Abstract
Background
Mycobacterium abscessus is an emerging pathogen causing severe pulmonary infections, particularly in individuals with underlying conditions, such as cystic fibrosis or chronic obstructive pulmonary disease. Macrolides, such as clarithromycin (CLR) or azithromycin (AZM), represent the cornerstone of antibiotherapy against the M. abscessus species. However, prolonged exposure to these macrolides can induce of Erm(41)-mediated resistance, limiting their spectrum of activity and leading to therapeutic failure. Therefore, inhibiting Erm(41) could thwart this resistance mechanism to maintain macrolide susceptibility, thus increasing the rate of treatment success. In our previous study, the Erm(41) methyltransferase was identified as a possible target enzyme of Cyclipostins and Cyclophostin compounds (CyC).
Methods
Herein, we exploited this feature to evaluate the in vitro activity of CLR and AZM in combination with different CyC via the checkerboard assay on macrolide-susceptible and induced macrolide-resistant M. abscessus strains selected in vitro following exposure CLR and AZM.
Results
Our results emphasize the use of the CyC to prevent/overcome Erm(41)‑induced resistance and to restore macrolide susceptibility.
Conclusion
This work should expand our therapeutic arsenal in the fight against a antibioticresistant mycobacterial species and could provide the opportunity to revisit the therapeutic regimen for combating M. abscessus pulmonary infections in patients, and particularly in erm(41)-positive strains.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12929-024-01091-w.
Keywords: Mycobacterium abscessus, Drug susceptibility, Macrolide, Resistance, Synergy testing, Erm(41)
Background
Mycobacterium abscessus is a rapid growing mycobacteria (RGM) causing pulmonary diseases in vulnerable individuals, such as cystic fibrosis (CF) or bronchiectasis patients [1–3]. These infections are particularly challenging to cure because M. abscessus is intrinsically resistant to most antibiotic classes and anti-tuberculosis drugs [4–8]. The current treatment consists in a multidrug regimen which involves an initial phase comprising a macrolide, usually clarithromycin (CLR) or azithromycin (AZM), associated with other antibiotics such amikacin (AMK), tigecycline, imipenem (IPM) or cefoxitin (CFX) for 3–12 weeks. This initial phase is followed by a continuation phase that comprises a macrolide, amikacin and one or several additional antibiotics (minocycline, clofazimine and moxifloxacin) depending on the severity of the infection, the tolerability of the regimen and the drug susceptibility profile of the strain [2, 9]. Macrolides are important in the context of polychemotherapy against NTMs. Despite this, the success rate of treatments using them against M. abscessus remains low (between 30 and 50%) [10, 11], particularly in the case of inducible macrolideresistant strains [12]. Macrolides target the large subunit of the bacterial ribosome and bind/occlude the nascent peptide exit tunnel, therefore inhibiting protein synthesis. However, prolonged exposure to macrolides, even at sub-inhibitory concentrations, can lead to the apparition of an inducible macrolide-resistant phenotype mediated by the Erm(41) methyltransferase [13–17]. This enzyme catalyzes the specific methylation of the adenine 2270 of the 23 S rRNA, which protects the ribosome from the macrolide activity. This mechanism can be observed in vitro, by exposing a susceptible strain to macrolides for up to 14 days instead of the classical 3–5 days [13, 15]. This intrinsic resistance mechanism occurs in M. abscessus subspecies M. abscessus as well as in M. abscessus subspecies M. bolletii, both expressing a functional Erm(41) enzyme, unlike M. abscessus subspecies M. massiliense which harbors a truncated version of the erm(41) gene [13, 18]. In this context, it is important to characterize the species within the abscessus complex, for example by using erm41 PCR, which allows both speciation within the complex and the determination of inducible resistance [9], hence its value as a complement to phenotypic tests [19, 20]. Thus, a yet unmet medical need would consist to develop new therapeutic approaches to counteract Erm(41)-induced resistance in M. abscessus [21, 22]. Similar strategies have been set up with the use of β-lactamase inhibitors to render M. abscessus strains susceptible to β-lactams [7, 23, 24] and highlight the importance and potential of molecules able to block drug resistance as attractive therapeutic adjuncts to reduce and/or circumvent resistance to antibiotics. Recently, the combination of rifabutin with CLR has been reported to suppress CLR resistance in erm(41)-expressing strains [24], whereby rifabutin acts as an inhibitor of erm(41) transcriptional induction, maintaining M. abscessus in a phenotypically susceptible state to CLR.
Cyclipostins and Cyclophostin analogues (CyC) represent attractive molecules with potent antibacterial activity against M. abscessus in vitro and inside infected macrophages with very low toxicity toward host cells [25–29]. Among these, the enolphosphate analogs CyC17 and CyC31 were the most active growth inhibitors of M. abscessus (against both smooth and rough variants) sharing minimal inhibitory concentrations (MIC) similar to those of imipenem or cefoxitin, often used in clinical settings [28, 29]. CyC compounds react with enzymes containing a catalytic serine and/or cysteine in their active site by forming an irreversible covalent bond [30–33]. This characteristic was exploited to identify their target enzymes using a competitive activity-based protein profiling (ABPP) approach [26–28, 30, 32–35]. In M. abscessus, 39 potential target enzymes of CyC17 were identified, most of which playing a role in lipid metabolism or cell wall synthesis [26]. In addition, 9 out of 39 have orthologs annotated as essential in the Mycobacterium tuberculosis genome [26]. Interestingly, 7 methyltransferases were also identified, including Erm(41). This finding highlights the potential of the CyC to thwart inducible macrolide resistance by blocking Erm(41), opening the possibility of exploiting these inhibitors as adjuncts molecules to suppress macrolide inducible resistance in the context of anti-M. abscessus chemotherapy.
In this study, we validated Erm(41) as an effective target of the CyC analogues and investigated their synergistic activity when given in combination with macrolides (CLR or AZM) as well as with other commonly used antibiotics (AMK, CFX and IPM) in susceptible and induced macrolide-resistant M. abscessus strains. Our results emphasize the potency of the CyC to restore the susceptibility of strains to macrolides, expanding our therapeutic arsenal against M. abscessus pulmonary diseases.
Methods
Antibiotics and compounds
Clarithromycin (Euromedex, France) and Azithromycin (Sigma Aldrich) were solubilized in 96% ethanol and dimethyl sulfoxide (DMSO), respectively. AMK, IPM and CFX was from Toku-E (France) and dissolved in water. The CyC analogues CyC17, CyC31,CyC8α and CyC8β were synthesized as described previously [32, 36]. Stock solutions (10 mM in DMSO) of the CyC compounds (purity of ≥ 95%) [27, 29] were stored at 4 °C.
Strain and bacterial culture
Escherichia coli DH10B cells used in cloning experiments were grown in Luria-Bertani broth (Invitrogen, Carlsbad CA, USA) or on agar plates at 37 °C. Transformants were selected on LB agar supplemented with 200 µg/mL hygromycin B (Toku-E). Mycobacterium smegmatis mc2155 groEL1ΔC strain [37] was grown in Middlebrook 7H9 (BD Difco) supplemented with 0.05% Tween 80 (Sigma-Aldrich, Saint-Quentin Fallavier, France) and 0.2% glycerol (Euromedex, France) (7H9-S) at 37 °C. For the M. abscessus CIP104536T [38] and M. massiliense (CIP 108297T) [39] S morphotypes, 7H9 was supplemented with 0.05% Tween 80, 0.2% glycerol and with 10% Oleic acid Albumin Dextrose Catalase (OADC) enrichment (BD Difco) (7H9-SOADC).
Expression and purification of recombinant Erm(41)
The MAB_2297 gene encoding Erm(41), was amplified by PCR using M. abscessus genomic DNA and the forward primer 5′GTATAACCATGGTTTCCGGCCAACGGTCGCGAC-3′ (NcoI site in bold) and reverse primer 5′-CATATTAAGCTTTGCGCCGCCTGATCACCAG-3′ (HindIII site in bold). The PCR product was cloned into the acetamide-inducible pMyC vector digested with NcoI and HindIII, as previously described [40], enabling the incorporation of a polyhistidine tag at the C-terminus of Erm(41). The integrity of the insert was confirmed by DNA sequencing (Eurofins Genomics). Approximately 200 ng of pMyC-erm(41) were electroporated into M. smegmatis mc2155 groEL1ΔC competent cells by using a Gene Pulser Xcell™ Electroporation System (BioRad, Marnes-la-Coquette, France) at 2500 V, 25 µF and 600 Ω. Recombinant clones were selected on 7H10 agar plates and used to inoculate 10 mL of 7H9-S supplemented with 50 µg/mL hygromycin at 37 °C under shaking at 180 rpm. After 3 days, the preparation was used to inoculate 2 L of culture medium for a large-scale production and the bacteria were grown at 37 °C under shaking until OD600nm value reached 1.5–2. Production of recombinant proteins was induced by adding acetamide (Sigma-Aldrich) to a final concentration of 0.5% (w/v) for 16 h at 37 °C. Cultures were harvested by centrifugation at 4500×g for 30 min at 4 °C. The bacterial pellets were resuspended in 30 mL CHES buffer (100 mM N-cyclohexyl-2-aminoethanesulfonic acid, pH 10, 150 mM NaCl and 1% Nlauroylsarcosine) (buffer A) and lysed using three passages through a French Press (Aminco, Silver Spring, MD, USA) at 1100 PSI. The supernatant was centrifuged for 30 min at 17,000×g and loaded onto a Ni2+-nitrilotriacetic acid (NTA) agarose gel column (Amersham Biosciences, UK). The column was extensively washed with buffer A to remove unspecific proteins and Erm(41) was eluted with buffer A containing 50 mM imidazole. Eluted fractions were collected and analyzed by SDS-PAGE followed by dialysis overnight against a solution of 100 mM CHES, pH 10, 150 mM NaCl and 0.05% Nlauroylsarcosine (buffer B) to remove imidazole. The final concentration of the recombinant protein was measured by determination of OD280nm using the molar extinction coefficient (ε = 32290 cm−1 M−1), concentrated by ultrafiltration to a final concentration of 1.5 mg/mL and stored at −80 °C.
