ABSTRACT
Unlocking the potential of broadly reactive coronavirus monoclonal antibodies (mAbs) and their derivatives offers a transformative therapeutic avenue against severe COVID-19, especially crucial for safeguarding high-risk populations. Novel mAb-based immunotherapies may help address the reduced efficacy of current vaccines and neutralizing mAbs caused by the emergence of variants of concern (VOCs). Using phage display technology, we discovered a pan-SARS-CoV-2 mAb (C10) that targets a conserved region within the receptor-binding domain (RBD) of the virus. Noteworthy, C10 demonstrates exceptional efficacy in recognizing all assessed VOCs, including recent Omicron variants. While C10 lacks direct neutralization capacity, it efficiently binds to infected lung epithelial cells and induces their lysis via natural killer (NK) cell-mediated antibody-dependent cellular cytotoxicity (ADCC). Building upon this pan-SARS-CoV-2 mAb, we engineered C10-based, Chimeric Antigen Receptor (CAR)-T cells endowed with efficient killing capacity against SARS-CoV-2-infected lung epithelial cells. Notably, NK and CAR-T-cell mediated killing of lung infected cells effectively reduces viral titers. These findings highlight the potential of non-neutralizing mAbs in providing immune protection against emerging infectious diseases. Our work reveals a pan-SARS-CoV-2 mAb effective in targeting infected cells and demonstrates the proof-of-concept for the potential application of CAR-T cell therapy in combating SARS-CoV-2 infections. Furthermore, it holds promise for the development of innovative antibody-based and cell-based therapeutic strategies against severe COVID-19 by expanding the array of therapeutic options available for high-risk populations.
Trial registration: ClinicalTrials.gov identifier: NCT04093596.
KEYWORDS: Non-neutralizing antibodies, infected cell-targeting, SARS-CoV-2, antibody-based therapy, cell therapy, CAR-T cells
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped RNA virus that belongs to the Coronaviridae group. This virus is known for causing the COVID-19 disease [1,2]. Since its outbreak in 2019, COVID-19 pandemic has been reported with millions of cases and deaths. Nowadays, thanks to the efforts from the global healthcare community, and worldwide vaccination programmes, the pandemic has been controlled. Even though global vaccination has induced immunity in the global population, there are different high-risk subgroups such as solid organ transplant recipients, patients with hematological disorders and immunocompromised patients, who are unable to acquire this immunity and are still at risk of COVID-19 disease with severe symptomatology [3–5].
An unprecedented effort by researchers around the world has resulted in the development of a spectrum of preventive and therapeutic approaches. Among them, several neutralizing antibodies directed against Spike protein have been developed and used to treat SARS-CoV-2 infection [6–9]. However, sustaining their effectiveness in the new variants of concern (VOCs) has been challenging because of the antibody escape mutations. It is therefore necessary to advance the development of immunotherapies that can effectively counter viral evolution. This could include the development of antibodies directed against highly conserved viral epitopes and able to target infected cells. Such antibodies and their derivatives hold promise for both antibody-based and cell-based therapeutic approaches. Most efforts for the development of protective anti-SARS-CoV-2 mAb have been focused thus far in targeting viral particles [6–9], while much less attention has been concentrated in targeting infected cells. This is important to take into consideration as antiviral antibodies display multiple Fc-mediated effector functions (i.e. antibody-dependent cellular cytotoxicity, ADCC; antibody-dependent cellular phagocytosis, ADCP, …) that enable them to kill infected cells [10]. In addition, neutralization does not always align with ADCC and some neutralizing antibodies such as Sotrovimab that display lower potency (high IC50) can still be efficient in reducing viral propagation [11]. Worthy of note, the assessment of the effector functions of anti-SARS-CoV-2 mAbs used in the clinic has mostly been done using Spike-expressing cells as a surrogate of infected cells [11,12]. However, the critical question of whether such antibodies effectively bind to and kill infected cells has been considerably understudied. It is therefore urgent to develop antibodies targeting highly conserved viral epitopes (known as “coldspots”) [13] and eliminating infected cells. This approach could emerge as a potent strategy against SARS-CoV-2 infection, poised to combat not only current variants but also future evolutions of the virus.
The discovery of anti-SARS-CoV-2 mAbs able to target infected cells holds therapeutic promise, with applications spanning diverse treatment modalities. They can be used as full-length molecules exploiting their Fc-mediated polyfunctionality. In addition, antibody fragments offer a pathway to develop new cell-based immunotherapies, notably Chimeric Antigen Receptor (CAR)-T cell therapy, which harnesses the combined strengths of antibodies and T-cells. In the recent years, CAR-T cell therapy has demonstrated remarkable success in the haematological cancers area. Encouraged by these achievements, there is growing interest in the development of CAR-T cell therapies for different diseases, including infectious diseases such as SARS-CoV-2 infection [14–17]. This enthusiasm is fuelled by recent insights suggesting that SARS-CoV-2-specific T-cell therapy may confer clinical benefits in severe cases of COVID-19 [18]. Additionally, evidence shows that CD8+ T cells contribute to survival in COVID-19 patients with hematologic cancers [19]. Given the absence of therapeutic options for high-risk populations vulnerable to SARS-CoV-2 infection, our aim was to pioneer novel therapeutic strategies aimed at eliminating infected cells, thereby aiding in the resolution of viral infection. These approaches could harness the diverse therapeutic potential of mAb and their derivative to create both antibody-based and cell-based interventions.
In the present work, we developed a potent pan-SARS-CoV-2 non-neutralizing mAb, called C10, which targets a conserved region within the receptor-binding domain (RBD) of the virus and effectively recognizes nearly all known VOCs. Importantly, C10 effectively binds to and lysates infected cells through ADCC leading to reduced viral titers. Leveraging this pan-SARS-CoV-2 mAb, we have engineered CAR-T cells capable of efficiently killing lung epithelial cells infected with the virus. Overall, our work identifies a pan-SARS-Cov-2 mAb able to target bona fide infected cells and provides a proof-of-concept for the potential use of CAR-T cell therapy in combating SARS-CoV-2 infections, especially in immunocompromised patients. Our findings also highlight the potential of non-neutralizing mAbs in fighting emerging infectious diseases and emphasize the promising prospects of repurposing them for therapeutic applications using various antibody formats.
Material & methods
Ethics
All human blood samples were fully anonymized and given with previous written consent of the donor. Samples were given under ethical approval of appropriate institutions.
Antibody selection
The human synthetic HuscI library in single-chain variable fragment (scFv) format is based on the sequence of the optimized 13R4 scFv [20] and is displayed on filamentous phages using the pHEN1 vector [21]. Diversity was introduced at positions [Aho numbering [22]] L38-40, L58, L69, L109, L111, L112, H39-40, H57, H59, H67, H69, and with eight 2–9 residue loops inserted between positions H110-134 of VH-CDR3 [23]. At each position the introduced diversity encoded a mix of five amino acids (Y, S, D, N, G), except when it resulted in a site of glycosylation (NxS/T), deamidation (NG), or isomerization (DG) where the introduced sequence was, depending on the surrounding framework sequences, QxS/T or NxA, QG or NA, EG or NA, respectively. The library diversity, as measured by the number of transformants, was 3 × 1010.
Antibody selection was performed by capture in 96-well immunoplates coated with Streptavidin then incubated with Tris-NTA-Biotin (Biotechrabbit #BR1001201) followed by his-tagged Wuhan Spike RBD (Sino Biological # 40592-V08H). The library (1012 cfu) was depleted twice using Streptavidin-coated plates, before the first round. Captured phages were eluted with 100 μL of freshly prepared elution buffer (50 mM Tris-HCl pH 8, 125 μg/mL trypsin and 1 mM CaCl2). These phages were used to prepare a new phage stock with TG1 bacteria and the KM13 helper phage [24]. After 3 rounds of panning, eluted phages were used to infect E. coli HB2151, plated and individual clones were used for monoclonal screening by Enzyme-linked immunosorbent assay (ELISA) using bacteria supernatant and scFv detection using HRP-coupled anti-myc 9E10 monoclonal antibody. After sequencing of the best binders, 7 VH sequences were selected, and a new library constructed by shuffling this pool of VH with the whole diversity of the original VL library using a Fab displaying vector derived from pHEN1. The resulting library was first panned as previously (R4), then the phage stock incubated with soluble his-tagged RBD in decreasing concentration (10 then 1 nM) and captured for 15 min on immunoplates coated with Streptavidin and incubated with Tris-NTA-Biotin. Eluted phages were used to infect E. coli HB2151, plated and bacteria supernatant containing Fab of individual clones were screened by ELISA using an anti-human Fab antibody (A0293-1ML; Sigma).
Antibody and antibody-fragment production
Full-length C8 and C10 mAbs were produced by Evitria (https://www.evitria.com/). They were produced in mammalian CHO cells upon transfection with the light chain and heavy chain variable sequences of C8 and C10 mAbs in a IgG1κ backbone. To obtain the C10 F(ab’)2 fragment, the recombinant C10 monoclonal IgG1 antibody was treated with IdeZ Protease (Promega; Cat# V8341) and purified using NAb™ Protein A Plus Spin Columns (Thermo Scientific #89952) following the manufacturers’ procedures. To produce the Fc-mutated C10 LALA-PG IgG1, which carries a silent Fc mutation via the Leu234Ala, Leu235Ala, and Pro329Gly (LALA-PG) substitutions in the Fc region [25], the VH and VL fragments were inserted into a production plasmid using Golden Gate assembly [25], the VH and VL fragments were inserted into a production plasmid using Golden Gate assembly. The VH and VL fragments were purified using the Macherey-Nagel PCR Clean-up and Gel Extraction kit. Following this, Golden Gate assembly was performed with the PEF-recipient-IgG-LALA-PG vector. The resulting clones were validated by PCR and confirmed through sequencing. Plasmids were then produced and used for transfection in HEK-293 T cells using the JET-PEI transfection agent. The IgG expression was driven by a human EF1 promoter, with both the light and heavy chains produced from the same transcript. An IRES sequence from ECMV facilitated the translation of the heavy chain. The supernatants from the transfected cells were collected, and the recombinant Fc-mutated antibodies were purified using recombinant Protein A agarose beads (ProteinMods).
Surface plasmon resonance (SPR)
SPR measurements were performed on a T200 apparatus (Cytiva) at 25°C in phosphate buffer saline buffer pH7.4 containing 0.05% of P20 surfactant (PBS-P+). The screening of Fabs (C10, G12, 2B6, 2B2, 2B3) or antibodies (G6, D12, E10, C8, D8) was performed by one injection of SARS-CoV-2-Spike-RBD-His protein (Sino-biological) at 50nM on Fab (15 nM) or IgG (20nM) captured on anti-Fab antibody (Anti-Fab IgG from IMab I5260). An anti-IL13RA2 Fab is taken as negative control.
For the antibodies C8, C10 and S309 (the parent antibody of Sotrovimab used as control), Single Cycle Kinetic (SCK) was used to evaluate more accurately the kinetic parameters of the interaction with the Spike variant trimeric proteins Wuhan, Delta, Omicron (from R&Dsystems). For this purpose, anti-Histidine antibody (R&D Biosystems) was immobilized on one flowcell of a CM5S sensorchip by amine coupling according to the manufacturer’s instructions (Cytiva). A flowcell control was prepared using the same chemistry without anti-his antibody. For SCK experiments, Spike protein was captured at 200–300RU on anti-his immobilized and five increasing concentrations of antibody were injected at 100 µL/min (injection time of 60s). After a dissociation of 400s in running buffer, sensor surface was regenerated using glycine-HCL pH1.7. All sensorgrams were corrected by subtracting the low signal of flowcell control and blank buffer injections. Kinetic parameters were evaluated using (T200 evaluation software 3.2.1). The sensorgrams were analysed using a Langmuir 1:1 interaction model for the screening of Fab and IgG, and a bivalent interaction model for SCK.
