Abstract
Growing evidence indicates that herpes simplex virus type 1 (HSV-1) acquires its final envelope in the trans-Golgi network (TGN). During the envelopment process, the viral nucleocapsid as well as the envelope and tegument proteins must arrive at this site in order to be incorporated into assembling virions. To gain a better understanding of how these proteins associate with cellular membranes and target to the correct compartment, we have been studying the intracellular trafficking properties of the small tegument protein encoded by the UL11 gene of HSV-1. This 96-amino-acid, myristylated protein accumulates on the cytoplasmic face of internal membranes, where it is thought to play a role in nucleocapsid envelopment and egress. When expressed in the absence of other HSV-1 proteins, the UL11 protein localizes to the Golgi apparatus, and previous deletion analyses have revealed that the membrane-trafficking information is contained within the first 49 amino acids. The goal of this study was to map the functional domains required for proper Golgi membrane localization. In addition to N-terminal myristylation, which allows for weak membrane binding, UL11 appears to be palmitylated on one or more of three consecutive N-terminal cysteines. Using membrane-pelleting experiments and confocal microscopy, we show that palmitylation of UL11 is required for both Golgi targeting specificity and strong membrane binding. Furthermore, we found that a conserved acidic cluster within the first half of UL11 is required for the recycling of this tegument protein from the plasma membrane to the Golgi apparatus. Taken together, our results demonstrate that UL11 has highly dynamic membrane-trafficking properties, which suggests that it may play multiple roles on the plasma membrane as well as on the nuclear and TGN membranes.
During herpes simplex virus type 1 (HSV-1) assembly, over 30 different proteins come together to form three major structures: the nucleocapsid, the glycoprotein-containing envelope, and the collection of proteins located between the capsid and envelope known as the tegument (30). While it is generally accepted that capsid assembly and genome packaging occur in the nucleus, the compartment(s) in which tegument and envelope are acquired is less well defined (30). As with other herpesviruses (e.g., pseudorabies virus, varicella-zoster virus, and human cytomegalovirus), the most recent model for HSV-1 envelopment suggests that assembled nucleocapsids are shuttled out of the nucleus by a budding-fusion event on the inner and outer nuclear membranes (respectively) and then travel through the cytoplasm as unenveloped capsids until reaching a trans-Golgi network (TGN)-derived vesicle (7, 12–14, 18, 31, 40, 43, 47). While at this site, nucleocapsids are thought to acquire their final lipid bilayer in a process that also results in the acquisition of the tegument and glycoproteins. The mature virions subsequently follow the secretory pathway out to the cell surface, where they are released into the extracellular medium.
Although several lines of evidence support this general model for HSV-1 envelopment, the specific molecular mechanisms by which the different components come together at the TGN have yet to be defined. In particular, almost nothing is known about how the tegument region of the virus is created during assembly. Understanding how this occurs is important because evidence suggests that the tegument proteins contain all of the functions required for budding at the TGN (23, 29). Therefore, determining how the tegument proteins travel to the same cellular compartment and interact to form a stable structure will provide insights into the overall mechanism of HSV-1 assembly. As a start to these investigations, we have been studying the trafficking properties of a small HSV-1 tegument protein, UL11.
The UL11 gene of HSV-1 encodes a 96-residue, myristylated protein (Fig. 1) that binds to the cytoplasmic face of internal membranes within infected cells (2, 21). By associating with these membranes, the UL11 protein is thought to play a role in the envelopment and egress of viral nucleocapsids (3, 22); however, the mechanism for this process has not been described. Previous studies have shown that UL11 localizes to both nuclear and cytoplasmic membranes in HSV-1-infected cells (2), but when expressed in the absence of other HSV-1 proteins, UL11 localizes predominantly to the Golgi apparatus (5). Mutational analyses have revealed that N-terminal myristylation and only the first 49 amino acids of UL11 are required for proper membrane binding and Golgi localization. Sequence alignment of several UL11 homologues (5) reveals that this region contains a conserved acidic cluster motif, similar to those found in membrane proteins that cycle between the plasma membrane and the Golgi apparatus (e.g., furin and cytomegalovirus gB) (6, 39, 41). The negatively charged amino acids of acidic clusters function by interacting with various components of the clathrin sorting machinery at the plasma membrane, endosomal, and Golgi compartments (9, 38, 42). Although the mechanism is poorly understood, this cycling process is typically dependent on the phosphorylation of a serine or threonine residue(s) within the acidic cluster. One exception, however, is the human immunodeficiency virus type 1 (HIV-1) Nef protein, which is a peripheral membrane protein that is recycled to the Golgi apparatus without requiring phosphorylation (26).
FIG. 1.
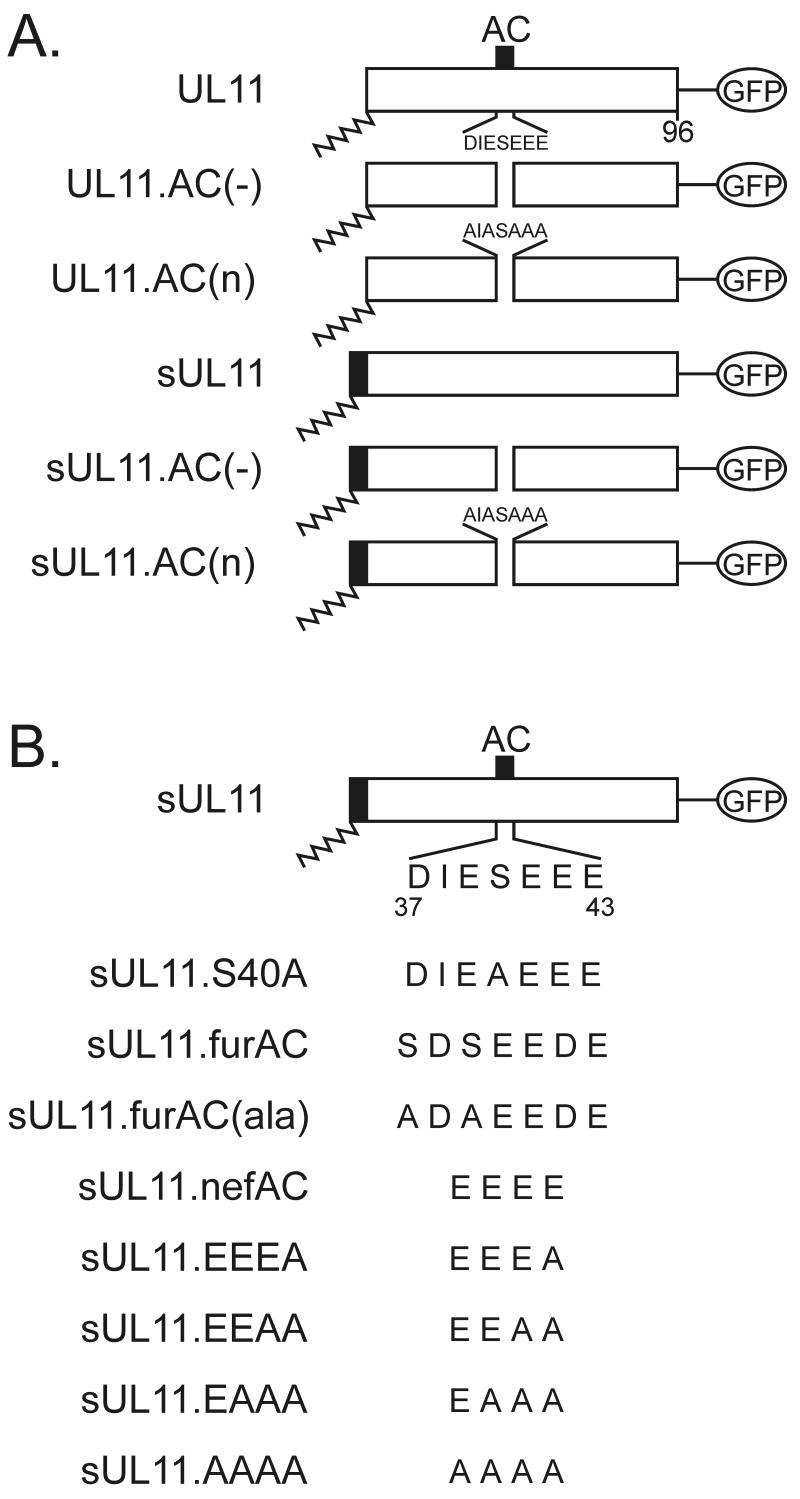
Mutational analysis of the acidic cluster region of UL11. (A) Inactivation of the wild-type sequence. The 96-amino-acid UL11 sequence (open rectangles) is shown with the position of its acidic cluster (AC) (solid black box). N-terminal myristylation (wavy line) is a natural modification of UL11 and was also provided by the first 10 amino acids of the Src oncoprotein (solid rectangle with wavy line). All constructs have the GFP protein (open oval) fused to their C termini. (B) Replacement with foreign acidic cluster sequences. The wild-type sequence of the UL11 acidic cluster is shown (residues 37 to 43). Wild-type and mutant acidic cluster sequences from furin and Nef were inserted in place of the UL11 sequence as shown. All constructs were expressed as UL11-GFP chimeras.
