By combining potent targeting of ALK oncoproteins and resistant mutants with brain penetrance and selectivity for ALK over TRK, the ALK inhibitor NVL-655 addresses key limitations of approved ALK inhibitors.
Abstract
Three generations of tyrosine kinase inhibitors (TKI) have been approved for anaplastic lymphoma kinase (ALK) fusion–positive non–small cell lung cancer. However, none address the combined need for broad resistance coverage, brain activity, and avoidance of clinically dose-limiting TRK inhibition. NVL-655 is a rationally designed TKI with >50-fold selectivity for ALK over 96% of the kinome tested. In vitro, NVL-655 inhibits diverse ALK fusions, activating alterations, and resistance mutations, showing ≥100-fold improved potency against ALKG1202R single and compound mutations over approved ALK TKIs. In vivo, it induces regression across 12 tumor models, including intracranial and patient-derived xenografts. NVL-655 inhibits ALK over TRK with 22-fold to >874-fold selectivity. These preclinical findings are supported by three case studies from an ongoing first-in-human phase I/II trial of NVL-655 which demonstrate preliminary proof-of-concept clinical activity in heavily pretreated patients with ALK fusion–positive non–small cell lung cancer, including in patients with brain metastases and single or compound ALK resistance mutations.
Significance: By combining broad activity against single and compound ALK resistance mutations, brain penetrance, and selectivity, NVL-655 addresses key limitations of currently approved ALK inhibitors and has the potential to represent a distinct advancement as a fourth-generation inhibitor for patients with ALK-driven cancers.
Introduction
The receptor tyrosine kinase anaplastic lymphoma kinase (ALK) can be constitutively activated by aberrant gene fusion, point mutation, and partial deletion to drive human cancers (1). Initially discovered as an oncogenic fusion in anaplastic large cell lymphoma (2), ALK fusions are present in 3% to 5% of non–small cell lung cancer (NSCLC; ref. 3), and oncogenic ALK has been detected in neuroblastomas, gliomas, inflammatory myofibroblastic tumors, and other cancers (4–8). More than 90 fusion partners have been reported for ALK (9). EML4 accounts for 84% of ALK fusions in NSCLC (10) with at least 15 known EML4–ALK breakpoint variants, the most common of which are variant 1 (v1) and variant 3 (v3) that conjoin ALK exon 20 with EML4 exon 13 and exon 6, respectively (11). Each fusion partner and breakpoint variant may differentially affect ALK subcellular localization; stability; transformation and metastatic capacity; drug sensitivity; and propensity to develop resistance mutations (12–17).
Six ALK tyrosine kinase inhibitors (TKI) spanning three generations (1G, 2G, and 3G) have been approved by one or more regulatory agencies around the globe for the treatment of patients with advanced or metastatic ALK fusion–positive (ALK+) NSCLC. As initial ALK TKI therapy, all six are efficacious with overall response rates of ∼70% to 80%, but each successive generation has achieved improved activity in the central nervous system (CNS) and median progression-free survival (mPFS; refs. 18–23). In phase III randomized studies against the 1G TKI crizotinib in TKI-naïve patients, benefit was achieved with the 2G TKIs alectinib (mPFS = 25.7 vs. 10.4 months; HR = 0.53), brigatinib (24.0 vs. 11.0 months; HR = 0.49), and ensartinib (25.8 vs. 12.7 months; HR = 0.51; approved in China only) and the 3G TKI lorlatinib (not estimable vs. 9.3 months; HR = 0.28) as assessed by independent review committees (20–23). Importantly, considerations related to resistance mutations, brain penetrance, or safety have limited the extent of clinical benefit from each of these TKIs.
The 1G ALK TKI crizotinib demonstrates a lack of potency against a broad spectrum of acquired secondary resistance mutations including ALKC1156Y, ALKI1171N/S/T, ALKF1174C/L/V, ALKL1196M, ALKG1202R, and ALKG1269A (24) and has minimal brain penetrance (25)—a major deficiency given that ∼30% to 40% of patients with ALK+ NSCLC have CNS metastases at diagnosis (26–28). Although 2G ALK TKIs can overcome some crizotinib-resistant ALK mutations and have demonstrated intracranial responses (24, 29–34), ALK resistance mutations causing disease relapse still occur, predominantly the G1202R solvent-front mutation detected in ∼35% of patients after progression on a 2G ALK TKI (33, 35).
The 3G ALK TKI lorlatinib is brain-penetrant and active against ALK single amino acid substitutions that confer resistance to 1G and 2G ALK TKIs (36–38). However, its efficacy following 2G ALK TKI therapy has been limited by the emergence of acquired resistance, including compound ALK resistance mutations identified in 25% to 50% of patients after progression on lorlatinib, with G1202R-containing compound mutations being the most prevalent and often resistant to all approved ALK TKIs (35, 36, 39). An investigational ALK TKI zotizalkib (TPX-0131) was designed to inhibit lorlatinib-refractory compound mutations (40), but its initial clinical study has been terminated (NCT04849273). At this time, effective treatment strategies for patients following disease relapse on lorlatinib remain limited. Lorlatinib has further been limited by a broad spectrum of CNS effects, reported to occur in 52% of patients (23, 41). These CNS effects include psychotic effects and changes in cognitive function, mood, speech, and mental status, which have been attributed to the inhibition of tropomyosin receptor kinase B (TRKB) in the CNS (41, 42). Inhibition of the TRK-family kinases, comprising TRKA, TRKB, and TRKC, may additionally cause weight gain, dizziness, and gait disturbance, which are also reported in patients receiving lorlatinib (23, 43–45).
Taken together, a need exists for a next-generation brain-penetrant ALK TKI that inhibits diverse ALK single and compound resistance mutations while sparing TRK. Here, we describe the discovery of NVL-655, an ALK TKI that addresses this need as evidenced by preclinical and preliminary proof-of-concept clinical activity.
Results
Design and Structure
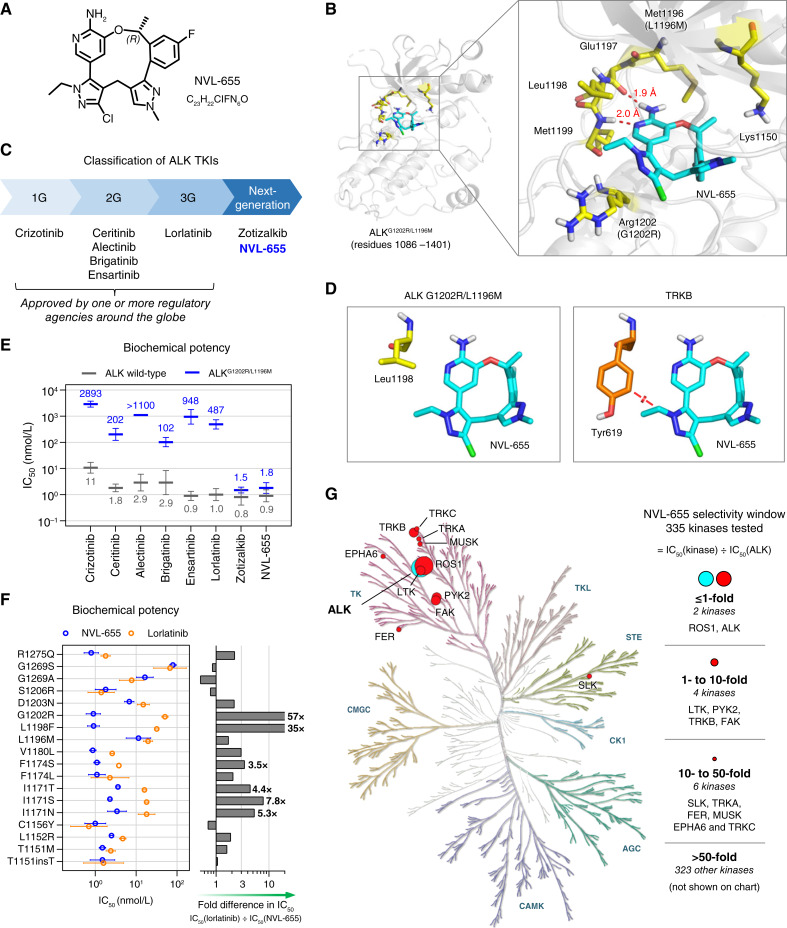
NVL-655 (Fig. 1A) is a macrocyclic small-molecule inhibitor that binds to the ALK ATP pocket. A crystal structure of NVL-655 bound to ALKG1202R/L1196M was obtained, which shows the aminopyridine moiety of NVL-655 forming two hydrogen bonds with Glu1197 and Met1199 in the hinge region (Fig. 1B; Supplementary Fig. S1A and S1B). The fluorophenyl ring fits into a recess in the floor of the binding pocket, and the methylpyrazole ring contacts the P-loop at the roof of the pocket. NVL-655 was designed to overcome the liabilities of approved and other investigational ALK TKIs (Fig. 1C) by addressing the following three medical needs.
Figure 1.
Design and biochemical activity of NVL-655. A, Chemical structure of NVL-655. B, X-ray structure of NVL-655 (cyan) in the binding pocket of ALKG1202R/L1196M, with key residues highlighted in yellow and hydrogen bonds in red. C, Classification of ALK TKIs into 1G, 2G, 3G, or next generation. D, Positioning of NVL-655 with respect to Leu1198 in the crystal structure of ALKG1202R/L1196M (left, yellow; PDB: 9GBE) or Tyr619 in TRKB (right, orange; PDB: 4AT3) based on α-carbon superposition. Red disc indicates a steric clash based on van der Waals overlap. E, Activity of eight TKIs against ALK (gray) and ALKG1202R/L1196M (blue) in biochemical assays. Geometric mean and SD are plotted, with numerical values of the means shown. “IC50 >” is treated as “IC50 =” for plotting. F, Biochemical activity of NVL-655 (blue) and lorlatinib (orange) against ALK with single amino acid substitution or insertion. Geometric mean and SD are plotted. Graph indicates relative potency to NVL-655. G, Kinome selectivity tree for NVL-655. Twelve kinases inhibited with IC50 within 50-fold of ALK are indicated.
Activity against ALK Single and Compound Mutations
The chemical structures of 1G, 2G, and 3G ALK TKIs have atoms that occupy the solvent-front region of the kinase ATP-binding pocket. Mutations in this region, particularly G1202R, have been hypothesized to cause a steric clash with these inhibitors, reducing affinity and thus limiting drug activity (36, 40, 46–48). NVL-655 was designed to minimize bulk in this region, potentially reducing steric clashes and preserving affinity for ALK mutants harboring G1202R (Fig. 1B).
Avoiding TRK Inhibition
There is a differentiating residue at ALK Leu1198, which corresponds to a sterically larger tyrosine residue in TRK (TRKA Tyr591, TRKB Tyr619, and TRKC Tyr619; Fig. 1D; refs. 49, 50). We took advantage of this residue and installed an N-ethylpyrazole on NVL-655 to avoid a negative interaction with the smaller ALK Leu1198 while clashing with the larger TRK tyrosine residue, which could result in preferential binding to wild-type (WT) and mutant ALK over TRK.
Brain Penetrance
The physicochemical properties of NVL-655 (Supplementary Fig. S1C) suggest that the compound has a good probability of brain penetrance. Specifically, topological polar surface area (TPSA) is a key correlate of free brain exposure, with lower TPSA values correlating with better CNS penetrance (51, 52). In comparison with lorlatinib, a compound with excellent CNS exposure (53), NVL-655 has a smaller TPSA (84 Å2 vs. 110 Å2). Based on this analysis, NVL-655 was predicted to have favorable CNS exposure.
Biochemical Activity
WT ALK
Eight ALK TKIs (crizotinib, ceritinib, alectinib, brigatinib, ensartinib, lorlatinib, zotizalkib, and NVL-655) were profiled in biochemical phosphorylation assays against the recombinant ALK domain. The assays were performed in the presence of 1 mmol/L ATP to simulate competitive inhibition against a physiologic ATP concentration. All TKIs including NVL-655 potently inhibited WT ALK (IC50 = 0.8–11 nmol/L; Fig. 1E; Supplementary Fig. S2A and S2B).
ALK Single Mutation
NVL-655 inhibited ALKT1151insT, ALKT1151M, ALKL1152R, ALKC1156Y, ALKI1171N, ALKI1171S, ALKI1171T, ALKF1174L, ALKF1174S, ALKV1180L, ALKL1198F, ALKG1202R, ALKD1203N, ALKS1206R, and ALKR1275Q potently (IC50 = 0.9–6.8 nmol/L) and inhibited ALKL1196M, ALKG1269A, and ALKG1269S moderately (IC50 = 11–79 nmol/L; Supplementary Fig. S2C). All of these have been identified as ALK TKI resistance mutations in NSCLC and/or gain-of-function mutations in neuroblastoma (24, 54, 55). NVL-655 was 57-fold more potent than lorlatinib against ALKG1202R (IC50 = 0.9 vs. 51 nmol/L; Fig. 1F), which represents the most common ALK resistance mutation in patients following 2G ALK TKI therapy (33, 35).
ALKG1202R/L1196M Compound Mutation
ALKG1202R/L1196M confers clinical resistance to lorlatinib and has been reported to confer resistance to 1G, 2G, and 3G ALK TKIs in vitro (35, 36). Indeed, 1G, 2G, and 3G ALK TKIs inhibited recombinant ALKG1202R/L1196M with IC50 > 100 nmol/L. By contrast, NVL-655 potently inhibited ALKG1202R/L1196M with an IC50 of 1.8 nmol/L (Fig. 1E; Supplementary Fig. S2A–S2D).
Kinome Selectivity
In a biochemical activity screen against 335 WT human kinases, NVL-655 selectively inhibited ALK with IC50 = 2.8 nmol/L (Fig. 1G; Supplementary Fig. S3A and S3B). Five kinases (ROS1, LTK, PYK, TRKB, and FAK) showed IC50 within 10-fold of ALK IC50, and six additional kinases (SLK, TRKA, FER, MUSK, EPHA6, and TRKC) showed IC50 within 50-fold of ALK IC50. Accordingly, NVL-655 had a >50-fold selectivity for ALK over 96% (323/335) of the kinome. TRKA, TRKB, and TRKC were inhibited with IC50 < 50-fold of ALK IC50 in this assay (Supplementary Fig. S3B). Because TRK proteins are key off-targets of concern, we further profiled them in biochemical and cell-based assays (see “ALK versus TRK Selectivity”).
