Abstract
Direct nanopore-based RNA sequencing can be used to detect posttranscriptional base modifications, such as N6-methyladenosine (m6A) methylation, based on the electric current signals produced by the distinct chemical structures of modified bases. A key challenge is the scarcity of adequate training data with known methylation modifications. We present Xron, a hybrid encoder–decoder framework that delivers a direct methylation-distinguishing basecaller by training on synthetic RNA data and immunoprecipitation (IP)-based experimental data in two steps. First, we generate data with more diverse modification combinations through in silico cross-linking. Second, we use this data set to train an end-to-end neural network basecaller followed by fine-tuning on IP-based experimental data with label smoothing. The trained neural network basecaller outperforms existing methylation detection methods on both read-level and site-level prediction scores. Xron is a standalone, end-to-end m6A-distinguishing basecaller capable of detecting methylated bases directly from raw sequencing signals, enabling de novo methylome assembly.
RNA modification plays essential roles in various biological processes, including stem cell differentiation and renewal, brain functions, immunity, aging, and cancer progression (D'Aquila et al. 2017; Sun et al. 2019; Qin et al. 2020; Boulias and Greer 2023). Among the various types of RNA modifications, N6-methyladenosine (m6A) is one of the most abundant versions and is involved in various biological processes including mRNA expression, splicing, nuclear exporting, translation efficiency, RNA stability, and miRNA processing (Boulias and Greer 2023). Accurate detection and quantification of m6A modifications is crucial for understanding their impact on gene regulation and cellular processes (Fu et al. 2014; Murakami and Jaffrey 2022).
High-throughput sequencing from Illumina, also known as sequencing by synthesis (SBS), identifies nucleotides through synthesis, leading to the loss of posttranscriptional information (Buermans and Den Dunnen 2014). Therefore, indirect methods are required to detect RNA modifications with SBS. These approaches first isolate the modified RNA and then conduct reverse transcription and cDNA sequencing to reveal the modifications. Two primary strategies are used to experimentally isolate RNA modifications. One type of approach involves immunoprecipitation (IP). Examples of methods using this approach include MeRIP-seq (Meyer et al. 2012), m6A-seq (Dominissini et al. 2012), PA-m6A-seq (Chen et al. 2015), m6A-CLIP/IP (Ke et al. 2015), miCLIP (Linder et al. 2015), m6A-LAIC-seq (Molinie et al. 2016), m6ACE-seq (Koh et al. 2019), and m6A-seq2 (Dierks et al. 2021). These methods rely on antibodies that target the modified ribonucleotide and enrich the RNA fragments with the target modified bases. The other type of approach is chemical-based detection. Examples of methods using this approach are Pseudo-seq (Carlile et al. 2014), AlkAniline-Seq (Marchand et al. 2018), MAZTER-seq (Garcia-Campos et al. 2019), m6A-REF-seq (Zhang et al. 2019), DART-seq (Meyer 2019), RBS-Seq (Khoddami et al. 2019), and m6A-SAC-seq (Hu et al. 2022). These techniques use chemical compounds or enzymes that selectively interact with the modified ribonucleotide, either cleaving or modifying the RNA reads to halt or disturb the reverse transcription process. This is followed by short-read cDNA sequencing, which identifies the RNA modifications by comparing the read ends of the cDNA or the base mismatches/deletions in cDNA. Although these methods were able to generate detailed maps of RNA modification sites, they all use external compounds which makes it hard to obtain the required single-base resolution. They also face other challenges and shortcomings including the limited availability of antibodies or compounds for specific modifications (Ryvkin et al. 2013), nonspecific antibody binding (Helm et al. 2019; McIntyre et al. 2020; Zhang et al. 2021), low single-nucleotide resolutions (Dominissini et al. 2012; Meyer et al. 2012), and, importantly, an inability to identify the exact location of a modification.
Direct RNA sequencing using nanopores offers a promising alternative (Garalde et al. 2018). An RNA molecule can be sequenced by measuring the intensity of the current flowing through the pore as the RNA molecules pass through it. Modified RNA nucleotides produce different signals than their unmodified counterparts, providing information about the modifications at the single-molecule read resolution (Jenjaroenpun et al. 2021; Leger et al. 2021). However, to detect specific modifications from subtle signal changes we need an optimized algorithm, which is normally obtained through supervised learning or a comparative approach (Wan et al. 2022). Unfortunately, current data are not immediately suitable for supervised learning due to the lack of experimental techniques for identifying the methylation state at the single-read resolution.
In vitro transcription (IVT) data, which are transcribed from either experimentally synthesized DNA sequences or native DNA (Liu et al. 2019; Jenjaroenpun et al. 2021), can provide reads that are either completely methylated or not methylated at all (all-or-none). However, the diversity of the sequence compositions in synthesized DNA data sets is limited due to constraints concerning the maximum DNA length that can be synthesized and the associated costs. In addition, the IVT data set lacks partially methylated reads with known methylation states. Although partially methylated reads can be generated by introducing a mixture of modified and canonical adenine during IVT, the location of methylation remains unknown because in such mixtures the RNA polymerase randomly selects adenine from either type during the transcription process. Models trained to identify modifications on all-or-none modified reads perform poorly on biological reads, which are usually sparsely methylated, regardless of the training feature used, such as basecalling error or signal difference (Liu et al. 2019; Zhong et al. 2023). Methods using such synthesized data sets include training a classifier to predict sequence segments (5-mers) given their corresponding nanopore raw signal segments (Gao et al. 2021) or features of these segments (Liu et al. 2019; Jenjaroenpun et al. 2021; Leger et al. 2021; Pratanwanich et al. 2021). The signal segments are extracted from the raw signal after performing basecalling and alignment, using models trained on canonical data (data with no methylation). As we show, the performance of such a classifier is limited since it is only trained on isolated short segments, losing contextual information. In addition, these models are trained solely on manually selected features including mean, standard deviation, and duration of isolated signal segments corresponding to five bases, which can lead to the loss of more detailed signal information. Recently, a new method, CHEUI, was trained using longer signal segments, yielding impressive results on IVT data (Acera Mateos et al. 2024). However, it suffers from overfitting when applied to real biological samples (Fig. 2; Hendra et al. 2022).
Figure 2.
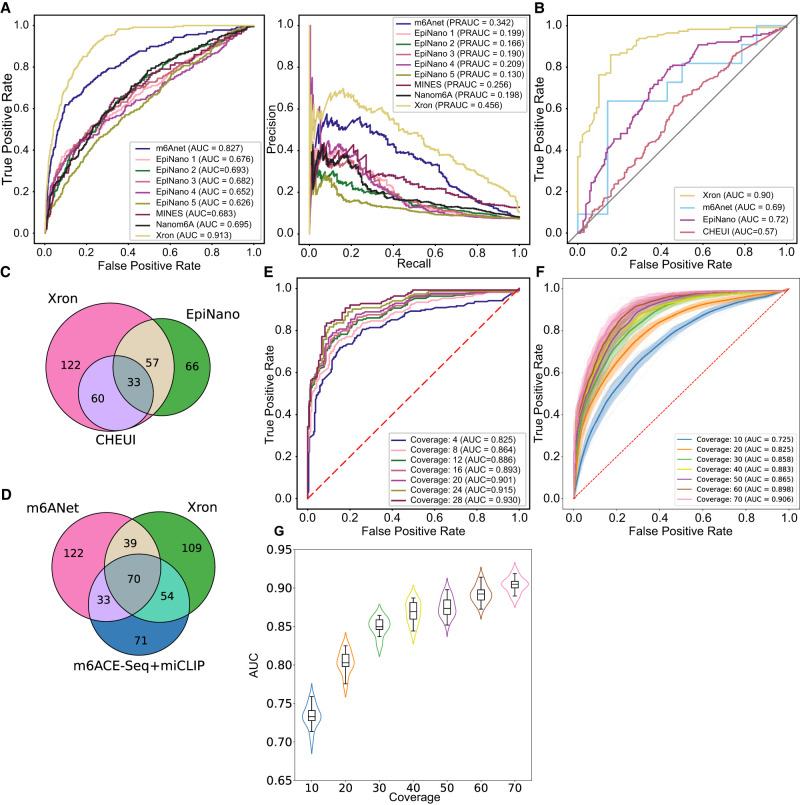
Comparison of Xron models across two different species. (A) ROC and PR curves of m6A prediction on human HEK293T cell line, produced by Xron and other models. (B) ROC curves produced by Xron and other models on yeast data. (C,D) Venn diagram showing the overlapping sites predicted by Xron and other methods on yeast (C) and HEK293T (D) data. (E) ROC curves produced by Xron for detecting m6A methylation in yeast data under different minimum sequence coverage thresholds. (F) ROC curves generated by Xron for detecting m6A methylation in down-sampled yeast data with different coverage. (G) Distribution of AUC score of Xron on down-sampled yeast data.
