Abstract
Common variable immunodeficiency (CVID) is the most prevalent primary immunodeficiency, marked by hypogammaglobulinemia, poor antibody responses, and increased infection susceptibility. The COVID-19 pandemic provided a unique opportunity to study the effects of prolonged viral infections on the immune responses of CVID patients. Here we use single-cell RNA-seq and spectral flow cytometry of peripheral blood samples before, during, and after SARS-CoV-2 infection showing that COVID-19 CVID patients display a persistent type I interferon signature at convalescence across immune compartments. Alterations in adaptive immunity include sustained activation of naïve B cells, increased CD21low B cells, impaired Th1 polarization, CD4+ T central memory exhaustion, and increased CD8+ T cell cytotoxicity. NK cell differentiation is defective, although cytotoxicity remains intact. Monocytes show persistent activation of inflammasome-related genes. These findings suggest the involvement of intact humoral immunity in regulating these processes and might indicate the need for early intervention to manage viral infections in CVID patients.
Subject terms: Primary immunodeficiency disorders, SARS-CoV-2, Computational biology and bioinformatics, Viral infection
Common variable immunodeficiency (CVID) is a prevalent primary immunodeficiency characterized by hypogammaglobulinemia, poor antibody responses, and enhanced susceptibility to infection. Here the authors examine CVID patients infected by SARS-CoV-2 and describe broad immune dysregulation, sustained type I IFN responses, altered B/T cell signaling, impaired NK cell differentiation, and persistent inflammasome activation.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), constituted a threat to public health worldwide (https://covid19.who.int/). Although many COVID-19 patients are asymptomatic or experience mild symptoms, some patients experience hyperinflammatory responses that lead to severe disease that can evolve to severe lung dysfunction, multiorgan system failure, and death1. In addition, persistent and often debilitating sequelae are increasingly recognized in convalescent individuals2.
In relation to the particular immune responses mounted by the host upon SARS-CoV-2 infection, a fast induction of SARS-CoV-2-specific CD4+ T cells in acute infection has been associated with a mild COVID-19 course and a rapid viral clearance3, whereas the lack of these specific CD4+ T cells was associated with severe COVID-194,5. Moreover, SARS-CoV-2-specific CD8+ T cell responses can develop rapidly during acute COVID-19, and the presence of these specific CD8+ T cells has been associated with better COVID-19 outcomes5,6. Regarding the humoral response, B cell memory rapidly evolves during SARS-CoV-2 infection7, and the production of neutralizing antibodies and especially the effectiveness of the anti-SARS-CoV-2 monoclonal antibodies demonstrate the effect of the humoral immune response on viral infection8–11. As part of the innate immune response, HLA-DRhiCD11chi inflammatory monocytes with an interferon-stimulated gene (ISG) signature are elevated in mild COVID-19, whereas severe COVID-19 is marked by the concurrence of neutrophil precursors, dysfunctional mature neutrophils, and HLA-DRlo monocytes12. Therefore, effective immune responses against SARS-CoV-2 require that the host’s immune system provides a balanced and coordinated response involving both innate and adaptive immune systems13,14.
In this context, patients with inborn errors of immunity (IEI) have demonstrated the role of type I interferon (IFN) in the control of the infection15. The prominent role of humoral memory in the antiviral protection, was underlined by the longer persistence of SARS-CoV-2 virus in patients with primary antibody deficiencies (PADs)16. Common variable immunodeficiency (CVID), the most common symptomatic PAD17, is characterized by hypogammaglobulinemia and poorly protective vaccine titers. Most CVID patients have recurrent severe infections, and a substantial proportion develop autoimmune and inflammatory features18,19. Besides, the classic defining antibody deficiency and B cell abnormalities, alterations in other immune compartments, including T cells and myeloid cells, have been described in CVID patients20–23.
The course and outcome of SARS-CoV-2 infection among patients with CVID in comparison with the general population has been a matter of debate. Most studies have reported a benign course of this particular viral infection in most CVID patients16,24–26, whereas one study found higher mortality rates among IEI patients including CVID patients27. In addition, it has been described that immunodeficient individuals, especially those with antibody deficiencies, show a persistent SARS-CoV-2 PCR positivity, as well as increased risks of both first infection and re-infection with SARS-CoV-2 among CVID individuals28. This situation provides the unique opportunity to investigate the effect of a persisting viral infection on the human immune system in the absence of a specific humoral response on a single-cell level.
Here, we performed a longitudinal study of peripheral blood samples from nine CVID patients collected prior, during and after SARS-CoV-2 infection. Using single-cell RNA-seq and spectral flow cytometry, we compared alterations of the immune system during viral infection and recovery of these patients in comparison with non-CVID cohorts29–31. Our analysis shows an increased type I interferon signature in almost all compartments, altered homeostasis of the entire adaptive immune system, and persistent inflammasome activation in monocytes.
Results
Disturbed homeostasis in both innate and adaptive immune compartments in COVID-19 CVID patients
Firstly, using single-cell RNA sequencing (scRNA-seq), we analyzed paired samples of peripheral blood mononuclear cells (PBMCs) from five CVID patients at three distinct stages of the SARS-CoV-2 infection: (1) baseline, before viral infection, (2) progression, during infection, and (3) convalescence, once the viral infection had been resolved and the patient was PCR negative (Fig. 1a and Supplementary Fig. 1a). All collected CVID patients were under regular immunoglobulin replacement therapy and displayed only mild symptoms during SARS-CoV-2 infection (for detailed patient description see Supplementary Data 1). We profiled a total of 70,643 single-cell transcriptomes of PBMCs from the CVID cohort (Supplementary Data 1).
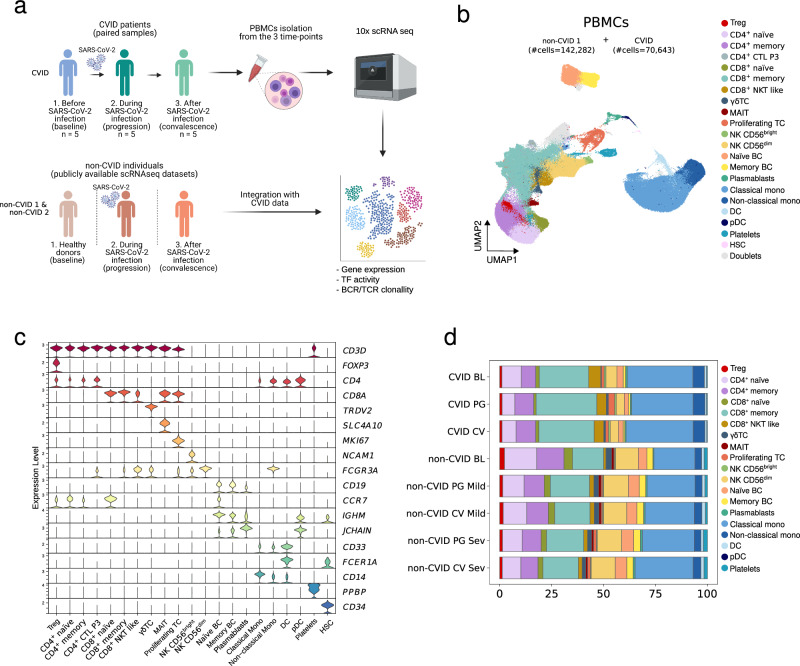
Fig. 1. Multi-stage single-cell analysis of PBMCs from CVID and non-CVID individuals with or without COVID-19.
a Overview of the participants included in this study, the samples collected and the data generated. Created in BioRender. Ballestar, E. (2024) https://BioRender.com/o27q186. b UMAP visualization showing different immune cell populations identified and cell-specific marker gene expression in the non-CVID 1 cohort (n = 6 for baseline, n = 5 for progression mild, n = 5 for progression severe, n = 5 for convalescence mild and n = 5 for convalescence severe) and in the CVID cohort (n = 5 paired samples at baseline, progression and convalescence). The number of captured cells in each cohort is indicated in brackets. The B cell compartment includes naïve B cells, memory B cells and plasmablasts. The T cell compartment includes CD4+ naïve T cells, CD4+ memory T cells, CD4+ cytotoxic CTL (only in sample P3), regulatory T cells (Treg), gamma-delta T (γδT) cells, CD8+ naïve T cells, CD8+ memory T cells, CD8+ NKT-like cells, mucosal-associated invariant T (MAIT) cells, and proliferating T cells. The NK cell compartment includes NK CD56bright and NK CD56dim cells. The myeloid cell compartment includes classical and non-classical monocytes, as well as conventional dendritic cells (cDC) and plasmacytoid dendritic cells (pDCs). In addition, we captured hematopoietic stem cells (HSC) and platelets. c Violin plots showing the gene expression levels of selected markers. d Stacked barplot showing cell cluster frequencies in each group. Source data for panels in this figure are provided in the Source Data Fig. 1 file.
The single-cell transcriptomes from the CVID cohort were processed and integrated with publicly available scRNA-seq datasets of PBMCs from non-CVID individuals without any known primary or secondary immunodeficiency (non-CVID 1 cohort), who were either negative or positive for SARS-CoV-2 (#cells = 142,282)29–31. This non-CVID 1 cohort included both non-CVID COVID-19 patients with mild symptoms (similar to patients in the CVID cohort) or severe symptoms (as a reference of a higher COVID-19-associated immune dysregulation), together with healthy individuals. As in our CVID cohort, non-CVID samples were classified in three stages: baseline, progression and convalescence. Samples from the non-CVID 1 cohort were selected to match our cohort of CVID patients in relation to sex, age and time of sample collection after symptoms onset (Supplementary Data 1a-1b and Supplementary Fig. 1a–g). The resulting integration of the CVID and non-CVID cohort 1 scRNA-seq datasets was used to create a ‘discovery object’. In parallel, to avoid potential biases associated with non-CVID samples due to technical differences and dataset origins, we integrated the single-cell transcriptomes from the CVID cohort with additional public scRNA-seq datasets of PBMCs from a cohort of individuals without known immunodeficiency (non-CVID 2 cohort) who were either negative or positive for SARS-CoV-2 (#cells = 270,448) (Supplementary Fig. 1h)32–35. The resulting scRNA-seq object (‘validation object’) was used to ensure that only genes exhibiting similar behavior across both non-CVID cohorts were selected for downstream analysis.
Our single-cell transcriptomic analysis identified several immune cell compartments at high resolution (Fig. 1b, c and Supplementary Fig. 1h, i). The comparison of cell proportions between CVID patients and non-CVID individuals (Fig. 1d and Supplementary Data 2), confirmed the expected reduction of memory B cells in CVID patients36. We also observed a significant reduction of regulatory T cells (Treg) and mucosal-associated invariant T (MAIT) cells at baseline in CVID patients, as previously described21,37,38. In addition, we detected a lower percentage of dendritic cells (DCs) in CVID patients compared with both mild or severe non-CVID individuals in convalescence (Fig. 1d and Supplementary Data 2), also previously described in CVID patients39.
