Abstract
More than 15% of human cancers have a viral etiology. In benign lesions induced by the small DNA tumor viruses, viral genomes are typically maintained extrachromosomally. Malignant progression is often associated with viral integration into host cell chromatin. To study the role of viral integration in tumorigenesis, we analyzed the positions of integrated viral genomes in tumors and tumor cell lines induced by the small oncogenic viruses, including the high-risk human papillomaviruses, hepatitis B virus, simian virus 40, and human T-cell leukemia virus type 1. We show that viral integrations in tumor cells lie near cellular sequences identified as nuclear matrix attachment regions (MARs), while integrations in nonneoplastic cells show no significant correlation with these regions. In mammalian cells, the nuclear matrix functions in gene expression and DNA replication. MARs play varied but poorly understood roles in eukaryotic gene expression. Our results suggest that integrated tumor virus genomes are subject to MAR-mediated transcriptional regulation, providing insight into mechanisms of viral carcinogenesis. Furthermore, the viral oncoproteins serve as invaluable tools for the study of mechanisms controlling cellular growth. Similarly, our demonstration that integrated viral genomes may be subject to MAR-mediated transcriptional effects should facilitate elucidation of fundamental mechanisms regulating eukaryotic gene expression.
Tumor virus infection represents an early and necessary event in the genesis of a variety of human cancers. However, viral infection alone is insufficient to produce a fully malignant phenotype. Progression to malignancy has been associated with the integration of viral DNA into the host genome (19, 56), but the mechanisms driving this association remain unclear. To investigate the hypothesis that malignant progression is influenced by the position of viral integration in host chromatin, we analyzed integrated oncogenic virus genomes in human tumors. The “high-risk” human papillomaviruses, HPV type 16 (HPV16) and HPV type 18 (HPV18) and hepatitis B virus (HBV) are the etiologic agents of human cervical and hepatocellular carcinoma, respectively, and as such contribute significantly to the worldwide cancer burden. Human T-cell leukemia virus type 1 (HTLV-1), a retrovirus, is associated with both adult T-cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), a nonneoplastic condition. Simian virus 40 (SV40) has served as a model for the study of basic mechanisms of viral carcinogenesis. However, the association of this tumor virus with human cancers is controversial. Preliminary reports link SV40 to mesothelioma, osteosarcoma, and brain tumors (7).
In mammalian cells, the nuclear matrix functions in gene expression and DNA replication (14). Nuclear matrix attachment regions (MARs) are DNA sequences that associate with the nuclear matrix or scaffold (12). MARs organize the eukaryotic genome into topologically independent transcriptional domains (38), augment the activity of nearby enhancers (20–22, 28), and can protect DNA transgenes from cellular silencing effects through local modifications of chromatin structure (30, 49). The association of MARs with eukaryotic transcriptional regulation suggests that these sequence elements may influence the expression of integrated tumor virus genomes.
To study the role of viral integration in tumorigenesis in vivo, we initially focused on tumors induced by HPV18. In HPV16-associated neoplasia, viral DNA may persist in mixed episomal and integrated forms (15, 33, 37, 50). In contrast, HPV18 genomes are reported to be exclusively integrated in HPV18-related malignancies (15, 37, 50). Tumors induced by this virus therefore represent an appropriate model system in which to study the role of integration in malignant progression. We developed an Alu-PCR technique to isolate cellular sequences flanking HPV18 integrants in cervical carcinoma.
To extend our analysis, we obtained cellular sequences flanking HPV16, HBV, SV40, and HTLV-1 integrations in tumor cells and cell lines from the public sequence databases. We determined the genomic position of each viral integration by BLAST similarity search (1) of the human genome and then defined the matrix association potential (MAP) of the integration site and surrounding 100 kb, using a mathematical algorithm that statistically weights various sequence motifs associated with cellular MARs, including ori signals, AT and TG richness, kinked and curved DNA, and topoisomerase II binding sites (48). We verified the algorithm's ability to accurately predict the positions of MARs in DNA sequences in separate experiments. We identified high-scoring regions (HSRs), DNA regions demonstrating a significant potential for attachment to the nuclear matrix, and determined the distance in kilobase pairs, D, from the viral integration point to the closest HSR. Dup and Ddown represent the distances from the upstream or downstream viral insertion point, IPup or IPdown, to the closest HSR upstream or downstream, respectively.
We calculated the corresponding mean distances to the closest HSR, dup and ddown, for viral integrations in both transformed and nonneoplastic cells. In some instances, viral integrations in transformed cells disrupted genes involved in the control of cellular growth, conferring a proliferative advantage to the host cell irrespective of viral gene expression; independent means were calculated for these integrations and for those that occurred outside known genes. The statistical significance of the observed mean distances was determined using Monte Carlo simulations to test the hypothesis that viral integration occurred randomly with respect to HSRs. We show that tumor virus integrations in transformed cells and cell lines are adjacent to cellular sequences identified as MARs. Viral integrations in nonneoplastic cells, and in tumor cells in which the integration disrupted genes involved in cellular growth control, show no association with these regions. MAR-mediated effects on the expression of integrated viral oncogenes might confer a proliferative advantage to a cell and its progeny, promoting tumor development.
MATERIALS AND METHODS
Tissue preparation and HPV detection.
Cervical tumors were obtained as part of a related project using protocols consistent with National Institutes of Health human studies guidelines. DNA was extracted by standard methods (44). HPV18-positive tumors were identified as previously described (2).
Isolation of HPV18 flanking sequences.