Cellular Activity
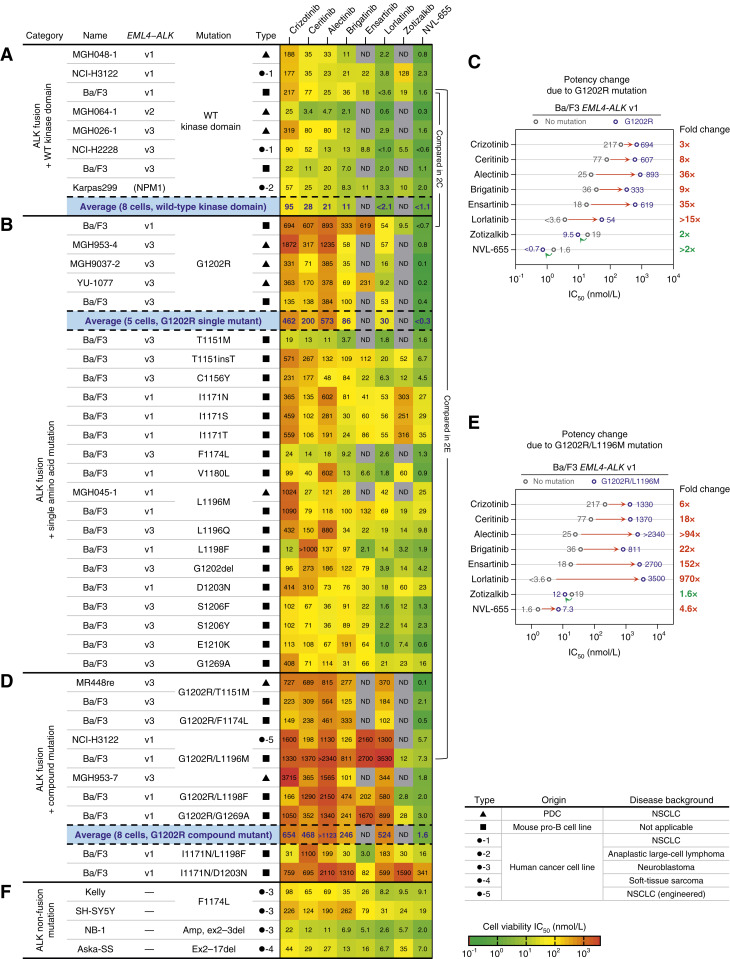
We profiled the activity of ALK TKIs, including NVL-655, in viability assays against 45 ALK-driven cell lines comprising nine patient-derived cell lines (PDC), seven human cancer cell lines, one engineered human cancer cell line, and 27 engineered Ba/F3 cell lines (Supplementary Figs. S4 and S5). The panel collectively represents a broad diversity of ALK alterations including two fusion partners (EML4 and NPM1); three EML4–ALK fusion breakpoint variants (v1, v2, and v3); two partial N-terminal deletions (ex2–3del and ex2–17del); 16 single point mutations (T1151M, C1156Y, I1171N, I1171S, I1171T, F1174L, V1180L, L1196M, L1196Q, L1198F, G1202R, D1203N, S1206F, S1206Y, E1210K, and G1269A); two small indels (T1151insT and G1202del); and seven compound mutations (G1202R/T1151M, G1202R/F1174L, G1202R/L1196M, G1202R/L1198F, G1202R/G1269A, I1171N/L1198F, and I1171N/D1203N).
ALK Fusion with the WT Kinase Domain
NVL-655 potently inhibited eight cell lines expressing WT ALK fusions with average IC50 < 1.1 nmol/L (Fig. 2A). NVL-655 exhibited an IC50 of 0.3 to 1.6 nmol/L against MGH048-1, MGH064-1, and MGH026-1 cell lines, which were established from TKI-naïve patients with NSCLC harboring EML4–ALK v1, v2, and v3 fusions, respectively (Supplementary Fig. S6A), and an IC50 of 2.0 nmol/L against the anaplastic large cell lymphoma cell line Karpas299 harboring the NPM1–ALK fusion. NVL-655 was on average more potent than any other ALK TKIs tested (Supplementary Fig. S6B). In ALK-driven human cancer cell lines treated with NVL-655 for 24 hours, we observed a reduction of cells in the S phase and an increase in caspase 3/7 activation. In a subset of the cell lines, we also observed an increase in the sub-G0–G1 population, annexin V staining, and propidium iodide (PI) staining (Supplementary Fig. S6C–S6E).
Figure 2.
NVL-655 inhibits the viability of ALK-driven cancer cell lines. A, Heat map showing the activity in cell viability assays against ALK fusion with the WT kinase domain. Karpas299 harbors the NPM1–ALK fusion, not EML4–ALK. B, Same as A but for ALK fusion with single amino acid mutation (substitution, insertion, or deletion). C, Plot showing the IC50 for ALK WT (gray) vs. G1202R (blue) in Ba/F3 EML4–ALK v1, along with the associated fold change in IC50. Red indicates an increase in IC50, whereas green indicates a decrease in IC50. D, Same as A but for ALK fusion with compound mutations. E, Same with C but for G1202R/L1196M. F, Same as A but for ALK nonfusion mutations. Potency values reflect geometric mean, and “IC50 <” and “IC50 >” are treated as “IC50 =” for plotting and heat map coloring. Errors, repeats, and maximal effects (Emax) are provided in Supplementary Figs. S5 and S6. Grayed-out entries indicate data not determined (ND). Average potencies (blue) reflect the geometric mean across the cell lines within each category. Amp, amplification.
ALK Fusion with Single Amino Acid Mutations
NVL-655 potently inhibited the growth of five cell lines harboring ALKG1202R fusions (IC50 = 0.1–0.8 nmol/L, average < 0.3 nmol/L; Fig. 2B), including three PDCs established after progressive disease on alectinib (MGH953-4 and YU-1077) or brigatinib (MGH9037-2; Supplementary Fig. S6A). NVL-655 was >14-fold more potent than zotizalkib, >100-fold more potent than lorlatinib, and >280-fold more potent than 1G and 2G ALK TKIs on average (Supplementary Fig. S6F). In Ba/F3 EML4–ALK v1 cells, introduction of G1202R mutation resulted in an 8- to 36-fold increase in the IC50 for 2G and 3G ALK TKIs but not NVL-655, consistent with its G1202R-accommodating design (Fig. 2C). Among ALK fusions with non-G1202R mutations, NVL-655 exhibited a potent IC50 (0.6–9.8 nmol/L) against ALKT1151M, ALKT1151insT, ALKC1156Y, ALKF1174L, ALKV1180L, ALKL1196Q, ALKL1198F, ALKG1202del, ALKS1206F, ALKS1206Y, and ALKE1210K and a moderate IC50 (16–35 nmol/L) against ALKI1171N, ALKI1171S, ALKI1171T, ALKL1196M, ALKD1203N, and ALKG1269A (Fig. 2B).
ALK Fusion with Compound Mutations
NVL-655 potently inhibited the growth of eight cell lines harboring diverse ALKG1202R compound mutations (IC50 = 0.1–7.3 nmol/L, average = 1.6 nmol/L; Fig. 2D) and was more potent than any other ALK TKIs (Supplementary Fig. S6G). MR448re (ALKG1202R/T1151M) and MGH953-7 (ALKG1202R/L1196M) cell lines, which were derived from patients with NSCLC after progressive disease on sequential 1G, 2G, and 3G ALK TKI treatment (39, 56–58), were highly sensitive to NVL-655 (IC50 = 0.1–1.8 nmol/L) but not FDA-approved ALK TKIs (IC50 > 100 nmol/L; Supplementary Fig. S6A). In Ba/F3 EML4–ALK v1 cells, introduction of the G1202R/L1196M compound mutation had a large impact on the IC50 of 2G and 3G ALK TKIs (18–970-fold increase) but a small impact on the IC50 of NVL-655 (4.6-fold increase), again consistent with the design of NVL-655 (Fig. 2E).
I1171N-based compound mutations including ALKI1171N/L1198F and ALKI1171N/D1203N have been reported in patients following relapse on lorlatinib (35, 36). In viability assays with engineered Ba/F3 cells, NVL-655 inhibited ALKI1171N/L1198F with a moderate IC50 of 16 nmol/L, which was 12-fold more potent than lorlatinib (IC50 = 183 nmol/L); however, NVL-655 and most other ALK TKIs showed limited activity against ALKI1171N/D1203N (Fig. 2D).
ALK Nonfusion Oncogenic Mutations
Besides fusion, ALK can be activated by point mutations or deletions in the extracellular domain (7, 59, 60). NVL-655 inhibited neuroblastoma cell lines Kelly (ALKF1174L), SH-SY5Y (ALKF1174L), and NB-1 (ALK amplification, ex2–3del), as well as a soft-tissue sarcoma cell line Aska-SS (ALK ex2–17del) with IC50 = 2 to 19 nmol/L (Fig. 2F).
Comparing Potency and Mutational Coverage of ALK TKIs
NVL-655 proved active against diverse oncogenic alterations, fusion partners, breakpoint variants, resistance mutations (Supplementary Fig. S7A), and disease backgrounds, with low IC50 values ranging from 0.1 to 35 nmol/L. We compared the IC50 of NVL-655 against seven other ALK TKIs across up to 45 cell lines, for a total of 285 pairwise comparisons. NVL-655 showed a lower IC50 than the comparator in the vast majority of comparisons (276/285 = 97%; Supplementary Fig. S7B). In the 9/285 cases in which NVL-655 showed a higher IC50, the IC50 difference was small, with a median of ∼1.3-fold only. By contrast, lorlatinib had a lower IC50 than the comparator in only 209/285 = 73% of comparisons (Supplementary Fig. S7C). In the 76/285 cases in which lorlatinib exhibited a higher IC50 than the comparator, most were a difference of >3-fold. This analysis indicated that among eight ALK TKIs tested, NVL-655 was the most potent inhibitor with the broadest coverage across ALK alterations.
Signaling Pathway Analysis
The effect of NVL-655 on ALK signaling was investigated in six cell lines representing diverse mutational categories: EML4–ALK v1, EML4–ALK v3 G1202R, EML4–ALK v3 G1202R/L1196M, EML4–ALK v3 G1202R/T1151M, and EML4–ALK v1 L1196M. In all these cellular models, NVL-655 suppressed phosphorylation of ALK and downstream effectors including ERK, AKT, and/or S6 in a dose-dependent manner at potencies consistent with cell viability measurements, supporting an on-target mechanism of action (Supplementary Fig. S8). NVL-655 induced PARP cleavage in MR448re, suggesting apoptosis. In PDCs harboring ALKG1202R single or compound mutations, NVL-655 showed maximal effects on pathway signaling at ≤10 nmol/L, whereas lorlatinib showed limited effects even at ≥100 nmol/L.
Further Evidence of On-target Activity
Consistent with the identification of ROS1 and LTK as targets in the kinome screen, NVL-655 potently inhibited Ba/F3 cells expressing oncogenic CD74–ROS1, CD74–ROS1G2032R, or CLIP1–LTK fusions (61) but not parental Ba/F3 cells cultured under IL3 (Supplementary Fig. S9). NVL-655 also did not inhibit ALK- and ROS1-independent cell lines including A549 (KRASG12S lung carcinoma) and A431 (EGFR-amplified epidermoid carcinoma; Supplementary Fig. S9). These observations suggest on-target activity and potentially limited general cytotoxicity.
ALK versus TRK Selectivity
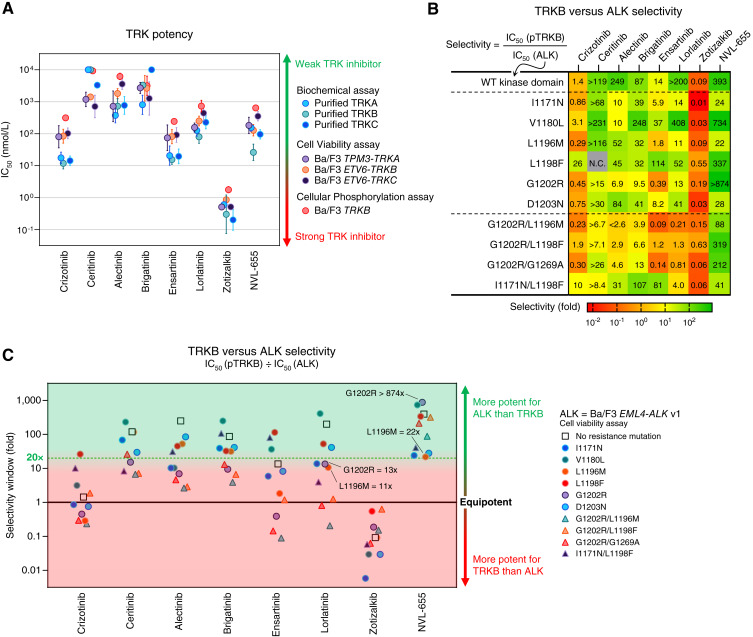
As avoidance of TRK inhibition was one of the main goals in designing NVL-655, we profiled the TRK inhibitory potency of ALK TKIs, including NVL-655, using (i) biochemical assays against purified TRKA, TRKB, and TRKC, (ii) cell viability assays against Ba/F3 cells expressing TPM3–TRKA, ETV6–TRKB, and ETV6–TRKC oncogenic fusions, and (iii) cellular phosphorylation assays against Ba/F3 TRKB cells stimulated with brain-derived neurotrophic factor (Fig. 3A; Supplementary Fig. S10A; ref. 49). Potency trends were highly consistent across the three TRK paralogs and three assay modalities, with correlation coefficients (R2) ranging from 0.87 to 0.98 (Supplementary Fig. S10B). Given the consistent results, we then selected cellular TRKB phosphorylation as the representative TRK assay because of its resemblance to ligand-mediated TRKB signaling in vivo. We calculated the selectivity window between TRKB inhibition and inhibition of diverse ALK oncogenes from cell viability assays. A selectivity window greater than 1 indicates stronger activity for ALK than TRKB, and a selectivity window less than 1 indicates stronger activity for TRKB than ALK (Fig. 3B and C).
Figure 3.
Selectivity for ALK vs. TRK. A, Activity against TRKA, TRKB, and/or TRKC in three assay modalities (biochemical, cell viability, and cellular phosphorylation assays). Geometric mean and SD are plotted. “IC50 >” and “IC50 <” are treated as “IC50 =” for plotting. B, Heat map showing the selectivity window calculated from cellular TRKB phosphorylation IC50 (Supplementary Fig. S10A) divided by Ba/F3 EML4–ALK v1 cell viability IC50 (Supplementary Fig. S4). “Selectivity <” and “selectivity >” are treated as “selectivity =” for heat map coloring. C, Graphical representation of the heat map in B. “Selectivity <” and “selectivity >” are treated as “selectivity =” for plotting. N.C., noncalculable.
