Abstract
Purpose
To investigate the osteogenic potential of human embryonic stem cell-derived exosomes (hESC-Exos) and their effects on the differentiation of human umbilical cord mesenchymal stem cells (hUCMSCs).
Methods
hESC-Exos were isolated and characterized using transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and Western blotting. hUCMSCs were cultured with hESC-Exos to assess osteogenic differentiation through alizarin red staining, quantitative PCR (qPCR), and Western blotting. miRNA profiling of hESC-Exos was performed using miRNA microarray analysis. In vivo bone regeneration was evaluated using an ovariectomized rat model with bone defects treated with exosome-loaded scaffolds.
Results
hESC-Exos significantly promoted the osteogenic differentiation of hUCMSCs, as evidenced by increased alizarin red staining and the upregulation of osteogenesis-related genes and proteins (ALP, RUNX2, OCN). miRNA analysis revealed that miR-21-5p is a key regulator that targets YAP1 and activates the Wnt/β-catenin signaling pathway. In vivo, hESC-Exos enhanced bone repair in ovariectomized rats, as demonstrated by increased bone mineral density and improved bone microarchitecture compared to those in controls.
Conclusion
hESC-Exos exhibit significant osteogenic potential by promoting the differentiation of hUCMSCs and enhancing bone regeneration in vivo. This study revealed that the miR-21-5p-YAP1/β-catenin axis is a critical pathway, suggesting that the use of hESC-Exos is a promising therapeutic strategy for bone regeneration and repair.
Keywords: human embryonic stem cells, exosomes, osteogenic differentiation, bone regeneration, miR-21-5p, YAP1
Graphical Abstract
Introduction
Fractures pose a substantial global health concern, impacting millions annually and giving rise to both immediate and long-term consequences for affected individuals.1 Osteoporosis is a skeletal disorder that significantly exacerbates the risk of fractures and is particularly prevalent among the aging population, notably among postmenopausal women.2 Statistically, osteoporotic fractures transpire every three seconds globally, with 1 in 3 women and 1 in 5 men over the age of 50 expected to experience such a fracture in their lifetime.3 This disorder arises from an imbalance between bone formation and resorption, the latter overshadowing the former. A leading contributor to this disequilibrium is postmenopausal estrogen deficiency,4,5 which results in deteriorated bone microarchitecture, heightened bone fragility, and increased fracture risk.6,7 Importantly, delayed bone healing is a consistent finding among osteoporotic patients, an observation well documented in both human and animal research.8,9
Additionally, hip fractures among the elderly raise serious health concerns, given their association with significant morbidity and an up to 30% one-year mortality rate.10 Fractures are more acute than other bone disorders, such as osteoarthritis, which is a chronic degenerative condition. While osteoarthritis leads to long-term pain and functional limitations, fractures have an immediate impact and can lead to chronic pain and disability if not managed appropriately.11 Osteoporotic patients further challenge orthopedic surgical procedures, particularly those involving traumatic bone defects, severe infections, and tumor resections.12 Economically, fractures incur hefty treatment and rehabilitation expenses, with indirect costs, such as lost work productivity, adding to the financial strain.13,14 Current treatments primarily involve autologous and allogeneic bone transplantation and scaffold materials fortified with growth factors or cells. Nevertheless, these modalities have limitations, emphasizing the need for more effective approaches.15
Exosomes, which are small extracellular vesicles secreted by cells, have gained significant attention in recent years due to their role in intercellular communication and potential therapeutic applications.16,17 Exosomes contain a variety of biomolecules, including proteins, lipids, and nucleic acids (mRNA, miRNA, and other noncoding RNAs). These components play essential roles in modulating cell behavior and physiological processes.18–20 Recent studies have shown that exosomes, especially those derived from mesenchymal stem cells (MSCs), have the potential to promote bone regeneration.21 MSC-derived exosomes contain specific miRNAs and proteins that can accelerate osteogenic differentiation, promote angiogenesis, and inhibit inflammatory responses, which are crucial for bone healing.22–24 In preclinical models, MSC-derived exosomes have shown promise in enhancing bone healing following fractures. This is attributed to their role in stimulating osteoblast proliferation and differentiation while inhibiting osteoclast activity.25,26 They also help modulate the inflammatory environment at the fracture site, thereby promoting faster and better-quality bone healing.27
Human embryonic stem cells (hESCs) are pluripotent cells capable of differentiating into almost all cell types in the human body.28 Exosomes derived from hESCs contain a specific set of proteins, lipids, and nucleic acids that reflect their origin. The molecular cargo of these exosomes often includes pluripotency markers and other molecules associated with the unique biology of hESCs.29,30 Due to their origination from pluripotent cells, the regenerative capacities of hESC-derived exosomes have been explored. Preliminary studies suggest that they might promote tissue repair and regeneration in various contexts, including lung and liver injuries.31,32 However, despite the positive therapeutic effects of bone marrow mesenchymal stem cells on tissue damage repair and their potential role in reversing aging and age-related osteogenic dysfunction,33 the role of hESC-Exos in bone regeneration remains relatively unexplored.
This study aimed to determine the potential of hESC-Exos to facilitate bone regeneration and promote osteogenesis in an osteoporotic animal model while simultaneously exploring their impact on the proliferation and differentiation of human umbilical cord mesenchymal stem cells (hUCMSCs) in vitro. Our findings underscore the significant osteogenic potential of hESC-Exos, suggesting that they are promising candidates for therapeutic interventions in osteoporotic bone defects.
Materials and Methods
Cell Culture
Human umbilical cord samples used for the isolation of hUCMSCs were obtained from healthy newborns, with written informed consent provided by the parents. The parents were fully informed about the purpose of the research and explicitly agreed to the use of these samples for scientific studies, including the isolation and culture of hUCMSCs. The study was approved by the Medical Ethical Committee of the Affiliated Hospital of Guangdong Medical University (Approval number: PJKT2023-024). All procedures were conducted in accordance with the Declaration of Helsinki. Immediately postdelivery, the umbilical cord was harvested and preserved on ice, and sample processing was initiated within 3 hours. Wharton’s jelly was meticulously isolated, dissected into 1–2 mm² fragments, and washed with antibiotic-containing phosphate-buffered saline (PBS). The tissue fragments were plated in 100 mm culture dishes and maintained in minimum essential medium (α-MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). The medium was refreshed after an initial 6 days and subsequently changed every 3 days. Upon reaching 70–80% confluence, the cells were detached utilizing TrypLE™ Express (Gibco, Grand Island, NY, USA) and replated in a 10 cm Petri dish at a density of 2000 cells/cm². Cells from passages 2–5 were used in the study.
hESCs were obtained from the National Engineering Research Center of Human Stem Cells (Changsha, Hunan, China), utilizing cell lines from passages 29 to 40. Cultivation was conducted under serum- and feeder-free conditions, employing mTeSR™ PLUS medium (STEMCELL Technologies, Vancouver, BC, CA), and the cells were housed on Matrigel (Corning, Brooklyn, NY, USA)-coated plates within a humidified incubator at 37°C and 5% CO2. After reaching ~80% confluence, the cells were passaged as clumps with ReLeSR™ Buffer (STEMCELL Technologies) and replated on Matrigel-coated plates using mTeSR™ PLUS medium.
Characterization of hUCMSCs
Flow cytometry analysis was performed to characterize the hUCMSCs. The cells were cultured in α-MEM supplemented with 10% FBS. Approximately 1×104 cells were exposed to a panel of specific antibodies: PE-conjugated anti-human CD45, PE-conjugated anti-human CD11b, PE-conjugated anti-human CD19, APC-conjugated anti-human CD73, PE-conjugated anti-human CD34, PE-conjugated anti-human CDHLA-DR, FITC-conjugated anti-human CD90, and PerCP-conjugated anti-human CD105 (all acquired from BD Biosciences, San Jose, CA, USA). After this incubation, flow cytometry was utilized for cell analysis.
Induction and Validation of the Multilineage Differentiation of hUCMSCs
hUCMSCs were subjected to differentiation into chondrogenic, adipogenic, and osteogenic lineages utilizing lineage-specific induction media. Osteogenic differentiation was promoted with osteogenesis induction medium (OIM) containing 10 mm β-glycerophosphate, 50 μM ascorbic acid, and 10−7 M dexamethasone. Adipogenic differentiation was initiated using adipogenesis induction medium (AIM) comprising 0.2 mm indomethacin, 0.1 mg/mL insulin, 1 mm 3-isobutyl-1-methylxanthine, and 10−6 M dexamethasone (all products from Sigma–Aldrich Corp., St. Louis, MO, USA). Chondrogenic differentiation was induced using chondrogenic differentiation medium (Gibco, Grand Island, NY, USA). The cells were cultured in these differentiation media for 14, 21, or 28 days. After the differentiation period, the cells were fixed in a 4% paraformaldehyde solution and then subjected to staining procedures using Alizarin Red (osteogenesis), Oil Red O (adipogenesis), and Alcian Blue (chondrogenesis) to validate successful differentiation.
