Abstract
Transposable elements (TEs) are repressed in plants through transcriptional gene silencing (TGS), maintained epigenetic silencing marks such as DNA methylation. However, the mechanisms by which silencing is first installed remain poorly understood in plants. Small interfering (si)RNAs and post-transcriptional gene silencing (PTGS) are believed to mediate the initiation of TGS by guiding the first deposition of DNA methylation. To determine how this silencing installation works, we took advantage of ÉVADÉ (EVD), an endogenous retroelement in Arabidopsis, able to recapitulate true de novo silencing with a sequence of PTGS followed by a TGS. To test whether PTGS is required for TGS, we introduce active EVD into RNA-DEPENDENT-RNA-POLYMERASE-6 (RDR6) mutants, an essential PTGS component. EVD activity and silencing are monitored across several generations. In the absence of PTGS, silencing of EVD is still achieved through installation of RNA-directed DNA methylation (RdDM). Our study shows that PTGS is dispensable for de novo EVD silencing. Although we cannot rule out that PTGS might facilitate TGS, or control TE activity, initiation of epigenetic silencing can take place in its absence.
Keywords: Silencing, Transposons, siRNAs, Epigenetics, Plants
Subject terms: Chromatin, Transcription & Genomics; Plant Biology
Synopsis
PTGS facilitates but is not required for the de novo epigenetic silencing of EVD, a proliferative retrotransposon in Arabidopsis.
PTGS and associated RDR6-RdDM promote homogeneous epigenetic silencing of 40–50 copies.
In absence of RDR6, silencing and installation of PolIV-RdDM at EVD regulatory sequences takes place stochastically.
TGS installation is independent of DNA methylation at EVD coding sequence and copy number.
PTGS facilitates but is not required for the de novo epigenetic silencing of EVD, a proliferative retrotransposon in Arabidopsis.

Introduction
Due to their mobile nature, transposable elements (TEs) pose a threat to genome integrity and can cause mutations compromising host fitness (McClintock, 1984; Bennetzen and Wang, 2014; Schubert and Vu, 2016; Bourque et al, 2018). TEs are mostly transcriptionally repressed across genomes through the action of epigenetic silencing mechanisms, limiting TE activity (Lippman et al, 2004; Allshire and Madhani, 2018).
In Arabidopsis thaliana, the best studied model for plants, DNA methylation (5-methylcytosine; 5mC) and histone H3 lysine-9 di-methylation (H3K9me2) cooperatively mediate transcriptional gene silencing (TGS) of TEs (Bernatavichute et al, 2008). Once established, 5mC and H3K9me2 patterns are propagated across generations to ensure transgenerational silencing of TEs. Maintenance of cytosine methylation depends on its sequence context, CG, CHG, or CHH (where H can be any nucleotide besides G). METHYLTRANSFERASE 1 (MET1) preserves 5mC in the CG context after each DNA replication cycle (Mathieu et al, 2007). Maintenance in CHG and CHH contexts occurs through a self-reinforcing loop with H3K9me2 (Chan et al, 2005; Du et al, 2012; Law et al, 2013; Du et al, 2014). In addition, DNA methylation deposition, particularly in the CHH context, can be mediated by the RNA-directed DNA methylation (RdDM) pathway. Canonical RdDM relies on the action of two plant specific RNA polymerases, PolIV and PolV. On the one hand, PolIV is recruited to TE loci through the histone mark H3K9me2. PolIV transcripts are converted into dsRNAs. These are further processed into 24-nt siRNA by DICER-LIKE 3 (DCL3), which are loaded into ARGONAUTE (AGO) 4/6-clade proteins. On the other hand, PolV is recruited to DNA-methylated TE loci. PolV provides the scaffold/target transcript for loaded AGO proteins, guiding the deposition of DNA methylation in all cytosine contexts (Law and Jacobsen, 2010; Kuhlmann and Mette, 2012; Matzke and Mosher, 2014; Erdmann and Picard, 2020). Given the dependency on PolIV-derived siRNAs, this branch of RdDM is also known as PolIV-RdDM.
While the maintenance of TE silencing relying on pre-existing heterochromatic marks is well described, the deposition of de novo silencing marks on active, proliferative transposable elements remains poorly understood. To gain insight into the initial molecular events by which plants recognize and silence active TEs, several studies have investigated host responses to environmentally, chemically, developmentally, or genetically induced TE reactivation (Teixeira et al, 2009; Slotkin et al, 2009; Mirouze et al, 2009; Reinders et al, 2009; Ito et al, 2011; Thieme et al, 2017). One of the best studied TEs in Arabidopsis is the Ty1/Copia long terminal repeat (LTR) retrotransposon ÉVADÉ (EVD; Copia93) (Mirouze et al, 2009). EVD is a functional, low copy TE in the reference Col-0 Arabidopsis ecotype and mainly regulated through CG methylation. Therefore, it can be released from silencing through loss of MET1 or the chromatin remodeler DECREASE IN DNA METHYLATION 1 (DDM1). Once lost, CG methylation cannot be reestablished, thus reactivated EVD remains active even after the reintroduction of functional (wild type) alleles of either MET1 or DDM1 and can rapidly increase in copy number all over the genome (Mathieu et al, 2007; Reinders et al, 2009; Mirouze et al, 2009). Owing to such property, EVD has quickly become a model system to study retrotransposon biology, TE bursts, and de novo silencing phenomena (Mirouze et al, 2009; Tsukahara et al, 2009; Marí-Ordóñez et al, 2013; Oberlin et al, 2017).
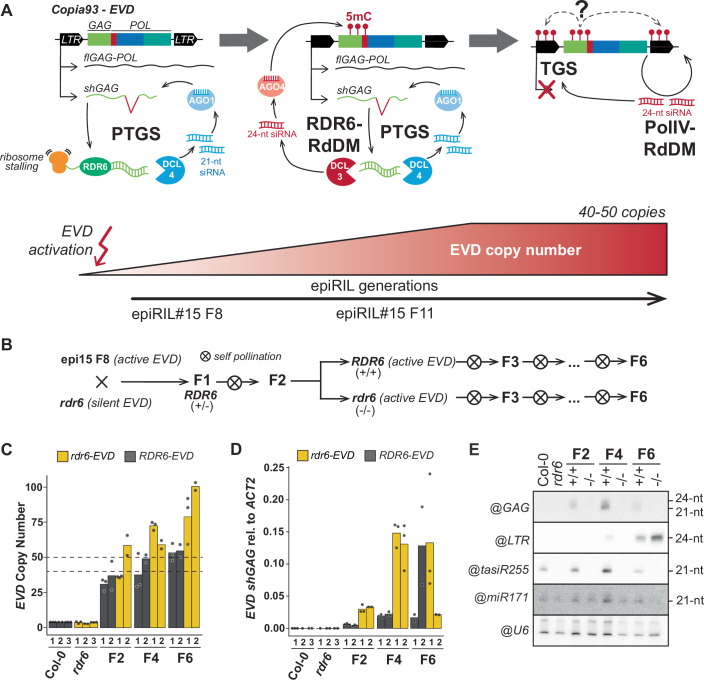
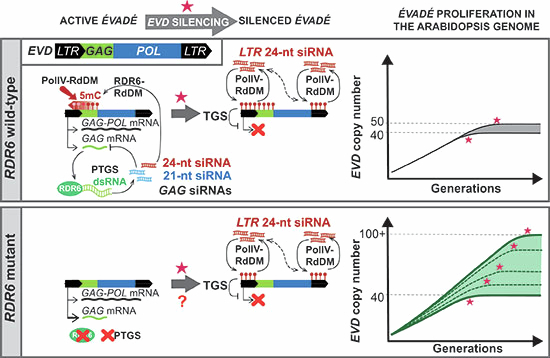
The EVD genome colonization and silencing cycle can be divided in well-defined stages. First, upon EVD reactivation, post-transcriptional gene silencing (PTGS) acts as initial host response (Fig. 1A) (Marí-Ordóñez et al, 2013; Oberlin et al, 2022). EVD PTGS is the result of its transcriptional and translational strategy to complete its transposition cycle. Due to an alternative splicing event, EVD produces two transcripts: (i) a full-length polycistronic mRNA (flGAG-POL) encoding for both its structural (Gag nucleocapsid) and catalytic (Pol) components; (ii) a short, Gag only transcript (shGAG), which is preferentially translated to generate the molar excess of Gag-to-Pol needed for the formation of virus-like particle (VLP) (Oberlin et al, 2017; 2022). A ribosome stalling event triggered during shGAG translation leads to the cleavage of the transcript. The resulting 3′ RNA fragment becomes a substrate for the RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) to produce double-stranded (ds)RNA, further processed by DICER-LIKE 4 (DCL4) into GAG-derived 21-nt siRNA (Oberlin et al, 2022). Albeit PTGS does reduce EVD GAG mRNA and protein levels, it does not prevent EVD transposition (Marí-Ordóñez et al, 2013; Oberlin et al, 2022). Next, EVD continues to increase its copy number across generations. When a threshold of 40–50 EVD copies per genome is reached, the excess of dsRNA produced by RDR6 is eventually processed by DCL3, giving rise to a population of GAG-derived 24-nt siRNAs (Fig. 1A). Loaded into AGO4-clade proteins, they guide the deposition of DNA methylation at EVD-GAG-coding sequences in a non-canonical RdDM pathway also known as RDR6-RdDM (Nuthikattu et al, 2013; Marí-Ordóñez et al, 2013). Finally, following GAG methylation, a switch from PTGS to transcriptional gene silencing (TGS) takes place. EVD TGS is characterized by the installation of PolIV-RdDM and the production of 24-nt siRNAs from EVD-LTR sequences, mediating DNA methylation of its regulatory sequences and the de novo TGS of new EVD copies (Fig. 1A) (Marí-Ordóñez et al, 2013).
Figure 1. Introgression and characterization of EVD in the rdr6 mutant background.
(A) Schematic representation of the three EVD silencing steps. Upon EVD reactivation, ribosome stalling during translation of EVD shGAG transcript triggers PTGS. 21-nt siRNA produced through RDR6 and DCL4 and loaded into AGO1. With increasing EVD copies across generations, the excess of dsRNA produced by RDR6 is processed by DCL3 to generate 24-nt siRNAs. Loaded into AGO4, shGAG siRNAs trigger DNA methylation (5mC) through RDR6-RdDM at GAG coding sequences without silencing. At 40–50 copies per genome, TGS is installed through Pol IV-RdDM, coincidentally with the appearance of DNA methylation and 24-nt siRNAs on the LTR sequences. (B) Crossing scheme to generate rdr6 mutant lines with active EVD. F2 plants were genotyped to select homozygous WT and mutant RDR6 lines, propagated through selfing until the F6 generation. (C) EVD copy number analysis by qPCR in RDR6-EVD and rdr6-EVD lines at generations F2, F4, and F6 derived from two independent F1s (biological replicates), using the EVD-GAG sequence as target. (D) qPCR analysis of shGAG expression normalized to ACT2 in EVD-RDR6 and EVD-rdr6 lines at generations 2, 4, and 6 derived from two independent F1s (biological replicates). In (C) and (D), each biological replicate, consistent of bulks of 8–10 plants, are represented for each genotype at each generation, dots show technical replicates. (E) RNA blot analysis of EVD siRNAs against GAG and LTR in RDR6 and rdr6 lines with active EVD at generations F2, F4, and F6. tasiR255 probe is used as control for RDR6 mutation, miR171 and snoRNA U6 are shown as loading controls. WT Col-0 and rdr6 with no reactivated EVD are shown as negative control for EVD activity. Source data are available online for this figure.
These observations have led to a model under which RDR6-RdDM is thought to contribute to the switch from PTGS to TGS by initiating the deposition of DNA methylation on EVD GAG-coding sequences. PTGS-derived siRNAs have been shown to play a role in restoring DNA methylation patterns in DNA methylation mutants (Teixeira et al, 2009; Nuthikattu et al, 2013; McCue et al, 2015) or during key developmental processes involving epigenetic reprogramming (Slotkin et al, 2009). Hence, RDR6-RdDM has been suggested as essential step to gap the transition from PTGS to TGS in the process of de novo transposon silencing (Marí-Ordóñez et al, 2013; Nuthikattu et al, 2013; McCue et al, 2015; Panda et al, 2016). More recently, using an EVD transgenic system, a mechanism for such transition has been proposed, under which AGO4, loaded with RDR6-derived siRNAs, interacts with PolII EVD transcripts at EVD loci to recruit PolV and downstream silencing components to ignite self-sustained PolIV-RdDM (Sigman et al, 2021). However, the requirement of PTGS for the initiation of TGS during a TE colonization event has never been experimentally tested. Furthermore, upon genome-wide loss of DNA methylation, the majority of reactivated TEs triggering RDR6-dependent PTGS are decayed TE-remnants incapable of transposition, while those intact enough to engage in translation do not (Oberlin et al, 2022). This has casted doubt on the universality of PTGS as a sensor of active TEs and initiator of epigenetic silencing during a TE colonization event.
In this study we address the necessity of PTGS and RDR6-RdDM in the process of de novo silencing of transposable elements. To achieve this, active EVD was introduced into RDR6 mutant plants to prevent RDR6-RdDM. EVD activity and silencing status were monitored over several generations in wild-type (WT) and rdr6 lines. While the mutation of RDR6 successfully prevented the production of siRNAs involved in RDR6-RdDM, silencing of EVD through installation of PolIV-RdDM in its LTRs was achieved in both, WT and mutant background. Therefore, PTGS is not essential for the installation of epigenetic silencing at active EVD copies. Given that EVD presents a unique case in triggering PTGS upon reactivation in the first place, we suggest that PTGS and RDR6-derived siRNA play a role in limiting EVD expression and transposition rather than initiating de novo silencing.
