ABSTRACT
Biocides and formulated biocides are used worldwide for an increasing number of applications despite tightening regulations in Europe and in the United States. One concern is that such intense usage of biocides could lead to increased bacterial resistance to a product and cross-resistance to unrelated antimicrobials including chemotherapeutic antibiotics. Evidence to justify such a concern comes mostly from the use of health care-relevant bacterial isolates, although the number of studies of the resistance characteristics of veterinary isolates to biocides have increased the past few years. One problem remains the definition of “resistance” and how to measure resistance to a biocide. This has yet to be addressed globally, although the measurement of resistance is becoming more pressing, with regulators both in Europe and in the United States demanding that manufacturers provide evidence that their biocidal products will not impact on bacterial resistance. Alongside in vitro evidence of potential antimicrobial cross-resistance following biocide exposure, our understanding of the mechanisms of bacterial resistance and, more recently, our understanding of the effect of biocides to induce a mechanism(s) of resistance in bacteria has improved. This article aims to provide an understanding of the development of antimicrobial resistance in bacteria following a biocide exposure. The sections provide evidence of the occurrence of bacterial resistance and its mechanisms of action and debate how to measure bacterial resistance to biocides. Examples pertinent to the veterinary field are used where appropriate.
BIOCIDE USAGE
Chemical biocides have been used for centuries for making water and foodstuff safe to consume, for treating wounds, and for preserving materials since well before the discovery of microorganisms. Today chemical biocides are heavily used in a wide range of applications and environments including the consumer product, water, wastewater, and food industries; goods manufacturing; the pharmaceutical industry; the health care and veterinary sectors; and the oil and gas industries (1). This wide range of applications reflects the versatility of biocide products for environmental disinfection, product preservation, and antisepsis (2). In Europe it is difficult to estimate the quantity of chemical biocides that are used in products or imported (1), although in 2006 the market for biocides was estimated to be €10 billion to €11 billion (1). It is, however, clear that the usage of chemical biocides is continuing to increase, particularly in consumer products. This increased usage may be partly due to consumers’ increased awareness of microbial contamination and infection. The rise in antibiotic resistance in bacteria might also have impacted on the usage of biocides, at least in the health care and veterinary settings (3). Widespread media coverage of issues of hospital cleanliness and “superbugs” have also contributed to better-informed customers, providing better marketing arguments for manufacturers and distributors of biocidal products (3). Alongside a better-informed public, the global increase in antimicrobial resistance in bacteria is forcing decision makers to tackle this growing issue. One of the recommended interventions is better hygiene and control of bacteria on surfaces in health care settings but also in animal husbandry (4).
In health care settings biocides are heavily used for the disinfection of environmental surfaces and medical devices and for antisepsis. The growing number of studies highlighting the presence, and at times persistence, of bacterial pathogens, including multidrug-resistant ones, on surfaces despite the use of decontamination (5–14) acknowledges that microorganisms can survive on surfaces and be transmitted to patients, staff, and inanimate objects (15), thus finally emphasizing the importance of controlling the microbial burden on surfaces. This newly found appreciation for controlling microbial pathogens on surfaces has led to an explosion of surface disinfection products and their marketing (16–18), contributing to a higher concentration of biocides eventually released in the environment.
The ability of biocides or biocidal products to decrease the microbial bioburden on surfaces is also highly relevant in animal husbandry, farm buildings, barns, equipment, and vehicles, where their use should contribute to reducing the spread of pathogens. This also includes their use to prevent infectious outbreaks from spreading from farms; for example, large quantities of biocides are being sprayed in the environment and on vehicles in an attempt to decrease the spread of animal viral diseases (19). The heavy use of biocidal products where heavy soiling is present, in particular, their use on vehicle wheels and undercarriages, deserves better scrutiny of its efficacy in preventing potential outbreaks.
The use of biocidal products also includes the disinfection of various environmental surfaces, antibiofouling, the preservation of building materials, and water and wastewater treatment. Biocides play an important role as food preservatives and for controlling microbial contaminants that may enter the food chain during food production. As mentioned previously, one growing area for biocide manufacturers is consumer products, including the preservation of cosmetics, but more recently, personal care products, household products, and textiles.
In Europe, the incorporation of biocides in products and the use of chemical biocides in general is heavily regulated (20), with the consequence that fewer biocides are available for manufacturers to use. This restriction on the number and type of chemical biocides available for manufacturers has, however, not reduced the number of biocidal products and biocide applications. On the contrary, awareness of the role of microorganisms in contamination, infection, or the production of odors, together with the growing threat of bacterial resistance to chemotherapeutic antibiotics, has resulted in the biocidal product market expanding. In Europe, the amounts of chemical biocides used per application is difficult to measure (1). Chemical biocides used in diverse applications eventually find their way to the environment (1). For example, high concentrations of triclosan have been found in river and wastewater effluents (1.4 to 40,000 ng/liter in surface water, up to 85,000 ng/liter in wastewater, and up to 133,000 μg/kg in biosolids from wastewater treatment plants) (20–23). There should be little doubt that chemical biocides even at a low concentration (i.e., sub-MIC level) will exert a selective pressure on microorganisms (18, 24–26), which should be monitored where biocidal products are heavily used (18, 27). The increase in the use of biocides and biocidal products might aggravate the possible link between biocide usage and emergence of antimicrobial resistance in bacteria (1, 3, 18, 28), although there is no doubt that overall biocide usage has brought immense benefit to human and animal health (1–3, 29).
This article explores reports of bacterial resistance to biocides and our current knowledge of the mechanisms of bacterial resistance. It also reflects on the effect of biocides’ interactions with bacteria that may lead to a change in susceptibility to antimicrobials. This article does not cover bacterial biofilms.
BIOCIDE RESISTANCE: A QUESTION OF DEFINITIONS
One of the main issues when dealing with bacterial resistance is the definition of “resistance.” This definition is linked with the test protocols to measure resistance, and these protocols are described later. There are many definitions of resistance to biocides, some of which describe only a small decrease in susceptibility (18, 28, 31–34). This contrasts with the definition of bacterial resistance to chemotherapeutic antibiotics, which reflects clinical resistance. With biocides the terms “resistance,” “tolerance,” “decreased susceptibility,” “reduced susceptibility,” “insusceptibility,” and “acquired reduced susceptibility” are used. Such diversity in terms reflects a lack of consensus within the scientific community and is contributing to a degree of confusion in our understanding of bacterial resistance to biocides. From a practical point of view, a bacterium surviving in a biocidal product is resistant to that product, whatever the concentration of biocide is in the product.
Many papers have used the term “reduced susceptibility,” which is based on the measurement of the MIC or the minimum bactericidal concentration. A biocide or biocide product at its in-use concentration may, however, still be effective (18, 35). One of the main difficulties is to determine what fold-difference in MIC or minimum bactericidal concentration reflects a change that will be significant in practice, i.e., a decrease in biocide effectiveness. This is likely to be biocide/biocidal product dependent.
From an academic perspective, other definitions of bacterial resistance have been used: (i) a bacterial strain that is not killed by a biocide concentration to which the majority of the bacterial species are susceptible and (ii) bacterial cells in a culture that survive biocide exposure that kills the majority of the bacterial population in that culture. This latest definition has been used mainly to identify specific mechanisms of biocide resistance in bacteria following stepwise exposure to a specific biocide.
Empirically, bacterial resistance to biocides has been labeled as intrinsic, a natural property of the bacterium, or acquired, following the acquisition of resistance genes or following mutations (36). These definitions still hold true, although the concept of transient resistance, following the expression of a mechanism(s) in response to a direct selective pressure, recognizes that the effect of a biocide on a bacterium may be more complex and short-lived as long as the biocide, exerting a selective pressure, is present (24, 25).
What appears to be more of a concern is the ability of a bacterium to become clinically resistant to an antibiotic(s) following exposure to a biocide/biocidal product. Such cross-resistance has been raised by the European Commission following reports from the Scientific Committee on Emerging and Newly Identified Health Risks (1, 37) and the Scientific Committee on Consumer Safety (38). The Biocidal Product Regulation (20), which regulates the commercialization of biocidal products on the European market, now mentions the potential issue of bacterial resistance and cross-resistance following biocide application. In the United States, the Federal Drug Administration (FDA) recently proposed several rules based on the concern about bacterial resistance linked to the use of certain chemical biocides (39). Demonstrating that a chemical biocide or a biocidal product will not give rise to resistance in bacteria is a question not only of definition but also of methodology.
OCCURRENCE OF BACTERIAL RESISTANCE TO BIOCIDES
Bacterial resistance to biocides and biocidal products has now been well documented in the literature, although examples are often anecdotal where a specific product was investigated. Biocides are a very diverse group of chemicals (1). Surprisingly, bacterial resistance has been studied with only a few biocides. For biocidal products, the formulation will help and hopefully optimize the delivery of the biocide(s) and/or negate some undesirable effects such as corrosiveness of surfaces, pungent smell, poor stability, or toxicity. Components of the formulations may also have a profound effect on biocide efficacy, either increasing or, on occasion, decreasing efficacy. In the peer-reviewed literature, formulations have rarely been studied in the past, although recently, several studies concerned the effect of formulated biocides on bactericidal efficacy (3, 18, 40, 41). Bacterial resistance has been investigated in vitro against several chemical classes, including phenolics (e.g., triclosan) (42–49), cationic biocides (e.g., chlorhexidine, quaternary ammonium compounds, particularly cetylpyridinium chloride, and benzalkonium chloride) (50–56), isothiazolinones (57), and more reactive biocides such as iodophors (58), alkylating agents (e.g., glutaraldehyde) (59–64), and several oxidizing compounds (65–68). Studies often differed in their methodology, rendering the comparison of results difficult (1, 18). Using realistic in vitro protocols to generate bacteria resistant to a specific biocide is not straightforward either (69). Investigations can generally be divided into four categories:
In vitro testing of bacterial resistance to a specific biocide, often involving training the bacteria to survive increasing concentrations of a biocide (46–48, 57, 69–73)
Studies reporting the isolation of environmental isolates resistant to specific biocides. These investigations principally concern environmental bacterial isolates from, for example, health care settings, manufacturing, and slaughterhouses and include biocides such as glutaraldehyde (59–62, 74), chlorine dioxide (65), chlorhexidine (75–79), triclosan (48, 80), quaternary ammonium compounds (79, 81–83), alcohol, and iodine (75)
Studies reporting the contamination of biocidal products principally used in health care settings and possible associations with infection outbreaks and pseudo-outbreaks (74, 84–89)
In situ studies reporting the impact on bacterial resistance of using specific biocidal products (90–93)
One criticism of in vitro studies is that they might not reflect the way bacteria encounter a biocide/biocidal product in practice (3, 69). For example, the use of stepwise training, i.e., the passaging of bacteria in increasing concentrations of a biocide, does not reflect conditions in situ. These studies have, however, yielded many insights on bacterial resistance mechanisms (46–48, 57, 69–71, 94). Another issue is that the development and nature of resistance to a biocide depend on the bacterial isolates investigated. Ciusa and colleagues (95) reported that bacterial strains from standard culture collection were not necessarily appropriate to study mechanisms of resistance to triclosan because they did not reflect the level and type of mutations observed with clinical isolates when exposed to bisphenol. This study also highlighted that valuable information is being learned through the study of large numbers of isolates (in this study 1,388 Staphylococcus aureus isolates were used) and questioned studies reporting the use of a single isolate (95).
