Abstract
USF1 and USF2 are basic helix-loop-helix transcription factors implicated in the control of cellular proliferation. In HeLa cells, the USF proteins are transcriptionally active and their overexpression causes marked growth inhibition. In contrast, USF overexpression had essentially no effect on the proliferation of the Saos-2 osteosarcoma cell line. USF1 and USF2 also lacked transcriptional activity in Saos-2 cells when assayed by transient cotransfection with USF-dependent reporter genes. Yet, there was no difference in the expression, subcellular localization, or DNA-binding activity of the USF proteins in HeLa and Saos-2 cells. Furthermore, Gal4-USF1 and Gal4-USF2 fusion proteins activated transcription similarly in both cell lines. Mutational analysis and domain swapping experiments revealed that the small, highly conserved USF-specific region (USR) was responsible for the inactivity of USF in Saos-2 cells. In HeLa, the USR serves a dual function. It acts as an autonomous transcriptional activation domain at promoters containing an initiator element and also induces a conformational change that is required for USF activity at promoters lacking an initiator. Taken together, these results suggest a model in which the transcriptional activity of the USF proteins, and consequently their antiproliferative activity, is tightly controlled by interaction with a specialized coactivator that recognizes the conserved USR domain and, in contrast to USF, is not ubiquitous. The activity of USF is therefore context dependent, and evidence for USF DNA-binding activity in particular cells is insufficient to indicate USF function in transcriptional activation and growth control.
USF is a family of evolutionarily conserved basic-helix-loop-helix-leucine zipper (bHLH-zip) transcription factors (8, 11, 17, 36) that interact with the DNA at symmetrical E boxes with the consensus sequence 5′ GGTCACGTGACC 3′ (2). In mammals, there are two ubiquitously expressed genes, USF1 and USF2, that play an essential role during embryonic development and also have pleiotropic effects in adult animals (10, 21, 37, 38).
One noticeable feature of the USF proteins is that they share with the Myc oncoproteins both a similar polypeptide structure and a similar DNA-binding specificity (2, 3, 14, 25). Yet, the cellular functions of USF and Myc seem quite different. For example, overexpression of c-myc, in collaboration with a second oncogene, is sufficient to trigger the complete transformation of primary embryo fibroblasts (20). Overexpression of USF can specifically abolish this transforming ability of c-Myc (22). Similarly, while c-Myc overexpression often contributes to the rapid proliferation of tumor cells, USF overexpression has instead been found to inhibit growth in a number of cancer cell lines (16, 22). Together, these observations suggest that USF and Myc play antagonistic roles in the control of mammalian cell proliferation.
The USF1 and USF2 polypeptides are very similar in their C-terminal regions, which contain the bHLH-zip domain, and consequently display identical dimerization and DNA-binding specificities (35, 36). Also extremely conserved between USF1 and USF2 is a small domain, termed the USF-specific region (USR), that is apparently unique to the USF proteins. The USR, which is located just upstream of the basic region, is essential for transcriptional activation by USF at promoters containing both a TATA box and an initiator element (23). The amino acid sequences of USF1 and USF2 are considerably more divergent in their N-terminal regions, which include in each case at least one other transcriptional activation domain (15, 23). The major USF species present in most tissues and cell lines is the heterodimer between USF1 and USF2. USF1 homodimers are less abundant, and USF2 homodimers are usually quite scarce (36, 37, 39).
Identification of cellular genes that are regulated by USF is complicated by the fact that most USF binding sites can also be recognized by other helix-loop-helix proteins, including all of the Myc (4, 14) and TFE3 (1, 7) family members. In addition, given their similarities but also their differences, various USF dimers may have both common and unique target genes (37). Among the proposed USF targets, it is noteworthy that many, including p53, transforming growth factor β2, and cyclin B1, are genes themselves involved in proliferation or cell cycle control (6, 27, 31).
While investigating the transcriptional and growth inhibitory properties of USF, we discovered that although the DNA-binding activity of USF was present in every cell line and tissue investigated, the transcriptional activities of the USF proteins varied. Here we report an analysis of the loss of USF function in the Saos-2 osteosarcoma cell line. The results of this analysis strongly suggest the existence of at least one additional protein, functioning as a specialized coactivator, that is absolutely required for USF function and, in contrast to USF itself, is not ubiquitously expressed.
MATERIALS AND METHODS
Cell culture and transfections.
The Saos-2 cell line was obtained from the American Type Culture Collection (Rockville, Md.). Both HeLa and Saos-2 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% donor calf serum (Sigma). HeLa cells were transfected by the calcium phosphate precipitation method as previously described (23). For Saos-2 cells, the transfection procedure was optimized by leaving the precipitate in contact with the cells for only 5 h in a 5% CO2 incubator. To compare results between the two cell lines, cotransfected plasmids included in each case 2.2 μg of pSV40-β-galactosidase as an internal control and the amounts of extracts used for electrophoretic mobility shift assay (EMSA) and Western blot analysis were normalized to equal β-galactosidase units. Luciferase and chloramphenicol acetyltransferase (CAT) assays were carried out as previously described (23).
For colony formation assay, the cells were transfected with 2 μg of pSV2neo and 6 μg of either pSG5, psvUSF1, or psvUSF2 (23, 24). After 3 weeks of selection in G418 (400 μg/ml), resistant colonies were stained with crystal violet and counted.
Plasmids.
Reporter plasmids were as described previously (24, 23). Derivation of the expression vectors for USF2ΔB, U2ΔN, U2Δ(7-186), U2ΔUSR, and G-U2(96-199) (Gal4-USF2) has also been previously reported (23). Construction of the USF2-VP16 expression vector, a generous gift from Howard Towle (University of Minnesota), is described in the work of Kayto et al. (12). Plasmid G-U1NS (Gal4-USF1) was constructed by inserting the end-filled NarI-to-ScaI fragment of the USF1 cDNA into the SmaI site of a pSG424 vector (29). The resulting Gal4-USF1 fusion protein therefore contained amino acids 80 to 169 of human USF1 downstream of the DNA-binding domain of Gal4 comprising amino acids 1 to 147. For construction of the U2ΔE5 mutant lacking amino acids 144 to 188 of USF2, restriction sites for NheI were introduced on each side of the exon 5 region in psvUSF2 by using the Transformer Site-Directed Mutagenesis kit (Clontech). The exon 5 region was then excised from the resulting plasmid by cleavage with NheI followed by intramolecular religation. For construction of pUSF2-TFE3, a DNA fragment encoding the bHLH-zip region of TFE3 (amino acids 130 to 234) was amplified by PCR using primers containing XhoI sites. The PCR product was then polymerized, cut with XhoI, and cloned into an XhoI-cut psvUSF2 vector. The same strategy was used to construct the E5-USF2 expression plasmid with a BamHI-restricted PCR fragment encoding amino acids 142 to 199 of USF2 that was cloned into a BamH1-cut psvUSF2 vector. The construction of all new plasmids was verified by DNA sequencing.
Construction and characterization of A-USF.
