Summary
During development, Shh attracts axons of spinal cord commissural neurons to the floor plate. Shh-mediated attraction of commissural axons requires the receptor Boc. How Boc regulates cytoskeletal changes in growth cones in response to Shh is not fully understood. To identify effectors of Boc in Shh-mediated axon guidance, we used BioID to screen for proteins in proximity to Boc. Top hits included members of the WAVE regulatory complex (WRC), which acts downstream of Rac1 to promote actin cytoskeleton assembly. Therefore, we hypothesized that the WRC is important for Shh-mediated growth cone turning. Using biochemical and cellular assays, we found that Boc directly interacts with the WRC and that this interaction can occur in live cells. Moreover, we found that knockdown of Nckap1 and Cyfip1/2, two subunits of the WRC, in commissural neurons, impairs axon attraction toward a Shh gradient. Our results demonstrate that the WRC is required for Shh-mediated axon attraction.
Subject areas: biological sciences, neuroscience, cell biology
Graphical abstract
Highlights
-
•
We used BioID to identify proteins in close proximity to the Shh receptor Boc
-
•
Boc interacts with the WAVE regulatory complex (WRC), an actin nucleation promoter
-
•
The WRC is expressed in spinal cord commissural neurons and colocalizes with Boc
-
•
Shh-mediated axon attraction requires Nckap1 and Cyfip1/2, two subunits of the WRC
Biological sciences; Neuroscience; Cell biology.
Introduction
Sonic hedgehog (Shh) is a morphogen that specifies cell fate during developmental patterning1 and functions as a guidance cue in axon pathfinding during nervous system development.2 In the developing spinal cord, Shh, secreted by the notochord and floor plate cells, forms a ventral-high dorsal-low gradient. The Shh gradient induces the fate of neural progenitors in the ventral spinal cord through a canonical transcription-dependent signaling pathway.1 In contrast, Shh attracts spinal cord commissural axons toward the floor plate through a non-canonical transcription-independent signaling pathway that acts locally at the growth cone.3 Shh signaling in the guidance of commissural axons requires the receptor Boc.4 Shh-mediated downstream signaling requires Smo activity,3,5 and Smo-dependent signaling leads to activation of Src-family kinases (SFKs) via β-arrestins.3,6
Signaling cascades in axon guidance promote remodeling of the growth cone actin cytoskeleton.7 Previously, we identified some of the effectors linking Shh signaling to the actin cytoskeleton. The mRNA-binding protein ZBP1 (Zipcode-binding protein 1), a downstream target of SFKs, is phosphorylated in response to Shh. Upon phosphorylation, it releases its cargo, β-actin mRNA, in the growth cone. The β-actin mRNA is then locally translated in the growth cone to produce β-actin protein, which facilitates polarized actin polymerization.8 In addition, the guanine nucleotide exchange factors (GEFs) Dock3 and 4 and their binding partners ELMO1 and 2 are also required for Shh-mediated axon guidance. Dock3/4 interacts with Boc through ELMO1/2, and the Dock/ELMO complex is released from Boc upon Shh stimulation. Dock/ELMO activates the Rho-family GTPase Rac1, which can remodel the growth cone actin cytoskeleton.9 However, how Boc regulates cytoskeletal remodeling in growth cones in response to Shh is still not fully understood.
To elucidate other effectors of Boc in Shh-dependent axon guidance, we screened for potential Boc-interacting proteins using a proximity-dependent biotin identification (BioID) assay. We found the WAVE regulatory complex (WRC) as a strong candidate for a Boc-interacting partner in the regulation of cytoskeletal dynamics. The WRC is a heteropentameric complex consisting of WAVE1/2/3 (or WASF1/2/3), CYFIP1/2 (or SRA-1/PIR121), NCKAP1/NCKAP1L [or NAP1(HEM2)/NAP1L(HEM1)], ABI1/2/3, and BRK1 (or HSPC300). WAVE1/2/3 are actin nucleation promoting factors, stimulating the activity of the Arp2/3 complex via their C-terminal VCA domain.10 The Arp2/3 complex nucleates branched actin filaments in lamellipodia, which are the actin-based membrane protrusions important for growth cone movement.11 WAVEs are usually in an inactive state in the WRC as its VCA domain is sequestered through intramolecular interactions.10,12 Various mechanisms, including the binding of GTP-bound Rac1 to CYFIP1/2, activate the WRC, leading to release of the sequestered VCA.12,13,14
The WRC has been shown to play a role in axon pathfinding. Previous work in Drosophila and Caenorhabditis elegans showed that the WRC is required for axon guidance and targeting.15,16,17 How various axon guidance receptors signal to the WRC is an area of active investigation. Several axon guidance receptors, including DCC, UNC5D, neogenin, Robo1, and Robo3, contain a conserved WRC interacting receptor sequence (WIRS), which directly interacts with a conserved surface formed by the CYFIP and ABI subunits.18 In axon guidance, this WIRS-dependent interaction has only been shown to be required for Slit-Robo1 mediated axon repulsion.19
In this study, we identified the WRC as a Boc-interacting partner and hypothesized that the WRC is required for Shh-mediated axon guidance. Using biochemical experiments, we show that Boc directly interacts with the WRC. We demonstrate that the WRC is expressed in spinal cord commissural neurons and that endogenous Boc and Cyfip2 interact in commissural neurons. Finally, we show that knockdown of Nckap1 or Cyfip1/2 prevents turning of commissural axons toward Shh in axon guidance assays, indicating that the WRC is required for Shh-dependent axon guidance.
Results
The WRC is a candidate BOC-interacting partner
To identify proteins that potentially interact with BOC, we performed a BioID experiment. We fused the biotin ligase, BirA, to human BOC so that proteins in close proximity to BOC could be biotinylated in cells. We incubated HEK293 T-Rex cells expressing FlagBirA∗-BOC or the FlagBirA∗ control with exogenous biotin and captured and then analyzed the biotinylated proteins by mass spectrometry.
After obtaining the list of peptides detected in two independent BOC BioID experiments (each containing two FlagBirA∗-BOC biological replicates), we removed the proteins that are promiscuous and/or common contaminants in mass spectrometry proximity-dependent biotinylation experiments. For this, proteins found in more than one-third (33%) of the mass spectrometry proximity-dependent biotinylation experiments reported in the Contaminant repository for affinity purification database20 were considered to be too frequent to be of interest and were thus removed from our list.
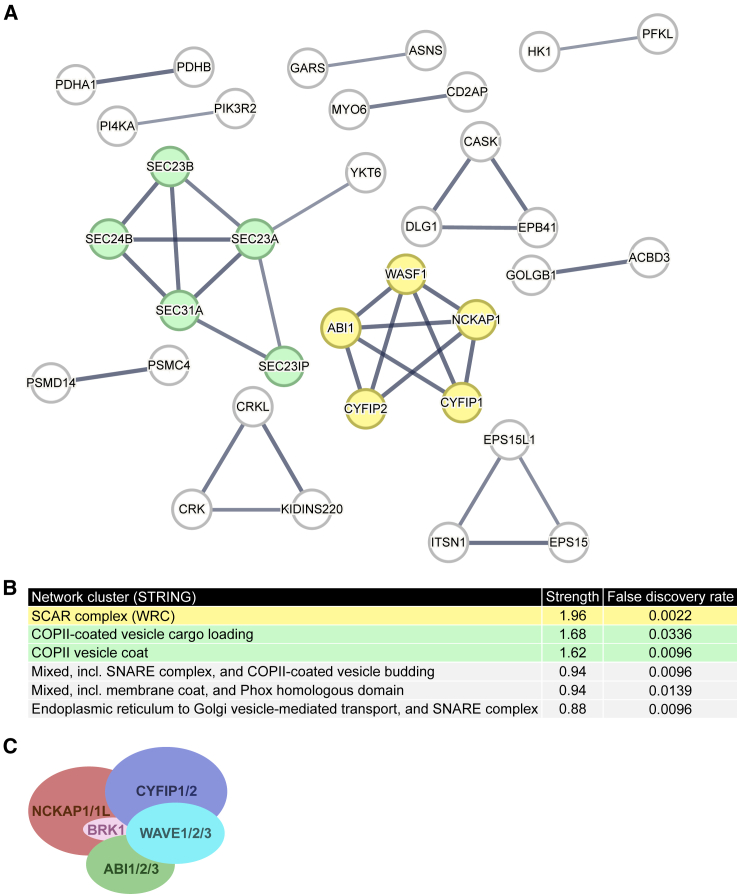
Following this, we focused our attention on the proteins that were represented by at least three peptides in each of the two FlagBirA∗-BOC BioID groups, while having no peptide in the four FlagBirA∗ control experiments. These criteria left us with 125 proteins (including BOC itself) as proteins proximal to BOC (Table S1). Of note, DOCK4, which we previously found to form a complex with Boc,9 was recovered in this BioID screen. To look for networks of potential BOC interactors, we performed a network cluster analysis of our 125 proteins (Figure 1A ; for brevity, only proteins forming a complex with at least one other protein found in the screen are shown). These 125 proteins have more interactions among themselves (protein-protein enrichment p value = 5.95e-10) than what would be expected for a random set of proteins of the same size. The top three ranking clusters were SCAR complex (WRC), COPII (coat protein complex II)-coated vesicle cargo loading, and COPII vesicle coat (Figure 1B). The COPII coat is responsible for the formation of vesicles from the endoplasmic reticulum, which is expected to be in proximity to transmembrane proteins such as BOC as they are anterogradely transported to the Golgi apparatus. The SCAR complex (WRC) ranked first in the network cluster analysis (strength = 1.96; FDR = 0.0022). Notably, we found five WRC proteins—CYFIP1, CYFIP2, NCKAP1, ABI1, and WASF1/WAVE1—which represent four of the five subunits of the WRC/SCAR complex in our screen (Figure 1C).
Figure 1.
The WRC is a candidate BOC-interacting partner
(A) Network cluster analysis using STRING of the 125 high-confidence proteins identified in the BOC BioID screen. Physical subnetworks are shown with the lines indicating that the proteins are part of a physical complex and line thickness indicating the strength of data support. Disconnected nodes were hidden, i.e., only proteins forming a complex with at least one other protein found in the BioID screen are shown. Protein subunits of the WRC are colored in yellow.
(B) List of enriched network clusters (STRING), sorted by cluster strength. Cluster strength: Log10(observed/expected). This measure describes how large the enrichment effect is. It is the ratio between (i) the number of proteins in the network that are annotated with a term and (ii) the number of proteins that are expected to be annotated with this term in a random network of the same size. False discovery rate describes how significant the enrichment is. Shown are p values corrected for multiple testing within each category using the Benjamini–Hochberg procedure.
(C) The five components of the WRC.
The WRC complex is a pentameric complex, and, in addition to the four subunits listed earlier, it also contains BRK1. BRK1 is a small protein (75 amino acids, 8.7 kDa) and is mostly buried inside the WRC complex,12 likely making it inaccessible to the biotin ligase of the BioID system, explaining why it was not identified by our BioID screen.
It is nonetheless remarkable that our BOC BioID screen detected four of five subunits of the WRC complex and that the WRC ranked first in the network cluster ontology analysis, suggesting that the WRC complex is a strong and bona fide interactor of Boc.
Boc interacts with the WRC
To confirm the interaction between BOC and the WRC, we performed co-immunoprecipitation (co-IP) experiments between Boc and the WRC components CYFIP2, NCKAP1, WAVE1, and ABI1, which correspond to the WRC components identified in the BOC BioID screen. We chose to use CYFIP2 and not CYFIP1, as it is the isoform more highly expressed in commissural neurons (Figure 4A). We expressed mouse Boc either with each individual WRC construct alone or with all the WRC constructs (CYFIP2-Flag, NCKAP1-Flag, WAVE1-Emerald, and ABI1-EGFP) together in HEK293T cells and immunoprecipitated Boc from the lysates with an anti-Boc antibody that specifically recognizes Boc (Figure S1). When Boc was co-expressed with all the WRC constructs, we detected CYFIP2, NCKAP1, WAVE1, and ABI1 in the immunoprecipitate (Figures 2A lane 6 and 2D lane 6; Figure S2). This indicates that mouse Boc interacts with the WRC and that the human BOC-WRC interaction discovered by the BioID data is conserved.