Lorlatinib had a >200-fold selectivity window for WT ALK, but this window was eroded to only 11- to 14-fold for ALKG1202R, ALKI1171N, and ALKL1196M (Fig. 3B and C). Lorlatinib has elicited tumor responses in patients harboring these single ALK mutants (62). Its relatively narrow selectivity window suggests that there may be nonnegligible TRKB inhibition at its clinically relevant concentration. Indeed, dose–response curves indicate 41% TRKB inhibition by lorlatinib at 640 nmol/L, a concentration required for ≥95% inhibition of ALK, ALKG1202R, and ALKL1196M (Supplementary Fig. S10C). Zotizalkib exhibited a ≤0.63-fold selectivity window for every ALK oncogene tested, indicating stronger activity against TRKB than ALK or ALK mutants (Fig. 3B and C).
NVL-655 was the most selective TKI for ALK over TRKB and was the only TKI to have a ≥22-fold window for ALK and every mutant ALK oncogene shown (Fig. 3B and C). This selectivity window enabled NVL-655 to avoid TRKB inhibition at an efficacious concentration; dose–response curves indicate only 2% TRKB inhibition by NVL-655 at 100 nmol/L, a concentration that provides ≥95% inhibition of ALK, ALKG1202R, ALKL1196M, and ALKG1202R/L1196M (Supplementary Fig. S10D). Orthogonal analyses using TRKA/B/C fusion viability assays were consistent with the findings from cellular TRKB phosphorylation assays (Supplementary Fig. S11A–S11C).
In Vivo Activity
Antitumor activity of NVL-655 was tested in 10 subcutaneous murine xenograft models, representing a wide diversity of fusion partners (EML4, HIP1, and STRN); EML4 breakpoint variants (v1 and v3); amino acid substitutions (I1171N, G1202R, G1202R/T1151M, G1202R/L1196M, and G1202R/G1269A); origins [PDCs, patient-derived xenografts (PDX), human cancer cell lines, and Ba/F3]; and indications (cholangiocarcinoma and NSCLC; Fig. 4A–D). NVL-655 was administered orally twice daily. Lorlatinib served as a control in some of these studies and, unless noted otherwise, was administered at 5 mg/kg twice daily, a precedented dosage (40) in which free steady-state trough exposure (Cmin,ss) approximates the reported free efficacious concentration (Ceff) of 125 nmol/L (63).
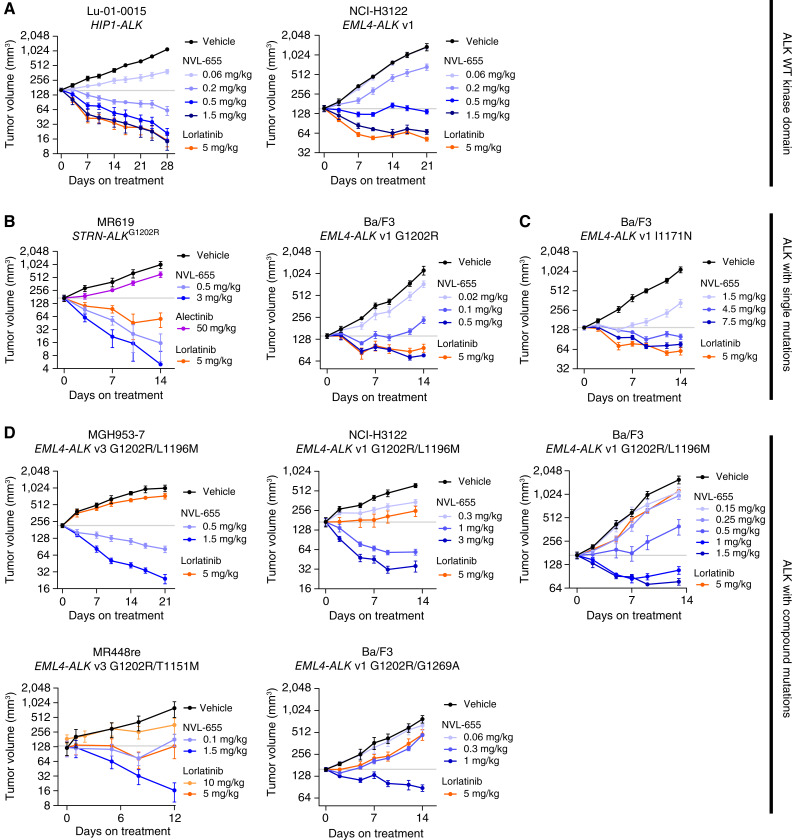
Figure 4.
NVL-655 inhibits subcutaneous ALK-driven tumor xenografts in mice. A–D, Change in tumor volume over time plotted as mean ± SEM for models harboring ALK fusion with a WT kinase domain (A), G1202R single mutation (B), I1171N single mutation (C), or G1202R compound mutations (D). Horizontal gray lines denote mean starting tumor volume of the vehicle group. All treatments were administered orally twice daily, except alectinib 50 mg/kg and lorlatinib 10 mg/kg, which were administered orally once daily. Each group contained 4–9 mice.
ALK Fusion with the WT Kinase Domain
NVL-655 at 1.5 mg/kg or lorlatinib at 5 mg/kg induced regression in xenograft models Lu-01-0015 (a TKI-naïve NSCLC PDX harboring HIP1–ALK) and NCI-H3122 (a human lung cancer cell line harboring EML4–ALK v1; Fig. 4A; Supplementary Fig. S12A and S12B). Western blot analysis of tumor samples confirmed ALK and S6 phosphorylation inhibition, consistent with an on-target mechanism of action, and revealed PARP cleavage, which suggests activation of apoptosis (Supplementary Fig. S12C and S12D). Lorlatinib at 5 mg/kg provided Cmin,ss = 120 to 134 nmol/L, matching the reported Ceff = 125 nmol/L (63), whereas NVL-655 at 1.5 mg/kg provided Cmin,ss = 5.75 to 14.3 nmol/L (Supplementary Fig. S12C and S12D). The Cmin,ss values of both compounds are expected to provide 100% inhibition of WT ALK fusion based on the in vitro cell viability assay, consistent with the observed regression in vivo.
ALK Fusion with Single Mutations
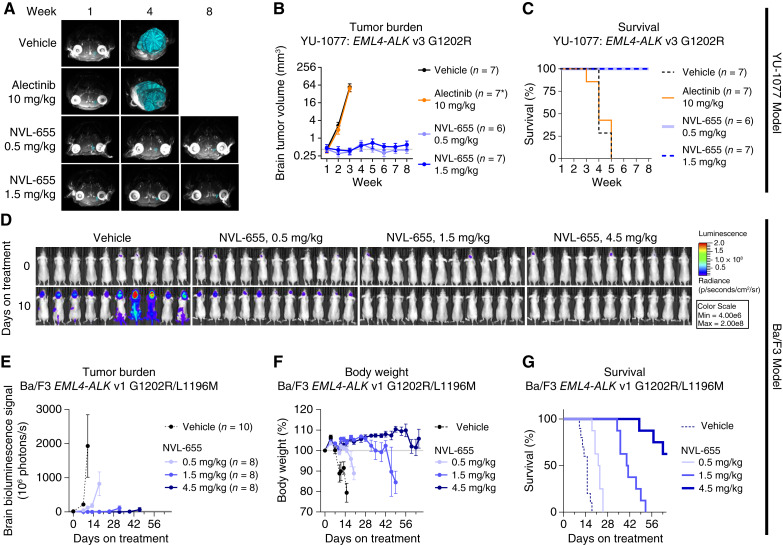
The MR619 model was derived from a patient with cholangiocarcinoma after progression on alectinib and harbored the STRN–ALK fusion with a G1202R mutation. MR619 xenografts were highly sensitive to NVL-655, with 0.5 mg/kg NVL-655 causing a 92% tumor volume decrease by day 14 (Fig. 4B; Supplementary Fig. S13A). NVL-655 at 0.5 mg/kg also induced tumor regression in a Ba/F3 EML4–ALK v1 G1202R xenograft model (Fig. 4B; Supplementary Fig. S13B).
In cell viability testing, I1171N was one of the least sensitive mutations for NVL-655, with IC50 = 27 nmol/L. Nevertheless, NVL-655 induced regression in the Ba/F3 EML4–ALK v1 I1171N model at 4.5 to 7.5 mg/kg (Fig. 4C; Supplementary Fig. S13C).
ALK Fusion with Compound Mutations
We profiled NVL-655 against five xenograft models with ALKG1202R compound mutations. Two of these—MGH953-7 (EML4–ALK v3 G1202R/L1196M) and MR448re (EML4–ALK v3 G1202R/T1151M)—were derived from patients with NSCLC following relapse on sequential 1G, 2G, and 3G ALK TKI treatments (Supplementary Fig. S6A). Two were generated from Ba/F3 cells expressing EML4–ALK v1 G1202R/L1196M or G1202R/G1269A, and the fifth model was generated from NCI-H3122 cells engineered to express EML4–ALK v1 G1202R/L1196M. These compound mutations were resistant to 1G, 2G, and 3G ALK TKIs in vitro (Fig. 2C), and lorlatinib treatment had limited to no activity in vivo (Fig. 4D). By contrast, NVL-655 at ≤1.5 mg/kg induced regression in all five models (Fig. 4D; Supplementary Fig. S14A–S14E). Western blot analysis of tumor samples confirmed inhibition of downstream signaling through the ERK, AKT, or STAT3 axes as well as upregulation of BIM, indicative of in vivo target inhibition and apoptosis activation (Supplementary Fig. S15A and S15B).
In all studies, we observed dose-dependent tumor inhibition by NVL-655 (Fig. 4A–D) with no significant body weight decrease (Supplementary Figs. S12–14), suggesting that NVL-655 was well tolerated.
Preclinical Intracranial Activity
NVL-655 exhibited an unbound brain-to-plasma partition coefficient (Kp,uu) of 0.16 in Wistar Han rats, measured at 1 hour after a single oral 10 mg/kg dosage. These Kp,uu values were comparable with that of lorlatinib (Kp,uu = 0.11), a TKI with high CNS penetration (49, 50, 53, 63).
YU-1077 is a NSCLC PDC harboring EML4–ALK v3 G1202R established after relapse on alectinib (Supplementary Fig. S6A; ref. 64). We implanted YU-1077 cells in the mouse brain and monitored tumor burden via MRI. The tumor grew ∼100-fold in size within 2 weeks after treatment initiation, and all vehicle-treated mice succumbed within 5 weeks, with a median overall survival (mOS) of 4 weeks (Fig. 5A–C; Supplementary Fig. S16A). Treatment with alectinib at 10 mg/kg once daily provided no antitumor activity. By contrast, NVL-655 at 0.5 and 1.5 mg/kg suppressed brain tumors and extended the survival of all mice to beyond the end of the study (week 8), representing a >2-fold increase in mOS. No tumor cells were detected by hematoxylin and eosin staining of brain slices in the NVL-655 treatment groups at week 3; however, MRI reconstruction indicated residual tumors of ∼0.5 mm3 throughout the study (Fig. 5B; Supplementary Fig. S16B).
Figure 5.
NVL-655 inhibits intracranial ALK-driven tumor xenografts in mice. A–D, NVL-655 inhibited tumor growth in the YU-1077 intracranial model. A, Superimposed MRI images from individual mice with brain tumor masses highlighted in cyan. B, Brain tumor volume over time based on MRI reconstruction, plotted as mean ± SEM. Horizontal gray line indicates initial tumor volume of the vehicle group. * indicates that six mice remained at the final treatment timepoint in the alectinib treatment group. C, Survival analysis. D–G, NVL-655 inhibited the Ba/F3 EML4–ALK v1 G1202R/L1196M luciferase intracranial model. D, Bioluminescence images indicating a change in tumor burden over 10 days of treatment. E, Brain bioluminescence over time plotted as mean ± SEM. Plotting for each treatment group stopped when the first animal was lost. Horizontal gray line indicates initial bioluminescence of the vehicle group. F, Body weight over time plotted as mean ± SEM. Horizontal gray line indicates initial body weight. G, Survival analysis. All treatment was administered orally twice daily, except alectinib 10 mg/kg, which was administered orally once daily.
We also intracranially implanted Ba/F3 cells coexpressing EML4–ALK v1 G1202R/L1196M and luciferase, which permitted tumor burden measurement via bioluminescence imaging. This tumor showed a ∼100-fold gain in brain bioluminescence signal within 10 days after treatment initiation and metastasized to peripheral organs (Fig. 5D and E), resulting in rapid weight decrease and loss of all vehicle-treated mice by day 18 with an mOS of 15 days (Fig. 5F and G). NVL-655 suppressed brain tumor growth in a dose-dependent manner and, at 4.5 mg/kg dosing, extended mOS to beyond the end of the study on day 65, representing a >4-fold survival improvement.
Preliminary Proof-of-concept Clinical Activity
Preliminary results from the ongoing phase I dose-escalation portion of the first-in-human ALKOVE-1 study provide preliminary clinical proof-of-concept for the design of NVL-655 (65). Preliminary clinical activity has been demonstrated in patients with ALK+ NSCLC, including those with brain metastases and single or compound ALK resistance mutations, with a favorable preliminary safety profile consistent with its ALK-selective, TRK-sparing design. Herein, we present three case studies from this trial in which intracranial and extracranial activities have been observed in heavily pretreated patients with ALK+ NSCLC, including two with detectable ALKG1202R compound mutations.
Case Study 1
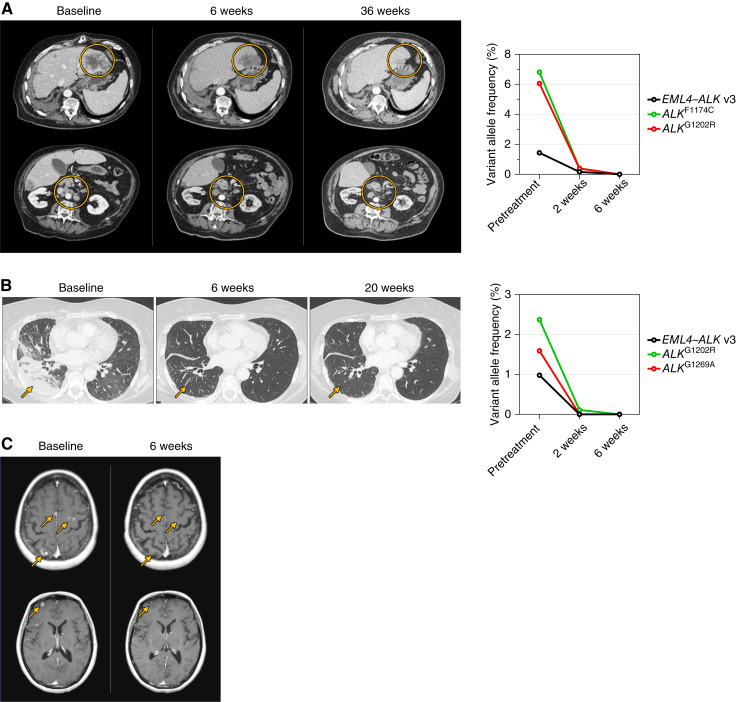
A 66-year-old patient diagnosed with stage IV lung adenocarcinoma with multiple lung, bone, and hepatic metastases had an EML4–ALK fusion identified in plasma ctDNA. The patient received alectinib followed by lorlatinib, each associated with tumor response and subsequent hepatic progression. The patient then received carboplatin and pemetrexed followed by docetaxel and nintedanib, with lung and hepatic progression as the best response to both treatments. Central ctDNA analysis performed at entry to the ALKOVE-1 study showed the EML4–ALK v3 fusion as well as a confirmed cis-allelic ALKG1202R/F1174C compound mutation. The patient initiated NVL-655 at 150 mg once daily and experienced immediate improvement in fatigue. Follow-up ctDNA analysis revealed complete clearance of the ALK fusion and mutant alleles by 6 weeks (Fig. 6A). The first on-treatment imaging at 6 weeks demonstrated a partial response (PR) by RECIST v1.1, including significant reduction in the size of liver metastases and mesenteric and retroperitoneal lymph nodes. As of the data cutoff, the patient has received NVL-655 for approximately 9 months with a confirmed PR (−64% by RECIST v1.1; Fig. 6A) and no reported toxicities.
