Abstract
Lung cancer is the leading cause of cancer mortality worldwide, yet there exists a limited view of the genetic lesions driving this disease. In this study, an integrated high-resolution survey of regional amplifications and deletions, coupled with gene-expression profiling of non-small-cell lung cancer subtypes, adenocarcinoma and squamous-cell carcinoma (SCC), identified 93 focal copy-number alterations, of which 21 span <0.5 megabases and contain a median of five genes. Whereas all known lung cancer genes/loci are contained in the dataset, most of these recurrent copy-number alterations are previously uncharacterized and include high-amplitude amplifications and homozygous deletions. Notably, despite their distinct histopathological phenotypes, adenocarcinoma and SCC genomic profiles showed a nearly complete overlap, with only one clear SCC-specific amplicon. Among the few genes residing within this amplicon and showing consistent overexpression in SCC is p63, a known regulator of squamous-cell differentiation. Furthermore, intersection with the published pancreatic cancer comparative genomic hybridization dataset yielded, among others, two focal amplicons on 8p12 and 20q11 common to both cancer types. Integrated DNA–RNA analyses identified WHSC1L1 and TPX2 as two candidates likely targeted for amplification in both pancreatic ductal adenocarcinoma and non-small-cell lung cancer.
Keywords: array comparative genomic hybridization, expression profiling, lung adenocarcinoma, squamous-cell lung carcinoma, TP73L
Lung cancer is the leading cause of cancer-related mortality in the United States, accounting for more than one-fourth of all cancer fatalities in 2004. Lung cancer is classified into two major subtypes, small-cell and non-small-cell lung cancer (NSCLC). NSCLC constitutes 75% of lung cancer cases and is subdivided further into three major histological subtypes: adenocarcinoma (AC), squamous-cell carcinoma (SCC), and large-cell carcinoma. The AC and SCC subtypes represent >85% of NSCLC cases. Although these NSCLC subtypes exhibit distinct pathological characteristics, the treatment approaches have remained generic and largely ineffective, despite advances in cytotoxic drugs, radiotherapy, and clinical management. For all stages of NSCLC, the 5-year survival rate has remained fixed at 15% for the last 15 years. The recent success of molecularly targeted therapies for a limited subset of cancer genotypes (1) has solidified the view that a more detailed knowledge of the spectrum of genetic lesions in lung cancer will, in turn, lead to meaningful therapeutic progress.
To date, the majority of lung cancer genetic studies have cataloged mutations or the promoter methylation status of known cancer genes, performed genome-wide loss-of-heterozygosity surveys, and applied comparative genomic hybridization (CGH) to audit regional copy-number alterations (CNAs) on metaphase chromosomes or small-scale bacterial artificial chromosome (BAC) arrays. These concerted efforts have identified a core set of lesions, including activating mutations in K-RAS and mutations that compromise p53 and Rb-pathway function (2). At the same time, the observed high number of recurrent chromosomal aberrations, particularly amplifications and deletions, suggests that only a small fraction of lung cancer genes has been identified. In particular, chromosomal CGH studies have revealed recurrent gains at 1q31, 3q25–27, 5p13–14, and 8q23–24 and deletions at 3p21, 8p22, 9p21–22, 13q22, and 17p12–13 (3–7). A recent array-CGH (aCGH) survey of known genes/loci using 348 BAC clones has confirmed recurrent chromosome-3p deletions and -3q gains and identified PIK3CA as a resident of the chromosome-3q amplicon (8).
Integrated CGH and expression profiling have emerged as effective entry points for cancer gene discovery, capable of providing a high-resolution view of the regional gains and losses throughout the cancer genome (9) and the associated copy-number-driven changes in gene expression (10, 11). In the microarray format, the resolution of CGH is dictated by the number and quality of mapped probes positioned along the genome (12). In this study, high-density gene-specific arrays were used to conduct high-resolution surveys of CNAs present in a collection of primary ACs and SCCs and of established NSCLC cell lines. Together with expression profiling, these datasets provide insights into the origins of, and genetic mechanisms driving, AC and SCC subtypes.
Materials and Methods
Cell Lines and Primary Tumors. All of the primary tumors were acquired from the Cooperative Human Tissue Network (Philadelphia) and the Brigham and Women's Hospital tissue bank (Boston) under an approved institutional protocol. The tumor histology was confirmed by a pathologist (M.J.Y.) before inclusion in this study. All of the cell lines were obtained from the American Type Culture Collection. The characteristics of the primary tumors and cell lines are detailed in Tables 2 and 3, respectively, which are published as supporting information on the PNAS web site. Three independent, normal RNA references isolated from adjacent, histologically normal lung tissues were used as the normal control for the expression analysis.
aCGH Profiling on Oligonucleotide and cDNA Microarrays. Genomic DNAs from cell lines and primary tumors were extracted according to manufacturer's instructions (Gentra Systems). Genomic DNA was fragmented and random-prime labeled as described in ref. 11 and http://genomic.dfci.harvard.edu/array_CGH.htm and hybridized to either human cDNA or oligonucleotide microarrays. The cDNA microarray contains 14,160 cDNA clones (Human 1 clone set, Agilent Technologies, Palo Alto, CA) with 13,281 genome-mappable clones, for which ≈11,211 unique map positions were defined (National Center for Biotechnology Information, Build 35). The median interval between mapped elements is 72.7 kb, 94.1% of intervals are <1 megabase (Mb), and 98.9% are <3 Mb. The oligonucleotide array contains 22,500 elements designed for expression profiling (Human 1A V2, Agilent Technologies), for which 16,097 unique map positions were defined (Build 35). The median interval between mapped elements is 54.8 kb, 96.7% of intervals are <1 Mb, and 99.5% are <3 Mb. Fluorescence ratios of scanned images of the arrays were calculated as the average of two paired arrays (dye swap), and the raw aCGH profiles were processed to identify statistically significant transitions in copy number by using a segmentation algorithm (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). In this study, significant copy-number changes are determined on the basis of segmented profiles only (for additional details on aCGH, expression profiling, quantitative PCR (QPCR) and FISH, see Supporting Materials and Methods).
