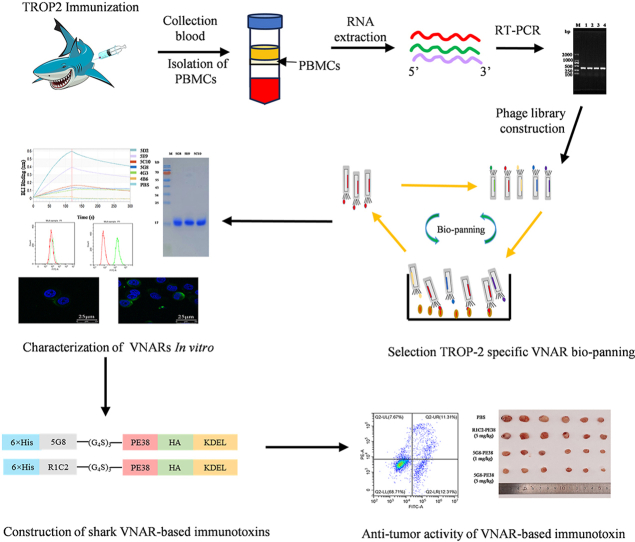
Abstract
TROP-2, a tumor-associated antigen, has been implicated in the progression of various epithelial tumors. Due to its favorable expression profile, TROP-2 has emerged as a promising target for antibody–drug conjugates (ADCs) based anti-tumor therapies. Although ADCs have shown efficacy in cancer treatment, their application in solid tumors is hindered by their high molecular weight, poor tumor penetration, and release of cytotoxic molecules. Therefore, a recombinant immunotoxin was developed based on a shark-derived variable domain of immunoglobulin new antigen receptor (VNAR) antibody. VNARs are only one-tenth the size of IgG antibodies and possess remarkable tissue penetration capabilities and high stability. In this study, a shark VNAR phage display library was created, leading to the identification of shark VNAR-5G8 that targets TROP-2. VNAR-5G8 exhibited a high affinity and cellular internalization ability towards cells expressing high levels of TROP-2. Epitope analysis revealed that VNAR-5G8 recognizes a hidden epitope consisting of CRD and TY-1 on TROP-2. Subsequently, VNAR-5G8 was fused with a truncated form of Pseudomonas exotoxin (PE38) to create the recombinant immunotoxin (5G8-PE38), which exhibited significant anti-tumor activity in vitro and in vivo. Overall, this study highlights the promise of 5G8-PE38 as a valuable candidate for cancer therapy.
Key words: Shark VNAR, Immunotoxins, TROP-2, Epitope identification, Anti-tumor, Pseudomonas exotoxin A, Nanobody, Breast cancer
Graphical abstract
A TROP2-binding nanobody (5G8) was identified from a VNAR phage display library. And an immunotoxin (5G8-PE38) was constructed, which showed effective anti-tumor activity.
1. Introduction
Human trophoblast cell surface antigen 2 (TROP-2) is a trans-membrane glycoprotein1 that exhibits higher expression levels in various tumor cells2,3. It regulates multiple signaling pathways and mediates tumor migration4. Over-expression of TROP-2 is associated with a poor prognosis of various malignancies, including breast cancer and non-small cell lung cancer (NSCLC)5,6. Therefore, TROP-2 represents a promising target for cancer treatment.
Antibody–drug conjugate (ADC)7 drugs targeting TROP-2 have been used for cancer therapy8. Sacituzumab Govitecan (SG) is the first ADC drug against TROP-2 approved for treating advanced triple-negative breast cancer9. Several clinical trials are ongoing to assess its efficacy in other cancer types and combination therapies10. In addition, similar approaches, such as Datopotamab deruxtecan and SKB264, are currently under clinical evaluation in patients with TROP-2 positive cancers11,12. However, these ADCs are based on IgG antibodies that have high molecular weights and poor tumor penetration. The release of toxins could lead to off-target toxicity and drug resistance.
To address the limitations of traditional ADC drugs, recombinant immunotoxins (RITs) have been increasingly attracting attention. RITs, similar to ADCs, are chimeric proteins consisting of a tumor-specific antibody variable region and a recombinant toxin protein13. Unlike traditional ADCs, the toxin portion of RITs is a protein toxin, such as Pseudomonas exotoxin A (PE) and its derivatives14. To reduce immunogenicity while maintaining the toxicity of PE, a smaller toxicity fragment known as PE38 has been created15,16 and utilized as a toxic ligand to develop various immunotoxins, such as Lumoxiti and LMB-10017,18. RITs are considered effective in reducing off-target effects and drug resistance. However, the antibody portion of recombinant immunotoxins primarily comprises single-chain variable fragments (ScFV) sourced from IgG antibodies, which exhibit lower stability and limited affinity19. Moreover, ScFv domains possess a hydrophobic interface and demonstrate low water solubility, resulting in a tendency for aggregation that compromises the efficacy of immunotoxins. Hence, there is an urgent need for innovative antigen-binding domains to drive the progress of immunotoxin development.
Nanobodies (Nbs) are variable domains of heavy chain antibodies sourced from camelids and sharks. Their compact size and extended complementary determining region (CDR) enable them to identify antigenic epitopes that traditional antibodies cannot access20. Due to these characteristics, nanobodies demonstrate significant promise for both diagnostic and therapeutic purposes21. Although the distinctive features of shark nanobodies (VNARs) are more pronounced, shark VNARs have not received as much attention as camel-derived nanobodies (VHHs). Shark VNAR possesses a unique V-region structure with a lower relative molecular weight than VHH, only 12 kDa22, exhibiting significant solid tumor tissue penetration ability23,24. Notably, the CDR3 loop of VNAR is longer, enabling them to recognize hidden antigenic epitopes25. The distinct internal environment of sharks, characterized by high urea content in their blood, results in exceptional chemical and thermal stability for VNARs26. Moreover, shark VNARs could be efficiently expressed in a wide range of host cells at low production cost27. Additionally, some small-sized sharks used as model animals for VNAR development, such as Chiloscyllium plagiosum, are abundant in resources and cost-effectiveness compared to alpacas. Thus, further exploration of shark VNARs can substantially enhance the expansion of bio-pharmaceutical resources and provide novel strategies for disease treatment.
In this study, several VNARs targeting the extracellular structural domain (ECD) of TROP-2 were obtained by screening a phage display library. VNAR-5G8 exhibited superior affinity and specificity for TROP-2 ECD. Subsequently, VNAR-5G8 was fused with the PE38 toxin to create the immunotoxin 5G8-PE38, and the anti-tumor activity of 5G8-PE38 was evaluated in vitro and in vivo. The results suggest that 5G8-PE38 is a potential candidate for cancer patients with high expression of TROP2.
2. Materials and methods
2.1. Immunization and library generation
Two C. plagiosum sharks were immunized with TROP-2-ECD (His27-Thr274) (Novoprotein, C07M). For the initial immunization, 250 μg of TROP-2-ECD was emulsified by mixing with complete Freund's adjuvant (Sigma–Aldrich, F5881) in equal proportions. The dorsal and lateral fin areas of the sharks were selected as the immunization sites. Subsequent immunizations utilized incomplete Freund's adjuvant (Sigma–Aldrich, F5506) mixed with TROP-2-ECD in equal proportions. The injection of TROP-2-ECD decreases with each round. Final immunization is performed by intravenous delivery of the antigenic protein.
Shark serum titers were measured every two weeks. Specifically, human TROP-2 extracellular structural domain protein of 1 μg/mL was coated on ELISA plates at 100 μL per well for overnight incubation at 4 °C. The wells of ELISA plates were washed three times with a plate washer equipped with PBST (0.1% Tween-20 in PBS, v/v). PBS containing 5% skimmed milk powder (MPBS, w/v) was added as a blocking solution for 1 h. Subsequently, the plate was washed, and gradient-diluted shark serum was added to the ELISA plate at 100 μL per well respectively to set up duplicate wells, and incubated for 1 h at 37 °C. The rabbit anti-shark poly-clonal antibody (produced in our laboratory)28 was diluted in MPBS (1:500) and added to each well. The plate was then incubated for 1 h at 37 °C. Then, the plate was washed, followed by the addition of HRP-labelled goat anti-rabbit IgG antibody (1:10,000 dilution, Millipore, AP156P), and incubated for 1 h (conditions were the same as above). After washing the plate, TMB (Solarbio, PR 1200) was added to each well and incubated for 15 min, then the reaction was terminated by the addition of H2SO4 at a concentration of 2 mol/L. The absorbance value was measured at 450 nm.
Five days after the last immunization, whole blood and spleen were collected. Total RNA was extracted using Trizol and reverse-transcribed, followed by PCR to amplify the VNAR coding sequences. The recombinant vector constructed by ligating the VNAR gene to pComb3xss was then electro-transformed into competent Escherichia coli TG1 to generate a library of VNARs, which was subsequently rescued by co-infection with the M13KO7 helper phage.
