Abstract
Alzheimer's disease (AD) is one of the most common neurodegenerative diseases. Its etiology and associated mechanisms are still unclear, which largely hinders the development of AD treatment strategies. Many studies have shown that dysregulation of energy metabolism in the brain of AD is closely related to disease development. Dysregulation of brain energy metabolism in AD brain is associated with reduced glucose uptake and utilization, altered insulin signaling pathways, and mitochondrial dysfunction. In this study, we summarized the relevant pathways and mechanisms regarding the dysregulation of energy metabolism in AD. In addition, we highlight the possible role of mitochondrial dysfunction as a central role in the AD process. A deeper understanding of the relationship between energy metabolism dysregulation and AD may provide new insights for understanding learning memory impairment in AD patients and in improving AD prevention and treatment.
Keywords: Alzheimer’s disease, Glucose metabolism, Glucose transporters, O-GlcNAc, Mitochondrial dysfunction, Mitochondrial autophagy, Mitochondrial genetics, β-Amyloid, Tau protein
Introduction
Alzheimer's disease (AD) is a neurological disorder characterized by progressive cognitive decline. AD is pathologically characterized by progressive neuronal and synaptic loss resulting from the deposition of amyloid-beta (Aβ) plaques and phosphorylated tau neurofibrillary tangles [1, 2]. Glucose is the main source of energy for the brain, producing adenosine triphosphate (ATP) through glycolysis, the tricarboxylic acid (TCA) cycle, and the electron transport chain (ETC). It is known to enter the brain from blood vessels via glucose transporter proteins (GLUTs) and requires insulin for optimal cellular utilization. Recent studies have shown that overexpression of glucose 6-phosphate dehydrogenase (G6PD), a rate-limiting enzyme in the pentose phosphate pathway, can save cognitive loss in double-transgenic APP/PS1 mouse models, which undoubtedly provides a new perspective for the treatment of AD. Previous studies have focused on enzymes involved in the production of Aβ or p-tau, and correcting dysregulation of sugar metabolism in the AD brain may also be an effective way to impede disease progression [3]. Much evidence suggests that brain glucose metabolism dysfunction, insulin resistance, and subsequent mitochondrial dysfunction play important roles in the progression of AD [4–6].
In this review, we summarized disordered glucose metabolism, impaired insulin signaling, mitochondrial dysfunctions, and their interactions with β-amyloid deposition and tau hyperphosphorylation in the AD brain [4, 6], highlighting the involvement of glucose transporter proteins, IRS/PI3K/Akt and its downstream signaling pathways and mitochondrial dysfunction including mitochondrial DNA mutations, abnormal mitochondrial fusion and division, altered mitochondrial dynamics, and oxidative stress in disease progression [4, 7, 8]. In general, they are interrelated and together affect the energy supply and metabolic balance of the brain. A deeper understanding of these mechanisms not only helps shed light on the pathophysiological processes of AD, but also provides important insights for developing new intervention strategies and treatments that ultimately improve patients' cognitive function and overall quality of life.
Search strategy
We searched PubMed using “Glucose metabolism” “Glucose transporters” “Mitochondrial dysfunction” “Mitochondrial autophagy” “Mitochondrial genetics” “β-Amyloid” “Tau protein” “Insulin resistance” “Oxidative stress” and “Alzheimer’s disease” as keywords and then excluded irrelevant articles based on the abstract. Finally, we reviewed the mechanism of brain energy metabolism dysregulation in AD. Most of the articles were published from 2015 to 2024.
Mechanisms related to glucose transport and metabolism in Alzheimer's disease
Although the brain makes up only 2% of the body overall, it uses 25% of the body's glucose. The high energy requirements of neurons are met primarily by the supply of d-glucose in the blood. The process of brain glucose metabolism involves both intracellular glucose metabolism and glucose transportation, and it involves multiple steps. The entry process of glucose into the brain requires transporter proteins. Energy requirements vary with brain activity, and deletion of major glucose transporter proteins leads to a reduction in brain glucose supply, which affects metabolic and other processes that depend on ATP production [9, 10]. Numerous studies have shown altered brain glucose transporter protein expression and function in AD brain.
2.1 Mechanisms of glucose transport in central nervous system
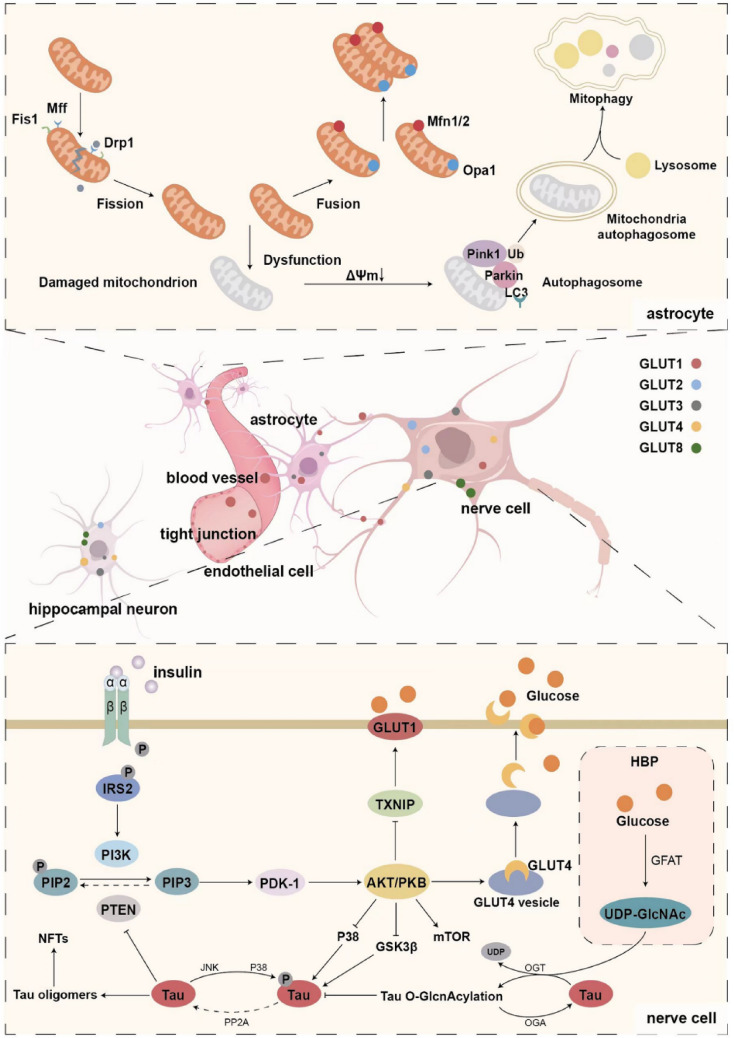
For optimal transport at various glucose concentrations and physiological demands, the brain expresses a family of transporter proteins from solute carrier 2 (SLC2), including GLUT1, GLUT2, GLUT3, GLUT4, GLUT8, and GLUT12 transporter proteins, as well as Na + -d-glucose cotransport proteins from the solute carrier 5 (SLC5) family, including sodium–glucose cotransporter 1 (SGLT1) [11]. Glucose transporter proteins are distributed in different locations and have different roles. The entry of glucose into the brain occurs primarily through the transporters GLUT1 and GLUT3 [10]. GLUT1 is located in capillary endothelial cells and astrocytes and is responsible for facilitating the passage of glucose through the blood–brain barrier and into astrocytes [11]. However, neurons do not express GLUT1, and the primary glucose transporter that facilitates glucose entrance into neurons is GLUT3. The density and distribution of GLUT3 in axons, dendrites, and neuronal bodies are related to local brain energy requirements, and low levels of GLUT3 are also detected in astrocytes and endothelial cells. Hypothalamic neurons contain GLUT2, which functions as a glucose sensor to control appetite, and it is believed that GLUT2 controls neurotransmitter release and synaptic activity in hippocampus neurons. Several insulin-sensitive transporters are present at low levels in the brain. GLUT4, an insulin-regulated glucose transporter primarily found in the hippocampus and amygdala, migrates from the cytoplasm to the cell membrane to help neurons absorb glucose, while GLUT8 is transferred to the rough endoplasmic reticulum in the cytoplasm after protein glycosylation to recycle available glucose [9–11]. In addition, GLUT12, a newly discovered glucose transporter, mainly exists in insulin-sensitive tissues. It is expressed more frequently in the frontal brain of Alzheimer's patients, according to recent studies, and the specific mechanism may be related to the deposition of Aβ [12]. (Fig. 1).
Fig. 1.