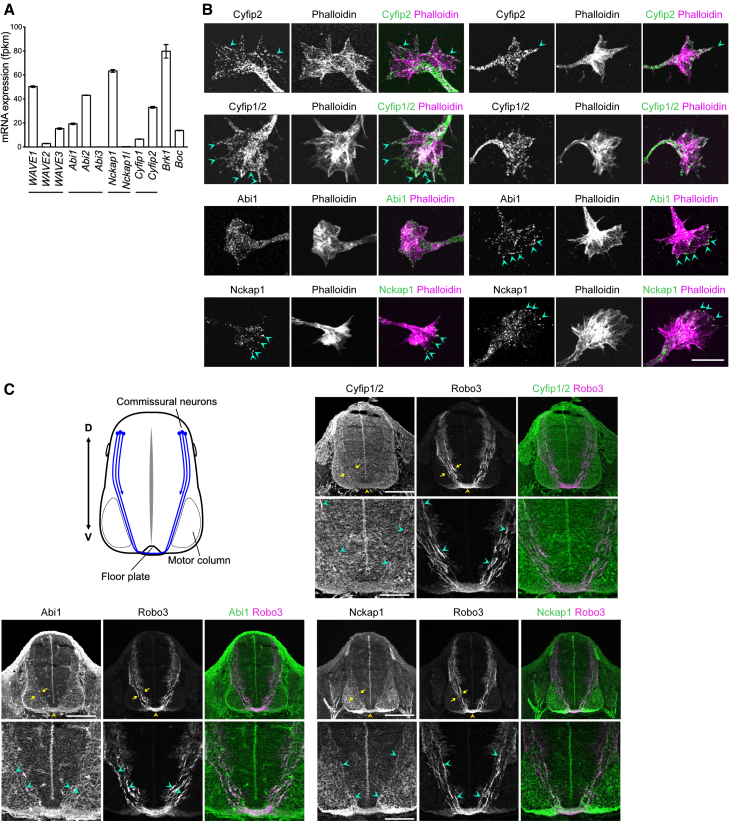
Figure 4.
The WRC is expressed in spinal cord commissural neurons
(A) The mean mRNA expression levels (±SEM) of the WRC components in dissociated rat commissural neurons (n = 3 independent experiments) from RNA sequencing of dissociated cultured embryonic rat commissural neurons (GSE268644; from Makihara et al.9). The expression level of Boc is included for comparison.
(B) Immunostaining of dissociated E13.5 rat commissural neurons for Cyfip2, Cyfip1/2, Abi1, and Nckap1. Two examples are shown for each antibody. F-actin was labeled with phalloidin. Cyan arrowheads indicate Cyfip2, Cyfip1/2, Abi1, or Nckap1 at the leading edge of the growth cone. Scale bar: 10 μm.
(C) Schematic representation of the trajectories of commissural axons in a spinal cord cross-section of E11.5 mouse. Commissural neurons located in the dorsal spinal cord project axons that are guided ventrally toward the floor plate. Immunostaining of Cyfip1/2, Abi1, or Nckap1 with the commissural neuron marker Robo3 in E11.5 mouse spinal cord sections. (Upper row) Yellow arrows delimitate the commissural axon tract. Yellow arrowhead indicates the floor plate. Scale bar: 200 μm. (Lower row) A zoom in of the ventral spinal cord is below each corresponding image. Cyan arrowheads indicate some commissural axons. Scale bar: 100 μm. See also Figures S4–S7.
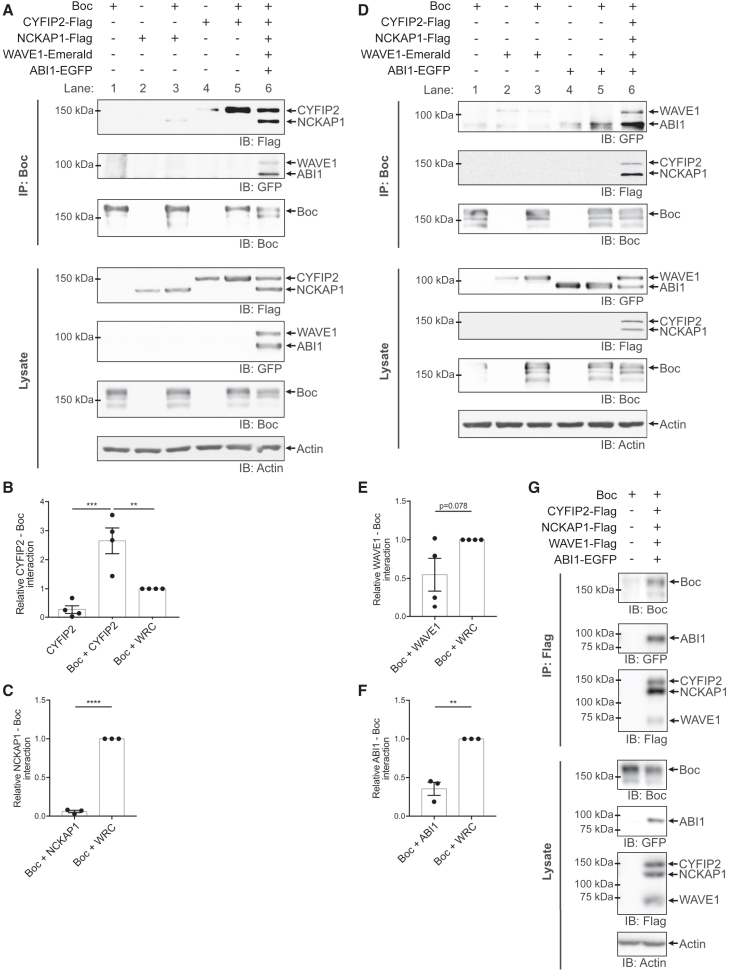
Figure 2.
Boc interacts with the WRC
(A and D) Boc and tagged WRC constructs were expressed in HEK293T cells as indicated. The lysates were immunoprecipitated with an anti-Boc antibody, and interacting proteins were analyzed through SDS-PAGE and western blot.
(B, C, E, F) The relative amount (mean ± SEM) of CYFIP2, NCKAP1, WAVE1, or ABI1 interacting with Boc was calculated by normalizing the amount of corresponding protein in the immunoprecipitate to its amount in the cell lysate and expressed relative to the “Boc + WRC” condition in each experiment. “WRC” refers to the condition in which NCKAP1, CYFIP2, WAVE1, and ABI1 were co-expressed. (B) One-way ANOVA (p = 0.0001), Tukey’s post hoc test, n = 4 independent experiments.
(C, E, F) Unpaired t tests, n = 3 independent experiments for (C) and (F) and n = 4 independent experiments for (E). ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(G) Boc and tagged WRC constructs were expressed in HEK293 cells as indicated. The lysates were immunoprecipitated with an anti-Flag antibody, and interacting proteins were analyzed through SDS-PAGE and western blot. Boc and ABI1-EGFP co-immunoprecipitated with CYFIP2-Flag/NCKAP1-Flag/WAVE1-Flag. See also Figures S1–S3.
We also tested whether individual WRC components could interact with Boc. When we co-expressed Boc with CYFIP2-Flag alone, we detected CYFIP2 in the immunoprecipitate (Figure 2A lane 5). The amount of CYFIP2 that co-immunoprecipitated with Boc was significantly higher than the weak non-specific binding we detected in our negative controls when CYFIP2 was expressed in the absence of Boc (Figure 2A lane 4; Figure 2B). This indicates that Boc and CYFIP2 can interact. Moreover, the amount of CYFIP2 interacting with Boc decreased when the other WRC components were expressed (Figures 2A lane 6 and 2B). This might be due to a conformational change in CYFIP2 when incorporated into the WRC, which may reduce the interaction between CYFIP2 and Boc.
On the other hand, when we co-expressed Boc with NCKAP1-Flag alone, we were barely able to detect NCKAP1 in the immunoprecipitate (Figure 2A lane 3). However, in the presence of the other WRC components, the amount of NCKAP1 that co-immunoprecipitated with Boc significantly increased (Figures 2A lane 6 and 2C). This suggests that the interaction between NCKAP1 and Boc is significantly enhanced when NCKAP1 is incorporated in the WRC. Similarly, when we co-expressed Boc with WAVE1-Emerald, we detected no specific Boc-WAVE1 interaction (Figure 2D lane 3; Figure S3). Like NCKAP1, the interaction of WAVE1 with Boc was enhanced by the expression of the other WRC components (Figures 2D lane 6 and 2E). When we co-expressed Boc with ABI1-EGFP alone, we detected Boc-ABI1 interaction (Figure 2D lane 5). Like NCKAP1 and WAVE1, the interaction of ABI1 with Boc was significantly enhanced by the expression of the other WRC components (Figures 2D lane 6 and 2F). Taken together, this suggests that NCKAP1, WAVE1, and ABI1 require the other WRC components to interact with Boc. Since CYFIP2 alone strongly interacts with Boc, it raises the possibility that the interaction between Boc and the WRC may occur through CYFIP1/2.
We also performed co-IP experiments in the other direction and immunoprecipitated the WRC components instead of Boc. We co-expressed Boc with the WRC constructs CYFIP2-Flag, NCKAP1-Flag, WAVE1-Flag, and ABI1-EGFP in HEK293 cells. We then immunoprecipitated the Flag-tagged proteins (CYFIP2, NCKAP1, and WAVE1) with an anti-Flag antibody and found that ABI1 and Boc were present in the immunoprecipitates (Figure 2G), demonstrating that Boc co-immunoprecipitates with the WRC. Thus, we detect a robust interaction between Boc and the WRC when we immunoprecipitate Boc or components of the WRC.
The interaction between Boc and the WRC occurs in live cells and is direct
To determine whether we could detect the interaction between Boc and the WRC in live cells, we used the NanoLuc Binary Technology (NanoBiT) assay to measure protein-protein interaction in live cells. We fused mouse Boc to the Large BiT subunit (LgBiT; 17.6kDa) to generate Boc-LgBiT and fused CYFIP2 to the Small BiT subunit (SmBiT; 11 amino acids) N-terminally and C-terminally to generate CYFIP2-SmBiT and SmBit-CYFIP2. If Boc and CYFIP2 interact, they bring the LgBiT and SmBiT subunits into close proximity to form a functional enzyme that generates a bright, luminescent signal in the presence of a nonlytic cell-permeable detection reagent. Expression of either Boc-LgBiT, CYFIP2-SmBiT, or SmBit-CYFIP2 alone (Figures 3A and 3B, left) in either HEK293 cells or COS7 cells did not generate a luminescent signal. However, when Boc-LgBiT was expressed together with CYFIP2-SmBiT (Figure 3A, left) or SmBit-CYFIP2 (Figure 3B, left), we detected a strong luminescent signal. This indicates that Boc interacts with CYFIP2 in live cells, consistent with our co-IP results demonstrating that CYFIP2 interacts with Boc (Figure 2A). We next co-expressed the WRC components NCKAP1-Flag, WAVE1-Emerald, ABI1-EGFP, and BRK1-Myc-Flag together with CYFIP2-SmBiT or SmBit-CYFIP2 and found that Boc and CYFIP2 also interact in live HEK293 and COS7 cells in the presence of the entire WRC (Figures 3A and 3B, right).
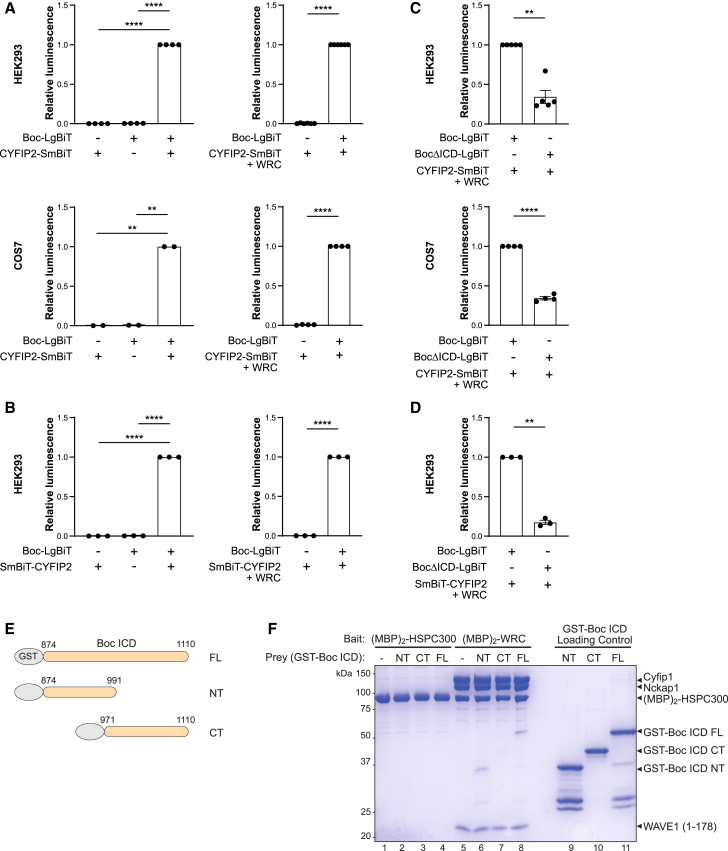
Figure 3.
