Abstract
Covering 2010–April 2024
There have been tremendous new discoveries and developments since 2010 in anticancer research based on marine cyanobacteria. Marine cyanobacteria are prolific sources of anticancer natural products, including the tubulin agents dolastatins 10 and 15 which were originally isolated from a mollusk that feeds on cyanobacteria. Decades of research have culminated in the approval of six antibody-drug conjugates (ADCs) and many ongoing clinical trials. Antibody conjugation has been enabling for several natural products, particularly cyanobacterial cytotoxins. Targeting tubulin dynamics has been a major strategy, leading to the discovery of the gatorbulin scaffold, acting on a new pharmacological site. Cyanobacterial compounds with different mechanisms of action (MOA), targeting novel or validated targets in a range of organelles, also show promise as anticancer agents. Important advances include the development of compounds with novel MOA, including apratoxin and coibamide A analogues, modulating cotranslational translocation at the level of Sec61 in the endoplasmic reticulum, largazole and santacruzamate A targeting class I histone deacetylases, and proteasome inhibitors based on carmaphycins, resembling the approved drug carfilzomib. The pipeline extends with SERCA inhibitors, mitochondrial cytotoxins and membrane-targeting agents, which have not yet advanced clinically since the biology is less understood and selectivity concerns remain to be addressed. In addition, efforts have also focused on the identification of chemosensitizing and antimetastatic agents. The review covers the state of current knowledge of marine cyanobacteria as anticancer agents with a focus on the mechanism, target identification and potential for drug development. We highlight the importance of solving the supply problem through chemical synthesis as well as illuminating the biological activity and in-depth mechanistic studies to increase the value of cyanobacterial natural products to catalyze their development.
We describe the pipeline of anticancer agents from marine cyanobacteria, highlighting critical steps from discovery towards development, including the identification of the molecular target and mechanism of action, and solving the supply problem.
1. Introduction
Natural products have played a key role in delivering drug candidates or provided the inspiration for the development of agents with new targets or new mechanisms of action, especially for infectious diseases and cancer.1–3 Historically, roughly 50% of all drugs are inspired by natural products and for cancer even 60%.3 Critical to the continuous success of natural products is the identification of biological sources with high biosynthetic potential and genetic diversity that translates into new chemical space and therefore therapeutic space, which has been increasingly realized by investigating marine sources that represent the greatest biodiversity on the planet. Through advances in genomics and target identification methodology, natural products are experiencing a renaissance, especially marine natural products that suffered most from the drawbacks of supply issue, structural complexity associated with difficulty in chemical synthesis and target identification.4–6 However, we have learned that novel biology exerted by these compounds justifies chemistry efforts, exemplified by the FDA approval of the sponge natural product (halichondrin B) inspired eribulin, requiring a 61-step synthesis.7 Requisites for successful drug development are the identification of novel chemical scaffolds with biological functions, a scalable total synthesis to solve the supply issue and explore structure–activity relationships (SAR) for further optimization, and the characterization of the mechanism of action (MOA) and the direct biological targets. The latter tends to be highly rewarding, since natural products point us to new ways to combat diseases, which translates into first-in-class agents.
Among marine organisms, cyanobacteria have become a validated source of drug leads and perhaps most successful organisms, with five current FDA approvals based on dolastatin 10 for the treatment of various cancers, accounting for 33% of FDA-approved marine drugs.8 Marine cyanobacteria's genetic diversity is associated with diverse and novel pharmacological space of the encoded compounds that are mainly modified peptides or peptide-polyketide hybrids. Cyanobacteria have also been demonstrated to produce most-potent-in-class natural products.9–11 Target discovery and mechanisms of action have been the catalyst to drive focused preclinical studies, to identify potential liabilities and rational application, and even combination therapy. Our review focuses on the recent progress in the discovery and development of anticancer agents from marine cyanobacteria, which usually required that structure determination was coupled with imperative chemical synthesis and in-depth mechanistic studies to recognize and maximize therapeutic opportunities. This review, covering the period from 2010 to April 2024, represents an extension of our group's 2015 review on general and representative biological targets and mechanisms of action of marine cyanobacterial compounds, now with an in-depth focus on anticancer activities and critical developmental issues and status.12 In contrast to other recent reviews focusing on structures and the vast array of biological activities,9,13–16 our review highlights promising anticancer activities of marine cyanobacterial compounds through the lens of potential drug development opportunities, with emphasis on target identification and mechanism of action, in addition to solving the supply issue. We present the most promising and well-characterized cyanobacterial compounds with distinct activity profiles, established synthetic strategies for obtaining material for rigorous preclinical assessment, classified compounds based on their MOA and target or target organelle, and application of enabling technologies for their development as ADCs.
2. Tubulin agents and their enablement as antibody-drug conjugates (ADCs)
2.1. Target engagement and mechanism of action
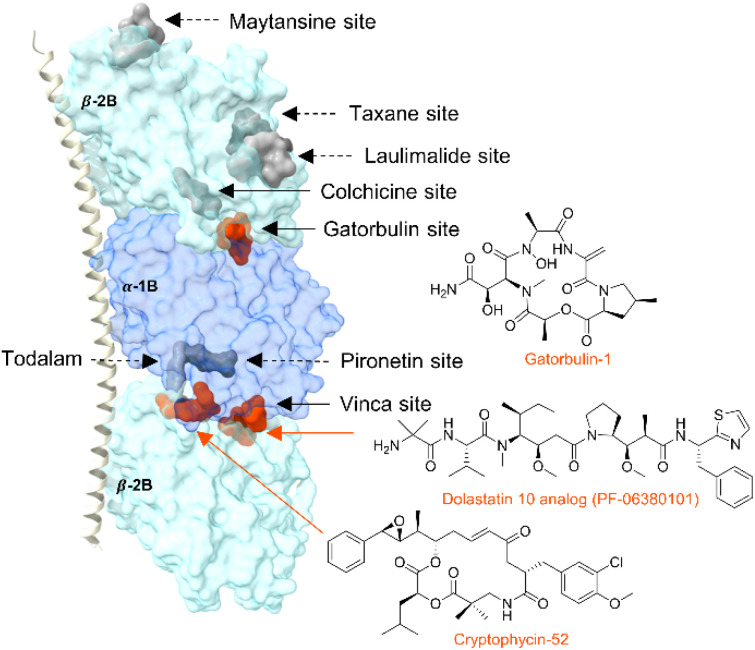
Tubulin-targeted chemotherapy has been highly successful, and the various marketed drugs are based on natural products scaffold, including cyanobacterial compounds.17,18 Tubulin heterodimers, composed of α-and β-subunits, form polarized polymers and these microtubules are involved in cellular structure, cell division and proliferation, motility, and intramolecular trafficking.17,19–21 Binding of pharmacological agents affects the tubulin dynamics and ultimately leads to anticancer activity.22 Tubulin agents are categorized based on their binding site at α/β-tubulin or the dimer interface and whether they stabilize or destabilize microtubules. Eight distinct sites are known (six of which lead to destabilization and two causing stabilization), including one of the first marine derived tubulin destabilizers, curacin A, which binds to the colchicine site based off biochemical data23,24 and the discovery of the seventh binding site targeted by the cyanobacterial microtubule-destabilizing depsipeptide, gatorbulin-1 (Fig. 1).25 The eighth binding site was recently discovered using computer-aided molecular design to create the first rationally designed tubulin binder to the interface between the maytansine site and the end of the pironetin pocket.26 The most well-known cyanobacterial tubulin agents in clinical use and advanced development include dolastatins 10 and 15 as well as the cryptophycins (Fig. 1), all of which are known to target the vinca site to interfere with tubulin assembly and destabilize microtubules.27–30 The recently discovered gatorbulins induce depolymerization targeting tubulin near the colchicine binding site at the interface between the α,β subunits.25,31 Target engagement in all cases leads to interference with tubulin dynamics, and one of the downstream effects is the disruption of mitotic spindle, leading to G2/M cell cycle arrest.32 These agents also produce a similar cytotoxicity profile in the NCI-60 cancer cell line screen, which indicates that they share a similar mechanism of action.25,33 However, despite these and other striking commonalities, the pharmacology appears to have left a distinct fingerprint and unique intricacies include drug-like properties. Targeting the gatorbulin site induces proteasome-mediated tubulin degradation,31 which is a unique characteristic compared with tubulin agents against other pharmacological sites.25
Fig. 1. Tubulin structure (PDBID: 5LA6) showing the known tubulin binding sites: maytasine (PDBID: 4TV8), taxane (zampanolide, 4I4T), laulimalide (4O4H), colchicine (plinabulin, 6S8K), gatorbulin (7ALR), pironetin (5LA6), todalam (todalam 4, 5SB3) and vinca (dolastatin 10 analogue, 4X1I and cryptophycin-52, 7LXB). Sites are indicated by a surface representation of the co-crystallized molecules (in parenthesis, when differing from the site name). Sites known to be targeted by marine cyanobacterial compounds are in orange.
2.2. Dolastatin 10
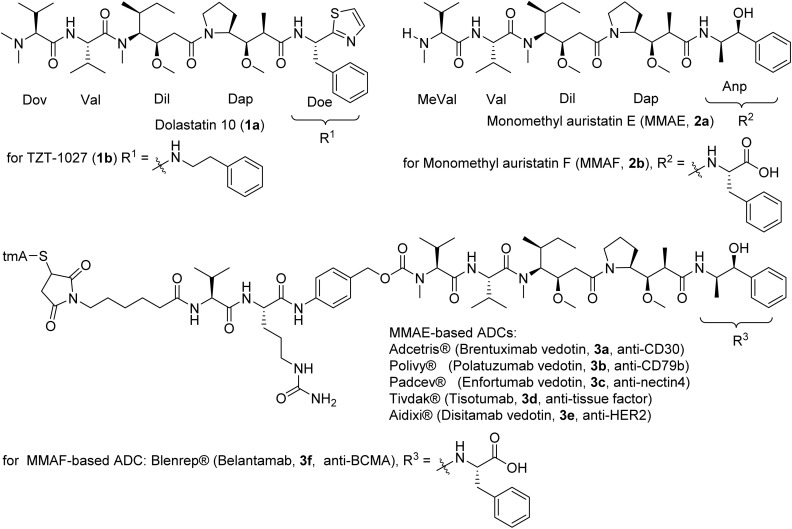
Dolastatin 10 (1a, Fig. 2) was originally isolated from the Indian Ocean sea hare Dolabella auricularia,34 but in 2001 the true producer was identified as a marine cyanobacterium, taxonomically classified in 2015 as a new genus Caldora penicillata.34–36 The compound (1a) has been isolated from cyanobacteria collected from Micronesia, Hawaii and Florida.34,35,37 Dolastatin 10 (1a) showed exquisite potency at the cellular level against cancer cells in the picomolar range (10–100 pM), notably with an IC50 of 0.03 nM for L1210 leukemia, 0.059 nM for small cell lung cancer NCI-H169, and 0.5 nM for human prostate cancer DU-145 cell lines.28,38–40 Dolastatin 10 reached Phase II clinical trials but was discontinued due to granulocytopenia and neutropenia at dose-limiting toxicities.41 The synthesis was achieved in various ways but always following a sequential elongation approach that was amenable to structure diversification, leading to TZT-1027 (1b) and monomethyl auristatin E (MMAE, 2a), which advanced to clinical trials (Fig. 2).42–44 Dolastatin 10 analogues did not progress further in clinical trials due to non-selectivity, but MMAE (2a) and monomethyl auristatin F (MMAF, 2b) were developed into the cytotoxic payload for antibody-drug conjugates (ADCs, 3a–3e, 3f) with tremendous success (Fig. 2).45–47
Fig. 2. Structures of dolastatin 10 (1a), and the synthetic analogues of monomethyl auristatins developed as ADCs approved by FDA for clinical use. The MMAF-based ADC has been withdrawn.
MMAE linked to the CD30 antibody via a protease-cleavable linker led to brentuximab vedotin (3a), approved in 2011 for the treatment of Hodgkin's lymphoma and anaplastic large cell lymphoma.48 (Fig. 2) The same cathepsin B-cleavable maleimidocaproyl-valyl-citrulline-p-aminobenzyl-carbamate linker ((mc-Val-Cit-PABC)-citrulline) was pivoted for polatuzumab vedotin (3b), where MMAE was linked to a CD79B-directed ADC that was approved in 2019 to treat non-Hodgkin lymphoma, chronic lymphocytic leukemia and refractory B-cell lymphoma.49 By utilizing an anti-nectin-4 monoclonal antibody, the ADC enfortumab vedotin (3c) (using the same payload and linker) was approved in 2019 to effectively target metastatic urothelial cancer.50
The success of these drugs prompted the launching of similar ADCs with the same payload and proven linker, differing only by the antibody, to target different cancers.51 In some instances, linkers were changed with the advances of linker technologies. MMAE-based tisotumab vedotin-tftv was the most recent FDA approved ADC (2021) with implications for recurrent or metastatic cervical cancer possessing disease progression during or post chemotherapeutic treatment.52 Belantamab mafodotin-blmf (3f) was the only MMAF-based FDA approved ADC (2020) as the first anti-B-cell maturation antigen therapy for patients with released or refractory multiple myeloma.53,54 The ADC contains a non-cleavable linker and an afucosylated Fc-engineered antibody.55 Later, approval was withdrawn due to inferior efficacy in progression-free survival of the patient.56Table 1 shows ADCs with dolastatin 10 analogues as the cytotoxic payload.8
Dolastatin 10 (1a) based ADCs approved or in clinical/preclinical trials.
| Trade name (ADC) | Company | Target (mAb mode) | Payload | Linker | FDA approved/clinical phase | Disease indication | Ref. |
|---|---|---|---|---|---|---|---|
| Adcetris® (brentuximab vedotin[BV], SGN-35) | Seattle Genetics/Takeda (Washington, U.S./Tokyo, Japan) | CD30 (cAC10 chimeric IgG1) | MMAE | Protease-cleavable linker, maleimidocaproyl-valine–citrulline-p-aminobenzyloxycarbonyl linker | 2011 | Relapsed/refractory Hodgkin lymphoma and anaplastic large cell lymphoma | 45 |
| Padcev® (enfortumab vedotin[EV], ASG-22ME, AGS-22M6E) | Seattle Genetics/Astella (Washington, U.S./Tokyo, Japan) | Nectin 4 (IgG1) | MMAE | Cleavable maleimidocaproyl-valine–citrulline-p-aminobenzyloxycarbonyl linker | 2019 | Locally advanced or metastatic urothelial cancer | 50 |
| Polivy® (polatuzumab vedotin-piiq[PV], DCDS4501, RG7596) | Roche (Basel, Switzerland) | CD79b (IgG1) | MMAE | Protease-cleavable maleimidocaproyl-valine–citrulline-p-aminobenzyloxycarbonyl linker | 2019 | Diffuse large B-cell lymphoma | 57 |
| Tisotumab vedotin (HuMax-TF-ADC) | Genmab (Vably, Denmark) | CD142 (Hu IgG1) | MMAE | Valine–citrulline peptide linker | 2021 | Multiple solid tumours | 52 and 58 |
| Disitamab vedotin (Aidixi) | RemeGene (California, U.S.) | HER2 | MMAE | Valine–citrulline peptide linker | 2021 | Multiple solid tumors | 59 |
| Telisotuzumab vedotin (ABBV-399) | AbbVie/Pierre Fabre (Illinois, U.S./Castres, France) | HGFR/cMet (Engineered IgG1) | MMAE | Valine–citrulline peptide linker | 2021 | Non-small cell lung cancer | 60 |
| ARX788 | Zhejiang Medicine/Ambrax (Zhejiang, China/California, U.S.) | HER2 | MMAF (Amberstatin 269) | Hydroxylamine-PEG4 | Phase III | Metastatic breast cancer/gastric cancer | 61 |
| BT8009 | Bicycle (Cambridge, U.K./Massachusetts, U.S.) | Nectin-4 (PVRL4) | MMAE | Valine–citrulline dipeptide linker | Phase II | Bladder and breast neoplasms, non-small cell lung and ovarian cancer, solid tumors | 62 |
| BT5528 | Bicycle (Cambridge, U.K./Massachusetts, U.S.) | EphA2 | MMAE | Valine–citrulline peptide linker | Phase II | Breast, gastric, and urothelial cancer, head/neck cancer, non-small cell lung, and ovarian cancer, solid tumors | 63 |
| Upifitamab rilsodotin (XMT-1536) | Mersana therapeutics (Massachusetts, U.S.) | NaPi2b & microtubules | Auristatin F-HPA | Poly-1-hydroxymethylethylene hydroxymethylformal | Phase II | Solid tumors | 64 and 65 |
| STI-6129 | Sorrento (California, USA) | CD38 | Duostatin 5 | Non-polyethylene glycol linker | Phase II | Relapsed/refractory systemic amyloidosis | 66 |
| RC-88 | RemeGen (China/U.S.) | Mesothelin | MMAE | Valine–citrulline peptide linker | Phase II | Solid tumors | 67 |
| A-166 | Klus Pharma (New Jersey, U.S.) | HER2 | Duostatin 5 | Stable protease-cleavable valine citrulline linker | Phase II | HER2-expressing cancer | 68 |
| MRG003 | Lepu Biopharma (China) | EGFR | MMAE | Valine–citrulline linker | Phase II | Non-small cell lung cancer | 69 |
| W0101 | Pierre Fabre (Paris, France) | IGF-R1 | Ab-auristatin | Noncleavable maleimidocaproyl (mc) linker | Phase II | Advanced or metastatic solid tumors | 70 |
| SolidAGS-16C3F | Astellas (Tokyo, Japan) | ENPP3 (Hu IgG1) | MMAF | Maleimidocaproyl linker | Phase II | Metastatic renal cell carcinoma | 71 |
| Ozuriftamab vedotin (BA-3021) | BioAtla (California, U.S.) | ROR2 | MMAE | Cleavable maleimidocaproyl-valyl-citrullinyl-p-aminobenzyloxycarbonyl linker | Phase II | NSCLC, triple – Breast cancer, and soft tissue sarcoma | 72 |
| Telisotuzumab vedotin (ABBV-399) | Abbvie (Illinois, U.S.) | c-Met | MMAE | Cleavable valine–citrulline peptide linker | Phase II | Solid tumors | 73 |
| Ladiratuzumab vedotin (SGN-LIV1A) | Seattle Genetics (Washington, U.S.) | LIV1 (Hz IgG1) | MMAE | Valine–citrulline peptide linker | Phase II | Breast cancer | 74 |
| CAB-AXL (BA-3011, Mecbotamab vedotin) | BioAlta (California, U.S.) | AXL | MMAE | Cleavable linker | Phase II | Solid tumor, non-small cell lung cancer, melanoma, sarcoma | 75 |
| ARX-517 | AmbRX (California, U.S.) | PSMA | MMAF (Amberstatin 269) | Hydroxylamine-PEG4 | Phase I | Prostate cancer | 76 |
| Cofetuzumab pelidotin (ABBV-647) | Abbvie (Illinois, U.S.) | PTK7 | Auristatin-0101 | Cleavable valine–citrulline based linker | Phase I | Non-small cell lung cancer | 77 |
| SGN-B6A | Pfizer (New York, U.S.) | Integrin beta-6 | MMAE | Valine–citrulline peptide linker | Phase I | Solid tumors | 78 |
| Zanidatamab zovodotin (ZW-49) | Zymeworks/BeiGene (BC, Canada/Beijing, China) | HER2 | Proprietary auristatin | Protease cleavable linker | Phase I | HER2 expressing cancer | 79 |
| ALT-P7 | 3Sbio/Alteogen (China/Korea) | HER2 & microtubules | MMAE | Cysteine-containing peptide motif | Phase I | Breast and gastric cancers | 80 |
Requisite for the success of the dolastatin 10-based ADC development was both the understanding of the biological and clinical relevance, as well as solving the supply issue by total synthesis.
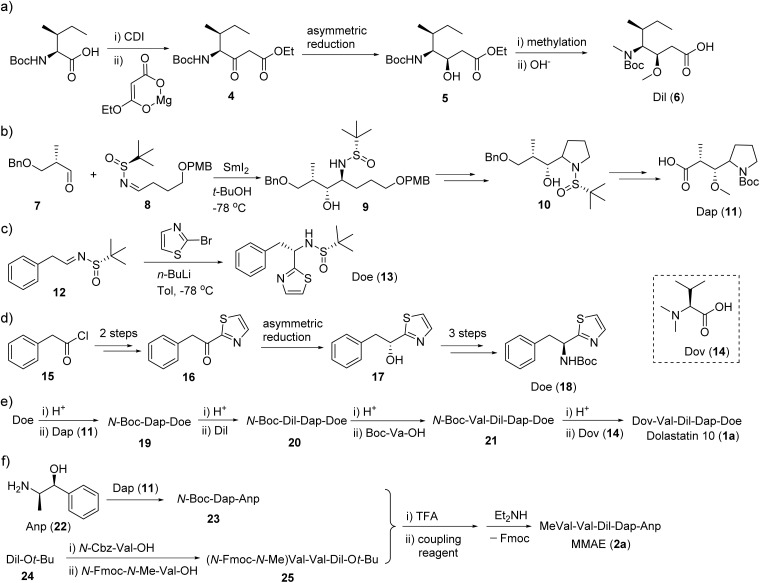
The total synthesis of dolastatin 10 (1a) always followed the sequential elongation approach using the five subunits. The difference between each method is the construct of the three key units, i.e., dolaisoleuine (Dil), dolaproine (Dap) and (S)-dolaphenine (Doe).81 The earlier synthesis of unit Dil and Dap was effected by using an Aldol reaction.82 A β-ketone ester approach was developed for the synthesis of the Dil fragment (Fig. 3a): the imidazole intermediate of isoleucine was reacted with magnesium enolate of ethyl hydrogen malonate to provide β-ketone ester.83,84 Sequential asymmetric reduction and methylation then gave the Dil fragment (Fig. 3a). Similarly, Genet followed the β-ketone ester approach to construct the Dap fragment.84 Wei developed a straightforward chiral N-sulfinyl imine method to generate both Dil and Doe fragments (Fig. 3b and c).85 However, when the Doe fragment was synthesized through a benzyl thiazolyl ketone, more than six steps were required (Fig. 3d).84 With the five subunits in hand, the synthesis of the final target started from Doe, and other units were installed in a stepwise manner by standard peptide coupling protocols (Fig. 3e). However, the general strategy of the total synthesis of MMAE (2a) is a little different: two subunits Dap-Anp and MeVal-Val-Dil were pre-synthesized and then coupled together to yield the final product MMAE (2a, Fig. 3f).86,87 The different strategies resulted from the different polarity of Dov and N-MeVal: the N,N-diMeVal is a polar moiety and its attachment at late-stage could avoid additional side reactions and a harsh purification process, while Fmoc protected MeVal does not face similar issues. Therefore a convergent approach was adopted in the synthesis of MMAE.88
Fig. 3. Synthesis steps (a–e) for dolastatin 10 (1a) and (f) MMAE (2a).
Nishio synthesized a series of dolastatin 10 derivatives with the replacement of Doe for versatile conjugations, and Mendelsohn modified the pyrrolidine ring (Dap unit) and synthesized a series of linear and macrocyclic dolastatin 10 analogues.89,90
2.3. Dolastatin 15
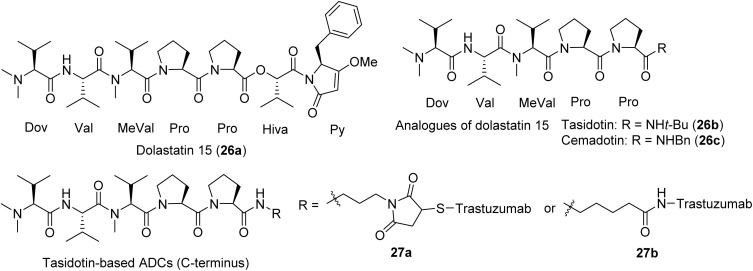
Dolastatin 15 (26a) was also first discovered from the shell-less mollusk D. auricularia by the Pettit group and only in 2020 published to be of proven cyanobacterial origin with its direct isolation from the marine cyanobacterium Symploca sp. by the Luesch group91 (Fig. 4). Dolastatin 15 possesses lower potency than dolastatin 10 in the nanomolar range (IC50 3–5 nM), and has been heavily investigated due to its action on tubulin.29,92 Dolastatin 15 is believed to target the vinca site, similar to dolastatin 10, although biochemical and cellular data also indicated differences.29,91,93 The dolastatin 15 analogues tasidotin (26b) and cemadotin (26c) (Fig. 4) have shown promising activity against solid tumors in preclinical models, and conjugation to trastuzumab on N- and C-terminus led to its evaluation as an ADC against human epidermal growth factor receptor (HER) 2+ ovarian cancer (Table 2).99 Only the ADCs derived from C-terminus maintained anti-tubulin activity (Fig. 4); the ones derived from N-terminus lost activity.100 Cemadotin (26c), a dolastatin 15 pentapeptide synthetic analogue, has been shown to cause tumor growth delays in vitro when conjugated through a thiazolidine linker to the fibronectin antibody (Fig. 4).81,82,101
Fig. 4. Dolastatin 15 (26a) and clinically evaluated analogues, including relevant ADCs that advanced to clinical trials.
Dolastatin 15 (26a) ADCs advanced to clinical trials.
| ADC (other names) | Company | Target | Clinical phase | Indication | Ref. |
|---|---|---|---|---|---|
| Tasidotin (26b, ILX651) | Genzyme (Massachusetts, U.S.) | Tubulin | Phase I | Advanced solid malignancies | 94 |
| Cemadotin (26c) | Hu and Hang | Tubulin | Phase I | Solid tumors | 95 |
| LU 103793 | BASF (Massachusetts, U.S.) | Tubulin | Phase II, discontinued | Non-small cell lung cancer, breast cancer, malignant melanoma | 96–98 |
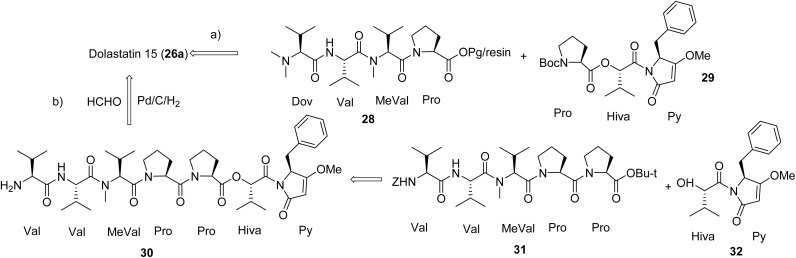
There are two main strategies published for the synthesis of dolastatin 15 (Fig. 5). Pettit's group used the [1 + 1] strategy through liquid solution method (Fig. 5a), first synthesizing the fragments Dov-Val-MeVal-Pro (28) and Pro-Hiva-Py (29), then fusing the two fragments aided by a coupling reagent to obtain the final product that was identical to the natural product.102,103 Akaji's group applied a similar strategy but used solid phase peptide synthesis (SPPS) to synthesize the same two fragments, in which a new coupling reagent, 2-chloro- 1,3-dimethylimidazolinium hexafluorophosphate (CIP), was utilized in multiple steps.104 However, the Jouin group applied a slightly different strategy (Fig. 5b), first synthesizing two fragments 2Val-MeVal-2Pro (30) and Hiva-Py (32), then fusing these two fragments into the linear peptide (Val to Py).105 The methylation of the end Val of the linear peptide with reductive hydrogenation using the system Pd/C/H2/HCHO provided the proposed product. However, their product did not match the synthesized natural product from Pettit's group but was comparable to the isolated natural product dolastatin 15 (26a). The Jouin group proposed the discrepancies were due to the lack of purity from the originally isolated compound and the lack of diastereomeric purity of an intermediate from Pettit's synthesis of dolastatin 15.105 The more purified synthetic product had slightly better activity than the isolated dolastatin 15 against the U.S. National Cancer Institute's P388 lymphocytic leukemia cell line (ED50 of 0.48 ng mL−1vs. 2.4 ng mL−1).105 Tasidotin (26b) and cemadotin (26c) are simplified analogues of dolastatin 15 (26a) and their total synthesis was straightforward, starting from its proline precursor.94,106
Fig. 5. Synthetic strategies (a and b) for dolastatin 15 (26a) that is also applicable to tasidotin (26b) and cemadotin (26c).
2.4. Cryptophycins
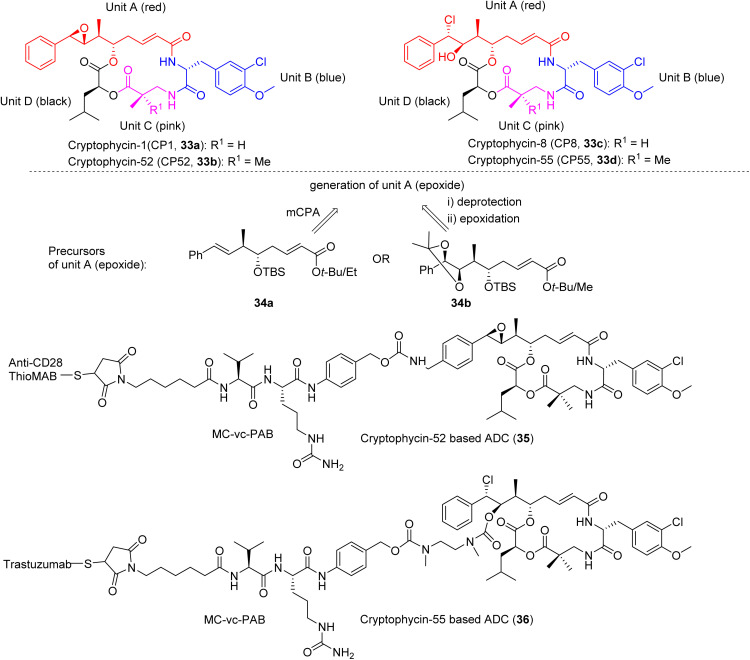
The super potent microtubule disruptors cryptophycins were originally isolated from terrestrial cyanobacteria as antifungal agents in 1990, and later characterized as anticancer agents by the Moore group in 1997.27,107 Subsequently, hundreds of analogues were either isolated or synthesized, many from the Moore group.108–110 Cryptophycins are presumably also produced by marine cyanobacteria, evidenced by cryptophycin-24 (arenastatin A) that was isolated from the cyanobacteria-harboring marine sponge Dysidea arenaria.111–113 The most abundant naturally occurring cryptophycin-1, demonstrated spectacular potency with IC50 values ranging from low-nanomolar to picomolar against many cancer cell lines (IC50 4.58 pM, KB cells; 7.63 pM, LoVo; 0.4 nM, C6 glioma; and 0.8 nM, HepG2) and was an ineffective substrate of P-glycoprotein (Pgp) in multidrug resistant cell lines.108,109,114,115 Cryptophycin-52 (HeLa IC50 0.011 nM) was selected as a clinical candidate but failed in Phase II clinical trials due to neurological toxicity (DLT 1.5 mg m−2) and limited efficacy.116–118 However, due to the exquisite potency, cryptophycins were predestined to be recognized as an attractive cytotoxic payload for ADCs, which led to their resurrection110,119 (Fig. 6). Met-Val-Cit-PABC was used as the linker to thiomab and trastuzumab as antibodies for cryptophycins-52 and -55 based ADCs, respectively.108,119,120 Cryptophycin-55-based ADC exhibited nanomolar cytotoxicity in HER2+ tumor cells (IC50 0.58–1.19 nM) and anti-tumor activity in xenograft models at 10 mg kg−1 (SKOV3 and NCIeN87).119
Fig. 6. Selected structures and units of cryptophycins (33a–d), including unit A precursors (34a,b) and ADCs (35, 36).