IP data from assays such as m6ACE-seq and m6A-CLIP-seq relies on the use of antibodies (Schwartz et al. 2013; Ke et al. 2015; Linder et al. 2015). However, this strategy only provides the modification proportion for each reference transcriptomic position, i.e., a site-level modification rather than the modification state for each individual read (read-level). m6Anet (Hendra et al. 2022) employs multiple-instance learning (Amores 2013) to train a classifier using IP data leading to improved site-level accuracy. However, IP data misses many methylation sites, particularly in low-coverage regions (McIntyre et al. 2020). Additionally, due to nonspecific antibody binding, the methylation detection results obtained through IP experiments produced a false-positive rate of ∼11%, which can vary between studies (Ke et al. 2017; Garcia-Campos et al. 2019). m6Anet also requires a minimum coverage level of 20 reads for a site to be detected due to the way the model is trained. The training involves maximizing the probability of detecting at least one methylated read among the reads covering a known methylated site. Such coverage depth is not always available. Finally, as in the other existing models, m6Anet relies on a basecaller and segmentation tools that are trained on nonmodified reads (canonical reads).
In summary, previous approaches try to identify m6A sites using basecalling errors (Liu et al. 2019; Jenjaroenpun et al. 2021; Leger et al. 2021; Pratanwanich et al. 2021), by comparing between control samples (Leger et al. 2021; Abebe et al. 2022), trained on IVT data (Gao et al. 2021; Acera Mateos et al. 2024), or trained on noisy labels from IP data (Hendra et al. 2022). As we will show, the fact that they are only trained on one type of data limits their performance. This work aims to address these limitations by introducing a framework that integrates multiple data types to improve the identification of m6A sites in nanopore direct RNA sequencing.
Results
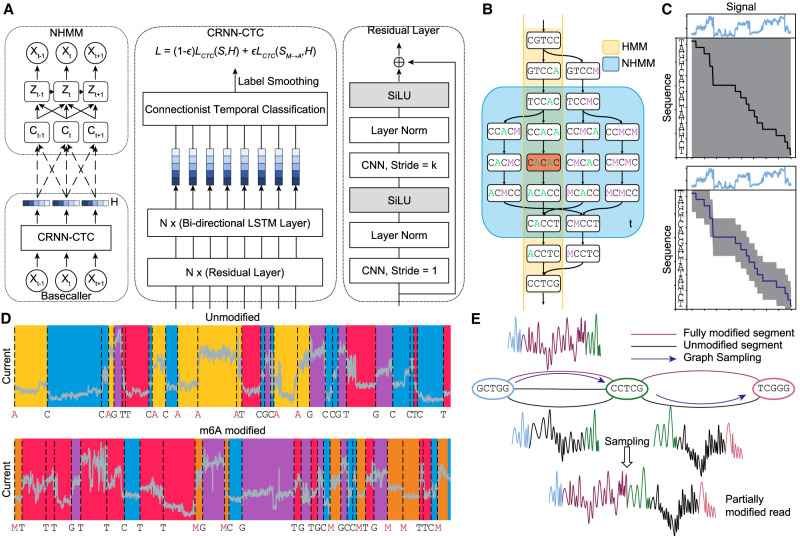
We present a method that takes a different approach by detecting methylation during the basecalling phase. We predict methylated bases directly from the current signal by training a methylation-distinguishing basecaller. To achieve this, we developed Xron, a hybrid encoder–decoder framework (Fig. 1). The encoder is a convolutional recurrent neural network (CRNN) encoding the observable signal into a k-mer representation. After it has been trained and fine-tuned, the CRNN serves as a methylation-distinguishing basecaller for new data. The decoder is a nonhomogeneous hidden Markov model (NHMM), which serves as a generative model for achieving signal segmentation and alignment when preparing the training data set. Applying the NHMM, we created a partially methylated data set to train the CRNN and produce a methylation-distinguishing basecaller. The CRNN is then fine-tuned using IP data, further enhancing the basecaller's generalizability (Supplemental Fig. S2). This framework enables us to obtain a highly accurate methylation-distinguishing basecaller by exploiting both IVT data and IP data, rather than using just one type of data (Supplemental Table S1). This approach outperforms all previous methods on synthesized and biological samples and provides a comprehensive, end-to-end solution for methylation base detection (Table 1; Fig. 2A,B; Supplemental Fig. S4).
Figure 1.
Schematics of Xron model and the data augmentation process through cross-linking and sampling. (A) Xron consists of two parts: a NHMM and a CRNN with a connectionist temporal classification (CTC) decoder. (B) Comparison between HMM and NHMM. The transition matrix of a HMM (yellow) encodes the whole Markov chain of k-mers, while the transition matrix of the NHMM (blue) at time t only encodes the Markov chain of the five nearby k-mers given the predicted k-mer (shown in red) at time t. The Markov chain is also expanded to include the k-mers with all combinations of the A and M (m6A) bases. We create partially methylated reads using data augmentation, first segmenting the signal and then cross-linking the reads and their corresponding signal in silico. To achieve this, we design a novel NHMM that can be trained to conduct signal segmentation in a semisupervised fashion on modified reads, even when lacking methylation labels. The NHMM is trained using the forward–backward algorithm with its transition matrix conditioned on a canonical basecalled sequence and its alignment, thus giving the maximum likelihood estimation of the model parameters regarding the methylation base. The Viterbi path of the NHMM gives the alignment between the current signal and sequence. Following the signal segmentation process performed with the NHMM, the NHMM was used to create a training data set with partially methylated reads and their true labels for methylation detection training by augmenting all-or-none modified reads. (C) The transition process of the NHMM is constrained by the neural network's output, leading to a smaller probability space and making it easier for the model to find the optimal alignment. (D) The NHMM is trained in a semisupervised manner on IVT data sets, including fully modified, unmodified, and partially modified reads. It provides accurate signal segmentation results for both unmodified and modified sequences. (E) In silico read cross-linking. The fully modified or unmodified reads are first broken into segments at the invariant k-mers to form a signal-k-mer graph, whose nodes are k-mers and whose edges are signal segments. Then, a partially methylated read is sampled from the k-mer signal graph.
Table 1.
Reported performance of m6A modification identification achieved by existing works
| Method | AUC-ROC | |||
|---|---|---|---|---|
| Read-levela | Site-levela | Yeast KOb | Humanc | |
| EpiNano (2019) (Liu et al. 2019) | – | 0.90 | 0.680 | – |
| ELIGOS (2021) (Jenjaroenpun et al. 2021) | – | 0.756 | 0.287 (F1) | – |
| Nanocompore (2021) (Leger et al. 2021) | – | – | 0.18 (F1) | – |
| Nanom6A (2021) (Gao et al. 2021) | – | 0.97 | 0.71 | – |
| CHEUI (2022) (Acera Mateos et al. 2024) | 0.806 | 0.92 | – | – |
| m6Anet (2022) (Hendra et al. 2022) | 0.90 | 0.94 | – | 0.83 |
| Xron (this work) | 0.93 | >0.99 | 0.90 | 0.91 |
Bold values: P < 0.001 (***). aThese results were reported on the IVT data set (Liu et al. 2019), in which single-read m6A modifications were known.
bYeast ime4Δ knockout data set from Liu et al. (2019).
cHuman HEK293T cell data set from Chen et al. 2021.
