Fc-enhanced anti-CTLA-4 leverages novel FcγR-dependent mechanisms to potentiate antitumor immunity and overcomes limitations of conventional anti-CTLA-4, effectively treating poorly immunogenic and refractory cancers.
Abstract
Conventional immune checkpoint inhibitors (ICI) targeting cytotoxic T-lymphocyte antigen 4 (CTLA-4) elicit durable survival but primarily in patients with immune-inflamed tumors. Although the mechanisms underlying response to anti–CTLA-4 remain poorly understood, Fc-gamma receptor (FcγR) IIIA coengagement seems critical for activity, potentially explaining the modest clinical benefits of approved anti–CTLA-4 antibodies. We demonstrate that anti–CTLA-4 engineered for enhanced FcγR affinity leverages FcγR-dependent mechanisms to potentiate T-cell responsiveness, reduce intratumoral regulatory T cells, and enhance antigen-presenting cell activation. Fc-enhanced anti–CTLA-4 promoted superior efficacy in mouse models and remodeled innate and adaptive immunity versus conventional anti–CTLA-4. These findings extend to patients treated with botensilimab, an Fc-enhanced anti–CTLA-4 antibody, with clinical activity across multiple poorly immunogenic and ICI treatment–refractory cancers. Efficacy was independent of tumor neoantigen burden or FCGR3A genotype. However, FCGR2A and FCGR3A expression emerged as potential response biomarkers. These data highlight the therapeutic potential of Fc-enhanced anti–CTLA-4 antibodies in cancers unresponsive to conventional ICI therapy.
Significance:
This study reveals that Fc-enhanced anti–CTLA-4 harnesses novel mechanisms to overcome the limitations of conventional anti–CTLA-4, effectively treating poorly immunogenic and treatment-refractory cancers. Our findings support the development of a new class of immuno-oncology agents, capable of extending clinical benefit to patients with cancers resistant to current immunotherapies.
Introduction
Immune checkpoint inhibitors (ICI) blocking cytotoxic T-lymphocyte antigen 4 (CTLA-4), programmed cell death 1 (PD-1), and PD-L1 have revolutionized treatments of many cancers. However, responses to approved ICI agents occur in a minority of tumors, and ICI resistance remains a barrier to optimal clinical benefit. CTLA-4 was the first immune checkpoint described (1–3). It negatively regulates T-cell activation by modulating costimulatory signals within the immunologic synapse between antigen-presenting cells (APC) and T cells. During T-cell priming, CTLA-4 is upregulated on T cells to outcompete binding of costimulatory CD28 to CD80 and CD86, thereby impairing T-cell activation (4, 5). Furthermore, CTLA-4 is highly expressed on regulatory T cells (Treg) and modulates their suppressive functions (6).
In mouse tumor models, anti–CTLA-4 (αCTLA-4) boosts tumor-reactive T-cell activation and selectively eliminates CTLA-4–expressing intratumoral Tregs (7, 8) that impair antitumor immunity through mechanisms attributed to αCTLA-4 antibody interactions with Fcγ receptors (FcγR) on intratumoral effector cells. Consequently, the first approved αCTLA-4 antibody ipilimumab was developed with an unmodified IgG1 Fc region (9), as efficacy in mouse tumor models was partially dependent on Fc-mediated Treg depletion (8). However, this effect in humans remains controversial (10). In contrast to ipilimumab, tremelimumab, another approved αCTLA-4 antibody, was developed with an IgG2 Fc region to minimize effector functions (11, 12). This difference has further drawn into question the relevance of Treg depletion for the clinical activity of αCTLA-4, at least in patients with melanoma. We previously showed that αCTLA-4 enhances T-cell priming through an FcγR-dependent mechanism, independent of Treg depletion (13).
Clinically, the efficacy of antibodies dependent on Fc–FcγR coengagement can be shaped by differential binding affinities associated with FCGR2A (FcγRIIA) and FCGR3A (FcγRIIIA) polymorphisms (14). In the context of αCTLA-4 therapy, ipilimumab-treated patients with high neoantigen burden and the germline high-affinity FcγRIIIA allele [valine (V)158] variant exhibited a significant survival advantage over patients carrying the low-affinity FCGR3A allele [phenylalanine (F)158] variant (15). Furthermore, patient responses correlated with higher baseline circulating FcγRIIIA-expressing nonclassical monocytes (16) and intratumoral FcγRIIIA-expressing macrophages (17). Given the association between αCTLA-4 clinical response and FcγR coengagement, we developed botensilimab, an Fc-enhanced multifunctional αCTLA-4 antibody designed to improve FcγR-mediated effector functions. Here, we show that Fc-enhanced αCTLA-4 antibodies leverage novel FcγR-dependent mechanisms to remodel both innate and adaptive immunity and promote superior antitumor immunity compared with conventional αCTLA-4. These findings extend to humans treated with botensilimab, with clinical activity across multiple treatment-refractory cancers, including those that progressed on prior ICI therapy (18–20).
Results
Fc-Enhanced αCTLA-4 Remodels the Lymphoid and Myeloid Immune Compartments in the Tumor Microenvironment
Previous studies have demonstrated that murine αCTLA-4 activity depends on coengaging FcγR-dependent mechanisms to promote T-cell priming, Treg depletion, and myeloid activation (8, 13, 15, 21). We assessed whether an Fc-engineered murine IgG2b (mIgG2b) αCTLA-4 antibody, with enhanced FcγR binding (Supplementary Fig. S1A–S1F) through mutations at serine 239, alanine 330, and isoleucine 332 [S239D/A330L/I332E (DLE); αCTLA-4DLE; ref. 22], promotes superior eradication of established tumors and remodels the tumor microenvironment (TME) versus a conventional Fc-competent mIgG2b mouse αCTLA-4 antibody, both recognizing the same CTLA-4 epitope. An mIgG2b Fc region was selected for its functional comparability with human IgG1, based on the activating-to-inhibitory FcγR binding ratio (Supplementary Fig. S1G and S1H; Supplementary Tables S1 and S2), previously shown to predict effector activity in vivo (23). In contrast, the murine IgG2a (mIgG2a) Fc region demonstrated comparably stronger binding to murine FcγRs (Supplementary Fig. S1I and S1J) and a higher activating-to-inhibitory FcγR binding ratio (Supplementary Table S1) compared with the corresponding human IgG1 αCTLA-4 (Supplementary Table S2), making it less suitable to be used as a murine surrogate of a conventional IgG1 αCTLA-4 antibody.
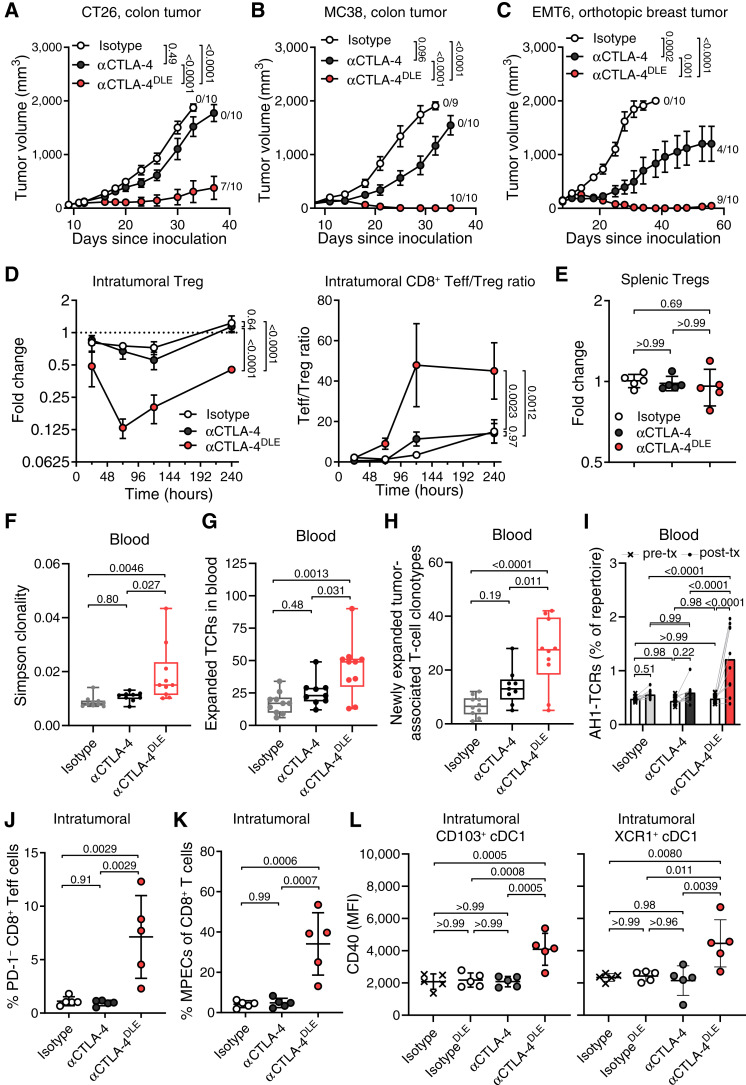
In CT26 and MC38 models of colorectal cancer exhibiting microsatellite stability (MSS) or microsatellite instability, respectively, and the EMT6 model of triple-negative breast cancer, αCTLA-4DLE promoted superior complete and long-term antitumor responses versus αCTLA-4 (Fig. 1A–C; Supplementary Fig. S2A–S2D). All mice experiencing complete CT26 regression resisted CT26 rechallenge regardless of antibody format (Supplementary Fig. S2E). Notably, αCTLA-4DLE efficacy was associated with significant intratumoral FOXP3+ Treg reduction up to 10 days posttreatment and increased the CD8/Treg ratio (Fig. 1D; Supplementary Fig. S3A) without affecting splenic Tregs (Fig. 1E; Supplementary Fig. S3B). CTLA-4 blockade with a conventional immunoglobulin format can remodel the intratumoral T-cell repertoire favorably but with limited antitumor activity (24, 25). In contrast, αCTLA-4DLE significantly increased peripheral T-cell receptor (TCR) clonality (Fig. 1F) and induced expansion of T cells in the periphery (Fig. 1G), and specifically tumor-associated T-cell clones (Fig. 1H), as determined by TCR sequencing of blood and tumor specimens. In addition, αCTLA-4DLE but not αCTLA-4 treatment induced a peripherally expanded and systemic antitumor T-cell response, as evidenced by an increased presence of AH1-specific (26) T-cell clones in blood after treatment (Fig. 1I). The improved antitumor immunity by αCTLA-4DLE correlated with increased intratumoral PD-1−CD8+ T effector (Teff) cells (Fig. 1J), Ki-67+CD8+ Teff cells (Supplementary Fig. S3C), granzyme B+ CD8+ Teff cells (Supplementary Fig. S3D), and CD62L−PD−1−Slamf7+CX3CR1−CD8+ memory precursor effector cell (MPEC) prevalence (Fig. 1K; Supplementary Fig. S3E). Multiplex immunofluorescence further showed augmented CD3+, CD4+, and CD8+ T-cell prevalence 9 days after αCTLA-4DLE versus αCTLA-4 and controls (Supplementary Fig. S3F).
Figure 1.
αCTLA-4DLE promotes superior antitumor immunity in tumor-bearing mouse models. Tumor growth in BALB/C mice bearing (A) subcutaneous CT26 tumors (n = 10 mice/group), (B) C57BL/6 mice bearing MC38 tumors (n = 9–10 mice/group), and (C) orthotopic EMT6 breast tumors (n = 10 mice/group) treated intraperitoneally with indicated antibodies. Fractions to the right of curves in A–C are CR over total tumor number. D, FOXP3+ Tregs relative to vehicle (dashed line) and tumor CD8+ Teff to Treg ratio, and (E) fold-change splenic FOXP3+ Tregs relative to isotype 72 hours posttreatment (n = 5 mice/time point) in CT26-bearing BALB/C mice treated with indicated antibodies. F, Changes in blood TCR Simpson clonality index, (G) expanded TCR clones in blood from pre- to posttreatment, (H) number of newly expanded tumor-associated T-cell clonotypes, and (I) total frequency of tumor-specific AH1 TCR clones in blood pretreatment (pre-tx) vs. posttreatment (post-tx) in CT26-bearing BALB/C mice (n = 9–10 mice/group) treated with indicated antibodies on days 0, 3, and 6. Pretreatment Blood samples and blood and tumor collected 11 days after initial dose. J, Percent intratumoral CD8+ Teff and (K) CD8+ MPECs by flow cytometry 10 days posttreatment in CT26-bearing BALB/C mice (n = 5 mice/group). L, CD40 mean fluorescence intensity (MFI) of intratumoral CD103+ or XCR1+ type 1 cDC1 in CT26-bearing BALB/C mice (n = 5/group) treated once with indicated antibodies. Tumors evaluated by flow cytometry 7 days posttreatment. Data represented as mean ± SEM (A–D) and mean ± SD (E and J–L). In box plots, center line, median; box limits, 25th and 75th percentile; whiskers, minimum and maximum values. Data analyzed by mixed-effects models with matched data (A–C) or two-way ANOVA (D and I) followed by the Tukey multiple comparisons test or one-way ANOVA followed by the Kruskal–Wallis test with Dunn correction (E–H and J–L).
We further confirmed our findings in mice challenged with staphylococcal enterotoxin B (SEB), eliciting a SEB-specific Vβ8+ TCR T-cell response (27). Consistent with tumor challenge data, αCTLA-4DLE increased Vβ8+ CD8+, CD8+ Teff, MPEC, and Ki-67+ and granzyme B+ CD8+ T-cell prevalence (Supplementary Fig. S4A–S4E) and reduced Tregs (Supplementary Fig. S4F), whereas αCTLA-4 induced no such changes. The expansion of T cells induced by αCTLA-4DLE was restricted to the Vβ8+ TCR repertoire of effector T cells, with no significant change in the nonspecific Vβ2+ T cells (Supplementary Fig. S4A–S4E). These data are consistent with our prior work demonstrating FcγR coengagement dependence for αCTLA-4–induced expansion and effector function of antigen-specific T cells (13).
Notably, the immune remodeling induced by αCTLA-4DLE was not limited to T cells. In CT26 tumor–bearing mice treated with αCTLA-4DLE, intratumoral CD103+ and XCR1+ type 1 conventional dendritic cells (cDC1) were also activated by αCTLA-4DLE but not αCTLA-4 (Fig. 1L). Upregulation of CD40 on cDC1 by αCTLA-4DLE (Fig. 1L) is consistent with improved T-cell priming, which is in line with recent work demonstrating that coengagement of immunoreceptor tyrosine-based activation motif–containing FcγRs by αCTLA-4 and subsequent downstream activation drives myeloid cells toward an activated, proinflammatory state (21).
Superior antitumor efficacy from αCTLA-4DLE versus αCTLA-4 was independent of bioavailability (Supplementary Fig. S5A and S5B). Although αCTLA-4 showed slightly higher maximum concentration exposure and half-life (Supplementary Fig. S5C), these attributes did not improve activity. Together, these results suggest that αCTLA-4DLE leverages novel FcγR-dependent mechanisms to induce antitumor immunity that correlates with improved cross-priming cDC1 activation, antigen-experienced CD8+ effector T-cell expansion, and reduced intratumoral Tregs.
Fc-Enhanced αCTLA-4 Improves Efficacy of Distinct Treatments
In a poorly immunogenic and aggressive syngeneic LSL-KrasG12D;Trp53R172H;Pdx-Cre (KPC) pancreatic tumor model comprised of both epithelial cancer cells and cancer-associated fibroblasts that mimics advanced refractory human pancreatic ductal adenocarcinoma (28), αCTLA-4DLE monotherapy performed comparably with or better than chemotherapy (clinically relevant gemcitabine, nab-paclitaxel, and cisplatin) and controlled 7 of 10 tumors when combined with chemotherapy (Supplementary Fig. S6A).
In immunotherapy-resistant B16F1.OVA melanoma cells, the combination of αCTLA-4DLE with αPD-1 [approved in melanoma (29)] and ovalbumin/TRP2 vaccine formulated with QS-21 and cytosine phosphoguanine adjuvants to improve antigen-specific T-cell priming (30, 31) significantly improved overall survival (OS) versus either therapy alone (Supplementary Fig. S6B). Furthermore, αCTLA-4DLE and αPD-1 combined with OT-1 transgenic CD8+ T cells recognizing ovalbumin significantly improved OS versus individual treatments (Supplementary Fig. S6C). Together, these studies highlight the broad potential of Fc-enhanced αCTLA-4 to improve antitumor immunity in poorly immunogenic and treatment-resistant tumors through diverse combinatorial strategies.
Botensilimab Is Fc-Engineered for Enhanced FcγR Coengagement and Signaling
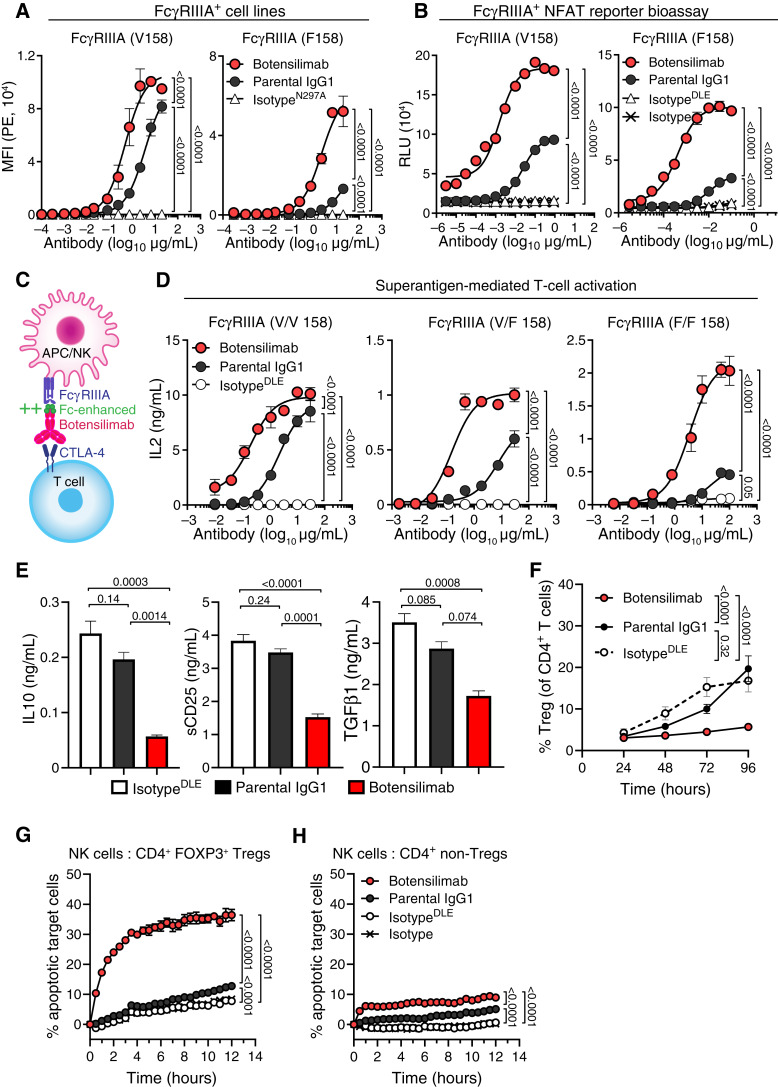
Conventional αCTLA-4 antibodies like ipilimumab were developed with an IgG1 Fc region, as antitumor activity in mouse models was partially dependent on Fc-mediated mechanisms (8, 32). However, these mechanisms have not been definitively shown in humans. To determine if αCTLA-4DLE–enhanced immune activity translated to humans, we designed botensilimab, a fully human IgG1 αCTLA-4 engineered with the same DLE mutations as mouse αCTLA-4DLE to improve FcγR-dependent functions versus wild-type (WT) human IgG1 αCTLA-4 (parental IgG1). Notably, Fc mutations of botensilimab did not affect its binding affinity to CTLA-4 nor its blockade of CTLA-4–ligand interactions (Supplementary Fig. S7). Botensilimab, the corresponding IgG1-WT, and ipilimumab demonstrated comparably high binding affinity with recombinant human CTLA-4-His and CTLA-4-Fc proteins (Supplementary Fig. S7A–S7C). Botensilimab did not bind to related CD28 family members, including B- and T-lymphocyte attenuator (BTLA), inducible T-cell costimulator (ICOS), CD28, or PD-1 (Supplementary Fig. S7D), indicative of expected CTLA-4 selectivity. Furthermore, botensilimab and IgG1-WT αCTLA-4 demonstrated comparable and complete CTLA-4 ligand blockade (Supplementary Fig. S7E) and CD28 pathway activation (Supplementary Fig. S7F and S7G). Although botensilimab, ipilimumab, and tremelimumab are distinct mAbs, they bind to similar epitopes on CTLA-4 (Supplementary Table S3), which significantly overlap with the binding site for the natural B7 ligands (33), consistent with these antibodies being complete blockers of CTLA-4–B7 ligand interactions (34–36).
