Summary
Multiple lines of evidence indicate that ankylosing spondylitis (AS) is a lymphocyte-driven disease. However, which lymphocyte populations are critical in AS pathogenesis is not known. In this study, we aimed to identify the key cell types mediating the genetic risk in AS using an unbiased functional genomics approach. We integrated genome-wide association study (GWAS) data with epigenomic and transcriptomic datasets of human immune cells. To quantify enrichment of cell type-specific open chromatin or gene expression in AS risk loci, we used three published methods—LDSC-SEG, SNPsea, and scDRS—that have successfully identified relevant cell types in other diseases. Natural killer (NK) cell-specific open chromatin regions are significantly enriched in heritability for AS, compared to other immune cell types such as T cells, B cells, and monocytes. This finding was consistent between two AS GWAS. Using RNA sequencing data, we validated that genes in AS risk loci are enriched in NK cell-specific gene expression. Using the human Space-Time Gut Cell Atlas, we also found significant upregulation of AS-associated genes predominantly in NK cells. We performed co-localization analyses between GWAS risk loci and genetic variants associated with gene expression (eQTL) to find putative target genes. This revealed four AS risk loci affecting regulation of candidate target genes in NK cells: two known loci, ERAP1 and TNFRSF1A, and two understudied loci, ENTR1 (SDCCAG3) and B3GNT2. Our findings suggest that NK cells may play a crucial role in AS development and highlight four putative target genes for functional follow-up in NK cells.
Keywords: ankylosing spondylitis, axial spondyloarthritis, natural killer (NK) cells, genome-wide association study (GWAS), transcriptomics, epigenomics, single-cell RNA-seq, immune-mediated disease, expression quantitative trait locus (eQTL), immune cells
Chiñas et al. gained new mechanistic insights into ankylosing spondylitis by integrating genetic data with epigenomic and transcriptomic datasets of peripheral immune cells and cells found in gut tissue. Their findings highlight natural killer cells as potential drivers of genetic susceptibility to the disease.
Introduction
Axial spondyloarthritis (axSpA) is a chronic inflammatory rheumatic disease characterized by inflammation of the spine and sacroiliac joints, with a proportion of persons with axSpA also presenting with arthritis in peripheral joints, uveitis, psoriasis (MIM: 177900), or inflammatory bowel disease (MIM: 266600).1 Historically, most genetic and pathogenetic studies in axSpA have been carried out in ankylosing spondylitis (AS [MIM: 106300]), a severe and well-characterized subtype of axSpA. The heritability of AS is high, with estimates ranging from between 40% and >90%.2 Two independent twin studies report heritability of >90%.3,4 The International Genetics of Ankylosing Spondylitis (IGAS) Consortium performed a genetic association study using the Immunochip, which ascertains a subset of risk loci in the genome that are relevant to immune-mediated diseases.5 IGAS reports a SNP-based heritability for AS of 32.7%, which is 1.6–2.4 greater than that for Crohn disease and psoriasis.6 In the genome-wide association study (GWAS) performed with UK Biobank data, in which an Affymetrix chip with genome-wide coverage was used, the SNP-based heritability for AS is estimated to be 69.1%, 3.3–3.8 greater than that for Crohn disease and psoriasis.7,8 Hence, AS has a strong genetic component, especially compared to other inflammatory diseases. Human leukocyte antigen B27 (HLA-B27) is the major risk allele for AS (odds ratio = 21.4).9 Additionally, GWASs have revealed >100 non-major histocompatibility complex (MHC) risk loci for AS, most of them implicating non-coding variants.5,6
Many immune cell types have been associated with axSpA. However, which ones are “driver” cell types actively contributing to the pathogenesis of the disease, as opposed to “bystanders” that become involved as a consequence of the disease, remain unclear. Studies leveraging genetic risk variants and their overlap with epigenomic and transcriptomic features variably suggested CD8+ T cells, CD4+ T cells, NK (natural killer) cells, monocytes, and gastrointestinal cells as potential mediators of AS genetic risk.10,11,12 However, these studies did not apply the new functional genomics datasets generated from human cells or the latest methodologies designed to integrate functional genomics with GWAS data. This new generation of methods takes advantage of the full range of SNPs examined in a GWAS (not just those surpassing the genome-wide significance threshold) and robustly control for genomic and linkage disequilibrium (LD) biases.13,14
For several immune-mediated diseases, these integrative functional genomics methods have successfully identified specific cell types as drivers of disease development. For example, for rheumatoid arthritis (RA [MIM: 180300]), multiple studies have found a significant enrichment of genetic risk in open or active chromatin regions (marking regulatory elements) specific for T cells.15,16,17 Both mouse and human studies corroborate the role of T cells as central players in RA pathogenesis.18,19 Similarly, for systemic lupus erythematosus (SLE [MIM: 152700]), studies have identified an enrichment of B cell-specific putative regulatory elements and gene expression in SLE risk loci,16,17 consistent with the well-established role of B cells in SLE pathogenesis.20 Hence, there is precedence that the integration of GWAS with functional genomics datasets can identify cellular drivers in inflammatory diseases with complex pathogenesis.
Here, we sought to investigate which immune cell populations could be drivers of AS development. We integrated GWAS summary statistics from two different AS cohorts with epigenomic and transcriptomic datasets of human leukocytes from peripheral blood and tissue using established methods that control for biases in genomic enrichment analyses. Our results bring forward NK cells as potential key drivers in the pathogenesis of AS.
Material and methods
GWASs
We used the GWAS Immunochip summary statistics from the IGAS Consortium. The IGAS study, led by Cortes et al.,5 performed high-density genotyping of 9,069 AS cases and 13,578 healthy controls. In addition, we used the GWAS summary statistics from the UK Biobank, which involved a case-control design with 1,185 AS cases and 419,276 controls, providing genome-wide coverage for AS susceptibility loci.21
We lifted the genomic positions of the genetic variants to genome build hg19 or hg38 according to the version compatible with subsequent analyses. Given the complexity and strong genetic association signals within the MHC region, we excluded variants located on chr6: 25 Mb–34 Mb.
We additionally used GWAS summary statistics for RA, Alzheimer disease, and SLE as positive control traits for which we know the disease-relevant immune cell types, and height as a negative control trait for which we do not expect immune cells to be relevant. The summary statistics for control traits were preprocessed by the Alkes Price laboratory. They included HapMap 3 (HM3) SNPs and SNPs that are in the 1000 Genomes Project, and they excluded the MHC region (chr6: 25 Mb–34 Mb). These summary statistics are available at https://alkesgroup.broadinstitute.org/.
Epigenomic and transcriptomic datasets
To identify cell-type-specific open chromatin regions in different immune cell types, we used the Calderon et al. study,16 in which the authors collected blood from four healthy subjects, sorted immune cell types, and generated chromatin accessibility profiles using assay for transposase accessible chromatin sequencing (ATAC-seq; GEO: GSE118189).
To find AS risk enrichment for cell-type-specific expression, we incorporated data from the study conducted by Gutierrez-Arcelus et al.,22 which involved low-input mRNA-seq data from sorted NK cells and six T cell subsets isolated from six healthy subjects (each with two replicates per cell type; GEO: GSE124731).
We used the Space-Time Gut Cell Atlas to identify cells exhibiting significant upregulation of disease-associated genes. This dataset includes single-cell (sc)RNA-seq profiling of 428,000 intestinal cells obtained from fetal (N = 16), pediatric (N = 8), and adult donors (N = 13). The dataset covers 11 different intestinal regions23 (https://www.gutcellatlas.org/).
Differential accessibility analysis
We used the counts of open chromatin consensus peaks called by Calderon et al.16 First, we transformed counts into reads per kilobase per million mapped reads (RPKM), then normalized by quantiles using the preprocess Core R package, and finally scaled to their log2 (normalized RPKM+1); thus, we account for differences in library size across samples and peak length variability. We pooled sorted samples into seven main immune cell types, aiming for a similar number of samples per cell type to avoid biases in the differential accessibility analyses: T cells (stimulated and unstimulated CD8+ T, unstimulated naive CD4 T, and memory CD4 T), B cells (stimulated and unstimulated bulk B cells, unstimulated memory and naive B cells), NK cells (stimulated and unstimulated mature NKs, unstimulated memory NKs, and immature NKs), monocytes (stimulated and unstimulated monocytes), plasmablasts (unstimulated plasmablasts), dendritic cells (DCs; unstimulated myeloid cells), and plasmacytoid DCs (unstimulated plasmacytoid DCs). The latter three cell types had fewer samples available; however, this did not impede our control trait Alzheimer disease to show significant heritability enrichment for myeloid DC-specific open chromatin regions, as expected.
