Abstract
Xenotransplantation may bridge the widening gap between the shortage of donor organs and the increasing number of patients waiting for transplantation. However, a major safety issue is the potential cross-species transmission of porcine endogenous retroviruses (PERV). This problem could be resolved if it is possible to produce pigs that do not contain replication-competent copies of this virus. In order to determine the feasibility of this, we have determined the number of potentially replication-competent full-length PERV proviruses and obtained data on their integration sites within the porcine genome. We have screened genomic DNA libraries from a Large White pig for potentially intact proviruses. We identified six unique PERV B proviruses that were apparently intact in all three genes, while the majority of isolated proviruses were defective in one or more genes. No intact PERV A proviruses were found in this pig, despite the identification of multiple defective A proviruses. Genotyping of 30 unrelated pigs for these unique proviruses showed a heterogeneous distribution. Two proviruses were uncommon, present in 7 of 30 and 3 of 30 pigs, while three were each present in 24 of 30 pigs, and one was present in 30 of 30 animals examined. Our data indicate that few PERV proviruses in Large White pigs are capable of productive infection and suggest that many could be removed by selective breeding. Further studies are required to determine if all potentially functional proviruses could be removed by breeding or whether gene knockout techniques will be required to remove the residuum.
Xenotransplantation offers the chance to bridge the growing gap between the numbers of patients requiring solid organ transplantation and the number of organs available. In addition, a number of clinical trials have been proposed or are ongoing using xenogeneic material for indications such as diabetes and hepatic support during acute liver failure (12, 34). While the potential benefits are considerable, there has been much public and scientific debate over the possible risk of zoonosis in xenotransplantation. The prolonged presence of an animal organ, together with systemic microchimerism in the recipient, presents an opportunity for the transmission of zoonotic agents (22, 28, 43, 49). In addition, the use of antirejection strategies, such as immunosuppression or complement inhibition, may lower the body's natural defenses against such pathogens (44). While it is possible to derive specific-or qualified pathogen-free pigs to exclude known exogenous pathogens with the potential for zoonotic spread from donor pig herds, there remain risks both from unknown agents and agents that are transmitted in the germ line. Concerns have been focused on porcine endogenous retrovirus (PERV), a gammaretrovirus with replication-competent human-tropic variants (33, 50), which is found in the germ line of all pig breeds examined to date (17). Other classes of PERV have been identified, including betaretroviruses (13, 32), and while more may remain to be found, to date, all of these appear to be defective.
Domestic pigs have up to 50 copies of PERV as measured using a pro probe on Southern blots (22). Three subgroups of PERV have been identified to date, PERV A, B, and C (1, 33). Although these subgroups were defined initially by molecular analysis, infection and pseudotyping experiments have shown that PERV A, B, and C use different, uncharacterized, cell receptors, with receptor usage being the method of formally classifying subgroups (45, 51). The gag and pol genes of all these subgroups are highly homologous, and they differ principally in the long terminal repeats (LTRs) and env hypervariable regions, the latter encoding putative cell receptor binding domains. PERV A and B are ubiquitous in domestic pigs; however, Large White pigs that do not have PERV C have been identified (6, 17). PERV A and B productively infect a wide range of cell types from both humans and other mammals, while PERV C has a more restricted tropism among the human cells examined, only infecting the HT1080 cell line nonproductively (45). PERVs released from pig tissues, such as peripheral blood mononuclear cells (PBMCs), endothelial cells, and pancreatic islets, have been shown to infect human cell lines, including 293 and endothelial cells (26, 45, 48, 50, 51).
Studies of humans treated with living pig tissue have not found any evidence of PERV infection (8, 11, 12, 14, 15, 23, 30, 31, 34, 38). These studies have included patients with a range of exposure routes to pig tissues, including skin grafts, transplantation with porcine pancreatic islets, and extracorporeal liver and splenic perfusion. Examination of multiple tissues in nonhuman primates transplanted with pig endothelial cells has also shown absence of in vivo infection with PERV (24, 25), although these primate cells are permissive for infection in vitro (5). However, two recent studies using SCID mouse models have shown evidence of PERV infection in multiple tissue compartments (10, 48). van der Laan et al. (48) showed infection of mouse cells in a SCID mouse transplanted with porcine pancreatic islets. The reason for the discrepancy between the results in humans and SCID mice is not clear, although it may reflect the highly compromised immune status of the SCID mouse or the fact that only PBMCs have been used to evaluate PERV status in human patients.
Despite the lack of evidence of PERV infection in humans, it would be desirable to use organ-source pigs that present the lowest possible zoonotic risk. Bosch et al. (6) found that most expressed PERV proviruses were defective, in common with endogenous retroviruses in other species, such as humans, mice, and cats (46). Although some pig-to-pig variation in proviral distribution has been found (6, 17), it has been speculated that some proviruses are likely to be common to all pigs. The functional status of these common proviruses will be the significant factor in determining safety. Provirus expression patterns vary between different cell types (1, 6) and may be altered due to cryptic stimuli during organ rejection. Cloning of pigs by nuclear transfer is being actively explored as a method to generate genetically modified pigs for xenotransplantation (4, 29, 35). It has been shown in other species that gene methylation status can alter during the nuclear reprogramming steps of this procedure (18–20), hence proviruses previously silenced by methylation may be reexpressed in this situation.
A systematic study is required to determine which proviruses are intact and what degree of pig-to-pig variance they display. We have taken a genomic approach to this problem and screened libraries for proviruses that have both gag and env sequences. These proviruses have then been subjected to further analysis to confirm that they have the potential to result in productive infection. Since Large White pigs negative for PERV C have been identified and our own studies have indicated a lack of full-length C proviruses in some Large White pigs analyzed to date (6) (D. Hart, personal communication), we have concentrated our analysis on PERV A and B. The study reported here indicates that although pigs carry approximately 50 proviruses, the number of potentially functional proviruses in the Large White pig is low and there is sufficient variation within one herd to remove most proviruses by breeding.
MATERIALS AND METHODS
Library preparation and screening.
DNA was extracted from porcine PBMCs and partially digested with Sau3AI according to the vector manufacturer's protocols. Libraries were constructed in BamHI-digested lambda Fix II or pSuperCos 1 vectors according to the manufacturer's instructions (Stratagene). The cosmid library was screened for PERV A, while the lambda library was screened for PERV B as described below.
For the cosmid library, three to fourfold genome equivalents were plated on LB agar plates supplemented with 50 μg of ampicillin/ml. Colonies were grown overnight at 30°C and lifted onto a Hybond N+ filter (Amersham Pharmacia Biotech) to create the master filter. The master filter was then replicated onto three other filters (sets A, B, and C) which were processed as described by in Sambrook et al. (37) and UV cross-linked on a Stratalinker cross-linker (120,000 μJ; Stratagene).
The triplicate replica filters (sets A, B, and C) were screened with probes labelled with [α-32P]dCTP, using the Megaprime random-primed labelling kit (Amersham Pharmacia Biotech) as follows. Filter set A was screened with a probe against PERV gag (sequence AF038600; bases 585 to 2159), while sets B and C were screened with a probe against PERV A env (sequence Y12238; bases 745 to 1101). Unincorporated nucleotide was removed by spin column chromatography on Sephadex G50 columns. The filters were prehybridized for 3 h at 65°C in hybridization buffer containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 20 mM NaH2PO4, 5× Denhardt's solution, 0.4% sodium dodecyl sulfate (SDS), and 500 μg of denatured salmon sperm DNA/ml. After addition of the probe, the filters were hybridized overnight. Filters were washed to a final stringency of 0.1× SSC–0.1% SDS at 65°C and either exposed for 24 to 48 h to X-ray film at −70°C with intensifying screens or for 1 to 5 h to a storage phosphor screen, followed by phosphorimaging on a Molecular Dynamics Storm phosphorimager. Phosphorimaged blots were printed onto acetate films prior to alignment on a light box with other relevant filters.
For the lambda Fix II library, a total of 1.5 × 106 PFU was plated on NZY agar plates according to the manufacturer's instructions (Stratagene). Duplicate filters were lifted using Hybond NX (Amersham Pharmacia Biotech) and denatured and neutralized as described previously (37). Filters were UV cross-linked as described above and prehybridized for 30 min in Quickhyb solution (Stratagene). Probes were labelled with [α-32P]dCTP using Prime-it II (Stratagene). The gag probe (AF038600; bases 1334 to 1813) was added to one filter, and the env probe (Y12239; bases 1387 to 1735) was added to the duplicate filter. The lambda library filters were hybridized and washed as described above for the cosmid library filters. The filters were exposed to X-ray film at −70°C for up to a week and developed using a Compact X4 automatic film processor (X-ograph Imaging Systems).
Clones that replicated on both the gag and env filters were then purified to homogeneity through multiple rounds of rescreening. Cosmid DNA was prepared using the Qiagen QIAwell system (Qiagen). Lambda DNA was prepared from liquid lysates as described by Ausubel et al. (2).
Primary analysis.
Cosmid and lambda clones were analyzed differently, as described below. This was a reflection of the lower yields of lambda DNA relative to cosmid DNA. Primary analysis of cosmid clones was performed by restriction fragment analysis, using SmaI and PvuII digests, to identify clones with identical restriction fragment fingerprints. Representative clones of each restriction fragment length polymorphism (RFLP) class were further analyzed by PCR of open reading frames (ORFs) as described below. Lambda clones were analyzed by restriction digestion with NotI, EcoRI, MslI, SmaI/NotI, or AvrII/MslI, followed by Southern blotting (37) onto Hybond NX (Amersham Pharmacia Biotech) and hybridization with either the 32P-labelled gag or PERV B env probes described above.
Secondary analysis.