In the present study, we have examined the roles of the acidic cluster and other sequences in the first half of UL11 in membrane trafficking. Using confocal microscopy, the acidic cluster was found to be required for the retrieval of UL11 from the plasma membrane to the Golgi apparatus but was not required for Golgi accumulation. Instead, Golgi accumulation was highly dependent upon three consecutive cysteines located near the N terminus of UL11. These cysteines, which we found to be required for the palmitylation of UL11, appeared to be essential for both membrane-targeting specificity and strong membrane binding. Thus, UL11 has dynamic trafficking properties, which suggests that it plays roles at multiple membrane compartments during viral replication.
MATERIALS AND METHODS
Cells.
A7 (human melanoma) cells, a gift from Gary Thomas (The Oregon Health Sciences University, Portland) were grown in Dulbecco's modified Eagle's medium (DMEM) (GIBCO) supplemented with 10% fetal bovine serum (FBS) and antibiotics (42). COS-1 (simian) cells were grown in DMEM supplemented with 3% FBS, 7% calf bovine serum, and antibiotics (44).
Construction of UL11 acidic cluster mutants.
The UL11 gene used for this study was obtained from the HSV-1 KOS genome and cloned into the Clontech pEGFP-N2 vector as previously described (5, 37). The pCMV.UL11.AC(−) construct was made by first PCR amplifying the UL11 gene from immediately downstream of the acidic cluster (AC)-coding sequence (nucleotide 129) to the 3′ end of the gene with a forward primer containing an AatII site and a reverse primer containing an EcoRI site. The resulting fragment was cut with AatII and ligated to another fragment of the UL11 gene that extends from 100 bp upstream of the UL11 start codon (−100) to a native AatII site that lies immediately upstream of the acidic cluster-coding sequence (+103). The ligation product, which represents the UL11 gene with a deletion in nucleotides 104 to 128, was then amplified using PCR with a forward primer (containing an SstI site) complementary to the sequence 100 bp upstream of the UL11 start codon and a reverse primer (containing an EcoRI site) complementary to the 3′ end of the UL11 gene. This product was then cut with SstI and EcoRI and cloned into the multiple cloning site (MCS) of the pEGFP-N2 vector, thereby producing a vector that encodes a UL11-green fluorescent protein (GFP) fusion protein.
To make the constructs in which the membrane-binding domain of the Src oncoprotein is fused to the N terminus of the UL11 protein [sUL11 and sUL11.AC(−)], a similar strategy was employed. The 256-bp SstI-AatII fragment cut from the previously described pSV.Src.HMG construct (5) was ligated to the AatII-EcoRI fragment of UL11 [derived from either pCMV.UL11 or pCMV.UL11.AC(−)]. The ligation products were PCR amplified, cut with SstI and EcoRI, and cloned into the MCS of the pEGFP-N2 vector in order to make the sUL11-GFP fusion protein.
The pCMV.CD4.UL11 and pCMV.CD4.UL11.AC(−) constructs were made by inserting the UL11-coding sequence in place of the last 87 bp of the human CD4 gene. To do this, a truncated CD4 gene [CD4T(−)], which encodes a protein that lacks the majority of the cytoplasmic tail, was cut from the pCMX.CD4T(−) vector (kindly provided by Chris Aiken, Vanderbilt University School of Medicine, Nashville, Tenn.) with HindIII and BamHI and ligated into the same sites in the pEGFP-N2 vector [pCD4T(−).GFP] (36). Next, the UL11-coding sequence was PCR amplified with a forward primer (containing a BglII site) that is complementary to the first 22 bp of the UL11 gene and a reverse primer (containing a NotI site) that is complementary to the last 20 bp of the UL11 gene. The resulting PCR fragment, which was cut with BglII and NotI, was used to replace the BamHI-NotI fragment from the pCD4T(−).GFP vector. In addition to removing a portion of the pEGFP-N2 MCS, this replacement removes the entire coding sequence of GFP.
All nucleotide substitutions within the UL11 acidic cluster-coding sequence were made by oligonucleotide-directed mutagenesis, as previously described (19). The UL11 gene was cloned into M13mp19, and the resulting recombinant phage genome was propagated to isolate single-stranded, uracil-containing template DNA. After mutagenesis, all mutants were subcloned from the M13 vector into pCMV.UL11 with SstI-EcoRI and into pCMV.sUL11 with MluI-EcoRI.
Other substitutions within UL11.
Mutations in the first half of UL11 (nucleotides 1 to 108) were made by a variety of methods. The Myr2sub.UL11 construct, in which the first 10 amino acids of Myr2.Gag (a myristylated form of Rous sarcoma virus Gag protein) (11, 44) were used to replace the first 10 residues of UL11, was made by PCR mutagenesis. In short, the sequence encoding the first 10 amino acids of Myr2.Gag (and ≈125 bp of noncoding upstream sequence) was PCR amplified and inserted in place of the first 30 bp of the UL11-coding sequence (and 100 bp of upstream noncoding sequence) using SstI and MluI (44). A similar strategy was employed to make the construct that encodes a protein that has the first 10 amino acids of Myr2.Gag attached to the N terminus of UL11 as an extension (Myr2ext.UL11). The Myr(−).UL11 construct, which serves as a negative control for the palmitylation experiments, has a glycine-to-alanine change at position 2 within the UL11 protein in order to abolish myristylation. To make this mutant, a Myr(−).UL11 gene was PCR amplified from the previously described pMyr(−).HMG construct, and the fragment was cloned into the pEGFP.N2 vector with SstI and EcoRI (5). The fUL11 construct, which is the positive control for the palmitylation studies, was made by attaching the sequence encoding the first 10 residues of the Fyn protein (which is both myristylated and palmitylated) as an extension from the 5′ end of the UL11-coding sequence (immediately upstream of the ATG). This sequence was PCR amplified out of a previously made pSV.Fyn.Gag construct (kindly provided by Leslie Parent, Pennsylvania State University College of Medicine, Hershey) and cloned into pCMV.sUL11 using the SstI and MluI enzymes. pCMV.UL11.CCC(−) was made by oligonucleotide-directed mutagenesis using an oligonucleotide coding for three alanines in place of the three consecutive N-terminal cysteines (residues 11 to 13).
Expression and metabolic labeling of UL11 mutants.
A7 or COS-1 cells were transfected by the calcium phosphate method, as previously described (8). To measure the apparent molecular mass and expression level of each UL11 derivative, at approximately 24 h posttransfection the cells were starved for 15 min in methionine-free DMEM and then metabolically labeled with 50 μCi (>1,000 Ci/mmol) of l-[35S]methionine for 2.5 h. The cells were then mixed with lysis buffer containing protease inhibitors (Sigma, product number P8340), and the UL11 chimeras were immunoprecipitated with a polyclonal anti-GFP serum (Clontech) (44).
Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide gels. The gels were dried, and radiolabeled proteins were detected by autoradiography with Kodak X-Omat AR5 film. Gels were exposed to film for 1 to 2 days.
To analyze UL11 phosphorylation in vivo, transfected COS-1 cells were metabolically labeled with [32P]orthophosphate at 24 h posttransfection. Cells were starved in phosphate-free DMEM (GIBCO) for 10 min at 37°C and then labeled with 250 μCi of [32P]orthophosphate (>8,500 Ci/mmol) for 6 to 16 h at 37°C. After medium was removed and the plates were washed once with Tris-buffered saline, the cells were mixed with lysis buffer containing protease inhibitors, 2 mM EDTA, 50 mM NaF, and 0.2 mM sodium vanadate (NaVO4). The UL11 proteins were then immunoprecipitated and analyzed by autoradiography as described above. Phosphorimager analysis was performed, and the phosphorylation efficiency was quantitated by dividing the amount of UL11 labeled with [32P]orthophosphate (phosphorylated protein) by the amount of UL11 labeled with [35S]methionine (total protein).