2.2. Enrichment of TROP-2-specific VNAR
Human TROP-2-ECD (His27–Thr274) protein was used to perform specific phage panning. In three rounds of panning, each well was coated with different concentrations of antigen for sequential reduction. TROP-2-ECD proteins were encapsulated in a decreasing gradient with concentrations of 10, 4, and 2 μg/mL, 200 μL per well. PBS was used as the negative control. After washing three times with PBST, the wells were blocked at 37 °C with PBS containing 3% (w/v) milk powder (or 5% BSA, alternating between different rounds of elution) for 1 h, and then washed three times with PBST. Approximately 1012 phages were added to the antigen-coated plates for incubation at 37 °C for 1 h. They were washed 10 times with PBST after the first round of elution. After two more rounds of elution, the plates were washed 20 times and the targeted TROP-2 phage was retained. The activated E. coli TG1 was added to the retained phage targeting TROP2 in plates, and phage infection was conducted by incubating at 37 °C for 30 min. The infected TG1 cells were harvested, and fresh TG1 were replenished, repeating this process three times. The coated plate was examined, and the number of clones that grew was recorded as the successfully infected TG1. For each bio-panning, enrichment was performed as described above and assessed by the bacterial population infected with the purified phage.
2.3. Production and purification of VNAR
The VNAR sequence was cloned into pET-28a(+) and transformed into E. coli BL21 (DE3) cells according to DNA sequencing. The colonies formed were pooled into 500 mL of LB medium containing 30 μg/mL kanamycin and incubated at 37 °C until OD600 reached 0.5. Cultures were induced with 0.1 mmol/L IPTG and incubated overnight at 16 °C to produce soluble proteins. The bacterial pellet was centrifuged and resuspended in an ice-cold lysis buffer. The resuspension was incubated on ice for 30 min, followed by sonication on ice. They were then centrifuged at 13,400×g for 30 min at 4 °C. Soluble VNAR containing His-tagged protein was purified from the cell lysate using Ni-NTA resin. Finally, VNAR was eluted with different concentrations of imidazole buffer and purified by exchanging the buffer to 1 × PBS on the Superdex 200 Increase 10/300 GL of size exclusion column (SEC) (Cytiva).
2.4. BLI-based affinity assay
Antibody affinity detection was performed using a forte BIO-Octet RED96e instrument (Sartorius). Biotinylated TROP-2 (NovoProtein, CY65) captured on SA (High Accuracy Streptavidin) biosensor. The hydrated SA sensor was equilibrated in assay buffer for 30 s and then allowed to bind to biotinylated TROP-2 for 45 s. After capturing the TROP2 protein, perform a second equilibration step for 60 s. The biosensor was bound to anti-TROP-2 VNAR (100 nmol/L) for 240–300 s and then dissociated for 240–300 s. Binding affinity was determined by analyzing binding curves generated in the Octet Data Analysis v12.0.2.3 software program. Controls were averaged and subtracted from the data before modeling, and dissociation constants (KD) were calculated.
2.5. ELISA for VNAR affinity and epitope binning
TROP-2-ECD at 1 μg/mL was coated onto 96-well ELISA plates overnight at 4 °C. The plates were blocked with MPBS at 37 °C for 2 h. VNARs were evaluated for affinity and MPBS was used as negative control. VNARs were serially diluted in a blocking buffer from 100 nmol/L to 10 pmol/L. After washing the ELISA plate with PBST, the bound VNARs were determined by an HA tag poly-clonal antibody conjugated with HRP (1:10,000 dilution, Abcam, ab1190) capable of reacting with TMB followed by OD450 value determination.
5G8, 5E9, and 5C10 (100 ng/well) were coated on the ELISA plates at 4 °C for 16 h. After washing the plates with PBST and blocking with 5% MPBS, TROP-2-ECD (1 μg/well) was added for 30 min at 37 °C. Following washing with PBST, 5E9 with the HA tag (100 ng/well) was introduced for 1 h at 37 °C. After 3rd washing with PBST (three times), the bound 5E9 was determined by the HA tag antibody conjugated with HRP (1:10,000 dilution, Abcam, ab1190) capable of reacting with TMB followed by OD450 value measurement.
2.6. Cell lines and cell culture
Human umbilical vascular endothelial cells (HUVEC), and human triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-468 were stocked and identified by STR analysis. They were grown as mono-layers in DMEM (Gibco, 11965092) medium supplemented with 10% FBS (PAN Biotech, ST30-3302), penicillin, and streptomycin (50 U/mL) at 37 °C in 5% CO2 and 80% relative humidity. Cells were passaged less than 2 months after each defrost and assessed for the presence of mycoplasma contamination.
2.7. Binding of VNARs to tumor cells
HUVEC, MDA-MB-231, and MDA-MB-468 cells were collected, washed with PBS, and incubated in FACS buffer (0.5% BSA in PBS). Expression of TROP-2 on HUVEC, MDA-MB-468, and MDA-MB-231 was verified using a rabbit anti-TROP-2 antibody (Cell Signaling Technology, 47866). The cells were incubated with FACS buffer containing 1 μg/mL of the TROP-2 antibody and VNARs for 30 min at 25 °C. They were then washed twice with FACS buffer and stained with CoraLite Plus 488-conjugated 6∗His-Tag monoclonal antibody (Proteintech, CL488-66005) for analysis by flow cytometry (FACS LSR II or FACS Verse, BD).
2.8. Internalization assay
MDA-MB-468 cells were prepared as described above for flow cytometry. Antibodies were bound to cells at 4 °C for 30 min. After twice washing with FACs buffer, the cells were placed at 37 °C to allow internalization for 15 min and 2 h, respectively. Internalization was estimated by the number of fluorescent cells. They were washed twice with FACS buffer and stained with the CoraLite Plus 488-conjugated 6∗His-Tag monoclonal antibody (Proteintech, CL488-66005). The difference in cell fluorescence intensity was calculated as fluorescence values indicative of internalization before and after 37 °C reaction versus that of 4 °C.
2.9. Immunofluorescence
MDA-MB-468 cells were cultured on tissue-treated cell sheets at 37 °C until fully appressed. Cells were fixed with 4% paraformaldehyde for 10 min and blocked with 1% BSA for 1 h. Then, 1 μg of VNAR and the TROP2 antibody were added and incubated at 4 °C overnight. After rinsing the cells with sterile PBS, the cells were sequentially incubated with Alexa Fluor 488-conjugated anti-His monoclonal antibody (dilution 1:500) and Alexa Fluor 488-conjugated anti-rabbit IgG (1:500, Abcam, ab150077). The nuclei were stained with DAPI. FITC fluorescence images were acquired using a confocal laser-scanning microscope (Leica, Germany), and the surface-stained images were merged to form a cytosolic staining map to localize the binding of VNAR to the surface of tumor cells.
2.10. Molecular weight determination
VNAR-5G8 was desalted and analyzed by LC–MS/MS29. LC separations were performed using an ACQUITY UPLC Protein BEH C4 Column, 300 Å, 1.7 μm, 2.1 mm × 50 mm, 1/pk (Waters) with a column temperature of 80 °C and an injection volume of 5 μL (5 μg). The mobile phase A was 0.1% TFA in water and the mobile phase B was 0.1% TFA in acetonitrile. The gradient was from 10% to 95% in 8 min; the flow rate was 0.3 mL/min; and the UV detection wavelength was 280 nm. MS experiments were carried out using a Xevo G2-XS Qtof instrument (Waters). The mass spectrometer was set up in positive ion scan mode with a primary resolution of 180,00 and a scan range of m/z 500 to 3500.
2.11. Disulfide bond analysis
VNAR-5G8 was desalted and NEM (N-ethylmethacrylamide) was used to seal free sulfhydryl groups. Trypsin was used for enzymatic digestion. The samples were incubated overnight at 37 °C. One sample was acidified to a final concentration of 1% FA for direct disulphide bond analysis. The other was reduced with DTT at 57 °C for 30 min, then alkylated with iodoacetamide for 30 min at room temperature, protected from light, and then acidified to a final concentration of 1% FA as a control sample. LC–MS/MS analysis conditions are the same as those for molecular weight determination.
2.12. Hydrogen-deuterium exchange mass spectrometry
HDX-MS30 experiments were performed using a high-resolution Xevo-G2XS QTof equipped with a nano-UPLC, a LEAP II autosampler, and HDX manager from Waters. Briefly, recombinant TROP-2 (1.2 mg/mL in PBS) and TROP-2 linked to VNAR (1:1 molar ratio) were used in this analysis. 5 μL of each sample was diluted (v/v) into 50 μL labeling buffer (100 mmol/L phosphate, 99.9% D2O, 0.5 mol/L TCEP, pH 7.0) at 20 °C to allow hydrogen-deuterium exchange. After 10–1000 s incubation, the exchange reaction was quenched and the peptic fragments were analyzed by QTof. Triplicated injections were performed for each exchange time. Mass assignment for each peptide without deuterium exchange was checked manually. Any assignment with a mass deviation >0.2 Da was removed. The relative fractional deuterium uptake for each residue amide proton at a given time was averaged and calculated following HD site equations, and the HDX difference between datasets was calculated by comparing the relative fractional uptake for each residue.