Results
Absence of RDR6 does not prevent the production of EVD-LTR 24-nt siRNAs
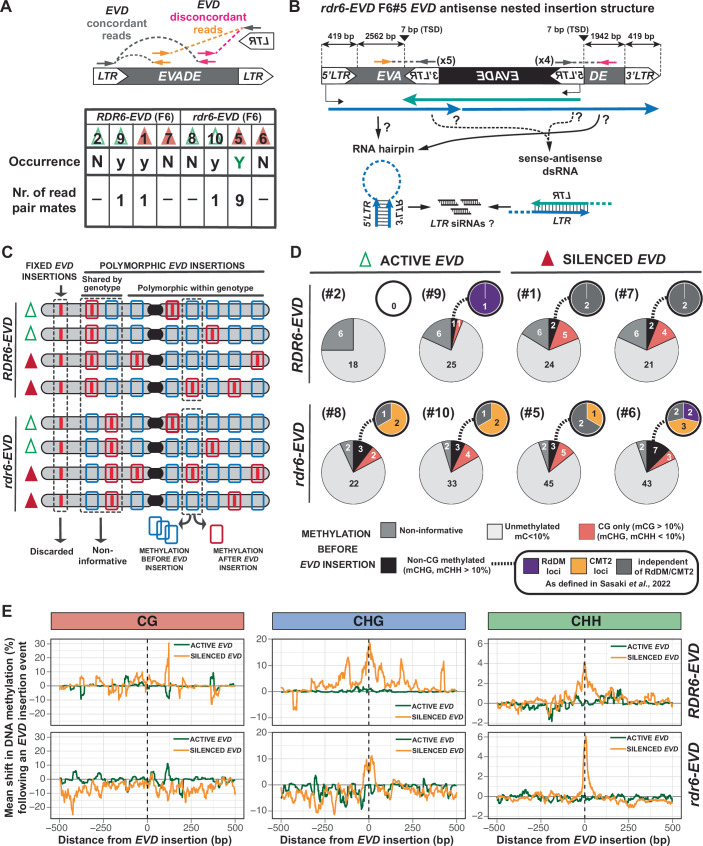
To investigate the role of PTGS and RDR6-RdDM in the initiation of canonical RdDM on EVD, and given that RDR6 is responsible for the production of EVD siRNA during the PTGS phase (Oberlin et al, 2017; 2022), rdr6-15 mutant plants were crossed to the 8th generation of the met1-derived epigenetic recombinant inbred line (epiRILs) number 15 (epi15 F8), a generation in which RDR6-RdDM has not yet been activated (Fig. 1B) (Marí-Ordóñez et al, 2013; Oberlin et al, 2022). This minimized the probability of introducing active EVD copies with GAG DNA methylation in the cross to rdr6. In the second generation (F2), two homozygous RDR6 wild-type (RDR6) and two mutant (rdr6) plants were selected by genotyping (hereafter referred to as RDR6-EVD and rdr6-EVD, respectively). They were allowed to self-pollinate in order to bulk-propagate two independent WT and two independent mutant populations until the 6th generation (F6), allowing EVD to colonize the genome (Fig. 1B). The silencing status of EVD in the two independent respective populations was monitored in bulks of 8 plants at generations F2, F4, and F6 by assessing the EVD copy number and expression levels through qPCR as well as the siRNA profile by RNA blot.
EVD copy numbers consistently increased across generations, confirming the inheritance of active, transposition-competent EVD copies from epi15 (Fig. 1C). EVD copies accumulated at a higher rate in rdr6-EVD background than in RDR6-EVD. The estimated copy number was consistently higher in rdr6-EVD than in RDR6-EVD plants, and the difference was significant in the F6, where loss of RDR6 activity allowed the accumulation of over 100 EVD copies (Fig. 1C). Consistent with an increasing copy number, EVD transcripts were also more abundant up to the 4th generation, particularly in rdr6-EVD. However, at the F6, large variations between biological replicates were observed in both RDR6-EVD and rdr6-EVD (Fig. 1D). This variation was observed consistently for the expression of both shGAG and flGAG-POL isoforms (Figs. 1D and EV1), suggesting a biological rather than a technical origin. The decrease and broad variation in EVD expression compared to the F4 (and despite the increase in copy number) suggested that TGS had started to take place. This was expected for the RDR6-EVD as the bulk of F6 individuals has already exceeded the 40–50 copy number limit, previously demonstrated to lead to TGS (Marí-Ordóñez et al, 2013). However, that result was unanticipated and intriguing for the rdr6-EVD lines and initiated the analysis of the potential EVD silencing mechanism by investigating the small RNA profile by RNA blots.
Figure EV1. EVD GAG-POL expression in RDR6 wild-type and mutant backgrounds.
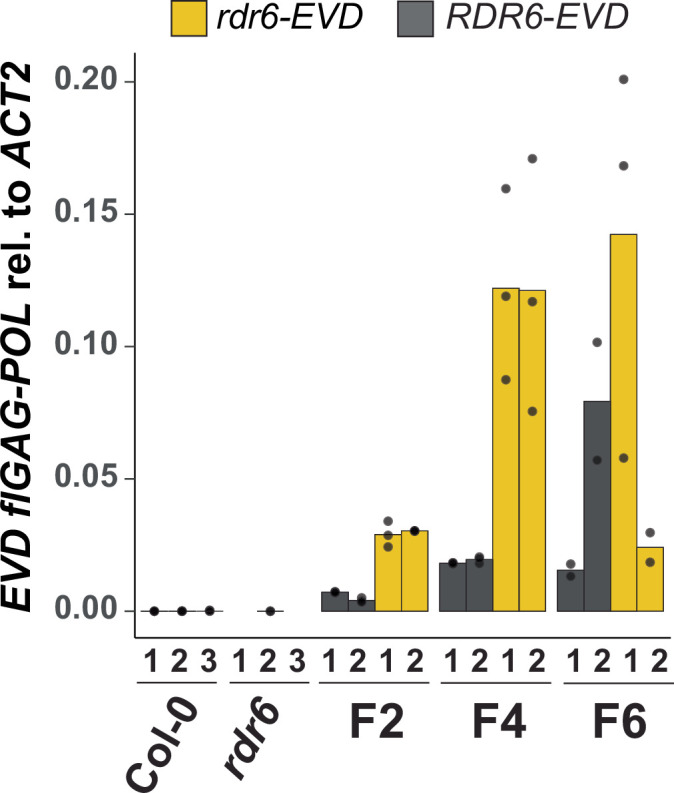
(A) qPCR analysis of flGAG-POL expression normalized to ACT2 in EVD-RDR6 and EVD-rdr6 lines at generations 2, 4, and 6 derived from two independent F1s (biological replicates). Each biological replicate, consistent of bulks of 8–10 plants, are represented for each genotype at each generation, dots show technical replicates. Source data are available online for this figure.
Although the transition of EVD from PTGS to TGS has been demonstrated to take place at the individual plant level, the detection of EVD-GAG and EVD-LTR siRNA in bulked material can also be used as proxy for the assessment of EVD silencing stage at each generation (Marí-Ordóñez et al, 2013). As expected from the dependency of GAG-derived siRNA on EVD expression, EVD-GAG siRNAs were detected in RDR6-EVD throughout generations, mirroring EVD expression levels (Fig. 1E). In these plants, 24-nt EVD-LTR siRNAs, involved in PolIV-RdDM and transcriptional silencing of EVD, were first detected in the F4 and increased in the F6 generation, coinciding with the decrease of EVD expression and GAG siRNAs (Fig. 1E). In contrast, as expected in the absence of RDR6, no RDR6-derived EVD-GAG siRNAs and trans-acting (ta)siRNAs were detectable in rdr6-EVD plants, EVD-LTR siRNAs were present in the F6 generation (Fig. 1E). Thus, the lack of PTGS and RDR6-derived EVD-GAG siRNA did not prevent the appearance of EVD-LTR 24-nt siRNAs, previously associated with EVD TGS.
RDR6-dependent EVD-GAG siRNAs are dispensable for EVD TGS
The presence of 24-nt EVD-LTR siRNAs, hallmark of successful PolIV-RdDM installation, indicated that silencing of EVD copies through TGS was likely taking place in both RDR6-EVD and rdr6-EVD backgrounds. Previous work had shown that EVD-GAG siRNAs (PTGS) and EVD-LTR 24-nt siRNAs (PolIV-RdDM) are mutually exclusive at the individual level. While both can be detected in bulks of plants, individuals with active or silenced EVD produce GAG or LTR siRNAs, respectively, as the transition to TGS has been shown to impact most if not all copies within one generation at the individual level (Marí-Ordóñez et al, 2013). The detection of both in RDR6-EVD bulks suggested that a subset of the individual plants successfully silenced EVD through TGS, pointing to a different silencing status in individual F6 plants within the bulked samples of both lines.
To investigate EVD silencing at the level of individual plants, and whether TGS had taken place in the absence of RDR6, ten RDR6-EVD and ten rdr6-EVD individuals from the F6 generations plants were selected. In RDR6-EVD, 24-nt EVD-LTR siRNAs were detected in all but two individuals, #2 and #9, where only EVD-GAG siRNAs were present (Fig. 2A). The presence of 24-nt EVD-LTR siRNAs was associated with a lower expression of EVD (Fig. 2B). Similarly, 24-nt EVD-LTR siRNAs were also detected in 6 out of 10 rdr6-EVD individuals (Fig. 2A) and the presence of 24-nt siRNAs correlated with a corresponding loss of EVD expression (Fig. 2B). Hence, silencing of EVD and production of associated LTR 24-nt siRNAs can take place in absence of RDR6-dependent GAG siRNAs.
Figure 2. Characterization of EVD silencing status in RDR6- and rdr6-EVD F6 individuals.
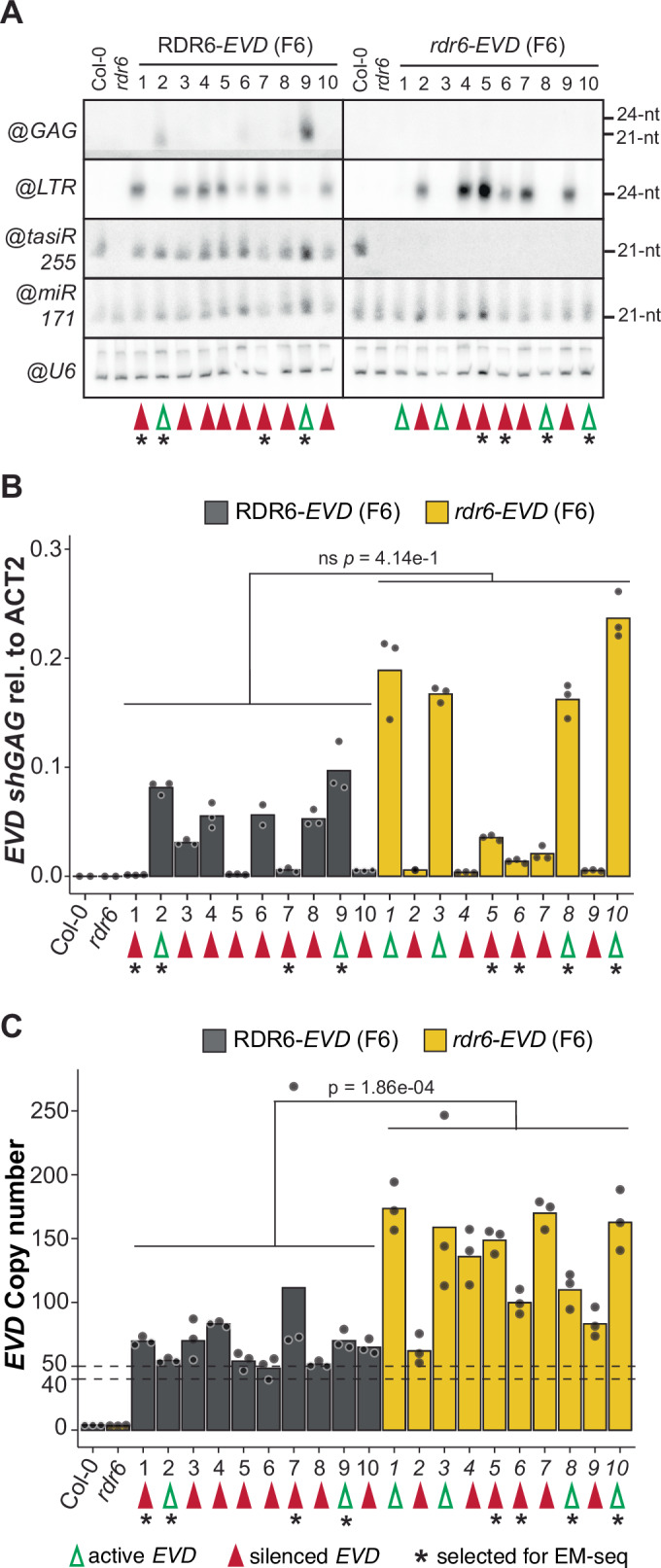
(A) RNA blot analysis of EVD siRNAs against GAG and LTR in 10 F6 individual plants of RDR6-EVD and rdr6-EVD lines. tasiR255 probe is used as control for RDR6 mutation, miR171 and snoRNA U6 are shown as loading controls. WT Col-0 and rdr6 with no reactivated EVD are shown as negative control for EVD activity. (B) Analysis of EVD shGAG expression of the same individuals investigated in A by qPCR, normalized to ACT2. (C) EVD copy number analysis by qPCR of the same individuals investigated in (A) and (B), using the EVD-GAG sequence as target. In (B) and (C), qPCR technical replicates for each sample are represented by dots. p-values for two-sided t-test between indicated samples are shown. Differences are considered statistically significant if p < 0.05 (5.00e−2) or non-significant (ns) if p ≥ 0.05. Green and red arrows indicate individuals with active and silenced EVD copies, respectively, selected individuals for subsequent EM-sequencing are indicated by an asterisk in (A–C). Source data are available online for this figure.
EVD silencing in the absence of RDR6 does not correlate with copy number
EVD switch to TGS has been experimentally established at around 40–50 copies in both met1 and ddm1 epiRILs, coinciding with the upper limit of natural variation for COPIA93 copies found within Arabidopsis ecotypes (Marí-Ordóñez et al, 2013; Quadrana et al, 2016). To assess whether a similar threshold applied in the absence of RDR6, we quantified EVD copy number in the same RDR6-EVD and rdr6-EVD F6 individuals used for EVD siRNAs and expression.
In agreement with the copy number threshold above which EVD TGS takes place, most RDR6-EVD individuals displayed homogenous copy number only slightly above this range. In rdr6-EVD plants, however, EVD silencing had taken place at a more variable copy number (Fig. 2C). Many plants displayed higher copy number, as previously observed in the bulk analysis (Fig. 1C); some individuals had switched to TGS at copy numbers just above the threshold (rdr6-EVD #2), while others did so at a copy number well above 100 (rdr6-EVD #4, 5, 7) (Fig. 2C). Nonetheless, EVD remained active in other individuals with copy numbers above 150 (rdr6-EVD #1, 3, 10) (Fig. 2C). Consequently, while PTGS seemed to facilitate the establishment of TGS in a reliable manner, in the absence of RDR6 activity, no clear copy number threshold for TGS installation was observed. Thus, in the rdr6 background, silencing of active EVD might be stochastic once the 40–50 copy number threshold is exceeded or depend on other factors not considered so far.
DNA methylation of EVD-GAG is dispensable for the transition to TGS
The copy number threshold is likely determined by the point at which the cumulative EVD expression from all new insertions causes DCL3 to process shGAG RDR6-derived dsRNA to initiate RDR6-RdDM (Marí-Ordóñez et al, 2013). Although EVD switch to TGS took place in absence of RDR6 and associated GAG siRNAs, we could not rule out the processing of shGAG transcripts by one of the other Arabidopsis RDR proteins, producing siRNA levels undetectable by Northern blots, but sufficient to induce the EVD-GAG DNA methylation believed to initiate PolIV-RdDM.