To detect palmitylation of the UL11 derivatives, transfected COS-1 cells (in 35-mm-diameter plates) were metabolically labeled with [3H]myristic acid (62.5 μCi/plate; 10 to 60 Ci/mmol) for 10 min at 37°C or with [3H]palmitic acid (156 μCi/plate; 30 to 60 Ci/mmol) for 60 min at 37°C. After these incubations, cells were mixed with lysis buffer containing protease inhibitors (Sigma, product number P8340), and the UL11 chimeras were immunoprecipitated with a polyclonal anti-GFP serum. Immunoprecipitated proteins were then mixed with sample buffer (β-mercaptoethanol was omitted to prevent release of palmitate) (16) and were separated by SDS-PAGE on 12% polyacrylamide gels. Gels were treated with Fluoro-Hance (Research Products Inc.) for 30 min prior to drying, and radiolabeled proteins were detected by autoradiography with Kodak X-Omat AR5 film. The gels were exposed to film for approximately 2 months.
Confocal microscopy of UL11 mutants.
A7 or COS-1 cells were transfected with the UL11-GFP chimeras as described above. At 12 to 36 h posttransfection, cells were washed once with Tris-buffered saline and immediately viewed using a Zeiss laser scanning microscope with a helium-argon laser (488-nm peak excitation).
To study the trafficking of the CD4.UL11 chimeras, a standard endocytosis assay was performed in A7 cells at 24 h posttransfection (39). The cells were washed twice with cold phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA) (Sigma), and anti-human CD4 monoclonal antibody (DAKO, product number M0716) was added at a 1:250 dilution in serum-free DMEM. The cells were incubated for 40 min at 4°C and then washed three times with cold PBS–3% BSA. Prewarmed DMEM supplemented with 10% FBS was added to the cells, and the plates were incubated at 37°C (5% CO2) to allow for endocytosis. At various times (0 min, 15 min, 30 min, 45 min, 1 h, and 2 h), the cells were fixed with either 3% paraformaldehyde (20 min, 4°C) or 95% ethanol–5% acetic acid (20 min, 0°C) and then permeabilized with 0.1% Triton X-100 (10 min, 23°C). A goat anti-mouse immunoglobulin G secondary antibody conjugated to fluorescein isothiocyanate was then added at a dilution of 1:75 in PBS–3% BSA. After being washed with PBS–3% BSA, cells were visualized by confocal microscopy as described above.
Membrane pelleting and quantitation of protein.
At 12 to 24 h posttransfection, A7 cells were washed twice with and harvested into NTE buffer (10 mM Tris, 100 mM NaCl, 1 mM Na2EDTA, pH 7.2). Intact cells were then pelleted for 5 min at 1,000 × g (4°C) and resuspended in 1 ml of hypotonic buffer (10 mM Tris, 0.2 mM MgCl2, pH 7.4). After 30 min (on ice), cells were lysed by Dounce homogenization (25 to 30 strokes), and lysates were centrifuged at 1,000 × g for 10 min to remove unbroken cells and nuclei. Postnuclear supernatants were then subjected to centrifugation at 100,000 × g for 40 min in a Beckman Optima TLX ultracentrifuge. Soluble and pellet fractions were collected and adjusted to 0.5% Triton X-100 in NTE buffer (32). The amount of UL11 protein (linked to GFP) present in each fraction was then quantitated using an Aminco-Bowman Series 2 fluorescence spectrophotometer with an excitation filter at 488 nm and an emission filter at 514 nm. The percentage of protein pelleted was calculated by dividing the amount in the pellet fraction (P) by the amount in both the soluble fraction (S) and pellet fraction [P/(P + S)]. In addition, soluble and pellet fractions were analyzed by Western blotting in order to confirm the fluorometric data.
RESULTS
Although the UL11 protein accumulates at the Golgi apparatus in both transfected and infected cells, the mechanism by which this occurs is still unclear. Moreover, it is unknown whether UL11 has a mechanism for direct Golgi targeting and retention or is capable of traveling to other cytoplasmic membranes from which it can be rapidly recovered and returned to the Golgi apparatus. Previous studies suggest that the first 49 amino acids of UL11 contain all of the necessary information for Golgi targeting (5), and a conserved acidic cluster within this region (Fig. 1) is similar to those found in membrane-spanning proteins that cycle between the plasma membrane and the Golgi apparatus. Therefore, we began this study by examining the importance of the acidic cluster sequence for Golgi apparatus targeting.
Mutational analysis of the acidic cluster.
To determine whether the acidic cluster is required for Golgi targeting, we examined the intracellular localization of two different mutants which lack this motif. One mutant, UL11.AC(−), contains a deletion of all seven residues (residues 37 to 43) from this region, while the other, UL11.AC(n), has the five aspartic and glutamic acid residues within the acidic cluster changed to alanines (Fig. 1A). To detect these mutants in living cells, GFP was linked to their C termini. A7 melanoma cells were used for these experiments because they have been used previously in other membrane-trafficking studies (26, 42), are easily transfected, and can be productively infected by HSV-1 (data not shown).
Expression of the wild-type and mutant UL11-GFP chimeras in A7 cells (Fig. 2A) and COS-1 cells (data not shown) verified that all constructs were expressed well and were of the expected molecular masses. Quantitation of protein band intensities by phosphorimager analysis revealed that both mutants are expressed three to five times better than wild-type UL11; however, the reason for this is not known.
FIG. 2.
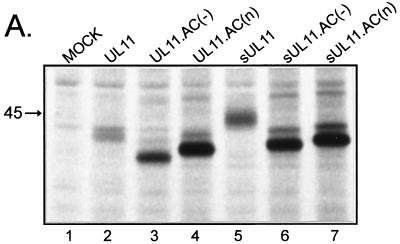
Expression of UL11 mutants with inactivated acidic clusters. (A) Biochemical analysis. A7 melanoma cells transfected with the indicated constructs were labeled for 2.5 h with l-[35S]methionine, and UL11-GFP chimeras were immunoprecipitated from cell lysates with a polyclonal antibody specific for GFP. Proteins were separated by SDS-PAGE and visualized by autoradiography. The position of the 45-kDa molecular mass marker is indicated. (B) Subcellular localization. Plasmids were transfected into A7 cells as in panel A, and UL11-GFP chimeras were visualized by live-cell confocal microscopy at approximately 18 h following transfection.
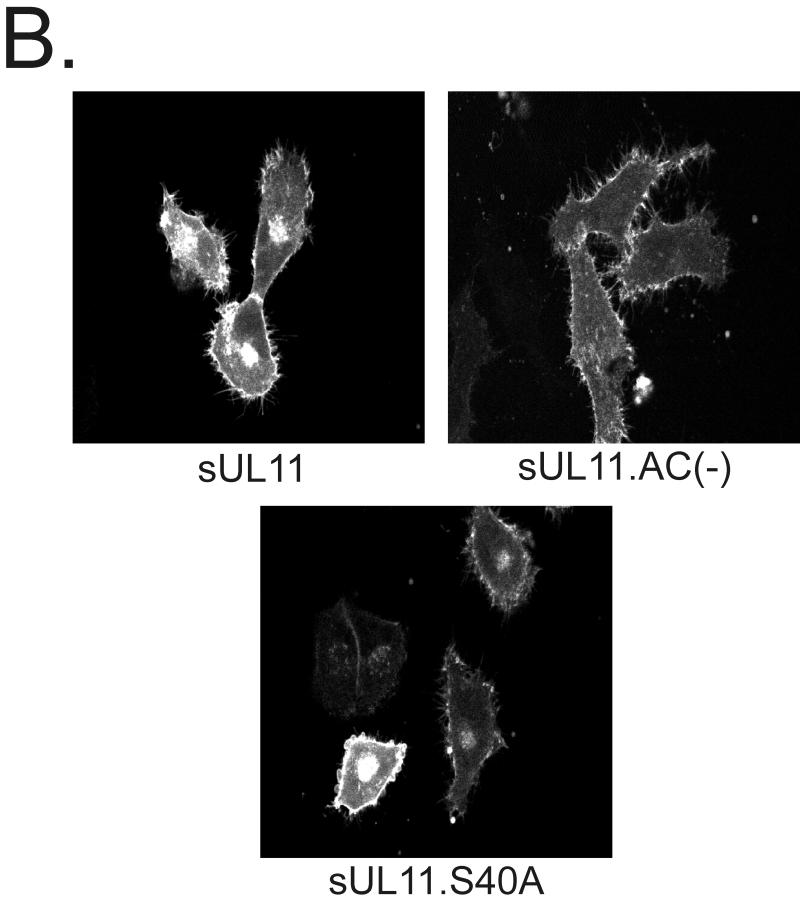
To determine where the acidic cluster mutants localize when transiently expressed in A7 cells, live-cell confocal microscopy was performed. If the acidic cluster is required for direct Golgi targeting and/or retention, then mutants lacking it should fail to accumulate at the Golgi apparatus. We found that both acidic cluster mutants [UL11.AC(−) and UL11.AC(n)] were targeted to the Golgi apparatus in a manner similar to that for the wild type (Fig. 2B, left panels); however, both mutants also appeared to accumulate on the plasma membrane. This suggests that the acidic cluster may be involved in retrieving UL11 from the plasma membrane rather than directly targeting it to the Golgi apparatus.