Next, we employed linear mixed-model regression to identify regions that exhibited differential accessibility between each cell type and the rest of the cell types. To account for potential donor-specific effects, we incorporated the donor ID variable as a random effect in our analysis.
For each cell type comparison, we tested peaks that had counts greater than the mean for that cell type in at least half of the samples. This yielded between 400,000 and 600,000 tested peaks depending on the cell type. To select the cell-type-specific open chromatin peaks for each cell type, we sorted open chromatin peaks by their t statistic and chose the positive top 10%.
Partitioned heritability enrichment analysis with LDSC-SEG
The LD score regression applied to specifically expressed genes (LDSC-SEG) version 1.0.1 method13 was applied to determine disease-relevant cell types for AS. Cell-type-specific open chromatin peaks were extended by 225 bp to each side to match the genomic coverage recommended by the LDSC-SEG authors. These annotations were then utilized as input for the partitioned heritability enrichment analysis by LDSC-SEG. To test whether the coefficient exceeds zero, LDSC-SEG utilizes a one-sided t test for computing regression p values. We used the baseline annotation version 1.2 provided by the Alkes Price lab for LDSC-SEG, comprising 75 background annotations. Additionally, we used all consensus peaks (N = 829,942) of Calderon et al. as the control annotation. Using other baselines or controls did not affect our results. We utilized SNP weight files derived from the HM3 project European population.
Analysis of cell-type-specific gene expression enrichment in risk loci using SNPsea
SNPsea analysis aimed to assess the association between risk SNPs and genes expressed specifically for a given cell type.24 We incorporated a curated list of risk SNPs for AS, compiled by Brown and Wordsworth,8 which includes genetic variants that have been associated with AS susceptibility. This list was derived from multiple AS studies conducted until 2017.
We utilized the expression data obtained from Gutierrez-Arcelus et al.22 The gene expression counts in this dataset were normalized to transcripts per million (TPM) and transformed to log2(TPM+1) values. To identify the genes with meaningful expression levels, we included those with log2(TPM+1) > 2 in at least 10 samples. SNPsea was then run for the normalized expression matrix and AS risk SNPs, using recombination intervals from Myers et al.,25 null SNPs from Lango Allen et al.,26 and the following parameters: --score single --slop 10000 --threads 2 --null-snpsets 0 --min-observations 100 --max-iterations 10000000.
Integration of GWAS with scRNA-seq with scDRS
We used the single-cell disease relevance score (scDRS) by combining scRNA-seq and GWAS to identify cells with significant upregulation of disease-associated genes, which are scored based on their strength of association with disease and are compared with null sets of genes present in the dataset.
As recommended by the scDRS authors, we first created disease-relevant gene sets using Multi-marker Analysis of GenoMic Annotation (MAGMA) version 1.10.27 We generated gene annotations with MAGMA, setting a window of 10 kb using the following parameters: --annotate window = 10,10 --snp-loc./g1000_eur/g1000_eur.bim --gene-loc./NCBI37.3/NCBI37.3.gene.loc. Then, we ran MAGMA using GWAS summary statistics for traits of interest with the following parameters: --bfile./magma_v1.10/g1000_eur/g1000_eur --pval GWAS.pval use = ‘SNP,P’ ncol = ‘N' --gene-annot./magma_v1.10/out/step1.genes.annot.
We ran scDRS using the disease-relevant gene sets from MAGMA and the expression data obtained from the Space-Time Gut Cell Atlas23 and corrected for biases by adding as covariates the number of genes expressed per cell and sample batch. Next, for visualization purposes and downstream analysis, we processed the single-cell dataset using Seurat,28 we performed integration across batches with Harmony,29 and we visualized cells in two dimensions with uniform manifold approximation and projection (UMAP). We labeled cells plotted in UMAP by the annotations defined by the Space-Time Gut Cell Atlas. Additionally, we colored cells by their scDRS score when cells passed the 0.20 false discovery rate (FDR) threshold.
Differential expression analyses
We used two differential expression analyses: low-input mRNA-seq and scRNA-seq. For the low-input mRNA-seq, data from Gutierrez-Arcelus et al.22 were analyzed to compare NK cells and T cell subsets from the peripheral blood of six healthy donors (each cell subset per donor in duplicate). The data were normalized using TPM and then log2 transformed. We tested genes with log2(TPM+1) > 2 in at least 10 samples. Linear mixed models (LMMs) were applied using the lme4 package in R, controlling for donor as a random effect. p values were adjusted for multiple testing using the FDR calculated by the qvalue R package. We then filtered significant genes within AS risk loci and used heatmaps to visualize the expression patterns.
For the scRNA-seq, pseudobulk expression data were generated by aggregating scRNA-seq data from NK cells and other immune cell types per donor. Counts per million normalization and log2 transformation were applied to the data before differential expression analysis. We tested genes with at least 5 counts in at least 20 donors. LMM was used, with donors included as a random effect. The analysis was conducted using the lme4 package in R, with p values adjusted for multiple testing using FDR calculated by the qvalue R package.
eQTL co-localization analysis
To select genomic loci for colocalization analysis, GWAS summary statistics were sorted by p values. Then, starting from the variant with the smallest p value, variants within a 50-kb window were removed. The process was repeated with the next most significant variant among the remaining variants until no variant with a p value below 5 × 10−5 was left. We performed colocalization analysis for GWAS studies against the eQTL (expression quantitative trait loci) Catalogue.30 We imported eQTL summary statistics from RNA-seq and microarray from Schmiedel et al.31 and Gilchrist et al.32 We fetched the summary statistics data using the tabix method with the seqminer R package (version 8.5). For each region tested, we included all biallelic SNPs that were ascertained in both the GWAS and eQTL study and performed the analysis only for genes within a window of ±500,000 bp from the GWAS top variant, and for which there was at least one eQTL passing the 5 × 10−5 p value threshold. Before merging GWAS and QTL data, the variant coordinates of the GWAS were lifted to the GRCh38 version of the reference genome using liftOver with the UCSC chain file. We used the coloc version 5.1.0.1 package33 in R version 4.1.0 to test for colocalization at each gene and dataset.
Each locus was plotted using plotgardener,34 and we recovered the LD of the top SNP in a given region in the GWAS dataset using the locuscomparer package.35 Then, we used plotgardener functions to display the regions near the lead variant and colored the genes tested using the posterior probability that the two traits share a causal variant (PP4).
Results
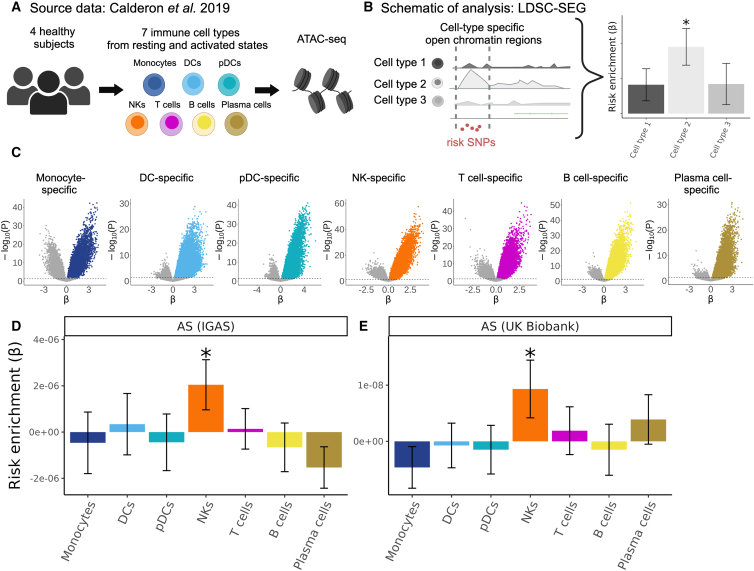
To assess which immune cell types might be mediating the genetic susceptibility to AS, we utilized a dataset of open chromatin profiles of immune cell subsets from peripheral blood of four healthy subjects16 (Figure 1A). Sorted cell subsets were analyzed using ATAC-seq with or without prior in vitro stimulation. For our study, we grouped the cells analyzed by Calderon et al. into seven main immune cell types: T cells, B cells, NK cells, plasmablasts, DCs, plasmacytoid DCs, and monocytes. We identified cell-type-specific open chromatin regions and assessed whether these were significantly enriched in AS genetic risk. We used the LDSC-SEG method13 to quantify enrichment of partitioned heritability in each of these cell-type-specific annotations (conceptual scheme in Figure 1B, data in Figure 1C) compared to baseline and control annotations, while taking into account the effects of LD. We excluded the MHC region from our analyses given the unusually high LD in this region and the fact that genetic associations with this locus are driven mostly by coding variants of the HLA-B gene. Using the Immunochip association study summary statistics from the IGAS Consortium,5 we found that NK cell-specific open chromatin regions were significantly enriched in genetic risk for AS (p = 0.026), while this was not the case for the other six immune cell types (Figure 1D).
