Abstract
The nonsegmented negative-strand RNA (NNS) viruses have a single-stranded RNA genome tightly encapsidated by the viral nucleocapsid protein. The viral polymerase transcribes the genome responding to specific gene-start and gene-end sequences to yield a series of discrete monocistronic mRNAs. These mRNAs are not produced in equimolar amounts; rather, their abundance reflects the position of the gene with respect to the single 3′-proximal polymerase entry site. Promoter-proximal genes are transcribed in greater abundance than more distal genes due to a localized transcriptional attenuation at each gene junction. In recent years, the application of reverse genetics to the NNS viruses has allowed an examination of the role of the gene-start and gene-end sequences in regulating mRNA synthesis. These studies have defined specific sequences required for initiation, 5′ modification, termination, and polyadenylation of the viral mRNAs. In the present report, working with Vesicular stomatitis virus, the prototypic Rhabdovirus, we demonstrate that a gene-end sequence must be positioned a minimal distance from a gene-start sequence for the polymerase to efficiently terminate transcription. Gene-end sequences were almost completely ignored in transcriptional units less than 51 nucleotides. Transcriptional units of 51 to 64 nucleotides allowed termination at the gene-end sequence, although the frequency with which polymerase failed to terminate and instead read through the gene-end sequence to generate a bicistronic transcript was enhanced compared to the observed 1 to 3% for wild-type viral mRNAs. In all instances, failure to terminate at the gene end prevented initiation at the downstream gene start site. In contrast to this size requirement, we show that the sequence between the gene-start and gene-end signals, or its potential to adopt an RNA secondary structure, had only a minor effect on the efficiency with which polymerase terminated transcription. We suggest three possible explanations for the failure of polymerase to terminate transcription in response to a gene-end sequence positioned close to a gene-start sequence which contribute to our emerging picture of the mechanism of transcriptional regulation in this group of viruses.
Vesicular stomatitis virus (VSV), the prototypic Rhabdovirus, has a nonsegmented negative-sense (NNS) RNA genome of 11,161 nucleotides (nt), tightly encapsidated by the viral nucleocapsid protein (N). The viral genome comprises a 50-nt leader region, five genes that encode N, phosphoprotein (P), matrix protein (M), glycoprotein (G), and large polymerase subunit protein (L), and a 59-nt trailer region, arranged in the order 3′ leader-N-P-M-G-L-trailer 5′. This encapsidated genome acts as the template for the viral polymerase, a complex of the P and L proteins (14) which is packaged in the virion (5).
On infection of a cell, the viral genome is first transcribed by the polymerase to yield a 47-nt leader RNA, which is neither capped nor polyadenylated (10, 11, 27), and five capped and polyadenylated mRNAs. These mRNAs are not produced in equimolar quantities; rather, their relative abundance decreases with distance from a single 3′-proximal polymerase entry site such that N > P > M > G > L (1, 4, 13, 51). This reflects a localized transcriptional attenuation at each gene junction where approximately 30% of polymerase molecules fail to transcribe the downstream gene following termination of the upstream gene (24). Thus, termination of transcription is a critical step in VSV gene expression, as initiation of a downstream gene is dependent on termination of transcription of the upstream gene (1, 4, 6, 7, 22).
The cis-acting signals essential for transcription include sequences within the leader region, the nontranscribed leader-N gene junction, and the first gene-start sequence (29, 33, 43, 55). In addition to the signals that constitute the 3′ promoter, polymerase activity is further regulated by a 23-nt conserved sequence 3′ …AUACUUUUUUU G/CA UUGUCNNUAG… 5′, present at each internal gene junction. Extensive analysis of this region has shown that termination and polyadenylation of the upstream mRNA require the AUACUUUUUUU and the first nucleotide of the nontranscribed intergenic dinucleotide (G/CA) (6, 7, 22, 45). Transcription of the downstream gene is influenced by the sequence and length of the intergenic dinucleotide, with discrimination against a U prior to the gene-start sequence (7, 44). The gene-start sequence (UUGUCNNUAG) is also required for initiation, and additionally it contains signals essential for capping and methylation of the nascent mRNA strand (45, 46).
In the present investigation we examined whether the spacing between the gene-start (defined as 3′ …UUGUCNNUAG… 5′) and gene-end (defined as 3′ …AUACUUUUUUUG/CA… 5′) sequences affected mRNA synthesis. We inserted a variety of transcriptional units that ranged in size from 41 to 1,098 nt between the leader region and the N gene. Results obtained from analysis of subgenomic replicons and infectious viruses show that a gene-end sequence must be a minimal distance from a gene-start sequence for efficient termination of transcription to occur.
MATERIALS AND METHODS
Plasmid construction and transfection.
Transcription plasmid pWT was designed to generate a VSV subgenomic replicon that contained the wild-type genomic 3′ leader and 5′ trailer regions surrounding a single transcriptional unit comprised of a fusion of the 3′ end of the N gene with the 5′ end of the L gene. This plasmid was identical to that described by Wertz et al. (52) except that the orientation of the VSV replicon with respect to the T7 promoter was reversed in order to generate a primary transcript of positive polarity. This was necessary because additional transcriptional units were cloned between the leader and N regions of pWT, and we previously demonstrated that bacteriophage T7 RNA polymerase was unable to efficiently transcribe through the negative-sense VSV intergenic region (53). The plasmid encoding the WT replicon was modified such that additional transcriptional units that ranged in size from 41 to 1,089 nt were inserted between the leader region and N gene (Fig. 1), using standard techniques (39). The inserted sequences are listed in Table 1. In addition, selected transcriptional units were incorporated into an infectious cDNA clone of VSV, pVSV1(+) (53). Plasmids expressing either VSV subgenomic replicons (34, 52) or the full-length VSV antigenome (53), together with plasmids encoding the VSV N, P, and L proteins, each under the control of T7 promoters, were transfected as described previously into baby hamster kidney (BHK-21) cells that were infected with a recombinant vaccinia virus, vTF7-3 (17), expressing T7 RNA polymerase.
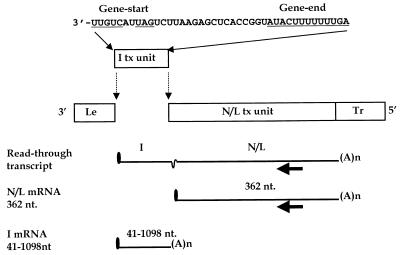
FIG. 1.
Structure of VSV subgenomic replicon and predicted mRNA products. The negative-sense VSV genome is represented diagrammatically. mRNA products are represented by solid lines. The filled arrow represents the oligonucleotide primer used in the primer extension reactions (Fig. 6). This primer anneals within the L gene to positive-sense RNA at positions 10925 to 10908 of the complete VSV genome sequence. I tx unit, inserted transcriptional unit; N/L tx unit, N/L transcriptional unit; Le, leader region; Tr, trailer region.
