Abstract
Cre recombinase is a phage-derived enzyme that has found utility for precise manipulation of DNA sequences. Cre recognizes and recombines pairs of loxP sequences characterized by an inverted repeat and asymmetric spacer. Cre cleaves and religates its DNA targets such that error-prone repair pathways are not required to generate intact DNA products. Major obstacles to broader applications are lack of knowledge of how Cre recognizes its targets, and how its activity is controlled. The picture emerging from high resolution methods is that the dynamic properties of both the enzyme and its DNA target are important determinants of its activity in both sequence recognition and DNA cleavage. Improved understanding of the role of dynamics in the key steps along the pathway of Cre-loxP recombination should significantly advance our ability to both redirect Cre to new sequences and to control its DNA cleavage activity in the test tube and in cells.
Keywords: Site-specific recombination, protein dynamics, DNA bending, auto-inhibition, DNA site selection
Introduction
The tyrosine family of site-specific DNA recombinases (Y-SSRs)[1–4] represent powerful tools for genome engineering – insertion, excision, or exchange of genes into precise locations in chromosomes. These enzymes (e.g., λ-integrase, Cre, and Flp recombinases) have proven useful for applications including construction of mouse libraries with nearly every gene under conditional control (http://www.creportal.org/) [5], lineage tracing in developing cells [6] and mapping of synaptic circuits in developing brains (Brainbow) [7]. Moreover, the therapeutic potential of Y-SSRs is illustrated by their use in excision of stably integrated proviral DNA [8,9], and for correcting a genomic inversion associated with Hemophilia A [10]. For in situ applications in eukaryotic cells, Y-SSRs have an advantage over gene editing systems that activate error-prone double-strand DNA repair pathways (as do TALENs, zinc finger nucleases, and CRISPR-Cas9 [11]), because many don’t require additional host-encoded factors and generate intact double-stranded DNA products [3,12,13].
Structural studies of Cre (Causes Recombination; Figure 1) and related tyrosine recombinases have provided high-resolution snapshots of steps along the recombination pathway [3,4,14,15], illuminating a role for both protein and DNA conformational changes (dynamics) in their function. The 343 amino acids of Cre comprise an N-terminal “core-binding” domain (CB) and a C-terminal “catalytic domain” (Cat) wherein resides the eponymous tyrosine residue, connected by an eleven residue linker. Cre and other Y-SSRs share structural and mechanistic similarity to type I-B topoisomerases (topo IB), which form a C-shaped clamp over duplex DNA and release strain by producing single-strand breaks via formation of a covalent DNA-protein phosphotyrosyl intermediate (reviewed in [16]). Conformational fluctuations in topo IB enzymes are found to be important for DNA binding, strand passage, and sensitivity to inhibitors [17]. An important distinction is that while topo IB enzymes function as monomers, Cre and other Y-SSRs require site-specific assembly of higher-order structures in which protein-protein contacts and changes in DNA topology modulate DNA cleavage activity [4,13,18].
Figure 1.
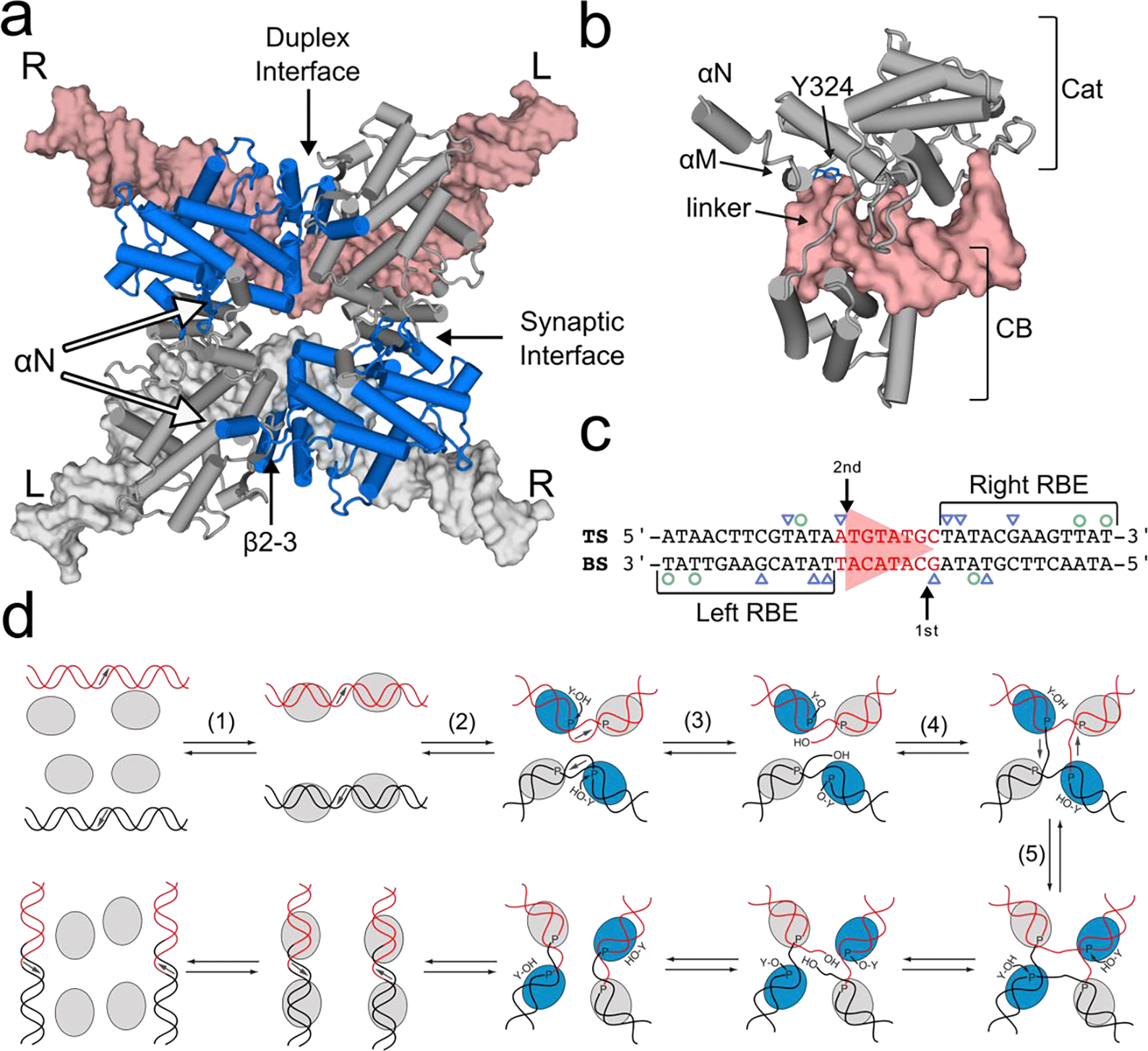
Structure and overall mechanism of Cre-loxP recombination. (a) Structure of the Cre-loxP synaptic complex (PDB 2HOI) comprises two homologous DNA duplexes and four Cre protomers. These antiparallel complexes exhibit approximate C2 symmetry, with the loxP DNA (pink, white) asymmetrically bent, and alternating protomers adopting “active” (blue) and “inactive” (grey) conformations as determined by protein-protein contacts in the synaptic and duplex interface. The C-terminal αN helix from each protomer makes a contact in trans with an adjacent protomer, generating a cyclic pattern of interactions. (b) Each Cre protomer (grey) forms a C-shaped clamp around DNA (pink), in which both the core binding (CB) and catalytic (Cat) domains bind DNA and are separated by a flexible linker. αM contains the catalytic Y324 (blue) and αN extends away from the DNA when bound. (c) The loxP DNA sequence comprises a pair of 13-bp inverted repeats (black; recombinase binding elements; RBEs) separated by an 8-bp asymmetric spacer (red, triangle). Arrows indicate the sites of preferred first and second cleavage on the “top” (TS) and “bottom” (BS) strands of each duplex. Base-specific major groove interactions are highlighted with blue triangles while minor groove interactions are denoted by green circles. (d) Overall mechanism of Y-SSR-mediated DNA recombination, from site selection (1), synapsis of doubly bound target sites (2), cleavage (3), strand exchange and ligation (4) to form a Holliday junction intermediate. In tetrameric complexes only one pair of protomers is thought to be active for DNA cleavage (blue), while a central isomerization step (5) interconverts active and inactive protomers and allows forward progression through the pathway.
