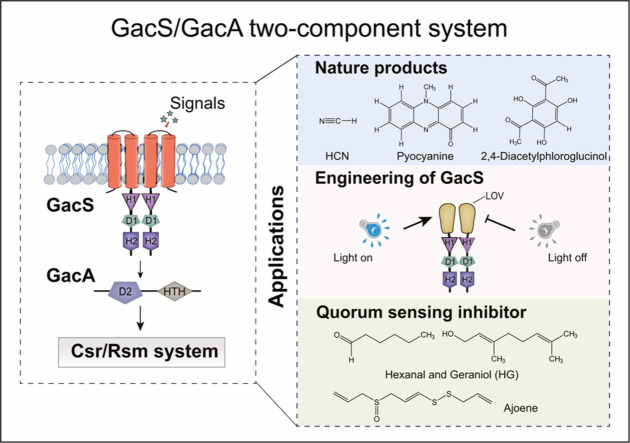
Abstract
The signal transduction system of microorganisms helps them adapt to changes in their complex living environment. Two-component system (TCS) is a representative signal transduction system that plays a crucial role in regulating cellular communication and secondary metabolism. In Gram-negative bacteria, an unorthodox TCS consisting of histidine kinase protein GacS (initially called LemA) and response regulatory protein GacA is widespread. It mainly regulates various physiological activities and behaviors of bacteria, such as quorum sensing, secondary metabolism, biofilm formation and motility, through the Gac/Rsm (Regulator of secondary metabolism) signaling cascade pathway. The global regulatory ability of GacS/GacA in cell physiological activities makes it a potential research entry point for developing natural products and addressing antibiotic resistance. In this review, we summarize the progress of research on GacS/GacA from various perspectives, including the reaction mechanism, related regulatory pathways, main functions and GacS/GacA-mediated applications. Hopefully, this review will facilitate further research on GacS/GacA and promote its application in regulating secondary metabolism and as a therapeutic target.
Keywords: GacS/GacA, Two-component system, Signaling regulatory network, Secondary metabolite, Antibiotic resistance
Graphical abstract
1. Introduction
Two-component system (TCS) is one of the most important tools by which bacteria recognize signaling molecules and regulate physiological responses as an adaptation to the environment [1,2]. The signal transduction mode of TCS is a conventional stimulus–response coupling mechanism [1,3]. In general, it involves the reaction of transfer of a phosphoryl group between two conserved components, namely, a histidine kinase protein (HK) containing a kinase core and a response regulatory protein (RR) containing a regulatory domain [4] (Fig. 1). TCS is also widespread in archaea and found in some eukaryotes [5].
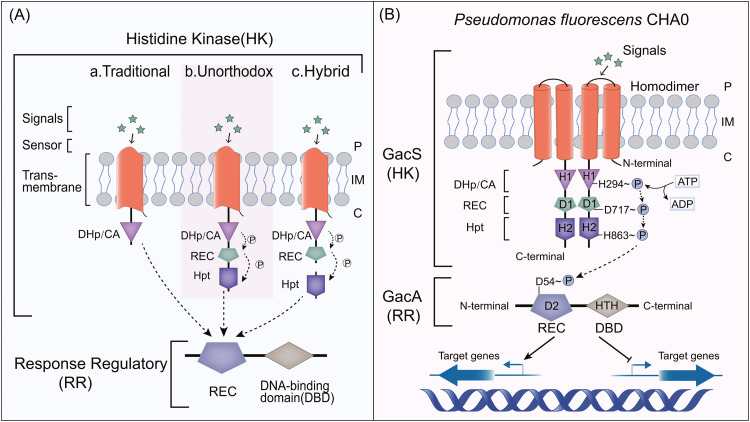
Fig. 1.
Signal-sensing and response mechanism of TCS. (A) Three types of HK: traditional HK, unorthodox HK and hybrid sensor HK [3,25,53,147]. (B) The domain composition of GacS/GacA. From top to bottom, GacS contains a periplasmic detection domain (GacSPD), two transmembrane helixes, HAMP (Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases) domain and three phosphotransfer domains: transmitter domain [a dimerization and histidine phosphotransfer (DHp) domain and a catalytic and ATP-binding (CA) domain], receiver (REC) domain and histidine phosphotransfer (Hpt) domain [6,53]. The GacSPD and HAMP domains are not shown, having been omitted for simplicity. GacA is a typical response regulator composed of the N-terminal REC and C-terminal helix-turn-helix (HTH) domain [147,148]. In Pseudomonas fluorescens CHA0, phosphorylated sites have been analyzed [28], [29], [30]. Phosphoryl group is indicated by “P”. P: Periplasm; IM: Inner Membrane; C: Cytoplasm.
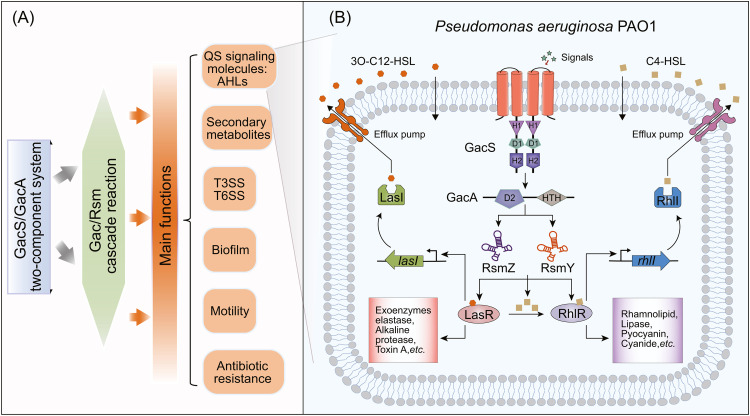
GacS/GacA is a TCS with the global regulation ability associated with environmental phenotypes [6,7]. Sensor kinase GacS and response regulator GacA (or their homologs) are widely present in various Gram-negative bacteria, such as Pseudomonas, Salmonella, Legionella and Vibrio, and have significant influence on the pathogenicity and virulence of pathogens [8], [9], [10]. GacS/GacA mainly regulates a series of physiological activities of bacteria through the Gac/Rsm signaling cascade pathway [6,11]. This pathway is evolutionarily conserved in different strains and plays different roles [12] (Fig. 2). Genomic and proteomic studies have identified many downstream pathways regulated by the response protein GacA, such as the production of quorum sensing (QS) signaling molecules (N-acyl homoserine lactones, AHLs) and secondary metabolites, participating in acute and chronic infection, biofilm formation, motility and antibiotic resistance [6,13,14].
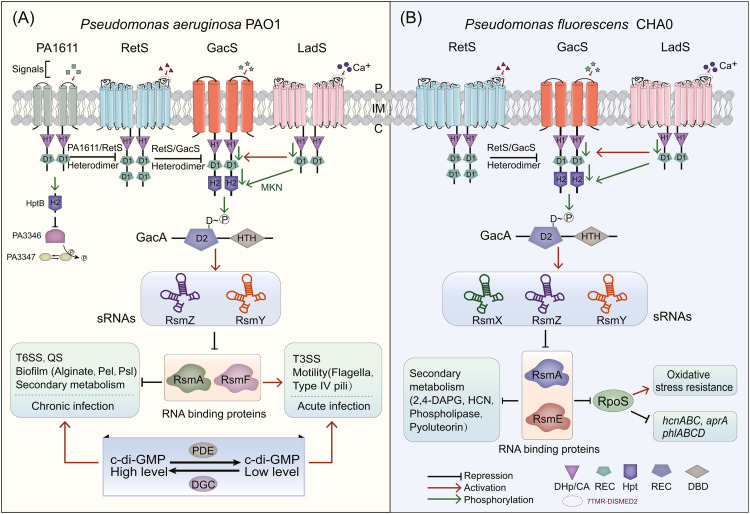
Fig. 2.
Signal transduction pathway of GacS/GacA in Pseudomonas. (A) Summary of Gac/Rsm signaling cascade pathway and regulatory phenotypes in Pseudomonas aeruginosa PAO1 [8,32,63,64,93]. (B) The regulation of the Gac/Rsm signaling cascade pathway in Pseudomonas fluorescens CHA0 [32]. MKN, multi-kinase network.
GacS/GacA is widely distributed and functionally diverse in microorganisms, but not in host mammals [15]. Importantly, its regulation of virulence and antibiotic resistance in pathogenic bacteria has motivated a number of related studies focused on it as a potential drug target [16], [17], [18]. In this review, we describe the GacS/GacA regulatory network from the perspectives of phosphorylation reaction mechanisms, related signaling pathways, functions and applications, with the aim of explaining the regulatory effects of this network on secondary metabolism and antibiotic resistance. This review should promote future research on GacS/GacA, especially GacS/GacA-mediated production and utilization of natural products, and addressing issues such as bacterial virulence and antibiotic resistance.
2. Reaction mechanism and homologous proteins of GacS/GacA
TCS is the link between bacteria sensing environmental signals and regulating their physiological behaviors [19]. The autophosphorylation of HK and the phosphorylation of RR are the main steps for TCS recognition and regulation [3,19]. HKs are signal-sensing proteins, which are usually membrane-bound, but some types are located in the cytoplasm [20]. Its function of autophosphorylation enables bacteria to continuously monitor environmental conditions, such as nutrients, temperature, pH, osmotic pressure and the presence of toxins [21]. Studies have provided detailed reviews of the signal transduction mechanism of TCS [3,[22], [23], [24]], and shown that HKs can be divided into three types [25] (1) traditional HK, (2) unorthodox HK, and (3) hybrid sensor HK (Fig. 1A).