Natural cryptophycins are composed of four units A–D (Fig. 6). Epoxide-containing (e.g., CP1, 33a or CP52, 33b) and chlorohydrin-containing (e.g., CP8, 33c or CP55, 33d) analogues represent two types of the most cytotoxic analogues. Unit A is the key fragment with respect to synthetic accessibility and bioactivity, i.e., it represents the most complex part for synthesis and the most important part in the determination of activity. The synthesis of unit A is challenging because it possesses four chiral centers, a labile epoxide and an unsaturated amide. Since the chlorohydrin could be readily obtained from the epoxide by transformation using acid-mediated ring opening (HCl), we here discuss the synthesis of epoxide-containing cryptophycins.121 The epoxide could be introduced at a late-stage by oxidation of the styrene moiety or at an early stage via protection by an acetonide that would be cleaved at a late-stage.121
A scalable and economically feasible synthesis was the key for rigorous biological evaluation in preclinical and clinical trials. Over 300 cryptophycins have been generated and tested, and, more recently, the biosynthesis was effectively constituted and chemoenzymatic synthesis were performed.122,123 However, the development of cryptophycin is attributed to its total synthesis.
There are many published methods for the synthesis of unit A. Generally, there are two strategies for the generation of the epoxide moiety, using both the styrene moiety approach and the acetonide moiety approach, as shown in Fig. 7.124–127 In the former approach, styrene was designed as the precursor for the epoxide moiety, which was generated by oxidation of styrene at a late-stage, while in the latter approach acetonide was designed as the precursor of epoxide moiety, generated by the deprotection-epoxidation sequence of the acetonide moiety.125,126 However, lower stereoselectivity was observed using the styrene approach (34a), i.e., the product was a mixture of diastereomers and the ratio of (R,R) and (S,S) varied from 1 : 1 to 2.5 : 1. This method is straightforward, as the diastereomers could be separated by HPLC but is only suitable for small-scale preparation.125 In the acetonide approach, the stereocenters were generated first and masked as acetonide and unmasked at a late-stage to produce an epoxide through another step. Though there are three more steps, this method could generate the (R,R) configuration, rendering it suitable for a scalable preparation of cryptophycins.126 In 2011, Sherman developed a novel chemoenzymatic approach to synthesize cryptophycin using the enzyme CrpD-M2 to incorporate a 2-hydroxy acid moiety (unit D) into cryptophycin analogues.128 The carboxy or amino terminus of 4-O-Me-phenylalanine (unit B) was frequently selected as the site of macrocyclization.
Fig. 7. Two strategies (a and b), for macrocyclization in the synthesis of cryptophycins.
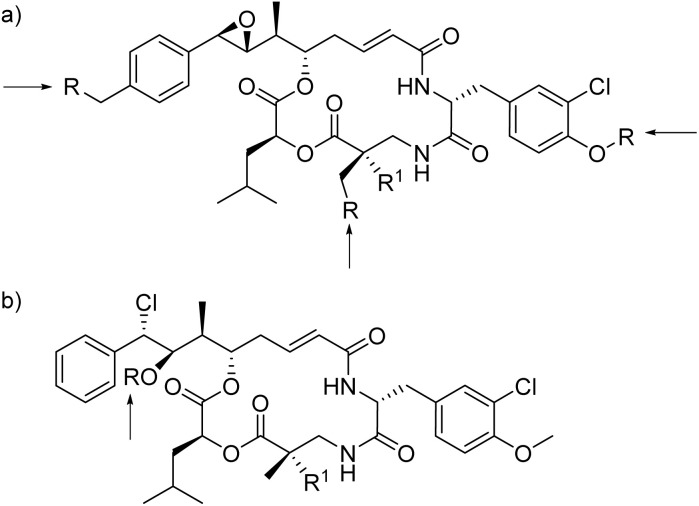
Due to the failure of cryptophycin-52 (33b) in Phase II clinical studies, ADCs have emerged as an important direction in the field of cryptophycin research (35, 36, Fig. 6).110,120,129 For the epoxide analogues (Fig. 8a), three sites were frequently chosen as a modification site, being the para-position of the aromatic ring of unit A, the OH site of unit B, and the methyl group of unit C. For the chlorohydrin analogue (Fig. 8b), the OH was derivatized to obtain ADCs.110
Fig. 8. Linker-antibody attachment sites for cryptophycin-based ADCs for (a) 35 and (b) 36 (R: derived moieties, R1: H or Me).
Recently, Sewald comprehensively reviewed the cryptophycins, including the fluorinated analogues and conjugates, biological activities, and SAR.108
2.5. Gatorbulins
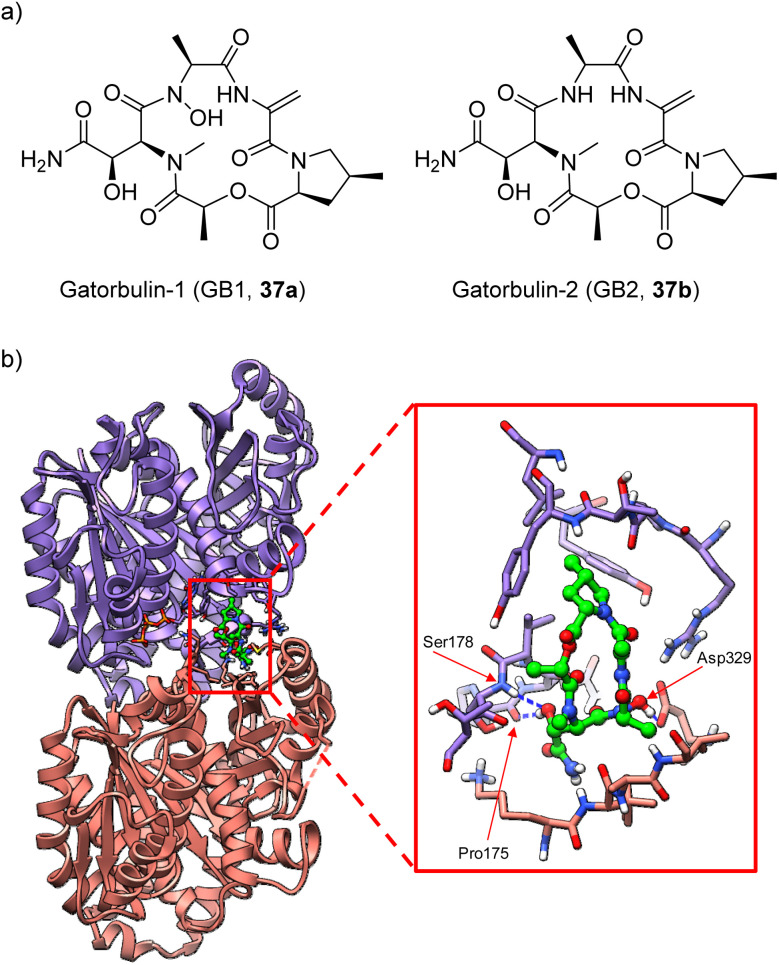
The gatorbulin family is the most recent addition to the microtubule-destabilizing agents derived from marine cyanobacteria. Gatorbulin-1 (GB1, 37a, Fig. 9) was described in 2021 from a Floridian Lyngbya confervoides by the Luesch group.25 The structure determination was achieved through standard 2D NMR coupled with 15N NMR, pinpointing the presence of a primary, secondary and tertiary amide as well as a hydroxamate group that was critical for the antiproliferative effects.25 This distinct cyclodepsipeptide of comparatively low molecular weight is a modified pentapeptide that displayed unique pharmacology, targeting a seventh tubulin pharmacological site, as revealed through multidimensional characterization in complementary mechanistic and phenotypic assays.25 Using fluorescent bona fide probes, it was found that GB1 could not displace fluorescent probes of eribulin (vinca site) or maytansine (maytansine/tip site) but competed successfully with a fluorescent probe for the colchicine site.31 This data initially suggested that GB1 might be the first peptide targeting the colchicine site; however, both the pharmacological data and mechanistic probing indicated that GB1 may not target the classic colchicine pocket.25 This hypothesis was successfully probed using macromolecular crystallography, culminating in the high-resolution structure determination of the α/β-tubulin-GB1 complex to reveal a new tubulin regulatory site near the colchicine site to modulate tubulin dynamics (Fig. 9).25,31 The tubulin-colchicine and tubulin-GB1 showed main differences in the β-tubulin loop T7 and α-tubulin loop T5, which imperatively change their conformations upon colchicine binding.31 GB1 binding precludes the concurrent ligand binding at the colchicine site, rationalizing the competition assay results and underscoring the distinct pharmacology of GB1.25,31
Fig. 9. (a) Structures of gatorbulin-1 (37a) and gatorbulin-2 (37b) and (b) crystal structure of the tubulin–gatorbulin-1 complex (PDBID: 7ALR). Purple: tubulin α-1B chain. Salmon: tubulin β-3 chain. Gatorbulin-1 carbons atoms are highlighted in green.
GB1 exhibited GI50 of 800 nM and IC50 of 300 nM in the primary screen against HCT116 colorectal cancer cells, without significantly affecting the cell viability of normal colon cells (IC50 > 10 μM).25 Of the cancer cells tested, GB1 was the most cytotoxic to colon cancer cells COLO205 (GI50 92 nM) and had strong activity in other cell lines, including melanoma (SK-MEL-5), ovarian (OVCAR-3), and prostate (DU-145).25 However, the activity of GB1 in Pgp and βIII-expressing HeLa cell lines was strongly attenuated, initially suggesting room for optimization.25 However, subsequent studies with MDCK cells stably transduced with the human efflux transporter MDR1/Pgp, showing similar permeability with and against transporter gradient, indicated that GB1 (44a) is a poor Pgp substrate.31
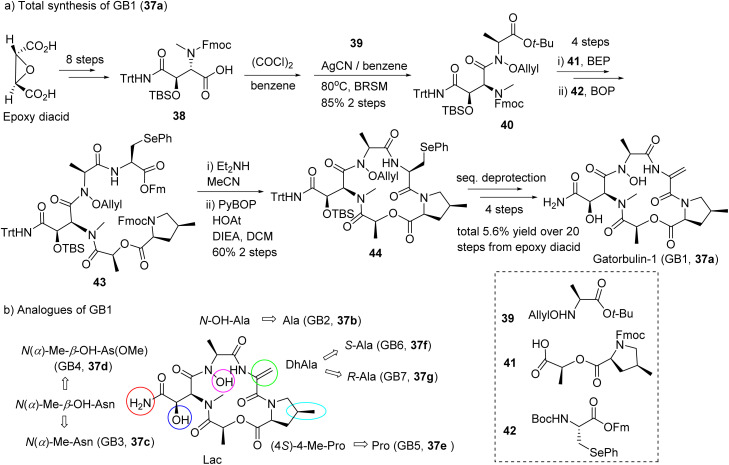
The co-isolated gatorbulin-2 (GB2, 37b), the N-deoxy derivative of GB1, lacked antiproliferative activity and provided preliminary SAR information. The structure of the α/β-tubulin-GB1 complex underscored that the N-hydroxy group shows wide-ranging interactions with loop 5's α- and β-tubulins.25 The synthesis of GB1 (37a) was achieved in 5.6% overall yield over 20 steps, solving the supply issue and established the basis for the synthesis of gatorbulins with simplified structures, GB2–7.31
The presence of almost exclusively unusual moieties (N-OH-Ala, DhAla, 4-MePro, OH-Asn) posed challenges for the total synthesis (Fig. 10a). Starting from trans-2,3-epoxysuccinic acid, the fully masked acid 38 was efficiently synthesized. Amidation of 38 by allyloxyamine 39 is a key step in this total synthesis, which was accomplished using the acid chloride method with AgCN as base. Fmoc–Fm pair was designed as the protection groups of amino and carboxy termini, respectively and Sec (Ph) was proposed as the precursor unit of DhAla. The PyBOP-mediated macrocyclization proceeded with good yield (60%). The Luesch group also synthesized six analogues (GB2–7, 37b–37g, Fig. 10b) applying a similar synthetic strategy.31
Fig. 10. Abbreviated synthesis (a) of GB1 (37a) and (b) the structures of gatorbulin analogues (GB2–7, 37b–37g) highlighting the structural modifications to the parent molecule.
GB1 (37a) has drug-like properties, including low molecular weight (<500 g mol−1), and is amenable to rapid synthetic modification. These characteristics increase the translational potential of gatorbulin-based, pharmacologically novel microtubule agents to not just become chemical probes but also drug candidates that complement the arsenal of current clinical and preclinical microtubule agents.
The SAR was systematically investigated (GB2–7, 37b–37g) at the biochemical and cellular level using GB1 (37a)-susceptible ovarian and cervical cancer cells.31 The hydroxamate moiety in the N-methyl-alanine residue is critical for activity. All other structural modifications present in GB1, including C-hydroxylation of asparagine, methylation at C-4 of proline, and sp2 hybridization in dehydro-alanine, were proven to be functionally relevant.31 Replacement of the primary amide with a methyl ester also resulted in reduced activity, indicating the intricate scaffold optimization by the GB1-producing cyanobacterium. Inhibition of tubulin polymerization in vitro and binding affinities correlated very well, translating into differential cellular efficacy. Using docking and molecular dynamics to evaluate the effects of the chemical simplification at the structural level, any changes resulted in loss of target interactions, although energetically modest. Similar to cevipabulin that targets two different sites on the tubulin dimer,31,130 GB1 promotes proteasome-mediated tubulin degradation but by an unknown mechanism, presumably distinct from that of cevipabulin.27 Comparison with GB1 (37a) indicated that cevipabulin binds to the same tubulin region although the binding mode is distinct.130 Cevipabulin almost exclusively interacts with α-tubulin, including nonexchangeable GTP.130 In contrast, GB1 (37a) makes extensive contact and hydrogen bonding with both α- and β-chains of tubulin.22 GB1–7 (37a–37g) showed excellent solubility and much higher than that of paclitaxel.31 Hepatic microsome stability was shown to be excellent, while human cytochrome P450s were not inhibited, and plasma binding was minimal with high free fractions.31 Passive permeability was predicted to be high based on parallel artificial membrane permeability assay (PAMPA) for GB1–6.31
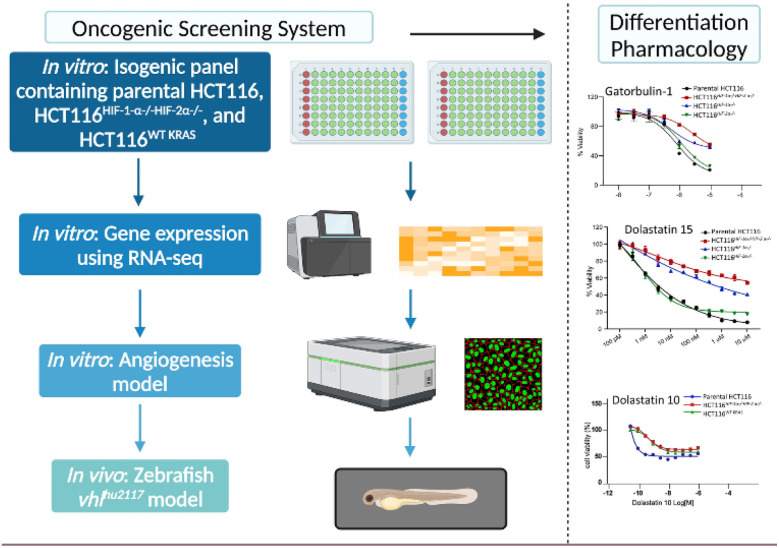
2.6. Repurposing as HIF inhibitors
Dolastatins 10 (1a) and 15 (26a) as well as gatorbulin-1 (37a) were discovered (or re-discovered) as agents with differential antiproliferative activity against hypoxia-inducible factor 1 (HIF-1) containing HCT116 cells, following deconvolution of initial differential selectivity for parental HCT116 cells versus dual HCT116HIF−1α−/−HIF−2α−/− knockouts.25,91,131 The screening system, described in 2016 to aid early-stage drug discovery by providing selectivity already in primary assays, enables the prioritization of agents that have a higher likelihood of activity and selectivity against solid tumors driven and characterized by oncogenic KRAS and hyperactivation of HIF (Fig. 11).131 It was demonstrated that microtubule agents belonged to the compounds with greatest differential, which was later proven for dolastatin 15 (26a) and gatorbulin-1 (37a).25,91 HIF inhibition appears to be a common downstream effect of tubulin-targeting agents as exemplified by multiple studies.25,91,131–133 The dual effect of tubulin-binding to interfere with tumor vascularization largely in endothelial cells, and HIF-inhibition to downregulate target genes such as VEGFA in growth factor secreting cells suggests that these compounds are also promising antiangiogenic agents, in addition to its primary utility as antimitotic agents.134,135 Downregulation of the HIF-target gene, VEGFA in parental HCT116 cells was observed in cancer cells for dolastatin 15 (26a) and gatorbulin-1 (37a) and both were effective in angiogenesis assay in vitro model systems using endothelial cells.25,91 Further, validation of HIF-regulated downstream effects by tubulin agents was demonstrated in vivo using a zebrafish model specifically in vhl mutants with activated (constitutively active) HIF signaling.91 Dolastatin 15 showed strong in vivo antiangiogenic effects concomitant with HIF target gene downregulation (Vegf and Egln3) that was revealed by in situ hybridization.91
Fig. 11. A multidimensional screening platform designed to identify preferential activity against cells with oncogenic KRAS mutations and downstream HIF hyperactivation in cellular and in vivo models.
The specific in vitro and in vivo effects on HIF signaling provided supportive rationale that these agents are expected to possess activity against solid tumors.136,137 As described above, dolastatin 10-based ADCs were originally approved for lymphomas but later also for bladder cancer (enfortumab vedotin).50 Tisotumab vedotin was designed to target tissue factor (TF) which is highly expressed in many solid tumors (ovarian, prostate, bladder, lung) for cervical and ovarian cancers.58 Ladiratuzumab vedotin was designed for metastatic triple negative/HR+/HER2-breast cancers are in different phases of clinical trials, while dolastatin 10-based ADCs undergo trials for ovarian cancer and other solid tumors (3a–3e, Table 1).74
3. Inhibitors of cotranslational translocation: Sec61 inhibitors
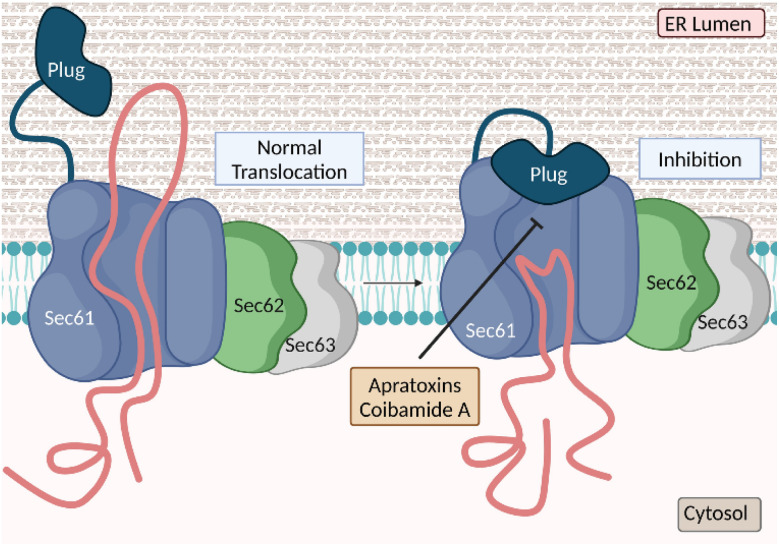
3.1. Mechanism of action
Cancer cells are usually characterized by overexpression and/or mutation of certain membrane proteins that function as pro-growth signaling receptors, depending on the tissue and cell type, and increased levels of secretion of ligands that act on these receptors.138,139 Both types of proteins, including receptor tyrosine kinases (RTKs) and growth factors, rely on a functional secretory pathway for proper localization on the cell membrane and extracellular milieu, respectively.140,141 Synthesis of proteins destined for secretion starts at cytosolic ribosomes and the firstly synthesized N-hydrophobic sequence (signal peptide) is recognized by the signal recognition particle (SRP) for subsequent docking at the SRP receptor in the endoplasmic reticulum (ER), where protein synthesis continues (Fig. 12).142 Cotranslational translocation is the first step in the secretory pathway and is initiated by protein insertion into the lumen of the ER.143 Insertion of the signal peptide is controlled by Sec61, the central protein translocation channel in the ER (Fig. 12).144 Targeting Sec61 may ultimately prevent ER translocation of all or a subset of proteins, depending on the specific compound and cellular context, which produces a potent antiproliferative effect.145 Two anticancer natural product classes from marine cyanobacteria, apratoxins146,147 and coibamides,148,149 have been demonstrated to act via this mechanism, by targeting Sec61.150–161 The different binding sites are hypothesized based on differential resistance to Sec61 mutants cells to apratoxin (45a) and coibamide A (52).145 (Fig. 13) Recently, high-resolution cryo-EM structures of seven inhibitors bound to chimeric human-yeast Sec61, including apratoxin F (45d), were characterized to bind to the lateral gate.145 These inhibitors have shown to induce different pharmacological fingerprints in the NCI-60 cell line screens.149,155,159,162,163
Fig. 12. Cotranslational translocation inhibition into the endoplasmic reticulum using Sec61 inhibitors.
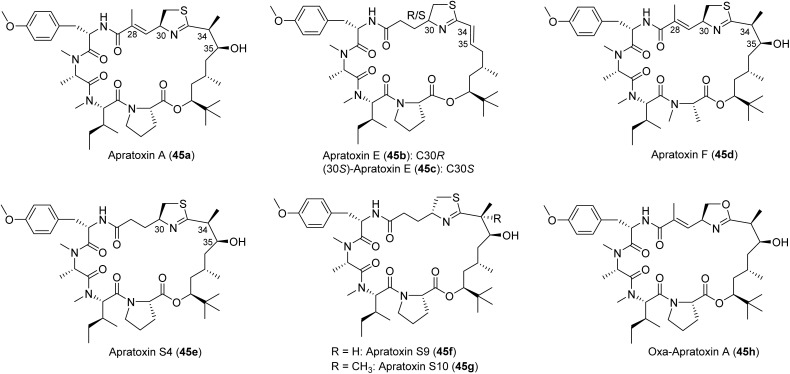
Fig. 13. Structures of selected natural apratoxins A (45a), E (45b), and F (45d), and other synthetic analogues (45C, 45e–45h).
3.2. Apratoxins
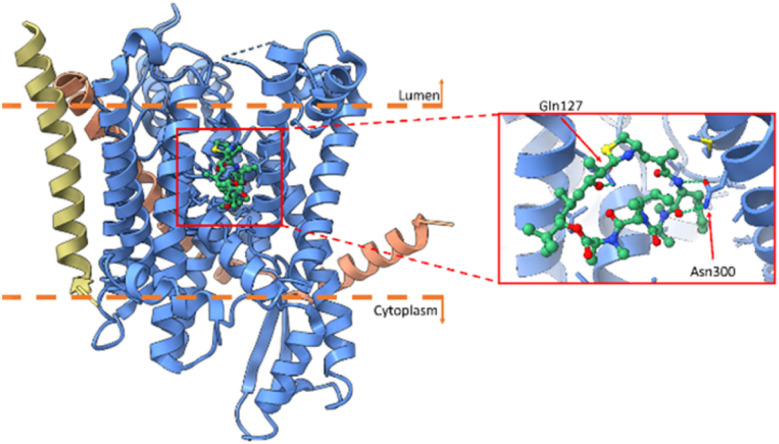
Following the discovery of natural apratoxin A (45a) the total synthesis was completed before the isolation of subsequent apratoxins, including apratoxins B and C Moore and co-workers,150 and subsequently apratoxins D–G151–153 by Gerwick and Luesch groups from Moorena bouillonii (recently reclassified as such).146,154,164 The total synthesis of its oxazoline analogues have been described.159,160,165,166 So far nine natural analogues have been discovered, with various SAR campaigns conducted to increase potency (Fig. 13). In 2015, Coltart reviewed the isolation, structure determination, and asymmetric total synthesis of natural apratoxins.167 The real interest in this compound class emerged after the mechanism of action was revealed, demonstrating apratoxin A an inhibitor of cotranslational translocation, a novel strategy to inhibit cancer cell growth at low nanomolar concentrations.156 The translocon Sec61 was shown to be the direct protein target of apratoxins, exemplified by using apratoxins A (45a) and F (45d) binding near the luminal plug domain (Fig. 12).145,157 The cryo-EM structure with F (45d) and other (non-cyanobacterial) Sec61 inhibitors suggested a common binding pocket for those Sec61 inhibitors (Fig. 14).145
Fig. 14. Cryo-EM structure of apratoxin F (45d) with Sec61 in blue (alpha), salmon (gamma) and gold (beta subunit) and apratoxin F in green (PDBID: 8DNZ).
In vivo studies with apratoxin S4 (45e) showed potent activity in a HCT116 colorectal cancer xenograft model.159 Daily dosing with 0.25 mg kg−1 produced a pronounced antitumor effect in mice with certain RTKs as pharmacodynamic markers shown to be selectively downregulated in the tumors.159 Pharmacokinetic studies for apratoxins A and S10 both showed enrichment in the pancreas and a favorable PK profile.147,168 Apratoxin S10 (45g), like apratoxin S4, lacked irreversible toxicity and at low doses apratoxin S10 showed efficacy in an orthotopic patient-derived xenograft (PDX) model for pancreatic cancer, which is the gold standard in the field.168 At a dosing schedule of 0.25 mg kg−1 every other day, apratoxin S10 inhibited the proliferation of pancreatic cancer cells in vivo, without inducing cytotoxicity, thus separating antiproliferative from cytotoxic activity.168 This effect may in part be attributed to the inhibition of growth factor and cytokine secretion from tumor-associated stromal cells that drives pancreatic cancer cell growth, suggesting that apratoxin S10 has an additional indirect effect on pancreatic cancer cells and therefore address a resistance mechanism.168,169 This finding on other cell types, coupled with the novel mechanism of action, opened up avenues for application beyond cancer.168,169 Apratoxin S10 was shown to inhibit the secretion of proangiogenic factors, VEGF-A and IL-6, and consequently evaluated for activity against cancer cells derived from highly vascularized tumors (renal, colon, neuroendocrine), where the compound showed 2000- to 5000-fold greater activity than FDA approved RTK inhibitors.169 The dual activity as inherent anticancer and antiangiogenic is particularly intriguing, as the compound may also address the resistance to antiangiogenic therapy through the inhibition of multiple proangiogenic pathways and inhibiting cancer cell growth simultaneously. The antiangiogenic activity in vitro has been determined for apratoxins A, S4 and S10 using human umbilical vein endothelial cells (HUVECs).155,163,169
To extend the scope of cancers that may be targeted by this compound class, apratoxin S4 (45e) was profiled against panels of cancer cells characterized by differential sensitivity to RTK inhibitors due to receptor mutations, oncogenic KRAS mutations, or activation of compensatory pathways.163 Apratoxin S4 (45e) was active at low-nanomolar to sub-nanomolar concentrations against panels of lung, head and neck, bladder, and pancreatic cancer cells, concomitant with downregulation of EGFR and other RTKs.163 Interestingly, the compound shows a differential substrate selectivity in cellular settings that was not anticipated based on biochemical studies.157 The selectivity was most pronounced in breast cancer cells, where apratoxin S4 preferentially downregulated HER3 over HER2. The activity of apratoxin S4 was also greater against estrogen receptor positive (ER+) and triple negative breast cancer (TNBC) cells than HER2+ breast cancer cells. The substrate selectivity appears to depend on the cellular context, since apratoxin A was shown previously to strongly downregulate HER2 in U2OS osteosarcoma cells.163 This apparent coupling or interplay of substrate selectivity and biological context is highly intriguing, and the molecular basis remains to be determined. Importantly, in contrast to known EGFR inhibitors, apratoxin S4 (45e) showed antiproliferative activity in mutant KRAS background, extending the application to addressing resistance to these selective targeted therapies.163
Apratoxins also differentially modulate cancer-related membrane proteins other than RTKs, including CUB-domain containing protein (CDCP1), a transmembrane protein linked to metastasis and invasion, forming a complex with EGFR to decrease cell adhesion.163 CDCP1 is also activated by KRAS but strongly downregulated by apratoxin S4 in a breast cancer cell type dependent manner. The fate of CDCP1 was monitored through pulldown experiments followed by proteomics, revealing the expected block in glycosylation by preventing cotranslational translocation and concomitant increase of the chaperone HSP70, presumably recruited due to misfolding of nonglycosylated CDCP1.163 Additionally, CDCP1 association with HUWE1 was increased in response to apratoxin S4 treatment. HUWE1 functions as an E3 ubiquitin ligase known to target many proteins, including BCL2-related MCL1, histones and DNA polymerases.170 Apratoxin S4 treatment presumably leads to HUWE1-mediated proteasomal degradation.163
SAR studies by alanine scanning pinpointed key amino acid residues, including the Tyr moiety and that the Pro can be replaced by N-Me-Ala, and that configurational changes of the Pro (l to d) are detrimental to the activity.159 Total synthesis led to the reassignment of the thiazoline configuration in apratoxin E (45b instead of 45c), which is different from all other natural apratoxins (Fig. 13). The significantly reduced activity of apratoxin E and dehydrated apratoxin A, indicated the importance of the hydroxy group in the Dtena unit. Importantly, re-engineering of the scaffold to remove the Michael acceptor in the modified cysteine (moCys) unit, further improved potency (apratoxin S4, 45e), indicating that the conjugated system was not involved in the mechanism of action.159 Furthermore, the unsaturated amide in apratoxin A was shown by mass spectrometry to be able to undergo conjugate (Michael) addition with thiol nucleophiles, presenting one potential liability and reason for the irreversible toxicity observed for apratoxin A.159 Collectively, these studies led to the prioritization of apratoxin A/E hybrids S4 (45e) and S9 (45f), with S9 being slightly more active.159 To avoid deactivation by dehydration and to improve stability, a gem-dimethyl was installed at C34 leading to the apratoxin S8 and S10 (45g, Fig. 13).160,169 All of these synthetic apratoxins demonstrated excellent in vivo activity and toxicity was managed through a carefully designed dosing schedule. However, the cytotoxicity of the oxazole analogue (45h) is much weaker than that of thiazoline analog.76
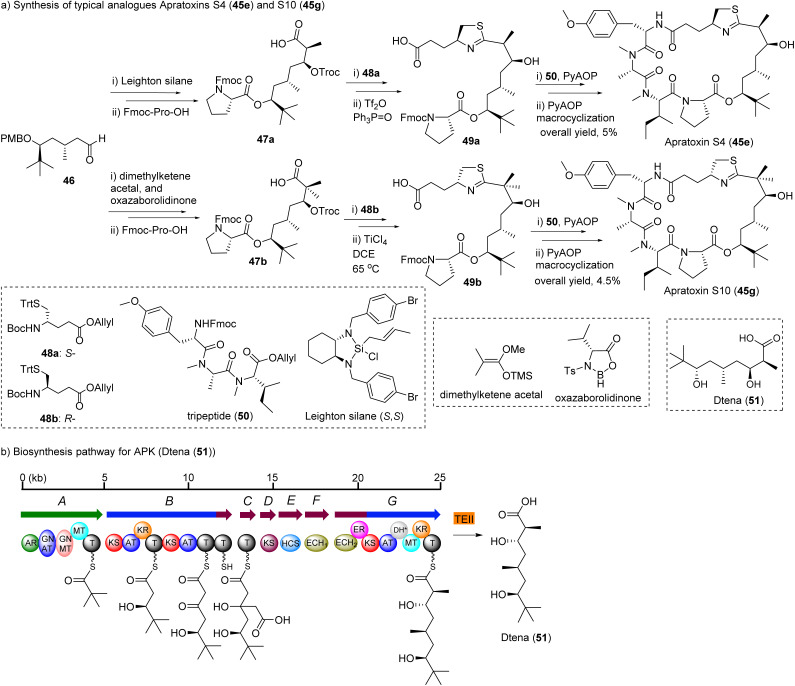
Among the synthetic analogues, apratoxin S4 and S10 are the promising candidates that have undergone most extensive biological evaluation.159,160,171 Their synthetic strategy is depicted in Fig. 15.