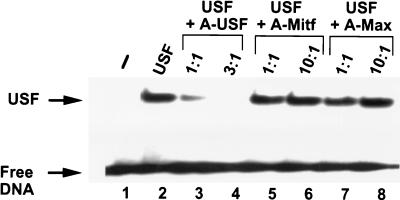
The USF dominant-negative mutant (A-USF) coding sequence was cloned as an NdeI-HindIII fragment into a pRc/CMV vector (Invitrogen) modified to contain an N-terminal hemagglutinin epitope (MYPYDVPDYA) and a new polylinker (pRc/CMV566). The protein sequence of A-USF is MAYPYDVPDYAHM-ASMTGGQQMGR-DPDE EEDDE EELE EDLENWIVQLSKIIPDCSMESTKSGQSKGGILSKACDYIQE LRQS N H RLSE E LQGLDQLQLDNDVLRQQVE DLKNKNLLLRAQLRHHGLEVVIKNDSN, where the bold letters are the acidic extension. The extent and specificity of the inhibition of USF DNA binding by A-USF were examined by EMSA (Fig. 1). This analysis revealed that one molar equivalent of A-USF inhibited USF DNA binding very strongly (more than fourfold) and three molar equivalents essentially abolished USF DNA binding. As a control, other bHLH-zip dominant-negative mutants containing the same acidic extension adjacent to the helix-loop-helix of Mitf (7, 9), or Max (4), were incapable of inhibiting USF DNA binding even at doses of 10 molar equivalents (Fig. 1). Natural USF1 dimers, at 4 μM in a buffer containing 12.5 mM phosphate (pH 7.4) and 150 mM KCl, have a melting temperature (Tm) of 41.7°C and a calculated stability of −9.3 kcal mol−1, while the heterodimers with A-USF display a Tm of 57.8°C and a stability of −13.9 kcal mol−1 (data not shown).
FIG. 1.
Inhibition of USF DNA binding by A-USF. The effect of different bHLH-zip dominant-negative mutants on USF DNA-binding activity was examined by EMSA. Binding reactions were performed under physiological conditions (12.5 mM phosphate [pH 7.4] and 150 mM KC1), and the reaction mixtures contained 10−11 M probe (17-bp double-stranded oligonucleotide containing a USF-specific binding site) alone (lane 1) or combined with 10−8 M recombinant USF1 (bHLH-zip domain) (lanes 2 to 8). When indicated, USF binding was challenged by the addition of the following dominant-negative mutants: A-USF at 1 × 10−8 or 3 × 10−8 M (lanes 3 and 4), A-Mitf at 1 × 10−8 or 1 × 10−7 M (lanes 5 and 6), or A-Max also at 1 × 10−8 or 1 × 10−7 M (lanes 7 and 8).
EMSA.
Mini-nuclear extracts from HeLa and Saos-2 cells were prepared in accordance with the procedure of Schreiber et al. (32). Standard DNA-binding reactions contained either 3 μg of protein from mini-nuclear extract or 1.5 μl of whole-cell extract prepared as for the luciferase assay, 1 μg of poly(dI-dC), and 0.1 ng of radiolabeled probe in 10-μl mixtures composed of 10 mM Tris-HCl (pH 7.9), 60 mM NaCl or KCl, 1 mM dithiothreitol, and 0.1% Triton X-100. Probes used for EMSA included a 150-bp DNA fragment or a 30- or 33-bp oligonucleotide, as indicated in the figure legends. When specified, the DNA-binding reaction mixtures also contained as a competitor DNA 10 ng of a 30-bp oligonucleotide containing or lacking the USF consensus binding site. After a 20-min incubation at 30°C, the reaction mixtures were supplemented with 2 μl of a 15% Ficoll solution and analyzed by electrophoresis on 4% acrylamide–0.2% bisacrylamide–22 mM Tris-borate (pH 8.3)–0.5 mM EDTA gels.
Antibodies and indirect immunostaining.
USF1- and USF2-specific rabbit polyclonal antibodies (35) were used for supershift analysis and immunostaining. USF polypeptide antibodies (Santa Cruz Biotechnology) were used for Western blot analysis. All procedures for immunostaining were carried out at room temperature. Transfected Saos-2 cells grown on coverslips were fixed in 4% formaldehyde for 15 min, washed three times with phosphate-buffered saline (PBS), permeabilized with 0.15% Triton X-100 in PBS, and washed three more times with PBS. After they were blocked with 1% bovine serum albumin (BSA) in PBS for 30 min, the cells were treated for 1 h with the primary antibodies diluted 1:200 in PBS containing 0.1% BSA, washed three times with PBS, and finally stained with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Cappel, West Chester, Pa.) diluted 1:1,000 in PBS containing 0.1% BSA. After counterstaining with propidium iodide, the coverslips were mounted on microscope slides by using 50% glycerol. Samples were examined and scanned at ×400 magnification with a Zeiss confocal laser scanning microscope. Composite images were assembled by using Adobe Photoshop and Canvas softwares.
RESULTS
USF is inactive in Saos-2 cells.
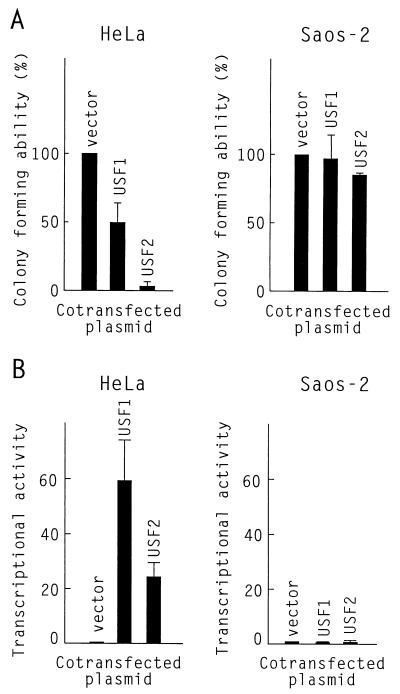
The growth inhibitory activity of the transcription factor USF in HeLa cells was demonstrated by a colony formation assay after cotransfection with a neomycin resistance gene (22). Overexpression of USF1 reduced by about 50% the number of neomycin-resistant colonies, while overexpression of USF2 essentially abolished colony formation. In contrast, USF1 or USF2 overexpression had essentially no effect on the colony-plating ability of Saos-2 cells (Fig. 2A).
FIG. 2.
USF1 and USF2 are both inactive in Saos-2 cells. (A) Colony formation assay. Plates of HeLa or Saos-2 cells were transfected with 2 μg of pSV2neo and 6 Εg of either psvUSF1, psvUSF2, or the pSG5 empty expression vector, as indicated. After G418 selection, colonies were stained with crystal violet and counted. The results shown are the average and standard deviation for colony numbers determined in three independent experiments. (B) Transient-transection assay. Plates of either HeLa or Saos-2 cells were transfected with 10 μg of the pU3MLLuc reporter plasmid and 9 μg of either pSG5, psvUSF1, or psvUSF2. Luciferase activities were determined 40 h after transfection. The results, expressed as fold activation, were averaged from a minimum of five independent experiments.
Since growth inhibition in HeLa cells requires the transcriptional activation domain of USF (22), this initial observation prompted us to examine the transcriptional activity of USF in Saos-2 cells. For this, we used the pU3MLLuc reporter, in which transcription of a luciferase gene is controlled by three USF binding sites upstream of the adenovirus major late minimum promoter (23). In Saos-2 cells, cotransfection of either USF1 or USF2 with the pU3MLLuc reporter did not enhance luciferase activity over the basal level obtained in the absence of exogenous USF. Yet, under the same conditions, transcription from the pU3MLLuc reporter in HeLa cells was stimulated about 60-fold by USF1 and 20-fold by USF2 (Fig. 2B). Taken together, these results suggest that exogenous USF was completely inactive in Saos-2 cells.