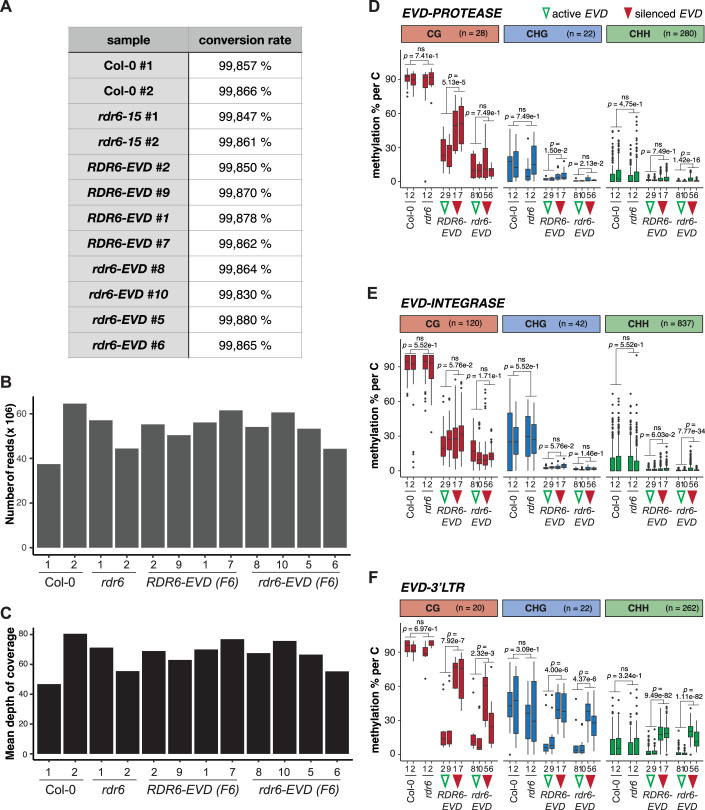
To corroborate the installation of PolIV-RdDM at EVD LTRs and address whether DNA methylation was independently of RDR6-generated siRNAs still deposited at EVD-GAG, or another coding region, EVD DNA methylation was assessed by whole-genome Enzymatic Methyl-sequencing (EM-seq) (Feng et al, 2020) on F6 individuals before and after the transition to TGS. We selected lines with active EVD (two RDR6-EVD and two rdr6-EVD individuals with high EVD expression and no 24-nt EVD-LTR siRNAs), as well as lines with silenced EVD (two RDR6-EVD and two rdr6-EVD individuals producing 24-nt EVD-LTR siRNAs and low EVD expression) (Fig. 2A,B; marked by asterisks). EM-seq on wild-type (Col-0) and rdr6 plants was performed as control for endogenous EVD methylation in the respective background. Overall, a 30–50X genome coverage with conversion rates above 99.8% was obtained for all samples (Fig. EV2A–C). Because short-read sequencing technology makes it challenging to estimate methylation levels along individual EVD insertions (~5 kb), EM-seq reads were mapped to a single fictitious EVD locus to estimate average methylation levels.
Figure EV2. EM-seq libraries general stats and EVD POL and 3′LTR methylation in RDR6- and rdr6-EVD lines.
(A) Cytosine-to-thymine conversion rates of the unmethylated chloroplastic DNA for each EM-seq library (see materials and methods for further information). (B) Number of paired-end fragments obtained in each EM-seq library. (C) Arabidopsis genome coverage in each EM-seq library. (D–F) EM-seq analysis of DNA methylation (as % per of methylated cytosines) in CG, CHG, and CHH contexts in WT Col-0, rdr6 and in RDR6- and rdr6-EVD F6 individuals with active and silenced EVD (marked with empty green and filled red arrowheads, respectively, numbers indicate same individuals as in Fig. 2) in: (D) EVD-Protease; (E) EVD-Integrase, and (F) EVD-5′LTR. In panels (D–F), n indicates the number of cytosines analyzed for each context per sample. In all boxplots: median is indicated by a solid bar, the boxes extend from the first to the third quartile and whiskers reach to the furthest values within 1.5 times the interquartile range. Dots indicate outliers, as data points outside of the above range. Wilcoxon rank sum test adjusted p-values between indicated groups of samples are shown. Differences are considered statistically significant if p < 0.05 (5.00e−2) or non-significant (ns) if p ≥ 0.05. Source data are available online for this figure.
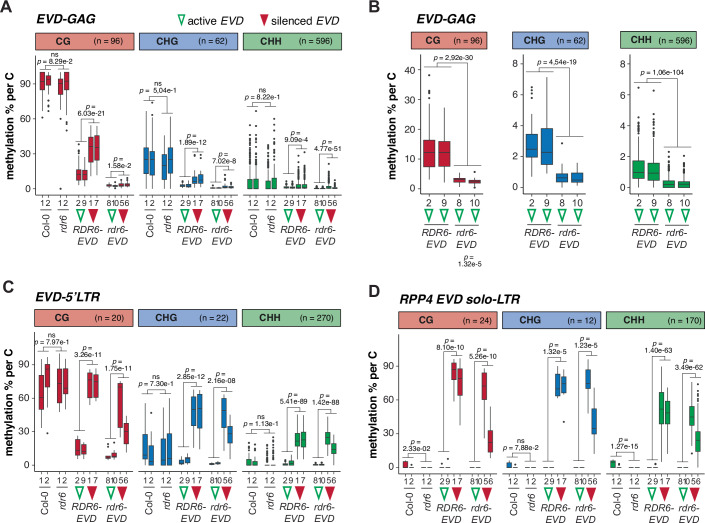
In RDR6-EVD with active EVD, low levels of GAG methylation in all cytosine contexts were observed. In contrast, the equivalent rdr6-EVD lines displayed near absence of DNA methylation (Fig. 3A,B). Furthermore, DNA methylation increased upon EVD silencing in RDR6-EVD but remained low after the EVD silencing in rdr6-EVD (Fig. 3A), indicating that absence of RDR6 was sufficient to abolish EVD-GAG DNA methylation, not only during EVD proliferation, but also after silencing. In addition, DNA methylation levels in rdr6-EVD lines remained low across other EVD coding regions in rdr6-EVD lines with active or silenced EVD, compared to RDR6-EVD (Fig. EV2D,E), ruling out that the switch to TGS is triggered through non-canonical-RdDM activity at other regions in the absence of RDR6.
Figure 3. DNA methylation of active and silenced EVD in RDR6- and rdr6-EVD lines.
EM-seq analysis of DNA methylation (as methylation % per cytosine) in CG, CHG, and CHH contexts in WT Col-0, rdr6 and in RDR6- and rdr6-EVD F6 individuals with active and silenced EVD (marked with empty green and filled red arrowheads, respectively, numbers indicate same individuals as in Fig. 2) in: (A) EVD-GAG; (B) EVD-GAG but only in RDR6- and rdr6-EVD F6 individuals with active EVD; (C) EVD-5′LTR; and (D) EVD solo-LTR in RPP4 (AT4G16869) promoter. In all panels, n indicates the number of cytosines analyzed for each context per sample. In all boxplots: median is indicated by a solid bar, the boxes extend from the first to the third quartile and whiskers reach to the furthest values within 1.5 times the interquartile range. Dots indicate outliers, as data points outside of the above range. Wilcoxon rank sum test adjusted p-values between indicated groups of samples are shown. Differences are considered statistically significant if p < 0.05 (5.00e−2) or non-significant (ns) if p ≥ 0.05. Source data are available online for this figure.
We next investigated DNA methylation at EVD-LTRs to confirm that presence of LTR 24-nt siRNA were bona fide indicators of PolIV-RdDM and the switch to EVD TGS. In both RDR6-EVD and rdr6-EVD lines with active EVD, DNA methylation at the LTRs was very low in all three contexts, while methylation levels were increased in those with silenced EVD (Fig. 3C). Furthermore, methylation levels at CHG and specifically at CHH were higher than those of the parental EVD copy in the wild-type Col-0 and rdr6 controls (Figs. 3C and EV2F), confirming the successful installation of PolIV-RdDM following the EVD burst, in contrast to RdDM-independent DNA methylation maintenance of EVD prior to its reactivation. To further validate that the switch to TGS through PolIV-RdDM installation had taken place, we inspected the methylation status of the EVD-derived solo-LTR present in the promoter of RECOGNITION OF PERONOSPORA PARASITICA 4 (RPP4, AT4G16860), referred to as RPP4 solo_LTR hereafter, which can be methylated in trans by EVD-LTR 24-nt siRNAs following an EVD de novo silencing event (Marí-Ordóñez et al, 2013). Indeed, EVD solo-LTR was only methylated in the RDR6-EVD and rdr6-EVD lines where EVD had been silenced (Fig. 3D).
Hence, the switch to TGS through PolIV-RdDM does neither require RDR6-RdDM nor the deposition of DNA methylation on EVD-GAG or other coding regions, to successfully silence EVD.
EM-seq paired-end sequencing data allows mapping of new EVD insertions
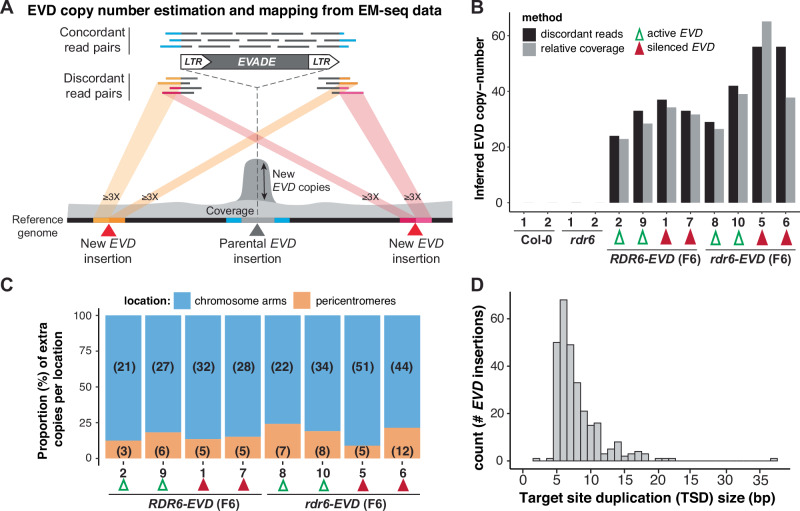
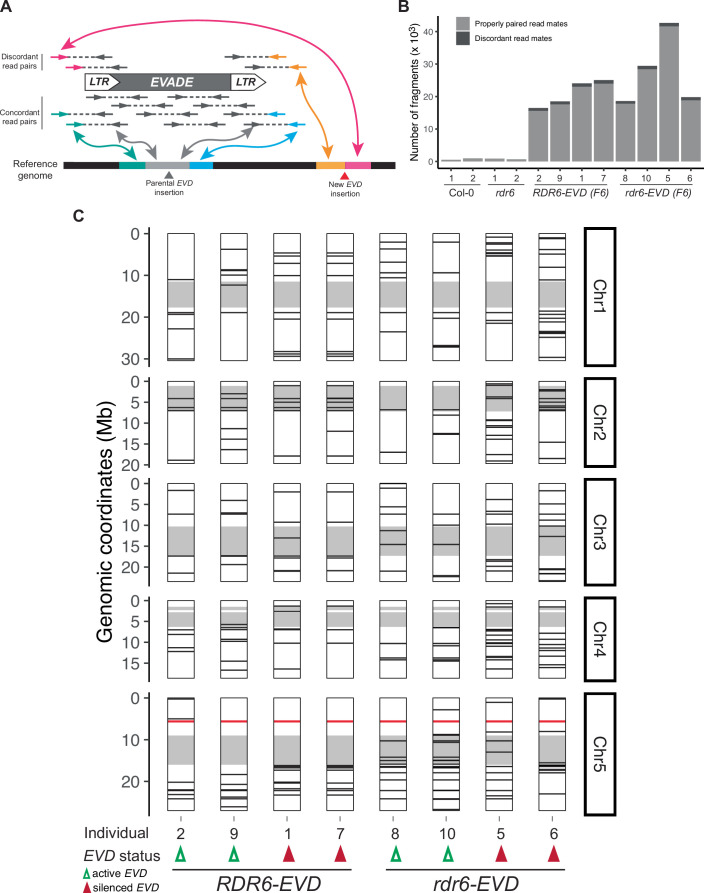
Once PolIV-RdDM is established, EVD silencing and associated deposition of DNA methylation impacts new copies genome-wide through the trans-activity of EVD LTR 24-nt siRNAs (Marí-Ordóñez et al, 2013). However, in EVD-silenced rdr6-EVD lines, LTR methylation levels were lower in the CG context for line #5 and in all contexts for line #6 than those in RDR6-EVD (Figs. 3C and EV2F). To investigate the homogeneity of DNA methylation at individual EVD insertions, and to ask whether it is influenced by the absence of RDR6-RdDM, we took advantage of discordant paired-read mates in our EM-seq data, where one of the read mates mapped to EVD LTRs and the other elsewhere in the genome, to identify and locate new EVD insertions (Gilly et al, 2014; Stuart et al, 2016; Quadrana et al, 2016; 2019) (Figs. 4A and EV3A) and to assess their methylation levels. Only new EVD insertions supported by three or more discordant paired-read mates from both of EVD LTRs were considered. Simultaneously, concordant paired-read mates mapping to EVD were used to independently estimate EVD copy numbers through the increase in EVD sequencing coverage due to additional insertions in RDR6- and rdr6-EVD relative to the Col-0 reference genome (Yoon et al, 2009; Quadrana et al, 2016) (Figs. 4A and EV3B).
Figure 4. Quantification of new EVD insertions in RDR6- and rdr6-EVD lines from EM-seq data.
(A) Schematic representation of the strategy used to quantify and map new EVD insertions from EM-seq data. EVD copy number was estimated using: (i) the increased EVD coverage of concordant paired read mates in EM-seq data, consequence of EVD transposition or, (ii) mapping new EVD insertions through discordant read pairs mapping to EVD and elsewhere in the genome. New insertions had to be supported by at least three discordant read pairs from each border to be considered. (B) Inferred EVD copy number using either relative coverage or discordant reads in WT Col-0, rdr6 and in RDR6- and rdr6-EVD F6 individuals with active and silenced EVD (marked with empty green and filled red arrowheads, respectively, numbers indicate same individuals as in Figs. 2 and 3). (C) Number and relative proportion (in %) of mapped new EVD insertions in pericentromeric or chromosomic arm locations in each indicated sample. Numbers of new EVD insertions at each location are indicated in brackets. (D) Histogram of the size (in bp) distribution of target site duplications at all new EVD insertions. Source data are available online for this figure.
Figure EV3. Mapping of new EVD insertions in RDR6- and rdr6-EVD lines from EM-seq data.