The presence of intact viral genes in lambda and cosmid isolates was examined by PCR across the gag, pol, and env ORF or by PCR of the ORF followed by protein truncation testing (PTT) to analyze the coding potential of the gag and env ORFs. Primers were designed to conserved sequences at the start and end of each ORF, based on published sequence data. PCR conditions were optimized on laboratory-generated plasmid clones of PERV C gag and PERV A, B, and C env which had been fully sequenced and which encoded full-length ORFs with >98% homology to the published prototype sequences. The pol PCR was optimized on DNA from PK15 cells.
For gag ORF PCR, the following primers were used: gag F (5′-GGATCCTAATACGACTCACTATAGGAACAGACCACCATGGGACAGACAGTGACTACC-3′) and gag R (5′-CCCTCCACCTTCAAAGTTAC-3′) (GenBank accession no. AF038600). Fifty-microliter reaction mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 150 nM each primer, 200 nM each deoxynucleoside triphosphate (dNTP), and 2.5 U of Amplitaq (PE Biosystems) with approximately 1 ng of cosmid or lambda template. Reaction mixtures were then cycled as follows: 95°C for 3 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min 50 s, followed by 10 min at 72°C. PCR was performed in an ABI2400 or ABI9600 thermocycler (PE Biosystems).
For pol ORF PCR, the primers used were: pol F (5′-GGATCCTAATACGACTCACTATAGGAACAGACCACCATGGGTGCCACAGGGCAAC-3′) and pol R (5′-GACCATTGTCTGACCCGATTA-3′) (accession no. AF038600). Fifty-microliter reaction mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 150 nM each primer, 200 nM each dNTP, and 2.5 U of Amplitaq (PE Biosystems) with approximately 1 ng of cosmid or lambda template. Reaction mixtures were then cycled as follows: 95°C for 3 min, followed by 30 cycles of 95°C for 40 s, 58°C for 30 s, 72°C for 3 min, and a final soak at 72°C for 10 min.
For env ORF PCR, the primers used were: env F (5′-GGATCCTAATACGACTCACTATAGGAACAGACCACCATGCATCCCACGTTAAGCCG-3′) and env R (5′-CGCTCTAGACTAAGCGTAGTCTGGGACGTCGTATGGGTAGAACTGGGAAGGGTAGAGGTCAGT-3′) (GenBank accession no. Y12238). Fifty-microliter reaction mixtures were prepared under the same conditions as described for gag (above) and then cycled as follows: 95°C for 3 min and then 30 cycles of 94°C for 1 min, 62°C for 1 min, 72°C for 2 min 10 s, followed by 10 min at 72°C.
The forward primer sequences shown for gag and env were modified so that they contained a T7 RNA polymerase promoter sequence, such that PCR products from clones giving approximately the predicted product sizes for gag and env could be further analyzed by PTT. Additionally, the stop codon in the env reverse primer was replaced with an in-frame hemagglutinin (HA) epitope tag, enabling detection of full-length products by Western blotting against the HA tag.
PTT (36) for ORFs was carried out using coupled transcription-translation in rabbit reticulocyte lysate (TNT T7 Rapid for PCR; Promega), with nonradioactive detection using biotinylated lysine tRNA. Each 25-μl reaction mixture contained 2.5 μl of unpurified PCR product, 1 μl of Transcend tRNA (Promega), and 20 μl of TNT T7 PCR Quick master mix (Promega) supplemented with 20 μM methionine. The reaction mixtures were incubated at 30°C for 90 min, and 2 μl was subjected to SDS-polyacrylamide gel electrophoresis on a 4 to 12% NuPAGE Bis-Tris gel (Novex/Invitrogen, Paisley, United Kingdom). Gels were semi-dry blotted to a Hybond enhanced chemiluminescence (ECL) membrane (Amersham Pharmacia Biotech). Biotinylated translation products were detected by blotting them with streptavidin-horseradish peroxidase (HRP) conjugate (1/3,750 dilution; Roche), followed by ECL detection (Amersham Pharmacia Biotech). Product sizes were assessed by reference to a translated gag or env plasmid clone of known sequence and a biotinylated protein molecular weight marker (Roche). HA epitope-tagged products were detected by blotting with high-affinity rat anti-HA antisera (1/500; Roche) and a secondary sheep anti-rat HRP conjugate (1/10,000; Sigma), followed by ECL detection.
Sequence analysis of cosmid and lambda clones.
Clones identified as having full-length gag, pol, and env genes together with full-length gag and env translation products were then sequenced by a combination of methods. Cosmid DNA and PCR products were directly sequenced using ABI PRISM Dye Terminator Cycle Sequencing on an ABI373 sequencer (Applied Biosystems), while some PCR products were subcloned into pCR2.1 (Invitrogen) prior to sequencing. The lambda clones were directly sequenced on a LICOR 4000 DNA sequencer (MWG Biotech) as follows: 800 ng of lambda DNA was prepared using a Thermo Sequenase fluorescence-labelled sequencing kit (Amersham), according to the manufacturer's instructions.
Analysis of PCR-negative cosmid clones.
Most clones that gave no PCR products for gag, pol, or env ORFs were subjected to Southern blotting to confirm that they were defective and/or contained deleted material. Southern blots of SmaI-digested cosmid DNA were probed with biotin-labelled oligonucleotides homologous to the region around the primer binding site (PBS) 5′-biotin, GCCTTTCATTTGGTGCGTTGGCCGGGAAATCCTCGCGACCACCCCTTACAC-3′ (nucleotide [nt] 661 to 711; accession no. AJ133817), and the 3′-end of env 5′-biotin, ACTGACCTCTAGCCTTCCCAGTTCTAAGATTAGAACTATTAACAAGACAA-3′ (nt 8121 to 8171; accession no. AJ133817). These probes were external to the probe sequences used in the initial library screening and to the gag and env PCR primer sets. Southern-blotted DNA was prehybridized at 40°C for 6 h in hybridization buffer (as described above) and then hybridized overnight at 40°C in hybridization buffer containing 27 nM biotin-labelled probe oligonucleotide. Blots were then washed in 1× SSC–0.1% SDS three times for 30 min at 42°C and then developed with streptavidin-HRP conjugate and ECL detection as described for the PTT above. It was assumed that, because of the sequence conservation seen about these regions in all known PERV variants, failure to bind these oligonucleotides indicated a provirus containing a deletion (deleted provirus).
Proviral flanking sequence analysis.
Proviral flanking sequence was obtained for all clones that were shown to contain apparently intact proviruses. Flanking sequence was generated by linker-mediated PCR between a primer in the PERV LTR and a primer in a linker ligated onto blunt-end-cut DNA (40). One-half microgram of PvuII-digested cosmid or lambda DNA was ligated to 60 pmol of double-stranded adaptor by use of 10 U of T4 DNA ligase (Invitrogen) in a 7-μl reaction mixture for 16 to 48 h at 15°C. The adaptor consisted of annealed upper-strand oligonucleotide (5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT-3′) and a 3′-amine-blocked lower-strand oligonucleotide (5′-ACCAGCCC-NH2-3′). One microliter of a 1:100 dilution of this reaction was used as template in a 25-μl PCR reaction mix containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 nM LTR-specific primer, 50 nM adaptor primer (5′-GTAATACGACTCACTATAGGGC-3′), 200 nM each dNTP, and 2.5 U of Amplitaq Gold (PE Biosystems). Reactions were cycled as follows: 94°C for 9 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min, and 72°C for 10 min. The LTR-specific primers used were in the U5 region, which is conserved among all known PERV variants; primer LTR-5′ (5′-GTGAACCCCATAAAAGCTGTC-3′) was used to isolate 5′-flanking sequence, while primer LTR-3′ (5′-GACAGCTTTTATGGGGTTCAC-3′) was used to isolate 3′-flanking sequence (GenBank accession no. AF038600 and AF038601).
Two PCR products were usually produced for each reaction, representing an LTR-internal proviral sequence and an LTR-flanking DNA sequence. Both products were either direct sequenced or gel purified with the Qiaquick Gel Extraction kit (Qiagen), cloned into pCR2.1-TOPO (Invitrogen), and then sequenced.
Analysis of provirus prevalence within a transgenic Large White pig herd.
Animals from Imutran Ltd's herd of pigs transgenic for the human complement regulator decay-accelerating factor (hDAF) were used to determine the incidence of the different proviruses. DNA was isolated from pig tissue by either a standard proteinase K-phenol-chloroform method (37) or by using Qiagen Genomic tips (Qiagen) according to the manufacturer's protocol. Based on sequence generated from the flanking PCR above, primer pairs were designed that would amplify between the PERV LTR and the adjacent flanking DNA, yielding a PCR product that would uniquely identify each provirus. Positive control reactions contained the lambda or cosmid clone the provirus was originally identified from. β-Globin (GenBank accession no. X86791) PCR was performed on each sample to confirm that the DNA was suitable for amplification, using ∼35 ng of template DNA in a 50-μl reaction mixture containing 150 nM each primer β-globin F1 (5′-GCAGATTCCCAAACCTTCGCAGAG-3′) and β-globin R1 (5′-TCTGCCCAAGTCCTAAATGTGCGT-3′), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 nM each dNTP, and 1.25 U of Amplitaq Gold (PE Biosystems). Reactions were cycled as follows: 95°C for 9 min, 30 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s, followed by 7 min at 72°C.
For the cosmid, 78 provirus, each reaction used ∼60 ng of template DNA in a 50-μl reaction mixture containing: 300 nM each primer 78ltrs (5′-GAACCCCATAAAAGCTGTCC-3′) and 78ltras (5′-GATCCTATGTTGGGTGCATTT-3′), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 nM each dNTP, and 2 U of Amplitaq Gold (PE Biosystems). Reactions were cycled as follows: 94°C for 9 min, followed by 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s, and a final 10 min at 72°C. The predicted product size was 323 bp based on sequence analysis.
