Abstract
Pleckstrin homology (PH) domains are found in numerous membrane-associated proteins and have been implicated in the mediation of protein-protein and protein-phospholipid interactions. Dynamin, a GTPase required for clathrin-dependent endocytosis, contains a PH domain which binds to phosphoinositides and participates in the interaction between dynamin and the βγ subunits of heterotrimeric G proteins. The PH domain is essential for expression of phosphoinositide-stimulated GTPase activity of dynamin in vitro, but its involvement in the endocytic process is unknown. We expressed a series of dynamin PH domain mutants in cultured cells and determined their effect on transferrin uptake by those cells. Endocytosis is blocked in cells expressing a PH domain deletion mutant and a point mutant that fails to interact with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. In contrast, expression of a point mutant with unimpaired PI(4,5)P2 interaction has no effect on transferrin uptake. These results demonstrate the significance of the PH domain for dynamin function and suggest that its role may be to mediate interactions between dynamin and phosphoinositides.
Dynamins are GTPases required for budding of clathrin-coated vesicles from the plasma membrane (reviewed in references 7, 22, 37, and 40) and also implicated in the internalization of caveolae (10, 25) and in vesicle budding from the trans-Golgi network (15). Two forms of dynamin have been partially characterized: neuronal specific dynamin I (DI), implicated in presynaptic vesicle recycling, and ubiquitous dynamin II (DII), believed to participate in receptor-mediated endocytosis. Dynamins contain three distinct domains: an N-terminal catalytic GTP-binding domain (around residues 1 to 300); a central pleckstrin homology (PH) domain (around residues 510 to 626), potentially involved in phospholipid and protein interactions; and a C-terminal proline/arginine-rich domain (PRD) (around residues 750 to 860) with several Src homology 3 binding motifs, which is required for targeting dynamin to the clathrin-coated pit (26, 30).
Numerous in vivo and in vitro studies have demonstrated that dynamin self-assembly and GTP hydrolysis are key elements of dynamin function. Dynamin mutants deficient in GTP binding and hydrolysis block the constriction of assembled clathrin-coated pits (38). Synaptosomes depolarized in the presence of GTPγS accumulate membrane-tethered coated vesicles with elongated necks surrounded by stacked dynamin collars (34). Dynamin “tubulates” liposomes upon binding to them and vesiculates them upon GTP addition (32, 33). Hence, GTP hydrolysis seems necessary for the scission of budding vesicles and for the disassembly of dynamin coils. Similar coiled structures can be generated from pure dynamin alone (13), provided that a region located between the PH domain and the PRD is not deleted (23). This region, termed the GTPase effector domain, appears to be necessary for the stimulation of enzymatic activity that accompanies dynamin self-association.
Several in vitro activators of dynamin GTPase activity have been identified, and it is likely that they stimulate activity by facilitating dynamin-dynamin interactions, either by providing a surface for self-association (as in the case of microtubules or phospholipid vesicles) or by directly cross-linking dynamin molecules (Grb2 and antidynamin antibodies). Until recently, the PRD was considered the sole site of interaction between dynamin and its GTPase activators, and indeed, deletion of the PRD results in loss of microtubule- and Grb2-stimulated GTPase activities (12, 18). However, stimulation of activity by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is nearly unaffected by this treatment, demonstrating that its interaction site resides elsewhere on the dynamin molecule (18). A growing body of evidence points to the PH domain as the likeliest phosphoinositide-binding site. PH domains are protein modules of approximately 120 amino acids with similar tertiary structures (8, 29). The expressed dynamin PH domain, like those of numerous other proteins, can bind to phosphoinositides in vitro (27, 42). Most importantly, dynamin mutants with deleted PH domains fail to bind phosphoinositides and consequently lose PI(4,5)P2-stimulated GTPase activity (27).
To examine the potential in vivo significance of the dynamin-phosphoinositide interaction we tested the effect of expression of dynamin PH domain mutants on transferrin uptake in Cos-7 cells. The same approach was used before to show that GTPase defective mutants inhibit endocytosis of host cells (11, 38). Those studies showed that mutant forms of DI are targeted to the coated pit region of cells that normally express only DII (HeLa and Cos-7), provided that the carboxyl-terminal domains of the mutant dynamins are intact. We report here that endocytosis is inhibited in cells expressing dynamin with a large deletion in the PH domain and in those expressing a point mutant which lacks PI(4,5)P2-stimulated GTPase activity. These are the first dynamin dominant-negative constructs with mutations outside the GTP-binding domain. Our results support the view that clathrin-dependent endocytosis may be subject to regulation by phosphoinositides and demonstrate the importance of the PH domain for normal dynamin function.