PCR amplification was performed with a sense or antisense biotinylated primer specific for HPV18-E6/E7, together with a sense or antisense primer complementary to the most highly conserved residues of the Alu repeat subfamilies Sb, Sb-1, Sb-2, Sc, Sp, Sq, Sx, and Alu-J (27). 3′ mismatches were incorporated to prevent amplification of 7SL RNA sequences. After purification of the PCR products over streptavidin-coated beads, a second PCR with an internal HPV primer was performed. An Alu-HPV18 hybrid plasmid was constructed from Alu consensus sequence pPD39 (4) and plasmid pHPV18 for use as a positive control. Normal genomic DNA and DNA extracted from an HPV16-positive cervical carcinoma served as negative controls. Cellular flanking sequences were reproducibly amplified in separate experiments. PCR conditions were as follows: 30 ng of tumor DNA or 30 pg of hybrid plasmid was amplified in a 50-μl PCR mixture containing 1× Expand buffer (Roche, Indianapolis, Ind.), 2.6 U of Expand High Fidelity enzyme (Roche), 2.0 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech, Piscataway, N.J.), and a 25 nM concentration of either R18Bio or F18Bio with either AluF or AluR. Thermocycling parameters were as follows: 3 min at 95°C; 30 s at 95°C, 15 s at 61°C, and 75 s at 72°C for a total of 34 cycles and a final 10 min at 72°C. The amplified biotinylated fragments were purified on Dynabeads-M280 Streptavidin (Dynal, Oslo, Norway). One microliter of the purified products was amplified in a second-round PCR with primers F18Int or R18Int and AluF or AluR. Second-round PCR conditions were identical to the first, but the cycles were limited to 25. A 5-μl volume of each second-round PCR product was electrophoresed in 1% agarose, transferred to a positively charged nylon membrane, and hybridized with digoxigenin-11-dideoxy-UTP-labeled (Roche) HPV18-specific oligonucleotide probe 18E6-3. Positive fragments were cloned into pCR 2.1-Topo vector (Invitrogen, Carlsbad, Calif.). Automated sequencing was performed using BigDye Terminator cycle sequencing (ABI Prism, Foster City, Calif.). DNA sequence analysis was performed with Wisconsin Package Version 10.1 (Genetics Computer Group, Madison, Wis.). PCR primer and probe sequences follow: R18Bio, 5′-Bio-TACTTGTGTTTCTCTGCGTC-3′; F18Bio, 5′-Bio-CCCTACAAGCTACCTGATCTG-3′; AluF, 5′-CCTGTAATCCCAGCACTTTG-3′; AluR, 5′-CCCAAAGTGCTGGGATTAC-3′; F18Int, 5′-ACAGTATACCCCATGCTGC-3′; R18Int, 5′-TTCAACGGTTTCTGGCAC-3′; and 18E6-3, 5′-CAGACTCTGTGTATGGAGACACATTGGAAAAACTAACTAA-3′.
Other viral flanking sequences.
Cellular sequences flanking single viral integrants, isolated by variety of methods from tumors, transformed cell lines, and nonneoplastic T cells, were obtained from the scientific literature and public sequence databases. The genomic position of each viral integration was determined by BLAST similarity search (version 2.1.2) of cellular flanking sequence against the nucleotide and high-throughput genome sequence databases. The maximum E value permissible for genomic localization of a viral integration was 10−25; the majority demonstrated perfect or near-perfect identity. No insertion point could be defined for HBV flanking sequences AJ222811 or AJ000498 or the SV40 integration in FRA7H (AF017104); these sequences were excluded from the analysis.
MAP analysis and proximity to viral insertions.
MAP was defined for the 100 kb of DNA sequence surrounding the integration site, when available, using Mar-Finder (http://www.futuresoft.org). MAP analysis was performed in 50-kb increments utilizing the default detection and clipping parameters (clip = 0.5 ρmax, where ρ = MAP). When viral integration occurred antiparallel to unfinished genomic sequence, MAP analysis was performed on the reverse complement of that sequence to facilitate statistical analysis. The minimum analyzed interval was 5 kb. The upstream and downstream IPs, IPup and IPdown, were the cellular nucleotides adjacent to the 5′ and 3′ ends, respectively, of the viral coding strand, where “coding strand” refers to the 5′-to-3′ nucleotide sequence indexed in the public sequence databases. When an integration occurred in oriented cellular sequence and the viral and cellular coding strands were antiparallel, the IPs and distances D were assigned with respect to the cellular “coding strand.” For each viral insertion, Dup and Ddown were calculated as the distance in kilobase pairs from IPup or IPdown to the closest HSR upstream or downstream, respectively, taking the endpoints of each analyzed interval as HSRs. When an IP occurred within an HSR, D = 0. Mean distances dup and ddown were calculated for the subsets of viral integrations listed (see Table 2).
TABLE 2.
Mean distances from viral IP to closest HSR (kb)a
| Subset | Virus | dup | ddown |
|---|---|---|---|
| a | HPV | 0.57 (n = 7, P < 0.005) | 7.3 (n = 10) |
| HBV | 0.00b(n = 3, P < 0.002) | 0.00 (n = 3, P < 0.002) | |
| SV40 | 10.2 (n = 2) | 0.66 (n = 3, P < 0.01) | |
| HTLV-1 in ATL | 0.09c(n = 3) | ||
| b | HPV + HBV in growth control genes | 11.6 (n = 7) | 5.2 (n = 8) |
| c | HTLV-1 in HAM/TSP | 5.9 (n = 2) | 12.8 (n = 8) |
dup and ddown give the mean distance from the viral IP to the closest HSR for the following subsets of viral integrations: a, integrations in tumors and transformed cell lines; b, integrations in tumors and cell lines that disrupted genes involved in the control of cellular growth; and c, integrations in nonneoplastic cells. P values are given when observed means are significantly different from values obtained under the assumption of random integration.
