Abstract
Although the vaccinia virus DNA polymerase is inherently distributive, a highly processive form of the enzyme exists within the cytoplasm of infected cells (W. F. McDonald, N. Klemperer, and P. Traktman, Virology 234:168–175, 1997). In the accompanying report we outline the purification of the 49-kDa A20 protein as a stoichiometric component of the processive polymerase complex (N. Klemperer, W. McDonald, K. Boyle, B. Unger, and P. Traktman, J. Virol. 75:12298–12307, 2001). To complement this biochemical analysis, we undertook a genetic approach to the analysis of the structure and function of the A20 protein. Here we report the application of clustered charge-to-alanine mutagenesis of the A20 gene. Eight mutant viruses containing altered A20 alleles were isolated using this approach; two of these, tsA20-6 and tsA20-ER5, have tight temperature-sensitive phenotypes. At the nonpermissive temperature, neither virus forms macroscopic plaques and the yield of infectious virus is <1% of that obtained at the permissive temperature. Both viruses show a profound defect in the accumulation of viral DNA at the nonpermissive temperature, although both the A20 protein and DNA polymerase accumulate to wild-type levels. Cytoplasmic extracts prepared from cells infected with the tsA20 viruses show a defect in processive polymerase activity; they are unable to direct the formation of RFII product using a singly primed M13 template. In sum, these data indicate that the A20 protein plays an essential role in the viral life cycle and that viruses with A20 lesions exhibit a DNA− phenotype that is correlated with a loss in processive polymerase activity as assayed in vitro. The vaccinia virus A20 protein can, therefore, be considered a new member of the family of proteins (E9, B1, D4, and D5) with essential roles in vaccinia virus DNA replication.
Vaccinia virus, the prototypic member of the poxvirus family, displays a great deal of genetic and physical autonomy from the host. The virus replicates solely within the cytoplasm of the host, and the 192-kb genome is thought to encode most if not all of the functions required for genome replication, gene expression, and virion morphogenesis. The centerpiece of the replication apparatus is the E9 DNA polymerase, which displays significant homology to the α and δ families of eucaryotic replicative polymerases as well as the polymerases encoded by herpesviruses. We and others have characterized the polymerase both genetically and biochemically (4, 5,9, 10, 12, 29–31, 36, 38, 39, 41). Temperature-sensitive (ts) alleles, mutator and anti-mutator alleles, and mutants conferring resistance to aphidicolin, phosphonoacetic acid, and cytosine arabinoside have been isolated and studied. The polymerase has been overexpressed and purified and shown to have both polymerase and proofreading exonuclease activities. We have also shown that the enzyme is inherently distributive in vitro, being able to catalyze the addition of <10 nucleotides (nt) per binding event when moderate levels of salt (40 mM NaCl) or divalent cations (8 mM MgCl2) are present (31). In sharp contrast, the cytoplasmic lysates of infected cells are able to catalyze the addition of as many as 7,000 nt in a single binding event under the same reaction conditions (29). We demonstrated that the protein(s) responsible for conferring processivity on the viral polymerase was present in extracts prepared from infected cells in which only early proteins were present but not in extracts prepared from uninfected cells. We also demonstrated that the vaccinia virus processivity factor had a native molecular mass of 45 kDa. In the accompanying paper, we describe our identification of the A20 protein as a stoichiometric component of the processive form of the polymerase. We show that A20 and DNA polymerase copurify through six chromatographic steps, that their physical interaction can be confirmed by coimmunoprecipitation, and that overexpression of both A20 and DNA polymerase leads to a corresponding increase in the levels of processive polymerase activity (24).
In this report, we describe a genetic analysis of A20 aimed at furthering our understanding of the structure of this protein and its role(s) in vivo. The use of conditionally lethal mutants as genetic tools for the study of vaccinia virus has been well established (8). Four complementation groups of ts mutants exhibiting a DNA− phenotype have been described; these contain lesions in the E9 DNA polymerase, the D5 DNA-independent nucleoside triphosphatase (NTPase), the B1 protein kinase, and the D4 uracil DNA glycosylase (16, 17, 33, 34, 36, 37, 41). Other proteins implicated in genome replication have been identified biochemically or by sequence analysis. These include the I3 single-strand DNA binding protein (SSB), the H6 topoisomerase, the A50 DNA ligase, the F2 dUTPase, the J3 thymidine kinase, the A48 thymidylate kinase, the F4:I4 ribonucleotide reductase, and the A22 resolvase (19, 40). The roles played by some of the latter group have been characterized using drug-resistant mutants, deletion mutants, or inducible recombinants.
Unfortunately, no mutants with lesions in the A20 gene were isolated in the initial mutant collections prepared by the Condit and Ensinger laboratories (6, 7, 13, 14). We therefore undertook a reverse genetic approach and subjected the A20 gene to targeted mutagenesis in an attempt to isolate a conditionally lethal allele. These efforts were successful, and we report here that the phenotype of the ts mutants we generated confirms that the A20 protein plays an essential role in viral DNA replication and that its disruption compromises the production of processive DNA polymerase activity.
(A preliminary report of this work was presented at the XIIIth International Poxvirus Workshop, Montpellier, France, September 2000.)
MATERIALS AND METHODS
Materials.
Restriction endonucleases, Taq DNA polymerase, T4 DNA ligase, calf intestinal phosphatase, pancreatic RNase, Escherichia coli DNA polymerase I, and DNA molecular weight standards were purchased from either Roche Molecular Biochemicals (Indianapolis, Ind.) or New England Biolabs, Inc. (Beverly, Mass.) and used as specified by the manufacturer. DNase I was obtained from Cooper Biochemicals, Inc. (West Chester, Pa.). [35S]methionine, 32P-labeled nucleoside triphosphates (NTPs), and [methyl-3H]thymidine were purchased from New England Nuclear Corporation (Boston, Mass.). Lipofectamine Plus, Geneticin (G418 sulfate), and 14C-labeled protein molecular weight markers were acquired from GIBCO-BRL Life Technologies (Gaithersburg, Md.) and used as specified.
Cells and viruses.
BSC40 African green monkey kidney cells and mouse L cells were maintained as monolayer cultures in Dulbecco modified Eagle medium (DMEM) (GIBCO-BRL, Gaithersburg, Md.) containing 5% fetal calf serum. Viral stocks of wild-type (wt) vaccinia virus (strain WR), ts42, ts17, and ts2 and the A20 mutants described below were prepared by ultracentrifugation of infected cytoplasmic extracts through a 36% sucrose cushion. Titers of all viral stocks were obtained by plaque assays performed on BSC40 cells. The permissive and nonpermissive temperatures for analysis of ts mutants were 31.5 and 40°C, respectively.
Mutagenesis and cloning of the vaccinia virus A20 gene.