Figure 1.
Human NK cell-specific open chromatin regions are enriched in AS genetic risk
(A) Calderon et al. study design. Peripheral blood cells from four healthy subjects were sorted into immune cell populations that we grouped in silico into seven cell types (see material and methods). Assay for transposase accessible chromatin using sequencing (ATAC-seq) was performed with and without prior in vitro activation.
(B) Graphical representation of LDSC-SEG analysis: identification of cell-type-specific annotations (in our case, open chromatin regions), followed by the integration with GWAS summary statistics to obtain a risk enrichment coefficient β and p value.
(C) Volcano plots showing results of differential accessibility analyses for each cell type compared to the other cell types. Colored dots indicate open chromatin peaks in the top decile of the t statistic for each cell type, which were used for LDSC-SEG analysis.
(D and E) Bar graphs displaying the AS genetic risk enrichment coefficient β and block jackknife SE for cell-type-specific open chromatin accounting for control peaks and baseline annotations. Summary statistics from the International Genetics of Ankylosing Spondylitis (IGAS) Consortium (D) and UK Biobank (E) GWASs were used.
∗p < 0.05.
We validated this finding in a GWAS with genome-wide genotyping using the summary statistics for AS from the UK Biobank. With this GWAS, we confirmed that open chromatin regions specific for NK cells were significantly enriched in AS heritability (p = 0.034; Figure 1E). To evaluate the reliability of our results, we included four control traits that have been extensively examined in similar studies integrating GWAS with functional genomics.13,15,16,17 As expected, RA presented the highest enrichment for T cell-specific open chromatin regions (p = 0.0018), Alzheimer disease (MIM: 104300) for myeloid DC (p = 0.00018), and SLE for B cells (p = 0.0015). We selected body height as a negative control trait, anticipating no significant enrichment for immune cells, a prediction that was confirmed by our data (all p > 0.1, Figure S1). The AS heritability enrichment results did not change for any of the two GWAS when cell-type-specific open chromatin regions were identified by controlling for stimulation status (Figure S2). Collectively, these epigenomic analyses suggest that AS risk alleles are preferentially located in regions that may influence gene regulation in NK cells.
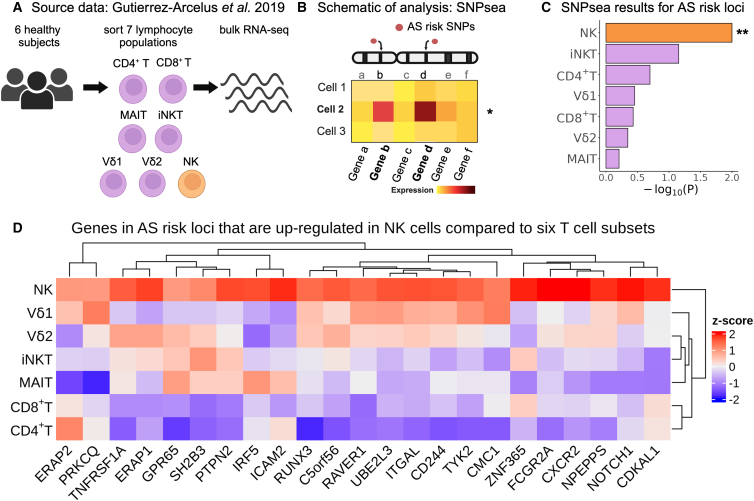
To corroborate these findings using an alternative experimental approach, we used our previously published RNA-seq dataset of sorted peripheral CD4+ T cells, CD8+ T cells, mucosal-associated invariant T cells (MAIT), invariant NK T cells (iNKT), γδ T cells expressing Vδ1 T cell receptor (TCR) chain (Vd1), γδ T cells expressing Vδ2 TCR chain (Vd2), and NK cells (each in duplicate from six healthy donors; Figure 2A).22 We also applied an alternative computational method to validate our findings, SNPsea, which quantifies enrichment of cell-type-specific gene expression in risk loci for a given trait (conceptual scheme in Figure 2B) by employing a non-parametric statistical method to calculate empirical p values through comparison with sets of null SNPs.24 We used SNPsea as an independent method that only uses risk loci that pass the genome-wide significance threshold, as opposed to LDSC-SEG, which uses all the summary statistics for a given GWAS and hence also incorporates information from genetic variants that do not pass the genome-wide significance threshold. For this analysis, we used the AS risk SNPs reported by Brown and Wordsworth in 2017, which were curated from multiple AS genetic studies (Table S1).8 This list includes the lead SNPs from AS risk loci that reached genome-wide significance in the IGAS and/or UK Biobank studies. SNPsea analysis revealed a significant enrichment of NK cell-specific gene expression in AS risk loci (p = 0.01), which was not observed in the other lymphocyte subsets included in the dataset (Figure 2C).
Figure 2.
NK cells show enrichment of cell-type-specific expression of AS-associated genes
(A) Gutierrez-Arcelus et al. study design. Peripheral blood cells from six healthy subjects were sorted into NK cells (orange) and six T cell populations (purple): CD4+ T, CD8+ T, MAIT, iNKT, and two γδ T cell populations. Bulk RNA-seq was performed on two replicates per sample.
(B) Graphical representation of the SNPsea method illustrating the integration of gene expression profiles with risk loci obtained from GWAS.
(C) Bar graphs showing −log10(p value) for enrichment of cell-type-specific expression of genes in AS risk loci using SNPsea. ∗∗p < 0.01.
(D) Heatmap showing expression levels for genes in AS risk loci that were significantly upregulated in NK cells compared to six T cells subsets. Expression levels are scaled by row.
We then performed a differential expression analysis comparing NK cells with the six T cell subsets (Table S2). Genes in AS risk loci with significant upregulation in NK cells are presented in Figure 2D. Two of these genes, RUNX3 (MIM: 600210) and TBX21 (MIM: 604895), encode transcription factors with important roles in lymphocytes. TNFRSF1A (MIM: 191190) encoding tumor necrosis factor (TNF) receptor 1 has a well-established association with AS that has been validated by multiple studies.36,37,38 FCGR2A (MIM: 146790) codes for the low-affinity Fcγ receptor IIA, an activating receptor involved in orchestrating immune response. Less-studied genes included NPEPPS (MIM: 606793), which encodes a puromycin-sensitive aminopeptidase, and LNPEP (MIM: 151300), which encodes a zinc-dependent aminopeptidase. Both genes are paralogs of ERAP1 (MIM: 606832) and belong to the MHC class I antigen processing and presentation pathway, along with other known AS risk genes.39 Collectively, the results of our second integrative analysis indicate that several genes within AS risk loci are highly expressed in NK cells relative to T cells, providing additional support for the emerging hypothesis that AS risk alleles exert their effects, at least in part, via NK cells.