The study of environmental isolates rather than standard culture collection strains yields important and probably more relevant information in terms of expressed mechanisms of resistance. Such investigations reiterate that bacteria can express multiple mechanisms at the same time and that some mechanisms responsible for a stable biocide-resistant phenotype are still unknown. Martin et al. (96) described a vegetative Bacillus subtilis endoscope washer isolate with stable resistance to the in-use concentration of chlorine dioxide and hydrogen peroxide, but also to peracetic acid (96, 97). Although this isolate is a good biofilm producer, the mechanisms responsible for the observed level of resistance to these oxidizing agents have not all been identified (96). Other studies that have isolated bacteria from environments where antimicrobials are heavily used identified a decrease in biocide susceptibility (81, 82, 95, 98) in some but not all isolates when compared to counterpart bacteria from standard culture collection (95, 98).
Studies reporting bacterial growth in biocidal products and subsequent infections or pseudo-infections have been very helpful in identifying the risks associated with some products and practices (89). Reported incidents often result from the inappropriate application of a product or the inappropriate preparation of a product, including the use of contaminated tap water, topping up of stock solutions, use of diluted products or inappropriate dilution, and inappropriate storage conditions. Some microorganisms, notably Pseudomonas spp., Burkholderia spp., and atypical mycobacteria can, however, contaminate the stock solution of a product because of their intrinsic resistance to the product (89). The preconceived idea that bacterial resistance occurs more readily in less reactive biocides such as phenolics (e.g., triclosan) and cationic biocides (e.g., chlorhexidine) rather than reactive ones such as alkylating and oxidizing agents does not hold true. For example, there have been many studies on atypical mycobacterial (Mycobacterium chelonae) resistance to 2% glutaraldehyde, which is used for the high-level disinfection of medical devices (59–64). It was speculated that these bacteria arose from a decrease in the effective concentration of glutaraldehyde (i.e., <2%) (60). Fisher et al. (99) reported the presence of glutaraldehyde-resistant atypical mycobacteria associated with endoscope reprocessing systems. Outbreaks of M. chelonae linked to endoscope reprocessing using glutaraldehyde have been described since 1991 (100). The more recent nosocomial outbreaks of Mycobacterium abscessus subsp. massiliense in Brazil, however, identified an isolate that was resistant to both 2% glutaraldehyde and first-line antimycobacterial antibiotics, highlighting the existence of cross-resistance mechanisms that remain to be identified (74).
Studies of the effect of biocidal product applications on emerging bacterial resistance in the community or health care settings remain scarce. These studies usually highlight the difficulty in data interpretation, notably in relation to the definition of “bacterial resistance.” The few in situ studies nevertheless provide interesting insight on the long-term usage of selected biocidal products. Two studies from Cole and colleagues failed to show any cross-resistance between antibiotics and antibacterial wash products (91, 92). Likewise, Aiello et al. (90) failed to show any statistically significant correlation between the use of triclosan-containing product and reduced susceptibility to antibiotics. A study of benzalkonium chloride-containing product usage in households, however, found a correlation between elevated QAC MIC and bacterial resistance to antibiotics (93).
There should be no doubt that bacteria have a great ability to survive biocide exposure and that the inappropriate use or preparation of biocidal products can result in bacterial resistance. The reporting of cross-resistance between biocides and unrelated chemicals such as chemotherapeutic antibiotics is increasing as scientists focus more on this possibility.
MECHANISMS OF BACTERIAL RESISTANCE
Biocides and biocidal products induce stress on the bacterial cell. In response, a bacterium expresses several mechanisms to prevent the detrimental effect caused by a biocide. These mechanisms aim to decrease the biocide concentration sufficiently that it is no longer damaging to the bacterial cells and include the ability of the bacterium to repair damages. If damage cannot be repaired efficiently or worsens, for example, because of high metabolic activity, the bacterial cell will be committed to a lethal pathway (Table 1). By some accounts that the maintenance of the cytoplasmic pH is key in that pathway (101, 102). Overall, our understanding of the bacterial mechanisms in place to decrease the susceptibility of a bacterium to biocides has improved, but they remain poorly studied. There is no doubt that bacteria have a plethora of mechanisms at their disposal and that often several mechanisms contribute together to the observed resistance phenotype. Our understanding of the effect of biocide interaction with bacteria and especially the stress response effect on gene expression remains poor. Examples given in the literature are often anecdotal. Understanding and measuring the expression of mechanisms following a biocide or biocidal product interaction with a bacterium has become important because it underlies the principle of the observed transient phenotypic changes in bacteria and the cross-resistance mechanisms between antimicrobials.
TABLE 1.
Levels of biocide interactions with a bacterial cell
| Exposure | Interactions | Types of damage | Events |
|---|---|---|---|
| Short exposure | Disruption of the transmembrane PMF leading to an uncoupling of oxidative phosphorylation and inhibition of active transport across the membrane | Reversible | |
| Inhibition of respiration or catabolic/anabolic reactions | |||
| Prolonged exposure | Disruption of metabolic processes | Reversible | |
| Disruption of replication | |||
| Loss of membrane integrity resulting in leakage of essential intracellular constituents (K+, inorganic phosphate, pentoses, nucleotides and nucleosides, proteins) | Imbalance of pHi | Irreversible | |
| Coagulation of intracellular materials | Commitment to cell death (autocidal pathway) | ||
| Lysis | Cell death |
Mechanisms that Decrease the Concentration of Antimicrobials in Bacteria
Bacteria can use several mechanisms to decrease the lethal or inhibitory concentration of a biocide. Biocides have multiple target sites against the bacterial structure and as such they are often regarded as nonspecific. The sum of the damage caused to multiple target sites and the importance of the target sites defines whether the interaction will lead to a lethal or inhibitory effect (Table 1) (3, 101–105). Decreasing a damaging concentration of a biocide/biocidal product will enable the target bacteria to survive. It should be recognized that a low concentration (sub- MIC) of a biocide will affect the bacteria and, notably, trigger mechanisms to further decrease the biocide concentration. It is now well established in in vitro laboratory experiments but also in practice that a low concentration of a biocide will give rise to bacteria that are less susceptible to the biocide, enabling at times the survival of the bacteria in products (60, 89, 96).
Furthermore, biocides are used in complex formulations (i.e., the biocidal product) in practice, yet the effect of a biocidal product on bacterial resistance is not often tested (3, 18, 41, 76, 81). Excipients such as surfactants, chelators, and wetting agents may have a direct effect on the bacterial cell structure and increase the efficacy of a biocide. Arguably, there is sometimes incompatibility between a biocide and an excipient, effectively reducing the bactericidal activity of the product.
Reducing biocide penetration
The effect of bacterial cell structure to prevent or reduce the penetration of antimicrobials has been well established, notably with bacterial endospores (106), Gram-negative bacteria, and mycobacteria (103, 104). The presence of the lipopolysaccharide layer in Gram-negative bacteria has been well documented for its role in decreasing the activity of several membrane active agents such as quaternary ammonium compounds and biguanides. Evidence of the role of lipopolysaccharide in decreasing the activity of a membrane active agent has often been indirect with the use of permeabilizing agents such as chelators and the use of bacterial protoplasts (103, 105, 107, 108). Genetic alterations of the bacterial membrane with, for example, transposon mutagenesis have also provided some important information on biocide/bacterial cell interactions (109). In mycobacteria, in the presence of mycolic acid associated with the arabinogalactan/arabinomannan cell wall, the lipid-rich outer cell wall is responsible for the lack of penetration of many antimicrobials (61, 104, 110–113). Likewise, porins have been shown to play an important role in the activity of glutaraldehyde and ortho-phthalaldehyde in mycobacteria (114). Reducing the expression of porins has been associated with reduced biocide and antibiotic efficacy (115, 116). Changes in bacterial cell membrane and cell wall composition following biocide exposure have been associated with a reduction in biocide activity (94, 115–120). Membrane alterations include membrane protein composition (57, 115, 121, 122), fatty acids (115, 123–127), and phospholipid content (128). A change in membrane potential has also been associated with a decrease in biocide susceptibility in Pseudomonas aeruginosa (129).
Efflux pumps
Efflux pumps, which are widespread in bacteria, contribute to decreasing the concentration of antimicrobials that penetrate the bacterial cells. The effect of active efflux on antimicrobial activity has been particularly well documented in S. aureus (130–139), P. aeruginosa (140–145), Escherichia coli (46, 82, 94, 146–149), Salmonella enterica serovar Typhimurium (150, 151), and Acinetobacter baumannii (116, 152). Five main classes of efflux pumps have been reported (Fig. 1) (153; 160): the drug/metabolite transporter superfamily, the major facilitator superfamily, the ATP-binding cassette family, the resistance-nodulation-division family, and the multidrug and toxic compound extrusion family.
FIGURE 1.
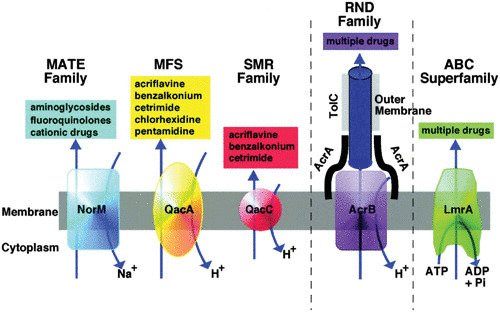
Diagrammatic comparison of the five families of efflux pumps (reproduced from reference 153). MATE, multidrug and toxic compound extrusion; MFS, major facilitator superfamily; SMR, •••; RND, resistance-nodulation-division; ABC, ATP-binding cassette.
The ability of efflux pumps alone to confer resistance to biocides/biocidal products is questionable, and it is likely that efflux pumps are part of several mechanisms used by a bacterium to survive biocide/biocidal product exposure (3, 83, 155). Some studies investigating triclosan claimed, however, that efflux was responsible for high-level resistance to the bisphenol (142, 144). Studies of bacterial isolates from environments where antimicrobials are heavily used, notably biguanides and QAC, have identified a high prevalence of efflux genes (e.g., qacA/B, norA, nor B, smr) in isolates that showed a decreased susceptibility to biocides (77–79, 82, 135).
Efflux can be induced by some antimicrobials (153, 156, 161). The expression of an efflux pump can increase following antimicrobial exposure, not necessarily by inducing the efflux pumps but by affecting global gene regulators, notably marA and soxS (46, 162). The effect of triclosan on bacteria has been particularly well studied with regard to efflux (46, 49, 140–142, 163, 164). In S. enterica serovar Typhimurium, overexpression of efflux results in decreased antimicrobial susceptibility (162–165). Overexpression of efflux pumps resulting in decreased biocide efficacy has also been described in Stenotrophomonas maltophilia with the overexpression of SmedEF (166); in E. coli with the overexpression of acrAB, marA, or soxS (46, 49, 162); and in Campylobacter jejuni overexpressing CmeB (167). The extent of efflux pumps and their role in bacteria are continuously evolving in the literature. Triggering overexpression of efflux in bacteria following biocide exposure is a concern that is debated later in this article.