Clustered charge-to-alanine mutagenesis was performed on multiple regions of the vaccinia virus A20 gene as summarized in Fig. 1. Mutations were introduced by overlap PCR (11). To construct each allele, two sets of primer pairs were used to amplify the targeted region: (i) an upstream 5′ primer (UN) and a 3′ primer which introduces the complement of the mutation (A20-XUP) and (ii) a 5′ primer which introduces the mutation (A20-XDN) and a downstream 3′ primer (DN). Both UN and DN contain BamHI sites at their 5′ termini. PCRs performed with these primer pairs and the WR genome as a template yielded products that overlapped by 17 nt. A mixture of these products then served as the template for a second round of PCR performed with corresponding upstream 5′ (UN) and downstream 3′ (DN) primers. The final product was gel purified, digested with BamHI, and cloned into pUC/neo, a plasmid containing the neomycin resistance gene under the control of the vaccinia virus P7.5 constitutive promoter (42). All constructs were subjected to DNA sequence analysis to confirm the insertion of the engineered mutations and the absence of spurious mutations. The primers used were as follows (the nucleotides changed to direct the alanine substitutions are boldfaced, and the BamHI sites used for cloning are underlined): A20-1, (i) UA (5′ ATGGATCCAGTCTATCATCGACAC 3′) and A20-1UP (5′ TTTGCTACCGCAGCATAATAATCAGATATTGACG 3′), (ii) A20-1DN (5′ TATGCTGCGGTAGCAAATAAACCGTTTAATAT 3′) and DA (5′ GCGGATCCAATATACATGAACGAG 3′); A20-2, (i) UA and A20-2UP (5′ ACTTGCCATAGCAGCTGGTATTTGAAAAGAGT 3′), (ii) A20-2DN (5′ CAGCTGCTATGGCAAGTGCGTGTAACAAAGT 3′) and DA; A20-3, (i) UB (5′ ATGGATCCGAGACGTCAATATCTG 3′) and A20-3UP (5′ ATGTGGCTGCCGCTATTTCAATTTCTAAAT 3′), (ii) A20-3DN (5′ AATAGCGGCAGCCACATTATTTGACGACGA 3′) and DB (5′ TAGGATCCTCGTCCTATAGTGTCT 3′); A20-4, (i) UB and A20-4UP (5′ ATAACGCGGCGGCAAATAATGTGTCTTCCT 3′), (ii) A20-4DN (5′ ATTTGCCGCCGCGTTATACTCTATTATAGAACG 3′) and DB; A20-ER, (i) UB and A20-ERUP (5′ AAAGAGGCTGCTATAATAGAGTATAACT 3′), (ii) A20-ERDN (5′ ATTATAGCAGCCTCTTTTGATGATAAATT 3′) and DB; A20-5, (i) UB and A20-5UP (5′ GAAATGCAGCAGCAAAAGAGCGTTCTATAA 3′), (ii) A20-5DN (5′ TTTTGCTGCTGCATTTCCAAAAATATCCAT 3′) and DB; A20-5ER, (i) UB and A20-5ERUP (5′ TGCAGCAGCAAAAGAGGCTGCTATAATAGAGTATAACT 3′), (ii) A20-5ERDN (5′ ATTATAGCAGCCTCTTTTGCTGCTGCATTTCCAAAAATATCCAT 3′) and DB; A20-6, (i) UC (5′ CAGGATCCTAGTGAGTACGGATTA 3′) and A20-6UP (5′ TTGCCGCTGCTACTGCTACAAATGTATGTTCTC 3′), (ii) A20-6DN (5′ AGCAGTAGCAGCGGCAAACACATTTTCCATTTT 3′) and DC (5′ TAGGATCCAATACCCTAACGGAG 3′); A20-7, (i) UC and A20-7UP (5′ TTATTGCTGCTGCTACAGTTCCGGATTTAT 3′), (ii) A20-7DN (5′ TGTAGCAGCAGCAATAAAAAACCAATCAGC 3′) and DC; A20-8, (i) UC and A20-8UP (5′ TTTGCTATTGCTGCTGCTACAGTTCCGGATTTAT 3′), (ii) A20-8DN (5′ AGCAGCAGCAATAGCAAACCAATCAGCATTTGA 3′) and DC.
FIG. 1.
Clustered charge-to-alanine mutagenesis of the vaccinia virus (strain WR) A20R gene. Altered alleles of A20R encoding proteins in which the highlighted charged residues were changed to alanine were generated as described in Materials and Methods. The endogenous A20 allele was then replaced with the mutant allele through transient dominant selection. Of the eight mutant viruses (vA20-1, -2, -3, -5, -ER5, -6, -7, and -8) generated using this procedure, two (tsA20-ER5 and tsA20-6) exhibited temperature-sensitive phenotypes. We were unable to isolate viruses containing mutant alleles 4 or ER, suggesting that these mutations might be lethal and unable to support virus viability.
Transient dominant selection and isolation of mutants.
The endogenous A20 allele in the viral genome was replaced with the mutant A20 alleles by transient dominant selection. Dishes (diameter, 35 mm) of BSC40 cells were infected with wt vaccinia virus at a multiplicity of infection (MOI) of 0.03 PFU/cell at 37°C. At 3 h postinfection (hpi), cells were transfected individually with 5 μg of a pUC/neo-A20 mutant construct by using the Lipofectamine Plus reagent and were shifted to 31.5°C. To select for neomycin-resistant virus generated by plasmid incorporation, Geneticin (G418) was added to the cells at 18 hpi to a final concentration of 3 mg/ml. Transfectants were harvested at 48 hpi, and neomycin-resistant viruses were isolated by two sequential rounds of plaque purification in the presence of G418. Plasmid integration was confirmed by PCR amplification of the neo gene. Subsequent plaque purifications were performed in the absence of G418 so that resolution of the tandemly duplicated A20 alleles, with the accompanying loss of the intervening neo gene, could occur. Loss of neo was confirmed by PCR. Determination of which plaques lacking neo retained the wt A20 allele and which had acquired the mutant A20 allele was accomplished by amplification and DNA sequence analysis of the A20 locus. Plaques identified as lacking neo and containing the mutant sequence were subjected to subsequent rounds of plaque purification until all progeny plaques lacked neo and contained the mutant A20 allele. These plaques were then expanded, and viral stocks were prepared. All rounds of infection were performed at 31.5°C.
Determination of 24-h viral yields.
Confluent BCS40 cells were infected with either wt virus, tsA20-6, tsA20-ER5, vA20-1, vA20-2, vA20-3, vA20-5, vA20-7, or vA20-8 at an MOI of 5 and incubated at either 31.5 or 40°C. At 24 hpi, infected cells were harvested by scraping, collected by sedimentation, and resuspended in a constant volume of 10 mM Tris, pH 9.0. Cells were disrupted by three cycles of freeze-thawing and two 15-s bursts of sonication. Viral yields were then determined by plaque assays performed at 31.5°C.
Analysis of viral protein synthesis by metabolic labeling.
Confluent 35-mm plates of BSC40 cells were infected with wt virus or tsA20-6 at an MOI of 10 at either 31.5 or 40°C. At the indicated times postinfection (2, 4, 6.5, and 8.5 hpi), cells were rinsed with prewarmed methionine-free DMEM and then incubated with the same medium containing 100 μCi of [35S]methionine/ml. After 45 min, the cells were harvested by scraping, collected by sedimentation, and resuspended in an equal volume of phosphate-buffered saline (PBS, comprising 140 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4 [pH 7.4]). Protein sample buffer (final concentrations, 1% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 10% glycerol, 25 mM Tris [pH 6.8]) was added to an equivalent aliquot of each sample, and cells were thoroughly disrupted by heating at 100°C. Proteins were resolved by electrophoresis on an SDS–10% polyacrylamide gel, and radiolabeled proteins were visualized by autoradiography.
Quantitation of viral DNA accumulation.
Dishes (diameter, 35 mm) of BSC40 cells were infected with either wt virus, tsA20-6, or tsA20-ER5 at an MOI of 5 and were maintained at 31.5 or 40°C. At 3, 6, 9, 12, and 24 hpi, cells were harvested by scraping, collected by sedimentation, washed once with PBS, and resuspended in 300 μl of loading buffer (10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–1 M ammonium acetate). Cells were then disrupted by three cycles of freeze-thawing. Lysates were diluted with an additional 450 μl of loading buffer, and a 25-μl aliquot of each sample was applied, in duplicate, to a Zeta probe membrane using a Bio-Dot microfiltration apparatus (both from Bio-Rad, Richmond, Calif.). Samples were denatured (with 1.5 M NaCl and 0.5 M NaOH) and washed twice (with 10× SSC) in situ. The membrane was then hybridized with a 32P-labeled nick-translated probe representing the HindIII E and F fragments of the vaccinia virus genome. After the membrane was washed and air dried, it was exposed to a phosphor screen overnight and data were acquired using the Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The data were then quantitated using ImageQuant software (Molecular Dynamics) and plotted using SigmaPlot (SSPS, Chicago, Ill.) software.
Determination of [3H]thymidine incorporation in tsA20-infected cells.