Expression, subcellular localization, and DNA-binding activity of USF in Saos-2 cells.
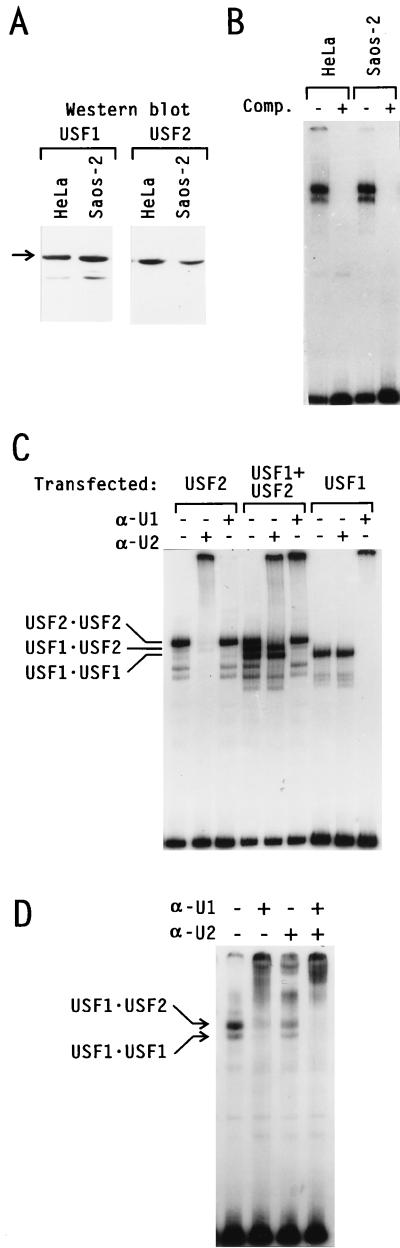
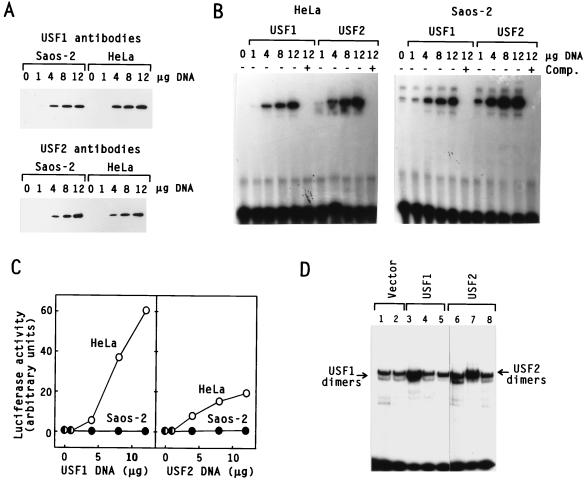
We next investigated whether there were differences in the expression, subcellular localization, or DNA-binding activity of USF that could account for the differences observed between Saos-2 cells and HeLa cells. As shown in Fig. 3A, Western blot analysis of endogenous USF in nuclear extracts from HeLa and Saos-2 cells prepared under identical conditions revealed comparable USF1 and USF2 polypeptide levels in both cell lines. EMSA analysis also demonstrated similar USF DNA-binding activities in the two cell lines (Fig. 3B). A supershift assay using rabbit polyclonal antibodies specific to either USF1 or USF2 (Fig. 3C) indicated that the major USF species in Saos-2 cells were the USF1-USF2 heterodimers; USF1 homodimers were less abundant, and USF2 homodimers were almost completely absent (Fig. 3D). This distribution is identical to that previously characterized in many other cell types, including HeLa (30, 35, 39, 36).
FIG. 3.
Expression of USF in Saos-2 cells. (A and B) Nuclear extracts from HeLa and Saos-2 cells prepared under identical conditions were used to compare the endogenous levels of USF1 and USF2 present in both cell lines by Western blot analysis (A) and EMSA (B). (C) Control for the specificity of the USF-specific antibodies in supershift analysis. DNA binding reaction mixtures were assembled with nuclear extracts from HeLa cells transiently transfected with either USF1, USF2, or both, as indicated. Prior to electrophoresis, the protein-DNA complexes were incubated with polyclonal antibodies specific to USF1 (α-U1) or USF2 (α-U2), as indicated above each lane. (D) Antibody supershift analysis of endogenous USF in Saos-2 nuclear extracts. Migration of the major USF dimers is indicated.
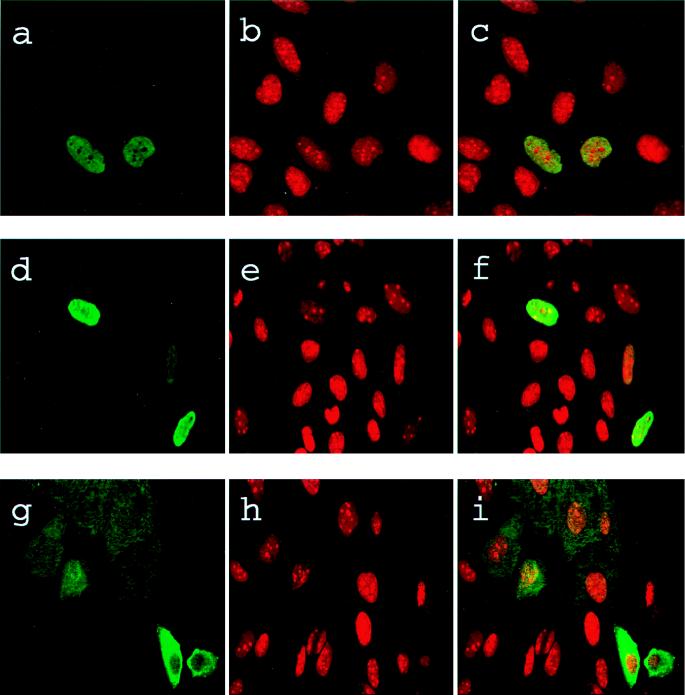
To examine the subcellular localization of USF in Saos-2 cells, transiently transfected cells were stained with USF1- or USF2-specific antibodies and fluorescein-conjugated secondary antibodies and then counterstained with propidium iodide and visualized by confocal microscopy. As illustrated in Fig. 4, staining of both USF1 and USF2 localized predominantly in the nuclei with nucleoli exclusion. In contrast, cells transfected with U2ΔBΔUSR, a USF2 mutant lacking both known nuclear localization signals (23), showed predominantly cytoplasmic staining. These experiments indicated that the loss of USF activity in Saos-2 cells was not related to an altered subcellular localization and that, at least for USF2, the same protein domains were required for nuclear localization in Saos-2 as in HeLa cells.
FIG. 4.
Subcellular localization of USF in Saos-2 cells. Cells were transfected with either USF2 (a to c), USF1 (d to f), or the U2ΔBΔUSR mutant (23) that lacks both of the USF2 nuclear localization signals (g to i). The transiently transfected cells were stained with USF1- or USF2-specific antibodies and fluorescein isothiocyanate-conjugated secondary antibodies (green) and counterstained with propidium iodide (red). Shown are confocal microscopy images that demonstrate the colocalization of the wild-type USF1 or USF2 proteins with DNA in the nuclei (c and f), while the nuclear localization mutant remained in the cytoplasm (i).