Enzymatic degradation
Some bacteria can produce enzymes that degrade biocides. The presence of catalase and superoxide dismutase, for example, has been shown to decrease bacterial susceptibility to oxidizing agents (66, 168). The production of enzymes alone conferring resistance to a biocide is, however, doubtful. This would suggest that enzymatic activity is high and that enzymes are not themselves affected by the biocide. It is more likely that the production of detoxifying enzymes contributes to the battery of mechanisms available to the bacteria to survive biocide injuries (96).
Other examples of enzymatic activity conferring decreased susceptibility to a biocide include the parabens (169, 170), aldehydes (171), and metallic ions. In the latter case the ions are reduced to the inactive metal (34).
Physiological and Metabolic Changes
Bacterial metabolism can be associated with antimicrobial efficacy in that bacteria with a high metabolism are more susceptible to antimicrobials than those with no metabolic activity (172). Exposure of a bacteria to a physical or chemical process, such as a biocide/biocidal product, results in a mixed population of dead, injured, and uninjured bacteria. In the food industry, the recovery of injured bacteria is considered essential (173). This is not so when the efficacy of a biocide treatment is measured. Standard efficacy tests do not consider the effect of the recovery media and incubation conditions post-biocide treatment. The impact of resuscitated injured bacteria following treatment has been exemplified by the dual use of traditional plate counting on a rich nonselective recovery media such as tryptone soy agar and the use of the Bioscreen microbial growth analyzer, which measures bacterial growth in liquid (174). The ability of a bacterium to repair injuries is likely to play an important role when resistance is considered. As shown in Table 1, initial damage caused by a biocide is reversible. In practice, where incubation conditions posttreatment favor recovery from injury, repairs can be visualized with an extended lag phase (173). Biocide exposure has, however, been linked to a decreased growth rate and extended lag phase in bacteria (172, 175–177) because of a direct action of the biocide on the bacterial cells, although in many studies the ability of bacteria to repair injuries was not considered. Change in metabolic pathways has been particularly well exemplified with S. enterica exposure to triclosan. The bisphenol at a low concentration has been shown to target specifically the enoyl acyl carrier reductase in bacteria, which affects fatty acid lipid synthesis in the target bacteria (47, 177–179). Webber et al. (180) showed that S. enterica could alter its metabolic pathway to produce pyruvate and fatty acids following triclosan exposure. This change was part of a “triclosan resistance network” involving the expression of distinct mechanisms (180). Curiao and colleagues reported similar findings, evoking multiple pathways in the adaptation of S. enterica to triclosan and other biocides such as chlorhexidine and benzalkonium chloride (181).
Codling and colleagues (109) showed that in Serratia marcescens, the disruption of biosynthetic and metabolic pathways of the bacterium increased bacterial susceptibility to a QAC. A change in metabolic processes following exposure to biocides has also been observed in other bacteria, including S. aureus (182) and P. aeruginosa (183). The full impact of a change in metabolic pathways on decreasing biocide/biocidal product efficacy has not been assessed, nor has the reproducibility of such a change when exposed to specific antimicrobials. At present, such observations have been bacteria/biocide specific.
Mutations
Mutations in bacteria are by nature random but can be driven by the continuous presence of a selective pressure, notably the presence of antimicrobials. Although it is widely recognized that the presence of chemotherapeutic antibiotics will drive target site mutations, there are far fewer examples with biocides. Mutations resulting from a biocide exposure have been mainly described with triclosan in several bacteria (43, 178, 180, 184–190). Mutations are linked to the use of triclosan at a low concentration and concern the enoyl-acyl reductase carrier protein (47, 179, 186, 191–193). In Salmonella biocide exposure resulted in mutations included de-repression of multidrug efflux pump AcrAB-TolC, and rpoA, which controls the RNA polymerase α-subunit (190). Interestingly, the investigation of 1,388 S. aureus clinical isolates’ response to triclosan exposure identified several mutations that were similar among the isolates (Fig. 2) but not comparable to those observed with the standard culture collection strain (95). Recent observations questioned the choice of a culture collection strain to study biocide resistance. Standard strains have been widely used in biocide resistance studies with the comparability of results between studies in mind. From Ciusa et al. (95), it appears that standard strains and clinical isolates do not behave the same way. Stepwise training protocols that rely on passaging bacteria in increasing concentrations of biocides have yielded bacteria with decreased susceptibility or resistance to a given biocide but may be criticized for not reflecting real-world conditions (3). Many in vitro studies have investigated genetic changes in standard culture collection strains following biocide exposure. The work of Ciusa et al. (95) addresses the appropriateness of this approach and would favor the use of environmental isolates that have been exposed to a biocide/biocidal product.
FIGURE 2.
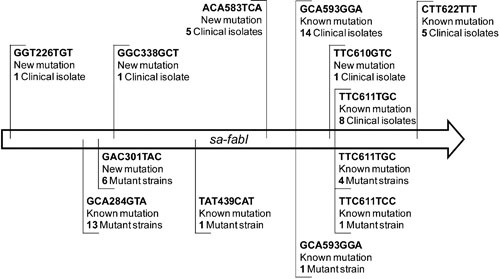
Schematic map of mutations in the Staphylococcus aureus fabI (sa-fabI) and Staphylococcus haemolyticus fabI (sh-fabI) genes. Mutations in sa-fabI are reported on a schematic map. Mutations detected in clinical isolates are mapped above the sequence, while mutations selected in vitro are shown below the sequence. (Reproduced from reference 95.)
INDUCTION OF GENE EXPRESSION CONFERRING BACTERIAL RESISTANCE
Biocide exposure even at a low concentration produces a stress on the target bacteria, even if the bacteria are intrinsically resistant to the biocide. The bacterial response to the stress will lead to a change in gene expression (24, 109, 116, 155, 162–167, 181, 194–196), particularly that of regulatory genes (46, 49, 68, 149, 162, 180, 197). The concentration of a biocide that is available to interact with the target bacteria is thus paramount (2, 3, 105, 198, 199), since a low, nonlethal concentration will not kill the bacterium but will undoubtedly produce a stress response. An indication of stress response is given by investigating the bacterial growth curve in the presence of a biocide at different concentrations. Increased lag phase or decreases in bacterial doubling time are indicators of a bacteria/biocide interaction and may reflect the induction/expression of mechanisms enabling the bacteria to decrease the toxicity of the biocide (28, 72, 172) and, as mentioned, allow earlier repair of injuries. Some bacterial mechanisms that play a role in decreasing the susceptibility to biocides are controlled by global regulators such as soxS and marA (46, 49, 146, 162). Antibiotic resistance mechanisms are also controlled by the same regulators (168, 200), which leads to the concern that biocide exposure can trigger antibiotic resistance. The induction of gene expression of global regulators leading to the expression of several mechanisms in bacterial resistance might not, however, be particularly problematic for the use of biocidal products since such expression might be transient. Some studies have shown that a decrease in bacterial susceptibility to biocides and sometimes to antibiotics was transient and only observed in the presence of the biocide (24, 25, 155).
As mentioned earlier, the efficient repair of sublethal injuries may play an important role in bacterial survival of biocide exposure. However, bacteria’s ability to repair damage following biocide exposure has received little attention (195, 201, 202). In E. coli polyhexamethylene biguanide alters the expression of several genes, notably rhs, involved in repairing nucleic acid (202). The involvement of effective DNA repair mechanisms has been proposed to explain the high-level resistance of an environmental isolate of B. subtilis to several oxidizing agents (96). Efficient DNA repair mechanisms enable Deinococcus radiodurans to survive ionizing and UV radiation and exposure to chemicals that damage nucleic acid (203). In Lactobacillus pentosus, strains that have adapted to sublethal concentrations of antimicrobials overexpressed ribosomal proteins and glutamyl tRNA synthetase, which was interpreted as a response to damaged proteins directly caused by the antimicrobial exposure (195).
CROSS-RESISTANCE
Exposure to a biocide/biocidal product can lead to a stress response involving the expression of global gene regulators and ultimately the expression of nonspecific mechanisms enabling bacterial survival (116, 155, 156, 162, 181, 190, 200, 204–211). The link between biocide usage and antibiotic resistance has led to many discussions with conflicting evidence; some studies support a link, while others fail to identify any cross-resistance (1, 24, 25, 48, 73, 78, 79, 81, 82, 90–93, 98, 196, 212–220). Where cross-resistance between biocide exposure and antibiotic resistance was identified, suggested common resistance mechanisms included overexpression of efflux (18, 82, 153, 156, 161), changes in bacterial cell wall permeability (115, 117), and changes in bacterial metabolism (180). Differences in protocols to (i) grow test bacteria, (ii) expose test bacteria to the biocide/biocidal product, and (iii) measure resistance to biocides and antibiotics contribute to differences in reported observations of the biocide’s effect on antibiotic resistance (18). Although the evidence is mainly in vitro based, the few in situ studies conducted also reported conflicting information about the association between the usage of biocidal products at home and an increase in antibiotic resistance among environmental isolates (89–93). It is worth noting, however, the study from Duarte and colleagues (74) reporting a postsurgical outbreak of a M. abscessus subsp. massiliense-resistant clone resistant to 2% glutaraldehyde and resistant to frontline antimycobacterial antibiotics.
There should be no doubt that bacteria have the capacity to express mechanisms that will lead to decreased susceptibility to both biocides/biocidal products and chemotherapeutic antibiotics. The question remains as to how commonly cross- resistance occurs in practice and what triggers emerging resistance in the first place. For example, efflux can easily be triggered in bacteria, not only by biocides but by a wide range of stimuli, such as spices and essential oil. (221).
MEASURING BACTERIAL RESISTANCE
One of the most important aspects of biocide/biocidal product resistance is how to measure bacterial resistance and cross-resistance. This has become even more pressing with the publication in Europe of the Biocidal Product Regulation (20), which asks manufacturers to demonstrate that their biocidal product will not cause emerging bacterial resistance. Likewise, in North America the FDA (39) issued a final rule on the safety and effectiveness of antibacterial soaps, effectively banning the use of certain biocides for that application. Rules concerning benzalkonium chloride, benzethonium chloride, and chloroxylenol, biocides that are commonly used in several products, deferred. One major issue for manufacturers is that neither the Biocidal Product Regulation nor the FDA indicate what appropriate tests should be conducted to demonstrate the safety of biocidal products where bacterial resistance is concerned.
