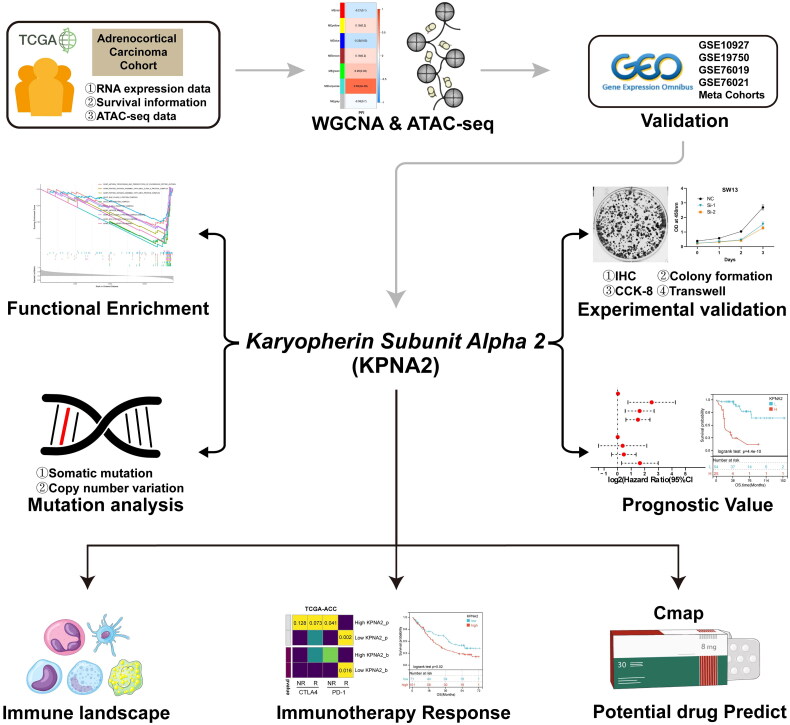
Abstract
Purpose
Adrenocortical carcinoma (ACC) afflicts both pediatric and adult populations and is characterized by dismal prognosis and elevated mortality. Given the inconsistent therapeutic benefits and significant side effects associated with the conventional chemotherapy agent, mitotane, and the nascent stage of immunotherapy and targeted treatments, there is an urgent need to identify novel prognostic biomarkers and therapeutic targets in ACC.
Methods
Utilizing multi-omic datasets from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO), we employed Weighted Gene Co-expression Network Analysis (WGCNA), Cox regression, Receiver Operating Characteristic (ROC) curves, and survival analyses to sift for potential prognostic biomarkers. We subsequently validated these findings through immunohistochemistry and cell assays, and delved into the biological role of KPNA2 in ACC through functional enrichment analysis, mutational landscape, and immune cell infiltration.
Results
A total of 77 progression-associated genes with aberrant chromosomal accessibility were discerned within the TCGA-ACC dataset. By integrating ROC and Cox regression from GEO datasets, KPNA2 emerged as an independent risk factor portending poor outcomes in ACC. ATAC-seq analysis revealed attenuated chromatin accessibility of KPNA2 in cases with unfavorable prognosis. Immunohistochemistry corroborated elevated KPNA2 expression, which was linked to enhanced proliferation and invasion. Elevated KPNA2 levels were found to activate oncogenic pathways while simultaneously suppressing immunological responses. Immune infiltration analysis further revealed a decrement in CD8+ T-cell infiltration in KPNA2-high cohorts.
Conclusion
This study demonstrates the clinical and biological significance of KPNA2 in ACC and suggests that KPNA2 could serve as a promising biomarker for predicting prognosis and immunotherapeutic responses in pediatric and adult ACC patients.
Keywords: Adrenocortical carcinoma, ATAC-seq, KPNA2, prognosis, immunotherapy
Introduction
Adrenocortical carcinoma (ACC) is a rare endocrine malignancy with an annual incidence rate of 0.5–2 cases per million adults and 0.2–0.3 cases per million children globally [1]. The overall 5-year survival rate for ACC is a mere 35%, with stage 4 patients experiencing less than a 10% 5-year survival rate [2]. Various therapeutic approaches for ACC encompass surgical resection, radiation therapy, chemotherapy, and targeted therapies [3,4]. For patients diagnosed at an early stage, surgical resection is typically the treatment of choice, especially when the tumor has not metastasized to other organs [5]. However, the efficacy of treatment may be compromised due to the tumor’s inherent resistance to conventional chemotherapy, particularly in advanced stages [6]. Recent years have seen intensified efforts in exploring the molecular mechanisms and biological characteristics of ACC in order to develop more therapy options, such as immunotherapy and targeted drugs [4,5]. Despite these advancements, the application of immunotherapy in ACC is still fraught with challenges due to inconsistent therapeutic outcomes, tumor heterogeneity, immune escape mechanisms, and adverse side effects [7,8]. Therefore, the identification of a novel biomarker for ACC could facilitate improved clinical management for therapeutic response and prognosis.
The heterogeneity of ACC serves as a pivotal determinant for its unpredictable prognosis and limitations in treatment modalities [9]. Multi-omics, an interdisciplinary approach amalgamating genomics, transcriptomics, proteomics, and metabolomics, offers an intricate exploration of ACC’s tumor heterogeneity at various molecular layers [10,11]. This promises to facilitate the identification of novel biomarkers. Within the multi-omics framework, Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) stands as a crucial methodology. It enables the identification of cancer-related genomic regions, including chromatin changes induced by mutations, oncogenes, tumor suppressor genes, and transcription factor binding sites [12,13]. By leveraging ATAC-seq, we can perform differential accessibility analysis to explore which protein-coding genes in ACC are influenced by the accessibility of non-coding regulatory elements. This not only aids in identifying potential biomarkers for ACC but also unveils prospective targets for immunotherapies and pharmacological interventions
In the present study, we identified Karyopherin Subunit Alpha 2 (KPNA2) as a potential biomarker for ACC. Previous research indicates that KPNA2 functions as a nuclear import protein, primarily involved in the regulation of protein transport between the cell nucleus and the cytoplasm. It plays a critical role in the processes of cancer cell growth and metastasis [14]. Numerous studies have demonstrated that aberrant overexpression of KPNA2 is closely associated with the progression, and therapy resistance of various cancers [15–17]. However, to date, there has been no reported research specifically investigating the role of KPNA2 in ACC.
Materials and methods
Data collection and processing
The baseline characteristics of the datasets employed in the present study are in Supplementary Table S1. To initiate this study, we downloaded RNA-seq data, corresponding mutation data, and clinical information for the adrenocortical carcinoma cohort from The Cancer Genome Atlas (TCGA-ACC) via the UCSC Xena platform [18]. We utilized the R packages clusterProfiler (version 4.8.1) [19] and org.Hs.eg.db (version 3.17.0) [20] to convert Ensembl IDs to SYMBOL IDs in the RNA-seq data. For ATAC-seq data, we employed the raw count matrix, normalized count matrix, and bigWig files acquired from the TCGA. From TCGA-ACC ATAC-seq, we selected 14 samples with matching RNA-seq data and clinical information for differential accessibility peak (DAP) analysis. As validation sets, we harvested four ACC sample datasets for both adults and pediatric cases from the GEO database [21–23]: Adults (GSE19750, GSE10927) and Pediatrics (GSE76019, GSE76021). Batch effects were corrected using the ComBat function in the sva R package (version 3.48.0) [24].
WGCNA and key module identification
Weighted Gene Co-expression Network Analysis (WGCNA) was employed for network-based gene filtering, aimed at detecting markers with specific attributes, such as progression. To delineate potential high co-expression gene clusters, we used the WGCNA package (version 1.72-1) [25] to construct a gene co-expression network for the TCGA-ACC. To obtain a more comprehensive analysis, we employed the ‘one-step’ method in WGCNA analysis, incorporating mRNA expression data (n = 19,563) from the TCGA-ACC dataset. To meet the conditions of a scale-free network, we determined the optimal soft-thresholding power (β = 16) and transformed the adjacency matrix into a topological overlap matrix (TOM). Additionally, we calculated the corresponding dissimilarity (1-TOM) and identified PFI-associated modules using the dynamic tree cutting method.