BSC40 cells were infected with either wt virus, tsA20-6, or tsA20-ER5 at an MOI of 5 and were maintained at either 31.5 or 40°C. At the indicated times postinfection (1.5, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 7, and 8 hpi), cells were rinsed with serum-free DMEM and then incubated with DMEM containing 10 μCi of [methyl-3H]thymidine/ml (83.7 Ci/mmol). After 30 min of labeling, the medium was removed and cells were rinsed with PBS and then treated in situ with ice-cold NP-40 lysis buffer (150 mM NaCl, 20 mM Tris [pH 7.8], 1.5 mM MgCl2, 0.65% NP-40). This treatment disrupts the plasma membrane but leaves the nuclei intact and adherent to the tissue culture dish. The lysis buffer was gently removed from the dish, and the cytoplasmic nucleic acids were precipitated by adding cold trichloroacetic acid (TCA) to 10%. Precipitates were collected on GF/C glass fiber filters (Whatman, Inc., Maidstone, Kent, England) and washed extensively with ice-cold 10% TCA. [3H]thymidine incorporation was determined by scintillation counting after the addition of Ready Protein liquid scintillation cocktail (Beckman Coulter, Fullerton, Calif.).
Determination of steady-state levels of the E9 polymerase and the A20 protein.
Confluent 35-mm dishes of BSC40 cells were infected with either wt virus, tsA20-6, or tsA20-ER5 at an MOI of 5. At 2, 4, and 8 hpi, cells were harvested by scraping and collected by sedimentation. Cells were washed once with PBS, resuspended in 1 mM Tris (pH 9.0), and disrupted by three cycles of freeze-thawing. Extracts were then fractionated on an SDS–10% polyacrylamide gel electrophoresis (PAGE) gel, and proteins were transferred electrophoretically to nitrocellulose filters (Schleicher & Schuell, Keene, N.H.). The blot was incubated with polyclonal sera directed against the A20 protein (24) or the E9 polymerase (28) (primary sera used at a 1:500 dilution) and subsequently with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:15,000 to 1:20,000) (Bio-Rad). Immunoreactive proteins were visualized by chemiluminescence using the Pierce (Rockford, Ill.) Super Signal reagents.
Preparation of vaccinia virus cell extracts for in vitro replication assays.
BSC40 cells and mouse L cells were infected with either wt virus, ts42 (E9 polymerase mutant), tsA20-6, tsA20-ER5, ts17 (D5 NTPase mutant), or ts2 (B1 kinase mutant) at an MOI of 15 PFU/cell and were maintained at either 31.5 or 40°C. Hydroxyurea (10 mM) was added to the medium when the inoculum was removed at 30 min postadsorption and was present until the cells were harvested at 6 hpi. Cells were scraped, collected via sedimentation, washed in isotonic buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA), and incubated in hypotonic buffer (10 mM Tris [pH 8.0], 10 mM KCl, 5 mM EDTA) for 10 min. Cells were then broken open with a Dounce homogenizer, and lysates were sedimented at low speed to remove nuclei and unbroken cells. The extract was further clarified by a second sedimentation at 16,000 × g for 20 min at 4°C. Glycerol was added to 12%, and aliquots were stored at −80°C until use.
Singly primed M13 replication assay.
A primed template was constructed as described previously (31). Briefly, a 24-mer oligonucleotide primer (5′ CGCCAGGGTTTTCCCAGTCACGAC 3′) was annealed to ssM13mp18 at a 20:1 molar ratio. Extracts prepared from cells infected with either wt virus or ts42, tsA20-6, tsA20-ER5, ts17, or ts2 at the permissive or nonpermissive temperature were prepared as described above. These extracts were then assayed for processive DNA polymerase activity in reaction mixtures containing 10 mM Tris-Cl (pH 7.5), 40 mg of bovine serum albumin/ml, 4% glycerol, 0.1 mM EDTA, 5 mM dithiothreitol, 8 mM MgCl2, 25 fmol of primed M13 DNA, 750 ng of E. coli SSB, 60 μM (each) dCTP, dGTP, and dATP, and 20 μM [α-32P]TTP (5 μCi/nmol). Reaction mixtures were preincubated with two of the four dNTPs (dCTP and dGTP) for 3 min at 30°C, and primer extension was initiated by addition of the remaining two dNTPs. Reaction mixtures were then incubated for 15 min, and reactions were stopped by addition of an equal volume of 1% SDS–40 mM EDTA. Primer extension products were fractionated on a 0.8% agarose gel containing 0.125 μg of ethidium bromide/ml; the gel was cast and run in 1× TBE (50 mM Tris, 50 mM boric acid, 1 mM EDTA). The agarose gel was then dried and exposed to Kodak MR film (Eastman Kodak, Rochester, N.Y.) for autoradiography.
Computer analysis.
Autoradiographic films were scanned with a SAPHIR scanner (Linotype Hell Co, Hauppauge, N.Y.) and adjusted using Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.) or Canvas 6.0 (Deneba Systems, Miami, Fla.) software. Plaque assay results were photographed using an AlphaImager (Alpha Innotech, San Leandro, Calif.) digital camera. Figures were labeled using Canvas 6.0 and printed using a Kodak 8670 dye sublimation printer.
RESULTS
To conduct a genetic analysis of how the A20 protein functions in the viral life cycle, we chose to generate altered alleles of A20 by clustered charge-to-alanine mutagenesis. This technique has been used successfully in our laboratory and in those of others to generate ts mutants of vaccinia virus and gain insight into gene function (11, 20, 21). The experimental rationale is that clusters of charged residues are apt to lie on the surface of the protein. Therefore, substitutions in these regions are likely to modulate such properties as protein-protein or protein-DNA interactions rather than to disrupt global protein folding. Our goal was to isolate viruses in which the endogenous allele of A20 had been replaced with a mutant allele that conferred a conditionally lethal phenotype.
Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: insertion of the altered alleles into the endogenous A20 locus by transient dominant selection.
We selected multiple regions of the vaccinia virus protein (Fig. 1) for mutagenesis, based on the presence of clusters of charged residues. Using overlap PCR, we generated constructs of the A19- A20-A21 region into which the nucleotides necessary to encode alanine substitutions had been introduced. We then utilized transient dominant selection to isolate viruses in which the mutant A20 allele had replaced the endogenous allele (18). Briefly, the mutant alleles were cloned into the pUC/neo plasmid, which contains the neomycin resistance gene under the control of a constitutive vaccinia virus promoter (26). Cells infected with wt virus were transfected with these plasmids and maintained at 31.5°C in the presence of G418. To select for viruses in which the plasmid had become incorporated into the genome, two rounds of plaque purification were performed in the presence of G418. In these viruses, the drug resistance gene is flanked by the endogenous and exogenous A20 alleles. Upon removal of G418 selection, the inherently unstable tandem duplication can resolve, and recombination occurs between the two A20 alleles. This recombinational resolution leads to the excision of the neo gene and the retention of either the wt or the mutant copy of the A20 gene. Loss of the neo gene was analyzed by PCR, and those plaques that had in fact retained only the desired mutant allele of A20 were identified by DNA sequence analysis.
We attempted to generate 10 mutant viruses using the procedure described above. vA20-1, -2, -3, -ER5, -5, -6, -7, and -8 were isolated and found to be viable at 31.5°C. We were unable to isolate viruses containing targeted mutations in region 4 or in the ER region adjacent to region 5. In each of these two cases, we analyzed more than 20 resolved progeny generated from multiple neo parental viruses; all were found to be wt at the A20 locus. We concluded that the particular mutations generated in these regions of the A20 protein are lethal to the virus at 31.5°C.
vA20-ER5 and vA20-6 are temperature sensitive as determined by plaque assay and single-step growth experiments.
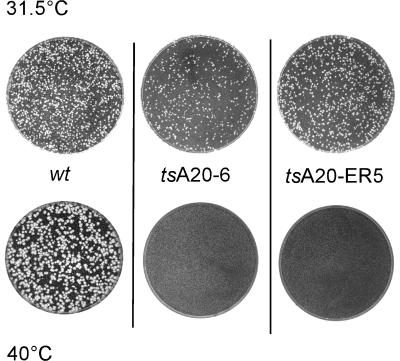
The eight mutants isolated were further characterized and tested for a ts phenotype. First, the viral stocks were titrated in parallel at 31.5 and 40°C. Two mutants, vA20-ER5, in which residues 185 to 191 were changed from ERSFDDK to AASFAAA (mutated nucleotides are boldfaced), and vA20-6, in which residues 265 to 269 were changed from KVKKK to AVAAA, were found to be temperature sensitive. Both mutants were unable to form plaques at 40°C but formed plaques of normal size when titrations were performed at 31.5°C (Fig. 2). These two mutants were henceforth designated tsA20-ER5 and tsA20-6. The plaques formed by the other mutants were comparable to those seen with wt virus at both temperatures (data not shown).
