Abstract
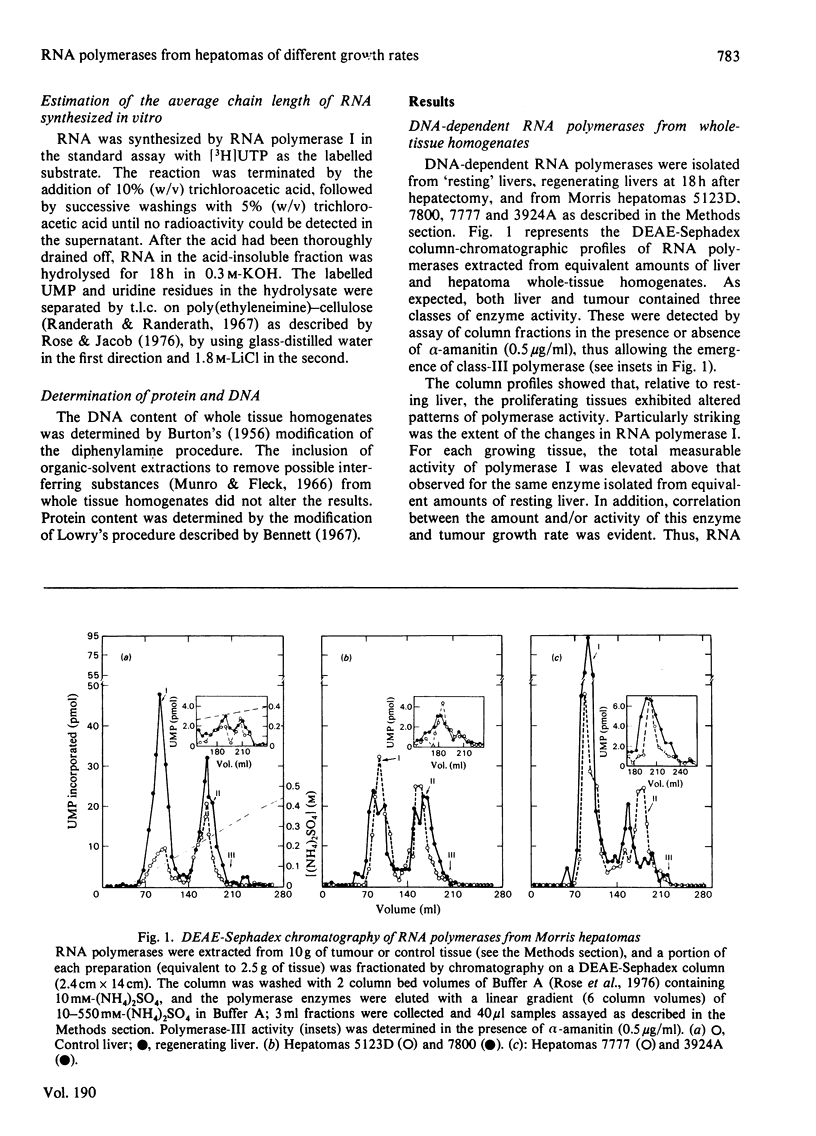
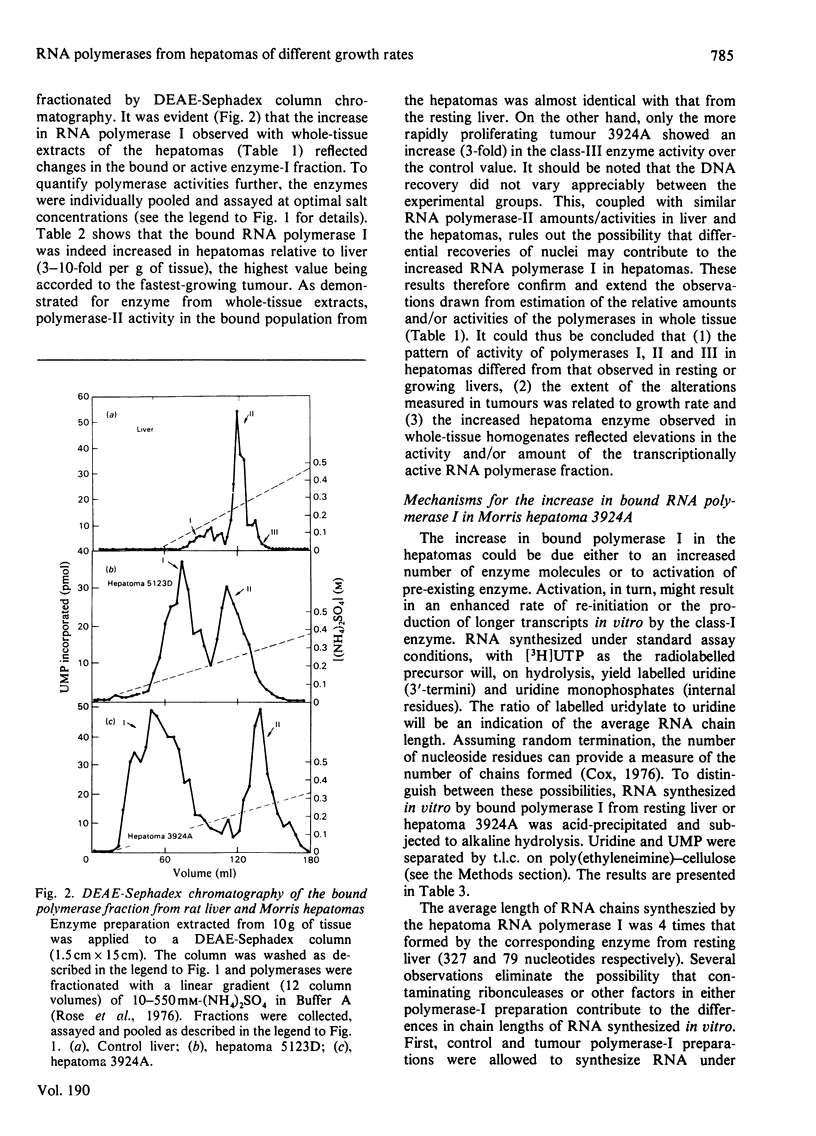
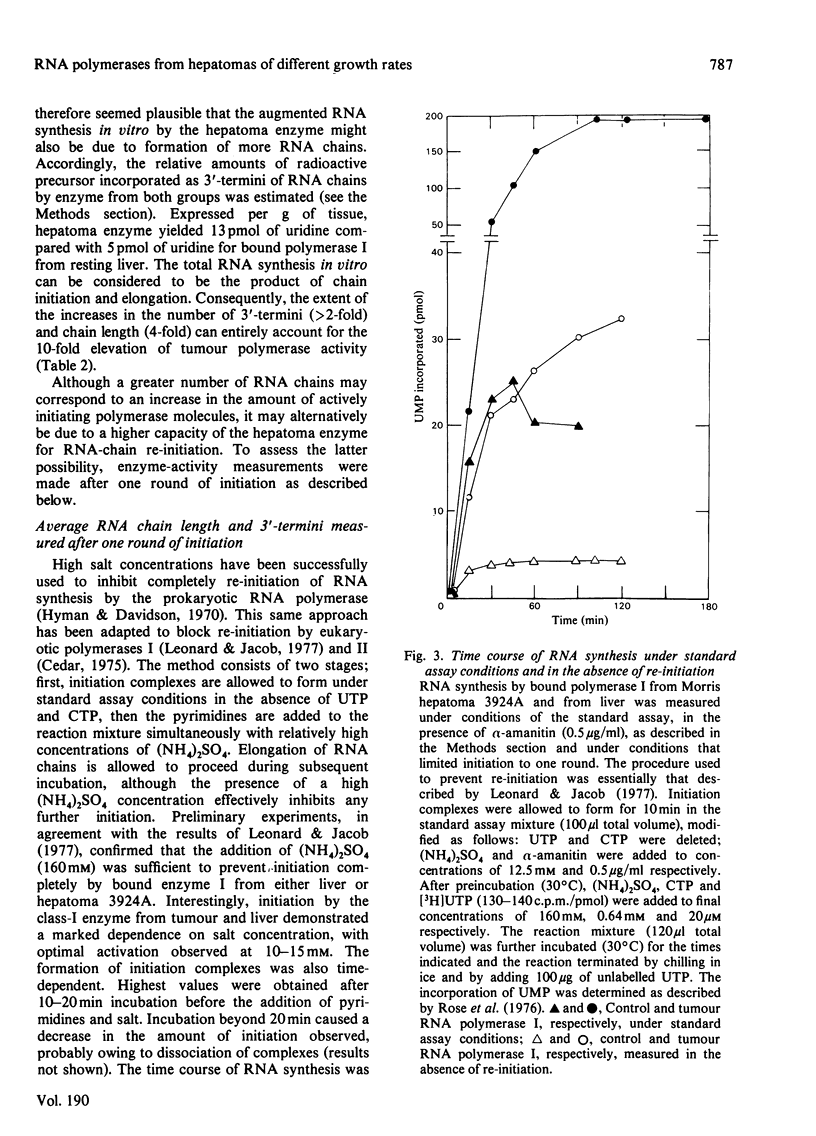
The amount and/or activity of DNA-dependent RNA polymerase I, Ii and III from resting liver, regenerating liver and a series of Morris hepatomas (5123D, 7800, 7777, 3924A) were determined after extraction of the enzymes from whole tissue homogenates and subsequent fractionation by DEAE-Sephadex column chromatography. When compared with resting liver, the tumours exhibited a characteristic enzyme pattern in which polymerase I, but not II, was increased. The increase in RNA polymerase I was proportional to the tumour growth rates. Alterations in polymerase III were confined to the most rapidly proliferating hepatomas. By contrast, all classes of RNA polymerase were found to be increased during liver regeneration. Relative to resting liver, the fastest growing tumour, 3924A, exhibited the highest activities and/or amounts of RNA polymerase I (8-fold) and III (5-fold) per g of tissue. These alterations in the tumour RNA polymerases were reflected in corresponding increases in the transcriptionally active (bound or chromatin-associated) enzyme population. The mechanisms underlying the augmented synthesis of RNA in vitro by bound polymerase I from hepatoma 3924A were elucidated by product analysis. The results indicated that, relative to liver RNA polymerase I, the tumour enzyme produced more nascent RNA chains and elongated these chains at a faster rate. The number of 3'-termini, as measured by incorporation into uridine, was higher in the hepatoma even under conditions which prevented re-initiation. suggesting increased amount of transcriptionally active RNA polymerase I in the tumour.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSCH H., BYVOET P., SMETANA K. The nucleolus of the cancer cell: a review. Cancer Res. 1963 Mar;23:313–339. [PubMed] [Google Scholar]
