Abstract
The serine esterase factor D of the complement system was purified from outdated human plasma with a yield of 20% of the initial haemolytic activity found in serum. This represented an approx. 60 000-fold purification. The final product was homogeneous as judged by sodium dodecyl sulphate/polyacrylamide-gel electrophoresis (with an apparent mol.wt. of 24 000), its migration as a single component in a variety of fractionation procedures based on size and charge, and its N-terminal amino-acid-sequence analysis. The N-terminal amino acid sequence of the first 36 residues of the intact molecule was found to be homologous with the N-terminal amino acid sequences of the catalytic chains of other serine esterases. Factor D showed an especially strong homology (greater than 60% identity) with rat 'group-specific protease' [Woodbury, Katunuma, Kobayashi, Titani, & Neurath (1978) Biochemistry 17, 811-819] over the first 16 amino acid residues. This similarity is of interest since it is considered that both enzymes may be synthesized in their active, rather than zymogen, forms. The three major CNBr fragments of factor D, which had apparent mol.wts. of 15 800, 6600 and 1700, were purified and then aligned by N-terminal amino acid sequence analysis and amino acid analysis. By using factor D labelled with di-[1,3-14C]isopropylphosphofluoridate it was shown that the CNBr fragment of apparent mol.wt. 6600, which is located in the C-terminal region of factor D, contained the active serine residue. The amino acid sequence around this residue was determined.
Full text
PDF



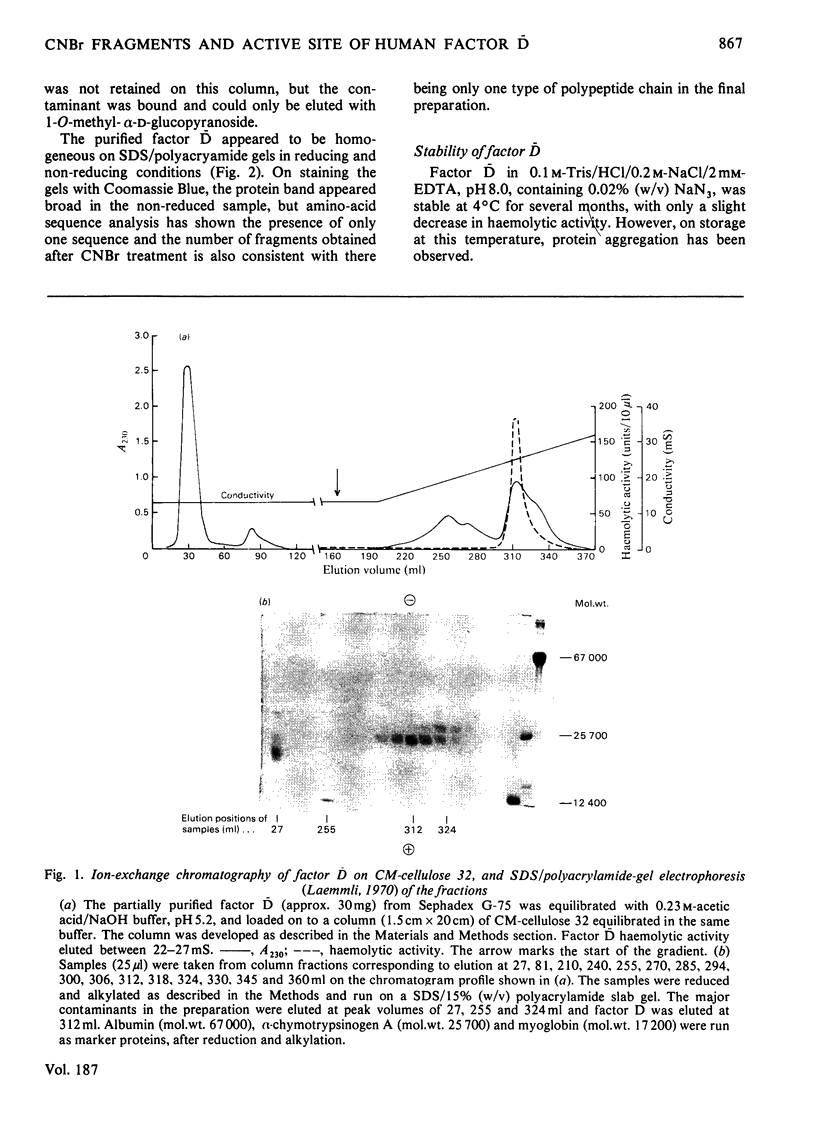
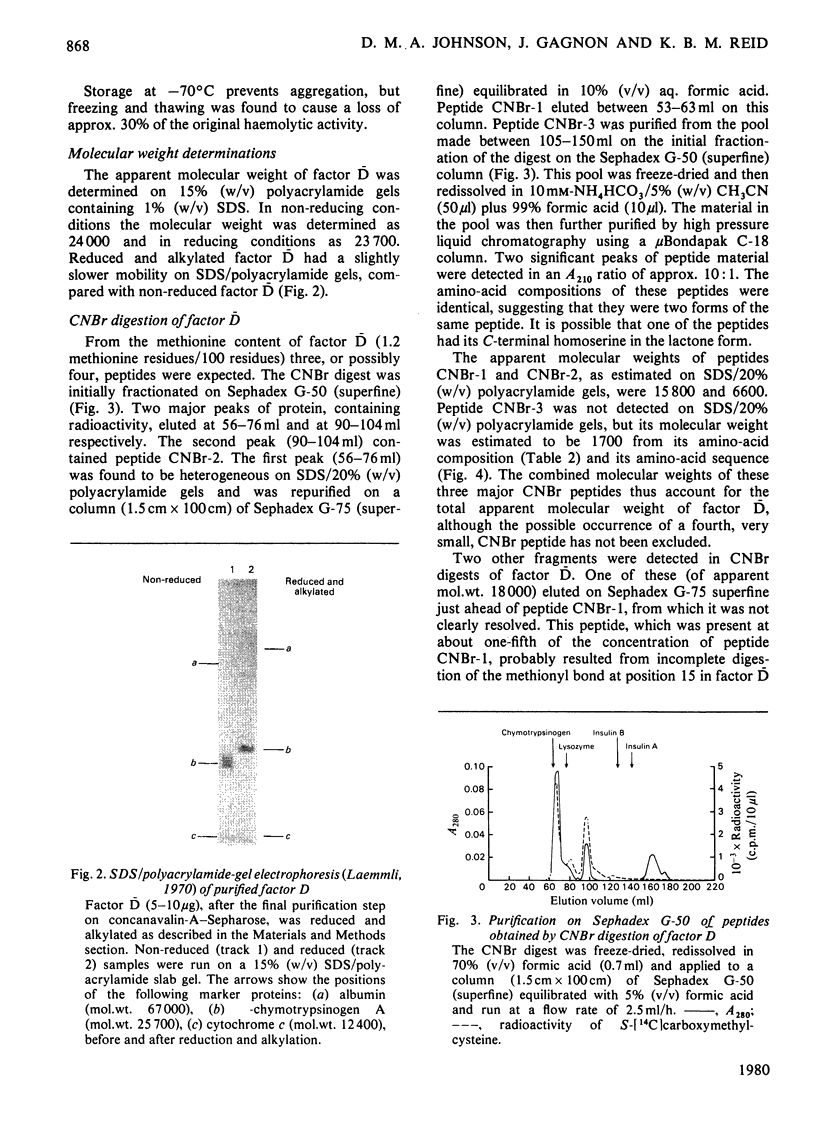





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austen K. F. Homeostasis of effector systems which can also be recruited for immunologic reactions. J Immunol. 1978 Sep;121(3):793–805. [PubMed] [Google Scholar]