FcγRIIIA is critical for IgG1-WT αCTLA-4 activity, including ipilimumab (13, 15). Consistent with IgG antibody Fc-binding properties (15, 37), botensilimab demonstrated superior binding to cells expressing human FcγRIIIA versus IgG1-WT αCTLA-4 (Fig. 2A). Remarkably, botensilimab demonstrated superior binding to both high- (V158) and low-affinity (F158) FcγRIIIA variants (Supplementary Fig. S8A) compared with IgG1-WT αCTLA-4 (Fig. 2A), ipilimumab (Supplementary Fig. S8B), or tremelimumab (Supplementary Fig. S8C). The enhanced FcγRIIIA binding by botensilimab correlated with potent and superior FcγRIIIA signaling in high- (V158) and low-affinity (F158) FcγRIIIA variants versus IgG1-WT αCTLA-4 (Fig. 2B) or ipilimumab (Supplementary Fig. S8D and S8E). These data differentiate botensilimab versus first-generation αCTLA-4 in engaging and eliciting FcγRIIIA signaling, even in cells expressing low-affinity FcγRIIIA, suggesting that botensilimab could benefit a broader patient population versus conventional αCTLA-4 therapy. Additionally, both botensilimab and IgG1-WT αCTLA-4 showed potent FcγRIIA signaling, but botensilimab generated greater maximal signals (Supplementary Fig. S8F and S8G). Botensilimab also exhibited improved binding to cell-expressed human FcγRIIB (Supplementary Fig. S8H) and FcγRIIA R/R 131 (Supplementary Fig. S8I) versus IgG1-WT αCTLA-4, whereas binding to FcγRIIA H/H 131 (Supplementary Fig. S8J) and FcγRI (Supplementary Fig. S8K) was comparable.
Figure 2.
Botensilimab enhances T-cell responsiveness independent of FcγRIIIA allele status and reduces Treg frequency. A, Binding of botensilimab, IgG1 variant of botensilimab (parental IgG1), or aglycosylated IgG1N297A isotype negative control antibody to CHO cells expressing FcγRIIIA V158 or F158 by flow cytometry. MFI, mean fluorescence intensity. B, Signaling through FcγRIIIA V158 and FcγRIIIA F158 in Jurkat cells expressing FcγRs upstream of an NFAT-dependent luciferase reporter, cocultured with CTLA-4–expressing cells and botensilimab, parental IgG1, IgG1DLE isotype, or IgG1 isotype. Luciferase expression shown as relative light units (RLU). C, Schematic depicting botensilimab binding to CTLA-4–expressing T cells and coengaging FcγRIIIA-expressing APC or (NK cells to create an immune synapse). D, IL2 secretion from SEA-stimulated healthy donor PBMC from FcγRIIIA homozygote V/V 158, FcγRIIIA heterozygote V/F 158, and FcγRIIIA homozygote F/F 158 donors treated with botensilimab, parental IgG1, or IgG1DLE isotype. E, IL10, soluble CD25 (sCD25), and TGFβ1 secretion from SEA-stimulated PBMC treated with 5 μg/mL botensilimab, parental IgG1, or IgG1DLE. FcγRIIIA heterozygote V/F 158 donor shown. F, Treg (CD3+CD4+CD25+FOXP3+) frequencies from SEA-stimulated healthy donor PBMCs treated with 5 μg/mL of botensilimab, parental IgG1, or IgG1DLE by flow cytometry (n = 4 donors). G, Dye-labeled and caspase 3/7–stained primary CD4+FOXP3+ Tregs or (H) conventional CD4+ T cells cocultured at a 1:1 ratio with FcγRIIIA-expressing NK-92 cells treated with indicated antibodies. Values measured by live imaging using confocal microscopy (n = 3). Data are represented as mean ± SEM (A, B, and D–G). Data analyzed by two-way ANOVA (A, B, D, and F–H) or one-way ANOVA (E), all followed by the Tukey multiple comparisons test.
Despite enhanced Fc-effector function, botensilimab demonstrated significantly reduced binding to C1q, the recognition molecule of the classical complement system, versus IgG1-WT αCTLA-4 (Supplementary Fig. S8L). Consequently, botensilimab did not promote complement-dependent cytotoxicity against CTLA-4–expressing cells, unlike IgG1-WT αCTLA-4 (Supplementary Fig. S8M). Lack of botensilimab complement binding and complement-dependent cytotoxicity is consistent with the Fc A330L point mutation that abrogates C1q binding (22), suggesting that botensilimab could avoid some immune-related adverse events (AE) such as complement-mediated hypophysitis associated with conventional αCTLA-4, including ipilimumab (38).
Botensilimab Enhances FcγR-Dependent T-cell Response
We previously established that FcγRIIIA coengagement on APCs by αCTLA-4 improves the T-cell−APC immune synapse, further enhancing TCR signaling strength and duration (13). To model the T-cell−APC immune synapse (Fig. 2C), human peripheral blood mononuclear cell (PBMC) cultures were stimulated with staphylococcal enterotoxin A (SEA) superantigen to cross-link APC MHC class II and a specific Vβ subset of TCRs to promote CD28 and CD86 coengagement within the immune synapse (39). Here, botensilimab was superior to IgG1-WT αCTLA-4 in inducing IL2 secretion (Fig. 2D). Notably, botensilimab enhanced T-cell responsiveness independent of FcγRIIIA genotype and induced robust IL2 secretion using APCs expressing only low-affinity FcγRIIIA, whereas IgG1-WT αCTLA-4 induced only weak responses, consistent with its lower FcγRIIIA binding affinity. Moreover, botensilimab, but not IgG1-WT αCTLA-4, significantly reduced immune-suppressive IL10, soluble CD25, and TGFβ1 from SEA-stimulated PBMCs (Fig. 2E). Together, these data further support that FcγR coengagement by αCTLA-4 antibodies plays a critical role in driving T-cell responsiveness and suggest that αCTLA-4 with enhanced FcγR binding can improve therapeutic efficacy.
Botensilimab Reduces the Frequency of Tregs
Although αCTLA-4 can deplete intratumoral Tregs (15, 32) and reduce Treg stability (40) in mouse models, the impact of ipilimumab on human Tregs in the clinic remains unsettled (10). In SEA-stimulated human PBMCs, botensilimab significantly reduced CD3+CD4+CD25+FOXP3+ Treg expansion (Fig. 2F) and maintained significantly higher conventional CD3+CD4+CD25−FOXP3− cell prevalence compared with IgG1-WT αCTLA-4 and isotype control (Supplementary Fig. S9A). Interestingly, the reduced prevalence of Tregs in botensilimab-treated, SEA-stimulated cultures was not attributed to depletion. Cultures treated with botensilimab 96 hours after SEA stimulation did not show a reduction in Tregs (Supplementary Fig. S9B), despite the presence of FcγRIIIA (CD16+)-expressing NK cells (Supplementary Fig. S9C) and high CTLA-4 expression in Tregs versus CD4+ non-Tregs or CD8+ T cells in culture (Supplementary Fig. S9D–S9F). This suggests that the presence of effector cells capable of depletion is insufficient to reduce Treg numbers and/or CTLA-4 is inadequately expressed on the cell surface on Tregs in this assay. Instead, the reduced Treg expansion is consistent with reduced TGFβ1 secretion in botensilimab-treated PBMC cultures.
To assess antibody-dependent cellular cytotoxicity, we cocultured primary human CD4+FOXP3+ Tregs or conventional CD4+ T cells with FcγRIIIA (V158)-expressing NK-92 cells and observed that botensilimab killed Tregs significantly better than IgG1-WT αCTLA-4 (Fig. 2G), consistent with improved intratumoral CD8+ effector T-cell/Treg ratios observed in αCTLA-4DLE–treated mice (Fig. 1D; Supplementary Fig. S3B). In contrast, botensilimab and IgG1-WT αCTLA-4 minimally affected conventional CD4+FOXP3− T cells (Fig. 2H), consistent with their lower cell surface CTLA-4 expression (41).
Botensilimab Activates Myeloid Cells
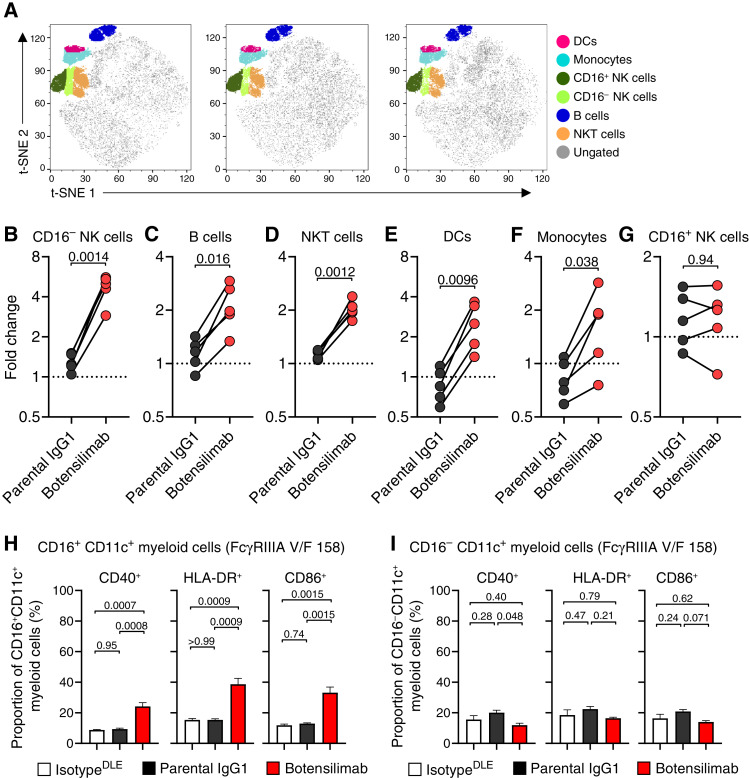
In superantigen-stimulated human PBMCs treated with botensilimab, we further analyzed the non–T-cell fraction (Fig. 3A; Supplementary Fig. S10A and S10B) and observed a significant increase in the frequency of CD16− NK cells (Fig. 3B), B cells (Fig. 3C), NK T cells (NKT; Fig. 3D), dendritic cells (DC; Fig. 3E), and monocytes (Fig. 3F). Interestingly, CD16+ NK cells did not change (Fig. 3G), suggesting that botensilimab preferentially expanded cytokine-producing regulatory NK cells that could affect T-cell polarization (42) rather than cytotoxic CD16+ NK effector cells.
Figure 3.
Botensilimab enhances the frequency of activated myeloid, NK, B, and NKT cells. A, Global t-distributed stochastic neighbor embedding (t-SNE) maps of total live immune cells (CD45+) after stimulation with SEA in the presence of 5 μg/mL botensilimab, parental IgG1, or IgG1DLE isotype. PBMC from five FcγRIIIA V/F 158 heterozygote healthy donors were used. Six phenotypically distinct clusters identified by individual phenotypic markers by flow cytometry within concatenated total live immune cells analyzed. Log2 fold changes in (B) CD16− NK, (C) B, (D) NKT, (E) DC, (F) monocyte, and (G) CD16+ NK cell counts in each immune cell cluster between samples treated with botensilimab and parental IgG1, compared with IgG1DLE isotype (n = 5/group; data are paired). H, Activated CD16+CD11c+ (I) and CD16−CD11c+ myeloid cell frequency determined by CD40, HLA-DR, and CD86 expression by flow cytometry. Representative data from an FcγRIIIA V/F 158 heterozygote donor. Data are represented as mean ± SEM (H and I). Data analyzed with a two-tailed paired t test (B–G) or one-way ANOVA followed by a Tukey multiple comparisons test (H and I).
Prior studies in tumor-bearing mouse models demonstrated that FcγR coengagement by αCTLA-4 can promote myeloid activation and type I IFN signaling (21), similar to αCTLA-4DLE enhancing intratumoral DC activation (Fig. 1L). However, translation to humans has not been previously established. Botensilimab significantly increased activated CD40− and human leukocyte antigen (HLA)-DR-expressing and CD86-expressing CD16+CD11c+ myeloid cells (Fig. 3H), consistent with its improved FcγRIIIA binding and signal induction (Fig. 2A and B). Notably, botensilimab-induced myeloid activation was independent of FcγRIIIA allele status, as similar increases in activated CD16+CD11c+ myeloid cell frequency were observed in donors homozygous for the low-affinity FcγRIIIA (F/F 158; Supplementary Fig. S10C). Although conventional IgG1 αCTLA-4 (Fig. 2A and B) and ipilimumab (Supplementary Fig. S8F, S8H, and S8I) engage FcγRIIIA, we did not observe significantly increased CD16+CD11c+ myeloid cell activation in donors expressing high-affinity (V/F 158; Fig. 3H) or low-affinity (F/F 158; Supplementary Fig. S10C) FcγRIIIA after treatment with conventional IgG1 αCTLA-4. Notably, the increased frequency of activated CD11c+ myeloid cells by botensilimab was specific to the CD16+ subset, as no significant changes were observed in the activated CD16−CD11c+ myeloid cell subset (Fig. 3I; Supplementary Fig. S10D). Together, these data suggest that enhanced FcγRIIIA signaling by botensilimab is important for potentiating T-cell and myeloid-cell activation, consistent with our previous reports that blockade of FcγRIIIA, but not other FcγRs, impairs T-cell responsiveness by αCTLA-4 (13).
Pharmacokinetics and Toxicity of Botensilimab in Cynomolgus Monkeys
To inform dose selection and safety monitoring strategies for clinical trials, the pharmacokinetics (PK), immunogenicity, and toxicology profiles of botensilimab were evaluated in nonhuman primates. Groups of male (n = 16) and female (n = 16) cynomolgus monkeys were administered weekly intravenous botensilimab at 5, 25, or 75 mg/kg/dose for up to 13 weeks followed by a 4-week recovery period to assess reversibility, persistence, or delayed occurrence of toxic effects. There were no botensilimab-related changes observed in food consumption, ophthalmic examinations, ECG, blood pressure, heart rate, respiration, neurologic examinations, hematology, coagulation, urinalyses, gross evaluation, or organ weight. Mortality occurred in one male at 75 mg/kg/dose that was euthanized because of poor clinical signs. Based on anatomic pathology findings, morbidity was attributed primarily to acute systemic inflammation and organ damage, characterized by perivascular/vascular infiltrates and neutrophilic infiltration in multiple organs, including the aorta, prostate, liver, and spleen. A high titer of antidrug antibodies was also observed. Botensilimab-related decreases in body weight (up to 11.6% and 21.3% for mid- and high-dose groups, respectively) were observed in males and females at ≥25 mg/kg/dose versus control. All other animals survived until necropsy. Histopathologic findings were observed in multiple organs in a dose-dependent manner (Supplementary Table S4). Botensilimab-related microscopic changes were observed in the blood vessels lining the periphery or within the parenchyma of numerous organs that were considered adverse but were reversible.
A noncompartmental analysis showed dose-proportional PK. Systemic exposure (AUC0–last and Cmax) to botensilimab increased dose-proportionally from 5 to 75 mg/kg in both sexes on days 1 and 85. After single (day 1) or repeated (day 85) intravenous administration of botensilimab to male and female monkeys, the median time to reach maximum concentration (Tmax) values for botensilimab was between 0.3 and 1.2 hours after the start of injection. The median half-life (T1/2) values for botensilimab ranged from 100.3 to 214.7 hours. Antidrug antibodies (ADA) were detected in a subset of animals, and the concentration–time points believed to be impacted by ADA were excluded from the noncompartmental assessment. Serum PK parameter estimates are shown in Supplementary Table S5. Under the conditions of this study, the highest nonseverely toxic dose was 25 mg/kg weekly, which is at least 48 times greater than the extrapolated human equivalent dosage of 1 mg/kg every 6 weeks reported to demonstrate antitumor activity in humans (43).
Botensilimab Effectively Treats Multiple Human Cancers
We have previously reported clinical results from an ongoing expanded phase I trial (NCT03860272) evaluating the safety and efficacy of botensilimab alone or combined with balstilimab [αPD-1 (functionally comparable to nivolumab); refs. 43, 44]. The results described herein focus on a subset of 211 patients with advanced solid cancers from this study with available biomarker data treated at any dose and schedule including botensilimab monotherapy and in combination with balstilimab, enrolled between April 1, 2019, and March 27, 2023 (Supplementary Fig. S11; baseline demographics and disease characteristics, Supplementary Table S6). The median patient age was 59 years (range, 19–83 years), with 57% female and 43% male. Seventy-eight (37%) patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 0, and 133 (63%) patients had a performance status of 1. The population was heavily pretreated with a median of 3 (range, 1–16) prior lines of therapy. Of the 211 patients, 63 (29%) had received prior PD-(L)1 inhibitors, 7 of whom also received prior CTLA-4 inhibition. In the dose-escalation portion of the study (3 + 3 design), patients with advanced solid tumors were treated with botensilimab monotherapy at dose levels starting at 0.1 mg/kg and escalating up to 3 mg/kg, administered intravenously every 3 weeks or every 6 weeks, or in combination with balstilimab 3 mg/kg administered intravenously every 2 weeks, for up to 2 years as previously described by Bullock and colleagues (43). Together with clinical efficacy and safety data, the optimal efficacious dosage of botensilimab when combined with balstilimab was determined to be 1 to 2 mg/kg every 6 weeks (43).
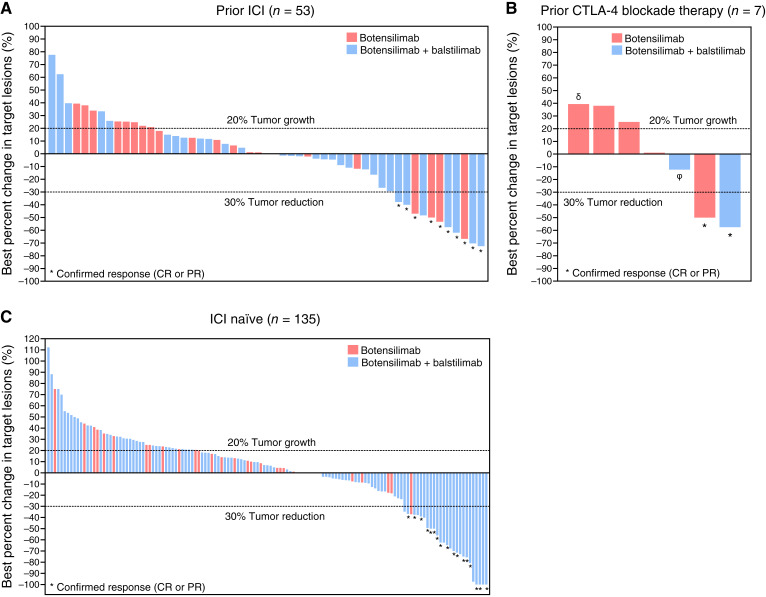
Botensilimab alone or combined with balstilimab elicited clinical activity across multiple cancer types, including immunotherapy-resistant and refractory tumors (18–20), such as metastatic MSS colorectal cancer (43), recurrent platinum-refractory/resistant ovarian carcinoma, αPD-(L)1–relapsed/refractory non–small cell lung cancer (18), various sarcomas (18), and ICI-relapsed/refractory melanoma including patients relapsed/refractory to conventional CTLA-4 inhibition (Supplementary Table S7; Supplementary Table S8; Fig. 4). Confirmed objective responses were observed in 28 of the 211 patients all with solid tumor treated at any dose level or regimen with available biomarker data [13.3%; 95% confidence interval (CI), 9.0–18.6], including two complete responses (CR). Additionally, 81 patients (38.4%) experienced stable disease (SD; ≥6 weeks), resulting in a disease control rate (DCR), defined as a best response of SD or better of 51.7% (95% CI, 44.7–58.6) for at least 6 weeks, 38.9% (95% CI, 32.2–45.8) for 12 weeks or more, and 19.9% (95% CI, 14.7–25.9) for 24 weeks or more (Supplementary Table S7). The median duration of response was not reached.
