Abstract
Proteins to be transported into the nucleus are recognized by members of the importin-karyopherin nuclear transport receptor family. After docking at the nuclear pore complex (NPC), the cargo-receptor complex moves through the aqueous pore channel. Once cargo is released, the importin then moves back through the channel for new rounds of transport. Thus, importin and exportin, another member of this family involved in export, are thought to continuously shuttle between the nuclear interior and the cytoplasm. In order to understand how nuclear transporters traverse the NPC, we constructed functional protein fusions between several members of the yeast importin family, including Pse1p, Sxm1p, Xpo1p, and Kap95p, and the green fluorescent protein (GFP). Complexes containing nuclear transporters were isolated by using highly specific anti-GFP antibodies. Pse1-GFP was studied in the most detail. Pse1-GFP is in a complex with importin-α and -β (Srp1p and Kap95p in yeast cells) that is sensitive to the nucleotide-bound state of the Ran GTPase. In addition, Pse1p associates with the nucleoporins Nsp1p, Nup159p, and Nup116p, while Sxm1p, Xpo1p, and Kap95p show different patterns of interaction with nucleoporins. Association of Pse1p with nucleoporins also depends on the nucleotide-bound state of Ran; when Ran is in the GTP-bound state, the nucleoporin association is lost. A mutant form of Pse1p that does not bind Ran also fails to interact with nucleoporins. These data indicate that transport receptors such as Pse1p interact in a Ran-dependent manner with certain nucleoporins. These nucleoporins may represent major docking sites for Pse1p as it moves in or out of the nucleus via the NPC.
Macromolecules move between the nucleus and the cytoplasm via aqueous channels spanning the nuclear envelope, termed nuclear pore complexes (NPCs). Transported molecules include proteins that move from the cytoplasm into the nucleus, RNAs that move outward to the cytoplasm, and proteins that shuttle back and forth. Thus, the processes of macromolecular import and export are intimately connected.
In general, transport in or out of the nucleus begins with recognition of the transported cargo by its cognate nuclear transport “receptor”. Proteins destined for the nuclear interior contain nuclear localization sequences (NLSs). The best characterized NLSs are from simian virus 40 T antigen and nucleoplasmin (44). Proteins containing these so-called “classical” NLSs are recognized in the cytoplasm by a heterodimeric receptor termed importin (or karyopherin) (29, 65). The NLS is bound by the smaller importin-α subunit, which interacts with the larger importin-β subunit for docking at the NPC and subsequent passage into the nucleus (11, 18, 31, 32, 36, 56, 81). In some cases, importin-β appears to bind and transport cargoes without importin-α (34, 40).
Although many NLS-containing proteins use importin-α/β to enter the nucleus, others do not contain the classical NLS and do not interact with importin-α/β. Instead, they interact with different import receptors that are members of a family of proteins related to importin-β. For example, the mRNA-binding protein hnRNPA1 contains a novel NLS that binds to transportin for its nuclear import (10, 24, 64). Transportin is one of several importin-β-like proteins that have no corresponding α-like partner, bind cargo directly, and dock at and move through the NPC (reviewed in reference 84).
The exit of proteins (and at least some RNA/protein complexes) from the nucleus appears to occur in a manner reciprocal to protein import, as illustrated by the human immunodeficiency virus Rev protein. Once inside the nucleus, Rev binds to Rev response element-containing RNAs and moves out of the nucleus (19). Rev and other similarly exported proteins contain a short stretch of leucine-rich amino acids, now termed the nuclear export signal (NES), that mediates their nuclear export (19, 26). The phenomenon of NES-dependent export led to the identification of export “receptors,” e.g., mammalian exportin and yeast Xpo1p/Crm1p, that bind NESs (22, 25, 61, 77). Exportins are also members of the importin-β family. Related export receptors for tRNAs have recently been identified (4, 33a, 50).
A general model is that cargoes move into or out of the nucleus complexed with their receptor. Once the cargo-receptor complex has reached its proper destination (i.e., the nucleoplasm or cytoplasm), the cargo dissociates and the transport receptors recycle for new rounds of transport. In support of this view, some importin-β proteins have been shown to cycle between the nucleus and the cytoplasm (38, 48, 77). In doing so, β proteins not only interact with their respective cargoes but also with proteins of the NPC (75).
In addition to the β proteins, the GTPase Ran and its regulators are central to the movement of macromolecules through the NPC. Ran is found in both the nucleus and the cytoplasm, whereas the Ran GTPase-activating protein (GAP) functions in the cytoplasm (7, 14, 35, 54) and the Ran GTP exchange factor (GEF) in the nucleus (3, 9, 60). This asymmetric distribution of the RanGAP and GEF with respect to the nuclear envelope has led to models of how molecules move in a vectorial manner between the nucleus and the cytoplasm (e.g., references 30, 45, and 53). According to one model, cytoplasmic RanGAP means that the Ran-GDP concentration would be high in the cytoplasm. The nuclear location of Rcc1 (the Ran GEF) would cause the concentration of Ran-GTP to be high in the nucleus. These distinct nucleotide-bound states of Ran are proposed to promote the binding and/or release of cargo from its particular carrier in the proper compartment and thus allow recycling of the receptors (12, 22, 30, 39, 49, 67). Recent reports suggest that importin-β can move through the NPC without binding cargo (46). However, the precise mode of Ran action remains controversial.
The NPC is a complex array of proteins spanning the double membrane bilayer of the nuclear envelope. The complete composition of the mammalian NPC is not known but, due in part to the sequencing of the yeast genome, the composition of the simpler yeast NPC is almost completely known (17). However, how macromolecules pass through the NPC channel remains a mystery. Blot overlay experiments have shown interactions between importins and various nucleoporins (Nups) in yeast and mammalian cells. Many Nups contain repeats of FXFG and/or GLFG. These repeats have been suggested to function as β-binding sites, and solution binding assays have provided some support for this hypothesis (2, 37, 56, 67, 69). However, these in vitro binding experiments with recombinant proteins may not adequately reproduce the specificity of the β-Nup interaction. Complexes containing importins and nucleoporins isolated from Xenopus cells indicate more-specific interactions (75) with certain nucleoporins on both the nucleoplasmic and cytoplasmic NPC surface.
Taken together, macromolecular transport in or out of the nucleus begins with binding of the transported cargo to the correct importin or exportin. After docking at the NPC, the cargo-receptor complex somehow moves through the nuclear pore channel. Thus, loading and unloading of cargoes and the interactions of cargo-receptor complexes with the NPC are the defining events of nuclear transport.
To understand how the cargo-receptor complexes move through the NPC, we have undertaken studies with the yeast Saccharomyces cerevisiae. The S. cerevisiae genome encodes at least 14 proteins with some similarity to importin-β (59). These proteins and their metazoan relatives compose the importin-β superfamily. The cell thus appears to have evolved an array of related carrier proteins to deliver diverse cargoes into and out of the nucleus. The relatedness of the carriers may reflect interactions with common transport factors such as Ran, whereas their diversity may reflect their ability to distinguish cargoes and nucleoporins. In order to address how importin-βs move through the NPC, we have studied the interactions between several yeast importin-β family members and nucleoporins by creating yeast strains where the only functional version of a particular nuclear transport receptor is fused to green fluorescent protein (GFP). Using highly specific anti-GFP antibodies, we have isolated and characterized complexes containing members of the importin family. In doing so, we have found that different importin-βs associate with distinct nucleoporins. Further, by analyzing one family member, Pse1p, in detail in mutants that affect the nucleotide-bound state of Ran, we show that interactions with distinct nucleoporins are Ran dependent.
