Abstract
Membrane proteins targeted to the plasma membrane are first transported from the endoplasmic reticulum (ER) to the Golgi apparatus. This study explored the mechanisms controlling plasma membrane trafficking of the boric acid channel AtNIP5;1 from the ER. Imaging-based screening using transgenic Arabidopsis identified six mutants in which GFP-NIP5;1 was localized in the ER in addition to the plasma membrane. Genetic mapping and whole-genome resequencing identified the responsible gene in four among the six mutants as KAONASHI3 (KNS3)/SPOTTY1/IMPERFECTIVE EXINE FORMATION. Among the plasma membrane-localized proteins tested, NIP5;1 and its homolog NIP6;1 were retained in the ER of the kns3 mutants. Our genetic analysis further discovered that two homologs of KNS3, KNSTH1 and KNSTH2, were also involved in the ER exit of NIP5;1. In Arabidopsis protoplasts and tobacco leaves, mCherry-fused KNS3 localized to the ER and Golgi, whereas KNSTH2 localized to the ER. The cytosolic C-terminal tail of KNS3 contains amino acids important for Golgi-to-ER trafficking. Furthermore, the ER-to-Golgi trafficking of KNS3 depended on KNSTH1 and KNSTH2, and the accumulation of these three proteins in Arabidopsis roots depended on each other. We propose that KNS3, KNSTH1, and KNSTH2 function as a cargo-receptor complex mediating the ER exit of NIP5;1.
Keywords: Arabidopsis thaliana, boric acid channel, cargo receptor complex, endoplasmic reticulum exit, KNS3/SPOT1/IEF, membrane trafficking
KAONASHI3 (KNS3)/SPOTTY1/IMPERFECTIVE EXINE FORMATION and its two homologs function cooperatively in the endoplasmic reticulum-to-plasma membrane traffic of boric acid channels as a potential cargo-receptor complex in Arabidopsis thaliana .
Introduction
Membrane proteins destined for the plasma membrane (PM) are co-translationally inserted into the membrane of the endoplasmic reticulum (ER) and then transported through the Golgi stacks and the trans-Golgi network to the PM. Trafficking between the ER and Golgi apparatus involves Coat Protein I (COPI)- and COPII-coated vesicles. COPI-coated vesicles mediate retrograde trafficking between the Golgi stacks and cis-Golgi to the ER, whereas COPII-coated vesicles mediate trafficking from the ER exit sites to the Golgi. COPII comprises five proteins, SAR1, SEC23, SEC24, SEC13, and SEC31. Among these components, SEC24 directly interacts with the signals exposed on the cytosolic side of cargo proteins. This behavior makes SEC24 an important factor in cargo selection for COPII vesicles (Béthune and Wieland, 2018; Brandizzi, 2018). Cargo proteins do not necessarily have signals that are recognized by SEC24. Cargo receptors interact with cargo proteins and SEC24 to facilitate the ER exit of the cargo proteins (Gomez-Navarro and Miller, 2016; Béthune and Wieland, 2018).
In plants, there is limited information on the cargo receptors for trafficking between the ER and Golgi apparatus. p24 proteins interact with the K/HDEL receptor ER RETENTION DEFECTIVE2 and facilitate the retrograde transport of ER RETENTION DEFECTIVE2 and K/HDEL ligands from the Golgi apparatus to the ER in Arabidopsis thaliana (Montesinos et al., 2014; Pastor-Cantizano et al., 2017, 2018). CORNICHON HOMOLOG (CNIH)1 functions as a possible cargo receptor for trafficking the sodium transporter OsHKT1;3 from the ER to the Golgi apparatus in rice (Oryza sativa) (Rosas-Santiago et al., 2015, 2017). AtCNIH1 and AtCNIH4 are essential for sorting and trafficking Glutamate Receptor-like3.3 from the ER to the PM in Arabidopsis (Wudick et al., 2018). PpCNIH2 functions as a cargo receptor important for ER exit and polar localization of the auxin efflux transporter PINA in the protonema cells of the moss Physcomitrium patens (Yáñez-Domínguez et al., 2023).
Boron (B) is an essential element for plant growth and is necessary for the structure and function of cell walls by cross-linking pectin in the rhamnogalacturonan II region (Funakawa and Miwa, 2015). Nodulin 26-like Intrinsic Protein (NIP)5;1 is a boric acid channel localized in the PM of root cells, including epidermal and endodermal cells. NIP5;1 is involved in B uptake under low-B conditions. Arabidopsis NIP5;1 T-DNA insertion mutants show severely reduced root and shoot growth owing to defects in B uptake under low-B conditions (Takano et al., 2006). NIP5;1 shows polar localization in the PM toward the soil side in root epidermal and endodermal cells, and this polar localization is important for the efficient transport of B in roots (Alassimone et al., 2010; Takano et al., 2010; Wang et al., 2017). NIP5;1 belongs to the NIP subfamily of major intrinsic proteins (aquaporins; Maurel et al., 2015; Jothi and Takano, 2023). In the NIP subfamily, there are two other boric acid channels, NIP6;1 and NIP7;1, which are important for B transport under low-B conditions. NIP6;1 is expressed in the nodal regions of shoots, especially in the phloem region of vascular tissues, and is required for the distribution of B toward young developing shoot tissues under low-B conditions. T-DNA insertion mutants of NIP6;1 exhibited reduced expansion of young rosette leaves under low-B conditions (Tanaka et al., 2008). NIP7;1 is expressed in the developing anthers and is involved in pollen development (Li et al., 2011). Compared with observations in the wild type (WT), T-DNA insertion mutants of NIP7;1 exhibited shorter siliques, a higher number of aborted seeds, and morphological defects in pollen grains under low-B conditions (Routray et al., 2018).
In previous work, we screened Arabidopsis mutants in which GFP-NIP5;1 showed aberrant localization and identified six mutant lines showing ER localization of GFP-NIP5;1 (Uehara et al., 2014). In the present study, we aimed to use these mutants to understand the mechanism underlying the ER exit of boric acid channels. Using genetic mapping and whole-genome resequencing, we identified KAONASHI3 (KNS3)/SPOTTY1/IMPERFECTIVE EXINE FORMATION and its homologs as the genes responsible for the mutant phenotype. We showed that KNS3 is localized in the ER and Golgi, and identified three amino acids in the C-terminal tail of KNS3 important for its Golgi-to-ER trafficking. We also showed that two homologs of KNS3 are important for ER-to-Golgi trafficking of KNS3 and that the accumulation of these proteins is dependent on each other. We propose that KNS3 and its two homologs function as a cargo-receptor complex for the ER exit of boric acid channels.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotypes Col-0 and Ler were from our laboratory stocks. The T-DNA insertion mutants kns3-2 (SALK_041228), kns3-3 (SALK_061320), knsth1-1 (SALK_027378), knsth1-2 (SALK_106609), knsth2-1 (SAIL_731_H03), and knsth2-3 (SAIL_670_H01) were obtained from the Arabidopsis Biological Resource Center. The kns3-2 knsth1-1, kns3-2 knsth2-1, and knsth1-1 knsth2-1 double mutants, and the kns3-2 knsth1-1 knsth2-1 triple mutant were generated by crossing. The nip5;1-1 mutant (SALK_122287) and ProNIP5;1 (–5ʹ untranslated regions):GFP-NIP5;1/nip5;1-1 transgenic plants were described previously (Takano et al., 2006; Tanaka et al., 2011).
Seeds were surface sterilized with 1% (w/v) NaClO for 10 min and rinsed with ultrapure water. For imaging of GFP-NIP5;1, plants were grown on solid medium [1.51 mM NaH2PO4, 0.26 mM Na2HPO4, 1.5 mM MgSO4, 2.0 mM Ca(NO3)2, 3.0 mM KNO3, 10.3 μM MnSO4, 1.0 μM ZnSO4, 1.0 μM CuSO4, 130 nM CoCl2, 24 nM (NH4)6Mo7O24, and 50 μM FeNa-EDTA] (Takano et al., 2005) containing 1% (w/v) sucrose, 1% (w/v) gellan gum, and 30 µM B for 4–10 d in growth chambers at 22 °C under fluorescent lamps (approximately 90 µmol m–2 s–1) with long-day conditions (16 h/8 h light/dark cycle). For phenotypic analysis and B determination in the reproductive growth stage, plants were cultivated in a hydroponic culture system (Takano et al., 2001) with liquid medium (Takano et al., 2005) containing 30 μM B for 10–12 d and then transferred to liquid medium containing 1 μM or 100 μM B for a further 15–30 d of growth under fluorescent lamps (approximately 110 µmol m–2 s–1) with long-day conditions. To observe pollen grains, plants were grown on Rockwool and vermiculite in pots and supplied with 1/1000 diluted Hyponex solution (Hyponex, Japan) once a week.
Genetic mapping and whole-genome resequencing
For genetic mapping, lines 1–3, 10–6, 14–3, and 15–2 (Col-0 background) were crossed with the Ler ecotype to obtain selfed F2 seeds. Genomic DNA was extracted from F2 plants exhibiting a mutant phenotype of GFP-NIP5;1. For rough mapping, simple sequence length polymorphism markers from the TAIR marker database and Cereon database (http://www.arabidopsis.org) were used as follows: map1-3M, map1-13.8M, and map1-24.4M for chromosome 1; map2-3.5M and nga168 for chromosome 2; nga162 and map3-18M for chromosome 3; NGA8 for chromosome 4; 2.5M and map5-19.9M for chromosome 5 (Supplementary Table S1). The whole-genome resequencing method has been described previously (Uehara et al., 2014).
Subcellular localization
The coding sequences of mCherry, KNS3, and KNSTH2 were amplified by PCR using specific primers (Supplementary Table S2). The PCR products were assembled using the In-Fusion technique (Clontech) into the pUB-DEST vector (Grefen et al., 2010) digested by XhoI for agroinfiltration into Nicotiana benthamiana (tobacco) leaves or into the pUGW2C vector (Nishimura et al., 2015) digested by PmeI and SpeI for polyethylene glycol (PEG)-mediated transformation into Arabidopsis mesophyll protoplasts. Alanine substitutions in the C-terminal regions of KNS3 and KNSTH2 were performed by inverse PCR using specific primers and vectors containing mCherry-KNS3 and mCherry-KNSTH2 as templates (Supplementary Table S2). The ends of the linear PCR products were fused using the In-Fusion technique. The 35S promoter:ST-YFP plasmid (Ito et al., 2018) was provided by Yoko Ito. The 2 × 35S promoter:Man1-GFP and GFP-HDEL plasmids (G-gb and ER-gb; Nelson et al., 2007) were provided by the Arabidopsis Biological Resource Center.
PEG-mediated protoplast transformation was performed as described previously (Maekawa et al., 2014). Agroinfiltration was performed as described previously (Yasuda et al., 2014) with some modifications. Agrobacterium was transformed with constructs using the heat shock method and then grown in Yeast Extract Peptone medium. The Agrobacterium cells were collected and resuspended with infiltration buffer containing 150 µM acetosyringone.
Immunofluorescence was performed as previously described (Yoshinari et al., 2019) with a few modifications. The specimens were incubated with the primary antibody [rabbit polyclonal anti-binding immunoglobulin protein (BiP) antibody (1:6000, Agrisera), chicken polyclonal anti-PIN FORMED 2 (PIN2) antibody (1:1000, Agrisera), rabbit polyclonal anti-PENTRATION3 (PEN3) antibody (1:1000, Agrisera), and rabbit polyclonal anti-PM intrinsic protein 2 (PIP2) antibody (1:1000, Agrisera) diluted in the blocking buffer] and secondary antibody [goat anti-rabbit IgG antibody conjugated with DyLight 549 (1:500; Thermo Fisher Scientific) and goat anti-chicken IgG antibody conjugated with CF568 (1:1000; Biotium)]. The specimens were counterstained with DAPI (2 μg ml–1) for 20 min at room temperature and washed four times with ultrapure water. Finally, the specimens were mounted using SlowFade Gold antifade solution (Thermo Fisher Scientific).