Schematic diagram of brain glucose transport, uptake and utilization-related mechanisms, and mitochondrial dynamics
Diminished expression of GLUT1 in the blood–brain barrier (BBB) is linked to diminished glucose transport in the early stages of AD [13]. The specific mechanism may be the accumulation of toxic soluble amyloid-beta-peptide (sAß) in the brain and plasma of AD patients, which reduces the expression of GLUT1 through the insulin/Akt/TXNIP axis, thus promoting BBB dysfunction [14]. The study discovered that decreased GLUT3 levels in AD patients' brain tissue hampered nerve cells' ability to absorb glucose [15]. Meanwhile, a 20% decrease of GLUT3 in the neurons of elderly AD mice is age related, while the total GLUT3 level is not affected in young AD mice [16]. It is possible that the primary cause of the decreased glucose absorption is that Aβ works by preventing GLUT3 vesicles from fusing with the plasma membrane [17].
Studies also demonstrated that the lack of glucose transport and metabolism in AD might be the result of impaired insulin signaling, which may be due to dysfunction of insulin-sensitive GLUTs. The young and elderly 3xTg-AD mice had considerably decreased total GLUT4 levels compared to age-matched non-Tg mice, by approximately 20% and 35%, respectively [16]. According to animal studies, insulin stimulation causes GLUT4 to be transported to the rat hippocampus plasma membrane, where it can enhance spatial memory and encourage glycolysis [18]. The PI3-K and MAPK pathways are required for GLUT4 translocation to the plasma membrane. Activation of protein kinase B (Akt/PKB) via the PI3-K pathway results in GLUT4 translocation [19]. Aβ(1–42) oligomers have no effect on GLUT1 or GLUT3, but they impair hippocampus insulin signaling, diminish GLUT4 translocation, and cause cognitive decline and hippocampal hypometabolism [20]. It is possible that upregulating GLUT4 mediates insulin's effect on memory [21, 22]. Hippocampal GLUT4 translocation was elevated by memory training, while GLUT4 blockage hindered memory acquisition [23].
Mitochondrial fission: Drp1 is transported from cytoplasm to mitochondria with the help of Fis1, Mff, etc. Drp1 then assembles and forms the oligomer ring structure, resulting in mitochondrial division through membrane shrinkage by GTP hydrolysis. Mitochondrial fusion: Two Mfn isomers (Mfn1/2) mediate outer membrane fusion, and Opa1 mediates inner membrane fusion to elongate mitochondria. Mitochondrial autophagy: Damaged mitochondrial membrane potential leads to depolarization, and the pathway of PINK1 into the inner mitochondrial membrane is blocked, resulting in stable accumulation of PINK1 in the outer mitochondrial membrane. E3 ubiquitin ligase is activated by phosphorylation of Parkin and its substrate ubiquitin molecule (Ub) at the Ser65 site (pSer65-Ub). The protein of the LC3 interaction domain in the mitochondrial outer membrane is the receptor of autophagy, and its binding causes extensive ubiquitination. After the recruitment of the autophagy receptor, the membrane of the autophagy begins to extend and eventually close, forming a double-membrane structure of the autophagy, and then fuses with the lysosome to form the autophagy lysosome, in which the damaged mitochondria are degraded.
Distribution of glucose transporters: The blood–brain barrier is mainly formed by the close connection of capillary endothelial cells, which are covered by continuous basement membrane, pericytes, and astrocyte terminal protrusions. Glucose transporters mediate D-glucose transport across specific membranes.
The insulin–insulin receptor (IR)–insulin receptor substrate (IRS)–PI3K pathway induces the phosphorylation and activation of PDK-1 and AKT. Activated AKT has many downstream effects: AKT phosphorylates the AKT substrate AS160, which controls the transport of GLUT4 to cell membranes; Inhibition of TXNIP by direct phosphorylation promotes plasma membrane localization of glucose transporter GLUT1; AKT inhibits Tau kinases such as GSK3β and p38, thereby limiting Tau hyperphosphorylation. Tau monomers also promote normal insulin signaling by inhibiting PTEN and thus inhibiting the conversion of PIP3 to PIP2. Intracellular glucose can enter the hexose amine biosynthetic pathway (HBP) and generate OGT substrate UDP-GlcNAc. And the glutamine-fructose-6-phosphate aminotransferase (GFAT) is the way for the speed limit of enzymes. O-GlcNAc transferase (OGT) and O-GlcNAc hydrolase (OGA) promote the installation and removal of O-GlcNAc on tau proteins, respectively, thereby regulating tau phosphorylation.
2.2 Overview of glucose utilization and metabolism
Upon entering the cell, glucose undergoes irreversible phosphorylation by hexokinase (HK) to become glucose 6-phosphate (G6P). G6P is then processed primarily through glycolysis, the pentose phosphate (PPP) pathway, glycogen generation, and other metabolic pathways. Glycolysis produces nicotinamide adenine dinucleotide (NADH), which is reoxidized in the mitochondrial ETC together with flavin adenine dinucleotide (FADH2) produced during the TCA cycle. The process of electron coupling of NADH and FADH2 with molecular oxygen to produce ATP is called oxidative phosphorylation [24].
The early and consistent feature of AD is a significant reduction in glucose utilization. The changes in glucose metabolism in AD patients are mainly related to the decrease of metabolic complex, the change of enzymes involved in glucose metabolism, the decrease of aerobic glycolysis, and the decrease of oxidative phosphorylation [24]. Specifically, the pyruvate dehydrogenase complex (PDHC), α-ketoglutarate dehydrogenase complex (KDHC), and enzymes in the electron transport chain (ETC), such as succinate dehydrogenase (Complex II) and cytochrome c oxidase (complex IV), exhibit reduced activity in AD patients [25]. Additionally, changes were seen in the glucose metabolism-related enzymes in AD, with decreased levels of hexokinase (HK) in both transgenic mouse models of amyloidosis and postmortem brain tissue of AD patients [26]. Activation of pyruvate dehydrogenase kinase 1 (PDK1) and lactate dehydrogenase A(LDHA) increases resistance to Aβ toxicity and causes a decrease in oxidative phosphorylation. Decreased expression of PDK1 was found in postmortem brain tissue of AD patients [27]. Santangelo found elevated lactate and decreased pyruvate in the brain of AD patients, suggesting that they rely more on anaerobic glycolysis for energy production, and that age-related loss of aerobic glycolysis may accelerate the pathology of AD [28].
2.3 Hexosamine biosynthetic pathway (HBP)
In addition to the above glucose utilization, approximately 3–5% of intracellular glucose enters the hexosamine biosynthesis pathway (HBP) and produces the OGT substrate uridine 50-diphosphate-n-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is the donor of protein O-linked beta-N-acetylglucosamine (O-GlcNAc) modification which is a post-translational modification that regulates a variety of cellular processes. O-GlcNActransferase (OGT) and O-GlcNAcase (OGA) promote the installation and removal of O-GlcNAc on proteins, respectively. The glycosylation of O-GlcNAc is mainly regulated by intracellular UDP-GlcNAc level, which varies with the availability of glucose in cells, so it is also known as the sensor of intracellular glucose metabolism [29]. (Fig. 1).
As a result, tau and other brain proteins have lower O-GlcNAcylation levels in AD brains. This is most likely because the brain's glucose metabolism is impaired. Both in vitro and in vivo, O-GlcNAcylation controlled the phosphorylation of tau in a site-specific way. O-GlcNAcylation on tau modulates local conformational changes that slow down fibrinogenic aggregation and counteract glycogen synthase kinase 3β (GSK-3β) induced phosphorylation [30]. Reduced tau O-GlcNAcylation was found to lead to abnormal hyperphosphorylation of tau in animal models [31].In the presence of tau pathology, an increase in O-GlcNAc can lead to a decrease both in β-amyloid peptide levels and amyloid plaque deposition [32]. It has been previously shown that O-GlcNAcylation increases the processing of non-amyloid α-secretase, leading to increased levels of neuroprotective soluble amyloid protein procurer α (sAPPα) fragments and decreased Aβ secretion [33]. Thus, O-GlcNAcylation may also be a molecular link between glucose dysregulation and β-amyloid pathology in AD. Furthermore, a multitude of research has demonstrated that O-GlcNAc also has significant impacts on mitochondria. O-GlcNAcylation controls the shape, dynamics, and oxidative stress of mitochondria [34]. Examples of these proteins include the respiratory chain complex, Milton, dynamin-related protein 1 (DRP1), and peroxisome proliferator-activated receptor-γ coactivator (PGC)−1 α ([35, 36]. In AD neurons, significant reductions in overall O-GlcNAcylation are strongly associated with mitochondrial defects, and OGA inhibitors can restore these mitochondrial abnormalities and stop cell death by raising O-GlcNAcylation levels [37].