Competition experiments
Competition experiments were performed to obtain information on the C10 binding regions. The first antibody (C10) was immobilized on the surface of CM5 chips (Cytiva) through standard amine coupling; RBD was then flowed to form the RBD/antibody complex, and shortly thereafter, the second antibody (Tixacevimab, Cilgavimab or Bebtelovimab) was injected. All the competition experiments were performed at 25°C on a Biacore 8 K instrument (Cytiva) using 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% Tween-20 as running buffer.
Docking and structural approaches
The C10 variable fragment was modelled according to the canonical structure method with the programme RosettaAntibody [26] as previously described [27]. Docking was performed using RosettaDock v3.1 as previously described [28]. In summary, C10 model was docked to Wuhan RBD experimental structure (PDBid: 6m17). Amongst the thousands of computationally generated complexes, the decoy in better agreement with experimental data (no cross competition with hACE2, Bebtelovimab, Cilgavimab and Tixacevimab and binding to all SARS-CoV-2 variants until Omicron XBB.1.5 and other RBD mutants) was selected and further refined by computational docking. The selected model was subjected to a 350 ns molecular dynamics (MD) simulation to adjust the local geometry and verify that the structure was energetically stable. MD was performed with GROMACS [29]. The system was initially set up and equilibrated through standard MD protocols: proteins were centred in a triclinic box, 0.2 nm from the edge, filled with SPCE water model and 0.15 m Na+ Cl- ions using the AMBER99SB-ILDN protein force field. Energy minimization was performed in order to let the ions achieve a stable conformation. Temperature and pressure equilibration steps, respectively at 310 K and 1 Bar, of 100 ps each were completed before performing the full MD simulations with the above-mentioned force field. MD trajectory files were analysed after removal of Periodic Boundary Conditions. The stability of each simulated complex was verified by root mean square deviation and visual analysis. The structures of C10 bound to SARS-CoV-2 Spike trimer was built using PyMOL software (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) PDB ID 6VXX served as a basis for the “three down” conformation; PDB ID 6VYB for the “one RBD up” conformation. Conformations not directly available (e.g. trimer with “three RBD up”) were obtained by structural alignment and repetition of the appropriate conformation in S monomer structures (e.g. monomeric “RBD up”) using PyMol software suite.
Human blood samples, primary cells purification and CAR-T generation
Healthy donor blood buffy coats were obtained from the local reference blood bank: Banc de Sang i Teixits, (BST, Barcelona, Spain; #HCB/2022/0137) and Etablissement Français du Sang (EFS, Montpellier, France; #21PLER2018-0069), according to local regulation. Human peripheral blood mononuclear cells (PBMCs) were obtained from buffy coats by density-gradient centrifugation (Lypmhoprep, StemCell Technologies), and were cultured in RPMI Medium 1640 (1X) (Gibco, 31870-025), 10% fetal bovine serum (FBS) (Sigma, F9665-500ML), penicillin/streptomycin (100 U/mL and 100 μg/mL, Gibco # 15140122).
Healthy donor NK cells were purified from human PBMCs by negative selection using a NK magnetic isolation kit (Miltenyi Biotech #130-092-657). Isolated NK cells were cultured in RPMI Medium 1640 (1X) (Gibco, 31870-025), 10% FBS (Sigma, F9665-500ML), penicillin/streptomycin and IL-2 (100 IU/mL; Miltenyi Biotec).
Healthy donor T cells were obtained from buffy coats by density-gradient centrifugation (Lymphoprep, StemCell technologies) using the RosetteSep™ Human T-cell Enrichment Cocktail (Stemcell technologies, 15061) to isolate T cells from whole blood by negative selection. The purified T cells were cultured in R10 Cell Medium, consisting in RPMI Medium 1640 (1X) (Gibco, 31870-025), 10% FBS (Sigma, F9665-500ML), penicillin/streptomycin, and IL-2 (50 IU/mL; Miltenyi Biotec). Cells were then activated and expanded for 4 days using beads conjugated with CD3 and CD28 mAbs (Dynabeads, Gibco, 11131D). The following day, T cells were transduced with the lentivirus encoding for the different CAR-T cells. Cells were expanded over 9–10 days, counting cells and media feeding daily. Finally, cells were frozen until used in the experiments.
Cell line culture
Human embryonic kidney cells (293 T HEK cells) (HEK 293T-ATCC-CRL-1575TM) and A549 cell (ATCC #CCL-185) were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, cat #31966) supplemented with 10% FBS, sodium pyruvate and antibiotics (penicillin–streptomycin). SARS-CoV-2 Spike-expressing cells (293GS and A549GS) were generated as follows. Briefly, a chimeric Gaussia luciferase construct designed for internal membrane binding was cloned into a third-generation lentivirus system. Next, the Wuhan spike protein under a T2A sequence for autoprolytic cleavage, was cloned into the same lentivirus plasmid. The resulting lentiviral vectors were then used to transduce 293 and A549 cells. After several passages, the cell lines were clonally isolated and selected based on Gaussia luciferase expression. They were further analysed using anti-Spike antibody staining and flow cytometry. Finally, the selected clones were passaged for over 40 days to evaluate transgene stability. Generated 293GS and A549GS cells were maintained in the same medium as parental cells. Jurkat cell line was cultured in RPMI media supplemented with 10% FBS and antibiotics (penicillin/streptomycin). Vero E6 cells (African green monkey kidney cells) were obtained from ECACC (#Vero C1008, ECACC 85020206) and maintained in DMEM supplemented with 10% heat-inactivated FBS and penicillin/streptomycin. Human pulmonary Alveolar A549-hACE2 cells were obtained from original A549 cells (ECACC #86012804) transduced with a lentiviral vector expressing human ACE2 receptor (manufactured by FlashTherapeutics company, Toulouse, France) and sorted by flow cytometry for having more than 80% hACE2 on their surface. The sorted A549-hACE2 cells were maintained in RPMI supplemented with 10% heat inactivated FBS, 1% sodium pyruvate, 0.5% HEPES and antibiotics (penicillin/streptomycin). Calu-3 cell line (EP-CL-0054, Elabscience Biotechnology Inc) was cultivated in the presence of DMEM (Sigma #D6429), 10% FBS, penicillin/streptomycin, 25 mM Hepes (Sigma #H0887). All cell lines were cultivated at 37°C / 5% CO2.
Virus production, purification and titration
SARS-CoV-2 strains used (detailed in the Supplementary Table 1) are: Wuhan, sequence: EPI_ISL_410720 (GISAID ID); D614G, sequence EPI_ISL_414631 (GISAID ID); ALPHA (UK), sequence EPI_ISL_1138411; BETA (South African), sequence EPI_ISL_1138745; DELTA, sequence EPI_ISL_3030060; GAMMA (Brasil), sequence EPI_ISL_2280284; Omicron, sequence EPI_ISL_8353353; Omicron BA.2, sequence EPI_ISL_9879476; Omicron BA.5, sequence EPI_ISL_13017789; Omicron XBB1.5, sequence EPI_ISL_16319988. These strains were supplied by the Institute Pasteur in the frame of a project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement #653316.
The virus strains were propagated in Vero E6 cells with DMEM containing 2.5% FBS at 37°C-5% CO2 and were harvested 72 h post-inoculation. Virus stocks were harvested and stored at −80°C as described [30]. The titers were determined by means of a plaque assay on a monolayer of Vero E6 cells as previously described [31]. Briefly, 100 μL from infected cell culture supernatants were titrated using 12 serial dilutions. The plaque-forming unit (PFU) values were determined by scoring the wells displaying cytopathic effects from 6 replicates/condition. The virus titer was determined as the number of PFU/mL, and multiplicity of infection (MOI) was the PFU/cell ratio.
Microscopy immunolabeling and imaging
Vero E6, A549-hACE2 and Calu-3 cells were cultured and infected with SARS-CoV-2 as described above. Cells were incubated with primary antibodies (C8 or C10) at different concentrations (ranging from 1 µg/mL to 50 µg/mL) during 1 h (4°C in PBS + 2% FBS) and then incubated with an Alexa Fluor® 647 anti-human IgG Fc secondary antibody (diluted 1:200; clone M1310G05 from BioLegend #410713) or goat anti-human IgG F(ab’)2 (1:200; polyclonal from Invitrogen # AB_429704) for 45 min. After washing, nuclei were labelled with Hoechst dye (1:5000; Molecular Probes). Images were acquired using a CellDiscoverer 7 LSM900 Airyscan2 microscope (ZEISS) equipped with a 10X (x2) and 20X (x2) objectives. Image analysis was performed using ZEN lite. Live-cell imaging was performed with time-lapse acquisition every 5 min over a 10-hour period (121 images) to visualize the cytotoxic capacity of C10-CAR-T cells. Infected cells were labelled in green (through the use of NeoGreen virus, a fluorescent SARS-COV-2 molecular clone) [32] and effector cells in red and blue (through the use of anti-CD45-APC antibody (Clone HI30, BioLegend) and Hoechst dye at 1:5000, respectively).
Real-time cytotoxicity assay
The cytotoxicity assay was performed using an xCELLigence real-time cell analyser (RTCA) System (Agilent Technology). Impedance-based RTCA was used for label-free and real-time monitoring of cytolysis activity. The cell index (CI) based on the measured cell-electrode impedance was used to measure cell viability.
For cytotoxic activity against Spike-expressing cells, basal readout was performed by addition of 50 μL of DMEM supplemented with 10% FBS, afterwards 1 × 104 target cells were seeded in E-plate 16 (Agilent, ref: 5469830001) and incubated for 16–24 h at 37 °C/5%CO2 in 50 μL DMEM supplemented with 10% FBS to allow their attachment. After the attachment period, CAR-T cells were added on top in 100 μL of RPMI-1640 supplemented with 10% FBS in an effector to target ratio 5:1.
For cytotoxic activity against infected lung epithelial cells, Calu-3 were used. Basal readout was performed by addition of 200 µL of DMEM supplemented with 10% FBS. Afterwards, Calu-3 were seeded at a density of 1,7 × 104 cells per well and grown for 72 h at 37°C/5% CO2 in 200 µL DMEM supplemented with 10% FBS to allow their attachment and growing. After the attachment period, Calu-3 were infected with SARS-CoV-2 (MOI 10−4) in DMEM without FBS during two hours. Afterwards, the medium was replaced by 80 µL of fresh RPMI-1640 supplemented with 10% FBS. Seven hours later, un-transduced cells (UTD) or SARS-CoV-2-directed CAR-T cells were added on top of the RTCA unit in 80 µL of RPMI-1640 at 5:1 ratio (effector:target). For ADCC assessment, primary NK cells were added at 1:1 (effector:target) ratio. NK cells were previously incubated with the C10 mAb, the F(ab’)2 fragment of C10 or the mutated C10-LALA-PG (at 10 µg/ml) for 1 h. The impedance signals were recorded for 96–120 h every 20-minute intervals.
Consecutive cytotoxicity assays
To evaluate the persistence in the cytotoxicity of CAR-T lymphocytes targeting the SARS-CoV-2 S protein, target cells and CAR-T lymphocytes were seeded at a 5:1 ratio in 48-well plates. After 48 h, double the initial number of target cells was added to the same well (rechallenge), repeating this process until reaching the fourth rechallenge. The final fourth rechallenge was conducted in the xCELLigence system to evaluate differences in the cytotoxicity activity of the different CAR-T lymphocytes. After the last rechallenge, the expression of markers associated with T-cell exhaustion was evaluated by flow cytometry. During the course of the experiment, at each rechallenge timepoint, a constant volume of cell culture was collected to label the cells with CD3 or CD45, in order to assess the variation of number of T lymphocytes during these cytotoxicity assays.
