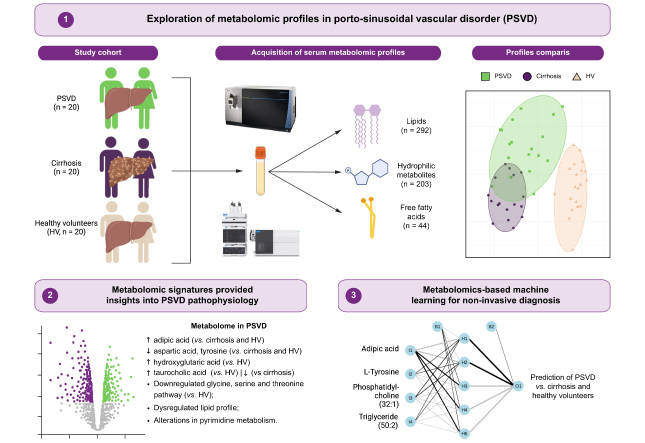
Abstract
Background & Aims
Porto-sinusoidal vascular disorder (PSVD) is a rare and diagnostically challenging vascular liver disease. This study aimed to identify distinct metabolomic signatures in patients with PSVD or cirrhosis to facilitate non-invasive diagnosis and elucidate perturbed metabolic pathways.
Methods
Serum samples from 20 healthy volunteers (HVs), 20 patients with histologically confirmed PSVD or 20 patients with cirrhosis were analyzed. Metabolites were measured using liquid chromatography-mass spectrometry. Differential abundance was evaluated with Limma’s moderated t-statistics. Artificial neural network and support vector machine models were developed to classify PSVD against cirrhosis or HV metabolomic profiles. An independent cohort was used for validation.
Results
A total of 283 metabolites were included for downstream analysis. Clustering effectively separated PSVD from HV metabolomes, although a subset of patients with PSVD (n = 5, 25%) overlapped with those with cirrhosis. Differential testing revealed significant PSVD-linked metabolic perturbations, including pertubations in taurocholic and adipic acids, distinguishing patients with PSVD from both HVs and those with cirrhosis. Alterations in pyrimidine, glycine, serine, and threonine pathways were exclusively associated with PSVD. Machine learning models utilizing selected metabolic signatures reliably differentiated the PSVD group from HVs or patients with cirrhosis using only 4 to 6 metabolites. Validation in an independent cohort demonstrated the high discriminative ability of taurocholic acid (AUROC 0.899) for patients with PSVD vs. HVs and the taurocholic acid/aspartic acid ratio (AUROC 0.720) for PSVD vs. cirrhosis.
Conclusions
High-throughput metabolomics enabled the identification of distinct metabolic profiles that differentiate between PSVD, cirrhosis, and healthy individuals. Unique alterations in the glycine, serine, and threonine pathways suggest their potential involvement in PSVD pathogenesis.
Impact and implications:
Porto-sinusoidal vascular disorder (PSVD) is a vascular liver disease that can lead to pre-sinusoidal portal hypertension in the absence of cirrhosis, with poorly understood pathophysiology and no established treatment. Our study demonstrates that analyzing the serum metabolome could reveal distinct metabolic signatures in patients with PSVD, including alterations in the pyrimidine, glycine, serine and threonine pathways, potentially shedding light on the disease's underlying pathways. These findings could enable earlier and non-invasive diagnosis of PSVD, potentially reducing reliance on invasive procedures like liver biopsy and guiding diagnostic pathways.
Keywords: PSVD, cirrhosis, ACLD, metabolomics, portal hypertension, non-cirrhotic portal hypertension
Graphical abstract
Highlights:
-
•
We identified distinct metabolic signatures that discriminate PSVD from cirrhosis and healthy controls.
-
•
A computational biology approach revealed alterations in the pyrimidine, glycine, serine and threonine pathways exclusively in patients with PSVD.
-
•
A machine learning-derived signature could reliably distinguish PSVD from healthy controls or cirrhosis using 4 to 6 metabolites.
-
•
These metabolomic perturbations might characterize pathophysiologically relevant pathways in PSVD.
-
•
Analyzing metabolomic profiles may allow for earlier non-invasive diagnosis of PSVD and guide diagnostic pathways.
Introduction
Porto-sinusoidal vascular disorder (PSVD) is a rare condition that encompasses vascular liver diseases with alterations of the periportal and sinusoidal vasculature often associated with pre-sinusoidal portal hypertension in the absence of cirrhosis.1,2 The clinical course of PSVD is linked with the severity of associated portal hypertension3 and patients occasionally require liver transplantation.4 Usually, patients with PSVD have preserved liver function, a low liver stiffness measurement (LSM),5,6 and often a normal or only slightly elevated hepatic venous pressure gradient (HVPG).3,[5], [6], [7], [8], [9] Intrahepatic venovenous communications, a less pronounced liver-surface nodularity (compared to cirrhosis), and specific alterations in the periportal area are among the commonly observed radio-morphological characteristics in PSVD.10,11 However, diagnosis and differentiation from cirrhosis remain challenging. First, diagnosis of PSVD requires an invasive liver biopsy of adequate quality,12 as well as evaluation by an expert pathologist. Second, patients usually remain asymptomatic for a long time and often present at late disease stages after portal hypertension-associated complications have already developed.3 Currently, non-invasive tests – especially the combination of LSM and spleen stiffness measurement – are increasingly being studied for non-invasive PSVD detection.13,14 However, no biomarkers have so far been proven sufficiently accurate to diagnose PSVD without performing a liver biopsy.
Metabolomics offers an approach to measure and analyze endogenous or exogenous metabolites (e.g., sugars, amino acids, organic acids, nucleotides, acylcarnitines, or lipids) in different biological samples.15 This includes both the identification and quantification of all (non-targeted metabolomics profiling) or specified (targeted metabolomics profiling) metabolites that constitute an individual’s metabolome.16 Two previous studies have examined plasma metabolomics in patients defined by the old criteria for “idiopathic non-cirrhotic portal hypertension” (INCPH).17,18 They identified ∼30 metabolites differentiating INCPH from cirrhosis and healthy volunteers (HVs).17 Based on a 5-metabolite signature including an acyl-carnitine, a bile acid, a fatty acid, a lysophosphatidylethanolamine, and sphingomyelin, patients with INCPH could be discriminated from patients with cirrhosis and HVs.18 However, these results remain to be validated. Considering the potential of metabolomics to shed light on the pathophysiology of PSVD and to differentiate PSVD from cirrhosis non-invasively, this study aimed to investigate the metabolome of patients with PSVD and compare it to those with cirrhosis and HVs.