When an IP occurred within an HSR, D = 0.
Upstream and downstream.
Validation of MAP algorithm.
The ability of the MAP algorithm to accurately predict DNA sequences that attach to the nuclear matrix was verified by analysis of genomic regions containing MARs characterized in vivo or in vitro. When the default detection parameters were used, there was excellent correlation between HSRs and known MARs in the serpin gene cluster (41), the apolipoprotein B gene (32), the immunoglobulin κ enhancer (54), the immunoglobulin heavy-chain μ enhancer (12), and intron 13 of topoisomerase I (43). Recognition of the MAR in intron 2 of topoisomerase I (43) and the corticosteroid-binding globulin gene (41) intronic MAR improved when the clip was < 0.4 ρmax, where ρ = MAP.
Statistical methods.
We performed Monte Carlo simulations to test the hypothesis that viral integration occurred randomly with respect to HSRs. For each subset of viral integrations, we chose insertion points at random from the genomic intervals in which the in vivo integrations occurred and calculated the mean distance dup or ddown to the nearest HSR for each of 10,000 independent simulations. P gives the fraction of simulated mean distances less than or equal to the observed mean; its value thus estimates the probability of the observed mean under the assumption that viral integration occurs randomly with respect to HSRs.
Database deposition.
Cellular sequences flanking integrated HPV18 in cervical carcinoma are available under GenBank accession numbers AF339133, AF339134, AF339135, AF339136, AF339137, AF339138, and AF339139.
RESULTS
We isolated cellular sequences flanking integrated HPV18 genomes from five of nine cervical tumors studied, including squamous-cell carcinoma, adenocarcinoma, and small-cell cervical cancers (Fig. 1). Sequences flanking HPV16, HBV, SV40, and HTLV-1 integrations, isolated from both neoplastic and nonneoplastic viral pathologies, were obtained from the scientific literature and public sequence databases, for a total of 54 viral flanking sequences. After BLAST localization, we determined the MAP of the genomic region surrounding each viral integration site. We identified HSRs, DNA regions demonstrating a significant potential for attachment to the nuclear matrix, and determined the distance, D, in kilobase pairs from the viral integration point to the closest HSR (Fig. 2). Dup and Ddown represent the distances from the upstream or downstream viral insertion point, IPup or IPdown, to the closest HSR upstream or downstream, respectively.
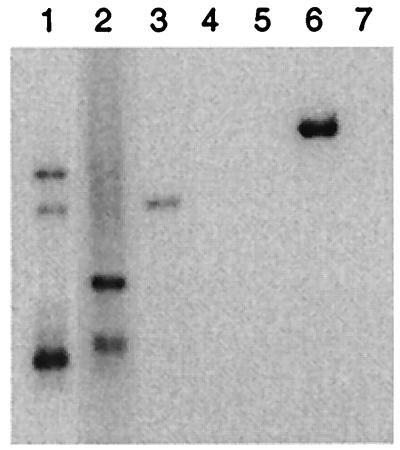
FIG. 1.
Southern hybridization of cellular sequences flanking integrated HPV18 in cervical carcinoma. Alu-PCR products were hybridized with an HPV18-specific oligonucleotide probe. Lanes 1 and 2, HPV18(+) cervical squamous-cell carcinoma; lane 3, HPV18(+) cervical adenocarcinoma; lane 4, HPV16(+) cervical squamous-cell carcinoma; lane 5, normal human genomic DNA; lane 6, Alu-HPV18 hybrid plasmid (2.3 kb); and lane 7, negative control. Tumors 1 and 2 harbored multiple HPV18 integrations.
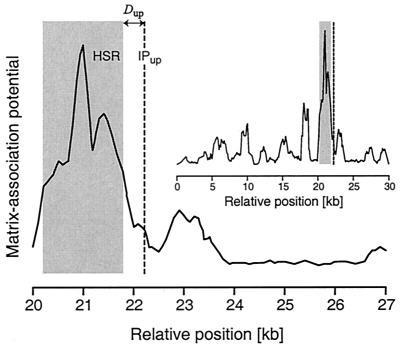
FIG. 2.
MAP of the genomic region surrounding an HPV18 integration in a cervical carcinoma. The HSR, defined using a mathematical algorithm that weights sequence motifs associated with cellular MARs, demonstrates a significant potential for attachment to the nuclear matrix. Dup is the distance from the IPup to the HSR. In this instance, the position of the downstream viral IP is unknown. The inset shows the full 30-kb analyzed interval, which contains a single HSR. In our study, MAP and HSR boundaries were defined using the MAR prediction algorithm's default detection and clipping parameters. The MAP of the genomic region illustrated here is shown with unclipped peaks for clarity.
Accession numbers of viral flanking sequences, highest-scoring BLAST homologies, and corresponding distances D to the closest HSR are given in Table 1. The mean distance between successive HSRs in the analyzed intervals surrounding the viral integration sites was 10.2 kb; no significant difference was observed between the mean distances in transformed cells (9.7 kb) and nonneoplastic cells (11.9 kb). The observed mean distances to the closest HSR, dup and ddown, for viral integrations in transformed and nonneoplastic cells are given in Table 2.
