Abstract
Tandem repeats (TRs) play important roles in genomic variation and disease risk in humans. Long-read sequencing allows for the accurate characterization of TRs; however, the underlying bioinformatics perspectives remain challenging. We present otter and TREAT: otter is a fast targeted local assembler, cross-compatible across different sequencing platforms. It is integrated in TREAT, an end-to-end workflow for TR characterization, visualization, and analysis across multiple genomes. In a comparison with existing tools based on long-read sequencing data from both Oxford Nanopore Technology (ONT, Simplex and Duplex) and Pacific Bioscience (PacBio, Sequel II and Revio), otter and TREAT achieve state-of-the-art genotyping and motif characterization accuracy. Applied to clinically relevant TRs, TREAT/otter significantly identify individuals with pathogenic TR expansions. When applied to a case-control setting, we replicate previously reported associations of TRs with Alzheimer's disease, including those near or within APOC1 (P = 2.63 × 10−9), SPI1 (P = 6.5 × 10−3), and ABCA7 (P = 0.04) genes. Finally, we use TREAT/otter to systematically evaluate potential biases when genotyping TRs using diverse ONT and PacBio long-read sequencing data sets. We show that, in rare cases (0.06%), long-read sequencing from coverage drops in TRs, including the disease-associated TRs in ABCA7 and RFC1 genes. Such coverage drops can lead to TR misgenotyping, hampering the accurate characterization of TR alleles. Taken together, our tools can accurately genotype TRs across different sequencing technologies and with minimal requirements, allowing end-to-end analysis and comparisons of TRs in human genomes, with broad applications in research and clinical fields.
Roughly 30% of the human genome consists of tandem repeats (TRs) characterized by one or more repeat motifs that are defined by their consecutive repetition (Hannan 2018). This repetitive pattern often leads to DNA instability, facilitating not only expansions and contractions of the repeating motif sequence, but also allelic diversity within the sequence (Pearson et al. 2005; Lynch et al. 2008). Several definitions of TRs have been introduced based on the motif length and size variability, including microsatellites, minisatellites, and macrosatellites. Microsatellites (or short tandem repeats, STRs) are the most abundant TRs in the human genome, are characterized by a repetitive motif of <6 bp, and tend to cluster in noncoding regions of the genome (Subramanian et al. 2003). Minisatellites are characterized by a repetitive motif with a size ranging 7–100 bp, and they are highly enriched in the telomeric regions of the genome (Linthorst et al. 2020). Macrosatellites are characterized by larger TRs units (>100 bp), and are enriched in the telomeric and centromeric portions of the genome (Dumbovic et al. 2017).
TRs can disrupt gene-expression regulation and contribute to over 40 neurological disorders (McMurray 2010; Hannan 2018; Khristich and Mirkin 2020). Pathogenic TR expansions, surpassing critical lengths, are linked to conditions like spinocerebellar ataxias, Huntington's disease, Fragile-X syndrome, amyotrophic lateral sclerosis (ALS), and myotonic dystrophy (McMurray 2010; Khristich and Mirkin 2020; Stevanovski et al. 2022). For instance, Fragile-X syndrome results from a GGC repeat expansion in the FMR1 gene, with affected individuals having up to 4000 copies compared to <50 in healthy individuals (Yu et al. 1991). Similarly, ALS is caused by an intronic hexa-nucleotide repeat expansion (GCCCCG) in the C9orf72 gene, exceeding a critical length of more than 200 copies (DeJesus-Hernandez et al. 2011). Beyond diseases-causing, TRs have been also identified as risk factor for complex human diseases: for example, the intronic TR in the ABCA7 gene is associated with a 4.5-fold increased risk of Alzheimer's disease (AD) when the TR exceeds 5720 bp (On Behalf of the BELNEU Consortium et al. 2018; De Roeck et al. 2019).
Traditionally, the evaluation of TR lengths and sequences has been challenging. Conventional methods, such as repeat-primed polymerase chain reaction (RP-PCR) and Southern blot assays, are time-consuming and limited in detecting TRs within PCR-based boundaries. Short-read sequencing approaches offer an alternative, but their limited read lengths often fail to span repetitive regions effectively. Despite heuristic methods and statistical modeling (Gymrek et al. 2012; Gelfand et al. 2014; Kristmundsdóttir et al. 2017; Bakhtiari et al. 2018; Dolzhenko et al. 2019; Eslami Rasekh et al. 2021), accurately assessing clinically relevant TRs remains difficult. The advent of long-read sequencing, particularly with Pacific Bioscience's (PacBio) High Fidelity (HiFi) and Oxford Nanopore Technology's (ONT) Duplex technology (10–20 kb on average, >99% accuracy) (Wenger et al. 2019; Sereika et al. 2022), has significantly improved TR evaluation by providing long and accurate sequencing fragments.
Characterizing TRs with long-read sequencing technology currently has two main limitations. First, there is the need to characterize TRs across different (long-read) sequencing technologies and data-types (Chiu et al. 2021; Masutani et al. 2023; Dolzhenko et al. 2024). This is critically important given the growing long-read sequencing initiatives aiming to comprehensively assess TRs in large genomic data sets (Gustafson et al. 2024), spanning both population-wide and clinical contexts. For example, some existing tools are constrained by predefined TR databases, hindering the identification of new TR features such as novel motif sequences (Ren et al. 2023); other tools are technology and data-type-dependent (Dolzhenko et al. 2024), or do not produce generalizable multisample outputs (Chiu et al. 2021; Masutani et al. 2023).
Second, there is a lack of comprehensive studies that have investigated potential biases when sequencing TRs. For example, DNA methylation has been previously shown to impact basecalling accuracy in long-read sequencing data (Gouil and Keniry 2019; Amarasinghe et al. 2020; Liu et al. 2021). Similarly, the formation of secondary structures due to TRs could impact enzyme efficiency (e.g., polymerase or nanopores) (Mirkin 2007), potentially reducing read quality and sequencing throughput in current long-read sequencing technologies. Furthermore, some technologies require the alignment of noisy reads to generate high-quality consensus sequences, which might be more difficult in case of repetitive regions. These problems may impact genotyping accuracy and lead to incorrect assessments of allele-sequences, including disease-associated TRs in patients.
Here, we present TREAT (Tandem REpeat Annotation Toolkit), a unified workflow for characterizing TRs across multiple genomes, cross-compatible with diverse long-read technologies and data-types (e.g., read-alignments and de novo assemblies). TREAT employs a novel generic targeted local assembler, otter, that can adapt to different sequencing chemistries to accurately characterize TRs. We benchmarked TREAT and otter with currently available tools for TR genotyping (PacBio's TRGT and LongTR) (Dolzhenko et al. 2024; Ziaei Jam et al. 2024) in terms of genotyping accuracy, motif identification, and running performances. We then showcase TREAT and otter applicability in a population-, clinical-, and case-control setting. Finally, we performed a systematic analysis of ∼864K genome-wide TRs in CHM13 reference genome to evaluate sporadic coverage drops that can affect TR genotyping accuracy. We did so using the well-characterized HG002 genome based on long-read sequencing data from ONT (Duplex and Simplex), HiFi, and non-HiFi data from PacBio's Revio and Sequel II instruments.
Results
Cross-compatible workflow for characterizing tandem repeats with otter and TREAT
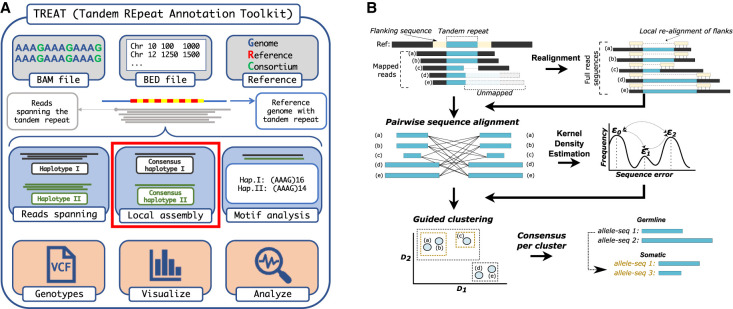
We present otter and TREAT, two bioinformatic tools that enable TR characterization across different long-read sequencing technologies and data-types with minimal input requirements (Fig. 1). Otter is a stand-alone generic targeted local assembler for long-read sequencing data, which automatically adapts to sequencing error-rates and coverage levels per target region. TREAT integrates otter to enable end-to-end unified workflow for de novo motif characterization and downstream analysis, including TR visualization, outlier-based, and case-control comparisons (see Methods; Fig. 1A). Both tools require sequencing data aligned to a reference genome (BAM files), the reference genome used (FASTA file), and the coordinates of the regions of interest (chromosome, start and end positions encoded in a BED file). TREAT/otter outputs a multisample gVCF (Genomic Variant Call Format) file reporting genotyped alleles, their size and relative repeat content (motif and number of copies), of each TR in each sample.