- Bennett T. P. Membrane filtration for determining protein in the presence of interfering substances. Nature. 1967 Mar 18;213(5081):1131–1132. doi: 10.1038/2131131a0. [DOI] [PubMed] [Google Scholar]
- Benz E. W., Jr, Getz M. J., Wells D. J., Moses H. L. Nuclear RNA polymerase activities and poly(A)-containing mRNA accumulation in cultured AKR mouse embryo cells stimulated to proliferate. Exp Cell Res. 1977 Aug;108(1):157–165. [PubMed] [Google Scholar]
- Büning J. Changes in RNA synthesis of endogenous "free" and template-engaged RNA polymerases in isolated nuclei of highly synchronized coleopteran embryos. Dev Biol. 1978 May;64(1):130–139. doi: 10.1016/0012-1606(78)90065-9. [DOI] [PubMed] [Google Scholar]
- Cedar H. Transcription of DNA and chromatin with calf thymus RNA polymerase B in vitro. J Mol Biol. 1975 Jun 25;95(2):257–269. doi: 10.1016/0022-2836(75)90394-0. [DOI] [PubMed] [Google Scholar]
- Chambon P. Eukaryotic nuclear RNA polymerases. Annu Rev Biochem. 1975;44:613–638. doi: 10.1146/annurev.bi.44.070175.003145. [DOI] [PubMed] [Google Scholar]
- Cox R. F. Quantitation of elongating form A and B RNA polymerases in chick oviduct nuclei and effects of estradiol. Cell. 1976 Mar;7(3):455–465. doi: 10.1016/0092-8674(76)90176-8. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Lin C. Y., Chen Y. M., Nagao R. T., Key J. L. Enhancement of soybean RNA polymerase I by auxin. Proc Natl Acad Sci U S A. 1975 Jan;72(1):69–72. doi: 10.1073/pnas.72.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Tata J. R. Differential activation of free and template-engaged RNA polymerase I and II during the resumption of development of dormant Artemia gastrulae. Dev Biol. 1977 Jun;57(2):293–304. doi: 10.1016/0012-1606(77)90216-0. [DOI] [PubMed] [Google Scholar]