- Bridgen P. J., Cross G. A., Bridgen J. N-terminal amino acid sequences of variant-specific surface antigens from Trypanosoma brucei. Nature. 1976 Oct 14;263(5578):613–614. doi: 10.1038/263613a0. [DOI] [PubMed] [Google Scholar]
- Davis A. E., 3rd, Zalut C., Rosen F. S., Alper C. A. Human factor D of the alternative complement pathway. Physicochemical characteristics and N-terminal amino acid sequence. Biochemistry. 1979 Nov 13;18(23):5082–5087. doi: 10.1021/bi00590a010. [DOI] [PubMed] [Google Scholar]
- Fearon D. T., Austen K. F., Ruddy S. Properdin factor D: characterization of its active site and isolation of the precursor form. J Exp Med. 1974 Feb 1;139(2):355–366. doi: 10.1084/jem.139.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Direct microsequence analysis of polypeptides using an improved sequenator, a nonprotein carrier (polybrene), and high pressure liquid chromatography. Biochemistry. 1978 May 30;17(11):2124–2133. doi: 10.1021/bi00604a016. [DOI] [PubMed] [Google Scholar]
- Klapper D. G., Wilde C. E., 3rd, Capra J. D. Automated amino acid sequence of small peptides utilizing Polybrene. Anal Biochem. 1978 Mar;85(1):126–131. doi: 10.1016/0003-2697(78)90282-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesavre P. H., Hugli T. E., Esser A. F., Müller-Eberhard H. J. The alternative pathway C3/C5 convertase: chemical basis of factor B activation. J Immunol. 1979 Aug;123(2):529–534. [PubMed] [Google Scholar]
- Lesavre P. H., Müller-Eberhard H. J. Mechanism of action of factor D of the alternative complement pathway. J Exp Med. 1978 Dec 1;148(6):1498–1509. doi: 10.1084/jem.148.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Lachmann P. J., Halbwachs L., Hobart M. J. Haemolytic diffusion plate assays for factors B and D of the alternative pathway of complement activation. Immunochemistry. 1976 Apr;13(4):317–324. doi: 10.1016/0019-2791(76)90341-4. [DOI] [PubMed] [Google Scholar]