Reaction conditions for all remaining primer sets described below were as follows: ∼20 ng of template DNA in a 50-μl reaction mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 150 nM each primer, 200 nM each dNTP, and 1.25 U of Amplitaq Gold (PE Biosystems).
Primer sets and cycle parameters used were as follows: Provirus 35121i, primers FLPR3 (5′-CATGACGGCAACTCCTGAAG-3′) and revltr1 (5′-GTGAACCCCATAAAAGCTGTC-3′) under the following cycle parameters: 95°C for 9 min, and then 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min, followed by 10 min at 72°C. The predicted product size was 455 bp based on sequence analysis.
Provirus 21321. primers 21f2 (5′-GGTCTGTGACCTACTCCATA-3′) and ltr1 (5′-GACAGCTTTTATGGGGTTCAC-3′) under the following cycle parameters: 95°C for 9 min, and then 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min, followed by 10 min at 72°C. The predicted product size was 518 bp based on sequence analysis.
Provirus 73414i, primers 73-4r-3 (5′-GTCATGCCGTTCATTTGGGA-3′) and revltr1 under the following cycle parameters: 95°C for 9 min, and then 40 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min, followed by 10 min at 72°C. The predicted product size was 388 bp based on sequence analysis.
Provirus 73414ii, primers 73-3f-3 (5′-AGCAACAGAATTGAAGTCAG-3′) and revltr1 under the same cycle parameters as described above for 73414i. The predicted product size was 438 bp based on sequence analysis.
Provirus 310518-5′, the primers 31f8 (5′-GGATGGAGACTATGCTCAGC-3′) and ltr1 under the following cycle parameters: 95°C for 9 min, and then 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min, followed by 10 min at 72°C. The predicted product size was 1,641 bp based on sequence analysis.
Provirus 310518-3′, the primers 31f2 (5′-AACTCATGATAAATGAAACACC-3′) and revltr1 under the same cycle parameters as described above for 310518-5′. The predicted product size was 815 bp based on sequence analysis.
RESULTS
Isolation of proviruses containing both gag and env sequences.
The screening strategy used was designed to rapidly exclude defective proviruses. To this end, only clones that gave signals for both gag and env probes were analyzed further. Proviruses that were missing either the entire gag gene or the VRA and VRB portions of env were thus excluded from further analysis. Clones with more subtle deletions or rearrangements were eliminated at later stages as described below.
The cosmid library was screened for PERV A proviruses by use of a PERV A env probe, and the lambda library was screened for PERV B proviruses by use of a PERV B env probe. Both libraries were also screened with PERV gag probes as described above. Both libraries yielded substantially more gag-positive colonies than env positives, partly reflecting the non-PERV subtype selectivity of the gag probes and in the case of the cosmid library the larger size of the gag probe used. Eighty-four clones were identified from the cosmid library as gag and env A positive, of which 78 were recovered. Of the 78 clones which were recovered, 74 were were purified to homogeneity through further rounds of screening, whereas sequence analysis showed that the other four clones did not contain an insert. From the lambda library, 40 plaques were initially isolated, of which four were found to be either gag or env B negative upon the second round of screening and were discarded. The remaining 36 positive plaques were purified to homogeneity. Multiple clones derived from each initial plaque were analyzed to ensure that any instability of the insert would not affect the results. The lambda library was also screened for the presence of PERV A; however, no PERV A env positives were associated with gag positives, suggesting an absence of full-length intact PERV A (data not shown). As a result, this was not pursued any further.
The doubly positive cosmid clones were then subjected to restriction fragment analysis to identify duplicate clones as shown in Table 1. Of the 74 clones analyzed, 54 were unique and 20 were duplicates of other clones. Where there were two or more cosmid clones with identical RFLP patterns, two or more representatives of each were analyzed to confirm results.
TABLE 1.
Analysis of gag and env double-positive clones obtained from a PERV A-screened cosmid library from the Large White pig
| Clone no.a | RFLP patternb | Presence of indicated probe by indicated test
|
Defect and probe | |||||
|---|---|---|---|---|---|---|---|---|
| gag PCR | gag PTT | pol PCRc | env PCR | env PTT | env HA PTT | |||
| 1 | +1 | CDg | +l | + | TPe | − | Cloning deletion | |
| 3 | U | − | − | − | Yes | |||
| 4 | U | + | FLd | + | − | Yes, env | ||
| 5 | U | − | +s | − | Yes, gag, pol, and env | |||
| 6 | U | − | + | + | TP | Yes, env | ||
| 7 | +11 | − | + | + | TP | Yes, gag and env | ||
| 9 | U | − | + | + | TP | Yes, gag and env | ||
| 10 | U | − | +l | − | Yes, gag and env | |||
| 12 | U | − | − | − | Yes | |||
| 13 | +6 | − | +l | − | Yes, gag and env | |||
| 15 | U | − | + | + | TP | Yes, gag and env | ||
| 18 | U | + | NDf | + | + | TP | Yes, env | |
| 19 | U | − | − | Yes | ||||
| 21 | U | + | ND | − | − | Yes, pol and env | ||
| 22 | U | − | − | − | Yes | |||
| 24 | U | CD | +l | − | Cloning deletion | |||
| 25 | U | + | + | + | TP | Yes, env | ||
| 27 | U | − | + | − | Yes, gag and env | |||
| 29 | U | − | +l | − | Yes, gag and env | |||
| 30 | U | − | − | − | Yes, gag and env | |||
| 31 | U | − | +l | − | Yes, gag and env | |||
| 34 | U | − | +l | − | Yes, gag and env | |||
| 35 | U | − | + | + | TP | Yes, gag and env | ||
| 36 | U | CD | ND | ND | Cloning deletion | |||
| 38 | U | − | − | − | Yes, gag and env | |||
| 39 | U | − | − | − | Yes, gag and env | |||
| 41 | U | − | − | − | Yes, gag and env | |||
| 43 | U | − | − | − | Yes, gross deletion | |||
| 45 | U | − | +l | − | Yes, gag and env | |||
| 46 | U | + | TP | + | + | FL | + | Yes, gag |
| 47 | U | − | − | − | No insert | |||
| 48 | U | + | FL | + | + | TP | − | Yes, env |
| 49 | U | + | FL | + | + | TP | − | Yes, env |
| 53 | U | − | − | − | Yes, gag and env | |||
| 54 | +1 | − | + | + | TP | Yes, gag and env | ||
| 56 | U | CDh | − | − | Cloning deletion | |||
| 57 | U | − | + | + | TP | Yes, env | ||
| 58 | U | + | FL | + | + | TP | − | Yes, env |
| 59 | U | + | ND | +l | − | Yes, env | ||
| 60 | U | + | FL | + | + | TP | − | Yes, env |
| 61 | U | + | FL | + | + | TP | − | Yes, env |
| 62 | U | − | − | − | Yes, env | |||
| 64 | U | CD | +l | − | Cloning deletion | |||
| 66 | U | − | +l | − | Yes, gag and env | |||
| 67 | U | CD | +l | − | Cloning deletion | |||
| 69 | U | − | + | − | Yes, gag and env | |||
| 70 | U | − | − | − | Yes, gag and env | |||
| 74 | U | − | +l | − | Yes | |||
| 77 | U | − | +l | − | Yes, gag and env | |||
| 78 | U | + | FL | + | + | FL | + | No intact |
| 79 | +1 | − | − | − | Yes, gag and env | |||
| 82 | U | − | + | − | Yes, gag and env | |||
| 83 | U | + | FL | + | + | TP | − | Yes, env |
| 84 | U | − | + | − | Yes, gag and env | |||
Clones were numbered sequentially on initial picking.
Unique (U) clones are indicated. Where duplicate clones were identified, the number of such duplicates is noted.
+, a product of the predicted size was obtained; + with an l or s, a respectively larger or smaller product was observed in the absence of the predicted product.
FL, full-length protein was translated.
TP, truncated protein was produced.
ND, not done.
CD, gag sequence was adjacent to vector sequence as a result of cloning deletions. Such clones are recorded as defective only if negative results were obtained for the env gene.
In the case of clone 56, the clone was truncated but negative PCR results were obtained for pol and env. This is recorded as a cloning deletion as it was not analyzed further.
As the yield of DNA from the lambda preparations was much lower than that from the cosmid clones, lambda clones were subjected to Southern blot analysis using the gag and env probes described in Materials and Methods. The results of the analysis of the lambda clones are shown in Table 2. Southern blotting showed that the majority of PERV B lambda clones possessed a variant RFLP with respect to the published PERV B sequence, suggesting they were potentially defective or deleted.
TABLE 2.