MATERIALS AND METHODS
Construction of yeast strains expressing GFP-tagged importins.
To replace PSE1 with an engineered gene encoding Pse1-GFP, we subcloned a ClaI-KpnI fragment containing the PSE1 open reading frame (ORF) fused in frame to the S65T V163A mutant of GFP and the NUF2 3′ UTR (42) from pPS1069 (74) into the nonreplicating plasmids pRS304 and pRS306 (76). The resulting plasmids, pPS1538 (TRP1) and pPS1539 (URA3), were linearized with NsiI or BstEII, and the linear DNA was transformed into PSY580 [(MATa ura3-52 leu2Δ1 trp1Δ63 GAL+) and mutant strains rna1-1 and prp20-1. The prp20-1 (PSY713 [45]) and rna1-1 (PSY714 [14]) strains result from backcrosses with FY86 (PSY581) and FY23 (PSY580). FY86 and FY23 are isogenic haploid S288c strains that are auxotrophic for different amino acids with opposite mating types (82). Similarly, to replace SXM1, we subcloned a SalI-KpnI fragment containing the SXM1 gene fused to GFP from pPS1117 (74) into pRS304, and the resulting plasmid pPS1541 was linearized with NsiI and transformed into PSY580. For XPO1, an ApaI-BamHI fragment containing XPO1-GFP (pKW470 [77]) was subcloned into pRS304, linearized with XcmI, and transformed into PSY580. To replace the KAP95 gene, we generated pJK169, which contains a 3′ fragment of the KAP95 gene amplified by PCR with the primers GATATTGCCTATGAGCTCG and GGctcgagCTAAGGATAATTGACGCTTC. The resulting PCR product was digested with SacI and XhoI and ligated into the similarly cut pPS967 (41). The resulting plasmid featured the last 1,689 bp of the KAP95 ORF fused in frame to the S65T V163A mutant GFP and the NUF2 3′ UTR. pJK169 was linearized with AgeI and transformed into an S288c-derived diploid strain. Transformants were selected for Ura+ prototrophy and checked by fluorescence microscopy for expression of Kap95p-GFP. The diploid strain was sporulated, and the functionality of Kap95p-GFP was assessed by determining the viability of the Ura+ spores. Of 12 tetrads dissected, 11 had four viable spores and, in all cases, the Ura+ phenotype segregated 2:2. All Ura+ spores exhibited nuclear rim signal by fluorescence microscopy. Furthermore, immunoblot analysis with a polyclonal antibody raised against the Kap95p protein indicated that, in Ura+ cells, there was no expression of the unfused Kap95p protein. Therefore, we conclude that the Kap95p-GFP protein is functional. Because the Kap95p-GFP protein is functional, pJK169 was transformed into the haploid strain PSY685 (MATa ura3-52 his3Δ200 trp1Δ63 leu2Δ1) to make PSY1227.
Antibodies.
To raise the anti-GFP polyclonal antibody, recombinant GFP was purified from an Escherichia coli-overproducing strain and injected into rabbits. Briefly, pJK83 a plasmid for the expression of 6×His-tagged GFP (S65T; V163A mutant) was transformed into XL1-Blue bacteria. To generate pJK83, the ORF of GFP was amplified by PCR with BamHI-linked oligonucleotides. The resulting PCR product was digested with BamHI and ligated into the bacterial expression vector pQE9 (Qiagen). 6×His-GFP was prepared by using Ni-NTA agarose resin (Qiagen) as directed by the manufacturer. The protein was injected into New Zealand White rabbits, and anti-GFP antibody was affinity purified from raw serum with GFP covalently attached to NHS-derivatized Sepharose (Pharmacia). GFP-specific antibodies were eluted with first 100 mM glycine (pH 2.5) and then 100 mM triethylamine (pH 11.5) and neutralized. Eluted antibodies were dialyzed against phosphate-buffered saline (PBS) and stabilized with 10 mg of bovine serum albumin (BSA) per ml and 0.05%. NaN3, with a final antibody concentration of 1 mg/ml.
Recombinant GST-Gsp1p was used to immunize rabbits, and the antiserum was affinity purified against immobilized Gsp1p (45). The generation of Pse1p- and Kap95p-specific antibodies was as described previously (45, 74). Srp1p-specific antibodies were provided by D. Gorlich (Heidelberg, Germany); Nup159p-specific antibodies were provided by C. Cole (Dartmouth, N.H.); Nsp1p-specific antibodies were provided by M. Stewart (Cambridge, United Kingdom); Nup116p-, Nup145p-, and Gle2-specific antibodies were provided by S. Wente (St. Louis, Mo.); and Pom152p-specific monoclonal antibody (MAb) 118C3 was provided by M. Rout (New York, N.Y.).
Immunoprecipitations.
Lysates for immunoprecipitations were usually prepared from 50-ml cultures grown at 25°C in yeast extract-peptone-dextrose (YEPD) medium to a cell density of 107 cells/ml. Wild-type cells transformed with pPS814 (a gift from Jim Haseloff, Medical Research Council, Cambridge, United Kingdom) encoding a fusion of GFP to β-galactosidase were grown in selective medium. Cells were washed in PBS (137 mM NaCl, 1.76 mM KH2PO4, 5.4 mM Na2HPO4, 2.7 mM KCl; pH 7.2) and resuspended in 1 ml of lysis buffer (PBS plus 3 mM MgCl2, 1 mM EDTA, and 0.5% Triton X-100) with protease inhibitors added (0.5 mM phenylmethylsulfonyl fluoride [PMSF]; 2.5 μg each of leupeptin, chymostatin, antipain, pepstatin A, and aprotinin per ml [all from Sigma]). In some cases, radioimmunoprecipitation assay (RIPA) buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in 150 mM NaCl and 50 mM Tris-HCl [pH 8.0]) with protease inhibitors was used. To lyse cells, a one-third volume of glass beads (425 to 600 μm in diameter) was added, and the samples were cooled on ice and treated three times mechanically for 1 min each cycle with a Mini-Bead Beater (Advanced Laboratory Research, Inc., Franklin, Mass.) at maximum speed. Lysates were clarified by centrifugation for 10 min at 12,000 × g at 4°C. To remove all cell debris, the supernatants were transferred into fresh tubes and centrifuged again. Protein concentrations of the lysates were determined with a protein assay kit (Bio-Rad). Equal amounts of protein corresponding to approximately 25 ml of culture were incubated with 7.5 μl of anti-GFP–beads for 30 min at 4°C. The beads were washed three times in lysis buffer and two times in lysis buffer without Triton X-100. Bound proteins were eluted with SDS buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol [DTT], 2% SDS, 0.1% bromophenol blue, 10% glycerol) and separated on an 8 or 12% gel by SDS-polyacrylamide gel electrophoresis (PAGE), followed by transfer to nitrocellulose membrane (Protan; Schleicher & Schuell). Membranes were temporarily stained with Ponceau-S (Sigma) to confirm equal loading. Silver-staining analyses of the corresponding samples revealed the GFP fusion to be the major component, with additional bands present at lower levels, some of which changed in a Ran-dependent manner (data not shown). To generate anti-GFP-beads, we incubated 200 μl of protein G-Sepharose (50% slurry; Pharmacia) with 100 μl of affinity-purified anti-GFP antibodies (1 mg/ml) in PBS with 10 mg of BSA per ml for 15 min at 25°C. The beads were washed three times with PBS and then resuspended in PBS plus 0.05% NaN3 to a final volume of 200 μl.