Identification and validation of prognostic genes
To identify key genes closely related to ACC progression, we employed the timeROC R package (version 0.4) [26] to compute the Area under curve (AUC) for assessing the predictive capacity of genes in candidate modules. In the TCGA-ACC cohort, genes with AUC > 0.7 were subjected to univariate Cox regression analysis to identify prognostic genes for overall survival. Subsequently, these results were further validated in GEO cohorts. We employed the R package survminer (version 0.4.9) (https://CRAN.R-project.org/package=survminer) and set the minimum group sample size to be greater than 30%. The optimal cutoff value for KPNA2 was calculated, dividing patients into high and low expression groups. Subsequently, we used the R package survival (version 3.5–5) (https://CRAN.R-project.org/package=survival) to analyze prognostic differences between the two groups. The log-rank test was applied to assess the significance of prognostic differences between samples in different groups [27].
Functional enrichment
Spearman correlation analysis was employed to evaluate the association between KPNA2 expression and all other genes. The clusterProfiler R package (version 4.8.1) [19] was used for functional enrichment analysis to identify significantly enriched terms related to Gene Ontology (GO).
Immunohistochemistry (IHC)
According to the protocol outlined in our previous studies [28], a brief description is provided below. ACC and adrenal adenoma tissue samples used in this study were sourced from Huizhou Central Hospital, with ethical approval granted by its ethics committee (No. KYLL2023105). All patients provided informed consent prior to the collection of their tissue samples. Samples were fixed in 4% paraformaldehyde prior to paraffin embedding. Tissue sections of 4 µm thickness were treated with 1% H2O2 solution, then blocked with non-immune goat serum. Sections were incubated with primary antibodies overnight at 4 °C, followed by a 30-minute incubation with biotinylated secondary antibodies at room temperature. Final scores were calculated by summing the percentages of positively stained cells and their staining intensities. Scoring was as follows: 0 (0%), 1 (1%–10%), 2 (11%–50%), 3 (>50%) for cell percentages; and 0 (negative), 1 (weak), 2 (moderate), 3 (strong) for staining intensity [29]. An anti-KPNA2 antibody (Immunoway, YT5691) was utilized.
Cell transient transfection
According to the protocol outlined in our previous studies [28], a brief description is provided below. The present study employed two human ACC cell lines, SW13 and NCI-H295R. The cells were cultured in DMEM (BC-M-005, Bio-Channel) and DMEM/F12 medium (BC-M-002, Bio-Channel), both supplemented with 10% fetal bovine serum (BC-SE-FBS07, Bio-Channel), and maintained in a humidified incubator at 37 °C with 5% CO2. According to the manufacturer’s instructions, negative control (NC) and KPNA2 siRNA (Genepharma, Suzhou, China) were transfected into ACC cells using GP-transfect-Mate (Genepharma, Suzhou, China). Plates were incubated for 48 h before total protein was harvested for Western Blot analysis. The siRNA sequences were showed in Table S2.
Western Blot
According to the protocol outlined in our previous studies [28], a brief description is provided below. Cells were harvested and lysed in RIPA buffer containing protease inhibitors. The resulting protein samples were separated by SDS-PAGE and transferred to PVDF membranes, which were blocked using 5% non-fat milk. Membranes were incubated with a primary anti-KPNA2 antibody (YT5691, Immunoway) and anti-β-actin (20536-1-AP, Proteintech), both at a 1:2000 dilution, followed by incubation with a secondary antibody (SA00001-2, Proteintech) at a 1:5000 dilution. Membranes were then washed thrice with PBST for 10 min each and exposed. β-actin served as a normalization control, and band intensities were quantified using Image J software.
Cell assays
According to the protocol outlined in our previous studies [28], a brief description is provided below.
For the CCK8 proliferation assay, approximately 4 × 103 transiently transfected cells were allocated to each well of a 96-well plate containing 100 μL of culture medium. Optical density at 450 nm was gauged 2 h post-addition of a 1:9 CCK8 solution at time intervals of 2, 24, 48, and 72 h using a spectrophotometer.
In the clonogenic assay, cells were plated in 6-well plates at a density of 1000 cells/well and incubated at 37 °C in a 5% CO2 atmosphere for a fortnight. Subsequent to dual PBS washes, cells were fixated with 4% paraformaldehyde for a quarter-hour and then stained with 1% crystal violet for 20 min at ambient temperature. The resulting colonies were enumerated, and the assay was performed in triplicate.
To evaluate invasive potential, a transwell assay was utilized. Around 4 × 104 transfected cells were seeded into the upper chamber containing serum-free medium, while the lower chamber was supplemented with complete medium. Following a 48-hour incubation under standard culture conditions, cells were PBS-washed, fixated in paraformaldehyde, and stained with 0.1% crystal violet. Subsequently, the stained cells were microscopically inspected and quantified.
Landscape of ACC mutations
The ‘maftools’ R package (version 2.16) [30] was used to calculate tumor mutational burden (TMB) for each patient. To investigate whether genomic mutations differed between high and low KPNA2 expression groups, a mutation waterfall plot was generated, visualizing the top 20 significantly mutated genes (SMGs) in ACC using the maftools and ComplexHeatmap R packages (version 2.16) [31]. Copy number variation (CNV) waterfall plots of the top 10 amplified and deleted chromosomal segments in ACC were also produced. Chi-square tests were employed to examine differences in CNV between KPNA2 expression subgroups, and Wilcoxon tests were conducted to evaluate differences in KPNA2 expression levels among mutated subgroups.
Assessment of immune cell infiltration
Immune scores of TCGA-ACC and GSE76019 were evaluated using the ‘IOBR’ R package (version 0.99.9) [32]. Scores for 22 types of immune cell infiltration across five algorithms were obtained. Spearman correlation analysis was employed to assess the relationship between KPNA2 expression and various immune cell scores.
Prediction of immunotherapy response and targeted drug efficacy
We employed the Subclass Mapping (Submap) algorithm [33] to predict responses to immune checkpoint blockade (ICB) therapy. We analyzed transcriptomic expression patterns between patient groups with differing KPNA2 expression levels and divergent immunotherapy responses. A p-value less than 0.05 was considered indicative of significant similarity between the subclasses. We then curated a collection of four immunotherapy datasets, namely Braun [34], GSE78220 [35], GSE91061 [36], and PRJNA482620 [37], from the Tiger database [38]. Anti-PD-1 immunotherapy samples were isolated and categorized based on optimal KPNA2 expression cut-off values for survival analysis, aiming to investigate the prognostic utility of KPNA2 expression in anti-PD-1 immunotherapy. Additionally, we utilized the Connectivity Map (CMap) [39], a data-driven systemic approach for identifying relationships among genes, chemical substances, and biological conditions, to identify potential compounds targeting ACC-associated pathways. Further specificity analyses were conducted using the CMap tool to elucidate mechanisms of action (MoA) and drug targets.
Statistical analysis
Data were analyzed and visualized using R version 4.3.1. A subset of the data visualization was performed using the Sanger Box bioinformatics analysis online tool [40]. Statistical analyses of immunohistochemistry and cellular experimental data were carried out using GraphPad Prism 8.0 software with a copyright license. The Wilcoxon rank-sum test and Kruskal–Wallis test were utilized for comparing differences between two or more groups. All p-values are two-sided, with statistical significance denoted as *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Identification of ACC progression-related gene modules through WGCNA
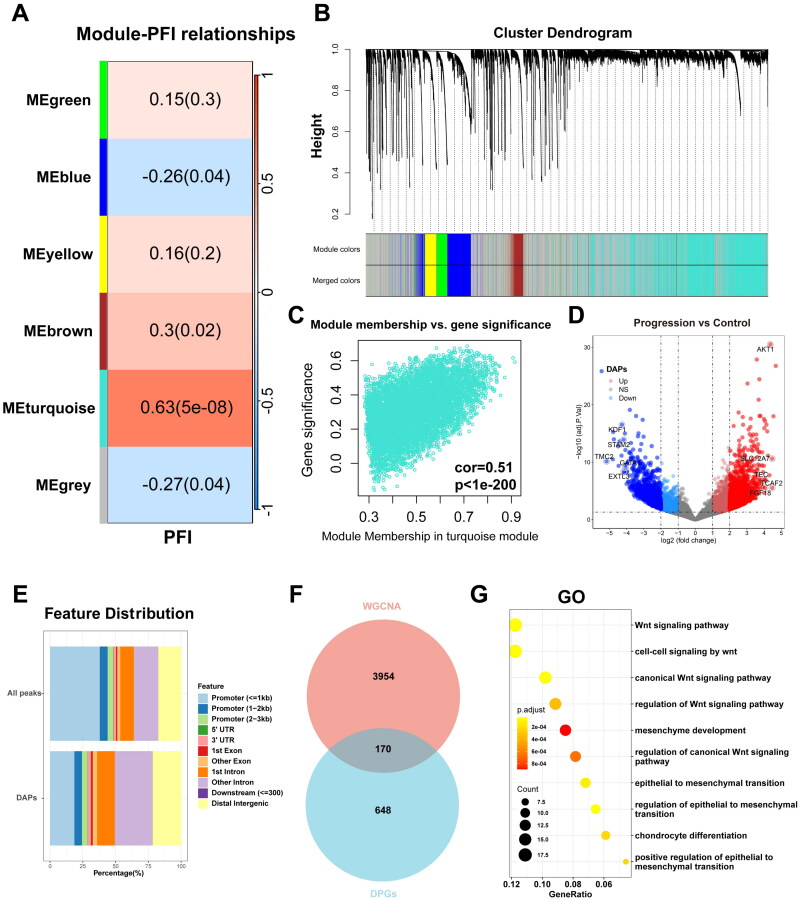
The workflow of the current study is depicted in Figure 1. Initially, we clustered the transcriptome sequencing dataset of 79 samples from TCGA-ACC based on median progression-free interval (PFI) times (Figure S1A). Subsequently, we performed WGCNA using 0.2 and 16 as the module merging threshold and minimum module size, respectively (Figure S1B). A heatmap was utilized to explore the relationships between the identified gene modules and PFI, resulting in six distinct gene modules (Figure 2A,B). Notably, the turquoise gene module exhibited a strong correlation with ACC progression (r = 0.63, p = 5e-08, Table S3). Additionally, in the Gene Significance vs Module Memberships plot, turquoise module genes displayed consistent results (r = 0.51, p = 1e-200) (Figure 2C). Consequently, after eliminating genes lacking statistical significance, we identified the turquoise module genes as those most highly correlated with ACC progression.