Cre catalyzes recombination between a pair of homologous 34 bp loxP sites via a series of steps involving (Figure 1d): (1) recognition and binding of a pair of Cre molecules to its target site containing two inverted repeats (recombinase binding elements, RBEs) separated by an asymmetric 8 bp “spacer”, (2) antiparallel “synapsis” of a pair of Cre dimers on loxP sites (Cre2-loxP) to form a stable tetrameric (Cre2-loxP)2 structure, (3) cleavage of two opposing DNA strands to produce an intermediate with two Cre protomers covalently attached to a 3’-phosphate via a tyrosyl linkage, followed by (4) strand exchange and re-ligation to form a four-way DNA Holliday junction (HJ) intermediate. A central isomerization step (5) that changes the geometry of the HJ intermediate is thought to determine which pair of protomers is active for DNA cleavage, thereby determining the direction by which the Holliday junction is resolved [1,3,13,19].
Despite much success in application of Y-SSRs to problems in molecular and cell biology, barriers to their broader application remain. Two major barriers to their broader application are that the specificity determinants of recombinases for their target sites, and the underlying mechanisms that control recombination outcome are not sufficiently understood to enable effective retargeting to new genomic sites [4,13,20–24]. It is probable that both target site specificity and outcome of the enzymatic reaction are governed by structural properties throughout the recombination pathway, implicating poorly understood protein and DNA dynamics. Here we highlight dynamic properties of the Cre-loxP and other Y-SSR systems and suggest that future exploration of these dynamics will allow us to better understand and apply Y-SSRs in gene editing applications.
DNA site recognition
Complementarity between the shape and electrostatic properties of proteins and their target DNA is critical to achieving sequence specificity. Specificity is achieved by a combination of “direct readout” of DNA bases by amino acid functional groups, and “indirect readout” arising from sequence-specific differences in the shape of the DNA helix [25]. Moreover, specificity requires proteins to overcome the long appreciated kinetic search problem of finding a set of unique “cognate” DNA sequences in a large background of “non-cognate” sequences with similar structures. Postulated solutions to this site search problem invoke three-dimensional diffusion, one-dimensional sliding, hopping, and direct transfer between strands [26–28], all of which require that protein-DNA interactions reflect a balance between loose association for efficient sequence scanning and high affinity binding for achieving specificity [29]. Consistent with these ideas, single molecule tracking experiments in cells show that DNA binding proteins (DBPs) spend most of their time bound to DNA [30] in a “non-specific” manner. Molecular strategies for achieving this balance between loose and tight association are suggested to include the presence of flexible inter-domain linkers in multidomain proteins [31], or negatively charged disordered regions [32].
Structural and biochemical studies have provided an incomplete picture of specificity determinants in Cre-loxP recognition. Crystal structures have shown that upon binding to an RBE each Cre protomer buries ~5000 Å2 in the protein-DNA interface but makes just a few direct contacts to the bases (Figure 1c) [4,14,33]. Nevertheless, high-throughput selection, screening and sequencing of recombined DNA libraries show that recombination is strongly dependent on nucleotide positions throughout the 34 bp site [34–37]. These observations implicate indirect readout (shape recognition) as a major player in Cre-mediated DNA recombination, bringing focus to the complementary structural and dynamic properties of Cre and its target site, loxP.
A high-resolution structure of Cre in the absence of DNA is not currently available, but several pieces of information provide important clues to how it achieves its function. Cre is monomeric in the absence of DNA, and superposition of two-dimensional 1H-15N NMR spectra of full-length Cre and of the isolated CB and Cat domains indicates that they fold independently [38], while the linker likely provides flexibility that would enable reorientation relative to each other. This flexibility could be expected to allow the protein to bind in different modes featuring both transient weak, and long-lived high affinity complexes with DNA, and to enable effective sequence scanning [31]. NMR 15N spin relaxation experiments sensitive to fast motions (< 10−9 s) have revealed dynamics associated with its function [38]. Large amplitude fluctuations are observed in the Cat domain for two loops: the J-K loop, which connects helices that play a role in contacting DNA; and the β2–3 loop, which bears one of the active site residues and forms part of the protein-protein interface that differs between active and inactive protomers in tetrameric assemblies (Figure 1). Upon monomer binding to a single RBE the J-K loop motions are dampened while the β2–3 loop remains flexible. Unexpectedly, the C-terminal region of the protein corresponding to the αN helix is rigid in the absence of DNA, indicating that it is not extended into solution to capture a neighboring protomer (Figure 1b). Additional NMR experiments with a C-terminal deletion or with a paramagnetic spin-labeled C-terminus revealed that in free protein the C-terminal sequence docks in cis, blocking the DNA binding site of the catalytic domain. The functional consequences of this cis-docking mode on target sequence scanning and activation are considered below.
Likewise, there is no direct experimental information on the structure of the Cre-free loxP DNA sequence, but there is abundant evidence that its structural features are important determinants to the efficiency and outcome of recombination reactions. Minimally, because protein-protein interactions are extensive in tetrameric complexes and differ between active and inactive protomers (Figure 1a), the ability to form these interactions depends on the bending propensity of the loxP site. A loxP half-site bound to Cre shows deformations from standard B-form, including widening of the major groove and narrowing of the minor groove at positions adjacent to the spacer [39]; some of these structural deviations are well aligned with predictions based on sequence [40,41]. These deformations become more pronounced in dimeric and tetrameric assemblies. The loxP spacer is observed to bend asymmetrically, with a higher degree of deformation on the AT-rich side [33] and this has been associated with favoring first cleavage to occur at the opposite end of the spacer region (on the bottom strand) [18,33]. Given that there are very few contacts between Cre and the spacer, it seems likely that the bias towards bottom strand cleavage is related to inherent bending of the spacer itself. MD simulations of free and Cre-bound loxP are consistent with the idea that local variation in flexibility and groove dimensions play roles in both site recognition and strand cleavage order [42]. High-resolution structural studies of free Cre and of loxP DNA would help to test these expectations.