Results
Identification of Known and Previously Uncharacterized CNAs in the NSCLC Genome. Forty-four tumors, frozen at the time of the initial resection, verified to be ACs (n = 18) or SCCs (n = 26), and possessing >70% tumor cellularity were subjected to genome-wide CGH profiling (Table 2). In addition to the primary tumors, CGH and expression profiling was performed on a panel of NSCLC cell lines (Table 3). All primary tumors and 14 NSCLC cell lines were analyzed by using an oligonucleotide array platform with a median resolution of 54.8 kb (13), and the remaining 20 NSCLC cell lines were previously interrogated by using a cDNA array platform with a median resolution of 72.7 kb (11). Eleven cell lines were analyzed by both platforms, revealing a high level of concordance between the two datasets (correlation coefficient of 0.88–0.95 in altered regions). Additionally, as demonstrated in ref. 13, the higher resolution of the oligonucleotide array platform relative to cDNA arrays uncovered additional focal CNAs and revealed greater structural detail of each CNA (see Fig. 5, which is published as supporting information on the PNAS web site; data not shown).
CGH profiles were generated to identify CNAs as described in ref. 11 (see also Materials and Methods and supporting information). These profiles revealed a NSCLC genome that is highly rearranged, harboring large numbers of distinct copy-number aberrations (CNAs = 319), many of which exhibited a high degree of structural complexity. Some of these CNAs are recurrent across different samples, allowing for the definition of a minimal common region (MCR) of gain/amplification or loss/deletion. The total number of MCRs is 220. To facilitate the identification of those MCRs that might have strong pathogenetic relevance (referred to as “high-priority MCRs”), we apply a set of criteria that include the occurrence in at least one tumor sample, the presence of at least one high-amplitude event (log20.8), and recurrence in at least two samples (see Supporting Materials and Methods). There are 93 of these high-priority MCRs: 74 amplifications and 19 deletions with a median size of 1.53 Mb.
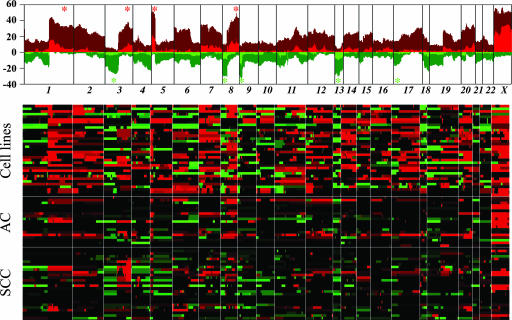
Upon comparison with existing NSCLC genomic data derived from chromosomal CGH, this high-priority MCR list was found to contain all known regional gains and losses, albeit with much finer definition. Specifically, our dataset included the known gains at 1q31, 3q25–27, 5p13–14, and 8q23–24 as well as the known deletions at 3p, 8p22, 9p21–22, 13q22, and 17p12–13 (Fig. 1). Virtually all the genes implicated in the pathogenesis of NSCLC were contained within the high-confidence MCRs, including p16INK4A and RB1 tumor-suppressor genes and MYC, EGFR, and KRAS2 oncogenes (see Table 4, which is published as supporting information on the PNAS web site). The whole short arm of chromosome 3 was consistently lost, with a peak recurrence at ≈50 Mb (29 of 79 samples, 36%). Segmentation did not identify obvious homozygous deletions, which would point to a specific target in this recurrently deleted region of 3p; however, this region contains RASSF1, TUSC2, SEMA3B, and FHIT, genes that have shown loss of heterozygosity in NSCLC (7) (Fig. 1; and see Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Genomic profiles of primary lung AC and squamous carcinomas and lung cancer cell lines. (Upper) Recurrence of chromosomal alterations. Integer-value recurrence of CNAs in segmented data (y axis) is plotted for each probe evenly aligned along the x axis in chromosome order. Dark red or green bars denote gain or loss of chromosome material, and bright red or green bars represent probes within regions of amplification or deletion (see Materials and Methods). Asterisks identify the most frequent region of gains (red) and losses (green), as reported in ref. 7. (Lower) Heat-map plot showing discrete CNAs within all samples, with the x axis representing probes ordered by genomic map positions and the y axis representing individual samples. Red represents chromosomal gain/amplification, and green denotes chromosomal loss/deletion.
The most notable feature of the high-priority MCR dataset was a subset (21 of 93) of highly focal MCRs that spanned <0.5 Mb and possessed a median of only 5 genes. From a total of 120 genes (of 139 genes in these 0.5-Mb MCRs) represented on the Affymetrix U133 Plus 2.0 microarray, 53 (≈40%) showed expression, further delimiting the number of potential cancer-relevant gene candidates. Among this subset of genes, several have already been established as having a role in cancer (e.g., ERBB3) or homology to known cancer genes (a PTEN-related molecule that is in a region of deletion). Additionally, three members of the cyclin family were present in two different MCRs (cyclins M3, M4, and J), and BRD9, a bromodomain gene with potential functional relatedness to BRD4, a gene located within the high-priority MCRs and involved in virus-induced cellular transformation (Table 1; and see Discussion). These findings highlight the potential for focused, high-yield cancer gene discovery.