If the acidic cluster is required for plasma membrane retrieval, then UL11 chimeras containing a plasma membrane-targeting signal but no acidic cluster should accumulate only on the plasma membrane. Because previous studies have demonstrated that the first 10 residues of the Src oncoprotein are sufficient for targeting heterologous proteins to the plasma membrane (45), this peptide was attached as an extension to the N termini of the acidic cluster mutants of UL11 (Fig. 1A). Unlike the wild-type Src-UL11 chimera (sUL11), which localizes to both the plasma membrane and Golgi apparatus, the mutants [sUL11.AC(−) and sUL11.AC(n)] localized to the plasma membrane but failed to accumulate at the Golgi apparatus, as would be expected if the acidic cluster is required for recovery from the plasma membrane (Fig. 2B, right panels). In addition, both mutants also localized (to various degrees) to what appear to be dispersed cytoplasmic vesicles, which suggests either that UL11 contains an endocytosis signal(s) or that Src targets the mutants to some non-plasma membrane locations.
UL11 can direct an integral plasma membrane protein to the Golgi apparatus.
To test whether UL11 contains signals that are sufficient for endocytosis, it or the AC(−) version was inserted in place of the cytoplasmic tail domain of human CD4 (Fig. 3A). By providing UL11 with a surrogate transmembrane and extracellular domain, it became possible to assay for endocytosis using established protocols (39). As a control, a CD4 mutant which has no cytoplasmic tail domain was also constructed [CD4T(−)]. All four constructs produced proteins of the expected size; however, both of the CD4-UL11 chimeras were expressed at levels that were lower (≈50%) than wild type (Fig. 3B).
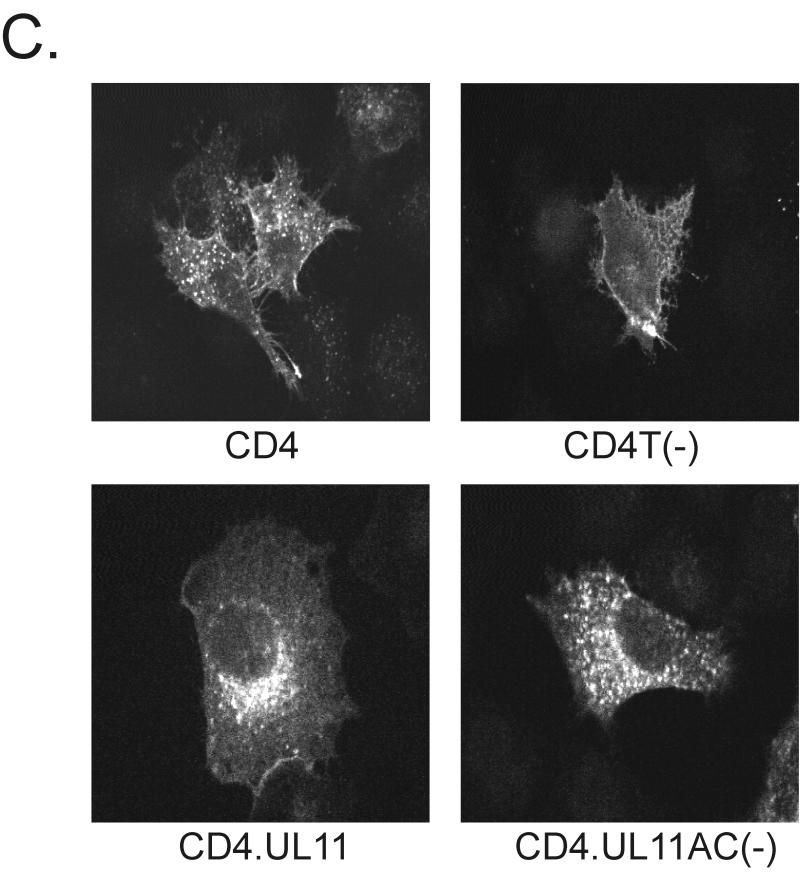
FIG. 3.
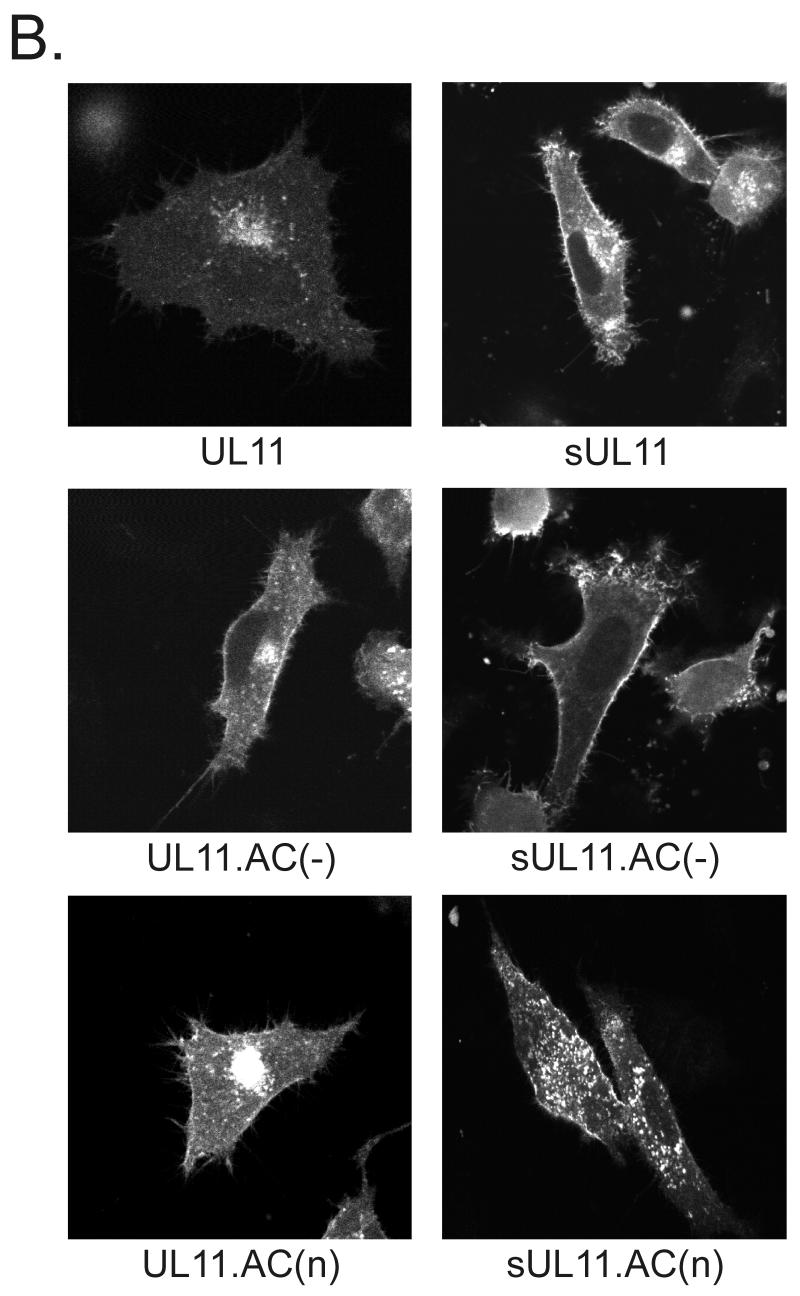
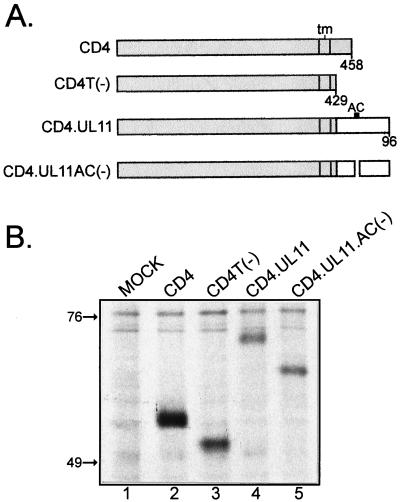
Recovery of CD4-UL11 chimeras from the plasma membrane. (A) CD4-UL11 constructs. The 458-amino-acid human CD4 protein (shaded rectangle) contains a large extracellular domain, a hydrophobic transmembrane domain (tm), and a short cytoplasmic domain. The wild-type UL11 sequence or the acidic cluster (AC) deletion mutant (open rectangles) was attached to the CD4 protein in place of the last 29 residues of the cytoplasmic tail, but in this case, GFP was not included. (B) Biochemical analysis. A7 cells were transfected and metabolically labeled with [35S]methionine for 2.5 h. The CD4 derivatives were immunoprecipitated with a monoclonal antibody specific for CD4, separated by SDS-PAGE, and visualized by autoradiography. The positions of molecular mass markers (in kilodaltons) are indicated. (C) Subcellular localization. A7 cells transfected with the indicated constructs were incubated with a monoclonal antibody specific for CD4 for 40 min on ice. After excess antibody was washed away, the cells were shifted to 37°C for 60 min to allow for endocytosis. To detect internalized antibody, the cells were fixed, permeabilized, stained with fluorescein isothiocyanate-labeled secondary antibody, and visualized by confocal microscopy.