Analysis of gag and env double-positive clones obtained from a PERV B-screened lambda library from the Large White pig
| Original plaque identification (ID) | Clone IDa | Southern blot datab | Presence of indicated probe by indicated test
|
Defect and probe | ||||
|---|---|---|---|---|---|---|---|---|
| gag PCR | gag PTT | pol PCR | env PCR | env PTT | ||||
| 2.1 | 21321 | PERV B Var | + | FLc | + | + | FL | No intact |
| 2.2. | 22332 | ND | + | TPd | NDe | + | FL | Yes, gag |
| 2.4 | 2441 | PERV B Var | + | FL | + | + | FL | No, intact |
| 2.5 | 25321 | PERV B Var | + | TP | + | + | TP | Yes, gag and env |
| 2.6 | 2622 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 2.7 | 27244 | PERV B Var | + | TP | + | + | FL | Yes, gag |
| 2.8 | 28321 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 3.1 | 31211 | PERV B Var | + | TP | + | + | FL | Yes, gag |
| 3.2 | 3211 | PERV B Var | + | TP | + | + | TP | Yes, gag and env |
| 3.3 | 3321 | PERV B | + | TP | ND | + | FL | Yes, gag |
| 3.4 | 34321 | PERV B Var | + | TP | ND | + | FL | Yes, gag |
| 3.5 | 35121 | PERV B | + | FL | + | + | FL | No intact |
| 3.6 | 3611 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 3.7 | 3711 | PERV B | − | − | − | Yes | ||
| 3.8 | 3821 | PERV B | + | TP | ND | + | ND | Yes, gag |
| 3.9 | 39111 | PERV B | + | FL | ND | + | TP | Yes, env |
| 3.10 | 310518 | PERV B | + | FL | + | + | FL | No, intact |
| 3.12 | 312146 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 4.1 | 411 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 4.3 | 436 | PERV B Var | + | ND | + | − | Yes, env | |
| 4.4 | 44111 | PERV B Var | + | FL | ND | + | TP | Yes, env |
| 4.5 | 45411 | PERV B | + | FL | + | + | TP | Yes, env |
| 5.1 | 5112 | PERV B Var | + | FL | + | + | TP | Yes, env |
| 5.2 | 52111 | PERV B | + | TP | ND | + | TP | Yes, gag and env |
| 6.1 | 61121 | PERV B Var | + | TP | ND | + | ND | Yes, gag |
| 6.2 | 62321 | PERV B | + | TP | + | + | FL | Yes, gag |
| 6.4 | 6411 | PERV B | + | TP | ND | + | ND | Yes, gag |
| 6.5 | 65321 | PERV B Var | + | FL | + | + | FL | No, intact |
| 7.1 | 7111 | PERV B Var | − | ND | − | Yes, gag and env | ||
| 7.2 | 7211 | PERV B Var | − | ND | − | Yes, gag and env | ||
| 7.3 | 73414 | PERV B Var | + | FL | + | + | FL | No, intact |
| 7.4 | 74411 | PERV B Var | + | FL | + | + | TP | Yes, env |
| 7.5 | 75321 | PERV B Var | − | ND | − | Yes, gag and env | ||
| 8.1 | 81111 | PERV B Var | + | TP | + | + | TP | Yes, gag and env |
| 8.2 | 82222 | PERV B Var | + | TP | + | + | TP | Yes, gag and env |
| 8.3 | 8311 | PERV B Var | + | TP | ND | + | TP | Yes, gag and env |
From each original plaque picked, multiple plaques were analyzed, the clone ID is the number of the clone that was analyzed in the greatest detail and was the least defective.
PERV B, the restriction pattern obtained was identical to that of the published PERV B provirus Y17013; PERV B Var, a variant RFLP pattern was observed, indicating that the proviral sequence was either variant from the published data or had a deletion somewhere in the proviral sequence.
FL, full-length protein was translated.
TP, truncated protein was produced.
ND, not done.
Secondary analysis.
Clones that gave both gag and env signals upon primary library screening were then analyzed by PCR or PCR followed by PTT, which identified clones with intact ORFs. The result of the PCR was regarded as positive if the clone gave a PCR product of approximately the size expected for the intact ORF based on published PERV sequences. If a clone was defective for either gag, pol, or env, analysis of the clone was terminated, because the provirus was deemed to be defective. In some cases, this means that results are not available for gag, pol, and env PCR. However, the results in Tables 1 and 2 demonstrate that sufficient data were obtained to conclude that such clones did contain defective proviruses.
gag.
The gag PCR showed major differences between the PERV A-screened cosmid library and the PERV B-screened lambda library. While 32 of 36 lambda clones gave full-length gag PCR products, only 13 of 54 unique cosmid clones yielded full-length products (Tables 1 and 2). Sequencing from the cosmid multiple cloning site (MCS) showed that in a number of cases this was a result of genomic DNA being cloned into the vector within the proviral sequence (termed cloning deletions in Table 1). However, for the majority of clones that gave no gag PCR product, sequence adjacent to the MCS was not PERV sequence, indicating that no cloning deletion had occurred. No gag products that were smaller than expected, which could have represented proviruses with internal deletions, were detected from either library.
PTT of apparently full-length gag genes from cosmid clones showed that 8 of 13 clones had intact gag ORFs and 1 of 13 had a truncated ORF, while 4 of 13 clones were not tested as they had already been excluded from analysis on the basis of a truncated env gene (Table 1).
In contrast to the cosmid clone results, only 11 of 32 lambda clones had an intact gag ORF and 20 of 34 had a truncated gag ORF. One clone of 32 was not analyzed by PTT as it had previously been found to be defective in gag (Table 2 and Fig. 1A).
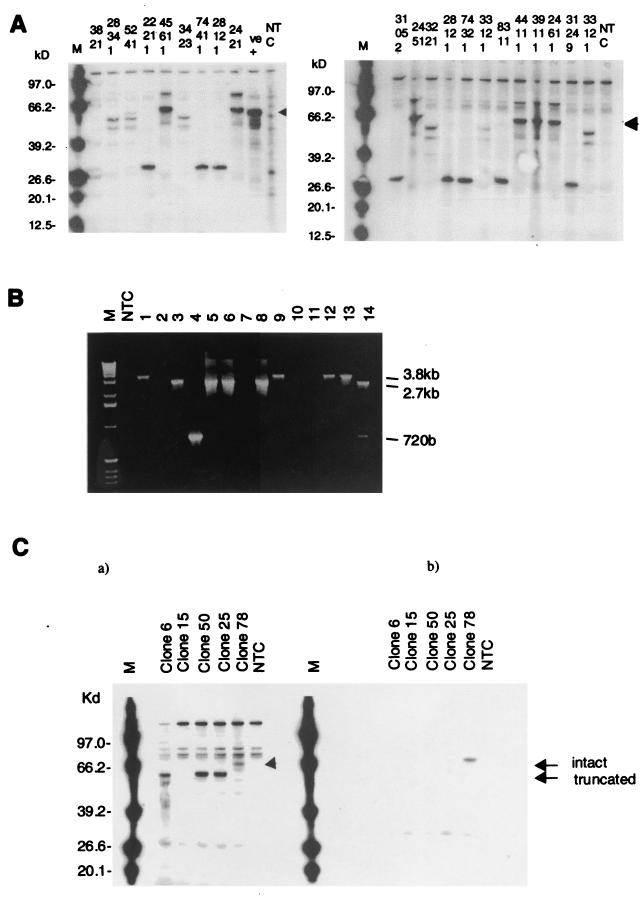
FIG. 1.
Sample results for the PCR and PTT assays for gag, pol, and env ORFs. PCR and PTT were carried out as described in Materials and Methods for gag, pol, and env. Selected results are shown. Full results are shown in Tables 1 and 2. (A) gag analysis. The size of the full-length product (arrowhead), at ∼60 kDa, is as predicted from translation of the sequence (59.5 kDa) and is the same size as the positive control. Full-length products were obtained from lambda clones 45611, 2421, 2451, 44111, 39111, and 24611. Products obtained from other clones were smaller than this and were recorded as truncated proteins; the provirus would therefore be defective. M, biotinylated protein marker (Roche); +ve, control sequenced full-length PERV C gag plasmid clone; NTC, no template control. (B) pol analysis. pol PCR on cosmid clones gave the following amplification products: negative (lanes 2, 10, and 11); 2.7-kb products as predicted for the pol ORF (lanes 3, 5, 6, and 8); ∼3.8-kb products, larger than the predicted product (lanes 1, 9, 12, and 13); ∼720-bp product, smaller than the predicted product (lane 5); and both 2.7-kb and ∼720-bp products (lane 14). (C) env analysis. Results are shown for five cosmids. (a) Conventional PTT shows that cosmid 78 yielded an ∼74-kDa product as predicted for the 73-kDa full-length PERV B env, with the ∼1-kDa epitope tag (arrow, intact). Clones 6, 25, and 50 gave truncated products at ∼63 kDa (arrow, truncated), whereas clone 15 gave a product of <20 kDa that is not visible on this gel. (b) Epitope-tagged PTT showed a strong signal only from clone 78, at ∼73 kDa, confirming the presence of the full-length protein identified in Fig. 1C, panel a. Signal is not present from products generated from the other clones, confirming the presence of a premature stop codon.
pol.
The pol PCR primers were designed to amplify an ∼2.7-kb product, based on sequence alignments with the protoype PERV sequence. Thirty-seven of 54 unique cosmid clones gave pol PCR products, while 16 of 54 clones gave no pol PCR products; 1 of 54 clones was not analyzed as it had been truncated in cloning (Table 1). A smaller ∼720-bp product was amplified from some cosmids and was the sole product from cosmid 5 (Fig. 1B). Sequencing of this product showed >96% homology over three regions to the PERV pol gene, with two internal deletions. There was homology from the pol ATG codon at base 2307 to base 2496 of PERV (accession no. AF038600), then a deletion, and homology from bases 3721 to 4051, then a deletion, and homology from base 4872 to the end of the PCR product at base 5070, giving a sequenced product with a size of 718 bp. This small product did not encode a functional Pol protein, and clones giving such PCR products were regarded as defective. Some clones also gave a larger than predicted 3.8-kb product. The 3.8-kb product could potentially encode a functional Pol protein, but this was not investigated further as work on gag and env showed clones giving this product were defective in at least one other gene. As shown in Fig. 1B, a few cosmid clones gave both the predicted full-length product and the 718-bp deleted product. In these cases, the larger fragment was assumed to be associated with the provirus under study due to the possibility that there were two or more proviruses or proviral fragments on the cosmid inserts.
Of the 18 of 36 lambda clones subjected to pol PCR, only one did not give the expected 2.7-kb product (Table 2). The remaining 18 lambda clones had previously been identified as being defective in Gag or Env and were not subjected to pol PCR.
env.