Figure 4.
Clinical response to botensilimab monotherapy and combination with balstilimab in patients with advanced solid cancers. Waterfall plot of maximal percentage change from baseline in sum of tumor target lesion diameters for patients treated with botensilimab monotherapy or in combination with balstilimab (A) who progressed on prior ICI [αPD-(L)1 and/or αCTLA-4] therapy (n = 53), (B) received prior αCTLA-4 therapy [ipilimumab, QL1706 (a mixture of αPD-1 IgG4 and αCTLA-4 IgG1; δ) or ALPN-202 (a dual PD-L1/CTLA-4 blocker and CD28 costimulator; ϕ; n = 7], or (C) received no prior ICI therapy (n = 135). Only patients treated with 1, 2, 3 mg/kg, or 150 mg of botensilimab monotherapy (red bar) or in combination with 3 mg/kg or 450 mg balstilimab (blue bar) are shown. Tumor reduction was assessed according to RECIST 1.1 criteria. Lower dotted line demarcates tumor reduction of 30%. * Indicates confirmed responses as of March 27, 2023.
Remarkably, responses were also observed in patients who had progressed on prior αPD-(L)1 or conventional αCTLA-4 therapies (Supplementary Table S7; Fig. 4A and B), with confirmed objective responses in 10 of 63 patients with available biomarker data (15.9%; 95% CI, 7.9–27.3), including one CR. Additionally, 28 patients (44.4%) had SD, resulting in a 60.3% DCR (95% CI, 47.2–72.4) at 6 weeks or more, 42.9% DCR (95% CI, 30.5–56.0) at 12 weeks or more, and 27.0% DCR (95% CI, 16.6–39.7) at 24 weeks or more (Supplementary Table S7). Notably, among the seven patients who received prior αCTLA-4 therapy, five had metastatic melanoma and were previously treated with ipilimumab. Of these five patients, two demonstrated a confirmed partial response (PR) and one achieved SD to botensilimab or botensilimab in combination with balstilimab (Fig. 4B). Similarly, in ICI-naïve patients, 18 of 148 patients (12.2%; 95% CI, 7.4–18.5) showed confirmed objective responses and a DCR of 37.2% (95% CI, 29.4–45.5) for 12 weeks or more (Supplementary Table S7; Fig. 4C). The combination of botensilimab and balstilimab elicited deeper responses and DCR versus botensilimab monotherapy (Supplementary Table S7). This is particularly noteworthy, as responses occurred in patients refractory to prior αPD-(L)1 and in tumor types that historically exhibited limited responses to conventional ICI therapy (Supplementary Table S8). Consistent with our preclinical data demonstrating αCTLA-4DLE efficacy in immunotherapy-resistant tumors, these data suggest that botensilimab modifies the molecular characteristics of the TME of these poorly immunogenic and ICI-refractory tumors to enhance responsiveness to subsequent T cell–targeting immunotherapies.
Botensilimab Avoids Complement-Mediated Toxicity
The clinical safety profile of botensilimab, both as monotherapy and in combination with balstilimab, has been reported previously (43). A summary of grade 3 or higher treatment-related AEs is summarized in Supplementary Table S9. The most common grade 3 or greater treatment-related AE was “diarrhea/colitis,” which comprised the preferred terms diarrhea, colitis, and enteritis. While acknowledging the limitations of cross-trial comparisons, the safety profile is generally consistent with other approved αCTLA-4 therapies, depending on dose and schedule (9, 45). However, a notable exception is the absence of hypophysitis in patients treated with botensilimab monotherapy (Supplementary Table S9), which is an immune-related AE sometimes associated with ipilimumab (46) and thought to be partially mediated by complement activation (38). This suggests that botensilimab avoids some of the limitations associated with conventional αCTLA-4 therapy, consistent with the A330L point mutation in botensilimab that precludes complement binding. Interestingly, only one patient treated with the combination of botensilimab and balstilimab experienced hypophysitis, which may be attributed to αPD-1 therapy (47). However, continued monitoring of hypophysitis incidence in larger patient cohorts treated with botensilimab is warranted to confirm these findings.
Botensilimab Enhances Peripheral T-cell Activation
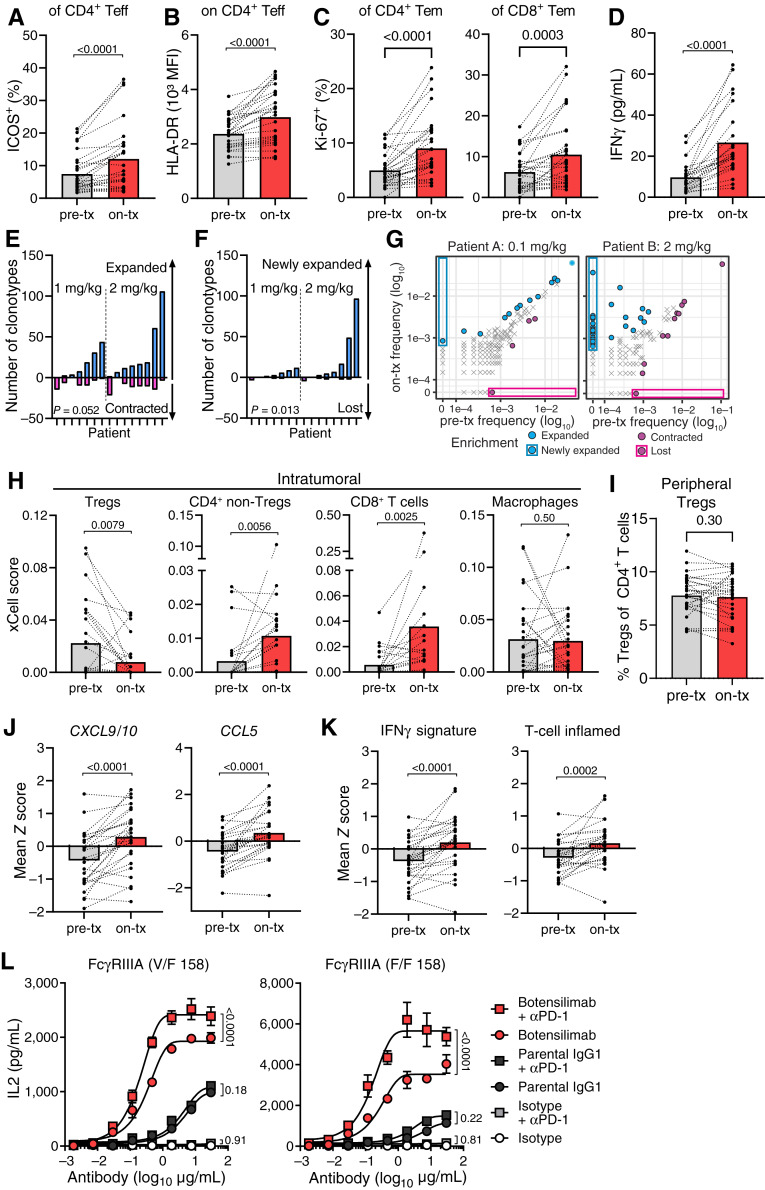
ICOS, HLA-DR, and IFNγ with increased CD4+ T-cell expression serve as pharmacodynamic markers of ipilimumab activity (48, 49). However, clinical responses remain limited to inflamed tumors like melanoma (50). To assess the pharmacodynamic effects of botensilimab, we performed flow cytometry analysis of peripheral blood (Supplementary Fig. S12), which revealed markers of T-cell activation following treatment. Although total CD4+ or CD8+ peripheral T cells remained unchanged with botensilimab monotherapy (Supplementary Fig. S13A), we observed significant dose-dependent increases in peripheral ICOS+ T-cell frequency, including conventional CD4+ (Supplementary Fig. S13B and S13C), CD4+ effector (Fig. 5A), CD4+ central memory, CD4+ effector memory, and CD45RA+ effector memory (Supplementary Fig. S13D) T cells 7 days posttreatment. Botensilimab also increased frequency and mean fluorescence intensity of HLA-DR–expressing CD4+ T cells, including central and effector memory T cells (Supplementary Fig. S13E–S13G), and effector T cells (Fig. 5B) and frequency of Ki-67+ CD4+ and CD8+ T-cell subsets (Fig. 5C; Supplementary Fig. S13H and S13I). Additionally, botensilimab increased PD-1+ CD4+ effector T-cell frequency (Supplementary Fig. S13J). Consistent with peripheral T-cell activation, botensilimab dose-dependently increased serum IFNγ (Fig. 5D; Supplementary Fig. S13K and S13L). Furthermore, given that active reshaping of the peripheral TCR repertoire is a key pharmacodynamic αCTLA-4 effect, associated with clinical efficacy (24, 51), we sequenced the CDR3 TCRβ chains in T cells from patient pretreatment blood and 3 to 4 weeks after botensilimab treatment. Consistent with enhanced TCR clonality and de novo expansion in tumor-bearing mouse models and peripheral T-cell activation in patients, botensilimab increased existing (Fig. 5E) and newly expanded T-cell clonotypes (Fig. 5F and G) in some patients; however, the small number or abundance of the newly expanded clones did not significantly impact the overall clonality (Supplementary Fig. S13M).
Figure 5.
Botensilimab enhances activated T-cell prevalence, reduces intratumoral Tregs, and upregulates genes associated with T cell–inflamed tumors in patients with advanced solid cancers. A–G, Pretreatment (pre-tx) and on-treatment (on-tx) blood samples from patients treated with 1 or 2 mg/kg botensilimab monotherapy. PBMC analyzed by flow cytometry for the (A) frequency of ICOS+ and (B) HLA-DR mean fluorescence intensity (MFI) on CD4+ Teff (CXCR3+) and (C) frequency of Ki-67+ CD4 and CD8 effector memory (Tem, CD45RO+CCR7−) T-cell subsets (n = 28; on-tx: 7 days after first dose). D, Plasma IFNγ in pre- and on-tx samples (n = 23; on-tx: 24 hours after first dose). Number of (E) expanded vs. contracted and (F) newly expanded vs. lost T-cell clonotypes in pre-tx vs. on-tx blood by differential abundance analysis between baseline (pre-tx; cycle 1 day 1) and 3–4 weeks postdose (n = 15; pre-tx: cycle 1 day 1; on-tx: 3–4 weeks after first dose). P values compare expanded vs. contracted T-cell clonotypes in E and F. G, T-cell clonotype abundance in two representative patients treated with 0.1 or 2 mg/kg botensilimab every 3 weeks. CDR3 sequencing of human TCRβ chains performed using immunoSEQ. H, Intratumoral cell type enrichment scores calculated using xCell for Tregs, CD4+ non-Tregs, CD8+ T cells, and macrophages as determined from RNA-seq of pre-tx and on-tx tumor biopsies from patients treated with 1 or 2 mg/kg botensilimab monotherapy every 3 or 6 weeks ± balstilimab every 2 weeks (n = 26; on-tx: cycle 2 day 1 for every 6-week cohort, or cycle 3 day 1 for every 3-week cohort). I, Percent peripheral Treg (CD4+, CD127low/−, CD25+) subsets (n = 28; on-tx: 7 days after first dose) analyzed by flow cytometry from patients treated with 1 or 2 mg/kg botensilimab monotherapy. J, Intratumoral CXCL9 and CXCL10 and CCL5 gene expression and (K) IFNγ and T cell–inflamed gene expression signatures (53) as determined from RNA-seq of pre-tx and on-tx tumor biopsies (n = 26). L, IL2 secretion from SEA-stimulated PBMC from FcγRIIIA heterozygote V/F 158 and FcγRIIIA homozygote F/F 158 donors treated with botensilimab, parental IgG1, or IgG1DLE isotype, alone ± αPD-1 (balstilimab). Paired data points with group mean (A–D and H–K) or mean ± SEM (L). Data analyzed with the two-tailed Wilcoxon matched-paired t test (A–F and H–K) or two-way ANOVA with the Tukey multiple comparisons test (L).
Botensilimab Remodels the TME of Refractory Tumors
We further assessed the ability of botensilimab monotherapy or combination with balstilimab to remodel the TME favorably in patients with poorly immunogenic and ICI-refractory tumors. Consistent with enhanced Fc-effector function, cell type enrichment analysis of bulk RNA sequencing (RNA-seq) from botensilimab-treated patients showed significant intratumoral Treg reduction (Fig. 5H) with a corresponding increase in conventional CD4+ and CD8+ T cells but no change in intratumoral macrophages. In line with these observations, chromogenic IHC of pretreatment and on-treatment tumor biopsies (Supplementary Fig. S14A–S14D) after botensilimab monotherapy or botensilimab plus balstilimab showed reduced FOXP3+ intratumoral Tregs with corresponding increased CD8+ T cells (Supplementary Fig. S14E–S14G). However, in contrast to that previously reported for ipilimumab (52), botensilimab did not impact the frequency of peripheral Tregs (Fig. 5I). These findings are consistent with αCTLA-4DLE–treated mouse tumor models (Fig. 1D, J, and K; Supplementary Fig. S3) and further differentiate the pharmacodynamic effects of botensilimab from those of ipilimumab (10).
Notably, the increased CD8+ T cells and reduced intratumoral Tregs correlated with significantly increased chemokine gene expression (CXCL9, CXCL10, and CCL5; Fig. 5J) and increased IFNG and T-cell inflamed signatures (Fig. 5K) associated with αPD-1 (pembrolizumab) response in patients with melanoma and head and neck squamous cell carcinoma (53). In superantigen-stimulation assays using PBMCs poorly responsive to αPD-1, we found that botensilimab alone strongly potentiated T-cell activation and, more notably, showed superior combination activity with αPD-1 versus IgG1-WT αCTLA-4 (Fig. 5L). Together, these data underscore that botensilimab enhances immune cell signatures and favorably remodels the TME in tumors refractory to conventional immunotherapy.
Botensilimab Efficacy Is Independent of CD16V158F and Neoantigen Burden
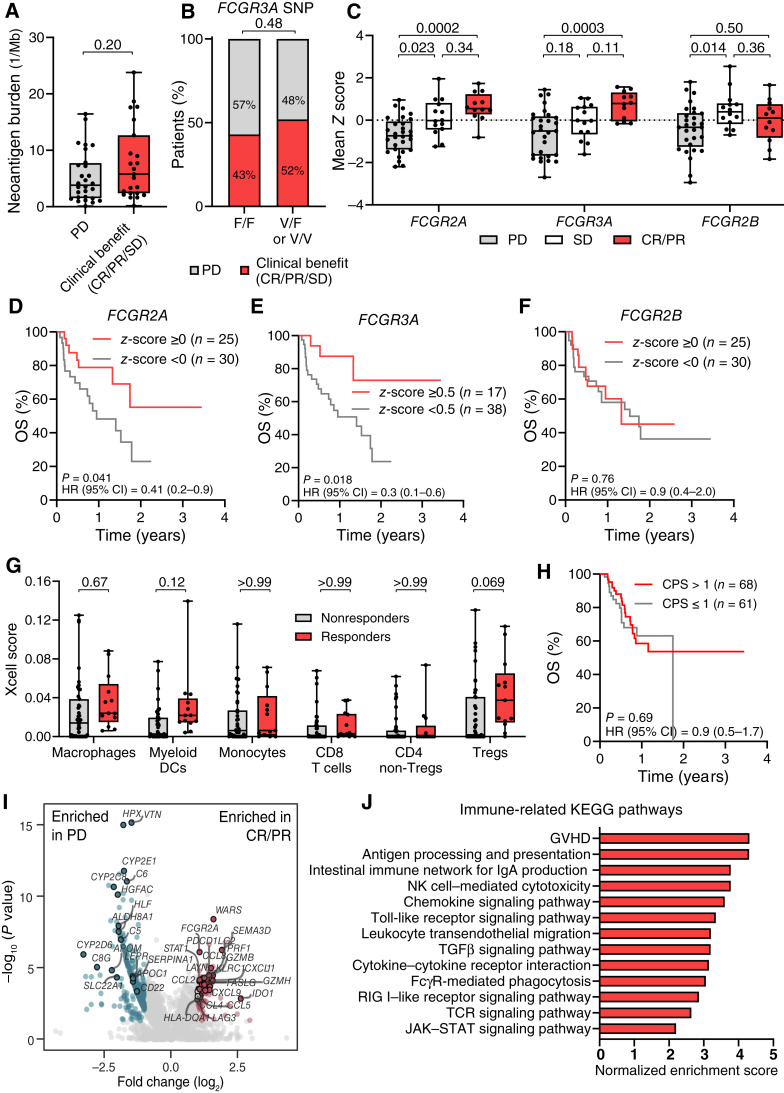
High neoantigen burden (54, 55) and FCGR3A (CD16)-V158F SNPs (15) are predictive markers of clinical response to ipilimumab in patients with advanced melanoma. However, botensilimab alone or botensilimab plus balstilimab was effectively independent of neoantigen burden (Fig. 6A; Supplementary Fig. S15A and S15B) or FCGR3AV158F allele status (Fig. 6B; Supplementary Fig. S15C and S15D), across multiple tumor types, including ICI-refractory tumors. These data are consistent with botensilimab engaging both the high- and low-affinity FcγRIIIA variants with higher affinity and potency versus conventional αCTLA-4 (Fig. 2A and B; Supplementary Fig. S8A) and with the murine botensilimab surrogate αCTLA-4DLE, promoting antitumor activity in poorly immunogenic tumor models (Supplementary Fig. S6).
Figure 6.
Clinical response to botensilimab is independent of neoantigen burden or FcγR polymorphism in patients with advanced solid cancers. Clinical benefit by (A) tumor neoantigen burden (TNB) at pretreatment and (B) FCGR3A genotype in patients treated with botensilimab monotherapy (TNB: n = 35; FCGR3A genotype: n = 33) or botensilimab plus balstilimab (TNB: n = 78; FCGR3A genotype: n = 71). PD, progressive disease. TNB by whole-exome sequencing with predicted HLA binding affinity <500 nmol/L (NetMHCpan). Clinical benefit defined as patients with CR, PR, or SD for ≥12 weeks per RECIST 1.1. C,FCGR2A, FCGR3A, and FCGR2B gene expression from pretreatment tumor biopsy by RNA-seq from patients treated with botensilimab monotherapy ± balstilimab (n = 55). Mean z-scores calculated from log2-scaled transcripts per million expression counts. Survival correlation with (D) FCGR2A, (E) FCGR3A, and (F) FCGR2B gene expression in pretreatment tumor biopsies. G, Intratumoral cell type fractions from pretreatment tumor biopsies by gene set enrichment analysis of bulk RNA-seq data by xCell. H, Kaplan–Meier survival by PD-L1 positivity by IHC using a CPS cutoff of 1 (n = 129). I, Volcano plot in which each dot represents one gene, and (J) immune-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched among differentially expressed genes between responders and patients with PD, using bulk RNA-seq of pretreatment tumors. In box plots, center line, median; box limits, 25th–75th percentile; whiskers, minimum and maximum values. Data analyzed with two-tailed Mann–Whitney test (A), Fisher exact test (B), and two-way ANOVA followed by the Tukey test (C) or Šídák (G) multiple comparisons test. Survival distributions compared by the log-rank test with HR and CI indicated (D–G). Differential gene expression and gene set enrichment were performed using a Benjamini–Hochberg adjustment (I and J).
FCGR Expression Predicts Response to Botensilimab
As Fc:FcγR coengagement is critical for αCTLA-4 activity (13, 15, 16), we hypothesized that botensilimab efficacy would be associated with activating FcγR expression. Notably, patients with CR or PR to botensilimab monotherapy or plus balstilimab had significantly higher FCGR2A and FCGR3A gene expression versus patients progressing on therapy, whereas FCGR2B gene expression did not predict efficacy (Fig. 6C). Higher mean FCGR2A (Fig. 6D) and FCGR3A (Fig. 6E), but not FCGR2B (Fig. 6F), gene expression correlated with better OS. In contrast, analysis of publicly available datasets (56–58), albeit from studies of patients with metastatic melanoma, revealed that FCGR2A and FCGR3A gene expression did not correlate with OS or response to αPD-1 (nivolumab or pembrolizumab) monotherapy (Supplementary Fig. S16A–S16D), αCTLA-4 (ipilimumab) monotherapy (Supplementary Fig. S16E), or the combination of αPD-1 and αCTLA-4 (Supplementary Fig. S16F and S16G). Collectively, these findings align with botensilimab’s enhanced FcγR binding and underscore the significance of Fc:FcγR coengagement for αCTLA-4 efficacy, including botensilimab. Moreover, these data imply that FCGR2A and FCGR3A could be predictive response biomarkers to botensilimab, warranting further investigation.