MIC determination has often been used as a marker for resistance (18, 28, 69–71, 183, 219), although the validity of MIC to measure bacterial resistance has been questioned (1, 3, 18) mainly since in practice, biocides are often used at concentrations exceeding the MIC (120) and biocides are used as part of a formulation whose ingredients will impact on product efficacy (3, 18, 76). MIC could, however, be used as a trend indicator (3, 18, 25, 28, 76, 198, 199, 222, 223). It is thus unfortunate that some studies measure an increase in bacterial resistance in terms of MIC (28, 224). Other studies have used a prevalue biocide concentration above which the environmental isolates were considered to be resistant. For example, Lavilla Lerma and colleagues used a threshold biocide concentration of 0.025 μg/ml at which any bacterial growth was considered bacterial resistance to the biocide (81). Furthermore, resistance has sometimes been defined as a small increase (e.g., 2-fold) in MIC. This definition remains questionable, especially when using a standard protocol such as a broth microdilution method (225) because a 2-fold change might only reflect a 1-dilution difference. The determination of changes in minimum biocidal concentration might be more appropriate, because this indicates a change in the lethal effect of the biocide (18, 223). Some studies have looked at a change in inactivation kinetics. Such protocols, although very useful because they determine the ability of a biocide/biocidal product to kill target bacteria over time, are very cumbersome and time-consuming and would not be able to be used routinely (3, 18).
The determination of a change in the susceptibility profile to chemotherapeutic antibiosis is somewhat easier to perform because the protocols used can follow well-established standards that provide clear guidance but also breakpoints for selected bacteria/antibiotics (226, 227). It is, however, clear that the clinical significance should be reported rather than reporting a mere change in the antibiotic zone of inhibition.
Measuring a change in the susceptibility profile to determine a prediction of the risk associated with biocidal product usage is acceptable if the exposure of the target bacterium to the biocidal product is realistic, i.e., if it reflects in situ exposure of bacteria with the biocidal products, encompassing dilution of the product upon usage if necessary, extended contact time for residual activity, etc. (18, 25). The test bacterial inoculum preparation needs to be strictly controlled to ensure reproducibility of the assay. When a significant change (here significant means a ≥10-fold change) in susceptibility profile is recorded (25, 222), the nature of this change, whether transient or permanent, needs to be established (18, 25, 223).
A protocol to predict the change in susceptibility profile of target bacteria following exposure to a biocide/biocidal product has been proposed (18). The use of such a protocol established the effect of various biocidal products on S. enterica (223), E. coli, and S. aureus (25). In these studies, triclosan was used as a positive control (25, 223). Triclosan is the most studied biocide in terms of interaction with bacteria. Studies have repeatedly show amended bacterial susceptibility profile to triclosan and antibiotics following exposure to the bisphenol. (23, 25, 228).
CONCLUSIONS
Biocidal products are useful compounds to control microbial contamination and kill pathogens. A biocidal concentration that will not kill the target bacteria will, regardless of the method of application, cause a stress response, which will lead to the expression of mechanisms that enable bacterial survival (3, 18, 28, 173). The concentration of biocide available to interact with the target bacteria is thus paramount. In the veterinary field, the presence of organic matter at the point of the biocidal product application, contributes to reduce the efficacy of the product, and therefore, cleaning the animate or inanimate surface prior to use of the biocidal product should be indicated but might not be practical.
Biocidal products are heavily used for veterinary applications, notably in animal husbandry, disinfection of udders in dairy animals, and in fish farming (1). Despite an increasing use of biocidal products, information related to the occurrence of bacterial resistance in these environments remains scarce (55, 73, 150, 229, 230). Nevertheless, with the growing knowledge and evidence of bacterial resistance in environments where biocides/biocidal products are heavily used, environmental surveillance has been timidly proposed to study the potential spread and occurrence of resistant bacteria (27, 73, 77).
With an increase in biocidal product usage, emerging bacterial resistance is possible, but to date, the risk associated with biocidal product usage has not been measured, mainly because of the lack of standard protocols. The only protocol to date has not yet been widely used against a small number of bacteria (18, 25, 223). The reproducibility of the data obtained may also depend on the bacterial species investigated (24). Such a predictive protocol also relies on using appropriate test parameters that reflect the biocidal product usage in practice (25, 223). Overall, investigating the biocidal effect on bacterial resistance should be welcome because it provides a better understanding of the biocide-bacteria interactions and should contribute to the development of more performant and safer biocidal products. This is particularly pertinent with the increased usage of biocidal products and the usage conditions in animal husbandry.
REFERENCES
- 1.Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). 2009. The antibiotic resistance effect of biocides. http://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf. Accessed January 2017.
- 2.Maillard J-Y. 2005. Usage of antimicrobial biocides and products in the healthcare environment: efficacy, policies, management and perceived problems. Ther Clin Risk Manag 1:340–370. [PMC free article] [PubMed] [Google Scholar]
- 3.Maillard J-Y, Denyer SP. 2009. Emerging bacterial resistance following biocide exposure: should we be concerned? Chim Oggi 27:26–28. [Google Scholar]
- 4.O’Neill J. 2016. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. HM Government, London, United Kingdom. [Google Scholar]
- 5.Otter JA, Yezli S, French GL. 2011. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol 32:687–699 10.1086/660363. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, Dougan G. 2010. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol 76:6895–6900 10.1128/AEM.00718-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476 10.1002/jmv.21237. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Boyce JM, Potter-Bynoe G, Chenevert C, King T. 1997. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect Control Hosp Epidemiol 18:622–627 10.1086/502213. [DOI] [PubMed] [Google Scholar]
- 9.Bhalla A, Pultz NJ, Gries DM, Ray AJ, Eckstein EC, Aron DC, Donskey CJ. 2004. Acquisition of nosocomial pathogens on hands after contact with environmental surfaces near hospitalized patients. Infect Control Hosp Epidemiol 25:164–167 10.1086/502369. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Vonberg RP, Kuijper EJ, Wilcox MH, Barbut F, Tüll P, Gastmeier P, van den Broek PJ, Colville A, Coignard B, Daha T, Debast S, Duerden BI, van den Hof S, van der Kooi T, Maarleveld HJ, Nagy E, Notermans DW, O’Driscoll J, Patel B, Stone S, Wiuff C, European C difficile-Infection Control Group, European Centre for Disease Prevention and Control (ECDC). 2008. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect 14(Suppl 5):2–20 10.1111/j.1469-0691.2008.01992.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130–138 10.1186/1471-2334-6-130. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fawley WN, Wilcox MH. 2001. Molecular epidemiology of endemic Clostridium difficile infection. Epidemiol Infect 126:343–350 10.1017/S095026880100557X. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talon D. 1999. The role of the hospital environment in the epidemiology of multi-resistant bacteria. J Hosp Infect 43:13–17 10.1053/jhin.1999.0613. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Hota B. 2004. Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin Infect Dis 39:1182–1189 10.1086/424667. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheeseman KE, Denyer SP, Hosein IK, Williams GJ, Maillard J-Y. 2009. Evaluation of the bactericidal efficacy of three different alcohol hand rubs against 57 clinical isolates of S. aureus. J Hosp Infect 72:319–325 10.1016/j.jhin.2009.04.018. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Williams GJ, Denyer SP, Hosein IK, Hill DW, Maillard J-Y. 2009. Limitations of the efficacy of surface disinfection in the healthcare setting. Infect Control Hosp Epidemiol 30:570–573 10.1086/597382. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Siani H, Cooper C, Maillard J-Y. 2011. Efficacy of “sporicidal” wipes against Clostridium difficile. Am J Infect Control 39:212–218 10.1016/j.ajic.2011.01.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Maillard J-Y, Bloomfield S, Coelho JR, Collier P, Cookson B, Fanning S, Hill A, Hartemann P, McBain AJ, Oggioni M, Sattar S, Schweizer HP, Threlfall J. 2013. Does microbicide use in consumer products promote antimicrobial resistance? A critical review and recommendations for a cohesive approach to risk assessment. Microb Drug Resist 19:344–354 10.1089/mdr.2013.0039. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Department for Environment, Food & Rural Affairs. 2012. Controlling disease in farm animals. https://www.gov.uk/guidance/controlling-disease-in-farm-animals. Accessed January 2017.
- 20.Pedrouzo M, Borrull F, Marcé RM, Pocurull E. 2009. Ultra-high-performance liquid chromatography-tandem mass spectrometry for determining the presence of eleven personal care products in surface and wastewaters. J Chromatogr A 1216:6994–7000 10.1016/j.chroma.2009.08.039. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Kumar KS, Priya SM, Peck AM, Sajwan KS. 2010. Mass loadings of triclosan and triclocarbon from four wastewater treatment plants to three rivers and landfill in Savannah, Georgia, USA. Arch Environ Contam Toxicol 58:275–285 10.1007/s00244-009-9383-y. [DOI] [PubMed] [Google Scholar]
- 22.Wilson B, Chen RF, Cantwell M, Gontz A, Zhu J, Olsen CR. 2009. The partitioning of triclosan between aqueous and particulate bound phases in the Hudson River Estuary. Mar Pollut Bull 59:207–212 10.1016/j.marpolbul.2009.03.026. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Scientific Committee on Consumer Safety. 2010. Opinion on triclosan antimicrobial resistance. http://ec.europa.eu/health//sites/health/files/scientific_committees/consumer_safety/docs/sccs_o_054.pdf. Accessed January 2017.
- 24.Knapp L, Rushton L, Stapleton H, Sass A, Stewart S, Amezquita A, McClure P, Mahenthiralingam E, Maillard J-Y. 2013. The effect of cationic microbicide exposure against Burkholderia cepacia complex (Bcc); the use of Burkholderia lata strain 383 as a model bacterium. J Appl Microbiol 115:1117–1126 10.1111/jam.12320. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Wesgate R, Grasha P, Maillard J-Y. 2016. Use of a predictive protocol to measure the antimicrobial resistance risks associated with biocidal product usage. Am J Infect Control 44:458–464 10.1016/j.ajic.2015.11.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Oggioni MR, Furi L, Coelho JR, Maillard JY, Martínez JL. 2013. Recent advances in the potential interconnection between antimicrobial resistance to biocides and antibiotics. Expert Rev Anti Infect Ther 11:363–366 10.1586/eri.13.16. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Cookson B. 2005. Clinical significance of emergence of bacterial antimicrobial resistance in the hospital environment. J Appl Microbiol 99:989–996 10.1111/j.1365-2672.2005.02693.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Maillard J-Y. 2007. Bacterial resistance to biocides in the healthcare environment: should it be of genuine concern? J Hosp Infect 65(Suppl 2):60–72 10.1016/S0195-6701(07)60018-8. [DOI] [PubMed] [Google Scholar]
- 29.Siani H, Maillard J-Y. 2015. Best practice in healthcare environment decontamination. Eur J Clin Microbiol Infect Dis 34:1–11 10.1007/s10096-014-2205-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Chapman JS. 1998. Characterizing bacterial resistance to preservatives and disinfectants. Int Biodeter Biodeg 41:241–245 10.1016/S0964-8305(98)00025-0. [DOI] [Google Scholar]
- 31.Chapman JS, Diehl MA, Fearnside KB. 1998. Preservative tolerance and resistance. Int J Cosmet Sci 20:31–39 10.1046/j.1467-2494.1998.171733.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Hammond SA, Morgan JR, Russell AD. 1987. Comparative susceptibility of hospital isolates of Gram-negative bacteria to antiseptics and disinfectants. J Hosp Infect 9:255–264 10.1016/0195-6701(87)90122-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Russell AD. 2003. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect Dis 3:794–803 10.1016/S1473-3099(03)00833-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Cloete TE. 2003. Resistance mechanisms of bacteria to antimicrobial compounds. Int Biodeter Biodegrad 51:277–282 10.1016/S0964-8305(03)00042-8. [DOI] [Google Scholar]
- 35.Dettenkofer M, Wenzler S, Amthor S, Antes G, Motschall E, Daschner FD. 2004. Does disinfection of environmental surfaces influence nosocomial infection rates? A systematic review. Am J Infect Control 32:84–89 10.1016/j.ajic.2003.07.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Poole K. 2002. Mechanisms of bacterial biocide and antibiotic resistance. J Appl Microbiol 92(Suppl):55S–64S 10.1046/j.1365-2672.92.5s1.8.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). 2010. Research strategy to address the knowledge gaps on the antimicrobial resistance effects of biocides. http://ec.europa.eu/health/scientific_committees/emerging/docs/scenihr_o_028.pdf. Accessed January 2017.