For the env PCR, 18 of 54 cosmid clones gave a PCR product of the predicted size of the ORF at ∼2 kb, 35 of 54 gave no PCR product, and one was not tested (Table 1 and Fig. 1C). No smaller PCR products representing env internal deletions were identified.
PTT was performed on the env PCR products shown in Fig. 1C. Only 2 of 18 cosmid clones gave full-length translation products for Env, which was confirmed by carboxy-terminal epitope tagging PTT as described in Materials and Methods, and is illustrated for clone 78 (Fig. 1C). Sequence analysis of cosmid clone 78, which also gave a full-length gag product, revealed that this was in fact a PERV B clones (see Fig. 3).
FIG. 3.
Sequence alignment of predicted Env proteins from full-length proviruses. Sequencing of env genes of apparently intact proviruses confirmed the presence of a full-length Env ORF, as predicted by the PTT results. Comparison of the proviral env sequence showed a high degree of homology (>99%) with the prototype PERV B sequence Y12239 at both the nucleotide and amino acid levels. The single sequence labelled 2441 is the consensus sequence obtained from clone 65321, 2441, and 310518 env sequences. Sequencing of env from lambda clones 73414 and 310518 gave only a single env consensus sequence, despite the two different 3′ LTRs that were present in these clones, suggesting that they were either identical or that one provirus did not have an intact env gene (GenBank accession no. AY056024 to -28).
Subsequent sequencing of the 3′ end of the env PCR product for clone 25 showed a CGA to TGA, Arg-to-stop mutation at amino acid 578, consistent with the ∼64-kDa product seen in Fig. 1C. The product for clone 15 contained multiple base insertions throughout the sequence, causing multiple frameshifts, and no significant ORF was present. The env genes for cosmid clones 6 and 50 were not sequenced.
Sixteen of 18 lambda PCR products gave truncated and therefore nonfunctional Env translation products. Sequence analysis of a number of cosmid isolates, including cosmid clones 15 and 25 (Fig. 1C), demonstrated that they were PERV A as expected.
In contrast to the results obtained from the cosmid library, 31 of 34 clones from the lambda library gave full-length PCR env products and only 3 of 34 yielded no product (Table 2). Thirteen of 31 PCR-positive clones gave full-length Env translation products on PTT, 10 of 31 gave truncated products, and 8 were not analyzed, as they had already been observed to be Gag defective.
To summarize, where the clones were amplifiable by one or more sets of primers but gave no product for one or more of the other ORFs, it was presumed that the remaining proviral sequences were defective and analysis was not continued further. By these criteria, no intact PERV A clones were identified from the cosmid library: of the 54 unique clones, only one gave full-length PTT products for Gag and Env, together with an appropriately sized pol PCR product. Sequence analysis of this clone, cosmid 78, showed that it was indeed PERV B. From the lambda library, six PERV B clones were identified as being potentially full length, clones 21321, 2441, 35121, 310518, 65321, and 73414. Analysis of these seven clones was continued further.
Analysis of the clones that did not give PCR products.
A number of the gag- and env-positive cosmid clones isolated did not amplify the gag, pol, or env primer sets. To exclude the possibility that these represented major sequence variants on the published PERV proviruses and hence were not amplified by the PCR primers used, these were further analyzed in one of two ways. (i) Sequencing from the cloning sites into the insert was used to determine if the provirus had been truncated during the cloning process. Six of 54 cosmid clones had internal proviral sequence adjacent to the cosmid cloning site due to such deletions (Table 1). No definitive analysis of these clones could therefore be completed. (ii) Southern blots of SmaI-digested cosmid DNA were probed with biotin-labelled oligonucleotides homologous to the region around the PBS and env LTR junction. It was assumed that, due to the sequence conservation seen within these regions in all known PERV variants, failure to bind these oligonucleotides would indicate a deleted provirus. Most of these clones showed no signal from either of these probes, indicating that they had deletions at both ends (Table 3). The remainder had deletions either at one end or the other. Cosmid clone 74 showed signals for both PBS and env LTR; however, sequence data showed a deletion of the 3′ end of the env gene (not shown).
TABLE 3.
Analysis by Southern blot of cosmid library clones that failed to give PCR products for gag, pol, or enva
| Clone | PBS band size (kb) | env band size (kb) | Comments |
|---|---|---|---|
| 12 | 0.5, 0.65 | − | No env signal: defective |
| 13 | − | No signal for PBS gag or env LTR: defective | |
| 19 | 1.2, 2.5 | 1.2 | env and PBS signal on the same fragment; therefore, deleted |
| 30 | − | No signal for PBS gag or env LTR: defective | |
| 43 | 2.3 | + | env and PBS signal on the same fragment; therefore, deleted |
| 53 | − | No signal for PBS gag or env LTR: defective | |
| 59 | >6.0b | − | No env signal: defective |
| 62 | >6.0b | − | No env signal: defective |
| 66 | >6.0b | − | No env signal: defective |
| 69 | − | No signal for PBS gag or env LTR: defective | |
| 70 | − | No signal for PBS gag or env LTR: defective | |
| 84 | 1.3 | + | env and PBS signal on the same fragment; therefore, deleted |
Cosmid DNA was digested with SmaI, electrophoresed, and subjected to Southern blotting. Blots were probed with a biotin-labelled probe against either PBS gag or env LTR, followed by detection of probe with steptavidin-HRP conjugate and ECL detection as described in Materials and Methods.
Product was too large to determine size accurately.
Analysis of flanking sequence and proviral prevalence.
Sequence data was obtained for the LTR-flanking DNA junction for each unique, intact clone identified from the cosmid and lambda libraries by use of the linker-mediated PCR method. For most clones 3′-flanking sequence was obtained (cosmid 78 and lambda clones 65321, 2441, 73414, and 35121). For lambda clone 21321, 5′-flanking sequence was obtained and for 310518 both 3′-and 5′-flanking sequences were identified. Sequence comparisons of a portion of the LTR-flanking sequences obtained by this method are shown in Fig. 2.
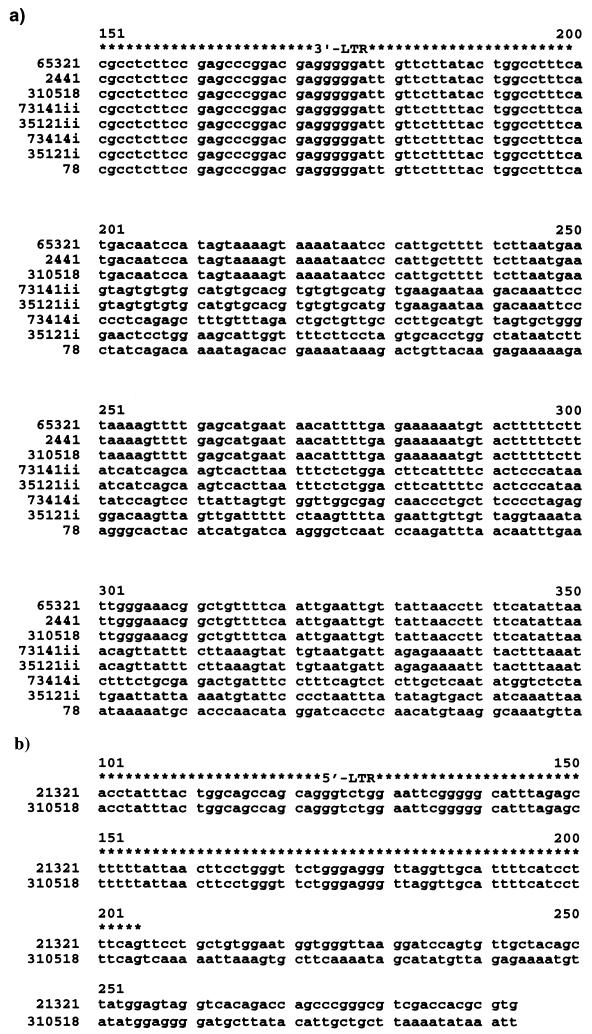
FIG. 2.
Analysis of proviral flanking sequences. Flanking sequence was obtained for all clones by the linker-mediated PCR method. For most clones 3′-flanking sequence was obtained (cosmid 78 and lambda clones 65321, 2441, 73414, and 35121) (GenBank accession no. AY056030 to -32 and AY056034 to -35), for lambda clone 21321 5′-flanking sequence (GenBank accession no. AY056033) was obtained, and for 310518 both 3′ and 5′-flanking sequences were obtained (GenBank accession no. AY056029). Sequence alignments were performed with the Wisconsin GCG Sequence analysis package, version 9 Pileup program. Pileups of a portion of either the 3′-LTR (a) or the 5′-LTR (b) flanking junctions are shown above. Asterisks above the sequence denote the U3(a) or U5 (b) region; sequence beyond this is flanking sequence.
Lambda clones 65321, 2441, and 310518 contained a common flanking sequence, confirming that they represented a single duplicated provirus. Lambda clones 73414 and 35121 both gave two different LTR-3′-flanking sequences (termed 73414i and 73414ii and 35121i and 35121ii, respectively); 73414ii and 35121ii were identical, while the other sequences showed no similarity (Fig. 2).
As the method used to generate the data was direct PCR or sequencing, the presence of two proviruses on a single clone means that we cannot distinguish between 73414 and 35121. These clones may represent a total of three infectious proviruses, complementary defective proviruses, or a combination of these possibilities. We treated them as intact proviruses and performed further analysis on them to determine their prevalence.
The 3′-flanking sequence of cosmid 78 was not related to any of the flanking sequences identified from the lambda clones.