Figure 1.
Schematic workflow of TREAT and otter. (A) TREAT workflow, highlighting the required inputs, the main features, and the main outputs of the tool. The red box highlights the main genotyping engine based on otter. (B) The main algorithmic steps of otter, a novel targeted local assembler for long-read sequencing data.
Otter and TREAT enable accurate characterization of both PacBio and ONT long-read data
We benchmarked TREAT and otter with TRGT and LongTR (Dolzhenko et al. 2024; Ziaei Jam et al. 2024), currently available tools to characterize TRs in long-read sequencing data. We compared: (1) genotyping accuracy, i.e., the accuracy of the predicted allele-sequences for a TR, (2) motif characterization accuracy, and (3) computational resources. We varied different long-read sequencing technologies (PacBio Sequel II and Revio, ONT Simplex and Duplex) as well as different coverages (5×, 10×, 15×, 20×, 25×, and 30×) of HG002 (Jarvis et al. 2022). We focused on a set of 161,382 TRs from PacBio's repeat catalog (see Methods). Predicted TR alleles were compared to the expected alleles based on the HG002 T2T assembly (see Methods).
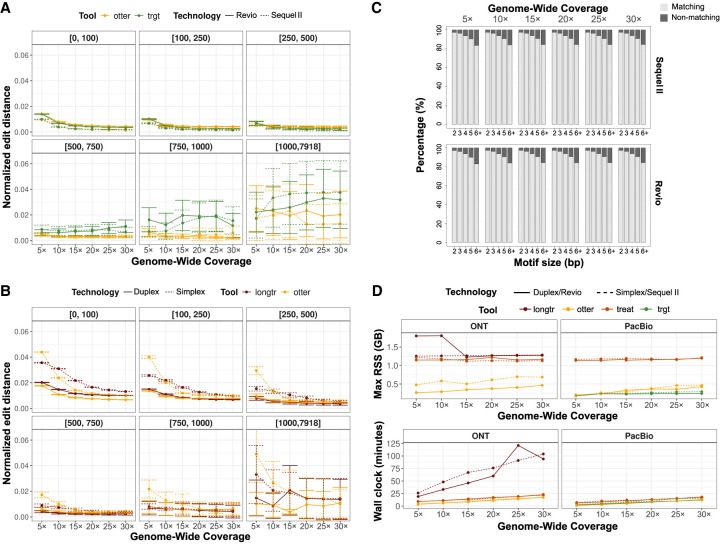
In PacBio data, we found comparable genotyping accuracy between otter (TREAT genotyping engine) and TRGT, for both Sequel II and Revio data sets, although otter generated more accurate genotypes for larger TRs (e.g., >500 bp), achieving average error-rates of 0.2%–2.5%, compared to 0.6%–3.8% of TRGT. Both methods were more accurate when increasing the coverage, although this was less pronounced for larger TRs (>500 bp). Notably, genotyping accuracy for both otter and TRGT was higher for PacBio's Sequel II data in comparison with Revio data (Fig. 2A; Supplemental Results). In ONT data, otter was generally more accurate than LongTR although differences for large TRs were less clear. For both tools, we observed better accuracies for Duplex data in comparison to Simplex data (Fig. 2B; Supplemental Results). Altogether, our benchmark across all tools revealed that PacBio led to more accurate genotypes for TRs <500 bp, with PacBio and ONT having similar performances for TRs ranging 500–1000 bp, and ONT leading to more accurate genotypes for TRs ≥1000 bp (see Fig. 2A,B; Supplemental Results).
Figure 2.
Benchmarking between TREAT/otter, TRGT, and LongTR. (A) Genotyping accuracy of otter and TRGT on PacBio Sequel II and Revio data, stratified by TR size, and sequencing depth. (B) Genotyping accuracy of otter and LongTR on ONT Simplex and Duplex data, stratified by TR size and sequencing depth. (C) Motif identification accuracy of TREAT and TRGT on PacBio Sequel II and Revio data, showing the overlap of matching motifs, stratified by motif size and sequencing depth. (D) Memory usage and running time of otter, TREAT, TRGT, and LongTR, across technologies and sequencing coverages.
The above observations remain when using different distance metrics and partitioned by different TR-types. For example, we observed similar performances when using the raw edit distance and correlation between observed and expected allele sizes (Supplemental Fig. S1; Supplemental Results). Furthermore, we found that TRs characterized by dinucleotide repeat motifs were on average less accurate than TRs with longer motifs (Supplemental Fig. S2). The fraction of alleles perfectly genotyped (i.e., with an edit distance of 0), compared to expected alleles, increased with higher coverage across all technologies and tools (Supplemental Fig. S3), with Sequel II data having the largest fraction of alleles perfectly matched, and ONT Simplex having the least. In PacBio Sequel II and Revio data, TRGT generated a slightly higher fraction of perfectly matched alleles with respect to otter (max difference 2.8%). In ONT data, otter outperformed LongTR in all settings.
Similarly, TREAT, which makes use of TR-genotypes from otter, achieved similar motif characterization accuracy relative to TRGT (Fig. 2C). In the GRCh38 reference genome, the motifs of the 161K TRs were mostly dinucleotide (49%), followed by tetranucleotide (22%) and 16+ bp motifs (11%) (Supplemental Fig. S4). Because LongTR does not directly report the identified TR motifs, we compared TR motifs between TREAT and TRGT. On average, TREAT identified the same motifs as TRGT in 96% of cases (Fig. 2C), and this did not change for different technologies (Sequel II or Revio) or different coverages. We observed a higher concordance in motif detection between tools for shorter motifs (Fig. 2C). When looking at the motifs identified by TREAT on the GRCh38 reference genome, these matched known TR annotations in 91% of the cases.
Finally, we evaluated the computational performances of otter (stand-alone), TREAT (integrated workflow with otter), TRGT and LongTR. When using four threads, TRGT and otter had similar run-time performances, while both were slightly faster than the integrated workflow from TREAT (Fig. 2D). On the other hand, for the ONT data, otter and TREAT were faster than LongTR. In terms of memory consumption, performances were comparable between TREAT and LongTR, while otter and TRGT used significantly less memory (Fig. 2D). When evaluating the multithreading capabilities in TREAT, we saw that when increasing the number of threads to 6, 8, 10, and 12, the running times decreased by 1.3-, 1.5-, 1.6-, and 1.8-fold (on average across the different technologies), compared to four CPU threads (Supplemental Fig. S5).
In addition to the high-quality HiFi data, PacBio can output non-HiFi data, i.e., reads that did not pass PacBio's internal HiFi quality thresholds, and that constitute a significant fraction of all sequenced data (45% in HG002). We explored whether integrating both HiFi and non-HiFi data could improve otter’s capability to accurately characterize TR allele-sequences. Because Revio uses a subset of these non-HiFi reads (those with at least 90% read quality) to improve throughput and accuracy via DeepConsensus (Baid et al. 2023), we performed this analysis only for Sequel II data. We found that non-HiFi data improved accuracy across all TR lengths. Specifically, when integrating non-HiFi reads of at least 85%–90% read quality, genotyping accuracy improved by nearly twofold (Supplemental Fig. S6).