Figure 6.
Preliminary clinical activity of NVL-655. A, Left, Representative CT images demonstrating a confirmed PR to NVL-655 in a patient with EML4–ALK v3 fusion–positive lung adenocarcinoma with ALKG1202R/F1174C compound resistance mutation after prior alectinib, lorlatinib, and chemotherapy. The patient received NVL-655 at 150 mg once daily. Yellow circles indicate liver metastases (top) and mesenteric and retroperitoneal lymph nodes (bottom) that decreased in size over the course of treatment. Right, Analysis of ctDNA indicated clearance of ALK fusion and mutation alleles. Not detected is imputed as 0% for graphing. B, Left, Representative CT images demonstrating a confirmed PR to NVL-655 in a patient with EML4–ALK v3 fusion–positive lung adenocarcinoma with ALKG1202R and ALKG1269A resistance mutations after prior alectinib, brigatinib, lorlatinib, and chemotherapy. The patient received NVL-655 at 150 mg once daily. Yellow arrows indicate lung metastases that became barely appreciable. Right, Analysis of ctDNA indicated clearance of ALK fusion and mutation alleles. Not detected is imputed as 0% for graphing. C, Representative images of MRI demonstrating a confirmed CNS CR to NVL-655 in a patient with EML4–ALK v1 fusion–positive lung adenocarcinoma without baseline detectable resistance mutation after prior crizotinib, ceritinib, lorlatinib, and chemotherapy. The patient received NVL-655 at 50 mg once daily. Yellow arrows indicate multiple brain metastases at baseline which completely disappeared after 6 weeks of therapy.
Case Study 2
A 62-year-old patient was diagnosed with stage IV lung adenocarcinoma harboring an EML4–ALK fusion. The patient was treated with alectinib, brigatinib, and lorlatinib sequentially, followed by chemotherapy with carboplatin, pemetrexed, and bevacizumab. Following disease progression, the patient was enrolled into the ALKOVE-1 study. Baseline central ctDNA analysis detected an EML4–ALK v3 fusion as well as ALKG1202R and ALKG1269A mutations. A cis-allelic configuration could not be confirmed as the two codons reside on different exons and are not covered by the same sequencing reads. NVL-655 was initiated at 150 mg once daily. Follow-up ctDNA analysis demonstrated complete clearance of the ALK fusion and mutant alleles by 6 weeks (Fig. 6B). Imaging after 6 weeks of therapy showed a PR (−82% by RECIST 1.1) with marked reduction of disease burden in the lungs. As of the data cutoff, the patient continues on NVL-655 at approximately 5 months with an ongoing confirmed PR (Fig. 6B) and has tolerated treatment well without grade ≥ 3 toxicity.
Case Study 3
A 47-year-old patient diagnosed with stage IV lung adenocarcinoma with multiple lung, hepatic, and infradiaphragmatic lymph node metastases had tumor ALK protein expression detected by IHC. The patient received—in sequence—crizotinib with CNS and hepatic progression at 1 month, cisplatin and pemetrexed with hepatic progression, ceritinib requiring dose reductions due to hepatic toxicity and with eventual CNS progression, and lorlatinib with initial CNS complete response (CR) and systemic PR followed by multifocal CNS progression and adrenal oligoprogression. Next-generation sequencing of the lorlatinib-refractory adrenal tumor showed EML4–ALK v1, with no known ALK resistance mutation. The patient then received datopotamab deruxtecan on a clinical trial. Upon multifocal CNS progression with the emergence of new brain metastases, the patient was enrolled in the ALKOVE-1 study and initiated NVL-655 at 50 mg once daily. Central ctDNA analysis did not detect ALK alterations in plasma, although we cannot exclude the possibility that such alterations were present but not detected. Imaging at week 6 demonstrated a CNS CR with complete resolution of multiple parenchymal brain metastases (Fig. 6C) and an overall response of PR (−31% of target lesions). Therapy was briefly interrupted because of grade 3 transaminase elevations without total bilirubin elevation that resolved after 2 weeks. NVL-655 was then restarted at the same dose. The patient remains on therapy for approximately 9 months as of the data cutoff with a confirmed overall PR (−50% by RECIST v1.1) and a sustained CNS CR without recurrence of transaminase elevations.
Estimated TRKB Inhibition in Humans
The steady-state maximum plasma-free drug concentrations were 483 nmol/L for lorlatinib (100 mg once daily; ref. 23) and 40 nmol/L for NVL-655 (150 mg once daily; ref. 65). These concentrations correspond to 31% and 3% estimated TRKB inhibition, respectively, based on in vitro dose–response curves (Supplementary Fig. S17). This pharmacokinetic (PK) analysis supports the potential of NVL-655 to avoid TRKB inhibition relative to lorlatinib at similar recommended dosages in humans and is consistent with findings in mice administered a single 10 mg/kg oral dose of lorlatinib or NVL-655 (Supplementary Fig. S17).
Discussion
ALK TKIs exemplify generational improvements in drug discovery and belong to a growing list of targeted therapies, including BCR–ABL, EGFR, and ROS1 inhibitors (49, 66, 67), that have traced parallel paths in multigenerational drug development. A common theme among these programs is the identification of medical needs left by earlier-generation therapies and the subsequent drug design effort to address those needs. Earlier-generation therapies have been limited by a number of factors, such as innate or acquired drug resistance, toxicity, and suboptimal PK properties, including inadequate brain penetration, which may collectively render the drugs ineffective or necessitate dose modifications that eventually lead to disease progression. By bridging the gap left by its predecessors, a next-generation therapy can re-induce tumor responses and may ultimately supplant an earlier-generation therapy as the standard of care due to improved response durability, CNS activity, and tolerability, as exemplified by the development of 3G EGFR TKI osimertinib in EGFR-mutant lung cancers (67).
Progress has been evident across the three generations of approved ALK TKIs. Compared with the 1G TKI crizotinib, 2G TKIs such as alectinib, brigatinib, and ensartinib offer enhanced potency, coverage of certain resistance mutations, and brain penetrance that altogether resulted in increased mPFS (20–22, 68, 69). The 3G TKI lorlatinib covers a wider mutational spectrum while achieving high brain penetrance, translating to a lower HR versus crizotinib than what had been achieved with 2G TKIs (23). These data have established 2G and 3G ALK TKIs as the preferred first-line therapy in advanced ALK+ NSCLC (70). For patients experiencing disease relapse on initial therapy with a 2G ALK TKI, lorlatinib represents a subsequent option as it has induced tumor responses in patients with ALK single mutations (38). However, clinical experiences have underscored two noteworthy deficiencies for lorlatinib. First, in relapsed patients with preexisting ALK single mutations, subsequent lorlatinib therapy can result in the development of compound ALK mutations refractory to most or all approved ALK TKIs. Second, CNS effects attributed to TRK inhibition have been observed in patients treated with lorlatinib (41, 42), consistent with its limited in vitro selectivity for ALK single mutations, in particular G1202R, over TRK. Motivated by these medical needs, NVL-655 was designed to surpass the limitations of existing ALK TKIs by combining potent activity against ALK single and compound mutations with high brain penetrance and selectivity over TRK.
NVL-655 is active against a diverse set of ALK-mutant oncoproteins, including ALKG1202R single and compound mutations, for which it was specifically designed. The potency against ALKG1202R solvent-front mutation is noteworthy, as (i) it is the most common on-target resistance mutation following relapse on standard 2G ALK TKIs (33, 35); (ii) although lorlatinib has demonstrated clinical efficacy against ALKG1202R (38), neurologic and other toxicities may necessitate dose reductions (71); and (iii) ALKG1202R compound mutations are recurrent after sequential 2G and 3G ALK TKIs and are particularly refractory, with no subsequent options for effective targeting (35, 39). NVL-655 was the most potent among eight approved or investigational ALK TKIs in nearly all the cell lines and ALK mutations tested. This in vitro activity translated into antitumor activity across diverse ALK+ xenograft models, including those derived from patients who had relapsed on 2G and/or 3G therapies and those harboring single or compound resistance mutations. Concordant with these results, NVL-655 has demonstrated activity in patients who have experienced disease progression on multiple ALK TKIs including lorlatinib, with or without single or compound ALK resistance mutations.
NVL-655 is also active against the less common ALK resistance mutations I1171N/S/T, L1196M, and G1269A, albeit with a reduced potency compared with its activity against WT ALK fusions. Hypotheses have been proposed based on computational studies for how these mutations confer resistance to other ALK TKIs. The ALKI1171N/S/T αC-helix mutations are proposed to disrupt the hydrophobic regulatory spine, electrostatically alter the binding pocket, and increase ATP affinity, shifting the kinase into an active state (72–74). The ALKL1196M gatekeeper mutation is proposed to increase steric hindrance and/or affect the activation loop and its interaction with the αC-helix, disrupting ligand binding (75–77), and the ALKG1269A mutation is proposed to increase steric hindrance (75,78). Resistance to ALK TKIs can be mediated by on-target or bypass mechanisms (79). NVL-655 has reduced activity against the ALKI1171N/D1203N compound mutation, which has been observed in patients progressing on lorlatinib (35). Additionally, as an ALK-selective TKI, NVL-655 is not predicted to prevent bypass resistance mechanisms that have been observed with other ALK TKIs, such as MET amplification and BRAFV600E mutation (80).
ALK + NSCLC has a high incidence of brain metastases (25–28), and the CNS represents a recurrent site of disease relapse on existing ALK TKIs, including 2G and 3G TKIs (30, 37, 81, 82). Once patients experience intracranial progression on lorlatinib, options to reestablish CNS disease control are extremely limited. In addition, some primary brain tumors are driven by ALK (6). Consistent with our design, NVL-655 was efficacious in intracranial models of ALKG1202R and ALKG1202R/L1196M and induced an intracranial CR in a patient (case study 3, no detected ALK resistance mutation) who had experienced intracranial progression on multiple ALK TKIs including lorlatinib.
To spare TRK and thereby minimize TRK-associated neurologic toxicities, NVL-655 presents an N-ethyl substituent that is positioned to clash with TRKA Tyr591 and TRKB/C Tyr619 while accommodating the smaller Leu1198 at the analogous position in ALK. This differential interaction might explain the observed selectivity over TRK and contribute to the broader kinome selectivity, as 60% of kinases harbor a Tyr at this position (50). Due in part to this feature, NVL-655 inhibited ALK, ALKG1202R, and ALKG1202R compound mutations 88-fold to >874-fold more potently than TRKB in vitro. This favorable potency differential should enable selection of a clinical NVL-655 dose that can strongly inhibit ALK oncoproteins in tumors without significant inhibition of TRK in the nervous system. Indeed, in the patient case studies reported here, NVL-655 induced responses without accompanying CNS effects attributed to off-target TRK inhibition. However, further study in more patients and with longer follow-up is needed to determine the extent—or the lack thereof—of neurologic adverse events; this is being assessed in the ongoing phase I/II clinical trial.
Despite the remarkable progress in targeting ALK fusions in lung cancer, important medical needs persist. NVL-655 has been developed to fulfill these needs and offers potential best-in-class features that integrate broad single and compound ALK mutation coverage, CNS penetrance, and selectivity aimed at minimizing clinical toxicities. These features, together with the encouraging early reports of clinical activity and safety in the ongoing first-in-human ALKOVE-1 trial, suggest that NVL-655 has the potential to improve outcomes for patients with ALK-driven cancers.
Methods
Compounds
Reference compounds were purchased from commercial sources: crizotinib (Combi-Blocks #QE-4059; MedChemExpress #HY-50878; Selleckchem #S1068), ceritinib (Ark Pharm Inc. #A-1739; Selleckchem #S7083; Anhui Senrise Technology #E120465), alectinib (Advanced ChemBlocks #I-9209; MedChemExpress #HY-13011; Selleckchem #S2762; Chemgood #C-1109), brigatinib (Advanced ChemBlocks # L13974; AstaTech #43213; MedChemExpress #HY-12857; Selleckchem #S8229), ensartinib (MedChemExpress #HY-103714; Wuhan Yongcan Biotechnology #184321; Selleckchem #S2934), lorlatinib (Ark Pharm Inc. #AK175603; eNovation Chemicals #Y0975931; Selleckchem #S7536; MedChemExpress #HY-12215; Chemgood #C-1260), zotizalkib (Pharma Resources; Shanghai Biochempartner #BCP42131), palbociclib (MedChemExpress #HY-50767), and Z-VAD-FML (MedChemExpress #HY-16658). NVL-655 was identified through a medicinal chemistry effort to balance potent activity against ALK and a range of ALK resistance mutations with avoidance of TRK inhibition. Synthesis of NVL-655 can be found in the International Patent Application Publication WO 2021/226269, Example 73. Calculator Plugins were used for structure property prediction and calculation, Marvin 19.20.0, 2019, Chemaxon (http://www.chemaxon.com): logD7.4, TPSA, and predicted basic pKa.
X-ray Crystallography
Protein Production for Crystallography
For structure analysis, a crystallization system was developed for ALKG1202R/L1196M, in which the kinase domain was fused to an N-terminal TEV-cleavable His-GST fusion tag. The protein was produced as previously published (83). In brief, the gene was expressed in baculovirus-infected insect cells, and the protein was purified by a three-step procedure comprising affinity and size exclusion chromatography. Protein for crystallization was concentrated to 10 mg/mL in crystallization buffer (500 mmol/L NaCl, 5.00 %v/v glycerol, 2 mmol/L dithiothreitol, and 20 mmol/L Tris-HCl pH 8).
