Abstract
Importance
Understanding the microbial diversity within the gastrointestinal tract of wild Korean water deer (KWD; Hydropotes inermis argyropus) is essential for gaining insights into their health and ecological interactions.
Objective
This study aims to isolate and identify aerobic and anaerobic bacterial species in the feces of wild KWD.
Methods
Fecal samples were collected from 55 wild KWD of varying age and sex. Aerobic bacteria were cultured at 37°C for 24–48 h under standard conditions, whereas anaerobic bacteria were cultured at 37°C for 48–72 h in an anaerobic environment. Bacterial identification was conducted using DNA extraction and polymerase chain reaction amplification targeting the 16S rRNA gene.
Results
The predominant aerobic bacteria identified belonged to the Firmicutes (58.18%) and Proteobacteria (41.82%) phyla, with Escherichia coli (31.82%) and Bacillus cereus (31.82%) being the most common species. Among anaerobic bacteria, most belonged to the Firmicutes (71.03%), Proteobacteria (27.10%), and Fusobacteriota (1.87%) phyla, with Paraclostridium bifermentans (28.97%) and E. coli (22.43%) being the most prevalent species. Other frequently identified anaerobic species were Fusobacterium varium, Lactococcus garvieae, Terrisporobacter glycolicus, Enterococcus faecalis, and Clostridium sporogenes.
Conclusions and Relevance
Our findings indicate a diverse microbial community in the feces of water deer, offering valuable insights into their gut microbiota and its potential implications for health and ecology.
Keywords: Hydropotes inermis argyropus, fecal microbiota, aerobic bacteria, anaerobic bacteria, 16S rRNA gene
INTRODUCTION
The Korean water deer (KWD) (Hydropotes inermis argyropus) is a significant and intriguing species within the Cervidae family [1]. Native to the Korean peninsula and parts of China, this herbivorous mammal thrives in marshy environments as wetland vegetation ensures food supply [2]. Despite its importance in the ecosystem, limited knowledge exists about the gut microbiota of KWD. These microorganisms are associated with deer health, nutrition, and habitat adaptations. The gastrointestinal tract of herbivorous mammals is a complex ecosystem with diverse microbial communities essential for digestion, metabolism, and immune function [3]. The fecal microbiota, which comprises numerous microorganisms, plays a crucial role in nutrient processing, maintaining host health, and defending against pathogens [4]. Understanding the composition and dynamics of the gut microbiome in KWD is crucial for comprehending their dietary adaptations, health status, and potential ecological impact [5]. Aerobic and anaerobic bacteria are essential microbial components, each serving different metabolic and protective functions. Aerobic bacteria rely on oxygen for survival, oxidize complex carbohydrates and proteins, produce vital vitamins, and offer protection against harmful microbes. Anaerobic bacteria thrive in oxygen-free environments, playing a pivotal role in fermenting plant materials, generating short-chain fatty acids (SCFAs), and maintaining gut health [6]. These bacteria assist in breaking down complex plant polysaccharides into absorbable nutrients, enabling herbivores to adapt to their specialized diets [7]. The gut microbiota influences host physiology and immune response, highlighting its broader ecological significance [4]. While the gut microbiota of various deer species, specifically European and North American species, such as red deer (Cervus elaphus) and white-tailed deer (Odocoileus virginianus), have been extensively studied [8,9], a notable gap exists in the literature concerning the gut microbial communities of Asian deer species, particularly the KWD. This lack of information limits our understanding of their gut health, dietary adaptations, and potential responses to environmental changes. Investigating the bacterial diversity in KWD feces provides insights into their specific digestive physiology, possible symbiotic relationships with particular bacterial taxa, and the ecological importance of their microbial inhabitants [10]. Furthermore, determining KWD fecal microbiota composition could aid public health and wildlife conservation efforts by providing insights into zoonotic risks and the spread of pathogens among domestic animals, wildlife, and humans [11].
This study aimed to isolate and identify aerobic and anaerobic bacterial species in the feces of wild KWD. This study represents the first comprehensive analysis of aerobic and anaerobic bacterial communities in the feces of wild KWD, establishing a foundational dataset for future research on wildlife microbiology and conservation.
METHODS
Ethics approval
The Institutional Animal Care and Use Committee at Chungbuk National University (CBNU) in the Republic of Korea approved this research (approved number: CBNUA-24-0024-01). All experimental procedures involving animals were conducted in strict accordance with applicable guidelines and regulations.
Sample collection
Fifty-five fecal samples, with an average weight of 10 g, were aseptically collected from the rectum of wild KWD at the Wildlife Center of Chungbuk, South Korea, between June and October 2023, based on availability and ethical considerations. The samples were obtained from male and female deer of varying ages. All animals were rescued and showed no signs of infectious diseases. The samples were carefully collected in sterile containers to avoid contamination and promptly transferred on ice to the Laboratory of Veterinary Laboratory Medicine at CBNU, South Korea. Upon arrival, the samples were immediately prepared for bacterial culturing.