Enzyme-linked immunosorbent assay (ELISA)
Co-culture supernatants were analysed for cytokine production (IFNγ and IL-2) by ELISA, following the manufacturer’s instructions (DuoSet Elisa, R&D Systems, Abington, UK). ELISA plates (NUNC-IMMUNO Maxisorp, Thermofisher, Waltham, USA) were coated (100 μl/well) with the capture antibody diluted at the working concentration in PBS at room temperature overnight. Then, plates were washed and blocked by adding 300 μL PBS containing 1% BSA to each well and incubated at room temperature for 1 h. The ELISA assay procedures were performed after sample dilution in Reagent Diluent. Briefly, 100 μL of sample or standards diluted in Reagent Diluent were added per well and incubated 2 h at room temperature. After three wash steps with 400 μL Wash Buffer, 100 μL of the Detection Antibody, diluted in Reagent Diluent, were added to each well and incubated 2 h at room temperature. After three wash steps, 100 μL of the working dilution of Streptavidin-HRP were added to each well and incubated for 20 min at room temperature. Again, after three wash steps, 100 μL of the working dilution of Substrate Solution were added to each well and incubated for 20 min at room temperature. Then, 50 μL of Stop Solution were added to each well and the plate was gently mixed. The optical density of each well was immediately determined using Epoch microplate spectrophotometer (BioTek Instrumentals, Inc., Winooski, VT, USA).
Flow cytometry
During the T-cell expansion, the following mAbs were employed: anti-CD4-PerCP (BD Biosciences, 555348), anti-CD8-AlexaFluor700 (BD Biosciences, 345773). CARs were detected using Biotin-SP AffiniPure Goat Anti-Mouse IgG, F(ab’)2 fragment (Jackson ImmunoResearch, 115-065-072) and Streptavidin-PE (eBioscience, 12-4317-87). In the case of the T-cell exhaustion panel, the following antibodies were employed: anti-PD1-APC (BD Biosciences, 558694), anti-CTLA4-PE (BD Biosciences, 557301), anti-LAG3-PeCy7 (eBioscience, 25-2239-42) and anti-CD8-APC H7 (BD Biosciences, 566855). In order to evaluate total T-cell proliferation an anti-CD3-BV510 antibody (BD Biosciences, 564713) was used.
For the detection of Spike expression, a rabbit mAb against the Spike protein was employed, followed by a staining with a mouse-anti rabbit IgG-FITC secondary antibody. Samples were run through the fluorescence-activated cell sorting flow cytometer Attune, and data were analysed using the FlowJo Software.
Luminiscence assay
For luminescence determination the Pierce™ Gaussia Luciferase Glow Assay Kit (ThermoFischer Scientific) was employed. A549GS or 293GS cells (1× 104) were plated in a white 96 flat well plate (ThermoFischer Scientific). Cells were lysed and coelenterazine was added to the cell lysate. After a 10-minute period incubation, luminescence was read using Synergy HT spectrophotometer (Biotek, Vermont, USA).
Transcriptomic studies
RNA was extracted from the different CAR-T constructs and UTD. Total RNA was then purified from 5 × 105 cells using the miRNeasy Micro (Qiagen GmbH) following the manufacturer's instructions. The quantification of all RNA samples was performed using the Nanodrop One (Thermo Fisher Scientific), and RNA integrity was evaluated using the Agilent 2100 Bioanalyser system. RNA expression profiling was performed using CAR-T Characterization Panel (770 genes covering the core pathways and cellular phenotypes that are known to predict CAR-T activity). Gene expression analysis was conducted with the NanoString technology. Briefly, 5 μL/sample containing 50 ng of total RNA was combined with the nCounter® reporter CodeSet (3 μL) and nCounter® capture ProbeSet (2 μL) along with hybridization buffer (5 μL) for an overnight hybridization reaction at 65°C. The reaction was then cooled to 4°C, the samples were purified, immobilized on a cartridge, and the data were assessed using the nCounter SPRINT. All expression data was reviewed using NanoString® nSolverTM Analysis software 4.0. The up-regulated genes were classified into the main 5 categories proposed by NanoString: Activation, Phenotype, Metabolism, Persistence, and Exhaustion.
Quantification of cytokines and chemokines secreted by infected cells (Vero E6, A549-hACE2 and Calu-3)
Secretion of soluble cytokines and chemokines was assayed from cell-free culture supernatants using bead-based immunoassays (LegendPlex, Biolegend). Two assays were used allowing the quantification of IL-1β, Il-6, TNF-α, CXCL10, IFN-λ1, IL-8, IL-12p70, IFN-α2, IFN-λ2/3, GM-CSF, IFN-β, IL-10, IFN-γ (LegendPlex Human anti-virus response panel, Biolegend #740390) and IL-8, CXCL10, CCL11, CCL17, CCL2, CCL5, CCL3, CXCL9, CXCL5, CCL20, CXCL1, CXCL11, CCL4 (LegendPlex Human proinflammatory chemokines panel, Biolegend #740985). Immunoassays plates were read on a Novocyte (Agilent Technologies) flow cytometer and data were analysed with the appropriate Biolegend LEGENDplexTM software (https://legendplex.qognit.com).
Statistics
Statistical analyses were performed using Prism software version 10.2.3 (GraphPad). Simple group comparisons were performed using the paired t-test. Multiple group comparisons were performed using Tukey’s multiple comparisons test. Significance was assigned as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Results
Identification and characterization of anti-SARS-CoV-2 Spike antibodies by phage display
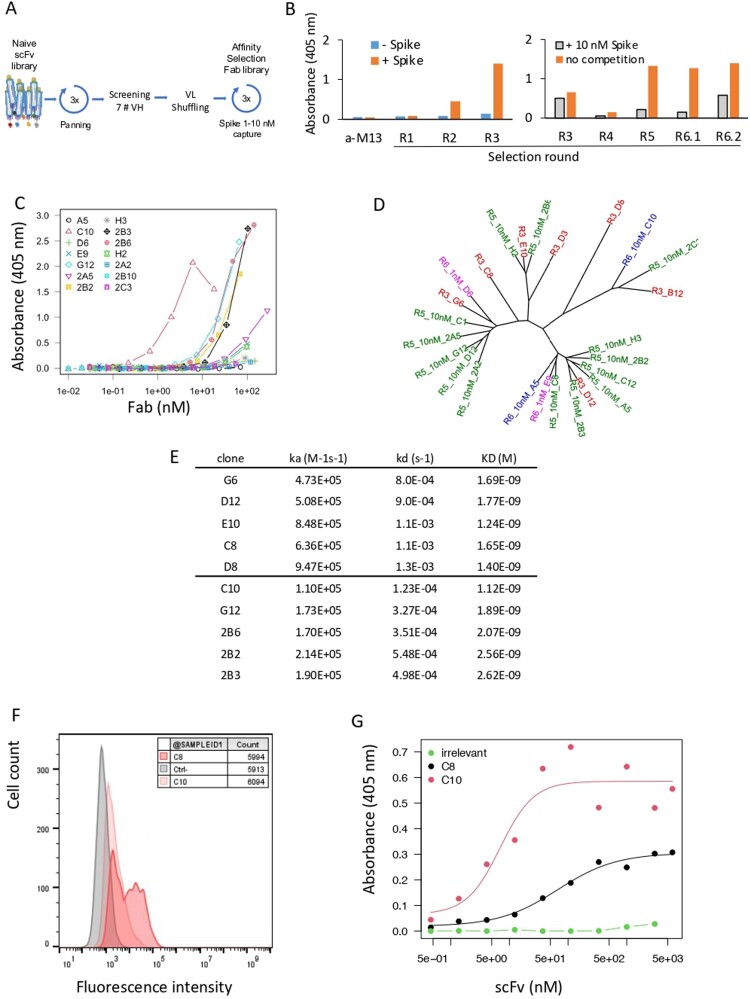
To select antibodies against SARS-CoV-2-Spike, a large naive synthetic scFv library was used for three consecutive rounds of biopanning on recombinant Wuhan Spike RBD captured by its poly-Histidine tag in microtiter plates (Figure 1(A)). After 2 and 3 rounds of panning the library was strongly enriched in RBD-positive clones, as shown by the positive signal by ELISA (Figure 1(B), left). Ninety-five clones were tested for binding and the best 7 clones (B12, C8, D3, D8, D12, E10, G6, referred to as parental clones) with different VH sequences were conserved. These 7 VH were shuffled with the whole VL library of a diversity of 4 × 105, and the resulting library in a fragment antibody (Fab) format was displayed on phages and sorted in solution by capture in decreasing concentrations of RBD (Figure 1(A)). To evaluate the efficiency of the process, pool of phages eluted after each round were tested for binding by ELISA in presence or absence of 10 nM competing soluble RBD. If binding is inhibited by the soluble RBD, this means that the Fabs expressed on phages are significantly complexed and likely have a nM affinity. Whereas the library before shuffling was only weakly inhibited (R3, 22%), after shuffling and selection all the libraries were strongly inhibited (R4 to R6.2, 67–88%) suggesting that most of the phages contained Fab with high affinities (Figure 1(B), right). In particular, the library after 2 rounds at 10 nM (R6.1) was inhibited by 88% but the final round at 1 nM (R6.2) was presumably too stringent and resulted in a decrease in the average affinity (R6.2, 59%). Individual clones from R5, R6.1 and R6.2 were tested by ELISA as soluble Fabs, quantified and ranked using dose–response curves (Figure 1(C)). Four clones isolated from R5 showed a strong binding with an EC50 around 20–50 nM (2B3, 2B2, 2B6, G12), and one high affinity clone with an EC50 around 2 nM (C10) was isolated from R6.1. No interesting clone was isolated from R6.2 as suggested by the decrease in the competition experiment in Figure 1(B). These 5 clones were derived from 4 different parental VH (B12, D12, E10, G6) as shown in the phylogenic tree in Figure 1(D). We then measured the affinity of these 5 clones by SPR by capturing them to the chip and applying the monomeric RBD in the flow to avoid avidity effects. In parallel we also tested the parental clones, except B12 and D3 which were no more active after re-cloning from phages. In these conditions all the affinities are comparable in the nM range (1.1–2.6 nM) but with different profiles. The non-matured parental clones (C8, D8, D12, E10, G6) showed an association rate faster than 5 × 105 M−1s−1 and a moderate dissociation rate around 10−3 s−1, whereas the matured Fab (2B3, 2B2, 2B6, G12, C10), showed slightly slower association rates around 2 × 105 M−1s−1 and slower dissociation rates in the 10−4 s−1 range (Figure 1(E)). The 10 clones were produced and purified as human IgG1 and tested by flow cytometry for binding to HEK293 cells stably expressing full-length Wuhan Spike at their surface. All the IgGs recognized the RBD presented in its native conformation at the cell surface but with different efficiencies (Supplementary Figure 1, Figure 1(F)). In particular, C8 clone was very efficient and strongly stained the cell line, despite being a non-matured antibody with a moderate affinity (Figure 1(F)). Based on their sequences, cytometry and SPR binding properties, we chose 2 clones for the design of CAR constructs. More precisely, C8 for its high binding to Spike expressing cells (Figure 1(F)), and C10 for its high affinity by SPR (Figure 1(E)). However, we first verified their binding capacity to RBD by ELISA in the scFv format used in CAR and showed activities comparable to the Fab format, C10 showing a much lower EC50 (7 nM) than C8 clones (67 nM) (Figure 1(G)). Finally, we employed SPR to assess the binding affinity of C8 and C10 mAbs to the trimeric form of the Spike protein (as shown in Supplementary Figure 2). Our results demonstrate effective binding of both mAbs to the trimeric Spike forms of the Wuhan strain and the Delta variant. However, C10 displayed a ten-fold higher affinity compared to C8. When tested against the Omicron variant, the C8 clone failed to recognize the trimer, while C10 exhibited very high affinity, exceeding 80 pM. Interestingly, C10 affinity for the Wuhan strain Spike trimer was significantly lower than that of clone S309 (the parent antibody of Sotrovimab), with Kd values of 0.2 and 0.04 nM, respectively. However, for the Delta variant, the affinities became comparable (0.05 and 0.09 nM for C10 and S309, respectively), and, against the Omicron variant, C10 demonstrated a 3.5-fold better affinity compared to S309 (80 pM vs. 280 pM). In conclusion, our evaluation using the trimeric Spike protein revealed that C10 exhibits comparable binding affinities to the S309 clone. However, C10 demonstrates a significantly greater resilience against mutations present in the Delta and Omicron variants.