(A) Schematic representation of the strategy used to map new EVD insertions using discordant read mates from EM-seq. (B) Number of fragments from concordant (properly paired) and discordant read mates mapping to EVD in each of the EM-seq libraries. (C) Genomic location of new EVD insertions mapped through discordant read pair mates in EM-seq data in the Arabidopsis genome. Parental EVD (AT5G17125) location is indicated with a red line. New EVD insertions are marked with black lines. Pericentromeric regions in each of Arabidopsis five chromosomes are marked in gray. See table provided in corresponding source data for precise chromosome coordinates of each new insertion. Source data are available online for this figure.
While no new EVD insertions were obtained in Col-0 and rdr6 controls, estimation methods yielded consistent increased EVD copy numbers in lines where EVD had proliferated (Fig. 4B). As shown above (Fig. 2C), more EVD insertions were found in rdr6-EVD than in RDR6-EVD (Fig. 4B). In all lines, 75% or more of new insertions were mapped to chromosome arms (Figs. 4C and EV3C), as expected from the integration preference of EVD into gene-rich regions (Quadrana et al, 2019). Furthermore, most new EVD insertions caused short sequence duplications, known as target site duplications (TSD), of 5–8 bp (Fig. 4D). This falls within the TSD range expected for LTR-RTE in plants, specifically a 5-bp TSDs as obtained here for EVD (Quadrana et al, 2016; Orozco-Arias et al, 2019; Jedlicka et al, 2019; Roquis et al, 2021). Therefore, the new EVD insertions mapped from EM-seq data displayed features of bona fide new EVD transposition events.
We noticed that EVD copy numbers quantified from EM-seq data were lower than the quantification by qPCR. This discrepancy might originate from inaccuracy of qPCR quantifications, including potential amplification of EVD extra-chromosomal cDNA, or from limited discordant read-mates coverage in the EM-seq data. Nonetheless, defined insertions identified in all samples allowed to investigate methylation levels at individual LTRs of new EVD insertions.
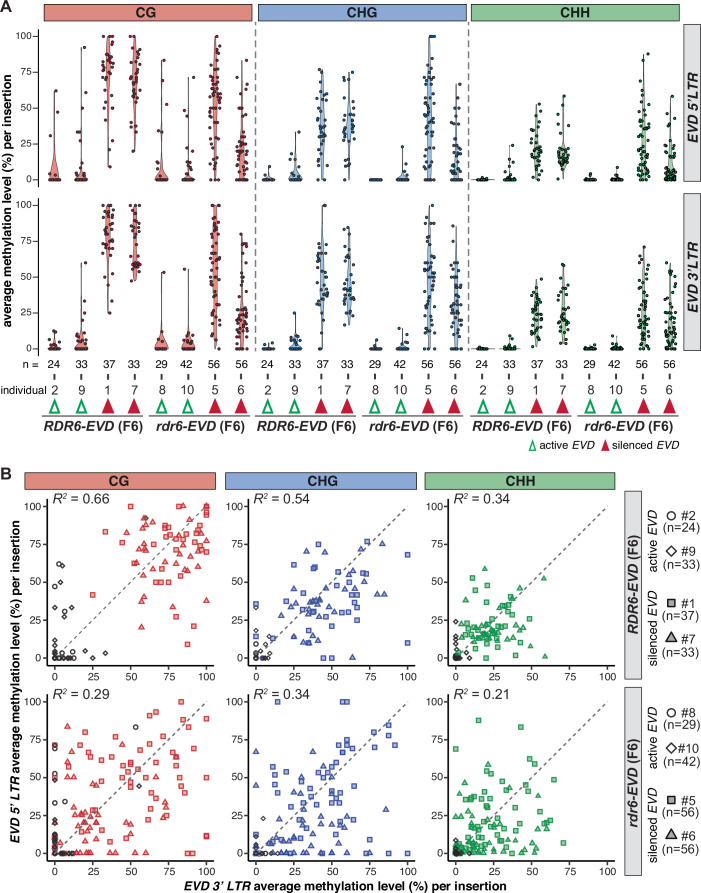
DNA methylation is not homogeneously deposited across new EVD insertions
As observed in our global analysis of EVD DNA methylation (Fig. 3), both LTRs of individual new EVD insertions gained DNA methylation in all contexts in EVD-silenced lines compared to those with active EVD. However, the LTR DNA methylation levels of individual EVD insertions were less homogeneous than expected, displaying a broad range in all contexts. Notwithstanding, we observed that the levels of DNA methylation in the three contexts were more heterogenous in rdr6-EVD lines than in the equivalent RDR6-EVD ones (Fig. 5A). This was most remarkable in the CG context, where most insertions in RDR6-EVD lines with silenced EVD displayed CG methylation levels above 50%, whereas several insertions in rdr6-EVD display lower or no methylation, especially in the rdr6-EVD line #6. A similar trend was observed in the CHG and CHH contexts, where some insertions displayed lower or no methylation in the rdr6-EVD lines after the switch to PolIV-RdDM (Fig. 5A).
Figure 5. LTR DNA methylation of individual new EVD insertions in RDR6- and rdr6-EVD lines.
(A) 5′ and 3′LTR average DNA methylation levels in each cytosine context for individual new EVD insertions in RDR6- and rdr6-EVD F6 individuals with active and silenced EVD. (B) Correlation between 5′ and 3′LTR average DNA methylation levels in each cytosine context for individual EVD insertions in RDR6- and rdr6-EVD F6 individuals. R2 indicates correlation coefficient between 5′ and 3′ LTR methylation levels within individual EVD insertions. Dashed line shows R2 = 1. Empty black symbols indicate EVD copies from individuals with active EVD and color-filled symbols from individuals with silenced EVD. In all panels, number of EVD insertions analyzed per individual correspond to those indicated in Fig. 4C. In all panels, n indicates the number of EVD insertions analyzed per sample. Source data are available online for this figure.
To investigate if the observed variation was the result of different methylation levels of individual insertions or the two LTRs from the same insertion being independently methylated, the correlation between 5′ and 3′ LTR DNA methylation for each insertion was assessed. While in the RDR6-EVD lines methylation between the two LTRs displayed a weak but positive correlation in the three contexts, in absence of RDR6 such correlation was lower for all three contexts (Fig. 5B). Again, this effect was more pronounced for CG methylation. Although we observed variation in RDR6-EVD silenced lines between 5′ and 3′ LTR methylation for a given insertion, in most cases DNA methylation remained above 50% in both LTRs. However, in rdr6-EVD lines, differences in CG methylation between LTRs of the same insertion were more pronounced, with several EVD insertions displaying high CG levels in one LTR but not the other. No bias for preferent methylation of either 5′ or 3′ LTR was observed (Fig. 5B).
Thus, in absence of RDR6 DNA methylation levels were not only less homogenous between de novo silenced EVD insertions (Fig. 5A), but also between LTRs of the same insertion (Fig. 5B). This could be a consequence of the absence of potential priming for the switch to TGS provided by EVD-GAG DNA methylation but might also reflect a difference in the timing of the switch. EVD-LTR 24-nt siRNAs were already detected at the 4th generation in RDR6-EVD but not in rdr6-EVD (Fig. 1E). Thus, in the later, EVD likely had been under PolIV-RdDM for less generations. This might be the case especially in the rdr6-EVD #6 line, where RPP4 solo_LTR methylation levels, which depends on EVD-LTR-derived 24-nt siRNAs, are also lower than in the other EVD-silenced lines (Fig. 3D). Nonetheless, the gain of DNA methylation at most new EVD insertions further supports that PolIV-RdDM gets installed at EVD-LTRs in the absence of RDR6-RdDM.
RdR6-RdDM is insufficient for EVD silencing in the absence of PolIV-RdDM
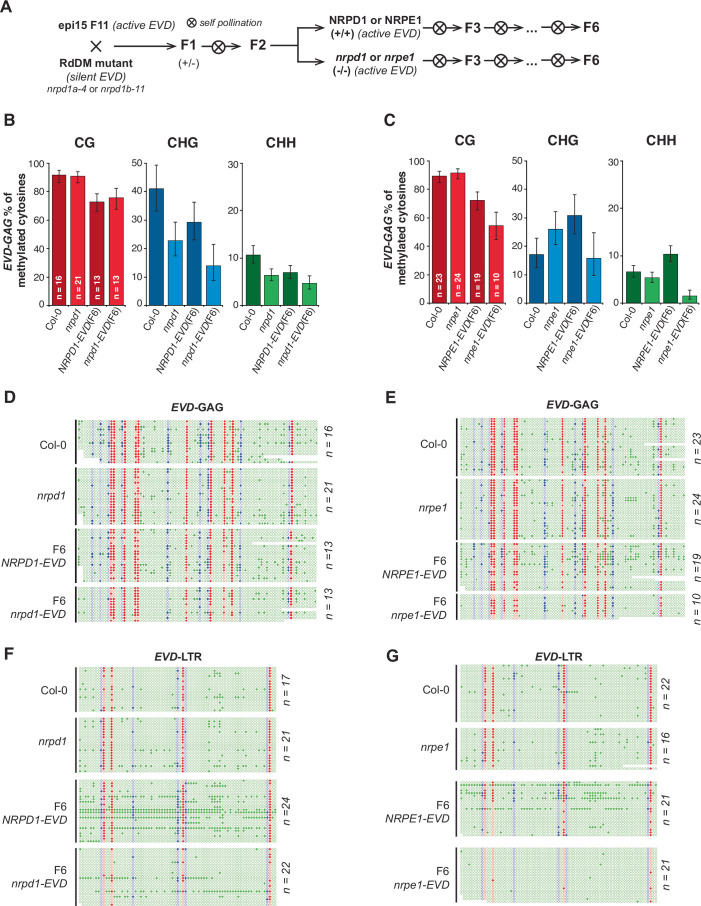
The above results indicated that, independently of RDR6-RdDM and EVD-GAG methylation, EVD silencing likely took place through the initiation of PolIV-RdDM at EVD-LTRs. While both PolIV and PolV are essential for PolIV-RdDM, only PolV is required for RDR6-RdDM, if siRNAs are provided through PTGS (Nuthikattu et al, 2013; McCue et al, 2015; Taochy et al, 2019; Sigman et al, 2021). Hence, to test the dependency of EVD TGS in PolIV-RdDM and, at the same time, explore if RDR6-RdDM could lead to TGS independently of PolIV-RdDM, we introduced active EVD in the mutants nrpd1 (PolIV largest subunit mutant) and nrpe1 (PolV largest subunit mutant), following the same strategy as for rdr6. This time, however, to ensure the presence of EVD-GAG methylation before the loss of RdDM, an epi15 F11 generation plant, already undergoing RDR6-RdDM (Marí-Ordóñez et al, 2013) (Fig. 1A), was used as EVD donor. Again, wild-type and mutant plants were selected in the F2 and propagated to F6 (Fig. EV4A).
Figure EV4. BS-PCR analysis of EVD-GAG DNA methylation levels in RdDM mutants.
(A) Crossing scheme to generate nrpd1- and nrpe1-EVD lines. F2 plants were genotyped to select homozygous WT and mutant lines for each background and propagated through selfing until the F6 generation. (B, C) % methylated cytosines by bisulfite -PCR DNA methylation analysis at EVD-GAG sequences in the F6 generation of NRPD1-EVD (B) and NRPE1-EVD (C) lines, in both WT (darker shade) and mutant (lighter shade) backgrounds. Col-0, nrpd1 and nrpe1 were used as controls. Error bars represent 95% confidence Wilson score intervals of the % of methylated cytosines (C) in each context (CG, CHG, CHH). (D, E) Dot-plot representation of bisulfite-PCR sequencing data for the F6 generation of NRPD1-EVD (D) and NRPE1-EVD (E) lines, in both WT and mutant backgrounds, at EVD-GAG sequences. Col-0 and nrpd1 or nrpe1 were used as control for the endogenous parental EVD copies. (F, G) Dot-plot representation of bisulfite-PCR sequencing data for the F6 generation of NRPD1-EVD (F) and NRPE1-EVD (G) lines, in both WT and mutant backgrounds, at EVD-LTR sequences. Col-0 and nrpd1 or nrpe1 were used as control for the endogenous parental EVD copies. In all dot-plots, filled circle represent methylated, empty circles unmethylated cytosines in the CG (red), CHG (blue), and CHH (green) context. Source data are available online for this figure.
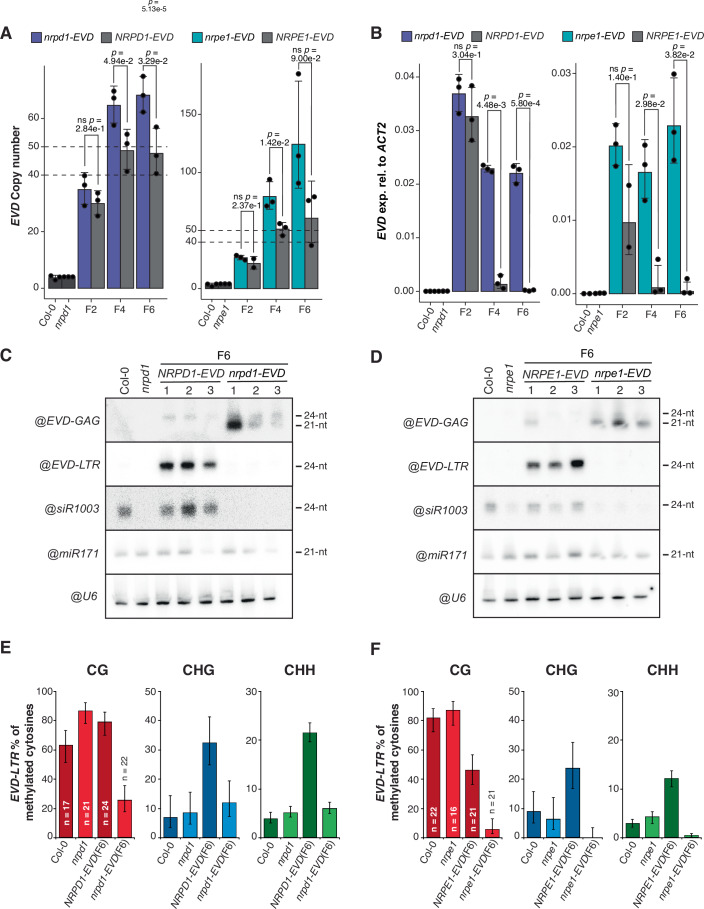
While lines carrying wild-type or mutant alleles were indistinguishable with respect to EVD copy number or expression in the F2 (Fig. 6A,B), EVD reached higher copy numbers, surpassing the 40–50 copy number threshold, in the following generations of both mutants (nrpd1-EVD and nrpe1-EVD) than in the lines carrying the corresponding wild-type alleles (NRPD1-EVD and NRPE1-EVD) (Fig. 6A). Similarly to the results obtained with RDR6-EVD lines, once the threshold was reached in lines with wild-type alleles, EVD expression was reduced. However, in both mutant backgrounds, EVD remained transcriptionally active (Fig. 6B), indicating that TGS was not installed in the absence of PolIV-RdDM. Furthermore, investigation of EVD small RNA patterns in three independent bulks of plants at the F6 generation confirmed the presence of EVD-LTR 24-nt siRNAs in both NRPD1- and NRPE1-EVD lines. On the contrary, in nrpd1- and nrpe1-EVD lines, only EVD-GAG siRNAs were detected (Fig. 6C,D), consistent with EVD expression triggering PTGS.
