Abstract
Cancer has a high mortality rate across the globe, and tissue biopsy remains the gold standard for tumor diagnosis due to its high level of laboratory standardization, good consistency of results, relatively stable samples, and high accuracy of results. However, there are still many limitations and drawbacks in the application of tissue biopsy in tumor. The emergence of liquid biopsy provides new ideas for early diagnosis and prognosis of tumor. Compared with tissue biopsy, liquid biopsy has many advantages in the diagnosis and treatment of various types of cancer, including non-invasive, quickly and so on. Currently, the application of liquid biopsy in tumor detection has received widely attention. It is now undergoing rapid progress, and it holds significant potential for future applications. Around now, liquid biopsies encompass several components such as circulating tumor cells, circulating tumor DNA, exosomes, microRNA, circulating RNA, tumor platelets, and tumor endothelial cells. In addition, advances in the identification of liquid biopsy indicators have significantly enhanced the possibility of utilizing liquid biopsies in clinical settings. In this review, we will discuss the application, advantages and challenges of liquid biopsy in some common tumors from the perspective of diverse systems of tumors, and look forward to its future development prospects in the field of cancer diagnosis and treatment.
Subject terms: Cancer, Cell biology
Introduction
Cancer is the second major cause of death in the world and is a major worldwide public health problem. Early detection and appropriate therapy are crucial for cancer patients to enhance their prognosis and enhance their chances of survival.1 Currently, the golden standard for tumor diagnosis is still tissue biopsy. Although tissue biopsy can definitively diagnose tumors and their subtypes, tissue biopsy is difficult to collect and, as an invasive test, it is prone to cause damage to patients and is not convenient for continuous monitoring of the disease progression.2 As tumors are sometimes hard to detect early, it is difficult to use tissue biopsies to accurately detect tumors at an early stage in the diseases.
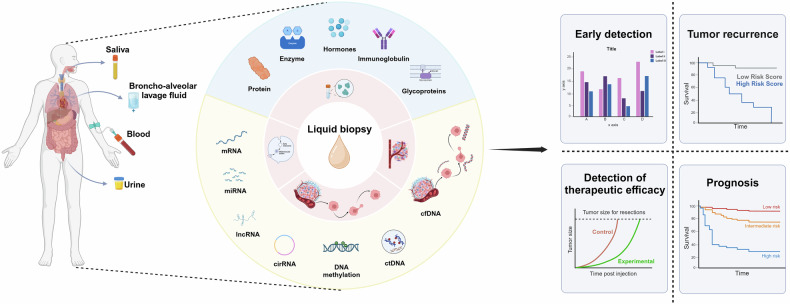
Liquid biopsy is a mini-invasive sample collection method that focuses on blood or body secretions for the detection of molecular alterations, tumor cells, and metabolites.3,4 Compared to tissue biopsies, liquid biopsies provide a role in early screening. Common specimens for liquid biopsy are blood and urine.5 Therefore, liquid biopsies are easier to perform than tissue biopsies and are virtually non-invasive to the patient,5,6 which makes liquid biopsies have the potential for continuous monitoring of tumor progression.7 Several molecular markers can be detected by liquid biopsy, such as circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), tumor-derived extracellular vesicles (EVs), tumor-educated platelets (TEPs), and circulating free RNA (cfRNA).7,8 Currently, more studies focus on the detection of CTCs, ctDNA and exosomes. In this paper, we will introduce various liquid biopsy molecular markers and summarize the current applications of liquid biopsy in various tumor systems from different systems.
The research history of liquid biopsy
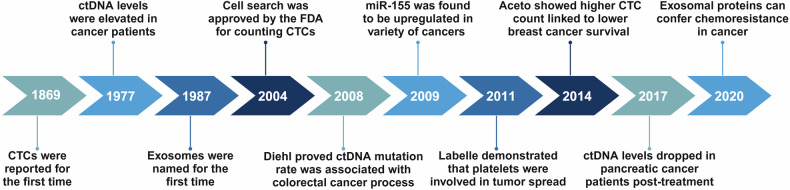
The development of liquid biopsy has gone through four main phases: the period of scientific exploration (before the 1990s), the period of scientific development (1990s), the period of industrial growth (2000–2010), and the period of industrial outbreak (2010-present) (Fig. 1).
Fig. 1.
History of liquid biopsy. Timeline of the research history and milestone events of study on liquid biopsy. CTCs Circulating tumor cells, ctDNA Circulating tumor DNA, FDA Food and Drug Administration. Created with BioRender.com
During the period of scientific exploration, several scholars have discovered the existence of CTCs, cfDNA and extracellular vehicles (EVs). In 1869, Australian physician Thomas Ashworth found cells similar to tumor cells in the blood of a recently deceased tumor patient.9 In 1948, Mandel and Metais made the groundbreaking discovery of the existence of unbound nucleic acid molecules in plasma.10 In 1967, Wolf obtained the first electron micrographs of EVs.11 In 1983, Stahl and Johnstone’s laboratory suggested that exosomes are discharged from EVs that had merged with the cell membrane through multivesicular structures.12 In addition, a study conducted by Leon et al. in 1977 revealed that levels of plasma free DNA were much elevated in individuals with tumors compared to those in the healthy population. This led to the hypothesis that free DNA is linked to the presence of tumors.13 In the period of scientific progress, CTC was initially isolated from blood in 1998 and was proven to correlate with pathologic staging, and it has only since been employed in the clinic.14 Additionally, in 1994, PCR was used to identify the first KRAS mutation in pancreatic cancer patients’ blood cfDNA, and the results were consistent with those found in tumor tissue.15 In 1996, Raposo provided evidence that EVs possess biological activity. It has been discovered that immune cells’ EVs can present antigens.16 Liquid biopsy indicators were discovered to be useful in the diagnosis of a variety of cancers during this time of industrial expansion. In patients with metastatic breast cancer, the quantity of CTCs prior to therapy was found to be an independent predictor of both overall survival and progression-free survival in 2005.17 Diehl F. et al. followed up on the ctDNA of 18 patients with bowel cancer in 2008 and used the BEAMing technique to identify hotspot mutations in genes like TP53, APC, KRAS, and PIK3CA. They discovered that the rate of ctDNA mutations changed over the course of treatment and that the trend of the change was positively correlated with both the tumor load and the CEA concentration.18 Several liquid biopsy markers were included into oncology guidelines and given the go-ahead for clinical use during the industrial boom. The use of ctDNA to identify EGFR mutations for concurrent Erizar diagnosis was authorized by the European Medicines Agency (EMA) in 2014, hence initiating the official clinical usage of ctDNA. According to the 2015 Chinese Expert Consensus on Blood EGFR Gene Mutation Testing in Non-Small Cell Lung Cancer (NSCLC), which was published in the Chinese Medical Journal, ctDNA from the blood (plasma) specimen can be used for evaluation if the tumor specimen cannot be assessed for EGFR gene status.19 And the use of CTC testing for prognostic assessment in breast cancer was addressed by AJCC recommendations in 2018.20 In 2019, CTC was included into the 2019 CSCO Breast Cancer Treatment Guidelines.21 More recently, in 2023, CTC entered the Chinese Technical Guidelines for Integrated Cancer Therapy (CACA).
Molecular markers of liquid biopsy
In this section we focus on several liquid biopsy biomarkers currently in use (Fig. 2). And summarizes the comparison of different liquid biopsy markers (Tables 1–5).
Table 2.
ctDNA detection techniques
| Detection Methods | Detection Principle | Role | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|
| ddPCR | DNA amplification, sample microtitration and reading of the starting concentration of target molecules by fluorescence signaling | Detection of single nucleotide variants, quantification of nucleotides | High sensitivity and specificity, relatively low cost for specific DNA detection, short time to achieve absolute quantification of target molecules, suitable for long-term monitoring of patients with known mutations | It cannot process a large amount of sequence information at the same time and can only amplify known sequences. | 98 |
| NGS | Sequence information is read after DNA amplification using signals emitted at base insertion into the DNA strand with the help of chemical markers | Whole exome sequencing (WES) as well as whole genome sequencing (WGS), detection of nucleotide variants | Large amount of sequence information can be processed at the same time, detection time is not long, suitable for patient screening of unknown mutations, lower cost compared to ddPCR for large amount of DNA detection | The sensitivity and specificity are not as good as ddPCR. | 99,100 |
Table 3.
Exosome detection techniques
| Technology | Mechanisms | Advantage | Disadvantage | Reference |
|---|---|---|---|---|
| differential centrifugation | Separation of substances of different sizes and densities by centrifugal force | The extraction method is simple, widely applicable, does not introduce additional markers, can handle a certain dose of sample, low cost, and does not contaminate exosomes | Cumbersome, time-consuming and labor-intensive, and the structure of the exosome may be destroyed. | 101 |
| filtration | Utilizes ultrafiltration membranes to selectively allow molecules or particles smaller than the membrane pore size to pass through. | High exosome recovery, simple handling, no introduction of additional markers | Poor ability to separate exosomes, time consuming, contamination of exosomes | 101 |
| polymer precipitation | and exosomes bind to each other, forming a complex, and then exosomes are separated by centrifugation | High recovery rate, easy to operate, can handle a large number of samples | Contamination of exosomes, low recovery purity, easy to damage the integrity of the exosome membrane | 101 |
| immunomagnetic bead method | Magnetic separation of exosomes using magnetism after specific binding of antibody-coated magnetic beads to exosome markers | High specificity, high purity of isolated exosomes, average difficulty in getting started | Takes time, costly, can’t handle large numbers of samples, tends to change exosome function | 101 |
| Chromatography (volumetric exclusion chromatography) | Separation of exosomes by continuous movement in different phases, taking advantage of the differences in the partitioning, adsorption and desorption properties of the components of the mixture between the stationary and mobile phases | High recovery rate, high purity, short time-consumption, low cost, simple operation, not easy to change the function of exosomes, no need for a large number of samples to isolate exosomes, can handle a large number of samples | Exosomes are diluted during the isolation process and may need to be subsequently concentrated, with potential contaminants that may contaminate the sample | 101 |
| microfluidic | Isolation of exosomes by methods such as exosome size or surface-specific markers | Good ability to isolate exosomes, high recovery of exosomes, no need for a large number of samples to isolate exosomes, fast isolation speeds | Costly to operate and maintain, requires specialized equipment, training prior to use, not able to process large quantities of samples | 57,101 |
Table 4.
RNA detection methods
| Technology | Mechanisms | Advantage | Disadvantage | Reference |
|---|---|---|---|---|
| RNA fluorescence in situ hybridization (RNA-FISH) | Hybridization signals were observed using fluorescence microscopy after binding to the target RNA with a fluorescent probe complementary to the target RNA sequence | High sensitivity and specificity, multi-color detection, relatively simple and time-consuming operation, tissue morphology can be maintained for detection | High sample requirements, need to ensure RNA integrity, need specialized equipment and probes, high cost, limited accuracy of quantification | 102 |
| RT-PCR (reverse transcription PCR) | PCR amplification after reverse transcription of RNA to cDNA | It is highly sensitive and specific, suitable for the detection of a wide range of RNAs, less time consuming and more accurate. | Complexity of operation, susceptibility to contamination by foreign products, expensive equipment and reagents | 103 |
| Northern Blotting | Complexity of operation, susceptibility to contamination by foreign products, expensive equipment and reagents | High sensitivity and specificity, quantitative detection of RNA compared to RNA-FISH | Time-consuming, more complex operations, high sample requirements, need to ensure RNA integrity, need specialized equipment and probes, higher costs | 104 |
| in situ hybridization | The principle is similar to RNA-FISH, but labeled using markers such as radioisotopes, biotin, digoxin, etc., and finally visualized by radioactive autoradiography, immunohistochemistry, etc. | Both DNA and RNA can be detected at a moderate cost | Not as accurate as RNA-FISH, multiple hybridizations are not as simple as RNA-FISH, can only capture RNA from cells at a certain time point | 105 |
| RNA microarray | Hybridization of RNA by immobilizing a large number of probes on a microarray | High throughput, accurate quantification and good reproducibility. | Can only detect highly expressed RNAs and cannot cover the full range of RNAs, especially lncRNAs. cost is high and affected by experimental complexity. | 106 |
| RNA sequencing | Direct sequencing of RNA molecules using high-throughput sequencing technology | Detects all RNAs, capable of deep sequencing with high sensitivity and specificity | Costly, requires advance removal of rRNAs | 106 |
Fig. 2.
Flowchart of applying liquid biopsy in cancers. Applications of liquid biopsies and types of biomarkers for liquid biopsies. Created with BioRender.com
Table 1.
Separation and detection of CTCs
| Cellular Assay Methods | Cell Characterization/Detection Principles | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Automatic scanning of fluorescence microscopes | Telomerase-specific replication-selective adenovirus expressing GFP (green fluorescent protein) | The assay is simple, detects a wide range of tumor cells, and does not require CTC enrichment. | Lack of large sample tests, relatively time-consuming and complicated procedures. | 93 |
| CTC-iChip | NA | No need to enrich CTC | Higher cost and only 39% CTC detection rate | 94 |
| Subtractive enrichment (SE) and immunostaining-FISH (iFISH) | Polyploid with chromosome 8 | Effective removal of leukocytes and erythrocytes, less loss of CTCs, significantly less time-consuming than traditional fish methods, and simultaneous detection of protein expression of multiple tumor markers specifically on CTCs | The development of relevant techniques is still in its infancy, and acute infectious lesions and benign space-occupying lesions may lead to false positives | 94,95 |
| Parsortix PC1 system | Physical method detection, microfluidic devices | Several multi-center clinical studies have demonstrated its ability to capture and collect CTCs | Processing is quite slow. | 96 |
| Cellsearch | EpCAM protein, immunomagnetic enrichment, fluorescent labeling | Considered the gold standard for CTC detection | Detection of EpCAM+ cells only, not applicable for some CTCs lacking EpCAM expression, e.g., GBM | 97 |
Table 5.
Comparison of different liquid biopsy markers
| Form | ctDNA | CTCs | Exosome |
|---|---|---|---|
| source | Blood, urine, saliva, synovial fluid, cerebrospinal fluid, etc. | Blood, cerebrospinal fluid, urine, etc. | Blood, urine, cerebrospinal fluid, ascites, pleural fluid, etc. |
| scale | Nanoscale (DNA fragments) | cellular level | Nanoscale (40–150 nm) |
| information load | Can carry information on multiple genetic variants | Complete genetic information, including genome, transcriptome, epigenetic variation | Carrying proteins, RNA and many other biomolecules |
| clinical significance | Early screening, companion diagnosis, prognostic assessment, MRD testing | Prognostic assessment, drug sensitivity prediction, drug resistance mechanism studies | Early diagnosis, prognostic assessment, drug response monitoring |
| stability | Relatively low (short half-life) | high | High (phospholipid bilayer protection) |
| rarity | High (especially in early-stage tumors) | high | moderate (interference from other vesicles in body fluids) |
| heterogeneity | low | High (large variation between CTCs) | moderate |
| Difficulty of isolation and purification | moderate | High (not yet standardized) | High (technically complex) |
| background noise | Moderate (normal cfDNA interference) | low | Moderate (interference from other vesicles in body fluids) |
| Difficulty of standardization | moderate | high | high |
| technical difficulty | Moderate (relies on high-sensitivity detection technology) | High (complex enrichment, identification techniques) | Medium (dependent on specific detection techniques) |
| operating difficulty | low | High (multi-step operation) | moderate |
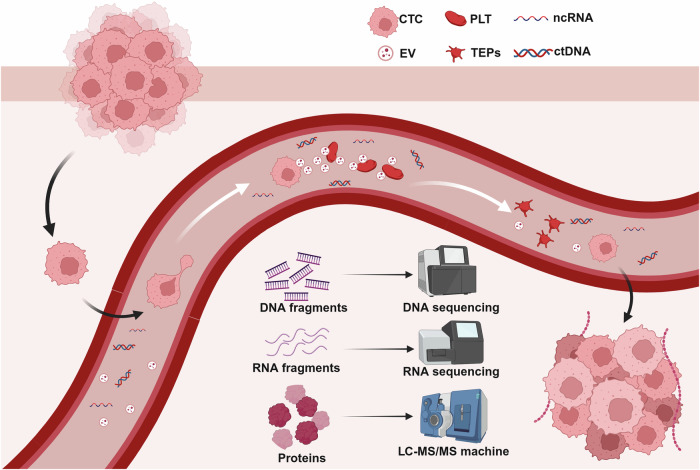
Circulating tumor cells (CTCs)
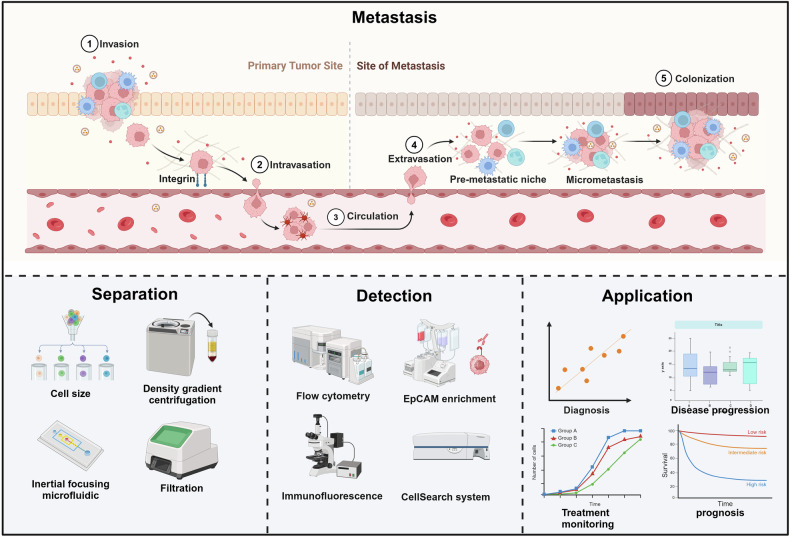
In 1869, Ashworth et al. first reported CTCs in the circulation of patients, which laid an important foundation for the study of CTCs. CTCs are cells released from primary and metastatic tumors that are shed into the blood or lymphatic vessels of cancer patients and circulate in the peripheral blood22 (Fig. 3). Although the proportion of CTCs in the blood is low, almost 1 CTCs is found per 1 million leukocytes, and most CTCs die in the peripheral blood in 1–2.5 h.23,24 However, in recent years, a large number of studies have demonstrated that the level of CTCs is associated with cancer development, especially playing an important role in the metastatic process of cancer,25 and these confirm that CTCs are an important biomarker. Therefore, CTCs have the potential to become an effective tool for cancer diagnosis, providing information for clinical decision-making and clinical research.26,27 A key challenge currently faced is how to isolate and collect CTCs more accurately, and the rapid development of technology has further facilitated the clinical application of CTCs.28 With technological advances and innovations, CTCs counts are associated with tumor status and higher accuracy. Studies have shown that higher levels of CTCs counts are associated with reduced progression-free survival and overall survival.29,30 For example, in 2014, Ramirez et al. demonstrated that in blood samples from breast cancer patients, an increased count of CTCs was found to be associated with a significant reduction in progression-free survival. As a result, the detection of CTCs has gained increasing attention as one of the important biomarkers for liquid biopsy. Due to the extremely low number of CTCs, it is high sensitivity advanced techniques to efficiently capture and detect CTCs that are necessary. Currently, methods used for the detection or isolation of CTCs are constantly being improved and have greatly increased in complexity and sensitivity.31 There are traditional methods such as density gradient centrifugation, inertial focusing, and filtration based on biophysical properties such as size, deformability, etc.32. There are also methods for the detection of cells by the expression of specific markers, epithelial cell adhesion molecule (EpCAM), vimentin, and N-cadherin, such as EpCAM enrichment, immunomagnetic separation, and microfluidic devices.33 Among them, the CellSearch® method is currently the only method authorized by the FDA to monitor the number of CTCs in blood samples.34 Even though these methods have a variety of shortcomings (Table 1), they have played a significant role in promoting research on the detection and clinical value of CTCs. CTCs, as an almost noninvasive test, will play an increasingly important value in the diagnosis, detection, and prognosis of tumors in the future.
Fig. 3.
Liquid biopsy markers—CTCs. The metastasis, separation detection and application of CTCs. Created with BioRender.com
Circulating tumor DNA (ctDNA)
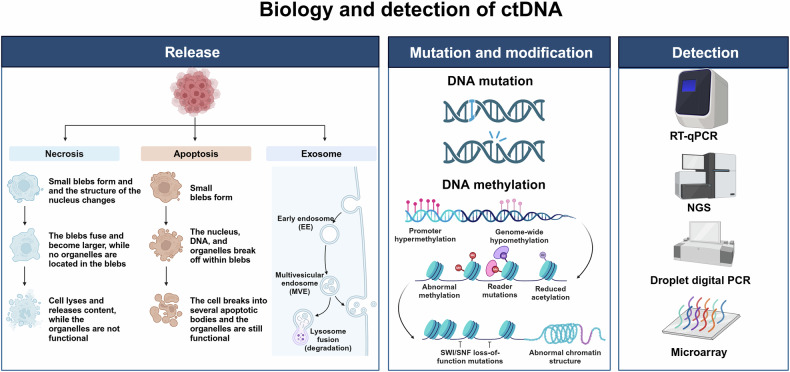
Circulating tumor DNA (ctDNA) can be extracted from the bloodstream and originates from the tumor. It is a type of circulating extracellular nucleic acids (cfDNA).35 CfDNA is primarily derived from normal leukocytes and stromal cells. However, in 1977, Leon et al. found that plasma-free DNA levels were significantly higher in patients with advanced tumors than in healthy individuals suggesting that cfDNA may also be derived from tumor cells.13 CtDNA only accounts for a small fraction of cfDNA, approximately 0.1–1.0% of its total36 (Fig. 4).
Fig. 4.
Liquid biopsy markers—ctDNA. CtDNA is usually actively secreted by tumor cells or released into the circulatory system during the apoptosis or necrosis of tumor cells. Mutations and methylation of ctDNA are often used as detection indicators. Created with BioRender.com
Similar to CTCs, ctDNA has traditionally been obtained from blood, but ctDNA can also be isolated by obtaining ascites, pleural fluid, urine, and cerebrospinal fluid (CSF). CfDNA is primarily derived from normal leukocytes and stromal cells, and ctDNA can dynamically respond to the state of the tumor at a given point in time. Compared with cfDNA, it has been shown that ctDNA base fragments in cancer patients are shorter than cfDNA, which is about 20–50 base pairs, making it less affected by intra-tumor heterogeneity.37 On the other hand, ctDNA has a shorter half-life, which is a prerequisite for its ability to be used as a real-time tumor biomarker, and it is these two characteristics of ctDNA that give it a distinct advantage when compared with traditional biopsy markers. The prognostic significance of ctDNA in cancer progression and its response to treatment has been described in recent years.38,39 It has been found that ctDNA levels are elevated in the serum of patients with pancreatic cancer (PC) and appear to decrease after treatment.13 In addition, the current clinical application often detects the mutation of target genes within ctDNA, for example, Diehl F and his team analyzed the serum ctDNA of 18 colorectal cancer patients and found hotspot mutated genes, such as APC, KRAS, TP53, and PIK3CA. And the mutation rate of ctDNA is related to its therapeutic process.18 Gene mutation can often trigger the imbalance of oncogenes and oncogenes, and then lead to cancer, so the mutation detection of ctDNA is of great significance for cancer detection. Abnormal DNA methylation also plays a key role in cancer development. In many tumors, an imbalance in DNA methylation usually precedes tumor formation and contributes to the early diagnosis of tumors.40 The detection of ctDNA has become increasingly sophisticated with technological advances, such as real-time quantitative polymerase chain reaction, digital droplet PCR (ddPCR), sanger sequencing, and next-generation sequencing (NGS).41–43 In the future, ctDNA assays will be widely used in new therapies to appropriately monitor the dynamics of tumor load and the cancer progression or prognosis.
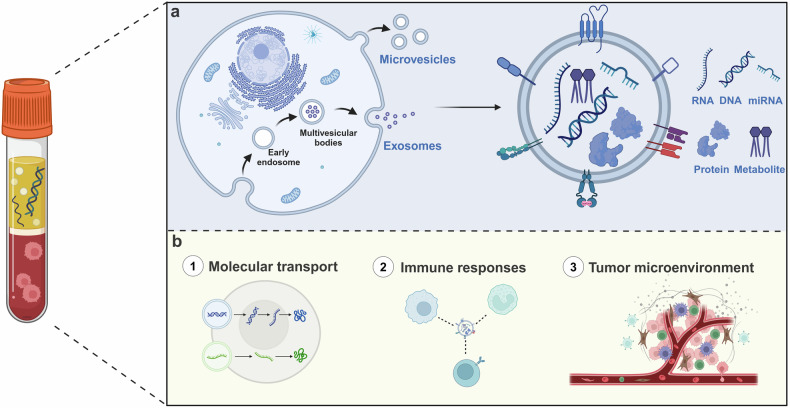
Exosomes
In 1987, Johnstone first named the vesicles released by sheep reticulocytes as exosomes.44 Exosomes are a subtype of extracellular vesicles that originate from endosomes produced by trap buds in the membranes of multivesicular bodies and are released outside the cell after the fusion of multivesicular endosomes with the cell membrane45 (Fig. 5). The other two major subtypes of extracellular vesicles are microvesicles and apoptotic vesicles whose categorization is based primarily on size and cellular origin. The three main subtypes of exosomes have received much attention in recent years.46 Exosomes can be detected in blood, saliva, urine, and other fluids, engaging in a variety of biological processes such as molecular transport, intercellular communication, and immune responses. In addition, it has been found that exosomes are key components of the tumor microenvironment and play an important role in cancer progression.47 While exosomes have unique advantages in the field of liquid biopsy, on the one hand, they are well stabilized, and on the other hand, they are more representative in describing the information of tumor cells.48 In recent years, exosomal products, such as nucleic acids, proteins, lipids, and metabolites have gradually become a focus of research in the field of cancer, for example, exosomal non-coding RNAs (ncRNAs) have been shown to provide important reference value in the diagnosis and treatment of cancer patients. The upregulation of exosomes miR-1246, miR-4644, miR-3976, and miR-4306 can be used as highly sensitive biomarkers in prostate cancer patients.49 In addition, exosomal lncRNA H19 was found to be upregulated in serum expression in bladder cancer patients, suggesting that exosomal lncRNAs have a potential role as important diagnostic markers.50 Due to their unusually large variety and number, exosomal proteins have also received extensive attention in recent years.51 Exosomal proteins have a regulatory role in the formation of the cancer microenvironment, tumor progression, and metastasis.52,53 In addition, exosomal proteins can also mediate chemoresistance in cancer treatment, and a recent study showed that plasma gelatin (pGSN), an isoform of GSN protein secreted by chemoresistant ovarian cancer cells, can be delivered to exosomes and activate α5β1 integrin. This leads to an increase in hypoxia-inducible factor 1 subunit α, which in turn promotes chemoresistance and survival of ovarian cancer cells.54 In view of the fact that exosomes are one of the markers of a liquid biopsy and their important clinical applications, it is particularly important to isolate and detect them efficiently and accurately. In recent years, such approaches as Reverse Transcription-Polymerase Chain Reaction (RT-PCR), genome sequencing, and proteomics are often available for the detection of exosomal content.55,56 Techniques such as differential ultracentrifugation, size-based separation, immunomagnetic separation, and microfluidics are commonly used for exosome isolation.57 In the future, with the development of technology and multidisciplinary fusion, exosome, one of the markers of liquid biopsy, will be more closely integrated with clinical applications, especially cancer detection.
Fig. 5.