The overexpression levels and integrities of exogenous USF1 and USF2 proteins produced by transient transfection with different amounts of expression vectors in HeLa and Saos-2 cells were examined by Western blotting (Fig. 5A) and EMSA (Fig. 5B) and found to be similar. These titration experiments also served to rule out the possibility that the inactivity of USF in Saos-2 cells could be due to a particularly strong self-squelching effect. When expressed at very high levels, many transcriptional activators squelch activated transcription (28). This squelching phenomenon is thought to be due to the sequestration in solution of transcriptional components, preventing their interaction at gene promoters. However, decreasing the concentration of overexpressed USF1 or USF2 in Saos-2 cells had no effect on the activity of the cotransfected reporter, while in HeLa cells reporter activity decreased with decreasing USF concentrations (Fig. 5C). These results were inconsistent with a strong squelching phenomenon causing the inactivity of USF in Saos-2 cells.
FIG. 5.
Expression of exogenous USF in HeLa and Saos-2 cells. (A to C) Cells were transiently transfected by using the pU3MLLuc reporter and various amounts of the USF1 or USF2 expression vector. The same extracts were used to examine the expression level and integrity of the overexpressed USF proteins by Western blot analysis (A) and EMSA (B) and to quantitate transcriptional activities by the luciferase assay (C). EMSA analysis was carried out with a radiolabeled 30-bp oligonucleotide containing the USF consensus binding site. Unlabelled competitor oligonucleotide (Comp.) was added as indicated. (D) Control for the overexpression of USF in stably transfected Saos-2 cells. G418-resistant Saos-2 colonies transfected with pSV2neo and the indicated USF expression vector, as described for Fig. 2, were individually expanded and processed for mini-nuclear extract preparation. EMSA was carried out with a radiolabeled 33-bp oligonucleotide containing the adenovirus major late USF binding site. The migration of complexes containing exogenous USF1 and USF2 homodimers is indicated.
We also verified that the lack of growth inhibition by USF in Saos-2 cells observed in the colony formation assay was not due to a lack of expression in the stable transfection assay. Several G418-resistant colonies from Saos-2 plates transfected with pSV2neo and either USF1 or USF2 were individually expanded, and nuclear extracts were prepared. In different colonies, EMSA revealed weak to very strong overexpression of the transfected USF gene in comparison to the endogenous levels observed in colonies transfected with the empty vector (Fig. 5D). Thus, the inability of USF to affect the colony-plating ability of Saos-2 cells was not due to a lack of expression in stably transfected cells. Instead, it probably reflected the inability of the USF proteins to activate transcription in this particular cell line.
Saos-2 USF can mediate transcriptional activation by the varicella-zoster virus IE62 protein.
Although endogenous and ectopically expressed USF extracted from Saos-2 cells bound DNA in vitro (Fig. 3B and 5B), none of the experiments described above could rule out the possibility that DNA binding by USF was prevented in vivo, for instance, by some labile posttranslational modification or by interaction with a repressor. To examine the DNA-binding ability of Saos-2 USF in vivo, we took advantage of the known interaction between USF and the viral IE62 protein. In HeLa and other cell lines, IE62 is a very strong transcriptional activator at promoters containing a single USF binding site. This activation by IE62 utilizes either endogenous USF or ectopic USF1 or USF2 and can be inhibited by cotransfection of a USF dominant-negative mutant (24). Cooperation with IE62 requires the DNA-binding domain of USF as well as other USF domains involved in transcriptional activation (24).
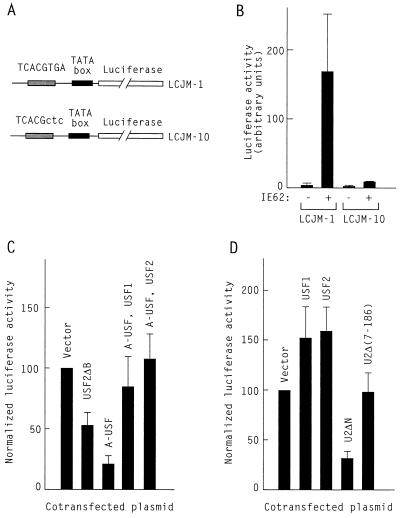
To determine whether IE62 could activate transcription in Saos-2 cells in cooperation with endogenous USF, cells were transfected with either the LCJM-1 reporter plasmid that contains the varicella-zoster virus DNA polymerase gene promoter driving luciferase or the related LCJM-10 plasmid in which a 3-bp mutation has been introduced in the USF binding site (24) (Fig. 6A). IE62 activated transcription from the LCJM-1 reporter in Saos-2 cells nearly 40-fold over the basal level, while mutation of the USF site in the LCJM-10 reporter essentially abolished IE62 transactivation (Fig. 6B). This result demonstrated that Saos-2 cells contained proteins that interacted with the USF binding site in plasmid LCJM-1 and mediated activation by IE62 through that site. To verify that these endogenous proteins were USF itself, we used USF dominant-negative mutants. Cotransfection of mutant USF2ΔB, which lacks the basic region of USF2, reduced transcriptional activation by IE62 in Saos-2 cells to 50% of the activity observed with IE62 alone (Fig. 6C). It is important to note that under the same conditions, inhibition by USF2ΔB in HeLa cells was usually more pronounced, reducing activation by IE62 to less than 20% of the control (24). However, this quantitative difference may simply reflect the lesser transfection efficiency for Saos-2 cells in comparison to that for HeLa cells. We therefore turned to a more potent dominant-negative mutant of USF, A-USF. In this construct, the basic region of the bHLH-zip domain of USF1 was replaced with an acidic sequence (EEEDDEEELEELE), which greatly stabilizes the heterodimers between A-USF and USF (18, 19, 26). Consequently, A-USF is a very efficient inhibitor of USF DNA binding (Fig. 1). Cotransfection of A-USF reduced transactivation by IE62 in Saos-2 cells to about 20% of the control value, demonstrating that endogenous USF was indeed an essential cooperating partner for activation by IE62 in Saos-2 cells, just as in HeLa cells. Transcriptional activation by IE62 in the presence of A-USF was restored by cotransfecting either USF1 or USF2, indicating that, as also true in HeLa cells, IE62 could cooperate with either of these transcription factors to activate transcription through USF sites in Saos-2 cells (Fig. 6C). These experiments demonstrate that both endogenous and ectopically expressed USF proteins were active in specific DNA binding in Saos-2 cells.
FIG. 6.
Transcriptional activation by IE62 in Saos-2 cells. (A) Schematic representation of the reporter constructs. LCJM-1 contains the varicella-zoster virus DNA polymerase promoter that drives transcription of a luciferase reporter gene. LCJM-10 is identical to LCJM-1 except for the indicated 3-bp substitution in the USF binding site. (B) Saos-2 cells were transfected with 10 μg of the indicated reporter plasmid and 0.4 μg of either the IE62 expression vector pCMV62 or the corresponding empty vector. Luciferase activities were determined 45 h after transfection. (C) Effect of USF dominant-negative mutants on IE62 activity in Saos-2 cells. Cotransfected plasmids included 10 μg of LCJM-1, 0.4 μg of pCMV62, and 5 μg each of the indicated USF expression vectors. The results of the luciferase assay were normalized relative to the activity obtained with IE62 alone. (D) Domains of USF required for cooperation with IE62 in Saos-2 cells. Cotransfected plasmids included 10 μg of LCJM-1, 0.4 μg of pCMV62, and 5 μg of the indicated expression vectors. The results of the luciferase assay were normalized relative to the activity obtained with IE62 alone.