Figure 6. Characterization of EVD proliferation and silencing in RdDM mutants.
(A) EVD copy number analysis by qPCR in NRPD1-EVD and NRPE1-EVD lines, in both WT (gray) and mutant (colored) backgrounds, at generations F2, F4, and F6 derived from three independent F1s (biological replicates), using the EVD-GAG sequence as target. (B) qPCR analysis of shGAG expression normalized to ACT2 in NRPD1-EVD and NRPE1-EVD lines, in both WT (gray) and mutant (colored) backgrounds, at generations F2, F4, and F6 derived from three independent F1s (biological replicates). In (A) and (B), biological replicates (bulks of 8–10 plants each) are individually represented by dots. Error bars show standard error of the mean in (A) and (B). p-values for two-sided t-test between indicated samples are shown. Differences are considered statistically significant if p < 0.05 (5.00e−2) or non-significant (ns) if p ≥ 0.05. (C, D) RNA blot analysis of EVD siRNAs against GAG and LTR in the F6 generation of 3 independent WT and mutant lines of NRPD1-EVD (C) and NRPE1-EVD (D). WT Col-0 and nrpd1 or nrpe1 with no reactivated EVD are shown as negative control for EVD activity. siR1003 probe is used as control for NRPD1 and NRPE1 mutations, miR171 and snoRNA U6 are shown as loading controls. (E, F) % methylated cytosines (C) by bisulfite-PCR DNA methylation analysis at EVD-LTR sequences in the F6 generation of NRPD1-EVD (E) and NRPE1-EVD (F) lines, in both WT (darker shade) and mutant (lighter shade) backgrounds. Col-0, nrp1d and nrpe1 were used as controls. n: number of clones analyzed. Error bars represent 95% confidence Wilson score intervals of the % of methylated cytosines (C) in each context (CG, CHG, CHH). Source data are available online for this figure.
Methylation of EVD was assessed in F6 bulks by Sanger sequencing of PCR amplicons from bisulfite-treated DNA (BS-PCR). Although this PCR-based method does not discern between integrated and extra-chromosomal DNA (ecDNA) integration intermediates in lines with active EVD, it allowed us to estimate overall EVD methylation levels. Regarding EVD-GAG, CG methylation was high in both wild type and mutant lines, as expected according to MET1 maintenance and previous exposure to RDR6-RdDM (Fig. EV4B–E). CHG and CHH methylation was present in both WT and mutant lines, albeit higher in NRPD1- and NRPE1-EVD lines than in their mutant equivalents (Fig. EV4B–E), probably due to the increase in methylation previously observed after the installation of TGS (Fig. 3A). Both nrpd1- and nrpe1-EVD lines displayed similar CHG methylation levels likely inherited from the epi15 F11 and maintained independently of siRNAs. However, CHH methylation was higher in nrpd1-EVD than in nrpe1-EVD lines (Fig. EV4B–E), in agreement with the absence of RDR6-RdDM in nrpe1 but not in nrpd1 mutants. Therefore, RdD6-RdDM seems to still be depositing CHH methylation in the nrpd1-EVD lines.
We next investigated the methylation status of EVD-LTR. As anticipated from the different epigenetic regulation of EVD before and after its mobilization and silencing, the presence of 24-nt siRNAs in NRPD- and NRPE-EVD lines correlated with increased DNA methylation levels, where CHG and CHH methylation levels surpassed those found in WT and mutant controls (Figs. 6E,F and EV4F,G). Furthermore, no loss of EVD DNA methylation, relative to Col-0, was observed neither in nrpd1 nor nrpe1 control samples, confirming the absence of PolIV-RdDM regulation at the parental EVD insertion (Fig. 6E,F). In contrast, in nrpd1- and nrpe1-EVD lines, DNA methylation in all contexts remained low in conformity with the absence of EVD-LTR 24-nt siRNAs and TGS (Fig. 6E,F). However, we noticed that DNA methylation in all contexts was higher in nrpd1-EVD than in nrpe1-EVD, suggesting that continuous RDR6-RdDM activity might cause weak DNA methylation in EVD-LTR. Still, as the BS-PCR used here amplifies both integrated and ecDNA EVD copies, a similar EM-seq strategy as used for the EVD-rdr6 F6 individuals will be required in the future to obtain LTR methylation levels of individual insertions in nrpd1- and nrpe1-EVD lines. Nonetheless, in the absence of PolIV-RdDM, EVD remained transcriptionally active up to the F6 generation despite the continuous action of PTGS and, in the case of nrpd1-EVD, RDR6-RdDM. Hence, PolIV-RdDM is required for de novo TGS initiation independently of RDR6-RdDM.
EVD antisense nested insertions and transposition into RdDM loci are potential TGS initiation events in absence of RdR6-RdDM
Given that PolIV-RdDM requires pre-existing epigenetic marks for its recruitment, EVD-LTR 24-nt siRNAs might be a consequence of a preceding EVD silencing event rather than the cause. To gain further insights into the trigger of EVD TGS in the absence of RDR6-RdDM, we investigated presumed triggers of TGS in the rdr6-EVD lines.
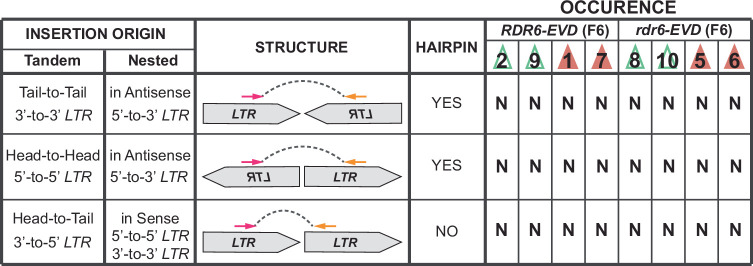
First, we attempted to explore the potential presence of LTR hairpins as the initial source of LTR siRNAs. TE-derived hairpins can arise as the result of genomic rearrangements between TE sequences or tandem/nested insertions and drive the silencing of homologous TEs (Slotkin et al, 2005). To do so, we initially examined the EM-seq data in search for discordant reads where the mate pairs will both map to EVD-LTR but in opposite orientations (Fig. EV5). We did not succeed to identify any read pair mate in such configuration (Fig. EV5). However, detecting LTR hairpins through such approach is technically limited by the insert size of the EM-seq library (~300–700 bp) as any pair of EVD-LTRs in antisense orientation further away than that will not be captured by such strategy with the EM-seq data. Therefore, as we could not detect the presence of two LTRs in close proximity, we expanded the search to paired read mates mapping to LTR and to antisense EVD sequences to identify potential nested antisense insertion configurations (Fig. 7A). Although such read pair mates were found in several individuals (Fig. 7A), we set a threshold of at least 2 read mates at each LTR to confidentially identify EVD nested insertions within itself and discard sequencing artifacts. Using this strategy, only one antisense nested insertion of EVD into itself was found in the rdr6-EVD #5 individual, in which EVD had been silenced, but not in any other sample (Fig. 7A,B). In the resulting EVD locus, the LTR of the initial insertion are in antisense to the LTR of the incoming EVD insertion. With the LTRs being 2562 and 1942 bp apart, this configuration can potentially result in LTR hairpin formation by transcription from the initial insertion into the antisense 3′LTR from the nested one or all the way through the antisense insertion to generate a second hairpin (Fig. 7B). Alternatively, convergent transcription driven by the 5′LTR of each of the insertions might result in the formation of dsRNA (Fig. 7B). Both cases can potentially trigger LTR siRNAs to initiate EVD TGS (Fig. 7B) and will require further investigation. However, such EVD antisense nested insertion was only confidentially found in one of the two silenced rdr6-EVD lines.
Figure EV5. Summary table for the search of close proximity sense-antisense EVD-LTR events.
Putative origin of LTR hairpins as consequence of EVD transposition in tandem or nested configurations next to the scheme of the strategy used to find them using discordant read mates from EM-seq where both read mates map to EVD-LTR. The table indicates the occurrence and number of read mates found in each sample. Arbitrary threshold of at least three paired read mates was set to confidentially identify two LTRs in close proximity. N: non-occurrence, y: presence of at least one discordant paired read mate, Y: presence of discordant paired read mates and above threshold for confident calling.
Figure 7. Genetic and epigenetic characterization of EVD insertion sites.
(A) Schematic representation of the strategy used to identify EVD antisense nested insertions within itself from EM-seq discordant paired read mates mapping to EVD-LTR and elsewhere on EVD coding sequence in antisense to the LTR. The table indicates the occurrence and number of read mates found in each sample. Arbitrary threshold of at least two paired read mates by LTR was set to confidentially identify an EVD nested insertion. N: non-occurrence, y: presence of at least one discordant paired read mate, Y: presence of discordant paired read mates and above threshold for confident presence of nested insertion. (B) Scheme of EVD antisense nested insertion structure based on EVD discordant reads in the sample rdr6-EVD F6 #5. Distance between LTRs as well as the length of the target-site duplication (TSD) caused by the insertion are indicated in base-pairs (bp). Colored arrows represent potential EVD transcripts within the locus. Putative sources of small RNAs are depicted below. (C) Schematic representation of methodology used to infer the methylation status of EVD landing sites prior to transposition using the EM-seq data. EVD insertions are represented by red lines. Red boxes indicate regions with new and polymorphic EVD insertions. Blue boxes represent the same region without the EVD insertion. (D) Pie charts of the distribution of insertions within non-methylated, CG-only methylated and non-CG methylated regions within each sample. Inlets represent the classification of non-CG methylated regions into RdDM-, CMT2-dependent or independent of both. Number of insertions in each category is indicated within the chart. (E) Metaplot of the mean shift in DNA methylation following a EVD insertion by genotype and methylation context depending on EVD silencing status. Source data are available online for this figure.
As an alternative trigger, we next investigated the local DNA methylation status of new EVD insertion sites. Transposition into a pre-existing RdDM locus might suffice to initiate PolIV-RdDM on an EVD insertion and spread to other copies through the action of 24-nt siRNAs. Therefore, we aimed to determine the methylation status of EVD landing sites prior to transposition in order to investigate if integration within methylated loci was a common feature in individuals with silenced EVD. However, extensive DNA methylation variation has been reported for the met1-epiRILs (Reinders et al, 2009). As the epiRIL#15 was used as the parental line carrying active EVD to generate the RDR6- and rdr6-EVD lines (Fig. 1B), their methylomes might differ from that of the wild-type. Hence, we used the EM-seq data to assess the methylation of individual landing sites for new EVD insertions by examining their DNA methylation in individuals without EVD at the corresponding locations (Fig. 7C). Fixed EVD insertions, present in all individuals sequenced, were discarded as no information about the methylation status of the region prior to insertion could be obtained. Only polymorphic EVD insertions were considered for the analysis (Fig. 7C). In addition, as absence of RDR6 has been shown to impact DNA methylation (Stroud et al, 2013), the methylation levels at regions without new EVD insertions were only assessed from individuals of the same genotype. Therefore, the methylation at positions with EVD insertions shared by all individuals of the same genotype were considered non-informative (Fig. 7C). Furthermore, their presence in all active and silenced EVD individuals within a given genotype likely disqualified them as the triggers of TGS. Regions were classified as unmethylated (5mC < 10%), mCG only (mCG > 10%, mCHG and mCHH < 10%) or non-CG methylated (mCHG, mCHH > 10%). In all samples, most new EVD insertions happened at unmethylated locations. EVD transposition events at CG and non-CG methylated loci were rare but also found in individuals with active or silenced EVD (Fig. 7D), suggesting that such events were not determinant of EVD silencing status. However, as CHH methylation can be maintained by PolIV-RdDM or independently of small RNAs through CMT2 activity, only insertions in PolIV-RdDM-dependent loci might led to 24-nt siRNAs production. Although the dependency of mCHH on PolIV-RdDM and CMT2 on a given locus might not be conserved in the RDR6- and rdr6-EVD lines given their epiRIL origin, we further subclassify non-CG methylated positions into RdDM, CMT2 or RdDM/CMT2-independent. Pathway assignment was based on the annotation performed by Sasaki and colleagues (Sasaki et al, 2022), who used a large methylome dataset from Arabidopsis mutants (Stroud et al, 2013) to determine locus-specific DNA methylation dependencies on different silencing pathways. No clear correlation between insertion of EVD in a putative PolIV-RdDM locus and its silencing was observed, as EVD insertions in RdDM loci were observed in an EVD active RDR6-EVD individual and only in one EVD silenced rdr6-EVD plant (#6) (Fig. 7D). Incidentally, such individual did not carry the EVD antisense nested insertion (Fig. 7A) and remains possible that those insertions triggered the switch to TGS in the rdr6-EVD plant #6.
Nonetheless, these analyses are not conclusive as the sample size is too small. Further small RNA, epigenetic characterization and genome sequencing of lines with active and silenced EVD will be required to find whether insertions within specific epigenetic landscape or EVD insertion configurations are responsible for initiating the epigenetic silencing of all new EVD insertions in absence of RDR6-RdDM.
EVD transposition and silencing has limited impact on local DNA methylation
Lastly, despite EVD preferential insertion within non-methylated regions (Fig. 7E), changes in local chromatin in response to transposition, and eventually leading to TGS, might had occurred. To additionally investigate if EVD transposition led to alterations in local DNA methylation, and whether those were influenced by the epigenetic status of EVD, we compared methylation levels at EVD insertion sites and their surroundings with and without EVD and before/after switch to TGS. Globally, no gains of DNA methylation in any context were observed around the insertion sites upon EVD transposition in plants with active EVD. However, methylation in CHG and CHH, but not in CG, increased at those positions following EVD transition to TGS (Fig. 7E). Although we noticed that CHG and CHH gains reached further away in RDR6 than in rdr6 lines, these did not spread far from the insertion site, likely reflecting the role of RdDM not only maintaining DNA methylation but also at preventing it from spreading outside of silenced loci boundaries (Zemach et al, 2013; Li et al, 2015).