Applying Xron to identify m6A methylation on direct RNA sequencing data sets
Xron performs methylation-distinguishing basecalling, outputting methylated bases directly from the raw sequencing signal emitted from the nanopore. Its neural network basecaller is trained on an augmented partially methylated data set and then fine-tuned using IP data. We tested Xron on three public direct RNA sequencing data sets: an IVT data set (Liu et al. 2019), a yeast data set (Liu et al. 2019), and a human embryonic kidney cells (HEK293T) data set (Hendra et al. 2022).
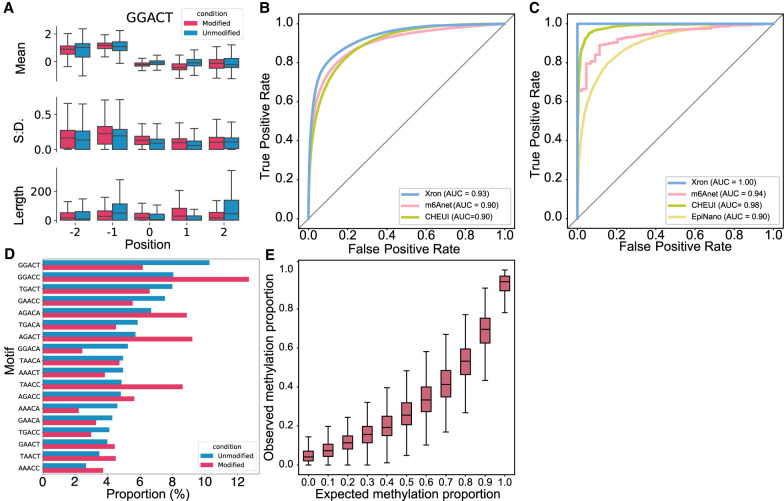
The IVT data set (Liu et al. 2019) was synthesized from artificially designed sequences followed by IVT. The data set contains either fully methylated or fully unmethylated reads. Signal intensity shows differences around the center base of the k-mer between modified and unmodified sites (Fig. 3A; Supplemental Fig. S1). The sequences are designed to contain all 5-mers, including the most common k-mer (GGACT) and all 18 DRACH motifs (Fig. 3A,B).
Figure 3.
Evaluation of the m6A detection results obtained for synthesized IVT RNA reads and stoichiometry prediction. (A) Box plot comparing the distribution of the mean, standard deviation, and length for the signal segmented by NHMM with 5232 modified sites and 18,464 unmodified sites for the GGACT motif. Horizontal lines show the median, the box denotes the interquartile range, and the whiskers extend to 1.5 times the interquartile range. Points beyond this range are considered outliers and are removed from the plot. (B,C) ROC curves of Xron against m6Anet and CHEUI for read-level (B) and site-level (C) m6A modification predictions. (D) Bar plot showing the relative proportion of DRACH 5-mer motif for 84,919 modified and 179,717 unmodified positions. (E) Box plot showing the m6A ratio predicted by Xron with different proportions of IVT control and IVT m6A RNA mixing.
The yeast data set (Liu et al. 2019) contains direct RNA sequencing reads from two strains, a wild-type strain, and a “ime4Δ” knockout strain, in which IME4 was deleted. The deletion of IME4 results in the complete elimination of m6A bases, making it a negative control. The yeast data set contains three independent biological replicates for each strain. Two were used in this study; the first replicate was used for training, and the second was used for evaluation.
The human HEK293T cell data set (Hendra et al. 2022) contains direct RNA-seq data from the HEK293T cell line (Pratanwanich et al. 2021), with methylation sites identified by m6ACE-seq (Koh et al. 2019) and miCLIP data (Linder et al. 2015) on the same cell line. The data set contains three replicates, and we used the first replicate to evaluate the method. (See Methods for details about replicates and data sets used for training and evaluation.)
The Arabidopsis data set (Parker et al. 2020) contains direct RNA sequencing reads from wild-type Arabidopsis (Col-0), mutants (vir-1) defective in m6A writer, and VIR-complemented lines. We used the three replicates of the wild-type line to evaluate the method.
Xron accurately identifies m6A sites
To evaluate the performance of Xron, we applied Xron that is fine-tuned on yeast data to direct RNA sequencing data derived from the human HEK293T cell line (Pratanwanich et al. 2021). Although Xron is pretrained using human IVT reads (see Methods), no human methylation information is used during training since all human reads are canonical. To validate the model, we used the m6A sites detected by m6ACE-seq and miCLIP from the human HEK293T cell line as the true labels during evaluation, following previous work (Hendra et al. 2022). We used the m6A sites identified by m6ACE-seq and miCLIP as positive samples and the other sites with the same 5-mer as negative samples. Xron achieved the best area under the receiver operating characteristic curve (AUC-ROC) of 0.91 (Fig. 2A; Supplemental Fig. S5A) compared with those of EpiNano (0.69) and m6Anet (0.83) and the best precision–recall (PR) AUC of 0.456 (Fig. 2A; Supplemental Fig. S5B) compared to m6Anet (0.342) and MINES (0.256; Lorenz et al. 2020).
Xron is sensitive to IME4 knockouts
In addition, we also evaluated Xron on a yeast data set using a ime4Δ knockout Saccharomyces cerevisiae strain where the m6A modification was completely eliminated (Schwartz et al. 2013) as the control data set, following a previous study (Liu et al. 2019). We used the second replicate sample of the data set for evaluation, as we had fine-tuned Xron on a subset of the first replicate. We treated the m6A sites in the wild-type strain as modified sites and the same sites in the ime4Δ knockout strain as unmodified sites. We compared Xron with other models for predicting modified/unmodified sites. Xron achieved an AUC-ROC score of 0.90 (Fig. 2B) on this task, providing a 21% increase over the second-best model, EpiNano (0.72). To fairly compare with other models that may not have been exposed to the yeast data set, we evaluated the performance of an Xron model fine-tuned on the human HEK293T cell line on yeast data and obtained similar accuracy (Supplemental Fig. S3A).
Xron detects more methylation sites and achieves high accuracy under low-coverage settings
As m6Anet intrinsically requires a minimum coverage of at least 20 to obtain site methylation predictions. This results in a much smaller sample size (11 sites detected). In the same setting, Xron yields 171 sites with a minimum coverage of 20 on the yeast data set, which results in higher AUC-ROC accuracy than m6Anet (0.90 vs. 0.69). In total, Xron detects 272 sites reported in the IP data, compared to the 156 sites detected by EpiNano and the 93 sites detected by CHEUI (Fig. 2C). Sites detected by Xron also show higher support from the IP technique (124) compared to m6Anet (107) in the HEK293T cell line (Fig. 2D). While different methods identify various m6A methylation sites, many sites are detected exclusively by one method. This observation aligns with previous reports (Koh et al. 2019; Hendra et al. 2022).
We next tested if including more low-coverage sites by setting different minimum sequencing coverage thresholds would influence the prediction accuracy of Xron (Fig. 2E). We found that increasing the read coverage yielded superior site-level methylation prediction accuracy, increasing from a 0.825 AUC-ROC score for a minimum read coverage level of 4 to a 0.930 AUC-ROC score with a minimum read coverage level of 28. This suggests that with higher sequencing depth, Xron can further enhance the precision and accuracy of methylation detection. Meanwhile, Xron outperforms other models by a large margin even when setting the minimum read coverage level to 4, with AUC 14% more than the second-best model, EpiNano (0.825 vs. 0.72). Furthermore, to evaluate Xron's performance in low-coverage regions, we down-sampled the reads to limit the maximum coverage at each site to a range of 10–70. Xron achieved an accuracy of 0.725 with maximum coverage of 10, outperforming other models with full data (Fig. 2F,G).