TABLE 1.
Genomic localizations of integrated tumor virus genomes in tumor cells, tumor cell lines, and nonneoplastic cells and the distance D in base pairs from viral IP to closest HSRa
| Cell type or lines | Virus | Source | Accession no. or reference | Localization | Dup | Ddown |
|---|---|---|---|---|---|---|
| Tumor cells | HPV18 | CxCa | AF339133 | AC008870 | 690 | |
| HPV18 | CxCa | AF339134 | AC008072 | 168 | ||
| HPV18 | CxCa | AF339135 | AC040933 | 0 | ||
| HPV18 | CxCa | AF339136 | AC013415 | 417 | ||
| HPV16 | CxCa | M33610 | AC007982 | 0 | ||
| HPV16 | CxCa | L43000 | AC068359 | 888 | ||
| HPV18 | CxCa | AF339137 | AC027364 | 554 | ||
| HPV18 | CxCa | AF339138 | AC087590 | 2,612 | ||
| HPV16 | CxCa | M33611 | AC007982 | 3,738 | ||
| HPV16 | CxCa | M33613 | AF152363 | 6,215 | ||
| HPV16 | CxCa | M33612 | AC007282 | 12,314 | ||
| HPV16 | CxCa | M33616 | AL133377 | 15,459 | ||
| HPV16 | CxCa | L42999 | AL033527 | 12,794 | ||
| HPV16 | CxCa | M33614 | U66722 | 12,445 | ||
| HPV16 | CxCa | 51 | AC026075 | 3,940 | ||
| HPV16 | CxCa | M59869 | AL136528b | 32,221 | ||
| HPV16 | CxCa | M59869 | AL136528b | 4,566 | ||
| HPV16 | CxCa | 13 | AC004518 | 10,637 | ||
| HPV16 | CxCa | 13 | AC004518 | 24,766 | ||
| HPV16 | CxCa | 51 | AC005325 | 10,746 | ||
| HPV18 | CxCa | AF339139 | AB025285 | 4,007 | ||
| HBV | HCC | AJ000499 | AC022525 | 0 | ||
| HBV | HCC | M27097c | AL049696 | 0 | ||
| HBV | HCC | M16635 | AC026982 | 0 | ||
| HBV | HCC | AJ000499 | AC022525 | 0 | ||
| HBV | HCC | M27097c | AL049696 | 0 | ||
| HBV | HCC | M16635 | AC026982 | 0 | ||
| HBV | HCC | K02716 | AC011323 | 347 | ||
| HBV | HCC | K02717 | AC011323 | 2,940 | ||
| HBV | HCC | S76119 | AF128893 | 1,642 | ||
| HBV | HCC | 52 | AC079341 | 38 | ||
| HBV | HCC | 52 | AC079341 | 356 | ||
| Tumor cell line | HPV16 | SiHa | AF001600 | AL158191 | 1,849 | |
| HPV16 | SiHa | AF001599 | AL356416 | 2,870 | ||
| HPV45 | IC-4 | AJ242956 | AC010145 | 16,071 | ||
| HPV45 | IC-4 | AJ242956 | AC010145 | 2,949 | ||
| HBV | HUH2-2 | M11225 | AC005203 | 10,931 | ||
| HBV | HUH2-2 | M11225 | AC005203 | 258 | ||
| SV40 | VA13 | D00846 | AC012178 | 20,454 | ||
| SV40 | VA13 | D00847 | AC012178 | 1,291 | ||
| SV40 | AG34 | U08313 | AC009075 | 0 | ||
| SV40 | AG34 | U08313 | AC009075 | 440 | ||
| SV40/Ad5 | H13.1 | X71401 | AL049569 | 248 | ||
| HTLV-1 | MT-4 | S80210 | AC036142 | 266 | ||
| HTLV-1 | MT-4 | S80215 | AL136981 | 0 | ||
| HTLV-1 | MT-4 | S80213 | AC021942 | 0 | ||
| HTLV-1 | MT-4 | S80212 | AC007222 | 26,450 | ||
| Nonneoplastic cells | HTLV-1 | HAM/TSP | AY003898 | AL163209 | 4,862 | |
| HTLV-1 | HAM/TSP | AY003897 | AC016195 | 5,193 | ||
| HTLV-1 | HAM/TSP | AY003891 | AC011753 | 2,079 | ||
| HTLV-1 | HAM/TSP | AY003890 | AL023806 | 0 | ||
| HTLV-1 | HAM/TSP | AY003892 | AC016941 | 56,400 | ||
| HTLV-1 | HAM/TSP | AY003888 | AL512422 | 8,995 | ||
| HTLV-1 | HAM/TSP | AY003886 | AL163204 | 6,965 | ||
| HTLV-1 | HAM/TSP | AY003894 | AC012559 | 13,317 | ||
| HTLV-1 | HAM/TSP | AY003893 | AC010552 | 2,182 | ||
| HTLV-1 | HAM/TSP | AY003899 | AL132712 | 13,933 |
CxCa, cervical carcinoma; HCC, hepatocellular carcinoma. When an IP occurred within an HSR, D = 0.
E > 10−25, but both flanks localized to the same clone.
Clip = 0.4 (see Materials and Methods).
Viral integrations in transformed cells.