2.4 Insulin signaling
Insulin signaling plays a crucial role in regulating glucose metabolism in nerve cells. Numerous studies have shown that insulin freely penetrates the blood–brain barrier from the bloodstream to control the metabolism of glucose in the brain, promotes neuronal growth, regulates the release and uptake of catecholamines, and controls the distribution and expression of GABA (gamma-aminobutyric acid) [38, 39]. Insulin resistance (IR) is a pathological state when cells' ability to respond to insulin is reduced, resulting in a biological effect that is less than expected from a given quantity of insulin [39]. Alterations in insulin signaling may hasten the aging of the brain, impact neuron plasticity, and potentially cause neurodegeneration. These changes are also responsible for insulin resistance. A fludeoxyglucose-18 (FDG) positron emission tomography (PET) study has shown that reduced local glucose metabolism is linked to insulin resistance, especially in the medial temporal lobe [40].
2.4.1 Altered insulin signaling and β-amyloid
Impaired insulin signaling has been linked to Aβ deposition in the brain, which can be harmful to synapses and contribute to memory loss in Alzheimer's disease [41]. Insulin regulates the metabolism of APP to maintain the equilibrium between the anabolism and catabolism of Aβ [42]. Insulin accelerates APP/Aβ trafficking from the trans-Golgi network, a key cellular location for Aβ synthesis, to the plasma membrane, thereby drastically reducing intracellular accumulation of Aβ, according to in vitro studies [43]. Research on animals indicates that a brain insulin shortage could result in higher production of Aβ, and insulin resistance increases the production of Aβ in the brain via altered insulin signal transduction, increased β-secretase, and γ-secretase activities [44], and autophagosome accumulations [45, 46]. Insulin-like growth factors (IGF), IGF receptors, insulin receptor substrate-1 (IRS-1) [47], and insulin-stimulated GLUT4 [48] are some of the IR components that are important in controlling APP processing, because they increase GSK-3β activity and decrease protein kinase B activity. It has also been shown that brain insulin and IGF-1 receptor dysfunction can also lead to Aβ aggregation and synaptic loss in the brain and that elevated levels of Aβ can in turn antagonize the binding of insulin and IGF-1 to their corresponding receptors, exacerbating neuronal resistance to insulin or IGF-1 [49].
The impaired insulin signaling pathway can affect the deposition of Aβ, while the accumulation of Aβ will further deepen the impaired insulin signaling pathway. Aβ can competitively bind insulin receptors and induce insulin resistance [50], inhibit autophosphorylation of insulin receptors, and also decrease insulin receptor levels and activity [51, 52], specifically on dendrites bound by soluble Aβ oligomers [51]. Additionally, it has been discovered that Aβ oligomers decide abnormal TNFα/JNK activation and IRS-1 inhibition in both in vitro and in vivo scenarios [51, 53], which induces tau hyperphosphorylation and altered insulin signaling.
On the other hand, the main Aβ degradation peptidase is insulin-degrading enzyme (IDE), which is controlled by insulin levels [54]. Mice lacking the IDE gene showed increased amounts of Aβ in the brain [55]. On the other hand, in APP transgenic mice, overexpression of IDE decreased brain Aβ levels and slowed or stopped Aβ plaque formation [56]. Insulin elevates IDE protein levels through the phosphatidylinositol-3-kinase (PI3K) pathway [57], and research demonstrates that in the brains of AD patients and APP transgenic mice, lower IDE was linked with defective insulin signaling (decreased PI3K subunit P85). Additionally, IDE serves as a mediator between AD and insulin resistance. In the mice brain, diet-induced insulin resistance promoted the production of Aβ, which correlated with enhanced γ-secretase activity and reduced IDE activity [58].
2.4.2 Altered insulin signaling and tau protein
Altered insulin signaling pathway might lead to abnormal phosphorylation of the tau protein. Tau protein controls intracellular signaling and maintains the integrity of microtubules [59], and the aggregation of hyperphosphorylated tau leads to neuronal dysfunction and degeneration in AD [60]. It has been demonstrated that by triggering the PI3 kinase (PI3-K) signaling pathway [61], insulin secretion and action decrease glycogen synthase kinase 3 (GSK-3β), a tau protein phosphorylation that has been extensively addressed [62]. Therefore, by blocking PI3-K/AKT and increasing GSK-3β activity, compromised insulin or IGF-1 signaling pathways can encourage tau phosphorylation [63].
In Alzheimer’s pathology, there is a vicious circle between insulin resistance, Tau pathology, and Aβ pathology. Impaired insulin signaling induces deposition of Aβ and hyperphosphorylation of tau, and those above pathological changes will further exacerbate insulin signaling impairment. The correlation between insulin signaling and Aβ has been discussed above. Tau deletion causes an impaired hippocampus insulin response due to decreased activity of IRS-1 and phosphatase and tensin congeners on chromosome 10 (PTEN), which negatively controls the PI3-K/AKT pathway. PTEN inhibits insulin signaling and reduces its lipid phosphatase activity [64]. Researchers believe that the pathologic loss of tau function promotes brain insulin resistance, which leads to cognitive and metabolic impairment in AD patients [65].
3 Mitochondrial dysfunction in Alzheimer's disease
Mitochondria produce ATP through oxidative phosphorylation (OXOPHOS) using reducing agents such as reducing NADH and FADH. ATP is necessary for maintaining neuronal function, transmitting neuronal information, and developing strong neural connections in learning and memory. Numerous studies have shown widespread mitochondrial abnormalities in the brains of AD patients. Below, we will analyze the mechanisms and effects of mitochondrial dysfunction in AD in four key areas, including gene level (mtDNA methylation, mutations, and non-coding RNA), calcium signaling, mitochondrial metabolism, and kinetic changes.
3.1 Gene level
The nuclear and mitochondrial genomes both affect mitochondrial activity [8] as most mitochondria proteins are encoded by the nucleus [66] and directed to the mitochondria by mitochondria-targeting sequences. Epigenetic modification of nuclear genes encoding mitochondrial proteins affects mitochondrial homeostasis. In addition, abnormal modification of mitochondrial DNA (mtDNA) can also lead to the development of diseases through mitochondrial dysfunction.
3.1.1 mtDNA methylation
In epigenetic processes, DNA methylation is probably the most easily understood epigenetic adaptation and the most common DNA modification. Stoccoro et al. found that mtDNA methylation was reduced by about 25% in blood samples of AD patients compared to D-loop methylation levels in the control group [67]. Blanch et al. found increased mtDNA methylation in the D-loop of the inner olfactory cortex of Braak stage I–II and III–IV AD [68]. The production of 5-hydroxymethylcytosine (5hmC), a crucial step in the demethylation of DNA, is facilitated by the proteins belonging to the 10–11 translocation (TET) family, which oxidize 5-methylcytosine (5mC) to 5hmC [69]. Previous small sample studies have shown elevated mitochondrial methylated cytosine-derived bases, and 5hmC levels in the superior temporal gyrus and the middle temporal gyrus of preclinical AD and advanced AD [70]. Recent studies have shown that with loss of TET function, AD-related pathology increases and that regulation of the methylome of TET enzymes, particularly TET1, may contribute to AD pathogenesis [71]. Single-carbon metabolism polymorphisms of MTRR 66 > G and DNMT3a −448A > G were significantly correlated with D-loop mtDNA methylation [72].
3.1.2 MtDNA mutation and damage
The accumulation of somatic mtDNA mutations affects mitochondrial function, and leads to AD progression [73]. Krishnan et al. demonstrated an increase in neurons expressing a mitochondrial biochemical defect-cytochrome-c oxidase (COX) deficit, which was well reported as a hallmark of mtDNA dysfunction in the hippocampus in sporadic AD patients relative to age-matched controls [74]. Mitochondrial dysfunction caused by oxidative damage may be related to mtDNA damage. In AD, there is more brain oxidative damage [75]. Previous research in the parietal cortex of AD patients demonstrated an extremely significant threefold increase in oxidative damage to mtDNA and a minor significant increase to nuclear DNA, confirming that mtDNA is especially vulnerable to oxidative damage [76]. However, another study has reported that the significant increase in the frequency of hippocampal mtDNA mutations in early AD patients may be related to the accumulation of replication errors rather than an immediate result of oxidative damage [77].
Research on AD patients and animal models indicates that early in the neurodegenerative process, unrepaired oxidative damage to nuclear and mtDNA may accumulate. It has been demonstrated that AD-related DNA repair dysfunction increases neurodegeneration. Base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), double-strand break repair (DSBR), and direct reversal are the primary DNA repair methods [78]. DSBR consists of both non-homologous end junction (NHEJ) and homologous recombination (HR). Compared to normal aging, AD brain cells have more extensive damage to the BER and DSBR pathways [79]. Haploid defect of DNA base excision repair enzyme is a key enzyme in the bit error rate of neurons, and the reduction of this enzyme can lead to the aggravation of AD characteristics in 3xTgAD mice, inducing neuronal dysfunction, cell death, impairment of memory, and synaptic plasticity [80]. DNA damage is a major activator of adenosine diphosphate-ribose polymerase-1 (PARP-1), an enzyme that uses nicotinamide adenine dinucleotide(NAD+) as a cofactor to catalyze post-translational modification of proteins. PARP-1 activation affects cytochrome oxidase IV protein levels, oxygen consumption, and membrane potential, leading to a cell bioenergy crisis that leads to cell death [81, 82]. Poly-ADP-ribose (PAR) protein accumulated and PARP1 activity increased in brain tissue samples from AD patients' susceptible areas. PARP1 is a unique signaling pathway that triggers programmed cell death [83]. The lack of NAD + in AD may be due to enhance oxidative stress-mediated DNA damage which leads to PARP1 activation, and finally caused mitochondrial dysfunction. There is a relationship between inflammation, mitochondrial dysfunction, and the pathophysiology of AD. AD pathogenesis can result from oxidative damage and lipid peroxide destruction of mtDNA, which can start inflammatory signals in astrocytes and set off cascade reactions [84].
