Abstract
Background
Diabetic kidney disease (DKD) is a global health issue. Epigenetic changes play an important role in the pathogenesis of this disease.
Summary
DKD is currently the leading cause of kidney failure worldwide. Although much is known about the pathophysiology of DKD, the research field of epigenetics is relatively new. Several recent studies have demonstrated that diabetes-induced dysregulation of epigenetic mechanisms alters the expression of pathological genes in kidney cells. If these changes persist for a long time, the so-called “metabolic memory” could be established. In this review, we highlight diabetes-induced epigenetic modifications associated with DKD. While there is a substantial amount of literature on epigenetic changes, only a few studies describe the underlying molecular mechanisms. Detailed analyses have shown that epigenetic changes play an important role in known pathological features of DKD, such as podocyte injury, fibrosis, accumulation of extracellular matrix, or oxidative injury, all of which contribute to the pathophysiology of disease. The transforming growth factor-β plays a key role as it is involved in all-mentioned epigenetic types of regulation.
Key Messages
Epigenetic is crucial for the development and progression of DKD, but the detailed molecular mechanisms have to be further analyzed more in detail.
Keywords: Diabetic kidney disease, Epigenetic, Histone modifications, DNA methylation, miRNA
Introduction
Dynamic gene regulation, such as chemical modifications of mRNA [1], 3D chromatin structure [2], or epigenetics [3], impacts transcript turnover and translation. Epigenetic alterations play an important role in the pathogenesis of diabetic kidney disease (DKD). Epigenetics affect gene expression without changing the DNA sequence itself and regulate numerous physiological and pathophysiological processes. The regulation of epigenetic mechanisms is influenced by the environment [3, 4]. Epigenetic modifications create a kind of “cellular memory” that can be transient, permanent, or heritable through cell divisions or across generations [3, 4].
The major types of epigenetic regulation are DNA methylation, post-translational histone modifications, and non-coding RNAs (ncRNAs) such as long ncRNAs, small interfering RNAs (siRNAs), or micro RNAs (miRNAs) [3, 4]. In recent decades, several epigenetic changes that drive disease have been described. However, the detailed mechanisms are still not fully understood.
Clinical trials and experimental models showed that patients with diabetes mellitus (DM) exhibit long-term complications despite a relatively short period of poor glucose control many years ago [5, 6]. Al-Dabet et al. [7] used a mouse model of hyperglycemia reversal where they treated mice with STZ (streptozotocin), an animal model for type 1 DM (T1DM). A subgroup of mice received a SGLT2 (sodium dependent glucose co-transporter 2) inhibitor for 6 weeks, while blood glucose levels remain high in another subgroup [7]. Their data showed that the mice in the SGLT2i group had lower albuminuria compared to the diabetic STZ group without the inhibitor, but higher albuminuria compared to the non-diabetic control animals [7]. RNAseq analyses demonstrated that diabetes changed the expression of several hundred genes (291 were upregulated, 746 were downregulated) compared with the control, and the reduction of blood glucose in the SGLT2i group had only marginal effects on the diabetes-induced, altered gene expression (271 upregulated, 274 downregulated) [7]. This phenomenon of persistence of the toxic effects of hyperglycemia is called metabolic memory [5].
For several years, many research groups have studied the molecular mechanisms of metabolic memory, particularly the role of epigenetics, to better understand DKD pathophysiology. Comprehending epigenetic modifications in DKD provides valuable insights into potential therapeutic targets, enabling the translation of these findings into clinical practice. Hyperglycemia, a hallmark of diabetes, can result in epigenetic changes affecting the expression of harmful genes that worsen kidney damage [5, 6].
The importance of epigenetics in modern medicine extends beyond DKD. It offers a framework for understanding how environmental factors influence gene expression and contribute to various diseases. This knowledge guides disease prevention, diagnosis, and treatment strategies. Epigenetic biomarkers could be developed for early detection of diseases, while epigenetic therapies could be explored for their potential to reverse harmful gene expression patterns. Given the potential of epigenetic research to transform our understanding and treatment of diseases like DKD, there is an urgent need for more comprehensive studies in this field.
The aim of this article was to review the literature on epigenetic hallmarks that contribute to the pathophysiology of DKD. We will focus on the impact of histone modifications but will also provide information on the role of DNA methylation and miRNAs.
Overview of DKD Pathophysiology
DKD develops in 25–40% of patients with DM and occurs in patients with T1DM and type 2 DM (T2DM) [8, 9]. In 2021, 11% of the global world population suffered from DM [8, 9]. DKD is the leading cause of kidney failure worldwide with growing prevalence for decades. This disease not only impairs the quality of living of the affected person but is also an enormous economic problem [8, 9]. Unfortunately, DKD can be silent for a long time in patients, making an early diagnosis difficult. The average time of diagnosis of kidney injury is about 7–10 years after onset of DM [8, 9].
DKD is characterized by significant structural changes in kidney morphology. One of the earliest events in DKD are modifications in the thickness and stiffness of the glomerular basement membrane due to glomerular hyperfiltration and direct alteration of collagen metabolism [10, 11]. This is followed by several pathological changes such as mesangial expansion, podocyte injury, and tubulointerstitial fibrosis [11]. Mesangial expansion occurs due to protein leakage, inflammation, and the accumulation of damaged tissue as well as growth stimulation of mesangial cells by high glucose, angiotensin II, and other factors [11–13]. Progressive mesangial destruction leads to glomerulosclerosis [11]. Podocyte injury is a key factor in the development of DKD. Hyperglycemia, oxidative stress, and inflammation impact podocytes, causing foot process effacement, actin rearrangement, increased tight junctions, slit diaphragm abnormalities, and apoptosis [11, 14]. Ultimately, fibrosis and atrophic kidney tissue develop, often through the process of epithelial-to-mesenchymal transition (EMT) [11]. EMT is characterized by the loss of epithelial markers such as E-cadherin, and induction of mesenchymal markers such as α-smooth muscle actin (α-SMA), vimentin, and N-cadherin [15].
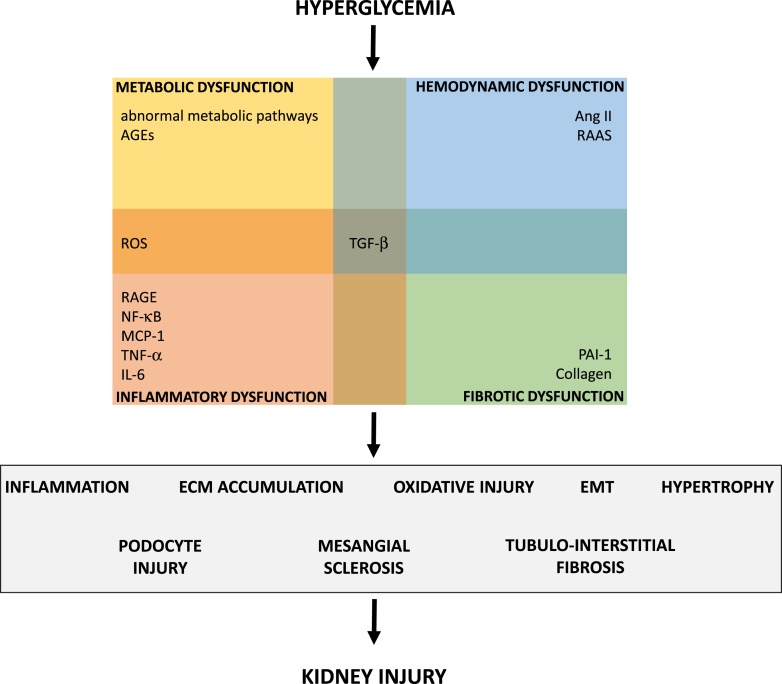
Hyperglycemia is the primary cause of pathological events in DKD, alongside hypertension [8, 11]. Hyperglycemia triggers various metabolic pathways and abnormal glucose metabolism, including the polyol pathway, hexosamine pathway, protein kinase C pathway, and the accumulation of advanced glycation end-products (AGEs) [8, 16, 17]. AGEs in turn induce the production of reactive oxygen species (ROS), leading to endothelial dysfunction [8, 16]. AGEs also activate the receptor for AGEs (RAGE) in podocytes and endothelial cells, as well as the NF-κB (nuclear factor-κB) pathway in podocytes [17–20]. RAGE is a multiligand pattern recognition receptor that triggers inflammation [17]. NF-κB signaling promotes inflammation and fibrosis through adhesion molecules and pro-inflammatory cytokines such as monocyte chemoattractant protein-1 (MCP-1), tumor necrosis factor (TNF-α), and interleukin (IL-6) [11, 16]. Pro-inflammatory mediators recruit inflammatory cells such as macrophages, that, in turn, produce further cytokines, ROS, and proteases, all contributing to kidney damage [8]. Elevated levels of ROS are also produced through hyperglycemia-mediated mitochondrial dysfunction and activation of NADPH oxidases (NOX) [10, 11, 16]. ROS play an important role in lipid oxidation and thus contribute to tissue damage [11, 16], causing foot process effacement, p53 activation and apoptosis, as well as protein kinase C activation that contributes to mesangial expansion [11]. Another pathological key mechanism in DKD is oxidative stress which can be caused by hyperglycemia, AGEs, and obesity via the upregulation of NF-κB, IL-1β, and IL-6 [11, 16]. In DKD, activation of the renin-angiotensin-aldosterone system (RAAS) induces mitogen-activated protein kinase (MAPK), transforming growth factor (TGF)-β, fibrosis, vascular endothelial growth factor (VEGF), NF-κB, ROS production, and inflammation via MCP-1, epidermal growth factor receptor (EGFR), and plasminogen activator inhibitor-1 (PAI-1), as well as other pro-inflammatory and profibrotic mediators [11, 16, 17]. Several growth factors are involved in DKD pathogenesis, with TGF-β being the best studied [8]. The profibrotic cytokine TGF-β is a multifunctional regulator that affects various cell types, including kidney cells [5, 21]. TGF-β acts via multiple signaling pathways such as Smad or MAPK. In kidney injury, upregulated TGF-β causes ECM (extracellular matrix) production, which leads to glomerulosclerosis as well as tubulointerstitial fibrosis, cellular hypertrophy, apoptosis, and podocyte foot process effacement. TGF-β also takes part in the process of EMT [5, 21]. Figure 1 summarizes the pathological consequences of chronic hyperglycemia contributing to DKD.