Although higher FCGR2A and FCGR3A expression could indicate greater tumor immune infiltration, such as with FcγRIIIA+ macrophages/monocytes associated with ipilimumab response (15, 16), response to botensilimab as monotherapy or in combination with balstilimab was independent of higher intratumoral macrophages, monocytes, or myeloid DC populations inferred from gene expression analyses (Fig. 6G; Supplementary Fig. S17A). However, higher FCGR2A (z-score ≥ 0; Supplementary Fig. S17B–S17E) and FCGR3A (z-score ≥ 0.5; Supplementary Fig. S17F–S17I) expression associated with improved survival, correlated with increased myeloid cells in the TME, suggesting that increased FCGR2A and FCGR3A gene expression in responders is partially dependent on myeloid cell infiltration. Consistent with their expression on myeloid cells, FCGR2A and FCGR3A expression showed a strong positive correlation (Supplementary Fig. S17J). However, neither gene’s expression correlated significantly with CD8+ T-cell enrichment scores (Supplementary Fig. S17K and S17L). Furthermore, unlike patients treated with conventional αCTLA-4 and/or αPD-1 (50, 59), response to botensilimab monotherapy or combination therapy was independent of baseline CD8+ and conventional CD4+ T-cell infiltration inferred from gene expression data (Fig. 6G; Supplementary Fig. S17A), further distinguishing the pharmacologic activity of botensilimab from ipilimumab. Moreover, in a much larger patient population, OS was independent of PD-L1 positivity (combined positive score cutoff of 1; Fig. 6H).
Exploratory transcriptomic profiling of responders to botensilimab monotherapy or combination therapy showed significant enrichment in genes associated with immune-related pathways such as WARS, GZMB, PRF1, and LAYN, chemokines such as CXCL9, CXCL10, CXCL11, and CCL5, immune signaling pathways such as STAT1, and semaphorins SEMA3A/D versus progressors (Fig. 6I). By contrast, genes associated with complement, immune proteases, leukocyte differentiation, and metabolism were enriched in progressors (Fig. 6I). Furthermore, using Kyoto Encyclopedia of Genes and Genomes pathway analyses, immune-related pathways such as T-cell receptor and JAK/STAT signaling, chemokine signaling, cytokine–cytokine receptor interaction, and those mediated by FcγR-expressing cell subsets such as antigen processing and presentation, FcγR-mediated phagocytosis, NK cell–mediated cytotoxicity, and Toll-like receptor signaling were significantly enriched in responders (Fig. 6J), supporting transcriptomic data. Further confirmation in larger randomized trials is required.
Discussion
Durable long-term survival is a hallmark of successful αCTLA-4 therapy. However, despite remarkable efficacy in mouse models, patient tumor response rates to conventional αCTLA-4 are relatively low and limited largely to immune-inflamed tumors (60). Although mechanisms for human αCTLA-4 response remain poorly characterized and controversial, recent evidence suggests that FcγR coengagement is a major contributor (13, 15, 21) and could explain the modest efficacy of conventional αCTLA-4. Furthermore, a recent study highlighted that ipilimumab modifies immune gene signatures predicting αPD-1 response in patients with advanced melanoma (56). However, given the low conventional ICI efficacy in colder tumors like MSS colorectal cancer (61), ipilimumab’s ability to favorably remodel the TME may be limited to immunologically active tumors like melanoma (56).
Here, we show that αCTLA-4 antibodies engineered for enhanced binding to activating FcγRs leverage novel FcγR-dependent mechanisms to potentiate T-cell responsiveness, reduce intratumoral Treg content, and enhance APC activation, independent of the FcγR genotype. We observed potent efficacy and favorable TME remodeling in distinct mouse models versus conventional αCTLA-4 and further demonstrated that these effects translate to patients treated with botensilimab across multiple treatment-refractory cancers including those that progressed on prior αPD-(L)1 or αCTLA-4 therapy (18).
Although efficacy studies in tumor-bearing mouse models suggested that αCTLA-4 activity was partly dependent on Fc-mediated intratumoral Treg depletion (8, 32), those studies utilized mouse IgG2a backbones that poorly predict human IgG1 performance and might not represent human conventional IgG1 αCTLA-4 effects. These findings have increased interest in developing Fc-enhanced αCTLA-4. We used the mouse IgG2b Fc backbone as a surrogate for conventional IgG1 αCTLA-4 and demonstrated suboptimal tumor control versus mouse Fc-enhanced αCTLA-4, αCTLA-4DLE. Notably, αCTLA-4DLE, but not WT-IgG2b αCTLA-4, promoted deeper responses across multiple tumor models, associated with favorable TME remodeling, including reduced Tregs, increased MPECs, and activated cDC1.
We previously reported that the ability of αCTLA-4 to enhance T-cell responsiveness independent of Treg depletion depended on APC-expressed FcγRIIIA coengagement (13), suggesting that CTLA-4 blockade alone was insufficient for optimal T-cell activation. Concordant with the kinetic segregation model of TCR signaling (62), we propose that αCTLA-4 with increased FcγRIIIA affinity, for example, botensilimab, enhances CTLA-4 blockade and T-cell activation by improving T-cell:APC immune synapse quality and prolonging the dwell time between CTLA-4–expressing T cells and FcγR-expressing APCs versus IgG1 αCTLA-4 (13).
In patients with metastatic melanoma, higher FcγRIIIA+ macrophage frequency (17) or FCGR3A polymorphisms with increased IgG1 binding capacity (15) correlate with improved OS in response to ipilimumab. Such data suggested Treg depletion, an unproven mechanism in humans (10). Our data demonstrate that conventional IgG1 αCTLA-4 depletes human Tregs poorly, even in the presence of high-affinity FcγRIIIA-expressing NK cells. In contrast, botensilimab mediates robust CTLA-4–expressing Treg killing and intratumoral Treg reduction in patients. Furthermore, botensilimab, but not conventional αCTLA-4, reduced Treg expansion in vitro, consistent with reduced TGFβ1 secretion, suggesting that Fc-enhanced anti–human CTLA-4 reduces Treg stability, corroborating with observations from mouse models (40). Instead of Treg modulation, our data suggest that suboptimal responses to conventional αCTLA-4 in patients expressing low-affinity FcγRIIIA could be attributed to poor T-cell activation. By comparison, botensilimab promoted robust T-cell activation independent of FCGR3A polymorphism, consistent with improved high- and low-affinity FcγRIIIA binding, and clinical responses independent of FCGR3A status, further distinguishing it from ipilimumab. Whether neoantigen burden predicts botensilimab efficacy requires further study.
Beyond T-cell modulation, myeloid cell FcγR coengagement by αCTLA-4 mouse IgG2a and signaling downstream of FcγR activation enhance myeloid activation and antitumor immunity in mice (21). We demonstrated that these Fc-dependent mechanisms effectively translate to humans, but only with Fc-enhanced, not with conventional αCTLA-4. Notably, botensilimab-induced myeloid activation was limited to FcγRIIIA-expressing myeloid cells. Prior mouse studies demonstrated that the myeloid compartment effects stemmed from direct FcγR coengagement, not Treg depletion, further highlighting a Treg-independent mechanism for αCTLA-4 efficacy (21). Increased monocyte, B cell, NK cell, and NKT cell prevalence, unique to botensilimab versus IgG1-WT αCTLA-4, further supports enhanced FcγR engagement in shaping the immune response. However, the full functional significance remains to be elucidated.
Botensilimab also induced intratumoral gene expression of cytotoxic T cell–recruiting cytokines predictive of ICI efficacy in patients (63). Although major sources of CXCL9 and CXCL10 are tumor-resident macrophages (63), we did not observe any changes in macrophage levels after botensilimab treatment, suggesting that the increase may be due to FcγRIIIA+ macrophage activation. A retrospective analysis of patients with metastatic melanoma treated with αPD-1 plus αCTLA-4 revealed a correlation between response and higher FcγRIIIA+ macrophage density and elevated CXCL9, CXCL10, and CXCL11 gene expression (17). However, the specific chemokine source remains undefined. Notably, botensilimab also induced expression of IFNG and T-cell inflamed signatures previously described as predictive of αPD-1 response (53), further underscoring its ability to remodel the TME to be more responsive to PD-1 blockade.
Although predictive biomarkers of human αCTLA-4 response are emerging, botensilimab response seems independent of known response signatures for ipilimumab alone or ipilimumab plus αPD-1, including tumor neoantigen burden (55), FCGR3A allele status (15), and intratumoral CD8+ T-cell infiltration (59). However, we observed strong correlations between intratumoral FCGR2A and FCGR3A gene expression and OS, consistent with the importance of FcγR coengagement. Additional studies of FcγRIIA+ and FcγRIIIA+ immune cell subsets are required.
This study has some limitations. The transcriptomic correlates are exploratory and lack a control group and robust sample size. We could not correlate pharmacodynamic responses with clinical outcomes because of limited posttreatment biopsy availability. Validation is needed in larger randomized trials to determine if these results represent predictive versus prognostic biomarkers. Although this study highlights novel and clinically relevant activity of botensilimab across many tumor types, direct comparison of immune correlates and response biomarkers for conventional αCTLA-4 is difficult as these data are largely acquired from melanoma cases. A limitation of the clinical data including safety and efficacy reported in this article is the inclusion of only patients with available biomarker data. This selection introduces a potential bias, resulting in findings that may not be generalizable to all patients in the trial. Additionally, confirmation of response rates across individual tumor types is pending evaluation of the full trial cohort.
In summary, we provide compelling preclinical and clinical evidence that enhancing FcγR engagement is a promising strategy to expand the clinical benefit of αCTLA-4 therapy and overcome some of the limitations of conventional ICI. These findings reveal an actionable pathway to improve treatment outcomes and extend survival to patient populations historically unresponsive to conventional ICI.
Methods
Cell Lines
The CT26 colorectal carcinoma and EMT6 mammary carcinoma cell lines were obtained from ATCC and maintained in RPMI 1640 supplemented with 10% heat-inactivated FBS and 1% each of penicillin and streptomycin (pen/strep). The MC38 cell line (64) was maintained in DMEM supplemented with 10% FBS and 1% each of pen/strep, L-glutamine, nonessential amino acids, N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid, and sodium pyruvate. B16F1.OVA (65) was maintained in RPMI supplemented with 10% FBS and 1% each of pen/strep and L-glutamine. All cell lines were kept in an incubator at 37°C and 5% CO2. Chinese hamster ovary (CHO) cells genetically engineered to express human FcγRI, FcγRIIA H/H131, FcγRIIA R/R131, FcγRIIB, FcγRIIIA F/F158, or FcγRIIIA V/V 158 and CHO cells expressing recombinant mouse FcγRI, FcγRIIB, FcγRIII, or FcγRIV were obtained from Collection de Cultures de Microorganismes, Institut Pasteur (37). Jurkat cells engineered to express human FcγRIIA (H/H 131), FcγRIIA (R/R 131), FcγRIIIA (V/V 158), or FcγRIIIA (F/F 158) with an NFAT-dependent firefly luciferase reporter gene were obtained from Promega. CHO cells engineered to express human CTLA-4 (GenScript) were cultured in F12K medium supplemented with 10% heat-inactivated FBS. Jurkat cells engineered to express human CTLA-4 (Jurkat-CTLA-4) at the cell surface were generated by transduction of intracellular domain–truncated CTLA-4 as described (66). Jurkat-CTLA-4 cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS. All cell lines were kept in an incubator at 37°C and 5% CO2. All cell lines were routinely tested for the presence of Mycoplasma.
Antibodies
Botensilimab (Fc-enhanced fully human anti–CTLA-4; IgG1κ), an unmodified IgG1κ variant of botensilimab (parental IgG1), and balstilimab (anti–PD-1; IgG4) were discovered using a proprietary mammalian display technology, Retrocyte Display (67), and produced using recombinant DNA technology in a CHO mammalian cell expression system. Botensilimab is engineered with amino acid substitutions DLE in the Fc region (22). The full sequence of botensilimab (68) and balstilimab (69) are publicly available. Ipilimumab was obtained from Myoderm (now Myonex). Human IgG1κ, IgG1DLE, and IgG4 and mouse IgG2bDLE variants of an antibody (VRC01) selective for the CD4-binding site of the HIV type 1 envelope protein, for which the sequence is also publicly available (RRID: AB_2491019), were used as isotype control antibodies. All Fc variants were produced in-house. The sequences of the variable regions of the heavy and light chains of αCTLA-4 clone 9D9 (RRID: AB_10949609) were used to generate surrogate antibodies with the constant regions of mouse IgG2a (mIgG2a) and mIgG2b and mutated variants with amino acid substitutions S245D/S336L/I338E (mIgG2aDLE and mIgG2bDLE; Fc-enhanced) or N297A (mIgG2aN297A; Fc-silent). InVivoPlus grade isotype control (clone MPC-11, mIgG2b) and mouse reactive anti–PD-1 (RMP1-14; Rat IgG2a, κ) were obtained from Bio X Cell.
Clinical Trial Design
The C-800-01 study (NCT03860272) is an open-label, phase I, multicenter study evaluating the safety, tolerability, PK, and pharmacodynamic profiles of botensilimab alone or in combination with anti–PD-1 (balstilimab) in patients with advanced solid tumors. Patients were enrolled at 14 sites across the United States. The study initially followed a 3 + 3 dose-escalation design and subsequently expanded to include separate indication and dose cohorts. Botensilimab was given every 3 or 6 weeks, starting at a dose level 0.1 up to 3 mg/kg, administered intravenously for up to 2 years, or in combination with balstilimab 3 mg/kg every 2 weeks administered intravenously for up to 2 years, or the combination of botensilimab 150 mg every 6 weeks with balstilimab 450 mg every 3 weeks for up to 2 years. All patients included in the clinical trial were subjected to written informed consent and consented prior to study enrollment. Select indication cohorts are still open for enrollment in the C-800-01 study.
Study Oversight
The C-800-01 study and collection and use of patient tumor and blood samples were conducted in compliance with the Declaration of Helsinki and International Conference on Harmonization Guidelines for Good Clinical Practice and were approved by the institutional review board (IRB) at each participating site: The Angeles Clinic and Research Institute, a Cedars-Sinai Affiliate; Beth Israel Deaconess Medical Center; City of Hope Comprehensive Cancer Center; Columbia University Medical Center; Dana-Farber Cancer Institute; HonorHealth Research & Innovation Institute; MD Anderson Cancer Center; Memorial Sloan Kettering Cancer Center; Providence Portland Cancer Center; Saint John’s Cancer Institute; University of Colorado; University of Miami Sylvester Comprehensive Cancer Center; University of Southern California Norris Comprehensive Cancer Center; and The University of Texas Health Science Center at San Antonio. All patients provided written informed consent prior to study enrollment.
Patient Characteristics
This article includes assessment of blood and tumor samples from patients with advanced solid tumors treated with 0.1, 0.3, 1, or 2 mg/kg botensilimab monotherapy every 3 weeks or every 6 weeks, or the combination of botensilimab with 3 mg/kg balstilimab every 2 weeks, or the combination of 150 mg botensilimab every 6 weeks with 450 mg balstilimab every 3 weeks, as part of an ongoing clinical trial sponsored by Agenus Inc (ClinicalTrials.gov Identifier: NCT03860272; Supplementary Fig. S11). All patient biospecimens were collected and used in compliance with the Agenus Inc C-800-01 study protocol, the Declaration of Helsinki, and International Conference on Harmonization Guidelines for Good Clinical Practice, with approval from institutional review boards at all sites and written informed consent obtained from all patients. Key inclusion criteria include patients ages 18 years or older with measurable disease per RECIST 1.1 and an ECOG performance status of 0 or 1 with adequate end-organ function. Prior ICI [anti–PD-(L)1 or anti–CTLA-4] was permitted. Patients must have had a confirmed diagnosis of metastatic or a locally advanced solid tumor for which no standard therapy was available or for which standard therapies previously failed. Key exclusion criteria include prior immune-related adverse events [aside from grade 1 or 2 rash per Common Terminology Criteria for Adverse Events (CTCAE) v.5] that required steroids for more than 7 days, steroids, or other immunosuppression within 1 week of starting study treatment and active or prior autoimmune disease requiring systemic treatment within 2 years with disease-modifying agents or steroids. Prior brain metastases were allowed if treated and off steroids for 10 days. Baseline demographics and tumor types are shown in Supplementary Table S6.
Clinical Endpoints
For biomarker analyses, clinical benefit was defined as patients who had a confirmed CR, PR, or SD for ≥12 weeks per RECIST 1.1. Imaging was performed every 6 weeks (±3 days) from the first treatment dose. Safety was assessed by evaluating AEs graded according to the NCI CTCAE version 5.0, vital signs (blood pressure, heart rate, and temperature); physical examinations; 12-lead electrocardiogram; ECOG performance status; and clinical laboratory assessments. AEs were coded according to the Medical Dictionary for Regulatory Activities. The severity of AEs was graded using NCI CTCAE.
Mouse In Vivo Tumor Studies
C57BL/6 and BALB/C mice were purchased from The Jackson Laboratory. KPC mice were obtained from the NCI Mouse Repository. Procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Agenus Inc following the guidance of the Public Health Services (PHS) Policy or The Translational Genomics Research Institute and conform to the relevant regulatory standards.
For CT26 and MC38 tumor studies, 5 × 104 cells and 1 × 105 cells, respectively, were suspended in 100 μL PBS and injected subcutaneously in the flank. Following engraftment, mice were randomized and treated via intraperitoneal administration with a single dose (10 or 100 µg) of mouse anti–CTLA-4 (mIgG2b or mIgG2bDLE) or isotype control antibodies as indicated, 7 (MC38) or 9 (CT26) days after tumor inoculation. Average tumor volume at the time of treatment was 63 mm3 (Fig. 1A), 120 mm3 (Fig. 1D–H; Supplementary Fig. S2D), 125 mm3 (Fig. 1I–L), or 61 mm3 (Supplementary Fig. S2B) for CT26 and 82 mm3 (Fig. 1B) or 73 mm3 (Supplementary Fig. S3B–S3F) for MC38 models. Mice cured of CT26 tumors were rechallenged with 1 × 106 CT26 cells tumors in the contralateral flank 180 days after initial tumor implantation and compared with concurrent tumor growth in CT26-naïve control mice to assess tumor-specific immunity. For pharmacodynamic studies in the CT26 and MC38 models, the tumor, spleen, and blood were collected posttreatment at the time points detailed in the figure legends and processed as previously described (13). For EMT6 tumor studies, 2 × 105 cells were suspended in 50 μL PBS and injected in the fourth mammary fat pad with 50% Matrigel. Following engraftment (mean tumor size: 62 mm3), mice were randomized and treated via intraperitoneal administration with three weekly doses of 100 μg mouse anti–CTLA-4 (mIgG2b or mIgG2bDLE) or isotype control antibodies starting 5 days after tumor inoculation. Differences in the proportion of complete responders versus nonresponders were assessed using the χ2 test on a 2 × 2 contingency table.
For pancreatic ductal adenocarcinoma models, murine pancreatic tumors were harvested from tumor-bearing KPC (NCI Mouse Repository) mice, minced using surgical scissors into small pieces, suspended in DMEM, mixed with Matrigel, and injected subcutaneously into the flanks of C57BL/6 mice. Treatment with chemotherapy was initiated when tumors reached ∼100 mm3 and was repeated 4 days later. Chemotherapy-treated mice received intraperitoneal administration of gemcitabine (70 mg/kg) and cisplatin (4 mg/kg) and intravenous administration of nab-paclitaxel (25 mg/kg). Therapeutic or isotype antibodies were administered intraperitoneally at 100 μg (αCTLA-4DLE) twice a week (treatment days were days 1, 4, 8, 11, 15, and 18 with day 1 being the first day of chemotherapy treatment).