Liquid biopsy markers – exosome. a The formation process of exosomes and the main detection contents such as RNA, DNA, miRNA, proteins, and metabolite. b The role of exosome in tumor progression. Created with BioRender.com
Tumor educated-platelets
When it comes to platelets, what often first comes to mind is their hemostatic and thrombotic role, however, the fact is that platelets are gradually being recognized as mediators of malignant disease.58 As the second most abundant cell in the peripheral blood, they play a role in hematological processes, such as wound healing, atherosclerosis, vascular growth regulation, and angiogenesis.59 In the 1800s Reiss et al. first reported that high platelet counts were associated with malignancy and that host-tumor interactions activate the coagulation cascade in many types of cancers, and since then, more relevant evidence has suggested a link between platelet counts and cancer.60,61 It has been found that platelet deposition is positively correlated with mortality in patients with cancer, and it is considered to be the second most common cause of cancer deaths.62 In addition, there is a unique type of platelet that is often used as a biomarker for liquid biopsies and has received much attention in recent years. It is a type of platelet that is isolated from tumor patients but exhibits a different RNA and protein profile, named TEPs63 (Fig. 6). Studies have shown the involvement of TEPs in the progression and spread of a variety of solid tumors. Specifically spliced TEP RNA markers can provide specific information on tumor presence, location, and molecular features, but the exact mechanisms require further research.64 While there are no present clinical applications for TEPs, numerous studies have explored the potential clinical uses of TEPs, providing valuable insights. Tumor platelets exert a bidirectional influence, causing platelets to consistently absorb proteins, nucleic acids, vesicles, and granules from tumors. This process results in alterations to the RNA and protein expression profiles of the platelets.65 Platelets possess several advantages as a component of liquid biopsy. They exhibit stability and ease of collection, as they may be readily obtained through low-speed centrifugation. Furthermore, the genetic material contained within platelets is relatively durable.66 Due to the limited lifespan of platelets, the composition of TEP can accurately indicate the current condition of the tumor, allowing for real-time surveillance of the tumor. Further investigation is required to fully understand the precise mechanism, but the spliced TEP RNA markers have the potential to offer precise details regarding the presence, location, and molecular features of tumors.64 Present research on platelets in persons with tumors has primarily concentrated on mRNA and lncRNA. Numerous studies have demonstrated the capability of RNA sequencing analysis to distinguish between tumor patients and those who are in good health.67 In 2022, Ye et al. discovered four specific long-stranded non-coding RNA (lncRNA) markers associated with colorectal cancer (CRC) that are found in platelets. These markers include LNCAROD, SNHG20, LINC00534, and TSPOAP-AS1. The expression levels of these lncRNAs were markedly increased in both platelets and serum samples from individuals diagnosed with colorectal cancer. This finding strongly indicates that these lncRNAs hold promising diagnostic value.68 A gene expression database specifically designed for platelet-based disease research was established in 2022. We anticipate that this database will significantly enhance the investigation of platelet liquid biopsies.69 Currently, the understanding of the mechanisms involving platelet RNA is incomplete, and the use of TEPs for tumor treatment is still in the conceptual phase, necessitating further extensive research.
Fig. 6.
Liquid biopsy markers—TEPs. The formation process and the detection of TEPs. CTC circulating tumor cell, EV extracellular vehicle, PLT platelet, TEPs tumor educated-platelets. Created with BioRender.com
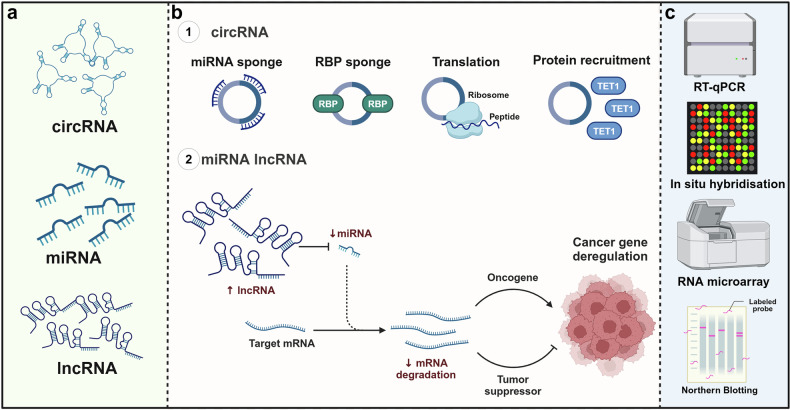
miRNA and lncRNA
Non-coding RNAs are diverse and play different functions and roles from coding RNAs in the cell. Initially, there was little understanding of non-coding RNAs, which had been considered to have a limited impact on tumorigenesis and development and were called spam-free RNAs. In recent years, numerous studies have demonstrated that non-coding RNAs play important roles in the development of different types of cancers.70 With further research, several non-coding RNAs have been used as biomarkers for liquid biopsies in cancer71 (Fig. 7).
Fig. 7.
Liquid biopsy markers—RNA. a Types of ncRNA. b The role of ncRNA. c The detection methods for ncRNA. Created with BioRender.com
miRNAs, a small (18–23 nt) single-stranded RNA molecule involved in post-transcriptional gene regulation, belong to the subclass of non-coding RNAs. It reduces the stability of mRNAs and inhibits gene expression by binding to 3′ untranslated region recognition sites.72 miRNA is the most widely studied factor in cancer research and the most studied ncRNA in liquid biopsies. miR-21 and miR-155 have been found to be up-regulated in a variety of cancers and may be able to become a promising cancer liquid biopsy marker.73 In recent years, more and more methods have been used for miRNA detection, such as qPCR, hybridization chain reaction, rolling circle amplification, and strand displacement amplification. These methods have greatly aided the study of miRNA, particularly in understanding its two primary features: abundance and tissue stability. These properties could potentially be advantageous in the future for developing non-invasive biomarkers for patients with tumors.
Currently, the second most abundant source of ncRNAs evaluated in cancer liquid biopsies is lncRNAs. lncRNAs are non-protein-coding transcripts more than 200 nt in length, which have a wide range of biological roles.74 For example, they regulate the transcription of genes, influence miRNA regulation of target genes, and, through their interactions with proteins affect the function and stability of proteins. Some lncRNAs can also regulate the cell cycle, which in turn affects cell proliferation and differentiation.75 Studies have shown that lncRNAs may be implicated in the development of cancer in relation to their ability to regulate key cancer-associated transcriptional activators.76 Because of their tissue-specific expression patterns, they may contribute to tumor heterogeneity.77
Several known cancer-related lncRNAs are overexpressed in the serum and plasma of cancer patients, enabling them to be promising biomarker candidates for non-invasive diagnosis.78,79 For example, it has been found that lncRNA can mediate pancreatic ductal adenocarcinoma (PDAC), which can be used as a liquid biopsy biomarker for PDAC.80 Hu and his team have found that lncRNA H19 can be used as a potential biomarker for the adjuvant diagnosis of lung cancer, because of its significant elevation in the plasma of patients with lung cancer.81 Although a large number of lncRNAs have been identified in recent years, the specific functions of some lncRNAs and the role they play in cancer are still unknown, so we need to pay close attention to the study of lncRNAs in the future, to fully evaluate its feasibility and accuracy as a liquid biopsy for cancer. Currently, there are abundant studies on lncRNA-based diagnostic and prognostic models.82–84 For example, one study discovered m6A immune-associated lncRNA risk models that can accurately forecast prognosis, immunological status, and treatment response in bladder cancer.82 And a study utilized overlapping long non-coding RNAs (lncRNAs) to create a signature of lncRNAs linked with cuproptosis. This signature can be employed to forecast the prognosis and determine the effectiveness of immune checkpoint blockade (ICB) therapy in individuals diagnosed with hepatocellular carcinoma.84 Despite the lack of clinical studies on the subject, there is no doubt that the modeling of biomarkers using miRNA and lncRNA is a crucial area of development in liquid biopsy.
CircRNA
Circular RNAs (circRNAs) are a distinct type of RNA molecules that possess a distinctive closed loop structure and do not code for proteins (Fig. 7). The initial documentation of circRNAs may be traced back to a 1971 investigation on potato spindle tuber disease. During this study, circRNAs were not yet recognized as a distinct concept, and scientists provisionally referred to them as a “virus-like” RNA with low molecular weight that has the ability to self-replicate.85 In 1976, Sanger et al. isolated this RNA and subjected it to different nuclease enzymes. They discovered that these RNAs were not easily broken down by most nuclease enzymes, indicating that they likely have a looped structure. This is because looped RNAs lack free ends at the 5′ and 3′ termini, making them less recognizable and degradable by nuclease enzymes. Sanger employed radioactive labeling to directly visualize the closed loop structure of virus-like RNAs. The RNA ends were labeled and it was seen that these ends were not labeled under both in vivo and in vitro circumstances, providing additional confirmation of the circRNA.86 The investigations conducted by Memczak et al. in 2013 and Hansen et al. in 2013 were significant contributions to the field of cyclic RNA research. These studies systematically have shown the extensive occurrence and significance of cyclic RNAs in human cells and tissues.87 Presently, scientists have discovered that circRNAs possess a multitude of biological roles, such as acting as miRNA sponges, controlling the splicing of precursor mRNAs, facilitating transcription, regulating their own stability and location through binding to RBPs (RNA-binding proteins), and encoding functional proteins, among others.88 circRNAs are not directly detectable by selective purification procedures that rely on polyA tails due to their absence of a typical polyA tail. Scientists have utilized several techniques like RT-PCR, RNAseq, northern hybridization, and high-throughput sequencing to detect circRNAs. This was achieved by developing primers that target specific reverse splice sites of circRNAs. Because of the inherent characteristics of circRNA, RNA exonuclease is unable to effectively degrade it, while linear RNA can be selectively broken down by RNA exonuclease for the purpose of enrichment.89 circRNAs can function as either proto-oncogenes or oncogenes in cancer, depending on the specific pathways they are connected with. One instance is circHIPK3, which can enhance the growth and movement of cancer cells by activating the miR-124/STAT3 pathway. STAT3 is a transcription factor that is linked to multiple oncogenes and the process of cell proliferation. The circHIPK3 molecule indirectly enhances the activation of the STAT3 signaling pathway by preventing the inhibitory effect of miR-124 on STAT3. This, in turn, controls the malignant activity of tumor cells.90 Studies have demonstrated that the circRNA ITCH functions as an oncogene in multiple types of cancer. The circ-ITCH molecule has the ability to bind to miRNAs, specifically miR-7, miR-17, and miR-214, resulting in an indirect control over the expression of its target genes. These microRNAs (miRNAs) and their target genes potentially play a role in many signaling pathways associated with tumors, including the Wnt/β-catenin system and the PI3K/AKT pathway.91 Aberrant expression of circ-ITCH can potentially facilitate tumor growth by disrupting the equilibrium of these pathways. It has been discovered that circ-ITCH is down-regulated as an oncogene in ovarian cancer, prostate cancer, glioma, and gastric cancer.92 To summarize, circRNAs contribute to the development of tumors by facilitating cell proliferation, avoiding growth inhibitors, increasing invasion and metastasis, inducing angiogenesis, disrupting cellular energy regulation, and fostering inflammation.
Technology for the detection of liquid biopsy markers
As previously stated, liquid biopsy markers primarily include CTCs, ctDNA, exosomes, free miRNA, lncRNA, circRNA, proteins, and so on, which are detected in various ways but share some similarities. CTCs detection necessitates enrichment of CTCs, which are subsequently labeled with particular antibodies or fluorescent dyes. These markers can bind to specific antigens on the surface of circulating tumor cells, generating visible fluorescence signals under a microscope. Physical separation methods and antigen–antibody conjugation methods are the most common approaches for enriching CTCs. Traditional physical separation methods involve separating cells based on screening parameters such as cell size, density, or charge. Traditional antigen-antibody binding approaches for identifying CTCs are primarily achieved by the CellSearch system, which is based on the principle of EpCAM to trap tumor cells.93–97 The primary objective of ctDNA detection is to identify specific mutations. Plasma DNA is concentrated and identified by using advanced technologies such as digital PCR (dPCR) and NGS.98–100 The identification of exosomes involves the enrichment of exosomes and subsequent analysis of their constituents. In this context, our primary focus is on the enrichment process. The main techniques employed for this purpose include differential centrifugation, filtration, polymer precipitation, immunomagnetic beads, chromatography (specifically volumetric exclusion chromatography), and the relatively new microfluidic technology.57,101 The methods used for RNA detection encompass RNA-FISH, RT-PCR, Northern Blotting, RNA Sequencing, RNA Microarray, In Situ Hybridization, and various other techniques.102–106 Proteins can be identified using western blot and mass spectrometry techniques. The subsequent tables provide a comparison of the principles linked to each technique, as well as their respective benefits and drawbacks (Tables 1–5).
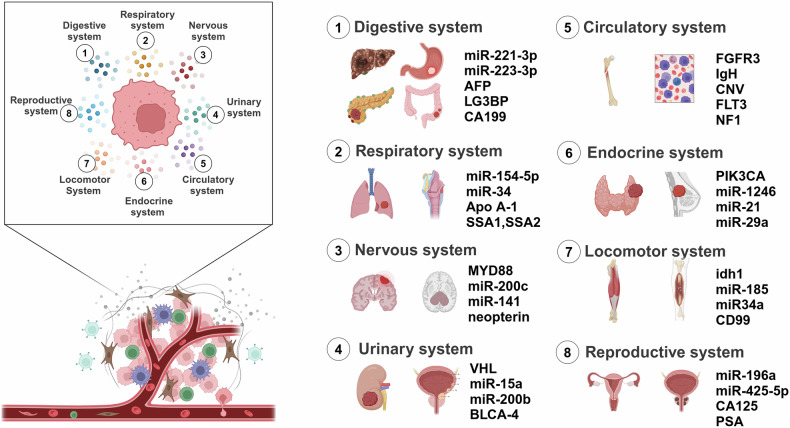
Liquid biopsy in systemic tumors
In this section we summarize the application of liquid biopsy in eight systems of tumors (Fig. 8).
Fig. 8.
Liquid biopsy biomarkers of systemic tumors. Application of liquid biopsy in tumors of different systems and some examples of biomarkers. Created with BioRender.com
Digestive systems
The digestive system concentrates on the use of liquid biopsy in hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), CRC, pancreatic cancer (PC) and gastric cancer (GC) (Table 6).
Table 6.
Liquid biopsy in digestive system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| HCC | miR-221-3p, miR-223-3p, miR-10b5p, miR-21-5p | Plasma exosome | up | Early diagnostic biomarker | 108 | |
| LG3BP, PIGR | Serum exosome | up | Early diagnostic biomarker | 110 | ||
| ECE1, HOXA1, cle11a, AK055957, PFKP, EMX1 methylation | Plasma | up | Early diagnostic biomarker | 112 | ||
| cfDNA | Plasma | up |
Early diagnostic biomarker, Efficacy monitoring biomarker |
115 | ||
| CTCs | Peripheral blood | up |
Early diagnostic biomarker, Tumor recurrence biomarker |
114 | ||
| Mixed CTCs, Mesenchymal CTCs | Peripheral blood | up |
Early diagnostic biomarker, Disease progression biomarker |
116 | ||
| CCA | microRNA-21, microRNA-221 | Plasma | up | Early diagnostic biomarker | 122 | |
| hTERT, CK19 | Peripheral blood | up | Prognostic biomarker | 123 | ||
| Osteopontin (OPN) | Serum | down | MMP1, MMP10, CXCR4 |
Efficacy monitoring biomarker, Prognostic biomarker |
124 | |
| MMP-7 | Serum | up | Early diagnostic biomarker | 127 | ||
| CYFRA 21-1 | Serum | up |
Early diagnostic biomarker, Disease progression biomarker |
129 | ||
| Osteopontin (OPN) | Serum | up | Efficacy monitoring biomarker | 130 | ||
| CTCs | Peripheral blood | up |
Tumor aggressiveness biomarker, Prognostic biomarker |
131 | ||
| cfDNA mutation | Bile | up | Prognostic biomarker | 132 | ||
| CRC | microRNA-203 | Serum exosome | up | M2-TAM | Prognostic biomarker | 135 |
| microRNA-21 | Plasma exosome | up |
Tumor recurrence biomarker, Prognostic biomarker |
137 | ||
| miR-17-5p, miR-92a-3p | Serum exosome | up |
Early diagnostic biomarker, Disease progression biomarker |
138 | ||
| miR-25-3p | Serum exosome | up | Tumor aggressiveness biomarker | 139 | ||
| miR-196b-5p | Serum exosome | up | STAT3 | Efficacy monitoring biomarker | 147 | |
| miR-301a, miR-23a | Serum exosome | UP | Early diagnostic biomarker | 140 | ||
| miR-19a | Serum exosome | up | Tumor recurrence biomarker | 142 | ||
| QSOX1 | Serum exosome | down | Early diagnostic biomarker | 145 | ||
| ctDNA | Plasma | up | Efficacy monitoring biomarker | 151 | ||
| ctDNA | Plasma | up |
Tumor recurrence biomarker, Prognostic biomarker |
152 |
Hepatocellular carcinoma (HCC)
In the diagnosis of HCC, alpha-fetoprotein (AFP) is detected as a classical tumor marker in most patients with HCC, but low expression of AFP in some patients with HCC is detrimental to the detection of HCC by AFP. Because HCC exhibits substantial tumor heterogeneity, neither AFP nor liver biopsy currently fulfills the clinical requirements for early diagnosis or prognosis assessment.107 Therefore, it is necessary and meaningful to search for alternative ways of detecting HCC.
Several liquid biopsy markers can be used for early diagnosis of hepatocellular carcinoma. On the one hand, it was found to be feasible to co-detect AFP with miRNA, and the diagnostic ability of patients with low AFP expression can be improved (AUC: 0.80, specificity: 95%, accuracy: 81%) by the combined detection of AFP and miRNAs (including miR-221-3p, miR-223-3p, miR-10b5p, and miR-21-5p).108 On the other hand, searching for other more effective protein markers may be an effective way to improve early diagnostic ability. For example, the exosomal proteins LG3BP and PIGR can promote the transformation, invasion, and proliferation of tumor cells, which are associated with a poor prognosis, and they show greater diagnostic ability as biomarkers compared to AFP.109,110 As a marker released into the peripheral blood by tumors, cfDNA is usually not used for screening purposes since there is minimal necrosis of tumor cells in the early stages, and only a small amount of ctDNA is released into the bloodstream.111 However, a recent study has shown that the methylation properties of ctDNA have great potential in the early diagnosis of tumors. Researchers identified six optimal methylated DNA markers (MDMs), including ECE1, HOXA1, cle11a, AK055957, PFKP, and EMX1, and performed phase I and phase II clinical validation, finding them to be highly AUC (0.96), sensitive (95%) and specific (92%) in the diagnosis of HCC.112 Expert consensus on early screening strategies for liver cancer in China incorporates cfDNA whole genome sequencing into the whole process of early liver cancer screening.113 CTCs are malignant cells that undergo epithelial-mesenchymal transition (EMT) in the primary tumor. Qi et al used the CanPatrol™ CTCs enrichment technology in 112 patients with HCC, and the positive rate exceeded 90% even for early-stage disease.114 In addition to the early diagnosis of tumors, liquid biopsy is also beneficial for patient treatment as well as prognosis. For example, ctDNA, mentioned above, is not only involved in the early diagnosis of tumors but can also be used as an indicator of the efficacy of tumor radiotherapy. Patients with high pre-radiotherapy ctDNA expression tended to have more advanced disease and larger tumors, and after radiotherapy, patients with low ctDNA expression had significantly better prognostic tumor response, intrahepatic non-failure rate, and local control (LC) rate (p = 0.017, p = 0.035, and p = 0.006, respectively).115 In addition to the detection of the number of CTCs, the form of CTCs is also an important test. It was found that the ratio of mixed CTCs to mesenchymal CTCs can be used to discriminatie metastatic HCC patients with non-metastatic patients (AUC: 0.861).116 Compared to mixed CTCs, mesenchymal CTCs have a greater potential for invasion and metastasis. Bai et al. found that high expression of the CXCR4 protein was more common in mixed CTCs, which may be associated with CTCs progression and metastasis.117 And the Guidelines for the Diagnosis and Treatment of Primary Liver Cancer in China suggest that CTCs testing can serve as a novel clinical tool for predicting prognosis and evaluating the effectiveness of treatment for liver cancer. In conclusion, the multiple markers of liquid biopsy can compensate for the inability to detect patients with low AFP expression and play a role in treatment as well as prognosis.
Cholangiocarcinoma (CCA)
The tumor’s stealthy growth seriously jeopardizes their early discovery, preventing patients from accessing potentially curative treatments.118 Additionally, the patient’s fragile and advanced illness state increases the danger of bleeding and peritoneal seeding, and the tiny amount of tissue retrieved might not be sufficient for confirmation by cytology or histology.119 For these reasons, liquid biopsy is essential for both the prognosis and diagnosis of cholangiocarcinoma.
The main markers that have been studied in cholangiocarcinoma (CCA) include cfDNA, CTCs, and miRNA. Compared with healthy control specimens, miR-21 and miR-221 showed significant overexpression in the plasma of patients, and higher circulating miR-21 expression was associated with poorer prognosis in ICCA.120 However, the current study found that high expression of miR-21 and miR-221 was not only detected in CCA but also in HCC and other liver diseases.121,122 Therefore, it is possible that the combination of miR-21 and miR-221 with other markers may be useful for the detection of CCA. For example, high levels of cytokeratin-19 (CYFRA 21-1), MMP-7, osteoblasts, periostin, and IL-6 can be detected in the serum of patients with CCA, which may be helpful for further diagnosis of CCA.123–130 In addition to miRNAs, CTCs is an important marker in liquid biopsy of CCA. High expression of CTCs is associated with strong tumor aggressiveness and short survival, and thus evaluation of CTCs may help identify CCA patients at risk of early death.131 Unlike miRNAs and CTCs, which are detected in blood, cfDNA can be detected in the bile of CCA patients, and tumor recurrence and prognosis can be inferred mainly by detecting single-nucleotide variants, insertions, and deletions of cfDNAs, but not their expression.132,133
Colorectal cancer (CRC)
Colorectal cancer is a complex illness characterized by numerous genetic or somatic changes, and it is identified in less than half of cases when it is locally advanced.134 Thus, the implementation of liquid biopsies is necessary to enhance the accuracy of colorectal cancer diagnosis and to forecast the advancement of the disease.
miRNAs have a crucial role in various aspects like tumorigenesis, proliferation, metastasis, and drug resistance in CRC. For example, high expression of miR-193a and miR25-3p, miR-17-5p and miR-92a-3p, miR-21, and miR-203 promotes liver metastasis by inducing vascular permeability/angiogenesis.135–139 Therefore, miRNAs have the potential to serve as an effective liquid biopsy marker. Several scholars have studied miRNAs and found that a variety of miRNAs, such as miR-23a, miR-301a,140 as well as miR-17-92a and miR-19a141,142 are significantly overexpressed in the blood of tumor-bearing patients and are predictive of early tumorigenesis as well as tumor aggressiveness. Consequently, some miRNAs can distinguish CRC patients from the population and help in the early diagnosis of CRC. As for CTCs, patients with colorectal cancer had higher CTCs counts than those with colorectal polyps (P < 0.001).143 And CTCs counts were positively correlated with CRC disease stage, with sensitivities ranging from 89 to 97% across the range of disease severity.144 However, not all liquid biopsy markers are present in the form of high expression in patients’ blood. The exosomal cargo protein QSOX1 is significantly reduced in the blood of tumor patients compared with healthy human controls while Glypican-1 (GPC1) is significantly increased in exosomes, and a series of recent studies have suggested that dysregulation of exosomal proteins could serve as a promising novel biomarker for the early diagnosis and non-invasive risk stratification of CRC.145 At present the monitoring of single extracellular vesicles (SEV) is also helpful in the diagnosis of colorectal cancer. A study has developed a new sensor that combines a DNA aptamer capable of explicitly binding to SEV surface proteins with a single microbead capable of immunoadsorbing EVs, allowing for the direct and rapid monitoring of SEV. Clinical trials have shown that it is able to detect exosomes directly from 2 μL plasma samples, and indicated that cancer patients have higher levels of CD63, EpCAM double-positive exosomes than healthy controls.146
In addition to the early diagnosis of tumors, the observation of the efficacy of tumor therapy and the prognosis of survival are important purposes of liquid biopsy. Up-regulation of miR-196b-5p in patients with CRC promotes chemoresistance to 5-FU.147 Besides, high expression of CTCs in patients’ blood is often a marker of high tumor recurrence rate and poor prognosis. The results of a study that performed CTCs counts on treatment days 1 and 15 showed that patients with high CTCs counts at baseline had worse overall survival (p < 0.001).148 In addition, the detection of CTCs surface markers such as thymidylate synthase and excision repair protein RAD23 homolog B can help to predict chemo-/radiotherapy resistance in patients.149 According to the Chinese Expert Consensus on Clinical Detection of Molecular Markers for Colorectal Cancer, CTCs could be effective for early screening, prognosis, and efficacy assessment of the disease.150 CtDNA has been shown to be useful in detecting the efficacy of surgery and chemotherapy and to play a role in the prediction of tumor recurrence. In patients receiving chemotherapy, downregulation of ctDNA is a predictor of response to treatment.151 Conversely, upregulation of ctDNA after surgery predicts a higher five-year risk of recurrence and poorer overall survival.152 Also, it is encouraging to note that studies have found a high degree of concordance between ctDNA mutations detected in the bloodstream and those found in biopsies of tumor tissues,153 suggesting that liquid biopsies may be able to play an even greater role in the future.
Pancreatic cancer (PC)
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of PC and accounts for more than 90% of PC154. The biology of PDAC is highly diverse and intricate, and its diversity is seen as a primary factor contributing to its resistance to therapies. Tumor heterogeneity is present not only across different patients (inter-tumor heterogeneity), but also within the same tumor (intratumor heterogeneity). Additionally, there is temporal heterogeneity caused by changes in PDAC over time and during treatment.155 Consequently, the early detection and monitoring of tumor development in PDAC via tissue biopsy is difficult. As a result, liquid biopsy holds significant research value in the diagnosis of PDAC and other related areas.
In the early diagnosis of PC, the number of CTCs can be effectively distinguished between PC patients and healthy controls, which has a high specificity (96.4%) but insufficient sensitivity (75.0%).156 Expert consensus of Oncology Committee of Chinese Medical Association in early diagnosis and treatment of pancreatic cancer states that CTCs can be used as a marker for early diagnosis and differential diagnosis of pancreatic cancer.157 Compared to CTCs, circulating epithelial cells (CECs) had a better performance in early diagnosis, with 77.8% patients showing detectable CECs, while only 15.8% of controls had detectable CECs.158 In early diagnosis, ctDNA relies heavily on the detection of its mutations. Since KRAS mutations are the most common genetic alterations in pancreatic cancer, and are present in more than 90% of patients, several scholars have investigated the use of KRAS mutations in liquid biopsy. It was found that detecting KRAS mutations by ctDNA alone had poor sensitivity (35.2%), accuracy (51.0%), and AUC (0.683).159 This may be due to the coexistence of KRAS mutations in a variety of other tumors.160 Therefore, the diagnostic power of ctDNA mutations can be effectively enhanced by combining ctDNA mutations with other markers, e.g., ctDNA mutations in combination with proteins,161 ctDNA mutations in combination with CA19-9, etc.29. Of these, the combination with CA199 had significantly higher sensitivity (78%) and specificity (91%).29 Compared to ctDNA mutations, methylation of ctDNA showed a stronger potential in early diagnosis, and methylation of ADAMTS1 and BNC1 performed well in the early diagnosis of PDAC in terms of its sensitivity (97.4%), specificity (91.6%), and AUC (0.95).162 Although CA19-9 is a classical tumor marker, it lacks specificity in early diagnosis as CA19-9 lacks tumor specificity. Therefore, monitoring CA19-9 in combination with other markers can help to improve the specificity of PC diagnosis. One study found that 66.10% of miRNA had better diagnostic value compared to CA19-9 by analyzing a variety of miRNAs.163 Expert consensus on the molecular diagnosis of early-stage pancreatic cancer (2023 edition) recommends miRNA combinations as markers for early-stage precision diagnosis of pancreatic cancer to provide guidance to clinicians. Moreover, miRNAs in combination with CA19-9 may have better application value.164 When combined with CA19-9, the AUC can be significantly increased compared to CA199 alone.165 In extracellular vesicles, the difference in extracellular vesicle long RNA levels had a very high AUC (0.949) in early diagnosis43.166 According to CACA TECHNICAL GUIDELINES FOR HOLISTIC INTEGRATIVE MANAGEMENT OF CANCER, the combination of CTCs, ctDNA, exosomes, microRNAs, etc., with CA19-9 can improve the accuracy of PC diagnosis. However, its widespread use in the clinic needs to be supported by high-quality clinical research.