- Hirsch J., Martelo O. J. Phosphorylation of rat liver ribonucleic acid polymerase I by nuclear protein kinases. J Biol Chem. 1976 Sep 10;251(17):5408–5413. [PubMed] [Google Scholar]
- Hyman R. W., Davidson N. Kinetics of the in vitro inhibition of transcription by actinomycin. J Mol Biol. 1970 Jun 14;50(2):421–438. doi: 10.1016/0022-2836(70)90202-0. [DOI] [PubMed] [Google Scholar]
- Jacob S. T. Mammalian RNA polymerases. Prog Nucleic Acid Res Mol Biol. 1973;13:93–126. doi: 10.1016/s0079-6603(08)60101-4. [DOI] [PubMed] [Google Scholar]
- Jacob S. T., Sajdel E. M., Munro H. N. Different responses of soluble whole nuclear RNA polymerase and soluble nucleolar RNA polymerase to divalent cations and to inhibition by alpha-amanitin. Biochem Biophys Res Commun. 1970 Feb 20;38(4):765–770. doi: 10.1016/0006-291x(70)90647-9. [DOI] [PubMed] [Google Scholar]
- Jaehning J. A., Stewart C. C., Roeder R. G. DNA-dependent RNA polymerase levels during the response of human peripheral lymphocytes to phytohemagglutinin. Cell. 1975 Jan;4(1):51–57. doi: 10.1016/0092-8674(75)90133-6. [DOI] [PubMed] [Google Scholar]
- Jänne O., Bullock L. P., Bardin C. W., Jacob S. T. Early androgen action in kidney of normal and androgen-insensitive (tfm/y) mice. Changes in RNA polymerase and chromatin template activities. Biochim Biophys Acta. 1976 Feb 5;418(3):330–343. doi: 10.1016/0005-2787(76)90295-1. [DOI] [PubMed] [Google Scholar]
- Lea M. A., Morris H. P., Weber G. Comparative biochemistry of hepatomas. VI. Thymidine incorporation into DNA as a measure of hepatoma growth rate. Cancer Res. 1966 Mar;26(3):465–469. [PubMed] [Google Scholar]
