Abstract
In contrast to Escherichia coli, where all tRNAs have the CCA motif encoded by their genes, two classes of tRNA precursors exist in the Gram-positive bacterium Bacillus subtilis. Previous evidence had shown that ribonuclease Z (RNase Z) was responsible for the endonucleolytic maturation of the 3′ end of those tRNAs lacking an encoded CCA motif, accounting for about one-third of its tRNAs. This suggested that a second pathway of tRNA maturation must exist for those precursors with an encoded CCA motif. In this paper, we examine the potential role of the four known exoribonucleases of B.subtilis, PNPase, RNase R, RNase PH and YhaM, in this alternative pathway. In the absence of RNase PH, precursors of CCA-containing tRNAs accumulate that are a few nucleotides longer than the mature tRNA species observed in wild-type strains or in the other single exonuclease mutants. Thus, RNase PH plays an important role in removing the last few nucleotides of the tRNA precursor in vivo. The presence of three or four exonuclease mutations in a single strain results in CCA-containing tRNA precursors of increasing size, suggesting that, as in E.coli, the exonucleolytic pathway consists of multiple redundant enzymes. Assays of purified RNase PH using in vitro-synthesized tRNA precursor substrates suggest that RNase PH is sensitive to the presence of a CCA motif. The division of labor between the endonucleolytic and exonucleolytic pathways observed in vivo can be explained by the inhibition of RNase Z by the CCA motif in CCA-containing tRNA precursors and by the inhibition of exonucleases by stable secondary structure in the 3′ extensions of the majority of CCA-less tRNAs.
INTRODUCTION
Transfer RNAs (tRNAs) are synthesized as longer precursor molecules that require maturation of both their 5′ and 3′ ends. Maturation of the 5′ side is catalysed by the quasi-universally conserved endonucleolytic ribozyme, RNase P (1), while maturation of the 3′ side can be either endonucleolytic or exonucleolytic. The best studied paradigm of the exonucleolytic pathway is that of Escherichia coli. Here, any one of at least four redundant enzymes, RNases T, PH, D and II (and possibly PNPase), can produce mature tRNA 3′ ends by degrading the 3′ extension (2), generally thought to result from cleavage of the primary transcript on the downstream side of the tRNA by endoribonuclease RNase E (3,4). Of the four enzymes involved in the exonucleolytic pathway of tRNA maturation in E.coli, RNase T and RNase PH are considered the most important (2). In the case of RNase T, this is thought to be because of the enzyme's ability to remove nucleotides close to stable secondary structures, such as the tRNA acceptor arm (5).
The endonucleolytic pathway of tRNA maturation is catalysed by the ubiquitous enzyme RNase Z, the structure of which was recently solved (6). About half of the sequenced bacteria contain an RNase Z ortholog (7). RNase Z cleaves tRNA precursors just downstream of the discriminator base [nucleotide (nt) 73 in the standard tRNA numbering system] in most cases, yielding a tRNA ready for addition of the CCA motif by nucleotidyl transferase (8–15). In many organisms, it has been shown that the CCA motif inhibits cleavage by RNase Z, thus preventing futile cycles of CCA addition and removal (14,16). The number of tRNA precursors with an encoded CCA motif varies widely in bacteria, from Thermotoga maritima which has just one such tRNA (17), to E.coli, where 100% of tRNAs have an encoded CCA motif. It was recently shown that RNase BN of E.coli, involved in the removal of erroneously incorporated nucleotides by nucleotidyl transferase or in the maturation of certain phage T4 tRNAs lacking a CCA (18), and previously thought to be a fifth enzyme of the exonucleolytic tRNA maturation pathway (2), is in fact an ortholog of RNase Z (19). We have shown that RNase Z is responsible for the endonucleolytic maturation of CCA-less tRNA precursors in Bacillus subtilis, which constitute about one-third (27 out of 86) of its tRNAs. The remaining tRNAs are correctly matured in vivo in a strain partially depleted for RNase Z (20), suggesting that a second maturation pathway must exist in B.subtilis for CCA-containing tRNA precursors. In this paper, we show that RNase PH is an important component of this exonucleolytic pathway.
MATERIALS AND METHODS
Strains
The strains used in this study are listed in Table 1. Substitution of part or all of ribonuclease coding sequences with antibiotic resistance markers have been described: ΔpnpA::cm (21), Δrph::spc (22), Δrnr::spc (23), ΔyhaM::pm (24) and Δrnr::tc (22). The isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac:rnz construct was also described previously (20). Strains with more than one ribonuclease mutation were constructed by transformation of RNase mutant strains with chromosomal DNA obtained from a strain bearing the desired mutation.