For chemoresistance in PC, a variety of liquid biopsy markers can be useful. Although CTCs counts may not be effective in predicting chemotherapy efficacy,167,168 detection of CTCs molecular features can help predict therapeutic efficacy, such as CXC-motif chemokine receptor 4 (CXCR4).169,170 Compared to CTCs, ctDNA has been more extensively studied in the detection of chemotherapy treatment. On the one hand, the probability of detectable ctDNA in the blood of patients receiving neoadjuvant chemotherapy is dramatically reduced.171 On the other hand, a decrease in cfDNA mutant allele fraction (MAF) predicts a response to chemotherapy, and drug-resistant patients show an increase in ctDNA MAF during the course of disease progression.172 Various ncRNAs such as miR-20a-5p and miR-373-3p have been found to be associated with chemotherapy resistance173,174 and have potential as indicators to monitor therapeutic efficacy. However, current studies on ncRNAs and EVs in chemoresistance have focused on mechanistic studies175 and more clinical studies are needed for validation.
In the prognostic prediction of PC, the positivity of CTCs was associated with poor prognosis in patients with PDAC.176,177 The KRAS mutation in ctDNA was found to be significantly associated with the prognosis of the patients.178 Mutated patients have a tendency to relapse early and have a significantly lower overall survival, and recurrence-free survival, as compared to unmutated patients.179 Multiple miRNAs were combined in one study, and the score model constructed could be used to predict 5-year OS in patients, which was lower in patients with higher risk scores.180 Similarly, the combined diagnosis of multiple markers in EVs (EV-CK18 mRNA, EV-CD63 mRNA, EV-miR-409, cfDNA concentration, and CA19-9) in the monitoring of PDAC metastasis has a favorable efficacy (accuracy of 84%, sensitivity of 78%, specificity of 88%, AUC of 0.85) due to conventional imaging.181
Although CA19-9 is a commonly used tumor marker, there are still 10% of patients who do not synthesize CA199, which is detrimental to the diagnosis of PC. Since the synthesis of CA19-9 is affected by common variants in the fucosyltransferase (FUT) enzymes FUT3 and FUT2, the combination of CA199 with FUT significantly improved the AUC (0.84-0.92).182 Measurement of the associated glycan DUPAN-2 is useful in individuals unable to synthesize CA19-9. A recent study found that the accuracy of early pancreatic cancer blood tests (CA19-9 and DUPAN-2) was improved when monitored by measuring the FUT2/FUT3 genotype subgroups and combining CA199 with DUPAN-2.183 Therefore, the detection of FUT added to patients with low CA19-9 expression may contribute to a more effective diagnosis of pancreatic cancer.
Gastric cancer (GC)
The primary indications of gastric cancer are nonspecific and typically involve dyspepsia, which is indicative of peptic ulcers. Patients and doctors sometimes overlook these symptoms, and a physical examination reveals no evident anomaly, or solely the presence of blood in the stool.184 Hence, it is imperative to discover novel and more efficient approaches for early detection of stomach cancer.
In early diagnosis, CTCs were found in 90.5% of patients. The sensitivity and specificity rates for detecting CTCs were 85.3% and 90.3%, respectively, among patients with gastric cancer and healthy individuals. Furthermore, it exhibits enhanced sensitivity in detecting advanced gastric cancer patients.185 Research has shown that the amount of cfDNA in the plasma of patients with stomach cancer is higher compared to healthy individuals.186 When comparing CTCs to cfDNA, it is found that cfDNA has a greater sensitivity (96.67%) and specificity (94.11%) in the early detection of gastric cancer. Additionally, it has an AUC value of 0.9914.187 In recent times, various methods have been developed to identify methylation in cfDNA for the purpose of early detection. These techniques offer a high level of accuracy (>90%) in terms of specificity, however their sensitivity is comparatively lower.188,189 Hence, there remains ample opportunity for enhancement. Certain circular RNAs (cirRNAs) have the potential to be utilized for early diagnosis.190 By combining various cirRNAs to create a prediction model, it is possible to more accurately distinguish between patients and healthy individuals.191 Moreover, the use of many miRNAs can be employed for the prompt detection of gastric cancer, exhibiting an impressive area under the curve (AUC) value of 0.9299.192 Furthermore, it was discovered that the levels of serum exosomal protein TRIM3 were notably decreased in patients with gastric cancer compared to individuals without the disease.193
Liquid biopsy can also reveal cancer progression. Several studies have indicated that CTCs are linked with GC stage, and the amount of CTCs is higher in patients with high stage than in individuals with low stage.194,195 CTCs was discovered in 96% of metastatic gastric cancer patients,196 and the number of CTCs was considerably higher in patients with GC distant organ metastases than in healthy controls and non-metastatic patients.197 The plasma cfDNA was demonstrated to show an elevated trend in its concentration with the progression of gastric cancer.198 And the serum cfDNA expression level of patients with stages III-IV was significantly higher than that of patients with stageI.199 The role of miRNAs in gastric cancer development has been identified, for example, down-regulation of either miR-17-5p or miR-4742-5p significantly inhibits GC cell proliferation, invasion, and metastasis,200,201 and HULC promotes ubiquitous cell invasion and migration through the Wnt/βcatenin signaling pathway,202 However, there is currently more mechanistic research and a lack of clinical data to validate the results. Upregulation of exosome hsa_circ_0015286 was found to be closely associated with tumor size, clinical stage, and lymph node metastasis, with an AUC of 0.778, a sensitivity of 82.1%, and a specificity of 65.7% in gastric cancer.203
During GC treatment, both CTCs and cfDNA have been found to be useful in predicting efficacy during ICB treatment. Immune checkpoint blockade therapy efficacy can be predicted by analyzing the number and type of CTCs and CTCs-PD-L1 expression.204 CfDNA, on the other hand, can be used to predict therapeutic efficacy by detecting microsatellite instability (MSI) in GC,205 For chemotherapy, ncRNAs have been mentioned more often, on the one hand, multiple miRNAs (miR100, miR-34a, miR-23a, miR-30a, let- 7g, miR-342, miR-16, miR-181, miR-1, and miR-34) were found to correlate with chemo-sensitivity through data prediction,206 and on the other hand, some ncRNAs were confirmed to be associated with chemo-sensitivity through basic research. For example, miR-30a with cisplatin chemotherapy,207 hsacirc_004413, miR-145-5p, circCPM with 5-FU resistance.208,209 Therefore, ncRNA may be useful for chemotherapy efficacy prediction, which needs to be supported by more clinical data. After undergoing surgical treatment, the expression level of serum exosomal LncRNAH19 was significantly reduced compared with the preoperative level, and its AUC for diagnosing GC was up to 0.849, with a sensitivity and specificity of 74.36% and 83.95%, respectively, and its expression level was significantly correlated with the TNM stage.210
For patient prognosis, the OS as well as PTS of patients after treatment showed a significant negative correlation with CTCs and ctDNA,211–213 and the detection of cfDNA levels was helpful in predicting the recurrence of patients.214 Methylation levels of the cfDNA genes such as RASSF1A, SOX17, and wi −1 were significantly correlated with reduced PFS as well as OS.215
Respiratory system
For the application of liquid biopsy in the respiratory system, we focus on lung cancer, laryngeal squamous cell carcinoma (LSCC), and nasopharyngeal cancer (Table 7).
Table 7.
Liquid biopsy in respiratory system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| LC | cfDNA methylation | Plasma | up | Early diagnostic biomarker | 217 | |
| CDO1, HOXA9, AJAP1, PTGDR, UNCX, MARCH11 methylation | Serum, Pleural effusion, Ascites | up | Early diagnostic biomarker, Prognostic biomarker | 218 | ||
| RASSF1A, CDKN2A, DLEC1 methylation | Plasma | up | Early diagnostic biomarker | 219 | ||
| ctDNA | Plasma | up | Efficacy monitoring biomarker | 222 | ||
| CTCs | Peripheral blood | up | Early diagnostic biomarker | 225 | ||
| let-7i-3p, miR-154-5p | Serum | down | Early diagnostic biomarker | 226 | ||
| miRNA | Plasma exosome | up | Early diagnostic biomarker | 228 | ||
| SSA1,SSA2 | Serum, Plasma | up | MMP-9 | Early diagnostic biomarker, Tumor aggressiveness biomarker | 221 | |
| LSCC | ctDNA | Plasma, Saliva | up | Early diagnostic biomarker | 240 | |
| ctDNA methylation | Plasma | up | Early diagnostic biomarker, Disease progression biomarker, Prognostic biomarker, Disease progression biomarker | 242 | ||
| CTCs | Peripheral blood | up | Early diagnostic biomarker | 237 | ||
| CTCs | Peripheral blood | up | Prognostic biomarker, Efficacy monitoring biomarker | 238 | ||
| CTCs | Peripheral blood | up | Prognostic biomarker | 239 | ||
| miR-21 | Serum exosome | up | Early diagnostic biomarker | 245 | ||
| miR-155 | Plasma | up | Early diagnostic biomarker | 246 | ||
| miRNA-130a | Plasma | down | Disease progression biomarker | 247 | ||
| miR-632 | Serum | up | Early diagnostic biomarker, Prognostic biomarker | 248 | ||
| Microbiota | Mouthwash | Early diagnostic biomarker | 236 | |||
| NPC | EBV DNA | Plasma | up | Early diagnostic biomarker, Tumor recurrence biomarker | 251 | |
| EBV DNA | Plasma | up | Efficacy monitoring biomarker | 253 | ||
| EBV DNA methylation | Saliva | up | Early diagnostic biomarker | 254 | ||
| EBV microRNA | Serum | up | Early diagnostic biomarker | 258 |
Lung cancer
The high mortality rate of lung cancer is mainly due to the late detection and diagnosis of lung cancer and the fact that most lung cancer patients show signs of metastasis at the time of symptom onset, leading to a decrease in the overall survival rate of lung cancer.3 Therefore, early diagnosis and early treatment are effective measures to reduce the mortality rate of primary lung cancer patients. In screening for lung cancer, ctDNA plays a role as a class of liquid biopsy markers in the diagnosis, treatment, and prognosis of the disease. Firstly, not only the expression of ctDNA is upregulated in lung cancer patients, but also its methylation level is upregulated in early-stage lung cancer, so ctDNA may be used as an effective marker for screening early-stage tumors.216–219 The exosome, which is currently popular in liquid biopsies, has likewise been found to serve as a liquid biopsy biomarker for lung cancer. In particular, exosomal proteins, a variety of proteins like SAA1, SAA2, Apo A-1, etc., have been found to be abnormally expressed in lung cancer patients and are considered to be potential markers for the early detection of lung cancer.220,221 Although CTCs do not play a significant role in early cancer screening, the number of CTCs detected does correlate strongly with tumor efficacy and prognosis.222–225 This idea was well confirmed in a recent study, in which patients with high CTCs counts before or after treatment had a significantly worse prognosis than those with low CTCs.224 The CSCO Small Cell Lung Cancer Diagnostic and Treatment Guidelines state that tracking CTCs can assist in accurately determining the disease’s clinical stage, which will help in selecting the best course of action, directing each patient’s unique course of care, keeping an eye on the tumor’s metastasis and recurrence, assessing the effectiveness of the treatment, and forecasting the prognosis for survival. miRNAs, as a prognostic biomarker for lung cancer, have also become an important component of liquid biopsies for lung cancer.226 In addition, miRNAs have been found to be involved in a variety of pathogenetic processes in cancer, such as proliferation, migration, and drug resistance.227,228 Therefore, miRNAs have the potential to become an effective biomarker for understanding tumor progression as well as treatment efficacy. In addition to this, the amount of ctDNA also reflects the different stages of lung cancer, and the detection rate of ctDNA rises with tumor stage, with ctDNA detected in 100% of plasma specimens from patients with stage II-IV NSCLC.229 Moreover, the expression of ctDNA is highly correlated with the volume and size of the tumors, and thus ctDNA detection may be synergistic with imaging, and more helpful in understanding the course of the patient’s disease. The 2021 IASLC NSCLC Liquid Biopsy Consensus states that plasma ctDNA can be considered a useful tool for genotyping newly diagnosed patients with advanced NSCLC, and that the results are often complementary to those from tissue analysis.230 Also, ctDNA mutations have been found to be of some significance in lung cancer, but their mutations are not associated with early screening of tumors but rather tend to guide the selection of treatment regimens. Since it has been found that drug-resistant recurrence in many patients is associated with mutations in ctDNA, ctDNA testing may be used as an adjunctive means of detecting therapeutic efficacy and providing more rational clinical drug use.222,231
Laryngeal squamous cell carcinoma (LSCC)
Laryngeal squamous cell carcinoma (LSCC) is the second most common cancer of the respiratory system after lung cancer.232 Due to the lack of early disease indicators, the diagnosis is typically made at a late stage. 40% of patients are diagnosed with lymph node metastases and have a bad outcome.233 Currently, imaging and tissue biopsy are the predominant diagnostic techniques of head and neck squamous cell carcinoma (HNSCC). However, imaging tools make it difficult to detect micrometastases and persistent lesions in the early stages. Because different metastatic lesions might arise in diverse tumor genetic landscapes, a single tissue sample cannot adequately capture tumor heterogeneity.234 As a result, clinical detection strategies to improve early identification and prolong survival of HNSCC are critical.
Classical CTCs as well as ctDNA have been shown to be associated with LSCC. Current studies have shown that ctDNA can be detected in the plasma and saliva of patients with early and advanced disease and that the amount of ctDNA is higher in patients with advanced and metastatic cancers than in patients with early-stage disease.235 A recent study found that ecological dysregulation of the oral microbiome is a key hallmark of LSCC and that LSCC can be identified by detecting microbiota in mouthwash, which provides a novel model for liquid biopsy of LSCC.236
And a series of studies have found that liquid biopsies have great potential for predicting the treatment efficacy and prognosis of patients. For CTCs, in addition to its early diagnostic role, it can also be used for treatment efficacy testing. CTCs counts are significantly reduced in tumor patients after treatment, and CTCs-negative patients have improved survival compared to CTCs-positive patients.237,238 Patients with high preoperative CTCs expression have a worse postoperative prognosis, and reduced CTCs values have been associated with an improved response to treatment.239. CtDNA may be associated with tumor recurrence and can appear prior to recurrence, which plays a predictive role.240 In addition, hypermethylation of ctDNA has been shown to correlate with tumor stage,241 and patients who exhibit high methylation levels early in life have a higher risk of death.242 Many miRNAs have been found to be dysregulated in cancers such as LSCC and are associated with tumor progression, and therefore miRNAs have received more attention in liquid biopsies for LSCC.243,244 To date, several miRNAs have been found to be highly expressed in the plasma of LSCC patients,245,246 and are strongly correlated with tumor size, advanced stage, and LNM.246 In addition, the expression of miRNAs such as miR130a and miR-632 has been associated with OS and DFS.247,248 LncRNA expression has been significantly correlated with the occurrence of LNM, advanced T-classification, and clinical stage, and may serve as a useful indicator of laryngeal cancer development.245
Nasopharyngeal cancer
Nasopharyngeal cancer is a malignant tumor of the respiratory system, which is often associated with EBV infection, and its symptoms are nonspecific and difficult to detect at an early stage.249 Because of the high correlation between nasopharyngeal cancer and EBV infection, EBV detection plays a very important role in liquid biopsy of nasopharyngeal cancer, and the circulating free EBV DNA tends to have the greatest role in early detection of nasopharyngeal cancer.250 By detecting the copy number of circulating free EBV (cfEBV) DNA, not only can it reflect the tumor load of patients, but also can be used for the prognosis prediction of metastatic nasopharyngeal cancer.251–253 Moreover, it has been found that the methylation of EBV DNA is significantly increased in the saliva of nasopharyngeal cancer patients, which suggests that it may be relevant to the detection of nasopharyngeal cancer.254 In addition, the detection of cfEBV DNA has shown other detection values, some scholars have found that the use of cfEBV DNA to guide routine imaging can effectively improve the detection efficiency and reduce the cost of detection.255 There is still much room for exploration of EBV in liquid biopsy of nasopharyngeal carcinoma. EBV-associated proteins such as EBNA1, EBER1, EBER2, etc. have been found to be useful in the diagnosis of nasopharyngeal cancer.256,257 Besides EBV-associated assays, various exosomal miRNAs have been found to be increased in the blood of patients with nasopharyngeal cancer, and anti-miRNA oligonucleotides (antagomiR) have a greater potential to become a therapeutic approach for nasopharyngeal cancer.258,259
Nervous system
In this part, we mainly introduce the application of liquid biopsy in gliomas as well as central nervous system lymphomas (Table 8).
Table 8.
Liquid biopsy in neverous system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| Glioblastoma | CTCs | Peripheral blood | up | Early diagnostic biomarker, Efficacy monitoring biomarker | 269 | |
| ctDNA mutation | CSF | up | Early diagnostic biomarker | 260 | ||
| ctDNA H3K27M mutation | CSF | up | Early diagnostic biomarker, Efficacy monitoring biomarker | 272 | ||
| ctDNA methylation | CSF | up | Early diagnostic biomarker | 268 | ||
| MCPH1 methylation | Serum | up | Early diagnostic biomarker, Efficacy monitoring biomarker | 274 | ||
| miR-320, miR-574-3p | Serum exosome | up | Early diagnostic biomarker | 276 | ||
| miRNA | CSF | up | Early diagnostic biomarker | 277 | ||
| PCNSL | MYD88, CARD11, CD79 mutation | CSF | up | Early diagnostic biomarker | 280 | |
| MYD88 | CSF | up | Early diagnostic biomarker | 284 | ||
| miR-200c, miR-141 | CSF exosome | down | ATP1B3, DYNC1H1, MATR3, NUCKS1, ZNF638, NUDT4, RCN2, GNPDA1, ZBTB38, DOLK | Early diagnostic biomarker, Efficacy monitoring biomarker | 289 | |
| SPP1, MARCKS, NPM1, VIM | CSF exosome | up | Early diagnostic biomarker | 291 | ||
| IL-10, sIL-2R | CSF | UP | Early diagnostic biomarker | 292 | ||
| IL-10 | CSF | up | Early diagnostic biomarker | 293 | ||
| Neopterin | CSF | UP | Early diagnostic biomarker | 295 |
Gliomas
Gliomas are the most prevalent primary malignant brain tumors in adults. Glioblastomas are highly malignant, with an average survival of 14.6 months.260,261 Early diagnosis of gliomas and therapeutic testing are therefore important for patients. The principal tool for monitoring gliomas is conventional magnetic resonance imaging, which has problems in separating true progression (TP) from pseudoprogression.262 As a result, more reliable and sensitive approaches are required to assess tumor response and evolution. Currently, liquid biopsy of gliomas involves specimens from blood and cerebrospinal fluid.
Firstly, for the early diagnosis of tumors, as EpCAM is widely expressed on the surface of CTCs derived from cancer cells, most CTCs detect cells targeting EpCAM, but EpCAM is not present in GBM cells.263 Thus, it has been suggested that circulating brain tumor cells are detected by GBM-specific expression of CD14, CD16, etc.264 In comparison to blood, CTCs in the cerebrospinal fluid are more readily identifiable and distinguishable from other cells,265 which may result from the presence of the blood-brain barrier and the more complex cellular composition of blood. According to NCCN Clinical Practice Guidelines in Oncology, Version 3.2020 on Central Nervous System Cancers, CTCs improve tumor cell detection and efficacy evaluation sensitivity.266 Secondly, the detection of ctDNA is also of diagnostic significance for gliomas, and studies conducted by several scholars have demonstrated that the sensitivity and accuracy of tumor ctDNA detection in cerebrospinal fluid is better than that in plasma compared with blood.261,267 In addition to mutations of ctDNA, its methylation can be used in the detection of cerebrospinal fluid, and the detection of ctDNA methylation can help analyze the subtypes of gliomas.268 Also, liquid biopsy can monitor the tumor progression. First is the classical CTCs, Various studies have illustrated that the number of CTCs does not only correlate with tumor progression, as well as prognosis.264,269 CTCs identification techniques may be taken into consideration for the evaluation of meningeal metastases, according to the Chinese Guidelines for Integrated Diagnosis and Treatment of Tumors—Metastatic Tumors of the Central Nervous System. In the detection of ctDNA, the detection of target mutations has received more attention. The detection of mutations can predict the degree of tumor malignancy.260 Liquid biopsy can be also predictive for the treatment and prognosis of gliomas. CTCs may correlate with tumor resistance.270 And the detection of ctDNA mutations can monitor the response to drug therapy.260,271 CtDNA mutations can also be used to select appropriate targeted therapeutic drugs, which is more conducive to the rational use of medication to improve the efficacy of treatment.272 Detection of ctDNA methylation in serum has revealed that the serum markers can reflect the characteristics of tissues and can effectively differentiate between gliomas and other malignant tumors, which can help in the diagnosis of gliomas as well as in the prediction of their prognosis.273,274 In addition to ctDNA in circulating tumor nucleic acids, miRNA is also a point of detection. Although miRNAs have advantages such as easy identification, their faster degradation leads to hindrance in the detection process. However, when miRNAs are incorporated into extracellular vesicles like exosomes, their degradation process is impeded, making them more stable and easier to detect.275 Detection of miRNAs in exosomes therefore currently appears to be positive in various aspects of the diagnosis of gliomas, for example, RNA RNU6-1 has been recognized as an identifying biomarker for GBM.276 Cerebrospinal fluid has been shown to be a source of GBM-specific 9 miRNAs.277 In addition, the detection of exosomal proteins has also proved to be promising for research.275 90% of GBM patients have at least one protein differently expressed in their exosomes, including EGFR, EGFRvIII, podoplanin, and IDH1.278 Furthermore, chloride intracellular channel 1 identified in exosomes enhances GBM growth and invasiveness, and is associated with poor prognosis.279 Currently, detecting changes in protein levels in body fluids or tissues is the most commonly used diagnostic method for the diagnosis, treatment, and prognosis of gliomas.
Primary central nervous system lymphoma (PCNSL)
Unlike other lymphomas, primary CNS lymphoma is not easily recognized and responded to by immune cells due to the blood-brain barrier and is therefore considered an “immune-privileged (IP)” lymphoma.280 Thus, timely diagnosis and treatment are crucial for improving patient prognosis and survival. Due to the difficulty of sampling tissue biopsies, liquid biopsies have the potential to be used in conjunction with radiologic features in the diagnosis of PCNSL.281 Currently, ctDNA is the most frequently discussed liquid biopsy for CNS lymphomas, but several studies have failed to find a relationship between the number of ctDNAs and the diagnosis of lymphomas, etc. More attention has been paid to ctDNA mutations such as MYD88, CARD11, CD79B, etc.280,282,283. Among them, MYD88 is the most well-researched, and it has been classified as a diagnostic marker for PCNSL.280
Several studies have demonstrated that detection of the MYD88 mutation in cerebrospinal fluid or plasma not only allows for the early diagnosis of PCNSL but also helps in the prediction of efficacy and drug resistance of chemotherapy and other therapeutic measures.284 Currently, the technology of ctDNA detection is constantly being updated, and a new rapid genotyping system (GeneSoC) based on microfluidic thermocycling technology with RT-PCR has recently made it possible to greatly reduce the detection time compared with the previous NGS and droplet digital PCR,285,286 which is more conducive to intraoperative detection and monitoring of the therapeutic efficacy.284,287 Liquid biopsy of ctDNA can reduce the impact of spatial heterogeneity of the tumor compared with tissue biopsy, and a recent study found that liquid biopsy detects ctDNA mutations earlier than tissue biopsy, so liquid biopsy of ctDNA has great potential for clinical application in PCNSL.288
In addition to the most attention in ctDNA, miRNAs have also been found to be useful as monitoring markers for PCNSL.289 There is a lack of research on miRNA compared to ctDNA, and miRNAs are currently mainly detected in exosomes due to the greater stability of miRNAs.290 The expression levels of miRNAs such as miR-200c and miR-141 etc. can be used to diagnose PCNNSL as well as to monitor the efficacy of chemotherapy.289 In addition to miRNAs, a variety of phosphoproteins associated with PCNNSL in cellular vesicles, including SPP1, MARCKS, NPM1, and VIM, have the potential to be used as markers of PCNNSL.291 Some inflammatory factors, such as CSF neopterin, the interleukin (IL)-10, CXCL13, etc. have been found to be up-regulated in the cerebrospinal fluid of PCNSL patients.280,292–294 Moreover, CSF neopterin has been found to be significantly higher in PCNSL patients than in patients with other brain tumors and pseudo-inflammatory encephalopathies, and thus neopterin levels may help to differentiate PCNSL from other CNS tumors.295
Urinary system
The liquid biopsy in urology has been focused on the following four tumors, including renal cell carcinoma (RCC), bladder cancer (BLCA), Wilms’ tumor (WT), and uroepithelial carcinoma (Table 9).
Table 9.