Patients and methods
Patient cohort & study design
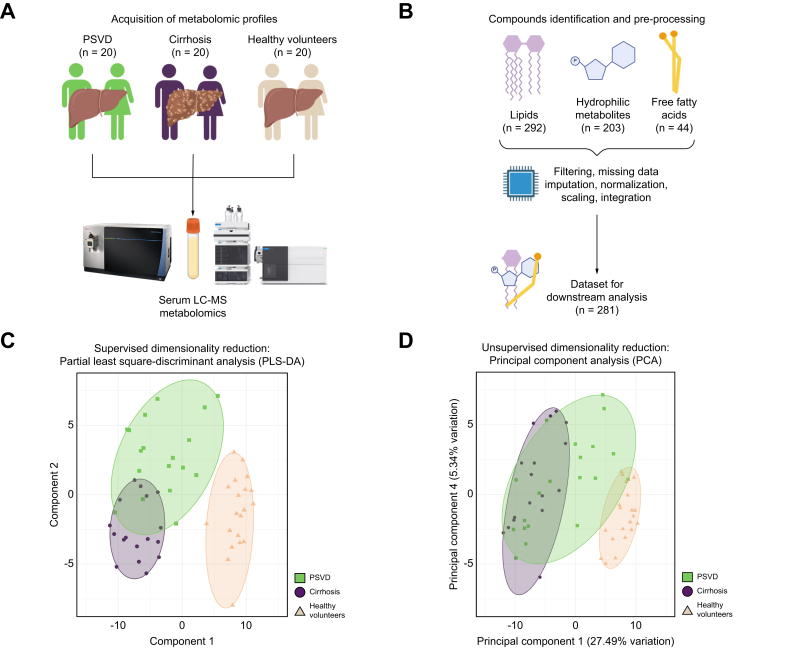
Twenty patients with histologically confirmed PSVD participating in the prospective Vienna Vascular Liver Disease Study (VALID study, ClinicalTrials.gov Identifier: NCT03541057), and 20 patients with cirrhosis from the Vienna Cirrhosis Study (VICIS, NCT03267615) were evaluated in this cross-sectional cohort study (Fig. 1A). As a control group, 20 HVs. were included. The PSVD group included patients with different presumed main etiologies (e.g., drug-induced or genetic). Patients with cirrhosis were matched using propensity scores including age, sex, BMI, MELD (model for end-stage liver disease) score, and history of hepatic decompensation, while HVs. were matched for age and sex. In patients with PSVD, clinical and laboratory data and data on LSM were collected at the time of histological confirmation of PSVD. All samples were collected after an overnight fasting period. For validation experiments, we investigated the cohort previously published by the group at Hospital Clinic Barcelona.18 In parallel, patients with cirrhosis were matched by gender, signs of portal hypertension and liver function, and HVs. were matched by age and gender. After outlier removal based on clustering (see “Metabolomics data analysis” chapter), the cohort consisted of 32 patients with PSVD, 30 patients with cirrhosis, and 33 HVs.
Fig. 1.
Study design.
(A) Study design and patient groups included in the study. (B) Metabolomics workflow. (C) Supervised dimensionality reduction, with group variable supplied for clustering. The leave-one-out method was used in validation with n folds = 1, 10 repeats, and five components used for testing. (D) Unsupervised dimensionality reduction based on unbiased whole-metabolomics profiles with removal of the low-variance features. In panels C and D, sample grouping is reflected by between-sample distance, while color-coded areas are provided for visual convenience.
Definitions
Diagnosis of PSVD was established according to a recent consensus statement.19 Specifically, PSVD was diagnosed based on a liver biopsy of adequate quality to rule out cirrhosis in the presence of either one clinical sign specific for portal hypertension or one histological lesion specific for PSVD. Specific clinical signs of portal hypertension included the presence of varices or portosystemic collaterals on endoscopy or cross-sectional imaging or a history of portal-hypertensive bleeding. Specific histological findings included obliterative portal venopathy (OPV) or nodular regenerative hyperplasia (NRH). Alternatively, the diagnosis of PSVD was established in a patient without cirrhosis on liver biopsy presenting with both an unspecific clinical sign of portal hypertension and an unspecific histological sign for PSVD. Unspecific clinical signs of portal hypertension included the presence of ascites, thrombocytopenia and splenomegaly ≥13 cm. Unspecific histological signs included portal tract abnormalities, architectural disturbances, non-zonal sinusoidal dilatation and mild perisinusoidal fibrosis.
Cirrhosis was diagnosed in the presence of clinically significant portal hypertension (CSPH, HVPG ≥10 mmHg) and LSM ≥15 kPa.
Patients with current or a history of occlusive portal vein thrombosis or hepatocellular carcinoma were excluded.
Histological work-up
Liver biopsy specimens were obtained by percutaneous liver biopsy (aspiration biopsy) or transjugular liver biopsy (aspiration or the TruCut biopsy), as previously described,20,21 and were required to have a length of ≥20 mm and/or include ≥6 portal tracts and/or be considered adequate for exclusion of cirrhosis by an expert liver pathologist.19 Biopsy specimens were evaluated by expert liver pathologists for the presence/absence of cirrhosis or histological features of PSVD.
Sample pre-processing and metabolomics data acquisition
A detailed description of sample pre-processing and metabolomics data acquisition for hydrophilic metabolites, lipids, and fatty acids can be found in the supplementary methods. In summary, metabolomics analyses were performed using reversed-phase liquid chromatography coupled to a high mass resolution Tribrid Orbitrap mass spectrometer for lipidomics and free fatty acid analysis and to a triple quadrupole mass spectrometer for hydrophilic metabolomics. For lipidomics analysis, lipids were extracted following the MTBE method.22 Studied lipophilic metabolites included phospholipids (phosphatidylcholine, phosphatidylethanolamine, phosphatidyl-serine, phosphatidylglycerol), sphingolipids (sphingomyelin, glucosylceramide, ceramide), glycerolipids (diacylglycerol, triacylglycerol), cholesteryl esters, and free cholesterol.
For measuring hydrophilic metabolites, hydrophilic small-molecule metabolites were extracted using ice-cold 80% methanol. Separation of small-molecule metabolites was achieved by ion-pairing reversed-phase chromatography coupled with dynamic MRM-based targeted tandem mass spectrometry analysis using the Agilent 6470 triple quadrupole mass spectrometer operated in negative electrospray ionization mode. Absolute quantification was based on an isotopically labeled internal standard mixture and external calibration curves. Metabolites analyzed by this method included acylcarnitines, amino acids and amino acid derivatives, biogenic acids and carboxylic acids, cofactors, nucleosides/nucleobases, nucleotides, phenyl acids, sugar derivatives, sugar phosphates, and vitamins. Finally, free fatty acids were measured using derivatization with 2-hydrazinoquinoline in acetonitrile. Absolute quantification was based on external calibration and isotopically labeled internal standardization.
Samples of the validation cohort were pre-processed and serum metabolomic profiles were acquired as previously described.18
Metabolomics data analysis
Peak finding and alignment were performed with Mass Hunter and TraceFinder software. The threshold for missing values was set as 10%, and metabolites with higher numbers of missing values were removed. Based on the removed metabolites detectable in at least one sample, we performed a missing pattern analysis (Supplementary Data 2). We imputed missing values for metabolites below the 10% threshold using the k-nearest neighbors’ algorithm in POMA (v.1.10.0).23 Outlier analysis was performed using PomaOutliers based on the formula Q3 + 3 ∗ IQR, and three identified metabolomic profiles (all within the cirrhosis group) were removed from the differential analysis but still used for machine learning prediction. The values were log-transformed and Pareto-scaled as previously described.24
Next, metabolomics datasets (lipids, hydrophilic metabolites, and fatty acids) were merged (Fig. 1B). Exploratory analysis was performed using principal component analysis (PCAtools, v. 2.12.0) for unsupervised exploration and partial least squares-discriminant analysis for clustering estimation with patient group considerations. Principal components were prioritized based on their ability to group and separate PSVD metabolic profiles from other groups. Differential metabolite abundance was tested using the Limma method (limma, v. 3.56.2)25 and with Benjamini Hochberg’s adjustment of p values. padjusted of <0.05 and logFC ≥|0.5| were used to identify significantly dysregulated metabolites. Functional analysis of metabolite groups was performed using the hypergeometric test of MetaboAnalystR (v. 4.0)26 against the KEGG database. A false discovery rate <25% was used as a threshold for significantly dysregulated pathways.
HVPG measurement
HVPG measurements were performed at the Vienna Hepatic Hemodynamic Lab according to a standardized and published protocol.21 HVPG was calculated as the mean difference between the wedged hepatic vein pressure and the free hepatic vein pressure after three measurements.27
Transient elastography
LSM was performed using FibroScan® (Echosens, Paris, France) by experienced operators, as previously described.28,29 The M and XL probes were used according to the manufacturer's recommendations. Only reliable measurements according to published criteria were considered for this study.30
Ethics
This study was approved by the ethics committees of the Medical University of Vienna (No. 1928/2017 and 1262/2017) and Universitat de Barcelona. All patients and HVs provided written informed consent to use their data and samples.