Nonhydrolyzable GMP-PNP (85% pure; Sigma) was dissolved in 50 mM Tris-HCl (pH 7.5), resulting in a 100 mM stock solution. Then 1 mM GMP-PNP and 1 mM MgCl2 were incubated with freshly prepared lysates for 10 min at 25°C, followed by incubation with anti-GFP beads for 30 min at 4°C.
Immunoblots.
To detect proteins, membranes were incubated for 1 h at 25°C with antibodies diluted in PBS–2.5% milk powder–0.05% Tween, followed by horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch Laboratories) and detection with enhanced chemiluminescence (Amersham Corp.). To analyze GFP-tagged proteins, we diluted the affinity-purified antibodies as follows: GFP-specific antibodies, 1:5,000; Pse1p-specific antibodies, 1:4,000; Kap95p-specific antibodies, 1:500; Srp1p-specific antibodies, 1:5,000; Nup159p-specific antibodies, 1:10,000; Nsp1p-specific antibodies, 1:1,000; Nup116p-specific antibodies, 1:1,000; and Pom152p-specific MAb 118C3, 1:5. To detect Ran/Gsp1p we used a 1:1,000 dilution of the Ran/Gsp1p-specific antibodies in PBS, 2.5% milk powder, and 0.01% Tween.
Immunofluorescence.
Cells were spheroplasted with 300 μg of Zymolase (100T; ICN) in solution P (0.1 M K2HPO4-KH2PO4 [pH 6.5], 1.2 M sorbitol) plus 25 mM DTT, washed in solution P, placed on slides coated with 0.3% polylysine, and permeabilized by the addition of 0.5% Nonidet P-40 in solution P for 5 min. Cells were blocked for 1 h with 5 mg of BSA per ml in 0.1 M Tris (pH 9.0), 150 mM NaCl, and 0.3% Triton X-100, followed by overnight incubation with a 1:2,000 dilution of MAb 414 (Babco) and a 1:100 dilution anti-Nop1p antibodies. Cells were washed two times with 0.1 M Tris (pH 9.0)–150 mM NaCl (Buffer A) and two times with 0.1 M Tris (pH 9.5)–100 mM NaCl–50 mM MgCl (Buffer B) and incubated for 1 h with mouse-specific Texas Red-conjugated secondary antibodies (Jackson Immunoresearch Laboratories) in a 1:1,000 dilution. Samples were washed two times in Buffer A and two times in Buffer B, and chromatin was stained with 1 μg of 4′6-diamidino-2-phenylindole (DAPI) for 5 min in Buffer B, followed by two washes with Buffer B.
GFP microscopy.
A Nikon Optiphot-2 epifluorescence microscope and a ×100 Plan APO 1.4 NA DIC objective were used to observe the yeast cells. A Chroma filter number 41018 (excitation HQ470/40, dichroic Q495LP, emission HQ 500LP; Chroma Technology Corp., Brattleboro, Vt.), together with a Princeton Instruments Micromax camera equipped with a Kodak KAF 1400 chip (1,317 × 1,035, 6.8 × 6.8 μm pixels; Princeton Instruments), operated by Metamorph Imaging software (Universal Imaging Corp., West Chester, Pa.), was used to capture the GFP images. The final figures were produced by using Adobe Photoshop without further manipulation.
RESULTS
The main focus of the following experiments is the dynamic behavior of the importin-β-like protein Pse1p. Pse1p is an essential protein implicated in the import of ribosomal proteins and certain transcription factors into the nucleus and perhaps also in the export of mRNAs from the nucleus (40, 43a, 69, 72, 74). Moreover, its function is at least in part redundant with that of the nonessential Kap123p, as evidenced by the synthetic lethality occurring between pse1-1ts and Δkap123 mutants (74).
In order to study the dynamics of Pse1p and some of its relatives, we have employed the following general strategy. Gene fusions were created that encode the entire protein of interest fused to a bright derivative of GFP (43) so that their distribution in cells can be observed directly by fluorescence microscopy without perturbations introduced by cell fixation. These were then introduced into the yeast genome so as to completely replace their wild-type counterparts, making the GFP fusion protein the only functional version in the cell. One of the strengths of this approach is that the distributions and interactions of the proteins are being studied at their normal levels and are not impacted by possible artifacts of overexpression from multicopy plasmids or competition from endogenous wild-type protein. Moreover, since the proteins remain fully functional, as evidenced by their ability to substitute for the wild-type version, the GFP moiety does not appear to alter their functions.
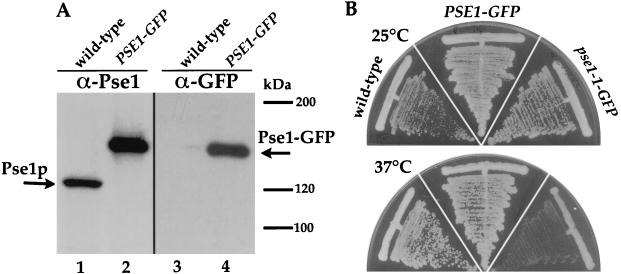
Replacement of PSE1 by a chimeric gene encoding a functional Pse1-GFP fusion protein is described in Materials and Methods. An immunoblot with anti-Pse1p antibodies confirms that the only version of Pse1p present in these strains is the GFP fusion. Pse1p is present in wild-type cells as a protein of the predicted molecular size of 121 kDa (Fig. 1A, lane 1). However, in lysates from cells where the endogenous PSE1 is replaced by PSE1-GFP, the anti-Pse1p antibody recognizes only a protein of 150 kDa (Fig. 1A, lane 2), which corresponds to the predicted molecular size of Pse1p plus GFP. This is confirmed by immunoblots probed with anti-GFP antibodies. No cross-reacting protein is present in the wild-type cells (Fig. 1A, lane 3), whereas the 150-kDa Pse1-GFP is observed in the strain containing the corresponding gene fusion as the only version of PSE1 (Fig. 1A, lane 4). Confirmation that Pse1-GFP is completely functional is illustrated in Fig. 1B; cells containing Pse1-GFP grow in a manner identical to the wild-type PSE1 cells at both 25 and 37°C.
FIG. 1.