Upregulation of BCR-related genes and persistent activation of naïve B cells with increased differentiation into CD21low B cells in COVID-19 CVID patients
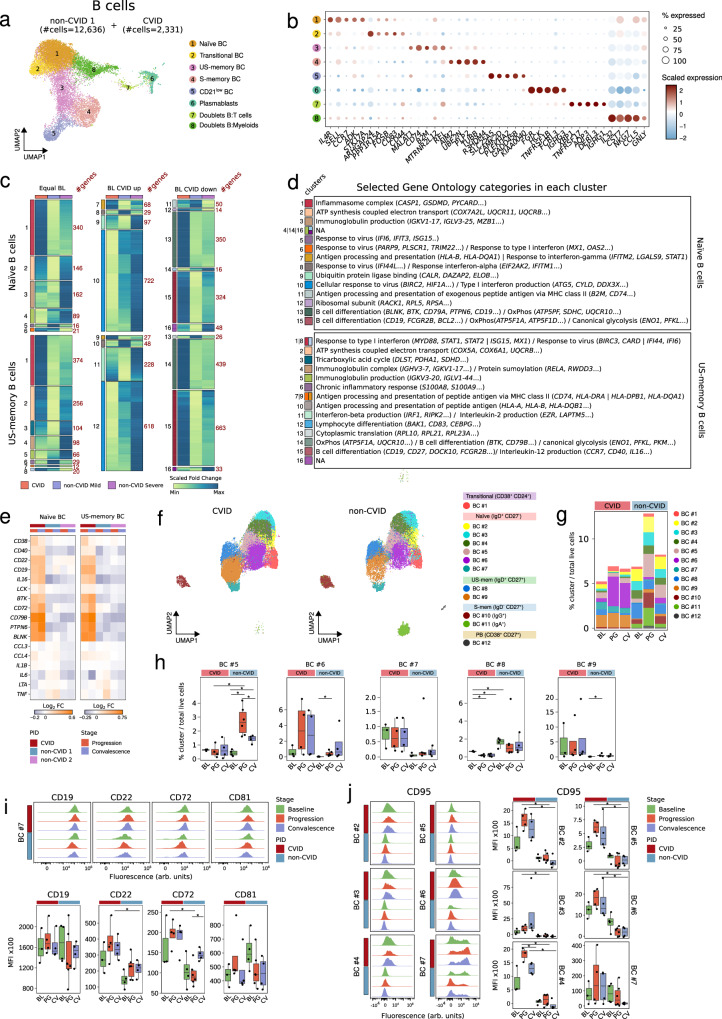
CVID patients are characterized by profound defects in B-cell-mediated immune responses. Consistent with this knowledge, none of the patients produced SARS-CoV-2-specific IgA antibodies and only one had detectable specific IgG antibodies after SARS-CoV-2 infection (Supplementary Data 1a). None of the CVID patients were positive for anti-type I IFNs autoantibodies previously described in patients with life-threatening COVID-1940. Reclustering of the B cell compartment annotated in Fig. 1b, comprising 2331 cells from CVID patients and 12,636 cells from non-CVID donors, allowed us to identify transitional, naïve, unswitched memory (US-memory), switched memory (S-memory), CD21low B cells subsets, and plasmablasts (Fig. 2a and b, and Supplementary Fig. 2a). Two additional clusters of doublets were detected but not used for downstream analysis. Furthermore, we performed a reclustering of the B cell compartment from the ‘validation object’, comprising 30,691 cells from non-CVID 2 cohort (Supplementary Fig. 2b and c). Because of the severe reduction of S-memory B cells and plasmablasts (Supplementary Data 2), we focused on the transcriptional alterations of naïve and US-memory B cells upon SARS-CoV2 infection. As we did not investigate SARS-CoV-2 specific responses, the results reflect global alterations in B cells during the viral infection and recovery. After excluding the two CVID patients lacking B cells, we calculated differentially expressed genes (DEGs) between baseline and progression in the CVID cohort or in the non-CVID 1 cohort (Supplementary Data 3). Then, we selected DEGs in the CVID cohort together with those DEGs in the non-CVID 1 cohort that were validated in the non-CVID 2 cohort. We then clustered these DEGs based on their expression profile during SARS-CoV-2 infection (Fig. 2c). They were grouped in three different categories comprising genes that exhibit either similar (BL equal), upregulated (BL CVID up), or downregulated (BL CVID down) baseline expression levels in CVID patients compared to non-CVID individuals (Fig. 2c). Among the different enriched gene ontology (GO) categories in the different clusters of DEGs (Supplementary Data 4), we focused on those related to immunological features (see Methods) and B cell biology (Fig. 2d). Thus, we observed that genes associated with type I IFN response, which play a role in the immune activity against viruses and other pathogens41–43, were already upregulated at baseline in the B cell compartment of CVID compared to non-CVID (Supplementary Fig. 2d) and showed less upregulation during COVID-19 progression, especially in US-memory B cells: clusters 1 and 8 (Fig. 2c, d and Supplementary Fig. 2d). At convalescence, however, the upregulation of type I IFN response-related genes was still maintained in CVID patients in contrast to non-CVID individuals (Supplementary Fig. 2d). On the other hand, we found genes related to B cell differentiation and regulation of the B cell receptor (BCR) signaling pathway in clusters of DEGs that were less expressed in CVID compared to non-CVID at baseline (Supplementary Fig. 2d) and become upregulated during COVID-19 progression and convalescence in CVID patients compared to non-CVID individuals: clusters 13 and 15 in naïve B cells, and cluster 14 and 15 in US-memory B cells (Fig. 2c, d and Supplementary Fig. 2d). Hence, we inspected the expression of BCR-related genes along the viral infection in more detail. We found a higher and more persistent transcriptional upregulation of several genes encoding members of the BCR signaling pathway, both activators and repressors, in the naïve and US-memory B cell compartment of COVID-19 CVID patients compared to COVID-19 non-CVID individuals (Fig. 2e and Supplementary Fig. 2e). This upregulation included BCR signaling pathway activator genes such as CD19, CD81, CD79B, BTK and BLNK, and also BCR inhibitory genes such as PTPN6 (which encodes the tyrosine phosphatase SHP-1), CD22 and CD72 (Fig. 2e and Supplementary Fig. 2e). In this line, among the dysregulated pathways identified through cell-cell communication inference analysis using CellChat44 (Supplementary Data 6), we observed that the CD22 signaling pathway, a negative regulator of BCR responses45,46, exhibited higher activity in naïve B cells from CVID patients compared to non-CVID individuals during viral infection (Supplementary Fig. 2f).
Fig. 2. Upregulation of BCR-related genes and persistent activation of naïve B cells with increased differentiation into CD21low B cells in COVID-19 CVID patients.
a UMAP visualization of B cell subpopulations, with cell numbers for CVID and non-CVID1 cohorts indicated. b Dot plot showing gene expression per B cell subpopulation; circle size denotes the percentage of expressing cells and colors indicate scaled average expression. (c) Heatmaps showing the fold change (baseline vs progression) of DEGs in CVID cohort, and non-CVID 1 cohort validated with non-CVID 2 cohort in selected B cell subsets. Genes are grouped by baseline expression comparison (equal, upregulated, or downregulated in CVID vs. non-CVID). Gene clusters are labeled on the left, and cluster sizes are indicated on the right. d Table of selected significantly enriched GO categories for clusters from (c), listing selected genes; clusters without significant enrichment are noted as NA. e Heatmaps showing fold change (baseline vs. progression/convalescence) of selected DEGs in CVID, non-CVID 1, and non-CVID 2 cohorts, showing only COVID-19 mild samples; color indicates Log2(Fold-change). f UMAP visualization of B cell clusters from spectral flow cytometry analysis. g Stacked barplot of B cell cluster frequency, calculated as cell counts per cluster over total live cells. h Box plots of selected B cell cluster frequencies. i Histograms and box plots of mean fluorescence intensity (MFI) for selected BCR-related proteins in cluster BC#7. (j) Histograms and box plots of CD95 protein levels in naïve B cells. Panels h, i, and j show biological replicates for CVID (BL: n = 3, PG: n = 4, CV: n = 4) and non-CVID controls (BL: n = 4, PG: n = 6, CV: n = 4). Two-sided t-tests were used (*p-value < 0.05). Box plot whiskers denote minimum and maximum values (excluding outliers), with the box spanning Q1 to Q3 and a horizontal line for the median. BL baseline, PG progression, CV convalescence. Source data are provided in Source Data Fig. 2 file.
We performed transcription factor (TF) activity analysis using decoupleR47, and observed changes in the activity of several TFs during SARS-CoV-2 infection in both CVID and non-CVID patients (Supplementary Fig. 2g). Notably, the activity of the B cell master regulator PAX5, which regulates the expression of genes critical to B cell identity, including components of the BCR such as CD19, CD79A, CD72 and BLK48, increased in CVID patients during infection and convalescence, in contrast to non-CVID individuals where PAX5 activity decreases (Supplementary Fig. 2h). Additionally, the TFs associated with the canonical NF-κB signaling pathway (p50, encoded by NFKB1 and p65, encoded by RELA), exhibited significantly a higher increment of activity in non-CVID individuals compared to CVID patients during infection. Of note, the canonical NF-κB signaling pathway displayed a higher activity in CVID patients compared to non-CVID at baseline (Supplementary Fig. 2h), possibly due to a chronic inflammatory background that characterizes many of these patients.
Following the observation of the dysregulated BCR related signature, we analyzed the BCR repertoires of naïve and US-memory B cells in our dataset at all stages of SARS-CoV-2 infection. The inspection of the dynamics of the VDJ usage upon SARS-CoV-2 infection indicates that the proportion of B cells expressing the IGHV3-53 gene, which has been described as the most frequently used IGHV gene among SARS-CoV-2 neutralizing antibodies49,50, was higher in COVID-19 non-CVID individuals than in COVID-19 CVID patients at both progression and convalescence stages (Supplementary Fig. 2i). Similar profiles were observed for the IGHV gene usage of other SARS-CoV-2 spike-targeting antibodies49,51, such as IGHV1-46, IGHV3-23 or IGHV5-51 among others (Supplementary Fig. 2i) suggesting suboptimal B cell selection in CVID patients.
We conducted further analysis to explore the phenotypic alterations in the B cell compartment using spectral cytometry on PBMCs (Supplementary Fig. 1j) collected from an additional cohort of CVID patients (paired samples) and non-CVID individuals, at baseline, COVID-19 progression and convalescence. This additional cohort was independent of the cohort analyzed by scRNA-seq (Supplementary Data 1). Through unsupervised cluster analysis using 33 surface antigens (Supplementary Data 7), we defined 12 main cell clusters within the B cell compartment (Fig. 2f and Supplementary Fig. 2j). Unsupervised cluster analysis showed that naïve B cells (IgD+CD27-) exhibited significant heterogeneity and were further divided into six main subclusters (Fig. 2f and Supplementary Fig. 2j). Within the naïve B cell compartment, we found a higher proportion of cells from cluster BC#7, corresponding with CD21low naïve B cells (Supplementary Fig. 2j), in CVID samples compared to non-CVID samples at all stages of infection (Fig. 2g and h). In addition, although not reaching significance due to group heterogeneity, we found a higher proportion of cells from cluster BC#6 (Fig. 2g and h), corresponding with CD86highHLA-DRhigh activated naïve B cells (Supplementary Fig. 2j), at progression and convalescence in CVID compared to non-CVID. On the contrary, we observed a lower percentage of cells from cluster BC#5 in CVID compared to non-CVID during SARS-CoV-2 infection and convalescence (Fig. 2g and h). Interestingly, cluster BC#5 is partly negative for CD21, and negative for CD27 and may reflect a small subset of reactive CD21lowCD27neg B cells not fully compatible with the CD21lowCD11chighT-bethigh B cell phenotype52,53 associated with viral infection. US-memory (IgD+CD27+) B cells were categorized into two clusters, with cells from clusters BC#8 and BC#9 exhibiting a lower and higher percentage, respectively, in CVID compared to non-CVID samples (Figs. 2g and h). As expected in CVID patient samples, we observed a diminished presence of both S-memory (IgD-CD27+) B cells and plasmablasts (CD38high CD27+) (Fig. 2f, g and Supplementary Fig. 2k). Finally, the spectral cytometry dataset allowed us to validate at the protein level the results from the scRNA-seq analysis regarding the upregulation of several BCR-related genes in the B cell compartment of CVID patients. We detected higher protein levels of CD19, CD22, CD72 and CD81 at progression in the CD21low cluster BC#7 in CVID compared to non-CVID (Fig. 2i). In this regard, the higher abundance of this cluster BC#7 in the CVID group (Figs. 2g and h) might partly explain the higher levels of BCR-related genes and the increased CD22 related cell-cell communication observed in CVID samples compared to non-CVID samples in the transcriptomic analysis. In addition, we found that the entire naïve B cell compartment displayed higher levels of CD95, the Fas receptor, together with lower levels of CXCR5 in the subsets BC#5 and BC#6 from CVID patients compared to non-CVID (Fig. 2j and Supplementary Fig. 2l).
Altogether, following SARS-CoV-2 infection, the B cell compartment of CVID patients exhibits an impaired activation of the canonical NF-κB signaling pathway and a dysregulated expression of several components of the BCR signaling pathway, possibly mediated by a higher activity of PAX5. In addition, CVID patients show a sustained type I IFN response signature at convalescence. Together with the described phenotypic alterations, these findings suggest a persistent activation of naïve B cells with increased differentiation into T-bethighCD21low B cells. However, the specific triggers and pathogenesis of these changes in the general B-cell compartment remain to be identified.
Defective increase of Th1-, but higher frequency of PD-1hiTIGIThi memory cells during and after SARS-CoV-2 infection in COVID-19 CVID patients
Next, we analyzed T cells from both CVID patients and non-CVID individuals at all stages of SARS-CoV-2 infection. After reclustering the T cell compartment previously annotated in Fig. 1b, we identified 17 clusters which comprised 46,094 cells from CVID patients and 66,194 cells from non-CVID donors (Fig. 3a and Supplementary Fig. 3a). Furthermore, we performed a reclustering of the T cell compartment from the validation object, comprising 178,483 cells from non-CVID 2 cohort (Supplementary Fig. 3b). The T cell compartment included CD4+ T cells (naïve, central memory and effector memory), CD8+ T cells (naïve, central memory and effector memory), together with γδT cells, MAIT cells, CD4+ cytotoxic, and proliferating T cells (Fig. 3a and b, and Supplementary Fig. 3a–d). One cluster of doublets was detected and excluded, and we focused on naïve, central memory (TCM) and effector memory (TEM) CD4+ and CD8+ T cells for downstream analysis.