Liquid biopsy in urologic system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| RCC | ctDNA | Plasma | up | Prognostic biomarker, Disease progression biomarker | 308 | |
| cfDNA | Plasma | up | Early diagnostic biomarker, Prognostic biomarker | 309 | ||
| TP53 mutation | Plasma | up | Prognostic biomarker | 310 | ||
| VHL, BAP1, PBRM1 mutation | Plasma | up | Prognostic biomarker | 311 | ||
| ctDNA methylation | Urine, Plasma | up | Early diagnostic biomarker | 299 | ||
| miR-21-5p, miR-150-5p, miR-145-5p, miR-146a-5p | Serum | up | Early diagnostic biomarker | 300 | ||
| miR-328-3p | Urine | down | Prognostic biomarker | 301 | ||
| miR-122-5p, miR-206 | Serum | up | Prognostic biomarker | 302 | ||
| miR-15a | Urine | up | Early diagnostic biomarker, Prognostic biomarker | 313 | ||
| MiR-30a-5p methylation | Urine | up | Early diagnostic biomarker, Prognostic biomarker | 303 | ||
| miR-210, miR-1233 | Serum exosome | up | Early diagnostic biomarker | 315 | ||
| has-mir-149-3p, has-mir-424-3p | Plasma exosome | up | Early diagnostic biomarker | 316 | ||
| has-mir-92a-1-5p | Plasma exosome | down | Early diagnostic biomarker | 316 | ||
| BLCA | CTCs | Peripheral blood | up | Early diagnostic biomarker | 340 | |
| CTCs | Peripheral blood | up | Efficacy monitoring biomarker | 345 | ||
| CTCs | Peripheral blood | up | Prognostic biomarker | 341 | ||
| p16(INK4a) methylation | Serum | up | Early diagnostic biomarker | 324 | ||
| APC, GSTP1, TIG1 methylation | Serum | up | Prognostic biomarker | 325 | ||
| p14ARF methylation | Plasma | up | Tumor recurrence biomarker | 326 | ||
| CDH13 methylation | Serum | up | Prognostic biomarker | 327 | ||
| ctDNA VAF | Plasma | up | Efficacy monitoring biomarker | 348 | ||
| ctDNA VAF | Plasma | up | Disease progression biomarker | 349 | ||
| ctDNA | Plasma | up | Efficacy monitoring biomarker, Tumor recurrence biomarker | 350 | ||
| miR-19a | Plasma | up | PTEN | Early diagnostic biomarker | 328 | |
| miR-200b | Plasma | up | Early diagnostic biomarker | 329 | ||
| miR-92, miR-33 | Plasma | down | Early diagnostic biomarker | 329 | ||
| miR-663b | Plasma exosome | up | Ets2 deterrence factor | Early diagnostic biomarker | 333 | |
| BLCA-4 | Urine | up | Early diagnostic biomarker | 334 | ||
| MCM5 | Urine | up | Early diagnostic biomarker | 335 | ||
| WT | miR-124-3p/miR-9-3p/miR-218-5p/miR-490-5p/miR-1538 | Serum | up | Early diagnostic biomarker | 549 | |
| TP53 mutation | Plasma, Serum, Urine | up | Tumor recurrence biomarker | 358 | ||
| ctDNA | Serum | up | Early diagnostic biomarker, Prognostic biomarker | 359 | ||
| Hyaluronidase | Urine | up | Early diagnostic biomarker | 361 | ||
| Basic fibroblast growth factor (bFGF) | Urine | up | Early diagnostic biomarker, Prognostic biomarker | 362 | ||
| UC | cfDNA | Plasma | up | Early diagnostic biomarker | 364 | |
| ctDNA mutation | Plasma | up | Efficacy monitoring biomarker | 365 | ||
| ctDNA methylation | Urine | up | Early diagnostic biomarker | 367 | ||
| TERT mutation, ONECUT2 methylation | Urine | up | Early diagnostic biomarker | 371 | ||
| miR-1343-5p, miR-6087 | Serum | up | Early diagnostic biomarker | 372 | ||
| miR-141 | Serum | up | Early diagnostic biomarker | 373 | ||
| miR-151b | Serum | up | Prognostic biomarker | 374 |
Renal cell carcinoma (RCC)
RCC is one of the most common malignant tumors which is the main type of kidney cancer. It is difficult to diagnose RCC in its early stages and is now prone to recurrence after surgery as well as radiotherapy, hence earlier diagnosis of RCC is required. RCC cancers take a lengthy period (up to 50 years) to evolve from their initial genetic changes to clinical symptoms. Although little histologic modifications are detectable in the comparable histologically normal renal tissues of individuals with renal tumors, epigenetic alterations have accumulated in this noncancerous renal tissue, indicating their potential application in early identification by liquid biopsy.296 Liquid biopsy can be used as an auxiliary test for early diagnosis of RCC, and the main biomarkers include CTCs, ctDNA, miRNA, and so on.297
In the early diagnosis of RCC, the detection of CTCs is less frequently concerned. And CTCs were detected in 100% of samples evaluated in patients with metastatic clear cell renal cell carcinoma (ccRCC) but not in healthy controls.298 In addition, in the differential diagnosis of RCC, ctDNA plays a role in detecting its methylation, and plasma cfDNA has been found to have 300 differentially methylated regions, which is effective in the diagnosis of RCC by detecting the methylation.299 Studies on miRNAs as liquid biopsy markers have shown that the combination of multiple miRNAs has high sensitivity and specificity in the diagnosis of RCC and helps to differentiate it from benign renal tumors.300–302 For example, four microRNA (miR-21-5p, miR-150-5p, miR-145-5p, and miR-146a-5p) panels were produced, and the AUC of the panels was 0.938 (95% CI: 0.889–0.971; sensitivity: 90.79%, specificity: 93.75%).300 Similar to ctDNA, methylation of miRNAs is also beneficial for the diagnosis and differentiation of RCC.303 In liquid biopsy of RCC, some scholars have found that in addition to CTCs, cfDNA, and cfRNA, some other biomarkers are also involved in the diagnosis of RCC, such as some metabolites, plasma proteins, and other biomarkers, which are also involved in the diagnosis of RCC, but there are fewer research reports, that require more in-depth exploration.304–307
Additionally, CK+CTCs are frequently detected and the number of them correlates with disease progression.298 miRNAs have been found to be associated with the grading and staging of RCC as well as distant metastasis. There are increased serum miR-122-5p and miR-206 levels in patients with metastatic diseases. In addition, miR-122-5p levels were associated with grade.302
More studies have found that liquid biopsy can be used for the treatment monitoring and prognosis prediction of RCC. With the development of genetic testing technology, studies on ctDNA and miRNA have been more focused in liquid biopsy. First, cfDNA content and fragment length play a role in prognostic prediction of RCC, with shorter cfDNA fragments significantly associated with shorter PFS and postoperative ctDNA associated with prognosis only in patients with metastatic RCC but not in those without metastasis.308–310 In the case of ctDNA, mutations in ctDNA continue to be of great interest in liquid biopsies of RCC, with several studies detecting a variety of mutations in ctDNA and miRNA. For ctDNA, its mutations remain of great interest in liquid biopsies of RCC, and several mutation sites have been detected in several studies, including VHL, brca1-associated protein 1 (BAP1), recombinant polybrominated gene 1 (PBRM1), TP53, ATM, and others, with the most common mutated genes being VHL.308,310–312 Its mutations correlate with prognosis, e.g., patients with high cfDNA concentrations and TP53 mutations have the worst PFS, whereas patients with low cfDNA and no mutations in TP53 have a longer PFS (p = 0.004).310 Mutation detection of ctDNA helps to predict the efficacy of ICI and TKI therapy, and the frequency of ctDNA mutations is significantly reduced after surgery.312 miR-15a has been regarded as a possible key molecule for liquid biopsy of RCC because it not only identifies benign tumors as well as RCC but also correlates with RCC postoperative prognosis(specificity:98.1%,sensitivity:100%, AUC: 0.955).313 The number of mixed CTCs in the metastasis and no-metastasis groups at 12 months postoperatively was significantly different from the number of mixed CTCs preoperatively, suggesting that the risk of recurrence or metastasis correlates with dynamic changes in the count of CTCs.314 Given that miRNAs are more stable in the exosomes, many studies have begun to target miRNAs in the exosomes, and a great deal of potential exists for their clinical application.315,316 Moreover, some circRNAs, lncRNAs, and piRNAs have been considered for liquid biopsy in RCC.317–320
Bladder cancer (BLCA)
Bladder cancer is a highly heterogeneous malignancy. BLCA can present as non-muscle-invasive bladder cancer (NMIBC), muscle-invasive bladder cancer (MIBC), or metastatic disease events, each characterized by distinct molecular drivers.321 Currently, invasive cystoscopy and tissue biopsy remain the gold standard for BLCA identification and surveillance. However, this method suffers from drawbacks such as sampling bias, invasiveness, and difficulty in sampling deep tumors, which limits its use in mass screening.322
In the early diagnosis of BLCA, CTCs and miRNA are the main liquid biopsy markers. In BLCA, CTCs can be quantified by detecting folate receptor-alpha and can be diagnostic for BLCA (sensitivity: 82.14%, specificity: 61.9%).323 The role of ctDNA methylation in liquid biopsy has received much attention. Various ctDNAs such as p16 DNA, APC, GSTP1, TIG1, etc. have been shown to be hypermethylated in patients with BLCA,324,325 and there is a positive correlation between the frequency of methylation and the stage, so the methylation of ctDNA may be used as a biomarker for the diagnosis of BLCA.326,327 miRNAs in plasma and exosomes have been widely studied as potential biomarkers and therapeutic targets. Firstly, in blood, miR-19a, miR-99a, miR-200b, miRNA-373, and other miRNAs have been shown to be expressed differently in the blood of BLCA patients than in healthy people, which is a potential biomarker for BLCA.328,329 When multiple miRNAs are integrated for combined diagnosis, they show high accuracy in early diagnosis and differential diagnosis of BLCA.330,331 A study has constructed logistic regression modeling that predicts diagnosis with 89% accuracy in detecting the presence or absence of BLCA, 92% accuracy in distinguishing invasive BLCA from other cases, and 100% accuracy in distinguishing MIBC from controls.329 In exosomes, miRNAs have been found to be associated with tumor progression and metastasis, and similar to the miRNA alterations detected in the bloodstream, exosomal miRNAs also play a role in the diagnosis of BLCA and in predicting the prognosis.332,333 Compared to blood, urine testing is mainly focused on protein and exfoliative cytology. Most of the proteins are detected by ELISA, such as the expression of BLCA-4(sensitivity: 93%, specificity: 97%, AUC: 0.9607), MCM5(sensitivity: 75.6%, specificity: 71.1%), etc., to assist in the early diagnosis of BLCA.334,335 On the other hand, for exfoliated cells, not only cell surface markers, such as Cytokeratin 17, can be used to identify tumor cells for the diagnosis of BLCA,336 but exfoliated cell DNA, including TERT promoter mutations(specificity: 100.00%, sensitivity: 46.67%) and FGFR3 mutations, are the most common mutations in somatic cells, which can be used to detect BLCA noninvasively and to monitor recurrence.337,338
For the detection of tumor progression, the presence of CTCs has also been associated with metastasis of BLCA, and CTCs have been shown to predict metastasis in NMIBC and to identify those at high risk of recurrence.339–341 Also, in MIBC, there is a higher level of CTCs, again demonstrating the correlation between CTCs and tumor muscle infiltration.339 Although ctDNA is rarely expressed in blood, it has been found to be superior compared to histology in reacting with advanced tumor load, for example. And plasma ctDNA has a high concordance with genes detected in tumor tissue.342
Liquid biopsies can also be used for therapeutic monitoring and prognosis prediction in BLCA. CTCs has been shown to be associated with disease recurrence and poor prognosis in several studies. After clinical treatment, CTCs-positive patients have worse progression-free survival, CSS, and OS (sensitivity: 35%, specificity: 97%).340,343,344 CTCs can be used to assess the efficacy of cisplatin-based chemotherapy, PDL1 immunotherapy, etc., and can help to better predict the efficacy of treatments.345,346 CTCs-positive patients have higher rates of cancer-related mortality and disease recurrence compared to CTCs-negative patients. And CTCs-positive patients who received neoadjuvant chemotherapy (n = 22) survived longer than those who were not CTCs-positive (n = 48).345
Mutations of ctDNA are another concern in addition to the DNA methylation hotspot. A variety of genetic mutations have been found to be present in the blood of BLCA patients with potential as prognostic markers. For example, FGFR3 and PI3KCA mutations are significantly associated with recurrence of the disease, and the number of genomic alterations has been correlated with response to immunotherapy.347–349 A clinical trial has found a strong correlation between ctDNA Variant Allel Frequency (VAF) and treatment duration, clinical activity, PFS, and OS. Compared with patients with dVAF ≥ 0, patients with lower mean VAF had a significantly better PFS and OS.348
In addition to this, ctDNA expression levels are a valid indicator, and it has been demonstrated that liquid biopsy results are detected earlier than imaging at the time of tumor recurrence, thus making ctDNA a potent prognostic marker for the patients.350 Whether it is chemotherapy, radiotherapy, immunotherapy, or cystectomy, ctDNA testing can respond to disease progression after treatment as well as detect treatment efficacy, suggesting that it may be possible to improve the treatment regimen for better therapeutic efficacy by continuous monitoring of ctDNA.351–353
However, compared to blood monitoring, although significant progress has been made in urine biomarkers and urocytology monitoring, their sensitivity and specificity are low, and thus their application in low-grade tumors is partially limited.322,354
Wilms’ tumor (WT)
Wilms’ tumor (WT), which often occurs in children, is the main type of renal tumor in children and currently has a recurrence rate of up to 15%.355 The majority of cases of WT are disseminated, caused by mutations in somatic cells that are often limited to tumor tissue, and the tumors are very genetically heterogenous.356 Liquid biopsy may play a role in its early screening as well as therapeutic monitoring, which may help in tumor treatment as well as reducing recurrence.
In blood, liquid biopsies are performed mainly by monitoring miRNA and ctDNA. Among them, several miRNAs have been found to serve as markers for WT diagnosis and to play a role in the differential diagnosis of WT from other tumors (accuracy: 97.5%, sensitivity: 99.8%, specificity: 94.7%).357 Mutations in ctDNA have been found to allow for the early identification of WT and the OS of ctDNA-positive patients is poorer than that of ctDNA-negative patients.358,359 Urine, as the other main sample for liquid biopsy, is less effective in the diagnosis and differentiation of WT than blood. For example, ctDNA was detected in the serum for 82% patients, but in the urine for 26% patients.359 However, proteomic monitoring of urine specimens acts as an important class of molecular markers in liquid biopsies. For example, neuron-specific enolase, basic fibroblast growth factor (bFGF), and hyaluronidase are enriched in the urine of patients with nephroblastoma and can be used as indicators for the diagnosis of WT.360,361 In addition, in the monitoring of the therapeutic efficacy of WT as well as in the determination of the prognosis, several protein biomarkers, such as transgenic specific enolase (NSE), hyaluronic acid (HA), hyaluronan-stimulating activity (HSA), and hyaluronidase, etc. can be used as assays to assist in the judgment.360,362
Uroepithelial carcinoma (UC)
Uroepithelial carcinoma (UC) can be divided into uroepithelial bladder cancer (UBC) and upper uroepithelial carcinoma (UTUC), UBC has been introduced in detail in the section of BLCA, so this section will mainly focus on UTUC. UTUC is known to be a particularly aggressive form of uroepithelial carcinoma, usually diagnosed at an advanced stage and posing serious therapeutic difficulties due to its anatomical location and the potential for early lymphatic and hematogenous dissemination.363
The main biomarkers for liquid biopsy in UTUC are ctDNA, miRNA, protein, etc. For ctDNA, the main focus is on its fragment size, mutation, and methylation. Plasma cfDNA fragment size correlates with UTUC and may be helpful in the diagnosis of UTUC (AUC: 0.72).364 Monitoring of ctDNA mutations and methylation in urine has revealed that both can diagnose UTUC (sensitivity: 96%, specificity: 88%),365–367 and predict the prognosis of the patients(sensitivity: 86.5%, specificity: 94.7%).368–370 Moreover, the combined detection of methylation and mutation can better monitor UTUC with higher sensitivity and specificity (sensitivity: 94.0%, specificity: 93.1%, AUC: 0.96).371 Multiple miRNAs have been identified as diagnostic markers for UTUC, and some miRNAs were found to be predictive of UTUC prognosis.372–374 For example, miR-151b was able to differentiate between two groups of UTUC patients with significant differences in tumor progression probability (p = 0.006) and cancer-specific survival probability (p = 0.034).374 In addition, a variety of proteins in plasma and urine may be useful for the detection of UTUC. For example, plasma phosphorylated protein 1 and urine FXYD3 can effectively identify patients with early UTUC, facilitating rapid screening for UC.375,376 Also, the survival prognosis of UTUC patients can be predicted by proteins, such as albumin-globulin ratio (AGR) and hemoglobin levels.377 Certain proteins, such as serum iron-regulated proteins and GDF-15 levels, have been associated with the progression and invasion of UTUC.378
Circulatory system
In the circulatory system, we will mainly discuss the application of liquid biopsies in myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML), lymphomas, and multiple myeloma (MM) (Table 10).
Table 10.
Liquid biopsy in circulatory system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| MDS/AML | ctDNA | Plasma | up | Disease progression biomarker | 382 | |
| ctDNA | Serum | up | Prognostic biomarker | 389 | ||
| cfDNA | Plasma | up | Disease progression biomarker | 383 | ||
| cfDNA mutation | Plasma, Serum | up | Disease progression biomarker | 385 | ||
| ctDNA methylation | Peripheral blood | up | Early diagnostic biomarker, Efficacy monitoring biomarker | 384 | ||
| Lymphoma | ctDNA | Serum | up | Disease progression biomarker, Efficacy monitoring biomarker | 392 | |
| ctDNA mutation | Peripheral blood | up | Disease progression biomarker | 395 | ||
| ctDNA | Peripheral blood | up | Early diagnostic biomarker, Efficacy monitoring biomarker | 396 | ||
| ctDNA mutation | Peripheral blood | up | Efficacy monitoring biomarker | 397 | ||
| ctDNA | Plasma | up | Efficacy monitoring biomarker | 399 | ||
| MM | CTCs | Peripheral blood | up | Early diagnostic biomarker | 403 | |
| ctDNA | Plasma | up | Disease progression biomarker | 407 | ||
| cfDNA | Plasma | up | Early diagnostic biomarker | 408 | ||
| ctDNA mutation | Serum | up | Disease progression biomarker | 405 | ||
| FGFR3, KMT2C, MAML2, ZFHX4 mutation | Peripheral blood | up | Efficacy monitoring biomarker | 409 |
Myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML)
Because of the analogous mutation profiles of MDS and AML genes and the characteristic transformation of MDS to AML, which occurs in ~30% of MDS, we have discussed MDS/AML jointly when discussing liquid biopsies of circulatory tumors. The reason why ctDNA is able to dominate liquid biopsies of MDS/AML is due to the influence of the following conditions. There are multiple genetic mutations in MDS/AML, and the correlation between ctDNA in the blood and the variants detected by bone marrow biopsy is high.379,380 In the plasma of AML patients, sequence variants and copy number variants of ctDNA are highly consistent with the results of the bone marrow biopsy.379,381 Also, due to the inherent heterogeneity of the tumors, the full information about the tumors may not be obtained at the time of the bone marrow puncture. The ctDNA test can be used to detect chromosomal aberrations and to detect the course of the disease in patients with MDS.382
Studies have shown that the concentration and integrity of DNA in patients with acute leukemia are higher than in healthy controls, and that relapse is associated with a significant increase in DNA integrity, so plasma DNA integrity may be a potential biomarker for detecting leukemia progression.383 Methylation of ctDNA has also received attention. AML can be reliably distinguished from the healthy population by detecting ctDNA methylation in the peripheral blood of AML patients.384
In addition, ctDNA mutations have been associated with the detection of therapeutic response. ctDNA in the blood of patients with MDS dynamically responds to the tumor load during treatment and demonstrates mutations and karyotypic abnormalities in MDS. Thus, ctDNA may respond to dynamic changes in myelogenetic abnormalities.385 Several studies have synthesized and discussed the detection of ctDNA in the plasma of patients with MDS and AML.386,387 After demethylation therapy or chemotherapy, ctDNA was detected in the peripheral blood of patients in complete remission/complete rheological recovery (CR/CRi) with a lower mean number of VAFs, and mutations were negatively correlated with longer progression-free survival (PFS) and overall survival (OS).388 And ctDNA methylation has been shown to correlate with tumor status after treatment. After treatment with azacitidine, AML patients showed a rapid decrease in peripheral blood ctDNA methylation levels.384
And for tumor recurrence, the rate of recurrence is higher in ctDNA-positive patients than in negative patients.389 As MDS progresses to AML, mutations in ctDNA can be detected in plasma earlier than changes in cell morphology.388
Lymphoma
Diffuse large B-cell lymphoma is the most common lymphoma and is an aggressive heterogeneous lymphoma. Multiple studies have shown that ctDNA can detect lymphoma-associated genetic alterations with 95% concordance with tissue biopsy results.390,391 CtDNA has been shown to be useful for disease progression and predicts progression (positive predictive value of 88.2% and negative predictive value of 97.8%) earlier than conventional imaging.392 CtDNA mutations in plasma are significantly downregulated after treatment,392,393 and their mutations are associated with event-free survival (EFS) and OS.394 To further improve the sensitivity of the analysis, ctDNA detection methods are being actively investigated, including phase variant enrichment and detection sequencing, CAPP-seq, and others.391,393,395 In Hodgkin’s lymphoma (HL), it has been demonstrated that ctDNA levels in HL plasma are higher than in the healthy population,396 and have correlated with radiologically detected findings, i.e., tumor volume.397,398 Its plasma ctDNA has been determined to be a reliable source of tumor DNA for identifying mutations in HL.397,399 During the research process of ctDNA mutations, it has been found that ctDNA mutation correlates with efficacy testing and prognosis of various therapies, including chemotherapy, immunotherapy, etc.399–401 Plasma ctDNA concentration before treatment could independently predict clinical outcome, and patients with particularly poor prognosis after radical immunochemotherapy could be identified by plasma ctDNA monitoring during treatment.400 In combination with PET/CT, ctDNA levels were found to correlate with total metabolic tumor volume detected by PET/CT,398 and ctDNA values were correlated with disease progression and survival.399 Therefore, the mutual complementarity of the two assays is conducive to a more accurate determination of treatment efficacy and risk of recurrence, etc.
Multiple myeloma (MM)
MM is characterized by an intricate array of genetic and epigenetic alterations that result in the malignant conversion of plasma cells. It is a hematologic disease that cannot be cured and exhibits significant variation in both space and time.402 Detection of CTCs in the serum of MM patients revealed that CTCs were higher in MM patients. Mutation detection of CTCs showed good agreement in the degree of mutation matching between MM cells in BM and CTCs in blood, and was 95% concordance in copy number alterations at the chromosome arm lFel.403,404 And in extramedullary (EM) plasmacytoma samples, an 87% concordance was found between the mutational profiles of EM tumor cells and CTCs, with the highly concordant mutations suggesting that CTCs may be responsible for the development of EM.403 Despite the good concordance of mutations in CTCs and MM cells, there are still some inconsistencies, which may indicate that a combination of the two tests may be more helpful in the diagnosis of MM. CfDNA is the most commonly used marker for MM liquid biopsy. It was found that changes in ctDNA levels preceded those of other serum markers, such as FLC.405,406 Therefore, ctDNA may be an earlier predictor of disease progression like CTCs. In addition, ctDNA can also be used to detect the progression of MM. Firstly, ctDNA levels were detected to show a sustained elevation during MM progression and the number was correlated with tumor load parameters such as the percentage of infiltration with bone marrow plasma cells.407,408 Moreover, serial analysis of the mutational status of cfDNA helps to assess the efficacy of treatment, and because EM is difficult to perform routine biopsies, mutation detection of ctDNA may be more of an examination advantage in the diagnosis as well as prognostic assessment of patients with EM.407 Studies have been conducted on cfDNA mutations and drug therapy, and it was found that ctDNA mutations such as DIS3, FGFR3, KMT2C, MAML2, and ZFHX4 mutations may predict resistance to certain therapies.409 Recently, a study demonstrated that the detection of ctDNA mutations to select the corresponding selective inhibitors for treatment has promising efficacy,410,411 further suggesting that liquid biopsy has great potential for use in the diagnosis and treatment of MM.
Endocrine system
The application of liquid biopsy in endocrine system tumors mainly includes thyroid cancer (TC) and breast cancer (BRCA) (Table 11).
Table 11.
Liquid biopsy in endocrine system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| TC | CTCs | Peripheral blood | up | Early diagnostic biomarker | 421 | |
| CTCs | Peripheral blood | up | Efficacy monitoring biomarker | 422 | ||
| ctDNA mutation | Plasma | up | Efficacy monitoring biomarker | 425 | ||
| ctDNA | Plasma | up | Disease progression biomarker | 424 | ||
| BRAFV600E mutation | Plasma | up | Efficacy monitoring biomarker | 426 | ||
| RET M918T mutation | Plasma | up | Prognostic biomarker | 427 | ||
| BRAF(T1799A) mutation | Plasma | up | Disease progression biomarker | 428 | ||
| PIK3CA mutation | Peripheral blood | up | Prognostic biomarker | 429 | ||
| SLC5A8, SLC26A4 methylation | Plasma | up | BRAF | Early diagnostic biomarker | 418 | |
| ctDNA methylation | Serum | up | Early diagnostic biomarker, Tumor recurrence biomarker | 430 | ||
| miR-146b-5p, miR-21a-5p | Plasma exosome | up | Early diagnostic biomarker | 420 | ||
| miR-29a | Serum exosome | down | Early diagnostic biomarker, Prognostic biomarker | 431 | ||
| BRCA | CTCs | Peripheral blood | up | Prognostic biomarker, Efficacy monitoring biomarker | 452 | |
| TP53, PIK3CA, ESR1 mutation | Plasma | up | Efficacy monitoring biomarker | 436 | ||
| ctDNA mutation | Plasma | up | Efficacy monitoring biomarker, Tumor recurrence biomarker | 437 | ||
| PIK3CA mutation | Plasma | up | Early diagnostic biomarker, Efficacy monitoring biomarker, Tumor recurrence biomarker | 438 | ||
| miR-1246, miR-21 | Plasma exosome | up | Early diagnostic biomarker | 441 | ||
| miR-101, miR-372 | Serum exosome | up | Early diagnostic biomarker | 442 | ||
| miR-106a-3p, miR-106a-5p, miR-20b-5p, miR-92a-2-5p | Serum exosome, Plasma exosome | up | Early diagnostic biomarker | 443 |
Thyroid cancer (TC)
TC is the most common endocrine malignancy. The majority of this tumor originates from epithelial tissue and includes differentiated thyroid carcinoma (DTC), poorly differentiated thyroid carcinoma (PDTC), anaplastic thyroid cancer (ATC).412 The differentiated type of TC, which usually exhibits an indolent clinical behavior and has a good prognosis, can be further classified into papillary thyroid carcinoma (PTC) (85–90%), follicular thyroid carcinoma (FTC) (5–10%), and Hürthle cell carcinoma (HCTC) (3%) cancers.413,414 The incidence of TC has been increasing globally over the past 30 years.412 Currently, rapidly evolving liquid biopsy techniques offer unique advantages in the diagnosis and prognostic testing of this disease. The following section focuses on liquid biopsy techniques related to TC.
First, for the detection of CTCs, in addition to enrichment based on its physical properties, immunoaffinity-based enrichment methods are now more commonly used, including assays for anti-EpCAM, tumor-specific cell surface antigens, cytokeratins (CKs), and other stem cell or mesenchymal markers.415,416 Of these, the CellSearch® method is currently the only method that is method authorized by the FDA to monitor the number of CTCs in blood samples.34 It has been shown that the number of CTCs is significantly increased in patients with DTC and that their number is proportional to the tumor stage at diagnosis, suggesting that CTCs may have a strong correlation with this tumor. CTCs values were significantly higher in TC patients compared to controls and the number of CTCs correlated with initial tumor stage.417 In addition to this, methylation of ctDNA has been recognized as a promising biomarker. One study found that the SLC5A8 and SLC26A4 genes have higher methylation expression in patients with TC.418 It has been shown that exosomal miRNAs are promising diagnostic markers for TC, being more resistant to the proteolytic activity of RNAases and more stable than free miRNAs in body fluids.419 Therefore, the detection of miRNAs in the exosomes seems to be more relevant. It was found that miR-146b-5p and miR-21a-5p were significantly elevated in the cellular exosomes of PTC patients compared to patients with benign multinodular disease, but no significant difference was found when free miRNAs in the blood were analyzed.420 This further demonstrates the superiority of exosomal miRNA detection testing for early diagnosis of TC.