TABLE 1.
Sequences engineered between the leader region and the N gene
| Construct | Inserted sequencea |
|---|---|
| 41 | 3′-UUGUCAUUAGUCUUAAGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 46 | 3′-UUGUCAUUAGUCUUAAGAUUUGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 51 | 3′-UUGUCAUUAGUCUUAAGUGUUAAUUUGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 56 | 3′-UUGUCAUUAGUCUUAAGAAUAUUGUUAAUUUGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 56SL1 | 3′-UUGUCAUUAGUUUUCUAAUGACAAAUCUUAAGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 56SL2 | 3′-UUGUCAUUAGUCUUAAGCCCCCCUUUGGGGGGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 56SL3 | 3′-UUGUCAUUAGUCUUAAGAAUAUUGUUACCCCCCAAAGGGGGGUAUACUUUUUUUGA-5′ |
| 56SL4 | 3′-UUGUCAUUAGUCUUAAGCCCCCCUUUGAAGAGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 56SL5 | 3′-UUGUCAUUAGUCUUAAGCUCUUCUUUGGGGGGAGCUCACCGGUAUACUUUUUUUGA-5′ |
| 60 | 3′-UUGUCAUUAGUCUUAAGAGCUCUACCGUAUGACAUUGGUAUUCCGGUAUACUUUUUUUGA-5′ |
| 65 | 3′-UUGUCAUUAGUCUUAAGAUUUGAGCUCUACCGUAUGACAUUGGUAUUCCGGUAUACUUUUUUUGA-5′ |
| 70 | 3′-UUGUCAUUAGUCUUAAGUGUUAAUUUGAGCUCUACCGUAUGACAUUGGUAUUCCGGUAUACUUUUUUUGA-5′ |
| 108 | 3′-UUGUCAUUAG-stuffer sequence derived from φX174-AUACUUUUUUUGA-5′ |
| 132 | 3′-UUGUCAUUAG-stuffer sequence derived from φX174-AUACUUUUUUUGA-5′ |
| 382 | 3′-UUGUCAUUAG-stuffer sequence derived from φX174-AUACUUUUUUUGA-5′ |
| 1098 | 3′-UUGUCAUUAG-sequence encoding GFP and stuffer from φX174-AUACUUUUUUUGA-5′ |
Presented negative-sense 3′-5′. The gene-start (UUGUCAUUAG) and gene-end (AUACUUUUUUUGA) sequences are indicated in bold. Potential stem-loop structures are indicated by underlining each half of the stem, taking advantage of potential G-U base pairings; SL4 and SL5 were designed to form in the positive and negative senses, respectively. φX174, sequences derived from bacteriophage φX174; GFP, green fluorescent protein-coding region derived from pGreen Lantern (GIBCO/BRL).
Analysis of RNA synthesis from subgenomic replicons.
RNA synthesis was examined by direct metabolic labeling of cells 15 h posttransfection. Cells were exposed for 6 h to [3H]uridine (33 μCi/ml) in the presence of actinomycin D (10 μg/ml). Cells were harvested, and total cytoplasmic RNA was extracted and analyzed as described previously (34, 52). Where possible, the abundance of the different mRNAs was determined as follows. Prior to agarose-urea gel electrophoresis, RNAs were annealed with oligo(dT) followed by incubation with RNase H as described elsewhere (6). This allowed the removal of the heterogeneous polyadenylate tails and resulted in the resolution of discrete bands that were quantitated by densitometry. Fluorogrammed gels were exposed to preflashed films that were scanned using a Howtek Scanmaster 3 and analyzed using DNA Quantity One software on a PDI model 320i densitometer.
Primer extension was used to map the position of the 5′ ends of the mRNAs and positive-sense replication products, using Superscript II reverse transcriptase (GIBCO/BRL) and an oligonucleotide primer that annealed to positive-sense RNA in the L gene at positions 10925 to 10908 of the complete VSV genome sequence (Fig. 1). Primer extensions were carried out as follows. Total cellular RNA obtained from a 60-mm-diameter dish 23 h posttransfection was isolated as described above and then incubated with RQ1 DNase (Promega Corporation, Madison, Wis.) to digest contaminating plasmid DNA. The resultant RNAs were purified using RNeasy columns (Qiagen Inc., Valencia, Calif.) and incubated a second time with RQ1 DNase. Following repurification, approximately one-sixth the total RNA obtained from a 60-mm-diameter dish was incubated with 50 pmol of oligonucleotide primer. This template-primer mix was heated to 100°C for 1 min and then placed directly on ice. Reverse transcriptions were performed at 50°C, using the buffer and deoxynucleoside triphosphate concentrations recommended by the manufacturer, in the presence of 2.5 μCi [35S]dATP (1,250 Ci/mmol). One-tenth of this reaction was analyzed by electrophoresis on a denaturing 6% polyacrylamide gel.
Characterization of recombinant viruses.
Recombinant VSV was recovered from cDNA clones that had been engineered to express an additional transcriptional unit, as described below. Recovered viruses were purified and amplified in BHK-21 cells as described elsewhere (53). Viral RNA synthesis was analyzed following infection of cells at a multiplicity of 3, by exposure to [3H]uridine in the presence of actinomycin D (10 μg/ml) 3 h postinfection for 4 h. Cells were harvested, and total cytoplasmic RNA was extracted and analyzed as described above.
RESULTS
Construction and characterization of a subgenomic replicon with two transcriptional units.
A wild-type subgenomic replicon, WT, consisting of a fusion of the 3′-terminal 210 nt with the 5′-terminal 265 nt of the VSV genome such that it encoded one mRNA, was engineered to contain a second transcriptional unit. A 41-nt transcriptional unit composed of the conserved gene-start and gene-end sequences surrounding an 18-nt polylinker was inserted at the leader-N gene junction to create WT+I-41 (Fig. 1). RNA synthesis directed by this bicistronic replicon was examined as described in Materials and Methods. The results of a typical experiment are shown in Fig. 2. The major RNA synthetic event programmed by replicon WT (Fig. 2, lane 1) was transcription of an mRNA, the N/L mRNA that was characterized previously (52, 55). This mRNA migrated as a broad band that resolved to a single discrete product when exposed to RNase H after annealing to oligo(dT), to remove polyadenylate tails, which indicated that it was polyadenylated (Fig. 2, compare lanes 1 and 2). The minor slower-migrating product (observed in lane 2) was previously identified as the genomic and antigenomic replication products of replicon WT (52, 55).