Table 1.
B.subtilis strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W168 | Wild-type | |
| SSB1030 | ΔpnpA::cm | This work |
| BG457 | Δrph::spc | This work |
| BG488 | Δrph::spc ΔpnpA::cm | This work |
| CCB020 | Δrph::spc ΔpnpA::cm Pspac:rnz | This work |
| CCB021 | Δrnr::spc | This work |
| YhaMd | ΔyhaM:ery | Gift from Y. Sadaie |
| BG393 | Δrph::cm Δrnr::spc | This work |
| BG395 | Δrph::cm ΔyhaM::pm | This work |
| BG503 | Δrnr::tc Δrph::spc ΔyhaM::pm | (22) |
| BG505 | ΔpnpA::cm Δrnr::tc Δrph::spc ΔyhaM::pm | This work |
Northern blots
The hybridization conditions for northern blots using oligonucleotide probes specific for each tRNA are described previously (20).
Purification of B.subtilis RNase PH
The B.subtilis rph gene was amplified by PCR using oligos CC075 and CC076 and cloned between the EcoRI and HindIII sites of pET28a (Stratagene). The sequences of all oligonucleotides used in the study are reported in Table 2. C-terminal histidine-tagged RNase PH was purified from E.coli BL21 CodonPlus (Stratagene) cells on a 1 ml Ni-NTA agarose (Qiagen) column according to the protocol described for RNase Z by Pellegrini et al. (20), except that elution was by pH-gradient from 50 mM sodium phosphate, pH 6.0, 0.3 M NaCl to 50 mM sodium acetate, pH 4.0 and 0.3 M NaCl.
Table 2.
Oligonucleotides used in this study
| Oligo | Sequence (5′→3′) |
|---|---|
| CC075 | AATTGAATTCGTACATACAATCGGAGGTAAAACATGAG |
| CC076 | AATTAAGCTTTTTCAGTTCTGGCAGAGAATCACC |
| CC079 | CATTTAAGCTTCCAAGCGG |
| CC107 | CATTGGAGCTTCCAAGCGG |
| CC201 | AAGTTATGCCGGCAAGAGGACTTGAA |
| CC202 | ATTAATACGACTCACTATAGCGGATGTGGCGGAATTGGCAGA |
| CC203 | GGTGGTGTTCTACCACTGAACTAC |
| CC204 | ATTAATACGACTCACTATAGGGGCCATAGCTCAGCTGGGGAGA |
| CC205 | ACTGGAGGATTCAGAGAATGAAACC |
| CC206 | ATTAATACGACTCACTATAGGCGGCATAGCCAAGTGGTAA |
| CC207 | TCCTGTGTGTCTGCCAATTCCACCA |
| CC208 | CAGTTACGGAGAGCAAGGGATTCGA |
| CC209 | AAATTATGCGGATGAAGGGACTTGAACCCC |
| CC214 | CAGTGGCGGAGACGAAGGGATTCGAA |
| CC215 | AAATGGTGCGGATGAAGGGACTTGAACC |
| HP62 | AACACAGCATGCCTGAACTACTTCCGC |
| HP294 | GGTAATAGTCGACTTACAATAGACTTGAATATTATATC |
| HP560 | ATTAATACGACTCACTATAGCTTCCATAGCTCAGCAGGTAG |
| HP563 | ATTAATACGACTCACTATAGCCGGTGTAGCTCAATTGGTAG |
Restriction sites are underlined.
Synthesis of tRNA precursors in vitro
32P-labelled tRNA precursors were synthesized by T7 RNA polymerase (Stratagene) in vitro using PCR fragments as templates as described previously (20). The following oligonucleotide pairs were used: trnI-Thr, HP560/HP62; trnI-Thr (+6, −CCA), HP560/CC079; trnI-Thr (+6, +CCA), HP560/CC107; trnB-Thr, HP563/HP294; trnB-Thr (+6, −CCA), HP563/CC201; trnD-Ser, CC198/CC199; trnD-Ser (+6, −CCA), CC198/CC208; trnD-Ser (+6, +CCA), CC198/CC214; trnB-Leu1, CC202/CC203; trnD-Leu1 (+6, −CCA), CC202/CC209; trnD-Leu1 (+6, +CCA), CC202/CC215; trnSL-Ala1, CC204/CC205; trnD-Cys, CC206/CC207. Transcripts were purified from unincorporated nucleotides on G50 spin columns (Amersham Biosciences).