Also, various liquid biopsy markers can be used to detect TC progression as well as treatment prognosis. And in the FTC typing of DTC, a research team found that CTCs was more common in malignant patients,421 while all benign patients were negative (specificity 100%, sensitivity 46%). So, CTCs may also be used as an indicator to judge the benignity or malignancy of a tumor. Furthermore, in a study by Qiu et al., it was found that metastasis was more likely to occur when the number of isolated CTCs was 5 or more (sensitivity: 64.3%, specificity: 83.8%), whereas isolation of seven or more CTCs predicted a poor prognosis in response to radioiodine treatment (sensitivity: 73.7%, specificity: 69.6%).422 Some studies have found that in the metastatic PTC patients mutations in RET and BRAF genes are more common and predict poor prognosis.423 Also, mutations in NRAS and TP53 were found to possibly accelerate tumor progression in DTC and ATC patients.424
In addition, ctDNA is significantly better than other imaging tests and protein marker tests in predicting the prognosis of TC. Several studies have shown that the detection rate of specific gene mutations in cfDNA is associated with overall survival and poor prognosis of TC, such as PIK3CA, BRAFV600E, RETM918T, and BRAFT1799A.424–428 Mutations in cfPIK3CA are also associated with poor prognosis in patients with ATC.429 Another study found that patients with recurrent TC have a higher positive rate (70%) of serum DNA methylation.430 Similarly, one study testing CTCs in patients with medullary thyroid cancer (MTC) both preoperatively and postoperatively, there were similar conclusions, which suggest that CTCs may be a good prognostic monitoring indicator.425 The same can be said for serum exosomes in predicting patient prognosis. One study found that PTC patients with high serum exosomal miR-29a levels had significantly greater OS and RFS than those with low exosomal miR-29a expression levels, suggesting that exosomal miR-29a levels may be associated with PTC recurrence.431 In addition, miR-146b, and miR-222 have also been shown to be potential markers of PTC recurrence.432 And a large number of miRNAs have also been found and reported, such as miR-16-2-3p, miR-223-5p, miR-130a-3p, miR182-5p, etc. have expressed their advantages in different aspects, which can provide some references for the diagnosis of clinical diseases.433
Breast cancer (BRCA)
Breast cancer (BRCA) is one of the most common malignant tumors among women in China, and its incidence rate has long been in the first place with the leading cause of tumor death among middle-aged women.434 Early screening and early diagnosis of this disease are extremely important because early metastatic cases are curable, while distant metastatic cases are currently considered incurable. Tissue biopsy and immunohistochemistry, the gold standard techniques for conventional BRCA treatment, have limited detection rates because of the high heterogeneity of the tumor, while liquid biopsy has unique advantages in this area, which provides sustainable and personalized medical treatment for patients.435
CtDNA has become a hotspot for scientists because of its high ability to assess tumor heterogeneity.435 Point mutations are one of the more common types of mutations in ctDNA, with genes such as TP53, PIK3CA, and ESR1 being the hotspot mutated genes in BRCA identified to date.436,437 By evaluating PIK3CA mutations in the plasma of BRCA patients, Beaver et al. found that the sensitivity of this method for detecting early BC was 93.3%, and the specificity of this gene reached 100%.438 Meanwhile the gene has been approved as a biomarker for the Pl3K inhibitor alpelisib in Europe and the United States.439 In addition to this, ctDNA methylation can also be used for early diagnosis as well as differentiation of breast cancer. A study has shown that the detection of ctDNA methylation biomarkers is highly accurate in the early diagnosis of breast cancer patients (AUC: 0.889, sensitivity: 100%, specificity: 75%).440 It has been reported that the expression levels of plasma exosomes miR-21(AUC:0.69), miR-1246(AUC:0.69), and serum exosomes miR-101, and miR-372 are significantly elevated in BRCA patients, suggesting that the above exosomal miRNAs could be used as a potential biomarker for the diagnosis of early-stage BRCA.441,442 By comparing the expression levels of circulating exosomal miRNAs in blood samples from 32 healthy volunteers and 32 BRCA patients, Li et al. found that the levels of four exosomal miRNAs (miR-106a-5p, miR-19b-3p, miR-20b-5p, and miR-92a-3p) were significantly elevated in sera of BRCA patients (sensitivity: 87%, specificity: 89, AUC: 0.937), and found that three plasma-derived exosomal miRNAs (miR-106a-3p, miR-106a-5p and miR-92a-2-5p) levels were elevated compared to healthy volunteers(sensitivity:82%, specificity:79%, AUC:0.889).443 It is suggested that the above seven circulating exosomal miRNAs are expected to be biomarkers for early diagnosis of BRCA. Among them, the expression of miR-106a-5p was higher than that of healthy volunteers both in plasma and serum, suggesting that exosomal miR-106a-5p has the potential to be a specific biomarker for early diagnosis of BRCA. In a recent study, a droplet digital ExoELISA method was developed for the detection of GPC-1(+) exosomes in clinical samples from healthy individuals, patients with benign breast cancer, patients with breast cancer, and post-breast cancer patients. The microtitre digital ExoELISA method demonstrated unprecedented accuracy and high specificity in exosome quantification with a detection limit of at least 10 exosomes per microliter. The results showed significantly higher GPC-1 expression in tumor-derived exosomes compared to normal and benign breast disease samples, and higher levels of GPC-1(+) exosomes in breast cancer patients than in healthy controls and patients with benign breast disease.444 For CTCs detection, the most effective CTCs test for BRCA is still the FDA-approved CellSearch® method.445 This method is an automated immunomagnetic enrichment method based on the EpCAM, which has a high degree of sensitivity and specificity. In addition, chip-based microfluidic methods are currently being developed.446 The main principle is to retain and isolate CTCs based on their different sizes and deformability from other blood components as blood flows within a microfluidic chip.447 It has been reported that the number of CTCs is positively correlated with the tumor stage,448 and that when CTCs are counted at a higher number, the worse the prognosis of the patient is,30 and the more prone to metastasis, which suggests that CTCs is a good prognostic indicator. According to the NCCN Clinical Practice Guidelines in Oncology, Version 3.2022, patients who have continuously increased CTCs levels following three weeks of first-line treatment have worse OS and PFS.449 A research team dynamically monitored ctDNA, CTCs, and CA15-3 levels in 30 patients with advanced BRCA. The results showed that changes in patient-specific ctDNA levels during treatment correlated most strongly with efficacy, and had higher sensitivity (90%) compared with CA15-3 and CTCs.450 Moreover, increased ctDNA levels indicated disease progression on average 5 months earlier than clinical imaging.450 In addition, a variety of exosomal miRNAs are associated with an increased risk of BRCA development and shorter survival. In addition, it has been found that genes such as miR-23a, miR-5100, miR-19b-3p, and miR-21 are involved in the process of EMT,451 which is also of clinical significance for the prognostic detection of tumors. We can also detect the number of CTCs in patients after taking treatment to reflect the clinical efficacy. For example, Nakamura et al. analyzed the relationship between the effect of chemotherapy and the change in the number of CTCs in metastatic BRCA and found that after 1 cycle of chemotherapy, the number of CTCs decreased by more than 90% compared with the number of CTCs before chemotherapy, 85.7% of the patients were in complete remission or partial remission; and for the patients who did not have a decrease or even an increase in the number of CTCs after chemotherapy, there were 63.6% of the patients who had progression of the disease.452 According to the Chinese Society of Clinical Oncology’s (CSCO) Breast Cancer Guidelines 2022, CTCs can partially mimic solid tumors and be employed in addition to genetic sequencing, pathological diagnosis, and disease surveillance.453 For patients with advanced BRCA, ctDNA is a sensitive and specific biomarker for monitoring tumor load, and changes in its levels suggest changes in tumor load and thus reflect drug efficacy. Although ctDNA has made large and substantial advances in BRCA and the applications of ctDNA are expanding,454 the information provided by ctDNA is usually limited to the assessment of disease burden and the presence or absence of genomic mutations. Further applications and tests remain to be explored.
In addition, lncRNAs and circRNAs have the potential to be diagnostic and prognostic liquid biopsy biomarkers for BRCA. LncRNAs can regulate transcription by binding to enhancer regions. LncRNAs are also involved in the binding of HOTAIR genes to the histone modification complexes PRC2 and LSD1, promoting H3K27 histone methylation and H3K4 demethylation, leading to target gene shutdown and promoting BRCA metastasis.455 As for circRNAs, their high stability, abundance, and specific expression make them have considerable clinical potential as new biomarkers. They are mainly involved in the regulation of cell survival, proliferation, and invasion through the MAPK/AKT signaling pathway. Some studies have found that in tumors hsa_circ_0017650 and hsa_circ_0017536 are less expressed in tumor tissues, and it is hypothesized that these two circRNAs may have a tumor suppressor effect in BRCA.456 In addition to the above conventional liquid biopsy techniques, other biomarkers for different manifestations of tumors are also developing, such as exosomal phosphorylated proteins, DNA methylation products, etc. The most appropriate liquid biopsy method for different manifestations of tumors can be adopted in order to provide an early reference for the diagnosis of the disease.
Locomotor system
Primary malignant bone tumors have a high morbidity and mortality rate in children and adolescents, so accurate and effective screening and diagnostic tools are particularly important. However, the invasive nature of routine tissue biopsy makes it impossible to repeat the sampling during patient prognostic monitoring, and patients are unable to receive sustainable precision medicine. Liquid biopsy, which is currently in the spotlight, overcomes the shortcomings of tissue biopsy and provides a new way of thinking for the diagnosis and prognostic monitoring of malignant tumors. Next, we mainly focus on several common bone tumors, including osteosarcoma (OS), chondrosarcoma (CS), and ewing’s sarcoma (ES), to provide a brief introduction to liquid biopsy techniques. However, since studies related to osteosarcoma and chondrosarcoma are also extremely limited, this section focuses on ewing’s sarcoma (Table 12).
Table 12.
Liquid biopsy in motor system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| EWSR1 Translocation | Plasma | up | Efficacy monitoring biomarker | 460 | ||
| ctDNA | Plasma | up | Prognostic biomarker | 462 | ||
| EWSR1 fusion sequence | Plasma | up | Efficacy monitoring biomarker | 458 | ||
| miR-34b | Plasma | down | Tumor aggressiveness biomarker | 472 | ||
| miR-143/145 | Plasma | down | FSCN1 | Early diagnostic biomarker | 473 | |
| miR34a | Plasma | down | Disease progression biomarker, Efficacy monitoring biomarker | 475 |
For bone tumors of mesenchymal origin, the ability of conventional assays to detect CTCs is extremely limited due to the obvious tumor heterogeneity and the lack of classical tumor markers, and there is also a lack of relevant studies. However, in a recent study, a team confirmed the presence of CTCs in ES patients based on the immunodetachment of CD99+ tumor cells and magnetic beads, followed by molecular analysis to detect specific fusion transcripts from chromosomal translocations,457 which provides a new way of detecting CTCs. Nevertheless, the potential clinical significance of CTCs is unclear and needs to be further explored. In patients with ES, the expression of CD99+ and the presence of chromosomal translocations are two of the most important features of the tumor.457 In particular, the detection of fusion genes such as EWSR1-FLI1, which is frequently present, is the gold standard for the diagnosis of ES and can be detected using genomic fusion sequences.458
More studies focus on detection of tumor progression and prediction of prognosis. The detection of ctDNA also has certain clinical significance, as some researchers have analyzed the ctDNA levels of IDH1 mutants in preoperative and postoperative patients with CS and found that the levels of ctDNA were correlated with different tumor grades, and a significant decrease in ctDNA levels was found in most patients who underwent surgical resection.459 While in OS patients, ctDNA levels also underwent similar changes after treatment.460,461 In addition, an increase in chromosome 8q has been observed in the ctDNA of patients with OS, which may be associated with a poorer prognosis.462 In addition, tumor cell exosomes have received much attention because they carry some of the functional proteins and genes of their “parent” tumor cells and may also play an important role in the pathogenesis, diagnosis, and treatment of primary bone tumors. Micro-fractionated ultracentrifugation is one of the most widely used methods in routine assays and is also a gold standard for exosome isolation.463 In the past few years, many new integrated microfluidic platforms have been developed for analyzing exosome levels, quantifying disease-specific subpopulations, and characterizing exosomal proteins and RNA at the histological level.464–467 Compared to conventional methods, emerging microfluidic platforms significantly reduce sample volume, reagent consumption, and separation time, while greatly improving separation recoveries and exosome quality levels for higher specificity.468,469 However, research in this area is still in its infancy and has great potential for development. Especially for osteosarcoma (OS), miRNA is highly suggestive of disease staging, metastasis, and therapeutic efficacy.470 As well, most of the studies to date have focused mainly on miRNA of OS, followed by CS and ES, for example, it was found that high expression of miR-135b, miR-150, miR-542-5p, and miR-652 may be associated with the onset and progression of OS.471 While down-regulation of miR-34b may be associated with metastasis of tumors in OS patients. Compared with non-metastatic patients, miR-34b plasma expression levels were significantly lower in metastatic patients.472 In CS, multiple miRNAs such as miR-20, miR-96, miR-100, miR-125b, miR-136 and other genes have significantly altered expression levels in both cell lines and tumor samples.473 And in a recent study, 17 key miRNAs were found to be involved in regulating the formation and growth of chondrosarcoma.474 Also, miR34a was found to be a potential biomarker for the development of ES.475 And miR-185 was shown to be involved in the formation and survival of ES cells.476
Reproductive system
In this section, we focus on the application of liquid biopsy in the following five reproductive system tumors, including cervical cancer (CC), endometrial carcinoma (EC), ovarian cancer (OC), prostate cancer (PCa), seminoma (Table 13).
Table 13.
Liquid biopsy in reproductive system cancers
| Cancer | Liquid biomarker | Origin | Tendency | Downstream target | Function | Reference |
|---|---|---|---|---|---|---|
| Cervical cancer | CTCs | Peripheral blood | up | Prognostic biomarker | 490 | |
| ccfHPV-DNA | Plasma | Efficacy monitoring biomarker | 478 | |||
| HOTAIR, PVT1, XLOC_000303, AL592284.1 | Plasma | up | Early diagnostic biomarker | 482 | ||
| miR-21, -25, -29a, -200a, -486-5p | Serum | up | Early diagnostic biomarker, Disease progression biomarker | 483 | ||
| miR-196a | Serum | up | Disease progression biomarker, Prognostic biomarker | 484 | ||
| miR-425-5p | Serum | up | Prognostic biomarker | 485 | ||
| ESR1, ERBB2 mutation | Plasma | up | Efficacy monitoring biomarker | 486 | ||
| Endometrial cancer | CTCs | Peripheral blood | up | Early diagnostic biomarker | 498 | |
| CK-20 | Peripheral blood | up | Tumor aggressiveness biomarker, Tumor recurrence biomarker | 550 | ||
| CTNNB1, KRAS, PTEN, PIK3CA | Plasma | up | Tumor recurrence biomarker, Efficacy monitoring biomarker | 500 | ||
| DNA methylation | Urine | up | Early diagnostic biomarker | 509 | ||
| Ovarian Cancer | claudin | Serum exosome | up | Early diagnostic biomarker | 523 | |
| miR-1307, miR-375 | Serum exosome | up | Early diagnostic biomarker | 524 | ||
| Prostatic carcinoma | CTCs | Peripheral blood | up | Prognostic biomarker | 530 | |
| cfDNA mutation | Serum | up | Early diagnostic biomarker | 531 | ||
| cfDNA | Plasma | up | Efficacy monitoring biomarker, Prognostic biomarker | 532 | ||
| miR-21 | Serum | up | Efficacy monitoring biomarker | 538 | ||
| miR-141, miR-146b-3p, miR-194 | Serum | up | Prognostic biomarker | 539 |
Cervical cancer (CC)
Cervical cancer (CC) is the fourth most common cancer among women worldwide, and HPV infection is the main cause of CC patients. Conventional screening and diagnostic tools are easily rejected because of their invasiveness, and as a non-invasive test, liquid biopsy may be an alternative and complementary tool to conventional screening and diagnosis. The following is a brief introduction to several common liquid biopsy methods.477
As an extremely important part of liquid biopsy, ctDNA is also clinically significant in patients with CC. Plasma ctDNA levels are significantly higher in patients with CC than in healthy controls and are strongly correlated with FIGO tumor stage, histologic grading, depth of infiltration, and lymphatic metastasis.478 Multiple studies have confirmed the potential use of this assay in clinical practice.479 And because of the high association between CC and human papillomavirus (HPV), it is also feasible to screen by detecting HPV cfDNA in the blood. In the detection of circulating HPV cfDNA, a magnetic bead-based HPV genotyping assay (E7-MPG) is an alternative method that is more accurate and significantly more sensitive (96.1%) than conventional dPCR.480 On the other hand, non-coding RNAs also have important roles in CC development and contribute to the early diagnosis of CC.481 Among them, lncRNAs and miRNAs have the most important roles. As a kind of lncRNA, HOX transcript antisense intragenic RNA (HOTAIR) is highly expressed in CC patients and promotes the proliferation and migration of tumor cells. Its combination with three other lncRNA (i.e., PVT1, AL592284.1, and XLOC_000303) significantly increased the positive predictive value (88%) and negative predictive value (84%) of CC.482 Meanwhile, combined testing of multiple miRNAs is often practiced in CC patients and can be more effective than individual testing. Jia et al. identified five serum miRNAs (i.e., miR-21, −25, −29a, −200a, and −486-5p) based on genome-wide miRNA sequencing and quantitative PCR (qPCR) validation, and the combination of these tests can differentiate CC patients from healthy controls.483
Moreover, studies have shown that genes such as miR-196a, miR-425-5p, and others have also been shown to be associated with the proliferation and migration of tumor cells in CC.484,485 In addition, another strategy for implementing liquid biopsy in CC detection involves the identification of somatic nucleotide variants (SNVs) in cancer driver genes.486–488 Tian et al. used an allelic fraction deviation (AFD) algorithm for evaluation and found that the value of AFD was positively correlated with the prognostic degree of patients.489 Initially, a phase III randomized clinical trial demonstrated that CTCs counts can be used as a predictive biomarker to guide the treatment of cervical cancer.490 Current strategies for the detection and isolation of CTCs in CC similarly rely on the physical and morphological characterization of the cells as well as on the identification and quantification of HPV oncogenes and epithelial markers through the use of molecular and/or immunofluorescence procedures.491–493 Compared to the detection of other solid tumors, there are fewer techniques related to the detection of CTCs in CC, and these methods have not yet been recognized.494
Endometrial carcinoma (EC)
Endometrial carcinoma (EC) is the most common cancer of the female reproductive tract, and its incidence is increasing year by year,495,496 which seriously affects the quality of life for female patients. Presently, there are no diagnostic techniques available for detecting EC in the general population. Endometrial sample carries the risk of causing discomfort, bleeding, infection, and uterine perforation. Additionally, in up to 25% of cases, a biopsy may not provide enough information to make a diagnosis.497Therefore, as an alternative sampling method to traditional tissue biopsy, liquid biopsy has been a boon for early diagnosis in female patients due to its non-invasive nature. In the following, we provide a brief introduction to liquid biopsy methods mainly from the aspects of CTCs, ctDNA, miRNA, and extracellular vesicles.
First, CTCs were also found in patients with endometrial cancer and there was a high correlation between CTCs and EC.498 The detection rate of CTCs in the peripheral blood of patients with EC was 7–75%.499 This discrepancy may be related to the population characteristics, CTCs detection techniques, and the number of patients studied. Furthermore, ctDNA is clinically important for EC patients. It has been found that at least one ctDNA mutation can be detected in the peripheral blood of 94% EC patients, which mainly occurs in the CTNNB1, KRAS, PTEN, and PIK3CA genes.500 In a clinical trial, it was found that miRNA expression levels also differed between healthy adults and EC patients, such as miR-15b(AUC: 0.768), miR-27a(AUC: 0.813), and miR-223(AUC: 0.768) were differentially expressed between endometrial cancer patients and healthy individuals, which is important for improving the diagnosis of endometrial cancer.501Another study showed that CTCs were detected in patients with stage III and IV EC or in close proximity to the tumor, but not in patients with early-stage or recurrence, so the prognostic significance of CTCs for patients with EC is still controversial.502
And the number of ctDNAs found in the blood increases with the progression of the disease.503 Moreover, it has been demonstrated that arch-related mutations (DNMT3A and TET2 genes) may be associated with poor prognosis in endometrial cancer patients, and DNMT3A mutations are more likely to be detected in EC patients in particular.504 The simple method of extraction and the ability to store for a long time prior to analysis make the detection of ctDNAs expected to be a potential marker for the diagnosis of EC. Moreover, a variety of miRNAs have been associated with EC tumorigenesis, invasion, and metastasis. miR-183-5p, miR-429, and miR-146a-5p expression were found to be up-regulated by liquid biopsy examination of saline lavage fluid from patients with endometrial cancer after saline infusion ultrasound intrauterine scintigraphy (SIS) procedure, whereas miR-296-5p and miR-204-5p were decreased.505 In addition, studies of EVs have found that elevated levels of membrane-bound protein A2 in EVs correlate with high-risk histology, grading, staging, and recurrence risk of EC, suggesting that it may have a role to play in disease surveillance, recurrence, and early disease detection.506
In addition to the more mainstream liquid biopsy techniques described above, additional novel assays for the detection of liquid biopsies continue to be developed. For example, for the detection of messenger RNA molecules (mRNA) in plasma samples.507 However, relatively few studies have been conducted on this substance, probably due to its poor stability, easy degradation, and low levels of circulation, which makes it more difficult to detect and analyze.508 The detection of DNA methylation in urine may also provide an attractive noninvasive testing strategy for early screening of asymptomatic EC patients.509
Ovarian cancer (OC)
Because there are no noticeable symptoms in the initial phases of the disease and no practical tests to detect it, almost 70% of patients with ovarian cancer (OC) are diagnosed in the advanced stages (Stages III and IV). Additionally, biomarkers like CA-125, which are specific to OC, are not sensitive or specific enough for regular screening.510 Thus, it is crucial to find new OC liquid biopsy markers.
The first is for classical CTCs, which are detected in OC patients by microbeads covered with the epithelial marker MOC31 or a mixture of cytokeratin and epidermal growth factor receptor (EGFR).511,512 The FDA approved the CellSearch® system (Menarini Silicon system, Italy) as the routine gold standard platform for CTCs isolation in clinical practice. Secondly, for ctDNA, the main methods currently used to identify ctDNA in the blood of OC patients are quantitative PCR, ddPCR, whole genome sequencing, and next-generation sequencing, which identify qualitative and quantitative alterations of ctDNA, such as gene fusions, aberrant DNA methylation, tumor-specific variants (TSV), copy number variants, and chromosomal instability.513 Up to now, most ctDNA identification techniques have focused on TP53 mutations in patients with high-grade plasmacytoid ovarian cancer (HGSOC).514 The use of ctDNA in OC reflects tumor heterogeneity more accurately than other assays and its shorter half-life makes it more precise compared to CA-125.515,516 However, the accuracy of ctDNA samples may be affected by its short half-life and low abundance in torrent blood (<0.5% of total cfDNA). Therefore, ctDNA analysis requires higher sensitivity techniques to minimize false negatives.517 On the other hand, in OC patients miRNAs are synthesized and activated faster than mRNAs and proteins, with a longer half-life.518 Therefore, miRNA may be more suitable for early OC detection.519 Several scholars have conducted studies on this topic, and in general, the current study found that lower overall survival in OC patients was mostly associated with the up-regulation of miR-21, miR-221, miR-141, and miR-429 and downregulation of miR-200c, miR-1290, miR-145, miR-199a, and miR-148a.520 In most of the studies, miRNA microarrays or NGS have been used to evaluate the miRNAs isolated from patients with ovarian cancer. In contrast to NGS, microarrays are more efficient and less cost-effective, but NGS has the potential to recognize novel miRNAs.507 In addition to miRNAs, lncRNAs and circRNAs have the potential to be diagnostic and prognostic liquid biopsy biomarkers of OC.513 CircRNAs are more stable in the peripheral circulation due to their specific covalent closure structure that makes them more resistant to destruction by RNase.521 It has been found that the expression of circRNAs in the differences between primary and metastatic sites of OC, may be associated with OC progression. As for lncRNAs, there are only partial data suggesting that clinical progression in OC patients is associated with lncRNA expression levels (XIST, H19, LSINCT5, AB073614, HOTAIR, CCAT2, and ANRIL). The diagnostic sensitivity and specificity of lncRNAs in these individuals have not yet been fully established, and no lncRNAs are licensed for therapeutic use, as their specific feasibility remains to be investigated.507 In addition, it was reported that the total exosome concentration was elevated in serum samples of OC patients, and exosomes of OC patients can carry a large number of miRNAs, therefore, exosomal proteins and miRNAs are the main indicators of exosomes and OC-related studies.522 Exosomal secreted proteins can be used as predictive or diagnostic indicators of ovarian cancer, e.g., it is seen in plasma and circulating exosomes of patients with OC overexpression of claudin-4 and can be used to monitor tumor progression.523 And the combined detection of exosomal miRNAs with the routine serum tumor biomarkers CA125 and HE4 can improve the detection rate of OC.524 In addition to this, thrombocytosis has been associated with increased cancer risk and shorter survival, especially in ovarian cancer. Because tumor cells are able to transcriptionally reprogram TEPs through multiple mechanisms,525 RNA sequencing of TEPs has become the latest component of liquid biopsy for tumor detection. It is also highly specific in the identification of OC and has been validated as a good test in different races and populations.
Prostate cancer (PCa)
Prostate cancer (PCa) is the second leading cause of cancer-related deaths in men, but the survival rate of PC improves significantly after appropriate treatment, so early screening and diagnosis of this disease is of great importance.526 Currently, prostate-specific antigen (PSA) and transrectal ultrasound-guided biopsy are mostly used in clinical practice, but the former is not a specific marker for tumors with low sensitivity and specificity, while the latter often causes rejection due to its invasiveness.527 In contrast, liquid biopsy shows its superiority here, and we will introduce the relevant methods from the following aspects.
Firstly, for the detection of CTCs, in addition to using physical properties, it can also be isolated using its biological properties, such as antibody-antigen interactions.528 Detection of the number of CTCs in the blood by flow cytometry predicts the prognosis of metastatic desmoplasia-resistant prostate cancer (mCRPC). Briefly, it means that when the number of detected CTCs is higher, the tumor load is higher and the survival prognosis of the patient is worse. And high CTCs phenotypic heterogeneity was also associated with poorer survival outcomes in mCRPC. According to Prostate Cancer, Version 4.2023, NCCN Clinical Practice Guidelines in Oncology, AR-V7 expression on CTCs can help CRPC patients treated with abiraterone/enzalutamide make decisions about their next course of treatment.529 In terms of CTCs phenotype, Lindsay et al. demonstrated that Ki67 and vimentin expression in CTCs correlates with poor prognosis in mCRPC.530 Second, plasma ctDNA was found to be a potential clinical marker for the early detection of prostate cancer, and its concentration can come to differentiate between malignant disease and benign hyperplasia of the prostate. Ekkehard Schutz et al. analyzed ctDNA using whole-genome amplification and found differences in the number of ctDNA sequence reads in the 100 kbp interval between patients with PCa and the healthy population.531 Similar to CTCs, ctDNA can be used to assess the prognosis of cachectic-resistant prostate cancer (CRPC). Analysis of 663 plasma samples from 140 patients with CRPC showed that ctDNA was associated with poor survival prognosis.532 In addition to testing for specific ctDNAs, quantitative characterization of ctDNA can be used as a less invasive and more reliable prognostic biomarker, especially for DNA methylation. Hypermethylation of glutathione-s-transferase P1 (GSTP1) is the most common epigenetic alteration in PCa, and methylation-specific PCR (MSPCR) for this substance has high sensitivity and specificity to differentiate between normal and neoplastic states.533 On the other hand, in comparison to the coding genes described above, noncoding RNAs have a unique advantage due to their high tissue and staging specificity for the disease.534 miRNAs, in turn, have become one of the most widely studied small noncoding RNAs due to their remarkable stability in body fluids.535–537 Serum miR-21 has been reported to be a very useful biomarker. Moreover, it was found that serum miR-21 levels were positively correlated with serum PSA levels in patients with hormone-refractory prostate cancer (HRPC).538 Thus, it was concluded that MiR-21 has the potential to serve as a marker and predictor of hormone-refractory disease transformation. Detection of miRNAs in the serum of patients with and without tumor recurrence revealed statistically significant differences in the expression of miR-141, miR-146b-3p, and miR-194 between the recurrence and no recurrence groups (P < 0.05). It was hypothesized that the three had potential as biomarkers for predicting disease progression, as they were elevated in PCa patients who subsequently experienced relapse.539 In addition to this, the clinical significance of some lncRNAs in PCa patients has been gradually emphasized. Among them, a variety of lncRNAs, including PCAT1, PCGEM1, SChLAP1, and PCAT6, have expressed their advantages in different aspects of PCa and so on, which can provide some references for the diagnosis of clinical diseases.540–543 In addition, there are currently major limitations in the development of exosomes in cells due to technological development and associated cost issues. Logozzi et al. found that plasma levels of PSA-expressing EVs were higher in PCa patients than in healthy subjects.544 Del Re found that AR-V7 variants in the RNAs of EVs predicted response to ARSI.545 However, another study suggests that EVs may be less predictive than CTCs, which contain a higher amount of AR-V7.546 Overall, further experimental studies are needed for exosome detection in PCa patients. In addition to routine liquid biopsies, other tests such as protein biomarkers, e.g., PCA3, PSA glycosylation, and DNA methylation biomarkers, etc., are also included. Different tests have different preferences, but they offer great promise for personalized treatment strategies for PCa in the future.
Seminoma
Seminoma is a common malignant tumor in men of reproductive age. However, routine diagnosis is a multi-step process with poor specificity. Liquid biopsy techniques can provide some early warnings to patients. Dora Raos et al. used pyrophosphate sequencing to assess liquid biopsy cfDNA methylation and compared it with samples from healthy volunteers. It was found that cfDNA methylation of OCT3/4, KITLG, and MAGEC2 can be used as potential non-invasive epigenetic biomarkers in liquid biopsies to some extent, but the conclusion still requires further experiments in larger populations. In addition to this, the results of the experiments will inevitably be affected because cancer-specific cfDNA methylation may be masked by cfDNA methylation in healthy cells.547
Conclusion and perspective
Tissue biopsy remains the gold standard for tumor diagnosis due to its high level of laboratory standardization, good consistency of results, relatively stable samples, and high accuracy of results. However, there are some drawbacks to tissue biopsy, such as the fact that it is invasive, so the part with the highest risk of complications cannot be sampled, and it is difficult to repeat the sampling, making it unsuitable for regular testing and treatment evaluation. Additionally, the tumor information obtained is heavily influenced by the heterogeneity of the sample and can only reflect the information of the sampling site, among other things. As a result, exploring new screening modalities is beneficial to patient therapy and prognosis. Liquid biopsy is now undergoing rapid progress, however its application in clinical practice is still limited. Compared to standard examination methods, various advantages have been established, including little invasiveness, low risk, multiple repeat sampling, suitability for dynamic monitoring, and the ability to mitigate the effect of tumor heterogeneity to some extent. However, there are several limitations to liquid biopsy, such as a lack of laboratory standardization, which weakens the consistency of test findings from different laboratories, the high requirements for sample manipulation, the need to increase accuracy, and so on.548
Liquid biopsy holds significant potential for future applications, although it also presents several areas that require enhancement. One key limitation in the practical use of liquid biopsy is the need to isolate, purify, and detect the markers involved in the monitoring process. Hence, it is imperative to prioritize the advancement of novel detection technologies and analysis platforms, with the establishment of standardized operating procedures and unified data analysis, in order to enhance the accuracy of liquid biopsy in future development. Furthermore, it might be attempted to integrate with the swiftly advancing artificial intelligence, which has the potential to become a more effective way of detection. However, in terms of clinical application, it is important to note that a liquid biopsy can only provide information about specific molecules or biomarkers, and cannot fully capture the complex nature of a disease.548 Therefore, liquid biopsy cannot completely replace tissue biopsy. Instead, the two methods work together to offer a more comprehensive understanding of the biological aspects of tumors. In order to implement liquid biopsy in a widespread manner in clinical settings, it is necessary to conduct extensive clinical trials, standardize the processes for enriching the samples, and establish consistent methods for downstream analysis.