Bacterial culture
Each sample was inoculated into Mueller-Hinton broth (Oxoid, United Kingdom) to analyze aerobic and anaerobic bacteria in the fecal samples. The broth containing aerobic bacteria was incubated at 37°C for 24 h in an aerobic environment to promote the growth of oxygen-dependent bacterial species. The broth for anaerobic bacteria was incubated at 37°C for 48–72 h in an anaerobic environment (oxygen-free environment) using an anaerobic jar equipped with a BD GasPak system (Becton, Dickinson and Company, USA) to increase the growth of bacteria that thrive without oxygen. After initial incubation, the samples were streaked onto sheep blood agar plates (KisanBio, Korea) via sterile inoculating loops to further isolate and cultivate bacterial colonies. The aerobic plates were incubated at 37°C for 24–48 h under aerobic conditions, while the anaerobic plates were incubated at 37°C for 48–72 h under anaerobic conditions. After the respective incubation periods, colonies with unique morphologies were selected from the aerobic and anaerobic plates and then subcultured onto fresh sheep blood agar plates using the same techniques to purify the cultures, ensuring each plate cultivated a single type of bacteria. The resulting pure colonies were prepared for DNA extraction.
DNA extraction and polymerase chain reaction (PCR) amplification
The genomic DNA was extracted from pure cultured isolates using a MagPurix Pathogen/Viral NAs B kit and the MagPurix 12s automated nucleic acid purification system (Zinexts Life Science, Taiwan), following the manufacturer’s instructions and protocol. The extracted DNA was stored at −80°C until PCR analysis.
For PCR analysis, the 16S rRNA gene from the isolated DNA was amplified using bacterial universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GG TTA CCT TGT TAC GAC TT-3′). A total volume of 50 µL used for the PCR consisted of 25 µL of 2× PCRBIO Ultra Mix Red (PCR Biosystems, UK), 2 µL of each forward and reverse primer (10 μM), 16 µL of distilled water, and 5 µL of template DNA. The PCR amplification conditions were as follows: 95°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 40 sec. A final extension step was performed at 72°C for 10 min. The PCR products were then separated on a 1.5% agarose gel and visualized under UV light after staining with EcoDye (BIOFACT, Korea).
Sequencing
The nucleotide sequences of the PCR products were analyzed using SolGent (Korea). Subsequently, bioinformatics tools and databases, such as Basic Local Alignment Search Tool, were used to compare and identify bacterial species by assessing the sequence similarities with known reference sequences available in public databases.
RESULTS
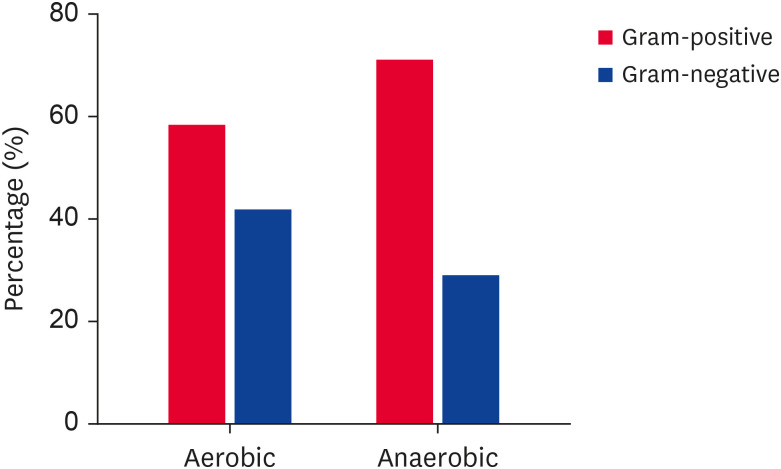
The study used 217 bacterial isolates obtained from the feces of wild KWD. These isolates included 110 aerobic bacterial isolates from 14 species and 107 anaerobic bacterial isolates belonging to 10 species. Of the aerobic isolates, 58.18% were gram-positive bacteria, and 41.81% were gram-negative bacteria. The anaerobic isolates were 71.03% gram-positive bacteria and 28.97% gram-negative bacteria (Fig. 1).
Fig. 1. Percentages of gram-negative and gram-positive bacteria isolated from the feces of wild Korean water deer (Hydropotes inermis argyropus).
