ABSTRACT
Metals and metalloids have been used alongside antibiotics in livestock production for a long time. The potential and acute negative impact on the environment and human health of these livestock feed supplements has prompted lawmakers to ban or discourage the use of some or all of these supplements. This article provides an overview of current use in the European Union and the United States, detected metal resistance determinants, and the proteins and mechanisms responsible for conferring copper and zinc resistance in bacteria. A detailed description of the most common copper and zinc metal resistance determinants is given to illustrate not only the potential danger of coselecting antibiotic resistance genes but also the potential to generate bacterial strains with an increased potential to be pathogenic to humans. For example, the presence of a 20-gene copper pathogenicity island is highlighted since bacteria containing this gene cluster could be readily isolated from copper-fed pigs, and many pathogenic strains, including Escherichia coli O104:H4, contain this potential virulence factor, suggesting a potential link between copper supplements in livestock and the evolution of pathogens.
INTRODUCTION
Metal compounds are widely used in livestock production because they are necessary supplements and play a very important role as essential trace elements that are part of the nutritional requirements of most animal species. However, copper and zinc compounds are also added to feed in larger concentrations for achieving additional beneficial effects. Therefore, we will focus this chapter on copper and zinc; their use and indications; the concerns related to the selection of resistance, toxicity, and environmental pollution and policies; and the alternatives and new developments regarding their use. Compounds derived from the nonessential metal arsenic, such as roxarsone, which have been used in livestock for feed supplementation in some countries around the world, will not be discussed in great detail in this chapter.
USE OF METAL COMPOUNDS IN AGRICULTURAL PRODUCTION: REQUIREMENTS AND RECOMMENDATIONS
Copper
Copper is an essential trace element that is necessary for the human body because it has major functions regarding fetal growth and early postnatal development, hemoglobin synthesis, maturation of the connective tissues, proper nerve function and bone development, and inflammatory processes. Therefore, most animal species are required to have copper in their feed and exhibit copper deficiency symptoms if copper is insufficient. Copper is also expected to interact with other compounds in the feed (phytates, fiber fraction, zinc, iron, calcium, ascorbic acid, molybdenum, and sulfur), and therefore the concentrations added to feed are increased to ensure that the necessary amount is available for absorption by the target animal species. This is also dependent on the animal’s intestinal tract, and these interactions influence the absorption, as extensively reviewed by Suttle (1).
An excess of copper can also be detrimental and lead to toxicity manifested as anemia and liver dysfunction. Release of copper from the liver may lead to vascular hemolysis and death. The toxicity of copper depends on speciation, with Cu(I) being more toxic then Cu(II). Certain animal species such as sheep and young calves are sensitive to excess copper (2). Copper might also exert a selection pressure on the gut microbiota and influence the composition of the gut flora. Furthermore, the use of therapeutic concentrations might select for resistance, which is addressed later in this chapter. Requirements for copper in animal feed depend on the target species, the age group and the feed composition (2).
Copper is contained in a number of plants and seeds in varying concentrations and may become concentrated with the use of copper compounds in agriculture, but the availability of copper in feed is not well known. For food-producing animals and small pet animals such as rabbits, the copper background levels in complete feeds are rather low and in the range of 5 (salmon feed) to 15 (rabbit feed) mg Cu/kg feed, and higher values are observed in feed for dairy cows, containing about 20 mg Cu/kg feed. Supplementation of animal feed is common practice, and the current limits for supplementation (3) were revised by the European Union in 2016 (Table 1) (4).
TABLE 1.
New proposed maximum limits of copper in complete animal feedh
| Target species, animal category | Ra | 1.5 × R | Background | NPMCb | CAMCb |
|---|---|---|---|---|---|
| Chickens for fattening, reared for laying | 8 | 12 | 10 | 25 | 25 |
| Laying hens, breeder hens | 8 | 12 | 10 | 25 | 25 |
| Turkeys for fattening, 0–8 weeks of age | 10 | 15 | 10 | 25 | 25 |
| Turkeys for fattening, from 8 weeks of age onward | 6 | 9 | 10 | 25 | 25 |
| Other poultry | 8 | 12 | 10 | 25 | 25 |
| Piglets, weaned | 8 | 12 | 10 | 25 | 170 |
| Pigs for fattening | 6 | 9 | 10 | 25 | 25 |
| Sows | 10 | 15 | 10 | 25 | 25 |
| Calves, milk replacer | 10 | 15 | 5 | 15c | 15 |
| Cattle for fattening | 8.8h | 13.2 | 10 | 30d | 35 |
| Dairy cows | 8.8–13.2h | 13.2–19.8 | 10 | 30 | 35 |
| Sheep | 7h | 10.6 | 10 | 15e | 15 |
| Goats | 7–22h | 10.6–33 | 10 | 35 | 25 |
| Horses | 8.8h | 13.2 | 10 | 25 | 25 |
| Rabbits | 5 | 7.5 | 10 | 25 | 25 |
| Salmonids | 5 | 7.5 | 10 | 25 | 25 |
| Other fish | 8 | 12 | 10 | 25 | 25 |
| Crustaceans | 50 | 50 | |||
| Dogs | 12 | 15.6f | 10 | 25 | 25 |
| Cats | 10 | 13f | 10 | 25 | 25 |
R, requirement.