Confocal image acquisition was performed using a Leica TCS SP8 equipped with a ×40 water-immersion lens or a ×63 oil-immersion lens. The laser excitation/spectral detection bandwidths were 405/420–470 nm for DAPI; 488/500–530 nm for GFP; 488/520–550 nm for YFP; 552/580–650 nm for mCherry, DyLight 549, and CF 568; and 552/650–700 nm for FM4-64. The image contrast was adjusted using ImageJ and Fiji software (Schindelin et al., 2012). Pearson correlation coefficients were calculated using Fiji/ImageJ with the PSC co-localization plugin (French et al., 2008).
Measurement of boron concentration in plant tissues
The roots, rosette leaves, and shoot apices (~1.5 cm from the top of the plants) were harvested from three or four independent plants grown hydroponically under long-day conditions. The samples were dried, weighed, and digested using concentrated nitric acid (FUJIFILM Wako Chemicals, Osaka, Japan) for B determination. The samples were dissolved in 0.3 M nitric acid. The B concentration was determined using a curcumin assay (Mohan and Jones, 2018).
Preparation of microsomal proteins and western blot analysis
The anti-KNS3, anti-KNSTH1, and anti-KNSTH2 antibodies were purchased from Sigma-Aldrich. Peptides [CNDHTSLKGGHAHS for KNS3 (68% purity), CALSGDGVLPRGEFHPLAA for KNSTH1 (80% purity), CSAPYEKTSHAHERPITN for KNSTH2 (85% purity)] were synthesized and used to immunize rabbits. Eight weeks after immunization, approximately 60 ml of serum was collected and affinity purified using these peptides.
Preparation of microsomal proteins and western blotting were performed as previously described (Yoshinari et al., 2019) with a few modifications. Root tissues (0.2–0.5 g) were lysed in 2–3 ml of homogenization buffer. Protein samples were heated at 70 °C for 10 min in SDS sample buffer containing 10% (v/v) 2-mercaptoethanol. Proteins were loaded on to a Bolt 4–12% Bis-Tris gel (Thermo Fisher Scientific) and transferred to a polyvinylidene fluoride membrane using a semi-dry transfer technique (Trans-Blot system; Bio-Rad). Rabbit anti-KNS3 polyclonal antibody (1:5000), rabbit anti-KNSTH1 polyclonal antibody (1:2000), and rabbit anti-KNSTH2 polyclonal antibody (1:5000) were diluted in Can Get Signal Solution 1 (Toyobo) and used as primary antibodies. An anti-rabbit IgG antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch) and diluted at 1:1 000 000 in Can Get Signal Solution 2 (Toyobo) was used as the secondary antibody. Protein signals were detected using a chemiluminescence imaging system (FUSION SOLO.7S.EDGE; Vilber, Germany). The membrane-bound proteins were stained with Coomassie Brilliant Blue (ATTO) after detection.
Statistical analysis
When data were found to be normally distributed, either Dunnett’s or Tukey’s multiple comparison test was conducted. When data were found not to be normally distributed, a non-parametric test (Mann–Whitney U test) was used. Prism 8 software (version 8.4.2; GraphPad Software, San Diego, CA, USA) was used for these analyses.
Multiple sequence alignment and phylogenetic analysis
Multiple sequence alignments of KNS3 and its orthologous amino acid sequences were performed using Clustal Omega (EMBL-EBI), and a phylogenetic tree was constructed using MEGA X (Kumar et al., 2018). The reliability of the topology was examined using the bootstrap method, which generated a bootstrap probability of 1000 replications at each interior branch of the tree.
Results
Mutations in KNS3 cause defects in the endoplasmic reticulum exit of NIP5;1
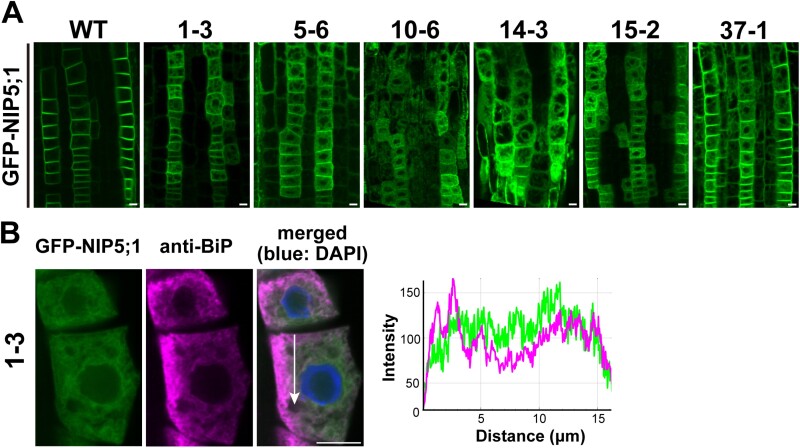
NIP5;1 is localized in the PM of the root cap and in root epidermal and endodermal cells in a polar manner toward the soil side (Takano et al., 2010). To understand the intracellular transport mechanism of NIP5;1, we performed genetic screening using an ethyl methanesulfonate-mutagenized population of GFP-NIP5;1 transgenic Arabidopsis plants (proNIP5;1 (Δ5ʹ untranslated regions):GFP-NIP5;1 in nip5;1-1). We screened approximately 40 000 M2 seedlings using a fluorescence microscope and identified three mutant lines showing GFP-NIP5;1 intracellular aggregation (Uehara et al., 2014). Additionally, we identified six mutant lines (1–3, 5–6, 10–6, 14–3, 15–2, and 37–1) in which GFP-NIP5;1 showed a network-like distribution in the cytoplasm in addition to localization in the PM of the root epidermal cells (Fig. 1A). To test for allelism, we crossed six mutant lines. In the F1 plants of line 1–3 × line 10–6, line 1–3 × line 15–2, line 10–6 × line 14–3, line 10–6 × line 15–2, and line 14–3 × line 15–2, GFP-NIP5;1 was localized in the ER and PM (Supplementary Fig. S1A). However, in the F1 plants of line 1–3 × line 37–1, line 5–6 × line 15–2, line 5–6 × line 37–1, and line 10–6 × line 37–1, GFP-NIP5;1 was localized only in the PM. These results suggested that lines 1–3, 10–6, 14–3, and 15–2 were allelic to each other and lines 5–6 and 37–1 were not allelic with these four lines.
Fig. 1.
Identification of mutants in which GFP-NIP5;1 was localized at the endoplasmic reticulum and plasma membrane. (A) GFP-NIP5;1 in the root epidermal cells of a wild-type (WT) plant and ethyl methanesulfonate mutants. (B) Left panel: immunofluorescence in root epidermal cells of line 1–3. GFP-NIP5;1 (green), endogenous binding immunoglobulin protein (BiP) detected by anti-BiP antibody (magenta), and nuclei stained with 2 μg ml‐ DAPI (blue) are shown. Right panel: intensity profiles of GFP-NIP5;1 (green) and anti-BiP (magenta), obtained using ImageJ software, along a line (white arrow) in the merged image. Plants were grown on solid medium containing 30 μM B for 7–10 d. Scale bar=10 μm.
To confirm the localization of GFP-NIP5;1 in lines 1–3, 10–6, 14–3, and 15-2, we performed immunofluorescence using an antibody against an ER-resident protein, BiP (Benghezal et al., 2000). BiP was distributed as a network in the cytoplasm, and line plot analysis showed an overlap with GFP-NIP5;1 in line 1–3 (Fig. 1B). The ER localization, in addition to PM localization, indicated that GFP-NIP5;1 was partially retained in the ER of the mutant lines.
Next, to identify the causative gene of the mutant phenotype in lines 1–3, 10–6, 14–3, and 15–2, we performed genetic mapping and whole-genome sequencing. We outcrossed these four mutants (Col-0 background) with the Ler ecotype and analyzed the phenotypes of the F2 plants. Approximately 25% of the population showed ER retention of GFP-NIP5;1, indicating that the phenotype was caused by a single recessive mutation. Genotyping of 25–35 F2 plants showing ER retention with 10 simple sequence length polymorphism markers showed a linkage to a marker located on the lower arm of chromosome 5 (Supplementary Fig. S1B; Supplementary Table S1).
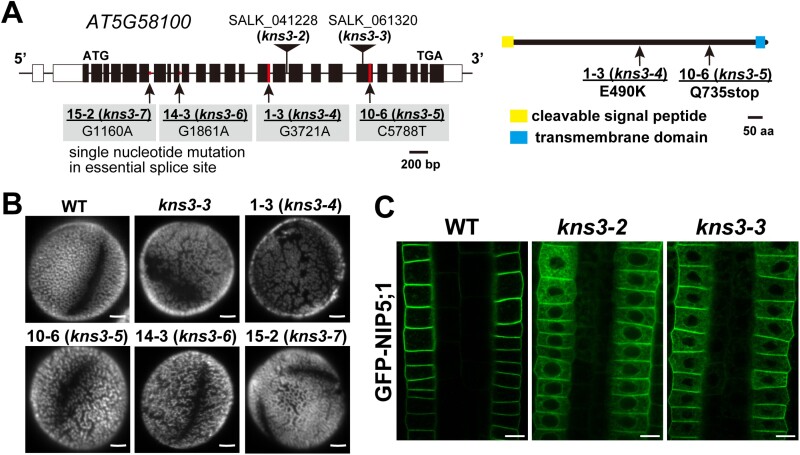
We performed whole-genome sequencing of the four lines using the SOliD platform (Applied Biosystems). The read sequences were mapped to the A. thaliana Col-0 genome (TAIR10.0). Single-nucleotide polymorphisms (SNPs) 6420, 3489, 8753, and 9239 were identified in lines 1–3, 10–6, 14–3, and 15–2, respectively. Among the genes containing SNPs, only At5g58100 was selected based on the following criteria: the type of SNP was (i) C to A or G to T, which are typical substitutions caused by ethyl methanesulfonate, (ii) located on chromosome 5, and (iii) common in the four allelic lines. We identified a missense mutation of the glutamic acid codon to a lysine codon (E490K) in exon 15 in line 1–3, a glutamine codon to a stop codon (Q735stop) in exon 20 in line 10–6, and a single nucleotide substitution in the essential splice site (G/A) at exon 6 downstream in line 15–2 and at exon 9 downstream in line 14–3 (Fig. 2A).
Fig. 2.
At5G58100 (KNS3) is the causative gene for mutants with defective NIP5;1 localization. (A) The exon–intron structure of the KNS3 gene (At5G58100). Filled boxes, open boxes, and thick bars indicate exons, untranslated regions, and introns, respectively. The topology of KNS3 was predicted by the Philius program. Positions for T-DNA insertion in the kns3-2 and kns3-3 mutants and point mutations in line 1–3 (kns3-4), 10–6 (kns3-5), 14–3 (kns3-6), and 15–2 (kns3-7) are indicated. (B) Pollen grains of wild-type (WT) plants and kns3 mutants. Pollen grains were stained with 0.001% Auramine O. Plants were grown in pots with vermiculite supplied with 1/1000 diluted Hyponex solution. (C) GFP-NIP5;1 in the root epidermal cells of KNS3 T-DNA insertion mutants. Plants were grown on solid medium containing 30 μM B for 7–10 d. Scale bars=2.5 μm (B) and 10 μm (C).