MATERIALS AND METHODS
Materials.
Phosphatidylcholine and PI(4,5)P2 were from Calbiochem. Monoclonal antibodies against residues 45 to 358 of DI were from Transduction Laboratories. Antipeptide polyclonal antibodies against residues 607 to 624 (anti-PH domain) (31) and polyclonal antibodies raised against intact rat brain DI were a generous gift from Thomas Südhof, University of Texas Southwestern Medical Center. Protease inhibitors, taxol, and GTP were from Sigma. [γ-32P]GTP was from Amersham. Glutathione-Sepharose was from Pharmacia Biotech Inc. Fluorescein-labeled transferrin and rhodamine-labeled goat anti-rabbit antibodies were from Molecular Probes. Restriction endonucleases were from New England Biolabs.
Mutagenesis.
Starting with pCMV96-7 (31) (rat dynamin Iaa in a mammalian expression vector), a PH domain deletion mutant (DI ΔPH) and two point mutants (DI K535M and DI K561M) were generated by the overlap extension procedure (14). Amino acids 541 to 618 were deleted in DI ΔPH. To obtain the DI N272 construct, a BglII site was inserted 5′ of methionine 272 in a PCR amplification; the amplified DNA was substituted for the region encoding the N terminus of dynamin. The integrity of the constructs was verified by sequencing.
Transfection and receptor-mediated endocytosis assay.
The mammalian expression vectors were transfected into coverslip-plated Cos-7 cells by using Lipofectamine from Gibco BRL according to the manufacturer’s instructions. Typical transfection efficiencies were 10 to 20%. At 48 h after transfection the cells were starved for 1 h in serum-free Dulbecco modified Eagle medium (DMEM). They were then labeled for 15 min with 20 μg of fluorescein-labeled transferrin per ml, rinsed briefly in phosphate-buffered saline, fixed with 3% paraformaldehyde for 25 min, permeabilized for 5 min with −20°C acetone, and immunostained with the antiserum against purified rat DI and rhodamine-labeled goat anti-rabbit immunoglobulin G secondary antibodies. Digital fluorescence images were acquired with a Photometrics (Tucson, Ariz.) cooled charge-coupled device (CCD) (384- by 576- by 14-bit Thompson chip) mounted on a Zeiss Axiovert 135 epifluorescence microscope.
Expression of DI mutants in Sf9 cells.
Wild-type DI, DI K535M, and DI K561M constructs were excised from the pCMV5 vector by BglII (5′) and partial SmaI (3′) digestion and ligated into the corresponding sites of the pBacPAK8 plasmid (Clontech). The resulting plasmids were cotransfected with Bsu36I-digested BacPAK6 viral DNA into Sf9 cells to produce recombinant baculoviruses which were then amplified. Typical batches of extracts were prepared by infecting 50 ml of Sf9 cells at 106 cells/ml (grown in IPL-41 medium with 10% fetal calf serum and 1% pleuronic acid) with recombinant baculoviruses at a multiplicity of infection of 10. After 64 h, the cells were harvested by centrifugation at 1,000 × g for 10 min and washed three times with phosphate-buffered saline. Cell pellets were resuspended in ice-cold solution containing 0.1 M 4-morpholineethanesulfonic acid (MES) (pH 7.0), 1 mM EGTA, 1 mM MgSO4, 1 mM dithiothreitol, and a range of protease inhibitors: 0.2 mM phenylmethylsulfonyl fluoride and 10 mg each of Na-benzoyl-l-arginine methyl ester, Na-p-tosyl-l-arginine methyl ester, Na-p-tosyl-l-lysine chloromethyl ketone, leupeptin, and pepstatin A per liter (buffer A). Cells were homogenized with a Dounce homogenizer with a 0.003-in. clearance, and then the homogenate was passed through a 271/2-gauge needle. The homogenate was centrifuged at 1,000 × g for 15 min, and the low-speed supernatant was centrifuged at 140,000 × g for 1 h. As monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, approximately 70% of expressed dynamin and dynamin mutants was present in the high-speed supernatants. Typically, levels of expression of wild type, DI K535M, and DI K561M were approximately 14, 18, and 35% of total extracted protein, respectively.
Other proteins.
Tubulin was purified according to the procedure of Williams and Lee (41), but MES instead of 1,4-piperazinediethanesulfonic acid (PIPES) buffer was used. For GTPase activation and binding experiments, tubulin was polymerized with taxol at a twofold molar excess to tubulin dimer. Grb2 was expressed in Escherichia coli as a fusion protein with glutathione S-transferase (GST) and purified on glutathione-Sepharose.