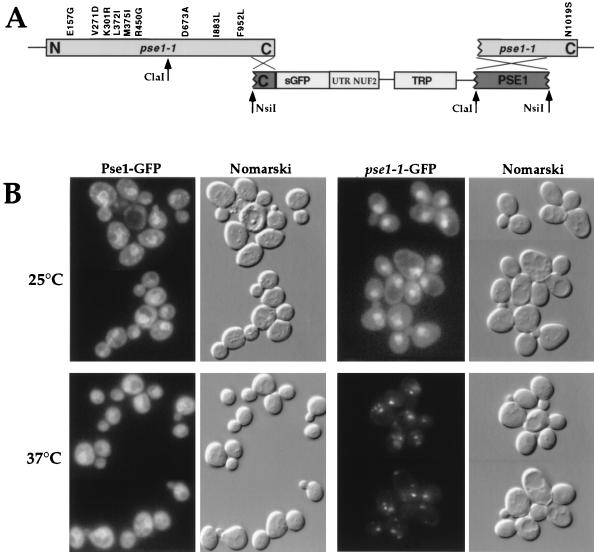
Expression of Pse1p, GFP-tagged Pse1p, and growth characteristics of wild-type, PSE1-GFP, and pse1-1–GFP strains. (A) Equal amounts of yeast lysates (20 μg of total protein) from a wild-type strain (PSY580) and a strain where PSE1 is replaced by PSE1-GFP were separated on an 8% gel by SDS-PAGE, transferred to nitrocellulose, and incubated with Pse1p-specific antibodies (74) (lanes 1 and 2) or with GFP-specific antibodies (lanes 3 and 4). (B) Growth of a wild-type yeast strain, a strain where PSE1 is replaced by PSE1-GFP, and a pse1-1 mutant strain where the C terminus of pse1-1 is replaced by the C-terminal portion of PSE1-GFP. Cells were streaked on YEPD plates and incubated for 60 h at 25 or 37°C.
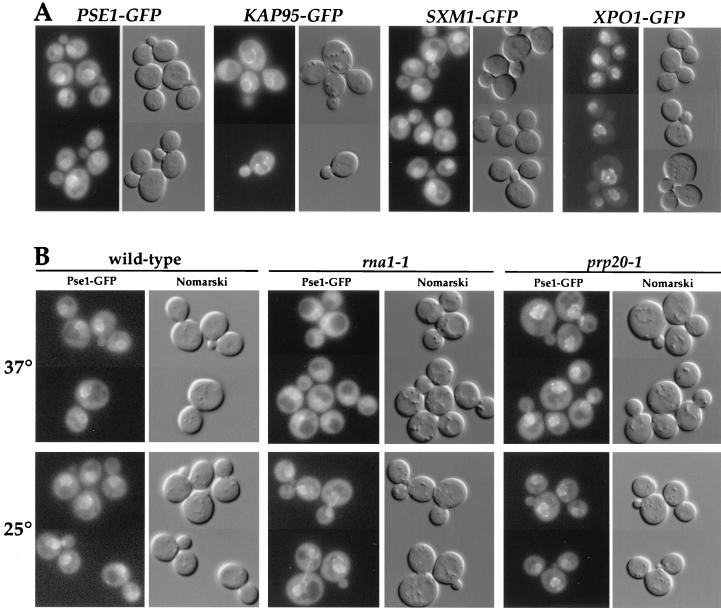
In order to compare the behavior of Pse1p to other importin-β-like proteins, strains were similarly constructed that contained only the GFP-tagged versions of SXM1, XPO1/CRM1, and KAP95. In each case, we confirmed that fusion proteins of the correct size were made and were functional in that their presence resulted in normal cell growth at all temperatures (data not shown). Fluorescence microscopy of cells grown at 25°C revealed a similar localization of Pse1-GFP, Sxm1-GFP, and Kap95-GFP. The three importin-βs are located at the nuclear envelope (Fig. 2A, first three panels), with some protein present in the cytoplasm and the nucleus as well. This distribution is consistent with the idea that these proteins may move between the nuclear interior and the cytoplasm, with a rate-limiting step occurring at the NPC. Sxm1-GFP, compared to Pse1-GFP and Kap95-GFP, shows more intranuclear accumulation. In contrast, Xpo1-GFP at steady state is restricted to the nucleus and the nuclear envelope (Fig. 2A, last panel). Unlike the other three proteins, Xpo1p is proposed to function only in export and thus may spend less time in the cytoplasm (77).
FIG. 2.
Intracellular localization of importin-β homologues. (A) Cells with PSE1, SXM1, KAP95, or XPO1 replaced by GFP-tagged versions were grown in selective media at 25°C and analyzed by fluorescence microscopy. (B) Localization of GFP-tagged Pse1p in wild-type, rna1-1, and prp20-1 mutant cells. Cells were grown at 25°C in selective media and shifted for 60 min to 37°C. All of the cells were transferred onto slides and examined immediately.
Interaction of Pse1-GFP with other importins.
In order to analyze interactions of Pse1p with other transport factors and the NPC, we isolated Pse1-GFP from cell lysates by using anti-GFP specific antibodies. Highly specific anti-GFP antibody was raised and affinity purified with recombinant GFP. The resulting antibodies were coupled to Sepharose beads for adsorption of the GFP fusion proteins from cell extracts.
Conditions were established for efficient solubilization of the GFP fusion proteins. In order to efficiently extract Pse1-GFP, we found that detergent must be present in the lysis buffer. A comparison of the amount of Pse1-GFP present in lysates prepared by glass bead lysis by using buffer without detergents, buffer with 0.5% Triton X-100, or RIPA buffer revealed only a minor fraction of the total Pse1-GFP extracted when no detergent was present in the buffer (data not shown). In contrast, the amount of Pse1-GFP in lysates prepared with 0.5% Triton X-100 represented a significant fraction of the total and did not increase when the more stringent RIPA buffer was used. Therefore, all experiments were carried out by lysing cells in buffer containing 0.5% Triton X-100.
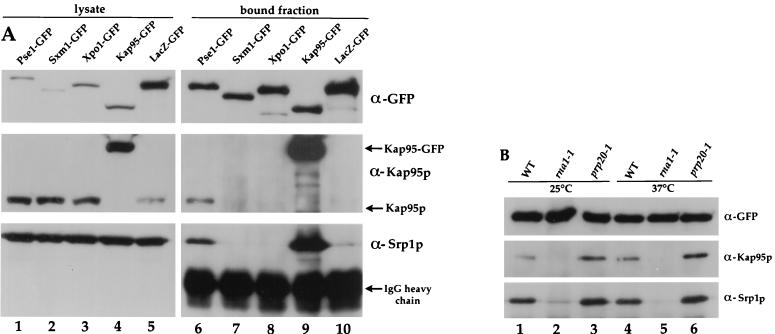
Pse1-GFP, Sxm1-GFP, Xpo1-GFP, and Kap95-GFP were isolated intact from cell lysates by immunoprecipitation with GFP-specific antibodies coupled to protein G-Sepharose. All tested GFP-tagged proteins were efficiently precipitated, including a negative control of LacZ-GFP (Fig. 3A, top panels). The bound fractions analyzed represent approximately 25-fold more than that analyzed for total lysates.
FIG. 3.
Expression and immunoprecipitation of GFP-tagged importin-βs. (A) Equal amounts of yeast lysates (10 μg of total protein) from strains with PSE1 (lane 1), SXM1 (lane 2), XPO1 (lane 3), or KAP95 (lane 4) replaced with GFP-tagged versions and wild-type cells expressing LacZ-GFP (lane 5) were separated on an 8% gel by SDS-PAGE and transferred onto nitrocellulose. Anti-GFP beads were used to precipitate complexes from lysates. PSE1 (lane 6), SXM1 (lane 7), XPO1 (lane 8), KAP95 (lane 9), and LacZ-GFP (lane 10) containing complexes and corresponding to 250 μl of yeast lysate (approximately 1 mg/ml) were separated and transferred to membranes as well. The top row shows a blot probed with GFP-specific antibodies. A second blot with identical samples was cut in half, and the upper portion was probed with Kap95p-specific antibodies, and the bottom half was probed with Srp1p-specific antibodies. (B) Genomic PSE1 was replaced by PSE1-GFP in wild-type, rna1-1, and prp20-1 strains. Cells were grown at 25°C in liquid culture, and half of the cells were shifted for 90 min to 37°C. Pse1-GFP was immunoprecipitated with anti-GFP beads, and the bound fraction was probed with GFP-, Kap95p-, and Srp1p-specific antibodies.