To assay for endocytosis, A7 cells expressing the CD4 derivatives were treated at 4°C with a monoclonal antibody raised against the human CD4 protein, shifted to 37°C to allow for internalization of the antibody-CD4 complexes, and then fixed, permeabilized, and stained with a fluorescently labeled anti-mouse serum (39). Unlike wild-type CD4, which was efficiently internalized and incorporated into vesicles, the CD4T(−) mutant remained largely localized to the plasma membrane (Fig. 3C). When wild-type UL11 was inserted in place of the CD4 cytoplasmic tail domain, the chimera was efficiently internalized and targeted to a perinuclear compartment reminiscent of the Golgi apparatus. In contrast, the CD4.UL11.AC(−) construct did not accumulate at the Golgi apparatus, although it was internalized from the plasma membrane (Fig. 3C). Thus, it appears that UL11 indeed contains endocytic signals and that the role of the acidic cluster is to direct the molecule back to the Golgi apparatus after being removed from the plasma membrane.
Lack of a role of phosphorylation in UL11 transport.
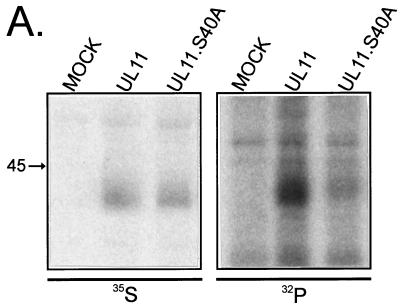
Recycling of various cellular and viral glycoproteins (e.g., furin and varicella-zoster virus gE) to the Golgi apparatus is dependent upon phosphorylation of a serine or threonine residue within an acidic cluster (1, 38, 42). Inhibition of phosphorylation often results in the accumulation of protein in the endosomal compartment and prevents recycling to the Golgi apparatus. To establish whether or not UL11 is phosphorylated on the serine located within its acidic cluster, in vivo phosphorylation experiments were performed. While both wild-type UL11 and a serine point mutant (UL11.S40A) were expressed well (Fig. 4A, left panel), metabolic labeling with [32P]orthophosphate demonstrated that UL11 was efficiently phosphorylated and that UL11.S40A was not (Fig. 4A, right panel). Quantitation of the bands by phosphorimager analysis revealed that mutating serine 40 to an alanine reduces phosphorylation by 60% ± 7% (n = 4) compared to wild-type UL11. These data suggest that serine 40 represents a major site of phosphorylation in UL11.
FIG. 4.
Phosphorylation of UL11. (A) Biochemical analysis. Duplicate plates of COS-1 cells were transfected with the indicated constructs. Approximately 18 h later, cells were labeled with either [35S]methionine for 2.5 h or [32P]orthophosphate for 16 h. The labeled proteins were immunoprecipitated with anti-GFP serum, subjected to SDS-PAGE analysis, and visualized by autoradiography. (B) Subcellular localization. The indicated constructs were transfected into A7 cells, and 18 h later, the sUL11-GFP chimeras were visualized by confocal microscopy.
To determine whether phosphorylation within the acidic cluster is required for UL11 recycling, the serine point mutant was expressed as a chimera with the Src peptide (Fig. 1B, sUL11.S40A) and confocal microscopy was performed. Similar to wild-type sUL11, the serine point mutant appeared to efficiently recycle to the Golgi apparatus (Fig. 4B). Furthermore, when expressed without the Src peptide on its N terminus, the serine point mutant behaves the same as the wild type (data not shown). Together, these results suggest that phosphorylation within the acidic cluster is not needed for UL11 recycling, as is also the case for the Nef protein (26)
Acidic clusters from other proteins can substitute for that of UL11.
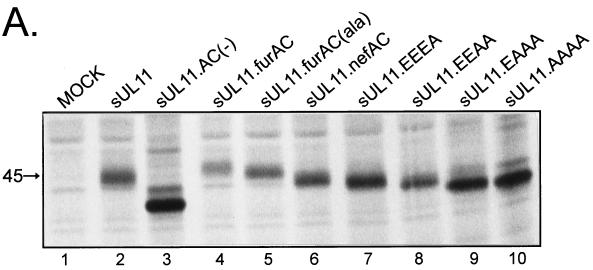
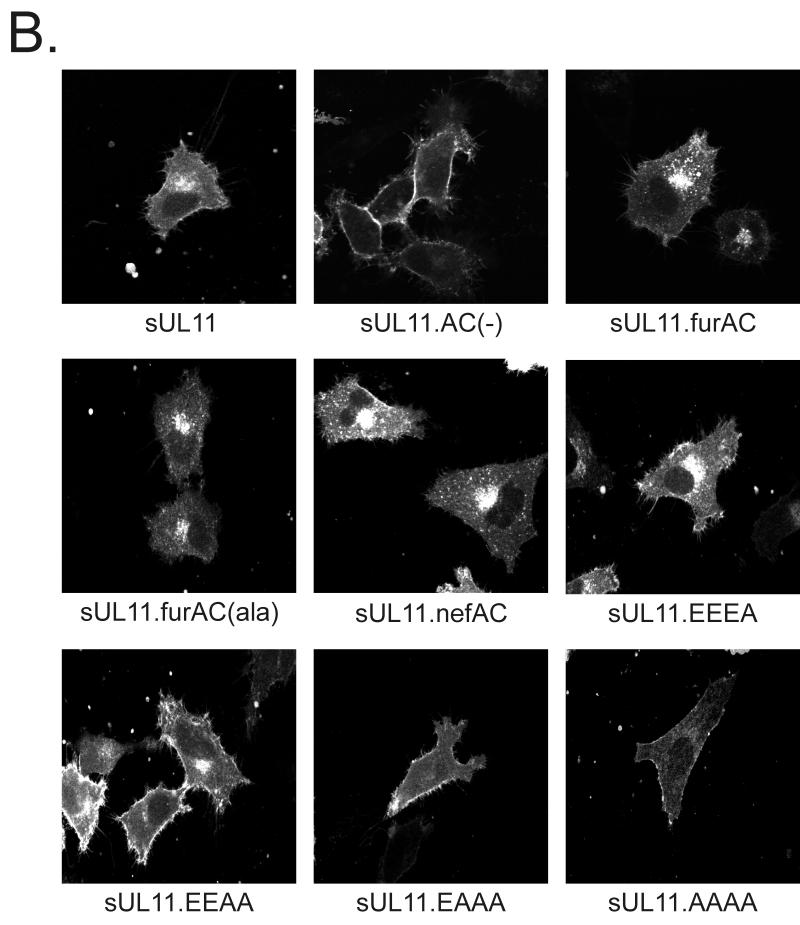
If the acidic cluster is needed for retrieval of UL11 from the plasma membrane, then our mutants should regain the ability to return to the Golgi apparatus when well-characterized acidic clusters from other recycled proteins are inserted into UL11. The acidic clusters of the furin and HIV-1 Nef proteins (SDSEEDE [42] and EEEE [26], respectively) were added to UL11 in the same position as the native acidic cluster, and the mutants were expressed as chimeras with the Src peptide (Fig. 1B). Resolution of these chimeras by SDS-PAGE revealed that all were expressed well and were of the expected sizes (Fig. 5A, lanes 2 to 4 and 6). Similar to sUL11, both chimeras were found to efficiently recycle to the Golgi apparatus when sent to the plasma membrane via Src (Fig. 5B, sUL11.furAC and sUL11.nefAC). Moreover, both were localized to the Golgi apparatus in the absence of the Src peptide (data not shown), which is expected since acidic clusters do not appear to be involved in direct Golgi targeting (see above). In addition, these chimeras provide further evidence that UL11 recycling is not regulated by phosphorylation within the acidic cluster, since the Nef acidic cluster (Fig. 1B, EEEE), which contains no serines or threonines, and a derivative of the furin acidic cluster which lacks serines (Fig. 1B, ADAEEDE) both enabled UL11 to be recovered from the plasma membrane (Fig. 5B).