With the ability of Xron to detect methylation in low-coverage regions or even at the single-read-level, we were able to check the read-level statistics of methylated k-mers. A comparison of the read-wise and site-wise relative frequency of methylated k-mers in yeast, human, and Arabidopsis shows differences in k-mer profiles across species. Site-wise counting treats multiple reads at one site as a single occurrence, while read-wise counts k-mer occurrence for each read and each site separately (Supplemental Fig. S7A–E). For yeast, the most frequently used motifs AGACA, GGACA, AGACT, and GGACT from the read-wise counting are also the most widely used motifs from the site-wise counting. But in human cell lines and Arabidopsis, read-wise counting indicates the most frequently used motif is different than the previously reported site-wise most “frequently” used motif, which is indicated by the site-wise counting. Motif GAACA in human cell lines has the highest (>17%) relative frequency in the read-wise count, exceeding the previously reported most methylated motif GGACT (∼12%), but it only possesses <8% relative frequency in the site-wise count while GGACT has >12% relative frequency. Motif TAACT in Arabidopsis has the highest (≈15%) relative frequency in the read-wise count, but drops to <10% in the site-wise count. The variation in k-mer profiles across different species offers an ideal scenario for assessing the generalizability of Xron. When comparing the Xron model fine-tuned on yeast and human data sets with different k-mer profiles, we found they give similar accuracy on yeast, human, and Arabidopsis data sets (Fig. 2A,B; Supplemental Fig. S3A–C).
Xron achieves nearly optimal site-level prediction on a synthesized RNA data set
We evaluated Xron on a synthesized RNA IVT data set (Liu et al. 2019) obtained from a different replicate than the training data set (see Methods). In this data set, the true methylation modifications were known for each position in each read, as the reads were either from a fully modified or a fully unmodified run. Our model achieved an AUC-ROC of 0.93 on the single-read-level prediction task (Fig. 3C), in which the model has to predict m6A bases or A bases for each read at DRACH sites identified by previous antibody IP experiments (Schwartz et al. 2013). Our model outperforms the second-best read-level model (m6Anet) by 3% (0.93 vs. 0.90) and achieves an almost optimal AUC-ROC of >0.99 for site-level prediction (Fig. 3D), outperforming the second-best site-level model (CHEUI) by nearly 2% (≈1 vs. 0.98).
Xron provides m6A stoichiometry
By aligning the reads to the reference genome and piling up the single-read m6A modification predictions for different sites, Xron can predict site-level m6A modification stoichiometry, i.e., the fraction of modified bases at a site. We evaluated this ability using a synthetic data set.
The data set was a mixture created by randomly sampling reads from fully modified or unmodified IVT data sets (Liu et al. 2019) with specific mixture proportions, which included 0%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, and 100%. We calculated the model-predicted m6A proportion as the number of m6A bases called per site divided by the total number of reads aligned to this site. The median relative modification proportion followed the same trend as the expected methylation proportion. The trend in stoichiometry level was successfully recovered (Fig. 3E).
Xron achieved high accuracy on SQK-RNA004 data
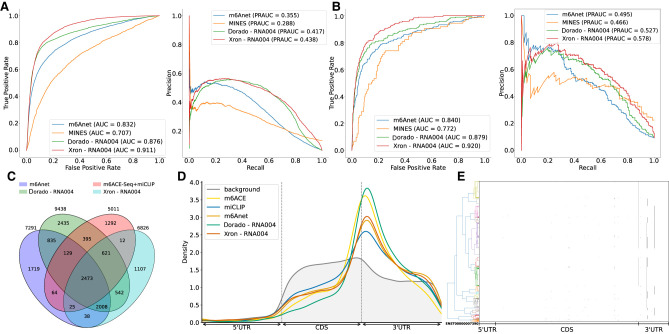
We trained an Xron model on a HEK293T cell line data set from the SG-NEx project, generated using the SQK-RNA004 direct RNA sequencing chemistry, a recently released sequencing kit that offers a higher sequencing rate and presumably better accuracy. Xron achieved an AUC of 0.91 and a PR-AUC of 0.438 for all sites (Fig. 4A), and an AUC of 0.92 and a PR-AUC of 0.578 for dense sites (Fig. 4B), surpassing the Oxford Nanopore Technologies m6A basecaller Dorado and other methods tested on the SQK-RNA002 data set in the same HEK293T cell line. A larger number of detected sites were mutually agreed upon by Xron and Dorado and were also supported by IP methods compared to the SQK-RNA002 data set on the same cell line, where most of the sites are detected by only one method (Figs. 4C, 2C,D). Modified sites detected from SQK-RNA004 data are enriched in the 3′ end of the coding sequence along the transcript coordinates, as expected for m6A (Fig. 4D).
Figure 4.
m6A detection on SQK-RNA004 data set. (A) ROC and PR curve of Xron on SQK-RNA004 data against Dorado. Results of m6Anet and MINES from SQK-RNA002 data on the same HEK293T cell line are also plotted for comparison. (B) Comparison of ROC and PR curves for Xron and Dorado on 2070 dense sites where neighboring modification sites exist within five bases. (C) Venn diagram showing the overlapping sites predicted by Xron and other methods on the HEK293T cell line. (D) Coordinate distribution of the m6A methylated sites predicted by five methods against the background distribution of all DRACH sites. Only sites with at least 20 coverage were chosen. (E) Clustering plot showing the modification of the TSR3 (ENSG00000007520) mRNA transcript over 780 reads. A modification is called if the predicted probability is >0.9 and is marked with a green dot.
Clustering analysis shows asynchronous modification
Xron enables direct access to read-level modification information, allowing us to examine the modification states across multiple sites within each read. Genes that have at least two m6A modification sites and with at least 500 coverage reads were selected. We found asynchronous modification states around the end of the coding sequence (CDS) and in the 3′-UTR region among these reads (Fig. 4E; Supplemental Fig. S8), where m6A methylation does not occur synchronously but in a combinatory pattern. For instance, in the TSR3 gene transcript (ENST00000007390.2) at positions 1041, 1096, 1105, and 1151, all 16 possible combinations of modification status at these four sites were observed with varying frequencies. This pattern suggests a complex regulatory mechanism based on m6A methylation.
Xron performs consistent basecalling on m6A-modified data sets
To compare the performance of Xron as a basecaller with a canonical basecaller, we evaluated the basecalling accuracy of Xron and compared it with that of the Guppy ONT basecaller (Table 2; Supplemental Table S2). We evaluated the basecall quality achieved on three data sets: the synthesized IVT RNA data set, the S. cerevisiae yeast data set, and the human HEK293T cell line data set, considering both modified and unmodified reads. When comparing the identity rate, only reads with potential modified sites are taken into account. For the synthesized IVT RNA and yeast data sets, we used the second replicate, which was not used as training data. Xron suffers less (or no) accuracy drop on data sets with m6A modifications. It exhibited no performance loss on data sets with methylation compared to the control data set. On the other hand, Guppy showed performance decreases on all three data sets with methylation compared to its performance on the unmodified control data sets, including a 14.47% drop in the identity rate on the synthesized reads and a 7.55% drop in the identity rate on the HEK293T reads. Guppy also shows a larger context bias for k-mers from DRACH motifs, comparing to Xron on the HEK293T reads (Supplemental Fig. S6), explaining the identity rate drop on basecalling m6A-modified reads.
Table 2.
Accuracy comparison between Xron and Guppy on three different data sets and their control data sets
| Condition | Model | Identity rate (%)(↑) | Identity rate change (%) |
|---|---|---|---|
| IVT Control | Xron | 87.35 | – |
| Guppy | 92.75 | – | |
| IVT m6A | Xron | 88.48 | 1.13 |
| Guppy | 78.28 | −14.47 | |
| Yeast ime4Δ KO | Xron | 83.42 | – |
| Guppy | 92.50 | – | |
| Yeast | Xron | 83.96 | 0.54 |
| Guppy | 91.94 | −0.56 | |
| HEK293T METTL3 KO | Xron | 85.91 | – |
| Guppy | 93.19 | – | |
| HEK293T | Xron | 87.12 | 1.21 |
| Guppy | 85.64 | −7.55 |
The identity rate (%) was defined as the number of matched bases in the query sequence divided by the number of bases in the reference sequence (the higher the better). All reported rates are mean values among the aligned reads.
Discussion
Several computational methods (Liu et al. 2019; Gao et al. 2021; Jenjaroenpun et al. 2021; Leger et al. 2021; Acera Mateos et al. 2024) have been used to detect m6A methylation. These methods require accurate training data, usually obtained using synthesized RNA reads containing the modification of interest, obtained through experimental methods such as m6ACE-seq or miCLIP, or from a comparative analysis against control data. However, these methods exhibit a performance drop when they are applied to other data sets, implying the existence of overfitting. In addition, these methods usually can only provide site-level methylation, losing read-level resolution. We developed an end-to-end m6A modification detection system for nanopore direct RNA sequencing and were among the first to create an m6A-distinguishing base caller. Our system, Xron, includes an NHMM model for k-mer decoding and a neural network basecaller. By employing data augmentation and semisupervised learning, we constructed an NHMM that is capable of performing accurate signal sequence alignment and introduced a novel training data set for m6A methylation detection. The training pipeline established in our work facilitates supervised basecaller training without necessitating complex feature engineering and using both IVT and IP data available to overcome overfitting.