Fig. 1.
Overview of hyperglycemia-induced factors contributing to the pathogenesis of DKD. AGEs, advanced glycation end-products; TGF, transforming growth factor; ROS, reactive oxygen species; Ang II, angiotensin II; RAAS, renin-angiotensin-aldosterone system; PAI-1, plasminogen activator inhibitor-1; RAGE, receptor of AGEs; NF-κB, nuclear factor-κB; MCP-1, monocyte chemoattractant protein-1; TNF-α, tumor necrosis factor-α; IL, interleukin.
Post-Translational Histone Modifications in DKD
The nucleosome consists of the four core histone proteins H2A, H2B, H3, and H4 [22, 23]. These proteins can be post-translational modified such as acetylated, methylated, phosphorylated, ubiquitinylated, or SUMOylated, which occurs primarily on the amino terminal ends of the histones [22, 23]. The modification-induced alterations of charge and structure of the histone tails cause chromatin remodeling and promote or inhibit the recruitment of transcriptional complexes, respectively, effector proteins [22, 23].
Post-Translational Histone Methylation in DKD
The addition of methyl groups mostly occurs on histone H3 or H4 sidechain nitrogen atoms of lysine (K) residues and is catalyzed by histone lysine methyltransferases (KMT) [24, 25]. It is a dynamic and reversible process. The removal of methyl groups is mediated by demethylases [24, 25]. Methylation of lysine exists in three possible states: mono-, di-, or tri-methylated and can be activating or repressing, depending on the final position of the methyl group [24, 25]; the di- and trimethylation at H3K4 enhances the expression of the respective genes, and methylation of H3K9, H3K27 as well as H4K20 represses gene expression [26]. Up to now, more than 50 lysine methyltransferases (writers) have been characterized that can be divided into two main groups: Suppressor of variegation, Enhancer of Zeste, Trithorax (SET) domain-containing KMTs and non-SET domain-containing KMTs (DOT1L-like proteins) [25]. Histone lysine demethylases (erasers) can be classified into amino oxidase homolog lysine demethylase 1 (KDM1) and JmjC domain-containing histone demethylases (such as KDM 2/3/4/5/6 etc.) [26]. Tables 1 and 2 give a short overview of histone methylases and demethylases, respectively, that are associated with DKD, and their substrates.
Table 1.
| Writers | Alias | Substrate(s) | Methylation |
|---|---|---|---|
| KMT1A, B, E, F | SUV39H1/2, SETDB1/2 | H3K9 | Di-/trimethylation |
| KMT2H | ASH1L | H3K4, H3K9, H4K20 | Mono-/di-/trimethylation |
| KMT4 | DOT1L | H3K79 | Mono-/di-/trimethylation |
| KMT5A | SETD8 | H4K20 | Monomethylation |
| KMT6B, A | EZH1/2 | H3K27 | Mono-/di-/trimethylation |
| KMT7 | SETD7, SET7/9 | H3K4 | Dimethylation |
| KMT8A | PRDM2 | H3K9 | Mono-/di-/trimethylation |
| KMT8D | PRDM8 | H3K9 | Dimethylation |
| KMT8E, F | PRDM3/16 | H3K9 | Monomethylation |
KMT, lysine methyltransferase.
Table 2.
| Erasers | Alias | Substrate(s) | Methylation |
|---|---|---|---|
| KDM1A | LSD1 | H3K4 | Mono-/dimethylation |
| KDM1B | LSD2 | H3K4, H3K9 | Mono-/dimethylation |
| KDM3A-C | JHDM2A/B, JMJD1C | H3K9 | Mono-/dimethylation |
| KDM4A-D | JMJD2A-D | H3K9, H3K36, H1.4K26 | Di-/trimethylation |
| KDM5A-D | JARID1A-D | H3K4 | Di-/trimethylation |
| KDM6A, B | UTX, JMJD3 | H3K27 | Di-/trimethylation |
| KDM7A, B | JHDM1D, PHF8 | H3K9, H3K27, H4K20 | Mono-/dimethylation |
KDM, lysine demethylases.
Several studies show the influence of post-translational histone methylation on the pathogenesis of DKD. Wang et al. [29] identified key methylation sites that may contribute to the pathogenesis of DKD, noting that diabetic kidney tissue exhibits 121 hypermethylated and 579 hypomethylated sites [29].
Komers et al. [30] analyzed epigenetic changes in mouse (OVE26) and rat STZ models of T1DM. They observed an increase in the activating H3K4me2 mark and a decrease of H3K27me3 in the tested genes of diabetic animals. Regarding the trimethylation of H3K27, they also investigated the expression of KDM6A and enhancer of zeste homolog 2 (EZH2), finding that KDM6A histone demethylase expression was increased in the kidneys of diabetic mice, while EZH2 expression remained unchanged compared to the controls [30]. In contrast, diabetic rats exhibited altered EZH2 expression without changes in KDM6A, indicating species-specific mechanisms of H3K27me3 loss in DKD [30]. Chen et al. [31] demonstrated that pharmacological prevention of KDM6A upregulation (that was also detected in patients with DKD) improved diabetes-induced kidney abnormalities and dysfunctions [31]. Additionally, the inhibition of KMT5A associated with reduced H4K20me1 [32], the upregulation of SET7/9 and changes in H3K4 methylation [33–36] as well as Dot1l and H3K79me2 [37] are involved in epigenetic dysregulation in DKD.
Impact of TGF-β1 on Histone Methylation in DKD
The upregulation of TGF-β1 in kidney cells is involved in cellular pathophysiological events in kidney disease. Sun et al. [38] showed that TGF-β1-induced expression of CTGF (connective tissue growth factor), Collagen I (Col I), and PAI-1 is associated with increased levels of the activating methylation marks H3K4me1/2/3 in high glucose-treated rat mesangial cells compared to the control. The changes in methylation marks at the promotors of CTGF, COL I, and PAI-1 genes are accompanied by a TGF-β1-mediated increase in the expression of the methyltransferases SET7/9 [38]. Congruently, the levels of the repressive marks H3K9me2/3 were decreased at the promotors of these genes under high glucose, but detailed mechanisms were not mentioned [38]. TGF-β1-mediated activation of SET7/9, associated with decreased H3K9 methylation and increased levels of the activating methylation at H3K4, also regulates p21 gene expression [39], which is involved in glomerular cell hypertrophy and senescence [7, 40–42].
Bai et al. [43] demonstrated the involvement of the histone lysine demethylase KDM6A in TGF-β1-mediated injury. The upregulation of this demethylase was associated with an increased expression of TGF-β, vimentin, and N-cadherin. Further analyses showed an enhanced methylation of the promotor region of E-cadherin, leading to a reduced E-cadherin expression [43].
Other analyses showed that TGF-β1 influences the methylation of H3K27 caused by dysregulation of EZH2, KDM6A, and KDM6B [44–47], leading to elevated expression of TGF-β1 target genes [45, 47] and genes of the TGF-β signaling pathway [46]. Additionally, alterations in H3K27 methylation were associated with an enhanced expression of Wnt1, which takes part in insulin sensitivity [44]. Jia et al. [45] showed that upregulation of KDM6A is involved in the TGF-β1 mediated hypertrophy of mesangial cells.
Histone Methylation and Podocyte Injury in DKD
Majumder et al. [48] showed that EZH2 and demethylases KDM6A- and KDM6B-mediated H3K27me3 regulation are involved in podocyte injury in diabetes. Deletion of EZH2 from the podocytes of mice, associated with reduced H3K27 trimethylation, increased the risk for glomerular disease. In contrast, inhibition of histone demethylases KDM6A and KDM6B by GSK-J4 in diabetic db/db mice (a mouse model for human T2DM) ameliorated albuminuria, accumulation of glomerular matrix, podocyte foot process effacement and glomerular increase of α-SMA. They also analyzed kidneys from patients with glomerular sclerosis and showed a decrease of H3K27me3 in glomerular cells and tubuli. In podocytes, the progressive loss of H3K27me3 correlates with the degree of glomerular injury [48]. EZH2 and, accordingly, H3K27me3 are also involved in diabetic podocyte injury caused by oxidative stress [49]. Inhibition of EZH2 expression in rats by DZNep (3-deazaneplanocin A, a histone methylation inhibitor) treatment caused reduced H3K27me3, higher glomerular thioredoxin-interacting protein (TxnIP; involved in cellular stress response as inhibitor of the antioxidant protein thioredoxin) expression and enhanced oxidative stress [49]. Moreover, the data show that depletion of EZH2 in diabetic rats goes along with changes in podocyte structure like vacuolization and foot process effacement [49]. In vitro cell culture studies demonstrated that EZH2 regulates H3K27me3 at the gene promotor of TxnIP via the trans-acting transcription factor paired box 6 (Pax6) [49].