Isolation and Characterization of hESC-Exos
hESCs were maintained in mTeSRTM PULS medium. Every 48 hours, conditioned media enriched with exosomes were harvested. The collected supernatants were subjected to sequential centrifugation to exclude dead cells and cellular debris: initial centrifugation at 300 × g for 10 minutes, followed by centrifugation at 2000 × g for 10 minutes, and finally centrifugation at 10,000 × g for 30 minutes, all at 4°C. The resulting supernatant was subjected to ultracentrifugation (Beckman Coulter, Brea, CA, USA) for 1 hour at 100,000 ×g and 4°C. The obtained pellet was rinsed with PBS, followed by another round of ultracentrifugation under identical conditions. The harvested hESC-Exos were quantified using a BCA protein assay (Beyotime, Shanghai, China). To identify the isolated exosomes, Western blotting was used to evaluate the expression of typical exosome markers.
Exosome size and concentration were assessed via NTA using the Zetaview PMX 110 system (Particle Metrix, Meerbusch, Germany) in conjunction with ZetaView 8.04.02 software (Particle Metrix, Meerbusch, Germany). Exosomes were diluted in 1× PBS (dilution factor: 1:1000) and introduced into the sample holder. NTA measurements were captured and evaluated at 11 distinct positions. Prior to analysis, the ZetaView system was calibrated with 110 nm polystyrene beads.
For TEM visualization, exosomes were fixed in 2% paraformaldehyde (PFA) for 5 minutes. The exosome solution was deposited onto an EM grid, allowing a 1-minute incubation. Next, the exosomes were stained with a 1% uranyl acetate (UA) solution for 1 min atop the grid. Superfluous UA was carefully absorbed with filter paper, and the grids were air-dried under ambient conditions. Exosome morphology and integrity were evaluated using TEM (JEOL, Tokyo, Japan).
Labeling and Visualization of hESC-Exos in hUCMSCs
hESC-Exos were labeled using a PKH-67 kit (Sigma–Aldrich Corp., St. Louis, MO, USA) according to the manufacturer’s guidelines. In brief, the exosomes were exposed to 500 μL of dilution C solution and 4 μL of PKH-26 dye solution and incubated for 5 minutes at room temperature in the dark. The staining reaction was halted by adding 500 μL of 1% bovine serum albumin. After centrifugation at 100,000 × g for 70 minutes, the labeled exosomes were resuspended in 200 μL of cold PBS. hUCMSCs were cultured with these labeled exosomes in a 48-well plate for 48 hours. Subsequently, the cells were fixed with 4% paraformaldehyde, stained with phalloidin (PHA) for 1 hour, and counterstained with DAPI for 5 minutes. The samples were visualized using fluorescence microscopy (Olympus, Japan).
Western Blot Analysis
Cells were lysed utilizing RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) to obtain endogenous proteins from cell lysates. Subsequently, protein concentrations were determined with a BCA protein assay kit (Beyotime, Shanghai, China). Equal quantities of proteins (20 μg) were separated on a 10% SDS‒PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane.
For chemiluminescence detection, membranes were blocked with 5% fat-free milk at room temperature for 1–2 hours, followed by an overnight incubation with primary antibodies at 4°C. After three washes with TBST (Tris-buffered saline with Tween 20) for 5 minutes each, the membrane was exposed to horseradish peroxidase-conjugated secondary antibodies for 1 hour. After an additional three washes with TBST (10 minutes each), the protein bands were visualized utilizing a chemiluminescence detection reagent (Merck Millipore, Billerica, MA, USA) and captured on a CCD camera (Azure Biosystems C500, Dublin, CA, USA).
For fluorescence detection, the membranes were blocked with Azure Fluorescent Blot Blocking Buffer (Azure Biosystems, Dublin, CA, USA), subjected to identical incubation and washing steps as described previously, washed with TBS (Tris-buffered saline), and visualized under an Odyssey Clx camera (Azure Biosystems, Dublin, CA, USA). All the Western blot data were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
The primary antibodies used for Western blot analysis were as follows: anti-CD81 (#ab109201, Abcam), anti-CD63 (#ab134045, Abcam), anti-CD9 (#ab92726, Abcam), anti-α-tubulin (#sc-32293, Santa Cruz), anti-β-actin (#sc-47778, Santa Cruz), anti-Osteri (#ab22552, Abcam), anti-OPN (#ab8448, Abcam), anti-β-catenin (#8480, CST), anti-DVL2 (#ab277903, Abcam), anti-DVL3 (#ab290351, Abcam), anti-YAP1 (#386123, ZEN-BIOSCIENCE), anti-RUNX2 (#860139, ZEN-BIOSCIENCE), anti-ALP (#R23427, ZEN-BIOSCIENCE), and anti-OCN (#514636, ZEN-BIOSCIENCE).
Cell Transfection
hUCMSCs were plated in 6-well plates at a density of 1×105 cells/well one day before transfection. The transfection process was performed using RNAiMAX transfection reagent (Life Technologies, Carlsbad, CA, USA) and 100 pmol/mL microRNA mimic according to the manufacturer’s guidelines. The medium was replenished 8 hours posttransfection. Cells were harvested 48 hours posttransfection, and transfection efficiency was assessed using qRT‒PCR.
Luciferase Reporter Assay
Transfection was performed using Lipofectamine 2000 reagents (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocols. Briefly, luciferase reporter vectors, including pmir-report-YAP1 wt, were transfected into HEK-293T cells along with miR-21-5p or miR-NC (nontargeting control mimic). After 24 hours of transfection, the cells were lysed using passive lysis buffer, and the fluorescence intensity was evaluated with the Dual-Luciferase Reporter Assay System (Life Technologies, Carlsbad, CA, USA).
siRNA-Mediated Knockdown of YAP1 in hUCMSCs
hUCMSCs were subjected to siRNA-mediated knockdown using YAP1-specific siRNA (siYAP1) purchased from GenePharma (Shanghai, China). For transfection, Lipofectamine™ RNAiMAX reagent (Thermo Fisher Scientific, USA) was used according to the manufacturer’s instructions. After 24 hours posttransfection, the culture medium was replaced with either standard growth medium or OIM. The cells were then harvested at predetermined time points for subsequent analyses and experimental procedures.
Antagomir-21-5p Treatment
hUCMSCs were cultured in serum-free medium and treated with 100 nM antagomir-21-5p or antagomir-NC (control) in a humidified incubator with 5% CO2 for 6 hours. After treatment, the supernatant was removed, and the cells were further cultured in osteogenic induction medium for differentiation.
Icg-001 Treatment
Following the manufacturer’s instructions, we prepared a 10 mm stock solution of Icg-001 using DMSO. This stock solution was added to the osteogenic induction medium to achieve a final concentration of 25 μM. The medium was replaced every three days to ensure consistent exposure to the inhibitor.
Quantitative RT‒PCR
To quantify gene expression at the mRNA level, total RNA was extracted from cells utilizing TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The extracted RNA was then transcribed into cDNA using a first-strand cDNA synthesis kit (Vazyme, Nanjing, China). qRT‒PCR was conducted using SYBR Premix EX Taq™ II (Vazyme, Nanjing, China) according to the manufacturer’s guidelines. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and small U6 RNA served as internal reference genes for mRNA and miRNA, respectively. The primers used for qRT‒PCR are listed in Supplementary Table 1. The relative expression of genes was calculated using the 2−ΔΔCt method.34 The data are shown as the mean ± standard deviation (SD) from three replicate experiments.
miRNA Microarray Analysis
Total RNA was isolated from hESC-Exos for miRNA analysis utilizing the miRNeasy Serum/Plasma Kit (QIAGEN, Dusseldorf, Germany) according to the manufacturer’s protocol. The concentration of the retrieved total RNA was determined using a Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), and its integrity was evaluated via an Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA, USA). A small RNA library, prepared from approximately 100 ng of total RNA, was constructed using the VAHTS Small RNA Library Prep Kit for Illumina® (Vazyme Biotech), involving the ligation of specific adapters to the 3’ and 5’ termini of miRNA, siRNA, and piRNA molecules, followed by reverse transcription, PCR amplification, and purification of the PCR products. The library quality and concentration were evaluated using an Agilent 2100 Bioanalyzer implemented with a DNA 1000 chip. Quantification for sequencing was precisely determined using the KAPA Biosystems Library Quantification Kit (Kapa Biosystems, Inc., Woburn, MA, USA) based on qRT‒PCR. Subsequently, the libraries were diluted, pooled equimolarly, and subjected to clustering. Single-end sequencing, featuring a 50 bp read length, was performed on all the samples.
