Abstract
In the past 15 years, ambient ionization techniques have witnessed a significant incursion into the field of mass spectrometry imaging, demonstrating their ability to provide complementary information to MALDI. Matrix-assisted laser desorption electrospray ionization (MALDESI) is one such technique that has evolved since its first demonstrations with ultraviolet lasers coupled to Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometers to extensive use with infrared lasers coupled to Orbitrap-based mass spectrometers. Concurrently, there have been transformative developments of this imaging platform due to the high level of control the principal group has retained over the laser technology, data acquisition software (RastirX), instrument communication, and image processing software (MSiReader). This review will discuss the developments of MALDESI since its first laboratory demonstration in 2005 to the most recent advances in 2021.
Keywords: MALDESI, Mass Spectrometry Imaging, IR Lasers, MSiReader, FTMS, Ambient Ionization, Direct Analysis
INTRODUCTION
The ability to ionize biomolecules directly from a sample has led to an exponential increase in the use of mass spectrometry imaging (MSI) techniques (Bodzon-Kulakowska & Suder, 2016; Buchberger et al., 2018; Palmer, Trede, & Alexandrov, 2016) to effectively address a diverse range of biological questions. Spatially resolved mass spectra acquired across a sample enable the visualization of sample-related ions using a heatmap, which can be fundamental to gain a more complete understanding of the underlying biology as location can be an important indicator of the roles that a given molecule plays in its system (Kumar et al., 2002; Simpson et al., 2000). This information is invaluable for studying the change in function or dysfunction, of a diseased or environmentally affected sample. It has become a complementary technique in many ways to augment information gained by alternative imaging approaches including confocal microscopy and magnetic resonance imaging (MRI).
The principles of MSI were first published over 5 decades ago using secondary ion mass spectrometry (SIMS) (Castaing & Slodzian, 1962; Liebl, 1967), which remains relevant today (Massonnet & Heeren, 2019; Muddiman, Gusev & Hercules, 1995). In static SIMS, a high energy ion beam is focused on the sample surface at very high lateral resolutions of 50 nm to 1 μm. The energy transfer from the primary ions results in the ejection of secondary ions from the top layer of the sample. This was first demonstrated on elements and has since been demonstrated to effectively ionize molecules as quasi-molecular ions ([M+H+]+ and [M-H+]−) or generate fragments thereof. High spatial resolution (<100 nm) is regularly achieved with SIMS, which makes it one of the best imaging methods for samples that need high resolution and can withstand substrate application. Significant fragmentation, particularly of larger molecules, complicates identification and quantification remains challenging; however, recent work using dynamic SIMS (with a sputtering primary ion beam instead of discrete primary ion beam) have shown that it is achievable (Keren et al., 2019).
The most widely used MSI technique today is matrix-assisted laser desorption ionization (MALDI) (Karas et al., 1987), which was first demonstrated for biological ion imaging in 1994 (Spengler, Hubert, & Kaufmann, 1994). MALDI is a soft ionization technique that allows for the analysis of small and macromolecules, such as peptides and proteins (Chaurand, Schwartz, & Caprioli, 2004). An ultraviolet (UV) laser excites a UV-absorbing matrix which desorbs molecules, eventually ions, in a rising plume of material (Jaskolla & Karas, 2011; Karas, Glückmann, & Schäfer, 2000). Some recent innovations have added an additional ionization step using a second laser, leading to the moniker MALDI-2, which increased the diversity of molecules beyond those typically found using traditional UV-MALDI (Soltwisch et al., 2015; Ellis et al., 2017). Micron level spatial resolution has been achieved using UV lasers for desorption in MALDI which has helped drive the MSI field toward cellular level resolution with sub-cellular spatial resolution recently reached; however, this is not routinely employed in most tissue imaging applications using MALDI and related techniques such as MALDI-2 (Buchberger et al., 2018; Korte et al., 2015; Feenstra et al., 2017; Dueñas et al., 2017; Niehaus et al., 2019). Many reviews have examined the advancement of MALDI technology and its applications in proteomics, lipidomics, and metabolomics (Baker, Han, & Borchers, 2017; Bodzon-Kulakowska & Suder, 2016; Schulz et al., 2019). Despite its widespread use, MALDI is not without its associated drawbacks. One concern is the high vacuum requirement, which inherently limits MALDI analyses to non-volatile species (Schwartz, Reyzer, & Caprioli, 2003). These issues have given rise to alternative MSI approaches operating at ambient conditions, including atmospheric pressure (AP) MALDI (Kompauer, Heiles, & Spengler, 2017; Laiko, Baldwin, & Burlingame, 2000; Li, Shrestha, & Vertes, 2007). Furthermore, application and co-crystallization heterogeneity of the ion producing matrix can affect spatial location and quantification, as well as producing highly abundant background peaks that can limit the sensitivity when imaging low m/z metabolites (Chaurand et al., 2011). Other ambient or atmospheric pressure MSI techniques were developed to minimize sample preparation and matrix-associated drawbacks.
Due in part to the limitations associated with MALDI and SIMS, the field of emerging ionization techniques blossomed, specifically those focused on MSI, and matured over the past two decades with innovation still occurring annually. Space in this review does not allow for extensive examination of each of these techniques, but they are listed in the Table 1 above with some of their most significant references. Several of the most common techniques are based to some degree on electrospray ionization (ESI) (Perez et al., 2019). ESI was demonstrated as a viable method for gas-phase transfer of large biomolecules for MS analysis in 1989 by Fenn and coworkers (Fenn et al., 1989; Yamashita & Fenn, 1984). ESI generates multiply-charged ions allowing for the analysis of large and small biomolecules, paving the way for the development of the most well-characterized ambient MSI technique: desorption electrospray ionization (DESI) (Takáts et al., 2004; Yin et al., 2019). In DESI, an electrospray plume is directed to the surface of the sample and secondary analyte ions are then generated by the interaction of the charged droplets with tissue molecules and/or the gas phase ions of electrospray hitting the tissue surface with high kinetic energy (Venter, Sojka, & Cooks, 2006). DESI is carried out under atmospheric conditions and does not require an endogenous or exogenous sample matrix. However, absolute quantification, even when using standards, is difficult as DESI does not consistently remove a specific amount of tissue (Vismeh et al., 2012).
Table 1.
Major emerging atmospheric pressure ionization techniques from 2000 to 2019.
| Name | Abbreviation | First Publication | Spatial Resolution (Range) | Attributes | References |
|---|---|---|---|---|---|
| Desorption electrospray ionization | DESI | 2004 | 5 – 500 μm | One of the most universal ambient techniques due to its ability to analyze small to large biomolecules | (Takats et al., 2004), (Harris et al., 2008), (Costa & Cooks, 2008), (Takats et al., 2005). (Ifa et al., 2007), (Pasilis et al., 2008), (Manincke et al., 2008) |
| Electrospray-assisted laser desorption ionization | ELDI | 2005 | 20 – 200 μm | Enabled the imaging of proteins by combining the resolved desorption with a laser with the multiple charging capacity of electrospray ionization | (Huang et al., 2006), (Guo et al., 2007), (Peng et al., 2007) |
| Easy ambient sonic spray ionization | EASI | 2006 | 150 μm - 10 μm | Based on sonic spray ionization (SSI) it enables positive and negative ionization simultaneously from an unbalanced distribution of cations and anions and with help from compressed qas | (Haddad et al., 2006), (Haddad et al., 2008), (Takats et al., 2003), (Sorensen et al., 2008J |
| Matrix-assisted laser desorption electrospray ionization | MALDESI | 2006 | 50 – 200 μm | Combines spatially resolved laser desorption like MALDI with ESI ionization | (Sampson et al.. 2006) |
| Laser ablation electrospray ionization | LAESI | 2007 | 40 – 350 μm | Similar to IR-MALDESI, use of an IR wavelength allows for minimal sample preparation for biological samples | (Nemes & Vertes, 2007), (Nemes et al., 2008), (Nemes et al., 2009), (Shrestha & Vertes, 2009). (Nemes et al., 2010) |
| Rapid evaporative ionization | REI | 2009 | 500 pm-2 μm | Used during operation to determine tissue composition by analyzing gas-phase ions from electrosurgical tools | (Balog et al., 2013), (Schafer et al., 2009), (Balog et al., 2010), (Ifa & Eberlin, 2016), (Balog et al., 2015) |
| tiquid-microjunetion surface sampling probe | LMJ-SSP | 2008 | 250 – 650 μm | Operates under similar principles as LESA with improved spatial resolution | (Van Berkel et al., 2007), (Kertesz & Van Berkel, 2010), (Van Berkel et al., 2009) |
| Liquid extraction surface analysis | LESA | 2010 | 1–1,5 μm | Decouples the desorption and ionization steps for orthogonal imaging of biological macromolecules | (Kertesz & Van Berkel, 2009), (Eikel et al., 2011), (Sarsby et al. 2014), (Swales et al,. 2014) |
| Nanospray desorption electrospray ionization | nanoDESl | 2010 | 2.5 – 200 μm | Allows for greater spatial resolution than traditional DESI and decouples the desorption and ionization steps | (Roach et al.. 2010a), (Laskin et al., 2012), (Roach et al., 2010b) |
| Air flow-assisted desorption electrospray ionization | AFADESI | 2011 | 160 – 500 μm | Increased effective ionization efficiency by combining air flow assisted ionization (AFAI) with DESI | (He et al., 2011), (Luo et al., 2013) |
| Plasma-assisted laser desorption ionization | PALDI | 2011 | 60 μm | An energy absorbing matrix aids in laser desorption before orthogonal DART ionization | (Feng et al., 2014), (Chang et al., 2013) |
| Laser ablation direct analysis in real time | LADI | 2017 | 50 – 126 μm | Removes the need for sample preparation by relying on laser desorption without matrix and DART ionization | (Fowble et al., 2017), (Fowble & Musah, 2019) |
| Atmospheric pressure nanoparticle- and plasma assisted laser desorption ionization | AP-nanoPALDI | 2017 | 1 −2.9 μm | Source geometry allows for simultaneous monitoring of the optical image during NIR laser desorption | (Kim et al.,2017), (Kim et al., 2018) |
| Direct analysis in real-time | DART | 2005 | Not really imaging | The grandfather technique for other ionization methods relying on Penning ionization, namely APCI processes | (Cody et al., 2006), (Jones et al., 2006), (Fernandez et al., 2006), (Pierce et al.. 2007), (Shelley et al., 2009), (Cody, 2009) |
| Desorption atmospheric pressure chemical ionization | DAPCI | 2006 | Not really imaging | “gaseous reagent ions generated by atmospheric pressure corona discharge are directed on to condensed-phase samples, causing desorption and ionization of the neutral target molecules” | (Williams et al., 2006), (Song & Cooks, 2006) |
| Desorption atmospheric pressure photon ionization | DAPPI | 2007 | Not really imaging | Easily photoionizes desorbed molecules as either radical cations or protonated molecules | (Haapala et al., 2007), (Luosujarvi et al., 2008) |
| Atmospheric pressure solids analysis probe | ASAP | 2005 | Not really imaging | Molecules with low polarity can be ionized with little sample preparation | (McEwen et al., 2005), (McEwen et al., 2007) |
| Dielectric barrier discharge ionization | DBDI | 2007 | No reported resolutions | (Sonnenfeld et al., 2001), (Na et al., 2007a), (Na et al., 2007b) | |
| Plasma-assisted desorption/ionization | PADI | 2007 | 1 μm | Generates ions using nonthermal atmospheric glow discharge with a lower operative voltage and hiqher current than DART | (Ratcliffs et al., 2007], (Bowfield et al., 2012) |
| Low-temperature plasma ionization | LTP | 2008 | 50 – 250 μm | Allows for portable MS systems, suitable for human skin as the temperature never exceeds 30*C | (Cody, 2009), (Harper et al., 2008), (Jackson et al., 2010), (Ma et al., 2009) |
| Flowing afterglow-atmospheric pressure glow discharge | FA-APGDI | 2008 | 20 μm | Allows for the analysis of polymer surfaces without any sample preparation | (Andrade et al., 2008), (Andrade et al., 2006), (Shelley et al., 2008) |
| Infrared laser-assisted desorption electrospray ionization | IR-LADESI | 2008 | No reported imaging | (Takats et al., 2003), (Harris et al., 2012) | |
| Secondary electrospray ionization | SESI | 2000 | Not really imaging | Highly sensitive for small volatile molecules and good method for charging neutral gaseous molecules | (Wu et al., 2000), (Martinez-Lozano et al., 2009), (Steiner et al., 2003), (Dillonet al., 2010), (Chen et al., 2007c) |
| Extractive electrospray ionization | EESI | 2006 | Not suitable for imaging | Stream of neutral volatile molecules interacts with ESI stream to analyze the most volatile molecules | (Chen et al., 2006), (Zhu et al., 2009), (Chen et al., 2007), (Law et al., 2010) |
| Neutral desorption EESI | ND-EESI | 2007 | Not suitable for imaqinq | Overcomes EESI speed and sampling limitations by combining neutral desorption with EESI | (Chen et al.. 2007a). (Chen et al.. 2007b), (Chen et al., 2009), (Williams & Scrivens, 2008), (Gu et al., 2010) |
| Fused droplet ESI | FD-ESI | 2002 | Not suitable for imaqinq | Overcomes contamination of surfactants and other interferents that normally plague ESI | (Chang et al., 2002), (Fan et al., 2005), (Pan et al., 2007) |
| Paper spray ionization | PSI | 2010 | No imaging examples | Allows for combination with other current analytical paper-based techniques and allows for non-laboratory uses including mass spectrometer miniaturization | (Wang et al., 2010), (Liu et al., 2010) |
One significant sub-category of ESI-based ambient MSI approaches, employ electrospray post-ionization. Secondary electrospray ionization (SESI) allows for analysis of volatiles by relying on the interaction of gas phase molecules with the charged droplets of an electrospray plume (Wu, Siems, & Hill, 2000; Zhu et al., 2010). Laser desorption with electrospray post-ionization techniques were then developed to analyze non-volatiles and to provide spatial information. These methods can be further sub-categorized by resonant and off-resonant laser desorption. Off-resonance techniques were first demonstrated with electrospray-assisted laser desorption ionization (ELDI) in 2005 (Shiea et al., 2005). In ELDI, the laser energy is primarily absorbed by the analyte, which leads to sample vaporization into the gas-phase where the molecules are then post-ionized by ESI (Huang et al., 2006; Lin et al., 2007). While multiple studies were carried out in the years after its initial publication, it became evident that resonant techniques have distinct advantages for the analysis of biological samples. In fact, in several studies published under the ELDI banner, the likely ionization mechanisms were more closely aligned with resonant laser desorption (Huang et al., 2010; Peng et al., 2007; Peng et al., 2010).
In resonant techniques, laser energy is absorbed by a matrix and some material is ablated as these neutral molecules are then post-ionized by ESI. The focus of this comprehensive review is to articulate the development and application of an innovative resonant technique and ionization method for the direct analysis and MSI of a diverse array of biological samples. Matrix-assisted laser desorption electrospray ionization (MALDESI) has been summarized in part by Bokhart (Bokhart & Muddiman, 2016). MALDESI was first demonstrated in the laboratory in 2005, where a common MALDI matrix and UV laser (337 nm) was used to desorb neutral biomolecules with subsequent post-ionization by ESI (Sampson, Hawkridge, & Muddiman, 2006). While the matrix was always thought to be essential for MALDESI to work, the matrix is not limited to the organic acid matrices that UV-MALDI typically uses. The matrix must be appropriately selected to absorb the wavelength of the laser being used (Robichaud et al., 2013), just as in IR-MALDI which uses water, glycerol and succinic acid as common matrices (Nelson et al., 1989; Nordhoff et al., 1994; Berkenkamp, Karas & Hillenkamp, 1996; Berkenkamp et al., 1997). Desorbed biomolecules are largely neutral species (neutral / ion ratio > 99%) that encounter the charged droplets of an ESI plume positioned orthogonally to their desorption and then experience the ionization mechanisms that have been characterized in ESI (Dixon & Muddiman, 2010). In this review the history of the MALDESI platform will be traced from its inception to 2021, spanning 15 years (the teenage years) and 4 generations of development (Figure 1).
Figure 1.
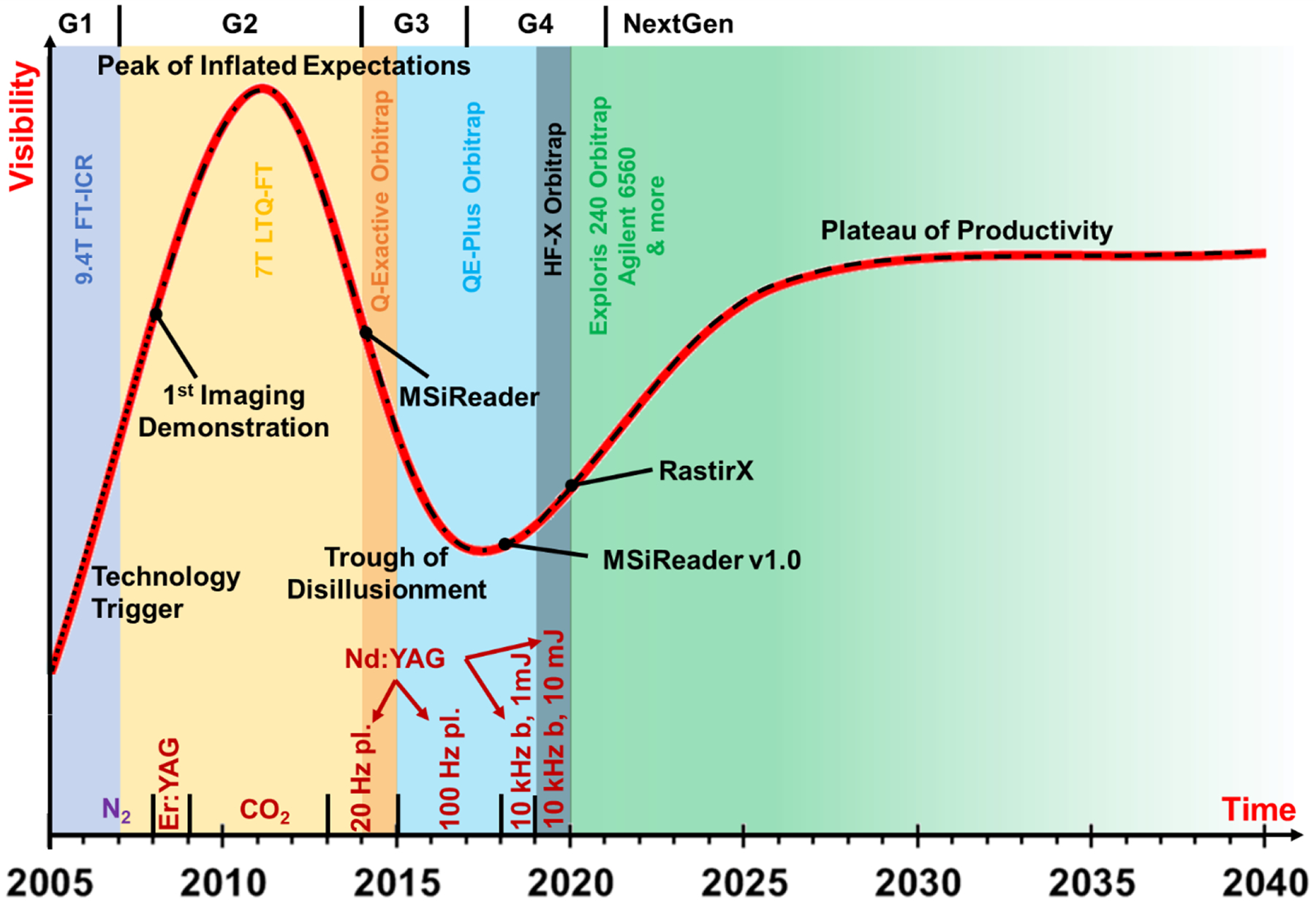
The MALDESI ‘Hype Cycle.’ The chart above tracks MALDESI from its development (‘Technology Trigger’) to the future plateau of productivity. The color-coded background specifies which mass spectrometry platform the source was coupled to for the years it covers on the horizontal axis. The small, dotted line represents the pre-imaging direct analysis and fundamentals, while the dotted and dashed line shows the optimal mixture of imaging, direct analysis and continued focus on fundamentals that characterizes MALDESI’s present and future. Then, once the fundamentals have been reasonably understood, imaging/direct analysis are the focuses during the dashed line to demonstrate its versatility and productivity. Key moments in the lifespan of MALDESI are shown on the chart, including laser technology at the bottom. The ‘Trough of Disillusionment’ was the result of wrestling with technological limitations, deepening the fundamental understanding of the technique and streamlining source engineering to advance the technique by lowering barriers to adoption by other labs.
SOURCE DEVELOPMENT
The original MALDESI source consisted of a 337 nm pulsed nitrogen laser, directed by a simple optical train to focus on the sample through a UV fused silica convex lens (Sampson et al., 2006) (Figure 2A). Orthogonal to the laser beam, a syringe pump provided the electrospray plume and directed the ions into an FT-ICR mass spectrometer with a 9.4 Tesla superconducting magnet. Shortly after the introduction of the original source, the manual linear XYZ sample positioning stage was replaced with a computer controlled motorized stage, marking a major step forward for MSI (Sampson, Hawkridge, & Muddiman, 2008b). A charge-coupled device (CCD) camera was positioned above the stage enabling real-time visualization of the ablation of the target sample. The combination of the camera and computer controlled motorized stage formed the foundation for the Rastir motion control software (see Software for Mass Spectrometry Imaging section) that allowed users to select the sample region for analysis with the smallest rectangle that encapsulated the region of interest (Robichaud et al., 2013). The stage then moved to the first spot and was synchronized with laser desorption for optimal scanning speeds using a motion control system (Figure 3). These were not trivial advances as they essentially required for that entire MSI platform to be built with a novel ionization source on an instrument not built for such integration. These changes were eventually implemented with a new source coupling, the Thermo LTQ-FT MS (Figure 2B). For the next 7 years numerous fundamental and biological studies were performed using that setup (see Diverse Array of Human Health and Disease Applications section).
Figure 2.