We also analyzed EZH2 and H3K27me3 in diabetic podocyte injury. Our data showed that diabetic conditions decreased the expression of EZH2 and accordingly the trimethylation of H3K27 in mouse podocytes in vitro and in vivo in db/db mice [50]. Suppression of EZH2 in cultured podocytes was associated with increased levels of TGF-β1 and SNAI1 (a transcription factor that regulates the expression of E-cadherin [15]) as well as the cyclin-dependent kinase inhibitor p27Kip1 [50]. We assume that the induction of p27Kip1 in murine cultured podocytes results in cell cycle arrest and cellular hypertrophy [51]. Additionally, downregulation of EZH2 activated RAGE, the transcription factor NF-κB and subsequently TNF-α expression [50, 51]. Moreover, our data [52] demonstrated that EZH2 and the trimethylation of H3K27 are also involved in hypoxia responses, which is a key pathophysiological event in diabetic disorders. Both, incubation of murine differentiated podocytes at 10% O2 and housing mice at 10% O2 significantly reduced EZH2 expression associated with a decrease of H3K27me3 in podocytes [52]. Additionally, pharmacological inhibition of HIFs (hypoxia inducible factor) diminished EZH2 and H3K27me3 in podocytes in vitro and in mice [52].
Lin et al. [53] found that dysregulated KDM6A contributed to podocyte injury in DKD. Cell culture experiments with murine podocytes showed that high glucose treatment of podocytes increased the KDM6A level and decreased the expression of nephrin (combined with an enhanced DNA methylation at the nephrin [NPHS1] promotor) [53]. Overexpression of KDM6A in podocytes reduced the expression of nephrin, WT1, and the trimethylation of H3K27 [53]. Moreover, GSK-J4 treatment ameliorated diabetic-induced albuminuria and normalized podocyte morphology. The knockout of KDM6A in diabetic STZ mice corrected features of DKD like GBM thickness and glomerular sclerosis [53].
Histone Methylation and Oxidative Stress in DKD
As mentioned above, epigenetic dysregulation induced TxnIP expression in podocytes, leading to oxidative stress. Similar data were published by De Marinis et al. [54] for mesangial cells. They found an increased expression of TxnIP in diabetic Sur1-E1506KD/D mice (a mouse model that develops diabetes and early signs of DKD) and in the glomeruli of db/db mice associated with higher methylation of H3K4 and reduced H3K27me3 [54].
Epigenetic histone modifications are involved in lipid metabolism [55]. High levels of 12/15 lipoxygenase (12/15-LO) in diabetes increased oxidized lipid products that cause oxidative stress and subsequently glomerular dysfunction and fibrosis [55]. These oxidized lipid products induced the translocation of SET7 to the promotors of profibrotic genes and consequently raised the methylation of H3K4me1/3 in rat mesangial cells [55].
Several studies provide evidence that post-translational histone methylation influences the pathogenesis of DKD. Key findings include alterations in histone methylation marks, such as H3K4me2 and H3K27me3, and species-specific changes in histone-modifying enzymes like EZH2, KDM6A, and SET7/9. These modifications affect gene expression linked to kidney dysfunction, hypertrophy, podocyte injury, and oxidative stress, highlighting their role in the epigenetic regulation of DKD.
Post-Translational Histone Acetylation in DKD
The acetylation/deacetylation of histone proteins is catalyzed by histone lysine acetyltransferases (HATs), respectively, histone lysine deacetylases (HDACs) [56]. In humans, HATs are particularly members of Gcn5 (Gcn5-related N-acetyltransferases)/PCAF (p300/CBP-associated factor) family, MYST family (with Tat-interactive protein 60 kD [Tip60], monocyticleukaemia zinc finger protein [MOZ], monocytic leukemic zinc finger-related factor [MORF], or human acetylase binding to ORC1 [HBO1]) or p300/CBP family [56, 57]. HDACs are divided into 4 classes: class I Rpd3-like proteins with HDAC1/2/3/8, class II Hda1-like proteins with HDAC4/5/6/7/9/10, class III Sir2-like proteins (silent information regulator sirtuin [SIRT] 1–7) and class 4 with HDAC11 [56, 58]. HATs and HDACs are mostly found in a complex with other proteins [56, 58].
Kidney biopsies from patients with DKD showed an upregulation of HDAC6 in tubular cells and infiltrated macrophages that are round to the diluted tubules [59]. Another study of podocytes from diabetic mice and in kidney biopsies from patient with podocytopathies demonstrated the involvement of SIRT6 in podocyte injury [60]. Studies on diabetic mice indicated that the deletion of SIRT6 in podocytes worsens podocyte injury and proteinuria [60].
Reddy et al. [61] analyzed epigenetic modifications in kidney glomeruli of diabetic mice. They revealed that changes in histone methylation and histone acetylation often occurred simultaneously [61]. The db/db diabetic mice showed a decrease in repressive histone marks H3K9me2/3 and H3K27me3, while the activating methylation marks H3K36me3 and H3K4me1, as well as the acetylation of H3K9, were increased [61]. Analyses of histone modification enzymes indicated the upregulation of the HATs Tip60, MOZ, and MORF, as well as of HDAC7 and HDAC9, and of the H3K4 histone methylases Setd7 and Setd4 [61]. Studies on mesangial cells cultured under diabetic conditions showed that the changes in histone methylation/acetylation were accompanied by the upregulation of RAGE, PAI-1, and MCP-1 [61].
Role of TGF-β1 in Dysregulated Histone Acetylation in DKD
Several studies have demonstrated a diabetes-induced dysregulation of HDACs associated with TGF-β-mediated kidney damage, including podocyte injury [62], ECM accumulation [63–67], fibrosis [63, 66–68], or EMT [64, 65, 69]. Members of the PCAF family are also involved in TGF-β1-associated epigenetic dysregulation. Malek et al. [70] displayed increased levels of PCAF/CBP in diabetic STZ rats, which were associated with the activation of NF-κB and TGF-β signaling and an increase of H2AK5Ac, H2BK5Ac, H3K18Ac, and H4K8Ac [70]. Yuan et al. [71] showed a role of p300/CBP in the activation of TGF-β target genes like PAI-2 and p21 in mesangial cells and in glomeruli from diabetic mice, associated with an increased acetylation of H3K9/14 [71]. Active p300 associated with the acetylation of H3K4 induced the synthesis of Col I during kidney fibrosis under diabetic conditions [72].
Histone Acetylation and Oxidative Stress in DKD
Changes in histone acetylation also contribute to oxidative stress in DKD. Lazar et al. [73] showed that p300/CBP is involved in the increase of Nox and the production of ROS in diabetic STZ mice via increased activation of H3K27Ac [73]. Liu et al. [74] found that diabetes-mediated upregulation of HDAC9 is involved in ROS generation and inflammation in podocytes. Moreover, altered histone acetylation affected the expression of nephrin and podocin in these cells contributing to the injury of podocytes [74].
The review of the literature shows that diabetic conditions lead to increased activating of acetylation marks, alongside with dysregulation of HDACs and HATs. These changes are associated with inflammation, oxidative stress, and TGF-β1-mediated kidney damage.
DNA Methylation in DKD
In mammalians, there are five DNA methyltransferases (DNMTs): DNMT1, DNMT2, DNMT3a, DNMT3B, DNMT3L. DNA methylation is often associated with transcriptional repression. It happens mainly at CpG sites, where a cytosine nucleotide is followed by a guanine nucleotide (5′ to 3′ direction) [75, 76]. CpG sites are mostly located in the promotor regions of genes but also in the gene body [76]. DNA demethylation is conducted by demethylases, mainly members of the ten-eleven translocation (TET) family (TET1, TET2, and TET3) [77].
Role of CpG Sites in the Progression of DKD
Numerous studies indicate that changes in DNA methylation, particularly in genes that are already known to be associated with kidney disease, contribute to the progression and clinical manifestation of DKD [78–92]. Interestingly, several of these analyses have identified CpG sites in the TXNIP gene [10, 82, 88, 90–93]. For example, Smyth et al. [82] conducted a study involving individuals with T1DM to assess differences in blood-derived DNA methylation between individuals with DKD and those without kidney disease. In meta-analyses of DKD in T1DM, they identified 32 differentially methylated CpGs in blood associated with DKD, mostly hypomethylated [82]. Seven of the CpGs overlapped with predicted transcription factor binding sites. Furthermore, methylation of 21 of the 32 CpGs was predictive of kidney failure development [82]. Li et al. [88] investigated the association between DNA methylation at CpG sites in peripheral blood and kidney function decline in patients with T2DM, identifying 48 CpGs associated with the rate of estimated glomerular filtration rate decline [88]. Notably, both studies [82, 88] observed higher DNA methylation levels at cg17944885, which correlated with the expression of a zinc finger protein and the progression of DKD [82, 88]. In another study, Gluck et al. [93], using micro-dissected human kidney tubule samples, identified 65 CpGs correlated with the degree of kidney fibrosis [93]. Both, Gluck et al. [93] and Li et al. [88] proposed the potential of DNA methylation biomarkers in predicting kidney function decline in diabetes, emphasizing the utility of incorporating DNA methylation data for early identification of high-risk patients [88, 93].
Changes in DNA Methylase Activity in DKD
Literature review reveals studies addressing the role of DNA methylases in kidney disease. Al-Dabet et al. [7], in a study using a mouse model of reversible hyperglycemia, observed persistent high expression of p21 in tubular cells despite lowering blood glucose levels, due to reduced methylation of the p21 promotor via suppression of DNMT1 and DNMT3B [7]. Gondaliya et al. [94] demonstrated an interaction between miRNA and DNA methylation in regulating gene expression in kidney tubular epithelial cells. Under conditions mimicking DKD (AGEs, high glucose, and angiotensin II), miR-29b downregulation led to an upregulation of DNMT3A, DNMT3B, and DNMT1. This was associated with an increased expression of fibronectin and KIM-1 in these cells [94].