FIG. 5.
Expression of UL11 chimeras having foreign acidic cluster sequences. (A) Biochemical analysis. A7 cells were transfected with the indicated constructs and labeled with [35S]methionine for 2.5 h. The labeled proteins were immunoprecipitated with anti-GFP serum, subjected to SDS-PAGE analysis, and visualized by autoradiography. (B) Subcellular localization. Constructs were transfected into A7 cells, and 18 h later, the UL11-GFP derivatives were visualized by confocal microscopy.
Given the simplicity of the four consecutive glutamic acid residues in the Nef acidic cluster, it was of interest to see whether still-shorter sequences were sufficient for recycling of UL11 (Fig. 1B). Surprisingly, an acidic cluster consisting of only two glutamic acidic residues (Fig. 5B, EEAA) was able to direct UL11 from the plasma membrane to the Golgi apparatus, although the efficiency was reduced relative to more acidic motifs. No recycling was seen with a single acidic residue (Fig. 5B, EAAA).
Targeting information is not found within the first 10 residues of UL11.
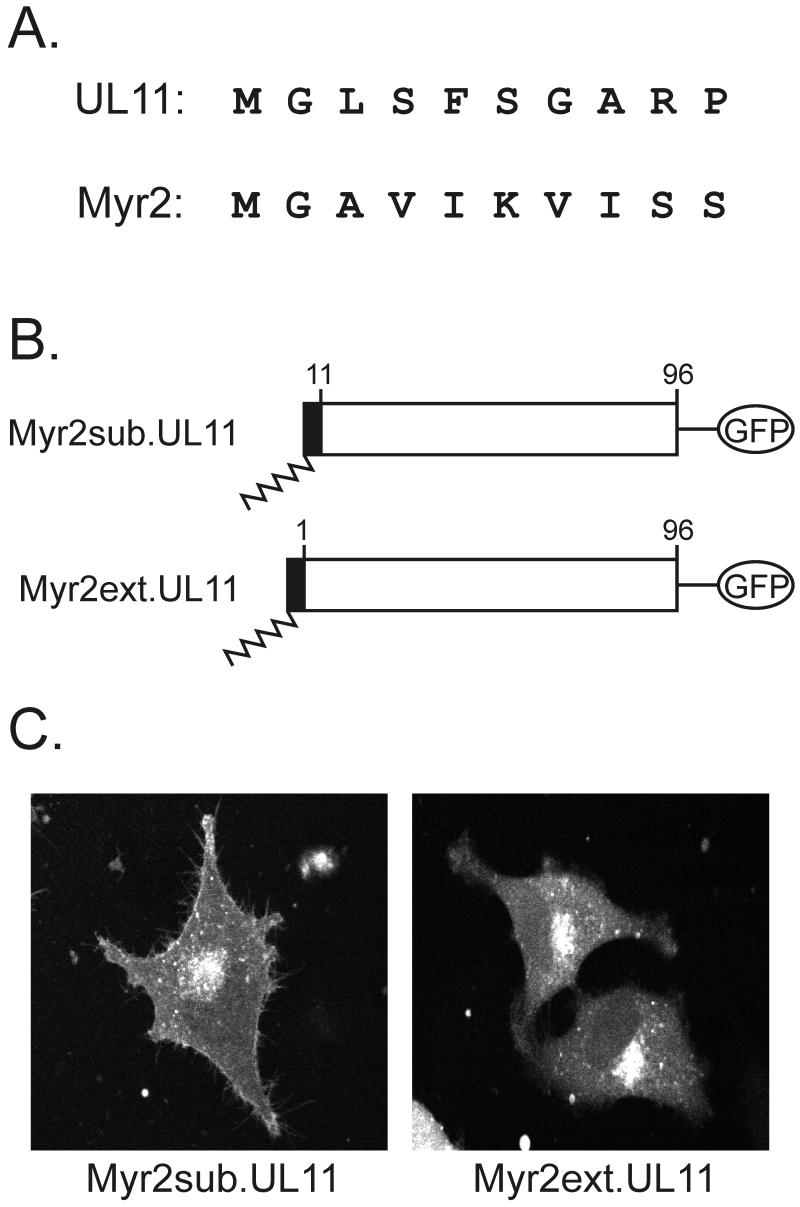
If the acidic cluster is required only for recovery from the plasma membrane, then what in the first half of UL11 is responsible for Golgi-specific targeting? It is well established that myristate alone is insufficient for stable or specific membrane interactions (25, 27) and that additional information must be provided by the amino acids within the polypeptide sequence. For instance, in the case of the Src protein, basic residues near the N terminus provide stabilizing interactions with acidic phospholipids found in the plasma membrane (27). Although UL11 contains only one basic residue near its N terminus (Fig. 6A), we were interested in determining whether its first 10 amino acids contain direct Golgi-targeting information. However, because myristylation itself requires certain N-terminal amino acids, it is not possible to alter this region without potentially eliminating myristylation (27). Therefore, to preserve this modification, a foreign myristylation signal, Myr2 (Fig. 6A), which lacks specific membrane-targeting information (11, 44) was inserted as either a substitution (Myr2sub.UL11) or an extension (Myr2ext.UL11) from the N terminus of UL11 (Fig. 6B). In spite of the drastic changes made in the N-terminal sequence of UL11 (Fig. 6A), both chimeras accumulated with high efficiency on the Golgi-derived membranes (Fig. 6C). Thus, the first 10 residues of UL11 do not appear to contain the direct Golgi targeting information, although myristylation is required (5).
FIG. 6.
Analysis of the first 10 residues of UL11. (A) Sequence comparison. The first 10 amino acids of the UL11 protein and a myristylated form of the Rous sarcoma virus Gag protein are shown. The glycine residue at the second position is the site of myristylation for both proteins, but otherwise the sequences are highly divergent. (B) Myr2-UL11 chimeras. The first 10 residues of the Myr2.Gag protein (black rectangle) and its associated myristate (wavy line) were either substituted in place of the first 10 residues of UL11 (Myr2sub.UL11) or attached to the N terminus as an extension (Myr2ext.UL11). Both constructs have the GFP protein (open oval) fused to their C termini. (C) Subcellular localization. The constructs were transfected into A7 cells, and 18 h later, the UL11-GFP chimeras were visualized by live-cell confocal microscopy.
Targeting is dependent on palmitylation at N-terminal cysteines.
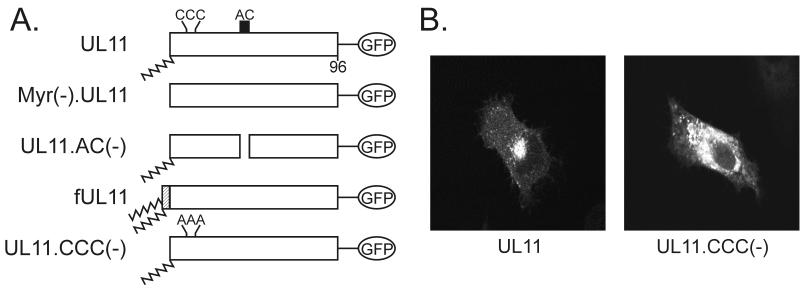
To locate the region that is responsible for Golgi accumulation, extensive substitution analysis was performed on residues 11 to 37 of UL11. Specifically, the following substitutions were introduced: R9A, CCC11,12,13AAA, R14A, NN15,16AA, LI18,19AA, D21A, D22A, E24A, H31A, D32A, F33A, and D34A. While the majority of these substitutions had little or no effect on Golgi accumulation (data not shown), replacement of the three N-terminal cysteines (residues 11 to 13) with alanines (Fig. 7A) had a profound effect on the ability of UL11 to specifically localize to the Golgi apparatus. In contrast to the wild type, which accumulates efficiently in the Golgi apparatus, the triple cysteine mutant [UL11.CCC(−)] appeared to reside in membranes dispersed throughout the cytoplasm (Fig 7B). This result suggests that the three cysteines are involved in directing UL11 specifically to the Golgi apparatus.
FIG. 7.