Table 1. High-confidence MCRs in lung ACs and SCCs, spanning <0.5 Mb.
| MCR recurrence
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MCRs
|
Gains/losses
|
Amplifications/deletions
|
||||||||
| Cytogenetic band | Position, Mb | Size, Mb | Maximum/minimum value | No. of transcripts | % | Tumors | Cell lines | Tumors | Cell lines | Candidate genes |
| Gains/amplifications | ||||||||||
| 1p36.32–1p36.32 | 2.37–2.47 | 0.10 | 2.92 | 2 | 22 | 9 | 9 | 3 | 0 | PEX10, RER1 |
| 1p34.3–1p34.3 | 37.82–38.13 | 0.32 | 2.16 | 11 | 18 | 7 | 8 | 1 | 1 | FLJ31434, YRDC, FLJ45459 |
| 1q32.2–1q32.2 | 206.17–206.24 | 0.06 | 1.14 | 3 | 39 | 15 | 17 | 1 | 4 | LAMB3 |
| 2q11.2–2q11.2 | 96.58–96.95 | 0.27 | 1.39 | 8 | 35 | 14 | 16 | 4 | 4 | CNNM3, CNNM4, SEMA4C |
| 2q31.1–2q31.1 | 170.19–170.32 | 0.13 | 2.67 | 4 | 28 | 11 | 13 | 1 | 1 | PPIG |
| 5p15.33–5p15.33 | 0.53–0.92 | 0.38 | 1.99 | 5 | 60 | 27 | 24 | 6 | 15 | BRD9 |
| 5q31.3–5q31.3 | 140.55–140.66 | 0.12 | 1.59 | 11 | 19 | 6 | 10 | 2 | 4 | PCDHB11, PCDHB12, PCDHB13 |
| 8p12–8p11.22* | 38.24–38.45* | 0.21 | 1.98 | 2 | 35 | 17 | 12 | 6 | 4 | FGFR1, WHSC1L1, LETM2 |
| 10q24.1–10q24.1 | 97.52–98.11 | 0.49 | 1.96 | 8 | 16 | 3 | 10 | 1 | 1 | CCNI |
| 10q26.3–10q26.3 | 135.24–135.24 | 0.01 | 1.04 | 1 | 11 | 1 | 8 | 1 | 3 | C10orf94 |
| 12q13.2–12q13.2 | 54.72–54.84 | 0.12 | 1.98 | 8 | 29 | 10 | 14 | 2 | 2 | ERBB3 |
| 14q32.13–14q32.13 | 93.66–94.03 | 0.37 | 1.15 | 9 | 29 | 6 | 18 | 1 | 4 | KIAA1622 |
| 16q22.2–16q22.2 | 69.52–69.87 | 0.26 | 1.4 | 2 | 18 | 6 | 9 | 1 | 4 | Hs.368781 |
| 18q12.1–18q12.1 | 26.83–27.31 | 0.48 | 1.1 | 6 | 19 | 9 | 7 | 2 | 1 | DSC1, DSC2, DSG2 |
| 19q13.33–19q13.33 | 55.52–55.68 | 0.16 | 0.73 | 8 | 17 | 5 | 9 | 1 | 1 | SPIB |
| 20q11.21–20q11.21 | 29.66–29.88 | 0.22 | 1.08 | 5 | 42 | 13 | 23 | 1 | 8 | BCL2L1, TPX2 |
| Losses/deletions | ||||||||||
| 7q34–7q34 | 142.28–142.44 | 0.16 | -1.39 | 4 | 13 | 7 | 4 | 2 | 1 | LOC154761 |
| 11q11–11q11† | 55.1–55.18 | 0.08 | -4.06 | 6 | 23 | 13 | 6 | 7 | 1 | OR4C11, OR4C6, OR4V1P† |
| 13q12.11–13q12.11 | 19.14–19.56 | 0.41 | -3.51 | 2 | 34 | 14 | 14 | 1 | 7 | HSMPP8, PSPC1 |
| 13q32.2–13q32.2 | 97.47–97.9 | 0.42 | -3.25 | 6 | 29 | 13 | 11 | 1 | 8 | FARP1 |
| 21p11.2–21p11.1 | 9.93–10.08 | 0.15 | -1.33 | 5 | 17 | 9 | 6 | 1 | 1 | TPTE |
The numbers of primary tumors or cell lines with gain or loss, and amplification or deletion, are listed. MCR recurrence is denoted as percentage of the total dataset. Number of transcripts is based on Build 35 of the National Center for Biotechnology Information. Only the known genes within the boundaries have been included. In bold face are the MCRs verified by RT-PCR and/or FISH (Figs. 3 and 7–10 and data not shown). The genes listed are among the subset of genes within the MCRs showing expression in the Affymetrix Plus 2.0.