Figure 1.
The flowchart of this study.
Figure 2.
Identification of progression-related genes with abnormal chromosomal accessibility in ACC. (A) Heatmap delineating gene module correlations with PFI. (B) Hierarchical gene clustering at the optimized soft-threshold. (C) Scatter plot displaying correlations within the turquoise module. (D) ATAC-seq volcano plot contrasting differential peaks in control and progression groups. (E) ChIPseeker-annotated bar chart of peak percentages. (F) Venn diagram showcasing overlap between WGCNA turquoise module genes and differential peaks. (G) GO analysis by progression-related genes with abnormal chromosomal accessibility.
Identification of aberrantly accessible differential peaks associated with ACC progression using ATAC-seq
Utilizing ATAC-seq as one of the multi-omics technologies, we explored the tumor heterogeneity in ACC and sought to identify aberrantly accessible differential peak genes associated with the progression of ACC. We performed a differential peak analysis on the ATAC-seq data from the aforementioned TCGA-ACC samples, categorized based on their median PFI values. TCGA-ACC consisted of 4 samples in the Control group and 10 in the Progression group, resulting in the identification of 3120 DAPs (Figure 2D, adjPval < 0.05, | log2 FC | > 2). Through peak-to-gene mapping, we identified 810 Differential Peak Genes (DPGs). Further annotation using the ChIPseeker package revealed that the percentage of distal elements, defined as non-promoter elements, was higher in DAPs compared to ALL peaks. This indicates a stronger specificity in distal elements’ response to ACC progression (Figure 2E). By intersecting genes from the turquoise module with DPGs, we identified 170 overlapping genes related to ACC disease progression (Figure 2F, Table S4) and conducted GO analysis (Figure 2G, Table S5). We found that these genes primarily enriched in the Wnt signaling pathway.
Identification of predictive biomarkers for ACC progression
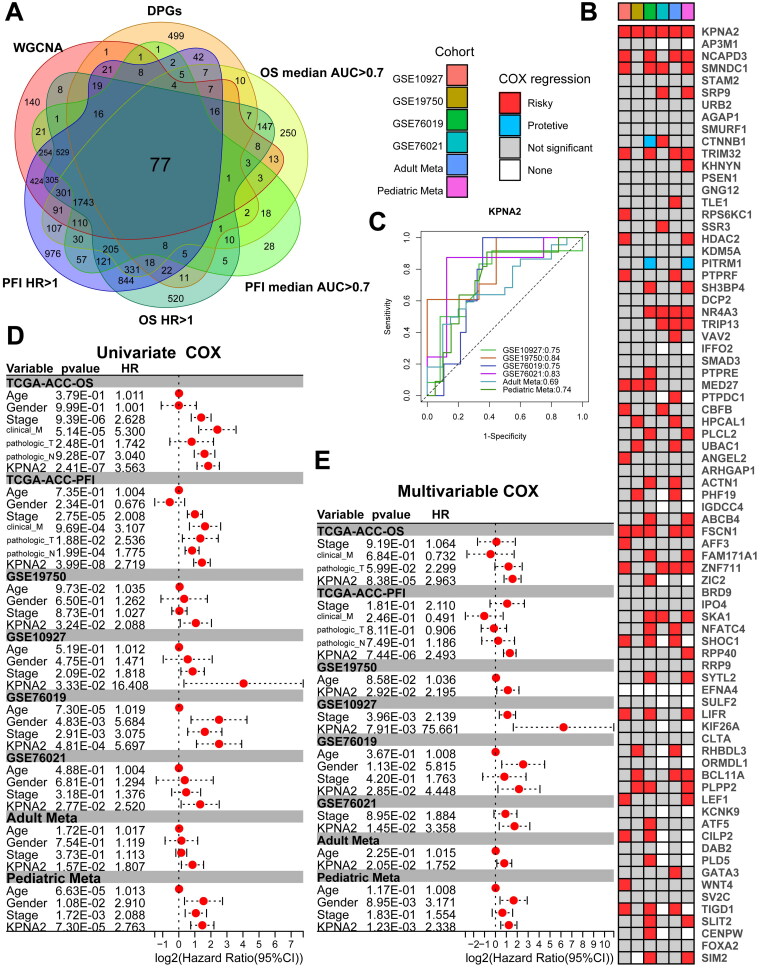
To identify key biomarkers within the aberrantly accessible gene set associated with ACC progression, we performed time-dependent univariate Cox regression analysis on all genes in the TCGA-ACC transcriptome sequencing dataset. These analyses were stratified by both median Overall Survival (OS) time and median PFI. We filtered for genes with an AUC greater than 0.7 and a Hazard Ratio (HR) greater than 1. The intersection with the ACC-related gene set yielded 77 candidate genes (Figure 3A). Subsequently, we leveraged the GEO database to obtain adult (GSE19750, GSE10927) and pediatric (GSE76019, GSE76021) ACC datasets and performed batch correction (Figure S2A–D). The adult datasets were prognostically anchored on OS, while the pediatric datasets were based on Event-Free Survival (EFS). We generated a total of six distinct validation cohorts. Upon conducting univariate Cox regression analyses across these cohorts for the previously identified set of 77 genes, we discerned that only KPNA2 consistently emerged as a significant prognostic risk factor across all cohorts (Figure 3B). Not only did KPNA2 display strong predictive power for adverse prognosis in ROC analysis (Figure 3C), but it also emerged as an independent prognostic factor for ACC patients in both univariate and multivariate Cox regression models after adjusting for other clinical characteristics (Figure 3D,E). Consequently, we postulate that KPNA2 could be a promising biomarker for ACC.
Figure 3.
Identification of KPNA2 as an ACC biomarker. (A) Venn diagram illustrating the overlap among WGCNA-derived genes, DPGs, TCGA univariate COX, and genes with AUC > 0.7. (B) Heatmap depicting univariate COX of intersecting genes in GEO datasets. (C) AUC metrics for KPNA2 across GEO datasets. (D, E) Univariate and multivariate regression assessing KPNA2 in TCGA and GEO cohorts.