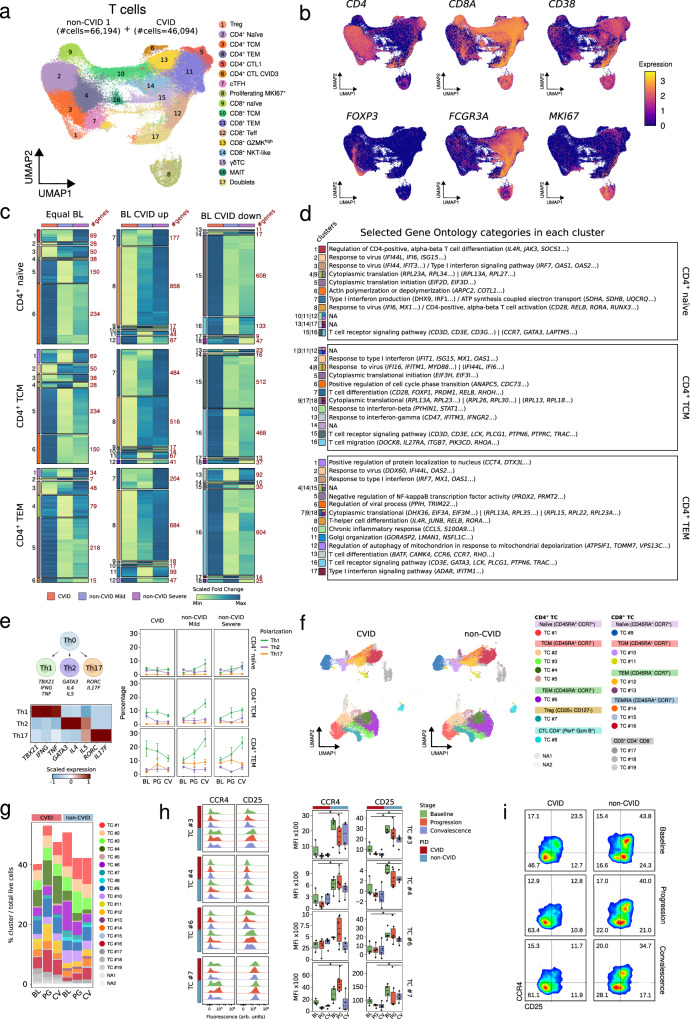
Fig. 3. Defective increase of Th1-, but higher frequency of PD-1hiTIGIThi memory cells during and after SARS-CoV-2 infection in COVID-19 CVID patients.
a UMAP visualization of T cell subsets, with cell numbers for CVID and non-CVID 1 cohorts indicated. b UMAPs showing expression of selected T cell genes; scale indicates expression levels. c Heatmaps showing fold change (baseline vs progression) of DEGs in CVID cohort, and non-CVID 1 cohort validated with non-CVID 2 cohort, for selected CD4+ T cell subsets. Genes are grouped by baseline expression comparison (equal, upregulated, or downregulated in CVID vs non-CVID). Gene clusters are labeled on the left, and cluster sizes are indicated on the right. d Table of selected significant GO categories for each cluster in c. Clusters without significant enrichment are marked as NA. e Scheme of T cell polarization into Th1/Th2/Th17 with related-gene expression and subset proportions by PID, infection stage, and COVID-19 severity. Mean and standard error bars reflect biological replicates: CVID (BL: n = 5, PG: n = 5, CV: n = 5) and the non-CVID control group (BL: n = 6, PG_Mild: n = 4, CV_Mild: n = 5, PG_Severe: n = 5, CV_Severe: n = 5). f UMAP of T cell clusters from spectral flow cytometry analysis. g Stacked barplot of T cell cluster frequencies, calculated as cell counts per cluster over total live cells. h Histograms and box plots showing levels of selected proteins in CD4+ T subsets by PID and infection stage. Dots represent biological replicates from different subjects in the CVID group (BL: n = 3, PG: n = 4, CV: n = 4) and the non-CVID control group (BL: n = 4, PG: n = 6, CV: n = 4). Two-sided t-test was used (*p-value < 0.05). i Density plot of CCR4/CD25 in cluster TC#3. Box plot whiskers denote minimum and maximum values (excluding outliers), with the box spanning Q1 to Q3 and a horizontal line for the median. BL baseline, PG progression, CV convalescence. Source data are provided in Source Data Fig. 3.
In these particular T cell subsets, we performed the same analysis as described above for B cells calculating DEGs between baseline and progression in each cohort (Supplementary Data 3). Regarding CD4+ T cells (Fig. 3c and d), among other GO categories (Supplementary Data 4), we found that those related to T cell receptor signaling were significantly enriched in clusters of genes upregulated in CVID patients during SARS-CoV-2 infection, although those genes were less expressed at baseline compared to non-CVID individuals: clusters 15 and 16 in CD4+ naïve T cells, cluster 15 in CD4+ TCM cells, and cluster 16 in CD4+ TEM cells (Fig. 3d and Supplementary Fig. 3e). Similarly, genes related to T cell migration were lower at baseline but increased more during progression in CD4+ TCM cells from CVID patients (cluster 16 in CD4+ TCM cells). Similar to the B cell compartment, type I IFN signature in the CD4+ T cells is already increased at baseline (Supplementary Fig. 3e), increases less strongly during infection especially evident in clusters 2, 3, 7, and 8 in CD4+ naïve T cells, clusters 2 and 8 in CD4+ TCM cells, and clusters 2, 3, and 17 in CD4+ TEM cells, but persists in CVID patients compared to non-CVID at convalescence (Supplementary Fig. 3e).
Furthermore, we performed cell-cell communication inference analysis. Among the identified dysregulated pathways (Supplementary Data 6), we focused on the CD40L/CD40 pathway due to its relevance in T cell biology, and we observed that the cell crosstalk between CD4+ TCM and B cells exhibited an impaired CD40L/CD40 signaling in CVID patients (Supplementary Fig. 3f).
Subsequently, we assessed CD4+ T cell polarization by analyzing the expression of TBX21 (Tbet), IFNG (IFN-γ), and TNF (TNF-α) for Th1 polarization, GATA3, IL4, and IL5 for Th2 polarization, or RORc (RORγ) and IL17F for Th17 polarization (Fig. 3e). Naïve CD4+ T cells did not show a clear increase in the Th1 phenotype during SARS-CoV-2 infection. In contrast, despite the higher Th1 phenotype observed at baseline in CVID samples, which is line with our previous observations reporting a skewed memory CD4+ T cell differentiation toward Th1 in complicated CVID patients54, both CD4+ TCM and CD4+ TEM of CVID patients showed a defective increase of Th1 cells at progression and convalescence stages compared to non-CVID individuals (Fig. 3e). However, IFNG expression was preserved at higher levels in CVID compared to non-CVID during SARS-CoV-2 infection, especially in the CD4+ TEM cell subset (Supplementary Fig. 3g), probably due to the initial skewed Th1 phenotype that characterizes CVID patients.
To further investigate phenotypic changes in the T cell compartment, we used spectral cytometry on PBMCs (Supplementary Fig. 1j) from the additional sets of CVID and non-CVID samples. Unsupervised cluster analysis using 35 surface antigens (Supplementary Data 7), identified 8 main cell clusters of CD4+ T cells, 8 clusters of CD8+ T cells and 3 clusters of CD4/CD8neg T cells (Fig. 3f and Supplementary Fig. 3g). In the CD4+ T cell compartment, we observed that the TCM cluster TC#3 is increased during SARS-CoV-2 infection in non-CVID individuals compared to CVID patients (Fig. 3g and Supplementary Fig. 3h). Conversely, the cluster TC#4, corresponding with CD4+TIGIThighPD1high TCM cells (Supplementary Fig. 3g), is more abundant across all stages of SARS-CoV-2 infection in CVID patients compared to non-CVID individuals potentially reflecting an exhausted phenotype (Fig. 3g and Supplementary Fig. 3h). Additionally, clusters TC#6, corresponding to the TEM subset, and TC#7, representing Tregs (CD25+CD127-), were more prevalent in non-CVID samples at baseline compared to CVID patients. However, both populations decreased during SARS-CoV-2 infection and convalescence in non-CVID compared to CVID (Fig. 3g and Supplementary Fig. 3h). In addition, we observed that cells from clusters TC#3 and TC#4, cluster TC#6, and cluster TC#7, expressed lower levels of CCR4 and CD25 in CVID patients compared to non-CVID individuals at all stages (Fig. 3h). Interestingly, within the TC#3 subset, which exhibited a more evident difference in the expression of both CCR4 and CD25 between CVID and non-CVID groups, the proportion of CCR4+CD25+ cells, potentially possessing a regulatory phenotype, was higher in non-CVID individuals than in CVID patients at baseline (Fig. 3i). In addition, this proportion decreased in both groups during SARS-CoV-2 infection and convalescence (Fig. 3i).
Taken together, our findings show a persistent type I IFN signature in CD4+ T cells at convalescence in CVID patients compared with non-CVID. Furthermore, we observed that during SARS-CoV-2 infection, the CD4+ T cell compartment of patients with CVID exhibits a defective increase of Th1 polarization, a higher abundance of TIGIThi PD-1hi central memory cells, a compromised CD40L/CD40 receptor/ligand interaction, and an impaired expression of CD25 and CCR4 especially on CD4+ TCM cells within the cluster TC#3.
Strong clonal expansion of cytotoxic CD8+ T effector compartment in COVID-19 CVID patients
In the CD8+ T cell compartment, we also observed distinct patterns of DEGs between baseline and progression in COVID-19 CVID patients and non-CVID individuals that were enriched in different GO categories (Fig. 4a, b and Supplementary Data 4). Naïve CD8+ T cells from CVID patients showed upregulation of genes related to TCR signaling (cluster 17), and reduced upregulation of genes related to antiviral response and type I IFN compared to mild COVID-19 non-CVID samples (clusters 6, 7, 8, and 9) during progression (Fig. 4a, b and Supplementary Fig. 4a). These patterns were also visible in CD8+ TCM cells (clusters 6, 8 and 9), but were clearly distinct in CD8+ TEM cells where antiviral response, type I IFN response and, remarkably, the cytotoxic gene signatures were more strongly upregulated in CVID patients compared to non-CVID individuals (clusters 1, 3, 12, 16, and 17) (Fig. 4a, b and Supplementary Fig. 4a), possibly due to a lower expression of cytotoxic-related genes at baseline in CVID patients compared to non-CVID (Supplementary Fig. 4a). As previously observed for B cells and CD4+ T cells, CD8+ T cells displayed persistent type I IFN signature in CVID patients compared to non-CVID at convalescence (Supplementary Fig. 4a).
Fig. 4. Strong clonal expansion of cytotoxic CD8+ T effector compartment in COVID-19 CVID patients.
a Heatmaps showing fold-change (baseline vs progression) of DEGs in CVID cohort, and non-CVID 1 cohort validated with non-CVID 2 cohort, for selected CD8+ T cell subsets. Genes are grouped by baseline expression comparison (equal, upregulated, or downregulated in CVID vs non-CVID). Gene clusters are labeled on the left, and cluster sizes are indicated on the right. b Table of selected significant GO categories for each cluster in (a). Clusters without significant enrichment are marked as NA. c Box plots showing ‘cytotoxic response’ score for selected CD8+ T cell subsets in CVID, non-CVID 1, and non-CVID 2 cohorts. Dots correspond to biological replicates from different subjects in the CVID group (BL: n = 5, PG: n = 5, CV: n = 5), the non-CVID 1 control group (BL: n = 6, PG_Mild: n = 4, CV_Mild: n = 5, PG_Severe: n = 5, CV_Severe: n = 5) and the non-CVID 2 control group (BL: n = 14, PG_Mild: n = 11, CV_Mild: n = 8, PG_Severe: n = 8, CV_Severe: n = 5). d Histograms and box plots showing levels of selected proteins in CD8+ T cell subsets, stratified by PID status and infection stage. Dots correspond to biological replicates from different subjects in the CVID group (BL: n = 3, PG: n = 4, CV: n = 4) and the non-CVID control group (BL: n = 4, PG: n = 6, CV: n = 4). Two-sided t-test was used (*p-value < 0.05). e UMAP depicting TCR clonal expansion in CVID patients by SARS-CoV-2 infection stages. Stacked barplot of TCR clonal expansion percentages across cell types and stages is indicated. Box plot whiskers denote minimum and maximum values (excluding outliers), with the box spanning Q1 to Q3 and a horizontal line for the median. BL baseline, PG progression, CV convalescence. Source data for panels are available in Source Data Fig. 4.
Furthermore, our cell-cell communication analysis indicated that, among the identified dysregulated pathways (Supplementary Data 6), prostaglandin and CLEC signaling pathways, which have been described as regulators of T cell function and activity55,56, are more active in CVID patients than non-CVID individuals during viral infection (Supplementary Fig. 4b).
It has been previously described that severe COVID-19 patients displayed a higher proportion of activated CD8+CD16+ (FCGR3A) T cells with increased cytotoxic functions associated with fatal outcomes of these patients33. Based on this previous study, our single-cell analysis allowed us to identify a FCGR3A+ cytotoxic T cell subpopulation (CD38+ MKI67+ FCGR3A+ CM T cells) that was highly expanded during SARS-CoV-2 infection in severe COVID-19 non-CVID individuals in comparison with mild COVID-19 non-CVID individuals or COVID-19 CVID patients (Supplementary Fig. 4c). This indicates that the expansion of this pathogenic cytotoxic T cell population is a feature of severe COVID-19 patients and not of mild COVID-19 individuals regardless of their CVID status.
Spectral cytometry analysis of the CD8+ T cell compartment showed that clusters TC#11, belonging to the CD8+ TCM subset, and TC#12, corresponding with the CD8+ TEM subset, were more abundant in CVID samples than in non-CVID samples (Fig. 3g and Supplementary Fig. 4d). This TEM cluster TC#12, highly prevalent in CVID patients, may correspond to a previously described subset of CD8+ TEM cells with features of exhaustion and activation57. In addition, the highly cytotoxic CD8+ TEMRA subset TC#16 is especially abundant in some CVID patients during infection compared to non-CVID individuals (Fig. 3g and Supplementary Fig. 4d). Furthermore, we performed differential protein expression analysis. Among the identified dysregulated proteins (Supplementary Data 7), we focused on some relevant molecules in CD8+ T cell biology. For instance, we found that the homing receptor CCR5 displayed lower levels in the CD8+ TEM clusters TC#11, TC#13, and CD8+ TEMRA clusters TC#14 and TC#16, together with higher levels of the cytotoxic molecules granzyme B and perforin in CVID patients compared to non-CVID individuals (Fig. 4d).
Finally, we conducted TCR analysis in CVID patients throughout SARS-CoV-2 infection. In contrast to the CD4+ T cell compartment, where no subpopulation exhibited the presence of clones larger than 20 cells at any stage (except for CD4+ CTL), the CD8+ T cell compartment showed the highest level of TCR clonal expansion (Fig. 4e). We also calculated the Shannon entropy index to assess the diversity of the TCR repertoire in each T cell cluster (Supplementary Fig. 4e). CD8+ naïve and CD8+ TCM cells did not exhibit an increase in hyperexpanded clones at progression (Fig. 4e) and showed no changes in TCR diversity (Supplementary Fig. 4e). In contrast, CD8+ TEM, CD8+ Teff, and CD8+ GZMKhigh cells, which displayed a higher proportion of hyperexpanded clones at progression (Fig. 4e), also displayed a decline in TCR diversity during SARS-CoV-2 infection (Supplementary Fig. 4e), potentially indicating the emergence of expanded clones. Interestingly, proliferating MKI67+ cells exhibited a higher proportion of hyperexpanded clones at progression (Fig. 4e) with no clear decline in TCR diversity (Supplementary Fig. 4e), suggesting a broad, not clonotype-specific expansion in this particular cell subpopulation.