MCR in 8p, which was subject to further fine mapping (see text)
The MCR at 11q11 has recently been shown by Sebat et al. (33) to be a copy number polymorphism (ORF511, chromosome 11q11)
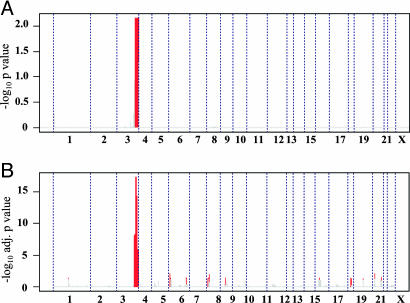
Common and Distinct Genomic Features in AC and SCC. Previous analyses of the NSCLC genome with low-resolution chromosomal or BAC aCGH have consistently shown that the only region of the genome differentiating SCCs from ACs was 3q26–29 (3–7). With our higher-resolution platform, capable of detecting previously unrecognized focal CNAs (see above), we sought to determine whether there exist additional genomic events that are characteristic of either SCC or AC. Surprisingly, the genomic profiles of AC and SCC were highly overlapping, such that neither supervised nor unsupervised clustering of the global CGH profiles was able to classify these tumors according to their histopathological subtypes (Fig. 1 and data not shown). Next, we asked whether there are significant regional differences between AC and SCC subtypes. To this end, we designed a permutation test to compare the incidence of events in each primary tumor subtype and to estimate the significance (see Supporting Materials and Methods). This permutation test identified only one region of gain/amplification on the long arm of chromosome 3, from 180 to 199 Mb, corresponding to 3q26–29, that was significantly targeted in the SCCs (Fig. 2A). Therefore, despite strikingly distinct histological presentations, SCC and AC are remarkably similar on the genomic level and are likely driven by many of the same oncogenes and tumor-suppressor gene mutations.
Fig. 2.
Chromosome 3q, from 180 to 199 Mb, is the only genomic region that shows a significant difference between AC and SCC by both aCGH (A) and expression (B) profiling. On both plots, x-axis coordinates represent probes ordered by genomic map positions, from chromosome 1 to chromosome X. (A) Probes significantly gained/amplified, comparing SCC and AC primary tumors, on aCGH. The y axis represents the -log10 P value of the permutation test. Probes presenting a P < 0.05 are plotted in red, and probes with a P > 0.05 are plotted in gray. No probes were significantly lost/deleted after comparison between SCC and AC primary tumors. (B) Genomic regions significantly enriched for differentially expressed probes comparing SCC and AC primary tumors. The y axis represents the -log10 adjusted P values of Fisher's exact test for enrichment. Red lines highlight regions with a P < 0.05, adjusted for multiple testing, and probes with an adjusted P > 0.05 are in gray.
We further hypothesize that the defined MCR on 3q harbors gene(s) driving the SCC phenotype and that such resident target(s) can be up-regulated by mechanisms other than gene amplification because, although common among SCCs, gain/amplification of 3q from 180 to 199 Mb is not present in all cases of SCC (54% in our samples and between 50% and 85% in the literature) (3–7). Thus, by comparing expression patterns of probes residing within the 3q180- to 199-Mb region, we would identify genes that are consistently overexpressed in SCC versus AC, regardless of copy number status, and such genes might have a higher probability of playing a critical role in driving the SCC phenotype. To this end, a one-way ANOVA and a post hoc Bonferroni test using all the probes (n = 166, corresponding to 106 genes) residing within the 180- to 199-Mb boundaries on 3q (see Supporting Materials and Methods) identified a subset of genes showing significant overexpression in SCCs versus ACs and were overexpressed, even in the absence of gene copy-number gains, on 3q in SCCs: p63, claudin 1, phosphatidylinositol glycan class X, and discs large homologue 1 (see Table 5, which is published as supporting information on the PNAS web site). The p63 gene is most notable, given its seminal role in squamous-tissue development and its links to squamous-cancer subtypes (14–17).
To further corroborate the above finding, we analyzed the global expression-profile data for the same NSCLC samples by using the program sam (significance analysis for microarray data) (18). A total of 297 probes were found to be significantly different between SCCs and ACs, based on a q value (false discovery rate) cut-off of 0.05 (19). Of this list of differentially expressed genes, we asked whether their differential expression was driven by underlying copy-number events. To this end, we mapped all 297 differentially expressed probes to their corresponding genomic positions and, using a 10-Mb moving-window analysis across the genome and applying a Fisher's exact test (see Supporting Materials and Methods), we identified those genomic regions that were significantly enriched for differentially expressed probes. As shown in Fig. 2B, this global analysis again identified 3q 180–199 Mb as the genomic region whose resident genes are most significantly enriched from among the list of differentially expressed genes between AC and SCC. Similar results were also obtained when we used a published lung cancer expression-profile dataset (20) (data not shown). In conclusion, our integrated aCGH and expression analyses strongly implicate a limited number of genes residing within 3q as key drivers of the SCC histopathological phenotype.
Cross-Tumor-Type Genomic Comparisons. The remarkable degree of overlap between lung cancer subtypes prompted a comparison with our recently generated high-resolution genomic profile of another lethal epithelial tumor type, pancreatic ductal AC (PDAC). Although the majority of the defined CNAs are distinct between lung cancer and PDAC, the intersection between these two datasets did reveal 17 shared loci in addition to expected common genomic alterations such as KRAS, c-MYC and INK4a/ARF (Table 4). Thus, cross-tumor-type comparison holds the potential to serve as a filter for CNAs that harbor targets that are potentially relevant to multiple cancer types.