Preparation and Characterization of Exosome-Loaded Natural Polymer Scaffolds
The scheme for the preparation of the natural polymer-derived scaffold is shown in Supplementary Figure 1. Chitosan (Chi, Sigma–Aldrich Corp, St. Louis, MO, USA) was dissolved in 2% acetic acid at a concentration of 1% (wt%) and then homogeneously mixed with type I collagen solution (ColI, 1.5%, MingRang Biotechnology, Sichuan, China) at a volume ratio of 5:1 with magnetic stirring. The mixed solutions were then poured into either rectangular molds or 384-well plates at a volume of 100 μL per well and freeze-dried for 24 hours in a freeze-drying machine (Scientz Biotechnology, Ningbo, China). After drying, a composite scaffold composed of Chi and Col I was obtained and designated Chi/Col I. To improve the stability and mechanical strength of the composite scaffold, Chi/Col I scaffolds were further crosslinked with oxidized chondroitin sulfate (oCS), which was prepared and characterized as described previously.35 Briefly, the Chi/ColI scaffolds were immersed in 10 mg/mL oCS solution (dissolved in distilled water) and incubated overnight with gentle shaking to ensure full interaction between oCS and Chi or Col I. Thereafter, the scaffolds were thoroughly rinsed with distilled water and freeze-dried. The resulting scaffolds were denoted as [Chi/ColI]-oCS and preserved in a dry environment.
To load these scaffolds with a consistent quantity of 200 μg of exosomes, exosome solutions were adjusted to a concentration of 20 μg/μL. A volume of 10 μL of this exosome solution was then added to each scaffold. To view the microstructure and distribution of exosomes in the scaffolds, the original and exosome-loaded scaffolds were spun coated with platinum-palladium using an Ion Sputter E-100 coating system (Hitachi High-Technologies, Japan) and detected with a field emission scanning electron microscope (FE-SEM, S-4800, Hitachi High-Technologies) under ambient (air) laboratory conditions.
Animal Experiments
All animal procedures were approved by the Ethical Review Committee of Guangdong Medical University and were carried out under sterile conditions (Approval number: GDY2202200). All experiments complied with the Chinese Law on Laboratory Animal Welfare (Guideline for Ethical Review of Animal Welfare, Standard number: GB/T 35892–2018), the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the Helsinki Declaration of the World Medical Association. A total of 46 female Sprague‒Dawley (SD) rats (9 weeks old) were procured from Changsheng Biological (Benxi City, Liaoning, China). After one week of acclimatization, the rats were randomly divided into two groups: a sham group of three rats and an ovariectomized (OVX) group of 43 rats. The rats were anesthetized by intraperitoneal injection of sodium thiopental. For the OVX group, bilateral ovariectomies were conducted through double dorsolateral incisions, resulting in surgical exposure and removal of both ovaries. In the sham group, a similar procedure was followed, but equivalent amounts of fat tissue adjacent to the ovaries were removed. After surgery, the OVX rats were allowed to recover for 2 weeks in individual cages. To confirm the establishment of the osteoporosis model, femur samples were collected from both the OVX and sham groups (three rats for each group) and subjected to X-ray evaluation to confirm the successful establishment of the osteoporosis animal model.
The bone defect model was established following a previously published method with minor modifications.36 In brief, OVX rats were anesthetized by an injection of 4% avertin (1 mL/100 g), and bone defects with a diameter of 1.5 mm and depth of 3 mm were created in the knee joint using an electric drill. The rats were then randomly assigned to four groups (8 rats/group): (1) the control group; (2) the [Chi/ColI]-oCS group; (3) the [Chi/ColI]-oCS/hESCs-Exo group (200 μg); and (4) the [Chi/ColI]-oCS/hUCMSCs-Exo group (200 μg). For groups 2 to 4, exosome-loaded scaffolds were implanted directly into the bone defects. At 6 or 8 weeks postsurgery, all rats were euthanized via excessive isoflurane inhalation, and the femurs were harvested for further analysis.
Microcomputed Tomography Analysis
The left femurs of the mice were analyzed using microcomputed tomography (μCT) to assess their structural properties. The harvested femurs were fixed in 4% paraformaldehyde (PFA) for 72 hours and then scanned using a microcomputed tomography system (Scanco Medical, vivaCT 80, Switzerland) at a voltage of 70 kV and a current of 114 μA. Cross-sectional images were used for 3D trabecular bone histomorphometric analysis, with a region of interest (ROI) representing 5% of the femoral length situated 100 µm below the growth plate. Data from this ROI, including bone mineral density (BMD), trabecular thickness (Tb. Th), trabecular separation (Tb. Sp), trabecular number (Tb. N), and bone volume fraction (BV/TV), were analyzed.
Statistical Analysis
Data analysis was conducted using GraphPad Prism 8.0 software (GraphPad, La Jolla, CA, USA). The results are presented as the mean ± standard deviation (SD). Two groups were compared using the unpaired Student’s t test, while multigroup comparisons utilized one-way ANOVA, followed by Tukey’s post hoc test. A p value less than 0.05 (*P < 0.05) was considered to indicate statistical significance.
Results
Characterization of hUCMSCs
Observation of the morphology of the hUCMSCs was conducted under a light microscope after 3 days of primary isolation and cultivation, revealing a characteristic spindle-like shape (Supplementary Figure 2A). To evaluate the multilineage differentiation potential of hUCMSCs, osteogenic, adipogenic, and chondrogenic induction media were utilized. Successful differentiation into osteogenic, adipogenic, and chondrogenic lineages was demonstrated through staining with Alizarin Red, Oil Red O, and Alcian Blue, respectively (Supplementary Figure 2B–2D). Flow cytometry analysis was employed to further validate the identity of the isolated hUCMSCs and ensure their adherence to international MSC standards through the assessment of specific surface marker expression. The analysis revealed that a substantial majority (>96%) of cells expressed the MSC characteristic markers CD105, CD90, and CD73, while a minority (<2%) expressed CD45, CD34, CD11b, CD19, and HLA-DR, which are markers typically associated with hematopoietic and immune cells (Supplementary Figure 2E). Consequently, morphological observations, multilineage differentiation potential, and specific surface marker expression collectively confirmed the successful identification of hUCMSCs, substantiating their applicability as stem cells for subsequent experiments.
Isolation and Characterization of hESC-Exos
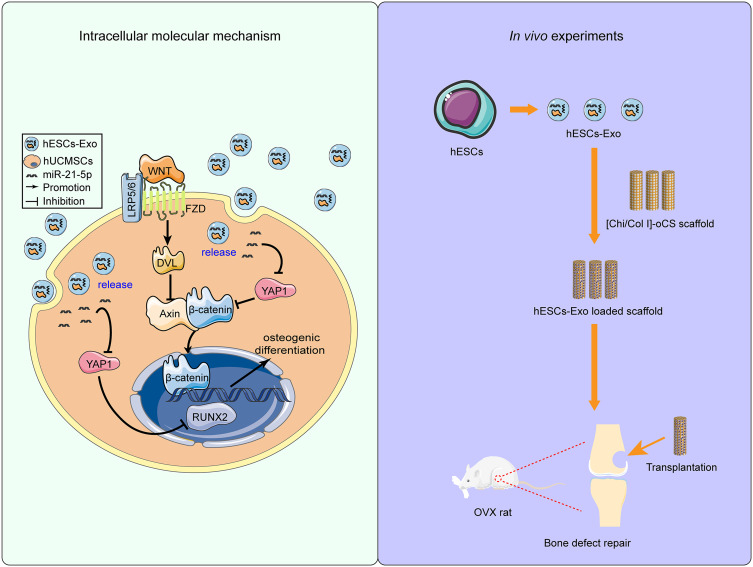
To isolate hESC-Exos from the conditioned medium of hESCs, we used ultrafast differential centrifugation. Concentrations of the hESC-Exos, ranging from 2.8 to 4.3 mg/mL, were quantified by the BCA method. Multiple experiments were conducted to identify and validate the characteristics of hESC-Exos. TEM revealed a spherical microvesicle structure of hESC-Exos, with diameters spanning approximately 50 nm to 200 nm (Figure 1A). NTA was performed to analyze the content and size distribution of the precipitate, revealing an average microvesicle diameter of approximately 164.5 nm ± 63.7 nm (Figure 1B). Furthermore, Western blot analysis revealed the expression of the exosomal surface markers CD81, CD63, and CD9 on hESC-Exos, which were not expressed on hESCs (Figure 1C). Collectively, these findings confirmed that the isolated hESC-Exos expressed characteristic exosome-specific markers and maintained the desired size and morphology.
Figure 1.
Characterization and identification of hESC-Exos. (A) Morphology of hESC-Exos visualized via TEM. The blue arrows indicate exosomes of different sizes. (B) Particle size distribution of hESC-Exos, as measured by NTA. (C) Surface markers of hESC-Exos, as determined by Western blotting.