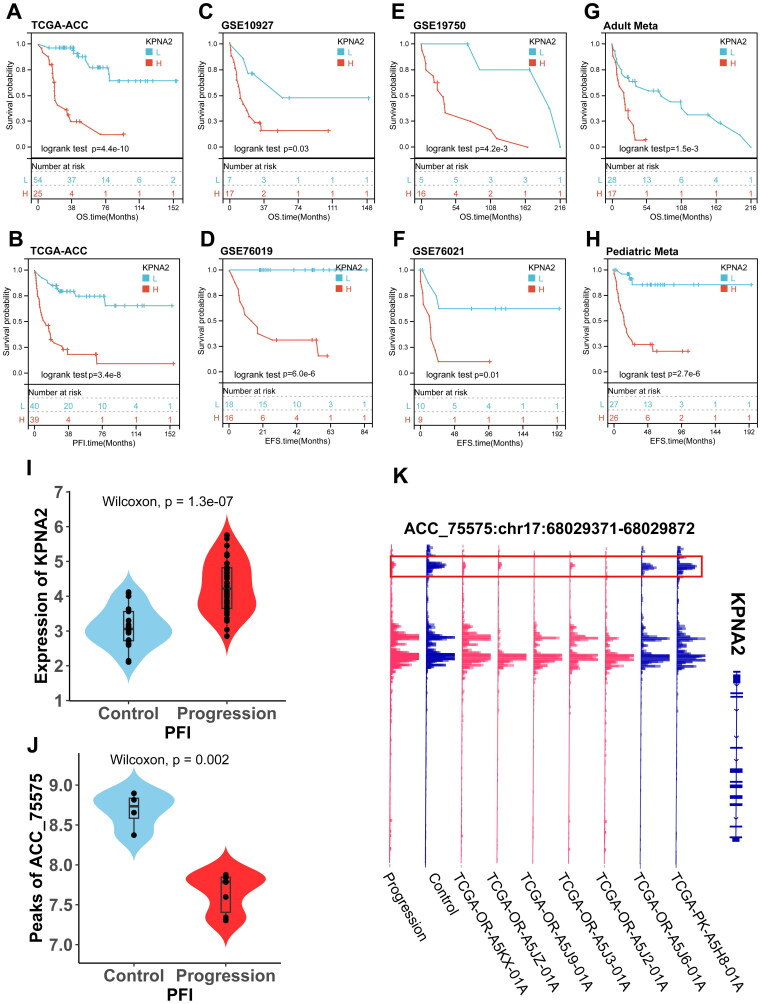
Furthermore, we performed Kaplan-Meier survival analyses within these cohorts. The results indicated that patients with ACC who exhibited elevated levels of KPNA2 expression manifested a significant trend towards poorer outcomes (Figure 4A–H). Additionally, we observed that in the TCGA-ACC cohort, KPNA2 expression levels were markedly higher in the Progression group compared to the Control group (p = 1.1e-07) (Figure 4I). In the ATAC-seq, the peak values corresponding to KPNA2 displayed variations in the Progression group. The Integrative Genomics Viewer (IGV) indicated a significant reduction in chromatin accessibility at the Distal Intergenic region corresponding to ACC-75575 (p = 0.002) (Figure 4J,K). Collectively, these findings suggest that elevated expression of KPNA2 portends adverse prognostic implications in multiple adult and pediatric ACC cohorts and may be associated with aberrant chromatin accessibility.
Figure 4.
KPNA2 Exhibits abnormal chromosomal accessibility and serves as a prognostic risk factor in ACC. (A–H) Kaplan–Meier survival analyses for KPNA2 across TCGA-ACC and GEO datasets. (I) Violin plot illustrating KPNA2 expression disparities between control and progression cohorts in TCGA-ACC. (J) Violin plot depicting ATAC-seq-derived differential peaks associated with KPNA2 in TCGA-ACC. (K) ATAC-seq gene track plot, with KPNA2-associated differential peaks accentuated in red frames.
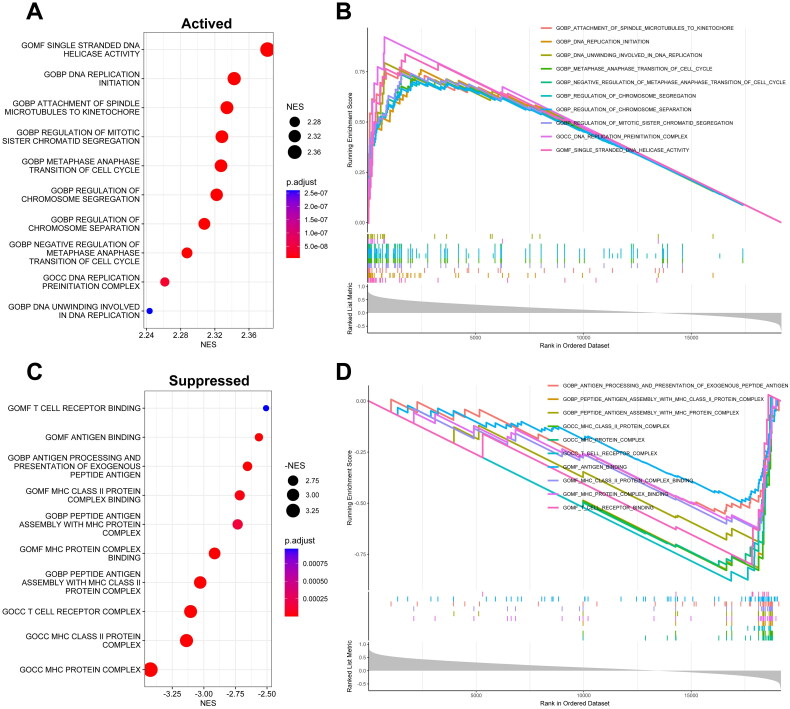
Functional enrichment analysis of KPNA2
To explore the biological functions of KPNA2, we employed Gene Set Enrichment Analysis (GSEA). As depicted in Figure 5, we selected the top 10 terms for both activation and inhibition based on the absolute values of the Normalized Enrichment Scores (NES). Notably, the activation set included terms related to cell proliferation such as ‘DNA Replication Initiation,’ ‘DNA Unwinding Involved in DNA Replication,’ and ‘DNA Replication Preinitiation Complex’ (Figure 5A,B, Table S6). Conversely, the inhibition set comprised immune-related terms such as ‘T Cell Receptor Complex,’ ‘T Cell Receptor Binding,’ ‘Antigen Binding,’ and ‘MHC Protein Complex’ (Figure 5C,D, Table S6). Based on these findings, KPNA2 may contribute to the malignant progression of ACC by activating pathways involved in tumor cell proliferation and growth, while suppressing processes related to antigen presentation and T cell activation.
Figure 5.
Functional Enrichment profiling of KPNA2. (A and B) Top 10 gene ontology (GO) terms depicting activation, ranked by normalized Enrichment score (NES). (C and D) top 10 GO terms indicating suppression, likewise ranked by NES.
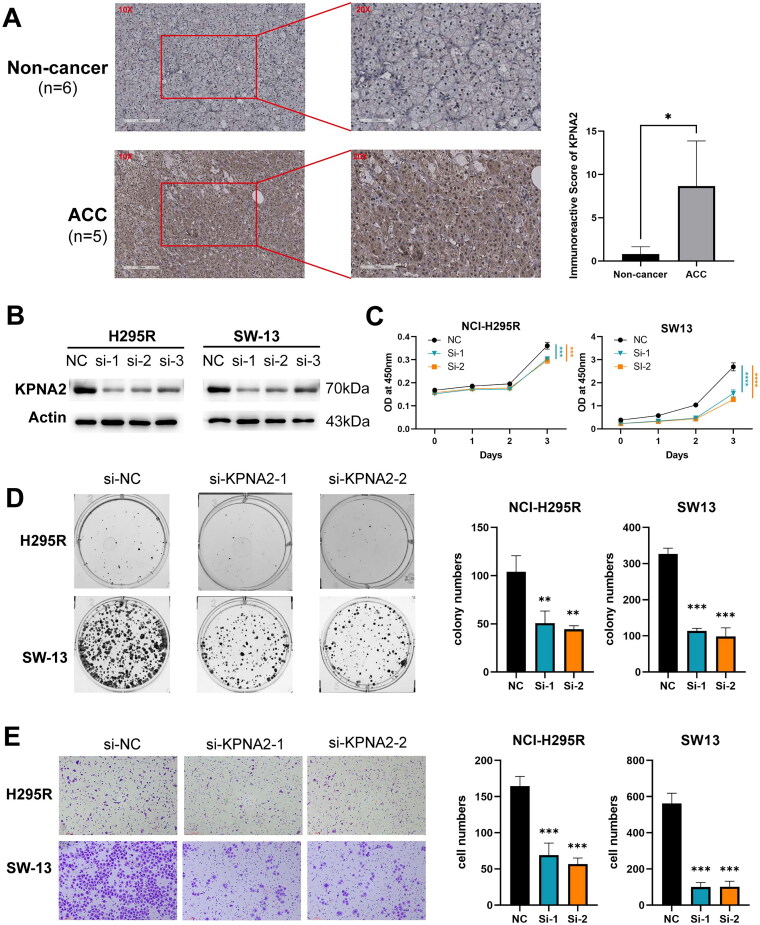
Experimental validation of KPNA2’s role in ACC
To further investigate the impact of KPNA2 on the phenotypic behavior of ACC cells, we performed a series of experimental analyses. To ascertain the expression profile of KPNA2 in ACC, clinical samples were collected and subjected to immunohistochemistry. Representative images of KPNA2 immunohistochemical staining are presented in Figure 6A. KPNA2 expression was predominantly localized in the cell membranes and cytoplasm of adrenal cells. Notably, the expression levels of KPNA2 protein were significantly higher in the ACC group compared to the non-cancerous group (p < 0.05). Moreover, we employed loss-of-function assays to validate the role of KPNA2 in ACC cells. As demonstrated in Figure 6B, siRNA1 and siRNA2 effectively knocked down KPNA2 expression in both SW13 and NCI-H295R cell lines. CCK-8 growth curves indicated that the downregulation of KPNA2 significantly inhibited the proliferation of SW13 and NCI-H295R ACC cells (Figure 6C). Colony formation assays further revealed that the downregulated KPNA2 led to a marked reduction in the number of cellular colonies formed by SW13 and NCI-H295R cells (Figure 6D). Additionally, transwell assays demonstrated that knockdown of KPNA2 substantially suppressed the invasiveness of ACC cells (Figure 6E). In summary, KPNA2 is overexpressed in ACC and promotes proliferation and invasion of ACC cells.