The interaction between Boc and the WRC occurs in live cells and is direct
(A–D) The NanoBiT structural complementation reporter system was used to detect interaction between Boc and CYFIP2 in live cells. Boc was fused to the Large BiT (LgBiT) subunit (Boc-LgBiT), and CYFIP2 was fused to the Small BiT (SmBiT) subunit either N-terminally (CYFIP2-SmBiT) or C-terminally (SmBiT-CYFIP2) and expressed in HEK293 or COS7 cells. When the two proteins interact, a luminescent signal is generated in the presence of substrate.
(A) Expression of Boc-LgBiT and CYFIP2-SmBit alone or (B) expression of Boc-LgBiT and SmBit-CYFIP2 alone does not generate a luminescent signal. (Left) When Boc-LgBiT is expressed with CYFIP2-SmBiT (A) or SmBit-CYFIP2 (B), they interact to generate a luminescent signal. (Right) Boc-LgBiT also interacts with CYFIP2-SmBiT (A) or SmBit-CYFIP2 (B) in the presence of co-expressed WRC components NCKAP1-Flag, WAVE1-Emerald, ABI1-EGFP, and BRK1-Myc-Flag (“+WRC”). Data are represented as mean ± SEM; error bars representing the SEM are too small to be visible. (Left) Repeated measures one-way ANOVA with Dunnett’s multiple comparison test, (right) paired t test. n = 2–6 independent experiments. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(C) Boc lacking an intracellular domain (ICD), BocΔICD-LgBiT, has a significantly lower interaction with CYFIP2-SmBiT and (D) SmBiT-CYFIP2 compared to full-length Boc-LgBiT in the presence of the WRC components NCKAP1-Flag, WAVE1-Emerald, ABI1-EGFP, and BRK1-Myc-Flag. Data are represented as mean ± SEM. Paired t test, n = 3–5 independent experiments. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(E) Schematic of GST-Boc ICD constructs.
(F) Coomassie-blue-stained SDS-PAGE gel showing MBP pull-down between purified (MBP)2-tagged HSPC300 or WRC and the indicated purified GST-Boc ICD proteins. Only GST-Boc ICD FL and GST-Boc ICD NT are pulled down by (MBP)2-WRC.
To test whether the interaction between Boc and CYFIP2 is mediated by the intracellular domain (ICD) of Boc, we generated BocΔICD-LgBiT, where the entire ICD of Boc is absent and replaced by LgBiT. We found that BocΔICD-LgBiT has a dramatically lower interaction with CYFIP2-SmBiT (Figure 3C) and SmBiT-CYFIP2 (Figure 3D) compared to full-length Boc-LgBiT, in the presence of the other WRC components. This demonstrates that the interaction between Boc and the CYFIP2 requires the intracellular domain of Boc.
Given that the WRC is the major protein complex identified from our BOC BioID screen, it suggests that the WRC is in very close proximity to Boc and that the interaction between Boc and the WRC may be direct. To test if Boc directly interacts with the WRC, we performed pull-down assays using purified recombinant proteins and previously established protocols.18 The N-terminus of the HSPC300 (BRK1) subunit was fused with two maltose-binding proteins, (MBP)2, which can immobilize HSPC300 either alone as (MBP)2-HSPC300 or incorporated into the WRC as (MBP)2-WRC, to amylose beads to facilitate pull-down assays. We performed pull-down assays with purified GST-tagged full-length Boc ICD, an N-terminal portion of the ICD (Boc ICD NT) and a C-terminal portion of the ICD (Boc ICD CT) (Figure 3E). Immobilized (MBP)2-HSPC300 alone was unable to pull down purified Boc ICD (Figure 3F lanes 1–4), demonstrating that HSPC300 alone cannot interact with the Boc ICD and that there is no non-specific binding of the Boc ICD to the beads or the MBP tag in the pull-down assay. In contrast, immobilized (MBP)2-WRC pulled down both Boc ICD FL and Boc ICD NT (Figure 3F lanes 6 and 8), but not Boc ICD CT (Figure 3F lane 7). This shows that the WRC directly interacts with the Boc ICD and that this interaction is mediated by the N-terminal sequence of the ICD.
The WRC is expressed in spinal cord commissural neurons
Boc is a receptor for Shh in axon guidance and is expressed in spinal cord commissural neurons.4 We hypothesized that the WRC acts downstream of Boc in Shh-mediated axon guidance. To determine which WRC components are expressed in commissural neurons, we used RNA sequencing of dissociated rat spinal cord commissural neurons9 (GSE268644). We found that all components of the WRC are expressed in commissural neurons and that expression levels of WAVE1, Abi2, Nckap1, and Cyfip2 are higher compared to their other homologs (Figure 4A).
Next, to determine whether the WRC components are localized to the growth cone, we immunostained dissociated spinal cord commissural neurons for Cyfip2, Cyfip1/2 (using an antibody that recognizes both Cyfip1 and Cyfip2), Abi1, and Nckap1. The CYFIP2, Cyfip1/2, Abi1, and Nckap1 antibodies that we used have been previously validated in knockout or knockdown cells and used for immunofluorescence staining.21,22,23,24,25 We further validated the specificity of these antibodies by immunostaining cells expressing tagged CYFIP2, Cyfip1, and NCKAP1 (Figures S4–S6), western blotting of cells expressing tagged Cyfip1/CYFIP2 proteins and of Cyfip1/2 KO cells (Figure S5), and immunostaining of dissociated commissural neurons knocked down for Cyfip2 and Nckap1 (Figures S4 and S6).
When we immunostained commissural neurons for Cyfip2, Cyfip1/2, Abi1, and Nckap1, we observed that Cyfip2, Cyfip1/2, Abi1, and Nckap1 are expressed throughout the growth cone (Figure 4B). We found that the WRC components are localized to the leading edge of the growth cone in some, but not all growth cones (Figure 4B, cyan arrowheads), and that this leading edge localization, when present, could be along part of the leading edge, discontinuous, or punctate. WRC immunostaining was also present at the tips of filopodia. The Cyfip2 and Cyfip1/2 immunostaining present throughout the growth cone and in the axon, particularly in the central region of the growth cone, is consistent with Cyfip1/Cyfip2 immunostaining in Xenopus retinal growth cones and rat hippocampal neuron growth cones, where Cyfip1/Cyfip2 is predominantly localized to the central region of the growth cone and the axon, with weaker immunostaining in the periphery and leading edge.23,26 The punctate discontinuous immunostaining pattern of the WRC at the growth cone leading edge that we observe is also reminiscent of the WRC localization in hippocampal neuron growth cones.26,27 Live cell imaging of Cyfip2 in Xenopus retinal growth cones has also revealed that Cyfip2 accumulation at the leading edge of growth cones is dynamic, transient, and often limited to only some parts of the leading edge.23 This illustrates that the WRC may not be always present along the entire leading edge of the growth cone. Thus, the varied localization patterns we observe may reflect the dynamic and transient nature of WRC accumulation at the growth cone leading edge. Moreover, in addition to being a component of the WRC, Cyfip2 also associates with ribonucleoprotein particles (RNPs) along the axon and in the central domain of the growth cone; its association with the WRC occurs primarily in the growth cone periphery.23 Therefore, Cyfip2 only partially colocalizes with other components of the WRC, such as Nckap1, and the colocalization is predominantly in the growth cone periphery.23 The variation in immunostaining patterns of the WRC components may also reflect differences between the affinities of the antibodies, their epitopes, and how well the epitopes are preserved during the fixation and labeling procedures. Overall, we find that Cyfip2, Cyfip1/2, Abi1, and Nckap1 are present throughout the growth cones of commissural neurons, including the leading edge.
To confirm the expression of the WRC components in vivo, we examined mouse embryonic spinal cord at E11.5, a stage where commissural axons are being guided toward the floor plate by Shh. We immunostained embryonic spinal cord sections for Cyfip1/2, Abi1, and Nckap1, together with the commissural neuron marker Robo3. Cyfip1/2, Abi1, and Nckap1 are all expressed throughout the spinal cord. They also colocalize with Robo3-expressing commissural axon tracts (Figure 4C; Figure S7), confirming that Cyfip1/2, Abi1, and Nckap1 are expressed in commissural neurons in vivo during the stage where commissural axons are guided toward the floor plate by Shh.
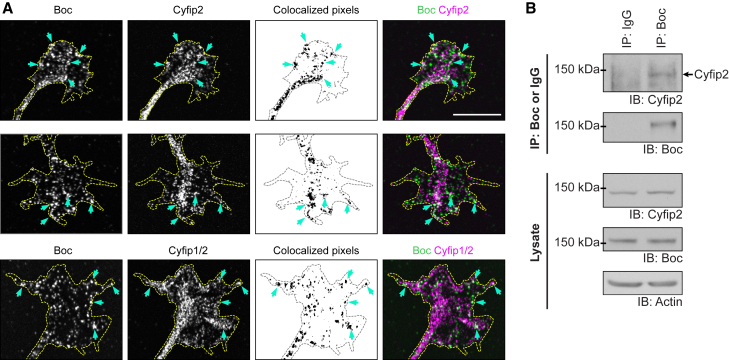
Cyfip2 colocalizes with and interacts with Boc in spinal cord commissural neurons
Given that the WRC is expressed in commissural neurons, we next determined if the WRC can interact with Boc in commissural neurons. We immunostained for Boc in commissural neurons using an antibody that has been previously verified for immunostaining,28 together with Cyfip2 (Figure S4) or Cyfip1/2 (Figure S5). Co-immunostaining of Boc and Cyfip2 in commissural neurons showed that they are both present in the growth cone and each have a punctate distribution. They partly colocalize, with some Boc puncta colocalizing with Cyfip2 (Figure 5A; cyan arrows). This is particularly prominent for the brighter Boc puncta. Similarly, Boc and Cyfip1/2 also partly colocalize, with some Boc puncta colocalizing with Cyfip1/2 (Figure 5A; cyan arrows). Together this supports an interaction between endogenous Boc and the WRC in growth cones.
Figure 5.
Cyfip2 colocalizes with and interacts with Boc in spinal cord commissural neurons
(A) Co-immunostaining of Boc with Cyfip2 or Cyfip1/2 in commissural neuron growth cones. Scale bar: 10 μm. Cyan arrows indicate colocalization between Boc and Cyfip2 or Boc and Cyfip1/2.
(B) Lysates from spinal cord commissural neurons were immunoprecipitated with an anti-Boc antibody or IgG. Co-immunoprecipitated proteins were analyzed through SDS-PAGE and western blot. Endogenous Cyfip2 co-immunoprecipitates with Boc. See also Figures S4–S8.
To test whether endogenous Boc can interact with the WRC in commissural neurons, we immunoprecipitated endogenous Boc from the lysates of dissociated commissural neurons. We detected Cyfip2 in the immunoprecipitate when the lysates were immunoprecipitated with an anti-Boc antibody but not when an IgG control antibody was used, indicating that endogenous Boc interacts with Cyfip2 in commissural neurons (Figure 5B; Figure S8).