Fig. 15. Abbreviated synthesis (a) of apratoxin S4 (45e), S10 (45g), and (b) biosynthesis pathway for apratoxin polyketide unit Dtena (51).
The moCys precursor units 48a, 48b were synthesized from cysteine through the amino aldehyde, Wittig reaction and reduction by NaBH4. However, the product was obtained in only 35% yield.159 An alternative method to construct moCys fragment was established, starting from glutamic acid derivative, and applied to the total synthesis of apratoxin E and its epimer.166 This method is straightforward and more efficient.
Aldehyde 46 was a common intermediate for all analogues. It was synthesized from pivalaldehyde by d-proline catalyzed aldol reaction in ten steps.160 The chiral center of acid 47a was constructed by crotylation of aldehyde 46 using commercially available material by Leighton's silica (S,S)-18.172 The product was easily obtained in high yields with excellent enantiomeric selectivity by simple mixing of both starting materials and keeping the mixture at −20 °C for some time.160 However, the gem-dimethyl acids 47b were synthesized using Kiyooka reaction through a boron auxiliary, oxazaborolidine.160,173 The formation of the thiazoline ring in 49a, and 49b from its open-chain precursor was a key step and for the mono-methyl (S4) or non-methyl analogues (S7).160 Kelly method (Ph3P(O)/Tf2O) provided products in 90% yield, but for gem-dimethyl analogues (S8 and S10) were only provided in low yield (5–10%).160 However, modified Kelly method (TiCl4/1,2-dichloroethane/heating) resulted in satisfactory yields (50–60%). PyAOP was selected as the coupling reagent of acid 49 with tripeptide 50. PyAOP also was used for final step macrocyclization, and good yields were obtained for all final targets. Other analogues were synthesized by parallel strategy of the total synthesis of apratoxin S4/S10. Each analogue can be synthesized from two fragments, tripeptide 50 and thiazole-containing carboxylic acid 49. An additional series of apratoxins by replacing the moCys unit with amino acids, has also been described without in-depth biological studies.174 Towards establishing a convenient hybrid chemistry–synthetic biology to access apratoxins, the key moiety of the apratoxin skeleton, Dtena (51, Fig. 15), was heterologously expressed in high yield (9.7 mg L−1).175
3.3. Coibamides
In 2008, Gerwick and McPhail isolated coibamide A (52, revised structure) a potent cytotoxin with antiproliferative activity against the NCI-60 cancer cell line panel, from the filamentous cyanobacterium Leptolyngbya sp.148 Coibamide A showed cell loss within S phase and increase in the G1 phase with little change in G2/M phase of the cell cycle.148 In 2013, the Ishmael group described coibamide A to possibly induce mTOR-independent autophagy in both mouse embryonic fibroblasts (MEFs) and human glioblastoma cell types, with morphological and biological characteristics of cell death in various cell lines.176 Coibamide A was more recently found to have a similar mechanism of action as apratoxins and the same molecular target, Sec61. Like apratoxins, coibamide A inhibits VEGFA and decreases expression of VEGFR2, as indicative of Sec61 inhibitors.177 However, as discussed above, comparative analysis of resistance mutations indicated differences in binding and pharmacological consequences, including cell type sensitivities.176,177 Coibamide A, originally, demonstrated potent cytotoxicity against MDA-MB-231 (GI50 2.8 nM), SNB-75 (GI50 7.6 nM), HL-60(TB) (GI50 7.4 nM), and LOX IMVI (GI50 7.4 nM) with good selectivity for breast, CNS, colon, and ovarian cancer cells.148 In a more recent study, these lariat depsipeptides demonstrated excellent inhibition of cell growth of six breast cancer cell lines in the nanomolar EC50 range, with MDA-MB-231 being the most sensitive (EC50 7.42 nM).158 In an effort to further define the sensitivity of coibamide A to breast cancer types, including triple negative breast cancer, HER and ErbB proteins indicated the depsipeptide suppressed all four HER protein expression and EGFR, exhibiting cell death as a function of exposure and concentration.148 However, endogenous HER2 was partially resistant to coibamide A (similarly shown for apratoxins)163 within the same concentrations of all HER proteins, indicating broad acting Sec61 inhibitors are not equally effective across all HER proteins,148,178 consistent with data for apratoxins.163
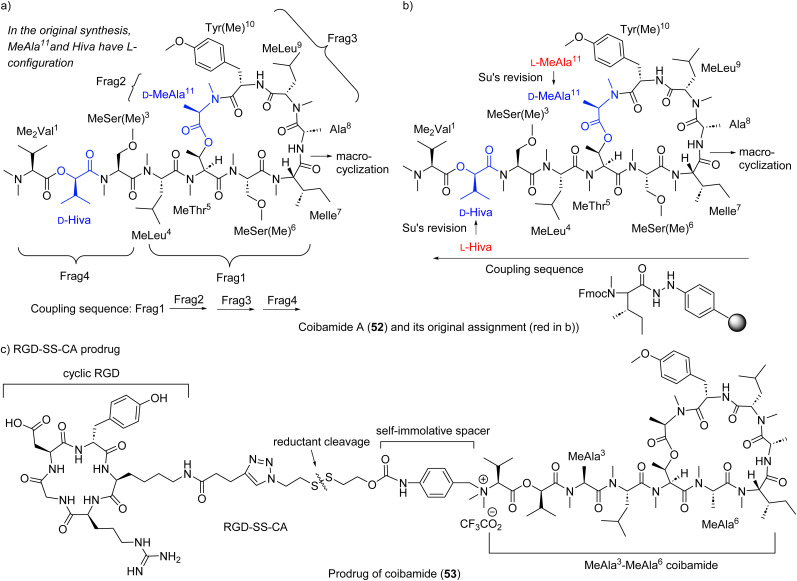
Yao group completed the total synthesis of the proposed structure in 2014.179 Using a liquid-phase and [(4 + 1) + 3 + 3] strategy, they constructed each fragment (Fig. 16a), fusing fragments 1, 2 and 3 sequentially, then preforming macrocyclization between N-Me-lle and Ala to obtain the macrocyclic core. Appending fragment 4 to the cyclic core provided the final target molecule. Unfortunately, the NMR spectra of synthetic coibamide were inconsistent with those of natural coibamide A, prompting a structural reassignment. Adopting Fmoc-based SPPS (Fig. 16b), the Su group also synthesized the originally proposed structure of coibamide A, substantiating Yao's result of the mismatch.180 Synthesis of two diastereomers with highest likelihood of representing natural coibamide A led to the unambiguous structural revision at MeAla (to d-MeAla) and Hiva (to d-Hiva) (Fig. 16b).180 In Su's SPPS method, the same site between N-Me-lle and Ala was chosen for macrocyclization. Starting from N-Me-lle on aryl hydrazide resin, they assembled amino acids via the sequence as shown in Fig. 16b, starting from N-Me-lle to Hiva, then MeAla to Ala to obtain linear precursors. Macrocyclization between N-Me-lle and Ala provided final targets.
Fig. 16. Synthetic strategies (a and b) for proposed coibamide A (52) and (c) for its RGD conjugate, and Su's structure revision as shown in (b): red represents the original assignment and blue the reassignment.
Su's group accessed a series of coibamide A analogues, as well as the MeAla3-MeAla6 coibamide, replacing Ser hydroxy and amino acids, respectively (Fig. 16c). The MeAla3-MeAla6 coibamide showed almost equal cytotoxicity against breast cancer cell line MDA-MB-231 compared with that of coibamide A.161 Furthermore, the MeAla3-MeAla6 coibamide exhibited better efficacy against tumor growth than coibamide A in vivo.161 Based on this slightly simplified analogue, authors designed and synthesized a stimuli-responsive peptide-drug conjugate (PDC) RGD-SS-CA as a prodrug (RGD: arginyl-glycyl-aspartic acid), which showed desirable drug release under stimuli and greater or similar cytotoxicity than coibamide A both in vitro and in vivo.181 In 2022, Oishi group generated series of analogues of coibamide A at MeThr5-MeAla11 site and Tyr(Me)10 site using a similar Fmoc-SPPS method and synthetic sequence as Su's method but the amide between Tyr(Me)10 and MeLeu9 was selected as the site for macrocyclization.182 A biphenyl (Bph) mimetic substituting Tyr(Me) was the most potent for the growth inhibition of A549 cells (IC50 0.06 μM).182
4. SERCA inhibitors
4.1. Mechanism of action
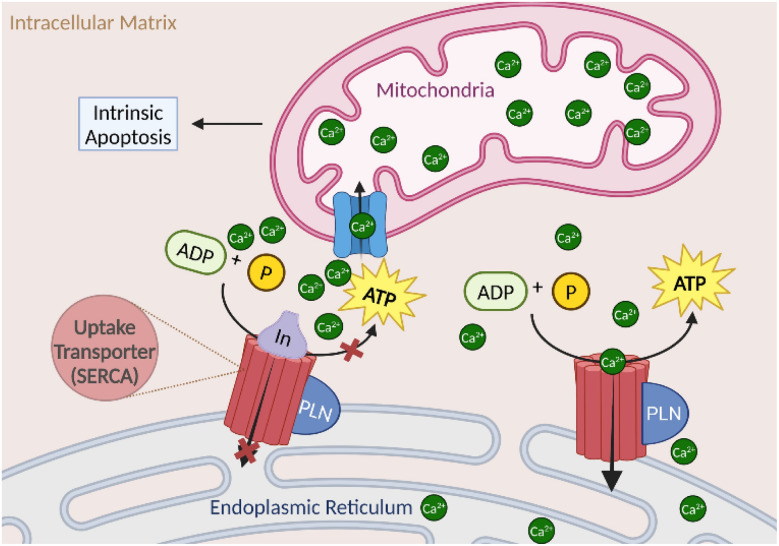
Calcium regulation intersects various cellular processes, including cancer metastasis and proliferation, and directly induces and regulates cell death.183 Thus the modulation of organelle Ca2+ homeostasis is a key target for treatment against cancer progression.183 The sarcoplasmic reticulum calcium ATPase (SERCA) is an endoplasmic reticulum 10 transmembrane calcium transporter protein found in all eukaryotic cells.184 The transporter family consists of SERCA1-3, with SERCA1α being the best characterized, and transfers calcium from the cytosol of the cell to the lumen of the ER.185 SERCA maintains the majority of calcium homeostasis, therefore potential therapeutic target development is advantageous for diseases with calcium dysregulation.185 The calcium cascade within the cell is initiated with the entry of calcium into the cell via a voltage-gated calcium channel or other calcium transporter, releasing calcium into the cytoplasm. SERCA then uptakes the calcium into the ER in an ATP-dependent manner to regulate the concentration of calcium in the membrane.184 Calcium concentration is regulated via small endogenous molecules and leak channels.186 When SERCA is inhibited, the concentration of calcium in the ER drops below 500 μM, initiating mitochondrial unfolded protein response (UPR) with the uptake of calcium into the mitochondria. Thus prolonged Ca2+ exposure leading to ER stress and subsequent mitochondrial calcium uptake leads to activation of pro-apoptotic signaling as shown (Fig. 17).187 The two marine natural products, iezoside (54a) (IC50 6.7 nM in HeLa; Ki 7.1 nM) and biselyngbyaside (62a) (IC50 0.1 μg mL−1 in HeLa; Ki 19 nM), were isolated and characterized as potent SERCA1α inhibitors.188–190
Fig. 17. Mechanism of action of sarcoplasmic reticulum calcium ATPase (SERCA) leading proapoptotic signaling.
4.2. Iezosides
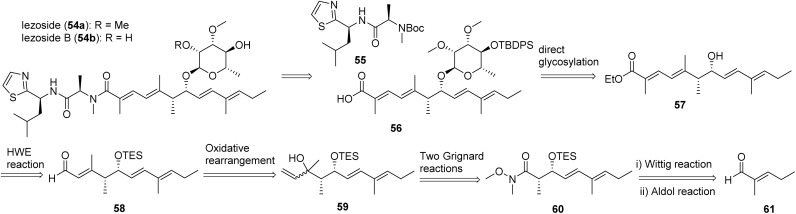
Iezoside (54a, Fig. 18) was recently isolated and characterized as a rare peptide-polyketide hybrid glycoside from Ie Island, Okinawa, Japan and identified from the marine cyanobacterium Leptochromothrix valpauliae by the Suenaga group.188 Iezoside features a sugar moiety and multiple double bonds and it also showed strong inhibitory activity against SERCA with comparable Japanese Foundation for Cancer Research 39 (JFCR39) cell lines profiles to the most well-known SERCA inhibitor thapsigargin from Thapsia plants and a calcium transporter ionophore A23187 also known as calcimycin.191–193 Validation of the target was conducted using the morphological changes that thapsigargin and cyclopiazonic (CA) induced upon HeLa cells, as well as the effect of the glycoside on the concentration of cytosolic Ca2+ in Fura-2-treated HeLa cells.188 SERCA1α was confirmed as the target, with Ki of 7.1 nM, making it the most potent marine natural product SERCA1α inhibitor to date.188 Iezoside (54a) exhibited potent antiproliferative activity against HeLa cells in cytotoxicity studies (IC50 6.8 ± 0.3 nM vs. 650 ± 82 nM) with an increase in G1 phase and a decrease in S phase cells at 10 nM, reflecting a cell cycle delay rather than cell cycle arrest due to moderate cell cycle distribution variation.188 More recently, iezoside (54a) and the demethylated analogue iezoside B (54b), were isolated by the Luesch group from Loggerhead Key in Florida from a marine cyanobacteria mixture composed of mainly Dichothrix sp. and Lyngbya sp.194 Additional validation of the bioactivity of iezoside and biological characterization of iezoside B were conducted showing cytotoxicity against non-small cell lung cancer cell line A549 (IC50 1.5 and 3.0 μM) and cervical cancer cell line HeLa (IC50 1.0 and 2.4 μM), providing preliminary SAR.155
Fig. 18. The structures of iezoside (54a) and iezoside B (54b), and synthetic strategy for iezoside.
The synthetic strategy of iezoside (54a) is depicted in Fig. 18, in which it was detached into two primary building blocks 55 and 56. Amine 55 is a thiazole-containing dipeptide. The alcohol 57 was glycosylated to afford 56 successfully in the presence of carbonyl group. From known aldehyde 56, via a series of sequential reactions including Wittig reaction, aldol reaction, Grignard reactions, oxidative rearrangement, and Horner–Wadsworth–Emmons (HWE) reaction, compound 57 was synthesized.188
4.3. Biselyngbyasides
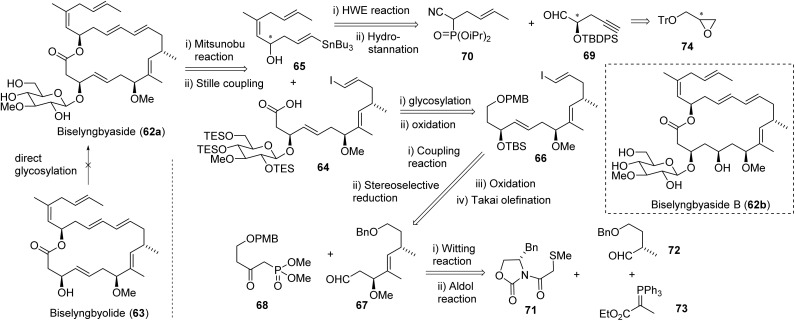
Biselyngbyaside (62a, BLS), an 18-membered ring macrolide, was isolated from Lyngbya sp. in 2009 by the Suenaga group.190 Similar to iezosides, structural features include sugar moieties and multiple double bonds. The macrolide exhibited cytotoxicity against HeLa S3 (IC50 100 ng mL−1) and an average GI50 of 600 nM against the JFCR39 human cancer cell line panel, with specific selectivity to glioblastoma SNB-78 cell line (GI50 36 nM) and the non-small cell lung cancer cell line NCI H522 (GI50 67 nM).189,190 The JFCR39 profile was similar to that of known SERCA inhibitors, with enzyme assays against SERCA1 and 2 confirming biselyngbyaside strongly inhibited SERCA1α (Ki ∼ 10 nM).189 Later studies described the isolation and biological activity of biselyngbyaside B (62b), which is less potent than BLS.195 Additional natural analogues (C–F) were isolated from the Lyngbya sp. and tested for potency against HeLa and HL60 cell lines.195,196
The Suenaga group reported the first total synthesis of BLS (62a, Fig. 19) in 2017 based on their successful total synthesis of biselyngbyolide (63) in 2016.197,198 They found that direct glycosylation of biselyngbyolide (63) at a late-stage failed to produce BLS (62) and proposed to install the sugar moiety before the carbonyl group was formed (Fig. 19). For the macrocycle formation, building blocks 64 and 65 were conjoined via Mitsunobu reaction, and then a Stille coupling between stannane of 65 and vinyl iodide of 64 closed the linear precursor. The glycosylation of free sec-OH of 66 was completed by trichloroacetimidate-activated glycoside donor, differing from the installation of a sugar moiety of iezoside. Further two-step oxidation of primary alcohol of 66 provided 64. The coupling reaction of 67 with 68, and then stereoselective reduction with BH3·SMe in the presence of chiral boron auxiliary (R)-Me-CBS (Corey-Bakshi-Shibata), followed by oxidation and Takai olefination to provide 66. Wittig reaction and Aldol reaction were involved when used building blocks 73, 72, and 71 to construct 67. For the construction of building block 65, the Ando's type phosphonate 70 with nitrile group was selected for HWE olefination, which afforded good yield and selectivity for E-configuration.
Fig. 19. The synthetic strategy for biselyngbyaside (62a).
While SERCA inhibitors may have gained momentum as potential agents against cancer, the major practical concern has been the difficulties associated with the synthesis of their complex structures for large scale synthesis. The synthesis of BLS still remains a laborious multistep methodology, while the lack of diversity in the structure scaffold with target specific inhibitory activity remains a challenge. The recent discovery of the potent cyanobacterial iezosides provides more feasibility in addressing both of these challenges.
5. Mitochondrial cytotoxins and extrinsic apoptosis inducers
5.1. Mechanism of action
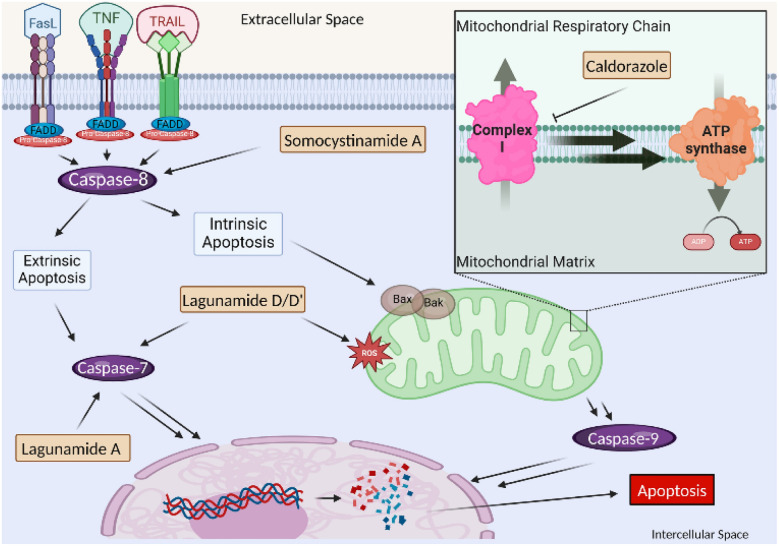
As the mitochondria are responsible for several critical cell functions, such as apoptosis regulation and oxidative phosphorylation, cancer pathology involving the organelle is a prime target for therapeutics.199,200 Mitochondrial cytotoxins target functional pathways resulting in the induction of tumor cell apoptosis (Fig. 20). Apoptosis is guided by the intrinsic and extrinsic pathways, both involving the activation of caspases.201 The intrinsic pathway is mediated by intracellular signals from an array of positive or negative non-receptor-mediated catalysts, producing a cascade effect to lose mitochondrial membrane potential and initiate the release of pro-apoptotic enzymes to execute the caspase cascade.202,203 Oxidative stress through various enhancers, like lagunamides (75a–e), increases signaling to induce an accelerated effect on caspase cascade induction.204 In contrast, the extrinsic pathway often involves pro-death signaling from outside of the cell via CD8-positive cytotoxic T or Natural Killer lymphocytes to death receptors on the cell membrane, including TNF-related apoptosis-inducing ligand (TRAIL), tumor necrosis factor (TNF), and Fas receptors.205 Caspase-8 is a gateway inducer of both intrinsic and extrinsic apoptotic pathways as a mediator of cell death and inflammation, which is targeted by the somocystinamide compound class (106b).206,207 Caspase-mediated mitochondrial cytotoxins can disrupt the membrane potential and cause overexpression of radical oxygen species, as well as upregulate/downregulate pro/anti-apoptotic signaling.208 This includes respiratory chain inhibitors targeting oxidative phosphorylation via interrupting glycolysis, terminating the austerity of tumor cell lines, such as caldorazole (90).209–211 Targeting apoptosis is one of the most successful non-surgical remedies and recognized as an effective universal target for treatment.201
Fig. 20. Mitochondrial cytotoxins within the cell and the downstream affects, including the caspases of the intrinsic apoptotic pathways and various signaling proteins to induce arrest or oxidative stress.
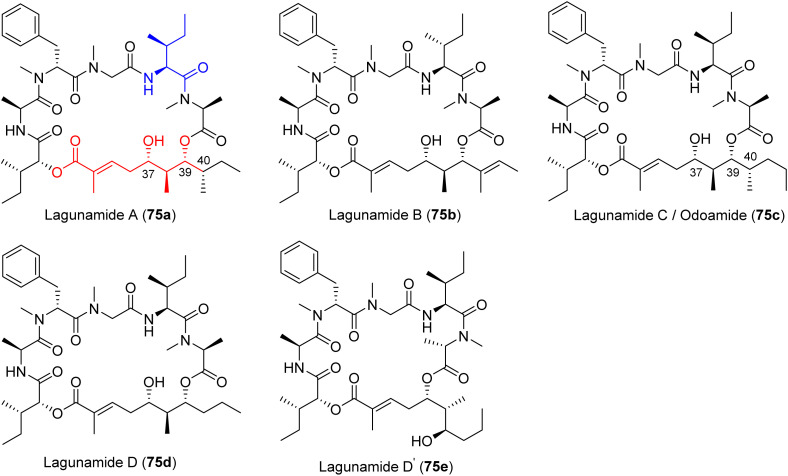
5.2. Lagunamides and odoamide
The lagunamides are cyclic dipeptides that were isolated from the cyanobacterium Lyngbya majuscula by the Tan and Luesch groups.212–214 The compounds are structurally related to aurilides and kulokekahilide-2.212,215–217 Lagunamide A (75a) induced morphological changes to A549 cells after a 24 h treatment, including shrinkage, pseudopodia retraction, karyopyknosis, and chromatin condensation, as well as nuclear cracking and DNA released into the cytoplasm via transmission electron microscopy (TEM).208 Further biological characterization of lagunamide A (75a) indicated the overproduction of ROS and mitochondrial membrane potential disruption, as well as upregulation of pro-apoptotic Bcl-2 proteins and downregulation of the anti-apoptotic Bcl-2 proteins, specifically Mcl-1.208 Lagunamide C (75c) was isolated in 2011 by the Tan group and indicated nanomolar cytotoxic potency against murine leukemia (P388; IC50 2.24 nM), lung carcinoma (A549; IC50 2.4 nM), prostate cancer (PC3; IC50 2.6 nM), ileocecal colorectal adenocarcinoma (HCT8; IC50 2.1 nM), and ovarian (SK-OV; IC50 4.5 nM) cancer cell lines.214 Recently, the structural revision of lagunamide C (75c) by the Kigoshi group revealed the lagunamide C is the identical to odoamide.218 Odoamide (75c) is structurally similar to lagunamide A (75a), differing only by a single methylene, and was isolated from the Okinawan cyanobacterium Okeania sp. by the Teruya group in 2016.219 Biological characterization of lagunamide D (75d), and D′ (75e, Fig. 21) indicated activation of caspases-3/7 within 6 h of treatment as well as mitochondrial rearrangement without fragmentation, exhibiting effects of both apoptotic pathways.204 The exact mechanism in which lagunamide D affects the mitochondria is yet to be elucidated, despite their ability to induce apoptosis.208,213,220 However, as a member of the aurilide class, the likely target could be prohibitin-1 (PHB1).221 PHB1 has a role in cell proliferation as a negative regulator and is localized to the inner membrane of the mitochondria.222 This is mediated through the stimulation of optic atrophy 1 (OPA1), which leads to the induction of mitochondrial fragmentation and cell death via apoptosis.221 However, a functional genomics driven approach using lagunamide D revealed that the depsipeptide might exert its affects through proteostasis modulation or having a chemical genetic interaction with the proteasome pathway.204 This was revealed through an unbiased chemogenomic RNAi screening. A targeted approach indicated the mechanism worked through mitochondrial network rearrangement, not mitochondrial fragmentation.204 Preliminary biological studies indicated potent cytotoxicity against HeLa S3 cells (IC50 26.3 nM). The later synthesized odoamide exhibited nanomolar potency against A549 cells (IC50 2.1 nM).219,223,224
Fig. 21. Mitochondrial cytotoxins from marine cyanobacteria structures of lagunamides (75a–e) and odamide (75c).
Ye and Xu finished the first total synthesis of lagunamide A (75a, Fig. 21) in 2012.225 In 2013, Wei achieved the synthesis using different strategies for the generation of the α-hydroxy acid unit and macrocyclization.226 In 2018, Kazmaier applied Matteson homologation as the major tool to finish the total synthesis of lagunamide A (75a).227 Oishi completed the total synthesis of odoamide (75c) in 2016 and then synthesized a series of analogues in 2018.223,224 The emphasis of the total synthesis of lagunamide or odoamide has been the construction of α-hydroxy acid fragment and macrocyclization (Fig. 21).
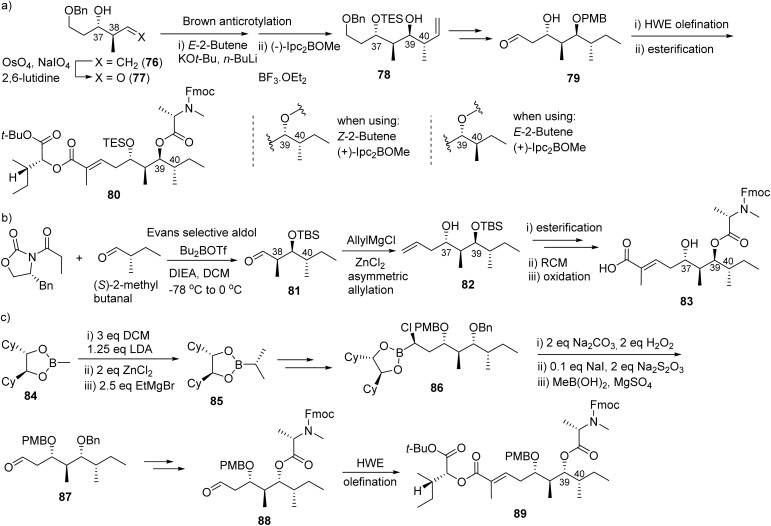
Fig. 22 shows the different strategies for the synthesis of the hydroxy acid fragment. In the synthesis of the proposed lagunamide A, Ye group found the configurational assignment for C39 and C40 was problematic and used the known chiral olefin as a starting point to furnish chiral carbons C37 and C38.225 Its aldehyde derivative, obtained by oxidative cleavage, afforded chiral carbon C39(S) and C40(S) through Brown anticrotylation in which E-2-butene/(−)-Ipc2BOMe assembly was used (Fig. 22a).225 Another two C40 epimers (C39(R)-C40(S) and C39(S)-C40(R)) were also synthesized using different assembly of butene and Ipc2BOMe (Fig. 22a). From these three parallel building blocks, they generated the corresponding diester fragments via HWE reaction and esterification and completed three analogues of lagunamides. By comparison, the correct structure of natural lagunamide A was determined to be the C39(R)-C40(S) isomer; however, the original assignment was C39(S)-C40(S).
