Abstract
The orchestration of our genes heavily relies on coordinated communication between enhancers and promoters, yet the mechanisms behind this dynamic interplay during active transcription remain unclear. Here, we investigated enhancer-promoter (E-P) interactions in relation to transcriptional bursting in mouse embryonic stem cells using sequential DNA/RNA/immunofluorescence–fluorescence in situ hybridization analyses. Our data reveal that the active state of specific genes is characterized by specific proximities between different genomic regions and the accumulation of transcriptional regulatory factors. Mathematical simulations suggest that an increase in local viscosity could potentially contribute to stabilizing the duration of these E-P proximities. Our study provides insights into the association among E-P proximity, protein accumulation, and transcriptional dynamics, paving the way for a more nuanced understanding of gene-specific regulatory mechanisms.
Active state during transcriptional bursting associates with proximities between genomic regions and protein accumulation.
INTRODUCTION
The transcription of cell type–specific genes is governed by enhancer regions that often are distant from the gene promoters. Enhancer regions are rich in binding sites for transcription factors that are expressed in a cell type–specific manner. For an enhancer to coordinate the cell-specific information instructed via transcription factors to a cognate promoter, it must be physically located near a promoter region. For example, a clear correlation between enhancer-promoter (E-P) proximity and transcriptional activity has been demonstrated by live-cell imaging in Drosophila (1). Conversely, several cases have been reported in which no apparent correlation between E-P proximity and transcriptional activity has been observed, highlighting the complexity of E-P interactions (2–6). These findings are compatible with the transcription hub model, which posits that E-P communication occurs through transcriptional regulatory factor condensates or clusters and does not necessitate direct contact between the E-P pair (7). Meanwhile, recent micrococcal nuclease chromatin capture (micro-C) analyses have uncovered clear interactions between E-P pairs just downstream of the transcription start site, at the location of the +1 nucleosome (8). However, the temporal aspects of these interactions remain undetermined.
Building upon our understanding of E-P interactions, studies have also elucidated the dynamic nature of transcriptional processes. Imaging and sequencing-based analyses have revealed that transcription is a dynamic process characterized by stochastic switches between active states, where RNA is continuously synthesized, and inactive states, where little to no synthesis occurs. This phenomenon, universally observed across various species, cell types, and genes, is commonly referred to as transcriptional bursting (9, 10). Transcriptional bursting plays a critical role in regulating both the level and cell-to-cell heterogeneity in gene expression. Recent findings indicate that during the active state, coactivators, such as bromodomain-containing protein 4 (BRD4)—which binds to acetylated histones at active enhancer regions—and the large subunit of RNA polymerase II (RPB1), form clusters in the spatial proximity of the gene locus (11). In addition, the gene region exhibits slower mobility in the active state (11, 12). The observations from recent studies suggest that genomic interactions, including E-P interactions, may undergo dynamic changes that regulate transcriptional dynamics. However, knowledge gaps remain in our understanding of the relationship between these dynamically changing E-P interactions and transcriptional dynamics.
To elucidate the relationship between transcriptional activity and the organization of transcriptional hubs—including genomic region interactions and the accumulation of transcription-related factors—acquiring multimodal data with preserved spatial information is essential. Recent advancements in spatial multiomic technologies have facilitated this, enabling the analysis of numerous genomic regions, transcriptional activities, proteins, and posttranslational modifications (PTMs) within their spatial contexts (13). Notable among these are oligo-based methods that offer comprehensive insights across these dimensions (14–17). Nevertheless, the requirement for cell fixation in spatial multiomics poses challenges for real-time analysis of the dynamic interplay between genomic interactions and the formation of transcriptionally active clusters.
Advances in techniques like Hi-C, in tandem with spatial omic technologies, have revolutionized our understanding of the cell’s higher-order genomic structures (18). The intergenomic interaction frequency map obtained from Hi-C can be used to estimate not only the higher-order genomic structure of the target genomic region but also its dynamics (19). Therefore, by combining the data obtained by spatial omic technologies and these analysis methods, it may be possible to investigate the relationship between the dynamics of higher-order genomic structure and transcriptional activity.
In this study, we used both sequential (seq) DNA/RNA/immunofluorescence (IF) fluorescence in situ hybridization (seq-DNA/RNA/IF-FISH) analyses and computational polymer simulations. The seq-DNA/RNA/IF-FISH analyses allowed us to visualize transcriptional activity states, higher-order genomic structures, transcriptional regulatory factors, and PTMs at the level of single alleles within individual mouse embryonic stem (ES) cells.
RESULTS
Multiplexed imaging of chromatin structure and transcriptional activity
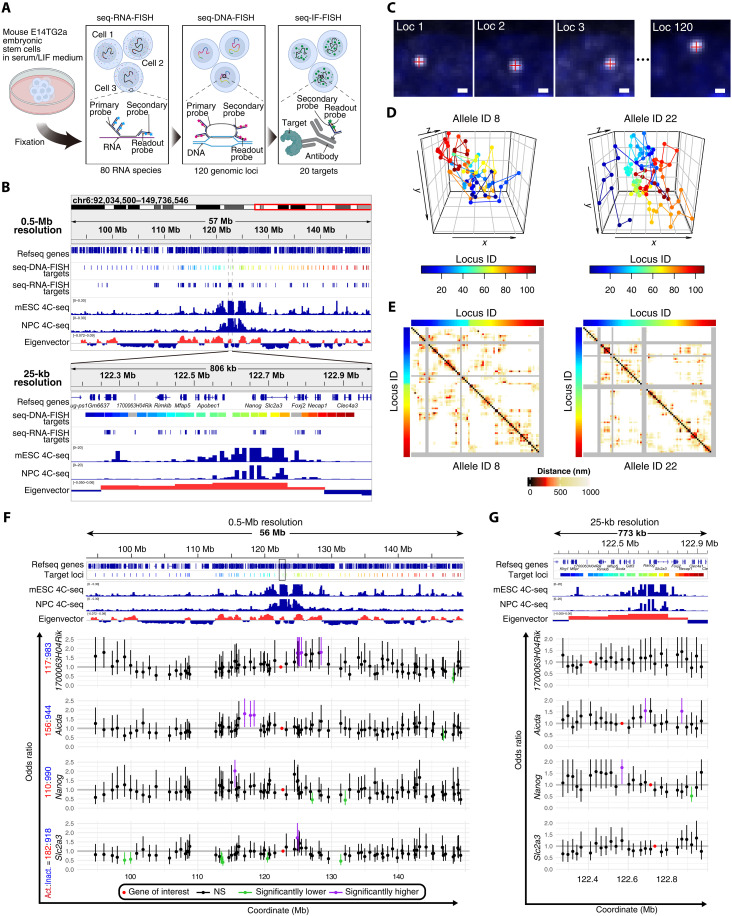
To explore the relationship between transcriptional dynamics and genomic interactions, we used imaging-based seq-DNA/RNA/IF-FISH analyses. This innovative technology combines seq-RNA-FISH, seq-DNA-FISH, and seq-IF-FISH on the same sample, thereby offering a comprehensive view of transcriptional activity, genomic interactions, and transcriptional regulatory factors/PTMs at single-allele resolution within individual cells (Fig. 1A) (14).
Fig. 1. Transcriptional activity state-specific genomic proximities.
(A) Diagram of seq-DNA/RNA/IF-FISH analysis. (B) Target regions for seq-DNA/RNA-FISH in this study, with regions for seq-DNA-FISH and seq-RNA-FISH, and genomic interaction data shown at a 0.5-Mb and 25-kb resolutions, excluding gray-highlighted regions due to quality checks. (C) seq-DNA-FISH fluorescent spots examples. Scale bars, 500 nm. (D) 3D chromatin traces of specific alleles. The color code at the bottom of the right corresponds to the target genomic regions in (B) at 0.5-Mb resolution. (E) Distance matrices for specific alleles. The gray regions indicate areas where fluorescent spots were not detected. Color codes on the top and left margins correspond to the 0.5-Mb resolution target genomic regions in (B). (F) Odds ratios compare genomic proximity frequencies by transcriptional state at 0.5 Mb, with error bars showing 95% confidence intervals (CIs). Red, purple, and green dots denote regions containing the gene of interest, regions with the lower limit of the 95% CI exceeding 1, and regions with the upper limit of the 95% CI below 1, respectively. Black dots represent regions not categorized as above. The count of alleles where the gene of interest was active (Act.) or inactive (Inact.) is listed beside the gene name. (G) Odds ratios calculated similar to (F) but for a 25-kb resolution. The target region at a 25-kb resolution is marked by an unfilled black rectangle in (F). Notation and sample sizes follow the same format as in (F). mESC, mouse embryonic stem cell; NPC, neural progenitor cell. The 4C-seq data use Nanog as bait (21). The eigenvector was calculated on the basis of Hi-C data from mouse ES cells cultured in serum/LIF conditions (24). For the eigenvector, please refer to Materials and Methods. NS, not significant.
The original seq-FISH technology used a “Primary-Probe Direct” detection method, where a primary probe, equipped with an adapter sequence for binding a fluorescence-tagged readout probe, is first hybridized to the target molecule (fig. S1A). This process makes the target molecule visible but requires synthesizing specialized readout probes, often entailing additional purification steps like high-performance liquid chromatography (HPLC), thus incurring considerable costs. In contrast, we opted for the “Secondary-Probe Mediated” approach used by other chromatin tracing technologies (16, 17). This method introduces a nonfluorescent secondary probe between the primary and readout probes, reducing the need for multiple unique readout probes and allowing their reuse across multiple targets. While this necessitates the preparation of many nonfluorescent secondary probes, the readout probes are only prepared in quantities corresponding to the fluorescent channels used—three in this study: Alexa488, ATTO565, and Alexa647—thus streamlining the process (fig. S1, B and C; see Materials and Methods).
We focused our investigation on the Nanog gene, a key regulator of pluripotency, in mouse ES cells cultured in a medium containing leukemia inhibitory factor (LIF) and serum (serum/LIF medium). In this condition, where pluripotency is maintained, transcriptional bursting at the Nanog locus has been previously observed (20). 4C-seq analyses of mouse ES cells and neural progenitor cells have shown that the Nanog locus can interact with genomic regions in a mouse ES cell–specific manner (Fig. 1B) (21). Although these distal interactions are relatively weak compared to the proximal regions of Nanog and occur across multiple A compartments (Fig. 1B), the functional consequences of these interactions on transcriptional bursting remain unclear. To elucidate the functional consequences of these distal interactions, we subjected genomic regions—both including and excluding these domains—to seq-DNA-FISH analyses at intervals of approximately 0.5 Mb. In addition, we subjected regions for seq-DNA-FISH analysis at 25-kb intervals within an approximately 750-kb region surrounding the Nanog locus (Fig. 1B). This 750-kb region largely comprises a singular topologically associating domain (TAD) (fig. S2). In total, we analyzed 120 genomic loci spanning approximately 60 Mb for seq-DNA-FISH analysis (Fig. 1, A and B).
Within the 60-Mb range targeted for seq-DNA-FISH analyses, we also chose 80 highly expressed genes in mouse ES cells as targets for seq-RNA-FISH (Fig. 1B; see Materials and Methods). Furthermore, to correlate the epigenetic state, transcription, and higher-order genomic structure, we targeted 20 types of entities that include transcriptional regulatory factors and histone PTMs, notably enriched in Nanog-interacting regions, and were investigated using seq-IF-FISH (see subsequent sections) (movie S1).
Analysis and data quality validation of seq-DNA-FISH experiments
In our study, two separate seq-FISH experiments were conducted, yielding data from 611 cells and 1100 alleles in the first replicate and 335 cells and 580 alleles in the second. Through seq-DNA-FISH analysis, we pinpointed the coordinates of gene region foci, facilitating measurements of distances between genomic loci (Fig. 1C). We observed low detection frequencies in some targeted regions. To ensure robust analysis, we excluded genomic regions where detection efficiency fell below half of the overall median (48.3%), focusing on 109 loci with higher detection rates for further investigation (fig. S3A; see Materials and Methods). The average detection efficiency was recorded at 70.51 ± 0.06% for the first replicate and 66.71 ± 0.08% for the second (fig. S3, A and B). Constructing two-dimensional (2D) distance matrices from the individual alleles’ 3D coordinates showed a degree of diversity in the higher-order genomic structures, aligning with prior studies, as illustrated by examples of distance matrices derived from two alleles (Fig. 1, D and E) (22, 23).
To assess the seq-DNA-FISH data’s quality, we evaluated interaction frequencies using set distance cutoffs at 25-kb and 0.5-Mb resolutions, comparing these to Hi-C data from mouse ES cells cultured in serum/LIF medium (24). Optimal correlation with Hi-C data was achieved at cutoffs of 350 and 950 nm for 25-kb and 0.5-Mb resolutions, respectively (fig. S3, C to E). While physical “contacts” or “interactions” are often defined using a cutoff of 150 nm or less, the larger cutoffs used in our study may also capture higher-order chromatin structures in addition to direct contacts between specific genomic regions. Furthermore, our data demonstrated that the localization precision for determining specific genomic regions, quantified as the variation in labeling the same spots across different cycles compared to the initial labeling, was substantially smaller (i.e., more precise) than the cutoff values used (fig. S3F). Therefore, we used the proximity frequencies calculated with 350- and 950-nm cutoffs for 25-kb and 0.5-Mb resolution data, respectively, to examine the relationship between the proximity of specific genomic regions and transcriptional activity.
Consistency was observed in the distribution of median spatial distances and proximity frequencies across replicates (fig. S4, A and B), with seq-DNA-FISH proximity frequencies showing a positive correlation with Hi-C data at corresponding resolutions (r > 0.75; fig. S4, C and D). Moreover, a sample size of approximately 500 suggested that proximity frequency closely mirrors that of the broader population (r > 0.95) (fig. S4, E and F), underscoring the seq-DNA-FISH data’s reliability. Unless otherwise specified, the results presented hereafter are based on data from the first replicate.