2.13. Molecular modeling and molecular dynamics (MD) simulation
The amino acid sequence of TROP-2-ECD was obtained from the NCBI database. The 3D structural model of TROP-2-ECD with VNAR-5G8 was constructed by AlphaFold231. The docking program pyDockWEB32 was employed to molecular dock the antigen TROP-2-ECD with VNAR-5G8, where the docking site was selected regarding deuterium relative absorbance data from HDX-MS. MD simulations were performed by a CUDA-accelerated version of the AMBER18 pmemd engine (pmemd. cuda)33. The ff19SB protein force field34 and the OPC water mode35 were used to describe the system. The simulated TROP-2-ECD-5G8 complex structure was based on the molecular docking results described above, where the corresponding complex structure with the lowest binding energy prediction was selected. During the simulations, the covalent bonding of hydrogen atoms was restricted using the SHAKE algorithm with an integration36 time step of 2 fs. Long-range electrostatic forces were handled using the Particle Mesh Ewald (PME)37 method of treatment with the mesh spacing set to 1 Å. Short-range non-bonding interactions were truncated at 12 Å. The temperature and pressure of the system were controlled at 310.15 K and 1.013 bar, respectively, using a Langevin thermostat and Berendsen constant pressure. Trajectory snapshots were saved at 100 ps intervals.
2.14. Construction of immunotoxins
The expressed sequence of VNAR immunotoxin was designed based on the cDNA sequences of 5G8 and the truncated P. exotoxin A (Gene ID: 877850), in which the type I region of the PE was truncated16 and the REDLK sequence of the protein tail was changed to KDEL, which was ligated with an HA tag. In addition, another VNAR immunotoxin was designed as the negative control, the sequence is based on the VNAR (R1C2) constructed from the neocoronavirus spiny protein38 and the P. exotoxin designed in the same way as the 5G8 fusion toxin. These two expression sequences were optimized based on E. coli codon preferences, and then the sequences of the enzymatic cleavage sites Nco I and Not I were added to the end of the sequence. The DNA fragment containing the target gene and the cleavage site was sequenced for confirmation.
2.15. Cytotoxicity assay
HUVEC and MDA-MB-468 cells were seeded into 96-well plates. 5G8, 5G8-PE38, and R1C2-PE38 (100 pmol/L–100 nmol/L) at different concentrations were incubated for 72 h at 37 °C in a 5% CO2 atmosphere. Afterwards, 5 mg/mL of MTT solution was added to each well and incubated at 37 °C for 6 h. The formazan was solubilized by DMSO, and its absorbance at 570 nm was measured.
2.16. Cell apoptosis assay
An annexin V-FITC apoptosis detection kit (Absin, abs50001) was used to analyze apoptosis using flow cytometry according to the manufacturer's instructions. Briefly, HUCEC and MDA-MD-468 cells were seeded (5 × 105 cells/mL) in a 6-well plate and incubated with either 5G8-PE38, R1C2-PE38, or vehicle for 48 h. Cells were collected, washed with PBS, and resuspended in 1 × binding buffer (300 μL) before adding annexin V-FITC (5 μL) and PI (5 μL). After incubation for 5 min in the dark, the log fluorescence values of annexin V-FITC and PI were shown on the X and Y axes, respectively.
2.17. Tumor inoculation and treatment schedule
Female BALB/c nude mice (4-week-old) were subcutaneously injected with 5 × 106 MDA-MB-468 cells. When the tumor volume reached 100 mm3, PBS, R1C2-PE38 (5 mg/kg), 5G8-PE38 (1 mg/kg), and 5G8-PE38 (5 mg/kg) were injected into mice through the tail vein. The animals received care in compliance with the guidelines for the Care and Use of Laboratory Animals. This study was approved by the ethical committee of Ocean University of China. All animal treatments and experiments were performed according to the Institutional Animal Care and Use guidelines39. Tumor volume changes were calculated by measuring the long and short diameters of mouse tumors every three days using an accurate vernier caliper. Tumor volume calculation formula Eq. (1):
| TV = (Length × Width2)/2 | (1) |
TV is the tumor volume, length is the length of the tumor, and width is the width of the tumor. The harvested organs (liver, spleen, and kidney) were routinely paraffinized for safety histological examinations (H&E staining)40. Tumor tissues were subjected to immunohistochemistry and immunofluorescence experiments41.
2.18. Statistical analysis
All experiments were replicated at least three times, and representative data or pooled data from repeat experiments were recorded. Statistical analysis was performed using GraphPad Prism 5.0. Two-way ANOVA with Tukey's multiple comparisons was performed to assess the differences in tumor burden (tumor volume and tumor weight). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 were considered statistically significant.
3. Results
3.1. Identification of VNARs against TROP-2
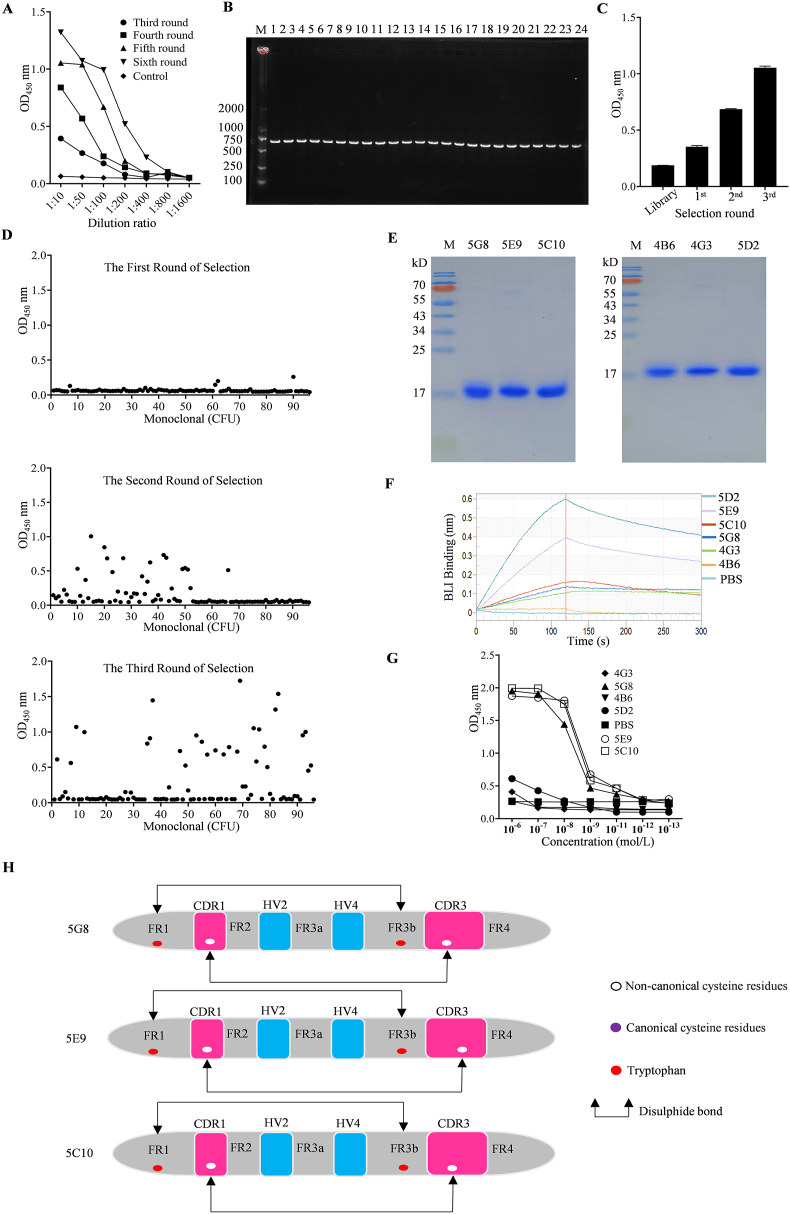
Two C. plagiosum sharks were subjected to six rounds of immunization with recombinant human-derived ECD of TROP-2 (His27–Thr274). The serum titer of shark specific to TROP-2-ECD was 1:800 (Fig. 1A). Subsequently, VNAR encoding gene fragments were amplified by PCR leading to a final library size of 1.28 × 108 independent clones. The PCR analysis of 24 randomly clones was performed and the insertion rate was determined to be 100% (Fig. 1B). Meanwhile, a high diversity of phage libraries was demonstrated by the large amino acid differences in the sequencing results of 24 clones (Supporting Information Fig. S1). After three rounds of bio-pannning using TROP-2-ECD, poly-clonal ELISA results showed a great enrichment of TROP-2-specific VNAR phage (Fig. 1C). Among the 288 monoclonal clones, sequences with OD450 values exceeding 0.8 were initially selected based on monoclonal ELISA results (Fig. 1D). Subsequently, antibody sequences with diverse amino acid compositions in the CDR3 region were identified through sequence comparison. Finally, six sequences (5G8, 5E9, 5C10, 4B6, 4G3, and 5D2) were selected for further analysis. The above six VNARs fused with a His tag at the C-terminus were expressed in the E. coli BL21 (DE3) strain using the pET28a vector. Ni + His Trap and gel filtration columns were used to purify them (Fig. 1E). BLI assays displayed that the KD values of VNARs 5G8, 5E9, and 5C10 were 8.73, 2.30, and 1.51 nmol/L, respectively (Fig. 1F and Supporting Information Table S1). Consistently, ELISA assays showed that they had good affinities to TROP-2 with EC50 values of 5.15, 1.68, and 2.808 nmol/L, respectively (Fig. 1G). Sequence analysis exhibited that they belong to type II shark VNARs and have different CDR3 domains (Fig. 1H).