Figure 1.
Antibody discovery. (A) Antibody selection against SARS-CoV-2-Spike. A large naive synthetic scFv library was used for three consecutive rounds of biopanning on recombinant Wuhan Spike RBD captured by its poly-Histidine tag in microtiter plates. (B) ELISA of pool of phages (1011 phages/well) eluted after each round of selection. Phages were detected using an HRP-coupled anti-M13 monoclonal antibody (Sino Biological #11973-MM05T-H). Bars represent the absorbance obtained by ELISA. (Left): Initial selection of the naive library (rounds R1, R2, R3). Absorbance of the ELISA on Spike RBD-coated (orange) or non-coated (blue) plates. (Right): Pool of phages selected after light-chain shuffling and selection (R4 to R6.2). Absorbance of the indicated phage stocks in an ELISA on RBD-coated plates (orange). Absorbance obtained with phage stocks first incubated with 10 nM of Spike RBD, then captured on RBD-coated plates (grey). (C) Binding capacity of 14 soluble selected Fabs recognizing Spike RBD determined by ELISA. (D) Phylogenic tree of selected clones. Parental clones isolated from R3, matured clones isolated from R5, R6.1 and R6.2 are depicted in red, green, blue and purple, respectively. (E) Affinity of 10 selected clones to monomeric RBD measured by SPR. (F) Binding of C8 and C10 IgGs to HEK293 cells stably expressing Wuhan Spike at the cell surface. C8 and C10 mAbs were used at 10 µg/mL and detected using an AF647-coupled anti-human antibody. Grey histogram: isotype antibody control. (G) Dose-response binding to RBD of purified scFv C8 and C10, assayed by ELISA.
Identification of a broadly reactive coronavirus antibody
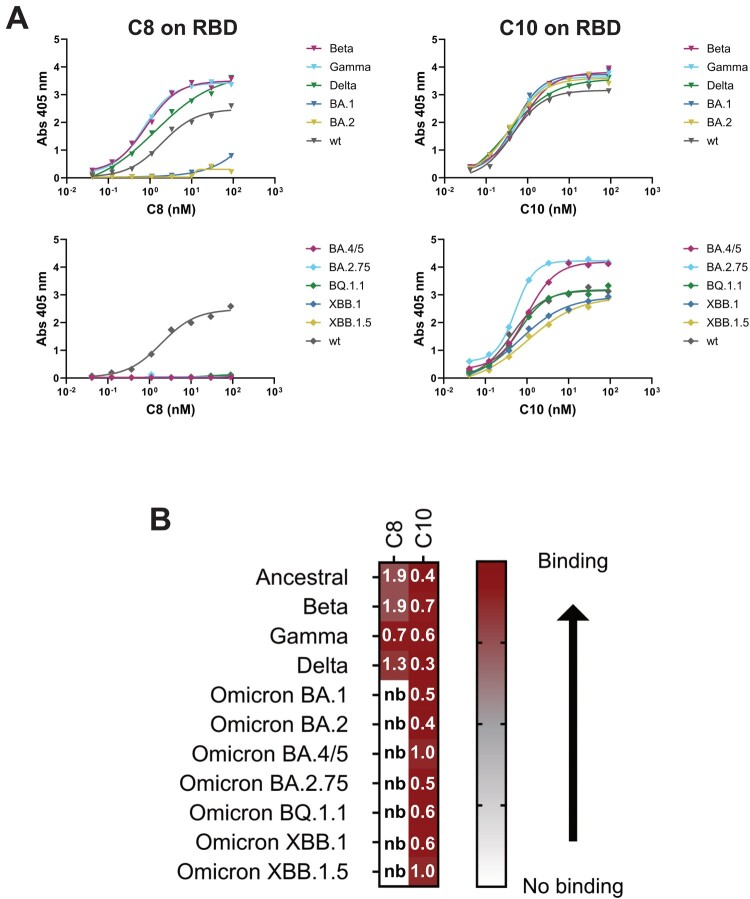
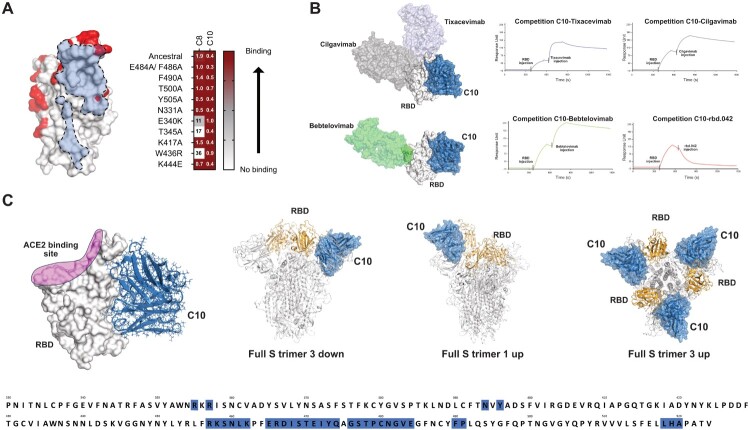
A good candidate for SARS-CoV-2 targeting must be able to bind all the circulating VOCs. C8 and C10 binding was assessed to multiple SARS-CoV-2 RBDs from VOCs spanning from the original Wuhan strain to more recent Omicrons (Figure 2). C10 mAb is able to bind all the RBDs tested, instead C8 fails to bind all Omicron (BA, BQ, and XBB) VOCs (Figure 2(A,B)). Binding results on C10 suggest that its epitope is located in a RBD conserved region. In order to obtain information on C10 epitope, binding competition with commercial mAbs, which epitopes are known, were performed using SPR. In addition, RBDs point mutants were used to discover if C10 binding was affected by mutations in specific regions. C10 binds to all the RBDs mutants tested (Figure 3(A) and Supplementary Figure 3) and does not compete for RBD binding with clinically previously used mAbs Bebtelovimab, Cilgavimab, Tixacevimab (Figure 3(B)). Interestingly, it competes with the neutralizing mAb rbd.042 that recognizes a coldspot of the SARS-CoV-2 Spike glycoprotein [13] (Figure 3(B)).
Figure 2.
Identification of a broadly reactive coronavirus antibody. (A-B) Binding capacity of C8 and C10 mAbs to multiple SARS-CoV-2 VOCs. (A) Antibody binding on RBD-SD1 variants, assessed by ELISA. (B) Heatmap representation of ELISA data from panel A. EC50 values are reported in nanomolar (nb = no binding).
Figure 3.
Docking model of C10 mAb binding to Spike RBD. (A) Binding capacity of C8 and C10 mAb to SARS-CoV-2 mutated RBDs shown as heatmap (right; EC50 values are reported in nanomolar). C10 fingerprint on RBD with mutated residues tested in binding assays (left). Residues mutated in the RBDs tested are shown in red. Light blue is the C10 fingerprint (6Å). (B) C10 mAb competition with clinically previously used mAbs and the neutralizing mAb rbd.042 for RBD binding (right). C10 antibody was immobilized on chip; RBD was then flowed to form the RBD/antibody complex, and shortly thereafter, Tixacevimab, Cilgavimab, Bebtelovimab or rbd.042 mAbs were injected. The binding event detected at the final step for Tixacevimab, Cilgavimab and Bebtelovimab, indicates that these second antibodies injected have a different epitope compared to the first (immobilized) antibody; instead, no binding, meaning competition, is detected for rbd.042. Structural representation of RBD with bound C10 and different Abs used for cross-competition experiments (left). (C) RBD surface representation with bound C10 based on the obtained with the docking model (left). C10 is shown in blue and ACE2 binding site is shown on RBD in pink. The putative epitope recognized by C10 (blue) encompasses amino acid residues dispersed across various regions within the RBD, predominantly situated in a region near the Subdomain 1 (SD1). C10 epitope accessibility on Spike trimer based on the docking model (right).
Experimental information on C10 binding to different RBDs mutants/variants and on C10 cross-competition with other anti-RBD antibodies with known epitope, were used to build a docking model for C10 binding to SARS-CoV-2 RBD [28,33,34]. Figure 3(C) shows the RBD surface representation with bound C10 based on the obtained docking model. C10 is shown in blue and ACE2 binding site is shown on RBD in pink. The putative epitope recognized by C10 (also shown in blue on the RBD sequence) encompasses amino acid residues dispersed across various regions within the RBD, predominantly situated between residues 457 to 491 (Figure 3(C)). C10 binding model was analysed in the context of SARS-CoV-2 full S trimer. We observed that C10 mAb makes clashes with adjacent Spike monomers when all the 3 RBDs are in “down” conformation. However, no clashes were observed when the RBDs are in “Up” conformation (Figure 3(C)). Overall, the docking model is in accordance with the cross-competition data on tested anti-RBD mAbs with known epitopes. It fits in a RBD region with no mutations (as C10 binds all the RBD mutants/variants tested).