Transcriptional activity state–specific genomic interactions
Using our seq-RNA-FISH data, we visualized individual RNA molecules. In regions where transcription is actively occurring, numerous nascent RNAs accumulate, resulting in the detection of bright fluorescence spots. In this study, bright spots detected near specific alleles, which were more intense than those from single RNA molecules, were identified as “transcriptional spots.” We used these spots as an indication that the gene is in an active state (see Materials and Methods). Upon calculating the proportion of transcriptionally active alleles for each gene, we found that only about 10% of the genes (8 of 80) exhibited an average active allele proportion exceeding 20% (fig. S5A). This observation is in accord with prior studies that suggest that most genes exhibit transcriptional bursting (10, 25). Furthermore, concurrent transcriptional activity, indicated by the presence of transcriptional spots within the same allele, was noted in less than 10% of gene pairs (fig. S5, B to E).
We then analyzed the proximity frequency matrix from seq-DNA-FISH data and categorized this matrix by transcriptional activity states to explore differences in genomic proximity frequencies during active and inactive states. By assessing the odds ratio for proximity between a gene and a genomic region based on its transcriptional state, for genes with over 100 alleles in an active state, we determined the likelihood of a gene being closer in either state (see Materials and Methods) (Fig. 1, F and G). Our analysis at both 0.5-Mb and 25-kb resolutions identified regions that significantly differ in their interaction with specific genes depending on whether the gene is in an active or inactive state (Fig. 1, F and G, and fig. S6A). Notably, the pattern of proximity changes varied by gene; for example, significant differences were observed at 0.5 Mb but not at 25-kb resolution for 1700063H04Rik and Slc2a3. In contrast, for Aicda and Nanog, changes in proximity frequency were significant across both proximal (25-kb resolution) and distal regions (0.5-Mb resolution).
To infer the biological implications of regions with significantly altered odds ratios, we plotted the enrichment levels and the values of various epigenomic-related factors, including chromatin openness as analyzed by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), H3K27ac (histone H3 lysine 27 acetylation), H3K27me3 (histone H3 lysine 27 trimethylation), H3K4me3 (histone H3 lysine 4 trimethylation), H3K9me3 (histone H3 lysine 9 trimethylation), BRD4 (a transcriptional coactivator that binds acetylated histones), ESRRB (estrogen related receptor beta, a transcription factor) and YY1 (Yin Yang 1, a transcription factor), SMC1A (structural maintenance of chromosomes 1A, a cohesin subunit), and CTCF (CCCTC-binding factor, a TAD boundary binding factor), as well as eigenvector values from Hi-C data (fig. S7A). At the 25-kb resolution, none of the factors showed significant differences. This finding suggests that regions with significant proximity changes in specific transcriptional states may not have a clear association with the localization of specific epigenomic factors or may reflect the limited number of genes analyzed in this study at the 25-kb resolution. In contrast, at the 0.5-Mb resolution, we observed significant differences with higher-order chromatin structure–related factors such as CTCF and SMC1A, as well as transcription factors ESRRB and YY1. This suggests a relationship between transcription-related proximity and these factors, even when analyzing data derived from bulk chromatin immunoprecipitation sequencing (ChIP-seq) or Hi-C analyses from many cells. This indicates that the localization of these epigenomic factors may vary at the single-cell or single-allele level. It has been reported that CTCF and cohesin dynamically interact with higher-order chromatin structures (26). In addition, transcription factors themselves are known to dynamically bind and dissociate (27, 28). These findings suggest that these factors might play a role in the proximity of distal regions in a transcription state–specific manner at a 0.5-Mb resolution.
While the cutoffs of 350 nm (25-kb resolution) and 950 nm (0.5-Mb resolution) are larger than the commonly used contact threshold (~150 nm), regions with significantly increased or decreased proximity frequencies at these cutoffs showed significant differences in the distribution of distances to the gene of interest (fig. S7B). Regions with significantly increased proximity frequencies were significantly closer than those with decreased proximity frequencies and vice versa. In addition, even when using a 150-nm cutoff, significant differences were observed at the 25-kb resolution (fig. S7C). However, at the 0.5-Mb resolution, significant differences were only observed with a 400-nm cutoff and not with a 350-nm cutoff (fig. S7C). These findings demonstrate that proximity frequencies with specific genes vary according to the transcriptional state, highlighting the dynamic nature of genomic architecture in relation to gene expression.
Relationship between cellular state and genomic proximities
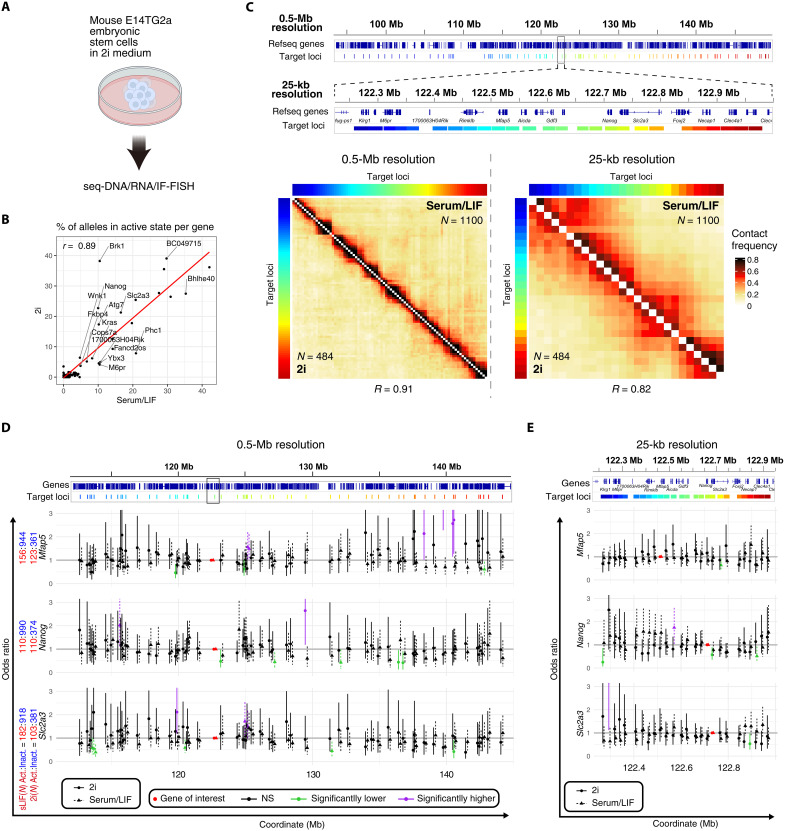
Higher-order genomic structure changes during cellular differentiation (24). In mouse ES cells, a notable reduction in cell-to-cell heterogeneity in gene expression and up-regulation of pluripotency marker genes, including Nanog, have been noted when cultured in medium containing mitogen-activated protein kinase kinase and glycogen synthase kinase 3 β inhibitors (2i medium), as opposed to a serum/LIF medium (29). To explore the relationship between these changes in cellular states and genomic proximities surrounding the Nanog locus, we cultured mouse ES cells in 2i medium, conducted seq-DNA/RNA/IF-FISH analyses, and obtained data from 262 cells and 484 alleles in a single replicate (Fig. 2A). Similar to the serum/LIF condition, spot detection was carried out, and it was confirmed that the 109 loci analyzed under serum/LIF conditions were detected with an efficiency of more than 50%. Furthermore, the average detection efficiency was recorded at 71.15 ± 0.08%. An increase in transcriptionally active states for genes including Nanog under 2i conditions was noted, aligning with prior research (Fig. 2B) (10, 20).
Fig. 2. Relationship between cellular state and higher-order genomic structure.
(A) Schematic representation of mouse ES cells cultured in 2i medium and subjected to seq-DNA/RNA/IF-FISH analysis. (B) Proportion of cells exhibiting transcriptional bursting (burst frequency) in mouse ES cells cultured in serum/LIF and 2i media. Serum/LIF: N = 1100; 2i: N = 484. Overall, high correlation values were observed, although the expression levels of certain genes varied substantially depending on the culture conditions. For instance, it is well-known that Nanog expression increases under 2i conditions; our study confirmed a corresponding increase in burst frequency in 2i culture. (C) Comparative analysis of proximity frequency for mouse ES cells cultured in different media. Serum/LIF: N = 1100; 2i: N = 484. Pearson correlation coefficients are provided for each comparison. (D) Odds ratios comparing proximity frequencies with the target gene region, based on the transcriptional activity state of the gene at a 0.5-Mb resolution. Error bars denote the 95% CI. Data for serum/LIF medium are consistent with those presented in Fig. 1F. Red, purple, green, and black dots indicate regions containing the target gene, regions with the lower limit of the 95% CI above 1, regions with the upper limit of the 95% CI below 1, and regions not fitting these descriptions, respectively. The counts of alleles with the gene of interest in active (Act.) or inactive (Inact.) states are listed next to the gene names. “sLIF” abbreviates serum/LIF. (E) Odds ratios for proximity frequencies with gene regions of interest (ROIs) at a 25-kb resolution, following the same notation and sample sizes as in (D). Data for serum/LIF medium are consistent with those presented in Fig. 1G.
Moreover, Hi-C data from mouse ES cells cultured in 2i (30) or serum/LIF medium (24) showed a high correlation (r > 0.9), with seq-DNA-FISH data also demonstrating a positive correlation (r > 0.8) (Fig. 2C and figs. S3, C to E, and S4), suggesting that the overall higher-order genomic structure remains similar across these conditions. To further investigate this, we calculated the odds ratio for genes in active states, with more than 100 alleles, under 2i conditions. We identified regions that significantly interact with specific genes in either active or inactive states (highlighted in purple or green, respectively) at both 0.5-Mb and 25-kb resolutions (Fig. 2, D and E). Although regions with significant differences in interaction frequency between active and inactive states were identified under both serum/LIF and 2i conditions, the specific regions showing significant changes did not overlap between these conditions, suggesting that the regions of significant interaction frequency changes due to transcriptional activity states vary according to the culturing condition (e.g., near the 140-Mb region in Mfap5 data or around the 130-Mb region in Nanog data).
To infer the biological implications of these regions with significantly altered odds ratios, we plotted either the enrichment levels or the values of various epigenomic-related factors, including chromatin openness as analyzed by ATAC-seq, H3K27ac, H3K27me3, H3K4me3, H3K9me3, BRD4, ESRRB, YY1, SMC1A, and CTCF (fig. S7D). Under 2i conditions, unlike serum/LIF conditions, no significant difference was observed for any of these factors at either 25-kb or 0.5-Mb resolution. This suggests that regions with significant proximity changes in specific transcriptional states may not have a clear association with the localization of specific epigenomic factors or may reflect the limited number of significantly altered regions. In addition, under 2i conditions, gene expression heterogeneity between cells is known to be reduced compared to serum/LIF conditions (29), which may account for the lack of significant differences in higher-order genomic structure between active and inactive states of specific genes.
These findings suggest that, even under 2i conditions, certain regions interact more frequently or rarely depending on the transcriptional state of specific genes, underscoring the dynamic relationship between cellular state, higher-order genomic structure, and gene expression.
Accumulation of transcriptional regulatory factors near genes in an active state
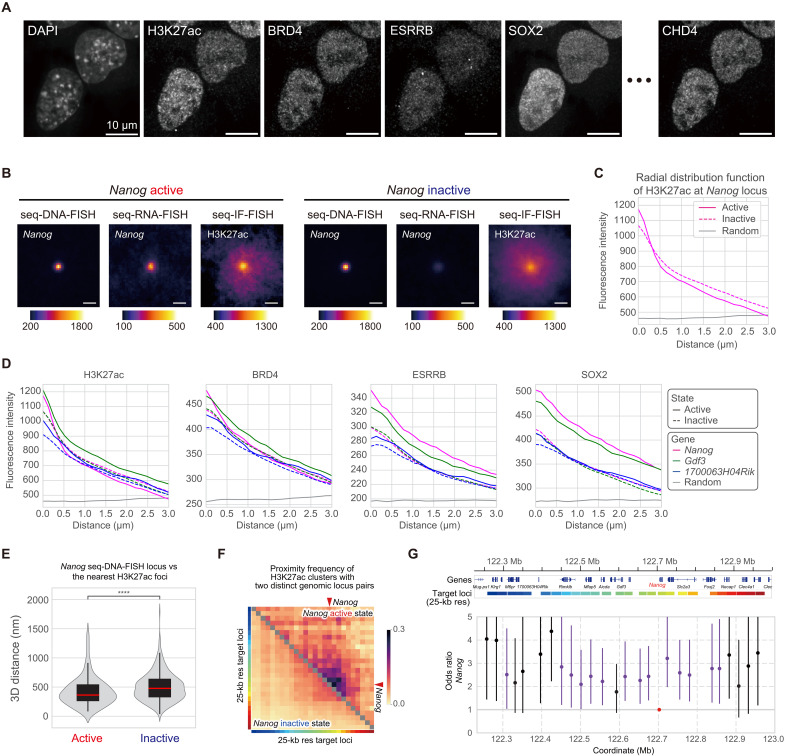
To elucidate the spatial relationship between the accumulation and localization of transcriptional regulatory factors/PTMs and transcriptional activity, we analyzed seq-IF-FISH data (Fig. 3A and fig. S8A). To avoid potential artifacts, we excluded regions with low relative fluorescence intensities compared to negative controls, focusing our investigation on 10 different proteins and PTMs (KDM1A, TCF3, SIN3A, OTX2, CHD4, ESRRB, MED12, BRD4, SOX2, and H3K27ac; fig. S8, A to C). To validate the seq-IF-FISH data, we measured fluorescence intensities at the coordinates of specific genomic regions determined by seq-DNA-FISH and compared these with ChIP-seq data, as previously reported (14). While seq-DNA-FISH can pinpoint the localization of specific genomic regions as single spots, enabling subvoxel-level precision, seq-IF-FISH involves numerous target molecules dispersed throughout the nucleus, making accurate quantification within a single voxel challenging. Nevertheless, at a 1-Mb resolution, we observed a positive correlation between the fluorescence intensities of transcriptional regulatory factors/PTMs at the 3D coordinates of specific genomic loci, as determined by seq-DNA-FISH, and the ChIP-seq enrichment data (fig. S8, D to G; see Materials and Methods). This suggests that our methods can effectively quantify the degree of protein accumulation at specific genomic loci.
