Abstract
Recently, Deep Learning (DL) models have shown promising accuracy in analysis of medical images. Alzeheimer Disease (AD), a prevalent form of dementia, uses Magnetic Resonance Imaging (MRI) scans, which is then analysed via DL models. To address the model computational constraints, Cloud Computing (CC) is integrated to operate with the DL models. Recent articles on DL-based MRI have not discussed datasets specific to different diseases, which makes it difficult to build the specific DL model. Thus, the article systematically explores a tutorial approach, where we first discuss a classification taxonomy of medical imaging datasets. Next, we present a case-study on AD MRI classification using the DL methods. We analyse three distinct models-Convolutional Neural Networks (CNN), Visual Geometry Group 16 (VGG-16), and an ensemble approach-for classification and predictive outcomes. In addition, we designed a novel framework that offers insight into how various layers interact with the dataset. Our architecture comprises an input layer, a cloud-based layer responsible for preprocessing and model execution, and a diagnostic layer that issues alerts after successful classification and prediction. According to our simulations, CNN outperformed other models with a test accuracy of 99.285%, followed by VGG-16 with 85.113%, while the ensemble model lagged with a disappointing test accuracy of 79.192%. Our cloud Computing framework serves as an efficient mechanism for medical image processing while safeguarding patient confidentiality and data privacy.
Keywords: Smart healthcare, Blockchain, Internet of things, Security & privacy, Patient data, Challenges
Subject terms: Computational platforms and environments, Health care, Biomedical engineering, Computer science
Introduction
In recent years, the healthcare industry has become increasingly interested in the latest technologies. Cloud computing (CC)1 and Deep Learning (DL)2 are two main domains that have revolutionized the healthcare ecosystem, having a great amount of potential in providing effective and innovative solutions to the challenges faced by healthcare professionals. One such example and challenge is the early treatment and diagnosis of Alzheimer’s Disease (AD), which is a neurological disorder that has affected millions of people worldwide. CC allows users to process and access data, storing data and applications on remote servers via Internet. The ability of healthcare providers to store and process enormous amounts of data in an economical and scalable manner due to CC has completely changed the healthcare sector3. However, significant security and privacy risks are associated with using CC in healthcare, especially when dealing with sensitive patient data4. Today, healthcare services make use of drones to deliver medical aid to remote places to extend the medical facility5,6.
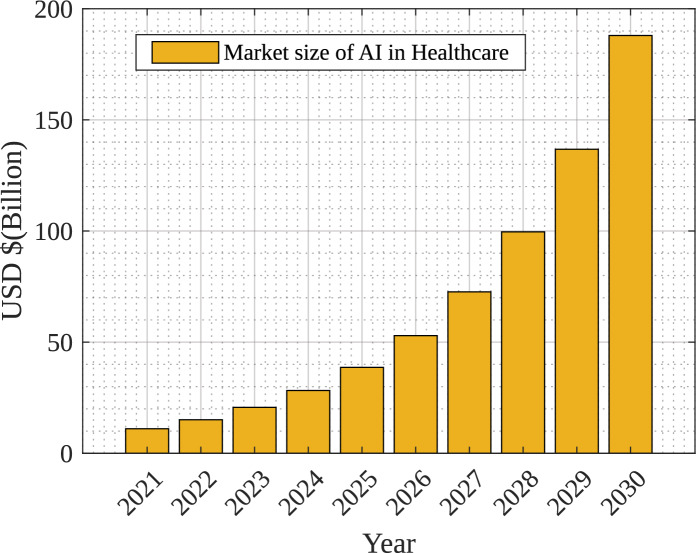
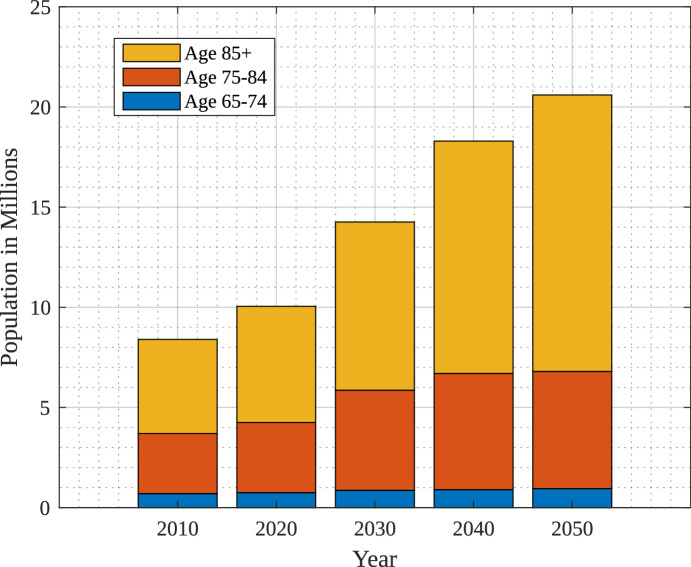
The impact of AI in 2022 was estimated at around 15 billion USD and it is expected to be 187 billion USD by the year 2030 which is a 36% compound annual growth rate. Figure 1 shows the growth of AI in healthcare over the year7. The use of digital technologies and enhanced care of patients are the main factors to boost the growth of market size in the healthcare domain. There have been observations on DL-based solutions over the years for diagnosis that have enhanced the trust of the medical community in primary research8. Moreover, explainable AI has provided a new dimension to verify and provide explanations for the decision made9. The projected number of people in different age groups in the U.S. suffering from Alzheimer’s Disease from the year 2010 to 2050 are shown in Fig. 2, as per study at10. Figure 2 also indicates drastic growth of Alzheimer’s Disease from 4.7 million to 13.8 million after the age of 85 years because of social and environmental conditions.
Figure 1.
Market size of AI in healthcare domain7.
Figure 2.
Impact of Alzheimer on age10.
DL is a promising approach that has demonstrated impressive performance in a number of areas, including Speech Recognition, Medical Image Classification, and Natural Language Processing11–14. Consequently, there has been a lot of interest in using DL models in healthcare in recent years. DL models in healthcare can help with early disease detection, effective diagnosis, and individualized treatment15. The early detection and diagnosis of AD is one of the most promising uses of DL in healthcare.
The field of medical imaging is progressing very fast with the developments presented for 2019 and beyond giving us interesting insights on both technology adoption and market dynamics. Ultrasound imaging, a critical aspect of diagnostic procedures, accounted for a large share in the market of 20.6% across globes as at 2019. North America was also prominent in this period by having a leading market portion of 42.1%, thereby emphasizing the pioneering nature of the region’s healthcare innovation16. An important trend is that Artificial Intelligence (AI) is being integrated into medical imaging, and it is anticipated to hit $4.3 billion by 2026 with regard to estimates. The global shift towards these diagnostic imaging processes is expected to reach 7.6 billion by 2030. Additionally, X-ray systems had dominated the market at around 30% in terms of its total sales in the year 2019 while multi-slice CT scanners had taken up about three quarters of their own markets back in 2018. The financial landscape valued this total at $3,199 million in FY2020 setting pace for notable valuations anticipated for MRI scanners’ markets in FY2027 or PET scanners market by FY2019 respectively. Moreover, the push towards advanced imaging techniques is mirrored in the growth predictions for nuclear medicine imaging and medical imaging software, poised for significant growth by 2026. The Asia-Pacific region, not to be outdone, is projected to experience a 5.9% CAGR in the medical imaging market between 2018 and 2023, reflecting a global tilt towards more sophisticated diagnostic capabilities. Teleradiology emerges as a pivotal technology, with its market projected to reach $10.8 billion by 2026. Amidst these advancements, General Electric’s market presence, with a 15.2% share in 2019, illustrates the competitive nature of the industry. Despite these technological strides, challenges such as the 20% of breast cancers undetected by mammography alone highlight the ongoing need for innovation and improvement in imaging modalities, including the promising growth of Optical Coherence Tomography (OCT) systems, expected to ascend at a CAGR of 10.4% from 2021 to 2028. These developments not only reflect the current state but also chart a course for the future of medical imaging, underscoring a blend of technological advancements, market shifts, and the perpetual aim of enhancing diagnostic accuracy and patient care.
Dataset images need to be processed into the platform that supports high computational environment. Hence, a platform for the storing, processing, and analysis of massive datasets is provided by CC that has transformed image-based categorization analysis in the healthcare system. CC offers a scalable and economical alternative for processing, storing, and analyzing enormous numbers of medical images in light of the growing digitization of medical data and the explosion in high-resolution medical imaging. Healthcare organizations can use the enormous computing capacity of the cloud to speed up challenging image classification tasks, such as locating cancers, lesions, or anomalies in radiological scans. Healthcare providers can drastically cut processing time by outsourcing resource-intensive computations to the cloud, enabling quicker diagnostic and treatments17.
Additionally, the collaborative nature of the cloud enables seamless data exchange and inter-institutional collaboration, allowing medical professionals to access and examine images from a distance. This has significant benefits for telemedicine and second-opinion settings where experts can share their opinion irrespective of regional limitations18. Advanced ML and AI techniques can also be used with cloud-based image analysis. These algorithms are capable of learning from various datasets and progressively increasing the precision. Hence, they can spot patterns and anomalies in medical images. The healthcare sector can increase diagnostic precision, boost patient outcomes, and streamline the entire healthcare workflow by utilizing the power of CC19. Therefore, CC is the best option for healthcare providers to store and process massive amount of patient data because of its scalability and everywhere accessibility. Using the given data, DL models can be trained on cloud platforms to find patterns and forecast outcomes20. As the CC resources are widely available so, DL models especially suited for CC in the healthcare industry must be developed21.
CC has completely changed how data is processed and stored22. CC has made it possible for healthcare workers to communicate effectively, share medical data, and monitor patients from a distance. With the help of the Cloud and DL the image classifications can be done in the optimal and fastest way. In this study we have focused on application of DL models for the early identification and diagnosis of AD in the healthcare industry. Alzheimer’s Disease (AD) is a degenerative neurological condition that impairs elderly people’s memory and cognitive abilities. A lot of patient information is needed for the diagnosis and treatment of AD, including medical histories, blood tests, and brain imaging studies. There is a need for effective and precise tools to analyze and derive valuable insights from the growing amount of healthcare provider data. A subset of ML called DL has demonstrated well in addressing this demand and has been used in a number of healthcare fields, including AD23.
In the field of categorizing AD, Machine Learning (ML) and DL have become crucial tools, providing previously unattainable insights into the complicated patterns of neuroimaging, clinical, and other medical-related data24. Support Vector Machines (SVMs) and Random Forests, ML approaches, have demonstrated proficiency in extracting discriminative features from neuroimaging modalities like Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET) scans. These techniques work well in lower-dimensional areas when the retrieved characteristics indicate distinct neuroanatomical differences linked to various illness phases. In addition to this, the development of DL has also played important advances in the classification of AD. Convolution Neural Networks (CNN) have demonstrated a special aptitude for identifying spatial patterns in neuroimaging data, catching minute structural anomalies that would challenge conventional analysis. The architecture’s innate capacity to preserve context from earlier time steps gives them the sensitivity to spot early alterations in brain activity linked to diseases. Utilizing Transfer Learning (TL), which allows pre-trained DL models, has also proved a good option for the identification of AD from the available datasets25.
For both medical professionals and patients, accurate classification of medical images using cutting-edge image analysis algorithms has important consequences26,27. The accurate classification of images serves as a powerful tool for decision-making for clinicians, enhancing their clinical knowledge with quantitative and data-driven insights28,29. By identifying small patterns and anomalies that can be invisible to the human eye, physicians can improve diagnostic accuracy by utilizing ML and DL models30. This is especially important in fields like radiology, where proper classification of medical pictures can help early discovery of the diseases, allowing prompt treatment and better patient health31. Additionally, automated image analysis lessens the cognitive burden on doctors, enabling them to devote more time for tailored patient care and nuanced interpretations as opposed to lengthy routine picture assessments32. Doctors may confidently diagnose illnesses, create personalized treatment plans, and track disease development with increased precision by incorporating computational models into their workflow.
Accurate medical picture classification benefits patients by earlier identification, more potent therapies, earlier start of suitable treatments, all-around improved healthcare experiences, and possibly preventing disease progression in its early stages. Advanced image analysis can also reduce diagnostic ambiguity, which reduces patient anxiety since people are better able to make educated decisions about their healthcare when information is disseminated quickly and accurately33. As medical technology develops, patients gain from a healthcare ecosystem where clinical knowledge34 and AI-powered image analysis work together to provide optimized care routes35. This environment promotes patient population trust and well-being36. For the classification challenge of the medical images for Alzheimer’s Disease, three DL models are analyzed and implemented using CNN, Visual Geometry Group (VGG-16), and the Ensembled model, in this study. The topic of early identification and detection of AD is addressed by the proposed architecture and algorithm.
Research contributions
This research offers insights into the current status of DL and CC research in healthcare. Through a comprehensive review of existing literature, it uncovers research gaps. By utilizing MRI images, the study enhances AD diagnosis and prognosis, potentially leading to faster and more precise treatment.
The paper compares the performance of three distinct DL models namely: CNN, VGG-16, and Ensembled model for classification and prediction task of Alzheimer’s Disease. This evaluation offers important insights into the applicability and efficicny of various models for medical image analysis.
Simulation results show that CNN and VGG-16 models perform better on accuracy and predictions for medical image analysis investigations.
The remaining work is organized as follows. Section “Related work” provides a literature survey of the existing states of the art. Section “Classification of medical imaging datasets” consists of the classification of diseases and their related database available in the public domain for research purposes. Section “Case study: Alzheimer’s MRI classification using DL methods” provides architecture and algorithm for the proposed case study on AD using DL and transfer learning, Sect. “Simulation results” provides the result and comparative analysis of CNN, VGG-16, and Ensembled model on different datasets. The final section “Conclusion” provides the summary of the comprehensive analysis of the different datasets.
Related work
This section presents a details analysis of the literature survey mentioned in the Table 1 with existing state-of-the-art approaches. In recent years, DL techniques for medical image processing have attracted increasing amounts of interest for medical practitioners. For various medical imaging modalities, such as ultrasound and MRI, researchers have presented a variety of DL architectures. In the field of healthcare Nancy et al.37 proposed a smart healthcare system that integrates Internet-of-Things (IoT) and a cloud-based monitoring system for the prediction of heart disease using DL. The suggested method predicts heart disease with a 92.8% accuracy rate. The authors conclude that the suggested approach can effectively predict heart disease and can be used in real-time healthcare monitoring analysis. An approach for a cloud-based encrypted communication system for diagnosing tuberculosis (TB) was presented by Ahmed et al.38. The proposed system seeks to protect the privacy and security of medical images while enabling efficient and accurate diagnosis using DL techniques. Authors in39 propose a hybrid Deep Learning network to enhance the classification of Mild Cognitive Impairment (MCI) from brain MRI scans. The model integrates the Swin Transformer, Dimension Centric Proximity Aware Attention Network (DCPAN), and Age Deviation Factor (ADF) to improve feature representation. By combining global, local, and proximal features with dimensional dependencies, the network achieves better classification results. The experimental evaluation on the ADNI dataset shows the model’s effectiveness, with an accuracy of 79.8%, precision of 76.6%, recall of 80.2%, and an F1-score of 78.4%. Furthermore, some studies have also focused on the security aspects of images as well. For instance, Gupta et al.40 presents a DL approach for enhancing the security of multi-cloud healthcare systems that outperforms other methods in terms of detection accuracy and false alarm rate. Qamar et al.41 proposed a DL-based method for analyzing healthcare data at the same time ensuring cloud-based security that achieves high accuracy in classifying healthcare data and can be used as an effective tool for healthcare data analysis with cloud-based cybersecurity.
Table 1.
Comparative analysis of the existing state of the art.
| References | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Pros | Cons |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gupta et al.55 | Y | N | N | N | N | N | Y | Y | Y | Y | Blockchain enabled system for early classification and detection of monkey-pox with the help of transfer learning on skin lesion dataset | Increased latency and bandwidth while accessing blockchain |
| Akhtar et al.56 | Y | N | N | N | N | Y | Y | Y | Y | N | Internet of medical thing based healthcare monitoring system that uses improved advanced feature set RNN | System requires high bandwidth and require large space to store patients data that is not secure |
| Nancy et al.37 | Y | Y | Y | N | N | Y | N | N | N | Y | A healthcare monitoring system for remote monitoring of patients health and real-time analysis using IOT and Cloud | To handle IOT data requires higher bandwidth |
| Ahmed et al.38 | Y | Y | Y | N | Y | Y | Y | N | Y | Y | A Perceptual Encryption method, applicable for both the images i.e. color and grayscale, improves robustness against different attacks | An assumption of a cloud computation server that is private, which may not be applicable in all settings |
| Gupta et al.40 | Y | N | N | Y | Y | Y | Y | N | N | N | The hierarchical model integrates seamlessly with the healthcare network’s hierarchical structure | Increased risk of malicious agents stealing or altering sensitive patient data |
| Qamar et al.41 | Y | N | N | N | N | N | Y | N | Y | Y | Utilizes DL-based classification and feature selection to analyze EHR data with a focus on cyber security | Additional complexity and potential security risks |
| Simeone et al.42 | Y | N | N | Y | Y | N | N | N | Y | Y | Cloud-based platform for worker health monitoring in hazardous manufacturing environments | The potential cost and complexity for implementation |
| Bolhasa-ni et al.57 | Y | N | N | Y | N | N | N | N | N | N | Comprehensive analysis of the possible uses of DL in IoT-based healthcare systems | Data Privacy and Security, increased risk of data breaches |
| Cotroneo et al.44 | Y | N | N | N | N | N | Y | Y | Y | Y | Yields comparable or superior results to manual clustering, which requires significant human effort and expertise | Requires high hardware requirements |
| Motwani et al.45 | Y | Y | N | N | N | Y | N | Y | Y | Y | Smart monitoring architecture monitors chronic patients in real-time and predicts Emergency, Alert, Warning, and Normal scenarios equally well locally and in the cloud | May require high costs for implementation and maintenance |
| Aazam et al.58 | Y | N | N | N | N | N | Y | N | Y | Y | Explored the use of ML in healthcare applications with edge computing | Challenges in ensuring interoperability between different devices and systems, which can limit the ability to scale and deploy such solutions on a larger scale |
| Hossain et al.46 | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Adequate for parallelization | Need for a reliable and high-speed internet connection |
| Praveen et al.59 | Y | Y | Y | N | N | Y | Y | N | Y | Y | OGSO-DNN is an energy-efficient illness detection and clustering approach for IoT-based sustainable healthcare systems | Scalability, Data privacy and security can be an issue |
| Shah et al.60 | Y | Y | N | Y | Y | Y | N | Y | Y | Y | Improves data accuracy and processing speed in IoT environments | Requires high bandwidth and robust infrastructure |
| Yan et al.50 | Y | N | N | N | N | N | Y | N | Y | Y | RSIF framework enhances healthcare data access in the cloud for users and service providers | May require a significant amount of computational resources |
| Tuli et al.52 | Y | N | N | Y | Y | Y | Y | Y | Y | Y | HealthFog: a portable, cost-effective solution for heart disease diagnosis using ensemble DL | Requires high compute resources for training and prediction |
| Durga et al.61 | Y | N | N | N | N | N | N | N | N | N | Explored algorithms for enhancing IoT-based healthcare systems in this study | High Complexity and Computation time |
1-Accuracy, 2-Sensitivity, 3-Specificity, 4-Latency, 5-Bandwidth Utilization, 6-Robustness, 7-Security, 8-Fault Tolerant, 9-Efficiency, 10-Reliability.
In risky manufacturing environments, Simeone et al.42 offers an AI-based cloud platform for monitoring risk. According to the authors, their platform can significantly lower the frequency of workplace accidents and increase employee productivity43. Cotroneo et al.44 proposed a DL-based technique to improve the analysis of software failures in CC systems. The authors described a DL-based method for identifying and categorizing software errors in CC systems using CNNs. In order to monitor patients in real-time and provide individualized recommendations, Motwani et al.45 proposed the Smart Patient Monitoring and Recommendation framework. A real-world dataset was used to evaluate the proposed framework, and the results demonstrated that it can accurately forecast patient conditions and offer useful personalized recommendations, thereby enhancing patient care. A DL-based pathology detection system for smart, connected healthcare was proposed by Hossain et al.46. The system uses images taken with a smartphone and sent to a cloud-based server for analysis to make a diagnosis of skin cancer. The accuracy of the suggested system was 87.31%. Ghaffar et al.47 presented a comparative analysis on COVID-19 detection for chest X-ray images. The authors used pretrained models like ResNet, DenseNet, and VGG to improve diagnostic accuracy. The study further builds upon these findings by providing a detailed comparative analysis of models such as MobileNet, EfficientNet, and InceptionV3, emphasizing their high accuracy rates in classifying COVID-19 infections, thus contributing valuable insights into effective methodologies for rapid and reliable disease detection in clinical practice. Illakiya and Karthik48 focused on the significant role of DL algorithms in medical image processing, particularly in helping radiologists to diagnose accurately disease. Their research reviews 103 articles to evaluate deep learning methods like CNNs, RNNs, and Transfer Learning for detecting Alzheimer’s Disease (AD) using neuroimaging modalities such as PET and MRI. The study highlights the effectiveness of these models in the detection of AD, while also emphasizing the need for a more detailed analysis of the progression from Mild Cognitive Impairment (MCI) to AD. Kang et al.49 address the challenge of overfitting in 3D CNN models for AD diagnosis due to limited labeled training samples. They propose a three-round learning strategy combining transfer learning and generative adversarial learning. This involves training a 3D Deep Convolutional Generative Adversarial Network (DCGAN) on sMRI data, followed by fine-tuning for AD versus cognitively normal (CN) classification, and transferring the learned weights for Mild Cognitive Impairment (MCI) diagnosis. The model achieves notable accuracies of 92.8% for AD versus CN, 78.1% for AD versus MCI, and 76.4% for MCI versus CN.