C10 mAb binds to SARS-CoV-2 infected cells and mediates cell killing through ADCC
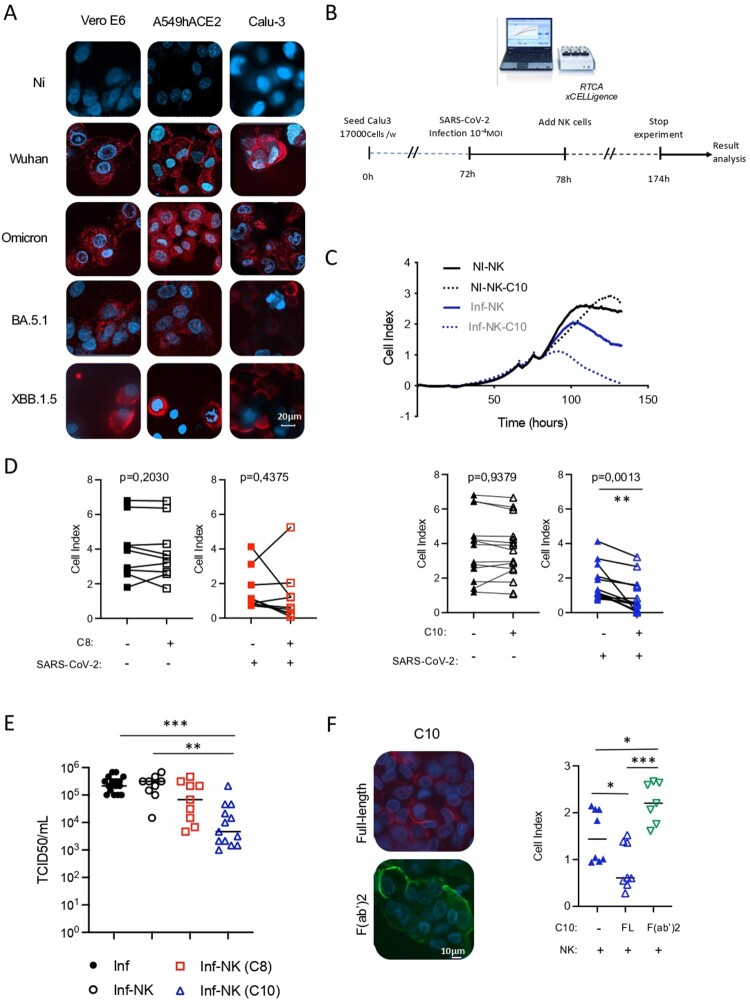
We assessed in parallel the functional properties of C8 and C10 mAbs. We studied their neutralization potential as well as their ability to target infected cells, both properties being important for countering viral spread. Neither C8 nor C10 displayed neutralizing capacity (Supplementary Figure 4A). Nevertheless, both mAbs demonstrated binding to Vero cells infected with SARS-CoV-2, albeit with different efficiencies, with C10 showing superior performance over C8, as determined by dose–response experiments (Supplementary Figure 4B). Both mAbs were able to target Vero cells as well as lung epithelial cells (A549-hACE2 and Calu-3) infected with different VOCs (Supplementary Figure 4C, Figure 4(A)). Based on these observations, we next assessed the cytotoxic capacity of C8 and C10 mAbs. To this end, we performed a cytotoxic assay based on the measure of electrical current using RTCA xCELLigence, which allows the determination of a cell index based on the electric impedance produced by adherent target cells (Figure 4(B,C)). We set a co-culture of effector cells (primary NK cells) and target cells (SARS-CoV-2-infected Calu-3 lung epithelial cells) in the presence or in the absence of mAbs. We showed that C10 mediates the killing of SARS-CoV-2 infected lung epithelial cells through NK-mediated ADCC (Figure 4(C,D)). The cell killing capacity of C8 was reduced compared to C10, as it was only observed in 3 out of 9 experiments, without reaching statistical significance. This might be the result of the lower binding of C8 to SARS-CoV-2-infected cells compared to C10 (as observed in dose response experiments Supplementary Figure 4B)), which likely accounts for its lack of ADCC activity in most of the tests conducted. This aligns with previous studies, which demonstrate that efficient ADCC requires sustained and robust binding of antiviral antibodies to viral antigens on the cell surface, and that stronger binding correlates with more effective killing activity [35]. Simultaneously, we evaluated the release of viral particles by infected target cells co-cultured with mAb-armed NK cells. Notably, a significant decrease in viral titer was observed in supernatants from infected cells co-cultured with C10-armed NK cells, whereas no such decrease was seen with C8-armed NK cells (Figure 4(E)). This highlights the capacity of the C10 mAb to eliminate infected cells and contribute to the decrease of viral propagation.
Figure 4.
Targeting capacity of SARS-CoV-2 infected cells by C8 and C10 mAbs. (A) Binding of C10 mAb to Vero E6 and lung epithelial cells infected with different SARS-CoV-2 VOCs. Non-infected cells (NI) were used as control. (B) Schematic representation of cytotoxicity assay timeline. (C) Graphical representation of cell index data obtained through the RTCA xCELLigence over-time, upon co-culture of non-infected cells (NI) or SARS-CoV2-infected Calu-3 cells (Inf) with NK cells armed or not with the C10 mAb (NK-C10 and NK, respectively). (D) ADCC capacity of C8 and C10 mAbs using infected lung epithelial cells as targets and primary NK cells as effector cells. Values are from 9 (C8) and 13 (C10) independent measurements. Significance was assessed using a paired t-test. (E) Viral titers in supernatants of infected cells in the presence of C8- and C10-armed NK cells. Values are from 13 (C8) and 9 (C10) independent measurements. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (F) Assessment of the binding to SARS-CoV-2-infected cells (left) and the cytotoxic capacity (right) of the F(ab’)2 fragment of C10 mAb as compared to the full-length (FL) C10 mAb. Values are from 4 independent measurements done in duplicate.
We next investigated whether the infected cell-killing capacity of the C10 mAb was dependent on its Fc fragment. To address this, we generated the F(ab’)2 fragment of C10 mAb and compared its cytotoxic activity to that of the full-length antibody. While the C10 F(ab’)2 fragment bound efficiently to SARS-CoV-2-infected cells, similar to the full-length C10 mAb (Figure 4(F), left), it did not exhibit any cytotoxic effect against infected cells (Figure 4(F), right). Additionally, we conducted ADCC assays using an Fc-silenced C10 mAb, which was engineered with the LALA-PG mutation [25] to significantly reduce Fc receptor binding. Like the C10 F(ab’)2 fragment, the C10-Fc-LALA-PG mutant did not show any cytotoxic activity against SARS-CoV-2-infected cells (Supplementary Figure 5). Overall, these findings show that the cytotoxic effect of the C10 mAb is Fc-dependent.
Development of second-generation CAR-T cells targeting SARS-CoV-2 spike protein
Based on the efficient binding capacity of C8 and C10 mAbs to infected cells, we next sought to repurpose these non-neutralizing antibodies for CAR-T cell therapy. To this aim we developed second-generation CAR-T cells from their sequences. Four different CAR designs were generated, using C8 or C10 scFv sequences combined with either a CD28 or 4-1BB intracellular co-stimulation (named C8-28z/C10-28z and C8-BBz/C10-BBz, respectively) (Figure 5(A)). Lentiviral vectors encoding for the different CAR designs were used to transduce healthy donor T cells to determine that they were properly expressed. Following T-cell activation with CD3/CD28 stimulation, we next assessed CAR-T cell proliferation during a 10 d course as an indicator of T-cell fitness. T cells expressing the different CAR molecules demonstrated a similar proliferation compared to control un-transduced T cells (UTD) (Figure 5(B)). Last, we aimed to detect CAR expression on T cell surface by flow cytometry staining after 10-day proliferation. For that, cells were stained using mAbs that specifically detect the scFv region of the CAR molecule. Sample acquisition revealed expression of all four CAR molecules on the surface of human T cells (Figure 5(C)). Altoghether, these results show that the designed CAR constructs can be properly expressed on human T cells without affecting their fitness.
Figure 5.
Development of second-generation CAR-T cells from C8 and C10 mAbs. (A) Design of different transgenes encoding for second-generation CAR molecules using two C8 and C10 scFv sequences, and two different costimulatory domains, CD28 and 4-1BB (named C8-28z/C10-28z and C8-BBz/C10-BBz, respectively). (B) Graph shows CAR-T cell proliferation during 10 days after stimulation with CD3/CD28 Dynabeads and IL-2, assessed by flow cytometry. (C) Flow cytometry plots showing CAR surface expression of the different constructs on human T cells.
Evaluation of CAR-T cell activity against SARS-CoV-2 spike-expressing cells
We next sought to evaluate the functionality of the different CAR-T cells we developed. To this end, we established a cell line model expressing the Spike protein on the cell surface. This model served as an initial platform to assess CAR-T cell activity, circumventing the stringent constraints associated with handling SARS-CoV-2 infected cells. To this end, we generated HEK293T and A549 cell lines expressing Gaussia Pierce Luciferase and the Spike protein from the Wuhan strain (Supplementary Figure 6A), named 293GS and A549GS, respectively. Both cell lines maintain the expression of Gaussia Luciferase (Supplementary Figure 6B) and express the Spike protein on the cell surface for at least 40 days. Of note, 293GS cells display a higher expression of the Spike protein on cell surface (Supplementary Figure 6C).
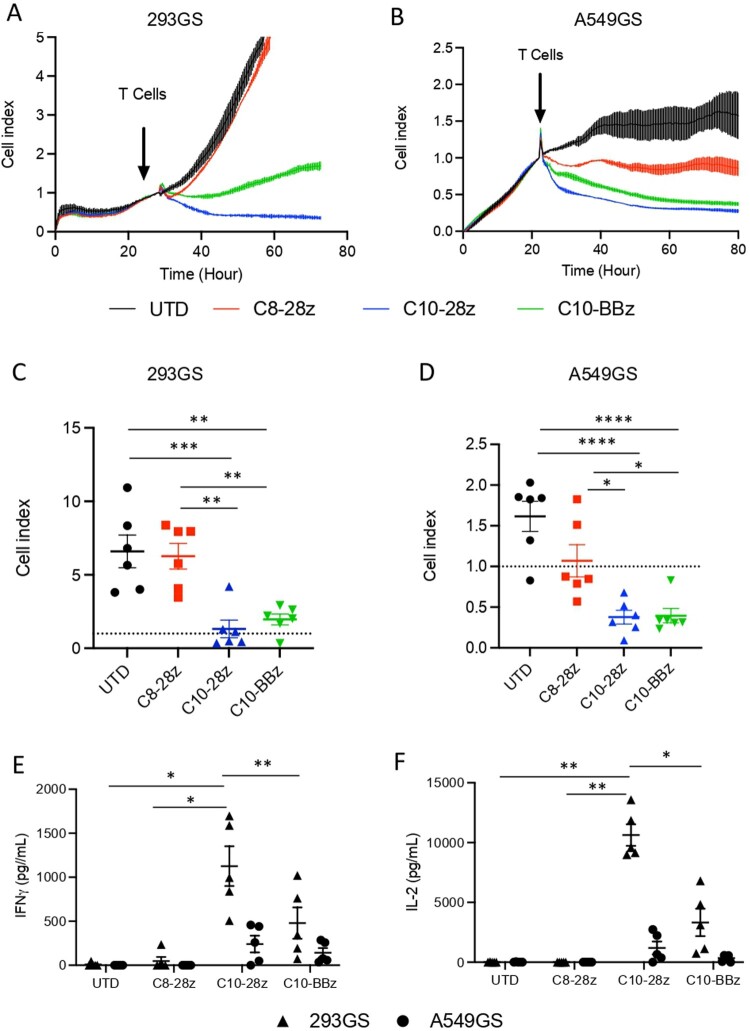
Subsequently, we performed a cytotoxic assay based on the measure of electrical current using RTCA xCELLigence, as above mentioned. We set a co-culture of CAR-T cells and target cells (293GS and A549GS) expressing the Spike protein at a 5:1 ratio. The results show a decrease in the cell index growth over time when target cells are co-cultured with CAR-T cells derived from the C10 scFv sequence, while C8-28z CAR-T does not have significant effect on target cell growth (Figure 6(A–D)). The C8-BBz CAR-T cell construct showed inefficient proliferation capacity (Figure 5(B)) and did not respond to target cells. Therefore, it was excluded from further experiments. Repeated experiments using T cells derived from different donors demonstrate that C10-28z and C10-BBz CAR-T cells recognize and eliminate the Spike-expressing cell lines 293GS and A549GS (Figure 6(C,D)). The analysis of the IFNγ and IL-2 release in cell culture supernatants revealed that UTD and C8-28z CAR-T cells do not produce detectable amounts of cytokines after 24 h of co-culture. On the contrary, C10-derived CAR-T cells exhibit substantial cytokine production, with notably higher secretion levels observed in C10-28z CAR-T cells compared to C10-BBz. This difference is particularly pronounced when co-cultured with 293GS cells (Figure 6(E,F)), suggesting that the Spike-expression level is key for CAR-T cell activation. These results prove that C10-derived CAR-T cells can be activated and perform a cytotoxic activity after encountering target cells. They also show that C10-28z CAR-T cells have a more potent activation than C10-BBz CAR-T cells.