Cohort statistics and machine learning models
Statistical analyses were performed using R 4.3.0 (R Core Team, R Foundation for Statistical Computing, Vienna, Austria). Continuous variables were reported as mean ± standard deviation or median (interquartile range), while categorical variables were reported as the proportion of patients with/without a certain characteristic. Student’s t test was used for group comparisons of normally distributed variables, and the Mann-Whitney U test for non-normally distributed variables. Group comparisons of categorical variables were performed using either Pearson's Chi-squared (X2) or Fisher’s exact test, as applicable. Details on metabolomics data processing are outlined above.
Machine learning models were employed for binary classification tasks in pairwise comparisons (PSVD vs. HV and PSVD vs. cirrhosis) with the Caret package. Model development was performed in two steps:
-
1.
Feature selection: We employed Naïve Bayes (klaR, v. 1.7-2) for recursive feature elimination, where differentially regulated metabolites of the PSVD group were used as inputs. It included all metabolites of PSVD vs. cirrhosis (n = 7); to reduce the dimensionality of the dataset for the moderate number of observations, only the top 20% significant metabolites of PSVD vs. HVs were employed (n = 22). The feature selection was performed in repeated cross-validation with five repeats, 10-fold each. Variable identification was performed considering the highest accuracy of a final model with <10 metabolites.
-
2.
Classification: We used the leave-one-out method with n = 1 in two models using selected features as an input: artificial neural network (ANN)31 with one hidden layer and polynomial support vector machine.32 The hyperparameter optimization was the following: hidden layer size from 5 to 10, decay from 0.1 to 2.5 for ANN; cost from 10 to 1,000, degree from 1 to 3, and scale from 0.1 to 1 for support vector machine. Prediction specificity, sensitivity, and Kappa values of final models were used for comparison. Importance metrics were extracted for each feature in the final models to compare feature contributions for the prediction. The minimization of overfitting in ANN has been considered by employing only one hidden layer and the abovementioned penalizing parameters for model cost and decay.
For validation experiments, normalized metabolite concentrations, or ratios of several such concentrations, were used for discriminatory analyses with the pROC package.33 Specifically, the validation cohort dataset was split into PSVD vs. cirrhosis and PSVD vs. HVs, where the patient group was defined as a response value, and the metabolite concentration (or ratio) as a predictor.
Results
Patient cohort description
Clinical, instrumental, and histological characterization
The median ages of the PSVD, cirrhosis, and HV groups were 51.0 (36.3-55.0), 53.0 (44.0-60.1), and 52.2 (39.7-58.9) years, while 13 (65.0%), 16 (80.0%), and 13 (65.0%) were male, respectively (Table 1). Patients with PSVD or cirrhosis had median MELD scores of 8 (7-10) vs. 8 (8-10) (p = 0.699) and a comparable incidence of previous hepatic decompensation (6 [30.0%] vs. 8 [40.0%], p = 0.740). Child-Pugh stages (A: 14 [70.0%] vs. 16 [80.0%], p = 0.715) and platelet counts (82 [63-117] vs. 84 [70-113], p = 0.642) were similar between the two groups. Expectedly, median LSM (7.8 [5.9-8.8] vs. 30.2 [23.9-67.5] kPa, p <0.001) and HVPG (5[4], [5], [6], [7], [8] vs. 18[13], [14], [15], [16], [17], [18], [19] mmHg, p <0.001) were significantly lower in patients with PSVD, whereas median serum albumin levels were higher (42.7 [39.8-44.5] vs. 39.5 [36.8-40.7] g/dl, p = 0.009). Specific signs of portal hypertension (varices and/or portosystemic collaterals) were present in all patients in both groups, while non-specific clinical signs (ascites, thrombocytopenia, and/or splenomegaly) were present in 95% of patients with PSVD.
Table 1.
Characteristics of patients with PSVD or cirrhosis and HVs.
| PSVD (n = 20) | Cirrhosis (n = 20) | HVs (n = 20) | p value | |
|---|---|---|---|---|
| Age, years | 51.0 (36.3-55.0) | 53.0 (44.0-60.1) | 52.2 (39.7-58.9) | 0.414 |
| Male sex | 13 (65.0%) | 16 (80.0%) | 13 (65.0%) | 0.699 |
| BMI, kg/m2 | 25.6 (22.9-28.4) | 25.9 (25.0-27.7) | - | 0.791 |
| History of decompensation | 6 (30.0%) | 8 (40.0%) | - | 0.740 |
| LSM, kPa | 7.8 (5.9-8.8) | 30.2 (23.9-67.5) | - | <0.001 |
| HVPG, mmHg | 5 (4-8) | 18 (13-19) | - | <0.001 |
| Child-Pugh score, points | 6 (5-7) | 6 (5-6) | - | 0.522 |
| Child-Pugh stage A | 14 (70.0%) | 16 (80.0%) | - | 0.715 |
| Child-Pugh stage B/C | 6 (30.0%) | 4 (20.0%) | - | |
| MELD | 8 (7-10) | 8 (8-10) | - | 0.699 |
| Platelet count, G/L | 82 (63-117) | 84 (70-113) | - | 0.642 |
| Albumin, g/dl | 42.7 (39.8-44.5) | 39.5 (36.8-40.7) | - | 0.009 |
| Spleen length, cm | 16.0 (14.5-17.5) | 14.0 (12.0-15.0) | 0.040 | |
| Specific clinical signs of portal hypertension | 20 (100%) | 20 (100%) | - | - |
| Varices | 19 (95.0%) | 19 (95.0%) | - | 1.000 |
| Portosystemic collaterals | 18 (90.0%) | 15 (75.0%) | - | 0.405 |
| Unspecific clinical signs of portal hypertension | 19 (95.0%) | 19 (95.0%) | - | 1.000 |
| Ascites | 2 (10.0%) | 8 (40.0%) | - | 0.068 |
| Thrombocytopenia | 16 (80.0%) | 18 (90.0%) | - | 0.661 |
| Splenomegaly | 18 (90.0%) | 14 (70.0%) | - | 0.235 |
HV, healthy volunteers; HVPG, hepatic venous pressure gradient; LSM, liver stiffness measurement; PSVD, porto-sinusoidal vascular disorder; Student’s t test, Mann-Whitney U test, Pearson's Chi-squared test, Fisher’s exact test. Bold value represents p <0.05.
Histological findings among patients with PSVD included OPV in 11 (55%), NRH in 3 (15%), portal tract abnormalities in 19 (95%), architectural disturbances in 16 (80%), non-zonal sinusoidal dilatation in 14 (70%) and mild perisinusoidal fibrosis in 10 (50%), summing up to 13 (65%) patients with specific histological features, while all patients had at least one unspecific feature. Liver biopsy was performed in 8/20 patients (40.0%) with cirrhosis, confirming the diagnosis of cirrhosis in all patients.
Etiological factors for PSVD included azathioprine exposure in 11 (55.0%), retroperitoneal fibrosis in 2 (10.0%), and cisplatin-based chemotherapy, HIV, antiphospholipid syndrome, acromegaly, myasthenia gravis, and systemic lupus erythematosus in one patient each (5.0%). No etiological factors were found in three young patients (18-20 years, 15.0%). Etiologies of cirrhosis included alcohol-related liver disease (8 patients, 40.0%), viral hepatitis (7, 35.0%), and non-alcoholic fatty liver disease (5, 25.0%).
Patient group separation based on whole-metabolomics profiles
Of the 539 identified serum metabolites, 256 (47.5%) were excluded during pre-processing, and 74 (13.7%) were complemented with imputation (Fig. 1B, Supplementary Data 1). Specifically, 226 metabolites (41.9%) were removed based on the 10% missing value threshold, and 30 metabolites (5.6%) were removed due to constant abundance or outlier concentrations (see “Metabolomics Data Analysis” chapter). The remaining 283 metabolites were used for downstream analysis. Missing pattern analysis of metabolites showed flavin adenine dinucleotide was predominantly absent in PSVD metabolomes, while fumaric acid non-detection was linked to cirrhosis and HV groups (Fig. S3).