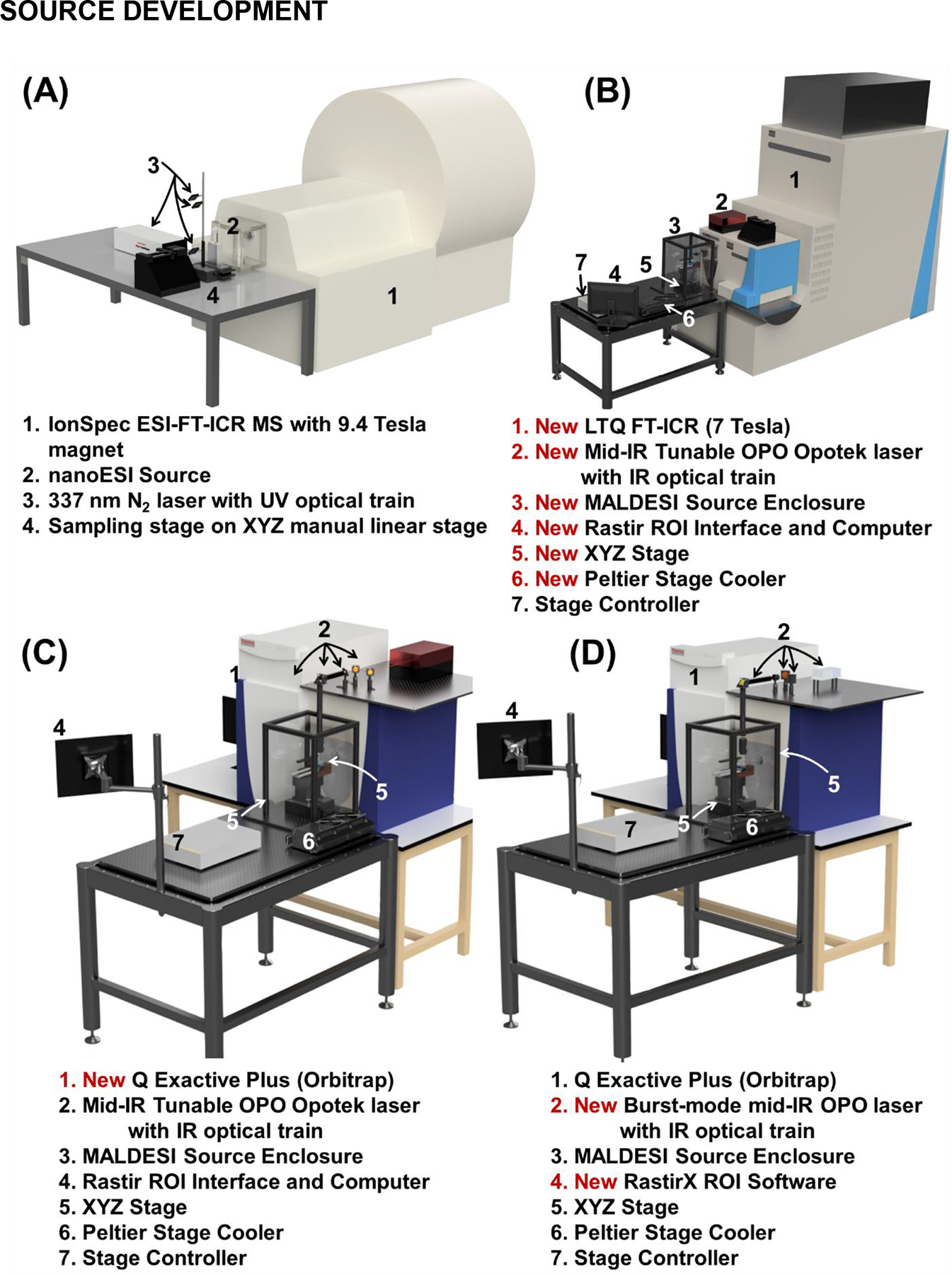
The MALDESI source from its initial demonstration in 2005 (Generation 1, A) to early development from 2007–13 (Generation 2, B) to Orbitrap coupling (Generation 3, C) to the current generation (Generation 4, D) in 2020. The Generation 4 (D) MALDESI source has been coupled to an Exploris 240 (Orbitrap, which will likely be the main platform for the completely re-engineered and improved NextGen MALDESI source.
Figure 3.
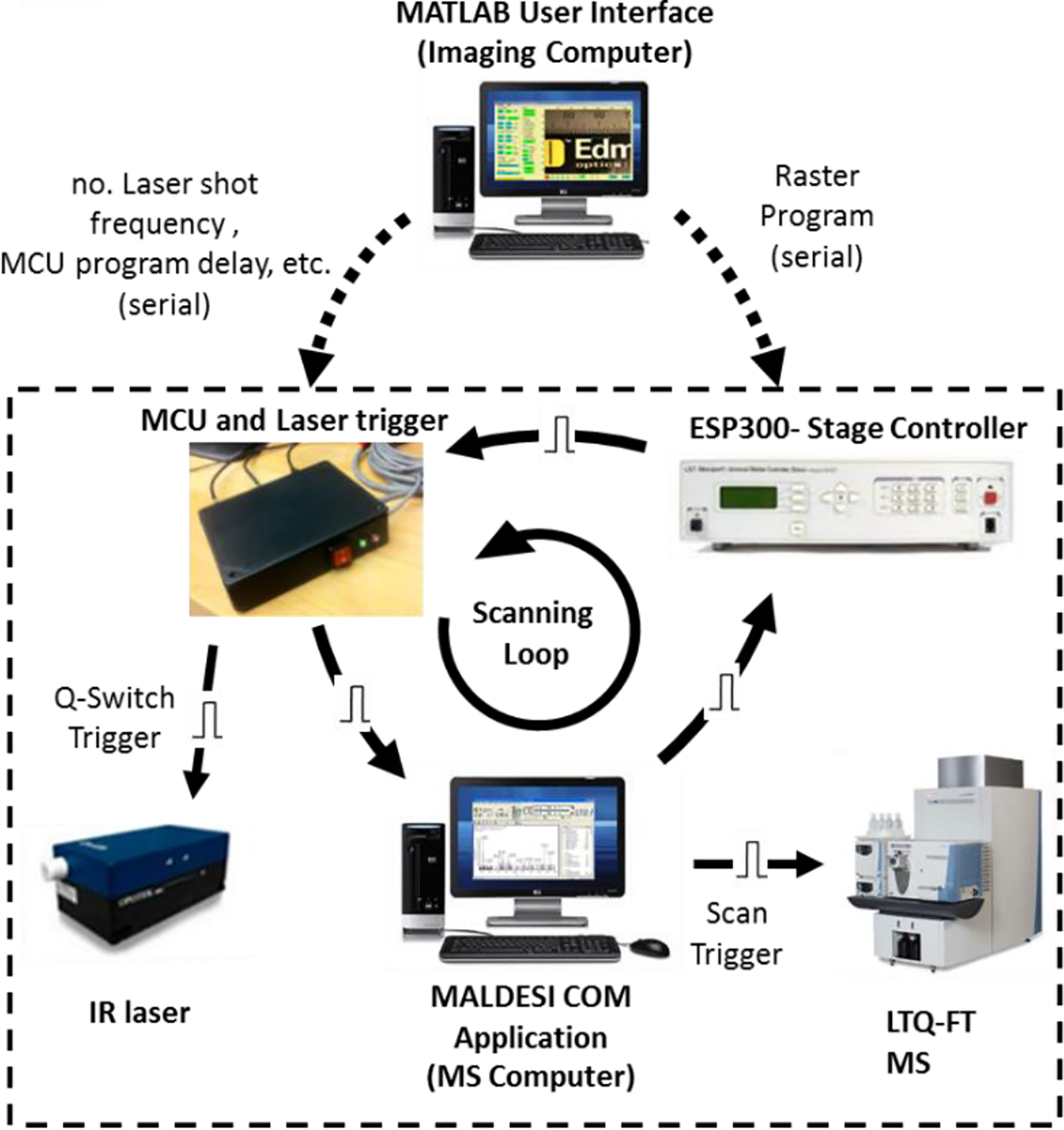
Communication between the different components of the MALDESI imaging source. Scanning parameters are selected in the Matlab user interface and sent to the stage controller and the MALDESI controller box. Communication between components in the dashed box (scanning loop) is then initiated. Communication sequence for one pixel (one cycle of scanning loop). Adapted by permission from Robichaud et al. (2013), copyright 2012 American Society for Mass Spectrometry. Adapted with permission from Robichaud et al. J. Am. Soc. Mass Spectrom 2013, 24, 1, 92–100. © 2013 American Chemical Society.
In the next generation (Figure 2C), the source was interfaced with a Q Exactive MS with Orbitrap mass analyzer (Barry et al., 2014; Meier, Garrard, & Muddiman, 2014), which provided accurate mass measurements available in previous couplings to Fourier transform (FT) MS (Barry, Robichaud, & Muddiman, 2013), but with faster acquisition rates due to reduced transient times. These characteristics made the Orbitrap platform the logical choice for high quality MSI data. Some features of the Orbitrap, namely automatic gain control (AGC), were turned off to match the pulsed nature of MALDESI ion generation.
The MALDESI source saw significant improvements in laser technology, deep characterization of the next Orbitrap-based MS in the Q Exactive series (Figure 2D), and fundamental improvements in the two main software components involved in experiments. Each development led to significant steps forward in the complexity of samples and hypotheses that could be tested using MALDESI. The combination of a high rep-rate mid-IR laser and an improved enclosure allowed for significant growth in biological applications of MALDESI as an imaging technique. Furthermore, an updated stage (Figure 2B) with a Z-dimension adjustment enabled the analysis of irregular samples that could not be readily cryosectioned.
More recent source development (~2017–2020) was focused largely on laser and software development (Figure 2D). Both of those developments are covered in more detail in their appropriate sections on Lasers and Software, respectively.
LASER DEVELOPMENT
Initial demonstrations of MALDESI were performed with UV lasers with traditional energy absorbing matrices. After successful demonstration with UV lasers (Sampson, Hawkridge, & Muddiman, 2007), the use of a mid-IR laser for desorption of cytochrome c with succinic acid proved to set the course for all future generations of MALDESI development (Figure 4) (Sampson, Murray, & Muddiman, 2009). The use of a 2940 nm laser was particularly important because of its ability to excite the O-H stretching mode of water, meaning that biological tissues could be analyzed without adding additional matrices necessary at UV wavelengths. Furthermore, liquid samples of many different kinds could be readily analyzed.
Figure 4.
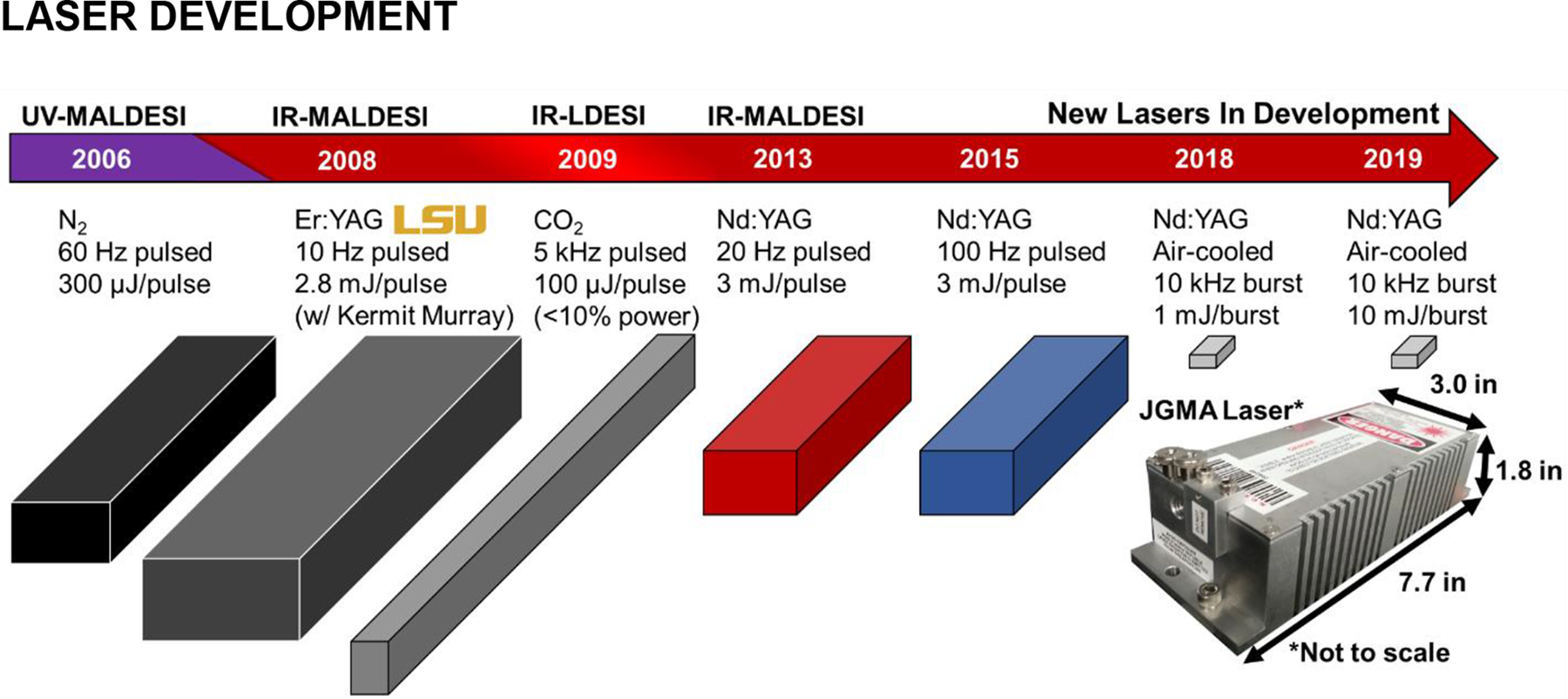
The development and application of different lasers for MALDESI. Their 3D footprints and key features are illustrated above to highlight the improvements in size and characteristics to make a more robust and agile system.
Knowing that the use of an ice layer as an energy absorbing matrix had been demonstrated before for IR-MALDI (Nelson et al., 1989; Nordhoff et al., 1994; Berkenkamp, Karas & Hillenkamp, 1996; Berkenkamp et al., 1997; Römpp et al., 2013), IR-MALDESI experiments began to incorporate ice as a matrix soon after the switch to 2940 nm lasers. An early design of experiments (DOE) evaluating the ice matrix showed an increase in the abundance of tissue-related ions and greater reproducibility compared to tissues relying solely on their endogenous water content. To achieve an appropriate ice layer the enclosure is purged with nitrogen before the sample stage is cooled to −8°C and the humidity is increased again to condense a thin ice layer over the tissue. The enclosure is purged with nitrogen again and a humidistat connected to a nitrogen gas supply keeps the relative humidity low enough (10% ± 2%) to maintain the same ice thickness without growth or defrosting throughout the experiment.
While the transition from UV to mid-IR lasers offered many benefits in sampling versatility, spatial resolution suffered. Single-cell resolution in IR-MALDESI is technically achievable using an oversampling method (Nazari & Muddiman, 2015) or with cell dispersion to get isolated singular cells (Xi et al., 2020) but to overcome natural limitations of the wavelength and move closer to single-cell resolution without oversampling, the geometry of the laser path was updated to include optics that produced a spot size closer to the diffraction-limited spot size, the theoretical highest resolution. The optical system increased spatial resolution from 150 μm to 50 μm by including an adjustable iris to trim the output into a near perfect gaussian beam profile, a beam expander to reduce beam divergence to provide smaller and circular ablation spots, and an aspheric focusing lens that corrected any spherical aberration that would hinder the spot size from reaching its diffraction limited size (Bokhart et al., 2017).
Even with the increased spatial resolution, commercially available mid-IR lasers left much to be desired. Most are expensive, flashlamp-pumped, and tunable optical parametric oscillator (OPO) lasers. A significant effort was made to develop fit-for-purpose improved laser technology that solved technical drawbacks, including periodic flashlamp maintenance and extensive water-cooling systems as well as decreased cost. A novel miniature, conductively cooled OPO laser was successfully built and characterized (Ekelöf et al., 2018). This diode-pumped, passively Q-switched solid-state Nd:OPO laser converts 1064-nm emission into 2970 nm light emitted for laser desorption. In each burst, the laser generated between 1 to 15 energy equivalent pulses in ~1 ms/burst at a maximum burst rate of 4 Hz. Laser control software and communications were created so users could program the number of pulses per burst. In diode-pumped passively Q-switched lasers, the length of the driving current in response to a trigger ensures that the chosen number of pulses are generated. The readout of a photodiode output signal, synchronous with the 1064 nm laser pulse, was fed back into the microcontroller to ensure the selected pulses-per-burst were consistently fired no matter the thermal state of the laser head and diode-pump array. Using this new laser and communication system, a full-body neonatal mouse imaging analysis demonstrated a 2× improvement in spatial resolution compared to the previous Opotek Nd:YAG OPO mid-IR tunable laser. The higher pulse rate of this laser also provided a way to achieve tighter synchronization with the mass spectrometer and thus improved the scan rate significantly. The new laser technology platform set the foundation for future laser development.
EVIDENCE FOR IONIZATION MECHANISM
Initial experiments performed on ubiquitin, bradykinin, angiotensin I, and glucagon illustrated that the MALDESI technique does in fact result in ESI-like ionization instead of MALDI-like ionization as evidenced by the observation of multiply-charged ions similar to those obtained using nanoESI (Figure 5). This approach was extended to the sampling of liquid droplets, with the same observed ionization patterns (Figure 5), which would be an important foundation for work in future generations (Sampson, Hawkridge, & Muddiman, 2008a).
Figure 5.
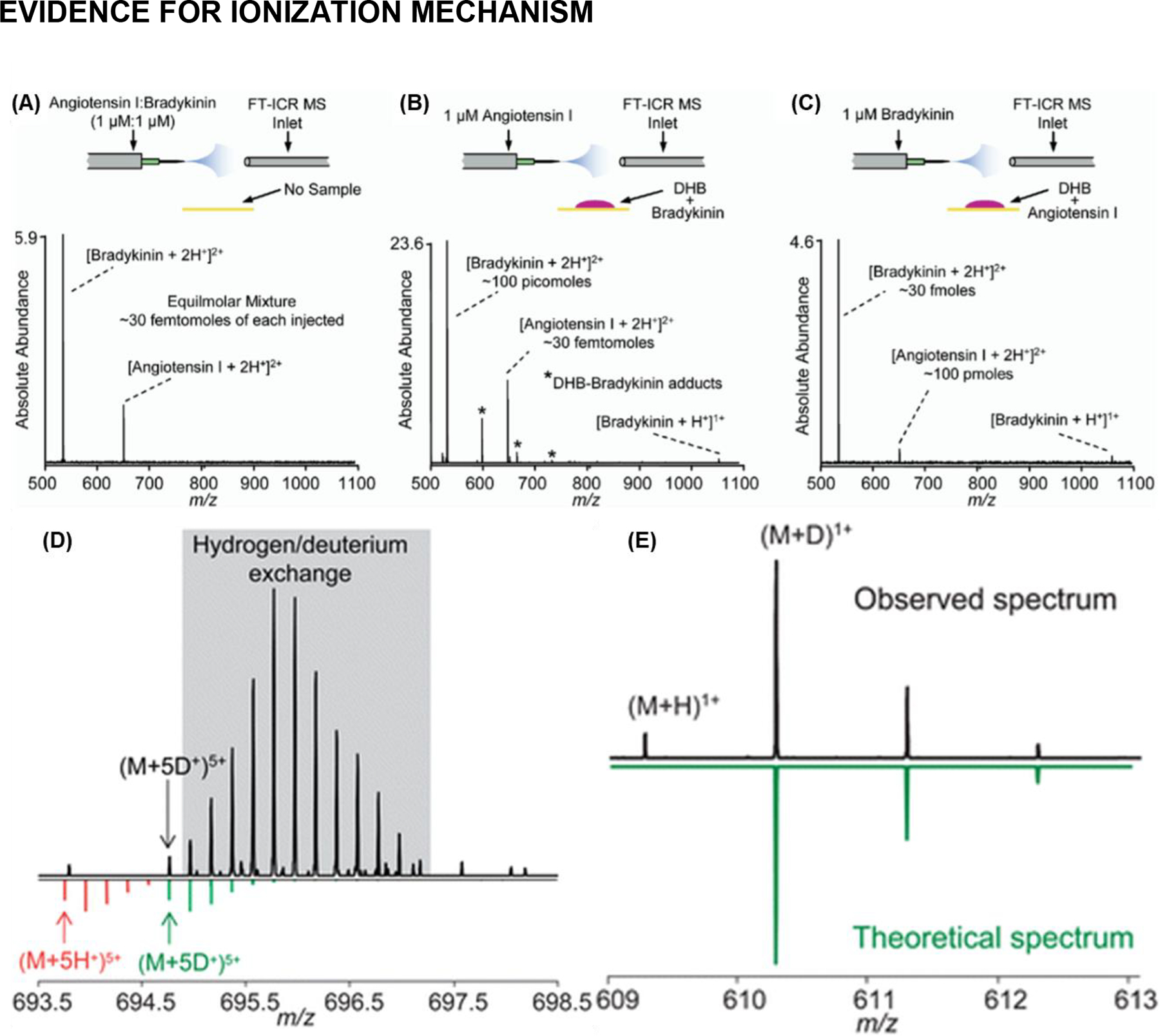
Evidence of ESI-like ionization in MALDESI. (A) NanoESI FT-ICR mass spectrum of an equimolar mixture of angiotensin I and bradykinin. (B) MALDESI of bradykinin and simultaneous nanoESI of angiotensin I. (C) MALDESI of angiotensin I and simultaneous nanoESI of bradykinin. Adapted with permission from Sampson et al., J. Am. Soc. Mass Spectrom, 2006, 17, 12, 1712–1716, © 2006 American Society for Mass Spectrometry. Published by Elsevier B.V. (D) The observed mass spectrum of the 5+ charge state of brain natriuretic peptide-32 (BNP-32); the gray region indicates portions of the isotopic distribution of the (M+5H+)5+ charge state of BNP-32; the green isotopic distribution is a theoretical isotopic distribution of the (M+5D+)5+ charge state of BNP-32. The extensive region for hydrogen/deuterium exchange resulted in follow up experiments illustrated in (E). On top the mass spectrum obtained while electrospraying deuterated solvents though Remote Analyte Sampling, Transport and Ionization Relay (RASTIR) source (Dixon et al., 2008) (Note: the original meaning here of RASTIR is different from the later developed Rastir imaging software); the most abundant peak is the monoisotopic peak for reserpine in the (M+D+)1+ form. On the right is the reflected theoretical mass spectrum of reserpine as a (M+D+)1+ ion. Adapted with permission from Dixon and Muddiman, Analyst, 2010, 135, 5, 880–882, © 2010 Analyst.
An early study looking at the ionization mechanisms involved in MALDESI used remote sampling (Dixon et al., 2008) to decouple laser desorption from the orthogonal ESI plume which would turn into a much larger area of interest in the field of MSI than was realized at the time. Using deuterated solvents doped in the electrospray Dixon and Muddiman were able to demonstrate the electrospray ionization was indeed the primary ionization mechanism in MALDESI (Figure 5) (Dixon & Muddiman, 2010). Deuterated solvents clearly shifted observed mass spectra (i.e., [M+D+]) relative to proton adduction, with the observed distribution nearly identical to theoretical spectra.