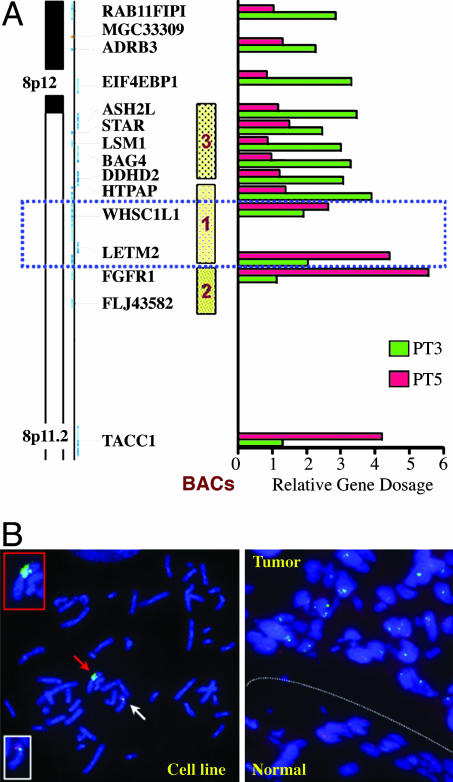
One of the shared loci was a focal amplification at 8p12–p11.2 (position 37.84–39.72 Mb) encompassing FGFR1, a cancer-relevant gene not previously implicated in lung cancer. Detailed mapping of the 8p12–p11.2 amplicon by real-time QPCR narrowed the MCR to 0.14 Mb in size, as defined by two informative primary tumor cases, PT3 and PT5 (Fig. 3A and data not shown). Because QPCR analysis clearly positioned the FGFR1 gene outside the telomeric boundary, this focal MCR contained only two annotated genes, WHSC1L1 and LETM2 (Fig. 3A). Because previous studies have implicated FGFR1 as the prime target of the 8p amplicon in other cancer types (21), interphase FISH was performed to verify the amplicon boundaries in several informative samples. FISH on the six primary tumors and two cell lines showing the amplification confirmed the presence of a high-copy-number amplicon at 8p (Fig. 3 A and B; and see Figs. 7–10, which are published as supporting information on the PNAS web site). Consistent with QPCR data, FISH with a BAC outside the MCR and including FGFR1 on PT3 revealed only two copies (Fig. 10 A and D), providing clear evidence that FGFR1 is not amplified in this sample. The integration of DNA copy number and RNA expression data was used to cull bystanders from true targets of the amplicon (10). Both gene-expression profiling and RT-QPCR (see Fig. 11, which is published as supporting information on the PNAS web site, and data not shown) demonstrated consistent overexpression of WHSC1L1. The other MCR resident gene, LETM2, did not show consistent overexpression in the presence of gene amplification. RAB11FIP1 and FGFR1, two genes positioned external to the telomeric and centromeric boundaries of the MCR, respectively, showed an expression pattern consistent with their placement outside the amplicon MCR.
Fig. 3.
Verification and boundary delimitation of the 8p12–p11.2 amplicon. (A) Real-time PCR verification of the 8p amplicon and its boundaries. Two primary tumors were used to delimit the boundaries, PT3 and PT5. The dotted blue rectangle includes the genes involved in the amplicon. (B Left) Metaphase FISH analysis of the cell line NCI-H1703, showing multiple green signals on one derivative chromosome (red arrow and Upper Inset). Green signal, BAC 2. A normal copy of chromosome 8 is also evident (white arrow and Lower Inset), showing two signals for BAC 2 (green) and two signals for the control BAC (RP11–138J2), red signal. (Right) Interphase FISH analysis on a section from the primary tumor PT3, comparing adjacent normal and tumor tissues. Note multiple, high-intensity signals with BAC 1 (green) in the tumor part and single or double signals with BAC 1 in the normal part of the section. Control, BAC (RP11–138J2), red signal.
To evaluate further the relevance of FGFR1 versus WHSC1L1 in lung cancer, the biological impact of small interfering RNA (siRNA)-mediated knockdown of each target was assessed in cell lines with and without 8p amplification (NCI-H1703 and NCI-H1395, respectively). For all genes, RT-QPCR documented >70% knockdown after siRNA pool transduction (data not shown). In soft agar assays with NCI-H1703, siRNA-mediated knockdown of WHSC1L1 resulted in 50% reduction in the number of H1703 soft agar colonies, whereas nearly complete FGFR1 depletion had no impact on colony formation in soft agar. As expected, knockdown of these two genes had no effect on NCI-H1395 colony formation. The copy-number-driven expression data, coupled with knockdown studies, supports the argument against a role for FGFR1 in lung cancers harboring an 8p amplicon and points to WHSC1L1 as a potential target of this amplification event.
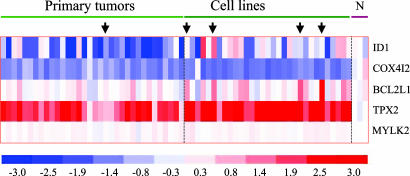
Another amplicon shared in the NSCLC and PDAC datasets mapped to 20q11.2 harboring BCL2L1 (previously BCL-xL), a known oncogene implicated in multiple cancer types. This amplicon was detected in one primary lung AC, one adenosquamous cell line, one SCC cell line and two AC cell lines. These samples together delimited the MCR to 220 kb, spanning positions 29.66–29.88 Mb and containing five genes: ID1, COX4I2, BCL2L1, TPX2, and MYLK2. Although BCL2L1 indeed exhibited modestly elevated expression, TPX2 was the only gene showing high-level copy-number-driven expression in most lung cancer cell lines and primary tumors tested, when compared with RNA derived from normal lung (Fig. 4). These findings suggest that, in addition to a known oncogene BCL2L1, TPX2 is a potential candidate oncogene targeted for amplification in both lung and pancreas cancers.
Fig. 4.