Figure 1.
Shark immunization and screening of VNARs against TROP-2. (A) Determination of serum titers after immunization. (B) PCR analysis of randomly picked clones from the original library. (C) Poly-clonal ELISA identification of phages specifically recognizing TROP-2. (D) VNARs were identified from 288 clones bound specifically to TROP-2 after three rounds of bio-panning by monoclonal ELISA. (E) SDS-PAGE analysis of 5G8, 5E9, 5C10, 4B6, 4G3 and 5D2. (F) Affinity analysis of six positive sequences screened for TROP-2 by BLI. (G) ELISA analysis of VNARs 5G8, 5E9, and 5C10 for binding to TROP-2. (H) Sequence structures of three high-affinity VNARs targeting TROP-2. Data in (A), (C), and (G) are reported as the mean ± SD, n = 3 per group.
3.2. Cellular binding and internalization of VNAR-5G8
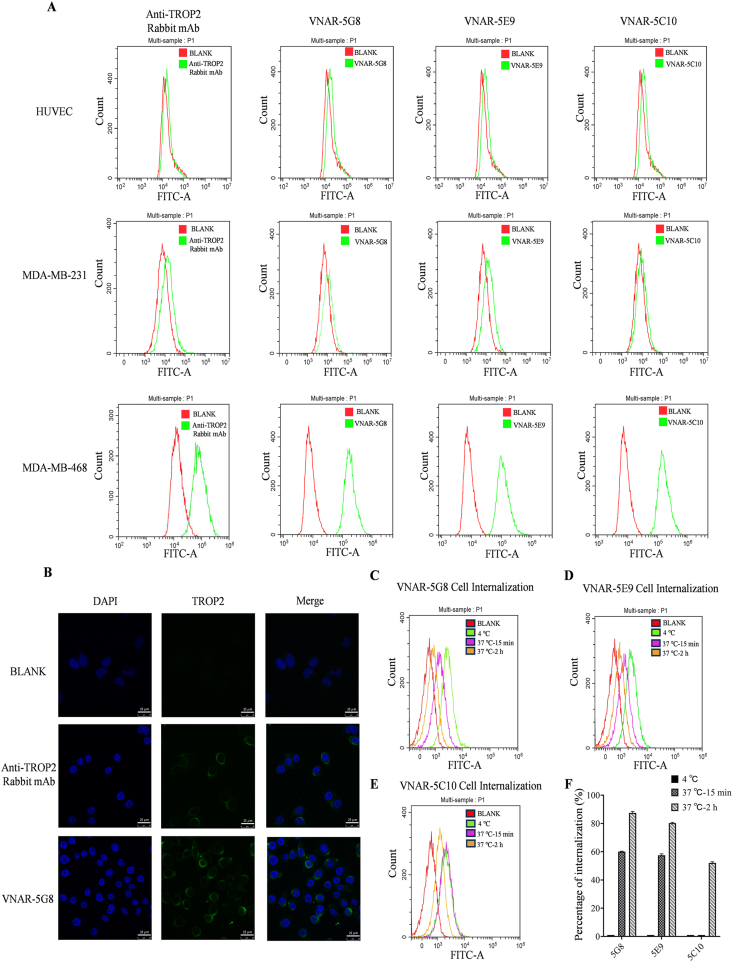
To evaluate the cell-binding capability of the VNARs, flow cytometry was performed. Firstly, expression of TROP-2 on the human umbilical vein endothelial cells (HUVEC), triple-negative breast cancer cell line, MDA-MB-468, and MDA-MB-231 were verified by a rabbit anti-TROP-2 monoclonal antibody. The results showed that the expression of TROP-2 was negative in HUVEC and MDA-MB-231 cells, whereas was positive in MDA-MB-468 cells. This demonstrated shark VNAR 5G8, 5E9, and 5C10 were bound to MDA-MB-468 cells but not to HUVEC and MDA-MB-231 cells (Fig. 2A), indicating that these VNARs possess comparable selectivity for the cell line. Immunofluorescence data further verified the above observation (Fig. 2B and Supporting Information Fig. S2). Then, the internalization of VNAR in MDA-MB-468 cells was examined using flow cytometry, and the internalization efficiencies for 5G8, 5E9, and 5C10 were 88%, 80% and 52%, respectively (Fig. 2C–F). Collectively, 5G8 was chosen for anti-tumor studies and its amino acid sequence is shown in Supporting Information Fig. S3.
Figure 2.
The results of flow cytometry and immunofluorescence from VNARs bonded to various cells. (A) Flow cytometry results of HUVEC, MDA-MB-231, and MDA-MB-468 cells bonded to a rabbit antibody against TROP-2 and VNARs (5G8, 5E9, and 5C10). (B) Confocal immunofluorescence images of MDA-MB-468 cells stained by the rabbit antibody against TROP-2 and VNARs 5G8 (scale bar = 25 μm). (C–E) Flow cytometric detection of internalized VNARs (5G8, 5E9, and 5C10) in MDA-MB-468 cells. (F) Internalized percentages of the three VNARs were calculated (mean ± SD, n = 3 in all data).
3.3. Quality analysis of VNAR-5G8
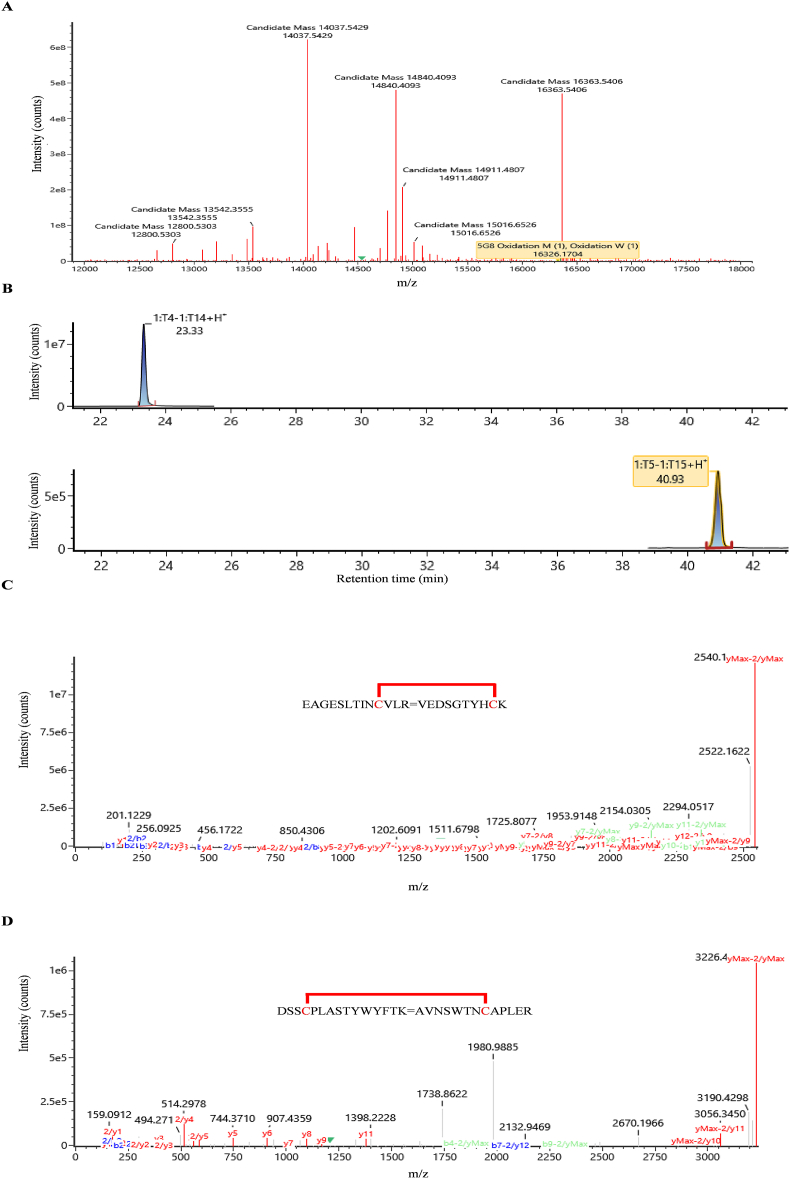
To further evaluate the quality of 5G8, physicochemical characterization experiments were carried out. Specifically, its molecular weight was determined to be 16,326.1704 Da (Fig. 3A and Supporting Information Fig. S4). The locations of the disulfide bond were identified by trypsinzing the VNAR followed by a subsequent reduction reaction. After enzymatic digestion, two sets of disulfide bond-linked polypeptide fragments with retention times of 23.33 and 40.93 min were observed (Fig. 3B). Secondary mass spectrometric analysis revealed the formation of two pairs of disulfide bonds with molecular weight of 2540.1717 Da (EAGESLTINCVLR/VEDSGTYHCK) and 3226.4530 Da (DSSCPLASTYWYFTK/AVNSWTNCAPLER), respectively (Fig. 3C and D). These results demonstrate the capability of our E. coli expression system to accurately produce the VNAR protein.