Figure 6.
Experimental validation of KPNA2. (A) Immunohistochemical staining micrographs accompanied by semi-quantitative KPNA2 analysis. (B) Western blot assessment of KPNA2 expression in SW13 and H295R. (C) CCK8 assay elucidating the proliferative potential of SW13 and H295R. (D) Clonogenic assay demonstrating the colony-forming abilities of SW13 and H295R. (E) Transwell migration assay quantifying invasion capabilities of SW13 and H295R. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
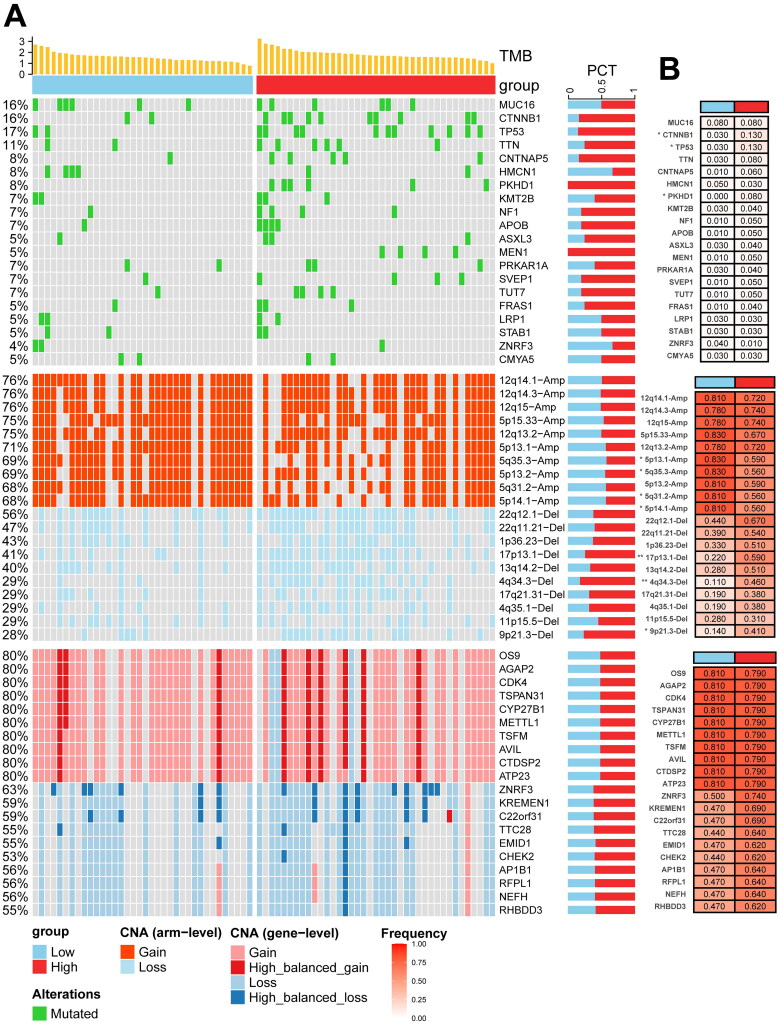
Mutational landscape in relation to KPNA2 expression in ACC
To further elucidate the role of KPNA2 in ACC from a multi-omics perspective, we examined the top 20 SMGs as well as the top 10 gain and loss CNVs at both arm-level and gene-level (Figure 7A). We then stratified these analyses by KPNA2 expression levels (Figure 7B). Firstly, ACC samples with elevated KPNA2 expression exhibited a significantly increased TMB (p = 0.0054) (Figure S3A). To investigate the impact of KPNA2 expression on tumor heterogeneity in ACC, we conducted inter-subgroup analyses focusing on the aforementioned SMGs and CNVs. Our results revealed that the KPNA2-high expression subgroup exhibited a higher prevalence of mutations in CTNNB1, TP53, and PKHD1 compared to the KPNA2-low expression subgroup (p < 0.05).
Figure 7.
Genomic analysis related to KPNA2. (A) Integrative landscape illustrating the interplay between KPNA2 expression, TMB, SMGs, and CNV. (B) Comparative mutational analysis, highlighting variations in SMGs and CNV across distinct KPNA2 expression subgroups. *p < 0.05; **p < 0.01; ***p < 0.001.
Interestingly, in CNVs at the arm-level, the KPNA2-low expression group exhibited a higher gain in 5p13.1, 5q35.3, 5q31.2, and 5p14.1, whereas the KPNA2-high expression group showed a higher loss in 17p13.1, 4q34.3, and 9p21.3 (all p < 0.05). However, no discernible differences were observed at the gene-level CNVs. Additionally, we explored KPNA2 expression differences under varying mutational statuses within SMGs and CNVs (Figure S3B–D). Overall, higher KPNA2 expression was associated with a more pronounced mutational landscape.
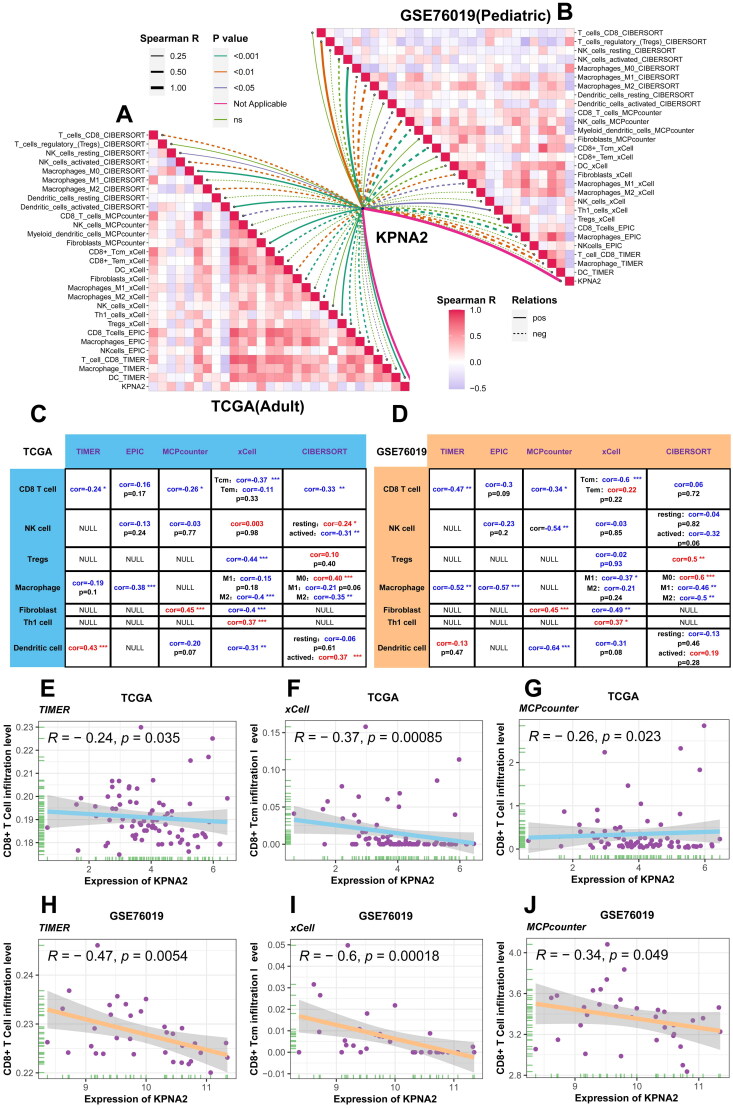
Correlation analysis of KPNA2 expression and immune cell infiltration
Building on our GSEA findings, which indicated a strong correlation between KPNA2 expression and tumor-immune pathways, we utilized datasets from TCGA-ACC and GSE76019 to represent adult and pediatric ACC populations, respectively, for the analysis of tumor immune cell infiltration (Figure 8A,B, Tables S7 and S8). Subsequently, we summarized the Spearman correlation analyses between KPNA2 expression and immune cell infiltration scores generated from TIMER, EPIC, MCPcounter, xCell, and CIBERSORT algorithms in both adult and pediatric ACC (Figure 8C,D). Specifically, in TIMER (TCGA-ACC: r = −0.24, p = 3.5 × 10−2; GSE76019: r = −0.47, p = 5.4 × 10−3), xCell (TCGA-ACC: r = −0.37, p = 8.5 × 10−4; GSE76019: r = −0.6, p = 1.8 × 10−4), and MCPcounter (TCGA-ACC: r = −0.26, p = 2.3 × 10−2; GSE76019: r = −0.34, p = 4.9 × 10−2) algorithms, a negative correlation was observed between CD8+ T-cell infiltration and KPNA2 expression in both adult and pediatric ACC (Figure 8E–J). Given that CD8+ T-cells generally play a tumor-killing role and their increased infiltration is often considered indicative of a favorable prognosis [41], we hypothesize that elevated KPNA2 expression may lead to adverse outcomes by suppressing the infiltration of CD8+ T-cells in the immune microenvironment of ACC.