Figure 6.
Evaluation of second generation CAR-T cell activity against Spike-expresing cells. (A,B) Graphical representation of cell index data obtained through the RTCA xCELLigence over-time. (C, D) Grouped analysis of CAR-T cell cytotoxicity against 293GS and A549GS cell lines at 48 h. Values are from 6 independent experiments. (E, F) Quantification of IL-2 and IFNγ at 24 h post co-culture of CAR-T cells with target cells. Values are from 5 independent measurements. Significance was assigned as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
CAR-T cells maintain their activity upon continuous stimulation by SARS-CoV-2 spike protein
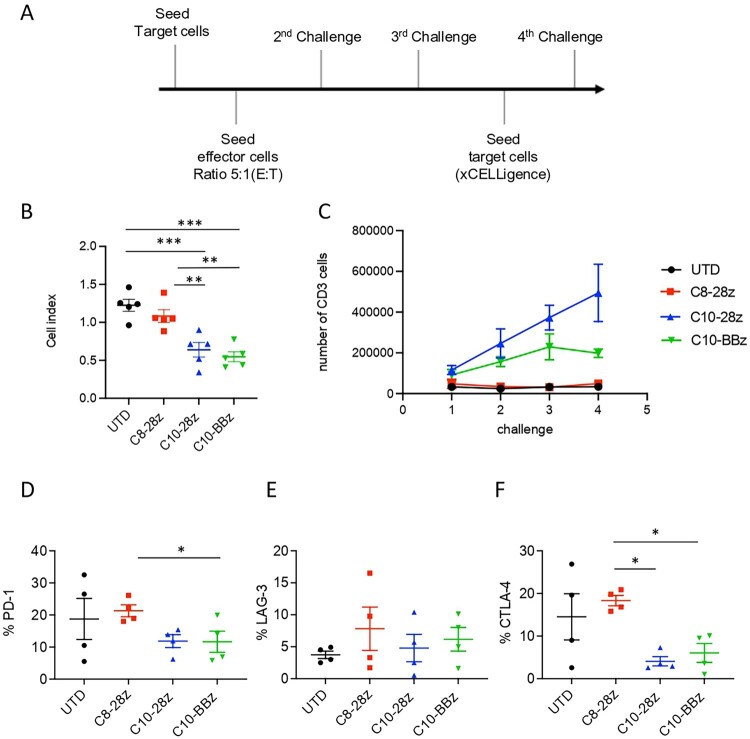
We next investigated C8- and C10-derived CAR-T-cell persistence to evaluate if they could maintain their activity when encountering continuous stimulation by the targeted antigen. To address this issue, we designed a rechallenge experiment based on sequential co-cultures of CAR-T cells with Spike-expressing cells (Figure 7(A)). We evaluated whether the cytotoxic capacity of CAR-T cells was compromised after four consecutive challenges. Results show that even after facing a continuous stimulation, C10-28z and C10-BBz CAR-T cells maintain their cytotoxicity towards target cells (Figure 7(B)). We also determined by flow cytometry the proliferation of CAR-T cells during consecutive challenges, measuring the total count of CD3+ cells. Results show that C10-derived CAR-T cells, but not C8-derived, maintain their proliferation throughout the sequential challenges. It is also important to note that C10-28z CAR-T cells exhibit a bigger expansion rate than the C10-BBz construct (Figure 7(C)). Moreover, we evaluated CAR-T cells exhaustion profile by assessing the expression of exhaustion markers such as LAG-3, CTLA-4 and PD-1 by flow cytometry. Results show a lower expression CTLA-4 and PD-1 exhaustion markers in C10-28z CAR-T cells as compared to C8-28z CAR-T cells (Figure 7(D–F)). Overall, the findings show a notably higher expansion rate and a reduced exhaustion phenotype in C10-derived CAR-T cells when compared to those derived from the C8 mAb.
Figure 7.
Evaluation of in vitro CAR-T cell persistence. (A) Schematic representation of rechallenge experiment time course. CAR-T cells were first exposed to target cells (ratio 5:1) and allowed to engage and eliminate the targets over a period of 24 h. Post initial challenge, CAR-T cells were re-isolated and re-challenged with a fresh batch of target cells three times (2nd, 3rd and 4th challenge). (B) Graph shows grouped analysis of CAR-T cell cytotoxicity during 4th challenge. Values are from 5 independent measurements. (C) Proliferation of CAR-T cells during the rechallenge experiment evaluated by total count of CD3+ T cells by flow cytometry. Values are from 4 independent measurements. (D-F) Exhaustion profile of the different CAR-T cells after 4 challenges evaluated by flow cytometry staining of PD-1, CTLA-4 and LAG-3 exhaustion markers. Values are from 4 independent measurements. Significance was assigned as follows: ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
CAR-T cells have cytotoxic activity against SARS-CoV-2-infected lung epithelial cells
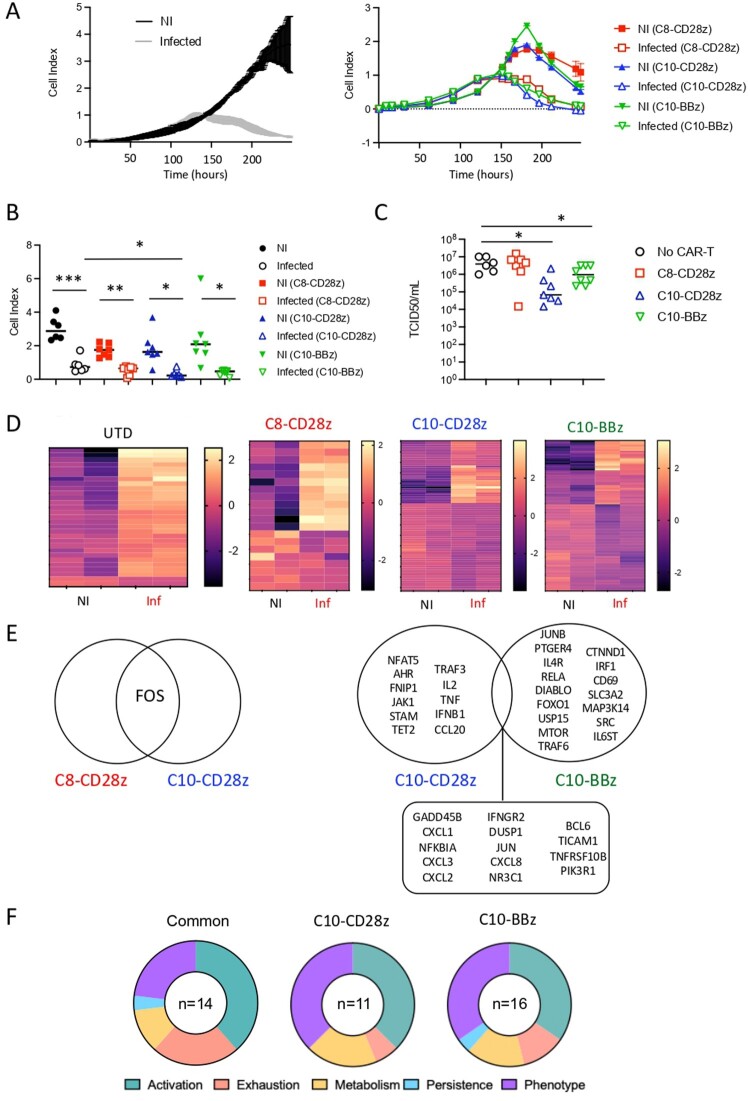
We next examined the cytotoxic potential of the different CAR-T cells against lung epithelial cells infected with SARS-CoV-2. This was important to asses as these cells secrete numerous cytokines and chemokines (Supplementary Figure 7) [36] that might affect CAR-T cell efficacy. Notably, Calu-3 cells exhibited a stronger inflammatory profile, both in the quantity and diversity of cytokine and chemokine secretion (Supplementary Figure 7). Therefore, this cell line was chosen for assessing the cytotoxic activity of CAR-T cells to mimic the hyperinflammatory environment associated with SARS-CoV-2 infection. As previously described [37,38], we observed a pronounced virus-induced cytopathic effect in lung-epithelial infected cells in the absence of CAR-T cells (Figure 8(A), left panel). However, despite this cytopathic effect reducing the window for assessing the cytotoxic potential of CAR-T cells, a significant decrease in the cell index growth over time was observed when Calu-3 infected cells were co-cultured with C10-28z CAR-T cells (Figure 8(A), right panel-8B). The cytotoxic capacity of C10-derived CAR-T cells was also visualized using cutting-edge imaging approaches (Supplementary Video 1). We assessed in parallel the release of viral particles by infected target cells co-cultured with the different CAR-T cells. Notably, a significant decrease in the viral titer was observed in supernatants recovered from infected cells co-cultured with both types of C10-derived CAR-T cells (C10-28z CAR-T and C10-BBz). In contrast, C8-28z CAR-T cells showed no effect (Figure 8(C)), as the viral titers measured were similar to those observed in the absence of CAR-T cells. These findings corroborate the impact of C10-28z CAR-T cells on cell index growth and infected cell-killing capacity. Thus, our results demonstrate the cytotoxic activity of C10-derived CAR-T cells against SARS-CoV-2-infected lung epithelial cells. Furthermore, they underscore the superior cytotoxic efficacy of C10-derived CAR-T cells against target cells, consistent with our observations in Spike-expressing cells.
Figure 8.
Evaluation of second generation CAR-T cells activity against infected cells and transcriptomic analysis of CAR-T cells. (A) Graphical representation of cell index data obtained through the RTCA xCELLigence over-time. Cell index data of non-infected (NI) and infected cells in the absence (left) and in the presence (right) of CAR-T cells. (B) Grouped analysis of CAR-T cells cytotoxicity against infected lung epithelial cells (Calu-3) at 48 h. (C) Viral titers in supernatants of Calu-3 infected cells in the absence and in the presence of C8- and C10-derived CAR-T cells. Values are from 6–7 independent experiments. (D) Heatmap representation of upregulated and downregulated genes in UTD, C8- and C10-derived CAR-T cells cultured with either non- infected or SARS-CoV-2 infected cells. Two independent experiments. (E) Venn diagram of upregulated genes in C8- and C10-derived CAR-T cells cultured with SARS-CoV-2 infected cells and not present in UTD cells upon co-culture with infected cells. (F) Number and principal biological pathways of differentially expressed genes in C10-derived CAR-T cells when exposed to infected cells, compared to the exposure to non-infected cells. Significance was assigned as follows: ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
The observed disparity in cytotoxic efficacity between C8- and C10-derived CAR-T cells prompted us to further characterize the different CAR-T cells, in an attempt to identify molecular mechanisms potentially involved in the enhancement of cytotoxic activity. To this end, we performed transcriptomic analysis using the NanoString nCounter technology. This approach allowed us to identify differentially expressed genes of the different CAR-T cells. As depicted in Figure 8(D), the heatmap representation of upregulated and downregulated genes shows a completely distinct transcriptional profile between C8- and C10-derived CAR-T cells co-cultured with infected cells. Notably, as compared to UTD co-cultured with infected cells, C10-derived CAR-T cells upregulated multiple genes associated with T-cell activation (PI3 K, JUN and NFκBIA as well as IL-2 and IFNGR2) and antiviral immune responses (IFNB, CXCL8, CXCL2, CXCL3), distinguishing them from C8-derived CAR-T cells (Figure 8(E)). Moreover, differentially expressed gene (DEG) analysis revealed specific signatures for each intracellular co-stimulation molecule, in particular genes involved in T-cell phenotype (AHR vs IL-4R and IL6ST) and persistence (Figure 8(F)). These observations show that specific SARS-CoV-2-CAR-T cell gene signatures are associated with successful response. The list of genes upregulated or downregulated and their fold increase is shown in Supplementary Tables 2–7.