Supervised dimensionality reduction clearly separated metabolic profiles between PSVD and HV groups (Fig. 1C). Unsupervised reduction mostly aligned with these findings, highlighting one patient with PSVD with an HV-like metabolome (Fig. 1D). However, the separation between PSVD and cirrhosis was less distinct, with five patients with PSVD in close proximity to the cirrhosis group in the supervised scenario (Fig. 1C). These five patients were further evaluated as a distinct PSVD subgroup later. Principal components (PC)-1 and PC4 best separated PSVD and cirrhosis (Fig. S1). PC1 was influenced by metabolites linked to oxidative phosphorylation and fatty acid beta-oxidation, while PC4 was driven by aspartic acid and beta-oxidation metabolites. PC1 was a key separator of patient etiology groups, while PC4 mainly explained the intra-group variance in patient metabolomes, separating samples within each group but not linked to similarities between patient groups. Both PCs featured several triacylglycerides as robust negative markers (Supplementary Data 2).
Pathophysiological analyses based on differentially expressed metabolites
Differential analysis reveals alterations in metabolite abundance in PSVD and cirrhosis
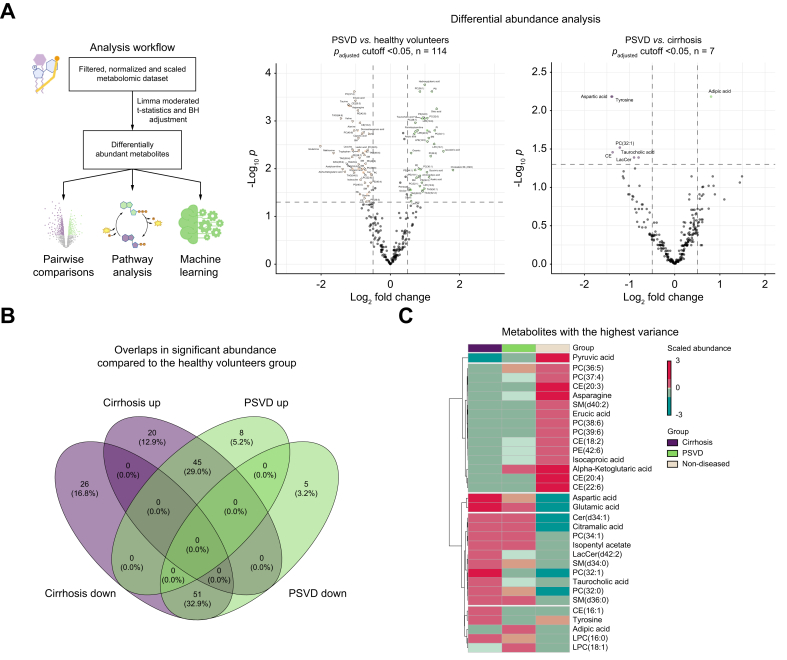
Differential testing identified distinct metabolic signatures for the PSVD and cirrhosis groups compared to the HV group (Fig. 2A). In PSVD, 114 metabolites were significantly altered, with cholesterol M (-H2O), isovaleric acid, and hydroxyglutaric acid being the most overrepresented and taurine, asparagine, erucic acid, and valine being the most downregulated (Supplementary Data 3). A pairwise comparison of PSVD vs. cirrhosis revealed seven differentially abundant metabolites (Supplementary Data 4). Adipic acid was upregulated in PSVD, while tyrosine, aspartic, and taurocholic acids, along with several lipids, were upregulated in cirrhosis.
Fig. 2.
Differential abundance of metabolites between the study groups.
(A) Volcano plots demonstrating upregulation (right direction) or downregulation (left direction) of metabolites in the respective comparison. The presented p values and log2 fold change were obtained using the limma’s moderated t-test with Benjamini Hochberg's adjustment, where significance thresholds were: padjusted < 0.05, log2 fold change > |0.5|. (B) Venn diagram showing overlaps in significant metabolites between PSVD or cirrhosis and non-diseased profiles. (C) Heatmap showing highest up- and downregulated metabolites, abundance values were scaled and average values for each group are presented. PSVD, porto-sinusoidal vascular disease.
Comparing patients with PSVD or cirrhosis to HVs identified major overlaps in downregulated (51 metabolites, 32.9%) and upregulated (45, 29%) directions (Fig. 2B). PSVD-specific alterations included upregulation of adipic acid, cholesterol M (-H2O), aminoadipic acid, xanthosine, and others, and downregulation of isoleucine, glycine, and threonine (Supplementary Data 5).
Unsupervised clustering of high-variance metabolites revealed five clusters (Fig. 2C). Cluster 1 included only pyruvic acid, downregulated in both disease groups compared to HVs, with stronger downregulation in cirrhosis. Cluster 2 included metabolites slightly upregulated in PSVD or less downregulated than in cirrhosis, such as alpha-ketoglutaric acid and phosphatidylcholine (36:5). Cluster 3 included strongly upregulated aspartic and glutamic acids in cirrhosis, with less pronounced upregulation in PSVD. Cluster 4 included metabolites less abundant in HV, such as taurocholic and citramalic acid. Cluster 5 included metabolites with unclear patterns, such as tyrosine, downregulated in patients with PSVD compared to those with cirrhosis or HVs, and adipic acid, upregulated only in PSVD.
Differential testing and high-variance metabolite exploration indicate that adipic and taurocholic acids are key determinants in PSVD metabolomic profiles, with phosphatidylcholines, cholesteryl esters, and sphingomyelins being prominent high-variance molecules.
We further compared metabolomes of the subset of five patients with PSVD with similarities to cirrhosis profiles (Fig. 1C, hereinafter referred to as PSVDsubgroup) to the rest of the patients with PSVD. Notably, all had one specific histological feature of PSVD (OPV in 4 patients and NRH in 1 patient) and at least one unspecific feature. Interestingly, 15 differentially abundant metabolites were identified: aconitic acid and 12 tri-/diglycerides were upregulated, while two phosphatidylcholines were downregulated in the PSVDsubgroup (Fig. S2, Supplementary Data 6A). Histologically confirmed steatosis (10-30%) was observed in 4/5 patients in this subgroup, and two had type 2 diabetes mellitus (Supplementary Data 6B). PSVDsubgroup patients had higher total serum triglyceride concentrations than other patients with PSVD (Fig. S2).
PSVD is linked to impairments in pathways associated with amino acid metabolism
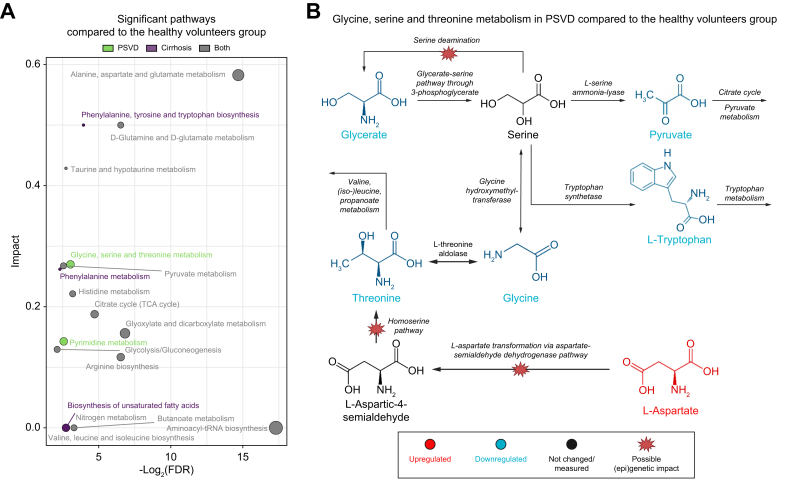
Functional analysis of differential metabolites identified altered metabolic pathways in PSVD (Supplementary Data 7). The most perturbed significant pathways distinguishing patients with PSVD and HVs were alanine, aspartate, and glutamate metabolism; D-Glutamine and D-glutamate metabolism; and taurine and hypotaurine metabolism pathways (Fig. 3A). Pyrimidine metabolism was also impacted: despite downregulation in L-glutamine, N-carbamoyl-L-aspartate, and orotate were overrepresented (Fig. S4).