Hence, the enhancement of detection technology and the integration of liquid biopsy markers or the amalgamation of liquid biopsy with other detection methods could potentially facilitate the advancement and utilization of liquid biopsy technology. Ultimately, liquid biopsy has garnered significant attention and investigation, despite certain remaining research deficiencies. Nevertheless, it holds immense clinical utility.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (82002751), Medical Science and Technology Project of Henan Province (SBGJ202102139), Henan Province Outstanding Young Talent Project in Health Science and Technology Innovation for Young and Middle-aged People (YQRC2023020) and Outstanding Youth Foundation of Henan Province (222300420071), Key scientific research project plan of colleges and universities in Henan Province (24A320048), and Key project of Henan Natural Science Foundation (242300421195).
Author contributions
JW, TS, and LM were responsible for the study’s design. LM and HG drafted the manuscript. YZ and ZL performed the tables and prepared the figures, while CW and JB meticulously collected the related references. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Liwei Ma, Huiling Guo.
Contributor Information
Liwei Ma, Email: maliwei@zzu.edu.cn.
Ting Sun, Email: sunting@zzu.edu.cn.
Jianwei Wei, Email: fccweijw@zzu.edu.cn.
References
- 1.Crosby, D. et al. Early detection of cancer. Science375, eaay9040 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Vaidyanathan, R., Soon, R. H., Zhang, P., Jiang, K. & Lim, C. T. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab. Chip19, 11–34 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Li, W. et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol. Cancer21, 25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casagrande, G. M. S., Silva, M. O., Reis, R. M. & Leal, L. F. Liquid biopsy for lung cancer: up-to-date and perspectives for screening programs. Int. J. Mol. Sci.24, 2505 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pantel, K. & Alix-Panabières, C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med.16, 398–406 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Fu, Y., Zhang, Y. & Khoo, B. L. Liquid biopsy technologies for hematological diseases. Med. Res. Rev.41, 246–274 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Lone, S. N. et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer21, 79 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikanjam, M., Kato, S. & Kurzrock, R. Liquid biopsy: current technology and clinical applications. J. Hematol. Oncol.15, 131 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvis, M. M., Romero, C. S., Bueno, T. O. & Teng, Y. Toward a new era for the management of circulating tumor cells. Adv. Exp. Med. Biol.1286, 125–134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandel, P. & Metais, P. Nuclear acids in human blood plasma. C. R. Seances. Soc. Biol. Fil.142, 241–243 (1948). [PubMed] [Google Scholar]
- 11.Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol.13, 269–288 (1967). [DOI] [PubMed] [Google Scholar]
- 12.Pan, B. T. & Johnstone, R. M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell33, 967–978 (1983). [DOI] [PubMed] [Google Scholar]
- 13.Leon, S. A., Shapiro, B., Sklaroff, D. M. & Yaros, M. J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res.37, 646–650 (1977). [PubMed] [Google Scholar]
- 14.Alix-Panabières, C. & Pantel, K. Liquid biopsy: from discovery to clinical application. Cancer Discov.11, 858–873 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Sorenson, G. D. et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomark. Prev.3, 67–71 (1994). [PubMed] [Google Scholar]
- 16.Raposo, G. et al. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med.183, 1161–1172 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnov, D. A. et al. Global gene expression profiling of circulating tumor cells. Cancer Res.65, 4993–4997 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, J., Lu, A. D., Zhang, L. P., Zuo, Y. X. & Jia, Y. P. Study of clinical outcome and prognosis in pediatric core binding factor-acute myeloid leukemia. Zhonghua. Xue. Ye. Xue. Za. Zhi.40, 52–57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giuliano, A. E., Edge, S. B. & Hortobagyi, G. N. Eighth edition of the AJCC cancer staging manual: breast cancer. Ann. Surg. Oncol.25, 1783–1785 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Huang, X. & Yin, Y. M. [Updates of Chinese society of clinical oncology (CSCO) guideline for breast cancer in 2018]. Zhonghua. Yi. Xue. Za. Zhi.98, 1213–1217 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Yu, M., Stott, S., Toner, M., Maheswaran, S. & Haber, D. A. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol.192, 373–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lianidou, E. S., Strati, A. & Markou, A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit. Rev. Clin. Lab Sci.51, 160–171 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Agashe, R. & Kurzrock, R. Circulating tumor cells: from the laboratory to the cancer clinic. Cancers12, 3065 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salu, P. & Reindl, K. M. Advancements in circulating tumor cell research: bridging biology and clinical applications. Cancers16, 1213 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen, S. J. et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol.26, 3213–3221 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Marcuello, M. et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med.69, 107–122 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Qiu, J. et al. Refining cancer management using integrated liquid biopsy. Theranostics10, 2374–2384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sefrioui, D. et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer117, 1017–1025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med.351, 781–791 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Vidlarova, M. et al. Recent advances in methods for circulating tumor cell detection. Int. J. Mol. Sci.24, 3902 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo, G. I. et al. The role of dielectrophoresis for cancer diagnosis and prognosis. Cancers14, 198 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozar, T. et al. Preclinical and clinical evaluation of magnetic-activated cell separation technology for CTC isolation in breast cancer. Front Oncol.10, 554554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrik, J. et al. Circulating tumor cells in colorectal cancer: detection systems and clinical utility. Int. J. Mol. Sci.23, 13582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y. Z., Kong, S. N., Liu, Y. P., Yang, Y. & Zhang, H. M. Can liquid biopsy based on ctDNA/cfDNA replace tissue biopsy for the precision treatment of EGFR-mutated NSCLC? J. Clin. Med.12, 1438 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campos-Carrillo, A. et al. Circulating tumor DNA as an early cancer detection tool. Pharm. Ther.207, 107458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underhill, H. R. et al. Fragment length of circulating tumor DNA. PLoS Genet12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov.7, 1394–1403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia, N. et al. Association of emergence of new mutations in circulating tumuor DNA during chemotherapy with clinical outcome in metastatic colorectal cancer. BMC Cancer21, 845 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Z. et al. Liquid biopsies for cancer: from bench to clinic. MedComm4, e329 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannigan, B. et al. Liquid biopsy assay for lung carcinoma using centrifuged supernatants from fine-needle aspiration specimens. Ann. Oncol.30, 963–969 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Olmedillas-López, S., Olivera-Salazar, R., García-Arranz, M. & García-Olmo, D. Current and emerging applications of droplet digital PCR in oncology: an updated review. Mol. Diagn. Ther.26, 61–87 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Ståhlberg, A. et al. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat. Protoc.12, 664–682 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L. & Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem.262, 9412–9420 (1987). [PubMed] [Google Scholar]
- 45.Witwer, K. W. & Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles8, 1648167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Niel, G. et al. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol.23, 369–382 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Zhou, Y., Zhang, Y., Gong, H., Luo, S. & Cui, Y. The role of exosomes and their applications in cancer. Int. J. Mol. Sci.22, 12204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han, Q. F. et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol. Cancer21, 207 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J. et al. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol.145, 102860 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Wang, J., Yang, K., Yuan, W. & Gao, Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med. Sci. Monit.24, 9307–9316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keerthikumar, S. et al. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol.428, 688–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, X. et al. Extracellular vesicle packaged LMP1-activated fibroblasts promote tumor progression via autophagy and stroma-tumor metabolism coupling. Cancer Lett.478, 93–106 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asare-Werehene, M. et al. The exosome-mediated autocrine and paracrine actions of plasma gelsolin in ovarian cancer chemoresistance. Oncogene39, 1600–1616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mu, Y. et al. Study of serum exosome miRNA as a biomarker for early onset adult ouclar myastthenia gravis. Gene896, 148034 (2024). [DOI] [PubMed] [Google Scholar]
- 56.Chen, Y. et al. Exosomal derived miR-1246 from hydroquinone-transformed cells drives S phase accumulation arrest by targeting cyclin G2 in TK6 cells. Chem. Biol. Interact.387, 110809 (2024). [DOI] [PubMed] [Google Scholar]
- 57.Chen, J. et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng. Biotechnol.9, 811971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franco, A. T., Corken, A. & Ware, J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood126, 582–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Italiano, J. E. Jr. & Battinelli, E. M. Selective sorting of alpha-granule proteins. J. Thromb. Haemost.7, 173–176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tranum, B. L. & Haut, A. Thrombocytosis: platelet kinetics in neoplasia. J. Lab. Clin. Med.84, 615–619 (1974). [PubMed] [Google Scholar]
- 61.Bailey, S. E., Ukoumunne, O. C., Shephard, E. A. & Hamilton, W. Clinical relevance of thrombocytosis in primary care: a prospective cohort study of cancer incidence using English electronic medical records and cancer registry data. Br. J. Gen. Pr.67, e405–e413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi, C. et al. P-selectin-mediated adhesion between platelets and tumor cells promotes intestinal tumorigenesis in Apc(Min/+) mice. Int. J. Biol. Sci.11, 679–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Y. et al. Application of tumor-educated platelets as new fluid biopsy markers in various tumors. Clin. Transl. Oncol.25, 114–125 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Best, M. G., Wesseling, P. & Wurdinger, T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res.78, 3407–3412 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Nilsson, R. J. et al. Blood platelets contain tumor-derived RNA biomarkers. Blood118, 3680–3683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stratz, C. et al. Micro-array profiling exhibits remarkable intra-individual stability of human platelet micro-RNA. Thromb. Haemost.107, 634–641 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Best, M. G. et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell28, 666–676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye, B. et al. A panel of platelet-associated circulating long non-coding RNAs as potential biomarkers for colorectal cancer. Genomics114, 31–37 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Zou, D. et al. PltDB: a blood platelets-based gene expression database for disease investigation. Bioinformatics38, 3143–3145 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Zhang, M., Dang, P., Liu, Y., Qiao, B. & Sun, Z. Noncoding RNAs in pyroptosis and cancer progression: Effect, mechanism, and clinical application. Front Immunol.13, 982040 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toden, S. & Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer126, 351–360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S. & Ghaffari, S. H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol.234, 5451–5465 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Tili, E., Croce, C. M. & Michaille, J. J. miR-155: on the crosstalk between inflammation and cancer. Int Rev. Immunol.28, 264–284 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Boon, R. A., Jaé, N., Holdt, L. & Dimmeler, S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol.67, 1214–1226 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Bridges, M. C., Daulagala, A. C. & Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol.220, e202009045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmitt, A. M. & Chang, H. Y. Long noncoding RNAs in cancer pathways. Cancer Cell29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramón, Y. C. S., Segura, M. F. & Hümmer, S. Interplay between ncRNAs and cellular communication: a proposal for understanding cell-specific signaling pathways. Front Genet.10, 281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen, Q. et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol. Lett.12, 1361–1366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin, Q. et al. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep.39, 2644–2652 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takahashi, K. et al. Long non-coding RNAs in epithelial-mesenchymal transition of pancreatic cancer. Front Mol. Biosci.8, 717890 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu, D. et al. Peripheral blood-based DNA methylation of long non-coding RNA H19 and metastasis-associated lung adenocarcinoma transcript 1 promoters are potential non-invasive biomarkers for gastric cancer detection. Cancer Control28, 10732748211043667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng, Z. H. et al. m6A-immune-related lncRNA prognostic signature for predicting immune landscape and prognosis of bladder cancer. J. Transl. Med.20, 492 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin, T. LncRNA DRAIR is a novel prognostic and diagnostic biomarker for gastric cancer. Mamm. Genome.32, 503–507 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Zhang, G., Sun, J. & Zhang, X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci. Rep.12, 11325 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diener, T. O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology45, 411–428 (1971). [DOI] [PubMed] [Google Scholar]
- 86.Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA73, 3852–3856 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hentze, M. W. & Preiss, T. Circular RNAs: splicing’s enigma variations. EMBO J.32, 923–925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peng, D. et al. CircRNA: an emerging star in the progression of glioma. Biomed. Pharmacother.151, 113150 (2022). [DOI] [PubMed] [Google Scholar]
- 89.Liang, Y. et al. A brief review of circRNA biogenesis, detection, and function. Curr. Genomics22, 485–495 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang, Y., Liu, Q. & Liao, Q. CircHIPK3: a promising cancer-related circular RNA. Am. J. Transl. Res.12, 6694–6704 (2020). [PMC free article] [PubMed] [Google Scholar]
- 91.Liu, T., Huang, T., Shang, M. & Han, G. CircRNA ITCH: insight into its role and clinical application prospect in tumor and non-tumor diseases. Front. Genet.13, 927541 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su, K., Yi, Q., Dai, X. & Liu, O. Circular RNA ITCH: an emerging multifunctional regulator. Biomolecules12, 359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kojima, T. et al. A simple biological imaging system for detecting viable human circulating tumor cells. J. Clin. Investig.119, 3172–3181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao, F. et al. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget7, 71330–71340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu, B. et al. Comprehensive atlas of circulating rare cells detected by SE-iFISH and image scanning platform in patients with various diseases. Front. Oncol.12, 821454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rushton, A. J., Nteliopoulos, G., Shaw, J. A. & Coombes, R. C. A review of circulating tumour cell enrichment technologies. Cancers13, 970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Andree, K. C., van Dalum, G. & Terstappen, L. W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol.10, 395–407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kojabad, A. A. et al. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol.93, 4182–4197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin, C., Liu, X., Zheng, B., Ke, R. & Tzeng, C. M. Liquid biopsy, ctDNA diagnosis through NGS. Life11, 890 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mandlik, J. S., Patil, A. S. & Singh, S. Next-generation sequencing (NGS): platforms and applications. J. Pharm. Bioallied Sci.16, S41–s45 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sidhom, K., Obi, P. O. & Saleem, A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int. J. Mol. Sci.21, 6466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meyer, C., Garzia, A. & Tuschl, T. Simultaneous detection of the subcellular localization of RNAs and proteins in cultured cells by combined multicolor RNA-FISH and IF. Methods118-119, 101–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Freeman, W. M., Walker, S. J. & Vrana, K. E. Quantitative RT-PCR: pitfalls and potential. Biotechniques26, 112–122 (1999). 124-115. [DOI] [PubMed] [Google Scholar]
- 104.He, S. L. & Green, R. Northern blotting. Methods Enzymol.530, 75–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Urbanek, M. O., Nawrocka, A. U. & Krzyzosiak, W. J. Small RNA detection by in situ hybridization methods. Int. J. Mol. Sci.16, 13259–13286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hong, M. et al. RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol.13, 166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou, Z. et al. Liquid biopsy in hepatocellular carcinoma. Methods Mol. Biol.2695, 213–225 (2023). [DOI] [PubMed] [Google Scholar]
- 108.Ghosh, S. et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int. J. Cancer147, 2934–2947 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Jiang, S. S. et al. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J. Transl. Med.12, 273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arbelaiz, A. et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology66, 1125–1143 (2017). [DOI] [PubMed] [Google Scholar]
- 111.von Felden, J. et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene40, 140–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kisiel, J. B. et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology69, 1180–1192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prospective suRveillance for very Early hepatoCellular cARcinoma(PreCar) expert panel. [Expert consensus on early screening strategies for liver cancer in China]. Zhonghua. Gan. Zang. Bing. Za. Zhi.29, 515–522 (2021). [DOI] [PubMed]
- 114.Qi, L. N. et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res.78, 4731–4744 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Park, S., Lee, E. J., Rim, C. H. & Seong, J. Plasma cell-free DNA as a predictive marker after radiotherapy for hepatocellular carcinoma. Yonsei Med. J.59, 470–479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen, J., Cao, S. W., Cai, Z., Zheng, L. & Wang, Q. Epithelial-mesenchymal transition phenotypes of circulating tumor cells correlate with the clinical stages and cancer metastasis in hepatocellular carcinoma patients. Cancer Biomark.20, 487–498 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Bai, T. et al. Circulating tumor cells and CXCR4 in the prognosis of hepatocellular carcinoma. Transl. Cancer Res.9, 1384–1394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Izquierdo-Sanchez, L. et al. Cholangiocarcinoma landscape in Europe: diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J. Hepatol.76, 1109–1121 (2022). [DOI] [PubMed] [Google Scholar]
- 119.Valle, J. W. et al. Biliary cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol.27, v28–v37 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Wang, Y. et al. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene533, 389–397 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Yan, Q. et al. The serum MicroRNA signatures for pancreatic cancer detection and operability evaluation. Front. Bioeng. Biotechnol.8, 379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Correa-Gallego, C. et al. Circulating plasma levels of MicroRNA-21 and MicroRNA-221 are potential diagnostic markers for primary intrahepatic cholangiocarcinoma. PLoS ONE11, e0163699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leelawat, K. et al. Prognostic relevance of circulating CK19 mRNA in advanced malignant biliary tract diseases. World J. Gastroenterol.18, 175–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou, K. Q. et al. Circulating osteopontin per tumor volume as a prognostic biomarker for resectable intrahepatic cholangiocarcinoma. Hepatobiliary Surg. Nutr.8, 582–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Julich-Haertel, H. et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol.67, 282–292 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Xu, H. et al. Elevation of serum KL-6 mucin levels in patients with cholangiocarcinoma. Hepatogastroenterology55, 2000–2004 (2008). [PubMed] [Google Scholar]
- 127.Leelawat, K., Sakchinabut, S., Narong, S. & Wannaprasert, J. Detection of serum MMP-7 and MMP-9 in cholangiocarcinoma patients: evaluation of diagnostic accuracy. BMC Gastroenterol.9, 30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kobayashi, S., Werneburg, N. W., Bronk, S. F., Kaufmann, S. H. & Gores, G. J. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology128, 2054–2065 (2005). [DOI] [PubMed] [Google Scholar]
- 129.Huang, L. et al. Serum CYFRA 21-1 in biliary tract cancers: a reliable biomarker for gallbladder carcinoma and intrahepatic cholangiocarcinoma. Dig. Dis. Sci.60, 1273–1283 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Loosen, S. H. et al. Elevated levels of circulating osteopontin are associated with a poor survival after resection of cholangiocarcinoma. J. Hepatol.67, 749–757 (2017). [DOI] [PubMed] [Google Scholar]
- 131.Yang, J. D. et al. Circulating tumor cells are associated with poor overall survival in patients with cholangiocarcinoma. Hepatology63, 148–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shen, N. et al. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep.42, 549–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goyal, L. et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov.7, 252–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zygulska, A. L. & Pierzchalski, P. Novel diagnostic biomarkers in colorectal cancer. Int. J. Mol. Sci.23, 852 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takano, Y. et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget8, 78598–78613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Teng, Y. et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun.8, 14448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tsukamoto, M., Iinuma, H., Yagi, T., Matsuda, K. & Hashiguchi, Y. Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology92, 360–370 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Fu, F., Jiang, W., Zhou, L. & Chen, Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol.11, 221–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zeng, Z. et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun.9, 5395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Karimi, N. et al. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J. Chin. Med Assoc.82, 215–220 (2019). [DOI] [PubMed] [Google Scholar]
- 141.Concepcion, C. P., Bonetti, C. & Ventura, A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J.18, 262–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsumura, T. et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer113, 275–281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang, C., Zhuang, W., Hu, Y. & Zhu, L. Clinical significance of peripheral circulating tumor cell counts in colorectal polyps and non-metastatic colorectal cancer. World J. Surg. Oncol.16, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsai, W. S. et al. Novel circulating tumor cell assay for detection of colorectal adenomas and cancer. Clin. Transl. Gastroenterol.10, e00088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ganig, N. et al. Proteomic analyses of fibroblast- and serum-derived exosomes identify QSOX1 as a marker for non-invasive detection of colorectal cancer. Cancers13, 1351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang, F., Zhang, Y., Chen, D., Zhang, Z. & Li, Z. Single microbead-based fluorescent aptasensor (SMFA) for direct isolation and in situ quantification of exosomes from plasma. Analyst146, 3346–3351 (2021). [DOI] [PubMed] [Google Scholar]
- 147.Ren, D. et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget8, 49807–49823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Camera, S. et al. Prognostic value of the pace of tumor progression as assessed by serial (18)F-FDG PET/CT scan and liquid biopsy in refractory colorectal cancer: the Coriolan trial. Cancers12, 2752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Troncarelli Flores, B. C. et al. Molecular and kinetic analyses of circulating tumor cells as predictive markers of treatment response in locally advanced rectal cancer patients. Cells8, 641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Colorectal Cancer Expert Committee of Chinese Society of Clinical Oncology (CSCO). [Consensus of Chinese experts on clinical detection of molecular markers of colorectal cancer]. Zhonghua. Wei Chang Wai Ke Za Zhi24, 191–197 (2021). [DOI] [PubMed]
- 151.Osumi, H., Shinozaki, E., Yamaguchi, K. & Zembutsu, H. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci. Rep.9, 17358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tie, J. et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. Int. J. Cancer148, 1014–1026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.(!!! INVALID CITATION !!!)..