Assay of RNase PH activity
RNase PH assays were performed in a 50 μl reaction volume with 6–8 ng/μl (200–300 nM) labelled precursor RNA and 60 ng/μl protein (38 nM hexamer) in 50 mM Tris, pH 8.0, 2.5 mM MgCl2, 30 mM NaCl, 10 mM K2HPO4 and 0.1 mM DTT at 37°C. Five microlitre aliquots were withdrawn at the times indicated and reactions were stopped by the addition of 5 μl 95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol. Samples were run on 5% polyacrylamide/7 M urea gels.
RESULTS
Short precursors of CCA-containing tRNAs accumulate specifically in the absence of RNase PH in vivo
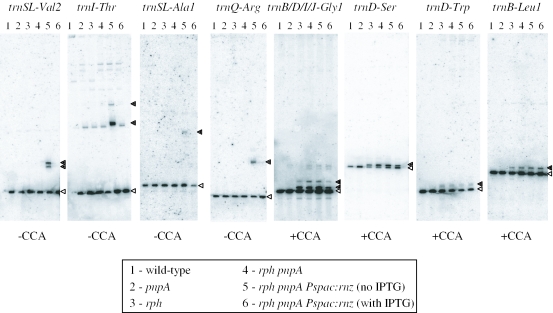
Only two of the 3′–5′ exoribonucleases potentially involved in tRNA maturation in E.coli are present in B.subtilis, RNase PH and PNPase, encoded by the rph and pnpA genes, respectively (25–27). Both are phosphorolytic enzymes. To determine whether either of these enzymes might be involved in tRNA maturation in B.subtilis, we examined the in vivo processing of eight individual tRNAs in rph and pnpA single deletion mutants and in an rph pnpA double mutant by northern blot analysis (Figure 1). Four of the tRNAs chosen for study had an encoded CCA and four did not. We also combined the rph pnpA double mutant with a construct where the gene coding for RNase Z (rnz) was placed under IPTG-dependent control and isolated total RNA, in the presence and absence of IPTG. Two classes of tRNAs clearly emerged from the northern blot analysis: CCA-less tRNAs accumulated as long precursors in the rph pnpA rnz strain grown in the absence of IPTG (lane 5), but in no other strain, while CCA-containing tRNAs accumulated as short precursors in any strain that lacked RNase PH (lanes 3–6). Most of the precursors that accumulated in the absence of RNase PH were 1–4 nt longer than the mature tRNA, while those that accumulated in the absence of RNase Z correspond to full-length precursors ending in transcription terminators (20). Thus, CCA-less tRNAs are processed by RNase Z, while maturation of CCA-containing tRNAs involves RNase PH. Although no overlap is apparent between the two pathways in the northern blot shown in Figure 1, we believe a small portion of B.subtilis tRNAs can be processed by either pathway (see Discussion). The pnpA mutation had no effect on tRNA maturation, either on its own or in combination with rph and rnz.
Figure 1.
Northern blots of total B.subtilis RNA in various ribonuclease mutants probed for individual tRNAs. Genotypes are listed in Table 1. The tRNAs probed are indicated by their gene names above each blot. tRNAGly1 is encoded by four individual genes trnB, trnD, trnI and trnJ. Precursor species are indicated by closed triangles; mature species by open triangles. The presence (+CCA) or absence (−CCA) of an encoded CCA motif is indicated below the blot for each tRNA species.
Multiple redundant exoribonucleases are involved in the maturation of B.subtilis CCA-containing tRNA precursors
A significant level of CCA-containing tRNA maturation still occurred in the absence of RNase PH, suggesting that at least one other enzyme is involved in the processing of these tRNAs. Only two other exoribonucleases have been identified thus far in B.subtilis, RNase R and YhaM, encoded by the rnr and yhaM genes, respectively (23,24). To determine whether either of these two ribonucleases played a role in the maturation of CCA-containing tRNA precursors, we performed northern blots of total RNA isolated from rnr and yhaM deletion mutants, alone or in combination with the rph mutation. The rnr and yhaM mutations had little effect on the maturation of the eight tRNAs tested, either alone or combined with the rph mutation (Figure 2, lanes 1–4). In strains with three exoribonuclease mutations (rph rnr yhaM), a small increase in precursor size was seen for trnD-Ser (compare lanes 4 and 5) and this effect was seen in varying degrees with other CCA-containing tRNA species, with minor variations in the relative importance of the different enzymes for specific tRNAs (data not shown). In strains with all four exoribonuclease mutations, a very clear increase in the size of the precursor fragments occurred, also specifically in tRNAs with encoded CCA motifs (Figure 2, lanes 5 and 6). As precursor size tends to increase with an accumulation of exonuclease mutations the data suggest that, as in E.coli, multiple redundant RNases appear to be involved in an exonucleolytic pathway of tRNA maturation.