The domains of USF required for interaction with IE62 in Saos-2 cells were investigated by examining the effect of cotransfecting different USF constructs. Cotransfection of either USF1 or USF2 did not significantly alter LCJM-1 transcription from the level observed with IE62 alone, indicating that endogenous USF levels were already sufficient to support full activation. Cotransfection of U2ΔN, a mutant containing the bHLH-zip domain of USF2 but lacking all N-terminal sequences, strongly inhibited IE62 activity (Fig. 6D). This demonstrated that other domains of USF2, besides those necessary for dimerization and DNA binding, were required for IE62 interaction. One of these IE62-interacting domains was located within the USR, since mutant U2Δ(7-186), which contains only the USR and bHLH-zip domains of USF2, was nearly as efficient at supporting IE62 activation as the full-length USF proteins (Fig. 6D). Since the USR is one of the most conserved USF domains, this result was consistent with the ability of IE62 to cooperate equally well with USF1 and USF2.
Activity of USF-VP16 fusion proteins in Saos-2 cells.
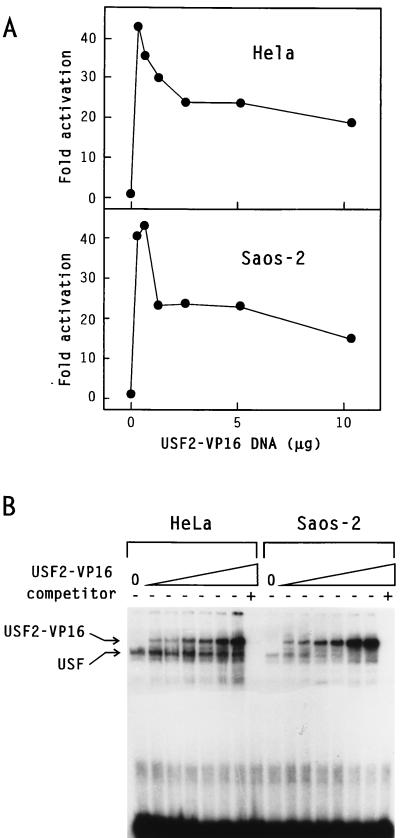
To determine whether exogenous USF could not only bind DNA in Saos-2 cells but also assemble into functional transcription complexes, we investigated the activity of different fusion proteins containing the DNA-binding domain of USF2 and the VP16 transcriptional activation domain. As shown in Fig. 7B, a hybrid protein containing the VP16 activation domain fused downstream of the leucine zipper of USF2 (12) was expressed at similar levels in HeLa and Saos-2 cells. In HeLa cells, USF2-VP16 efficiently activated transcription of the pU3MLLuc reporter gene when present at low concentrations. Greater expression levels, however, were inhibitory, presumably as a result of self-squelching (28). Interestingly, activation of pU3MLLuc transcription by USF2-VP16 in Saos-2 cells closely resembled that observed in HeLa cells, with a similar amplitude and a similar squelching at elevated concentrations (Fig. 7A). Identical results were obtained for fusion constructs containing the VP16 activation domain inserted either at the N-terminus or in the middle of USF2 (data not shown). Taken together, these experiments demonstrated that the transcriptional deficiency of Saos-2 cells was not a general phenomenon, since transactivators such as IE62 and USF2-VP16 were active in this cell line. Also, the DNA-binding domain of USF was clearly functional in Saos-2 cells, since activation by USF2-VP16 required the presence of USF binding sites in the reporter gene (data not shown). Thus, the inactivity of the wild-type USF proteins in Saos-2 cells was not caused by their inability to interact with the promoter DNA but by their inability to subsequently activate transcription.
FIG. 7.
Transactivation by USF2-VP16 in HeLa and Saos-2 cells. HeLa or Saos-2 cells were transfected with 10 μg of the pU3MLLuc reporter plasmid and variable amounts of the USF2-VP16 expression vector, as indicated. (A) Transcriptional activity. The results of the luciferase assay were averaged from two independent experiments and are expressed as fold activation over the basal level in the absence of USF2-VP16. (B) Levels of USF2-VP16 expression. The DNA-binding activity of USF2-VP16 was monitored by EMSA using a radiolabelled 33-bp oligonucleotide containing the USF consensus binding site. The migration of complexes containing endogenous USF or the USF2-VP16 protein is indicated.
The USR is responsible for the inactivity of USF2 in Saos-2 cells on initiator-containing reporters.
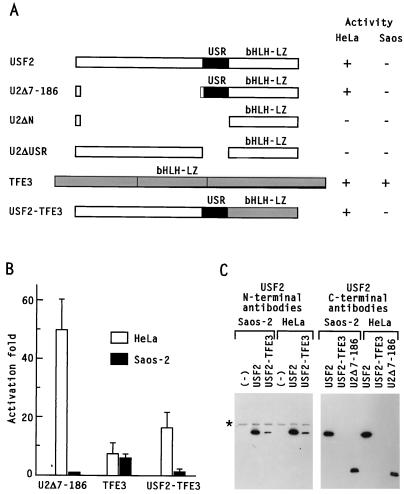
A series of deletion mutants and hybrid proteins was used to investigate the domains responsible for the inactivity of USF2 in Saos-2 cells. In the case of reporter genes containing an initiator element, previous studies had demonstrated that the USR of USF1 and USF2 functioned as an autonomous, specialized activation domain (23). In agreement with these earlier results, a construct containing only the USR and DNA-binding domains of USF2 (U2Δ7-186) was found to strongly activate the pU3MLLuc reporter in HeLa cells, while a USF2 mutant lacking only the USR (U2ΔUSR) was inactive (Fig. 8A). Like wild-type USF2, U2Δ7-186 was completely inactive in Saos-2 cells (Fig. 8B). This result narrowed down the possible domains responsible for USF2 loss of function in Saos-2 cells to either the USR or the C-terminal dimerization and DNA-binding domain. To discriminate between these last two possibilities, a domain swapping experiment was performed with the related bHLH-zip transcription factor TFE3 (1). Overexpression of wild-type TFE3 activated transcription from the pU3MLLuc reporter to similar levels in HeLa and Saos-2 cells, confirming that the transcriptional defect of Saos-2 cells was specific to USF (Fig. 8B). A hybrid protein composed of the N-terminal domain of USF2 and the DNA-binding domain of TFE3 displayed strong activity in HeLa cells (Fig. 8B), even though its expression level was lower than that of wild-type USF2 (Fig. 8C). In contrast, the same USF2-TFE3 fusion protein was completely inactive in Saos-2 cells (Fig. 8B). This result demonstrated that the DNA-binding domain of USF2 was dispensable for cell type specificity and that inactivity in Saos-2 cells was instead associated with the USR-containing N-terminal region.
FIG. 8.