FIG. 2.
tsA20-ER5 and tsA20-6 do not form plaques at the nonpermissive temperature. Equivalent numbers of PFU of wt virus, tsA20-ER5, or tsA20-6 were used to infect confluent 60-mm dishes of BSC40 cells. Cells were maintained at either 31.5 or 40°C for 48 h, after which plates were stained with crystal violet and compared for plaque formation.
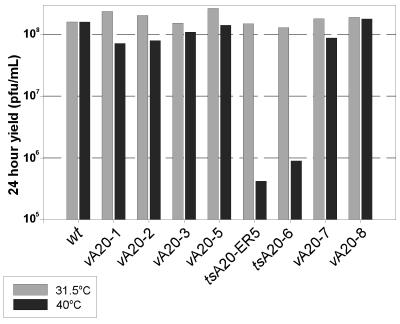
Second, single-step growth experiments were performed with all of the mutants isolated. Confluent monolayers of BSC40 cells were infected at both 31.5 and 40°C (at an MOI of 5), and the 24-h viral yield from this single round of infection was then quantitated (Fig. 3). Cells infected with tsA20-6 at 40°C produced 2 orders of magnitude less virus than parallel cultures maintained at 31.5°C; a comparable result was obtained from infections performed in L cells (data not shown). tsA20-ER5-infected cultures showed a 2.5-log-unit decrease in yield at 40°C relative to the yield obtained at 31.5°C. wt-infected cultures, and those infected with the viruses containing the other mutant A20 alleles, produced comparable yields of virus at both temperatures. Thus, the A20-1, -2, -3, -5, -7, and -8 mutations appeared to have no impact on the infectious cycle, and the viruses containing these alleles were therefore not subjected to further analysis.
FIG. 3.
tsA20-6 and tsA20-ER5 show a temperature-sensitive defect in the production of infectious virus from a single round of infection. Confluent dishes of BSC40 cells were infected with either wt virus, vA20-1, vA20-2, vA20-3, vA20-5, tsA20-ER5, tsA20-6, vA20-7, or vA20-8 at an MOI of 5 and were maintained at either 31.5 or 40°C for 24 h. Cells were then harvested, and the viral yield was determined in plaque assays performed at 31.5°C.
The data described above indicated that mutation of the A20 locus could have a profound impact on the ability of vaccinia virus to mediate a productive infection. Confirmation that the temperature sensitivity exhibited upon acquisition of the A20-6 allele was indeed linked to this allele was obtained from marker rescue studies. Briefly, transfection of a plasmid encoding the wt A20 allele (but not the corresponding vector alone) could “rescue” the ts phenotype by recombination with the tsA20-6 genome (data not shown). These data confirmed that the phenotype of tsA20-6 is due to the altered A20 allele and not to any other changes within the viral genome. Coinfections with wt virus were also performed, and quantitation of the viral yield indicated that both tsA20 alleles have a recessive phenotype (data not shown). Taken together, the data in Fig. 2 and 3 indicate that the protein encoded by the vaccinia virus A20 gene plays an essential role in the viral life cycle.
tsA20-6-infected cells fail to synthesize late viral proteins at the nonpermissive temperature.
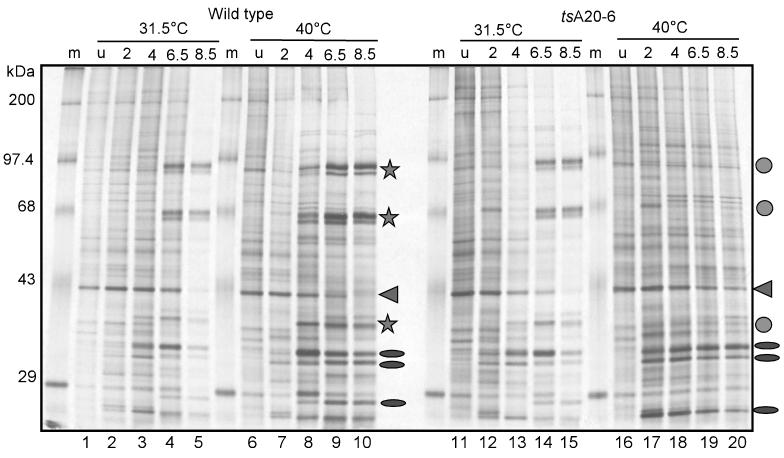
Both tsA20-6 and tsA20-ER5 show a profound temperature sensitivity that causes a severe arrest in the infectious cycle. To determine which stage of the vaccinia virus life cycle was affected in these ts mutants, we first examined the temporal profile of protein synthesis in cells infected with wt virus or tsA20-6 at the permissive and nonpermissive temperatures. After infection at an MOI of 10, BSC40 cells were metabolically labeled with [35S]methionine for 45 min at various times postinfection; total cell lysates were then resolved by SDS-PAGE and visualized by autoradiography (Fig. 4). The expression pattern seen in cells infected with tsA20-6 at the permissive temperature is virtually identical to that seen in wt-infected cells (Fig. 4; compare lanes 12 to 15 with lanes 2 to 5). However, a clear defect is seen in cells infected with tsA20-6 at the nonpermissive temperature. Although the expression of early proteins appears to be normal (Fig. 4; compare lanes 17 to 20 [tsA20-6] to lanes 7 to 10 [wt]), late viral proteins are not expressed (compare lanes 19 and 20 with lanes 8 to 10, 14, and 15). Thus, at the nonpermissive temperature, tsA20-6 is apparently able to enter cells and direct the expression of early mRNAs and proteins. However, the life cycle arrests prior to the expression of late genes. This arrest could reflect a defect in secondary uncoating, DNA replication, or intermediate or late gene expression.
FIG. 4.
Synthesis of late viral proteins is abolished during nonpermissive infections performed with tsA20-6. Confluent 35-mm dishes of BSC40 cells were infected with either wt virus or tsA20-6 at an MOI of 10 and maintained at either 31.5 or 40°C. Cells were metabolically labeled with [35S]Met for 45 min prior to being harvested at the times indicated. Uninfected cells (u) were analyzed in parallel at both temperatures. Cell extracts were fractionated on an SDS–10% PAGE gel, and the nascent polypeptides were visualized by autoradiography. Ovals and stars represent characteristic early and late viral proteins, respectively. Circles indicate the late proteins that are absent in lanes 19 and 20. The arrowhead indicates a cellular protein whose synthesis declines during vaccinia virus infections.
DNA replication is severely impaired in tsA20-ER5- and tsA20-6-infected cells.