CyC and Erm(41) interaction
To validate the inhibitory interaction between CyC analogues and Erm(41), 10 µg of Erm(41) in 50 µL was incubated for 30 min with different CyC inhibitors at a molar excess xI of 100 in buffer B at 37 °C with shaking. Next, Erm(41) pretreated or not with the CyC analogues were further incubated with 5 µM ActivX TAMRA-FP probe (Thermo Fisher Scientific) for 1 h at room temperature in the darkness. The reaction was stopped by adding 5X Laemmli reducing buffer followed by boiling and proteins were separated by 12% SDS-PAGE. Subsequently, TAMRA FP-labeled proteins were detected by fluorescent gel scanning (TAMRA: λex 557 nm, λem 583 nm) using the Cy®3 filter of a ChemiDoc MP Imager (Bio-Rad) before staining the gels with Coomassie Brilliant Blue dye. Densitometric analyses were performed using the ImageLab™ software version 5.0 (Bio-Rad) to determine the fluorescence intensity content per sample. All raw data were exported as CSV files, imported in the R studio software (The R Project for Statistical Computing, version 4.2.1) and graphs were plotted with the ggplot2 package (version 3.4.0). Histogram represented the mean ± standard deviation of two independent replicates.
Molecular docking in silico
The 3D models structures of Erm(41) (UniProt accession no. B1MAV7) were built with the Phyre2 web portal for protein modeling, prediction and analysis using Erm(E) structure from Saccharopolyspora erythraea (PDB 6NVM) and Erm(38) structure from M. smegmatis (PDB 7F8B) as structural templates [41]. The flexible side chain method, where the ligand (i.e., CyC17) is joined in an arbitrary conformation with the target [i.e., Erm(41) protein] and then modeled as a fully flexible side chain in the AutoDock/Vina simulation, was used [42].
The PyMOL Molecular Graphics System (version 1.4, Schrödinger, LLC) was used as working environment with an in-house version of the AutoDock/Vina PyMOL plugin to perform in silico molecular docking with the CyC17 [43, 44]. A model structure file was generated for the CyC17 molecule, and its geometry was refined using the Avogadro open-source program (version 1.1.1. “http://avogadro.openmolecules.net/”). The box size used for the receptor was chosen to fit the whole protein and to allow nonconstructive binding positions, and was further refined to the inhibitor binding site in Erm(41).
Mass spectrometry
Global mass analyses were determined on purified Erm(41) (15 mg/ml in 100 mM CHES, pH 10, 150 mM NaCl and 0.05% Nlauroylsarcosine buffer) pre-incubated or not with CyC17 at a molar excess, xI, of 100. Ten µL of each protein sample ± CyC17 were desalted on ZipTip C4 (Millipore, Molsheim, France) and eluted by 3 µL of sinapinic acid matrix solution in 0.3% TFA/CH3CN (50:50 v/v). One µL of the latter sample was analyzed on a MALDI TOF-TOF Bruker Ultraflex III spectrometer (Bruker Daltonics, Wissembourg, France) controlled by the Flexcontrol 3.0 package (Build 51). This instrument was used at a maximum accelerating potential of 25 kV and was operated in linear mode using the m/z range from 20,000 to 100,000 (LP_66kDa_method) as already described [30].
Erm(41) quantification
Proteomic studies on macrolide-resistant strains were performed from 20 mL of M. abscessus cultures. Cells were washed twice in PBS (Phosphate Buffer Saline) solution pH 7 and lysed by mechanical disruption on a BioSpec Beadbeater in PBS containing 8 M urea. Quantification of Erm(41) was performed using Liquid chromatography-mass spectrometry (LC-MS/MS) analysis using an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (ThermoFisher Scientific, San Jose, CA) online with an Ultimate 3000RSLCnano chromatography system (ThermoFisher Scientific, Sunnyvale, CA). MS was performed using a data-independent acquisition (DIA) mode. Quantification was based on relative label-free intensity (LFQ) calculated using the DIA-NN 1.8 algorithm [45]. Main DIA-NN output file was further filtered and the LFQ intensity was calculated using our DIAgui package at 1% q value (https://github.com/marseille-proteomique/DIAgui) [46]. Relative quantification was calculated using fold changes of LFQ intensity under CLR or AZM exposure relative to LFQ intensity in non-exposure condition.
The full mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the PRIDE partner repository (https://www.ebi.ac.uk/pride/login) with the dataset identifiers PXD055560.
Preparation of anti-Erm(41) immune serum
Mouse anti-Erm(41) antibodies were produced as follows. A His-tagged version of Erm(41) was prepared as described above. Purified Erm(41) was subcutaneously injected into five BALB/c mice (Janvier, France) (20 µg per mouse) with incomplete Freund’s adjuvant (1/1, v/v) on days 1, 28, and 57. One week after days 28 and 57, blood samples were obtained from the retroorbital plexus, centrifuged and stored at −20 °C until use. Mouse experiments were performed according to institutional and national ethical guidelines (Agreement n˚783223; approved by the Ministry of Higher Education and Research with APAFIS#11465-2016111417574906v4).
Synergistic assay
Synergistic activities were tested in 96-well microplates by the checkerboard assay in Middlebrook 7H9-SOADC broth and using microdilution method [47]. MIC of the antibiotics were determined in 96 well flat-bottom Nunclon Delta Surface microplates with lid (Thermo-Fisher Scientific, ref. 167008) using optical density to assess growth. Log-phase bacteria were diluted to a cell density of 5 × 106 cells/mL in 7H9-SOADC medium. Then, 100 µL of this bacterial suspension (5 × 105 cells final per well) was added to each well containing 100 µL of the 7H9-SOADC medium, serial two-fold dilutions of the selected antibiotics or controls to a final volume of 200 µL. In addition, wells containing 200 µL of 7H9-SOADC medium only were used as sterility/background controls, whereas wells containing 100 µg/mL kanamycin (Euromedex) was used as positive control for growth inhibition. Plates were incubated at 37 °C for 4 to 5 days. The optical density (OD) at 600 nm was quantified using a Tecan Spark 10 M™ multimode microplate reader (Tecan Group Ltd., France). Relative growth units were defined as: RGU% = (test well OD600nm/mean OD600nm of growth control wells) ×100. For each condition, three biological replicates were performed. The MIC of each drug was defined as the lowest drug concentration that inhibited more than 90% of the bacterial growth.
The fractional inhibitory concentration index (FICI) is the reference parameter for quantifying the interaction between two antibiotics in a combination [48]. The fractional inhibitory concentration (FIC) of antibiotic A is defined as the MIC of antibiotic A in the combination divided by the MIC of antibiotic A alone, and vice versa for the FIC of antibiotic B. The FICI value was obtained by the sum of the FICA and FICB. A FICI value of less than 0.5 is considered synergistic; between 0.5 and 4, the effect is considered indifferent; when greater than 4, the interaction is antagonistic [48]. All FICI presented correspond to the minimal FICI obtained. All the results were exported as CSV files, imported in the R studio software and graphs were plotted with the ggplot2 package.
Induction of macrolide resistance in M. abscessus strains
Macrolide resistance was induced in the M. abscessus S reference strain following exposure to CLR or AZM for 14 days. Briefly, M. abscessus S strain was used to inoculate 10 mL of 7H9SOADC at an OD600 nm of 0.05 in absence/presence of 0.1 µg/mL CLR or 0.5 µg/mL AZM corresponding to values ~ 10 times lower than the respective MICs. After 5 days of incubation at 37 °C with shaking (50 rpm), 0.2 mL of the bacterial suspensions were used to inoculate 10 mL of fresh culture medium with or without CLR/AZM, and after 5 additional days of incubation, 0.2 mL of the latter preparation was used to re-inoculate 10 mL of fresh culture medium with or without CLR/AZM. The MIC of CLR or AZM was determined before macrolide exposure and during the induction phase at 5, 10, and 14 days. Susceptibility testing was performed in Middlebrook 7H9-SOADC broth using the microdilution method. MIC of CLR and AZM were determined in 96-well flat-bottom Nunclon Delta Surface microplates with lid using the same method, as described in the previous section. The MIC of each drug was defined as the lowest drug concentration that inhibited more than 90% of the bacterial growth. For each condition, three biological replicates were performed.