Fig. 3. Accumulation of transcriptional regulatory factors near genes in an active state.
(A) Nuclear localization images of various proteins and PTMs obtained by seq-IF-FISH. Scale bars, 10 μm. Additional images are in fig. S8A. (B) Median signals of seq-DNA-FISH for the Nanog locus, seq-RNA-FISH for Nanog, and seq-IF-FISH for H3K27ac, centered at Nanog seq-DNA-FISH foci, separated by active and inactive states. Sample sizes: Nanog active (N = 110), inactive (N = 990). Scale bars, 1 μm. (C) Radial distribution function of the H3K27ac seq-IF-FISH signal for the median IF signal in (B). (D) Radial distribution function of H3K27ac, BRD4, ESRRB, and SOX2 seq-IF-FISH signals centered at Nanog, Gdf3, and 1700063H04Rik seq-DNA-FISH foci, shown for active and inactive states. Sample sizes: Nanog active (N = 110), inactive (N = 990); Gdf3 active (N = 217), inactive (N = 883); 1700063H04Rik active (N = 117), inactive (N = 983). (E) 3D distance distribution between the Nanog seq-DNA-FISH locus and the nearest H3K27ac foci, separated by Nanog’s active and inactive states. The red line in the box plot shows the median. Statistical significance determined by Wilcoxon test. ****P ≤ 0.0001. (F) Colocalization frequency matrix showing H3K27ac clusters proximal to genomic locus pairs within 350 nm. Color coding on heatmap margins corresponds to seq-DNA-FISH regions at 25-kb resolution, linked to gene regions detailed in (G). Red arrowheads indicate the genes of interest. (G) Odds ratios comparing the frequency of regions containing the gene of interest, specific genomic regions, and H3K27ac foci within 350-nm proximity, based on gene activity. Error bars show 95% CIs. Red, purple, and black dots indicate regions with the target gene, regions where the 95% CI lower limit exceeds 1, and regions not categorized as such, respectively.
We recognize the challenge in quantifying accumulation at resolutions below 1 Mb due to the diffraction limit. Therefore, we examined the distribution of seq-IF-FISH signals at the spatial coordinates of specific genes determined by seq-DNA-FISH. To do this, we extracted seq-IF-FISH images containing the spatial coordinates of specific genes determined by seq-DNA-FISH and generated median intensity projection images (Fig. 3B). Furthermore, we calculated the radial distribution function for these images (Fig. 3C). The results suggested that the H3K27ac signal at the Nanog gene region tends to have higher fluorescence intensity in the central region when Nanog is in an active state. Next, we examined the radial distribution function of the seq-IF-FISH data for the Nanog, Gdf3, and 1700063H04Rik gene regions. The findings suggested that transcription-related factors and PTMs such as H3K27ac, BRD4, ESRRB, and SOX2 tend to accumulate more around gene regions that are in an active state (Fig. 3D and fig. S9A). Moreover, when compared to the radial distribution function in random nuclear regions, the fluorescence intensity of these factors was consistently higher near these gene regions, regardless of whether they were in an active or inactive state. This suggests that these gene-containing regions belong to A compartments (Fig. 1B and fig. S2B), where transcription-related factors are more likely to localize compared to the rest of the nuclear space. Nonetheless, the tendency for transcription-related factors to accumulate even more in specific gene regions during their active state is intriguing.
Consistent with previous reports in both live and fixed cells, transcription-related factors and RNA polymerase II are known to cluster near actively transcribing genes (11, 31–33). Similarly, when the images of H3K27ac were magnified, the fluorescence intensity was observed as foci within the nucleus (fig. S9B). Using seq-IF-FISH data, we measured the distances from specific genes to their nearest foci (see Materials and Methods) and found that, in the active state of Nanog, 1700063H04Rik, and Gdf3, H3K27ac foci were significantly closer to these genes (Fig. 3E and fig. S9, C and D). Furthermore, we observed distinct differences between the active and inactive states in the colocalization frequency of H3K27ac foci with two distinct genomic locus pairs, defined as being within 350 nm of each other, especially in regions close to the target genes (figs. S3, F and G, and S9, E and F). These findings suggest that several transcription-related factors tend to accumulate around genes in their active state and that H3K27ac foci are more likely to be in proximity to active genes associated with specific characteristic genomic regions.
Transcriptional regulation by dynamic interaction of distal E-P
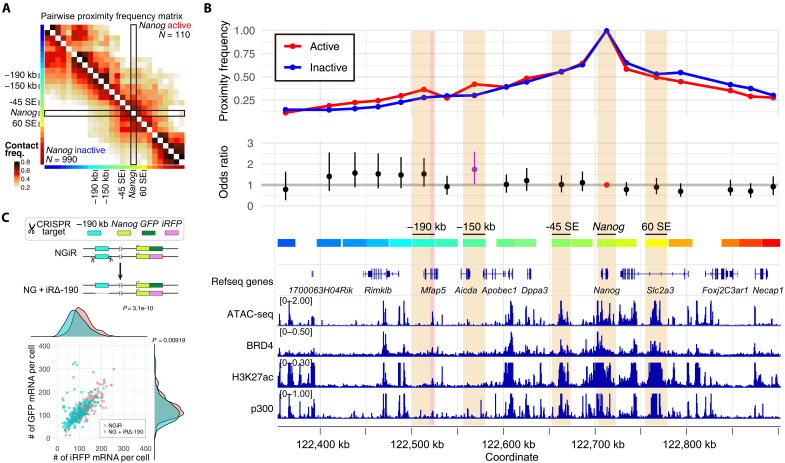
To elucidate how genomic proximities contribute to transcriptional activity, we focused on Nanog. A comparative analysis of proximity frequency matrices according to the transcriptional activity states of Nanog revealed little difference in proximity with known super-enhancers (SEs) of Nanog, such as −45 SE and 60 SE, depending on the transcriptional state of Nanog (Fig. 4, A and B) (34). This observation suggests that interaction frequencies with these SEs do not significantly influence Nanog’s transcriptional activity shifts during its transcriptional bursting.
Fig. 4. Transcriptional regulation through dynamic E-P proximity.
(A) Proximity frequency matrix for Nanog when in active or inactive transcriptional states. Sample sizes for active (inactive) states of Nanog are denoted as N = 110 (990). Color codes on the left and bottom of the heatmap correspond to the seq-DNA-FISH target regions at a 25-kb resolution, with details on their correlation with gene regions provided in (B). (B) Outline of the proximity frequency with the Nanog region based on its transcriptional activity state. The top depicts proximity frequencies with the Nanog gene region in active versus inactive states, relating to the section marked by the black frame in (A). The middle panel presents the odds ratio, comparing proximity frequencies with the Nanog gene region across different transcriptional states of Nanog, identical to the data shown in Fig. 1G, replicated here for enhanced reader comprehension. The bottom illustrates the localization of epigenomic factors near the Nanog locus, highlighting open chromatin domains (as identified by ATAC-seq data) and the presence of BRD4, H3K27ac, and p300 in the Mfap5 gene body region, indicative of enhancer-like properties. (C) Allele-specific single-molecule RNA-FISH analysis in NGiR and NG + iRΔ-190 cell lines. NGiR: N = 196; NG + iRΔ-190: N = 198. P values were determined using a two-sided Wilcoxon rank sum test.
However, we observed an overall increase in proximity frequency between the Nanog gene region and its upstream regions during active transcription states compared to inactive ones, with the region at −150 kb being particularly notable for this rise (Fig. 4, A and B). This specific −150-kb region exhibited a distinctly pronounced increase in interaction frequency with the Nanog gene during transcriptionally active states compared to inactive ones. While not statistically significant, regions beyond −190 kb upstream also demonstrated a trend toward more frequent interactions with Nanog during its active states (Fig. 4B). Despite the lack of typical enhancer markers like H3K27ac, BRD4, and histone acetyltransferase p300, as well as open chromatin signals in the −150-kb region, these markers were notably present in the −190-kb region. This led us to hypothesize that the −190-kb region might act as an enhancer for Nanog, prompting further investigation (Fig. 4B).
To investigate the functional role of the −190-kb region in Nanog transcription, we attempted its deletion via genome editing (Fig. 4C and fig. S10). Using the NGiR cell line, where GFP and iRFP were knocked into the Nanog gene (10), we established a cell line with the −190-kb region deleted from the Nanog-iRFP allele, termed NG + iRΔ-190 (fig. S10). Quantification of transcriptional activity for each allele using single-molecule RNA FISH (smRNA-FISH) revealed a significant decrease in the number of Nanog-iRFP mRNA molecules originating from the allele lacking the −190-kb region (Fig. 4C). This suggests that the −190-kb region functions as one of the enhancers contributing to Nanog expression.
Dynamics of E-P communication
Alterations in genomic proximities, including those between enhancers and promoters (E-P proximities), associated with transcriptional activity, may affect the local dynamics of both promoters and enhancers. Our previous work has demonstrated that the Nanog locus exhibits reduced mobility when in an active state (11, 12). In addition, a growing body of evidence suggests that inhibiting transcription can enhance genomic dynamics, potentially stabilizing specific higher-order genomic configurations in a transcription-dependent manner (35, 36).
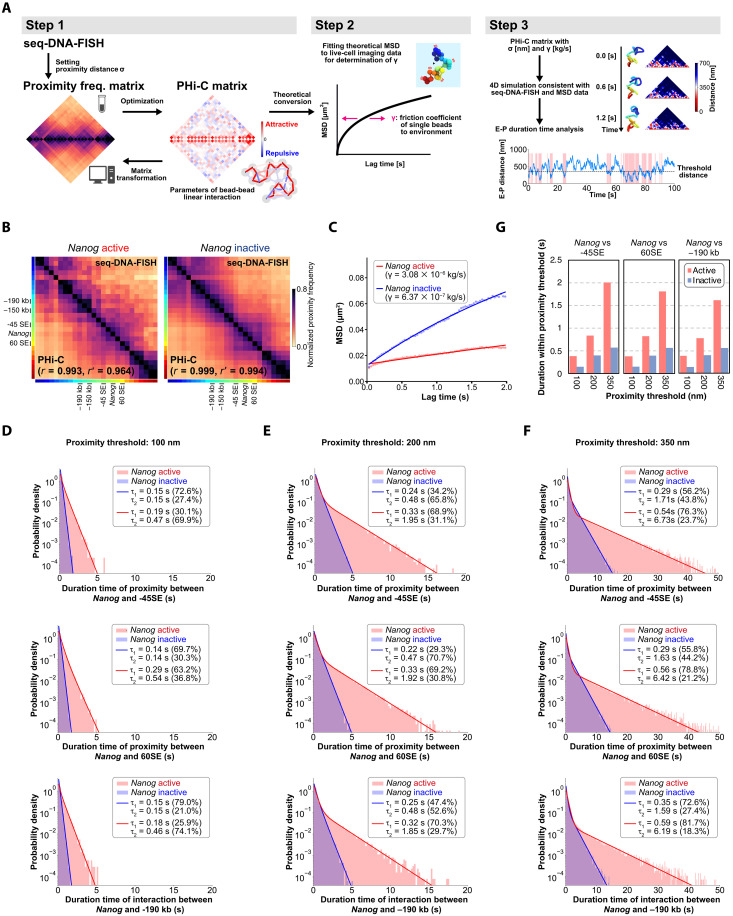
To explore this further, we used a three-step computational strategy incorporating polymer modeling (PHi-C) (37, 38), seq-DNA-FISH data, and live-cell imaging (Fig. 5A). The PHi-C polymer model features a PHi-C matrix that represents linear attractive/repulsive interaction parameters between beads. Transforming an input proximity frequency matrix into the PHi-C matrix with high pairwise correlation effectively reconstructs a polymer model that mirrors the input data. In the first step, we generated the PHi-C matrix using the PHi-C optimization algorithm, inputting the proximity frequency matrix obtained from seq-DNA-FISH with the proximity distance σ. Next, we theoretically transformed the PHi-C matrix to estimate the mean square displacement (MSD) for a specific genomic region. However, this theoretical MSD curve includes an element of uncertainty due to an unknown parameter, γ, which influences the curve’s shape over time and represents the friction coefficient of individual beads within their environment. By fitting this theoretical MSD to experimental data from live-cell imaging, we can estimate the value of γ. This adjustment ensures that the PHi-C polymer model’s 4D (3D + time) features align with both seq-DNA-FISH and live-cell imaging data. Thus, in the final step, we numerically analyze the E-P interaction dynamics by conducting Brownian dynamic simulations of the PHi-C polymer model.
Fig. 5. Estimation of higher-order genomic structural dynamics for Nanog transcriptional state by PHi-C.
(A) Integration of seq-DNA-FISH and live-cell imaging into a polymer model via PHi-C. Step 1: seq-DNA-FISH data is transformed into a proximity matrix by setting proximity distance σ and then optimized into a PHi-C matrix. Step 2: The PHi-C matrix is theoretically converted into MSD for specific genomic regions, with curve shape adjusted by the parameter γ (representing bead friction coefficient) based on live-cell imaging data. Step 3: Analysis of E-P dynamics through Brownian dynamic simulations, integrating seq-DNA-FISH and live-cell imaging data within the PHi-C model. (B) Reconstruction of higher-order genome architecture using PHi-C, informed by a proximity frequency matrix generated from seq-DNA/RNA-FISH data. The top right represents the seq-DNA-FISH data, while the bottom left showcases the proximity frequency matrix as estimated through PHi-C. Proximity frequency matrices are distinctly displayed, based on Nanog’s active and inactive states. (C) MSD of the Nanog locus. MSD values from PHi-C were fitted to the experimental data for the Nanog region (11) to determine the friction coefficient γ. See Materials and Methods for more details. (D) Distribution of the duration of interaction between Nanog and −45 SE, between Nanog and 60 SE, and between Nanog and −190 kb at a proximity threshold of 100 nm. (E) Distribution of the duration of interaction between Nanog and −45 SE, between Nanog and 60 SE, and between Nanog and −190 kb at a proximity threshold of 200 nm. (F) Distribution of the duration of interaction between Nanog and −45 SE, between Nanog and 60 SE, and between Nanog and −190 kb at a proximity threshold of 350 nm. (G) Duration of closeness below the proximity threshold between Nanog and −45 SE, between Nanog and 60 SE, and between Nanog and −190 kb.