TGF-β1 and DNA Methylation in DKD
Similar to histone modifications, DNA methylation is associated with TGF-β1-mediated injury in DKD. Oba et al. [95] showed that TGF-β1 is demethylated in primary mesangial cells from diabetic db/db mice, associated with a decreased activity of DNMT1 [95]. Yang et al. [96] demonstrated upregulation of TGF-β1 in diabetic rats due to TET2-mediated demethylation of the TGF-β1 promoter contributing to fibrotic changes and exacerbating DKD pathology.
DNA Methylation and Podocyte Injury in DKD
Literature indicates that alterations in DNA methylation contribute to podocyte dysfunction in DKD [53, 97–99]. Treatment of human immortalized podocytes with high glucose in vitro alters DNA methylation and gene expression patterns. Methylation profiling identified 337 hypomethylated genes that were upregulated, while only two genes were hypermethylated and downregulated [98].
Several publications found in the course of this review showed a connection between DNA methylation and the expression of nephrin [53, 97, 99]. Hayashi et al. [97] showed that the expression of the transcription factor Kruppel-like factor 4 (KLF4) is reduced in mouse and human podocytes, which was associated with diminished nephrin expression and proteinuria [97]. Lin et al. [53] explored the KDM6A – KLF10 interrelationship in podocyte injury, revealing that the KDM6A-induced suppression of nephrin and WT1 expression in podocytes was associated with upregulation of its downstream effector KLF10 and recruitment of DNMT1 [53]. Zhang et al. [99] highlighted the role of DNMT1 in podocytes, showing increased DNMT1 levels under diabetic conditions correlating with reduced expression of slit diaphragm proteins like nephrin [99].
The previous findings highlight that changes in DNA methylation, particularly at specific CpG sites, play a crucial role in the progression of DKD and the decline in kidney function in individuals with diabetes. Alterations in DNA methyltransferase activity (e.g., DNMT1, DNMT3A, and DNMT3B) are associated with the regulation of genes involved in podocyte injury (affecting key proteins such as nephrin, with evidence suggesting the involvement of transcription factors), as well as kidney fibrosis and dysfunction, including TGF-β1-mediated dysregulation.
Dysregulation of miRNAs in DKD
Non-coding RNAs are regulatory molecules that act through various mechanisms and can interact with each other to build complex and dynamic networks. They are divided into small ncRNAs with a length of up to 200 nucleotides (such as miRNAs or siRNAs), long ncRNAs (lncRNAs) with more than 200 nucleotides, and circular RNAs (circRNAs) [100]. Here, we will focus on the dysregulation of miRNAs in DKD. MiRNAs are negative regulators of the expression of most genes encoded by the human genome that controls gene expression at the post-transcriptional level through incomplete base pairing with the 3′-untranslated region (UTR) of target messenger RNAs [101, 102]. MiRNAs not only act in cells but also have paracrine effects and affects cellular interactions [103]. That means that miRNAs are also present and highly stable in body fluids like serum, plasma, or urine and thus easily detectable serving as a diagnostic tool [101, 102]. Several studies address the dysregulation of miRNA expression in DKD using urine, serum or plasma samples from patients with diabetic disorders. A short overview of detected miRNAs and their dysregulation in DKD are listed in Table 3.
Table 3.
Dysregulated miRNAs in DKD
| Analyzed material from patients | Detected miRNAs | ||
|---|---|---|---|
| Pooled urine samples from patients with DKD | miR-126, miR-155, miR-29b | ↑ | [104] |
| Urinary exosomes from patients with T2DM | miR-15b, miR-34a, miR-636, miR-133b, miR-342, miR-30a, miR-514a-5p, miR-451a, miR-126-3p, miR-214, miR-503, miR-21-5p, let-7e-5p, miR-23b-3p, miR-155-5p, miR-28-3p, miR-425-5p, miR-4491, miR-2117, miR-4507, miR-5088-5P, miR-1587, miR-219a-3p, miR-5091, miR-498, miR-4687-3p, miR-516b-5p, miR-4534, miR-1275, miR-5007-3p, miR-4516, miR-4270, miR‐4756, miR-615-3p | ↑ | [105–112] |
| Urine samples from pediatric patients with T1DM | miR-377 | ↑ | [113] |
| Serum from patients with T2DM with DKD | miR-145-5p, miR-874-3p, miR-101-3p, miR-99b, miR-122 | ↑ | [114, 115] |
| Serum from patients with T1DM with DKD | miR-518-3p, miR-34a-5p, miR-126-5p, miR-425-5p, miR-618, miR-139-5p | ↑ | [116] |
| Plasma from patients with T1DM with DKD | miRNA-21, miR-21-3p and miR-378-3p | ↑ | [117, 118] |
| Urinary samples from patients with DKD | miR-27b-3p, miR-1228-3p | ↓ | [119] |
| Urine samples from pediatric patients with T1DM | miR-216a | ↓ | [113] |
| Urinary exosomes from patients with T2DM with DKD | miR-21-3p, miR-4792, miR-375, miR-1268a, miR-501-5p, miR-582, miR-30b-5p, miR-125b-5p, miR-663a, miR-3137, miR-3147 | ↓ | [106–108, 110, 112] |
| Urinary exosomes from patients with T1DM with DKD | miR-30e-5p | ↓ | [120] |
| Serum samples from patients with DKD | miR-23a-3p, miR-26a-5p, miR-27a-3p | ↓ | [121] |
| Serum from patients with T2DM with DKD | miR-20a, miR-486 | ↓ | [115] |
| Plasma from patients with T1DM with DKD | miR-16-5p, miR-29a-3p | ↓ | [118] |
↑, upregulation; ↓, downregulation; DKD, diabetic kidney disease; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus.
Currently, numerous miRNAs involved in the pathogenesis of DKD have been described, but targets or functions of only a fraction of these are known [103]. Some studies have analyzed the molecular mechanisms of miRNAs more in detail. Zanchi et al. [122] detected an upregulation of miR-184 in Zucker diabetic fatty (ZDF) rats, showing that tubular miR-184 expression is induced by albumin and is associated with collagen accumulation [122]. Interestingly, the upregulation of miR-184 is epigenetically regulated by DNA demethylation and histone lysine acetylation [122]. Lee et al. [123] analyzed the role of miR-146a in DKD, demonstrating that the downregulation of miR-146a negatively correlates with albuminuria and glomerular damage in patients with T2DM. Animal studies demonstrated that the reduction of miR-146a causes mesangial sclerosis and foot process effacement [123]. Chen et al. [124] have shown that miR-296 regulates the expression of SGLT2 [124]. Wang et al. [125] found that miR-92b-3p, a direct target of Smad7, is one of many miRNAs regulated by AGEs [125].
miRNAs in Oxidative Stress and Inflammation
Several publications describe a role for miRNAs in oxidative stress and inflammation. Sheng et al. [126] demonstrated that miR-23a-3p is involved in inflammatory and fibrotic processes that cause tubular injury in DKD. Targeting miR-23a-3p in HK-2 cells in vitro influenced the expression of IL-6, TNF-α, and fibronectin [126]. Another study showed that miR-140-5p plays a role in ROS generation and inflammation with miR-140-5p being reduced in kidney biopsies from patients with DKD [127]. The authors demonstrated that miR-140-5p targets Toll-like receptor (TLR 4) and accordingly regulates NF-κB signaling [127]. miR-25 is involved in the regulation of oxidative stress as direct interaction partner of NOX4 [128]. Wang et al. [129] showed that the upregulation of miR-155 in podocytes drives the expression of inflammatory molecules as MCP-1, TNF-α, IL-1β, IL-6 via SIRT1 downregulation. They tested the role of miR-155 in vivo, finding that inhibition of miR-155 in an animal model reduces the albumin-creatinine ratio and inflammatory processes, reversing kidney pathological changes as foot process effacement [129].
TGF-β1 and miRNAs in DKD
Numerous publications underline the importance of miRNAs in TGF-β1-mediated injury in the pathogenesis of DKD, as summarized in Table 4.
Table 4.