3.1.3 Non-coding RNA (miRNA)
In mitochondria, non-coding RNAs from the nucleus or mitochondrial genome involved in epigenetic regulation mainly include miRNA, small interfering RNA (siRNA), Piwi-interacting RNA (piRNA), long non-coding RNAs(lncRNA), and circular RNA(circRNA). Among them, miRNAs regulate gene expression by inhibiting or degrading the level of target messenger RNA (mRNAs) after transcription. In vitro studies have shown that amyloid-oligo-β42 (OAβ42) can stimulate the production of mitochondrial tumor necrosis factor-α(TNF-α), thereby elevating the degree of miR-34a expression, which decreases five important proteins' expression in mitochondrial ETC through base pair complementarity [85], thus leading to mitochondrial dysfunction. Moreover, miR-34a negatively regulates the expression of B-cell lymphoma-2(Bcl2) by directly targeting the 3’-untranslated region(3’UTR) of Bcl2, an anti-apoptotic protein mainly located in mitochondria, whose expression is known to protect cell viability by inhibiting apoptosis induced by caspase-9 [86]. In AD transgenic mice, downregulation of Bcl2 leads to caspase3 activation and neurodegeneration [87, 88]. John et al. concluded and analyzed that miR-15b, miR-23a/b, miR-107, miR-132, miR-143, and miR-330 having neuroprotective effects of preventing mitochondrial dysfunction and Aβ formation were down-regulated in neurons of AD patients. However, miR-15a, miR-34a, miR-126, miR-181a, miR-210-3p, miR-425, and miR-424 are elevated in AD, which can lead to the loss of mitochondrial structure and function and play a pathological role in AD [89]. Some of these miRNAs act on mitochondria and regulate mitochondrial biogenesis (miR-23a/23b), kinetics (miR-140, miR-195), and mitochondrial autophagy (miR-101) [90].
3.2 Calcium signaling
In AD, there is a close relationship between calcium ion signaling and its homeostasis, ATP production, and mitochondrial dysfunction. As mentioned above, after being transported via GLUTs, glucose can be converted into ATP through glycolysis and TCA cycle thereby meeting the primary energy requirements of the brain and maintaining the normal operation of neuronal signals and other non-signaling activities. Among them, intracellular Ca2+ buffering is one of the most important ATP-dependent neuronal mechanisms [91]. Intracellular Ca2 + is a common second messenger that is essential for regulating many aspects of neuronal function, including action potential, learning and memory, neuron development and differentiation, and synaptic plasticity [92]. Two ATPases, sarco endoplasmic reticulum Ca2+ ATPase(SERCA) and plasma membrane Ca2+ ATPase(PMCA) of the substrate, respectively, produce ATP into the endoplasmic reticulum Ca2+ pump and pump out of the cell [93]. In the absence of an adequate supply of ATP, intracellular Ca2+ levels may increase, resulting in certain effects or loss of function of the cell, leading to neurological susceptibility [94]. Meanwhile, recent studies have found that calcium homeostasis regulatory protein(CALHM)2 V136G mutation (rs232660) is significantly correlated with AD [95]. CALHM is a voltage-dependent calcium channel with a quaternary transmembrane structure and is also an important channel mediating cellular ATP release, which is activated by the decrease of extracellular Ca2+ level and plasma membrane depolarization [96, 97]. V136G mutation leads to loss of CALHM2 ATP release function and impaired synaptic plasticity in astrocytes, and CALHM2V136G mutation makes mice prone to cognitive decline in old age [95]. Thus, this could result in a destructive cycle of Ca2+ and ATP that together promote AD disease progression.
Mitochondrial Ca2+ homeostasis is essential for mitochondrial function and ATP production [98]. Mitochondrial Ca2+ imbalance triggers mitochondrial dynamic(transport, fission, fusion) changes and mitochondrial autophagy, thus leading to cell death in AD [99–101]. The main mechanism may be that amyloid deposition and tau hyperphosphorylation have neurotoxic effects on mitochondrial Ca2+ homeostasis [98]. Additionally, hyperphosphorylated tau protein disrupts neuronal circuits, opens voltage-gated Ca2 + channels (VGCCs), and depletes nuclear Ca2 + , all of which have an impact on Ca2 + homeostasis [102–104]. Aβ has the ability to create holes in the plasma membrane or over-activate channels, which permits a significant amount of Ca2 + to enter the cytoplasm from the extracellular space. On the other hand, studies have also confirmed that elevated mitochondrial Ca2+ levels are associated with plaque deposition and neuronal death in mouse models of AD [105]. It has been found that in vivo mitochondrial Ca2+ overload involving toxic extracellular Aβ oligomers is prevented by blocking mitochondrial Ca2+ mono-transporters. Therefore, the reduction of neuronal mitochondrial Ca2+ by inhibiting mitochondrial Ca2+ mono-transporters could serve as a new potential therapeutic target for the treatment of AD [105].
3.3 Mitochondrial metabolism and kinetic changes
Changes in mitochondrial metabolism and dynamics have been closely linked to various diseases (e.g., AD and Parkinson's disease), and it is critical to study mitochondrial morphology and dynamics, because they directly affect cellular health and function. The main dynamic activities of mitochondria are fusion (two organelles merging into one), fission (a single organelle splitting into two), transport (directed movement within the cell), and autophagy (targeted destruction through the autophagy pathway). In mammals, mitochondria fusion is mediated by three GTPases: Mitofusin1 (Mfn1), Mitofusin2 (Mfn2), and opticnerveatrophyprotein1 (Opa1). Mfn1 and Mfn2 mediated the mitochondrial outer membrane fusion; Opa1 collaborates with Mfn1/2 and other proteins mediated intimal fusion and ridge remodeling [106, 107]. As a complement to fusion, mitochondrial division is equally important for cellular and organismal physiology. Mitochondrial division is mediated by dynein-associated protein 1(Drp1), a large GTPase that mediates mitochondrial fission through the fission factor (Fis1) or mitochondrial fission factor (Mff) receptor expressing in the outer membrane of mitochondria [108]. The possibility exists that Drp1 phosphorylation is linked to the regulation mechanism of mitochondrial fission [109]. Another method of maintaining mitochondrial mass is mitochondrial autophagy, which is primarily mediated by phosphatase and tensin homolog-induced putative kinase 1(Pink1) and E3 ubiquitin ligase (Parkin) [110]. Increased reactive oxygen species (ROS) levels and mitochondrial membrane depolarization tend to accumulate Pink1 on the mitochondrial outer membrane, further phosphorylates protein including the recruitment and activation of Parkin, and reduce oxidative stress by mediating autophagy signaling and autophagy receptors’ activation [108, 111] (Fig. 1).
Previous research has demonstrated a significant decrease in the protein expression levels of the big kinetic protein-related GTPases involved in fission and fusion in the AD brain, including DRP1, Opa1, Mfn1, and Mfn2, while Fis1 is significantly increased [110, 112]. Aβ can lead to increased mitochondrial fragmentation and decreased neuronal fusion [113], while in APP mice, reducing Drp1 can reduce Aβ generation and mitochondrial dysfunction, improving mitochondrial biogenesis and synaptic activity [114]. In addition, Drp1 and glycogen synthase kinase 3β (GSK3β) are related to the pathogenesis of AD. Studies have demonstrated that Drp1Ser693 is the phosphorylation site of GSK3β [109]. Therefore, GSK3β in AD neurons can be mediated by the Drp1 phosphorylation of mitochondria.
A key function of PINK1 is to maintain a balance between mitochondrial fission and fusion. While PINK1 suppression can cause mitochondrial fusion, PINK1 overexpression can encourage mitochondrial fission [115]. PINK1 provides protection against oxidative stress, proteasome activity, endoplasmic reticulum stress, and neurodegeneration caused by mitochondrial malfunction [5]. A decrease in mitochondrial levels was found in 12 month old APP mice, along with a decrease in PINK1 and telomerase reverse transcriptase (TERT) levels involved in mitochondrial autophagy [116]. Activation of the Adrb2/Akt/Pink1 signaling pathway could improve mitochondrial autophagy in mice, and alleviate mitochondrial dysfunction, and tau pathology [117].