Early attempts at understanding energy deposition in MALDESI were completed using cocaine (Rosen et al., 2015). This subject was then revisited with an increased focus and depth to quantify the degree of “softness” of IR-MALDESI with direct comparison to a conventional ESI source. In this study, the internal energy depositions on common sample types were determined using the survival yield method (Gabelica & De Pauw, 2005) with a series of para-substituted benzyl pyridiniums (BPs) as thermometer ions (Figure 6) (Tu & Muddiman, 2019). IR-MALDESI generates ions with comparable internal energy distributions to those obtained with conventional ESI in analysis of both liquid and solid samples, corroborating the view that the softness of IR-MALDESI is equivalent to ESI. Data from this study indicated that an externally deposited ice layer slightly increases the internal energy when compared to the endogenous water in animal tissues.
Figure 6.
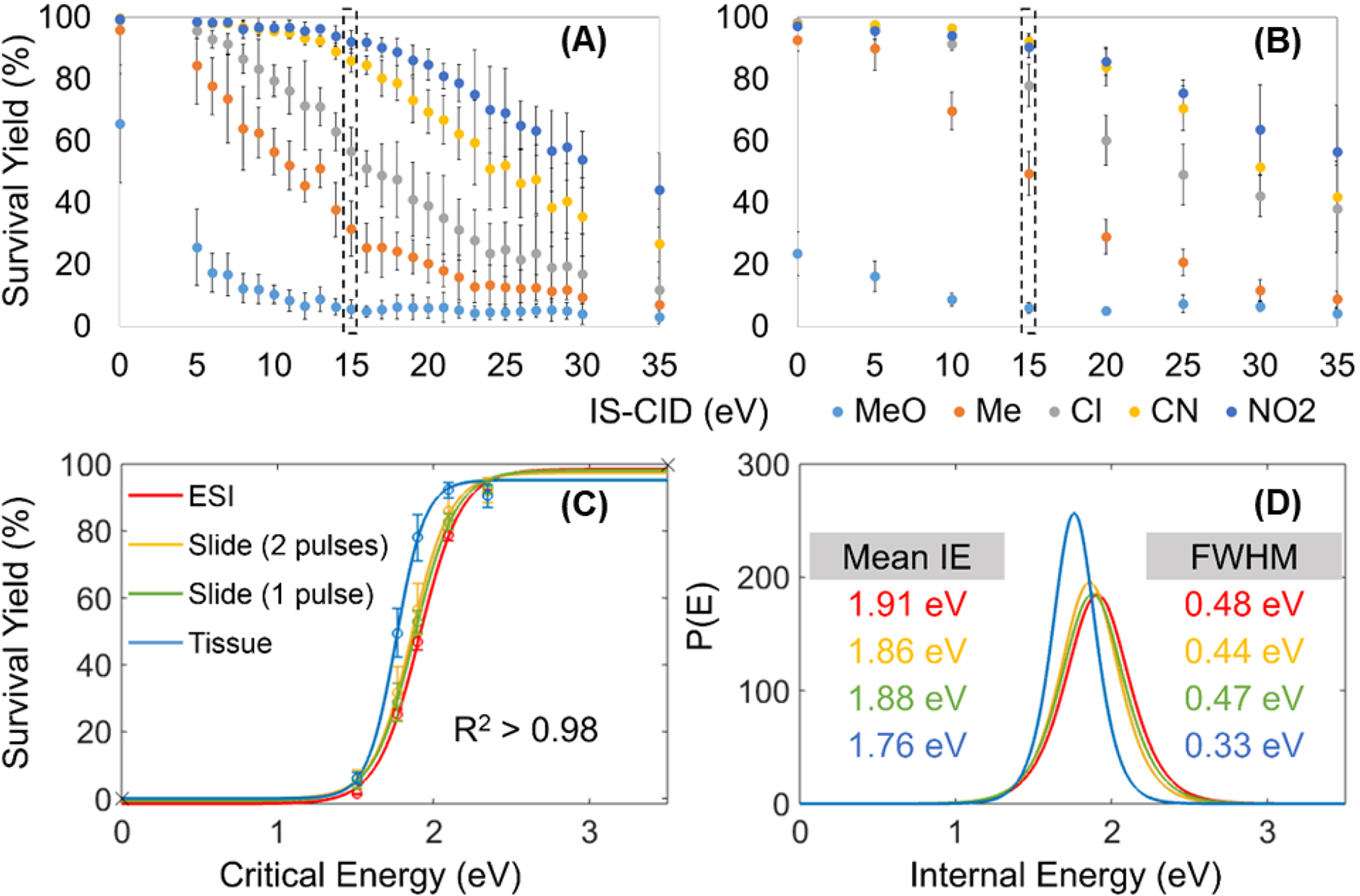
Survival yields of five BP cations versus IS-CID energies for IR-MALDESI analysis of (A) sprayed slides and (B) tissue sections on sprayed slides. The comparison of (C) Boltzmann’s sigmoidal curves and (D) Internal Energy distributions between ESI and IR-MALDESI was made at 15 eV IS-CID. Adapted with permission from Tu and Muddiman, J. Am. Soc. Mass Spectrom 2019, 30, 11, 2380–2391 © 2019 American Chemical Society.
INCREASING MOLECULAR COVERAGE AND SIGNAL ABUNDANCE IN MALDESI
From the very first iteration of MALDESI there have been several aspects that can govern the outcomes of a given experiment. Due to the many variables and interactions, a global optimization of the MALDESI source was performed using a design of experiments (DOE) (Hecht, Oberg, & Muddiman, 2016) to determine the optimal parameters of stage height, ESI-inlet distance, sample-inlet distance, ESI flow rate, plate voltage, laser fluence and repetition rate for IR-MALDESI (Barry & Muddiman, 2011). Another DOE was carried out to evaluate the presence of an ice layer over a tissue sample, where ice was shown to improve desorption plume dynamics, thereby increasing tissue ion abundances even for multiple laser pulses in one location (Robichaud, Barry, & Muddiman, 2014). The effect of electrospray solvent composition was also briefly evaluated in 2015 (Barry et al., 2015) before a more extensive analysis in 2019 (Bagley, 2019). Each of these studies were designed to increase the depth of biological information that can be obtained using IR-MALDESI.
The DOE did not resolve every factor that could affect ion abundances. For example, to achieve synchronization between the laser trigger and MS acquisition from each position the ions need to accumulate in the radio frequency-only storage trap (C-trap) for a constant and sufficient time (injection time, IT) prior to injection into the Orbitrap analyzer. The goal of the study was to decrease the total amount of time the C-trap remained open since sensitivity decreases the longer it is open due to signal decay from space charge effects from the ambient background ions. The effect of C-trap accumulation time on ion abundance was characterized (Rosen et al., 2015) before experimental method optimization based on time-resolved ion yields (Figure 7) (Ekelöf & Muddiman, 2018). The C-trap IT was decreased to improve analyte signals by incorporating an IR laser with a 100-Hz repetition rate to reduce the IT from >100 ms when using a 20-Hz IR laser to 30 ms, which resulted in increased ion abundance by up to 3 orders of magnitude for analytes ranging from xenobiotics to endogenous lipids (Figure 7A, 7B). The decrease of C-trap accumulation time was achieved concurrently with optimization of the timing between the laser ablation event and the start of ion collection. Previously, the instrument operated in the “handshake mode” which introduced a variable delay of up to 100 ms between trigger and response, requiring the injection time to be 110 ms for 2 laser pulses. The “low-latency handshake mode” provided by Thermo Fisher Scientific, and the home-designed external circuits, enabled the mass spectrometer to be precisely synchronized to a laser trigger within microseconds. Several tissue-specific ions acquired using incremental IT following laser ablation were measured to determine the signal rise time. Using the novel low-latency mode, 75 ms of IT after two 20-Hz laser pulses was suggested as the optimum MSI setting, yielding 2× signal abundances and improved repeatability (Figure 7C, 7D). These improvements at faster laser firing rates, and lower IT, were a large driving force in developments in laser technology to operate at effective repetition rates higher than 100 Hz (see Laser Development section).
Figure 7.
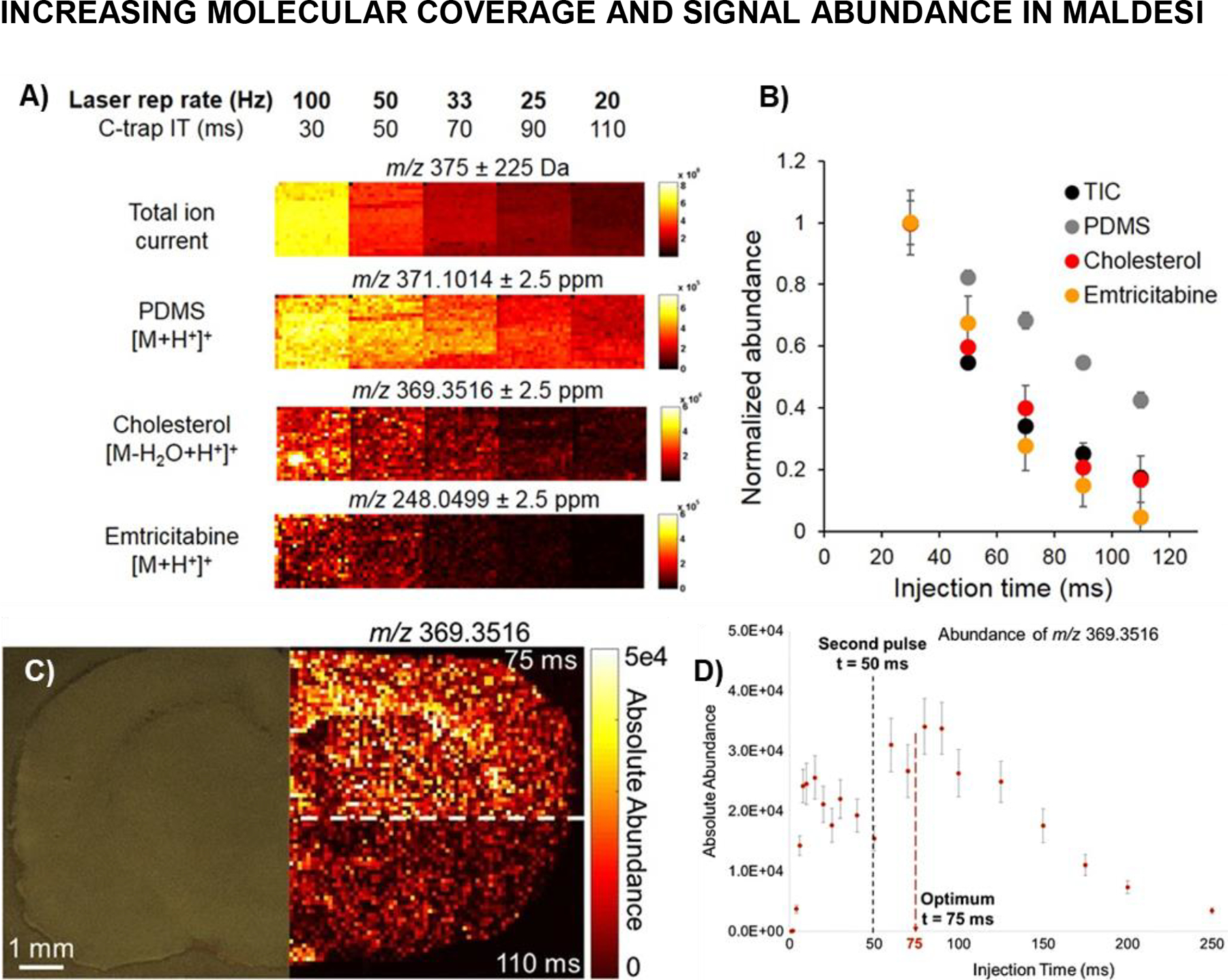
Dependence of Q Exactive Plus C-trap accumulation time on ambient and tissue-specific ion abundance during IR-MALDESI MSI analysis of ARV-incubated human cervical tissue. (A) MSI ion maps indicate increased ion abundance as C-trap inject time is reduced for: total ion current (150—600 m/z); ambient polydimethylcyclosiloxane (PDMS); endogenous cholesterol; and xenobiotic emtricitabine (FTC). (B) Normalized ion abundance for each analyte, including 95% confidence limit, showing 2–100 fold increase when reducing accumulation time from 110 ms (previous limiting conditions with 20 Hz laser) to 30 ms (100 Hz laser). Adapted with permission from Rosen et al., 2015, Anal Chem, 87, 20, 10483–10490, © 2015 American Chemical Society. (C) MSI of cholesterol in rat brain to illustrate the improved ion abundances when inject time was lowered from previously found optimum for two pulses firing at 20 Hz (A,B). (D) Graph showing the ion abundance dynamics of cholesterol with lines showing the timing of laser pulses and optimum resultant injection time. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Analytical and Bioanalytical Chemistry IR-MALDESI method optimization based on time-resolved measurement of ion yields, M Ekelöf, DC Muddiman © 2018.
Like most MSI techniques, MALDESI must rely on HRAM to resolve similar m/z and relative abundances in isotopic envelopes (spectral accuracy) for elemental composition determination. It was crucial to understand how spectral accuracy (e.g., carbon and sulfur counting) changed with absolute ion abundance in the mass analyzer to determine thresholds in ion abundances required to accurately estimate the number of atoms in a molecule (Khodjaniyazova et al., 2018). Two experimentally determined optimal conditions were established for three arbitrarily defined mass windows of small (100 < MW (Da) < 400), medium (400 < MW (Da) < 900), and large (1000< MW (Da) < 1500) compounds: (1) optimum absolute 13C1, 15N1, and 34S1 ion abundance for accurate carbon, nitrogen, and sulfur counting, and (2) thresholds for the absolute monoisotopic ion abundances required for high spectral accuracy. These findings informed untargeted MSI analyses of healthy and diseased tissues to confirm the presence of the analyte of interest based on carbon, nitrogen and sulfur counting, as well as all future untargeted studies (Pace & Muddiman, 2020).
Efforts to improve the value of HRAM experimental data often revolve around expanding or deepening coverage for a class of molecules. Deepening coverage in MALDESI is not as simple as repeated sampling of pooled samples, available to techniques such as LC-MS, because laser ablation results in complete tissue removal from each voxel. Using a technique known as polarity-switching, the metabolome and lipidome coverage was deepened on the ovarian cancer model at the cost of losing 2× spatial resolution in the x-dimension (Nazari & Muddiman, 2016). In a polarity switching experiment, every other voxel is analyzed in either positive or negative ionization mode, with a delay between each voxel as the ESI stabilizes. Solvent compositions were evaluated for optimal analysis in both modes, with the optimal electrospray solvent being 50:50 MeOH/H2O with 1 mM acetic acid (Muddiman et al., 1996). Although polarity switching does result in less spatial resolution in the x-direction (i.e. 100 μm turns into 200 μm), the y-resolution stays the same. For a given region of tissue, there is now information about more metabolites as different classes of molecules ionize with different efficiencies in each polarity. Some molecules that would have been completely unseen in positive mode are now seen in the negative mode voxels, and vice versa. Polarity switching provides the same information as doing sequential positive and negative analyses performed on serial sections, while offering a 42% reduction in analysis time. It also saves sample preparation time, eliminates bias from using different samples for positive and negative polarity in separate experiments, and is the best option for precious samples with limited availability. The polarity switching technique has been demonstrated for several diverse sample types (Cochran et al., 2015; Kalmar et al., 2020).
Even while great strides have been made in improving the understanding and application of MALDESI, fuller characterization was needed to understand the areas for growth in experimental and data analysis performance. Part of this included revisiting a partial determination of optimal ESI parameters for mammalian tissue samples completed during the early characterization of MALDESI and performing a comprehensive study to determine the effects electrospray parameters have on lipid profiles during IR-MALDESI analyses (Bagley, Ekelöf, & Muddiman, 2020). A liquid chromatography pump was used to create solvent gradients for H2O:ACN and H2O:MeOH solvent systems to determine the proper solvent system and composition for lipid studies in positive and negative polarities. A series of 12-minute gradients were run across the entire solvent composition range (5–95%) for multiple applied voltages as rat liver tissue was analyzed. Then, 25-minute gradients were run across tighter gradient ranges selected from the short gradients. More optimal settings were found in both polarities that increased lipidome coverage, abundance of detected lipids, and decreased background peaks so that future studies can go deeper into the metabolome. With the new electrospray settings, a 3-fold increase in abundance and 15% increase in coverage in positive mode was achieved, and a 1.5-fold abundance increase, and a 10% coverage increase in negative mode.
SOFTWARE FOR MASS SPECTROMETRY IMAGING
The high volume of high-resolution accurate mass (HRAM) data collected using IR-MALDESI coupled to an Orbitrap mass spectrometer, necessitated better MSI analysis software. MSiReader was first developed as vendor-neutral software in 2013 and allowed the MSI community to perform several data analysis and interrogations on their own data (Figure 8) (Robichaud et al., 2013). At the time of publication there were only two other free MSI software options: Datacube Explorer (Klinkert et al., 2014) and Biomap (Stoeckli et al., 2002). Datacube Explorer limited mass resolution to 0.1 Da and Biomap only had 16-bits of dynamic range. Although the field has grown to offer more options, MSiReader has remained a significant contributor as it is free and open-source software in the MSI community (Buchberger et al., 2018).
Figure 8.
MSiReader supports and is driven by input from the MSI community. The MSiReader application programming interface also provides a structured framework for implementing new features and algorithms.
The second-generation release of MSiReader v1.0 (Bokhart et al., 2018) grew to meet the community needs for data analysis using diverse sampling techniques around the world. Regardless of the technique used to acquire MS data, many users share the need to process MSI data without compromising its integrity by limiting the dynamic range or resolution, which is particularly important when processing HRAM data. In MSiReader v1.0, new features allowed for multiple data sets to be analyzed simultaneously without having to merge files, which could significantly increase the time and tedium often associated with data analysis (Figure 8). Furthermore, the MSiQuantification tool allowed users to define the regions associated with their absolute quantification calibration curve and then calculate the concentration of desired analyte(s) across the tissue. Absolute quantification is possible with IR-MALDESI due to the complete ablation at each pixel in the image resulting in voxels with known dimensions. MSiReader was also updated to appropriately parse polarity switching data for several different polarity-switching patterns, so that data analysis in both modes could be more swiftly accomplished on the same data file upload. An optical image tool was added in response to experimental improvements, namely for whole-body imaging sets, that has applications for every type of imaging. An optical image of the analyzed tissue can be incredibly informative when overlaid with MSI data. Therefore, a tool was created in MSiReader v1.0 that enables an image overlay with a user-defined transparency scale bar. While basic translation, rotation, and scaling tools are provided for image overlays, the IR-MALDESI platform captures an inherently co-registered optical image for each run. Regardless of the experiment, HRAM MSI data must be constantly evaluated for mass measurement accuracy and spectral accuracy. In that vein, MSiReader v1.0 offers several quality assurance features that generate heatmaps and histograms of MMA and spectral accuracy for a given m/z across the image file. As of this writing, there have been 1.5 downloads per day since 2013 with an estimated 1300 unique users resulting in nearly 400 citations to MSiReader (Robichaud et al, 2013; Bokhart et al., 2017b).
MSiReader saw many developments in its 3rd generation that improved data analysis (msireader.wordpress.ncsu.edu). The MSiCorrelation tool (Ekelöf et al., 2018) generates batch images for an m/z list (imported or internally generated by MSiReader) and can also correlate these images against a reference, assigning a rank or measure of similarity to each. The reference can be a molecular image or any graphical file that Matlab can import (e.g., an optical image). Four algorithms are provided to score images including the Structural Similarity Index (SSIM), which was originally developed for video processing and compression (Wang et al., 2004). In contrast to Minkowski metrics (e.g., mean squared error) and Pearson correlation, SSIM is not a pixel-by-pixel comparison, rather luminance, contrast, and structure are considered, separately mimicking the human vision system. With SSIM, MSiCorrelation can process several hundred thousand images in a few hours to find ions having spatial distribution similar to a molecule of interest using an ordinary desktop computer, allowing for more rapid and deeper data analysis (Ekelöf et al., 2018). Coupled with MSiReader’s principal component analysis tool (MSiPCA) a robust and efficient visualization and analysis workflow is realized for large imaging data sets. The PCA tool is interactive, linking component space as visualized in a biplot with MSiReader’s spatial heatmaps. SSIM is an effective pre-filter for reducing very large search spaces that would otherwise exhaust memory prior to performing principal component analysis. Other notable features implemented during the 3rd generation are additional normalization options, new filtering options, more efficient m/z resampling, 3D visualization, ROI management, and perceptually linear colormaps optimized for color vision deficiency. MSiReader continues to evolve to support IR-MALDESI source development and the analysis of biological systems and rises to meet the needs and suggestions from the entire MSI community.
Similarly, the software in which the user defines the region to be raster scanned by IR-MALDESI, denoted Rastir, was also developed to meet experimental needs. Rastir is the software responsible for user control over imaging parameters prior to analysis (Figure 9). Communication with a video camera allows users to verify the region of interest for analysis and edit experimental parameters such as scan lines and spot-to-spot distance. These settings are stored and run once communication with the Arduino microcontroller has been established. Throughout the experiment the Arduino and Rastir communicate to ensure MS analysis, stage motion, and laser firing are properly synchronized. For the majority of MALDESI’s history, analyses were limited to the smallest possible rectangle that could fit around the sample. Rectangular ROIs resulted in significant off-tissue analysis time that provided no extra biological information and wasted time. The original platform was updated in the 3rd generation to allow for arbitrary ROI selection in which the user creates a polygon that allows for minimal off-sample analysis to limit biological degradation and decrease the amount of unwanted off-tissue material injected into the MS (Figure 10). Each arbitrary ROI in RastirX is saved after creation and can be repeated for multiple analyses, with MS data easily co-registered in MSiReader with the saved optical images of the sample for each analysis. This new version, RastirX (Garrard et al., 2020), was used in several projects (Bagley et al., 2020; Khodjaniyazova et al., 2019) and MSiReader was updated to accept such data files. It will serve as the platform for future analysis of irregularly shaped samples not well served by current ROI tools in both 2- and 3-D imaging.
Figure 9.