TGF-β-related dysregulation of miRNAs in DKD
| miRNA | In vivo detection | Supposed mechanisms | |
|---|---|---|---|
| let-7d | TGF-β1 decreases let-7d in NRK-52E cells -> Col I ↑, E-cadherin ↓ | [130] | |
| let-7d | ↓ in glomeruli of STZ-treated mice | TGF-β1 decreases let-7d in mouse mesangial cells -> TGF-R1 ↑ | [131] |
| miR-34a-5p | ↑ in db/db mice | Upregulation of miR-34a-5p -> SIRT1 ↓ -> TGF-β1 ↑, fibronectin ↑ -> kidney fibrosis | [132] |
| miR-130b | ↓ in glomeruli of STZ-treated mice | TGF-β1 decreases miR-130b in mouse mesangial cells -> TGF-R1 ↑, Col4A1 ↑, Col12A1 ↑, CTGF ↑, PAI-1 ↑ | [131] |
| miR-143/145 | ↑ in glomeruli of db/db mice | TGF-β1 -> SMAD signaling or mTOR -> miR-143/145 ↑ in human podocytes -> WT-1 ↓, Zonula-1 ↓, Col I ↑, α-SMA ↑ | [133] |
| miR-154-5p | ↑ in glomeruli of STZ and high-fat diet rats | miR-154-5p ↑ in rat mesangial cells -> TGF-β1/SMAD3 ↑ -> cell proliferation ↑ | [134] |
| miR-192, miR-200b/c | ↑ in glomeruli of STZ-treated and db/db mice | miR-192 in mouse mesangial cells ↑-> miR-200b/c ↑ -> TGF-β1 ↑ -> Col IA2 ↑, Col IVA1 ↑ | [135, 136] |
| miR-199b-3p | ↓ STZ mice | Downregulation of miR-199b-3p in HK-2 cells -> KDM6A ↑ -> E-cadherin ↓, TGF-β1 ↑, vimentin ↑, N-cadherin ↑ | [43] |
| miR-204 | ↓ in patients with DKD | miR-204 knockdown in mice -> TGF-β1 ↑, Col IA1 ↑, MCP-1 ↑ | [137] |
| miR-205 | ↓ in db/db mice | TGF-β1 downregulates miR-205 via HDAC2 -> fibronectin ↑, Col I ↑, SNAI1 ↑, CTGF ↑ | [66] |
| miR-342-3p | ↓ in db/db mice | miR-342-3p downregulation in mouse mesangial cells -> TGF-β1 ↑, fibronectin ↑, Col IV ↑, PTEN ↓ via SOX6 ↑ | [138] |
| miR-378 | ↓ in db/db mice | TGF-β1 reduces miR-378 in NRK-52E cells -> Col I, Col IV, α-SMA | [139] |
| miR-378 | ↓ in db/db mice | TGF-β1 decreases miR-378 in primary human mesangial cells -> mesangial cell expansion and proliferation | [139] |
| miR-1207-5p | miR-1207-5p upregulation in mesangial cells -> TGF-β1 ↑, PAI-1 ↑, fibronectin ↑ | [140] |
↑, upregulation; ↓, downregulation; Col, collagen; STZ, streptozotocin; TGF-R1, transforming growth factor-β receptor 1; CTGF, connective tissue growth factor; PAI, plasminogen activator inhibitor; WT-1, Wilms’ tumor 1; mTOR, mechanistic Target of Rapamycin; α-SMA, smooth muscle actin; DKD, diabetic kidney disease; HDAC, histone deacetylase; PTEN, Phosphatase and Tensin homolog; SIRT1, sirtuin 1; KDM6A, lysine demethylase 6A; MCP-1, monocyte chemoattractant protein-1; SOX, SRY-Box Transcription Factor.
Translational Relevance of miRNAs
The translational relevance of miRNAs in kidney disease has been explored in recent studies. Baker et al. [141] performed a tissue-specific approach to analyze miRNA expression profiles across four different types of kidney disease. Their data showed that distinct miRNAs are differentially expressed in glomeruli or proximal tubuli in each type of kidney disease providing an excellent basis for the development of biomarkers [141]. Other studies have already tested miRNAs as therapeutic targets in animal models of DKD [142, 143]. Kölling et al. [142] analyzed miRNA profile in diabetic kidneys and found miR-21 as the most highly upregulated, causing mesangial cell hypertrophy and changes in podocyte phenotype. To investigate the therapeutic potential of silencing miR-21 in ameliorating DKD, they antagonized miR-21 in STZ mice [142]. Silencing miR-21 improved kidney function and reduced pathological changes such as albuminuria, mesangial expansion, podocyte loss, fibrosis, and inflammation [142]. Putta et al. [143] identified miR-192 as a critical regulator of kidney fibrosis in DKD. To test the translational relevance of this miRNA, they treated STZ mice with an inhibitor of miR-192, resulting in a reduction of kidney fibrosis by normalization of the expression of profibrotic genes and upregulation of ant-fibrotic genes [143].
Experimental evidence has shown that numerous miRNAs are involved in the pathogenesis of DKD through mechanisms such as inflammation, fibrosis, oxidative stress, ECM accumulation, and TGF-β1-mediated injury. Some studies have also demonstrated the therapeutic potential of targeting these miRNAs to improve kidney function and slow disease progression.
Conclusion
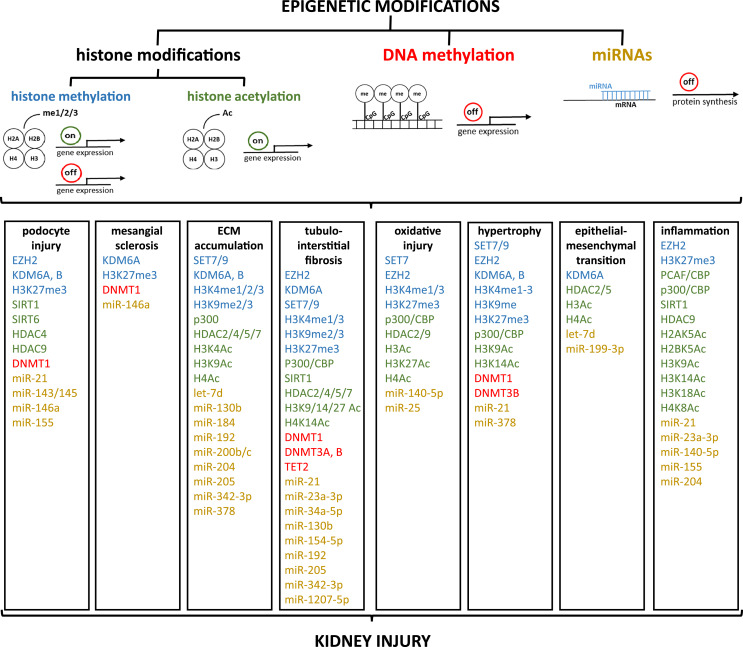
The literature review showed that epigenetic dysregulation contributes to the pathophysiology of DKD, as illustrated in Figure 2. Mostly, different epigenetic mechanisms, and often a combination of epigenetic modifications, drive kidney injury. But it seems that epigenetic dysregulation can be species and pathway specific. Since epigenetic modifications are dynamic, it is challenging to determine whether they are a cause or a consequence of diseases. On one hand, epigenetic modifications can predispose individuals to certain diseases by affecting the regulation of genes involved in critical biological processes. On the other hand, diseases themselves can induce epigenetic changes. Chronic conditions like hyperglycemia can lead to alterations in the epigenome, which may exacerbate the disease or contribute to its progression.
Fig. 2.
Schematic illustration of epigenetic changes found during the literature review, assigned to the respective pathological pathways associated with kidney injury. me, methylation; Ac, acetylation; miR, mi RNA; ECM, extracellular matrix; EZH2, enhancer of zeste homolog 2; KDM, lysine demethylases; SIRT, silent information regulator sirtuin; HDAC, histone lysine deacetylases; DNMT, DNA methyltransferases; SET, Suppressor of variegation, Enhancer of Zeste, Trithorax; TET, ten-eleven translocation; CBP, CREB-binding protein; PCAF, p300/CBP-associated factor.
Only a few studies emphasize the role of epigenetic changes in the pathogenesis of DKD. However, understanding the detailed molecular mechanisms is crucial. Additionally, the literature review identified only one publication that addresses the role of the sex in epigenetic signature. Recent research has clearly illuminated that molecular differences between male and female individuals exist. Loeffler et al. [144] showed that the sex causes differences in the development and the progression of DKD, as well as kidney failure [144]. Therefore, it is important to focus more on sex-related epigenetic changes in the pathogenesis of DKD. Epigenetics has a transformative impact on modern medicine, particularly in understanding the pathophysiology of disease. It enables more personalized and effective therapeutic approaches. Moreover, epigenetic markers can serve as biomarkers for early diagnosis, prognosis, and monitoring of diseases. Additionally, because epigenetic modifications are reversible, they are attractive targets for new therapies. Thus, epigenetics in medicine holds promise for preventing or halting the development of DKD.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was not supported by any sponsor or funder.
Author Contributions
M.L. and G.W. reviewed the literature, wrote, and edited the manuscript.
Funding Statement
This study was not supported by any sponsor or funder.