The formation of advanced glycation end products (AGEs) in cells and tissues is a normal aspect of aging, but it accelerates in AD, according to transmission electron microscopy [118]. AGEs can influence mitochondrial dynamics by affecting the fusion–fission balance, significantly increasing mitochondrial fission in AD. The influence of AGEs could be attributed to the fact that AGEs up-regulate the expression of fission proteins Drp1 and Fis1, and down-regulate the expression of fusion proteins Mfn1, Mfn2, and Opa1 [7], and through ROS, AGEs are also implicated in aberrant APP processing and the generation of Aβ [119]. In a study using transgenic tau mice, mitochondrial malfunction was also discovered. This included decreased cytochrome oxidase activity, elevated amounts of H2O2 and lipid peroxidation, and decreased levels of mitochondrial ATP. In addition, these mice had reduced fusion proteins (Mfn2, Mfn1, and Opa12) [120], suggesting that phosphorylated tau protein can induce mitochondrial abnormalities.
Overall, the primary causes of mitotic autophagy defects in AD are generally attributed to the production and accumulation of Aβ and phosphorylated tau proteins [7], the abnormal interaction of Aβ and phosphorylated tau proteins with Drp1 and volt-dependent anion channels (VDAC), and decreased levels of PINK1 and parkin1 [108].
4 Translational therapy
In recent years, there has been a gradual increase in the number of clinical studies addressing the mechanisms of glucose metabolism in AD patients, with researchers focusing on dietary interventions (e.g., intermittent fasting [121] or ketogenic diet [122]) and pharmacological therapies (glucose-lowering medications, medications to improve insulin sensitivity, etc.), to explore whether these can attenuate or slow down the progression of AD. For example, liraglutide, a glucagon-like peptide-1 (GLP-1) analog, restores BBB glucose transport [123], and is also an insulin sensitizer with neuroprotective properties, which can ameliorate insulin resistance in patients with AD [124]. Newer glucose-lowering drugs, such as dipeptidyl peptidase 4 (DPP-4) inhibitors, glucagon-like peptide-1 receptor agonists (GLP-1RAs), and sodium–glucose cotransporter-2 (SGLT2) inhibitors, may also help to prevent or delay the onset of dementia through a variety of mechanisms, including reducing insulin resistance and oxidative stress, and attenuating amyloid deposition and tau phosphorylation [125].
State-of-the-art therapeutic approaches to mitochondrial dysfunction in AD are emerging as a hot topic of research and are currently focused on several key areas. Current mainstream therapeutic targets, anti-amyloid antibodies (e.g., Aducanumab, Lecanemab, and Donanemab) have been used to reduce brain amyloid load in AD patients [126]. Meanwhile, research on tau proteins is rapidly advancing (anti-p-tau217 antibody), inhibiting the aberrant phosphorylation of tau proteins, thereby slowing the formation of neurofibrillary tangle [127]. Targeting Aβ or tau can also reduce mitochondrial damage at the cellular level [126].
Meanwhile, multiple therapeutic strategies are currently being investigated to improve mitochondrial function and reduce oxidative stress in AD. Metformin has been found to have a positive effect on improving cognition in patients with cognitive impairment, which may be related to improved mitochondrial metabolism and insulin signaling, as well as activation of AMPK, modulation of microglial cell phenotypes, and an increase in autophagy in the brain [128, 129]. Some other mitochondria-targeted antioxidants, such as MitoQ, SS-31, SkQ, MitoApocynin, MitoTEMPO, and MitoVitE, can reduce ROS levels and oxidative stress [130]. Mitochondrial division inhibitors (e.g., Mdivi-1) help reduce neuronal damage and improve cognitive function by modulating the activity of the dynamin-related protein Drp1 [131]. Some small molecule drugs, such as Urolithin A and Mito Q, are effective in modulating the mitochondrial autophagy process and attenuating oxidative stress damage to neuronal cells [132, 133]. There is also a trend toward gene therapy, particularly the use of clustered regularly interspaced palindromic repeats (CRISPR)-associated protein 9 (Cas9) technology to correct defective mitochondrial genes for the treatment of AD [134].
4 Conclusion
Reduced expression and dysfunction of GLUTs in AD might impair glucose uptake in neurons. Decreased complexes involved in glucose metabolism, enzyme changes, decreased aerobic glycolysis and oxidative phosphorylation, and impaired insulin signaling pathway may impair glucose utilization and metabolism. At the same time, mitochondrial dysfunction is also an important factor that interacts with the reduction of glucose utilization. Impaired energy metabolism during AD is intrinsically linked to the loss of mitochondrial structural and functional integrity; in addition, dysregulation of glucose metabolism and mitochondrial dysfunction are related to amyloid deposition and tau hyperphosphorylation in AD, whether as primary or secondary events, which can provide a variety of new therapeutic targets. An increasing number of studies have begun to focus on the application of regulating glucose metabolism, insulin signaling pathway, and mitochondrial function in AD treatment strategies. We hope that this review can provide a reference for intervention studies on the dysregulation of energy metabolism in AD.
Author contributions
Material preparation, data collection, and analysis were performed by Zhao Yang and Yuan Yue. The first draft of the manuscript was written by Yuan Yue and Zhao Gang. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China under Grant No. 81600924.
Data availability
Not applicable.
Declarations
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
References
- 1.Soria Lopez JA, González HM, Léger GC (2019) Alzheimer’s disease. Handb Clin Neurol 167:231–255. 10.1016/B978-0-12-804766-8.00013-3 [DOI] [PubMed] [Google Scholar]
- 2.Grundke-Iqbal I, Iqbal K, Tung YC et al (1986) Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci 83:4913–4917. 10.1073/pnas.83.13.4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Correas AG, Olaso-Gonzalez G, Roca M et al (2024) Glucose 6-phosphate dehydrogenase overexpression rescues the loss of cognition in the double transgenic APP/PS1 mouse model of Alzheimer’s disease. Redox Biol 75:103242. 10.1016/j.redox.2024.103242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Zhao F, Ma X et al (2020) Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener 15:30. 10.1186/s13024-020-00376-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewanjee S, Chakraborty P, Bhattacharya H et al (2022) Altered glucose metabolism in Alzheimer’s disease: role of mitochondrial dysfunction and oxidative stress. Free Radic Biol Med 193:134–157. 10.1016/j.freeradbiomed.2022.09.032 [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA, Halliwell B (2019) Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci 20:148–160. 10.1038/s41583-019-0132-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pradeepkiran JA, Reddy PH (2020) Defective mitophagy in Alzheimer’s disease. Ageing Res Rev 64:101191. 10.1016/j.arr.2020.101191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quirós PM, Mottis A, Auwerx J (2016) Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 17:213–226. 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- 9.Szablewski L (2017) Glucose transporters in brain. In Health and in Alzheimer’s Disease. J Alzheimers Dis JAD 55:1307–1320. 10.3233/JAD-160841 [DOI] [PubMed] [Google Scholar]
- 10.Kyrtata N, Emsley HCA, Sparasci O et al (2021) A systematic review of glucose transport alterations in Alzheimer’s Disease. Front Neurosci 15:626636. 10.3389/fnins.2021.626636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koepsell H (2020) Glucose transporters in brain in health and disease. Pflugers Arch 472:1299–1343. 10.1007/s00424-020-02441-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil-Iturbe E, Solas M, Cuadrado-Tejedo M et al (2020) GLUT12 expression in brain of mouse models of Alzheimer’s Disease. Mol Neurobiol 57:798–805. 10.1007/s12035-019-01743-1 [DOI] [PubMed] [Google Scholar]
- 13.Winkler EA, Nishida Y, Sagare AP et al (2015) GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat Neurosci 18:521–530. 10.1038/nn.3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Curran GL, Min PH et al (2022) Amyloid beta peptides inhibit glucose transport at the blood–brain barrier by disrupting insulin-Akt pathway in Alzheimer’s Disease. Pharmacol Toxicol. 10.1101/2022.11.21.517280 [Google Scholar]
- 15.Simpson IA, Chundu KR, Davies-Hill T et al (1994) Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer’s disease. Ann Neurol 35:546–551. 10.1002/ana.410350507 [DOI] [PubMed] [Google Scholar]