Figure 3.
Molecular weight and disulfide bond formation of VNAR-5G8 were revealed by LC–MS/MS. (A) Molecular weight mass profile of VNAR-5G8. (B) Extracted ion chromatogram (XIC) of two pairs of disulfide bonds formed in VNAR-5G8. (C) Mass spectra of the VNAR-5G8 fragments with disulfide bond formation between FR1 and FR3. (D) Mass spectra of the VNAR-5G8 fragments with disulfide bond formation between CDR1 and CDR3.
3.4. Epitope analysis and molecular docking
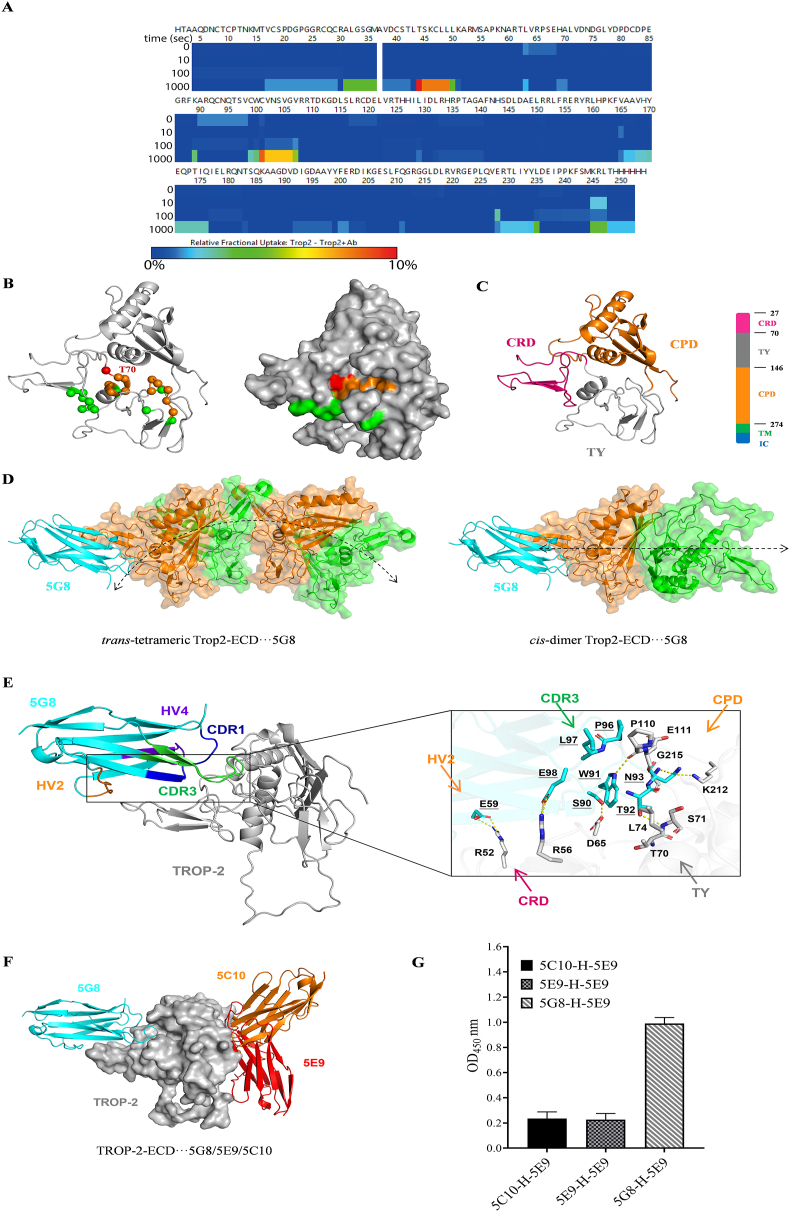
HDX-MS was utilized to analyze the binding epitope of 5G8 on TROP-2. The data collected from both TROP-2 and TROP-2+5G8 are high-quality as depicted by the peptide coverage map for TROP-2. A total of 146 peptides were obtained with 98.4% coverage and 7.44 overlap (Supporting Information Fig. S5). The heat map illustrated the differences in the relative uptake fraction of deuterium between TROP-2 and TROP-2+5G8, which became more pronounced with a longer labeling time (Fig. 4A). Notably, two representative peptides, TSKCLL and CVNSVGV (Supporting Information Fig. S6), exhibited the most significant disparities in relative uptake fractions of deuterium between TROP-2 and TROP-2+5G8. These outcomes revealed that the binding site of VNAR-5G8 relative to TROP-2 resided in a concealed epitope constituted by peptides CRD (T70–L75) and TY (C127) (Fig. 4B). In contrast, Sacituzumab targets a stretch peptide (Q237–Q252) of the CPD structural domain42, highlighting the notable differences in binding epitope and structure between Sacituzumab and VNAR-5G8.
Figure 4.
HDX-MS and molecular dynamics simulation of TROP-2 in the presence of VNAR-5G8. (A) The heat map shows the difference in relative fractional uptake of deuterium between TROP-2 and TROP-2+5G8 by HDX-MS. (B) 3D projection of the regions with the most significant difference in relative fractional uptake of deuterium. The residues colored in red carried the largest difference (∼10%), the residues colored in brown carried the mild difference (∼5%), and the residues in green carried no difference (<1%). (C) Schematic diagram of TROP-2-ECD. TROP-2-ECD contains an ectodomain (27–274), consisting of a cysteine-rich domain (CRD), thyroglobulin type-1 domain (TY-1) and cysteine-poor domain (CPD), trans-membrane (TM), and cytoplasmic regions. The cartoon represents an ectodomain of TROP-2, including CRD (red), TY-1 domain (gray), and CPD (yellow), with all disulfide bonds shown as sticks. (D) Binding position of 5G8 on the trans-tetrameric and cis-dimer of TROP-2-ECD. The assembly direction of TROP-2-ECD is represented by a double-arrowed line. (E) Detailed view of TROP-2-ECD binding model with VNAR-5G8. VNAR-5G8 and TROP-2-ECD are represented as cyan, and grey cartoons. Key residues are indicated as cyan and grey rods. Hydrogen bonds are shown as yellow dashed lines. Among them, HV4 of VNAR is purple, HV2 is orange, CDR3 is green, and CDR1 is blue. (F) Schematic representation of the structural overlay of the TROP-2-ECD···5G8, TROP-2-ECD···5E9, and TROP-2-ECD···5C10 composite systems. TROP-2-ECD, 5G8, 5E9, and 5C10 are denoted as grey surfaces, cyan, red and orange cartoons, respectively. (G) Double antibody sandwich ELISA was used to detect the epitope competition of VNAR binding to TROP-2 for the three shark VNARs. The horizontal and vertical axes represent the concentration and OD450 value of VNAR, respectively (mean ± SD, n = 3).