NPMC, newly proposed total maximum contents of copper in complete feed (4); CAMC, currently authorized maximum content of copper in complete feed (3).
Because copper in milk-based diets shows a high bioavailability and does not contain substantial amounts of copper antagonists, the background level (5 mg/kg) is not added to the allowance (15 mg/kg).
Considering cattle feeding on pasture, a potentially high amount of copper antagonists in forage is taken into account.
NPMC is limited by the Mineral Tolerance of Animals (MTL); dietary concentrations above 15 mg/kg could provoke chronic copper poisoning (CCP).
No requirement data available; the allowance (12 and 10 mg/kg for dogs and cats, respectively) is therefore only multiplied by 1.3 to consider different bioavailability of copper supplements.
Adjusted from dry matter to complete feed with 88% dry matter.
Source: reference 4.
Copper additives
The list of copper compounds authorized for copper supplementation in feed is extensive and includes cupric acetate, monohydrate; basic cupric carbonate, monohydrate; cupric chloride, dihydrate; cupric methionate; cupric oxide; cupric sulfate, pentahydrate; cupric chelate of amino acid hydrate; copper lysine sulfate; cupric chelate of glycine hydrate; copper chelate of hydroxy analogue of methionine; dicopper chloride trihydroxide; and copper bislysinate. These authorized compounds fall under European Economic Community (EEC) number E4 (3). Most copper-containing compounds show bioavailability similar to copper sulfate for all animal species. However, cupric oxide and cupric carbonate (and cuprous iodide in poultry) showed a lower and more variable relative bioavailability (4).
Therapeutic use of copper
Copper may be prescribed for copper deficiency and is normally administered orally using premixes in feed, or it may be administrated using injectable solutions. Additionally, copper has antimicrobial properties, and copper sulfate may be used in footbaths as a fungicide for the control of foot-rot in cattle and sheep (5 to 10% solution) (5). Copper has also been used as a growth promoter, even though the efficacy of this practice has not been very well demonstrated. Growth promotion might depend on the feed antagonists present and relate to improved appetite or digestibility in pigs and poultry (1).
Zinc
Similar to copper, zinc is also an essential trace element and therefore is necessary to comply with the nutritional requirements of livestock. Zinc is part of 10% of all proteins and contributes to their tertiary structure and the function of enzymes, and therefore its role is essential in diverse organs and systems. For example, zinc is needed for glucose and lipid metabolism, cell proliferation, embryogenesis, and systems related to the nervous and immune systems.
Symptoms of zinc deficiency can be observed in most animal species (6). Known examples of this are paraqueratosis in swine (7) and growth deficiencies in feathering and skeletal development in poultry (8). Zinc is an essential trace element, and zinc deficiency might be related to malabsorption of this metal compound. Similarly, Friesian cows and bull terrier dogs have been shown to have genetic syndromes related to zinc malabsorption (9, 10).
Most of the absorption of zinc occurs in the small intestine, and feed components such as phytates may hamper the absorption in monogastric species, while low molecular weight binding ligands such as citrate, picolinate, EDTA, and amino acids such as histidine and glutamate were shown to increase the absorption of zinc (11).
Zinc is mainly metabolized in the liver, and the main zinc deposit is in bone. Measurement of zinc in bone may be used to determine zinc status or utilization, while measurement of zinc in hair may be used for diagnostic purposes. Zinc concentrations in plasma are not reliable because zinc binds extensively to proteins.
Since zinc is mainly excreted in feces, it is the variable zinc excretion that together with absorption maintains zinc homeostasis in the body. These processes also influence the seepage of zinc from manure into the environment. Zinc absorption and bioavailability are influenced by the feed composition; not only the plant component but also high concentrations of other metals such as copper, nickel, and iron may lead to zinc deficiency. Therefore, formulations of feed need to take into account other feed components that might interfere with zinc absorption and bioavailability.