Expression analyses of genes residing in the chromosome-20q11 amplicon. Heat-map representation of the expression level of each of the five genes, ID1, COX4I2, BCL2L1, TPX2, and MYLK2, in the amplicon from Affymetrix analysis. When multiple Affymetrix probes for a gene were present, the median value among the probes was used. Each column represents an individual sample, and each row represents a gene. N, normal lung tissue. The color intensity on the heat map correlates with the intensity of the expression, with red representing overexpression and blue indicating reduced expression. The arrows indicate the samples that present amplification.
Discussion
In light of the few validated oncogenes and tumor-suppressor genes linked to NSCLC pathogenesis (2) and the limited therapeutic options for this disease, the definition of 93 focal MCRs based on our genome-wide high-resolution aCGH analysis provides a rational starting point for productive gene-discovery efforts in the lung cancer research field. The identification, based on integrated DNA–RNA analyses, of candidate targets with plausible or known links to cancers resident within these focal CNAs reinforces the validity of our approach.
On the global level, the ability to define, at high resolution, the compendium of genomic events in NSCLC permitted an unbiased comparison of the genomes of AC and SCC histological subtypes, revealing 3q26–29 as the only genomic signature for SCC. It is notable that this region has been reported to be the most common and an early genetic alteration in SCC of the head and neck, a group of tumors that share similar developmental, histological, and pathogenetic features with SCCs of the lung (22). One potential explanation for this similarity is that these two histological subtypes of lung cancer are derived from the same lung stem/precursor cell and that only a few unique genetic alterations are sufficient to confer either an AC or a SCC phenotype. Indeed, there is some experimental evidence that the alveolar type-II cell is a pluripotential stem cell involved in the genesis of human AC and SCC (23). Our results suggest that, in a subset of lung SCCs, overexpression of genes residing within 3q, mediated either by amplification or by other mechanisms, could selectively induce a squamous-cell phenotype against the backdrop of a genetic background that is otherwise common between ACs and SCCs. Interestingly, among the genes that did show overexpression in SCC samples, irrespective of copy-number changes, was p63. Overexpression of p63 has been reported in several SCCs (14, 15), and mutations in p63 have been reported in human genetic disorders affecting ectodermal development (16). In the mouse, p63 deficiency leads to profound defects in, or frank loss of, the entire spectrum of epithelial tissues (14). Conditional transgenic mice expressing p63 isoforms in the epithelial lining of the bronchioles in the lung developed severe squamous metaplasia (17). These findings support the view that p63 exerts a critical role in maintaining the proliferative capacity of the epidermal-cell population as well as driving an epithelial stratification program (14). Based on our data and on the features of the genes overexpressed in 3q, it is tempting to speculate that AC and SCC arise from a common cellular origin and are driven to a malignant endpoint by common genetic and biological mechanisms.
Comparison of the NSCLC dataset with the recently published PDAC dataset (7) proved useful on two levels. First, despite their epithelial nature and several shared cancer gene mutations, the distinctive nature of the lung and pancreas datasets further underscored the high degree of similarity between the AC and SCC CNA profiles (data not shown). Second, these comparisons and the search for commonly targeted loci provided an opportunity to identify loci with strong cancer relevance among the large number of CNAs identified. The chromosome-8p amplicon, shared by the NSCLC and PDAC datasets, has also been detected in carcinomas of the breast, prostate, and bladder and in T cell lymphomas (21, 24). Whereas FGFR1 resides within this larger CNA and has been considered the prime candidate target of this amplification, detailed QPCR and FISH mapping and expression analysis excluded the FGFR1 gene. In addition, sequence analysis of exons encoding the juxtamembrane and kinase domains of FGFR1 failed to reveal any mutations (data not shown). These findings in lung cancer are consistent with recent studies in breast cancer that show FGFR1 does not play a pathogenetic role in breast cancer cells harboring amplification of this 8p locus (24), thereby pointing to other resident gene(s) as the true target of this 8p12–p11.2 amplicon.
The gene that appears to be the most likely candidate target for this amplicon is WHSC1L1, on the basis of the physical mapping data, copy-driven expression patterns, and functional-assay results. Additionally, data from the literature strongly point to this gene as causally involved in hematological and solid tumors. WHSC1L1 is involved in a chromosomal translocation in acute myeloid leukemia, t(8;11)(p11.2;p15), that preserves all of the domains in WHSC1L1, excluding one PWWP domain (25). In addition, amplification of WHSC1L1 has been demonstrated in breast cancer (26). Several members of the SET2 family of histone lysine methyltransferases, to which this gene belongs, have roles in cancer (27). In particular, the two homologues of WHSC1L1, NSD1 and WHSC1, have been implicated in acute myeloid leukemia and multiple myeloma, respectively (28, 29). Together, these data provide a compelling case for WHSC1L1 as a lung cancer oncogene and as a prime target for amplification in NSCLC.