Nckap1 and Cyfip1/2 are required for Shh-mediated growth cone turning
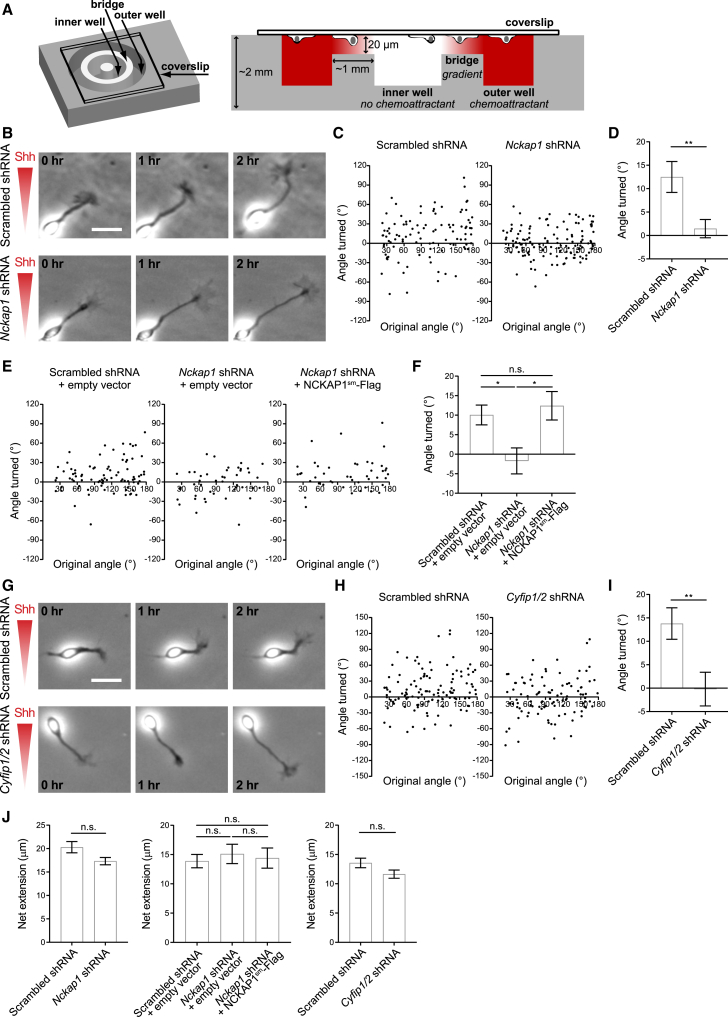
Since Boc interacts with the WRC and the WRC promotes actin cytoskeleton assembly, we next tested whether the WRC is required for Shh-mediated attraction of commissural axons in vitro. Knockdown of any one component of the WRC leads to degradation of the rest of the components.22,29,30,31 Therefore, knockdown of one component of the WRC is sufficient to ablate WRC function. To knock down the WRC in commissural neurons, we targeted either Nckap1, since only a single homolog is expressed in commissural neurons (Figure 4A), or Cyfip1/2, due to the strong Boc-CYFIP2 interaction detected in both our co-IP (Figures 2A, 2B, and 5B) and NanoBiT experiments (Figures 3A–3D). We generated shRNAmir (short hairpin RNA with microRNA scaffold) against rat Nckap1, Cyfip1, and Cyfip2 mRNAs. We evaluated their efficiency by co-expressing each shRNAmir or control scrambled shRNA with its target-tagged expression plasmids in HEK293T cells. The shRNAmirs targeted against Nckap1, Cyfip1, and Cyfip2 reduced their target-tagged protein expression by 91%, 77%, and 98%, respectively (Figure S9), and were also able to reduce endogenously expressed Cyfip2 and Nckap1 by ∼40%–80% in commissural neurons (Figures S4C, S6C, and S10). Consistent with previous measurements of knockdown efficiency in commissural neurons,9,32 we found that western blotting more accurately measured knockdown levels (∼80% for Nckap1) compared to immunofluorescence (∼40% for Nckap1), due to less background signal and ease of discrimination of the signal of the protein band of interest from any cross-reactive proteins.
After validation of the shRNAmirs, we electroporated commissural neurons isolated from E13.5 rat spinal cords either with control scrambled shRNAmir or with shRNAmir against Nckap1 or Cyfip1/2. Nckap1 or Cyfip1/2 knockdown did not affect overall neuron morphology nor axon length (Figure S11). We then assessed the ability of these axons to respond to a Shh gradient using a Dunn chamber axon turning assay3 (Figure 6A). A stable gradient of a chemoattractant is generated in the Dunn chamber, and we monitor and measure the turning response of axons exposed to that gradient. Positive angles turned represent attraction of axons up the gradient. Axons of commissural neurons electroporated with control scrambled shRNAmir showed a bias toward positive angles turned (Figures 6B and 6C) and turned toward high concentrations of Shh with a mean angle turned of ∼13° (Figure 6D). However, shRNA knockdown of Nckap1 in commissural neurons completely abolished the ability of axons to turn toward Shh, showing no net turning (Figures 6B–6D). This indicates that Nckap1 is required for commissural axons to turn toward Shh in vitro.
Figure 6.
Nckap1 and Cyfip1/2 are required for Shh-mediated growth cone turning
(A) Dunn chamber schematic from top (left) and side (right) views from Yam et al. 2009.20 The inner well is filled with media containing no chemoattractant, whereas the outer well is filled with media containing chemoattractant. Diffusion of the chemoattractant from the outer well to the inner well forms a gradient of the chemoattractant over the bridge region. The neurons cultured on a coverslip and exposed to the gradient in the bridge region are imaged.
(B and G) Time-lapse imaging of commissural neurons electroporated either with scrambled shRNA or shRNA targeting Nckap1 or Cyfip1/2 and exposed to a Shh gradient in a Dunn chamber. Axons of commissural neurons electroporated with scrambled shRNA turned toward high concentrations of Shh, whereas axons of Nckap1 or Cyfip1/2 knockdown neurons did not change their direction of growth. The Shh gradient increases along the y axis in the images. Scale bar: 20 μm.
(C) Scatterplots of the angle turned versus the original angle between the axons and the direction of the Shh gradient for neurons under the indicated conditions. Positive angles represent turning of axons toward the Shh gradient.
(D) The mean angle turned (±SEM) of axons of commissural neurons in a Shh gradient. Nckap1 knockdown inhibits the turning of axons up a Shh gradient. Welch’s t test. n = 100 and n = 134 axons for scrambled and Nckap1 shRNA electroporated commissural neurons, respectively.
(E) Scatterplots of the angle turned vs. the original angle and (F) the mean angle turned (±SEM) of axons of commissural neurons in a Shh gradient under the indicated conditions. Expression of NCKAP1sm-Flag rescues the inhibitory effect of Nckap1 knockdown on the turning of axons up a Shh gradient. One-way ANOVA, n = 84, n = 46, and n = 45 axons for scrambled shRNA + empty vector, Nckap1 shRNA + empty vector, and Nckap1 shRNA + NCKAP1sm-Flag electroporated commissural neurons, respectively.
(H) Scatterplots of the angle turned vs. the original angle and (I) the mean angle turned (±SEM) of axons of commissural neurons in a Shh gradient under the indicated conditions. Cyfip1/2 knockdown inhibits the turning of axons up a Shh gradient. Unpaired t test, n = 114 and n = 101 axons for scrambled and Cyfip1/2 shRNA electroporated commissural neurons, respectively.
(J) Net extension (±SEM) of the axons during the 2 h exposure to a Shh gradient. Mann-Whitney test, n = 100 and n = 134 axons for scrambled and Nckap1 shRNA electroporated commissural neurons, respectively. Kruskal-Wallis test, n = 84, n = 46, and n = 45 axons for scrambled shRNA + empty vector, Nckap1 shRNA + empty vector, and Nckap1 shRNA + NCKAP1sm-Flag electroporated commissural neurons, respectively. Mann-Whitney test, n = 114 and n = 101 axons for scrambled and Cyfip1/2 shRNA electroporated commissural neurons, respectively. Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant. See also Figures S9–S11.
Then, we tried to rescue the inhibitory effect of Nckap1 knockdown on the turning of commissural axons toward Shh by electroporating commissural neurons with an shRNA-resistant form of NCKAP1 (NCKAP1sm-Flag) together with Nckap1 shRNA. Expression of NCKAP1sm-Flag completely rescued the turning response of commissural axons toward a Shh gradient (Figures 6E and 6F). This indicates that the effect of Nckap1 knockdown on blocking the ability of axons to turn toward Shh is not an off-target effect.
If the WRC is required for commissural axons to turn to Shh, knockdown of another WRC component should also block Shh-mediated axon guidance. Indeed, we found that shRNA knockdown of Cyfip1/2 in commissural neurons also blocked the ability of axons to turn toward Shh (Figures 6G–6I), indicating that Cyfip1/2 is also required for commissural axons to turn toward Shh. In addition, neither Nckap1 nor Cyfip1/2 knockdown affected the net extension of axons during the experiment (Figure 6J). This is consistent with our finding that the total axon length is not affected by Nckap1 or Cyfip1/2 knockdown (Figure S11) and indicates that the inhibition of axon turning to Shh when Nckap1 or Cyfip1/2 were knocked down was not due to insufficient axon growth. Together, these results show that the WRC is a downstream effector of Boc required for growth cone turning toward Shh.
Discussion
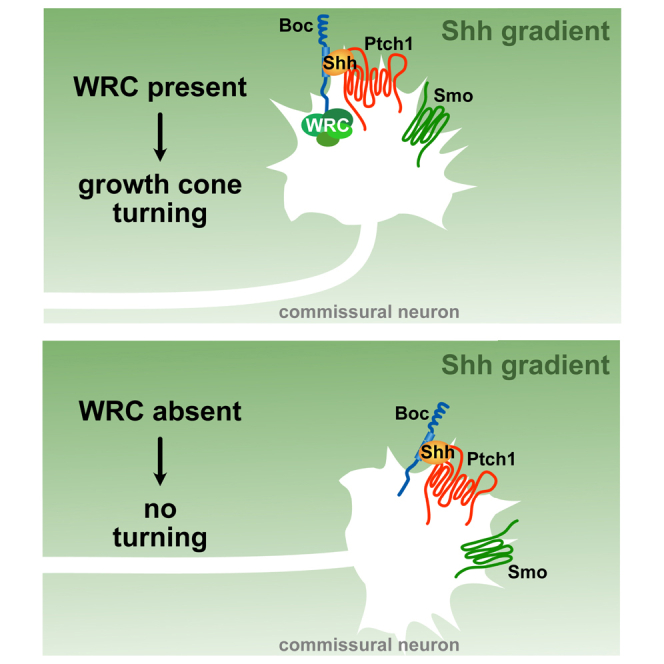
In this study, we identify the WRC as an effector downstream of Boc in Shh-mediated axon guidance. Boc directly interacts with the WRC, and knockdown of Nckap1 or Cyfip1/2 prevents turning of commissural axons toward Shh, demonstrating that the WRC is required for Shh-mediated axon guidance. We propose that the WRC promotes directional actin polymerization in growth cones in response to Shh, enabling growth cone turning.
Shh does not induce axon growth in vitro,3,5 consistent with the phenotype of Smo conditional and Boc−/− mice that show misrouted, but not shorter, commissural axons.4,5 Given that Shh does not induce axon growth, only axon turning3,5 and we do not find any effect of Nckap1 or Cyfip1/2 knockdown on axon growth (Figure 6J; Figure S11), we conclude that the WRC acts in axon turning, but not elongation, downstream of Shh/Boc signaling.
Regulation of the WRC in axon guidance
The WRC can be activated by the binding of GTP-bound Rac1 to CYFIP1/2.12,13,33 Boc interacts with Dock/ELMO, which is a GEF for Rac1, and Shh signaling in axon guidance activates Rac1 in commissural neurons in a Dock-dependent manner.9 Therefore, Shh may activate the WRC through Rac1.
WRC activity can also be regulated by phosphorylation.34 Shh signaling in axon guidance activates SFKs.3 SFK activation could facilitate activation of the WRC since Src, an SFK, phosphorylates WAVE1 at Tyr125, which contributes to WAVE1 activation. This is predicted to occur by the release of the VCA sequestration of WAVE.12,35 In addition, the Boc intracellular domain interacts with the non-receptor tyrosine kinase Abelson (ABL1),36 which can phosphorylate WAVE2 at Tyr150 (corresponding to Tyr151 in WAVE1 and WAVE3), thereby activating the WRC, again predicted to occur by the release of VCA sequestration of WAVE.12,37,38 ABL also phosphorylates ABI1/2 at Tyr148, Tyr155, Tyr248, and Tyr285, and this may also contribute to the activation of the WRC.39
It is also possible that the binding of the WRC to a receptor can regulate the WRC. For receptors that bind to the WRC through the WIRS motif, the binding of the intracellular domain of these receptors to the WRC has variable effects on WRC activity: some intracellular domains enhance Rac1-mediated WRC activation, whereas others have no effect on activity or are slightly inhibitory.18 It remains to be determined whether the binding of Boc to the WRC directly affects the activity of the WRC.
Other potential Boc-interacting proteins of interest identified by BioID
Our BOC BioID experiments, in addition to identifying the WRC as a Boc-interacting complex and identifying DOCK4, which we previously found to form a complex with Boc,9 also identified other proteins in proximity to Boc that might be important for Shh-/Boc-mediated axon guidance (Table S1). One of them is TRIO, a GEF for Rac1, reported to act downstream of DCC in netrin-mediated axon guidance of commissural neurons.40 It is noteworthy that it could also function downstream of Shh/Boc-mediated axon guidance.