Yan et al.50 proposed a framework for DL-based concurrent processing and storage of healthcare data in a distributed CC setting51. The framework that has been proposed aims to address the difficulties involved in the processing and real-time analysis of massive amounts of healthcare data. An evaluation of the proposed framework using a prototype system reveals that it is capable of processing and storing large volumes of health data effectively. Tuli et al.52 proposed “HealthFog” for the automatic diagnosis of heart disease by integrating the IoT and fog computing environment. HealthFog uses an ensemble DL model to classify heart diseases based on ECG signals, with this system has improved the accuracy and efficiency of heart disease diagnosis53. Illakiya et al.54 present a novel Adaptive Hybrid Attention Network (AHANet) for the early detection of AD using brain MRI. AHANet integrates Enhanced Non-Local Attention (ENLA) for capturing global spatial and contextual information and Coordinate Attention for extracting local features. Additionally, an Adaptive Feature Aggregation (AFA) module effectively fuses these features, enhancing the network’s performance. Trained on the ADNI dataset, AHANet achieves a remarkable classification accuracy of 98.53%, outperforming existing methods in AD detection.
Novelty of the work
The article outlines a ground-breaking method for utilizing DL in healthcare, which focuses on medical image processing, specifically for MRI data-based AD diagnosis. In an effort to increase the accuracy of Alzheimer’s diagnosis. This study also conducts a thorough analysis of DL algorithms and CC. The article also discusses the various healthcare imaging databases analysis. For the experimental analysis, the three models-CNN, VGG-16, and an ensemble model have been analyzed for classification by carefully choosing an Alzheimer’s dataset. The recommendation of a novel design with three essential layers-the input layer, the cloud layer for preprocessing and model implementation, and the diagnostic layer for thorough categorization and notification generation is a standout addition. This layered approach improves model comprehension and provides guidelines for upcoming implementations in the domain of Healthcare systems. Beyond model performance, the research presents a ground-breaking CC framework that revolutionizes medical image processing while putting the needs of patient privacy and data confidentiality first. This study is positioned as a leading contribution to the breakthroughs in DL in the healthcare ecosystem.
Classification of medical imaging datasets
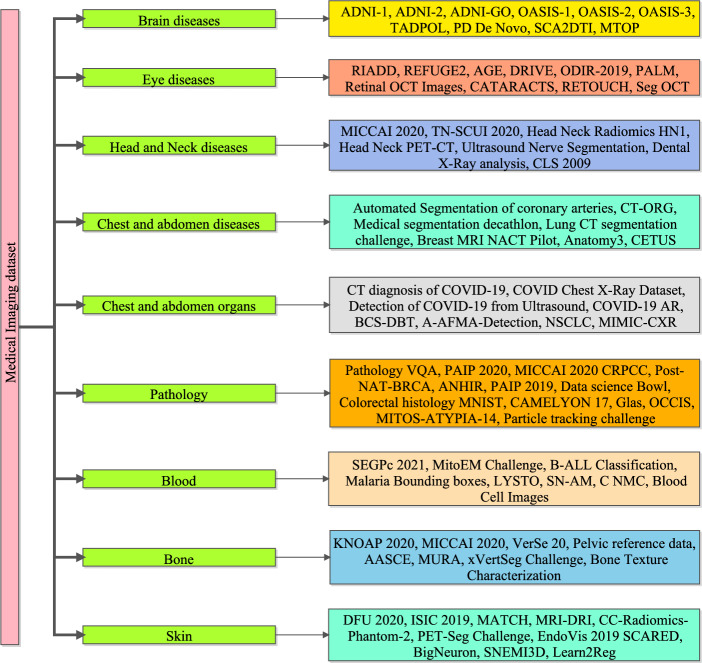
An overview of all the image datasets, associated diseases, and techniques/technologies used for retrieving medical data is presented. [NOTE: All the presented datasets in the work are public and links are provided in the appendix. As of April 4, 2024, the links provided for accessing the datasets were verified to be functional. However, it is important to note that the authors cannot be held responsible for any changes, removal, or updates to the datasets as these links were sourced from the Internet. Any alterations to the dataset or additional information not accessible through these links are beyond the control and responsibility of the authors. Users are advised to verify the availability and integrity of the datasets independently]. The subsections provide vital information about the techniques/technologies and information regarding the associated disease or considered body parts. Medical imaging datasets are essential for the evolution of diagnostic and prognostic techniques in the healthcare area. Figure 3 presents the taxonomy of medical imaging data sets such as brain disease, eye diseases, head and neck disease, chest and abdomen, pathology, blood, bone, and skin database. These datasets include a wide range of medical images taken from various clinical contexts, such as X-rays images, MRI, CT, and ultrasound images. Medical imaging datasets are useful because they allow for the development and testing of cutting-edge algorithms, predictive models, and AI-based applications that improve medical diagnosis, treatment planning, and patient care. Medical imaging datasets are significant because they can also provide researchers and healthcare practitioners with a plethora of visual information about a patient’s internal anatomical structures, physiological processes, and potential problems. This knowledge allows for the early detection and accurate characterization of a wide range of medical problems, including, but not limited to, malignancies, cardiovascular diseases, neurological disorders, and orthopedics pathologies etc. The following section discusses the various datasets available for the various known diseases.
Figure 3.
Taxonomy of medical imaging dataset.
Associated diseases
This section contains necessary information related to the diseases mentioned in all medical image datasets.
Alzheimer’s disease
AD is a brain disorder that gradually affects the brain and results in memory, thinking, and loose the ability to work normally. As the disease progresses, individuals with Alzheimer’s may have difficulty with everyday activities, eventually losing the ability to recognize and communicate with people. The main cause of Alzheimer’s is under research, but it is believed to involve a complex interaction between genetics, lifestyle factors, and environmental factors62.
Parkinson’s diseases
Parkinson’s disease is a neurological condition that causes a chronic, progressive movement disorder. It results from the decline of dopamine-producing neurons in a particular region of the brain, which lowers dopamine levels63. Tremors, stiffness, slowness of movement, and issues with balance and coordination are some signs of Parkinson’s disease64. Non-motor symptoms such as cognitive decline and mood disorders can also be brought on by the disease. There is currently no cure for Parkinson’s disease, which typically develops gradually and gets worse over time65.
Spinocerebellar ataxia type II
The central nervous system is impacted by the uncommon genetic disorder known as Spinocerebellar ataxia type II. It is brought on by a mutation in the ATXN2 gene, which causes the cerebellum and other areas of the brain to deteriorate. Progressive movement issues, such as poor coordination, balance issues, slurred speech, and muscle stiffness, are a hallmark of SCA2. Additionally, it can cause psychological and cognitive symptoms like anxiety, depression, and memory issues. SCA2 is inherited in an autosomal dominant manner, which means that all it takes for a person to be affected by the condition is for one copy of the mutated gene to be inherited from one parent. SCA2 currently has no known treatment options; instead, efforts are made to control the illness’s symptoms66.
Mild traumatic brain injury
Concussion, also referred to as MTBI is a type of brain injury brought on by a blow to the head or body. Although the injury is usually not life-threatening, it is still referred to as “mild” because it can still result in many of physical and mental symptoms, including dizziness, confusion, memory issues, and mood swings. These symptoms may need medical attention and rehabilitation and can last for days, weeks, or even months67.
AMD
Age-related Macular Degeneration is referred to as AMD. It is a chronic eye condition that damages the macula, the retina region that controls central vision. AMD can result in a progressive loss of vision, making it challenging to read, drive, or identify people68.
DR
Diabetic Retinopathy, also known as DR, is an eye condition associated with diabetes that can cause blindness69. The high level of sugar in the blood affects the retina blood vessels leak and constrict. If untreated, DR is a progressive condition that can result in vision loss. In order to stop or slow the progression of DR, routine eye exams and blood sugar control are crucial70.
Glaucoma
It is a disease that damages the optic nerve which causes vision loss or blindness. The optic nerve transmits visual information from eye to the brain. This damage often occurs due to increased pressure in the eye, which can lead to vision loss and, ultimately blindness if left untreated. Glaucoma can come in a variety of eye forms, such as primary angle-closure, open-angle, and normal-tension glaucoma71.
Pathologic myopia
Pathologic myopia is a severe form of myopia (nearsightedness) characterized by a refractive error of at least − 6 diopters and associated structural changes in the eye. In pathologic myopia, the eye elongates, causing thinning and stretching of the retina, which can lead to various complications such as retinal detachment, choroidal neovascularization, and macular atrophy72.
Techniques/Technologies
In the medical field, different types of medical imaging technologies or techniques are used to capture images of the human body for treatment purposes and diagnosis. Each modality has its own strengths and weaknesses, and the particular medical issue being identified or treated determines which modality should be used.
DWI
Diffusion-weighted Imaging (DWI), is a type of medical imaging that creates images by utilizing the characteristics of water molecules in tissues. It is commonly used in MRI and is sensitive to changes in the microstructure of tissues, such as those caused by stroke, inflammation, or tumors73. DWI is particularly useful for detecting early signs of stroke and for monitoring the response to treatment74.
PT
The term “positron emission tomography” (PT) is an imaging technique that uses radioactive material to measure and visualize the changes in the metabolic process. Different tracers are used to monitor the flow, absorption, and chemical composition of blood. It is often used as a medical and research tool to detect tumor imaging75.
T1 and T2
T1 and T2 are MRI modalities that provide different types of tissue contrast. T1-weighted images have a shorter echo and repetition time, which makes them sensitive to changes in tissue composition, particularly in fat content. On the other hand, T2-weighted images have a longer echo and repetition time, which makes them sensitive to changes in tissue water content and can highlight fluid-filled structures such as cysts or edema76.
MRI FLAIR
Fluid Attenuated Inversion Recovery (FLAIR) is an MRI pulse sequence used to suppress fluids such as Cerebrospinal Fluid (CSF) and highlight pathological tissues with high water content, such as tumors, inflammation, and demyelination lesions. It is commonly used in neuroimaging to visualize subtle abnormalities in the brain and spinal cord77.
CT
Computed Tomography (CT) medical imaging produces comprehensive internal images of the body using X-rays. It is frequently used to identify and track many illnesses, including tumors, internal bleeding, and bone fractures78. Multiple X-ray images of that area of the body taken by the CT scanner from various angles are combined to form a complex cross-sectional image of the body component being investigated79.
Ultrasound
The medical imaging method known as ultrasound uses high-frequency sound waves to produce pictures of the organs and tissues of the body. The internal organs, tissues, and blood flow can be seen using this non-invasive, painless procedure. While ultrasound is frequently used in obstetrics to see the growing fetus, it can also be used to diagnose and track a number of medical conditions in other areas of the body, including the abdomen, pelvis, and heart80.
Computed radiography
Digital radiographic images are created using Computed Radiography (CR), a medical imaging technique that employs Photostimulable Phosphor plates. A digital image that can be viewed on a computer is created in CR by scanning an imaging plate with a laser beam after it has been exposed to X-rays. For its superior image quality and adaptability, CR is widely used in medical imaging, particularly in radiography. Digital radiography (DR), which provides quicker image acquisition and lower radiation exposure for patients, has largely replaced it81.
MR
By mixing radio waves and a potent magnetic field, the medical imaging method known as magnetic resonance produces detailed images of the body’s internal components. Frequently used to visualize the brain, spine, joints, and soft tissues and is particularly helpful in diagnosing conditions that might not be visible on other medical imaging techniques, like X-rays or CT scans. For some patients, MR imaging offers a safer alternative because it doesn’t expose them to ionizing radiation82.
Coronary CT angiography
In coronary CT Angiography (CTA), Computed Tomography (CT) is a non-invasive imaging method used to see the coronary arteries that provide blood to the heart. Contrast dye is injected into a vein and then circulated through the coronary arteries in this procedure. A 3D image of the coronary arteries is created by reconstructing images of the arteries taken by the CT scanner as the dye travels through them83. Utilizing this method, coronary artery disease, blockages, and other heart conditions can be identified or assessed84.
Radiotherapy structure set
A radiotherapy structure set is a collection of anatomical structures that are identified on medical images, such as organs or tumors, and which are typically used in the planning and administration of radiation therapy for the treatment of cancer. The structure set makes it easier to precisely direct radiation towards the tumor while minimizing damage to healthy cells and organs85.
PET
Positron Emission Tomography (PET) is known as PET. It is a type of medical imaging procedure that creates 3D images of bodily functions using radioactive materials called radiotracers. PET scans are frequently used to identify and treat cancer as well as other heart and brain diseases86.
Endoscopy
It involves the insertion of a long thin tube with a camera on the other end to examine internal organs or tissues. It is frequently used to check the joints, respiratory system, and digestive system. Endoscopy can also be used to obtain tissue samples for biopsies and perform minor surgical procedures87.
General microscopy
Using a microscope to view and examine samples that cannot be seen with the unaided eye, such as microorganisms, cells, tissues, and small structures, is known as general microscopy. It involves using a microscope to enlarge the image of the sample being examined in order to examine its features and characteristics in greater detail. Several disciplines, including biology, medicine, material science, and forensics, among others, can benefit from general microscopy88.
Electron microscope
An electron microscope that uses a beam of electrons for illumination to create an image of a sample. There are two variants of it one uses a transmission process and the other uses a scanning process. By colliding with the sample’s atoms, the electrons in the beam generate signals that can be picked up and used to build images with extremely high levels of magnification and resolution. Numerous scientific disciplines, such as materials science, biology, and nanotechnology, use electron microscopes89.
Datasets associated with the classification of brain diseases
The objective of the AD Neuroimaging Initiative (ADNI) is to understand how the disease develops and to find biomarkers for its early diagnosis and treatment. The ADNI study is divided into several phases, such as ADNI-1, ADNI-2, ADNI-3, and ADNI-GO (Grand Opportunity)90,91. Based on their cognitive abilities, the study participants can be divided into three groups: those with normal cognition (NC), mild cognitive impairment (MCI), and AD. Early Mild Cognitive Impairment (EMCI) and Later Mild Cognitive Impairment (LMCI) are two subgroups of MCI. An assortment of Neuroimaging datasets, such as structural and functional MRI scans, as well as data on demographics and clinical conditions, can be found in the Open Access Series of Imaging Studies (OASIS). Three datasets, referred to as OASIS-1, OASIS-2, and OASIS-392–94, have been made available by the OASIS project and are primarily used for research on AD. However, researchers have also used these datasets for related tasks like functional area segmentation. Table 2 depicts an overview of the datasets and difficulties associated with classifying brain diseases.
Table 2.
An overview of the datasets and difficulties associated with classifying brain diseases.
| Dataset | Year | Techniques | Associated disease | Classification |
|---|---|---|---|---|
| ADNI-190 | 2004 | T1, T2, DWI and PT | Alzheimer’s | NC, MCI and AD |
| ADNI-291 | 2015 | T1, T2, DWI and PT | Alzheimer’s | NC, EMCI, LMCI and AD |
| ADNI-GO90 | 2009 | T1, T2, DWI and PT | Alzheimer’s | NC, MCI and AD |
| OASIS 192 and OASIS 293 | 2007,2009 | T1 | Alzheimer’s | NC and AD |
| OASIS 394 | 2019 | T1, T2, PT and MRI FLAIR | Alzheimer’s | NC, AD |
| TADPOLE95 | 2017 | T1, T2, DWI and PT | Alzheimer’s | NC, MCI, AD |
| PD De Novo96 | 2019 | T1 | Parkinson’s | NC and PD |
| SCA2 DTI97 | 2018 | T1 and DWI | Spinocerebellar ataxia type II | NC and SCA2 |
| MTOP98 | 2016 | T1 and DWI | Mild traumatic brain injury | Healthy, category I or category II |
An extensive, global initiative to advance the comprehension, detection, and care of AD and similar diseases is the TADPOLE (Trajectory Analysis Project for AD) dataset95. The dataset contains clinical and imaging information from people who have been given the detection of Mild Cognitive Impairment (MCI), AD, or cognitive normality (CN). PD De Novo is a dataset of individuals who were newly infected with Parkinson’s disease (PD) and followed for up to three years to track the progression of the disease96. The dataset includes clinical assessments, imaging data, genetic information, and biospecimen samples from participants, and is intended to help researchers better understand the early stages of PD and develop new treatments. Some datasets concentrate on diagnosing mild traumatic brain injury and others on Spinocerebellar Ataxia type II (SCA2)97.
Datasets associated with eye diseases
Retinal Image Analysis for Multi-disease Detection (RIADD) is a public dataset that has a total of 2,740 retinal fundus images99. The dataset has been specifically intended for the development of algorithms that can detect and categorize various common retinal illnesses, including retinopathy caused by diabetes, glaucoma, and macular degeneration caused by age. Retinal Fundus Glaucoma Challenge 2019 (REFUGE2 ) is a public dataset consisting of retinal fundus images of both healthy and glaucomatous eyes100 . It was introduced as part of a challenge to develop ML algorithms for automated glaucoma diagnosis. The AGE dataset is related to the disease glaucoma. It is a dataset consisting of retinal fundus images collected from patients with glaucoma as well as healthy individuals101. The dataset is intended for use in developing and testing algorithms for automated glaucoma detection and diagnosis based on retinal fundus images. In the Paediatric Automated Lamellar Keratoplasty Missions (PALM ) dataset102, 1159 color scans and their segmentation masks show the optic nerve head (ONH) and macula, two ophthalmology-related regions of interest. The retinal OCT (Optical Coherence Tomography) pictures dataset is a collection of retinal OCT images used to train and evaluate DL models for the detection of retinal illnesses such as macular degeneration, diabetic retinopathy, and glaucoma103,104. A collection of retinal images used to identify cataracts is called the CATARACTS dataset. The dataset includes eyes with and without cataracts, and it is made up of retinal images taken with a slit-lamp camera. Retinal Optical Coherence Tomography (OCT)105 scans with Diabetic Macular Edema (DME) make up the Seg OCT (DME) dataset, a medical imaging collection that is openly accessible106. Table 3 depicts an overview of eye disease datasets along with challenges.
Table 3.
An overview of datasets and challenges related to tasks involving eye diseases.
| Dataset | Year | Techniques | Associated disease |
|---|---|---|---|
| RIADD99 | 2020 | FP | AMD, DR |
| REFUGE2100 | 2020 | FP | Glaucoma |
| AGE101 | 2019 | OCT | Closure glaucoma |
| DRIVE107 | 2019 | FP | Vessel extraction |
| ODIR-2019108 | 2019 | FP | AMD, glaucoma, diabetes, cataract, hypertension |
| PALM102 | 2019 | FP | Pathologic myopia |
| Retinal OCT Images103104 | 2018 | OCT | DR |
| CATARACTS105 | 2017 | Video | Surgery tools detection |
| RETOUCH109 | 2017 | OCT | Fluid segmentation |
| Seg OCT(DME)106 | 2015 | OCT | Diabetic macular edema |
Datasets associated with head and neck diseases
The MICCAI 2020 HECKTOR challenge dataset is a multi-institutional dataset of head and neck CT scans. It includes CT scans of 198 patients with head and neck tumors, along with annotations for Organs at Risk (OARs) and Gross Tumor Volumes (GTVs)110. The dataset is intended for use in developing and evaluating algorithms for automated segmentation of OARs and GTVs in head and neck CT scans. TN-SCUI 2020 is a dataset for thyroid nodule ultrasound image classification and segmentation111 . The dataset consists of 5949 ultrasound images of thyroid nodules. The Head Neck Radiomics HN1 dataset is a collection of head and neck cancer patients’ medical images and clinical data. The dataset includes CT, MRI, and PET-CT images of the patient’s head and neck region, as well as clinical information such as tumor stage and treatment outcomes. The Head Neck PET-CT dataset consists of Positron Emission Tomography (PET) and computed tomography (CT) images of the head and neck region, which are used for diagnosis and treatment planning of head and neck cancer. The dataset includes images from 58 patients, with each patient having a PET and CT scan112,113 .
The Ultrasound Nerve Segmentation dataset is a medical imaging dataset used for nerve segmentation tasks. It contains ultrasound images of the neck area of patients, with the goal of segmenting nerve structures in the images. The Dental X-Ray Analysis 1 Dataset is a publicly available dataset of dental X-ray images for the task of dental pathology detection. The dataset contains 5 different classes of dental pathologies, including Dental caries, Dental fillings, Dental implants, Dental infections, and Dental fractures. The CLS 2009 dataset is a medical image dataset of the carotid bifurcation. It contains 200 contrast-enhanced CT Angiography (CTA) scans of the neck region, including the carotid bifurcation, from 100 patients113–116 . Table 4 depicts an overview of challenges and datasets related to head and neck diseases.
Table 4.
An overview of the challenges and datasets related to head and neck diseases.
| Dataset | Year | Techniques | Emphasis |
|---|---|---|---|
| MICCAI 2020: HECKTOR110 | 2020 | CT, PT | Tumors originating in the head and neck area |
| TN-SCUI 2020111 | 2020 | Ultrasound | Diagnosis of nodules in the thyroid gland |
| Head Neck Radiomics HN1112 | 2019 | CT | Malignant growths in the head and neck |
| Head Neck PET-CT113 | 2017 | CT, PT | Tumor |
| Ultrasound Nerve Segmentation114 | 2016 | Ultrasound | Nerve |
| Dental X-Ray Analysis115 | 2015 | Computed Radiography | Caries |
| CLS 2009116 | 2009 | CT Angiography | Carotid bifurcation |
Datasets associated with chest and abdomen
A medical imaging dataset called the automated Segmentation of Coronary Arteries contains heart CT Angiography (CTA) images for the segmentation of coronary arteries. The dataset consists of 101 CTA heart images with an average of 291 slices per image. A collection of computed tomography (CT) volumes with various organ segmentations is called the CT-ORG dataset. It consists of 1,010 volumetric images of the thoracic and abdominal regions of the human body, with segments of the liver, pancreas, kidneys, stomach, gallbladder, aorta, and portal vein in each volume. The Medical Segmentation Decathlon (MSD) dataset consists of ten distinct medical image segmentation tasks for a variety of imaging modalities, including microscopy, CT, and MRI. Segmenting brain tumors, heart, liver, lungs, pancreas, prostate, and other organs are just a few of the tasks117,118.