Analysis of antigen-binding epitopes plays a crucial role in the development of antibody-based therapeutics. TROP-2-ECD contains an N-terminal cysteine-rich domain (CRD), a thyroglobulin type-1 domain (TY), and a C-terminal cysteine-deficient domain (CPD) (Fig. 4C)42. Previous studies showed that TROP-2-ECD is exhibited as an oligomer on the cell surface and can be assembled through cis- or trans-multimers to perform its biological functions43. To gain a deeper understanding of the molecular binding mode between VNAR-5G8 and TROP-2-ECD, HDX-MS data is used to perform molecular dynamics (MD) simulations. Interestingly, 5G8 binds to both assembly pathways of TROP-2-ECD which may prevent the latter from aggregating intracellularly or intercellularly, thereby retaining the normal function of TROP-2 (Fig. 4D). This phenomenon was not found in Sacituzumab. Meanwhile, MD simulations showed that the CDR3 of 5G8 was embedded in the hidden space epitope formed by CRD and TY, and some residues of HV2 were also involved in its binding to TROP-2-ECD. The binding stability of VNAR-5G8 to TROP-2-ECD is maintained by multiple pairs of residue interactions (Fig. 4E). R52 and R56 of TROP-2-ECD form salt bridges with E59 in the HV2 region and E98 in the CDR3 region of VNAR, respectively. The residue pairs at the interface between TROP-2-ECD and the CDR3 region (D65|S90, S71|T92, P110|W91, K212|N93, and G215|N93) mainly form hydrogen bonds. In addition, the residue pairs T70|T92, L74|W91, P110|L97, P110|P96, and E111|P96 mainly form hydrophobic interactions to stabilize the system (Supporting Information Table S2). The binding modes of other VNARs to TROP-2-ECD were also investigated, demonstrating that 5G8 has a different binding epitope compared to 5E9 and 5C10 (Fig. 4F and G). These findings provide valuable information on the intricate interactions between VNAR-5G8 and TROP-2-ECD, shedding light on its potential mechanisms of action.
3.5. Anti-tumor activity of 5G8 based immunotoxins
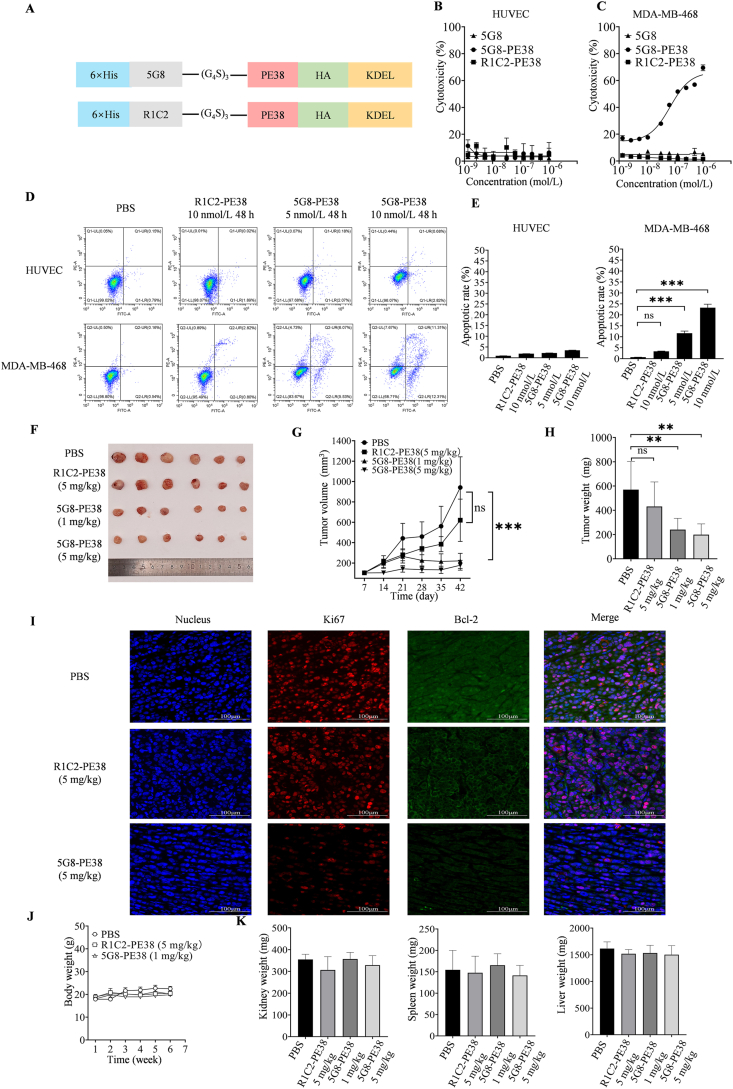
To explore the potential application of VNAR-5G8 in cancer treatment, we constructed an immunotoxin by fusing 5G8 with PE38 using a G4S linker (Fig. 5A). 5G8-PE38 was successfully expressed and purified showing a molecular weight of approximately 60 kDa (Supporting Information Fig. S7A). As a negative control, immunotoxin-based on an unrelated VNAR (R1C2) was also constructed (Fig. 5A). ELISA analysis showed that 5G8-PE38 had a high affinity for TROP-2 that was not affected by PE38 (Fig. S7B). For MTT assay, 5G8 was non-toxic to both HUVEC and MDA-MB-468 cells. Meanwhile, 5G8-PE38 did not exhibit significant cytotoxicity towards HUVEC cells which expressed low levels of TROP-2 (Fig. 5B). As expected, 5G8-PE38 displayed significant cytotoxicity against MDA-MB-468 cells with high expression of TROP-2 (IC50 = 6.69 nmol/L; Fig. 5C). Finally, the impact of 5G8-PE38 on MDA-MB-468 cell apoptosis was determined. As shown in Fig. 5D and E, early and late apoptosis rates of MDA-MB-468 cells treated with 5G8-PE38 for 48 h were significantly higher than that of the control, whereas R1C2-PE38 showed no effect in the cases.
Figure 5.
In vitro and in vivo activities of VNAR-5G8 fused with PE38. (A) Schematic view of 5G8-PE38 construction. (B) Cytotoxicity of 5G8-PE38 in HUVEC cells. (C) Cytotoxicity of 5G8-PE38 in MDA-MB-468 cells. (D) Apoptosis of HUVEC and MDA-MB-468 cells treated with PBS, R1C2-PE38, and 5G8-PE38 for 24 h determined by flow cytometry. (E) Statistical plot of 5G8-PE38 induced apoptosis in HUVEC and MDA-MB-468 cells. (F) Tumor tissues were stripped from mice at the end of treatment. (G) Treatment timeline in mice receiving 5G8-PE38 and relative tumor volumes (n = 6 mice per group). (H) Tumor weight following treatment. (I) Representative immunofluorescence images of MDA-MB-468 tumor sections stained for DAPI (4′,6-diamidino-2-phenylindole) (DAPI, blue), Bcl-2 (green) and Ki-67 (red) proliferation marker (Scale bar = 100 μm). (J) Body weight following treatment (n = 6 mice per group). (K) Liver, spleen, and kidney weights following treatment. (n = 6 mice per group). Data are reported as the mean ± SD, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
In vivo experiments were also performed to evaluate the therapeutic efficacy of 5G8-PE38 in the MDA-MB-468 tumor xenograft mouse model. The mice were treated with different doses of 5G8-PE38 (1 and 5 mg/kg). Tumor tissues (Fig. 5F), volumes (Fig. 5G), and weights (Fig. 5H) were recorded. The results demonstrated the anti-tumor efficacy of 5G8-PE38. In addition, immunofluorescence staining of tumor tissues showed a significant decrease in Ki67 and Bcl-2 in 5G8-PE38 treated animals (Fig. 5I). In terms of safety, no obvious difference was found in the mass and H&E morphology of the liver, spleen, and kidney (Fig. 5J and K, Supporting Information Fig. S8).
4. Discussion
In recent years, substantial progress has been made in the development of ADCs44. In addition to HER2, TROP-2 has become another promising target, such as Sacituzumab Govitecan, the world's first ADC targeting TROP-2 for treating metastatic triple-negative breast cancer and locally advanced or metastatic urothelial carcinoma (UC)11. However, ADCs are still limited by their high molecular weights and inability to penetrate solid tumor45. Approximately 0.1% of the administered ADC dose reaches tumor cells, while the majority is metabolized by normal cells, resulting in unwanted off-target toxicity46. These issues are affected by various factors, including cell death triggered by target-dependent and independent mechanisms from ADCs bonded to the cell surface, premature release of cytotoxic payload to the circulation, and non-specific endocytosis resulting from the instability of linker-carriers45.
On the other hand, most toxic ligands in ADCs are small molecule compounds. To overcome the shortcomings of small molecule compounds, a novel therapeutic strategy has emerged for immunotoxins by fusing protein toxins with antibody variable regions, such as those used for cancer treatment by targeting CD22 and HER217,47,48. Lumoxiti is a cytotoxic agent against CD22 to treat hairy cell leukemia19, while trastuzumab-derived single-chain antibodies (ScFv) were utilized to design immunotoxins against HER2-positive cancer. HER2-specific ScFv was also fused with DFF40 toxin as a novel immunotoxin47. However, there are existing immunotoxins that are also based on intact IgG-type antibodies and their ScFv, whose tendency to self-aggregate and the possibility of unintended fusion of the VH/VL structural domains reduces the affinity and affects the functionality of the immunotoxin. The VNAR has a molecular weight much lower than IgG and better penetration to solid tumors. It naturally lacks Fc-mediated off-target effects caused by binding to FcγRs. VNARs are more specific than IgG and can reduce cytotoxicity caused by non-specific endocytosis42.