Quantifying the transcriptome-wide modification rates is one of the key challenges in methylation detection. From the read-level methylation states given by Xron, the modification stoichiometry for each site can be obtained. Meanwhile, our method does not require a high minimum coverage depth, which is essential for detecting methylation in low-expression regions. Comparative methods detect methylation by analyzing data from different conditions (Leger et al. 2021; Pratanwanich et al. 2021). While Xron does not require a control sample to detect methylation, it can facilitate the use of a control sample by comparing the same site across samples. In addition, compared to other methods where the model performance is influenced by aspects such as basecalling algorithms, accuracy in the alignment of the reference sequence to signal, and segmentation of the raw signal, Xron reads out methylation information directly from the raw signal. More training data on different experimental protocols and different organisms will likely further improve the accuracy of Xron and other supervised approaches, while the training framework of Xron can easily adopt these additional training data into the fine-tuning pipeline.
As a basecaller, Xron achieves a consistent identity rate among methylation and unmethylation data sets. Although there is a performance gap in terms of identity rate between Xron and the basecaller Guppy, this is likely due to the different neural network architecture used. In future research, it would be beneficial to investigate various neural network structures since previous studies have shown that alterations to the CRNN architecture can yield enhanced basecalling accuracy. For example, Guppy uses QuartzNet (Kriman et al. 2020), a neural network designed initially for speech recognition. SACall (Huang et al. 2022) employs an attention mechanism, while RODAN (Neumann et al. 2022) integrated squeeze-and-excitation (Hu et al. 2018) layers into a base convolutional neural network (CNN).
Currently, the NHMM takes only raw signal as its input. This has several advantages, including being easy to train and having a closed-form solution for parameter estimation. However, additional input features can be added to the NHMM, including the encoded representation from the neural network base caller. The strategy used by NHMM can also help provide more accurate signal segmentation in other downstream current-based applications, such as postbasecalled sequence correction (e.g., Nanopolish by Simpson et al. [2017]). We leave this as future work. Xron was used to detect m6A modification; however, our framework is suitable for training a basecaller for detecting any natural posttranscription modification, including DNA methylation such as 5mC and other types of RNA modification. Xron can also be retrained to detect artificial modifications at a single-molecule level, such as detecting modifications introduced in small noncoding RNA (Shi et al. 2022).
Methods
Xron is trained using both IVT and IP data sets to obtain better performance. It was first trained on a surrogated IVT data set and then fine-tuned on IP data. To make efficient fine-tuning and to avoid overfitting to the all-or-none methylated reads in IVT data when training with the long current signal, we create partially methylated reads using data augmentation, first segmenting the signal and then cross-linking the reads and its corresponding signal in silico. To achieve this, we design a novel NHMM that can be trained to conduct signal segmentation in a semisupervised fashion on modified reads, even when lacking methylation labels. The NHMM is trained using the forward–backward algorithm with its transition matrix conditioned on a canonical basecalled sequence and its alignment, thus giving the maximum a posteriori estimation of the model parameters regarding methylation base. The Viterbi path of the NHMM gives the alignment between the current signal and sequence. Following the signal segmentation process with the NHMM, we prepared a partially methylated data set through data augmentation, splicing the fully methylated and unmethylated segments. Training on this augmented data set diminishes the inductive bias of the model on partially methylated reads when training with entirely methylated or nonmethylated reads. We then trained an end-to-end methylation detection basecaller on the augmented data set, and it achieved high-accuracy methylation base detection at a single-read resolution. We further improved the basecaller by applying a fine-tuning procedure on IP data with label smoothing to obtain a more accurate basecalling model. Finally, we benchmarked different m6A detection methods on three data sets, including a synthetic IVT data set, a yeast data set, and a human HEK293T cell line, demonstrating that Xron yields accurate methylation-aware basecalls and generalizes to different species.
NHMM trained using semisupervised learning
We design a hybrid framework to conduct signal segmentation and alignment when methylated bases are present. A homogeneous HMM (we refer to this model as an HMM throughout the remainder of this paper for convenience), as employed in the Nanopolish preprocessing tool (Simpson et al. 2017), faces challenges when applied to sequences with methylation bases. The absence of ground truth for the methylation states in each basecalled sequence prevents supervised HMM training. However, training the HMM unsupervised, using only signal and reference genome, is difficult due to the high noise contained in nanopore sequencing signals, the long lengths of the electrical signals, and the similar signal levels between certain k-mers and their methylated counterparts. Additionally, totally unsupervised training is not necessary as we already have the canonical basecalled sequence with alignment given by the canonical basecaller and the reference genome. Although the signals are error-prone in the methylated region, they still provide a general sketch of the sequence. Thus, instead of performing unsupervised learning with the HMM, we develop a semisupervised training process using an NHMM, where we use the basecalled canonical sequence as a prior when building the transition chain backbone in the NHMM. In contrast with an HMM possessing a homogeneous transition matrix that remains constant over time t, an NHMM possesses a nonhomogeneous transition matrix that depends on the external variables and varies over time t, allowing the use of dynamic control for the transition process. Various NHMMs have been used in meteorology (Hughes et al. 1999) and economics (Meligkotsidou and Dellaportas 2011; Neumann et al. 2022) by constructing transition matrices that depend on time-varying covariates, such as seasonality (Hughes et al. 1999) or economic cycle indicators (Meligkotsidou and Dellaportas 2011). In our case, the base probabilities along time t predicted by an existing canonical basecaller (a base caller trained to predict only canonical bases) are used as the time covariates of the transition matrix. This approach enables the model to concentrate on the section of the Markov chain guided by the predicted base probability (Fig. 1C), rather than dealing with the entire chain as is required in unsupervised learning using HMM, which is more challenging and error-prone.
NHMM for methylated sequence segmentation and alignment
The NHMM represents the input sequence of raw current signals as X = (x1, …, xT) for a given k-mer sequence Z = (z1, …, zT) inside a nanopore over the sequencing duration T. Each signal point xt represents a normalized current value, while zt is a variable indicating the k-mer at time t. The transition matrix of the NHMM is constrained on the basecalled sequence and its alignment given by the canonical basecaller. More specifically, suppose we are given the base probability matrix H = (h1, …, hT) ∈ ℝB×T, where B is the number of bases and is the probability of base b at time t, which is obtained from an existing canonical neural network basecaller (Fig. 1A; Graves et al. 2006; Teng et al. 2018). From the base probability matrix H, we extract the most probable basecalled sequence and its corresponding alignment A(t) which aligns the signal point time t to sequence index τ, giving t → τ. After correcting the basecalled sequence with the reference genome, we construct a reference k-mer sequence C by sliding a window of size k (in our case, k = 5) across the basecalled sequence, moving one base at a time. Each windowed segment forms a k-mer and is added to the sequence . From now on, to simplify the notation, we use ct to denote the corresponding k-mer at time t after transitioning through alignment cA(t). All time offsets of the k-mer sequence reside in the sequence domain, meaning ct−1 refers to cA(t)−1. Finally, we derived the k-mer transition matrix Ψ from k-mer sequence C; for details, see the next section. Then, the likelihood of observing an electrical signal X is given by
| (1) |
Here, Z is the hidden state representing the underlying k-mer sequence, zt is the k-mer at time t, and cA(t) is the corrected k-mer representation at time t acquired from the canonical neural network output H (Fig. 1A). T is the maximum time stamp for a given sequence segment. m is the window size for the k-mers to be considered. P(x|z) is the emission probability of the signal x given the k-mer z, as modeled by a Gaussian distribution.