Figure 2.
Northern blots of total B.subtilis RNA in exoribonuclease mutants probed for individual tRNAs. Genotypes are listed in Table 1. The tRNAs probed are indicated by their gene names above each blot. tRNAGly1 is encoded by four individual genes trnB, trnD, trnI and trnJ. Precursor species are indicated by closed triangles; mature species by open triangles. The presence (+CCA) or absence (−CCA) of an encoded CCA motif is indicated below the blot for each tRNA species; (+/−CCA) indicates that, for a given tRNA, some genes have an encoded CCA, while others do not.
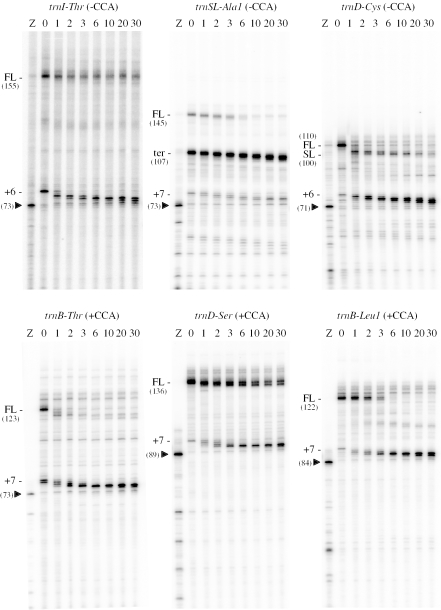
RNase PH degrades 3′ extensions in vitro and is arrested 3–4 nt from the discriminator base
To further analyse the role of RNase PH in the maturation of CCA-containing precursors, we overproduced and purified C-terminal histidine-tagged B.subtilis RNase PH in E.coli. We then measured the rate of RNase PH degradation of the 3′ extensions of various B.subtilis tRNA precursors, synthesized by T7 RNA polymerase in vitro, some of which possess an encoded CCA motif and some which lack it. Some tRNA precursor preparations had additional products besides the designed full-length transcript, which provided additional information about the ability of RNase PH to traverse secondary structures (see Discussion). A significant portion of T7 RNA polymerase paused or terminated 6 or 7 nt downstream from the 3′ end of all tRNAs synthesized. In addition, a natural transcription terminator occurs 35 nt before the 3′ end of the trn-Ala1 transcript and a non-native stem–loop is present 10 nt from the end of the trnD-Cys transcript, which also caused T7 RNAP to pause or terminate 6–7 nt downstream.
The degradation kinetics of the different tRNA precursors in vitro were highly dependent on the enzyme-to-substrate ratio and the presence of secondary structure in the 3′ extensions. At near stoichiometric RNase PH-to-substrate ratios, RNase PH rapidly degraded both classes of tRNA precursor to 1 nt from the discriminator base where it remained stalled for periods characteristic for each tRNA, but independent of the CCA motif (data not shown). At limiting enzyme concentrations, RNase PH first degraded the 3′ extensions to within 6 or 7 nt of the discriminator base with the accumulation of few intermediates, indicative of a processive mode of degradation (Figure 3). For trnSL-ala1 and trnD-cys, the accumulated intermediates were attributed to the secondary structures which caused both the termination of T7 transcription in the synthesis of the substrate and a block to RNase PH processivity. At 6–7 nt from the discriminator base, RNase PH action became more distributive, and precursors progressively shortened by 1 nt were visible, culminating in a significant accumulation of species 3 or 4 nt from the discriminator base. The trnB-Thr, trnD-Ser and trnB-Leu1 precursors, all of which possess a CCA motif, were degraded primarily to +4 or +3 (where the discriminator base is defined as 0) at the end of the 30 min experiment, while the CCA-less precursors trnI-Thr, trnSL-AlaI and trnD-Cys were reproducibly degraded 1 nt further (+3 or +2) in the same time period. This suggested that RNase PH might be sensitive to the presence of the CCA motif to some extent.
Figure 3.