Overall, these results indicate distinct effects on the different CD8+ T cell subsets during viral infection and convalescence phase. Most remarkably, the cytotoxic CD8+ T cell compartment in CVID patients show a strong and specific clonal expansion upon SARS-CoV-2 infection, together with a general larger CD8+ T effector compartment compared to non-CVID individuals. In addition, CD8+ T cells from CVID patients display lower levels of the receptor CCR5, but increased levels of the cytotoxic molecules granzyme B and perforin during SARS-CoV-2 infection in the entire CD8+ memory T cell compartment, and at convalescence in the highly cytotoxic CD8+ TEMRA subset TC#16.
Impaired differentiation, preserved cytotoxic and increased immune checkpoint molecules by NK cells from COVID-19 CVID patients
We next examined the NK cell compartment, which plays key roles during SARS-CoV-2 infection58–60. To gain deeper insights, we performed reclustering of the NK cells previously annotated in Fig. 1b from both the CVID cohort (2,677 cells) and the non-CVID 1 cohort (16,566 cells) (Fig. 5a). According to existing literature in the field60, we accurately annotated six NK cell subpopulations, namely NK CD56bright, inflamed CD56dim, cytokine CD56dim, HLA-DRhigh CD56dim and CD56dim, together with an additional population that we annotated as SAP+ CD56dim NK cells (Fig. 5a and b, and Supplementary Fig. 5a and b). Furthermore, we performed a reclustering of the NK cell compartment from the ‘validation object’, comprising 23,615 cells from the non-CVID 2 cohort (Supplementary Fig. 5c), and we successfully identified the same abovementioned NK cell subpopulations (Supplementary Fig. 5c and d). Among these cell subsets, only NK CD56bright, CD56dim, and HLA-DRhigh CD56dim clusters were well represented in the CVID cohort (see Methods), thus becoming the primary focus for downstream analysis.
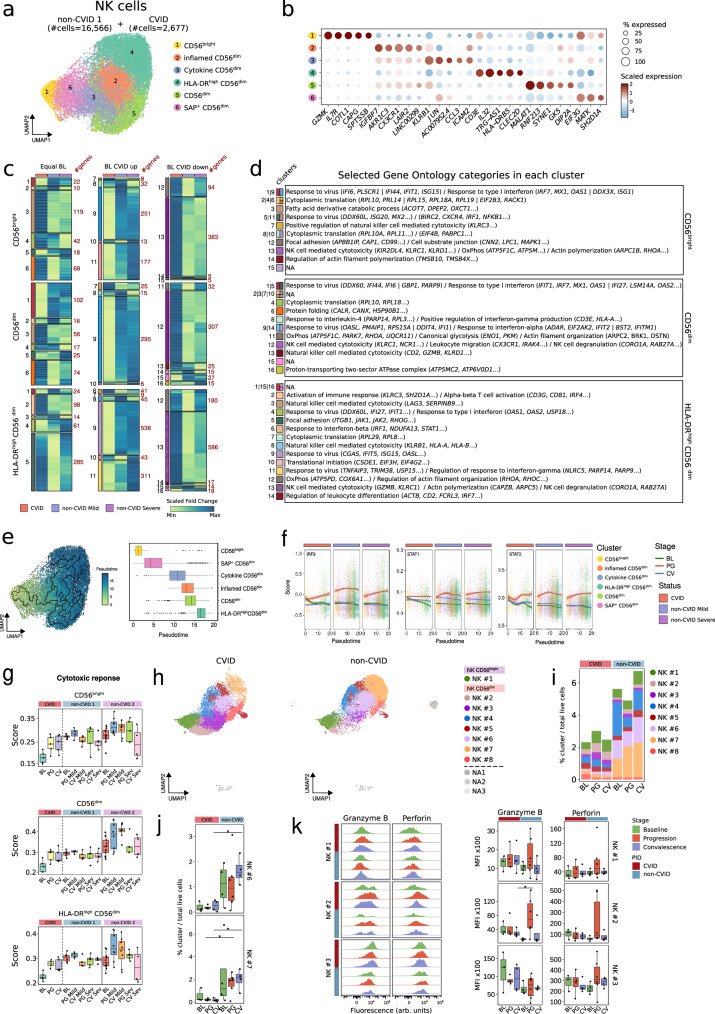
Fig. 5. Impaired differentiation, preserved cytotoxic and increased immune checkpoint molecules by NK cells from COVID-19 CVID patients.
a UMAP showing NK cell clusters, with cell numbers for CVID and non-CVID 1 cohorts indicated. b Dot plot showing gene expression per NK cell subpopulation; circle size denotes the percentage of expressing cells and colors indicate scaled gene expression. c Heatmaps showing fold-change (baseline vs progression) of DEGs in CVID cohort, and non-CVID 1 cohorts validated with non-CVID 2 cohort, for selected NK cell subsets. Genes are grouped by baseline expression comparison (equal, upregulated, or downregulated in CVID vs non-CVID). Gene clusters (left) and cluster sizes (right) are labeled. d Table of selected significant GO categories for each cluster in c. NA indicates clusters lacking enrichment. e UMAP and box plots showing cell trajectory of NK cells. Scale refers to pseudotime trajectory. f Enrichment scores for IRF9, STAT1 and STAT2 in single-cells by NK cell subset. g Box plots showing ‘cytotoxic response’ score for selected NK cell subsets in CVID, non-CVID 1, and non-CVID 2 cohorts. Dots correspond to biological replicates from different subjects in CVID (BL: n = 5, PG: n = 5, CV: n = 5), non-CVID1 (BL: n = 6, PG_Mild: n = 4, CV_Mild: n = 5, PG_Severe: n = 5, CV_Severe: n = 5) and non-CVID 2 control group (BL: n = 14, PG_Mild: n = 11, CV_Mild: n = 8, PG_Severe: n = 8, CV_Severe: n = 5). h UMAP of NK cells from spectral flow cytometry analysis. i Stacked barplot of NK cell cluster frequencies, calculated as cell counts per cluster over total live cells. j Box plots showing frequencies of selected NK cell clusters. k Histograms and box plots showing granzyme B and perforin levels in selected NK cells. Panels j and k show biological replicates for CVID (BL: n = 3, PG: n = 4, CV: n = 4) and non-CVID controls (BL: n = 4, PG: n = 6, CV: n = 4). Two-sided t-tests were used (*p-value < 0.05). Box plot whiskers denote minimum and maximum values (excluding outliers), with box spanning Q1 to Q3 and horizontal line for the median. BL baseline, PG progression, CV convalescence. Source data for panels are provided in the Source Data Fig. 5 file.
Subsequently, we performed the same analysis as described above for B and T cells by classifying DEGs (Supplementary Data 3) into three categories (Fig. 5c and d). Among the different enriched GO categories (Supplementary Data 4), we observed that, similar to B and T cells, response to type I IFN was upregulated at baseline, less prominently enriched during SARS-CoV-2 infection in the three NK cell populations analyzed (clusters 1, 5, 9 and 11 in CD56bright NK cells, clusters 1, 5, 9 and 14 in CD56dim NK cells, and clusters 4, 9 and 11 in HLA-DRhigh CD56dim NK cells) (Fig. 5c and d), but persisted at convalescence especially in the HLA-DRhigh CD56dim NK cell subset from CVID patients compared to non-CVID (Supplementary Fig. 5e). In this regard, when we defined cell differentiation trajectories across the different NK cell subpopulations identified (Fig. 5e) and represented the scored expression of the regulons controlled by the TFs of the type I IFN pathway—specifically, STAT1, STAT2, and IRF961— along pseudotime (Fig. 5f), we observed that the activation of these TFs appeared to be less robust in CVID patients compared to non-CVID individuals, particularly in CD56bright NK cells, being only partially recovered in CD56dim and HLA-DRhigh CD56dim NK cells (Fig. 5f). On the other hand, several GO categories related to NK cell mediated cytotoxicity and degranulation were significantly enriched in clusters containing genes that become more upregulated in CVID patients than in non-CVID individuals during progression (clusters 7 and 13 in CD56bright NK cells, clusters 12 and 13 in CD56dim NK cells, and cluster 13 in HLA-DRhigh CD56dim NK cells) (Fig. 5c and d). In this regard, the bona fide cytotoxic genes GZMB and PRF1 showed increased expression during SARS-CoV-2 infection in CVID patients, which is partially maintained at convalescence (Fig. 5g and Supplementary Fig. 5g). This increased upregulation of cytotoxicity-related genes in CVID samples during infection occurs mostly in lowly expressed genes at baseline in CVID (Supplementary Fig. 5e and f).
Considering the established role of NK cells in modulating fibrosis in various tissues62–64, and the dysfunctional antifibrotic activity of NK cells in severe COVID-19 patients60, we analyzed the expression of a profibrotic gene set (AREG, DUSP2, ZFP36L2, CXCR4, and TSC22D3) in the NK compartment during SARS-CoV-2 infection. As previously60, we observed a tendency toward a higher profibrotic signature in the three examined NK cell subsets of severe COVID-19 non-CVID individuals compared to mild COVID-19 non-CVID individuals (Supplementary Fig. 5h). Notably, this elevated profibrotic signature was not observed in CVID patients following SARS-CoV-2 infection (Supplementary Fig. 5h).
Spectral cytometry analysis of PBMCs (Supplementary Fig. 1j) in the additional cohort of CVID and non-CVID individuals at different stages of SARS-CoV-2 infection, and unsupervised cluster analysis using 35 surface antigens (Supplementary Data 7), identified 1 cell cluster of CD56bright NK cells and 7 NK clusters of CD56dim (Fig. 5h and Supplementary Fig. 5i). Moreover, CVID patients exhibited similar proportions of CD56bright compared to non-CVID, but an overall reduction in the frequencies of CD56dim NK cells affecting especially more differentiated subsets, such as NK#6 and NK#7 (Fig. 5i, j and Supplementary Fig. 5j). In general, we observed similar levels of the cytotoxicity-related proteins granzyme B and perforin in the NK cell compartment of CVID and non-CVID individuals at all stages of SARS-CoV-2 infection (Fig. 5k). In addition, among the significantly differentially expressed proteins (Supplementary Data 8), we focused on some of those with a previously described role in NK cell biology, such as CCR565,66, CD3867, TIGIT68 and TIM-369,70. In this regard, we detected lower levels of CCR5, together with higher levels of CD38, TIGIT and TIM-3 in CVID compared to non-CVID (Fig. 5k, and Supplementary Fig. 5k and 5l).
All these data suggest that the differentiation of advanced stages of CD56dim NK cells is impaired in CVID patients in comparison with non-CVID donors. However, this is not associated with a decreased cytotoxicity as NK cells of CVID patients strongly upregulate cytotoxicity-related gene sets during infection and convalescence despite reduced expression at baseline. This conclusion is corroborated by the similar amounts of the cytotoxic molecules granzyme B and perforin in CVID and non-CVID individuals at the protein level. In contrast, the homing receptor CCR5 displays lower expression, and CD38, TIGIT and TIM-3 show higher levels in CVID patients compared to non-CVID individuals indicating increased expression of immune checkpoint molecules.
Persistent activation of inflammasome genes in COVID-19 CVID monocytes
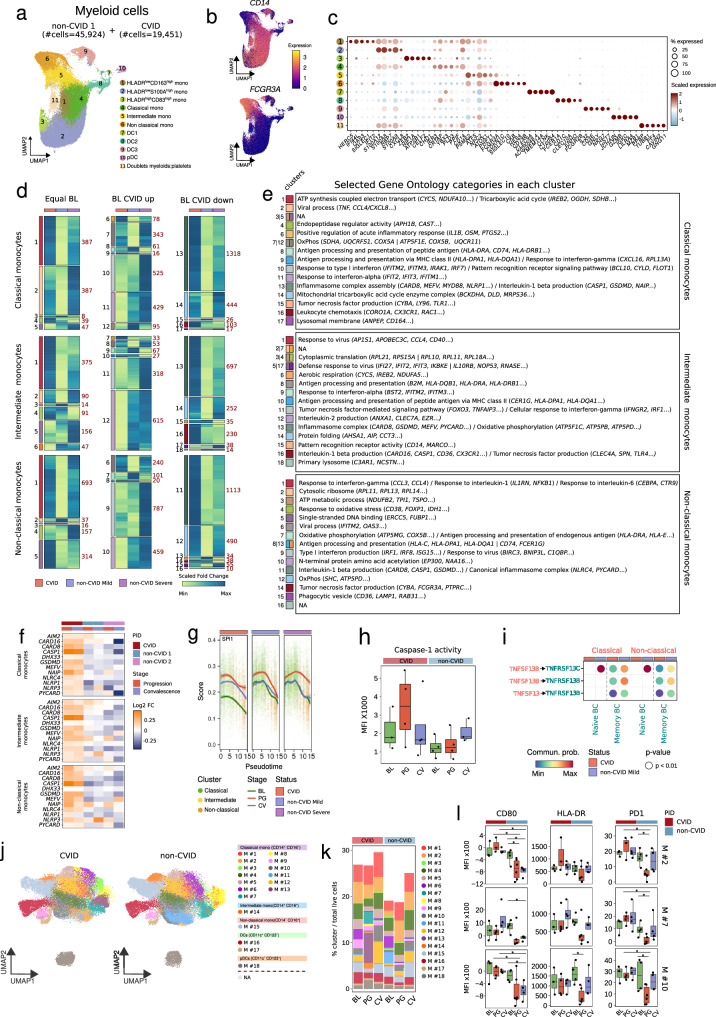
Severe COVID-19 patients display functional and phenotypic alterations in monocytes12,71–73. Hence, after reclustering the myeloid cells annotated in Fig. 1b, we analyzed the three main subsets of monocytes within the myeloid compartment (classical, intermediate and non-classical) during SARS-CoV-2 infection and recovery (Fig. 6a–c and Supplementary Fig. 6a). Furthermore, we annotated three additional clusters of monocytes, namely HLA-DRlowCD163high, HLA-DRlowS100Ahigh and HLA-DRhighCD83high monocytes, to put our analysis in context with existing literature12 (Fig. 6a–c, and Supplementary Fig. 6a and b). Reclustering of the myeloid cell compartment from the validation object, comprising 25,740 cells from non-CVID 2 cohort (Supplementary Fig. 6c), rendered the same abovementioned myeloid cell subpopulations (Supplementary Fig. 6c and d). Similarly to other cell compartments, we calculated DEGs and grouped them based on their expression profiles during SARS-CoV-2 infection (Fig. 6d). Subsequently, we performed GO category enrichment analysis for the identified DEG clusters to gain insights into their functional characteristics (Fig. 6d and e).