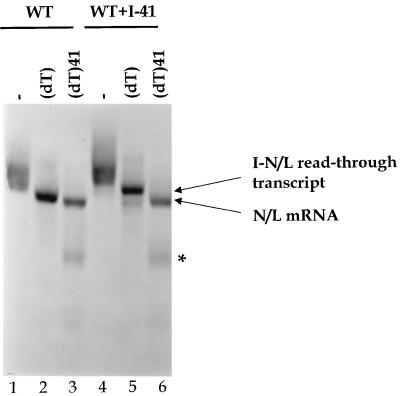
FIG. 2.
Identification of the RNAs synthesized by subgenomic replicons. Cells were infected with vTF7-3, transfected with cDNAs for subgenomic replicon WT (lanes 1 to 3) or WT+I-41 (lanes 4 to 6) along with plasmids encoding the VSV N, P, and L proteins, and exposed to [3H]uridine (33 μCi/ml) in the presence of actinomycin D (10 μg/ml) as described in Materials and Methods. Cytoplasmic extracts were prepared, and RNAs were annealed with the indicated strand- and sequence-specific oligonucleotides prior to exposure to RNase H as described in Materials and Methods. The resultant RNAs were analyzed by electrophoresis on agarose-urea gels and visualized by fluorography as described in Materials and Methods. Lanes 1 and 4, no oligonucleotide; lanes 2 and 5, oligo(dT); lanes 3 and 6, oligo(dT) and a negative-sense oligonucleotide designed to anneal to the entire I-41 mRNA and the first 10 nt of the N/L mRNA. Under the conditions used, this latter oligonucleotide, which was designed to anneal to the entire I-41 mRNA and the first 10 nt of the N/L mRNA, appears to mediate some cleavage of the N/L mRNA to yield a product of approximately 200 nt (∗). This presumably reflects inefficient hybridization of this large oligonucleotide at a suboptimal site within the N/L mRNA which results in its cleavage by RNase H.
The major product transcribed by replicon WT+I-41 migrated as a broad band slightly slower than the N/L mRNA of replicon WT (Fig. 2, compare lanes 1 and 4). This product was shown to be polyadenylated by cleavage with RNase H after annealing to oligo(dT) to yield two faster-migrating species (Fig. 2, lane 5). The minor faster-migrating product comigrated with the deadenylated N/L mRNA of replicon WT (Fig. 2, compare lanes 2 and 5), indicating that a small amount of the N/L mRNA was transcribed from WT+I-41. The major product was larger than the deadenylated N/L mRNA, suggesting that it was a transcript comprising a read-through of the inserted I-41 mRNA into the N/L mRNA. Consistent with this, discrete monocistronic I-41 mRNA was not detected. To examine whether the larger RNA was an I-41-N/L read-through transcript, RNAs were exposed to RNase H after annealing to oligo(dT) and an oligonucleotide designed to anneal to the entire I-41 mRNA and the first 10 nt of the N/L mRNA (Fig. 2, lanes 3 and 6). A major product that migrated slightly faster than the deadenylated N/L mRNA was observed for WT+I-41 (Fig. 2, lane 6). This product comigrated with the cleavage product of the N/L mRNA extracted from replicon WT-transfected cells (Fig. 2, compare lanes 6 and 3). These data demonstrated that the major product of transcription from WT+I-41 was a bicistronic read-through transcript of the I-41 mRNA covalently linked to the N/L mRNA (I-41-N/L read-through), indicating that the VSV polymerase failed to efficiently terminate mRNA synthesis at the gene-end sequence of I-41. This observation was striking, as previously we and others demonstrated that the AUACUUUUUUUG/C found at each intergenic junction was sufficient for termination of mRNA synthesis and that read-through of a wild-type gene junction occurred only 1 to 3% of the time (6, 7, 21, 22, 24, 41, 42). We therefore suspected that the inefficient termination observed at the I-41 gene end was a consequence of the short distance between the gene-start and gene-end sequences.
Effect on RNA synthesis of altering the spacing between the gene-start and gene-end sequences.
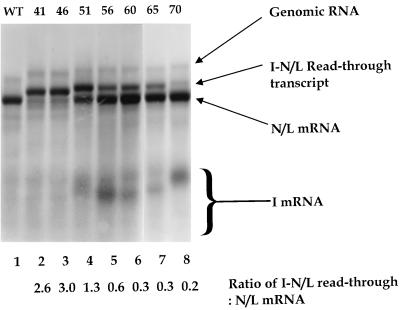
To examine the effect of the spacing between the gene-start and gene-end sequences on mRNA synthesis, the distance between these two conserved elements was increased. Using standard cloning techniques, the first transcriptional unit of WT+I-41 was gradually enlarged from 41 up to 1,098 nt (Table 1). The effects of these insertions on RNA synthesis were examined as described in Materials and Methods, and the results are shown in Fig. 3. Levels of replication were indistinguishable from those of replicon WT, as evidenced by the slowest-migrating product observed in each lane (Fig. 3). The nature and abundance of the mRNA products varied according to the size of the first transcriptional unit. Replicon WT+I-46 behaved similarly to WT+I-41, predominantly directing the synthesis of an I-46-N/L read-through transcript, with only a small amount of the discrete N/L mRNA synthesized (Fig. 3, lane 3). Increasing the spacing between the gene-start and gene-end sequences a further 5 nt produced replicon WT+I-51, which directed the synthesis of an I-51-N/L read-through transcript, but significantly more discrete N/L mRNA was synthesized (Fig. 3, lane 4). The discrete I-51 mRNA was difficult to detect owing to its small size and low uridine content, although on the exposure of the autoradiograph shown here, a faint product was observed (Fig. 3, lane 4). When the inserted transcriptional unit was increased in size a further 5 nt to yield WT+I-56, monocistronic I-56 mRNA was observed and more N/L mRNA was produced than the I-56-N/L read-through transcript (Fig. 3, lane 5). Increasing the size of the transcriptional unit to 60 or 65 nt had little discernible effect on the relative levels of transcription of the inserted mRNA, the N/L mRNA, or the I-N/L read-through (Fig. 3, compare lane 5 with lanes 6 and 7). However, increasing the transcriptional unit to 70 nt resulted in a further decrease in the proportion of the I-N/L read-through transcript synthesized (Fig. 3, lane 8). Increasing the size of the transcriptional unit up to 1,098 nt had no further effect on production of the discrete I mRNA, the N/L mRNA, or the products of polymerase read-through to generate bicistronic transcripts (data not shown). These data thus demonstrated that the spacing between the gene-start and gene-end sequences was an important determinant of whether a transcript is efficiently terminated at a wild-type gene-end sequence.
FIG. 3.