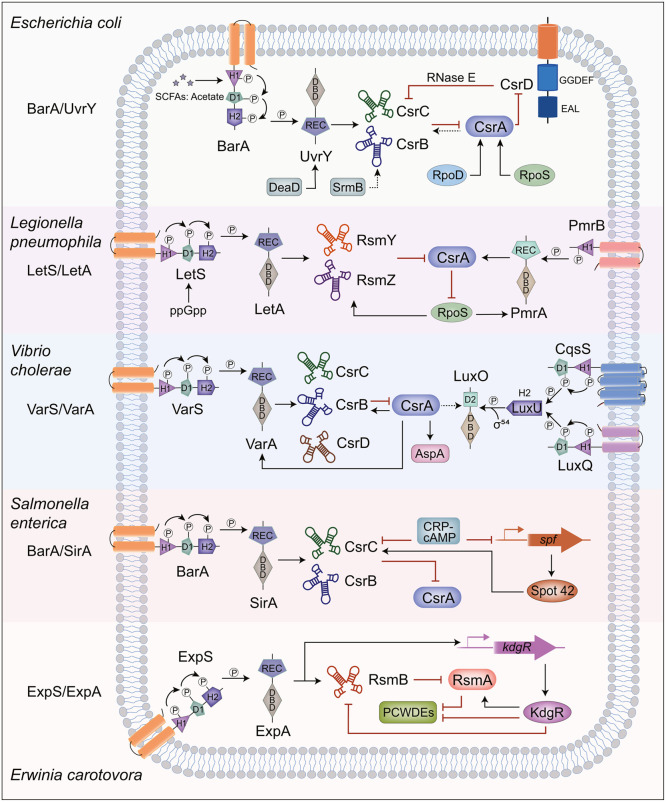
Intensive research has focused on GacS/GacA since the 1990s. GacA was originally named a global activator (abbreviated as Gac) for antibiotic and cyanide synthesis. Studies using genetic analysis and biochemical experiments showed that GacA is the homologous response regulator of GacS, and the two form a set of TCS [26,27]. According to the characteristics of its domains, GacS belongs to the unorthodox HK group, and Fig. 1B shows the structural composition of GacS/GacA. In Pseudomonas fluorescens CHA0, the key sites of GacS/GacA causing autophosphorylation and conformational changes have been revealed [28] (Fig. 1B). Detecting unknown signals through GacSPD (Periplasmic detection) domain, GacS autophosphorylates at His294 residue (H1), after which the phosphoryl groups are transferred to Asp717 (D1) and His863 (H2) in turn. After that, the phosphoryl group is transferred to residue Asp54 (D2) in the REC (Receiver) domain of GacA [28], [29], [30]. Phosphorylated RR is sometimes dephosphorylated by HK [19]. After sensing environmental signals, GacS/GacA can regulate the expression of downstream genes to adapt to changes in the environment [21]. GacS/GacA is highly conserved in Gram-negative bacteria and is prevalent in Pseudomonas [12,31]. More functions of GacS/GacA have also been found. We describe its functions in Section 4. Its homologs have been identified in other genera, and some have different names (Table 1). Fig. 3 briefly summarizes the regulatory pathways of homologous proteins in several widely studied pathogenic bacteria, including BarA/UvrY of Escherichia coli, ExpS/ExpA of Erwinia carotovora, LetS/LetA of Legionella pneumophila, BarA/SirA of Salmonella enterica and VarS/VarA of Vibrio cholerae.
Table 1.
Study on GacS/GacA and its homologous TCSs, including downstream regulated Csr/Rsm system members and major regulatory phenotypes.
| Species | GacS/GacA homologs | RNA binding proteins | Regulated sRNAs | Major regulatory phenotypes |
| Pseudomonas aeruginosa | GacS/GacA [71,78,83,97] | RsmA, RsmF | RsmY, RsmZ | AHLs, virulence, biofilm, motility, metabolism, T3SS, T6SS |
| Pseudomonas fluorescens | GacS/GacA[[35], [40], [41], [68]] | RsmA, RsmE | RsmX, RsmY, RsmZ | Metabolism, protease, phospholipase, RpoS, motility |
| Pseudomonas syringae | GacS (LemA)/GacA[[26], [42], [153], [154]] | RsmA1–5 | RsmX1–5, RsmY, RsmZ | Growth, syringomycin, motility, alginate, virulence, metabolism, QS |
| Escherichia coli | BarA/UvrY [[27], [155], [156]] | CsrA | CsrB, CsrC | Metabolism, motility, biofilm, stress resistance, virulence, QS |
| Legionella pneumophila | LetS/LetA[[152], [157], [158], [159]] | CsrA | RsmY, RsmZ | Cytotoxicity, virulence, motility, cell morphology, stress response, T4SS |
| Vibrio cholerae | VarS/VarA [[160], [161], [162], [163]] | CsrA | CsrB, CsrC, CsrD | QS, biofilm, virulence, cell shape |
| Salmonella enterica | BarA/SirA[[149], [164], [165], [166]] | CsrA | CsrB, CsrC | Virulence, motility, metabolism, stress survival, biofilm |
| Pectobacterium wasabiae (Erwinia carotovora) | ExpS/ExpA[[150], [167], [168], [169], [170]] | RsmA | RsmB | Extracellular enzymes, virulence, flagella, HarpinEcc, T2SS, T6SS, PCWDEs |
| Azotobacter vinelandii | GacS/GacA[[118], [171], [172]] | RsmA | RsmZ1–7, RsmY | Alginate, PHB, ARs, flagella |
| Pseudomonas chlororaphis | GacS/GacA [[173], [174], [175]] | RsmA | RsmX, RsmY, RsmZ | QS, biofilm, HCN, PHZ, LPS, trehalose, siderophore, motility |
| Acinetobacter baumannii | GacS/GacA [[136], [137]] | — | — | Virulence, biofilms, motility, PAA |
| Dickeya oryzae | TzpS/TzpA [146] | RsmA | RsmB | Zeamines, virulence, biofilms, motility |
The list of regulated phenotypes is not complete and is meant to suggest the conservation as well as breadth of regulation. Species are listed in the order mentioned in the text. AHLs, N-acyl-homoserine lactones; T3SS, type III secretion system; QS, quorum sensing; PCWDE, plant cell wall-degrading enzyme; PHB, polyhydroxybutyrate; AR, alkylresorcinol; HCN, hydrogen cyanide; PHZ, phenazine; LPS, lipopolysaccharide; PAA, phenylacetic acid.
Fig. 3.
Signal transduction regulation pathway of GacS/GacA homologous proteins. In Escherichia coli, Erwinia carotovora, Legionella pneumophila, Salmonella enterica and Vibrio cholerae, GacS/GacA homologous proteins sense and respond to signals, and activate the Csr/Rsm system in downstream pathways [12,[149], [150], [151], [152]]. The lines indicate the interactive relationship: Arrow, activation; T bar, repression; Dotted line, predicted interaction.
3. Summary of GacS/GacA regulatory networks
3.1. Gac/Rsm signaling cascade pathway
Initially, it was thought that the GacS/GacA global response regulator controls gene expression only at the transcriptional level. However, it was later found that the GacS/GacA system mainly regulates the Rsm/Csr (Carbon storage regulator) system at the post-transcriptional level [32,33]. There are two components in the Rsm/Csr pathway: RNA binding proteins (RsmA/CsrA families) and noncoding small RNAs (sRNAs). RsmA/CsrA families are regulators of gene expression [12]. The Gac/Rsm signaling cascade pathway includes the GacS/GacA and Rsm post-transcriptional regulatory system [31]. Signaling molecules stimulate the transfer of phosphoryl groups in GacS/GacA. Phosphorylated GacA can bind to a conserved palindromic upstream activating sequence (GacA box) to activate the transcription of sRNAs, thus triggering Rsm cascade reaction [34,35]. These sRNAs have complex stem-loop structures containing GGA motifs, and they have multiple RNA binding protein sites [33]. This means that sRNAs can modulate the activation/inhibition impact of RNA binding proteins to target genes [36]. Therefore, Gac/Rsm pathway can regulate multiple phenotype-related genes at the post-transcriptional level [36].
In Pseudomonas aeruginosa, GacS/GacA directly controls the expression of two sRNAs, RsmY and RsmZ [35] (Fig. 2A). There are differences in the transcriptional efficiency of RsmY and RsmZ, and the expression level of RsmY is about double that of RsmZ [35]. RNA binding proteins, RsmA and RsmF, in P. aeruginosa regulates protein synthesis and/or mRNA stability by specifically binding to the GGA motif in the 5'-UTR (untranslated region) of the target mRNAs [36]. High levels of RsmY and RsmZ can also isolate RsmA/RsmF from target mRNAs to relieve the regulatory effect of RsmA/RsmF and promote/inhibit the expression of target genes [37]. Each sRNA may play a different role in controlling RsmA/RsmF activities [38,39]. In P. fluorescens, GacA regulates the transcription of three sRNAs: RsmX, RsmY and RsmZ [40]. These sRNAs jointly regulate metabolism (HCN, 2,4-DAPG and exoprotease) by relieving the inhibitory effects of the RNA binding proteins RsmA and RsmE [34,40,41] (Fig. 2B). In Pseudomonas syringae, the Gac/Rsm pathway is more complex. Specifically, GacA can activate and regulate the expression of seven sRNAs (RsmX1–5, RsmY and RsmZ) [42]. These regulatory relationships make GacS/GacA a core activator/suppressor of many phenotypes.
3.2. Regulation of the Gac/Rsm pathway
In Pseudomonas, the Gac/Rsm pathway can be affected by factors in other pathways [43]. Although only GacS-P can phosphorylate GacA, other HKs can also affect the process of GacA phosphorylation through GacS [44,45]. For example, hybrid sensor HKs, LadS and RetS, can regulate GacS and influence the Gac/Rsm pathway (Fig. 2). Hybrid sensor HKs are characterized by the presence of additional REC domains at the C-terminus. Moreover, the C-terminus of RetS has two REC domains, while LadS has only one. The signal binding domains of LadS and RetS consist of a seven-transmembrane (7TMR) region and a periplasmic sensor domain (diverse intracellular signaling module extracellular 2, DISMED2) [46,47]. DISMED2 is responsible for sensing signal molecules and promoting HK phosphorylation. 7TMR-DISMED2 was identified in carbohydrate-binding proteins and it was predicted that this domain could sense a variety of glycans in the environment [48]. Wang et al. found that mucin glycans were the signal molecules of RetS and inhibited the activity of GacS [49]. However, mucin glycans are heterogeneous and diverse, while the mechanisms of binding between mucin glycans and RetS-DISMED2 are unknown.