Fig. 3.
Functional analysis of metabolic sets.
(A) Overlaps in significantly altered pathways in PSVD or cirrhosis compared to non-diseased profiles. Color code indicates significance of a pathway in PSVD, cirrhosis, or both. The over-representation was tested using the hypergeometric test, and significant pathways were those with padjusted <0.05. Color code indicates significance of a pathway in PSVD, cirrhosis, or both. (B) Metabolic map of glycine, serine and threonine metabolic pathways, differentially regulated only in PSVD compared to non-diseased profiles. Color code indicates downregulated (blue) and upregulated (yellow) metabolites, scaled between -1-1. PSVD, porto-sinusoidal vascular disease.
Glycine, serine, and threonine metabolic pathways were uniquely linked to PSVD, but not to cirrhosis (Supplementary Data 8). Perturbations of this pathway were followed by the significant downregulation of glycine and threonine, as well as pyruvate and L-tryptophan (Fig. 3B). These changes in L-glutamine as a precursor aligned with the reduction in alpha-ketoglutarate, as they are both members of the glutaminolysis pathway (Fig. 2C). Upregulation in N-carbamoyl-L-aspartate and orotate may indicate dysregulation of uridine metabolism, as the other molecule linked to its exchange, deoxyuridine, was also found to be upregulated in PSVD (Fig. S4).
These alterations in amino acid turnover and their crosslinks to the tricarboxylic acid cycle and pyrimidine synthesis may provide a first glimpse at pathophysiologically relevant metabolic disturbances in PSVD.
Metabolomics for non-invasive diagnosis of PSVD
Machine learning models for metabolite-based PSVD identification
Machine learning models were employed to prioritize metabolites and assess their diagnostic value (Fig. S5A). Recursive feature elimination identified four metabolites for classifying PSVD vs. cirrhosis and six for distinguishing patients with PSVD from HVs (Table 2).
Table 2.
Performance of machine learning models.
| Prediction | Prioritized parameters | Model | Sensitivity | Specificity | K |
|---|---|---|---|---|---|
| PSVD vs. HV |
|
ANN | 0.95 | 1 | 0.95 |
| SVM | 1 | 0.952 | 0.95 | ||
| PSVD vs. cirrhosis |
|
ANN | 0.8 | 0.9 | 0.7 |
| SVM | 0.833 | 0.773 | 0.6 |
ANN, artificial neural network; HVPG, hepatic venous pressure gradient; LSM, liver stiffness measurement; PH, portal hypertension; PSVD, porto-sinusoidal vascular disorder; SVM, support vector machine; K, Cohen's Kappa. Bold value represents p <0.05.
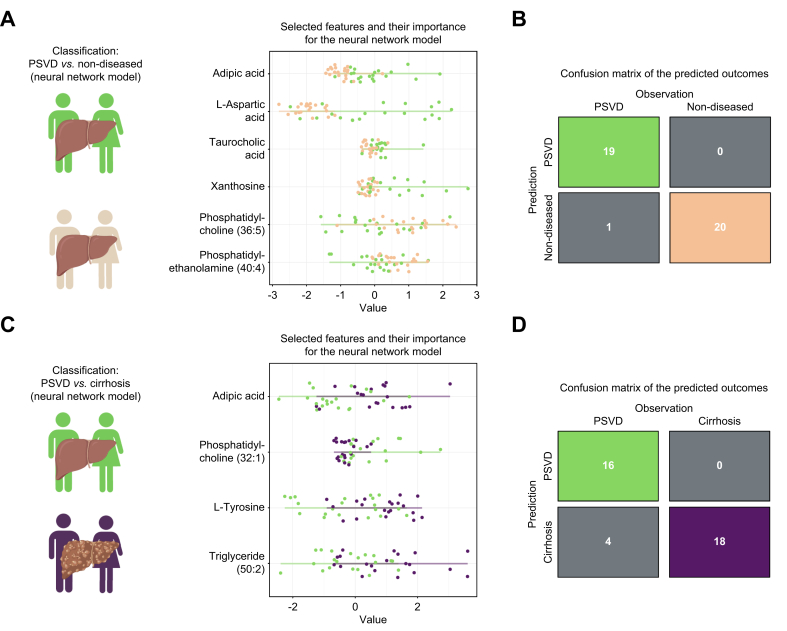
The ANN model for PSVD vs. HV classification used adipic acid, L-aspartic acid, taurocholic acid, xanthosine, phosphatidylcholine (36:5), and phosphatidylethanolamine (40:4) (Fig. 4A). It correctly classified all HVs and misclassified only one patient with PSVD (Fig. 4B), achieving a specificity of 1 and sensitivity of 0.95.
Fig. 4.
Class prediction based on significantly dysregulated metabolites using artificial neural networks.
(A) Features selected for the model between PSVD and non-diseased metabolic profiles. (B) Confusion matrix showing the difference between real and predicted classes between PSVD and non-diseased patients. (C) Prioritized features for classification between PSVD and cirrhosis profiles. (D) Confusion matrix for patients with PSVD and cirrhosis. Feature importance values represent their impact on the classification of each patient (separate dots) in a model with optimal hyperparameters. The features are sorted from the most important (top) to the least important for the prediction (bottom). Confusion matrices are color-coded according to the patient groups, with gray indicating discordance between real and predicted values. The features were prioritized via repeated cross-validation with n = 5 repeats and n = 10 folds using caret's naive Bayes algorithm.
The ANN for PSVD vs. cirrhosis used adipic acid, phosphatidylcholine (32:1), L-tyrosine, and triglyceride (50:2) (Figs 4C and S5B). It misclassified PSVD in four cases and cirrhosis in two (Fig. 4D), with a sensitivity of 0.8 and specificity of 0.9 (Table 2).
Confirmation of the diagnostic importance of taurocholic and L-aspartic acids in the Barcelona cohort
A comparison of patient characteristics between the Barcelona and Vienna cohorts can be found in Table S1. Importantly, both cohorts were comparable with regard to liver disease severity.
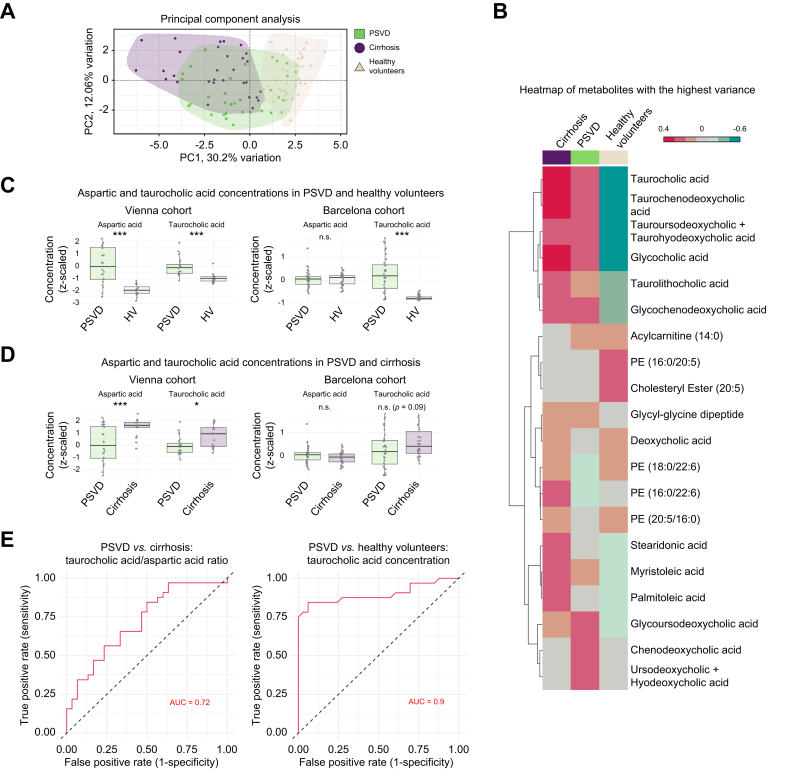
The first two PCs, mainly comprising lipids and taurocholic acid derivatives, explained the most variance in the Barcelona cohort. As in the Vienna dataset, there was a clear separation between the cirrhosis and HV groups, with some patients with PSVD clustering among other groups (Fig. 5A).