RNA extraction and reverse transcription
RNA was prepared from 109M. abscessus cells at different time points 1, 2, 5, 6, 7, 10, 11, 12 and 14 days. The bacteria were harvested and frozen at −80 °C beforehand. Pellet was resuspended in homogenization solution supplemented of 2.5% 1-Thioglycerol (v/v) and then lysed by mechanical disruption on a BioSpec Beadbeater. Total RNA was purified using Maxwell® 16 miRNA Tissue Kit (Promega) according to the manufacturer’s instructions with an extra TURBO DNase (Invitrogen) digestion step to eliminate the contaminating DNA. Finally, the RNA quality was assessed by Tapestation system (Agilent). To obtain cDNAs, GoScript™ Reverse Transcription System protocol (Promega) was used. The final mix contained 1 µg total RNA, 0.5 µg random primers (Promega), 4 µL GoScript™ 5X Reaction Buffer (Promega), 2 µL of 25 mM MgCl2, 1 µL 40 mM dNTP and 1 µL GoScript™ Reverse Transcriptase (Promega).
Quantitative PCR analysis
Quantitative real-time PCR (qPCR) analyses were performed on a CFX96 real-time system (Bio-Rad). The reaction volume was 15 µL, and the final concentration of each primer was 0.5 µM. Specific primers used for qPCR are the following: erm(41) forward 5′CTCAGGGGAGTTCGTTGTGG-3′ and reverse 5′-CCGCTATCCGGACATCTTCC-3′ and rrs (MAB_r5051) forward 5′-CATGGTGAGTGGTGCAAAGC-3′ and reverse 5′AGTCTGGGCCGTATCTCAGT-3′. The cycling parameters of the qPCR were 98 °C for 2 min, followed by 45 cycles of 98 °C for 5 s, 64 °C for 10 s, and 72 °C for 1 s. A final melting curve from 65 °C to 95 °C was added to determine the specificity of the amplification. To determine the amplification kinetics of each product, the fluorescence derived from the incorporation of SYBERGreen into the double-stranded PCR products was measured at the end of each cycle using the Sso Advanced Universal SYBRGreen Supermix kit (Bio-Rad, France). The results were analyzed using Bio-Rad CFX Maestro software, version 2.3 (Bio-Rad, France). The 16 S RNA gene (rrs) was used as a reference for normalization. For each point, a technical duplicate was performed. All the results were exported as CSV files, imported in the R studio software and graphs were plotted with the ggplot2 package.
Results
Characterization of the CyC-Erm(41) interaction
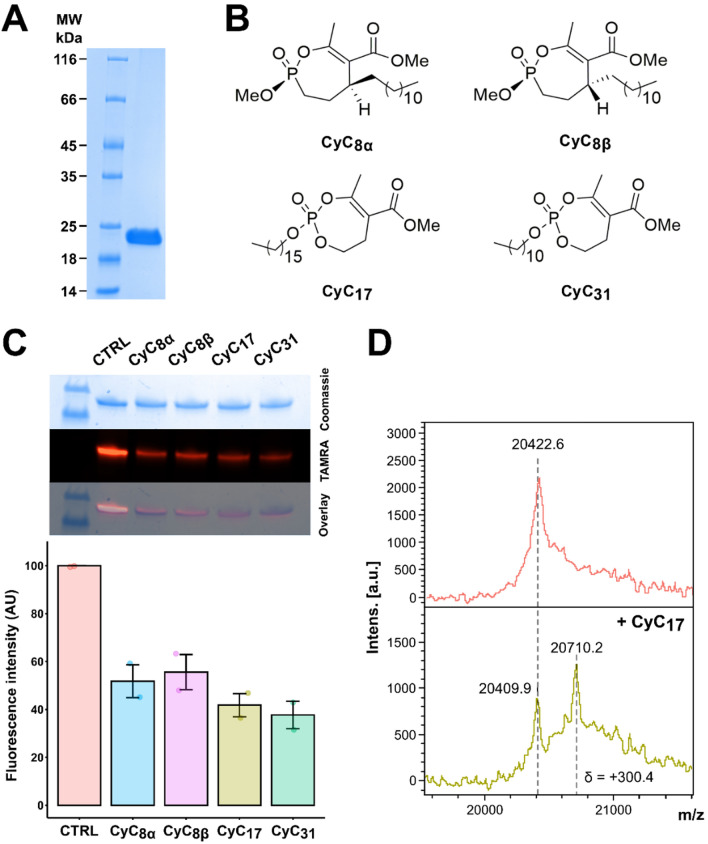
In a previous study, competitive ABPP conducted with the CyC17 inhibitor identified the Erm(41) methyltransferase responsible for inducible macrolide resistance as a possible biological target in M. abscessus at a permutation false discovery rate of 5% [26]. To investigate whether other CyC analogues displaying activity against M. abscessus interact and inhibit Erm(41), the erm(41) gene (MAB_2297) was cloned within a homemade pMyC vector in frame with a 6xHis-tag, allowing strong expression of Erm(41)-His6 in the presence of acetamide [49]. The recombinant protein (theoretical mass ~ 22 400 Da) was produced in M. smegmatis mc2155 groEL1ΔC, and purified to homogeneity by nickel affinity, leading to around 20 mg of pure recombinant protein per liter of culture. Purity and the molecular weight were checked by 12% SDS-PAGE and global mass spectrometry (Fig. 1).
Figure. 1.
Biochemical characterization of the CyC-Erm(41) interaction. A Protein purity assessed by SDS-PAGE. Ten micrograms of protein were loaded onto a 12% polyacrylamide gel and stained with Coomassie brilliant blue G-250 solution. MW, molecular weight standards (5 µg; Euromedex). B Chemical structures of the CyC analogues used in this study. C Equal amounts of Erm(41) were pre-treated with CyC8α. CyC8β. CyC17 and CyC31 , incubated with TAMRA-FP, separated by SDS-PAGE, and visualized by Coomassie Blue staining (top) or in-gel fluorescence (middle). Fluorescence intensity was quantified using the ImageLab™ software. The TAMRA-FP without CyC (CTRL) was arbitrarily placed at 100 AU of fluorescence intensity. D Global mass modification of Erm(41) alone (upper panel) or pre-incubated with CyC 17 (lower panel) as determined using an Ultraflex III mass spectrometer (Bruker Daltonics) in linear mode with the LP_66kDa method
Considering the structure and mechanism of action of the CyC analogues on catalytic serine or cysteine residues, a chemically relevant fluorophosphonate (FP) probe bearing a fluorophore (i.e., rhodamine for TAMRA-FP) and exhibiting a similar mode of action [33, 50] was selected and tested in competitive inhibition tests to assess the covalent interaction between Erm(41) and various CyC (Fig. 1B). Purified Erm(41) was first incubated with either CyC17 and CyC31, two potent inhibitors of extracellularly-growing M. abscessus; or CyC8α and CyC8β, only active against intracellularly-growing M. abscessus [26, 28, 29]. The Erm(41)-CyC complexes were further co-incubated for 1 h with TAMRA-FP and equal amounts of protein were separated by SDS-PAGE and visualized by Coomassie staining (Fig. 1C, upper panel) and ingel fluorescence for TAMRA detection (Fig. 1C, middle panel). In each case, pre-treatment with the CyC molecules resulted in a significant decrease in the fluorescence intensity vs. the control, indicating that the reaction with the TAMRA probe was impeded in the presence of Erm(41)-CyC adducts. Comparison of the fluorescence intensity between the four CyC-Erm(41) complexes and the control condition without CyC, showed a decrease in fluorescence of ~ 50–60% (Fig. 1C). Surprisingly, incubation with higher concentrations of CyC, up to a molar excess xI of 200 related to 1 mol of Erm(41), did not further reduce the fluorescence intensity (data not shown). Taken together, these results suggest that CyC8α, CyC8β, CyC17 and CyC31 directly interact with Erm(41), confirming that this methyltransferase is an effective target of these inhibitors. The fact that the CyC-mediated fluorescence inhibition of was incomplete, remains however unexplained.
To further confirm this interaction, the CyC17-Erm(41) complex obtained above was subjected to MALDI-TOF mass spectrometry (Fig. 1D), which allows to track mass changes if the CyC inhibitor is covalently bound to the protein. Total mass results showed the presence of two peaks: a first peak (20,409.9 ± 3 Da) corresponds to the unmodified protein, and a second one (20,710 Da) to the CyC17-bound Erm(41) adduct. It is noteworthy, however, that the observed 300.4-Da mass shift increment in global mass was 146.08 Da lower than the expected CyC17 theoretical molecular mass of 446.28 Da (Fig. 1D). This mass difference is consistent with previous studies with the thioesterase TesA [31], the antigen 85 complex [33] and the hydrolase HsaD [30] where rearrangement of the covalently bound CyC17 inhibitors occurred, resulting in the loss of the methyl 2-acetyl-4-hydroxybutanoate moiety to reach a stable thermodynamic state. Overall, these results therefore support the formation of a covalent and irreversible Erm(41)-CyC complex.