Figure 8.
Correlation between KPNA2 and immune cell infiltration. (A and B) Correlation graphs of KPNA2 with immune (C and D) comprehensive summary detailing the statistical significance of the association between KPNA2 and immune cell infiltration levels in TCGA-ACC and GSE76019. (E–J) Scatter plots generated through TIMER, xCell, and MCPcounter algorithms to elucidate the correlation between KPNA2 expression and CD8+ T-cell infiltration scores in TCGA-ACC and GSE76019.
Immunotherapy and potential drug targets of KPNA2 in ACC
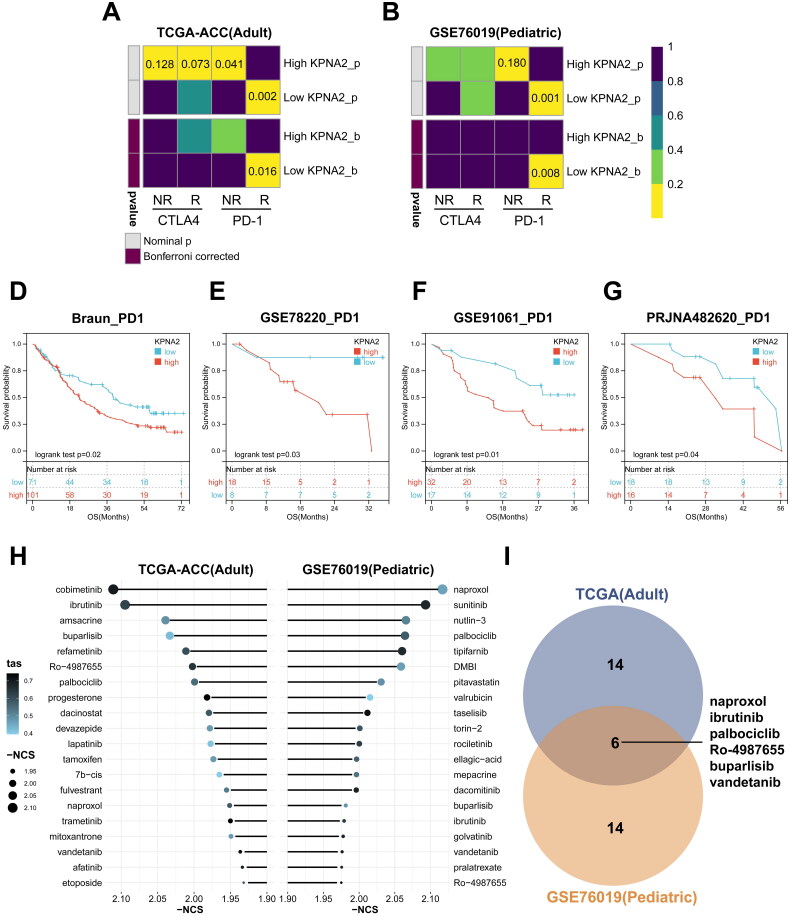
Following the discovery of the potential association between KPNA2 and the immune microenvironment in ACC, we investigated its utility as a biomarker for immunotherapy. CTLA-4 and PD-1, common targets for immunotherapy, inherently suppress autoimmunity, thereby preventing the immune system from killing cancer cells [42]. To delve deeper into the role of KPNA2 in immunotherapy for both adult and pediatric ACC, we initially employed SubMap analysis on TCGA-ACC and GSE76019 datasets. We found that adult and pediatric ACC patients with low KPNA2 expression demonstrated significant expression similarity to anti-PD-L1 responsive cohorts within SubMap (p < 0.05; Figure 9A,B). This suggests that patients with lower KPNA2 expression may be more sensitive to anti-PD-1 therapies compared to those with higher expression, while showing no significant response to anti-CTLA-4 therapies. Subsequently, we sourced four immunotherapy datasets—Braun, GSE78220, GSE91061, PRJNA482620—from the Tiger database, and selected anti-PD-1 therapy samples for survival analysis. The results indicated that patients in the high KPNA2 expression group had significantly poorer prognoses (Figure 9D–G), suggesting limited benefits from anti-PD-1 therapy in these individuals.
Figure 9.
Exploration of immunotherapy responses and Identification of potential drug targets associated with KPNA2. (A and B) Contingency tables delineating the relationship between immunotherapy responses and KPNA2 expression clusters, as stratified by the Submap algorithm in TCGA-ACC and GSE76019 cohorts. (D–G) Kaplan–Meier survival curves evaluating OS across KPNA2 expression subcategories within multiple anti-PD1 cohorts (Braun, GSE78220, GSE91061, PRJNA482620). (H) Bubble plot representing the results of cmap analysis. (I) Venn diagram illustrating the intersection of significant findings derived from cmap analysis.
Moreover, we employed the CMap database to identify compounds that could potentially target KPNA2-associated pathways in ACC. According to Normalized Connectivity Scores (NCS), we selected the top 20 compounds from both TCGA-ACC and GSE76019 datasets (Figure 9H). Mechanism-of-action (MOA) analysis (Figure S4A,B) revealed six potential ACC therapeutic agents—naproxol, ibrutinib, palbociclib, Ro-4987655, buparlisib, and vandetanib—that could potentially target KPNA2. Collectively, our findings indicate that KPNA2 serves as a potential biomarker for immunotherapy and as a drug target in ACC.
Discussion
ACC is a highly malignant tumor, characterized by its propensity for metastasis and resistance to standard therapies. Many patients are diagnosed at advanced stages, resulting in poor prognosis [2,43]. Mitotane is currently the only approved chemotherapeutic agent for treating ACC, primarily used in cases where surgical resection is not feasible or when recurrence or metastasis occurs post-surgery. However, its therapeutic efficacy is generally slow, varies among individuals, and is accompanied by significant side effects [4]. Immunosuppressive agents and targeted therapies, as emerging directions for ACC treatment, are still in clinical trials and face challenges such as inconsistent efficacy, intense side effects, and drug resistance [2,8]. Therefore, the identification of a biomarker that can predict the prognosis and immunotherapeutic response in ACC patients is of paramount importance. In this study, using bioinformatics, immunohistochemistry, and in vitro experiments, we identified KPNA2 as a gene associated with ACC progression and found that it has robust predictive power for immunotherapeutic responses.
After identifying a gene set associated with ACC progression through WGCNA and ATAC-seq, we performed GO analysis and discovered a significant enrichment of the Wnt/β-catenin signaling pathway within this gene set. Aberrant activation of the Wnt/β-catenin pathway can drive tumorigenesis by promoting cellular proliferation, survival, and migration [44]. Numerous studies have shown that inhibiting the Wnt/β-catenin pathway can suppress the proliferation and growth of ACC cells, consequently slowing tumor development. Research by Morgan K Penny et al. found that targeting the oncogenic Wnt/β-catenin signaling pathway could disrupt ECM expression and impact ACC tumor growth [45]. Rottlerin, a natural plant polyphenol, has been shown to inhibit cell proliferation and induce apoptosis in ACC cell lines and xenograft models [46]. Additionally, Niclosamide can downregulate the expression of β-catenin and inhibit the levels of epithelial-mesenchymal transition mediators [47].