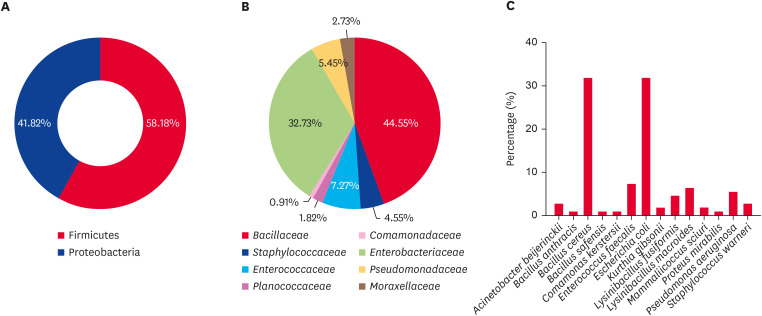
The aerobic isolates were predominantly from the phylum Firmicutes (58.18%), with Proteobacteria accounting for the remaining 41.82% (Fig. 2A). Within Firmicutes, the Bacillaceae family was the most prevalent, constituting 44.55% of the aerobic isolates (Fig. 2B). Bacillus cereus was the most commonly isolated species in the Bacillaceae family, representing 31.82% of all aerobic isolates. Other notable species in this family included Bacillus anthracis (0.91%), Lysinibacillus fusiformis (4.55%), Lysinibacillus macroides (6.36%), and Bacillus safensis (0.91%) (Fig. 2C). The presence of B. anthracis is particularly noteworthy due to its potential as a zoonotic pathogen. The Staphylococcaceae family accounted for 4.55% of the aerobic isolates, with Staphylococcus warneri and Mammaliicoccus sciuri at 2.73% and 1.82%, respectively. Enterococcus faecalis, the sole representative of the Enterococcaceae family, constituted 7.27% of the total aerobic isolates. Kurthia gibsonii from the Planococcaceae family accounted for 1.82% of all the isolates. The Enterobacteriaceae family was the most prevalent within the phylum Proteobacteria, constituting 32.73% of the aerobic isolates. Escherichia coli was the dominant species within this family, representing 31.82% of the aerobic isolates, while Proteus mirabilis accounted for 0.91%. Pseudomonas aeruginosa, a member of the Pseudomonadaceae family, comprised 5.45% of the isolates. The Comamonadaceae family was represented by a single isolate of Comamonas kerstersii (0.91%), and the Moraxellaceae family was represented by Acinetobacter beijerinckii (2.73%).
Fig. 2. Percentages of aerobic bacteria at the (A) phylum, (B) family, and (C) species level from the feces of wild Korean water deer (Hydropotes inermis argyropus).
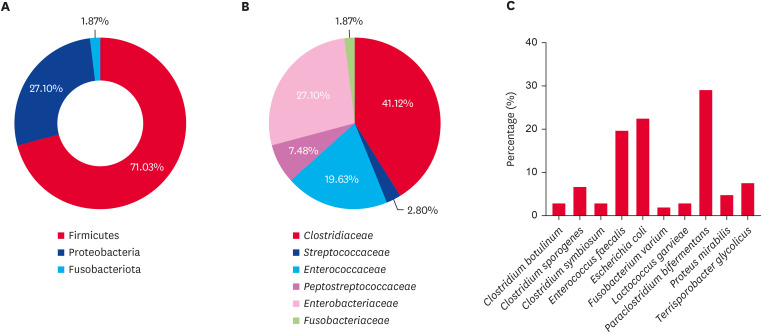
Under anaerobic conditions, most isolates belonged to the phylum Firmicutes (71.03%), followed by Proteobacteria (27.10%) and Fusobacteriota (1.87%) (Fig. 3A). Within Firmicutes, the Clostridiaceae family was the most common, accounting for 41.12% of the anaerobic isolates (Fig. 3B). Paraclostridium bifermentans, the most frequently isolated species within this family, constituted 28.97% of the total anaerobic isolates. Other species included Clostridium symbiosum (2.80%), Clostridium botulinum (2.80%), and Clostridium sporogenes (6.54%) (Fig. 3C). The Enterococcaceae family, represented solely by E. faecalis, comprised 19.63% of the anaerobic isolates. Another notable family within Firmicutes was Peptostreptococcaceae, with Terrisporobacter glycolicus accounting for 7.48% of the isolates. The Streptococcaceae family included Lactococcus garvieae, which comprised 2.80% of the anaerobic isolates. Within the phylum Proteobacteria, the Enterobacteriaceae family was the most prevalent, with E. coli as the dominant species, accounting for 22.43% of the anaerobic isolates, while P. mirabilis accounted for 4.67%. The phylum Fusobacteriota consisted of the family Fusobacteriaceae, comprising 1.87% of the anaerobic isolates, with Fusobacterium varium being the identified species.
Fig. 3. Percentages of anaerobic bacteria at the (A) phylum, (B) family, and (C) species level from the feces of wild Korean water deer (Hydropotes inermis argyropus).
The identities of the isolated strains were confirmed by sequencing and comparison with the National Center for Biotechnology Information database. Each strain showed high query coverage and similarity, typically ranging from 99% to 100%, ensuring accurate identification. The obtained sequences were submitted to the GenBank database and can be accessed under the accession numbers listed in Tables 1 and 2.