TREAT's unified workflow enables characterization of diverse tandem repeats
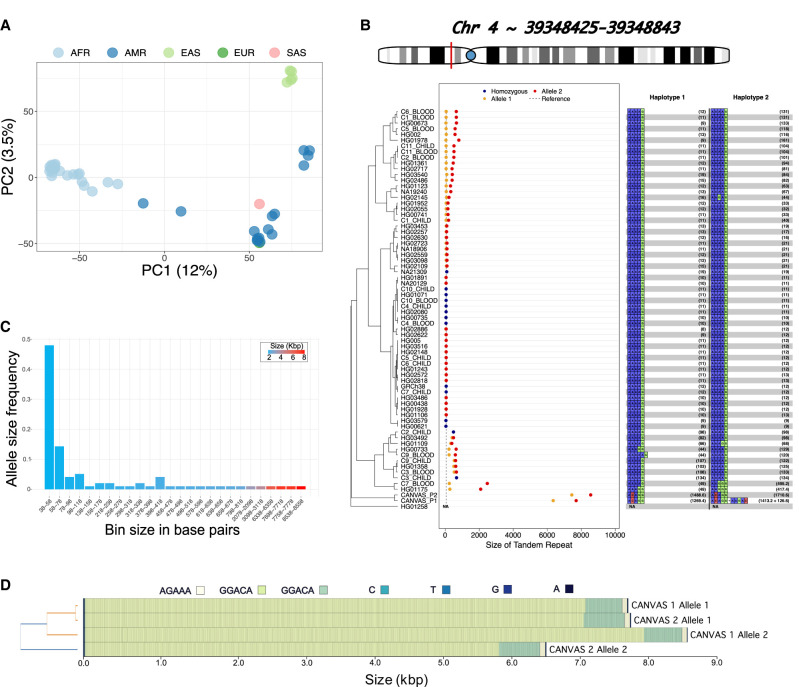
We applied TREAT's unified workflow to characterize TRs in a population and clinical setting. First, we genotyped the set of 161K TRs in 47 genomes from the Human Pangenome Research Consortium (HPRC) (Wang et al. 2022), for which PacBio HiFi data were available. We then extracted the top 20% most variable TRs (N = 32,208, based on the coefficient of variation) (see Methods), and performed a principal component analysis (PCA) (Fig. 3A) on the joint allele sizes (i.e., the sum of the maternal and paternal alleles). We found that PC1 explained 12% of the total variance and genetically represented the African-American axis, while PC2 explained 3.5% of variance and corresponded to the American-Asian axis. The explained variance was similar to that of a PCA including 40/47 matching samples and 30,544 random common (minor allele frequency >10%) single-nucleotide polymorphisms (SNPs; PC1: 14%, PC2: 4%) (Supplemental Fig. S7).
Figure 3.
TREAT visualization and analysis modules. (A) The PCA of the ancestry-based analysis based on the 20% most variable TRs across 47 HPRC genomes. (B) The main TR is in the RFC1 gene. y-axis: individuals, x-axis: TR size (in bp). Blue dots: smaller allele, orange dots: larger allele, red dots: homozygous genotypes. Dashed line: the allele in the reference genome GRCh38. The right side of the plot reports, for each sample and each allele, the motif and relative number of copies. The TR lengths of the two CANVAS patients were identified as significant outliers compared to the length-distribution of 47 samples from the HPRC. (C) The distribution of allele sizes for the TR in RFC1 gene. (D) Motif representation in CANVAS patients, as produced with MotifScope (Zhang et al. 2024).
We then used TREAT's outlier analysis to detect and score extreme TR expansions or contractions of 35 clinically relevant TRs (Supplemental Table S1) in 47 genomes from the HPRC, as well as two Dutch CANVAS patients and 10 parent–offspring duos (see Methods; Salazar et al. 2023; van de Pol et al. 2023). The two CANVAS patients were previously characterized to harbor expansions in the intronic TR in RFC1 (van de Pol et al. 2023). For all individuals, PacBio HiFi data were generated with Sequel II instrument. In total, we identified 30 instances where the TR length in certain samples were significantly different from the distribution of TR lengths across all 69 genomes. The most significant deviations were observed for the two CANVAS patients in the TR intronic of RFC1 gene (P < 2 × 10−16 for both patients) (Fig. 3B–D). The joint allele size for these samples was 78- and 89-fold higher than the median TR size across all 69 genomes. Significant TR expansions were also found in the TR in ATXN8 gene (HG01123 sample, P < 2 × 10−16) (Supplemental Fig. S8), and in DMD gene (HG02622 sample, P = 6.90 × 10−3) (Supplemental Fig. S9). In the TR intronic of RFC1 gene, we also observed a significant heterozygous expansion in one parent of the parent–offspring duos (P = 1.7 × 10−3 and P = 5.18 × 10−11, respectively, for the short and long alleles) (Fig. 3B). Unexpectedly, the child reported a homozygous nonexpanded genotype, suggesting a misassembly or an allele dropout.
Finally, we applied TREAT to characterize unique TRs that are present in CHM13 reference genome but absent in GRCh38 across the 47 HPRC genomes. We first curated a set of ∼864K genome-wide TRs in the CHM13 reference genome (see Methods). We evaluated genotyping accuracy by applying TREAT/otter to CHM13-aligned long-read data sets of HG002 (PacBio's Revio and Sequel II as well as ONT's Duplex and Simplex). We observed similar performances as those observed when using ∼161K TRs from GRCh38 (see Supplemental Fig. S10; Supplemental Results). These results showcase otter and TREAT's ability to de novo characterize TRs across different reference genomes, and without prior knowledge of TR motif composition. Based on a CHM13-to-GRCH38 liftOver procedure, we found 1017 unique TRs present in CHM13 and absent in GRCh38, 37% of which overlapped coding sequences (Supplemental Methods; Supplemental Table S2). We used TREAT/otter to characterize these TRs across the 47 HPRC genomes and found a mean TR size of 129 bp (median = 45 bp), mainly composed of trinucleotide motifs (42%), followed by homopolymers (26%), and 6+ nt motifs (22%) (Supplemental Fig. S11).
Tandem repeats may be sensitive to coverage dropouts in long-read sequencing
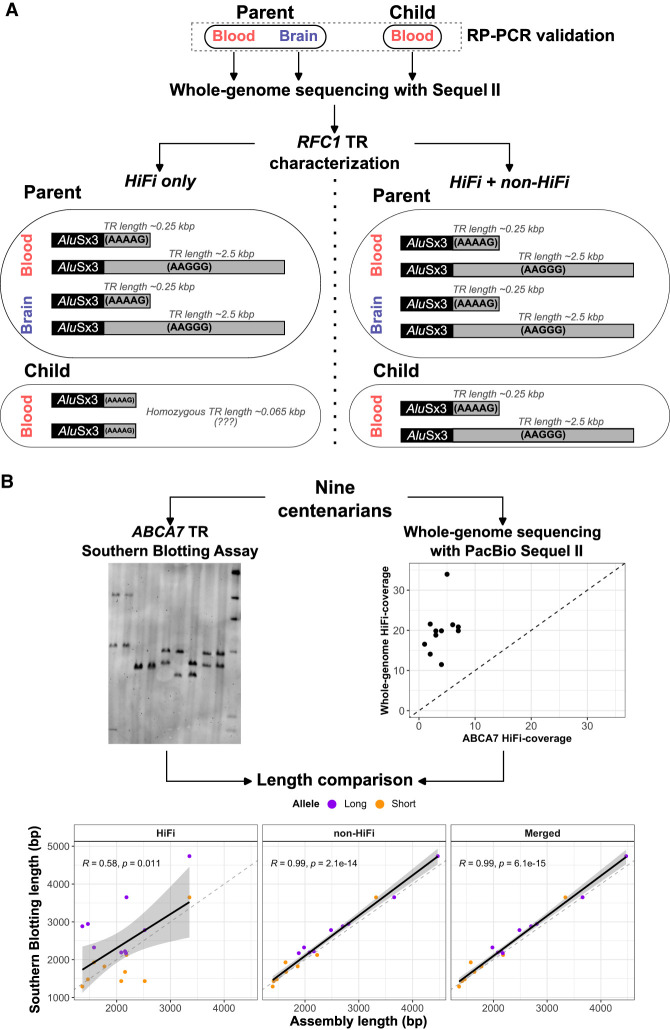
A closer investigation of PacBio long-read data revealed unexpected drops of coverage in clinical TRs, consequently leading to misgenotyping of disease-associated TRs. One example is the CANVAS-associated intronic TR in RFC1, where the most common allele consists of an (AAAAG)11 motif, with a total size of ∼55 bp. In CANVAS patients, the TR can range from 2 to 10 kbp in total length (Fig. 3B–D; Supplemental Results). In one parent–child duo, we found that the parent harbored an expanded heterozygous version of the TR: a shorter allele with a total length of 244 bp with the (AAAAG)50 motif; and a longer allele with a total length of 2.49 kbp, composed primarily of the (AAGGG)490 motifs (Fig. 4A). Long-read sequencing of brain tissue from the same individual (PacBio Sequel II) confirmed these results, although the longer allele was further expanded by 180 bp (36 additional motif-copies), suggesting a somatic expansion in the brain relative to blood (Fig. 4A). However, long-read data from the child yielded a homozygous allele-sequence of 63 bp with the (AAAAG)12 motif (Fig. 4A). This was unexpected as at least one of the two allele-sequences from the parent should be inherited in the child. A closer analysis of HiFi long-read-pileup strongly supported this genotype. However, we observed an abnormal coverage drop in both the parent and child for this TR, which was alleviated when including non-HiFi data (Supplemental Results). After merging HiFi and non-HiFi data of the child, TREAT/otter correctly assembled the expanded allele-sequence at 2.65 kbp in size with (AAGGG) > 374. Penta-repeat-primed PCR (RP-PCR) confirmed that both parent and child harbored repeat expansions separately composed of the (AAAAG) and (AAGGG) motifs (Supplemental Fig. S12). Therefore, HiFi data alone failed to capture this expanded allele-sequence, which was recoverable when including the non-HiFi data.