Constructing a transition matrix from the basecalled sequence and its alignment
We loosely constrain the transition matrix at time t in the nonhomogeneous HMM by using the base prediction output H derived from a canonical basecaller, thereby using the segmentation results provided by the basecaller in an error-tolerant manner (Fig. 1B). By calculating the most probable path from H, we can obtain both the basecalled sequence and the alignment between each base within the most probable path and the sequencing time t. Following this, we correct the basecalled sequence using the reference genome, and we also make appropriate revisions to the alignment to address the deletion or insertion errors in the basecalled sequence. We transform the corrected sequence into a k-mer sequence C = {ct:t = 1, …, T}, incorporating the k bases surrounding each base in the basecalled sequence; then, this k-mer sequence is reformatted into transition matrices Ψ = {ψt:t = 1, …, T} by including at most m transitions, where each ψt is the temporal transition matrix at time t. During the process of constructing the k-mer sequence C from H, the basecalled RNA sequence is corrected by aligning it to a reference genome through the following steps:
For mismatched bases, we replace the bases in the k-mer with the reference bases.
For insertions/deletions in the basecalled sequences that are smaller than five bases, we determine the new signal alignment boundary of the inserted/deleted bases by evenly merging/splitting the signal boundaries of nearby bases; i.e., we redistribute the occupancy of the inserted bases to the nearby bases and allocate occupancy for the deleted bases from the nearby bases.
We skip the sequence segments with insertions and deletions that are larger than five bases for quality control purposes.
The transition matrix Ψ is then constrained by C, masking out the irrelevant transition paths so that only transition paths that are likely to occur at time t are retained. To more clearly see what these temporal transition matrices stand for, let be the transition probability from k-mer i to k-mer j given constraint k-mers ci from a time window with a width of at most m, i.e., from to . At the start and end of sequence, the window size is less than k due to boundary constraints. In comparison with the transition matrix ϕi,j = P(zt = i|zt−1 = j) of a homogeneous HMM, the transition matrix now changes over time t:
| (2) |
where ⊗ is the tensor product operation, denotes elementwise multiplication, ei is a one-hot vector where only the ith element is 1, and ϕi,j is the transition matrix in which ϕi,j = 1 if the transition from k-mer i to k-mer j is valid (otherwise, it is 0). For example, AAACT to AACTA is valid, while AAACT to ACTCC is not, as we only allow 1 base step. is the k-mer transition matrix from the k-mer sequence described above; it is a binary value matrix indicating the k-mer transition i → j at time t, where 1 denotes a possible transition and 0 represents an impossible transition.
We construct the transition matrix from m nearby k-mers instead of only the k-mer at time t from k-mer sequence C because the base probability predicted by the canonical basecaller is not exact due to the CTC loss used (Graves et al. 2006; Teng et al. 2018) and the insertion/deletion errors in the sequence, nor is it totally correct due to the previously unseen modified bases. Thus, we allow the NHMM to explore the alignment space in two ways. First, at each time point, the transition matrix of the NHMM is restricted to the current transition probability and the m nearby transition probabilities, where m is a hyperparameter (Eq. 2). This is done to make sure that the final alignment output by the NHMM is not too far away from the given the alignment from canonical basecalling but still allows for exploration within the m-base window. Second, the transition path of the underlying Markov chain is broadened to encompass all possible modified counterparts for each k-mer along the path (Fig. 1C). As an example, AACGT is extended to include four alternative k-mers with modified bases, AACGT (the original k-mer), AMCGT, MACGT, and MMCGT, leading to expanded paths. After the transition matrix is constructed for all the time points, the NHMM is then trained using the expectation–maximization (EM) algorithm (Baum et al. 1970) until it converges (Supplemental Fig. S2B).
Preparing the training data with data augmentation and read sampling
All-or-none methylated reads exhibit either complete methylation of all adenine (A) bases or none at all, whereas in actual biological samples, methylation typically occurs less frequently and is more sporadically distributed. To prevent the neural network from overfitting to all-or-none methylation reads, we create a training data set containing partially methylated reads with labels. This is accomplished by dividing the signals from the all-or-none modified reads into smaller segments and subsequently splicing them together. The corresponding sequences are recombined according to their alignment with the signal, as provided by the NHMM. Merging the signals generated from distinct k-mers at their junction points can result in substantial discrepancies between the combined signal and the actual signal obtained from a real sequencing run. To avoid such deviations caused by k-mer mismatches, we ensure that the preceding and succeeding k-mers at the joint sections are identical. For instance, we can merge the signal segments with basecalled sequences such as GGMCGTTCXXX and XXXCGTTCTAG to form GGMCGTTCTAG. To achieve this, we define nonmethylatable k-mers as k-mers without adenine (CGTTC in the example). They have the same sequencing signal distributions in both modified and unmodified reads, making them suitable for use as joint anchors. We employ the trained NHMM to decode both the canonical and fully modified reads in the training IVT data set, using the base probability prediction from the canonical basecaller as described before. The alignment between the sequence and signal is established through a Viterbi path, which assigns each signal point to its corresponding k-mer (Fig. 1D). Each read is subsequently divided into segments at nonmethylatable k-mers. These segments are used to construct a k-mer signal graph, where each node represents an invariant k-mer. Each edge corresponds to a signal segment whose aligned sequence begins and ends at the respective k-mers of the connected nodes (Fig. 1E). We then perform a random walk on the graph, choosing the next edge via an -greedy sampling strategy with an upper confidence bound (UCB) (Sutton and Barto 2018), as used in the multiarmed bandit algorithm, to ensure maximum diversity in the sampling sequence (see Algorithm 1 in Supplemental Material).
Data processing
Acquisition and processing of direct RNA sequencing data sets
All data sets used in this study are acquired from Liu et al. (2019), Jenjaroenpun et al. (2021), Workman et al. (2019), Hendra et al. (2022), and Chen et al. (2021). We obtained both replicates (replicate 1 and replicate 2) from the EpiNano-synthesized IVT RNA data set (Liu et al. 2019) and the only single replicate from the ELIGOS-synthesized IVT RNA data set (Jenjaroenpun et al. 2021). Both of these data sets contain fully modified reads and unmodified control reads. We also obtained all the NA12878 IVT RNA reads from the Oxford Nanopore human reference data set repository: https://github.com/nanopore-wgs-consortium/NA12878/blob/master/RNA.md (Workman et al. 2019). For the yeast data set, we obtained all three replicates of the wild strain and ime4-knockout strain (ime4Δ) (Liu et al. 2019). Reads are extracted if mapped to m6A-modified RRACH sites previously identified by antibody IP (Schwartz et al. 2013). For the human HEK293T cell line, we obtained two replicates (replicate 1 and replicate 2) of the wild-type human HEK293T cell (Hendra et al. 2022) to evaluate models. Following a previous study (Hendra et al. 2022), we used the reference transcriptome and its genome annotation provided by SG-NEx project: https://github.com/GoekeLab/sg-nex-data (Chen et al. 2021). We used the same m6A DRACH sites in the m6Anet paper (Hendra et al. 2022), which were originally identified by m6ACE-seq and miCLIP experiments (Linder et al. 2015; Koh et al. 2019). We also obtained the first replicate of the wild-type cell line, generated using the SQK-RNA004 sequencing kit from the SG-NEx data repository v5.0.1 (Chen et al. 2021). Currently, there is only one replicate of this data set available. Therefore, we split the data set randomly by reads for training and evaluation purposes. For the Arabidopsis data set, we obtained three wild-type replicates (Col0-1 to Col0-3) from Parker et al. (2020). We used the TAIR10 reference transcriptome (cDNA) and genome from Ensembl: https://plants.ensembl.org/Arabidopsis_thaliana/Info/Index. All replicates in the data sets are biological replicates, which are independent biological samples sequenced using the same direct RNA nanopore sequencing protocol. As for synthesized IVT reads, RNA replicates were transcribed from synthesized DNA reads with different sequences. See the sections below for details on replicates used for training and evaluating. All SQK-RNA002 samples were generated using the Nanopore R9.4.1 flow cell, except for the human IVT data, which came from the R9.4 flow cell. The only significant difference between the two flow cells is the slightly improved yield in the R9.4.1. SQK-RNA004 samples were generated using the FLO-PRO004RA flow cell (Chen et al. 2021).