Osteogenic Differentiation of hUCMSCs Facilitated by hESC-Exos
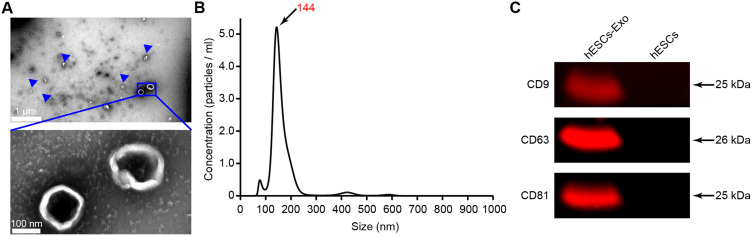
We demonstrated the uptake of PKH67-labeled hESC-Exos by hUCMSCs. Over a coincubation period of 48 hours, there was a discernible increase in the green fluorescence intensity, suggesting the progressive internalization of the labeled exosomes within the cytoplasm of the hUCMSCs. As a cellular reference, blue fluorescence, indicating nuclear staining with DAPI, and red fluorescence, indicating cytoskeletal staining, indicate contextual localization of the assimilated exosomes. These observations confirmed the ability of hUCMSCs to internalize hESC-Exos, hinting at a conceivable cellular interplay between the exosomes and their recipient cells (Figure 2A).
Figure 2.
hESC-Exos promote the osteogenic differentiation of hUCMSCs. (A) Uptake of hESC-Exos by hUCMSCs over 48 hours. The green fluorescence indicates PKH-67-labeled hESC-Exos. The hUCMSC nuclei were distinctly illuminated by blue fluorescence through DAPI staining, whereas cellular F-actin, which was stained with FITC-phalloidin, was visualized as red fluorescence. (B) Alizarin red staining of hUCMSCs after 14 days of treatment with exosomes. (C) Quantitative analysis of alizarin red staining. (D) mRNA expression levels of ALP, RUNX2, OCN, and OPN were assessed by qRT‒PCR. (E) The protein levels of OSX, ALP, OPN, RUNX2, and OCN in hUCMSCs were measured by Western blotting and statistically analyzed using ImageJ normalization, and the quantified protein levels were normalized to those of α-tubulin. The data are presented as the means ± SDs; n=3. *P<0.05, **P<0.01, ***P<0.001, compared to the control group; ‘ns’ indicates no significant difference.
Treatment with hESC-Exos notably augmented the osteogenic differentiation potential of hUCMSCs. Culturing hUCMSCs in osteogenic induction differentiation medium enriched with hESC-Exos elicited a conspicuous increase in alizarin red staining and activity, which was evident after 14 days of osteogenic induction. A concentration of 20 μg/mL of hESC-Exos had a particularly pronounced osteogenic effect compared to that of the other groups (Figure 2B and C). Moreover, hUCMSCs treated with 20 μg/mL hESC-Exos for 3 days exhibited upregulated mRNA expression levels of early osteogenesis-associated genes, namely, ALP and RUNX2. A similar pattern was observed for the late osteogenesis-related gene OCN after treatment with 20 μg/mL for 7 days. Importantly, the group treated with 20 μg/mL hESC-Exos exhibited more pronounced upregulation of osteogenesis-associated genes than both the control group and the group treated with 10 μg/mL hESC-Exos (Figure 2D). Additionally, hESC-Exo treatment increased the protein expression of OSX, ALP, OPN, RUNX2, and OCN and notably increased the protein levels of these osteogenic markers in the hESC-Exo-treated group (Figure 2E). Collectively, these findings indicate that hESC-Exos can significantly promote the osteogenic differentiation of hUCMSCs in a concentration-dependent manner.
miRNA Profiling and Validation in hESC-Exos
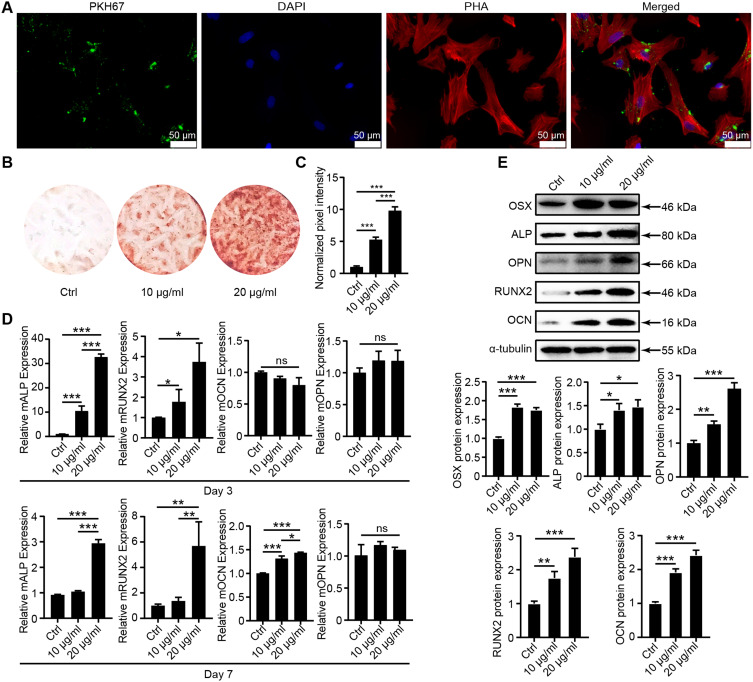
An extensive exploration of the miRNA expression profile in hESC-Exos was undertaken utilizing a miRNA microarray, revealing a varied assortment of miRNAs encased within the exosomes (Figure 3A). Subsequent sequencing data analysis, oriented toward identifying miRNAs on the premise of their abundance, revealed the top 13 miRNAs (Figure 3B). To substantiate the sequencing results, total RNA was isolated from hESC-Exos, and qRT‒PCR was used to determine the expression levels of 12 miRNAs, which were previously underscored for their high abundance in the sequencing data. Remarkably, miR-21-5p and miR-221-3p were more than 1000-fold more abundant in hESC-Exos than was miR-144-3p (Figure 3C). This intriguing pattern suggests that hESC-Exos might exert their biological effects by transporting miR-21-5p and miR-221-3p to target cells. Nevertheless, comprehensive experiments are needed to elucidate the definitive roles and intricate mechanisms associated with these miRNAs in hESC-Exos.
Figure 3.
Profiling and validation of hESC-Exo miRNAs. (A) Global miRNA expression levels standardized after miRNA microarray assessment. (B) A tabulation of the most abundant miRNAs identified through sequencing. (C) Validation of selected miRNAs via qRT‒PCR utilizing total RNA extracted from hESC-Exos. (D) Alizarin red staining of hUCMSCs after 14 days of treatment with hESC-Exos or antagomir-21-5p. “+” indicates treatment applied; “−” indicates no treatment.
To further explore the functional role of miR-21-5p in hESC-Exo-mediated bone regeneration in vitro, we used antagomir-21-5p to specifically silence miR-21-5p expression. Compared with that in the control group, the mineralization capacity of the hUCMSCs in the antagomir-21-5p group was significantly diminished (Figure 3D). These findings suggest that miR-21-5p is a crucial effector molecule in the osteogenic promotion facilitated by hESC-Exos in hUCMSCs.
Role of miR-21-5p in Enhancing Osteogenic Differentiation of hUCMSCs Induced by hESC-Exos
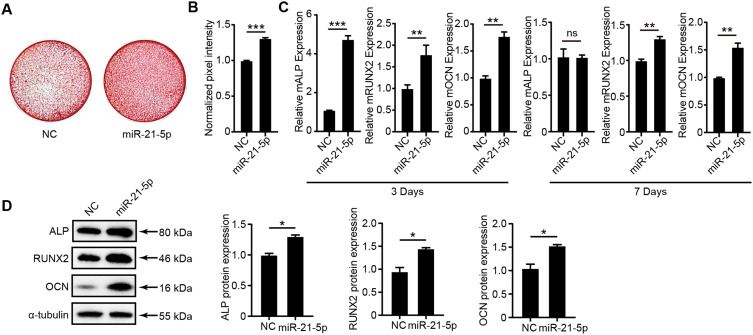
To elucidate the contribution of miR-21-5p to the osteogenic differentiation mediated by hESC-Exos, hUCMSCs were transfected with miR-21-5p mimics. After transfection, a notable increase in miR-21-5p expression was observed within 8 hours. Subsequently, we examined the potential influence of miR-21-5p on osteogenic differentiation in hUCMSCs. Alizarin red staining revealed that compared with NC treatment, miR-21-5p mimic treatment notably intensified osteogenic differentiation (Figure 4A and B). Complementary qRT‒PCR analyses on day 3 revealed increased mRNA expression of ALP, RUNX2, and OCN in the miR-21-5p mimic cohort. By day 7, the RUNX2 and OCN mRNA concentrations remained distinctly elevated in the miR-21-5p mimic group relative to those in the mimic NC group (Figure 4C). Western blot assays further confirmed a marked increase in the protein expression levels of ALP, RUNX2, and OCN in the miR-21-5p mimic group (Figure 4D). Taken together, these findings underscore the pivotal role of miR-21-5p in bolstering the osteogenic differentiation of hUCMSCs orchestrated by hESC-Exos.