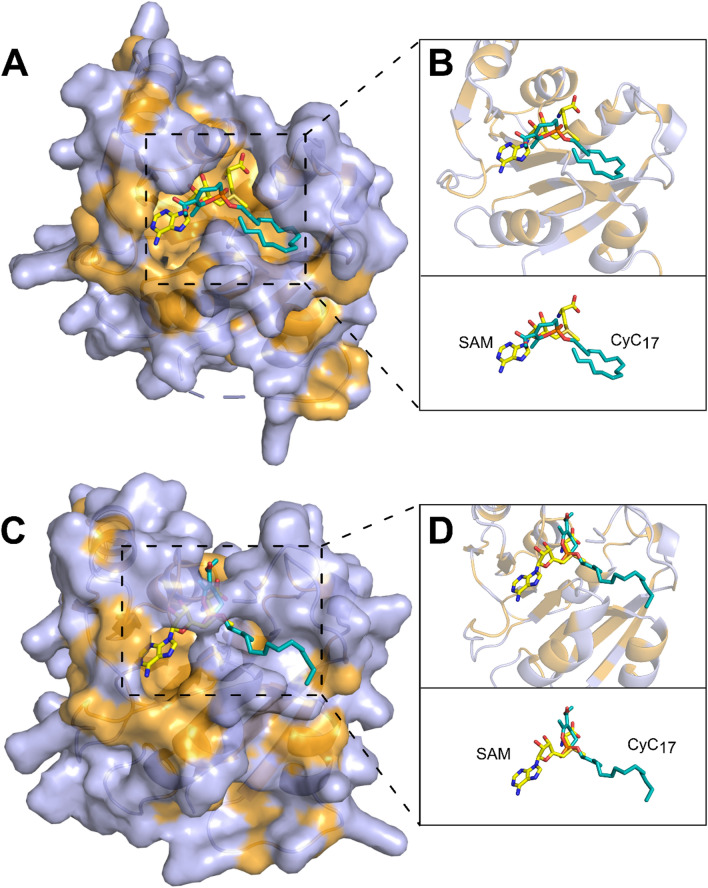
To identify the potential binding site of the CyC into the protein, we generated three dimensional structural models of Erm(41) based on the Saccharopolyspora erythraea Erm(E) (PDB id: 6NVM) and on the M. smegmatis Erm(38) (PDB id: 7F8B) crystal structures, Erm(E) and Erm(38) share 28% and 30% sequence identity with Erm(41), respectively [51, 52]. Both models which are based on high-resolution 3D structures (1.75–2.25 Å), share sequence coverage of 92% with Erm(41), ensuring a proper orientation of the amino acid side chains (Fig. 2). Although these two models generated slightly different 3D structures (RSMD 1.12 Å), both unraveled an Y-shaped catalytic pocket able to accommodate the S-adenosyl-L-methionine (SAM) co-factor. SAM is the methyl donor for Erm(41) to methylate the 23 S rRNA at the adenine 2270 [53, 54]. The Erm(41) pocket comprises mostly hydrophobic and positively-charge residues at its entrance, facilitating the interaction with the negatively-charged rRNA, for the methyltransferase reaction.
Fig. 2.
Molecular docking of CyC 17 in the active site of Erm(41). Model structures of Erm(41) generated using the Phyre2 web portal for protein modeling and based on A , B Erm(E) structure (PDB id: 6NVM) or C , D Erm(38) structure (PDB id: 7F8B). B , D Catalytic pocket view in presence of CyC17 and SAM co-factor. Hydrophobic residues (alanine, leucine, isoleucine, valine, tryptophan, tyrosine, phenylalanine, proline and methionine) are highlighted in orange. The inhibitor and SAM cofactor are in stick representation with the following atom color-code: oxygen red; phosphorus orange; carbon cyan for CyC 17 and yellow for SAM. Structures were drawn with PyMOL Molecular Graphics System (version 1.4, Schrödinger, LLC)
The docking of CyC17 in the Erm(41) pocket generated from the two structural models revealed that this CyC would adopt a productive orientation that could prevent the positioning of SAM in active site (Fig. 2B and D). Taken together, these 3D models along with the biochemical data not only validate the ability of CyC17 to interact with Erm(41), but importantly, suggest that its location inside the catalytic pocket prevents the accommodation of SAM, rendering the enzyme inactive.
Combined efficacity of the CyC with other drugs on macrolide-susceptible strains
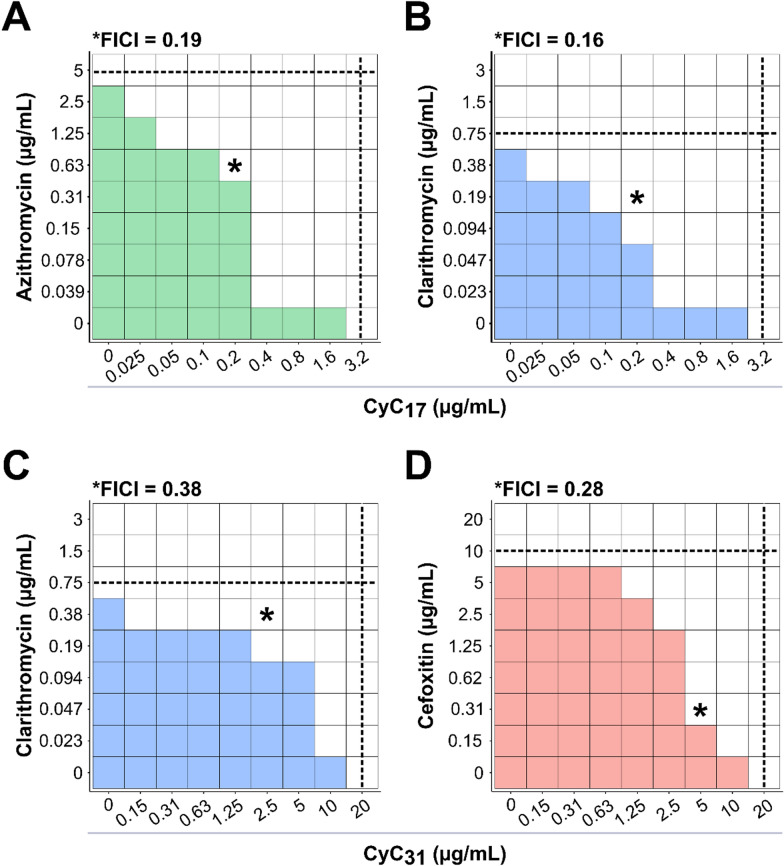
We next investigated the in vitro interaction of the CyC in association with a panel of drugs used in clinical settings: CLR, AZM, AMK, IPM and CFX. The CyC17 and CyC31 which are the best inhibitors of extracellularly-growing M. abscessus [26] have been selected and tested with CLR and AZM using the checkerboard assay. The MIC of each compound tested alone was 5 µg/mL for AZM, 0.75 µg/mL for CLR, 3.2 µg/mL for CyC17 and 20 µg/mL for CyC31. The association of AZM with CyC17 showed a clear synergistic effect with a fractional inhibitory concentration index (FICI) of 0.19 (Fig. 3A). In this context, the best combination displayed MICAZM and MICCyC17 values that were 8-fold (0.63 µg/mL) and 16-fold lower (0.2 µg/mL) than that of AZM and CyC17 alone, respectively. Also, the association of CLR with CyC17 and CyC31 showed a synergistic effect with a FICI of 0.16 and 0.38 respectively (Fig. 3B and C). In this context, the best combination displayed MICCLR and MICCyC17 values that were 8-fold (0.094 µg/mL) and 32-fold lower (0.025 µg/mL) than that of CLR and CyC17 alone, respectively. About CyC31-CLR association, the MICCLR and MICCyC31 values were 4-fold (0.19 µg/mL) and 128-fold lower (0.15 µg/mL) than that of CLR and CyC31 alone, respectively. Regarding the CyC31-AZM association, no effect could be measured on either compound for every combination, with calculated FICI between 0.63 and 2.5, the interaction was qualified as indifferent (Figure S1).
Figure. 3.
Synergistic activity of CyC-antibiotic combinations against M. abscessus. Schematic checkerboard representation of association between A azithromycin or B clarithromycin and CyC 17 and between C clarithromycin or D cefoxitin and CyC31. The growth is represented by the colored boxes. Black dotted lines represent the respective MIC of the drug (horizontal) and the CyC (vertical). The calculated fractional inhibitory concentration index (FICI) of the best combination is indicated on top of each representation and also by a black star. The smallest FICI obtained for a combination is taken as the FICI of the association. Data are representative of three independent biological replicates
We next evaluated the in vitro activity of AMK, IPM or CFX in association with CyC17 or CyC31. The MIC values of AMK, IPM and CFX alone were 10 µg/mL, > 20 µg/mL and 10 µg/mL, respectively, in agreement with published values [55]. As observed above with AZM and CLR, the combination of CFX with CyC31 showed a synergistic effect, with a FICI of 0.28 (Fig. 3D), resulting in a 32-fold (0.31 µg/mL) and 4-fold (5 µg/mL) decrease in MIC values of CFX and CyC31 respectively. For the other combinations, including CyC17-AMK, CyC31-AMK, CyC17-IPM, CyC31-IPM and CyC17-CFX, the FICI values were between 0.51 and 2.0, considered as indifferent interactions (Figure S1). However, it should be noted that in the CyC31-AMK association, the presence of CyC31 allowed a 2‑fold decrease in MICAMK in 0.15–1.25 µg/mL interval and a 4‑fold decrease in MICAMK in 2.5–10 µg/mL interval.