Dynamics associated with DNA binding and sequence scanning
The observation that in free Cre its C-terminal region blocks the DNA binding site of the Cat domain [38] raises questions about the functional consequences of this “auto-inhibited” conformation. As conspicuous as is this auto-inhibited state, upon forming a C-clamp structure around a DNA double helix the C-terminus is displaced from the DNA binding interface and extends in a disordered fashion into solution, wherein it is available to dock on a neighboring protomer [38,39]. On purely thermodynamic grounds, the presence of a conformation with blocked or weakened DNA binding affinity would be expected to reduce the overall affinity of Cre for both cognate and non-cognate DNA (Figure 2). As the C-terminus appears only to block the Cat domain, the CB domain could be expected to remain available for transient DNA binding enabling the enzyme to exhibit enhanced rates of DNA scanning. The idea that auto-inhibition could enhance sequence searching in Cre recombinase is supported by studies on DNA scanning by other sequence-specific DNA binding proteins [32,43,44].
Figure 2.
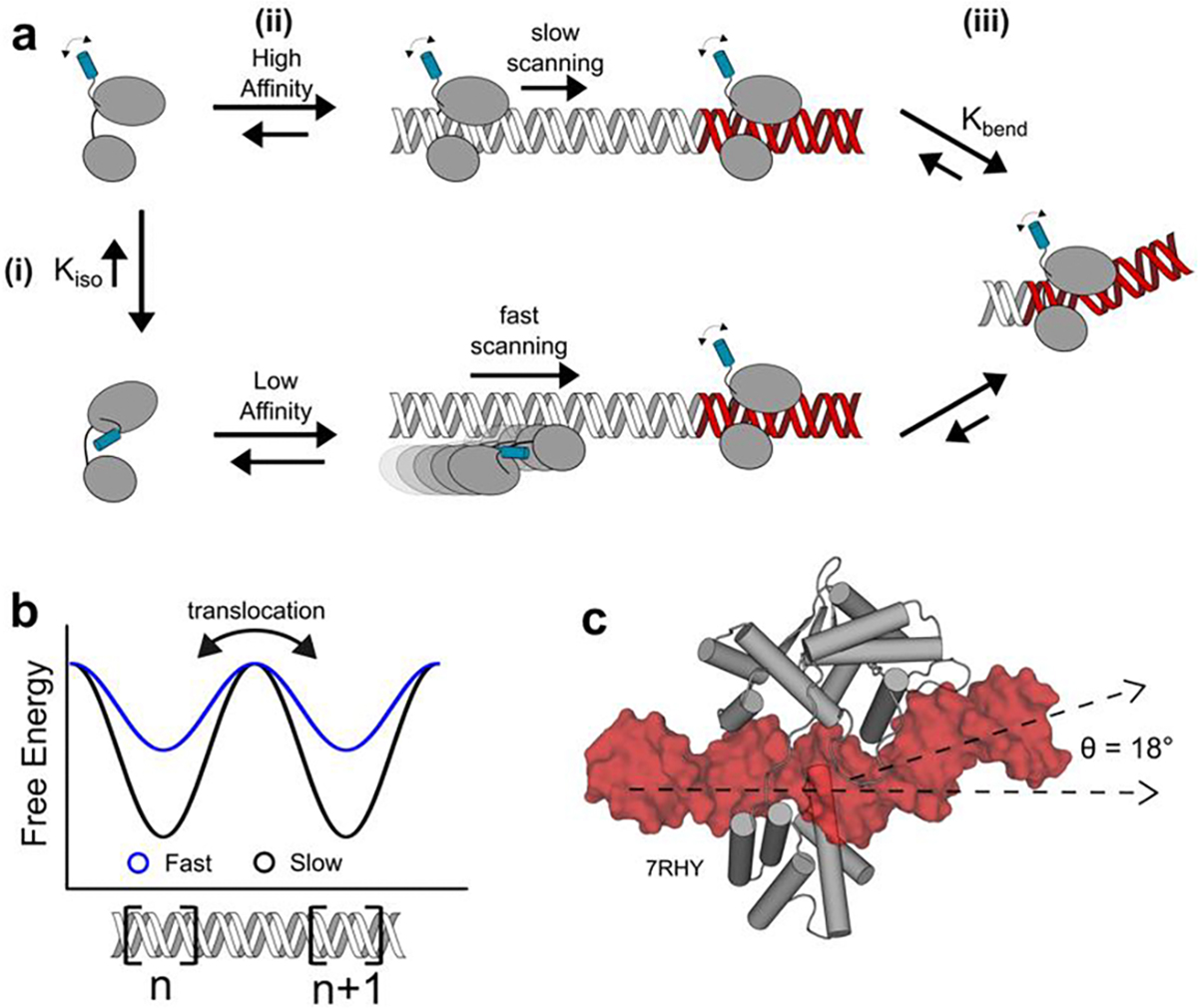
Mechanism of DNA scanning and site selection by Cre recombinase. (a) Equilibrium KIso to an autoinhibited state involving the C-terminus of Cre (blue) allows for two modes of DNA binding (i). Binding by the uninhibited conformation leads to a high affinity, slow scanning complex (top), whereas DNA binding by the autoinhibited state of the protein produces a low affinity, fast scanning complex (ii). Upon reaching a loxP sequence, Cre induces complex-stabilizing conformational changes including DNA bending (iii). (b) Hypothetical free energy landscape of DNA scanning. Low-affinity, fast scanning complexes have smaller energy barriers when translocating between adjacent DNA binding sites, n and n+1, compared to high affinity, slow scanning complexes. (c) Binding of a Cre protomer to an RBE (PDB 7RHY) forms a specific complex featuring an 18° bend in the DNA, along with stabilizing conformational changes in the protein.
Protein and DNA dynamics are also implicated in the observation that site-specific binding of Cre to loxP is accompanied by distortion of the DNA through bending and alternation of groove dimensions (Figure 2 b) [39]. loxP sequences exhibit low GC content, with TA base steps being amenable to protein-induced DNA distortions compared to other sequences with greater GC content [45,46]. Differences in the structures of Cre bound as monomers, dimers and tetramers to DNA [39], implicate protein dynamics in achieving site-specific recognition. The likely importance of binding-coupled conformational changes in site recognition by Cre is underscored by the observation that the protein makes very few base-specific contacts to the 34 bp loxP site (Figure 1). Indeed, single molecule force spectroscopy experiments have shown that when deformation of DNA is disallowed by applying tension, DNA binding proteins that induce bending upon target site binding have decreased specificity [47,48].
Protein-protein interactions regulate Cre
Fluctuating protein-protein interactions (PPIs) are at the crux of Cre function. Upon binding DNA, Cre may capture a second protomer via trans-docking by the C-terminal αN helix, burying ~2600 Å2 of protein-protein surface area. A thus assembled Cre2-loxP dimer can then synapse with another such dimer by both donating and accepting a C-terminal trans docking interaction, burying an additional 8000 Å2 of protein-protein surfaces across the synapse. Although each protomer makes similar contacts with its RBE [13,14], synaptic complexes are two-fold, not four-fold symmetric (Figure 1), and the protein-protein interfaces differ between the duplex and synaptic interfaces. These differences reveal that the PPIs are malleable and able to dynamically sample multiple conformations.