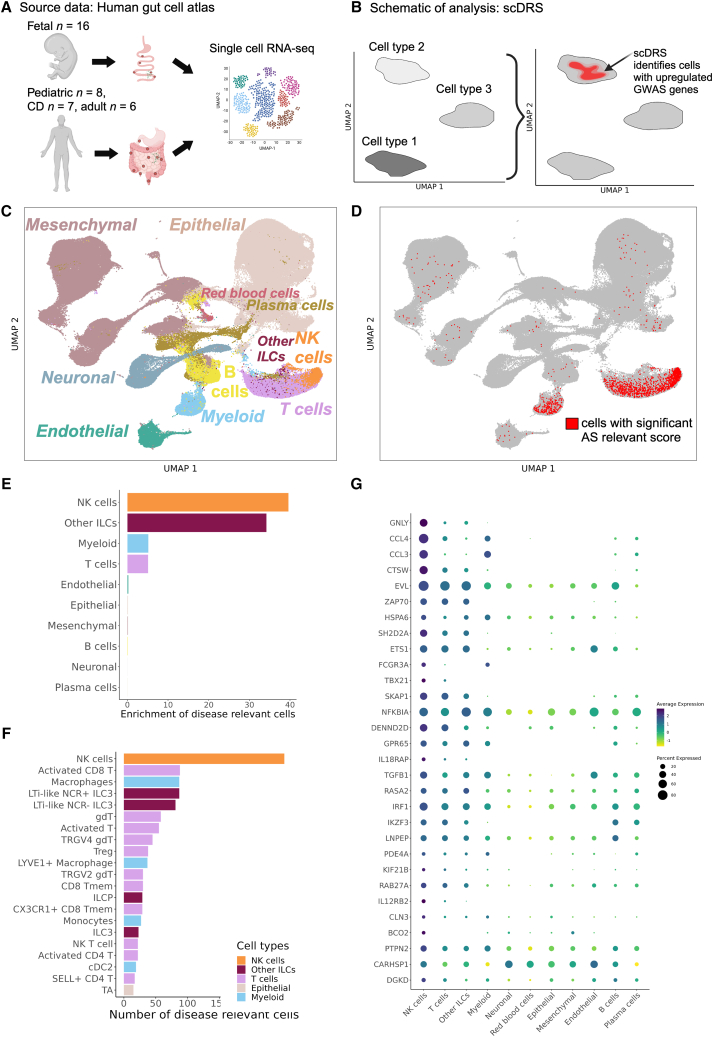
The transcriptomic phenotype of immune cells commonly differs between blood and tissue.40,41 Hence, in addition to analyzing peripheral blood as in the previous analyses, we sought to evaluate disease-relevant cell subsets from a tissue relevant for AS. We used the human Space-Time Gut Cell Atlas,23 which includes scRNA-seq data for samples from various locations of fetal (N = 16), pediatric (N = 8), and adult (N = 13, including 6 healthy subjects and 7 subjects with Crohn disease) intestine (Figure 3A). We applied the scDRS method,14 which identifies cells that overexpress a significant proportion of genes implicated by GWAS, weighted on their strength of association with disease, compared to null sets of control genes in the dataset (conceptual scheme in Figure 3B). This method is similar to LDSC-SEG in that it uses information from both risk loci that pass the genome-wide significance threshold and those that do not. The Space-Time Gut Cell Atlas investigators identified the following broad cell types: mesenchymal, epithelial, endothelial, neuronal, myeloid, red blood cells, B cells, plasmablasts, T cells, NK cells, and other innate lymphoid cells (ILCs) (Figure 3C). scDRS identified 1,852 cells with significantly enriched expression of AS GWAS genes (20% FDR; Figure 3D). Of these, 765 were T cells, 264 were myeloid cells, 320 were NK cells and 319 were other ILCs. Normalized for cell type abundance in the dataset, NK cells showed the highest enrichment (39-fold), followed by other ILCs (34-fold), T cells (5-fold), and myeloid cells (5-fold; Figure 3E). In contrast, non-immune cell types exhibited a depletion of disease-relevant cells relative to their abundance in the entire dataset (Figure S3). We then used the fine-grained annotations of the Space-Time Gut Cell Atlas to identify the particular cell subsets that had significant expression enrichment of AS-associated genes. This revealed NK cells as the most abundant (N = 320), followed by LTi-like natural cytotoxicity receptor-positive (NCR+) ILC3 cells (N = 147), activated CD8+ T cells (N = 132), macrophages (N = 130), LTi-like NCR− ILC3 cells (N = 112), γδ T cells (N = 94), and other T cells, ILCs, and myeloid subsets (Figure 3F). At lower FDR thresholds, few cells had a significant disease-relevant score (457 at 10% FDR and 20 at 5% FDR), but NK cells remained the top-enriched cell type (Figure S4). Genes in AS risk loci with high expression in gut NK cells include GNLY (MIM: 188855), CCL4 (MIM: 182284), and CCL3 (MIM: 182283) (Figure 3G). In addition, by comparing the MAGMA-scored genes used for scDRS analysis with the genes within 250 kb of the risk SNPs used in the SNPsea analysis, we identified eight AS-associated genes that are upregulated in NK cells at 5% FDR in both peripheral blood lymphocytes and immune cell types from the gut (Table S3). These genes are FCGR3A (MIM: 146740), SLAMF7 (MIM: 606625), TBX21, APOBR (MIM: 605220), NOTCH1 (MIM: 190198), RUNX3, IL18R1 (MIM: 604494), and GPR65 (MIM: 604620). Using the control traits specified earlier, we confirmed T cells as the main disease-relevant cell type for RA and monocytes for Alzheimer disease (Figure S2). No significant disease-relevant cells were identified for height (as expected) and for SLE, which could mean that B cells in the gut are in a state not pertinent to SLE or that the dataset lacked sufficient power to detect an association for this disease (Figure S2). In summary, our analyses indicate that tissue-resident NK cells exhibit significant expression of AS-associated genes.
Figure 3.
Human gut single-cell atlas reveals significant upregulation of AS-associated genes in NK cells
(A) Generation of the Space-Time Gut Cell Atlas, with samples from fetal, pediatric, and adult subjects.
(B) Graphical representation of the scDRS method, which integrates GWAS risk genes with single-cell data to identify disease-relevant cells.
(C) Visualization of the Space-Time Gut Cell Atlas data using UMAP on the top 20 principal components from 1,997 variable genes from the scRNA-seq expression matrix.
(D) Same UMAP visualization as in (C). Cells with significant scDRS score (20% FDR) are colored in red.
(E) Bar graph showing enrichment of scDRS significant cells per cell type (cell-type percentage within scDRS significant cells over cell-type percentage in whole dataset).
(F) Bar graph showing the number of significant scDRS cells for each cell type using the fine-grained annotations from the Space-Time Gut Cell Atlas. Cell populations with at least 15 significant scDRS cells are shown.
(G) Scaled average expression levels and percentage of cells expressing a given gene for 50 genes associated with AS that had significant upregulation (5% FDR) in NK cells compared to the other cell types. Genes are sorted by multiplying their MAGMA score (strength of association with AS) by their average level of expression in NK cells.
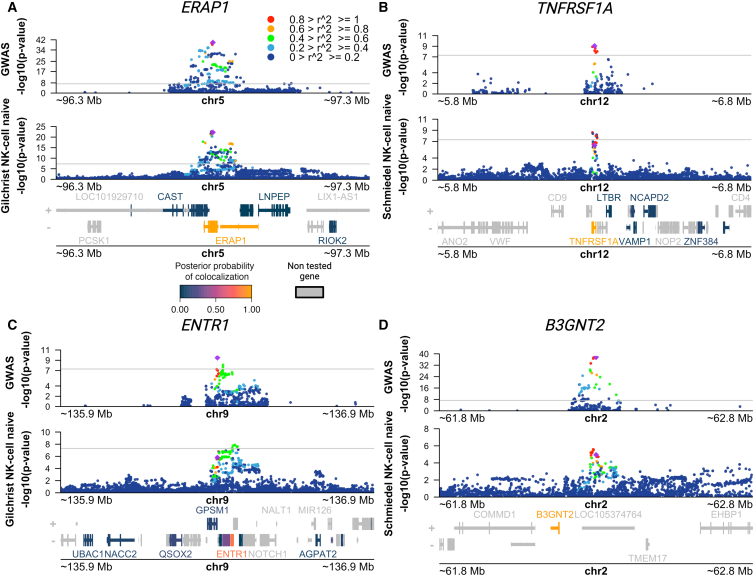
Lastly, we sought to find putative target genes for AS risk variants in NK cells. To this end, we performed co-localization analyses between AS GWAS risk loci and genetic variants associated with gene expression (eQTLs) using coloc.33 We leveraged eQTL summary statistics from the eQTL Catalogue,30 drawing upon data from a study on the transcriptomic profiling of peripheral NK cells from 91 genotyped individuals,31 as well as a microarray QTL study that profiled NK cells from 245 genotyped individuals.32 We found four AS risk loci with genome-wide significance (p < 5 × 10−8) and a high posterior probability (>0.8) of sharing a causal variant with an NK cell eQTL (PP4, Table 1; Figure 4). An additional 10 loci with suggestive AS association p values (3.56 × 10−5 < p < 5.40 × 10−8) showed evidence of co-localization with NK cell eQTLs for 18 genes (PP4 > 0.75; Table 1). Within the genome-wide significant loci we identified the established target genes ERAP1 and TNFRSF1A, as well as the putative target genes ENTR1 (MIM: 618289)(also known as SDCCAG3) and B3GNT2 (MIM: 605581), which have been studied less.