Discussion
The emergence of immune evasive SARS-CoV-2 VOCs underscores the urgent need for the development of novel therapeutic options against severe COVID-19, in particular for individuals at high-risk. The development of broadly reactive coronavirus mAbs and derived mAb-based molecules might help to overcome the diminished efficacy of neutralizing mAbs due to evolving antigenic landscape of SARS-CoV-2. Here, we described a non-neutralizing pan-SARS-CoV-2 mAb targeting a conserved region of SARS-CoV-2 that recognizes all tested VOCs and effectively binds to infected cells. This leverages the cytotoxic potential of this mAb, either in full-length format (through ADCC) or through its use for the generation of CAR-T cells for clearance of SARS-CoV-2-infected lung epithelial cells.
Our findings identify C10 as a pan-SARS-CoV-2 mAb with remarkable binding efficiency to RBD proteins harbouring various mutations. Its ability to recognize cells infected by diverse VOCs underscores its specificity towards a highly conserved region of the virus. Consistent with this notion, both our molecular docking model and cross-competition studies employing the rdd.042 mAb [13], suggest that C10 targets an epitope in a coldspot of the RBD predominantly situated between residues 457 to 491, near subdomain 1 (SD1). To further elucidate the precise epitope recognized by C10 mAb, future endeavours will necessitate obtaining the crystal structure, enabling a comprehensive characterization of its binding interactions.
In agreement with previous studies, our work highlights the therapeutic potential of non-neutralizing mAbs targeting conserved regions [12,39,40]. Notably, investigations have revealed that non-neutralizing antibodies generated in vaccinated individuals have been shown to mediate protection [40]. In addition, several non-neutralizing mAbs isolated from either vaccinated individuals or individuals that recovered from SARS-CoV-2 infection, have been show to mediate protection in mouse and hamster models of SARS-CoV-2 infection [12,39]. These protective effects manifest through a spectrum of mechanisms, including Fc-independent and Fc-dependent pathways, underlining that multiple mechanism of action such as ADCC and ADCP are involved in mAb-mediated protection [12,39,41]. Similar to C10 mAb, such mAbs were directed to highly conserved regions (coldspots). This underscores the rationale behind utilizing non-neutralizing mAbs in the development of innovative therapeutic strategies [42]. These approaches might capitalize on the multifaceted mechanisms of action inherent in antiviral mAbs and their derivatives, which can play a pivotal role in controlling and clearing infected cells.
Many studies investigating the Fc-mediated antiviral effect of non-neutralizing anti-SARS-CoV-2 mAbs have relied on reporter systems measuring FcγR binding [39] and/or the use of recombinant cells expressing the Spike protein [12]. Worthy of note, our research breaks new ground by demonstrating the ADCC capacity of C10 mAb against bona fide infected cells, which is much closer to the pathological conditions. Similarly, previous studies reporting the cytotoxic capacity of cell therapies based on antibody derivatives, such as the use of bi-specific T-cell engagers [43], NK-CAR cells [44], CAR-macrophages [45] and conventional CAR-T cells [46], are also based on the use of non-infected Spike-expressing cells, which do not take into account the virus-induced cytopathic effect. In addition, none of these studies addressed the capacity of the different antibody-engineered cells to control viral replication. Importantly, our findings demonstrate the cytotoxic capacity of CAR-T cells against SARS-CoV-2- infected cells. Our observations hold dual significance. Firstly, they demonstrated that CAR-T-cell mediated killing of lung-infected cells effectively reduce viral titers. Secondly, they reveal that CAR-T-cells retain their cytotoxic capacity in the strong pro-inflammatory environment associated with SARS-CoV-2-infected lung epithelial cells.
The observed difference in cytotoxic efficacy between CAR-T cells derived from C8 and C10 mAbs underscores the intricate interplay of effective antigen binding, antigen expression levels, and the selection of signal transduction molecules. Studies have shown that the binding affinity of a CAR scFv can significantly impact T cell signalling strength, which, in turn, influences CAR-T cell function [47–49]. Specifically, affinity alterations achieved through mutagenesis of complementary-determining regions or by targeting different epitopes on the same antigen have been demonstrated to modulate T-cell responses. Thus, the specific epitope targeted by C10 can indeed play a crucial role in determining the effectiveness of the interaction (i.e. it may benefit from targeting a particularly immunogenic or accessible region on the S protein). In this context, it is particularly noteworthy the contrast in efficacy between C8- and C10-derived CAR-T cells employing the same intracellular co-stimulation (CD28), with the latter displaying robust cytotoxic activity while the former had negligible impact on target cell killing. This discrepancy aligns with the disparate binding capacities of the two mAbs (with C10 exhibiting superior binding). Moreover, the choice of intracellular co-stimulation further influences outcomes; C10-derived CAR-T cells employing either CD28 or 4-1BB intracellular co-stimulation yielded different results, with CD28 co-stimulation proving more efficacious. Notably, this enhanced efficacy correlates with distinct transcription profiles. While C8-derived CAR-T cells fail to exhibit enhanced gene expression in the presence of infected cells compared to UTD-CAR-T cells, both C10-derived CAR-T cells display increased expression of genes associated with immune cell activation. Nevertheless, each intracellular co-stimulation molecule elicits unique transcriptional signatures, particularly in genes related to T-cell phenotype and persistence, in agreement with previous studies in the context of anticancer CAR-T cell-based therapeutic approaches [50–53]. These differences underscore the importance of tailored approaches in the development of effective antiviral CAR-T cell therapies, necessitating meticulous consideration of antigen binding properties and intracellular signalling cascades to optimize therapeutic outcomes.
Our study underscores the potential use of CAR-T cell-based therapies to combat coronavirus infections. While adoptive T-cell therapies are undergoing clinical trials [18], CAR-T cells are increasingly recognized for their potential in fighting infectious diseases, including SARS-CoV-2 [17,54]. However, this approach still faces numerous limitations, including challenges related to the acute nature of SARS-CoV-2 infection. This hinders the use of autologous cells due to the time required for CAR-T cell generation. Therefore, the generation of “off-the-shelf” Universal CAR-T might help overcome several drawbacks of conventional CAR-T cells [55]. First, they might allow the immediate availability of a stored and ready to use T-cell product usable for any patient. Second, it might allow to overcome the suboptimal amounts of T-cells available in immunocompromised patients and/or due to the lymphopenia associated with SARS-CoV-2 infection. Third, it might help reducing the extremely high costs associated with autologous patient cell-derived CAR-T cells. However, allogeneic CAR-T approaches pose two major challenges: the risk of graft-versus-host disease (GvHD) and the potential for rapid elimination of the allogeneic T cells by the host immune system. To mitigate these risks, sophisticated genetic engineering techniques are being developed. This includes the use of gene editing tools like CRISPR to create “universal” CAR-T cells by knocking out HLA class I and II molecules or the T cell receptor (TCR). Additionally, protein retention systems can prevent TCR assembly on the T-cell surface, further reducing the risk of GvHD [56,57]. Such allogeneic CAR-T cells, maintaining cytotoxic potential while mitigating alloreactivity, are currently under development [58,59] and clinical testing [60] (https://clinicaltrials.gov/study/NCT04093596). Addressing immune rejection in allogeneic CAR-T therapy also involves combining genetic engineering with other therapeutic strategies, such as immune checkpoint modulation and co-infusion with regulatory cells [56,57].
Other challenges are also associated with CAR-T cell therapy, including cytopenia, poor trafficking and cytokine syndrome release. Cytopenia can be addressed through several tailored approaches, depending on the underlying cause, specific type and severity of the cytopenia, and the patient’s overall condition [61]. These strategies include the administration of growth factors, such as G-CSF for neutropenia and erythropoiesis-stimulating agents for anemia, as well as blood and platelet transfusions. Additionally, reducing infection risk is crucial. Corticosteroids and/or immunosuppressive agents can also be employed for managing both immune-mediated cytopenia and cytokine release syndrome. Enhancing the trafficking and persistence of CAR-T cells is critical for optimizing CAR-T cell therapy. Several strategies can be employed, including engineering CAR-T cells to secrete cytokines that promote immune cell recruitment or to co-express homing receptors that guide them to target tissues. Additionally, upregulating adhesion molecules can improve CAR-T cell interactions with endothelial cells, enhancing their infiltration into sites of disease [62–64]. These and other approaches might allow improved cell trafficking and lifelong persistence of the treatment. Finally, in the particular context of SARS-CoV-2 infection, approaches to counteract the hyperinflammation associated with both the viral infection and CAR-T cell therapy may need to be considered. In this regard, several FDA-approved drugs have demonstrated efficacy in inhibiting cytokine release from CAR-T cells and attenuating SARS-CoV-2-mediated lethal inflammation [65].
The efficient targeting and killing of infected cells by pan SARS-CoV-2 mAbs might also be leveraged in other cell-based therapeutic approaches, such as allogenic CAR-NK cells. These cells have been shown to be well tolerated without evidence of GvHD [66]. Moreover, antibody fragments from potent anti-SARS-CoV-2 mAbs that specifically target infected cells, like C10, can be used to develop different antibody-based therapeutic products such as bi-specific antibodies and T-cell engagers. These engagers are able to target infected cells and recruit cytotoxic immune cells to sites of infection. Such compounds have shown therapeutic efficacy in cancer patients and may circumvent the challenges associated with genetically modifying immune cells at a reduced cost [67]. Finally, the utilization of infected cell-targeting, pan SARS-CoV-2 mAbs in their full-length format, either unmodified or engineered to enhance ADCC, could offer another therapeutic avenue to reduce viral burden. Given that the severity of COVID-19 is frequently associated with its respiratory symptoms, in addition to systemic administration, the direct delivery of potent mAbs through inhalation warrants consideration. Indeed, inhalation of a combination of anti-SARS-CoV-2 mAbs has been demonstrated to be safe and resulted in a higher proportion of participants experiencing complete resolution of respiratory symptoms [68]. This underscores the potential utility of inhaled protective antibodies in the management of COVID-19.
In conclusion, we discover a pan SARS-CoV-2 mAb that binds to a conserved epitope and targets infected cells. This mAb offers numerous potential applications in both antibody-based and cell-based therapeutic approaches. This advancement significantly expands the therapeutic options available to combat the evolving antigenic landscape of SARS-CoV-2 and overcome the inefficacies of available antibody-based therapeutic approaches.
Supplementary Material
Acknowledgements
We thank the BSL3 research laboratory CEMIPAI (CNRS UAR3725) and her scientific director, Dr. Delphine Muriaux, for providing help and advice, and the Centre de Ressources Biologiques (CRB) (CHU, Montpellier, France) for providing the different VOCs. We thank the Transcriptomic Core Facility of the Institute in Regenerative Medicine and Biotherapy, CHU-INSERM-UM Montpellier where the applied Biosystem-Affymetrix microarrays were processed. We thank Cecile Monzo and Carole Crozet that are in charge of the Nanostring Sprint Profiler platform in the Institute for Neurosciences Montpellier (INM), for providing assistance in the transcriptomic studies. We thank the Montpellier Proteomics Platform (PPM-PP2I, BioCampus Montpellier) facilities where the SPR experiments were carried out.