Previously, loss-of-function mutants of At5g58100 have been reported as kns3 (Suzuki et al., 2008), spotty1 (Dobritsa et al., 2011), and imperfective exine formation (Wang et al., 2021), which have defects in outer pollen wall formation. To confirm that At5g58100 is the causative gene for ER retention of NIP5;1, we acquired kns3-2 and kns3-3 T-DNA insertion lines (Fig. 2A) and compared their phenotypes. As expected, Auramine O staining showed a collapsed reticulate exine structure on the pollen surface in all four allelic lines and kns3-3 (Fig. 2B). We then introduced the GFP-NIP5;1 construct into the kns3-2 and kns3-3 mutants and observed the fluorescence of GFP-NIP5;1 in root epidermal cells. GFP-NIP5;1 was localized in the ER and PM of the kns3-2 and kns3-3 mutants (Fig. 2C), in a pattern similar to that observed in the four allelic lines (Fig. 1A). Collectively, these results establish that At5g58100 is the causative gene for ER retention of NIP5;1. The four allelic lines, 1–3, 10–6, 14–3, and 15–2, were named kns3-4, kns3-5, kns3-6, and kns3-7, respectively.
KNS3 functions in the endoplasmic reticulum exit of specific plasma membrane proteins, including NIP5;1 and NIP6;1
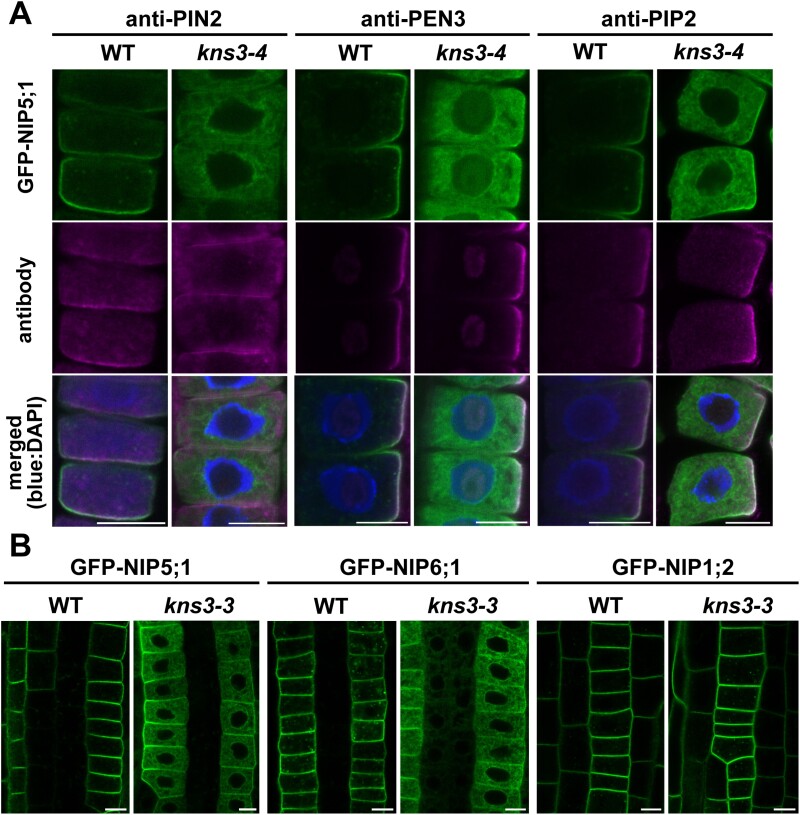
To examine whether KNS3 specifically affects the intracellular transport of NIP5;1, we investigated the localization of an auxin transporter, PIN2 (Abas et al., 2006), an ATP-binding cassette transporter, PEN3/PLEIOTROPIC DRUG RESISTANCE8 (Langowski et al., 2010), and an aquaporin, PIP2 (Maurel et al., 2015), in kns3-4 and the WT (Col-0). In the WT, immunofluorescence with anti-PIN2, anti-PEN3, and anti-PIP2 antibodies showed the co-localization of these proteins with GFP-NIP5;1 in the PM of epidermal cells. In kns3-4 mutant cells, these three proteins were localized in the PM, whereas GFP-NIP5;1 accumulated in the ER (Fig. 3A). These results indicate that KNS3 is not required for the trafficking of PIN2, PEN3, and PIP2 to the PM.
Fig. 3.
KNS3 functions in the trafficking of specific plasma membrane proteins, including NIP5;1 and NIP6;1. (A) Immunofluorescence of PIN2, PEN3, and PIP2 in the root epidermal cells of the wild-type (WT) and kns3-4 mutant. GFP-NIP5;1 (green), endogenous PIN2, PEN3, and PIP2 detected by antibodies (magenta), and nuclei stained with 2 μg ml DAPI (blue) are shown. (B) GFP-NIP5;1, GFP-NIP6;1, and GFP-NIP1;2 expressed under the control of the NIP5;1 promoter in the WT and kns3-3 background. Plants were grown on solid medium containing 30 μM B for 7–10 d. Scale bar=10 μm.
We also examined the localization of NIP6;1 and NIP1;2, which belong to the NIP subfamily. To examine their localization in the root epidermis, we used plants expressing GFP-NIP6;1 and GFP-NIP1;2 under the control of the NIP5;1 promoter. In root epidermal cells, GFP-NIP6;1 was localized to the PM in WT but to the ER and PM in kns3-3, similar to GFP-NIP5;1. However, GFP-NIP1;2 was localized in the PM of both WT and kns3-3 mutants (Fig. 3B). Therefore, ER retention in the kns3 mutant was observed only for NIP5;1 and its close homolog NIP6;1 among the PM-localized proteins tested.
It has been reported that NIP5;1 is localized in the PM in a polar manner toward the soil side in epidermal cells and that polar localization is important for the efficient transport of B in roots (Wang et al., 2017). Therefore, we investigated whether KNS3 functions in the polar trafficking of NIP5;1. In optical longitudinal sections at the center of the WT root, we observed GFP-NIP5;1 in the soil-side PM domain but not in the stele-side PM domain in the epidermal cells (Supplementary Fig. S2). Comparison with the signal of FM4-64, a lipophilic styryl dye that stains the PM, confirmed its polar localization (Supplementary Fig. S2). Although intracellular signals were observed, GFP-NIP5;1 in the PM of epidermal cells of kns3-3 (Supplementary Fig. S2) showed a polar localization. These results indicate that KNS3 is involved in ER exit but not in polar trafficking to the PM.
Defects in silique development but not pollen surface structure of kns3 mutants are partially rescued by higher boron supply
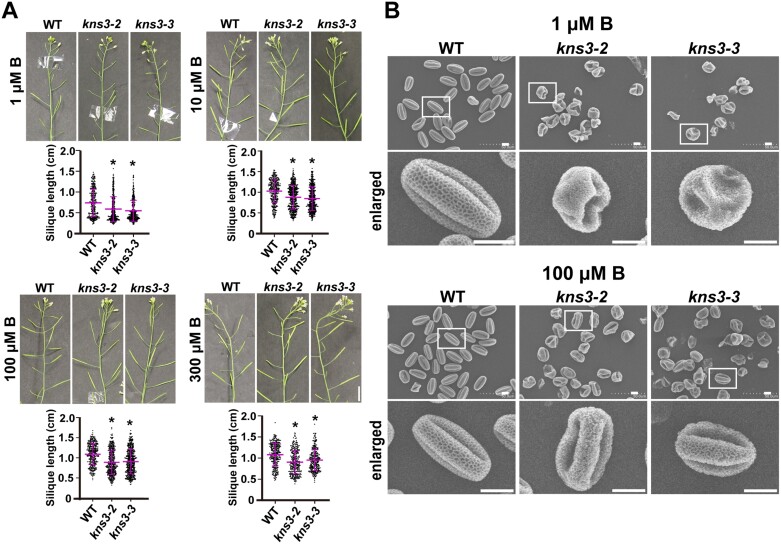
Given that GFP-NIP5;1 and GFP-NIP6;1 showed significant ER retention in the kns3 mutants (Figs 2C, 3B), B transport was possibly affected in these mutants. To investigate the effect of B conditions on the phenotypes of the kns3 mutants, kns3-2, kns3-3, nip5;1-1, a NIP5;1 T-DNA insertion mutant (Takano et al., 2006), and WT plants were grown on solid medium with low to sufficient (0.03, 0.3, 3, and 30 µM) B concentrations for 7 d. The kns3 mutants did not show any significant differences from the WT in terms of shoot fresh weight or root length under different B conditions, whereas both values were significantly reduced in nip5;1-1, as reported previously (Supplementary Fig. S3A) (Takano et al., 2006). Subsequently, kns3-3, nip6;1-1, a NIP6;1 T-DNA insertion mutant (Tanaka et al., 2008), and WT plants were grown in a hydroponic system with low (0.1 µM), moderately low (1 µM), and sufficient (100 µM) B concentrations. As previously reported, the nip6;1-1 mutant showed reduced expansion of young rosette leaves and loss of apical dominance under low B conditions but normal growth under sufficient B conditions (Tanaka et al., 2008) (Supplementary Fig. S3B, C). In contrast to the nip6;1-1 mutant, the kns3 mutants grew normally with no visible growth defects until flowering under these conditions (Supplementary Fig. S3B, C). However, the siliques of kns3-2 and kns3-3 mutants were shorter than those of the WT in low to moderately high B conditions: silique lengths of kns3-2 were 80, 85, 82, and 84% of those of the WT in 1, 10, 100, and 300 μM B, respectively, and those of kns3-3 were 75, 82, 83, and 88% of those of the WT, respectively (Fig. 4A). Although the siliques of kns3 mutants were shorter than those of the WT under all conditions tested, the difference decreased as the B supply was increased. We then analyzed the pollen structure of kns3-2 and kns3-3 by scanning electron microscopy in plants grown with 1 µM and 100 µM B supply. Under both conditions, all pollen grains had a collapsed exine structure, in contrast to the reticulate pattern of the WT pollen surface (Fig. 4B).
Fig. 4.
Defects in silique development but not pollen surface structure of kns3 mutants were partially rescued by a high B supply. (A) Siliques of wild-type (WT), kns3-2, and kns3-3 plants grown hydroponically with 30 µM B for 10–12 d and then with 1, 10, 100, or 300 μM B for 30 d. The lengths of 191–386 siliques from three plants were measured. Data represent the mean ±SD. Asterisks indicate mutants that showed significant differences from the WT (*P<0.01; Mann–Whitney U test). (B) Scanning electron micrographs of pollen grains from WT and kns3 mutants grown hydroponically with 30 µM B for 10–12 d and then with 1 μM or 100 μM B for 20 d. More than 50 pollen grains were observed, and representative images are shown. Scale bars=1 cm (A) and 10 μm (B).
To investigate the possible alteration of B transport in the kns3 mutants, WT and kns3 mutants were grown hydroponically with 30 μM B for 10–12 d and then transferred to low (1 µM) or sufficient (100 µM) B conditions for an additional 15 d. B concentrations in the roots, rosette leaves, and shoot apices of kns3-2 and kns3-3 mutants were not significantly different from those of the WT under both low and sufficient B conditions (Supplementary Fig. S3D). It is likely that the boric acid channels remaining in the PM of the cells of the kns3 mutants sufficiently supported the uptake and translocation of B. It is possible that B transport in flowers is affected and that factors other than B are also involved in the phenotypes of pollen and siliques in the kns3 mutants.