GTPase assays.
GTPase activities were measured by the release of 32Pi from [γ-32P]GTP (16) after incubation at 37°C in buffer A containing 1 mM MgGTP. The reaction times varied from 5 to 15 min, depending on dynamin or activator concentrations, to ensure that less than 15% of GTP was hydrolyzed. In some experiments (as indicated in the figure legends) buffer A contained 0.1 M NaCl. Dynamin concentrations used for obtaining specific activities (as in Fig. 4 and 5) were estimated by scanning Coomassie blue-stained gels of Sf9 cell extracts, using electrophoresed and stained bovine serum albumin to generate a standard curve.
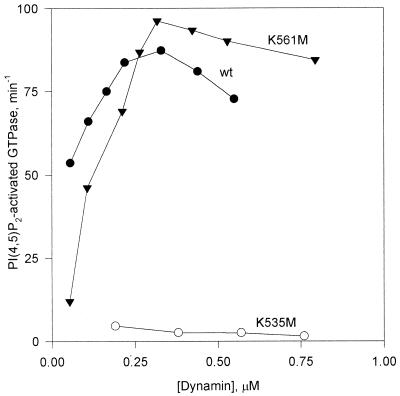
FIG. 4.
Specific PI(4,5)P2-stimulated GTPase activities of wild-type (wt) and mutant dynamins as a function of dynamin concentration. Specific activities were calculated by dividing moles of Pi released per minute by the dynamin concentration in each assay. Activities in the absence of PI(4,5)P2 were subtracted from each data point. Experiments were performed in buffer A containing 0.1 M NaCl. The PI(4,5)P2 concentration was 5 μM.
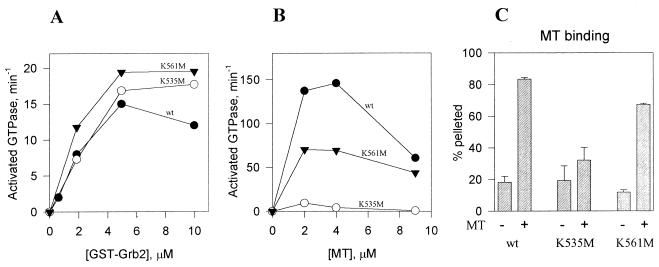
FIG. 5.
Interactions of dynamin mutants with GST-Grb2 and microtubules. (A) GTPase activation by GST-Grb2. The assays were performed in buffer A containing 0.1 M NaCl. (B) Effect of microtubules (MT) on GTPase activity. Assays were carried out in buffer A without added salt. (C) Binding of expressed dynamins to microtubules. Wild-type (wt) and mutant dynamins were subjected to a cosedimentation assay with microtubules (5 μM tubulin dimer). Means and standard errors of triplicate measurements are shown. Concentrations of wt DI, DI K561M, and DI K535M were, respectively, 0.33, 0.95, and 0.76 μM for panel A, 0.14, 0.47, and 0.39 μM for panel B, and 0.8, 0.8, and 1.14 μM for panel C.
Microtubule binding assay.
Samples of Sf9 cell extracts containing equal concentrations of expressed dynamin mutants were mixed with microtubules in buffer A, incubated for 15 min at room temperature, and centrifuged at 140,000 × g for 15 min. The pellets were resuspended in the original volume, and the amount of dynamin in the supernatants and pellets was determined by scanning densitometry of Coomassie blue-stained SDS-polyacrylamide gels.
Other methods.
Phospholipid vesicles were prepared as mixtures of 10% PI(4,5)P2 and 90% phosphatidylcholine as previously described (18). Cos-7 cells were grown in DMEM with 10% calf serum and penicillin-streptomycin from Gibco BRL. The protein concentration was determined as described by Bradford (4) with bovine serum albumin as a standard. SDS-polyacrylamide gel electrophoresis was carried out according to the method of Laemmli (17) as modified by Matsudaira and Burgess (21). Immunoblot analysis was carried out by the method of Towbin et al. (35) as described previously (39).
RESULTS
Expression of mutant dynamins.