- 38.Scientific Committee on Consumer Safety (SCCS). 2010. Opinion on triclosan antimicrobial resistance. http://ec.europa.eu/health//sites/health/files/scientific_committees/consumer_safety/docs/sccs_o_054.pdf. Accessed January 2017.
- 39.U.S. Food and Drug Administration. 2016. Safety and effectiveness of consumer antiseptics; topical antimicrobial drug products for over-the-counter human use. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm517478.htm. Accessed January 2017. [PubMed]
- 40.Lavilla Lerma L, Benomar N, Casado Muñoz MC, Gálvez A, Abriouel H. 2015. Correlation between antibiotic and biocide resistance in mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces throughout meat chain production. Food Microbiol 51:33–44 10.1016/j.fm.2015.04.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Cowley NL, Forbes S, Amézquita A, McClure P, Humphreys GJ, McBain AJ. 2015. Effects of formulation on microbicide potency and mitigation of the development of bacterial insusceptibility. Appl Environ Microbiol 81:7330–7338 10.1128/AEM.01985-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasatsu M, Shimizu K, Noguchi N, Kono M. 1993. Triclosan-resistant Staphylococcus aureus. Lancet 341:756 10.1016/0140-6736(93)90526-M. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem 273:30316–30320 10.1074/jbc.273.46.30316. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Bamber AI, Neal TJ. 1999. An assessment of triclosan susceptibility in methicillin-resistant and methicillin-sensitive Staphylococcus aureus. J Hosp Infect 41:107–109 10.1016/S0195-6701(99)90047-6. [DOI] [PubMed] [Google Scholar]
- 45.Randall LP, Cooles SW, Piddock LJ, Woodward MJ. 2004. Effect of triclosan or a phenolic farm disinfectant on the selection of antibiotic-resistant Salmonella enterica. J Antimicrob Chemother 54:621–627 10.1093/jac/dkh376. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.McMurry LM, Oethinger M, Levy SB. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett 166:305–309 10.1111/j.1574-6968.1998.tb13905.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.McMurry LM, McDermott PF, Levy SB. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother 43:711–713. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cottell A, Denyer SP, Hanlon GW, Ochs D, Maillard JY. 2009. Triclosan-tolerant bacteria: changes in susceptibility to antibiotics. J Hosp Infect 72:71–76 10.1016/j.jhin.2009.01.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Curiao T, Marchi E, Viti C, Oggioni MR, Baquero F, Martinez JL, Coque TM. 2015. Polymorphic variation in susceptibility and metabolism of triclosan-resistant mutants of Escherichia coli and Klebsiella pneumoniae clinical strains obtained after exposure to biocides and antibiotics. Antimicrob Agents Chemother 59:3413–3423 10.1128/AAC.00187-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adair FW, Geftic SG, Gelzer J. 1971. Resistance of Pseudomonas to quaternary ammonium compounds. II. Cross-resistance characteristics of a mutant of Pseudomonas aeruginosa. Appl Microbiol 21:1058–1063. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell AD. 2002. Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. J Appl Microbiol 92(Suppl):121S–135S 10.1046/j.1365-2672.92.5s1.12.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Chapman JS. 2003. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeter Biodegrad 51:271–276 10.1016/S0964-8305(03)00044-1. [DOI] [Google Scholar]
- 53.Stickler DJ. 1974. Chlorhexidine resistance in Proteus mirabilis. J Clin Pathol 27:284–287 10.1136/jcp.27.4.284. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillespie MT, May JW, Skurray RA. 1986. Plasmid-encoded resistance to acriflavine and quaternary ammonium compounds in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 34:47–51 10.1111/j.1574-6968.1986.tb01346.x. [DOI] [Google Scholar]
- 55.Randall LP, Cooles SW, Sayers AR, Woodward MJ. 2001. Association between cyclohexane resistance in Salmonella of different serovars and increased resistance to multiple antibiotics, disinfectants and dyes. J Med Microbiol 50:919–924 10.1099/0022-1317-50-10-919. [DOI] [PubMed] [Google Scholar]
- 56.Romão CMCPA, Faria YN, Pereira LR, Asensi MD. 2005. Susceptibility of clinical isolates of multiresistant Pseudomonas aeruginosa to a hospital disinfectant and molecular typing. Mem Inst Oswaldo Cruz 100:541–548 10.1590/S0074-02762005000500015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Winder CL, Al-Adham IS, Abdel Malek SM, Buultjens TE, Horrocks AJ, Collier PJ. 2000. Outer membrane protein shifts in biocide-resistant Pseudomonas aeruginosa PAO1. J Appl Microbiol 89:289–295 10.1046/j.1365-2672.2000.01119.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.O’Rourke E, Runyan D, O’Leary J, Stern J. 2003. Contaminated iodophor in the operating room. Am J Infect Control 31:255–256 10.1067/mic.2003.13. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Griffiths PA, Babb JR, Bradley CR, Fraise AP. 1997. Glutaraldehyde-resistant Mycobacterium chelonae from endoscope washer disinfectors. J Appl Microbiol 82:519–526 10.1046/j.1365-2672.1997.00171.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.van Klingeren B, Pullen W. 1993. Glutaraldehyde resistant mycobacteria from endoscope washers. J Hosp Infect 25:147–149 10.1016/0195-6701(93)90107-B. [DOI] [PubMed] [Google Scholar]
- 61.Manzoor SE, Lambert PA, Griffiths PA, Gill MJ, Fraise AP. 1999. Reduced glutaraldehyde susceptibility in Mycobacterium chelonae associated with altered cell wall polysaccharides. J Antimicrob Chemother 43:759–765 10.1093/jac/43.6.759. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Fraud S, Maillard J-Y, Russell AD. 2001. Comparison of the mycobactericidal activity of ortho- phthalaldehyde, glutaraldehyde and other dialdehydes by a quantitative suspension test. J Hosp Infect 48:214–221 10.1053/jhin.2001.1009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Walsh SE, Maillard J-Y, Russell AD, Hann AC. 2001. Possible mechanisms for the relative efficacies of ortho-phthalaldehyde and glutaraldehyde against glutaraldehyde-resistant Mycobacterium chelonae. J Appl Microbiol 91:80–92 10.1046/j.1365-2672.2001.01341.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Nomura K, Ogawa M, Miyamoto H, Muratani T, Taniguchi H. 2004. Antibiotic susceptibility of glutaraldehyde-tolerant Mycobacterium chelonae from bronchoscope washing machines. Am J Infect Control 32:185–188 10.1016/j.ajic.2003.07.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Martin DJH, Denyer SP, McDonnell G, Maillard J-Y. 2008. Resistance and cross-resistance to oxidising agents of bacterial isolates from endoscope washer disinfectors. J Hosp Infect 69:377–383 10.1016/j.jhin.2008.04.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Greenberg JT, Demple B. 1989. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. J Bacteriol 171:3933–3939 10.1128/jb.171.7.3933-3939.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA 87:6181–6185 10.1073/pnas.87.16.6181. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dukan S, Touati D. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol 178:6145–6150 10.1128/jb.178.21.6145-6150.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh SE, Maillard J-Y, Russell AD, Catrenich CE, Charbonneau DL, Bartolo RG. 2003. Development of bacterial resistance to several biocides and effects on antibiotic susceptibility. J Hosp Infect 55:98–107 10.1016/S0195-6701(03)00240-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Tattawasart U, Maillard J-Y, Furr JR, Russell AD. 1999. Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J Hosp Infect 42:219–229 10.1053/jhin.1999.0591. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Thomas L, Maillard J-Y, Lambert RJW, Russell AD. 2000. Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a “residual” concentration. J Hosp Infect 46:297–303 10.1053/jhin.2000.0851. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Thomas L, Russell AD, Maillard J-Y. 2005. Antimicrobial activity of chlorhexidine diacetate and benzalkonium chloride against Pseudomonas aeruginosa and its response to biocide residues. J Appl Microbiol 98:533–543 10.1111/j.1365-2672.2004.02402.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Molina-González D, Alonso-Calleja C, Alonso-Hernando A, Capita R. 2014. Effect of sub-lethal concentrations of biocides on the susceptibility to antibiotics of multi-drug resistant Salmonella enterica strains. Food Control 40:329–334 10.1016/j.foodcont.2013.11.046. [DOI] [Google Scholar]
- 74.Duarte RS, Lourenço MCS, Fonseca LS, Leão SC, Amorim EL, Rocha IL, Coelho FS, Viana-Niero C, Gomes KM, da Silva MG, Lorena NS, Pitombo MB, Ferreira RM, Garcia MH, de Oliveira GP, Lupi O, Vilaça BR, Serradas LR, Chebabo A, Marques EA, Teixeira LM, Dalcolmo M, Senna SG, Sampaio JL. 2009. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol 47:2149–2155 10.1128/JCM.00027-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wisplinghoff H, Schmitt R, Wöhrmann A, Stefanik D, Seifert H. 2007. Resistance to disinfectants in epidemiologically defined clinical isolates of Acinetobacter baumannii. J Hosp Infect 66:174–181 10.1016/j.jhin.2007.02.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Bock LJ, Wand ME, Sutton JM. 2016. Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect 93:42–48 10.1016/j.jhin.2015.12.019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Liu Q, Zhao H, Han L, Shu W, Wu Q, Ni Y. 2015. Frequency of biocide-resistant genes and susceptibility to chlorhexidine in high-level mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MuH MRSA). Diagn Microbiol Infect Dis 82:278–283 10.1016/j.diagmicrobio.2015.03.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Hijazi K, Mukhopadhya I, Abbott F, Milne K, Al-Jabri ZJ, Oggioni MR, Gould IM. 2016. Susceptibility to chlorhexidine amongst multidrug-resistant clinical isolates of Staphylococcus epidermidis from bloodstream infections. Int J Antimicrob Agents 48:86–90 10.1016/j.ijantimicag.2016.04.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Conceição T, Coelho C, de Lencastre H, Aires-de-Sousa M. 2015. High prevalence of biocide resistance determinants in Staphylococcus aureus isolates from three African countries. Antimicrob Agents Chemother 60:678–681 10.1128/AAC.02140-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lear JC, Maillard J-Y, Dettmar PW, Goddard PA, Russell AD. 2002. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources. J Ind Microbiol Biotechnol 29:238–242 10.1038/sj.jim.7000320. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Lavilla Lerma L, Benomar N, Gálvez A, Abriouel H. 2013. Prevalence of bacteria resistant to antibiotics and/or biocides on meat processing plant surfaces throughout meat chain production. Int J Food Microbiol 161:97–106 10.1016/j.ijfoodmicro.2012.11.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Grande Burgos MJ, Fernández Márquez ML, Pérez Pulido R, Gálvez A, Lucas López R. 2016. Virulence factors and antimicrobial resistance in Escherichia coli strains isolated from hen egg shells. Int J Food Microbiol 238:89–95 10.1016/j.ijfoodmicro.2016.08.037. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Martínez-Suárez JV, Ortiz S, López-Alonso V. 2016. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front Microbiol 7:638 10.3389/fmicb.2016.00638. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanford JP. 1970. Disinfectants that don’t. Ann Intern Med 72:282–283 10.7326/0003-4819-72-2-282. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Prince J, Ayliffe GAJ. 1972. In-use testing of disinfectants in hospitals. J Clin Pathol 25:586–589 10.1136/jcp.25.7.586. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bridges K, Lowbury EJL. 1977. Drug resistance in relation to use of silver sulphadiazine cream in a burns unit. J Clin Pathol 30:160–164 10.1136/jcp.30.2.160. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klasen HJ. 2000. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 26:131–138 10.1016/S0305-4179(99)00116-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Reiss I, Borkhardt A, Füssle R, Sziegoleit A, Gortner L. 2000. Disinfectant contaminated with Klebsiella oxytoca as a source of sepsis in babies. Lancet 356:310 10.1016/S0140-6736(00)02509-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 89.Weber DJ, Rutala WA, Sickbert-Bennett EE. 2007. Outbreaks associated with contaminated antiseptics and disinfectants. Antimicrob Agents Chemother 51:4217–4224 10.1128/AAC.00138-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aiello AE, Marshall B, Levy SB, Della-Latta P, Larson E. 2004. Relationship between triclosan and susceptibilities of bacteria isolated from hands in the community. Antimicrob Agents Chemother 48:2973–2979 10.1128/AAC.48.8.2973-2979.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cole EC, Addison RM, Rubino JR, Leese KE, Dulaney PD, Newell MS, Wilkins J, Gaber DJ, Wineinger T, Criger DA. 2003. Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J Appl Microbiol 95:664–676 10.1046/j.1365-2672.2003.02022.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Cole EC, Addison RM, Dulaney PD, Leese KE, Madanat HM, Guffey AM. 2011. Investigation of antibiotic and antibacterial susceptibility and resistance in Staphylococcus form the skin of users and non-users of antibacterial wash products in home environments. Int J Microbiol Res 3:90–96 10.9735/0975-5276.3.2.90-96. [DOI] [Google Scholar]
- 93.Carson RT, Larson E, Levy SB, Marshall BM, Aiello AE. 2008. Use of antibacterial consumer products containing quaternary ammonium compounds and drug resistance in the community. J Antimicrob Chemother 62:1160–1162 10.1093/jac/dkn332. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alonso-Calleja C, Guerrero-Ramos E, Alonso-Hernando A, Capita R. 2015. Adaptation and cross-adaptation of Escherichia coli ATCC 12806 to several food-grade biocides. Food Control 56:86–94 10.1016/j.foodcont.2015.03.012. [DOI] [Google Scholar]
- 95.Ciusa ML, Furi L, Knight D, Decorosi F, Fondi M, Raggi C, Coelho JR, Aragones L, Moce L, Visa P, Freitas AT, Baldassarri L, Fani R, Viti C, Orefici G, Martinez JL, Morrissey I, Oggioni MR, BIOHYPO Consortium. 2012. A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int J Antimicrob Agents 40:210–220 10.1016/j.ijantimicag.2012.04.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Martin DJH, Wesgate RL, Denyer SP, McDonnell G, Maillard J-Y. 2015. Bacillus subtilis vegetative isolate surviving chlorine dioxide exposure: an elusive mechanism of resistance. J Appl Microbiol 119:1541–1551 10.1111/jam.12963. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Bridier A, Le Coq D, del Pilar Sanchez-Vizuete M, Aymerich S, Meylheuc T, Maillard J-Y, Thomas V, Dubois-Brissonnet F, Briandet R. 2012. Biofilms of a Bacillus subtilis endoscope WD isolate that protect Staphylococcus aureus from peracetic acid. PLoS One 7:e44506 10.1371/journal.pone.0044506. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lear JC, Maillard J-Y, Dettmar PW, Goddard PA, Russell AD. 2006. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources: susceptibility to antibiotics and other biocides. Int Biodeter Biodegrad 57:51–56 10.1016/j.ibiod.2005.11.002. [DOI] [Google Scholar]
- 99.Fisher CW, Fiorello A, Shaffer D, Jackson M, McDonnell GE. 2012. Aldehyde-resistant mycobacteria bacteria associated with the use of endoscope reprocessing systems. Am J Infect Control 40:880–882 10.1016/j.ajic.2011.11.004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarado CJ, Stolz SM, Maki DG, Centers for Disease Control (CDC). 1991. Nosocomial infection and pseudoinfection from contaminated endoscopes and bronchoscopes—Wisconsin and Missouri. MMWR Morb Mortal Wkly Rep 40:675–678. [PubMed] [Google Scholar]
- 101.Denyer SP, Stewart GSAB. 1998. Mechanisms of action of disinfectants. Int Biodeter Biodegrad 41:261–268 10.1016/S0964-8305(98)00023-7. [DOI] [Google Scholar]
- 102.Maillard J-Y. 2002. Bacterial target sites for biocide action. J Appl Microbiol 92(Suppl):16S–27S 10.1046/j.1365-2672.92.5s1.3.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Denyer SP, Maillard J-Y. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. J Appl Microbiol 92(Suppl):35S–45S 10.1046/j.1365-2672.92.5s1.19.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Lambert PA. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol 92(Suppl):46S–54S 10.1046/j.1365-2672.92.5s1.7.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leggett MJ, Schwarz JS, Burke PA, Mcdonnell G, Denyer SP, Maillard J-Y. 2015. Resistance to and killing by the sporicidal microbicide peracetic acid. J Antimicrob Chemother 70:773–779 10.1093/jac/dku445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Munton TJ, Russell AD. 1970. Effect of glutaraldehyde on protoplasts of Bacillus megaterium. J Gen Microbiol 63:367–370 10.1099/00221287-63-3-367. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Ayres HM, Payne DN, Furr JR, Russell AD. 1998. Effect of permeabilizing agents on antibacterial activity against a simple Pseudomonas aeruginosa biofilm. Lett Appl Microbiol 27:79–82 10.1046/j.1472-765X.1998.00397.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Codling CE, Jones BV, Mahenthiralingam E, Russell AD, Maillard J-Y. 2004. Identification of genes involved in the susceptibility of Serratia marcescens to polyquaternium-1. J Antimicrob Chemother 54:370–375 10.1093/jac/dkh351. [PubMed] [DOI] [PubMed] [Google Scholar]
- 110.Walsh SE, Maillard J-Y, Russell AD, Hann AC. 2001. Possible mechanisms for the relative efficacies of ortho-phthalaldehyde and glutaraldehyde against glutaraldehyde-resistant Mycobacterium chelonae. J Appl Microbiol 91:80–92 10.1046/j.1365-2672.2001.01341.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.McNeil MR, Brennan PJ. 1991. Structure, function and biogenesis of the cell envelope of mycobacteria in relation to bacterial physiology, pathogenesis and drug resistance; some thoughts and possibilities arising from recent structural information. Res Microbiol 142:451–463 10.1016/0923-2508(91)90120-Y. [DOI] [PubMed] [Google Scholar]
- 112.Broadley SJ, Jenkins PA, Furr JR, Russell AD. 1995. Potentiation of the effects of chlorhexidine diacetate and cetylpyridinium chloride on mycobacteria by ethambutol. J Med Microbiol 43:458–460 10.1099/00222615-43-6-458. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Fraud S, Hann AC, Maillard J-Y, Russell AD. 2003. Effects of ortho-phthalaldehyde, glutaraldehyde and chlorhexidine diacetate on Mycobacterium chelonae and Mycobacterium abscessus strains with modified permeability. J Antimicrob Chemother 51:575–584 10.1093/jac/dkg099. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Svetlíková Z, Skovierová H, Niederweis M, Gaillard J-L, McDonnell G, Jackson M. 2009. Role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde-based disinfectants and drugs. Antimicrob Agents Chemother 53:4015–4018 10.1128/AAC.00590-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tattawasart U, Maillard JY, Furr JR, Russell AD, Russell AD. 2000. Outer membrane changes in Pseudomonas stutzeri resistant to chlorhexidine diacetate and cetylpyridinium chloride. Int J Antimicrob Agents 16:233–238 10.1016/S0924-8579(00)00206-5. [DOI] [PubMed] [Google Scholar]
- 116.Fernández-Cuenca F, Tomás M, Caballero-Moyano FJ, Bou G, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual Á, Spanish Group of Nosocomial Infections (GEIH) from the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC) and the Spanish Network for Research in Infectious Diseases (REIPI), Spanish Group of Nosocomial Infections GEIH from the Spanish Society of Clinical Microbiology and Infectious Diseases SEIMC and the Spanish Network for Research in Infectious Diseases REIPI. 2015. Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. J Antimicrob Chemother 70:3222–3229. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Tattawasart U, Hann AC, Maillard J-Y, Furr JR, Russell AD. 2000. Cytological changes in chlorhexidine-resistant isolates of Pseudomonas stutzeri. J Antimicrob Chemother 45:145–152 10.1093/jac/45.2.145. [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Braoudaki M, Hilton AC. 2005. Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan. Int J Antimicrob Agents 25:31–37 10.1016/j.ijantimicag.2004.07.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Pagès JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903 10.1038/nrmicro1994. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656 10.1128/MMBR.67.4.593-656.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gandhi PA, Sawant AD, Wilson LA, Ahearn DG. 1993. Adaptation and growth of Serratia marcescens in contact lens disinfectant solutions containing chlorhexidine gluconate. Appl Environ Microbiol 59:183–188. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brözel VS, Cloete TE. 1994. Resistance of Pseudomonas aeruginosa to isothiazolone. J Appl Bacteriol 76:576–582 10.1111/j.1365-2672.1994.tb01655.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Jones MV, Herd TM, Christie HJ. 1989. Resistance of Pseudomonas aeruginosa to amphoteric and quaternary ammonium biocides. Microbios 58:49–61. [PubMed] [PubMed] [Google Scholar]