The USR activation domain is responsible for the inactivity of USF2 in Saos-2 cells. Cotransfection experiments were carried out in HeLa and Saos-2 cells by using the pU3MLLuc reporter (10 μg) and different USF2 (5 μg) or TFE3 (15 μg) expression vectors. Reporter gene activity was determined 40 to 42 h after transfection. (A) Schematic representation of the USF constructs. (B) The results of the luciferase assays, expressed as fold activation over the levels determined in the presence of the corresponding empty vector, were averaged in each case from a minimum of three independent experiments. (C) Expression of USF2 and USF2-TFE3 constructs in HeLa and Saos-2 cells was compared in one set of extracts by Western blot analysis using peptide antibodies specific to either the N or C terminus of USF2. The asterisk indicates the migration of a USF-unrelated cross-reacting protein.
Taken together, the results of this mutational analysis strongly suggest that the inactivity of USF2 on the pU3MLLuc reporter in Saos-2 cells resulted from the inability of the USR to function as an initiator-dependent activation domain. Since the USR is highly conserved between USF1 and USF2, loss of USR function in Saos-2 cells accounted for the inactivity of both USF1 and USF2.
USF1 and USF2 contain activation domains that are active in Saos-2 cells.
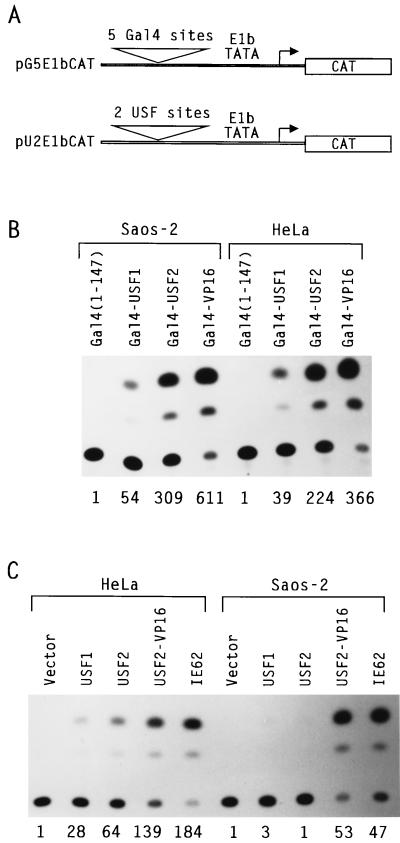
The USR is a specialized, initiator-dependent activation domain that does not function in the context of Gal4 fusion proteins (23). However, additional activation domains have also been identified in both USF1 and USF2 that can, at least in HeLa cells, function as Gal4 fusions (13, 15, 23). To investigate the activity of these more classical USF domains in Saos-2 cells, we carried out cotransfections experiments with the pG5E1bCAT reporter (Fig. 9A) and various Gal4 fusion proteins. As illustrated in Fig. 9B, control transfections with Gal4-VP16 revealed similar activities of this potent transactivator on the pG5E1bCAT reporter in both HeLa and Saos-2 cells. Interestingly, significant transcriptional activity was also detected in both cell lines for Gal4-USF2 and Gal4-USF1 hybrid proteins. This result demonstrated that the coactivators required for the classical activation domains of USF1 and USF2 were present in Saos-2 cells.
FIG. 9.
Context-dependent activity of the classical activation domains of USF1 and USF2 in HeLa and Saos-2 cells. (A) Schematic representation of the reporter plasmids. (B) Activity of Ga14-USF fusions in HeLa and Saos-2 cells. Cotransfections were carried out with 10 μg of pG5E1bCAT and 5 μg of the indicated Ga14 expression vectors. Transcriptional activities were determined by CAT assay. An autoradiogram from a representative experiment is shown, with quantitation indicated below each lane as the ratio of CAT activity observed in each case to the basal level observed with Ga14(1-147) alone. (C) Transactivation of the pU2E1bCAT reporter in HeLa and Saos-2 cells. Cotransfections were carried out with pU2E1bCAT (10 μg) and expression vectors for USF1 (9 μg), USF2 (9 μg), USF2-VP16 (0.32 μg), or IE62 (0.5 μg), as indicated, and analyzed by the CAT assay as described for panel B.
The existence of USF activation domains that were functional in Saos-2 cells suggested that the USF inactivity could be promoter dependent. To explore this possibility, we next compared the activities of USF1 and USF2 in HeLa and Saos-2 cells by using a reporter gene containing the same E1b core promoter present in the reporter used with the Gal4-USF fusion proteins (pU2E1bCAT) (Fig. 9A). Control transfections demonstrated that this reporter was strongly activated by USF2-VP16 as well as by IE62 in both HeLa and Saos-2 cells. In contrast, the natural USF1 and USF2 proteins again demonstrated transcriptional activity in HeLa cells exclusively and not in Saos-2 cells (Fig. 9C). Since the activity of the Gal4-USF fusions indicated a similar availability of coactivators in both cell lines, this observation suggested that the activity of the classical activation domains of USF1 and USF2 in their natural context was somehow inhibited in Saos-2 cells.
Essential role of the USR in transcriptional activation by USF in the absence of an initiator element.
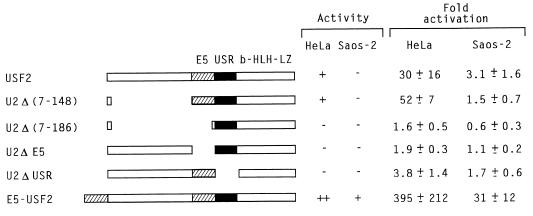
Earlier studies in HeLa cells had mapped the USF2 exon 5-encoded region as the activation domain essential both for activation by Gal4-USF2 and for activation of pU2E1bCAT by wild-type USF2 (23). Yet, the inability of USF2 to activate pU2E1bCAT in Saos-2 cells suggested additional requirements besides the presence of this exon 5 domain. These requirements were investigated by testing additional mutants (Fig. 10). In HeLa cells, an N-terminal deletion mutant of USF2 containing only the exon 5, USR, and DNA-binding domains [construct U2Δ(7-148)] (Fig. 10) was even more active than the wild-type protein in stimulating transcription of the pU2E1bCAT reporter. However, further deletion of the exon 5 region, as in construct U2Δ(7-186), abolished activity. A USF2 mutant lacking only the exon 5 region (construct U2ΔE5) proved also completely inactive. These results confirmed the importance of the exon 5 activation domain for transactivation of E1b-driven reporters. Furthermore, since the U2Δ(7-186) and the U2ΔE5 mutants both contained an intact USR domain and were fully active with the pU3MLLuc reporter (reference 23 and data not shown), this analysis also confirmed the specificity of the USR activation domain for promoters that contained an initiator element.
FIG. 10.
In its natural context, the classical activation domain of USF2 is controlled by the USR. Cotransfections were carried out in HeLa and Saos-2 cells by using the pU2E1bCAT reporter and the various USF mutants depicted on the left of the figure. The results, expressed as fold activation, were calculated from a minimum of three independent experiments. E5, exon 5-encoded activation domain (hatched boxes).