Table 1. Identification of aerobic bacterial isolates by nucleotide BLAST search of NCBI database using 16S rRNA gene sequences from feces of wild Korean water deer (Hydropotes inermis argyropus).
| Serial No. | Isolate No. (strain designation) | Identified species | Length of sequences (bp) | Query coverage | Identity match | GenBank accession No. of corresponding sequence | GenBank accession No. of studied strains |
|---|---|---|---|---|---|---|---|
| 1 | CBNU_2B1 | Lysinibacillus macroides | 1,362 | 100% | 99.93% | MT197307 | PP600166 |
| 2 | CBNU_54B1 | Lysinibacillus fusiformis | 1,394 | 100% | 100% | MN543848 | PP600167 |
| 3 | CBNU_18B1 | Bacillus cereus | 1,388 | 100% | 100% | MT545090 | PP594158 |
| 4 | CBNU_41B2 | Bacillus anthracis | 1,384 | 100% | 99.93% | CP054816 | PP594159 |
| 5 | CBNU_50B1 | Bacillus safensis | 1,393 | 100% | 100% | MT533923 | PP660955 |
| 6 | CBNU_55B1 | Staphylococcus warneri | 1,406 | 100% | 100% | MT642942 | PP600205 |
| 7 | CBNU_9B1 | Mammaliicoccus sciuri | 1,389 | 100% | 100% | MT550814 | PP600220 |
| 8 | CBNU_16B2 | Enterococcus faecalis | 1,401 | 100% | 100% | MT611694 | PP600130 |
| 9 | CBNU_45B1 | Kurthia gibsonii | 1,386 | 100% | 99.93% | MN960342 | PP598129 |
| 10 | CBNU_5B1 | Comamonas kerstersii | 1,372 | 100% | 100% | PP600214 | PP600214 |
| 11 | CBNU_7B | Proteus mirabilis | 1,382 | 100% | 100% | PP600216 | PP600216 |
| 12 | CBNU_23B2 | Escherichia coli | 1,343 | 100% | 100% | MN557058 | PP597403 |
| 13 | CBNU_8B2 | Pseudomonas aeruginosa | 1,378 | 100% | 99.93% | MT646431 | PP600217 |
| 14 | CBNU_37B2 | Acinetobacter beijerinckii | 1,362 | 100% | 100% | JN644620 | PP598609 |
BLAST, Basic Local Alignment Search Tool; NCBI, National Center for Biotechnology Information.
Table 2. Identification of anaerobic bacterial isolates by nucleotide BLAST search of NCBI database using 16S rRNA gene sequences from feces of wild Korean water deer (Hydropotes inermis argyropus).
| Serial No. | Isolate No. (strain designation) | Identified species | Length of sequences (bp) | Query coverage | Identity match | GenBank accession No. of corresponding sequence | GenBank accession No. of studied strains |
|---|---|---|---|---|---|---|---|
| 1 | CBNU_1NB1 | Paraclostridium bifermentans | 1,348 | 100% | 99.78% | MN758863 | PP600132 |
| 2 | CBNU_12NB1 | Clostridium symbiosum | 1,365 | 100% | 99.85% | LC515566 | PP660969 |
| 3 | CBNU_30NB1 | Clostridium botulinum | 1,348 | 100% | 100% | MF040752 | PP661238 |
| 4 | CBNU_11NB1 | Clostridium sporogenes | 1,358 | 100% | 100% | MT356160 | PP600302 |
| 5 | CBNU_10NB1 | lactococcus garvieae | 1,413 | 100% | 99.65% | MF354518 | PP594165 |
| 6 | CBNU_21NB1 | Enterococcus faecalis | 1,399 | 100% | 100% | MT611694 | PP600303 |
| 7 | CBNU_40NB1 | Terrisporobacter glycolicus | 1,345 | 100% | 100% | OQ147126 | PP775802 |
| 8 | CBNU_23NB3 | Escherichia coli | 1,381 | 100% | 100% | CP146681 | PP600137 |
| 9 | CBNU_35NB1 | Proteus mirabilis | 1,327 | 99% | 99.92% | MW217053 | PP600133 |
| 10 | CBNU_55NB2 | Fusobacterium varium | 1,346 | 100% | 100% | LR134390 | PP600301 |
BLAST, Basic Local Alignment Search Tool; NCBI, National Center for Biotechnology Information.
DISCUSSION
This study provides a comprehensive understanding of the fecal microbiota composition of wild KWD. The bacterial community encompasses a wide variety of aerobic and anaerobic bacteria. The coexistence of beneficial and potentially harmful bacterial species highlights the complex microbial ecosystem within the digestive system of these herbivorous animals.
The analysis of aerobic bacterial diversity in the feces of wild KWD provides valuable insights into the gut microbiota. This investigation identified aerobic bacterial isolates, mostly belonging to the phyla Firmicutes and Proteobacteria (Fig. 2A). Previous studies conducted in Europe [12] and Asia [13] extensively documented these phyla in studies involving the gastrointestinal microbiota of deer species. Firmicutes play a vital role in the digestion and absorption of nutrients in herbivores, such as the KWD, by breaking down intricate plant polysaccharides into fermentable sugars [14,15].