Figure 4.
The role of miR-21-5p in promoting the osteogenic differentiation of hUCMSCs. (A) Alizarin red staining illustrating osteogenic differentiation in hUCMSCs posttreatment with miR-21-5p mimics or NC mimics (Ctrl) over an 8-hour span. (B) Quantitative assessment of alizarin red staining intensity via Photoshop software. (C) mRNA expression levels of ALP, RUNX2, and OCN determined by qPCR. (D) The protein levels of ALP, RUNX2, and OCN in hUCMSCs were quantified by Western blotting. The analysis was anchored by ImageJ normalization, with protein quantifications standardized against α-tubulin levels. The data are presented as the means ± SDs; n=3; *P<0.05, **P<0.01, ***P<0.001, compared to the NC group; ‘ns’ indicates no significant difference.
Influence of miR-221-3p on the Osteogenic Differentiation of hUCMSCs
After transfection of miR-221-3p mimics into hUCMSCs, a notable increase in calcification deposition was detected compared to that in the NC-transfected control group, as evidenced by alizarin red staining after 14 days of osteogenic induction (Supplementary Figure 3A and B). qRT‒PCR analyses revealed a significant increase in the mRNA levels of ALP and OCN at both 3 and 7 days postosteogenic induction in the miR-221-3p transfection group, while the RUNX2 mRNA levels remained unaffected (Supplementary Figure 3C). In contrast, Western blot analyses did not reveal any significant differences in the protein expression of ALP, RUNX2, or OCN between the miR-221-3p-transfected and NC groups after the 14-day osteogenic induction period (Supplementary Figure 3D). These findings suggest that miR-221-3p may modulate calcification deposition during the osteogenic differentiation of hUCMSCs at the mRNA level; however, its influence on protein expression warrants further exploration.
Mechanistic Role of miR-21-5p in Regulating Osteogenic Differentiation via YAP1 Downregulation
Based on the sequencing results above, we hypothesized that hESC-Exos might chiefly regulate the osteogenic differentiation of hUCMSCs through the conveyance of miR-21-5p. Computational target identification for miRNAs was performed by employing the web-based algorithms TargetScan and miRanda. Notably, YAP1 not only achieved a relatively high score among the projected results but also exhibited an association with the osteogenic differentiation-initiating transcription factor RUNX2.37 However, the underlying mechanism, particularly whether miR-21-5p mediates the osteogenic differentiation of hUCMSCs by targeting YAP1, remains to be elucidated (Figure 5A).
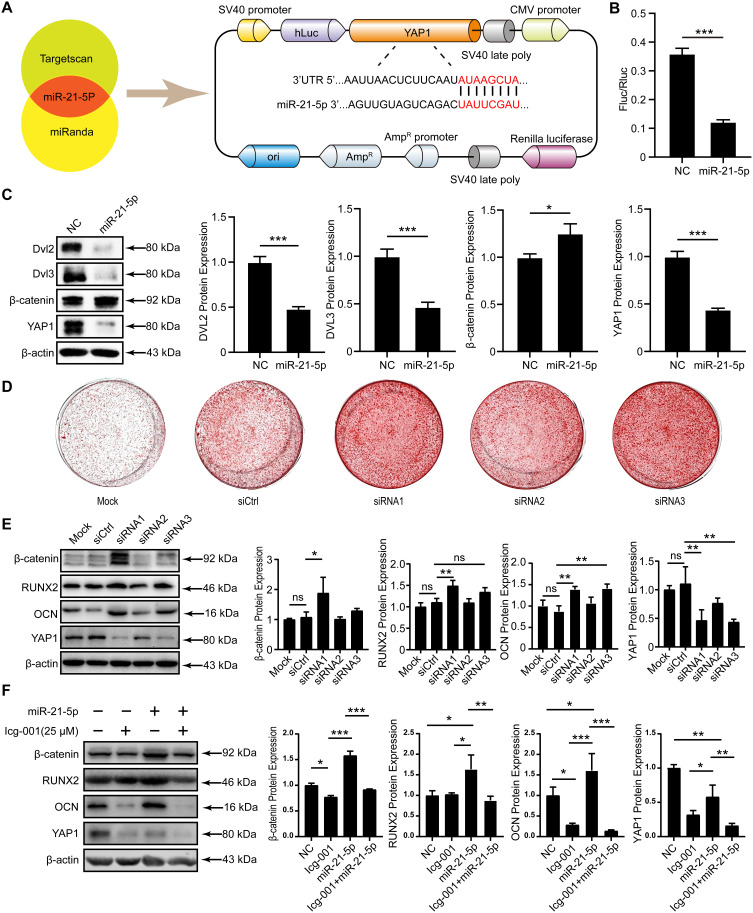
Figure 5.
Investigation of miR-21-5p targeting of YAP1 and its subsequent influence on the osteogenic differentiation of hUCMSCs. (A) The TargetScan and miRanda online databases were used to predict potential miR-21-5p target genes and construct plasmids harboring the YAP1 3’UTR. (B) Cotransfection of the plasmid and miR-21-5p into 293T cells, followed by verification of the targeting relationship between miR-21-5p and YAP1 via a dual-luciferase reporter assay. (C) The protein expression levels of DVL2, DVL3, β-catenin, and YAP1 in hUCMSCs were quantified by Western blotting and statistically analyzed by ImageJ normalization, and the protein levels were quantified by normalization to the level of β-actin. (D) Representative images of alizarin red staining following 14 days of osteogenic induction demonstrating increased calcification in the siRNA-treated group compared to the siCtrl group. (E) Western blot analysis of β-catenin, RUNX2, OCN, and YAP1 protein levels in hUCMSCs, with quantification normalized to β-actin using ImageJ software. (F) Western blot analysis and quantification of β-catenin, RUNX2, OCN, and YAP1 protein levels in hUCMSCs, normalized to β-actin. The data are presented as the means ± SDs; n=3; *P<0.05, **P<0.01, ***P<0.001 relative to the NC group or siCtrl group; “ns” indicates no significant difference.
To verify the direct targeting of YAP1 by miR-21-5p, we transfected 293T cells with luciferase reporter constructs containing the putative 3ʹUTR of YAP1. A marked decrease in luciferase activity after incubation with miR-21-5p indicated a strong interaction between YAP1 and miR-21-5p (Figure 5B). Following miR-21-5p transfection into hUCMSCs and induction of osteogenic differentiation for 14 days, Western blotting revealed pronounced YAP1 downregulation in the miR-21-5p mimic group. Concurrently, the levels of β-catenin, which is integral to the Wnt pathway, increased, while the levels of Dvl2 and Dvl3, which are markers of alternative Wnt pathway activation, decreased substantially (Figure 5C).
In further experiments, to ascertain whether the influence of miR-21-5p on the osteogenic differentiation of hUCMSCs was mediated by YAP1, knockdown experiments were conducted using a YAP1-specific siRNA (siYAP1). After one day of siYAP1 exposure, the culture medium was replaced with MEM-Alpha. Subsequently, the medium was transitioned to OIM, and the cells were induced for 14 days. Alizarin red staining revealed that compared with siCtrl, siRNA substantially enhanced osteogenic differentiation (Figure 5D). Western blot analysis demonstrated a substantial reduction in YAP1 protein levels in the siYAP1 group, accompanied by a corresponding increase in β-catenin and RUNX2 levels (Figure 5E). These findings indicate that miR-21-5p might enhance the osteogenic differentiation of hUCMSCs by activating the Wnt pathway via YAP1 downregulation.
To further confirm whether the osteogenic effects of miR-21-5p are mediated through WNT pathway activation, hUCMSCs transfected with the miR-21-5p mimic were treated with osteogenic induction medium, either with or without the WNT pathway inhibitor Icg-001 (25 μM). Western blot analysis revealed that in the Icg-001-treated group, the protein levels of RUNX2, OCN, and YAP1 were significantly elevated, whereas β-catenin levels were markedly reduced, indicating successful inhibition of the WNT pathway (Figure 5F). These results strongly indicate that the osteogenic promotion effect of miR-21-5p on hUCMSCs is mediated primarily through the activation of the WNT signaling pathway.