The chromosome-20 amplicon shared by NSCLC and PDAC contained five genes. However, only BCL2L1 and TPX2 showed copy-number-driven expression. The identification of BCL2L1 suggests that gene amplification is one of the mechanisms driving BCL2L1 activation in NSCLC. The relevance of BCL2L1 amplification and overexpression in the development of NSCLC is strengthened by previous studies establishing a critical oncogenic role for BCL2L1 in PDAC (30). Interestingly, TPX2 emerges from our analysis as a strong candidate targeted for amplification and overexpression in NSCLC and PDAC. TPX2 is required for targeting aurora-A kinase to the spindle apparatus. Elevated expression of aurora A has been reported in breast, bladder, colon, ovarian, and pancreatic cancers and correlates with chromosomal instability and clinically aggressive disease. However, no instances of specific amplification or overexpression of aurora kinase A have been reported so far in NSCLC, a finding consistent with the absence of aurora kinase A gene amplification in our dataset. Manda et al. (31) have demonstrated that this gene is overexpressed in lung cancer tissue, compared with normal lung. Additionally, using the program oncomine (32), we compared the expression level of TPX2 in different cancer types with the corresponding levels in normal tissues. Lung SCCs and ACs, lung small-cell carcinomas (20), and prostate and hepatocellular carcinoma showed significant overexpression of TPX2, compared with the respective normal tissues. Intriguingly, in our expression data, a high correlation existed between TPX2 and Aurora kinase A expression (r = 0.7801, P < 0.001) and even more with many other genes involved in spindle formation and mitotic progression, for example, Bub1 (r = 0.93, P < 0.001), CDC20 (r = 0.93, P < 0.001), and Aurora kinase B (r = 0.90, P < 0.001). The amplification of TPX2 and the correlation of its expression with genes involved in spindle formation and progression through the cell cycle suggest a possible critical role for TPX2 in lung and pancreas carcinogenesis.
In conclusion, by using gene-specific CGH platforms, custom bioinformatics tools, and integration of expression profiles, we have identified many recurrent amplifications and deletions in the NSCLC genome. The high degree of NSCLC genomic complexity, the recurrent nature of these lesions, and preliminary functional characterization of resident genes support the view that a large number of important oncogenes and tumor-suppressor genes remain to be identified, opening potential therapeutic and diagnostic opportunities for this dismal disease.
Supplementary Material
Acknowledgments
We thank Drs. Ruben Carrasco, Elizabeth Maher, Ergun Sahin, Omar Kabbarah, Mariela Jaskelioff, and Aram Hezel for helpful discussions and manuscript revisions and Chris Leo, Melissa Donovan, and Ilana Perna for superb technical advice and support. aCGH profiles were generated at the Arthur and Rochelle Belfer Cancer Genomic Center at Dana–Farber Cancer Institute. G.T. is supported by a grant from The Fund to Cure Myeloma and a Specialized Program of Research Excellence Multiple Myeloma Career Development Award. K.K.W. is supported by National Institutes of Health (NIH) Grant K08AG 2400401, the Sidney Kimmel Foundation for Cancer Research, and the Joan Scarangello Foundation to Conquer Lung Cancer. L.C. is supported by NIH Grants R01 CA099041 and U01-CA084313-07. R.A.D. is an American Cancer Society (ACS) Research Professor and an Ellison Medical Foundation Senior Scholar and is supported by a grant from the ACS and NIH Grants U01-CA084313-07 and R01 CA084628-12.
Author contributions: G.T., K.-K.W., C.B., Y.Z., D.B.K., L.C., and R.A.D. designed research; G.T., K.-K.W., G.M., C.B., B.F., D.B.K., Y.Z., A.P., M.J.Y., A.J.A., E.S.M., Z.Y., and H.J. performed research; C.B., B.F., Y.Z., and D.B.K. contributed new reagents/analytic tools; G.T., K.-K.W., C.B., Y.Z., and D.B.K. analyzed data; and G.T., K.-K.W., G.M., B.F., Y.Z., D.B.K., L.C., and R.A.D. wrote the paper.
Abbreviations: AC, adenocarcinoma; CGH, comparative genomic hybridization; aCGH, array-CGH; CNAs, copy-number alterations; Mb, megabase; MCR, minimal common region; NSCLC, non-small-cell lung cancer; PDAC, pancreatic ductal AC; QPCR, quantitative PCR; SCC, squamous-cell carcinoma.
References
- 1.Pao, W. & Miller, V. A. (2005) J. Clin. Oncol. 23, 2556-2568. [DOI] [PubMed] [Google Scholar]
- 2.Minna, J. D., Roth, J. A. & Gazdar, A. F. (2002) Cancer Cell 1, 49-52. [DOI] [PubMed] [Google Scholar]
- 3.Bjorkqvist, A. M., Husgafvel-Pursiainen, K., Anttila, S., Karjalainen, A., Tammilehto, L., Mattson, K., Vainio, H. & Knuutila, S. (1998) Genes Chromosomes Cancer 22, 79-82. [PubMed] [Google Scholar]