Neither the REC nor the Hpt (Histidine phosphotransfer) domain is required for RetS to inhibit the GacS/GacA signaling pathway. Instead, it directly forms heterodimers with GacS to prevent the transfer of phosphoryl groups from GacS to GacA [50], [51], [52], [53]. RetS downregulates the signal transduction function of GacS in three ways: it obtains phosphoryl groups from GacS-P, exerts phosphatase activity to accelerate the dephosphorylation of GacS-P, and inhibits the autophosphorylation of GacS [54,55]. These mechanisms allow RetS to inhibit the transcription of RsmY and RsmZ by interfering with GacS/GacA activity, thereby regulating the expression of target genes. These mechanisms prevent GacA from being phosphorylated, resulting in the inactivation of sRNA transcription. When the expression of sRNA decreases, the inhibitory function of RsmA is no longer restricted. Moreover, RsmA can inhibit the transcription of RetS and relieve the inhibitory effect of RetS on the Gac/Rsm signaling cascade pathway [56]. PA1611, which is another hybrid sensor HK of P. aeruginosa, can also interacts with RetS to relieve the inhibitory effect of RetS on the Gac/Rsm pathway [57] (Fig. 2A). Its binding mode may be similar to that of RetS/GacS [57]. PA1611 has a higher affinity with RetS, which can help release GacS from the GacS/RetS heterodimer to form a PA1611/RetS heterodimer [57,58]. This released GacS forms homodimers and activates the Gac/Rsm pathway. Moreover, PA1611 overexpression is consistent with the phenotypic data of retS deletion mutant, indicating that PA1611/RetS heterodimer blocks the inhibitory effect of RetS on GacS [58]. In addition, the PA1611 pathway can regulate the Gac system through HptB [59]. HptB is a single-domain Hpt protein, which transmits phosphoryl groups from PA1611 to PA3346-PA3347 [59]. However, the regulation of sRNA by HptB may be independent of GacS/GacA, which is affected by cell growth conditions [60,61]. Moreover, a conserved membrane protein, CmpX (PA1775), was identified as a regulator of PA1611 expression in GacS/GacA regulatory networks [62]. These complex regulators allow the Gac/Rsm signaling cascade to more precisely regulate the expression of target genes.
Unlike RetS and PA1611, which regulate signaling cascade pathways through protein–protein interactions, LadS works with GacS/GacA to form a multi-component signal transduction system with a primitive phosphoryl cascade [63]. When signal Ca2+ binds to the DISMED2 domain, its kinase function is activated [63]. Subsequently, the H1 and D1 domains of LadS and the H2 domain of GacS form a multi-kinase network to enhance the transfer of a phosphoryl group from GacS to GacA and induce regulation of the Gac/Rsm pathway [64]. Currently, although LadS has been reported to sense Ca2+, the signals sensed by the other three HKs (GacS, PA1611 and RetS) remain unclear [25].
4. Main functions of GacS/GacA in regulatory pathways
4.1. Regulating the production of AHLs
Through QS systems, microorganisms regulate physiological behavior to greatly improve environmental adaptability [65]. QS signal molecules, also called autoinducers, are intermediaries in communication between bacteria [66]. With increasing cell density, signal molecules can accumulate to high concentrations in the environment, resulting in the activated transcription of target genes to regulate various physiological behaviors and adapt to the changeable living environment [67]. Studies have shown that the expression of GacS/GacA is affected by QS. Because the expression of GacA is regulated by the growth process and peaks in the stationary phase [68], the expression of many genes targeted by GacS/GacA also improves significantly with increasing cell density [69]. Therefore, it is speculated that bacteria growing at high density secrete signal molecules that activate GacS/GacA [70], although such signal molecules remain largely unknown. In addition, Reimmann et al. found that GacS/GacA could regulate the synthesis of the QS signal molecules AHLs, thus indirectly participating in QS [71].
AHLs are produced by members of the AHL synthase families (including the LuxI, HdtS and LuxM families). Most AHL synthesis-related genes in Gram-negative bacteria belong to the LuxI family [72,73]. AHLs can regulate the physiological activities and behaviors of bacteria, such as the production of virulence factors (pyocyanin, cyanide and lipase), and activate the transcription of RpoS [71]. There are two typical AHLs in P. aeruginosa. One is N-3-oxo-dodecanoyl-l-homoserine lactone (3O-C12-HSL), which is synthesized and regulated by the LasR/LasI QS system. The other is N-butanoyl-l-homoserine lactone (C4-HSL), the synthesis of which depends on the RhlR/RhlI QS system [74], [75], [76], [77]. Reimmann et al. found that deletion of the gene gacA in P. aeruginosa PAO1 delayed and reduced the production of C4-HSL, RhlR and LasR [71]. In addition, Kay et al. studied the regulatory mechanism of GacS/GacA on AHLs and found that the deletion of rsmY and rsmZ impaired C4-HSL production, and that the same phenomenon occurred in the strain ΔgacA [78]. GacA can regulate the transcription of RsmY and RsmZ, so it is concluded that GacS/GacA regulates the expression of AHLs through the Rsm pathway. Moreover, compared with the wild-type, rsmY/rsmZ deletion mutants exhibited decreases in the extracellular products regulated by QS [78]. The regulatory effects of GacS/GacA on AHL synthesis make it indirectly participate in QS networks [71] (Fig. 4B). Meanwhile, GacS/GacA is a two-component system that can sense signaling molecules and directly participate in bacterial communication. GacS/GacA is not a simple linear signal transduction pathway in the regulatory network of bacteria and its complete regulatory networks in QS require further study.
Fig. 4.
Main functions of GacS/GacA regulation. (A) Via its global regulatory ability, GacS/GacA participates in a variety of important regulatory functions. (B) GacS/GacA is involved in the synthesis of AHLs in Pseudomonas aeruginosa PAO1[[78], [121]].
Chancey et al. revealed the complexity of the GacS/GacA regulatory pathway in biocontrol strain Pseudomonas aureofaciens 30–84 and showed that GacS/GacA and AHLs worked together to control gene expression [79]. Transcriptomic data of Pseudomonas chlororaphis 30–84 showed that the gacA gene controled the expression of genes involved in phenazine antibiotic synthesis (phzXYFABCD and phzO) [80,81]. Meanwhile, GacS/GacA can regulate the expression of the AHL synthesis gene phzI [79]. This means that the Gac/Rsm pathway can directly regulate the production of phenazine antibiotics or indirectly regulate it through AHL molecules. In short, the regulatory effects of GacS/GacA on AHLs make its signal regulation network more complex. Studying the interaction between different signal pathways can provide a deeper understanding of how bacteria adapt to complex and changeable environmental conditions.
4.2. GacS/GacA is responsible for the production of secondary metabolites and extracellular enzymes
GacS/GacA can control the expression of hundreds of genes and positively regulate the production of various secondary metabolites through sRNAs [15]. Wei et al. explored the global impact of gacA mutation on the transcriptome and metabolome of P. aeruginosa M18, and found that GacA significantly affected the transcription of about 15% (839) of annotated genes in its genome [14]. Many genes of extracellular enzymes and secondary metabolites, such as siderophores (pyoverdine and pyochelin) and virulence factors, are activated by GacS/GacA [[14], [82]].
Virulence factors, such as pyocyanine (phz), HCN (hcnABC) and 2,4-DAPG (phl), play important roles in improving fitness and infection rate [83]. GacS/GacA can regulate the synthesis of virulence factors by controlling various transcriptional and post-transcriptional factors [84]. In the animal pathogen P. aeruginosa, GacS/GacA was found to be one of the main systems regulating the expression of virulence factors, and to rely on the Rsm post-transcriptional system in order to function [83,85]. Moreover, in the plant pathogen P. syringae, GacA/GacS also controls the expression of virulence genes through Rsm, and its pathway is more complex [42]. There are seven sRNAs and five RsmA homologous proteins in the Rsm system to regulate the infection process [86]. In biocontrol bacteria P. fluorescens CHA0, GacA was found to positively regulate the biosynthesis of biocontrol factors [87]. Deletion of gacS or gacA greatly reduces the expression of genes required for HCN, 2,4-DAPG and other virulence factors, leading to loss of the ability to protect plants from pathogen invasion in mutant strains [30,31,87]. Moreover, P. fluorescens In5 can produce the antifungal cyclic lipopeptides (CLPs) nunamycin and nunapeptin, but this is not the case in strain ΔgacA, resulting in this mutant being unable to inhibit the growth of fungi and oomycetes [88].
Microorganisms are an important resource of secondary metabolites and enzymes, and many strains are ideal producers of enzymes [89]. Besides participating in the synthesis of virulence factors, Sacherer et al. confirmed that gacA was also essential for producing two extracellular enzymes (protease and phospholipase C) in P. fluorescens [90]. In Dickeya dadantii (Erwinia chrysanthemi 3937), the production of pectin lyase, protease and cellulase by strain ΔgacA decreased due to the increased inhibitory effect of RsmB/RsmA on extracellular enzyme expression [91]. Therefore, as an indispensable part of secondary metabolites and extracellular enzymes, GacS/GacA and its regulatory pathway can be used as targets to develop inhibitors of animal or plant pathogens and inducers of biocontrol bacteria [92].
4.3. GacS/GacA mediates secretory systems and acute/chronic infection
Pathogenic bacteria maintain acute and chronic infection by regulating the alternate functions of the type III secretory system (T3SS) and type VI secretory system (T6SS) [93]. The process can be affected by GacS/GacA, hybrid sensor kinases that influence the Gac/Rsm pathway, and the secondary messenger c-di-GMP [57,94]. GacS/GacA can regulate the expression of the secretory systems of pathogenic bacteria, including T3SS and T6SS [93,95]. P. aeruginosa infection readily occurs in cystic fibrosis (CF) cases [96]. Patients with CF initially exhibit acute infection and later develop chronic respiratory diseases due to expression of the secretory systems [96]. GacS/GacA can negatively regulate T3SS related to acute infection and positively regulate T6SS related to chronic infection through the Rsm system [49,97] (Fig. 2A). Moreover, many phenotypes related to acute and chronic virulence have been shown to be inversely regulated by RsmA/RsmF [39]. Specifically, low levels of RsmA/RsmF can upregulate the expression of genes associated with chronic infection, while high levels of RsmA/RsmF are conducive to the expression of acute virulence-related factors [98].