Fig. 22. Comparison of different strategies (a–c) for the synthesis of hydroxy acid fragment.
Generally, Wei applied Evans's selective aldol reaction of Evans (R)-oxazolidinone and (S)-2-methylbutanal to establish the three chiral centers, C38-40, of the hydroxy acid building block (Fig. 22b), similar to the strategy Ye used in the primary effort.225,226 Asymmetric allylation with allylmagnesium chloride generated the chiral carbon C37, in which the diastereoselectivity was improved to 90 : 10 when ZnCl2 was selected as an additive (Fig. 22b). Furthermore, Wei applied the ring closing metathesis (RCM) reaction to furnish the unsaturated acid side chain for this fragment. Creatively, Kazmaier applied Matteson homologation to establish the four chiral centers of the hydroxy acid fragment (Fig. 22c).227 For the macrolactamization, it seems that the site between Ala and Hmpa (2-hydroxy-3-methylpentanoic acid) provided better cyclization efficiency than the site between Gly and Ile.
Oishi achieved the total synthesis of odoamide (75c) and some analogues by establishing the stereogenic center, C38–C39, using a method similar to that of Ye's and Wei's.223–226 Instead of Evans selective aldol reaction, Oishi used Mukaiyama aldol reaction to generate the chiral center at C37.223
Oishi chose the site between Ala and Hmpa as macrocyclization site. However, serious epimerization problems at isoleucine were encountered because isoleucine was installed prior to the tetrapeptide.
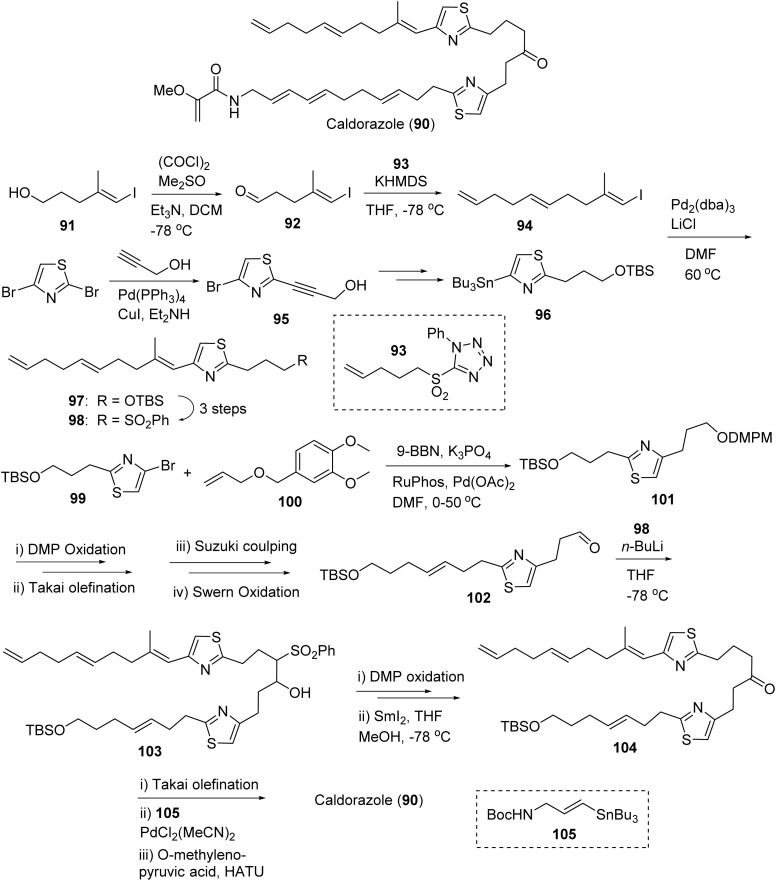
5.3. Caldorazole
Caldorazole (90, Fig. 23) was isolated from Caldora sp. off the coast of Tomuruzaki in Ishigaki Island, Japan in 2022 by the Suenaga group.228 The polyketide contains two thiazole rings, a terminal olefin and an O-methylenolpyruvamide moiety (90, Fig. 23). The compound showed potent cytotoxic activity (IC50 < 100 nM) against several solid tumor cell lines (HeLa IC50 23 nM; CaSki IC50 68 nM; HT1080 IC50 74 nM).228 The potential as a phosphoenolpyruvate mimic was confirmed via selectivity in the presence of 2-deoxy-d-glucose (2DG), which suppresses glycolysis in the mitochondrial respiratory chain complex 1.228 Selectivity for glucose-suppressed conditions in tumor cells was confirmed with caldorazole showing no toxicity against the normal cell line WI38 in the presence of 2DG. Caldorazole did not show selectivity against HeLa S3Mer− cells, which are highly selective for DNA alkylating agents; thus the mechanism of cytotoxicity via DNA alkylation was not relevant to caldorazole.228 Biakamides, a sponge isolated natural products, are a similar structure class with similar mechanism to caldorazole in inhibition of mitochondrial respiration making them both attractive agents for developing therapies that can potentially cause “nutrient starvation” in cancer cells.229,230
Fig. 23. Structures and total synthesis of caldorazole (90).
The Suenaga group and their co-workers completed the total synthesis of caldorazole (90) in 2023 as depicted in Fig. 23.211 The authors' preliminary convergent synthetic strategy is to disassemble the molecule into three parts at the sites of amide and middle keto and the ketone will be formed by sulfone coupling. The synthesis of the left side building block was started with Swern oxidation of iodide alcohol 91. The aldehyde was subjected to Julia-Kocienski olefination with sulfone 93 to afford vinyl iodide 94 (E/Z 4 : 1). The selective alkynylation of 2,4-dibromothiazole with Sonogashira cross-coupling to provide alkyne 95, which was transformed into stannane 96via reduction of alkyne by TsNHNH2 and stannylation by Bu3SnCl. Stille coupling between 94 and 96 gave TBS ether 97. Then its sulfide (by PhSSPh/n-Bu3P) was oxidized to sulfone 98. Suzuki–Miyaura cross coupling of 99 with the hydroboration product of allyl ether 100 afforded protected ether 101. The elongation of the TBS ether side of 101 was completed by sequential deprotection, DMP oxidation, Takai olefination and Suzuki coupling with the corresponding product being converted into aldehyde 102 by Swern oxidation. It is noteworthy that a 10 : 1 E/Z olefination ratio was achieved. Sulfone coupling of 102 and 98 produced β-OH sulfone 103, which was subjected to oxidation and desulfonylation to provide keto compound 104. The sequential deprotection of TBS group of 104, oxidation, Takai olefination, Suzuki coupling (with 105) and the coupling with O-methylenolpyruvic acid afforded the final product caldorazole 90 in 0.47% overall yield.211
5.4. Somocystinamide A and laucysteinamide A
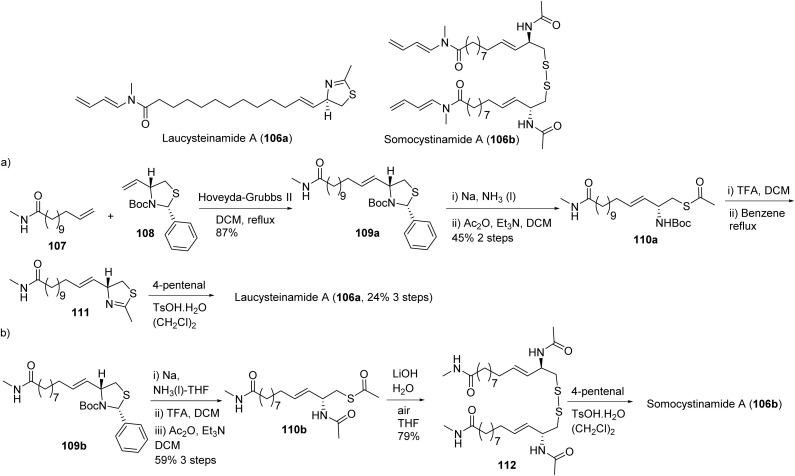
Somocystinamide A (106b, Fig. 24) was isolated by the Gerwick group from a cyanobacteria mixture Lyngbya majuscula/Schizothrix sp. in 2002 from Somosomo, Fiji.231 This novel lipopeptide of mixed PKS/NRPS biosynthetic origin was mildly cytotoxic to mouse neuro-2a neuroblastoma cells (IC50 1.4 μg mL−1) originally,231 then later revealed potent cytotoxicity against leukemia (Jurkat IC50 3 nM; CEM IC50 14 nM) and lung carcinoma (A549 IC50 46 nM) and good cytotoxicity against breast carcinoma (MCF-7 IC50 210 nM), neuroblastoma (NB7 IC50 819 nM), and prostate carcinoma (PC3 IC50 970 nM).206 Further characterization revealed somocystinamide A activates programmed cell death via caspase-8 as well as apoptotic activity in tumors resistant to death receptor-mediated killing and Fas-mediated apoptosis via plasma membrane lipid compartment modifications.206 Together, the mechanistic studies revealed the unique method in which ScA exhibits both an angiogenetic effect and inhibition of tumor cell progression. (Fig. 24, 106b).206
Fig. 24. Structures of laucysteinamide A (106a) and somocystinamide A (106b) and the synthetic strategies (a and b) for the two compounds.
Laucysteinamide A (106a, Fig. 24), monomeric analogue of somocystinamide A, was isolated from Caldora penicillata in the Northern Mariana Islands by the Gerwick group.232 The lipopeptide exhibited weak cytotoxicity, compared to somocystinamide A, against lung cell carcinoma (H460) with an IC50 of 11 μM, with the synthetic equivalent of laucysteinamide A exhibiting cytotoxicity against H460 at 20 μM.233
Using the their improved protocols for the total synthesis of somocystinamide (106b), Gerwick and co-workers completed the total synthesis of laucysteinamide A (106a) in 2021 (Fig. 24, b).233,234 The available unsaturated compound 107 and 108 were subjected to olefin cross metathesis catalyzed by Hoveyda-Grubbs II catalyst to afford thiazolidine 109a. Birch reduction followed by acylation of 109a provided thioester 110a. The thiazoline compound 111 was prepared by reflux to induce dehydrative cyclization of the Boc-cleaved 110a. The condensation of 111 with 4-pentenal using TsOH as a catalyst afforded the final product laucysteinamide A (106a) in 9.4% overall yield. The authors applied the improved protocols to the total synthesis of somocystinamide (106b, Fig. 24) and obtained a better overall yield. The thioester 110a was prepared from 109a using the same protocols for 110a. The basic hydrolysis of thioester of 110b in the presence of air made the dimerization provided somocystinamide A (106b) with an overall yield of 16.1%.233,234
6. Epigenetic modulators
6.1. Mechanism of action
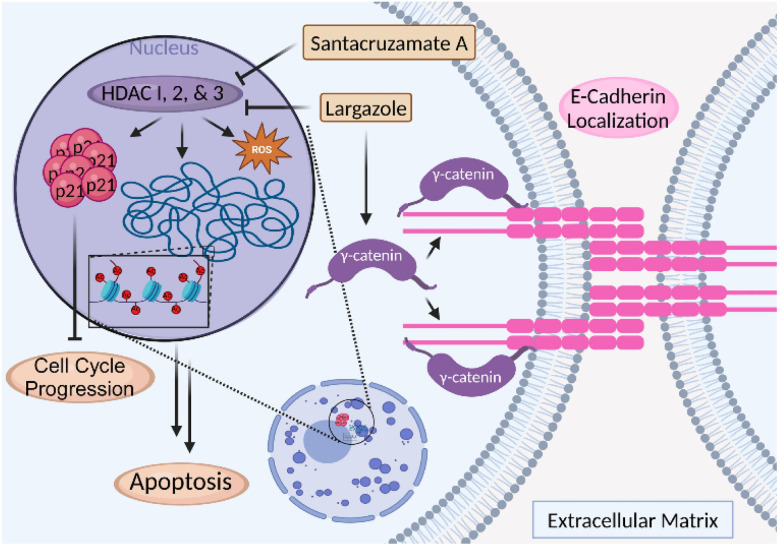
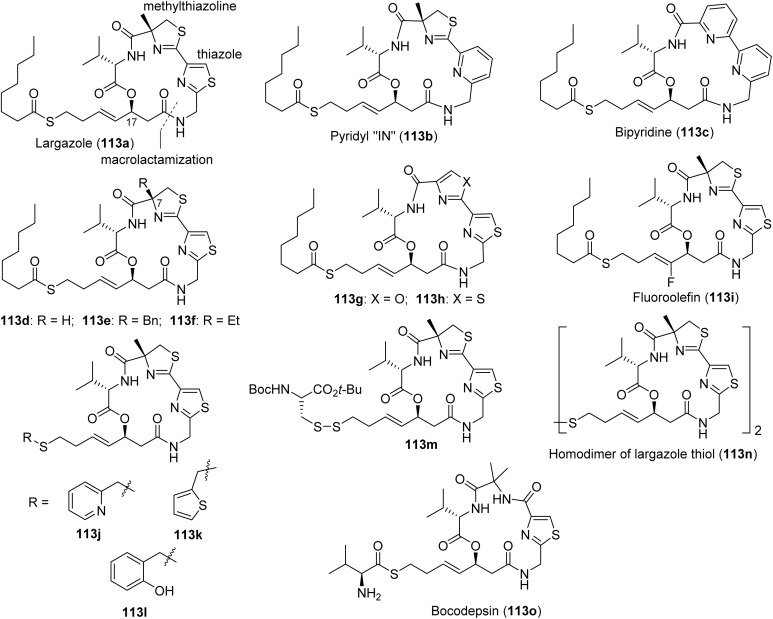
Inhibition of histone deacetylases (HDACs) is a validated anticancer strategy. Several drugs that target zinc-dependent isoforms (HDAC1–11) with different selectivity profiles have been approved for the treatment of various lymphomas (T-cell lymphomas, multiple myeloma) but have not shown significant clinical efficacy against solid tumors.235,236 HDAC inhibition leads to hyperacetylation of histones, open chromatin structure and activation of gene expression, including genes encoding cell cycle inhibitors and pro-apoptotic proteins, which mainly or at least partially account for the anticancer activities of pharmacological HDAC inhibitors.237 Recent reviews have highlighted the importance of these targets for therapeutic applications.235,238 (Fig. 25) To date, two HDAC inhibitors have been identified from marine cyanobacteria: (1) largazole (122a, Fig. 26), a thioester prodrug that liberates a zinc-binding thiol group upon activation, and (2) santacruzamate A (130, Fig. 30), containing a hydroxamate functionality as the moiety with Zn2+ affinity.239–242
Fig. 25. Class I HDAC inhibitors regulate histone deacetylation through HDAC 1, 2, and 3, therefore activating gene expression, but also possess cytoplasmic (non-nuclear) targets that promote anticancer activity, including modulation of the E-cadherin complex and consequent cell–cell adhesion.
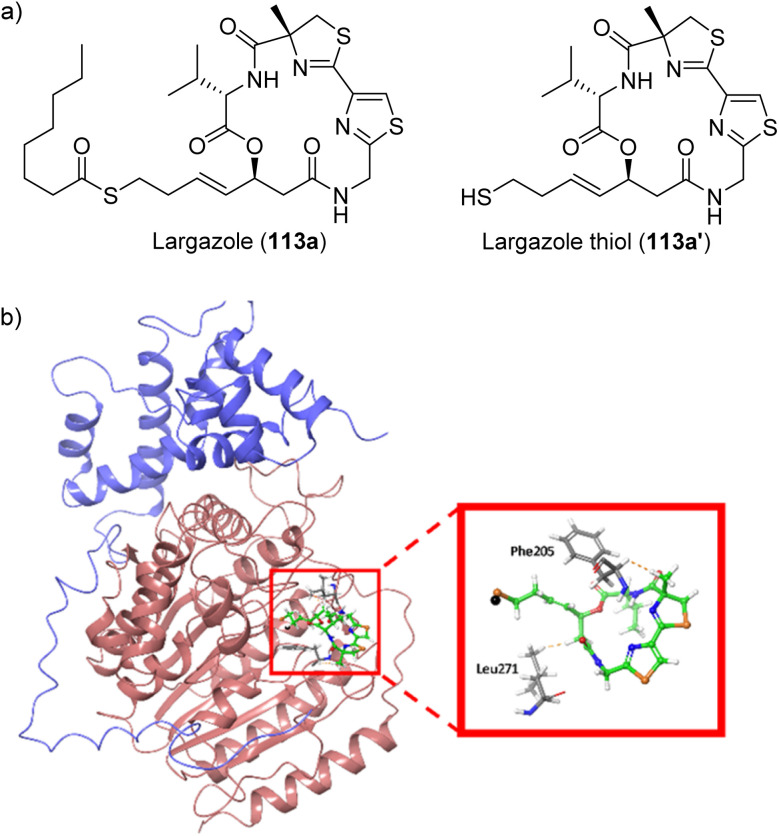
Fig. 26. (a) Structures of largazole (prodrug) and its active species (largazole thiol) and (b) molecular docking of largazole thiol (113a′) to HDAC1 crystal structure (PBDID: 5ICN).
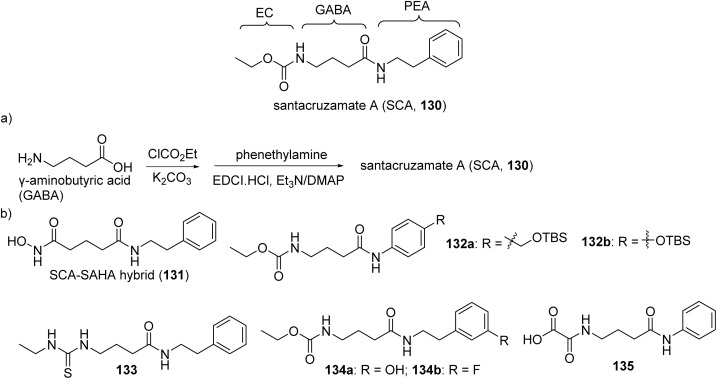
Fig. 30. Parent structure of santacruzamate A (130), (a) synthesis and (b) analogues.
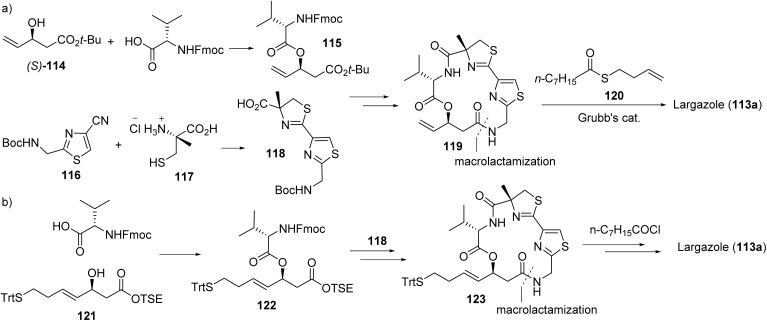
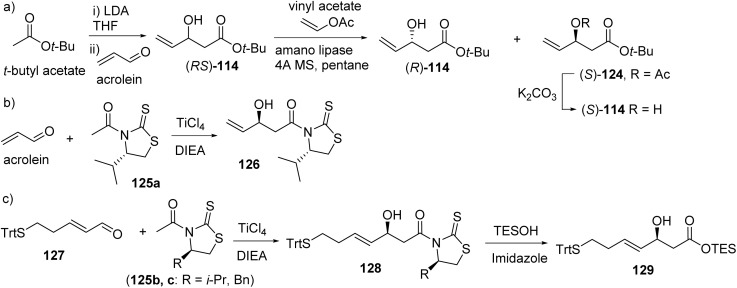
6.2. Largazole
In 2012, Hong and Luesch reviewed the discovery, general synthetic strategies, cytotoxicity, HDAC isoform selectivity profile, HDAC8-largazole thiol crystal structure, mechanism of action, and SAR for largazole (113a).243 Initial biological evaluation indicated nanomolar potency on human bone cancer (U2Os IC50 55 nM) and mouse normal breast/fibroblast cells (NMuMG IC50 122 nM/NIH3T3 IC50 480 nM), with superior potency for the breast cancer cell line MDA-MB-231 (IC50 7.7 nM).241 Bowers et. al. characterized the HDAC selectivity as HDACs 1, 2, and 3 selective over HDAC6.239 Importantly, largazole (113a) showed solid tumor activity in a HCT116 colorectal cancer xenograft mouse model at 5 mg kg−1, correlating with the histone hyperacetylation status at that dose, so that the antitumor effect was attributed to gene expression changes due to HDAC inhibition.244 Although to a lesser extent than tubulin agents dolastatins 10 and 15 (Sections 2.2, and 2.3), largazole also showed greater efficacy in HCT116 cells containing oncogenic KRAS and selectivity for HCT116 cells containing functional HIF transcription factors.131 Largazole phenocopied the colgate 1 zebrafish mutant and inhibited HIF-induced angiogenesis in vivo in vhl mutant zebrafish, but also to a lesser degree than dolastatin 15 (Section 2.3).131
Largazole (113a) is a prodrug that is activated by protein-assisted hydrolysis to generate largazole thiol (113a′) that is optimized to access the zinc within the active site of the HDAC to give its potent inhibitory effect, similar to the active (reduced) form of FDA approved drug FK228 (romidepsin).245 Largazole thiol (113a′) exhibited potent, sub-nM activity against class I isoforms HDAC 1, 2, and 3 (Ki of 0.07, 0.07, 0.17 nM), and to a much lesser extent class IIb isoform HDAC6 (Ki 25 nM).239 For comparison, FK288 exhibited Ki values of 0.12, 0.14, 0.28, and 35 nM against HDAC1, 2, 3, and 6, crowning largazole thiol (113a′) as the more potent HDAC inhibitor.239 The molecular docking of largazole thiol to HDAC1 crystal structure has provided a foundation in which these inhibitors interact with HDAC (Fig. 26). The largazole thiol binds to the zinc deep within the active cleft of the zinc finger, forcing the overall metal coordination geometry to be almost tetrahedral.243 Additionally, the macrocyclic portion exists outside of the convergent region of all HDACs, providing a base to induce isoform selectivity via SAR of the amino acid residues on the ring.243 Comprehensive profiling against all eleven zinc-dependent HDAC isoforms underscored the HDAC1–3 selectivity.243 Largazole (113a) exhibits greater antineoplastic effects than largazole thiol (113a′) on malignant melanoma cells (IC50 45 nM–315 nM vs. IC50 360 nM–2600 nM), highlighting the importance of the prodrug for cellular efficacy.239
Largazole (113a) was shown to both induce the expression of the tumor suppressor E-cadherin and also to modulate the composition of the E-cadherin complex in triple negative breast cancer cells (TNBCs) by increasing the content of γ-catenin, promoting transport of E-cadherin to the cell surface to mediate cell–cell interaction and reduce invasion.246,247 Reduced or lack of E-cadherin expression and improper cytoplasmic localization (if expressed) are characteristic feature and causative for the invasiveness of TNBCs, and largazole reversed both TNBC features to “normalize” the cellular phenotype to a less invasive state.246,248 This dual effect of largazole on gene expression and modulation of protein–protein interaction translated into mouse models.246 TNBC line tumors ectopically expressing GFP-tagged E-cadherin demonstrated the proper cell surface localization upon treatment with largazole ex vivo using excised tumors or by systemic administration in vivo (10 mg kg−1).246 The activity was enhanced in combination with dexamethasone which acts by preventing CDCP1 cleavage and formation of the invasive cleaved (cCDCP1) form.246 The cooperative effects with glucocorticoids indicated that largazole (113a), or HDAC inhibitors in general, could be effective in this biological context, potentially providing the basis for combination therapy clinical trials for TNBC.246
Largazole (113a) was shown to demonstrate potent and selective cytotoxicity towards a number of lung cancer cell lines, inhibiting cell proliferation within an IC50 range of 0.077 to 0.57 μM in wild type or mutant EGFR cell lines. The 16-HBE normal human bronchial epithelial cells exhibited less potency at more than 10 μM.249 Largazole was shown to arrest the cell cycle at G1 phase with high concentrations of treatment causing activation of caspase 9 with the downregulation of pro-caspase 9, indicating a role in the intrinsic apoptotic pathway with the cleavage of PARP using Annexin V/PI staining and flow cytometry analysis.249 The upregulation of p21 as demonstrated in A549 cells further supports the conclusion that largazole induces G1 phase arrest.249
More recently, largazole (113a) was found to be bioavailable in the brain and can reach in vivo concentrations sufficient to inhibit glioblastoma cell proliferation in vitro.250 At 50 mg per kg ip, largazole treatment for 12 h induced neuroprotective and cancer-related gene expression changes, extending the applicability of largazole to neurodegenerative diseases and brain cancers.250 Global gene expression profiling of largazole and Ingenuity Pathway Analysis (IPA) of RNA sequencing data sets revealed upregulation of neuronal transcription factor Pax6 and Oprm1.250 In addition to its role in neurogenesis and neuronal plasticity, Pax6 is known to suppress proliferation, invasion and colony formation of glioblastoma cells.251 Pax6 expression also inversely correlates with tumor grade. The data implicates Pax6 as a relevant indirect largazole target to mediate both beneficial activities. Oprm1 is a μ-opioid receptor shown to induce neuroprotective activity via the mTOR signaling against β-amyloid peptide neurotoxicity and is the target for morphine.252
The therapeutic potential of largazole (113a, Fig. 27) to treat cancer and other diseases triggered the synthesis of analogues, including different prodrugs like hydroxamates, ketones and disulfides (113b–113n, Fig. 27)247,253–263 The pyridyl “IN” analogue (113b) showed similar potency against 797 NUT midline carcinoma cell line.264 Bipyridine analogue (113c), series of C7 and thiazole analogues (113d–113h), and the fluoroolefin analogue (113i) exhibited similar potency against HDAC1 or HDAC8.259,265,266 Zinc-binding group analogues (113j–113m), including different disulfide prodrugs (113m), were also synthesized to modulate activity profiles and change the modes and timing of activation, providing opportunities for conjugation to targeting agents, and to address potential liabilities or efficacy issues due to the rapid protein-assisted thioester hydrolysis observed for largazole during extensive pharmacokinetic studies in rats.244,247,260,262,267 The disulfide homodimer (113n) generates two equivalents of largazole thiol and therefore has superior in vitro potency on a molar basis. Other structural modifications were aimed at tuning the class I HDAC isoform selectivity profiles (HDACs 1–3, 8), which was achieved by replacing the valine residue with other amino acids (Phe, Tyr, His, Asp, and lower homologues of Lys). These efforts led to largazole analogues without significant HDAC8 inhibition and where HDAC2 activity was dialed down (Phe), resulting in an HDAC1,3 inhibitor with 5.9-fold selectivity for HDAC1 over HDAC2. Replacing Val with His resulted in the most HDAC1-selective isoform inhibitor with 7.0- and 5.5-fold selectivity over HDAC2 and HDAC3, respectively.257 This data suggests that the largazole scaffold can be a template for the design of next-generation isoform selective inhibitors. One largazole-based thioester prodrug with simplified core structure, bocodepsin (OKI-179, 113o), has reached the clinic so far. Designed through a lead candidate optimization program to enhance isoform selectivity and oral bioavailability, its thiol retained class I selectivity HDACs (IC50 1.2 nM for, HDAC1; 2.4 nM for HDAC2, 2.0 nM for HDAC3, and 47 nM for HDAC8) over class IIb HDACs, including HDAC6 and 10 (IC50 47 nM, 2.8 nM); and class IV HDAC11 (IC50 2.3 nM).268,269 Recently, a first-in-human phase 1b/2 trial was conducted using bocodepsin (113o) as a treatment for advanced solid tumors and concluded in prolonged disease stabilization as the first orally bioavailable, Class I targeting HDAC inhibitor.270,271
Fig. 27. Structures of selected largazole analogues (113a–o) designed and synthesized for SAR studies.
For the synthesis of largazole (113a), most groups chose the site between β-hydroxy acid and thiazole fragment for macrocyclization, which offers less steric hindrance. Two main convergent strategies were developed for the total synthesis of largazole (113a). The major difference between these two strategies is the timing of incorporation of the pendant side chain and pharmacophore. In one strategy (Fig. 28), the allylic cyclic core 119 was synthesized first from vinyl hydroxy acid 114, valine, thiazolyl nitrile 116 and α-methylcysteine 117.264,272–274 Then thiol 120 was fused to the cyclic core using a cross metathesis reaction to complete the final target. This strategy is amenable to the synthesis of diverse analogues using various building blocks, 114, 116, 117, Val and 120. However, in the second strategy (Fig. 28b), the trityl-masked thiol (121) was first esterified with Fmoc-protected valine.239,275,276 Subsequently, the cyclic core was synthesized from this fragment, the valine and thiazoyl-thiazoline moiety (118). The n-octanoyl chloride was coupled with unmasked thiol group to provide final product (113a). Strategy depicted in Fig. 28a is suitable for the late-stage diversification of the side chain and installation of various prodrugs.
Fig. 28. Two main strategies (a and b) for the total synthesis of largazole (113a).
Hydroxy acid (114, Fig. 28) is the key building block in both synthetic strategies (Fig. 28a and b). The Nagao auxiliary masked hydroxy acid (126, Fig. 29b) induced a direct reaction without the cleavage of the auxiliary.240 Aldol reaction between acrolein and enolate from t-butyl acetate provided a racemic allylic hydroxy ester (RS)-114 (Fig. 29a).265,272,273,275 The (S)-enantiomer of 114 could be separated from the (R)-114 when the (S)-114 was selectively protected by allyl acetate under enzymatic resolution (amano lipase). Base-catalyzed deacetylation afforded (S)-hydroxy acid (114). However, the hydroxy acid 114 is sensitive to basic condition and prone to dehydration. From this point of view, the asymmetric selective aldol reactions (Fig. 29b and c) using Nagao auxiliaries (125a–c) has advantages over the method in Fig. 28a.240
Fig. 29. The synthesis of hydroxy acid moiety ((S)-114) using different strategies (a–c).