We used seq-DNA-FISH proximity frequency data with a resolution of 25 kb as input and performed a PHi-C analysis with σ set to 350 nm. Through this approach, PHi-C allows us to estimate higher-order genomic structures and dynamics from the input proximity frequency matrices obtained from seq-DNA/RNA-FISH data, in both the transcriptionally active and inactive states of Nanog (Fig. 5B and fig. S11A). To better approximate the model to actual physical scales, we fitted the theoretical function of the PHi-C model to our experimental MSD data for the Nanog region (11), thereby estimating the value of the friction coefficient γ (kilograms per second; Fig. 5C). We found that the active state exhibited a fivefold higher friction coefficient compared to the inactive state (Nanog active state: 3.08 × 10−6 kg/s; Nanog inactive state: 6.37 × 10−7 kg/s). In physics, an elevated friction coefficient does not merely suggest a higher level of viscosity in the conventional sense of “thickness” or “stickiness” of a liquid. Instead, it indicates a state where various interactions within the solvent or medium slow down the movement of target molecules, or restrict it, due to increased resistance. Thus, this heightened friction coefficient implies a higher level of such “viscous” interactions between the Nanog region and its surrounding environment, affecting the mobility and interaction dynamics of genomic elements (Fig. 5C and movies S2 and S3).
To verify the similarity between the higher-order genomic structures inferred by PHi-C and those determined by seq-DNA-FISH, we compared the distribution of median distance matrices obtained from seq-FISH and PHi-C (using 1000 randomly sampled structures inferred from PHi-C) and found a high correlation between them (fig. S12A). Furthermore, we conducted a similar comparison by sampling 1000 structures along the time series in PHi-C, which also showed a high correlation (fig. S12, B to D). While the ability of PHi-C to infer dynamic structures has been demonstrated in previous research (37), this study did not experimentally validate the higher-order genomic dynamics predicted by PHi-C. However, these results suggest that the PHi-C analysis based on the proximity frequency matrix from seq-DNA-FISH has the potential to estimate those dynamics.
Consistent with the transcription hub model, our data suggest that various transcriptional regulatory factors tend to accumulate near active genes (Fig. 3). This fivefold increase in the friction coefficient suggests that protein accumulation influences the dynamics not only of the gene in the active state but also of other genomic loci (movies S2 and S3) (39). Recent studies using micro-C have revealed characteristic interactions between the +1 nucleosome downstream of the transcription start sites of enhancers and promoters (8). These nucleosomes are inferred to be in remarkably close proximity (<100 s nm), suggesting that such states are maintained for a certain extent of time. To elucidate the proximity dynamics between the Nanog locus and −45 SE, we measured the duration of closeness using 100, 200, and 350 nm as proximity thresholds in the Brownian dynamic simulation of the PHi-C polymer model. At each of these thresholds, we found that the duration of interactions within the threshold distance was more than doubled in the active state compared to the inactive state (Fig. 5, D to G). Notably, not only was this true for −45 SE, but the durations also increased for other surrounding enhancer regions like −190 kb and 60 SE (Fig. 5, D to G). Consequently, it can be posited that, in the active state of Nanog, sustained close proximities with multiple enhancer regions induce continuous transcriptional activity.
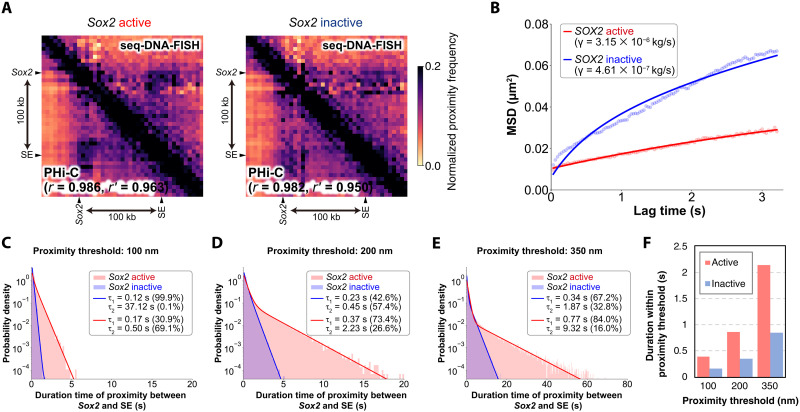
We also observed that the dynamics of Sox2 are less mobile in the active state (11). Reanalysis of chromatin tracing data for the Sox2 region (40) revealed that the proximity duration with a downstream SE located 100 kb downstream more than doubled during the active state (Fig. 6 and fig. S11B).
Fig. 6. Estimation of higher-order genomic structural dynamics for Sox2 transcriptional state by PHi-C.
(A) Proximity frequency matrix derived from 5-kb resolution chromatin tracing data (40) for Sox2 transcriptional activity, with a threshold set at 150 nm (17), used to estimate higher-order genome architecture through PHi-C. The top right shows the seq-DNA-FISH data, and the bottom left depicts the proximity frequency matrix as estimated by PHi-C. (B) MSD of the Sox2 locus. MSD values from PHi-C were fitted to the experimental data for the Sox2 region (11) to determine the friction coefficient γ. Further details can be found in Materials and Methods. (C) Distribution of the duration of interaction between Sox2 and SE at a proximity threshold of 100 nm. (D) Distribution of the duration of interaction between Sox2 and SE at a proximity threshold of 200 nm. (E) Distribution of the duration of interaction between Sox2 and SE at a proximity threshold of 350 nm. (F) Duration of closeness below the proximity threshold between Sox2 and SE.
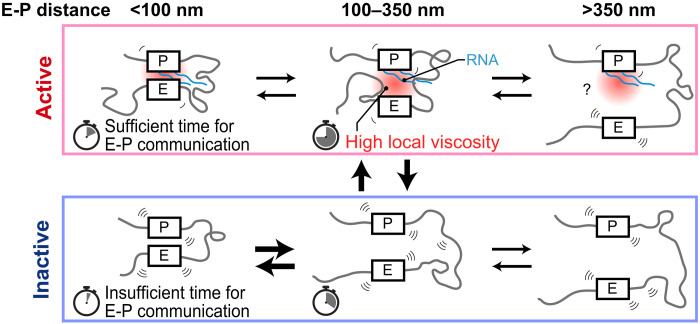
In summary, our research suggests that within specific gene regions, alterations in genomic interactions coupled with an increase in local viscosity due to the formation of transcription hubs serve to prolong the duration of E-P interactions in the active state. Regarding the estimation of local viscosity changes using PHi-C, several issues remain, such as the use of data from numerous fixed cells obtained by seq-FISH, the difference in cell lines used in live-cell imaging data and those used in seq-FISH, and the inability to consider the structural diversity between alleles detected in seq-FISH. Nevertheless, the insights obtained from this study offer a nuanced understanding of the mechanistic intricacies governing transcriptional regulation (Fig. 7).
Fig. 7. Transcriptional model incorporating increased E-P proximity durations specific to the active state.
In the active state, specific genomic regions come into close proximity, along with the accumulation of proteins and RNAs. This may result in a higher-viscosity environment around the gene, extending the duration of E-P interactions. In the inactive state, viscosity is lower, leading to shorter E-P interactions compared to the active state. Timer icons represent the average duration of each state.
DISCUSSION
In this study, we applied sequential DNA/RNA/IF-FISH techniques to investigate the genomic proximities and the local accumulation of transcriptional regulatory factors in the active states of specific genes. Our findings reveal that proximities between a given gene region and other genomic regions significantly change with the transcriptional activity state of that gene. In addition, proteins tend to accumulate more near genes when they are transcriptionally active compared to their inactive state. Furthermore, mathematical simulations informed by seq-DNA-FISH and live-cell imaging data suggest an increase in viscosity around actively transcribing genes, which prolong E-P interaction times (Fig. 7).
Our results find additional support in the model proposed by Zuin et al. (41), who accounted for the nonlinear relationship between E-P interaction frequencies and RNA expression levels through E-P communication–mediated transcriptional bursting. Notably, our findings, with 100 and 200 nm as thresholds, showed average proximity durations of approximately 0.4 and 0.8 s, aligning well with the E-P interaction timescales described by Zuin et al. (41). Despite this consistency, we observed substantial variability in physical distances between genomic regions (fig. S6B). This suggests that several factors, such as the gene region’s higher-order genomic structure, E-P genomic distance, and promoter activity strength, contribute to the lack of a straightforward correlation between transcriptional activation and E-P proximity frequency (Fig. 7) (2–6).
Building on these observations, our findings lend principal support to the transcriptional hub model (7). In our model, changes in genomic proximities and viscosity lead to the formation of hubs approximately 350 nm in size. Our model also accommodates recent observations that reported E-P interactions at distances of less than hundreds of nanometers (Fig. 7) (8).
Our data suggest that E-P proximity frequency alone may not suffice to fully explain functional relationships with transcriptional activity (Figs. 5 and 6). For instance, while deletion of the SE −45 SE significantly reduces Nanog transcription levels, we observed no significant change in interaction frequency between −45 SE and Nanog across different transcriptional states (Fig. 4B) (34). However, our simulations suggest an increase in the duration of interactions between Nanog and its enhancer regions, including −45 SE, in the active state, underscoring the importance of extended interaction times for efficient E-P communication. This has implications for the recruitment of transcription factors and coactivators, as well as Mediator complex assembly (42). The limitations of static methodologies, such as Hi-C and FISH, in capturing these dynamic processes highlight the need for advanced imaging techniques that can provide high-temporal-resolution imaging in multiple colors (<50-ms intervals).
There are several limitations to estimating the dynamic properties of target genomic regions using PHi-C. For instance, our estimation of local viscosity changes using PHi-C relies on data from many fixed cells obtained through seq-DNA-FISH. In addition, the live-cell imaging data used differ from the cell lines used in seq-FISH, and the structural diversity between alleles detected by seq-DNA-FISH is not accounted for. Furthermore, our simulations did not consider the loop extrusion effect by cohesin. Loop extrusion is considered an important driving force in the formation of higher-order chromatin structures in mammals, and many models of higher-order genomic structure incorporate this effect (43). However, several unresolved issues remain regarding the dynamic nature of loop extrusion within the nucleus and its precise role in chromatin organization, making it an active area of research. Therefore, in this study, we did not include the effects of loop extrusion in our simulations. This decision is based on the complexity and current limitations in understanding the dynamic behavior of loop extrusion at the single-cell level. Incorporating loop extrusion into future models will be crucial for more accurately simulating chromatin behavior. We believe that future research integrating loop extrusion effects is necessary to provide a more comprehensive understanding of chromatin organization.
Our simulations aim to capture the broad changes in chromatin dynamics between transcriptionally active and inactive states, using viscosity as a surrogate for the overall landscape of molecular crowding and interactions. However, the model cannot fully account for protein-protein interactions, active transcription, and other intracellular processes that might influence chromatin behavior. While viscosity serves as a useful parameter for capturing general differences in chromatin dynamics, it should not be interpreted as the sole driving factor behind these changes. Nonetheless, it is evident that the behavior of specific genomic regions changes depending on the transcriptional activity state (11, 12), and understanding the causes of such behavioral changes could provide insights into the mechanisms controlling transcriptional bursting.
MATERIALS AND METHODS
Cell lines
The E14tg2a cell line (AES0135, Riken Cell Bank, Japan) was cultured at 37°C and 5% CO2, on either laminin-511 (LN511) (BioLamina, Stockholm, Sweden) or a gelatin-coated dish. Two types of mediums were used: serum/LIF medium, which included Dulbecco’s modified Eagle’s medium (Wako, Osaka, Japan, 197-16275), 15% fetal bovine serum (GE Healthcare, Little Chalfont, UK, SH30396.03), 0.5 mM monothioglycerol solution (Wako, 195-15791), 1× minimum essential medium (MEM) nonessential amino acids (Wako, 139-15651), 2 mM l-alanyl-l-glutamine solution (Wako, 016-21841), LIF (1000 U/ml; Wako, 195-16053), and gentamicin (20 μg/ml; Wako, 078-06061); and 2i medium, which contained the same ingredients as the serum/LIF medium along with 3 μM CHIR99021 (Cayman Chemical, Ann Arbor, MI, USA, 13122) and 1 μM PD0325901 (Chemscene, Monmouth Junction, NJ, USA, CS-0062).
C57BL/6J mouse ES cells (Bruce 4 C57BL/6J, male; EMD Millipore, Billerica, MA) and other knock-in derivatives, including the NGiR cell line (160329-B6-2 GFP/iRFP+4) (10) and NG + iRΔ-190 cell line, were cultured under the same conditions, using either LN511 or a gelatin-coated dish and either serum/LIF or 2i medium.
Plasmids
In the present study, plasmids were engineered using conventional molecular biology techniques, and we used the following three plasmid variants. The sequences of these plasmids can be accessed through the respective URLs provided. For CRISPR-mediated gene editing, two CRISPR vectors were used: eSpCas9-EF-49_10_L1 (available at https://benchling.com/s/seq-nS1bLoWAlYFDGQ2DtaiK?m=slm-pCIkvw6Miu0Vhto7abmZ) and eSpCas9-EF-49_10_R1 (accessible at https://benchling.com/s/seq-McPdsNwm54kANzquOwyv?m=slm-7jSZnlfEwwcawOARrJ7e). In addition, the mTagBFP2 expression vector, pCAG-mTagBFP2, was used (Addgene plasmid #122373; available at https://addgene.org/122373/). pCAG-mTagBFP2 can be sourced from Addgene (Watertown, MA, USA).