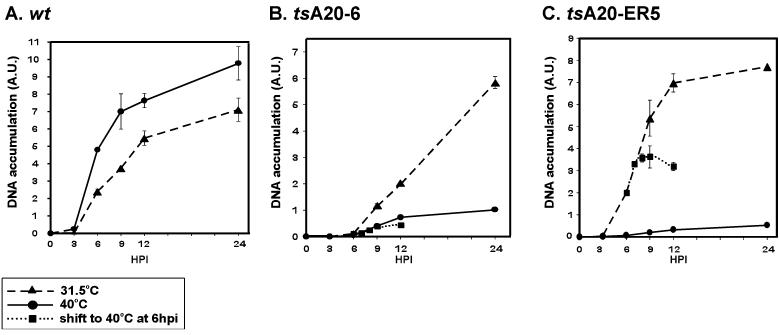
To establish if the block in tsA20-6 and tsA20-ER5 infections occurs at the stage of DNA replication, the accumulation of viral genomes during the infectious cycle was quantitated. Cells were infected with either wt virus, tsA20-ER5, or tsA20-6 and maintained at either 31.5 or 40°C. At 3, 6, 9, 12, and 24 hpi, cells were harvested and aliquots of total cellular extracts were subjected to DNA dot blot hybridization analysis. Data were quantitated on a phosphorimager and are shown graphically in Fig. 5. When cells were infected with wt virus, the profile of viral DNA accumulation was similar at both temperatures, with somewhat more DNA being accumulated at 40°C (Fig. 5A). At 9, 12, and 24 hpi, levels of DNA accumulated at 40°C relative to those at 31.5°C were 186, 139, and 137%, respectively. This was not true in cells infected with tsA20-ER5 or tsA20-6. At the permissive temperature, the profile seen after tsA20-ER5 infection was similar to that observed with wt virus (Fig. 5C): DNA accumulation increased after 3 hpi and began to level off after 12 hpi. In tsA20-6-infected cells, DNA accumulation was not seen until 6 hpi and continued without leveling off for the duration of the 24-h infection (Fig. 5B). Most importantly, cells infected at 40°C with either ts mutant showed a severe impairment in the accumulation of viral DNA (Fig. 5B and C). At 24 hpi, the ratio of DNA accumulation in cells infected with tsA20-6 at 40°C to that in cells infected at 31.5°C was 17%; in cells infected with tsA20-ER5 this ratio was 7%. These data provide compelling evidence that the lesions in the tsA20 viruses compromise viral DNA replication.
FIG. 5.
DNA replication is defective at the nonpermissive temperature in cells infected with tsA20-ER5 or tsA20-6. Confluent monolayers of BSC40 cells were infected with either wt virus (A), tsA20-6 (B), or tsA20-ER5 (C) and maintained at 31.5°C (dashed line) or 40°C (solid line). In some cases, cells were infected at 31.5°C and then shifted to 40°C at 6 hpi (dotted line). At the indicated time points, cells were harvested and lysed, and equivalent amounts of sample were spotted in duplicate onto a Zeta probe membrane. Accumulation of viral DNA was monitored by dot blot hybridization using a radiolabeled nick-translated probe representing the HindIII E and F fragments of the vaccinia virus genome. Signals were quantitated using a Storm PhosphorImager, and a graphic representation was prepared using SigmaPlot software.
To confirm that the defect is indeed intrinsic to DNA replication and does not affect the prerequisite stage of secondary uncoating, we performed shift-experiments. Infections were allowed to initiate at 31.5°C and were then shifted to 40°C at 6 hpi, a point at which the infectious cycle has progressed past uncoating and DNA replication has already begun. Indeed, as shown in Fig. 4 (lane 14), late gene expression is already robust at this time in cells infected with tsA20-6 at 31.5°C. We examined the accumulation of viral DNA at 7, 8, and 9 hpi in these shifted cultures (Fig. 5B and C). In both tsA20-6- and tsA20-ER5-infected cells, further DNA accumulation was curtailed, indicating that the mutations have a direct impact on DNA replication. In cells infected with tsA20-6, in which little DNA has accumulated by 6 hpi, the levels of viral DNA seen at 7, 8, and 9 hpi in the shifted cultures paralleled those seen in cells maintained at 40°C for the entire experiment. In cells infected with tsA20-ER5, there appeared to be a 1-h lag during which replication continued after the shift to the nonpermissive temperature; after 7 hpi, however, no additional viral DNA accumulated.
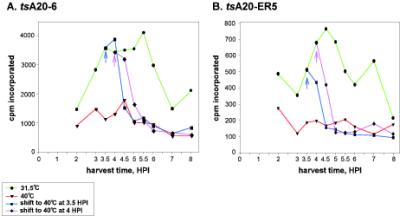
To investigate the block in DNA replication in more detail, we examined the early stages of viral DNA replication by monitoring the incorporation of [3H]thymidine in tsA20-6- and tsA20-ER5-infected cells. Cells were infected with the ts mutants and either maintained at 31.5 or 40°C or shifted from 31.5 to 40°C at 3.5 or 4 hpi. The incorporation of [3H]thymidine into cytoplasmic nucleic acids during sequential 30-min periods of pulse-labeling was then determined. For tsA20-6-infections maintained at 31.5°C, thymidine incorporation showed a broad peak from 3.5 to 5.5 hpi and then declined (Fig. 6A). For tsA20-ER5 infections, a narrower peak of thymidine incorporation occurred between 4 and 5 hpi (Fig. 6B). This general profile of thymidine incorporation is similar to what has been reported by us and others for wt-infected cells (17, 32, 35); the decline in thymidine incorporation at a time when DNA replication continues at a robust rate is attributed to feedback inhibition of the viral thymidine kinase as well as changes in the intracellular nucleotide pools (22, 40). Both ts mutants showed impaired thymidine incorporation at the nonpermissive temperature. These data support our conclusion that these ts mutants are defective in DNA replication. When cultures were infected with tsA20-6 at 31.5°C and then shifted to 40°C, thymidine incorporation began to decline 30 min after the cultures were shifted and had dropped to the levels seen with the 40°C-only cultures by 60 to 90 min postshift. In tsA20-ER5-infected cultures, a decrease in thymidine incorporation was seen immediately after the shift, and incorporation was reduced to that seen in the 40°C-only cultures by 60 min postshift. These data confirm that both of the tsA20 mutants have profound defects in viral DNA replication.
FIG. 6.
[3H]thymidine incorporation is impaired in cells infected with tsA20-ER5 and tsA20-6 at the nonpermissive temperature. Cells were infected with either tsA20-6 (A) or tsA20-ER5 (B) and incubated at either 31.5 or 40°C. In some cases, infection was initiated at 31.5°C and then cultures were shifted to 40°C at 3.5 or 4 hpi. At various times postinfection, cells were incubated for 30 min with DMEM containing 10 μCi of [methyl-3H]thymidine/ml prior to being harvested under conditions which leave the nuclei intact and adherent to the culture dish. Cytoplasmic nucleic acids were concentrated by acid precipitation and immobilized on glass fiber filters. [3H]thymidine incorporation was quantitated by liquid scintillation counting. Data shown in panels A and B were obtained in independent assays; therefore, the use of different scales on the ordinate is not significant.
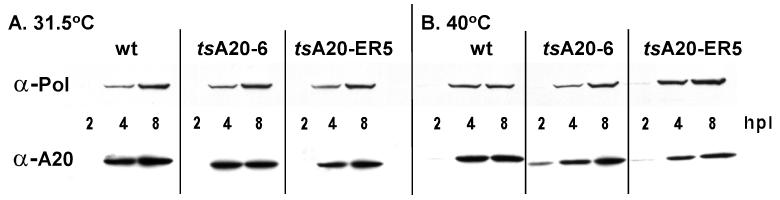
Both the A20 and polymerase proteins accumulate normally during nonpermissive tsA20 infections in BSC40 cells.
Having established that defects in the A20 protein cause a DNA− phenotype, we were interested in determining what aspect(s) of A20's functioning was impaired in these ts alleles. To investigate whether the proteins encoded by the tsA20-6 and tsA20-ER5 alleles were thermolabile, we prepared extracts from BSC40 cells at 2, 4, and 8 h after infection with either wt virus, tsA20-6, or tsA20-ER5 at the permissive or nonpermissive temperature. Aliquots were resolved by SDS-PAGE, transferred to nitrocellulose filters, and probed with the anti-A20 serum (Fig. 7). The temporal profiles of A20 accumulation were comparable in all of the infections, indicating that the A20 proteins encoded by the ts alleles are not thermolabile. The A20 protein was first detected at 4 hpi at 31.5°C and at 2 hpi at 40°C. Interestingly, the A20 proteins encoded by the two mutant alleles had slightly faster electrophoretic mobilities than the wt protein (Fig. 7A; compare 4- and 8-hpi lanes for wt virus, tsA20-6, and tsA20-ER5). We were also interested in determining whether the lesions in the A20 protein might cause destabilization of the E9 DNA polymerase, which could in turn compromise DNA replication. The upper half of the same blot was therefore probed with the anti-DNA polymerase antibody. The levels of polymerase at the nonpermissive and permissive temperatures were equivalent in cells infected with wt virus or with either of the ts mutants. Since neither the level of the A20 protein nor that of the DNA polymerase was affected in nonpermissive tsA20-6 and tsA20-ER5 infections, the defect in these infections appears to be at the level of protein function rather than at the level of synthesis or stability.