Hybrid sensor kinases RetS, PA1611 and LadS are involved in the regulatory effects of Gac/Rsm on pathogen infectivity [45,57]. GacS is necessary to activate the phenotype associated with chronic persistent infection [8]. Unlike GacS, RetS accelerates the process of acute infection by inhibiting the Gac/Rsm pathway [50], [51], [52]. It inhibits T6SS while activating T3SS [44,52,60]. In contrast, Ca2+-activated LadS leads to pathogens transitioning from acute to chronic infection by regulating the Gac/Rsm pathway [45]. Moreover, the LadS-mediated Gac/Rsm pathway also endows the strain with antibiotic tolerance by slowing down the growth rate of P. aeruginosa [63]. This reflects an original strategy that, in different living environments, pathogens sense environmental signals through complex multi-component signaling transduction systems to control their infectious state and maximize the rate of successful infection.
In addition, the secondary messenger c-di-GMP relying on the Gac/Rsm pathway participates in switching the productive states of T3SS and T6SS by changing its level [requiring diguanylate cyclase (DGC) or phosphodiesterase (PDE)] [93,94] (Fig. 2A). c-di-GMP is synthesized by DGC and hydrolyzed by PDE [99]. In P. aeruginosa, RetS is based on the Gac/Rsm pathway to exert effect. The retS deletion mutant shows a high level of c-di-GMP in the cell, which promotes T6SS-mediated chronic infection [93]. In contrast, a low level of c-di-GMP can promote the expression of virulence factors required for acute infection. In addition, the T3SS/T6SS conversion induced by c-di-GMP depends on two sRNAs, RsmY and RsmZ, and the DGC enzyme is strictly inhibited by RsmA [93].
4.4. GacS/GacA participates in biofilm formation
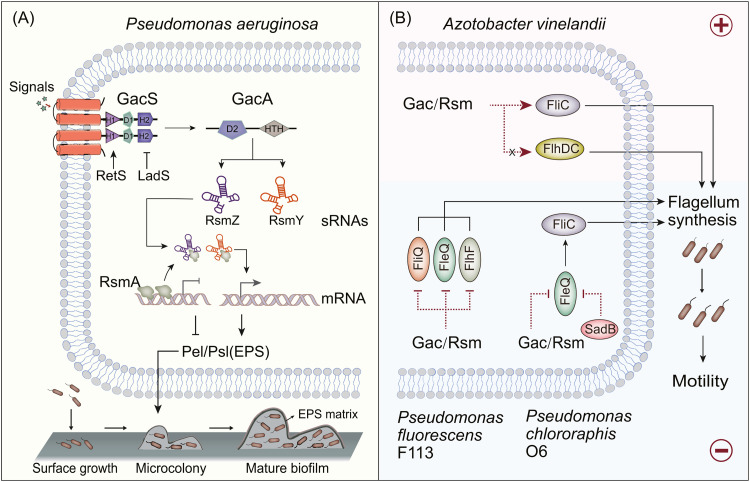
Biofilms are composed of microbial communities growing on surfaces and extracellular polymeric substance (EPS) [100]. EPS contains polysaccharides, proteins and DNA [101]. Alginate, Pel, Psl and other polysaccharides are determinants of the structural stability of biofilms [102]. If bacteria cannot produce these polysaccharides, they exhibit severe defects in biofilm development. Biofilm formation confers greater resistance to environmental challenges, antibiotic penetration and the host immune system [103]. The GacS/GacA regulatory network is necessary for biofilm formation in P. aeruginosa [104], [105], [106], [107] (Fig. 5A). Parkins et al. found that the biofilm formation of ΔgacA was reduced to one-tenth of its typical level in P. aeruginosa PA14 [83]. GacS has a GacSPD domain composed of 126 residues [108]. Ahmad et al. analyzed the GacSPD function in P. aeruginosa PAK and found that strain PAKgacSΔPD showed a similar phenotype to PAKΔgacS in terms of biofilm formation, and that the biofilm thickness was reduced to one-quarter that in the PAK strain [108]. Further functional analysis showed that this effect was accompanied by a change of transcription of the sRNAs RsmY/RsmZ [108]. Moreover, the polysaccharide Pel/Psl was found to be directly negatively regulated by RsmA [109]. Therefore, high levels of RsmZ/RsmY can bind RsmA, upregulate the expression of Psl/Pel and promote biofilm formation. Furthermore, the regulation of GacS by RetS/LadS has been demonstrated in studies on biofilm regulation by the Gac/Rsm pathway [45,53]. Analyzing the mechanism by which GacS/GacA regulates biofilm formation can help solve the side effects caused by biofilm, especially their mediation of chronic infection and antibiotic resistance, and provide solutions for overcoming these in clinical practice. An overview of bacterial antibiotic resistance mediated by biofilm formation through GacS/GacA is provided in Section 4.6.
Fig. 5.
Regulatory effects of GacS/GacA on biofilm formation and motility. (A) In Pseudomonas aeruginosa, GacS/GacA positively regulates the synthesis of main matrix structure polysaccharides, Psl and Pel, through the Rsm pathway to promote biofilm formation [53,104,106]. (B) In Pseudomonas fluorescens F113 and Pseudomonas chlororaphis O6, Gac/Rsm negatively regulates flagellum synthesis [110,115]. In Azotobacter vinelandii, Gac/Rsm positively regulates flagellum synthesis to promote motility [118].
4.5. GacS/GacA regulates bacterial motility
Motility is one of the most crucial characteristics of Pseudomonas and enables bacteria to settle in niches and gain a competitive advantage [110]. Three types of movements have been described in Pseudomonas, namely, swimming, swarming and twitching [110,111]. Navazo et al. confirmed that these three motility-related phenotypes in P. fluorescens F113 are negatively regulated by GacS/GacA[110]. In addition, Martinez-Granero et al. found that the strain ΔgacS produced greater swimming haloes [112]. There are two flagellum systems in P. fluorescens F113, mainly regulated by FleQ and FlhDC [113]. Gac/Rsm reduces the expression of the flagellum regulatory gene fleQ, thereby reducing bacterial motility [114]. This inhibitory effect was shown to disappear in strains overexpressing RsmA and RsmE [114]. In P. chlororaphis O6, GacS also negatively regulates flagellum formation and cell motility [115]. At the stationary phase of strain ΔgacS, the expression of three genes (fleQ, fliQ and flhF) involved in flagellum formation was found to increase, resulting in an elongated cell shape and a two-fold increase in flagellum number [115]. Therefore, GacS/GacA can be used as a negative regulator to regulate flagellum formation and cell morphology, then affect motility.
Notably, GacS/GacA exhibits the species difference in the regulation of the types and mechanisms of motility (Fig. 5B). In P. syringae pv. tomato DC3000, GacS/GacA positively regulates swimming and swarming movement, while RsmA negatively regulates swarming and does not regulate swimming [116]. This reveals the variability of the Gac/Rsm pathway in regulating motility. Compared with Pseudomonas, Azotobacter vinelandii has unusual peritrichous flagellation, and FlhDC is the master regulator of flagellum synthesis [117]. López-Pliego et al. found that the Gac/Rsm pathway might not regulate the FlhDC of A. vinelandii, but positively regulated FliC, thereby promoting swimming [118]. However, the targets of the Gac/Rsm pathway and whether there is direct or indirect regulation of flagellum synthesis remain to be determined. Moreover, on the swarming edge, gacS and gacA are spontaneously lost to help bacteria adapt to the changing growth conditions [119].
4.6. GacS/GacA mediates antibiotic resistance
In recent decades, the overuse of antibiotics has led to the emergence of multidrug-resistant bacteria [120]. The main known mechanisms behind antibiotic resistance include the overexpression of efflux pumps, production of antibiotic-modified enzymes, modification of antibiotic-targeted sites and biofilm formation [121]. Brinkman et al. found that the deletion of gacS could reduce the minimum inhibitory concentration (MIC) of P. aeruginosa PAK on gentamicin, amikacin and chloramphenicol [122]. This showed that GasS/GacA could regulate the antibiotic resistance of bacteria [122,123].
Bacteria in biofilm have a unique mechanism for tolerating antibiotics. It has been proven that GacS/GacA is involved in antibiotic resistance in bacteria by increasing biofilm formation [83]. GacS/GacA can participate in the resistance against three different families of antibiotics, represented by tetracycline, tobramycin and ciprofloxacin, through RsmA/RsmZ [120]. Under their sub-minimum inhibitory concentrations (sub-MIC), biofilm formation regulated by Gac/Rsm is induced to increase antibiotic resistance [124]. In P. aeruginosa PA14, deletion of gacS was found to lead to reduced biofilm formation and increased sensitivity to antibiotics [125]. However, when characterizing the strain ΔgacS, phenotypic variation occurred in the biofilm and resulted in the formation of small colony variants (SCVs) [126]. SCVs have thicker biofilm and higher antibiotic resistance level than wild-type and parental strain ΔgacS [125]. Moreover, when the gene gacS was added back into SCVs, bacteria resumed their normal phenotype, which reflects a special strategy of adaptation to the environment [127,128]. In general, GacS/GacA affects antibiotic resistance by mediating biofilm formation. This is one of the reasons why microbial infections are so difficult to control.
Studies have shown that bacterial motility can also affect antibiotic resistance [129]. Meanwhile, GacS/GacA can regulate the motility of bacteria [119]. Therefore, in theory, GacS/GacA could interfere with antibiotic resistance by regulating bacterial motility. However, no studies on this have been reported. Although swarming motility is a primary stage of mature biofilm formation [130], the mechanism by which motility confers antibiotic resistance may differ from this. In contrast to bacteria within biofilms, swarming cells are unprotected in a complex extracellular matrix but metabolically very active, while still exhibiting multiple drug-resistant phenotypes [129]. Therefore, the GacS/GacA-mediated model of bacterial motility regulation causing the antibiotic resistance has important research value and warrants further study.