Discussion
PTGS is required for GAG methylation but dispensable for the TGS of EVD
Post transcriptional gene silencing mediated by RDR6-dependent siRNAs has been hypothesized to be the initiating step of TE epigenetic silencing, in particular EVD (Teixeira et al, 2009; Marí-Ordóñez et al, 2013; Nuthikattu et al, 2013; McCue et al, 2015; Panda et al, 2016; Sigman et al, 2021). In this study, by investigating the silencing fate of active EVD in wild type and mutant RDR6 backgrounds, we show that, although DNA methylation deposited in the EVD-GAG sequence is a consequence of PTGS through RDR6-RdDM and might contribute to its final silencing, in absence of RDR6 and the associated EVD-GAG siRNAs, transcriptional gene silencing is still achieved through the installation of PolIV-RdDM without any prior DNA methylation at EVD coding sequences. Therefore, PTGS and gene body DNA methylation are dispensable for de novo EVD silencing.
Our results add further evidence supporting that PTGS can direct the deposition of DNA methylation (Wu et al, 2012; Nuthikattu et al, 2013; McCue et al, 2015; Taochy et al, 2019). However, RDR6-RdDM is neither necessary nor sufficient to for TGS initiation, which requires PolIV-RdDM at regulatory sequences. Previous work has shown that an immobile EVD overexpression transgenic system under the control of the CaMV 35S promoter (35S:EVD), despite triggering the same PTGS response as the endogenous EVD, does not transition to TGS in WT plants (Marí-Ordóñez et al, 2013; Oberlin et al, 2022). This is in line with the observation that in endogenous loci and transgenes triggering strong PTGS and undergoing RDR6-RdDM, DNA methylation is unable to spread from transcribed regions to regulatory sequences to set TGS (Taochy et al, 2019).
PTGS might facilitate the installation of EVD TGS
Nonetheless, given the consistent installation of silencing in presence of PTGS, limiting EVD proliferation beyond 40–50 copies, and the loss of LTR DNA methylation homogeneity between as well as within newly silenced EVD insertions, RDR6-RdDM likely sensitizes EVD loci to facilitate TGS initiation, explaining the uniformity of EVD copy number at which RdDM is installed at EVD, compared to the stochasticity observed in rdr6 mutants. Introduction of either active EVD or 35S:EVD in dcl2 dcl4 double mutants, results in the switch to TGS at 20–30 EVD copies and silencing of 35S:EVD coupled with the production of 24-nt siRNAs from their promoters (Marí-Ordóñez et al, 2013). Hence, in the case of EVD, enhancing RDR6-RdDM by promoting DCL3 activity upon RDR6 dsRNA products in the absence of DCL2 and DCL4, expedites EVD switch from PTGS to TGS. Here, despite the lack of PolIV-RdDM, a low level of DNA methylation at EVD-LTR was found in RDR6-RdDM competent nrpd1-EVD plants. Given the close proximity of GAG to 5′ regulatory sequences, strong or prolonged GAG RDR6-RdDM might result in enough methylation adjacent to the promoter to recruit PolIV-RdDM as previously suggested (Marí-Ordóñez et al, 2013; Sigman et al, 2021). This situation may be favored for endogenous EVD. Although both endogenous EVD and 35S:EVD produce high RDR6-dependent siRNA levels, in contrast to the ubiquitously expressed 35S promoter, EVD-LTR drives expression in only a few cells (Marí-Ordóñez et al, 2013), where intracellular siRNAs levels might be high enough to promote DNA methylation spreading into the nearby LTR. However, as the BS-PCR used here to measure EVD DNA methylation in polIV and polV mutant backgrounds does not provide information about the methylation status of individual insertions and it captures both integrated and ecDNA EVD copies, a similar EM-seq strategy as used for the EVD-rdr6 F6 individuals will have to be applied in the future. This will allow to obtain a better resolution of the LTR methylation levels of individual insertions in nrpd1- and nrpe1-EVD lines to gain further insights in the role of RDR6-RdDM in priming or sensitizing EVD insertions for the switch to TGS.
PTGS potentially acts as an EVD copy number control system
Alternatively, although not mutually exclusive with the priming role of RDR6-RdDM in the switch to TGS, we propose that PTGS triggered during translation can act as a mechanism to regulate EVD proliferation until the switch to TGS takes place. Mutations in RDR6 lead to increased EVD shGAG mRNA, protein and VLP levels, resulting in increased transposition (this study and (Lee et al, 2021; Oberlin et al, 2022)). Although the mechanisms triggering shGAG ribosome stalling and cleavage has not yet been elucidated, given that PTGS is not commonly triggered by most TEs undergoing translation in Arabidopsis (Oberlin et al, 2022), it is possible that EVD hijacks the PTGS pathway to mediate copy number control (CNC) and regulate its own transposition rate. A variety of CNC factors have been found to be self-encoded by TEs, such as peptides to inhibit VLP formation or antisense ORFs to hamper reverse transcription, as means to minimize genomic damage on their host caused by over proliferation (Matsuda and Garfinkel, 2009; Cottee et al, 2021). However, EVD also reached high copy numbers in nrpd1- and nrpe1-EVD lines, despite the continuous action of PTGS. A full understanding of the impact of PTGS in EVD transposition and host recognition and defense mechanisms will require further investigation of EVD activity in genetic backgrounds defective for both PTGS and RdDM.
PolIV-RdDM is essential for EVD control and de novo silencing
Recruitment of RdDM to EVD-LTRs independently of RDR6-RdDM, together with the absence of EVD TGS in either nrpd1 or nrpe1 backgrounds, indicates that PolIV-RdDM is essential for de novo EVD silencing during its genome colonization. Although absence of RdDM has little impact in the reactivation of silenced TEs in the Arabidopsis genome (He et al, 2021; 2022), a role for RdDM in controlling TE propagation has been previously shown for the heat-responsive LTR-retrotransposon (LTR-RTE) ONSEN, for which proliferation and copy number following induction is increased in NRPD1 mutants (Ito et al, 2011; Matsunaga et al, 2012; Hayashi et al, 2020; Niu et al, 2022). Furthermore, genetic variation in the RdDM components RDR2 and NRPE1 has been associated with variation in TE content within Arabidopsis natural populations (Baduel et al, 2021; Sasaki et al, 2022; Jiang et al, 2023). Therefore, PolIV-RdDM might play a major role in de novo TE silencing besides its function as a DNA methylation maintenance pathway.
Coincidentally, RdDM at long, young, and potentially functional TEs, operates at their edges (Zemach et al, 2013; Stroud et al, 2013; 2014), similarly to the patterns found at EVD after the transition to TGS independently of PTGS. As mentioned previously, most LTR-RTEs intact enough to be translation-competent do not trigger PTGS when transcriptionally active (Oberlin et al, 2022). Hence, the phenomena of RdDM installation at flanking LTRs in absence of PTGS observed here for EVD might represent a more general mechanism of de novo silencing of LTR-RTEs.
Potential mechanisms of TGS initiation in absence of PTGS
In absence of PTGS and the initial deposition of DNA methylation within the EVD-GAG sequence, the mechanism(s) triggering or initiating epigenetic silencing on EVD remain obscure as we have not identified a common cause. Apart from TE activity, several mechanisms have been shown to recruit RdDM or trigger the deposition of DNA methylation. Some of which have been explored in this work and will be further discussed here as they could explain the stochasticity of silencing observed.
For example, double-stranded DNA breaks (DSBs) have been shown to trigger the production of siRNAs and promote the deposition of DNA methylation at the borders of the break points in Arabidopsis (Wei et al, 2012; Schalk et al, 2016; 2017; Du et al, 2022). In addition, little is known about the chromatin landscape of newly integrated TE copies, which might contain histone variants or histone modifications predisposing for DSB-induced silencing (Yelagandula et al, 2014; Lorković et al, 2017; Osakabe et al, 2018; Bourguet et al, 2021). However, no changes in DNA methylation were observed around new insertion sites in individuals with active EVD. Moreover, such mechanism would imply that new insertions become individually silenced as they integrate, which is incompatible with the low levels of DNA methylation in lines with active EVD and its coordinated increase once the switch to TGS takes place. A significant number of simultaneous transposition events generating excessive DNA damage might be required to elicit a silencing response. Nonetheless, EVD does remain active in some individuals despite a high copy number and expression levels.
Given the arbitrary copy number at which TGS took place in rdr6-EVD lines, silencing might therefore depend on more stochastic events. Due to trans-activity of EVD-LTR 24-nt siRNAs, the initiation of RdDM in one or few EVD copies might suffice to spread silencing across all new active insertions. RNA hairpins resulting of TE tandem or nested insertions and acting as source of 24-nt siRNAs has been shown to mediate the epigenetic silencing of the Mutator TE in maize (Slotkin et al, 2003). Though this mechanism was previously deemed unlikely to trigger EVD TGS in RDR6 wild-type backgrounds (Marí-Ordóñez et al, 2013), increased transposition upon absence of PTGS might enhance the chances of hairpin formation. We have indeed detected event of an antisense EVD nested insertion into itself, potentially leading to the formation of LTR hairpins, in a rdr6-EVD individual where EVD has switched to TGS. However, we did not test if such EVD locus did become a source of siRNAs to initiate EVD switch to TGS. Nonetheless, RNA hairpins are generally processed into several siRNA sizes (Zilberman et al, 2004; Fusaro et al, 2006), and LTR siRNAs in such individual were predominantly 24-nt long.
Alternatively, and analogously to TE silencing triggered by transposition within piRNA clusters in Drosophila melanogaster (Brennecke et al, 2007; Goriaux et al, 2014; Guida et al, 2016), increased transposition might increase the probability of integration events within pre-existing heterochromatin domains, subjecting one or few EVD copies to RdDM. However, no increase in absolute or relative pericentromeric insertions was observed in rdr6-EVD lines that had switch to EVD TGS and, although EVD insertions within regions with non-CG methylation were observed, those were not exclusive to individuals that had switch to TGS. Transposition within putative RdDM loci were observed in one rdr6-EVD individual with silenced EVD but as well in one RDR6-EVD individual with active EVD. Therefore, although transposition events leading to the formation of EVD loci with the potential to trigger small RNAs independently of RDR6 or within RdDM loci can occur, a more detailed study with increased population size, better characterization of the chromatin environment and small RNAs in EVD landing sites will be required in the future to properly explore if transposition within certain chromatin environments or loci already subjected to RdDM can initiate EVD TGS.
In addition, the repetitiveness of EVD during a transposition burst, might led to changes in the three-dimensional chromatin organization forming, or bringing EVD to, chromatin interaction clusters associated with silencing (Grob et al, 2014; Grob and Grossniklaus, 2019). Furthermore, such repetitiveness might cause genome instability through non-allelic homologous recombination events (Sammarco et al, 2022) that trigger the silencing of EVD. Future investigation of the presence of genome rearrangements and chromatin conformation changes before and after EVD switch to TGS should help elucidate whether such changes are taking place and are indeed linked to de novo EVD silencing.
Several of the above mechanisms might be favored by the presence of LTRs. Although single LTRs from exogenous TEs have been shown to trigger RdDM when transformed into Arabidopsis (Fultz and Slotkin, 2017), they are found as duplicated sequences at both ends of intact LTR-RTEs. Such arrangement might help the formation of LTR hairpins through tandem (either in head-to-head or tail-to-tail configurations) or nested EVD insertions, as observed for multicopy T-DNA events (Neve et al, 1997; Buck et al, 1999). Moreover, EVD-LTRs being twice the number of EVD insertions might increase non-allelic recombination rates between LTRs. Furthermore, in Arabidopsis, it has been shown that the structure of a locus can influence its epigenetic fate using a transgenic selection marker flanked by tandemly duplicated promoter sequences, mimicking an LTR-RTE configuration. Such transgenic locus can be found in either active or silenced state (epialleles). The silenced epiallele, characterized by the be presence of 24-nt siRNA from the repeated regions (like EVD), has been shown to induce silencing of the active one in an RdDM-dependent manner when both are crossed with each other (Scheid et al, 2003; Foerster et al, 2011; Bente et al, 2021). Deletion of the duplicated region at the end of the transgene from the silent epiallele lead to a decrease in its trans-silencing ability, while its removal from the active epiallele strongly impaired its capacity to become silenced (Bente et al, 2021). Hence, this type of sequence configuration might contribute to spread and stabilize TGS across EVD insertions once one of them becomes targeted by RdDM.
Finally, RdDM has been implicated in antiviral defense against Geminiviruses, plant single-stranded circular DNA viruses replicating in the nucleus (Al-Kaff et al, 1998; Raja et al, 2008). Linear and circular extra chromosomal DNA (ecDNA and eccDNA, respectively), products of reverse transcription, have been detected upon expression off several LTR-RTEs, including EVD (Lanciano et al, 2017; Mann et al, 2022; Niu et al, 2022). In absence of PTGS, EVD ecDNA and eccDNA content has been shown to increase in plant tissues (Lee et al, 2020; Zhang et al, 2023). Although it remains to be determined whether RdDM can act on TE ecDNA or eccDNA, they harbor the potential to become RdDM targets for the initiation of TGS on integrated EVD copies.
In conclusion, our study reveals that, while PTGS activity contributes to limit TE proliferation ensuring the faithful setting of TGS, PTGS alone is insufficient to explain the de novo initiation of epigenetic silencing of an active proliferative TE in the Arabidopsis genome. TGS initiation in absence of PTGS is more stochastic and, although we have not identified a common trigger event, some of the potential causes identified here will increase the chance of occurring as more EVD copies accumulate in the genome. Besides those, several of the above-mentioned phenomena, including those not investigated here, alone or in combination, could contribute to de novo silencing of EVD and TEs, and deserve future investigation to gain insights into the mechanisms of defense against genomic parasites.