FIG. 7.
The A20 and E9 (polymerase) proteins accumulate to normal levels during nonpermissive infections with tsA20-ER5 and tsA20-6. Cells were infected with either wt virus, tsA20-ER5, or tsA20-6 at an MOI of 5 and were incubated at either 31.5 or 40°C. At the indicated times postinfection, cells were harvested and disrupted by three cycles of freeze-thawing. Aliquots of the extracts were fractionated on an SDS–10% PAGE gel and transferred to a nitrocellulose filter. The filter was cut in half at the 68-kDa marker; the top half of the blot was probed with a polyclonal anti-DNA polymerase (α-Pol) serum, and the bottom half was probed with a polyclonal anti-A20 serum.
Extracts prepared from cells infected with tsA20-6 or tsA20-ER5 do not direct the processive conversion of singly primed M13 DNA to the RFII form.
There is, as yet, no in vitro system for poxvirus DNA replication. Nor are assays available to dissect replication in vivo, beyond the ability to monitor [3H]thymidine incorporation, accumulation of viral DNA, and resolution of replication intermediates into monomeric genomes. We have, however, previously described the ability of cytoplasmic extracts of wt-infected cells to direct processive DNA synthesis on a singly primed M13 template in vitro (29). This assay was the basis of our conclusion that processive synthesis required both the E9 polymerase and at least one other early viral protein. Activity was abolished when cells were infected with a mutant encoding a temperature-sensitive DNA polymerase (ts42). Recombinant polymerase, however, could be used to reconstitute processive activity when added back to these polymerase-deficient extracts but not when added to similarly prepared extracts of uninfected cells. The accompanying paper (24) demonstrates that extracts prepared from cells overexpressing both the A20 protein and DNA polymerase show a significant increase in processive polymerase activity over that in extracts from cells in which only the polymerase is overexpressed. These data provide provocative evidence that A20 and DNA polymerase together might constitute the processive enzyme.
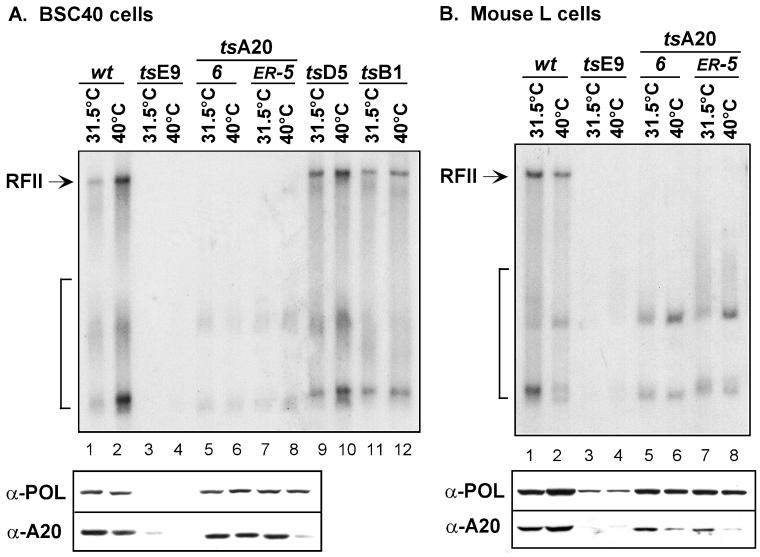
As an alternative approach to analyzing the role of A20 in the generation of processive polymerase activity, we examined the abilities of cytoplasmic extracts of cells infected with tsA20-6 or tsA20-ER5 to form RFII in the same in vitro assay. BSC40 cells or mouse L cells were infected with either wt virus, ts42 (DNA polymerase mutant), tsA20-6, tsA20-ER5, ts17 (D5 NTPase mutant), or ts2 (B1 kinase mutant) and maintained at either the permissive or the nonpermissive temperature. Cytoplasmic extracts were prepared from these infected cells as described in Materials and Methods, and these extracts were then tested for the ability to direct RFII formation using a singly primed M13 template in vitro. After a 3-min preincubation reaction that allowed replication complexes to assemble at the primer-template junction, synchronous DNA synthesis was initiated and allowed to proceed for 15 min at 30°C. These reactions were performed in the presence of 8 mM MgCl2, under which conditions the free E9 polymerase is highly distributive and is unable to copy the >7-kB template in the time allotted (31). Reaction products were fractionated on a TBE-agarose gel and visualized by autoradiography. Results are shown in the upper panels of Fig. 8; both the short products generated during distributive synthesis and the full-length RFII product generated during processive synthesis are indicated. Consistent with previous results (29), extracts prepared from BSC40 or L cells infected with ts42 (E9 polymerase mutant) at either the permissive or nonpermissive temperature failed to catalyze RFII formation (Fig. 8, lanes 3 and 4). The polymerase encoded by this mutant is extremely thermolabile, and no active enzyme can be detected in vitro (28). Even the short products diagnostic of distributive synthesis are absent in these reactions. Of significant importance to the present work is the observation that extracts prepared from BSC40 or L cells infected with tsA20-6 or tsA20-ER5 also failed to form RFII (Fig. 8, lanes 5 to 8). This defect was seen with extracts prepared from cells infected at either temperature and indicates that cells infected with these A20 mutants produce no processive polymerase activity that can be assayed in vitro. It is worth noting that the short products generated by the free polymerase acting in a distributive mode are present in these reactions.
FIG. 8.
Extracts prepared from cells infected with tsA20-ER5 or tsA20-6 cannot direct the processive synthesis of RFII in a singly primed M13 replication assay. BSC40 cells (A) and mouse L cells (B) were infected at an MOI of 15 with wt virus (lanes 1 and 2), tsE9 (lanes 3 and 4), tsA20-6 (lanes 5 and 6), tsA20-ER5 (lanes 7 and 8), tsD5 (lanes 9 and 10), or tsB1 (lanes 11 and 12) and maintained at 31.5°C (lanes 1, 3, 5, 7, 9, and 11) or 40°C (lanes 2, 4, 6, 8,10, and 12). At 6 hpi, infected cells were harvested, and cytoplasmic extracts were prepared as described in Materials and Methods. These extracts (2.0 μg) were then assayed in reaction mixtures containing 25 fmol of primed M13 ssDNA, 750 ng of E. coli SSB, 60 μM (each) dATP, dGTP, and dCTP, 20 μM [α-32P]TTP, and 8 mM MgCl2. After 15 min of incubation at 30°C, the reactions were stopped, and DNA products were fractionated on a 0.8% TBE-agarose gel containing ethidium bromide and visualized by autoradiography. The arrow indicates the RFII product; the bracket indicates the short DNA products that result from distributive polymerase activity. Aliquots of the same extracts were also subjected to immunoblot analysis with antisera directed against the DNA polymerase (α-POL) or A20 protein (α-A20). The immunoblot is aligned underneath the corresponding samples of the autoradiograph illustrating RFII formation.
The experiment for which results are shown in Fig. 7 indicated that the A20 proteins encoded by tsA20-6 and tsA20-ER5 were not thermolabile and accumulated to wt levels during nonpermissive infections performed in BSC40 cells. We wanted to confirm that the inability of the cytoplasmic lysates prepared from tsA20-infected cells to direct processive RFII formation was not due, however, to the absence of soluble DNA polymerase or A20 protein in these lysates. Therefore, the cytoplasmic lysates prepared from both the BSC40 and L cells were resolved by SDS-PAGE and transferred to a nitrocellulose filter. The top half of the filter was developed with an anti-DNA polymerase serum, and the bottom half was developed with an anti-A20 serum. Results are shown in the lower panels of Fig. 8; the immunoblots are aligned with the autoradiograph illustrating RFII formation. Both the DNA polymerase and the A20 protein were present at wt levels in lysates prepared from BSC40 or L cells infected with tsA20-6 or tsA20-ER5 at 31.5°C (Fig. 8; compare lanes 5 and 7 with lanes 1 and 2 in both panels). Therefore, the absence of processive polymerase activity in these lysates cannot reflect a loss of the A20 and polymerase proteins per se but rather reflects their inability to collaborate in forming RFII. When lysates were prepared from BSC40 cells infected at 40°C with tsA20-6, equivalent levels of the DNA polymerase and A20 appeared to be present (Fig. 8A, lane 6). There was a specific diminution, however, in the levels of A20 seen in some cultures infected at 40°C: BSC40 cells infected with tsA20-ER5 (Fig. 8A, lane 8) and L cells infected with tsA20-ER5 or tsA20-6 (Fig. 8B, lanes 6 and 8). As expected, polymerase was significantly depleted in extracts prepared from ts42-infected cells (Fig. 8, lanes 3 and 4); a corresponding diminution in the levels of A20 was also observed. The latter was unexpected and suggests that A20 might be either unstable or less soluble in the presence of the defective ts42-encoded DNA polymerase.