Dynamics are also implicated in controlling DNA cleavage activity by Cre. Single Cre protomers bound to DNA lack cleavage activity [49], and in tetrameric complexes, only alternating protomers adopt an “active” conformation (Figure 1). Comparison of the active site structures of Cre-loxP complexes at different points during assembly to that of a type IB topoisomerase [50] reveal variation in the position of a lysine that serves a general acid, and the tyrosine nucleophile, K201 and Y324 in Cre (Figure 3). These residues are critical for cleavage activity and their misarrangement in lower-order oligomers is consistent with Cre’s lack of topoisomerase-like activity [49]. These variations in the active site structure of Cre during assembly and between protomers in the tetrameric complex are linked to differences in the PPIs. Various sets of PPIs stabilize Cre-loxP complexes while maintaining an “inactive” conformation, while a unique set of PPIs is responsible for producing an “active” conformation for only one protomer on each duplex (Figure 3). The requirement of a unique set of PPIs to activate Cre may provide a mechanism to avoid unproductive DNA cleavage activity. As well, the necessity of conformational changes in both protein and DNA substrate to generate the active conformation is likely to be a source of specificity for Cre and other recombinases, as only certain DNA substrates will adopt conformations that allow these critical PPIs to form.
Figure 3. Protein-protein contacts regulate Cre activity.
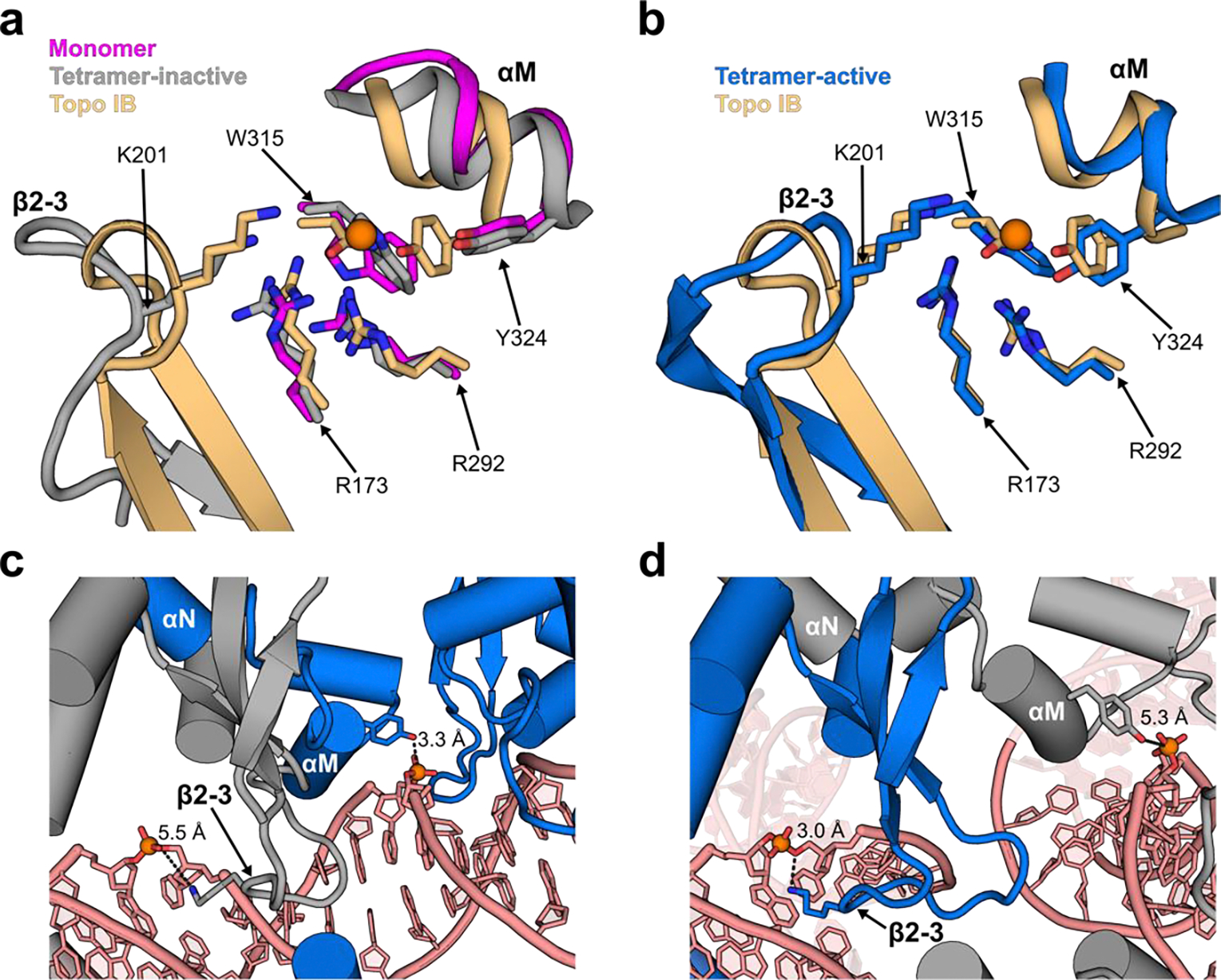
(a) Superposition of active site residues from a Cre monomer bound to a half-site (magenta, 7RHY), the inactive protomer from a synaptic complex (grey, 2HOI) and the DNA-free topoisomerase IB from Deinococcus radiodurans (gold, 2F4Q) reveals mis-positioning of K201 and Y324 relative to the scissile phosphate in the inactive states (orange sphere). (b) By comparison, the active protomer from the synaptic complex (blue) aligns well to the constitutively active topoisomerase IB structure. (c) The duplex interface within the synaptic complex of Cre-loxP displays packing of the β2–3 of the inactive protomer (grey) against the αM helix of the active protomer (blue). This properly positions Y324 of the active protomer, while displacing K201 of the inactive protomer away from the scissile phosphate. (d) The synaptic interface exhibits a different orientation of β2–3 and αM in the two protomers. In this interface the β2–3 of the active protomer (blue) invades the minor groove of loxP and positions K201 near the scissile phosphate for catalysis. With the loss of contact from β2–3 of the active protomer, the αM helix and Y324 of the inactive protomer (grey) are more distant from the scissile phosphate, not poised for nucleophilic attack.
Dynamics and synapse heterogeneity
In addition to playing dominant roles in site selection and control over protomer cleavage activity, protein and DNA dynamics are also likely to directly influence recombination outcomes. As two Cre2-loxP “dimer” complexes synapse to form tetrameric complexes, dynamic equilibria between at least two alternative conformations of the dimer may have deterministic consequences on the recombination pathway (Figure 4). Of particular interest is how these dynamic processes govern the bias for first cleavage in Cre mediated recombination.
Figure 4. Heterogeneity in Cre-loxP synaptic assembly.