- 154.Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M. & Maitra, A. Pancreatic cancer: advances and challenges. Cell186, 1729–1754 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stosic, K. et al. A comprehensive review of the potential role of liquid biopsy as a diagnostic, prognostic, and predictive biomarker in pancreatic ductal adenocarcinoma. Cells13, 3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ankeny, J. S. et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br. J. Cancer114, 1367–1375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Early Diagnosis and Treatment Group, the Oncology Committee of Chinese Medical Association. [Expert consensus of oncology committee of Chinese medical association in early diagnosis and treatment of pancreatic cancer]. Zhonghua Zhong Liu Za Zhi42, 706–712 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Rhim, A. D. et al. EMT and dissemination precede pancreatic tumor formation. Cell148, 349–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang, R. et al. Diagnostic and prognostic values of KRAS mutations on EUS-FNA specimens and circulating tumor DNA in patients with pancreatic cancer. Clin. Transl. Gastroenterol.13, e00487 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med.25, 1928–1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cohen, J. D. et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA114, 10202–10207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Eissa, M. A. L. et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenet.11, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wnuk, J., Strzelczyk, J. K. & Gisterek, I. Clinical value of circulating miRNA in diagnosis, prognosis, screening and monitoring therapy of pancreatic ductal adenocarcinoma-a review of the literature. Int. J. Mol. Sci.24, 5113 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mann, D. V., Edwards, R., Ho, S., Lau, W. Y. & Glazer, G. Elevated tumour marker CA19-9: clinical interpretation and influence of obstructive jaundice. Eur. J. Surg. Oncol.26, 474–479 (2000). [DOI] [PubMed] [Google Scholar]
- 165.Dittmar, R. L. et al. Plasma miRNA biomarkers in limited volume samples for detection of early-stage pancreatic cancer. Cancer Prev. Res.14, 729–740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu, S. et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut69, 540–550 (2020). [DOI] [PubMed] [Google Scholar]
- 167.Yeo, D. et al. Exploring the clinical utility of pancreatic cancer circulating tumor cells. Int. J. Mol. Sci.23, 1671 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim, H. et al. Clinical significance of circulating tumor cells after chemotherapy in unresectable pancreatic ductal adenocarcinoma. Transl. Oncol.16, 101321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fusi, A. et al. Expression of chemokine receptors on circulating tumor cells in patients with solid tumors. J. Transl. Med.10, 52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gardner, K. P., Tsai, S., Aldakkak, M., Gironda, S. & Adams, D. L. CXCR4 expression in tumor associated cells in blood is prognostic for progression and survival in pancreatic cancer. PLoS ONE17, e0264763 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Groot, V. P. et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin. Cancer Res.25, 4973–4984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wei, T. et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol. Cancer Ther.18, 196–203 (2019). [DOI] [PubMed] [Google Scholar]
- 173.Lu, H. et al. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy. Biosci. Rep.39, BSR20181374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.van der Sijde, F. et al. Serum miR-373-3p and miR-194-5p are associated with early tumor progression during FOLFIRINOX treatment in pancreatic cancer patients: a prospective multicenter study. Int. J. Mol. Sci.22, 10902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mikamori, M. et al. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci. Rep.7, 42339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Song, B. G. et al. Detection of circulating tumor cells in resectable pancreatic ductal adenocarcinoma: a prospective evaluation as a prognostic marker. Front. Oncol.10, 616440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nitschke, C. et al. Characterization of RARRES1 expression on circulating tumor cells as unfavorable prognostic marker in resected pancreatic ductal adenocarcinoma patients. Cancers14, 4405 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ako, S. et al. Plasma KRAS mutations predict the early recurrence after surgical resection of pancreatic cancer. Cancer Biol. Ther.22, 564–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guo, S. et al. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br. J. Cancer122, 857–867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kandimalla, R. et al. Identification of Serum miRNA signature and establishment of a nomogram for risk stratification in patients with pancreatic ductal adenocarcinoma. Ann. Surg.275, e229–e237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang, Z. et al. A multianalyte panel consisting of extracellular vesicle miRNAs and mRNAs, cfDNA, and CA19-9 shows utility for diagnosis and staging of pancreatic ductal adenocarcinoma. Clin. Cancer Res.26, 3248–3258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dbouk, M. et al. Diagnostic performance of a tumor marker gene test to personalize serum CA19-9 reference ranges. Clin. Cancer Res.29, 4178–4185 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ando, Y. et al. Using tumor marker gene variants to improve the diagnostic accuracy of dupan-2 and carbohydrate antigen 19-9 for pancreatic cancer. J. Clin. Oncol.42, 2196–2206 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.López, M. J. et al. Characteristics of gastric cancer around the world. Crit. Rev. Oncol. Hematol.181, 103841 (2023). [DOI] [PubMed] [Google Scholar]
- 185.Tang, L. et al. Diagnostic accuracy of circulating tumor cells detection in gastric cancer: systematic review and meta-analysis. BMC Cancer13, 314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park, J. L. et al. Quantitative analysis of cell-free DNA in the plasma of gastric cancer patients. Oncol. Lett.3, 921–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim, K. et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann. Surg. Treat. Res.86, 136–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ren, J. et al. Genome-scale methylation analysis of circulating cell-free DNA in gastric cancer patients. Clin. Chem.68, 354–364 (2022). [DOI] [PubMed] [Google Scholar]
- 189.Ko, K. et al. Methylation status and long-fragment cell-free DNA are prognostic biomarkers for gastric cancer. Cancer Med.10, 2003–2012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ma, S. et al. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int.21, 44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Roy, S. et al. Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol. Cancer21, 42 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xu, Y. et al. Clinical role of miR-421 as a novel biomarker in diagnosis of gastric cancer patients: a meta-analysis. Medecine101, e29242 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fu, H. et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res.37, 162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ito, H. et al. Long-term prognostic impact of circulating tumour cells in gastric cancer patients. World J. Gastroenterol.22, 10232–10241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huang, X. et al. Clinicopathological and prognostic significance of circulating tumor cells in patients with gastric cancer: a meta-analysis. Int. J. Cancer136, 21–33 (2015). [DOI] [PubMed] [Google Scholar]
- 196.Negishi, R. et al. Transcriptomic profiling of single circulating tumor cells provides insight into human metastatic gastric cancer. Commun. Biol.5, 20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hiraiwa, K. et al. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann. Surg. Oncol.15, 3092–3100 (2008). [DOI] [PubMed] [Google Scholar]
- 198.Zhong, Y. et al. Plasma cfDNA as a potential biomarker to evaluate the efficacy of chemotherapy in gastric cancer. Cancer Manag Res.12, 3099–3106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qian, C. et al. Alu-based cell-free DNA: a novel biomarker for screening of gastric cancer. Oncotarget8, 54037–54045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bae, W. J. et al. miR-4742-5p promotes invasiveness of gastric cancer via targeting Rab43: An in vitro study. Biochem. Biophys. Res. Commun.613, 180–186 (2022). [DOI] [PubMed] [Google Scholar]
- 201.Yifei, S., Chunxiao, H. & Dinuo, L. MiR-17-5p inhibits the proliferation and metastasis of gastric cancer cells by targeting PTEN protein. Alter. Ther. Health Med.28, 23–29 (2022). [PubMed] [Google Scholar]
- 202.Cai, Y. et al. YncRNA PTCSC3 and lncRNA HULC Negatively affect each other to regulate cancer cell invasion and migration in gastric cancer [Retraction]. Cancer Manag. Res.13, 8003–8004 (2021). [DOI] [PMC free article] [PubMed]
- 203.Zheng, P., Gao, H., Xie, X. & Lu, P. Plasma exosomal hsa_circ_0015286 as a potential diagnostic and prognostic biomarker for gastric cancer. Pathol. Oncol. Res.28, 1610446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cheng, B. et al. Enumeration and characterization of circulating tumor cells and its application in advanced gastric cancer. Onco Targets Ther.12, 7887–7896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Willis, J. et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin. Cancer Res.25, 7035–7045 (2019). [DOI] [PubMed] [Google Scholar]
- 206.Azimi, M., Totonchi, M. & Ebrahimi, M. Determining the role of MicroRNAs in self-renewal, metastasis and resistance to drugs in human gastric cancer based on data mining approaches: a systematic review. Cell J.24, 1–6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Abbasi, A., Hosseinpourfeizi, M. & Safaralizadeh, R. All-trans retinoic acid-mediated miR-30a up-regulation suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Life Sci.307, 120884 (2022). [DOI] [PubMed] [Google Scholar]
- 208.Zhou, F. et al. The regulation of hsacirc_004413 promotes proliferation and drug resistance of gastric cancer cells by acting as a competing endogenous RNA for miR-145-5p. PeerJ10, e12629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fang, L. et al. Circular CPM promotes chemoresistance of gastric cancer via activating PRKAA2-mediated autophagy. Clin. Transl. Med.12, e708 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhou, H., Shen, W., Zou, H., Lv, Q. & Shao, P. Circulating exosomal long non-coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J. Int. Med. Res.48, 300060520934297 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Matsusaka, S. et al. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in patients with advanced gastric cancer. Cancer Sci.101, 1067–1071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yu, P. et al. Application of circulating tumor cells and circulating free DNA from peripheral blood in the prognosis of advanced gastric cancer. J. Oncol.2022, 9635218 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Normando, S. R. C. et al. Circulating free plasma tumor DNA in patients with advanced gastric cancer receiving systemic chemotherapy. BMC Clin. Pathol.18, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lan, Y. T. et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget8, 3009–3017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Karamitrousis, E. I. et al. Prognostic Role of RASSF1A, SOX17 and Wif-1 promoter methylation status in cell-free DNA of advanced gastric cancer patients. Technol. Cancer Res. Treat.20, 1533033820973279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nicolazzo, C. et al. True conversions from RAS mutant to RAS wild-type in circulating tumor DNA from metastatic colorectal cancer patients as assessed by methylation and mutational signature. Cancer Lett.507, 89–96 (2021). [DOI] [PubMed] [Google Scholar]
- 217.Vrba, L. et al. DNA methylation biomarkers discovered in silico detect cancer in liquid biopsies from non-small cell lung cancer patients. Epigenetics15, 419–430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ooki, A. et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin. Cancer Res.23, 7141–7152 (2017). [DOI] [PubMed] [Google Scholar]
- 219.Yang, Z. et al. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv. Clin. Exp. Med.28, 355–360 (2019). [DOI] [PubMed] [Google Scholar]
- 220.Kang, S. M. et al. The Haptoglobin β chain as a supportive biomarker for human lung cancers. Mol. Biosyst.7, 1167–1175 (2011). [DOI] [PubMed] [Google Scholar]
- 221.Sung, H. J. et al. Identification and validation of SAA as a potential lung cancer biomarker and its involvement in metastatic pathogenesis of lung cancer. J. Proteome Res.10, 1383–1395 (2011). [DOI] [PubMed] [Google Scholar]
- 222.Cabanero, M. & Tsao, M. S. Circulating tumour DNA in EGFR-mutant non-small-cell lung cancer. Curr. Oncol.25, S38–s44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Revelo, A. E. et al. Liquid biopsy for lung cancers: an update on recent developments. Ann. Transl. Med.7, 349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Li, R. Y. & Liang, Z. Y. Circulating tumor DNA in lung cancer: real-time monitoring of disease evolution and treatment response. Chin. Med. J.133, 2476–2485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang, Z. et al. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology21, 519–525 (2016). [DOI] [PubMed] [Google Scholar]
- 226.Huang, J. et al. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed. Res. Int.2014, 364316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rodríguez, M. et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer53, 713–724 (2014). [DOI] [PubMed] [Google Scholar]
- 228.Rabinowits, G., Gerçel-Taylor, C., Day, J. M., Taylor, D. D. & Kloecker, G. H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer10, 42–46 (2009). [DOI] [PubMed] [Google Scholar]
- 229.Szczerba, B. M. et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature566, 553–557 (2019). [DOI] [PubMed] [Google Scholar]
- 230.Rolfo, C. et al. Liquid biopsy for advanced NSCLC: a consensus statement from the International Association for the study of Lung Cancer. J. Thorac. Oncol.16, 1647–1662 (2021). [DOI] [PubMed] [Google Scholar]
- 231.Lee, D. H. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): the road to a success, paved with failures. Pharm. Ther.174, 1–21 (2017). [DOI] [PubMed] [Google Scholar]
- 232.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 233.Iacob, R. et al. Liquid biopsy in squamous cell carcinoma of the esophagus and of the head and neck. Front. Med.9, 827297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yang, W. Y. et al. Liquid biopsy in head and neck squamous cell carcinoma: circulating tumor cells, circulating tumor DNA, and exosomes. Expert Rev. Mol. Diagn.20, 1213–1227 (2020). [DOI] [PubMed] [Google Scholar]
- 235.Payne, K. et al. Circulating tumor DNA as a biomarker and liquid biopsy in head and neck squamous cell carcinoma. Head. Neck40, 1598–1604 (2018). [DOI] [PubMed] [Google Scholar]
- 236.Yu, S. et al. Oral-microbiome-derived signatures enable non-invasive diagnosis of laryngeal cancers. J. Transl. Med.21, 438 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kawada, T. et al. Circulating tumor cells in patients with head and neck squamous cell carcinoma: feasibility of detection and quantitation. Head. Neck39, 2180–2186 (2017). [DOI] [PubMed] [Google Scholar]
- 238.Nichols, A. C. et al. Detection of circulating tumor cells in advanced head and neck cancer using the cell search system. Head. Neck34, 1440–1444 (2012). [DOI] [PubMed] [Google Scholar]
- 239.Rizzo, M. I. et al. Detection of circulating tumor cells in patients with laryngeal cancer using screen cell: comparative pre- and post-operative analysis and association with prognosis. Oncol. Lett.19, 4183–4188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang, Y. et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med.7, 293ra104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sanchez-Cespedes, M. et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res.60, 892–895 (2000). [PubMed] [Google Scholar]
- 242.Schröck, A. et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin. Chem.63, 1288–1296 (2017). [DOI] [PubMed] [Google Scholar]
- 243.Reddy, K. B. MicroRNA (miRNA) in cancer. Cancer Cell Int.15, 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yu, X. & Li, Z. The role of microRNAs expression in laryngeal cancer. Oncotarget6, 23297–23305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wang, J. et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol.31, 148 (2014). [DOI] [PubMed] [Google Scholar]
- 246.Wang, J. L., Wang, X., Yang, D. & Shi, W. J. The expression of MicroRNA-155 in plasma and tissue is matched in human laryngeal squamous cell carcinoma. Yonsei Med. J.57, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Powrózek, T. et al. miRNA-130a significantly improves accuracy of SGA nutritional assessment tool in prediction of malnutrition and cachexia in radiotherapy-treated head and neck cancer patients. Cancers10, 294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cao, Y. C. et al. Serum miR-632 is a potential marker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Acta Otolaryngol.140, 418–421 (2020). [DOI] [PubMed] [Google Scholar]
- 249.Hsu, C. L., Chang, Y. S. & Li, H. P. Molecular diagnosis of nasopharyngeal carcinoma: past and future. Biomed. J. 100748, 10.1016/j.bj.2024.100748 (2024). [DOI] [PubMed]
- 250.Tan, R. et al. Clinical utility of Epstein-Barr virus DNA and other liquid biopsy markers in nasopharyngeal carcinoma. Cancer Commun.40, 564–585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lo, Y. M. et al. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res.59, 5452–5455 (1999). [PubMed] [Google Scholar]
- 252.Chan, K. A. et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med.378, 973 (2018). [DOI] [PubMed]
- 253.Hsu, C. L. et al. Plasma Epstein-Barr virus DNA concentration and clearance rate as novel prognostic factors for metastatic nasopharyngeal carcinoma. Head Neck34, 1064–1070 (2012). [DOI] [PubMed] [Google Scholar]
- 254.Zheng, X. H. et al. Saliva biopsy: detecting the difference of EBV DNA methylation in the diagnosis of nasopharyngeal carcinoma. Int. J. Cancer153, 882–892 (2023). [DOI] [PubMed] [Google Scholar]
- 255.Wu, C. F. et al. Liquid biopsy posttreatment surveillance in endemic nasopharyngeal carcinoma: a cost-effective strategy to integrate circulating cell-free Epstein-Barr virus DNA. BMC Med.19, 193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sheu, L. F. et al. Enhanced malignant progression of nasopharyngeal carcinoma cells mediated by the expression of Epstein-Barr nuclear antigen 1 in vivo. J. Pathol.180, 243–248 (1996). [DOI] [PubMed] [Google Scholar]
- 257.Murono, S. et al. Detection of Epstein-Barr virus in nasopharyngeal carcinoma by in situ hybridization and polymerase chain reaction. Laryngoscope107, 523–526 (1997). [DOI] [PubMed] [Google Scholar]
- 258.Wong, A. M., Kong, K. L., Tsang, J. W., Kwong, D. L. & Guan, X. Y. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer118, 698–710 (2012). [DOI] [PubMed] [Google Scholar]
- 259.Stenvang, J., Petri, A., Lindow, M., Obad, S. & Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence3, 1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhao, Z. et al. Applications of cerebrospinal fluid circulating tumor DNA in the diagnosis of gliomas. Jpn. J. Clin. Oncol.50, 325–332 (2020). [DOI] [PubMed] [Google Scholar]
- 261.Borba, L. A. B., Passos, G. & Oliveira, I. Liquid biopsy and tumor DNA/RNA detection in the cerebrospinal fluid of patients diagnosed with central nervous system glioma—a review article. Surg. Neurol. Int.14, 183 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Li, K. et al. Imaging and liquid biopsy for distinguishing true progression from pseudoprogression in gliomas, current advances and challenges. Acad. Radiol.31, 3366–3383 (2024). [DOI] [PubMed] [Google Scholar]
- 263.Thege, F. I. et al. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab Chip14, 1775–1784 (2014). [DOI] [PubMed] [Google Scholar]
- 264.Sullivan, J. P. et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov.4, 1299–1309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wick, M., Gross, C. C., Isenmann, S. & Strik, H. Cytology of cerebrospinal fluid: standards, importance and modern methods. Nervenarzt87, 1276–1281 (2016). [DOI] [PubMed] [Google Scholar]
- 266.Nabors, L. B. et al. Central nervous system cancers, version 3.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw.18, 1537–1570 (2020). [DOI] [PubMed] [Google Scholar]
- 267.De Mattos-Arruda, L. et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun.6, 8839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Orzan, F. et al. Liquid biopsy of cerebrospinal fluid enables selective profiling of glioma molecular subtypes at first clinical presentation. Clin. Cancer Res.29, 1252–1266 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Macarthur, K. M. et al. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res.74, 2152–2159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Müller, C. et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med.6, 247ra101 (2014). [DOI] [PubMed] [Google Scholar]
- 271.Anfossi, S., Babayan, A., Pantel, K. & Calin, G. A. Clinical utility of circulating non-coding RNAs—an update. Nat. Rev. Clin. Oncol.15, 541–563 (2018). [DOI] [PubMed] [Google Scholar]
- 272.Wierzbicki, K. et al. Targeting and therapeutic monitoring of H3K27M-mutant glioma. Curr. Oncol. Rep.22, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med.6, 224ra224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ghodsi, M., Shahmohammadi, M., Modarressi, M. H. & Karami, F. Investigation of promoter methylation of MCPH1 gene in circulating cell-free DNA of brain tumor patients. Exp. Brain Res.238, 1903–1909 (2020). [DOI] [PubMed] [Google Scholar]
- 275.Westphal, M. & Lamszus, K. Circulating biomarkers for gliomas. Nat. Rev. Neurol.11, 556–566 (2015). [DOI] [PubMed] [Google Scholar]
- 276.Manterola, L. et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol.16, 520–527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Akers, J. C. et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget8, 68769–68779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Shao, H. et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med.18, 1835–1840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Setti, M. et al. Extracellular vesicle-mediated transfer of CLIC1 protein is a novel mechanism for the regulation of glioblastoma growth. Oncotarget6, 31413–31427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Hiemcke-Jiwa, L. S. et al. Molecular analysis in liquid biopsies for diagnostics of primary central nervous system lymphoma: review of literature and future opportunities. Crit. Rev. Oncol. Hematol.127, 56–65 (2018). [DOI] [PubMed] [Google Scholar]
- 281.Baraniskin, A. & Schroers, R. Modern cerebrospinal fluid analyses for the diagnosis of diffuse large B-cell lymphoma of the CNS. CNS Oncol.3, 77–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Akhter, A. et al. Differential expression of Toll-like receptor (TLR) and B cell receptor (BCR) signaling molecules in primary diffuse large B-cell lymphoma of the central nervous system. J. Neurooncol.121, 289–296 (2015). [DOI] [PubMed] [Google Scholar]
- 283.Chapuy, B. et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood127, 869–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yamaguchi, J. et al. Rapid detection of the MYD88 L265P mutation for pre- and intra-operative diagnosis of primary central nervous system lymphoma. Cancer Sci.114, 2544–2551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Hiemcke-Jiwa, L. S. et al. The use of droplet digital PCR in liquid biopsies: a highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid. Hematol. Oncol.36, 429–435 (2018). [DOI] [PubMed] [Google Scholar]
- 286.Zorofchian, S. et al. Detection of the MYD88 p.L265P mutation in the CSF of a patient with secondary central nervous system lymphoma. Front. Oncol.8, 382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Fontanilles, M. et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget8, 48157–48168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hou, Y., Zi, J., Liu, S., Ge, Q. & Ge, Z. Mutational profiling of circulating tumor DNA and clinical characteristics in lymphoma: based on next generation sequencing. Mol. Carcinog.62, 200–209 (2023). [DOI] [PubMed] [Google Scholar]
- 289.Hu, Y. et al. Exosomal miR-200c and miR-141 as cerebrospinal fluid biopsy biomarkers for the response to chemotherapy in primary central nervous system lymphoma. Discov. Oncol.14, 205 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mathieu, M., Martin-Jaular, L., Lavieu, G. & Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol.21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 291.Deng, Y. et al. Phosphoproteome analysis of cerebrospinal fluid extracellular vesicles in primary central nervous system lymphoma. Analyst148, 3594–3602 (2023). [DOI] [PubMed] [Google Scholar]
- 292.Ikeguchi, R., Shimizu, Y., Shimizu, S. & Kitagawa, K. CSF and clinical data are useful in differentiating CNS inflammatory demyelinating disease from CNS lymphoma. Mult. Scler.24, 1212–1223 (2018). [DOI] [PubMed] [Google Scholar]
- 293.Sasagawa, Y., Akai, T., Tachibana, O. & Iizuka, H. Diagnostic value of interleukin-10 in cerebrospinal fluid for diffuse large B-cell lymphoma of the central nervous system. J. Neurooncol.121, 177–183 (2015). [DOI] [PubMed] [Google Scholar]
- 294.Shao, J. et al. High level of IL-10 in cerebrospinal fluid is specific for diagnosis of primary central nervous system lymphoma. Cancer Manag Res.12, 6261–6268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Viaccoz, A. et al. CSF neopterin level as a diagnostic marker in primary central nervous system lymphoma. Neuro Oncol.17, 1497–1503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kubiliute, R. & Jarmalaite, S. Epigenetic biomarkers of renal cell carcinoma for liquid biopsy tests. Int. J. Mol. Sci.22, 8846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Li, M. et al. Liquid biopsy at the frontier in renal cell carcinoma: recent analysis of techniques and clinical application. Mol. Cancer22, 37 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Bade, R. M. et al. Development and initial clinical testing of a multiplexed circulating tumor cell assay in patients with clear cell renal cell carcinoma. Mol. Oncol.15, 2330–2344 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nuzzo, P. V. et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med.26, 1041–1043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Chen, X. et al. Identification of a four-microRNA panel in serum for screening renal cell carcinoma. Pathol. Res. Pract.227, 153625 (2021). [DOI] [PubMed] [Google Scholar]
- 301.Di Meo, A. et al. Prognostic urinary miRNAs for the assessment of small renal masses. Clin. Biochem.75, 15–22 (2020). [DOI] [PubMed] [Google Scholar]
- 302.Heinemann, F. G. et al. Serum miR-122-5p and miR-206 expression: non-invasive prognostic biomarkers for renal cell carcinoma. Clin. Epigenet.10, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Outeiro-Pinho, G. et al. MicroRNA-30a-5p(me): a novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J. Exp. Clin. Cancer Res.39, 98 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Di Meo, A. et al. Searching for prognostic biomarkers for small renal masses in the urinary proteome. Int. J. Cancer146, 2315–2325 (2020). [DOI] [PubMed] [Google Scholar]
- 305.Xu, W. et al. Plasma KIM-1 is associated with recurrence risk after nephrectomy for localized renal cell carcinoma: a trial of the ECOG-ACRIN Research Group (E2805). Clin. Cancer Res.27, 3397–3403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sato, T. et al. Accurate quantification of urinary metabolites for predictive models manifest clinicopathology of renal cell carcinoma. Cancer Sci.111, 2570–2578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wang, Z. et al. UPLC-MS-based urine untargeted metabolomic analyses to differentiate bladder cancer from renal cell carcinoma. BMC Cancer19, 1195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Yamamoto, Y. et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci.110, 617–628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Salinas-Sánchez, A. S. et al. Clinical value of perioperative levels of DNA and mRNA in plasma of patients with renal cell carcinoma. Transl. Oncol.14, 100999 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Del Re, M. et al. The amount of DNA combined with TP53 mutations in liquid biopsy is associated with clinical outcome of renal cancer patients treated with immunotherapy and VEGFR-TKIs. J. Transl. Med.20, 371 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Bacon, J. V. W. et al. Plasma circulating tumor DNA and clonal hematopoiesis in metastatic renal cell carcinoma. Clin. Genitourin. Cancer18, 322–331.e322 (2020). [DOI] [PubMed] [Google Scholar]
- 312.Koh, Y. et al. Early dynamics of circulating tumor DNA predict clinical response to immune checkpoint inhibitors in metastatic renal cell carcinoma. Int. J. Urol.29, 462–469 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mytsyk, Y. et al. MicroRNA-15a expression measured in urine samples as a potential biomarker of renal cell carcinoma. Int. Urol. Nephrol.50, 851–859 (2018). [DOI] [PubMed] [Google Scholar]
- 314.Wang, Z. L. et al. Dynamic changes of different phenotypic and genetic circulating tumor cells as a biomarker for evaluating the prognosis of RCC. Cancer Biol. Ther.20, 505–512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhang, W. et al. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur. Urol. Focus4, 412–419 (2018). [DOI] [PubMed] [Google Scholar]
- 316.Xiao, C. T., Lai, W. J., Zhu, W. A. & Wang, H. MicroRNA derived from circulating exosomes as noninvasive biomarkers for diagnosing renal cell carcinoma. Onco Targets Ther.13, 10765–10774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Guo, R. et al. LncRNA RCAT1 promotes tumor progression and metastasis via miR-214-5p/E2F2 axis in renal cell carcinoma. Cell Death Dis.12, 689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Liu, H. et al. circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics10, 10791–10807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Li, Y. et al. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol. Med.21, 381–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Zhao, C. et al. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J. Urol.37, 1639–1647 (2019). [DOI] [PubMed] [Google Scholar]
- 321.Piao, X. M., Cha, E. J., Yun, S. J. & Kim, W. J. Role of exosomal miRNA in bladder cancer: a promising liquid biopsy biomarker. Int. J. Mol. Sci.22, 1713 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Li, S. et al. Blood-based liquid biopsy: insights into early detection, prediction, and treatment monitoring of bladder cancer. Cell Mol. Biol. Lett.28, 28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Qi, F. et al. Quantitation of rare circulating tumor cells by folate receptor α ligand-targeted PCR in bladder transitional cell carcinoma and its potential diagnostic significance. Tumour Biol.35, 7217–7223 (2014). [DOI] [PubMed] [Google Scholar]
- 324.Valenzuela, M. T. et al. Assessing the use of p16(INK4a) promoter gene methylation in serum for detection of bladder cancer. Eur. Urol.42, 622–628 (2002). [DOI] [PubMed] [Google Scholar]
- 325.Ellinger, J. et al. Hypermethylation of cell-free serum DNA indicates worse outcome in patients with bladder cancer. J. Urol.179, 346–352 (2008). [DOI] [PubMed] [Google Scholar]
- 326.Domínguez, G. et al. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin. Cancer Res.8, 980–985 (2002). [PubMed] [Google Scholar]
- 327.Lin, Y. L., Sun, G., Liu, X. Q., Li, W. P. & Ma, J. G. Clinical significance of CDH13 promoter methylation in serum samples from patients with bladder transitional cell carcinoma. J. Int. Med. Res.39, 179–186 (2011). [DOI] [PubMed] [Google Scholar]
- 328.Feng, Y. et al. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J. Exp. Clin. Cancer Res.33, 67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Adam, L. et al. Plasma microRNA profiles for bladder cancer detection. Urol. Oncol.31, 1701–1708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Jiang, X. et al. Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget7, 36733–36742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Jiang, X. et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int. J. Cancer136, 854–862 (2015). [DOI] [PubMed] [Google Scholar]
- 332.Liang, Z., Liu, L., Gao, R., Che, C. & Yang, G. Downregulation of exosomal miR-7-5p promotes breast cancer migration and invasion by targeting RYK and participating in the atypical WNT signalling pathway. Cell Mol. Biol. Lett.27, 88 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Yin, X. et al. Exosomal miR-663b targets Ets2-repressor factor to promote proliferation and the epithelial-mesenchymal transition of bladder cancer cells. Cell Biol. Int.44, 958–965 (2020). [DOI] [PubMed] [Google Scholar]
- 334.Cai, Q. et al. Urine BLCA-4 exerts potential role in detecting patients with bladder cancers: a pooled analysis of individual studies. Oncotarget6, 37500–37510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Roupret, M. et al. Diagnostic accuracy of MCM5 for the detection of recurrence in nonmuscle invasive bladder cancer followup: a blinded, prospective cohort, multicenter European study. J. Urol.204, 685–690 (2020). [DOI] [PubMed] [Google Scholar]
- 336.Southgate, J., Harnden, P. & Trejdosiewicz, L. K. Cytokeratin expression patterns in normal and malignant urothelium: a review of the biological and diagnostic implications. Histol. Histopathol.14, 657–664 (1999). [DOI] [PubMed] [Google Scholar]
- 337.Hosen, M. I. et al. Urinary TERT promoter mutations are detectable up to 10 years prior to clinical diagnosis of bladder cancer: evidence from the Golestan Cohort Study. EBioMedicine53, 102643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hernández, S. et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J. Clin. Oncol.24, 3664–3671 (2006). [DOI] [PubMed] [Google Scholar]
- 339.Haga, N. et al. Increase in circulating tumor cells in invasive bladder cancer after transurethral resection of bladder tumor. Anticancer Res.40, 4299–4307 (2020). [DOI] [PubMed] [Google Scholar]
- 340.Gazzaniga, P. et al. Circulating tumor cells detection has independent prognostic impact in high-risk non-muscle invasive bladder cancer. Int. J. Cancer135, 1978–1982 (2014). [DOI] [PubMed] [Google Scholar]
- 341.Gazzaniga, P. et al. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: a CellSearch analysis. Ann. Oncol.23, 2352–2356 (2012). [DOI] [PubMed] [Google Scholar]
- 342.Vandekerkhove, G. et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat. Commun.12, 184 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Raimondi, C., Gradilone, A. & Gazzaniga, P. Circulating tumor cells in early bladder cancer: insight into micrometastatic disease. Expert Rev. Mol. Diagn.14, 407–409 (2014). [DOI] [PubMed] [Google Scholar]
- 344.Zhang, Z. et al. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: a meta-analysis of 30 published studies. Oncotarget8, 59527–59538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Beije, N. et al. Circulating tumour cells to drive the use of neoadjuvant chemotherapy in patients with muscle-invasive bladder cancer. ESMO Open7, 100416 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Anantharaman, A. et al. Programmed death-ligand 1 (PD-L1) characterization of circulating tumor cells (CTCs) in muscle invasive and metastatic bladder cancer patients. BMC Cancer16, 744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med.379, 1754–1765 (2018). [DOI] [PubMed] [Google Scholar]