Six cellular sequences upstream of high-risk HPV integrations, isolated from five independent cervical tumors, were analyzed. Although the mean distance between successive HSRs in the genomic intervals in which the HPV integrations occurred was almost 10 kb, six of six in vivo integrations localized within approximately 1/2 kb of an upstream HSR. The mean distance to an HSR upstream, dup, in HPV-associated tumors, together with that observed in the cervical carcinoma cell line SiHa, which harbors a single HPV16 integration (34), was 0.57 kb (n = 7, P < 0.005). HPV downstream flanks, however, showed no significant correlation with HSRs (ddown = 7.3 kb, n = 10). The difference between the HPV upstream and downstream means, dup and ddown, was statistically significant (P < 0.004).
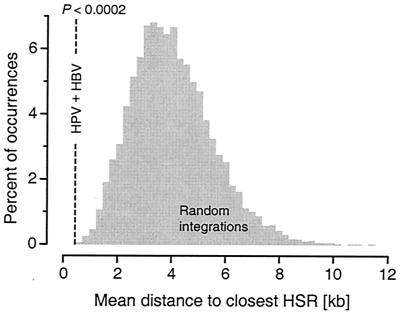
Similarly, HBV integrations in hepatocellular carcinoma cells occurred within HSRs (dup = 0.0 kb, n = 3, P < 0.002 and ddown = 0.0 kb, n = 3, P < 0.002). Taken together, the mean distance dup to the closest HSR for HPV and HBV integrations in tumor cells was 0.401 kb (n = 10, P < 0.0002). Random integrations simulated in those same genomic intervals occurred an average of 4 kb from an HSR (Fig. 3).
FIG. 3.
Mean distance to an HSR, dup, for high-risk HPV and HBV integrations in tumor cells and the distribution of means in Monte Carlo simulations. The distribution was computed under the assumption that viral integration occurs randomly with respect to HSRs. P gives the fraction of simulated mean distances less than or equal to the observed mean; here, P < 0.0002.
Although our sample size for SV40 integrations in transformed cell lines was small, the results were particularly intriguing. Here, SV40 downstream, but not upstream, flanking sequences were adjacent HSRs (ddown = 0.66 kb, n = 3, P < 0.01 and dup = 10.2 kb, n = 2). Interestingly, the SV40 oncogenes, large and small T antigens, are transcribed from the minus strand of this bidirectionally transcribed virus. Our results suggest that the effect of a nearby MAR on the expression of an integrated viral genome is directional and is manifest when a MAR is 5′ on the viral coding strand.
Viral integrations that disrupted growth control genes.
Some HPV and HBV integrations disrupted genes involved in the control of cellular growth, including TP73, ERBB2, MYCN, TERT, RARB, and CCNA2 (10, 16, 34, 39, 45, 52). The integrations in ERBB2, MYCN, and TERT occurred in intronic, untranslated exonic, and regulatory cellular sequences, respectively. Increased expression of the virally disrupted gene was demonstrated for MYCN, RARB, and CCNA2 (16, 45, 53). The insertional activation of host genes, by either direct mutation or transcriptional activation, is reminiscent of cellular transformation induced by the nonacutely transforming retroviruses; here, viral integration promotes tumorigenesis irrespective of viral gene expression. Interestingly, the positions of HPV and HBV integrations in cellular growth control genes showed no association with HSRs (dup = 11.6 kb, n = 7 and ddown = 5.2 kb, n = 8).
HTLV-1 proviral integrations in nonneoplastic and transformed cells.
Finally, HTLV-1 integrations in nonneoplastic T cells showed no correlation with HSRs (dup = 5.9 kb, n = 2 and ddown = 12.8 kb, n = 8). Several studies of HTLV-1 integration in adult T-cell leukemia have been published, but few flanking sequences have been deposited in the public databases. Our analysis of HTLV-1 insertions in malignant cells was therefore limited to the ATL cell line MT-4. MT-4 harbors multiple HTLV-1 integrations and expresses high levels of HTLV-1 Tax protein (26). Although HTLV-1 proviral insertions in nonneoplastic cells showed no association with HSRs, three of the four integrations isolated from this transformed cell line (8) were immediately adjacent to HSRs (d = 0.09 kb).
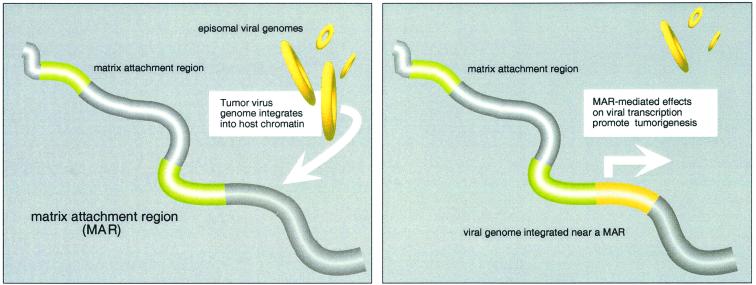
Thus, tumor virus integrations in transformed cells and cell lines were adjacent to cellular sequences identified as MARs (Fig. 4). Viral integrations in nonneoplastic cells, and in tumor cells in which the integration disrupted genes involved in cellular growth control, showed no association with these regions.
FIG. 4.
Integrated tumor virus genomes in transformed cells and cell lines are adjacent to MARs. Viral integrations in nonneoplastic cells, and in tumor cells in which the integration disrupted genes involved in cellular growth control, show no association with these regions. MAR-mediated effects on the expression of integrated viral oncogenes might confer a proliferative advantage to a cell and its progeny, promoting tumor development.