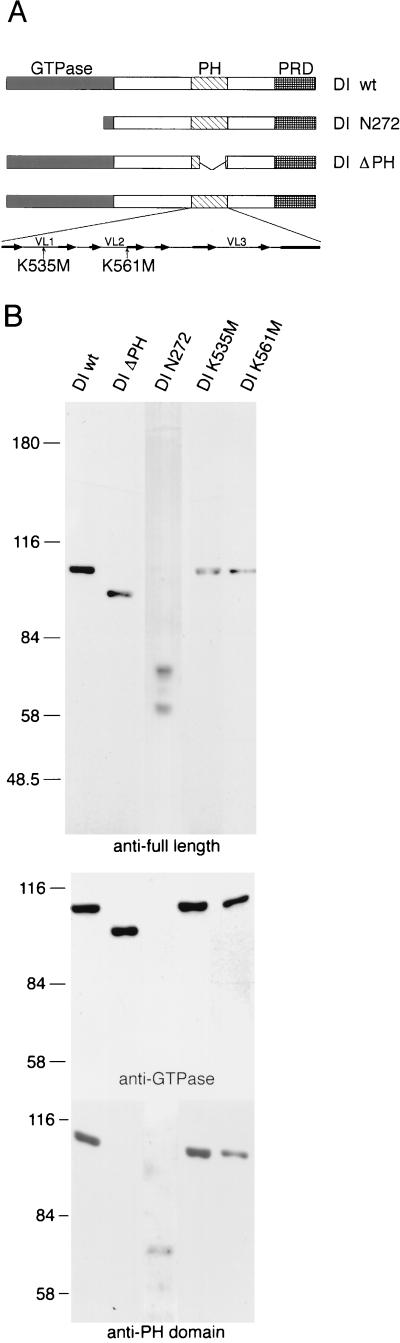
Three mutants were generated to evaluate the importance of the dynamin PH domain in endocytosis: DI ΔPH, which lacks most of the PH domain (residues 541 to 618 deleted), and two point mutants, DI K535M and DI K561M, with lysine 535 or 561 replaced by methionine. DI ΔPH was shown to lack PI(4,5)P2-stimulated GTPase activity but to retain Grb2-stimulated activity (27). Our choice of point mutations was based on nuclear magnetic resonance spectroscopy data (27, 42) (see Discussion). We also expressed, as controls, wild-type DI and a deletion mutant, DI N272, lacking most of the GTP-binding domain. Both proteins were previously tested in endocytosis assays (11); overexpression of wild-type DI did not interfere with receptor-mediated endocytosis, while DI N272 blocked transferrin uptake in host cells. A diagram of the expressed mutant dynamin constructs is shown in Fig. 1A.
FIG. 1.
Characterization of expressed dynamin mutants. (A) Diagram of the expressed mutant dynamin constructs. The PH domain structural elements (8) are shown in the bottom expansion, in which the arrows represent β-sheet strands, and the rectangle represents an α-helix. VL, variable loop. (B) Immunoblots of transfected Cos cell extracts, obtained with antidynamin antibodies raised against intact rat brain DI (anti-full length), against a synthetic peptide corresponding to residues 607 to 624 (anti-PH domain), and against residues 45 to 358 (anti-GTPase). The construct transfected is indicated above each lane. Numbers on the left are molecular masses, in kilodaltons. The 60-kDa band recognized by the antibodies in DI N272 preparations is probably a degradation product. wt, wild type.
The identity of expressed proteins was verified by immunoblotting (Fig. 1B). All five expressed proteins are recognized by antiserum generated against full-length rat DI. As expected, DI ΔPH is not recognized by antibodies directed against the PH domain, and DI N272 is not recognized by antibodies against the GTP-binding domain. Mutants DI K535M and DI K561M were recognized by all three antidynamin antibodies. Expressed proteins migrated on SDS gels with expected apparent molecular masses (97 kDa for wild-type DI, DI K535M, and DI K561M; 90 kDa for DI ΔPH; and 72 kDa for DI N272). The protein of approximately 60 kDa that was recognized by anti-full-length-dynamin antibodies in DI N272 cell extract was probably a degradation product of DI N272. Since this band was consistently observed despite the presence of protease inhibitors, it is possible that some degradation occurs in vivo. However, this control had the expected dominant-negative effect (see Fig. 2).