A collection of lung lung-CT scans makes up the Lung CT Segmentation Challenge dataset. Automated lung segmentation from CT scans is a crucial step in the diagnosis and treatment of many lung diseases, including cancer and emphysema. This project was designed to develop and evaluate algorithms for this task. A medical imaging dataset called the Breast MRI NACT Pilot is made up of breast MRI scans from breast cancer patients who received Neoadjuvant Chemotherapy (NACT) treatment. The data set might contain MRI images of the breast from various patients annotated for various breast structures, including breast tissue, tumor, and surrounding anatomy, 514 for example. Anatomy 3 (Year: 2015): This dataset focuses on the segmentation of organs like the liver, lung, kidney, aorta, and trachea and includes both MR and CT scans. Developing algorithms to precisely segment these organs from MR and CT images, which are frequently used for various diagnostic and treatment planning purposes, may be a challenge. The data set might contain MRI and CT scans from various patients that have annotations for the relevant organs. CETUS 2014 (Year: 2014): This dataset focuses on the segmentation of the heart and utilizes ultrasound imaging. The challenge may involve developing algorithms to accurately segment the heart from ultrasound images, which are commonly used for cardiac imaging. The dataset may include ultrasound images from different patients with annotations for different cardiac structures such as the left ventricle, right ventricle, and myocardium119–122. Table 5 depicts an overview of the datasets and challenges for tasks that involve organ segmentation in the chest and abdomen.
Table 5.
Overview of datasets and challenges for tasks involving segmentation of organs in the chest and abdomen.
| Dataset | Year | Techniques | Considered body parts |
|---|---|---|---|
| Automated Segmentation of coronary arteries117 | 2020 | Coronary CT angiography | Coronary arteries |
| CT-ORG118 | 2019 | CT | Liver, lung, kidney and bladder |
| Medical segmentation decathlon119 | 2019 | MR, CT | Liver, lung, heart, pancreas and colon |
| Lung CT segmentation challenge120,123 | 2017 | Radiotherapy structure set | Lung and heart |
| Breast MRI NACT pilot121 | 2016 | MR | Breast |
| Anatomy3124 | 2015 | MR, CT | Liver, lung, kidney, aorta and trachea |
| CETUS122 | 2014 | Ultrasound | Heart |
Datasets associated with segmentation of organs in chest and abdomen regions
The COVID-19 CT Diagnosis dataset is a collection of CT scan data from patients who tested positive and negative for the virus125–127. Studies have employed CNNs and other machine learning models to extract features from chest X-rays and CT scans, achieving significant improvements in diagnostic accuracy. For instance, authors in128 demonstrated the efficacy of using CNNs in combination with recurrent neural networks (RNNs) for automated disease detection, underscoring the potential of hybrid models in enhancing diagnostic capabilities.
In order to generate the dataset, 2973 CT scans from 1173 individuals, including 888 COVID-19 positive cases and 285 COVID-19 negative cases, were obtained. The COVID Chest X-Ray Dataset is a collection of chest X-ray scans for cases that tested positive for COVID-19, for normal cases, and for instances that tested positive for other kinds of pneumonia. The dataset is comprised of 2 parts: Metadata, Which contains the patient’s data, and Image files, Which include the chest X-ray scans in Portable Network Graphics (PNG) format. There are a total of 12,721 images in the collection. COVID-19 AR, a dataset introduced in 2020, combined CT and CR imaging techniques to facilitate organ segmentation in patients with COVID-19. BCS-DBT, a dataset focused on digital breast tomosynthesis, was introduced in the same year with an emphasis on breast cancer segmentation129–133.
A-AFMA-Detection, a 2020 dataset, utilized ultrasound imaging to detect amniotic fluid in the abdomen region. In 2019, the NSCLC-RadiomicsInterobserver1 dataset was introduced, focusing on NSCLC and utilizing a radiotherapy structure set for organ segmentation. The MIMIC-CXR dataset, introduced in 2019, utilized electronic health records and report data for chest image analysis. The NSCLC-Radiomics-Interobserver1 dataset is a collection of medical images and annotations for NSCLC patients. The dataset includes CT images of the chest region for 30 NSCLC patients, with annotations for various regions of interest (ROIs) within each image. The MIMIC-CXR (Medical Information Mart for Intensive Care Chest X-ray) dataset contains radiology reports and chest X-ray images. More than 65,000 patients’ chest X-ray images and corresponding reports totaling more than 350,000 are included. The patient’s associated demographic and clinical information, as well as frontal and lateral views of the chest, are all included in the dataset134–136. Table 6 depicts an overview of Datasets and Challenges in tasks related to the Segmentation of Organs in the Chest and abdomen regions.
Table 6.
An overview of datasets and challenges in tasks related to the segmentation of organs in the chest and abdomen regions.
| Dataset | Year | Techniques | Emphasis |
|---|---|---|---|
| CT Diagnosis of COVID-19129 | 2020 | CT | COVID-19 |
| COVID Chest X-Ray Dataset130 | 2020 | CR | COVID-19 |
| Detection of COVID-19 from Ultrasound131 | 2020 | Ultrasound | COVID-19 |
| COVID-19 AR132 | 2020 | CT and CR | COVID-19 |
| BCS-DBT133 | 2020 | Digital breast tomosynthesis | Breast cancer |
| A-AFMA-Detection134 | 2022 | Ultrasound | Amniotic fluid detection |
| NSCLC-Radiomics-Interobserver1135 | 2019 | Radiotherapy structure set | Non-small cell lung cancer |
| MIMIC-CXR136 | 2019 | Electronic health record and report | Chest image analysis |
Datasets related with image analysis in Pathology
Pathology VQA (2020): This dataset was created especially for pathology-related visual question-answering (VQA) tasks. It involves responding to inquiries about pathology images, which necessitates comprehending the visual information contained in the images and doing so137.
PAIP 2020 (2020): In order to analyze and spot patterns and features in pathology images related to this particular type of cancer, this dataset specifically focuses on colorectal cancer138.
MICCAI 2020 CRPCC (2020): To investigate the synergy between radiology and pathology in disease diagnosis, this dataset combines radiology and pathology data for classification tasks139.
Post-NAT-BRCA (2019): The analysis of changes in cell morphology and structure following treatment is made possible by this dataset, which focuses on cell images specifically for studying the effects of post-neoadjuvant therapy in breast cancer140.
ANHIR (2019): This dataset was created for pathology image registration, which entails aligning and registering images from various sources or time points to allow for additional analysis and comparison141.
PAIP 2019 (2019): This dataset emphasizes the segmentation of liver tumors and tumor burden, offering information for the creation of algorithms and models for the precise segmentation of liver tumors and the estimation of tumor burden142.
Data Science Bowl 2018: The goal of this dataset, which contains images of cells, was to test participants’ ability to create algorithms for cell detection and classification tasks143.
Colorectal Histology MNIST (2018): This data which is based on patch-based analysis of colorectal pathology, can be used to examine and spot patterns in histology images of colorectal cancer144.
CAMELYON 17 (2017): This dataset focuses on breast cancer metastases and provides information for creating models and algorithms for identifying and classifying metastatic lesions in breast cancer pathology images145.
Glas (2015): This dataset, which is based on patches of colorectal cancer pathology, offers information for examining and identifying features in patches of colorectal cancer pathology images146.
OCCIS (2014): Data for studying cell morphology and structure in relation to cervical cancer screening is provided by this dataset, which consists of overlapping images of cervical cells147.
MITOS-ATYPIA-14 (2014): This dataset focuses on the identification of mitotic cells with the goal of creating algorithms for the precise identification of these crucial markers of cell division and proliferation148.
Particle Tracking Challenge (2012): This dataset involves particle tracking in microscopy images, providing data for developing algorithms and models for accurately tracking particles and analyzing their dynamics149.
Table 7 depicts an overview of Datasets and difficulties associated with image analysis in Pathology.
Table 7.
An Overview of Datasets and difficulties associated with image analysis in Pathology.
| Dataset | Year | Emphasis |
|---|---|---|
| Pathology VQA137 | 2020 | Visual question answering |
| PAIP 2020138 | 2020 | Colorectal cancer |
| MICCAI 2020 CRPCC139 | 2020 | Classification combining radiology and pathology data |
| Post-NAT-BRCA140 | 2019 | Cell |
| ANHIR141 | 2019 | Pathology image registration |
| PAIP 2019142 | 2019 | Liver cancer and tumor burden segmentation |
| Data Science Bowl143 | 2018 | Cell |
| Colorectal Histology MNIST144 | 2018 | Patch-based analysis of colorectal pathology |
| CAMELYON 17145 | 2017 | Breast cancer metastases |
| Glas146 | 2015 | Colorectal cancer pathology patch |
| OCCIS147 | 2014 | Overlapping cervical cell |
| MITOS-ATYPIA-14148 | 2014 | Mitotic |
| Particle Tracking Challenge149 | 2012 | Particle |
Datasets associated with the analysis of blood-related images
Medical datasets connected to blood-related image analysis have proven to be useful in furthering both research and therapeutic applications. These datasets typically consist of a collection of digital photographs of blood samples that have been annotated with information such as cell kinds, morphologies, and potential disease markers. The use of these datasets has aided in the development of advanced ML algorithms capable of accurately identifying and categorizing blood components. Researchers have made substantial advances in the early diagnosis of diseases such as leukemia, anemia, and different blood-borne infections by exploiting these databases. The diversity of these datasets, which include a wide range of blood properties, ensures a solid training basis for AI algorithms, contributing to a better knowledge of hematological illnesses. Such data-driven approaches have also resulted in more tailored treatment strategies, ultimately improving overall patient care quality. Below are various examples of the same.
SegPc 2021—Myeloma Plasma Cell: The analysis of myeloma plasma cells, a subset of white blood cells essential to the growth of multiple myeloma, a plasma cell cancer, is the main goal of the SegPc 2021 dataset. This dataset can be used for tasks like classifying, quantifying, and segmenting myeloma plasma cells in images related to blood150.
MitoEM Challenge 2020—Mitochondria: The analysis of mitochondria, significant organelles in charge of generating energy in cells, is the main focus of the MitoEM Challenge dataset. The understanding of mitochondrial function and dysfunction in various diseases can be aided by using this dataset for tasks like segmentation, detection, and classification of mitochondria in blood-related images151.
B-ALL Classification 2019—Leukemic Blasts in an Immature Stage: The analysis of leukemic blasts in an early stage is the main focus of the B-ALL Classification dataset. Blood cancer of the B-ALL (B-cell Acute Lymphoblastic Leukaemia) variety mainly affects lymphoid cells. This dataset can help with the development of B-ALL diagnostic and prognostic tools by being used for tasks like classifying, detecting, and quantifying leukemic blasts in blood-related images151.
Malaria Bounding Boxes 2019—Cells in Blood: The analysis of blood cells infected with malaria parasites is done using the Malaria Bounding Boxes dataset. Millions of people worldwide suffer from malaria, a mosquito-borne illness, and accurate identification and quantification of infected blood cells is essential for both diagnosis and treatment. This dataset can be used for tasks like object recognition, blood-related image segmentation, and classification of infected blood cells152.
LYSTO 2019—Lymphocytes: The analysis of lymphocytes, a category of white blood cells that is crucial to the immune response, is the main focus of the LYSTO dataset. The understanding of lymphocyte function in various diseases and conditions can be improved with the help of this dataset, which can be used for tasks like the segmentation, classification, and quantification of lymphocytes in images related to blood153.
SN-AM Dataset 2019—Stain Normalization: For the analysis of stain normalization methods in images involving blood, the SN-AM Dataset was created. In order to address staining variability across various samples and laboratories, stain normalization is a crucial preprocessing step in medical image analysis. For image analysis tasks involving blood, this dataset can be used to evaluate and benchmark stain normalization techniques154.
C NMC Dataset 2019—Classification of Immature Leukemic Cells: The immature leukemic cells, which are abnormal white blood cells that are indicative of leukemia, are the main focus of the C NMC Dataset. This dataset can be used for tasks like categorization, detection, and quantification of immature leukemic cells in images of blood as well as for the creation of automated tools for leukemia diagnosis and monitoring154.
Blood Cell Images 2018—Cells in Blood: Red blood cells, white blood cells, and platelets are just a few of the different types of blood cells included in the Blood Cell Images dataset. This dataset can be used for tasks like classifying, quantifying, and segmenting blood cells in images of the blood. It can also be a useful tool for researching blood cell morphology and comprehending various blood-related diseases and conditions155.
Table 8 depicts an overview of Datasets and challenges related to the Analysis of blood-related images
Table 8.
An overview of datasets and challenges related to the analysis of blood-related images.
| Dataset | Year | Emphasis |
|---|---|---|
| SegPc 2021150 | 2020 | Myeloma plasma cell |
| MitoEM challenge151 | 2020 | Mitochondria |
| B-ALL Classification156 | 2019 | Leukemic blasts in an immature stage |
| Malaria bounding boxes152 | 2019 | Cells in blood |
| LYSTO153 | 2019 | Lymphocytes |
| SN-AM dataset154 | 2019 | Stain normalization |
| C NMC dataset154 | 2019 | Classification of immature leukemic cells |
| Blood cell images155 | 2018 | Cells in blood |
Datasets associated with the analysis of bone-related images
Medical datasets connected to bone-related image processing are a valuable asset in the domains of orthopedics and radiography. These datasets frequently include a wide range of imaging modalities, such as X-rays, CT scans, and MRI images depicting diverse bone structures and accompanying disorders. Annotations in these databases may include information about fractures, osteoporosis, bone cancers, and other orthopedic diseases. Healthcare professionals can accomplish more exact diagnoses, prognoses, and treatment planning by using ML and AI algorithms trained on these massive datasets. The availability of such datasets has fueled innovation in the creation of automated diagnostic systems, saving time and perhaps reducing human error in analyzing complex bone pictures. As a result, these data-driven techniques are changing the landscape of bone health care by encouraging early intervention and leading to better patient outcomes. Some examples of the same have been showcased here.
KNOAP 2020—MR and CR Knee Osteoarthritis: The MR and CR images used to analyze knee osteoarthritis are the primary focus of the KNOAP 2020 dataset. This dataset can be used for tasks like segmenting, classifying, and quantifying features related to knee osteoarthritis, and it can help with the creation of early detection and monitoring methods for this degenerative joint disease157.
MICCAI 2020 RibFrac Challenge 2020—CT Detection and Classification of Rib Fractures: The CT images used in the MICCAI 2020 RibFrac Challenge dataset are intended for the detection and classification of rib fractures. This dataset can be used for tasks like fracture localization, classification, and detection, and it can aid in the creation of automated tools for managing and assessing rib fractures158.
VerSe 20 2020—CT Vertebra: The VerSe 20 dataset is dedicated to the CT-based analysis of vertebrae. This dataset can be used for tasks like categorizing, quantifying, and segmenting vertebral structures and abnormalities. It can also be used to help with the diagnosis and planning of treatments for a variety of conditions and diseases involving the spine159.
Pelvic Reference Data 2019—CT Pelvic Images: For use in a variety of pelvic-related image analysis tasks, the Pelvic Reference Data dataset offers CT images of the pelvic anatomy. This dataset can be used to perform operations on the pelvic structures, such as segmentation, registration, and measurement, and it can help with the creation of automated tools for pelvic imaging analysis in clinical and academic settings160.
AASCE 19 2019—CR Spinal Curvature: The CR images used for the analysis of spinal curvature are the main focus of the AASCE 19 dataset. This dataset can be used for tasks like segmentation, measurement, and classification of abnormal spinal curvature, which can help us understand conditions and deformities affecting the spine161.
MURA 2018—CR Abnormality in Musculoskeletal: The MURA dataset includes different musculoskeletal injuries and abnormalities, including those involving bones. This dataset can be a helpful tool for researching musculoskeletal disorders and injuries because it can be used for tasks like the localization, classification, and detection of abnormalities in CR images162.
xVertSeg Challenge 2016—CR Fractured Vertebrae: The CR images of fractured vertebrae are the focus of the xVertSeg Challenge dataset. This dataset can be used for tasks like segmenting, classifying, and localizing vertebral fractures. It can also help with the creation of automated tools for managing and assessing vertebral fractures163.
Bone Texture Characterization 2014—CR Bone Crisps: Using computed radiography (CR) images, the Bone Texture Characterization dataset is intended for the analysis of bone crisps. This dataset can be used for tasks like bone crisp classification, feature extraction, and texture analysis, which can help us understand the characteristics of bone texture and how they relate to bone health164.
Table 9 depicts an overview of Datasets and Challenges Related to the Analysis of bone-related images.
Table 9.
An overview of datasets and challenges related to the analysis of bone-related images.
| Dataset | Year | Techniques | Emphasis |
|---|---|---|---|
| KNOAP 2020157 | 2020 | MR and CR | Knee osteoarthritis |
| MICCAI 2020 RibFrac Challenge158 | 2020 | CT | Detection and classification of rib fractures |
| VerSe 20159 | 2020 | CT | Vertebra |
| Pelvic reference data160 | 2019 | CT | Pelvic images |
| AASCE 19161 | 2019 | CR | Spinal curvature |
| MURA162 | 2018 | CR | Abnormality in musculoskeletal |
| xVertSeg Challenge163 | 2016 | CR | Fractured vertebrae |
| Bone texture characterization164 | 2014 | CR | Bone crisps |
Datasets associated with skin and animal image analysis tasks
The fields of dermatology, veterinary medicine, and wildlife biology all place high importance on medical datasets linked to skin analysis and animal image analysis activities. Images of various dermatological disorders, including melanoma, psoriasis, and eczema, are frequently included in skin-related datasets, allowing the creation of AI-driven diagnostic tools that support early detection and treatment planning. Animal picture databases also offer important information about the morphology, behavior, and health of both domestic and wild animals. These could be information from monitoring wildlife for conservation and demographic studies, or veterinary medical imaging for disease diagnosis. By incorporating ML models that have been trained on such a wide range of datasets, healthcare, and ecology researchers are encouraged to take a multidisciplinary approach, opening up new possibilities for better medical diagnosis, individualized treatment plans, and greater animal species knowledge. With the help of technological improvements, the merging of skin and animal picture collections represents a cutting-edge frontier in scientific inquiry into ecology and medicine. Here are a few of its instances.
DFU 2020: This dataset focuses on the detection of foot ulcers in diabetic patients. Foot ulcers are a common complication in diabetic patients and early detection is crucial for timely intervention and prevention of serious complications such as infections and amputations. The emphasis of this dataset is on developing image analysis techniques using RGB imaging to accurately detect foot ulcers from medical images, which can aid in early diagnosis and effective management of diabetic foot ulcers165.
ISIC 2019: This dataset is focused on the classification of 9 different skin diseases, including melanoma, benign nevi, and dermatofibroma, among others. The emphasis of this dataset is to develop image analysis techniques using RGB imaging for accurate and automated classification of skin diseases, which can assist dermatologists in clinical decision-making and improve patient outcomes. Early and accurate classification of skin diseases can aid in timely treatment and management, and potentially prevent the progression of malignant skin conditions166.
MATCH 2020: This dataset focuses on tumor tracking in markerless lung CT images. The emphasis of this dataset is on developing advanced image analysis techniques using CT imaging to track lung tumors in dynamic medical images, which can aid in radiation therapy planning, assessment of tumor response to treatment, and monitoring disease progression. Accurate tumor tracking can improve treatment efficacy and minimize radiation exposure to healthy tissues167.
MRI-DIR: This dataset focuses on multi-modality registration with phantom images in MR and CT images. The emphasis of this dataset is on developing image registration techniques using MR and CT imaging to align different imaging modalities for accurate fusion and analysis of medical images. Accurate multi-modality registration can improve image-guided interventions, treatment planning, and diagnosis by providing a comprehensive view of anatomical and functional information from different imaging modalities168.
CC-Radiomics-Phantom-2: This dataset focuses on assessing feature variability using phantom images in CT imaging. The emphasis of this dataset is on developing image analysis techniques to assess the variability of radiomic features extracted from phantom images, which can help in evaluating the robustness and reproducibility of radiomics-based image analysis methods. Understanding feature variability in phantom images is crucial for ensuring the reliability and generalizability of radiomics-based models in clinical practice169.
PET-Seg Challenge 2016: This dataset focuses on phantom registration and research using CT and PET imaging. The emphasis of this dataset is on developing image registration techniques to accurately align CT and PET images of phantoms, which are used for quality assurance and calibration purposes in PET imaging. Accurate phantom registration can ensure the accuracy and reliability of PET imaging results, which are widely used for cancer diagnosis, staging, and treatment response assessment170.
EndoVis SCARED 2019: This dataset focuses on depth estimation from endoscopic data. The emphasis of this dataset is on developing computer vision and image processing techniques to accurately estimate the depth information from endoscopic images or videos. Accurate depth estimation can aid in 3D reconstruction, virtual reality visualization, and surgical planning during minimally invasive surgeries, improving the safety and efficacy of these procedures171.
BigNeuron 2016: This dataset focuses on general microscopy for animal neuron reconstruction. The emphasis of this dataset is on developing image analysis techniques to reconstruct the complex 3D structures of animal neurons from microscopy images. Accurate neuron reconstruction can provide insights into neural networks and connectivity, aiding in understanding brain function and neurological diseases172.
SNEMI3D 2013: This dataset focuses on the segmentation of neurites from electron microscopy images. The emphasis of this dataset is on developing advanced image segmentation techniques to accurately segment neurites, which are the elongated projections of nerve cells, from electron microscopy images. Accurate neurite segmentation is crucial for studying the morphology and connectivity of neurons, understanding neural circuits, and investigating neurological disorders173.
Learn2Reg 2020: This dataset focuses on image registration in medical imaging using MR and CT images. The emphasis of this dataset is on developing image registration techniques to align and fuse MR and CT images for various medical imaging tasks such as image-guided interventions, treatment planning, and disease diagnosis. Accurate image registration can improve the accuracy and precision of medical image analysis, aiding in better patient care and treatment outcomes174.
Table 10 depicts an overview of the Datasets and challenges related to Skin, phantom, and animal image analysis tasks.
Table 10.