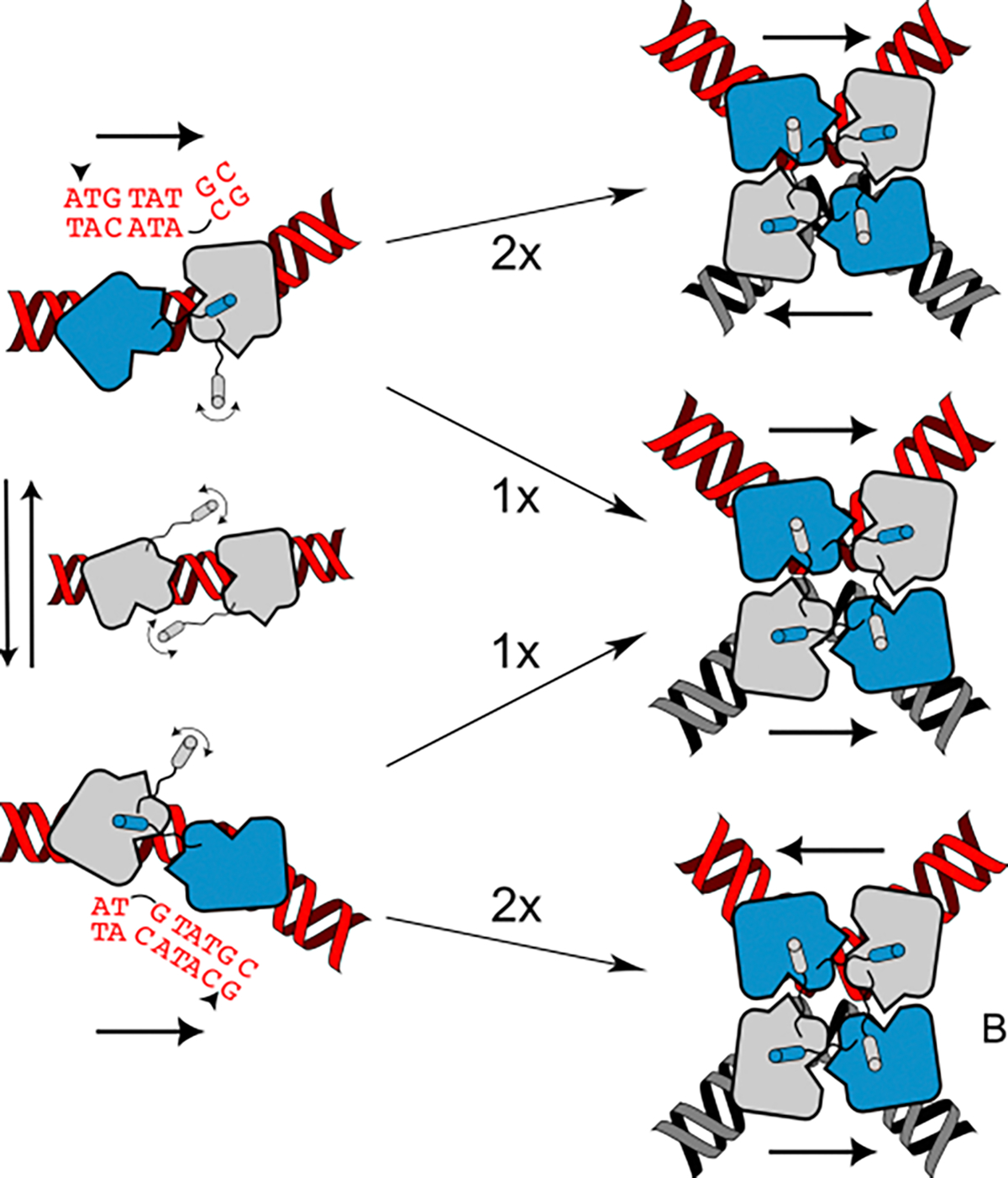
(Left) Cre2-loxP dimers may exchange between conformations in which the protomer on the left RBE donates its αN to the neighboring protomer, and is primed (blue) for cleavage of the top strand, or bent in the opposite direction, favoring bottom strand cleavage by the protomer on the right RBE. Assembly of two like dimers results in anti-parallel synapsis (with respect to the loxP sequence) and subsequent activation of the protomers (blue) for the corresponding first cleavage on the top or bottom strands. Synapsis between dimers with opposite bends results in a parallel tetramer, which due to lack of proper base pairing and possible steric hinderance during recombination is assumed to be unproductive.
In reaction with loxP DNA substrates, Cre preferentially cleaves the bottom-strand (BS) first [51,52]. A variety of experiments using systematic and random variation of the loxP spacer have shown that sequence variants can alter the thermodynamics of synapsis, cleavage bias, and recombination efficiency of Cre-mediated recombination [51–54]. Since formation of productive PPIs by Cre requires large deformations in the DNA substrate, it follows that such mutations may alter the flexibility of pre-synaptic complexes, disfavoring formation of productive conformations.
Bias for BS cleavage is proposed to arise from preferential synapsis of Cre2-loxP dimers with the substrate bent in a direction that primes the protomer bound to the right RBE for cleavage of the BS, whereas dimers bent in the opposite direction prime the protomer on the left RBE for cleavage of the TS (Figure 4) [33,51]. Synapsis between two similarly bent dimers would produce tetramers that are antiparallel with respect to the asymmetric spacer, that are poised for TS or BS cleavage, while synapsis of dimers with opposite bends would generate parallel complexes that are likely dead-end products [51,55–57]. Equal populations of TS and BS dimers would be expected to result in assembly of equal probability of assembling TS, BS and parallel synaptic complexes, whereas a bias in the dimer population favoring either BS or TS cleavage would favor those complexes and disfavor formation of dead-end products. As mentioned above, intrinsic properties of the DNA substrate, and its ability to adopt the conformations necessary to proceed with assembly and recombination [42], are likely important determinants. Thus, understanding the structure and dynamics of Cre2-loxP complexes preceding synapsis, and the mechanisms by which a dynamic equilibrium of dimeric conformations is produced, may be essential for understanding the order of stand cleavage and recombination efficiency.
Dynamics of synaptic tetrameric complexes
Cre and other Y-SSRs exhibit conformational shifts within the synaptic complex before and in conjunction with the first cleavage event [39,58–61]. Synapsis of two Cre2-loxP dimers into a tetrameric complex establishes new protein-protein interactions (PPIs) that activate the enzyme for DNA cleavage [51,53]. Structural evidence shows that synapsis is accompanied by additional DNA bending and rigidification of regions implicated in recombination activity [33,62,63]. Of particular interest is the β2–3 loop which contains active site residue K201. In synaptic complexes the β2–3 loop from inactive protomers pack against the αM helix of the adjacent protomer in the duplex interface (Figure 3), helping to position the neighboring Y324 for cleavage; cleavage-primed protomers display a β2–3 conformation closer to the scissile phosphate, also properly positioning K201 (Figure 1d) [33]. Analysis of structure variability within the cryo-EM data of pre-cleavage CreK201A-loxP tetramers revealed a continuum of DNA bending angles that are coupled to restructuring of the β2–3 loop and thus priming Cre for DNA cleavage (Figure 5). In addition to contributing to activity in synaptic complexes, it seems likely that this structural variability in synaptic complexes provide insight into the types of dynamics that must take place during the downstream steps of Holliday junction isomerization of and resolution.