From the 5′-flanking sequence obtained, clones 21321 and 310518 were different, but because 5′-flanking sequence was not obtained for the other clones, it was not possible to determine if 21321 was the same provirus as the 73414-35121 group or cosmid 78. However, although the patterns of prevalence in pigs were identical for 35121 and 21321 (Table 4), there are a number of nucleotide and amino acid differences in Env that confirm that they represent different proviruses (Fig. 3).
TABLE 4.
Prevalence of six potentially full-length PERV B proviruses in 30 unrelated Large White pigsa
| Pig no. | Provirus tested
|
||||||
|---|---|---|---|---|---|---|---|
| 78 | 35121i | 310518-3′ | 310518-5′ | 21321 | 73414i | 73414ii | |
| H370 | − | ++ | ++ | ++ | ++ | − | ++ |
| H225 | − | ++ | ++ | ++ | ++ | − | ++ |
| G838 | − | ++ | ++ | ++ | ++ | − | ++ |
| H768 | − | − | ++ | ++ | − | − | ++ |
| G759 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| G597 | − | ++ | ++ | ++ | ++ | − | ++ |
| G585 | − | ++ | ++ | ++ | ++ | − | ++ |
| G473 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| G116 | − | ++ | ++ | ++ | ++ | − | ++ |
| F934 | − | ++ | ++ | ++ | ++ | − | ++ |
| F894 | − | − | − | − | − | − | ++ |
| F800 | − | ++ | ++ | ++ | ++ | ++ | ++ |
| F748 | − | ++ | ++ | ++ | ++ | − | ++ |
| E765 | − | − | ++ | ++ | − | − | ++ |
| D681 | − | ++ | ++ | ++ | ++ | − | ++ |
| H744 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| H800 | − | ++ | ++ | ++ | ++ | − | ++ |
| H679 | − | − | ++ | ++ | − | ++ | ++ |
| H338 | ++ | − | − | − | − | − | ++ |
| H863 | − | ++ | ++ | ++ | ++ | − | ++ |
| H522 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| H888 | − | − | ++ | ++ | − | − | ++ |
| H874 | − | ++ | − | − | ++ | − | ++ |
| H539 | − | ++ | ++b | − | ++ | − | ++ |
| H791 | − | ++ | − | − | ++ | − | ++ |
| H765 | − | ++ | − | − | ++ | ++ | ++ |
| H858 | − | ++ | − | − | ++ | − | ++ |
| H440 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| H588 | − | ++ | ++ | ++ | ++ | − | ++ |
| G768 | ++ | ++ | ++ | ++ | ++ | − | ++ |
| Total | 7/30 | 24/30 | 24/30 | 23/30 | 24/30 | 3/30 | 30/30 |
PCR screening for the presence of each provirus was carried out as described in Materials and Methods. ++, provirus was present in the pig, −, provirus was not present.
This result showed a discrepancy between the 3′- and 5′-flanking LTR for provirus 310518. It is possible that this provirus may exist as two alleles, one with a partial deletion at the 5′ LTR.
Primer pairs were designed for each of these unique LTR-flanking DNA junction sequences to analyze 30 unrelated pigs for the presence of each of the proviruses. The herd from which the test animals were selected was Imutran Ltd's closed genetic herd of pigs transgenic for the human complement regulator hDAF. These pigs have previously been used in Imutran Ltd's xenotransplantation work (7). Thirty animals were selected to exclude siblings and closely related animals and to include as many ancestors (different parents and grandparents) as possible. This selection of animals on pedigree increased the number of variable genotypes tested compared to a random selection of animals, because commonality by descent was minimized. Suitability of the DNA for PCR amplification was confirmed by using porcine β-globin primers, and the results are summarized in Table 4.
The PERV cosmid 78 provirus was present in 7 of 30 pigs; 23 of 30 pigs were negative for this provirus.
Provirus 73414i was present in 3 of 30 pigs, while provirus 73414ii was in 30 of 30 pigs. Provirus 35121i was present in 24 of 30 pigs. A smaller ∼200-bp fragment was produced from lambda clone 35121, the positive control, which was also visible in some of the pig PCRs. As this did not interfere with the assay, it was disregarded and was not sequenced to determine its origin.
For provirus 310518, independent PCRs were performed for the 5′ and 3′ flanks. These gave concordant results in 29 of 30 pigs; 23 of 30 pigs were positive for the provirus and 6 of 30 were negative. Pig H539 was positive for the 3′-LTR-flanking junction and negative for the 5′-LTR-flanking junction, thus giving an ambiguous result. An attempt to use the primers in the flanking DNA to PCR amplify the preintegration site failed, giving an irrelevant PCR product which was sequenced to reveal an amplified porcine repeat sequence (data not shown). Provirus 21321 was present in 24 of 30 pigs.
env sequence analysis.
Sequencing of env genes on all clones containing proviruses that appeared intact as shown in Tables 1 and 2 showed that each contained a full-length Env ORF, confirming the PTT results. Comparison of the proviral env with published PERV sequences showed a high degree of homology (>99%) with the published PERV B sequence Y12239 at both the nucleotide and amino acid levels (Fig. 3). The single sequence labelled 2441 is the consensus sequence obtained from clones 65321, 2441, and 310518 env sequences. Sequencing of env from lambda clones 73414 and 310518 gave only a single env consensus sequence from each, despite the two different 3′ LTRs that were present in these clones.
The receptor-interacting VRA and VRB regions in the middle of the Env protein showed no variation from the prototype sequence, although there were a few polymorphisms between the env genes that confirmed that they represented different proviruses (Fig. 3).
In summary, all clones were found to be intact in all three ORFs, as confirmed by PTT and sequencing. Sequence analysis of the clones with published PERV variants showed high levels of homology between them, as expected.
DISCUSSION
While extensive analysis of patients treated with living porcine tissue has shown that none of these patients has been infected by PERV, the future of porcine xenotransplantation may be dependent on using pigs with the smallest possible load of PERV proviruses. As a first step toward the objective of selecting such pigs, we have identified a number of potentially full-length, functional PERV proviruses and isolated their flanking sequences to enable their chromosomal locations to be identified. In addition, we have analyzed their prevalence in 30 Large White pigs transgenic for the human complement regulator hDAF.
Our strategy was designed to identify full-length proviral inserts encoding potentially infectious PERV A and B viral subtypes from a cosmid and a lambda library, respectively. Both libraries were derived from DNA of the same pig. These clones were coscreened by using both gag and env probes to increase the probability of isolating full-length proviruses and to rapidly eliminate grossly defective proviruses. Doubly positive clones were then screened using either restriction fragment analysis alone (cosmid library) or restriction fragment analysis with Southern blotting (lambda library) to identify unique and duplicate clones and to confirm the presence of proviral sequences. Subsequently, clones were screened by PCR and PTT to identify clones containing full-length Gag and Env ORFs, and each provirus was sequenced to confirm they were intact. Sequence flanking the proviral integration site of apparently intact proviruses was isolated and used to design primer pairs to screen pigs for the presence of each provirus. The data are summarized in Tables 1 and 2.
From the original cosmid-screening filters, only 10 to 12% of clones that gave gag-positive signals were also env positive (data not shown). This is due mainly to the nonvariant specificity of the gag probe, which would have detected all PERV variants, compared to the env probes, which were variant specific. We attempted to differentially screen for PERV A and B, using probes in the VRA and VRB regions that showed the most difference between the two. Although the PERV A and B probes showed specificity on Southern blots, they cross-hybridized to some extent under the conditions employed for cosmid screening. Consequently, it is likely that we would have detected proviruses with less homology to the published PERV A and B sequences. Such sequence variants have been identified by other groups (27, 51). Additionally, we detected a PERV B provirus in the PERV A-screened cosmid library, which presumably reflects some degree of probe cross-reactivity.
Most of the proviruses had pol sequences that were amplifiable by PCR (Fig. 1 and 2). We identified some polymorphism in this product, obtaining fragments of three different sizes. Sequencing revealed the smallest product to be a pol-related fragment with multiple deletions, proviruses giving this fragment would be defective. One product was of the size predicted for published PERV pol sequences, while the other fragment was larger than the published ones. Sequencing the ends of both of these products showed that they both had >96% pol homology and could both potentially encode functional pol, although the larger products were not fully sequenced since proviruses with this pol sequence were defective in either gag or env. Clone 78, the only provirus in the cosmid library that gave both-full length gag and env ORFs by PTT, gave a pol product of the predicted size from published pol genes.
On sequencing the env gene of the provirus on cosmid 78, it was apparent that it was a PERV B provirus (Fig. 2), despite the fact that it was identified using a PERV A env probe. The reason for this is not clear, as we did not identify the other PERV B proviruses that were found in the lambda library from the cosmid library. Conversely, the clone 78 PERV B provirus was not isolated from the lambda library. This possibly reflects the fact that the PERV A env probe could cross-hybridize with PERV B env to some extent. It is also possible that as filters were washed in bulk, the part of the library screening filter that clone 78 was isolated from was not subjected to a stringent enough wash after hybridization, hence cross-reactivity was observed. The failure to identify the cosmid 78 PERV B provirus in the lambda library is cause for more concern. It is possible due to the screening strategy used that lambda clones that contained the cosmid 78 provirus were all truncated during library construction and despite the three to four genome equivalents of library that were screened, the provirus would not be identified by this approach.
Unlike the results of the pol PCR assay, in the gag and env PCR screen only a relatively small fraction of clones isolated from the cosmid library yielded products (Table 1). This is unlikely to be due to small sequence changes that prevented PCR primer extension, as all published PERV A-, B-, and C-related proviruses have highly homologous sequences around the start and stop codons of all three viral genes. Additionally, we have successfully used a single primer pair to amplify gag and env genes of all currently known PERV variants under the low target complexity used here (not shown). It is likely that failure to produce a PCR product is related to gross mutations or deletions around the PBSs. This was supported by the Southern blot data shown in Table 3, where most of the clones that failed to yield PCR products failed to bind probes encompassing either the PBS or env LTR junction.