Integration and Surface Characterization of Exosome-Loaded Scaffolds
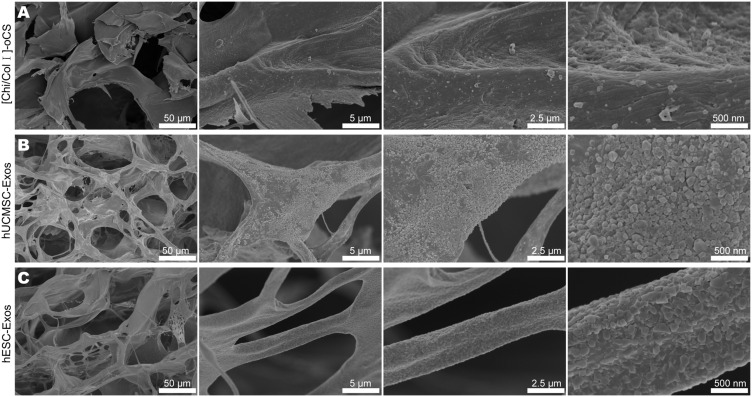
To assess the potential of exosomes for in vivo bone defect repair, we integrated two distinct exosome types into individual scaffolds. Specifically, we soaked [Chi/Col I]-oCS scaffolds in exosome solutions to ensure complete absorption before performing scanning electron microscopy (SEM) to analyze their surface topography. Notably, all the scaffolds maintained a well-interconnected 3D pore structure after freeze-drying. However, it is worth noting that a more uniform and smooth surface structure was found for the pure [Chi/Col I]-oCS scaffold (Figure 6A). In contrast, a somewhat rougher surface structure with homogenous and dense nanoparticles was observed on both the hUCMSC-Exo- and hESC-Exo-loaded scaffolds (Figure 6B and C). These nanoparticles exhibited a typical sphere-shaped structure with a diameter of approximately 150 nm, indicating successful exosome loading.
Figure 6.
SEM characterization of [Chi/Col I]-oCS scaffolds laden with exosomes. (A) SEM image highlighting the surface morphology of the pure [Chi/Col I]-oCS scaffold. (B) SEM image of the [Chi/Col I]-oCS scaffold postloaded with hUCMSC-Exos. (C) SEM image of the surface topography of the [Chi/Col I]-oCS scaffold postloaded with hESC-Exos.
Vivo Efficacy of hESC-Exos in Bone Defect Repair
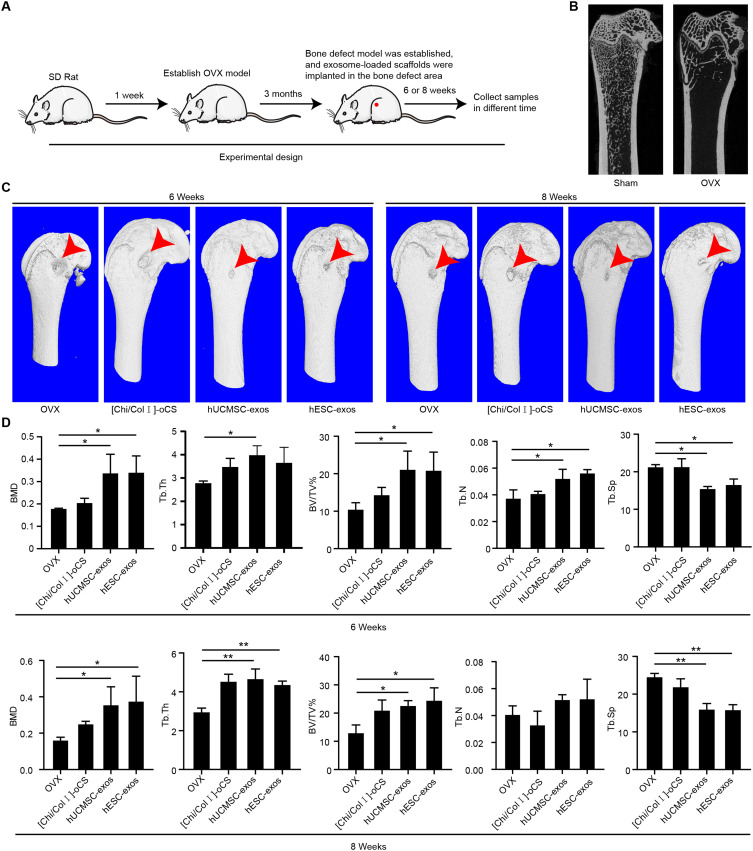
To elucidate the therapeutic potential of hESC-Exos for bone repair in vivo, a series of animal experiments were conducted (Figure 7A). Three months postbilateral ovariectomy, a substantial reduction in cancellous bone was observed in the OVX group, indicating the successful induction of osteoporosis (Figure 7B). Wang et al demonstrated that hUCMSC-Exos, when coated with hydrogels, effectively promoted rat bone defect healing;38 thus, hUCMSC-Exos were selected as a control in this study. At 6 and 8 weeks following bone defect repair, 3D reconstructed images revealed notable differences in bone defect repair among the different groups (Figure 7C). Both the hESC-Exo- and hUCMSC-Exo-loaded scaffolds exhibited enhanced bone healing, and no significant differences were found between them. Furthermore, both the hESC-Exo and hUCMSC-Exo groups displayed elevated BMD, BV/TV, Tb.Th, and Tb.N values in comparison to those of the OVX and [Chi/Col I]-oCS groups, while the Tb.Sp values were lower (Figure 7D). These findings demonstrated the efficacy of hESC-Exos in treating bone defects in vivo.
Figure 7.
Therapeutic effects of hESC-Exos on bone defect repair in OVX rats. (A) Schematic representation of the animal experimental design. (B) Micro-CT detection of femoral spongy bone content three months after bilateral ovary removal. The sham group, which underwent identical adipose tissue removal adjacent to the ovaries, served as a control. (C) Three-dimensional microCT reconstruction of each group’s bone defect site at 6 and 8 weeks postrepair, with arrows indicating the location of the bone defects. (D) Quantitative analyses of BMD, BV/TV, Tb.N, Tb.Th, and Tb.Sp for each group at 6 and 8 weeks following bone defect repair. The data are presented as the means ± SDs; n=3; *P<0.05, **P<0.01, compared to the OVX group.
Discussion
Postmenopausal osteoporosis, which is predominantly characterized by osteoporotic defects, remains a considerable therapeutic challenge. Existing treatments such as bisphosphonates, calcitonin, and selective estrogen receptor modulators primarily emphasize the prevention and management of osteoporosis.39 Autogenous bone grafting, although regarded as the gold standard for bone defect treatment, has inherent limitations, such as donor site morbidity and limited bone supply.40,41 These constraints underscore the urgent need for innovative strategies that not only bolster bone regeneration but also inhibit bone loss and minimize side effects. In this context, our research demonstrated the exceptional osteogenic potential of hESC-Exos both in vitro and in vivo.
Stem cell therapy has been lauded for its immense potential in bone tissue repair and regeneration, with osteoporosis being a primary target.42,43 Among stem cell types, hUCMSCs are of particular significance due to their advantages, including easy accessibility, noninvasive procurement, robust immunomodulatory properties, and enhanced self-renewal capabilities.44,45 Therefore, focusing on the osteogenic differentiation of hUCMSCs seems to be a promising approach for clinical bone tissue repair. Our findings emphasize the pronounced osteogenic differentiation of hUCMSCs, which is further potentiated by hESC-Exos, as indicated by the expression of key markers such as ALP, RUNX2, and OCN.
Exosomes have recently been recognized as critical modulators of particular cellular functions, chiefly owing to their capacity to transfer bioactive proteins and miRNAs.46 The Wnt/β-catenin pathway, which encompasses extracellular signals and membrane, cytoplasmic, and nuclear segments, is integral in this context. Predominantly, Wnt proteins mediate extracellular signals, while the membrane segment principally consists of Wnt receptors such as FZD (specifically, the sevenfold transmembrane receptor, Frizzled protein) and LRP5/6. The cytoplasmic segment notably includes β-catenin, DVL, and AXIN. When Wnts are recognized by FZD and LRP5/6, the “destruction complex”, which loses its ability to degrade β-catenin, is recruited to the cell membrane through interaction with FZD. Consequently, induced β-catenin translocates to the nucleus, activating the transcription of target genes.47 Our study revealed a complex mechanism following the internalization of hESC-Exos by hUCMSCs. On the one hand, miR-21-5p in hESC-Exos, through a sponge effect, targets YAP1 to inhibit its expression, subsequently promoting the activity of the Wnt/β-catenin pathway, which in turn leads to an increase in the osteogenic differentiation ability of the cells. On the other hand, reduced expression of YAP1 might directly facilitate the transcriptional activity of RUNX2, thereby promoting the osteogenic differentiation process. Specifically, miR-21-5p has been previously shown to influence osteogenic differentiation.48 An increase in the expression of miR-21-5p in hUCMSCs led to an increase in the expression of osteogenic markers and enhanced calcification.