Figure 4.
Coverage drops in TR in RFC1 and ABCA7 genes, associated with CANVAS and AD. (A) The genotyped TR alleles and relative motif characterization in a parent–child duo using only HiFi data, and HiFi + non-HiFi data. Long-read data from the brain of the parent were also available. Adding non-HiFi data rescued the missing allele in the child. (B) The comparison between experimentally validated alleles in the TR intronic of ABCA7 gene, and genotyped alleles based on HiFi data alone, and HiFi + non-HiFi. Experimental validation of TR alleles was performed with Southern blot assay, and was available for nine individuals for which long-read data were also available. When adding non-HiFi data, we could recover the expanded alleles in the child that were missed by HiFi data alone.
We observed similar situations of abnormal coverage drops in PacBio data in a separate intronic TR in ABCA7, previously associated with AD. We experimentally validated the lengths of this TR using Southern blotting in a subset of nine centenarians for which long-read sequencing was performed (Supplemental Fig. S13; Supplemental Methods). The local HiFi coverage for these individuals ranged 1–7× (Fig. 4B; Supplemental Results). The correlation between experimentally validated alleles and HiFi-based alleles was 0.58 (Pearson's correlation) (Fig. 4B). However, the inclusion of non-HiFi data increased read support by fourfold to an average coverage of 22×. As a result, the correlation with experimentally validated allele sizes increased to 0.99 (Fig. 4B). These results highlight standing challenges of characterizing TRs with long-read sequencing data, and suggest systematic biases of long-read sequencing in certain genomic regions.
The above observations motivated us to systematically characterize genome-wide coverage drops of TRs across long-read sequencing technologies. We did this by investigating coverage drops in the curated set of ∼864K genome-wide TRs in the CHM13 reference genome, using both PacBio and ONT long-read data sets of HG002 at ∼38× coverage (see Supplemental Results and Methods). The average TR length in this curated set was 93 bp, with motifs being mostly 16+ bp motifs (23%), followed by dinucleotide (18%), tetranucleotide (14%), and homopolymers (13%) (Supplemental Fig. S4). For each TR, we defined the coverage ratio by dividing the local TR coverage versus global genome-wide coverage. We found the average coverage ratio to be 1.01, 1.02, 0.99, and 1.03, respectively, for Sequel II, Revio, ONT Simplex, and Duplex technologies. This indicated generally no unexpected coverage drops in TRs (Supplemental Fig. S14A). However, 486 (0.06%) unique TRs had ratios below 0.25 (i.e., a fourfold lower coverage than expected based on the global average coverage), of which 454 (93%) were present in the HG002 T2T reference assembly (Supplemental Table S3). The majority of the low-coverage TRs (294/454, 65%) overlapped gene annotations, potentially leading to misgenotyping that may impact biological interpretation. Furthermore, we observed that some of these TRs were within 5 kbp of each other, suggesting that coverage drops can extend across multi-kbp regions. Overall, we observe significantly more low-coverage TRs in PacBio data sets compared to ONT (OR = 9.4, P-value < 2 × 10−16, Fisher's exact test), with N = 437 TRs (89%) being specific to PacBio data sets. Moreover, 22% of these TRs (N = 98) had low coverage in both Sequel II and Revio data sets, suggesting potential systematic challenges in both technologies (Supplemental Fig. S14B–G). This included the intronic TR in ABCA7, previously associated with AD. The average number of non-HiFi reads in these TRs was 10, indicating that although reads were generated for these TRs, most were flagged as low-quality during HiFi data generation.
Within the ONT data sets, we observe significantly more low-coverage TRs in the Duplex data set relative to the Simplex data set (OR = 2.6, P-value = 1.76 × 10−3, Fisher's exact test).
We characterized the sequences of all low-coverage TRs to investigate potential characteristic features. When comparing the 454 low-coverage TRs with the remaining of ∼864K genome-wide TRs, we found that low-coverage TRs were longer (P-value = 8.68 × 10−14; 493 bp longer on average) and harbored higher GC-content (P-value = 2.28 × 10−50; 17.4% higher on average). A comparison of dinucleotide content revealed that AG, CC, CG, CT, and GG dinucleotides were significantly enriched in the low-coverage TRs (Supplemental Fig. S14H,I). Moreover, we found that G-quadruplex DNA secondary structures (G4s) were more likely to occur in low-coverage TRs (P-value = 2.48 × 10−45; 3.76% higher) (Supplemental Fig. S14H; Supplemental Methods).
Comparing tandem repeats across multiple genomes in a case-control setting
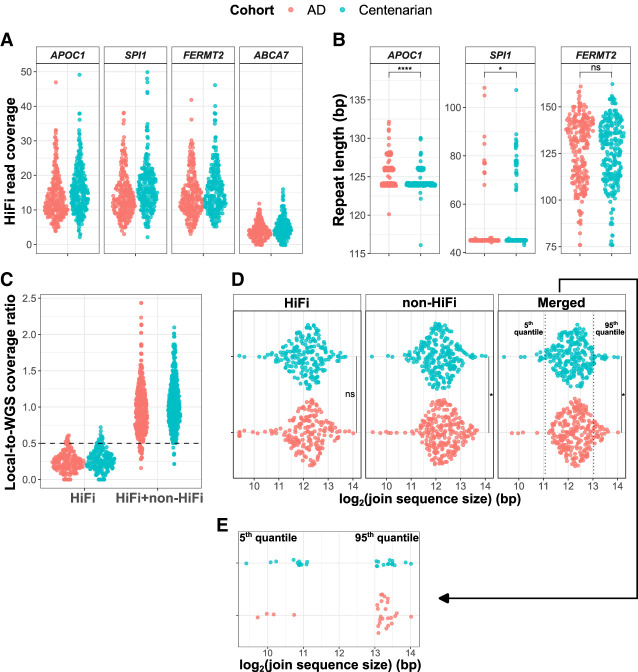
With the acquired knowledge about possible allele dropouts in TRs, we used TREAT/otter in a case-control setting to replicate the association of four TRs that were previously shown to associate with AD risk (Table 1, Supplemental Table S4). We did so by using a set of 246 AD patients (mean age = 67.9 ± 9.8, 70% females) and N = 248 cognitively healthy centenarians (mean age = 101.2 ± 2.5, 70% females) that were sequenced with PacBio Sequel II instrument (Methods; Supplemental Fig. S15; Salazar et al. 2023). Across all 494 genomes, we observed a median coverage (HiFi data) of 14, 15, 14, and 4, respectively, for the TRs in APOC1, SPI1, FERMT2, and ABCA7 (Fig. 5A). The combined allele size (i.e., the sum of the maternal and paternal alleles) of the TR nearby APOC1 (Chr 19:44921096–44921134) was significantly expanded in AD patients compared to cognitively healthy centenarians (beta = 0.38, P = 2.63 × 10−9) (Table 1; Fig. 5B). In contrast, the short allele of the TR within SPI1 gene was significantly contracted in AD patients compared to cognitively healthy centenarians (beta = −0.03, P = 6.5 × 10−3) (Table 1; Fig. 5B). The direction of effect of these TRs was in line with the original studies (Guo et al. 2023; Wang et al. 2023). We could not replicate the association of the TR within FERMT2 (beta = 0.01, P = 0.27, short allele) (Table 1; Fig. 5B).