Globally, these results suggest that the CyC exert a positive effect when associated with other antibiotics, notable with AZM, CLR and CFX with FICI ranging between 0.16 and 0.38. These drug combinations reduced by at least a 4-fold the MIC of the antibiotics.
Induction of macrolide resistance anderm(41) expression
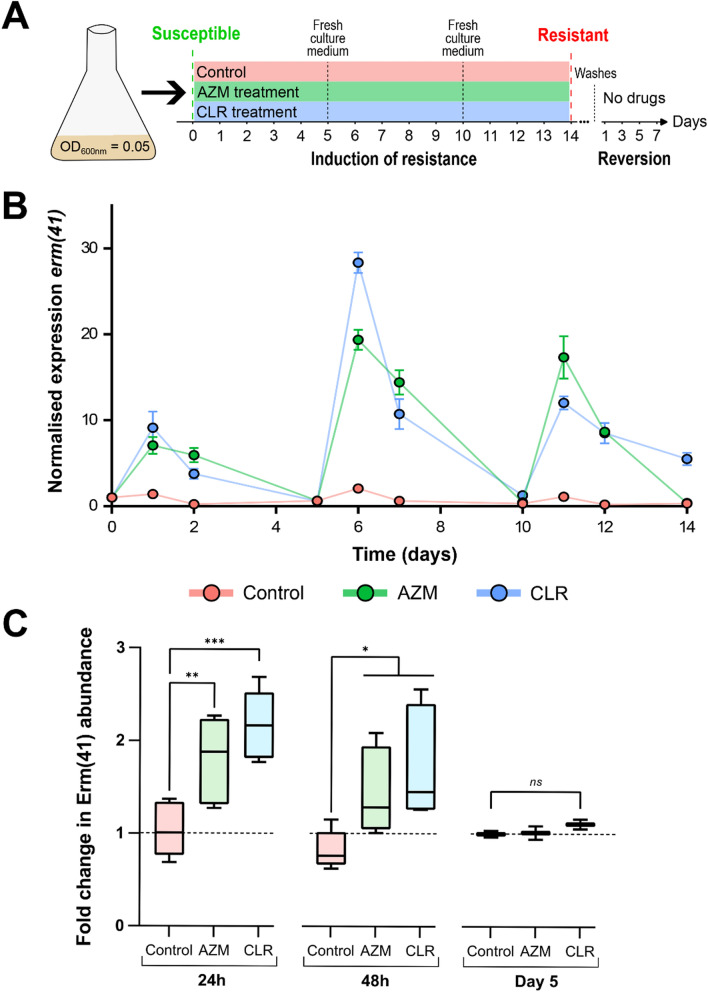
The validation of Erm(41) as a target of the CyC inhibitors prompted us to evaluate their capacity to block the Erm(41)-mediated inducible macrolide resistance. To do so, M. abscessus was first pre-exposed to CLR or AZM for 14 days, as previously reported [13–17]. To avoid the generation of spontaneous resistant mutant in rrl gene (MAB_r5052), coding for rRNA 23 S which is the main target of macrolides, M. abscessus was incubated with sub-inhibitory macrolide concentrations corresponding to 0.1×MIC, i.e., 0.1 µg/mL and 0.5 µg/mL for CLR and AZM, respectively (Fig. 4A). The inducible macrolide resistance was followed by MIC measurements, Western blotting, global proteomic analysis and determination of the erm(41) transcriptional levels, as reported [13–15]. Determination of MIC was assessed after 5, 10 and 14 days of treatment (Table 1). As expected, the prolonged treatment with CLR or AZM increased both the MICCLR and MICAZM values up to 50times (≥ 40 µg/mL) and 17-times (> 100 µg/mL), respectively, after 14 days for both CLR-treated and AZM-treated strains. As noticed earlier [17], the MIC of AZM increased more rapidly than the one of CLR. These results confirm that pre-exposure of M. abscessus to sub-MIC concentrations of CLR or AZM induces macrolide resistance. Conversely, when performed on M. massiliense which lacks a functional Erm(41) protein [13], no resistance was observed (MICAZM = 1.03 ± 0.10 µg/mL; MICCLR = 1.45 ± 0.02 µg/mL) even after 14 days of treatment, which validates the macrolide induction protocol.
Fig. 4.
Induction of macrolide resistance. A Schematic protocol of induction of macrolide resistance and reversion in M. abscessus strains B Expression of erm(41) transcripts was quantified by RT-qPCR during the induction of macrolide resistance in M. abscessus. The amount of transcript in the untreated condition (control) at day 0 is taken as reference. Data represent the mean of normalized expression of two biological replicates. C Quantification of Erm(41) abundance by mass spectrometry. The graph shows the variations in fold change values for Erm(41) abundance after 24 h, 48 h and 5 days of induction of macrolide resistance in M. abscessus . In each case, the untreated control strain was taken as reference with a fold change of 1. Statistical analysis was done using a non-parametric Mann–Whitney test with Prism 8.0 (Graphpad Inc): * p value < 0.05; ** p value < 0.01; *** p value < 0.001
Table 1.
Resistance levels to macrolides following exposure of M. abscessus to CLR or AZM
| MIC (µg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Growth conditions | Before exposition | Day 5 | Day 10 | Day 14 | Reversion | |||||
| CLR | AZM | CLR | AZM | CLR | AZM | CLR | AZM | CLR | AZM | |
| Control (Untreated) | 0.9 ± 0.5 | 5.8 ± 1.6 | 0.8 ± 0 | 6.9 ± 1.4 | 0.3 ± 0.1 | 4.5 ± 1.5 | 0.6 ± 0.2 | 6.0 ± 2.0 | ND | ND |
| CLR | 0.9 ± 0.5 | 5.8 ± 1.6 | 3.1 ± 1.7 | 47 ± 17 | 9.4 ± 3.4 | > 100 | > 40 | > 100 | 0.8 ± 0.4 | 4.1 ± 1.2 |
| AZM | 0.9 ± 0.5 | 5.8 ± 1.6 | 3.6 ± 2.1 | 63 ± 0 | 8.3 ± 3.2 | > 100 | 40 ± 11 | > 100 | 0.7 ± 0.2 | 3.6 ± 1.1 |
The MICs are given in µg/mL. For the reversion experiments, resistant mutants obtained after 14 days of induction were washed with fresh medium, grown without antibiotic for 7 day and susceptibility testing was performed to check the capacity of the strains to revert to a susceptible phenotype. The MIC presented in the table correspond to the mean ± SD of the MIC obtained from three independent biological replicates. ND not determined
In parallel, transcriptional expression of erm(41) was monitored during the 14 days of exposure to CLR or AZM. The erm(41) mRNA levels were quantified by reverse transcription PCR at day 1, 2, 5, 6, 7, 10, 11, 12 and 14, with drug renewal every 5 days (Fig. 4A). As shown in Fig. 4B, subMIC macrolide concentrations induced erm(41) expression with a peek at 24 h, followed by a progressive decrease until day 5. Antibiotic renewal significantly increased the mRNA level of erm(41) for both macrolides although following a similar erm(41) gene expression kinetic (rapid induction at day 6 and progressive decrease up to day 10). These results demonstrate the importance to renew macrolides during the induction of resistance phase in order to maintain high erm(41) expression levels. Next, M. abscessus lysates from cultures exposed to CLR or AZM for 24 h and 48 h, respectively, were probed using Erm(41)-specific murine antibodies. However, we were unable to detect the protein in these samples despite several attempts (data not show). Thus, Erm(41) detection was done by analyzing the complete proteome of CLR- and AZM-treated M. abscessus cultures after 24 h, 48 h and 5 days of exposure. Statistical differential analysis with the non-treated M. abscessus proteome showed the presence of Erm(41) protein in the AZM- and CLR-treated M. abscessus proteome at 24 h and 48 h but not at 5 days of treatment to macrolides (Fig. 4C). These results confirm the RT-qPCR results, but also show a weak overexpression of the protein with a fold-change of 1.7–2.1 at 24 h and 1.3–1.6 at 48 h compared to the proteome of non-treated cultures. The presence of the Erm(41) at 24 and 48 h, together with the low level of erm(41) transcripts and its disappearance at day 5, suggests that this protein may be produced in low scale and is probably very unstable.
To evaluate whether inducible macrolide resistance mechanism is reversible, macrolide-resistant strains were washed with fresh 7H9-SOADC medium and grown in culture medium without antibiotic for 7 days [17]. Then, susceptibility testing was performed to check the capacity of the strains to revert to a susceptible phenotype (Table 1, Reversion condition). Whatever the macrolide used for the induction of resistance, MIC of AZM and CLR measured after 7 days of culture medium without any antibiotic were identical to those before macrolide induction, confirming that this resistance phenomenon is reversible. Moreover, no mutation was found in rrl is AZM- or CLR-treated strains (data not show), thus excluding the emergence of a genetically-acquired resistance mechanism.