Degradation kinetics of tRNA 3′ extensions by RNase PH in vitro. The full-length transcript is indicated by FL. The T7 RNA polymerase pause/termination site downstream from the tRNA 3′ end is indicated by +6 or +7. The transcription terminator (ter) of trnSL-Ala1 and stem–loop (SL) of trnD-Cys are also shown. The sizes of the various species are given in parentheses. The full-length transcripts are larger than the natural 3′ ends of the transcripts in vivo and were chosen for their suitability for oligonucleotide hybridization for template amplification by PCR. The position of the discriminator base (arrowhead) was identified for each tRNA species by cleavage of the precursor by 14 ng/μl RNase Z (lane Z) for 15 min as described by Pellegrini et al. (20). For those tRNA precursors containing a CCA motif, short variants were made lacking a CCA motif, permitting cleavage by RNase Z. Incubation times are given in minutes. The zero minute time point is in the absence of RNase PH.
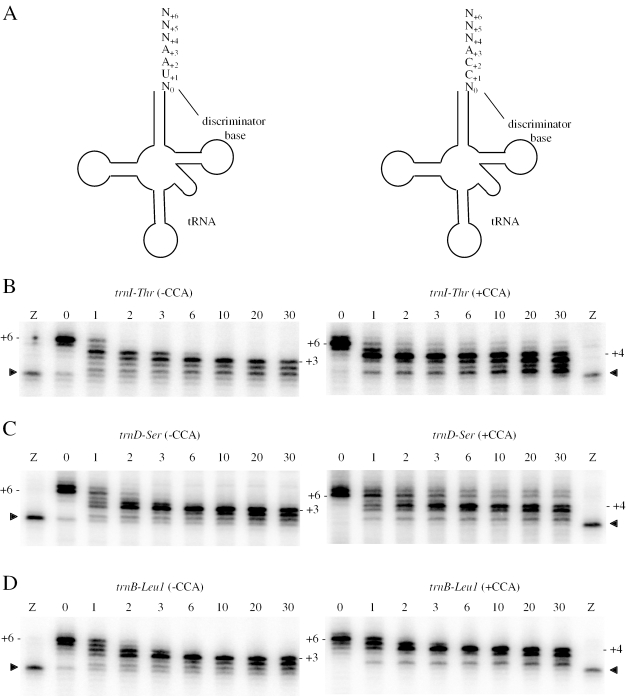
RNase PH is slow to remove the last nucleotide 3′ to the CCA motif in vitro
To determine whether the CCA motif had an effect on the degradation kinetics of RNase PH independently of sequence variation in either the tRNA or the 3′ extension, we synthesized short variants of the trnI-Thr, trnD-Ser and trnB-Leu1 tRNA precursors, with and without a CCA sequence, and compared their degradation rates in vitro at limiting RNase PH concentrations. The precursors were designed to terminate 6 nt (+6) downstream from the discriminator base and the species with and without CCA differed from each other by substituting UA for the CC dinucleotide of the CCA motif. As was observed for the longer tRNA precursors, the first major pause by RNase PH was at +3 for CCA-less precursors, while for the CCA-containing variants, the enzyme first stopped at +4, before eventually moving to +3, the position required to yield a functional tRNA (Figure 4). This suggests that RNase PH can indeed sense the presence of the CCA motif when it gets close to the acceptor stem and, for reasons that are as yet unclear, hesitates before removing the +4 nt specifically from CCA-containing tRNA precursors. Purified native RNase PH behaves identically in vitro (data not shown), showing that this degradation pattern is not an artefact due to the His tag. RNase PH arrest at +4 in vitro is independent of nucleotide identity; A, C or U occupy this position in the three tRNA precursors tested.
Figure 4.
Effect of CCA motif on RNase PH degradation kinetics. (A) tRNA precursor substrates showing positions of nucleotides relative to discriminator base (N0). All precursor tRNAs were designed to have 6 nt (+6) 3′ extensions. The CCA-less variants have a UAA motif instead of the CCA. (B) Degradation kinetics of trnI-Thr tRNA precursors, with and without a CCA motif, by RNase PH in vitro. Similar reactions are shown for (C) trnD-Ser and (D) trnB-Leu1. The arrowhead indicates the position of the discriminator base of the tRNA, identified by cleaving the −CCA version of the tRNA precursor with RNase Z (lane Z). Incubation times are given in minutes. The zero minute time point is in the absence of RNase PH. The position of the major stalling site is indicated as +3 or +4 to the right of each autoradiogram.
DISCUSSION
Previous evidence had suggested the existence of at least two pathways for tRNA maturation in B.subtilis: an RNase Z-dependent endonucleolytic pathway limited to CCA-less tRNA precursors and a second pathway for those containing a CCA motif. In this paper, we show that, while long 3′ extensions of CCA-containing tRNA precursors can be matured by multiple redundant exoribonucleases in vivo, RNase PH plays a major role in degrading the last few nucleotides before the acceptor stem. We show that, similar to RNase Z, which is inhibited by the presence of the CCA motif at the end of the acceptor stem, RNase PH also appears to be sensitive to this sequence and is surprisingly slow to remove the last nucleotide to generate the functional tRNA.