References
- 1. Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022;38(2):182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han MH, Issagulova D, Park M. Interplay between epigenome and 3D chromatin structure. BMB Rep. 2023;56(12):633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kanherkar RR, Bhatia-Dey N, Csoka AB. Epigenetics across the human lifespan. Front Cell Dev Biol. 2014;2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y. Modern epigenetics methods in biological research. Methods. 2021;187:104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58(3):443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kato M, Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol. 2019;15(6):327–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Dabet MM, Shahzad K, Elwakiel A, Sulaj A, Kopf S, Bock F, et al. Reversal of the renal hyperglycemic memory in diabetic kidney disease by targeting sustained tubular p21 expression. Nat Commun. 2022;13(1):5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jung CY, Yoo TH. Pathophysiologic mechanisms and potential biomarkers in diabetic kidney disease. Diabetes Metab J. 2022;46(2):181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gembillo G, Ingrasciotta Y, Crisafulli S, Luxi N, Siligato R, Santoro D, et al. Kidney disease in diabetic patients: from pathophysiology to pharmacological aspects with a focus on therapeutic inertia. Int J Mol Sci. 2021;22(9):4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohandes S, Doke T, Hu H, Mukhi D, Dhillon P, Susztak K. Molecular pathways that drive diabetic kidney disease. J Clin Invest. 2023;133(4):e165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qaz M, Sawaf, H, Ismail, J, Qazi, H, Vachharajani, T. Pathophysiology of diabetic kidney disease. EMJ Nephrol. 2022;10(1):102–13. [Google Scholar]
- 12. Wolf G, Haberstroh U, Neilson EG. Angiotensin II stimulates the proliferation and biosynthesis of type I collagen in cultured murine mesangial cells. Am J Pathol. 1992;140(1):95–107. [PMC free article] [PubMed] [Google Scholar]
- 13. Ziyadeh FN, Sharma K, Ericksen M, Wolf G. Stimulation of collagen gene expression and protein synthesis in murine mesangial cells by high glucose is mediated by autocrine activation of transforming growth factor-beta. J Clin Invest. 1994;93(2):536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolf G, Chen S, Ziyadeh FN. From the periphery of the glomerular capillary wall toward the center of disease: podocyte injury comes of age in diabetic nephropathy. Diabetes. 2005;54(6):1626–34. [DOI] [PubMed] [Google Scholar]
- 15. Naber HP, Drabsch Y, Snaar-Jagalska BE, ten Dijke P, van Laar T. Snail and Slug, key regulators of TGF-β-induced EMT, are sufficient for the induction of single-cell invasion. Biochem Biophys Res Commun. 2013;435(1):58–63. [DOI] [PubMed] [Google Scholar]
- 16. Sugahara M, Pak WLW, Tanaka T, Tang SCW, Nangaku M. Update on diagnosis, pathophysiology, and management of diabetic kidney disease. Nephrology. 2021;26(6):491–500. [DOI] [PubMed] [Google Scholar]
- 17. Tuttle KR, Agarwal R, Alpers CE, Bakris GL, Brosius FC, Kolkhof P, et al. Molecular mechanisms and therapeutic targets for diabetic kidney disease. Kidney Int. 2022;102(2):248–60. [DOI] [PubMed] [Google Scholar]
- 18. Ruster C, Bondeva T, Franke S, Forster M, Wolf G. Advanced glycation end-products induce cell cycle arrest and hypertrophy in podocytes. Nephrol Dial Transpl. 2008;23(7):2179–91. [DOI] [PubMed] [Google Scholar]
- 19. Ruster C, Bondeva T, Franke S, Tanaka N, Yamamoto H, Wolf G. Angiotensin II upregulates RAGE expression on podocytes: role of AT2 receptors. Am J Nephrol. 2009;29(6):538–50. [DOI] [PubMed] [Google Scholar]
- 20. Bondeva T, Ruster C, Franke S, Hammerschmid E, Klagsbrun M, Cohen CD, et al. Advanced glycation end-products suppress neuropilin-1 expression in podocytes. Kidney Int. 2009;75(6):605–16. [DOI] [PubMed] [Google Scholar]
- 21. Loeffler I, Wolf G. Transforming growth factor-β and the progression of renal disease. Nephrol Dial Transpl. 2014;29(Suppl 1):i37–45. [DOI] [PubMed] [Google Scholar]
- 22. Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. [DOI] [PubMed] [Google Scholar]
- 23. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al Ojaimi M, Banimortada BJ, Othman A, Riedhammer KM, Almannai M, El-Hattab AW. Disorders of histone methylation: molecular basis and clinical syndromes. Clin Genet. 2022;102(3):169–81. [DOI] [PubMed] [Google Scholar]
- 25. Yu C, Zhuang S. Histone methyltransferases as therapeutic targets for kidney diseases. Front Pharmacol. 2019;10:1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DʼOto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1(2):34–40. [PMC free article] [PubMed] [Google Scholar]
- 27. Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor-Papadimitriou J, Burchell JM. Histone methylases and demethylases regulating antagonistic methyl marks: changes occurring in cancer. Cells. 2022;11(7):1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang YZ, Xu WW, Zhu DY, Zhang N, Wang YL, Ding M, et al. Specific expression network analysis of diabetic nephropathy kidney tissue revealed key methylated sites. J Cell Physiol. 2018;233(10):7139–47. [DOI] [PubMed] [Google Scholar]
- 30. Komers R, Mar D, Denisenko O, Xu B, Oyama TT, Bomsztyk K. Epigenetic changes in renal genes dysregulated in mouse and rat models of type 1 diabetes. Lab Invest. 2013;93(5):543–52. [DOI] [PubMed] [Google Scholar]
- 31. Chen H, Huang Y, Zhu X, Liu C, Yuan Y, Su H, et al. Histone demethylase UTX is a therapeutic target for diabetic kidney disease. J Physiol. 2019;597(6):1643–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu L, Zhong Z, Gu J, Nan K, Zhu M, Miao C. ets1 associates with KMT5A to participate in high glucose-mediated EndMT via upregulation of PFN2 expression in diabetic nephropathy. Mol Med. 2021;27(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Guo Y, Zeng W, Huang L, Pang Q, Nie L, et al. ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am J Physiol Ren Physiol. 2014;306(8):F916–925. [DOI] [PubMed] [Google Scholar]
- 34. Sharma N, Sankrityayan H, Kale A, Gaikwad AB. Role of SET7/9 in the progression of ischemic renal injury in diabetic and non-diabetic rats. Biochem Biophys Res Commun. 2020;528(1):14–20. [DOI] [PubMed] [Google Scholar]
- 35. Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, et al. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transpl. 2010;25(6):1811–7. [DOI] [PubMed] [Google Scholar]
- 36. Surse VM, Gupta J, Tikoo K. Esculetin induced changes in Mmp13 and Bmp6 gene expression and histone H3 modifications attenuate development of glomerulosclerosis in diabetic rats. J Mol Endocrinol. 2011;46(3):245–54. [DOI] [PubMed] [Google Scholar]
- 37. Zhang L, Chen L, Gao C, Chen E, Lightle AR, Foulke L, et al. Loss of histone H3 K79 methyltransferase Dot1l facilitates kidney fibrosis by upregulating endothelin 1 through histone deacetylase 2. J Am Soc Nephrol. 2020;31(2):337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21(12):2069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, Li C, Li X, Cui P, Li Q, Guo Q, et al. Involvement of histone lysine methylation in p21 gene expression in rat kidney in vivo and rat mesangial cells in vitro under diabetic conditions. J Diabetes Res. 2016;2016:3853242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolf G, Schroeder R, Zahner G, Stahl RA, Shankland SJ. High glucose-induced hypertrophy of mesangial cells requires p27(Kip1), an inhibitor of cyclin-dependent kinases. Am J Pathol. 2001;158(3):1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shankland SJ, Wolf G. Cell cycle regulatory proteins in renal disease: role in hypertrophy, proliferation, and apoptosis. Am J Physiol Ren Physiol. 2000;278(4):F515–529. [DOI] [PubMed] [Google Scholar]
- 42. Zahner G, Wolf G, Ayoub M, Reinking R, Panzer U, Shankland SJ, et al. Cyclooxygenase-2 overexpression inhibits platelet-derived growth factor-induced mesangial cell proliferation through induction of the tumor suppressor gene p53 and the cyclin-dependent kinase inhibitors p21waf-1/cip-1 and p27kip-1. J Biol Chem. 2002;277(12):9763–71. [DOI] [PubMed] [Google Scholar]
- 43. Bai S, Xiong X, Tang B, Ji T, Li X, Qu X, et al. hsa-miR-199b-3p prevents the epithelial-mesenchymal transition and dysfunction of the renal tubule by regulating E-cadherin through targeting KDM6A in diabetic nephropathy. Oxid Med Cell Longev. 2021;2021:8814163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang B, Ji G, Naeem H, Wang J, Kantharidis P, Powell D, et al. The use of targeted next generation sequencing to explore candidate regulators of TGF-β1ʼs impact on kidney cells. Front Physiol. 2018;9:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jia Y, Reddy MA, Das S, Oh HJ, Abdollahi M, Yuan H, et al. Dysregulation of histone H3 lysine 27 trimethylation in transforming growth factor-β1-induced gene expression in mesangial cells and diabetic kidney. J Biol Chem. 2019;294(34):12695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Estaras C, Fueyo R, Akizu N, Beltran S, Martinez-Balbas MA. RNA polymerase II progression through H3K27me3-enriched gene bodies requires JMJD3 histone demethylase. Mol Biol Cell. 2013;24(3):351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hung PH, Hsu YC, Chen TH, Ho C, Lin CL. The histone demethylase inhibitor GSK-J4 is a therapeutic target for the kidney fibrosis of diabetic kidney disease via DKK1 modulation. Int J Mol Sci. 2022;23(16):9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Majumder S, Thieme K, Batchu SN, Alghamdi TA, Bowskill BB, Kabir MG, et al. Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J Clin Invest. 2018;128(1):483–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siddiqi FS, Majumder S, Thai K, Abdalla M, Hu P, Advani SL, et al. The histone methyltransferase enzyme enhancer of zeste homolog 2 protects against podocyte oxidative stress and renal injury in diabetes. J Am Soc Nephrol. 2016;27(7):2021–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liebisch M, Wolf G. AGE-induced suppression of EZH2 mediates injury of podocytes by reducing H3K27me3. Am J Nephrol. 2020;51(9):676–92. [DOI] [PubMed] [Google Scholar]