Analysis of the clones isolated from the lambda and cosmid library showed that there were significant differences between the clones from both libraries. Results for PERV B isolated from the lambda library showed some clear differences from the PERV A clones from the cosmid library as described above (compare Tables 1 and 2). First, most clones gave PCR products for both gag and env, indicating that more of the PERV B clones were intact at the gross sequence level. Approximately 50% of these clones gave full-length gag or env translation products as determined by PTT. Six clones gave both gag and env translation products, all of which gave an appropriately sized pol PCR product. Analysis of the flanking sequences from lambda clones 65321, 2441, and 310518 demonstrated that they had a common flanking sequence and represented a single provirus. Lambda clones 73414 and 35121 each gave two different LTR-3′-flanking sequences (termed 73414i and 73414ii and 35121i and 35121ii, respectively); 73414ii and 35121ii were identical, whereas 73414i and 35121i sequences showed no similarity. The 21321 5′-flanking sequence was not the same as the 310518 sequence, indicating that this represented another provirus. Thus, we identified five unique PERV B proviruses from the lambda library together with one PERV B clone from the cosmid library on the basis of their differing flanking sequences (Fig. 2). These flanking sequences were not the same as those identified by Deng et al. (9). It is also unclear if the flanking sequences isolated by Deng et al. were associated with a full-length proviral insert (9). Only one env sequence was obtained from clones 73414 and 35121, indicating that there were either two identical env sequences or that one provirus was defective and yielded no env product. The six env sequences showed that they have a high (>99%) degree of homology with the prototype PERV B sequence Y12239 at both the amino acid level (Fig. 3) and the DNA level (not shown).
As described above, we did not detect any intact PERV A proviruses in this pig. These differences between the results for PERV A and B cannot be explained simply by differences between clones isolated from the lambda and cosmid libraries, as the cosmid library contained multiple defective PERV A proviruses and we isolated an intact PERV B provirus, clone 78. Due to the larger insert size of the cosmid library, it is less likely that proviruses would be truncated during library construction than those used for the lambda library. Krach et al. (21) were able to isolate an infectious PERV A clone from PK15 cells, a cell type that is known to express high levels of infectious PERV A and B. There are two main possibilities to account for the absence of infectious PERV A in this pig. Data derived by coculture of pig aortic endothelial cells with permissive cells for replication suggest that PERV infectivity differs between different pigs (D. Cunningham et al. unpublished data). This implies that the number of infectious proviruses is low, as Krach et al. (21) have also suggested, and that there is significant diversity between individuals, as we have found (see below). Alternatively, full-length proviruses encoding infectious PERV A may be absent from this pig and infectious PERV A may be generated by recombination with other PERV sequences present during infection cycles. Wilson et al. (51) demonstrated that passage of their infectious virus PERV-NIH through human cells generated clones that when sequenced were found to be recombinants of PERV A and PERV C.
Retroviruses are diploid, and virions can undergo complex recombinations involving more than one internal template (switch displacement/assimilation or forced copy choice model) during reverse transcription (16). During this process, packaged defective A genomes could undergo recombination with subgroup B genomes to generate an infectious subgroup A virus. Recombinations of this form are known to be required for the generation of polytropic murine leukemia virus (42) and the subgroup B feline leukemia virus (41). It is also possible that, due to the relatively poor superinfection resistance conferred by PERV Env expression in PERV-infected cells, some of the proviruses examined by Krach et al. (21) were derived through reinfection cycles in the long-established PK15 cell line. It is possible that pig tissues or primary cell lines may differ in this respect.
The number of intact, unique proviruses identified here is broadly in agreement with other data on numbers of intact proviruses identified in Large White pigs. Bosch et al. (6) analyzed PERV RNA from Large White pigs and found 11 different transcripts, of which only 4 had intact env genes. No data were published on gag or pol genes. Additionally, other nonexpressed proviruses may have been present in this pig. We currently do not know if the six unique proviruses we have identified are expressed in Large White pigs and which, if any, correspond to the proviral transcripts noted by Bosch et al. (6). Analysis of the LTRs of these proviruses is currently under way. Krach et al. (21) identified one PERV A and two PERV B infectious proviruses from their screen of a genomic library of PK15 cells. We do not currently know if the six proviruses identified in this work are infectious.
A caveat of our screening approach is that if there are two or more proviral sequences on the same clone, we cannot determine if either is intact, due to cross-complementation in both the PCR and sequencing reactions. As noted above, both clones 73414 and 35121 appear to be contiguous and may each contain two defective, but complementary, proviruses based on the results of LTR analysis. This question will be answered by further work on these two clones. Additionally, as discussed above, viral complementation and recombination in other species have been shown to lead to the emergence of infectious virus and may be the origin for infectious PERV A in some pigs. In addition to removing full-length functional proviruses, it may be necessary to examine the possibility that recombination between defective proviruses may generate infectious viruses after infection. Further studies on the relative expression rates of defective genomes and how efficiently they are copackaged is needed to address this point.
We used LTR-flanking sequence to design primer pairs to determine the prevalence of the six proviruses within 30 unrelated Large White hDAF pigs. As shown in Table 4, three patterns of prevalence were observed: 73414ii was present in all pigs examined; 35121i, 310518, and 21321 were common, but some pigs did not have them; and cosmids 78 and 73414i were uncommon. This pattern may reflect either founder effects within the herd or the fact that the more common proviruses were integrated farther back in the evolution of the domestic pig. Ancient proviruses in the founder animals will be present in all offspring, while more recently integrated proviruses may show a more sparse distribution. The degree of heterogeneity between pigs is similar to that noted for PERV C (6,17). Interestingly, sequence analysis of the LTR-flanking sequences showed that lambda clones 73414 and 35121 are contiguous, sharing LTR-flanking sequence 73414ii, while 35121i and 73414i were unique to each respective lambda clone. This indicates that there are three almost concatameric PERV proviruses at this locus, implying a preferred integration site. Preferred targets for retroviral integration have been suggested previously. Shih et al. (39) identified that within these targets, independent integration events can occur at the same nucleotide base.
We have demonstrated that of the approximately 50 PERV proviruses enumerated using a pro probe (22), only six unique intact PERV B proviruses and no intact PERV A proviruses were present in the genome of the Large White pig studied. Thus, the majority of proviruses are defective in one or more of the viral genes. No PERV C env sequences were detectable in this pig by PCR. However, there inevitably remain a large number of proviruses that have one or two ORFs, which may complement each other. It is possible that some of the PERV proteins may have functions, such as protection against reinfection with exogenous PERV viruses, perhaps by providing superinfection resistance (3) or by Fv-type restriction mechanisms (47).
Regardless of speculation, these data indicate that it would be straightforward to remove proviruses 78 and 73414i by breeding. Whether provirus 73414ii can be removed by breeding will require a survey of a larger number of pigs to determine if there is variation at this locus. The recent developments in pig cloning (4, 29, 35) using nuclear transfer may also enable the genetic knockout of the more ubiquitous proviruses, whereas breeding out would necesarily be a more protracted process. The remaining proviruses show sufficient variation to indicate that a breeding strategy to remove them should be be possible. In this regard 2 of the 30 pigs examined (F894 and H338) had much lower proviral numbers. However, it is recognized that selection purely on the basis of these five proviruses may fix other proviruses that are not represented in this pig but are present in other pigs. Consequently, a breeding strategy will require detailed analysis, using a similar strategy, of the proviruses present in a number of founder pigs. This will ensure that all intact proviruses present in the founder herd are identified.
ACKNOWLEDGMENTS
This work was funded by Imutran Ltd., a Novartis Pharma company.
Thanks are due to Grahame McKenzie for the cosmid and lambda libraries and Eleanor Lees-Jones for the identification of unrelated and outbred Large White pigs.