5. Controlling the Gac/Rsm pathway and maximizing its application value
5.1. Environmental factors affect the Gac/Rsm pathway
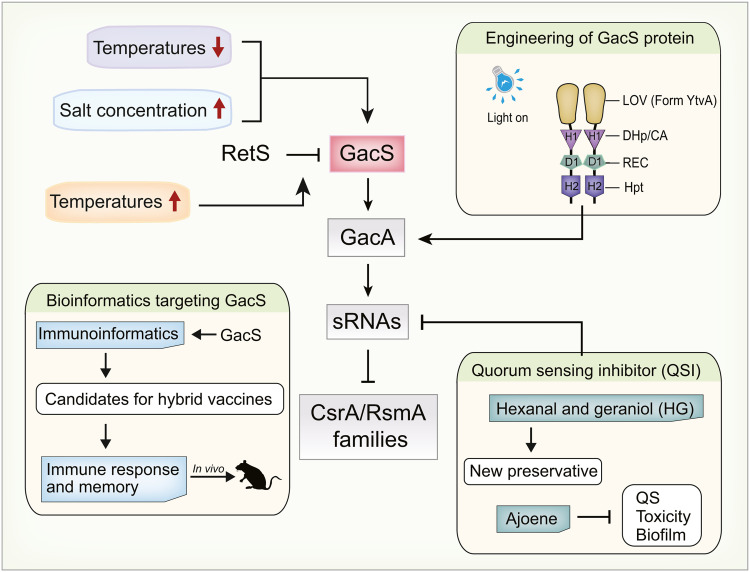
The Gac/Rsm pathway can regulate the secondary metabolism and antibiotic resistance of bacteria. If the expression of the Gac/Rsm system can be modulated by environmental factors, the production of secondary metabolism products or enzymes can be altered. Studies have shown that environmental factors have a significant effect on the expression of TCS. In Pseudomonas putida PCL1445, the production of cyclic lipopeptides putisolvin I and II was shown to be regulated by GacS/GacA [131]. Moreover, Dubern et al. showed that the expression of gacS/gacA was strongly induced at a low temperature (11°C) and high concentration of NaCl (1 M) (Fig. 6), resulting in the increased production of putisolvins [132]. Furthermore, Humair et al. tested the potential effect of temperature on two auxiliary sensors, LadS and RetS, and found that the temperature sensitivity mainly depended on RetS, which inhibits the Gac/Rsm pathway at high temperatures [133]. Compared with the findings at a standard incubation temperature of 30°C, P. fluorescens CHA0 was shown to only produce a small number of HCN and antibiotic compounds on rich solid medium at 35°C. Its sensing mechanism may involve changes in membrane fluidity, making the heterodimer RetS/GacS more robust at 35C [133]. Physiological and environmental conditions are important factors controlling the Gac/Rsm pathway. Therefore, studies on how to regulate the GacS/GacA pathway by optimizing the relevant temperature and salinity conditions to promote the maximum production of secondary metabolites could be a focus of future research.
Fig. 6.
Application of targeted Gac/Rsm pathway. The information obtained from Section 5 [132,133,135,138,141,143].
5.2. Engineering of GacS protein
GacS is the upstream initiator of the Gac/Rsm pathway. Significantly, the sensor domain of GacS can be replaced by an existing photosensor domain to realize the engineering modification of the optogenetic system [134]. Cheng et al. replaced two transmembrane regions and the HAMP (Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases) domain of GacS in P. aeruginosa with the light-oxygen-voltage (LOV) blue light sensor domain of the photosensitive protein YtvA of Bacillus subtilis [135] (Fig. 6). These light-regulated derivatives of GacS can only be regulated by blue light, while still retaining kinase activity, which can reversibly stimulate the global Gac/Rsm signaling pathway. More importantly, the new GacS photosensitive protein can transform acute infection to chronic infection and control the expression of virulence factors of P. aeruginosa. This further expands the precise control of bacterial pathogenicity.
In TCSs related to the virulence of Acinetobacter baumannii, GacS/GacA plays an important role in the regulation of antibiotic sensitivity and virulence mechanism [136,137]. Considering that prevention is the best strategy for dealing with A. baumannii infection, Smiline et al. used immunoinformatic methods to predict novel vaccine peptide candidates of GacS protein against multidrug-resistant strains [138]. This study suggested the use of five antigenic peptides as hybrid vaccine candidates. The GacS vaccine peptide data and immunoinformatic methods available in A. baumannii provide a good reference for the study of GacS in other strains. The application of bioinformatic databases and methodologies for the engineering of GacS protein increases the possibility of finding new methods with minimal trial and error. However, the process of developing vaccine peptides is complex, involving a need to design chimeric vaccine constructs and perform in vivo experiments to further confirm the immune response and memory (Fig. 6). With the continuous progress of research, bioinformatic databases and methods should continue to provide valuable insights into the targeting of GacS/GacA to overcome infections by pathogenic bacteria.
5.3. Quorum sensing inhibitor working through Gac/Rsm cascade pathways
The reaction initiated by QS signaling molecules can not only directly promote the expression of virulence factors, but also enable bacteria to resist antibacterial compounds, such as by forming biofilms [139]. Meanwhile, Quorum sensing inhibitor (QSI) can reduce the virulence of bacteria and biofilm formation, providing promising biotechnological applications [140]. Gac/Rsm is a key element in the QS signaling cascade regulatory pathway and a promising target for QSI (Fig. 6). For example, the QSI combination of hexanal and geraniol has been proved to significantly inhibit the expression of gacS/gacA in P. fluorescens and can be further developed as a new preservative for agricultural products [141]. In addition to the TCS, sRNA in the Gac/Rsm cascade pathway is also a target for inhibitors. Ajoene is a small sulfur-rich molecule and is the main QSI in garlic [142]. Experimental evidence provided by Jakobsen et al. revealed that ajoene exerted its own QSI activity by reducing the sRNA expression of RsmY and RsmZ in P. aeruginosa [143]. Overall, ajoene plays a role upstream of the QS pathway by targeting the Gac/Rsm cascade, inhibiting QS, toxicity and biofilm formation. However, these results require further studies, especially for determining the specific mechanism by which ajoene affects sRNA.
6. Future perspectives
TCS is critical for cell growth and adaptation to complex environmental changes. GacS/GacA is a TCS with global regulatory capacity that functions through the Gac/Rsm signaling cascade pathway [31]. The highlights of this review include (1) an emphasis on the studied GacS/GacA regulatory pathways and functions, particularly in Pseudomonas; and (2) summarizing Gac/Rsm pathway-mediated applications, which deserve more attention. In recent years, the application of targeted GacS/GacA has emerged, and some research strategies are worthy of further exploration. Because the global regulatory capacity of GacS/GacA is known to confer some secondary metabolic advantages, it is highly recommended that the corresponding pathways are directly and rationally modified to obtain a large amount of required product. The regulation of virulence and antibiotic resistance by GacS/GacA makes it a potential target for drug therapy [144,145]. Photogenetic modification of GacS protein has been applied to study pathogen–host (P. aeruginosa–Caenorhabditis elegans) interactions [135]. By introducing pathogens into hosts, we can further understand the virulence, host susceptibility and infection mechanism of pathogens, which should provide a way of studying antibiotics and new targets of infection. Notably, GacS/GacA is conserved and functionally diverse in different strains. Insight into the mechanism and regulatory pathway of GacS/GacA can provide ideas for the development of natural products and drugs.
Although some pathways and functions of GacS/GacA have been revealed, the GacS/GacA regulatory networks are not complete and require further supplementation and improvement. Finding the precise types of signaling molecules that GacS, RetS and LadS can recognize is a great challenge in this field. Latour summarized GacS-related content from a signaling molecule perspective and presented the progress made in characterizing GacS signaling and new strategies. And his review greatly assists future inquiry into the families and structures of the exact signals sensed by GacS/GacA [6]. The signals sensed by GacS can directly regulate the Gac/Rsm pathways, while the signals sensed by RetS and LadS can interfere with GacS/GacA by changing its phosphorylation [64]. To date, most studies have indicated that the function of the GacS/GacA system mainly depends on the Gac/Rsm signaling cascade pathway. After finding the signal molecules, it is also necessary to verify whether the signal transduction pathway acts directly through Gac/Rsm. Moreover, GacS/GacA homologous proteins in other strains are also continuously being identified. Chen et al. identified a homologous protein in Dickeya oryzae and named it TzpS/TzpA [146]. In addition, sRNAs and RNA binding proteins of the Rsm/Csr system are still being found [38,116]. The regulatory function of GacS/GacA in particular requires further exploration. For example, there is still a lack of evidence of an effect of regulating motility on antibiotic resistance. As research continues to intensify, we believe it will become easier to explore the physiological processes and regulatory networks in which GacS/GacA participates. Finally, the existing GacS/GacA-mediated applications are only at the laboratory test stage, and it is necessary to design in vivo experiments. The results of these future works should be of great significance for understanding the mechanism of bacterial regulation and identifying new applications targeting GacS/GacA.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (42176108, 31870023), the Young Taishan Scholars Program of Shandong Province (tsqn202103029) and the Fundamental Research Funds for the Central Universities (201941009).