- 124.Méchin L, Dubois-Brissonnet F, Heyd B, Leveau JY. 1999. Adaptation of Pseudomonas aeruginosa ATCC 15442 to didecyldimethylammonium bromide induces changes in membrane fatty acid composition and in resistance of cells. J Appl Microbiol 86:859–866 10.1046/j.1365-2672.1999.00770.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Guérin-Méchin L, Dubois-Brissonnet F, Heyd B, Leveau JY. 1999. Specific variations of fatty acid composition of Pseudomonas aeruginosa ATCC 15442 induced by quaternary ammonium compounds and relation with resistance to bactericidal activity. J Appl Microbiol 87:735–742 10.1046/j.1365-2672.1999.00919.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Guérin-Méchin L, Dubois-Brissonnet F, Heyd B, Leveau JY. 2000. Quaternary ammonium compound stresses induce specific variations in fatty acid composition of Pseudomonas aeruginosa. Int J Food Microbiol 55:157–159 10.1016/S0168-1605(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 127.Tkachenko O, Shepard J, Aris VM, Joy A, Bello A, Londono I, Marku J, Soteropoulos P, Peteroy-Kelly MA. 2007. A triclosan-ciprofloxacin cross-resistant mutant strain of Staphylococcus aureus displays an alteration in the expression of several cell membrane structural and functional genes. Res Microbiol 158:651–658 10.1016/j.resmic.2007.09.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Boeris PS, Domenech CE, Lucchesi GI. 2007. Modification of phospholipid composition in Pseudomonas putida A ATCC 12633 induced by contact with tetradecyltrimethylammonium. J Appl Microbiol 103:1048–1054 10.1111/j.1365-2672.2007.03346.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Bruinsma GM, Rustema-Abbing M, van der Mei HC, Lakkis C, Busscher HJ. 2006. Resistance to a polyquaternium-1 lens care solution and isoelectric points of Pseudomonas aeruginosa strains. J Antimicrob Chemother 57:764–766 10.1093/jac/dkl011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Lyon BR, Skurray R. 1987. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 51:88–134. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tennent JM, Lyon BR, Midgley M, Jones IG, Purewal AS, Skurray RA. 1989. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J Gen Microbiol 135:1–10. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Littlejohn TG, Paulsen IT, Gillespie MT, Tennent JM, Midgley M, Jones IG, Purewal AS, Skurray RA. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett 74:259–265 10.1111/j.1574-6968.1992.tb05376.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Leelaporn A, Paulsen IT, Tennent JM, Littlejohn TG, Skurray RA. 1994. Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J Med Microbiol 40:214–220 10.1099/00222615-40-3-214. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Heir E, Sundheim G, Holck AL. 1998. The Staphylococcus qacH gene product: a new member of the SMR family encoding multidrug resistance. FEMS Microbiol Lett 163:49–56 10.1111/j.1574-6968.1998.tb13025.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Heir E, Sundheim G, Holck AL. 1999. The qacG gene on plasmid pST94 confers resistance to quaternary ammonium compounds in staphylococci isolated from the food industry. J Appl Microbiol 86:378–388 10.1046/j.1365-2672.1999.00672.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Rouch DA, Cram DS, DiBerardino D, Littlejohn TG, Skurray RA. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol 4:2051–2062 10.1111/j.1365-2958.1990.tb00565.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Huet AA, Raygada JL, Mendiratta K, Seo SM, Kaatz GW. 2008. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 154:3144–3153 10.1099/mic.0.2008/021188-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Schindler BD, Kaatz GW. 2016. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist Updat 27:1–13 10.1016/j.drup.2016.04.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Santos Costa S, Viveiros M, Rosato AE, Melo-Cristino J, Couto I. 2015. Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus. BMC Microbiol 15:232 10.1186/s12866-015-0572-8. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 45:428–432 10.1128/AAC.45.2.428-432.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chuanchuen R, Narasaki CT, Schweizer HP. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J Bacteriol 184:5036–5044 10.1128/JB.184.18.5036-5044.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. 2007. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J Bacteriol 189:7600–7609 10.1128/JB.00850-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schweizer HP. 1998. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother 42:394–398. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chuanchuen R, Karkhoff-Schweizer RR, Schweizer HP. 2003. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am J Infect Control 31:124–127 10.1067/mic.2003.11. [PubMed] [DOI] [PubMed] [Google Scholar]
- 145.Morita Y, Murata T, Mima T, Shiota S, Kuroda T, Mizushima T, Gotoh N, Nishino T, Tsuchiya T. 2003. Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother 51:991–994 10.1093/jac/dkg173. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.Moken MC, McMurry LM, Levy SB. 1997. Selection of multiple-antibiotic-resistant (mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother 41:2770–2772. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nishino K, Yamaguchi A. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol 183:5803–5812 10.1128/JB.183.20.5803-5812.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lomovskaya O, Lewis K. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA 89:8938–8942 10.1073/pnas.89.19.8938. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pagès JM. 2008. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets 9:750–759 10.2174/138945008785747824. [PubMed] [DOI] [PubMed] [Google Scholar]
- 150.Randall LP, Cooles SW, Coldham NG, Penuela EG, Mott AC, Woodward MJ, Piddock LJ, Webber MA. 2007. Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J Antimicrob Chemother 60:1273–1280 10.1093/jac/dkm359. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Webber MA, Randall LP, Cooles S, Woodward MJ, Piddock LJ. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 62:83–91 10.1093/jac/dkn137. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Rajamohan G, Srinivasan VB, Gebreyes WA. 2010. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother 65:228–232 10.1093/jac/dkp427. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402 10.1128/CMR.19.2.382-402.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Noguchi N, Suwa J, Narui K, Sasatsu M, Ito T, Hiramatsu K, Song JH. 2005. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med Microbiol 54:557–565 10.1099/jmm.0.45902-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Sánchez MB, Decorosi F, Viti C, Oggioni MR, Martínez JL, Hernández A. 2015. Predictive studies suggest that the risk for the selection of antibiotic resistance by biocides is likely low in Stenotrophomonas maltophilia. PLoS One 10:e0132816 10.1371/journal.pone.0132816. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176 10.1080/07853890701195262. [PubMed] [DOI] [PubMed] [Google Scholar]
- 157.Brown MH, Paulsen IT, Skurray RA. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol 31:394–395 10.1046/j.1365-2958.1999.01162.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Borges-Walmsley MI, Walmsley AR. 2001. The structure and function of drug pumps. Trends Microbiol 9:71–79 10.1016/S0966-842X(00)01920-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.Poole K. 2001. Multidrug resistance in Gram-negative bacteria. Curr Opin Microbiol 4:500–508 10.1016/S1369-5274(00)00242-3. [DOI] [PubMed] [Google Scholar]
- 160.Poole K. 2002. Outer membranes and efflux: the path to multidrug resistance in Gram-negative bacteria. Curr Pharm Biotechnol 3:77–98 10.2174/1389201023378454. [PubMed] [DOI] [PubMed] [Google Scholar]
- 161.Buffet-Bataillon S, Tattevin P, Maillard J-Y, Bonnaure-Mallet M, Jolivet-Gougeon A. 2016. Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol 11:81–92 10.2217/fmb.15.131. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Bailey AM, Constantinidou C, Ivens A, Garvey MI, Webber MA, Coldham N, Hobman JL, Wain J, Woodward MJ, Piddock LJ. 2009. Exposure of Escherichia coli and Salmonella enterica serovar Typhimurium to triclosan induces a species-specific response, including drug detoxification. J Antimicrob Chemother 64:973–985 10.1093/jac/dkp320. [PubMed] [DOI] [PubMed] [Google Scholar]
- 163.Randall LP, Cooles SW, Coldham NG, Penuela EG, Mott AC, Woodward MJ, Piddock LJ, Webber MA. 2007. Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J Antimicrob Chemother 60:1273–1280 10.1093/jac/dkm359. [PubMed] [DOI] [PubMed] [Google Scholar]
- 164.Webber MA, Randall LP, Cooles S, Woodward MJ, Piddock LJ. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 62:83–91 10.1093/jac/dkn137. [PubMed] [DOI] [PubMed] [Google Scholar]
- 165.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856 10.1111/j.1462-5822.2005.00671.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 166.Sánchez P, Moreno E, Martinez JL. 2005. The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother 49:781–782 10.1128/AAC.49.2.781-782.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pumbwe L, Randall LP, Woodward MJ, Piddock LJV. 2004. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J Antimicrob Chemother 54:341–347 10.1093/jac/dkh331. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Demple B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon: a review. Gene 179:53–57 10.1016/S0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 169.Hutchinson J, Runge W, Mulvey M, Norris G, Yetman M, Valkova N, Villemur R, Lepine F. 2004. Burkholderia cepacia infections associated with intrinsically contaminated ultrasound gel: the role of microbial degradation of parabens. Infect Control Hosp Epidemiol 25:291–296 10.1086/502394. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Valkova N, Lépine F, Valeanu L, Dupont M, Labrie L, Bisaillon JG, Beaudet R, Shareck F, Villemur R. 2001. Hydrolysis of 4-hydroxybenzoic acid esters (parabens) and their aerobic transformation into phenol by the resistant Enterobacter cloacae strain EM. Appl Environ Microbiol 67:2404–2409 10.1128/AEM.67.6.2404-2409.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kümmerle N, Feucht HH, Kaulfers PM. 1996. Plasmid-mediated formaldehyde resistance in Escherichia coli: characterization of resistance gene. Antimicrob Agents Chemother 40:2276–2279. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gomez Escalada M, Russell AD, Maillard J-Y, Ochs D. 2005. Triclosan- bacteria interactions: single or multiple target sites? Lett Appl Microbiol 41:476–481. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Wu VCH. 2008. A review of microbial injury and recovery methods in food. Food Microbiol 25:735–744 10.1016/j.fm.2008.04.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 174.Lambert RJW, van der Ouderaa M-LH. 1999. An investigation into the differences between the Bioscreen and the traditional plate count disinfectant test methods. J Appl Microbiol 86:689–694 10.1046/j.1365-2672.1999.00712.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 175.Brown MRW, Williams P. 1985. Influence of substrate limitation and growth phase on sensitivity to antimicrobial agents. J Antimicrob Chemother 15(Suppl A):7–14 10.1093/jac/15.suppl_A.7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 176.Wright NE, Gilbert P. 1987. Influence of specific growth rate and nutrient limitation upon the sensitivity of Escherichia coli towards chlorhexidine diacetate. J Appl Bacteriol 62:309–314 10.1111/j.1365-2672.1987.tb04925.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.Gomez Escalada M, Harwood JL, Maillard J-Y, Ochs D. 2005. Triclosan inhibition of fatty acid synthesis and its effect on growth of E. coli and Ps. aeruginosa. J Antimicrob Chemother 55:879–882 10.1093/jac/dki123. [PubMed] [DOI] [PubMed] [Google Scholar]
- 178.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532 10.1038/28970. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR, Slabas AR, Rice DW, Rafferty JB. 1999. Molecular basis of triclosan activity. Nature 398:383–384 10.1038/18803. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Webber MA, Coldham NG, Woodward MJ, Piddock LJV. 2008. Proteomic analysis of triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 62:92–97 10.1093/jac/dkn138. [PubMed] [DOI] [PubMed] [Google Scholar]
- 181.Curiao T, Marchi E, Grandgirard D, León-Sampedro R, Viti C, Leib SL, Baquero F, Oggioni MR, Martinez JL, Coque TM. 2016. Multiple adaptive routes of Salmonella enterica Typhimurium to biocide and antibiotic exposure. BMC Genomics 17:491 10.1186/s12864-016-2778-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Seaman PF, Ochs D, Day MJ. 2007. Small-colony variants: a novel mechanism for triclosan resistance in methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 59:43–50 10.1093/jac/dkl450. [PubMed] [DOI] [PubMed] [Google Scholar]