In the process chemistry study, the Luesch group adopted the method from Fig. 29a, with notable critical modifications.277 Only 0.5 eq. K2CO3 was used in the large-scale preparation and the dehydration was alleviated significantly. When the thioester (120) was installed at the late-stage, the final product largazole (113a) was contaminated with catalyst (brownish product). This problem was solved by the assembly separation using normal-phase and reversed-phase C18 chromatography. For the synthesis of the thiazoline-thiazole moiety (118), most groups proceeded in a similar manner.
6.3. Santacruzamate A
Santacruzamate A (130, Fig. 30) is an achiral cytotoxin isolated from the cyanobacterium Symploca sp. collected in Coiba National Park off the Pacific coast of Panama by the Gerwick and Balunas groups. The compound possesses structural similarities to the clinically approved HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, vorinostat).242 Preliminary bioactivity indicated selectivity for HDAC2 at an IC50 of 0.119 nM and cytostatic activity against cutaneous T-cell lymphoma (Hut-78 GI50 1.4 μM) and colorectal cancer (HCT116 GI50 29.4 μM).242 In an independent study, the HDAC2 inhibitory activity of santacruzamate A (130) was found to block nuclear translocation of PD-L1, presumably by enhancing the acetylation status of Lys263, leading to reprogramming of the expression of immune-response related genes.242 Consequently, santacruzamate A was found to enhance the antitumor activity of PD-1 blockage using anti-PD-1 antibodies in a MC38 syngeneic mouse tumor model.278 This effect was not observed in immunocompromised nude mice, supporting the hypothesis of T-cell dependent combinatorial effect, opening up new therapeutic opportunities for HDAC2 inhibitors.9
The chemical structure of santacruzamate A (SCA, 130, Fig. 30a) is simple and consists of three sections: an ethyl carbamate (EC) terminus, a γ-aminobutyric acid (GABA) linker and a phenethylamine (PEA) cap. The original total synthesis of SCA was reported by Balunas, alongside the isolation work.242 As shown in Fig. 30a, coupling of the N-terminal and C-terminal of GABA with the ethyl chloroformate and PEA, respectively, provided product SCA (130). Starting from monomethyl glutarate, Balunas also synthesized the SCA-SAHA hybrid (131, Fig. 30b). In 2016, following similar protocols, Balunas synthesized 40 analogues of SCA by alteration of EC, GABA and PEA with different moieties, respectively.279 Unfortunately, all analogues, including the SCA (both natural and synthetic) and SCA-SAHA hybrid (131), were inactive against HDAC2 enzyme as well as HCT116 cells. Only two PEA-altered analogues 132a, 132b showed moderate to weak activity against MCF-7 with micromolar GI50. However, using similar synthetic strategy as Balunas, Wen and Rodriquez developed more potent analogues of SCA in 2015 and 2017 against HCT116 cell line with IC50 values in the micromolar range at 48 h and 72 h (133, 134a,b and 135, Fig. 30b).280,281
7. Proteasome inhibitors
7.1. Mechanism of action
Multiple myeloma (MM) is malignant plasma cells that metastasize as osteolytic bone legions without the formation of new bone, with proteasome inhibitors as typical treatment for the incurable disease.282 Proteasomes are large protein complexes working with polymerized ubiquitin to degrade mutated or misfolded intracellular proteins in a metabolic depleting fashion. Ubiquitin targets damaged proteins, initiates a 3-step cascade to produce a tagged protein with a polyubiquitin chain, in which the proteasome then passes the protein through its 20S core to be degraded into small oligopeptides (Fig. 31).283 Inhibition of proteasomes are generally well received for clinical use with minimal side effects, with the FDA approval of bortezomib in 2003 for multiple myeloma and later the semi-synthetic carfilzomib, derived from epoxomicin, for refractory multiple myeloma in 2012.283 Both inhibit the chymotrypsin-like, trypsin-like, and caspase-like subunits of the 20S proteasome (β5, β2, β1) and are given in triple combination therapy with an immunomodulatory drug and dexamethasone. Other therapies, like the γ-lactam-β-lactone natural product salinosporamide A, target the 20S proteasome as well and is currently in a phase III trial under the name marizomib as a treatment for patients with recently diagnosed glioblastoma.284 However, the plasma cells of MM are highly resistant, leading to patient serial relapse post treatment with no clear resistant mechanism defined.285 Precision therapy via ADCs are the imminent treatment for resistant cells, with the FDA approval of the B-cell maturation antigen (BCMA) targeted ADC belantamab mafodotin (Bela) in 2020. The marine cyanobacterial natural product carmaphycin is in development by linking carmaphycin B analogues with the antibody trastuzumab (Fig. 32).286–288
Fig. 31. Proteasome degradation mechanism with inhibitors acting on 20S, stabilizing proteins that upregulate apoptotic signals and downregulate proteins that induce cell proliferation.
Fig. 32. Structures of the natural products carmaphycin A (136a) and B (136b) and two ADCs containing a carmaphycin B-based payload (137a–b).
7.2. Carmaphycins
Carmaphycins A and B (136a,b, Fig. 32) were first isolated from Symploca sp. from Curacao in 2012 by the Gerwick group and feature an α,β-epoxyketone warhead with potential for ADC preparation via a methionine sulfoxide or methionine sulfone (A and B).287 Both target the β5 subunit of the 20S proteasome with an IC50 of 2.5 and 2.6 nM, respectively.287 Cytotoxicity assays for carmaphycin A (136a) and B (136b) initially resulted in strong potency against H-460 (EC50 9 and 6 nM) and HCT116 (IC50 19 and 43 nM) cancer cell lines. In the NCI 60 cell-line panel, most GI50 values ranged between 1 and 50 nM, with specific antiproliferative effects in CNS, lung, and colon tumor cell lines with specific sensitivities to mutants of KRAS or p53 cell lines.287 With little effect on total growth inhibition at higher concentrations, the α,β-epoxyketones signify the need for modification to the natural product warhead for drug development, as was done for the approved drugs carfilzomib and epoxomicin with similar warheads.289
The key feature of carmaphycins is the α,β-epoxyketone and the key issue in the total synthesis was the construction of this fragment (Fig. 33). The Gerwick group completed the first total synthesis and reported it along with the isolation and structure determination.287 In their initial work, Gerwick used the typical procedure (straight way) for the synthesis of the epoxyketone fragment (139): the combination of a Grignard reaction of Leu Weinreb amide and oxidation with oxone provided the α,β-epoxyketone fragment 139 as major product (Fig. 33, route a).287 The yield for these two steps was 26% and the diastereomeric selectivity was 1.7 : 1.290 For ADC preparation, a different protocol was followed for the synthesis of epoxyketone 139, through alcohol intermediates 140, 141 (route b), and the total yield for these three steps was 60–80%.288,291 However, only 27% yield for this three-step reaction was reported by Zhou.291 With epoxyketone 139 in hand, general peptide coupling with dipeptide 142 (commercially available) gave carmaphycin A (136a), which was further oxidized to carmaphycin B (136b) with oxone. Similar to published synthetic routes, a series of analogues were synthesized as well as ADCs of carmaphycin A.288,292,293 Development of carmaphycin ADCs resulted in the coupling of a cleavable and non-cleavable payload of DBCO-C6-acid to carmaphycin B analogues with the eventual conjugation to the antibody trastuzumab (137b and 137a, respectively, Fig. 32).287 The synthesis of a novel ADC with the natural product scaffold was successful, but biological evaluation revealed decreased potency compared to the antibody itself and significantly less potency than the payload MMAE.288
Fig. 33. Abbreviated synthesis of carmaphycins (136a–b) and analogue 142 for SAR studies.
8. Macrocyclic membrane-targeting agents
8.1. Mechanism of action
Therapeutics targeting membranes dominate the druggable sphere as more than 60% of targets are membrane proteins, with macrocyclic agents as an exciting class of druggable entities.294 But targets are not necessarily proteins. Bacterial membrane inhibitors, such as the soil bacteria metabolites lysocin E and daptomycin, target predominantly phospholipid classes like menaquinone and phosphatidylglycerol, respectively.295 The eukaryotic lipid membrane targets include ergosterol (fungi) and cholesterol (mammalian cells), with various natural product inhibitors with a wide structure diversity as described by Nishimura and Matsumori in a 2020 review.296 The frontline defense of polyene antifungal agents, like amphotericin B, complex ergosterol in the plasma membrane, disrupting membrane fluidity and integrity by permeabilizing the fungal membrane to inhibit cellular functions.297 The marine cyanobacterial polyhydroxylated macrolide amantelide A (143a) has a similar mechanism to polyene antifungals, with the additional ability to promote the polymerization of actin in cell-free systems.298,299 The marine sponge helix forming product polytheonamide B works as an ion channel and methyl-β-cyclodextrin as an acute cholesterol depleting agent, and the natural product theonellamides, the bicyclic peptide, targets both ergosterol and cholesterol. Cholesterol plays an essential role in the mechanical stability of the membrane during division and reduces shear force stress in cell separation. It thus could represent a prime target for cancer therapeutics as cancer cells restructure their membrane to avoid apoptosis, ensure proliferation, and resist treatment300,301 (Fig. 34).
Fig. 34. Natural macrocyclic agents targeting eukaryotic sterol containing membranes.
8.2. Amantelides and other polydroxylated macrolides
The isolation of amantelides A and B were described by the Luesch group from a gray cyanobacterium collected from Tumon Bay, Guam.298 The mode of action of amantelide A (143a, Fig. 35) has been studied through a combination of yeast knockout mutants and chemogenomics, as well as targeted biophysical and biochemical methods.299 Amantelide A preferentially binds to sterol-containing membranes based on 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine(POPC)-based liposome studies, although the interaction is not completely dependent on sterols; however, the binding affinity increased with sterols compared with POPC liposomes alone.299 In contrast to other sterol-targeting compounds, amantelide A does not require β-configuration of the 33-OH group; binding occurred equally to cholesterol and epi-cholesterol containing POPC membranes.299 Interaction with cholesterol-containing membranes is the likely cause of rapid cell death of mammalian cells. In yeast, the (ergosterol-containing) membrane was determined to be the major functionally relevant target, causing pore formation and cell death.299 The mode of interaction and lipid recognition on the molecular level remains to be investigated. Interestingly, C33-acetyl analogue, amantelide B (143b, Fig. 35), also binds to POPC and the presence of sterols decreased binding, providing clear SAR based on only two compounds.298 The lack of activity of amantelide B (143b) suggests that acylation might serve as a self-protective mechanism of the native organism.
Fig. 35. Structures of amantelides (143a–c) and the related cyanobacterial macrolides, bastimolides (144a,b), caylobolides (145a,b), palstimolide A (146) and nulapolide (147).
Other related macrolides from marine cyanobacteria such as bastimolide A (144a), and B (144b), palstimolide A (146), and caylobolides (145a, 145b) with similar structural scaffold (Fig. 35) may have a comparable, membrane-mediated mechanism of action with to-be-determined affinity for membrane components.302–307 It has been challenging to deduce the absolute configuration of the stereocenters, also hampering synthetic accessibility so far. Preliminary SAR for amantelide A (143a) and B (143b) suggests that minor changes drastically influence recognition and binding, indicating selectivity can likely be tuned. However, amantelides also bind to membranes in a sterol-independent manner.298
In addition, chemogenomic profiling has suggested that actin modulation might also play a role in the mechanism of action of amantelide A.299 Amantelides A (143a) and B (143b) both appeared to directly affect actin dynamics, promoting polymerization to a minor extent in vitro. Since both compounds have the same effect in biochemical assays and membrane integrity is quickly compromised in cells treated amantelide A (143a) before actin interaction can occur, (1) actin binding is most likely not functionally relevant and may not occur in cells, and (2) the direct effects on sterol-membranes and actin can be separated, as is the case for amantelide B (143b). Compound activity, however, could potentially be modulated by actin as the cytoskeleton is associated with the membrane.299
To serve as an antifungal agent, amantelide A (143a) is too non-selectively cytotoxic against fungi and mammalian cells (HT29 IC50 870 nM; HeLa IC50 870 nM). However, the modulation of sterol-selectivity might lead to an antifungal if cholesterol affinity can be dialled out.298 For cancer applications, it may require targeted delivery to cancer cells to spare normal cells, or the differential biological context for cancer and normal cells beyond sterols can be exploited. Nevertheless, target identification has provided the basis for further evaluation of this compound class.
There are several publications about the total synthesis of polydroxylated macrolides, but selectively constructing multiple chiral centers in this type of molecules is very challenging. Achieving an excellent stereochemical control by enantioselective organocatalytic halogenation, Quintard group completed the synthesis of the C15–C27 fragment of bastimolide A (144a, Fig. 36).308 The total synthesis of bastimolide B (144b, Fig. 36) was completed by Aggarwal group in 2022 as showed in Fig. 36.309 The key strategy for success was the combined application of iterative boronic ester homologation with metal carbenoids and metal catalyzed alkene hydroboration and diboration. Excellent remote stereocenter control was achieved in the construction of acyclic backbone of bastimolide B (144b). Fig. 36 depicts the retrosynthetic analysis and stepwise synthesis of the authors' work.
Fig. 36. Total synthesis of bastimolide B (144b) with the (a) retrosynthetic analysis, (b) synthesis of the tail, and (c) synthesis of the macrocyclic core and coupling of the two fragments.
9. Chemosensitizing agents
Resistance to chemotherapeutic agents in cancer treatment has been reported for most drugs used to treat lethal cancers and contributes to the failure of therapy and the cause of tumor recurrence.310 Typical drug resistance involve the tumor microenvironment (hypoxia, vascular viscosity, and reduced glucose supply), progression pathways (PI3K/Akt, MAPK, and Wnt pathways), drug influx and efflux throughout the cell (acidic pH and limited solute carrier transporters), and impaired apoptosis (enhanced DNA repair via Wnt signaling).310 Chemosensitizing agents discussed here fall into two groups: (1) inherently cytotoxic but do not display the requisite selectivity or inappropriately used, including compounds that act on actin dynamics grassypeptolide A (165), lyngbyabellin A (166) and dolastatin 12 (167), and (2) compounds that are weakly or non-cytotoxic but enhance the sensitivity to cytotoxins and to inducers of apoptosis (doscadenamides (175a,b; 176a,b)).311–315
9.1. Overcoming Nrf2-mediated drug resistance: actin agents and grassypeptolides
Similar to microtubules, the actin cytoskeleton is critical for many processes, including cell division and migration.21 However, actin agents, including promoters of G-actin polymerization into F-actin or microfilament depolymerizers, have not been successful as stand-alone agents which can likely be due to cardiac and respiratory toxicities associated with non-specific binding to actin.311,316,317 However, nuances including different binding sites exist that might lead to distinct pharmacological consequences.318 Actin also influences signaling pathways and actin related protein suggested by the identification of ACTR10 in a genomic siRNA screen for Nrf2 inhibitors.319,320 In a small molecule screen, the cyanobacterial actin agents lyngbyabellin A (166) and dolastatin 12 (167) were also identified as inhibitors of Nrf2 signaling, a stress response pathway that is linked to drug resistance (Fig. 37).311,313,320 Nrf2 is an important regulator of detoxification and electron exchange via antioxidant response elements (ARE) gene expression and Phase II detoxification enzyme regulation, critical for cell survival and suppression of carcinogenesis in early stage cancer diagnosis.318,321 However, overactivation of Nrf2 within cancer cells can induce the overexpression of metabolic enzymes, contributing to the metabolic reprogramming to avoid apoptosis, thus installing a cancer resistance mechanism.318,321 Grassypeptolide A (165) was identified in the same screen as an Nrf2 inhibitor and also sensitized cancer cells to chemotherapeutic tubulin agent vinblastine, revealing a potential opportunity for combination therapy at low doses.314,320 Grassypeptolide A (165) isolated from the marine cyanobacterium Lyngbya confervoides has modest (low micromolar) antiproliferative activity and induces p21 and p27 and decreases cyclin D1 expression, presumably leading to the observed G1 cell cycle arrest.314 These compound classes with unrelated mechanisms would probably benefit from detailed mechanistic studies and could be resurrected for use as agents for combination therapy (Fig. 38).
Fig. 37. Structures of grassypeptolide A (165), lyngbyabellin A (166), and dolastatin 12 (167).
Fig. 38. NRF2 pathway and indirect inhibition to overcome drug resistance.
The total synthesis of grassypeptolide A (165) is depicted in Fig. 39.322,323 The precursors of thiazoline ring, thioacylating agents 169, and 170 were activated by nitrobenzotriazole and fused into peptide chain (168) to afford subunit 171. The installation of O-TBS-Ser was mediated by PyAOP and achieved in 88% yield, but yields were low when other coupling reagents were used (EDCI, HATU, BOPCl, CMPI). A similar problem was encountered in the synthesis of fragment 172. The esterification of Maba to Pla residue (alcohol) resulted in low yields when mediated by EDCI (DCC)/DMAP and CMPI. However, the acyl chloride method provided 90% yield. The convergent synthesis in which subunits 172 and 171 were jointed between Maba and Thr through amidation, and the site between Pro and N-Me-Phe-thn-ca was chosen for macrolactamization, which proceeded smoothly. However, the macrolactonization failed between Maba and Pla and initiated intramolecular cyclization between the Pro and N-Me-Phe-thn-ca to form the hydroxy acid. The two thiazoline rings were formed simultaneously when the primary alcohols of thioamide were activated with DAST, following TBS deprotection. However, the secondary alcohol of Thr must be protected (by Troc) in the reaction of bis(thiazoline) ring formation as the free hydroxy would react to form the oxazoline at Thr.
Fig. 39. Total synthesis of grassypeptolide A (165).
9.2. Synergy with TRAIL: doscadenamides
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) binds to death receptors DR4 and DR5, leading to selective cancer cell death.324 However, recombinant TRAIL protein and agents targeting the TRAIL pathway lacked sufficient efficacy in the clinic due to developing resistance to TRAIL.325 Quorum sensing (QS) activators in Gram-negative bacteria also have eukaryotic targets with cancer relevance.326 Mutant studies revealed that doscadenamide A (175a, Fig. 40b) binds to LasR to activate quorum sensing in Pseudomonas aeruginosa, similar to N-(3-oxododecanoyl)-l-homoserine lactone (C12) that was reported to synergize with TRAIL to induce apoptosis in cancer cells.315 Doscadenamides were first reported from the marine cyanobacterium Moorena bouillonii by the Luesch group.315 Similarly, the doscadenamide scaffold (Fig. 40a) can act as a non-homoserine lactone template for combination with TRAIL or TRAIL pathway activators based on model studies in triple negative MDA-MB-231 breast cancer cells at 50 μM for most analogues.327 Synthesis of the natural doscadenamides (175a) and a strategic set of analogues (175b–c, 175b) (Fig. 40a and b) solved the supply problem and enabled the rigorous probing of the SAR.315,327
Fig. 40. Natural and synthetic doscadenamides (a) showing generic structure with side chains and (b) structures of selected key analogues with enhanced activity and (c) synthetic route for doscadenamide A (175a).
Biological testing using various Gram-negative bacteria and TNBC cells (MDA-MB-231) revealed that side chain Moya2 (see 175a in Fig. 40b) is predominantly responsible for both QS and TRAIL-sensitizing activity.327 Specifically, doscadenamides F (175b, natural product) and S4 (175c, synthetic) with partial or complete saturation of the two acyl side chains, respectively, and doscadenamides S10–S12 (synthetic), lacking one of the acyl chains (Moya1), were most effective in sensitizing to TRAIL.327 These compounds reduced the MDA-MB-231 cancer cell viability and induced PARP cleavage as an indicator of apoptosis. The data suggest that the scaffold may be further tuned to enhance the activity and, with the synthetic method in hand, further rapid diversification can be achieved rather easily.327
The total synthesis of doscadenamide A is depicted in Fig. 40c. 7-octynoic acid (Oya) was regio- and stereo-selectively methylated via its oxazolidinone derivative (R-177) to afford (2R)-methyl-7-octynoic acid (Moya, R-178). The (5S)-pyrrolinone 180 was synthesized by the condensation of N-Fmoc-L-Lyn(Boc)-OH with Meldrum's acid, followed by thermolysis and methylation.328 The key step for this total synthesis was the acylation of 180, which was accomplished by treatment of the PFP ester of 178 (179) with n-BuLi as base at −50 °C to afford intermediate 176a (doscadenamide S10).315 Using standard deprotection and coupling protocols, final product doscadenamide A (175a) was successfully synthesized from 176 and 178. Using a parallel strategy, a series of analogues was created to determine the effect of the saturation status of both acyl chains or the necessity of the presence to have one or the other chain (Fig. 40a).
Although the eukaryotic target has not yet been elucidated, the fact that the l-homoserine lactone (C12) and doscadenamides target the same receptor in bacteria and exhibit similar effects in human cancer cells suggest that they may share the same target and/or mechanism in eukaryotic systems, which might provide opportunities to explore cancer vulnerabilities for rational combination therapy. However, the potency of doscadenamides is very low (25 to 50 μM) hence these agents are merely proof-of-concept compounds at this stage.327
10. Antimetastatic agents
Several cyanobacterial compounds that do not have inherent cytotoxic anticancer effects have shown to inhibit proteases and modulate GPCRs that mediate cell migration and invasion.329,330 The cellular effects, mainly assessed in invasive breast cancer cells, suggest their potential as antimetastatic agents.331 Proteases are also components of the tumor microenvironment and protease inhibitors might also indirectly affect cancer cells through inhibition of aberrant protein processing in different cell types. As described in Section 3.2, apratoxins also modulate the tumor microenvironment; however, they also have inherent antiproliferative effects that are not observed with the compounds discussed in this section (Fig. 41).
Fig. 41. Protease inhibitors with various targets.
10.1. Protease inhibitors
Drug development efforts for protease inhibitors based on natural products, including those of marine cyanobacterial origin, was recently reviewed.332 While the focus has been mainly on pulmonary diseases, certain serine proteases play a role in cancer progression and metastasis and are targeted by marine cyanobacterial compounds.9,332 One such protease is human neutrophil elastase (HNE) which has mainly been linked to pulmonary inflammatory conditions, but also cancer progression, cell migration and metastasis.333 HNE has broad substrate specificity and overactivity leads to degradation of extracellular matrix, cleavage of inflammatory mediators and induction of cytokines and growth factors. Dysfunction of kallikreins (KLKs), a group of 15 family members, has also been impacted in cancer and many other disorders.334 Among them, KLK7 was recently shown to be targeted by the same class of cyclodepsipeptides as HNE inhibitors.335
The serine proteases, chymotrypsin-like human neutrophil elastase (HNE), kallikreins (KLK), and trypsin-like enzymes are broad spectrum mediators of biological processes with multiple targets, including cytokine receptors and EGFR for HNE, PAR receptors for KLK, and trypsin, plasmin, and matriptase for trypsin-like proteases.336 The aspartic proteases include cathepsin D and E which are both biomarkers of cancer (particularly cathepsin D) and induce downstream proliferation of cancer cells after entered the cell from the lysosomes.337,338 BACE1 is an aspartic protease involved with poor prognosis and overexpression in tumor progression.339 ADAM9 and 10 are metalloproteases that promote tumor progression and metastasis, with effect on therapeutic resistance and poor diagnosis via ADAM9 (Fig. 41).340
10.2. Chymotrypsin-like serine protease inhibitors: symplostatin 5, molassamide, lyngbyastatin 7, and tutuilamide A
Among the over 100 members of the 3-amino-6-hydroxy-2-piperidone (Ahp)-containing cyclodepsipeptides with 19-membered core ring structure, only a few have been rigorously characterized and evaluated for potential therapeutic applications (Fig. 42), including symplostatin 5 (181d), molassamide (181b), lyngbyastatin 7 (Lbs7, 181a) and tutuilamide A (TtaA, 181i).341 While Ahp unit is the classical pharmacophore for the serine protease-inhibitory activity, the adjacent 2-amino-2-butenoic acid (Abu) drives the selectivity towards chymotrypsin-like enzymes (elastase), especially HNE and KLK7, leading to nanomolar to sub-nanomolar activity at the enzyme level (IC50). Selectivity is presumably determined by hydrogen bonding of the Abu unit, as previously seen with the stabilization of the ethylidene unit of Abu via CH/π interactions and the interaction of the Abu-containing inhibitor FR901277 towards the porcine pancreatic elastase.342,343 Specifically, under identical conditions, lyngbyastatin 7 (181a) and tutuilamide A (181i) inhibited HNE with IC50 of 0.85 nM and 0.73 nM, respectively, and human KLK7 with IC50 of 3.1 nM and 5.0 nM, respectively.335 Docking studies of lyngbyastatin 7 (181a) and tutuilamide A (181i) to HNE showed that both interact with the enzymes similar to other Ahp-Abu elastase inhibitors (Fig. 43).335 Additionally, docking models of lyngbyastatin 7 and tutuilamide A against KLK7 revealed the presence of key interactions between Asn192 and Phe218, amino acids that are not conserved between other KLK homologues.335 This could indicate the reason for the observed selectivity of KLK7 by both compounds.
Fig. 42. Structures of lyngbyastatin 7 (181a), and symplostatin 5 (181d) and naturally occurring and synthetic analogues of lyngbyastatin 7 with side chain modifications.
Fig. 43. Molecular docking of lyngbyastatin 7 (181a) (Lbs7: green) and tutuilamide A (181i) (TtaA: blue) to (a) HNE (PDBID: 3Q76) and (b) KLK7 (PDBID: 2QXI).
Molassamide (181b) first reported from the assemblages of Dichothrix utahensis, by the Luesch group, as a representative member of the Ahp-Abu cyclodepsipeptide family was shown to prevent CD40 proteolytic processing in biochemical assays at 1 μM and in MDA-MB-231 breast cancer cells at 10 μM, when exogenously adding 100 nM HNE.344 CD40 is a membrane protein and member of the tumor necrosis factor receptor family that is a known extracellular HNE substrate, which regulates NF-κB signaling to promote expression of genes encoding breast cancer cell migratory factors, including ICAM-1, which is a cell surface receptor that regulates cell–cell adhesion.345 Molassamide (181b) not only attenuated ICAM1 gene expression but also inhibited HNE-induced membrane-bound ICAM-1 protein cleavage, therefore preventing soluble ICAM-1 (sICAM-1) release from the cell surface receptor. These molecular changes translated into phenotypic effects, i.e., inhibition of migration specifically of HNE-induced MDA-MB-231 cells but not basal migration.346
Lyngbyastatin 7 and tutuilamide A have been synthesized and the side chain diversified, using a different strategy than that previously used for the generation of somamide A (181c).335 The new strategy allowed the late-stage modulation of drug-like properties by introducing changes in the pendant side chain (Fig. 44). The Shioiri group completed the total synthesis of somamide A (181c, Fig. 44a) in 2002, as the first member of this Abu-containing Ahp-cyclodepsipeptides.347 The Ahp unit was viewed as glutamate-derived alcohol and Abu was derived from Thr. Considering the risk of epimerization, they chose the site at Abu/norvaline; in their synthetic strategy, the precursor 182a was composed of three fragments 184a, 185a and 186 and the side chain was installed into fragment 184a in advance (Fig. 44a). Abu was synthesized from Thr through oxidation by Martin's sulfurane and Ahp was synthesized by biomimetic strategy from 5-OH norvaline via its aldehyde (Fig. 44b). In order to build a macrocyclic core-based library for SAR studies, the Luesch group developed a new strategy, constructing the macrocyclic core first and installing the side chain at late-stage (Fig. 44c).348,349 This new strategy made it is possible to synthesize a small library of lyngbyastatin 7 (181a) with diverse side chains (Fig. 44a). In the same fashion, the Luesch group accomplished the total synthesis of tutuilamide A (181i) using this convergent approach via only five steps from the macrocycle 183 (Fig. 44c).335
Fig. 44. The different synthetic strategies for (a) somamide (181c), (b) the Abu/Ahp unit, and (c) lyngbyastatin 7 (181a) and tutuilamide A (181i).
10.3. Trypsin-like serine protease inhibitors: kempopeptins
The serine protease inhibitors kempopeptins are also Ahp-containing cyclodepsipeptides produced from a tentative Lyngbya species, compared with those in Section 10.2, favoring trypsin-like enzymes. The Lys-bearing kempopeptins B and C (188a,b, Fig. 45) discovered from a cyanobacterium in the Florida Keys differ only by the nature of the halogen atom (Br vs. Cl) at the N,O-diMe-Tyr moiety.350,351 Kempopeptin C (188b, chlorinated) showed 3- to 7-fold higher potency at the enzyme level in direct comparison to kempopeptin B with IC50 values of 0.19 μM (trypsin), 0.36 μM (plasmin) and 0.28 μM (matriptase).351 Differences in potency might not only be attributed to steric hindrance within the binding pocket from the Br atom, reducing binding affinity, but also to differential electronic effects. Binding of kempopeptin C (188b) to the active site of matriptase (Fig. 45) at 1 to 10 μM prevented the cleavage of membrane targets of matriptase, including CDCP1 and desmoglein-2 (Dsg-2), as shown in vitro using human recombinant proteins.351 Kempopeptin C (188b, 10 μM) also attenuated CDCP1 cleavage in MDA-MB-231 cells upon exogenous addition of recombinant human matriptase. At the same concentration, a significant inhibitory effect on cell migration was detected, presumably due to inhibition of trypsin-like serine proteases.
Fig. 45. (a) Structures of kempopeptins B (188a) and C (188b) and (b) docking of kempopeptin C into matriptase (PDBID: 2GV7).
When the Abu unit is replaced with a basic residue (Lys, Arg), then the selectivity of the scaffold changes to preferentially inhibit trypsin, plasmin and matriptase, which recognize the X–Y–Z-sequence in substrates. Particularly plasmin and matriptase have been implicated in cancer progression and metastasis because of their processing of cancer-relevant membrane proteins. Despite the wealth of known cyanobacterial serine protease inhibitors specifically targeting these enzymes as substrate mimics, including micropeptins,352,353 there have been limited studies to probe cellular activity and phenotypic consequences in the context of cancer.354
10.4. Aspartic protease inhibitors: grassystatins
Among aspartic proteases, particularly cathepsin D has been linked to cancer, as a biomarker for aggressive breast cancer with poor prognosis but also therapeutic target.337 Cathepsin D controls several signaling cascades, including plasminogen activators and cysteine cathepsins B and L that lead to metastasis and invasion. Specifically, plasminogen activators are inhibited by PAI-1 which is one target of cathepsin D, while another proteolytic cathepsin D target, cystatin C (Cys-C), inhibits cysteine proteases that play a role in cancer progression.355
The pepstatin-like aspartic protease inhibitors of the grassystatin class (189a, 189b, Fig. 46) and certain tasiamides, are linear peptides with the statin or statin-derived pharmacophore that potently inhibit cathepsin D and related proteases with differential selectivity for cathepsin E.356–359 A small subset has been investigated for cellular activity against cathepsin D-mediated molecular changes and migration using MDA-MB-231 breast cancer cells, which are characterized by high levels of cathepsin D and secreted PAI-1 and Cys-C.357 Grassystatin A (189a, Fig. 46) was the most potent cathepsin D inhibitor, while grassystatin F (189c) was used in proof-of-concept studies and inhibited migration, in contrast to grassystatin D (189b, Fig. 46), supporting in vitro SAR results that underscore the importance of the N-terminal, N,N-diMe amino acid.356,357 Selectivity for cathepsin D over cathepsin E may be enhanced by phenyl-statin-containing pharmacophore as in tasiamides B and F and further tuning in other sites based on molecular modeling results (Fig. 47).358,359
Fig. 46. Chemical structures of grassystatins A (189a), D (189b) and F (189c) with retrosynthetic strategy.