Figure 5. Pre-cleavage dynamics in Tyr-recombinase synaptic complexes.
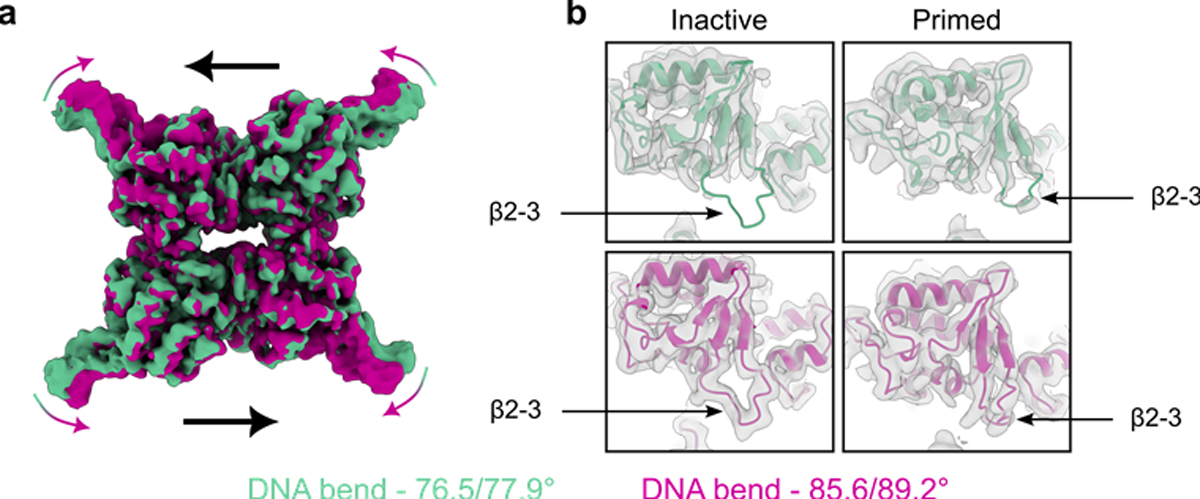
(a) Overlay of electron density maps from first (green) and last (violet) frames of 3D variability analysis of the CreK201A-loxP pre-cleavage synaptic complex.[45] (b) Models of inactive and active Cre protomers fit to the electron density maps of the CreK201A-loxP complex in a, highlighting the change in density and restructuring of the β2–3 loop region. Panels a and b were adapted from [45].
Holliday junction formation and isomerization
Conformational isomerization of the asymmetric Cre-loxP Holliday junction (HJ) intermediate is thought to exchange the active and inactive protomer pairs, interconverting TS and BS complexes, and determining the direction of resolution to duplex DNA [13,64–66]. Formation of the HJ intermediate requires a series of rearrangements in the tetrameric synaptic complex: coordinated cleavage of a pair of strands by active protomers, exchange of the resulting pair of 5’-hydroxyl-bearing single strands across the synapse, and reversal of the 3’-phophotyrosine linkages (Figure 1d, steps 3–4). Mechanistic details are scarce regarding dynamics in these steps [51,66], but macroscopic similarity of the structures of synaptic and HJ complexes suggests that necessary protein and DNA rearrangements are local.
Application of single molecule fluorescence microscopy experiments that are sensitive to dynamics and distance provided insights into the kinetics of steps in the recombination pathway [57]. In a long DNA duplex containing two loxP sites, with FRET donor/acceptor pairs positioned to be able to distinguish TS and BS complexes, experiments with recombination deficient Cre mutants allowed detection of particles with either low or high FRET states. The use of wild-type protein resulted in particles with intermediate FRET, suggesting that the high and low FRET states are being rapidly averaged on a timescale faster the 100 ms exposure time of the camera [57]. Remarkably, this timescale is similar to that measured for isomerization of protein-free Holliday junctions [67]. One interpretation is that the protein is just a passenger and neither constrains nor enhances this intrinsic dynamic behavior of the DNA four-way junction. Alternatively, the protein may play a role in steering dynamics towards productive isomerization.
Knowledge of the structure and dynamics of HJ isomerization could help reveal the role of Cre in the process. In crystals, Cre [65,66] and related Y-recombinases [68,69] adopt a nearly planar “open” conformation wherein the HJ arms splay out in a nearly cruciform fashion. By contrast, naked HJs favor a “closed” conformation, resulting in a pair of coaxial stacks. Open HJ conformations may represent the transition state between stacked isomers and are observed at low ionic strength and in the absence of cofactors [67,70]. Loop-closure kinetics experiments suggest that Cre-loxP tetrameric complexes can adopt non-planar intermediates different from the planar structures seen in crystals [71]. Rotation about the Cre-loxP synaptic interface to generate tetrahedral-like structures is also thought to be stable for the same reason closed-junctions exhibit an interhelical rotation [71]. Thus, Cre binding to loxP under physiological conditions may stabilize open-like structures that help shepherd HJs along the recombination pathway, making use of intrinsic junction dynamics to remodel protein-protein interfaces.
Cre-mediated recombination in eukaryotic cells
Given that Cre is an enzyme of bacteriophage origin, it bears considering how its biophysical properties, which evolved for function in bacteria, enable powerful genome editing in eukaryotic cells. Remarkably, Cre features sequence elements that have been shown to enable its active transport into the eukaryotic nucleus [72]. Moreover, experiments with mammalian chromosomes have shown that while recombination efficiency decreases with genetic distance, it can nevertheless occur when that distance is maximized [73]. Various experiments indicate that nucleosomal packaging presents a barrier to Cre-mediated recombination: increased recombination is observed in mouse spermatids wherein DNA becomes exposed upon chromatin reorganization [74]; increased recombination is observed during the S phase of the cell cycle of synchronized HeLa cells, when DNA is exposed for replication [75]; and recombination frequency with evolved recombinases is strongly correlated with epigenetic marks of active transcription [76]. Nevertheless, there is also evidence that Cre and related recombinases can effectively compete with chromatin for access to its target sites [77]. These observations call for insightful experiments to better understand mechanisms of site selection, synapsis, strand exchange and resolution in the much more complex environment in the eukaryotic nucleus.
Conclusion
Molecular tools to edit genomic DNA in a precise and efficient manner would tremendously advance our understanding of biology and could potentially provide targeted therapy. Structural studies of Cre-mediated DNA recombination have provided exquisite snapshots of steps along the recombination pathway. Those structures have illuminated discrete conformational changes associated with DNA site recognition and protein activation, while providing an incomplete picture of the specificity determinants. With the rapidly expanding list of natural and engineered Cre-like enzymes capable of editing target DNA, effective tools now seem within reach. Improved understanding of the role of dynamics in the key steps along the Cre-loxP recombination pathway should significantly advance that objective.