RNAs synthesized by genomic analogs with increasingly longer first transcriptional units. The fluorogram shows an agarose-urea gel of actinomycin D-resistant RNAs synthesized in cells transfected with the indicated VSV genomic analog and the VSV N, P, and L support plasmids. RNAs were labeled, harvested, purified, annealed with oligo(dT), and exposed to RNase H as described in Materials and Methods. Replication products, the N/L mRNA, and the I-N/L read-through transcript are indicated. The I mRNA (which varied in size) is indicated by a bracket (see text for details). Replicons were named according to the size of the inserted transcriptional unit (indicated above the lanes). To indicate the efficiency with which polymerase terminated transcription at the I gene-end sequence, the molar ratio of the I-N/L read-through transcript to the N/L mRNA was determined as described in the text. These values are shown below the lanes.
Owing to the small size and low U content of the I mRNAs, it was not possible to accurately quantify their abundance directly. In addition, the pH 3.0 agarose-urea gels separate RNAs both on the basis of size and charge (28), resulting in unpredictable mobility, particularly of these small RNAs. However, the relative proportions of the I-N/L read-through transcript versus the N/L mRNA act as an indirect measure of the efficiency with which polymerase terminated mRNA synthesis in response to the I gene-end sequence. For each replicon, the ratio of the I-N/L read-through to N/L mRNA was determined as described in Materials and Methods, and these values are indicated (Fig. 3). Ratios greater than 1.0 indicate that the read-through transcript was produced in molar excess over the N/L mRNA. The close proximity of the I-N/L read-through and the N/L mRNA resulted in an overassessment of the abundance of the less intense product, such as the N/L mRNA of WT+I-41 and WT+I-46, and the read-through transcript of WT+I-70 (Fig. 3, lanes 2, 3, and 8, respectively). However, the ratios given are consistent with those obtained following quantitation of total RNA synthesized in a 23-h time period as assayed by primer extension (see below).
Effects on viral transcription of insertion of an additional transcriptional unit between leader and the N gene.
To determine whether the data obtained with subgenomic replicons would be the same when examined during transcription from infectious viruses, selected transcriptional units were inserted into an infectious cDNA clone of VSV, pVSV1(+). Recombinants corresponding to WT+I-41, WT+I-60, WT+I-1098, and three additional recombinants with 108-, 132-, or 382-nt transcriptional units were engineered into pVSV1(+), and the ability to rescue infectious virus examined. Infectious viruses were readily recovered from all cDNAs except the recombinant corresponding to WT+I-41. Virus was not recovered from pVSV1(+)41 from 35 transfections under conditions that routinely allowed rescue of virus from other engineered cDNAs. Extrapolation of the data obtained from analysis of RNA synthesis by subgenomic replicon WT+I-41 suggest that this is likely a consequence of the inability of such an engineered virus to synthesize discrete monocistronic N mRNA. A read-through transcript of the 41-nt inserted transcriptional unit and the N mRNA may affect translation of N protein because the positive-sense copy of the I gene-end sequence (UAUGAAAAAAA) would now provide the first methionine codon of the read-through transcript. This methionine residue would be in the −1 reading frame with respect to that of the N protein AUG. That pVSV1(+)41 did not contain mutations that prevented recovery of infectious virus was demonstrated by pVSV1(+)60 and pVSV1(+)1098, in which the additional sequences were directly cloned into the transcriptional unit of pVSV1(+)41 and infectious virus was recovered.
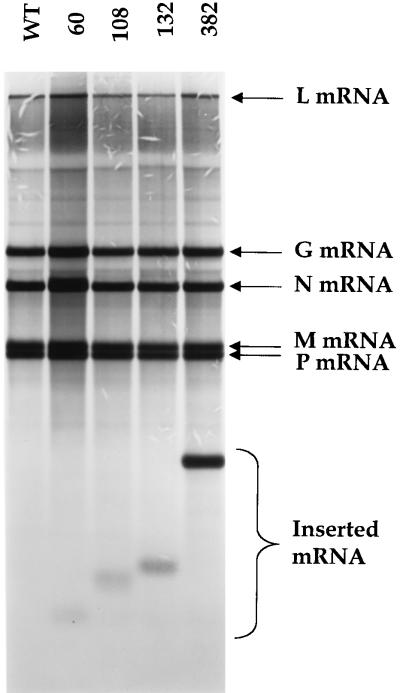
The recombinant viruses were characterized by examining the products of RNA synthesis in infected cells, as described in Materials and Methods. The data shown in Fig. 4 demonstrate that each of the recovered viruses produced an additional mRNA that corresponded to the size of the additional transcriptional unit [data not shown for rVSV1(+) 1098]. Recombinant virus rVSV1(+)60 synthesized an I-60-N read-through transcript (Fig. 4), consistent with the abundance of the I-60-N/L read-through transcript produced by subgenomic replicon WT+I-60 (Fig. 3, lane 6). For rVSV1(+)108, -132, -382, and -1098, the abundance of the I mRNA relative to the N mRNA was quantitated. The I mRNA was produced in a 1.3:1 to 1.7:1 molar ratio compared to the N mRNA. These data are consistent with the sequential and polar transcription of VSV mRNAs whereby promoter-proximal genes are transcribed in greater abundance than more distal genes. In addition, these data demonstrate that the level of attenuation observed at the I-N gene junction ranged from 24 to 41%, which is consistent with the approximate 30% attenuation reported at the N-P, P-M, and M-G gene junctions (24). Taken together, these findings support our observations obtained using the subgenomic replicons, which demonstrate that there is a minimal size requirement for efficient termination of a VSV mRNA.
FIG. 4.
Fluorogram of an agarose-urea gel of actinomycin D-resistant mRNAs synthesized in BHK-21 cells infected with recombinant VSV viruses that contained an additional transcriptional unit inserted at the leader-N gene junction. RNAs were labeled, harvested, purified, annealed with oligo(dT), and exposed to RNase H as described in Materials and Methods. The VSV N, P, M, G, and L mRNAs are identified. The inserted mRNA (which varied in size) is indicated by a bracket (see text for details). Recombinant viruses were named according to the number of nucleotides inserted at the leader-N gene junction (indicated above the lanes).
Effects of sequence and structure on termination.
The experiments described above indicated that the VSV polymerase was unable to efficiently terminate mRNA synthesis in response to a gene-end sequence of a transcriptional unit less than 46 nt in length. However, the polymerase terminated mRNA synthesis in response to the gene-end sequence of transcriptional units that were 5 nt longer (WT+I-51), and it terminated mRNA synthesis with greater efficiency when the transcriptional unit was 10 nt longer (WT+I-56 [Fig. 3, lane 5]). The transcriptional unit present in WT+I-41 contained an 18-nt polylinker sequence. This presented the possibility that the palindromic nucleotides within this sequence might influence polymerase behavior, and that only when these nucleotides were interrupted could termination occur. To explore this possibility, we examined whether primary RNA sequence with a potential to form an RNA secondary structure could influence the minimal size requirement for termination by generating subgenomic replicons whose I mRNA sequences had the potential to adopt specific secondary structures.