To comprehensively evaluate the therapeutic potential of hESC-Exos in vivo, a series of animal experiments were performed. The significant reduction in cancellous bone three months postbilateral ovariectomy in OVX rats reaffirmed the induction of osteoporosis, consistent with our preliminary hypotheses. We aimed to further explore the effectiveness of hydrogel-coated hUCMSC-Exos in promoting rat bone defect healing, as highlighted in a previous study.38 Importantly, 3D reconstructed images of rat femurs after bone defect repair revealed significant differences in the healing processes among the different experimental groups. Although both the hESC-Exo and hUCMSC-Exo groups exhibited enhanced bone repair, the differences were not significant, and a closer examination highlighted the unique advantages associated with hESC-Exos. These advantages are rooted in the fact that hESCs, the source of hESC-Exos, possess remarkable characteristics, including the ability to indefinitely pass through. Consequently, exosomes derived from hESCs maintain a high degree of uniformity in terms of quality and properties. On the other hand, exosomes derived from hUCMSCs might face challenges in maintaining consistent quality and characteristics due to individual variations in umbilical cord-derived material and potential differences between batches. As a result, exosomes derived from hESCs offer distinct advantages, making them more promising for clinical applications.
The regulatory influence of miR-21-5p on YAP1, as delineated in our study, resonates with past research pointing to the multifaceted roles of YAP and TAZ in osteoblast differentiation.49 In a paradigm shift, YAP and TAZ act as nuclear relays, translating mechanical signals from extracellular matrix rigidity and cell morphology into biochemical cues.50 An interaction between YAP and RUNX2, as evidenced in our work and others, appears pivotal in regulating mesenchymal stem cell osteogenic differentiation, potentially influenced by external pathways such as Wnt signaling.51–53
Our findings highlighted the prominent role of miR-21-5p in augmenting the osteogenic differentiation of hUCMSCs, revealing a novel miR-21-5p-YAP1/β-catenin axis that is a promising arena for activating the Wnt signaling pathway, which is crucial for the osteogenic differentiation of hUCMSCs.
Lineage decisions emanating from mesenchymal stem cells into osteogenic and adipogenic spectra are finely modulated, as detailed by studies elucidating the crucial role of SOX2 in these processes.51 Further research has shed light on how pathways such as Wnt signaling are modulated by other effectors, accentuating the nuanced nature of these interactions.54–56 However, our study has several limitations that should be addressed in future work. First, we utilized only one type of mesenchymal stem cell (hUCMSC) for the in vitro experiments, which may limit the generalizability of our findings. Future studies should consider examining the effects of hESC-Exos on other types of mesenchymal stem cells (eg, bone marrow-derived MSCs) and primary osteoblasts to evaluate whether the observed osteogenic effects are consistent across different cell types. This broader analysis could provide a more comprehensive understanding of the therapeutic potential of hESC-Exos in bone tissue engineering. Additionally, while our in vivo experiments demonstrated that hESC-Exos directly enhance bone healing at the site of bone defects in mice, the long-term safety and efficacy of using hESC-Exos in clinical settings remain to be investigated. Future research should include extended in vivo studies with larger animal models to assess the durability of bone repair and to monitor for potential adverse effects.
In the realm of tissue repair, exosomes have been proven to be potent vectors, as multiple studies have substantiated.57,58 Prior research emphasizing the efficacy of hydrogels combined with hUCMSC-derived exosomes in repairing bone defects in rats sets the stage for our exploration.38 We elucidated the synergistic effect of hESC-Exos and [Chi/Col I]-oCS, culminating in a composite scaffold that significantly enhances bone formation.
Conclusion
In conclusion, our study underscores the crucial role of hESC-Exos in promoting the osteogenic differentiation of hUCMSCs. We demonstrated both the in vitro and in vivo efficacy of hESC-Exos, particularly in enhancing bone healing in an osteoporotic rat model, and provided mechanistic insights into the function of miR-21-5p. Our findings highlight the importance of the miR-21-5p-YAP1/β-catenin axis, a key regulatory pathway that activates Wnt signaling and drives the osteogenic differentiation of hUCMSCs. Overall, our findings suggest that targeting the miR-21-5p-YAP1/β-catenin axis through hESC-Exos represents a novel strategy for enhancing osteogenic differentiation. This pathway offers a promising direction for the development of exosome-based therapies for bone defects, warranting further exploration and optimization in future research.
Acknowledgments
This work was supported by the Affiliated Hospital of Guangdong Medical University “Clinical Medicine +” CnTech Coconstruction Platform Foundation (No. CLP2021A001), the Competitive Allocation Project of Special Funds for Science and Technology Development in Zhanjiang City (No. 2022A01198), and the High-Level Talent Research Start-Up Fund of the Affiliated Hospital of Guangdong Medical University (No. GCC2022014).
Disclosure
The author(s) report no conflicts of interest in this work.
References
- 1.Clynes MA, Harvey NC, Curtis EM, Fuggle NR, Dennison EM, Cooper C. The epidemiology of osteoporosis. Br Med Bull. 2020;133(1):105–117. doi: 10.1093/bmb/ldaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu HY, Wu AT, Tsai CY, et al. The balance between adipogenesis and osteogenesis in bone regeneration by platelet-rich plasma for age-related osteoporosis. Biomaterials. 2011;32(28):6773–6780. doi: 10.1016/j.biomaterials.2011.05.080 [DOI] [PubMed] [Google Scholar]
- 3.Sozen T, Ozisik L, Basaran NC. An overview and management of osteoporosis. Eur J Rheumatol. 2017;4(1):46–56. doi: 10.5152/eurjrheum.2016.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Go M, Shin E, Jang SY, Nam M, Hwang GS, Lee SY. BCAT1 promotes osteoclast maturation by regulating branched-chain amino acid metabolism. Exp Mol Med. 2022;54(6):825–833. doi: 10.1038/s12276-022-00775-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu R, Zeng Q, Xia C, et al. Fractions of Shen-Sui-Tong-Zhi formula enhance osteogenesis via activation of beta-Catenin signaling in growth plate chondrocytes. Front Pharmacol. 2021;12:711004. doi: 10.3389/fphar.2021.711004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah FA, Ruscsak K, Palmquist A. 50 years of scanning electron microscopy of bone-a comprehensive overview of the important discoveries made and insights gained into bone material properties in health, disease, and taphonomy. Bone Res. 2019;7:15. doi: 10.1038/s41413-019-0053-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377(9773):1276–1287. doi: 10.1016/S0140-6736(10)62349-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng S, Zhou C, Yang H, et al. Melatonin accelerates osteoporotic bone defect repair by promoting osteogenesis-angiogenesis coupling. Front Endocrinol. 2022;13:826660. doi: 10.3389/fendo.2022.826660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCann RM, Colleary G, Geddis C, et al. Effect of osteoporosis on bone mineral density and fracture repair in a rat femoral fracture model. J Orthop Res. 2008;26(3):384–393. doi: 10.1002/jor.20505 [DOI] [PubMed] [Google Scholar]
- 10.Downey C, Kelly M, Quinlan JF. Changing trends in the mortality rate at 1-year post hip fracture - a systematic review. World J Orthop. 2019;10(3):166–175. doi: 10.5312/wjo.v10.i3.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White DK, Tudor-Locke C, Felson DT, et al. Do radiographic disease and pain account for why people with or at high risk of knee osteoarthritis do not meet physical activity guidelines? Arthritis Rheum. 2013;65(1):139–147. doi: 10.1002/art.37748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manoochehri H, Ghorbani M, Moosazadeh Moghaddam M, Nourani MR, Makvandi P, Sharifi E. Strontium doped bioglass incorporated hydrogel-based scaffold for amplified bone tissue regeneration. Sci Rep. 2022;12(1):10160. doi: 10.1038/s41598-022-14329-0 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Geraerds A, Haagsma JA, de Munter L, Kruithof N, de Jongh M, Polinder S. Medical and productivity costs after trauma. PLoS One. 2019;14(12):e0227131. doi: 10.1371/journal.pone.0227131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singaram S, Naidoo M. The physical, psychological and social impact of long bone fractures on adults: a review. Afr J Prim Health Care Fam Med. 2019;11(1):e1–e9. doi: 10.4102/phcfm.v11i1.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Liu Q, Guo H, Liang J, Zhang Y. Overview of physical and pharmacological therapy in enhancing bone regeneration formation during distraction osteogenesis. Front Cell Dev Biol. 2022;10:837430. doi: 10.3389/fcell.2022.837430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahan S, Mukherjee S, Ali S, et al. Pioneer role of extracellular vesicles as modulators of cancer initiation in progression, drug therapy, and vaccine prospects. Cells. 