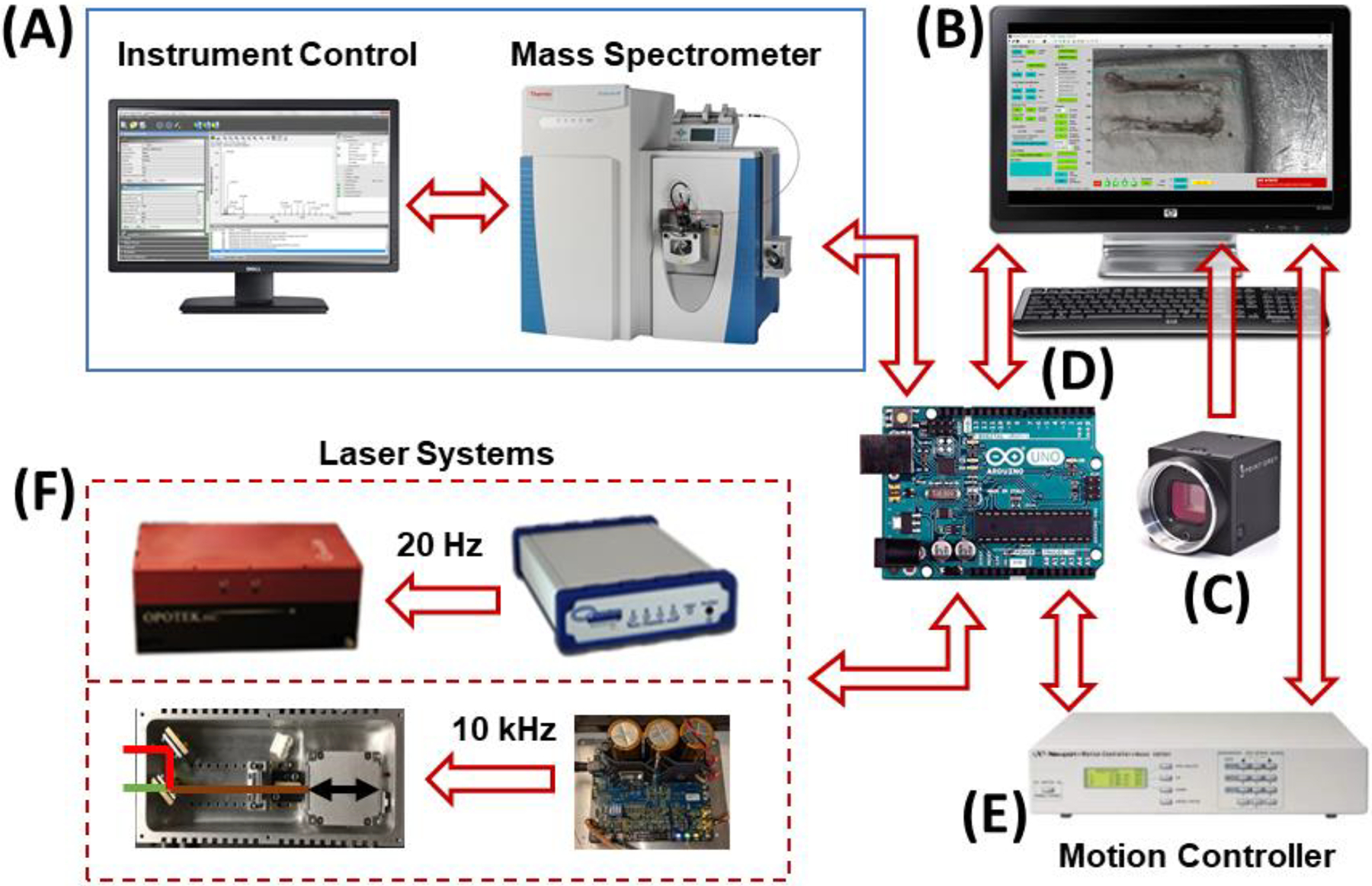
IR-MALDESI System Components. (A) RastirX is agnostic with respect to the mass spectrometer, provided there are two “handshake” signals: one to trigger scan acquisition and another indicating that the instrument is ready to acquire a scan. The voltage and polarity of these signals for various instruments is easily accommodated with opto-coupled relays and simple microprocessor (D) code changes. Mass Spectrometers interfaced to date include: Thermo Fisher Scientific LQT-FT-ICR Ultra, Q Exactive, Q Exactive Plus, Q Exactive HF-X, and Exploris 240, and Agilent 6560 IM-QTOF. (B) The RastirX user interface computer is any Windows PC with Matlab R2014a or later and the Image Processing and Image Acquisition toolboxes. RAM and HD (or SSD) requirements are modest – 8GB and 250GB, respectively. At least 3 USB ports are needed for communication with the video camera (C), microcontroller (D) and motion control system (E). (C) A video camera with fixed focal length lens. 4K DCI resolution is desirable although any USB camera recognized by the Image Acquisition toolbox will work, as will any webcam. (D) An Arduino Uno microcontroller for synchronization of the laser, stage controller, and mass spectrometer. A very simple custom shield has been built for interfacing TTL I/O pins with the mass spectrometer. (E) A motion controller. Currently a Newport ESP300 is connected to a USB serial port on the user interface PC (B) to send commands and report current position and system status. TTL level signals are sent to and received from the microcontroller (D). (F) A mid-IR laser. Two laser systems are shown: a 20 Hz pulse rate Opotek Q-switched, tunable laser (2700 – 3100 nm wavelength) along with a Quantum Composers Sapphire 9200 pulse generator for precision triggering (upper), and a 10 kHz pulse rate JGMA laser (2970 wavelength) with a DM-100 power supply/pulse generator (lower). A menu selection in the RastirX interface is used to indicate which laser is installed. Adapted with permission from Garrard et al., J. Am. Soc. Mass Spectrom 2020, 31, 12, 2547–2552 © 2020 American Chemical Society.
Figure 10.
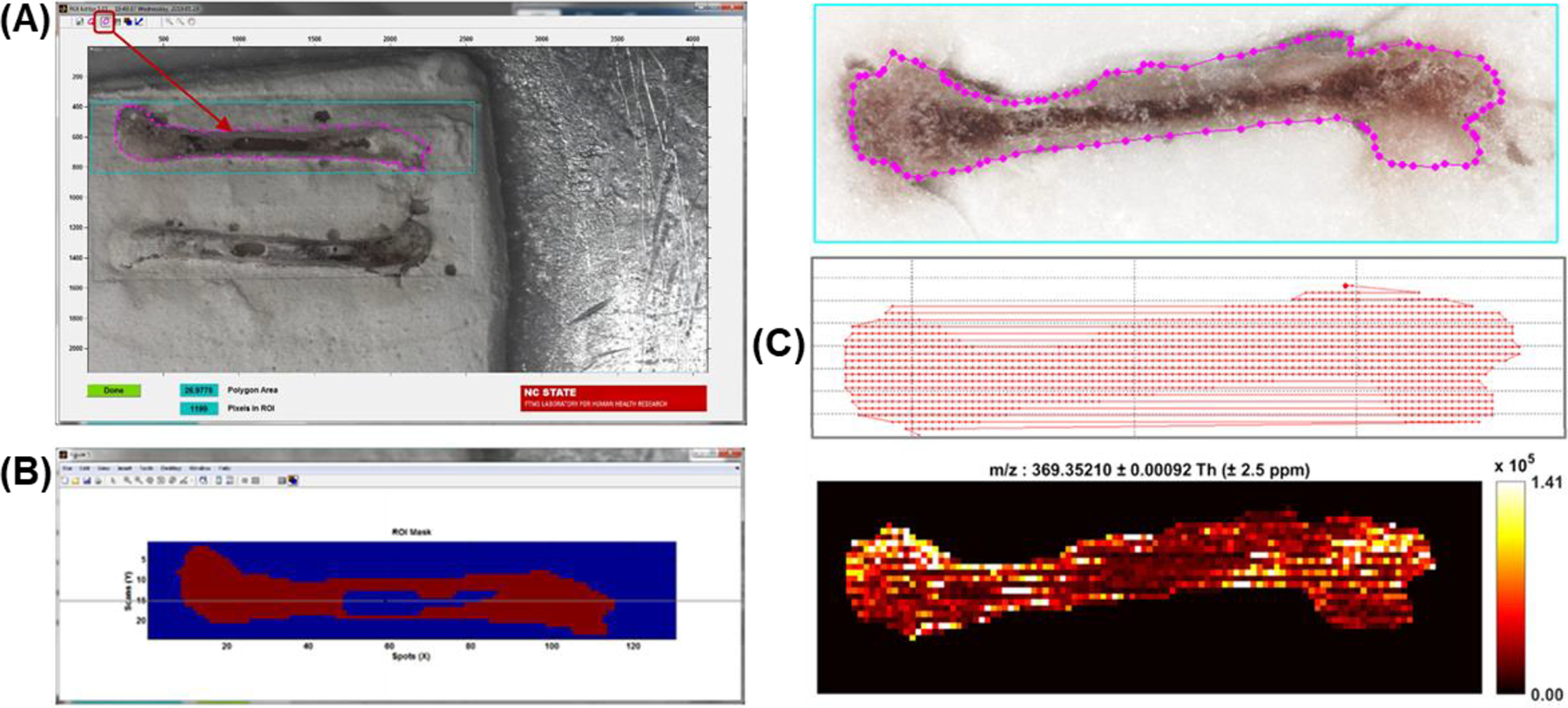
Demonstration of the value of RastirX on mouse bones. (A) ROI Editor polygon tool can be used to draw any closed polygon inside the rectangular ROI to delimit the area of the sample that is imaged. The polygon can be edited and moved after it is drawn. When the ROI Editor is launched, the last saved polygon can be recovered, or the user can draw a new one. (B) Further refinement of the ROI can be made using the mask editor and mouse to include or exclude any pixel in the rectangular ROI. (C) Motion path and the pixels that are imaged can be plotted by RastirX. Adapted with permission from Garrard et al., J. Am. Soc. Mass Spectrom 2020, 31, 12, 2547–2552 © 2020 American Chemical Society.
DIVERSE ARRAY OF HUMAN HEALTH AND DISEASE APPLICATIONS
MALDESI has been providing MSI for diverse human health applications from the beginning. Many of the earliest applications centered on drug distribution of cancer or HIV drugs, and adherence to treatment. One of the first forays in human health focused MSI using IR-MALDESI involved imaging the cancer drug lapatinib in liver tissue and comparing it with UV-MALDI MSI, where it was demonstrated to be an appropriate alternative to UV-MALDI (Barry et. al., 2015). The next study looked at the spatial distribution and abundance of several HIV drugs in cervical tissues where the concentration of drugs was compared against homogenized samples analyzed using LC-MS/MS (Figure 11). Again, IR-MALDESI proved to be a viable alternative to traditional MS and MSI techniques by providing roughly the same sensitivity as LC-MS/MS analyses.
Figure 11.
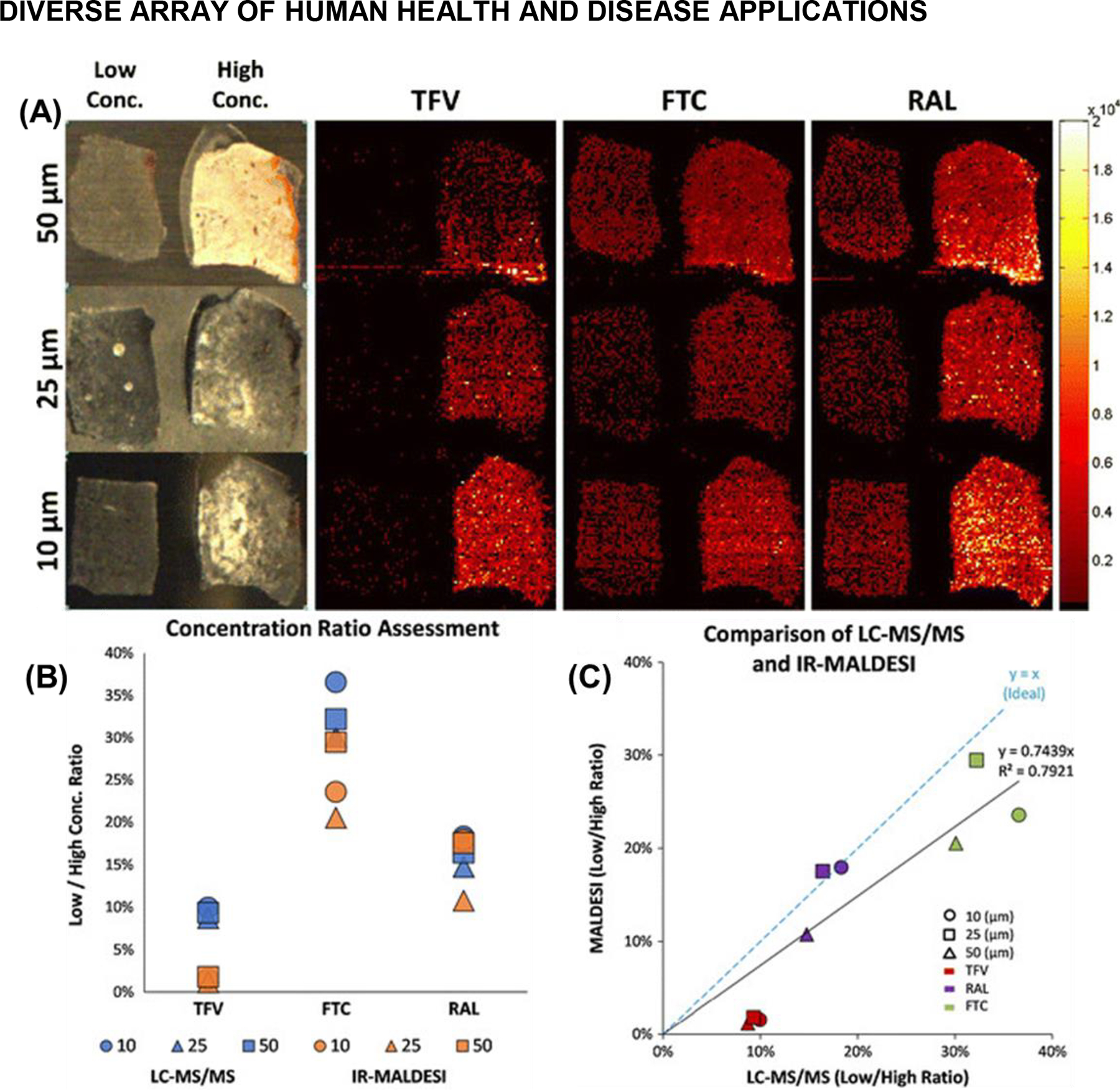
IR-MALDESI MSI analysis of cervical tissues incubated in either a low or high concentration of three HIV drugs: emtricitabine (FTC), tenofovir (TFV), and raltegravir (RAL). (A) Three different tissue thicknesses were investigated (10, 25, and 50 μm). The ion maps for all three drugs at each tissue thickness are shown on the same intensity scale to highlight relative differences in abundance. (B) Plot of the low to high concentration ratios of all three drugs across the three tissue thicknesses that were investigated. (C) Plot of the data from both methods (LC-MS/MS versus IR-MALDESI). A slope near 1 indicates relatively good agreement between the results from the LC-MS/MS and IR-MALDESI MSI experiments. Adapted with permission from Barry et al., J. Am. Soc. Mass Spectrom 2014, 25, 12, 2038–2047 © 2014 American Chemical Society.
Since IR-MALDESI ablates probed regions of the sample completely, it is well-suited for quantitative imaging analyses. The first demonstration was the quantitative analysis of antiretroviral drug-incubated tissues (Bokhart et al., 2015). A structural analogue to each antiretroviral was sprayed onto the glass specimen slide before tissue mounting for normalization, which reduced voxel-to-voxel variability and is the fundamental technique for achieving relative quantification (Figure 12). Absolute quantification was demonstrated for the analysis of several antiretrovirals in human cervical tissues and compared to traditional absolute quantification on an LC-MS/MS platform. Absolute quantitative MSI was achieved by spotting SIL calibration standards onto the tissue matrix to produce a spatial calibration curve that accounts for tissue-matrix effects. IR-MALDESI showed comparable results to LC-MS/MS analyses (Figure 12). This was later replicated for glutathione in hen ovarian tissue (Nazari et al., 2019).
Figure 12.
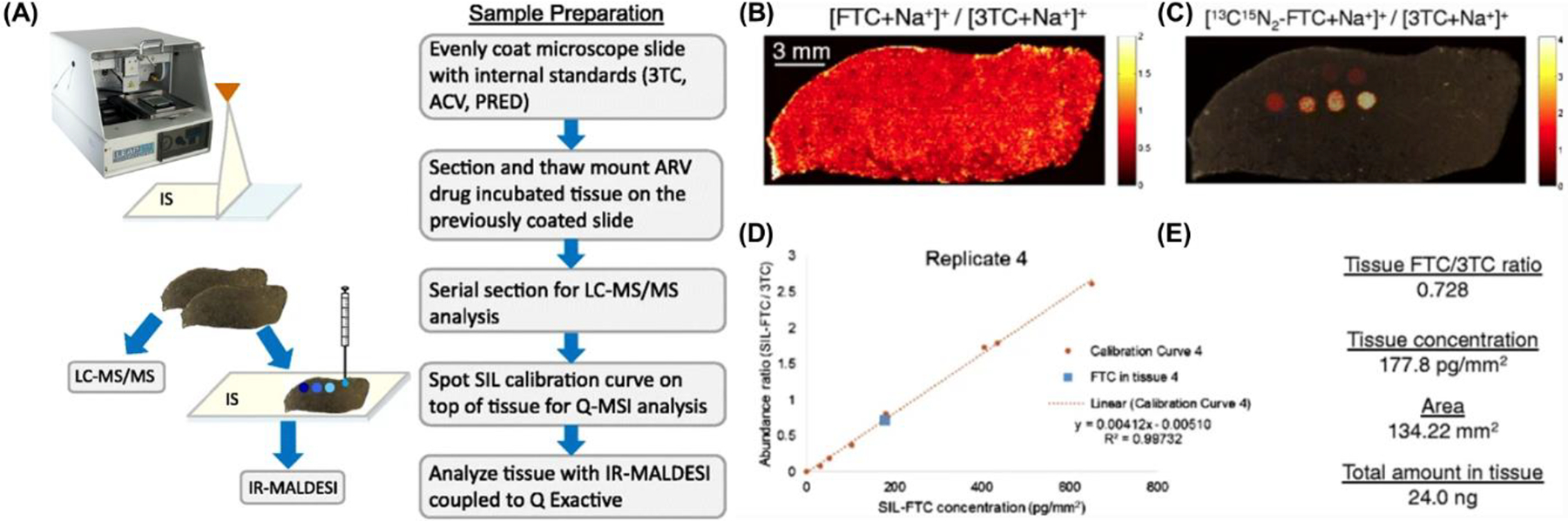
Summary for quantitative IR-MALDESI MSI of FTC in tissue. (A) Workflow for quantitative IR-MALDESI MSI. (B) Ion map of [FTC+Na+]+ / [3TC+Na+]+ representing abundance of incubated FTC in the tissue section. The average ratio was 0.728. (C) Ion map of [13C15N2-FTC+Na+]+ / [3TC+Na+]+ representing the calibration curve at 0, 0.25, 0.5, 1, 2, 4, 6, and 8 μg/mL solution. (D) Resulting calibration curve generated from 13C15N2-FTC showing good linearity with R 2 = 0.9973. The calculated tissue concentration was near the center of the calibration range. (E) Summary of values used to generate the total amount of drug present in tissue section. Using the average ratio and the equation of the calibration curve returns a value of the FTC concentration in tissue in picograms per square millimeter. Using the area of the tissue, the total amount of FTC in the section was determined to be 24.0 ng. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Analytical and Bioanalytical Chemistry Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using matrix-assisted laser desorption electrospray ionization, MT Bokhart, E Rosen, C Thompson, C Sykes, AD Kashuba, DC Muddiman © 2015.
With other early studies looking at HIV drug distributions in incubated tissues, IR-MALDESI pivoted to provide HIV clinicians with a useful tool to determine the adherence of patients to their drug regimen. The longitudinal profiles of various antiretrovirals (ARVs) would provide evidence of daily adherence over roughly a month with a temporal resolution of ~8 hours. Recent experiments have re-examined IR-MALDESI’s effectiveness on detecting such ARVs in hair that has been cosmetically treated. Even after harsh treatments such as relaxers or bleach, most ARVs were still detectable, proving IR-MALDESI is a robust system for determining adherence to ARV regimens (Figure 13). By using IR-MALDESI, clinicians can get an accurate picture of patient drug adherence without drawing blood or plasma.
Figure 13.
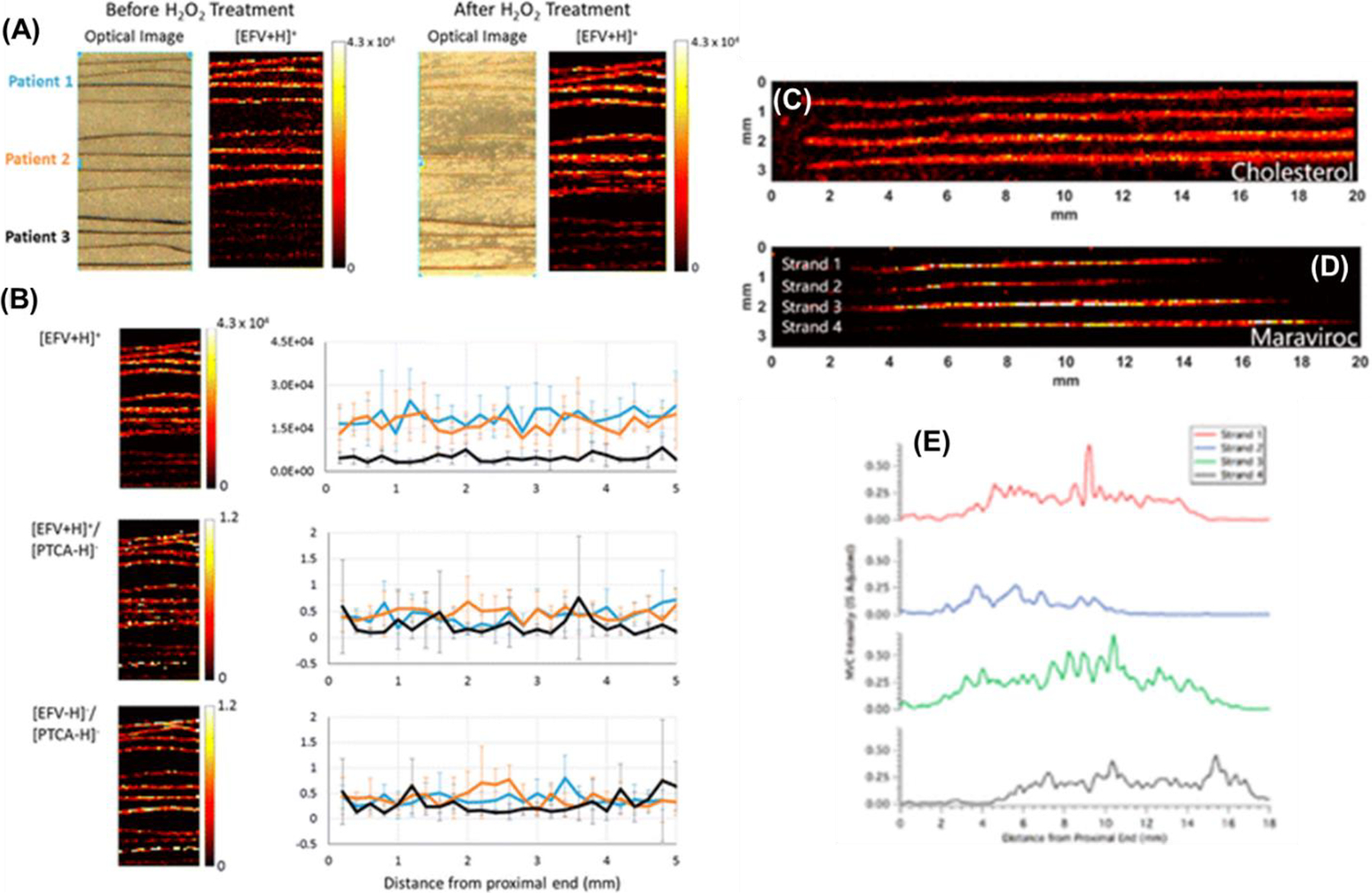
MSI of antiretrovirals in hair to evaluate drug adherence. (A) IR-MALDESI response to [EFV + H]+ from strands (n = 4) of three dosed patients before (left) and after (right) oxidation of melanin by H2O2 indicating no significant degradation in response to EFV. (B) Top panel: ion map of [EFV + H]+ (left) and average longitudinal profile (right) for each of three patients, indicating a 4-fold difference in response to accumulated EFV in hair. Middle panel: Normalization of [EFV + H]+ response by [PTCA – H]− results in similar longitudinal profiles for each of three patients to fixed-dose intake of EFV. Bottom panel: Comparative normalization approach for EFV, matching ionization mechanisms ([EFV – H]−/PTCA – H]−). Adapted with permission from Rosen et al., Anal Chem, 2016, 88, 2, 1336–1344, © 2015 American Chemical Society. (C) Ion image corresponding to cholesterol. (D) Ion image from the same hair strands corresponding to maraviroc (corrected by IS). (E) Longitudinal profiles showing the intensity of maraviroc along the length of the hair strands in panel D. Each of the profiles were smoothed by moving average (n = 3). The top profile corresponds to the top strand in the image. Adapted with permission from Gilliland et al., Anal Chem, 2019, 91, 16, 10816–10822 © 2019 American Chemical Society.