Acknowledgements
TDB and MJB acknowledge support from NIH T32GM086252. JSM received support from NIH T32GM118291. Research on Cre recombinase in the MPF laboratory was supported by NIH grant R01GM122432.
References
- 1.Craig NL: The Mechanism of Conservative Site-Specific Recombination. Annu Rev Genet 1988, 22:77–105. [DOI] [PubMed] [Google Scholar]
- 2.Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A: Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res 1998, 26:391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grindley NDF, Whiteson KL, Rice PA: Mechanisms of site-specific recombination. Annu Rev Biochem 2006, 75:567–605. [DOI] [PubMed] [Google Scholar]
- 4.Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F: Cre Recombinase and Other Tyrosine Recombinases. Chem Rev 2016, 116:12785–12820. [DOI] [PubMed] [Google Scholar]
- 5.Perry MN, Smith CM, Onda H, Ringwald M, Murray SA, Smith CL: Annotated expression and activity data for murine recombinase alleles and transgenes: the CrePortal resource. Mamm Genome 2022, 33:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian X, Zhou B: Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J Biol Chem 2021, 296:100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW: Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450:56–62. [DOI] [PubMed] [Google Scholar]
- 8.Rojo-Romanos T, Karpinski J, Millen S, Beschorner N, Simon F, Paszkowski-Rogacz M, Lansing F, Schneider PM, Sonntag J, Hauber J, et al. : Precise excision of HTLV-1 provirus with a designer-recombinase. Mol Ther 2023, 31:2266–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar I, Hauber I, Hauber J, Buchholz F: HIV-1 proviral DNA excision using an evolved recombinase. Science 2007, 316:1912–1915. [DOI] [PubMed] [Google Scholar]
- 10.Lansing F, Mukhametzyanova L, Rojo-Romanos T, Iwasawa K, Kimura M, Paszkowski-Rogacz M, Karpinski J, Grass T, Sonntag J, Schneider PM, et al. : Correction of a Factor VIII genomic inversion with designer-recombinases. Nat Commun 2022, 13:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu JH, Davis KM, Liu DR: Chemical Biology Approaches to Genome Editing: Understanding, Controlling, and Delivering Programmable Nucleases. Cell Chem Biol 2016, 23:57–73. [DOI] [PubMed] [Google Scholar]
- 12.Fogg PCM, Colloms S, Rosser S, Stark M, Smith MCM: New Applications for Phage Integrases. J Mol Biol 2014, doi: 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Duyne GD: Cre Recombinase. In Mobile DNA III. Edited by Lambowitz AM, Gellert M, Chandler M, Craig NL, Sandmeyer SB, Rice PA. American Society of Microbiology; 2015:119–138. [Google Scholar]
- 14.Van Duyne GD: A Structural View of Cre-loxP Site-Specific Recombination. Annu Rev Biophys Biomol Struct 2001, 30:87–104. [DOI] [PubMed] [Google Scholar]
- 15.Van Duyne GD, Landy A: Bacteriophage lambda site-specific recombination. Mol Microbiol 2024, n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoeffler AJ, Berger JM: DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys 2008, 41:41–101. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi DT, Gadelle D, Agama K, Kiselev E, Zhang H, Yab E, Petrella S, Forterre P, Pommier Y, Mayer C: Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states. Nat Commun 2022, 13:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibb B, Gupta K, Ghosh K, Sharp R, Chen J, Duyne GDV: Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res 2010, 38:5817–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoess RH, Abremski K: The Cre-lox Recombination System. In Nucleic Acids and Molecular Biology 4. Springer, Berlin, Heidelberg; 1990:99–109. [Google Scholar]
- 20.Gaj T, Sirk SJ, Barbas III CF: Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol Bioeng 2014, 111:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdanove AJ, Bohm A, Miller JC, Morgan RD, Stoddard BL: Engineering altered protein–DNA recognition specificity. Nucleic Acids Res 2018, 46:4845–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt LT, Schneider A, Posorski J, Lansing F, Jelicic M, Jain M, Sayed S, Buchholz F, Sürün D: Quantification of evolved DNA-editing enzymes at scale with DEQSeq. Genome Biol 2023, 24:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt LT, Paszkowski-Rogacz M, Jug F, Buchholz F: Prediction of designer-recombinases for DNA editing with generative deep learning. Nat Commun 2022, 13:7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoersten J, Ruiz-Gómez G, Lansing F, Rojo-Romanos T, Schmitt LT, Sonntag J, Pisabarro MT, Buchholz F: Pairing of single mutations yields obligate Cre-type site-specific recombinases. Nucleic Acids Res 2022, 50:1174–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS: Origins of Specificity in Protein-DNA Recognition. Annu Rev Biochem 2010, 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg OG, Winter RB, von Hippel PH: Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 1981, 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 27.von Hippel PH, Berg OG: Facilitated target location in biological systems. J Biol Chem 1989, 264:675–678. [PubMed] [Google Scholar]
- 28.Halford SE, Marko JF: How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res 2004, 32:3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zandarashvili L, Esadze A, Vuzman D, Kemme CA, Levy Y, Iwahara J: Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc Natl Acad Sci 2015, 112:E5142–E5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stracy M, Schweizer J, Sherratt DJ, Kapanidis AN, Uphoff S, Lesterlin C: Transient non-specific DNA binding dominates the target search of bacterial DNA-binding proteins. Mol Cell 2021, 81:1499–1514.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuzman D, Polonsky M, Levy Y: Facilitated DNA Search by Multidomain Transcription Factors: Cross Talk via a Flexible Linker. Biophys J 2010, 99:1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Bigman LS, Greenblatt HM, Yu B, Levy Y, Iwahara J: Negatively charged, intrinsically disordered regions can accelerate target search by DNA-binding proteins. Nucleic Acids Res 2023, doi: 10.1093/nar/gkad045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo F, Gopaul DN, Duyne GDV: Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc Natl Acad Sci USA 1999, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missirlis PI, Smailus DE, Holt RA: A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination. BMC Genomics 2006, 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bessen JL, Afeyan LK, Dančík V, Koblan LW, Thompson DB, Leichner C, Clemons PA, Liu DR: High-resolution specificity profiling and off-target prediction for site-specific DNA recombinases. Nat Commun 2019, 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamauchi Y, Matsukura H, Motone K, Ueda M, Aoki W: Evaluation of a library of loxP variants with a wide range of recombination efficiencies by Cre. PLOS ONE 2022, 17:e0276657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cautereels C, Smets J, De Saeger J, Cool L, Zhu Y, Zimmermann A, Steensels J, Gorkovskiy A, Jacobs TB, Verstrepen KJ: Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts. Nat Commun 2024, 15:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unnikrishnan A, Amero C, Yadav DK, Stachowski K, Potter D, Foster MP: DNA binding induces a cis-to-trans switch in Cre recombinase to enable intasome assembly. Proc Natl Acad Sci 2020, 117:24849–24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP: Mechanisms of Cre recombinase synaptic complex assembly and activation illuminated by Cryo-EM. Nucleic Acids Res 2022, 50:1753–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R: DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res 2013, 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Chiu T-P, Rohs R: Predicting DNA structure using a deep learning method. Nat Commun 2024, 15:1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abi-Ghanem J, Samsonov SA, Pisabarro MT: Insights into the preferential order of strand exchange in the Cre/loxP recombinase system: impact of the DNA spacer flanking sequence and flexibility. J Comput Aided Mol Des 2015, 29:271–282. [DOI] [PubMed] [Google Scholar]