Another GEF that we found in proximity to Boc is Intersectin 1 (ITSN1; Figure 1A). ITSN1 is a GEF for the Rho-family GTPase Cdc42. ITSN1 physically associates with EphB2, a receptor for B-ephrin ligands. EphB2 activates the GEF activity of ITSN1 in cooperation with neural Wiskott-Aldrich syndrome protein (N-WASP), which in turn activates Cdc42.41 The authors suggested that this mechanism is important for dendritic spine morphogenesis. Interestingly, in Itsn1 mutant mice, intercortical tracts, such as the corpus callosum, ventral hippocampal, and anterior commissures, fail to cross the midline.42 This phenotype is noteworthy given the role that Shh/Boc-mediated signaling plays in midline attraction of spinal commissural axons.5,43
Additional interesting candidates in proximity to Boc are CRK, CRKL, and KIDINS220, which form a network cluster (Figure 1A). CRK (CRK proto-oncogene, adaptor protein, also known as CRK-II), together with CRKL (CRK-like), are members of an adapter protein family that bind tyrosine-phosphorylated proteins. KIDINS220 (kinase-D-interacting substrate 220, also known as ankyrin repeat-rich membrane spanning [ARMS]) is a scaffold protein at the cell membrane and interacts with CRKL.44 CRK and CRKL regulate cell adhesion, spreading and migration.45,46 Interestingly, CRK and CRKL can regulate cell adhesion and migration through their ability to interact with various members of the DOCK family.47,48 Given the similarities between cell migration and axon guidance mechanisms, this raises the possibility that CRK, CRKL, and KIDINS220 might work together to modulate DOCK activity in Shh/Boc-mediated axon guidance.
Whether TRIO, ITSN1, CRK, CRKL, and KIDINS220 are true interactors of Boc and whether they play a role in Shh/Boc-mediated axon guidance will be an exciting avenue for future research.
The WRC can function downstream of diverse axon guidance receptors
The WRC component ABI1 has been suggested to be required for UNC-6/UNC-40-mediated attraction of AVM and PVM neurons.17 The WRC is also an effector of Slit/Robo1-mediated axon repulsion at the midline.19 Robo1 contains a WIRS motif, the conserved sequence found to directly interact with a surface formed by CYFIP and ABI18 and directly binds to the WRC. The WIRS is required for Robo1 to mediate axon repulsion.19 The netrin-1 receptors DCC, neogenin, and UNC5D and the NELL2 receptor Robo3 also have a WIRS motif, although in each of these cases it is unknown whether their WIRS motifs are required for their function as axon guidance receptors.18 In contrast, Boc does not have a WIRS and is able to directly interact with the WRC.
Some membrane proteins including HPO-30 and retrolinkin also do not contain the WIRS and directly interact with the WRC,49,50 indicating that there are binding motifs other than the WIRS that mediate interaction with the WRC. HPO-30, a claudin superfamily member, directly interacts with the WRC and acts with the dendrite branching receptor DMA-1 to mediate dendrite branching in PVD neurons of C. elegans.50 Retrolinkin, a neuronal endosomal membrane protein, recruits the WRC through its direct interaction with CYFIP2.49 The intracellular domain of Boc does not share clear regions of homology with HPO-30 and retrolinkin, suggesting that Boc may interact with the WRC through a unique binding mechanism.
In this study, we found that the WRC is an effector of non-canonical Shh signaling to mediate growth cone turning toward Shh. Understanding how different axon guidance receptors signal to the WRC will shed light on how the WRC might integrate signals from different receptors and act as a common downstream effector.
Limitations of the study
Ideally, an in vivo analysis of the requirement of the WRC in Shh-mediated axon guidance would complement our study. However, the WRC is required for Slit/Robo1-mediated axon repulsion. Thus, the phenotype that we would observe in a WRC knockout mouse (such as Wave1−/−, which is not embryonically lethal unlike most of the other knockout mice of WRC components) or a mouse with a conditional deletion of one component of the WRC could not be attributed to a specific role for the WRC in Shh-mediated axon guidance.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Frédéric Charron (frederic.charron@ircm.qc.ca).
Materials availability
All reagents generated in this study are available upon request to the lead contact.
Data and code availability
-
•
The BioID data have been deposited at ProteomeXchange and is publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Dr. Hideto Takahashi for generously providing ChromPure goat IgG, Dr. Jia-Jia Liu for generously providing the Cyfip2-Myc (mouse) and Nckap1-Flag (mouse) plasmids, Dr. Giorgio Scita for providing the anti-Abi1 antibody, and Dr. Klemens Rottner for facilitating the sharing of reagents. We thank Kevin Zhang, Dr. Sabrina Schlienger, Dr. Julien Ferent, and Dr. Shirin Makihara for technical assistance. This study was funded by the Canadian Institutes of Health Research grants FDN334023 and PJT-180647 (to F.C.) and MOP-84273 (to S.A.), the Canada Foundation for Innovation grants CFI33768 and CFI NeuroBasis 39794 (to F.C.), and the National Institutes of Health grant R35 GM128786 (to B.C.). F.C. holds the Canada Research Chair in Developmental Neurobiology.
Author contributions
Conceptualization, N.B., S.A., B.C., P.T.Y., and F.C.; methodology, N.B., J.F.M., S.L., S.A., P.T.Y., and F.C.; investigation, N.B., J.F.M., R.S., K.S.A., S.L., Y.L., D.A.K., K.Z., A.S., and F.C.; resources, A.S. and T.S.; writing—original draft, N.B., P.T.Y., and F.C.; writing—review and editing: N.B., S.A., B.C., P.T.Y., and F.C; funding acquisition, S.A., B.C., and F.C.; supervision, B.C., P.T.Y., and F.C.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Boc | R&D Systems | Cat# AF2385; RRID: AB_2066909 |
| Goat polyclonal anti-Robo3 | R&D Systems | Cat# AF3076; RRID: AB_2181865 |
| Mouse monoclonal anti-β-actin (clone AC-15) | Sigma-Aldrich | Cat# A5441; RRID: AB_476744 |
| Mouse monoclonal anti-Abi1 (clone W8.3) | Gift from Dr Giorgio Scita (Innocenti et al.21) | N/A |
| Mouse monoclonal anti-Flag (clone M2) | Sigma-Aldrich | Cat# F3165; RRID: AB_259529 |
| Rabbit polyclonal anti-Cyfip1/2 | Steffen et al.21 | Anti-Sra-1A (2240) |
| Rabbit polyclonal anti-CYFIP2 | Abcam | Cat# ab79716; RRID: AB_10673468 |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific (Invitrogen) | Cat# A-11122; RRID: AB_221569 |
| Rabbit polyclonal anti-Nckap1 | Steffen et al.21 | 2391-C |
| Rabbit polyclonal anti-Nckap1 | Steffen et al.21 | 4952-B |
| Donkey anti-goat IgG 647 | Jackson ImmunoResearch Laboratory | Cat# 705-605-147; RRID: AB_2340437 |
| Donkey anti-mouse IgG 488 | Molecular Probes | Cat# A21202; RRID: AB_141607 |
| Donkey anti-rabbit IgG 488 | Molecular Probes | Cat# A21206; RRID: AB_2535792 |
| Donkey anti-rabbit IgG Cy3 | Jackson ImmunoResearch Laboratory | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey anti-goat IgG (HRP) | Jackson ImmunoResearch Laboratory | Cat# 705-035-147; RRID: AB_2313587 |
| Goat anti-mouse IgG (HRP) | Jackson ImmunoResearch Laboratory | Cat# 115-035-003; RRID: AB_10015289 |
| Goat anti-rabbit IgG (HRP) | Jackson ImmunoResearch Laboratory | Cat# 111-035-003; RRID: AB_2313567 |
| ChromPure Goat IgG | Jackson ImmunoResearch Laboratory | Cat# 005-000-003; RRID: AB_2336985 |
| Bacterial and virus strains | ||
| DH5α | Thermo Fisher Scientific (Invitrogen) | Cat# 18265-017 |
| Arctic Express DE3 RIL | Agilent | Cat# 230193 |
| BL21 (DE3)TIR | Sigma-Aldrich | Cat# B2935 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Human Sonic Hedgehog (C24II) N-Terminus | R&D | Cat# 1845-SH |
| Recombinant Human Sonic Hedgehog Protein, High Activity | R&D | Cat# 8908-SH |
| Bovine serum albumin (BSA) | MultiCell | Cat# 500-0206 |
| Bovine serum albumin (BSA) IgG-free, Protease-free | Jackson ImmunoResearch Laboratory | Cat# 001-000-161 |
| Phalloidin-TRITC | Sigma-Aldrich | Cat# P1951 |
| Alexa Fluor™ 488 Phalloidin | Thermo Fisher Scientific (Invitrogen) | Cat# A12379 |
| Mowiol 4-88 | Sigma-Aldrich | Cat# 81381 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11 873 580 001 |
| PhosSTOP™ | Roche | Cat# 04 906 837 001 |
| Poly-L-lysine solution (molecular weight 70,000–150,000, concentration: 0.01%) | Sigma-Aldrich | Cat# P4707 |
| DAPI | Sigma-Aldrich | Cat# D95964 |
| Protein A/G-PLUS Agarose beads | Santa Cruz | Cat# sc-2003 |
| Lipofectamine™ 2000 | Thermo Fisher Scientific (Invitrogen) | Cat# 11668019 |
| Lipofectamine 3000 | Life Technologies | Cat# L3000-015 |
| Glutathione Sepharose beads | Cytiva | Cat# 17075605 |
| Amylose beads | NEB | Cat# E8021L |
| SOURCE Q15 | Cytiva | Cat# 17094702 |
| Superdex 200 Increase 10/300 GL (24 mL) | Cytiva | Cat# 28990944 |
| Superdex 75 Increase 10/300 GL, 24mL | Cytiva | Cat# 29148721 |
| L-glutathione reduced, 99% | Fisher | Cat# 50-112-6940 |
| Biotin | Sigma | Cat# B4501-1G |
| Streptavidin Sepharose high performance | GE Healthcare | Cat# 17-5113-01 |
| Sequencing Grade Modified Trypsin | Promega | Cat# V5111 |
| Critical commercial assays | ||
| In-Fusion® Snap Assembly Kit | Takara Bio Inc | Cat# 638945 |
| Nano-Glo® Live Cell Assay System | Promega | Cat# N2012 |
| Deposited data | ||
| Commissural neuron RNAseq dataset | Makihara et al.9 | GSE268644; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE268644 |
| BOC BioID dataset (ProteomeXchange) | This paper | Accession number PXD041651, MassIVE MSV000091742. https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD041651 |
| Contaminant repository for affinity purification database | Mellacheruvu et al.20 | http://www.crapome.org/ |
| Experimental models: Cell lines | ||
| HEK293T cells | Gift from Dr Audrey Claing | N/A |
| HEK293 cells | Gift from Dr Marc Tessier-Lavigne | N/A |
| Cyfip1/2 KO #3 B16-F1 cells | Schaks et al. 201814 | N/A |
| COS7 cells | ATCC® CRL-1651™ | N/A |
| Sf9 cells in Sf-900™ | Thermo Fisher Scientific - Invitrogen | Cat# 11496015 |
| High Five™ (Tni) Cells | Expression Systems | Cat# 94-002F |
| Flp-in T-REx 293 cell | Invitrogen | Cat# R78007 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: BocAP−2 (Boctm2Rsk) | Gift from Dr. Benjamin Allen (Zhang et al.51) | MGI: 5000229 |
| Rat: ARS/Sprague Dawley | Charles River (St. Constant, Canada) | N/A |
| Recombinant DNA | ||
| mEmerald-WASP1-C-14 | Gift from Dr. Michael Davidson | Addgene #54314; RRID: Addgene_54314 |
| pCAGGS-mBoc | Fabre et al.52 | N/A |
| pCAGGS-NCKAP1-Flag | This paper | N/A |
| pCAGGS-NCKAP1sm-Flag (shRNA resistant) | This paper | N/A |
| pcDNA3.1(+)Myc-Cyfip2 | Gift from Dr. Jia-Jia Liu (Xu et al.49) | N/A |
| pcDNA6.2-GW/EmGFP-miR | Invitrogen | K493600 |
| pcDNA6.2-GW/EmGFP-miR (Nckap1) | This paper | N/A |
| pcDNA6.2-GW/EmGFP-miR (Cyfip1) | This paper | N/A |
| pcDNA6.2-GW/EmGFP-miR (Cyfip2) | This paper | N/A |
| pcDNA5 FRT/TO FLAGBirA∗-hBOC | This paper | N/A |
| pCMV-FLAG-CYFIP2 | MRC PPU Reagents and Services | DU23294 |
| pCAGGS-CYFIP2-Flag | This paper | N/A |
| pCMV-Tag2B-Nap1(Flag tagged) | Gift from Dr. Jia-Jia Liu (Xu et al.49) | N/A |
| pEGFP-Abi1 | Innocenti et al.21 | Addgene #74905; RRID: Addgene_74905 |
| pEGFP-C2-Cyfip1 | Steffen et al.22 | N/A |
| pFlag-NAP1-CMV-2 | Gift from Dr. Jean-François Côté (generated by Yamamoto et al.53). | N/A |
| pcDNA3-Human WAVE1-Flag | Westphal et al.54 | N/A |
| pCMV6-Entry-BRK1(HSPC300)-Myc-DDK(Flag) | Origene | Cat# RC200804 |
| pBit1.1-C-TK-LgBit, pBit2.1-C-TK-SmBit and pBit2.1-N-TK-SmBit (NanoBiT® PPI MCS Starter System Vectors) | Promega | Cat# N2014 |
| pCAGGs-mBoc-LgBiT | This paper | N/A |
| pCAGGs-mBoc-ΔICD-LgBiT | This paper | N/A |
| pCAGGs-CYFIP2-SmBiT | This paper | N/A |
| pCAGGs-SmBiT-CYFIP2 | This paper | N/A |
| GST-mBoc FL (Codon optimized) in pGEX-Tev vector | This paper | pKA037 |
| GST-mBoc NT (Codon optimized) in pGEX-Tev vector | This paper | pKA038 |
| GST-mBoc CT (Codon optimized) in pGEX-Tev vector | This paper | pKA034 |
| (MBP)2-HSPC300 in pMalC2Tev vector | Chen et al.18 | pDK118 |
| MBP-WAVE 1–178 in pMalC2Tev vector | Chen et al.18 | pDK120 |
| MBP-Abi2 1–158 in pMalC2Tev vector | Chen et al.18 | pDK075 |
| His6-Sra1 in pAV5a vector | Chen et al.18 | pDK116 |
| Nap1 in pAV5a vector | Chen et al.18 | pDK149 |
| Software and algorithms | ||
| Prism 7 for Mac OSX and Prism 8 for Windows | Graphpad | https://www.graphpad.com |
| ImageJ | NIH | https://imagej.net |
| Volocity 6.0 | Quorum Technologies | https://www.volocity4d.com/download |
| Image Lab | BioRad | http://www.bio-rad.com/ja-jp/product/imagelab-software |
| BLOCK-iT™ RNAi Designer | Thermo Fisher Scientific | https://rnaidesigner.thermofisher.com/rnaiexpress/ |
| Affinity Designer | Serif (Europe) Ltd. | https://affinity.serif.com/en-us/designer/ |
| STRING | STRING CONSORTIUM 2023 | https://string-db.org/ |
| ProHits (Protein High-throughput Solution) | Mount Sinai Hospital | https://www.prohitsms.com/Prohits_download/list.php |
| Other | ||
| Dunn Chamber | Hawksley | DC-100 |
| White 96-well cell culture-treated flat-bottom microplates | Falcon | Cat# 353296 |
| Pierce C18 spin tips | Thermo Scientific | Cat# 84850 |
Experimental model and study participant details
Animals
All animal work was performed in accordance with the Canadian Council on Animal Care Guidelines and approved by the IRCM Animal Care Committee. Wildtype C57BL/6 mice and Boc−/− mice51 (BocAP−2, MGI: 5000229, a gift from Benjamin Allen) were maintained in the IRCM specific pathogen-free animal facility in static microinsulator cages, with up to five mice per cage at a temperature of 20°C–24°C and 40–70% humidity. Embryonic day 0 (E0) was defined as midnight of the night before a plug was found. Tissue from E11.5 embryos of either sex (not determined) was used for experiments. Staged pregnant female Sprague-Dawley rats were obtained from Charles River (St. Constant, Canada and New York, USA). E13.5 embryos of both sexes (not determined) were randomly used for immunostaining and primary dissociated neuron culture. Given that many embryos need to be rapidly pooled to obtain enough material for primary dissociated neuron culture, we cannot determine the sex of the embryos. Thus we are unable to determine if sex might influence the results of our study.