The importance of analyzing samples without chemical derivatization was demonstrated on label-free metabolomic studies of expanded metabolic classes. First, neurotransmitters lower than 400 m/z pose a challenge in MALDI-MSI due to significant matrix interference in the low mass range, requiring additional methods such as chemical derivatization to increase the molecular weight of metabolites above the interfering peaks. Unfortunately, these methods are inherently selective and prevent deep coverage in untargeted studies. IR-MALDESI first demonstrated deeper coverage in the analysis of rat brains exposed to the highly abundant flame retardant, tetrabromobisphenol A (Bagley et al., 2018). This study showed the ability to detect neurotransmitters without chemical derivatization as well as model changes in their distribution after exposure to flame retardants. In a follow-up study, both neurotransmitters and their pathway-related metabolites were investigated in rat placenta tissue (Pace et al., 2020) (Figure 14). This method detected 49 neurotransmitters and metabolites without chemical derivatization, 40% higher than previously reported MALDI analyses with chemical derivatization (35 neurotransmitters & metabolites) (Shariatgorji et al., 2019).
Figure 14.
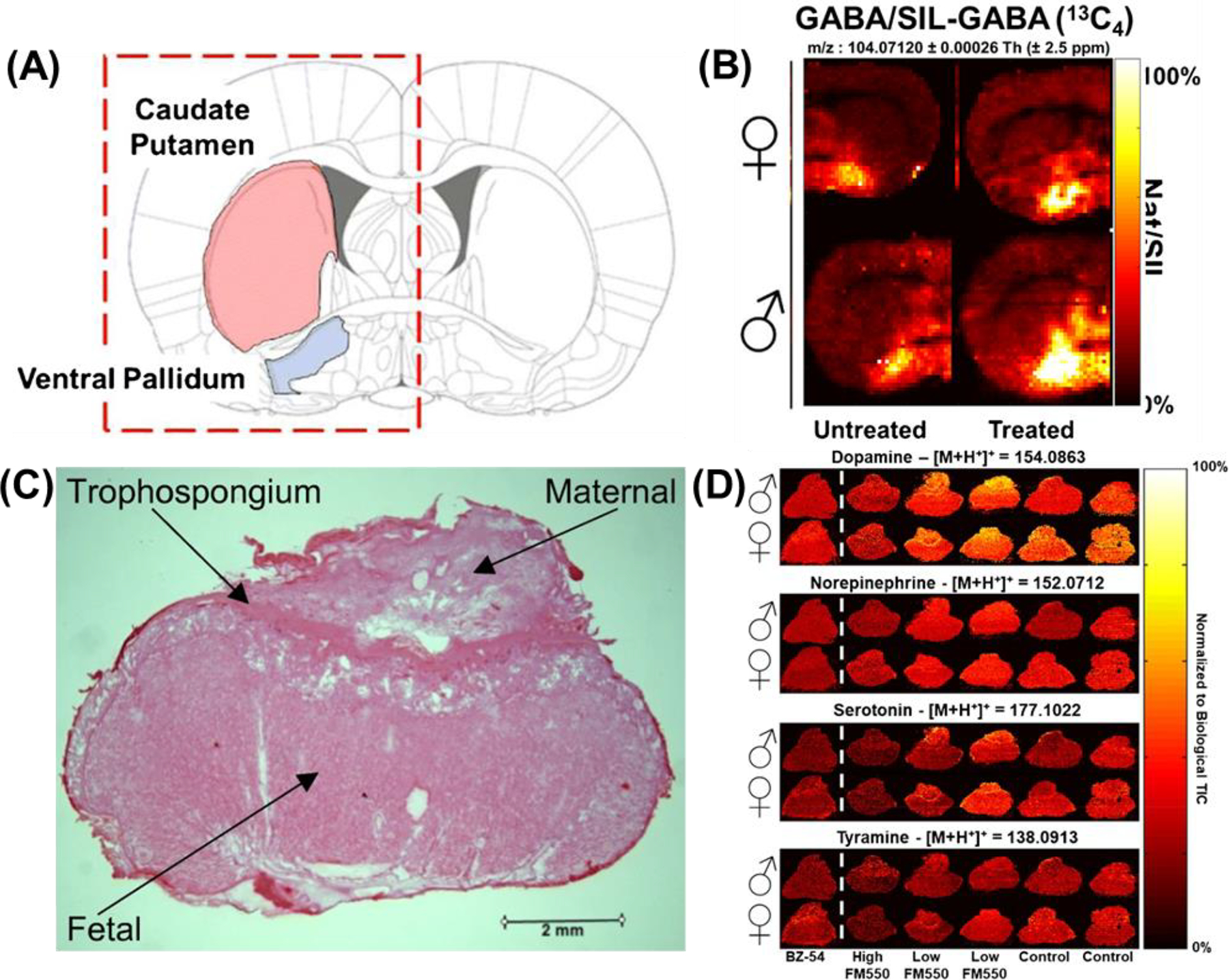
Underivatized neurotransmitters were analyzed in rat brains and rat placenta. Ion abundance in the (A) caudate putamen for GABA represented as (B) ion heat maps (m/z 104.0710) normalized to their SIL isotopes. The inset abundance charts demonstrate an example of the difference between bright and dark voxels in the images. The top row of each image set are female samples, the bottom row are males. The left columns are untreated rats, the right columns are treated rats. The red dashed box shows the region of the brain sample that was analyzed. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Analytical and Bioanalytical Chemistry IR-MALDESI mass spectrometry imaging of underivatized neurotransmitters in brain tissue of rats exposed to tetrabromobisphenol A, MC Bagley, M Ekelöf, K Rock, H Patisaul, DC Muddiman © 2018. (C) Optical image of the rat placenta depicting the maternal, trophospongium, and fetal regions. (D) Spatial distributions of neurotransmitters in placenta tissue as a function of exposure. All four of these neurotransmitters are localized across the whole placenta. While dopamine and norepinephrine appear to be unaffected by exposure levels, serotonin and tyramine have lower normalized abundance in the BZ-54 and high FM550 exposure groups compared with the control group. The BZ-54, high FM550, low FM550, and control exposure groups received a total of 1000 mg BZ-54, 110 mg FM550, 100 mg FM550, and 0 mg FM550, respectively. Dotted white line is used to separate BZ-54 from FM550 exposed tissues. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Analytical and Bioanalytical Chemistry Analysis of neurotransmitters in rat placenta exposed to flame retardants using IR-MALDESI mass spectrometry imaging, CL Pace, B Horman, H Patisaul, DC Muddiman © 2020.
Derivatization is also necessary for MALDI analyses of N-linked glycans. First, the sialic acid residues must be stabilized by esterification, otherwise they dissociate from glycans due to their labile nature. The negative charge on the carboxylic acid of sialic acid predisposes glycans to negative mode ionization, where MALDI has inherent low sensitivity. Using IR-MALDESI N-linked glycans, with sialic acids intact, were ionized without chemical derivatization in both positive (4 glycans) and negative (12 glycans, Figure 15) ionization modes (Pace & Muddiman, 2020).
Figure 15.
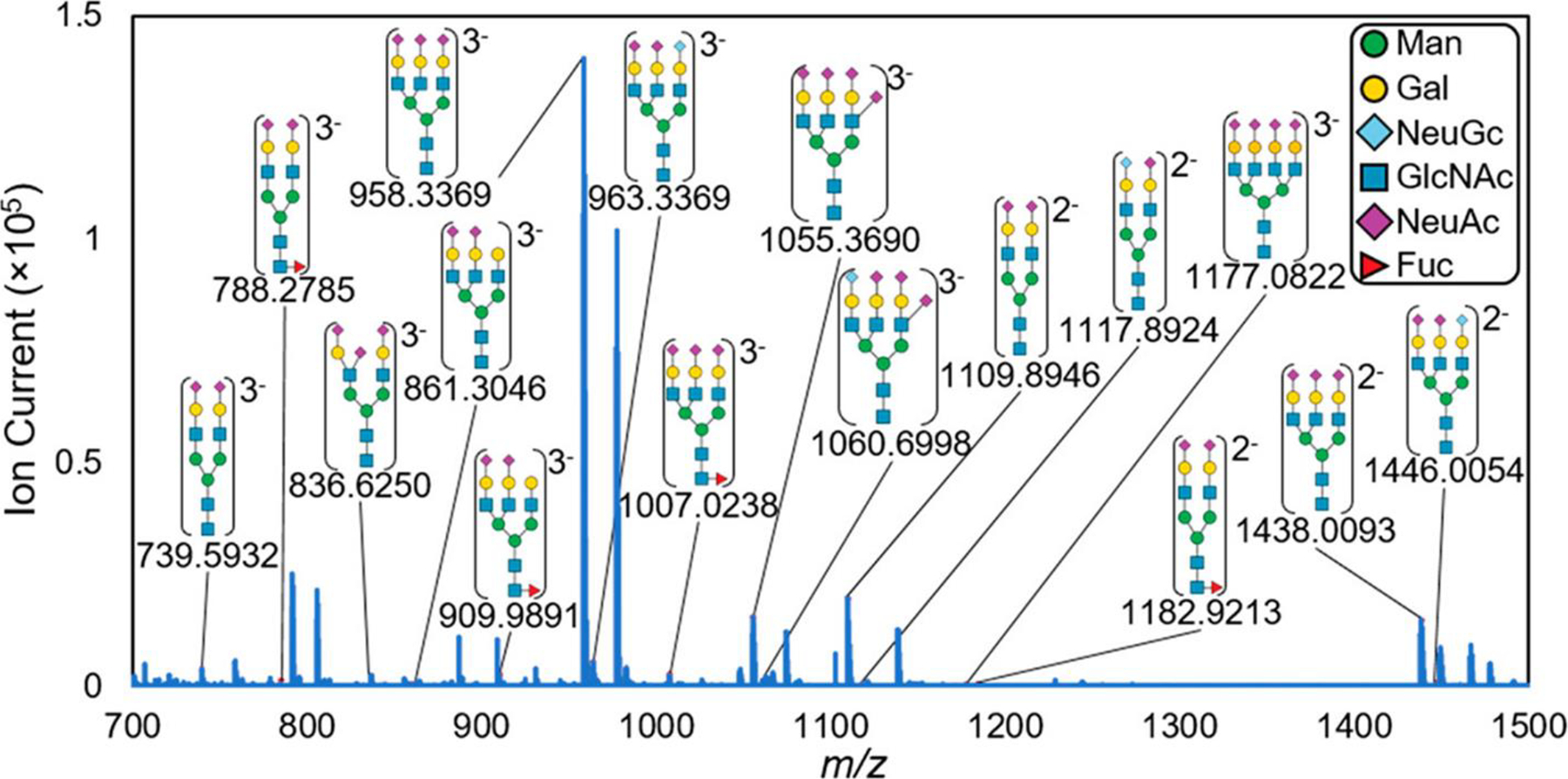
IR-MALDESI mass spectrum of N-linked glycans in negative mode with their experimental m/z shown below each putative identification. These N-linked glycans were detected as glycosylamines, which is discussed in more detail below. Putative structures were assigned based on accurate mass and literature-based characterizations and are heavily composed of sialic acid residues (purple diamonds). Adapted with permission from Pace and Muddiman, J. Am. Soc. Mass Spectrom 2020, 31, 8, 1759–1762 © 2020 American Chemical Society.
In the past few years, the MALDESI MSI focus on human health has been broadened to include not only the human and mammalian tissues discussed in the preceding paragraphs, but also plants particularly in the study highlighted below in Figure 16. IR-MALDESI MSI provided key pieces of evidence challenging the dogma that artemisinin, an anti-malarial compound found naturally in Artemisia annua, was only found in glandular trichomes. In fact, it was demonstrated through several different techniques, including MSI, that the artemisinin pathway is found in non-glandular trichome cells, which offers some hope that artemisinin, the most important ingredient in one of the most effective anti-malarial treatments, might eventually be naturally produced in A. annua. Since then, there have been several published studies analyzing plants with human health implications (Jaiswal et al., 2020; Bagley et al., 2020).
Figure 16.
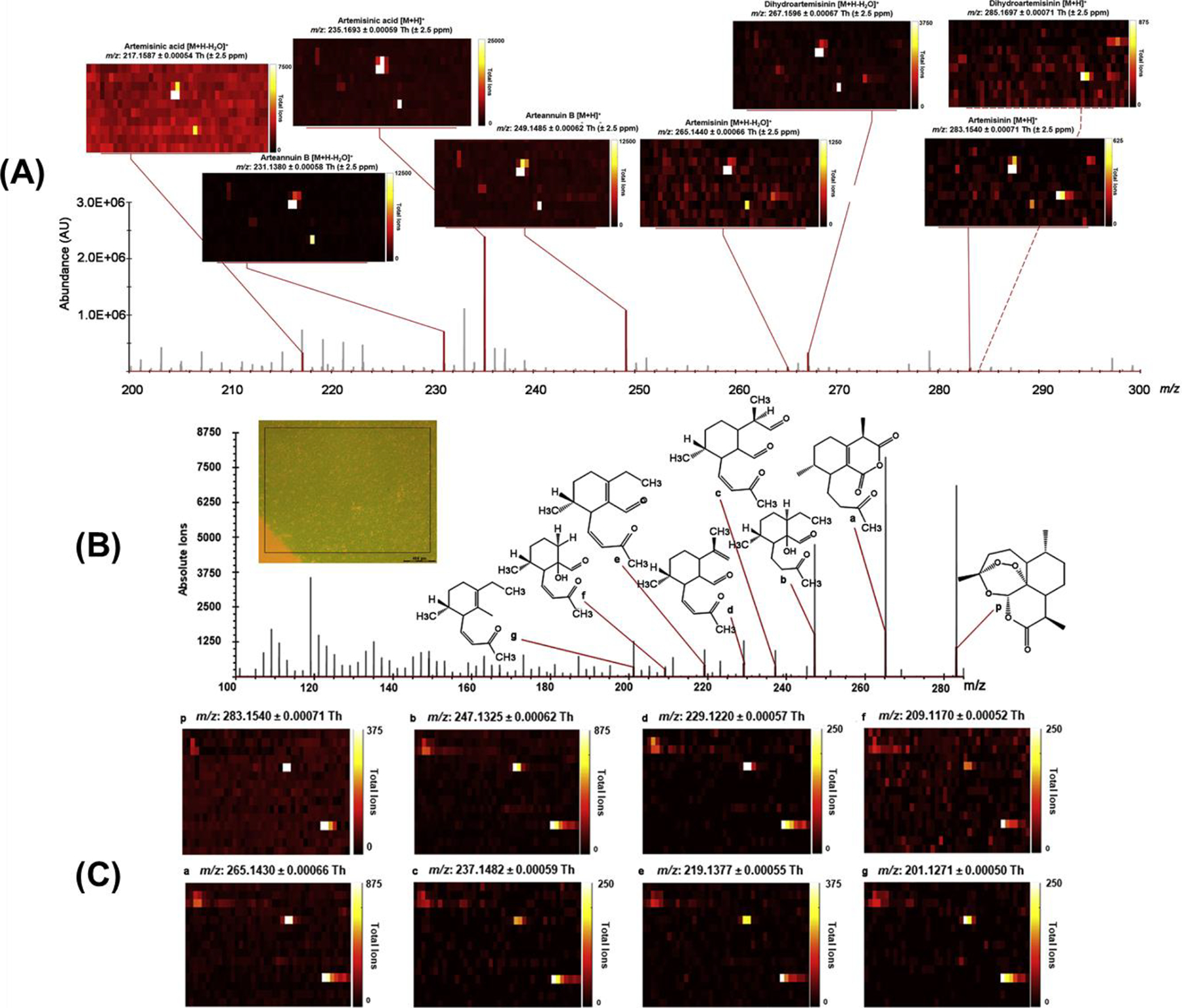
IR-MALDESI Analyses of Glandular Trichome-free Leaves. (A) IR-MALDESI ion abundance map from 200 to 300 Th demonstrates the localization of artemisinin and its related metabolites in glandular trichome (GT)-free leaves. Ions are highlighted in red and are shown in the text above the heatmaps. From left to right: artemisinic acid [M+H-H2O]+, arteannuin B [M+H-H2O]+, artemisinic acid [M+H]+, arteannuin B [M+H]+, artemisinin [M+H-H2O]+, dihydroartemisinin [M+H-H2O]+, artemisinin [M+H]+, dihydroartemisinin [M+H]+. The ion abundance map for dihydroartemisinin [M + H]+ is connected by dashed red lines, which indicates that it colocalizes with several pixel groups of artemisinin [M+H]+. (B) Spectra of fragments associated with MS/MS fragmentation of the artemisinin precursor (283.1540 m/z) and monitored by PRM were obtained from artemisinin in the leaf section of A. annua shown in the insert. Fragment peaks identified by analysis of the artemisinin standard are highlighted with red lines. All m/z values are reported with ±2.5 ppm specificity. (C) Ion distribution heatmaps correlate by letter to the artemisinin fragments. The sampling area shown in the distribution heatmaps is correlated to the area within the box on the leaf section shown in (B), which lacks glandular trichomes. Adapted with permission from Judd et al., Mol. Plant, 2019, 12, 5, 704–714, © 2019 Elsevier B.V.
ANALYSIS OF CHALLENGING SAMPLES WITH ALTERNATIVE MS-FRIENDLY MATRICES
The agility of IR-MALDESI has largely been due to the laser desorption wavelength, which simply needs water in or on the sample to desorb the tissue. However, there are times when ice matrix formation is not possible or fresh-frozen samples cannot be cryosectioned and mounted directly onto the specimen slide. Agarose was one of the first matrices to be required due to sample considerations, namely the need to rapidly analyze fixed seedlings while minimizing dehydration that could change metabolite profiles (Bagley et al., 2020). Agarose was the appropriate matrix because it remained moist while immobilizing the seedling. Furthermore, it enabled SIL-based relative quantification by doping the agarose with SIL-indoleacetic acid, which mirrored the abundance and %RSD of SIL sprayed compounds used for previous quantification in MALDESI studies (Bagley et al., 2020).
While agarose was an appropriate substrate for sampling seedlings, a different sample substrate was designed for whole-body zebrafish imaging, which serves as a translational model for understanding human diseases due to similarities in their genomes. For typical whole-body zebrafish MSI, embedding is commonly used to maintain tissue integrity during sectioning. However, adult male zebrafish pose a significant challenge for cryosectioning due to their large air-filled swim bladder and absence of ovaries to provide support to the abdominal wall. Therefore, gender and life stage impact the quality of whole-body sectioning making consistency in longitudinal studies across sexes difficult to maintain. To this end, IR-MALDESI successfully imaged the lipidome and metabolome (Stutts et al., 2020) of adult male zebrafish embedded in a 5% carboxymethyl cellulose + 10% gelatin solution, by adapting a sectioning method previously described by Nelson et al. (Nelson et al., 2013).
A third sampling substrate was needed to immobilize and maintain the structural integrity of bones even during cryosectioning. One of the gold standards for preparation of bone tissues prior to MSI is decalcification (Stout & Teitelbaum, 1976), where fresh bone is soaked in various solvents (e.g., 10% hydrochloric acid) and kept there for days and sometimes even weeks. During this process, all bone minerals are removed, making bone softer and thus easier to cut in thin slices. Traditionally, there are only two MSI sources compatible with analyses of bone tissues: MALDI and SIMS. Both methods are performed on decalcified bones, which causes chemical and physical changes to the bone. Therefore, developing a derivatization free MSI method for direct analysis of fresh bones was critical for detection of biochemical signatures unique to healthy and disease-affected bones. Seven different embedding materials were evaluated for maintaining structural integrity during analysis, before settling on Plaster of Paris. A CMC/gelatin mixture, agarose, liquid nails, thin-set mortar, water, and alginate were each deemed inappropriate embedding material as they either modified the bone, did not freeze properly, or suppressed signals of bone-specific ions (Figure 17). After method development, healthy and stroke-affected bones from mice were imaged and putatively identified 826 and 669 tissue-specific features in healthy and stroke-affected humeri (Khodjaniyazova et al., 2019), demonstrating deeper coverage than other methods.
Figure 17.