References
- 1.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Gao R., Mack T.R., Stock A.M. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschiazzo A., Trajtenberg F. Two-component sensing and regulation: how do histidine kinases talk with response regulators at the molecular level? Annu. Rev. Microbiol. 2019;73:507–528. doi: 10.1146/annurev-micro-091018-054627. [DOI] [PubMed] [Google Scholar]
- 4.Gao R., Stock A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuichet K., Cantwell B.J., Zhulin I.B. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 2010;13:219–225. doi: 10.1016/j.mib.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latour X. The evanescent GacS signal. Microorganisms. 2020;8 doi: 10.3390/microorganisms8111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan Q., Lopes L.D., Shaffer B.T., Kidarsa T.A., Vining O., Philmus B., Song C., Stockwell V.O., Raaijmakers J.M., McPhail K.L., Andreote F.D., Chang J.H., Loper J.E. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio. 2018;9 doi: 10.1128/mBio.01845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gooderham W.J., Hancock R.E. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009;33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier J.D., Jones M.K., Thiaville P., Joseph J.L., Swain R.A., Krediet C.J., Gulig P.A., Teplitski M., Wright A.C. Role of GacA in virulence of Vibrio vulnificus. Microbiol. (Read.) 2010;156:3722–3733. doi: 10.1099/mic.0.043422-0. [DOI] [PubMed] [Google Scholar]
- 10.Heeb S., Haas D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 11.Ferreiro M.D., Gallegos M.T. Distinctive features of the Gac-Rsm pathway in plant-associated Pseudomonas. Environ. Microbiol. 2021;23:5670–5689. doi: 10.1111/1462-2920.15558. [DOI] [PubMed] [Google Scholar]
- 12.Vakulskas C.A., Potts A.H., Babitzke P., Ahmer B.M., Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 2015;79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortet P., Fochesato S., Bitbol A.-.F., Whitworth D.E., Lalaouna D., Santaella C., Heulin T., Achouak W., Barakat M. Evolutionary history expands the range of signaling interactions in hybrid multikinase networks. Sci. Rep. 2021;11:11763. doi: 10.1038/s41598-021-91260-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei X., Huang X., Tang L., Wu D., Xu Y. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J. Bacteriol. 2013;195:3387–3400. doi: 10.1128/JB.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez A.F., Barba-Ostria C., Silva-Jiménez H., Georgellis D. Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 2016;18:3210–3226. doi: 10.1111/1462-2920.13397. [DOI] [PubMed] [Google Scholar]
- 16.Gotoh Y., Eguchi Y., Watanabe T., Okamoto S., Doi A., Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010;13:232–239. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Bem A.E., Velikova N., Pellicer M.T., Baarlen P., Marina A., Wells J.M. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem. Biol. 2015;10:213–224. doi: 10.1021/cb5007135. [DOI] [PubMed] [Google Scholar]
- 18.Fihn C.A., Carlson E.E. Targeting a highly conserved domain in bacterial histidine kinases to generate inhibitors with broad spectrum activity. Curr. Opin. Microbiol. 2021;61:107–114. doi: 10.1016/j.mib.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zschiedrich C.P., Keidel V., Szurmant H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krell T., Lacal J., Busch A., Silva-Jiménez H., Guazzaroni M.-.E., Ramos J.L. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 21.Ishii E., Eguchi Y. Diversity in sensing and signaling of bacterial sensor histidine kinases. Biomolecules. 2021:11. doi: 10.3390/biom11101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung K., Fried L., Behr S., Heermann R. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 2012;15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Casino P., Rubio V., Marina A. The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Jacob-Dubuisson F., Mechaly A., Betton J.-.M., Antoine R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018;16:585–593. doi: 10.1038/s41579-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu C., Sun D., Zhu J., Liu W. Two-component signal transduction systems: a major strategy for connecting input stimuli to biofilm formation. Front. Microbiol. 2018;9:3279. doi: 10.3389/fmicb.2018.03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich J.J., Kinscherf T.G., Kitten T., Willis D.K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernestig A.K., Melefors O., Georgellis D. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 2001;276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 28.Haas D., Keel C. Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- 29.Haas D., Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 30.Zuber S., Carruthers F., Keel C., Mattart A., Blumer C., Pessi G., Gigot-Bonnefoy C., Schnider-Keel U., Heeb S., Reimmann C., Haas D. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 2003;16:634–644. doi: 10.1094/MPMI.2003.16.7.634. [DOI] [PubMed] [Google Scholar]
- 31.Lapouge K., Schubert M., Allain F.H., Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 32.Sonnleitner E., Haas D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 2011;91:63–79. doi: 10.1007/s00253-011-3332-1. [DOI] [PubMed] [Google Scholar]
- 33.Kusmierek M., Dersch P. Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr. Opin. Microbiol. 2018;41:58–67. doi: 10.1016/j.mib.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Humair B., Wackwitz B., Haas D. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 2010;76:1497–1506. doi: 10.1128/AEM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brencic A., McFarland K.A., McManus H.R., Castang S., Mogno I., Dove S.L., Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babitzke P., Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Brencic A., Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen K.H., Diaz M.R., Gode C.J., Wolfgang M.C., Yahr T.L. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol. 2018;200 doi: 10.1128/JB.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janssen K.H., Diaz M.R., Golden M., Graham J.W., Sanders W., Wolfgang M.C., Yahr T.L. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J. Bacteriol. 2018;200 doi: 10.1128/JB.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kay E., Dubuis C., Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Zhang B., Wu H., Wu X., Yan Q., Zhang L.Q. Pleiotropic effects of RsmA and RsmE proteins in Pseudomonas fluorescens 2P24. BMC Microbiol. 2020;20:191. doi: 10.1186/s12866-020-01880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreiro M.D., Behrmann L.V., Corral A., Nogales J., Gallegos M.T. Exploring the expression and functionality of the rsm sRNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 2021;18:1818–1833. doi: 10.1080/15476286.2020.1871217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francis V.I., Stevenson E.C., Porter S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Ventre I., Goodman A.L., Vallet-Gely I., Vasseur P., Soscia C., Molin S., Bleves S., Lazdunski A., Lory S., Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent F., Round A., Reynaud A., Bordi C., Filloux A., Bourne Y. Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ. Microbiol. 2010;12:1775–1786. doi: 10.1111/j.1462-2920.2010.02264.x. [DOI] [PubMed] [Google Scholar]
- 47.Jing X., Jaw J., Robinson H.H., Schubot F.D. Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins. 2010;78:1631–1640. doi: 10.1002/prot.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anantharaman V., Aravind L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics. 2003;4:34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang B.X., Wheeler K.M., Cady K.C., Lehoux S., Cummings R.D., Laub M.T., Ribbeck K. Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. Curr. Biol. 2021;31:90–102. doi: 10.1016/j.cub.2020.09.088. e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laskowski M.A., Kazmierczak B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 2006;74:4462–4473. doi: 10.1128/IAI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodman A.L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia Y., Xu C., Wang D., Weng Y., Jin Y., Bai F., Cheng Z., Kuipers O.P., Wu W. YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2020 doi: 10.1128/AEM.02171-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan Kaler K.M., Nix J.C., Schubot F.D. RetS inhibits Pseudomonas aeruginosa biofilm formation by disrupting the canonical histidine kinase dimerization interface of GacS. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francis V.I., Waters E.M., Finton-James S.E., Gori A., Kadioglu A., Brown A.R., Porter S.L. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 2018;9:2219. doi: 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mancl J.M., Ray W.K., Helm R.F., Schubot F.D. Helix cracking regulates the critical interaction between RetS and GacS in Pseudomonas aeruginosa. Structure. 2019;27:785–793. doi: 10.1016/j.str.2019.02.006. e785. [DOI] [PubMed] [Google Scholar]
- 56.Corley J.M., Intile P., Yahr T.L. Direct inhibition of RetS synthesis by RsmA contributes to homeostasis of the Pseudomonas aeruginosa Gac/Rsm signaling system. J. Bacteriol. 2022;204 doi: 10.1128/jb.00580-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong W., Chen L., Zhao J., Shen T., Surette M.G., Shen L., Duan K. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 2013;88:784–797. doi: 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- 58.Bhagirath A.Y., Pydi S.P., Li Y., Lin C., Kong W., Chelikani P., Duan K. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 2017;3:162–175. doi: 10.1021/acsinfecdis.6b00153. [DOI] [PubMed] [Google Scholar]
- 59.Hsu J.L., Chen H.C., Peng H.L., Chang H.Y. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bordi C., Lamy M.C., Ventre I., Termine E., Hachani A., Fillet S., Roche B., Bleves S., Mejean V., Lazdunski A., Filloux A. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jean-Pierre F., Tremblay J., Deziel E. Broth versus surface-grown cells: differential regulation of RsmY/Z small RNAs in Pseudomonas aeruginosa by the Gac/HptB system. Front. Microbiol. 2016;7:2168. doi: 10.3389/fmicb.2016.02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhagirath A.Y., Somayajula D., Li Y., Duan K. CmpX affects virulence in Pseudomonas aeruginosa through the Gac/Rsm signaling pathway and by modulating c-di-GMP levels. J. Membr. Biol. 2018;251:35–49. doi: 10.1007/s00232-017-9994-6. [DOI] [PubMed] [Google Scholar]
- 63.Broder U.N., Jaeger T., Jenal U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016;2:16184. doi: 10.1038/nmicrobiol.2016.184. [DOI] [PubMed] [Google Scholar]