Dissociated commissural neuron culture
Primary commissural neuron cultures were prepared as described previously and maintained at 5% CO2 and 37°C in a humidified incubator.55 Briefly, commissural neurons were isolated from the dorsal fifth of E13.5 rat neural tubes. They were plated in Neurobasal medium supplemented with 10% heat-inactivated FBS and 2 mM GlutaMAX (Life Technologies 35050-061). After ∼18–20 h, the medium was changed to Neurobasal supplemented with 2% B27 (Life Technologies 17504-044) and 2 mM GlutaMAX. Dissociated commissural neuron cultures were used for experiments after 2 days of culture in vitro. For Dunn chamber experiments, electroporated commissural neurons were plated at 300,000–400,000 cells/well in a six-well plate on acid-washed PLL-coated 18 mm square #3D coverslips (Assistent, Germany). For immunostaining, commissural neurons were plated at 35,000 cells/well in a 24-well plate on acid-washed PLL-coated 12 mm round #1D coverslips. For biochemical experiments, commissural neurons were plated at ∼5.8 × 106 cells/plate on PLL-coated 60 mm plates (Falcon 353002).
Cell lines
HEK293T cells (derived from female), HEK293 cells (derived from female) and COS7 cells (derived from male) were maintained in DMEM +10% FBS + penicillin/streptomycin (Invitrogen) at 37°C in a 5% CO2 humidified incubator. Cyfip1/2 KO #3 B16-F1 cells (derived from male) were from Schaks et al.14 The cell lines have not been authenticated and have not been tested for mycoplasma contamination.
Method details
Antibodies
The following antibodies were used: anti-Boc (R&D Systems AF2385; 1:2000 for WB, 1:100 for IF), anti-Flag M2 (Sigma F3165; 1:2000 for WB, 1:1000 for IF), anti-GFP (Invitrogen A11122; 1:2000 for WB), anti-GFP (Invitrogen A11120; 1:1000 for IF), anti-β-actin (Sigma A5441; 1:5000 for WB), anti-Robo3 (R&D Systems AF3076; 1:200 for IHC), anti-CYFIP2 (Abcam 79716; 1:1000 for WB, 1:100 for IF, 1:50 for IHC), anti-Nckap1 (2391-C)22 (1:10 for IF, 1:5 for IHC), anti-Nckap1 (4952-B)22 (1:3000 for WB), anti-Abi121 (1:20 for IF and IHC), anti-Cyfip1/2 (anti-Sra-1A, 2240)22 (1:100 for IF and IHC), donkey-anti-mouse IgG 488 (Molecular Probes A21202), donkey-anti-rabbit IgG 488 (Molecular Probes A21206), donkey-anti-goat IgG 647 (Jackson ImmunoResearch Laboratory 705-605-147), donkey anti-mouse IgG Cy3 (Jackson ImmunoResearch Laboratory 715-165-151), donkey anti-rabbit IgG Cy3 (Jackson ImmunoResearch Laboratory 711-165-152), donkey anti-mouse IgG 647 (Jackson ImmunoResearch Laboratory 715-605-151), donkey anti-goat HRP (Jackson ImmunoResearch Laboratory 705-035-147), goat anti-rabbit HRP (Jackson ImmunoResearch Laboratory 111-035-003), goat anti-mouse HRP (Jackson ImmunoResearch Laboratory 115-035-003), ChromPure goat IgG (Jackson ImmunoResearch Laboratory 005-000-003). All the secondary antibodies for immunostaining were used at 1:1000 and all the secondary antibodies for Western blotting were used at 1:10000 except for the donkey anti-goat HRP which was used at 1:5000.
Plasmids
pCAGGS-mBoc was generated by our lab.52 mEmerald-WASP1-C-14 was a gift from Dr. Michael Davidson (Addgene plasmid #54314; http://n2t.net/addgene:54314; RRID:Addgene_54314). pEGFP-Abi1 was a gift from Dr. Giorgio Scita (Addgene plasmid #74905; http://n2t.net/addgene:74905; RRID:Addgene_74905).21 pCMV5-FLAG3-CYFIP2 was from MRC PPU Reagents and Services (DU23294). CYFIP2-Flag was subcloned from pCMV5-FLAG3-CYFIP2-Flag to pCAGGS using In-Fusion Cloning Technology (In-Fusion Snap Assembly Kit: Takara Bio Inc, cat# 638945) to generate pCAGGS-CYFIP2-Flag. pFLAG-CMV-2-NAP1 plasmid was a gift from Dr. Jean-François Côté (generated by Yamamoto et al.53). NAP1-Flag-WT was subcloned from pFLAG-CMV-2-NAP1 to pCAGGS (in Kpn1 at the multiple cloning site (MCS)) using In-Fusion to generate pCAGGS-NCKAP1-Flag. pcDNA3.1(+)Myc-Cyfip2 (mouse) and pCMV-Tag2B-Nap1 (Flag tagged) (mouse) were gifts from Dr. Jia-Jia Liu.49 pEGFP-C2-Cyfip1 (mouse) was generated by Steffen et al.22 pcDNA3-Human WAVE1-Flag was a gift from Dr. Greg Bashaw (generated by Westphal et al.54). pCMV6-Entry-BRK1(HSPC300)-Myc-DDK(Flag) was obtained from Origene (cat# RC200804).
pCAGGS-NCKAP1sm-Flag, the shRNA resistant form of human NCKAP1, was made by subcloning NAP1-Flag-WT from pFLAG-CMV-2-NAP1 to pCAGGS (into the Kpn1 site) to have better expression in commissural neurons, along with making silent mutations in NCKAP1, using In-Fusion Cloning Technology. The three silent mutations were at human NCKAP1 bp 328–330 (L110) from TTG to TTA, bp 331–333 (L111) from CTG to CTC, and bp 334–336 (N112) from AAT to AAC.
The pBit1.1-C-TK-LgBiT, pBit2.1-C-TK-SmBiT and pBit2.1-N-TK-SmBiT vectors were included in the NanoBiT PPI MCS Starter System Vectors (Promega, cat# N2014). pCAGGS-mBoc-LgBiT was made by subcloning a fusion of mBoc (full length) amplified from pCAGGS-mBoc and the C-terminal LgBiT (including the LgBiT linker) amplified from pBit1.1-C-TK-LgBiT into the Not1 site within the MCS of pCAGGS, using In-Fusion Cloning Technology. pCAGGs-mBoc-ΔICD-LgBiT was made by subcloning a fusion of mBoc, without the intracellular domain, amplified from pCAGGS-mBoc, and the C-terminal LgBiT (including the LgBiT linker) amplified from pBit1.1-C-TK-LgBiT, into the Not1 site within the MCS of pCAGGS, using In-Fusion Cloning Technology. In the final protein expressed, the mBoc intracellular domain is replaced by C-terminal LgBiT (including the LgBiT linker).
pCAGGS-CYFIP2-SmBiT was made by subcloning a fusion of CYFIP2 amplified from pCAGGS-CYFIP2-Flag and the C-terminal SmBiT (including the SmBiT linker) amplified from pBit2.1-C-TK-SmBiT into the Not1 site within the MCS in pCAGGS, using In-Fusion Cloning Technology. pCAGGS-SmBiT-CYFIP2 was made by subcloning a fusion of CYFIP2 amplified from pCAGGS-CYFIP2-Flag and the N-terminal SmBiT (including the SmBiT linker) amplified from pBit2.1-N-TK-SmBiT into the Not1 site within the MCS of pCAGGS using In-Fusion Cloning Technology.
BioID
HEK293 T-Rex Flp-In cells stably expressing pcDNA5 FRT/TO FLAGBirA∗ or pcDNA5 FRT/TO FLAGBirA∗-hBOC were generated using the Flp-In system as previously described.56 After selection in DMEM +10% FBS, 1% penicillin/streptomycin and 100 μg/mL Hygromycin B, 20 × 15 cm plates of subconfluent (60%) cells were incubated for 24 h in complete media supplemented with 1 μg/mL tetracycline (Sigma) and 50 μM biotin (Sigma). Cells were collected and pelleted (5 plates/pellet) at 500×g for 3 min. Cell pellets were washed twice with PBS and dried pellets were snap frozen. Pellets were lysed in 10 mL of modified RIPA lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, protease inhibitor cocktail, 250U Benzonase) at 4°C for 1 h, then sonicated (30 s at 35% power, Sonic Dismembrator 500; Fisher Scientific) to disrupt visible aggregates. The lysate was centrifuged at 21,000×g for 30 min. Clarified supernatants were incubated with 30 μL packed, preequilibrated streptavidin-Sepharose beads (GE) at 4°C for 3 h. Beads were collected by centrifugation (3000 rpm, 2 min), washed six times with 50 mM ammonium bicarbonate pH 8.3, and digested with trypsin (Promega) at 37°C for 18 h. The supernatant containing the tryptic peptides was collected, desalted with C18 tips (ThermoFisher Scientific) and lyophilized. Peptides were resuspended in 0.1% formic acid and 1/6th of the sample was analyzed per mass spectrometry run.