Fig. 6. Persistent activation of inflammasome genes in COVID-19 CVID monocytes.
a UMAP showing myeloid cell clusters, with cell numbers for CVID and non-CVID 1 cohorts indicated. b UMAPs showing expression of CD14 and FCGR3A; scale indicates expression levels. c Dot plot showing gene expression per myeloid cluster; circle size denotes the percentage of expressing cells and colors indicate average expression. d Heatmaps showing fold-change (baseline vs progression) of DEGs in CVID cohort, and non-CVID 1 cohort validated with non-CVID 2 cohort, across selected myeloid subsets. Genes are grouped by baseline expression comparison (equal, upregulated, or downregulated in CVID vs non-CVID). Gene clusters (left) and cluster sizes (right) are labeled. e Table of selected significant GO categories for each cluster in d. NA indicates clusters lacking enrichment. f Heatmaps showing fold-change (baseline vs progression/convalescence) of selected DEGs in CVID, non-CVID1, and non-CVID2 cohorts, showing only COVID-19 mild samples; color indicates Log2(fold-change). g SPI1 regulon enrichment score in single-cells by NK subset. h Box plots depicting caspase-1 activity in CVID and non-CVID samples across SARS-CoV-2 infection stages. Dots correspond to biological replicates from different subjects in the CVID group (BL: n = 3, PG: n = 4, CV: n = 4) and the non-CVID control group (BL: n = 4, PG: n = 4, CV: n = 3). i Cell-cell communication analysis of BAFF and APRIL signaling pathways during SARS-CoV-2 infection. Sender cells and ligands in red, receiver cells and receptors in green. j UMAP showing myeloid clusters from spectral flow cytometry analysis. k Stacked barplot of myeloid cluster frequencies, calculated as cell counts per cluster over total live cells. l Box plots showing the MFI of selected proteins in selected monocyte clusters. Dots correspond to biological replicates from different subjects in CVID group (BL: n = 3, PG: n = 4, CV: n = 4) and non-CVID control group (BL: n = 4, PG: n = 6, CV: n = 4). Two-sided t-test was used (*p-value < 0.05). Box plot whiskers denote minimum and maximum values (excluding outliers), with the box spanning Q1 to Q3 and a horizontal line for the median. BL baseline, PG progression, CV convalescence. Source data for panels are provided in the Source Data Fig. 6 file.
Among the enriched GO categories identified (Supplementary Data 4), like in all other cell compartments, we observed increased baseline expression of type I IFN response genes in all three monocyte compartments (Supplementary Fig. 6e), reduced upregulation during progression in CVID patients compared to non-CVID individuals: clusters 10 and 11 in classical monocytes, and clusters 9 in intermediate and non-classical monocytes (Fig. 6d and e). In contrast to the other immune compartments analyzed, this upregulation did not persist in monocytes from CVID patients compared to non-CVID at convalescence (Supplementary Fig. 6e). Remarkably, upregulated genes in monocytes from COVID-19 CVID patients displayed significant enrichment in categories linked to acute inflammation, IL-1β production, and inflammasome complex assembly. This enrichment was evident across classical monocytes (clusters 6 and 13), intermediate monocytes (clusters 13 and 16), and non-classical monocytes (cluster 11) (Fig. 6d and e). In this regard, key inflammasome-related genes, including CASP1 (encoding caspase-1), GSDMD (encoding gasdermin), MEFV (encoding pyrin), NLRP1, NLRC4 and CARD8, were upregulated in classical, intermediate, and non-classical monocytes from CVID patients throughout the disease progression, contrasting with individuals from both non-CVID cohorts (Fig. 6f). This pattern was also observed in HLA-DRlow CD163high, HLA-DRlow S100Ahigh and HLA-DRhigh CD83high monocytes (Supplementary Fig. 7a and b). The upregulation of these genes, together with other inflammasome-related genes such as PYCARD and NAIP, persisted at the convalescence stage in the monocytes from CVID patients, in contrast to non-CVID individuals (Fig. 6f, and Supplementary Fig. 6f, 7a and b).
Next, we analyzed the TF activities within the specified monocyte subsets. Among the different TFs that show differences in their activity during SARS-CoV-2 infection, we noticed a more pronounced activation of PU.1 (encoded by SPI1) in monocytes from CVID patients compared to their non-CVID counterparts, persisting across both progression and convalescence stages. This observation was further supported when we defined cell differentiation trajectories across the different monocyte subpopulations identified (Supplementary Fig. 6g) and represented the scored expression of the regulons controlled by PU.1 along pseudotime (Fig. 6g and Supplementary Fig. 7c).
To confirm the potential dysregulation of inflammasome function in monocytes of COVID-19 CVID patients, we stimulated PBMCs from an additional cohort comprising both non-CVID individuals and CVID patients at different stages—before, during, and after SARS-CoV-2 infection—with the inflammasome activator nigericin. Measurement of caspase-1 activity in the CD14+ monocyte compartment revealed that, in the course of progression, COVID-19 CVID patients exhibited a higher inflammasome activity compared to non-CVID individuals (Fig. 6h). This increased inflammasome activation after nigericin stimulation was not detected during convalescence, despite the elevated expression of inflammasome genes.
Furthermore, cell-cell communication analysis showed a reduced crosstalk between classical and non-classical monocytes with naïve and memory B cells affecting both the APRIL and BAFF signaling pathways in CVID patients compared to non-CVID individuals during SARS-CoV-2 infection (Fig. 6i).
Utilizing unsupervised clustering analysis with data from 33 surface antigens (Supplementary Data 7), we characterized the myeloid compartment (Supplementary Fig. 1j) in the additional cohort of CVID and non-CVID individuals at different stages of SARS-CoV-2 infection. Our analysis revealed 13 distinct cell clusters of classical monocytes (CD14+CD16-), one cluster of intermediate monocytes (CD14+CD16+), one cluster of non-classical monocytes (CD14-/lowCD16+), 2 clusters of dendritic cells (DCs), and 1 cluster of plasmacytoid dendritic cells (pDCs) (Fig. 6j, k and Supplementary Fig. 6h). Moreover, we observed a higher expression of proteins associated with antigen presentation, such as CD80 and HLA-DR, and higher levels of PD-1, implicated in the suppression of antiviral responses, in CVID patients compared to non-CVID individuals at progression (Fig. 6l).
All these results indicate that upregulation and persistent activation of specific inflammasome genes is a feature of the monocytic compartment in COVID-19 CVID patients, although we could detect increased caspase-1 activity only during the viral infection. In addition, the crosstalk between monocytes and B cells through BAFF and APRIL pathways is disrupted in CVID patients during progression. Finally, monocytes from CVID patients show increased antigen presenting and costimulatory molecules but also higher PD-1 levels during SARS-CoV-2 infection than non-CVID individuals.
Discussion
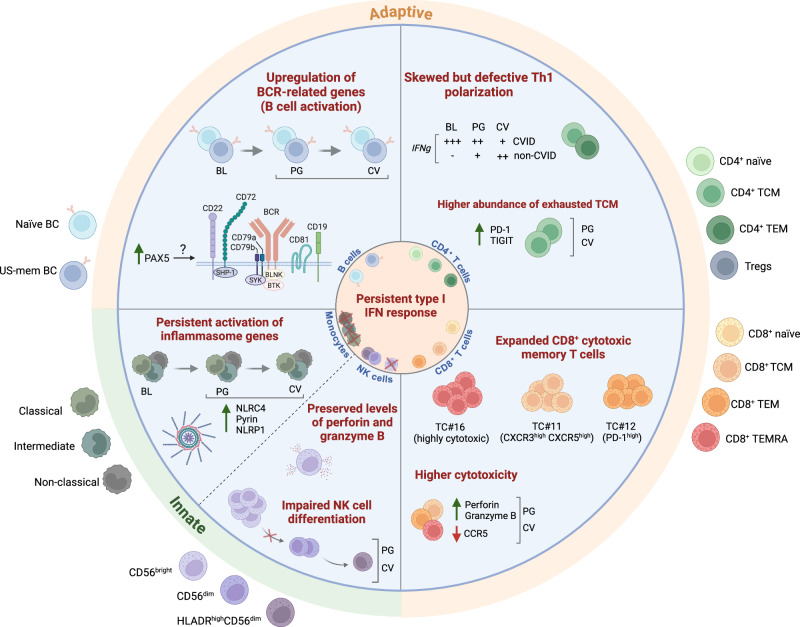
Our results show a broad dysregulation of both the innate and the adaptive immunity systems during SARS-CoV-2 infection and subsequent recovery in CVID patients, which gives us a unique insight into the effects of a viral challenge on the different immune cells in the absence of a proper humoral response, and in patients who face the infection with a pre-established immune-dysregulated background. An important clinical feature in this context is the long-lasting positivity to SARS-CoV-2 in most of our COVID-19 CVID patients, a characteristic also described by others74. We hypothesized that the poor humoral response contributes, at least in part, to the viral persistence, leading to an altered innate and adaptive immune activation and homeostasis even beyond infection into convalescence, as elucidated in this study. Thus, we observed a persistent type I IFN response at convalescence in all the immune compartments analyzed, except for monocytes. Following SARS-CoV-2 infection, we observed that CVID patients, compared to non-CVID individuals, displayed increased expression of BCR signaling-related genes in B cells, and persistent general activation of naïve B cells and expansion of CD21low B cells. Additionally, TCR signaling-related genes were upregulated in CD4+ T cells; however, this was combined with signs of exhaustion of the central memory compartment and a defective increase in Th1 response in CD4+ T cells, leading to an even reduced abundance of Th1 cells during convalescence compared to non-CVID COVID-19 patients. Despite this, there was high expression of IFNG during infection, likely due to the highly skewed Th1 phenotype at baseline that characterizes CVID patients54. CD8+ T cells show a strong clonal expansion of effector memory T cells, with increased expression of cytotoxicity-related genes. This was also preserved in NK cells despite a differentiation defect and elevated expression of immune checkpoint molecules. In monocytes, the upregulation of inflammasome-related genes in CVID patients with detectable increase in caspase-1 activity was the most remarkable finding (all summarized in Fig. 7).
Fig. 7. Overview of the dysregulated immune homeostasis in COVID-19 CVID patients.
Scheme depicting the altered immune homeostasis in CVID patients upon SARS-CoV-2 infection and recovery. BL baseline, PG progression, CV convalescence. Created in BioRender. Ballestar, E. (2024) https://BioRender.com/j59o518.
Many of the transcriptional alterations detected in COVID-19 CVID patients were persistent at the convalescence stage. The sustained immune activation including the type I IFN pathway, CD4+ T cell exhaustion, increased clonal expansion of cytotoxic CD8+ T cells, and inflammasome activity in monocytes has raised major concerns in the context of SARS-CoV-2 infection as some of these changes are associated with an increased acute pathology75. While this has not been the observation in most CVID patients, it is tempting to speculate how much the persistent alterations in the context of an ineffective clearance of viral infections might contribute to chronic immune dysregulation in complex CVID patients, as similar immunological alterations have been described also in these patients without overt viral infection57,76,77. This highlights the importance of studying the effect of disturbed humoral immunity on overall immune homeostasis in the context of SARS-CoV-2 infection and subsequent recovery.
Early type I IFN responses are important to control viral replication, and an impaired type I IFN response, therefore, leads to severe COVID-1975. Our results indicate that type I IFN response is very similar in COVID-19 CVID patients compared to mild COVID-19 non-CVID individuals at the progression stage, rendering type I IFN response independent from an intact humoral response at this stage of the infection. The persistent type I IFN response during convalescence might be one of the drivers of CD4+ T cell exhaustion, increased cytotoxic differentiation of CD8+ T cells, and the inflammasome activity in monocytes discussed below78.
The underlying poor specific humoral response was confirmed by the severely impaired anti-SARS-CoV-2 IgG and IgA antibody response, and a poor selection of BCR repertoires associated with SARS-CoV-2 neutralizing antibodies. In association with the absence of specific antibody response and general expansion of memory and plasmablasts which was observed in non-CVID individuals, we detected a persistent activation of “naïve” and US-memory B cells evidenced by the upregulation of BCR signaling-related genes, both activators and inhibitors, and the persistent expansion of a CD86+ “naïve” B cell cluster (BC#6) in some CVID patients. We detected an impaired increase of the canonical NF-κB signaling in these cells, a phenomenon observed already in CVID patients, independently of a known acute viral infection79. Analyzing the activity of additional dysregulated TF activities identified in this study could provide insights into the defective B cell responses observed in CVID patients, as a reference for future research in the field.
Reduced expression during SARS-CoV-2 infection of CXCR5, a chemokine receptor involved in B cell migration to B cell follicles80,81 and for appropriate B cell localization within the light zone of germinal centers82, might reflect and increased extrafollicular at the expense of a GC response which would be underlined by the increased expansion of CD21low B cells.