Table 1.
Replication of TRs previously associated with Alzheimer's disease
| TRs previously associated with AD | ||||
|---|---|---|---|---|
| Region | Chr 19:44921096–44921134 | Chr 11:47775208–47775243 | Chr 19:1049436–1050066 | Chr 14:52832909–52832938 |
| Gene | APOC1 | SPI1 | ABCA7 | FERMT2 |
| Best model | Joint alleles | Short alleles | Joint alleles | Short alleles |
| Beta (OR) | 0.38 (1.46) | −0.03 (0.97) | 8.63 × 10−5 (1.01) | 0.01 (1.01) |
| P-value | 2.6 × 10−9 | 6.5 × 10−3 | 0.041 | 0.27 |
| Original study | 38014121 | 37745545 | 29589097 | 37745545 |
| Original OR | NA | −0.01 (0.99) | 4.5 | 0.01 (1.01) |
| Original model | Longer allele | Joint alleles | Individuals with alleles >5720 bp | Joint alleles |
| Original method | Logistic regression | Mixed linear models | Fisher's exact | Mixed linear models |
| Original P-value | 4.3 × 10−10 | NA | 0.008 | NA |
| Original samples | 1489 AD versus 1492 controls | 6328 AD versus 6580 controls | 275 AD versus 177 controls | 6328 AD versus 6580 controls |
| Data type | Short-read sequencing | Short-read sequencing | Southern blot | Short-read sequencing |
Region: genomic coordinates of the TR with respect to GRCh38; Gene: the closest gene as reported in the original publications; Best model: model that yielded the most significant association, in our comparison: short allele, long allele, or joint alleles size; Beta (OR): effect size and relative Odds Ratio with respect to AD: an increased TR size leads to increased AD risk for positive estimates; P-value: P-value of association. We used logistic regression models using TR size (short allele, long allele, and combined allele size) as predictor for AD case-control status, using 246 AD patients (cases) and 248 cognitively healthy centenarians (controls); Original study: the PubMed ID of the original study; Original OR: the odds ratio as reported in the original study; Original model: model used for association in the original study; Original method: method used for association in the original study; Original P-value: the P-value reported in the original study; Original samples: the number of AD cases and controls used in the original study; Data type: the data on which the association were identified.
(TR) Tandem repeat, (AD) Alzheimer's disease.
Figure 5.
Replication of the association with AD of TRs in APOC1, SPI1, FERMT2, and ABCA7. (A) The coverage distribution of the four TRs in AD patients and cognitively healthy centenarians. (B) The TR size difference between AD patients and cognitively healthy centenarians in APOC1, SPI1, and FERMT2. For the associations, we used logistic regression models using the TR size as predictor for AD case-control status. (C) HiFi and combined HiFi + non-HiFi coverage distribution of the TR intronic of ABCA7 gene. (D) Comparison of the joint allele size of ABCA7 TR between AD cases and cognitively healthy centenarians, respectively, using HiFi data, non-HiFi data, and the merged data set of HiFi and non-HiFi. (E) Number of AD cases and cognitively healthy centenarians in the lower 5th quantile and upper 95th quantile. Quantiles were defined based on the distribution of the joint TR allele size in the centenarians. We tested for the differential enrichment of AD and centenarians in each quantile with Fisher's exact tests.
For the intronic TR in ABCA7, we found significant expansions in AD cases after integrating non-HiFi data (beta = 8.63 × 10−5, P = 0.04, joint allele size) (Fig. 5C,D). We note that 22 samples were omitted due to reduced coverage levels even after integrating HiFi and non-HiFi data. We then identified TR size boundaries in the centenarian controls corresponding to the 5th and 95th percentiles of the joint TR allele sizes (2.2 kbp and 8.4 kbp, respectively). The number of centenarians with a TR size lower than the 5th percentile was threefold higher than that of AD cases (1-tailed Fisher's exact test P = 0.023, OR = 3.2) (Fig. 5E), and the number of AD cases with a TR size larger than the 95th percentile was twofold higher than that of centenarians (1-tailed Fisher's exact test P = 0.04, OR = 2.0) (Fig. 5E). Given the difficulties in correctly assessing the allele-sequences of this TR, we cannot exclude that additional samples suffer from allelic dropouts, especially for the larger expanded allele-sequences.
Discussion
In this study, we provide novel contributions to better characterize TRs with long-read sequencing data. First, we present our novel tools, otter and TREAT, that provide a unified workflow to accurately characterize TRs using both PacBio and ONT data sets. This enabled us to characterize genome-wide TRs in patients with neurodegenerative diseases and genomes from the HPRC. Second, we show that in rare instances, long-read sequencing technologies can suffer from abnormal coverage drops in TRs due to potential systematic challenges, particularly in PacBio's HiFi technology. These coverage drops can lead to TR misgenotyping, as we observed in CANVAS and AD-associated TRs. Finally, we applied TREAT/otter to a case-control setting and replicated TRs previously associated with AD across 494 long-read sequenced AD patients and cognitively healthy centenarian genomes.
Our benchmark of otter and TREAT highlighted state-of-the-art performances of our tools in terms of TR genotyping and motif identification accuracy. We showed that otter, TREAT, and other existing tools provide generally accurate characterizations of TRs on both PacBio and ONT data sets, and with improved accuracies at higher sequencing coverages. Across technologies, our benchmark revealed that PacBio leads to generally more accurate genotypes for relatively smaller TRs, with PacBio and ONT having similar performances for TRs ranging 500–1000 bp, and ONT leading to more accurate genotypes for larger TRs. These results remained when using other distance metrics as well as in a similar benchmark using the CHM13 reference genome and a larger set of genome-wide TRs.
Our systematic analysis of coverage drops revealed that overall, coverage drops of TRs are rare (0.6%), and do not impact the overall genotyping performances of TREAT/otter and other tools. However, our analysis relied on HG002, a highly homozygous genome sequenced at high coverage (38×). Hence, TR coverage drops may be more prevalent in other (low-coverage) genomes that harbor expanded TR sequences, especially those with GC-rich sequences. TRs with coverage drops were often large (>500 bp), high in GC-content, and with higher densities of predicted G4s. G4s have been previously reported to reduce polymerase efficiency (Lago et al. 2021). As PacBio's HiFi technology relies on multiple successful passes of a DNA polymerase in a circular DNA template (Wenger et al. 2019), we speculate that the interference of G4s might reduce the number of passes in the circular template, possibly leading to lower quality reads (non-HiFi reads). Altogether, incidents of TR coverage drops were enriched in PacBio's Revio and Sequel II data sets, and to a lower extent in ONT's Duplex and Simplex data sets, with ONT Simplex suffering the least. Although rare, we showed and experimentally validated that coverage drops in TRs can occur at clinically relevant TRs, requiring extra attention when characterizing these TRs. To this end, we showed that local versus global coverage ratio is an effective way to identify such problematic regions, and that for PacBio, these regions can be (in part) rescued by adding noisier non-HiFi data, as shown for the TRs in ABCA7 and RFC1 genes.
TREAT and otter can be used to genotype and characterize potentially any type of repetitive sequences. However, this remains challenging for very large TRs spanning several kilobases, for example, those in telomeric and centromeric regions of the genome. We also note that regions where sequencing error-rates exceed interallele dissimilarities may still be difficult to genotype. As the error rate in ONT Simplex data is relatively higher than PacBio and ONT Duplex, this is likely driving the lower genotyping accuracy observed in ONT Simplex. These limitations are not only specific to TREAT and otter, but extend to other existing tools. With newer sequencing technologies bringing longer read lengths (e.g., ONT ultra-long reads), together with more complete reference genome assemblies, it might become possible to genotype any satellite region (micro-, mini-, and macrosatellites) in the genome with TREAT and otter.