Moreover, we filtered out KPNA2 from this gene set as the most predictive biomarker for ACC prognosis and as a potential drug target. KPNA2 is a nuclear transport protein belonging to the karyopherin protein family. It plays a crucial role in the molecular transport process between the cell nucleus and the cytoplasm [48,49]. Its primary function is to shuttle proteins containing nuclear localization signals from the cytoplasm to the nucleus to participate in nuclear biological processes such as gene transcription, DNA repair, and cell cycle regulation [50] Through GO analysis, we found that KPNA2 significantly activates pathways related to cell replication and cell cycle progression. In subsequent experiments, we also discovered that KPNA2 promotes ACC cell proliferation and metastasis. Some research indicates that KPNA2 is overexpressed in multiple types of cancer and promotes tumor progression both in vitro and in vivo, correlating with poor patient prognosis. For example, studies have shown that KPNA2 is associated with shorter overall survival in lung adenocarcinoma and that its overexpression enhances the migratory ability of lung adenocarcinoma cells [51,52]. Similarly, Altan et al. found that KPNA2 promotes gastric cancer progression and poor patient prognosis through the activation of the Wnt/β-catenin signaling pathway [53]. Additionally, in ovarian and colorectal cancers, KPNA2 facilitates tumor progression by participating in the AKT signaling pathway [54,55]. Hence, it is evident that the roles and mechanisms of KPNA2 vary across different types of cancer.
Tumor heterogeneity serves as a critical determinant of both prognosis and therapy response in ACC. Variations in mutations across different cells can lead to disparities in cell growth, proliferation, and signaling pathways, thereby contributing to heterogeneity. Genomic mutational analysis can elucidate the landscape of gene mutations within the tumor [56]. In the present study, we found significant differences in TP53 and CTNNB1 mutations among the KPNA2 expression subgroups, both of which have been confirmed to be associated with the occurrence and progression of ACC [57–59]. Our findings indicate that the expression levels of KPNA2 in ACC are significantly correlated to various degrees with TMB, SMGs, and CNV, suggesting that KPNA2 is a predictor of higher TMB, with potential implications for immunotherapeutic responsiveness [60].
The tumor immune microenvironment, comprising immune cells, cytokines, chemokines, and immune checkpoint molecules, plays a pivotal role in cancer onset, progression, metastasis, and therapy response. It dictates how the immune system interacts with cancer cells, thus affecting their survival, proliferation, and migration [61]. In our analysis, we observed that KPNA2 significantly inhibits immune response-related pathways. Research has demonstrated that increased nuclear transporter KPNA2 contributes to tumor immune evasion by enhancing PD-L1 expression in pancreatic ductal adenocarcinoma (PDAC) [16]. In addition, multiple algorithms indicate that KPNA2 expression negatively correlates with CD8+ T cells in both adult and pediatric datasets. Submap analysis revealed that low expression of KPNA2 is significantly correlated with a potential PD-L1 immune response, while high expression of KPNA2 in the immunotherapy cohort suggests a poor prognosis. The results indicate that patients with low KPNA2 expression, coupled with upregulated immune checkpoints and increased infiltration of CD8+ T cells, are most likely to benefit from immunotherapy. Consequently, KPNA2 possesses potential prognostic value for immunotherapeutic interventions.
In addition to existing treatments, exploring the combination of Mitotane and KPNA2 inhibitors presents a promising direction for research. Mitotane, a specific anticancer drug used for treating ACC, is an isomer of dichlorodiphenyltrichloroethane (DDT) and demonstrates direct cytotoxic effects on adrenal tissues, though its exact mechanism of action is not fully understood [62]. However, the efficacy of Mitotane as a monotherapy in ACC is hampered by its variable outcomes and significant side effects [63,64]. Combining Mitotane with other drugs is a critical avenue for ACC treatment, but recent clinical trial results have been less than satisfactory [65,66]. KPNA2 inhibitors have already shown some pre-clinical promise in breast cancer and colorectal cancer [15,17]. In our future research, we plan to investigate the combined effect of KPNA2 inhibitors and Mitotane in ACC.
While the role of KPNA2 has been explored in various other cancers, its specific impact on ACC has been largely uncharted until now, thereby filling a critical knowledge gap in the existing literature. It is important, however, to acknowledge certain limitations inherent in our research. Firstly, although we have validated our findings through publicly available databases, the sample size of ACC specimens obtained for this study remains limited, necessitating further validation from a more expansive dataset. Secondly, while our data analysis has identified potential agents for targeting KPNA2, ongoing work involves a more exhaustive series of cellular and other experimental assays aimed at confirming the efficacy and mechanistic pathways of these candidate compounds.
Conclusion
In conclusion, our study introduces KPNA2 as a novel biomarker for ACC, offering positive implications for prognostic risk assessments and shaping future directions in the development of targeted therapeutics and immunomodulatory interventions for ACC patients.
Supplementary Material
Acknowledgment
We would like to express our gratitude to all the contributors of the public datasets.
Funding Statement
This research was supported by grants from the National Natural Science Foundation of China (Grant no. 82003271). The Guangzhou Planned Project of Science and Technology (Grant no. 2023A04J1269). Medical Science and Technology Research Fund of Guangdong Province (Grant no. B2021317). Huizhou High Level Hospital Construction Science and Technology Special Project (Grant No. 2022CZ010004).
Authors contributions
Jianming Lu, Jiahong Chen, and Zhong Dong played instrumental roles in the conceptualization and design of the study. Bioinformatics analysis was conducted by Jianming Lu, Yihao Chen, Yongcheng Shi, and Fengping Liu. The collection of clinical samples was undertaken by Jiahong Chen and Zhong Dong. Supervision of the research was carried out by Jianming Lu, Zhong Dong, Xiaohui Ling, Junhong Deng and Weide Zhong. Manuscript preparation was done by Jiahong Chen, Yihao Chen and Xiaohui Ling, while experimental validation was achieved by Chuanfan Zhong, Shumin Fang, Shanshan Mo and Yihao Chen. All authors made substantial contributions to the article and granted approval for its submission.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statements
The public data used in this study has been described in the Materials and Methods.