In the T cell compartment, at baseline, we observed the previously reported CD4+ Th1 bias in CVID patients with the CD21low phenotype and immune dysregulation54. However, despite this bias towards Th1, we identified a compromised Th1 polarization in CVID patients during and especially after SARS-CoV-2 infection, potentially impacting proper antiviral responses. Particularly, the CD4+ TCM cell compartment from CVID patients was strongly affected by the infection. While there is an expansion of the partially CCR4+CD25+ cluster TC#3, a subset potentially exhibiting regulatory characteristics, during infection in non-CVID individuals, this was absent in CVID patients and replaced by a strong shift towards TIGIT+PD-1+ T cells. In combination with an impaired CD40/CD40L interaction during SARS-CoV-2 infection, the increased expression of immune checkpoint molecules on CD4+ TCM cells is compatible with a dysfunctional TCM compartment as it has been described for HCV infection83 and may potentially impact the proper activation of immune subsets such as B cells and monocytes and the induction of a GC response. The impaired upregulation of CD40L on CD4+ T cells from CVID patients has been previously reported84–86.
In the CD8+ T cell compartment, we identified a partly clonal expansion of effector memory CD8+ T cells and transcriptional and flow cytometry analysis revealed a significant upregulation of cytotoxic genes like GZMB and PRF1 in CVID samples compared to non-CVID samples. It has been described how SARS-CoV-2-triggered complement activation creates an inflammatory milieu that drives differentiation of CD16+ cytotoxic T cells with high immunopathogenic potential33. Our analysis indicates that severe COVID-19 patients display a significant increase in the proportion of that pathogenic T cell subset, which was not observed in mild COVID-19 patients with independence of their PID status.
Similar to CD8+ T cells, we observed an increased signature of cytotoxicity-related genes in NK cells from CVID patients. This increased cytotoxicity signature was observed independently of the infection status and despite impaired differentiation into more mature CD56dim NK cell populations. A reduction in circulating NK cells in CVID has been previously reported87, but the altered differentiation status has not been well described and requires further investigation. Similarly, the function of NK cells in CVID remains to be fully understood, as we observed higher levels of the CD38 protein, which is associated with NK cell activation and mediates cytotoxic responses88, on one hand, and high levels of the inhibitory molecules TIGIT and TIM-3, which have been associated with exhaustion in NK cells from COVID-19 patients89,90, on the other. Finally, we examined the monocyte compartment of CVID patients in the context of COVID-19. Altered monocyte activation has been described in CVID patients22. Upon SARS-CoV-2 infection, these immunodeficient individuals displayed an enhanced and persistent gene upregulation of several components of different inflammasomes complexes, including pyrin (MEFV), NLRP1-CARD8 and NLRC4/IPAF-NAIP in the monocytic compartment, while there was no clear differential expression observed for AIM2 and NLRP3 related inflammasomes. It is only recently that the activation of the inflammasome via CARD8 and NLRP1 viral sensing in the context of mammalian effector-triggered immunity (ETI) has been described for rhinovirus, SARS-CoV2, and others91. NLRC4 activation in SARS-CoV-2 infection is reported to contribute to the upregulation of CD8692, which may also explain the observed upregulation of CD80 in our CVID patients. Conversely, the role of monocyte infection by SARS-CoV-2 via anti-spike antibody-mediated phagocytosis93, which induces NLRP3 and AIM2 inflammasomes activation, might be less significant in antibody-deficient patients who are unable to produce anti-spike antibodies. Furthermore, our findings indicate a more pronounced and sustained activation of the transcription factor PU.1 in monocytes derived from CVID patients compared to their non-CVID counterparts which might contribute to the increased transcriptional regulation of the IL1B gene and inflammasome activation as previously described in myeloid cells94,95. The proinflammatory transcriptome observed in the myeloid compartment of COVID-19 CVID patients persisted at the convalescent stage, although our investigation by triggering inflammasome activity with nigericin did not elicit increased caspase-1 activity at this time. This is compatible with a prolonged inflammasome priming, but not inflammasome activation. This phenomenon might be also attributed to suboptimal activation of the CARD-containing sensors NLRP1, NLRC4, and CARD8 related-inflammasomes by nigericin, allowing for other triggers in the context of a persisting viral infection to cause a sustained inflammasome activity also during convalescence. Future studies will determine whether poor viral clearance in immunocompromised patients contributes to chronic immune activation even in the absence of overt infection as it is observed in CVID.
It is surprising that the sustained SARS-CoV-2 positivity and the associated persistent type I IFN response, preserved or increased cytotoxicity in NK cells and CD8+ T cells respectively, and inflammasome activation observed here in the COVID-19 CVID patients, did not cause a more severe course of the disease. As this clinical outcome has also been reported in patients with agammaglobulinemia and other CVID patients, who similarly experienced prolonged yet not more severe illness96–98, it remains to be understood what protected most of these patients from severe COVID-19. It is plausible that the expansion and enhanced cytotoxic response in the CD8+ T cell compartment, along with the skewed Th1 polarization (associated with higher levels of IFN-γ), and with the observed robust type I IFN response in these CVID patients, might have contributed to the control of the viral infection. Nonetheless, while the defective humoral response in CVID patients may not significantly increase the severity of COVID-19, and might be beneficial due to the impaired production of anti-IFN autoantibodies observed in these patients40, it appears to elevate the risks of both prolonged infection and possibly subsequent reinfection28.
In summary, our investigation detected a strong general impact of the absence of humoral immunity during and subsequent to SARS-CoV-2 infection on the innate and adaptive immune system in patients with CVID. Our study provides valuable insights into the persistent immune dysregulation observed in CVID patients following SARS-CoV-2 infection, yet there are limitations that warrant consideration. The small cohort size limits the generalizability of our findings and restricts the statistical power needed to detect subtle immune variations across different CVID presentations. Although longitudinal single-cell RNA-seq and spectral flow cytometry provided a high-resolution view of immune alterations, the results may need further validation in larger CVID cohorts and with other viral infections to assess their specificity and reproducibility.
Our hypothesis, displayed in Fig. 7, is that the preserved type I IFN and cytotoxic response allows patients with agammaglobulinemia and CVID to finally control novel viruses like SARS-CoV2 without a severe course in most patients. The severely impaired humoral immunity seems, however, to predispose these patients to a longer persistence of the virus and thereby to prolonged immune stimulation, although this needs to be confirmed in future studies with larger cohorts. This was suggested by the persisting type I IFN response which might in turn contribute to the observed prolonged broad T and B cell activation, increased exhaustive phenotype of CD4+ central memory T cells, expression of cytotoxic signature in CD8+ and NK cells and inflammasome activation in monocytes. The decreasing Th1 signature during the course of the viral infection in patients naturally prone to type 1 immunity was remarkable. This reduction might be related to the decreased CD40L/CD40 communication, and likely reflects the increasing exhaustion of the CD4+ T cell compartment in the context of chronic viral infections, as has been suggested in other viral infections83 and also for bacterial infections in complex CVID77. These findings open new avenues for exploring triggers and pathomechanisms of the immune dysregulation in complex CVID patients and prompt further investigations in other patient cohorts with impaired humoral immunity.
Methods
Patients and ethics approval
Human blood samples were collected from CVID patients diagnosed according to the European society of immune deficiencies (ESID) criteria99,100. Samples were collected before (baseline), during (progression) and after (convalescence) SARS-CoV-2 infection. They were collected at the Medical Center-University of Freiburg, Germany. The CVID patients in our cohort were all infected between April 2020 and May 2021, when the wild type strain and the alpha variant of SARS-CoV-2 were predominant in Germany. SARS-CoV-2 infection diagnosis was based on nasopharyngeal PCR. All patients included in the study had a longstanding diagnosis of CVID (age range 32-70 years old). All CVID patients but CVID4 belonged to the complex form of CVID with signs of immune dysregulation listed in the Supplementary Data 1a. The EUROclass distribution showed mostly low B cells or an expansion of CD21low B cells as it is expected in this clinical subgroup of patients, especially when they are older as the B cell numbers in this subgroup often decrease. Thus, the patients were typical CVID patients. The non-CVID donors analyzed were infected between January 2020 and April 2021, also when the wild-type strain and the alpha variant were predominant. In addition, none of the CVID patients included in the study, nor the reference non-CVID individuals, received anti-SARS-CoV-2 vaccination (Supplementary Data 1a). At the time of infection of the patients included in this study, no known antiviral was available, and various drugs, including HCQ and ivermectin, were administered under compassionate use before their efficacy or lack thereof was demonstrated (Supplementary Data 1a). All donors received oral and written information about the possibility that their blood would be used for research purposes, and any questions that arose were answered. The study was approved by the local Ethics Committees of the participating center (507/16; 282/11). Prior to sample collection, donors signed a consent form approved by the Ethics Committee of their corresponding center, which adhered to the principles set out in the WMA Declaration of Helsinki. There was no compensation to participants.
Sample collection
PBMCs were obtained from peripheral blood by Ficoll gradient using Lymphocyte Isolation Solution. Once PBMCs had been isolated, all samples were cryopreserved and stored at −150 °C. In the case of the publicly available datasets of non-CVID individuals, data from samples at baseline, progression or convalescence stages were used but they did not correspond with paired samples.
Single-cell capture
Cryopreserved PBMCs were thawed rapidly in a 37 °C water bath, then slowly diluted in pre-warmed growth medium, centrifuged, resuspended in PBS + 0.04% BSA and filtered with a 40 um Bel-art Flowmi strainer. In order to optimize single-cell reagents and to minimize batch effects, PBMC samples from different donors were pooled. Cells from different donors were counted and concentration adjusted to load 50,000 cells on the 10X-Genomics Chromium Station. In cases where PBMCs from different donors were pooled, a fraction was taken to isolate genomic DNA for genotyping, using Illumina Infinium Global Screening Array, and the other fraction was used to generate single-cell gel beads-in-emulsion (GEMs). Genomic DNA was isolated from PBMCs for genotyping using a Maxwell® 16 Blood DNA Purification Kit from Promega following the manufacturer’s instructions.
Library generation and sequencing
Libraries were constructed following the manufacturer’s protocol for the Chromium Next GEM Single Cell V(D)J Reagent Kits v1.1 with Feature Barcode technology for Cell Surface Protein (10X Genomics Rev E), but with two amendments: the amount of SI primer was doubled, and the number of PCR cycles was set at 7. Samples were sequenced using the Illumina NovaSeq 6000, where cellular gene expression, as well as B cell and T cell clonality were simultaneously profiled. We targeted ∼300 M raw reads per sample (∼60,000 raw reads per cell) with cycle numbers of 100 for read 1, 100 for read 2, and 100 for the index read.
Single-cell data alignment, quantification, and quality control
For the CVID samples, the single-cell transcriptome data were aligned and quantified by Cell Ranger v3.1 using GRCh38 (Ensembl 93) concatenated to the SARS-Cov-2 genome as a reference. Pooled donor samples were deconvolved using Souporcell101, which yielded a genotype variant that allowed donor identity to be matched across samples. Cells that could not be explained by a single genotype were considered doublets and removed before analysis. Additionally, the two cohorts of PBMCs from non-CVID donors were obtained from publicly available scRNA-seq datasets of individuals without any known immunodeficiency (Supplementary Data 1).
Cells with the following features were filtered out: more than 20% mitochondrial gene content, more than 7500 counts per cell, fewer than 500 genes, or more than 5000 genes. Additionally, genes expressed in fewer than three cells were removed. Downstream analysis was performed using the Seurat R package102 (version 4.3.0.1).
Deconvolution of donors in pooled scRNA-seq samples
Pooled scRNA-seq samples containing cells from multiple individuals were demultiplexed using souporcell. Briefly, the algorithm identifies genotypic differences between single cells by variant calling aligned reads using STAR103 and generating a VCF (Variant Call Format) file using Freebayes (Marth 2012). Souporcell was run with the following command for all samples (argument $1 corresponds to sample ID, and N is the number of multiplexed individuals in a sample): /software/singularity-v3.5.1/bin/singularity exec./souporcell.sif./souporcell_pipeline.py -i./cellranger302_count_$1_GRCh38-1_2_0/possorted_genome_bam.bam -b./cellranger302_count_$1_GRCh38-1_2_0/filtered_feature_bc_matrix/barcodes.tsv -f./refdata-cellranger-GRCh38-1.2.0/fasta/genome.fa -t 8 -o souporcell_result_$1 -k N --skip_remap True --common_variants./filtered_2p_1kgenomes_GRCh38.vcf, where the last VCF file with common variants was downloaded as per instructions in https://github.com/wheaton5/souporcell with the following command: wget --load-cookies /tmp/cookies.txt “https://docs.google.com/uc?export=download&confirm =$(wget-quiet-save-cookies/tmp/cookies.txt-keep-session-cookies-no-check-certificate ‘ https://docs.google.com/uc?export=download&id=13aebUpEKrtjliyT9rYzRijtkNJVUk5F_’-O-|sed-rn’s/. *confirm = ([0-9A-Za-z_]+).*/\1\n/p’)&id=13aebUpEKrtjliyT9rYzRijtkNJVUk5F_”-Ocommon_variants_grch38.vcf & & rm-rf/tmp/cookies.txt.
Furthermore, to find out the exact donor identity of each donor’s barcode cluster in souporcell results, the cells in parallel have undergone genotyping using Illumina Infinium Global Screening Array. In order to unambiguously identify every individual in the pooled samples, each donor’s variants were separated from the pooled VCF and each single-donor VCF was matched to the genotype data using PLINK104. This software matches each souporcell sample with the genotype data giving a concordance ratio (based on the similarity of the variants) that allows us to distinctly identify each sample with each donor ID.
