Abstract
A study has been made of e.p.r. signals due to Mo(V) in reduced sulphite oxidase (EC 1.8.3.1) from chicken liver. Reduction by SO3(2-), or photochemically in the presence of a deazaflavin derivative, produces spectra indistinguishable from one another. Three types of spectra from the enzyme were distingusihed and shown to correspond to single chemical species, since they could be simulated at both 9 and 35 GHz by using the same parameters. These were the low-pH form of the enzyme, with gav. 1.9805, the high-pH form, with gav. 1.9681 and a phosphate complex, with gav. 1.9741. The low-H form shows interaction with a single exchangeable proton, with A(1H)av. (hyperfine coupling constant) = 0.98 mT, probably in the form of an MoOH group. Parameters of the signals are compared with those for signals from xanthine oxidase and nitrate reductase. The signal from the phosphate complex of sulphite oxidase in unique among anion complexes of Mo-containing enzymes in showing no hyperfine coupling to protons. There is no evidence for additional weakly coupled protons or nitrogen nuclei in the sulphite oxidase signals. The possibility is considered that the enzymic mechanism involves abstraction of a proton and two electrons from HSO3- by a Mo = O group in the enzyme.
Full text
PDF

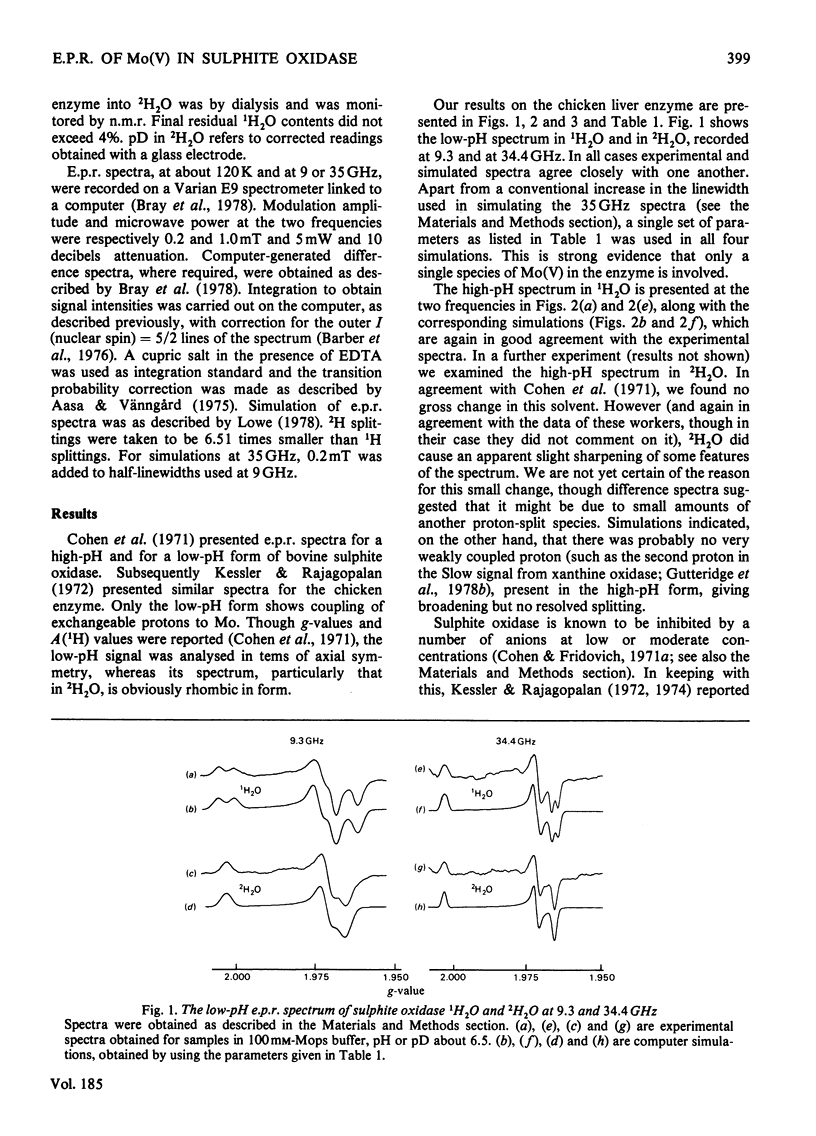



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barber M. J., Bray R. C., Lowe D. J., Coughlan M. P. Studies by electron-paramagnetic-resonance spectroscopy and stopped-flow spectrophotometry on the mechanism of action of turkey liver xanthine dehydrogenase. Biochem J. 1976 Feb 1;153(2):297–307. doi: 10.1042/bj1530297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray R. C., Barber M. J., Lowe D. J. Electron-paramagnetic-resonance spectroscopy of complexes of xanthine oxidase with xanthine and uric acid. Biochem J. 1978 Jun 1;171(3):653–658. doi: 10.1042/bj1710653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray R. C., Vänngård T. "Rapidly appearing" molybdenum electron-paramagnetic-resonance signals from reduced xanthine oxidase. Biochem J. 1969 Oct;114(4):725–734. doi: 10.1042/bj1140725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H. J., Fridovich I., Rajagopalan K. V. Hepatic sulfite oxidase. A functional role for molybdenum. J Biol Chem. 1971 Jan 25;246(2):374–382. [PubMed] [Google Scholar]
- Guiard B., Lederer F. The "b5-like" domain from chicken-liver sulfite oxidase: a new case of common ancestral origin with liver cytochrome b5 and bakers' yeast cytochrome b2 core. Eur J Biochem. 1977 Mar 15;74(1):181–190. doi: 10.1111/j.1432-1033.1977.tb11379.x. [DOI] [PubMed] [Google Scholar]