Shark VNAR-5G8, which targets TROP2, exhibits suitable affinity and high internalization efficiency. Through the searches of literature and databases (including NCBI, PDB, IMGT, INDI, and SYNBIP)49, 50, 51, 52, 5G8 is the first reported shark VNAR known to target TROP2, as well as the first shark VNAR employed in immunotoxin construction. At the same time, the binding epitope of VNAR-5G8 to TROP2-ECD was revealed for the first time, adding a novel dimension to our understanding. The binding site of the marketed drug Sacituzumab Govitecan is localized to the stretch peptide in the TROP-2 CPD (Q237–Q252)42. However, shark VNAR-5G8 developed a significantly difference from Sacituzumab Govitecan in terms of antigenic epitopes. The T70 amino acid residue in TROP2 is the primary binding site, followed by S71 and L74 binding sites, respectively. In the CDR3 region of VNAR-5G8, T92, and W91 form residue pairs with T70, S71, and L74 in TROP2, respectively, as the main binding mode. Structural analysis revealed that this region does not hinder the tumor-specific protein cleavage of TROP-2 and does not have a cytotoxic effect on tumor cells53. VNAR-5G8 alone did not effectively suppress tumor cell proliferation. Recent studies have shown that TROP-2 molecules cluster on the cell membrane of tumor cells42, and the binding region of VNAR-5G8 to TROP-2 at the site of TROP-2 cluster formation may affect TROP-2 aggregation, thereby limiting tumor cell metastasis. The above results prove that nanoscale structural characteristics of shark VNARs enable deeper epitope binding, which may have more functional applications in tumor therapy than conventional IgG-based monoclonal antibodies.
In this study, we constructed an immunotoxin by combining VNAR-5G8 with PE38. 5G8-PE38 not only retains the specificity of VNAR but also utilizes the functional properties of PE38. This immunotoxin demonstrated both in vitro and in vivo anti-tumor activities. The genetic engineering approach used to prepare the fusion toxin offers advantages in terms of convenience and batch stability compared to the traditional chemical coupling method. In future experiments, it will be interesting to explore the fusion of other toxins with VNAR and provide new strategies for cancer treatment.
5. Conclusions
In this study, we designed, constructed and characterized a novel shark nanobody-based immunotoxin targeting TROP2. Briefly, we constructed a shark VNAR phage display library and identified shark VNAR-5G8 targeting TROP-2. VNAR-5G8 exhibits strong affinity and internalization towards cells expressing high levels of TROP-2, and it can recognize spatially hidden epitopes of the TROP2 protein. These properties are advantageous for the development of nanobody-based anti-tumor drugs. Subsequently, VNAR-5G8 was fused with a truncated form of P. exotoxin A (PE38) to create a recombinant immunotoxin (5G8-PE38). 5G8-PE38 displayed promising anti-tumor activity in vitro and in vivo. Overall, this study highlights the potential of 5G8-PE38 as a valuable candidate for cancer treatment.
Author contributions
Xiaozhi Xi: Writing – original draft, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Yanqing Wang: Software, Methodology, Formal analysis, Data curation. Guiqi An: Software, Methodology, Data curation. Shitao Feng: Methodology, Data curation. Qiumei Zhu: Formal analysis. Zhongqiu Wu: Data curation. Jin Chen: Writing – review & editing, Formal analysis, Data curation. Zhicheng Zuo: Software. Qiang Wang: Writing – review & editing. Ming-Wei Wang: Writing – review & editing, Supervision. Yuchao Gu: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Investigation, Funding acquisition, Conceptualization.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This research was supported by the Qingdao Marine Science and Technology Center (No. 2022QNLM030003-4 and 8-01, China), the Program of National Natural Science Foundation of China (No. 82273846), and the Taishan Scholars Program (No. tsqn202211058, China). We also would like to thank Professor Wei Deng of Soochow University for his help in hydrogen-deuterium exchange mass spectrometry (HDX-MS) and data analysis.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.08.023.
Appendix A. Supporting information
The following is the Supporting Information to this article:
References
- 1.Shvartsur A., Bonavida B. Trop2 and its overexpression in cancers: regulation and clinical/therapeutic implications. Genes Cancer. 2015;6:84–105. doi: 10.18632/genesandcancer.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chou J., Trepka K., Sjöström M., Egusa E.A., Chu C.E., Zhu J., et al. TROP2 expression across molecular subtypes of urothelial carcinoma and enfortumab vedotin-resistant cells. Eur Urol Oncol. 2022;5:714–718. doi: 10.1016/j.euo.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDougall A.R., Tolcos M., Hooper S.B., Cole T.J., Wallace M.J. Trop2: from development to disease. Dev Dyn. 2015;244:99–109. doi: 10.1002/dvdy.24242. [DOI] [PubMed] [Google Scholar]
- 4.Liu X., Zhou T., Wang Y., Pei M., Wang G., Chu W., et al. TROP2 as patient-tailoring but not prognostic biomarker for breast cancer. Onco Targets Ther. 2022;15:509–520. doi: 10.2147/OTT.S354048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen Y., Ouyang D., Zou Q., Chen Q., Luo N., He H., et al. A literature review of the promising future of TROP2: a potential drug therapy target. Ann Transl Med. 2022;10:1403. doi: 10.21037/atm-22-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakach E., Sacks R., Kalinsky K. Trop2 as a therapeutic target in breast cancer. Cancers (Basel) 2022;14:5936. doi: 10.3390/cancers14235936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J., Duan Q., Ran C., Fu T., Liu Y., Tan W. Recent progress of aptamer–drug conjugates in cancer therapy. Acta Pharm Sin B. 2023;13:1358–1370. doi: 10.1016/j.apsb.2023.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H., Fang X., Tuhin I.J., Tan J., Ye J., Jia Y., et al. CAR T cells equipped with a fully human scFv targeting Trop2 can be used to treat pancreatic cancer. J Cancer Res Clin Oncol. 2022;148:2261–2274. doi: 10.1007/s00432-022-04017-x. [DOI] [PubMed] [Google Scholar]
- 9.Syed Y.Y. Sacituzumab Govitecan: first approval. Drugs. 2020;80:1019–1025. doi: 10.1007/s40265-020-01337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furlanetto J., Marmé F., Loibl S. Sacituzumab Govitecan: past, present and future of a new antibody–drug conjugate and future horizon. Future Oncol. 2022;18:3199–3215. doi: 10.2217/fon-2022-0407. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y., Yuan X., Tian Q., Huang X., Chen Y., Pu Y., et al. Preclinical profiles of SKB264, a novel anti-TROP2 antibody conjugated to topoisomerase inhibitor, demonstrated promising antitumor efficacy compared to IMMU-132. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.951589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okajima D., Yasuda S., Maejima T., Karibe T., Sakurai K., Aida T., et al. Datopotamab Deruxtecan, a novel TROP2-directed antibody–drug conjugate, demonstrates potent antitumor activity by efficient drug delivery to tumor cells. Mol Cancer Ther. 2021;20:2329–2340. doi: 10.1158/1535-7163.MCT-21-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weerakkody L.R., Witharana C. The role of bacterial toxins and spores in cancer therapy. Life Sci. 2019;235 doi: 10.1016/j.lfs.2019.116839. [DOI] [PubMed] [Google Scholar]
- 14.Havaei S.M., Aucoin M.G., Jahanian-Najafabadi A. Pseudomonas exotoxin-based immunotoxins: over three decades of efforts on targeting cancer cells with the toxin. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.781800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green D.S., Husain S.R., Johnson C.L., Sato Y., Han J., Joshi B., et al. Combination immunotherapy with IL-4 Pseudomonas exotoxin and IFN-α and IFN-γ mediate antitumor effects in vitro and in a mouse model of human ovarian cancer. Immunotherapy. 2019;11:483–496. doi: 10.2217/imt-2018-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mai P.T., Lim D., So E., Kim H.Y., Duysak T., Tran T.Q., et al. Constitutive expression of a cytotoxic anticancer protein in tumor-colonizing bacteria. Cancers (Basel) 2023;15:1486. doi: 10.3390/cancers15051486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhillon S. Moxetumomab pasudotox: first global approval. Drugs. 2018;78:1763–1767. doi: 10.1007/s40265-018-1000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W., Tai C.H., Liu X., Pastan I. Anti-mesothelin immunotoxin induces mesothelioma eradication, anti-tumor immunity, and the development of tertiary lymphoid structures. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2214928119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weatherill E.E., Cain K.L., Heywood S.P., Compson J.E., Heads J.T., Adams R., et al. Towards a universal disulphide stabilised single chain Fv format: importance of interchain disulphide bond location and vL-vH orientation. Protein Eng Des Sel. 2012;25:321–329. doi: 10.1093/protein/gzs021. [DOI] [PubMed] [Google Scholar]