- 16.Sancheti H, Akopian G, Yin F et al (2013) Age-dependent modulation of synaptic plasticity and insulin mimetic effect of lipoic acid on a mouse model of Alzheimer’s disease. PLoS ONE 8:e69830. 10.1371/journal.pone.0069830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uemura E, Greenlee HW (2001) Amyloid beta-peptide inhibits neuronal glucose uptake by preventing exocytosis. Exp Neurol 170:270–276. 10.1006/exnr.2001.7719 [DOI] [PubMed] [Google Scholar]
- 18.McNay EC, Ong CT, McCrimmon RJ et al (2010) Hippocampal memory processes are modulated by insulin and high-fat-induced insulin resistance. Neurobiol Learn Mem 93:546–553. 10.1016/j.nlm.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura T (2023) The mechanisms of glucose transporter type 4 translocation regulated by insulin receptor signaling. Nihon Yakurigaku Zasshi Folia Pharmacol Jpn. 10.1254/fpj.22106 [DOI] [PubMed]
- 20.Pearson-Leary J, McNay EC (2012) Intrahippocampal administration of amyloid-β(1–42) oligomers acutely impairs spatial working memory, insulin signaling, and hippocampal metabolism. J Alzheimers Dis JAD 30:413–422. 10.3233/JAD-2012-112192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sano H, Eguez L, Teruel MN et al (2007) Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab 5:293–303. 10.1016/j.cmet.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 22.Pearson-Leary J, Jahagirdar V, Sage J, McNay EC (2018) Insulin modulates hippocampally-mediated spatial working memory via glucose transporter-4. Behav Brain Res 338:32–39. 10.1016/j.bbr.2017.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson-Leary J, McNay EC (2016) Novel roles for the insulin-regulated glucose transporter-4 in hippocampally dependent memory. J Neurosci Off J Soc Neurosci 36:11851–11864. 10.1523/JNEUROSCI.1700-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han R, Liang J, Zhou B (2021) Glucose metabolic dysfunction in neurodegenerative diseases-new mechanistic insights and the potential of hypoxia as a prospective therapy targeting metabolic reprogramming. Int J Mol Sci 22:5887. 10.3390/ijms22115887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoffner JM (1997) Oxidative phosphorylation defects and Alzheimer’s disease. Neurogenetics 1:13–19. 10.1007/s100480050002 [DOI] [PubMed] [Google Scholar]
- 26.Cuadrado-Tejedor M, Vilariño M, Cabodevilla F et al (2011) Enhanced expression of the voltage-dependent anion channel 1 (VDAC1) in Alzheimer’s Disease transgenic mice: an insight into the pathogenic effects of amyloid-β. J Alzheimers Dis 23:195–206. 10.3233/JAD-2010-100966 [DOI] [PubMed] [Google Scholar]
- 27.Newington JT, Rappon T, Albers S et al (2012) Overexpression of pyruvate dehydrogenase kinase 1 and lactate dehydrogenase A in nerve cells confers resistance to amyloid β and other toxins by decreasing mitochondrial respiration and reactive oxygen species production. J Biol Chem 287:37245–37258. 10.1074/jbc.M112.366195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santangelo R, Giuffrida ML, Satriano C et al (2021) β-amyloid monomers drive up neuronal aerobic glycolysis in response to energy stressors. Aging 13:18033–18050. 10.18632/aging.203330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuzwa SA, Vocadlo DJ (2014) O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem Soc Rev 43:6839–6858. 10.1039/C4CS00038B [DOI] [PubMed] [Google Scholar]
- 30.Cantrelle F-X, Loyens A, Trivelli X et al (2021) Phosphorylation and O-GlcNAcylation of the PHF-1 epitope of tau protein induce local conformational changes of the c-terminus and modulate tau self-assembly into fibrillar aggregates. Front Mol Neurosci 14:661368. 10.3389/fnmol.2021.661368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Iqbal K, Grundke-Iqbal I et al (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci U S A 101:10804–10809. 10.1073/pnas.0400348101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuzwa SA, Shan X, Jones BA et al (2014) Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic tau/APP mutant mice. Mol Neurodegener 9:42. 10.1186/1750-1326-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobsen KT, Iverfeldt K (2011) O-GlcNAcylation increases non-amyloidogenic processing of the amyloid-β precursor protein (APP). Biochem Biophys Res Commun 404:882–886. 10.1016/j.bbrc.2010.12.080 [DOI] [PubMed] [Google Scholar]
- 34.Tan EP, Villar MT, E L, et al (2014) Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function. J Biol Chem 289:14719–14730. 10.1074/jbc.M113.525790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu H, Pekkurnaz G, Falk J et al (2021) FHL2 anchors mitochondria to actin and adapts mitochondrial dynamics to glucose supply. J Cell Biol 220:e201912077. 10.1083/jcb.201912077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gawlowski T, Suarez J, Scott B et al (2012) Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J Biol Chem 287:30024–30034. 10.1074/jbc.M112.390682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinho TS, Correia SC, Perry G et al (2019) Diminished O-GlcNAcylation in Alzheimer’s disease is strongly correlated with mitochondrial anomalies. Biochim Biophys Acta Mol Basis Dis 1865:2048–2059. 10.1016/j.bbadis.2018.10.037 [DOI] [PubMed] [Google Scholar]
- 38.Neth BJ, Craft S (2017) Insulin resistance and Alzheimer’s Disease: bioenergetic linkages. Front Aging Neurosci 9:345. 10.3389/fnagi.2017.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sędzikowska A, Szablewski L (2021) Insulin and insulin resistance in Alzheimer’s Disease. Int J Mol Sci 22:9987. 10.3390/ijms22189987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willette AA, Bendlin BB, Starks EJ et al (2015) Association of insulin resistance with cerebral glucose uptake in late middle-aged adults at risk for Alzheimer Disease. JAMA Neurol 72:1013–1020. 10.1001/jamaneurol.2015.0613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao W-Q, Lacor PN, Chen H et al (2009) Insulin receptor dysfunction impairs cellular clearance of neurotoxic oligomeric a{beta}. J Biol Chem 284:18742–18753. 10.1074/jbc.M109.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burillo J, Marqués P, Jiménez B et al (2021) Insulin resistance and diabetes mellitus in Alzheimer’s disease. Cells 10:1236. 10.3390/cells10051236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasparini L, Gouras GK, Wang R et al (2001) Stimulation of beta-amyloid precursor protein trafficking by insulin reduces intraneuronal beta-amyloid and requires mitogen-activated protein kinase signaling. J Neurosci Off J Soc Neurosci 21:2561–2570. 10.1523/JNEUROSCI.21-08-02561.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Son SM, Song H, Byun J et al (2012) Altered APP processing in insulin-resistant conditions is mediated by autophagosome accumulation via the inhibition of mammalian target of rapamycin pathway. Diabetes 61:3126–3138. 10.2337/db11-1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Son SM, Song H, Byun J et al (2012) Accumulation of autophagosomes contributes to enhanced amyloidogenic APP processing under insulin-resistant conditions. Autophagy 8:1842–1844. 10.4161/auto.21861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devi L, Alldred MJ, Ginsberg SD, Ohno M (2012) Mechanisms underlying insulin deficiency-induced acceleration of β-amyloidosis in a mouse model of Alzheimer’s disease. PLoS ONE 7:e32792. 10.1371/journal.pone.0032792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Q-L, Yang F, Rosario ER et al (2009) Beta-amyloid oligomers induce phosphorylation of tau and inactivation of insulin receptor substrate via c-Jun N-terminal kinase signaling: suppression by omega-3 fatty acids and curcumin. J Neurosci Off J Soc Neurosci 29:9078–9089. 10.1523/JNEUROSCI.1071-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao J, Yamashita H, Qiao L, Friedman JE (2000) Decreased Akt kinase activity and insulin resistance in C57BL/KsJ-Leprdb/db mice. J Endocrinol 167:107–115. 10.1677/joe.0.1670107 [DOI] [PubMed] [Google Scholar]
- 49.Pandini G, Pace V, Copani A et al (2013) Insulin has multiple antiamyloidogenic effects on human neuronal cells. Endocrinology 154:375–387. 10.1210/en.2012-1661 [DOI] [PubMed] [Google Scholar]
- 50.Xie L, Helmerhorst E, Taddei K et al (2002) Alzheimer’s beta-amyloid peptides compete for insulin binding to the insulin receptor. J Neurosci Off J Soc Neurosci 22:RC221. 10.1523/JNEUROSCI.22-10-j0001.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao W-Q, De Felice FG, Fernandez S et al (2008) Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J Off Publ Fed Am Soc Exp Biol 22:246–260. 10.1096/fj.06-7703com [DOI] [PubMed] [Google Scholar]
- 52.Ling X, Martins RN, Racchi M et al (2002) Amyloid beta antagonizes insulin promoted secretion of the amyloid beta-protein precursor. J Alzheimers Dis JAD 4:369–374. 10.3233/jad-2002-4504 [DOI] [PubMed] [Google Scholar]