We know that importin-β should bind importin-α. Therefore, as a control for our experimental conditions, we probed the immunoprecipitates for the presence of importin-α by immunoblotting with affinity-purified antibody, raised against Srp1p, the yeast homologue of importin-α. As predicted, a significant portion of the total Srp1p remained associated with Kap95-GFP (Fig. 3A, lane 9). No Srp1p associated with Sxm1-GFP, Xpo1-GFP, or LacZ-GFP. However, a reproducibly smaller amount of Srp1p was always observed in association with Pse1-GFP (Fig. 3A, lane 6). Moreover, when the same precipitates were probed with affinity-purified antibody against Kap95p, it was also present in association with Pse1-GFP. These results suggest that Pse1p may bind directly to Srp1p/Kap95p or be part of larger complex that perhaps also contains NPC binding partners, a topic which will be addressed below. The observation that other importin-βs do not display a similar interaction suggests that this association with Srp1p-Kap95p is specific to Pse1p.
Ran regulators affect the behavior of Pse1p.
In yeast cells, the two major Ran/Gsp1p regulators are encoded by PRP20 and RNA1. Prp20p corresponds to the nuclear-localized GTP exchange factor for Gsp1p (20). Temperature-sensitive prp20-1 mutants result in loss of function of Prp20p, a block in nuclear transport, and presumably a decrease in the ratio of nuclear Ran-GTP to Ran-GDP (1, 23). Rna1p corresponds to the cytoplasmically located GAP for Gsp1p (5, 8, 14). The use of temperature-sensitive rna1-1 mutants results in loss of function of Rna1p, a block in nuclear transport and presumably a corresponding increase in the ratio of Ran-GTP to Ran-GDP in the cell (14, 35). In order to directly examine the effect on the distribution of Pse1-GFP of the nucleotide-bound state of Gsp1p in cells, we replaced the genomic PSE1 gene with PSE1-GFP in rna1-1 and prp20-1 strains. Both strains remained temperature sensitive after the gene replacement (data not shown).
The intracellular distribution of Pse1-GFP in the mutant cells was examined by fluorescence microscopy at 25°C and following a shift to the nonpermissive temperature of 37°C. In rna1-1 cells, some of the nuclear envelope-associated GFP fluorescence was lost, with a corresponding increase in the cytoplasmic fluorescence (Fig. 2B, middle panel), suggesting less association of Pse1-GFP with the nuclear envelope. In contrast, in prp20-1 cells most of the Pse1-GFP fluorescence was concentrated at the nuclear rim (Fig. 2B, right panel), a finding consistent with an increase in the association of Pse1-GFP with the NPC. The total amount of Pse1-GFP was not altered in any of the mutant backgrounds when compared to the wild-type cells (data not shown). The apparent high concentration of Pse1-GFP at the nuclear envelope in prp20-1 cells suggests that the nucleotide-bound state of Ran/Gsp1p plays a role in Pse1p’s docking at or release from the NPC as it moves in or out of the nucleus.
The association of Pse1-GFP with the Srp1p/Kap95p complex is disrupted in rna1-1 cells. Lysates were prepared from rna1-1 cells at either permissive or nonpermissive temperature, and Pse1p-interacting proteins were assessed by immunoprecipitation with anti-GFP antibodies as described above. In both cases, the amount of coprecipitating Srp1p/Kap95p was significantly decreased (Fig. 3B, lanes 2 and 5) compared to extracts prepared from wild-type cells (Fig. 3B, lanes 1 and 4). On the other hand, little effect on the interaction of Pse1-GFP with importins was observed for extracts prepared from prp20-1 cells at either temperature (Fig. 3B, lanes 3 and 6). If anything, the amount of bound Kap95p/Srp1p increased slightly in prp20-1 cells. Moreover, under any conditions that generated a high concentration of Ran/Gsp1p-GTP, a decrease in the level of binding of Srp1p/Kap95p to Pse1-GFP was observed. That this effect is mediated by the nucleotide-bound state of Ran is supported by the following observations (data not shown). First, the addition of the nonhydrolyzable GTP-analogue GMP-PNP to extracts decreased the amount of Srp1p associated with Pse1-GFP. Second, the overexpression of a mutant Ran/Gsp1p stabilized in the GDP-bound form (T26N) as opposed to overexpression of mutant Ran/Gsp1p stabilized in the GTP-bound form (G21V) (71, 83) also increased the amount of Srp1p associated with Pse1-GFP. Ran-GTP is known to dissociate the importin-α/β complex (12, 30, 67). Dissociation could also release Srp1p/Kap95p from Pse1p or perhaps from a common binding site shared with Pse1p at the NPC.
Interactions between importins and proteins of the NPC.
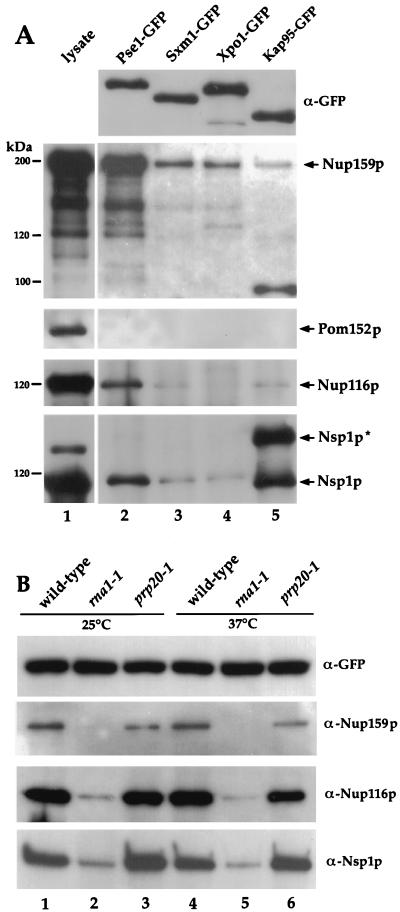
To examine interactions between members of the importin-β family and proteins of the NPC, we probed equal amounts of the immunoprecipitated Pse1-GFP, Sxm1-GFP, Xpo1-GFP, and Kap95-GFP complexes with antisera specific for various nucleoporins. Both Kap95-GFP and Pse1-GFP complexes contained significant amounts of the nucleoporin Nsp1p, whereas little Nsp1p was present in association with Sxm1-GFP or Xpo1-GFP (Fig. 4A). As we have previously reported, we consistently observe enrichment of a higher-molecular-weight protein (which we designate Nsp1p*) that reacts with the affinity-purified anti-Nsp1p antibody in association with Kap95p but not with Pse1p (80). The nature of this presumably modified form of Nsp1p is not known, but treatment with phosphatase does not affect its mobility on the SDS gel (data not shown).
FIG. 4.