Crystallization and Structure Analysis
Crystals of ALK in complex with NVL-655 were grown by mixing protein solution (5.5 mg/ mL + 2 mmol/L NVL-655) with reservoir solution [16%–22% (w/v) polyethylene glycol 3350 and 0.15 mol/L buffer solution pH 7.0–7.5 at a 1:1 ratio] using the sitting drop vapor diffusion method at 293 K. Before flash freezing in liquid nitrogen, crystals were cryoprotected by being immersed in reservoir solution supplemented with 20% (v/v) polyethylene glycol 200. Diffraction data of the complex were collected at the Swiss Light Source. The crystals belonged to space group P 21 21 21 containing one molecule per asymmetric unit. The structure was solved to a final resolution of 1.58 Å. The phase information necessary to determine and analyze the structure was obtained by molecular replacement (2) using a previously solved structure of ALK as a search model. Subsequent model building and refinement were performed according to standard protocols using CCP4 and COOT (3). Ligand parametrization and generation were carried out using CORINA (4). The water model was built using the “find waters2” algorithm of COOT, followed by refinement using REFMAC (5) and checking all waters using the validation tool of COOT. Full data collection, processing, and refinement statistics for the structure of NVL-655 bound to human ALK are given in Supplementary Fig. S1A. Data collection and model statistics are shown in Supplementary Fig. S1A. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 9GBE).
Computational Modeling
TRKB [PDB: 4AT3 (84)] was superimposed onto the X-ray structure of NVL-655 in ALKG1202R/L1196M using the Schrodinger Maestro binding-site alignment protocol to illustrate the proximity between NVL-655 and TRKBY619. NVL-655 was not separately docked onto TRKB. All residue numbering is based on the canonical amino acid sequence entries on the UniProt database: ALK (Q9UM73), TRKA (P04629-1), TRKB (Q16620-1), TRKC (Q16288-1), ROS1 (P08922), and LTK (P29376-1).
Biochemical Kinase Activity Assay
The activity of purified kinases was measured using the PhosphoSens assay (AssayQuant Technologies; ref. 85). For testing site 1A, test compounds were dissolved in DMSO to 100-fold over the desired concentration and dispensed at 250 nL into a 384-well plate in a threefold dilution series. A 12.5 µL solution containing 2 mmol/L ATP with 26 µmol/L fluorogenic peptide substrate AQT0101 (AssayQuant Technologies #AQT0101; paired with ALK and ALKG1202R/L1196M) or AQT0104 (AssayQuant Technologies #AQT0104; paired with TRKA, TRKB, and TRKC) in buffer [50 mmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 0.01% Brij-35, 0.5 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), and 10 mmol/L MgCl2] was added to the plate. The reaction was triggered by the addition of a 12.5 µL solution containing 3 nmol/L ALK (Carna #08-518), 10 nmol/L ALKG1202R/L1196M (SignalChem #NP20-118G), 1 nmol/L TRKA (BPS Bioscience #40280), 3 nmol/L TRKB (SignalChem #N17-11G), or 0.5 nmol/L TRKC (BPS Bioscience #40282) kinase domains in buffer (50 mmol/L HEPES pH 7.5, 0.01% Brij-35, 2% glycerol, 0.4 mg/mL BSA, 0.5 mmol/L EGTA, and 10 mmol/L MgCl2). The final concentrations were 1 mmol/L ATP, 13 µmol/L AQT0101 or AQT0104, 0.5 to 5 nmol/L kinase (1.5 nmol/L ALK, 5 nmol/L ALKG1202R/L1196M, 0.5 nmol/L TRKA, 1.5 nmol/L TRKB, or 0.5 nmol/L TRKC), 50 mmol/L HEPES pH 7.5, 0.01% Brij-35, 1% glycerol, 0.2 mg/mL BSA, 0.5 mmol/L EGTA, and 10 mmol/L MgCl2.
For testing site 1B, test compounds were dissolved in 10% DMSO in water to 10-fold over the desired concentration and dispensed at 2.5 µL into a 384-well plate in a threefold dilution series. Aqueous buffer was added to each well such that the final volume was 20 µL, and the final concentrations were 15 µmol/L AQT0101 (AssayQuant Technologies #AQT0101), 1 mmol ATP, 1 mmol DTT, 0.5 mmol EGTA, 50 mmol/L HEPES pH 7.5, 10 mmol/L MgCl2, 0.01% Brij-35, and 0.5 to 5 nmol/L kinase: 5 nmol/L ALKT1151insT (SignalChem # A19-12G), 0.5 nmol/L ALKT1151M (SignalChem # A19-12BG), 0.5 nmol/L ALKL1152R (SignalChem # A19-12CG), 0.5 nmol/L ALKC1156Y (SignalChem # A19-12DG), 0.5 nmol/L ALKI1171N (Carna # 08-566), 0.5 nmol/L ALKI1171S (Carna # 08-569), 1 nmol/L ALKI1171T (Carna # 08-590), 1 nmol/L ALKF1174L (SignalChem # A19-12EG), 0.5 nmol/L ALKF1174S (SignalChem # A19-12FG), 1.5 nmol/L ALKV1180L (Carna # 08-570), 1 nmol/L ALKL1196M (SignalChem # A19-12GG), 1 nmol/L ALKL1198F (Carna # 08-571), 1 nmol/L ALKG1202R (Carna # 08-544), 1.5 nmol/L ALKD1203N (Carna # 08-583), 1 nmol/L 5 nmol/L ALKS1206R (SignalChem # A19-12IG), ALKG1269A (SignalChem # A19-12JG), 0.5 nmol/L ALKG1269S (SignalChem # A19-12KG), or 2 nmol/L ALKR1275Q (SignalChem # A19-12LG).
For both testing sites, the plate was sealed, and the fluorescence signal was recorded using a plate reader at λemission = 485 nm every few minutes for up to 240 minutes at 30°C. The change in fluorescence intensity over time during the initial, linear phase of the reaction is the initial velocity. IC50 was calculated from the plot of initial velocity versus log (inhibitor concentration) regressed to the four-parameter logistic equation.
Kinase Panel Screening
Inhibition of kinase activity was measured using radiolabeled [γ-33P]-ATP (Reaction Biology). A solution containing [γ-33P]-ATP was mixed with NVL-655 (100 nmol/L or 1 μmol/L), each of the 335 kinases, and the corresponding kinase substrate, with ATP at the concentration close to the Michaelis–Menten constant (KM). The reaction was stopped after 1 hour, and the incorporation of 33P was quantified using a scintillation counter. Inhibition was measured by the residual activity of 33P. Based on the 335-kinase screen, 31 kinases showing >50% inhibition were selected for focused IC50 determination using the same assay at 10 concentrations of NVL-655 ranging from 3 μmol/L to 0.09 nmol/L. IC50 was determined from the plot of residual activity against log (inhibitor concentration) regressed to a four-parameter logistic equation. Kinome map illustration was reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com; ref. 86).
Cell Culture
The MGH026-1, MGH048-1, and MGH064-1 cell lines were developed from malignant pleural effusions of TKI treatment–naïve patients with NSCLC harboring EML4–ALK rearrangement. The MGH045-1, MGH953-4, MGH9037-2, and MGH953-7 cell lines were developed from a surgical biopsy of pleural tumor (for MGH045-1 only) or from malignant pleural effusions (for the remaining three cell lines) of patients with NSCLC with EML4–ALK rearrangement who relapsed on prior ALK TKI treatments as indicated in Supplementary Fig. S6A. Prior to model generation, the patients signed written informed consent to participate in a Dana-Farber/Harvard Cancer Center Institutional Review Board–approved protocol, providing permission for research to be performed on their samples. Cell lines were cultured in RPMI1640 + 10% FBS or DMEM + 10% FBS and were sequenced to confirm the presence of ALK rearrangement and/or mutation. Additional authentication was performed by SNP fingerprinting.
After receiving written informed consent from the patent, the MR448re PDC was established from ascitic fluid of a patient with NSCLC following relapse on sequential crizotinib, alectinib, brigatinib, and lorlatinib treatment. Ascitic mononuclear cells were isolated by Ficoll centrifugation and cultured in “TCM” media containing DMEM/F-12, GlutaMAX, 10% FBS, 0.4 μg/mL hydrocortisone, 8.4 ng/mL cholera toxin, 24 μg/mL adenine, and 5 μmol/L Y-27632 (a ROCK inhibitor; Selleckchem # S1049). After stable cancer cell growth was obtained in vitro, culture media were transitioned to DMEM + 10% FBS. The presence of driver alterations was confirmed by RT-PCR and Sanger sequencing.
After receiving written informed consent from the patent, the YU-1077 PDC was established from the pleural effusion of a patient with NSCLC after progression on alectinib and during subsequent ceritinib treatment for 1 month. The patient’s pleural effusion samples were centrifuged at 500× g for 10 minutes at 25°C and resuspended in PBS. Cells were separated with Ficoll-Paque PLUS solution as per protocol. The interface containing the mononuclear cells was washed twice in Hank’s Balanced Salt Solution and plated on collagen IV–coated plates in RPMI medium supplemented with 10% FBS. Sanger sequencing was performed to determine whether cells maintained patient characteristics. FACS staining of EpCAM confirmed PDCs with more than 99% cancer purity. Sanger sequencing confirmed EML4–ALK v3 and G1202R mutations.
Ba/F3 cells were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan, and were maintained in RPMI1640 + 10% FBS in the presence of IL3 (10 ng/mL). Genes encoding human EML4–ALK v1 (WT, G1202R, G1202R/L1196M, G1202R/L1198F, G1202R/G1269A, T1151instT, C1156Y, I1171N, I1171S, I1171T, V1180L, L1196M, L1196Q, L1198F, G1202del, D1203N, S1206F, S1206Y, E1210K, G1269A, I1171N/L1198F, or I1171N/D1203N), CD74–ROS1 (WT, G2032R, or L2086F), CLIP1–LTK, TPM3–TRKA, ETV6–TRKB, ETV6–TRKC, or TRKB (full-length) were synthesized, cloned into a retroviral vector with a puromycin-resistant marker, and packaged into retroviral particles. The virus was used to infect Ba/F3 cells. Stable cell lines were selected by IL3 withdrawal and with puromycin for at least 7 days. The polyclonal culture was used in assays directly, or monoclonal cultures were established through limiting dilution before being used in assays. Successful transformants were confirmed by Sanger sequencing and Western blot. All cells were confirmed to express the full desired protein. Although rendering Ba/F3 CD74–ROS1 and ETV6–TRKC cells IL3-independent, the genes underwent a spontaneous C-terminal frameshift past the kinase domain that was deemed inconsequential to the activity or sensitivity of the kinase domains.
Alternatively, Ba/F3 cells were purchased from DSMZ and cultured in DMEM + 10% FBS in the presence of IL3 (0.5 ng/mL) and were infected with lentiviral constructs according to the manufacturer’s protocol (Thermo Fisher Scientific # K4955) to express EML4–ALK v3 (WT, G1202R, G1202R/T1151M, G1202R/F1174L, T1151M, or F1174L). Infected Ba/F3 cells were selected in the presence of blasticidin (14 mg/mL) and IL3 (0.5 ng/mL) followed by IL3 withdrawal. EML4–ALK fusion and kinase domain mutations were confirmed by Sanger sequencing.
A431, A549, Aska-SS, Karpas299, Kelly, NB-1, NCI-H2228, NCI-H3122, and SH-SY5Y cell lines were obtained at Pharmaron, except for the Western blot experiment (Supplementary Fig. S8) in which NCI-H3122 was purchased from ATCC. Cell lines were cultured in the following media: A431 (DMEM + 10% FBS), A549 (F12K + 10% FBS), Aska-SS (DMEM + 20% FBS), Karpas299 (RPMI + 10% FBS), Kelly (RPMI + 10% FBS + 1% GlutaMAX), NB-1 (RPMI + 10% FBS), and SH-SY5Y (1:1 mixture of F12K:EMEM + 10% FBS). NCI-H3122 was transduced with the EML4–ALK v1 G1202R/L1196M construct (same as used for Ba/F3), selected for 2 weeks on puromycin, expanded monoclonally, and confirmed by Sanger sequencing and Western blot.
All cells were maintained at 37°C with 5% CO2. All cells were inspected for Mycoplasma contamination using ABM PCR Mycoplasma Detection Kit (Cat #G238) or the Lonza MycoAlert Detection Kit (LT07-710) at a frequency of at least once per 3 months, with the following exceptions. YU-1077 cells were inspected for Mycoplasma contamination prior to experimentation using Myco-Read Mycoplasma Polymerase Chain Reaction (PCR) Detection Kit (Cat #SMD0171). MR448re cells were not tested for Mycoplasma contamination but were not maintained in culture for more than 2 months after establishment or thawing.
Cell Viability Assay
For testing site 2A, 2,000 to 5,000 cells (MGH026-1, MGH048-1, MGH064-1, MGH045-1, MGH953-4, MGH9037-2, and MGH953-7) were plated in triplicate into 96-well plates. Cells were incubated with CellTiter-Glo (Promega) after 5-day drug treatment, and luminescence was measured using a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices, LLC). GraphPad Prism (GraphPad Software) was used to graphically display data and determine IC50 values by a nonlinear regression model utilizing a four-parameter analytic method.
For testing site 2B, cells (MR448re and engineered Ba/F3 EML4–ALK v3) were tested in cell viability assays in 96-well plates using the CellTiter-Glo reagent (G7570, Promega) after a 72-hour (Ba/F3) or 144-hour (MR448re) treatment. Data were normalized to DMSO vehicle wells, and the IC50 was determined using GraphPad Prism.
For testing site 2C, about 3,000 YU-1077 cells were seeded in 96-well plates in growth media and incubated overnight at 37°C before adding serially diluted inhibitors. Cells were incubated at 37°C for 72 hours before performing CellTiter-Glo (G924C, Promega) following the manufacturer’s protocol. Dose–response curves and IC50 values were calculated using GraphPad Prism 10 (GraphPad Software, Inc.).
For testing site 2D, cells (A431, A549, Aska-SS, Karpas299, Kelly, NB-1, NCI-H2228, NCI-H3122, SH-SY5Y, and engineered NCI-H3122 and Ba/F3 EML4–ALK v1 cells) were seeded into 384-well plates, and test compounds were added in a threefold dilution series in complete culture medium containing 10% FBS. For cell viability of parental Ba/F3 cells supplemented with IL3, 10 ng/mL IL3 was used. After a 72-hour incubation with the inhibitor, cell viability was measured using the CellTiter-Glo reagent (Promega). Untreated wells served as negative controls (no inhibition), whereas wells treated with high concentrations of the nonspecific kinase inhibitor staurosporine served as positive controls (full inhibition). IC50 was calculated from percent inhibition and log (inhibitor concentration) using four-parameter logistic regression.