For models of subcutaneous melanoma used to evaluate combination with cancer vaccines, 5 × 104 B16F1.OVA cells, suspended in 100 μL PBS, were injected subcutaneously in C57BL/6 mice. Three days after tumor implantation, mice were vaccinated subcutaneously with a recombinant heat shock protein 70-peptide complex comprising synthetic long ovalbumin and melanoma-associated antigen TRP2 epitopes in QS-21 saponin and cytosine phosphoguanine adjuvants. Vaccination was administered on days 3, 8, and 13. Mice treated with immune checkpoint blockade (ICB) therapy, received intraperitoneal administration of Fc-enhanced anti–CTLA-4 (mIgG2bDLE; 50 μg) weekly beginning on day 3 for 3 weeks (treatment days: days 3, 8, and 13) and anti–PD-1 (200 μg) biweekly beginning on day 3 for 2 weeks (treatment days: day 3, 6, 10, and 13). Survival was determined based on tumor volume (>2,000 mm3 considered death), body weight (>20% loss considered death), or due to animal incapacitation or morbidity.
For models involving adoptive cell therapy, 1.5 × 105 B16F1.OVA cells, suspended in 100 μL PBS, were injected subcutaneously into the flank of CD90.1 C57BL/6 transgenic mice. OT-1–specific transgenic CD8+ T cells harvested from OT-1 mice (purchased from The Jackson Laboratory) and expanded with IL2 for 5 days were administered intravenously 8 days after tumor implantation. Mice treated with immune checkpoint blockade therapy received intraperitoneal administration of Fc-enhanced anti–CTLA-4 (mIgG2bDLE; 50 μg) weekly beginning on day 3 for 3 weeks and anti–PD-1 (200 μg) biweekly beginning on day 3 for 3 weeks.
For graphical considerations, in graphs that show mean tumor growth, a value of 2,000 mm3 was given to tumors of mice in which tumors exceed 2,000 mm3 (mice are euthanized when tumors reach 2,000 mm3); a mean is plotted if the group has at least one mouse alive. For statistical analysis, all individual values up to the day in which all groups have at least one mouse alive, and when showing individual values, are included. In all experiments, tumors were measured twice weekly using Vernier calipers and volumes calculated using the formula length × width2 × 0.52 (CT26, MC38, and B16.F1 models) or length × width2 × 0.5 (EMT6).
Mouse In Vivo SEB Superantigen Studies
C57BL/6 mice were injected once intraperitoneally with 150 μg SEB superantigen (Toxin Technologies) and 100 μg mouse anti–CTLA-4 (mIgG2b or mIgG2bDLE) or corresponding isotype control antibodies as previously described (13). Blood was collected 6 days posttreatment and analyzed by flow cytometry as described (13, 27). All the procedures related to animal handling, care, and treatment in this study were performed according to guidelines approved by the IACUC of Agenus Inc following the guidance of the PHS Policy.
PK of Mouse Anti–CTLA-4 Antibodies
BALB/C mice were treated intraperitoneally with a dose of 10, 30, or 100 µg of anti–CTLA-4 (mIgG2b or mIgG2bDLE) antibodies. Antibodies were administered to groups of four mice per dose cohort per collection schedule. Blood was collected by submandibular venous puncture in K2EDTA-coated tubes at 5 minutes and 1, 3, 6, 9, 24, 48, 72, 96, 168 (7 days), 336 (14 days), and 504 hours (21 days) after antibody administration to prepare plasma for analysis. All the procedures related to animal handling, care, and treatment in this study were performed according to guidelines approved by the IACUC of Agenus Inc following the guidance of the PHS Policy.
Anti–CTLA-4 antibody concentrations were measured using a bespoke ELISA. Recombinant mouse CTLA-4-His (2 µg/mL) immobilized on an ELISA plate was incubated with a dose range (5.6 × 10−7–0.10 µg/mL) of soluble anti–CTLA-4 antibodies. Sample plates were washed 5 times and antibody binding detected using a secondary peroxidase-conjugated AffiniPure goat anti–mouse IgG (H+L) antibody, activated by the addition of tetramethylbenzidine (TMB) substrate, and the resulting optical density measured at 450 nm using an EnVision Multimode plate reader (PerkinElmer). A noncompartmental analysis was performed using Phoenix WinNonlin 64 version 8.3.3.33 (Certara, Inc.).
Binding Kinetics by Surface Plasmon Resonance
Affinity of botensilimab to human CTLA-4 was determined using a Biacore T200 instrument (Cytiva) equipped with a CM5 sensor chip. Botensilimab was immobilized on the CM5 sensor chip by amine coupling chemistry. To collect kinetic binding data, increasing concentrations ranging from 0.9 to 30 nmol/L of recombinant human CTLA-4 His-tagged (Sino Biological) or Fc-fusion (ACROBiosystems) were injected over the flow cells at a flow rate of 30 μL/minute and at a temperature of 25°C. The complex was allowed to associate for 180 seconds and dissociate for 600 seconds. Binding affinities to FcγRs were determined using a Biacore T200 or a Biacore 8K+ instrument (Cytiva). A CM5 chip was immobilized with an anti-His antibody (Invitrogen), and the different His-tagged human FcγRIIB, FcγRIIIA F158, and V158 and mouse FcγRIIB and FcγRIV captured on the chip. For binding to FcγRIIIA, botensilimab and the parental IgG1 were injected over the flow cells at concentrations ranging from 0.9 to 60 nmol/L. Ipilimumab and tremelimumab were injected over the flow cells at concentrations ranging from 0.94 to 60 nmol/L (FcγRIIIA V158) and from 7.81 to 8 µmol/L (FcγRIIIA F158). For binding to FcγRIIB, botensilimab and the parental IgG1 were injected over the flow cells at concentrations ranging from 15.6 to 1 µmol/L, ipilimumab at concentrations ranging from 7.81 nmol/L to 8 µmol/L, and tremelimumab at concentrations ranging from 120 nmol/L to 8 µmol/L.
For binding to mouse FcγRIV, αCTLA-4 mouse antibodies (mIgG2a, mIgG2bDLE, and mIgG2b) were injected at concentrations ranging from 1.88 to 60 nmol/L. For binding to mouse FcγRIIB, antibodies were injected at concentrations ranging from 125 nmol/L to 8 µmol/L. The antibodies were injected at a flow rate of 30 or 50 μL/minute at a temperature of 25°C. The chip surface was regenerated after each cycle with 10 mmol/L glycine pH 2.0. Binding kinetics or steady state analyses were carried out using Biacore Insight Evaluation software (Cytiva 4.0.8.20368) or Biacore T200 Evaluation software (GE Healthcare version 3.1).
Binding Specificity by ELISA
To demonstrate specificity of botensilimab for CTLA-4, binding to CTLA-4 and related CD28 family proteins CD28, ICOS, BTLA, and PD-1 was evaluated by ELISA. Recombinant CD28 family member proteins were immobilized to an assay plate overnight at 4°C, blocked with StartingBlock Blocking Buffer (Thermo Fisher Scientific) for 15 minutes, and incubated with a dose range (1.37–1,000 ng/mL) of botensilimab or Fc-enhanced isotype control (IgG1DLE) antibody for 1 hour. Binding was detected using a human IgGκ light chain secondary antibody linked to horseradish peroxidase (HRP) and visualized with 3,3′,5,5′-TMB substrate. Absorbance data were collected at 450 nm using an EnVision Multimode plate reader (PerkinElmer).
Cell-Based Ligand Blocking Assay
To assess CTLA-4 ligand binding, CD80-Fc and CD86-Fc proteins (R&D Systems) were conjugated to Alexa Fluor 647 (AF647) dye (Invitrogen) using the AF647 labeling kit (Life Technologies), as per manufacturer’s instructions. CHO cells, genetically engineered to stably express human CTLA-4 at the cell surface (GenScript), were incubated with a dose range of botensilimab, an unmodified IgG1 variant of botensilimab (parental IgG1), or corresponding isotype control antibodies (human IgG1 and human IgG1DLE) antibodies for 30 minutes at 4°C. Cells were washed and subsequently incubated with AF647 fluorescently labeled CD80-Fc or CD86-Fc proteins for 30 minutes at 4°C. Following incubation, CD80-Fc and CD86-Fc binding was analyzed by flow cytometry. A CTLA-4 ligand blocking bioassay (Promega) was used to further characterize the potential of the anti–CTLA-4 antibodies to block CTLA-4–ligand interactions and promote T-cell receptor and CD28 activation independent of FcγR coengagement. In this system, a human Jurkat T-cell line engineered to constitutively express cell-surface CTLA-4, together with a luciferase reporter gene under the control of a native IL2 promoter (IL2-luc) which responds to TCR and CD28 activation, was treated with titrated concentrations of botensilimab, parental IgG1, or corresponding isotype control antibodies (human IgG1 and human IgG1DLE), together with Raji B cells that endogenously expressed CD80 and CD86, and a plasma membrane–expressed anti–CD3 mAb fragment to trigger TCR activation. Following a 6-hour incubation, luciferase activity was determined using a Bio-Glo Luciferase Assay System (Promega).
Fc–FcγR Cellular Binding
To assess mouse Fc–FcγR binding, CHO cell lines genetically engineered to express mouse FcγRI, FcγRIIB, FcγRIII, and FcγRIV were incubated with titrated concentrations of mIgG2a, mIgG2aDLE, mIgG2b, mIgG2bDLE, and mIgG2bN297A variants of αCTLA-4 (clone 9D9) antibodies for 1 hour at 4°C. Antibody binding was detected using an excess of AF647-conjugated F(ab′)2 fragment of goat anti–mouse Fab/F(ab′)2 IgGs (Jackson ImmunoResearch) and analyzed by flow cytometry. Median fluorescent intensity values were used to visualize the binding.
To evaluate human Fc–Fcγ receptor binding, CHO cell lines genetically engineered to express human FcγRI, FcγRII-R/R 131, FcγRII-H/H 131, FcγRIIB, FcγRIIIA-V/V 158, and FcγRIIIA-F/F158 were incubated with titrated concentrations of botensilimab, an IgG1 variant of botensilimab (parental IgG1), ipilimumab, or an isotype IgG1.N297A (aglycosylated) negative control antibody cross-linked with an equimolar concentration of anti–light chain F(ab′)2 fragment (Jackson ImmunoResearch). Following a 1-hour incubation, binding of the antibodies was detected by flow cytometry using a R-PE–conjugated goat anti–human IgG Fcγ fragment–specific secondary antibody (Jackson ImmunoResearch).
FcγR Signaling Assay
CTLA-4–expressing Jurkat cells (Promega) or CTLA-4–expressing CHO cells (GenScript) were incubated with increasing concentrations of botensilimab, an unmodified IgG1 variant of botensilimab (parental IgG1), or corresponding isotype control (human IgG1 and human IgG1DLE) antibodies. Effector Jurkat cells engineered to express FcγRIIA H/H131, FcγRIIA R/R131, FcγRIIIA F/F158, or FcγRIIIA V/V 158 with an NFAT-dependent firefly luciferase reporter gene (Promega) were added at an effector-to-target cell ratio of 2.5:1. Following a 20-hour incubation at 37°C, NFAT-luciferase activity was determined using a Bio-Glo Luciferase Assay System (Promega) and an EnVision Multimode plate reader (PerkinElmer).
PK, Toxicity, and Immunogenicity of Botensilimab in Cynomolgus Macaques
In a 13-week toxicity study, cynomolgus monkeys (n = 16/sex; 4 dose groups) were injected once weekly for up to 13 weeks by intravenous injection with 0 (formulation buffer, vehicle), 5, 25, and 75 mg/kg/dose botensilimab and following a 4-week recovery period. Dosing levels were based on the results from a 4-week toxicity study, in which the indicated no observed adverse effect level was identified as 30 mg/kg. Animals were approximately 4 to 10 years of age and with body weights ranging from 3.6 to 7.4 kg in males and 2.4 to 6.4 kg in females at dosing initiation. A complete dosing-phase necropsy was conducted on day 92 and a recovery-phase necropsy conducted on day 120. Criteria for evaluation included viability (morbidity/mortality), clinical observations, body weight, food consumption, ophthalmic examinations, safety pharmacology (ECG, blood pressure, heart rate, respiration, and neurologic examinations), clinical pathology (hematology, serum chemistry, coagulation, and urinalyses), immunogenicity, gross (necropsy) evaluation, organ weight, histopathologic evaluation, and toxicokinetic analysis.
Blood samples for hematology, clinical chemistry, and coagulation were collected in K2EDTA tubes, plain tubes with separating gel, and sodium citrate tubes, respectively. The samples were chilled, mixed gently, and centrifuged following blood collections. The resultant plasma was frozen immediately over dry ice and stored in a freezer set to maintain −80°C. Serum samples were prepared from blood collected at predose and 5 and 15 minutes and 1, 4, 8, 24, 36, 48, 72, 96, and 144 hours postdose on study days 1 and 85. The samples were analyzed for drug concentrations using a validated immunoassay analytic procedure (dynamic range = 30–1,000 ng/mL). A noncompartmental model was performed using Certara WinNonlin. A three-tiered process was used to detect, confirm, and titer antibodies against botensilimab (ADA) using a validated analytic method. Serum samples were prepared from blood collected predose on study days 1, 29, and 85. The assay method utilized an electrochemiluminescent bridging immunoassay. In brief, samples were diluted 1 to 50 with acetic acid followed by neutralization by tris base before the addition of a mixture of biotinylated- and ruthenylated-AGEN1181. A streptavidin-coated plate was used to capture immune complexes prior to wash and addition of read buffer. The concentration of botensilimab for the confirmatory assay was set at 2 µg/mL.
The 13-week toxicity study was performed at WuXi AppTec Co., Ltd. in accordance with IACUC guidelines, adhered to the US FDA Good Laboratory Practice Regulations 21 CFR 58, animal care welfare regulations and guidelines set forth by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International, the Guide for the Care and Use of Laboratory Animals of the National Research Council (2011), and the People’s Republic of China, Ministry of Science and Technology, “Regulations for the Administration of Affairs Concerning Experimental Animals” 2017.
Human Subjects and Samples
Human whole blood was obtained from healthy, consenting, and anonymized adult volunteers through Research Blood Components, LLC and PBMC isolated by density centrifugation using Ficoll-Paque Plus (Thermo Fisher Scientific). All blood collection followed American Association of Blood Banks guidelines and was approved by the New England Independent Review Board (120160613). Blood samples from patients with cancer were obtained after they provided consent on IRB-approved protocols. Pre- and posttreatment blood samples from patients with advanced solid cancer (n = 37) treated with 0.1, 0.3, 1, or 2 mg/kg botensilimab monotherapy every 3 weeks or every 6 weeks (NCT03860272) were obtained by venipuncture. All patients provided written informed consent. Blood samples were subjected to PBMC isolation using a lymphoprep density gradient centrifugation in SepMate tubes and cells cryopreserved until sample testing. On-treatment PBMCs were collected 7 days after treatment.
Pretreatment and on-treatment plasma was prepared from blood collected from patients with advanced solid cancer (n = 32) treated with 0.1, 0.3, 1, or 2 mg/kg botensilimab monotherapy every 3 weeks or every 6 weeks. All patients provided written informed consent. Blood for plasma was collected in NaHep collection tube and processed immediately at the clinical sites. On-treatment plasma was collected 24 hours after treatment. Plasma aliquots were stored at −80°C until testing. IFNγ cytokine levels in plasma were tested using the Meso Scale Discovery V-PLEX Plus Proinflammatory Panel 1 electrochemiluminescence assay (K15049G) performed by Precision for Medicine.
FcγR Genotyping
For nonclinical assays, genomic DNA was extracted from human PBMC samples using the QIAamp DNA blood mini kit (Qiagen) and the concentrations measured using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). FCGR2A and FCGR3A polymorphisms were identified by PCR, using custom-designed primers for FCGR2A (forward primer: CCCTTGGAATCTATCCTTACA; reverse primer: GACTACCTATTACCTGGGA) and FCGR3A (forward primer: GTTCAAGGAGGAAGACCC; reverse primer: CTTGAGTGATGGTGATGTTCA). The PCR products were checked by electrophoresis using a 1.5% agarose gel containing 0.5 μg/μL ethidium bromide (Sigma-Aldrich) and FcγR genotyping confirmed by Sanger sequencing of PCR products using standard approaches. Sequence traces were assembled and analyzed to identify genomic alterations using the Benchling software package (Benchling).
For clinical samples, genomic DNA was isolated from blood using QIAamp DNA Midi Kit (including proteinase K, RNase, and lysis steps) and subjected to TaqMan genotyping. A measure of 25 ng of genomic DNA was amplified using a ProFlex PCR System with two TaqMan genotyping assays designed by Q2S|EA Genomics to detect SNPs rs1801274 (FCGR2A) and rs396991 (FCGR3A). All patients provided written informed consent. Data acquisition and initial calls were performed using the ViiA 7 or QuantStudio instrument (Thermo Fisher Scientific). Each run included control samples with known genotypes representing all three genotypes (AA, AB, and BB) for the SNPs of interest and a no template control (NTC) lacking the DNA template. Cluster plots were visually examined to ensure that the results for control samples within each plate clustered as expected and that test sample genotype results were segregated into clearly defined clusters. Clinical specimens (and control samples) were run in technical duplicates. For FCGR3A, if either or both PCR replicates had an undetermined or discordant result, the genotype was tested using PCR amplification and Sanger sequencing using FCGR3A-specific primers designed by Q2S|EA Genomics. Blood samples from patients with cancer were obtained after they provided consent on IRB-approved protocols.
Primary T Cell and APC Coculture Stimulation Assays
Human PBMCs, isolated from healthy donor whole blood, were cultured at 1 × 105 cells/well in 96-well round bottom tissue culture plates (Corning). Cells were stimulated with 10 ng/mL of superantigen SEA peptide (Toxin Technology, Inc.) and treated with titrated or fixed concentrations of soluble botensilimab, an IgG1 variant of botensilimab (parental IgG1), or isotype control (IgG1DLE) antibodies alone or in combination with a fixed dose of anti–PD-1 (balstilimab) or an IgG4 isotype control antibody. After 4 days, IL2, IL10, soluble CD25, and TGFβ1 levels in culture supernatants were analyzed using a bead-based luminescent amplification assay (AlphaLISA, PerkinElmer) or Luminex bead array (R&D Systems). The frequencies of Tregs and non-Tregs were assessed longitudinally at 12, 24, 48, 72, and 96 hours by flow cytometry from cultures treated with 10 ng/mL of SEA peptide and 5 µg/mL of the indicated antibodies. To evaluate the potential for Treg depletion in SEA-stimulated cultures, antibodies were added 96 hours post-SEA stimulation, and Treg frequencies were assessed 24 hours later. Tregs were defined as viable CD45+ CD3+ CD4+ CD25+ FOXP3+ cells, whereas non-Tregs were defined as viable CD45+ CD3+ CD4+ CD25− cells. The frequencies of CD16− and CD16+ NK cells (CD3− CD56+), B cells (CD3−, CD19+), NKT cells (CD3+ CD56+), DC (CD3− CD19− CD14− CD11c+ HLA-DR+), and monocytes (CD3− CD19− CD14+ HLA-DR+) from five SEA-stimulated, FcγRIIIA V/F 158 healthy donors at 96 hours were assessed by flow cytometry. Expression of CD40, HLA-DR, and CD86 was assessed on CD16+ CD11c+ and CD16− CD11c+ myeloid cells by flow cytometry.
Treg Depletion Assay
Tregs (CD4+ CD127low CD25+) and non-Tregs (CD4+ CD25−) were isolated from human healthy donor PBMCs using the EasySep Human CD4+ CD127low CD25+ Regulatory T Cell Isolation Kit as per the manufacturer’s protocol (STEMCELL Technologies). T-cell identity was confirmed with FOXP3 expression and FOXP3 epigenetic methylation pattern. Tregs were expanded for 11 days in X-VIVO 15 Media supplemented with 5% human AB serum (Gemini), 100 ng/mL rapamycin (Sigma), ImmunoCult Human CD3/CD28 T Cell Activator (STEMCELL Technologies), and 100 ng/mL human IL2 (R&D Systems). CD4+ non-Tregs were expanded for 11 days in X-VIVO 15 Media supplemented with 20 ng/mL human IL2 and human CD3/CD28 (ImmunoCult). T cells were stained with CellTrace Far Red and CellEvent Caspase-3/7 Green kits as per the manufacturer’s protocol (Thermo Fisher Scientific) and cocultured with NK-92 cells engineered to express FcγRIIIA (34) at an effector cell:target cell ratio of 1:1.3, with 10,000 T cells and 13,000 NK-92 cells per well. Cells were treated with 10 µg/mL of botensilimab, an IgG1 variant of botensilimab (parental IgG1), Fc-engineered isotype control antibody (IgG1DLE), or IgG1 isotype control antibodies for 16 hours at 37°C in 5% CO2. Antibody-dependent cellular cytotoxicity of T cells was captured using a high-content confocal microscope (ImageXpress Micro, Molecular Devices). Images were taken every 30 minutes for 16 hours, analyzed using MetaXpress image analysis software (Molecular Devices).