Interestingly, a small deletion removing only the USR abolished the activity of USF2 in HeLa cells on the pU2E1bCAT reporter despite the presence of an intact exon 5 domain (mutant U2ΔUSR) (Fig. 10). This result demonstrated that, within its normal context, activity of the exon 5 activation domain required the presence of the USR. Since the USR domain was found to be inactive in Saos-2 cells, its absolute requirement for activity of the exon 5 domain also explained the inactivity of USF2 in Saos-2 cells with the pU2E1bCAT reporter.
In wild-type USF2, the exon 5 and USR domains are normally adjacent. It was therefore important to establish whether this positioning played a role in the USR requirement for activity of the exon 5 domain. For this, we constructed a mutant in which an additional exon 5 domain was inserted at the N terminus of USF2, in a position expected to be out of the control of the USR. The resulting construct, E5-USF2, was found functional in Saos-2 cells, where its activity was similar to that of natural USF2 in HeLa cells (Fig. 10). This result confirmed the intrinsic activity of the exon 5 domain in Saos-2 cells. It also suggested that, in wild-type USF2, one of the USR functions is to trigger a conformational change that exposes the adjacent exon 5 activation domain. As expected, E5-USF2, with two active exon 5 domains, displayed in HeLa cells a greatly enhanced activity as compared to that of wild-type USF2 (Fig. 10).
DISCUSSION
The results presented in this report indicate a loss of USF function in the Saos-2 cell line. In HeLa cells, the USF proteins function as transcriptional effectors and, when overexpressed, inhibit proliferation. These two activities of USF are in all likelihood related. Indeed, effective growth inhibition by USF2 requires all functional domains of the protein, including those directly implicated in transcriptional activation (22, 22a). Therefore, it seems likely that the inability of USF to inhibit the growth of Saos-2 cells is a direct consequence of its transcriptional inactivity.
Several possible explanations for the absence of USF activity in Saos-2 cells were ruled out by our analysis. For instance, we showed that the USF proteins properly localize in the nucleus of Saos-2 cells (Fig. 4), where they are furthermore fully active in DNA binding (Fig. 3 and 5). This latter property is also demonstrated by the ability of both endogenous and exogenous USF to mediate transcriptional activation by IE62 in a USF binding site-dependent fashion (Fig. 6). It is further illustrated by the activity in Saos-2 cells of the USF2-VP16 fusion protein, which necessitates its specific interaction with the promoter DNA. The inactivity of both USF1 and USF2 in Saos-2 cells on all promoters tested is not due to a deficiency of general transcription factors and/or coactivators in this cell line since several other transcription factors were as active in Saos-2 cells as in HeLa cells. Activation domains that functioned similarly in both HeLa and Saos-2 cells included those of VP16, TFE3, and IE62, as well as the classical activation domains of USF1 and USF2. The transcriptional defect of Saos-2 cells is therefore not of a general nature but is instead specific to USF.
Given that, in Saos-2 cells, USF is capable of specific interaction with its cognate binding sites in gene promoters and contains at least one functional activation domain, its complete inactivity on all reporters tested indicates that this activation domain must be masked. Two different models can be considered to account for these observations. First, a corepressor, present in Saos-2 but not HeLa cells, could interact with promoter-bound USF and interfere with the function of its activation domain. Alternatively, a specific coactivator necessary for USF activity could be either absent or inactivated in Saos-2 cells. Although our results cannot formally exclude the repressor model, several observations seem inconsistent with this idea. First, one would expect that the large amounts of exogenous USF produced in transient-transfection assays would eventually titrate out the putative repressor and unmask the activity. Second, one would imagine that the interaction of a repressor with DNA-bound USF would also prevent activation by at least some of the USF fusion proteins. Yet, in all cases tested, insertion of an additional activation domain (e.g., that of VP16) at different locations within USF2 restored transcriptional activity in Saos-2 cells. It seems therefore more likely that the transcriptional activity of USF1 and USF2 is controlled by interaction of a specialized coactivator that recognizes the conserved USR domain and is somehow inactivated in Saos-2 cells.
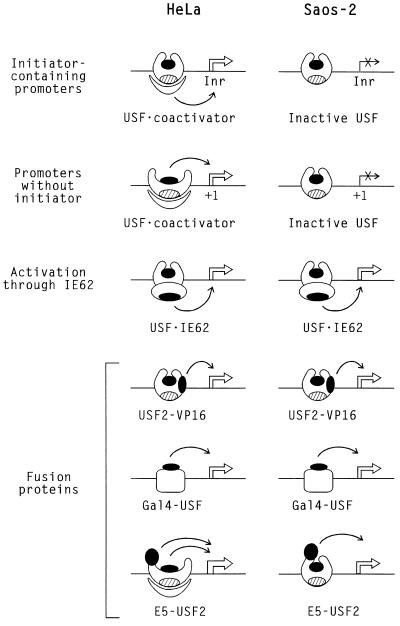
As depicted schematically in Fig. 11, the specific coactivator model is also entirely consistent with the importance of different domains for transcriptional activation of different reporter genes by USF2. In the presence of an initiator element, the USR was found both necessary and sufficient for USF2 activity in HeLa cells. This result is perfectly understandable if the cofactor mediates the stimulatory effect of USF by interacting with or perhaps recruiting initiator-binding proteins. In the absence of an initiator element, cofactor binding may trigger a conformational change in USF that exposes the classical activation domain. This would explain the dual requirement in this case for both the USR and the USF2 exon 5 activation domains. Finally, the USR-dependent coactivator model is also consistent with the context-independent activity of the USF-VP16 and Gal4-USF fusion proteins (Fig. 11).
FIG. 11.
Specific coactivator model for the cell-type-dependent activity of transcription factor USF. USF and related proteins are schematically represented with classical activation domains indicated by black ovals and the USR indicated by hatched ovals. In HeLa cells, association of a specific cofactor (crescent) interacting with the USR domain mediates the transcriptional activation by USF of initiator-containing promoters. In the case of promoters lacking an initiator, binding of the cofactor triggers a conformational change that exposes the other activation domain of USF. In Saos-2 cells, the absence of the cofactor, or its inability to recognize the USR due to altered posttranslational modification, is responsible for the complete inactivity of native USF proteins. Interaction of IE62 (oval) bypasses the requirement for the cellular coactivator. This model also explains the activity in Saos-2 cells of mutants such as USF2-VP16 and E5-USF2 in which the additional activation domains are independent of proper USR function.
Is the coactivator simply missing in Saos-2 cells? While this is certainly the simplest and perhaps most likely possibility, it is also conceivable that posttranslational modifications could either permit or prevent the USF-cofactor interaction. In that case, HeLa and Saos-2 cells would differ in the expression of the cognate modifying enzymes. Finally, it is important to note that the repressor and cofactor models of USF function are not necessarily exclusive. USF activity may well be controlled in different cells by both mechanisms. A similar regulation by both corepressors and coactivators is well documented in the case of the hormone receptors (33). Whatever the mechanism involved in the cell type-dependent activity of USF, the observations reported here strongly suggest the existence of at least one cellular factor that can simultaneously affect the activity of all USF proteins. This cellular cofactor could be very similar to the viral IE62 protein, which functions equally well with USF1 or USF2 and also recognizes the USR (Fig. 6D).