Methods
Reagents and tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number | |
|---|---|---|---|
| Experimental Models | |||
| A. thaliana Col-0 (WT) | (Alonso et al, 2003) | NASC ID: N60000; ABRC ID: CS60000 | |
| A. thaliana rdr6-15 | (Fahlgren et al, 2006) | NASC ID: N879578; ABRC ID: CS879578 (SAIL_617_H07) | |
| A. thaliana epi15 F8 | (Reinders et al, 2009) | N/A | |
| A. thaliana epi15 F8 x rdr6-15 | (Oberlin et al, 2022) | N/A | |
| A. thaliana epi15 F11 | (Marí-Ordóñez et al, 2013) | N/A | |
| A. thaliana nrpd1a-4 (nrpd1) | (Pontier et al, 2005) | NASC ID: N583051; ABRC ID: SALK_083051 | |
| A. thaliana nrpd1b-11 (nrpe1) | (Pontier et al, 2005) | NASC ID: N529919; ABRC ID: SALK_029919 | |
| A. thaliana epi15 F11 x nrpd1 | This work | N/A | |
| A. thaliana epi15 F11 x nrpe1 | This work | N/A | |
| Oligonucleotides and other sequence-based reagents | |||
| PCR/qPCR | |||
| Use | Target | Sequence 5′ → 3′ | Notes |
| Copy number/Expresion levels | ACT2 F | GCACCCTGTTCTTCTTACCG | |
| Copy number/Expresion levels | ACT2 R | AACCCTCGTAGATTGGCACA | |
| Copy number | EVD-GAG F | TTTGACCCGCGTGTTTGAAG | |
| Copy number | EVD-GAG R | AATCTTCGGGTCAAGCGTTC | |
| Copy number | EVD-IN F | CCGGAGAACAAAGAAGCAAGC | |
| Copy number | EVD-IN R | AATGTGCGGTTCTTGGTTGG | |
| Expression levels | shGAG F | GTTGGTTGCTACATCCACACCT | |
| Expression levels | shGAG R | TTTTCCCGTCTCAATATCCGGATT | |
| BiS-PCR | EVD-LTR F | GGATATGTATTATAAGAGAGAGTGGGTYGAATATATG | |
| BiS-PCR | EVD-LTR R | TTTATAARCATAAAAACATAATCTTATRCTCTAATACCATA | |
| BiS-PCR | EVD-3′GAG F | GTGTTTGAAGTGGAAGAAGGYGATTAAYGAATTAA | |
| BiS-PCR | EVD-3′GAG R | ATAACCCRACTTAACTTTRCTCCTCATAAATTTCTTAAA | |
| Genotyping | SAIL_LB3 | tagcatctgaatttcataaccaatctcgatacac | T-DNA BP primer for genotyping rdr6-15 |
| Genotyping | rdr6-15 LP | TGAATCCATTCCTGAACAAGC | mut rdr6-15: BP x LP |
| Genotyping | rdr6-15 RP | CAATGCAACCTCATCTTGGATG | WT RDR6: LP x RP |
| Genotyping | SALK_LBa1 | TGGTTCACGTAGTGGGCCATCG | T-DNA BP for genotyping nrpd1a-4 and nrpd1b-11 |
| Genotyping | NRPD1B-11 LP | CAAAGTGGTGATGCATGGAGG | mut nrpd1b-11: BP x LP |
| Genotyping | NRPD1B-11 RP | ATGTAAATTTTGGAAGTCGGC | WT NRPD1A-11: LP x RP |
| Genotyping | NRPD1A-4 LP | GCACGGGTTCGAATACGGG | mut nrpd1a-4: BP x LP |
| Genotyping | NRPD1A-4 RP | GTATCTGACACCGCGGACTC | WT NRPD1A-4: LP x RP |
| Probes | |||
| Use | Target | Sequence 5′ → 3′ | Notes |
| Northern blots | EVD-LTR F | ATGATGCTCGAGAGTGCGACAAGATCGATGTAGGT | PCR probe |
| Northern blots | EVD-LTR R | TACAATTCCGCATATTCTTTCATGGTATCAGAGCATA | PCR probe |
| Northern blots | EVD-GAG F | TAAGTCAAGAAGACTTAGAGTTTA | PCR probe |
| Northern blots | EVD-GAG R | AAGAAACTCATGAGGAGCAAAGT | PCR probe |
| Northern blots | siR1003 | ATGCCAAGTTTGGCCTCACGGTCT | Oligo probe |
| Northern blots | tasi255 | TACGCTATGTTGGACTTAGAA | Oligo probe |
| Northern blots | miR171 | GATATTGGCGCGGCTCAATCA | Oligo probe |
| Northern blots | U6 | AGGGGCCATGCTAATCTTCTC | Oligo probe |
| Chemicals, Enzymes and other reagents | |||
| TRIzol™ | Thermo Fisher Scientific | #15596018 | |
| DNase I | Thermo Fisher Scientific | #EN0521 | |
| RevertAid Reverse Transcription Kit | Thermo Fisher Scientific | #K1622 | |
| KAPA SYBR FAST qPCR Master Mix (2X) | Sigma-Aldrich/Merck | #KK4602 | |
| 2X NovexTM TBE-Urea sample buffer | Thermo Fisher Scientific | #LLC6876 | |
| Proto+Urea protein and sequencing gel sytem | National Diagnostics | EC-830 + EC-835 + EC-840 | |
| Hybond-NX Nylon membrane | Cytivia | #RPN303T | |
| 1-methylimidazol | Merk | #M50834 | |
| EDC (N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride) | Merk | #E7750 | |
| [α-32P]-dCTP | Hartmann Analytic | #SRP-305 | |
| [γ-32P]-ATP | Hartmann Analytic | #SRP-501 | |
| Prime-it II Random Primer Labelling Kit | Agilent | #300385 | |
| T4 Polynucleotide kinase | Thermo Fisher Scientific | #EK0031 | |
| illustra Microspin G-50 columns | Cytivia | #27533001 | |
| PerfectHyb | Sigma-Aldrich/Merck | #H7033 | |
| DNeasy plant kit | Qiagen | #69204 | |
| NEBNext® Enzymatic Methyl-seq Kit | NEB | #M7634 | |
| EZ DNA Methylation-Gold Kit | Zymo Research | #D5005 | |
| CloneJET PCR Cloning Kit | Thermo Fisher Scientific | #K1231 | |
| Software | |||
| The code developed for this publication can be found here: | https://github.com/GregViallefond/Trasser2024/ | ||
| Excel (16.86) | Microsoft | N/A | |
| R (4.4.1) | R Core Team; https://www.R-project.org | N/A | |
| ggplot2 (3.5.1) | (Wickham, 2016) https://cloud.r-project.org/web/packages/ggplot2/index.html | N/A | |
| TrimGalore (0.6.2) | https://github.com/FelixKrueger/TrimGalore | N/A | |
| Bismark (v0.24.2) |
(Krueger and Andrews, 2011) |
N/A | |
| SAMtools (1.20) | (Li et al, 2009) https://github.com/samtools/samtools | N/A | |
| bedtools (2.28) | https://github.com/arq5x/bedtools2 | N/A | |
| picard (3.2.0) | https://broadinstitute.github.io/picard/ | N/A | |
| Kismeth | (Gruntman et al, 2008) https://katahdin.girihlet.com/kismeth/revpage.pl | N/A | |
| Other | |||
| QuantStudio 5 Real-Time PCR System | Thermo Fisher Scientific | A28575 | |
| Typhoon FLA 9500 | GE Healthcare | N/A | |
| NovaSeq SP | Illumina | N/A | |
Plant material and growth conditions
Plants were grown on soil at 21 °C, in 16 h light/8 h dark cycle with an LED light intensity of 85 µM m−2 s−1. After germination, seedlings were transplanted at a rate of 1 plant per pot. Plants were genotyped by PCR as soon as a tip of a young rosette leaf could be harvested (see Reagents and Tools table for a list of primers used). Inflorescences (closed flower buds, no visible petals) were harvested, flash frozen in liquid nitrogen and stored at −70 °C. Cauline leaves used for EVD copy number quantification were harvested on ice and immediately stored at −70 °C.
Estimation of EVD expression and copy number by qPCR
Total RNA was extracted from 6 to 8 closed inflorescences ground to a fine powder by TRIzol™ according to manufacturer’s instructions and precipitated in 1 volume of cold isopropanol. Total RNA was treated with RNase-free DNase I and cDNA synthesis performed using the RevertAid Reverse Transcription Kit with random hexamer primers. qPCR reactions were performed in a total volume of 10 μl using the KAPA SYBR FAST qPCR Master Mix (2X) with low ROX reference dye according to the manufacturer’s instruction and run on a QuantStudio 5 Real-Time PCR System. Each reaction was performed in 2–3 technical replicates. Relative expression was calculated as fold change of the ratio of target of interest and ACT2 (AT3G18780). For EVD copy number estimations, DNA was extracted from frozen ground cauline leaves using Edwards buffer (200 mM Tris pH 8, 200 mM NaCl, 25 mM EDTA, 0.5% (v/v) SDS). Samples were precipitated in 1 volume of 70% ethanol and diluted at 1/100 in double-distilled water. EVD copy number was estimated by absolute quantification from the ΔΔCt EVD and ACT2 levels, normalized by their inherent copy numbers of two in WT plants, respectively. Oligonucleotides used are listed in the Tools and Reagents table. Data analysis was done in Excel (Microsoft) and R. Plots were generated using ggplot2. Statistical significance was calculated using pairwise Student’s t-test. No blinding was performed.
Small RNA blot analysis
Total RNA was extracted from 6 to 8 closed inflorescences ground to a fine powder by TRIzol™ according to manufacturer’s instructions and precipitated in 1 volume of cold isopropanol. 10–20 μg of total RNA were mixed with an equal volume of 2X Novex™ TBE-Urea sample buffer and resolved on a 17.5% National Diagnostics polyacrylamide-ureal gel according to manufacturer’s instructions, followed by electroblotting on a Hybond-NX Nylon membrane and chemical crosslinking (12.5 M 1-methylimidazol, 31.25 mg/mL EDC (N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride)) according to (Pall and Hamilton, 2008). PCR probes were labeled with [α-32P]-dCTP using the Prime-it II Random Primer Labelling Kit and purified on illustra Microspin G-50 columns according to manufacturer’s instructions. Oligo probes were labeled using [γ-32P]-ATP, using T4 Polynucleotide kinase according to manufacturer’s instructions and purified on illustra Microspin G25 columns. Hybridization was performed in PerfectHyb hybridization buffer for PCR probes, or in Church buffer (7% SDS, 0.5 M Na2HPO4/NaH2PO4 pH 7.2, 1 mM EDTA) for oligonucleotide probes. Following overnight hybridization at 42 °C in a rotary oven, membranes were washed three times at 50 °C with 2X SCC, 0.1% (v/v) SDS. Detection was performed with a Typhoon FLA 9500. Oligonucleotide used for probes are listen in the Tools and Reagents table.
Methylation analysis by EM-Seq
DNA was extracted from closed inflorescences from single plants, using the DNeasy plant kit according to manufacturer’s instructions. Libraries were prepared using the NEBNext® Enzymatic Methyl-seq Kit following the manufacturer’s instructions. Sequencing was performed on an Illumina NovaSeq SP to produce paired-end reads of 150 bp. Sequenced reads were quality filtered and adapter trimmed using TrimGalore version 0.6.2. Enzymatic-converted reads were aligned to the TAIR10 Arabiopsis genome using Bismark version v0.24.2 with bismark (--unmapped --ambiguous --maxins 700) PCR/optical duplicates were removed using deduplicate_bismark and the resulting BAM files were further processed to generate cytosine files bismark_methylation_extractor (--paired-end --cytosine_report --CX_context --comprehensive --no_overlap). Weighted average methylation (Schultz et al, 2012) was calculated and for cytosines laying on each EVD domain. Conversion rates were assessed by calculating the average methylation level for reads mapping to the unmethylated chloroplast (Cokus et al, 2008). Statistical testing was done in R using the function wilcox_test from the rstatix package. P-values were adjusted for multiple testing using Benjamini & Hochberg. Meaning of significance: ****p < 10−4, ***p < 10−3, **p < 0.01, *p < 0.05, ns p ≥ 0.05. No blinding was performed.
EVD copy number estimation and mapping of new insertions from EM-seq
Coverage-based copy number estimation
Depth of coverage at every base between the 5′UTR and 3′UTR of reference EVD insertion (Chr5:5630399-5634902), as well as on two ~10 kb intervals upstream and downstream of EVD (Chr5:5619977-5629277, Chr5:5636011-5645311) was calculated for each paired-end BAM using samtools depth (-aa). The ratio between mean depth of coverage on EVD and mean depth of coverage on the upstream and downstream intervals was calculated for each sample. The mean inverse ratio from Col-0 and rdr6-15 samples only carrying the reference insertion were used as a normalization factor in every other sample.
Discordant read mate pairs-based mapping of EVD insertions
For the identification of new EVD insertions, an existing protocol for the detection of TE insertions from short-read sequencing data, was adapted for EM-seq datasets (Baduel et al, 2021). Each pair of fastq files containing unmapped mates was remapped in single-end (-s) mode using bismark (--local for both mates, --pbat for second in pair) to the TAIR10 genome, as well as to the EVD DNA sequence extracted from TAIR10 (Chr5:5629976-5635312) with parameters --local -D 20 -R 3 -N 1 for both mates, --pbat for second in pair. Since multi-mapping reads are automatically removed by bismark, part of the 3′ LTR from the extracted EVD sequence was masked using bedtools maskfasta (Chr5:5635050:5635156) to prevent reads from mapping exactly to both LTRs, leading to some missmapped reads but maximizing coverage. Fragments for which one mate mapped onto the extracted EVD sequence and the other to any other position in the genome were considered as discordant and candidate reads for identification of de novo insertions. Cases where a single read would map to both EVD and another genomic location were discarded.
Candidate reads were extracted using bedtools bamtobed and loaded into R for processing. A simple scheme was used to identify potential insertions: reads that mapped outside of EVD were sorted by chromosome and start coordinates, and then put into clusters based on their proximity. The directionality (relative to the reference genome) of their component reads was deduced from their strandness and their pairing (first or second in pair). The directionality of candidate reads mapping around a bona fide non-reference insertion is known as they should all point towards the insertion site. Therefore, valid clusters (comprised of m + n reads) were considered those for which the first m reads (upstream of the insertion site) pointed toward the 3′ end of the reference chromosome and the remaining n reads (downstream of the insertion site) pointed towards the 5′ end of the reference chromosome (m ≥ 3, n ≥ 3), from those the insertion site was inferred as located between the end coordinate of the mth read and the start coordinates of the m+1st read. Similarly, paired-end mates of the first m reads (or last n) should all share the same directionality and additionally, the first m and the last n reads display opposite directionality. Any cluster that did not adhere to these rules was discarded. Within each cluster, the directionality of reads mapping onto EVD was used to assess the strandness of the insertion, and from it each read’s LTR-of-origin inferred.
For each remaining cluster, presence and size of target-site-duplication (TSD) was established by inspecting the cigar strings of the mth and m+1st reads. If the mth read was soft-clipped on its 5′ end and the m+1st read was soft-clipped on its 3′ end, and both reads overlapped each other (which was always true), then a TSD was deemed present and the size of the overlap taken as the size of the TSD. 91% of clusters showed such a pattern.
Identification of nested/tandem insertions was done similarly. In short pairs of reads mapping discordantly onto the EVD sequence, with at least one mate overlapping the 5′/3′ LTR were retrieved. For each sample, such reads and their mapping coordinates were then visually inspected.
Assessment of DNA methylation levels in new EVD insertions
For each identified new insertion, the associated cluster of reads was retrieved and LTR-of-origin determined individually for every read. In each sample, the appropriate BAM file was subsampled so it would only include the appropriate clustered reads containing LTR-of-origin information using picard FilterSamReads. For each sample, cluster and LTR-of-origin, a cytosine report was obtained from the subsampled BAM as specified before and a single methylation level for each cytosine context was obtained using weighted average methylation as described above was calculated and for cytosines laying on the each individual LTR.