Fig. 47. Molecular docking of grassystatin A (189a) into cathepsin E (PDBID: 4PEP).
The first total synthesis of grassystatin A (189a, Fig. 46) was reported in 2012.360 In the retrosynthetic strategy, grassystatin A was disassembled into two sub-fragments at the site Leu/Asn site (190, 191, Fig. 46).360 Choosing amidation for fragment coupling provided versatility with respect to analogue generation through building different sub-fragments 190 and 191. Purification by silica gel chromatography led to decomposition; however, the pure final product 189a was obtained in 45% yield by directly submitting the crude product to HPLC for purification. More recently, the Hunter group designed and synthesized a series of fluorinated analogues of grassystatin A using SPPS.361
11. GPCR modulators
G-Protein Coupled Receptors (GPCRs) are a diverse group of membrane receptors that function within most physiological processes across the body. This includes regulation, growth, and homeostasis, with multiple avenues for dysregulation, such as causing an assortment of neurodegenerative disorders, cardiovascular diseases and cancer.362 Due to the vast effect of the proteins within the physiological functions as well as their downstream affects, GPCRs can affect tumor cell invasion and metastasis, promote favorable conditions for tumor survival by enhancing the tumor microenvironment, and affect abnormal cell growth and survival.363 Recently, cellular profiling of marine cyanobacterial natural products identified selective GPCR agonists and antagonists, including modulators of cancer-relevant GPCRs.364
11.1. Brintonamides
Brintonamides were isolated by the Luesch group from intertidal cyanobacterial mats mainly from Oscillatoriaceae and characterized as modified peptides with dual activity targeting protease and GPCRs (Fig. 48).365 Using functional, β-arrestin assays, brintonamides C–E (192c–e, Fig. 49) have been found to target CCR10, OXTR, SSTR3, TACR2 (antagonism) and CXCR7 (agonism) with differential selectivity profiles depending on the nature of the N-terminal residue that confers the depsipeptide character.365 Only brintonamide D (192d, Fig. 48) exhibited sub-micromolar activity against one of the GPCRs, namely CCR10, while others were in the micromolar range.365 The C–C motif chemokine receptor 10 (CCR10) selectivity and link of CCR10 to metastasis prompted targeted studies to assess the antimetastatic potential of brintonamides A–E (192a–e, Fig. 49) along with synthetic analogues that were accessible through standard methodology.365
Fig. 48. Activity of brintonamide D (192d) as a CCR10 antagonist and potential downstream effects. Only effects on proliferation and on migration as an indicator of metastasis have been studied so far.
Fig. 49. Brintonamides A–D (192a–d) (a) chemical structures and (b) synthesis.
CCR10 is associated with cutaneous T-cell lymphoma, T-cell mediate skin inflammation and metastasis. A putative binding model for the interaction of brintonamide D with CCR10 is depicted in Fig. 50. CCR10 is the receptor for ligand CCL27. Since the invasive MDA-MB-231 breast cancer cells are TNBCs with high CCR10 expression, the antimetastatic potential was assessed in this cell type using cell proliferation and migration assays upon stimulation with CCL27 under serum-starved condition.366 Brintonamide D dose-dependently inhibited specifically the CCL-27 induced proliferation (10% viability reduction) and more pronouncedly inhibited migration, which was functionally linked to CCR10 antagonism because the compound phenocopied effects of an siRNA targeting CCR10 but not other GPCRs.365
Fig. 50. Molecular docking of brintonamide D (192d) into CCR10 homology model (PDBIDs: 5LWE, 5UIW, 4MBS, 4RWD, 4N6H, 3OE9).
Interestingly, all brintonamides were also inhibitors of the serine protease KLK7, although in high micromolar range and without appreciable SAR and cellular activity against this target.
The total synthesis of brintonamide A (192a, Fig. 49b) was accomplished by general SPPS protocol (Fig. 49), in which Fmoc-l-Pro was attached to CTC resin first, and then other amino acids were attached sequentially (C → N). The cleavage from the CTC resin, followed by sequential methylation of the C-terminal Pro and acylation of the other Pro by Hmpa provided the final product, brintonamide A (192a). Further esterification of brintonamide A by cinnamide acid using Yamaguchi method afforded brintonamide D (192d).365 Other analogues were synthesized using same protocol and provided critical insight into the SAR.
12. Perspective & conclusion
Marine cyanobacteria are a promising source of new drug candidates, particular for cancer, as validated by the approval of several ADCs with cyanobacteria-based cytotoxic payloads. As demonstrated in the review, many compounds from these sources represent new chemical scaffolds that target novel pharmacological space (Fig. 51). In other cases, biological targets may be identical to those of compounds from other microbial, including terrestrial sources; however, the potency is oftentimes superior or binding sites differ. Even though the pharmacophore may be similar to those from other sources, the specific decoration or modifications modulate selectivity profiles. Cyanobacterial protecting group chemistry that keeps compounds in a latent condition may also be unique or characteristic. Natural products research oftentimes leads to the elucidation of a structurally intriguing compound. However, the structural beauty (to a natural product chemist) is not enough to trigger developmental efforts. Translating the chemical diversity into therapeutic potential requires target identification, rigorous pharmacological profiling, and mechanistic studies, which can guide towards suitable biomedical applications. The identification of novel biology ultimately justifies and drives the tedious and sometimes seemingly brute-de-force chemistry, which should never be the major hurdle given the expense of drug development at other stages that is larger by magnitudes. The medicinal chemistry campaigns are not as elaborate as for non-natural products due to the structural complexity. They may also not have to be as extensive since compounds are already fairly optimized by nature for physiologically relevant targets. Adaption to human targets and physiology may require tweaking selectivity or dialing out liabilities that may lead to off-target toxicity. Oftentimes the starting point provided by nature is good enough, assuming intellectual property protection concerns can be addressed to ensure development.367 Fundamentally, both the supply problem and novel pharmacological action need to be addressed in order to differentiate the compounds from other agents in development or in the clinic. For almost all compounds discussed in this review, the key biology (target) and chemistry (supply) issues have been solved. Some compounds may not have drug-like properties; however, they could provide the basis for the design of compounds with similar pharmacology that are more amenable for development. The target space of cyanobacterial agents is tremendously broad for cancer alone. While this review has focused on cancer, the same approach can be applied to other disease indications, particularly infectious diseases, where natural products always have been playing and will continue to play a dominant role.368
Fig. 51. Overview of marine cyanobacterial anticancer agents and their various targets.
Lastly, we have only seen the tip of the iceberg of specialized metabolites from marine cyanobacteria, as genome or metagenome sequencing suggest that the biosynthetic potential is oftentimes 10-fold higher than what is observed at the chemical level.369 Translating biosynthetic potential into screenable and therefore actionable compounds through synthetic biology is another exciting avenue in the field and for drug discovery.353,370,371 Furthermore, enabling technologies such as antibody conjugation and other emerging technologies will guarantee that we maximize the potential of these evolutionarily optimized compounds.372–374
13. Conflicts of interest
H. L. is cofounder of Oceanyx Pharmaceuticals, Inc. that has licensed patents and patent applications related to apratoxins and optioned patents based on largazole.
Acknowledgments
Anticancer drug discovery from marine cyanobacteria in the authors' laboratory is supported by the NIH grant R01CA172310 (H. L.), and the Debbie and Sylvia DeSantis Chair professorship (H. L.). E. K. E. is supported by NIGMS grant T32GM136583: Chemistry–Biology Interface Training Program at the University of Florida. We thank Gustavo Seabra (University of Florida) for preparing docking figures. Pathway figures were created using BioRender.
Biographies
Biography
Hendrik Luesch.

Hendrik Luesch received his Diplom in Chemistry at the University of Siegen (Germany) in 1997 and his PhD with Professor Richard Moore at the University of Hawaii at Manoa in 2002. After postdoctoral studies at The Scripps Research Institute with Professor Peter Schultz, he joined the faculty at the University of Florida in 2005, where he is leading a comprehensive marine natural products program with chemistry- and genomics-based discovery platforms. Since 2024 he holds a joint appointment at Duke-NUS Medical School in Singapore. Research highlights include the discovery and characterization of dolastatin 10, apratoxins, largazole, and gatorbulins from marine cyanobacteria.
Biography
Emma K. Ellis.

Emma Ellis received her BSc in Biochemistry from Oklahoma Baptist University in 2022. After graduation, she joined the Department of Medicinal Chemistry at the University of Florida as a predoctoral candidate. She was selected as a trainee in the UF Chemistry–Biology Interface Predoctoral Training Program (T32) and is mentored by Professor Hendrik Luesch. Her research is focused on the isolation and characterization of novel metabolites from marine cyanobacteria, including synthesis and biological evaluation of lead compounds with pharmacological relevance for the treatment of cancer.
Biography
Qi-Yin Chen.

Qi-Yin Chen received BSc and MS in Chemistry from Henan Normal University (1997) and Central South University (2000), respectively, and PhD in Organic Chemistry in 2005 from the Institute of Chemistry (CAS). He completed postdoctoral training with Professors Yujiro Hayashi (Tokyo University of Science), Alan Katritzky (UF), and Hendrik Luesch (UF). He currently is a Research Associate Professor and serves as the leader for Synthetic Chemistry Core at Center for Natural Products, Drug Discovery and Development (CNPD3) at UF. His research focuses on the synthesis of bioactive marine natural products, and small-molecule drug leads. He has co-authored over 40 peer-reviewed articles.
Biography
Ranjala Ratnayake.

Ranjala Ratnayake obtained her BSc (1996) and MPhil (2002) from the Open University in Sri Lanka. She obtained her PhD at the University of Queensland, Australia with Professor Robert J Capon in 2007. She completed postdoctoral trainings at the National Cancer Institute (NCI) in the Molecular Targets Development Program (MTDP), in Frederick, Maryland and at UF. She is currently a Research Associate Professor in the Department of Medicinal Chemistry at UF. She has co-authored over 50 peer-reviewed research articles in small molecule characterization, assay development, screening, and mechanistic studies of natural products including dolastatin 15, apratoxins, gatorbulins, lagunamides and largazole.
15 References
- Atanasov A. G. Zotchev S. B. Dirsch V. M. Supuran C. T. Nat. Rev. Drug Discovery. 2021;20:200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler J. A. Curr. Protoc. Pharmacol. 2019;86:e67. doi: 10.1002/cpph.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J. Cragg G. M. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- Liang X. Luo D. Luesch H. Pharmacol. Res. 2019;147:104373. doi: 10.1016/j.phrs.2019.104373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. Kong G. Liu M. Jiang M. Cheng D. Chen F. Green Synth. Catal. 2022;3:243–258. doi: 10.1016/j.gresc.2022.07.007. [DOI] [Google Scholar]
- Li G. Peng X. Guo Y. Gong S. Cao S. Qiu F. Front. Chem. 2021;9:761609. doi: 10.3389/fchem.2021.761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou K. C. Pan S. Shelke Y. Rigol S. Bao R. Das D. Ye Q. Proc. Natl. Acad. Sci. U. S. A. 2022;119:e2208938119. doi: 10.1073/pnas.2208938119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Approved Marine Drugs, https://www.marinepharmacology.org/approved, accessed December 15, 2023
- Tan L. T. Phyo M. Y. Molecules. 2020;25:2197. doi: 10.3390/molecules25092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpine R. Sieber S. Curr. Res. Biotechnol. 2021;3:65–81. doi: 10.1016/j.crbiot.2021.03.001. [DOI] [Google Scholar]
- Haque N. Parveen S. Tang T. Wei J. Huang Z. Mar. Drugs. 2022;20:528. doi: 10.3390/md20080528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Reyes L. A. Luesch H. Nat. Prod. Rep. 2015;32:478–503. doi: 10.1039/C4NP00104D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Bañuelos B. Durán-Riveroll L. M. Rangel-López E. Pérez-López H. I. González-Maya L. Molecules. 2022;27:4814. doi: 10.3390/molecules27154814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Zhang T. Yao J. Lu J. Liu Z. Ding L. Eur. J. Med. Chem. 2020;201:112473. doi: 10.1016/j.ejmech.2020.112473. [DOI] [PubMed] [Google Scholar]
- Nawaz T. Gu L. Fahad S. Saud S. Jiang Z. Hassan S. Harrison M. T. Liu K. Khan M. A. Liu H. El-Kahtany K. Wu C. Zhu M. Zhou R. Food Energy Secur. 2023;12:e495. doi: 10.1002/fes3.495. [DOI] [Google Scholar]
- D'Agostino P. M. Nat. Prod. Rep. 2023;40:1701–1717. doi: 10.1039/D3NP00011G. [DOI] [PubMed] [Google Scholar]
- Kaul R. Risinger A. L. Mooberry S. L. J. Nat. Prod. 2019;82:680–685. doi: 10.1021/acs.jnatprod.9b00105. [DOI] [PubMed] [Google Scholar]
- Dall'Acqua S. Curr. Top. Med. Chem. 2014;14:2272–2285. doi: 10.2174/1568026614666141130095311. [DOI] [PubMed] [Google Scholar]
- Akhmanova A. Steinmetz M. O. Nat. Rev. Mol. Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Mühlethaler T. Gioia D. Prota A. E. Sharpe M. E. Cavalli A. Steinmetz M. O. Angew. Chem., Int. Ed. 2021;60:13331–13342. doi: 10.1002/anie.202100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M. Koenderink G. H. Nat. Rev. Mol. Cell Biol. 2019;20:38–54. doi: 10.1038/s41580-018-0067-1. [DOI] [PubMed] [Google Scholar]
- Kumar B. Kumar R. Skvortsova I. Kumar V. Curr. Cancer Drug Targets. 2017;17:357–375. doi: 10.2174/1568009616666160928110818. [DOI] [PubMed] [Google Scholar]
- Blokhin A. V. Yoo H. D. Geralds R. S. Nagle D. G. Gerwick W. H. Hamel E. Mol. Pharmacol. 1995;48:523–531. [PubMed] [Google Scholar]
- Steinmetz M. O. Prota A. E. Trends Cell Biol. 2018;28:776–792. doi: 10.1016/j.tcb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Matthew S. Chen Q.-Y. Ratnayake R. Fermaintt C. S. Lucena-Agell D. Bonato F. Prota A. E. Lim S. T. Wang X. Díaz J. F. Risinger A. L. Paul V. J. Oliva M. Á. Luesch H. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2021847118. doi: 10.1073/pnas.2021847118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Peña H. Abel A.-C. Shevelev M. Prota A. E. Pieraccini S. Horvath D. Biomolecules. 2023;13:285. doi: 10.3390/biom13020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D. Himes R. H. Moore R. E. Wilson L. Jordan M. A. Biochemistry. 1997;36:12948–12953. doi: 10.1021/bi971302p. [DOI] [PubMed] [Google Scholar]
- Bai R. Petit G. R. Hamel E. Biochem. Pharmacol. 1990;39:1941–1949. doi: 10.1016/0006-2952(90)90613-P. [DOI] [PubMed] [Google Scholar]
- Bai R. Friedman S. J. Pettit G. R. Hamel E. Biochem. Pharmacol. 1992;43:2637–2645. doi: 10.1016/0006-2952(92)90153-A. [DOI] [PubMed] [Google Scholar]
- Eren E. Watts N. R. Sackett D. L. Wingfield P. T. J. Biol. Chem. 2021;297:101138. doi: 10.1016/j.jbc.2021.101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.-Y. Ratnayake R. Hortigüela R. Seabra G. M. Cameron M. D. Fernando Díaz J. Ángela Oliva M. Luesch H. Bioorg. Med. Chem. 2023;95:117506. doi: 10.1016/j.bmc.2023.117506. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Vicente J. J. Cancers. 2021;13:5650. doi: 10.3390/cancers13225650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck S. L. Collins J. M. Doroshow J. H. Mol. Cancer Ther. 2010;9:1451–1460. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit G. R. Kamano Y. Herald C. L. Tuinman A. A. Boettner F. E. Kizu H. Schmidt J. M. Baczynskyj L. Tomer K. B. Bontems R. J. J. Am. Chem. Soc. 1987;109:6883–6885. doi: 10.1021/ja00256a070. [DOI] [Google Scholar]
- Luesch H. Moore R. E. Paul V. J. Mooberry S. L. Corbett T. H. J. Nat. Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- Engene N. Tronholm A. Salvador-Reyes L. A. Luesch H. Paul V. J. J. Phycol. 2015;51:670–681. doi: 10.1111/jpy.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit G. R. Kamano Y. Herald C. L. Fujii Y. Kizu H. Boyd M. R. Boettner F. E. Doubek D. L. Schmidt J. M. Chapuis J.-C. Michel C. Tetrahedron. 1993;49:9151–9170. doi: 10.1016/0040-4020(93)80003-C. [DOI] [Google Scholar]
- Maki A. Mohammad R. Raza S. Saleh M. Govindaraju K. D. Pettit G. R. Al-Katib A. Anticancer Drugs. 1996;7:344–350. doi: 10.1097/00001813-199605000-00016. [DOI] [PubMed] [Google Scholar]
- Turner T. Jackson W. H. Pettit G. R. Wells A. Kraft A. S. Prostate. 1998;34:175–181. doi: 10.1002/(SICI)1097-0045(19980215)34:3<175::AID-PROS4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kalemkerian G. P. Ou X. Adil M. R. Rosati R. Khoulani M. M. Madan S. K. Pettit G. R. Cancer Chemother. Pharmacol. 1999;43:507–515. doi: 10.1007/s002800050931. [DOI] [PubMed] [Google Scholar]
- Singh S. B. J. Nat. Prod. 2022;85:666–687. doi: 10.1021/acs.jnatprod.1c01135. [DOI] [PubMed] [Google Scholar]
- Tomioka K. Kanai M. Koga K. Tetrahedron Lett. 1991;32:2395–2398. doi: 10.1016/S0040-4039(00)79932-1. [DOI] [Google Scholar]
- Miyazaki K. Kobayashi M. Natsume T. Gondo M. Mikami T. Sakakibara K. Tsukagoshi S. Chem. Pharm. Bull. 1995;43:1706–1718. doi: 10.1248/cpb.43.1706. [DOI] [PubMed] [Google Scholar]
- Maderna A. Doroski M. Subramanyam C. Porte A. Leverett C. A. Vetelino B. C. Chen Z. Risley H. Parris K. Pandit J. Varghese A. H. Shanker S. Song C. Sukuru S. C. K. Farley K. A. Wagenaar M. M. Shapiro M. J. Musto S. Lam M.-H. Loganzo F. O'Donnell C. J. J. Med. Chem. 2014;57:10527–10543. doi: 10.1021/jm501649k. [DOI] [PubMed] [Google Scholar]
- Senter P. D. Sievers E. L. Nat. Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- Gao G. Wang Y. Hua H. Li D. Tang C. Mar. Drugs. 2021;19:363. doi: 10.3390/md19070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Y.-T. Mayes P. A. Acharya C. Zhong M. Y. Cea M. Cagnetta A. Craigen J. Yates J. Gliddon L. Fieles W. Hoang B. Tunstead J. Christie A. L. Kung A. L. Richardson P. Munshi N. C. Anderson K. C. Blood. 2014;123:3128–3138. doi: 10.1182/blood-2013-10-535088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco J. A. Cerveny C. G. Meyer D. L. Mixan B. J. Klussman K. Chace D. F. Rejniak S. X. Gordon K. A. DeBlanc R. Toki B. E. Law C.-L. Doronina S. O. Siegall C. B. Senter P. D. Wahl A. F. Blood. 2003;102:1458–1465. doi: 10.1182/blood-2003-01-0039. [DOI] [PubMed] [Google Scholar]
- Dornan D. Bennett F. Chen Y. Dennis M. Eaton D. Elkins K. French D. Go M. A. T. Jack A. Junutula J. R. Koeppen H. Lau J. McBride J. Rawstron A. Shi X. Yu N. Yu S.-F. Yue P. Zheng B. Ebens A. Polson A. G. Blood. 2009;114:2721–2729. doi: 10.1182/blood-2009-02-205500. [DOI] [PubMed] [Google Scholar]
- Challita-Eid P. M. Satpayev D. Yang P. An Z. Morrison K. Shostak Y. Raitano A. Nadell R. Liu W. Lortie D. R. Capo L. Verlinsky A. Leavitt M. Malik F. Aviña H. Guevara C. I. Dinh N. Karki S. Anand B. S. Pereira D. S. Joseph I. B. J. Doñate F. Morrison K. Stover D. R. Cancer Res. 2016;76:3003–3013. doi: 10.1158/0008-5472.CAN-15-1313. [DOI] [PubMed] [Google Scholar]
- López de Sá A. Díaz-Tejeiro C. Poyatos-Racionero E. Nieto-Jiménez C. Paniagua-Herranz L. Sanvicente A. Calvo E. Pérez-Segura P. Moreno V. Moris F. Ocana A. J. Hematol. Oncol. 2023;16:118. doi: 10.1186/s13045-023-01519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Drug Evaluation and Research (CDER), FDA, accessed March 13, 2024 [Google Scholar]
- Trudel S. Lendvai N. Popat R. Voorhees P. M. Reeves B. Libby E. N. Richardson P. G. Anderson L. D. Sutherland H. J. Yong K. Hoos A. Gorczyca M. M. Lahiri S. He Z. Austin D. J. Opalinska J. B. Cohen A. D. Lancet Oncol. 2018;19:1641–1653. doi: 10.1016/S1470-2045(18)30576-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J. T. W. Harris P. W. R. Brimble M. A. Kavianinia I. Molecules. 2021;26:5847. doi: 10.3390/molecules26195847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos M. A. Hungria V. T. M. Radinoff A. Delimpasi S. Mikala G. Masszi T. Li J. Capra M. Maiolino A. Pappa V. Chraniuk D. Osipov I. Leleu X. Low M. Matsumoto M. Sule N. Li M. McKeown A. He W. Bright S. Currie B. Perera S. Boyle J. Roy-Ghanta S. Opalinska J. Weisel K. Lancet Haematol. 2023;10:e801–e812. doi: 10.1016/S2352-3026(23)00243-0. [DOI] [PubMed] [Google Scholar]
- GSK provides an update on Blenrep (belantamab mafodotin-blmf) US marketing authorization | GSK US, https://us.gsk.com/en-us/media/press-releases/gsk-provides-an-update-on-blenrep-belantamab-mafodotin-blmf-us-marketing-authorization/, accessed September 20, 2024
- Sehn L. H. Herrera A. F. Flowers C. R. Kamdar M. K. McMillan A. Hertzberg M. Assouline S. Kim T. M. Kim W. S. Ozcan M. Hirata J. Penuel E. Paulson J. N. Cheng J. Ku G. Matasar M. J. J. Clin. Oncol. 2020;38(2):155–165. doi: 10.1200/JCO.19.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagen Inc., Open Label Phase 2 Study of Tisotumab Vedotin for Locally Advanced or Metastatic Disease in Solid Tumors, https://clinicaltrials.gov/, accessed March 13, 2024
- Deeks E. D. Drugs. 2021;81:1929–1935. doi: 10.1007/s40265-021-01614-x. [DOI] [PubMed] [Google Scholar]
- Goldman J. W. Horinouchi H. Cho B. C. Tomasini P. Dunbar M. Hoffman D. Parikh A. Blot V. Camidge D. R. J. Clin. Oncol. 2022;40:9013. doi: 10.1200/JCO.2022.40.16_suppl.9013. [DOI] [Google Scholar]
- Yu W. Ramprasad M. P. Pal M. Chen C. Paruchuri S. Skidmore L. Knudsen N. Allen M. Buechler Y. PLoS One. 2023;18:e0284198. doi: 10.1371/journal.pone.0284198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd G. E. Scott H. Chen L. van Rietschoten K. Ivanova-Berndt G. Dzionek K. Brown A. Watcham S. White L. Park P. U. Jeffrey P. Rigby M. Beswick P. J. Med. Chem. 2022;65:14337–14347. doi: 10.1021/acs.jmedchem.2c00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G. Brown A. Mudd G. Huxley P. Van Rietschoten K. Pavan S. Chen L. Watcham S. Lahdenranta J. Keen N. Mol. Cancer Ther. 2020;19:1385–1394. doi: 10.1158/1535-7163.MCT-19-1092. [DOI] [PubMed] [Google Scholar]
- Bodyak N. D. Mosher R. Yurkovetskiy A. V. Yin M. Bu C. Conlon P. R. Demady D. R. DeVit M. J. Gumerov D. R. Gurijala V. R. Lee W. McGillicuddy D. Park P. U. Poling L. L. Protopova M. Qin L. Stevenson C. A. Ter-Ovanesyan E. Uttard A. Xiao D. Xu J. Xu L. Bergstrom D. A. Lowinger T. B. Mol. Cancer Ther. 2021;20:896–905. doi: 10.1158/1535-7163.MCT-20-0183. [DOI] [PubMed] [Google Scholar]
- Yurkovetskiy A. V. Bodyak N. D. Yin M. Thomas J. D. Clardy S. M. Conlon P. R. Stevenson C. A. Uttard A. Qin L. Gumerov D. R. Ter-Ovanesyan E. Bu C. Johnson A. J. Gurijala V. R. McGillicuddy D. DeVit M. J. Poling L. L. Protopopova M. Xu L. Zhang Q. Park P. U. Bergstrom D. A. Lowinger T. B. Mol. Cancer Ther. 2021;20:885–895. doi: 10.1158/1535-7163.MCT-20-0166. [DOI] [PubMed] [Google Scholar]
- Li L. Hau A. Tong W. Lau M. Zhu T. Fells K. Wei A. Li X. Deng D. Sun Y. Kovacs E. Khasanov A. Yan Z. Zhu P. Zhou H. Ji H. Li H. Zhang H. Cancer Res. 2020;80:LB–227. [Google Scholar]
- Li Q. Wang L. Zhang J. Zhao G. Liu Z. Ma X. Jiang J. Clin. Transl. Sci. 2023;16:1232–1242. doi: 10.1111/cts.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Liu R. Gao S. Li W. Chen Y. Meng Y. Liu C. Jin W. Wu J. Wang Y. Hao Y. Yi S. Qing Y. Ge J. Hu X. npj Breast Cancer. 2023;9:28. doi: 10.1038/s41523-023-00522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F. Wang F. Shi Y.-R. Guo Y. Shu X.-L. Pan S. Qu S.-H. Zhang P. Jiang Y. Xu M.-J. Lei K. Qu S. Lei F. Lv X. Xiang Y.-Q. Xu R.-H. Ann. Oncol. 2023;34:S559. doi: 10.1016/j.annonc.2023.09.2006. [DOI] [Google Scholar]
- Akla B. Broussas M. Loukili N. Robert A. Beau-Larvor C. Malissard M. Boute N. Champion T. Haeuw J.-F. Beck A. Perez M. Dreyfus C. Pavlyuk M. Chetaille E. Corvaia N. Mol. Cancer Ther. 2020;19:168–177. doi: 10.1158/1535-7163.MCT-19-0219. [DOI] [PubMed] [Google Scholar]
- Kollmannsberger C. Choueiri T. K. Heng D. Y. C. George S. Jie F. Croitoru R. Poondru S. Thompson J. A. Oncologist. 2021;26:182–e361. doi: 10.1002/onco.13628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. L. Adkins D. Lorch J. H. Thomas J. S. Grewal J. S. J. Clin. Oncol. 2023;41:TPS6107. doi: 10.1200/JCO.2023.41.16_suppl.TPS6107. [DOI] [Google Scholar]