An overview of the datasets and challenges related to skin, phantom, and animal image analysis tasks.
| Dataset | Year | Techniques | Emphasis |
|---|---|---|---|
| DFU 2020165 | 2020 | RGB | Detection of foot ulcers in diabetic patients |
| ISIC 2019166 | 2019 | RGB | Classification of 9 diseases |
| MATCH167 | 2020 | CT | Tumor tracking in lung |
| MRI-DIR168 | 2018 | MR, CT | Multi-modality registration with phantom |
| CC-Radiomics-Phantom-2169 | 2018 | CT | Feature assessment with phantom |
| PET-Seg Challenge170 | 2016 | CT, PET | Phantom registration and research |
| EndoVis 2019 SCARED171 | 2019 | Endoscopy | Depth estimation from endoscopic data |
| BigNeuron172 | 2016 | General microscopy | Animal neuron reconstruction |
| SNEMI3D173 | 2013 | Electron microscope | Segmentation of neurites |
| Learn2Reg174 | 2020 | MR, CT | Image registration |
Case study: Alzheimer’s MRI classification using DL methods
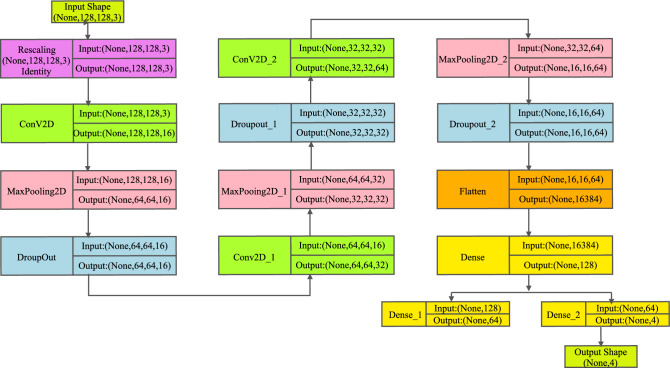
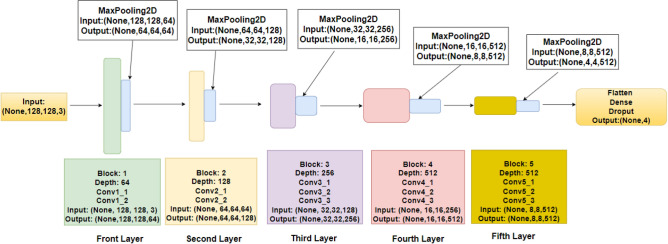
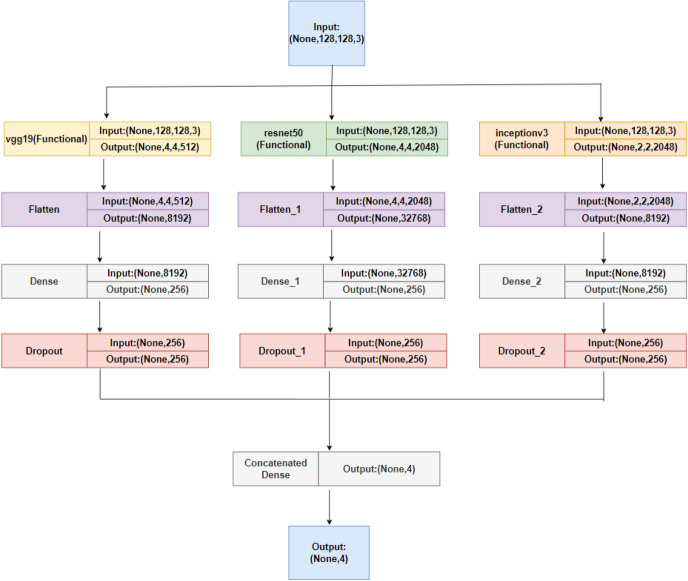
DL techniques, notably CNNs and pre-trained models such as VGG-16, have been used successfully in the field of neuroimaging to categorize AD using MRI images175. CNNs, a sort of deep neural network primarily built for image processing, excel at extracting features from high-dimensional input176. They recognize complicated patterns in data using convolutional layers, pooling, and fully linked layers, making them excellent for MRI image processing. VGG-16, a CNN variation, is pre-trained on millions of images, generating a robust feature detection capability. Due to its resilience and depth (16 layers), it can record subtle patterns in MRI scans, greatly helping to diagnose AD and separate it from other types of dementia. However, the fundamental difficulty with DL models is the requirement for large volumes of training data to avoid overfitting and ensure accurate predictions. Obtaining a large volume of labeled data in the field of medical imaging, particularly in the context of AD, can be difficult due to privacy concerns and the time-consuming nature of the labeling procedure. To remedy this, consider using ensemble ML approaches. Ensemble learning is a method in which numerous models or ’learners’ are developed and strategically merged to tackle a specific computer intelligence problem. Combining CNN or VGG-16 output, for example, with other ML algorithms helps improve and refine the decision-making process, enhancing the accuracy and robustness of the final model. Furthermore, by efficiently learning from minority classes, ensemble methods can help address class imbalance concerns that are frequently seen in medical datasets. To summarize, the use of DL algorithms such as CNN, VGG-16, and ensemble ML approaches has promising promise in Alzheimer’s MRI Classification, potentially improving diagnostic precision and allowing for early therapies. The following sections elucidate the dataset used in this study and present the results achieved through the various methodologies mentioned above.
Dataset used
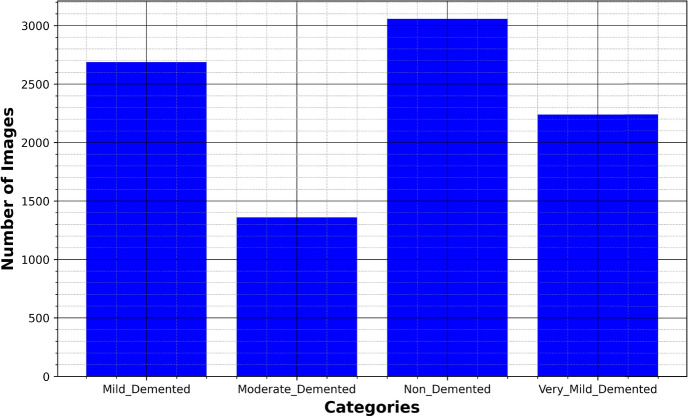
The disease taken into consideration is AD and the dataset taken into consideration is the Alzheimer’s MRI dataset. Figure 4 presents the distribution of different images in the dataset.
Figure 4.
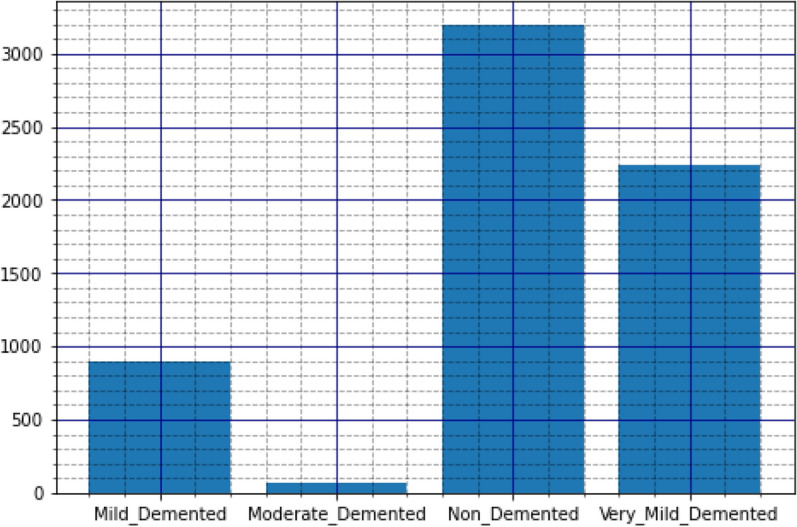
Original dataset distribution.
The dataset includes a total of 6400 images, which have all been resized to 128 × 128 pixels. The dataset consists of mild, moderate demented, non-demented, and mild demented images consisting of 896 images, Moderate Demented (64 images), Non-Demented (3200 images), and Very Mild Demented (2240 images). The images were preprocessed to remove noise and artifacts and then resized to 128 × 128 pixels for consistency. Each image is labeled with a class label (1–4) corresponding to the severity of dementia, as determined by a clinical expert.
The dataset has a class imbalance problem in case of Moderate Demented and Mild Demented cases. Thus, we apply different data augmentation techniques to address the imbalance. Initially, we generated extra images by basic augmentation like rotation, flipping, scaling, and translation transformations. In particular, for each minority class image , the techniques are applied as follows.
Rotation: Each image is rotated by leading to new images .
Flipping: Horizontal H(x) and vertical V(x) images flips are applied, to create additional images.
Scaling: Images are resized by factors leading to scaled images .
Translation: The images were translated with a shift along the x and y axes by pixels, resulting in as the translated images.
With the basic techniques, we generated 3584 new images for Mild Demented cases, and 256 new images for moderate demented. However, we removed images which are similar with 50% subtraction on augmentation, and generated finally 2688 total images in mild demented, and 320 images in moderate demented.
Next, we used the Generative Adversarial Network (GAN) model on the moderate demented case177. GANs are models that consist of a generator G and a discriminator D, which both play a minimax game. The generator , usually given some noise input z with parameters , aims to produce realistic-looking images. In contrast, the discriminator , which works with either natural or fake data x with parameters , is trained to discriminate between these two classes. The objective function is as follows.
| 1 |
Applying GANs to the moderate demented case, we were able to generate high-quality synthetic images that closely resemble real images in this category. After applying GAN, we increased the number of Moderate Demented images from 320 to 1360. Further, resampling technique using Synthetic Minority Over-sampling Technique (SMOTE) is applied to generate new synthetic samples by interpolating the moderate demented class samples. Mathematically, given a sample in the minority class and one of its k-nearest neighbors, , a new synthetic sample is created, where is a random number drawn from the interval [0, 1], that defines a linear space between the sample and . We further did undersampling on the Non-demented class to mitigate its domination over moderate demented class, resulting in 3057 images in the non-demented class. Figure 5 shows the final dataset distribution after applying the balancing techniques.
Figure 5.
Modified dataset distribution post balancing.
The purpose of this dataset is to aid in the development of computer-aided diagnosis systems for AD. The dataset provides a valuable resource for researchers interested in developing DL models to identify patterns in MRI images associated with the disease. The dataset has several potential applications in medical research and clinical practice. Various ML and DL models trained on this dataset could be used to automate the diagnosis of dementia based on MRI images, which could reduce the time and cost associated with traditional diagnostic methods. So considering this dataset, various DLs are applied on this dataset for classification and prediction into the necessary classes of dementia. Subsection “Methodology” give depicts the methodology portion.
Methodology
After identifying the necessary dataset following models were applied and trained on this dataset and appropriate results and graphs were obtained. The following are the models:
CNN: We used a CNN model to classify AD based on preprocessed MRI data. The model was implemented using Keras with a TensorFlow backend. The following steps were taken to construct the model: A Rescaling layer was added to the model with a scaling factor of 1./255 and the input shape of IMG HEIGHT x IMG WIDTH x 3. This layer helps to scale down the pixel values of the input images to a range between 0 and 1, which can improve model performance. Figure 6 depicts how data is distributed and transformed amongst different layers that include Convolutional178, Pooling179, Dense180, and Dropout layers181 in CNN182. A 2D convolutional layer was added with 16 filters, a kernel size of (3, 3), padding of ’same’, ReLU activation function, and He normal kernel initializer. This layer helps to extract features from the input images by sliding a filter window over the image and computing dot products between the filter and local image patches. A max-pooling layer was added with a pool size of (2, 2). This layer helps to down-sample the feature maps produced by the convolutional layer by taking the maximum value in each non-overlapping subregion. A dropout layer was added with a rate of 0.20. This layer randomly drops out a percentage of the neurons in the preceding layer during training to prevent overfitting. Steps were repeated twice with increasing numbers of filters (32 and 64) to further extract and down-sample features. The output from the convolutional layers was flattened to create a one-dimensional feature vector. A fully connected layer with 128 neurons, ReLU activation function, and He normal kernel initializer was added. This layer helps to map the high-dimensional feature vector to a lower-dimensional representation. Another fully connected layer with 64 neurons and ReLU activation function was added to further reduce the dimensionality. The output layer with 4 neurons and a softmax activation function was added to produce class probabilities for the input images.
VGG-16: The VGG-16 model is a deep CNN183. It is a popular model for image classification because of its relatively simple architecture. In this study, we used the VGG-16 model as a base model for our Alzheimer’s MRI classification task. Figure 7 indicates all the layers included in the model and input is transformed from one layer to another which includes many convolutional layers and required flatten and dropout layers. We used a pre-trained VGG-16 model with ImageNet weights to learn meaningful features from many images. We excluded the final fully connected layers by setting the included top parameter to False and added our own dense layers for classification. The pre-trained layers were frozen to prevent weight updates during training. We then added a flattened layer to the output of the base model, which converts the output tensor to a 1-dimensional tensor that can be passed to a fully connected layer. We added a dense layer with 256 neurons, ReLU activation function, and He normal kernel initializer. This was followed by a dropout layer with a rate of 0.20, which helps prevent inaccurate prediction by randomly dropping out some of the neurons during training. The output layer had 4 neurons and a softmax activation function for producing a probability distribution over the four classes. We then compiled the model with the Adam optimizer184, sparse categorical cross-entropy loss function185, and accuracy as the evaluation metric186.
Ensembled: We defined an ensemble model that combines three pre-trained DL models - VGG19, ResNet50187, and InceptionV3188 into a single model for image classification. VGG19 is selected due of its straightforward and deep architecture, which makes it highly effective for image classification tasks. VGG19’s uniform structure, consisting of multiple convolutional layers followed by fully connected layers, excels at extracting detailed and hierarchical features from images. This characteristic is particularly important in medical imaging, where subtle differences in brain scans need to be captured accurately to distinguish between various stages of Alzheimer’s disease. The simplicity of VGG19 also allows for easier interpretability and implementation, making it a reliable component of our ensemble.
Figure 6.
Architectural diagram of layers of CNN.
Figure 7.
Architectural diagram of layers of VGG-16.
ResNet50 is incorporated into our ensemble due to its innovative use of residual connections, which address the vanishing gradient problem that often hampers the training of deep neural networks. These residual connections enable ResNet50 to train much deeper networks without the risk of degradation in performance. This model’s strength in capturing intricate patterns and its robustness in training deep architectures enhance the ensemble’s ability to accurately classify Alzheimer’s stages.
InceptionV3 is chosen for its advanced architecture that includes Inception modules, which allow the network to handle multiple receptive fields within the same layer. This design enables the model to process and integrate information at different scales, which is crucial for analyzing the diverse patterns present in MRI images (Supplementary Information). InceptionV3’s efficiency in managing computational resources while maintaining high performance is another reason for its inclusion. Its capacity to perform complex feature extraction and its adaptability to various types of image data make it an excellent addition to our ensemble.
By combining these three models, our ensemble leverages the unique strengths of each: VGG19’s depth and simplicity, ResNet50’s ability to train deep networks effectively, and InceptionV3’s multi-scale feature extraction189. The ensemble model features improved performance in distinguishing the various stages of Alzheimer’s disease from MRI scans. In the ensemble, to prevent any kind of update on model weights, the model weights are frozen. By combining the models, the ensembled model can learn from the strengths of each model and improve overall performance. The input images are passed through each of the three models, and the output features are concatenated together. Figure 8 depicts all the layers of the ensembled model where input is passed through each model with necessary layers like flatten, dense, and dropout, and finally after concatenating all models necessary output is obtained. For training of our ensembled model, we employed the Adam optimizer, known for its adaptive learning rate capabilities which facilitate efficient convergence to optimal solutions. The initial learning rate was set to 0.001, a commonly used value that strikes a balance between rapid convergence and stable training. This learning rate was chosen based on preliminary experiments and fine-tuning, ensuring that the models in the ensemble could learn effectively without overshooting the loss function minima190. Additionally, to further enhance the model’s performance during the latter stages of training, we implemented a learning rate schedule that reduced the learning rate by a factor of 0.1 if the validation loss plateaued for five consecutive epochs.
Figure 8.
Architectural diagram of layers of ensembled model.
Regarding the dataset split, we divided the dataset into training, validation, and testing subsets to ensure accurate performance assessment and validation of the model. Specifically, 70% of the dataset was allocated for training. This subset was used to adjust the weights of the models based on the backpropagation algorithm and to learn the underlying patterns in the data. Another 15% of the dataset was set aside for validation. The remaining 15% of the dataset was reserved for testing. This test set was used to evaluate the final performance of the ensembled model after the training process was complete, providing a final assessment of how well the model generalizes to unseen data.
Table 11.
Comparison of deep learning models: VGG19, ResNet50, and InceptionV3.
| Feature | VGG19 | ResNet50 | InceptionV3 |
|---|---|---|---|
| Architecture type | Convolutional Neural Network (CNN) | Residual Neural Network (ResNet) | Convolutional Neural Network (CNN) |
| Depth | 19 layers | 50 layers | 48 layers |
| Key feature | Simple and deep | Residual connections to avoid vanishing gradient | Inception modules with dimensionality reduction |
| Convolution layers | 16 | 49 | 48 |
| Pooling layers | 5 | 1 | Multiple pooling operations within modules |
| Fully connected layers | 3 | 1 | 1 |
| Input image size | 224 × 224 | 224 × 224 | 299 × 299 |
| Advantages | Easy to understand and implement | Solves vanishing gradient problem, deeper networks | Efficient with complex data, good for transfer learning |
| Disadvantages | Large number of parameters | Computationally intensive | Complex architecture |
| Use cases | Image classification, feature extraction | Image recognition, object detection | Image classification, object detection, and more complex feature extraction |
With a total of 19 layers, VGG19 is a kind of CNN that is distinguished by its depth and simplicity. It consists of three fully connected layers, five pooling layers, and sixteen convolutional layers. This model has about 143 million parameters and can handle pictures. VGG19 is a good choice for image categorization and feature extraction applications because it is simple to comprehend and apply. Its numerous parameters, which need a substantial amount of processing power, can be a drawback. On the other hand, ResNet50 is a ResNet subclass that has 50 layers, 49 of which are convolutional layers and only one pooling layer. This subclass is deeper than others. In order to solve the vanishing gradient issue, it makes use of residual relationships, which permits the creation of deeper networks. This model can also process 224 × 224 images and has about 25.6 million parameters. The main benefit of ResNet50 is that it can manage deep networks well, which makes it perfect for object detection and picture recognition. Its primary disadvantage is that it requires a lot of computing. Another CNN, but one with a more intricate structure is InceptionV3. Its 48 layers include different pooling operations inside its modules and several convolutional layers. It includes Inception modules, which enable effective handling of complex data and dimensionality reduction. InceptionV3 has about 23 million parameters and can process larger input images up to 299 by 299 in size. The concatenated features are then passed through a dense layer with 256 neurons, with a ReLU activation function191 and a dropout layer with a rate of 0.20 to prevent inaccurate predictions. The outputs from each model are concatenated and passed through a final dense layer192 with a softmax activation function, which generates a probability distribution over the four classes of Alzheimer’s MRI images. The ensemble model is initialized with the categorical cross-entropy loss function193 and Adam optimizer. The purpose of using an ensemble model is to improve the accuracy and robustness194 of the model by combining the strengths of multiple models.
Subsection “Simulation results” presents the simulation results after applying all three models to the training and validation dataset. The simulation results consist of the accuracy and loss graphs of training and validation data. Finally, after that prediction tasks were performed and their results are also mentioned in the section.
Proposed architecture of cloud-based analysis of medical data
This architecture is intended for the classification and diagnosis of medical images. Figure 9 represents the proposed architecture which consists of three layers [NOTE: The cliparts used in the figure are freely available icons under the CCO license.]:
- Input layer: This layer takes the medical image datasets and the patient’s medical history if any, as an input. The datasets may contain X-rays, CT scans, MRI images, or other medical images. PMH (Patient Medical History) is a record of a patient’s past health conditions, medical treatments, surgeries, allergies, medications, and any other relevant medical information that may impact their current or future health. The dataset along with PMH (Patient Medical History) is passed to the cloud layer for further processing, through some gateway.
- Medical image dataset: Consists of Images obtained from various medical imaging modalities, of numerous diseases such as Alzheimer’s, Parkinson’s, etc.
- Cloud layer: In this layer image classification and prediction tasks are performed using DL models. These models are trained on large datasets to learn the patterns and features that are indicative of various medical conditions. The cloud layer may use several DL models such as CNN, VGG-16, etc. to process the medical images and extract features, and after the classification, i.e. categorize diseases based on their characteristic symptoms, and predict the likelihood of a person having that specific disease or health condition, the results are passed to diagnose layer.
- Data cleaning and reduction: Data cleaning and reduction are two essential steps in the process of data preparation for analysis. It involves the identification of errors and removing them and also the removal of any inconsistencies in data. Data reduction involves reducing the dataset while keeping the necessary data and information. Usually, this is done to speed up computation and enhance the performance of ML algorithms.
- Extracting feature: Extracting features is the process of extracting essential and relevant information from raw data. Feature extraction is frequently used to improve the performance of ML models by removing redundant or irrelevant data and lowering the dimensionality of the data, making it simpler and more quickly to process.
- Data classification and prediction: Training a model on a labeled dataset to identify patterns and make predictions on fresh, untainted data is known as data classification. Contrarily, making predictions typically involves training a model on a labeled dataset and applying it to unobserved and relatively new data. The aim of prediction is to precisely calculate the likelihood of a specific result or event using the information that is currently available.
Diagnose layer: This layer receives the predicted classification of the medical image from the cloud layer. The diagnose layer notifies the patient, doctor, and hospital if the predicted classification points to a medical condition. The alert may contain the predicted diagnosis, the probability of the diagnosis, and recommendations for further testing or treatment. The diagnose layer can also be integrated with EMR systems to maintain a record of the medical history of patients and share it with healthcare providers. Overall, this architecture provides a comprehensive solution for medical image classification and prediction, with the ability to alert healthcare providers and patients of potential medical conditions.
Gateway module: It acts as a bridge between different layers of the architecture, enabling data to be transferred between them. Here image encryption and decryption tasks are performed using the Advanced Encryption Standard (AES) algorithm in CBC mode. It first sets up a random 16-byte key and initialization vector, loads an image, and displays the original image using Matplotlib. Then it encrypts the image bytes using AES in CBC mode and saves the encrypted image to a new file. It also displays the encrypted image using Matplotlib. Next, it loads the encrypted image bytes from the file, decrypts the image bytes using AES in CBC mode, and creates a new image from the decrypted bytes. It displays the decrypted image using Matplotlib and saves it to a new file. Figure 10 depicts the entire process of encryption and decryption.
Figure 9.