We were able to replicate previously reported TRs associated with AD by comparing a cohort of AD patients and cognitively healthy centenarians. We acknowledge that these TRs were previously identified using different experimental methods (e.g., short-read sequencing, Southern blotting), and analyses strategies (logistic regressions, linear mixed models, Fisher's exact test) (On Behalf of the BELNEU Consortium et al. 2018; Guo et al. 2023; Wang et al. 2023). While this heterogeneity hampers the direct comparison of the effect size estimates, all associations we observed were in the same direction as the original studies. In particular, the TR intronic of ABCA7 was shown to carry an odds ratio for AD of 4.5 when one allele was expanded >5.7 kbp (On Behalf of the BELNEU Consortium et al. 2018). Similarly, we observed that individuals carrying larger allele-sequences were significantly associated with AD. However, in our cohort, the effect was mainly driven by cognitively healthy centenarians having a shorter joint allele size (i.e., more AD-protection), rather than AD cases having a more expanded TR sizes. While we cannot exclude that we have missed some expanded genotypes due to allele dropouts, the centenarians that we included were previously shown to be enriched with the protective alleles in the majority of SNPs associated with AD (Tesi et al. 2024).
In summary, otter and TREAT are flexible and accurate bioinformatics tools compatible with different sequencing platforms and requiring minimal input requirements, that enable end-to-end analysis and comparisons of TRs in human genomes with broad applications in research and clinical fields.
Methods
TREAT
The main analysis is the assembly analysis, which uses otter for TR genotyping, and is followed by TR content characterization (identification of motif and number of copies) on the individual TR alleles. In addition to the assembly analysis, TREAT implements a reads analysis (Fig. 1A). Here, TR genotyping is performed using an iterative clustering framework based on TR sizes (Supplemental Methods). This is followed by TR content characterization, which is done on all individual reads (Supplemental Methods; Supplemental Results). This analysis may be preferred when information from all reads is needed, for example, for performing a multiple sequence alignment, or when studying somatic instability.
In all cases, TR content characterization is performed with pytrf (https://github.com/lmdu/pytrf). When multiple motif annotations for the same sequence are found by pytrf, a consensus representation of the repeat content is generated. Briefly, if the fraction of sequence annotated with a given motif is >95%, then the relative motif is regarded as the best motif describing the TR. In case two or more motifs are found, each describing a portion of the sequence, then the intersection is calculated by intersecting the motif-specific start and end positions. If the intersection is <90%, then the motifs and the relative number of copies are combined. For example, for sequence TGTGTGTGTGTGTGGAGAGAGAGAGAGA, pytrf identifies (1) seven copies of TG (ranging positions 1–14, 50% of the sequence covered), and (2) seven copies of GA (ranging positions 15–28, 50% of the sequence covered). In this case, the combined sequence annotation will be TG + GA, repeated 7 + 7 times (see Supplemental Methods).
TREAT's analysis module consists of an outlier-detection framework, and a case-control analysis. The outlier-detection scores extreme variations in TR allele sizes across a set of samples. Outliers are detected using a normalized distance that quantifies how far each allele size is from the median allele size, scaled by the variability of the data (Supplemental Methods). A P-value for each individual is then calculated by comparing each data point's distance to a χ2 distribution. The case-control analysis employs logistic regression models to compare allele sizes (short allele, long allele, and joint allele size) between cases and controls.
Otter: a stand-alone, fast, local assembler
Otter is a generic stand-alone method for generating fast local assemblies of a given region or genotyping whole-genome de novo assemblies. Otter in the main genotyping engine of TREAT assembly analysis. Briefly, given a region of interest, otter uses the htslib library to identify spanning reads (region of interest is fully contained in the reads) and nonspanning reads (only partially contained) in a given BAM file, and extracts the corresponding subsequence per read based on their alignment (Fig. 1B; Bonfield et al. 2021). When a reference genome is provided, it will perform local read-realignments on nonspanning reads if it detects a clipping-signal, which can indicate suboptimal mappings to due highly divergent sequences (Fig. 1B). This is done by aligning (using WFA2-lib alignment library) (Marco-Sola et al. 2023) the flanking sequences of a region (100 bp by default, modifiable with “‐‐flank-size” parameter) derived from the reference genome onto each read, which are then used to recalibrate the corresponding subsequence of the region of interest. Recalibrated nonspanning reads are reclassified as spanning if both flanking sequences are successfully aligned with a minimum length and sequence similarity (by default, 90% sequence similarity, modifiable with “‐‐min-sim” parameter). In the context of TRs, this realignment procedure often correctly recalibrates the alignments of TRs with major length and/or motif-composition differences relative to a reference genome.
Otter identifies unique allele-sequences by clustering spanning reads via pairwise-sequence alignment (Fig. 1B; Supplemental Methods). To manage high somatic variation and/or sequencing errors, otter estimates local baseline error-rates per region using a Gaussian-kernel density estimator. This produces a one-dimensional distribution of spanning pairwise-sequence distances. In single homozygous allele-sequences, the distribution is unimodal centered at 0. With multiple allele-sequences, the distribution is multimodal, where peaks represent sequence errors between reads from different allele-sequences. Otter identifies these peaks and performs hierarchical clustering, stopping when distances exceed the densest peak, partitioning reads into initial clusters. This procedure is followed by a curation step to ensure sufficient read support, adapting to local coverage (Fig. 1B). If no maximum number of alleles (α) is enforced, otter outputs all clusters. Otherwise, clusters below the coverage threshold are merged, and if clusters exceed α, hierarchical clustering continues until α clusters remain. Otter then generates a final consensus sequence per cluster via pseudo-partial order alignment procedure of spanning and nonspanning reads inspired from Ye and Ma (2016).
Genomes included for testing
HPRC
Publicly available PacBio long-read HiFi data of 47 individuals from the HPRC were downloaded (https://github.com/human-pangenomics/HPP_Year1_Data_Freeze_v1.0?tab=readme-ov-file) (Wang et al. 2022). For the well-characterized HG002 genome (Jarvis et al. 2022), we also downloaded data generated with ONT (Duplex and Simplex chemistries) and PacBio Revio technologies. Finally, we generated long-read sequencing data for HG002 using the PacBio Sequel II instrument across three SMRT cells, keeping both HiFi and non-HiFi data. ONT data were aligned to the reference genomes (GRCh38 and CHM13) using minimap2 (2.21-r1071, specifying -x map-ont) (Li 2018). PacBio data were aligned using pbmm2 (1.9.0, specifying –preset CCS and –preset SUBREADS, respectively, for HiFi and non-HiFi data) (Wenger et al. 2019).
100-plus Study cohort and Alzheimer Dementia Cohort
For the replication of TRs previously associated with AD, we used HiFi sequencing (Sequel II) data from the blood DNA of N = 246 patients with AD from the Amsterdam Dementia Cohort (ADC) (van der Flier and Scheltens 2018; Salazar et al. 2023), and N = 248 cognitively healthy centenarians from the 100-plus Study cohort (Holstege et al. 2018; Salazar et al. 2023). Ten cognitively healthy centenarians were sequenced as a trio, including the blood-derived DNA from the centenarian, the brain-derived DNA from the centenarian and blood-derived DNA from a child of the centenarian. The combined set of a centenarian and child is referred to as parent–child duo throughout the manuscript. Sequencing data preprocessing was conducted as previously described (Supplemental Methods; Salazar et al. 2023). Long-read sequencing data for these individuals is available on the Alzheimer Genetics Hub (AGH, https://alzheimergenetics.org/) upon submission of a research project proposal through the contact form (https://alzheimergenetics.org/contact/).
CANVAS patients
We used the HiFi data (Sequel II) of two patients diagnosed with CANVAS (Cerebellar ataxia with neuropathy and vestibular areflexia syndrome), caused by a TR expansion in RFC1 gene (van de Pol et al. 2023).