2022;11(3):490. doi: 10.3390/cells11030490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo MH, Lee AR, Moon KS. Characteristics of extracellular vesicles and preclinical testing considerations prior to clinical applications. Biomedicines. 2022;10(4):869. doi: 10.3390/biomedicines10040869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurunathan S, Kang MH, Song H, Kim NH, Kim JH. The role of extracellular vesicles in animal reproduction and diseases. J Anim Sci Biotechnol. 2022;13(1):62. doi: 10.1186/s40104-022-00715-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasser C, Alikhani VS, Ekstrom K, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan SHS, Wong JRY, Sim SJY, et al. Mesenchymal stem cell exosomes in bone regenerative strategies-a systematic review of preclinical studies. Mater Today Bio. 2020;7:100067. doi: 10.1016/j.mtbio.2020.100067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang W, Han B, Hai Y, Sun D, Yin P. Mechanism of action of mesenchymal stem cell-derived exosomes in the intervertebral disc degeneration treatment and bone repair and regeneration. Front Cell Dev Biol. 2021;9:833840. doi: 10.3389/fcell.2021.833840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Cao H, Hua W, et al. Mesenchymal stem cell-derived extracellular vesicles for bone defect repair. Membranes. 2022;12(7):716. doi: 10.3390/membranes12070716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Wu S, Liu W, Wang L, Dong M, Niu W. microRNAs delivered by small extracellular vesicles in MSCs as an emerging tool for bone regeneration. Front Bioeng Biotechnol. 2023;11:1249860. doi: 10.3389/fbioe.2023.1249860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerami MH, Khorram R, Rasoolzadegan S, et al. Emerging role of mesenchymal stem/stromal cells (MSCs) and MSCs-derived exosomes in bone- and joint-associated musculoskeletal disorders: a new frontier. Eur J Med Res. 2023;28(1):86. doi: 10.1186/s40001-023-01034-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Mai Z, Cui L, Zhao X. Engineering exosomes and biomaterial-assisted exosomes as therapeutic carriers for bone regeneration. Stem Cell Res Ther. 2023;14(1):55. doi: 10.1186/s13287-023-03275-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hade MD, Suire CN, Suo Z. Mesenchymal stem cell-derived exosomes: applications in regenerative medicine. Cells. 2021;10(8):1959. doi: 10.3390/cells10081959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosseini K, Lekholm E, Ahemaiti A, Fredriksson R. Differentiation of human embryonic stem cells into neuron, cholinergic, and glial cells. Stem Cells Int. 2020;2020:8827874. doi: 10.1155/2020/8827874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belair C, Sim S, Kim KY, et al. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J Cell Biol. 2019;218(8):2564–2582. doi: 10.1083/jcb.201811148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi Y, Qiao X, Liu Q, et al. Systemic proteomics and miRNA profile analysis of exosomes derived from human pluripotent stem cells. Stem Cell Res Ther. 2022;13(1):449. doi: 10.1186/s13287-022-03142-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montay-Gruel P, Zhu Y, Petit B, et al. Extracellular vesicles for the treatment of radiation-induced normal tissue toxicity in the lung. Front Oncol. 2020;10:602763. doi: 10.3389/fonc.2020.602763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang N, Li X, Zhong Z, et al. 3D hESC exosomes enriched with miR-6766-3p ameliorates liver fibrosis by attenuating activated stellate cells through targeting the TGFbetaRII-SMADS pathway. J Nanobiotechnol. 2021;19(1):437. doi: 10.1186/s12951-021-01138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong L, Chen B, Zhang J, et al. Human ESC-sEVs alleviate age-related bone loss by rejuvenating senescent bone marrow-derived mesenchymal stem cells. J Extracell Vesicles. 2020;9(1):1800971. doi: 10.1080/20013078.2020.1800971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi: 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- 35.Zhao M, Li L, Zhou C, et al. Improved stability and cell response by intrinsic cross-linking of multilayers from collagen I and oxidized glycosaminoglycans. Biomacromolecules. 2014;15(11):4272–4280. doi: 10.1021/bm501286f [DOI] [PubMed] [Google Scholar]
- 36.Zhao R, Chen S, Zhao W, et al. A bioceramic scaffold composed of strontium-doped three-dimensional hydroxyapatite whiskers for enhanced bone regeneration in osteoporotic defects. Theranostics. 2020;10(4):1572–1589. doi: 10.7150/thno.40103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peruzzi B, Cappariello A, Del Fattore A, Rucci N, De Benedetti F, Teti A. c-Src and IL-6 inhibit osteoblast differentiation and integrate IGFBP5 signalling. Nat Commun. 2012;3:630. doi: 10.1038/ncomms1651 [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Wang J, Zhou X, et al. A new self-healing hydrogel containing hucMSC-derived exosomes promotes bone regeneration. Front Bioeng Biotechnol. 2020;8:564731. doi: 10.3389/fbioe.2020.564731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng N, Dai J, Cheng X, et al. Porous CaP/silk composite scaffolds to repair femur defects in an osteoporotic model. J Mater Sci Mater Med. 2013;24(8):1963–1975. doi: 10.1007/s10856-013-4945-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migliorini F, Cuozzo F, Torsiello E, Spiezia F, Oliva F, Maffulli N. Autologous bone grafting in trauma and orthopaedic surgery: an evidence-based narrative review. J Clin Med. 2021;10(19):4347. doi: 10.3390/jcm10194347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gholami L, Nooshabadi VT, Shahabi S, et al. Extracellular vesicles in bone and periodontal regeneration: current and potential therapeutic applications. Cell Biosci. 2021;11(1):16. doi: 10.1186/s13578-020-00527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antebi B, Pelled G, Gazit D. Stem cell therapy for osteoporosis. Curr Osteoporos Rep. 2014;12(1):41–47. doi: 10.1007/s11914-013-0184-x [DOI] [PubMed] [Google Scholar]
- 43.Hao ZC, Wang SZ, Zhang XJ, Lu J. Stem cell therapy: a promising biological strategy for tendon-bone healing after anterior cruciate ligament reconstruction. Cell Prolif. 2016;49(2):154–162. doi: 10.1111/cpr.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Omar R, Beroud J, Stoltz JF, Menu P, Velot E, Decot V. Umbilical cord mesenchymal stem cells: the new gold standard for mesenchymal stem cell-based therapies? Tissue Eng Part B Rev. 2014;20(5):523–544. doi: 10.1089/ten.TEB.2013.0664 [DOI] [PubMed] [Google Scholar]
- 45.Abbaszadeh H, Ghorbani F, Derakhshani M, Movassaghpour A, Yousefi M. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles: a novel therapeutic paradigm. J Cell Physiol. 2020;235(2):706–717. doi: 10.1002/jcp.29004 [DOI] [PubMed] [Google Scholar]
- 46.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Xiao Q, Xiao J, et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3. doi: 10.1038/s41392-021-00762-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian F, Zhao C, Qu J, et al. Icariin attenuates titanium particle-induced inhibition of osteogenic differentiation and matrix mineralization via miR-21-5p. Cell Biol Int. 2018;42(8):931–939. doi: 10.1002/cbin.10957 [DOI] [PubMed] [Google Scholar]
- 49.Dupont S, Morsut L, Aragona M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 50.Tang Y, Feinberg T, Keller ET, Li XY, Weiss SJ. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat Cell Biol. 2016;18(9):917–929. doi: 10.1038/ncb3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo E, Basu-Roy U, Gunaratne PH, et al. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013;3(6):2075–2087. doi: 10.1016/j.celrep.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng Y, Wu A, Li P, et al. Yap1 regulates multiple steps of chondrocyte differentiation during skeletal development and bone repair. Cell Rep. 2016;14(9):2224–2237. doi: 10.1016/j.celrep.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 53.Ying J, Wang P, Zhang S, et al. Transforming growth factor-beta1 promotes articular cartilage repair through canonical Smad and Hippo pathways in bone mesenchymal stem cells. Life Sci. 2018;192:84–90. doi: 10.1016/j.lfs.2017.11.028 [DOI] [PubMed] [Google Scholar]
- 54.Huo KL, Yang TY, Zhang WW, Shao J. Mesenchymal stem/stromal cells-derived exosomes for osteoporosis treatment. World J Stem Cells. 2023;15(3):83–89. doi: 10.4252/wjsc.v15.i3.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Y, Li X, Zhang Q, et al. Exosome-guided bone targeted delivery of Antagomir-188 as an anabolic therapy for bone loss. Bioact Mater. 2021;6(9):2905–2913. doi: 10.1016/j.bioactmat.2021.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Liu X, Li H, et al. Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway. Stem Cell Res Ther. 2016;7(1):136. doi: 10.1186/s13287-016-0391-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Guan J, Niu X, et al. Exosomes released from human induced pluripotent stem cells-derived MSCs facilitate cutaneous wound healing by promoting collagen synthesis and angiogenesis. J Transl Med. 2015;13:49. doi: 10.1186/s12967-015-0417-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Chopp M, Meng Y, et al. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J Neurosurg. 2015;122(4):856–867. doi: 10.3171/2014.11.JNS14770 [DOI] [PMC free article] [PubMed] [Google Scholar]