- Moore G. W., Goodman M. Alignment statistic for identifying related protein sequences. J Mol Evol. 1977 Apr 29;9(2):121–130. doi: 10.1007/BF01732744. [DOI] [PubMed] [Google Scholar]
- Schreiber R. D., Pangburn M. K., Lesavre P. H., Müller-Eberhard H. J. Initiation of the alternative pathway of complement: recognition of activators by bound C3b and assembly of the entire pathway from six isolated proteins. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3948–3952. doi: 10.1073/pnas.75.8.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim R. B., Porter R. R., Reid K. B., Gigli I. The structure and enzymic activities of the C1r and C1s subcomponents of C1, the first component of human serum complement. Biochem J. 1977 May 1;163(2):219–227. doi: 10.1042/bj1630219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis J. E., Schrohenloher R. E., Stroud R. M. Human factor D of the alternative complement pathway: purification and characterization. J Immunol. 1977 Jul;119(1):337–342. [PubMed] [Google Scholar]
- Woodbury R. G., Katunuma N., Kobayashi K., Titani K., Neurath H., Anderson W. F., Matthews B. W. Covalent structure of a group-specific protease from rat small intestine. Appendix: crystallographic data for a group specific protease from rat intestine. Biochemistry. 1978 Mar 7;17(5):811–819. doi: 10.1021/bi00598a010. [DOI] [PubMed] [Google Scholar]
- de Haën C., Neurath H., Teller D. C. The phylogeny of trypsin-related serine proteases and their zymogens. New methods for the investigation of distant evolutionary relationships. J Mol Biol. 1975 Feb 25;92(2):225–259. doi: 10.1016/0022-2836(75)90225-9. [DOI] [PubMed] [Google Scholar]