In some human cancer cell lines, we observed a biphasic dose–response behavior. We attributed the first dose response to on-target growth inhibition caused by ALK inhibition and the second dose response to off-target cytotoxicity caused by other pathways beyond ALK. In such cases, only the first IC50 (Supplementary Fig. S4) and its associated plateau (Emax; Supplementary Fig. S5) are reported and indicate on-target ALK inhibition.
Western Blot and Cellular Phosphorylation Assays
For testing site 3A, cells (NCI-H3122, MGH045-1, MGH953-4, and MGH953-7) were treated for 6 hours. Total protein lysates were analyzed by Western blotting with the following antibodies (all from Cell Signaling Technology): pALK Y1282/1283 (#9687), pALK Y1604 (#3341), ALK (#3633), pAKT S473 (#4060), AKT (#4691), pERK1/2 T202/Y204 (#9101), ERK1/2 (#9102), pS6 S240/244 (#5364), S6 (#2217), and β-actin (#4970).
For testing site 3B, MR448re cells were treated for 6 hours and were analyzed by Western blotting using the following antibodies (all from Cell Signaling Technology except for β-actin): pALK Y1604 (#3341), ALK (#3333), pAKT S473 (#4060), AKT (#4961), pERK1/2 T202/Y204 (#9101), ERK1/2 (#9102), pS6 S235/S236 (#4858), S6 (#2217), cleaved PARP Asp214 (#9541), and β-actin antibody (Sigma-Aldrich #A1978).
For testing site 3C, YU-1077 cells were treated for 6 hours and were analyzed by Western blotting using the following antibodies (all from Cell Signaling Technology): pALK Y1604 (#3341), ALK (#3363), pAKT S473 (#9271), AKT (#9272), pERK1/2 T202/Y204 (#9101), ERK1/2 (#9102), pS6 S235/S236 (#4858), and S6 (#2217). Cell lysates were centrifuged at 1,300 rpm for 20 minutes at 4°C, and the resultant supernatants were transferred to new tubes. The protein concentration was quantified using a Bradford assay (Bio-Rad). Equal amounts of protein were separated by SDS-PAGE and transferred to the polyvinylidene difluoride membrane. The immunoblots were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Statistical analyses were performed using GraphPad Prism 10 software (GraphPad Software, Inc.). Cell line experiments were independently repeated more than three times with technical and biological repeats in each condition.
For testing site 3D, Ba/F3 TRKB cells were seeded into 384-well plates, and test compounds were added in a threefold dilution series in full culture medium + 10% FBS. Cells were stimulated with 100 ng/mL brain-derived neurotrophic factor for 20 minutes. TRK phosphorylation was measured using the phospho-TRKA (Tyr674/675)/phospho-TRKB (Tyr706/Tyr707) AlphaLISA reagent (PerkinElmer #ALSU-PTRKAB). Untreated wells served as negative controls (no inhibition), whereas wells treated with high concentrations of the nonspecific kinase inhibitor staurosporine served as positive controls (full inhibition). IC50 was calculated from percent inhibition and inhibitor concentration using four-parameter logistic regression.
Cell Cycle and Apoptosis Assays
Cell-cycle analysis was performed using the FITC BrdU Flow Kit according to the manufacturer’s instructions (BD Pharmingen # 559619). Briefly, NCI-H3122, Karpas299, and NB-1 cells were treated with NVL-655 or palbociclib for 24 hours followed by the FITC BrdU solution for 2 hours. Cells were washed, fixed and permeabilized, and treated with DNase, RNase A, and then 7-aminoactinomycin D (7-AAD). Cells were analyzed by flow cytometry (iQue3) with the following gating: sub-G0–G1 phase (low 7-AAD), G0–G1 phase (7-AAD+, BrdU−), S phase (7-AAD+/++, BrdU+), and G2–M phase (7-AAD++, BrdU−).
Caspase 3/7 activation was measured using Caspase-Glo 3/7 Assay System according to the manufacturer’s instructions (Promega # G8092) after treatment of NCI-H3122, Karpas299, and NB-1 cells with NVL-655 or Z-VAD-FMK for 24 hours. Annexin V and PI staining was performed using the Dead Cell Apoptosis Kits according to the manufacturer’s instructions (Invitrogen # V13242). Briefly, NCI-H3122 and NB-1 cells were treated with NVL-655 or Z-VAD-FMK for 24 hours. Cells were washed, treated with FITC annexin V and PI, and analyzed by flow cytometry (iQue3) with the following gating: early apoptosis (annexin V+, PI−) and late apoptosis (annexin V+, PI+).
Subcutaneous Xenograft Studies
MGH953-7 Model
All animal studies were conducted in accordance with the guidelines as published in the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) of Massachusetts General Hospital. Xenografts were implanted subcutaneously into the flanks of female athymic nude (Nu/Nu) mice aged 6 to 8 weeks. Mice were maintained in laminar flow units in sterile filter-top cages with Alpha-Dri bedding. Mice were randomized into groups once the tumors had attained a volume of 150 mm3. The treatment groups were treated twice a day at approximately 9-hour/15-hour intervals with drug solution dissolved in acid water (lorlatinib) or drug solution dissolved in 20% hydroxypropyl-β-cyclodextrin (HP-β-CD; NVL-655) by oral gavage. Tumor volumes were measured twice weekly and calculated using the formula mm3 = 0.52 × L × W2. The numbers of mice at the beginning of the study were n = 7 (vehicle), n = 8 (lorlatinib), n = 8 (NVL-655, 0.5 mg/kg), and n = 9 (NVL-655, 1.5 mg/kg). On day 17 and day 21 timepoints, the numbers of mice were n = 6 (vehicle), n = 7 (lorlatinib), n = 8 (NVL-655, 0.5 mg/kg), and n = 8 (NVL-655, 1.5 mg/kg) due to mouse loss.
MR448re and MR619 Models
All animal procedures and studies were performed in accordance with the approved guidelines for animal experimentation by the Ethics Committee at University Paris Sud (CEEA 26, Project 2020_074_27871) following EU regulation. Animals were housed under pathogen-free conditions with food and water ad libitum. For each model, cancer cells were grafted subcutaneously (1 graft/mouse) in 8-week-old female Swiss nude (MR448re) or male NOD/SCIDγ mice (MR619; Charles River Laboratories) until tumor volume reached 80 to 200 mm3 for the efficacy study or 250 to 400 mm3 for the pharmacodynamics (PD) study. The animals were then randomized for treatment. Treatment was administered once or twice a day (at 8:00 and 18:00 hours), with dosing skipped on weekends. Western blotting was performed using the following antibodies (all from Cell Signaling Technology except for β-actin): pALK Y1604 (#3341), ALK (#3333), pAKT S473 (#4060), AKT (#4961), pERK1/2 T202/Y204 (#9101), ERK1/2 (#9102), pS6 S235/S236 (#4858), S6 (#2217), BIM (#2933), and β-actin antibody (Sigma #A1978). MR619 and MR448re PDCs harbored the corresponding driver fusions and mutations as confirmed by RT-PCR and Sanger sequencing. PDX models were subjected to whole-exome sequencing and whole-transcriptome sequencing for complete characterization.
Lu-01-0015 Model
All experimental procedures were performed according to and with approval from the guidelines of the IACUC of Wuxi AppTec following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Briefly, 6- to 8-week-old female Balb/c nude mice were implanted subcutaneously into the right flank with Lu-01-0015 tumor slices (∼30 mm3) with Matrigel. In the efficacy study, after tumors grew to an average tumor volume of about 162 mm3, mice (n = 5 per group) were randomized and administered vehicle (20% HP-β-CD), lorlatinib, or NVL-655 by oral gavage twice daily (12-hour intervals). In a separate PK and PD study, after tumors grew to an average tumor volume of about 411 mm3, mice received a single dose or twice daily × 5 days of vehicle, lorlatinib, or NVL-655, and tumor and blood were collected at 1 and 12 hours after the last dose. Western blotting was performed using the following antibodies (all from Cell Signaling Technology): pALK Y1604 (#3341), ALK (#3633), and β2-microglobulin (#12851).
NCI-H3122 Models and Ba/F3 Models
All the procedures related to animal handling, care, and the treatment in this study was performed according to guidelines approved by the IACUC of Pharmaron following the guidance of the AAALAC. Briefly, 6- to 8-week-old female Balb/c nude mice were implanted subcutaneously into the right flank with 2 × 106 (NCI-H3122 or NCI-H3122 EML4–ALK v1 G1202R/L1196M) or 1 × 106 (Ba/F3 EML4–ALK v1 G1202R, I1171N, G1202R/L1196M, or G1202R/G1269A) tumor cells. In the efficacy studies, after tumors reached an average tumor volume of approximately 154 to 170 mm3, mice were randomized and administered vehicle (20% HP-β-CD), lorlatinib, or NVL-655 by oral gavage twice daily (12-hour intervals). In separate PK and PD studies, after tumors reached an average tumor volume of approximately 420 to 450 mm3, mice received a single dose or twice daily × 5 days of vehicle, lorlatinib, or NVL-655, and tumor and blood were collected at 1 and/or 12 hours after the last dose. Western blotting was performed using the following antibodies (all from Cell Signaling Technology): pSTAT3 (#9145), STAT3 (#12640), GAPDH (#97166), pALK Y1604 (#3341), ALK (#3633), pS6 (#4858), S6 (#2217), cleaved PARP Asp214 (#5625), PARP (#9532), and β2-microglobulin (#12851).
P values were calculated using the Wilcoxon rank-sum test for tumor volume changes between the first day and the indicated day of treatment in each pairwise comparison. The calculation was performed using an open-access web tool (87).
Kp,uu and Intracranial Studies
Kp,uu
All the procedures related to animal handling, care, and the treatment in these studies were performed according to guidelines approved by the IACUC of Pharmaron following the guidance of AAALAC. NVL-655 was formulated as 1 mg/mL solution in 20% HP-β-CD in deionized water. NVL-655 was administered as a single oral dose to male Wistar Han rats (n = 3 each) at 10 mg/kg. After 1 hour, brain and plasma samples were collected, and brain samples were homogenized in PBS pH 7.4. Brain and plasma samples were precipitated by acetonitrile and centrifugation (4,700 rpm, 15 minutes). Total drug concentrations in the supernatants were quantified by LC/MS-MS. Unbound fractions were determined using rapid equilibrium dialysis. Kp,uu was calculated as the ratio between the unbound drug concentration in the brain and unbound drug concentration in the plasma.
Ba/F3 EML4–ALK v1 G1202R/L1196M Intracranial Model
All the procedures related to animal handling, care, and the treatment in these studies were performed according to guidelines approved by the IACUC of Pharmaron following the guidance of AAALAC. Ba/F3 EML4–ALK G1202R/L1196M cells were transduced with viral particles containing the firefly luciferase gene and a neomycin-resistant marker. Infected cells were selected on neomycin, and monoclonal cultures were established through limiting dilution. Successful transformants were confirmed by Sanger sequencing and bioluminescence. For the in vivo study, 5 × 104 Ba/F3 EML4–ALK G1202R/L1196M luciferase cells were stereotactically implanted into the right forebrains of 6- to 8-week-old female Balb/c nude mice. After 7 days, mice were randomized based on mean bioluminescence signal into four groups of n = 10 mice each and received vehicle or NVL-655. Bioluminescence and body weight were measured at regular intervals until the end of the study (65 days after treatment start) or until animals met the criteria for euthanasia.
YU-1077 Model
All animal experiments were performed in compliance with the standard animal care conditions by and with approval from the IACUC at Yonsei University College of Medicine. Mouse cages were limited to a maximum of five animals per cage and checked daily for cage cleanliness and sufficient food/water. Briefly, 10,000 YU-1077 cells (diluted in 5 µL PBS) were stereotactically injected into the brain of 7-week-old male Balb/c nude mice. Mice were treated with NVL-655 orally twice daily for 2 weeks. Tumors were evaluated with MRI 1 week after implantation and every subsequent week after treatment with NVL-655. All MRI scans were evaluated volumetrically using OsiriX Lite software (Pixmeo SARL).
Clinical Study
ALKOVE-1 (NCT05384626) is a first-in-human, tumor-agnostic, phase I/II trial of NVL-655 in patients with solid tumors harboring ALK rearrangements or activating ALK mutations. All patients provided signed written informed consent for participation. The study was conducted in accordance with the ethical principles of Good Clinical Practice and the Declaration of Helsinki and was approved by the institutional review board/independent ethics committee at each participating site. Case studies are reported as of the data cutoff date of December 21, 2023. The ctDNA next-generation sequencing analysis was conducted with plasma samples collected at cycle 1 day 1 (C1D1, predose), C1D15, and C3D1 using the Guardant360 assay (Guardant Health). Allelic configuration (cis vs. trans) was determined for samples with ≥2 ALK mutations on the same exon as previously described (39). For ALKG1202R/F1174C, the two mutant codons are in close enough proximity and can be covered by the same sequencing reads, revealing the presence of a cis-allelic configuration.
Data Availability
No datasets were generated or analyzed in this study.
Supplementary Material
Supplementary Figure S1. Chemistry/X-ray. Supplementary Figure S2. Biochemical activity of ALK TKIs. Supplementary Figure S3. Kinome profiling of NVL-655. Supplementary Figure S4. Potency in cell viability assay. Supplementary Figure S5. Maximal effect (Emax) in cell viability assay. Supplementary Figure S6. Cellular activity of NVL-655. Supplementary Figure S7. Comparing NVL-655 and lorlatinib potency. Supplementary Figure S8. Pathway analysis in cell lines. Supplementary Figure S9. Further evidence for on-target activity of NVL-655. Supplementary Figure S10. TRK activity. Supplementary Figure S11. Selectivity for ALK versus TRK using additional assays. Supplementary Figure S12. In vivo studies for ALK fusion with a wild-type kinase domain. Supplementary Figure S13. In vivo studies for ALK fusion with G1202R or I1171N single mutations. Supplementary Figure S14. In vivo studies for ALK fusion with G1202R compound mutations. Supplementary Figure S15. Pharmacodynamic analysis in G1202R compound mutation models. Supplementary Figure S16. Body weight and H&E staining for the YU-1077 intracranial study. Supplementary Figure S17. Estimating TRKB inhibition in humans and mice.