- 64.Chambonnier G., Roux L., Redelberger D., Fadel F., Filloux A., Sivaneson M., de Bentzmann S., Bordi C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLos Genet. 2016;12 doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abisado R.G., Benomar S., Klaus J.R., Dandekar A.A., Chandler J.R. Bacterial quorum sensing and microbial community interactions. mBio. 2018;9 doi: 10.1128/mBio.02331-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S., Bassler B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whiteley M., Diggle S.P., Greenberg E.P. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laville J., Voisard C., Keel C., Maurhofer M., Defago G., Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakatsu Y., Matsui H., Yamamoto M., Noutoshi Y., Toyoda K., Ichinose Y. Quorum-dependent expression of rsmX and rsmY, small non-coding RNAs, in Pseudomonas syringae. Microbiol. Res. 2019;223-225:72–78. doi: 10.1016/j.micres.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Bertani I., Venturi V. Regulation of the N-acyl homoserine lactone-dependent quorum-sensing system in rhizosphere Pseudomonas putida WCS358 and cross-talk with the stationary-phase RpoS sigma factor and the global regulator GacA. Appl. Environ. Microbiol. 2004;70:5493–5502. doi: 10.1128/AEM.70.9.5493-5502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reimmann C., Beyeler M., Latifi A., Winteler H., Foglino M., Lazdunski A., Haas D. The global activator GacA of Pseudomonas aeruginosa PAO1 positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 72.Parsek M.R., Val D.L., Hanzelka B.L., Cronan J.E., Jr, Greenberg E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z., Nair S.K. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012;21:1403–1417. doi: 10.1002/pro.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schuster M., Lostroh C.P., Ogi T., Greenberg E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soto-Aceves M.P., Cocotl-Yanez M., Servin-Gonzalez L., Soberon-Chavez G. The Rhl quorum-sensing system is at the top of the regulatory hierarchy under phosphate-limiting conditions in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2021:203. doi: 10.1128/JB.00475-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kariminik A., Baseri-Salehi M., Kheirkhah B. Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immunol. Lett. 2017;190:1–6. doi: 10.1016/j.imlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 77.Schuster M., Greenberg E.P. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 78.Kay E., Humair B., Denervaud V., Riedel K., Spahr S., Eberl L., Valverde C., Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chancey S.T., Wood D.W., Pierson L.S., 3rd Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 1999;65:2294–2299. doi: 10.1128/AEM.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang D., Lee S.H., Seeve C., Yu J.M., Pierson L.S., 3rd, Pierson E.A. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiologyopen. 2013;2:505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu J.M., Wang D., Ries T.R., Pierson L.S., 3rd, Pierson E.A. An upstream sequence modulates phenazine production at the level of transcription and translation in the biological control strain Pseudomonas chlororaphis 30-84. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0193063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng X., de Bruijn I., van der Voort M., Loper J.E., Raaijmakers J.M. The Gac regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 2013;5:608–619. doi: 10.1111/1758-2229.12061. [DOI] [PubMed] [Google Scholar]
- 83.Parkins M.D., Ceri H., Storey D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 84.Mhedbi-Hajri N., Malfatti P., Pedron J., Gaubert S., Reverchon S., Van Gijsegem F. PecS is an important player in the regulatory network governing the coordinated expression of virulence genes during the interaction between Dickeya dadantii 3937 and plants. Environ. Microbiol. 2011;13:2901–2914. doi: 10.1111/j.1462-2920.2011.02566.x. [DOI] [PubMed] [Google Scholar]
- 85.Balasubramanian D., Schneper L., Kumari H., Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ge Y., Lee J.H., Liu J., Yang H.W., Tian Y., Hu B., Zhao Y. Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant Pathol. 2019;20:1217–1236. doi: 10.1111/mpp.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blumer C., Heeb S., Pessi G., Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christiansen L., Alanin K.S., Phippen C.B.W., Olsson S., Stougaard P., Hennessy R.C. Fungal-associated molecules induce key genes involved in the biosynthesis of the antifungal secondary metabolites nunamycin and nunapeptin in the biocontrol strain Pseudomonas fluorescens In5. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.01284-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen X.-.L., Wang Y., Wang P., Zhang Y.-.Z. Proteases from the marine bacteria in the genus Pseudoalteromonas: diversity, characteristics, ecological roles, and application potentials. Mar. Life Sci. Technol. 2020;2:309–323. doi: 10.1007/s42995-020-00058-8. [DOI] [Google Scholar]
- 90.Sacherer P., Defago G., Haas D. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 1994;116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 91.Yang S., Peng Q., Zhang Q., Yi X., Choi C.J., Reedy R.M., Charkowski A.O., Yang C.H. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937) Mol. Plant Microbe Interact. 2008;21:133–142. doi: 10.1094/MPMI-21-1-0133. [DOI] [PubMed] [Google Scholar]
- 92.Zhang B., Zhang Y., Liang F., Ma Y., Wu X. An extract produced by Bacillus sp. BR3 influences the function of the GacS/GacA two-component system in Pseudomonas syringae pv. tomato DC3000. Front. Microbiol. 2005;10 doi: 10.3389/fmicb.2019.02005. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moscoso J.A., Mikkelsen H., Heeb S., Williams P., Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 94.Jimenez P.N., Koch G., Thompson J.A., Xavier K.B., Cool R.H., Quax W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Malley M.R., Chien C.F., Peck S.C., Lin N.C., Anderson J.C. A revised model for the role of GacS/GacA in regulating type III secretion by Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2020;21:139–144. doi: 10.1111/mpp.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Winstanley C., O’Brien S., Brockhurst M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Valentini M., Gonzalez D., Mavridou D.A., Filloux A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018;41:15–20. doi: 10.1016/j.mib.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 98.Marden J.N., Diaz M.R., Walton W.G., Gode C.J., Betts L., Urbanowski M.L., Redinbo M.R., Yahr T.L., Wolfgang M.C. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Valentini M., Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 101.Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 102.Ryder C., Byrd M., Wozniak D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan J., Bassler B.L. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rasamiravaka T., Labtani Q., Duez P., El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015 doi: 10.1155/2015/759348. (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chambers J.R., Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21:39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mikkelsen H., Sivaneson M., Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 107.Moradali M.F., Ghods S., Rehm B.H.A. Lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ali-Ahmad A., Fadel F., Sebban-Kreuzer C., Ba M., Pelissier G.D., Bornet O., Guerlesquin F., Bourne Y., Bordi C., Vincent F. Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2017;7:11262. doi: 10.1038/s41598-017-11361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Irie Y., Starkey M., Edwards A.N., Wozniak D.J., Romeo T., Parsek M.R. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Navazo A., Barahona E., Redondo-Nieto M., Martinez-Granero F., Rivilla R., Martin M. Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2009;2:489–498. doi: 10.1111/j.1751-7915.2009.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feng T., Han Y., Li B., Li Z., Yu Y., Sun Q., Li X., Du L., Zhang X.-.H., Wang Y. Interspecies and intraspecies signals synergistically regulate Lysobacter enzymogenes twitching motility. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.01742-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez-Granero F., Rivilla R., Martin M. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 2006;72:3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barahona E., Navazo A., Garrido-Sanz D., Muriel C., Martinez-Granero F., Redondo-Nieto M., Martin M., Rivilla R. Pseudomonas fluorescens F113 can produce a second flagellar apparatus, which is important for plant root colonization. Front. Microbiol. 2016;7:1471. doi: 10.3389/fmicb.2016.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martinez-Granero F., Navazo A., Barahona E., Redondo-Nieto M., Rivilla R., Martin M. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS ONE. 2012;7:e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J.S., Kim Y.H., Anderson A.J., Kim Y.C. The sensor kinase GacS negatively regulates flagellar formation and motility in a biocontrol bacterium, Pseudomonas chlororaphis O6. Plant Pathol. J. 2014;30:215–219. doi: 10.5423/PPJ.NT.11.2013.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ferreiro M.D., Nogales J., Farias G.A., Olmedilla A., Sanjuan J., Gallegos M.T. Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2018;31:525–536. doi: 10.1094/MPMI-09-17-0232-R. [DOI] [PubMed] [Google Scholar]
- 117.Leon R., Espin G. flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiol. (Read.) 2008;154:1719–1728. doi: 10.1099/mic.0.2008/017665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lopez-Pliego L., Lara-Flores N., Molina-Romero D., May-Compan G., Carreno-Lopez R., Nunez C.E., Castaneda M. The GacS/A-Rsm pathway positively regulates motility and flagella synthesis in Azotobacter vinelandii. Curr. Microbiol. 2021;79:17. doi: 10.1007/s00284-021-02695-3. [DOI] [PubMed] [Google Scholar]
- 119.Song C., Kidarsa T.A., van de Mortel J.E., Loper J.E., Raaijmakers J.M. Living on the edge: emergence of spontaneous gac mutations in Pseudomonas protegens during swarming motility. Environ. Microbiol. 2016;18:3453–3465. doi: 10.1111/1462-2920.13288. [DOI] [PubMed] [Google Scholar]
- 120.Bhagirath A.Y., Li Y., Patidar R., Yerex K., Ma X., Kumar A., Duan K. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y.Y., Feng T., Wang Y. The role of bacterial signaling networks in antibiotics response and resistance regulation. Mar. Life Sci. Technol. 2022 doi: 10.1007/s42995-022-00126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brinkman F.S., Macfarlane E.L., Warrener P., Hancock R.E. Evolutionary relationships among virulence-associated histidine kinases. Infect. Immun. 2001;69:5207–5211. doi: 10.1128/IAI.69.8.5207-5211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tierney A.R., Rather P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019;14:533–552. doi: 10.2217/fmb-2019-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Linares J.F., Gustafsson I., Baquero F., Martinez J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davies J.A., Harrison J.J., Marques L.L., Foglia G.R., Stremick C.A., Storey D.G., Turner R.J., Olson M.E., Ceri H. The GacS sensor kinase controls phenotypic reversion of small colony variants isolated from biofilms of Pseudomonas aeruginosa PA14. FEMS Microbiol. Ecol. 2007;59:32–46. doi: 10.1111/j.1574-6941.2006.00196.x. [DOI] [PubMed] [Google Scholar]