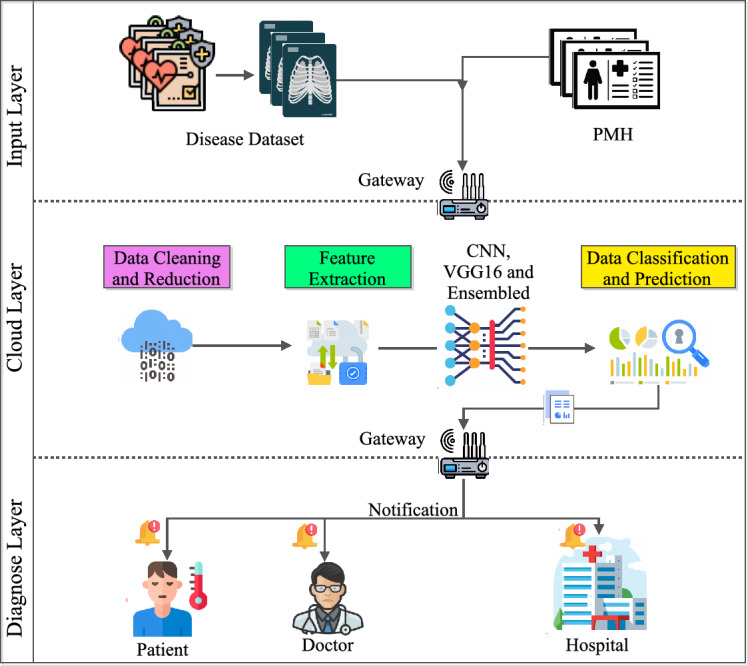
Proposed architecture.
Figure 10.
Encryption and decryption of images.
The proposed cloud-based architecture for medical data analysis plays a vital role in population health management. This is made possible by an efficient process of data gathering, integration, and analysis- a critical process in identifying at-risk populations for focused interventions. The architecture commences with the input Layer, which aggregates comprehensive health data, including datasets on medical images and patient medical history (PMH). It is through such extensive data collection that forms the core of population health management. It collects various data to ensure it gets all the necessary details and information for processing at later stages. The collected data is then processed in the cloud Layer. Advanced DL models like CNN, VGG-16, and ensembled models in this layer would be used to analyze the medical images and extract relevant features. The use of DL models in the cloud layer would be essentially crucial because population-based health management deals with large-scale data. Medical conditions can be easily detected early, and interventions made using the cloud layer through accurate classification and prediction of the likelihood of their occurrences, which forms an essential task in effective population health management.
The cleaning and reduction of data processes in the cloud layer ensure that the data used in the analysis is accurate and relevant, hence improving the reliability of the predictions by the DL models. The architecture retains high data quality by reducing errors and inconsistencies, and it also reduces the dimensionality of the data-all important in making correct predictions regarding at-risk populations. Additional data is optimized by feature extraction, where it identifies the most relevant information from the raw datasets. This increases the efficiency and effectiveness of the DL model in dealing with the data’s volume in less time and more accurately. The extracted features from this massive dataset would enable the system to detect subtle signs of health risks, hence early detection of at-risk people.
Data classification and prediction abilities of the cloud layer help the system to identify the populations at risk. The DL models learn to detect patterns associated with different health conditions by training on labeled datasets. When applied to new data, these models can predict the likelihood of an individual developing a specific condition, thus enabling healthcare providers to focus their attention and resources on those who are most at risk. This predictive capability is central to population health management, as it allows for the proactive management of health issues before they become more serious and widespread.
Algorithm 1.
The proposed architecture
The diagnose Layer translates these predictions into actionable insights, alerting the healthcare provider and patient of potential health risks based on detailed diagnostic reports, with recommendations for further testing or treatment. The correct information delivered to the right people at the right time ensures timely and targeted interventions. The diagnose layer further passes any such insights to Electronic Medical Record (EMR) systems to make them available across the healthcare continuum. The gateway module ensures that data transfer between the layers is secure because the data is encrypted with AES encryption, automatically securing patient data and compliance with regulations. These securities are essential in retaining trust and protecting sensitive health information, two essential elements in effective population health management.
Algorithm 1 depicts the architectural approach of the entire process, which takes input as the medical image dataset() and patient medical history(PMH) which is sent to , and as an output, respective entity get notification alerts such as patient, doctor, and hospital. This algorithm is a representation of the entire process that aims to process medical data generate a diagnosis report and send notification alerts to the doctor and patients. Following is the breakdown of the steps mentioned above in the algorithm:
Preprocess (Dataset): This is a function that processes the raw medical data set to make it suitable for analysis. This could involve tasks such as data cleaning, normalization, feature extraction, or other transformations that make the data more usable.
Preprocesses (PMH): Similarly, this function processes the patient’s previous medical history (PMH) to make it usable in the pipeline. This could involve tasks such as converting unstructured data into structured data, removing irrelevant information, or extracting key features.
Cloudlayer: This step involves sending the processed data sets to a cloud-based layer for further processing. This could involve tasks such as running the data through a DL model or other ML/DL algorithms to generate a prediction.
DL-CNN (Ds1, Ds2): This refers to the CNN that is used to generate predictions from the medical data set and previous medical history.
Rclassification: This refers to the predicted diagnosis classification generated by the CNN.
Classification (Y): This step involves using a classifier algorithm to generate a diagnosis report based on the predicted classification generated by the CNN.
Prediction (Rclassification, Y): This step generates the diagnosis report based on the results of the classification algorithm.
Diagnoselayer: This step involves sending the diagnosis report to another layer for further processing. This could involve tasks such as storing the report in a database, generating visualizations, or other analyses.
Send-Alert (Report): This step involves sending alert notifications to relevant entities based on the diagnosis report. In this case, the algorithm loops through the entities Eh, Ep, and Ed and sends alerts to each entity as necessary. Overall, this algorithm represents a typical DL pipeline for medical diagnosis that involves preprocessing data sets, using DL algorithms to generate predictions, and sending alerts to relevant entities based on the results.
Cross validation techniques
While constructing an ML model, we provide initial data to train the model and subsequently provide some unknown data to test the accuracy and check how effectively the model is performing against the unseen data. A model that performs well on unseen data, displaying consistency and accurately predicting a wide range of input data, is considered stable. However, this is not always the case. Machine learning models can be unstable, necessitating an evaluation of their stability. This is where Cross Validation becomes relevant, which asses the ML model’s accuracy on unseen data i.e. how the results of statistical analysis are used to generalize to the concept of the dataset to be independent. The need for cross-validation is very important, because if we have developed a ML model to address a particular problem, and after training it on a specific dataset, we find that its accuracy on the training data is around 98%. This does not imply that your model has been trained effectively and is the best choice due to its high accuracy. A high accuracy on the training data does not guarantee that the model will perform well on unseen data. A model with high training accuracy might have overfit the training data, meaning it has learned to capture even the smallest variations in the training data, rather than generalizing well to new, unseen data. When such an overfitted model is exposed to new data, its performance might drop significantly. Also, On the other hand, if a model has low training accuracy, it might not have learned the underlying patterns in the data well enough. This can result in poor performance not only on the training set but also on new, unseen data. This scenario is known as underfitting. So, in general, training accuracy is important, but it is not the only indicator of a good model which make essential to model’s performance on a separate test set or using cross-validation to ensure that it can generalize well to new data. To handle such issues we use cross-validation, some commonly used cross-validation techniques are listed below:
- Holdout validation: This technique is used in ML to evaluate the performance of a model. It involves splitting the dataset into two subsets: a training set and a test set. The model is trained on the training set and then evaluated on the test set to estimate its performance on unseen data. If is the complete data set consisting of k samples. we split into two subset training set and testing set each consist of k training and testing set. The models is trained on , once the model is trained we evaluate its performance on to estimate generalization error. Let be the true label for sample k and be the predicted label by the model. The performance metric, such as accuracy, can be calculated as:
where I is the indicator function that returns 1 if the predicted label matches the true label and 0 otherwise.2 - Leave-one-out cross-validation: This technique used to estimate the performance of an ML model dataset is divided into k subsets, where k is the number of instances in the dataset. For each iteration, one instance is held out as the validation set, and the model is trained on the remaining k-1 instances. This process is repeated k times, with each instance used once as the validation set. The performance of the model is then evaluated by averaging the performance metrics (such as accuracy, error rate, etc.) obtained in each iteration. It provides an unbiased estimate of the model’s performance, as each instance is used for both training and validation. The error rate is defined as:
L is the loss function used to measure the difference between the actual target value and the predicted target value . The sum is taken over all instances in the dataset. This techniques is generally used when working with small dataset.3 - K-fold cross-validation: The basic idea behind K-Fold Cross-Validation is to divide the dataset into K subsets, or folds, of approximately equal size. The model is then trained and evaluated K times, each time using a different fold as the test set and the remaining folds as the training set. Evaluate the model on the test set and calculate the evaluation metric (e.g., accuracy, precision, recall, etc.) and save the evaluation metric for fold k. Finally, Calculate the average evaluation metric across all K folds to obtain the final performance estimate of the model.
Where E is the average evaluation matrix across all k folds and is the evaluation matrix for fold k.4 Stratified K-fold cross-validation: This technique used in ML to address the issue of class imbalance in datasets. It is an extension of the traditional K-Fold Cross-Validation method, where the dataset is divided into k subsets (folds) while ensuring that each fold has the same proportion of target variable classes as the entire dataset. This helps in producing more reliable and unbiased estimates of model performance, especially when dealing with imbalanced datasets.
Class balancing methods
When the number of samples in each class is roughly equal, then most of the ML algorithms perform best, as they are designed to maximize the accuracy and minimize the error. When the dataset consists of imbalanced classes, then the ML algorithm exhibits biases toward some classes. In such cases, the model may achieve high accuracy by predicting the majority class but fail to capture the minority class. For instance, if 98% of the data from the bank transaction belongs to the majority class, i.e., valid transactions, and only 2% transactions are fraudulent, and your model predicts every transaction as valid without checking it, you still get 98% accuracy. So, the basic classification models like logistic regression or decision trees may struggle to identify and classify minority class data points correctly. To handle this problem, class balancing techniques are used, some of the widely used techniques are listed below:
- Random under-sampling: It involves reducing the number of instances in the majority class to match the number of instances in the minority class. This helps prevent the model from being biased towards the majority class and improves its ability to generalize to new data. If is a dataset if k samples, where is the number of samples of the majority class and is the number of samples of the minority class, then the imbalance ratio can be defined as:
Determine the desired number of samples in the balanced , randomly select instances from the majority class, and now Combine the randomly selected instances from the majority class with all instances from the minority class to create the balanced dataset. Finally train the machine learning model on the balanced dataset.5 - Random over-sampling: This method involves randomly selecting instances from the minority class and duplicating them until the class distribution is balanced. If is a dataset if k samples and c distinct classes and is the number of samples of the majority class and is the number of samples of the minority class, then imbalance ratio is defined as:
Now randomly select some new samples from equivalent to - and replace them with new randomly selected ones. Now is balanced.6 - Synthetic minority oversampling technique: This technique uses class imbalance in datasets by generating synthetic samples for the minority class. It creates synthetic samples that are similar to existing minority class samples, thereby increasing the representation of the minority class in the dataset. This helps to balance the class distribution and improve the performance of machine learning models, especially in scenarios where the minority class is underrepresented. Let x be the set of minority class samples and is the total number of minority class samples, then for each sample in x compute k-nearest neighbor and randomly select one of the k-nearest neighbors for each .
where is the generated synthetic samples and is a random value between 0 and 1. Repeat this process until the desired balance between the minority and majority classes is achieved.7 Near miss: This technique addresses class imbalance by selecting a subset of samples from the majority class that are close to the minority class samples. The goal is to reduce the imbalance between classes while maintaining the separability between classes. It is an under-sampling technique that uses distance to make the majority class equal to the minority class. Each sample in the minority class, calculate its distances to the samples in the majority class and for each sample in the minority class, select the “nearest” samples from the majority class based on a euclidean distance. Now, based on the specific variant of never-miss, different criteria are used to select the samples from the majority class. The “Near Miss” algorithm is a simple yet effective way to address class imbalance by focusing on the samples that are most relevant to the minority class.
Simulation results
This subsection presents the analysis of results on the medical dataset when CNN, VGG-16, and Ensemble model are used.
CNN: Fig. 11 shows the loss in training and validation data, whereas Fig. 12 shows the accuracy in training and validation data. The model was initially trained on the preprocessed MRI data for 50 epochs with a batch size of 64 using the Adam optimizer. The validation dataset was used to assess model performance and prevent inaccurate predictions on testing data. The Y-axis of the graph indicates loss and X-axis indicates the number of epochs in Fig. 11. In Fig. 12 the Y-axis determines accuracy and X-axis determines the number of epochs. We can observe from the figures that if the model is trained on more epochs the model provides better accuracy with loss as quite as low, This helps us to do classification tasks appropriately. Figure 13 indicates the prediction tasks performed after training the model on the dataset. Here X-Axis indicates the actual form of the disease and Y-Axis indicates the predicted form; if the actual and predicted form match, then the predicted result will be indicated in green color text, and if not then the predicted result will be indicated in red color text. Our study successfully addresses the prediction tasks and achieved high accuracy in our analysis, and achieved 99.285% test accuracy.
VGG-16: Fig. 14 shows the loss in training and validation data, Fig. 15 shows the accuracy on training and validation data. For observation, we trained the model using the fit function with a batch size of 64 and 20 epochs while keeping the pre-trained layer weights fixed. The Y-axis of the graph indicates accuracy and X-axis indicates the number of epochs in Fig. 14. In Fig. 15 the Y-axis determines loss and X-axis determines the number of epochs. As the model is much more complex because it includes many convolutional layers and due to its deeper architecture, the model was trained on fewer epochs compared to CNN, but the model still performed better on 20 epochs, if we increase the number of epochs we can achieve better accuracy. In Fig. 15, the initial value of loss is very high for few epochs because the model is adjusting its weights to better fit the training data. As the training progresses the model converges to a better solution and reduces the loss drastically. Figure 16 indicates the prediction tasks performed after training the model on the dataset. Here X-Axis indicates the actual form of the disease and Y-Axis indicates the predicted form; if the actual and predicted form match, then the predicted result will be indicated in green color text, and if not then the predicted result will be indicated in red color text. Our study successfully addresses the prediction tasks and achieved high accuracy in our analysis, and achieved 85.113% test accuracy.
Ensembled: For the Ensembled Model, Fig. 17 depicts the loss on training data and validation data and Fig. 18 indicates the accuracy on training data and validation data. The ensemble model is compiled with the categorical cross-entropy loss function, the Adam optimizer, and accuracy as the evaluation metric with 10 epochs with a batch size of 64. The ensemble model is more complex due to the combination of multiple models that includes VGG19, Resnet, and Inceptionv3, the model is trained on fewer epochs as it requires high additional computational resources. However the accuracy results is not as good compared to CNN and VGG-16 models, but with higher epochs, better accuracy can be achieved with lower value of loss. Figure 19 depicts the prediction results, but we can clearly observe from the figure that there are many wrong predictions obtained as compared to CNN and VGG-16 models, even if accuracy is higher there are chances of many wrong predictions. In this setup, we achieved 79.192% test accuracy.
Figure 11.
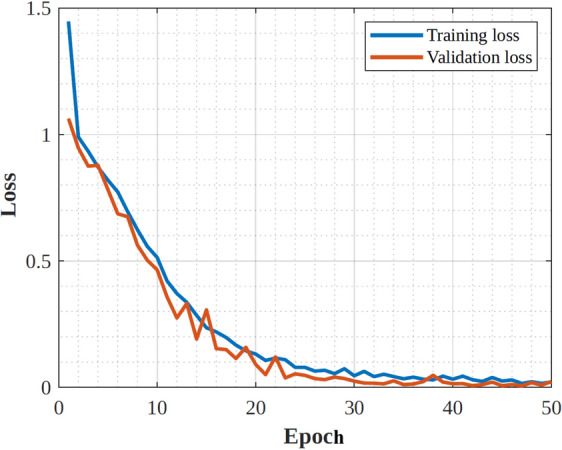
CNN model training and validation loss.
Figure 12.
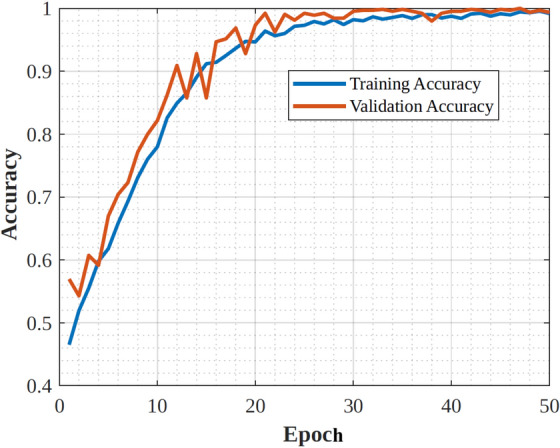
CNN model training and validation accuracy.
Figure 13.
Prediction results for CNN model.
Figure 14.
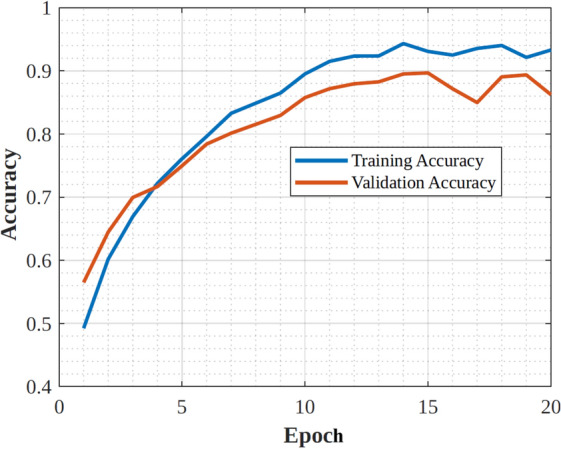
VGG-16 model training and validation accuracy.
Figure 15.
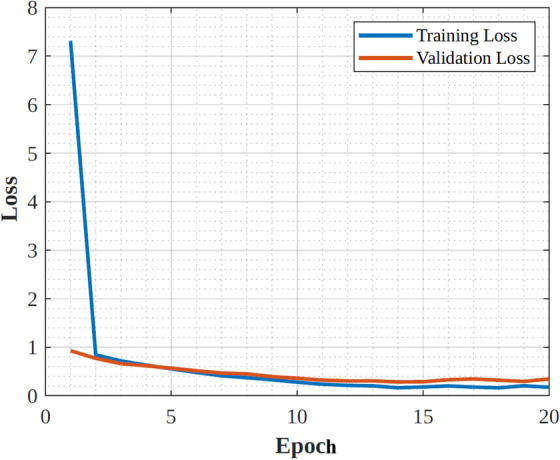
VGG-16 model training and validation loss.
Figure 16.
Prediction result for VGG-16 model.
Figure 17.
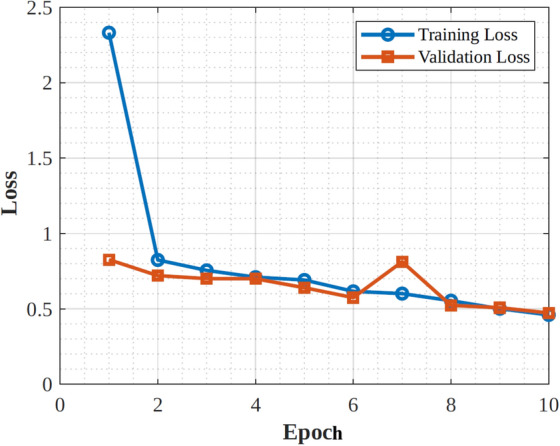
Ensembled model training and validation loss.
Figure 18.
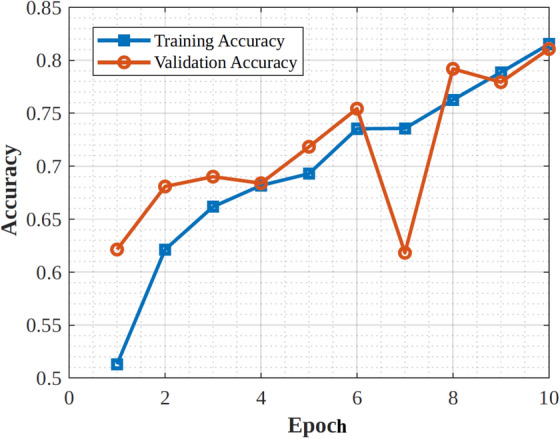
Ensembled model training and validation accuracy.
Figure 19.
Prediction result for ensembled model.