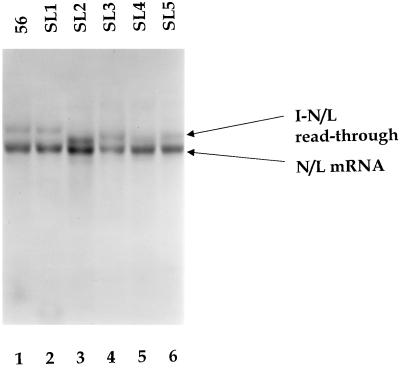
Working with WT+I-56, we generated five additional replicons designed to introduce RNA stem-loop structures (SL1 to SL5) and/or homopolymeric tracts (Table 1) and examined the effects of these alterations on transcription. WT+I-56SL1 behaved indistinguishably from the WT+I-56 (Fig. 5, compare lanes 1 and 2), whereas WT+I-56SL2 appeared to synthesize slightly higher levels of the read-through transcript than WT+I-56 (Fig. 5, compare lanes 1 and 3), suggesting that the potential to form a stem-loop structure in the I-56 mRNA or its encoding template reduced the efficiency of termination at the gene junction immediately downstream. The low-pH agarose-urea gels separate RNAs on the basis of base composition as well as size (28), which presumably explains the mobility shift observed for the I-N/L read-through transcript of WT+I-56SL2. As SL2 reduced termination and SL1 was indistinguishable from replicon WT, we examined whether the proximity of the potential stem-loop structure to the gene-end sequence influenced termination. A potential stem-loop structure was introduced adjacent to the gene-end sequence (WT+I-56SL3), and the effects on RNA synthesis were examined. Only minor differences in the proportions of the N/L mRNA and the I-N/L read-through transcript were observed in WT+I-56SL3 compared with WT+I-56SL2 (Fig. 5, lane 4), suggesting that the proximity of the potential stem-loop structure to the gene end was not a major determinant of the efficiency of termination.
FIG. 5.
RNAs synthesized by genomic analogs in which the first transcriptional unit or its mRNA product was designed such that a portion of it could adopt a stem-loop structure. A fluorogram of an agarose-urea gel of actinomycin D-resistant RNAs synthesized in cells transfected with the indicated VSV genomic analog and the VSV N, P, and L support plasmids is shown. RNAs were labeled, extracted, annealed with oligo(dT), and exposed to RNase H as described in Materials and Methods. The N/L mRNA and the I-N/L read-through transcript are shown. The sequences of the inserted transcriptional units and the potential stem-loop structures are shown in Table 1.
The sequences of the I regions of replicons WT+I-56SL1-3 could adopt stem-loop structures in both the positive and negative strands. To examine whether the reduction in termination observed for WT+I-56SL2 was a consequence of a potential structure in either the positive or negative strand, two additional replicons, WT+I-56SL4 and WT+I-56SL5, were generated. By virtue of G-U base pairs, WT+I-56SL4 and WT+I-56SL5 should adopt a stem-loop structure in only the positive and only the negative strand, respectively. The quantity of the I-N/L read-through versus N/L mRNA suggested that termination was slightly more efficient in WT+I-56SL4 than WT+I-56SL5 (Fig. 5, compare lanes 5 and 6). However, these differences were relatively minor, and neither WT+I-56SL4 nor WT+I-56SL5 appeared to affect termination as much as WT+I-56SL2. While WT+I-56SL2 displayed a reduction in termination efficiency, this analysis showed that failure of polymerase to terminate mRNA synthesis in response to a gene-end sequence was influenced considerably less by either the primary RNA sequence, or the potential to adopt a stem-loop structure, than the size of the transcriptional unit.
Analysis of the positive-sense products of transcription by primer extension.
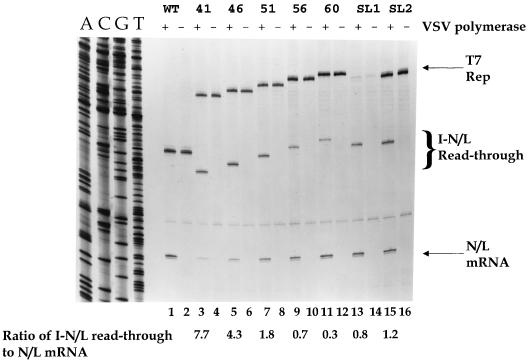
To accurately quantitate the abundance of the positive-sense products of transcription, primer extension analyses were performed using an oligonucleotide that annealed to positive-sense VSV RNA at residues 10925 to 10909 of the complete VSV genome sequence. As predicted, extension of this primer by reverse transcriptase (RT) on RNAs extracted from WT-transfected cells yielded a product of 192 nt from extension on the N/L mRNA (Fig. 6, lane 1), which was absent when the plasmid expressing the L polymerase subunit was omitted from the transfection (Fig. 6, lane 2). For technical reasons, namely, that T7 RNA polymerase fails to reliably transcribe a negative-sense copy of a VSV intergenic junction at 37°C (53), replicons were generated as positive-sense transcripts. Thus, the plus-strand genomic replication products could not be readily distinguished from the primary T7 transcript (Fig. 6, lanes 1 and 2). For each of the replicons that contained an additional transcriptional unit, positive-sense products corresponding to the N/L mRNA, the I-N/L read-through, and the primary T7 transcripts were observed. The abundance of the N/L mRNA and the I-N/L read-through varied between the different replicons (Fig. 6, lanes 3 to 12). The general trend in relative levels of the monocistronic N/L mRNA versus the I-N/L read-through agreed with that observed by agarose-urea gel analysis (Fig. 2 to 5). As the size of the I mRNA increased, the abundance of the read-through transcript decreased and the quantity of the N/L mRNA increased. However, by primer extension analysis it was possible to detect the small quantity of N/L mRNA produced from WT+I-41 and WT+I-46 (Fig. 6, lanes 3 to 6), which was barely detectable by agarose-urea gel analysis (Fig. 3, lanes 2 and 3).
FIG. 6.
Primer extension analysis of the positive-sense RNAs synthesized by subgenomic replicons with increasingly longer first transcriptional units. A polyacrylamide gel of the primer extension products obtained from total cytoplasmic RNAs extracted from BHK-21 cells that were transfected 23 h earlier with the indicated genomic analog and either the VSV N, P, and L support plasmids (+) or the VSV N and P support plasmids (−) is shown. The oligonucleotide primer annealed to positive-sense RNA at positions 10925 to 10908 of the complete VSV genome sequence (Fig. 1). Reverse transcriptions were performed as described in Materials and Methods. A sequence ladder of replicon WT sequenced with the same primer is shown for reference. Replicons were named according to the number of nucleotides present in the inserted transcriptional unit or its potential to adopt a stem-loop structure (Table 1). As a measure of the efficiency with which polymerase terminated at the I gene-end sequence, the molar ratio of the I-N/L read-through transcript to the N/L mRNA was determined as described in the text, and these values are indicated below the lanes. Significantly less T7 transcript was detected for replicon WT+I-56SL1; however, the proportions of the I-N/L read-through transcript and the N/L mRNA observed were consistent with those observed following agarose-urea gel analysis (Fig. 5). Products of primer extension on the initial positive-sense T7 transcripts and the replicated antigenomes are indicated (T7 and Rep, respectively).