- 126.Proctor R.A., von Eiff C., Kahl B.C., Becker K., McNamara P., Herrmann M., Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 127.Malone J.G. Role of small colony variants in persistence of Pseudomonas aeruginosa infections in cystic fibrosis lungs. Infect. Drug Resist. 2015;8:237–247. doi: 10.2147/IDR.S68214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schniederjans M., Koska M., Haussler S. Transcriptional and mutational profiling of an aminoglycoside-resistant Pseudomonas aeruginosa small-colony variant. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lai S., Tremblay J., Deziel E. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009;11:126–136. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 130.Shrout J.D., Chopp D.L., Just C.L., Hentzer M., Givskov M., Parsek M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 131.Dubern J.F., Lagendijk E.L., Lugtenberg B.J.J., Bloemberg G.V. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J. Bacteriol. 2005;187:5967–5976. doi: 10.1128/JB.187.17.5967-5976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dubern J.-.F., Bloemberg G.V. Influence of environmental conditions on putisolvins I and II production in Pseudomonas putida strain PCL1445. FEMS Microbiol. Lett. 2006;263:169–175. doi: 10.1111/j.1574-6968.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 133.Humair B., González N., Mossialos D., Reimmann C., Haas D. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J. 2009;3:955–965. doi: 10.1038/ismej.2009.42. [DOI] [PubMed] [Google Scholar]
- 134.Lindner F., Diepold A. Optogenetics in bacteria-applications and opportunities. FEMS Microbiol. Rev. 2022;46 doi: 10.1093/femsre/fuab055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cheng X., Pu L., Fu S., Xia A., Huang S., Ni L., Xing X., Yang S., Jin F. Engineering Gac/Rsm signaling cascade for optogenetic induction of the pathogenicity switch in. ACS Synth. Biol. 2021;10:1520–1530. doi: 10.1021/acssynbio.1c00075. [DOI] [PubMed] [Google Scholar]
- 136.Cerqueira G.M., Kostoulias X., Khoo C., Aibinu I., Qu Y., Traven A., Peleg A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014;210:46–55. doi: 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 137.De Silva P.M., Kumar A. Signal transduction proteins in Acinetobacter baumannii: role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019;10:49. doi: 10.3389/fmicb.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smiline Girija A.S. Delineating the immuno-dominant antigenic vaccine peptides against gacS-sensor kinase in Acinetobacter baumannii: an in silico investigational approach. Front. Microbiol. 2020;11:2078. doi: 10.3389/fmicb.2020.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saurav K., Costantino V., Venturi V., Steindler L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar. Drugs. 2017;15 doi: 10.3390/md15030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kalia V.C., Purohit H.J. Quenching the quorum sensing system: potential antibacterial drug targets. Crit. Rev. Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 141.Yu H., Liu Y., Yang F., Xie Y., Guo Y., Cheng Y., Yao W. The combination of hexanal and geraniol in sublethal concentrations synergistically inhibits quorum sensing in Pseudomonas fluorescens-in vitro and in silico approaches. J. Appl. Microbiol. 2022 doi: 10.1111/jam.15446. [DOI] [PubMed] [Google Scholar]
- 142.Jakobsen T.H., van Gennip M., Phipps R.K., Shanmugham M.S., Christensen L.D., Alhede M., Skindersoe M.E., Rasmussen T.B., Friedrich K., Uthe F., Jensen P.Ø., Moser C., Nielsen K.F., Eberl L., Larsen T.O., Tanner D., Høiby N., Bjarnsholt T., Givskov M. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jakobsen T.H., Warming A.N., Vejborg R.M., Moscoso J.A., Stegger M., Lorenzen F., Rybtke M., Andersen J.B., Petersen R., Andersen P.S., Nielsen T.E., Tolker-Nielsen T., Filloux A., Ingmer H., Givskov M. A broad range quorum sensing inhibitor working through sRNA inhibition. Sci. Rep. 2017;7:9857. doi: 10.1038/s41598-017-09886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hirakawa H., Kurushima J., Hashimoto Y., Tomita H. Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics. 2020:9. doi: 10.3390/antibiotics9100635. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goswami M., Espinasse A., Carlson E.E. Disarming the virulence arsenal of Pseudomonas aeruginosa by blocking two-component system signaling. Chem. Sci. 2018;9:7332–7337. doi: 10.1039/c8sc02496k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen Y., Li Y., Zhu M., Lv M., Liu Z., Chen Z., Huang Y., Gu W., Liang Z., Chang C., Zhou J., Zhang L.-.H., Liu Q. The GacA-GacS type two-component system modulates the pathogenicity of Dickeya oryzae EC1 mainly by regulating the production of zeamines. Mol. Plant Microbe Interact. 2022 doi: 10.1094/MPMI-11-21-0292-R. [DOI] [PubMed] [Google Scholar]
- 147.Fadel F., Bassim V., Francis V.I., Porter S.L., Botzanowski T., Legrand P., Perez M.M., Bourne Y., Cianférani S., Vincent F. Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS. Structure. 2022 doi: 10.1016/j.str.2022.06.002. [DOI] [PubMed] [Google Scholar]
- 148.Willett J.W., Crosson S. Atypical modes of bacterial histidine kinase signaling. Mol. Microbiol. 2017;103:197–202. doi: 10.1111/mmi.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.El Mouali Y., Esteva-Martinez G., Garcia-Pedemonte D., Balsalobre C. Differential regulation of CsrC and CsrB by CRP-cAMP in Salmonella enterica. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.570536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Broberg M., Lee G.W., Nykyri J., Lee Y.H., Pirhonen M., Palva E.T. The global response regulator ExpA controls virulence gene expression through RsmA-mediated and RsmA-independent pathways in Pectobacterium wasabiae SCC3193. Appl. Environ. Microbiol. 2014;80:1972–1984. doi: 10.1128/AEM.03829-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vakulskas C.A., Pannuri A., Cortés-Selva D., Zere T.R., Ahmer B.M., Babitzke P., Romeo T. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol. Microbiol. 2014;92:945–958. doi: 10.1111/mmi.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Faucher S.P., Shuman H.A. Small regulatory RNA and Legionella pneumophila. Front. Microbiol. 2011;2:98. doi: 10.3389/fmicb.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kitten T., Kinscherf T.G., McEvoy J.L., Willis D.K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 154.Liu J., Yu M., Ge Y., Tian Y., Hu B., Zhao Y. The RsmA RNA-binding proteins in Pseudomonas syringae exhibit distinct and overlapping roles in modulating virulence and survival under different nutritional conditions. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.637595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Camacho M.I., Alvarez A.F., Chavez R.G., Romeo T., Merino E., Georgellis D. Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J. Bacteriol. 2015;197:983–991. doi: 10.1128/JB.02325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zere T.R., Vakulskas C.A., Leng Y., Pannuri A., Potts A.H., Dias R., Tang D., Kolaczkowski B., Georgellis D., Ahmer B.M., Romeo T. Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hammer B.K., Tateda E.S., Swanson M.S. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 158.Sahr T., Bruggemann H., Jules M., Lomma M., Albert-Weissenberger C., Cazalet C., Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mendis N., McBride P., Saoud J., Mani T., Faucher S.P. The LetA/S two-component system regulates transcriptomic changes that are essential for the culturability of Legionella pneumophila in water. Sci. Rep. 2018;8:6764. doi: 10.1038/s41598-018-24263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wong S.M., Carroll P.A., Rahme L.G., Ausubel F.M., Calderwood S.B. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 1998;66:5854–5861. doi: 10.1128/IAI.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lenz D.H., Miller M.B., Zhu J., Kulkarni R.V., Bassler B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 162.Butz H.A., Mey A.R., Ciosek A.L., Payne S.M. Vibrio cholerae CsrA directly regulates varA to increase expression of the three nonredundant Csr small RNAs. mBio. 2019;10 doi: 10.1128/mBio.01042-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lemos Rocha L.F., Peters K., Biboy J., Depelteau J.S., Briegel A., Vollmer W., Blokesch M. The VarA-CsrA regulatory pathway influences cell shape in Vibrio cholerae. PLos Genet. 2022;18 doi: 10.1371/journal.pgen.1010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Altier C., Suyemoto M., Ruiz A.I., Burnham K.D., Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 2000;35:635–646. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 165.Teplitski M., Goodier R.I., Ahmer B.M. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int. J. Med. Microbiol. 2006;296:449–466. doi: 10.1016/j.ijmm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 166.Teplitski M., Al-Agely A., Ahmer B.M.M. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiol. (Read.) 2006;152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- 167.Eriksson A.R., Andersson R.A., Pirhonen M., Palva E.T. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 1998;11:743–752. doi: 10.1094/MPMI.1998.11.8.743. [DOI] [PubMed] [Google Scholar]
- 168.Hyytiainen H., Montesano M., Palva E.T. Global regulators ExpA (GacA) and KdgR modulate extracellular enzyme gene expression through the RsmA-rsmB system in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 2001;14:931–938. doi: 10.1094/MPMI.2001.14.8.931. [DOI] [PubMed] [Google Scholar]
- 169.Cui Y., Chatterjee A., Chatterjee A.K. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant Microbe Interact. 2001;14:516–526. doi: 10.1094/MPMI.2001.14.4.516. [DOI] [PubMed] [Google Scholar]
- 170.Valente R.S., Xavier K.B. The Trk potassium transporter is required for RsmB-mediated activation of virulence in the phytopathogen Pectobacterium wasabiae. J. Bacteriol. 2016;198:248–255. doi: 10.1128/JB.00569-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Castaneda M., Sanchez J., Moreno S., Nunez C., Espin G. The global regulators GacA and sigma(S) form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 2001;183:6787–6793. doi: 10.1128/JB.183.23.6787-6793.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hernandez-Eligio A., Moreno S., Castellanos M., Castaneda M., Nunez C., Muriel-Millan L.F., Espin G. RsmA post-transcriptionally controls PhbR expression and polyhydroxybutyrate biosynthesis in Azotobacter vinelandii. Microbiol. (Read.) 2012;158:1953–1963. doi: 10.1099/mic.0.059329-0. [DOI] [PubMed] [Google Scholar]
- 173.Li J., Yang Y., Dubern J.F., Li H., Halliday N., Chernin L., Gao K., Camara M., Liu X. Regulation of GacA in Pseudomonas chlororaphis strains shows a niche specificity. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0137553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Y., Wang Z., Bilal M., Hu H., Wang W., Huang X., Peng H., Zhang X. Enhanced fluorescent siderophore biosynthesis and loss of phenazine-1-carboxamide in phenotypic variant of Pseudomonas chlororaphis HT66. Front. Microbiol. 2018;9:759. doi: 10.3389/fmicb.2018.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kang B.R., Anderson A.J., Kim Y.C. Hydrogen cyanide produced by Pseudomonas chlororaphis O6 is a key aphicidal metabolite. Can. J. Microbiol. 2019;65:185–190. doi: 10.1139/cjm-2018-0372. [DOI] [PubMed] [Google Scholar]