In contrast, we generated a spontaneous macrolide-resistant mutant strain by using a high concentration of macrolide (10× MIC), displaying high level of resistance to CLR (MIC > 40 µg/mL) and AZM (MIC > 100 µg/mL). This mutant carries an adenine-to-guanine replacement at position 2271 in rrl avoiding the macrolide to interact with the 23 S rRNA (Figure S2), as already reported in M. abscessus clinical isolates [56, 57].
CyC-macrolide association on macrolide-resistant strains
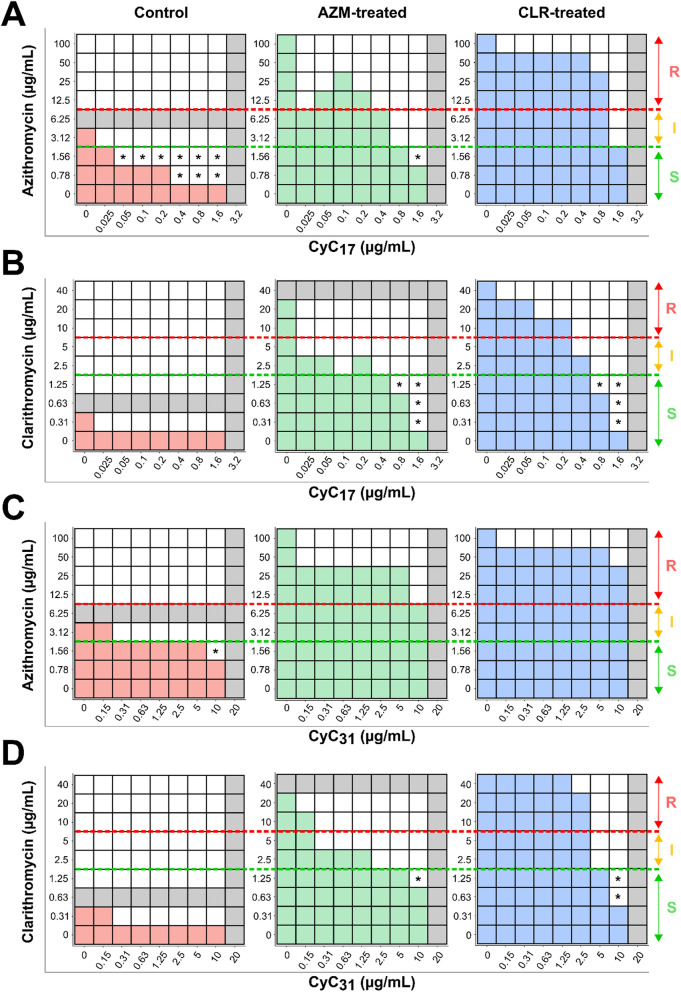
The ability of the CyC to interact in vitro with purified Erm(41) protein suggest that these inhibitors could antagonize the Erm(41) activity and, consequently, restore susceptibility to macrolides. To test this hypothesis, we evaluated in vitro the association of AZM and CLR with CyC17 or CyC31 using the checkerboard assay with reversible resistant strains generated as described below (Fig. 5, Table S1 and S2). For susceptibility breakpoints, we followed the Clinical and Laboratory Standards Institute (CLSI) guidelines [58].
Fig. 5.
Activity of CyC-macrolides association on macrolide-resistant M. abscessus strains. Schematic checkerboard representation of association between A azithromycin or B clarithromycin and CyC 17 , and between C azithromycin or D clarithromycin and CyC 31 using WT (Control), AZM- and CLR-treated strains. Bacterial growth is represented by colored boxes, and the corresponding MIC values for macrolides and CyC are shown in gray. Based on CLSI guidelines, the green dotted lines represent the maximum macrolide concentration for which the strain is considered susceptible, and the red dotted lines correspond to the minimum concentration for which the strain is considered resistant. If bacterial growth is above the red line, the phenotype is considered resistant to macrolides, below the green line, the phenotype is considered susceptible, and between the two lines, the phenotype is intermediate. The new susceptible macrolides conditions have been indicated by a black star. S susceptible, I intermediate and R resistant
The control strain, corresponding to the untreated condition, showed already an intermediate AZM sensitivity profile (MICAZM = 6 µg/mL). The association of CyC17 with AZM resulted in a 3- and 7-fold decrease in MICAZM in the 0.05–0.2 µg/mL (MICAZM = 2 µg/mL) and in the 0.4–1.6 µg/mL (MICAZM = 0.8 µg/mL) CyC17 concentration range, respectively. Similarly, a 2-fold reduction in MICAZM (MICAZM = 3 µg/mL) was reached in presence of CyC31 in the 0.31–5 µg/mL concentration range, the strain becoming fully susceptible from 10 µg/mL of CyC31. Although the addition of CyC17 and CyC31 with CLR did not modify the CLR susceptibility profile of the control strain, their presence led to a 2-fold reduction in CMICLR.
As reported above resistant strains displayed a MICAZM >100, MICCLR >40 µg/mL, MICCyC17 = 3.2 µg/mL and MICCyC31 = 20 µg/mL, respectively.
For the CLR-treated strain, two combinations were able to restore susceptibility to macrolides. The association of CLR with CyC17 leads to a gradual decrease of MICCLR until an intermediate susceptibility profile at 0.4 µg/mL, corresponding to CyC17’s MIC/8. The strain become fully susceptible at CyC17’s MIC/4 concentration with a fold change reduction of 40 (MICCLR = 1 µg/mL), and even reached a fold change in its MICCLR up to 133 (MICCLR = 0.3) at CyC17’s MIC/2. Similarly, 5 µg/mL (i.e., MIC/4) CyC31 allowed to dramatically decrease the MICCLR (3 µg/mL) by 13-fold. When using 10 µg/mL (i.e., CyC31’s MIC/2) the strain became fully susceptible to CLR with a fold change of 66 in its MICCLR.
In the case of AZM-treated strains, three combinations were able to restore susceptibility to macrolides. The AZM association with 0.8 µg/mL (MIC/4) of CyC17 allowed to reach a MICAZM value of 3 µg/mL (33-fold decrease), which is considered as ‘intermediate’. In the presence of 1.6 µg/mL (MIC/2) of CyC17 a 50-fold decrease in MICAZM was reached (MICAZM=2 µg/mL) and the strain was considered susceptible. With the CyC31, regardless the concentration used, the AZM-treated strain remained resistant, even if a 7-fold decrease in MICAZM was obtained at a CyC31’s MIC/2. Regarding CLR susceptibility to this AZM-treated strain, the most efficient associations was the combination of CLR with CyC17 or CyC31. The addition of low concentration of CyC17 corresponding to MIC/64 decreased drastically MICCLR until an intermediate level. The use of CyC17’s MIC/4 concentration (0.8 µg/mL) fully restored CLR susceptibility. Similarly, the addition of CyC31 together with CLR resulted in an 8-fold decrease in MICCLR for CyC31 concentrations ranging from 0.31 to 1.25 µg/mL. Finally, 10 µg/mL of CyC31 (MIC/2) rendered the strain susceptible to CLR.
On the other hand, when testing the association of AZM and CLR with the CyC17 on M. massiliense, no effect was observed with respect to the MICs of AZM (0.625–1.25 µg/mL), CLR (0.093–0.188 µg/mL) or the CyC17 (3.2 µg/mL). Aligning with these results, the activity of CyC17 and CyC31 with macrolides against the macrolide-resistant spontaneous mutant carrying an irreversible point mutation in rrl gene did not influence the susceptibility profile to CLR or AZM (Table S3). Overall, the results based on M. massiliense and the rrl mutant suggest that the potentiating effect of the CyC for macrolides is only effective in the case of Erm(41)-dependent inducible resistance.
Discussion
M. abscessus is considered as the main pathogenic RGM in humans [59], responsible for a broad spectrum of infections ranging from mucocutaneous infections in immunocompetent individuals to severe pulmonary infections in patients with CF or chronic obstructive pulmonary disease [2, 3]. The natural resistance of M. abscessus to most conventional antibiotics, the poor clinical outcome and the lengthy regimens render treatments particularly difficult. In clinical practice, drug combinations are given in efforts to prevent the development of drug resistance during therapy and to optimize the efficacy of the treatments. In this context, the discovery of a new family of the multitargeted CyC inhibitors, acting specifically on mycobacteria with no toxicity toward mammalian cells, adds new hope for subsequent therapeutic developments [26–28]. Among the targets impacted by the CyC, we identified the Erm(41) a methyltransferase involved in the resistance induced by macrolides, which are the pillar of anti-M. abscessus therapy and whose inactivation often leads to a therapeutic end [1, 20]. Here, we investigated the interaction between the CyC and Erm(41) to bypass inducible macrolide resistance, thus opening the door to new therapeutic interventions.
Foremost, we showed that Erm(41) interacts with several CyC, including CyC17 and CyC31 which exhibit M. abscessus growth in vitro growth; and CyC8α and CyC8β, primarily acting against M. abscessus residing in the macrophage [26, 29]. Biochemical studies involving the TAMRA-FP probe that binds to serine hydrolases along with mass spectrometry and in silico molecular docking, suggest that the CyC analogues are covalently bound to the active site of Erm(41), thereby preventing the accommodation of SAM into Erm(41) the catalytic site, resulting in enzymatic blockage. The future high-resolution structure of Erm(41) and detailed description of the catalytic mechanism of the enzyme may lead to the development of other chemical entities capable to inhibit its enzymatic activity. Alternatively, the development of other classes of Erm(41)specific inhibitors or SAM derivatives may provide means to preserve high susceptibility levels of M. abscessus isolates to macrolides [60, 61].