- Leonard T. B., Jacob S. T. Alterations in DNA-dependent RNA polymerase I and II from rat liver by thioacetamide: preferential increase in the level of chromatin-associated nucleolar RNA polymerase IB. Biochemistry. 1977 Oct 4;16(20):4538–4544. doi: 10.1021/bi00639a032. [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Bullock L. P., Bardin C. W., Jacob S. T. Effect of medroxyprogesterone acetate and testosterone on solubilized RNA polymerases and chromatin template activity in kidney from normal and androgen-insensitive (Tfm/Y) mice. Biochemistry. 1978 Oct 31;17(22):4833–4838. doi: 10.1021/bi00615a034. [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Rose K. M., Jacob S. T. Evidence for the nuclear origin of RNA polymerases identified in the cytosol: release of enzymes from the nuclei isolated in isotonic sucrose. Biochem Biophys Res Commun. 1976 Sep 7;72(1):114–120. doi: 10.1016/0006-291x(76)90968-2. [DOI] [PubMed] [Google Scholar]
- Matsui T., Onishi T., Muramatsu M. Nucleolar DNA-dependent RNA polymerase from rat liver. 2. Two forms and their physiological significance. Eur J Biochem. 1976 Dec 11;71(2):361–368. doi: 10.1111/j.1432-1033.1976.tb11122.x. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Tantravahi R., Miller D. A., Yu L. C., Szabo P., Prensky W. Marked increase in ribosomal RNA gene multiplicity in a rat hepatoma cell line. Chromosoma. 1979 Feb 21;71(2):183–195. doi: 10.1007/BF00292822. [DOI] [PubMed] [Google Scholar]
- Munro H. N. The determination of nucleic acids. Methods Biochem Anal. 1966;14:113–176. doi: 10.1002/9780470110324.ch5. [DOI] [PubMed] [Google Scholar]
- Nowell P. C., Morris H. P., Potter V. R. Chromosomes of "minimal deviation" hepatomas and some other transplantable rat tumors. Cancer Res. 1967 Sep;27(9):1565–1579. [PubMed] [Google Scholar]
- Rose K. M., Jacob S. T. Poly(adenylic acid) synthesis in isolated rat liver mitochondria. Biochemistry. 1976 Nov 16;15(23):5046–5052. doi: 10.1021/bi00668a015. [DOI] [PubMed] [Google Scholar]