DISCUSSION
Here we demonstrate a significant correlation between the positions of diverse integrated tumor virus genomes and sequences identified as MARs. Where these viral integrations in tumor cells do not disrupt genes involved in the control of cellular growth, they lie near MARs, thereby providing strong evidence for an association with tumorigenesis. MAR-mediated effects on the expression of integrated viral oncogenes might confer a proliferative advantage to a cell and its progeny, promoting tumor development.
Due to their limited genetic content, the small tumor viruses depend on the host cell machinery for replication. The viral oncoproteins HPV16 and HPV18 E6/E7, SV40 T antigen, and HTLV-1 Tax target regulators of the cell cycle, including p53, Rb, and CBP/p300, to promote cellular proliferation and a stable environment for viral replication (7, 25). Because they functionally inactivate cellular tumor suppressors, the viral oncoproteins play an important role in the development of human cancers. However, only a small percentage of cells infected with oncogenic viruses progress to malignancy. In cervical carcinogenesis, malignant progression correlates with papillomaviral integration into host chromatin. Hepatocarcinogenesis, though incompletely understood, likely involves a complex interplay between chronic liver inflammation and the effects of HBV gene expression on hepatocellular growth control and resistance to apoptosis. Again, the integration of HBV DNA into host chromatin represents an important step in the development of hepatocellular carcinoma.
In HPV-related cancers, the integration of viral DNA into the human genome is believed to lead to transcriptional deregulation of the viral transforming genes E6 and E7. Integration frequently disrupts the viral E2 open reading frame (3, 46); loss of the E2 gene product relieves its repression of the HPV E6/E7 promoter, increasing expression of the viral oncoproteins (47). In the absence of integration, mutational inactivation of E2 has also been demonstrated to increase the transforming potential of HPV16 in vitro (42). However, no parallel mechanism explaining the role of integration in the development of cancers associated with other oncogenic viruses has been elucidated, suggesting that a more unifying mechanism explaining the role of integration in cellular transformation exists.
If integrated tumor virus genomes are subject to position effects of the type noted for endogenous (23) and therapeutic (36) transgenes, expression of integrated viral sequences might be maintained or increased in some cellular loci. Cells harboring such integrants would manifest higher levels of the viral oncoproteins, exhibit increased proliferative capacity, and undergo clonal expansion. Interestingly, variation in HPV16 E7 protein levels has been noted among subclonal populations of cervical epithelial cells harboring exclusively integrated HPV16 DNA (29). Despite disruption of the E2 open reading frame in all populations, significant variation in cellular E7 protein levels was demonstrated and in some cases equaled that seen in Caski, a cervical cancer cell line harboring an estimated 60 to 600 copies of HPV16 (34). The observed variation in E7 protein levels in independent populations of epithelial cells harboring integrated HPV16 might reflect position-mediated effects on the expression of the integrated viral oncogenes.
Some viral integrations disrupt genes involved in the control of cellular growth, indirectly conferring a proliferative advantage to the host cell. We show that, in the absence of such alterations, integrated DNA and RNA tumor viruses in tumors and tumor cell lines lie near cellular sequences identified as MARs. The proximity of viral integrants to cellular MARs could simply reflect enhanced recombigenicity in these genomic loci. However, the random distribution of viral integrants in nonneoplastic cells and in tumor cells with disruptions in growth control genes argues against an absolute mechanistic requirement for integrations within or near MARs.
A previous report located a single woodchuck hepatitis virus integration in a woodchuck hepatocellular carcinoma near a MAR (18). However, the ubiquity of MARs in mammalian genomes implies that analysis of a single viral integration site cannot establish an association between integrated viral genomes and MARs, let alone with tumorigenesis. Associations of viral integration sites with local AT richness (11) and polypurine or polypyrimidine (9) or topoisomerase I motifs (31) have also been reported. Although no correlation of topoisomerase I motifs with MARs has been demonstrated, AT richness and polypurine and polypyrimidine tracts are fundamental sequence characteristics of MARs; our study thus confirms and extends previous observations. We propose that the proximity of integrated viral genomes to cellular MARs demonstrated here represents a unifying mechanism underlying the role of viral integration in the development of diverse human cancers.
In this study, MARs were identified using a computational algorithm that statistically weights sequence motifs characteristic of MARs identified by traditional biochemical methods. These methods rely upon nuclear isolation, followed by separation of nuclear matrix and loop fractions and hybridization with specific probes (in vivo assay), or alternatively by reassociation of putative MARs to the purified nuclear matrix (in vitro assay). Because these assays are subject to experimental variability (17), many studies employ both in vivo and in vitro biochemical approaches to more definitively characterize putative MARs in DNA sequences. In our laboratory, we observed a strong correlation between the positions of MARs identified in vivo and/or in vitro and those predicted in silico. The concordance between the boundaries of experimentally determined MARs and HSRs typically averaged less than 100 or 200 bp over a 50-kb analyzed interval. Further, the MAR-Finder algorithm has been validated in several independent studies (24, 40). By providing data about the relative frequency of specific sequence motifs within each HSR, the use of a computational approach for the characterization of MARs in virally transformed cells may help to identify subsets or clusters of sequence motifs that typify those classes of MARs playing roles in the modulation of cellular gene expression, providing the beginnings of a resolution of these diverse regulatory elements at the sequence level.
Our data are consistent with the hypothesis that tumor virus genomes are differentially expressed when integrated in close proximity to MARs. Specific mechanisms for the transcriptional activation of viral oncogenes in host chromatin remain to be elucidated. Functionally diverse, MARs define chromatin domains, augment the activity of flanking enhancers, and protect transgenes from position effects. These sequence elements lack a consensus motif; MAR-binding proteins typically recognize structural features of these AT-rich DNA elements, which display enhanced flexibility and a propensity for base-unpairing and DNA bending and unwinding (5).