Microscopy
Images were acquired using a Nikon Ti-2 microscope with a CSU-W1 confocal unit (Yokogawa), a 100× Nikon Plan Apo λ oil-immersion objective lens (numerical aperture, 1.4), and an iXon Ultra electron-multiplying charge-coupled device (EMCCD, Andor Technology), operated using NIS-Elements software (ver. 5.11.01; Nikon). The microscope was also equipped with 405-, 488-, 561-, and 637-nm lasers (Andor Technology) and an ASI MS-2000 piezo stage (Applied Scientific Instrumentation). Z-stack images spanning 10 μm with 200-nm intervals (51 sections; 130 nm/pixel) were acquired.
Validation of the fluorescent detection method in sequential RNA-FISH
C57BL/6J murine ES cells were seeded at a density of 7.5 × 104 cells in four wells of an eight-well chambered cover glass with #1.5 glass, coated with LN511 (Cellvis, Sunnyvale, CA, USA, C8-1.5HN). Cells were fixed the following day with 4% paraformaldehyde (PFA) for a 10-min incubation. This was followed by three washes with 1× phosphate-buffered saline (PBS) without calcium and magnesium, a permeabilization step for 1 hour at −20°C in 70% ethanol, and air drying for 10 min. Subsequent treatment with 0.5% Triton X-100 in PBS was performed at room temperature for 15 min, after which the cells were washed three times with PBS. Cells were then blocked at room temperature for 15 min in a solution containing PBS, UltraPure bovine serum albumin (BSA; 10 mg/ml; Invitrogen, AM2616), 0.3% Triton X-100, 0.1% dextran sulfate (Nacalai Tesque, Kyoto, Japan, 03879-72), and sheared salmon sperm DNA (0.5 mg/ml; Invitrogen, AM9680). Following blocking, samples were washed with 2× SSC (Nacalai Tesque, Kyoto, Japan, 32146-91).
Hybridization was performed at 37°C for 12 hours in a humidified chamber using seq-RNA-FISH probe sets (see below) and a 10 nM poly-thymine locked nucleic acid (polyT LNA) oligonucleotide (table S1) in 50% hybridization buffer. This buffer consisted of 50% formamide (Wako, 066-02301), 2× SSC, and 10% (w/v) dextran sulfate. Posthybridization, samples were washed at room temperature for 30 min with a 55% wash buffer, followed by three rinses with 4× SSC.
Hybridization with either 100 nM readout probe or a mixture of secondary probe and readout probe was executed in 10% EC buffer [10% ethylene carbonate (Sigma-Aldrich, E26258), 10% dextran sulfate (Sigma-Aldrich, D4911), and 4× SSC] at room temperature for 20 min. The samples were then washed with 4× SSCT (4× SSC and 0.1% Triton X-100) and subsequently with 12.5% wash buffer (12.5% formamide, 2× SSC, and 0.1% Triton X-100) at room temperature for 30 s. Additional staining with 4′,6-diamidino-2-phenylindole (DAPI) solution [4× SSCT solution with DAPI (1:100; Dojindo, 340-07971)] was carried out for 30 s at room temperature. Samples were imaged under a microscope after the addition of an antibleaching buffer [50 mM tris-HCl (pH 8.0), 300 mM NaCl, 2× SSC, 3 mM trolox (Sigma-Aldrich, 238813), 0.8% d-(+)-glucose (Nacalai Tesque, Kyoto, Japan, 16806-25), 1000-fold diluted catalase (Sigma-Aldrich, C3155), and glucose oxidase (0.5 mg ml−1; Sigma-Aldrich, G2133)]. Postimaging, samples were treated for 2 min at room temperature with 55% wash buffer (55% formamide, 2× SSC, and 0.1% Triton X-100) to facilitate stripping, washed twice with 4× SSCT, and reprocessed from hybridization to imaging. Poststripping verification was confirmed by imaging the samples treated with DAPI solution and antibleaching buffer. Probe sequences used in this study are listed in data S1.
Readout and secondary probe design and synthesis
In this study, we generated 1 million 15-base probes using the numpy.random function in Python. From this pool, we selectively isolated those with 5′ ends starting with either adenine (A) or thymine (T) and a guanine-cytosine (GC) content within the range of 40 to 60%. Indices were crafted from human and mouse transcriptome data available from the National Center for Biotechnology Information (NCBI). The generated probes were then mapped to these indices in sequential fashion—first to the human and then to the mouse transcriptome—using Bowtie2 in its default mode (search for multiple alignments, report the best one). Probes failing to map were retained for further analysis. From these, a subset of 5000 was chosen and subjected to BLAST analysis via its web interface, sequentially against the human and mouse transcriptomes to eliminate those demonstrating high complementarity. Further refinement was carried out using blastn in BLAST+ to exclude probes with high complementarity (those matching 10 bases or more). Of the remaining probes, 227 were used as secondary probes and/or readout probes. Each secondary probe was designed to include sequences complementary to the primary probe, in addition to two identical readout probe binding sequences. The secondary probes were custom-synthesized and purified using an oligonucleotide purification cartridge (OPC) column by Eurofins. Readout probes tagged with Alexa 488 and Alexa 647 at their 5′ ends were synthesized by Thermo Fisher Scientific, while those labeled with ATTO565 at the 5′ end was synthesized by Sigma-Aldrich. All readout probes underwent purification through HPLC.
Primary probe design for sequential DNA/RNA-FISH
Target regions were selected on the basis of their interactions with Nanog in mouse ES cells. This was guided by evidence from 4C-seq data (fig. S2A) (21) as well as promoter-capture Hi-C analyses in mouse ES cells (44). Aside from the aforementioned Nanog-interactive regions, other regions of interest (ROIs) were set at approximately 0.5-Mb intervals to comprehensively cover the genome landscape. Furthermore, we also designated 28 regions at intervals of 25 kb. All target regions used for seq-DNA-FISH were cataloged in data S2.
Probe sequences were generated by selecting 35–nucleotide (nt) sections from target regions, focusing specifically on GC content that lies outside the 45 to 65% range. For the design of primary probes, the target genomic region was obtained from the unmasked and repeat-masked GRCm38/mm10 mouse genome FASTA files, downloaded from Ensembl release 102. Probe design was facilitated through Oligominer (https://github.com/beliveau-lab/OligoMiner) (45), using parameters set as “-l 35 -L 35 -g 45 -G 65 -t 37 -s 300 -S 5 -c 5 -C 1.” Subsequently, Bowtie2 was used to identify similar sequences with parameters specified as “-t -k 11 --local -D 20 -R 3 -N 0 -L 19 -i C,1 -S.” Following the initial probe design, alignment to the unmasked mouse genome was performed using Bowtie2 for off-target evaluation. Criteria for off-target hits were specified as any alignment with a minimum of 19 matched bases that resided outside the genomic coordinates of the designated target region. Probes yielding more than 10 off-target matches were excluded. Concurrently, probes were scrutinized against a BLAST database formulated from common repeating sequences in mammals. The FASTA file for “simple repeat” sequences restricted to “mammalia only” was sourced from Repbase (46). Probes with at least 19 matched bases to the repeat index were disregarded.
Subsequent to this, cross-hybridization candidates were eliminated using blastn. Probe pairs displaying at least 19 matched bases were discarded in the final probe selection process. The finalized probe sets were meticulously chosen to preserve probe specificity and to ensure a relatively uniform distribution of probes along the target sequence. Probes located near the central region of the target genomic region received preferential selection. For each genomic locus targeted, a range of 100 to 200 primary probes were used within a single target genomic region to visualize individual loci as diffraction-limited spots, as per seq-DNA-FISH methodology.
The primary probes for seq-DNA-FISH are composed of 35-nt sequences specific to the genomic region, accompanied by four identical 15-nt secondary probe binding sites. In addition, these probes have 20-nt primer binding sites situated at both the 5′ and 3′ termini. The 15-nt secondary probe binding sites are associated with 1 of the 40 sequential rounds used for diffraction-limited spot imaging (Fig. 1A). The target sequences for these probes are listed in data S3.
As fiducial markers, the primary probe targeting the repetitive sequence located in the 3632454L22Rik locus on the X chromosome was used. This fiducial probe is similarly constructed, containing 35-nt genomic-specific sequences, albeit flanked by three distinct 15-nt secondary probe binding sites. These probes also have a pair of 20-nt primer binding sites, identical to the previously described primary probes. The target sequence for the fiducial probe is AAGGAAGCCAGCTGTGGGTAAGGAAGCCAGCTGTG. Phosphorylated 5′ terminus of this fiducial probe was synthesized by Eurofins and purified via HPLC.
For seq-RNA-FISH primary probe design, target candidates were initially selected from among the regions previously defined for seq-DNA-FISH, specifically focusing on those that included a transcription start site. Further refinement was based on gene expression levels; only those regions exhibiting nonnegligible expression were included in the analysis. Expression levels were determined on the basis of CEL-seq2 data from serum/LIF cultured mouse ES cells (10), accessed at https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132591, considering genes with a unique molecular identifier count of 10 or more as suitable candidates for the seq-RNA-FISH analysis. The target sequences were selected referencing PaintSHOP (https://oligo.shinyapps.io/paintshop/) (47). Each targeted RNA was imaged with up to 48 primary probes per target RNA to visualize individual RNAs as diffraction-limited spots. The number of probes could vary for shorter genes, with the minimum being 15. These RNA-specific primary probes range from 30 to 37 nt in length and are flanked by three identical 15-nt secondary probe binding sites. They also contain 20-nt primer binding sites at both the 5′ and 3′ ends. The 15-nt secondary probe binding sites are associated with 1 of 27 sequential rounds for diffraction-limited spot imaging. The targeted sequences for these seq-RNA-FISH probes are listed in data S4.
Primary probe preparation for seq-DNA/RNA-FISH
Primary probes designed for seq-DNA/RNA-FISH, as described in the “Primary probe design for sequential DNA/RNA-FISH” section, were obtained as oligoarray complex pools from Twist Bioscience. These oligo pools served as templates and were amplified in accordance with previously reported methodologies (14). For seq-DNA-FISH primary probes, amplification from the oligo pool was conducted using seqDNA1F and seqDNA1R primers, using KOD One PCR Master Mix (TOYOBO, Japan). The resultant DNA was then purified using DNA Clean and Concentrator (Zymo Research). A second round of PCR amplification was executed using seqDNA2F and seqDNA2R primers, followed by in vitro transcription using the MEGAshortscript Kit (Thermo Fisher Scientific, AM1354). Reverse transcription was facilitated using the seqDNA_revT primer and Thermo Fisher Scientific Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific, EP0751). After reverse transcription, the single-stranded DNA probes were subjected to alkaline hydrolysis with 1 M NaOH at 65°C for 15 min to degrade the RNA templates, subsequently neutralized with 1 M acetic acid, and then ethanol precipitated. These amplified primary probes were resuspended in seq-DNA-FISH primary hybridization buffer, which consisted of approximately 1 nM per probe, 100 nM 3632454L22Rik fiducial marker probe, 40% formamide, 2× SSC, and 10% (w/v) dextran sulfate. If any precipitation was observed, then it was dissolved by heat treatment at 65°C for 15 min. The probes were stored at −20°C until use.
For seq-RNA-FISH primary probes, a similar amplification protocol was followed. However, the initial amplification from the oligo pool used seqRNA1F and seqRNA1R primers, while the second round of PCR used seqRNA2F and seqRNA2R primers. For reverse transcription, the seqRNA1F primer was used. All other procedures were analogous to those used for the seq-DNA-FISH primary probes. The oligonucleotides used are listed in table S1.
DNA-antibody conjugation
Genomic regions interacting specifically with the Nanog promoter region in mouse ES cells were identified as subjects for investigation, analyzed using enrichment analysis in ChIP-Atlas (48), based on findings previously published by de Wit et al. (21). Targets were selected on the basis of low P values and the availability of commercial antibodies not containing BSA or gelatin. A list of targeted antigens and the corresponding antibodies used in this study is provided in table S2.
The preparation of oligonucleotide DNA–conjugated primary antibodies was conducted as previously described (49). Briefly, antibodies (100 μg) underwent buffer exchange to PBS via 50-kDa Amicon Ultra Centrifugal Filters (Millipore, UFC505096) and were treated with 10 equivalents of PEGylated SMCC cross-linker [SM(PEG)2] (Thermo Fisher Scientific, 22102) diluted in anhydrous N,N′-dimethylformamide (Vector Laboratories, S4001005). Following incubation at 4°C for 2 hours, the solution was purified using 7K molecular weight cutoff Zeba Spin Desalting Columns. Concurrently, 300 μM 5′ thiol-modified 18-nt DNA oligonucleotides—composed of an AAA base sequence and a 15-base secondary probe binding sequence (Eurofins)—was reduced with 50 mM dithiothreitol in PBS at room temperature for 2 hours and purified using NAP5 columns (GE Healthcare, 17-0853-01). Maleimide-activated antibodies were then combined with 11 equivalents of the reduced form of the thiol-modified DNA oligonucleotides in PBS and incubated at 4°C overnight. The resulting DNA–primary antibody conjugates were washed four times with PBS and concentrated via 50-kDa Amicon Ultra Centrifugal Filters. The concentrations of the conjugated oligonucleotide DNA and antibodies were quantified using a BCA Protein Assay Kit (Thermo Fisher Scientific 23225) and NanoDrop (Thermo Fisher Scientific).
Functional validation of the conjugated antibodies was confirmed through SDS–polyacrylamide gel electrophoresis and Coomassie brilliant blue staining, which demonstrated the expected reduction in electrophoretic mobility due to oligonucleotide conjugation. Additional verification was conducted through immunofluorescence, using fluorophore-conjugated secondary antibodies that recognize the host species of each antibody; successful nuclear staining affirmed the functionality of the oligonucleotide-labeled antibodies.