- 348.Raja, R. et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res.24, 6212–6222 (2018). [DOI] [PubMed] [Google Scholar]
- 349.Shohdy, K. S. et al. Serial ctDNA analysis predicts clinical progression in patients with advanced urothelial carcinoma. Br. J. Cancer126, 430–439 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Birkenkamp-Demtröder, K. et al. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur. Urol.73, 535–540 (2018). [DOI] [PubMed] [Google Scholar]
- 351.de Kruijff, I. E. et al. Liquid biopsies to select patients for perioperative chemotherapy in muscle-invasive bladder cancer: a systematic review. Eur. Urol. Oncol.4, 204–214 (2021). [DOI] [PubMed] [Google Scholar]
- 352.Powles, T. et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature595, 432–437 (2021). [DOI] [PubMed] [Google Scholar]
- 353.Zhang, J. et al. Circulating tumor DNA analyses predict disease recurrence in non-muscle-invasive bladder cancer. Front. Oncol.11, 657483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Koguchi, D. et al. Diagnostic potential of circulating tumor cells, urinary MicroRNA, and urinary cell-free DNA for bladder cancer: a review. Int. J. Mol. Sci.23, 9148 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Zheng, H., Liu, J., Pan, X. & Cui, X. Biomarkers for patients with Wilms tumor: a review. Front. Oncol.13, 1137346 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Miguez, A. C. K. et al. Assessment of somatic mutations in urine and plasma of Wilms tumor patients. Cancer Med.9, 5948–5959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Schmitt, J. et al. Treatment-independent miRNA signature in blood of Wilms tumor patients. BMC Genom.13, 379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Treger, T. D. et al. Somatic TP53 mutations are detectable in circulating tumor DNA from children with anaplastic Wilms tumors. Transl. Oncol.11, 1301–1306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Madanat-Harjuoja, L. M. et al. Circulating tumor DNA as a biomarker in patients with stage III and IV Wilms tumor: analysis from a Children’s Oncology Group Trial, AREN0533. J. Clin. Oncol.40, 3047–3056 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Perotti, D. et al. Hallmark discoveries in the biology of Wilms tumour. Nat. Rev. Urol.21, 158–180 (2024). [DOI] [PubMed] [Google Scholar]
- 361.Stern, M., Longaker, M. T., Adzick, N. S., Harrison, M. R. & Stern, R. Hyaluronidase levels in urine from Wilms’ tumor patients. J. Natl. Cancer Inst.83, 1569–1574 (1991). [DOI] [PubMed] [Google Scholar]
- 362.Lin, R. Y., Argenta, P. A., Sullivan, K. M. & Adzick, N. S. Diagnostic and prognostic role of basic fibroblast growth factor in Wilms’ tumor patients. Clin. Cancer Res.1, 327–331 (1995). [PubMed] [Google Scholar]
- 363.Padullés, B. et al. Prognostic value of liquid-biopsy-based biomarkers in upper tract urothelial carcinoma. Int. J. Mol. Sci.25, 3695 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Nakano, K. et al. Fragmentation of cell-free DNA is induced by upper-tract urothelial carcinoma-associated systemic inflammation. Cancer Sci.112, 168–177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Blumendeller, C. et al. Use of plasma ctDNA as a potential biomarker for longitudinal monitoring of a patient with metastatic high-risk upper tract urothelial carcinoma receiving pembrolizumab and personalized neoepitope-derived multipeptide vaccinations: a case report. J. Immunother. Cancer9, e001406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Springer, S. U. et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife7, e32143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Ghoreifi, A. et al. A urine-based DNA methylation marker test to detect upper tract urothelial carcinoma: a prospective cohort study. J. Urol.209, 854–862 (2023). [DOI] [PubMed] [Google Scholar]
- 368.Ge, G. et al. Urothelial carcinoma detection based on copy number profiles of urinary cell-free DNA by shallow whole-genome sequencing. Clin. Chem.66, 188–198 (2020). [DOI] [PubMed] [Google Scholar]
- 369.Lu, H. et al. Aristolochic acid mutational signature defines the low-risk subtype in upper tract urothelial carcinoma. Theranostics10, 4323–4333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Chalfin, H. J. et al. Circulating tumor cell and circulating tumor DNA assays reveal complementary information for patients with metastatic urothelial cancer. Eur. Urol. Oncol.4, 310–314 (2021). [DOI] [PubMed] [Google Scholar]
- 371.Xu, Y. et al. A urine-based liquid biopsy method for detection of upper tract urinary carcinoma. Front. Oncol.10, 597486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Urabe, F. et al. Independent verification of circulating miRNA as diagnostic biomarkers for urothelial carcinoma. Cancer Sci.113, 3510–3517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Kriebel, S. et al. Analysis of tissue and serum microRNA expression in patients with upper urinary tract urothelial cancer. PLoS ONE10, e0117284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Montalbo, R. et al. Prognostic value of circulating microRNAs in upper tract urinary carcinoma. Oncotarget9, 16691–16700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Li, Y. et al. Identification of plasma secreted phosphoprotein 1 as a novel biomarker for upper tract urothelial carcinomas. Biomed. Pharmacother.113, 108744 (2019). [DOI] [PubMed] [Google Scholar]
- 376.Hsu, Y. P. et al. Instrument-free detection of FXYD3 using vial-based immunosensor for earlier and faster urothelial carcinoma diagnosis. ACS Sens.5, 928–935 (2020). [DOI] [PubMed] [Google Scholar]
- 377.Mori, K. et al. Prognostic value of preoperative blood-based biomarkers in upper tract urothelial carcinoma treated with nephroureterectomy: a systematic review and meta-analysis. Urol. Oncol.38, 315–333 (2020). [DOI] [PubMed] [Google Scholar]
- 378.Traeger, L. et al. Serum Hepcidin and GDF-15 levels as prognostic markers in urothelial carcinoma of the upper urinary tract and renal cell carcinoma. BMC Cancer19, 74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Rogers, A. et al. Relative increase in leukemia-specific DNA in peripheral blood plasma from patients with acute myeloid leukemia and myelodysplasia. Blood103, 2799–2801 (2004). [DOI] [PubMed] [Google Scholar]
- 380.Zhao, P. et al. Using circulating tumor DNA to monitor myelodysplastic syndromes status. Hematol. Oncol.37, 531–533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Ruan, M. et al. Targeted next-generation sequencing of circulating tumor DNA, bone marrow, and peripheral blood mononuclear cells in pediatric AML. Front. Oncol.11, 666470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Garcia-Gisbert, N. et al. Molecular and cytogenetic characterization of myelodysplastic syndromes in cell-free DNA. Blood Adv.6, 3178–3188 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Gao, Y. J. et al. Increased integrity of circulating cell-free DNA in plasma of patients with acute leukemia. Clin. Chem. Lab Med.48, 1651–1656 (2010). [DOI] [PubMed] [Google Scholar]
- 384.Božic, T. et al. Investigation of measurable residual disease in acute myeloid leukemia by DNA methylation patterns. Leukemia36, 80–89 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Suzuki, Y. et al. Peripheral blood cell-free DNA is an alternative tumor DNA source reflecting disease status in myelodysplastic syndromes. Cancer Sci.107, 1329–1337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Yao, C. Y. et al. Distinct mutation profile and prognostic relevance in patients with hypoplastic myelodysplastic syndromes (h-MDS). Oncotarget7, 63177–63188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Liu, L. P. et al. Early detection of molecular residual disease and risk stratification for children with acute myeloid leukemia via circulating tumor DNA. Clin. Cancer Res.30, 1143–1151 (2024). [DOI] [PubMed] [Google Scholar]
- 388.Xue, Y. et al. Applications of circulating tumor DNA in myelodysplastic syndromes and acute myeloid leukemia: promises and challenges. Front. Biosci. (Landmark Ed.).29, 86 (2024). [DOI] [PubMed] [Google Scholar]
- 389.Nakamura, S. et al. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood133, 2682–2695 (2019). [DOI] [PubMed] [Google Scholar]
- 390.Rossi, D. et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood129, 1947–1957 (2017). [DOI] [PubMed] [Google Scholar]
- 391.Scherer, F. et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med.8, 364ra155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Roschewski, M. et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol.16, 541–549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Lauer, E. M., Mutter, J. & Scherer, F. Circulating tumor DNA in B-cell lymphoma: technical advances, clinical applications, and perspectives for translational research. Leukemia36, 2151–2164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Kurtz, D. M. et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell178, 699–713.e619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Kurtz, D. M. et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol.39, 1537–1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Oki, Y. et al. Detection of classical Hodgkin lymphoma specific sequence in peripheral blood using a next-generation sequencing approach. Br. J. Haematol.169, 689–693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Desch, A. K. et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia34, 151–166 (2020). [DOI] [PubMed] [Google Scholar]
- 398.Roschewski, M., Rossi, D., Kurtz, D. M., Alizadeh, A. A. & Wilson, W. H. Circulating tumor DNA in lymphoma: principles and future directions. Blood Cancer Discov.3, 5–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Spina, V. et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood131, 2413–2425 (2018). [DOI] [PubMed] [Google Scholar]
- 400.Mutter, J. A. et al. Circulating tumor DNA profiling for detection, risk stratification, and classification of brain lymphomas. J. Clin. Oncol.41, 1684–1694 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Roemer, M. G. M. et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J. Clin. Oncol.36, 942–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Li, S., Zhang, E. & Cai, Z. Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma. Biomark. Res.11, 27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Garcés, J. J. et al. Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia34, 3007–3018 (2020). [DOI] [PubMed] [Google Scholar]
- 404.Mishima, Y. et al. The mutational landscape of circulating tumor cells in multiple myeloma. Cell Rep.19, 218–224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Rustad, E. H. et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica102, 1266–1272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Mithraprabhu, S. et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia31, 1695–1705 (2017). [DOI] [PubMed] [Google Scholar]
- 407.Mithraprabhu, S., Sirdesai, S., Chen, M., Khong, T. & Spencer, A. Circulating tumour DNA analysis for tumour genome characterisation and monitoring disease burden in extramedullary multiple myeloma. Int J. Mol. Sci.19, 1858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Gerber, B. et al. Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica103, e245–e248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Coffey, D. G. et al. High-throughput drug screening and multi-omic analysis to guide individualized treatment for multiple myeloma. JCO Precis. Oncol.5, PO.20.00442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Li, S. et al. Targeting the GCK pathway: a novel and selective therapeutic strategy against RAS-mutated multiple myeloma. Blood137, 1754–1764 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Giesen, N. et al. A phase 2 clinical trial of combined BRAF/MEK inhibition for BRAFV600E-mutated multiple myeloma. Blood141, 1685–1690 (2023). [DOI] [PubMed] [Google Scholar]
- 412.Brown, R. L., de Souza, J. A. & Cohen, E. E. Thyroid cancer: burden of illness and management of disease. J. Cancer2, 193–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Kure, S. & Ohashi, R. Thyroid Hürthle cell carcinoma: clinical, pathological, and molecular features. Cancers13, 26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Schlumberger, M. & Leboulleux, S. Current practice in patients with differentiated thyroid cancer. Nat. Rev. Endocrinol.17, 176–188 (2021). [DOI] [PubMed] [Google Scholar]
- 415.Bankó, P. et al. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol.12, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Feng, Z. et al. Circulating tumor cells in the early detection of human cancers. Int. J. Biol. Sci.18, 3251–3265 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Ehlers, M. et al. Increased numbers of circulating tumor cells in thyroid cancer patients. Horm. Metab. Res.50, 602–608 (2018). [DOI] [PubMed] [Google Scholar]
- 418.Zane, M. et al. Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAF(V600E): a non-invasive tool panel for early detection of thyroid cancer. Biomed. Pharmacother.67, 723–730 (2013). [DOI] [PubMed] [Google Scholar]
- 419.Liu, Y., Geng, H., Liu, X., Cao, M. & Zhang, X. A meta-analysis of circulating microRNAs in the diagnosis of papillary thyroid carcinoma. PLoS One16, e0251676 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Delcorte, O. et al. Two miRNAs enriched in plasma extracellular vesicles are potential biomarkers for thyroid cancer. Endocr. Relat. Cancer29, 389–401 (2022). [DOI] [PubMed] [Google Scholar]
- 421.Sato, T. et al. Circulating tumor cells detected by reverse transcription-polymerase chain reaction for carcinoembryonic antigen mRNA: distinguishing follicular thyroid carcinoma from adenoma. Surgery137, 552–558 (2005). [DOI] [PubMed] [Google Scholar]
- 422.Qiu, Z. L. et al. Circulating tumor cells correlate with clinicopathological features and outcomes in differentiated thyroid cancer. Cell Physiol. Biochem.48, 718–730 (2018). [DOI] [PubMed] [Google Scholar]
- 423.Yan, C., Huang, M., Li, X., Wang, T. & Ling, R. Relationship between BRAF V600E and clinical features in papillary thyroid carcinoma. Endocr. Connect.8, 988–996 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Allin, D. M. et al. Circulating tumour DNA is a potential biomarker for disease progression and response to targeted therapy in advanced thyroid cancer. Eur. J. Cancer103, 165–175 (2018). [DOI] [PubMed] [Google Scholar]
- 425.Ciampi, R. et al. Pre- and post-operative circulating tumoral DNA in patients with medullary thyroid carcinoma. J. Clin. Endocrinol. Metab.107, e3420–e3427 (2022). [DOI] [PubMed] [Google Scholar]
- 426.Lubitz, C. C. et al. Circulating BRAF(V600E) levels correlate with treatment in patients with thyroid carcinoma. Thyroid28, 328–339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Cote, G. J. et al. Prognostic significance of circulating RET M918T mutated tumor DNA in patients with advanced medullary thyroid carcinoma. J. Clin. Endocrinol. Metab.102, 3591–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Cradic, K. W. et al. Mutant BRAF(T1799A) can be detected in the blood of papillary thyroid carcinoma patients and correlates with disease status. J. Clin. Endocrinol. Metab.94, 5001–5009 (2009). [DOI] [PubMed] [Google Scholar]
- 429.Qin, Y. et al. Clinical utility of circulating cell-free DNA mutations in anaplastic thyroid carcinoma. Thyroid31, 1235–1243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Hu, S. et al. Detection of serum deoxyribonucleic acid methylation markers: a novel diagnostic tool for thyroid cancer. J. Clin. Endocrinol. Metab.91, 98–104 (2006). [DOI] [PubMed] [Google Scholar]
- 431.Wen, Q., Wang, Y., Li, X., Jin, X. & Wang, G. Decreased serum exosomal miR-29a expression and its clinical significance in papillary thyroid carcinoma. J. Clin. Lab. Anal.35, e23560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Lee, J. C. et al. Papillary thyroid cancer-derived exosomes contain miRNA-146b and miRNA-222. J. Surg. Res.196, 39–48 (2015). [DOI] [PubMed] [Google Scholar]
- 433.Toraih, E. A. et al. Diagnostic and prognostic performance of liquid biopsy-derived exosomal microRNAs in thyroid cancer patients: a systematic review and meta-analysis. Cancers13, 4295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J. Clin.66, 115–132 (2016). [DOI] [PubMed] [Google Scholar]
- 435.Freitas, A. J. A. et al. Liquid Biopsy as a Tool for the Diagnosis, Treatment, and Monitoring of Breast Cancer. Int. J. Mol. Sci.23, 9952 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Guttery, D. S. et al. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin. Chem.61, 974–982 (2015). [DOI] [PubMed] [Google Scholar]
- 437.Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med.7, 302ra133 (2015). [DOI] [PubMed] [Google Scholar]
- 438.Beaver, J. A. et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res.20, 2643–2650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Rugo, H. S. et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol.22, 489–498 (2021). [DOI] [PubMed] [Google Scholar]
- 440.Hai, L., Li, L., Liu, Z., Tong, Z. & Sun, Y. Whole-genome circulating tumor DNA methylation landscape reveals sensitive biomarkers of breast cancer. MedComm3, e134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Hannafon, B. N. et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res.18, 90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Eichelser, C. et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget5, 9650–9663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Li, M. et al. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat.170, 257–270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Liu, C. et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett.18, 4226–4232 (2018). [DOI] [PubMed] [Google Scholar]
- 445.Racila, E. et al. Detection and characterization of carcinoma cells in the blood. Proc. Natl Acad. Sci. USA95, 4589–4594 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Ozkumur, E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med.5, 179ra147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Hvichia, G. E. et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer138, 2894–2904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res.10, 6897–6904 (2004). [DOI] [PubMed] [Google Scholar]
- 449.Gradishar, W. J. et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw.20, 691–722 (2022). [DOI] [PubMed] [Google Scholar]
- 450.Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med.368, 1199–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 451.Famta, P. et al. Enigmatic role of exosomes in breast cancer progression and therapy. Life Sci.289, 120210 (2022). [DOI] [PubMed] [Google Scholar]
- 452.Nakamura, S. et al. Multi-center study evaluating circulating tumor cells as a surrogate for response to treatment and overall survival in metastatic breast cancer. Breast Cancer17, 199–204 (2010). [DOI] [PubMed] [Google Scholar]
- 453.Jiang, Z. et al. Chinese Society of Clinical Oncology (CSCO) breast cancer guidelines 2022. Transl. Breast Cancer Res.3, 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Klinge, C. M. Non-coding RNAs in breast cancer: intracellular and intercellular communication. Noncoding RNA4, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Tierno, D., Grassi, G., Zanconati, F., Dapas, B. & Scaggiante, B. Plasma circular RNAs as biomarkers for breast cancer. Biomedicines12, 875 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Benini, S. et al. Detection of circulating tumor cells in liquid biopsy from Ewing sarcoma patients. Cancer Manag Res.10, 49–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Krumbholz, M. et al. Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in Ewing sarcoma. Clin. Cancer Res.22, 4356–4365 (2016). [DOI] [PubMed] [Google Scholar]
- 459.Gutteridge, A. et al. Digital PCR analysis of circulating tumor DNA: a biomarker for chondrosarcoma diagnosis, prognostication, and residual disease detection. Cancer Med.6, 2194–2202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Klega, K. et al. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children With solid tumors. JCO Precis. Oncol.2018, PO.17.00285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.McBride, D. J. et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer49, 1062–1069 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Shulman, D. S. et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children’s Oncology Group. Br. J. Cancer119, 615–621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Momen-Heravi, F. et al. Current methods for the isolation of extracellular vesicles. Biol. Chem.394, 1253–1262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Fang, S. et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One12, e0175050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Liang, L. G. et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci. Rep.7, 46224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Vaidyanathan, R. et al. Detecting exosomes specifically: a multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal. Chem.86, 11125–11132 (2014). [DOI] [PubMed] [Google Scholar]
- 467.Sina, A. A. et al. Real time and label free profiling of clinically relevant exosomes. Sci. Rep.6, 30460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Gholizadeh, S. et al. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: current status and future directions. Biosens. Bioelectron.91, 588–605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Liga, A., Vliegenthart, A. D., Oosthuyzen, W., Dear, J. W. & Kersaudy-Kerhoas, M. Exosome isolation: a microfluidic road-map. Lab. Chip15, 2388–2394 (2015). [DOI] [PubMed] [Google Scholar]
- 470.Nugent, M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag. Res.6, 15–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Lulla, R. R. et al. Identification of differentially expressed MicroRNAs in osteosarcoma. Sarcoma2011, 732690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Tian, Q. et al. A causal role for circulating miR-34b in osteosarcoma. Eur. J. Surg. Oncol.40, 67–72 (2014). [DOI] [PubMed] [Google Scholar]
- 473.Urdinez, J. et al. The miR-143/145 cluster, a novel diagnostic biomarker in chondrosarcoma, acts as a tumor suppressor and directly inhibits Fascin-1. J. Bone Min. Res.35, 1077–1091 (2020). [DOI] [PubMed] [Google Scholar]
- 474.Parafioriti, A. et al. Expression profiling of microRNAs and isomiRs in conventional central chondrosarcoma. Cell Death Discov.6, 46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Sciandra, M. et al. Circulating miR34a levels as a potential biomarker in the follow-up of Ewing sarcoma. J. Cell Commun. Signal14, 335–347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Zhang, S., Li, D., Jiao, G. J., Wang, H. L. & Yan, T. B. miR-185 suppresses progression of Ewing’s sarcoma via inhibiting the PI3K/AKT and Wnt/β-catenin pathways. Onco Targets Ther.11, 7967–7977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Cafforio, P. et al. Liquid biopsy in cervical cancer: hopes and pitfalls. Cancers13, 3968 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Thangarajah, F. et al. Digital droplet PCR-based quantification of ccfHPV-DNA as liquid biopsy in HPV-driven cervical and vulvar cancer. J. Cancer Res. Clin. Oncol.149, 12597–12604 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Charo, L. M. et al. Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients. Mol. Oncol.15, 67–79 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Galati, L. et al. Detection of circulating HPV16 DNA as a biomarker for cervical cancer by a bead-based HPV genotyping assay. Microbiol. Spectr.10, e0148021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Tornesello, M. L. et al. The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front. Oncol.10, 150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Sun, W. et al. Four circulating long non-coding RNAs act as biomarkers for predicting cervical cancer. Gynecol. Obstet. Investig.83, 533–539 (2018). [DOI] [PubMed] [Google Scholar]
- 483.Jia, W. et al. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol.3, 851–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Liu, P., Xin, F. & Ma, C. F. Clinical significance of serum miR-196a in cervical intraepithelial neoplasia and cervical cancer. Genet. Mol. Res.14, 17995–18002 (2015). [DOI] [PubMed] [Google Scholar]
- 485.Sun, L. et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann. Clin. Biochem.54, 127–133 (2017). [DOI] [PubMed] [Google Scholar]
- 486.Page, K. et al. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin. Chem.63, 532–541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Bohers, E. et al. Somatic mutations of cell-free circulating DNA detected by next-generation sequencing reflect the genetic changes in both germinal center B-cell-like and activated B-cell-like diffuse large B-cell lymphomas at the time of diagnosis. Haematologica100, e280–e284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Chicard, M. et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin. Cancer Res.24, 939–949 (2018). [DOI] [PubMed] [Google Scholar]
- 489.Tian, X. et al. Dynamic analysis of circulating tumor DNA to predict prognosis and monitor therapeutic response in metastatic relapsed cervical cancer. Int. J. Cancer148, 921–931 (2021). [DOI] [PubMed] [Google Scholar]
- 490.Tewari, K. S. et al. Circulating Tumor Cells In Advanced Cervical Cancer: NRG Oncology-Gynecologic Oncology Group Study 240 (NCT 00803062). Mol. Cancer Ther.19, 2363–2370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Weismann, P. et al. The detection of circulating tumor cells expressing E6/E7 HR-HPV oncogenes in peripheral blood in cervical cancer patients after radical hysterectomy. Neoplasma56, 230–238 (2009). [DOI] [PubMed] [Google Scholar]
- 492.Obermayr, E. et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer10, 666 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Kiss, I., Kolostova, K., Pawlak, I. & Bobek, V. Circulating tumor cells in gynaecological malignancies. J. Buon25, 40–50 (2020). [PubMed] [Google Scholar]
- 494.Du, K. et al. Circulating tumor cells counting act as a potential prognostic factor in cervical cancer. Technol. Cancer Res. Treat.19, 1533033820957005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Constantine, G. D., Kessler, G., Graham, S. & Goldstein, S. R. Increased incidence of endometrial cancer following the women’s health initiative: an assessment of risk factors. J. Women’s. Health28, 237–243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Lortet-Tieulent, J., Ferlay, J., Bray, F. & Jemal, A. International patterns and trends in endometrial cancer incidence, 1978-2013. J. Natl. Cancer Inst.110, 354–361 (2018). [DOI] [PubMed] [Google Scholar]
- 497.Shen, Y., Shi, R., Zhao, R. & Wang, H. Clinical application of liquid biopsy in endometrial carcinoma. Med. Oncol.40, 92 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Kiss, I. et al. Correlation between disease stage and the presence of viable circulating tumor cells in endometrial cancer. Anticancer Res.38, 2983–2987 (2018). [DOI] [PubMed] [Google Scholar]
- 499.Bogani, G. et al. Detection of circulating tumor cells in high-risk endometrial cancer. Anticancer Res.35, 683–687 (2015). [PubMed] [Google Scholar]
- 500.Bolivar, A. M. et al. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: identification of plasma-based, tumor-associated mutations in early-stage patients. Mod. Pathol.32, 405–414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Wang, L. et al. Circulating microRNAs as a fingerprint for endometrial endometrioid adenocarcinoma. PLoS One9, e110767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Buscail, E. et al. Tumor-proximal liquid biopsy to improve diagnostic and prognostic performances of circulating tumor cells. Mol. Oncol.13, 1811–1826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Grant, B. M., Pugh, T. J. & Oza, A. M. Molecular monitoring in endometrial cancer-ready for prime time? Clin. Cancer Res.29, 305–308 (2023). [DOI] [PubMed] [Google Scholar]
- 504.He, D. et al. DNMT3A/3B overexpression might be correlated with poor patient survival, hypermethylation and low expression of ESR1/PGR in endometrioid carcinoma: an analysis of The Cancer Genome Atlas. Chin. Med J.132, 161–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Yang, J. et al. Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid. Int. J. Mol. Sci.24, 8683 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Urabe, F. et al. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Cell Physiol.318, C29–c39 (2020). [DOI] [PubMed] [Google Scholar]
- 507.Nakamura, K. et al. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol. Cancer15, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.S, E. L. A., Mäger, I., Breakefield, X. O. & Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov.12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 509.van den Helder, R. et al. Non-invasive detection of endometrial cancer by DNA methylation analysis in urine. Clin. Epigenet.12, 165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Karimi, F. et al. Liquid biopsy in ovarian cancer: advantages and limitations for prognosis and diagnosis. Med. Oncol.40, 265 (2023). [DOI] [PubMed] [Google Scholar]
- 511.Marth, C., Kisic, J., Kaern, J., Tropé, C. & Fodstad, Ø. Circulating tumor cells in the peripheral blood and bone marrow of patients with ovarian carcinoma do not predict prognosis. Cancer94, 707–712 (2002). [DOI] [PubMed] [Google Scholar]
- 512.Judson, P. L. et al. Preoperative detection of peripherally circulating cancer cells and its prognostic significance in ovarian cancer. Gynecol. Oncol.91, 389–394 (2003). [DOI] [PubMed] [Google Scholar]
- 513.Zhu, J. W., Charkhchi, P. & Akbari, M. R. Potential clinical utility of liquid biopsies in ovarian cancer. Mol. Cancer21, 114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Asante, D. B., Calapre, L., Ziman, M., Meniawy, T. M. & Gray, E. S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: ready for prime time? Cancer Lett.468, 59–71 (2020). [DOI] [PubMed] [Google Scholar]
- 515.Siena, S. et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann. Oncol.29, 119–126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 517.Zheng, X., Li, X. & Wang, X. Extracellular vesicle-based liquid biopsy holds great promise for the management of ovarian cancer. Biochim. Biophys. Acta Rev. Cancer1874, 188395 (2020). [DOI] [PubMed] [Google Scholar]
- 518.Wang, L., Zhao, F., Xiao, Z. & Yao, L. Exosomal microRNA-205 is involved in proliferation, migration, invasion, and apoptosis of ovarian cancer cells via regulating VEGFA. Cancer Cell Int.19, 281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Mateescu, B. et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med.17, 1627–1635 (2011). [DOI] [PubMed] [Google Scholar]
- 520.Konstantinopoulos, P. A., Lheureux, S. & Moore, K. N. PARP inhibitors for ovarian cancer: current indications, future combinations, and novel assets in development to target DNA damage repair. Am. Soc. Clin. Oncol. Educ. Book40, 1–16 (2020). [DOI] [PubMed] [Google Scholar]
- 521.Wang, M. et al. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci.13, 1497–1506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Zhou, W. et al. Serum exosomes from epithelial ovarian cancer patients contain LRP1, which promotes the migration of epithelial ovarian cancer cell. Mol. Cell Proteom.22, 100520 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Li, J. et al. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer9, 244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Su, Y. Y. et al. Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J. Ovar. Res.12, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Yazawa, H. et al. Hydrodynamics-based gene delivery of naked DNA encoding fetal liver kinase-1 gene effectively suppresses the growth of pre-existing tumors. Cancer Gene Ther.13, 993–1001 (2006). [DOI] [PubMed] [Google Scholar]
- 526.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48 (2023). [DOI] [PubMed] [Google Scholar]
- 527.Lilja, H., Ulmert, D. & Vickers, A. J. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat. Rev. Cancer8, 268–278 (2008). [DOI] [PubMed] [Google Scholar]
- 528.Sharma, S. et al. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv.36, 1063–1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Schaeffer, E. M. et al. Prostate cancer, version 4.2023, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw.21, 1067–1096 (2023). [DOI] [PubMed] [Google Scholar]
- 530.Lindsay, C. R. et al. Vimentin and Ki67 expression in circulating tumour cells derived from castrate-resistant prostate cancer. BMC Cancer16, 168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Schütz, E. et al. Chromosomal instability in cell-free DNA is a serum biomarker for prostate cancer. Clin. Chem.61, 239–248 (2015). [DOI] [PubMed] [Google Scholar]
- 532.Choudhury, A. D. et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight3, e122109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Han, X. Y. et al. A new mycobacterium species causing diffuse lepromatous leprosy. Am. J. Clin. Pathol.130, 856–864 (2008). [DOI] [PubMed] [Google Scholar]
- 534.Singh, N. et al. The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun.12, 7349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.O’Brien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol.9, 402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Sharova, E. et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br. J. Cancer114, 1362–1366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Matsuzaki, K. et al. MiR-30b-3p and miR-126-3p of urinary extracellular vesicles could be new biomarkers for prostate cancer. Transl. Androl. Urol.10, 1918–1927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Zhang, H. L. et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate71, 326–331 (2011). [DOI] [PubMed] [Google Scholar]
- 539.Selth, L. A. et al. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. Cancer109, 641–650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Liang, C. et al. Long non-coding RNA PCAT-1 in human cancers: a meta-analysis. Clin. Chim. Acta480, 47–55 (2018). [DOI] [PubMed] [Google Scholar]
- 541.Xue, Y. et al. Association between lncrna PCGEM1 polymorphisms and prostate cancer risk. Prostate Cancer Prostatic Dis.16, 139–144 (2013). [DOI] [PubMed] [Google Scholar]
- 542.Prensner, J. R. et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol.15, 1469–1480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Prensner, J. R. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet.45, 1392–1398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Logozzi, M. et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett.403, 318–329 (2017). [DOI] [PubMed] [Google Scholar]
- 545.De Giorgi, U., Conteduca, V., Scarpi, E. & Re: Marzia Del Re, Elisa Biasco, Stefania Crucitta, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal rna strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol 2017;71:680-7. Eur. Urol.73, e9–e10 (2018). [DOI] [PubMed] [Google Scholar]
- 546.Nimir, M. et al. Detection of AR-V7 in liquid biopsies of castrate resistant prostate cancer patients: a comparison of AR-V7 analysis in circulating tumor cells, circulating tumor RNA and exosomes. Cells8, 688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Raos, D. et al. cfDNA methylation in liquid biopsies as potential testicular seminoma biomarker. Epigenomics14, 1493–1507 (2022). [DOI] [PubMed] [Google Scholar]
- 548.Wang, K., Wang, X., Pan, Q. & Zhao, B. Liquid biopsy techniques and pancreatic cancer: diagnosis, monitoring, and evaluation. Mol. Cancer22, 167 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Murray, M. J. et al. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol. Biomark. Prev.24, 350–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Klein, A., Fishman, A., Zemer, R., Zimlichman, S. & Altaras, M. M. Detection of tumor circulating cells by cytokeratin 20 in the blood of patients with endometrial carcinoma. Gynecol. Oncol.78, 352–355 (2000). [DOI] [PubMed] [Google Scholar]