HMGA (previously known as HMG-I/Y) is a MAR-binding, nonhistone chromatin protein that displays a marked affinity for the bent DNA and AT tracts characteristic of MARs (5). Remarkably, the HPV18 E6/E7 promoter was recently reported to be under the control of an enhanceosome containing HMGA and the JunB/Fra-2 heterodimer (6). Enhanceosomes are cooperatively assembled nucleosome complexes that incorporate DNA-bending architectural proteins such as HMGA with sequence-specific transcription factors (35). Binding of the architectural protein to a specific DNA structural motif facilitates the recruitment of transcription factors and the formation of a stable, three-dimensional enhanceosome complex; the assembled enhanceosome induces a dramatic increase in the rate at which preiniatiation complexes assemble at a given promoter (55).
Thus, the integration of an HPV18 genome adjacent to a MAR might facilitate the assembly of the HMGA-containing enhanceosome on the viral promoter, driving the transcription of the integrated HPV oncogenes. Experiments in our laboratory are currently under way to investigate the influence of MARs on the activity of the HPV18 promoter and specifically on the assembly of HMGA-containing enhanceosomes. Other integrated DNA and RNA tumor viruses may be activated through similar MAR-mediated effects on transcriptional initiation.
ACKNOWLEDGMENTS
We acknowledge the contributions of our colleagues in preparation of the manuscript. We thank Connie P. Matthews and Jenn Ihle Koop for tissue acquisition; George Bonnet, Marija Helt, Andy McShea, Mike Barrett, and Brooks Shera for helpful discussions; and Denise Galloway, Steve Schwartz, and Keith Fournier for insightful comments on the manuscript. pHPV18 was a gift of H. zur Hausen.
This work was supported by grants NCI CA42792 (J.K.M.) and NIDCD DC03687 (C.A.S.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Shera K, Ihle J, Billman L, Goff B, Greer B, Tamimi H, McDougall J, Klingelhutz A. Telomerase activation in cervical cancer. Am J Pathol. 1997;151:25–31. [PMC free article] [PubMed] [Google Scholar]
- 3.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batzer M A, Alegria-Hartman M, Deininger P L. A consensus Alu repeat probe for physical mapping. Genet Anal Tech Appl. 1994;11:34–38. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 5.Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit Rev Eukaryot Gene Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- 6.Bouallaga I, Massicard S, Yaniv M, Thierry F. An enhanceosome containing the Jun B/Fra-2 heterodimer and the HMG-I(Y) architectural protein controls. HPV 18 transcription. EMBO Rep. 2000;1:422–427. doi: 10.1093/embo-reports/kvd091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butel J S. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 8.Cavrois M, Wain-Hobson S, Wattel E. Stochastic events in the amplification of HTLV-I integration sites by linker-mediated PCR. Res Virol. 1995;146:179–184. doi: 10.1016/0923-2516(96)80578-4. [DOI] [PubMed] [Google Scholar]
- 9.Choo K B, Chen C M, Han C P, Cheng W T, Au L C. Molecular analysis of cellular loci disrupted by papillomavirus 16 integration in cervical cancer: frequent viral integration in topologically destabilized and transcriptionally active chromosomal regions. J Med Virol. 1996;49:15–22. doi: 10.1002/(SICI)1096-9071(199605)49:1<15::AID-JMV3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Choo K B, Lee H H, Liew L N, Chong K Y, Chou H F. Analysis of the unoccupied site of an integrated human papillomavirus 16 sequence in a cervical carcinoma. Virology. 1990;178:621–625. doi: 10.1016/0042-6822(90)90366-y. [DOI] [PubMed] [Google Scholar]
- 11.Chou K S, Okayama A, Su I J, Lee T H, Essex M. Preferred nucleotide sequence at the integration target site of human T-cell leukemia virus type I from patients with adult T-cell leukemia. Int J Cancer. 1996;65:20–24. doi: 10.1002/(SICI)1097-0215(19960103)65:1<20::AID-IJC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 13.Cone R W, Minson A C, Smith M R, McDougall J K. Conservation of HPV-16 E6/E7 ORF sequences in a cervical carcinoma. J Med Virol. 1992;37:99–107. doi: 10.1002/jmv.1890370205. [DOI] [PubMed] [Google Scholar]
- 14.Cook P R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 15.Cullen A P, Reid R, Campion M, Lorincz A T. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dejean A, Bougueleret L, Grzeschik K H, Tiollais P. Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature. 1986;322:70–72. doi: 10.1038/322070a0. [DOI] [PubMed] [Google Scholar]
- 17.Donev R M. The type of DNA attachment sites recovered from nuclear matrix depends on isolation procedure used. Mol Cell Biochem. 2000;214:103–110. doi: 10.1023/a:1007159421204. [DOI] [PubMed] [Google Scholar]
- 18.D'Ugo E, Bruni R, Argentini C, Giuseppetti R, Rapicetta M. Identification of scaffold/matrix attachment region in recurrent site of woodchuck hepatitis virus integration. DNA Cell Biol. 1998;17:519–527. doi: 10.1089/dna.1998.17.519. [DOI] [PubMed] [Google Scholar]
- 19.Feitelson M A. Hepatitis B virus in hepatocarcinogenesis. J Cell Physiol. 1999;181:188–202. doi: 10.1002/(SICI)1097-4652(199911)181:2<188::AID-JCP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez L A, Winkler M, Forrester W, Jenuwein T, Grosschedl R. Nuclear matrix attachment regions confer long-range function upon the immunoglobulin mu enhancer. Cold Spring Harbor Symp Quant Biol. 1998;63:515–524. doi: 10.1101/sqb.1998.63.515. [DOI] [PubMed] [Google Scholar]