REFERENCES
- 1.Akiyoshi D E, Denaro M, Zhu H, Greenstein J L, Banerjee P, Fishman J A. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J Virol. 1998;72:4503–4507. doi: 10.1128/jvi.72.5.4503-4507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Ltd.; 1997. [Google Scholar]
- 3.Berkowitz R D, Goff S P. Point mutations in Moloney murine leukemia virus envelope protein: effects on infectivity, virion association, and superinfection resistance. Virology. 1993;196:748–757. doi: 10.1006/viro.1993.1532. [DOI] [PubMed] [Google Scholar]
- 4.Betthauser J, Forsberg E, Augenstein M, Childs L, Eilertsen K, Enos J, Forsythe T, Golueke P, Jurgella G, Koppang R, Lesmeister T, Mallon K, Mell G, Misica P, Pace M, Pfister-Genskow M, Strelchenko N, Voelker G, Watt S, Thompson S, Bishop M. Production of cloned pigs from in vitro systems. Nat Biotechnol. 2000;18:1055–1059. doi: 10.1038/80242. [DOI] [PubMed] [Google Scholar]
- 5.Blusch J H, Patience C, Takeuchi Y, Templin C, Roos C, Von Der H, Steinhoff G, Martin U. Infection of nonhuman primate cells by pig endogenous retrovirus. J Virol. 2000;74:7687–7690. doi: 10.1128/jvi.74.16.7687-7690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch S, Arnauld C, Jestin A. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J Virol. 2000;74:8575–8581. doi: 10.1128/jvi.74.18.8575-8581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozzi E, Bhatti F, Schmoeckel M, Chavez G, Smith K G, Zaidi A, Bradley J R, Thiru S, Goddard M, Vial C, Ostlie D, Wallwork J, White D J, Friend P J. Long-term survival of nonhuman primates receiving life-supporting transgenic porcine kidney xenografts. Transplantation. 2000;70:15–21. [PubMed] [Google Scholar]
- 8.Cunningham, D. A., C. Herring, X. M. Fernández-Suárez, A. J. Whittam, K. Paradis, and G. A. Langford. 2001. Trends in cardiology, in press. [DOI] [PubMed]
- 9.Deng Y M, Lee J H, Moran C, Jin J H, Tuch B E, Rawlinson W D. Mapping dispersed repetitive loci using semi-specific PCR cloning and somatic cell hybrid mapping. Nucleic Acids Res. 2000;28:E103. doi: 10.1093/nar/28.23.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Y M, Tuch B E, Rawlinson W D. Transmission of porcine endogenous retroviruses in severe combined immunodeficient mice xenotransplanted with fetal porcine pancreatic cells. Transplantation. 2000;70:1010–1016. doi: 10.1097/00007890-200010150-00004. [DOI] [PubMed] [Google Scholar]
- 11.Dinsmore J H, Manhart C, Raineri R, Jacoby D B, Moore A. No evidence for infection of human cells with porcine endogenous retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplantation. 2000;70:1382–1389. doi: 10.1097/00007890-200011150-00020. [DOI] [PubMed] [Google Scholar]
- 12.Elliott R B, Escobar L, Garkavenko O, Croxson M C, Schroeder B A, McGregor M, Ferguson G, Beckman N, Ferguson S. No evidence of infection with porcine endogenous retrovirus in recipients of encapsulated porcine islet xenografts. Cell Transplant. 2000;9:895–901. doi: 10.1177/096368970000900616. [DOI] [PubMed] [Google Scholar]
- 13.Ericsson T, Oldmixon B, Blomberg J, Rosa M, Patience C, Andersson G. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J Virol. 2001;75:2765–2770. doi: 10.1128/JVI.75.6.2765-2770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heneine W, Tibell A, Switzer W M, Sandstrom P, Rosales G V, Mathews A, Korsgren O, Chapman L E, Folks T M, Groth C G. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet. 1998;352:695–699. doi: 10.1016/S0140-6736(98)07145-1. [DOI] [PubMed] [Google Scholar]
- 15.Herring C, Cunningham D A, Whittam A J, Fernandez-Suarez X M, Langford G A. Monitoring xenotransplant recipients for infection by PERV. Clin Biochem. 2001;34:23–27. doi: 10.1016/s0009-9120(00)00187-9. [DOI] [PubMed] [Google Scholar]
- 16.Hu W S, Temin H M. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin H, Inoshima Y, Wu D. Expression of porcine endogenous retrovirus in peripheral blood leukocytes from ten different breeds. Transplant Infect Dis. 2000;2:11–14. doi: 10.1034/j.1399-3062.2000.020103.x. [DOI] [PubMed] [Google Scholar]
- 18.Kikyo N, Wolffe A P. Reprogramming nuclei: insights from cloning, nuclear transfer and heterokaryons. J Cell Sci. 2000;113:11–20. doi: 10.1242/jcs.113.1.11. [DOI] [PubMed] [Google Scholar]
- 19.Kono T. Nuclear transfer and reprogramming. Rev Reprod. 1997;2:74–80. doi: 10.1530/ror.0.0020074. [DOI] [PubMed] [Google Scholar]
- 20.Kono T, Obata Y, Yoshimzu T, Nakahara T, Carroll J. Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat Genet. 1996;13:91–94. doi: 10.1038/ng0596-91. [DOI] [PubMed] [Google Scholar]
- 21.Krach U, Fischer N, Czauderna F, Tonjes R R. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J Virol. 2001;75:5465–5472. doi: 10.1128/JVI.75.12.5465-5472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Tissier P, Stoye J P, Takeuchi Y, Patience C, Weiss R A. Two sets of human-tropic pig retrovirus. Nature. 1997;389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 23.Levy M F, Crippin J, Sutton S, Netto G, McCormack J, Curiel T, Goldstein R M, Newman J T, Gonwa T A, Banchereau J, Diamond L E, Byrne G, Logan J, Klintmalm G B. Liver allotransplantation after extracorporeal hepatic support with transgenic (hCD55/hCD59) porcine livers: clinical results and lack of pig-to-human transmission of the porcine endogenous retrovirus. Transplantation. 2000;69:272–280. doi: 10.1097/00007890-200001270-00013. [DOI] [PubMed] [Google Scholar]
- 24.Loss M, Arends H, Winkler M, Przemeck M, Steinhoff G, Rensing S, Kaup F J, Hedrich H J, Winkler M E, Martin U. Analysis of potential porcine endogenous retrovirus (PERV) transmission in a whole-organ xenotransplantation model without interfering microchimerism. Transplant Int. 2001;14:31–37. doi: 10.1111/j.1432-2277.2001.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 25.Martin U, Steinhoff G, Kiessig V, Chikobava M, Anssar M, Morschheuser T, Lapin B, Haverich A. Porcine endogenous retrovirus (PERV) was not transmitted from transplanted porcine endothelial cells to baboons in vivo. Transplant Int. 1998;11:247–251. doi: 10.1007/s001470050136. [DOI] [PubMed] [Google Scholar]
- 26.Martin U, Winkler M E, Id M, Radeke H, Arseniev L, Takeuchi Y, Simon A R, Patience C, Haverich A, Steinhoff G. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV) Xenotransplantation. 2000;7:138–142. doi: 10.1034/j.1399-3089.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- 27.Onions D, Hart D, Mahoney C, Galbraith D, Smith K. Endogenous retroviruses and the safety of porcine xenotransplantation. Trends Microbiol. 1998;6:430–431. doi: 10.1016/s0966-842x(98)01386-9. [DOI] [PubMed] [Google Scholar]
- 28.Onions D, Cooper D K, Alexander T J, Brown C, Claassen E, Foweraker J E, Harris D L, Mahy B W, Minor P D, Osterhaus A D, Pastoret P P, Yamanouchi K. An approach to the control of disease transmission in pig-to-human xenotransplantation. Xenotransplantation. 2000;7:143–155. doi: 10.1034/j.1399-3089.2000.00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry A C. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000;289:1188–1190. doi: 10.1126/science.289.5482.1188. [DOI] [PubMed] [Google Scholar]
- 30.Paradis K, Langford G, Long Z, Heneine W, Sandstrom P, Switzer W M, Chapman L E, Lockey C, Onions D, Otto E. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science. 1999;285:1236–1241. doi: 10.1126/science.285.5431.1236. [DOI] [PubMed] [Google Scholar]
- 31.Patience C, Patton G S, Takeuchi Y, Weiss R A, McClure M O, Rydberg L, Breimer M E. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet. 1998;352:699–701. doi: 10.1016/S0140-6736(98)04369-4. [DOI] [PubMed] [Google Scholar]
- 32.Patience C, Switzer W M, Takeuchi Y, Griffiths D J, Goward M E, Heneine W, Stoye J P, Weiss R A. Multiple groups of novel retroviral genomes in pigs and related species. J Virol. 2001;75:2771–2775. doi: 10.1128/JVI.75.6.2771-2775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 34.Pitkin Z, Mullon C. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif Organs. 1999;23:829–833. doi: 10.1046/j.1525-1594.1999.06444.x. [DOI] [PubMed] [Google Scholar]
- 35.Polejaeva I A, Chen S H, Vaught T D, Page R L, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares D L, Colman A, Campbell K H. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- 36.Roest P A, Roberts R G, Sugino S, van Ommen G J, den Dunnen J T. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum Mol Genet. 1993;2:1719–1721. doi: 10.1093/hmg/2.10.1719. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schumacher J M, Ellias S A, Palmer E P, Kott H S, Dinsmore J, Dempsey P K, Fischman A J, Thomas C, Feldman R G, Kassissieh S, Raineri R, Manhart C, Penney D, Fink J S, Isacson O. Transplantation of embryonic porcine mesencephalic tissue in patients with PD. Neurology. 2000;54:1042–1050. doi: 10.1212/wnl.54.5.1042. [DOI] [PubMed] [Google Scholar]
- 39.Shih C C, Stoye J P, Coffin J M. Highly preferred targets for retrovirus integration. Cell. 1988;53:531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- 40.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoye J P, Coffin J M. The dangers of xenotransplantation. Nat Med. 1995;1:1100. doi: 10.1038/nm1195-1100a. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi Y, Liong S H, Bieniasz P D, Jager U, Porter C D, Friedman T, McClure M O, Weiss R A. Sensitization of rhabdo-, lenti-, and spumaviruses to human serum by galactosyl(α1-3)galactosylation. J Virol. 1997;71:6174–6178. doi: 10.1128/jvi.71.8.6174-6178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi Y, Patience C, Magre S, Weiss R A, Banerjee P T, Le Tissier P, Stoye J P. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998;72:9986–9991. doi: 10.1128/jvi.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tonjes R R, Czauderna F, Kurth R. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J Virol. 1999;73:9187–9195. doi: 10.1128/jvi.73.11.9187-9195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Laan L J, Lockey C, Griffeth B C, Frasier F S, Wilson C A, Onions D E, Hering B J, Long Z, Otto E, Torbett B E, Salomon D R. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature. 2000;407:90–94. doi: 10.1038/35024089. [DOI] [PubMed] [Google Scholar]
- 49.Weiss R A, Magre S, Takeuchi Y. Infection hazards of xenotransplantation. J Infect. 2000;40:21–25. doi: 10.1053/jinf.1999.0604. [DOI] [PubMed] [Google Scholar]
- 50.Wilson C A, Wong S, Muller J, Davidson C E, Rose T M, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson C A, Wong S, Van Brocklin M, Federspiel M J. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol. 2000;74:49–56. doi: 10.1128/jvi.74.1.49-56.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]