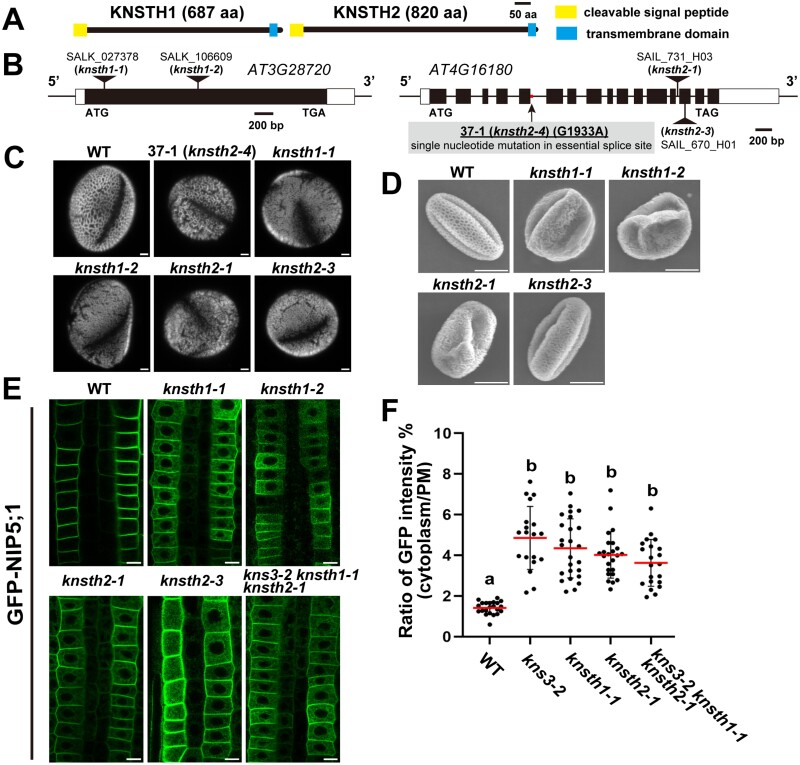
Two predicted homologs of KNS3 are also important for the endoplasmic reticulum exit of NIP5;1
To investigate the possible functional redundancy of KNS3, we searched for KNS3 homologs in the A. thaliana genome (TAIR10.0). A protein BLAST search in the National Center for Biotechnological Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that At3g28720.1 and At4g16180.2 have 26% and 28% identity with KNS3 (At5G58100.1), respectively. These two proteins were predicted to have an N-terminal signal peptide and a C-terminal transmembrane domain similar to those of KNS3 by the Philius program (Reynolds et al., 2008) (Fig. 5A) and the AlphaFold protein structure database (Varadi et al., 2022) (Supplementary Fig. S4B, C). Thus, we named At3g28720 and At4g16180 as KNS THREE HOMOLOGS (KNSTH) 1 and 2, respectively. Importantly, we found that mutant line 37–1 had a single-nucleotide mutation in the essential splice site (C/T) downstream of exon 5 in KNSTH2 (Fig. 5B). We named line 37–1 as knsth2-4. We observed a defective pollen wall structure in the knsth1-1 and knsth2-1 T-DNA insertion lines, similar to that in kns3 mutants (Fig. 5C, D). Moreover, the defective pollen wall was similarly observed in kns3-2 knsth1-1, kns3-2 knsth2-1, and knsth1-1 knsth2-1 double mutants, and in the kns3-2 knsth1-1 knsth2-1 triple mutant (Supplementary Fig. S5) We then introduced the GFP-NIP5;1 construct into the knsth1 and knsth2 mutants and observed fluorescence in root epidermal cells. GFP-NIP5;1 was localized in the ER and PM in knsth1-1, knsth1-2, knsth2-1, knsth2-3, and the kns3-2 knsth1-1 knsth2-1 triple mutant (Fig. 5E), as well as in kns3 mutants (Fig. 2C). The cytoplasm/PM ratios of the GFP-NIP5;1 signal were significantly higher in the kns3-2, knsth1-1, knsth1-2, and triple mutant plants than in the WT plants and were not significantly different among these mutants (Fig. 5F). GFP-NIP5;1 in the PM showed polar localization in the kns3-3 single mutant and the triple mutant, similar to that in the WT (Supplementary Fig. S2). These results suggest that KNS3 and its two homologs work together in the ER exit of NIP5;1 and pollen wall formation.
Fig. 5.
Two predicted homologs of KNS3 are also important for the endoplasmic reticulum exit of NIP5;1. (A) Topology of KNSTH1 and KNSTH2 containing an N-terminal signal peptide and a C-terminal transmembrane domain. (B) The exon–intron structures of KNSTH1 (At3G28720) and KNSTH2 (At4G16180). Positions for T-DNA insertions in knsth1-1 and knsth2-1, and a point mutation in line 37–1 (knsth2-4), are shown. (C) Pollen of wild-type (WT) and the knsth1 and knsth2 mutants. Pollen grains were stained with 0.001% Auramine O. (D) SEM images of pollen grains from WT, knsth1, and knsth2 mutants. Plants were grown in pots with vermiculite supplied with 1/1000 diluted Hyponex solution. (E) GFP-NIP5;1 in kns3, knsth1, and knsth2 single T-DNA insertion mutants and the triple mutant. Plants were grown on solid medium containing 30 μM B for 7–10 d. (F) Ratio of GFP intensity in the cytoplasm and plasma membrane. Dot plots show the distribution among 23 (WT), 23 (kns3-2), 25 (knsth1-1), 24 (knsth2-1), and 25 (kns3-2 knsth1-1 knsth2-1) cells from three or four independent primary roots. Data represent the mean (red lines) ±SD (bars). Different letters indicate significant differences based on Tukey’s test (P<0.01). Scale bars=2.5 μm (C) and 10 μm (D, E).
KNS3 is localized in the endoplasmic reticulum and Golgi, whereas KNSTH2 is mainly localized in the endoplasmic reticulum
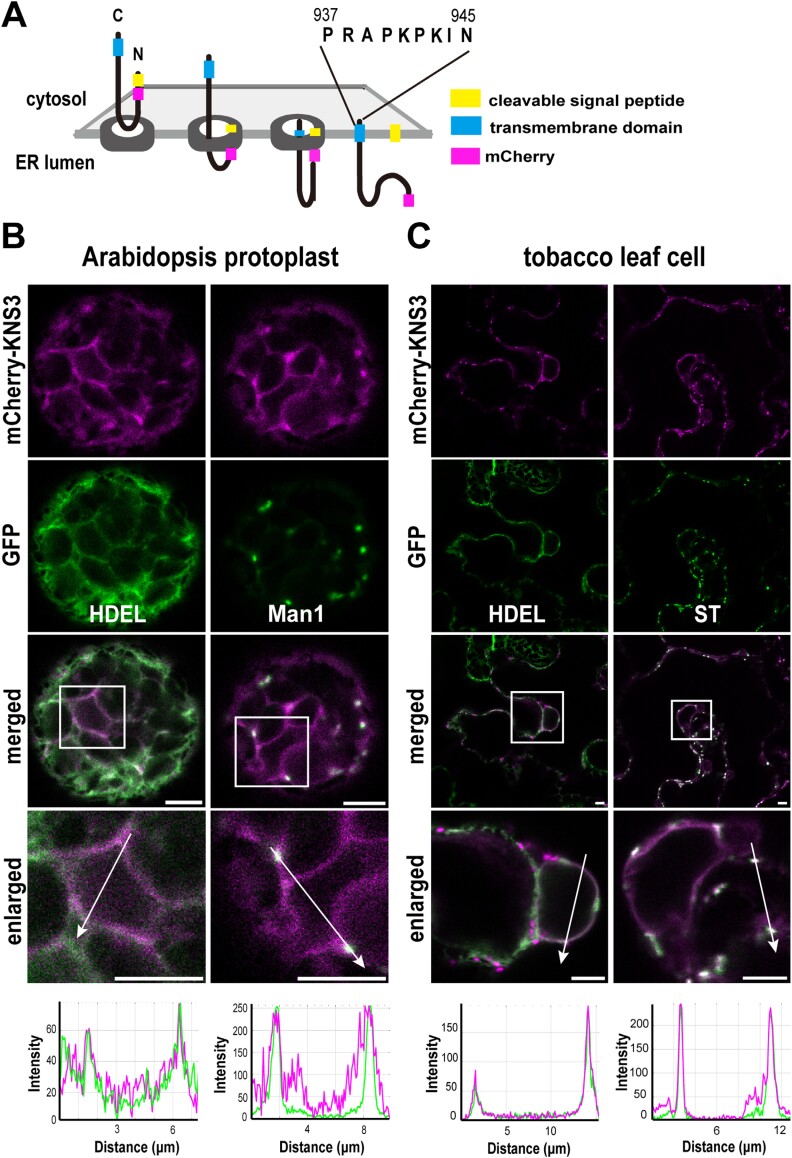
KNS3 was predicted to contain an N-terminal signal peptide and a C-terminal transmembrane domain (Fig. 2A; Supplementary Fig. S4A). It is probable that KNS3 is inserted into the ER membrane, its N-terminal soluble region is located in the ER lumen, and its short (nine amino acids) C-terminal tail is located in the cytosol. To investigate the intracellular localization of KNS3, we designed a ProUBQ10:mCherry-KNS3 construct, in which the mCherry sequence was located downstream of the signal peptide sequence (Fig. 6A). We examined KNS3 localization in protoplasts from the leaf mesophyll cells of WT Arabidopsis by PEG-mediated transformation. In protoplasts, mCherry-KNS3 showed ring- and network-like localization that overlapped with the ER marker GFP-HDEL, and showed punctate structures that co-localized with the Golgi marker Man1-GFP (Nebenführ et al., 1999) (Fig. 6B). We also introduced the mCherry-KNS3 construct into tobacco leaves by agroinfiltration. In the epidermal cells, mCherry-KNS3 was observed in ring-like and punctate patterns. In the ring-like structures, mCherry-KNS3 co-localized with the ER marker GFP-HDEL (Nebenführ et al., 2000) (Fig. 6C). In the punctate structures, mCherry-KNS3 co-localized with the trans-Golgi marker ST-YFP (Saint-Jore et al., 2002) (Fig. 6C). Line plot analysis confirmed co-localization. In summary, mCherry-KNS3 was detected in the ER and Golgi apparatus in both Arabidopsis protoplasts and tobacco leaf cells.
Fig. 6.
mCherry-KNS3 is localized in the endoplasmic reticulum and Golgi. (A) Topology of KNS3 with an N-terminal signal peptide and a transmembrane domain. In the mCherry-KNS3 construct, mCherry was inserted after the signal peptide. (B) mCherry-KNS3 and GFP-HDEL or Man1-GFP expressed in protoplasts from wild-type (Col-0) Arabidopsis leaf mesophyll cells using polyethylene glycol-mediated transformation. (C) mCherry-KNS3 and GFP-HDEL, or ST-YFP in N. benthamiana leaf epidermal cells after agroinfiltration. Scale bars=5 μm.