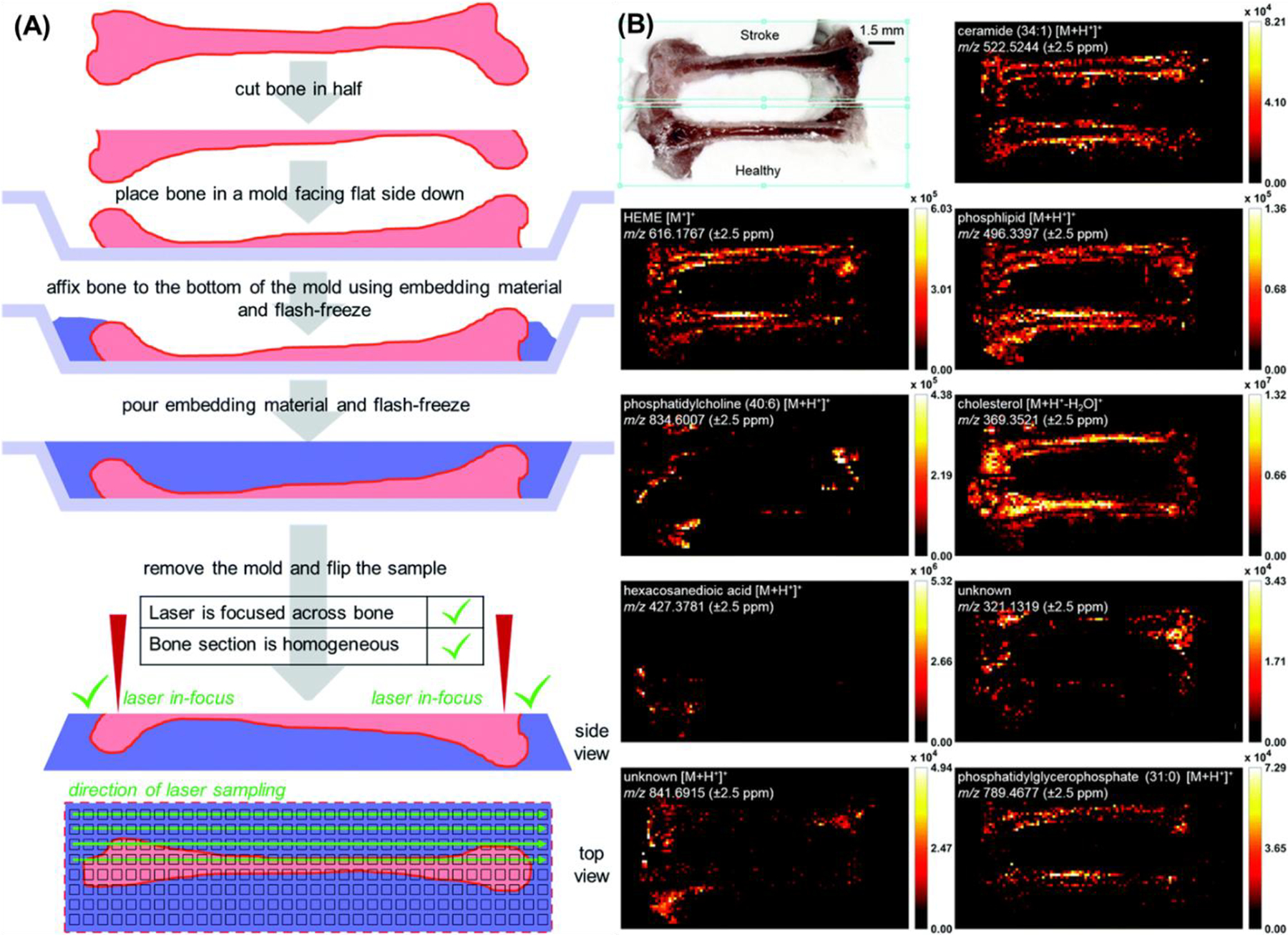
IR-MALDESI MSI of healthy and stroke-affected mouse humeri embedded in Plaster of Paris. (A) Flash-freeze fresh bone; cut bone in half using cryomicrotome; place trimmed bone in a mold facing flat side down; affix bone to the mold using embedding material and wait until the material sets; pour the rest of the embedding material and smooth out the top surface with a blade. Finally, a representation of the ROI and direction of laser sampling. (B) Optical image (+40% brightness, +40% contrast) and ion heatmaps. Features from MS1 scans were putatively annotated using METASPACE annotation engine. Scale bar is 1.5 mm for both the bone image and ion distributions. Adapted with permission from Khodjaniyazova et al., Anal Methods, 2019, 11, 46, 5929–5938 © 2019 The Royal Society of Chemistry.
NEW DIRECTIONS: THE NEXT PHASE OF MALDESI
One of the most recent improvements in IR-MALDESI was marked by pushing past traditional 2D and into 3D imaging. A commercially available over the counter (OTC) pharmaceutical in the form of multiple unit pellet systems (MUPS) was used as a model to demonstrate the feasibility of IR-MALDESI for 3D MSI (Figure 18) (Bai et al., 2020). Without the need of consecutive cryosectioning required in other methods, 3D IR-MALDESI MSI was performed by repeatedly collecting 2D images over the same region of interest (ROI). The depth resolution was investigated as a function of laser energy levels and density of ablated material and found to be 2.3 μm at reduced energy level (0.3 mJ/pulse). A follow up 3D MSI analysis on a region of interest of 15 × 15 voxels across 50-layers was performed on a half pill, with an analysis duration of 70 minutes. After the demonstration of an ablation-based 3D MSI on the hard sample, attention was shifted to imaging soft tissues which are much more complex modalities (Figure 19). A total of 594 lipids were putatively detected on a fresh frozen skin tissue in a pilot study and skin structured was resolved with differential lipids distributions in a 56-layer 3D MSI at x and y resolution of 50 μm and z resolution of 7 μm, which was comparable to the histological image.
Figure 18.
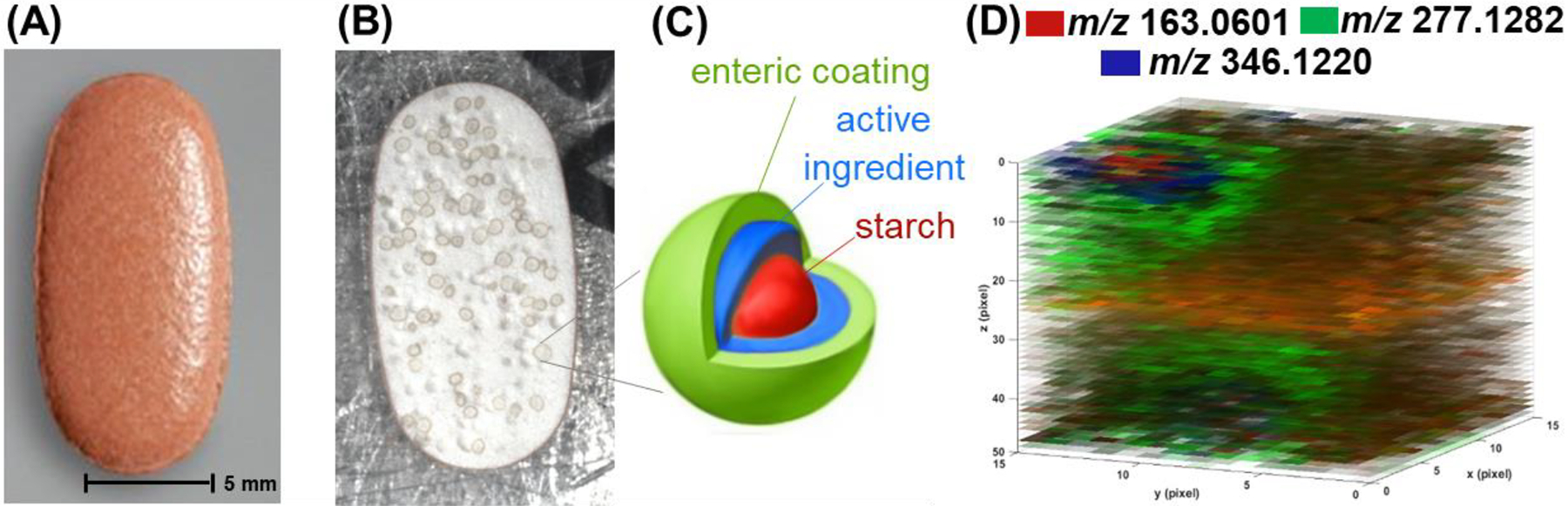
Three-dimensional demonstration of IR-MALDESI MSI performed on a pill. (A) Optical image of a full pill, where the z-resolution at different energy levels was determined. (B) Optical image of a pill trimmed in half for 2D and 3D MSI; small circles are due to MUPS formulation. (C) Schematic of a pellet and its components, where each color indicates different components. (D) Three-dimensional intensity maps for starch (m/z 163.0601), triethyl citrate (m/z 277.1282), and omeprazole (m/z 346.1220). Colocalization of three markers in the pill with laser spot size 80 μm and depth resolution 16.3 μm. Adapted with permission Bai et al., J. Am. Soc. Mass Spectrom 2020, 31, 2, 292–297 © 2020 American Chemical Society.
Figure 19.
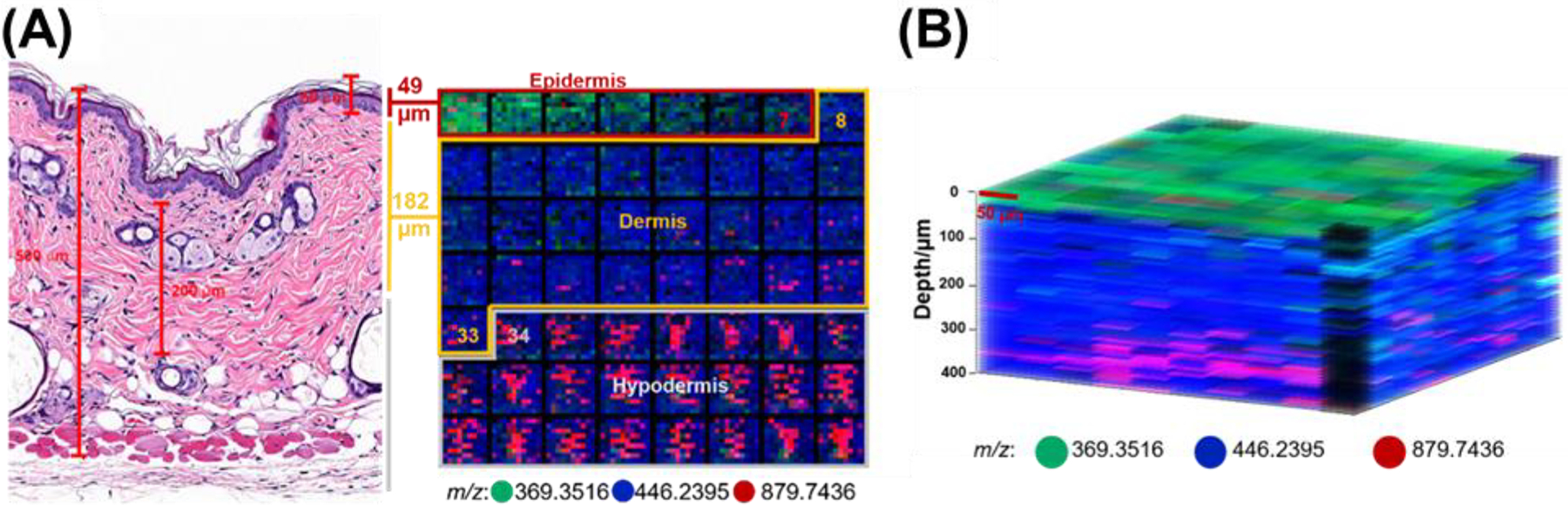
(A) Correlation of MSI images with H&E-stained histological image. Overlaid image of three putatively identified lipids (cholesterol at m/z 369.3516 in the green channel, S1P at m/z 446.2395 in the blue channel, TG at m/z 879.7436 in the red channel) shows a clear interface between the 7th and 8th layer as well as the 33rd and 34th layer. The estimated thickness of the epidermis and dermis agrees with the histological data. (B) 3D three-color overlaid image of cholesterol at m/z 369.3516 (green), S1P at m/z 446.2395 (blue), and TG at m/z 879.7436 (52:3) (red). The lateral resolution is 50 μm and the depth resolution 7 μm. The magenta color represents the concurrent presence of blue and red color. Adapted by permission from Springer Nature Customer Service Centre GmbH: Springer Analytical and Bioanalytical Chemistry Three-dimensional (3D) imaging of lipids in skin tissues with infrared matrix-assisted laser desorption electrospray ionization (MALDESI) mass spectrometry, H Bai, KE Linder, DC Muddiman © 2021.
An IR-MALDESI source was coupled with a drift tube ion mobility spectrometer-mass spectrometer (IMS-MS) to increase the coverage of isomeric and isobaric species (Figure 20) (Ekelöf et al., 2020). Ion mobility provides structural resolution of charged species prior to their detection in a mass analyzer. To demonstrate the range of applications possible with this novel platform, small molecules, lipids, carbohydrates, and intact proteins were directly analyzed from standards and/or biological samples. Undiluted Coca-Cola was directly analyzed to separate fructose and glucose (structural isomers, C6H12O6) as well as to absolutely quantify caffeine in the drink. The protein thioredoxin was directly analyzed with an observed charge-state distribution from 8+−12+. A proof-of-concept MSI experiment was performed on an oak leaf where isomeric flavonoids as well as multiply-charged ions were detected. Using MSiReader the collected IR-MALDESI IMS-MS data was analyzed, a significant step forward in the agility of this platform for the MSI community which has identified IMS as a promising new direction for MSI applications (Rivera et al., 2020; Laskin et al., 2020).
Figure 20.
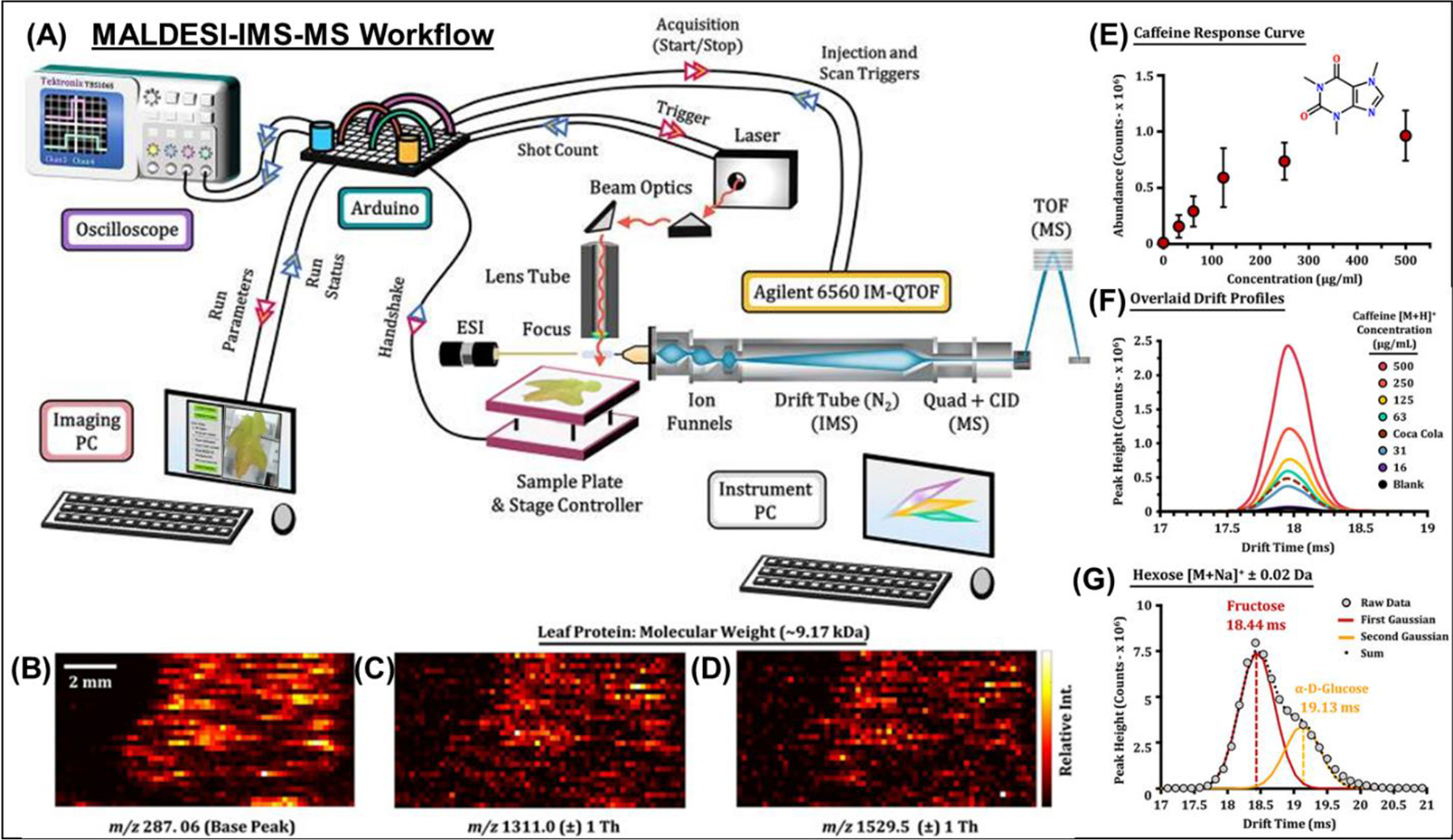
MALDESI-IMS-MS workflow. (A) A diagram is shown illustrating the major components of the IMS-QTOF MS platform and the annotated connections to the Arduino microcontroller board used to control each acquisition. (B,C,D) Raster experiment with consecutive line scans of a locally sourced oak leaf. After sample collection, spectra were acquired using IR-MALDESI-IMS-MS and exported to MSiReader for data visualization. Ion heatmaps for the base peak and highly charged features are illustrated with corresponding mass spectra. (E) Average abundance evaluations for the [M + H]+ signal of caffeine summed for per concentration level (or 50 IMS acquisitions) with error bars illustrating one standard deviation. (F) IMS drift time profiles for each caffeine concentration level, including a solvent blank and undiluted Coca-Cola. (G) The drift time profile of the hexose sugar [M + Na]+ ion acquired from the Coca-Cola sample. Two Gaussian peaks were fit to the profile illustrating CCS values consistent with fructose and glucose in an abundance ratio of 68:32. Adapted with permission Ekelöf et al., J. Am. Soc. Mass Spectrom 2020, 31, 3, 642–650 © 2020 American Chemical Society.
MALDESI’s versatility for everything from tissue to liquid analysis (Nazari et al., 2018), while enabling the soft ionization characteristics of ESI, made it a potential MS technique for high-throughput screening (HTS). While many HTS methods have relied on either UV/Vis or fluorescence spectroscopy for detection, significant drawbacks associated with these techniques have created interest in a label-free, sensitive, and specific method that is not limited by tagging efficiency or obfuscated by autofluorescence or non-specific tagging. Mass spectrometry offers all these characteristics; however, historical application for HTS has been limited due to the time requirements of HPLC-MS or UPLC-MS and the need for sample cleanup to remove nonvolatile compounds that could compromise the analysis. IR-MALDESI was demonstrated to be a viable, even desirable option for HTS studies by combining the benefits of traditional MS, without many of the limitations (Nazari et al., 2017). In principle, the contents of a 1536-well microtiter plate could be analyzed in ~30 minutes using IR-MALDESI. This can be accomplished without cleaning up samples to remove salts or buffers, as IR-MALDESI has shown to have high salt tolerance up to molar concentrations. A proof-of-concept study looking at the enzymatic activity of isocitrate dehydrogenase 1 (IDH1) was found to have a high Z-factor demonstrating that IR-MALDESI can separate true positives from false positives with a high degree of confidence (Nazari et al., 2017). Since then, another study using a custom-built IR-MALDESI HTS platform was used to analyze three more biochemical assays (IDH1, DGKζ, P300) and a pilot screen (IDH1) showing comparable data to traditional HTS methods, and the ability to decrease analysis time from 1.9 s/well to 100 ms/well (Pu et al., 2021).
While MALDESI has grown tremendously since its creation in 2006 (Figures 1,2,4 Table 2), there is more to accomplish regarding fundamental and experimental improvements. The next generation of development will include exploration of other mid-IR wavelengths to determine if there are benefits for surface sensitivity, depth resolution for 3D imaging, and spatial resolution. The current sample stage will be upgraded to a more accurate and precise planar XY stage that allows for faster scanning by shortening the raster time between spots. As 3D analysis becomes a larger focus of IR-MALDESI, it will be important to integrate a motorized Z axis that can keep the surface of the sample at the appropriate focal depth of the laser and enable appropriate sampling of surfaces with significant topography. To improve experimental data, the IR-MALDESI source will be coupled to more advanced MS systems including the Thermo Exploris 240, which would allow for greater resolution and higher scan rates, and the Agilent 6560 IM-TOF, which would allow the distinguishing of structural isomers. MSiReader will evolve in coordination with these improvements and with input from the entire MSI community. Taken together, these improvements will keep the IR-MALDESI platform at the cutting edge of ambient MSI, providing value to the MSI community and the broader scientific community.
Table 2.
Table with other innovations, characteristics and improvements made in IR-MALDESI categorized by generation. ‘Materials’ and ‘Molecules’ columns are not broken down by generation because they were more driven by biological interests and collaborations. This creates a valuable feedback loop where technology improvements can enable greater biological/molecular diversity and where biological/molecular needs necessitate improvements in technology. By the end of generation 4, and moving forward in NextGen, all materials and molecules can be analyzed.
| Generation | Acquisition Rate | Spot Size | Resolving Power | Other Innovations | Materials | Molecules |
|---|---|---|---|---|---|---|
| 1 | 0.5–1 scan/s (instrument limited) | 150 | >200,000 FWHM @ 200 m/z | Soft Tissues (e.g., Brain, Heart, Lung) Whole Body (e.g., Mouse, Zebrafish) Bone (w/o demineralization) Hair Skin (Human, Pig, Mouse) Plants with Medicinal Value OTC Formulated Pills/Counterfeit Drugs Cerebrospinal Fluid Plasma Cells (including single cell analysis) Paper-based Cell Culture Tumor Model Spheroids High Throughout / High Content Screening |
Neurotransmitters ARV Drugs Chemotherapy Drugs Drugs of abuse Lipids Polar Metabolites Bioactive Peptides Proteins Glycans Exposure (e.g., PFAS) |
|
| 2 | 1.2–1.5 scan/s (laser limited) | 100 | 50,000 FWHM @ 400 m/z | 1st Source Designed 1st Use of Infrared laser 1st Imaging Demonstration Global Optimization for IR-MALDESI MSiReader developed | ||
| 3 | 1.7 scan/s (instrument, stage limited) | 100–150 (50) | 140k @ 200 m/z | 1st Ice Matrix Demonstration Cellular Level MSI by Oversampling Quantitative Imaging Polarity Switching Whole-body imaging | ||
| 4 | 1.8 scan/s (instrument, stage limited) | 50–100 | 140k @ 200 m/z | 50 micron resolution demonstration 1st HTS Demonstration Injection Time Optimization MSiReader v1.0 developed Absolute Quantitative MSI Novel Burst-Mode Laser Digital Image Recognition 3D Imaging Demonstration Ion Mobility Demonstration MSI of Arbitrary Spatial Patterns | ||
| NextGen | ≥2 scan/s | ≤50 | 240k @ 200 m/z Ion Mobility |
ACKNOWLEDGEMENTS
The authors thank Crystal Lynn Pace, Anqi Tu, Hellena Bai and Jeff Enders for their assistance in preparing this manuscript. The authors gratefully acknowledge financial support from the National Institutes of Health (R01GM087964), the W.M. Keck Foundation, and North Carolina State University. This work was performed in part by the Molecular Education, Technology and Research Innovation Center (METRIC) at NC State University, which is supported by the State of North Carolina.