C1q Binding
C1q binding to botensilimab and an IgG1 variant of botensilimab (parental IgG1) was determined using ELISA. Plates were coated with increasing concentrations of the indicated anti–CTLA-4 antibodies followed by the complement component C1q (2 μg/mL). Bound C1q to the antibodies was detected using a biotinylated anti-C1q antibody, streptavidin–HRP, and the chromogen 3,3′,5,5′-TMB, and absorbance at 450 nm measured using an EnVision Multimode plate reader (PerkinElmer).
Complement-Dependent Cytotoxicity Assay
Jurkat-CTLA-4 cells were incubated with 20% normal human serum derived from three healthy donors, as a source of complement, and 10 μg/mL of botensilimab, an IgG1 variant of botensilimab (parental IgG1), or no antibody (PBS buffer) for 4 hours at 37°C and 5% CO2. As a positive control for confirming active complement, Raji cells endogenously expressing CD20 were incubated with normal human serum and 10 μg/mL of anti-CD20 antibody Rituxan. Cell viability was assessed postincubation by staining with 7-amino-actinomycin viability dye. Following a 10-minute incubation at 4°C, cell viability was assessed by flow cytometry to measure antibody-mediated complement-dependent cytotoxicity activity.
Whole-Exome Sequencing and Bulk RNA-seq
Genomic DNA and RNA from formalin-fixed, paraffin-embedded, tumor-containing tissue sections and matched blood samples were isolated and subjected to whole-exome paired-end sequencing and RNA-seq using the Personalis ImmunoID NeXT platform. All patients provided written informed consent. DNA/RNA extraction, library preparation, NovaSeq sequencing and data analysis were performed at Personalis, demonstrating accuracy and precision of their experimental protocols, with a sensitivity of 99% for single-nucleotide variants (3% false-positive rate) and 94% for indels (3% false-positive rate). The coverage was 150× for germline control blood samples and 300× for tumor samples. Somatic variants were called from the exome reads and the reference human genome using hs37d5 and Burrows–Wheeler Aligner and STAR alignment tools, and variants were called using MuTect, Verdict, and FusionCatcher. Neoantigen burden was defined as peptides/Mb that originate from tumor-specific nonsynonymous somatic variants (single-nucleotide variants and indels) with predicted HLA binding affinity <500 nmol/L (based on NetMHCpan prediction). FCGR2A, FCGR2B, and FCGR3A gene expression was assessed from pretreatment biopsies by RNA-seq from patients with advanced solid cancers treated with 1, 2, or 3 mg/kg botensilimab monotherapy (n = 12) or in combination with balstilimab (n = 38). Survival analysis was performed using Kaplan–Meier estimate with the clinical data cutoff on March 27, 2023. OS was censored at the last contact date unless a death was reported for a patient prior to the data cutoff date. The log-rank test was used to compare OS between groups with high (z-score > 0) and low (z-score ≤ 0) FCGR2A, FCGR2B, and FCGR3A gene expression. HR with associated two-sided CI was calculated using a Cox proportional hazards regression model, with a level of gene expression as a single covariate. IFNγ gene signature (53), T cell–inflamed gene signature (53), and CXCL9 and CXCL10, and CCL5 gene expression by RNA-seq were assessed from paired pretreatment and on-treatment tumor biopsies from patients with advanced solid cancers and treated intravenously with 0.3, 1, or 2 mg/kg botensilimab every 3 weeks or every 6 weeks in combination with 3 mg/kg balstilimab every 2 weeks. On-treatment biopsies were taken on cycle 2 day 1 for the every 6 weeks cohort or cycle 3 day 1 for the every 3 weeks cohort. A mean z-score was calculated from log2-scaled transcripts per million expression counts. To predict cell types from bulk RNA-seq expression data, cell-type enrichment analysis by xCell was performed as previously described (70). An R package for running xCell is available as an open-source code in a GitHub repository (https://github.com/dviraran/xCell). The RNA-seq expression counts (transcripts per million) were supplied to the model as a gene by sample matrix and the resulting predictions compared across pretreatment and on-treatment sample time points. For differential gene expression analysis, genes with <10 counts in >90% samples were filtered out. DESeq2 in R compared 13 responders (CR + PR) with 29 nonresponders (PD) with normal log2FC shrinkage. Gene set enrichment analysis employed the R library “fgsea” using C2-CP:KEGG signatures from the Broad Institute’s MSigDB. Signatures were curated postenrichment to eliminate duplicates and highlight statistically significant pathways (immune related) that were relevant to this study. The Benjamini–Hochberg method was used to adjust P values for both differential gene expression and gene set enrichment analysis. Tumor and blood samples from patients with cancer were obtained after they provided consent on IRB-approved protocols.
Additionally, we analyzed four published melanoma datasets in which pretreatment biopsy samples were subjected to RNA-seq to investigate the relationship between FCGR gene expression and response to ICI therapy. The datasets included Gide and colleagues (BioProject accession number PRJEB23709), Hugo and colleagues (BioProject accession number PRJNA312948), CheckMate 067 (BioProject accession number PRJNA923698), and CheckMate 064 (BioProject accession number PRJNA92369) cohorts. Raw data were processed with the nf-core rnaseq pipeline (doi: 10.5281/zenodo.1400710) using default parameters (71). In these studies, patients with metastatic melanoma were treated with αPD-1 therapy (pembrolizumab or nivolumab), αCTLA-4 therapy (ipilimumab), or the combination αPD-1 and αCTLA-4 therapies. Responders were defined as patients with a RECIST response of CR, PR, or SD of greater than 6 months with no progression and nonresponders as progressive disease or SD for less than or equal to 6 months before disease progression.
TCR Sequencing
TCR repertoire analysis on blood and tumor samples from CT26 tumor–bearing Balb/C mice treated intraperitoneally with 100 μg of Fc-enhanced anti–CTLA-4 (Clone 9D9, mIgG2bDLE), anti–CTLA-4 (Clone 9D9, IgG2b), or isotype control (clone MPC-11, mIgG2b) was performed by Adaptive Biotechnologies. This study was performed according to guidelines approved by the IACUC of Agenus Inc following the guidance of the PHS Policy. Briefly, when tumors reached an average volume of 130 mm3 (10 days postimplantation), mice were treated with the indicated antibodies 3 times a week for 1 week. Blood (100 μL) was collected prior to the first antibody treatment in a standard EDTA tube, frozen, and stored at −80°C. All animals were euthanized 11 days after first dose, and tumor and blood collected. Pretreatment blood and posttreatment blood and tumor samples were submitted for sequencing of the CDR3 regions of mouse TCRβ chains (immunoSEQ mouse TCRβ assay) by Adaptive Biotechnologies.
For TCR sequencing of clinical samples, genomic DNA from blood PBMCs was isolated from patients treated with 0.1, 1, or 2 mg/kg botensilimab every 3 weeks or every 6 weeks pretreatment (cycle 1 day 1) and 3 to 4 weeks postdose. All patients provided written informed consent. Blood samples from patients with cancer were obtained after they provided consent on IRB-approved protocols. Samples were subjected to high-throughput sequencing of the CDR3 variable chain of the TCR gene using the multiplex PCR-based Adaptive immunoSEQ platform and Adaptive immunoSEQ analysis pipeline. Downstream TCR sequencing analyses were performed using the identified productive TCRβ CDR3 amino acid sequence (referred to as “clonotype”) and included a number of unique clonotypes, their counts, and frequencies, Simpson clonality index, and differential abundance of T-cell clones between pre and posttreatment blood.
When indicated, samples were downsampled to allow for comparison. Differential abundance analysis was performed using an exact binomial test, and abundance (clonotype frequency) change was considered significant (expanded or contracted clonotype) if P value adjusted by the Benjamini–Hochberg method was lower or equal to 0.01. Only clones with a count >5 was included in this analysis. When determining the number of tumor-associated T-cell clonotypes that expanded peripherally in CT26 tumor–bearing mice, only TCRs that expanded in blood and were present in tumors posttreatment were included. Lastly, a previously published TCR CDR3β signature of AH1-specific T-cell clones was used to determine the frequency of AH1-specific (tumor-specific) clones in pre- and posttreatment blood samples (72).
Flow Cytometry
A detailed list of the flow cytometry antibodies used can be found in Supplementary Table S10. Cell surface and intracellular (FOXP3) flow cytometry analyses of both mouse and human samples were performed as previously described (13). Briefly, cells were stained with the indicated mAbs in flow buffer (PBS-BSA 0.5%, for all cell surface antigens) or permeabilization buffer (eBioscience FoxP3/Transcription Factor Staining Buffer Set for all intracellular antigens) for a minimum of 45 minutes on ice. The gating strategy used for tumor-bearing mouse studies is as follows: for the assessment of CD40 on CD103+ and XCR+ cDC1 cells, cDC1 cells were defined as viable CD45+ CD3− Nkp46− CD19− Ly6C− MHC-II+ CD11c+ cells; Tregs were defined as viable CD45+ CD19− F4/80− CD11b− Ly-6G− CD4+ CD8− CD25+ FOXP3+ cells; CD8 Teff cells were defined as viable CD45+ CD19− F4/80− CD11b− Ly-6G− CD4− CD8+ CD62L− PD-1− cells; and CD8 MPECs were defined as viable CD45+ CD19− F4/80− CD11b− Ly-6G− CD4− CD8+ CD62L− PD-1− Slamf7+ CX3CR1− cells. The gating strategy used for SEB-reactive CD8 T-cell subsets in mouse studies is as follows: CD8 T cells were defined as viable CD8+ Vβ8+ cells; Teff cells were defined as viable CD8+ Vβ8+ CD44+ CD62L−; and MPECs were defined as viable CD8+ Vβ8+ CD44+ CD62L− PD-1− Slamf7+ CX3CR1− cells. Nonspecific T-cell subsets were defined as viable cells expressing CD8 and Vβ2 markers.
All cytometric analysis was conducted on a BD Symphony instrument (BD Biosciences) or LSRFortessa instrument (BD Biosciences). Flow cytometry data were analyzed using FlowJo v.10.8.2 software (Tree Star LLC) with plug-ins for T-distributed stochastic neighbor embedding. For SEA-stimulated human PBMC assays, t-distributed stochastic neighbor embedding analysis was performed in 1,008,000 live CD45+ cells using equal sampling cell numbers among triplicate samples from seven SEA-stimulated healthy donors treated with 10 μg/mL of botensilimab, parental IgG1, or isotype control (IgG1DLE) antibodies for 4 days, with 3,000 iterations, a perplexity of 30, and learning rate (eta) of 70560 (73). The gating strategy used for T-cell subsets from SEA-stimulated PBMC cultures is summarized in Supplementary Fig. S9F. CD8 T cells were defined as viable CD45+ CD3+ CD8+ cells. Tregs were defined as viable CD45+ CD3+ CD4+ CD25+ FOXP3+ cells, whereas non-Tregs were defined as viable CD45+ CD3+ CD4+ CD25− FoxP3− cells. NK cells were defined as viable CD45+ CD3− CD19− CD14− CD56+ cells.
For clinical samples from patients treated with botensilimab monotherapy, PBMCs were thawed, stained with Zombie Yellow Fixable Viability Dye (BioLegend) for 20 minutes at 4°C, and subsequently preincubated for 10 minutes at 4°C with 1% human serum (Sigma-Aldrich) to block Fc receptors. Cells were then incubated for 25 minutes at 4°C with 20 µg/mL of balstilimab (anti–PD-1, Agenus Inc) antibody, washed, and incubated for 25 minutes at 4°C with 10 µg/mL of biotin α-IgG4 antibody (Life Technologies). A second step of Fc receptor blocking was performed for 10 minutes at 4°C and cells stained for 20 minutes at 4°C with a combination of appropriately titrated antibodies directed against CD3 (UCHT1, BioLegend), CD4 (RPA-T4, BioLegend), CD45RO (UCHL1, BioLegend), CCR7 (G043H7, BioLegend), CXCR3 (G025H7, BioLegend), CXCR5 (J252D4, BioLegend), HLA-DR (L243, BioLegend), CD8 (RPA-T8, BD Bioscience), ICOS (DX29, BD Bioscience), Ki-67 (Clone B56, BD Biosciences), and streptavidin (BioLegend). Subsequently, the cells were fixed and acquired on a BD LSRFortessa cytometer (BD Biosciences) at Precision For Medicine. All patients provided written informed consent. Data were analyzed using FlowJo v.10.8.2 software (Tree Star LLC). For gating, T cells were defined as single viable CD3+ lymphocytes. CD4 and CD8 T cells were defined as CD4+ CD8− T cells and CD4− CD8+ T cells, respectively. For the CD4 T-cell subset, naïve, central memory, effector memory, and terminally differentiated effector memory CD4+ T cells were defined as CCR7+ CD45RO−, CCR7+ CD45RO+, CCR7− CD45RO+, and CCR7− CD45RO− cells. Tregs were defined as CD127low/− CD25+. The gating strategy is summarized in Supplementary Fig. S12.
IHC and Immunofluorescent Staining
CD8, FOXP3, and CD68 chromogenic triplex IHC staining was performed on 4-μm formalin-fixed, paraffin-embedded tissue sections on pretreatment and on-treatment biopsies from patients with advanced solid cancer and treated with 2 mg/kg botensilimab every 3 weeks monotherapy (n = 5 patients), 1 mg/kg botensilimab every 6 weeks in combination with 3 mg/kg balstilimab every 2 weeks (n = 3), or 2 mg/kg botensilimab every 6 weeks in combination with 3 mg/kg balstilimab every 2 weeks (n = 5) using Ventana Discovery Ultra Autostainer (Roche Diagnostics) according to the manufacturer’s instructions. All patients provided written informed consent. On-treatment biopsies were taken on cycle 2 day 1 for the every 6 weeks cohort or cycle 3 day 1 for the every 3 weeks cohort. Briefly, slides were baked for 1 hour at 60°C and deparaffinized followed by heat-induced epitope retrieval in Ventana Ultra Cell Conditioning Solution 1 (pH 8.0; Cat. # 950-224) at 90°C for 64 minutes. The slides were stained with primary antibodies listed in Supplementary Table S10 in the following sequence: CD68 (ready to use) → CD8 (ready to use) → FoxP3 (working concentration 1.5 μg/mL). OmniMap anti-Ms HRP (Roche/Ventana) and OmniMap anti-Rb HRP (Roche/Ventana) multimer secondary reagents or DISCOVERY anti-Rb (Roche/Ventana) and DISCOVERY anti-NP (Roche/Ventana) hapten secondary reagents were used together with DISCOVERY Chromogen kits (Roche/Ventana) for the detection of primary antibodies. Heat denaturation (100°C, 8 minutes) in ULTRA CC2 buffer (Roche/Ventana) was performed prior to CD8 and FOXP3 staining to remove antibodies deposited in the previous cycles. Hematoxylin (Roche/Ventana) and bluing reagent (Roche/Ventana) were used for counterstaining. Stained slides were scanned using an Aperio AT Turbo brightfield scanner and reviewed by a certified pathologist. Quantitative image analysis was performed using Halo (v3.6.4134.166) platform with a DenseNet (v2) algorithm. Areas identified as necrotic and irrelevant to the tumor, alongside regions affected by staining artifacts, and tissue folds were excluded from the analysis. Hematoxylin and eosin (H&E)–stained slides from serial sections were used to improve tissue annotation.
PD-L1 protein expression was evaluated in pretreatment biopsy specimens from patients with advanced colorectal cancer (n = 87), ovarian cancer (n = 19), and sarcoma (n = 23), using PD-L1 IHC 28-8 pharmDx qualitative IHC assay (Dako/Agilent, Cat. # SK005) according to the manufacturer’s instructions. All patients provided written informed consent. H&E staining was performed prior to the PD-L1 detection, and tumor presence was examined by a certified pathologist. Tissue samples with <100 tumor cells per slide were considered nonevaluable. PD-L1 expression in all evaluable samples was assessed by tumor proportion score, combined proportional score (CPS), and %PD-L1+ immune cell scoring. Tumor nonrelevant areas (normal and necrotic tissues) and tissue and staining artefacts were excluded from the analysis. Tumor proportion score was calculated as a number of PD-L1+ tumor cells of total tumor cells. %PD-L1+ immune cell was calculated as the percentage of PD-L1+ immune cells of total immune cells. CPS score was calculated using the following formula:
Mouse tumors were embedded in FSC 22 Frozen Section Media (Leica Biosystems) and freshly frozen on dry ice/ethanol slurry. The embedded tumors were sectioned at 10 μm and transferred either on the superfrost glass slides (Thermo Fisher Scientific) for the histology and eosin staining or glass coverslips (22 × 22 mm, Electron Microscopy) for the PhenoCycler multiplex staining using the Leica CM3050 S cryostat (Leica Biosystems). Coverslips were handled using Dumont coverslip forceps (Fine Science Tools) throughout the staining process. Mouse tumors were obtained from studies performed according to guidelines approved by the IACUC of Agenus Inc following the guidance of the PHS Policy.
Multiplex immunofluorescence was performed as previously described using the Akoya Biosciences CODEX platform (74). Briefly, 10-μm fresh frozen tissue sections were thawed at room temperature for 15 minutes and fixed with acetone for 10 minutes followed by 3× washing with hydration buffer. The sections were further fixed for another 10 minutes using 1.6% paraformaldehyde followed by 3× washing in the hydration buffer. Sections were then equilibrated in the staining buffer for 30 minutes. The coverslip was then stained with an antibody cocktail of the antibodies (CD3, CD4, and CD8; Supplementary Table S11) to a volume of 200 μL at 1:200 dilution overnight at 4°C in a sealed humidity chamber. After multiple washes with staining buffer, fixation steps were performed using 1.6% paraformaldehyde, 100% methanol, and fixation kit provided by Akoya Biosciences followed by 3× PBS washing. Then, the coverslip was transferred to the storage buffer. The reporter plate was designed using the fluorescent dyes which were also conjugated with the complimentary barcodes to match with the barcodes in the primary antibodies. Imaging was performed using a Keyence BZ-X800 inverted fluorescence microscope equipped with a 20× Plan Apo λ NA 0.75 objective (Nikon), an Akoya CODEX microfluidics instrument, and CODEX instrument manager v1.29.3.6 (Akoya Biosciences). Light exposure times and the arrangement of cycles are outlined in Supplementary Table S11. At the conclusion of the CODEX multicycle reaction, images were processed using CODEX Processor v1.7.0.6. H&E staining was performed on the corresponding serial sections, and images were acquired in brightfield mode using a Keyence BZ-X800 microscope. H&E staining slides were reviewed by a board-certified pathologist (Julie Feldstein, MD) to confirm the tumor content and any other pathologic changes.
Statistics
Statistical analyses were done using Prism 10.0 (GraphPad Software) and P values calculated using one-way ANOVA, two-way ANOVA, a Students t test, Mann–Whitney test, or Fishers exact test where indicated. For the purpose of multiple comparison adjustment, P values were adjusted based on the underlying statistical test as indicated in the figure legends and text elsewhere. Briefly, for mixed-effects model and two-way ANOVAs, the Tukey or Šídák multiple comparisons test was used. For one-way ANOVAs, the Tukey multiple comparisons or Kruskal–Wallis test with Dunn or Benjamini, Krieger, and Yekutieli correction was used. For pairwise comparisons of two groups, a Students’ t, Mann–Whitney, or Wilcoxon test was used. Survival analysis was done using Kaplan–Meier estimate with the clinical data cutoff of March 27, 2023. OS was censored at the last contact date unless a death was reported for a patient prior to the data cutoff date. The log-rank test was used to compare OS between groups with high (z-score > 0) and low (z-score ≤ 0) FCGR2A, FCGR2B, and FCGR3A gene expression and PD-L1 positivity by IHC using a CPS cutoff of 1. HRs with associated two-sided CIs were calculated using a Cox proportional hazards regression model, with a level of gene expression as a single covariate. All P values were two-sided with a level of significance set at P < 0.05. Comparisons of the proportion of patients were assessed in a 2 × 2 contingency table and using a Fisher exact test.