Bisulfite-PCR sequencing based DNA methylation analysis
DNA was extracted from 6 to 8 closed inflorescences bulked from 4 plants ground to a fine powder, using the DNeasy plant kit according to manufacturer’s instructions. Bisulfite treatment was performed with the EZ DNA Methylation-Gold Kit using the manufacturer’s standard protocol. DNA fragments of interest were amplified by PCR. PCR reactions and primer design was conducted according to published recommendations (Tools and Reagents table, (Henderson et al, 2010)) and cloned using CloneJET PCR Cloning Kit. Single colonies were sequenced by Sanger sequencing. Sequences were analyzed using the browser-based Kismeth software. Wilson score intervals were used to find 95% confidence intervals for the percentages of methylated sites in each context. Results shown were obtained from two independent experiments.
Supplementary information
Acknowledgements
This work was supported by the Gregor Mendel Institute of the Austrian Academy of Sciences core funding attributed to AM-O and MN. We thank current and former members of the Marí-Ordóñez group as well as colleagues from the Gregor Mendel Institute and the Vienna BioCenter campus for useful discussions, ideas and feedback on the project and manuscript. We are grateful to the Vienna BioCenter Core Facilities (VBCF): NGS facility for EM-seq library preparation and sequencing, and the Plant Science Facility for assistance with plant work. We also thank the IMP Molecular Biology Service for providing reagents and Sanger sequencing. Lastly, we would like to acknowledge the VBC in-house COVID-19 testing facility for their efforts in providing a safe working environment during the pandemic.
Expanded view
Author contributions
Marieke Trasser: Conceptualization; Data curation; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Grégoire Bohl-Viallefond: Formal analysis; Investigation; Visualization; Methodology. Verónica Barragán-Borrero: Investigation; Methodology. Laura Diezma-Navas: Investigation. Lukas Loncsek: Formal analysis; Investigation. Magnus Nordborg: Supervision; Writing—review and editing. Arturo Marí-Ordóñez: Conceptualization; Resources; Data curation; Formal analysis; Supervision; Funding acquisition; Validation; Visualization; Writing—original draft; Project administration; Writing—review and editing.
Source data underlying figure panels in this paper may have individual authorship assigned. Where available, figure panel/source data authorship is listed in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00304-5.
Data availability
NGS data generated for this study are accessible at NCBI SRA under ID number PRJNA1111825. Accompanying code for this publication can be found here: https://github.com/GregViallefond/Trasser2024/.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00304-5.
Disclosure and competing interests statement
The authors declare no competing interests.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44319-024-00304-5.
References
- Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ (1998) Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113–2115 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Madhani HD (2018) Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 19:229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657 [DOI] [PubMed] [Google Scholar]
- Baduel P, Leduque B, Ignace A, Gy I, Gil J, Loudet O, Colot V, Quadrana L (2021) Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biol 22:138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Quadrana L, Colot V (2021) Plant transposable elements, methods and protocols. Methods Mol Biol 2250:157–169 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Plant Biol 65:505–530 [DOI] [PubMed] [Google Scholar]
- Bente H, Foerster AM, Lettner N, Scheid OM (2021) Polyploidy-associated paramutation in Arabidopsis is determined by small RNAs, temperature, and allele structure. PLoS Genet 17:e1009444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3:e3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet P, Picard CL, Yelagandula R, Pélissier T, Lorković ZJ, Feng S, Pouch-Pélissier M-N, Schmücker A, Jacobsen SE, Berger F et al (2021) The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation. Nat Commun 12:2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS et al (2018) Ten things you should know about transposable elements. Genome Biol 19:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103 [DOI] [PubMed] [Google Scholar]
- Buck SD, Jacobs A, Montagu MV, Depicker A (1999) The DNA sequences of T‐DNA junctions suggest that complex T‐DNA loci are formed by a recombination process resembling T‐DNA integration. Plant J 20:295–304 [DOI] [PubMed] [Google Scholar]
- Chan SWL, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Beckwith SL, Letham SC, Kim SJ, Young GR, Stoye JP, Garfinkel DJ, Taylor IA (2021) Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control. Nat Commun 12:5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Groth M, Feng S, Hale CJ, Li S, Vashisht AA, Gallego-Bartolome J, Wohlschlegel JA, Patel DJ et al (2014) Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell 55:495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Liu Y, Lu L, Shi J, Xu L, Li Q, Cheng X, Chen J, Zhang X (2022) Accumulation of DNA damage alters microRNA gene transcription in Arabidopsis thaliana. BMC Plant Biol 22:576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A et al (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151:167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann RM, Picard CL (2020) RNA-directed DNA methylation. PLoS Genet 16:e1009034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944 [DOI] [PubMed] [Google Scholar]
- Feng S, Zhong Z, Wang M, Jacobsen SE (2020) Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing Epigenetics Chromatin 13:42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster AM, Dinh HQ, Sedman L, Wohlrab B, Scheid OM (2011) Genetic rearrangements can modify chromatin features at epialleles. PLoS Genet 7:e1002331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz D, Slotkin RK (2017) Exogenous transposable elements circumvent identity-based silencing, permitting the dissection of expression-dependent silencing. Plant Cell 29:360–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic‐Hagan J, Ellacott GA, Watson JM, Wang M, Brosnan C, Carroll BJ et al (2006) RNA interference‐inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7:1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly A, Etcheverry M, Madoui M-A, Guy J, Quadrana L, Alberti A, Martin A, Heitkam T, Engelen S, Labadie K et al (2014) TE-Tracker: systematic identification of transposition events through whole-genome resequencing. BMC Bioinforma 15:377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriaux C, Théron E, Brasset E, Vaury C (2014) History of the discovery of a master locus producing piRNAs: the flamenco/COM locus in Drosophila melanogaster. Front Genet 5:257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Grossniklaus U (2019) Invasive DNA elements modify the nuclear architecture of their insertion site by KNOT-linked silencing in Arabidopsis thaliana. Genome Biol 20:1–15 [DOI] [PMC free article] [PubMed]
- Grob S, Schmid MW, Grossniklaus U (2014) Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol Cell 55:678–693 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinforma 9:371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida V, Cernilogar FM, Filograna A, Gregorio RD, Ishizu H, Siomi MC, Schotta G, Bellenchi GC, Andrenacci D (2016) Production of small noncoding RNAs from the flamenco locus is regulated by the gypsy retrotransposon of Drosophila melanogaster. Genetics 204:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Takehira K, Nozawa K, Suzuki T, Masuta Y, Kato A, Ito H (2020) ONSEN shows different transposition activities in RdDM pathway mutants. Genes Genet Syst 95:183–190 [DOI] [PubMed]
- He L, Huang H, Bradai M, Zhao C, You Y, Ma J, Zhao L, Lozano-Durán R, Zhu J-K (2022) DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat Commun 13:1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Zhao C, Zhang Q, Zinta G, Wang D, Lozano-Durán R, Zhu J-K (2021) Pathway conversion enables a double-lock mechanism to maintain DNA methylation and genome stability. Proc Natl Acad Sci USA 118:e2107320118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Chan SR, Cao X, Johnson L, Jacobsen SE (2010) Accurate sodium bisulfite sequencing in plants. Epigenetics 5:47–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115–119 [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Lexa M, Vanat I, Hobza R, Kejnovsky E (2019) Nested plant LTR retrotransposons target specific regions of other elements, while all LTR retrotransposons often target palindromes and nucleosome-occupied regions: in silico study. Mob DNA 10:50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Xu Y-C, Zhang Z-Q, Chen J-F, Niu X-M, Hou X-H, Li X-T, Wang L, Zhang YE, Ge S et al (2023) Forces driving transposable element load variation during Arabidopsis range expansion. Plant Cell 36:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann M, Mette MF (2012) Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol Biol 79:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S, Carpentier M-C, Llauro C, Jobet E, Robakowska-Hyzorek D, Lasserre E, Ghesquière A, Panaud O, Mirouze M (2017) Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet 13:e1006630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Maple Y-S, Dürr R, Dawson J, Tamim A, Genio S, del C, Papareddy R, Luo A, Lamb JC, Amantia S et al (2021) A transposon surveillance mechanism that safeguards plant male fertility during stress. Nat Plants 7:34–41 [DOI] [PubMed] [Google Scholar]
- Lee SC, Ernst E, Berube B, Borges F, Parent J-S, Ledon P, Schorn A, Martienssen RA (2020) Arabidopsis retrotransposon virus-like particles and their regulation by epigenetically activated small RNA. Genome Res 30:576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup 1000 Genome Project Data Processing (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM et al (2015) RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci USA 112:14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD et al (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471–476 [DOI] [PubMed] [Google Scholar]
- Lorković ZJ, Park C, Goiser M, Jiang D, Kurzbauer M-T, Schlögelhofer P, Berger F (2017) Compartmentalization of DNA damage response between heterochromatin and euchromatin is mediated by distinct H2A histone variants. Curr Biol 27:1192–1199 [DOI] [PubMed] [Google Scholar]
- Mann L, Seibt KM, Weber B, Heitkam T (2022) ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinforma 23:40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O (2013) Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45:1029–1039 [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Čaikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG Methylation. Cell 130:851–862 [DOI] [PubMed] [Google Scholar]
- Matsuda E, Garfinkel DJ (2009) Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci USA 106:15657–15662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga W, Kobayashi A, Kato A, Ito H (2012) The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol 53:824–833 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15:394–408 [DOI] [PubMed] [Google Scholar]
- McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801 [DOI] [PubMed] [Google Scholar]
- McCue AD, Panda K, Nuthikattu S, Choudury SG, Thomas EN, Slotkin RK (2015) ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J 34:20–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461:427–430 [DOI] [PubMed] [Google Scholar]
- Neve MD, Buck SD, Jacobs A, Montagu MV, Depicker A (1997) T‐DNA integration patterns in co‐transformed plant cells suggest that T‐DNA repeats originate from co‐integration of separate T‐DNAs. Plant J 11:15–29 [DOI] [PubMed] [Google Scholar]
- Niu X, Chen L, Kato A, Ito H (2022) Regulatory mechanism of a heat-activated retrotransposon by DDR complex in Arabidopsis thaliana. Front Plant Sci 13:1048957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK (2013) The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 162:116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin S, Rajeswaran R, Trasser M, Barragán‐Borrero V, Schon MA, Plotnikova A, Loncsek L, Nodine MD, Marí‐Ordóñez A, Voinnet O (2022) Innate, translation‐dependent silencing of an invasive transposon in Arabidopsis. EMBO Rep 23:e53400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin S, Sarazin A, Chevalier C, Voinnet O, Marí-Ordóñez A (2017) A genome-wide transcriptome and translatome analysis of Arabidopsistransposons identifies a unique and conserved genome expression strategy for Ty1/Copiaretroelements. Genome Res 27:1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Arias S, Isaza G, Guyot R (2019) Retrotransposons in plant genomes: structure, identification, and classification through bioinformatics and machine learning. Int J Mol Sci 20:3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe A, Lorković ZJ, Kobayashi W, Tachiwana H, Yelagandula R, Kurumizaka H, Berger F (2018) Histone H2A variants confer specific properties to nucleosomes and impact on chromatin accessibility. Nucleic Acids Res 46:7675–7685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ (2008) Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3:1077–1084 [DOI] [PubMed] [Google Scholar]
- Panda K, Ji L, Neumann DA, Daron J, Schmitz RJ, Slotkin RK (2016) Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation Genome Biol 17:170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19:2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui M-A, Guy J, Silveira AB, Engelen S, Baillet V, Wincker P et al (2019) Transposition favors the generation of large effect mutations that may facilitate rapid adaption Nat Commun 10:3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Silveira AB, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V (2016) The Arabidopsis thaliana mobilome and its impact at the species level. Elife 5:e15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P, Sanville BC, Buchmann RC, Bisaro DM (2008) Viral genome methylation as an epigenetic defense against geminiviruses. J Virol 82:8997–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BBH, Mirouze M, Ordóñez AM, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23:939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquis D, Robertson M, Yu L, Thieme M, Julkowska M, Bucher E (2021) Genomic impact of stress-induced transposable element mobility in Arabidopsis. Nucleic Acids Res 49:gkab828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammarco I, Pieters J, Salony S, Toman I, Zolotarov G, Placette CL (2022) Epigenetic targeting of transposon relics: beating the dead horses of the genome? Epigenetics 17:1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Gunis J, Reichardt-Gomez I, Nizhynska V, Nordborg M (2022) Conditional GWAS of non-CG transposon methylation in Arabidopsis thaliana reveals major polymorphisms in five genes. PLoS Genet 18:e1010345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk C, Cognat V, Graindorge S, Vincent T, Voinnet O, Molinier J (2017) Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci USA 114:E2965–E2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk C, Drevensek S, Kramdi A, Kassam M, Ahmed I, Cognat V, Graindorge S, Bergdoll M, Baumberger N, Heintz D et al (2016) DNA DAMAGE BINDING PROTEIN2 shapes the DNA methylation landscape. Plant Cell 28:2043–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid OM, Afsar K, Paszkowski J (2003) Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet 34:450–454 [DOI] [PubMed] [Google Scholar]
- Schubert I, Vu GTH (2016) Genome stability and evolution: attempting a holistic view. Trends Plant Sci 21:749–757 [DOI] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR (2012) ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet 28:583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman MJ, Panda K, Kirchner R, McLain LL, Payne H, Peasari JR, Husbands AY, Slotkin RK, McCue AD (2021) An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation. Nat Plants 7:1461–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D (2003) Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics 165:781–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D (2005) Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37:641–644 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, TanurdZiC M, Becker JOD, Feij OJA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152:352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R (2016) Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5:e20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taochy C, Yu A, Bouché N, Bouteiller N, Elmayan T, Dressel U, Carroll BJ, Vaucheret H (2019) Post-transcriptional gene silencing triggers dispensable DNA methylation in gene body in Arabidopsis. Nucleic Acids Res 47:9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF et al (2009) A role for RNAi in the selective correction of DNA methylation defects. Science 323:1600–1604 [DOI] [PubMed] [Google Scholar]
- Thieme M, Lanciano S, Balzergue S, Daccord N, Mirouze M, Bucher E (2017) Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol 18:134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461:423–426 [DOI] [PubMed]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Danielsen JMR, Yang Y-G, Qi Y (2012) A role for small RNAs in DNA double-strand break repair. Cell 149:101–112 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) Ggplot2: elegant graphics for data analysis. Springer
- Wu L, Mao L, Qi Y (2012) Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol 160:990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, Muthurajan UM, Nie X, Kawashima T, Groth M et al (2014) The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158:98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J (2009) Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 19:1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153:193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Mbodj A, Soundiramourtty A, Llauro C, Ghesquière A, Ingouff M, Slotkin RK, Pontvianne F, Catoni M, Mirouze M (2023) Extrachromosomal circular DNA and structural variants highlight genome instability in Arabidopsis epigenetic mutants. Nat Commun 14:5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14:1214–1220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data generated for this study are accessible at NCBI SRA under ID number PRJNA1111825. Accompanying code for this publication can be found here: https://github.com/GregViallefond/Trasser2024/.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00304-5.