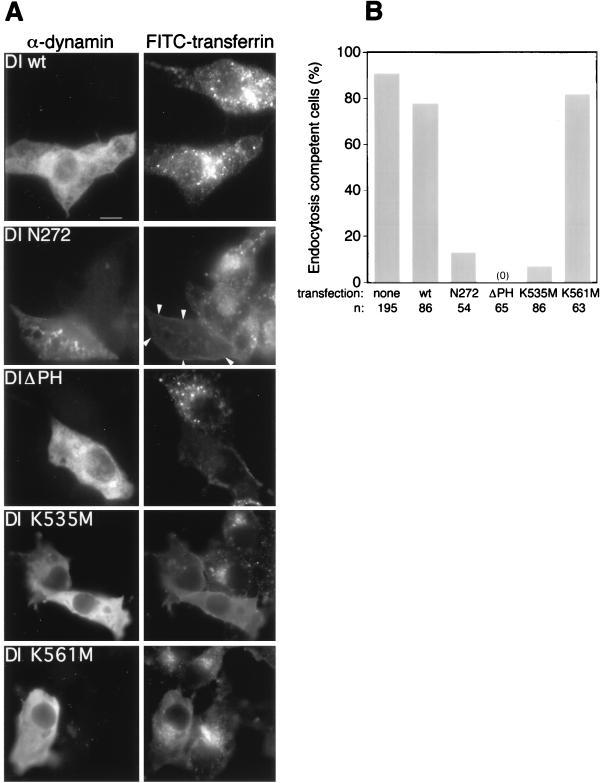
FIG. 2.
Effects of DI mutants on transferrin internalization. (A) Immunofluorescence images of cells. Left panels show the transfected cells identified by staining with antiserum against full-length DI (α-dynamin). Right panels show the fluorescence signals from cells which have internalized FITC-transferrin; arrowheads point to the cell transfected with DI N272. Bar, 10 μm. (B) Quantitative assessment of the endocytic poisoning potential of each construct. Cells considered endocytosis incompetent lacked any FITC-transferrin spots. wt, wild type.
Effect of mutant dynamin on endocytosis.
Our primary goal was to determine if mutations in the PH domain interfere with the ability of dynamin to function in endocytosis. Thus, we transiently transfected Cos-7 cells with the DI mutants described above and examined the ability of transfected cells to internalize transferrin. After a 15-min incubation with fluorescein isothiocyanate (FITC)-transferrin, the cells were processed for immunofluorescence with antibodies against full-length dynamin. In agreement with a previous report (11), staining was not evident in untransfected cells, allowing us to distinguish transfected from untransfected cells. Also consistent with previous observations (11), the mutant DI N272 gives a punctate staining pattern whereas the wild-type dynamin has a more diffuse distribution (Fig. 2A, left). Interestingly, mutants DI ΔPH, DI K561M, and DI K535M also have diffuse staining patterns, indicating a mainly cytosolic localization.
As expected, overexpression of wild-type DI did not affect the uptake of fluorescein-labeled transferrin, whereas uptake was essentially abolished in cells transfected with DI N272 (11) (Fig. 2A, right). Expression of DI ΔPH also blocked endocytosis in host cells. We next tested the effects on endocytosis of overexpression of the two PH domain point mutants, DI K535M and DI K561M. The mutant defective in phosphoinositide binding, DI K535M (see below) blocked transferrin uptake in host cells, whereas mutation of lysine 561, which does not interfere with phosphoinositide binding, did not yield a dominantly inhibitory dynamin. Only 7% of cells with expressed DI K535M (n = 86) were able to internalize transferrin, compared to 82% of cells with the DI K561M mutation (n = 63) (Fig. 2B).
In vitro properties of dynamin mutants.
The above results suggested to us that the inhibitory effect of DI K535M on endocytosis could be due to the inability of this mutant to interact with phosphoinositides. To test this possibility we overexpressed that mutant, as well as mutant DI K561M and wild-type DI, in Sf9 cells for biochemical characterization. As evident in Coomassie blue-stained gels (Fig. 3A), the expressed dynamins were by far the most abundant proteins of infected Sf9 cell extracts. In a typical preparation, bands corresponding to DI K535M, DI K561M, and wild-type dynamins accounted for 18, 35, and 14%, respectively, of the staining intensities of electrophoresed extracts. The identities of expressed dynamins were verified by immunoblotting, as in Fig. 1B. GTPase activation of wild-type and mutant dynamins by microtubules, Grb2, and PI(4,5)P2 were assayed by using extracts of infected Sf9 cells. Dynamins were not further purified because the extracts of uninfected cells had very low basal GTPase activities, which, most importantly, were unaffected by dynamin activators. Moreover, the estimated basal activities of wild-type and mutant dynamins in infected cell extracts were nearly identical to that of purified bovine brain dynamin (5 to 10 min−1) (18), indicating that they were neither stimulated nor inhibited significantly by endogenous factors.
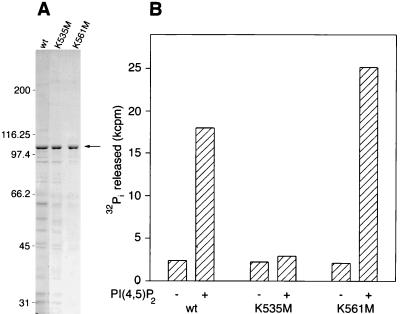
FIG. 3.