For clinical outcomes from the C-800-01 study (NCT03860272), data were evaluated as observed and presented, with no imputation method for missing values unless otherwise specified. Descriptive statistics were used in general to summarize trial results, including means, medians, ranges, and measures of variability. Qualitative variables were summarized by counts and percentages. The objective response rate (ORR) per investigator assessment was evaluated over the entire trial period according to RECIST 1.1. For a best overall response of a PR or CR, confirmation of the response was required. DCR was defined as a best response of a confirmed CR or PR or SD ≥6, 12, or 24 weeks.
Data Availability
RNA-seq data from patient tumor samples has been deposited to the Database of Genotypes and Phenotypes, under accession number phs003704.v1.p1. Deidentified individual participant clinical data, whole-exome sequencing data, and bulk RNA-seq data that underlie the results reported in this article are available for transfer upon request for academic use and within the limitations of the provided informed consent. Interested investigators can obtain and certify the data transfer agreement and submit requests to the principal investigator (D. Chand). Investigators who consent to the terms of the data transfer agreement, including, but not limited to, the use of these data only for research purposes, and to protect the confidentiality of the data and limit the possibility of identification of participants in any way whatsoever for the duration of the agreement, will be granted access. The raw DNA TCR sequence data have been deposited into the immuneACCESS project repository of the Adaptive Biotechnologies database (https://clients.adaptivebiotech.com/pub/delepine-2023-nm). The remaining data presented in the article are available within the article, Supplementary Information, and data files. No custom code has been used to produce data for this article.
Supplementary Material
This dataset contains the binding affinity measurements of the various mouse anti-CTLA-4 (αCTLA-4) antibodies to mouse Fc gamma receptors as determined by surface plasmon resonance. It also includes calculated activating to inhibitory (A/I) ratios for each antibody. This information is crucial for understanding the Fc-mediated effector functions of the αCTLA-4 antibodies used in the study, providing insight into their in vivo efficacy and mechanisms of action.
This dataset contains the binding affinity measurements of the botensilimab and the corresponding parental IgG1 anti-CTLA-4 antibody to human Fc gamma receptors as determined by surface plasmon resonance. It also includes calculated activating to inhibitory (A/I) ratios for each antibody.
This table presents a comparison of the binding epitopes of botensilimab, ipilimumab, and tremelimumab on human CTLA-4. It highlights the specific amino acid residues on CTLA-4 that are involved in the binding interface for each antibody. This comparison provides insight into binding characteristics of botensilimab relative to established anti-CTLA-4 therapies.
This dataset summarizes the toxicology findings from a 13-week repeat-dose study of botensilimab in cynomolgus monkeys. This comprehensive toxicology profile is crucial for assessing the safety of botensilimab, and is particularly relevant for predicting potential human toxicity.
This dataset presents the serum pharmacokinetic parameters of botensilimab in cynomolgus monkeys. The study includes both male and female monkeys, allowing for sex-based comparisons. These data are crucial for understanding botensilimab's behavior in the body and inform dose selection for clinical trials. The dataset provides insights into potential differences in drug metabolism between sexes and across different dose levels. Importantly, it also helps contextualize the toxicology findings, allowing for a more comprehensive assessment of the drug's safety and efficacy profile.
This dataset provides an overview of the baseline demographic characteristics of patients with advanced solid tumors who participated in the ongoing phase 1 trial of botensilimab. This demographic information is crucial for understanding the patient population studied and is valuable for interpreting the safety and efficacy outcomes of the study.
This dataset summarizes the clinical response and disease control rates of patients with advanced solid tumors treated in the ongoing phase 1 trial of botensilimab. These efficacy measures are crucial for assessing the therapeutic potential of botensilimab alone and in combination with balstilimab. Importantly, these data further highlight the therapeutic potential in patients who have progressed on prior immune checkpoint therapy, providing valuable insights into botensilimab's efficacy in this challenging-to-treat population.
This dataset summarizes the best overall response of each patient reported in the manuscript. It offers insights into clinical responses to botensilimab alone and in combination with balstilimab across various advanced solid tumor types. Importantly, it includes data on patients who progressed on prior conventional immune checkpoint therapy or have tumor types historically non-responsive to immunotherapy. By providing a patient-level view of treatment efficacy, this dataset allows for a nuanced understanding of how different tumor types and individual patients respond to botensilimab.
This dataset summarizes the grade 3 or higher treatment-related adverse events (TRAEs) observed in patients with advanced solid tumors treated in the ongoing phase 1 trial of botensilimab. This safety information is crucial for assessing the risk-benefit profile of botensilimab alone and in combination with balstilimab.
This key resource table provides a comprehensive list of all antibodies and proteins used in the study, thus allowing other researchers to accurately replicate or conduct similar studies using the same reagents.
This table provides technical information about the multiplex immunofluorescence studies performed using the Akoya Biosciences CODEX platform. This is crucial for the reproducibility and interpretation of the multiplex immunofluorescence results.
This dataset presents the binding characteristics of the mouse anti-CTLA-4 (αCTLA-4) antibodies to various mouse Fc gamma receptors (FcγRs). This information is crucial for understanding the Fc-mediated effector functions of the αCTLA-4 antibodies used in the study. The binding data help explain the mechanisms of action and potential efficacy differences between the various αCTLA-4 antibodies in mouse models.
This dataset presents comparative results demonstrating the enhanced efficacy of Fc-enhanced anti-CTLA-4 versus the parental anti-CTLA-4 antibody in various tumor-bearing mouse models. Data from multiple tumor-bearing mouse models, strengthens the generalizability of the findings across different tumor types. The individual mouse data allows for a detailed view of treatment responses, including, variability within treatment groups, timing and rate of tumor growth or regression and potential identification of exceptional responders or non-responders.
This dataset presents comparative results demonstrating the enhanced T cell modulation in the tumor microenvironment promoted by Fc-enhanced anti-CTLA-4 versus the parental anti-CTLA-4 antibody in two specific tumor-bearing mouse models: MC38 and CT26. These data demonstrate the superior ability of Fc-enhanced anti-CTLA-4 to promote T cell infiltration and Treg depletion in the tumor microenvironment. This information is crucial for understanding the mechanistic basis for the improved anti-tumor efficacy of Fc-enhanced anti-CTLA-4 antibodies.
This dataset presents comparative results demonstrating the superior ability of Fc-enhanced anti-CTLA-4 to potentiate antigen-specific T cell responses in vivo versus the parental anti-CTLA-4 antibody. The use of the Staphylococcal Enterotoxin B-challenged model in non-tumor bearing mice highlights the broader immunological effects of Fc-enhanced anti-CTLA-4 beyond the tumor microenvironment.
This dataset presents the pharmacokinetic (PK) profile of mouse Fc-enhanced anti-CTLA-4 antibody versus the parental anti-CTLA-4 antibody. The PK profile is important for interpreting the efficacy data, and further supports the conclusion that the enhanced anti-tumor effects are likely due to the engineered Fc-mediated functions rather than altered drug exposure or persistence in the body.
This dataset presents results demonstrating the broad combination activity of Fc-enhanced anti-CTLA-4 in various poorly immunogenic tumor-bearing mouse models. It demonstrates the potential of Fc-enhanced anti-CTLA-4 antibodies to improve outcomes in various combination treatment strategies, supporting their broad applicability in cancer immunotherapy.
This dataset provides detailed information on the binding characteristics and functional properties of botensilimab. This comprehensive dataset demonstrates botensilimab's high affinity and selectivity for CTLA-4, its ability to effectively block CTLA-4 interactions with its ligands comparable to that of the parental IgG1 anti-CTLA-4 antibody. Importantly, this data further supports that the superior functional attributes of botensilimab are due to the Fc-engineered region and not the variable region of the antibody.
This dataset provides a comprehensive analysis of the Fcγ receptor binding kinetics and Fc-mediated functions of botensilimab compared to other approved anti-CTLA-4 antibodies. These results highlight the unique Fc-mediated properties of botensilimab, particularly its enhanced engagement of activating Fcγ receptors without inducing complement-dependent cytotoxicity. This data further provides mechanistic insights into botensilimab's potential for enhanced immune modulation and anti-tumor efficacy.
This dataset presents results showing the effect of botensilimab on CD4+ non-regulatory T cells (non-Tregs) in stimulated human peripheral blood mononuclear cell (PBMC) cultures. It demonstrates that unlike conventional anti-CTLA-4, botensilimab treatment does not lead to a decrease in CD4+ non-Treg populations in stimulated human PBMC cultures. This information is crucial for understanding botensilimab's unique mechanism of action and its potential advantages over conventional IgG1 anti-CTLA-4 antibodies. Importantly, these results further support the hypothesis that under context of antigen-stimulation, botensilimab decreases the generation of Tregs rather than promoting depletion via antibody-dependent cellular cytotoxicity. This mechanism distinguishes botensilimab from conventional anti-CTLA-4 antibodies and may account for its superior T cell responsiveness and activation.
This dataset provides important insights into botensilimab's effects on the myeloid compartment of the immune system. The increase in CD11c+ HLA-DR+ myeloid cells, which likely represent antigen-presenting cells, suggests that botensilimab may enhance antigen presentation and T cell activation. The lack of effect on CD16- myeloid cells indicates a selective impact on specific myeloid subsets. These findings contribute to understanding botensilimab's unique mechanism of action, suggesting it may enhance anti-tumor immunity not only through effects on T cells but also by modulating myeloid cell populations.
This CONSORT flow diagram for the expanded phase I clinical trial of botensilimab is essential for assessing the validity and generalizability of the trial results. It allows readers to understand how many patients were initially considered, how many were actually treated, and how many contributed to the biomarker analyses. This transparency is essential for interpreting the strength of the evidence supporting botensilimab's efficacy and safety in advanced solid cancers.
This figure describes the flow cytometry gating strategy used for T cell immune profiling in cryopreserved peripheral blood mononuclear cells (PBMCs) from patients with advanced solid cancers treated with botensilimab. The gating strategy allows for a comprehensive analysis of various CD4+ T cell subpopulations.
This dataset presents additional evidence of botensilimab's immunological effects on peripheral T cell activation in patients with advanced solid cancers. It demonstrates dose-dependent increases in key T cell activation markers. These findings provide further insight into botensilimab's mechanism of action, offer potential biomarkers for treatment monitoring, and support its dose-response relationship. The data further help researchers and clinicians better understand botensilimab's immunomodulatory potential in cancer treatment.
This dataset presents analysis of intratumoral regulatory T cells (Tregs) and CD8+ T cells in tumor biopsies from patients with advanced solid cancers before and after botensilimab treatment. Importantly, these data further support and confirm the changes previously reported by deconvolution analyses of RNA sequencing data. This corroboration between protein-level staining and transcriptomic analyses provides robust evidence of botensilimab's effects on the tumor immune microenvironment, offering valuable insights into its mechanism of action and potential to enhance anti-tumor immunity.
This dataset shows that response to botensilimab, alone or with balstilimab, in patients with advanced cancers is independent of tumor neoantigen burden and FcγRIIIA allele status. This suggests that botensilimab's efficacy may be broad and not limited by these commonly used biomarkers typically associated with response to conventional immune checkpoint therapy. The data highlights a potential advantage of botensilimab over traditional immunotherapies, indicating it might be effective in a wider range of patients, including those who may not typically respond to standard checkpoint inhibitors.
This dataset examines the relationship between FCGR2A and FCGR3A gene expression and clinical response to anti-PD-1 or conventional anti-CTLA-4 therapies in patients with advanced melanoma. Analysis of multiple clinical trial datasets shows that response to these conventional immunotherapies is not associated with FCGR2A or FCGR3A expression levels. This contrasts with botensilimab's efficacy profile, supporting its unique Fc-dependent mechanism of action. The data further support that FCGR2A and FCGR3A expression could be predictive biomarkers of response to botensilimab, distinguishing it from conventional immunotherapies.
This dataset analyzes the relationship between FCGR2A and FCGR3A gene expression and immune cell type enrichment in tumor biopsies from patients treated with botensilimab alone or in combination with balstilimab. The analysis provides insights into the immune landscape associated with FCGR2A and FCGR3A expression in the tumor microenvironment, suggesting potential cellular mechanisms underlying botensilimab's efficacy. This data supports the role of FCGR2A and FCGR3A as potential biomarkers and helps elucidate how their expression may relate to specific immune cell infiltration patterns relevant to botensilimab's unique Fc-dependent mechanism of action.
Acknowledgments
The study was sponsored by Agenus Inc We are grateful to the patients who were recruited in our study and their families and study teams who participated in the C-800-01 clinical trial. We thank Vidur Patel and Sudesh Pawaria for assistance with mouse B16F1.OVA studies. We are grateful to the Cancer Discovery editorial team for their feedback.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
C. Delepine reports employment with Agenus Inc and ownership of stocks of the company. A. Tanne reports employment with and being a share holder of Agenus Inc at the time he worked on this project. H. Han reports grants from Agenus Inc during the conduct of the study. S. Bourdelais reports other support from Agenus Inc outside the submitted work. J.D. Waight reports personal fees from Agenus Inc during the conduct of the study. A. Sebastin-Yague reports personal fees from Agenus Inc and MiNK Therapeutics outside the submitted work. A. Slee reports employment with Agenus Inc. J.E. Grossman reports other support from Agenus Inc during the conduct of the study. N.S. Wilson reports a patent for https://patents.google.com/patent/US11013802B2/en issued and a patent for https://patents.google.com/patent/US10323091B2/en issued. D.D. Von Hoff reports grants from Agenus Inc during the conduct of the study; other support from multiple entities outside the submitted work; and being a consultant for Agenus Inc in the past but not currently when he worked on this project. J. Stebbing reports other support from Agenus Inc from 2022 till date during the conduct of the study; being editor-in-chief in Oncogene; being on SABs/advisory boards for Eli Lilly and Company, Agenus Inc, Celltrion, Greenmantle, vTv Therapeutics, APIM, Onconox, IO Labs, Clinical ink, Zephyr AI, BenevolentAI, Biozen, Pear Bio, Sable Bio, and LinkGevity; receiving consulting fees from Lansdowne Partners and Vitruvian; being a board member for Graviton Bioscience BV, etira Therapeutics, and Portage; and being on the board of BB Biotech Healthcare Trust PLC previously. T.J. Curiel reports personal fees and other support from Agenus Inc during the conduct of the study. R.B. Stein reports personal fees from Agenus Inc during the conduct of the study; personal fees from Samsara Biocapital, ImmunoGenesis, Flame Biosciences, PolyPid, MiroBio, Caliper Life Sciences, Codify Therapeutics, XOMA, The Ohio State University DDI, Washington University School of Medicine in St. Louis, and MiNK Therapeutics outside the submitted work; and a patent for Checkpoint Modulator Antibodies pending. No disclosures were reported by the other authors.
Authors’ Contributions
D. Chand, N.S. Wilson, and R.B. Stein: Conceived the project. D. Chand, D.A. Savitsky, J.D. Waight, D.L. Levey, and J.M. Patel: Conceptualized the experiments. M.K. Wilkens, S. Vincent, C. Galand, A. Tanne, H. Han, D.A. Savitsky, S. Ng, M. Marques, and D. Chand: Designed and performed mouse in vivo studies. D. Chand, S. Krishnan, D.A. Savitsky, M. Manrique, R.B. Gombos, R. Ward, and M.R. Costa: Performed cell-based assays. P. Garcia-Broncano, S. Krishnan, Y. Qin, and K.T. Soh: Performed flow cytometry. G. Mednick, A. Sebastian-Yague, and C. Delepine: Performed computational data analyses. B. Joshi and J.G. Keith: Performed multiplex imaging experiments and analyses. J. Ridpath, O. Huber, V. Venkatraman, M. Pupecka-Swider, and B. Morin: Performed surface plasmon resonance analyses. A. Slee: Designed and supervised the nonhuman primate studies. W. Wu, N.-P. Rudqvist, S. Bourdelais, and O. Udartseva: Contributed to data analyses. S.J. O’Day and J.E. Grossman: Led the clinical trial. D.D. Von Hoff, J.S. Buell, R.B. Stein, N.S. Wilson, J. Stebbing, and T.J. Curiel: Provided critical insights and intellectual input during the studies. D. Chand and T.J. Curiel: Wrote the manuscript. D. Chand: Supervised the project.