The Bacillaceae family is the most abundant within the phylum Firmicutes, with B. cereus as the primary species within this family (Fig. 2B and C). B. cereus is known for its wide range of functions, including the cycling of nutrients and the decomposition of organic materials in the environment [16,17]. Although B. cereus is commonly linked to foodborne illnesses, it can aid organic matter decomposition and enhance nutrient absorption in the deer gut. These results align with findings from earlier studies, which have shown that Bacillus species help maintain overall health and gut function by assisting in the breakdown of complex compounds [18,19]. L. macroides and L. fusiformis are notable species in the Bacillaceae family recognized for their capacity to break down intricate organic compounds [20]. The analysis identified B. anthracis in small amounts, indicating its minimal pathogenic effect under typical circumstances. B. safensis, a member of the Bacillaceae family, is known to withstand environmental stressors, suggesting varying environmental exposure in deer [21,22].
The Staphylococcaceae family, although less common, includes species such as S. warneri and M. sciuri. Although these species are typically harmless bacteria found on the skin and mucosal surfaces, they can sometimes lead to infections. Their detection in fecal samples indicates their temporary acquisition from environmental sources or through interaction with other animals. Previous studies have demonstrated Staphylococci in various environmental samples, suggesting that their presence in deer feces could be due to environmental exposure [2,23,24]. The family Enterococcaceae, represented by only E. faecalis, has a well-documented function in maintaining gut health. It helps balance the intestinal microbiota and inhibits the growth of harmful bacteria through competition and the production of bacteriocins. This observation is consistent with findings in domestic and wild herbivores, where Enterococcus species are crucial to gut health [25]. Although identified in only two strains, K. gibsonii from the family Planococcaceae demonstrated microbial diversity and the potential presence of less-studied bacterial groups in the deer gut.
The phylum Proteobacteria accounted for 41.82% of all aerobic isolates, with the family Enterobacteriaceae being ubiquitous. E. coli, the predominant species in this family, is commonly found in the digestive system of various mammals and plays a vital role in producing vitamin K and preventing the growth of harmful bacteria. Nevertheless, the widespread occurrence of this phenomenon also increases the likelihood of pathogenic strains that could have detrimental effects on the well-being of the deer population and their surroundings [26]. P. mirabilis, a less common member of the Enterobacteriaceae family, is recognized for its involvement in urinary tract infections across different hosts. Its presence in feces increases the possibility of temporary colonization or environmental contamination [27]. As a metabolically adaptable bacterium, P. aeruginosa belongs to the family Pseudomonadaceae and may thrive in various settings, such as soil, water, and host tissues. Its discovery in fecal samples suggests that deer interact with many habitats and are exposed to the environment [28]. As a member of the Moraxellaceae family, A. beijerinckii is renowned for its resistance to environmental stressors and ability to decompose organic matter. Their presence in fecal samples highlights deer contact with soil and plant material, enhancing gut microbiome diversity [29]. The aerobic bacterial population has become more complex following the identification of C. kerstersii, a single strain of the family Comamonadaceae. This bacterium is commonly observed in water and soil environments, indicating that deer may encounter aquatic ecosystems [30].
Overall, the fecal aerobic bacterial diversity in wild KWD was primarily due to Firmicutes and Proteobacteria, which indicated herbivorous dietary habits and environmental interaction. The presence of beneficial and potentially harmful bacteria emphasizes the complex nature of deer gut microbiota and its vital role in their overall health and ecology.
The feces of wild KWD contain an anaerobic bacterial community that plays a vital role in their digestion and overall health. Our study aimed to isolate anaerobic bacterial species, predominantly from the phylum Firmicutes (Fig. 3A). Previous studies have shown that Firmicutes play crucial roles in the digestion of plant materials and SCFA production needed for energy metabolism and gut health in herbivorous mammals [31].
The family Clostridiaceae was highly prevalent within the phylum Firmicutes (Fig. 3B). Among the anaerobic bacterial isolates, P. bifermentans, the most abundant species (Fig. 3C), is recognized for its ability to ferment various carbohydrates and proteins, resulting in the production of SCFAs such as butyrate, acetate, and propionate. These metabolites help preserve a healthy gut lining and serve as a host energy source [32]. Research on other herbivores, such as cows and horses, has also emphasized the prevalence of Clostridiaceae and their importance in fermentative digestion [33]. Additional notable species in Clostridiaceae are C. sporogenes, C. symbiosum, and C. botulinum. C. sporogenes produces butyrate, a SCFA with anti-inflammatory properties, and supports gut health. C. symbiosum and C. botulinum are known for their unique metabolic capabilities, although they are less common than other bacteria. C. botulinum is specifically recognized for its neurotoxicity, raising concerns over its potential as a pathogen, even in small amounts. These findings align with previous research on wildlife species, indicating that Clostridium can positively and negatively affect gut interactions [34,35].