References
- 1.Fassnacht M, Assie G, Baudin E, et al. Adrenocortical carcinomas and malignant phaeochromocytomas: ESMO-EURACAN clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(11):1476–1490. doi: 10.1016/j.annonc.2020.08.2099. [DOI] [PubMed] [Google Scholar]
- 2.Else T, Kim AC, Sabolch A, et al. Adrenocortical carcinoma. Endocr Rev. 2014;35(2):282–326. doi: 10.1210/er.2013-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berruti A, Grisanti S, Pulzer A, et al. Long-term outcomes of adjuvant mitotane therapy in patients with radically resected adrenocortical carcinoma. J Clin Endocrinol Metab. 2017;102(4):1358–1365. doi: 10.1210/jc.2016-2894. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Galindo C, Krailo MD, Pinto EM, et al. Treatment of pediatric adrenocortical carcinoma with surgery, retroperitoneal lymph node dissection, and chemotherapy: the children’s oncology group ARAR0332 protocol. J Clin Oncol. 2021;39(22):2463–2473. doi: 10.1200/JCO.20.02871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassnacht M, Dekkers O, Else T, et al. European Society of Endocrinology clinical practice guidelines on the management of adrenocortical carcinoma in adults, in collaboration with the European Network for the Study of Adrenal Tumors. Eur J Endocrinol. 2018;179(4):G1–G46. doi: 10.1530/EJE-18-0608. [DOI] [PubMed] [Google Scholar]
- 6.Else T, Williams AR, Sabolch A, et al. Adjuvant therapies and patient and tumor characteristics associated with survival of adult patients with adrenocortical carcinoma. J Clin Endocrinol Metab. 2014;99(2):455–461. doi: 10.1210/jc.2013-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgantzoglou N, Kokkali S, Tsourouflis G, et al. Tumor microenvironment in adrenocortical carcinoma: barrier to immunotherapy success? Cancers. 2021;13(8):13. doi: 10.3390/cancers13081798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karwacka I, Obołończyk Ł, Kaniuka-Jakubowska S, et al. The role of immunotherapy in the treatment of adrenocortical carcinoma. Biomedicines. 2021;9(2):98. doi: 10.3390/biomedicines9020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bougeard G, Renaux-Petel M, Flaman JM, et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33(21):2345–2352. doi: 10.1200/JCO.2014.59.5728. [DOI] [PubMed] [Google Scholar]
- 10.Olivier M, Asmis R, Hawkins GA, et al. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. 2019;20(19):4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasin Y, Seldin M, Lusis A.. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan F, Powell DR, Curtis DJ, et al. From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 2020;21(1):22. doi: 10.1186/s13059-020-1929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corces MR, Granja JM, Shams S, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362(6413):eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Z, Xu T, Bao L, et al. CREB3L1 promotes tumor growth and metastasis of anaplastic thyroid carcinoma by remodeling the tumor microenvironment. Mol Cancer. 2022;21(1):190. doi: 10.1186/s12943-022-01658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma A, Tang M, Zhang L, et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene. 2019;38(13):2405–2419. doi: 10.1038/s41388-018-0590-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhou KX, Huang S, Hu LP, et al. Increased nuclear transporter KPNA2 contributes to tumor immune evasion by enhancing PD-L1 expression in PDAC. J Immunol Res. 2021;2021:6694392–6694313. doi: 10.1155/2021/6694392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han F, Zhang L, Liao S, et al. The interaction between S100A2 and KPNA2 mediates NFYA nuclear import and is a novel therapeutic target for colorectal cancer metastasis. ONCOGENE. 2022;41(5):657–670. doi: 10.1038/s41388-021-02116-6. [DOI] [PubMed] [Google Scholar]
- 18.Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giordano TJ, Kuick R, Else T, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15(2):668–676. doi: 10.1158/1078-0432.CCR-08-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demeure MJ, Coan KE, Grant CS, et al. PTTG1 overexpression in adrenocortical cancer is associated with poor survival and represents a potential therapeutic target. Surgery. 2013;154(6):1405–1416; discussion 1416. doi: 10.1016/j.surg.2013.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto EM, Rodriguez-Galindo C, Choi JK, et al. Prognostic significance of major histocompatibility complex class II expression in pediatric adrenocortical tumors: a St. Jude and Children’s Oncology Group Study. Clin Cancer Res. 2016;22(24):6247–6255. doi: 10.1158/1078-0432.CCR-15-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langfelder P, Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanche P, Dartigues JF, Jacqmin-Gadda H.. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32(30):5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
- 27.Rich JT, Neely JG, Paniello RC, et al. A practical guide to understanding Kaplan-Meier curves. Otolaryngol Head Neck Surg. 2010;143(3):331–336. doi: 10.1016/j.otohns.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong C, Long Z, Yang T, et al. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int J Biol Sci. 2023;19(5):1543–1563. doi: 10.7150/ijbs.77133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J, Dong W, He H, et al. Autophagy induced by overexpression of DCTPP1 promotes tumor progression and predicts poor clinical outcome in prostate cancer. Int J Biol Macromol. 2018;118(Pt A):599–609. doi: 10.1016/j.ijbiomac.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Z, Eils R, Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 32.Zeng D, Ye Z, Shen R, et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12:687975. doi: 10.3389/fimmu.2021.687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshida Y, Brunet JP, Tamayo P, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PLOS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun DA, Hou Y, Bakouny Z, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26(6):909–918. doi: 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. CELL. 2017;168(3):542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. CELL. 2017;171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Chen AX, Gartrell RD, et al. Author Correction: immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(6):1022–1022. doi: 10.1038/s41591-019-0449-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Luo Z, Zhang D, et al. TIGER: a web portal of tumor immunotherapy gene expression resource. Genomics Proteomics Bioinformatics. 2022;21(2):337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malta TM, Sokolov A, Gentles AJ, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. CELL. 2018;173(2):338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen W, Song Z, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Leun AM, Thommen DS, Schumacher TN.. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20(4):218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25(21):9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fassnacht M, Terzolo M, Allolio B, et al. Combination chemotherapy in advanced adrenocortical carcinoma. N Engl J Med. 2012;366(23):2189–2197. doi: 10.1056/NEJMoa1200966. [DOI] [PubMed] [Google Scholar]
- 44.Nusse R, Clevers H.. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. CELL. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Penny MK, Lerario AM, Basham KJ, et al. Targeting oncogenic Wnt/β-catenin signaling in adrenocortical carcinoma disrupts ECM expression and impairs tumor growth. Cancers. 2023;15(14):3559. doi: 10.3390/cancers15143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, Wang M, Zhao X, et al. Rottlerin as a novel chemotherapy agent for adrenocortical carcinoma. Oncotarget. 2017;8(14):22825–22834. doi: 10.18632/oncotarget.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satoh K, Zhang L, Zhang Y, et al. Identification of niclosamide as a novel anticancer agent for adrenocortical carcinoma. Clin Cancer Res. 2016;22(14):3458–3466. doi: 10.1158/1078-0432.CCR-15-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radu A, Blobel G, Moore MS.. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci U S A. 1995;92(5):1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldfarb DS, Corbett AH, Mason DA, et al. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14(9):505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Kelley JB, Talley AM, Spencer A, et al. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11(1):63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang CI, Wang CL, Wu YC, et al. Quantitative proteomics reveals a novel role of karyopherin alpha 2 in cell migration through the regulation of vimentin-pErk protein complex levels in lung cancer. J Proteome Res. 2015;14(4):1739–1751. doi: 10.1021/pr501097a. [DOI] [PubMed] [Google Scholar]
- 52.Li XL, Jia LL, Shi MM, et al. Downregulation of KPNA2 in non-small-cell lung cancer is associated with Oct4 expression. J Transl Med. 2013;11(1):232. doi: 10.1186/1479-5876-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altan B, Yokobori T, Mochiki E, et al. Nuclear karyopherin-α2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis. 2013;34(10):2314–2321. doi: 10.1093/carcin/bgt214. [DOI] [PubMed] [Google Scholar]
- 54.Huang L, Zhou Y, Cao XP, et al. KPNA2 promotes migration and invasion in epithelial ovarian cancer cells by inducing epithelial-mesenchymal transition via Akt/GSK-3β/Snail activation. J Cancer. 2018;9(1):157–165. doi: 10.7150/jca.20879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang P, Zhao Y, Liu K, et al. Wip1 cooperates with KPNA2 to modulate the cell proliferation and migration of colorectal cancer via a p53-dependent manner. J Cell Biochem. 2019;120(9):15709–15718. doi: 10.1002/jcb.28840. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. NATURE. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durand J, Lampron A, Mazzuco TL, et al. Characterization of differential gene expression in adrenocortical tumors harboring beta-catenin (CTNNB1) mutations. J Clin Endocrinol Metab. 2011;96(7):E1206–11. doi: 10.1210/jc.2010-2143. [DOI] [PubMed] [Google Scholar]
- 58.Bonnet-Serrano F, Bertherat J.. Genetics of tumors of the adrenal cortex. Endocr Relat Cancer. 2018;25(3):R131–r152. doi: 10.1530/ERC-17-0361. [DOI] [PubMed] [Google Scholar]
- 59.Das R, Ghosh Chowdhury M, Raundal S, et al. Objective assessment of adrenocortical carcinoma driver genes and their correlation with tumor pyruvate kinase M2. GENE. 2022;822:146354. doi: 10.1016/j.gene.2022.146354. [DOI] [PubMed] [Google Scholar]
- 60.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–1492. doi: 10.1093/annonc/mdw168. [DOI] [PubMed] [Google Scholar]
- 62.Bedrose S, Daher M, Altameemi L, et al. Adjuvant therapy in adrenocortical carcinoma: reflections and future directions. Cancers. 2020;12(2):508. doi: 10.3390/cancers12020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Institute of Diabetes and Digestive and Kidney Diseases . LiverTox: clinical and research information on drug-induced liver injury. Bethesda: National Institute of Diabetes and Digestive and Kidney Diseases; 2012. [Google Scholar]
- 64.Puglisi S, Calabrese A, Basile V, et al. Mitotane concentrations influence the risk of recurrence in adrenocortical carcinoma patients on adjuvant treatment. J Clin Med. 2019;8(11):1850. doi: 10.3390/jcm8111850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debets P, Dreijerink KMA, Engelsman A, et al. Impact of EDP-M on survival of patients with metastatic adrenocortical carcinoma: a population-based study. Eur J Cancer. 2023;196:113424. doi: 10.1016/j.ejca.2023.113424. [DOI] [PubMed] [Google Scholar]
- 66.Tourneau CL, Hoimes C, Zarwan C, et al. Avelumab in patients with previously treated metastatic adrenocortical carcinoma: phase 1b results from the JAVELIN solid tumor trial. J Immunother Cancer. 2018;6(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public data used in this study has been described in the Materials and Methods.