To quantitate the abundance of the I-N/L read-through transcript relative to the N/L mRNA, the autoradiograph was scanned and the primer extension products were quantified as described in Materials and Methods. The relative proportion of the I-N/L read-through versus N/L mRNA acts as an indirect measure of the efficiency with which polymerase terminated mRNA synthesis at the I gene-end sequence. Failure to terminate transcription at the I gene-end sequence resulted in synthesis of a greater quantity of the I-N/L read-through transcript and decreased levels of the N/L mRNA. This was reflected in a higher numerical value for the ratio of the I-N/L read-through to N/L mRNA. Quantitation of the RNA products following agarose-urea gel electrophoresis (Fig. 3 and 5) was complicated by the similar migration of the N/L mRNA and the I-N/L read-through transcript and the greatly different uridine contents of the I and N/L mRNAs. The primer extension analysis allowed us to ascribe a more accurate value to the reduction of termination at the gene junction (Fig. 6). The I-N/L read-through transcript was more abundant than the N/L mRNA, as indicated by a ratio of >1.0, when the inserted transcriptional unit was less than 56 nt.
Analysis of polymerase behavior at the ignored gene junction.
Termination of transcription at a VSV gene junction involves multiple processes, including (i) reiterative transcription or polymerase slippage on the U7 tract to generate the polyadenylate tails and (ii) termination and release of the nascent transcript. Recently we have shown that in certain circumstances read-through RNAs contain additional A residues that result from polymerase slippage while copying the U7 tract of the intergenic junction (J. N. Barr and G. W. Wertz, unpublished data). The relationship between polymerase slippage and termination is complex and remains poorly understood. However, we sought to examine the behavior of polymerase at the ignored gene junctions of WT+I-41, WT+I-46, and WT+I-51 to determine whether any reiterative transcription occurred. The primer extension analysis described above showed that the majority of the read-through transcripts were homogeneous in length, indicating that they were faithful copies of the intergenic junction lacking additional A residues. However, if uncontrolled slippage on the U7 tract occurred at a low level, the primer extension products would have variable 5′ termini, which would make detection of these transcripts difficult. To assess whether a small proportion of the read-through transcripts contained nontemplated A residues at an intergenic junction, RNAs were amplified by RT-PCR, and the resultant cDNAs were cloned and sequenced. To avoid DNA contamination of the RT-PCR and to avoid contamination with the primary T7 transcript, subgenomic replicons were assembled into infectious particles that were used to infect cells expressing the VSV N, P, and L proteins, as described previously (34, 54). The resultant RNAs were reverse transcribed using Moloney murine leukemia virus RT and a primer that annealed in the L gene at positions 10925 to 10908 of the complete VSV genome sequence and were subsequently amplified by PCR. As a control, PCRs were performed on samples in which RT had been omitted from the reaction. These controls failed to generate products in the subsequent PCR, thus demonstrating that the products were RNA dependent (data not shown). A total of three independent PCRs were performed, and the resultant products were individually cloned and sequenced. The sequence through the intergenic region was examined from 53 separate clones and was shown to be a direct copy of the intergenic region 48 times. Of the remaining five sequences, four contained a single additional A residue, and one sequence had an expanded A run (23 A residues). These data thus confirmed that the VSV polymerase typically failed to slip on the seven U residues at the intergenic junction on these read-through transcripts.
DISCUSSION
Termination of transcription is a crucial step in the regulation of gene expression in VSV. Since the viral transcriptase enters the genome at a single site (13), expression of each downstream gene is dependent on termination of the preceding gene (1, 4, 6, 7, 22). We and others previously demonstrated that the gene-end sequence 3′ AUACUUUUUUUG/C 5′ functioned as an essential cis-acting signal for termination of mRNA synthesis (6, 22). However, as described here, the ability of polymerase to terminate mRNA synthesis in response to this element is diminished in transcriptional units of less than 70 nt and is almost completely abolished for transcriptional units of less than 51 nt.
Within the confines of this minimal size requirement for mRNA synthesis, we examined whether the primary RNA sequence or ability of the RNA to adopt a stem-loop structure influenced the efficiency with which polymerase utilized the gene-end sequence (Fig. 5). The I transcriptional unit of replicon WT+I-56 was altered such that potential stem-loop structures could form in either the template or nascent strand. The molar ratio of I-56-N/L read-through transcript to N/L mRNA was altered from 0.7 in WT+I-56 to 1.2 in WT+I-56SL2, indicating that the presence of a stem-loop structure in the I mRNA, or its encoding template, reduced the efficiency of termination at the I-N/L gene junction. However, for replicon WT+I-56SL1, the molar ratio of the I-N/L read-through to N/L mRNA was 0.8, only slightly different from that observed for WT+I-56 (Fig. 6). These data suggest that sequences outside the conserved gene junction can modulate the behavior of the polymerase at the gene junction but show that these effects were relatively minor. Consequently we conclude that the failure of the polymerase to terminate at the intergenic junction in WT+I-41 and WT+I-46 was predominantly a consequence of the size of the transcriptional unit, rather than its sequence.
How might such an effect be mediated? The components of the VSV transcription reaction are the template, the polymerase complex, and the product mRNA. Each of these components could influence the behavior of the polymerase at a gene-end sequence and thus may be responsible for the inefficient termination observed in transcriptional units of 51 nt or less.
The template for transcription by the VSV polymerase is the negative-sense RNA genome tightly encapsidated by the N protein (14). Available evidence indicates that the accessory P component of the polymerase can bind to the template prior to binding of the L core polymerase subunit (18, 31), though stronger binding occurs with a P-L complex (18). Binding sites for the P protein have been mapped to both the leader region (25) and the complement of the genomic trailer (23). As each of these binding sites spans approximately 20 nt (23, 25), it seems likely that the footprint of the fully assembled polymerase would be larger. The physical size of the polymerase complex might prevent either the recognition or use of the essential cis-acting signals for termination at a gene junction positioned less than 56 nt from a gene-start sequence.