- 51. Liebisch M, Bondeva T, Franke S, Daniel C, Amann K, Wolf G. Activation of the receptor for advanced glycation end products induces nuclear inhibitor of protein phosphatase-1 suppression. Kidney Int. 2014;86(1):103–17. [DOI] [PubMed] [Google Scholar]
- 52. Barth J, Loeffler I, Bondeva T, Liebisch M, Wolf G. The role of hypoxia on the trimethylation of H3K27 in podocytes. Biomedicines. 2023;11(9):2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin CL, Hsu YC, Huang YT, Shih YH, Wang CJ, Chiang WC, et al. A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction. EMBO Mol Med. 2019;11(5):e9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. De Marinis Y, Cai M, Bompada P, Atac D, Kotova O, Johansson ME, et al. Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney Int. 2016;89(2):342–53. [DOI] [PubMed] [Google Scholar]
- 55. Yuan H, Reddy MA, Deshpande S, Jia Y, Park JT, Lanting LL, et al. Epigenetic histone modifications involved in profibrotic gene regulation by 12/15-lipoxygenase and its oxidized lipid products in diabetic nephropathy. Antioxid Redox Signal. 2016;24(7):361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23(5):329–49. [DOI] [PubMed] [Google Scholar]
- 57. Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6(7):a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6(4):a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hou Q, Kan S, Wang Z, Shi J, Zeng C, Yang D, et al. Inhibition of HDAC6 with CAY10603 ameliorates diabetic kidney disease by suppressing NLRP3 inflammasome. Front Pharmacol. 2022;13:938391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun. 2017;8(1):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reddy MA, Sumanth P, Lanting L, Yuan H, Wang M, Mar D, et al. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int. 2014;85(2):362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang X, Liu J, Zhen J, Zhang C, Wan Q, Liu G, et al. Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int. 2014;86:712–25. [DOI] [PubMed] [Google Scholar]
- 63. Khan S, Jena G, Tikoo K. Sodium valproate ameliorates diabetes-induced fibrosis and renal damage by the inhibition of histone deacetylases in diabetic rat. Exp Mol Pathol. 2015;98(2):230–9. [DOI] [PubMed] [Google Scholar]
- 64. Xu Z, Jia K, Wang H, Gao F, Zhao S, Li F, et al. METTL14-regulated PI3K/Akt signaling pathway via PTEN affects HDAC5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. Cell Death Dis. 2021;12(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Noh H, Oh EY, Seo JY, Yu MR, Kim YO, Ha H, et al. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury. Am J Physiol Ren Physiol. 2009;297(3):F729–39. [DOI] [PubMed] [Google Scholar]
- 66. Zheng Z, Zhang S, Chen J, Zou M, Yang Y, Lu W, et al. The HDAC2/SP1/miR-205 feedback loop contributes to tubular epithelial cell extracellular matrix production in diabetic kidney disease. Clin Sci. 2022;136(3):223–38. [DOI] [PubMed] [Google Scholar]
- 67. Khan S, Jena G. Sodium butyrate, a HDAC inhibitor ameliorates eNOS, iNOS and TGF-β1-induced fibrogenesis, apoptosis and DNA damage in the kidney of juvenile diabetic rats. Food Chem Toxicol. 2014;73:127–39. [DOI] [PubMed] [Google Scholar]
- 68. Ma X, Wang Q. Short-chain fatty acids attenuate renal fibrosis and enhance autophagy of renal tubular cells in diabetic mice through the HDAC2/ULK1 Axis. Endocrinol Metab Seoul. 2022;37(3):432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yoshikawa M, Hishikawa K, Marumo T, Fujita T. Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by TGF-beta1 in human renal epithelial cells. J Am Soc Nephrol. 2007;18(1):58–65. [DOI] [PubMed] [Google Scholar]
- 70. Malek V, Sharma N, Gaikwad AB. Histone acetylation regulates natriuretic peptides and neprilysin gene expressions in diabetic cardiomyopathy and nephropathy. Curr Mol Pharmacol. 2019;12(1):61–71. [DOI] [PubMed] [Google Scholar]
- 71. Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-β1-mediated gene transcription in mesangial cells. Am J Physiol Ren Physiol. 2013;304(5):F601–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu H, Wu X, Qin H, Tian W, Chen J, Sun L, et al. Myocardin-related transcription factor A epigenetically regulates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2015;26(7):1648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lazar AG, Vlad ML, Manea A, Simionescu M, Manea SA. Activated histone acetyltransferase p300/CBP-related signalling pathways mediate up-regulation of NADPH oxidase, inflammation, and fibrosis in diabetic kidney. Antioxidants. 2021;10(9):1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu F, Zong M, Wen X, Li X, Wang J, Wang Y, et al. Silencing of histone deacetylase 9 expression in podocytes attenuates kidney injury in diabetic nephropathy. Sci Rep. 2016;6:33676. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75. Li S, Tollefsbol TO. DNA methylation methods: global DNA methylation and methylomic analyses. Methods. 2021;187:28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Greenberg MVC, Bourcʼhis D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. [DOI] [PubMed] [Google Scholar]
- 77. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81–92. [DOI] [PubMed] [Google Scholar]
- 78. Smyth LJ, Kilner J, Nair V, Liu H, Brennan E, Kerr K, et al. Assessment of differentially methylated loci in individuals with end-stage kidney disease attributed to diabetic kidney disease: an exploratory study. Clin Epigenetics. 2021;13(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Khurana I, Kaipananickal H, Maxwell S, Birkelund S, Syreeni A, Forsblom C, et al. Reduced methylation correlates with diabetic nephropathy risk in type 1 diabetes. J Clin Invest. 2023;133(4):e160959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6(1):20–8. [DOI] [PubMed] [Google Scholar]
- 81. Aldemir O, Turgut F, Gokce C. The association between methylation levels of targeted genes and albuminuria in patients with early diabetic kidney disease. Ren Fail. 2017;39(1):597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smyth LJ, Dahlstrom EH, Syreeni A, Kerr K, Kilner J, Doyle R, et al. Epigenome-wide meta-analysis identifies DNA methylation biomarkers associated with diabetic kidney disease. Nat Commun. 2022;13(1):7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bomsztyk K, Denisenko O, Wang Y. DNA methylation yields epigenetic clues into the diabetic nephropathy of Pima Indians. Kidney Int. 2018;93(6):1272–5. [DOI] [PubMed] [Google Scholar]
- 84. Lecamwasam A, Novakovic B, Meyer B, Ekinci EI, Dwyer KM, Saffery R. DNA methylation profiling identifies epigenetic differences between early versus late stages of diabetic chronic kidney disease. Nephrol Dial Transpl. 2021;36(11):2027–38. [DOI] [PubMed] [Google Scholar]
- 85. Marumo T, Hoshino J, Kawarazaki W, Nishimoto M, Ayuzawa N, Hirohama D, et al. Methylation pattern of urinary DNA as a marker of kidney function decline in diabetes. BMJ Open Diabetes Res Care. 2020;8(1):e001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cole E, Brown TA, Pinkerton KE, Postma B, Malany K, Yang M, et al. Perinatal exposure to environmental tobacco smoke is associated with changes in DNA methylation that precede the adult onset of lung disease in a mouse model. Inhal Toxicol. 2017;29(10):435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoshimoto N, Hayashi K, Hishikawa A, Hashiguchi A, Nakamichi R, Sugita-Nishimura E, et al. Significance of podocyte DNA damage and glomerular DNA methylation in CKD patients with proteinuria. Hypertens Res. 2023;46(4):1000–8. [DOI] [PubMed] [Google Scholar]
- 88. Li KY, Tam CHT, Liu H, Day S, Lim CKP, So WY, et al. DNA methylation markers for kidney function and progression of diabetic kidney disease. Nat Commun. 2023;14(1):2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chu AY, Tin A, Schlosser P, Ko YA, Qiu C, Yao C, et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8(1):1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen Z, Miao F, Braffett BH, Lachin JM, Zhang L, Wu X, et al. DNA methylation mediates development of HbA1c-associated complications in type 1 diabetes. Nat Metab. 2020;2(8):744–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen Z, Miao F, Paterson AD, Lachin JM, Zhang L, Schones DE, et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci USA. 2016;113(21):E3002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim H, Bae JH, Park KS, Sung J, Kwak SH. DNA methylation changes associated with type 2 diabetes and diabetic kidney disease in an east asian population. J Clin Endocrinol Metab. 2021;106(10):e3837–51. [DOI] [PubMed] [Google Scholar]
- 93. Gluck C, Qiu C, Han SY, Palmer M, Park J, Ko YA, et al. Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nat Commun. 2019;10(1):2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gondaliya P, Dasare A, Srivastava A, Kalia K. miR29b regulates aberrant methylation in In-Vitro diabetic nephropathy model of renal proximal tubular cells. PLoS One. 2018;13(11):e0208044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Oba S, Ayuzawa N, Nishimoto M, Kawarazaki W, Ueda K, Hirohama D, et al. Aberrant DNA methylation of Tgfb1 in diabetic kidney mesangial cells. Sci Rep. 2018;8(1):16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang L, Zhang Q, Wu Q, Wei Y, Yu J, Mu J, et al. Effect of TET2 on the pathogenesis of diabetic nephropathy through activation of transforming growth factor β1 expression via DNA demethylation. Life Sci. 2018;207:127–37. [DOI] [PubMed] [Google Scholar]
- 97. Hayashi K, Sasamura H, Nakamura M, Azegami T, Oguchi H, Sakamaki Y, et al. KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J Clin Invest. 2014;124(6):2523–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li Z, Chen H, Zhong F, Zhang W, Lee K, He JC. Expression of glutamate receptor subtype 3 is epigenetically regulated in podocytes under diabetic conditions. Kidney Dis. 2019;5(1):34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang L, Zhang Q, Liu S, Chen Y, Li R, Lin T, et al. DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int. 2017;92(1):140–53. [DOI] [PubMed] [Google Scholar]