PI(4,5)P2 stimulation of the GTPase activity of dynamins expressed in Sf9 cells. (A) Coomassie blue-stained gel of Sf9 cell extracts used for GTPase assays. The arrow indicates dynamin. wt, wild type. Numbers on the left are molecular masses, in kilodaltons. (B) GTPase activities of Sf9 cell extracts containing wild-type (wt) or mutant dynamins measured in the absence or presence of 5 μM PI(4,5)P2. Assays were carried out in buffer A containing additionally 0.1 M NaCl for 5 min. Dynamin concentrations (0.44 μM wt DI, 0.57 μM DI K535M, and 0.53 μM K561M) were estimated by densitometric scanning of Coomassie blue-stained gels (as in panel A), using electrophoresed bovine serum albumin to generate a standard curve. Typical GTPase activities of mock-infected Sf9 cell extracts were one-third to one-half of the basal values of dynamin-expressing extracts.
Figure 3B shows that DI K561M, like wild-type dynamin, is stimulated about 10-fold by PI(4,5)P2. This stimulation is on a level similar to that obtained for purified bovine brain dynamin (2, 18). In contrast, the GTPase activity of DI K535M is not affected by PI(4,5)P2. These results are consistent with the lipid binding experiments of Salim et al. (27) mentioned above. We and others have previously shown that, in the presence of microtubules or PI(4,5)P2-containing liposomes, specific GTPase activity increases cooperatively as a function of dynamin concentration (18, 36). This kinetic behavior is presumably due to dynamin self-association, which is facilitated by binding to surfaces provided by microtubules or liposomes. As shown in Fig. 4, wild-type dynamin and DI K561M display this characteristic cooperative increase in PI(4,5)P2-stimulated activity but DI K535M is not activated, even at high enzyme concentrations. The figure shows data obtained with 4 μM PI(4,5)P2, a concentration of lipid which is optimal for stimulation of wild-type and K561M dynamins. There was no evidence of PI(4,5)P2-stimulated GTPase activity of DI K535M over the range of lipid concentrations tested (0.5 to 10 μM).
We next asked whether PH domain point mutations affected dynamin’s interactions with microtubules or Grb2, two GTPase activators reported to interact with the C-terminal PRD (12). Grb2, a much less potent activator than PI(4,5)P2 or microtubules, stimulated the enzymatic activities of mutant and wild-type dynamins equally well (Fig. 5A) (27). This result indicates that the mutations altered neither the Grb2-binding site within the PRD (which overlaps the putative coated-pit targeting region of dynamin [26, 30]) nor the self-assembly associated with high enzymatic activity. Surprisingly, microtubule-stimulated GTPase activities of the PH domain point mutants were inhibited (Fig. 5B). Inhibition was more pronounced in DI K535M, which also has a much lower affinity for microtubules than do wild-type and DI K561M proteins (Fig. 5C). The physiological significance of this result is unclear because microtubules appear not to be involved in dynamin-dependent steps of endocytosis (24, 28). Also, unlike phosphoinositide- and Grb2-stimulated GTPase activities, microtubule-stimulated activity must be assayed under essentially salt-free conditions because the dynamin-microtubule interaction is abolished at physiological ionic strength. Apparently, lysines in the PH domain, along with arginines within the PRD, contribute to the charge-charge interactions that determine microtubule-dynamin binding.
DISCUSSION
The present results demonstrate the importance of the PH domain for dynamin function and support the hypothesis that interactions between the PH domain and phosphoinositides are critical for clathrin-dependent endocytosis. The most pertinent evidence for this is our finding that DI K535M, which is not stimulatable by PI(4,5)P2, is an inhibitor of transferrin uptake whereas mutant DI K561M, which behaves like wild-type dynamin in this regard, is not. A role for phosphoinositides in endocytosis was also implied by Bauerfeind et al. (3), who showed that synaptojanin, a phosphoinositide-5-phosphatase, forms a complex with dynamin and amphiphysin and localizes to the clathrin-coated pit. In a previous paper members of our group speculated that synaptojanin regulates dynamin by converting PI(4,5)P2 and PI(3,4,5)P3, which stimulate GTPase activity, to PI(4)P and PI(3,4)P2, which do not (2). Recently, an inactivating mutation in phosphatidylinositol 5-kinase, a key enzyme in the production of PI(4,5)P2 and PI(3,4,5)P3, was found to inhibit colony-stimulating factor 1 receptor-mediated endocytosis in NIH 3T3 cells (5).