Nucleoporins are present in importin-β–GFP complexes. (A) Yeast lysate from a strain expressing Pse1-GFP (lane 1) and precipitated proteins from strains where importin-β genes were replaced either by PSE1-GFP, SXM1-GFP, XPO1-GFP, or KAP95-GFP were separated by SDS-PAGE and analyzed by immunoblotting. Duplicates of identical blots were probed from top to bottom with GFP-specific antibodies, Nup159p-specific antibodies, Pom152p-specific antibodies, Nup116p-specific antibodies, and Nsp1p-specific antibodies. (B) Wild-type, rna1-1, and prp20-1 cells where genomic PSE1 was replaced by PSE1-GFP were grown at 25°C, and half of each culture was shifted for 90 min to 37°C. Identical samples with precipitated Pse1-GFP complexes were separated on an 8% gel by SDS-PAGE, transferred onto nitrocellulose, and incubated with GFP-, Nup159p-, Nup116p-, or Nsp1p-specific antibodies.
The Pse1-GFP complex also contains the nucleoporin Nup116p (Fig. 4A, lane 2). Only very minor amounts of Nup116p are present in association with Sxm1-GFP, Xpo1-GFP, and Kap95-GFP (Fig. 4A, lanes 3 to 5). Similarly, the Pse1-GFP complex is enriched for Nup159p/Rat7p (Fig. 4A, lane 2), whereas the other importin-β complexes contain only minor amounts of Nup159p/Rat7p (Fig. 4A, lanes 3 to 5).
The immunoprecipitates were also probed with antibodies to a number of other nucleoporins which were not found to be present in the complex. These include antibodies to Pom152p (Fig. 4A), Nup145p, Gle2p, Nup82p, and Nup1p (data not shown).
Consistent with the alterations in localization of Pse1-GFP at the nuclear envelope in rna1-1 and prp20-1 mutants, the interactions of Pse1p with nucleoporins are also affected in these mutants. The amount of Nsp1p, Nup116p, and Nup159p associated with Pse1-GFP is drastically reduced when immunoprecipitates are prepared from rna1-1 cells (Fig. 4B, lanes 2 and 5). In contrast, Pse1-GFP still displays interactions with Nsp1p, Nup116p, and Nup159p in extracts from prp20-1 cells (Fig. 4B, lanes 3 and 6), suggesting that the Pse1p-NPC complex is stable when the concentration of Ran-GDP is high but not when Ran-GTP predominates. Interestingly, the amount of Nup159p associated with Pse1-GFP did decrease somewhat in the prp20-1 strain compared to the wild type (Fig. 4B, lanes 3 and 6).
Mutations in PSE1 affect the localization of the mutant protein.
We have previously described temperature-sensitive alleles of PSE1 (74). Sequence analysis of the PCR-generated pse1-1 reveals 11 mutations corresponding to 10 amino acid changes and a G-to-A mutation 11 bp upstream of the ATG (see Fig. 5A). Five of the ten amino acid changes are located in the N-terminal region of Pse1p within the Ran-binding domain, as defined by homology among importin-βs (27).
FIG. 5.
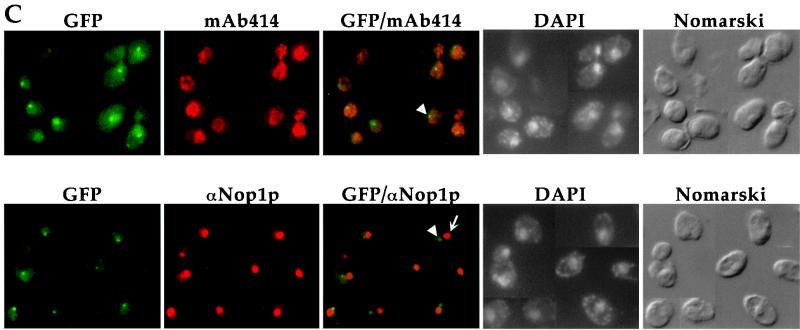
Intracellular localization of Pse1-1 protein. (A) The schematic (not drawn to scale) depicts pPS1538 containing a C-terminal fragment of PSE1 fused to GFP and the resulting arrangement after the replacement of the pse1-1 allele with the C-terminal portion of PSE1-GFP. Amino acid changes due to mutations are indicated. (B) Cells expressing Pse1-GFP or Pse1-1–GFP were grown at 25°C in selective media, and half of the cultures were shifted for 30 min to 37°C. Cells were transferred to slides and immediately subjected to microscopic analysis. The GFP signal from Pse1-1 cells was detected by a threefold longer exposure time than that from wild-type cells. (C) Cells expressing Pse1-1–GFP were probed with MAb 414 to visualize the nuclear envelope and with anti-Nop1p to visualize the nucleolus and then stained with DAPI to visualize their DNA after a shift to 37°C. Arrowheads indicate Pse1-1–GFP, and the smaller arrow indicates the nucleolus, as determined by localization of Nop1p.
In order to analyze further the nature of pse1-1, we created a strain where GFP was fused to the genomic pse1-1, thereby encoding Pse1-1–GFP. The integration strategy was such that all but the most 3′ mutation were preserved in the gene fusion (Fig. 5A). The resulting strain remains temperature-sensitive for growth (Fig. 1B).
Pse1-1–GFP was localized by fluorescence microscopy. At the permissive temperature of 25°C, more Pse1-1–GFP was observed inside the nucleus compared to cells bearing wild-type Pse1-GFP (Fig. 5B, top panels). After a shift to the nonpermissive temperature of 37°C, Pse1-1–GFP accumulated in small “dots” (Fig. 5B, lower panels). When cells were costained with an MAb that recognizes the repeat regions of nucleoporins (MAb 414), the Pse1-1 “dots” were often observed at the nuclear envelope (Fig. 5C, top panels). However, this localization is distinct from the nucleolus, as determined by costaining with anti-Nop1p antibodies (Fig. 5C, bottom panels). There was no concomitant alteration in the morphology of the nuclear envelope or fragmentation of the nucleoli, as determined by anti-Nop1p and DAPI staining.
Mutant Pse1-1p does not interact with Ran-GTP.
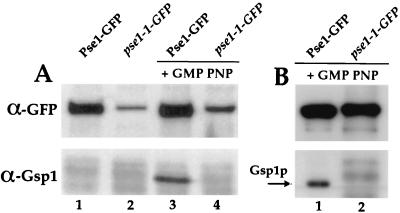
No Ran/Gsp1p was detectable in a complex with mutant Pse1-1–GFP, whereas wild-type Pse1-GFP could be isolated complexed to Ran/Gsp1p. Equal amounts of cell lysate from either wild-type or pse1-1 cells (grown at 25°C) were adsorbed to anti-GFP Sepharose and analyzed for the presence of Ran/Gsp1p. In the presence of the GTP analog GMP-PNP, no Ran/Gsp1p was bound by Pse1-1–GFP, whereas binding to wild-type Pse1-GFP was observed (Fig. 6A, lanes 3 and 4). This result was also seen for wild-type and mutant Pse1-GFP proteins when cells were shifted to 37°C (data not shown). As we observed previously, mutant Pse1-1p is produced at lower levels than wild-type Pse1p, and this is confirmed by the total amount of GFP fusion associated with the Sepharose beads (Fig. 6A, top panels). Thus, in order to ensure that we did not miss the presence of Ran/Gsp1p-GTP associated with Pse1-GFP, 10 times more pse1-1 lysate was also tested. Even under these conditions, no Ran/Gsp1p was present in the Pse1-1 immunoisolated complexes (Fig. 6B).
FIG. 6.