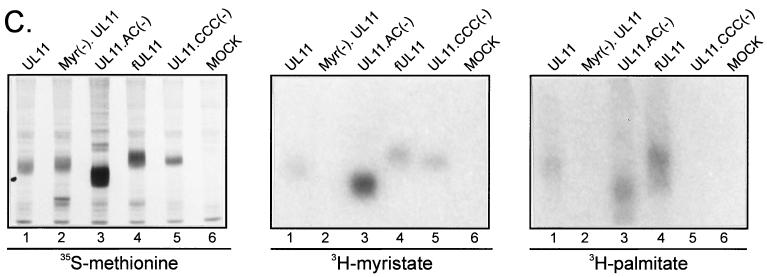
Palmitylation of UL11. (A) Mutational analysis of the CCC motif. The wild-type UL11 protein (top) is myristylated (wavy line) and contains the acidic cluster (AC) (residues 37 to 43, indicated by a black box) along with a cluster of three cysteines (residues 11 to 13). An N-terminal 10-amino-acid sequence from the Fyn protein (hatched box) is known to be sufficient for both myristylation and palmitylation (double wavy line). All constructs have the GFP protein (open oval) fused to their C termini. (B) Subcellular localization. The indicated constructs were transfected into COS-1 cells, and 18 h later, the UL11-GFP chimeras were visualized by confocal microscopy. (C) Biochemical analysis. Transfected COS-1 cells were labeled with either [35S]methionine for 2.5 h, [3H]myristic acid for 10 min, or [3H]palmitic acid for 60 min. Cell lysates were prepared, and UL11-GFP proteins were immunoprecipitated, mixed with sample buffer (without β-mercaptoethanol), resolved by SDS-PAGE, and visualized by autoradiography.
Cysteine residues near the N termini of peripherally bound membrane proteins often serve as sites of modification with palmitic acid. Addition of this 16-carbon fatty acid to the sulfhydryl group of cysteine residues (27) increases the protein's affinity for membranes and has been shown to be required for membrane binding specificity (46). Although the mechanism for this is not known, it is thought that palmitylation serves to lock proteins into the membrane compartment where palmitoyl acyl transferases (PATs) are located (27) (see Discussion).
To determine whether UL11 is palmitylated in vivo, transfected cells were metabolically labeled with [3H]palmitic acid. While the wild type appeared to be labeled efficiently with [3H]palmitic acid in vivo (Fig. 7C, right panel, lane 1), the construct that lacks all three cysteine residues was not (Fig. 7C, right panel, lane 5). Similar to the wild type, UL11.AC(−) and a construct that contains the first 10 residues of the myristylated and palmitylated Fyn protein (Fig. 7A, fUL11) both appear to be palmitylated (Fig. 7C, right panel, lanes 3 and 4), whereas a mutant that lacks the myristylation signal [Myr(−).UL11] and thus the ability to be palmitylated on membranes was not labeled to any detectable level (Fig. 7C, right panel, lane 2). As controls, cells expressing the different UL11 derivatives were also labeled with [35S]methionine and [3H]myristic acid. All constructs appeared to be expressed to levels equal to or greater than wild type (Fig. 7C, left panel) and all derivatives containing a myristylation signal were labeled efficiently with [3H]myristic acid (Fig. 7C, center panel). Taken together, these data demonstrate that loss of the three cysteines inhibits UL11 palmitylation in vivo and suggest that one or more of these cysteines represents the site of palmitylation within UL11.
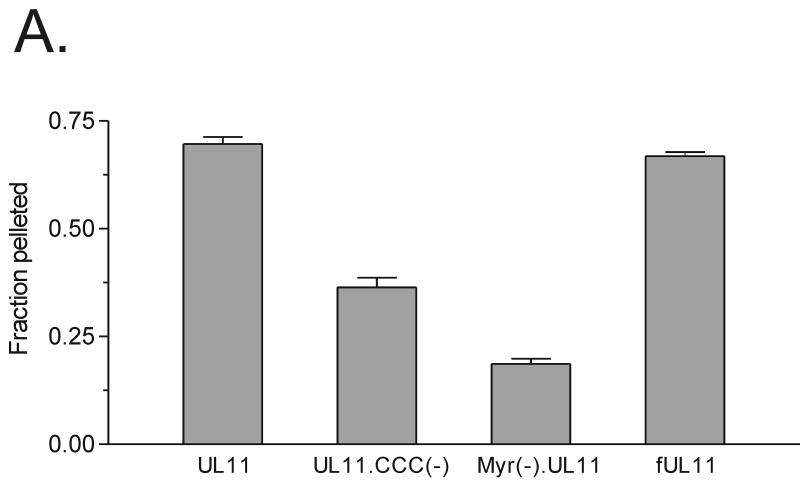
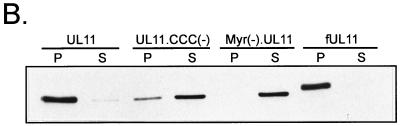
To determine whether or not palmitylation of UL11 affects the strength of membrane binding, standard membrane-pelleting experiments were performed (32). As shown in Fig. 8A, almost 70% of the wild-type UL11 and fUL11 protein (positive control) pelleted with cellular membranes, compared to only about 20% of the Myr(−).UL11 protein (negative control). This suggests that when UL11 contains both fatty acid modifications, it associates very strongly with membranes, and when it contains neither, it is essentially soluble. Interestingly, an intermediate phenotype is observed when UL11 is myristylated but not palmitylated. For instance, about 35% of UL11.CCC(−) pelleted with cellular membranes, suggesting that it binds to membranes with less affinity than wild-type UL11. Similar patterns were observed when the soluble and pellet fractions were directly analyzed by Western blot analysis (Fig. 8B) rather than indirectly by fluorometry (Fig. 8A). Taken together, these data demonstrate that palmitylation of UL11 is required for both membrane binding specificity and membrane binding strength.
FIG. 8.
Membrane binding of UL11 derivatives. Transfected A7 cells were harvested and washed in NTE buffer and then allowed to swell for 1 h in hypotonic buffer. Cells were disrupted by Dounce homogenization, and nuclei were removed by a low-speed spin. To separate membrane-bound molecules from soluble forms, the supernatants were subjected to centrifugation at 100,000 × g. The soluble (S) and pellet (P) fractions were collected and analyzed for the presence of UL11 protein by fluorometry (A) or Western blot analysis (B). The fraction pelleted was calculated by dividing the amount pelleted by the total amount of protein (soluble plus pellet). Error bars indicate standard deviations.
DISCUSSION
While the intracellular localization of many tegument proteins has been well described, little is known about how they are targeted to the compartment in which they accumulate. In this report, we have provided evidence that the tegument protein UL11 utilizes multiple pathways in order to efficiently accumulate in the Golgi apparatus. In particular, it appears that UL11 is first targeted directly to the Golgi in a process that requires palmitylation (primary targeting), then sent to the plasma membrane from the Golgi by some unknown mechanism (discussed below), and then recovered from the plasma membrane and targeted to the Golgi via the acidic cluster (secondary targeting). While the mechanism of acidic cluster-mediated trafficking has been well characterized in other recycled proteins, the relationship between palmitylation and membrane targeting is less understood.
Palmitylation and membrane targeting.
Most of the evidence linking palmitylation to targeting comes from studies in which disruption of a palmitylation signal within a membrane-associated protein leads to a decreased ability to bind to membranes and/or target to the correct compartment (for example, see reference 46). Although the mechanism for this is not known, two independent models have been proposed. In one model, proteins containing a single acyl group (e.g., myristate) are palmitylated in one compartment and then travel to other compartments through interactions with carrier proteins and receptors. Indeed, several palmitylated proteins (e.g., Ras and Src family kinases) are known to form interactions with both cytoplasmic and membrane-bound proteins and thus may rely on these interactions for their targeting specificity. The other model, known as the kinetic bilayer trapping model (35), proposes that proteins containing either a myristate or farsenyl moiety transiently associate with multiple intracellular membranes until reaching a compartment which contains a specific PAT. At this site, the protein is palmitylated and stably anchored as a result of the strong binding energy that a palmitate residue provides.
Although a few groups have reported the identification and purification of PATs from various mammalian cell lines, little is known about the localization and specificity of these enzymes (27). In support of the kinetic bilayer trapping model, one study reported purification of PATs that are enriched in the plasma membrane and that are responsible for the palmitylation of the plasma membrane-resident Src family kinases and Gα proteins (4, 10). In contrast, although several palmitylated proteins have been found to be localized to the TGN, no one has identified or purified PATs enriched in these membranes. Further characterization of the mechanism and cellular location of palmitylation will be important for a more complete understanding of both cellular and viral processes.
Many viruses are known to encode palmitylated proteins that play important roles in the assembly process. One particularly interesting example of a virus that does this is vaccinia virus. Of the six palmitylated proteins that this virus encodes, four are known to be important in either envelopment, egress, or cell-to-cell spread (15). In addition, the two palmitylated glycoproteins of Sindbis virus (17) and HSV-1 UL11 (characterized in this study) represent palmitylated proteins from other viruses that are known to function in assembly and envelopment. In all, it will be interesting to determine if other HSV-1 proteins are palmitylated and whether they are also important for viral assembly.