CCA-containing precursors a few nucleotides longer than the mature tRNA accumulated in rph mutant strains in vivo, for all of the tRNAs we examined. This is in sharp contrast to E.coli, where effects of the rph mutation were only visible on one of the eleven tRNA species examined, or upon precursor tRNA overproduction (2). The accumulation of precursor species in E.coli generally required removal of a second exoribonuclease, RNase T. The increased relative importance of RNase PH for removal of the last few nucleotides in B.subtilis in vivo may be due to the lack of an RNase T ortholog to provide this overlapping function.
The presence of multiple exonuclease mutations in one strain led to increased length of CCA-containing tRNA precursors in vivo, suggesting that, as in E.coli, multiple redundant enzymes are involved in the exonucleolytic pathway of tRNA maturation in B.subtilis. The quadruple deletion strain, rnr yhaM rph pnpA, grows slowly (doubling time of ∼60 min in rich medium). Whether the level of accumulation of tRNA precursors seen in this strain is sufficient to account for its slow growth phenotype remains to be seen. It is clear, however, that a significant level of tRNA maturation still occurs in the quadruple exoribonuclease mutant, suggesting that at least one other enzyme of this pathway remains at large. Preliminary experiments have suggested that recently identified ribonucleases encoded by the B.subtilis ykqC, ymfA genes (28) or the kapD gene, encoding a homolog of Homo sapiens 3′ hExo and Caenorhabditis elegans ERI-1 RNases (29,30), are not responsible for this activity (data not shown).
The size of the precursor tRNA that accumulated in vivo in strains lacking RNase PH in Figure 1 was dependent on the tRNA. The trnD-Ser precursor, for example, had only 1–2 extra nucleotides, whereas the trnB-Leu1 precursor was clearly a few nucleotides larger. Since these two tRNA precursors showed no difference in their degradation profiles by purified RNase PH in vitro (Figure 4), we presume this difference in precursor size observed in vivo is owing to variations in sensitivities of these tRNAs to the remaining processing enzyme(s).
The concentration of RNase PH (38 nM) necessary for the kinetic experiments, even though limiting relative to the substrate (200–300 nM), was quite high, presumably because of the relatively high Km of RNase PH [1 μM for the E.coli enzyme (31)] for tRNA. At limiting RNase PH concentrations, the first site of RNase PH arrest detected in vitro is 3–4 nt from the discriminator base. Curiously, relatively little of the +2 species is seen in the CCA-containing precursors compared to those without CCA (Figure 4), suggesting that if the CCA sequence is attacked by RNase PH, it has a tendency to remove all but the final cytosine (+1). Should this occur in vivo, the tRNA could be repaired by nucleotidyl transferase by adding CA to the C at +1. For the CCA-less precursors, significant quantities of the +3, +2 and +1 products are seen, whereas no discriminator product accumulates, i.e. degradation to nt 0. None of these products would be a substrate for nucleotidyl transferase activity, which adds either to the discriminator base or to C- or CC-ends. In this case, RNase Z would be required to cleave endonucleolytically at the discriminator base, generating a viable substrate for nucleotidyl transferase activity.
RNase PH hesitates before removing the +4 nt specifically from CCA-containing tRNA precursors in vitro. This is in agreement with results in E.coli that show that, while RNase PH can easily remove the +5 nt from tRNA precursors, it is significantly less efficient at removing the +4 nt than RNase T (32). The dynamics of removal of the last nucleotides of CCA-less tRNAs has not been addressed with E.coli RNase PH, mainly because this organism possesses only tRNAs with encoded CCA. Since RNase PH can degrade CCA-less precursors to +3, this would suggest that the phenomenon is sequence-, rather than position-dependent. This is not an artefact of the sequence we chose to replace the CCA motif in the experiment shown in Figure 4, since RNase PH behaves in the same manner when the CCA motif is replaced with a sequence other than UAA in the case of trnI-Thr (data not shown). It is not clear why a check-point at +4 for CCA-containing tRNAs would be a beneficial property of RNase PH or, indeed, whether this is a significant event in vivo where other enzymes are present. The distributive nature of RNase PH degradation close to the acceptor stem, also observed in the E.coli enzyme (33), would allow other, possibly more efficient enzymes, access to the substrate at each successive round of nucleotide removal. The active site of B.subtilis RNase PH is deep within the enzyme, at the base of a cleft that can accommodate the CCA motif and the first few base pairs of the acceptor stem (34). Thus, the potential for sensing the presence of the CCA motif clearly exists.