- 53.Bomfim TR, Forny-Germano L, Sathler LB et al (2012) An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer’s disease- associated Aβ oligomers. J Clin Invest 122:1339–1353. 10.1172/JCI57256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vekrellis V, Ye Z, Qiu WQ et al (2000) Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci Off J Soc Neurosci 20:1657–1665. 10.1523/JNEUROSCI.20-05-01657.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farris W, Mansourian S, Chang Y et al (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A 100:4162–4167. 10.1073/pnas.0230450100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leissring MA, Farris W, Chang AY et al (2003) Enhanced proteolysis of β-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron 40:1087–1093. 10.1016/S0896-6273(03)00787-6 [DOI] [PubMed] [Google Scholar]
- 57.Zhao L, Teter B, Morihara T et al (2004) Insulin-degrading enzyme as a downstream target of insulin receptor signaling cascade: implications for Alzheimer’s disease intervention. J Neurosci 24:11120–11126. 10.1523/JNEUROSCI.2860-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho L, Qin W, Pompl PN et al (2004) Diet-induced insulin resistance promotes amyloidosis in a transgenic mouse model of Alzheimer’s disease. FASEB J 18:902–904. 10.1096/fj.03-0978fje [DOI] [PubMed] [Google Scholar]
- 59.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW (1975) A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A 72:1858–1862. 10.1073/pnas.72.5.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ittner LM, Ke YD, Delerue F et al (2010) Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142:387–397. 10.1016/j.cell.2010.06.036 [DOI] [PubMed] [Google Scholar]
- 61.Hong M, Lee VM (1997) Insulin and insulin-like growth factor-1 regulate tau phosphorylation in cultured human neurons. J Biol Chem 272:19547–19553. 10.1074/jbc.272.31.19547 [DOI] [PubMed] [Google Scholar]
- 62.Tau Phosphorylation by GSK3 in different conditions. https://www.hindawi.com/journals/ijad/2012/578373/. Accessed 29 Dec 2022 [DOI] [PMC free article] [PubMed]
- 63.Doble BW, Woodgett JR (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 116:1175–1186. 10.1242/jcs.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gratuze M, Planel E (2017) Regulation of brain insulin signaling: a new function for tau. J Exp Med 214:2171–2173. 10.1084/jem.20170979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marciniak E, Leboucher A, Caron E et al (2017) Tau deletion promotes brain insulin resistance. J Exp Med 214:2257–2269. 10.1084/jem.20161731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mokranjac D, Neupert W (2005) Protein import into mitochondria. Biochem Soc Trans 33:1019–1023. 10.1042/BST20051019 [DOI] [PubMed] [Google Scholar]
- 67.Stoccoro A, Siciliano G, Migliore L, Coppedè F (2017) Decreased methylation of the mitochondrial D-loop region in late-onset Alzheimer’s disease. J Alzheimers Dis 59:559–564. 10.3233/JAD-170139 [DOI] [PubMed] [Google Scholar]
- 68.Blanch M, Mosquera JL, Ansoleaga B et al (2016) Altered mitochondrial DNA methylation pattern in Alzheimer disease-related pathology and in Parkinson disease. Am J Pathol 186:385–397. 10.1016/j.ajpath.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 69.Iacobazzi V, Castegna A, Infantino V, Andria G (2013) Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol Genet Metab 110:25–34. 10.1016/j.ymgme.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 70.Bradley-Whitman MA, Lovell MA (2013) Epigenetic changes in the progression of Alzheimer’s disease. Mech Ageing Dev 134:486–495. 10.1016/j.mad.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armstrong MJ, Jin Y, Vattathil SM et al (2023) Role of TET1-mediated epigenetic modulation in Alzheimer’s disease. Neurobiol Dis 185:106257. 10.1016/j.nbd.2023.106257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoccoro A, Tannorella P, Migliore L, Coppedè F (2020) Polymorphisms of genes required for methionine synthesis and DNA methylation influence mitochondrial DNA methylation. Epigenomics 12:1003–1012. 10.2217/epi-2020-0041 [DOI] [PubMed] [Google Scholar]
- 73.Corral-Debrinski M, Horton T, Lott MT et al (1994) Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics 23:471–476. 10.1006/geno.1994.1525 [DOI] [PubMed] [Google Scholar]
- 74.Krishnan KJ, Ratnaike TE, De Gruyter HLM et al (2012) Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer’s disease. Neurobiol Aging 33:2210–2214. 10.1016/j.neurobiolaging.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 75.Perry G, Nunomura A, Hirai K et al (2002) Is oxidative damage the fundamental pathogenic mechanism of Alzheimer’s and other neurodegenerative diseases? Free Radic Biol Med 33:1475–1479. 10.1016/S0891-5849(02)01113-9 [DOI] [PubMed] [Google Scholar]
- 76.Mecocci P, MacGarvey U, Beal MF (1994) Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol 36:747–751. 10.1002/ana.410360510 [DOI] [PubMed] [Google Scholar]
- 77.Hoekstra JG, Hipp MJ, Montine TJ, Kennedy SR (2016) Mitochondrial DNA mutations increase in early stage Alzheimer disease and are inconsistent with oxidative damage. Ann Neurol 80:301–306. 10.1002/ana.24709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeppesen DK, Bohr VA, Stevnsner T (2011) DNA repair deficiency in neurodegeneration. Prog Neurobiol 94:166–200. 10.1016/j.pneurobio.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou Y, Song H, Croteau DL et al (2017) Genome instability in Alzheimer disease. Mech Ageing Dev 161:83–94. 10.1016/j.mad.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sykora P, Misiak M, Wang Y et al (2015) DNA polymerase β deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res 43:943–959. 10.1093/nar/gku1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martire S, Fuso A, Mosca L et al (2016) Bioenergetic impairment in animal and cellular models of Alzheimer’s disease: PARP-1 inhibition rescues metabolic dysfunctions. J Alzheimers Dis 54:307–324. 10.3233/JAD-151040 [DOI] [PubMed] [Google Scholar]
- 82.Martire S, Mosca L, d’Erme M (2015) PARP-1 involvement in neurodegeneration: A focus on Alzheimer’s and Parkinson’s diseases. Mech Ageing Dev 146–148:53–64. 10.1016/j.mad.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 83.Strosznajder JB, Czapski GA, Adamczyk A, Strosznajder RP (2012) Poly(ADP-ribose) polymerase-1 in amyloid beta toxicity and Alzheimer’s disease. Mol Neurobiol 46:78–84. 10.1007/s12035-012-8258-9 [DOI] [PubMed] [Google Scholar]
- 84.Wilkins HM, Swerdlow RH (2016) Relationships between mitochondria and neuroinflammation: implications for Alzheimer’s disease. Curr Top Med Chem 16:849–857. 10.2174/1568026615666150827095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Russell AE, Doll DN (2016) TNF-α and beyond: rapid mitochondrial dysfunction mediates TNF-α-induced neurotoxicity. J Clin Cell Immunol 7:467. 10.4172/2155-9899.1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen M, Guerrero AD, Huang L et al (2007) Caspase-9-induced mitochondrial disruption through cleavage of anti-apoptotic BCL-2 family members. J Biol Chem 282:33888–33895. 10.1074/jbc.M702969200 [DOI] [PubMed] [Google Scholar]
- 87.Jian C, Lu M, Zhang Z et al (2017) miR-34a knockout attenuates cognitive deficits in APP/PS1 mice through inhibition of the amyloidogenic processing of APP. Life Sci 182:104–111. 10.1016/j.lfs.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 88.Wang X, Liu P, Zhu H et al (2009) miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull 80:268–273. 10.1016/j.brainresbull.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 89.John A, Kubosumi A, Reddy PH (2020) Mitochondrial MicroRNAs in aging and neurodegenerative diseases. Cells 9:1345. 10.3390/cells9061345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang R, Zhou H, Jiang L et al (2016) MiR-195 dependent roles of mitofusin2 in the mitochondrial dysfunction of hippocampal neurons in SAMP8 mice. Brain Res 1652:135–143. 10.1016/j.brainres.2016.09.047 [DOI] [PubMed] [Google Scholar]
- 91.Dienel GA (2019) Brain glucose metabolism: integration of energetics with function. Physiol Rev 99:949–1045. 10.1152/physrev.00062.2017 [DOI] [PubMed] [Google Scholar]
- 92.Cascella R, Cecchi C (2021) Calcium dyshomeostasis in Alzheimer’s disease pathogenesis. Int J Mol Sci 22:4914. 10.3390/ijms22094914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brini M, Carafoli E (2009) Calcium pumps in health and disease. Physiol Rev 89:1341–1378. 10.1152/physrev.00032.2008 [DOI] [PubMed] [Google Scholar]
- 94.Holahan MR, Tzakis N, Oliveira FA (2019) Developmental aspects of glucose and calcium availability on the persistence of memory function over the lifespan. Front Aging Neurosci 11:253. 10.3389/fnagi.2019.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liao Y, Wang Y, Tao Q-Q et al (2023) CALHM2 V136G polymorphism reduces astrocytic ATP release and is associated with depressive symptoms and Alzheimer’s disease risk. Alzheimers Dement J Alzheimers Assoc 19:4407–4420. 10.1002/alz.13366 [DOI] [PubMed] [Google Scholar]