REFERENCES
- Ai W, Nie H, Song S, Liu X, Bai Y, & Liu H. 2018. A versatile integrated ambient ionization source platform. J Am Soc Mass Spectrom 29:7:1408–1415. [DOI] [PubMed] [Google Scholar]
- Andrade FJ, Shelley JT, Wetzel WC, Webb MR, Gamez G, Ray SJ, & Hieftje GM. 2008. Atmospheric pressure chemical ionization source. 2. Desorption−Ionization for the direct analysis of solid compounds. Anal Chem 80:8:2654–2663. [DOI] [PubMed] [Google Scholar]
- Andrade FJ, Wetzel WC, Chan GC-, Webb MR, Gamez G, Ray SJ, & Hieftje GM. 2006. A new, versatile, direct-current helium atmospheric-pressure glow discharge. J Anal at Spectrom 21:11:1175–1184. [Google Scholar]
- Bagley MC, Ekelöf M, & Muddiman DC. 2020. Determination of optimal electrospray parameters for lipidomics in infrared-matrix-assisted laser desorption electrospray ionization mass spectrometry imaging. J Am Soc Mass Spectrom 31:2:319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley MC, Stepanova AN, Ekelöf M, Alonso JM, & Muddiman DC. 2020. Development of a relative quantification method for infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging of arabidopsis seedlings. Rapid Commun Mass Spectrom 34:6:e8616. [DOI] [PubMed] [Google Scholar]
- Bagley MC, Ekelöf M, Rock K, Patisaul H, & Muddiman DC. 2018. IR-MALDESI mass spectrometry imaging of underivatized neurotransmitters in brain tissue of rats exposed to tetrabromobisphenol A. Anal Bioanal Chem 410:30:7979–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley MC, Pace CL, Ekelöf M, Muddiman DC. 2020. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) mass spectrometry imaging analysis of endogenous metabolites in cherry tomatoes. Analyst 145:5516–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Khodjaniyazova S, Garrard KP, & Muddiman DC. 2020. Three-dimensional imaging with infrared matrix-assisted laser desorption electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 31:2:292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Linder KE, & Muddiman DC. 2021. Three-dimensional (3D) imaging of lipids in skin tissues with infrared matrix-assisted laser desorption electrospray ionization (MALDESI) mass spectrometry. Anal Bioanal Chem 413:2793–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TC, Han J, & Borchers CH. 2017. Recent advancements in matrix-assisted laser desorption/ionization mass spectrometry imaging. Curr Opin Biotech 43:62–69. [DOI] [PubMed] [Google Scholar]
- Balog J, Szaniszlo T, Schaefer K, Denes J, Lopata A, Godorhazy L, … Takats Z. 2010. Identification of biological tissues by rapid evaporative ionization mass spectrometry. Anal Chem 82:17:7343–7350. [DOI] [PubMed] [Google Scholar]
- Balog J, Kumar S, Alexander J, Golf O, Huang J, Wiggins T, … Takats Z. 2015. In vivo endoscopic tissue identification by rapid evaporative ionization mass spectrometry (REIMS). Angew Chem Int Ed 54:38:11059–11062. [DOI] [PubMed] [Google Scholar]
- Balog J, Sasi-Szabó L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, … Takáts Z. 2013. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med 5:194:194ra93. [DOI] [PubMed] [Google Scholar]
- Barry JA, Groseclose MR, Robichaud G, Castellino S, & Muddiman DC. 2015. Assessing drug and metabolite detection in liver tissue by UV-MALDI and IR-MALDESI mass spectrometry imaging coupled to FT-ICR MS. Int J Mass Spectrom 377:448–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JA, Robichaud G, Bokhart MT, Thompson C, Sykes C, Kashuba AD, & Muddiman DC. 2014. Mapping antiretroviral drugs in tissue by IR-MALDESI MSI coupled to the Q exactive and comparison with LC-MS/MS SRM assay. J Am Soc Mass Spectrom 25:12:2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JA, & Muddiman DC. 2011. Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments. Rapid Commun Mass Spectrom 25:23:3527–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JA, Robichaud G, & Muddiman DC. 2013. Mass recalibration of FT-ICR mass spectrometry imaging data using the average frequency shift of ambient ions. J Am Soc Mass Spectrom 24:7:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkenkamp S, Karas M, & Hillenkamp F. 1996. Ice as a matrix for IR-matrix-assisted laser desorption/ionization: mass spectra from a protein single crystal. Proc Natl Acad Sci USA 93:14:7003–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkenkamp S, Menzel C, Karas M, and Hillenkamp F. 1997. Performance of infrared matrix-assisted laser desorption/ionization mass spectrometry with lasers emitting in the 3 μm wavelength range. Rapid Commun Mass Spectrom 11:13:1399–1406. [Google Scholar]
- Bodzon-Kulakowska A, & Suder P. 2016. Imaging mass spectrometry: Instrumentation, applications, and combination with other visualization techniques. Mass Spec Rev 35:1:147–169. [DOI] [PubMed] [Google Scholar]
- Bokhart MT, & Muddiman DC. 2016. Infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging analysis of biospecimens. Analyst 141:18:5236–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhart MT, Manni J, Garrard KP, Ekelöf M, Nazari M, & Muddiman DC. 2017. IR-MALDESI mass spectrometry imaging at 50 micron spatial resolution. J Am Soc Mass Spectrom 28:10:2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba AD, & Muddiman DC. 2015. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal Bioanal Chem 407:8:2073–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhart MT, Nazari M, Garrard KP, & Muddiman DC. 2017b. MSiReader v1.0: Evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J Am Soc Mass Spectrom 29:1:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowfield A, Barrett DA, Alexander MR, Ortori CA, Rutten FM, Salter TL, … Bradley JW. 2012. Surface analysis using a new plasma assisted desorption/ionisation source for mass spectrometry in ambient air. Rev Sci Instrum 83:6:063503. [DOI] [PubMed] [Google Scholar]
- Buchberger AR, DeLaney K, Johnson J, & Li L. 2018. Mass spectrometry imaging: A review of emerging advancements and future insights. Anal Chem 90:1:240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing R, & Slodzian G. 1962. Microanalyse par emission ionique secondaire. J Microsc 1:395–410. [Google Scholar]
- Chang C, Li X, Bai Y, Xu G, Feng B, Liao Y, & Liu H. 2013. Graphene matrix for signal enhancement in ambient plasma assisted laser desorption ionization mass spectrometry. Talanta 114:54–59. [DOI] [PubMed] [Google Scholar]
- Chang D, Lee C, & Shiea J. 2002. Detecting large biomolecules from high-salt solutions by fused-droplet electrospray ionization mass spectrometry. Anal Chem 74:11:2465–2469. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Schwartz SA, & Caprioli RM. 2004. Peer reviewed: Profiling and imaging proteins in tissue sections by MS. Anal Chem 76:5:86 A–93 A. [PubMed] [Google Scholar]
- Chaurand P, Cornett DS, Angel PM, & Caprioli RM. 2011. From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol Cell Proteomics 10:2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wortmann A, & Zenobi R. 2007. Neutral desorption sampling coupled to extractive electrospray ionization mass spectrometry for rapid differentiation of biosamples by metabolomic fingerprinting. J Mass Spectrom 42:9:1123–1135. [DOI] [PubMed] [Google Scholar]
- Chen H, Venter A, & Cooks RG. 2006. Extractive electrospray ionization for direct analysis of undiluted urine, milk and other complex mixtures without sample preparation. Chem Commun 19:2042–2044. [DOI] [PubMed] [Google Scholar]
- Chen H, Yang S, Wortmann A, & Zenobi R. 2007. Neutral desorption sampling of living objects for rapid analysis by extractive electrospray ionization mass spectrometry. Angew Chem Int Ed Engl 46:40:7591–7594. [DOI] [PubMed] [Google Scholar]
- Chen H, Sun Y, Wortmann A, Gu H, & Zenobi R. 2007a. Differentiation of maturity and quality of fruit using noninvasive extractive electrospray ionization quadrupole time-of-flight mass spectrometry. Anal Chem 79:4:1447–1455. [DOI] [PubMed] [Google Scholar]
- Chen H, Sun Y, Wortmann A, Gu H, & Zenobi R. 2007b. Differentiation of maturity and quality of fruit using noninvasive extractive electrospray ionization quadrupole time-of-flight mass spectrometry. Anal Chem 79:4:1447–1455. [DOI] [PubMed] [Google Scholar]
- Chen H, Hu B, Hu Y, Huan Y, Zhou Z, & Qiao X. 2009. Neutral desorption using a sealed enclosure to sample explosives on human skin for rapid detection by EESI-MS. J Am Soc Mass Spectrom 20:4:719–722. [DOI] [PubMed] [Google Scholar]
- Cochran KH, Barry JA, Robichaud G, & Muddiman DC. 2015. Analysis of trace fibers by IR-MALDESI imaging coupled with high resolving power MS. Anal Bioanal Chem 407:3:813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran KH, Barry JA, Muddiman DC, & Hinks D. 2013. Direct analysis of textile fabrics and dyes using infrared matrix-assisted laser desorption electrospray ionization mass spectrometry. Anal Chem 85:2:831–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody RB. 2009a. Observation of molecular ions and analysis of nonpolar compounds with the direct analysis in real time ion source. Anal Chem 81:3:1101–1107. [DOI] [PubMed] [Google Scholar]
- Cody RB. 2009b. Observation of molecular ions and analysis of nonpolar compounds with the direct analysis in real time ion source. Anal Chem 81:3:1101–1107. [DOI] [PubMed] [Google Scholar]
- Costa AB, & Graham Cooks R. 2008. Simulated splashes: Elucidating the mechanism of desorption electrospray ionization mass spectrometry. Chem Phys Lett 464:1:1–8. [Google Scholar]
- Dillon LA, Stone VN, Croasdell LA, Fielden PR, Goddard NJ, & Paul Thomas CL. 2010. Optimisation of secondary electrospray ionisation (SESI) for the trace determination of gas-phase volatile organic compounds. Analyst. 135:2:306–314. [DOI] [PubMed] [Google Scholar]
- Dixon RB, Sampson JS, Hawkridge AM, & Muddiman DC. 2008. Ambient aerodynamic ionization source for remote analyte sampling and mass spectrometric analysis. Anal Chem 80:13:4994–5001. [DOI] [PubMed] [Google Scholar]
- Dixon RB, & Muddiman DC. 2010. Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst 135:5:880–882. [DOI] [PubMed] [Google Scholar]
- Dueñas ME, Essner JJ, & Lee YJ. 2017. 3D MALDI mass spectrometry imaging of a single cell: spatial mapping of lipids in the embryonic development of zebrafish. Sci Rep 7:14946:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, & Henion JD. 2011. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: The terfenadine example. Rapid Commun Mass Spectrom 25:23:3587–3596. [DOI] [PubMed] [Google Scholar]
- Ekelöf M, Manni J, Nazari M, Bokhart M, & Muddiman DC. 2018. Characterization of a novel miniaturized burst-mode infrared laser system for IR-MALDESI mass spectrometry imaging. Anal Bioanal Chem 410:9:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekelöf M, McMurtrie EK, Nazari M, Johanningsmeier SD, & Muddiman DC. 2017. Direct analysis of triterpenes from high-salt fermented cucumbers using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). J Am Soc Mass Spectrom 28:2:370–375. [DOI] [PubMed] [Google Scholar]
- Ekelöf M, & Muddiman DC. 2018. IR-MALDESI method optimization based on time-resolved measurement of ion yields. Anal Bioanal Chem 410:3:963–970. [DOI] [PubMed] [Google Scholar]
- Ekelöf M, Dodds J, Khodjaniyazova S, Garrard KP, Baker ES, & Muddiman DC. 2020. Coupling IR-MALDESI with drift tube ion mobility-mass spectrometry for high-throughput screening and imaging applications. J Am Soc Mass Spectrom 31:3:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekelöf M, Garrard KP, Judd R, Rosen EP, Xie D, Kashuba ADM, & Muddiman DC. 2018. Evaluation of digital image recognition methods for mass spectrometry imaging data analysis. J Am Soc Mass Spectrom 29:12:2467–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Soltwisch J, Paine MRL, Dreisewerd K, & Heeren RMA. 2017. Laser post-ionisation combined with a high resolving power orbitrap mass spectrometer for enhanced MALDI-MS imaging of lipids. Chem Comm 53:53:7246–7249. [DOI] [PubMed] [Google Scholar]
- Feenstra AD, Dueñas ME, & Lee YJ. 2017. Five micron high resolution MALDI mass spectrometry imaging with simple, interchangeable, multi-resolution optical system. J Am Soc Mass Spectrom 28:3:434–442. [DOI] [PubMed] [Google Scholar]
- Feng B, Zhang J, Chang C, Li L, Li M, Xiong X, … Liu H. 2014. Ambient mass spectrometry imaging: Plasma assisted laser desorption ionization mass spectrometry imaging and its applications. Anal Chem 86:9:4164–4169. [DOI] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, & Whitehouse CM. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 246:4926:64–71. [DOI] [PubMed] [Google Scholar]
- Fernández FM, Cody RB, Green MD, Hampton CY, McGready R, Sengaloundeth S, … Newton PN. 2006. Characterization of solid counterfeit drug samples by desorption electrospray ionization and direct-analysis-in-real-time coupled to time-of-flight mass spectrometry. Chem Med Chem 1:7:702–705. [DOI] [PubMed] [Google Scholar]
- Fideler J, Johanningsmeier SD, Ekelöf M, & Muddiman DC. 2019. Discovery and quantification of bioactive peptides in fermented cucumber by direct analysis IR-MALDESI mass spectrometry and LC-QQQ-MS. Food Chem 271:715–723. [DOI] [PubMed] [Google Scholar]
- Fowble KL, Teramoto K, Cody RB, Edwards D, Guarrera D, & Musah RA. 2017. Development of “Laser ablation direct analysis in real time imaging” mass spectrometry: Application to spatial distribution mapping of metabolites along the biosynthetic cascade leading to synthesis of atropine and scopolamine in plant tissue. Anal Chem 89:6:3421–3429. [DOI] [PubMed] [Google Scholar]
- Gabelica V, & De Pauw E. 2005. Internal energy and fragmentation of ions produced in electrospray sources. Mass Spectrom Rev 24:4:566–587. [DOI] [PubMed] [Google Scholar]
- Garrard K, Ekelöf M, Khodjaniyazova S, Bagley M, & Muddiman D. 2020. A versatile platform for mass spectrometry imaging of arbitrary spatial patterns. J Am Soc Mass Spectrom 31:12:2547–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland WM, Prince HMA, Poliseno A, Kashuba ADM, & Rosen EP. 2019. Infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging of human hair to characterize longitudinal profiles of the antiretroviral maraviroc for adherence monitoring. Anal Chem 91:16:10816–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Yang S, Li J, Hu B, Chen H, Zhang L, & Fei Q. 2010. Geometry-independent neutral desorption device for the sensitive EESI-MS detection of explosives on various surfaces. Analyst 135:4:779–788. [DOI] [PubMed] [Google Scholar]
- Guo H, Liu AH, Ye M, Yang M, & Guo DA. 2007. Characterization of phenolic compounds in the fruits of forsythia suspensa by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21:5:715–729. [DOI] [PubMed] [Google Scholar]
- Haapala M, Pól J, Saarela V, Arvola V, Kotiaho T, Ketola RA, … Kostiainen R. 2007. Desorption atmospheric pressure photoionization. Anal Chem 79:20:7867–7872. [DOI] [PubMed] [Google Scholar]
- Haddad R, Sparrapan R, & Eberlin MN. 2006. Desorption sonic spray ionization for (high) voltage-free ambient mass spectrometry. Rapid Commun Mass Spectrom 20:19:2901–2905. [DOI] [PubMed] [Google Scholar]
- Haddad R, Sparrapan R, Kotiaho T, & Eberlin MN. 2008. Easy ambient sonic-spray ionization-membrane interface mass spectrometry for direct analysis of solution constituents. Anal Chem 80:3:898–903. [DOI] [PubMed] [Google Scholar]
- Harper JD, Charipar NA, Mulligan CC, Zhang X, Cooks RG, & Ouyang Z. 2008. Low-temperature plasma probe for ambient desorption ionization. Anal Chem 80:23:9097–9104. [DOI] [PubMed] [Google Scholar]
- Harris GA, Nyadong L, & Fernandez FM. 2008. Recent developments in ambient ionization techniques for analytical mass spectrometry. Analyst 133:10:1297–1301. [DOI] [PubMed] [Google Scholar]
- Harris GA, Graf S, Knochenmuss R, & Fernández FM. 2012. Coupling laser ablation/desorption electrospray ionization to atmospheric pressure drift tube ion mobility spectrometry for the screening of antimalarial drug quality. Analyst 137:13:3039–3044. [DOI] [PubMed] [Google Scholar]
- He J, Tang F, Luo Z, Chen Y, Xu J, Zhang R, … Abliz Z. 2011. Air flow assisted ionization for remote sampling of ambient mass spectrometry and its application. Rapid Commun Mass Spectrom 25:7:843–850. [DOI] [PubMed] [Google Scholar]
- Hecht ES, Oberg AL, & Muddiman DC. 2016. Optimizing mass spectrometry analyses: a tailored review on the utility of design of experiments. J Am Soc Mass Spectrom 27:5:767–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Jhang S, Cheng C, Cheng S, & Shiea J. 2010. Effects of matrix, electrospray solution, and laser light on the desorption and ionization mechanisms in electrospray-assisted laser desorption ionization mass spectrometry. Analyst 135:4:759–766. [DOI] [PubMed] [Google Scholar]
- Huang M, Hsu H, Lee J, Jeng J, & Shiea J. 2006. Direct protein detection from biological media through electrospray-assisted laser desorption ionization/mass spectrometry. J Proteome Res 5:5:1107–1116. [DOI] [PubMed] [Google Scholar]
- Ifa DR, & Eberlin LS. 2016. Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem 62:1:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifa DR, Wiseman JM, Song Q, & Cooks RG. 2007. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI). Int J Mass Spectrom 259:1:8–15. [Google Scholar]
- Jackson AU, Garcia-Reyes J, Harper JD, Wiley JS, Molina-Díaz A, Ouyang Z, & Graham Cooks R. 2010. Analysis of drugs of abuse in biofluids by low temperature plasma (LTP) ionization mass spectrometry. Analyst 135:5:927–933. [DOI] [PubMed] [Google Scholar]
- Jaiswal YS, Yerke AM, Bagley MC, Ekelöf M, Weber D, Haddad D, … Williams LL. 2020. 3D imaging and metabolomic profiling reveal higher neuroactive kavalactone contents in lateral roots and crown root peels of piper methysticum (kava). GigaScience 9:9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolla TW, & Karas M. 2011. Compelling evidence for lucky survivor and gas phase protonation: The unified MALDI analyte protonation mechanism. J Am Soc Mass Spectrom 22:6:976–988. [DOI] [PubMed] [Google Scholar]
- Jones RW, Cody RB, & McClelland JF. 2006. Differentiating writing inks using direct analysis in real time mass spectrometry. J Forensic Sci 51:4:915–918. [DOI] [PubMed] [Google Scholar]
- Judd R, Bagley MC, Li M, Zhu Y, Lei C, Yuzuak S, … Xie D. 2019. Artemisinin biosynthesis in non-glandular trichome cells of artemisia annua. Mol Plant 12:5:704–714. [DOI] [PubMed] [Google Scholar]
- Kalmar JG, Oh Y, Dean RA, & Muddiman DC. 2020. Investigating host-pathogen meta-metabolic interactions of magnaporthe oryzae infected barley using infrared matrix-assisted laser desorption electrospray ionization mass spectrometry. Anal Bioanal Chem 412:1:139–147. [DOI] [PubMed] [Google Scholar]
- Karas M, Bachmann D, Bahr U, & Hillenkamp F. 1987. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom 78:53–68. [Google Scholar]
- Karas M, Glückmann M, & Schäfer J. 2000. Ionization in matrix-assisted laser desorption/ionization: Singly charged molecular ions are the lucky survivors. J Mass Spectrom 35:1:1–12. [DOI] [PubMed] [Google Scholar]
- Keren L, Bosse M, Thompson S, Risom T, Vijayaragavan K, McCaffrey E, … Angelo M. 2019. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv 5:10:eaax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz V, & Van Berkel GJ. 2010a. Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform. J Mass Spectrom 45:3:252–260. [DOI] [PubMed] [Google Scholar]
- Kertesz V, & Van Berkel GJ. 2010b. Liquid microjunction surface sampling coupled with high-pressure liquid Chromatography−Electrospray ionization-mass spectrometry for analysis of drugs and metabolites in whole-body thin tissue sections. Anal Chem 82:14:5917–5921. [DOI] [PubMed] [Google Scholar]
- Khodjaniyazova S, Nazari M, Garrard KP, Matos MPV, Jackson GP, & Muddiman DC. 2018. Characterization of the spectral accuracy of an orbitrap mass analyzer using isotope ratio mass spectrometry. Anal Chem 90:3:1897–1906. [DOI] [PubMed] [Google Scholar]
- Khodjaniyazova S, Hanne NJ, Cole JH, & Muddiman DC. 2019. Mass spectrometry imaging (MSI) of fresh bones using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Anal Methods 11:46:5929–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Lee SY, Kim H, Park J, Lim D, & Moon DW. 2018. Biomolecular imaging of regeneration of zebrafish caudal fins using high spatial resolution ambient mass spectrometry. Anal Chem 90:21:12723–12730. [DOI] [PubMed] [Google Scholar]
- Kim JY, Seo ES, Kim H, Park J, Lim D, & Moon DW. 2017. Atmospheric pressure mass spectrometric imaging of live hippocampal tissue slices with subcellular spatial resolution. Nature Comm 8:1:2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert I, Chughtai K, Ellis SR, & Heeren RMA. 2014. Methods for full resolution data exploration and visualization for large 2D and 3D mass spectrometry imaging datasets. Int J Mass Spectrom 362:40–47. [Google Scholar]
- Kompauer M, Heiles S, & Spengler B. 2017. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat Methods 14:1:90–96. [DOI] [PubMed] [Google Scholar]
- Korte AR, Yandeau-Nelson MD, Nikolau BJ, & Lee YJ. 2015. Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal BioAnal Chem 407:2301–2309. [DOI] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, … Snyder M. 2002. Subcellular localization of the yeast proteome. Genes Dev 16:6:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiko VV, Baldwin MA, & Burlingame AL. 2000. Atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem 72:4:652–657. [DOI] [PubMed] [Google Scholar]
- Laskin J, Heath BS, Roach PJ, Cazares L, & Semmes OJ. 2012. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem 84:1:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WS, Wang R, Hu B, Berchtold C, Meier L, Chen H, & Zenobi R. 2010. On the mechanism of extractive electrospray ionization. Anal Chem 82:11:4494–4500. [DOI] [PubMed] [Google Scholar]
- Li Y, Shrestha B, & Vertes A. 2007. Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal Chem 79:2:523–532. [DOI] [PubMed] [Google Scholar]
- Liebl H 1967. Ion microprobe mass analyzer. J Appl Phys 38:13:5277–5283. [Google Scholar]
- Lin S, Huang M, Chang H, & Shiea J. 2007. Using electrospray-assisted laser desorption/ionization mass spectrometry to characterize organic compounds separated on thin-layer chromatography plates. Anal Chem 79:22:8789–8795. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Manicke NE, Lin J, Cooks RG, & Ouyang Z. 2010. Development, characterization, and application of paper spray ionization. Anal Chem 82:6:2463–2471. [DOI] [PubMed] [Google Scholar]
- Loziuk P, Meier F, Johnson C, Ghashghaei HT, Muddiman DC. 2016. TransOmic analysis of forebrain sections in Sp2 conditional knockout embryonic mice using IR-MALDESI imaging of lipids and LC-MS/MS label-free proteomics. Anal Bioanal Chem 408:13:3453–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, He J, Chen Y, He J, Gong T, Tang F, … Abliz Z. 2013. Air flow-assisted ionization imaging mass spectrometry method for easy whole-body molecular imaging under ambient conditions. Anal Chem 85:5:2977–2982. [DOI] [PubMed] [Google Scholar]
- Luosujärvi L, Arvola V, Haapala M, JP, Saarela V, Franssila S, … Kauppila TJ. 2008. Desorption and ionization mechanisms in desorption atmospheric pressure photoionization. Anal Chem 80:19:7460–7466. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang S, Lin Z, Liu Y, Xing Z, Yang C, & Zhang X. 2009. Real-time monitoring of chemical reactions by mass spectrometry utilizing a low-temperature plasma probe. Analyst 134:9:1863–1867. [DOI] [PubMed] [Google Scholar]
- Manicke NE, Wiseman JM, Ifa DR, & Cooks RG. 2008. Desorption electrospray ionization (DESI) mass spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: Ionization, adduct formation, and fragmentation. J Am Soc Mass Spectrom 19:4:531–543. [DOI] [PubMed] [Google Scholar]
- Martínez-Lozano P, Rus J, Fernández de la Mora Gonzalo, Hernández M, & Fernández de la Mora Juan. 2009. Secondary electrospray ionization (SESI) of ambient vapors for explosive detection at concentrations below parts per trillion. J Am Soc Mass Spectrom 20:2:287–294. [DOI] [PubMed] [Google Scholar]
- Massonnet P, & Heeren RMA. 2019. A concise tutorial review of TOF-SIMS based molecular and cellular imaging. J Anal At Spectrom 34:11:2217–2228. [Google Scholar]
- McEwen C, & Gutteridge S. 2007. Analysis of the inhibition of the ergosterol pathway in fungi using the atmospheric solids analysis probe (ASAP) method. J Am Soc Mass Spectrom 18:7:1274–1278. [DOI] [PubMed] [Google Scholar]
- McEwen CN, McKay RG, & Larsen BS. 2005. Analysis of solids, liquids, and biological tissues using solids probe introduction at atmospheric pressure on commercial LC/MS instruments. Anal Chem 77:23:7826–7831. [DOI] [PubMed] [Google Scholar]
- Meier F, Garrard KP, & Muddiman DC. 2014. Silver dopants for targeted and untargeted direct analysis of unsaturated lipids via infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Rapid Comm Mass Spectrom 28:22:2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MSiReader. Retrieved from https://msireader.wordpress.ncsu.edu/ Accessed: February 2, 2021.