Role of additional signals in UL11 trafficking.
Although our studies on the acidic cluster have provided clues as to how UL11 trafficks within the cell, they have raised a number of further questions. For instance, does UL11 exit the Golgi apparatus by bulk secretory flow, or does it contain additional sequences that direct it out? Studies with furin have demonstrated that sorting signals within its cytoplasmic tail (e.g., YXXL and LI) interact with the clathrin sorting machinery at the Golgi apparatus to mediate vesicle formation and exocytosis at a rate that is greater than that provided by bulk flow (9, 38, 42). Because UL11 is a viral protein that is required for efficient nucleocapsid egress, it might also contain signals that direct it out of the Golgi (3). In fact, UL11 does contain a dileucine motif (LI) within the first half of the protein (residues 18 and 19) that may serve this purpose. Additionally, UL11 also contains three DXE motifs, which have the potential to accelerate export through the secretory pathway and increase protein expression on the cell surface (20, 24, 34)
In addition to exiting the Golgi apparatus, UL11 must be internalized from the plasma membrane in order to recycle back to the Golgi apparatus. Deletion of the acidic cluster results in an increase of protein on the plasma membrane, suggesting that the acidic cluster may play some role in endocytosis (Fig. 2B). However, because acidic cluster deletion mutants are still internalized when directly sent to the plasma membrane (Fig. 2B and 3C), we know that the acidic cluster is not the only endocytosis signal. Studies are under way to determine whether the LI motif is also involved in this process.
Role for UL11 trafficking in HSV-1-infected cells.
The observation that UL11 is able to recycle suggests that it may serve functions on membranes other than the TGN and nuclear membrane, where it has been previously observed in HSV-1-infected cells. For instance, it is possible that UL11 interacts with other HSV-1 proteins (or cellular proteins) on the plasma membrane and directs them to the proper site of envelopment. By doing this, UL11 may be serving to recruit one or more essential “budding” factors to this site. Although there is no direct evidence for this recruitment mechanism, data presented here and in other studies do suggest that UL11 interacts with something in HSV-1-infected cells. In particular, it has been observed that UL11 localizes primarily to the Golgi apparatus when expressed in the absence of other HSV-1 proteins but localizes to both the Golgi apparatus and the nuclear membrane in HSV-1-infected cells (2, 5). This difference provides evidence for an interaction of UL11 with either a viral protein(s) or a virally induced cellular protein(s). Based on a sequence alignment of different UL11 homologues, we predict that the nonconserved C terminus of UL11 is the region of the protein that is responsible for forming these interactions, but further experimentation is required to test this.
Although it is likely that UL11 trafficking is important for virus envelopment and/or egress, it is possible that recycling of UL11 serves some other purpose. For example, retrieval from the plasma membrane may simply represent a way in which UL11 remains concentrated in the Golgi apparatus and ensures its incorporation into assembling virions. Alternatively, UL11 trafficking may be required for the downregulation of other molecules (cellular or viral) from the cell surface. Indeed, the peripheral membrane protein HIV-1 Nef, which is recycled to the Golgi apparatus in a manner similar to that for UL11, binds to and downregulates both CD4 and class I major histocompatibility complex from the cell surface in HIV-1-infected cells (28, 33). Studies to determine whether UL11 performs similar functions are under way.
ACKNOWLEDGMENTS
We extend special thanks to Michael G. Fried (Department of Biochemistry and Molecular Biology, The Pennsylvania State University College of Medicine) for use of the fluorometer and for assistance with the fluorometric analyses and to Michael Brignati for discussions and careful review of the manuscript.
This work was supported in part by National Institutes of Health (NIH) grants to R.J.C. (CA42460 and CA72058) and to J.W.W. (CA47482). J.B.B. was partially supported by NIH training grant CA60395, and J.S.L was partially supported by a fellowship from the Life Sciences Consortium of Pennsylvania State University.
REFERENCES
- 1.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Jacob R J, Simmerman L, Roizman B. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthiaume L, Resh M D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J Biol Chem. 1995;270:22399–22405. doi: 10.1074/jbc.270.38.22399. [DOI] [PubMed] [Google Scholar]
- 5.Bowzard J B, Visalli R J, Wilson C B, Loomis J S, Callahan E M, Courtney R J, Wills J W. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J Virol. 2000;74:8692–8699. doi: 10.1128/jvi.74.18.8692-8699.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau A D, Del Rio T, Wolffe E J, Enquist L W. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J Virol. 1999;73:4372–4384. doi: 10.1128/jvi.73.5.4372-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittié A S, Thomas L, Thomas G, Tooze S A. Interaction of furin in immature secretory granules from neuroendocrine cells with the AP-1 adaptor complex is modulated by casein kinase II phosphorylation. EMBO J. 1997;16:4859–4870. doi: 10.1093/emboj/16.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunphy J T, Greentree W K, Manahan C L, Linder M E. G-protein palmitoyltransferase activity is enriched in plasma membranes. J Biol Chem. 1996;271:7154–7159. doi: 10.1074/jbc.271.12.7154. [DOI] [PubMed] [Google Scholar]
- 11.Erdie C R, Wills J W. Myristylation of Rous sarcoma virus Gag protein does not prevent replication in avian cells. J Virol. 1990;64:5204–5208. doi: 10.1128/jvi.64.10.5204-5208.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow H, Klupp B G, Fuchs W, Veits J, Osterrieder N, Mettenleiter T C. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosenbach D W, Hansen S G, Hruby D E. Identification and analysis of vaccinia virus palmitylproteins. Virology. 2000;275:193–206. doi: 10.1006/viro.2000.0522. [DOI] [PubMed] [Google Scholar]
- 16.Hausmann J, Ortmann D, Witt E, Veit M, Seidel W. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology. 1998;244:343–351. doi: 10.1006/viro.1998.9135. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones F, Grose C. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J Virol. 1988;62:2701–2711. doi: 10.1128/jvi.62.8.2701-2711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Zerangue N, Lin Y F, Collins A, Yu M, Jan Y N, Jan L Y. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 21.MacLean C A, Clark B, McGeoch D J. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol. 1989;70:3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- 22.MacLean C A, Dolan A, Jamieson F E, McGeoch D J. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J Gen Virol. 1992;73:539–547. doi: 10.1099/0022-1317-73-3-539. [DOI] [PubMed] [Google Scholar]
- 23.McLauchlan J, Rixon F J. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura N, Balch W E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 25.Peitzsch R M, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 26.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resh M D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 28.Rhee S S, Marsh J W. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J Virol. 1994;68:5156–5163. doi: 10.1128/jvi.68.8.5156-5163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rixon F J, Addison C, McLauchlan J. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J Gen Virol. 1992;73:277–284. doi: 10.1099/0022-1317-73-2-277. [DOI] [PubMed] [Google Scholar]
- 30.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 31.Sanchez V, Greis K D, Sztul E, Britt W J. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 34.Sevier C S, Weisz O A, Davis M, Machamer C E. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol Biol Cell. 2000;11:13–22. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahinian S, Silvius J R. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34:3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 36.Shaw A S, Amrein K E, Hammond C, Stern D F, Sefton B M, Rose J K. The Lck tyrosine protein kinase interacts with the cytoplasmic tail of the CD4 glycoprotein through its unique amino-terminal domain. Cell. 1989;59:627–636. doi: 10.1016/0092-8674(89)90008-1. [DOI] [PubMed] [Google Scholar]
- 37.Smith K O. Relationship between the envelope and the infectivity of herpes simplex virus. Proc Soc Exp Biol Med. 1964;115:814–816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- 38.Teuchert M, Schäfer W, Berghofer S, Hoflack B, Klenk H, Garten W. Sorting of furin at the trans-Golgi network: interaction of the cytoplasmic tail sorting signals with AP-1 Golgi-specific assembly proteins. J Biol Chem. 1999;274:8199–8207. doi: 10.1074/jbc.274.12.8199. [DOI] [PubMed] [Google Scholar]
- 39.Tugizov S, Maidji E, Xiao J, Pereira L. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J Virol. 1999;73:8677–8688. doi: 10.1128/jvi.73.10.8677-8688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Genderen I L, Brandimarti R, Torrisi M R, Campadelli G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 41.Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks M S, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan L, Molloy S S, Thomas L, Liu G, Xiang Y, Rybak S L, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 43.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolven A, Okamura H, Rosenblatt Y, Resh M D. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol Biol Cell. 1997;8:1159–1173. doi: 10.1091/mbc.8.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]