Data were acquired on an Easy nLC-1200 coupled to a Thermo QExactive HF mass spectrometer (ThermoFisher Scientific) operating in a top 20 mode. The mobile phase was composed of Buffer A (0.1% formic acid) and Buffer B (0.1% formic acid in 90% acetonitrile). Peptides were separated using a PepMap RSLC C18 2 μm, 75 μm × 50 cm column and a PepMap 100 C18 3 μm, 75 μm × 2 cm precolumn with a 2 h gradient of 11%–33% Buffer B. Data were analyzed using the trans-proteomic pipeline via the ProHits 2.0.0 software suite. Proteins were identified with a ProteinProphet cut-off of 0.85 (corresponding to 1% FDR). Four biological replicate acquisitions of control HEK293 T-Rex cells expressing the FLAGBirA∗ tag only were performed for comparative purposes with two independent Boc BioID experiments (each experiment containing two biological replicates of T-Rex cells expressing pcDNA5 FRT/TO FLAGBirA∗-hBOC).
To generate a list of high-confidence peptides from the BOC BioID experiments (Table S1), we removed proteins that are promiscuous and/or common contaminants in mass spectrometry proximity dependent biotinylation experiments: proteins found in more than one-third (33%) of the mass spectrometry proximity dependent biotinylation experiments reported in the Contaminant repository for affinity purification database20 were considered to be too frequent to be of interest and were thus removed from our list. Following this, we kept proteins that obtained at least three peptides in each of the two FlagBirA∗-BOC BioID groups, while having no peptide in the four FlagBirA∗ control experiments. These criteria left us with 125 proteins, which are listed in Table S1. Network cluster analysis was performed using STRING (string-db.org), with the following parameters: Only physical subnetworks are shown (thus the edges indicate that the proteins are part of a physical complex), line thickness indicates the strength of data support, minimum required interaction score was set to high confidence (0.700), and disconnected nodes were hidden (i.e., only proteins forming a complex with at least one other protein found in the screen are shown).
Recombinant protein purification
(MBP)2-tagged human HSPC300 and WRC was expressed and purified as previously described.13,18,57 Different GST-tagged mouse Boc intracellular domain (ICD) constructs were expressed in Arctic Express (DE3) RIL (Agilent) at 10°C or BL21 (DE3)T1R cells (Sigma) at 18°C overnight after induction with 1 mM IPTG. To improve expression, Boc ICD FL and NT used a codon-optimized sequence from Thermo Fisher. Proteins were purified by Glutathione Sepharose beads (Cytiva), followed by anion-exchange chromatography through a Source Q15 column and gel filtration chromatography through a Superdex Increase 75 or 200 column. All ion exchange and gel filtration chromatography steps were performed using columns from Cytiva on an ÄKTATM Pure protein purification system.
In vitro pull-down assays
MBP pull-down experiments followed previous procedures.18 Briefly, 40 pmol of (MBP)2-tagged proteins (bait) and 600–1000 pmol of GST-tagged proteins (prey) were mixed with 20 μL of amylose beads (New England Biolabs) in 1 mL of binding buffer [10 mM HEPES pH 7, 50 mM NaCl, 5% (w/v) glycerol, 0.05% (w/v) Triton X-100, and 5 mM beta-mercaptoethanol] at 4°C for 30 min. The beads were centrifuged and washed with 1 mL of binding buffer three times. Bound proteins were eluted with 40 μL of binding buffer supplemented with 2% (w/v) maltose and examined by SDS-PAGE and Coomassie blue staining.
Dunn chamber axon guidance assay and analysis
To quantify the growth cone turning of commissural neurons in response to gradients, we performed the Dunn chamber axon guidance assay as described previously.3 Briefly, commissural neurons were grown on PLL-coated square #3D coverslips as described above. The coverslips were assembled into a Dunn chamber. Gradients were generated in the Dunn chamber with 0.1–0.2 μg/mL Shh (R&D Systems, 1845-SH or 8908-SH) in the outer well. The behavior of commissural neurons was monitored by time-lapse phase contrast microscopy for 2 h at 37°C with a 10× Fluotar or 20× Fluotar objective on a Leica DMIRE2 inverted microscope (Leica, Germany) equipped with an MS-2000 XYZ automated stage (ASI, Eugene, OR). All images were collected on an Orca ER CCD camera (Hamamatsu) using Volocity (Improvision, Waltham, MA). The angle turned of the commissural axons was defined as the angle between the original direction of the axon and a straight line connecting the base of the growth cone from the first to the last time point of the assay period. Axons that were already growing parallel up the Shh gradient (defined as within a 20° angle with respect to the gradient) were excluded from the analysis because they were already growing up the Shh gradient.
shRNA generation and evaluation
shRNAs with a microRNA stem (shRNAmir) were designed against rat/mouse Nckap1, Cyfip1 and Cyfip2 mRNAs using BLOCK-iT RNAi Designer (ThermoFisher Scientific). Oligonucleotides encoding the sequences were ligated into the pcDNA6.2-GW/EmGFP-miR vector. The EmGFP-miR cassettes were then amplified by PCR and cloned into the pCAGGS vector. The efficiency and specificity of these shRNAmirs were evaluated in vitro using HEK293T cells co-transfected with tagged expression vectors (Figure S9).
shRNA target sequences used for knockdown are as below:
Nckap1 shRNA: 5′-ATGTCTGTGACTTGCTGAATA-3’
Cyfip1 shRNA: 5′-CACAACGTGTCTGCACCTTAT-3’
Cyfip2 shRNA: 5′-ATCGAGTATGCAGAACTCAAA-3’
Electroporation of commissural neurons
Neurons were electroporated with the Amaxa 96-well Shuttle using the P3 Primary Cell 96-well Nucleofector Kit (Lonza, Switzerland). For each electroporation in one well of a 96-well Nucleofector plate, 0.45–1 × 106 commissural neurons were electroporated with either 0.5–0.6 μg shRNA expression vector or 0.45 μg shRNA expression vector together with 0.25 μg NCKAP1sm-Flag or pCAGGS empty vector according to the manufacturer’s instructions. The electroporation was performed with the 96-DC-100 program. ∼80–90% of commissural neurons successfully expressed the vectors after electroporation.
Immunostaining
For immunostaining of spinal cord cross-sections, mouse embryos were collected at E11.5 and fixed in 4% PFA for 1 h at 4°C and washed with PBS. Fixed embryos were transferred to 30% sucrose in PBS at 4°C overnight, then embedded in OCT and frozen. 12 μm sections were cut from the frozen tissues and then immunostained. Immunostaining was performed by permeabilizing the spinal cord tissue sections with 0.3% Triton X-100 for 15 min followed by washing in PBS. Spinal cord tissue sections were then blocked with 5% donkey serum with 0.05% Triton X-100 in PBS for 1 h. The blocking solution was replaced with the primary antibody diluted in PBS with 1% donkey serum and 0.05% Triton X-100 and incubated overnight at 4°C. After washing, the samples were incubated with the secondary antibodies diluted in PBS with 1% donkey serum and 0.05% Triton X-100 for 1 h at room temperature. The samples were mounted in Mowiol.
For immunostaining of dissociated commissural neurons, cells were fixed for 15 min by adding an equal volume of 8% PFA in PBS (final concentration 4% PFA) to the cultures. COS7 cells were transfected with the indicated plasmids using Lipofectamine 3000 (Invitrogen) and cultured for 48 h. Cells were fixed for 15 min in 4% PFA. Commissural neurons and COS7 cells were then washed in PBS and permeabilized with 0.05% Triton X-100 in PBS for 1 min followed by washing in PBS. The cells were then blocked with 5% donkey serum or 10% BSA in PBS for 30 min - 1 h. The blocking solution was replaced with the primary antibodies diluted in 1% BSA (Jackson ImmunoResearch) in PBS. After washing, the samples were incubated with the secondary antibodies diluted in 1% BSA in PBS. F-actin was labeled by incubation with phalloidin-TRITC (Sigma-Aldrich; 1:150-1:250) or phalloidin-Alexa Fluor 488 (Life Tech A12379; 1:250) in 1% BSA in PBS. Nuclei were stained with DAPI (Sigma-Aldrich D9564; 1:10000). The samples were mounted in Mowiol.
Western blotting
Cells were lysed with LMB buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 1% NP-40, 10 mM MgCl2, 1 mM EDTA, 10% glycerol) or SLB buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40) with protease inhibitors (Roche 11873580001) and phosphatase inhibitors (Roche 04906837001) and boiled in SDS sample buffer (Laemmli buffer with β-mercaptoethanol) for 5 min. Protein samples were separated by SDS-PAGE and transferred to PVDF membrane. The membranes were blocked with 5% skim milk in TBST (0.01 M Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Tween 20), followed by primary antibody incubation in 1% skim milk in TBST. Secondary antibodies were conjugated to horseradish peroxidase and Western blots were visualized with chemiluminescence.
Brains from WT and Boc−/− E11.5 embryos were collected and lysed with RIPA lysis buffer (150 mM sodium chloride, 50 mM Tris-HCl pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with phosphatase inhibitors and protease inhibitors. Then, the protein concentration of the lysates was measured by Bradford assay. 20 μg of protein was used for SDS-PAGE followed by Western blotting.
Co-immunoprecipitation
HEK293T or HEK293 cells were transfected with expression vectors using Lipofectamine 2000 (Invitrogen) or Lipofectamine 3000 (Life Technologies L3000-015). 24–48 h after transfection, cells were serum-starved overnight in DMEM with 0.2–0.5% FBS and lysed with LMB lysis buffer when the immunoprecipitation was performed with the anti-Boc antibody (R&D Systems, AF2385), and SLB buffer with protease and phosphatase inhibitors when the immunoprecipitation was performed with the anti-FLAG antibody (Sigma, #F3165). For co-IP of endogenous proteins, commissural neurons were lysed with SLB buffer with protease and phosphatase inhibitors. 2–3 mg of HEK cell lysates or 0.9 mg of commissural neuron lysates in ∼750 μL LMB/SLB buffer with protease and phosphatase inhibitors were incubated with 2 μg antibody for HEK cell lysates or 0.5 μg antibody for commissural neuron lysates and Protein A/G-agarose beads (Santa Cruz Protein A/G PLUS-Agarose) for 4 h at 4°C (Boc antibody) or 1–2 h at 4°C (Flag M2 antibody) to capture the immunoprecipitated proteins. The beads were washed 3 times with LMB/SLB buffer and proteins binding to the beads were eluted by adding SDS sample buffer at 95°C for 5 min. The immunoprecipitated proteins were analyzed by SDS-PAGE and Western blotting.
Actin was used as a loading control to verify that equal amounts of lysates were used for the experiments. For the quantification in Figure 2, we measured the band intensities on the Western blots to determine the amount of protein present. We normalised the amount of the protein in the immunoprecipitate (ProteinAIP) to the amount of the protein in the cell lysate (ProteinAlysate), to account for any variation in protein expression between the different lysates. This measurement, ProteinAIP/ProteinAlysate, was then expressed relative to the “Boc + WRC” condition in each co-IP experiment, such that the relative interaction between ProteinA and Boc was set to “1” in the presence of the WRC, i.e., relative ProteinA-Boc interaction = [ProteinAIP/ProteinAlysate]conditionX/[ProteinAIP/ProteinAlysate]Boc+WRC.
Image acquisition
Images of immunostained dissociated commissural neurons were acquired using a Leica SP8 confocal microscope with a 63× objective. Image acquisition for spinal cord immunohistochemistry was done using an LSM700 confocal microscope (Carl Zeiss) with either a 10X or a 40× objective.
NanoBiT assays
NanoBiT assays were used to detect protein interactions in live cells using the Nano-Glo Live Cell Assay System (Promega, N2012). HEK293 cells were seeded at 14 000 cells/well and COS7 cells were seeded at 8 000 cells/well in white 96-well cell culture-treated flat-bottom microplates (Falcon, 353296). ∼24 h later, HEK293 and COS7 cells were transfected with the indicated LgBiT and SmBiT expression vectors using Lipofectamine 3000 (Life Technologies L3000-015). Each condition was always done in triplicate. 24 h after transfection, the media from each well was carefully changed to 80 μL/well of DMEM with 0.5% FBS. Then 20 μL of 5× Nano-Glo Live Cell Substrate (containing the cell-permeable furimazine substrate diluted in LCS dilution buffer) was added to each well. The plates were gently mixed and luminescence was read immediately using a Glomax Microplate Reader using the Glomax Luminescence Protocol with an integration time of 0.5 s. When confirmation of protein expression levels was needed, cells were then lysed with SLB lysis buffer with protease inhibitors and phosphatase inhibitors and analyzed by SDS-PAGE and Western blotting.