- 96.Structure and assembly of calcium homeostasis modulator proteins—PubMed. https://pubmed.ncbi.nlm.nih.gov/31988524/. Accessed 24 Dec 2023
- 97.Microglial Calcium Homeostasis Modulator 2: novel anti-neuroinflammation target for the treatment of neurodegenerative diseases|Neuroscience Bulletin. 10.1007/s12264-023-01153-3. Accessed 24 Dec 2023 [DOI] [PMC free article] [PubMed]
- 98.Calvo-Rodriguez M, Bacskai BJ (2021) Mitochondria and calcium in Alzheimer’s disease: from cell signaling to neuronal cell death. Trends Neurosci 44:136–151. 10.1016/j.tins.2020.10.004 [DOI] [PubMed] [Google Scholar]
- 99.Manczak M, Reddy PH (2012) Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum Mol Genet 21:2538–2547. 10.1093/hmg/dds072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manczak M, Calkins MJ, Reddy PH (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet 20:2495–2509. 10.1093/hmg/ddr139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease|Nature Neuroscience. https://www.nature.com/articles/s41593-018-0332-9. Accessed 1 Nov 2023 [DOI] [PMC free article] [PubMed]
- 102.Mahoney R, Ochoa Thomas E, Ramirez P et al (2020) Pathogenic Tau Causes a Toxic Depletion of Nuclear Calcium. Cell Rep 32:107900. 10.1016/j.celrep.2020.107900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Esteras N, Kundel F, Amodeo GF et al (2021) Insoluble tau aggregates induce neuronal death through modification of membrane ion conductance, activation of voltage-gated calcium channels and NADPH oxidase. FEBS J 288:127–141. 10.1111/febs.15340 [DOI] [PubMed] [Google Scholar]
- 104.Busche MA, Wegmann S, Dujardin S et al (2019) Tau impairs neural circuits, dominating amyloid-β effects, in Alzheimer models in vivo. Nat Neurosci 22:57–64. 10.1038/s41593-018-0289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Calvo-Rodriguez M, Hou SS, Snyder AC et al (2020) Increased mitochondrial calcium levels associated with neuronal death in a mouse model of Alzheimer’s disease. Nat Commun 11:2146. 10.1038/s41467-020-16074-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Friedman JR, Nunnari J (2014) Mitochondrial form and function. Nature 505:335–343. 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu S, Zhang X, Liu C et al (2021) Role of mitochondria in neurodegenerative diseases: from an epigenetic perspective. Front Cell Dev Biol 9:688789. 10.3389/fcell.2021.688789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Medala VK, Gollapelli B, Dewanjee S et al (2021) Mitochondrial dysfunction, mitophagy, and role of dynamin-related protein 1 in Alzheimer’s disease. J Neurosci Res 99:1120–1135. 10.1002/jnr.24781 [DOI] [PubMed] [Google Scholar]
- 109.Chou C-H, Lin C-C, Yang M-C et al (2012) GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS ONE 7:e49112. 10.1371/journal.pone.0049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pickrell AM, Youle RJ (2015) The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85:257–273. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du F, Yu Q, Yan S et al (2017) PINK1 signaling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer’s disease. Brain 140:3233–3251. 10.1093/brain/awx258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang X, Su B, Lee H et al (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci Off J Soc Neurosci 29:9090–9103. 10.1523/JNEUROSCI.1357-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kandimalla R, Reddy PH (2016) Multiple faces of dynamin-related protein 1 and its role in Alzheimer’s disease pathogenesis. Biochim Biophys Acta 1862:814–828. 10.1016/j.bbadis.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manczak M, Kandimalla R, Fry D et al (2016) Protective effects of reduced dynamin-related protein 1 against amyloid beta-induced mitochondrial dysfunction and synaptic damage in Alzheimer’s disease. Hum Mol Genet 25:5148–5166. 10.1093/hmg/ddw330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panchal K, Tiwari AK (2019) Mitochondrial dynamics, a key executioner in neurodegenerative diseases. Mitochondrion 47:151–173. 10.1016/j.mito.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 116.Manczak M, Kandimalla R, Yin X, Reddy PH (2018) Hippocampal mutant APP and amyloid beta-induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum Mol Genet 27:1332–1342. 10.1093/hmg/ddy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chai G-S, Wu J-J, Gong J et al (2022) Activation of β2-adrenergic receptor ameliorates amyloid-β-induced mitophagy defects and tau pathology in mice. Neuroscience 505:34–50. 10.1016/j.neuroscience.2022.09.020 [DOI] [PubMed] [Google Scholar]
- 118.Srikanth V, Maczurek A, Phan T et al (2011) Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol Aging 32:763–777. 10.1016/j.neurobiolaging.2009.04.016 [DOI] [PubMed] [Google Scholar]
- 119.Li X-H, Du L-L, Cheng X-S et al (2013) Glycation exacerbates the neuronal toxicity of β-amyloid. Cell Death Dis 4:e673. 10.1038/cddis.2013.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kandimalla R, Manczak M, Yin X et al (2018) Hippocampal phosphorylated tau induced cognitive decline, dendritic spine loss and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum Mol Genet 27:30–40. 10.1093/hmg/ddx381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kapogiannis D, Manolopoulos A, Mullins R et al (2024) Brain responses to intermittent fasting and the healthy living diet in older adults. Cell Metab 36:1668-1678.e5. 10.1016/j.cmet.2024.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fortier M, Castellano C-A, St-Pierre V et al (2021) A ketogenic drink improves cognition in mild cognitive impairment: results of a 6-month RCT. Alzheimers Dement J Alzheimers Assoc 17:543–552. 10.1002/alz.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gejl M, Brock B, Egefjord L et al (2017) Blood–brain glucose transfer in Alzheimer’s disease: effect of GLP-1 analog treatment. Sci Rep 7:17490. 10.1038/s41598-017-17718-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Watson KT, Wroolie TE, Tong G et al (2019) Neural correlates of liraglutide effects in persons at risk for Alzheimer’s disease. Behav Brain Res 356:271–278. 10.1016/j.bbr.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 125.Tang H, Shao H, Shaaban CE et al (2023) Newer glucose-lowering drugs and risk of dementia: a systematic review and meta-analysis of observational studies. J Am Geriatr Soc 71:2096–2106. 10.1111/jgs.18306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jucker M, Walker LC (2023) Alzheimer’s disease: from immunotherapy to immunoprevention. Cell 186:4260–4270. 10.1016/j.cell.2023.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang D, Zhang W, Ming C et al (2024) P-tau217 correlates with neurodegeneration in Alzheimer’s disease, and targeting p-tau217 with immunotherapy ameliorates murine tauopathy. Neuron 112:1676-1693.e12. 10.1016/j.neuron.2024.02.017 [DOI] [PubMed] [Google Scholar]
- 128.Battini V, Cirnigliaro G, Leuzzi R et al (2023) The potential effect of metformin on cognitive and other symptom dimensions in patients with schizophrenia and antipsychotic-induced weight gain: a systematic review, meta-analysis, and meta-regression. Front Psychiatry 14:1215807. 10.3389/fpsyt.2023.1215807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cao B, Rosenblat JD, Brietzke E et al (2018) Comparative efficacy and acceptability of antidiabetic agents for Alzheimer’s disease and mild cognitive impairment: A systematic review and network meta-analysis. Diabetes Obes Metab 20:2467–2471. 10.1111/dom.13373 [DOI] [PubMed] [Google Scholar]
- 130.Pszczołowska M, Walczak K, Miśków W et al (2024) Mitochondrial disorders leading to Alzheimer’s disease—perspectives of diagnosis and treatment. GeroScience 46:2977–2988. 10.1007/s11357-024-01118-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bhatti JS, Kaur S, Mishra J et al (2023) Targeting dynamin-related protein-1 as a potential therapeutic approach for mitochondrial dysfunction in Alzheimer’s disease. Biochim Biophys Acta BBA - Mol Basis Dis 1869:166798. 10.1016/j.bbadis.2023.166798 [DOI] [PubMed] [Google Scholar]
- 132.Hou Y, Chu X, Park J-H et al (2024) Urolithin A improves Alzheimer’s disease cognition and restores mitophagy and lysosomal functions. Alzheimers Dement J Alzheimers Assoc 20:4212–4233. 10.1002/alz.13847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bao X, Liu X, Wu Q et al (2023) Mitochondrial-targeted antioxidant MitoQ-mediated autophagy: a novel strategy for precise radiation protection. Antioxidants 12:453. 10.3390/antiox12020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chacko L, Chaudhary A, Singh B et al (2023) CRISPR-Cas9 in Alzheimer’s disease: therapeutic trends, modalities, and challenges. Drug Discov Today 28:103652. 10.1016/j.drudis.2023.103652 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.