The polymerase complex ostensibly comprises the viral P and L proteins. While several host cell proteins have been found associated with the L polymerase subunit (12), a role for these in RNA synthesis has yet to be demonstrated. One possible explanation for the failure of polymerase to terminate an mRNA within a limited distance of a gene-start sequence is that the polymerase complex itself may undergo a postinitiation modification event. This modification may be relatively subtle, such as an alteration to the phosphorylation status of a component of the complex, and/or could reflect the presence or absence of accessory factors. Precedent for modifications to a transcribing polymerase complex is provided by RNA polymerase II (pol II). Purification of initiating and elongating forms of yeast pol II provided evidence for the presence of a “mediator” at the initiation step, which is absent during polymerase elongation (47). Loss of this mediator during the transition to an elongating polymerase complex is accompanied by hyperphosphorylation of the carboxy-terminal domain of pol II. Whether analogous events occur during VSV transcription remains to be determined, but differences between initiating and elongating polymerase complexes may account for the inability of the polymerase to terminate mRNA synthesis in response to a gene-end sequence positioned close to a gene-start sequence.
In many systems, the nascent mRNA chain acts as a regulatory target during elongation and termination. Signals in the nascent strand can be transmitted either directly or indirectly to the elongating polymerase. For example, during transcription of the human immunodeficiency virus genome, the viral protein Tat interacts with the TAR element on the nascent RNA chain, and this is necessary to generate full-length viral transcripts (30). If the nascent mRNA strand influences the response of the VSV polymerase to a gene-end sequence, then it seems likely that this would be a feature conserved among all VSV mRNAs. Conserved elements are found at the 5′ and 3′ termini of VSV mRNAs, namely, 5′ AACAGNNAUC and UAUGAn 3′, respectively. Additionally, the 5′ terminus is modified by capping and methylation resulting in the structure 5′m7G(5′)ppp(5′)ApmApCpApGp (32, 37). Consequently, either the conserved sequence elements themselves or the modifications that occur could act as regulatory targets during transcription.
Consistent with this possibility, a recent analysis of the first three nucleotides of a VSV gene-start sequence demonstrated a role for these residues in the postinitiation events of capping and methylation and showed that initiation and correct 5′-end modification were separable events during mRNA synthesis (46). Transcripts that were not correctly modified at their 5′ termini lacked a poly(A) tail and were truncated, ranging in size from 40 to 200 nt. These observations led to the suggestion that polymerase processivity is linked to proper 5′-end modification and suggest a recognition event between the 5′ end of the nascent mRNA chain and the transcribing polymerase complex.
Capping of VSV mRNAs is unusual in that the ∝ and β phosphates of the 5′-5′ triphosphate bridge of the GpppA cap are derived from a presumed GDP donor (2, 3). This contrasts with capping of other eukaryotic mRNAs, where GMP derived from GTP is transferred by RNA guanylyltransferase to the 5′ diphosphate of an RNA chain in the nucleus. This unusual mechanism, combined with the cytoplasmic location of VSV RNA synthesis, has led to the suggestion that the VSV polymerase is responsible for capping its mRNAs, though direct evidence for this is lacking. However, short uncapped transcripts corresponding to the 5′ ends of VSV mRNAs cannot be chased into capped transcripts in in vitro transcription reactions (8, 26, 50), suggesting that capping itself is intimately linked with transcription elongation. Intriguingly, Piwnica-Worms and Keene reported previously that transcripts less than 37 nt long were not capped in vitro (36). These similarities raise the possibility that an mRNA must be a minimal size in order to be capped, and that this process is a prerequisite for mRNA termination.
In contrast to its role in capping, there is good evidence to suggest a direct role for the VSV polymerase in methylation of the cap structure of its transcripts (20, 49). Although under- and unmethylated VSV mRNAs can be generated (suggesting that methylation is not essential for transcription), transcription reactions performed in the presence of the methylation inhibitor S-adenosylhomocysteine produced VSV mRNAs that contained giant heterogeneous poly(A) (38), suggesting that methylation can influence 3′ end formation. This effect was attributed to be a consequence of failure to methylate the 5′ end of the downstream mRNA. However, it remains possible that this effect was mediated by failure to methylate the 5′ end of the nascent mRNA transcript.
The shortest transcripts observed in VSV-infected cells are the uncapped and nonpolyadenylated leader transcripts synthesized from the 3′ ends of the genomic and antigenomic viral RNAs (27). These transcripts are 47 and 45 nt in length for the positive- and negative-sense leader RNAs, respectively (10, 27). To date, neither specific initiation nor termination signals for the synthesis of discrete leader RNAs have been defined. While signals for leader RNA synthesis have not yet been defined, the sizes of the leader products are within the range identified in this study at which a low level of termination should occur, and read-through would be the predominant activity. However, leader RNA is produced in excess of N mRNA, with read-through of the leader-N gene junction being a relatively minor event, except in the case of a template-associated defect in the N protein (35), suggesting that this size requirement does not apply to the leader RNA. It is intriguing that two major differences between the nascent leader RNA transcript and that of a viral mRNA are that the leader RNA is not capped or polyadenylated. In this regard it would be of interest to determine whether generating a capped and/or polyadenylated leader RNA would influence polymerase behavior at the leader-N gene junction.
Excluding the positive- and negative-sense leader RNAs, the shortest capped and polyadenylated transcript synthesized by an NNS RNA virus is that generated from the 68-nt overlap at the M2-L gene junction of respiratory syncytial (RS) virus (9). Here the frequency with which the polymerase failed to terminate at the M2 gene-end sequence was approximately 10%. In RS virus the gene-end sequences vary. The ability of polymerase to terminate mRNA synthesis at a gene-end sequence is influenced by the primary sequence (S. B. Harmon, A. G. Megaw, and G. W. Wertz, unpublished data) and also by a trans-acting protein, M2-1, which enhances polymerase read-through at gene junctions (19). Based on the findings of this study, the close proximity of the L gene-start sequence to the M2 gene end may also contribute to the frequency with which polymerase reads through this junction. A recent study that reduced the spacing between the L gene-start signal and the M2 gene-end signal from 68 to 24 nt in an RS virus subgenomic replicon resulted in an increase in the abundance of transcripts that initiated at the L gene start (15). Based on the work described here, this would presumably be caused by a reduction in termination at the M2 gene end.
Overlapping gene junctions have been observed in other NNS viruses, such as sigma rhabdovirus (48), and the filoviruses Ebola virus and Marburg virus (16, 40). The work presented offers an explanation how these viruses efficiently transcribe the downstream mRNA. The efficiency with which polymerase terminates transcription in response to a gene-end sequence is reduced by its close proximity to an upstream gene-start sequence. The work presented here suggests that polymerase that initiates at the gene-start sequence within the upstream gene is unable to efficiently terminate transcription at the gene-end sequence of the 33-nt overlap found for sigma virus (48), and the 18- to 20-nt overlaps observed for Ebola and Marburg viruses (16, 40).