The inactivity of USF is not unique to Saos-2 cells. Preliminary studies in several cell lines suggested that a loss of USF function is not uncommon in transformed cells, especially among those derived from breast tumors (10a). These observations may have profound implications for the mechanisms of cancer progression. Indeed, our studies of embryonic fibroblasts suggested that the activity of USF could be essential in protecting cells against the tumorigenic potential of Myc (22). Myc overexpression, whether due to gene amplification, translocation, or increased message stability, is a common event in tumor progression that favors rapid proliferation (16). In other contexts, events leading to USF inactivation may well promote uncontrolled growth just like Myc deregulation does. While simultaneous inactivation of all alleles of the USF1 and USF2 genes is obviously unlikely to occur, the results described here strongly suggest the existence of a cofactor or modifying enzyme that is absolutely required for USF function. Thus, a complete loss of USF function could be brought about by the inactivation of a single gene, and such an event could play an essential role in tumorigenesis. The fact that Saos-2 cells are known to be deficient in both p53 and Rb activities (5, 34) raises the intriguing possibility that the inactivity of USF may come as a consequence of the loss of one of these two tumor suppressors. However, preliminary experiments indicated that neither p53 nor Rb overexpression could restore USF activity in Saos-2 cells, as determined by a transient-cotransfection assay (data not shown). Those results seem to rule out a direct involvement of either p53 or Rb in the cell-type-dependent activity of USF.
ACKNOWLEDGMENTS
We are grateful to Howard C. Towle for the USF2-VP16 construct, to Kuo Ooi for assisting in plasmid construction, and to Marilyn N. Szentirmay and Michael W. Dyke for critical reading of the manuscript.
This work was supported by grants G-1195 from the Robert A. Welch foundation, DMAD17-96-1-6221 from the Department of the Army, and CA79578 from the National Institutes of Health. Additional support was provided by institutional funds and by a postdoctoral fellowship from National Cancer Institute Training Grant CA09299 (T.L.). The confocal microscopy facility at the M.D. Anderson Cancer Center is supported by grant CA16672.
REFERENCES
- 1.Beckmann H, Su L-K, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer μE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 2.Bendall A S, Molloy P L. Base preference for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 5.Chandar N, Billing B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogswell J P, Godlevski M M, Bonham M, Bisis J, Babiss L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol Cell Biol. 1995;15:2782–2790. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher D E, Carr C S, Parent L A, Sharp P A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991;5:2342–2352. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- 8.Gregor P D, Sawadogo M, Roeder R G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 9.Hemesath T T, Steingrimsson E, McGill G, Hansen M J, Vaught J, Hodgkinson C A, Arnheiter H, Copeland N G, Jenkins N A, Fisher D E. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 10.Henrion A A, Vaulont S, Raymondjean M, Kahn A. Mouse USF1 gene cloning: comparative organization with the c-myc gene family. Mamm Genome. 1996;7:803–809. doi: 10.1007/s003359900241. [DOI] [PubMed] [Google Scholar]
- 10a.Ismail, P. M., and T. Lu. Unpublished observations.
- 11.Kaulen H, Pognonec P, Gregor P D, Roeder R G. The Xenopus B1 factor is closely related to the mammalian activator USF and is implicated in the developmental regulation of TFIIIA gene expression. Mol Cel Biol. 1991;11:412–424. doi: 10.1128/mcb.11.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kayto E N, Shih H M, Towle H C. Carbohydrate regulation of hepatic gene expression—evidence against a role for the upstream stimulatory factor. J Biol Chem. 1997;272:7525–7531. doi: 10.1074/jbc.272.11.7525. [DOI] [PubMed] [Google Scholar]
- 13.Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of an eukaryotic regulatory protein. Science. 1986;231:699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 14.Kerkhoff E, Bister K, Klempnauer K H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci USA. 1991;88:4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschbaum B J, Pognonec P, Roeder R G. Definition of the transcriptional activation domain of recombinant 43-kilodalton USF. Mol Cell Biol. 1992;12:5094–5101. doi: 10.1128/mcb.12.11.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koskinen P J, Alitalo K. Role of myc amplification and overexpression in cell growth, differentiation and death. Cancer Biol. 1993;4:3–12. [PubMed] [Google Scholar]
- 17.Kozlowski M T, Gan L, Venuti J M, Sawadogo M, Klein W H. Sea urchin USF: a helix-loop-helix protein active in embryonic ectoderm cells. Dev Biol. 1991;148:625–630. doi: 10.1016/0012-1606(91)90280-g. [DOI] [PubMed] [Google Scholar]
- 18.Krylov D, Echlin D, Kasai K, Arnheiter H, Taparowsky E, Vinson C. A general method to design dominant negative to bHLHXZip proteins that abolish DNA binding. Proc Natl Acad Sci USA. 1997;94:12274–12279. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Land H, Parada L F, Weinberg R A. Tumorigenic conversion of primary fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin Q, Luo X, Sawadogo M. Archaic structure of the gene encoding transcription factor USF. J Biol Chem. 1994;269:23894–23903. [PubMed] [Google Scholar]
- 22.Luo X, Sawadogo M. Antiproliferative properties of the USF family of transcription factors. Proc Natl Acad Sci USA. 1996;93:1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Luo, X. Unpublished observations.
- 23.Luo X, Sawadogo M. Functional domains of transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier J L, Luo X, Sawadogo M, Straus S E. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol. 1994;14:6896–6906. doi: 10.1128/mcb.14.10.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and Myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 26.Olive M, Krylov D, Echlin D R, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 27.Reisman D, Rotter V. The helix-loop-helix containing transcription factor USF binds to and transactivates the promoter of the p53 tumor suppressor gene. Nucleic Acids Res. 1993;21:345–350. doi: 10.1093/nar/21.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadowki I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski I, Ptashne M. A vector for expressing Ga14(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawadogo M, Van Dyke M W, Gregor P D, Roeder R G. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem. 1988;263:11985–11993. [PubMed] [Google Scholar]
- 31.Scholtz B, Kingsley-Kallesen M, Rizzino A. Transcription of the transforming growth factor-β2 gene is dependent on an E-box located between an essential cAMP resopnse element/activation transcription factor motif and the TATA box of the gene. J Biol Chem. 1996;271:32375–32380. doi: 10.1074/jbc.271.50.32375. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber E, Matthias P, Muhler M M, Schaffner W. Rapid detection of octamer binding proteins with min-extracts prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulman I G, Li C, Schwabe J W R, Evans R M. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 34.Shew J Y, Lin B T, Chen P L, Yang-Feng T L, Lee W H. C-terminal truncation of the retinoblastoma gene product leads to functional inactivation. Proc Natl Acad Sci USA. 1990;87:6–10. doi: 10.1073/pnas.87.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirito M, Walker S, Lin Q, Kozlowski M T, Klein W H, Sawadogo M. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 1992;2:231–240. [PMC free article] [PubMed] [Google Scholar]
- 36.Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirito M, Lin Q, Deng J M, Behringer R R, Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci USA. 1998;95:3758–3763. doi: 10.1073/pnas.95.7.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallet V S, Henrion A A, Bucchini D, Casado M, Raymondjean M, Kahn A, Vaulont S. Glucose-dependent liver gene expression in upstream stimulatory factor 2−/− mice. J Biol Chem. 1997;272:21944–21949. doi: 10.1074/jbc.272.35.21944. [DOI] [PubMed] [Google Scholar]
- 39.Viollet B, Lefrancois-Martinez A M, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]