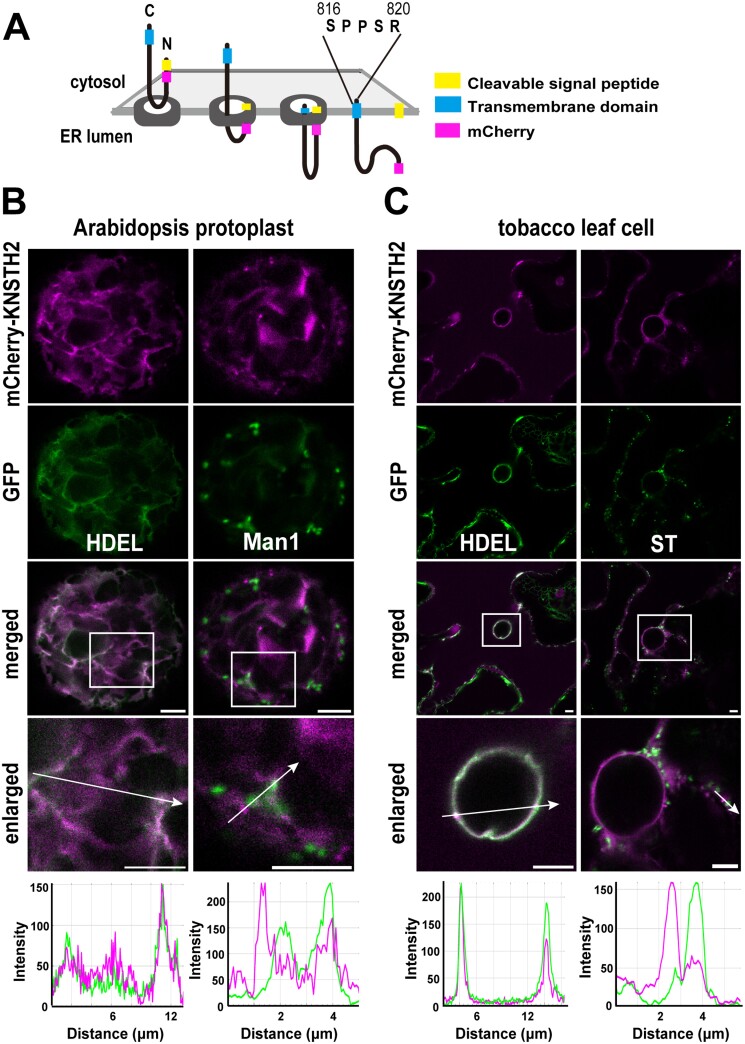
Next, we designed a ProUBQ10:mCherry-KNSTH2 construct, similar to the mCherry-KNS3 construct (Fig. 7A). We examined KNSTH2 localization in protoplasts from leaf mesophyll cells of Arabidopsis WT plants. In protoplasts, mCherry-KNSTH2 showed ring-like, network-like, and occasionally punctate localization. In the ring- and network-like structures, mCherry-KNSTH2 overlapped with GFP-HDEL (Fig. 7B). mCherry-KNSTH2 did not significantly overlap with Man1-GFP (Fig. 7B). In tobacco leaf epidermal cells, mCherry-KNSTH2 exhibited ring-like and occasional punctate structures. In the ring-like structures, mCherry-KNSTH2 overlapped with GFP-HDEL. mCherry-KNSTH2 did not co-localize with ST-YFP (Fig. 7C). In summary, mCherry-KNSTH2 was observed mainly in the ER of Arabidopsis protoplasts and tobacco leaf cells.
Fig. 7.
mCherry-KNSTH2 is mainly localized in the endoplasmic reticulum. (A) Topology of KNSTH2 with an N-terminal signal peptide and a transmembrane domain. In the mCherry-KNSTH2 construct, mCherry was inserted after the signal peptide. (B) mCherry-KNSTH2 and GFP-HDEL or Man1-GFP expressed in protoplasts from leaf mesophyll cells of wild-type (Col-0) Arabidopsis using polyethylene glycol-mediated transformation. (C) mCherry-KNSTH2 and GFP-HDEL or ST-YFP expressed in N. benthamiana leaf epidermal cells by agroinfiltration. Scale bars=5 μm.
We also designed a ProUBQ10:mCherry-KNSTH1 construct, similar to mCherry-KNS3 and mCherry-KNSTH2. However, the introduction of this construct into competent cells of various Escherichia coli strains resulted in the rare appearance of colonies. Plasmids from these rare colonies contained mutations that caused amino acid substitutions or deletions in KNSTH1. Therefore, the toxicity of KNSTH1 hampered analysis of its localization. We also introduced mCherry-KNSTH1 PCR products into the protoplasts of Arabidopsis leaf mesophyll cells (Lu et al., 2013). Although a control experiment using mCherry-HDEL PCR products showed mCherry signals, we failed to detect any fluorescence from mCherry-KNSTH1.
K941, K943, and I944 in the C-terminal tail of KNS3 are important for trafficking from the Golgi to the endoplasmic reticulum
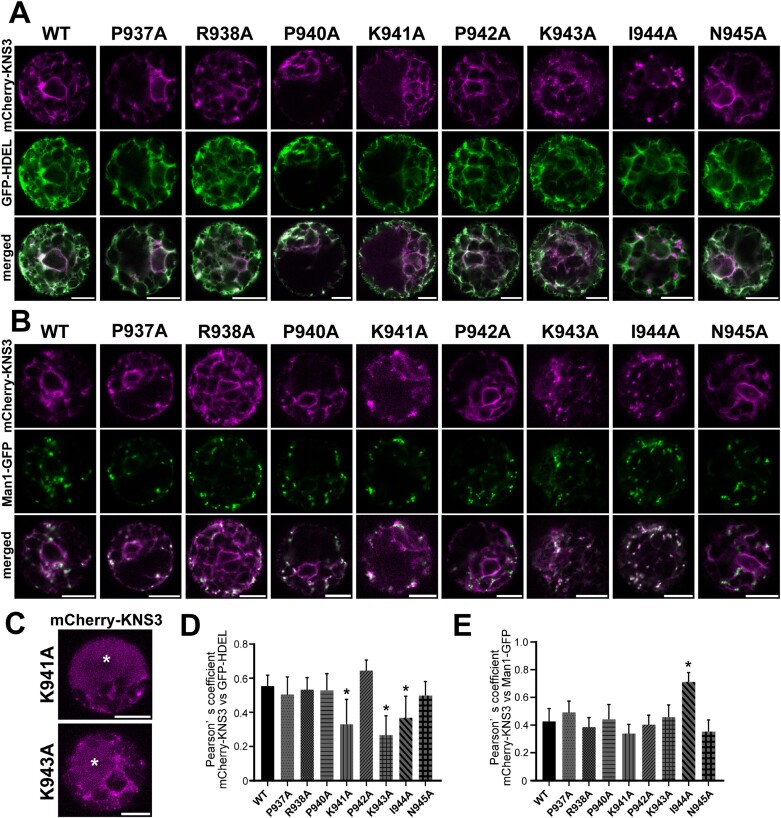
mCherry-KNS3 was localized in the ER and Golgi apparatus in both Arabidopsis protoplasts and tobacco leaf cells (Fig. 6). To explore the importance of amino acids in KNS3 for trafficking between the ER and Golgi apparatus, we performed alanine scanning mutagenesis of the cytosolic C-terminal tail (PRAPKPKIN). We expressed mCherry-KNS3 WT and variants with the ER marker GFP-HDEL or the Golgi marker Man1-GFP in protoplasts from Arabidopsis leaf mesophyll cells. mCherry-KNS3 WT, P937A, R938A, P940A, P942A, and N945A were localized in ring- and network-like structures co-labeled with the ER marker and in punctate structures co-labeled with the Golgi marker (Fig. 8A, B). mCherry-KNS3 K941A and K943A were also localized in ring- and network-like structures and punctate structures (Fig. 8A, B), whereas they occasionally showed diffuse mCherry signals in the cells (four of 20 protoplasts with K941A and four of 21 protoplasts with K943A) (Fig. 8C). Considering that leaf mesophyll protoplasts are largely occupied by vacuoles (for examples, refer to images of tonoplast or vacuolar labeling in Aluri and Büttner, 2007; Kang et al., 2012; Lee et al., 2013), we judged that the diffuse mCherry signals were in the vacuole. mCherry-KNS3 I944A was not observed in ring- or network-like structures but was mainly in punctate structures co-labeled with Man1-GFP (Fig. 8A, B). Pearson correlation coefficients between mCherry-KNS3 P937A, R938A, P940A, P942A, N945A, and GFP-HDEL (0.50–0.65) were similar to that of mCherry-KNS3 WT (0.56), while those of K941A, K943A, and I944A (0.27–0.37) were significantly lower (Fig. 8D). Pearson correlation coefficients between mCherry-KNS3 P937A, R938A, P940A, K941A, P942A, K943A, N945A and Man1-GFP (0.34–0.49) were similar to that of mCherry-KNS3 WT (0.43), while that of I944A (0.71) was significantly higher than that of the WT (Fig. 8E). We also expressed mCherry-KNS3 WT and variants with the ER marker GFP-HDEL or the Golgi marker ST-YFP in tobacco leaf epidermal cells. mCherry-KNS3 WT, P937A, R938A, P940A, P942A, and N945A were localized in the ER and Golgi apparatus (Supplementary Fig. S6A, B). Signals from mCherry-KNS3 K941A, K943A, and I944A were observed as punctate structures co-labeled with the Golgi marker (Supplementary Fig. S6B) and as diffuse signals (Supplementary Fig. S6A, B). Since leaf epidermal cells are largely occupied by vacuoles (Foresti et al., 2010; Scabone et al., 2011; Vieira et al., 2019; Wang et al., 2020), the diffuse mCherry signals were considered to be in the vacuole. The results are summarized in Supplementary Table S3. These results suggest that K941, K943, and I944 are important for the retrograde trafficking of KNS3 from the Golgi to the ER, and that the alanine-substituted variants were mistargeted to the vacuole from the Golgi.
Fig. 8.
K941, K943, and I944 in the C-terminal tail of KNS3 are important for trafficking from the Golgi to the endoplasmic reticulum. (A, B) mCherry-KNS3 wild type (WT) and variants expressed with GFP-HDEL (A) or Man1-GFP (B) in protoplasts of WT (Col-0) Arabidopsis leaf mesophyll cells. (C) mCherry-KNS3 K941A and K943A were expressed in protoplasts of WT (Col-0) Arabidopsis leaf mesophyll cells. Asterisks indicate the vacuole. Scale bars=10 μm. (D, E) Pearson correlation coefficients of mCherry-KNS3 WT and variants with GFP-HDEL (D) or Man1-GFP (E). Data represent the mean ±SD of 10–16 protoplasts. Asterisks denote significant differences between the WT and variants (*P<0.01; Dunnett’s test).
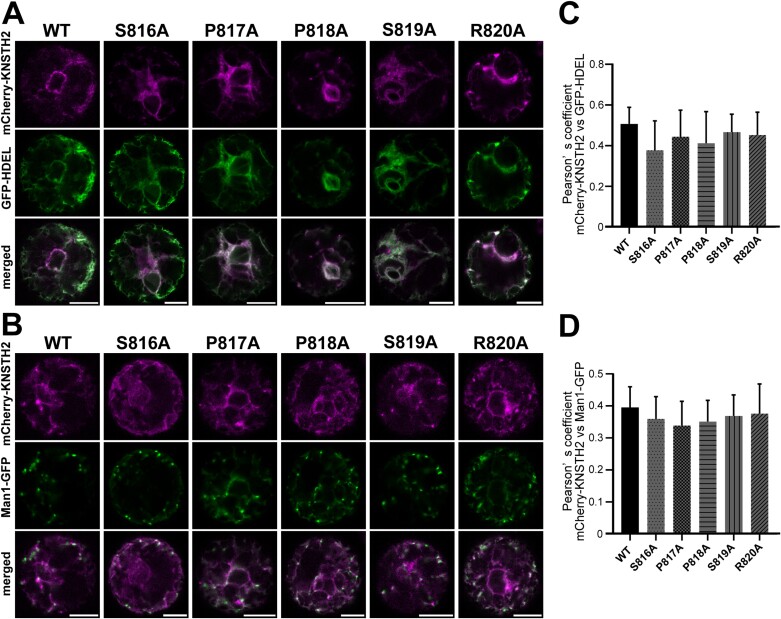
Mutations in the C-terminal tail do not affect the endoplasmic reticulum localization of KNSTH2
KNSTH2 was mainly localized in the ER in both Arabidopsis protoplasts and tobacco leaf cells (Fig. 7). To investigate the possible shuttling of KNSTH2 between the ER and Golgi and the involvement of its C-terminal tail in trafficking, we performed alanine scanning mutagenesis of the cytosolic C-terminal tail (SPPSR) of KNSTH2 (Fig. 7A). In Arabidopsis protoplasts, mCherry-KNSTH2 WT, S816A, P817A, P818A, S819A, and R820A showed ring- and network-like structures co-labeled with GFP-HDEL (Fig. 9A). These five variants and the WT hardly showed punctate localization and rarely co-localized with Man1-GFP (Fig. 9B). Pearson correlation coefficients between mCherry-KNSTH2 variants and GFP-HDEL (0.41–0.44) were similar to that of mCherry-KNSTH2 WT and GFP-HDEL (0.51) (Fig. 9C). Pearson correlation coefficients between mCherry-KNSTH2 variants and Man1-GFP (0.32–0.36) were similar to that of mCherry-KNSTH2 WT and Man1-GFP (0.39) (Fig. 9D). We also expressed mCherry-KNSTH2 WT and variants with GFP-HDEL or ST-YFP in tobacco leaf epidermal cells. Similar to mCherry-KNSTH2 WT, the variants co-localized well with the ER marker GFP-HDEL but not with the Golgi marker ST-YFP (Supplementary Fig. S7). In summary, mutations in the KNSTH2 C-terminal tail did not influence the ER localization of mCherry-KNSTH2 (Supplementary Table S3).