Mutant Pse1-1p does not interact with Ran/Gsp1p. (A) Cells expressing Pse1-GFP or Pse1-1–GFP were grown at 25°C. Lysates were split, and half of each lysate was incubated with 1 mM GMP-PNP. Pse1-GFP and pse1-1–GFP were immunoprecipitated, bound proteins were separated on a 12% gel by SDS-PAGE and transferred to nitrocellulose, and the membrane was cut in half. The top portion was probed with GFP-specific antibodies, and the bottom half was probed with Ran/Gsp1p-specific antibodies. (B) Lysates from Pse1-1–GFP and Pse1-GFP expressing cells were treated with GMP-PNP and subjected to immunoprecipitation. To match equal amounts of Pse1-GFP, 10-fold more pse1-1–GFP lysate was used for immunoprecipitation. The bound proteins were analyzed as described for panel A.
Mutant Pse1-1p fails to interact with importins and some proteins of the NPC.
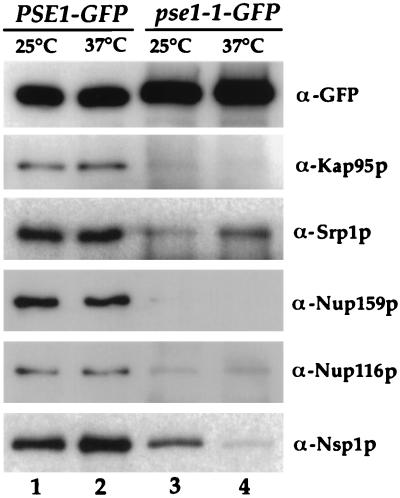
The ability of Pse1-1–GFP to interact with nucleoporins and importins compared to wild-type Pse1-GFP was examined at both 25 and 37°C. Compared to the wild-type Pse1-GFP, very low amounts of Srp1p/Kap95p were present in the immunoprecipitated Pse1-1–GFP complex (Fig. 7, compare lanes 1 and 2 to lanes 3 and 4) at either temperature. Similarly, the amount of copurifying Nup116p and Nup159p was significantly less for Pse1-1. At 25°C, some Nsp1p remained associated with the mutant Pse1-1–GFP, but at 37°C most Nsp1p was absent from the Pse1-1–GFP (Fig. 7, lanes 3 and 4).
FIG. 7.
Nucleoporins and importins present in Pse1-GFP and pse1-1–GFP complexes. Cells were grown at 25°C, and half of each culture was shifted for 1 h to 37°C. Pse1-GFP- and pse1-1–GFP-containing complexes were precipitated, and identical samples were separated on an 8% gel by SDS-PAGE and transferred to nitrocellulose. Blots were incubated with GFP-, Nsp1p-, Nup116p-, or Rat7/Nup159p-specific antibodies. One blot was cut in half; the upper half was incubated with Kap95p, and the bottom half was incubated with Srp1p-specific antibodies.
DISCUSSION
Elucidating how nuclear transporters such as Pse1p and other members of the importin-β superfamily move through the aqueous channel of the NPC provides the key to understanding the vectorial nature of nuclear transport. The asymmetric distribution of the major regulators of the Ran GTPase plays a central role in the transport process. The levels of Ran-GDP would be high in the cytoplasm due to the localization of the RanGAP (Rna1p) in the cytoplasm and at the NPC (14, 35), perhaps on its cytoplasmic face. Conversely, the levels of Ran-GTP would be high in the nucleus because of the nuclear-localized exchange factor, Rcc1 (or Prp20p in yeast cells) (60). Additional regulators reside in both compartments and further effect the nucleotide-bound state of Ran (e.g., references 15, 52, 55, 63, 73, and 79).
To understand further how macromolecules are transported in and out of the nucleus, we have studied in detail the protein interactions of an essential member of the importin-β family, Pse1p, in the yeast S. cerevisiae. We find that under conditions where the Ran nucleotide exchange factor, Prp20p, is inactive, Pse1p associates with the importin-α/β heterodimer and with certain nucleoporins. When the RanGAP, Rna1p, is inactive, these interactions are lost and less Pse1p appears to be localized at the nuclear envelope. Taken together, we suggest that these interactions reflect major NPC binding sites for certain nuclear transport receptors as they pass through the nuclear pore channel.
There are several possible explanations for how certain cargoes together with their transport receptors move between the nucleus and the cytoplasm. By one model, cargo loading would allow the receptor-cargo complex to move in the proper direction through the NPC (22, 32, 36, 45, 49). Once through, cargo unloading would result in a change in confirmation, resulting in recycling of the transport receptor for further rounds of transport (6, 12, 67). Cargo loading and unloading would be effected by the different nucleotide-bound states of Ran in the nucleus versus the cytoplasm. This model is supported by the observations that Ran-GTP dissociates cargo from importins, which would be expected to occur on the nucleoplasmic side of the pore (21, 30, 67). Furthermore, Ran-GTP stimulates binding of NES-bearing cargoes to exportins, which also would occur in the nucleus prior to their movement out (4, 22, 67). Additional experiments have also indicated the presence of Ran-GDP in complex with importin and cargo as it docks on the cytoplasmic side of the NPC during the first step of import (13).
A second possibility is that the nuclear transport receptors simply move continuously in both directions through the NPC independent of cargo and/or Ran. Support for this possibility comes from observations that importin-β mutants that do not bind Ran can still enter the nucleus (28, 46, 58). However, this may not accurately reflect the state of importin in the cell, where it may always be exposed to some form of Ran. Thus, in any model, the interaction of transport receptors with the NPC, as well as loading and unloading of cargoes, are the defining events of nuclear transport.
Several features of the yeast system have allowed us to develop a powerful approach to simultaneously assess the dynamics and the protein-protein interactions of transporters such as Pse1p under physiological conditions. First, we have taken advantage of the ability of yeast to efficiently express functional GFP fused to a number of proteins. We also substituted the GFP fusions for the normal copy of a particular gene, thus creating yeast strains where the only version of the protein of interest is the GFP fusion expressed at its wild-type levels. Finally, we have generated a highly specific polyclonal anti-GFP antibody that, when coupled to Sepharose, efficiently precipitates GFP fusion proteins from yeast cells. We can use these tools to observe the location of a particular protein without artifacts introduced by fixation or overexpression, and we can employ GFP as an effective affinity tag.
Using this approach, we have documented interactions between Pse1p and the NPC. We found that Pse1p localizes at the nuclear envelope and interacts with three nucleoporins, Nup159p, Nup116p and Nsp1p, under conditions where Ran-GTP should be low, e.g., in the prp20-1 mutant (1). When Ran-GTP is high, e.g., in an rna1-1 (14) mutant or in the presence of GMP-PNP, Pse1p is located in the cytoplasm and no longer binds to the three nucleoporins. These data are summarized in Fig. 8. Moreover, a mutant Pse1p that can no longer bind Ran does not efficiently localize at the NPC or bind to nucleoporins but does appear to be able to enter the nucleus.
FIG. 8.
Diagram summarizing Pse1p interactions. In wild-type cells or when Prp20p is inactive, Pse1p is at the NPC complexed to Nup159, Nup116, and Nsp1p.
One interpretation of these data is that Nup159, Nup116, and Nsp1p are part of a major docking site for Pse1p as it moves into the nucleus. Thus, when Ran-GDP is high in prp20-1, this might favor the NPC docked form. When Ran-GTP is high, this would prevent docking and Pse1p would remain cytoplasmic. This interpretation is supported by the observations that at least a portion of Nsp1p and Nup159p are located on the cytoplasmic side of the NPC (47, 70).