- 183.Abdel-Malek SM, Al-Adham IS, Winder CL, Buultjens TE, Gartland KM, Collier PJ. 2002. Antimicrobial susceptibility changes and T-OMP shifts in pyrithione-passaged planktonic cultures of Pseudomonas aeruginosa PAO1. J Appl Microbiol 92:729–736 10.1046/j.1365-2672.2002.01575.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 184.Parikh SL, Xiao G, Tonge PJ. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39:7645–7650 10.1021/bi0008940. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Chen Y, Pi B, Zhou H, Yu Y, Li L. 2009. Triclosan resistance in clinical isolates of Acinetobacter baumannii. J Med Microbiol 58:1086–1091 10.1099/jmm.0.008524-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Zhu L, Lin J, Ma J, Cronan JE, Wang H. 2010. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother 54:689–698 10.1128/AAC.01152-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Heath RJ, Li J, Roland GE, Rock CO. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J Biol Chem 275:4654–4659 10.1074/jbc.275.7.4654. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Slater-Radosti C, Van Aller G, Greenwood R, Nicholas R, Keller PM, DeWolf WE Jr, Fan F, Payne DJ, Jaworski DD. 2001. Biochemical and genetic characterization of the action of triclosan on Staphylococcus aureus. J Antimicrob Chemother 48:1–6 10.1093/jac/48.1.1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 189.Massengo-Tiassé RP, Cronan JE. 2008. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem 283:1308–1316 10.1074/jbc.M708171200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJV. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248 10.1093/jac/dkv109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Roujeinikova A, Levy CW, Rowsell S, Sedelnikova S, Baker PJ, Minshull CA, Mistry A, Colls JG, Camble R, Stuitje AR, Slabas AR, Rafferty JB, Pauptit RA, Viner R, Rice DW. 1999. Crystallographic analysis of triclosan bound to enoyl reductase. J Mol Biol 294:527–535 10.1006/jmbi.1999.3240. [PubMed] [DOI] [PubMed] [Google Scholar]
- 192.Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C. 1999. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol 290:859–865 10.1006/jmbi.1999.2907. [PubMed] [DOI] [PubMed] [Google Scholar]
- 193.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274:11110–11114 10.1074/jbc.274.16.11110. [PubMed] [DOI] [PubMed] [Google Scholar]
- 194.McCay PH, Ocampo-Sosa AA, Fleming GTA. 2010. Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 156:30–38 10.1099/mic.0.029751-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 195.Casado Muñoz MC, Benomar N, Ennahar S, Horvatovich P, Lavilla Lerma L, Knapp CW, Gálvez A, Abriouel H. 2016. Comparative proteomic analysis of a potentially probiotic Lactobacillus pentosus MP-10 for the identification of key proteins involved in antibiotic resistance and biocide tolerance. Int J Food Microbiol 222:8–15 10.1016/j.ijfoodmicro.2016.01.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 196.Casado Muñoz MC, Benomar N, Lavilla Lerma L, Knapp CW, Gálvez A, Abriouel H. 2016. Biocide tolerance, phenotypic and molecular response of lactic acid bacteria isolated from naturally-fermented Aloreña table to different physico-chemical stresses. Food Microbiol 60:1–12 10.1016/j.fm.2016.06.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 197.Jang H-J, Chang MW, Toghrol F, Bentley WE. 2008. Microarray analysis of toxicogenomic effects of triclosan on Staphylococcus aureus. Appl Microbiol Biotechnol 78:695–707 10.1007/s00253-008-1349-x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 198.Cerf O, Carpentier B, Sanders P. 2010. Tests for determining in-use concentrations of antibiotics and disinfectants are based on entirely different concepts: “resistance” has different meanings. Int J Food Microbiol 136:247–254 10.1016/j.ijfoodmicro.2009.10.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 199.Russell AD, McDonnell G. 2000. Concentration: a major factor in studying biocidal action. J Hosp Infect 44:1–3 10.1053/jhin.1999.0654. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Koutsolioutsou A, Peña-Llopis S, Demple B. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother 49:2746–2752 10.1128/AAC.49.7.2746-2752.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mokgatla RM, Gouws PA, Brözel VS. 2002. Mechanisms contributing to hypochlorous acid resistance of a Salmonella isolate from a poultry-processing plant. J Appl Microbiol 92:566–573 10.1046/j.1365-2672.2002.01565.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 202.Allen MJ, White GF, Morby AP. 2006. The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology 152:989–1000 10.1099/mic.0.28643-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 203.Slade D, Radman M. 2011. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191 10.1128/MMBR.00015-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Daniels C, Ramos JL. 2009. Adaptive drug resistance mediated by root-nodulation-cell division efflux pumps. Clin Microbiol Infect 15(Suppl 1):32–36 10.1111/j.1469-0691.2008.02693.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 205.Maseda H, Hashida Y, Konaka R, Shirai A, Kourai H. 2009. Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob Agents Chemother 53:5230–5235 10.1128/AAC.00631-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Walsh C, Fanning S. 2008. Antimicrobial resistance in foodborne pathogens: a cause for concern? Curr Drug Targets 9:808–815 10.2174/138945008785747761. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623 10.2165/11317030-000000000-00000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Oethinger M, Kern WV, Goldman JD, Levy SB. 1998. Association of organic solvent tolerance and fluoroquinolone resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother 41:111–114 10.1093/jac/41.1.111. [PubMed] [DOI] [PubMed] [Google Scholar]
- 209.Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183:3890–3902 10.1128/JB.183.13.3890-3902.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Fraise AP. 2002. Biocide abuse and antimicrobial resistance: a cause for concern? J Antimicrob Chemother 49:11–12 10.1093/jac/49.1.11. [PubMed] [DOI] [PubMed] [Google Scholar]
- 211.Langsrud S, Sidhu MS, Heir E, Holck AL. 2003. Bacterial disinfectant resistance: a challenge for the food industry. Int Biodeter Biodegrad 51:283–290 10.1016/S0964-8305(03)00039-8. [DOI] [Google Scholar]
- 212.Braoudaki M, Hilton AC. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J Clin Microbiol 42:73–78 10.1128/JCM.42.1.73-78.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Braoudaki M, Hilton AC. 2004. Low level of cross-resistance between triclosan and antibiotics in Escherichia coli K-12 and E. coli O55 compared to E. coli O157. FEMS Microbiol Lett 235:305–309 10.1111/j.1574-6968.2004.tb09603.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 214.Gilbert P, McBain AJ. 2003. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin Microbiol Rev 16:189–208 10.1128/CMR.16.2.189-208.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Russell AD. 2004. Bacterial adaptation and resistance to antiseptics, disinfectants and preservatives is not a new phenomenon. J Hosp Infect 57:97–104 10.1016/j.jhin.2004.01.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 216.Alonso-Hernando A, Capita R, Prieto M, Alonso-Calleja C. 2009. Comparison of antibiotic resistance patterns in Listeria monocytogenes and Salmonella enterica strains pre-exposed and exposed to poultry decontaminants. Food Control 20:1108–1111 10.1016/j.foodcont.2009.02.011. [DOI] [Google Scholar]
- 217.Weber DJ, Rutala WA. 2006. Use of germicides in the home and the healthcare setting: is there a relationship between germicide use and antibiotic resistance? Infect Control Hosp Epidemiol 27:1107–1119 10.1086/507964. [PubMed] [DOI] [PubMed] [Google Scholar]
- 218.Pumbwe L, Skilbeck CA, Wexler HM. 2007. Induction of multiple antibiotic resistance in Bacteroides fragilis by benzene and benzene-derived active compounds of commonly used analgesics, antiseptics and cleaning agents. J Antimicrob Chemother 60:1288–1297 10.1093/jac/dkm363. [PubMed] [DOI] [PubMed] [Google Scholar]
- 219.Lara HH, Ayala-Nunez NV, Turrent LDCI, Padilla CR. 2010. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J Microbiol Biotechnol 26:615–621 10.1007/s11274-009-0211-3. [DOI] [Google Scholar]
- 220.Peyrat MB, Soumet C, Maris P, Sanders P. 2008. Phenotypes and genotypes of Campylobacter strains isolated after cleaning and disinfection in poultry slaughterhouses. Vet Microbiol 128:313–326 10.1016/j.vetmic.2007.10.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 221.Gilbert P, McBain AJ, Bloomfield SF. 2002. Biocide abuse and antimicrobial resistance: being clear about the issues. J Antimicrob Chemother 50:137–139, author reply 139–140 10.1093/jac/dkf071. [PubMed] [DOI] [PubMed] [Google Scholar]
- 222.Lear JC, Maillard J-Y, Dettmar PW, Goddard PA, Russell AD. 2006. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources: susceptibility to antibiotics and other biocides. Int Biodeter Biodegrad 57:51–56 10.1016/j.ibiod.2005.11.002. [DOI] [Google Scholar]
- 223.Knapp L, Amézquita A, McClure P, Stewart S, Maillard J-Y. 2015. Development of a protocol for predicting bacterial resistance to microbicides. Appl Environ Microbiol 81:2652–2659 10.1128/AEM.03843-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sundheim G, Langsrud S, Heir E, Holck AL. 1998. Bacterial resistance to disinfectants containing quaternary ammonium compounds. Int Biodeter Biodegrad 41:235–239 10.1016/S0964-8305(98)00027-4. [DOI] [Google Scholar]
- 225.International Organization for Standardization. 2006. ISO: 20776-1. Clinical laboratory testing and in vitro diagnostic test systems: susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. Part 1. Reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases. British Standard Institute, London, United Kingdom. [Google Scholar]
- 226.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2014. Breakpoint tables for interpretation of MICs and zone diameters. Version 4.0. 2014. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf. Accessed January 2017.
- 227.Andrews JM, BSAC Working Party on Susceptibility Testing. 2009. BSAC standardized disc susceptibility testing method (version 8). J Antimicrob Chemother 64:454–489 10.1093/jac/dkp244. [PubMed] [DOI] [PubMed] [Google Scholar]
- 228.Saleh S, Haddadin RNS, Baillie S, Collier PJ. 2011. Triclosan: an update. Lett Appl Microbiol 52:87–95 10.1111/j.1472-765X.2010.02976.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.Gradel KO, Randall L, Sayers AR, Davies RH. 2005. Possible associations between Salmonella persistence in poultry houses and resistance to commonly used disinfectants and a putative role of mar. Vet Microbiol 107:127–138 10.1016/j.vetmic.2005.01.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 230.Chuanchuen R, Pathanasophon P, Khemtong S, Wannaprasat W, Padungtod P. 2008. Susceptibilities to antimicrobials and disinfectants in Salmonella isolates obtained from poultry and swine in Thailand. J Vet Med Sci 70:595–601 10.1292/jvms.70.595. [PubMed] [DOI] [PubMed] [Google Scholar]