In summary, we have shown a minimal size requirement for VSV mRNA synthesis. The identification of such a requirement suggests that (i) a physical constraint is placed on the polymerase by the close proximity of the gene end to the gene start which prevents termination, (ii) a modification occurs to the actively transcribing polymerase complex that is essential for recognition of the gene-end sequence, or (iii) the nascent mRNA strand acts as a regulatory target during transcription. Further experiments are under way to test these hypotheses.
ACKNOWLEDGMENTS
We acknowledge the members of the G. W. Wertz and L. A. Ball laboratories for critical reviews of the manuscript.
This work was supported by PHS grant AI12464 from NIAID to G.W.W.
REFERENCES
- 1.Abraham G, Banerjee A K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham G, Rhodes D P, Banerjee A K. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 3.Abraham G, Rhodes D P, Banerjee A K. Novel initiation of RNA synthesis in vitro by vesicular stomatitis virus. Nature. 1975;255:37–40. doi: 10.1038/255037a0. [DOI] [PubMed] [Google Scholar]
- 4.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltimore D, Huang A S, Stampfer M. Ribonucleic acid synthesis of vesicular stomatitis virus, II. An RNA polymerase in the virion. Proc Natl Acad Sci USA. 1970;66:572–576. doi: 10.1073/pnas.66.2.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr J N, Whelan S P, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr J N, Whelan S P, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanda P K, Banerjee A K. Identification of promoter-proximal oligonucleotides and a unique dinucleotide, pppGpC, from in vitro transcription products of vesicular stomatitis virus. J Virol. 1981;39:93–103. doi: 10.1128/jvi.39.1.93-103.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Olmsted R A, Spriggs M K, Johnson P R, Buckler-White A J. Gene overlap and site-specific attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1987;84:5134–5138. doi: 10.1073/pnas.84.15.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colonno R J, Banerjee A K. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell. 1978;15:93–101. doi: 10.1016/0092-8674(78)90085-5. [DOI] [PubMed] [Google Scholar]
- 11.Colonno R J, Banerjee A K. A unique RNA species involved in initiation of vesicular stomatitis virus RNA transcription in vitro. Cell. 1976;8:197–204. doi: 10.1016/0092-8674(76)90003-9. [DOI] [PubMed] [Google Scholar]
- 12.Das T, Mathur M, Gupta A K, Janssen G M, Banerjee A K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci USA. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson S U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 14.Emerson S U, Wagner R R. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearns R, Collins P L. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann H, Muhlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H D. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Lenard J. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J Virol. 1995;69:7718–7723. doi: 10.1128/jvi.69.12.7718-7723.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy R W, Harmon S B, Wertz G W. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J Virol. 1999;73:170–176. doi: 10.1128/jvi.73.1.170-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hercyk N, Horikami S M, Moyer S A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 21.Herman R C, Schubert M, Keene J D, Lazzarini R A. Polycistronic vesicular stomatitis virus RNA transcripts. Proc Natl Acad Sci USA. 1980;77:4662–4665. doi: 10.1073/pnas.77.8.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaac C L, Keene J D. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J Virol. 1982;43:241–249. doi: 10.1128/jvi.43.1.241-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 25.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazzarini R A, Chien I, Yang F, Keene J D. The metabolic fate of independently initiated VSV mRNA transcripts. J Gen Virol. 1982;58:429–441. doi: 10.1099/0022-1317-58-2-429. [DOI] [PubMed] [Google Scholar]
- 27.Leppert M, Rittenhouse L, Perrault J, Summers D F, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 28.Lerach H, Diamond D, Wozney J, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Pattnaik A K. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J Virol. 1999;73:444–452. doi: 10.1128/jvi.73.1.444-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marciniak R A, Calnan B J, Frankel A D, Sharp P A. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- 31.Mellon M G, Emerson S U. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27:560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyer S A, Abraham G, Adler R, Banerjee A K. Methylated and blocked 5′ termini in vesicular stomatitis virus in vivo mRNAs. Cell. 1975;5:59–67. doi: 10.1016/0092-8674(75)90092-6. [DOI] [PubMed] [Google Scholar]
- 33.Moyer S A, Smallwood-Kentro S, Haddad A, Prevec L. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol. 1991;65:2170–2178. doi: 10.1128/jvi.65.5.2170-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 35.Perrault J, Clinton G M, McClure M A. RNP template of vesicular stomatitis virus regulates transcription and replication functions. Cell. 1983;35:175–185. doi: 10.1016/0092-8674(83)90220-9. [DOI] [PubMed] [Google Scholar]
- 36.Piwnica-Worms H, Keene J D. Sequential synthesis of small capped RNA transcripts in vitro by vesicular stomatitis virus. Virology. 1983;125:206–218. doi: 10.1016/0042-6822(83)90074-0. [DOI] [PubMed] [Google Scholar]
- 37.Rhodes D P, Banerjee A K. 5′-terminal sequence of vesicular stomatitis virus mRNA's synthesized in vitro. J Virol. 1975;17:33–42. doi: 10.1128/jvi.17.1.33-42.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose J K, Lodish H F, Brock M L. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J Virol. 1977;21:683–693. doi: 10.1128/jvi.21.2.683-693.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 41.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert M, Lazzarini R A. In vivo transcription of the 5′-terminal extracistronic region of vesicular stomatitis virus RNA. J Virol. 1981;38:256–262. doi: 10.1128/jvi.38.1.256-262.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smallwood S, Moyer S A. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology. 1993;192:254–263. doi: 10.1006/viro.1993.1028. [DOI] [PubMed] [Google Scholar]
- 44.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stillman E A, Whitt M A. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svejstrup J Q, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg R D. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teninges D, Bras F, Dezelee S. Genome organization of the sigma rhabdovirus: six genes and a gene overlap. Virology. 1993;193:1018–1023. doi: 10.1006/viro.1993.1219. [DOI] [PubMed] [Google Scholar]
- 49.Testa D, Banerjee A K. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Testa D, Chanda P K, Banerjee A K. Unique mode of transcription in vitro by vesicular stomatitis virus. Cell. 1980;21:267–275. doi: 10.1016/0092-8674(80)90134-8. [DOI] [PubMed] [Google Scholar]
- 51.Villarreal L P, Breindl M, Holland J J. Determination of molar ratios of vesicular stomatitis virus induced RNA species in BHK21 cells. Biochemistry. 1976;15:1663–1667. doi: 10.1021/bi00653a012. [DOI] [PubMed] [Google Scholar]
- 52.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whelan S P, Ball L A, Barr J N, Wertz G T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan S P, Wertz G W. The 5′ terminal trailer region of vesicular stomatitis virus contains a position-dependent cis-acting signal for assembly of RNA into infectious particles. J Virol. 1999;73:307–315. doi: 10.1128/jvi.73.1.307-315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whelan S P J, Wertz G W. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]