Cell type identification and cluster annotation
Our scRNA-seq data from CVID patients (CVID cohort) was integrated with both non-CVID 1 and non-CVID 2 cohorts independently. The integration was performed by finding anchors between batches from the CVID cohort, and samples from the non-CVID cohort following the Seurat pipeline. Both objects were normalized (NormalizeData) considering counts per million (CPM), top 2000 highest variable genes were selected based on the vst Seurat algorithm (FindVariableFeatures) and data was scaled (ScaleData). Principal component analysis (PCA) was performed on the list of features used to integrate the cohorts (SelectIntegrationFeatures), the first 50 principal components (PCs) were used. Cell clustering was generated with the Louvain algorithm (FindNeighbors, FindClusters) and then Uniform Manifold Approximation and Projection (UMAP) was performed for visualization, the top 50 PCs were selected. VlnPlot of known canonical markers were used for visualization with Seurat.
Preliminary annotation for the obtained unsupervised clusters by Seurat was assigned by manual annotation with known cell-specific marker genes (Fig. 1b, c).
For a more detailed evaluation, subclustering analysis of B cells, T cells, NK cells and myeloid cells was performed following the same procedure as above. In that case the top 30 PCs were used for dimensional reduction and clustering. Additionally, for the CVID + non-CVID 2 cohort samples with a reduced number of cells were filtered out. Similarly, cell clusters were annotated based on known cell markers12,33,60. Common CVID cells in the two cohorts were used to validate cell cluster annotation for reproducibility. Dotplots of selected and specific genes per cluster were visualized with Seurat.
Differential gene expression analysis
Differentially expressed genes (DEGs) were calculated with FindMarkers using Wilcoxon Rank Sum test and Bonferroni correction. DEGs were calculated by comparing the different samples grouped by SARS-CoV-2 stage (baseline, progression, or convalescence), PID status (CVID or non-CVID), and cell cluster. In addition, we calculated DEGs comparing CVID BL vs non-CVID BL. Only DEGs in non-CVID 1 cohort that were validated in non-CVID 2 cohort (FDR ≤ 0.1 & Log2(fold change) sign equal in both non-CVID cohorts) were considered for downstream analysis. Only cell clusters containing a minimum of 10 cells from at least 3 different patients in each comparison group (based on PID, SARS-CoV-2 infection stage, and COVID-19 severity) were used for downstream analysis. For scRNA-seq analysis of the B cell compartment, only the three CVID donors with B cells (CVID1, CVID2 and CVID4) were used for DEGs calculation and downstream analysis. Sex-stratified analysis was not performed due to the limited size of the cohort.
Gene Ontology (GO) analysis
For each immune cell compartment, once DEGs were calculated between baseline and progression in the CVID cohort or in the non-CVID 1 cohort, we selected only those DEGs in non-CVID 1 cohort that were also validated in non-CVID 2 cohort. Afterward, we clustered these DEGs based on their expression profile during SARS-CoV-2 infection and grouped them in three different categories: ‘BL equal’, comprising genes that exhibit similar expression levels in CVID and non-CVID individuals at BL; ‘BL CVID up’, for genes that are upregulated in CVID patients compared with COVID-19 controls; ‘BL CVID down’, including genes that are downregulated in CVID patients compared to non-CVID individuals at BL. Next, we performed gene ontology (GO) category enrichment analysis in each of the gene clusters obtained. GO terms enriched in each cluster were obtained using the R package clusterProfiler105 (version 4.14.0). with the default parameters. Annotation Dbi R package “org.Hs.eg.db” (version 3.20.0) was used to map gene identifiers. All significantly enriched GO terms are collected in Supplementary Data 4. Among the entire list of significantly enriched GO categories, we selected those containing particular labels of immunological features. We selected common labels across compartments containing the following word patterns: “interleukin”, “interferon”, “necrosis”, “adaptive”, “inflamm”, “toll”,”Fc”, “pattern”, “chemo”, migration”, “glycolysis”, “tricarbo”, “fatty”, “electron”, and “oxidative phosphorylation”. Furthermore, in the case of specific immune cell compartments, the word patterns additionally used were: a) for B cells: “humoral”, “b cell”, “bcr”, “differentiation” and “immuno”, b) for T cells: “t cell”, “vir”, “cyto”, “kill”, “antigen”, “polarization”, “Th”, “differentiation” and “prolifer”, c) for NK cells: “natural” and “kill”, and d) for myeloid cells: “present”. To visualize the pattern of selected GO categories along all stages of SARS-CoV-2 in CVID and non-CVID individuals, we used AddModuleScore function. For score calculation, lists of genes from selected GO categories for each population were obtained from Gene Set Enrichment Analysis (GSEA) and AmiGO Gene Ontology. In order to correct for different cell numbers per patient group, we calculated gene expression scores in each single-cell and depicted the mean of these scores by PID status (CVID, non-CVID), stage (baseline, progression or convalescence) and cell population.
Cytotoxic and anti-fibrotic response analysis
Cytotoxic response was calculated with a list of leukocyte-mediated cytotoxic-related genes from the GSEA (GO:0001909). Likewise, pro-fibrotic activity was calculated with a gene list of profibrotic genes (AREG, DUSP2, ZFP36L2, CXCR4, and TSC22D3)60. The scores were obtained with AddModuleScore. Then, we calculated the mean of the obtained scores per each patient, stage (baseline, progression or convalescence) and cell population in order to correct potential differences in cell numbers.
CD4+ T cell polarization analysis
For the T helper cell polarization Th0, Th1, Th2, and Th17 cells were annotated within the CD4+ T cells population. TBX21, TNF, and IFNG were used as Th1 markers; GATA3, IL5, and IL4 were used as Th2 markers and RORC and IL17F as Th17 markers. Each cell was annotated according to the maximum cell score value obtained using AddModuleScore, with a defined threshold of an average expression value above 0.4. In case a cell did not reach the 0.4 expression threshold, it was considered as Th0. To correct cell differences by patient, the mean and the standard error of the Th cells percentage per stage and patient were calculated.
Transcription factor activity analysis
Inference of TF activities from expression values were calculated using decoupleR47, a curated resource of TFs and their targets compiled from various sources including the literature, ChIP-Seq peaks, in silico predictions, as well as gene expression data, was used to estimate TF activities from combined expression values of gene targets. We calculated TF activity at a single-cell resolution using the default parameters of run_ulm function. To address potential variations due to different cell numbers per patient, we conducted subsampling to ensure an equal number of cells across patients and stages. The mean scores from significant TFs (Supplementary Data 9) were visualized as a heatmap. Additionally, TF scores of subsampled single cells for selected TFs were depicted in box plots.
To define canonical NF-κB target genes that are specifically activated in B cells, we selected DEGs from the publicly available dataset GSE61608 that overlap with both RELA and NFKB1 regulons. These target genes were named as “NFKB1/RELA” regulon.
T cell receptor and B cell receptor clonality
Single-cell TCR and BCR data were processed with the Cell Ranger v3.1 vdj pipeline using GRCh38 as a reference. For BCR downstream analysis, we calculated the percentage of VDJ usage upon SARS-CoV-2 infection by each patient and stage, and represented it as a box plot.
TCR downstream analysis was performed using scRepertoire106 R package (version 1.10.1). For the TCR gene analysis in the CVID cohort, the absolute frequency of clonotypes per SARS-CoV-2 infection stage and patient was calculated by combineTCR and combineExpression, removing any cell barcode with more than 2 immune receptor chains. We determined expansion of clonotypes as: single equal to 1, small between 1 and 5, medium between 5 and 20, large between 20 and 100, and hyperexpanded between 100 and 500. The relative frequency of were calculated per SARS-CoV-2 infection stage and cluster, filtering out the cells without TCR. Clonal diversity was also calculated using clonalDiversity, showing the Shannon entropy index in box plots.
Pseudotime trajectory analysis
Monocle 3 package107,108 (version 1.3.1) was used to calculate pseudotime trajectories of myeloid and NK cells. The root nodes of the trajectories were selected as Classical Monocytes and CD56bright NK cells, respectively based on literature109. Genes differentially expressed across the trajectory were calculated using Moran’s I test (graph_test), only genes with positive value were selected.
Discriminant Regulon Expression Analysis (DoRothEA) database was used to identify regulons for IRF9, STAT1, STAT2, and SPI1, using only activator genes with A and B confidence scores. These gene sets were taken to calculate scores with AddModuleScore, and then visualize them based on the pseudotime order.
Statistical analysis
Statistical analyses were performed in R (v4.3.0). Differences in the representation of cell types was reported using two-tailed Welch’s t tests, comparing the proportion of cell types in each patient and cluster between the CVID and non-CVID cohorts.
Data visualization
Unsupervised subsamplings have been performed for all splitted UMAP visualizations, selecting the lowest number of cells per CVID status, COVID-19 severity, and infection stage.
For data visualization the R packages Seurat, ggplot2 (version 3.5.1), monocle3, and scRepertoire have been used. In box plots, whiskers correspond with the minimum and maximum values of the dataset. The box is drawn from Q1 to Q3 with a horizontal line to indicate the median.
Spectral cytometry
All antibodies used for staining are listed in Supplementary Data 7. One million cryopreserved PBMCs were thawed rapidly in a 37 °C water bath, then slowly diluted in pre-warmed growth medium, centrifuged, resuspended in 3 mL of PBS. The centrifugation steps after each wash were performed for 5 min at RT 300 g, for viability and intra-staining steps, and 400 g, for surface staining steps.
Cells were stained for viability (Supplementary Data 7) for 15 min at RT in the dark, then washed with 3 mL of FACSflow and centrifuged. After viability staining, cells were sequentially stained by incubating with “Post-Viability Mix” for 10 mins, “Chemokines Mix” for 10 mins, and “General Master Mix” for 20 mins. Then, cells were washed with FACSflow twice. Finally, for the myeloid + B cells cytometry panel, cells were incubated for 10 mins at RT with 300 μL of PBS + 1% PFA, washed with FACSflow, incubated with “PostPFA Mix” for 10 mins at RT and washed. In parallel, for the T + NK cells cytometry panel, we used Dako’s IntraStain (Agilent, K2311) protocol, following manufacturer’s guides. Cells were acquired on a Cytek Aurora 5-laser spectral flow cytometer.
Spectral cytometry analysis was performed using FlowJo v.10.10 for initial gating (viability, SSC-A/FSC-A doublets, and cell-lineage markers), and for the UMAP and Phenograph clustering (FlowJo plugins), using all protein markers as input parameters. UMAP calculation was made using 10 NN (nearest neighbors), 0.1 min_dist and 2 n_components. For Phenograph calculation k = 250 was set. Downstream analysis was performed using flowCore (version 2.18.0), ggplot2 (version 3.5.1), pheatmap (version 1.0.12), and ggpubr (version 0.6.0) R packages.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank the CERCA Program/Generalitat de Catalunya and the Josep Carreras Foundation for institutional support. This publication is part of the Human Cell Atlas:www.humancellatlas.org/publications. The project that gave rise to these results has received funding from “la Caixa” Foundation under the grant agreement LCF/PR/HR22/52420002, Spanish Ministry of Science and Innovation (grant number PID2020-117212RB-I00/AEI/10.13038/. 501100011033) (E.B.), by the Wellcome Trust Grant 206194 and 108413/A/15/D (R.V.-T.), Instituto de Salud Carlos III (ISCIII), Ref. AC18/00057, associated. with i-PAD project (ERARE European Union program) (E.B.), and the Chan Zuckerberg Initiative (grant 2020-216799) (R.V.-T. and E.B.). This publication has also been supported by the Unstoppable campaign of the Josep Carreras Leukemia Foundation. We are indebted to the donors for participating in this research. Some samples for this project were obtained from the CCI-Biobank, a partner biobank of the University Medical Center Freiburg and Medical Faculty “Center for Biobanking—FREEZE”. We thank the producers of all publicly available datasets utilized in this study and acknowledge their contribution in our research.
Author contributions
J.R.-U., R.V.-T., K.W. and E.B. conceived the study; J.R.-U., L.C. and G.G.-T. performed 10X Chromium experiments; J.R.-U., G.G.-T., R.H., T.P. and E.P. performed sample and library preparation; E.A.-L. and J.M. performed patient genotyping; J.R-U., J.C.-S., C.L.C., L.-F. H., and C.C.-F. performed computational analysis; J.R.-U., J.C.-S., C.L.C., L.-F. H., B.K., C.C.-F., G.G.-T., K.W., R.V.-T. and E.B. analyzed and interpreted the data; B.K., M.H., A.D. and K.W. contributed with clinical material and clinical interpretation of the results; J.R-U. and E.B. wrote the manuscript with contributions from J.C.-S., C.L.C., L.-F. H., B.K, K.W. and R.V-T.; R.V.-T., K.W. and E.B. co-directed the study. All authors read and accepted the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The chromium droplet-based scRNA-seq data in this study have been deposited in EGA database under accession code EGAD50000000543. Interactive visualizations of single-cell transcriptomic datasets from this study can be accessed via https://www.immunodeficiencycellatlas.org. Spectral flow cytometry files have been deposited in FlowRepository under accession code FR-FCM-Z8F8. The Whole Exome Sequencing (WES) data are not publicly available due to ethical and privacy regulations. Access to the data can be granted upon request by contacting the corresponding authors. Requests for access will be reviewed and responded to within 30 days, and any approved access will be governed by a Data Use Agreement, which may impose specific conditions on the permissible uses of the data. We used additional scRNA-seq data publicly-available datasets with the following accession numbers: HRA000150, CRA002497, GSE158055, E-MTAB-10026, GSE196456, GSE154567 and GSE175450. Source data are provided with this paper.
Code availability
The code used in the analyses can be found at https://github.com/BALLESTARLAB/CVID-COVID110.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Javier Rodríguez-Ubreva, Josep Calafell-Segura, Celia L. Calvillo, Baerbel Keller.
These authors jointly supervised this work: Roser Vento-Tormo, Klaus Warnatz, Esteban Ballestar.
Contributor Information
Javier Rodríguez-Ubreva, Email: jrodriguez@carrerasresearch.org.
Roser Vento-Tormo, Email: rv4@sanger.ac.uk.
Klaus Warnatz, Email: klaus.warnatz@uniklinik-freiburg.de.