Table 1.
Putative target genes identified by co-localization analysis between AS-associated loci and eQTLs in NK cells
| Lead GWAS variant | p value | GWAS | Putative target gene | Posterior probability of shared causal variant | Quantification method | eQTL study |
|---|---|---|---|---|---|---|
| rs27529 | 1.24E−40 | IGAS | ERAP1 | 0.99186 | microarray | Gilchrist et al. (2021)32 |
| rs6759298 | 2.07E−38 | IGAS | B3GNT2 | 0.97245 | RNA-seq | Schmiedel et al. (2018)31 |
| rs1128905 | 3.17E−10 | IGAS | ENTR1 | 0.8207 | microarray | Gilchrist et al. (2021)32 |
| rs1860545 | 8.66E−10 | IGAS | TNFRSF1A | 0.99673 | RNA-seq | Schmiedel et al. (2018)31 |
| rs11065898 | 5.41E−8 | IGAS | TMEM116 | 0.87563 | microarray | Gilchrist et al. (2021)32 |
| rs9619386 | 4.42E−7 | IGAS | UBE2L3 | 0.96397 | microarray | Gilchrist et al. (2021)32 |
| rs1250542 | 2.07E−6 | IGAS | ZMIZ1 | 0.87272 | microarray | Gilchrist et al. (2021)32 |
| rs1250542 | 2.07E−6 | IGAS | ZMIZ1 | 0.83268 | RNA-seq | Schmiedel et al. (2018)31 |
| rs952594 | 2.08E−6 | IGAS | APEH | 0.75689 | microarray | Gilchrist et al. (2021)32 |
| rs952594 | 2.08E−6 | IGAS | RBM6 | 0.91981 | RNA-seq | Schmiedel et al. (2018)31 |
| rs952594 | 2.08E−6 | IGAS | UBA7 | 0.94237 | microarray | Gilchrist et al. (2021)32 |
| rs952594 | 2.08E−6 | IGAS | UBA7 | 0.8851 | RNA-seq | Schmiedel et al. (2018)31 |
| rs6565217 | 2.82E−6 | IGAS | AC135050.3 | 0.91556 | RNA-seq | Schmiedel et al. (2018)31 |
| rs6565217 | 2.82E−6 | IGAS | STX4 | 0.94653 | RNA-seq | Schmiedel et al. (2018)31 |
| rs7191548 | 3.13E−6 | IGAS | EIF3CL | 0.7822 | microarray | Gilchrist et al. (2021)32 |
| rs7191548 | 3.13E−6 | IGAS | NPIPB8 | 0.97545 | microarray | Gilchrist et al. (2021)32 |
| rs7191548 | 3.13E−6 | IGAS | SGF29 | 0.85225 | microarray | Gilchrist et al. (2021)32 |
| rs7191548 | 3.13E−6 | IGAS | TUFM | 0.82246 | microarray | Gilchrist et al. (2021)32 |
| rs7191548 | 3.13E−6 | IGAS | TUFM | 0.95694 | RNA-seq | Schmiedel et al. (2018)31 |
| rs6583441 | 3.84E−6 | IGAS | IKZF1 | 0.96244 | RNA-seq | Schmiedel et al. (2018)31 |
| rs4690326 | 6.49E−6 | IGAS | DGKQ | 0.91883 | microarray | Gilchrist et al. (2021)32 |
| rs4690326 | 6.49E−6 | IGAS | DGKQ | 0.89989 | RNA-seq | Schmiedel et al. (2018)31 |
| rs4690326 | 6.49E−6 | IGAS | IDUA | 0.98988 | microarray | Gilchrist et al. (2021)32 |
| rs4690326 | 6.49E−6 | IGAS | SLC49A3 | 0.801 | RNA-seq | Schmiedel et al. (2018)31 |
| rs26481 | 9.17E−6 | UK Biobank | CAST | 0.95638 | microarray | Gilchrist et al. (2021)32 |
| rs26481 | 9.17E−6 | UK Biobank | ERAP1 | 0.95565 | microarray | Gilchrist et al. (2021)32 |
| rs2236167 | 3.57E−5 | IGAS | PPP2R3C | 0.9491 | microarray | Gilchrist et al. (2021)32 |
Figure 4.
Co-localization of AS risk loci and NK cell eQTLs points to putative target genes for AS risk variants
(A–D) Manhattan plots showing AS GWAS and NK cell eQTL −log10(p values) for SNPs within 500 kb of a lead GWAS SNP. The color of each SNP indicates its level of linkage disequilibrium (LD) between with the lead GWAS SNP (purple diamond). Genes in the region are colored according to their posterior probability of hypothesis four (PP4) (i.e., that the same causal variant is shared between AS and the eQTL for that gene).
(A) Manhattan plots identifying putative target gene ERAP1 using AS IGAS GWAS (top) and NK microarray gene eQTL data obtained from Gilchrist et al.32 (bottom).
(B) Manhattan plots identifying putative target gene TNFRSF1A using AS IGAS GWAS (top) and NK gene eQTL data obtained from Schmiedel et al.31 (bottom).
(C) Manhattan plots identifying putative target gene ENTR1 using AS IGAS GWAS (top) and NK microarray gene eQTL data obtained from Gilchrist et al.32 (bottom).
(D) Manhattan plots identifying putative target gene B3GNT2 using AS IGAS GWAS (top) and NK gene expression QTL (eQTL) data obtained from Schmiedel et al.31 (bottom). All QTL summary statistics taken from eQTL Catalog.
Discussion
In this study, we integrated epigenomic and transcriptomic datasets with AS genetic risk data to find candidate cellular drivers of AS pathogenesis. Our unbiased approach, applying three different methods to datasets from both peripheral blood and tissue, consistently identified NK cells as the dominant disease-relevant cell type. Specifically, we found that NK-specific open chromatin regions and NK-specific gene expression were significantly enriched for non-MHC AS genetic risk. This suggests that a significant portion of AS risk variants affects gene regulation in NK cells, pointing to NK cells as potential key mediators of AS pathogenesis.
NK cells have the ability to directly destroy target cells through cell lysis and in addition play a significant role in shaping immune responses by releasing cytokines. Previous studies support a role for NK cells in AS. Persons with AS with chronic subclinical intestinal inflammation were found to have an increased abundance of NKp44+ NK cells in their gut, and these cells were the major producers of interleukin-22 (IL-22) in the lamina propria, suggesting a possible role in tissue protection.42 One could speculate that dysfunctional NK cells “drive” AS development by contributing to intestinal inflammation, which is in line with the gut-joint axis hypothesis.43 Alternatively, NK cells may play a critical role through activities in spinal tissues. Cuthbert et al. studied entheseal immunology using discarded surgical specimens from persons with back pain (not axSpA) undergoing laminectomy and reported that NK cells are present in both entheseal soft tissue and peri-entheseal bone.44 We are not aware of any data assessing the presence of NK cells at spinal enthesis in subjects with AS or in the subchondral bone marrow in subjects with sacroiliitis.