- 4.Luk, C., Tsao, M. S., Bayani, J., Shepherd, F. & Squire, J. A. (2001) Cancer Genet. Cytogenet. 125, 87-99. [DOI] [PubMed] [Google Scholar]
- 5.Pei, J., Balsara, B. R., Li, W., Litwin, S., Gabrielson, E., Feder, M., Jen, J. & Testa, J. R. (2001) Genes Chromosomes Cancer 31, 282-287. [DOI] [PubMed] [Google Scholar]
- 6.Petersen, I., Bujard, M., Petersen, S., Wolf, G., Goeze, A., Schwendel, A., Langreck, H., Gellert, K., Reichel, M., Just, K., et al. (1997) Cancer Res. 57, 2331-2335. [PubMed] [Google Scholar]
- 7.Balsara, B. R. & Testa, J. R. (2002) Oncogene 21, 6877-6883. [DOI] [PubMed] [Google Scholar]
- 8.Massion, P. P., Kuo, W. L., Stokoe, D., Olshen, A. B., Treseler, P. A., Chin, K., Chen, C., Polikoff, D., Jain, A. N., Pinkel, D., et al. (2002) Cancer Res. 62, 3636-3640. [PubMed] [Google Scholar]
- 9.Kallioniemi, O. P., Kallioniemi, A., Piper, J., Isola, J., Waldman, F. M., Gray, J. W. & Pinkel, D. (1994) Genes Chromosomes Cancer 10, 231-243. [DOI] [PubMed] [Google Scholar]
- 10.Pollack, J. R., Sorlie, T., Perou, C. M., Rees, C. A., Jeffrey, S. S., Lonning, P. E., Tibshirani, R., Botstein, D., Borresen-Dale, A. L. & Brown, P. O. (2002) Proc. Natl. Acad. Sci. USA 99, 12963-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguirre, A. J., Brennan, C., Bailey, G., Sinha, R., Feng, B., Leo, C., Zhang, Y., Zhang, J., Gans, J. D., Bardeesy, N., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9067-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albertson, D. G. & Pinkel, D. (2003) Hum. Mol. Genet. 12, R145-R152. [DOI] [PubMed] [Google Scholar]
- 13.Brennan, C., Zhang, Y., Leo, C., Feng, B., Cauwels, C., Aguirre, A. J., Kim, M., Protopopov, A. & Chin, L. (2004) Cancer Res. 64, 4744-4748. [DOI] [PubMed] [Google Scholar]
- 14.McKeon, F. (2004) Genes Dev. 18, 465-469. [DOI] [PubMed] [Google Scholar]
- 15.Massion, P. P., Taflan, P. M., Jamshedur Rahman, S. M., Yildiz, P., Shyr, Y., Edgerton, M. E., Westfall, M. D., Roberts, J. R., Pietenpol, J. A., Carbone, D. P. & Gonzalez, A. L. (2003) Cancer Res. 63, 7113-7121. [PubMed] [Google Scholar]
- 16.Westfall, M. D. & Pietenpol, J. A. (2004) Carcinogenesis 25, 857-864. [DOI] [PubMed] [Google Scholar]
- 17.Koster, M. I., Kim, S., Mills, A. A., DeMayo, F. J. & Roop, D. R. (2004) Genes Dev. 18, 126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey, J. D. & Tibshirani, R. (2003) Proc. Natl. Acad. Sci. USA 100, 9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon, R., Richter, J., Wagner, U., Fijan, A., Bruderer, J., Schmid, U., Ackermann, D., Maurer, R., Alund, G., Knonagel, H., et al. (2001) Cancer Res. 61, 4514-4519. [PubMed] [Google Scholar]
- 22.Huang, Q., Yu, G. P., McCormick, S. A., Mo, J., Datta, B., Mahimkar, M., Lazarus, P., Schaffer, A. A., Desper, R. & Schantz, S. P. (2002) Genes Chromosomes Cancer 34, 224-233. [DOI] [PubMed] [Google Scholar]
- 23.Ten Have-Opbroek, A. A., Benfield, J. R., van Krieken, J. H. & Dijkman, J. H. (1997) Histol. Histopathol. 12, 319-336. [PubMed] [Google Scholar]
- 24.Ray, M. E., Yang, Z. Q., Albertson, D., Kleer, C. G., Washburn, J. G., Macoska, J. A. & Ethier, S. P. (2004) Cancer Res. 64, 40-47. [DOI] [PubMed] [Google Scholar]
- 25.Rosati, R., La Starza, R., Veronese, A., Aventin, A., Schwienbacher, C., Vallespi, T., Negrini, M., Martelli, M. F. & Mecucci, C. (2002) Blood 99, 3857-3860. [DOI] [PubMed] [Google Scholar]
- 26.Angrand, P. O., Apiou, F., Stewart, A. F., Dutrillaux, B., Losson, R. & Chambon, P. (2001) Genomics 74, 79-88. [DOI] [PubMed] [Google Scholar]
- 27.Schneider, R., Bannister, A. J. & Kouzarides, T. (2002) Trends Biochem. Sci. 27, 396-402. [DOI] [PubMed] [Google Scholar]
- 28.Jaju, R. J., Fidler, C., Haas, O. A., Strickson, A. J., Watkins, F., Clark, K., Cross, N. C., Cheng, J. F., Aplan, P. D., Kearney, L., et al. (2001) Blood 98, 1264-1267. [DOI] [PubMed] [Google Scholar]
- 29.Stec, I., Wright, T. J., van Ommen, G. J., de Boer, P. A., van Haeringen, A., Moorman, A. F., Altherr, M. R. & den Dunnen, J. T. (1998) Hum. Mol. Genet. 7, 1071-1082. [DOI] [PubMed] [Google Scholar]
- 30.Trauzold, A., Schmiedel, S., Roder, C., Tams, C., Christgen, M., Oestern, S., Arlt, A., Westphal, S., Kapischke, M., Ungefroren, H. & Kalthoff, H. (2003) Br. J. Cancer 89, 1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manda, R., Kohno, T., Matsuno, Y., Takenoshita, S., Kuwano, H. & Yokota, J. (1999) Genomics 61, 5-14. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., Barrette, T., Pandey, A. & Chinnaiyan, A. M. (2004) Neoplasia 6, 1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. (2004) Science 305, 525-528. [DOI] [PubMed] [Google Scholar]
- 34.Futreal, P. A., Coin, L., Marshall, M., Down, T., Hubbard, T., Wooster, R., Rahman, N. & Stratton, M. R. (2004) Nat. Rev. Cancer 4, 177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.