Acknowledgments
We thank the patients, caregivers, advocates, physicians, nurses, and research staff for their participation in the ongoing ALKOVE-1 clinical trial. All work presented here was solely funded by Nuvalent, Inc. J.J. Lin and A.N. Hata are supported by the R01 CA164273 grant from the NCI of the NIH independently and outside of the work presented here. A. Drilon is supported by the P30 CA008748 grant from the NCI of the NIH independently and outside of the work presented here.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
J.J. Lin reports personal fees from C4 Therapeutics, Blueprint Medicines, Mirati Therapeutics, AnHeart Therapeutics, CLaiM Therapeutics, Regeneron, Yuhan, Ellipses Pharma, Daiichi Sankyo, AstraZeneca, and Hyku Biosciences, grants and personal fees from Genentech, Nuvalent, Bayer, Elevation Oncology, Novartis, Turning Point Therapeutics, and Bristol Myers Squibb, grants, personal fees, and other support from Pfizer, grants from Relay Therapeutics, Roche, and Linnaeus Therapeutics, and personal fees and other support from Merus outside the submitted work. J.C. Horan reports employment with and ownership interest in Nuvalent, Inc., and a patent for US11667649 granted, a patent for WO 2023/196900 pending, and a patent for WO 2024/086634 pending. A. Tangpeerachaikul reports employment with and ownership interest in Nuvalent, Inc., and a patent for US11667649 granted, a patent for WO 2023/196910 pending, and a patent for and a patent for WO 2024/086634 pending. M.L. Johnson reports grants from Nuvalent during the conduct of the study, as well as grants and other support from AbbVie, Amgen, Arcus Biosciences, ArriVent Biopharma, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, Eli Lilly and Company, Fate Therapeutics, Genentech/Roche, Genmab, GlaxoSmithKline, Gritstone Oncology, Immunocore, Janssen, Merck, Mirati Therapeutics, Novartis, Pfizer, Revolution Medicines, Sanofi, and Takeda Pharmaceuticals, grants from Adaptimmune, Array BioPharma, Artios Pharma, Bayer, BeiGene, BerGenBio, BioAtla, Black Diamond, Calithera Biosciences, Carisma Therapeutics, City of Hope National Medical Center, Conjupro Biotherapeutics, Corvus Pharmaceuticals, Curis, CytomX, Dracen Pharmaceutical, Elicio Therapeutics, EMD Serono, EQRx, Erasca, Exelixis, Genocea Biosciences, Harpoon, Helsinn Healthcare, Hengrui Therapeutics, Hutchinson MediPharma, IDEAYA Biosciences, IGM Biosciences, Immuneering Corporation, Immunitas Therapeutics, IMPACT Therapeutics, Incyte, Kartos Therapeutics, LockBody Therapeutics, Loxo Oncology, Memorial Sloan Kettering, Merus, Mythic Therapeutics, NeoImmune Tech, Neovia Oncology, NextPoint Therapeutics, Numab Therapeutics, Nuvalent, OncoC4, Palleon Pharmaceuticals, PMV Pharmaceuticals, Rain Therapeutics, RasCal Therapeutics, Regeneron Pharmaceuticals, Relay Therapeutics, Ribon Therapeutics, Rubius Therapeutics, Seven and Eight Biopharmaceuticals, Shattuck Labs, Silicon Therapeutics, Summit Therapeutics, Syndax Pharmaceuticals, SystImmune, Taiho Oncology, TCR2 Therapeutics, Tempest Therapeutics, Theras, Tizona Therapeutics, Tmunity Therapeutics, Turning Point Therapeutics, Vividion, Vyriad, and Y-mAbs Therapeutics, and other support from Alentis Therapeutics, Biohaven Pharmaceuticals, D3 Bio Limited, Genmab, Gilead Sciences, Hookipa Biotech, Jazz Pharmaceuticals, ModeX Therapeutics, Molecular Axiom, Normunity, Novocure, Pyramid Biosciences, Seagen, Synthekine, Takeda Pharmaceuticals, and Zai Lab outside the submitted work. B. Besse reports other support from AbbVie, BioNTech SE, Bristol Myers Squibb, Chugai Pharmaceutical, CureVac AG, Daiichi Sankyo, F. Hoffmann-La Roche Ltd., PharmaMar, Regeneron, Sanofi Aventis, Turning Point Therapeutics, Eli Lilly and Company, Ellipses Pharma Ltd., Genmab, Immunocore, Janssen, MSD, Ose Immunotherapeutics, Owkin, Taiho oncology, BeiGene, Genmab A/S, GlaxoSmithKline, Janssen, Roche-Genentech, Sanofi, Takeda, Hedera Dx, and Springer Healthcare Ltd. during the conduct of the study. D.R. Camidge reports personal fees from Nuvalent, Pfizer, Takeda, and Roche during the conduct of the study. T. Fujino reports grants and personal fees from Nuvalent, Inc. during the conduct of the study; grants from Takeda Science Foundation, Eli Lilly Japan K.K., and Kinnate Biopharma Inc. outside the submitted work; and a patent for KU220115PCT pending. S. Yoda reports grants from Nuvalent, Inc. during the conduct of the study, as well as grants and personal fees from Nuvalent, Inc. and personal fees from Tango Therapeutics outside the submitted work. S. Mente reports employment with and ownership interest in Nuvalent, Inc., and a patent for US 11667649 granted. Y. Sun reports employment with and ownership interest in Nuvalent, Inc. N.E. Kohl reports personal fees from Nuvalent, Inc., during the conduct of the study; personal fees from M.D. Anderson Cancer Center, personal fees from ATPACAV LLC, outside the submitted work. J.R. Porter is a Board member and employee of and has ownership interest in Nuvalent, Inc., and a patent for WO 2023/196910 pending. M.D. Shair is a consultant/board member of and has ownership interest in Nuvalent, Inc. during the conduct of the study; has employment with Harvard University outside the submitted work; and reports a patent for US 11667649 granted. V.W. Zhu reports employment with and ownership interest in Nuvalent, Inc., and a patent for WO 2023/196910 pending. E. Felip reports other support from AbbVie, Amgen, AstraZeneca, Bayer, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly and Company, F. Hoffmann-La Roche, Genmab, Gilead, GSK, Janssen, Merck Serono, MSD, Novartis, Peptomyc, Pfizer, Regeneron, Sanofi, Takeda, Turning Point Therapeutics, Daiichi Sankyo, Genentech, Medical Trends, Medscape, PeerVoice, touchONCOLOGY, and Grifols outside the submitted work. B.C. Cho reports personal fees from Champions Oncology, Crown Bioscience, Imagen, PearlRiver Bio GmbH, Abion, BeiGene, Novartis, AstraZeneca, Boehringer Ingelheim, Roche, Bristol Myers Squibb, CJ Bioscience, CureLogen, Cyrus therapeutics, Ono Pharmaceutical, ONEGENE BIOTECHNOLOGY, Yuhan, Pfizer, Eli Lilly and Company, GI Cell, Guardant Health, HK inno.N, IMNEURUN Biosciences Inc., Janssen, Takeda, MSD, MedPacto, Blueprint Medicines, RandBio, Hanmi, Yonsei University Health System, Kanaph Therapeutic Inc., BridgeBio Therapeutics, Oscotec Inc., J INTS Bio, Therapex Co., Ltd., Gilead, Amgen, TheraCanVac Inc., Gencurix Inc., and Interpark Bio Convergence Corp., other support from DAAN Biotherapeutics, and grants from MOGAM Institute, LG Chem, Oscotec, Interpark Bio Convergence Corp., GI Innovation, GI Cell, Abion, AbbVie, AstraZeneca, Bayer, Blueprint Medicines, Boehringer Ingelheim, Champions Onocology, CJ Bioscience, CJ Blossom Park, Cyrus, Dizal Pharma, Genexine, Janssen, Eli Lilly and Company, MSD, Novartis, Nuvalent, Oncternal, Ono, Regeneron, Dong-A ST, BridgeBio Therapeutics, Yuhan, ImmuneOncia, Illumina, Kanaph Therapeutics, Therapex, J INTS BIO, Hanmi, CHA Bundang Medical Center, and Vertical Bio AG outside the submitted work. L. Friboulet reports grants from Nuvalent during the conduct of the study, as well as grants from Sanofi, Relay Therapeutics, and Amgen outside the submitted work. A.N. Hata reports grants and personal fees from Nuvalent during the conduct of the study, as well as grants and personal fees from Amgen and Pfizer, grants from BridgeBio, Bristol Myers Squibb, C4 Therapeutics, Eli Lilly and Company, Novartis, and Scorpion Therapeutics, and personal fees from Engine Biosciences, Oncovalent, TigaTx, and TOLREMO therapeutics outside the submitted work. H.E. Pelish reports employment with and ownership interest in Nuvalent, Inc., and a patent for US 11667649 granted, a patent for WO 2023/196910 pending, and a patent for WO 2024/086634 pending. A. Drilon reports personal fees from 14ner/Elevation Oncology, Amgen, AnHeart Therapeutics, AbbVie, ArcherDX, AstraZeneca, Bayer, BeiGene, BerGenBio, Blueprint Medicines, Boundless Bio, Bristol Myers Squibb, Chugai Pharmaceutical, EcoR1, EMD Serono, Entos, Exelixis, Helsinn, Hengrui Therapeutics, Ignyta/Genentech/Roche, InnoCare, Janssen, Loxo/Lilly, Merus, Monopteros, Monte Rosa, Novartis, Nuvalent, Pfizer, Prelude, Regeneron, Repare RX, Takeda/ARIAD/Millennium, Treeline Biosciences, TP Therapeutics, Tyra Biosciences, Verastem, Zymeworks, and MBrace and other support from Boehringer Ingelheim, Merck, and Puma during the conduct of the study; other support from Foundation Medicine, Teva, Taiho, GlaxoSmithKline, and PharmaMar and personal fees from Wolters Kluwer, and UpToDate Answers in CME, Applied Pharmaceutical Science, Inc., Axis, Clinical Care Options, Doc Congress, EPG Health, Harborside Nexus, i3 Health, IMEDEX, Liberum, Medendi, Medscape, Med Learning, Medtalks, MJH Life Sciences, MORE Health, Ology, OncLive, Paradigm, PeerView Institute, PeerVoice, Physicians’ Education Resources, Projects in Knowledge, Resources, Remedica Ltd., Research To Practice, RV More, Springer Healthcare, Targeted Oncology, Touch IME, and WebMD outside the submitted work; and a patent for selpercatinib osimertinib pending. No disclosures were reported by the other authors.
References
- 1. Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol 2016;27(Suppl 3):iii4–15. [DOI] [PubMed] [Google Scholar]
- 2. Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994;263:1281–4. [DOI] [PubMed] [Google Scholar]
- 3. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. [DOI] [PubMed] [Google Scholar]
- 5. Delsol G, Lamant L, Mariamé B, Pulford K, Dastugue N, Brousset P, et al. A new subtype of large B-cell lymphoma expressing the ALK kinase and lacking the 2; 5 translocation. Blood 1997;89:1483–90. [PubMed] [Google Scholar]
- 6. Guerreiro Stucklin AS, Ryall S, Fukuoka K, Zapotocky M, Lassaletta A, Li C, et al. Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun 2019;10:4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008;455:930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griffin CA, Hawkins AL, Dvorak C, Henkle C, Ellingham T, Perlman EJ. Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res 1999;59:2776–80. [PubMed] [Google Scholar]
- 9. Ou S-HI, Zhu VW, Nagasaka M. Catalog of 5′ fusion partners in ALK-positive NSCLC circa 2020. JTO Clin Res Rep 2020;1:100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ross JS, Ali SM, Fasan O, Block J, Pal S, Elvin JA, et al. ALK fusions in a wide variety of tumor types respond to anti-ALK targeted therapy. Oncologist 2017;22:1444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers (Basel) 2017;9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Armstrong F, Duplantier M-M, Trempat P, Hieblot C, Lamant L, Espinos E, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene 2004;23:6071–82. [DOI] [PubMed] [Google Scholar]
- 13. Heuckmann JM, Balke-Want H, Malchers F, Peifer M, Sos ML, Koker M, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682–90. [DOI] [PubMed] [Google Scholar]
- 14. Woo CG, Seo S, Kim SW, Jang SJ, Park KS, Song JY, et al. Differential protein stability and clinical responses ofEML4-ALK fusion variants to various ALK inhibitors in advancedALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791–7. [DOI] [PubMed] [Google Scholar]
- 15. Childress MA, Himmelberg SM, Chen H, Deng W, Davies MA, Lovly CM. ALK fusion partners impact response to ALK inhibition: differential effects on sensitivity, cellular phenotypes, and biochemical properties. Mol Cancer Res 2018;16:1724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin JJ, Zhu VW, Yoda S, Yeap BY, Schrock AB, Dagogo-Jack I, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol 2018;36:1199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang SS, Nagasaka M, Zhu VW, Ou S-HI. Going beneath the tip of the iceberg. Identifying and understanding EML4-ALK variants and TP53 mutations to optimize treatment of ALK fusion positive (ALK+) NSCLC. Lung Cancer 2021;158:126–36. [DOI] [PubMed] [Google Scholar]
- 18. Pfizer Inc . XALKORI (crizotinib) [package insert]. [cited 2024 Jan 11]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/202570s036lbl.pdf.
- 19. Novartis Pharmaceuticals Corporation . ZYKADIA (ceritinib) [package insert]. [cited 2024 Jan 11]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/205755s019lbl.pdf.
- 20. Genentech . ALECENSA (alectinib) [package insert]. [cited 2024 Jan 11]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/208434s012lbl.pdf.
- 21. Takeda Pharmaceuticals . ALUNBRIG (brigatinib) [package insert]. [cited 2024 Jan 11]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/208772s013lbl.pdf.
- 22. Horn L, Wang Z, Wu G, Poddubskaya E, Mok T, Reck M, et al. Ensartinib vs crizotinib for patients with anaplastic lymphoma kinase-positive non-small cell lung cancer: a randomized clinical trial. JAMA Oncol 2021;7:1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfizer Inc . LORBRENA (lorlatinib) [package insert]. [cited 2024 Jan 11]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/210868s004lbl.pdf.