- Muddiman DC, Gusev AI, Hercules DM. 1995. Application of secondary ion and matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for the quantitative analysis of biological molecules. Mass Spectrom Rev 14:6:383–429. [Google Scholar]
- Muddiman DC, Cheng X, Udseth HR, & Smith RD. 1996. Charge-state reduction with improved signal intensity of oligonucleotides in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 7:8:697–706. [DOI] [PubMed] [Google Scholar]
- Na N, Zhao M, Zhang S, Yang C, & Zhang X. 2007. Development of a dielectric barrier discharge ion source for ambient mass spectrometry. J Am Soc Mass Spectrom 18:10:1859–1862. [DOI] [PubMed] [Google Scholar]
- Na N, Zhang C, Zhao M, Zhang S, Yang C, Fang X, & Zhang X. 2007. Direct detection of explosives on solid surfaces by mass spectrometry with an ambient ion source based on dielectric barrier discharge. J. Mass Spectrom 42:8:1079–1085. [DOI] [PubMed] [Google Scholar]
- Nazari M, & Muddiman DC. 2015. Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Anal Bioanal Chem 407:8:2265–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari M, & Muddiman DC. 2016. Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 141:2:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari M, Bokhart MT, & Muddiman DC. 2016. Whole-body mass spectrometry imaging by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). JoVE 109:e53942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari M, Bokhart MT, Loziuk PL, & Muddiman DC. 2018. Quantitative mass spectrometry imaging of glutathione in healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 143:3:654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari M, Malico AA, Ekelöf M, Lund S, Williams GJ, & Muddiman DC. 2018. Direct analysis of terpenes from biological buffer systems using SESI and IR-MALDESI. Anal Bioanal Chem 410:3:953–962. [DOI] [PubMed] [Google Scholar]
- Nazari M, Ekelöf M, Khodjaniyazova S, Elsen NL, Williams JD, & Muddiman DC. 2017. Direct screening of enzyme activity using infrared matrix-assisted laser desorption electrospray ionization. Rapid Commun Mass Spectrom 31:22:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KA, Daniels GJ, Fournie JW, & Hemmer MJ. 2013. Optimization of whole-body zebrafish sectioning methods for mass spectrometry imaging. J Biomol Tech 24:3:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RW, Rainbow MJ, Lohr DE, & Williams P. 1989. Volatilization of high molecular weight DNA by pulsed laser ablation of frozen aqueous solutions. Science 246:4937:1585–1587. [DOI] [PubMed] [Google Scholar]
- Nemes P, & Vertes A. 2007. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem 79:21:8098–8106. [DOI] [PubMed] [Google Scholar]
- Nemes P, Woods AS, & Vertes A. 2010. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem 82:3:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, Barton AA, & Vertes A. 2009. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem 81:16:6668–6675. [DOI] [PubMed] [Google Scholar]
- Nemes P, Barton AA, Li Y, & Vertes A. 2008. Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization mass spectrometry. Anal Chem 80:12:4575–4582. [DOI] [PubMed] [Google Scholar]
- Niehaus M, Soltwisch J, Belov ME, & Dreisewerd K. 2019. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat Methods 16:9:925–931. [DOI] [PubMed] [Google Scholar]
- Nordhoff E, Kirpekar F, Karas M, Cramer R, Hahner S, Hillenkamp F, Kristiansen K, Roepstroff P, & Lezius A. 1994. Comparison of IR- and UV-matrix-assisted laser desorption/ionization mass spectrometry of oligodeoxynucleotides. Nucleic Acids Res 22:13:2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CL, Horman B, Patisaul H, & Muddiman DC. 2020. Analysis of neurotransmitters in rat placenta exposed to flame retardants using IR-MALDESI mass spectrometry imaging. Anal Bioanal Chem 412:15:3745–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CL & Muddiman DC. 2020. Direct analysis of native N-linked glycans by IR-MALDESI. J Am Soc Mass Spectrom 31:8:1759–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A, Trede D, & Alexandrov T. 2016. Where imaging mass spectrometry stands: Here are the numbers. Metabolomics 12:6:107. [Google Scholar]
- Pan CT, Shiea J, & Shen SC. 2007. Fabrication of an integrated piezo-electric micro-nebulizer for biochemical sample analysis. J Micromech Microengineering 17:3:659–669. [Google Scholar]
- Pasilis SP, Kertesz V, & Van Berkel GJ. 2008. Unexpected analyte oxidation during desorption electrospray ionization-mass spectrometry. Anal Chem 80:4:1208–1214. [DOI] [PubMed] [Google Scholar]
- Peng IX, Shiea J, Loo RRO, & Loo JA. 2007. Electrospray-assisted laser desorption/ionization and tandem mass spectrometry of peptides and proteins. Rapid Commun Mass Spectrom 21:16:2541–2546. [DOI] [PubMed] [Google Scholar]
- Peng IX, Shiea J, Loo RRO, & Loo JA. 2007. Electrospray-assisted laser desorption/ionization and tandem mass spectrometry of peptides and proteins. Rapid Commun Mass Spectrom 21:16:2541–2546. [DOI] [PubMed] [Google Scholar]
- Peng IX, Ogorzalek Loo RR, Margalith E, Little MW, & Loo JA. 2010. Electrospray-assisted laser desorption ionization mass spectrometry (ELDI-MS) with an infrared laser for characterizing peptides and proteins. Analyst 135:4:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CJ, Bagga AK, Prova SS, Yousefi Taemeh M, & Ifa DR. 2019. Review and perspectives on the applications of mass spectrometry imaging under ambient conditions. Rapid Commun Mass Spectrom 33:27–53. [DOI] [PubMed] [Google Scholar]
- Pierce CY, Barr JR, Cody RB, Massung RF, Woolfitt AR, Moura H, … Fernandez FM. 2007. Ambient generation of fatty acid methyl ester ions from bacterial whole cells by direct analysis in real time (DART) mass spectrometry. Chem Commun 8:807–809. [DOI] [PubMed] [Google Scholar]
- Pu F, Radosevich AJ, Sawicki J, Chang-Yen D, Talaty NN, Gopalakrishnan SM, Williams JD, Elsen NL. 2021. High-throughput label-free biochemical assays using infrared matrix assisted desorption electrospray ionization mass spectrometry. Anal Chem. in press doi: 10.1021/acs.analchem.1c00737. [DOI] [PubMed] [Google Scholar]
- Ratcliffe LV, Rutten FJM, Barrett DA, Whitmore T, Seymour D, Greenwood C, … McCoustra M. 2007. Surface analysis under ambient conditions using plasma-assisted desorption/ionization mass spectrometry. Anal Chem 79:16:6094–6101. [DOI] [PubMed] [Google Scholar]
- Rivera ES, Djambazova KV, Neumann EK, Caprioli RM, Spraggins JM. 2020. Integrating ion mobility and imaging mass spectrometry for comprehensive analysis of biological tissues: a brief review and perspective. J Mass Spectrom 55:ii-ii e4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, Laskin J, & Laskin A. 2010a. Molecular characterization of organic aerosols using nanospray-desorption/electrospray ionization-mass spectrometry. Anal Chem 82:19:7979–7986. [DOI] [PubMed] [Google Scholar]
- Roach PJ, Laskin J, & Laskin A. 2010b. Nanospray desorption electrospray ionization: An ambient method for liquid-extraction surface sampling in mass spectrometry. Analyst 135:9:2233–2236. [DOI] [PubMed] [Google Scholar]
- Robichaud G, Garrard KP, Barry JA, & Muddiman DC. 2013. MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 24:5:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G, Barry JA, & Muddiman DC. 2014. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom 25:3:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G, Barry JA, Garrard KP, & Muddiman DC. 2013. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom 24:1:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römpp A, Schäfer KC, Guenther S, Wang Z, Köstler M, Leisner A, Paschke C, Schramm T, & Spengler B. 2013. High-resolution atmospheric pressure infrared laser desorption/ionization mass spectrometry imaging of biological tissue. Anal Bioanal Chem 405:22:6959–6968. [DOI] [PubMed] [Google Scholar]
- Rosen EP, Bokhart MT, Nazari M, & Muddiman DC. 2015. Influence of C-trap ion accumulation time on the detectability of analytes in IR-MALDESI MSI. Anal Chem 87:20:10483–10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EP, Bokhart MT, Ghashghaei HT, & Muddiman DC. 2015. Influence of desorption conditions on analyte sensitivity and internal energy in discrete tissue or whole body imaging by IR-MALDESI. J Am Soc Mass Spectrom 26:6:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EP, Thompson CG, Bokhart MT, Prince HM, Sykes C, Muddiman DC, & Kashuba AD. 2016. Analysis of antiretrovirals in single hair strands for evaluation of drug adherence with infrared-matrix-assisted laser desorption electrospray ionization mass spectrometry imaging. Anal Chem 88:2:1336–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JS, Hawkridge AM, & Muddiman DC. 2008a. Development and characterization of an ionization technique for analysis of biological macromolecules: Liquid matrix-assisted laser desorption electrospray ionization. Anal Chem 80:17:6773–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JS, Hawkridge AM, & Muddiman DC. 2008b. Construction of a versatile high precision ambient ionization source for direct analysis and imaging. J Am Soc Mass Spectrom 19:10:1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JS, Hawkridge AM, & Muddiman DC. 2007. Direct characterization of intact polypeptides by matrix-assisted laser desorption electrospray ionization quadrupole fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 21:7:1150–1154. [DOI] [PubMed] [Google Scholar]
- Sampson JS, Hawkridge AM, & Muddiman DC. 2006. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom 17:12:1712–1716. [DOI] [PubMed] [Google Scholar]
- Sampson JS, Murray KK, & Muddiman DC. 2009. Intact and top-down characterization of biomolecules and direct analysis using infrared matrix-assisted laser desorption electrospray ionization coupled to FT-ICR mass spectrometry. J Am Soc Mass Spectrom 20:4:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DM, Creger Steve, Singla V, Kurulugama RT, Fjeldsted J, & Laskin J. 2020. Ion mobility-mass spectrometry imaging workflow. J Am Soc Mass Spectrom 31:12:2437–2442. [DOI] [PubMed] [Google Scholar]
- Sarsby J, Martin NJ, Lalor PF, Bunch J, & Cooper HJ. 2014. Top-down and bottom-up identification of proteins by liquid extraction surface analysis mass spectrometry of healthy and diseased human liver tissue. J Am Soc Mass Spectrom 25:11:1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer K, Dénes J, Albrecht K, Szaniszló T, Balog J, Skoumal R, … Takáts Z. 2009. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angewandte Chemie 48:44:8240–8242. [DOI] [PubMed] [Google Scholar]
- Schulz S, Becker M, Groseclose MR, Schadt S, & Hopf C. 2019. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr Opin Biotech 55:51–59. [DOI] [PubMed] [Google Scholar]
- Schwartz SA, Reyzer ML, & Caprioli RM. 2003. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: Practical aspects of sample preparation. J Mass Spectrom 38:7:699–708. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Fridjonsdottir E, Vallianatou T, Källback P, Katan L, … Andrén PE. 2019. Comprehensive mapping of neurotransmitter networks by MALDI–MS imaging. Nat Methods 16:10:1021–1028. [DOI] [PubMed] [Google Scholar]
- Shelley JT, Ray SJ, & Hieftje GM. 2008. Laser ablation coupled to a flowing atmospheric pressure afterglow for ambient mass spectral imaging. Anal Chem 80:21:8308–8313. [DOI] [PubMed] [Google Scholar]
- Shelley JT, Wiley JS, Chan GCY, Schilling GD, Ray SJ, & Hieftje GM. 2009. Characterization of direct-current atmospheric-pressure discharges useful for ambient desorption/ionization mass spectrometry. J Am Soc Mass Spectrom 20:5:837–844. [DOI] [PubMed] [Google Scholar]
- Shiea J, Huang M, HSu H, Lee C, Yuan C, Beech I, & Sunner J. 2005. Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids. Rapid Commun Mass Spectrom 19:24:3701–3704. [DOI] [PubMed] [Google Scholar]
- Shieh I, Lee C, & Shiea J. 2005. Eliminating the interferences from TRIS buffer and SDS in protein analysis by fused-droplet electrospray ionization mass spectrometry. J Proteome Res 4:2:606–612. [DOI] [PubMed] [Google Scholar]
- Shrestha B, & Vertes A. 2009. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem 81:20:8265–8271. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, & Wiemann S. 2000. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep 1:3:287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Müthing J, & Dreisewerd K. 2015. Mass spectrometry imaging with laser-induced postionization. Science 348:6231:211. [DOI] [PubMed] [Google Scholar]
- Song Y, & Cooks RG. 2006. Atmospheric pressure ion/molecule reactions for the selective detection of nitroaromatic explosives using acetonitrile and air as reagents. Rapid Commun Mass Spectrom 20:20:3130–3138. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld A, Tun TM, Zajíčková L, Kozlov KV, Wagner H-, Behnke JF, & Hippler R. 2001. Deposition process based on organosilicon precursors in dielectric barrier discharges at atmospheric Pressure—A comparison. Plasmas and Polymers 6:4:237–266. [Google Scholar]
- Sørensen MB, Aaslo P, Egsgaard H, & Lund T. 2008. Determination of D/L-amino acids by zero needle voltage electrospray ionisation. Rapid Commun Mass Spectrom 22:4:455–461. [DOI] [PubMed] [Google Scholar]
- Spengler B, Hubert M, & Kaufmann R. 1994. MALDI ion imaging with a new scanning UV-laser microprobe. Proceedings of the 42nd Annual Conference on Mass Spectrometry and Allied Topics 1041. [Google Scholar]
- Steiner WE, Clowers BH, Haigh PE, & Hill HH. 2003. Secondary ionization of chemical warfare agent simulants: Atmospheric pressure ion mobility time-of-flight mass spectrometry. Anal Chem 75:22:6068–6076. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Staab D, Staufenbiel M, Wiederhold K, & Signor L. 2002. Molecular imaging of amyloid β peptides in mouse brain sections using mass spectrometry. Anal Biochem 311:1:33–39. [DOI] [PubMed] [Google Scholar]
- Stout SD, & Teitelbaum SL. 1976. Histological analysis of undecalcified thin sections of archeological bone. Am J Phys Anthropol 44:2:263–269. [DOI] [PubMed] [Google Scholar]
- Stutts WL, Knuth MM, Ekelöf M, Mahapatra D, Kullman SW, & Muddiman DC. 2020. Methods for cryosectioning and mass spectrometry imaging of whole-body zebrafish. J Am Soc Mass Spectrom 31:4:768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swales JG, Tucker JW, Strittmatter N, Nilsson A, Cobice D, Clench MR, … Goodwin RJA. 2014. Mass spectrometry imaging of cassette-dosed drugs for higher throughput pharmacokinetic and biodistribution analysis. Anal Chem 86:16:8473–8480. [DOI] [PubMed] [Google Scholar]
- Takats Z, Nanita SC, Cooks RG, Schlosser G, & Vekey K. 2003a. Amino acid clusters formed by sonic spray ionization. Anal Chem 75:6:1514–1523. [DOI] [PubMed] [Google Scholar]
- Takáts Z, Wiseman JM, Gologan B, & Cooks RG. 2004. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306:5695:471. [DOI] [PubMed] [Google Scholar]
- Takáts Z, Wiseman JM, & Cooks RG. 2005. Ambient mass spectrometry using desorption electrospray ionization (DESI): Instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom 40:10:1261–1275. [DOI] [PubMed] [Google Scholar]
- Thompson CG, Rosen EP, Prince HMA, White N, Sykes C, de la Cruz G, … Kashuba ADM. 2019. Heterogeneous antiretroviral drug distribution and HIV/SHIV detection in the gut of three species. Sci Transl Med 11:499: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CG, Bokhart MT, Sykes C, Adamson L, Fedoriw Y, Luciw PA, … Rosen EP. 2015. Mass spectrometry imaging reveals heterogeneous efavirenz distribution within putative HIV reservoirs. Antimicrob Agents Chemother 59:5:2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu A, & Muddiman DC. 2019. Systematic evaluation of repeatability of IR-MALDESI-MS and normalization strategies for correcting the analytical variation and improving image quality. Anal Bioanal Chem 411:22:5729–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu A, & Muddiman DC. 2019. Internal energy deposition in infrared matrix-assisted laser desorption electrospray ionization with and without the use of ice as a matrix. J Am Soc Mass Spectrom 30:11:2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V, & King RC. 2009. High-throughput mode liquid microjunction surface sampling probe. Anal Chem 81:16:7096–7101. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V, Koeplinger KA, Vavrek M, & Kong AT. 2008. Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J Mass Spectrom 43:4:500–508. [DOI] [PubMed] [Google Scholar]
- Vandercruyssen K, D’Hondt M, Vergote V, Jansen H, Burvenich C, & De Spiegeleer B. 2014. LC–UV/MS quality analytics of paediatric artemether formulations. J Pharm Anal 4:1:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter A, Sojka PE, & Cooks RG. 2006. Droplet dynamics and ionization mechanisms in desorption electrospray ionization mass spectrometry. Anal Chem 78:24:8549–8555. [DOI] [PubMed] [Google Scholar]
- Vismeh R, Waldon DJ, Teffera Y, & Zhao Z. 2012. Localization and quantification of drugs in animal tissues by use of desorption electrospray ionization mass spectrometry imaging. Anal Chem 84:12:5439–5445. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu J, Cooks R, & Ouyang Z. 2010. Paper spray for direct analysis of complex mixtures using mass spectrometry. Angewandte Chemie 49:5:877–880. [DOI] [PubMed] [Google Scholar]
- Wang Z, Bovik AC, Sheikh HR, & Simoncelli EP. 2004. Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Processing 13:4:600–612. [DOI] [PubMed] [Google Scholar]
- Williams JP, Patel VJ, Holland R, & Scrivens JH. 2006. The use of recently described ionisation techniques for the rapid analysis of some common drugs and samples of biological origin. Rapid Commun Mass Spectrom 20:9:1447–1456. [DOI] [PubMed] [Google Scholar]
- Williams JP, & Scrivens JH. 2008. Coupling desorption electrospray ionisation and neutral desorption/extractive electrospray ionisation with a travelling-wave based ion mobility mass spectrometer for the analysis of drugs. Rapid Commun Mass Spectrom 22:2:187–196. [DOI] [PubMed] [Google Scholar]
- Wu C, Siems WF, & Hill HH. 2000. Secondary electrospray ionization ion mobility spectrometry/mass spectrometry of illicit drugs. Anal Chem 72:2:396–403. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fenn JB. 1984. Electrospray ion source. another variation on the free-jet theme. J Phys Chem 88:20:4451–4459. [Google Scholar]
- Yin R, Burnum-Johnson K, Sun X, Dey SK, & Laskin J. 2019. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat Protoc 14:12:3445–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Gamez G, Chen H, Chingin K, & Zenobi R. 2009. Rapid detection of melamine in untreated milk and wheat gluten by ultrasound-assisted extractive electrospray ionization mass spectrometry (EESI-MS). Chem Commun 5:559–561. [DOI] [PubMed] [Google Scholar]
- Zhu J, Bean HD, Kuo YM, & Hill JE. 2010. Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization mass spectrometry. J Clin Microbiol 48:12:4426–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]