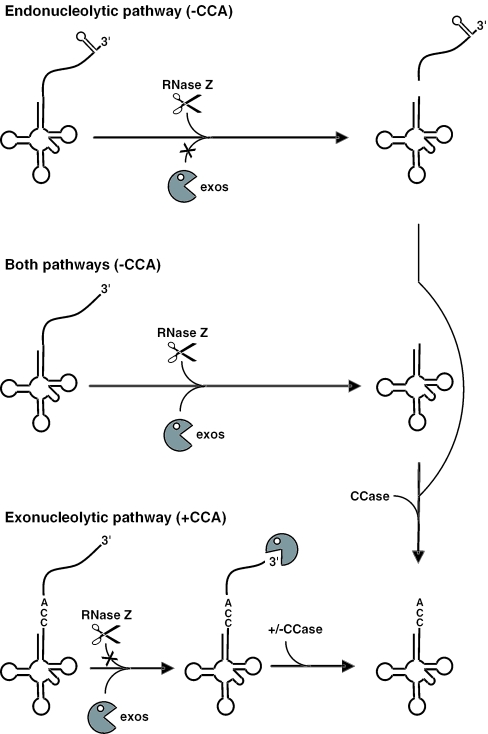
Although RNase PH is sensitive to the CCA motif in vitro, this only appears to concern the removal of the very last nucleotide 3′ to the CCA motif; 3′ extensions of CCA-less precursors are also degraded by RNase PH in vitro (Figures 3 and 4). What then accounts for the in vivo observation that the rph and other exonuclease mutations only affect the processing of CCA-containing tRNAs? This result could be explained by an inhibition of exonucleases by secondary structures downstream of the majority of CCA-less tRNAs, making these precursors substrates primarily for RNase Z (Figure 5). Out of the 27 CCA-less tRNA precursors in B.subtilis, 17 are predicted to have transcription terminators (13 species) or stable stem–loops (4 species) protecting their 3′ ends from exonucleolytic degradation (data not shown). A good example of the inhibitory effect of a terminator structure on RNase PH degradation in vitro was seen with the trnSL-Ala1 tRNA in Figure 3. We believe that the 10 remaining CCA-less tRNAs can be processed either endonucleolytically or exonucleolytically and would, thus, not be predicted to accumulate as precursors in the absence of only one pathway. Indeed, three CCA-less tRNAs (trnD-Cys, trnD-Thr and trnSL-Thr1) were identified by Pellegrini et al. (20) that were still correctly matured under conditions of RNase Z depletion. In their primary transcripts, these tRNAs are followed closely by other tRNA species and are expected to have short unprotected 3′ extensions upon processing of the 5′ side of the downstream tRNA by RNase P, permitting maturation either by the exonucleases or by RNase Z. Thus, the two pathways of tRNA maturation in B.subtilis are not likely to be fully mutually exclusive; 17 tRNAs are predicted to be processed primarily by RNase Z, 59 by exonucleases and 10 by either pathway.
Figure 5.
Model for 3′ maturation of tRNAs in B.subtilis. In the maturation of CCA-less tRNAs by the endonucleolytic pathway, exoribonucleases (pac-man symbol; exos) are inhibited (indicated by the X) by 3′ terminal structures and these tRNAs are thus primarily substrates of RNase Z (scissors). The 3′ trailer fragment generated by RNase Z is presumably eventually degraded by exonucleases in a much slower reaction. CCA-less tRNA precursors that are not protected by secondary structures can be degraded by either pathway. In the maturation of CCA-containing tRNAs, RNase Z activity is inhibited by the CCA motif, and the 3′ extension is degraded by exonucleases. Should part of the CCA sequence be removed by the exonuclease, it is repaired by nucleotidyl transferase (CCase).
Acknowledgments
We thank L. Bénard for critical reading of the manuscript, M. Deutscher, K. F. Jensen, N. Mathy, P. Stragier and F. Allemand for helpful discussion, and P. Régnier and E. Hajnsdorf for use of lab facilities. Purified native RNase PH was a kind gift from K. F. Jensen. This work was supported by funds from the CNRS (UPR 9073), Université Paris VII-Denis Diderot, PRFMMIP 2001/2003 and ACI Jeunes Chercheurs from the Ministère de l'Education Nationale. Work in the laboratory of DHB was funded by Public Health Service grant GM-48804 from the National Institutes of Health. Funding to pay for the Open Access publication charges for this article was provided by CNRS (UPR 9073).
Conflict of interest statement. None declared.