- 21.Fernández L A, Winkler M, Grosschedl R. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol Cell Biol. 2001;21:196–208. doi: 10.1128/MCB.21.1.196-208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrester W C, Fernandez L A, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gdula D A, Gerasimova T I, Corces V G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glazko G V, Rogozin I B, Glazkov M V. Comparative study and prediction of DNA fragments associated with various elements of the nuclear matrix. Biochim Biophys Acta. 2001;1517:351–364. doi: 10.1016/s0167-4781(00)00297-9. [DOI] [PubMed] [Google Scholar]
- 25.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 26.Jeang K-T, Derse D, Matocha M, Sharma O. Expression status of Tax protein in human T-cell leukemia virus type 1-transformed MT4 cells: recall of MT4 cells distributed by the NIH AIDS Research and Reference Reagent Program. J Virol. 1997;71:6277–6278. doi: 10.1128/jvi.71.9.6277-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jendraschak E, Kaminski W E. Isolation of human promoter regions by Alu repeat consensus-based polymerase chain reaction. Genomics. 1998;50:53–60. doi: 10.1006/geno.1998.5290. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 29.Jeon S, Allen-Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalos M, Fournier R E. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol Cell Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konopka A K. Compilation of DNA strand exchange sites for non-homologous recombination in somatic cells. Nucleic Acids Res. 1988;16:1739–1758. doi: 10.1093/nar/16.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy-Wilson B, Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5′ and 3′ boundaries of the gene. J Biol Chem. 1989;264:21196–21204. [PubMed] [Google Scholar]
- 33.Matsukura T, Koi S, Sugase M. Both episomal and integrated forms of human papillomavirus type 16 are involved in invasive cervical cancers. Virology. 1989;172:63–72. doi: 10.1016/0042-6822(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 34.Meissner J D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 35.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 36.Neff T, Shotkoski F, Stamatoyannopoulos G. Stem cell gene therapy, position effects and chromatin insulators. Stem Cells. 1997;15:265–271. doi: 10.1002/stem.5530150834. [DOI] [PubMed] [Google Scholar]
- 37.Park J S, Hwang E S, Park S N, Ahn H K, Um S J, Kim C J, Kim S J, Namkoong S E. Physical status and expression of HPV genes in cervical cancers. Gynecol Oncol. 1997;65:121–129. doi: 10.1006/gyno.1996.4596. [DOI] [PubMed] [Google Scholar]
- 38.Paulson J R, Laemmli U K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 39.Quade K, Saldanha J, Thomas H, Monjardino J. Integration of hepatitis B virus DNA through a mutational hot spot within the cohesive region in a case of hepatocellular carcinoma. J Gen Virol. 1992;73:179–182. doi: 10.1099/0022-1317-73-1-179. [DOI] [PubMed] [Google Scholar]
- 40.Rogozin I B, Glazko G V, Glazkov M V. Computer prediction of sites associated with various elements of the nuclear matrix. Brief Bioinform. 2000;1:33–44. doi: 10.1093/bib/1.1.33. [DOI] [PubMed] [Google Scholar]
- 41.Rollini P, Namciu S J, Marsden M D, Fournier R E. Identification and characterization of nuclear matrix-attachment regions in the human serpin gene cluster at 14q32.1. Nucleic Acids Res. 1999;27:3779–3791. doi: 10.1093/nar/27.19.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romanczuk H, Howley P M. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romig H, Ruff J, Fackelmayer F O, Patil M S, Richter A. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur J Biochem. 1994;221:411–419. doi: 10.1111/j.1432-1033.1994.tb18753.x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sastre-Garau X, Favre M, Couturier J, Orth G. Distinct patterns of alteration of myc genes associated with integration of human papillomavirus type 16 or type 45 DNA in two genital tumours. J Gen Virol. 2000;81:1983–1993. doi: 10.1099/0022-1317-81-8-1983. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 47.Shah K, Howley P M. Papillomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2077–2109. [Google Scholar]
- 48.Singh G B, Kramer J A, Krawetz S A. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stief A, Winter D M, Stratling W H, Sippel A E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989;341:343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- 50.Stoler M H, Rhodes C R, Whitbeck A, Wolinsky S M, Chow L T, Broker T R. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992;23:117–128. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- 51.Thorland E C, Myers S L, Persing D H, Sarkar G, McGovern R M, Gostout B S, Smith D I. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60:5916–5921. [PubMed] [Google Scholar]
- 52.Wang J, Chenivesse X, Henglein B, Brechot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Brechot C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene. 1992;7:1653–1656. [PubMed] [Google Scholar]
- 54.Whitehurst C, Henney H R, Max E E, Schroeder H W, Jr, Stuber F, Siminovitch K A, Garrard W T. Nucleotide sequence of the intron of the germline human kappa immunoglobulin gene connecting the J and C regions reveals a matrix association region (MAR) next to the enhancer. Nucleic Acids Res. 1992;20:4929–4930. doi: 10.1093/nar/20.18.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yie J, Senger K, Thanos D. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc Natl Acad Sci USA. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]