Our data do not exclude the possibility that the PH domain serves to promote a protein-protein interaction rather than (or in addition to) a protein-lipid interaction. At present the only known protein to bind to the dynamin PH domain is the βγ subunit complex of heterotrimeric G protein, which was shown to inhibit basal and PI(4,5)P2-stimulated GTPase activities (19). However, Gβγ interacts with the C-terminal α-helical portion of the PH domain (20), which is far removed from residue 535 in the three-dimensional structure (8, 9). The strongest evidence that dynamin PH domains interact with specific proteins was provided by Artalejo et al. (1). They showed that rapid endocytosis following stimulation of chromaffin cells is blocked by introduction of expressed DI PH domains but not by the DII PH domain, even though DI and DII are stimulated equally well by PI(4,5)P2 (18). If specific PH domain-binding proteins are eventually identified it will be of great interest to measure their affinities for dynamin mutants, such as K535M, which block clathrin-dependent endocytosis.
The in vitro interaction between dynamin’s PH domain and phosphoinositides has been explored by two different groups (27, 42). Using nuclear magnetic resonance spectroscopy and soluble PI(4,5)P2 head group analogs, they proposed different phospholipid binding sites on the PH domain. Thus, Zheng et al. (42) found that K561 (and other residues, but not K535) showed chemical shift differences upon 1-(α-glycerophosphoryl)-inositol 4,5-bisphosphate binding. In contrast, the data of Salim et al. (27) implicate K535 (and other residues, but not K561) in d-myo-inositol 1,4,5-trisphosphate binding; moreover, binding of PI(4,5)P2 liposomes to GST-PH domains was abolished by the K535M mutation but nearly unaffected by the K561M mutation. Consistent with the report by Salim et al. (27), our GTPase activation data (Fig. 4) indicate that in the context of the full-length protein, K535 is necessary for PIP2 binding, but K561 does not affect this interaction significantly.
As dynamin mediates several stages of the coated-pit cycle, it remains to be determined at which step the PH domain’s function is crucial. In this regard, further insights may be obtained from electron microscopy and in vitro endocytosis assays. Our data, together with a substantial body of additional circumstantial evidence (6), implicate phosphoinositides in clathrin-mediated endocytosis. However, while phosphoinositides are probably not inert membrane components during receptor-mediated endocytosis, details of their role remain to be clarified.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM55562-01A2, American Heart Association grant-in-aid 97G-111 (to B.B.), and National Institutes of Health training grant 2-T32 GM07062-22 (to M.A.).
We thank Hsin Chien Lin for assistance in molecular biological procedures and insightful discussions, Keng-Mean Lin for providing phospholipids, Irma Rodman for excellent technical assistance, and Jose Rizo-Rey for helpful discussions.
REFERENCES
- 1.Artalejo C R, Lemmon M A, Schlessinger J, Palfrey H C. Specific role for the PH domain of dynamin-1 in the regulation of rapid endocytosis in adrenal chromaffin cells. EMBO J. 1997;16:1565–1574. doi: 10.1093/emboj/16.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barylko B, Binns D, Lin K M, Atkinson M A L, Jameson D M, Yin H L, Albanesi J P. Synergistic activation of dynamin GTPase by GRB2 and phosphoinositides. J Biol Chem. 1998;273:3791–3797. doi: 10.1074/jbc.273.6.3791. [DOI] [PubMed] [Google Scholar]
- 3.Bauerfeind R, Takei K, De Camilli P. Amphiphysin I is associated with coated endocytic intermediates and undergoes stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem. 1997;272:30984–30992. doi: 10.1074/jbc.272.49.30984. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Davis J N, Rock C O, Cheng M, Watson J B, Ashmun R A, Kirk H, Kay R J, Roussel M F. Complementation of growth factor-dependent mitogenic signaling by a truncated phosphatidylinositol 4-phosphate 5-kinase. Mol Cell Biol. 1998;17:7398–7406. doi: 10.1128/mcb.17.12.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Camilli P, Emr S D, McPherson P S, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 7.De Camilli P, Takei K, McPherson P S. The function of dynamin in endocytosis. Curr Biol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 9.Fushman D, Cahill S, Lemmon M A, Schlessinger J, Cowburn D. Solution structure of the pleckstrin homology domain of dynamin by heteronuclear NMR spectroscopy. Proc Natl Acad Sci USA. 1995;92:816–820. doi: 10.1073/pnas.92.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henley J R, Krueger E W A, Oswald B J, McNiven M A. Dynamin-mediated internalization of caveolae. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herskovits J, Burgess C, Obar R, Vallee R. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–576. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herskovits J, Shpetner H, Burgess C, Vallee R. Microtubules and SRC homology 3 domains stimulate the dynamin GTPase via its C-terminal domain. Proc Natl Acad Sci USA. 1993;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinshaw J, Schmid S. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Jones S M, Howell K E, Henley J R, Cao H, McNiven M A. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- 16.Korn E D, Collins J H, Maruta H. Myosins from Acanthamoeba castellanii. Methods Enzymol. 1982;85:357–363. doi: 10.1016/0076-6879(82)85034-9. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lin H C, Barylko B, Achiriloaie M, Albanesi J P. Phosphatidylinositol (4,5)-bisphosphate-dependent activation of dynamins I and II lacking the proline/arginine-rich domains. J Biol Chem. 1997;272:25999–26004. doi: 10.1074/jbc.272.41.25999. [DOI] [PubMed] [Google Scholar]
- 19.Lin H C, Gilman A G. Regulation of dynamin I GTPase activity by G protein βγ subunits and phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1996;271:27979–27982. doi: 10.1074/jbc.271.45.27979. [DOI] [PubMed] [Google Scholar]
- 20.Liu J-P, Yajima Y, Li H, Ackland S, Akita Y, Stewart J, Kawashima S. Molecular interactions between dynamin and G-protein βγ-subunits in neuroendocrine cells. Mol Cell Endocrin. 1997;132:61–71. doi: 10.1016/s0303-7207(97)00120-2. [DOI] [PubMed] [Google Scholar]
- 21.Matsudaira P T, Burgess D R. SDS microslab linear gradient polyacrylamide gel electrophoresis. Anal Biochem. 1978;87:386–396. doi: 10.1016/0003-2697(78)90688-7. [DOI] [PubMed] [Google Scholar]
- 22.McClure S J, Robinson P J. Dynamin, endocytosis and intracellular signalling. Mol Membr Biol. 1996;13:189–215. doi: 10.3109/09687689609160598. [DOI] [PubMed] [Google Scholar]
- 23.Muhlberg A B, Warnock D E, Schmid S L. Domain structure and intramolecular regulation of dynamin. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda Y, Nakata T, Hirokawa N. Localization of dynamin: widespread distribution in neurons and association with membranous organelles. Neuroscience. 1993;55:113–127. doi: 10.1016/0306-4522(93)90459-s. [DOI] [PubMed] [Google Scholar]
- 25.Oh P, McIntosh D P, Schnitzer J E. Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J Cell Biol. 1998;141:101–114. doi: 10.1083/jcb.141.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto P M, Herskovits J S, Vallee R B. Role of basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 27.Salim K, Bottomley M J, Querfurth E, Zvelebil M J, Gout I, Scaife R, Margolis R L, Gigg R, Smith C I E, Driscoll P C, Waterfield M D, Panayotou G. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 28.Scaife R, Margolis R L. Dynamin coated tubular membrane invaginations induced by GTPγS in nerve terminals. J Cell Biol. 1990;111:3023–3033. doi: 10.1083/jcb.111.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 30.Shpetner H S, Herskovits J S, Vallee R B. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Sontag J-M, Fykse E M, Ushkaryov Y, Liu J-P, Robinson P J, Südhof T C. Differential expression and regulation of multiple dynamins. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- 32.Sweitzer S M, Hinshaw J E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 33.Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- 34.Takei K, McPherson P, Schmid S, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTPgammaS in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuma P, Collins C. Activation of dynamin GTPase is a result of positive cooperativity. J Biol Chem. 1994;269:30842–30847. [PubMed] [Google Scholar]
- 37.Urrutia R, Henley J R, Cook T, McNiven M A. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Bliek A, Redelmeier T, Damke M, Tisdale E, Meyerowitz E, Schmid S. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner M C, Barylko B, Albanesi J P. Tissue distribution and subcellular localization of mammalian myosin I. J Cell Biol. 1992;119:163–170. doi: 10.1083/jcb.119.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warnock D E, Schmid S L. Dynamin GTPase, a force-generating molecular switch. Bioessays. 1996;18:885–893. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- 41.Williams R C, Lee J C. Preparation of tubulin from brain. Methods Enzymol. 1982;85:376–385. doi: 10.1016/0076-6879(82)85038-6. [DOI] [PubMed] [Google Scholar]
- 42.Zheng J, Cahill S M, Lemmon M A, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the PH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]