E. faecalis, a member of the Enterococcaceae family, is a versatile facultative anaerobe capable of thriving in aerobic and anaerobic environments. This adaptability is crucial to regulating gut homeostasis. Furthermore, E. faecalis contributes to gut microbiota balance and possesses probiotic properties, including the production of bacteriocins, which inhibit the growth of harmful bacteria. Research on domestic animals and humans has consistently demonstrated the beneficial effects of Enterococcus species in the gut [25].
T. glycolicus, accounting for 7.48% of the anaerobic isolates, represents the Peptostreptococcaceae family, another significant group within Firmicutes. T. glycolicus is known for its fermentative metabolism, which aids the breakdown of complex polysaccharides and assists digestion in herbivores. The presence of this bacterium highlights the specialized fermentation activities occurring in the anaerobic environment of the herbivorous gut [36].
The presence of L. garvieae from the Streptococcaceae family highlights the wide range of lactic acid bacteria in the deer gut. L. garvieae is commonly linked with dairy products but is also found in the gastrointestinal tract, aiding lactic acid fermentation and gut health. Other animal studies have demonstrated the importance of lactic acid bacteria, such as Lactococcus species, in maintaining a healthy gut microbiota [37].
Phylum Proteobacteria, with the family Enterobacteriaceae comprising the majority of this group, and E. coli are the most prevalent species within this family (Fig. 3C). The adaptability and importance of E. coli in the gut microbiota is underscored by its ability to switch between aerobic and anaerobic existence. Although it is usually a commensal organism, certain E. coli strains can become pathogenic and pose health risks. Research on herbivores and other animals has also emphasized the significant impact of E. coli on gut health and disease [26]. P. mirabilis, another member of the Enterobacteriaceae family, is known to produce urease, which can modulate the pH and affect gut microbial balance through its alkalizing effect. Its presence in feces suggests potential environmental contamination or temporary colonization. Research on other animals has shown that Proteus species are also found in the gastrointestinal tract and can influence gut health through their metabolic activities [27].
The phylum Fusobacteriota, consisting of F. varium from the Fusobacteriaceae family, produces SCFAs such as butyrate and propionate during the fermentation of proteins and amino acids in the gastrointestinal tract. The presence of these microorganisms, even in low quantities, underscores the array of metabolic processes occurring within the deer gut. Research on humans and other animals suggests that Fusobacterium species play a role in maintaining gut health and contribute to inflammatory processes [38].
The feces of wild KWD contain diverse and well-suited microbial ecosystems comprising anaerobic bacteria. Firmicutes, Proteobacteria, and Fusobacteriota are involved in the fermentation of dietary fibers and proteins. These processes lead to the production of SCFAs, which offer advantages to the host. The microbial community in the KWD gut is crucial for nutrient absorption, energy production, and overall gut health. These findings support the dietary habits and ecological adaptability of the deer.
In addition, certain pathogenic bacteria identified in this study, including B. anthracis, C. botulinum, and E. coli, have zoonotic importance [39,40,41], as highlighted by the One Health perspective. These pathogens pose risks to both wildlife and human populations, emphasizing the need for continuous monitoring to prevent potential disease transmission between animals and humans.
This investigation relied on a robust culturing and molecular approach; however, each has limitations. Some species are fussy or unculturable under laboratory conditions; therefore, culture may not represent the entire microbial ecosystem. Although more detailed, molecular approaches may fail to distinguish between living and dead bacteria, influencing functional microbiota interpretations. Metagenomic sequencing and transcriptomics may help understand microbial community functions and host relationships. Identifying and functionally characterizing unculturable bacteria might improve our understanding of the microbial environment. This study did not consider seasonal microbial composition changes. Microbial communities change during the year based on deer food and health; therefore, the results are only snapshots. Future research should include more samples from other seasons and locations. This strategy would help explain spatial and temporal changes in the microbial flora and their impact on deer health.
In conclusion, this study examined the bacterial diversity in the feces of wild KWD using culture-based and molecular techniques. Firmicutes and Proteobacteria were the two most prevalent phyla identified in aerobic and anaerobic isolates. B. cereus was the most prevalent species among the aerobic bacteria, while P. bifermentans dominated the anaerobic isolates. The bacterial community in KWD is closely linked to their herbivorous diet, which is essential for digestion, nutrient absorption, and overall gut health. Future research should incorporate advanced molecular techniques such as metagenomic sequencing for a more comprehensive understanding of microbial communities and their functional roles.
ACKNOWLEDGMENTS
I would like to extend my sincere gratitude to Dr. Jongseung Kim, Veterinary Doctor and Jooyoung Park, Manager at the Wildlife Center of Chungbuk, for their invaluable assistance in the collection of fecal samples from Korean water deer for this study. Their expertise and dedication significantly contributed to the successful completion of our research.
Footnotes
Funding: This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Companion Animal Life Cycle Industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (322103-5).
Conflict of Interest: The authors declare no conflicts of interest.
Data Availability Statement: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
- Conceptualization: Islam MA, Kim S, Park S, Taili I, Jeong DH, Na KJ.