- 100. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. 2024;25(3):211–32. [DOI] [PubMed] [Google Scholar]
- 101. Franczyk B, Gluba-Brzozka A, Olszewski R, Parolczyk M, Rysz-Gorzynska M, Rysz J. miRNA biomarkers in renal disease. Int Urol Nephrol. 2022;54(3):575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Szeto CC. Urine miRNA in nephrotic syndrome. Clin Chim Acta. 2014;436:308–13. [DOI] [PubMed] [Google Scholar]
- 103. Mahtal N, Lenoir O, Tinel C, Anglicheau D, Tharaux PL. MicroRNAs in kidney injury and disease. Nat Rev Nephrol. 2022;18(10):643–62. [DOI] [PubMed] [Google Scholar]
- 104. Beltrami C, Simpson K, Jesky M, Wonnacott A, Carrington C, Holmans P, et al. Association of elevated urinary miR-126, miR-155, and miR-29b with diabetic kidney disease. Am J Pathol. 2018;188(9):1982–92. [DOI] [PubMed] [Google Scholar]
- 105. Eissa S, Matboli M, Aboushahba R, Bekhet MM, Soliman Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J Diabetes Complications. 2016;30(8):1585–92. [DOI] [PubMed] [Google Scholar]
- 106. Zapala B, Kaminska A, Piwowar M, Paziewska A, Gala-Bladzinska A, Stepien EL. miRNA signature of urine extracellular vesicles shows the involvement of inflammatory and apoptotic processes in diabetic chronic kidney disease. Pharm Res. 2023;40(4):817–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zang J, Maxwell AP, Simpson DA, McKay GJ. Differential expression of urinary exosomal MicroRNAs miR-21-5p and miR-30b-5p in individuals with diabetic kidney disease. Sci Rep. 2019;9(1):10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sinha N, Puri V, Kumar V, Nada R, Rastogi A, Jha V, et al. Urinary exosomal miRNA-663a shows variable expression in diabetic kidney disease patients with or without proteinuria. Sci Rep. 2023;13(1):4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhao Y, Shen A, Guo F, Song Y, Jing N, Ding X, et al. Urinary exosomal MiRNA-4534 as a novel diagnostic biomarker for diabetic kidney disease. Front Endocrinol. 2020;11:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li X, Xu R, Liu X, Xu L, Xue M, Cheng Y, et al. Urinary miR-3137 and miR-4270 as potential biomarkers for diabetic kidney disease. J Clin Lab Anal. 2020;34(12):e23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jia Y, Zheng Z, Yang Y, Zou M, Li J, Wang L, et al. MiR-4756 promotes albumin-induced renal tubular epithelial cell epithelial-to-mesenchymal transition and endoplasmic reticulum stress via targeting Sestrin2. J Cell Physiol. 2019;234(3):2905–15. [DOI] [PubMed] [Google Scholar]
- 112. Wang J, Tao Y, Zhao F, Liu T, Shen X, Zhou L. Expression of urinary exosomal miRNA-615-3p and miRNA-3147 in diabetic kidney disease and their association with inflammation and fibrosis. Ren Fail. 2023;45(1):2121929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. El-Samahy MH, Adly AA, Elhenawy YI, Ismail EA, Pessar SA, Mowafy ME, et al. Urinary miRNA-377 and miRNA-216a as biomarkers of nephropathy and subclinical atherosclerotic risk in pediatric patients with type 1 diabetes. J Diabetes Complications. 2018;32(2):185–92. [DOI] [PubMed] [Google Scholar]
- 114. Zahari Sham SY, Ng CT, Azwar S, Yip WK, Abdullah M, Thevandran K, et al. Circulating miRNAs in type 2 diabetic patients with and without albuminuria in Malaysia. Kidney Blood Press Res. 2022;47(2):81–93. [DOI] [PubMed] [Google Scholar]
- 115. Regmi A, Liu G, Zhong X, Hu S, Ma R, Gou L, et al. Evaluation of serum microRNAs in patients with diabetic kidney disease: a nested case-controlled study and bioinformatics analysis. Med Sci Monit. 2019;25:1699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Santos-Bezerra DP, Santos AS, Guimaraes GC, Admoni SN, Perez RV, Machado CG, et al. Micro-RNAs 518d-3p and 618 are upregulated in individuals with type 1 diabetes with multiple microvascular complications. Front Endocrinol. 2019;10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fouad M, Salem I, Elhefnawy K, Raafat N, Faisal A. MicroRNA-21 as an early marker of nephropathy in patients with type 1 diabetes. Indian J Nephrol. 2020;30(1):21–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Assmann TS, Recamonde-Mendoza M, Costa AR, Punales M, Tschiedel B, Canani LH, et al. Circulating miRNAs in diabetic kidney disease: case-control study and in silico analyses. Acta Diabetol. 2019;56(1):55–65. [DOI] [PubMed] [Google Scholar]
- 119. Conserva F, Barozzino M, Pesce F, Divella C, Oranger A, Papale M, et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of kidney fibrosis in diabetic nephropathy. Sci Rep. 2019;9(1):11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dieter C, Assmann TS, Costa AR, Canani LH, de Souza BM, Bauer AC, et al. MiR-30e-5p and MiR-15a-5p expressions in plasma and urine of type 1 diabetic patients with diabetic kidney disease. Front Genet. 2019;10:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ji JL, Shi HM, Li ZL, Jin R, Qu GT, Zheng H, et al. Satellite cell-derived exosome-mediated delivery of microRNA-23a/27a/26a cluster ameliorates the renal tubulointerstitial fibrosis in mouse diabetic nephropathy. Acta Pharmacol Sin. 2023;44(12):2455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zanchi C, Macconi D, Trionfini P, Tomasoni S, Rottoli D, Locatelli M, et al. MicroRNA-184 is a downstream effector of albuminuria driving renal fibrosis in rats with diabetic nephropathy. Diabetologia. 2017;60(6):1114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lee HW, Khan SQ, Khaliqdina S, Altintas MM, Grahammer F, Zhao JL, et al. Absence of miR-146a in podocytes increases risk of diabetic glomerulopathy via up-regulation of ErbB4 and notch-1. J Biol Chem. 2017;292(2):732–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chen S. circ_000166/miR-296 aggravates the process of diabetic renal fibrosis by regulating the SGLT2 signaling pathway in renal tubular epithelial cells. Dis Markers. 2022;2022:6103086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang LP, Geng JN, Sun B, Sun CB, Shi Y, Yu XY. MiR-92b-3p is induced by advanced glycation end products and involved in the pathogenesis of diabetic nephropathy. Evid Based Complement Alternat Med. 2020;2020:6050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sheng S, Zou M, Yang Y, Guan M, Ren S, Wang X, et al. miR-23a-3p regulates the inflammatory response and fibrosis in diabetic kidney disease by targeting early growth response 1. Vitro Cell Dev Biol Anim. 2021;57(8):763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Su J, Ren J, Chen H, Liu B. MicroRNA-140-5p ameliorates the high glucose-induced apoptosis and inflammation through suppressing TLR4/NF-κB signaling pathway in human renal tubular epithelial cells. Biosci Rep. 2020;40(3):BSR20192384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Fu Y, Zhang Y, Wang Z, Wang L, Wei X, Zhang B, et al. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am J Nephrol. 2010;32(6):581–9. [DOI] [PubMed] [Google Scholar]
- 129. Wang X, Gao Y, Yi W, Qiao Y, Hu H, Wang Y, et al. Inhibition of miRNA-155 alleviates high glucose-induced podocyte inflammation by targeting SIRT1 in diabetic mice. J Diabetes Res. 2021;2021:5597394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wang Y, Le Y, Xue JY, Zheng ZJ, Xue YM. Let-7d miRNA prevents TGF-β1-induced EMT and renal fibrogenesis through regulation of HMGA2 expression. Biochem Biophys Res Commun. 2016;479(4):676–82. [DOI] [PubMed] [Google Scholar]
- 131. Castro NE, Kato M, Park JT, Natarajan R. Transforming growth factor β1 (TGF-β1) enhances expression of profibrotic genes through a novel signaling cascade and microRNAs in renal mesangial cells. J Biol Chem. 2014;289(42):29001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Huang C, Yu J, Da J, Dong R, Dai L, Yang Y, et al. Dendrobium officinale Kimura and Migo polysaccharide inhibits hyperglycaemia-induced kidney fibrosis via the miRNA-34a-5p/SIRT1 signalling pathway. J Ethnopharmacol. 2023;313:116601. [DOI] [PubMed] [Google Scholar]
- 133. Tabei A, Sakairi T, Hamatani H, Ohishi Y, Watanabe M, Nakasatomi M, et al. The miR-143/145 cluster induced by TGF-β1 suppresses Wilmsʼ tumor 1 expression in cultured human podocytes. Am J Physiol Ren Physiol. 2023;325:F121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bian C, Luan Z, Zhang H, Zhang R, Gao J, Wang Y, et al. miR-154-5p affects the tgfβ1/smad3 pathway on the fibrosis of diabetic kidney disease via binding E3 ubiquitin ligase Smurf1. Oxid Med Cell Longev. 2022;2022:7502632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80(4):358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kato M, Dang V, Wang M, Park JT, Deshpande S, Kadam S, et al. TGF-β induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal. 2013;6(278):ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cheng Y, Wang D, Wang F, Liu J, Huang B, Baker MA, et al. Endogenous miR-204 protects the kidney against chronic injury in hypertension and diabetes. J Am Soc Nephrol. 2020;31(7):1539–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Jiang ZH, Tang YZ, Song HN, Yang M, Li B, Ni CL. miRNA-342 suppresses renal interstitial fibrosis in diabetic nephropathy by targeting SOX6. Int J Mol Med. 2020;45(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang B, Yao K, Wise AF, Lau R, Shen HH, Tesch GH, et al. miR-378 reduces mesangial hypertrophy and kidney tubular fibrosis via MAPK signalling. Clin Sci. 2017;131(5):411–23. [DOI] [PubMed] [Google Scholar]
- 140. Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One. 2013;8(10):e77468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Baker MA, Davis SJ, Liu P, Pan X, Williams AM, Iczkowski KA, et al. Tissue-specific MicroRNA expression patterns in four types of kidney disease. J Am Soc Nephrol. 2017;28(10):2985–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kolling M, Kaucsar T, Schauerte C, Hubner A, Dettling A, Park JK, et al. Therapeutic miR-21 silencing ameliorates diabetic kidney disease in mice. Mol Ther. 2017;25(1):165–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23(3):458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Loeffler I, Ziller N. Sex-related aspects in diabetic kidney disease-an update. J Clin Med. 2023;12(8):2834. [DOI] [PMC free article] [PubMed] [Google Scholar]