The IVT RNA data sets were obtained from the EpiNano project (Liu et al. 2019) from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE124309. The ELIGOS IVT RNA data sets were obtained from the ELIGOS Project (Jenjaroenpun et al. 2021) from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP166020. The yeast data sets (wild and ime4-knockout) were obtained from the EpiNano project (Liu et al. 2019) through the GEO database (GSE126213). The HEK293T cell lines data were obtained from the SG-NEx Project (Chen et al. 2021) through the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) (PRJEB40872). The Arabidopsis data were obtained through ENA (PRJEB32782). The SQK-RNA004 data were an early access data set obtained from the SG-NEx data repository v5.0.1.
Canonical basecalling and mapping
All reads in the training data set were basecalled using the Guppy 5.0.11 ONT basecaller (https://pypi.org/project/ont-pyguppy-client-lib/5.0.11/) and then mapped to the reference genome using minimap2 v2.24 (Li 2018) with the settings “-ax map-ont -uf ‐‐secondary=no ‐‐MD”. The mapped reads were then transferred to the BAM format using SAMtools 1.11.0 (Li et al. 2009). A canonical neural network basecaller with the same structure as the CRNN was then trained using the NA12878 IVT reads, and this basecaller was then used to produce the base probability prediction. This canonical basecaller is used as a starting model when we retrain it on the augmented IVT data and subsequently fine-tune it on the yeast data (Liu et al. 2019).
Training data sets
We randomly selected 300,000 canonical (unmodified) read chunks and 300,000 fully modified read chunks from replicate 1 of each of the two synthesized IVT RNA data sets (Liu et al. 2019; Jenjaroenpun et al. 2021), as well as the first 300,000 canonical read chunks from the Oxford Nanopore Human IVT reference data set (Workman et al. 2019) to construct the k-mer signal graph we described above. Reads were filtered out if the corresponding basecalled sequence was shorter than three bases, if the signal had a dwell time (the putative duration a k-mer remains in the pore) exceeding 2000 signal time points, if the basecalled sequence could not be aligned to the reference genome, or if a single base type comprised more than 60% of the basecalled sequence. This filtering process resulted in 228,983 canonical read chunks and 204,822 methylated read chunks from the first synthesized IVT data set (Liu et al. 2019), 195,161 canonical read chunks and 213,085 methylated read chunks from the second synthesized IVT data set (Jenjaroenpun et al. 2021), and 188,004 canonical read chunks from the Human IVT reference data set (Workman et al. 2019). Methylation sites identified by antibody IP (Schwartz et al. 2013), derived from the first replicate of the wild-type and the first replicate of the ime4Δ from the yeast data set (Liu et al. 2019) were used to create the fine-tuning data set. We regarded all sites from the wild-type strain as methylated and all sites from the ime4Δ strain as unmethylated. However, we considered these classifications noisy labels and used label smoothing during fine-tuning. Human HEK293T cell data set (Hendra et al. 2022) was not used for training and only used in the evaluation.
Evaluation data sets
All the accuracy evaluation data sets we used are sourced from previously published resources. These include a synthesized IVT data set (Liu et al. 2019), a yeast data set (Liu et al. 2019), and a human HEK293T cell data set (Hendra et al. 2022). We used the second replicate from both the synthesized IVT and yeast data sets, as we had already used the first replicate of these two data sets for training and fine-tuning, and we used the first replicate of the human HEK293T cell data set as it was not included in training. A subset of the human HEK293T cell data set containing 500 genes was randomly sampled from the original data set. For the yeast data, we assessed model performance based on the sites identified by m6A-seq (Schwartz et al. 2013) for the wild-type strain, and the ime4Δ strains where no methylation should be observed. For the evaluation on human data, following previous work (Hendra et al. 2022), we regarded the combined sites identified by m6ACE-seq (Koh et al. 2019) and miCLIP (Linder et al. 2015) as methylated sites, and other randomly selected sites with the DRACH motif as unmethylated sites.
Training and fine-tuning a m6A methylation-sensitive neural network basecaller
We used the partially modified reads sampled from the signal k-mer graph to retrain a canonical basecaller. Before performing retraining on the pretrained canonical basecaller, we reinitialized the parameters of the last fully connected hidden layer with random weights but kept the same standard deviation. We then retrained the model using a smaller learning rate (0.00001) than the usual learning rate (0.001). We fine-tuned our model on biological samples with m6A sites identified by antibody experiments (Liu et al. 2019), labeling the A base at each modified site as an m6A base for every read (Supplemental Fig. S2B). Since the bases at methylation sites are usually not methylated in every read, this approach would introduce many false-positive labels. To address this issue, we applied label smoothing to the CTC loss that was used to train the basecaller. A label sequence of length L was defined as S = {si:i = 1, 2, …, L}, and each si belonged to the set {A, C, G, T, M}. The base probability logit output H ∈ RT/K×N was a (T/K)-by-N matrix derived from the basecaller's CRNN, where K is the total number of strides (i.e., the number of steps the convolutional filter moves across the input at each operation), and N is the number of bases used for prediction plus 1 (a blank symbol). The altered CTC loss with label smoothing under a strength factor represented by was then defined as
| (3) |
where M stands for the m6A base, LCTC is the usual CTC loss, and SM→A is the sequence in which every m6A base is replaced with an A base. We set empirically for the fine-tuning process, with an expectation that the methylation label is correct with probability .
Software availability
Code for Xron is available at GitHub (https://github.com/haotianteng/xron) and as Supplemental Code. Xron is available under a GNU GENERAL PUBLIC LICENSE v3.0. Xron is built with Python 3.8 and PyTorch 1.12, and has been tested on PyTorch 1.13 and 2.0.
Supplemental Material
Acknowledgments
This work was supported in part by the US National Science Foundation (DBI-1937540, III-2232121), the US National Institutes of Health (R01HG012470), and by the generosity of Eric and Wendy Schmidt by recommendation of the Schmidt Futures program. We also thank the Pittsburgh Supercomputing Center for providing computational resources through the Bridges2 system. H.T. is supported by funding from Oxford Nanopore Technologies plc and the School of Computer Science, Carnegie Mellon University—the Joint CMU-Pitt PhD Program in Computational Biology (CPCB). We used ChatGPT to correct grammatical errors and improve the flow of early drafts of this manuscript. We thank Minh Hoang for reviewing the manuscript and offering valuable feedback. We thank Tim Massingham (XGenomes Corp.) for the helpful discussion on signal segmentation.
Author contributions: H.T., Z.B.-J., and C.K. conceived the study. H.T. and C.K. designed the Xron algorithm. H.T. implemented the Xron algorithm. H.T. ran the performed comparison and analysis. H.T. and M.S. prepared the training data. H.T., Z.B.-J., and C.K. wrote the initial draft. H.T., Z.B.-J., M.S., and C.K. refined the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278960.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
C.K. is a co-founder of Ocean Genomics, Inc. H.T. is supported by funding from Oxford Nanopore Technologies plc. M.S. is an employee of Oxford Nanopore Technologies plc.