- Gutteridge S., Tanner S. J., Bray R. C. Comparison of the molybdenum centres of native and desulpho xanthine oxidase. The nature of the cyanide-labile sulphur atom and the nature of the proton-accepting group. Biochem J. 1978 Dec 1;175(3):887–897. doi: 10.1042/bj1750887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart L. I., McGartoll M. A., Chapman H. R., Bray R. C. The composition of milk xanthine oxidase. Biochem J. 1970 Mar;116(5):851–864. doi: 10.1042/bj1160851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Jones H. P., Rajagopalan K. V. In vitro reconstitution of demolybdosulfite oxidase by a molybdenum cofactor from rat liver and other sources. J Biol Chem. 1977 Jul 25;252(14):4994–5003. [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V. Electron paramagnetic resonance of the tungsten derivative of rat liver sulfite oxidase. J Biol Chem. 1976 Sep 25;251(18):5505–5511. [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V. Tryptic cleavage of rat liver sulfite oxidase. Isolation and characterization of molybdenum and heme domains. J Biol Chem. 1977 Mar 25;252(6):2017–2025. [PubMed] [Google Scholar]
- Kessler D. L., Rajagopalan K. V. Hepatic sulfite oxidase. Effect of anions on interaction with cytochrome c. Biochim Biophys Acta. 1974 Dec 29;370(2):389–398. doi: 10.1016/0005-2744(74)90100-4. [DOI] [PubMed] [Google Scholar]
- Kessler D. L., Rajagopalan K. V. Purification and properties of sulfite oxidase from chicken liver. Presence of molybdenum in sulfite oxidase from diverse sources. J Biol Chem. 1972 Oct 25;247(20):6566–6573. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lowe D. J. Electron paramagnetic resonance in biochemistry. Computer simulation of spectra from frozen aqueous samples. Biochem J. 1978 Jun 1;171(3):649–651. doi: 10.1042/bj1710649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V., Hemmerich P. A photochemical procedure for reduction of oxidation-reduction proteins employing deazariboflavin as catalyst. J Biol Chem. 1977 Aug 25;252(16):5612–5614. [PubMed] [Google Scholar]
- Tsopanakis A. D., Tanner S. J., Bray R. C. pH-jump studies at subzero temperatures on an intermediate in the reaction of xanthine oxidase with xanthine. Biochem J. 1978 Dec 1;175(3):879–885. doi: 10.1042/bj1750879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. P., Bray R. C. Electron-paramagnetic-resonance studies on nitrate reductase from Escherichia coli K12. Biochem J. 1978 Jun 1;171(3):639–647. doi: 10.1042/bj1710639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. P. Oxidation--reduction potentials of molybdenum and iron--sulphur centres in nitrate reductase from Escherichia coli. Biochem J. 1979 Feb 1;177(2):757–759. doi: 10.1042/bj1770757. [DOI] [PMC free article] [PubMed] [Google Scholar]