Fig. 5.
Exploration of the validation dataset.
(A) Principal component analysis reveals a similar separation pattern as in the original study dataset: patients with PSVD show a distinct metabolic profile, while some of them overlap with other groups. (B) A heatmap of 20 metabolites with the highest variance demonstrates similar perturbations in taurocholic acid and its derivatives. Each column represents the group’s mean concentration. (C) Comparative analysis of the metabolites with the highest diagnostic performance between PSVD and healthy volunteers. (D) Comparative analysis of the metabolites with the highest diagnostic performance between PSVD and cirrhosis. For C and D, normalized and Z-scaled values were used for testing. Shapiro-Wilk test for normality was performed, following Welch two sample t-test (normal distribution) or Wilcoxon rank sum test (non-normal distribution), results of which are reported on the plots. Box plots represent medians, minimum and maximum concentrations within 1.5x the interquartile range. n.s.: p >0.05, ∗∗∗p ≤0.001. (E) Metabolites prioritized for the machine learning model in the Vienna cohort (see Fig. 4) showing the best discrimination between patients with PSVD or cirrhosis, and healthy volunteers in the Barcelona cohort. PSVD, porto-sinusoidal vascular disease.
Metabolites with the highest variance showed similar taurocholic acid perturbation as in the Vienna cohort: taurocholic acid and its derivatives were downregulated in HVs compared to patients with PSVD and upregulated in cirrhosis compared to PSVD (Fig. 2, Fig. 5C).
Although metabolite panels did not completely overlap between cohorts, L-aspartic acid, taurocholic acid, phosphatidylcholine (32:1), and L-tyrosine were detected in both. Taurocholic and L-aspartic acids showed consistent levels in PSVD across cohorts, with significant differences only seen in the HV cohorts (Fig. S6). Taurocholic acid concentrations were higher in patients with PSVD compared to HVs in both cohorts (p ≤0.001), while aspartic acid was upregulated only in PSVDVienna compared to HVVienna (p ≤0.001) (Fig. 5C). Taurocholic acid levels were further increased in cirrhosis compared to PSVD, significantly in the Vienna cohort (p = 0.01) and as a trend in the Barcelona cohort (p = 0.09) (Fig. 5D).
Taurocholic acid alone distinguished patients with PSVD from HVs with an AUROC of 0.899 (Table 3, Supplementary Data 10). The ratio of taurocholic acid to L-aspartic acid was the best predictor for PSVD vs. cirrhosis (AUROC = 0.720) and the second-best for PSVD vs. HV (AUROC = 0.645). These findings highlight the diagnostic potential of prioritized metabolites, warranting further evaluation in larger PSVD cohorts.
Table 3.
Discriminative performance of metabolite concentrations or their ratios in the Barcelona cohort.
| Comparison | Metabolite(s) | AUROC | 95% CI |
|---|---|---|---|
| vs. cirrhosis | |||
| PSVD vs. cirrhosis | Taurocholic acid/L-aspartic acid (ratio) | 0.720 | 0.592-0.847 |
| PSVD vs. cirrhosis | Tyrosine/L-aspartic acid (ratio) | 0.631 | 0.491-0.771 |
| PSVD vs. cirrhosis | Tyrosine/taurocholic acid (ratio) | 0.620 | 0.479-0.760 |
| PSVD vs. cirrhosis | L-aspartic acid | 0.605 | 0.463-0.747 |
| vs. HVs | |||
| PSVD vs. HV | Taurocholic acid | 0.899 | 0.812-0.985 |
| PSVD vs. HV | Taurocholic acid/L-aspartic acid (ratio) | 0.645 | 0.506-0.783 |
| PSVD vs. HV | L-tyrosine | 0.612 | 0.461-0.762 |
Only metabolites previously prioritized in the machine learning models in the Vienna cohort were assessed and reported when AUROC values >0.6 are observed. Values in bold indicate the best prediction for each comparison and are graphically displayed in Fig. 5E. HV, healthy volunteer; PSVD, porto-sinusoidal vascular disorder; Area-under-the-receiver-operator-characteristics-curve.
Discussion
This study aimed to showcase the potential of advanced high-throughput metabolomics analyses in gaining further insights into the pathophysiology of PSVD and in facilitating its non-invasive diagnosis.
Our study provides insights into potential pathophysiological mechanisms. Adipic acid was upregulated in PSVD compared to both non-diseased and cirrhosis groups. This dicarboxylic acid is not a primary human metabolic product nor a major fermentation product of the gut microbiome.34,35 Its increased urine concentration has been reported in type 2 diabetes.35,36 However, our discovery of its association with PSVD is a novel finding. As a xenobiotic in humans, it undergoes β-oxidation into smaller metabolites.37 Hence, we hypothesize that an unknown genetic or epigenetic factor in PSVD impacts mechanisms involved in adipic acid transformation (e.g., acyl-CoA synthetases, medium-chain acyl-CoA dehydrogenase, or crotonase).
Taurocholic acid was significantly downregulated in patients with PSVD in the Vienna cohort. Alterations in glycine, threonine, and serine metabolic pathways were unique to PSVD, potentially contributing to its pathobiology. Downregulation of pathway members such as pyruvate and L-tryptophan suggests reduced substrate availability for the tricarboxylic acid cycle, aligning with the downregulation of alpha-ketoglutarate and L-glutamine. Downregulation of the latter pathway, known as glutaminolysis, indicates that this alternative source of tricarboxylic acid cycle-substrates is not involved in compensation for cell energy balance impairments in PSVD.38 The upregulation of gene sets related to the tricarboxylic acid cycle enzymes, observed in previous RNA microarray studies, follows our findings and hints at the compensatory nature of such changes.39 Recent genomic findings describe a pathogenic variant in FCHSD1, an mTOR pathway member gene,40 potentially linked to these amino acid perturbations.41 Still, the characterization of these processes on a cellular level is lacking in PSVD and would require a multi-omics setup to combine our findings with tissue and cellular expression of the involved enzymes.
The upregulation of N-carbamoyl-L-aspartate and orotate suggests increased pyrimidine synthesis, hinting at a dysregulation in uridine metabolism. This may not impact energy production directly via the tricarboxylic acid cycle but is crucial for DNA and RNA synthesis.42 The impairment of pyrimidine turnover in PSVD warrants further investigation.
We deliberately included a heterogeneous PSVD and cirrhotic cohort to obtain generalizable results and avoid overfitting. While studying narrowly defined groups could yield more explicit metabolomic signatures, this approach risks non-reproducibility in external cohorts. Similarly, not limiting patients with cirrhosis to those undergoing liver biopsy avoided selection bias.
Recent studies have evaluated metabolomic signatures in patients diagnosed according to the previous INCPH criteria.17,18 One study compared metabolomic profiles of 33 patients with PSVD, 33 with cirrhosis, and 33 HVs,17 identifying 28 differentially abundant metabolites using ultra-performance liquid chromatography with mass spectrometry. These metabolites could differentiate PSVD from cirrhosis with an AUC of 0.99. Another model with 31 metabolites also differentiated patients with PSVD from HVs with similar accuracy, although the spectral peaks were not linked to specific metabolic compounds, limiting the generalizability of the results.17 Another study identified a five-metabolite signature that accurately discriminated patients with INCPH from those with cirrhosis and HVs, but these studies used the older INCPH criteria, which may not represent the current, broader PSVD patient population.18 As it is estimated that current PSVD criteria extend the number of patients by three times,3 the previous studies might not represent the current population. Nevertheless, when investigated for validation purposes, the dataset from Seijo et al. (2015)18 confirmed the diagnostic role of our prioritized markers, taurocholic and L-aspartic acids.