To achieve vendor-recommended concentrations for application, a mixture of 20 distinct antibodies was combined in a 1.5-ml tube. Subsequently, this mixture was brought to a final volume of 500 μl using PBS. DNA-primary antibodies were buffer-exchanged to PBS using 50-kDa Amicon Ultra Centrifugal Filters. Concentrations were verified using a BCA Protein Assay Kit and NanoDrop. These preparations were stored at −80°C until use.
seq-DNA/RNA/IF-FISH
Glass-bottom six-well plates (Cellvis, P06-1.5H-N) were equipped with cell culture inserts (ibidi, ib80209). Only the area within the cell culture inserts was treated with poly-d-lysine (Sigma-Aldrich, P6407) at a concentration of 0.1 mg/ml for 60 min. Following the treatment, the plates were air-dried overnight. The inserts were subsequently coated with LN511 at 37°C for 1 hour. E14tg2a ES cells, precultivated in either serum/LIF or 2i media, were seeded at a density of 2 × 104 cells per insert and incubated at 37°C with 5% CO2. The next day, cells were fixed with 4% PFA for 10 min, followed by three washes with PBS. Supernatants were discarded, and cells were treated with 70% ethanol at −20°C for 1 hour and air-dried for 10 min. Custom HybriWell Sealing Systems (Grace Bio Labs, Custom HBW13 FL_1, 13 mm in inner diameter × 0.25 mm in depth, 25 mm × 28 mm outer dimensions, 1.5-mm ports, Adhesive A12) were applied at this stage.
Cell permeabilization was initiated with a 15-min treatment of 0.5% Triton X-100 in PBS at room temperature, followed by three PBS washes. Blocking was carried out for 15 min at room temperature using a blocking solution that included UltraPure BSA (10 mg/ml; Invitrogen, AM2616), 0.3% Triton X-100, 0.1% dextran sulfate (Nacalai Tesque, 03879-72), and sheared salmon sperm DNA (0.5 mg/ml; Invitrogen, AM9680). DNA oligonucleotide–conjugated primary antibodies (table S2) were incubated in this blocking solution, with 100-fold diluted SUPERase In RNase Inhibitor (Invitrogen, AM2694), at 4°C overnight.
Subsequent to three 10-min washes with PBS, the samples underwent postfixation with freshly prepared 4% PFA in PBS for 5 min at room temperature. This was followed by six washes with PBS and an additional postfixation step using 1.5 mM BS(PEG)5 (Thermo Fisher Scientific, A35396) in PBS for 20 min at room temperature, quenched by 100 mM tris-HCl (pH 7.5) for 5 min. After washing with PBS, the samples were rinsed with 2× SSC (Nacalai Tesque, 32146-91). For hybridization, seq-RNA-FISH probe sets (1 nM each per probe) and a 10 nM polyT LNA oligonucleotide (table S1) were combined in a 50% hybridization buffer consisting of 50% formamide (Wako, 066-02301), 2× SSC, and 10% (w/v) dextran sulfate. The hybridization process was carried out at 37°C for 48 hours in a humidified chamber. To prevent dehydration, the port seals accompanying the HybriWell Sealing System were applied. Following hybridization, samples were rinsed with a 55% wash buffer at room temperature for 30 min and then washed three times with 4× SSC. For imaging, please refer to the “seq-DNA/RNA/IF-FISH imaging” section.
Following seq-RNA-FISH imaging, the specimens underwent a preparation process for seq-DNA-FISH primary probe hybridization. Initially, the specimens were washed using PBS and then subjected to a 1-hour incubation at 37°C in RNase A/T1 Mix (Thermo Fisher Scientific, EN0551), diluted 100 times. Subsequent to this step, the samples were subjected to three rinses in PBS and an additional three washes in a 50% denaturation buffer containing 50% formamide and 2× SSC; this was followed by a 15-min incubation at room temperature. The samples were then exposed to a 90°C heat treatment for 4.5 min in the 50% denaturation buffer while sealing the inlet and outlet of the custom chamber with accompanying seals from the HybriWell Sealing System.
Postheating, the samples were again rinsed with 2× SSC. The seq-DNA-FISH primary hybridization buffer, comprising approximately 1 nM per probe, a 100 nM 3632454L22Rik fiducial marker probe, 40% formamide, 2× SSC, and 10% (w/v) dextran sulfate, was applied and incubated at 37°C for 72 hours within a humidified chamber. To prevent dehydration, the HybriWell Sealing System’s holes were sealed with the provided port seals. Subsequent to the hybridization, the samples were washed with a 40% wash buffer containing 40% formamide, 2× SSC, and 0.1% Triton X-100, at room temperature for 15 min. This was followed by three additional washes in 4× SSC. Thereafter, the samples underwent further processing to “padlock” the primary probes. For this purpose, a 31-nt global ligation bridge (final concentration: 100 nM; table S1) was hybridized in a 20% hybridization buffer, composed of 20% formamide, 10% dextran sulfate (Sigma-Aldrich, D4911), and 4× SSC, at 37°C for 2 hours. Following this, the samples were washed three times for a total of 15 min with a 10% wash buffer, consisting of 10% formamide, 2× SSC, and 0.1% Triton X-100. The specimens were then incubated with a 20-fold diluted Quick Ligase in 1× Quick Ligase Reaction Buffer from the Quick Ligation Kit (New England Biolabs, M2200), supplemented with an additional 1 mM adenosine triphosphate (Takara Bio, 4041), at room temperature for 1 hour. This step was designed to facilitate the ligation reaction between the 5′ and 3′ ends of the seq-DNA-FISH primary probes. The samples were then subjected to a series of washes and incubations, including fixation and amine modification steps, to further stabilize the primary probes. Imaging for seq-DNAFISH and seq-IF-FISH was carried out as described in the subsequent section (see the “seq-DNA/RNA/IF-FISH imaging” section).
seq-DNA/RNA/IF-FISH imaging
The sequential imaging procedure was carried out on the basis of the methodology previously reported by Takei et al. (14). The fluidic delivery system used for this process also adhered to prior research (25). Briefly, an automated fluidic delivery system was constructed, comprising two multichannel fluidic valves (IDEX Health & Science, EZ1213-820-4) and a Hamilton syringe pump (Hamilton Company, 63133-01). Integration of the fluidic valves and syringe pump with homemade connectors, as well as the coordination with microscope imaging, was managed via a custom Python script. Samples were mounted on the microscope, and tubes were positioned to allow the flow of solutions and waste fluids into the apertures of HybriWell seals. A volume of 500 μl of DAPI solution was flowed through the system to stain cell nuclei, followed by the capture of 30 to 50 fields of view (FOVs). Subsequently, a 400-μl mixture of 100 nM secondary and readout probes was flowed through in 10% EC buffer (containing ethylene carbonate, dextran sulfate, and 4× SSC) and hybridized at room temperature for 20 min. This was followed by a brief room temperature wash, using 500 μl of 4× SSCT (4× SSC and 0.1% Triton X-100) and 500 μl of 12.5% wash buffer (12.5% formamide, 2× SSC, and 0.1% Triton X-100). Another 500 μl of DAPI solution (4× SSCT solution with DAPI at a 1:100 dilution; Dojindo, 340-07971) was flowed and incubated for 30 s at room temperature. An antibleaching buffer [50 mM tris-HCl (pH 8.0), 300 mM NaCl, 2× SSC, 3 mM trolox (Sigma-Aldrich, 238813), 0.8% d-(+)-glucose (Nacalai Tesque, 16806-25), 1000-fold diluted catalase (Sigma-Aldrich, C3155), and glucose oxidase (0.5 mg/ml; Sigma-Aldrich, G2133)] was then flowed through the system, followed by imaging 150 s later. Postimaging, 1.4 ml of 55% wash buffer (55% formamide, 2× SSC, and 0.1% Triton X-100) was applied for a 2-min stripping operation at room temperature. The samples were subsequently washed twice with 4× SSCT. These procedures were repeated for each set of readout and secondary probes. In the final round of seq-RNA-FISH, images were captured using readouts against polyT LNA oligonucleotide to stain all RNA. After seq-RNA-FISH, samples were removed from the microscope for further experimental procedures preceding seq-DNA-FISH imaging (refer to the “seq-DNA/RNA/IF-FISH” section). For seq-DNA-FISH and seq-IF-FISH imaging, FOVs previously captured during seq-RNA-FISH were used. During seq-DNA-FISH, a mixture of secondary and readout probes for fiducial marker staining (comprising three types: Alexa488, ATTO565, and Alexa647) was each added at 50 nM. After seq-DNA-FISH imaging, to avoid contamination with fiducial markers, the samples underwent another stripping procedure, and the fluidic delivery system was cleaned. If noticeable shifts in imaging position occurred, additional rounds were executed. At the end of the day, corrections were made for any nuclear deformation and misalignments caused by fluidic delivery.
Establishment of a cell line with deletion of the −190-kb region
In each well of 24-well gelatin-coated plates (Nunc), we plated 5 × 105 C57BL6J ES cells with knocked-in mNeonGreen (GFP) and iRFP670 (iRFP) in their respective Nanog alleles (referred to as NGiR) in 0.5 ml of 2i medium. Cells were cultured for 1 hour. Transfection complexes were prepared as follows: In one 1.5-ml tube, 25 μl of opti-MEM, 1 μl of P3000 reagent, 500 ng of eSpCas9-EF-49_10_L1, 500 ng of eSpCas9-EF-49_10_R1, and 300 ng of pCAG-mTagBFP2 (Addgene plasmid #122373) were combined. In another tube, 25 μl of opti-MEM and 1.8 μl of Lipofectamine 3000 were mixed well. The contents of both tubes were combined and incubated for 15 min before being added to the plated cells. The cells were cultured overnight, defining this day as day 0. The medium was replaced on day 1. On day 2, an approximate 1000 blue fluorescent protein (BFP)-positive cells, presumed to be successfully transfected, were sorted using a FACSAria III cell sorter (BD Biosciences, Franklin Lakes, NJ, USA) and plated on a 6-cm dish. The medium was replaced with fresh 2i medium on day 4. On day 7, 24 colonies were picked for downstream analysis and verification of gene targeting. PCR was performed using Δ-190_gPCR-F1 and Δ-190_gPCR-R1 primers. No cell lines with deletions on both alleles were obtained; thus, candidate lines presumed to have deletion in a single allele were selected for further analysis. For additional verification, regions surrounding the upstream and downstream CRISPR target sequences were amplified and sequenced using the following primers: Δ-190_gPCR-F2 and Δ-190_gPCR-R3 for the upstream region and Δ-190_gPCR-F3 and Δ-190_gPCR-R2 for the downstream region. In addition, the deleted region was amplified and sequenced using Δ-190_gPCR-F2 and Δ-190_gPCR-R2 primers through Sanger sequencing. The linkage between the deleted −190-kb region and the Nanog-iRFP knock-in allele was confirmed through smRNA-FISH using GFP/iRFP probes, as well as DNA-FISH using a probe specific to the -190-kb region. The confirmed cell line was designated as NG + iRΔ-190.
Single-molecule RNA FISH
Both NGiR and NG + iRΔ-190 cell lines were seeded onto eight-well chambered cover glasses coated with LN511 at a density of 7.5 × 104 cells per well in 2i media. The following day, cells were fixed by a 10-min incubation with 4% PFA and subsequently washed three times with PBS. The samples were then permeabilized in 70% ethanol at −20°C for 1 hour and air-dried for 10 min. For cellular membrane disruption, samples were treated with 0.5% Triton X-100 in PBS at room temperature for 15 min and washed again three times with PBS. Cells were blocked for 15 min at room temperature using a blocking solution containing PBS, UltraPure BSA (10 mg/ml; Invitrogen, AM2616), 0.3% Triton X-100, 0.1% dextran sulfate (Nacalai Tesque, Kyoto, Japan, 03879-72), and sheared salmon sperm DNA (0.5 mg/ml; Invitrogen, AM9680). Postblocking, the samples were rinsed with 2× SSC (Nacalai Tesque, Kyoto, Japan, 32146-91). Probes for mNeonGreen (GFP) and iRFP670 conjugated with CAL Fluor Red 590 and Quasar 670 (10), along with a 10 nM polyT LNA oligonucleotide, were hybridized in 30% hybridization buffer. The hybridization was carried out at 37°C for 12 hours in a humidified chamber. Following hybridization, samples were washed with a 35% wash buffer (35% formamide, 2× SSC, and 0.1% Triton X-100) at room temperature for 30 min and subsequently rinsed three times with 4× SSC. The cells were then stained for 30 s at room temperature with a DAPI staining solution (4× SSCT solution with DAPI at 1:100 dilution; Dojindo, 340-07971). Last, an antibleaching buffer containing 50 mM tris-HCl (pH 8.0), 300 mM NaCl, 2× SSC, 3 mM trolox (Sigma-Aldrich, 238813), 0.8% d-(+)-glucose (Nacalai Tesque, Kyoto, Japan, 16806-25), 1000-fold diluted catalase (Sigma-Aldrich, C3155), and glucose oxidase (0.5 mg/ml; Sigma-Aldrich, G2133) was added before imaging via microscopy.