Recent cloud schemes in the medical fields
CC in healthcare encompasses a multifaceted scheme aimed at revolutionizing various aspects of the industry and the same has been discussed in Table 12, which indicates the various usages of the CC schemes for healthcare ecosystems with its benefits, challenges, examples, and the numerous supporting AI algorithms. One prominent application is the adoption of cloud-based electronic health records (EHRs), enabling the secure storage, management, and sharing of patient health data. Additionally, cloud infrastructure facilitates health information exchange between different healthcare entities, ensuring seamless and secure data sharing. Moreover, cloud services offer robust solutions for healthcare data storage and backup, crucial for preserving sensitive medical information such as images and genomic data. Cloud replication of vital systems and data improves disaster recovery and business continuity plans and guarantees quick recovery in an emergency. The utilization of wearables and medical IoT devices expands the scope of healthcare by permitting telemedicine, population health management, and remote patient monitoring195. Cloud platforms use analytics and machine learning to mine large healthcare datasets and extract meaningful information for research, quality improvement, and clinical decision-making. Secure cloud-based communication tools enable virtual consultations and remote monitoring for telemedicine and virtual care platforms. Cloud-enabled data gathering and analysis helps population health management programs by assisting in the identification of populations that are at-risk and the implementation of focused interventions. Strict cybersecurity protocols are put in place to protect patient information and guarantee adherence to legal requirements. Self-service portals and mobile apps improve patient engagement by enabling people to quickly manage their appointments and access their health records. Furthermore, cloud-based solutions streamline the distribution, inventory control, and purchase of medical equipment and supplies, improving the efficiency of the healthcare supply chain. Moreover, real-time monitoring and predictive maintenance are made possible by the integration and analysis of data from various healthcare IoT devices, providing useful insights to enhance patient care results196. All things considered, cloud computing revolutionizes the healthcare sector by improving accessibility, efficiency, and quality of care delivery while protecting patient privacy and security. However, the role of the Transformer is also supportive to the healthcare industry. Originally intended for NLP, the Transformer architecture has found use in the healthcare industry, especially in cloud computing settings. This deep learning model effectively processes sequential input by means of self-attention techniques. The Transformer can be used for tasks like image analysis, predictive modeling, and patient monitoring in the healthcare industry, where data is frequently provided in sequences, such as medical images or patient records. Accurate predictions are made possible by effectively capturing complicated connections in data, particularly in situations with subtle patterns. Because of its parallelizability and scalability, it can be deployed in the cloud and process massive healthcare datasets effectively. All things considered, the Transformer design holds great potential for increasing data analysis, decision support, and eventually patient outcomes in the healthcare industry. Mathematically, the Transformer architecture can be represented as follows: Let X be the input sequence of data, where , and n is the length of the sequence. Each element represents a feature vector or a token in the sequence. Feed-forward neural networks and self-attention mechanisms are the mainstays of the Transformer design. The self-attention mechanism computes attention scores between all pairs of elements in the input sequence X, capturing the relevance or importance of each element with respect to others. Mathematically, the self-attention mechanism can be formulated as shown in Eq. 7:
| 8 |
where Q, K, and V are the query, key, and value matrices obtained by linear transformations of the input sequence X. represents the dimensionality of the key vectors. After scaling by the square root of the key vectors’ dimensionality and utilizing the dot product between the query and key vectors to calculate the attention scores, a softmax function is applied to produce a probability distribution. Ultimately, the result is calculated as the weighted sum of the value vectors, with the attention scores serving as the basis for the weight calculations. The output of the self-attention mechanism is sent via feed-forward neural networks to identify non-linear linkages and interactions in the data, following the computation of attention scores. A non-linear activation function, usually a Rectified Linear Unit (ReLU), comes after two linear transformations in a feed-forward neural network. The feed-forward neural network can be expressed mathematically as shown below in equation 8:
| 9 |
where , , , and are learnable parameters of the neural network. Positional encodings are another feature of the Transformer architecture that provide details on the placement or order of the elements in the input sequence. The input embeddings are enhanced with these positional encodings prior to being fed into feed-forward neural networks and the self-attention mechanism. All things considered, the Transformer architecture provides an effective framework for handling sequential healthcare data, which makes it ideal for a range of applications including clinical decision support, predictive modeling, medical image analysis, and patient monitoring in cloud healthcare systems.
Table 12.
Exploring cloud computing strategies in healthcare: benefits, challenges, examples and ML/DL usages.
| Cloud computing scheme | Benefits | Challenges | Examples/Platforms | AI (ML/DL) Algorithms roles |
|---|---|---|---|---|
| Cloud-based electronic health records197 | Real-time access to patient information, interoperability, automatic updates, reduced infrastructure costs | Data security and privacy concerns, interoperability challenges, regulatory compliance | Epic Systems, Cerner Corporation, Athenahealth | Natural Language Processing (NLP), Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), Support Vector Machines (SVM), Decision Trees |
| Health information exchange198 | Improved care coordination, streamlined workflows, support for population health management | Interoperability issues, data governance, privacy and security risks | CommonWell Health Alliance, Carequality, InterSystems HealthShare | Machine Learning, Natural Language Processing (NLP), Deep Learning, Graph Neural Networks (GNN), Ensemble Learning |
| Healthcare data storage and backup199 | Scalability, cost-effectiveness, data redundancy, disaster recovery | Data security and privacy, compliance with regulatory requirements, data migration challenges | Amazon Web Services (AWS), Microsoft Azure, Google Cloud Platform | Machine Learning, Deep Learning, Natural Language Processing (NLP), AutoML, Random Forests |
| Disaster recovery and business continuity200 | Minimize downtime, data loss prevention, scalability, cost savings | Data replication and synchronization, network latency, dependency on cloud service providers | VMware Site Recovery, Zerto, Druva Phoenix | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Time Series Forecasting, Autoencoder |
| Medical IoT and wearables201 | Real-time monitoring, personalized care, early detection of health issues | Data interoperability, device compatibility, security and privacy concerns, data integration complexity | Philips Healthcare, Qualcomm Life, Validic | Machine Learning, Deep Learning, Time Series Analysis, Anomaly Detection, Natural Language Processing (NLP) |
| Healthcare analytics and machine learning202 | Predictive analytics, personalized medicine, improved outcomes, cost reduction | Data quality and integrity, algorithm bias, interpretability of results, data privacy and security | Health Catalyst, SAS Analytics, IBM Watson Health | Supervised Learning, Unsupervised Learning, Deep Learning, Reinforcement Learning, Natural Language Processing (NLP) |
| Telemedicine and virtual care platforms203 | Improved access to care, convenience, reduced healthcare costs, patient engagement | Technological barriers (e.g., internet connectivity), reimbursement challenges, regulatory and licensure issues | Teladoc Health, Amwell, Doxy.me | Machine Learning, Deep Learning, Natural Language Processing (NLP), Computer Vision, Sentiment Analysis |
| Population health management204 | Disease prevention, care coordination, resource allocation, improved outcomes | Data integration and standardization, interoperability, privacy concerns, scalability | Arcadia, Cerner Population Health, Philips Wellcentive | Machine Learning, Deep Learning, Graph Analytics, Predictive Modeling, Clustering |
| Cybersecurity and compliance management205 | Data encryption, access controls, threat detection, regulatory compliance | Evolving cybersecurity threats, compliance with multiple regulations (e.g., HIPAA, GDPR), resource constraints | Palo Alto Networks, Fortinet, Microsoft Azure Security Center | Machine Learning, Deep Learning, Natural Language Processing (NLP), Anomaly Detection, Predictive Analytics |
| Patient engagement and self-service portal206 | Patient empowerment, improved communication, enhanced care coordination | User adoption and engagement, accessibility barriers, data security and privacy concerns | MyChart (Epic Systems), FollowMyHealth (Allscripts), Cerner HealtheLife | Machine Learning, Deep Learning, Natural Language Processing (NLP), Recommender Systems, Sentiment Analysis |
| Healthcare supply chain management207 | Cost savings, inventory optimization, supply chain visibility, improved patient care | Supply chain complexity, demand forecasting accuracy, inventory management challenges | Oracle Healthcare, SAP Integrated Business Planning, GHX | Machine Learning, Deep Learning, Time Series Analysis, Predictive Modeling, Optimization |
| Healthcare IoT data integration and analytics208 | Predictive maintenance, real-time monitoring | Latency and data security | IoT Health Monitoring System, Remote Patient Monitoring Platform, Philips Healthcare | Autoregressive Integrated Moving Average (ARIMA), Isolation Forest, One-Class SVM,Predictive models |
Comparative analysis of models
This section presents the comparison of all models that are implemented and trained on a dataset with 10 only epochs, as VGG-16 and Ensembled models require high computational resources, which makes the model too complex and takes more time to train the dataset. Figure 20 indicates training data accuracy and Fig. 21 indicates validation data accuracy of the 3 models. We considered Adam optimizer with a batch size of 64 and sparse categorical cross-entropy loss function. The Adam optimizer gives better performance and uses optimized gradient descent whereas the cross-entropy loss function provides better adjustment of model weights during training. We can observe from both the figures that VGG-16 provides better accuracy as compared to CNN and Ensemble model, but CNN achieved higher accuracy when trained for a higher number of epochs.
Figure 20.
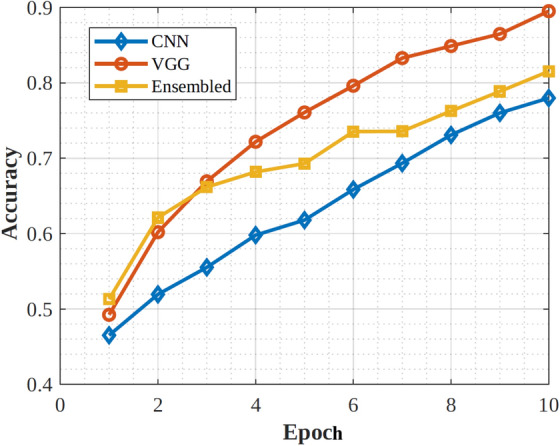
Comparison of training accuracy with 10 epoch.
Figure 21.
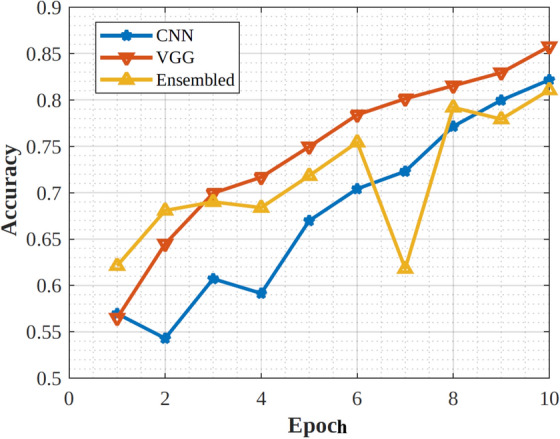
Comparison of validation accuracy with 10 epoch.
Table 13 shows the comparison based on the specific characteristics and requirements of MRI datasets, such as the need for robustness to variations in MRI data, the importance of interpretability in medical imaging, and the potential for improved performance through ensembling. Table 14 shows the observation on performance classification of model used by209,210 with proposed approach. This shows the per-class classification of the proposed model against the two baseline deep learning CNN model. The accuracy of the proposed model is 92.18% against the two model 78.0% and 91.20%.
Table 13.
Analysis metric for CNN, VGG16, Ensembled model.
| Factor | CNN | VGG16 | Ensembled model |
|---|---|---|---|
| Architecture | Custom deep learning specially designed for spatial data such as MRI | Predefined, 16 layers, 3X3 filter, and max-pooling layer that helps to learn hierarchical features from the input MRI images | Combination of models, possibly including CNNs, VGG16, and other architectures. |
| Performance metrics | F1-score = 96.5, Sensitivity = 96.5, Precision = 96.5 | F1-score = 96, Sensitivity = 97, Precision = 94.5 | F1-score = 98.5, Sensitivity = 98.7, Precision = 98.25 |
| Computational complexity | High when dealing with complex MRI dataset | Model depth contribute to high computational complexity | Depend on the number of base model and ensembled method |
| Training and inference time | Longer training time | Longer training time | Longer training time |
| Interpretability | Difficult to understand the specific features or patterns in the images that lead to those predictions. | Challenging to interpret how it makes decisions based on MRI images | More interpretable and depend on the base model. |
| Robustness | Robust due to hierarchical features of images | Robust due to ability to learn features from images | Potential for improved robustness |
Table 14.
Comparative Analysis metric of proposed model.
| Sarraf et al.210 | Islam et al.209 | Proposed | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ND | VMD | MiD | MoD | ND | VMD | MiD | MoD | ND | VMD | MiD | MoD | |
| Precision | 0.88 | 0.75 | 0.62 | 0.32 | 0.80 | 0.44 | 0.27 | 0.50 | 0.96 | 0.98 | 0.67 | 0.55 |
| Recall | 0.95 | 0.50 | 0.70 | 0.51 | 0.90 | 0.33 | 0.60 | 0.50 | 0.96 | 0.36 | 0.86 | 0.51 |
| F1-score | 0.91 | 0.60 | 0.62 | 0.42 | 0.82 | 0.38 | 0.25 | 0.50 | 0.96 | 0.60 | 0.75 | 0.50 |
| Support | 72.0 | 6.00 | 7.00 | 2.00 | 73.0 | 6.00 | 6.00 | 2.00 | 78.0 | 7.00 | 7.00 | 2.00 |
ND: Non Demented, VMD: Very mild Demented, MiD: Mild Demented, MoD: Moderate Demented.
Discussion
The VGG-16 neural network design has been used more frequently to classify medical images, demonstrating its significant utility in this particular area. VGG-16 has demonstrated success in extracting complex patterns and characteristics from medical pictures, including those related to various diseases and Alzheimer’s MRI Classification, thanks to its depth and usage of small convolutional filters.
Its application across diverse medical imaging tasks such as tumor detection, organ segmentation, and tissue classification demonstrates its adaptability and potency in handling complex visual data. Furthermore, the architecture’s ability to be fine-tuned allows it to be tailored to specific medical scenarios, resulting in enhanced performance compared to more generic models. The VGG-16 consistently succeeds in various medical image classification for Alzheimer’s MRI Classification endeavors despite some restrictions in computational efficiency and resource consumption, demonstrating its significant contribution to the development of AI-driven healthcare diagnostics and research. Using medical datasets offers a number of challenges that prevent their ideal application in clinical practice and healthcare research. First and foremost, accuracy depends on the quality and consistency of the data, which necessitates stringent pre-processing and standardization. Second, strict compliance with rules like HIPAA in the United States or GDPR in Europe is required due to the privacy and ethical issues surrounding patient information, which can restrict accessibility and collaboration. Thirdly, complex integration and alignment techniques are frequently needed to extract relevant insights from the heterogeneous nature of medical data, including different imaging modalities, textual reports, and genetic information. Last but not least, biased models that inaccurately represent the overall population may result from the dataset’s frequently unbalanced distribution of specific medical diseases. Together, these difficulties highlight the complexity of utilizing medical datasets, and they call for a careful, interdisciplinary strategy to get around them and realize the full promise of data-driven healthcare.
Statistical analysis of experiment results using T-test
In order to make sense of the data gathered during study and experimentation, statistical analysis is essential. The t-test is one approach that is frequently used to compare two sets of data. This article explores the fundamentals and uses of the t-test in statistical testing, offering a thorough explanation of how this technique is applied to examine experimental results related to medical imaging. The discussion here covers the t-test’s theoretical foundations, assumptions, several t-test varieties, and step-by-step directions for the test. To show how it might be applied and interpreted in the actual world, concrete examples and experiments have been investigated. A t-test is a statistical technique for making inferences about datasets. Its purpose is to measure whether the means of two groups exhibit significant differences while being linked to specific attributes. The t-test is a fundamental method within the realm of statistical hypothesis testing. This method is most appropriate when comparing two distinct groups, known as pairwise testing or comparison. The decision to employ a t-test hinges on two primary considerations: firstly, whether the groups being compared originate from the same or different groups, and secondly, whether the intention is to assess significance in a particular direction or both directions.
Paired T Test: If the groups come from the same population, a paired t-test should be employed (for instance, when comparing results from an experiment before and after).
Two sample T test: It is also known as an independent t-test, which should be advised to be used if the samples come from two distinct populations such as two different species or persons from two different locations.
One sample T-test : Use a t-test of one sample if only one set is being compared to a standard value (for instance, comparing a liquid’s acidity to pH 7 as the neutral value).
- T-tests with one and two tails: It is advised to use if two populations are different. To evaluate whether the mean of one population is greater or less than the other, a one-tailed t-test can be performed. Below is the two-sample t-test formula.
In Eq. (10), the is the value of t, and are denoted as the means of the two groups, which is being compared, and the is denoted as the pooled standard error of the two groups. The and are designated as the number of observations for each group. A high t-value signifies group mean difference is more important than the standard error of the pool, which indicates the more significant difference between the two groups. In the present study, we reject the null hypothesis and come to the conclusion that the two groups are distinct.10
To calculate the t-test the Mean Absolute error (MAE), as shown in the Eq. (11) has been computed for the medical image classification.
The MAE is represented as follows:
| 11 |
where
n: Total number of samples in the dataset
: Predicted value for the sample
: Actual (ground truth) value for the sample
This formula provides an average measure of the absolute differences between predicted and actual values, serving as an evaluation metric for regression tasks. Table 15 shows the statistical testing data of one tail and two tails for MAE values. Here the level of significance is 0.05, and hence, the NULL hypothesis is rejected here. The values that have been discussed here are lesser than the level of significance, i.e., less than the 0.05 level. The same is also described in Table 15 the ANN value has been calculated as one-tail values as 2.7E03 and two-tail as 5.42E03. Similarly, the values for the Random forest have been calculated as 6.41E03 for One-tail and 1.28E03 for two-tail. Likewise, the values of the KNN, SVM, and Residual Networks are found as 3.10E03, 4.23E03, and 2.40E03 for one-tail and the two-tail values are calculated as 3.36E02, 3.48E03, and 1.90E03.
Table 15.
T test statistical analysis for the MAE values.
| Schemes used for the comparison with our proposed model | One tail values | Two tail values |
|---|---|---|
| ANN | 2.70E03 | 5.42E03 |
| Random forest | 6.41E03 | 1.28E03 |
| KNN | 3.10E03 | 3.36E02 |
| SVM | 4.23E03 | 3.48E03 |
| Gradient boosting | 2.40E03 | 1.90E03 |
Conclusion
The study presents a tutorial approach on the use of DL for reliable CC in healthcare, using Alzheimer’s-related MRI scans as a test case. Our article covered an extensive review of literature, hand-picking the right dataset, employing various computational models like CNN, VGG-16, and an ensemble method, as well as creating our own unique architecture and algorithms. We also conducted simulations and engaged in prediction activities to test the efficacy of these models. The CNN model emerged as the top performer, boasting a test accuracy of an impressive 99.285%. On the other hand, VGG-16 secured a decent score with an 85.113% accuracy rate. The ensemble model trailed behind, achieving only 79.192% accuracy and displaying numerous false predictions, thereby showing its limitations in this particular predictive task. The results indicate that CNN based models show immense potential in MRI imaging. Moreover, our proposed architecture offers a secure and trustworthy blueprint for integrating CC into healthcare systems.
Future research could delve into how these computational models apply to other sets of medical data and evaluate their effectiveness in real-world clinical scenarios. Another promising avenue could be the exploration of encrypting medical images to further enhance privacy and security measures. In sum, our study paves the way for the fusion of dependable CC in healthcare settings and the progression of DL models in the realm of medical image analysis.
Supplementary Information
Acknowledgements
All authors read and approved the final manuscript.
Author contributions
Conceptualization, V.K.P., P.B., S.S., S.C., and S.A; methodology, S.A. and N.A., M.B.; software, S.A., V.K.P., S.S., S.C., M.B.; formal analysis, S.A. N.A., M.B., S.S., S.C.,; writing–original draft preparation, ALL AUTHORS WERE INVOLVED; writing–review and editing, ALL AUTHORS WERE INVOLVED HERE supervision, S.C. AND S.A.; All authors have read and agreed to the current version of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71358-7.
References
- 1.Prasad, V. K. & Bhavsar, M. D. Monitoring IAAS cloud for healthcare systems: Healthcare information management and cloud resources utilization. Int. J. E-Health Med. Commun. (IJEHMC)11(3), 54–70 (2020). [Google Scholar]
- 2.Celard, P. et al. A survey on deep learning applied to medical images: from simple artificial neural networks to generative models. Neural Comput. Appl.35(3), 2291–2323 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aruna, M., Arulkumar, V., Deepa, M. & Latha, G. C. P. Medical healthcare system with hybrid block based predictive models for quality preserving in medical images using machine learning techniques. In 2022 International Conference on Advanced Computing Technologies and Applications (ICACTA) 1–10 (2022). 10.1109/ICACTA54488.2022.9753355.
- 4.Verma, A. et al. Data localization and privacy-preserving healthcare for big data applications: Architecture and future directions. In Emerging Technologies for Computing, Communication and Smart Cities (eds Singh, P. K. et al.) 233–244 (Springer, Singapore, 2022). [Google Scholar]
- 5.Verma, A., Bhattacharya, P., Zuhair, M., Tanwar, S. & Kumar, N. VaCoChain: Blockchain-based 5G-assisted UAV vaccine distribution scheme for future pandemics. IEEE J. Biomed. Health Inf.26(5), 1997–2007. 10.1109/JBHI.2021.3103404 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Verma, A. et al. SanJeeVni: Secure UAV-envisioned massive vaccine distribution for COVID-19 underlying 6g network. IEEE Sens. J.23(2), 955–968. 10.1109/JSEN.2022.3188929 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Precedence research: Artificial Intelligence in Healthcare market by component, application and technology. https://www.precedenceresearch.com/artificial-intelligence-in-healthcare-market. Accessed: 2023-08-20
- 8.Prasad, V. K. et al. ABV-COVID: An ensemble forecasting model to predict availability of beds and ventilators for COVID-19 like pandemics. IEEE Access10, 74131–74151. 10.1109/ACCESS.2022.3190497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raswat, D. et al. Explainable AI for healthcare 5.0: Opportunities and challenges. IEEE Access10, 84486–84517. 10.1109/ACCESS.2022.3197671 (2022). [Google Scholar]
- 10.2020 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia, 16(3), 391–460 (2020) 10.1002/alz.12068 [DOI] [PubMed]
- 11.Mujahid, M. et al. An efficient ensemble approach for Alzheimerrsquo’s disease detection using an adaptive synthetic technique and deep learning. Diagnostics13(15), 2489. 10.3390/diagnostics13152489 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naseem, S. et al. DeepFert: An intelligent fertility rate prediction approach for men based on deep learning neural networks. IEEE Access11, 75006–75022. 10.1109/ACCESS.2023.3290554 (2023). [Google Scholar]
- 13.Rehman, A., Saba, T., Mujahid, M., Alamri, F. S. & ElHakim, N. Parkinsonrsquo’s disease detection using hybrid LSTM-GRU deep learning model. Electronics12(13), 2856. 10.3390/electronics12132856 (2023). [Google Scholar]
- 14.Hassan, A. et al. (eds) Federated Learning and AI for Healthcare 1–391 (IGI Global, Hershey, 2024). 10.4018/979-8-3693-1082-3. [Google Scholar]
- 15.Li, X. et al. Deep learning attention mechanism in medical image analysis: Basics and beyonds. Int. J. Netw. Dyn. Intell.2(1), 93–116. 10.53941/ijndi0201006 (2023). [Google Scholar]
- 16.Nabrawi, E., Alanazi, A. T. & Al Alkhaibari, E. Imaging in healthcare: A glance at the present and a glimpse into the future. Cureus15(3), e36111 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayan, V. et al. Enhance-Net: An approach to boost the performance of deep learning model based on real-time medical images. J. Sensors2023, 8276738 (2023). [Google Scholar]
- 18.Saraswat, D., Bhattacharya, P., Zuhair, M., Verma, A. & Kumar, A. AnSMart: A SVM-based anomaly detection scheme via system profiling in smart grids. In 2021 2nd International Conference on Intelligent Engineering and Management (ICIEM), London, United Kingdom, 417–422 (2021). 10.1109/ICIEM51511.2021.9445353
- 19.Patel, M., Prasad, V. K., Bhattacharya, P., Bhavsar, M. & Zuhair, M. Privacy preservation for big data healthcare management. In 2022 3rd International Conference on Intelligent Engineering and Management (ICIEM), 211–216 (IEEE, 2022)
- 20.Mewada, A., Gujaran, R., Prasad, V. K., Chudasama, V., Shah, A. & Bhavsar, M. Establishing trust in the cloud using machine learning methods. In Proceedings of First International Conference on Computing, Communications, and Cyber-Security (IC4S 2019) 791–805 (Springer, 2020)
- 21.Dhar, T., Dey, N., Borra, S. & Sherratt, R. S. Challenges of deep learning in medical image analysis-improving explainability and trust. IEEE Trans. Technol. Soc.4(1), 68–75. 10.1109/TTS.2023.3234203 (2023). [Google Scholar]
- 22.Verma, A. et al. FedRec: Trusted rank-based recommender scheme for service provisioning in federated cloud environment. Digit. Commun. Netw.9(1), 33–46. 10.1016/j.dcan.2022.06.003 (2023). [Google Scholar]
- 23.Manemann, S. M. et al. Predicting Alzheimer’s disease and related dementias in heart failure and atrial fibrillation. Am. J. Med.136(3), 302–307. 10.1016/j.amjmed.2022.11.010 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jothi, G., Inbarani, H. H., Azar, A. T. & Devi, K. R. Rough set theory with Jaya optimization for acute lymphoblastic leukemia classification. Neural Comput. Appl.31(9), 5175–5194. 10.1007/s00521-018-3359-7 (2019). [Google Scholar]
- 25.Kim, H. E. et al. Transfer learning for medical image classification: A literature review. BMC Med. Imaging22(1), 69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emary, E., Zawbaa, H. M., Hassanien, A. E., Schaefer, G. & Azar, A. T. Retinal vessel segmentation based on possibilistic fuzzy c-means clustering optimised with cuckoo search. In Neural Networks (IJCNN), 2014 International Joint Conference On 1792–1796 (IEEE, 2014)
- 27.Emary, E., Zawbaa, H. M., Hassanien, A. E., Schaefer, G. & Azar, A. T. Retinal blood vessel segmentation using bee colony optimisation and pattern search. In Neural Networks (IJCNN), 2014 International Joint Conference On 1001–1006 (IEEE, 2014)
- 28.Amiri, Z., Heidari, A., Navimipour, N. J., Unal, M. & Mousavi, A. Adventures in data analysis: A systematic review of deep learning techniques for pattern recognition in cyber-physical-social systems. Multimed. Tools Appl.83, 22909–22973. 10.1007/s11042-023-16382-x (2023).