Funding Statement
This work was supported by the consortium ACT4COVIDCellnex (220110FF, RAK200019FFA) and by EU-OPENSCREEN ERIC and has received funding from the European Union´s Horizon Europe research and innovation programme ISIDORe under grant agreement No 10146133.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cucchiari D, Egri N, Bodro M, et al. Cellular and humoral response after MRNA-1273 SARS-CoV-2 vaccine in kidney transplant recipients. Am J Transplant. 2021;21(8):2727–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertini CD, Khawaja F, Sheshadri A.. Coronavirus disease-2019 in the immunocompromised host. Clin Chest Med. 2023;44(2):395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spanjaart AM, Ljungman P, de La Camara R, et al. Poor outcome of patients with COVID-19 after CAR T-cell therapy for B-cell malignancies: results of a multicenter study on behalf of the European Society for Blood and Marrow Transplantation (EBMT) Infectious Diseases Working Party and the European Hematology Association (EHA) Lymphoma Group. Leukemia. 2021;35(12):3585–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto D, Sauer MM, Czudnochowski N, et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373(6559):1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox M, Peacock TP, Harvey WT, et al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat Rev Microbiol. 2023;21(2):112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Song G, Liu H, et al. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease. Immunity. 2023;56(3):669–686.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Campos-Mata L, Trinité B, Modrego A, et al. A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity. Nat Commun. 2024;15(1):1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu LL, Suscovich TJ, Fortune SM, et al. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018;18(1):46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruel T, Stéfic K, Nguyen Y, et al. Longitudinal analysis of serum neutralization of SARS-CoV-2 Omicron BA.2, BA.4, and BA.5 in patients receiving monoclonal antibodies. Cell Rep Med. 2022;3(12):100850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson NV, Wall SC, Kramer KJ, et al. Discovery and characterization of a pan-betacoronavirus S2-binding antibody. Structure. 2024;32(11):1893–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchini F, Crivelli V, Abernathy ME, et al. Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein. Sci Immunol. 2023;8(81):eade0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldini CR, Ellis GI, Riley JL.. CAR T cells for infection, autoimmunity and allotransplantation. Nat Rev Immunol. 2018;18(10):605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seif M, Einsele H, Löffler J.. CAR T cells beyond cancer: hope for immunomodulatory therapy of infectious diseases. Front Immunol. 2019;10:2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bupha-Intr O, Haeusler G, Chee L, et al. CAR-T cell therapy and infection: a review. Expert Rev Anti Infect Ther. 2021;19(6):749–758. [DOI] [PubMed] [Google Scholar]
- 17.Morte-Romea E, Pesini C, Pellejero-Sagastizábal G, et al. CAR immunotherapy for the treatment of infectious diseases: a systematic review. Front Immunol. 2024;15:1289303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papadopoulou A, Karavalakis G, Papadopoulou E, et al. SARS-CoV-2-specific T cell therapy for severe COVID-19: a randomized phase 1/2 trial. Nat Med. 2023;29(8):2019–2029. [DOI] [PubMed] [Google Scholar]
- 19.Bange EM, Han NA, Wileyto P, et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med. 2021;27(7):1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martineau P, Jones P, Winter G.. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol. 1998;280(1):117–127. [DOI] [PubMed] [Google Scholar]
- 21.Hoogenboom HR, Griffiths AD, Johnson KS, et al. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19(15):4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honegger A, Plückthun A.. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol. 2001;309(3):657–670. [DOI] [PubMed] [Google Scholar]
- 23.Robin G, Sato Y, Desplancq D, et al. Restricted diversity of antigen binding residues of antibodies revealed by computational alanine scanning of 227 antibody-antigen complexes. J Mol Biol. 2014;426(22):3729–3743. [DOI] [PubMed] [Google Scholar]
- 24.Kristensen P, Winter G.. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 1998;3(5):321–328. [DOI] [PubMed] [Google Scholar]
- 25.Lo M, Kim HS, Tong RK, et al. Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J Biol Chem. 2017;292(9):3900–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sircar A, Kim ET, Gray JJ.. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009;37(Web Server issue):W474-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedotti M, Simonelli L, Livoti E, et al. Computational docking of antibody-antigen complexes, opportunities and pitfalls illustrated by influenza hemagglutinin. Int J Mol Sci. 2011;12(1):226–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonelli L, Beltramello M, Yudina Z, et al. Rapid structural characterization of human antibody-antigen complexes through experimentally validated computational docking. J Mol Biol. 2010;396(5):1491–1507. [DOI] [PubMed] [Google Scholar]
- 29.Van Der Spoel D, Lindahl E, Hess B, et al. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. [DOI] [PubMed] [Google Scholar]
- 30.Lyonnais S, Hénaut M, Neyret A, et al. Atomic force microscopy analysis of native infectious and inactivated SARS-CoV-2 virions. Sci Rep. 2021;11(1):11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Assou S, Ahmed E, Morichon L, et al. The transcriptome landscape of the in vitro human airway epithelium response to SARS-CoV-2. Int J Mol Sci. 2023;24(15):12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rihn SJ, Merits A, Bakshi S, et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021;19(2):e3001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertoglio F, Meier D, Langreder N, et al. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat Commun. 2021;12(1):1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcotte H, Cao Y, Zuo F, et al. Conversion of monoclonal IgG to dimeric and secretory IgA restores neutralizing ability and prevents infection of Omicron lineages. Proc Natl Acad Sci U S A. 2024;121(3):e2315354120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruel T, Guivel-Benhassine F, Amraoui S, et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun. 2016;7:10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira EPV, da Silva Felipe SM, de Freitas RM, et al. Transcriptional profiling of SARS-CoV-2-infected calu-3 cells reveals immune-related signaling pathways. Pathogens. 2023;12(11):1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Garcia P, Fiorillo Moreno O, Zarate Peñata E, et al. From cell to symptoms: the role of SARS-CoV-2 cytopathic effects in the pathogenesis of COVID-19 and long COVID. Int J Mol Sci. 2023;24(9):8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu N, Wang W, Liu Z, et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11(1):3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark J, Hoxie I, Adelsberg DC, et al. Protective effect and molecular mechanisms of human non-neutralizing cross-reactive spike antibodies elicited by SARS-CoV-2 mRNA vaccination. bioRxiv. 2024;2024.02.28.582613. [DOI] [PubMed]
- 40.Rahman MO, Kamigaki T, Thandar MM, et al. Protection of the third-dose and fourth-dose mRNA vaccines against SARS-CoV-2 Omicron subvariant: a systematic review and meta-analysis. BMJ Open. 2023;13(12):e076892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiakolas AR, Kramer KJ, Wrapp D, et al. Cross-reactive coronavirus antibodies with diverse epitope specificities and Fc effector functions. Cell Rep Med. 2021;2(6):100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mader K, Dustin LB.. Beyond bNAbs: uses, risks, and opportunities for therapeutic application of non-neutralising antibodies in viral infection. Antibodies (Basel). 2024;13(2):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dogan M, Kozhaya L, Placek L, et al. Targeting SARS-CoV -2 infection through CAR-T -like bispecific T cell engagers incorporating ACE2. Clin Transl Immunology. 2022;11(10):e1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma MT, Badeti S, Chen C-H, et al. CAR-NK cells effectively target SARS-CoV-2-spike-expressing cell lines in vitro. Front Immunol. 2021;12:652223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu W, Lei C, Ma Z, et al. CAR macrophages for SARS-CoV-2 immunotherapy. Front Immunol. 2021;12:669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X, Kazanova A, Thurmond S, et al. Effective chimeric antigen receptor T cells against SARS-CoV-2. iScience. 2021;24(11):103295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Jiang S, Fang C, et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 2015;75(17):3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chmielewski M, Hombach A, Heuser C, et al. T cell activation by antibody-like immunoreceptors: increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J Immunol. 2004;173(12):7647–7653. [DOI] [PubMed] [Google Scholar]
- 49.Guedan S, Calderon H, Posey AD, et al. Engineering and design of chimeric antigen receptors. Mol Ther Methods Clin Dev. 2019;12:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salter AI, Ivey RG, Kennedy JJ, et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci Signal. 2018;11(544):eaat6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawalekar OU, O’Connor RS, Fraietta JA, et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. 2016;44(2):380–390. [DOI] [PubMed] [Google Scholar]
- 52.Priceman SJ, Gerdts EA, Tilakawardane D, et al. Co-stimulatory signaling determines tumor antigen sensitivity and persistence of CAR T cells targeting PSCA+ metastatic prostate cancer. Oncoimmunology. 2018;7(2):e1380764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong Q, Zhu Y-M, Zheng L-L, et al. Chimeric antigen receptor-T cells with 4-1BB co-stimulatory domain present a superior treatment outcome than those with CD28 domain based on bioinformatics. Acta Haematol. 2018;140(3):131–140. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Liu C, Fang Y, et al. Developing CAR-immune cell therapy against SARS-CoV-2: current status, challenges and prospects. Biochem Pharmacol. 2024;222:116066. [DOI] [PubMed] [Google Scholar]
- 55.Depil S, Duchateau P, Grupp SA, et al. “Off-the-shelf” allogeneic CAR T cells: development and challenges. Nat Rev Drug Discov. 2020;19(3):185–199. [DOI] [PubMed] [Google Scholar]
- 56.Ali A, DiPersio JF.. ReCARving the future: bridging CAR T-cell therapy gaps with synthetic biology, engineering, and economic insights. Front Immunol. 2024;15:1432799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Z, Wang Y, Jin X, et al. Universal CAR cell therapy: challenges and expanding applications. Transl Oncol. 2024;51:102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aparicio C, Acebal C, González-Vallinas M.. Current approaches to develop “off-the-shelf” chimeric antigen receptor (CAR)-T cells for cancer treatment: a systematic review. Exp Hematol Oncol. 2023;12(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamiya T, Wong D, Png YT, et al. A novel method to generate T-cell receptor-deficient chimeric antigen receptor T cells. Blood Adv. 2018;2(5):517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mailankody S, Matous JV, Chhabra S, et al. Allogeneic BCMA-targeting CAR T cells in relapsed/refractory multiple myeloma: phase 1 UNIVERSAL trial interim results. Nat Med. 2023;29(2):422–429. [DOI] [PubMed] [Google Scholar]
- 61.Jain T, Olson TS, Locke FL.. How I treat cytopenias after CAR T-cell therapy. Blood. 2023;141(20):2460–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhlmann A-S, Peterson CW, Kiem H-P.. Chimeric antigen receptor T-cell approaches to HIV cure. Curr Opin HIV AIDS. 2018;13(5):446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong M, Talluri S, Chen YY.. Advances in promoting chimeric antigen receptor T cell trafficking and infiltration of solid tumors. Curr Opin Biotechnol. 2023;84:103020. [DOI] [PubMed] [Google Scholar]
- 64.Pawlowski KD, Duffy JT, Gottschalk S, et al. Cytokine modification of adoptive chimeric antigen receptor immunotherapy for glioblastoma. Cancers (Basel). 2023;15(24):5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia L, Yuan L-Z, Hu Y-H, et al. A SARS-CoV-2-specific CAR-T-cell model identifies felodipine, fasudil, imatinib, and caspofungin as potential treatments for lethal COVID-19. Cell Mol Immunol. 2023;20(4):351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reina-Ortiz C, Giraldos D, Azaceta G, et al. Harnessing the potential of NK cell-based immunotherapies against multiple myeloma. Cells. 2022;11(3):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shanshal M, Caimi PF, Adjei AA, et al. T-cell engagers in solid cancers-current landscape and future directions. Cancers (Basel). 2023;15(10):2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maranda B, Labbé SM, Lurquin M, et al. Safety and efficacy of inhaled IBIO123 for mild-to-moderate COVID-19: a randomised, double-blind, dose-ascending, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. 2024;24(1):25–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.