HLA-B27 can bind to the killer cell immunoglobulin-like receptor (KIR) KIR3DL1 and affect the function of NK cells, including their ability to lyse cells.45,46 HLA-B27 homodimers can also bind KIR3DL2.47 Chan and colleagues showed an expansion of KIR3DL2+ NK and CD4+ T cells in persons with AS.48 Subsequent studies by the same group focused on CD4+ T cells demonstrating that KIR3DL2+ CD4+ T cells were major IL-17A producers.49 However, an expansion of KIR3DL2+ NK and T cells has not been observed in other axSpA cohorts.50,51 Multiple risk loci for AS include genes relevant for NK cell function, including KIR3DL1 (MIM: 604946), KIR2DS5 (MIM: 604956), KIR3DS1 (MIM: 620778), and KIR2DL5 (MIM: 605305).52,53 In another study, investigators co-cultured endoplasmic reticulum aminopeptidase 1 (ERAP1)-inhibited M1 macrophages with NK cells from persons with AS and found that the ERAP1 protective alleles in individuals led to decreased CD69 and CD107a on NK cells and a lower number of interferon-γ+ NK cells as compared to persons carrying non-protective alleles.54
Our findings do not rule out the involvement of other cell types in AS pathogenesis. Indeed, in the human Space-Time Gut Cell Atlas, we identified significant expression of AS-associated genes in T cell subsets and ILC subsets (Figures 3D–3F), which share transcriptional programs with NK cells.22,55 Indeed, it is likely that genetic risk to AS is mediated through multiple cell types, as is the case for other complex diseases such as multiple sclerosis, for which studies have found risk enrichment in open/active chromatin regions specific to both T and B cells.17 We and others have shown that eQTLs often exhibit impact in multiple cell types.56,57 Hence, determining the specific cell type through which a disease risk variant is exerting its pathogenic effects can be challenging.
Our co-localization analyses using two eQTL NK cell datasets identified four putative target genes for AS risk variants: ERAP1, TNFRSF1A, ENTR1 (also known as SDCCAG3) and B3GNT2. The importance of ERAP1 in AS risk is well established, and polymorphisms affecting its expression have been reported for multiple cell types, including macrophages, monocytes, T cells, induced pluripotent stem cells, fibroblasts, and immortalized B cells.30 Similarly, multiple studies have found significant associations between non-coding polymorphisms at or near TNFRSF1A and AS, including in European and east Asian populations.36,37 While there are multiple genes in this genomic locus, including PLEKHG6 (MIM: 611743), SCNN1A (MIM: 600228), and LTBR (MIM: 600979), our co-localization results suggest that TNFRSF1A, which encodes TNFR1, is the target gene of the causal variant in this locus, and its dysregulation can occur in NK cells. This is consistent with the therapeutic efficacy of TNF inhibitors in AS and the known function of TNF as a booster of the cytolytic capacity of NK cells.58 Interestingly, TNFRSF1A has been functionally linked to ENTR1, a less extensively studied putative target gene identified in this study. ENTR1, which encodes an endosome-associated trafficking regulator, is needed for TNFR expression on the cell surface.59 Lastly, B3GNT2 encodes an acetylglucosaminyltransferase enzyme that is a type II transmembrane protein. A recent study in a Taiwanese cohort demonstrated that a non-coding genetic variant near B3GNT2 is associated with AS susceptibility, and that B3GNT2 blood mRNA levels were negatively correlated with C-reactive protein, erythrocyte sedimentation rate, syndesmophyte formation, and the Bath AS functional index.60
While the four putative target genes identified here make sense in the context of AS and potential impact on NK cell function, they are not as numerous as we would have expected given the 32 genome-wide significant risk loci included in the co-localization analyses. However, similar challenges have been encountered with non-coding risk variants in other complex diseases, where only 20%–47% of risk variants co-localized with eQTLs.61,62 Our research, along with that of others, suggests that many regulatory effects might remain undetected due to their presence in cell states of activation or differentiation that have not been thoroughly explored.63,64,65 Moreover, the sample size of typical eQTL studies is likely insufficient to find the regulatory effects of most risk variants identified by GWAS.66 Hence, we believe that better-powered eQTL studies ascertaining multiple activation states in NK cells are needed to find additional target genes for AS risk variants.
Our study has several limitations. Due to a lack of published data we have an incomplete assessment of the spectrum of potentially relevant immune cell subsets and states, particularly those present in inflamed sacroiliac joints and spine. Consequently, if the real driver for AS pathogenesis is a cell subset or state that was not present in the analyzed datasets but has transcriptomic and epigenomic similarities to NK cells, then our results may suffer from a “guilt-by-association” bias. To our knowledge, current transcriptomic datasets profiling multiple immune cell types from persons with AS are limited to peripheral blood. When we applied scDRS to a recently published scRNA-seq dataset of 98,884 peripheral blood mononuclear cells from 10 subjects with AS and 29 healthy controls,67 we found no significant cells for the disease-relevant gene expression score (data not shown), possibly due to lack of power in the study for this type of analysis. Another limitation is that the reported nominal p values in the LDSC-SEG and SNPsea analyses would not have been significant if we had applied FDR correction, accounting for the seven tests performed per analysis. However, NK cells consistently showed marginal significance (defined as nominal p < 0.05) for heritability enrichment in cell-type-specific open chromatin regions across two independent GWAS and in the cell-type-specific expression analysis where NK cells were compared with T cell subsets using a curated list of genome-wide significant loci for AS. Additionally, the results from the Gut Cell Atlas confirmed NK cells as the top enriched expressors of AS-associated genes.
Our study encompassed a broad spectrum of immune cell states within the gastrointestinal tract and peripheral blood of healthy subjects and consistently pointed to NK cells. Since GWASs pinpoint genetic regions implicated in the onset of disease, including early stages when future patients are still asymptomatic, the study of samples from healthy subjects is relevant, despite the possibility that not all cell states are represented. Future investigations, particularly larger-scale studies of samples from blood and inflamed tissue from persons with AS including untreated subjects in the early phases of the disease, will be key to establish whether NK cells are indeed drivers of AS pathogenesis.
Data and code availability
All data and methods are publicly available as specified above. The code generated during this study is available at GitHub https://github.com/gutierrez-arcelus-lab/AS_RelevantCellTypes.
Acknowledgments
This study was supported by a seed grant from the Spondyloarthritis Research and Treatment Network (SPARTAN) and a microgrant from the Joint Biology Consortium (1P30AR070253-01). P.A.N. was supported by P30AR070253 and R01AR073201. J.E. was supported by NIH grant R21 AR076040-01 and an ASPIRE grant from Pfizer. M.G.-A. was supported by P30AR070253, the Arthritis National Research Foundation, the Lupus Research Alliance, and the Gilead Sciences Rheumatology Research Scholars Award. This work was supported by the Cell Discovery Network, a collaborative initiative funded by the Manton Foundation and the Warren Alpert Foundation at Boston Children’s Hospital. We thank Soumya Raychaudhuri and Kamil Slowikowski for guidance on implementing SNPsea, and Steven Gazal for guidance on implementing LDSC-SEG. We thank the Gutierrez-Arcelus and Nigrovic laboratories for feedback on this study. The scientific illustrations were created in BioRender.
Author contributions
Conceptualization, M.C., J.E., and M.G-A.; Methodology, M.C., D.F-S., V.R.C.A., V.E.N-C., and M.G-A.; Software, M.C., D.F-S., V.R.C.A., and V.E.N-C.; Formal Analysis, M.C., D.F-S., V.R.C.A., and V.E.N-C.; Resources, M.L., J.E., and M.G-A.; Writing - Original Draft, M.C., J.E., and M.G-A.; Writing - Review & Editing, M.C., V.R.C.A., P.A.N., J.E., and M.G-A.; Visualization, M.C. and M.G-A. Supervision, V.R.C.A., P.A.N., J.E., and M.G-A.; Funding Acquisition, P.A.N., J.E., and M.G-A.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100375.
Contributor Information
Joerg Ermann, Email: jermann@bwh.harvard.edu.
Maria Gutierrez-Arcelus, Email: mgutierr@broadinstitute.org.