- 24. Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aldea M, Besse B, Hendriks LEL. ALK inhibitors in ALK-positive NSCLC with central nervous system metastases. Eur Oncol Haematol 2020;16:18–21. [Google Scholar]
- 26. Gainor JF, Tseng D, Yoda S, Dagogo-Jack I, Friboulet L, Lin JJ, et al. Patterns of metastatic spread and mechanisms of resistance to crizotinib in ROS1-positive non-small-cell lung cancer. JCO Precis Oncol 2017;2017:PO.17.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ou S-HI, Zhu VW. CNS metastasis in ROS1+ NSCLC: an urgent call to action, to understand, and to overcome. Lung Cancer 2019;130:201–7. [DOI] [PubMed] [Google Scholar]
- 28. Pacheco JM, Gao D, Smith D, Purcell T, Hancock M, Bunn P, et al. Natural history and factors associated with overall survival in stage IV ALK-rearranged non-small cell lung cancer. J Thorac Oncol 2019;14:691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chow LQ, Barlesi F, Bertino EM, van den Bent MJ, Wakelee H, Wen PY, et al. Results of the ASCEND-7 phase II study evaluating ALK inhibitor (ALKi) ceritinib in patients (pts) with ALK+ non-small cell lung cancer (NSCLC) metastatic to the brain. Ann Oncol 2019;30:v602–3. [Google Scholar]
- 30. Gadgeel S, Peters S, Mok T, Shaw AT, Kim DW, Ou SI, et al. Alectinib versus crizotinib in treatment-naive anaplastic lymphoma kinase-positive (ALK+) non-small-cell lung cancer: CNS efficacy results from the ALEX study. Ann Oncol 2018;29:2214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Camidge DR, Kim D-W, Tiseo M, Langer CJ, Ahn M-J, Shaw AT, et al. Exploratory analysis of brigatinib activity in patients with anaplastic lymphoma kinase-positive non-small-cell lung cancer and brain metastases in two clinical trials. J Clin Oncol 2018;36:2693–701. [DOI] [PubMed] [Google Scholar]
- 32. Horn L, Wang Z, Wu G, Poddubskaya E, Mok T, Reck M, et al. Ensartinib vs crizotinib for patients with anaplastic lymphoma Kinase−Positive non–small cell lung cancer. JAMA Oncol 2021;7:1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koopman B, Groen HJM, Schuuring E, Hiltermann TJN, Timens W, den Dunnen WFA, et al. Actionability of on-target ALK resistance mutations in patients with non-small cell lung cancer: local experience and review of the literature. Clin Lung Cancer 2022;23:e104–15. [DOI] [PubMed] [Google Scholar]
- 34. Horn L, Infante JR, Reckamp KL, Blumenschein GR, Leal TA, Waqar SN, et al. Ensartinib (X-396) in ALK-positive non–small cell lung cancer: results from a first-in-human phase I/II, multicenter study. Clin Cancer Res 2018;24:2771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dagogo-Jack I, Rooney M, Lin JJ, Nagy RJ, Yeap BY, Hubbeling H, et al. Treatment with next-generation ALK inhibitors fuels plasma ALK mutation diversity. Clin Cancer Res 2019;25:6662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoda S, Lin JJ, Lawrence MS, Burke BJ, Friboulet L, Langenbucher A, et al. Sequential ALK inhibitors can select for lorlatinib-resistant compound ALK mutations in ALK-positive lung cancer. Cancer Discov 2018;8:714–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Solomon BJ, Bauer TM, Mok TSK, Liu G, Mazieres J, de Marinis F, et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: updated analysis of data from the phase 3, randomised, open-label CROWN study. Lancet Respir Med 2023;11:354–66. [DOI] [PubMed] [Google Scholar]
- 38. Shaw AT, Solomon BJ, Besse B, Bauer TM, Lin C-CC, Soo RA, et al. ALK resistance mutations and efficacy of lorlatinib in advanced anaplastic lymphoma kinase-positive non–small-cell lung cancer. J Clin Oncol 2019;37:1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shiba-Ishii A, Johnson TW, Dagogo-Jack I, Mino-Kenudson M, Johnson TR, Wei P, et al. Analysis of lorlatinib analogs reveals a roadmap for targeting diverse compound resistance mutations in ALK-positive lung cancer. Nat Cancer 2022;3:710–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murray BW, Zhai D, Deng W, Zhang X, Ung J, Nguyen V, et al. TPX-0131, a potent CNS-penetrant, next-generation inhibitor of wild-type ALK and ALK-resistant mutations. Mol Cancer Ther 2021;20:1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camidge DR, Shakespeare W, Iv H, Nagasaka M, Ou SI. Lorlatinib should not be considered as the preferred first-line option in patients with advanced ALK rearranged NSCLC. J Thorac Oncol 2021;16:528–31. [DOI] [PubMed] [Google Scholar]
- 42. Shaw AT, Bauer TM, de Marinis F, Felip E, Goto Y, Liu G, et al. First-line lorlatinib or crizotinib in advanced ALK-positive lung cancer. N Engl J Med 2020;383:2018–29. [DOI] [PubMed] [Google Scholar]
- 43. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Drilon A. TRK inhibitors in TRK fusion-positive cancers. Ann Oncol 2019;30:VIII23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu D, Flory J, Lin A, Offin M, Falcon CJ, Murciano-Goroff YR, et al. Characterization of on-target adverse events caused by TRK inhibitor therapy. Ann Oncol 2020;31:1207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hatcher JM, Bahcall M, Choi HG, Gao Y, Sim T, George R, et al. Discovery of inhibitors that overcome the G1202R anaplastic lymphoma kinase resistance mutation. J Med Chem 2015;58:9296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu X, Smaill JB, Ding K. Medicinal chemistry strategies for the development of kinase inhibitors targeting point mutations. J Med Chem 2020;63:10726–41. [DOI] [PubMed] [Google Scholar]
- 48. Chen C, He Z, Xie D, Zheng L, Zhao T, Zhang X, et al. Molecular mechanism behind the resistance of the G1202R-mutated anaplastic lymphoma kinase to the approved drug ceritinib. J Phys Chem B 2018;122:4680–92. [DOI] [PubMed] [Google Scholar]
- 49. Drilon A, Horan JC, Tangpeerachaikul A, Besse B, Ou S-HI, Gadgeel SM, et al. NVL-520 is a selective, TRK-sparing, and brain-penetrant inhibitor of ROS1 fusions and secondary resistance mutations. Cancer Discov 2023;13:598–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson TW, Richardson PF, Bailey S, Brooun A, Burke BJ, Collins MR, et al. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h] [2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem 2014;57:4720–44. [DOI] [PubMed] [Google Scholar]
- 51. Kelder J, Grootenhuis PD, Bayada DM, Delbressine LP, Ploemen JP. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res 1999;16:1514–9. [DOI] [PubMed] [Google Scholar]
- 52. Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 2000;43:3714–7. [DOI] [PubMed] [Google Scholar]
- 53. Bauer TM, Shaw AT, Johnson ML, Navarro A, Gainor JF, Thurm H, et al. Brain penetration of lorlatinib: cumulative incidences of CNS and non-CNS progression with lorlatinib in patients with previously treated ALK-positive non-small-cell lung cancer. Target Oncol 2020;15:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 2014;26:682–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mizuta H, Bigot L, Tangpeerachaikul A, Pelish H, Friboulet L. EP08.02-020 preclinical activity of NVL-655 in a patient-derived NSCLC model with lorlatinib-resistant ALK G1202R/T1151M mutation. J Thorac Oncol 2022;17:S406. [Google Scholar]
- 57. Fujino T, Nguyen L, Yoda S, Yu M, Mizuta H, Bigot L, et al. Preclinical activity of NVL-655 in patient-derived models of ALK cancers, including those with lorlatinib-resistant G1202R/L1196M compound mutation. Eur J Cancer 2022;174:S78–9. [Google Scholar]
- 58. Pailler E, Faugeroux V, Oulhen M, Mezquita L, Laporte M, Honoré A, et al. Acquired resistance mutations to ALK inhibitors identified by single circulating tumor cell sequencing in ALK-rearranged non–small-cell lung cancer. Clin Cancer Res 2019;25:6671–82. [DOI] [PubMed] [Google Scholar]
- 59. Okubo J, Takita J, Chen Y, Oki K, Nishimura R, Kato M, et al. Aberrant activation of ALK kinase by a novel truncated form ALK protein in neuroblastoma. Oncogene 2012;31:4667–76. [DOI] [PubMed] [Google Scholar]
- 60. Fleuren EDGG, Vlenterie M, Van Der Graaf WTAA, Hillebrandt-Roeffen MHSS, Blackburn J, Ma X, et al. Phosphoproteomic profiling reveals ALK and MET as novel actionable targets across synovial sarcoma subtypes. Cancer Res 2017;77:4279–92. [DOI] [PubMed] [Google Scholar]
- 61. Izumi H, Matsumoto S, Liu J, Tanaka K, Mori S, Hayashi K, et al. The CLIP1–LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature 2021;600:319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shaw AT, Solomon BJ, Chiari R, Riely GJ, Besse B, Soo RA, et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol 2019;20:1691–701. [DOI] [PubMed] [Google Scholar]
- 63. Shaw AT, Felip E, Bauer TM, Besse B, Navarro A, Postel-Vinay S, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee JB, Yu MR, Yun MR, Lee YW, Oh SY, Lee EJ, et al. Abstract 4022: preclinical intracranial activity of NVL-655 in an alectinib-resistant patient-derived model harboring EML4-ALK fusion with G1202R mutation. Cancer Res 2023;83:4022. [Google Scholar]
- 65. Lin JJ, Johnson M, Felip E, Ou S-HI, Besse B, Baik C, et al. Abstract B154: safety and preliminary activity of the selective ALK inhibitor NVL-655 in patients with ALK fusion-positive solid tumors. Mol Cancer Ther 2023;22:B154–4. [Google Scholar]
- 66. Rossari F, Minutolo F, Orciuolo E. Past, present, and future of Bcr-Abl inhibitors: from chemical development to clinical efficacy. J Hematol Oncol 2018;11:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 68. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, et al. Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med 2017;377:829–38. [DOI] [PubMed] [Google Scholar]
- 69. Camidge DR, Kim HR, Ahn M-J, Yang JCH, Han J-Y, Hochmair MJ, et al. Brigatinib versus crizotinib in ALK inhibitor–naive advanced ALK-positive NSCLC: final results of phase 3 ALTA-1L trial. J Thorac Oncol 2021;16:2091–108. [DOI] [PubMed] [Google Scholar]
- 70. National Comprehensive Cancer Network, Inc . NCCN clinical Practice guidelines in oncology (NCCN Guidelines®) for guideline name; 2024[cited 2024 Jan 25]. Available from:https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
- 71. Bauer TM, Felip E, Solomon BJ, Thurm H, Peltz G, Chioda MD, et al. Clinical management of adverse events associated with lorlatinib. Oncologist 2019;24:1103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 2014;20:5686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Balasundaram A, C Doss GP. Comparative atomistic insights on apo and ATP-I1171N/S/T in nonsmall-cell lung cancer. ACS Omega 2023;8:43856–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. He M, Li W, Zheng Q, Zhang H. A molecular dynamics investigation into the mechanisms of alectinib resistance of three ALK mutants. J Cell Biochem 2018;119:5332–42. [DOI] [PubMed] [Google Scholar]
- 75. Nagasundaram N, Wilson Alphonse CR, Samuel Gnana PV, Rajaretinam RK. Molecular dynamics validation of crizotinib resistance to ALK mutations (L1196M and G1269A) and identification of specific inhibitors. J Cell Biochem 2017;118:3462–71. [DOI] [PubMed] [Google Scholar]
- 76. Kay M, Dehghanian F. Exploring the crizotinib resistance mechanism of NSCLC with the L1196M mutation using molecular dynamics simulation. J Mol Model 2017;23:323. [DOI] [PubMed] [Google Scholar]
- 77. Chen J, Wang J, Zhu W. Mutation L1196M-induced conformational changes and the drug resistant mechanism of anaplastic lymphoma kinase studied by free energy perturbation and umbrella sampling. Phys Chem Chem Phys 2017;19:30239–48. [DOI] [PubMed] [Google Scholar]
- 78. Amin AD, Li L, Rajan SS, Gokhale V, Groysman MJ, Pongtornpipat P, et al. TKI sensitivity patterns of novel kinase-domain mutations suggest therapeutic opportunities for patients with resistant ALK+ tumors. Oncotarget 2016;7:23715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schneider JL, Lin JJ, Shaw AT. ALK-positive lung cancer: a moving target. Nat Cancer 2023;4:330–43. Available from: 10.1038/s43018-023-00515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shi R, Filho SNM, Li M, Fares A, Weiss J, Pham N-A, et al. BRAF V600E mutation and MET amplification as resistance pathways of the second-generation anaplastic lymphoma kinase (ALK) inhibitor alectinib in lung cancer. Lung Cancer 2020;146:78–85. [DOI] [PubMed] [Google Scholar]
- 81. Camidge DR, Kim HR, Ahn M-J, Yang JCH, Han J-Y, Hochmair MJ, et al. Brigatinib versus crizotinib in advanced ALK inhibitor–naive ALK-positive non–small cell lung cancer: second interim analysis of the phase III ALTA-1L trial. J Clin Oncol 2020;38:3592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Solomon BJ, Cappuzzo F, Felip E, Blackhall FH, Costa DB, Kim D-W, et al. Intracranial efficacy of crizotinib versus chemotherapy in patients with advanced ALK-positive non-small-cell lung cancer: results from PROFILE 1014. J Clin Oncol 2016;34:2858–65. [DOI] [PubMed] [Google Scholar]
- 83. Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 2011;19:679–90. [DOI] [PubMed] [Google Scholar]
- 84. Bertrand T, Kothe M, Liu J, Dupuy A, Rak A, Berne PF, et al. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol 2012;423:439–53. [DOI] [PubMed] [Google Scholar]
- 85. Shults MD, Imperiali B. Versatile fluorescence probes of protein kinase activity. J Am Chem Soc 2003;125:14248–9. [DOI] [PubMed] [Google Scholar]
- 86. Eid S, Turk S, Volkamer A, Rippmann F, Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics 2017;18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Enot DP, Vacchelli E, Jacquelot N, Zitvogel L, Kroemer G. TumGrowth: an open-access web tool for the statistical analysis of tumor growth curves. Oncoimmunology 2018;7:e1462431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Chemistry/X-ray. Supplementary Figure S2. Biochemical activity of ALK TKIs. Supplementary Figure S3. Kinome profiling of NVL-655. Supplementary Figure S4. Potency in cell viability assay. Supplementary Figure S5. Maximal effect (Emax) in cell viability assay. Supplementary Figure S6. Cellular activity of NVL-655. Supplementary Figure S7. Comparing NVL-655 and lorlatinib potency. Supplementary Figure S8. Pathway analysis in cell lines. Supplementary Figure S9. Further evidence for on-target activity of NVL-655. Supplementary Figure S10. TRK activity. Supplementary Figure S11. Selectivity for ALK versus TRK using additional assays. Supplementary Figure S12. In vivo studies for ALK fusion with a wild-type kinase domain. Supplementary Figure S13. In vivo studies for ALK fusion with G1202R or I1171N single mutations. Supplementary Figure S14. In vivo studies for ALK fusion with G1202R compound mutations. Supplementary Figure S15. Pharmacodynamic analysis in G1202R compound mutation models. Supplementary Figure S16. Body weight and H&E staining for the YU-1077 intracranial study. Supplementary Figure S17. Estimating TRKB inhibition in humans and mice.
Data Availability Statement
No datasets were generated or analyzed in this study.