Quantification and statistical analysis
Statistical analysis was performed with Prism 7 for Mac OSX or Prism 10 for Windows (GraphPad). All error bars represent SEM and asterisks (∗) indicate significance as follows: ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001, n.s. = not significant (p > 0.05). The statistical analysis used in each experiment and the definition of n are stated in the figure legends. All t-tests were two-tailed, except for those measuring protein levels after shRNA knockdown, where one-tailed t-tests were used.
Published: November 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111333.
Contributor Information
Patricia T. Yam, Email: patricia.yam@ircm.qc.ca.
Frédéric Charron, Email: frederic.charron@ircm.qc.ca.
Supplemental information
References
- 1.Dessaud E., McMahon A.P., Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 2.Yam P.T., Charron F. Signaling mechanisms of non-conventional axon guidance cues: the Shh, BMP and Wnt morphogens. Curr. Opin. Neurobiol. 2013;23:965–973. doi: 10.1016/j.conb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Yam P.T., Langlois S.D., Morin S., Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 2009;62:349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 4.Okada A., Charron F., Morin S., Shin D.S., Wong K., Fabre P.J., Tessier-Lavigne M., McConnell S.K. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- 5.Charron F., Stein E., Jeong J., McMahon A.P., Tessier-Lavigne M. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 6.Sauve R., Morin S., Yam P.T., Charron F. beta-arrestins Are Scaffolding Proteins Required for Shh-Mediated Axon Guidance. J. Neurosci. 2024;44 doi: 10.1523/JNEUROSCI.0261-24.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dent E.W., Gupton S.L., Gertler F.B. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harbor Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lepelletier L., Langlois S.D., Kent C.B., Welshhans K., Morin S., Bassell G.J., Yam P.T., Charron F. Sonic Hedgehog Guides Axons via Zipcode Binding Protein 1-Mediated Local Translation. J. Neurosci. 2017;37:1685–1695. doi: 10.1523/JNEUROSCI.3016-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makihara S., Morin S., Ferent J., Côté J.F., Yam P.T., Charron F. Polarized Dock Activity Drives Shh-Mediated Axon Guidance. Dev. Cell. 2018;46:410–425.e7. doi: 10.1016/j.devcel.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Kurisu S., Takenawa T. The WASP and WAVE family proteins. Genome Biol. 2009;10:226. doi: 10.1186/gb-2009-10-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omotade O.F., Pollitt S.L., Zheng J.Q. Actin-based growth cone motility and guidance. Mol. Cell. Neurosci. 2017;84:4–10. doi: 10.1016/j.mcn.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., Rosen M.K. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B., Chou H.T., Brautigam C.A., Xing W., Yang S., Henry L., Doolittle L.K., Walz T., Rosen M.K. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife. 2017;6 doi: 10.7554/eLife.29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaks M., Singh S.P., Kage F., Thomason P., Klünemann T., Steffen A., Blankenfeldt W., Stradal T.E., Insall R.H., Rottner K. Distinct Interaction Sites of Rac GTPase with WAVE Regulatory Complex Have Non-redundant Functions in Vivo. Curr. Biol. 2018;28:3674–3684.e6. doi: 10.1016/j.cub.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shakir M.A., Jiang K., Struckhoff E.C., Demarco R.S., Patel F.B., Soto M.C., Lundquist E.A. The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics. 2008;179:1957–1971. doi: 10.1534/genetics.108.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan R., Gohl C., Fleige A., Klämbt C., Bogdan S. Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila. Mol. Biol. Cell. 2011;22:4079–4092. doi: 10.1091/mbc.E11-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y., Quinn C.C. MIG-10 functions with ABI-1 to mediate the UNC-6 and SLT-1 axon guidance signaling pathways. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B., Brinkmann K., Chen Z., Pak C.W., Liao Y., Shi S., Henry L., Grishin N.V., Bogdan S., Rosen M.K. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhari K., Gorla M., Chang C., Kania A., Bashaw G.J. Robo recruitment of the Wave regulatory complex plays an essential and conserved role in midline repulsion. Elife. 2021;10 doi: 10.7554/eLife.64474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E.B., Di Fiore P.P., Carlier M.F., Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 22.Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T.E.B. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cioni J.M., Wong H.H.W., Bressan D., Kodama L., Harris W.A., Holt C.E. Axon-Axon Interactions Regulate Topographic Optic Tract Sorting via CYFIP2-Dependent WAVE Complex Function. Neuron. 2018;97:1078–1093.e6. doi: 10.1016/j.neuron.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kage F., Doring H., Mietkowska M., Schaks M., Gruner F., Stahnke S., Steffen A., Musken M., Stradal T.E.B., Rottner K. Lamellipodia-like actin networks in cells lacking WAVE regulatory complex. J. Cell Sci. 2022;135:jcs260364. doi: 10.1242/jcs.260364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toret C.P., D'Ambrosio M.V., Vale R.D., Simon M.A., Nelson W.J. A genome-wide screen identifies conserved protein hubs required for cadherin-mediated cell-cell adhesion. J. Cell Biol. 2014;204:265–279. doi: 10.1083/jcb.201306082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., Takenawa T., Kaibuchi K. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol. Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soderling S.H., Guire E.S., Kaech S., White J., Zhang F., Schutz K., Langeberg L.K., Banker G., Raber J., Scott J.D. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen B.L., Song J.Y., Izzi L., Althaus I.W., Kang J.S., Charron F., Krauss R.S., McMahon A.P. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunda P., Craig G., Dominguez V., Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Rogers S.L., Wiedemann U., Stuurman N., Vale R.D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steffen A., Faix J., Resch G.P., Linkner J., Wehland J., Small J.V., Rottner K., Stradal T.E.B. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell. 2006;17:2581–2591. doi: 10.1091/mbc.e05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlienger S., Yam P.T., Balekoglu N., Ducuing H., Michaud J.F., Makihara S., Kramer D.K., Chen B., Fasano A., Berardelli A., et al. Genetics of mirror movements identifies a multifunctional complex required for Netrin-1 guidance and lateralization of motor control. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding B., Yang S., Schaks M., Liu Y., Brown A.J., Rottner K., Chowdhury S., Chen B. Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. Nat. Commun. 2022;13:5444. doi: 10.1038/s41467-022-33174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza M.C. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin. Cell Dev. Biol. 2013;24:272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ardern H., Sandilands E., Machesky L.M., Timpson P., Frame M.C., Brunton V.G. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil Cytoskeleton. 2006;63:6–13. doi: 10.1002/cm.20101. [DOI] [PubMed] [Google Scholar]
- 36.Vuong T.A., Leem Y.E., Kim B.G., Cho H., Lee S.J., Bae G.U., Kang J.S. A Sonic hedgehog coreceptor, BOC regulates neuronal differentiation and neurite outgrowth via interaction with ABL and JNK activation. Cell. Signal. 2017;30:30–40. doi: 10.1016/j.cellsig.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart J.R., Gonzalez F.H., Kawai H., Yuan Z.M. c-Abl interacts with the WAVE2 signaling complex to induce membrane ruffling and cell spreading. J. Biol. Chem. 2006;281:31290–31297. doi: 10.1074/jbc.M602389200. [DOI] [PubMed] [Google Scholar]
- 39.Huang C.H., Lin T.Y., Pan R.L., Juang J.L. The involvement of Abl and PTP61F in the regulation of Abi protein localization and stability and lamella formation in Drosophila S2 cells. J. Biol. Chem. 2007;282:32442–32452. doi: 10.1074/jbc.M702583200. [DOI] [PubMed] [Google Scholar]
- 40.Briancon-Marjollet A., Ghogha A., Nawabi H., Triki I., Auziol C., Fromont S., Piche C., Enslen H., Chebli K., Cloutier J.F., et al. Trio mediates netrin-1-induced Rac1 activation in axon outgrowth and guidance. Mol. Cell Biol. 2008;28:2314–2323. doi: 10.1128/MCB.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irie F., Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat. Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- 42.Sengar A.S., Ellegood J., Yiu A.P., Wang H., Wang W., Juneja S.C., Lerch J.P., Josselyn S.A., Henkelman R.M., Salter M.W., Egan S.E. Vertebrate intersectin1 is repurposed to facilitate cortical midline connectivity and higher order cognition. J. Neurosci. 2013;33:4055–4065. doi: 10.1523/JNEUROSCI.4428-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Z., Makihara S., Yam P.T., Teo S., Renier N., Balekoglu N., Moreno-Bravo J.A., Olsen O., Chédotal A., Charron F., Tessier-Lavigne M. Long-Range Guidance of Spinal Commissural Axons by Netrin1 and Sonic Hedgehog from Midline Floor Plate Cells. Neuron. 2019;101:635–647.e4. doi: 10.1016/j.neuron.2018.12.025. [DOI] [PubMed] [Google Scholar]
- 44.Arevalo J.C., Pereira D.B., Yano H., Teng K.K., Chao M.V. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J. Biol. Chem. 2006;281:1001–1007. doi: 10.1074/jbc.M504163200. [DOI] [PubMed] [Google Scholar]
- 45.Lopez M.L., Lo M., Kung J.E., Dudkiewicz M., Jang G.M., Von Dollen J., Johnson J.R., Krogan N.J., Pawłowski K., Jura N. PEAK3/C19orf35 pseudokinase, a new NFK3 kinase family member, inhibits CrkII through dimerization. Proc. Natl. Acad. Sci. USA. 2019;116:15495–15504. doi: 10.1073/pnas.1906360116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders M.A., Basson M.D. p130cas but not paxillin is essential for Caco-2 intestinal epithelial cell spreading and migration on collagen IV. J. Biol. Chem. 2005;280:23516–23522. doi: 10.1074/jbc.M413165200. [DOI] [PubMed] [Google Scholar]
- 47.Nishihara H., Maeda M., Oda A., Tsuda M., Sawa H., Nagashima K., Tanaka S. DOCK2 associates with CrkL and regulates Rac1 in human leukemia cell lines. Blood. 2002;100:3968–3974. doi: 10.1182/blood-2001-11-0032. [DOI] [PubMed] [Google Scholar]
- 48.Sanders M.A., Ampasala D., Basson M.D. DOCK5 and DOCK1 regulate Caco-2 intestinal epithelial cell spreading and migration on collagen IV. J. Biol. Chem. 2009;284:27–35. doi: 10.1074/jbc.M808010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu C., Fu X., Zhu S., Liu J.J. Retrolinkin recruits the WAVE1 protein complex to facilitate BDNF-induced TrkB endocytosis and dendrite outgrowth. Mol. Biol. Cell. 2016;27:3342–3356. doi: 10.1091/mbc.E16-05-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou W., Dong X., Broederdorf T.R., Shen A., Kramer D.A., Shi R., Liang X., Miller D.M., Xiang Y.K., Yasuda R., et al. A Dendritic Guidance Receptor Complex Brings Together Distinct Actin Regulators to Drive Efficient F-Actin Assembly and Branching. Dev. Cell. 2018;45:362–375. doi: 10.1016/j.devcel.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W., Hong M., Bae G.U., Kang J.S., Krauss R.S. Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis. Model. Mech. 2011;4:368–380. doi: 10.1242/dmm.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fabre P.J., Shimogori T., Charron F. Segregation of ipsilateral retinal ganglion cell axons at the optic chiasm requires the Shh receptor Boc. J. Neurosci. 2010;30:266–275. doi: 10.1523/JNEUROSCI.3778-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto A., Suzuki T., Sakaki Y. Isolation of hNap1BP which interacts with human Nap1 (NCKAP1) whose expression is down-regulated in Alzheimer's disease. Gene. 2001;271:159–169. doi: 10.1016/s0378-1119(01)00521-2. [DOI] [PubMed] [Google Scholar]
- 54.Westphal R.S., Soderling S.H., Alto N.M., Langeberg L.K., Scott J.D. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langlois S.D., Morin S., Yam P.T., Charron F. Dissection and culture of commissural neurons from embryonic spinal cord. J. Vis. Exp. 2010;39 doi: 10.3791/1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coyaud E., Mis M., Laurent E.M.N., Dunham W.H., Couzens A.L., Robitaille M., Gingras A.C., Angers S., Raught B. BioID-based Identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 Ligase Substrates. Mol. Cell. Proteomics. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen B., Padrick S.B., Henry L., Rosen M.K. Biochemical reconstitution of the WAVE regulatory complex. Methods Enzymol. 2014;540:55–72. doi: 10.1016/B978-0-12-397924-7.00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The BioID data have been deposited at ProteomeXchange and is publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.