Web resources
Alkes Price Summary Statistics: https://alkesgroup.broadinstitute.org/
eQTL Catalog: https://www.ebi.ac.uk/eqtl/
MAGMA: https://cncr.nl/research/magma/
Space-Time Gut Cell Atlas: https://www.gutcellatlas.org/
Online Mendelian Inheritance in Man: https://www.omim.org/
UK Biobank Summary Statistics: https://pan.ukbb.broadinstitute.org/downloads
Supplemental information
References
- 1.Navarro-Compán V., Sepriano A., El-Zorkany B., van der Heijde D. Axial spondyloarthritis. Ann. Rheum. Dis. 2021;80:1511–1521. doi: 10.1136/annrheumdis-2021-221035. [DOI] [PubMed] [Google Scholar]
- 2.Díaz-Peña R., Castro-Santos P., Durán J., Santiago C., Lucia A. The Genetics of Spondyloarthritis. J. Pers. Med. 2020;10:151. doi: 10.3390/jpm10040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown M.A., Kennedy L.G., MacGregor A.J., Darke C., Duncan E., Shatford J.L., Taylor A., Calin A., Wordsworth P. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 1997;40:1823–1828. doi: 10.1002/art.1780401015. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen O.B., Svendsen A.J., Ejstrup L., Skytthe A., Harris J.R., Junker P. Ankylosing spondylitis in Danish and Norwegian twins: occurrence and the relative importance of genetic vs. environmental effectors in disease causation. Scand. J. Rheumatol. 2008;37:120–126. doi: 10.1080/03009740701824613. [DOI] [PubMed] [Google Scholar]
- 5.International Genetics of Ankylosing Spondylitis Consortium IGAS. Cortes A., Hadler J., Pointon J.P., Robinson P.C., Karaderi T., Leo P., Cremin K., Pryce K., Harris J., et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hübenthal M., et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchhouse C. Heritability of >2,000 traits and disorders in the UK Biobank. Neale lab. 2017 http://www.nealelab.is/blog/2017/9/15/heritability-of-2000-traits-and-disorders-in-the-uk-biobank [Google Scholar]
- 8.Brown M.A., Wordsworth B.P. Genetics in ankylosing spondylitis - Current state of the art and translation into clinical outcomes. Best Pract. Res. Clin. Rheumatol. 2017;31:763–776. doi: 10.1016/j.berh.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Reveille J.D., Zhou X., Lee M., Weisman M.H., Yi L., Gensler L.S., Zou H., Ward M.M., Ishimori M.L., Learch T.J., et al. HLA class I and II alleles in susceptibility to ankylosing spondylitis. Ann. Rheum. Dis. 2019;78:66–73. doi: 10.1136/annrheumdis-2018-213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Haynes K., Pennisi D.J., Anderson L.K., Song X., Thomas G.P., Kenna T., Leo P., Brown M.A. Epigenetic and gene expression analysis of ankylosing spondylitis-associated loci implicate immune cells and the gut in the disease pathogenesis. Genes Immun. 2017;18:135–143. doi: 10.1038/gene.2017.11. [DOI] [PubMed] [Google Scholar]
- 11.Yazar S., Alquicira-Hernandez J., Wing K., Senabouth A., Gordon M.G., Andersen S., Lu Q., Rowson A., Taylor T.R.P., Clarke L., et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science. 2022;376:eabf3041. doi: 10.1126/science.abf3041. [DOI] [PubMed] [Google Scholar]
- 12.Brown A.C., Cohen C.J., Mielczarek O., Migliorini G., Costantino F., Allcock A., Davidson C., Elliott K.S., Fang H., Lledó Lara A., et al. Comprehensive epigenomic profiling reveals the extent of disease-specific chromatin states and informs target discovery in ankylosing spondylitis. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2023.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M.J., Hou K., Dey K.K., Sakaue S., Jagadeesh K.A., Weinand K., Taychameekiatchai A., Rao P., Pisco A.O., Zou J., et al. Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 2022;54:1572–1580. doi: 10.1038/s41588-022-01167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calderon D., Nguyen M.L.T., Mezger A., Kathiria A., Müller F., Nguyen V., Lescano N., Wu B., Trombetta J., Ribado J.V., et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 2019;51:1494–1505. doi: 10.1038/s41588-019-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soskic B., Cano-Gamez E., Smyth D.J., Rowan W.C., Nakic N., Esparza-Gordillo J., Bossini-Castillo L., Tough D.F., Larminie C.G.C., Bronson P.G., et al. Chromatin activity at GWAS loci identifies T cell states driving complex immune diseases. Nat. Genet. 2019;51:1486–1493. doi: 10.1038/s41588-019-0493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee S., Webber C., Poole A.R. The induction of arthritis in mice by the cartilage proteoglycan aggrecan: roles of CD4+ and CD8+ T cells. Cell. Immunol. 1992;144:347–357. doi: 10.1016/0008-8749(92)90250-s. [DOI] [PubMed] [Google Scholar]
- 19.Rao D.A., Gurish M.F., Marshall J.L., Slowikowski K., Fonseka C.Y., Liu Y., Donlin L.T., Henderson L.A., Wei K., Mizoguchi F., et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542:110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caielli S., Wan Z., Pascual V. Systemic Lupus Erythematosus Pathogenesis: Interferon and Beyond. Annu. Rev. Immunol. 2023;41:533–560. doi: 10.1146/annurev-immunol-101921-042422. [DOI] [PubMed] [Google Scholar]
- 21.Pan-UKB team Pan-UK Biobank. 2020. https://pan.ukbb.broadinstitute.org
- 22.Gutierrez-Arcelus M., Teslovich N., Mola A.R., Polidoro R.B., Nathan A., Kim H., Hannes S., Slowikowski K., Watts G.F.M., Korsunsky I., et al. Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat. Commun. 2019;10:687. doi: 10.1038/s41467-019-08604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elmentaite R., Kumasaka N., Roberts K., Fleming A., Dann E., King H.W., Kleshchevnikov V., Dabrowska M., Pritchard S., Bolt L., et al. Cells of the human intestinal tract mapped across space and time. Nature. 2021;597:250–255. doi: 10.1038/s41586-021-03852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slowikowski K., Hu X., Raychaudhuri S. SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics. 2014;30:2496–2497. doi: 10.1093/bioinformatics/btu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 26.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerimov N., Hayhurst J.D., Peikova K., Manning J.R., Walter P., Kolberg L., Samoviča M., Sakthivel M.P., Kuzmin I., Trevanion S.J., et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 2021;53:1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175:1701–1715.e16. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilchrist J.J., Makino S., Naranbhai V., Sharma P.K., Koturan S., Tong O., Taylor C.A., Watson R.A., de Los Aires A.V., Cooper R., et al. Natural Killer cells demonstrate distinct eQTL and transcriptome-wide disease associations, highlighting their role in autoimmunity. Nat. Commun. 2022;13:4073. doi: 10.1038/s41467-022-31626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer N.E., Davis E.S., Wenger C.D., Deoudes E.M., Parker S.M., Love M.I., Phanstiel D.H. Plotgardener: cultivating precise multi-panel figures in R. Bioinformatics. 2022;38:2042–2045. doi: 10.1093/bioinformatics/btac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B., Gloudemans M.J., Rao A.S., Ingelsson E., Montgomery S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson S.I., Liu Y., Danoy P.A., Wu X., Thomas G.P., Jiang L., Sun L., Wang N., Han J., Han H., et al. Association of STAT3 and TNFRSF1A with ankylosing spondylitis in Han Chinese. Ann. Rheum. Dis. 2011;70:289–292. doi: 10.1136/ard.2010.133322. [DOI] [PubMed] [Google Scholar]
- 37.Karaderi T., Pointon J.J., Wordsworth T.W.H., Harvey D., Appleton L.H., Cohen C.J., Farrar C., Harin A., Brown M.A., Wordsworth B.P., Australo-Anglo-American Spondyloarthritis Consortium Evidence of genetic association between TNFRSF1A encoding the p55 tumour necrosis factor receptor, and ankylosing spondylitis in UK Caucasians. Clin. Exp. Rheumatol. 2012;30:110–113. [PubMed] [Google Scholar]