We investigated the synergistic potential of the CyC in combination with multiple antimicrobials active against M. abscessus. Several combinations were tested in vitro with CyC17 and CyC31, emphasizing optimal synergistic interactions between CyC17-AZM, CyC17-CLR and CyC31-CLR. Our data also highlighted the potent association between CyC31 and CFX, a drug largely used for the treatment against M. abscessus infections. Overall, this study confirms that the CyC can be used in association with other clinically relevant drugs against this species, displaying a clear improvement regarding the existing treatments, at least in vitro. Pre-clinical studies are now warranted to study the impact and efficacy of these treatment combinations in animal models.
Importantly, the CyCs are able to counteract macrolide resistance mediated by Erm(41) or remain active on spontaneous resistant mutants carrying a mutation in the rrl gene. This restoration of macrolide susceptibility is of prime interest and offers new prospects for the development of drug regimens.
In this study, we demonstrated that the Erm(41) protein is also able to interact with the CyC8(α,β) which are only active ex vivo inside infected macrophages [26]. These results may suggest that these CyC8(α,β) might also be able to restore macrolide susceptibility in resistant strains generated by a treatment to macrolides. To confirm this hypothesis, it will be necessary to test the association of CyC8(α,β) with AZM or CLR against macrophages infected with M. abscessus strain whose resistance has previously been induced.
Finally, our data confirmed the efficacy of the CyC to restore the CLR or AZM susceptibility on macrolide-resistant M. abscessus strain following Erm(41) induction. A major asset of the CyC relies on the fact that these molecules can target multiple enzymes, thus preventing the emergence of spontaneous resistant mutants. The work presented here describes a novel and unique aspect of this family of inhibitors by lowering/suppressing a mechanism that leads to drug in inactivation. To the best of our knowledge, CyC are the first inhibitors of the Erm(41) methyltransferase.
In addition, this work revealed that the synergy activity of the CyC in association with CFX should also be taken into consideration. Further experiments are, however, needed to better understand the relationship between the CFX and CyC compounds.
The recently studied CLR-Rifabutin combination showed also a synergistic effect able to block Erm(41) induction and so increased the efficiency of macrolide [24]. Thereby, blocking the main actor responsible for the resistance to macrolide by molecules unable to generate spontaneous mutants deserves more attention.
Conclusion
These results provide the opportunity to revisit the therapeutic regimen for combating M. abscessus pulmonary infections in CF patients, and particularly erm(41)-positive strains. Long-term prospects should help to expand our therapeutic arsenal in the fight against a particularly antibioticresistant mycobacterial species.
Supplementary Information
Acknowledgements
We acknowledge the Grégory Lemarchal and Vaincre la Mucoviscidose associations for their constant help and support since many years.
Abbreviations
- ABPP
Activity-based protein profiling
- AMK
Amikacin
- AZM
Azythromycin
- CF
Cystic fibrosis
- CLR
Clarithromycin
- CFX
Cefoxitin
- CyC
Cyclipostins and Cyclophostin analogues
- IPM
Imipenem
- FIC
Fractional inhibitory concentration
- FICI
Fractional inhibitory concentration index
- MIC
Minimal inhibitory concentration
- RGM
Rapid-growing mycobacteria
Author contributions
Conceptualization: J-FC, LK, J-LH and SC. Methodology and validation: MS, IP, PF, SA, LC, YD, CDS J-FC. Formal analysis: MS, IP, PF, SA, LC, YD, PS, CDS J-FC and SC. Investigation: MS, IP, PF, SA, LC, YD, J-FC. Writing—review and editing: MS, J-FC, LK, PS, VLM, J-LH and SC. Supervision: SC. Project administration: SC.
Funding
This work was supported by the Centre National de la Recherche Scientifique (CNRS), Aix-Marseille University (AMU), the Association Grégory Lemarchal and Vaincre la Mucoviscidose (projects N°RF20190502466 and RF20220503053), and the Agence Nationale de la Recherche (LipInTB project N°ANR-19-CE44-0011). M.S. was supported by a PhD fellowship from the Association Grégory Lemarchal and Vaincre la Mucoviscidose (project N°RF20190502466 and RF20220503053). Marseille Proteomics (marseille-proteomique.univ-amu.fr/) was supported by IBISA (Infrastructures Biologie Santé et Agronomie), the Canceropôle PACA, the Provence-Alpes-Côte d’Azur Région, the Institut Paoli-Calmettes, and the Fonds Européen de Développement Régional (FEDER).
Availability of data and materials
All data generated and analyzed during this study are included in this article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication and references
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/25/2024
The original online version of this article was revised: The family name of the author ‘Vincent Le Moigne’ in article xml has been corrected.
References
- 1.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. [DOI] [PubMed] [Google Scholar]
- 2.Cowman S, van Ingen J, Griffith DE, Loebinger MR. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J. 2019. 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 3.Martiniano SL, Nick JA, Daley CL. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med. 2022;43(4):697–716. [DOI] [PubMed] [Google Scholar]
- 4.Wallace RJ Jr., Brown-Elliott BA, Crist CJ, Mann L, Wilson RW. Comparison of the in vitro activity of the glycylcycline tigecycline (formerly GAR-936) with those of tetracycline, minocycline, and doxycycline against isolates of nontuberculous mycobacteria. Antimicrob Agents Chemother. 2002;46(10):3164–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. 2012;67(4):810–8. [DOI] [PubMed] [Google Scholar]
- 6.Maurer FP, Bruderer VL, Castelberg C, Ritter C, Scherbakov D, Bloemberg GV, Bottger EC. Aminoglycoside-modifying enzymes determine the innate susceptibility to aminoglycoside antibiotics in rapidly growing mycobacteria. J Antimicrob Chemother. 2015;70(5):1412–9. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre AL, Le Moigne V, Bernut A, Veckerle C, Compain F, Herrmann JL, Kremer L, Arthur M, Mainardi JL. Inhibition of the beta-lactamase bla(Mab) by avibactam improves the in vitro and in vivo efficacy of imipenem against Mycobacterium abscessus. Antimicrob Agents Chemother. 2017. 10.1128/aac.02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rominski A, Roditscheff A, Selchow P, Bottger EC, Sander P. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother. 2017;72(2):376–84. [DOI] [PubMed] [Google Scholar]
- 9.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash K, Parodi N, Strong A, Gee M, Smith T, Wallace RJ Jr. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol. 2015;53(4):1211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Bottger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis. 2020;71(4):905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax. 2016;71(Suppl 1):i1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi H, Kim SY, Kim DH, Huh HJ, Ki CS, Lee NY, Lee SH, Shin S, Shin SJ, Daley CL, Koh WJ. Clinical characteristics and treatment outcomes of patients with acquired macrolide-resistant Mycobacterium abscessus lung disease. Antimicrob Agents Chemother. 2017;61(10):10–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash KA, Brown-Elliott BA, Wallace RJ Jr. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53(4):1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi GE, Shin SJ, Won CJ, Min KN, Oh T, Hahn MY, Lee K, Lee SH, Daley CL, Kim S, Jeong BH, Jeon K, Koh WJ. Macrolide treatment for Mycobacterium abscessus and Mycobacterium massiliense infection and inducible resistance. Am J Respir Crit Care Med. 2012;186(9):917–25. [DOI] [PubMed] [Google Scholar]
- 15.Maurer FP, Castelberg C, Quiblier C, Bottger EC, Somoskovi A. Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J Antimicrob Chemother. 2014;69(6):1559–63. [DOI] [PubMed] [Google Scholar]
- 16.Schildkraut JA, Pennings LJ, Ruth MM, de Brouwer AP, Wertheim HF, Hoefsloot W, de Jong A, van Ingen J. The differential effect of clarithromycin and azithromycin on induction of macrolide resistance in Mycobacterium abscessus. Future Microbiol. 2019;14:749–55. [DOI] [PubMed] [Google Scholar]
- 17.Richard M, Gutierrez AV, Kremer L. Dissecting erm(41)-mediated macrolide-inducible resistance in Mycobacterium abscessus. Antimicrob Agents Chemother. 2020;64(2):10–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kook YH. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol Immunol. 2010;54(6):347–53. [DOI] [PubMed] [Google Scholar]
- 19.Harada T, Akiyama Y, Kurashima A, Nagai H, Tsuyuguchi K, Fujii T, Yano S, Shigeto E, Kuraoka T, Kajiki A, Kobashi Y, Kokubu F, Sato A, Yoshida S, Iwamoto T, Saito H. Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J Clin Microbiol. 2012;50(11):3556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux AL, Catherinot E, Soismier N, Heym B, Bellis G, Lemonnier L, Chiron R, Fauroux B, Le Bourgeois M, Munck A, Pin I, Sermet I, Gutierrez C, Veziris N, Jarlier V, Cambau E, Herrmann JL, Guillemot D, Gaillard JL. Comparing Mycobacterium massiliense and Mycobacterium abscessus lung infections in cystic fibrosis patients. J Cyst Fibros. 2015;14(1):63–9. [DOI] [PubMed] [Google Scholar]