Fig. 9.
Mutations in the C-terminal tail do not affect the endoplasmic reticulum localization of KNSTH2. (A, B) mCherry-KNSTH2 WT and variants expressed with GFP-HDEL (A) or Man1-GFP (B) in protoplasts extracted from wild-type (WT) (Col-0) Arabidopsis leaf mesophyll cells. (C, D) Pearson coefficients of mCherry-KNSTH2 WT and variants with GFP-HDEL (C) or Man 1-GFP (D). Data represent the mean ±SD of 10 or 11 protoplasts. No significant differences were observed (P>0.05; Dunnett’s test). Scale bar=10 μm.
KNSTH1 and KNSTH2 are important for the trafficking of KNS3 from the endoplasmic reticulum to the Golgi
To investigate whether KNS3 and its homologs affect the trafficking of each other, we expressed mCherry-KNS3 in protoplasts from the knsth1 and knsth2 mutants. In knsth1-1 or knsth2-1 cells, mCherry-KNS3 showed ring- and network-like structures and occasionally showed punctate structures, similar to those in the WT cells (Supplementary Fig. S8A, B). The Pearson correlation coefficients between mCherry-KNS3 and GFP-HDEL or Man1-GFP in knsth1-1, knsth2-1, and WT cells were not significantly different (Supplementary Fig. S8C, D). Subsequently, we expressed mCherry-KNSTH2 in protoplasts from the WT, kns3-3, and knsth1-1. mCherry-KNSTH2 showed ring- and network-like structures in kns3-3 and knsth1-1, similar to those in the WT (Supplementary Fig. S8E, F). The Pearson correlation coefficients between mCherry-KNSTH2 and GFP-HDEL or Man1-GFP in the WT, kns3-3, and knsth1-1 were not significantly different (Supplementary Fig. S8G, H). These results suggest that most KNS3 and KNSTH2 are localized in the ER, and the lack of their homologs does not affect steady-state localization.
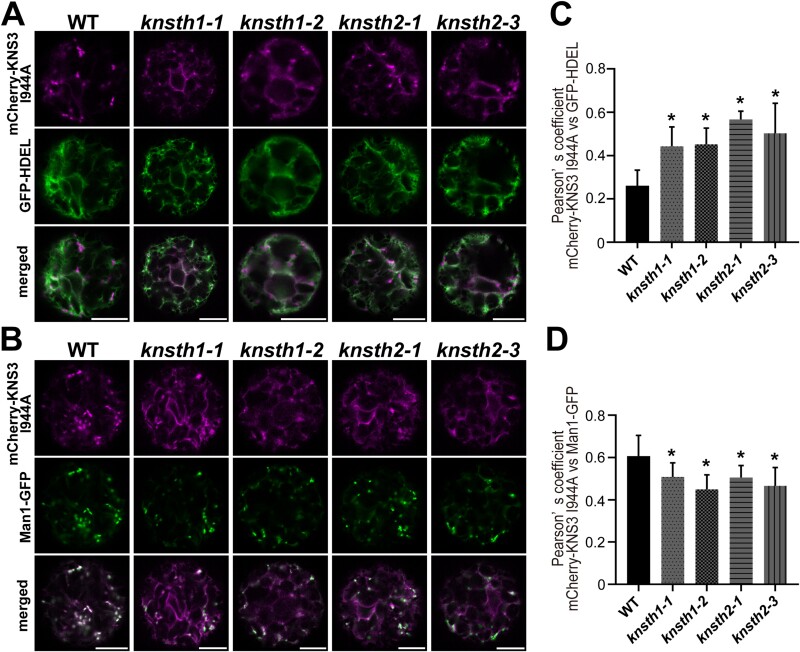
We then analyzed the localization of mCherry-KNS3 I944A, for which we showed an increased co-localization with the Golgi apparatus in Arabidopsis leaf mesophyll protoplasts (Fig. 8B, E). Owing to the defect in the mCherry-KNS3 I944A variant in retrograde trafficking from the Golgi to the ER, we were able to analyze the contributions of KNSTH1 and KNSTH2 in the ER-to-Golgi trafficking of KNS3. In contrast to the punctate localization seen in WT protoplasts, mCherry-KNS3 I944A showed both ER-like and punctate localization in knsth1-1, knsth1-2, knsth2-1, and knsth2-3 cells (Fig. 10A, B). The Pearson correlation coefficients between mCherry-KNS3 I944A and GFP-HDEL in the WT were approximately 0.3, whereas they were significantly higher, up to approximately 0.6, in knsth1-1, knsth1-2, knsth2-1, or knsth2-3 cells. (Fig. 10C). The Pearson correlation coefficients between mCherry-KNS3 I944A and Man1-GFP in the WT were approximately 0.6, whereas they were significantly lower, at approximately 0.5, in knsth1-1, knsth1-2, knsth2-1, or knsth2-3 cells (Fig. 10D). The results are summarized in Supplementary Table S3. These results indicated that KNSTH1 and KNSTH2 are important for the trafficking of KNS3 from the ER to the Golgi.
Fig. 10.
KNSTH1 and KNSTH2 are important for the trafficking of KNS3 from the endoplasmic reticulum to the Golgi. (A, B) mCherry-KNS3 I944A and GFP-HDEL (A) or Man1-GFP (B) in protoplasts from wild-type (WT) (Col-0), knsth1-1, knsth1-2, knsth2-1, and knsth2-3 Arabidopsis leaf mesophyll cells. Scale bars=10 μm. (C, D) Pearson correlation coefficients of mCherry-KNS3 I944A with GFP-HDEL (C) or Man1-GFP (D). Data represent the mean ±SD of 10–17 protoplasts. Asterisks denote significant differences between the WT and the mutant lines (*P<0.01; Dunnett’s test).
Accumulation of KNS3 and its two homologs is mutually dependent
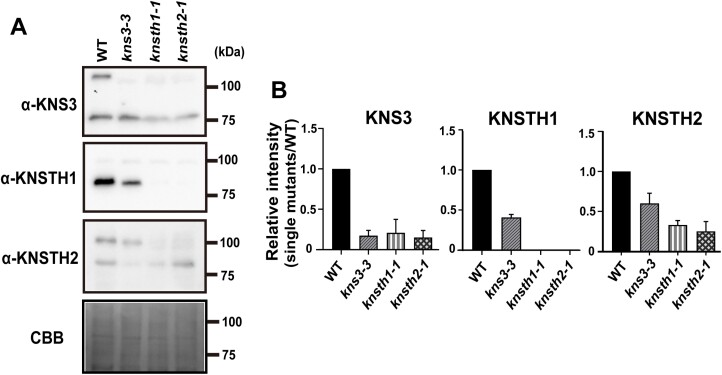
The rate of ER retention of GFP-NIP5;1 in the kns3-2 knsth1-1 knsth2-1 triple mutant was not significantly different from that in the single mutants (Fig. 5F). This result suggests that KNS3 shares a function with its two homologs in the ER exit of NIP5;1. To investigate the relation between KNS3 and its two homologs at the protein accumulation level, we quantified the protein in the roots of WT, kns3-3, knsth1-1, and knsth2-1 plants using newly generated anti-KNS3, KNSTH1, and KNSTH2 antibodies (Fig. 11A). The predicted molecular weights of KNS3, KNSTH1, and KNTH2 were 107.7, 76.5, and 92.4 kDa, respectively. In the microsome preparations, anti-KNS3 detected a clear band at approximately 120 kDa in the WT strain and a faint band at approximately 115 kDa in the kns3-3 mutant (Fig. 11A). The slight accumulation of the smaller protein was possibly due to translation from an aberrant mRNA containing a part of the T-DNA sequence (Fig. 2A). Anti-KNSTH1 detected clear bands at approximately 90 kDa in the WT but not in the knsth1-1 mutant. Anti-KNSTH2 detected clear bands at approximately 105 kDa in the WT and faint bands at approximately 105 kDa and 100 kDa in the knsth2-1 mutant. This faint expression was possibly due to translation from an mRNA in which the 13th intron, including the T-DNA sequence, was spliced out (Fig. 5B). These results indicated that the antibodies specifically detected the respective proteins. We then analyzed protein accumulation in the homolog mutants. The band intensities of KNS3 were approximately 20% and 15% in knsth1-1 and knsth2-1, respectively, compared with those in the WT (Fig. 11B). The band intensity of KNSTH1 was approximately 40% in kns3-3 and not detected in knsth2-1. The band intensities of KNSTH2 were approximately 60% and 30% in kns3-3 and knsth1-1, respectively (Fig. 11A, B). These results indicate that KNS3, KNSTH1, and KNSTH2 are unstable when one of the two homologs is not expressed.
Fig. 11.
The accumulation of KNS3 and its two homologs is mutually dependent. (A) Immunoblotting of KNS3, KNSTH1, and KNSTH2 proteins in microsomal fractions. wild-type (WT), kns3-3, knsth1-1, and knsth2-1 plants were grown on solid medium containing 30 μM B for 3 weeks. Approximately 60 plants were used as one sample for microsome preparation. Immunoblot analysis was performed using anti-KNS3, anti-KNSTH1, and anti-KNSTH2 antibodies. (B) Densitometric quantification. Data represent the mean ±SD of three independent experiments.
Discussion
Endoplasmic reticulum exit of NIP5;1 requires KNS3, KNSTH1, and KNSTH2
In our fluorescence imaging-based genetic screening, we identified six mutant lines (lines 1–3, 5–6, 10–6, 14–3, 15–2, and 37–1) in which GFP-NIP5;1 showed ER and PM localization (Fig. 1A). Using genetic mapping and whole-genome sequencing, we identified KNS3 as the gene responsible for the four allelic lines (line 1–3, 10–6, 14–3, and 15–2; Fig. 2A) and KNSTH2 as the gene responsible for line 37–1 (Fig. 5B). Retention of GFP-NIP5;1 in the ER was consistently observed in kns3 and knsth2 T-DNA insertion mutants (Figs 2C, 5E). We did not identify KNSTH1 from the forward-genetic study. However, protein BLAST showed that KNSTH1 has 26% identity with KNS3, and the topology of KNSTH1 was similar to that of KNS3 (Fig. 5A; Supplementary Fig. S4B). In knsth1 T-DNA insertion mutants, GFP-NIP5;1 showed ER and PM localization similar to that of the kns3 and knsth2 mutants (Fig. 5E). These results established that KNS3 and the newly described proteins KNSTH1 and KNSTH2 are required for the ER exit of NIP5;1. Phylogenetic analysis showed that these three proteins are well conserved in mosses, lycophytes, and angiosperms (Supplementary Table S4; Supplementary Fig. S9), suggesting conserved roles in plant physiology.