REFERENCES
- 1.Frank D.N., Adamidi C., Ehringer M.A., Pitulle C., Pace N.R. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., Deutscher M.P. Maturation pathways for E.coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 3.Li Z., Deutscher M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ow M.C., Kushner S.R. Initiation of tRNA maturation by RNase E is essential for cell viability in E.coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Pandit S., Deutscher M.P. 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li de la Sierra-Gallay I., Pellegrini O., Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 7.Condon C., Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garber R.L., Gage L.P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979;18:817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- 9.Hagenbuchle O., Larson D., Hall G.I., Sprague K.U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979;18:1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- 10.Castano J.G., Tobian J.A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase) J. Biol. Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- 11.Stange N., Beier H. A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J. 1987;6:2811–2818. doi: 10.1002/j.1460-2075.1987.tb02577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oommen A., Li X.Q., Gegenheimer P. Cleavage specificity of chloroplast and nuclear tRNA 3′-processing nucleases. Mol. Cell. Biol. 1992;12:865–875. doi: 10.1128/mcb.12.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S.J., Kang H.S. Purification and characterization of the precursor tRNA 3′-end processing nuclease from Aspergillus nidulans. Biochem. Biophys. Res. Commun. 1997;233:354–358. doi: 10.1006/bbrc.1997.6448. [DOI] [PubMed] [Google Scholar]
- 14.Nashimoto M. Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res. 1997;25:1148–1154. doi: 10.1093/nar/25.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer M., Schiffer S., Marchfelder A. tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry. 2000;39:2096–2105. doi: 10.1021/bi992253e. [DOI] [PubMed] [Google Scholar]
- 16.Mohan A., Whyte S., Wang X., Nashimoto M., Levinger L. The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA. 1999;5:245–256. doi: 10.1017/s1355838299981256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minagawa A., Takaku H., Takagi M., Nashimoto M. A novel endonucleolytic mechanism to generate the CCA 3′ termini of tRNA molecules in Thermotoga maritima. J. Biol. Chem. 2004;279:15688–15697. doi: 10.1074/jbc.M313951200. [DOI] [PubMed] [Google Scholar]
- 18.Asha P.K., Blouin R.T., Zaniewski R., Deutscher M.P. Ribonuclease BN: identification and partial characterization of a new tRNA processing enzyme. Proc. Natl Acad. Sci. USA. 1983;80:3301–3304. doi: 10.1073/pnas.80.11.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezraty B., Dahlgren B., Deutscher M.P. The RNase Z homologue encoded by Escherichia coli elaC gene is RNase BN. J. Biol. Chem. 2005;280:16542–16545. doi: 10.1074/jbc.C500098200. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Bechhofer D.H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oussenko I.A., Abe T., Ujiie H., Muto A., Bechhofer D.H. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oussenko I.A., Bechhofer D.H. The yvaJ gene of Bacillus subtilis encodes a 3′-to-5′ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J. Bacteriol. 2000;182:2639–2642. doi: 10.1128/jb.182.9.2639-2642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oussenko I.A., Sanchez R., Bechhofer D.H. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in Gram-positive bacteria. J. Bacteriol. 2002;184:6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craven M.G., Henner D.J., Alessi D., Schauer A.T., Ost K.A., Deutscher M.P., Friedman D.I. Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J. Bacteriol. 1992;174:4727–4735. doi: 10.1128/jb.174.14.4727-4735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luttinger A., Hahn J., Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 27.Mitra S., Hue K., Bechhofer D.H. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol. Microbiol. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- 28.Even S., Pellegrini O., Zig L., Labas V., Vinh J., Brechemmier-Baey D., Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominsky Z., Yang X.-C., Kaygun H., Dadlez M., Marzluff W. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy S., Wang D., Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 31.Kelly K.O., Deutscher M.P. Characterization of Escherichia coli RNase PH. J. Biol. Chem. 1992;267:17153–17158. [PubMed] [Google Scholar]
- 32.Li Z., Deutscher M.P. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem. 1994;269:6064–6071. [PubMed] [Google Scholar]
- 33.Cudny H., Deutscher M.P. 3′ processing of tRNA precursors in ribonuclease-deficient Escherichia coli. Development and characterization of an in vitro processing system and evidence for a phosphate requirement. J. Biol. Chem. 1988;263:1518–1523. [PubMed] [Google Scholar]
- 34.Harlow L.S., Kadziola A., Jensen K.F., Larsen S. Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding. Protein Sci. 2004;13:668–677. doi: 10.1110/ps.03477004. [DOI] [PMC free article] [PubMed] [Google Scholar]