The defect in RFII formation seen with extracts prepared from tsA20-infected cells provides strong evidence that mutation of A20 causes a specific defect in processive DNA polymerase activity. This in vitro reaction asks for nothing more complex than processive DNA synthesis: a free primer terminus is provided, and the template is single-stranded and kept free of secondary structure by the inclusion of E. coli SSB. Thus, auxiliary functions that might be involved in initiation or template unwinding are not required. The specificity of the defect seen with ts42, tsA20-ER5, and tsA20-6 is underscored by the results obtained with ts mutants carrying lesions in the D5 DNA-independent NTPase (ts17) (15–17, 35) or the B1 protein kinase (ts2) (33, 34). Extracts from BSC40 cells infected with ts17 or ts2 at either the permissive or nonpermissive temperature are able to catalyze RFII formation in vitro (Fig. 8A, lanes 9 to 12). Thus, although these mutants exhibit a ts DNA− phenotype in vivo, their defect is not manifested in vitro in this specific assay, which demands only the ability to elongate a primer in a processive fashion.
DISCUSSION
Using a biochemical approach, we identified the vaccinia virus A20 protein as a stoichiometric component of the processive form of the viral DNA polymerase (24). In this report, we undertook a complementary genetic analysis of A20 in order to better understand its structure and function in the viral life cycle. Using clustered charge-to-alanine mutagenesis, we generated 10 altered alleles of the A20 gene and attempted to isolate viruses containing each of these alleles in place of the endogenous A20 gene. We were successful in isolating eight such viruses (vA20-1, -2, -3, -5, -ER5, -6, -7, and -8). We were unable to isolate vA20-4 or vA20-ER and feel justified in concluding that these alleles are not competent to support a productive infectious cycle. Six of the other alleles (A20-1, -2, -3, -5, -7, and -8) supported virus viability and did not confer any apparent phenotype. Two alleles (A20-ER5 and A20-6), however, engendered profound temperature-sensitivity.
Since the sequence of the A20 gene does not show any significant homology to genes other than the orthologs found in other poxviruses, and since it does not contain any recognizable motifs, it was difficult to predict which mutations might alter the protein in a way that would perturb its function in vivo. One of the alleles that appeared to be lethal to the virus, A20-4 (177DDE179→AAA), affects a region that is somewhat conserved among chordopoxvirus genomes; these three charged residues are flanked by two invariant aromatic residues. It was also of interest to see that whereas the A20-ER allele (185ERSFDDK191→AASFDDK) appeared to be incompatible with virus viability, and the adjacent A20-5 allele (185ERSFAAA191) conferred no phenotype in vivo, the two in combination (A20-ER5) (185AASFAAA191) conferred a temperature-sensitive phenotype. This region is not highly conserved in the A20 homologs encoded by diverse poxviruses (see reference 24) (Fig. 2A), but the presence of charged residues at the position corresponding to 185ER186 is frequent. In essence, the A20-5 allele serves as a partial intragenic suppressor of the ER allele. The other allele that conferred a temperature-sensitive phenotype, A20-6 (265KVKKK269→AVAAA), affects a region that is fairly well conserved in the various poxvirus homologs. Both of the ts alleles encode proteins that are stable in vivo, which is unusual; in our experience, most proteins encoded by ts alleles are thermolabile.
Generation of two viruses in which mutation of the A20 allele conferred a dramatic temperature-sensitive phenotype allowed us to probe the role of this protein in the viral life cycle. At the permissive temperature, the infectious cycles directed by these ts viruses were comparable to those observed with wt virus. At the nonpermissive temperature, plaque formation was abolished and the viral yield produced in a 24-h infection (at an MOI of 5) was reduced by 2 to 2.5 orders of magnitude. The viral life cycle appeared to progress normally through the expression of early proteins but then arrested at the phase of DNA replication. Dot blot hybridization analysis of viral DNA accumulation, and quantitation of [3H]thymidine incorporation, indicated that DNA synthesis was largely abolished at the nonpermissive temperature. We observed a cessation of DNA synthesis when cultures that had been infected at 31.5°C were shifted to 40°C at a time when replication had already begun, confirming that the A20 lesions have a direct impact on ongoing DNA synthesis. They do not, instead, act indirectly by inhibiting a prerequisite step such as secondary uncoating. The cessation of synthesis in the shifted cultures was not immediate, however; a lag of 30 to 60 min was typical. The presence of this lag was somewhat surprising, since many mutants with defects in key replication proteins show a fast-stop phenotype, in which ongoing DNA synthesis ceases immediately when cultures are shifted to the nonpermissive temperature (17). The lag seen with both tsA20-ER5 and tsA20-6 suggests that the mutant A20 proteins might lose function slowly upon shift to the nonpermissive temperature and/or might manifest their defect only when a new round of synthesis begins on a fresh template. The mutant proteins are not thermolabile but instead show a functional thermosensitivity; perhaps A20 undergoes a conformational change or a loss of function when it dissociates from the template after helping the polymerase complete synthesis on a given template.
The observation that tsA20-ER5 and tsA20-6 had a profound and specific defect in viral DNA replication was fully consistent with our purification of A20 as a major component of the processive DNA polymerase complex. We therefore reasoned that the DNA− phenotype would reflect an inability of the A20/DNA polymerase complex to direct processive synthesis in vivo and that this deficit would be retained in vitro. This hypothesis was tested by examining the ability of extracts prepared from cells infected with tsA20-ER5 or tsA20-6 to convert a singly primed M13 template to the RFII product in a processive manner. These extracts were incompetent in this assay, unlike extracts prepared from wt-infected cells. No RFII product was seen using the extracts prepared at 31.5 or 40°C. This constitutive absence of activity is also characteristic of extracts prepared from cells infected with ts42, a virus with a lesion in the DNA polymerase gene (29). Presumably, although these mutant proteins retain sufficient function in vivo at 31.5°C, perhaps due to the stabilizing influence of other viral proteins or the cytoplasmic architecture, they do not manifest this function in vitro. This is particularly striking in the case of the tsA20 extracts, since we have shown that both the A20 and DNA polymerase proteins are stable and are indeed present within these extracts. Furthermore, the short products characteristic of distributive polymerase action are formed by the tsA20 extracts, and we have not observed any defect in the ability of these extracts to synthesize short stretches of DNA using an activated salmon sperm template (data not shown) (30). Thus, the A20-ER5 and A20-6 proteins are unable to perform some or all of the activities required in the M13 assay. Because of the relative simplicity of this assay, we presume that it is the interaction with the polymerase, the interaction with the DNA template, or a hypothetical interaction(s) with other modulatory proteins that is affected. Determination of what facet of its activity as a likely processivity factor is incapacitated by the lesions in tsA20-ER5 and tsA20-6 will be investigated in future studies.