These findings indicated that linear methods based on serum whole-metabolome features can partially differentiate profiles for PSVD. Advanced approaches, e.g. based on machine learning, are needed for more precise separation. Hence, our study identified two metabolite signatures that enabled the non-invasive differentiation between PSVD, cirrhosis, and health with high sensitivity and specificity. Our PSVD patient group included diverse etiological factors, so these signatures likely reflect conserved metabolic perturbations in PSVD.
Finally, we identified a subgroup of patients with PSVD and metabolomic profiles overlapping those of patients with cirrhosis, characterized by lipid-related alterations, including higher total triglyceride levels and histological signs of hepatic steatosis. Indeed, the observed dysregulated lipids were previously linked with MASLD and the metabolic syndrome, indicating that the co-existence of MASLD and vascular changes might define a specific subset of patients with PSVD, which requires further evaluation in larger PSVD cohorts.
Despite the promise of untargeted metabolomics, its primary application will likely remain research-oriented. Thus, our results should foster multicenter, prospective collaborations, utilizing targeted assays for the suggested candidate biomarkers to test their diagnostic value. This could lead to cost-effective, easily applicable tests for non-invasive PSVD diagnosis.
Our study has limitations. The small sample size reflects the rare nature of PSVD, limiting opportunities to subdivide the cohort to assess heterogeneity and impacting the performance of machine learning models. Despite using best practices to minimize overfitting, it remains a concern due to the small cohort size. Although comparable metabolite perturbations were observed in the Barcelona cohort, more extensive, prospective multicenter studies are needed to validate the models' accuracy.
The absence of paired liver metabolomic readouts and enzymatic activity measurements, linked to material-related challenges, means extrahepatic effects of cirrhosis and PSVD may drive some metabolomic changes. Additionally, technical differences between studies affected the overlap of detected metabolites, necessitating further studies to explore other markers predicted to perform well for PSVD identification.
Abbreviations
ANN, artificial neural network; HVs, healthy volunteers; HVPG, hepatic venous pressure gradient; INCPH, idiopathic non-cirrhotic portal hypertension; LSM, liver stiffness measurement; NRH, nodular regenerative hyperplasia; OPV, obliterative portal venopathy; PC, principal component; PSVD, porto-sinusoidal vascular disorder.
Financial support
This work was supported by a grant from the “Ärztekammer für Wien” (0023-WS2020) and the “Medical Scientific Fund of the Mayor of the City of Vienna” (784259/2021) awarded to G.S. G.S., L.B., B.Si., K.L., M.T., M.M., T.R., and B.Sc. received financial support by the Clinical Research Group MOTION, Medical University of Vienna, Vienna, Austria – a Clinical Research Group Programme project funded by the Ludwig Boltzmann Gesellschaft (Grant Nr LBG_KFG_22_32) with funds from the Fonds Zukunft Österreich.
Conflicts of interest
G.S. received travel support from Amgen. O.P. has nothing to disclose. J.J.L. has nothing to disclose. S.S. has nothing to disclose. J.I.S.-A. has nothing to disclose. N.M. has nothing to disclose. J.T.H. has nothing to disclose. K.W. has nothing to disclose. L.B. has nothing to disclose. B.Si. received travel support from AbbVie and Gilead. K.L. has nothing to disclose. B.M. has nothing to disclose. M.T. received grant support from Albireo, Alnylam, Cymabay, Falk, Gilead, Intercept, MSD, Takeda and UltraGenyx; honoraria for consulting from Abbvie, Albireo, Boehringer Ingelheim, BiomX, Falk, Genfit, Gilead, Hightide, Intercept, Jannsen, MSD, Novartis, Phenex, Pliant, Regulus, Siemens and Shire; speaker fees from Albireo, Bristol-Myers Squibb, Falk, Gilead, Intercept, MSD and Madrigal, as well as travel support from AbbVie, Falk, Gilead and Intercept. He is also a co-inventor on patents on the medical use of norUDCA/norucholic acid filed by the Medical University of Vienna. M.M. served as a speaker and/or consultant and/or advisory board member for AbbVie, Collective Acumen, Echosens, Gilead, Takeda, and W. L. Gore & Associates, and received travel support from AbbVie and Gilead. T.R. received grant support from AbbVie, Boehringer-Ingelheim, Gilead, MSD, Philips Healthcare, Pliant Pharmaceuticals, W.L. Gore & Associates, and Siemens, served as a speaker for AbbVie, Gilead, Gore, Intercept, Roche, MSD, and W.L. Gore & Associates, received consulting/advisory board fee from AbbVie, Bayer, Boehringer-Ingelheim, Gilead, Intercept, MSD, and Siemens, and travel support from Boehringer-Ingelheim, Gilead, and Roche. J.C.G.-P. received grant support from Gore & Associates, Astra Zeneca, Cook and Mallinckrodt. B.Sc. received travel support from AbbVie, AstraZeneca, Ipsen, and Gilead, grant support from AstraZeneca and Eisai and speaker’s honoraria from AstraZeneca and Eisai.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Study conception (G.S., O.P., T.R., and B.Sc.), data collection (all authors), metabolomics data acquisition and pre-processing (TH, NM), statistical analysis (G.S., O.P., and B.Sc.), drafting of the manuscript (G.S., O.P., T.R., and B.Sc.), validation cohort (J.J.L., S.S., J.C.G.-P.), review for important intellectual content and approval of the final manuscript (all authors).
Data availability statement
Pre-processed metabolomics data and scripts required to reproduce the figures are available via GitHub: https://github.com/xander-p/SEMPETSCH-PSVD-metabolomics.
Ethics approval statement
This study was approved by the ethics committee of the Medical University of Vienna (No. 1928/2017 and 1262/2017) as well as the Universitat de Barcelona. All patients and HV gave written informed consent to the use of their data and samples.
Acknowledgements
G.S., O.P., L.B., B.Si., L.H., M.J., M.M., T.R. and B.Sc. were supported by the Austrian Federal Ministry for Digital and Economic Affairs, the National Foundation for Research, Technology and Development, the Christian Doppler Research Association, and Boehringer Ingelheim.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101208.
Contributor Information
Juan-Carlos García-Pagán, Email: jcgarcia@clinic.cat.