Preprocessing of seq-DNA/RNA/IF-FISH imaging data
Imaging data from seq-DNA/RNA/IF-FISH were initially processed using Fiji software. Subsequently, each FOV was chronologically organized along the time axis. To correct uneven background illumination, a dark image subtraction was performed. To prevent pixel values from becoming zero, a constant value of 1 was added to the entire image set. Flat-field correction was applied to each fluorescence image by dividing the images by the normalized background illumination while retaining the intensity profiles of fluorescent spots. Subsequently, drift correction was executed on the basis of the DAPI images using ImageJ’s “Correct 3D drift” function (parameters: edge_enhance only = 0, lowest = 1, highest = 51, max_shift_x = 50, max_shift_y = 50, and max_shift_z = 30). Hyperstack images were subjected to minimum intensity projection along the z and time dimensions, followed by cropping to exclude xy regions containing zero values. Similarly, the hyperstack images were processed in the x and time dimensions to ensure the exclusion of yz regions containing zero values. Additional drift correction was performed based on DAPI images using Fiji’s “descriptor-based series registration (2d/3d + t)” (parameters: series_of_images = stack, brightness_of = Low, approximate_size = [5 px], type_of_detections = [Minima & Maxima], subpixel_localization = [3-dimensional quadratic fit], transformation_model = [Affine (3d)], images_are_roughly_aligned, number_of_neighbors = 3, redundancy = 1, significance = 3, allowed_error_for_ransac = 5, global_optimization = [All against first image (no global optimization)], range = 5, choose_registration_channel = 1, image = [Fuse and display], and interpolation = [Linear Interpolation]). Henceforth, voxel dimensions are x:y:z = 130:130:130 nm. At this point, only seq-DNA-FISH images detected with a 647-nm readout probe were extracted and subjected to average intensity projection along the time axis. Given that all images contain fiducial markers, this procedure emphasizes these fiducial regions, saved as Fiducial_enhance_stack.tif. Subsequently, maximum intensity projection was applied along the time axis to the 647-nm seq-DNA-FISH images, thereby generating an overlaid image of the detected foci, saved as seqDNAFISH_647_foci_enhance_stack.tif. Postdrift correction images were saved separately for each FOV, capture timing, and color channel.
Nuclear segmentation of seq-DNA/RNA/IF-FISH data
3D segmentation of the cell nuclei was performed using Cellpose (version 0.6.5) (50). Initially, nuclear images were opened and resampled to reduce the xy resolution by a factor of one-fifth. These images were further subjected to Gaussian blurring with an SD of 1.5 pixels. Using these preprocessed images, 3D segmentation was carried out using Cellpose. The original xy resolution was restored, and a series of morphological operations was executed: erosion with a radius of 1, dilation with a radius of 2, opening with a radius of 8, and another dilation with a radius of 4. These processed images were saved as ROIs. These provisional ROIs were then visually verified and further refined manually using Napari (DOI: 10.5281/zenodo.3555620). Nuclei touching the xy boundaries of the image were excluded from the analysis.
seq-DNA-FISH analysis
For the detection of fluorescent spots in seq-DNA-FISH, we used Big-FISH (version 0.6.2) (51). Fluorescent spots were initially detected using the “detection.detect_spots” function, with parameters set as follows: “return_threshold = True” and “voxel_size” and “spot_radius” both set to (130, 130, 130) and (240, 189, 189), respectively. Subsequent to this initial detection, subpixel fitting was executed using the “detection.fit_subpixel” function, maintaining the same voxel_size and spot_radius parameters. It is noteworthy that any detected spots not falling within the nuclear regions as delineated in the “Nuclear segmentation of seq-DNA/RNA/IF-FISH data” section were excluded from further analysis. Moreover, any spots with fluorescence intensities ranking outside the top 10 within individual cells were also omitted from the analysis. Subsequently, Fiducial_enhance_stack.tif was used for spot identification via Big-FISH. All detected spots within a 3-pixel radius were considered as fiducial markers. In different channel images, fiducials in close proximity (within 2 pixels) were associated. However, if two fiducial candidate spots in the same channel were located within 3 pixels of each other, both were excluded from consideration as fiducial markers due to potential calibration difficulties. The 647-nm channel was considered the fiducial, and the offset between fiducial marker candidates in the same capture series across three channels was calculated. Using the chromatic offset information from the closest neighboring fiducial marker, other spot positions were adjusted. These spots were subsequently verified using Napari, and any that were unequivocally identified as fiducial markers were excluded. Spot intensities were normalized to the median nuclear intensity, and spots below a value of 1.1 were omitted from analysis. Subsequently, only the top four brightest spots within each channel and each cell during each capture were selected for analysis, excluding the rest. Further, we applied aligner.find_all_chr from Jia et al. (52) to assess whether the fluorescent spots were located on the same chromosome, using parameters: cell_pts_input = cell_pts_input, gene_dist = gene_dist.values.astype(‘int’), bin_size = bin_size, nm_per_bp = 0.34, pixel_dist = 130.0, num_skip = 5, total_num_skip_frac = 0.8, norm_skip_penalty = false, stretch_factor = 1.01, init_skip_frac = 0.15, lim_init_skip = False, and max_iter = 2. For chromosomes appearing to have swapped connections in proximity, correction was performed by determining two centroids using k-means [KMeans(n_clusters = 2)], and chromosomes were reassigned to the nearest centroid. From the assigned fluorescent spots, median xyz coordinates were calculated for each chromosome. Distances from these median coordinates to each spot were computed. Outliers exceeding the threshold of average distance + 2 × SD (34.9 pixels) were excluded from the analysis. Target genomic regions with detection efficiency below half of the overall median detection efficiency were also excluded (fig. S3A). Furthermore, in the median distance matrix of replicate 1, data corresponding to three regions that displayed patterns distinctly divergent from those of replicate 2 were excluded from the analysis (fig. S3A). The finalized data of the seq-DNA-FISH fluorescent spots are listed in data S5.
seq-RNA-FISH analysis
For the spot detection in preprocessed seq-RNA-FISH images, we used Big-FISH. The analysis was conducted using the detection.detect_spots function with the parameters: return_threshold = True, voxel_size = (130, 130, 130), and spot_radius = (240, 189, 189). While the automatically determined threshold was often deemed sufficient, manual adjustments were made when necessary. On the basis of the set threshold, fluorescent spots were identified. Further, transcriptional foci were discerned using the detection.decompose_dense and detection.detect_clusters functions with a minimum requirement of three spots (nb_min_spots = 3). Subsequently, the presence or absence of these transcriptional foci was determined on the basis of the coordinates of the gene vicinity as ascertained through seq-DNA-FISH analysis. If the distance from the median coordinates of the specific chromosome’s seq-DNA-FISH spots was within 30 pixels, then transcription was considered to have occurred. The finalized transcriptional foci data are listed in data S6.
seq-IF-FISH analysis
Using the nuclear ROI determined through the “Nuclear segmentation of seq-DNA/RNA/IF-FISH data” section, we calculated the mean fluorescence intensity within cell nuclei. Only proteins/PTMs with a relative intensity value exceeding 1.5 when compared to the same channel’s negative control—captured without the addition of a readout probe and a secondary probe—were considered for analysis (fig. S8, B and C). Notably, histone deacetylase 3 and SMAD4, which exhibited localization patterns divergent from those typically obtained through general immunofluorescence, were excluded from the study (fig. S8, A and C).
In 1-Mb intervals, we examined the correlation between ChIP-seq enrichment levels and fluorescence intensity data obtained from seq-IF-FISH. Initially, 55 intervals of 1 Mb each were designated starting at chr6:94,672,507. However, segments numbered 9, 11, 16, 18, and 36 were excluded as the corresponding seq-DNA-FISH data were ultimately not analyzed (fig. S3A). Using genomeCoverageBed, ChIP-seq enrichment levels were calculated in terms of reads per million. The pertinent Bigwig files are listed in table S3. Within the seq-IF-FISH images, the mean fluorescence intensity was calculated in a spherical region with a radius of 3 pixels, centered on the 3D coordinates of specific genome regions as determined by seq-DNA-FISH analysis. When multiple seq-DNA-FISH targets were included within the predefined segments, this mean value was used. From these parameters, we calculated the z score as (mean fluorescence intensity of the sphere − mean nuclear intensity)/SD of nuclear intensity.
We generated the median intensity projection images of seq-IF-FISH centered at specific seq-DNA-FISH foci and calculated the radial distribution functions of fluorescence intensity from seq-IF-FISH projection images. First, we used the seq-DNA-FISH spot coordinates corresponding to the target locus to extract the relevant seq-DNA-FISH, seq-RNA-FISH, and seq-IF-FISH images in either the active or inactive gene state. The images were cropped to a 51 × 51–pixel area centered on the seq-DNA-FISH spots at a specific z focal plane. Next, we computed median intensity projection images separately for the active and inactive states from these cropped images. In addition, 1200 randomly selected images from the nucleus were cropped similarly, and median intensity projection images were generated. Subsequently, radial distribution functions were calculated using these median intensity projection images. The radial distribution functions were generated by calculating the average fluorescence intensity along radii extending from the center of the images [using the RadialProfile class from Astropy/Photutils 1.13.0 with default settings: astropy/photutils: 1.13.0 (zenodo.org)].
For the detection of H3K27ac foci, Big-FISH was used. Spots were identified using the detection.detect_spots function with parameters: threshold = 40, voxel_size = (130, 130, 130), and spot_radius = (200, 200, 200). Subpixel fitting was performed on these detected spots using the detection.fit_subpixel function, also with parameters: voxel_size = (130, 130, 130) and spot_radius = (200, 200, 200). The distance to the 3D coordinates of each target, as determined by seq-DNA-FISH analysis, was calculated. The entity closest to a specific genome coordinate was classified as the nearest clusters. The finalized H3K27ac foci data are listed in data S7.
Odds ratio calculation
To assess the association between transcriptional activity and the proximity of specific DNA regions, we computed the odds ratio using the formula: odds ratio = (ad)/(bc). Here, proximity is defined as occurring when the distance between two genomic regions is below a specific threshold (350 nm for 25-kb resolution data and 950 nm for 0.5-Mb resolution data). In this equation, a indicates the number of cells exhibiting both proximity and transcription, b is the number of cells with proximity but no transcription, c is the number of cells with transcription but no proximity, and d is the count of cells where neither transcription nor proximity is observed. Bootstrap methods were used to determine the 95% confidence intervals for these odds ratios. A genomic region was considered proximal to a gene-containing region if it was within 350 nm for data at a 25-kb resolution, and within 950 nm for a 0.5-Mb resolution, from the gene’s promoter.
The same formula was applied to investigate the relationship between the presence of H3K27ac foci and their proximity to two distinct genomic locus pairs, with proximity defined as being within 350 nm for 25-kb resolution data.
A/B compartment calculation
First, we used the .hic formatted file (4DNFI4OUMWZ8 in the 4DN data portal) from in situ Hi-C experiments on mouse ES cells (24). Next, using Straw (53), we extracted the Knight-Ruiz (KR)-normalized observed/expected matrix for chromosome 6 at 25-kb resolution. We then calculated the first eigenvector of the log2 ratio of the observed-to-expected matrix. Last, we flipped the eigenvector profile to ensure a positive correlation coefficient with the gene density profile (54).
Modeling of higher-order genomic structural dynamics using PHi-C
On the basis of seq-DNA/RNA-FISH data, we can generate the proximity frequency map with the 350-nm distance threshold according to a specific gene’s transcriptionally active/inactive state. We analyzed the genomic regions of Nanog and Sox2 from our data and multiplexed error-robust fluorescence in situ hybridization (MERFISH) data (40) with 25- and 5-kb resolutions, respectively. Then, we input the generated proximity frequency data into the PHi-C2 pipeline (38). First, the PHi-C pipeline reconstructs a proximity frequency map through optimization of the parameters of the PHi-C polymer model. Here, we used the default input parameters of the PHi-C2 optimization command. The output proximity frequency maps were in good agreement with the inputs (Figs. 5B and 6A and fig. S11).
Then, the PHi-C pipeline allows for calculating structure and dynamics using the optimized parameters of the PHi-C polymer model. However, the spatial and temporal scales of the polymer model are normalized in the units. To recover the model in actual spatial and temporal scales, we need two parameters: the proximity distance σ (meters) and the friction coefficient of a polymer segment γ (kilograms per second). We set σ = 350 (nanometers). To obtain the value of γ in active and inactive states, we used the MSD data of Nanog and Sox2 (11). First, we multiplied the MSD values by 1.5 to convert from two dimensions to three dimensions. Then, we fitted the value of γ using the following theoretical MSD of the PHi-C model (37)
where t represents the time, kB is the Boltzmann constant [= 1.380649 × 10−23 (m2 kg s−2 K−1)], T means the temperature set as 310 (K), Q is the orthogonal matrix of the optimized Laplacian matrix of the PHi-C polymer model, is the normalized eigenvalue of the pth mode, n is the matrix index of Nanog or Sox2 region, and ε represents the estimation error in position determination (55) and is also a fitted parameter. We used a SciPy function (scipy.optimize.curve_fit) in the fitting.
Analysis of duration time between E-P regions in PHi-C dynamic simulations
We obtained polymer models consistent with structures and dynamics in actual spatial and temporal scales based on the above PHi-C modeling to the proximity frequency matrices from seq-DNA/RNA-FISH data. Then, we calculated the duration times for which E-P regions (Nanog and −45 SE, 60 SE, −190 kb; Sox2 and SE) are close in PHi-C dynamic simulations (Figs. 5, D to G, and 6). In the PHi-C2 dynamics command, we set the step size parameter so that the integration time step corresponds to actual time Δt = 0.1 (s):kBTΔt /(γσ2), where γ is the fitted friction coefficient and σ = 350 (nm). In addition, we numerically integrated 105 steps corresponding to 104 s, for 100 different initial conformations. Last, we measured the duration times that E-P regions are in proximity threshold distances of 100, 200, and 350 (nm). First, we estimated the probability density of the duration times by the following two-component exponential model: . Here, τ1 and τ2 (>τ1) represent the characteristic duration times between E-P regions. Then, we calculated the average duration time by .
Visualization of PHi-C dynamic simulations
In the visualization (movies S2 and S3), we fixed the center of mass of polymer conformations to the origin and calculated the dynamics and distance maps within 50 s. We used Visual Molecular Dynamics (VMD) to visualize polymer dynamics (56).