- 20.Xi X., Xiao G., An G., Liu L., Liu X., Hao P., et al. A novel shark single-domain antibody targeting OGT as a tool for detection and intracellular localization. Front Immunol. 2023;14 doi: 10.3389/fimmu.2023.1062656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narbona J., Hernández-Baraza L., Gordo R.G., Sanz L., Lacadena J. Nanobody-based EGFR-targeting immunotoxins for colorectal cancer treatment. Biomolecules. 2023;13:1042. doi: 10.3390/biom13071042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y., Wang R., Liu L., Feng S., Xi X., Yu W., et al. Identification and characterization of shark VNARs targeting the Helicobacter pylori adhesin HpaA. Artif Cell Nanomed Biotechnol. 2023;51:509–519. doi: 10.1080/21691401.2023.2255635. [DOI] [PubMed] [Google Scholar]
- 23.Qin L., Ren Q., Lu C., Zhu T., Lu Y., Chen S., et al. Screening and anti-glioma activity of Chiloscyllium plagiosum anti-human IL-13Rα2 single-domain antibody. Immunology. 2023;170:105–119. doi: 10.1111/imm.13658. [DOI] [PubMed] [Google Scholar]
- 24.Clarke E., Stocki P., Sinclair E.H., Gauhar A., Fletcher E.J.R., Krawczun-Rygmaczewska A., et al. A single domain shark antibody targeting the transferrin receptor 1 delivers a TrkB agonist antibody to the brain and provides full neuroprotection in a mouse model of Parkinson's disease. Pharmaceutics. 2022;14:1335. doi: 10.3390/pharmaceutics14071335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao S., Chen X., He Z., Wu L., Xie X., Sun Z., et al. Colorimetric and surface-enhanced Raman scattering dual-mode lateral flow immunosensor using phage-displayed shark nanobody for the detection of crustacean allergen tropomyosin. J Hazard Mater. 2024;468 doi: 10.1016/j.jhazmat.2024.133821. [DOI] [PubMed] [Google Scholar]
- 26.Jiao S., Xie X., He Z., Sun Z., Wang Z., Zhang S., et al. Lateral flow immunochromatographic assay for competitive detection of crustacean allergen tropomyosin using phage-displayed shark single-domain antibody. J Agric Food Chem. 2024;72:1811–1821. doi: 10.1021/acs.jafc.3c07569. [DOI] [PubMed] [Google Scholar]
- 27.Leow H.C., Fischer K., Leow Y.C., Braet K., Cheng Q., McCarthy J. Cytoplasmic and periplasmic expression of recombinant shark VNAR antibody in Escherichia coli. Prep Biochem Biotechnol. 2019;49:315–327. doi: 10.1080/10826068.2019.1566145. [DOI] [PubMed] [Google Scholar]
- 28.Xiao G.K., Hao P.Y., Feng S.T., Liu X.C., Fan Y.H., Lv J.X., et al. Preparation and application of rabbit anti-striped bamboo shark IgNAR polyclonal antibody. Chin Mar Drugs. 2022;41:37–42. [Google Scholar]
- 29.Yang J., Zhou A., Li M., He Q., Zhou J., Crommen J., et al. Mimotope peptide modified pompon mum-like magnetic microparticles for precise recognition, capture and biotransformation analysis of rituximab in biological fluids. Acta Pharm Sin B. 2024;14:1317–1328. doi: 10.1016/j.apsb.2023.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi Y., Zhang M., Xue H., Yu R., Bao Y.O., Kuang Y., et al. Schaftoside inhibits 3CLpro and PLpro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19. Acta Pharm Sin B. 2022;12:4154–4164. doi: 10.1016/j.apsb.2022.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casadevall G., Duran C., Osuna S. AlphaFold2 and deep learning for elucidating enzyme conformational flexibility and its application for design. JACS. 2023;3:1554–1562. doi: 10.1021/jacsau.3c00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez-García B., Pons C., Fernández-Recio J. pyDockWEB: a web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013;29:1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 33.Lee T.S., Allen B.K., Giese T.J., Guo Z., Li P., Lin C., et al. Alchemical binding free energy calculations in AMBER20: advances and best practices for drug discovery. J Chem Inf Model. 2020;60:5595–5623. doi: 10.1021/acs.jcim.0c00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian C., Kasavajhala K., Belfon K.A.A., Raguette L., Huang H., Migues A.N., et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theor Comput. 2020;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 35.Izadi S., Anandakrishnan R., Onufriev A.V. Building water models: a different approach. J Phys Chem Lett. 2014;5:3863–3871. doi: 10.1021/jz501780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 37.Darden T., York D., Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10094. [Google Scholar]
- 38.Liu X., Wang Y., Sun L., Xiao G., Hou N., Chen J., et al. Screening and optimization of shark nanobodies against SARS-CoV-2 spike RBD. Antivir Res. 2024;226:105898. doi: 10.1016/j.antiviral.2024.105898. [DOI] [PubMed] [Google Scholar]
- 39.Song N., Guan X., Zhang S., Wang Y., Wang X., Lu Z., et al. Discovery of a pyrrole-pyridinimidazole derivative as novel SIRT6 inhibitor for sensitizing pancreatic cancer to gemcitabine. Cell Death Dis. 2023;14:499. doi: 10.1038/s41419-023-06018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker S., McDowall C., Sanchez-Perez L., Osorio C., Duncker P.C., Briley A., et al. Immunotoxin-αCD40 therapy activates innate and adaptive immunity and generates a durable antitumor response in glioblastoma models. Sci Transl Med. 2023;15:682. doi: 10.1126/scitranslmed.abn5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J., Xu L., Zhang X.Q., Liu X., Zhang Z.X., Zhu Q.M., et al. Discovery of a natural small-molecule AMP-activated kinase activator that alleviates nonalcoholic steatohepatitis. Mar Life Sci Technol. 2023;5:196–210. doi: 10.1007/s42995-023-00168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun M., Zhang H., Jiang M., Chai Y., Qi J., Gao G.F., et al. Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2. iScience. 2021;24 doi: 10.1016/j.isci.2021.103190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavšič M., Ilc G., Vidmar T., Plavec J., Lenarčič B. The cytosolic tail of the tumor marker protein Trop2—a structural switch triggered by phosphorylation. Sci Rep. 2015;5 doi: 10.1038/srep10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters J.C., Nickens D.J., Xuan D., Shazer R.L., Amantea M. Clinical toxicity of antibody drug conjugates: a meta-analysis of payloads. Invest New Drugs. 2018;36:121–135. doi: 10.1007/s10637-017-0520-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z., Li H., Gou L., Li W., Wang Y. Antibody–drug conjugates: recent advances in payloads. Acta Pharm Sin B. 2023;13:4025–4049. doi: 10.1016/j.apsb.2023.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarantino P., Ricciuti B., Pradhan S.M., Tolaney S.M. Optimizing the safety of antibody–drug conjugates for patients with solid tumours. Nat Rev Clin Oncol. 2023;20:558–576. doi: 10.1038/s41571-023-00783-w. [DOI] [PubMed] [Google Scholar]
- 47.Lu D., Guo Y., Hu Y., Wang M., Li C., Gangrade A., et al. Fusion of apoptosis-related protein Cytochrome c with anti-HER-2 single-chain antibody targets the suppression of HER-2+ breast cancer. J Cel Mol Med. 2021;25:10638–10649. doi: 10.1111/jcmm.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y., Wu Z., Pang Z., Zhang L., Song D., Liu F., et al. Manufacture and evaluation of a HER2-positive breast cancer immunotoxin 4D5Fv-PE25. Microb Cel Fact. 2023;22:100. doi: 10.1186/s12934-023-02115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lefranc M.P. Antibody informatics: IMGT, the international ImMunoGeneTics information system. Microbiol Spectr. 2014;2:10. doi: 10.1128/microbiolspec.AID-0001-2012. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Li F., Qiu W., Xu B., Li Y., Lian X., et al. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022;50:560–570. doi: 10.1093/nar/gkab926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deszyński P., Młokosiewicz J., Volanakis A., Jaszczyszyn I., Castellana N., Bonissone S., et al. INDI-integrated nanobody database for immunoinformatics. Nucleic Acids Res. 2022;50:1273–1281. doi: 10.1093/nar/gkab1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamble P.R., Rane S., Breed A.A., Joseph S., Mahale S.D., Pathak B.R. Proteolytic cleavage of Trop2 at Arg87 is mediated by matriptase and regulated by Val194. FEBS Lett. 2020;594:3156–3169. doi: 10.1002/1873-3468.13899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.