- 21.Blondiaux N, Moune M, Desroses M, Frita R, Flipo M, Mathys V, Soetaert K, Kiass M, Delorme V, Djaout K, Trebosc V, Kemmer C, Wintjens R, Wohlkonig A, Antoine R, Huot L, Hot D, Coscolla M, Feldmann J, Gagneux S, Locht C, Brodin P, Gitzinger M, Deprez B, Willand N, Baulard AR. Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420. Science. 2017;355(6330):1206–11. [DOI] [PubMed] [Google Scholar]
- 22.Laws M, Shaaban A, Rahman KM. Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol Rev. 2019;43(5):490–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubee V, Bernut A, Cortes M, Lesne T, Dorchene D, Lefebvre AL, Hugonnet JE, Gutmann L, Mainardi JL, Herrmann JL, Gaillard JL, Kremer L, Arthur M. Beta-lactamase inhibition by avibactam in Mycobacterium abscessus. J Antimicrob Chemother. 2015;70(4):1051–8. [DOI] [PubMed] [Google Scholar]
- 24.Aziz DB, Go ML, Dick T. Rifabutin suppresses inducible clarithromycin resistance in Mycobacterium abscessus by blocking induction of whiB7 and erm41. Antibiot (Basel). 2020;9(2):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavalier JF, Spilling CD, Durand T, Camoin L, Canaan S. Lipolytic enzymes inhibitors: A new way for antibacterial drugs discovery. Eur J Med Chem. 2021;209:112908. [DOI] [PubMed] [Google Scholar]
- 26.Madani A, Ridenour JN, Martin BP, Paudel RR, Abdul Basir A, Le Moigne V, Herrmann JL, Audebert S, Camoin L, Kremer L, Spilling CD, Cavalier JF. Cyclipostins and cyclophostin analogues as multitarget inhibitors that impair growth of Mycobacterium abscessus. ACS Infect Dis. 2019;5(9):1597–608. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen PC, Delorme V, Benarouche A, Martin BP, Paudel R, Gnawali GR, Madani A, Puppo R, Landry V, Kremer L, Brodin P, Spilling CD, Cavalier JF, Canaan S. Cyclipostins and Cyclophostin analogs as promising compounds in the fight against tuberculosis. Sci Rep. 2017;7(1):11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen PC, Madani A, Santucci P, Martin BP, Paudel RR, Delattre S, Herrmann JL, Spilling CD, Kremer L, Canaan S, Cavalier JF. Cyclophostin and cyclipostins analogues, new promising molecules to treat mycobacterial-related diseases. Int J Antimicrob Agents. 2018;51(4):651–4. [DOI] [PubMed] [Google Scholar]
- 29.Sarrazin M, Martin BP, Avellan R, Gnawali GR, Poncin I, Le Guenno H, Spilling CD, Cavalier JF, Canaan S. Synthesis and biological characterization of fluorescent cyclipostins and cyclophostin analogues: new insights for the diagnosis of mycobacterial-related diseases. ACS Infect Dis. 2022;8(12):2564–78. [DOI] [PubMed] [Google Scholar]
- 30.Barelier S, Avellan R, Gnawali GR, Fourquet P, Roig-Zamboni V, Poncin I, Point V, Bourne Y, Audebert S, Camoin L, Spilling CD, Canaan S, Cavalier JF, Sulzenbacher G. Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis. Febs J. 2023;290(6):1563–82. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen PC, Nguyen VS, Martin BP, Fourquet P, Camoin L, Spilling CD, Cavalier JF, Cambillau C, Canaan S. Biochemical and structural characterization of TesA, a major thioesterase required for outer-envelope lipid biosynthesis in Mycobacterium tuberculosis. J Mol Biol. 2018;430(24):5120–36. [DOI] [PubMed] [Google Scholar]
- 32.Point V, Malla RK, Diomande S, Martin BP, Delorme V, Carriere F, Canaan S, Rath NP, Spilling CD, Cavalier JF. Synthesis and kinetic evaluation of cyclophostin and cyclipostins phosphonate analogs as selective and potent inhibitors of microbial lipases. J Med Chem. 2012;55(22):10204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viljoen A, Richard M, Nguyen PC, Fourquet P, Camoin L, Paudal RR, Gnawali GR, Spilling CD, Cavalier JF, Canaan S, Blaise M. Cyclipostins and cyclophostin analogs inhibit the antigen 85 C from Mycobacterium tuberculosis both in vitro and in vivo. J Biol Chem. 2018;293(8):2755–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutta S, Malla RK, Bandyopadhyay S, Spilling CD, Dupureur CM. Synthesis and kinetic analysis of some phosphonate analogs of cyclophostin as inhibitors of human acetylcholinesterase. Bioorg Med Chem. 2010;18(6):2265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spilling CD. The chemistry and biology of cyclophostin, the cyclipostins and related compounds. Molecules. 2019;24(14):2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin BP, Vasilieva E, Dupureur CM, Spilling CD. Synthesis and comparison of the biological activity of monocyclic phosphonate, difluorophosphonate and phosphate analogs of the natural AChE inhibitor cyclophostin. Bioorg Med Chem. 2015;23(24):7529–34. [DOI] [PubMed] [Google Scholar]
- 37.Noens EE, Williams C, Anandhakrishnan M, Poulsen C, Ehebauer MT, Wilmanns M. Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1DeltaC expression strain. BMC Biotechnol. 2011;11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4(6):e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adekambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, Drancourt M. Amoebal coculture of Mycobacterium massiliense sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol. 2004;42(12):5493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakala N, Schue JC, Carriere M, Geerlof F, Canaan S. Evidence for the cytotoxic effects of Mycobacterium tuberculosis phospholipase C towards macrophages. Biochim Biophys Acta. 2010;1801(12):1305–13. [DOI] [PubMed] [Google Scholar]
- 41.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianco G, Forli S, Goodsell DS, Olson AJ. Covalent docking using autodock: two-point attractor and flexible side chain methods. Protein Sci. 2016;25(1):295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24(5):417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020;17(1):41–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerault MA, Camoin L, Granjeaud S. DIAgui: a Shiny application to process the output from DIA-NN. Bioinform Adv. 2024;4(1):vbae001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Run E, Arthur M, Mainardi JL. In vitro and intracellular activity of imipenem combined with rifabutin and avibactam against Mycobacterium abscessus. Antimicrob Agents Chemother. 2018;62(8):10–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52(1):1. [DOI] [PubMed] [Google Scholar]
- 49.Santucci P, Point V, Poncin I, Guy A, Crauste C, Serveau-Avesque C, Galano JM, Spilling CD, Cavalier JF, Canaan S. LipG a bifunctional phospholipase/thioesterase involved in mycobacterial envelope remodeling. Biosci Rep. 2018;38(6):BSR20181953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96(26):14694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stsiapanava A, Selmer M. Crystal structure of ErmE – 23S rRNA methyltransferase in macrolide resistance. Sci Rep. 2019;9(1):14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goh BC, Xiang X, Lescar J, Dedon PC. Crystal structure and functional analysis of mycobacterial erythromycin resistance methyltransferase Erm38 reveals its RNA-binding site. J Biol Chem. 2022;298(2):101571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skinner R, Cundliffe E, Schmidt FJ. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983;258(20):12702–6. [PubMed] [Google Scholar]
- 54.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253(4):618–32. [DOI] [PubMed] [Google Scholar]
- 55.Singh S, Bouzinbi N, Chaturvedi V, Godreuil S, Kremer L. In vitro evaluation of a new drug combination against clinical isolates belonging to the Mycobacterium abscessus complex. Clin Microbiol Infect. 2014;20(12):O1124-1127. [DOI] [PubMed] [Google Scholar]
- 56.Wallace RJ Jr, Meier A, Brown BA, Zhang Y, Sander P, Onyi GO, Bottger EC. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40(7):1676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipworth S, Hough N, Leach L, Morgan M, Jeffery K, Andersson M, Robinson E, Smith EG, Crook D, Peto T, Walker T. Whole-genome sequencing for predicting clarithromycin resistance in Mycobacterium abscessus. Antimicrob Agents Chemother. 2019. 10.1128/aac.01204-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woods GL, Brown-Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, Pfyffer GE, Ridderhof JC, Siddiqi SH, Wallace RJ Jr, Warren NG, Witebsky FG. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Wayne: Clinical and Laboratory Standards Institute; 2011. [PubMed] [Google Scholar]
- 59.Johansen MD, Herrmann JL, Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol. 2020;18(7):392–407. [DOI] [PubMed] [Google Scholar]
- 60.Foik IP, Tuszynska I, Feder M, Purta E, Stefaniak F, Bujnicki JM. Novel inhibitors of the rRNA ErmC’ methyltransferase to block resistance to macrolides, lincosamides, streptogramine B antibiotics. Eur J Med Chem. 2018;146:60–7. [DOI] [PubMed] [Google Scholar]
- 61.Fischer TR, Meidner L, Schwickert M, Weber M, Zimmermann RA, Kersten C, Schirmeister T, Helm M. Chemical biology and medicinal chemistry of RNA methyltransferases. Nucleic Acids Res. 2022;50(8):4216–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this article.