- Kujtan L. Subramanian J. Transl. Lung Cancer Res. 2023;12:1826–1829. doi: 10.21037/tlcr-23-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A. Cusmai A. Acquafredda S. Rinaldi L. Palmiotti G. Expert Opin. Invest. Drugs. 2022;31:495–498. doi: 10.1080/13543784.2022.2042252. [DOI] [PubMed] [Google Scholar]
- CAB Portfolio – BioAtla, https://www.bioatla.com/cabportfolio/, accessed March 13, 2024
- Shen J. Pachynski R. Nordquist L. Adra N. Bilen M. Aggarwal R. Reichert Z. Schweizer M. Xu D. Patel M. Zhang J. Shi W. L'Heureux D. Z. Tagawa S. Cancer Res. 2023;83:CT121. doi: 10.1158/1538-7445.AM2023-CT121. [DOI] [Google Scholar]
- Maitland M. L. Sachdev J. C. Sharma M. R. Moreno V. Boni V. Kummar S. Stringer-Reasor E. Lakhani N. Moreau A. R. Xuan D. Li R. Powell E. L. Jackson-Fisher A. Bowers M. Alekar S. Xin X. Tolcher A. W. Calvo E. Clin. Cancer Res. 2021;27:4511–4520. doi: 10.1158/1078-0432.CCR-20-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon R. P. Schmitt M. W. Jonas M. Franz C. E. Trueblood E. S. Yumul R. Westendorf L. Ryan M. C. Cancer Res. 2020;80:2906. doi: 10.1158/1538-7445.AM2020-2906. [DOI] [Google Scholar]
- Jhaveri K. Han H. Dotan E. Oh D.-Y. Ferrario C. Tolcher A. Lee K.-W. Liao C.-Y. Kang Y.-K. Kim Y. H. Hamilton E. Spira A. Patel N. Karapetis C. Rha S. Y. Boyken L. Woolery J. Bedard P. Ann. Oncol. 2022;33:S749–S750. doi: 10.1016/j.annonc.2022.07.589. [DOI] [Google Scholar]
- Park Y. H. Ahn H. K. Kim J.-Y. Ahn J. S. Im Y.-H. Kim S.-H. Lee S. CHUNG H.-S. Park S. J. J. Clin. Oncol. 2020;38:3551. [Google Scholar]
- Pettit G. R. Singh S. B. Hogan F. Lloyd-Williams P. Herald D. L. Burkett D. D. Clewlow P. J. J. Am. Chem. Soc. 1989;111:5463–5465. doi: 10.1021/ja00196a061. [DOI] [Google Scholar]
- Roux F. Maugras I. Poncet J. Niel G. Jouin P. Tetrahedron. 1994;50:5345–5360. doi: 10.1016/S0040-4020(01)80692-X. [DOI] [Google Scholar]
- Shioiri T. Hamada Y. Synlett. 2001;2001:184–201. doi: 10.1055/s-2001-10758. [DOI] [Google Scholar]
- Mordant C. Reymond S. Tone H. Lavergne D. Touati R. Ben Hassine B. Ratovelomanana-Vidal V. Genet J.-P. Tetrahedron. 2007;63:6115–6123. doi: 10.1016/j.tet.2007.03.036. [DOI] [Google Scholar]
- Zhou W. Nie X.-D. Zhang Y. Si C.-M. Zhou Z. Sun X. Wei B.-G. Org. Biomol. Chem. 2017;15:6119–6131. doi: 10.1039/C7OB01395G. [DOI] [PubMed] [Google Scholar]
- World Intellectual Property Organization, WO02088172A2, 2002
- Bajjuri K. M. Liu Y. Liu C. Sinha S. C. ChemMedChem. 2011;6:54–59. doi: 10.1002/cmdc.201000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit G. R. , Barkoczy J., US Pat., US5635483A, 1997
- Yokosaka S. Izawa A. Sakai C. Sakurada E. Morita Y. Nishio Y. Bioorg. Med. Chem. 2018;26:1643–1652. doi: 10.1016/j.bmc.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Akaiwa M. Martin T. Mendelsohn B. A. ACS Omega. 2018;3:5212–5221. doi: 10.1021/acsomega.8b00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake R. Gunasekera S. P. Ma J. J. Dang L. H. Carney T. J. Paul V. J. Luesch H. Eur. J. Biol. Chem. Res. 2020;21:2356–2366. doi: 10.1002/cbic.202000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg G. M., Kingston D. G. I. and Newman D. J., Anticancer Agents from Natural Products, CRC Press, Boca Raton, 1st edn, 2005 [Google Scholar]
- Lopus M. Mol. Cell. Biochem. 2013;382:93–102. doi: 10.1007/s11010-013-1721-8. [DOI] [PubMed] [Google Scholar]
- Ray A. Okouneva T. Manna T. Miller H. P. Schmid S. Arthaud L. Luduena R. Jordan M. A. Wilson L. Cancer Res. 2007;67:3767–3776. doi: 10.1158/0008-5472.CAN-06-3065. [DOI] [PubMed] [Google Scholar]
- Hu M. K. Huang W. S. J. Pept. Res. 1999;54:460–467. doi: 10.1034/j.1399-3011.1999.00130.x. [DOI] [PubMed] [Google Scholar]
- Marks R. S. Graham D. L. Sloan J. A. Hillman S. Fishkoff S. Krook J. E. Okuno S. H. Mailliard J. A. Fitch T. R. Addo F. Am. J. Clin. Oncol. 2003;26(4):336–337. doi: 10.1097/01.COC.0000020962.25210.6F. [DOI] [PubMed] [Google Scholar]
- Kerbrat P. Dieras V. Pavlidis N. Ravaud A. Wanders J. Fumoleau P. Eur. J. Cancer. 2003;39:317–320. doi: 10.1016/S0959-8049(02)00531-2. [DOI] [PubMed] [Google Scholar]
- Smyth J. Boneterre M. Schellens J. Calvert H. Greim G. Wanders J. Hanauske A. Ann. Oncol. 2001;12(4):509–511. doi: 10.1023/A:1011194910571. [DOI] [PubMed] [Google Scholar]
- Gianolio D. A. Rouleau C. Bauta W. E. Lovett D. Cantrell Jr W. R. Recio 3rd A. Wolstenholme-Hogg P. Busch M. Pan P. Stefano J. E. Kramer H. M. Goebel J. Krumbholz R. D. Roth S. Schmid S. M. Teicher B. A. Cancer Chemother. Pharmacol. 2012;70:439–449. doi: 10.1007/s00280-012-1925-8. [DOI] [PubMed] [Google Scholar]
- Newman D. J. Cragg G. M. Mar. Drugs. 2017;15:99. doi: 10.3390/md15040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casi G. Huguenin-Dezot N. Zuberbühler K. Scheuermann J. Neri D. J. Am. Chem. Soc. 2012;134:5887–5892. doi: 10.1021/ja211589m. [DOI] [PubMed] [Google Scholar]
- Pettit G. R. Herald D. L. Singh S. B. Thornton T. J. Mullaney J. T. J. Am. Chem. Soc. 1991;113:6692–6693. doi: 10.1021/ja00017a061. [DOI] [Google Scholar]
- Pettit G. R. Thornton T. J. Mullaney J. T. Boyd M. R. Herald D. L. Singh S.-B. Flahive E. J. Tetrahedron. 1994;50:12097–12108. doi: 10.1016/S0040-4020(01)89562-4. [DOI] [Google Scholar]
- Akaji K. Hayashi Y. Kiso Y. Kuriyama N. J. Org. Chem. 1999;64:405–411. doi: 10.1021/jo981055a. [DOI] [Google Scholar]
- Patino N. Frérot E. Galeotti N. Poncet J. Coste J. Dufour M.-N. Jouin P. Tetrahedron. 1992;48:4115–4122. doi: 10.1016/S0040-4020(01)92190-8. [DOI] [Google Scholar]
- Bai R. Edler M. C. Bonate P. L. Copeland T. D. Pettit G. R. Ludueña R. F. Hamel E. Mol. Pharmacol. 2009;75:218–226. doi: 10.1124/mol.108.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. E. Hirsch C. F. Sesin D. F. Flor J. E. Chartrain M. Fromtling R. E. Harris G. H. Salvatore M. J. Liesch J. M. Yudin K. J. Ind. Microbiol. 1990;5:113–123. doi: 10.1007/BF01573860. [DOI] [Google Scholar]
- Weiss C. Figueras E. Borbely A. N. Sewald N. J. Pept. Sci. 2017;23:514–531. doi: 10.1002/psc.3015. [DOI] [PubMed] [Google Scholar]
- Trimurtulu G. Ohtani I. Patterson G. M. L. Moore R. E. Corbett T. H. Valeriote F. A. Demchik L. J. Am. Chem. Soc. 1994;116:4729–4737. doi: 10.1021/ja00090a020. [DOI] [Google Scholar]
- Verma V. A. Pillow T. H. DePalatis L. Li G. Phillips G. L. Polson A. G. Raab H. E. Spencer S. Zheng B. Bioorg. Med. Chem. Lett. 2015;25:864–868. doi: 10.1016/j.bmcl.2014.12.070. [DOI] [PubMed] [Google Scholar]
- Kobayashi M. Aoki S. Ohyabu N. Kurosu M. Wang W. Kitagawa I. Tetrahedron Lett. 1994;35:7969–7972. doi: 10.1016/S0040-4039(00)78398-5. [DOI] [Google Scholar]
- Leao T. Castelão G. Korobeynikov A. Monroe E. A. Podell S. Glukhov E. Allen E. E. Gerwick W. H. Gerwick L. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3198–3203. doi: 10.1073/pnas.1618556114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggen M. Mossman C. J. Buck S. B. Nair S. K. Bhat L. Ali S. M. Reiff E. A. Boge T. C. Georg G. I. J. Org. Chem. 2000;65:7792–7799. doi: 10.1021/jo000767+. [DOI] [PubMed] [Google Scholar]
- Iacobazzi R. M. Annese C. Azzariti A. D'Accolti L. Franco M. Fusco C. La Piana G. Laquintana V. Denora N. ACS Med. Chem. Lett. 2013;4:1189–1192. doi: 10.1021/ml400300q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagaki J. Shi G. Miyauchi S. Murakami S. Yang Y. Anticancer Drugs. 2015;26:259–271. doi: 10.1097/CAD.0000000000000183. [DOI] [PubMed] [Google Scholar]
- Panda D. DeLuca K. Williams D. Jordan M. A. Wilson L. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9313–9318. doi: 10.1073/pnas.95.16.9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman M. J. Gandara D. R. Hausner P. Israel V. Thornton D. DeSanto J. Doyle L. A. Lung Cancer. 2003;39:197–199. doi: 10.1016/S0169-5002(02)00511-1. [DOI] [PubMed] [Google Scholar]
- Eggen M. Georg G. I. Med. Res. Rev. 2002;22:85–101. doi: 10.1002/med.10002. [DOI] [PubMed] [Google Scholar]
- Lai Q. Wu M. Wang R. Lai W. Tao Y. Lu Y. Wang Y. Yu L. Zhang R. Peng Y. Jiang X. Fu Y. Wang X. Zhang Z. Guo C. Liao W. Zhang Y. Kang T. Chen H. Yao Y. Gou L. Yang J. Eur. J. Med. Chem. 2020;199:112364. doi: 10.1016/j.ejmech.2020.112364. [DOI] [PubMed] [Google Scholar]
- Su D. Kozak K. R. Sadowsky J. Yu S.-F. Fourie-O'Donohue A. Nelson C. Vandlen R. Ohri R. Liu L. Ng C. He J. Davis H. Lau J. Del Rosario G. Cosino E. dela Cruz-Chuh J. Ma Y. Zhang D. Darwish M. Cai W. Chen C. Zhou H. Lu J. Liu Y. Kaur S. Xu K. Pillow T. H. Bioconjugate Chem. 2018;29:1155–1167. doi: 10.1021/acs.bioconjchem.7b00785. [DOI] [PubMed] [Google Scholar]
- Weiss C. Sammet B. Sewald N. Nat. Prod. Rep. 2013;30:924–940. doi: 10.1039/C3NP70022D. [DOI] [PubMed] [Google Scholar]
- Schmidt J. J. Khatri Y. Brody S. I. Zhu C. Pietraszkiewicz H. Valeriote F. A. Sherman D. H. ACS Chem. Biol. 2020;15:524–532. doi: 10.1021/acschembio.9b00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck Z. Q. Aldrich C. C. Magarvey N. A. Georg G. I. Sherman D. H. Biochemistry. 2005;44:13457–13466. doi: 10.1021/bi051140u. [DOI] [PubMed] [Google Scholar]
- Eißler S. Stoncius A. Nahrwold M. Sewald N. Synthesis. 2006;22:3747–3789. [Google Scholar]
- Bolduc K. L. Larsen S. D. Sherman D. H. Chem. Commun. 2012;48:6414–6416. doi: 10.1039/C2CC32417B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissler S. Nahrwold M. Neumann B. Stammler H.-G. Sewald N. Org. Lett. 2007;9:817–819. doi: 10.1021/ol063032l. [DOI] [PubMed] [Google Scholar]
- Mast C. A. Eißler S. Stončius A. Stammler H.-G. Neumann B. Sewald N. Chem.–Eur. J. 2005;11:4667–4677. doi: 10.1002/chem.200500282. [DOI] [PubMed] [Google Scholar]
- Ding Y. Rath C. M. Bolduc K. L. Håkansson K. Sherman D. H. J. Am. Chem. Soc. 2011;133:14492–14495. doi: 10.1021/ja204716f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras E. Borbély A. Ismail M. Frese M. Sewald N. Beilstein J. Org. Chem. 2018;14:1281–1286. doi: 10.3762/bjoc.14.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P. Yan W. Yang J. Cytoskeleton. 2024;81(6–7):255–263. doi: 10.1002/cm.21813. [DOI] [PubMed] [Google Scholar]
- Bousquet M. S. Ma J. J. Ratnayake R. Havre P. A. Yao J. Dang N. H. Paul V. J. Carney T. J. Dang L. H. Luesch H. ACS Chem. Biol. 2016;11:1322–1331. doi: 10.1021/acschembio.5b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabjeesh N. J. Escuin D. LaVallee T. M. Pribluda V. S. Swartz G. M. Johnson M. S. Willard M. T. Zhong H. Simons J. W. Giannakakou P. Cancer Cell. 2003;3:363–375. doi: 10.1016/S1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Carbonaro M. Escuin D. O'Brate A. Thadani-Mulero M. Giannakakou P. J. Biol. Chem. 2012;287:11859–11869. doi: 10.1074/jbc.M112.345587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangjee A. Pavana R. K. Ihnat M. A. Thorpe J. E. Disch B. C. Bastian A. Bailey-Downs L. C. Hamel E. Bai R. ACS Med. Chem. Lett. 2014;5:480–484. doi: 10.1021/ml4004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Chen J. Xiao M. Li W. Miller D. D. Pharm. Res. 2012;29:2943–2971. doi: 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. Recktenwald M. Hutt E. Fuller S. Briggs M. Goel A. Daringer N. Cancers. 2022;14:1259. doi: 10.3390/cancers14051259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. Ruan W. Bobrow B. Carmeliet P. Eltzschig H. K. Nat. Rev. Drug Discovery. 2024;23:175–200. doi: 10.1038/s41573-023-00848-6. [DOI] [PubMed] [Google Scholar]
- Fares J. Fares M. Y. Khachfe H. H. Salhab H. A. Fares Y. Signal Transduction Targeted Ther. 2020;5:1–17. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip H. Y. K. Papa A. Cells . 2021;10:659. doi: 10.3390/cells10030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z. Lovly C. M. Mol. Cancer . 2018;17:58. doi: 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witsch E. Sela M. Yarden Y. Physiology. 2010;25:85–101. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. K. Miller E. A. Genetics. 2013;193:383–410. doi: 10.1534/genetics.112.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y. Wilkinson B. M. Pool M. R. Biochim. Biophys. Acta, Mol. Cell Res. 2013;1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Itskanov S. Park E. Cold Spring Harbor Perspect. Biol. 2023;15(1):a041250. doi: 10.1101/cshperspect.a041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskanov S. Wang L. Junne T. Sherriff R. Xiao L. Blanchard N. Shi W. Q. Forsyth C. Hoepfner D. Spiess M. Park E. Nat. Chem. Biol. 2023;19:1063–1071. doi: 10.1038/s41589-023-01337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H. Yoshida W. Y. Moore R. E. Paul V. J. Corbett T. H. J. Am. Chem. Soc. 2001;123:5418–5423. doi: 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- Huang K.-C. Chen Z. Jiang Y. Akare S. Kolber-Simonds D. Condon K. Agoulnik S. Tendyke K. Shen Y. Wu K.-M. Mathieu S. Choi H. Zhu X. Shimizu H. Kotake Y. Gerwick W. H. Uenaka T. Woodall-Jappe M. Nomoto K. Mol. Cancer Ther. 2016;15:1208–1216. doi: 10.1158/1535-7163.MCT-15-0648. [DOI] [PubMed] [Google Scholar]
- Medina R. A. Goeger D. E. Hills P. Mooberry S. L. Huang N. Romero L. I. Ortega-Barría E. Gerwick W. H. McPhail K. L. J. Am. Chem. Soc. 2008;130:6324–6325. doi: 10.1021/ja801383f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter D. Paatero A. O. Kawaguchi S. Kazemi S. Serrill J. D. Kellosalo J. Vogel W. K. Richter U. Mattos D. R. Wan X. Thornburg C. C. Oishi S. McPhail K. L. Ishmael J. E. Paavilainen V. O. ACS Chem. Biol. 2020;15:2125–2136. doi: 10.1021/acschembio.0c00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H. Yoshida W. Y. Moore R. E. Paul V. J. Bioorg. Med. Chem. 2002;10:1973–1978. doi: 10.1016/S0968-0896(02)00014-7. [DOI] [PubMed] [Google Scholar]
- Tidgewell K. Engene N. Byrum T. Media J. Doi T. Valeriote F. A. Gerwick W. H. Chembiochem. 2010;11:1458–1466. doi: 10.1002/cbic.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew S. Schupp P. J. Luesch H. J. Nat. Prod. 2008;71:1113–1116. doi: 10.1021/np700717s. [DOI] [PubMed] [Google Scholar]
- Gutiérrez M. Suyama T. L. Engene N. Wingerd J. S. Matainaho T. Gerwick W. H. J. Nat. Prod. 2008;71:1099–1103. doi: 10.1021/np800121a. [DOI] [PubMed] [Google Scholar]
- Thornburg C. C. Cowley E. S. Sikorska J. Shaala L. A. Ishmael J. E. Youssef D. T. A. McPhail K. L. J. Nat. Prod. 2013;76:1781–1788. doi: 10.1021/np4004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H. Chanda S. K. Raya R. M. DeJesus P. D. Orth A. P. Walker J. R. Izpisúa Belmonte J. C. Schultz P. G. Nat. Chem. Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- Liu Y. Law B. K. Luesch H. Mol. Pharmacol. 2009;76:91–104. doi: 10.1124/mol.109.056085. [DOI] [PubMed] [Google Scholar]
- Paatero A. O. Kellosalo J. Dunyak B. M. Almaliti J. Gestwicki J. E. Gerwick W. H. Taunton J. Paavilainen V. O. Cell Chem. Biol. 2016;23:561–566. doi: 10.1016/j.chembiol.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Shi W. Lu D. Wu C. Li M. Ding Z. Li Y. Chen B. Lin X. Su W. Shao X. Xia Z. Fang L. Liu K. Li H. Biochem. Biophys. Res. Commun. 2021;547:52–58. doi: 10.1016/j.bbrc.2021.01.112. [DOI] [PubMed] [Google Scholar]
- Chen Q.-Y. Liu Y. Luesch H. ACS Med. Chem. Lett. 2011;2:861–865. doi: 10.1021/ml200176m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.-Y. Liu Y. Cai W. Luesch H. J. Med. Chem. 2014;57:3011–3029. doi: 10.1021/jm4019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G. Wang W. Ao L. Cheng Z. Wu C. Pan Z. Liu K. Li H. Su W. Fang L. J. Med. Chem. 2018;61:8908–8916. doi: 10.1021/acs.jmedchem.8b01141. [DOI] [PubMed] [Google Scholar]
- Luesch H. Paavilainen V. O. Nat. Prod. Rep. 2020;37:717–736. doi: 10.1039/C9NP00066F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W. Ratnayake R. Wang M. Chen Q.-Y. Raisch K. P. Dang L. H. Law B. K. Luesch H. Curr. Res. Pharmacol. Drug Discov. 2021;2:100053. doi: 10.1016/j.crphar.2021.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Forsyth C. J. J. Am. Chem. Soc. 2003;125:8734–8735. doi: 10.1021/ja036050w. [DOI] [PubMed] [Google Scholar]
- Ma D. Zou B. Cai G. Hu X. Liu J. O. Chem.–Eur. J. 2006;12:7615–7626. doi: 10.1002/chem.200600599. [DOI] [PubMed] [Google Scholar]
- Mao Z.-Y. Si C.-M. Liu Y.-W. Dong H.-Q. Wei B.-G. Lin G.-Q. J. Org. Chem. 2017;82:10830–10845. doi: 10.1021/acs.joc.7b01598. [DOI] [PubMed] [Google Scholar]
- Tarsis E. M. Rastelli E. J. Wengryniuk S. E. Coltart D. M. Tetrahedron. 2015;71:5029–5044. doi: 10.1016/j.tet.2015.05.047. [DOI] [Google Scholar]
- Cai W. Ratnayake R. Gerber M. H. Chen Q.-Y. Yu Y. Derendorf H. Trevino J. G. Luesch H. Invest. New Drugs. 2019;37:364–374. doi: 10.1007/s10637-018-0647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W. Chen Q.-Y. Dang L. H. Luesch H. ACS Med. Chem. Lett. 2017;8:1007–1012. doi: 10.1021/acsmedchemlett.7b00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz V. Bommert K. S. Kruk J. Schwinning D. Chatterjee M. Stühmer T. Bargou R. Bommert K. Sci. Rep. 2020;10:18419. doi: 10.1038/s41598-020-75499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. Pissarnitski D. A. Huang X. Palani A. Zhu Z. Greenlee W. J. Hyde L. A. Song L. Terracina G. Zhang L. Parker E. M. ACS Med. Chem. Lett. 2017;8:1002–1006. doi: 10.1021/acsmedchemlett.7b00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird J. W. A. Ng P. Y. Kubota K. Wang X. Leighton J. L. J. Am. Chem. Soc. 2002;124:7920–7921. doi: 10.1021/ja0264908. [DOI] [PubMed] [Google Scholar]
- Kiyooka S. Kaneko Y. Komura M. Matsuo H. Nakano M. J. Org. Chem. 1991;56:2276–2278. doi: 10.1021/jo00007a003. [DOI] [Google Scholar]
- Onda Y. Masuda Y. Yoshida M. Doi T. J. Med. Chem. 2017;60:6751–6765. doi: 10.1021/acs.jmedchem.7b00833. [DOI] [PubMed] [Google Scholar]
- Dhakal D. Kallifidas D. Chen M. Kokkaliari S. Chen Q.-Y. Paul V. J. Ding Y. Luesch H. Org. Lett. 2023;25:2238–2242. doi: 10.1021/acs.orglett.3c00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau A. M. Greenwood J. A. Löhr C. V. Serrill J. D. Proteau P. J. Ganley I. G. McPhail K. L. Ishmael J. E. PLoS One. 2013;8:e65250. doi: 10.1371/journal.pone.0065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrill J. D. Wan X. Hau A. M. Jang H. S. Coleman D. J. Indra A. K. Alani A. W. G. McPhail K. L. Ishmael J. E. Invest. New Drugs. 2016;34:24–40. doi: 10.1007/s10637-015-0303-x. [DOI] [PubMed] [Google Scholar]
- Kazemi S. Kawaguchi S. Badr C. E. Mattos D. R. Ruiz-Saenz A. Serrill J. D. Moasser M. M. Dolan B. P. Paavilainen V. O. Oishi S. McPhail K. L. Ishmael J. E. Biochem. Pharmacol. 2021;183:114317. doi: 10.1016/j.bcp.2020.114317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W. Qiu H.-B. Chen Y.-J. Xi J. Yao Z.-J. Tetrahedron Lett. 2014;55:6109–6112. doi: 10.1016/j.tetlet.2014.09.047. [DOI] [Google Scholar]
- Yao G. Pan Z. Wu C. Wang W. Fang L. Su W. J. Am. Chem. Soc. 2015;137:13488–13491. doi: 10.1021/jacs.5b09286. [DOI] [PubMed] [Google Scholar]
- Wu C. Cheng Z. Lu D. Liu K. Cheng Y. Wang P. Zhou Y. Li M. Shao X. Li H. Su W. Fang L. J. Med. Chem. 2021;64:991–1000. doi: 10.1021/acs.jmedchem.0c01387. [DOI] [PubMed] [Google Scholar]
- Kitamura T. Suzuki R. Inuki S. Ohno H. McPhail K. L. Oishi S. ACS Med. Chem. Lett. 2022;13:105–110. doi: 10.1021/acsmedchemlett.1c00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bong A. H. L. Monteith G. R. Biochim. Biophys. Acta, Mol. Cell Res. 2018;1865:1786–1794. doi: 10.1016/j.bbamcr.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Arbabian A. Brouland J.-P. Gélébart P. Kovàcs T. Bobe R. Enouf J. Papp B. BioFactors. 2011;37:139–149. doi: 10.1002/biof.142. [DOI] [PubMed] [Google Scholar]
- Periasamy M. Kalyanasundaram A. Muscle Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- Zhai X. Sterea A. M. El Hiani Y. Cells. 2020;9:1536. doi: 10.3390/cells9061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterková L. Kmoníčková E. Ruml T. Rimpelová S. J. Med. Chem. 2020;63:1937–1963. doi: 10.1021/acs.jmedchem.9b01509. [DOI] [PubMed] [Google Scholar]
- Kurisawa N. Iwasaki A. Teranuma K. Dan S. Toyoshima C. Hashimoto M. Suenaga K. J. Am. Chem. Soc. 2022;144:11019–11032. doi: 10.1021/jacs.2c04459. [DOI] [PubMed] [Google Scholar]
- Morita M. Ogawa H. Ohno O. Yamori T. Suenaga K. Toyoshima C. FEBS Lett. 2015;589:1406–1411. doi: 10.1016/j.febslet.2015.04.056. [DOI] [PubMed] [Google Scholar]
- Teruya T. Sasaki H. Kitamura K. Nakayama T. Suenaga K. Org. Lett. 2009;11:2421–2424. doi: 10.1021/ol900579k. [DOI] [PubMed] [Google Scholar]
- Sagara Y. Inesi G. J. Biol. Chem. 1991;266:13503–13506. doi: 10.1016/S0021-9258(18)92726-2. [DOI] [PubMed] [Google Scholar]
- Abbott B. J. Fukuda D. S. Dorman D. E. Occolowitz J. L. Debono M. Farhner L. Antimicrob. Agents Chemother. 1979;16:808–812. doi: 10.1128/AAC.16.6.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen D. C. Kean W. S. Verma A. Cole J. T. Watson W. D. Biol. Proced. Online. 2012;14:4. doi: 10.1186/1480-9222-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkaliari S. Luo D. Paul V. J. Luesch H. Mar. Drugs. 2023;21:378. doi: 10.3390/md21070378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M. Ohno O. Teruya T. Yamori T. Inuzuka T. Suenaga K. Tetrahedron. 2012;68:5984–5990. doi: 10.1016/j.tet.2012.05.038. [DOI] [Google Scholar]
- Watanabe A. Ohno O. Morita M. Inuzuka T. Suenaga K. Bull. Chem. Soc. Jpn. 2015;88:1256–1264. doi: 10.1246/bcsj.20150117. [DOI] [Google Scholar]
- Sato E. Sato M. Tanabe Y. Nakajima N. Ohkubo A. Suenaga K. J. Org. Chem. 2017;82:6770–6777. doi: 10.1021/acs.joc.7b00905. [DOI] [PubMed] [Google Scholar]
- Sato E. Tanabe Y. Nakajima N. Ohkubo A. Suenaga K. Org. Lett. 2016;18:2047–2049. doi: 10.1021/acs.orglett.6b00660. [DOI] [PubMed] [Google Scholar]
- Liu Y. Sun Y. Guo Y. Shi X. Chen X. Feng W. Wu L.-L. Zhang J. Yu S. Wang Y. Shi Y. Int. J. Biol. Sci. 2023;19:897–915. doi: 10.7150/ijbs.81609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Youle R. J. Annu. Rev. Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer C. M. Singh A. T. K. Int. J. Mol. Sci. 2018;19:448. doi: 10.3390/ijms19020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C. Fang M. Li Y. Li L. Wang X. Cell. 2000;102:33–42. doi: 10.1016/S0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan A. M. Neoplasia. 1999;1:5–15. doi: 10.1038/sj.neo.7900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D. Ratnayake R. Atanasova K. R. Paul V. J. Luesch H. Biochem. Pharmacol. 2023;213:115608. doi: 10.1016/j.bcp.2023.115608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro B. A. El-Deiry W. S. Nat. Rev. Clin. Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrasidlo W. Mielgo A. Torres V. A. Barbero S. Stoletov K. Suyama T. L. Klemke R. L. Gerwick W. H. Carson D. A. Stupack D. G. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2313–2318. doi: 10.1073/pnas.0712198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orning P. Lien E. J. Leukocyte Biol. 2021;109:121–141. doi: 10.1002/JLB.3MR0420-305R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Huang W. Li L. Sun X. Song S. Xu Q. Zhang L. Wei B.-G. Deng X. Mol. Pharm. 2016;13:3756–3763. doi: 10.1021/acs.molpharmaceut.6b00564. [DOI] [PubMed] [Google Scholar]
- Abdel-Naime W. A. Kimishima A. Setiawan A. Fahim J. R. Fouad M. A. Kamel M. S. Arai M. Mar. Drugs. 2020;18:555. doi: 10.3390/md18110555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer N. Cramer T. Drug Resistance Updates. 2011;14:191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y. Iwasaki A. Fujimura H. Kudo C. Kurisawa N. Ohno O. Suenaga K. J. Org. Chem. 2023;88:3208–3216. doi: 10.1021/acs.joc.2c03007. [DOI] [PubMed] [Google Scholar]
- Tripathi A. Puddick J. Prinsep M. R. Rottmann M. Tan L. T. J. Nat. Prod. 2010;73:1810–1814. doi: 10.1021/np100442x. [DOI] [PubMed] [Google Scholar]
- Luo D. Putra M. Y. Ye T. Paul V. J. Luesch H. Mar. Drugs. 2019;17:83. doi: 10.3390/md17020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A. Puddick J. Prinsep M. R. Rottmann M. Chan K. P. Chen D. Y.-K. Tan L. T. Phytochemistry. 2011;72:2369–2375. doi: 10.1016/j.phytochem.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Suenaga K. Mutou T. Shibata T. Itoh T. Fujita T. Takada N. Hayamizu K. Takagi M. Irifune T. Kigoshi H. Yamada K. Tetrahedron. 2004;60:8509–8527. doi: 10.1016/j.tet.2004.06.125. [DOI] [Google Scholar]
- Han B. Gross H. Goeger D. E. Mooberry S. L. Gerwick W. H. J. Nat. Prod. 2006;69:572–575. doi: 10.1021/np0503911. [DOI] [PubMed] [Google Scholar]
- Nakao Y. Yoshida W. Y. Takada Y. Kimura J. Yang L. Mooberry S. L. Scheuer P. J. J. Nat. Prod. 2004;67:1332–1340. doi: 10.1021/np049949f. [DOI] [PubMed] [Google Scholar]
- Hagimoto K. Tojo S. Teruya T. Yoshida M. Kigoshi H. Tetrahedron. 2024;154:133871. doi: 10.1016/j.tet.2024.133871. [DOI] [Google Scholar]
- Sueyoshi K. Kaneda M. Sumimoto S. Oishi S. Fujii N. Suenaga K. Teruya T. Tetrahedron. 2016;72:5472–5478. doi: 10.1016/j.tet.2016.07.031. [DOI] [Google Scholar]
- Tripathi A. Fang W. Leong D. T. Tan L. T. Mar. Drugs. 2012;10:1126–1137. doi: 10.3390/md10051126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. Murata A. Orihara T. Shirakawa T. Suenaga K. Kigoshi H. Uesugi M. Chem. Biol. 2011;18:131–139. doi: 10.1016/j.chembiol.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Signorile A. Sgaramella G. Bellomo F. De Rasmo D. Cells. 2019;8:71. doi: 10.3390/cells8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M. Sueyoshi K. Teruya T. Ohno H. Fujii N. Oishi S. Org. Biomol. Chem. 2016;14:9093–9104. doi: 10.1039/C6OB01583B. [DOI] [PubMed] [Google Scholar]