- Rose K. M., Ruch P. A., Morris H. P., Jacob S. T. RNA polymerases from a rat hepatoma. Partial purification and comparison of properties with corresponding liver enzymes. Biochim Biophys Acta. 1976 Apr 15;432(1):60–72. doi: 10.1016/0005-2787(76)90041-1. [DOI] [PubMed] [Google Scholar]
- Sajdel E. M., Jacob S. T. Mechanism of early effect of hydrocortisone on the transcriptional process: stimulation of the activities of purified rat liver nucleolar RNA polymerases. Biochem Biophys Res Commun. 1971 Nov 5;45(3):707–715. doi: 10.1016/0006-291x(71)90474-8. [DOI] [PubMed] [Google Scholar]
- Schwartz L. B., Sklar V. E., Jaehning J. A., Weinmann R., Roeder R. G. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J Biol Chem. 1974 Sep 25;249(18):5889–5897. [PubMed] [Google Scholar]
- Sebastian J., Mian F., Halvorson H. O. Effect of the growth rate on the level of the DNA-dependent RNA polymerases in Saccharomyces cerevisiae. FEBS Lett. 1973 Aug 15;34(2):159–162. doi: 10.1016/0014-5793(73)80782-3. [DOI] [PubMed] [Google Scholar]
- Tillyer C. R., Butterworth P. H. The relationship between the activities of different pools of RNA polymerases I and II during PHA-stimulation of human lymphocytes. Nucleic Acids Res. 1978 Jun;5(6):2099–2111. doi: 10.1093/nar/5.6.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann R., Roeder R. G. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc Natl Acad Sci U S A. 1974 May;71(5):1790–1794. doi: 10.1073/pnas.71.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. L. An improved method for the quantitative isolation of rat liver nuclear RNA polymerases. Biochim Biophys Acta. 1975 Jul 7;395(3):329–336. doi: 10.1016/0005-2787(75)90204-x. [DOI] [PubMed] [Google Scholar]
- Yu F. L. Increased levels of rat hepatic nuclear free and engaged RNA polymerase activities during liver regeneration. Biochem Biophys Res Commun. 1975 Jan 2;64(3):1107–1115. doi: 10.1016/0006-291x(75)90161-8. [DOI] [PubMed] [Google Scholar]