- 29.Jothi, G., Inbarani, H. H. & Azar, A. T. Hybrid tolerance rough set: PSO based supervised feature selection for digital mammogram images. Int. J. Fuzzy Syst. Appl. (IJFSA)3(4), 15–30 (2013). [Google Scholar]
- 30.Anter, A. M., Azar, A. T., Hassanien, A. E., El-Bendary, N. & ElSoud, M. A. Automatic computer aided segmentation for liver and hepatic lesions using hybrid segmentations techniques. In Federated Conference on Computer Science and Information Systems (FedCSIS 2013) (IEEE, 2013) (submitted, 2013).
- 31.Banu, P. K. N., Azar, A. T. & Inbarani, H. H. Fuzzy firefly clustering for tumour and cancer analysis. Int. J. Model. Ident. Control27(2), 92–103. 10.1504/IJMIC.2017.082941 (2017). [Google Scholar]
- 32.Azar, A. T., Banu, P. & Inbarani, H. H. PSORR-an unsupervised feature selection technique for fetal heart rate. In Modelling, Identification & Control (ICMIC), 2013 Proceedings of International Conference On 60–65 (IEEE, 2013).
- 33.Ahmad, S., Shakeel, I., Mehfuz, S. & Ahmad, J. Deep learning models for cloud, edge, fog, and IoT computing paradigms: Survey, recent advances, and future directions. Comput. Sci. Rev.49, 100568. 10.1016/j.cosrev.2023.100568 (2023). [Google Scholar]
- 34.Prasad, V. K. & Bhavsar, M. D. SLAMMP framework for cloud resource management and its impact on healthcare computational techniques. Int. J. E-Health Med. Commun. (IJEHMC)12(2), 1–31 (2021). [Google Scholar]
- 35.Uthiramoorthy, A., Bhardwaj, A., Singh, J., Pant, K., Tiwari, M. & Gonzáles, J. L. A. A comprehensive review on data mining techniques in managing the medical data cloud and its security constraints with the maintained of the communication networks. In 2023 International Conference on Artificial Intelligence and Smart Communication (AISC) 618–623 (2023). 10.1109/AISC56616.2023.10085161.
- 36.Hassan, A. et al. (eds) Lightweight Digital Trust Architectures in the Internet of Medical Things (IoMT) 1–448 (IGI Global, Hershey, 2024). 10.4018/979-8-3693-2109-6. [Google Scholar]
- 37.Nancy, A. A., Ravindran, D., Raj Vincent, P. D., Srinivasan, K. & Gutierrez Reina, D. IoT-cloud-based smart healthcare monitoring system for heart disease prediction via deep learning. Electronics11(15), 2292 (2022). [Google Scholar]
- 38.Ahmad, I. & Shin, S. A perceptual encryption-based image communication system for deep learning-based tuberculosis diagnosis using healthcare cloud services. Electronics11(16), 2514 (2022). [Google Scholar]
- 39.Illakiya, T. & Karthik, R. A dimension centric proximate attention network and Swin transformer for age-based classification of mild cognitive impairment from brain MRI. IEEE Access11, 128018–128031. 10.1109/ACCESS.2023.3332122 (2023). [Google Scholar]
- 40.Gupta, L. et al. Cybersecurity of multi-cloud healthcare systems: A hierarchical deep learning approach. Appl. Soft Comput.118, 108439 (2022). [Google Scholar]
- 41.Qamar, S. Healthcare data analysis by feature extraction and classification using deep learning with cloud based cyber security. Comput. Electr. Eng.104, 108406 (2022). [Google Scholar]
- 42.Simeone, A., Caggiano, A., Boun, L. & Grant, R. Cloud-based platform for intelligent healthcare monitoring and risk prevention in hazardous manufacturing contexts. Procedia CIRP99, 50–56 (2021). [Google Scholar]
- 43.Prasad, V. K., Bhavsar, M. D. & Tanwar, S. Influence of montoring: Fog and edge computing. Scalable Comput. Pract. Exp.20(2), 365–376 (2019). [Google Scholar]
- 44.Cotroneo, D., De Simone, L., Liguori, P. & Natella, R. Enhancing the analysis of software failures in cloud computing systems with deep learning. J. Syst. Softw.181, 111043 (2021). [Google Scholar]
- 45.Motwani, A., Shukla, P. K. & Pawar, M. Novel framework based on deep learning and cloud analytics for smart patient monitoring and recommendation (SPMR). J. Ambient Intell. Humaniz. Comput.14, 5565–5580. 10.1007/s12652-020-02790-6 (2021).
- 46.Hossain, M. S. & Muhammad, G. Deep learning based pathology detection for smart connected healthcare. IEEE Netw.34(6), 120–125 (2020). [Google Scholar]
- 47.Ghaffar, Z., Shah, P. M., Khan, H., Zaidi, S. F. A., Gani, A., Khan, I. A., Shah, M. A. & ul Islam, S. Comparative analysis of state-of-the-art deep learning models for detecting COVID-19 lung infection from chest X-ray images. arXiv:2208.01637 (2022).
- 48.Illakiya, T. & Karthik, R. Automatic detection of Alzheimer’s disease using deep learning models and neuro-imaging: Current trends and future perspectives. Neuroinformatics21(2), 339–364 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Kang, W., Lin, L., Sun, S. & Wu, S. Three-round learning strategy based on 3D deep convolutional GANs for Alzheimer’s disease staging. Sci. Rep.13(1), 5750 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, S., He, L., Seo, J. & Lin, M. Concurrent healthcare data processing and storage framework using deep-learning in distributed cloud computing environment. IEEE Trans. Ind. Inf.17(4), 2794–2801 (2020). [Google Scholar]
- 51.Prasad, V. K., Mehta, H., Gajre, P., Sutaria, V. & Bhavsar, M. Capacity planning through monitoring of context aware tasks at IaaS level of cloud computing. In Future Internet Technologies and Trends: First International Conference, ICFITT 2017, Surat, India, August 31-September 2, 2017, Proceedings 1, 66–74 (Springer, 2018).
- 52.Tuli, S. et al. HealthFog: An ensemble deep learning based smart healthcare system for automatic diagnosis of heart diseases in integrated IoT and fog computing environments. Futur. Gener. Comput. Syst.104, 187–200 (2020). [Google Scholar]
- 53.Prasad, V. K. et al. Federated learning for the internet-of-medical-things: A survey. Mathematics11(1), 151. 10.3390/math11010151 (2023). [Google Scholar]
- 54.Illakiya, T., Ramamurthy, K., Siddharth, M. V., Mishra, R. & Udainiya, A. AHANet: Adaptive hybrid attention network for Alzheimer’s disease classification using brain magnetic resonance imaging. Bioengineering10(6), 714. 10.3390/bioengineering10060714 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta, A., Bhagat, M. & Jain, V. Blockchain-enabled healthcare monitoring system for early monkeypox detection. J. Supercomput.79, 15675–15699. 10.1007/s11227-023-05288-y (2023). [DOI] [PMC free article] [PubMed]
- 56.Akhtar, M. M., Shatat, R. S. A., Shatat, A. S. A., Hameed, S. A. & Ibrahim, Alnajdawi S. IoMT-based smart healthcare monitoring system using adaptive wavelet entropy deep feature fusion and improved RNN. Multimed. Tools Appl.82(11), 17353–17390 (2023). [Google Scholar]
- 57.Bolhasani, H., Mohseni, M. & Rahmani, A. M. Deep learning applications for IoT in health care: A systematic review. Inf. Med. Unlocked23, 100550 (2021). [Google Scholar]
- 58.Aazam, M., Zeadally, S. & Flushing, E. F. Task offloading in edge computing for machine learning-based smart healthcare. Comput. Netw.191, 108019 (2021). [Google Scholar]
- 59.Praveen, K. et al. Deep learning based intelligent and sustainable smart healthcare application in cloud-centric IoT. Comput. Mater. Contin.66(2), 1987–2003 (2021). [Google Scholar]
- 60.Shah, P. M. et al. 2D-CNN based segmentation of ischemic stroke lesions in MRI scans. In Advances in Computational Collective Intelligence (eds Hernes, M. et al.) 276–286 (Springer, Cham, 2020). [Google Scholar]
- 61.Durga, S., Nag, R. & Daniel, E. Survey on machine learning and deep learning algorithms used in internet of things (IoT) healthcare. In 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC), 1018–1022 (IEEE, 2019).
- 62.Van Dyck, C. H. et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med.388(1), 9–21 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Hussain, S. S. et al. Classification of Parkinson’s disease in patch-based MRI of substantia nigra. Diagnostics13(17), 2827. 10.3390/diagnostics13172827 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shah, P. M., Zeb, A., Shafi, U., Zaidi, S. F. A. & Shah, M. A. Detection of Parkinson disease in brain MRI using convolutional neural network. In 2018 24th International Conference on Automation and Computing (ICAC), Newcastle Upon Tyne, UK, 1–6 (2018). 10.23919/IConAC.2018.8749023.
- 65.Dar, M. N., Akram, M. U., Yuvaraj, R., Khawaja, S. G. & Murugappan, M. EEG-based emotion charting for Parkinson’s disease patients using convolutional recurrent neural networks and cross dataset learning. Comput. Biol. Med.144, 105327 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Sousa, A. et al. On the identification of potential novel therapeutic targets for spinocerebellar ataxia type 1 (sca1) neurodegenerative disease using evoppi3. J. Integr. Bioinform.20(2), 20220056. 10.1515/jib-2022-0056 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhan, X. et al. Translational models of mild traumatic brain injury tissue biomechanics. Curr. Opin. Biomed. Eng.24, 100422. 10.1016/j.cobme.2022.100422 (2022). [Google Scholar]
- 68.Han, J. et al. Classifying neovascular age-related macular degeneration with a deep convolutional neural network based on optical coherence tomography images. Sci. Rep.12(1), 2232 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prasad, V. K., Nimavat, V., Trivedi, K. & Bhavsar, M. Utilizing deep learning methodology to classify diabetic retinopathy. In Inventive Communication and Computational Technologies (eds Ranganathan, G. et al.) 679–692 (Springer, Singapore, 2023). [Google Scholar]
- 70.Nagpal, D., Panda, S. N., Malarvel, M., Pattanaik, P. A. & Zubair Khan, M. A review of diabetic retinopathy: Datasets, approaches, evaluation metrics and future trends. J. King Saud Univ. Comput. Inf. Sci.34(9), 7138–7152. 10.1016/j.jksuci.2021.06.006 (2022). [Google Scholar]
- 71.Singh, L. K., Pooja, Garg H. & Khanna, M. Deep learning system applicability for rapid glaucoma prediction from fundus images across various data sets. Evol. Syst.13(6), 807–836 (2022). [Google Scholar]
- 72.Fang, H. et al. PALM: Open fundus photograph dataset with pathologic myopia recognition and anatomical structure annotation. Sci. Data11(1), 99 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan, H., Shah, P. M., Shah, M. A., ul Islam, S. & Rodrigues, J. J. P. C. Cascading handcrafted features and convolutional neural network for IoT-enabled brain tumor segmentation. Comput. Commun.153, 196–207. 10.1016/j.comcom.2020.01.013 (2020). [Google Scholar]
- 74.Orset, T., Royo, J., Santin, M. D., Pouget, P. & de Thiebaut, Schotten M. A new open, high-resolution, multishell, diffusion-weighted imaging dataset of the living squirrel monkey. Sci. Data10(1), 224 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKay, N. S. et al. Positron emission tomography and magnetic resonance imaging methods and datasets within the dominantly inherited alzheimer network (dian). Nat. Neurosci.26(8), 1449–1460 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanaat, A., Shiri, I., Ferdowsi, S., Arabi, H. & Zaidi, H. Robust-Deep: A method for increasing brain imaging datasets to improve deep learning models’ performance and robustness. J. Digit. Imaging35(3), 469–481 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar, P. S., Sakthivel, V., Raju, M. & Satya, P. Brain tumor segmentation of the FLAIR MRI images using novel ResUnet. Biomed. Signal Process. Control82, 104586 (2023). [Google Scholar]
- 78.Khan, M. et al. IoMT-enabled computer-aided diagnosis of pulmonary embolism from computed tomography scans using deep learning. Sensors23(3), 1471. 10.3390/s23031471 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdulkareem, K. H. et al. Automated system for identifying COVID-19 infections in computed tomography images using deep learning models. J. Healthc. Eng.2022, 5329014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Sahu, A., Das, P. K. & Meher, S. High accuracy hybrid CNN classifiers for breast cancer detection using mammogram and ultrasound datasets. Biomed. Signal Process. Control80, 104292 (2023). [Google Scholar]
- 81.Tyystjärvi, T., Virkkunen, I., Fridolf, P., Rosell, A. & Barsoum, Z. Automated defect detection in digital radiography of aerospace welds using deep learning. Weld. World66(4), 643–671 (2022). [Google Scholar]
- 82.Long, T. et al. Constructing a digital twin of the birdcage coil in an MR scanner by map matching: For radio frequency heating evaluation of implantable medical devices. IEEE Trans. Instrum. Meas.71, 1–9. 10.1109/TIM.2022.3212552 (2022). [Google Scholar]
- 83.Zhang, Y. et al. An end-to-end multimodal 3D CNN framework with multi-level features for the prediction of mild cognitive impairment. Knowl.-Based Syst.281, 111064. 10.1016/j.knosys.2023.111064 (2023). [Google Scholar]
- 84.Zeng, A. et al. ImageCAS: A large-scale dataset and benchmark for coronary artery segmentation based on computed tomography angiography images. Comput. Med. Imaging Graph.109, 102287 (2023). [DOI] [PubMed] [Google Scholar]
- 85.Bose, P. et al. Multimodal deep learning methods on image and textual data to predict radiotherapy structure names. BioMedInformatics3(3), 493–513 (2023). [Google Scholar]
- 86.Kwiecinski, J. et al. Latent coronary plaque morphology from computed tomography angiography, molecular disease activity on positron emission tomography, and clinical outcomes. Arterioscler. Thromb. Vasc. Biol.43, e279–e290 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang, J. et al. A benchmark dataset of endoscopic images and novel deep learning method to detect intestinal metaplasia and gastritis atrophy. IEEE J. Biomed. Health Inform.27(1), 7–16. 10.1109/JBHI.2022.3217944 (2023). [DOI] [PubMed] [Google Scholar]
- 88.Bilodeau, A. et al. Microscopy analysis neural network to solve detection, enumeration and segmentation from image-level annotations. Nat. Mach. Intell.4(5), 455–466 (2022). [Google Scholar]
- 89.Conrad, R. & Narayan, K. Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model trained on a diverse dataset. Cell Syst.14(1), 58–71 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mueller, S. G. et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. North Am.15(4), 869–877. 10.1016/j.nic.2005.09.008 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aisen, P. S. et al. Alzheimer’s disease neuroimaging initiative 2 clinical core: Progress and plans. Alzheimer’s Dement.11(7), 734–739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marcus, D. S. et al. Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci.19(9), 1498–1507 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Marcus, D. S., Fotenos, A. F., Csernansky, J. G., Morris, J. C. & Buckner, R. L. Open access series of imaging studies: Longitudinal MRI data in nondemented and demented older adults. J. Cogn. Neurosci.22(12), 2677–2684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.LaMontagne, P. J., Benzinger, T. L., Morris, J. C., Keefe, S., Hornbeck, R., Xiong, C., Grant, E., Hassenstab, J., Moulder, K., Vlassenko, A. G., et al. OASIS-3: Longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. MedRxiv, 2019–12 (2019).
- 95.Marinescu, R. V., Oxtoby, N. P., Young, A. L., Bron, E. E., Toga, A. W., Weiner, M. W., Barkhof, F., Fox, N. C., Eshaghi, A., Toni, T. et al. The Alzheimer’s disease prediction of longitudinal evolution (TADPOLE) challenge: Results after 1 year follow-up. arXiv preprint arXiv:2002.03419 (2020).
- 96.Tessa, C. et al. Central modulation of parasympathetic outflow is impaired in de novo Parkinson’s disease patients. PLoS ONE14(1), 0210324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mascalchi, M. et al. Histogram analysis of DTI-derived indices reveals pontocerebellar degeneration and its progression in SCA2. PLoS ONE13(7), 0200258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bellotti, R., Lombardi, A., Guaragnella, C., Amoroso, N., Tateo, A. & Tangaro, S. Mild traumatic brain injury outcome prediction based on both graph and K-NN methods. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Second International Workshop, BrainLes 2016, with the Challenges on BRATS, ISLES and mTOP 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 17, 2016, Revised Selected Papers 2, 271–281 (Springer, 2016).
- 99.Quellec, G., Lamard, M., Conze, P.-H., Massin, P. & Cochener, B. Automatic detection of rare pathologies in fundus photographs using few-shot learning. Med. Image Anal.61, 101660 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Orlando, J. I. et al. Refuge challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Med. Image Anal.59, 101570 (2020). [DOI] [PubMed] [Google Scholar]
- 101.Fu, H. et al. Age challenge: Angle closure glaucoma evaluation in anterior segment optical coherence tomography. Med. Image Anal.66, 101798 (2020). [DOI] [PubMed] [Google Scholar]
- 102.Baid, U., Baheti, B., Dutande, P. & Talbar, S. Detection of pathological myopia and optic disc segmentation with deep convolutional neural networks. In TENCON 2019—2019 IEEE Region 10 Conference (TENCON), 1345–1350 (2019). 10.1109/TENCON.2019.8929252.
- 103.Kermany, D., Zhang, K. & Goldbaum, M. Large dataset of labeled optical coherence tomography (OCT) and chest x-ray images. Mendeley Data3(10.17632), 170 (2018). [Google Scholar]
- 104.Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell172(5), 1122–1131 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Al Hajj, H. et al. CATARACTS: Challenge on automatic tool annotation for cataract surgery. Med. Image Anal.52, 24–41 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Chiu, S. J. et al. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed. Opt. Express6(4), 1172–1194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hemelings, R. et al. Artery-vein segmentation in fundus images using a fully convolutional network. Comput. Med. Imaging Graph.76, 101636 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Wang, J., Yang, L., Huo, Z., He, W. & Luo, J. Multi-label classification of fundus images with EfficientNet. IEEE Access8, 212499–212508. 10.1109/ACCESS.2020.3040275 (2020). [Google Scholar]
- 109.Bogunović, H. et al. Retouch: The retinal oct fluid detection and segmentation benchmark and challenge. IEEE Trans. Med. Imaging38(8), 1858–1874 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Andrearczyk, V. et al. Overview of the HECKTOR challenge at MICCAI 2020: Automatic head and neck tumor segmentation in PET/CT. In Head Neck Tumor Segmentation (eds Andrearczyk, V. et al.) 1–21 (Springer, Cham, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang, Q. et al. Thyroid nodule segmentation and classification in ultrasound images through intra-and inter-task consistent learning. Med. Image Anal.79, 102443 (2022). [DOI] [PubMed] [Google Scholar]
- 112.Aerts, H. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun.5, 4006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vallieres, M., Kay-Rivest, E., Perrin, L. J., Liem, X., Furstoss, C., Khaouam, N., Nguyen-Tan, P. F., Wang, C.-S. & Sultanem, K. Data from head-neck-PET-CT The Cancer Imaging Archive. Published (2017). [DOI] [PMC free article] [PubMed]
- 114.Wu, H., Liu, J., Wang, W., Wen, Z. & Qin, J. Region-aware global context modeling for automatic nerve segmentation from ultrasound images. In Proceedings of the AAAI Conference on Artificial Intelligence, Vol. 35(4), 2907–2915. 10.1609/aaai.v35i4.16397 (2021).