- 43.Kim J, Ahn D, Park C-J: FOXO4 Transactivation Domain Interaction with Forkhead DNA Binding Domain and Effect on Selective DNA Recognition for Transcription Initiation. J Mol Biol 2021, 433:166808. [DOI] [PubMed] [Google Scholar]
- 44.Krois AS, Dyson HJ, Wright PE: Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci 2018, 115:E11302–E11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Travers AA: The structural basis of DNA flexibility. Philos Trans R Soc Lond Ser Math Phys Eng Sci 2004, 362:1423–1438. [DOI] [PubMed] [Google Scholar]
- 46.Marin-Gonzalez A, Vilhena JG, Perez R, Moreno-Herrero F: A molecular view of DNA flexibility. Q Rev Biophys 2021, 54:e8. [DOI] [PubMed] [Google Scholar]
- 47.Losito M, Smith QM, Newton MD, Cuomo ME, Rueda DS: Cas12a target search and cleavage on force-stretched DNA. Phys Chem Chem Phys 2021, 23:26640–26644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Broek B, Noom MC, Wuite GJL: DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res 2005, 33:2676–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abremski K, Wierzbicki A, Frommer B, Hoess RH: Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J Biol Chem 1986, 261:391–396. [PubMed] [Google Scholar]
- 50.Patel A, Shuman S, Mondragón A: Crystal Structure of a Bacterial Type IB DNA Topoisomerase Reveals a Preassembled Active Site in the Absence of DNA*. J Biol Chem 2006, 281:6030–6037. [DOI] [PubMed] [Google Scholar]
- 51.Ghosh K, Lau C-K, Gupta K, Van Duyne GD: Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat Chem Biol 2005, 1:275–282. [DOI] [PubMed] [Google Scholar]
- 52.Lee L, Sadowski PD: Sequence of the loxP Site Determines the Order of Strand Exchange by the Cre Recombinase. J Mol Biol 2003, 326:397–412. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh K, Guo F, Duyne GDV: Synapsis of loxP Sites by Cre Recombinase. J Biol Chem 2007, 282:24004–24016. [DOI] [PubMed] [Google Scholar]
- 54.Lee G, Saito I: Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 1998, 216:55–65. [DOI] [PubMed] [Google Scholar]
- 55.Hamilton DL, Abremski K: Site-specific recombination by the bacteriophage P1 lox-Cre system: Cre-mediated synapsis of two lox sites. J Mol Biol 1984, 178:481–486. [DOI] [PubMed] [Google Scholar]
- 56.Huffman KE, Levene SD: DNA-sequence asymmetry directs the alignment of recombination sites in the FLP synaptic complex. J Mol Biol 1999, 286:1–13. [DOI] [PubMed] [Google Scholar]
- 57.Pinkney JNM, Zawadzki P, Mazuryk J, Arciszewska LK, Sherratt DJ, Kapanidis AN: Capturing reaction paths and intermediates in Cre- loxP recombination using single-molecule fluorescence. Proc Natl Acad Sci 2012, 109:20871–20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bebel A, Karaca E, Kumar B, Stark WM, Barabas O: Structural snapshots of Xer recombination reveal activation by synaptic complex remodeling and DNA bending. eLife 2016, 5:e19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zawadzki P, May PFJ, Baker RA, Pinkney JNM, Kapanidis AN, Sherratt DJ, Arciszewska LK: Conformational transitions during FtsK translocase activation of individual XerCD– dif recombination complexes. Proc Natl Acad Sci 2013, 110:17302–17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller AN, Xin Y, Boer S, Reinhardt J, Baker R, Arciszewska LK, Lewis PJ, Sherratt DJ, Löwe J, Grainge I: Activation of Xer-recombination at dif: structural basis of the FtsKγ–XerD interaction. Sci Rep 2016, 6:33357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grainge I, Lesterlin C, Sherratt DJ: Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res 2011, 39:5140–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP: Mechanisms of Cre recombinase synaptic complex assembly and activation illuminated by Cryo-EM. Nucleic Acids Res 2022, 50:1753–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo F, Gopaul DN, Van Duyne GD: Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 1997, 389:40–46. [DOI] [PubMed] [Google Scholar]
- 64.Lee L, Sadowski PD: Directional resolution of synthetic holliday structures by the Cre recombinase. J Biol Chem 2001, 276:31092–31098. [DOI] [PubMed] [Google Scholar]
- 65.Gopaul DN, Guo F, Van Duyne GD: Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J 1998, 17:4175–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh K, Lau CK, Guo F, Segall AM, Van Duyne GD: Peptide Trapping of the Holliday Junction Intermediate in Cre-loxP Site-specific Recombination*. J Biol Chem 2005, 280:8290–8299. [DOI] [PubMed] [Google Scholar]
- 67.McKinney SA, Déclais A-C, Lilley DMJ, Ha T: Structural dynamics of individual Holliday junctions. Nat Struct Biol 2003, 10:93–97. [DOI] [PubMed] [Google Scholar]
- 68.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T: A structural basis for allosteric control of DNA recombination by λ integrase. Nature 2005, 435:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Rice PA: The Role of the Conserved Trp330 in Flp-mediated Recombination. J Biol Chem 2003, 278:24800–24807. [DOI] [PubMed] [Google Scholar]
- 70.Lilley DMJ: Structures of helical junctions in nucleic acids. Q Rev Biophys 2000, 33:109–159. [DOI] [PubMed] [Google Scholar]
- 71.Shoura MJ, Giovan SM, Vetcher AA, Ziraldo R, Hanke A, Levene SD: Loop-closure kinetics reveal a stable, right-handed DNA intermediate in Cre recombination. Nucleic Acids Res 2020, 48:4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Y, Gagneten S, Tombaccini D, Bethke B, Sauer B: Nuclear targeting determinants of the phage P1 Cre DNA recombinase. Nucleic Acids Res 1999, 27:4703–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A: Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol 2000, 20:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR: Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci 2000, 97:13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hashimoto M, Taniguchi M, Yoshino S, Arai S, Sato K: S Phase-preferential Cre-recombination in mammalian cells revealed by HIV-TAT-PTD-mediated protein transduction. J Biochem (Tokyo) 2008, 143:87–95. [DOI] [PubMed] [Google Scholar]
- 76.Pandey S, Gao XD, Krasnow NA, McElroy A, Tao YA, Duby JE, Steinbeck BJ, McCreary J, Pierce SE, Tolar J, et al. : Efficient site-specific integration of large genes in mammalian cells via continuously evolved recombinases and prime editing. Nat Biomed Eng 2024, doi: 10.1038/s41551-024-01227-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwikardi M, Dröge P: Use of site-specific recombination as a probe of nucleoprotein complex formation in chromatin. Eur J Biochem 2001, 268:6256–6262. [DOI] [PubMed] [Google Scholar]