The defect in boron transport partially explains the defective fertility of kns3 mutants
The kns3 mutants showed defects in the ER exit of the boric acid channels NIP5;1 and NIP6;1 (Figs 2C, 3B). However, the kns3 mutants did not show similar B-deficient phenotypes to the nip5;1 or nip6;1 mutants (Supplementary Fig. S3), and the B concentrations in the roots, rosette leaves, and shoot apices of the mutants were not significantly different from those of the WT (Supplementary Fig. S3D). This was probably because a sufficient number of boric acid channels were still transported to the PM in the mutants. Additionally, we noticed that the siliques of kns3 mutants were shorter than those of the WT in the low B condition and were partially rescued by a normal to sufficient B supply (Fig. 4A). These results suggest that defective B transport in floral organs affects plant fertility. We also found that the pollen of kns3 mutants showed an abnormal exine structure, irrespective of B conditions (Fig. 4B). It is likely that KNS3 is involved in the ER exit of boric acid channels and other cargoes responsible for the construction of the exine structure. It was previously demonstrated that the loss of function of the COPII components SEC31B, SEC23A and D, and SAR1B and C resulted in defective exine structures on the pollen surface and male-sterile phenotypes in Arabidopsis (Zhao et al., 2016; Aboulela et al., 2018; Liang et al., 2020). These examples suggest that efficient ER exit is particularly important in tapetum cells, where massive secretion is required for pollen development.
KNS3 and its homologs function as a possible cargo-receptor complex for boric acid channels
KNS3, KNSTH1, and KNSTH2 are predicted to contain a short cytosolic C-terminal tail, a single transmembrane domain, and a large luminal ER region (Figs 2A, 5A; Supplementary Fig. S4). These structures are similar to those of some ER-Golgi cargo receptors, such as ER-Golgi intermediate compartment-53 (ERGIC-53)/lectin mannose-binding 1 (LMAN1) in mammals and yeast, and p24 proteins in mammals, yeast, and plants (Zhang et al., 2009; Pastor-Cantizano et al., 2016). Generally, cargo receptors help package cargo proteins into vesicles for subsequent trafficking. ERGIC-53/LMAN1 and p24 proteins transport glycoproteins and glycosylphosphatidylinositol-anchored proteins, respectively, from the ER to the Golgi apparatus (Zhang et al., 2009; Pastor-Cantizano et al., 2016). Based on the similarity in the protein structure and function of cargo proteins at the ER exit, we speculate that KNS3, KNSTH1, and KNSHT2 are cargo receptors for the ER exit of boric acid channels and other cargoes.
We observed that the rate of ER retention of GFP-NIP5;1 in the kns3-2 knsth1-1 knsth2-1 triple mutant was not significantly different from that in the single mutants (Fig. 5F). We also observed that the accumulation of KNS3, KNSTH1, and KNSTH2 was interdependent (Fig. 11B). Based on the ePlant database (Waese et al., 2017), KNS3, KNSTH1, and KNSTH2 are expressed ubiquitously in various tissues, but not in mature pollen (Honys and Twell, 2004; Nakabayashi et al., 2005; Schmid et al., 2005) (Supplementary Figs S10, S11, S12). These proteins likely function together in various cell types. Human ERGIC-53/LMAN1 forms a complex with MCFD2 (Zhang et al., 2003, 2005). Saccharomyces cerevisiae p24 proteins, including Erp1p, Erp2p, Emp24p, and Erv25p, form a heteromeric complex, and their accumulation is interdependent (Marzioch et al., 1999). Based on the analogy between these complexes, we hypothesize that KNS3 and its homologs function as a cargo-receptor complex. However, our co-immunoprecipitation assay using transgenic Arabidopsis expressing GFP-NIP5;1 and anti-KNS3/KNSTH1/KNSTH2 antibodies, and tobacco leaves expressing mCherry-KNS3 and GFP-NIP5;1 failed to detect interactions. This is probably because most of the GFP-NIP5;1 was localized in the PM, not in the ER. It will be important to test the direct interaction between the KNS3 complex and boric acid channels in the ER in future studies.
Next, we discuss the different roles of KNS3, KNSTH1, and KNSTH2 in the cargo-receptor complex. In Arabidopsis mesophyll protoplasts and tobacco leaf cells, mCherry-KNS3 localized to the ER and Golgi apparatus (Fig. 6).In contrast, the K941A and K943A variants localized to the Golgi and showed reduced ER localization (Fig. 8A, B, D), and the I944A variant localized to the Golgi but not to ring- and network-like structures in Arabidopsis protoplasts (Fig. 8A, B, E). The mCherry fluorescence derived from the K941A and K943A variants in Arabidopsis protoplasts and tobacco leaf cells and the I944A variant in tobacco leaf cells was also observed in the vacuoles (Fig. 8C; Supplementary Fig. S6A, B). These results suggest that the KxKI motif is important for Golgi-to-ER trafficking, and that defects in retrograde trafficking cause mistargeting of mCherry-KNS3 variants from the Golgi to the vacuole. In S. cerevisiae, the double lysine motif (KK) in the C-terminus of ERGIC-53/LMAN1, and the KxK motif in the C-terminal tail (-GKFFVKQKIL) of Erp1p, a member of the p24α subfamily, are important for retrograde trafficking from the Golgi to the ER (Schindler et al., 1993; Marzioch et al., 1999; Pastor-Cantizano et al., 2016). Based on this analogy, the cytosolic C-terminal KxKI motif in KNS3 may bind to the COPI complex for retrograde trafficking of KNS3 from the Golgi to the ER. This KxKI motif is conserved in KNS3 clade proteins from mosses to angiosperms (Supplementary Fig. S13). According to our working hypothesis, KNS3 is likely responsible for retrograde trafficking in the KNS3-KNSTH1-KNSTH2 complex (Supplementary Fig. S14).
KNSTH1 has a longer C-terminal tail (KRDRLFRNKRKQF) than that of KNS3 (PRAPKPKIN) or KNSTH2 (SPPSR). Previously, the double phenylalanine motif (FF) at the C-terminus of ERGIC-53/LMAN1 was shown to interact with the SEC23-SEC24 subcomplex of the COPII complex and to act as an ER-exit motif (Kappeler et al., 1997). The C-terminal tail of the p24γ subfamily contains a ΦF motif (where Φ represents a bulky hydrophobic residue), which presumably allows binding to a COPII component for ER exit (Pastor-Cantizano et al., 2016). These examples imply the possibility that KNSTH1 could bind to a COPII component with the ΦF motif in its C-terminal tail. The ΦF motif is conserved in the KNSTH1 clade proteins from mosses to angiosperms (Supplementary Fig. S13). Although we were unable to test this hypothesis due to the toxic effects of mCherry-KNSTH1 expression in bacterial and plant cells, KNSTH1 may be responsible for ER exit by binding to the COPII component in the KNS3-KNSTH1-KNSTH2–cargo complex (Supplementary Fig. S14).
In Arabidopsis mesophyll protoplasts and tobacco leaf cells, mCherry-KNSTH2 is localized mainly in the ER. Although we did not observe any changes in the localization of mCherry-KNSTH2 in the ER upon substitution of amino acids in the C-terminal tail (Fig. 9; Supplementary Fig. S7), we cannot rule out the possibility that KNSTH2 moves between the ER and Golgi, along with KNS3 and KNSTH1.
In conclusion, we revealed that KNS3, KNSTH1, and KNSTH2 are important for the ER exit of the boric acid channel NIP5;1 and possibly other proteins involved in the construction of the pollen wall structure. We propose that KNS3, KNSTH1, and KNSTH2 form a cargo-receptor complex that interacts with boric acid channels and other cargoes in the ER, transporting them to the Golgi by packaging them into COPII vesicles (Supplementary Fig. S14). In the Golgi apparatus, the cargo receptor complex separates from the cargo and returns to the ER via COPI vesicles. Subsequently, the boric acid channels and other cargo molecules move from the trans-Golgi network to the PM via the secretion pathway.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. At5g58100 (KNS3) is the causative gene for the ER retention of NIP5;1.
Fig. S2. GFP-NIP5;1 shows polar localization in mutants of KNS3 and its homologs.
Fig. S3. Growth of kns3 mutants under different B conditions.
Fig. S4. Topologies of KNS3, KNSTH1, and KNSTH2.
Fig. S5. Pollen structure in kns3, knsth1, and knsth2 multiple mutants.
Fig. S6. K941, K943, and I944 in the C-terminal tail of KNS3 are important for its trafficking from the Golgi to the ER.
Fig. S7. Mutations in the C-terminal tail of KNSTH2 do not affect its ER localization.
Fig. S8. Localization of mCherry-KNS3 and KNSTH2 is unchanged in single mutants of their homologs.
Fig. S9. Phylogenetic tree of KNS3 and its homologs.
Fig. S10. Expression pattern of KNS3.
Fig. S11. Expression pattern of KNSTH1.
Fig. S12. Expression pattern of KNSTH2.
Fig. S13. Multiple alignments of the amino acid sequences of the C-terminal tail of KNS3 and its homologs.
Fig. S14. A working hypothesis of the functions of KNS3, KNSTH1, and KNSTH2 in an ER-Golgi cargo-receptor complex.
Table S1. SSLP markers for rough mapping.
Table S2. Primers used in this research.
Table S3. Summary of the localization of mCherry-KNS3 and mCherry-KNSTH2 in Arabidopsis protoplasts and tobacco leaf cells.
Table S4. List of proteins collected using BLAST in the NCBI database.
Acknowledgements
We acknowledge Masahira Hattori and Kenshiro Oshima (University of Tokyo) for genome sequencing, Yoko Ito (Ochanomizu University) for the ST-YFP plasmid, and Arabidopsis Biological Resource Center (ABRC) for the GFP-HDEL and Man1-GFP plasmids. We thank Yuka Ohkita (Osaka Prefecture University) for skillful technical assistance, Motoaki Tojo (Osaka Metropolitan University) for technical guidance on SEM, and Keita Muro and Mayuki Tanaka (Osaka Metropolitan University) for the comments on the manuscript.
Contributor Information
Zhe Zhang, Graduate School of Life and Environmental Sciences, Osaka Prefecture University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka, 599-8531, Japan.
Shunsuke Nakamura, Graduate School of Agriculture, Hokkaido University, Kita 8, Nishi 5, Kita-ku, Sapporo, Hokkaido, 060-0808, Japan.
Arisa Yamasaki, Graduate School of Life and Environmental Sciences, Osaka Prefecture University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka, 599-8531, Japan.
Masataka Uehara, Graduate School of Agriculture, Hokkaido University, Kita 8, Nishi 5, Kita-ku, Sapporo, Hokkaido, 060-0808, Japan.
Shunsuke Takemura, Graduate School of Bioagricultural Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan.
Kohei Tsuchida, Graduate School of Bioagricultural Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan.
Takehiro Kamiya, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan.
Shuji Shigenobu, National Institute for Basic Biology, 38 Nishigonaka, Myodaiji-cho, Okazaki, 444-8585, Japan.
Katsushi Yamaguchi, National Institute for Basic Biology, 38 Nishigonaka, Myodaiji-cho, Okazaki, 444-8585, Japan.
Toru Fujiwara, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan.
Sumie Ishiguro, Graduate School of Bioagricultural Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan.
Junpei Takano, Graduate School of Life and Environmental Sciences, Osaka Prefecture University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka, 599-8531, Japan; Graduate School of Agriculture, Osaka Metropolitan University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka, 599-8531, Japan.
James Murray, Cardiff University, UK.