Evaluating otter and TREAT performances
Comparison with existing tools
We compared TREAT/otter to TRGT and LongTR (Dolzhenko et al. 2024; Ziaei Jam et al. 2024). For the comparison, we used the HG002 genome and a set of 161,382 TRs from PacBio's repeat catalog (version 0.3.0, available at https://github.com/PacificBiosciences/trgt/tree/main/repeats). We compared the tools’ genotyped alleles to the expected alleles from the T2T assembly of HG002. As metrics, we used (1) normalized edit distance, (2) raw edit distance, (3) allele size correlation between the observed and expected alleles, and (4) fraction of perfectly genotyped alleles. In addition, we evaluated motif identification accuracy, and computational resources.
TREAT/otter applications
We compared the performances of TREAT assembly and reads analyses by correlating the estimated TR allele sizes with each other (Supplemental Results). Then, we used TRs for a population stratification analysis: using the set of 161K TRs, we selected the top 20% most variable TRs based on the coefficient of variation (ratio of standard deviation to the mean TR joint allele size). Then we applied PCA based on the joint allele sizes. For 40/47 matching samples with SNP data from the 1000 Genomes Project (1000 Genomes Project Consortium 2015), we also performed PCA based on 30,544 randomly sampled common (minor allele frequency >10%) SNPs.
To evaluate clinical applicability, we applied the TREAT/otter outlier analysis module on the combined data set of 47 HPRC genomes plus the two CANVAS patients and the 10 parent–child duos. For this analysis, we focused on 35 clinically relevant TRs (Supplemental Table S1), that were previously associated with neurological diseases (McMurray 2010; On Behalf of the BELNEU Consortium et al. 2018; Khristich and Mirkin 2020). Finally, TREAT/otter case-control analysis module was used to replicate the association of four TRs that were previously associated with AD (On Behalf of the BELNEU Consortium et al. 2018; Guo et al. 2023; Wang et al. 2023). The commands used for the outlier and case-control analyses are available in Supplemental Methods.
Systematic analysis of allele dropouts in tandem repeats
Curated set of TRs in CHM13
We downloaded and curated repeat annotations for the CHM13 reference genome (version 2.0, https://github.com/marbl/CHM13) (Supplemental Methods). This curated data set counted 864,424 TRs genome-wide. We extracted the corresponding parental and maternal allele-sequences in HG002 for these TRs by aligning the HG002 T2T assembly (version 0.7) to CHM13 (Jarvis et al. 2022).
TRs unique to CHM13
We first genotyped the 864K TRs using otter in HG002 from different technologies (Sequel II, Revio, Simplex and Duplex), and at different coverage levels (5×, 10×, 15×, 20×, 25×, and 30×), and calculated the normalized edit distance between observed and expected TR alleles (Supplemental Results). We then focused on a set of TRs present in CHM13 and absent in GRCh38, and used TREAT/otter to characterize the repeat content of these TRs in 47 genomes from HPRC.
Evaluation of coverage drops in TRs
Using HG002 data from Sequel II, Revio, Simplex and Duplex technologies (∼30× coverage each), we calculated the ratio between local TR coverage and average global coverage. TRs where this ratio was <0.25 were regarded as low-coverage TRs. We then investigated sequence characteristics of low-coverage TR, including average size, dinucleotide content, and propensity to form G4s. For the latter, we used pqsfinder (v2.10.1) with “min_score = 20” parameter (Hon et al. 2017).
Consent statement
The Medical Ethics Committee of the Amsterdam UMC and Radboud UMC approved all studies. All participants and/or their legal representatives provided written informed consent for participation in clinical and genetic studies.
Software availability
Otter is written in C++ and the source code is freely available at https://github.com/holstegelab/otter.
TREAT is written in Python and R (for plots). The source code is freely available at https://github.com/holstegelab/treat along with example data sets, documentation, a dedicated Conda configuration file and a Docker image to ease the installation.
The source code of TREAT and otter are also available as Supplemental Code.
Supplemental Material
Acknowledgments
Part of the work in this manuscript was carried out on the Cartesius supercomputer, which is embedded in the Dutch national e-infrastructure with the support of SURF Cooperative. Computing hours were granted to H.H. by the Dutch Research Council (100plus: project# vuh15226, 15318, 17232, and 2020.030; Role of VNTRs in AD; project# 2022.31, Alzheimer's Genetics Hub project# 2022.38). N.T. is appointed at ABOARD and H.H., M.R., S.v.d.L. are recipients of ABOARD, a public-private partnership receiving funding from ZonMW (#73305095007) and Health∼Holland, Topsector Life Sciences and Health (PPP-allowance; #LSHM20106). This work is supported by a VIDI grant from the Dutch Scientific Counsel (#NWO 09150172010083) and a public-private partnership with TU Delft and PacBio, receiving funding from ZonMW and Health∼Holland, Topsector Life Sciences and Health (PPP-allowance), and by Alzheimer Nederland WE.03-2018-07. S.v.d.L. is recipient of ZonMW funding (#733050512). H.H. was supported by the Hans und Ilse Breuer Stiftung (2020), Dioraphte 16020404 (2014), and the HorstingStuit Foundation (2018). Acquisition of the PacBio Sequel II long-read sequencing machine was supported by the ADORE Foundation (2022). We are grateful to all the reviewers involved in the peer-review process for their comments which have largely improved our manuscript.
Author contributions: N.T., A.S., H.H., M.H., and M.R. conceived the presented idea; N.T., A.S., and Y.Z. performed the analyses; L.K., S.W., J.K., A.-F.S., M.P., and K.S. performed the sequencing and wet-lab experiments; E.-J.K. shared data from patients with TR expansions; N.T. and A.S. contributed to the first draft of the manuscript; Y.Z., S.v.d.L., M.H., E.-J.K., K.S., M.R., H.H., N.T., and A.S. contributed to the critical revision of the manuscript and the rebuttal.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279351.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
H.H. has a collaboration contract with Muna Therapeutics, PacBio, Neurimmune, and Alchemab. She serves on the scientific advisory boards of Muna Therapeutics and is an external advisor for Retromer Therapeutics. All other authors declare that they have no competing interests.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. 2020. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 21: 30. 10.1186/s13059-020-1935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baid G, Cook DE, Shafin K, Yun T, Llinares-López F, Berthet Q, Belyaeva A, Töpfer A, Wenger AM, Rowell WJ, et al. 2023. DeepConsensus improves the accuracy of sequences with a gap-aware sequence transformer. Nat Biotechnol 41: 232–238. 10.1038/s41587-022-01435-7 [DOI] [PubMed] [Google Scholar]
- Bakhtiari M, Shleizer-Burko S, Gymrek M, Bansal V, Bafna V. 2018. Targeted genotyping of variable number tandem repeats with adVNTR. Genome Res 28: 1709–1719. 10.1101/gr.235119.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield JK, Marshall J, Danecek P, Li H, Ohan V, Whitwham A, Keane T, Davies RM. 2021. HTSlib: C library for reading/writing high-throughput sequencing data. Gigascience 10: giab007. 10.1093/gigascience/giab007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R, Rajan-Babu I-S, Friedman JM, Birol I. 2021. Straglr: discovering and genotyping tandem repeat expansions using whole genome long-read sequences. Genome Biol 22: 224. 10.1186/s13059-021-02447-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roeck A, Van Broeckhoven C, Sleegers K. 2019. The role of ABCA7 in Alzheimer's disease: evidence from genomics, transcriptomics and methylomics. Acta Neuropathol 138: 201–220. 10.1007/s00401-019-01994-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, Deshpande V, Schlesinger F, Krusche P, Petrovski R, Chen S, Emig-Agius D, Gross A, Narzisi G, Bowman B, et al. 2019. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics 35: 4754–4756. 10.1093/bioinformatics/btz431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, English A, Dashnow H, De Sena Brandine G, Mokveld T, Rowell WJ, Karniski C, Kronenberg Z, Danzi MC, Cheung WA, et al. 2024. Characterization and visualization of tandem repeats at genome scale. Nat Biotechnol. 10.1038/s41587-023-02057-3 [DOI] [PubMed] [Google Scholar]