References
- Abebe JS, Price AM, Hayer KE, Mohr I, Weitzman MD, Wilson AC, Depledge DP. 2022. DRUMMER—rapid detection of RNA modifications through comparative nanopore sequencing. Bioinformatics 38: 3113–3115. 10.1093/bioinformatics/btac274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acera Mateos P, Sethi AJ, Ravindran A, Srivastava A, Woodward K, Mahmud S, Kanchi M, Guarnacci M, Xu J, Yuen ZWS, et al. 2024. Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications. Nat Commun 15: 3899. 10.1038/s41467-024-47953-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores J. 2013. Multiple instance classification: review, taxonomy and comparative study. Artif Intell 201: 81–105. 10.1016/j.artint.2013.06.003 [DOI] [Google Scholar]
- Baum LE, Petrie T, Soules G, Weiss N. 1970. A maximization technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann Math Stat 41: 164–171. 10.1214/aoms/1177697196 [DOI] [Google Scholar]
- Boulias K, Greer EL. 2023. Biological roles of adenine methylation in RNA. Nat Rev Genet 24: 143–160. 10.1038/s41576-022-00534-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans H, Den Dunnen J. 2014. Next generation sequencing technology: advances and applications. Biochim Biophys Acta 1842: 1932–1941. 10.1016/j.bbadis.2014.06.015 [DOI] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Lu Z, Wang X, Fu Y, Luo G-Z, Liu N, Han D, Dominissini D, Dai Q, Pan T, et al. 2015. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew Chem 127: 1607–1610. 10.1002/ange.201410647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Davidson NM, Wan YK, Patel H, Yao F, Low HM, Hendra C, Watten L, Sim A, Sawyer C, et al. 2021. A systematic benchmark of nanopore long read RNA sequencing for transcript level analysis in human cell lines. bioRxiv 10.1101/2021.04.21.440736 [DOI] [Google Scholar]
- D'Aquila P, Montesanto A, Mandalà M, Garasto S, Mari V, Corsonello A, Bellizzi D, Passarino G. 2017. Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell 16: 966–975. 10.1111/acel.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A, Sideri T, Varier RA, Brandis A, Stelzer Y, et al. 2021. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods 18: 1060–1067. 10.1038/s41592-021-01242-z [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. 2014. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 15: 293–306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu X, Wu B, Wang H, Xi F, Kohnen MV, Reddy AS, Gu L. 2021. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using nanopore direct RNA sequencing. Genome Biol 22: 22. 10.1186/s13059-020-02241-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A, et al. 2019. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell 178: 731–747.e16. 10.1016/j.cell.2019.06.013 [DOI] [PubMed] [Google Scholar]
- Graves A, Fernández S, Gomez F, Schmidhuber J. 2006. Connectionist temporal classification: labelling unsegmented sequence data with recurrent neural networks. In Proceedings of the 23rd International Conference on Machine Learning (ICML), Pittsburgh, PA, pp. 369–376. 10.1145/1143844.1143891 [DOI] [Google Scholar]
- Helm M, Lyko F, Motorin Y. 2019. Limited antibody specificity compromises epitranscriptomic analyses. Nat Commun 10: 5669. 10.1038/s41467-019-13684-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendra C, Pratanwanich PN, Wan YK, Goh WSS, Thiery A, Göke J. 2022. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods 19: 1590–1598. 10.1038/s41592-022-01666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shen L, Sun G. 2018. Squeeze -and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141. IEEE, Salt Lake City, UT. [Google Scholar]
- Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, Harada BT, Dai Q, Wei J, Zhang L, et al. 2022. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol 40: 1210–1219. 10.1038/s41587-022-01243-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Nie F, Ni P, Luo F, Wang J. 2022. SACall: a neural network basecaller for Oxford nanopore sequencing data based on self-attention mechanism. IEEE/ACM Trans Comput Biol Bioinf 19: 614–623. 10.1109/TCBB.2020.3039244 [DOI] [PubMed] [Google Scholar]
- Hughes JP, Guttorp P, Charles SP. 1999. A non-homogeneous hidden Markov model for precipitation occurrence. J R Stat Soc Ser C: Appl Stat 48: 15–30. 10.1111/1467-9876.00136 [DOI] [Google Scholar]
- Jenjaroenpun P, Wongsurawat T, Wadley TD, Wassenaar TM, Liu J, Dai Q, Wanchai V, Akel NS, Jamshidi-Parsian A, Franco AT, et al. 2021. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res 49: e7. 10.1093/nar/gkaa620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, Cairns BR. 2019. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci 116: 6784–6789. 10.1073/pnas.1817334116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CW, Goh YT, Goh WS. 2019. Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes. Nat Commun 10: 5636. 10.1038/s41467-019-13561-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriman S, Beliaev S, Ginsburg B, Huang J, Kuchaiev O, Lavrukhin V, Leary R, Li J, Zhang Y. 2020. Quartznet: deep automatic speech recognition with 1D time-channel separable convolutions. In 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), IEEE, pp. 6124–6128. IEEE, Barcelona, Spain. [Google Scholar]
- Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, et al. 2021. RNA modifications detection by comparative nanopore direct RNA sequencing. Nat Commun 12: 7198. 10.1038/s41467-021-27393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. 2019. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10: 4079. 10.1038/s41467-019-11713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz DA, Sathe S, Einstein JM, Yeo GW. 2020. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26: 19–28. 10.1261/rna.072785.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Ayadi L, Ernst FG, Hertler J, Bourguignon-Igel V, Galvanin A, Kotter A, Helm M, Lafontaine DL, Motorin Y. 2018. AlkAniline-seq: profiling of m7G and m3C RNA modifications at single nucleotide resolution. Angew Chem Int Ed Engl 57: 16785–16790. 10.1002/anie.201810946 [DOI] [PubMed] [Google Scholar]
- McIntyre AB, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, Mason CE. 2020. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci Rep 10: 6590. 10.1038/s41598-020-63355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meligkotsidou L, Dellaportas P. 2011. Forecasting with non-homogeneous hidden Markov models. Statist Comput 21: 439–449. 10.1007/s11222-010-9180-5 [DOI] [Google Scholar]
- Meyer KD. 2019. DART-seq: an antibody-free method for global m6A detection. Nat Methods 16: 1275–1280. 10.1038/s41592-019-0570-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinie B, Wang J, Lim KS, Hillebrand R, Lu Z-X, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. 2016. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat Methods 13: 692–698. 10.1038/nmeth.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Jaffrey SR. 2022. Hidden codes in mRNA: control of gene expression by m6A. Mol Cell 82: 2236–2251. 10.1016/j.molcel.2022.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D, Reddy AS, Ben-Hur A. 2022. RODAN: a fully convolutional architecture for basecalling nanopore RNA sequencing data. BMC Bioinf 23: 142. 10.1186/s12859-022-04686-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG. 2020. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife 9: e49658. 10.7554/eLife.49658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratanwanich PN, Yao F, Chen Y, Koh CW, Wan YK, Hendra C, Poon P, Goh YT, Yap PM, Chooi JY, et al. 2021. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat Biotechnol 39: 1394–1402. 10.1038/s41587-021-00949-w [DOI] [PubMed] [Google Scholar]
- Qin Y, Li L, Luo E, Hou J, Yan G, Wang D, Qiao Y, Tang C. 2020. Role of m6A RNA methylation in cardiovascular disease. Int J Mol Med 46: 1958–1972. 10.3892/ijmm.2020.4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang L-S. 2013. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19: 1684–1692. 10.1261/rna.036806.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhou T, Chen Q. 2022. Exploring the expanding universe of small RNAs. Nat Cell Biol 24: 415–423. 10.1038/s41556-022-00880-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14: 407–410. 10.1038/nmeth.4184 [DOI] [PubMed] [Google Scholar]
- Sun T, Wu R, Ming L. 2019. The role of m6A RNA methylation in cancer. Biomed Pharmacother 112: 108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
- Sutton RS, Barto AG. 2018. Reinforcement learning: an introduction, 2nd ed. MIT Press, Cambridge, MA. [Google Scholar]
- Teng H, Cao MD, Hall MB, Duarte T, Wang S, Coin LJM. 2018. Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience 7: giy037. 10.1093/gigascience/giy037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YK, Hendra C, Pratanwanich PN, Göke J. 2022. Beyond sequencing: machine learning algorithms extract biology hidden in nanopore signal data. Trends Genet 38: 246–257. 10.1016/j.tig.2021.09.001 [DOI] [PubMed] [Google Scholar]
- Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, et al. 2019. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16: 1297–1305. 10.1038/s41592-019-0617-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen L-Q, Zhao Y-L, Yang C-G, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo G-Z. 2019. Single-base mapping of m6A by an antibody-independent method. Sci Adv 5: eaax0250. 10.1126/sciadv.aax0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen T, Chen H-X, Xie Y-Y, Chen L-Q, Zhao Y-L, Liu B-D, Jin L, Zhang W, Liu C, et al. 2021. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat Methods 18: 1213–1222. 10.1038/s41592-021-01280-7 [DOI] [PubMed] [Google Scholar]
- Zhong Z-D, Xie Y-Y, Chen H-X, Lan Y-L, Liu X-H, Ji J-Y, Wu F, Jin L, Chen J, Mak DW, et al. 2023. Systematic comparison of tools used for m6A mapping from nanopore direct RNA sequencing. Nat Commun 14: 1906. 10.1038/s41467-023-37596-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.