Statistical analysis
The exact number, n, of data points and their representation (such as cells and independent experiments) and statistical tests used are indicated in the respective figure legends and in Results. All experiments were performed as two or more independent experiments. The same conclusions were obtained from each experiment. Statistical tests were performed in R software (The R Project for Statistical Computing, Vienna, Austria) and Python (Python Software Foundation, https://python.org/).
Acknowledgments
We thank Y. Ochiai for providing technical assistance. We extend our heartfelt gratitude to I. Hiratani (RIKEN, Japan) and H. Miura (RIKEN, Japan) for invaluable discussions about seq-DNA-FISH analysis. We also thank L. Cai (Caltech) for providing insights into the automation of seq-DNA/RNA/IF-FISH and S. Tsuda (Ansanga Lab) for indispensable support in the fabrication and control of the automation apparatus. In addition, we appreciate the critical feedback and insightful comments on the manuscript provided by M. Francois (University of Sydney) and I. Solovei (Ludwig-Maximilians-University of Munich), which greatly enriched the content of our work.
Funding: This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology to H.Oc. (JP21H05753, JP22H02609, JP22H04694, and JP24H02326), to H.Oh. (JP22K15084), to S.S. (JP23H04297), and to Y.O. (JP18H05527 and JP24H02323); CREST from the Japan Science and Technology Agency to H.Oc. and S.S. (JPMJCR23N3); NIG-JOINT to H.Oc. (76A2023, 15R2023, 69A2024, and 14R2024); the Medical Research Center Initiative for High Depth Omics, Kyushu University, Japan to H.Oc., H.Oh., and Y.O.; MEXT Promotion of Development of a Joint Usage/Research System Project: the Cooperative Research Project Program to H.Oc., H.Oh., and Y.O.; MEXT Promotion of Development of a Joint Usage/Research System Project: Coalition of Universities for Research Excellence Program (CURE) Grant Number: JPMXP1323015486 to H.Oc., H.Oh., and Y.O.; the RIKEN-Hiroshima University Science and Technology Hub Collaborative Research Program to H.Oc. and S.S.; the Takeda Science Foundation to H.Oc.; and The Mitsubishi Foundation to H.Oc.
Author contributions: Conceptualization: H.Oc. Methodology: H.Oh., S.S., H.Ow., T.F., K.H., S.O., T.Y., Y.O., and H.Oc. Investigation: H.Oh., S.S., H.Ow., and H.Oc. Visualization: H.Oh., S.S., and H.Oc. Funding acquisition: H.Oh., S.S., Y.O., and H.Oc. Project administration: H.Oc. Supervision: H.Oc. Writing—original draft: H.Oc. Writing—review and editing: H.Oh., S.S., Y.O., K.H., and H.Oc.
Competing interests: K.H. participated in this research while affiliated with Ansanga Lab, Suita, Japan. Now, his primary affiliation is with the Center for Information and Neural Networks (CiNet), National Institute of Information and Communications Technology (NICT), Suita, Osaka, Japan. In addition, he holds positions at the Life and Medical Sciences Area, Health Sciences Discipline, Kobe University, Kobe, Hyogo, Japan and the RIKEN Center for Biosystems Dynamics Research, Suita, Osaka, Japan. His involvement with Ansanga Lab is now in a secondary capacity. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Publicly available software and packages were used in this study.
Supplementary Materials
The PDF file includes:
Figs. S1 to S12
Tables S1 to S3
Legends for movies S1 to S3
Legends for data S1 to S7
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
Data S1 to S7
REFERENCES AND NOTES
- 1.Chen H., Levo M., Barinov L., Fujioka M., Jaynes J. B., Gregor T., Dynamic interplay between enhancer–promoter topology and gene activity. Nat. Genet. 50, 1296–1303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander J. M., Guan J., Li B., Maliskova L., Song M., Shen Y., Huang B., Lomvardas S., Weiner O. D., Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8, e41769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benabdallah N. S., Williamson I., Illingworth R. S., Kane L., Boyle S., Sengupta D., Grimes G. R., Therizols P., Bickmore W. A., Decreased enhancer-promoter proximity accompanying enhancer activation. Mol. Cell 76, 473–484.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heist T., Fukaya T., Levine M., Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 116, 15062–15067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinola S. M., Götz M., Bellec M., Messina O., Fiche J.-B., Houbron C., Dejean M., Reim I., Gizzi A. M. C., Lagha M., Nollmann M., Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 53, 477–486 (2021). [DOI] [PubMed] [Google Scholar]
- 6.L. I. G. Acuña, I. Flyamer, S. Boyle, E. T. Friman, W. A. Bickmore, Transcription decouples estrogen-dependent changes in enhancer-promoter contact frequencies and spatial proximity. bioRxiv 534720 [Preprint] (2023); https://biorxiv.org/content/10.1101/2023.03.29.534720. [DOI] [PMC free article] [PubMed]
- 7.Lim B., Levine M. S., Enhancer-promoter communication: Hubs or loops? Curr. Opin. Genet. Dev. 67, 5–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barshad G., Lewis J. J., Chivu A. G., Abuhashem A., Krietenstein N., Rice E. J., Ma Y., Wang Z., Rando O. J., Hadjantonakis A.-K., Danko C. G., RNA polymerase II dynamics shape enhancer–promoter interactions. Nat. Genet. 55, 1370–1380 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez J., Larson D. R., Transcription in living cells: Molecular mechanisms of bursting. Annu. Rev. Biochem. 89, 189–212 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Ochiai H., Hayashi T., Umeda M., Yoshimura M., Harada A., Shimizu Y., Nakano K., Saitoh N., Liu Z., Yamamoto T., Okamura T., Ohkawa Y., Kimura H., Nikaido I., Genome-wide kinetic properties of transcriptional bursting in mouse embryonic stem cells. Sci. Adv. 6, eaaz6699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohishi H., Shimada S., Uchino S., Li J., Sato Y., Shintani M., Owada H., Ohkawa Y., Pertsinidis A., Yamamoto T., Kimura H., Ochiai H., STREAMING-tag system reveals spatiotemporal relationships between transcriptional regulatory factors and transcriptional activity. Nat. Commun. 13, 7672 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochiai H., Sugawara T., Yamamoto T., Simultaneous live imaging of the transcription and nuclear position of specific genes. Nucleic Acids Res. 43, e127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandereyken K., Sifrim A., Thienpont B., Voet T., Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takei Y., Yun J., Zheng S., Ollikainen N., Pierson N., White J., Shah S., Thomassie J., Suo S., Eng C.-H. L., Guttman M., Yuan G.-C., Cai L., Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takei Y., Zheng S., Yun J., Shah S., Pierson N., White J., Schindler S., Tischbirek C. H., Yuan G.-C., Cai L., Single-cell nuclear architecture across cell types in the mouse brain. Science 374, 586–594 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Su J.-H., Zheng P., Kinrot S. S., Bintu B., Zhuang X., Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182, 1641–1659.e26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateo L. J., Murphy S. E., Hafner A., Cinquini I. S., Walker C. A., Boettiger A. N., Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner A., Boettiger A., The spatial organization of transcriptional control. Nat. Rev. Genet. 24, 53–68 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Feng C., Wang J., Chu X., Large-scale data-driven and physics-based models offer insights into the relationships among the structures, dynamics, and functions of chromosomes. J. Mol. Cell Biol. 15, mjad042 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochiai H., Sugawara T., Sakuma T., Yamamoto T., Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci. Rep. 4, 7125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit E., Bouwman B. A. M., Zhu Y., Klous P., Splinter E., Verstegen M. J. A. M., Krijger P. H. L., Festuccia N., Nora E. P., Welling M., Heard E., Geijsen N., Poot R. A., Chambers I., de Laat W., The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature 501, 227–231 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Bintu B., Mateo L. J., Su J.-H., Sinnott-Armstrong N. A., Parker M., Kinrot S., Yamaya K., Boettiger A. N., Zhuang X., Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn E. H., Pegoraro G., Brandão H. B., Valton A.-L., Oomen M. E., Dekker J., Mirny L., Misteli T., Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonev B., Cohen N. M., Szabo Q., Fritsch L., Papadopoulos G. L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A., Cavalli G., Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah S., Takei Y., Zhou W., Lubeck E., Yun J., Eng C.-H. L., Koulena N., Cronin C., Karp C., Liaw E. J., Amin M., Cai L., Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabriele M., Brandão H. B., Grosse-Holz S., Jha A., Dailey G. M., Cattoglio C., Hsieh T.-H. S., Mirny L., Zechner C., Hansen A. S., Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science 376, 496–501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Zhang Z., Li L., Chen B.-C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E., Tjian R., Liu Z., Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh T.-H. S., Cattoglio C., Slobodyanyuk E., Hansen A. S., Darzacq X., Tjian R., Enhancer–promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat. Genet. 54, 1919–1932 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolodziejczyk A. A., Kim J. K., Tsang J. C. H., Ilicic T., Henriksson J., Natarajan K. N., Tuck A. C., Gao X., Bühler M., Liu P., Marioni J. C., Teichmann S. A., Single Cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaughlin K., Flyamer I. M., Thomson J. P., Mjoseng H. K., Shukla R., Williamson I., Grimes G. R., Illingworth R. S., Adams I. R., Pennings S., Meehan R. R., Bickmore W. A., DNA methylation directs polycomb-dependent 3d genome re-organization in naive pluripotency. Cell Rep. 29, 1974–1985.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabari B. R., Dall’Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cisse I. I., Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Dong A., Saydaminova K., Chang H., Wang G., Ochiai H., Yamamoto T., Pertsinidis A., Single-molecule nanoscopy elucidates RNA polymerase II transcription at single genes in live cells. Cell 178, 491–506.e28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blinka S., Reimer M. H. Jr., Pulakanti K., Rao S., Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes. Cell Rep. 17, 19–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagashima R., Hibino K., Ashwin S. S., Babokhov M., Fujishiro S., Imai R., Nozaki T., Tamura S., Tani T., Kimura H., Shribak M., Kanemaki M. T., Sasai M., Maeshima K., Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germier T., Kocanova S., Walther N., Bancaud A., Shaban H. A., Sellou H., Politi A. Z., Ellenberg J., Gallardo F., Bystricky K., Real-time imaging of a single gene reveals transcription-initiated local confinement. Biophys. J. 113, 1383–1394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinkai S., Nakagawa M., Sugawara T., Togashi Y., Ochiai H., Nakato R., Taniguchi Y., Onami S., PHi-C: Deciphering Hi-C data into polymer dynamics. Nar Genom. Bioinform. 2, lqaa020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinkai S., Itoga H., Kyoda K., Onami S., PHi-C2: Interpreting Hi-C data as the dynamic 3D genome state. Bioinformatics 38, 4984–4986 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alshareedah I., Moosa M. M., Pham M., Potoyan D. A., Banerjee P. R., Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun. 12, 6620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang H., Zhu Q., Jussila A., Han Y., Bintu B., Kern C., Conte M., Zhang Y., Bianco S., Chiariello A. M., Yu M., Hu R., Tastemel M., Juric I., Hu M., Nicodemi M., Zhuang X., Ren B., CTCF mediates dosage- and sequence-context-dependent transcriptional insulation by forming local chromatin domains. Nat. Genet. 53, 1064–1074 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuin J., Roth G., Zhan Y., Cramard J., Redolfi J., Piskadlo E., Mach P., Kryzhanovska M., Tihanyi G., Kohler H., Eder M., Leemans C., van Steensel B., Meister P., Smallwood S., Giorgetti L., Nonlinear control of transcription through enhancer–promoter interactions. Nature 604, 571–577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vernimmen D., Bickmore W. A., The hierarchy of transcriptional activation: From enhancer to promoter. Trends Genet. 31, 696–708 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Chiariello A. M., Bianco S., Esposito A., Fiorillo L., Conte M., Irani E., Musella F., Abraham A., Prisco A., Nicodemi M., Physical mechanisms of chromatin spatial organization. FEBS J. 289, 1180–1190 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Novo C. L., Javierre B.-M., Cairns J., Segonds-Pichon A., Wingett S. W., Freire-Pritchett P., Furlan-Magaril M., Schoenfelder S., Fraser P., Rugg-Gunn P. J., Long-range enhancer interactions are prevalent in mouse embryonic stem cells and are reorganized upon pluripotent state transition. Cell Rep. 22, 2615–2627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beliveau B. J., Kishi J. Y., Nir G., Sasaki H. M., Saka S. K., Nguyen S. C., Wu C., Yin P., OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U.S.A. 115, E2183–E2192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao W., Kojima K. K., Kohany O., Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershberg E. A., Camplisson C. K., Close J. L., Attar S., Chern R., Liu Y., Akilesh S., Nicovich P. R., Beliveau B. J., PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat. Methods 18, 937–944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C., ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saka S. K., Wang Y., Kishi J. Y., Zhu A., Zeng Y., Xie W., Kirli K., Yapp C., Cicconet M., Beliveau B. J., Lapan S. W., Yin S., Lin M., Boyden E. S., Kaeser P. S., Pihan G., Church G. M., Yin P., Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Imbert A., Ouyang W., Safieddine A., Coleno E., Zimmer C., Bertrand E., Walter T., Mueller F., FISH-quant v2: A scalable and modular tool for smFISH image analysis. RNA 28, 786–795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia B. B., Jussila A., Kern C., Zhu Q., Ren B., A spatial genome aligner for resolving chromatin architectures from multiplexed DNA FISH. Nat. Biotechnol. 41, 1004–1017 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durand N. C., Robinson J. T., Shamim M. S., Machol I., Mesirov J. P., Lander E. S., Aiden E. L., Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miura H., Poonperm R., Takahashi S., Hiratani I., Practical analysis of Hi-C Data: Generating A/B compartment profiles. Methods Mol. Biol. 1861, 221–245 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Martin D. S., Forstner M. B., Käs J. A., Apparent subdiffusion inherent to single particle tracking. Biophys. J. 83, 2109–2117 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3
Legends for movies S1 to S3
Legends for data S1 to S7
Movies S1 to S3
Data S1 to S7