- 38.Sode J., Bank S., Vogel U., Andersen P.S., Sørensen S.B., Bojesen A.B., Andersen M.R., Brandslund I., Dessau R.B., Hoffmann H.J., et al. Genetically determined high activities of the TNF-alpha, IL23/IL17, and NFkB pathways were associated with increased risk of ankylosing spondylitis. BMC Med. Genet. 2018;19:165. doi: 10.1186/s12881-018-0680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitulano C., Tedeschi V., Paladini F., Sorrentino R., Fiorillo M.T. The interplay between HLA-B27 and ERAP1/ERAP2 aminopeptidases: from anti-viral protection to spondyloarthritis. Clin. Exp. Immunol. 2017;190:281–290. doi: 10.1111/cei.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meininger I., Carrasco A., Rao A., Soini T., Kokkinou E., Mjösberg J. Tissue-Specific Features of Innate Lymphoid Cells. Trends Immunol. 2020;41:902–917. doi: 10.1016/j.it.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Domínguez Conde C., Xu C., Jarvis L.B., Rainbow D.B., Wells S.B., Gomes T., Howlett S.K., Suchanek O., Polanski K., King H.W., et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science. 2022;376 doi: 10.1126/science.abl5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciccia F., Accardo-Palumbo A., Alessandro R., Rizzo A., Principe S., Peralta S., Raiata F., Giardina A., De Leo G., Triolo G. Interleukin-22 and interleukin-22-producing NKp44+ natural killer cells in subclinical gut inflammation in ankylosing spondylitis. Arthritis Rheum. 2012;64:1869–1878. doi: 10.1002/art.34355. [DOI] [PubMed] [Google Scholar]
- 43.Gracey E., Vereecke L., McGovern D., Fröhling M., Schett G., Danese S., De Vos M., Van den Bosch F., Elewaut D. Revisiting the gut–joint axis: links between gut inflammation and spondyloarthritis. Nat. Rev. Rheumatol. 2020;16:415–433. doi: 10.1038/s41584-020-0454-9. [DOI] [PubMed] [Google Scholar]
- 44.Cuthbert R.J., Fragkakis E.M., Dunsmuir R., Li Z., Coles M., Marzo-Ortega H., Giannoudis P.V., Jones E., El-Sherbiny Y.M., McGonagle D. Brief Report: Group 3 Innate Lymphoid Cells in Human Enthesis. Arthritis Rheumatol. 2017;69:1816–1822. doi: 10.1002/art.40150. [DOI] [PubMed] [Google Scholar]
- 45.Peruzzi M., Wagtmann N., Long E.O. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B∗2705. J. Exp. Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart-Jones G.B.E., di Gleria K., Kollnberger S., McMichael A.J., Jones E.Y., Bowness P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B∗2705. Eur. J. Immunol. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 47.Kollnberger S., Chan A., Sun M.-Y., Chen L.Y., Wright C., di Gleria K., McMichael A., Bowness P. Interaction of HLA-B27 homodimers with KIR3DL1 and KIR3DL2, unlike HLA-B27 heterotrimers, is independent of the sequence of bound peptide. Eur. J. Immunol. 2007;37:1313–1322. doi: 10.1002/eji.200635997. [DOI] [PubMed] [Google Scholar]
- 48.Chan A.T., Kollnberger S.D., Wedderburn L.R., Bowness P. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum. 2005;52:3586–3595. doi: 10.1002/art.21395. [DOI] [PubMed] [Google Scholar]
- 49.Ridley A., Hatano H., Wong-Baeza I., Shaw J., Matthews K.K., Al-Mossawi H., Ladell K., Price D.A., Bowness P., Kollnberger S. Activation-Induced Killer Cell Immunoglobulin-like Receptor 3DL2 Binding to HLA-B27 Licenses Pathogenic T Cell Differentiation in Spondyloarthritis. Arthritis Rheumatol. 2016;68:901–914. doi: 10.1002/art.39515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jansen D.T.S.L., Hameetman M., van Bergen J., Huizinga T.W.J., van der Heijde D., Toes R.E.M., van Gaalen F.A. IL-17-producing CD4+ T cells are increased in early, active axial spondyloarthritis including patients without imaging abnormalities. Rheumatology. 2015;54:728–735. doi: 10.1093/rheumatology/keu382. [DOI] [PubMed] [Google Scholar]
- 51.Larid G., Trijau S., Barral C., Lafforgue P., Pham T. Absence of overexpression of KIR3DL2 on CD4+ T cells and NK cells in patients with axial spondyloarthritis. Rheumatology. 2023;62:e114–e116. doi: 10.1093/rheumatology/keac546. [DOI] [PubMed] [Google Scholar]
- 52.Díaz-Peña R., Blanco-Gelaz M.A., Suárez-Alvarez B., Martínez-Borra J., López-Vázquez A., Alonso-Arias R., Brüges-Armas J., Vidal-Castiñeira J.R., López-Larrea C. Activating KIR genes are associated with ankylosing spondylitis in Asian populations. Hum. Immunol. 2008;69:437–442. doi: 10.1016/j.humimm.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Larrea C., Blanco-Gelaz M.A., Torre-Alonso J.C., Bruges Armas J., Suarez-Alvarez B., Pruneda L., Couto A.R., Gonzalez S., Lopez-Vázquez A., Martinez-Borra J. Contribution of KIR3DL1/3DS1 to ankylosing spondylitis in human leukocyte antigen-B27 Caucasian populations. Arthritis Res. Ther. 2006;8:R101. doi: 10.1186/ar1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babaie F., Mohammadi H., Salimi S., Ghanavatinegad A., Abbasifard M., Yousefi M., Hajaliloo M., Khalili Y., Zamanlou S., Safari R., et al. Inhibition of ERAP1 represses HLA-B27 free heavy chains expression on polarized macrophages and interrupts NK cells activation and function from ankylosing spondylitis. Clin. Immunol. 2023;248 doi: 10.1016/j.clim.2023.109268. [DOI] [PubMed] [Google Scholar]
- 55.Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., Colonna M., Immunological Genome Consortium Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez-Arcelus M., Ongen H., Lappalainen T., Montgomery S.B., Buil A., Yurovsky A., Bryois J., Padioleau I., Romano L., Planchon A., et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S., et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R., Jaw J.J., Stutzman N.C., Zou Z., Sun P.D. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 2012;91:299–309. doi: 10.1189/jlb.0611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neznanov N., Neznanova L., Angres B., Gudkov A.V. Serologically defined colon cancer antigen 3 is necessary for the presentation of TNF receptor 1 on cell surface. DNA Cell Biol. 2005;24:777–785. doi: 10.1089/dna.2005.24.777. [DOI] [PubMed] [Google Scholar]
- 60.Wang C.-M., Jan Wu Y.-J., Lin J.-C., Huang L.-Y., Wu J., Chen J.-Y. Genetic effects of B3GNT2 on ankylosing spondylitis susceptibility and clinical manifestations in Taiwanese. J. Formos. Med. Assoc. 2022;121:1283–1294. doi: 10.1016/j.jfma.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 61.Chun S., Casparino A., Patsopoulos N.A., Croteau-Chonka D.C., Raby B.A., De Jager P.L., Sunyaev S.R., Cotsapas C. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mu Z., Wei W., Fair B., Miao J., Zhu P., Li Y.I. The impact of cell type and context-dependent regulatory variants on human immune traits. Genome Biol. 2021;22:122. doi: 10.1186/s13059-021-02334-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutierrez-Arcelus M., Baglaenko Y., Arora J., Hannes S., Luo Y., Amariuta T., Teslovich N., Rao D.A., Ermann J., Jonsson A.H., et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet. 2020;52:247–253. doi: 10.1038/s41588-020-0579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umans B.D., Battle A., Gilad Y. Where Are the Disease-Associated eQTLs? Trends Genet. 2021;37:109–124. doi: 10.1016/j.tig.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Connally N.J., Nazeen S., Lee D., Shi H., Stamatoyannopoulos J., Chun S., Cotsapas C., Cassa C.A., Sunyaev S.R. The missing link between genetic association and regulatory function. Elife. 2022;11 doi: 10.7554/eLife.74970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mostafavi H., Spence J.P., Naqvi S., Pritchard J.K. Systematic differences in discovery of genetic effects on gene expression and complex traits. Nat. Genet. 2023;55:1866–1875. doi: 10.1038/s41588-023-01529-1. [DOI] [PubMed] [Google Scholar]
- 67.Alber S., Kumar S., Liu J., Huang Z.-M., Paez D., Hong J., Chang H.-W., Bhutani T., Gensler L.S., Liao W. Single Cell Transcriptome and Surface Epitope Analysis of Ankylosing Spondylitis Facilitates Disease Classification by Machine Learning. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.838636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and methods are publicly available as specified above. The code generated during this study is available at GitHub https://github.com/gutierrez-arcelus-lab/AS_RelevantCellTypes.