Author contributions
ZZ performed the majority of the experiments; SN, AS, MU, ST, and KT performed experiments; TK, SS, and KY analyzed the genome resequencing data; JT, SI, and TF designed the research and supervised the experiments; ZZ and JT wrote the manuscript; all the authors contributed to the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
This work is supported by a JSPS Grant-in-Aid (16K15082, 19H05763, 19H00934) and the NEXT program (GS001) to JT, and a JSPS Grant-in-Aid (19H05637) to TF.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Abas L, Benjamins R, Malenica N, Paciorek TT, Wiřniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C.. 2006. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nature Cell Biology 8, 249–256. [DOI] [PubMed] [Google Scholar]
- Aboulela M, Nakagawa T, Oshima A, Nishimura K, Tanaka Y.. 2018. The Arabidopsis COPII components, AtSEC23A and AtSEC23D, are essential for pollen wall development and exine patterning. Journal of Experimental Botany 69, 1615–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alassimone J, Naseer S, Geldner N.. 2010. A developmental framework for endodermal differentiation and polarity. Proceedings of the National Academy of Sciences, USA 107, 5214–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluri S, Büttner M.. 2007. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proceedings of the National Academy of Sciences, USA 104, 2537–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Wasteneys GO, Jones DA.. 2000. The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. The Plant Cell 12, 1179–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béthune J, Wieland FT.. 2018. Assembly of COPI and COPII vesicular coat proteins on membranes. Annual Review of Biophysics 47, 63–83. [DOI] [PubMed] [Google Scholar]
- Brandizzi F. 2018. Transport from the endoplasmic reticulum to the Golgi in plants: where are we now? Seminars in Cell and Developmental Biology 80, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Geanconteri A, Shrestha J, et al. 2011. A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiology 157, 947–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Gershlick DC, Bottanelli F, Hummel E, Hawes C, Denecke J.. 2010. A recycling-defective vacuolar sorting receptor reveals an intermediate compartment situated between prevacuoles and vacuoles in tobacco. The Plant Cell 22, 3992–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP.. 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nature Protocols 3, 619–628. [DOI] [PubMed] [Google Scholar]
- Funakawa H, Miwa K.. 2015. Synthesis of borate cross-linked rhamnogalacturonan II. Frontiers in Plant Science 6, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, Miller E.. 2016. Protein sorting at the ER–Golgi interface. Journal of Cell Biology 215, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR.. 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. The Plant Journal 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D.. 2004. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology 5, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Uemura T, Nakano A.. 2018. The Golgi entry core compartment functions as a COPII-independent scaffold for ER-to-Golgi transport in plant cells. Journal of Cell Science 131, 1–9. [DOI] [PubMed] [Google Scholar]
- Jothi M, Takano J.. 2023. Understanding the regulatory mechanisms of B transport to develop crop plants with B efficiency and excess B tolerance. Plant and Soil 487, 1–20. [Google Scholar]
- Kang H, Kim SY, Song K, Sohn EJ, Lee Y, Lee DW, Hara-Nishimura I, Hwang I.. 2012. Trafficking of vacuolar proteins: the crucial role of Arabidopsis vacuolar protein sorting 29 in recycling vacuolar sorting receptor. The Plant Cell 24, 5058–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler F, Klopfenstein DRC, Foguet M, Paccaud J-P, Hauri H-P.. 1997. The recycling of ERGIC-53 in the early secretory pathway. Journal of Biological Chemistry 272, 31801–31808. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski L, Růžička K, Naramoto S, Kleine-Vehn J, Friml J.. 2010. Trafficking to the outer polar domain defines the root-soil interface. Current Biology 20, 904–908. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jang M, Song K, Kang H, Lee MH, Lee DW, Zouhar J, Rojo E, Sohn EJ, Hwang I.. 2013. Functional identification of sorting receptors involved in trafficking of soluble lytic vacuolar proteins in vegetative cells of Arabidopsis. Plant Physiology 161, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Choi W, Wallace IS, Baudry J, Roberts DM.. 2011. Arabidopsis thaliana NIP7;1: an anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 50, 6633–6641. [DOI] [PubMed] [Google Scholar]
- Liang X, Li SW, Gong LM, Li S, Zhang Y.. 2020. COPII components Sar1b and Sar1c play distinct yet interchangeable roles in pollen development. Plant Physiology 183, 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chen X, Wu Y, Wang Y, He Y, Wu Y.. 2013. Directly transforming PCR-amplified DNA fragments into plant cells is a versatile system that facilitates the transient expression assay. PLoS One 8, e57171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Inada N, Yasuda S, Fukao Y, Fujiwara M, Sato T, Yamaguchi J.. 2014. The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiology 164, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJM, Solari RCE, Rowley A.. 1999. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Molecular Biology of the Cell 10, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L.. 2015. Aquaporins in plants. Physiological Reviews 95, 1321–1358. [DOI] [PubMed] [Google Scholar]
- Mohan T, Jones A.. 2018. Determination of boron content using a simple and rapid miniaturized curcumin assay. Bio-Protocol 8, e2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Pastor-Cantizano N, Robinson DG, Marcote MJ, Aniento F.. 2014. Arabidopsis p24δ5 and p24δ9 facilitate Coat Protein I-dependent transport of the K/HDEL receptor ERD2 from the Golgi to the endoplasmic reticulum. The Plant Journal 80, 1014–1030. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E.. 2005. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal 41, 697–709. [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA.. 2000. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiology 124, 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA.. 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiology 121, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A.. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Ishikawa S, Matsunami E, et al. 2015. New Gateway-compatible vectors for a high-throughput protein-protein interaction analysis by a bimolecular fluorescence complementation (BiFC) assay in plants and their application to a plant clathrin structure analysis. Bioscience, Biotechnology and Biochemistry 79, 1995–2006. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Bernat-Silvestre C, Marcote MJ, Aniento F.. 2018. Loss of Arabidopsis p24 function affects ERD2 trafficking and Golgi structure, and activates the unfolded protein response. Journal of Cell Science 131, jcs203802. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, García-Murria MJ, Bernat-Silvestre C, Marcote MJ, Mingarro I, Aniento F.. 2017. N-linked glycosylation of the p24 family protein p24δ5 modulates retrograde Golgi-to-ER transport of K/HDEL ligands in Arabidopsis. Molecular Plant 10, 1095–1106. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Montesinos JC, Bernat-Silvestre C, Marcote MJ, Aniento F.. 2016. p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253, 967–985. [DOI] [PubMed] [Google Scholar]
- Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS.. 2008. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Computational Biology 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Santiago P, Lagunas-Gómez D, Barkla BJ, Vera-Estrella R, Lalonde S, Jones A, Frommer WB, Zimmermannova O, Sychrová H, Pantoja O.. 2015. Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. Journal of Experimental Botany 66, 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Santiago P, Lagunas-Gomez D, Yáñez-Domínguez C, Vera-Estrella R, Zimmermannová O, Sychrová H, Pantoja O.. 2017. Plant and yeast cornichon possess a conserved acidic motif required for correct targeting of plasma membrane cargos. Biochimica et Biophysica Acta. Molecular Cell Research 1864, 1809–1818. [DOI] [PubMed] [Google Scholar]
- Routray P, Li T, Yamasaki A, Yoshinari A, Takano J, Choi WG, Sams CE, Roberts DM.. 2018. Nodulin intrinsic protein 7;1 is a tapetal boric acid channel involved in pollen cell wall formation. Plant Physiology 178, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C.. 2002. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. The Plant Journal 29, 661–678. [DOI] [PubMed] [Google Scholar]
- Scabone CM, Frigerio L, Petruccelli S.. 2011. A fluorescent reporter protein containing AtRMR1 domains is targeted to the storage and central vacuoles in Arabidopsis thaliana and tobacco leaf cells. Plant Cell Reports 30, 1823–1833. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carrera I, Frise E, et al. 2012. Fiji: an open-source platform for biological image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler R, Itin C, Zerial M, Lottspeich F, Hauri HP.. 1993. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. European Journal of Cell Biology 61, 1–9. [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU.. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Masaoka K, Nishi M, Nakamura K, Ishiguro S.. 2008. Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant and Cell Physiology 49, 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, Von Wirén N, Fujiwara T.. 2005. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proceedings of the National Academy of Sciences, USA 102, 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T.. 2010. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proceedings of the National Academy of Sciences, USA 107, 5220–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Wada M, Ludewig U, Schaaf G, Von Wirén N, Fujiwara T.. 2006. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. The Plant Cell 18, 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Yamagami M, Noguchi K, Hayashi H, Fujiwara T.. 2001. Preferential translocation of boron to young leaves in Arabidopsis thaliana regulated by the BOR1 gene. Soil Science and Plant Nutrition 47, 345–357. [Google Scholar]
- Tanaka M, Takano J, Chiba Y, Lombardo F, Ogasawara Y, Onouchi H, Naito S, Fujiwaraa T.. 2011. Boron-dependent degradation of NIP5;1 mRNA for acclimation to excess boron conditions in Arabidopsis. The Plant Cell 23, 3547–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T.. 2008. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. The Plant Cell 20, 2860–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara M, Wang S, Kamiya T, Shigenobu S, Yamaguchi K, Fujiwara T, Naito S, Takano J.. 2014. Identification and characterization of an Arabidopsis mutant with altered localization of NIP5;1, a plasma membrane boric acid channel, reveals the requirement for d-galactose in endomembrane organization. Plant and Cell Physiology 55, 704–714. [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, et al. 2022. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira V, Peixoto B, Costa M, Pereira S, Pisarra J, Pereira C.. 2019. N-linked glycosylation modulates Golgi-independent vacuolar sorting mediated by the plant specific insert. Plants 8, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, et al. 2017. ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. The Plant Cell 29, 1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhao X, Pang C, Zhou S, Qian X, Tang N, Yang N, Xu P, Xu X, Gao J.. 2021. IMPERFECTIVE EXINE FORMATION (IEF) is required for exine formation and male fertility in Arabidopsis. Plant Molecular Biology 105, 625–635. [DOI] [PubMed] [Google Scholar]
- Wang S, Yoshinari A, Shimada T, Hara-Nishimura I, Mitani-Ueno N, Ma JF, Naito S, Takano J.. 2017. Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. The Plant Cell 29, 824–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ma J, Jin X, Yue N, Gao P, Mai KKK, Wang X, Li D, Kang B, Zhang Y.. 2020. Three‐dimensional reconstruction and comparison of vacuolar membranes in response to viral infection. Journal of Integrative Plant Biology 63, 353–364. [DOI] [PubMed] [Google Scholar]
- Wudick MM, Portes MT, Michard E, et al. 2018. CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 360, 533–536. [DOI] [PubMed] [Google Scholar]
- Yáñez-Domínguez C, Lagunas-Gómez D, Torres-Cifuentes DM, Bezanilla M, Pantoja O.. 2023. A cornichon protein controls polar localization of the PINA auxin transporter in Physcomitrium patens. Development 150, dev201635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Sato T, Maekawa S, Aoyama S, Fukao Y, Yamaguchi J.. 2014. Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. Journal of Biological Chemistry 289, 15179–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari A, Hosokawa T, Amano T, Beier MP, Kunieda T, Shimada T, Hara-Nishimura I, Naito S, Takano J.. 2019. Polar localization of the borate exporter BOR1 requires AP2-dependent endocytosis. Plant Physiology 179, 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Cunningham MA, Nichols WC, et al. 2003. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nature Genetics 34, 220–225. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kaufman RJ, Ginsburg D.. 2005. LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. Journal of Biological Chemistry 280, 25881–25886. [DOI] [PubMed] [Google Scholar]
- Zhang YC, Zhou Y, Yang CZ, Xiong DS.. 2009. A review of ERGIC-53: its structure, functions, regulation and relations with diseases. Histology and Histopathology 24, 1193–1204. [DOI] [PubMed] [Google Scholar]
- Zhao B, Shi H, Wang W, Liu X, Gao H, Wang X, Zhang Y, Yang M, Li R, Guo Y.. 2016. Secretory COPII protein SEC31B is required for pollen wall development. Plant Physiology 172, 1625–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.