References
- 1. Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med 1995;182:459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995;3:541–7. [DOI] [PubMed] [Google Scholar]
- 3. Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 1995;270:985–8. [DOI] [PubMed] [Google Scholar]
- 4. Azuma M, Ito D, Yagita H, Okumura K, Phillips JH, Lanier LL, et al. B70 antigen is a second ligand for CTLA-4 and CD28. Nature 1993;366:76–9. [DOI] [PubMed] [Google Scholar]
- 5. Hathcock KS, Laszlo G, Dickler HB, Bradshaw J, Linsley P, Hodes RJ. Identification of an alternative CTLA-4 ligand costimulatory for T cell activation. Science 1993;262:905–7. [DOI] [PubMed] [Google Scholar]
- 6. Walker LSK. Treg and CTLA-4: two intertwining pathways to immune tolerance. J Autoimmun 2013;45:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996;271:1734–6. [DOI] [PubMed] [Google Scholar]
- 8. Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res 2013;1:32–42. [DOI] [PubMed] [Google Scholar]
- 9. Lonberg N, Korman AJ. Masterful antibodies: checkpoint blockade. Cancer Immunol Res 2017;5:275–81. [DOI] [PubMed] [Google Scholar]
- 10. Sharma A, Subudhi SK, Blando J, Scutti J, Vence L, Wargo J, et al. Anti-CTLA-4 immunotherapy does not deplete FOXP3+ regulatory T cells (Tregs) in human cancers. Clin Cancer Res 2019;25:1233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel TH, Brewer JR, Fan J, Cheng J, Shen Y-L, Xiang Y, et al. FDA approval summary: tremelimumab in combination with durvalumab for the treatment of patients with unresectable hepatocellular carcinoma. Clin Cancer Res 2024;30:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan S, Burt DJ, Ralph C, Thistlethwaite FC, Hawkins RE, Elkord E. Tremelimumab (anti-CTLA4) mediates immune responses mainly by direct activation of T effector cells rather than by affecting T regulatory cells. Clin Immunol 2011;138:85–96. [DOI] [PubMed] [Google Scholar]
- 13. Waight JD, Chand D, Dietrich S, Gombos R, Horn T, Gonzalez AM, et al. Selective FcγR co-engagement on APCs modulates the activity of therapeutic antibodies targeting T cell antigens. Cancer Cell 2018;33:1033–47.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol 2008;26:1789–96. [DOI] [PubMed] [Google Scholar]
- 15. Arce Vargas F, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell 2018;33:649–63.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romano E, Kusio-Kobialka M, Foukas PG, Baumgaertner P, Meyer C, Ballabeni P, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci U S A 2015;112:6140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee H, Ferguson AL, Quek C, Vergara IA, Pires daSilva I, Allen R, et al. Intratumoral CD16+ macrophages are associated with clinical outcomes of patients with metastatic melanoma treated with combination anti-PD-1 and anti-CTLA-4 therapy. Clin Cancer Res 2023;29:2513–24. [DOI] [PubMed] [Google Scholar]
- 18. Wilky B, El-Khoueiry A, Bullock A, Tsimberidou A, Mahadevan D, Margolin K, et al. 778 Botensilimab, a novel innate/adaptive immune activator, plus or minus balstilimab (anti-PD-1) in “cold” and I-O refractory metastatic solid tumors. J Immunother Cancer 2022;10:A810–10. [Google Scholar]
- 19. Bullock A, Fakih M, Gordon M, Tsimberidou A, El-Khoueiry A, Wilky B, et al. LBA-4 Results from an expanded phase 1 trial of botensilimab (BOT), a multifunctional anti-CTLA-4, plus balstilimab (BAL; anti-PD-1) for metastatic heavily pretreated microsatellite stable colorectal cancer (MSS CRC). Ann Oncol 2023;34:S178–9. [Google Scholar]
- 20. Wilky BA, Trent JC, Gordon MS, El-Khoueiry AB, Bullock AJ, Henick B, et al. Efficacy and safety of botensilimab (BOT) plus balstilimab (BAL) in patients (pts) with refractory metastatic sarcoma. Ann Oncol 2023;35:S1031–61. [Google Scholar]
- 21. Yofe I, Landsberger T, Yalin A, Solomon I, Costoya C, Demane DF, et al. Anti-CTLA-4 antibodies drive myeloid activation and reprogram the tumor microenvironment through FcγR engagement and type I interferon signaling. Nat Cancer 2022;3:1336–50. [DOI] [PubMed] [Google Scholar]
- 22. Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci U S A 2006;103:4005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science 2005;310:1510–2. [DOI] [PubMed] [Google Scholar]
- 24. Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med 2014;6:238ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudqvist N-P, Pilones KA, Lhuillier C, Wennerberg E, Sidhom J-W, Emerson RO, et al. Radiotherapy and CTLA-4 blockade shape the TCR repertoire of tumor-infiltrating T cells. Cancer Immunol Res 2018;6:139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci U S A 1996;93:9730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyahara Y, Khattar M, Schroder PM, Mierzejewska B, Deng R, Han R, et al. Anti-TCRβ mAb induces long-term allograft survival by reducing antigen-reactive T cells and sparing regulatory T cells. Am J Transplant 2012;12:1409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang L, TeSlaa T, Ng S, Nofal M, Wang L, Lan T, et al. Ketogenic diet and chemotherapy combine to disrupt pancreatic cancer metabolism and growth. Med 2022;3:119–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015;372:2521–32. [DOI] [PubMed] [Google Scholar]
- 30. Newman MJ, Wu JY, Gardner BH, Munroe KJ, Leombruno D, Recchia J, et al. Saponin adjuvant induction of ovalbumin-specific CD8+ cytotoxic T lymphocyte responses. J Immunol 1992;148:2357–62. [PubMed] [Google Scholar]
- 31. Miconnet I, Koenig S, Speiser D, Krieg A, Guillaume P, Cerottini J-C, et al. CpG are efficient adjuvants for specific CTL induction against tumor antigen-derived peptide. J Immunol 2002;168:1212–8. [DOI] [PubMed] [Google Scholar]
- 32. Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 2013;210:1695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He M, Chai Y, Qi J, Zhang CWH, Tong Z, Shi Y, et al. Remarkably similar CTLA-4 binding properties of therapeutic ipilimumab and tremelimumab antibodies. Oncotarget 2017;8:67129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gombos RB, Gonzalez A, Manrique M, Chand D, Savitsky D, Morin B, et al. Toxicological and pharmacological assessment of AGEN1884, a novel human IgG1 anti-CTLA-4 antibody. PLoS One 2018;13:e0191926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, et al. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci U S A 2017;114:E4223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee JY, Lee HT, Shin W, Chae J, Choi J, Kim SH, et al. Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy. Nat Commun 2016;7:13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009;113:3716–25. [DOI] [PubMed] [Google Scholar]
- 38. Iwama S, De Remigis A, Callahan MK, Slovin SF, Wolchok JD, Caturegli P. Pituitary expression of CTLA-4 mediates hypophysitis secondary to administration of CTLA-4 blocking antibody. Sci Transl Med 2014;6:230ra45. [DOI] [PubMed] [Google Scholar]
- 39. Levy R, Rotfogel Z, Hillman D, Popugailo A, Arad G, Supper E, et al. Superantigens hyperinduce inflammatory cytokines by enhancing the B7-2/CD28 costimulatory receptor interaction. Proc Natl Acad Sci U S A 2016;113:E6437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zappasodi R, Serganova I, Cohen IJ, Maeda M, Shindo M, Senbabaoglu Y, et al. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature 2021;591:652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valk E, Rudd CE, Schneider H. CTLA-4 trafficking and surface expression. Trends Immunol 2008;29:272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morandi B, Bougras G, Muller WA, Ferlazzo G, Münz C. NK cells of human secondary lymphoid tissues enhance T cell polarization via IFN-gamma secretion. Eur J Immunol 2006;36:2394–400. [DOI] [PubMed] [Google Scholar]
- 43. Bullock AJ, Schlechter BL, Fakih MG, Tsimberidou AM, Grossman JE, Gordon MS, et al. Botensilimab plus balstilimab in relapsed/refractory microsatellite stable metastatic colorectal cancer: a phase 1 trial. Nat Med 2024;30:2558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O’Malley DM, Oaknin A, Monk BJ, Selle F, Rojas C, Gladieff L, et al. Phase II study of the safety and efficacy of the anti-PD-1 antibody balstilimab in patients with recurrent and/or metastatic cervical cancer. Gynecol Oncol 2021;163:274–80. [DOI] [PubMed] [Google Scholar]
- 45. Fecher LA, Agarwala SS, Hodi FS, Weber JS. Ipilimumab and its toxicities: a multidisciplinary approach. Oncologist 2013;18:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jessel S, Weiss SA, Austin M, Mahajan A, Etts K, Zhang L, et al. Immune checkpoint inhibitor-induced hypophysitis and patterns of loss of pituitary function. Front Oncol 2022;12:836859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Di Dalmazi G, Ippolito S, Lupi I, Caturegli P. Hypophysitis induced by immune checkpoint inhibitors: a 10-year assessment. Expert Rev Endocrinol Metab 2019;14:381–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liakou CI, Kamat A, Tang DN, Chen H, Sun J, Troncoso P, et al. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci U S A 2008;105:14987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ng Tang D, Shen Y, Sun J, Wen S, Wolchok JD, Yuan J, et al. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol Res 2013;1:229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Subrahmanyam PB, Dong Z, Gusenleitner D, Giobbie-Hurder A, Severgnini M, Zhou J, et al. Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. J Immunother Cancer 2018;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robert L, Tsoi J, Wang X, Emerson R, Homet B, Chodon T, et al. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res 2014;20:2424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kavanagh B, O’Brien S, Lee D, Hou Y, Weinberg V, Rini B, et al. CTLA4 blockade expands FoxP3+ regulatory and activated effector CD4+ T cells in a dose-dependent fashion. Blood 2008;112:1175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Campbell KM, Amouzgar M, Pfeiffer SM, Howes TR, Medina E, Travers M, et al. Prior anti-CTLA-4 therapy impacts molecular characteristics associated with anti-PD-1 response in advanced melanoma. Cancer Cell 2023;41:791–806.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019;35:238–55.e6. [DOI] [PubMed] [Google Scholar]
- 58. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li F, Li C, Cai X, Xie Z, Zhou L, Cheng B, et al. The association between CD8+ tumor-infiltrating lymphocytes and the clinical outcome of cancer immunotherapy: a systematic review and meta-analysis. EClinicalMedicine 2021;41:101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Korman AJ, Garrett-Thomson SC, Lonberg N. The foundations of immune checkpoint blockade and the ipilimumab approval decennial. Nat Rev Drug Discov 2022;21:509–28. [DOI] [PubMed] [Google Scholar]
- 61. Chen EX, Jonker DJ, Loree JM, Kennecke HF, Berry SR, Couture F, et al. Effect of combined immune checkpoint inhibition vs best supportive care alone in patients with advanced colorectal cancer: the Canadian cancer trials group CO.26 study. JAMA Oncol 2020;6:831–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol 2006;7:803–9. [DOI] [PubMed] [Google Scholar]
- 63. House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. [DOI] [PubMed] [Google Scholar]
- 64. Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014;515:572–6. [DOI] [PubMed] [Google Scholar]
- 65. Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, et al. Expansion and activation of CD103+ dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 2016;44:924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakaseko C, Miyatake S, Iida T, Hara S, Abe R, Ohno H, et al. Cytotoxic T lymphocyte antigen 4 (CTLA-4) engagement delivers an inhibitory signal through the membrane-proximal region in the absence of the tyrosine motif in the cytoplasmic tail. J Exp Med 1999;190:765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Breous-Nystrom E, Schultze K, Meier M, Flueck L, Holzer C, Boll M, et al. Retrocyte Display® technology: generation and screening of a high diversity cellular antibody library. Methods 2014;65:57–67. [DOI] [PubMed] [Google Scholar]
- 68. Van Dijk M, Mundt CA, Ritter G, Schaer D, Wolchok JD, Merghoub T, et al. Anti-CTLA-4 antibodies and methods of use thereof. United States patent US 11013802. 2021. [Google Scholar]
- 69. Van Dijk M, Wilson NS, Mundt CA, Ritter G, Wolchok JD, Merghoub T, et al. Anti-PD-1 antibodies and methods of use thereof. United States patent US 10323091. 2019. [Google Scholar]
- 70. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38:276–8. [DOI] [PubMed] [Google Scholar]
- 72. Rudqvist N-P, Charpentier M, Lhuillier C, Wennerberg E, Spada S, Sheridan C, et al. Immunotherapy targeting different immune compartments in combination with radiation therapy induces regression of resistant tumors. Nat Commun 2023;14:5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Belkina AC, Ciccolella CO, Anno R, Halpert R, Spidlen J, Snyder-Cappione JE. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun 2019;10:5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schurch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 2020;182:1341–59.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This dataset contains the binding affinity measurements of the various mouse anti-CTLA-4 (αCTLA-4) antibodies to mouse Fc gamma receptors as determined by surface plasmon resonance. It also includes calculated activating to inhibitory (A/I) ratios for each antibody. This information is crucial for understanding the Fc-mediated effector functions of the αCTLA-4 antibodies used in the study, providing insight into their in vivo efficacy and mechanisms of action.
This dataset contains the binding affinity measurements of the botensilimab and the corresponding parental IgG1 anti-CTLA-4 antibody to human Fc gamma receptors as determined by surface plasmon resonance. It also includes calculated activating to inhibitory (A/I) ratios for each antibody.
This table presents a comparison of the binding epitopes of botensilimab, ipilimumab, and tremelimumab on human CTLA-4. It highlights the specific amino acid residues on CTLA-4 that are involved in the binding interface for each antibody. This comparison provides insight into binding characteristics of botensilimab relative to established anti-CTLA-4 therapies.
This dataset summarizes the toxicology findings from a 13-week repeat-dose study of botensilimab in cynomolgus monkeys. This comprehensive toxicology profile is crucial for assessing the safety of botensilimab, and is particularly relevant for predicting potential human toxicity.
This dataset presents the serum pharmacokinetic parameters of botensilimab in cynomolgus monkeys. The study includes both male and female monkeys, allowing for sex-based comparisons. These data are crucial for understanding botensilimab's behavior in the body and inform dose selection for clinical trials. The dataset provides insights into potential differences in drug metabolism between sexes and across different dose levels. Importantly, it also helps contextualize the toxicology findings, allowing for a more comprehensive assessment of the drug's safety and efficacy profile.
This dataset provides an overview of the baseline demographic characteristics of patients with advanced solid tumors who participated in the ongoing phase 1 trial of botensilimab. This demographic information is crucial for understanding the patient population studied and is valuable for interpreting the safety and efficacy outcomes of the study.
This dataset summarizes the clinical response and disease control rates of patients with advanced solid tumors treated in the ongoing phase 1 trial of botensilimab. These efficacy measures are crucial for assessing the therapeutic potential of botensilimab alone and in combination with balstilimab. Importantly, these data further highlight the therapeutic potential in patients who have progressed on prior immune checkpoint therapy, providing valuable insights into botensilimab's efficacy in this challenging-to-treat population.
This dataset summarizes the best overall response of each patient reported in the manuscript. It offers insights into clinical responses to botensilimab alone and in combination with balstilimab across various advanced solid tumor types. Importantly, it includes data on patients who progressed on prior conventional immune checkpoint therapy or have tumor types historically non-responsive to immunotherapy. By providing a patient-level view of treatment efficacy, this dataset allows for a nuanced understanding of how different tumor types and individual patients respond to botensilimab.
This dataset summarizes the grade 3 or higher treatment-related adverse events (TRAEs) observed in patients with advanced solid tumors treated in the ongoing phase 1 trial of botensilimab. This safety information is crucial for assessing the risk-benefit profile of botensilimab alone and in combination with balstilimab.
This key resource table provides a comprehensive list of all antibodies and proteins used in the study, thus allowing other researchers to accurately replicate or conduct similar studies using the same reagents.
This table provides technical information about the multiplex immunofluorescence studies performed using the Akoya Biosciences CODEX platform. This is crucial for the reproducibility and interpretation of the multiplex immunofluorescence results.
This dataset presents the binding characteristics of the mouse anti-CTLA-4 (αCTLA-4) antibodies to various mouse Fc gamma receptors (FcγRs). This information is crucial for understanding the Fc-mediated effector functions of the αCTLA-4 antibodies used in the study. The binding data help explain the mechanisms of action and potential efficacy differences between the various αCTLA-4 antibodies in mouse models.
This dataset presents comparative results demonstrating the enhanced efficacy of Fc-enhanced anti-CTLA-4 versus the parental anti-CTLA-4 antibody in various tumor-bearing mouse models. Data from multiple tumor-bearing mouse models, strengthens the generalizability of the findings across different tumor types. The individual mouse data allows for a detailed view of treatment responses, including, variability within treatment groups, timing and rate of tumor growth or regression and potential identification of exceptional responders or non-responders.
This dataset presents comparative results demonstrating the enhanced T cell modulation in the tumor microenvironment promoted by Fc-enhanced anti-CTLA-4 versus the parental anti-CTLA-4 antibody in two specific tumor-bearing mouse models: MC38 and CT26. These data demonstrate the superior ability of Fc-enhanced anti-CTLA-4 to promote T cell infiltration and Treg depletion in the tumor microenvironment. This information is crucial for understanding the mechanistic basis for the improved anti-tumor efficacy of Fc-enhanced anti-CTLA-4 antibodies.
This dataset presents comparative results demonstrating the superior ability of Fc-enhanced anti-CTLA-4 to potentiate antigen-specific T cell responses in vivo versus the parental anti-CTLA-4 antibody. The use of the Staphylococcal Enterotoxin B-challenged model in non-tumor bearing mice highlights the broader immunological effects of Fc-enhanced anti-CTLA-4 beyond the tumor microenvironment.
This dataset presents the pharmacokinetic (PK) profile of mouse Fc-enhanced anti-CTLA-4 antibody versus the parental anti-CTLA-4 antibody. The PK profile is important for interpreting the efficacy data, and further supports the conclusion that the enhanced anti-tumor effects are likely due to the engineered Fc-mediated functions rather than altered drug exposure or persistence in the body.
This dataset presents results demonstrating the broad combination activity of Fc-enhanced anti-CTLA-4 in various poorly immunogenic tumor-bearing mouse models. It demonstrates the potential of Fc-enhanced anti-CTLA-4 antibodies to improve outcomes in various combination treatment strategies, supporting their broad applicability in cancer immunotherapy.
This dataset provides detailed information on the binding characteristics and functional properties of botensilimab. This comprehensive dataset demonstrates botensilimab's high affinity and selectivity for CTLA-4, its ability to effectively block CTLA-4 interactions with its ligands comparable to that of the parental IgG1 anti-CTLA-4 antibody. Importantly, this data further supports that the superior functional attributes of botensilimab are due to the Fc-engineered region and not the variable region of the antibody.
This dataset provides a comprehensive analysis of the Fcγ receptor binding kinetics and Fc-mediated functions of botensilimab compared to other approved anti-CTLA-4 antibodies. These results highlight the unique Fc-mediated properties of botensilimab, particularly its enhanced engagement of activating Fcγ receptors without inducing complement-dependent cytotoxicity. This data further provides mechanistic insights into botensilimab's potential for enhanced immune modulation and anti-tumor efficacy.
This dataset presents results showing the effect of botensilimab on CD4+ non-regulatory T cells (non-Tregs) in stimulated human peripheral blood mononuclear cell (PBMC) cultures. It demonstrates that unlike conventional anti-CTLA-4, botensilimab treatment does not lead to a decrease in CD4+ non-Treg populations in stimulated human PBMC cultures. This information is crucial for understanding botensilimab's unique mechanism of action and its potential advantages over conventional IgG1 anti-CTLA-4 antibodies. Importantly, these results further support the hypothesis that under context of antigen-stimulation, botensilimab decreases the generation of Tregs rather than promoting depletion via antibody-dependent cellular cytotoxicity. This mechanism distinguishes botensilimab from conventional anti-CTLA-4 antibodies and may account for its superior T cell responsiveness and activation.
This dataset provides important insights into botensilimab's effects on the myeloid compartment of the immune system. The increase in CD11c+ HLA-DR+ myeloid cells, which likely represent antigen-presenting cells, suggests that botensilimab may enhance antigen presentation and T cell activation. The lack of effect on CD16- myeloid cells indicates a selective impact on specific myeloid subsets. These findings contribute to understanding botensilimab's unique mechanism of action, suggesting it may enhance anti-tumor immunity not only through effects on T cells but also by modulating myeloid cell populations.
This CONSORT flow diagram for the expanded phase I clinical trial of botensilimab is essential for assessing the validity and generalizability of the trial results. It allows readers to understand how many patients were initially considered, how many were actually treated, and how many contributed to the biomarker analyses. This transparency is essential for interpreting the strength of the evidence supporting botensilimab's efficacy and safety in advanced solid cancers.
This figure describes the flow cytometry gating strategy used for T cell immune profiling in cryopreserved peripheral blood mononuclear cells (PBMCs) from patients with advanced solid cancers treated with botensilimab. The gating strategy allows for a comprehensive analysis of various CD4+ T cell subpopulations.
This dataset presents additional evidence of botensilimab's immunological effects on peripheral T cell activation in patients with advanced solid cancers. It demonstrates dose-dependent increases in key T cell activation markers. These findings provide further insight into botensilimab's mechanism of action, offer potential biomarkers for treatment monitoring, and support its dose-response relationship. The data further help researchers and clinicians better understand botensilimab's immunomodulatory potential in cancer treatment.
This dataset presents analysis of intratumoral regulatory T cells (Tregs) and CD8+ T cells in tumor biopsies from patients with advanced solid cancers before and after botensilimab treatment. Importantly, these data further support and confirm the changes previously reported by deconvolution analyses of RNA sequencing data. This corroboration between protein-level staining and transcriptomic analyses provides robust evidence of botensilimab's effects on the tumor immune microenvironment, offering valuable insights into its mechanism of action and potential to enhance anti-tumor immunity.
This dataset shows that response to botensilimab, alone or with balstilimab, in patients with advanced cancers is independent of tumor neoantigen burden and FcγRIIIA allele status. This suggests that botensilimab's efficacy may be broad and not limited by these commonly used biomarkers typically associated with response to conventional immune checkpoint therapy. The data highlights a potential advantage of botensilimab over traditional immunotherapies, indicating it might be effective in a wider range of patients, including those who may not typically respond to standard checkpoint inhibitors.
This dataset examines the relationship between FCGR2A and FCGR3A gene expression and clinical response to anti-PD-1 or conventional anti-CTLA-4 therapies in patients with advanced melanoma. Analysis of multiple clinical trial datasets shows that response to these conventional immunotherapies is not associated with FCGR2A or FCGR3A expression levels. This contrasts with botensilimab's efficacy profile, supporting its unique Fc-dependent mechanism of action. The data further support that FCGR2A and FCGR3A expression could be predictive biomarkers of response to botensilimab, distinguishing it from conventional immunotherapies.
This dataset analyzes the relationship between FCGR2A and FCGR3A gene expression and immune cell type enrichment in tumor biopsies from patients treated with botensilimab alone or in combination with balstilimab. The analysis provides insights into the immune landscape associated with FCGR2A and FCGR3A expression in the tumor microenvironment, suggesting potential cellular mechanisms underlying botensilimab's efficacy. This data supports the role of FCGR2A and FCGR3A as potential biomarkers and helps elucidate how their expression may relate to specific immune cell infiltration patterns relevant to botensilimab's unique Fc-dependent mechanism of action.
Data Availability Statement
RNA-seq data from patient tumor samples has been deposited to the Database of Genotypes and Phenotypes, under accession number phs003704.v1.p1. Deidentified individual participant clinical data, whole-exome sequencing data, and bulk RNA-seq data that underlie the results reported in this article are available for transfer upon request for academic use and within the limitations of the provided informed consent. Interested investigators can obtain and certify the data transfer agreement and submit requests to the principal investigator (D. Chand). Investigators who consent to the terms of the data transfer agreement, including, but not limited to, the use of these data only for research purposes, and to protect the confidentiality of the data and limit the possibility of identification of participants in any way whatsoever for the duration of the agreement, will be granted access. The raw DNA TCR sequence data have been deposited into the immuneACCESS project repository of the Adaptive Biotechnologies database (https://clients.adaptivebiotech.com/pub/delepine-2023-nm). The remaining data presented in the article are available within the article, Supplementary Information, and data files. No custom code has been used to produce data for this article.