Esteban Ballestar, Email: eballestar@carrerasresearch.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54732-x.
References
- 1.Gavriatopoulou, M. et al. Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med.20, 493–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehandru, S. & Merad, M. Pathological sequelae of long-haul COVID. Nat. Immunol.23, 194–202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan, A. T. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep.34, 108728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, J. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature587, 270–274 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Rydyznski Moderbacher, C. et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell183, 996–1012.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng, Y. et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol.21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartley, G. E. et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol.5, eabf8891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin, M. J. et al. Intramuscular AZD7442 (Tixagevimab-Cilgavimab) for prevention of Covid-19. N. Engl. J. Med.386, 2188–2200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien, M. P. et al. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N. Engl. J. Med.385, 1184–1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P. et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med.384, 229–237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougan, M. et al. Bamlanivimab plus Etesevimab in Mild or Moderate Covid-19. N. Engl. J. Med.385, 1382–1392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell182, 1419–1440.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultze, J. L. & Aschenbrenner, A. C. COVID-19 and the human innate immune system. Cell184, 1671–1692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sette, A. & Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell184, 861–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science370, eabd4570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milito, C. et al. Clinical outcome, incidence, and SARS-CoV-2 infection-fatality rates in Italian patients with inborn errors of immunity. J. Allergy Clin. Immunol. Pract.9, 2904–2906.e2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidel, M. G. et al. The European Society for Immunodeficiencies (ESID) Registry working definitions for the clinical diagnosis of inborn errors of immunity. J. Allergy Clin. Immunol. Pract.7, 1763–1770 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Bonilla, F. A. et al. International Consensus Document (ICON): Common variable immunodeficiency disorders. J. Allergy Clin. Immunol. Pract.4, 38–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameratunga, R. Assessing disease severity in Common Variable Immunodeficiency Disorders (CVID) and CVID-Like disorders. Front. Immunol.9, 2130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman, Ea. L. et al. T cell phenotypes in patients with common variable immunodeficiency disorders: associations with clinical phenotypes in comparison with other groups with recurrent infections. Clin. Exp. Immunol.170, 202–211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azizi, G. et al. Circulating Helper T-Cell Subsets and Regulatory T Cells in Patients With Common Variable Immunodeficiency Without Known Monogenic Disease. J. Investig. Allergol. Clin. Immunol.28, 172–181 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Barbosa, R. R. et al. Monocyte activation is a feature of common variable immunodeficiency irrespective of plasma lipopolysaccharide levels. Clin. Exp. Immunol.169, 263–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cambronero, R., Sewell, W. A., North, M. E., Webster, A. D. & Farrant, J. Up-regulation of IL-12 in monocytes: a fundamental defect in common variable immunodeficiency. J. Immunol.164, 488–494 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Marcus, N. et al. Minor clinical impact of COVID-19 pandemic on patients with primary immunodeficiency in Israel. Front. Immunol.11, 614086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drabe, C. H. et al. Low morbidity in Danish patients with common variable immunodeficiency disorder infected with severe acute respiratory syndrome coronavirus 2. Infect. Dis.53, 953–958 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Cohen, B. et al. COVID-19 infection in 10 common variable immunodeficiency patients in New York City. J. Allergy Clin. Immunol. Pract.9, 504–507.e1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shields, A. M., Burns, S. O., Savic, S. & Richter, A. G. & UK PIN COVID-19 Consortium. COVID-19 in patients with primary and secondary immunodeficiency: The United Kingdom experience. J. Allergy Clin. Immunol.147, 870–875.e1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katzenstein, T. L. et al. Outcome of SARS-CoV-2 infection among patients with common variable immunodeficiency and a matched control group: A Danish nationwide cohort study. Front. Immunol.13, 994253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, J.-Y. et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol.21, 1107–1118 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Yu, K. et al. Dysregulated adaptive immune response contributes to severe COVID-19. Cell Res.30, 814–816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen, W. et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov.6, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephenson, E. et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med.27, 904–916 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georg, P. et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell185, 493–512.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoo, W. H. et al. Tracking the clonal dynamics of SARS-CoV-2-specific T cells in children and adults with mild/asymptomatic COVID-19. Clin. Immunol.246, 109209 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, C. et al. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep.34, 108590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehr, C. et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood111, 77–85 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Yesillik, S., Agrawal, S., Gollapudi, S. V. & Gupta, S. Phenotypic analysis of CD4+ Treg, CD8+ Treg, and Breg cells in adult common variable immunodeficiency patients. Int. Arch. Allergy Immunol.180, 150–158 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Arduini, S., Dunne, J., Conlon, N., Feighery, C. & Doherty, D. G. Mucosal-associated invariant T cells are depleted and functionally altered in patients with common variable immunodeficiency. Clin. Immunol.176, 23–30 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Yong, P. F. K. et al. Selective deficits in blood dendritic cell subsets in common variable immunodeficiency and X-linked agammaglobulinaemia but not specific polysaccharide antibody deficiency. Clin. Immunol.127, 34–42 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science370, eabd4585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol.15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, J. S. & Shin, E.-C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol.20, 585–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galbraith, M. D. et al. Specialized interferon action in COVID-19. Proc. Natl. Acad. Sci. USA.119, e2116730119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giovannone, N. et al. Galectin-9 suppresses B cell receptor signaling and is regulated by I-branching of N-glycans. Nat. Commun.9, 3287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao, A. et al. Galectin-9 binds IgM-BCR to regulate B cell signaling. Nat. Commun.9, 3288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv.2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Revilla-I-Domingo, R. et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J.31, 3130–3146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, M. et al. Structural basis of a shared antibody response to SARS-CoV-2. Science369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao, Y. et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B Cells. Cell182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren, X. et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell184, 1895–1913.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau, D. et al. Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation. Sci. Immunol.2, eaai8153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zurbuchen, Y. et al. Human memory B cells show plasticity and adopt multiple fates upon recall response to SARS-CoV-2. Nat. Immunol.24, 955–965 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unger, S. et al. The TH1 phenotype of follicular helper T cells indicates an IFN-γ-associated immune dysregulation in patients with CD21low common variable immunodeficiency. J. Allergy Clin. Immunol.141, 730–740 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Maseda, D., Ricciotti, E. & Crofford, L. J. Prostaglandin regulation of T cell biology. Pharmacol. Res.149, 104456 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Thebault, P. et al. The C-type lectin-like receptor CLEC-1, expressed by myeloid cells and endothelial cells, is up-regulated by immunoregulatory mediators and moderates T cell activation. J. Immunol.183, 3099–3108 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Klocperk, A. et al. Distinct CD8 T cell populations with differential exhaustion profiles associate with secondary complications in common variable immunodeficiency. J. Clin. Immunol.42, 1254–1269 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuri-Cervantes, L. et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol.5, eabd7114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maucourant, C. et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol.5, eabd6832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krämer, B. et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity54, 2650–2669.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol.14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ong, S. et al. Natural killer cells limit cardiac inflammation and fibrosis by halting eosinophil infiltration. Am. J. Pathol.185, 847–861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glässner, A. et al. Impaired CD4+ T cell stimulation of NK cell anti-fibrotic activity may contribute to accelerated liver fibrosis progression in HIV/HCV patients. J. Hepatol.59, 427–433 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Yang, M., Vanderwert, E., Kimchi, E. T., Staveley-O’Carroll, K. F. & Li, G. The important roles of natural killer cells in liver fibrosis. Biomedicines11, 1391 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brownlie, D. et al. Comparison of lung-homing receptor expression and activation profiles on NK cell and T cell subsets in COVID-19 and influenza. Front. Immunol.13, 834862 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlin, L. E., Hemann, E. A., Zacharias, Z. R., Heusel, J. W. & Legge, K. L. Natural killer cell recruitment to the lung during influenza a virus infection is dependent on CXCR3, CCR5, and Virus Exposure Dose. Front. Immunol.9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deaglio, S. et al. Human CD38 and CD16 are functionally dependent and physically associated in natural killer cells. Blood99, 2490–2498 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Stanietsky, N. et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA.106, 17858–17863 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ndhlovu, L. C. et al. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood119, 3734–3743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gleason, M. K. et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood119, 3064–3072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kvedaraite, E. et al. Major alterations in the mononuclear phagocyte landscape associated with COVID-19 severity. Proc. Natl. Acad. Sci. USA.118, e2018587118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vanderbeke, L. et al. Monocyte-driven atypical cytokine storm and aberrant neutrophil activation as key mediators of COVID-19 disease severity. Nat. Commun.12, 4117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brauns, E. et al. Functional reprogramming of monocytes in patients with acute and convalescent severe COVID-19. JCI insight7, e154183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan, M. et al. Persistent SARS-CoV-2 PCR positivity despite anti-viral treatment in immunodeficient patients. J. Clin. Immunol.43, 1083–1092 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sefik, E. et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park, J. et al. Interferon signature in the blood in inflammatory common variable immune deficiency. PLoS One8, e74893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perreau, M. et al. Exhaustion of bacteria-specific CD4 T cells and microbial translocation in common variable immunodeficiency disorders. J. Exp. Med.211, 2033–2045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science369, 718–724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keller, B. et al. Disturbed canonical nuclear factor of κ light chain signaling in B cells of patients with common variable immunodeficiency. J. Allergy Clin. Immunol.139, 220–231.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Förster, R. et al. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell87, 1037–1047 (1996). [DOI] [PubMed] [Google Scholar]
- 81.Ansel, K. M. et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature406, 309–314 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Allen, C. D. C. et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol.5, 943–952 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Ackermann, C. et al. HCV-specific CD4+ T cells of patients with acute and chronic HCV infection display high expression of TIGIT and other co-inhibitory molecules. Sci. Rep.9, 10624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodríguez-Ubreva, J. et al. Single-cell Atlas of common variable immunodeficiency shows germinal center-associated epigenetic dysregulation in B-cell responses. Nat. Commun.13, 1779 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thon, V. et al. Defective integration of activating signals derived from the T cell receptor (TCR) and costimulatory molecules in both CD4+ and CD8+ T lymphocytes of common variable immunodeficiency (CVID) patients. Clin. Exp. Immunol.110, 174–181 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brugnoni, D. et al. CD4+ cells from patients with Common Variable Immunodeficiency have a reduced ability of CD40 ligand membrane expression after in vitro stimulation. Pediatr. Allergy Immunol.7, 176–179 (1996). [DOI] [PubMed] [Google Scholar]
- 87.Ebbo, M. et al. Low circulating natural killer cell counts are associated with severe disease in patients with common variable immunodeficiency. EBioMedicine6, 222–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sconocchia, G. et al. CD38 triggers cytotoxic responses in activated human natural killer cells. Blood94, 3864–3871 (1999). [PubMed] [Google Scholar]
- 89.Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med.26, 1070–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li, M. et al. Elevated exhaustion levels of NK and CD8+ T cells as indicators for progression and prognosis of COVID-19 disease. Front. Immunol.11, 580237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barry, K., Murphy, C. & Mansell, A. NLRP1- A CINDERELLA STORY: a perspective of recent advances in NLRP1 and the questions they raise. Commun. Biol.6, 1274 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsukalov, I. et al. NFκB and NLRP3/NLRC4 inflammasomes regulate differentiation, activation and functional properties of monocytes in response to distinct SARS-CoV-2 proteins. Nat. Commun.15, 2100 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Junqueira, C. et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature606, 576–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ha, S.-D., Cho, W., DeKoter, R. P. & Kim, S. O. The transcription factor PU.1 mediates enhancer-promoter looping that is required for IL-1β eRNA and mRNA transcription in mouse melanoma and macrophage cell lines. J. Biol. Chem.294, 17487–17500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yashiro, T. et al. PU.1 and IRF8 modulate activation of NLRP3 inflammasome via regulating its expression in human macrophages. Front. Immunol.12, 649572 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morita, R. et al. COVID-19 relapse associated with SARS-CoV-2 evasion from CD4+ T-cell recognition in an agammaglobulinemia patient. iScience26, 106685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Speletas, M. et al. B cells and COVID-19: Lessons from patients with Agammaglobulinemia and the study of functional B-cell polymorphisms. J. Investig. Allergol. Clin. Immunol.32, 53–55 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Quinti, I. et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J. Allergy Clin. Immunol.146, 211–213.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bousfiha, A. et al. The 2017 IUIS phenotypic classification for primary immunodeficiencies. J. Clin. Immunol.38, 129–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ameratunga, R. et al. Comparison of diagnostic criteria for common variable immunodeficiency disorder. Front. Immunol.5, 415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods17, 615–620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res.9, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Björkström, N. K., Strunz, B. & Ljunggren, H.-G. Natural killer cells in antiviral immunity. Nat. Rev. Immunol.22, 112–123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calvillo C. L. & Calafell-Segura J. BALLESTARLAB/CVID-COVID. November-2024. 10.5281/zenodo.14046594 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The chromium droplet-based scRNA-seq data in this study have been deposited in EGA database under accession code EGAD50000000543. Interactive visualizations of single-cell transcriptomic datasets from this study can be accessed via https://www.immunodeficiencycellatlas.org. Spectral flow cytometry files have been deposited in FlowRepository under accession code FR-FCM-Z8F8. The Whole Exome Sequencing (WES) data are not publicly available due to ethical and privacy regulations. Access to the data can be granted upon request by contacting the corresponding authors. Requests for access will be reviewed and responded to within 30 days, and any approved access will be governed by a Data Use Agreement, which may impose specific conditions on the permissible uses of the data. We used additional scRNA-seq data publicly-available datasets with the following accession numbers: HRA000150, CRA002497, GSE158055, E-MTAB-10026, GSE196456, GSE154567 and GSE175450. Source data are provided with this paper.
The code used in the analyses can be found at https://github.com/BALLESTARLAB/CVID-COVID110.