During the course of this work, the laboratories of B. Moss and S. Fields published a yeast two-hybrid analysis of interactions among the full repertoire of vaccinia virus proteins (27). In this assay, the A20 protein was shown to interact with the D5 (dNTPase) and D4 (uracil DNA glycosylase) proteins, which have been shown to play essential roles in viral DNA replication (40). These observations are provocative and warrant further investigation. An interaction with the H5 protein was also observed. The abundant H5 phosphoprotein appears to have multiple roles in the viral life cycle. H5 has been shown to stimulate the transcription of late genes in vitro (25), and several mutant alleles of H5 confer a dominant, temperature-sensitive phenotype in vivo and cause an arrest at an early stage of virion morphogenesis (11). H5 is also a substrate for the B1 protein kinase, which has been shown to be important for viral DNA replication, and appears to relocate to replication factories as DNA synthesis begins (1–3). If H5 serves as a scaffolding protein for DNA synthesis, then an interaction with A20 might prove to be important.
Subsequent to this two-hybrid analysis, the Moss laboratory also undertook a clustered charge-to-alanine mutagenesis of the A20 gene; their findings, as well as ours, were reported at the XIIIth International Poxvirus Workshop (Montpellier, France, September 2000), and their work has already been published (23). The collections of A20 alleles generated in the two studies overlap, and our results are in general agreement with theirs regarding the phenotypes observed. Of note, our tsA20-ER5 virus corresponds to mutant 185 of Ishii and Moss, and for those assays performed in common, comparable evidence of a severe ts DNA− phenotype was obtained.
In sum, the results presented here and in the accompanying paper (24) identify the A20 protein as playing an essential role in vaccinia virus replication. Unraveling how the A20 protein interacts with DNA, the DNA polymerase, and other modulatory viral proteins to accomplish processive and faithful replication of the genome is of significant interest. These studies will enhance our understanding of vaccinia virus biology and should also provide insight into the broader question of how microorganisms and cells accomplish the universal task of DNA replication.
ACKNOWLEDGMENTS
This work was supported by a grant to P.T. from the NIH (AI 21758). K.B. is supported by an NRSA from the Public Health Service (AI 10428).
We thank R. Tether and B. Tchizhed for technical assistance and Nancy Klemperer for discussions at the initiation of this project.
REFERENCES
- 1.Beaud G, Beaud R. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J Gen Virol. 1997;78:3297–3302. doi: 10.1099/0022-1317-78-12-3297. [DOI] [PubMed] [Google Scholar]
- 2.Beaud G, Beaud R, Leader D P. Vaccinia virus gene H5R encodes a protein that is phosphorylated by the multisubstrate vaccinia virus B1R protein kinase. J Virol. 1995;69:1819–1826. doi: 10.1128/jvi.69.3.1819-1826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown N G, Nick Morrice D, Beaud G, Hardie G, Leader D P. Identification of sites phosphorylated by the vaccinia virus B1R kinase in viral protein H5R. BMC Biochem. 2000;1:2. doi: 10.1186/1471-2091-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Challberg M D, Englund P T. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J Biol Chem. 1979;254:7812–7819. [PubMed] [Google Scholar]
- 5.Challberg M D, Englund P T. The effect of template secondary structure on vaccinia DNA polymerase. J Biol Chem. 1979;254:7820–7826. [PubMed] [Google Scholar]
- 6.Condit R C, Motyczka A. Isolation and preliminary characterization of temperature sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 7.Condit R C, Motyczka A, Spizz G. Isolation, characterization and physical mapping of temperature sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- 8.Condit R C, Niles E G. Orthopoxvirus genetics. Curr Top Microbiol Immunol. 1990;163:1–39. doi: 10.1007/978-3-642-75605-4_1. [DOI] [PubMed] [Google Scholar]
- 9.DeFilippes F M. Effect of aphidicolin on vaccinia virus: isolation of an aphidicolin-resistant mutant. J Virol. 1984;52:474–482. doi: 10.1128/jvi.52.2.474-482.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFilippes F M. Site of the base change in the vaccinia virus DNA polymerase gene which confers aphidicolin resistance. J Virol. 1989;63:4060–4063. doi: 10.1128/jvi.63.9.4060-4063.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMasi J, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a temperature-sensitive mutant with a profound defect in morphogenesis. J Virol. 2000;74:2393–2405. doi: 10.1128/jvi.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl P L, Jones E V, Moss B. Homology between DNA polymerase of poxviruses, herpesviruses and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci USA. 1986;83:3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensinger M. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus. J Virol. 1982;43:778–790. doi: 10.1128/jvi.43.3.778-790.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ensinger M, Rovinsky M. Marker rescue of temperature-sensitive mutations of vaccinia virus WR: correlation of genetic and physical maps. J Virol. 1983;48:419–428. doi: 10.1128/jvi.48.2.419-428.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans E, Klemperer N, Ghosh R, Traktman P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J Virol. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans E, Traktman P. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J Virol. 1987;61:3152–3162. doi: 10.1128/jvi.61.10.3152-3162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans E, Traktman P. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- 18.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia A D, Moss B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol. 2001;75:6460–6471. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassett D E, Condit R C. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc Natl Acad Sci USA. 1994;91:4554–4558. doi: 10.1073/pnas.91.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassett D E, Lewis J I, Xing X, DeLange L, Condit R C. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- 22.Hruby D E. Inhibition of vaccinia virus thymidine kinase by the distal products of its own metabolic pathway. Virus Res. 1985;2:151–156. doi: 10.1016/0168-1702(85)90245-x. [DOI] [PubMed] [Google Scholar]
- 23.Ishii K, Moss B. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J Virol. 2001;75:1656–1663. doi: 10.1128/JVI.75.4.1656-1663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klemperer N, McDonald W, Boyle K, Unger B, Traktman P. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J Virol. 2001;75:12298–12307. doi: 10.1128/JVI.75.24.12298-12307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald W F, Crozel-Goudot V, Traktman P. Transient expression of the vaccinia virus DNA polymerase is an intrinsic feature of the early phase of infection and is unlinked to DNA replication and late gene expression. J Virol. 1992;66:534–547. doi: 10.1128/jvi.66.1.534-547.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald W F, Klemperer N, Traktman P. Characterization of a processive form of the vaccinia virus DNA polymerase. Virology. 1997;234:168–175. doi: 10.1006/viro.1997.8639. [DOI] [PubMed] [Google Scholar]
- 30.McDonald W F, Traktman P. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr Purif. 1994;5:409–421. doi: 10.1006/prep.1994.1059. [DOI] [PubMed] [Google Scholar]
- 31.McDonald W F, Traktman P. Vaccinia virus DNA polymerase: in vitro analysis of parameters affecting processivity. J Biol Chem. 1994;269:31190–31197. [PubMed] [Google Scholar]
- 32.McFadden G, Dales S. Biogenesis of poxviruses: preliminary characterization of conditional lethal mutants of vaccinia virus defective in DNA synthesis. Virology. 1980;103:68–79. doi: 10.1016/0042-6822(80)90126-9. [DOI] [PubMed] [Google Scholar]
- 33.Rempel R E, Anderson M K, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rempel R E, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roseman N A, Hruby D E. Nucleotide sequence and transcript organization of a region of the vaccinia virus genome which encodes a constitutively expressed gene required for DNA replication. J Virol. 1987;61:1398–1406. doi: 10.1128/jvi.61.5.1398-1406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sridhar P, Condit R C. Selection for ts mutations in specific vaccinia virus genes: isolation of a phosphonoacetic acid-resistant, temperature sensitive virus mutant. Virology. 1983;128:444–457. doi: 10.1016/0042-6822(83)90269-6. [DOI] [PubMed] [Google Scholar]
- 37.Stuart D T, Upton C, Higman M A, Niles E G, McFadden G. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J Virol. 1993;67:2503–2512. doi: 10.1128/jvi.67.5.2503-2512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taddie J A, Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: identification of point mutations conferring altered drug sensitivities and reduced fidelity. J Virol. 1991;65:869–879. doi: 10.1128/jvi.65.2.869-879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taddie J A, Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3′–5′ exonuclease domain. J Virol. 1993;67:4323–4336. doi: 10.1128/jvi.67.7.4323-4336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traktman P. Poxvirus DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 775–798. [Google Scholar]
- 41.Traktman P, Kelvin M, Pacheco S. Molecular genetic analysis of vaccinia virus DNA polymerase mutants. J Virol. 1989;63:841–846. doi: 10.1128/jvi.63.2.841-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traktman P, Liu K, Rollins R A, Jesty S A, Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]