- Kaneda M. Kawaguchi S. Fujii N. Ohno H. Oishi S. ACS Med. Chem. Lett. 2018;9:365–369. doi: 10.1021/acsmedchemlett.8b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. Chen B. Lei H. Wang Z. Liu Y. Xu Z. Ye T. Chem. Commun. 2012;48:8697–8699. doi: 10.1039/C2CC34187E. [DOI] [PubMed] [Google Scholar]
- Huang W. Ren R.-G. Dong H.-Q. Wei B.-G. Lin G.-Q. J. Org. Chem. 2013;78:10747–10762. doi: 10.1021/jo401687s. [DOI] [PubMed] [Google Scholar]
- Gorges J. Kazmaier U. Org. Lett. 2018;20:2033–2036. doi: 10.1021/acs.orglett.8b00576. [DOI] [PubMed] [Google Scholar]
- Ohno O. Iwasaki A. Same K. Kudo C. Aida E. Sugiura K. Sumimoto S. Teruya T. Tashiro E. Simizu S. Matsuno K. Imoto M. Suenaga K. Org. Lett. 2022;24:4547–4551. doi: 10.1021/acs.orglett.2c01566. [DOI] [PubMed] [Google Scholar]
- Kotoku N. Ishida R. Matsumoto H. Arai M. Toda K. Setiawan A. Muraoka O. Kobayashi M. J. Org. Chem. 2017;82:1705–1718. doi: 10.1021/acs.joc.6b02948. [DOI] [PubMed] [Google Scholar]
- Ishida R. Matsumoto H. Ichii S. Kobayashi M. Arai M. Kotoku N. Chem. Pharm. Bull. 2019;67:210–223. doi: 10.1248/cpb.c18-00587. [DOI] [PubMed] [Google Scholar]
- Nogle L. M. Gerwick W. H. Org. Lett. 2002;4:1095–1098. doi: 10.1021/ol017275j. [DOI] [PubMed] [Google Scholar]
- Zhang C. Naman C. B. Engene N. Gerwick W. H. Mar. Drugs. 2017;15:121. doi: 10.3390/md15040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. S. Zhang C. Glukhov E. Gerwick W. H. Suyama T. L. J. Nat. Prod. 2021;84:865–870. doi: 10.1021/acs.jnatprod.0c01317. [DOI] [PubMed] [Google Scholar]
- Suyama T. L. Gerwick W. H. Org. Lett. 2008;10:4449–4452. doi: 10.1021/ol8016947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Wang S. Chen J. Yu Z. Int. J. Med. Sci. 2019;16:424–442. doi: 10.7150/ijms.30154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-C. Sethy B. Liou J.-P. Front. Cell Dev. Biol. 2020;8:576391. doi: 10.3389/fcell.2020.576391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. Zhu L. Wang H. Tan Y. Yang Z. Yang L. Wan L. Bioorg. Med. Chem. 2021;52:116510. doi: 10.1016/j.bmc.2021.116510. [DOI] [PubMed] [Google Scholar]
- Peng X. Sun Z. Kuang P. Chen J. Eur. J. Med. Chem. 2020;208:112831. doi: 10.1016/j.ejmech.2020.112831. [DOI] [PubMed] [Google Scholar]
- Bowers A. West N. Taunton J. Schreiber S. L. Bradner J. E. Williams R. M. J. Am. Chem. Soc. 2008;130:11219–11222. doi: 10.1021/ja8033763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y. Taori K. Kim H. Hong J. Luesch H. J. Am. Chem. Soc. 2008;130:8455–8459. doi: 10.1021/ja8013727. [DOI] [PubMed] [Google Scholar]
- Taori K. Paul V. J. Luesch H. J. Am. Chem. Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- Pavlik C. M. Wong C. Y. B. Ononye S. Lopez D. D. Engene N. McPhail K. L. Gerwick W. H. Balunas M. J. J. Nat. Prod. 2013;76:2026–2033. doi: 10.1021/np400198r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. Luesch H. Nat. Prod. Rep. 2012;29:449–456. doi: 10.1039/C2NP00066K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Salvador L. A. Byeon S. Ying Y. Kwan J. C. Law B. K. Hong J. Luesch H. J. Pharmacol. Exp. Ther. 2010;335:351–361. doi: 10.1124/jpet.110.172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurek-George A. Cecil A. R. L. Mo A. H. K. Wen S. Rogers H. Habens F. Maeda S. Yoshida M. Packham G. Ganesan A. J. Med. Chem. 2007;50:5720–5726. doi: 10.1021/jm0703800. [DOI] [PubMed] [Google Scholar]
- Law M. Corsino P. Jahn S. Davis B. Chen S. Patel B. Pham K. Lu J. Sheppard B. Nørgaard P. Hong J. Higgins P. Kim J.-S. Luesch H. Law B. Oncogene. 2013;32:1316–1329. doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador L. A. Park H. Al-Awadhi F. H. Liu Y. Kim B. Zeller S. L. Chen Q.-Y. Hong J. Luesch H. ACS Med. Chem. Lett. 2014;5:905–910. doi: 10.1021/ml500170r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi S. Yashiro M. Takashima T. Nomura S. Noda S. Kawajiri H. Ishikawa T. Wakasa K. Hirakawa K. Br. J. Cancer. 2010;103:249–255. doi: 10.1038/sj.bjc.6605735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-C. Wen Z.-S. Qiu Y.-T. Chen X.-Q. Chen H.-B. Wei M.-M. Liu Z. Jiang S. Zhou G.-B. ACS Med. Chem. Lett. 2013;4:921–926. doi: 10.1021/ml400093y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Salvador-Reyes L. A. Elsadek L. A. Ratnayake R. Chen Q.-Y. Luesch H. ACS Chem. Neurosci. 2020;11:1937–1943. doi: 10.1021/acschemneuro.0c00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-H. Wu X. Tan F. Shi Y.-X. Glass T. Liu T. J. Wathen K. Hess K. R. Gumin J. Lang F. Yung W. K. A. J. Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- Xu C. Liu G. Ji H. Chen W. Dai D. Chen Z. Zhou D. Xu L. Hu H. Cui W. Chang L. Zha Q. Li L. Duan S. Wang Q. Mol. Med. Rep. 2018;18:4297–4302. doi: 10.3892/mmr.2018.9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers A. A. Greshock T. J. West N. Estiu G. Schreiber S. L. Wiest O. Williams R. M. Bradner J. E. J. Am. Chem. Soc. 2009;131:2900–2905. doi: 10.1021/ja807772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen D. J. Smith W. B. Haines B. E. Wiest O. Bradner J. E. Williams R. M. Bioorg. Med. Chem. 2015;23:5061–5074. doi: 10.1016/j.bmc.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Bubb J. M. Bowers A. A. Smith W. B. Paranal R. Estiu G. Wiest O. Bradner J. E. Williams R. M. Bioorg. Med. Chem. Lett. 2013;23:6025–6028. doi: 10.1016/j.bmcl.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Tu Z. Li H. Liu C. Li Z. Sun Q. Yao Y. Liu J. Jiang S. ACS Med. Chem. Lett. 2013;4:132–136. doi: 10.1021/ml300371t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Park H. Salvador L. A. Serrano P. E. Kwan J. C. Zeller S. L. Chen Q.-Y. Ryu S. Liu Y. Byeon S. Luesch H. Hong J. Bioorg. Med. Chem. Lett. 2014;24:3728–3731. doi: 10.1016/j.bmcl.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhansali P. Hanigan C. L. Perera L. Casero R. A. Tillekeratne L. M. V. Eur. J. Med. Chem. 2014;86:528–541. doi: 10.1016/j.ejmech.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaliti J. Al-Hamashi A. A. Negmeldin A. T. Hanigan C. L. Perera L. Pflum M. K. H. Casero R. A. Jr. Tillekeratne L. M. V. J. Med. Chem. 2016;59:10642–10660. doi: 10.1021/acs.jmedchem.6b01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Ratnayake R. Lee H. Shi G. Zeller S. L. Li C. Luesch H. Hong J. Bioorg. Med. Chem. 2017;25:3077–3086. doi: 10.1016/j.bmc.2017.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G. Di Fabio R. Ferrante L. Summa V. Botta M. ChemMedChem. 2017;12:1917–1926. doi: 10.1002/cmdc.201700563. [DOI] [PubMed] [Google Scholar]
- Bhansali P. Hanigan C. L. Casero R. A. Jr. Tillekeratne L. M. V. J. Med. Chem. 2011;54:7453–7463. doi: 10.1021/jm200432a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J. Qiu Y. Ma K. Yao Y. Wang Z. Li X. Zhang D. Tu Z. Jiang S. Tetrahedron. 2014;70:7763–7769. doi: 10.1016/j.tet.2014.05.078. [DOI] [Google Scholar]
- Clausen D. J. Smith W. B. Haines B. E. Wiest O. Bradner J. E. Williams R. M. Bioorg. Med. Chem. 2015;23:5061–5074. doi: 10.1016/j.bmc.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto J. A. Vaz E. Lepore I. Pöppler A.-C. Franci G. Álvarez R. Altucci L. de Lera Á. R. J. Med. Chem. 2010;53:4654–4667. doi: 10.1021/jm100244y. [DOI] [PubMed] [Google Scholar]
- Zhang B. Shan G. Zheng Y. Yu X. Ruan Z.-W. Li Y. Lei X. Mar. Drugs. 2019;17:333. doi: 10.3390/md17060333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. Salvador L. A. Sy S. K. B. Tang Y. Singh R. S. P. Chen Q.-Y. Liu Y. Hong J. Derendorf H. Luesch H. Mar. Drugs. 2014;12:1623–1640. doi: 10.3390/md12031623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Waschke B. C. Woolaver R. A. Chen Z. Zhang G. Piscopio A. D. Liu X. Wang J. H. Cancer Immunol. Res. 2019;7:1318–1331. doi: 10.1158/2326-6066.CIR-18-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Phillips J., Ungermannove D., Nasveschuk C. G. and Zhang G., US Pat., 20130203681A1, 2014
- Schreiber A. R. Kagihara J. A. Corr B. R. Davis S. L. Lieu C. Kim S. S. Jimeno A. Camidge D. R. Williams J. Heim A. M. Martin A. DeMattei J. A. Holay N. Triplett T. A. Eckhardt S. G. Litwiler K. Winkler J. Piscopio A. D. Diamond J. R. Cancers. 2023;16:91. doi: 10.3390/cancers16010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OnKure Therapeutics – Scientific Publications, https://onkuretherapeutics.com/pipeline/publications/, accessed October 26, 2024
- Seiser T. Kamena F. Cramer N. Angew. Chem., Int. Ed. 2008;47:6483–6485. doi: 10.1002/anie.200802043. [DOI] [PubMed] [Google Scholar]
- Nasveschuk C. G. Ungermannova D. Liu X. Phillips A. J. Org. Lett. 2008;10:3595–3598. doi: 10.1021/ol8013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y. Taori K. Kim H. Hong J. Luesch H. J. Am. Chem. Soc. 2008;130:8455–8459. doi: 10.1021/ja8013727. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K. Kulkarni S. Org. Lett. 2008;10:3907–3909. doi: 10.1021/ol8014623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q. Wang L.-P. Jiao X.-Z. Liu X.-Y. Wu Q. Xie P. J. Asian Nat. Prod. Res. 2010;12:940–949. doi: 10.1080/10286020.2010.510114. [DOI] [PubMed] [Google Scholar]
- Chen Q.-Y. Chaturvedi P. R. Luesch H. Org. Process Res. Dev. 2018;22:190–199. doi: 10.1021/acs.oprd.7b00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Nihira N. T. Bu X. Chu C. Zhang J. Kolodziejczyk A. Fan Y. Chan N. T. Ma L. Liu J. Wang D. Dai X. Liu H. Ono M. Nakanishi A. Inuzuka H. North B. J. Huang Y.-H. Sharma S. Geng Y. Xu W. Liu X. S. Li L. Miki Y. Sicinski P. Freeman G. J. Wei W. Nat. Cell Biol. 2020;22:1064–1075. doi: 10.1038/s41556-020-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromek S. M. deMayo J. A. Maxwell A. T. West A. M. Pavlik C. M. Zhao Z. Li J. Wiemer A. J. Zweifach A. Balunas M. J. Bioorg. Med. Chem. 2016;24:5183–5196. doi: 10.1016/j.bmc.2016.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. Lu W. Ma M. Liao J. Ganesan A. Hu Y. Wen S. Huang P. RSC Adv. 2014;5:1109–1112. doi: 10.1039/C4RA13889A. [DOI] [Google Scholar]
- Randino R. Gazzerro P. Mazitschek R. Rodriquez M. Bioorg. Med. Chem. 2017;25:6486–6491. doi: 10.1016/j.bmc.2017.10.026. [DOI] [PubMed] [Google Scholar]
- Roodman G. D. Leukemia. 2009;23:435–441. doi: 10.1038/leu.2008.336. [DOI] [PubMed] [Google Scholar]
- Nunes A. T. Annunziata C. M. Semin. Oncol. 2017;44:377–380. doi: 10.1053/j.seminoncol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulder T. A. M. Moore B. S. Angew Chem. Int. Ed. Engl. 2010;49:9346–9367. doi: 10.1002/anie.201000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimando A. G. Malerba E. Leone P. Prete M. Terragna C. Cavo M. Racanelli V. Front. Oncol. 2022;12:1–17. doi: 10.3389/fonc.2022.973836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham A. Drugs. 2020;80:1607–1613. doi: 10.1007/s40265-020-01404-x. [DOI] [PubMed] [Google Scholar]
- Pereira A. R. Kale A. J. Fenley A. T. Byrum T. Debonsi H. M. Gilson M. K. Valeriote F. A. Moore B. S. Gerwick W. H. ChemBioChem. 2012;13:810–817. doi: 10.1002/cbic.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaliti J. Miller B. Pietraszkiewicz H. Glukhov E. Naman C. B. Kline T. Hanson J. Li X. Zhou S. Valeriote F. A. Gerwick W. H. Eur. J. Med. Chem. 2019;161:416–432. doi: 10.1016/j.ejmech.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaliti J. Fajtová P. Calla J. LaMonte G. M. Feng M. Rocamora F. Ottilie S. Glukhov E. Boura E. Suhandynata R. T. Momper J. D. Gilson M. K. Winzeler E. A. Gerwick W. H. O'Donoghue A. J. Chem.–Eur. J. 2023;29:e202203958. doi: 10.1002/chem.202203958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin N. Kim K. B. Elofsson M. Meng L. Auth H. Kwok B. H. B. Crews C. M. Bioorg. Med. Chem. Lett. 1999;9:2283–2288. doi: 10.1016/S0960-894X(99)00376-5. [DOI] [PubMed] [Google Scholar]
- Zhou H.-J. Aujay M. A. Bennett M. K. Dajee M. Demo S. D. Fang Y. Ho M. N. Jiang J. Kirk C. J. Laidig G. J. Lewis E. R. Lu Y. Muchamuel T. Parlati F. Ring E. Shenk K. D. Shields J. Shwonek P. J. Stanton T. Sun C. M. Sylvain C. Woo T. M. Yang J. J. Med. Chem. 2009;52:3028–3038. doi: 10.1021/jm801329v. [DOI] [PubMed] [Google Scholar]
- Trivella D. B. B. Pereira A. R. Stein M. L. Kasai Y. Byrum T. Valeriote F. A. Tantillo D. J. Groll M. Gerwick W. H. Moore B. S. Chem. Biol. 2014;21:782–791. doi: 10.1016/j.chembiol.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte G. M. Almaliti J. Bibo-Verdugo B. Keller L. Zou B. Y. Yang J. Antonova-Koch Y. Orjuela-Sanchez P. Boyle C. A. Vigil E. Wang L. Goldgof G. M. Gerwick L. O'Donoghue A. J. Winzeler E. A. Gerwick W. H. Ottilie S. J. Med. Chem. 2017;60:6721–6732. doi: 10.1021/acs.jmedchem.7b00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. Flynn A. D. Annu. Rev. Biomed. Eng. 2016;18:51–76. doi: 10.1146/annurev-bioeng-092115-025322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R. M. Walker C. Epand R. F. Magarvey N. A. Biochim. Biophys. Acta, Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Matsumori N. Nat. Prod. Rep. 2020;37:677–702. doi: 10.1039/C9NP00059C. [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A. Rice L. B. Clin. Microbiol. Rev. 1999;12:501–517. doi: 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Reyes L. A. Sneed J. Paul V. J. Luesch H. J. Nat. Prod. 2015;78:1957–1962. doi: 10.1021/acs.jnatprod.5b00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsadek L. A. Matthews J. H. Nishimura S. Nakatani T. Ito A. Gu T. Luo D. Salvador-Reyes L. A. Paul V. J. Kakeya H. Luesch H. Chembiochem. 2021;22:1790–1799. doi: 10.1002/cbic.202000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra S. K. Biochim. Biophys. Acta, Rev. Cancer. 2008;1785:182–206. doi: 10.1016/j.bbcan.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Preta G. Front. Cell Dev. Biol. 2020;8:1–10. doi: 10.3389/fcell.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C.-L. Linington R. G. Balunas M. J. Centeno A. Boudreau P. Zhang C. Engene N. Spadafora C. Mutka T. S. Kyle D. E. Gerwick L. Wang C.-Y. Gerwick W. H. J. Org. Chem. 2015;80:7849–7855. doi: 10.1021/acs.joc.5b01264. [DOI] [PubMed] [Google Scholar]
- Shao C.-L. Mou X.-F. Cao F. Spadafora C. Glukhov E. Gerwick L. Wang C.-Y. Gerwick W. H. J. Nat. Prod. 2018;81:211–215. doi: 10.1021/acs.jnatprod.7b00917. [DOI] [PubMed] [Google Scholar]
- Keller L. Siqueira-Neto J. L. Souza J. M. Eribez K. LaMonte G. M. Smith J. E. Gerwick W. H. Molecules. 2020;25:1604. doi: 10.3390/molecules25071604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S. J. Antibiot. 2021;74:769–785. doi: 10.1038/s41429-021-00468-5. [DOI] [PubMed] [Google Scholar]
- MacMillan J. B. Molinski T. F. Org. Lett. 2002;4:1535–1538. doi: 10.1021/ol025759p. [DOI] [PubMed] [Google Scholar]
- Salvador L. A. Paul V. J. Luesch H. J. Nat. Prod. 2010;73:1606–1609. doi: 10.1021/np100467d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintard A. Sperandio C. Rodriguez J. Org. Lett. 2018;20:5274–5277. doi: 10.1021/acs.orglett.8b02213. [DOI] [PubMed] [Google Scholar]
- Fiorito D. Keskin S. Bateman J. M. George M. Noble A. Aggarwal V. K. J. Am. Chem. Soc. 2022;144:7995–8001. doi: 10.1021/jacs.2c03192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. Sadeghi S. Tabatabaeian H. Int. J. Mol. Sci. 2021;22:9451. doi: 10.3390/ijms22179451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H. Yoshida W. Y. Moore R. E. Paul V. J. Mooberry S. L. J. Nat. Prod. 2000;63:611–615. doi: 10.1021/np990543q. [DOI] [PubMed] [Google Scholar]
- Milligan K. E. Marquez B. L. Williamson R. T. Gerwick W. H. J. Nat. Prod. 2000;63:1440–1443. doi: 10.1021/np000133y. [DOI] [PubMed] [Google Scholar]
- Harrigan G. G. Yoshida W. Y. Moore R. E. Nagle D. G. Park P. U. Biggs J. Paul V. J. Mooberry S. L. Corbett T. H. Valeriote F. A. J. Nat. Prod. 1998;61:1221–1225. doi: 10.1021/np9801211. [DOI] [PubMed] [Google Scholar]
- Kwan J. C. Ratnayake R. Abboud K. A. Paul V. J. Luesch H. J. Org. Chem. 2010;75:8012–8023. doi: 10.1021/jo1013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. Matthew S. Chen Q.-Y. Kwan J. C. Paul V. J. Luesch H. Org. Lett. 2019;21:7274–7278. doi: 10.1021/acs.orglett.9b02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. McPhail K. L. Gross H. Goeger D. E. Mooberry S. L. Gerwick W. H. Tetrahedron. 2005;61:11723–11729. doi: 10.1016/j.tet.2005.09.036. [DOI] [Google Scholar]
- Marquez B. L. Watts K. S. Yokochi A. Roberts M. A. Verdier-Pinard P. Jimenez J. I. Hamel E. Scheuer P. J. Gerwick W. H. J. Nat. Prod. 2002;65:866–871. doi: 10.1021/np0106283. [DOI] [PubMed] [Google Scholar]
- Zhang D. D. Chapman E. Nat. Prod. Rep. 2020;37:797–826. doi: 10.1039/C9NP00061E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. L. Fu M. Drerup C. M. Gray R. S. Harty B. L. Ackerman S. D. O'Reilly-Pol T. Johnson S. L. Nechiporuk A. V. Barres B. A. Monk K. R. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9153–E9162. doi: 10.1073/pnas.1711088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J. H. Liang X. Paul V. J. Luesch H. ACS Chem. Biol. 2018;13:1189–1199. doi: 10.1021/acschembio.7b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Lu H. Bai Y. Cancer Med. 2019;8:2252–2267. doi: 10.1002/cam4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Liu Y. Wang Z. Xing X. Maguire A. R. Luesch H. Zhang H. Xu Z. Ye T. Chem.–Eur. J. 2013;19:6774–6784. doi: 10.1002/chem.201203667. [DOI] [PubMed] [Google Scholar]
- Liu H. Liu Y. Xing X. Xu Z. Ye T. Chem. Commun. 2010;46:7486–7488. doi: 10.1039/C0CC01200A. [DOI] [PubMed] [Google Scholar]
- Thorburn A. J. Thorac. Oncol. 2007;2:461–465. doi: 10.1097/JTO.0b013e31805fea64. [DOI] [PubMed] [Google Scholar]
- Trivedi R. Mishra D. P. Front. Oncol. 2015;5:69. doi: 10.3389/fonc.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K. Bassler B. L. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. Chen Q.-Y. Seabra G. M. Matthew S. Kwan J. C. Li C. Paul V. J. Luesch H. J. Nat. Prod. 2021;84:779–789. doi: 10.1021/acs.jnatprod.0c01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M. Kringelum H. Murray A. Tønder J. E. Org. Lett. 2006;8:2103–2106. doi: 10.1021/ol060500i. [DOI] [PubMed] [Google Scholar]
- Ruiz-Torres V. Encinar J. A. Herranz-López M. Pérez-Sánchez A. Galiano V. Barrajón-Catalán E. Micol V. Molecules. 2017;22:1037. doi: 10.3390/molecules22071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratspahić E. Freissmuth M. Gruber C. W. Trends Pharmacol. Sci. 2019;40:309–326. doi: 10.1016/j.tips.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar H. Hussain K. Soni A. Khan A. Hussain T. Chénais B. Molecules. 2021;26:247. doi: 10.3390/molecules26010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Luesch H. Nat. Prod. Rep. 2020;37:827–860. doi: 10.1039/C9NP00060G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman I. Hammes S. R. Steroids. 2018;133:96–101. doi: 10.1016/j.steroids.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S. Kryza T. Batra J. Clements J. Nat. Rev. Cancer. 2022;22:223–238. doi: 10.1038/s41568-021-00436-z. [DOI] [PubMed] [Google Scholar]
- Chen Q.-Y. Luo D. Seabra G. M. Luesch H. Bioorg. Med. Chem. 2020;28:115756. doi: 10.1016/j.bmc.2020.115756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom L. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Seo S. U. Woo S. M. Im S.-S. Jang Y. Han E. Kim S. H. Lee H. Lee H.-S. Nam J.-O. Gabrielson E. Min K. Kwon T. K. Cell Death Dis. 2022;13:1–13. doi: 10.1038/s41419-022-04581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadati T. Houben T. Bitorina A. Shiri-Sverdlov R. Cells. 2020;9:1679. doi: 10.3390/cells9071679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris F. Matafora V. Bachi A. J. Exp. Clin. Cancer Res. 2021;40:147. doi: 10.1186/s13046-021-01953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-W. Huang Y.-K. Kuo T.-T. Liu J.-P. Sher Y.-P. Int. J. Mol. Sci. 2020;21:7790. doi: 10.3390/ijms21207790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köcher S. Rey J. Bongard J. Tiaden A. N. Meltzer M. Richards P. J. Ehrmann M. Kaiser M. Angew. Chem., Int. Ed. 2017;56:8555–8558. doi: 10.1002/anie.201701771. [DOI] [PubMed] [Google Scholar]
- Nishio M. Umezawa Y. Hirota M. Takeuchi Y. Tetrahedron. 1995;51:8665–8701. doi: 10.1016/0040-4020(94)01066-9. [DOI] [Google Scholar]
- Nakanishi I. Kinoshita T. Sato A. Tada T. Biopolymers. 2000;53:434–445. doi: 10.1002/(SICI)1097-0282(20000415)53:5<434::AID-BIP7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Gunasekera S. P. Miller M. W. Kwan J. C. Luesch H. Paul V. J. J. Nat. Prod. 2010;73:459–462. doi: 10.1021/np900603f. [DOI] [PubMed] [Google Scholar]
- Wang W. Nag S. A. Zhang R. Curr. Med. Chem. 2015;22:264–289. doi: 10.2174/0929867321666141106124315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Paul V. J. Luesch H. Chembiochem. 2018;19:815–825. doi: 10.1002/cbic.201700627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa F. Shioiri T. Tetrahedron Lett. 2002;43:8673–8677. doi: 10.1016/S0040-4039(02)02178-0. [DOI] [Google Scholar]
- Luo D. Chen Q.-Y. Luesch H. J. Org. Chem. 2016;81:532–544. doi: 10.1021/acs.joc.5b02386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D. Luesch H. ACS Med. Chem. Lett. 2020;11:419–425. doi: 10.1021/acsmedchemlett.9b00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taori K. Paul V. J. Luesch H. J. Nat. Prod. 2008;71:1625–1629. doi: 10.1021/np8002172. [DOI] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Salvador L. A. Law B. K. Paul V. J. Luesch H. Mar. Drugs. 2017;15:290. doi: 10.3390/md15090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrir-Ilan E. Carmeli S. Tetrahedron. 2010;66:9194–9202. doi: 10.1016/j.tet.2010.09.067. [DOI] [Google Scholar]
- Thuan N. H. An T. T. Shrestha A. Canh N. X. Sohng J. K. Dhakal D. Front. Chem. 2019;7:2296–2646. doi: 10.3389/fchem.2019.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf A. Khalil A. A. Olatunde A. Khan M. Anwar S. Alafnan A. Rengasamy K. R. Pharmacol. Res. 2021;166:105521. doi: 10.1016/j.phrs.2021.105521. [DOI] [PubMed] [Google Scholar]
- Breznik B. Mitrović A. Lah T. T. Kos J. Biochimie. 2019;166:233–250. doi: 10.1016/j.biochi.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Kwan J. C. Eksioglu E. A. Liu C. Paul V. J. Luesch H. J. Med. Chem. 2009;52:5732–5747. doi: 10.1021/jm9009394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Law B. K. Paul V. J. Luesch H. J. Nat. Prod. 2017;80:2969–2986. doi: 10.1021/acs.jnatprod.7b00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Ratnayake R. Paul V. J. Luesch H. Bioorg. Med. Chem. 2016;24:3276–3282. doi: 10.1016/j.bmc.2016.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Li H. Jiang F. Wang Z. Zhang W. Bioorg. Med. Chem. 2022;57:116646. doi: 10.1016/j.bmc.2022.116646. [DOI] [PubMed] [Google Scholar]
- Yang S. Zhang W. Ding N. Lo J. Liu Y. Clare-Salzler M. J. Luesch H. Li Y. Bioorg. Med. Chem. 2012;20:4774–4780. doi: 10.1016/j.bmc.2012.05.077. [DOI] [PubMed] [Google Scholar]
- Lawer A. Nesvaderani J. Marcolin G. M. Hunter L. Tetrahedron. 2018;74:1278–1287. doi: 10.1016/j.tet.2017.12.033. [DOI] [Google Scholar]
- Zhang B. Li S. Shui W. Front. Chem. 2022;10:843502. doi: 10.3389/fchem.2022.843502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary P. K. Kim S. Cells. 2021;10:3288. doi: 10.3390/cells10123288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Reyes L. A. Engene N. Paul V. J. Luesch H. J. Nat. Prod. 2015;78:486–492. doi: 10.1021/np500931q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H. Gao B. Rezaei M. A. Kwan J. C. Li C. Ye T. Paul V. J. Luesch H. J. Med. Chem. 2018;61:6364–6378. doi: 10.1021/acs.jmedchem.8b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. Sun S. Lu X. Chen P. Chen C. Liang W. Peng C. Int. Immunopharmacol. 2017;51:124–130. doi: 10.1016/j.intimp.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Bekiari M., in Blue Planet Law: The Ecology of our Economic and Technological World, ed. M. da G. Garcia and A. Cortês, Springer International Publishing, Cham, 2023, pp. 237–252 [Google Scholar]
- Cock I. E. Cheesman M. J. Molecules. 2023;28:7127. doi: 10.3390/molecules28207127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. Hotta K. Deng Y. Yuan R. Quan S. Chen X. Microorganisms. 2021;9:2551. doi: 10.3390/microorganisms9122551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunach M. Guljamow A. Miguel-Gordo M. Dittmann E. Nat. Prod. Rep. 2024;41:347–369. doi: 10.1039/D3NP00045A. [DOI] [PubMed] [Google Scholar]
- Scherlach K. Hertweck C. Nat. Commun. 2021;12:3864. doi: 10.1038/s41467-021-24133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponziani S. Di Vittorio G. Pitari G. Cimini A. M. Ardini M. Gentile R. Iacobelli S. Sala G. Capone E. Flavell D. J. Ippoliti R. Giansanti F. Int. J. Mol. Sci. 2020;21:5510. doi: 10.3390/ijms21155510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Martyn A. P. Quinn R. J. Nat. Prod. Rep. 2022;39:2292–2307. doi: 10.1039/D2NP00038E. [DOI] [PubMed] [Google Scholar]
- Manochkumar J. Cherukuri A. K. Kumar R. S. Almansour A. I. Ramamoorthy S. Efferth T. Comput. Biol. Med. 2023;165:107425. doi: 10.1016/j.compbiomed.2023.107425. [DOI] [PubMed] [Google Scholar]