- 115.Besnard, C. et al. 3d analysis of enamel demineralisation in human dental caries using high-resolution, large field of view synchrotron x-ray micro-computed tomography. Mater. Today Commun.27, 102418. 10.1016/j.mtcomm.2021.102418 (2021). [Google Scholar]
- 116.Hameeteman, K. et al. Evaluation framework for carotid bifurcation lumen segmentation and stenosis grading. Med. Image Anal.15(4), 477–488 (2011). [DOI] [PubMed] [Google Scholar]
- 117.Han, D. et al. Deep learning analysis in coronary computed tomographic angiography imaging for the assessment of patients with coronary artery stenosis. Comput. Methods Programs Biomed.196, 105651. 10.1016/j.cmpb.2020.105651 (2020). [DOI] [PubMed] [Google Scholar]
- 118.Rister, B., Shivakumar, K., Nobashi, T. & Rubin, D. L. CT-ORG: CT volumes with multiple organ segmentations [dataset]. Cancer Imaging Arch. (2019).
- 119.Simpson, A. L., Antonelli, M., Bakas, S., Bilello, M., Farahani, K., Van Ginneken, B., Kopp-Schneider, A., Landman, B. A., Litjens, G., Menze, B. et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019).
- 120.Yang, J. et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med. Phys.45(10), 4568–4581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Newitt, D. & Hylton, N. Single site breast DCE-MRI data and segmentations from patients undergoing neoadjuvant chemotherapy. Cancer Imaging Arch., 2 (2016).
- 122.Bernard, O., et al. Challenge on endocardial three-dimensional ultrasound segmentation (CETUS). Proc. MICCAI Chall. Echocardiogr. Three Dimens. Ultrasound Segmentation (CETUS), 1–8 (2014).
- 123.Yang, J., Sharp, G., Veeraraghavan, H., van Elmpt, W., Dekker, A., Lustberg, T. & Gooding, M. Data from lung CT segmentation challenge. Cancer Imaging Arch. (2017).
- 124.Jimenez-del-Toro, O. et al. Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: Visceral anatomy benchmarks. IEEE Trans. Med. Imaging35(11), 2459–2475 (2016). [DOI] [PubMed] [Google Scholar]
- 125.Prasad, V. K., Tanwar, S. & Bhavsar, M. C2B-SCHMS: Cloud computing and bots security for COVID-19 data and healthcare management systems. In Proceedings of Second International Conference on Computing, Communications, and Cyber-Security: IC4S 2020, 787–797 (Springer, 2021).
- 126.Bhushan, B. et al. Towards a secure and sustainable internet of medical things (IoMT): Requirements, design challenges, security techniques, and future trends. Sustainability15(7), 6177. 10.3390/su15076177 (2023). [Google Scholar]
- 127.Vadiati, M. et al. A sustainable trend in COVID-19 research: An environmental perspective. Front. Environ. Sci.11, 1104679. 10.3389/fenvs.2023.1104679 (2023). [Google Scholar]
- 128.Shah, P. M., Ullah, F., Shah, D., Gani, A., Maple, C., Wang, Y., Shahid, Abrar, M. & Islam, S. U. Deep GRU-CNN model for COVID-19 detection from chest X-rays data. IEEE Access, 10, 35094–35105 (2022). 10.1109/ACCESS.2021.3077592. [DOI] [PMC free article] [PubMed]
- 129.Yang, X., He, X., Zhao, J., Zhang, Y., Zhang, S. & Xie, P. COVID-CT-dataset: A CT scan dataset about COVID-19. arXiv preprint arXiv:2003.13865 (2020).
- 130.Tartaglione, E., Barbano, C. A., Berzovini, C., Calandri, M. & Grangetto, M. Unveiling COVID-19 from chest x-ray with deep learning: A hurdles race with small data. Int. J. Environ. Res. Public Health17(18), 6933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Born, J., Brändle, G., Cossio, M., Disdier, M., Goulet, J., Roulin, J. & Wiedemann, N. POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS). arXiv preprint arXiv:2004.12084 (2020).
- 132.Desai, S. et al. Chest imaging representing a COVID-19 positive rural us population. Sci. Data7(1), 414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Buda, M., Saha, A., Walsh, R., Ghate, S., Li, N., Święcicki, A., Lo, J. Y. & Mazurowski, M. A. Detection of masses and architectural distortions in digital breast tomosynthesis: a publicly available dataset of 5060 patients and a deep learning model. arXiv preprint arXiv:2011.07995 (2020). [DOI] [PMC free article] [PubMed]
- 134.Fiorentino, M. C., Villani, F. P., Di Cosmo, M., Frontoni, E. & Moccia, S. A review on deep-learning algorithms for fetal ultrasound-image analysis. Med. Image Anal.83, 102629. 10.1016/j.media.2022.102629 (2023). [DOI] [PubMed] [Google Scholar]
- 135.Wee, L., Aerts, H., Kalendralis, P. & Dekker, A. Data from NSCLC-Radiomics-Interobserver1[data set]. Cancer Imaging Arch., 10 (2019).
- 136.Johnson, A. E. et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data6(1), 317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He, X., Zhang, Y., Mou, L., Xing, E. & Xie, P. PathVQA: 30000+ questions for medical visual question answering. arXiv preprint arXiv:2003.10286 (2020).
- 138.Kim, K. et al. PAIP 2020: Microsatellite instability prediction in colorectal cancer. Med. Image Anal.89, 102886. 10.1016/j.media.2023.102886 (2023). [DOI] [PubMed] [Google Scholar]
- 139.Li, J., Zhu, G., Hua, C., Feng, M., BasheerBennamoun, Li, P., Lu, X., Song, J., Shen, P., Xu, X., Mei, L., Zhang, L., Shah, S. A. A. & Bennamoun, M. A Systematic Collection of Medical Image Datasets for Deep Learning (2021).
- 140.Martel, A., Nofech-Mozes, S., Salama, S., Akbar, S. & Peikari, M. Assessment of residual breast cancer cellularity after neoadjuvant chemotherapy using digital pathology [data set]. Cancer Imaging Arch. (2019).
- 141.Borovec, J. et al. ANHIR: Automatic non-rigid histological image registration challenge. IEEE Trans. Med. Imaging39(10), 3042–3052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim, Y. J. et al. PAIP 2019: Liver cancer segmentation challenge. Med. Image Anal.67, 101854. 10.1016/j.media.2020.101854 (2021). [DOI] [PubMed] [Google Scholar]
- 143.Caicedo, J. C. et al. Nucleus segmentation across imaging experiments: The 2018 data science bowl. Nat. Methods16(12), 1247–1253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamida, A. B. et al. Deep learning for colon cancer histopathological images analysis. Comput. Biol. Med.136, 104730 (2021). [DOI] [PubMed] [Google Scholar]
- 145.Litjens, G. et al. 1399 H &E-stained sentinel lymph node sections of breast cancer patients: The CAMELYON dataset. GigaScience7(6), 065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu, Y. et al. Recent advances of deep learning for computational histopathology: Principles and applications. Cancers14(5), 1199. 10.3390/cancers14051199 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tareef, A. et al. Automatic segmentation of overlapping cervical smear cells based on local distinctive features and guided shape deformation. Neurocomputing221, 94–107. 10.1016/j.neucom.2016.09.070 (2017). [Google Scholar]
- 148.Roux, L. Mitosis atypia 14 grand challenge. Mitosis atypia 14 grand challenge (2014).
- 149.Meijering, E., Dzyubachyk, O. & Smal, I. Chapter nine—Methods for cell and particle tracking. In Imaging and Spectroscopic Analysis of Living Cells Methods in Enzymology Vol. 504 (ed. Conn, P. M.) 183–200 (Academic Press, Cambridge, 2012). 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 150.Gupta, A. et al. SegPC-2021: A challenge dataset on segmentation of multiple myeloma plasma cells from microscopic images. Med. Image Anal.83, 102677. 10.1016/j.media.2022.102677 (2023). [DOI] [PubMed] [Google Scholar]
- 151.Wei, D., Lin, Z., Franco-Barranco, D., Wendt, N., Liu, X., Yin, W., Huang, X., Gupta, A., Jang, W.-D., Wang, X. et al. MitoEM dataset: Large-scale 3D mitochondria instance segmentation from EM images. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part V 23, 66–76 (Springer, 2020). [DOI] [PMC free article] [PubMed]
- 152.Manku, R. R., Sharma, A. & Panchbhai, A. Malaria Detection and Classificaiton (2020).
- 153.Jiao, Y., van der Laak, J., Albarqouni, S., Li, Z., Tan, T., Bhalerao, A., Ma, J., Sun, J., Pocock, J., Pluim, J. P. W., Koohbanani, N. A., Bashir, R. M. S., Raza, S. E. A., Liu, S., Graham, S., Wetstein, S., Khurram, S. A., Watson, T., Rajpoot, N., Veta, M. & Ciompi, F. LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset (2023). [DOI] [PubMed]
- 154.Gupta, A. & Gupta, R. SN-AM dataset: White blood cancer dataset of B-ALL and MM for stain normalization. Cancer Imaging Arch. (2019).
- 155.Tran, T., Kwon, O.-H., Kwon, K.-R., Lee, S.-H., Kang, K.-W. Blood cell images segmentation using deep learning semantic segmentation. In 2018 IEEE International Conference on Electronics and Communication Engineering (ICECE), 13–16 (2018). 10.1109/ICECOME.2018.8644754.
- 156.Pan, Y., Liu, M., Xia, Y. & Shen, D. Neighborhood-correction algorithm for classification of normal and malignant cells. In ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging (eds Gupta, A. & Gupta, R.) 73–82 (Springer, Singapore, 2019). [Google Scholar]
- 157.Knapp, P., Weishuhn, L., Pizzimenti, N. & Markel, D. C. Risk factors for manipulation under anaesthesia after total knee arthroplasty. Bone Joint J.102–B(6 Supple A), 66–72. 10.1302/0301-620X.102B6.BJJ-2019-1580.R1 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Jin, L. et al. Deep-learning-assisted detection and segmentation of rib fractures from CT scans: Development and validation of fracnet. EBioMedicine62, 103106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tahtabasi, M. et al. The prognostic value of vertebral bone density on chest CT in hospitalized COVID-19 patients. J. Clin. Densitom.24(4), 506–515. 10.1016/j.jocd.2021.07.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yorke, A., McDonald, G., Solis, D. & Guerrero, T. Pelvic reference data. Cancer Imaging Arch. (2019).
- 161.Zhou, L. & Zhang, J. Advances of area-wise distributed monitoring using long gauge sensing techniques. Sensors19(5), 1038. 10.3390/s19051038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rajpurkar, P., Irvin, J., Bagul, A., Ding, D., Duan, T., Mehta, H., Yang, B., Zhu, K., Laird, D., Ball, R. L. et al. MURA: Large dataset for abnormality detection in musculoskeletal radiographs. arXiv preprint arXiv:1712.06957 (2017).
- 163.Schubl, S. D. et al. Temporal bone fracture: Evaluation in the era of modern computed tomography. Injury47(9), 1893–1897. 10.1016/j.injury.2016.06.026 (2016). [DOI] [PubMed] [Google Scholar]
- 164.Song, Y., Cai, W., Zhang, F., Huang, H., Zhou, Y. & Feng, D. D. Bone texture characterization with fisher encoding of local descriptors. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) 5–8 (IEEE, 2015).
- 165.Cassidy, B. et al. The DFUC 2020 dataset: Analysis towards diabetic foot ulcer detection. touchREVIEWS Endocrinol.17(1), 5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Codella, N., Rotemberg, V., Tschandl, P., Celebi, M. E., Dusza, S., Gutman, D., Helba, B., Kalloo, A., Liopyris, K., Marchetti, M. et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC). arXiv preprint arXiv:1902.03368 (2019).
- 167.Mueller, M. et al. The markerless lung target tracking AAPM grand challenge (MATCH) results. Med. Phys.49(2), 1161–1180 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ger, R. B. et al. Synthetic head and neck and phantom images for determining deformable image registration accuracy in magnetic resonance imaging. Med. Phys.45(9), 4315–4321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Latifi, K., Ullah, G., Gillies, R. & Moros, E. Credence cartridge radiomics phantom CT scans with controlled scanning approach. TCIA (2018).
- 170.Farahani, K. et al. Computational challenges and collaborative projects in the NCI quantitative imaging network. Tomography2(4), 242–249. 10.18383/j.tom.2016.00265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Allan, M., Mcleod, J., Wang, C., Rosenthal, J. C., Hu, Z., Gard, N., Eisert, P., Fu, K. X., Zeffiro, T., Xia, W., Zhu, Z., Luo, H., Jia, F., Zhang, X., Li, X., Sharan, L., Kurmann, T., Schmid, S., Sznitman, R., Psychogyios, D., Azizian, M., Stoyanov, D., Maier-Hein, L. & Speidel, S. Stereo Correspondence and Reconstruction of Endoscopic Data Challenge. arXiv:2101.01133 (2021).
- 172.Acciai, L., Soda, P. & Iannello, G. Automatic neuron tracing using a locally tunable approach. In 2016 IEEE 29th International Symposium on Computer-Based Medical Systems (CBMS) 130–135 (2016). 10.1109/CBMS.2016.48.
- 173.Arganda-Carreras, I., Seung, S., Vishwanathan, A. & Berger, D. SNEMI 3D: 3D Segmentation of Neurites in EM Images (2013).
- 174.Dalca, A., Hu, Y., Vercauteren, T., Heinrich, M., Hansen, L., Modat, M., De Vos, B., Xiao, Y., Rivaz, H., Chabanas, M., et al.Learn2Reg-the challenge. Zenodo (2020).
- 175.Fathi, S. et al. A deep learning-based ensemble method for early diagnosis of Alzheimer’s disease using MRI images. Neuroinformatics22(1), 89–105 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Saraswat, D. et al. Secure 5G-assisted UAV access scheme in IoBT for region demarcation and surveillance operations. IEEE Commun. Standards Mag.6(1), 58–66. 10.1109/MCOMSTD.0001.2100057 (2022). [Google Scholar]
- 177.Shah, P. M. et al. DC-GAN-based synthetic X-ray images augmentation for increasing the performance of EfficientNet for COVID-19 detection. Expert Syst.39(3), 12823. 10.1111/exsy.12823 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang, Q., Xiao, J., Tian, C., Chun-Wei Lin, J. & Zhang, S. A robust deformed convolutional neural network (CNN) for image denoising. CAAI Trans. Intell. Technol.8(2), 331–342 (2023). [Google Scholar]
- 179.Nirthika, R., Manivannan, S., Ramanan, A. & Wang, R. Pooling in convolutional neural networks for medical image analysis: A survey and an empirical study. Neural Comput. Appl.34(7), 5321–5347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Faisal, M., Leu, J.-S., Avian, C., Prakosa, S. W. & Köppen, M. DFNet: Dense fusion convolution neural network for plant leaf disease classification. Agron. J.116, 826–838 (2023). [Google Scholar]
- 181.Iparraguirre-Villanueva, O. et al. Text prediction recurrent neural networks using long short-term memory-dropout. Indones. J. Electr. Eng. Comput. Sci29, 1758–1768 (2023). [Google Scholar]
- 182.Salehi, A. W. et al. A study of CNN and transfer learning in medical imaging: Advantages, challenges, future scope. Sustainability15(7), 5930. 10.3390/su15075930 (2023). [Google Scholar]
- 183.Shah, S. R. et al. Comparing inception V3, VGG 16, VGG 19, CNN, and ResNet 50: A case study on early detection of a rice disease. Agronomy13(6), 1633. 10.3390/agronomy13061633 (2023). [Google Scholar]
- 184.Truong, T. X. et al. A new approach based on TensorFlow deep neural networks with ADAM optimizer and GIS for spatial prediction of forest fire danger in tropical areas. Remote Sens.15(14), 3458. 10.3390/rs15143458 (2023). [Google Scholar]
- 185.Mohamad Almustafa, K., Kumar Sharma, A. & Bhardwaj, S. STARC: Deep learning algorithms’ modelling for structured analysis of retina classification. Biomed. Signal Process. Control80, 104357. 10.1016/j.bspc.2022.104357 (2023). [Google Scholar]
- 186.Luong, H. H. et al. Fine-tuning VGG16 for Alzheimer’s disease diagnosis. In Complex, Intelligent and Software Intensive Systems (ed. Barolli, L.) 68–79 (Springer, Cham, 2023). [Google Scholar]
- 187.Umamaheswari, S., Vishal, N. R., Pragadesh, N. R. & Lavanya, S. Performance analysis of resnet50 architecture based pest detection system. In 2023 9th International Conference on Advanced Computing and Communication Systems (ICACCS), Vol. 1, 578–583 (2023). 10.1109/ICACCS57279.2023.10112802
- 188.Ahmed, M., Afreen, N., Ahmed, M., Sameer, M. & Ahamed, J. An inception V3 approach for malware classification using machine learning and transfer learning. Int. J. Intell. Netw.4, 11–18. 10.1016/j.ijin.2022.11.005 (2023). [Google Scholar]
- 189.Thayumanasamy, I. & Ramamurthy, K. Performance analysis of machine learning and deep learning models for classification of Alzheimer’s disease from brain MRI. Traitement du Signal39(6), 1961–1970. 10.18280/ts.390608 (2022). [Google Scholar]
- 190.Hu, Z., Li, Y., Wang, Z., Zhang, S. & Hou, W. Conv-Swinformer: Integration of CNN and shift window attention for Alzheimer’s disease classification. Comput. Biol. Med.164, 107304. 10.1016/j.compbiomed.2023.107304 (2023). [DOI] [PubMed] [Google Scholar]
- 191.Mao, T. & Zhou, D.-X. Rates of approximation by ReLU shallow neural networks. J. Complex.79, 101784 (2023). [Google Scholar]
- 192.Sreenivasu, S. et al. Dense convolutional neural network for detection of cancer from CT images. BioMed Res. Int.222, 1293548 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 193.Dan, Z., Zhao, Y., Bi, X., Wu, L. & Ji, Q. Multi-task transformer with adaptive cross-entropy loss for multi-dialect speech recognition. Entropy24(10), 1429. 10.3390/e24101429 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Obayya, M. et al. Artificial intelligence driven biomedical image classification for robust rheumatoid arthritis classification. Biomedicines10(11), 2714. 10.3390/biomedicines10112714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Trivedi, C. et al. A transformative shift toward blockchain-based IoT environments: Consensus, smart contracts, and future directions. Secur. Privacy6(5), 308. 10.1002/spy2.308 (2023). [Google Scholar]
- 196.Hassan, A. et al. (eds) Pioneering Smart Healthcare 5.0 with IoT, Federated Learning, and Cloud Security 1–352 (IGI Global, Hershey, 2024). 10.4018/979-8-3693-2639-8. [Google Scholar]
- 197.Gong, Z. et al. Smart urban planning: Intelligent cognitive analysis of healthcare data in cloud-based IoT. Comput. Electr. Eng.110, 108878. 10.1016/j.compeleceng.2023.108878 (2023). [Google Scholar]
- 198.Dixon, B. E. Chapter 1: introduction to health information exchange. In Health Information Exchange (ed. Dixon, B. E.) 3–20 (Academic Press, Cambridge, 2023). 10.1016/B978-0-323-90802-3.00013-7. [Google Scholar]
- 199.Ranchal, R. et al. Disrupting healthcare silos: Addressing data volume, velocity and variety with a cloud-native healthcare data ingestion service. IEEE J. Biomed. Health Inform.24(11), 3182–3188. 10.1109/JBHI.2020.3001518 (2020). [DOI] [PubMed] [Google Scholar]
- 200.Rizvi, S., Ayres, J., Pensyl, J. & Ihnat, M. Developing data protection and recovery plan for healthcare IoT domain. J. Comput. Sci. Coll.39(3), 210–225 (2023). [Google Scholar]
- 201.Surantha, N., Atmaja, P., David & Wicaksono, M. A review of wearable internet-of-things device for healthcare. Procedia Comput. Sci.179, 936–943 (2021). 10.1016/j.procs.2021.01.083. 5th International Conference on Computer Science and Computational Intelligence 2020
- 202.Desai, F. et al. HealthCloud: A system for monitoring health status of heart patients using machine learning and cloud computing. Internet Things17, 100485. 10.1016/j.iot.2021.100485 (2022). [Google Scholar]
- 203.Mbunge, E., Batani, J., Gaobotse, G. & Muchemwa, B. Virtual healthcare services and digital health technologies deployed during coronavirus disease 2019 (COVID-19) pandemic in South Africa: A systematic review. Global Health J.6(2), 102–113. 10.1016/j.glohj.2022.03.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.López-Martínez, F., Núñez-Valdez, E. R., García-Díaz, V. & Bursac, Z. A case study for a big data and machine learning platform to improve medical decision support in population health management. Algorithms13(4), 102. 10.3390/a13040102 (2020). [Google Scholar]
- 205.Thomasian, N. M. & Adashi, E. Y. Cybersecurity in the internet of medical things. Health Policy Technol.10(3), 100549. 10.1016/j.hlpt.2021.100549 (2021). [Google Scholar]
- 206.Singh, A., Joshi, S. & Domb, M. Embedded conversational AI, chatbots, and NLP to improve healthcare administration and practices. In 2023 2nd International Conference on Automation, Computing and Renewable Systems (ICACRS) 38–45 (2023). 10.1109/ICACRS58579.2023.10404985
- 207.Furstenau, L. B. et al. Resilience capabilities of healthcare supply chain and supportive digital technologies. Technol. Soc.71, 102095. 10.1016/j.techsoc.2022.102095 (2022). [Google Scholar]
- 208.Dang, V. A., Vu Khanh, Q., Nguyen, V.-H., Nguyen, T. & Nguyen, D. C. Intelligent healthcare: Integration of emerging technologies and internet of things for humanity. Sensors23(9), 4200. 10.3390/s23094200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Islam, J. & Zhang, Y. Brain MRI analysis for Alzheimer’s disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inform.5, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sarraf, S. & Tofighi, G. Classification of Alzheimer’s Disease Structural MRI Data by Deep Learning Convolutional Neural Networks (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.