- Data curation: Islam MA, Kim S, Na KJ.
- Formal analysis: Islam MA, Na KJ.
- Funding acquisition: Na KJ.
- Investigation: Islam MA, Kim S, Na KJ.
- Methodology: Islam MA, Kim S, Na KJ.
- Project administration: Na KJ.
- Resources: Islam MA, Kim S, Na KJ.
- Supervision: Na KJ.
- Validation: Islam MA, Na KJ.
- Visualization: Islam MA, Na KJ.
- Writing - original draft: Islam MA, Na KJ.
- Writing - review & editing: Islam MA, Islam O, Kim S, Jeong DH, Na KJ.
References
- 1.Schilling AM, Rössner GE. The (sleeping) beauty in the beast–a review on the water deer, Hydropotes inermis. Hystrix Ital J Mammal. 2017;28(2):121–133. [Google Scholar]
- 2.Bakker ES, Pagès JF, Arthur R, Alcoverro T. Assessing the role of large herbivores in the structuring and functioning of freshwater and marine angiosperm ecosystems. Ecography. 2016;39(2):162–179. [Google Scholar]
- 3.Mackie RI. Mutualistic fermentative digestion in the gastrointestinal tract: diversity and evolution. Integr Comp Biol. 2002;42(2):319–326. doi: 10.1093/icb/42.2.319. [DOI] [PubMed] [Google Scholar]
- 4.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279(1):70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen J, Lara-Gutiérrez J, Stocker R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol Rev. 2021;45(4):1–16. doi: 10.1093/femsre/fuaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Megonigal JP, Hines ME, Visscher PT. In: Biogeochemistry. Schlesinger WH, editor. Elsevier-Pergamon; 2004. Anaerobic metabolism: linkages to trace gases and aerobic processes; pp. 317–424. [Google Scholar]
- 7.Cholewińska P, Czyż K, Nowakowski P, Wyrostek A. The microbiome of the digestive system of ruminants - a review. Anim Health Res Rev. 2020;21(1):3–14. doi: 10.1017/S1466252319000069. [DOI] [PubMed] [Google Scholar]
- 8.Delgado ML, Singh P, Funk JA, Moore JA, Cannell EM, Kanesfsky J, et al. Intestinal microbial community dynamics of white-tailed deer (Odocoileus virginianus) in an agroecosystem. Microb Ecol. 2017;74(2):496–506. doi: 10.1007/s00248-017-0961-7. [DOI] [PubMed] [Google Scholar]
- 9.Menke S, Heurich M, Henrich M, Wilhelm K, Sommer S. Impact of winter enclosures on the gut bacterial microbiota of red deer in the Bavarian Forest National Park. Wildl Biol. 2019;2019(1):1–10. [Google Scholar]
- 10.Yan J, Wu X, Wang X, Shang Y, Zhang H. Uncovering the fecal bacterial communities of sympatric sika deer (Cervus nippon) and wapiti (Cervus canadensis) Animals (Basel) 2022;12(18):2468. doi: 10.3390/ani12182468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peixoto RS, Harkins DM, Nelson KE. Advances in microbiome research for animal health. Annu Rev Anim Biosci. 2021;9(1):289–311. doi: 10.1146/annurev-animal-091020-075907. [DOI] [PubMed] [Google Scholar]
- 12.Pacheco-Torres I, Hernández-Sánchez D, García-De la Peña C, Tarango-Arámbula LA, Crosby-Galván MM, Sánchez-Santillán P. Analysis of the intestinal and faecal bacterial microbiota of the Cervidae family using 16S next-generation sequencing: A review. Microorganisms. 2023;11(7):1860. doi: 10.3390/microorganisms11071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan Y, Yang H, Han S, Feng L, Wang T, Ge J. Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express. 2017;7(1):212. doi: 10.1186/s13568-017-0517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dearing MD, Kohl KD. Beyond fermentation: Other important services provided to endothermic herbivores by their gut microbiota. Integr Comp Biol. 2017;57(4):723–731. doi: 10.1093/icb/icx020. [DOI] [PubMed] [Google Scholar]
- 15.White BA, Lamed R, Bayer EA, Flint HJ. Biomass utilization by gut microbiomes. Annu Rev Microbiol. 2014;68(1):279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 16.Majed R, Faille C, Kallassy M, Gohar M. Bacillus cereus biofilms—same, only different. Front Microbiol. 2016;7:1054. doi: 10.3389/fmicb.2016.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ. Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol. 2020;128(6):1583–1594. doi: 10.1111/jam.14506. [DOI] [PubMed] [Google Scholar]
- 18.Luise D, Bosi P, Raff L, Amatucci L, Virdis S, Trevisi P. Bacillus spp. probiotic strains as a potential tool for limiting the use of antibiotics and improving the growth and health of pigs and chickens. Front Microbiol. 2022;13:801827. doi: 10.3389/fmicb.2022.801827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G, Shen K, Yu R, Wu Q, Yan Q, Chen W, et al. Probiotic (Bacillus cereus) enhanced growth of Pengze crucian carp concurrent with modulating the antioxidant defense response and exerting beneficial impacts on inflammatory response via Nrf2 activation. Aquaculture. 2020;529:735691 [Google Scholar]