Bernhard Scheiner, Email: bernhard.scheiner@meduniwien.ac.at.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.De Gottardi A., Rautou P.E., Schouten J., et al. Porto-sinusoidal vascular disease: proposal and description of a novel entity. Lancet Gastroenterol Hepatol. 2019;4:399–411. doi: 10.1016/S2468-1253(19)30047-0. [DOI] [PubMed] [Google Scholar]
- 2.De Gottardi A., Sempoux C., Berzigotti A. Porto-sinusoidal vascular disorder. J Hepatol. 2022;77:1124–1135. doi: 10.1016/j.jhep.2022.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Wöran K., Semmler G., Jachs M., et al. Clinical course of porto-sinusoidal vascular disease is distinct from idiopathic noncirrhotic portal hypertension. Clin Gastroenterol Hepatol. 2022;20:e251–e266. doi: 10.1016/j.cgh.2020.11.039. [DOI] [PubMed] [Google Scholar]
- 4.Magaz M., Giudicelli-Lett H., Nicoară-Farcău O., et al. Liver transplantation for porto-sinusoidal vascular liver disorder: long-term outcome. Transplantation. 2023;107:1330–1340. doi: 10.1097/TP.0000000000004444. [DOI] [PubMed] [Google Scholar]
- 5.Seijo S., Reverter E., Miquel R., et al. Role of hepatic vein catheterisation and transient elastography in the diagnosis of idiopathic portal hypertension. Dig Liver Dis. 2012;44:855–860. doi: 10.1016/j.dld.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Sharma P., Agarwal R., Dhawan S., et al. Transient elastography (Fibroscan) in patients with non-cirrhotic portal fibrosis. J Clin Exp Hepatol. 2017;7:230–234. doi: 10.1016/j.jceh.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuppalanchi R., Mathur K., Pyko M., et al. Liver stiffness measurements in patients with Noncirrhotic portal hypertension-the devil is in the details. Hepatology. 2018;68:2438–2440. doi: 10.1002/hep.30167. [DOI] [PubMed] [Google Scholar]
- 8.Bissonnette J., Généreux A., Côté J., et al. Hepatic hemodynamics in 24 patients with nodular regenerative hyperplasia and symptomatic portal hypertension. J Gastroenterol Hepatol. 2012;27:1336–1340. doi: 10.1111/j.1440-1746.2012.07168.x. [DOI] [PubMed] [Google Scholar]
- 9.Da B.L., Surana P., Kapuria D., et al. Portal pressure in non-cirrhotic portal hypertension: to measure or not to measure. Hepatology. 2019 doi: 10.1002/hep.30862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lampichler K., Semmler G., Wöran K., et al. Imaging features facilitate diagnosis of porto-sinusoidal vascular disorder. Eur Radiol. 2022 doi: 10.1007/s00330-022-09132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valainathan S.R., Sartoris R., Elkrief L., et al. Contrast-enhanced CT and liver surface nodularity for the diagnosis of porto-sinusoidal vascular disorder: a case-control study. Hepatology. 2022 doi: 10.1002/hep.32367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stift J., Semmler G., Walzel C., et al. Transjugular aspiration liver biopsy performed by hepatologists trained in HVPG measurements is safe and provides important diagnostic information. Dig Liver Dis. 2019;51:1144–1151. doi: 10.1016/j.dld.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Furuichi Y., Moriyasu F., Taira J., et al. Noninvasive diagnostic method for idiopathic portal hypertension based on measurements of liver and spleen stiffness by ARFI elastography. J Gastroenterol. 2013;48:1061–1068. doi: 10.1007/s00535-012-0703-z. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira-Silva J., Gaspar R., Liberal R., et al. Splenic-hepatic elastography index is useful in differentiating between porto-sinusoidal vascular disease and cirrhosis in patients with portal hypertension. Dig Liver Dis. 2022 doi: 10.1016/j.dld.2022.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes E., Wilson I.D., Nicholson J.K. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Seijo S., Lozano J.J., Alonso C., et al. Metabolomics discloses potential biomarkers for the noninvasive diagnosis of idiopathic portal hypertension. Am J Gastroenterol. 2013;108:926–932. doi: 10.1038/ajg.2013.11. [DOI] [PubMed] [Google Scholar]
- 18.Seijo S., Lozano J.J., Alonso C., et al. Metabolomics as a diagnostic tool for idiopathic non-cirrhotic portal hypertension. Liver Int. 2016;36:1051–1058. doi: 10.1111/liv.12972. [DOI] [PubMed] [Google Scholar]
- 19.De Gottardi A., Rautou P.E., Schouten J., et al. Porto-sinusoidal vascular disease: proposal and description of a novel entity. Lancet Gastroenterol Hepatol. 2019;4:399–411. doi: 10.1016/S2468-1253(19)30047-0. [DOI] [PubMed] [Google Scholar]
- 20.Stift J., Semmler G., Wöran K., et al. Comparison of the diagnostic quality of aspiration and core-biopsy needles for transjugular liver biopsy. Dig Liver Dis. 2020;52:1473–1479. doi: 10.1016/j.dld.2020.08.028. [DOI] [PubMed] [Google Scholar]
- 21.Reiberger T., Schwabl P., Trauner M., et al. Measurement of the hepatic venous pressure gradient and transjugular liver biopsy. J Vis Exp. 2020 doi: 10.3791/58819. [DOI] [PubMed] [Google Scholar]
- 22.Matyash V., Liebisch G., Kurzchalia T.V., et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellano-Escuder P., Gonzalez-Dominguez R., Carmona-Pontaque F., et al. POMAShiny: a user-friendly web-based workflow for metabolomics and proteomics data analysis. Plos Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Berg R.A., Hoefsloot H.C.J., Westerhuis J.A., et al. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong J., Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;34:4313–4314. doi: 10.1093/bioinformatics/bty528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiberger T., Ferlitsch A., Payer B.A., et al. Non-selective beta-blockers improve the correlation of liver stiffness and portal pressure in advanced cirrhosis. J Gastroenterol. 2012;47:561–568. doi: 10.1007/s00535-011-0517-4. [DOI] [PubMed] [Google Scholar]
- 28.Reiberger T., Ferlitsch A., Payer B.A., et al. Noninvasive screening for liver fibrosis and portal hypertension by transient elastography—a large single center experience. Wiener Klinische Wochenschrift. 2012;124:395–402. doi: 10.1007/s00508-012-0190-5. [DOI] [PubMed] [Google Scholar]
- 29.Semmler G., Wöran K., Scheiner B., et al. Novel reliability criteria for controlled attenuation parameter assessments for non-invasive evaluation of hepatic steatosis. United Eur Gastroenterol J. 2020;8:321–331. doi: 10.1177/2050640619900820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boursier J., Zarski J.P., de Ledinghen V., et al. Determination of reliability criteria for liver stiffness evaluation by transient elastography. Hepatology. 2013;57:1182–1191. doi: 10.1002/hep.25993. [DOI] [PubMed] [Google Scholar]
- 31.Venables W.N., Ripley B.D. 4th ed. Springer; New York, NY: 2002. Modern applied statistics with S. [Google Scholar]
- 32.Kuhn M. Building predictive models inRUsing thecaretPackage. J Stat Softw. 2008;28 doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 33.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruss K.M., Chen H., Liu Y., et al. Host-microbe co-metabolism via MCAD generates circulating metabolites including hippuric acid. Nat Commun. 2023;14:1–12. doi: 10.1038/s41467-023-36138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu S., Liu S., Li H., et al. Identification of gut Microbiota and metabolites signature in patients with irritable bowel syndrome. Front Cell Infect Microbiol. 2019;9 doi: 10.3389/fcimb.2019.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villarreal-Perez J.Z., Villarreal-Martinez J.Z., Lavalle-Gonzalez F.J., et al. Plasma and urine metabolic profiles are reflective of altered beta-oxidation in non-diabetic obese subjects and patients with type 2 diabetes mellitus. Diabetol Metab Syndr. 2014;6:129. doi: 10.1186/1758-5996-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strittmatter C.S., Eggers J., Biesgen V., et al. Insights into the degradation of medium-chain-length dicarboxylic acids in cupriavidus necator H16 reveal β-oxidation differences between dicarboxylic acids and fatty acids. Appl Environ Microbiol. 2022;88 doi: 10.1128/AEM.01873-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoo H.C., Yu Y.C., Sung Y., et al. Glutamine reliance in cell metabolism. Exp Mol Med. 2020;52:1496–1516. doi: 10.1038/s12276-020-00504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernández-Gea V., Campreciós G., Betancourt F., et al. Co-expression gene network analysis reveals novel regulatory pathways involved in porto-sinusoidal vascular disease. J Hepatol. 2021;75:924–934. doi: 10.1016/j.jhep.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Shan J., Megarbane A., Chouchane A., et al. Genetic predisposition to porto-sinusoidal vascular disorder: a functional genomic-based, multigenerational family study. Hepatology. 2023;77:501–511. doi: 10.1002/hep.32735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le T.T., Ziemba A., Urasaki Y., et al. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J Lipid Res. 2013;54:1044–1057. doi: 10.1194/jlr.M034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pre-processed metabolomics data and scripts required to reproduce the figures are available via GitHub: https://github.com/xander-p/SEMPETSCH-PSVD-metabolomics.