Another possibility is that the different nucleoporin interactions represent distinct steps as Pse1p moves in or out of the nucleus. For instance, the association with Nsp1p may be the import docking step favored by Ran-GDP. Conversely, the binding to Nup159p might reflect movement of Pse1p out of the nucleus, which might require interaction of Pse1p with Ran-GTP once it has entered the nucleus. Thus, in prp20-1 where Ran-GTP would be lower, the interaction of Pse1p with Nup159p is slightly lower compared to wild-type cells. These ideas are supported by the fact that nsp1 mutants have a block in nuclear import (57, 70), whereas nup159/rat7 mutants have a block in nuclear export (at least of export of mRNAs (16)). However, our data do not address which nucleoporins are in direct contact with Pse1p as opposed to part of a larger complex.
We also created strains expressing only functional Sxm1-GFP, Xpo1-GFP, and Kap95-GFP. Interestingly, we find that the pattern of nucleoporin interactions differs for different importin-β family members. All three importin-βs bind Nup159p, but to a much lesser extent than that observed for Pse1p. Only marginal binding to Nup116p is observed for all three as well. Sxm1p and Xpo1p also show only a marginal level of Nsp1p binding. However, as we previously reported, Kap95p binds very strongly to Nsp1p and to a higher-migrating form of Nsp1p (80). These differences may indicate that not all importin-βs take the same route through the NPC. If this is the case, differential interactions between importin-βs and Nups may contribute to their vectorial movement.
Others have assessed interactions between some yeast importin-βs and Nups. For example, Kap95p was shown to bind Nup1p, Nsp1p, Nup145p, Nup100p, Nup159p, Nup116p, and Nup57p in blot overlay experiments (37, 47, 67, 69). Similarly, Sxm1p bound Nsp1p, Nup1p, and Nup159p (68). However, it is difficult to ascertain the biological significance of these interactions as they were carried out with isolated proteins displayed on blots and did not take into account the distribution of the various Nups on the NPC in vivo. Moreover, they do not display the Ran-dependent binding that we have observed. In a more physiological study with Xenopus oocytes, Impβ was found to interact with three nucleoporins in a Ran-dependent manner (75). Two of these, TPR and Nup153, reside on the nuclear surface of the NPC (62, 66, 78). It is proposed that these interactions reflect a terminal import step prior to release of cargo from Impβ (75). Impβ also interacted with Nup358, which lies on the cytoplasmic fibers extending from the NPC (56, 85, 86). This may reflect an initial docking step. It is not yet clear if yeast NPCs contain such elaborate cytoplasmic fibers.
In addition to nucleoporin interactions, we detected an association of Srp1p/Kap95 with Pse1p and not with other importin-βs. This interaction was also dissipated by Ran-GTP. There are at least two explanations for these observations. One is that there is a direct interaction between Pse1p and Srp1p/Kap95p. The second is that the interaction is indirect and reflects a common binding site for the two nuclear transporters at the NPC. The interaction of both Kap95p and Pse1p with Nsp1p suggests that this may be the site of their interaction.
In sum, we have developed a strategy for simultaneously assessing the in vivo dynamics and biochemical interactions of a nuclear transport receptor. In doing so, we have been able to document Ran-dependent interactions between Psep1p and the NPC. These data further our understanding of how macromolecules may transit between the nuclear interior and the cytoplasm.
ACKNOWLEDGMENTS
We especially thank Laura Davis, Michael Rout, Susan Wente, Dirk Gorlich, Chuck Cole, and Ed Hurt for providing us with numerous antibodies. We thank the members of the Silver laboratory and Anita Corbett for comments on the manuscript.
This work was supported by grants from the National Institutes of Health to P.A.S. and by Postdoctoral Fellowships from the Deutsche Forschungsgemeinschaft and the Japanese Science Foundation to M.S. and T.T., respectively. M.D. was supported by an NIH Training Grant in Biophysics to Harvard University, and J.K. was supported by an NIH Training Grant to The Dana Farber Cancer Institute.
REFERENCES
- 1.Aebi M, Clark M W, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- 2.Aitchison J D, Blobel G, Rout M P. Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 3.Amberg D C, Fleischmann M, Stagljar I, Cole C N, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arts G J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 5.Becker J, Melchior F, Gerke V, Bischoff F R, Ponstigl H, Wittinghoffer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff F R, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff F R, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 10.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi N C, Adam E J, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi N C, Adam E J H, Visser G D, Adam S A. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi N C, Adam S A. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbett A H, Koepp D M, Schlenstedt G, Lee M S, Hopper A K, Silver P A. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett A H, Silver P A. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 16.Del Priore V, Heath C, Snay C, MacMillan A, Gorsch L, Dagher S, Cole C. A structure/function analysis of Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae. J Cell Sci. 1997;110:2987–2999. doi: 10.1242/jcs.110.23.2987. [DOI] [PubMed] [Google Scholar]
- 17.Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 18.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 19.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann M, Clark M W, Forrester W, Wickens M, Nishimoto T, Aebi M. Analysis of yeast prp20 mutations and functional complementation by the human homologue RCC1, a protein involved in the control of chromosome condensation. Mol Gen Genet. 1991;227:417–423. doi: 10.1007/BF00273932. [DOI] [PubMed] [Google Scholar]
- 21.Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- 22.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 23.Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 24.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 26.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 29.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 30.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 31.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 32.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 33.Gorsch L C, Dockendorff T C, Cole C N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Hellmuth K, Lau D M, Bischoff F R, Kunzler M, Hurt E, Simos G. Nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 35.Hopper A K, Traglia H M, Dunst R W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- 37.Iovine M K, Watkins J L, Wente S R. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iovine M K, Wente S R. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahana J A, Schlenstedt G, Geiser J R, Evanchuck D M, Hoyt A, Silver P A. The yeast dynactin complex is involved in partitioning the mitotic spindle between the mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahana J A, Schnapp B J, Silver P A. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahana J A, Silver P A. Use of A. victoria green fluorescent protein to study protein dynamics in vivo. In: Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley and Sons, Inc.; 1996. pp. 9.6.13–9.6.19. [Google Scholar]
- 43a.Kaffman A, Rank N M, O’Shea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 45.Koepp D M, Wong D H, Corbett A H, Silver P A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraemer D M, Strambio-de-Castillia C, Blobel G, Rout M P. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- 48.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 49.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 51.Lee M S, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 52.Lounsbury K M, Beddow A L, Macara I G. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- 53.Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 54.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mutvei A, Dihlmann S, Herth W, Hurt E C. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation. Eur J Cell Biol. 1992;59:280–295. [PubMed] [Google Scholar]
- 58.Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 59.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 60.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromatin condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 62.Pante N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paschal B M, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 65.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 67.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 68.Rosenblum J S, Pemberton L F, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 70.Schlenstedt G, Hurt E, Doye V, Silver P A. Reconstitution of nuclear protein transport with semi-intact yeast cells. J Cell Biol. 1993;123:785–798. doi: 10.1083/jcb.123.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlenstedt G, Saavedra C, Loeb J D, Cole C N, Silver P A. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+ RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Gorlich D, Ponstingl H, Bischoff F R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schlenstedt G, Wong D H, Koepp D M, Silver P A. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 78.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 79.Taura T, Krebber H, Silver P A. A member of the Ran-binding protein family, Yrb2, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 82.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 83.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P A. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 85.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 86.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]