- 20.Ahsan N, Shimizu M. Lysinibacillus species: their potential as effective bioremediation, biostimulant, and biocontrol agents. Rev Agric Sci. 2021;9:103–116. [Google Scholar]
- 21.Azeem MA, Shah FH, Ullah A, Ali K, Jones DA, Khan MEH, et al. Biochemical characterization of halotolerant Bacillus safensis PM22 and its potential to enhance growth of maize under salinity stress. Plants (Basel) 2022;11(13):1721–1742. doi: 10.3390/plants11131721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satomi M, La Duc MT, Venkateswaran K. Bacillus safensis sp. nov., isolated from spacecraft and assembly-facility surfaces. Int J Syst Evol Microbiol. 2006;56(Pt 8):1735–1740. doi: 10.1099/ijs.0.64189-0. [DOI] [PubMed] [Google Scholar]
- 23.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos B, Rosalino LM, Palmeira JD, Torres RT, Cunha MV. Antimicrobial resistance in commensal Staphylococcus aureus from wild ungulates is driven by agricultural land cover and livestock farming. Environ Pollut. 2022;303:119116. doi: 10.1016/j.envpol.2022.119116. [DOI] [PubMed] [Google Scholar]
- 25.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 26.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 27.Drzewiecka D. Significance and roles of Proteus spp. bacteria in natural environments. Microb Ecol. 2016;72(4):741–758. doi: 10.1007/s00248-015-0720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novik G, Savich V, Kiseleva E. An insight into beneficial Pseudomonas bacteria. Microbiol Agric Human Health. 2015;1(5):73–105. [Google Scholar]
- 29.Dahal U, Paul K, Gupta S. The multifaceted genus Acinetobacter: from infection to bioremediation. J Appl Microbiol. 2023;134(8):1–18. doi: 10.1093/jambio/lxad145. [DOI] [PubMed] [Google Scholar]
- 30.Rong K, Delport J, AlMutawa F. Comamonas kerstersii bacteremia of unknown origin. Case Rep Infect Dis. 2022;2022(1):1129832. doi: 10.1155/2022/1129832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H, Yeo S, Arellano K, Kim HR, Holzapfel W. In: Probiotics and Prebiotics in Animal Health and Food Safety. Di Gioia D, Biavati B, editors. Springer International Publishing AG; 2018. Role of the gut microbiota in health and disease; pp. 35–62. [Google Scholar]
- 32.Macfarlane GT, Macfarlane S. Physiological and nutritional factors affecting synthesis of extracellular metalloproteases by Clostridium bifermentans NCTC 2914. Appl Environ Microbiol. 1992;58(4):1195–1200. doi: 10.1128/aem.58.4.1195-1200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bomba A, Jonecová Z, Gancarčíková S, Nemcová R. In: Gastrointestinal Microbiome. Ouwehand AC, Vaughan EE, editors. CRC Press; 2006. The gastrointestinal microbiota of farm animals; pp. 381–400. [Google Scholar]
- 34.Guo P, Zhang K, Ma X, He P. Clostridium species as probiotics: potentials and challenges. J Anim Sci Biotechnol. 2020;11(24):24. doi: 10.1186/s40104-019-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Songer JG. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev. 1996;9(2):216–234. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pahalagedara AS, Flint S, Palmer J, Brightwell G, Luo X, Li L, et al. Non-targeted metabolomic profiling identifies metabolites with potential antimicrobial activity from an anaerobic bacterium closely related to Terrisporobacter species. Metabolites. 2023;13(2):252–263. doi: 10.3390/metabo13020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinderola G, Ouwehand A, Salminen S, von Wright A. Lactic Acid Bacteria: Microbiological and Functional Aspects. 5th ed. CRC Press; 2019. p. 744. [Google Scholar]
- 38.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson CJ, Kracalik IT, Ross N, Alexander KA, Hugh-Jones ME, Fegan M, et al. The global distribution of Bacillus anthracis and associated anthrax risk to humans, livestock and wildlife. Nat Microbiol. 2019;4(8):1337–1343. doi: 10.1038/s41564-019-0435-4. [DOI] [PubMed] [Google Scholar]
- 40.Rasetti-Escargueil C, Lemichez E, Popoff MR. Public health risk associated with botulism as foodborne zoonoses. Toxins (Basel) 2019;12(1):1–24. doi: 10.3390/toxins12010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García A, Fox JG, Besser TE. Zoonotic enterohemorrhagic Escherichia coli: a one health perspective. ILAR J. 2010;51(3):221–232. doi: 10.1093/ilar.51.3.221. [DOI] [PubMed] [Google Scholar]