- Dumbovic G, Forcales S-V, Perucho M. 2017. Emerging roles of macrosatellite repeats in genome organization and disease development. Epigenetics 12: 515–526. 10.1080/15592294.2017.1318235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami Rasekh M, Hernández Y, Drinan SD, Fuxman Bass JI, Benson G. 2021. Genome-wide characterization of human minisatellite VNTRs: population-specific alleles and gene expression differences. Nucleic Acids Res 49: 4308–4324. 10.1093/nar/gkab224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand Y, Hernandez Y, Loving J, Benson G. 2014. VNTRseek—a computational tool to detect tandem repeat variants in high-throughput sequencing data. Nucleic Acids Res 42: 8884–8894. 10.1093/nar/gku642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q, Keniry A. 2019. Latest techniques to study DNA methylation. Essays Biochem 63: 639–648. 10.1042/EBC20190027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MH, Lee W-P, Vardarajan B, Schellenberg GD, Phillips-Cremins J. 2023. Polygenic burden of short tandem repeat expansions promote risk for Alzheimer's disease. medRxiv 10.1101/2023.11.16.23298623 [DOI] [Google Scholar]
- Gustafson JA, Gibson SB, Damaraju N, Zalusky MP, Hoekzema K, Twesigomwe D, Yang L, Snead AA, Richmond PA, De Coster W, et al. 2024. High-coverage nanopore sequencing of samples from the 1000 Genomes Project to build a comprehensive catalog of human genetic variation. Genome Res (this issue) 34: 2061–2073. 10.1101/gr.279273.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Golan D, Rosset S, Erlich Y. 2012. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res 22: 1154–1162. 10.1101/gr.135780.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ. 2018. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19: 286–298. 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- Holstege H, Beker N, Dijkstra T, Pieterse K, Wemmenhove E, Schouten K, Thiessens L, Horsten D, Rechtuijt S, Sikkes S, et al. 2018. The 100-plus Study of cognitively healthy centenarians: rationale, design and cohort description. Eur J Epidemiol 33: 1229–1249. 10.1007/s10654-018-0451-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon J, Martínek T, Zendulka J, Lexa M. 2017. Pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 33: 3373–3379. 10.1093/bioinformatics/btx413 [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Formenti G, Rhie A, Guarracino A, Yang C, Wood J, Tracey A, Thibaud-Nissen F, Vollger MR, Porubsky D, et al. 2022. Semi-automated assembly of high-quality diploid human reference genomes. Nature 611: 519–531. 10.1038/s41586-022-05325-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristich AN, Mirkin SM. 2020. On the wrong DNA track: molecular mechanisms of repeat-mediated genome instability. J Biol Chem 295: 4134–4170. 10.1074/jbc.REV119.007678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristmundsdóttir S, Sigurpálsdóttir BD, Kehr B, Halldórsson BV. 2017. popSTR: population-scale detection of STR variants. Bioinformatics 33: 4041–4048. 10.1093/bioinformatics/btw568 [DOI] [PubMed] [Google Scholar]
- Lago S, Nadai M, Cernilogar FM, Kazerani M, Domíniguez Moreno H, Schotta G, Richter SN. 2021. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat Commun 12: 3885. 10.1038/s41467-021-24198-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linthorst J, Meert W, Hestand MS, Korlach J, Vermeesch JR, Reinders MJT, Holstege H. 2020. Extreme enrichment of VNTR-associated polymorphicity in human subtelomeres: genes with most VNTRs are predominantly expressed in the brain. Transl Psychiatry 10: 369. 10.1038/s41398-020-01060-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Rosikiewicz W, Pan Z, Jillette N, Wang P, Taghbalout A, Foox J, Mason C, Carroll M, Cheng A, et al. 2021. DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation. Genome Biol 22: 295. 10.1186/s13059-021-02510-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, et al. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci 105: 9272–9277. 10.1073/pnas.0803466105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Sola S, Eizenga JM, Guarracino A, Paten B, Garrison E, Moreto M. 2023. Optimal gap-affine alignment in O(s) space. Bioinformatics 39: btad074. 10.1093/bioinformatics/btad074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani B, Kawahara R, Morishita S. 2023. Decomposing mosaic tandem repeats accurately from long reads. Bioinformatics 39: btad185. 10.1093/bioinformatics/btad185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. 2010. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 11: 786–799. 10.1038/nrg2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. 2007. Expandable DNA repeats and human disease. Nature 447: 932–940. 10.1038/nature05977 [DOI] [PubMed] [Google Scholar]
- On Behalf of the BELNEU Consortium, De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van den Bossche T, Cras P, Vandenberghe R, De Deyn PP, et al. 2018. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease. Acta Neuropathol 135: 827–837. 10.1007/s00401-018-1841-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Edamura KN, Cleary JD. 2005. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6: 729–742. 10.1038/nrg1689 [DOI] [PubMed] [Google Scholar]
- Ren J, Gu B, Chaisson MJP. 2023. vamos: variable-number tandem repeats annotation using efficient motif sets. Genome Biol 24: 175. 10.1186/s13059-023-03010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar A, Tesi N, Knoop L, Pijnenburg Y, Van Der Lee S, Wijesekera S, Krizova J, Hiltunen M, Damme M, Petrucelli L, et al. 2023. An AluYb8 retrotransposon characterises a risk haplotype of TMEM106B associated in neurodegeneration. medRxiv 10.1101/2023.07.16.23292721 [DOI] [Google Scholar]
- Sereika M, Kirkegaard RH, Karst SM, Michaelsen TY, Sørensen EA, Wollenberg RD, Albertsen M. 2022. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat Methods 19: 823–826. 10.1038/s41592-022-01539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovski I, Chintalaphani SR, Gamaarachchi H, Ferguson JM, Pineda SS, Scriba CK, Tchan M, Fung V, Ng K, Cortese A, et al. 2022. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci Adv 8: eabm5386. 10.1126/sciadv.abm5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L. 2003. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol 4: R13. 10.1186/gb-2003-4-2-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesi N, Van Der Lee S, Hulsman M, Van Schoor NM, Huisman M, Pijnenburg Y, Van Der Flier WM, Reinders M, Holstege H. 2024. Cognitively healthy centenarians are genetically protected against Alzheimer's disease. Alzheimers Dement 20: 3864–3875. 10.1002/alz.13810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pol M, O'Gorman L, Corominas-Galbany J, Cliteur M, Derks R, Verbeek NE, van de Warrenburg B, Kamsteeg E-J. 2023. Detection of the ACAGG repeat motif in RFC1 in two Dutch Ataxia families. Mov Disord 38: 1555–1556. 10.1002/mds.29441 [DOI] [PubMed] [Google Scholar]
- van der Flier WM, Scheltens P. 2018. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis 62: 1091–1111. 10.3233/JAD-170850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Antonacci-Fulton L, Howe K, Lawson HA, Lucas JK, Phillippy AM, Popejoy AB, Asri M, Carson C, Chaisson MJP, et al. 2022. The Human Pangenome Project: a global resource to map genomic diversity. Nature 604: 437–446. 10.1038/s41586-022-04601-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dombroski BA, Cheng P-L, Tucci A, Si Y-Q, Farrell JJ, Tzeng J-Y, Leung YY, Malamon JS, Alzheimer's Disease Sequencing Project, et al. 2023. Structural variation detection and association analysis of whole-genome-sequence data from 16,905 Alzheimer's Diseases Sequencing Project subjects. medRxiv 10.1101/2023.09.13.23295505 [DOI] [Google Scholar]
- Wenger AM, Peluso P, Rowell WJ, Chang P-C, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, et al. 2019. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 37: 1155–1162. 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Ma ZS. 2016. Sparc: a sparsity-based consensus algorithm for long erroneous sequencing reads. PeerJ 4: e2016. 10.7717/peerj.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, et al. 1991. Fragile X genotype characterized by an unstable region of DNA. Science 252: 1179–1181. 10.1126/science.252.5009.1179 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hulsman M, Salazar A, Tesi N, Knoop L, Van Der Lee S, Wijesekera S, Krizova J, Kamsteeg E-J, Holstege H. 2024. MotifScope: a multi-sample motif discovery and visualization tool for tandem repeats. bioRxiv 10.1101/2024.03.06.583591 [DOI] [PubMed] [Google Scholar]
- Ziaei Jam H, Zook JM, Javadzadeh S, Park J, Sehgal A, Gymrek M. 2024. LongTR: genome-wide profiling of genetic variation at tandem repeats from long reads. Genome Biol 25: 176. 10.1186/s13059-024-03319-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.