Toxicity to zinc is low in general, and tolerance to high levels of zinc has been observed. However, ruminants, especially young and pregnant animals, are more susceptible to the effects of excess zinc compared to pigs or poultry. Zinc toxicity has been observed in steers and heifers, but tolerance seems to be higher in dairy cows. Sheep may develop symptoms of excess zinc in their diet, manifesting as symptoms ranging from weakness and jaundice to abortions and stillbirth. Horses, especially young and pregnant animals, are among the most sensitive to excess zinc in the diet and react with lameness and osteochondrosis. Pigs are among the most zinc-tolerant animal species, and it has been shown that even though they can exhibit symptoms of toxicity, it depends on the exposure time to excess zinc diet and the relation of other components of the diet such as calcium. Therefore, in some countries, high zinc concentrations are allowed for therapeutic purposes (12).
Zinc is naturally occurring in agricultural produce, but the levels vary greatly. The content of zinc in natural products depends on the zinc concentration in the soil and the conditions that might influence zinc uptake by plants. Some zinc may also be added due to deposition on plants, soil contamination, and processing. Similar to copper, the available concentrations in raw materials vary and are relatively low in relation to the animal requirements. Therefore, zinc supplementation to animal feed is common (12). However, supplementation has been at relatively high levels, and several organizations have indicated that these levels could be lowered to reduce the impact on environment (12, 13).
For requirements in different animal species and age groups as well as current and proposed supplementation levels, please consult the data in Table 2 (12).
TABLE 2.
New proposed maximum limits of zinc in complete feedf
| Target species, animal category | Ra | 1.5 × R | Background | NPMCb | CAMCc |
|---|---|---|---|---|---|
| Chickens for fattening, reared for laying | 40–50 | 60–75 | 30 | 100 | 150 |
| Laying hens, breeder hens | 45 | 67.5 | 30 | 100 | 150 |
| Turkeys for fattening, 0–8 weeks of age | 70 | 105 | 30 | 120 | 150 |
| Turkeys for fattening, from 8 weeks of age onward | 50 | 75 | 30 | 120 | 150 |
| Other poultry | 40, 60, 60 | 80 | 30 | 100 | 150 |
| Piglet below 11 kg weight | 100 | 150 | 30 | 150 | 150 |
| Piglet, weaned above 11 kg weight | 80 | 120 | 30 | 150 | 150 |
| Pigs for fattening | 60 | 90 | 30 | 100 | 150 |
| Sows | 50–100 | 75–150 | 30 | 150 | 150 |
| Calves, milk replacer | 40 | 60 | 30 | 100 | 200 |
| Cattle for fattening | 35d | 53 | 30 | 100 | 150 |
| Dairy cows, dairy heifer | 44d | 66 | 30 | 100 | 150 |
| Sheep | 40d | 60 | 30 | 100 | 150 |
| Goats | 31d | 47 | 30 | 100 | 150 |
| Horses | 44d | 66 | 30 | 100 | 150 |
| Rabbits | 70 | 105 | 50 | 150 | 150 |
| Salmonids | 50 | 75 | 60 | 150 | 200 |
| Other fish | 20 | 30 | 60 | 100 | 200 |
| Dogs | 100e | 70 | 150 | 250 | |
| Cats | 75e | 70 | 150 | 250 |
R, requirement.
NPMC, newly proposed total maximum contents of zinc in complete feed (13).
CAMC, currently authorized maximum content of zinc in complete feed (3).
Adjusted from dry matter to complete feed with 88% dry matter.
Allowance, taken as 1.5 times the requirement.
Source: reference 13.
Zinc additives
For supplementation of feed, a number of additives are utilized, including zinc oxide, zinc chloride monohydrate, zinc sulfate monohydrate, zinc sulfate heptahydrate, zinc chelate of amino acids hydrate, zinc acetate dihydrate, and zinc lactate trihydrate, which fall under EEC number E6 (3). In regard to therapeutic use, the products used are similar but require a prescription if regulation permits. In the European Union, this usage has been allowed in some countries but it will be discontinued because the Committee for Veterinary Medicine Products at the European Medicines Agency has recommended that market authorizations be withdrawn for existing products, and new products will be refused largely due to environmental risks and acknowledgement of the risk for of coselection (14).
Therapeutic use of zinc
Zinc may be used in some countries under prescription for treatment and metaphylaxis of diarrheal disease in young animals such as postweaning piglets. Zinc supplements are normally given in the form of premixes used in feed or drinking water. The most relevant zinc forms for this practice are zinc oxide, zinc chloride, and zinc sulfate. For example, in Denmark and Belgium, veterinarians have until recently prescribed zinc oxide supplementation of up to 2,500 ppm for weaning pigs for up to 14 days after weaning, which is much more than the maximum concentration allowed in pig feed—250 ppm in the European Union, with the usual concentrations ranging between 50 and 125 ppm (15–17). Similarly, in other countries around the world, the therapeutic use of zinc is relatively widespread (18), and doses for therapeutic use are between 1,000 and 3,000 ppm for up to 2 to 3 weeks (19). This is contradictory to the fact that these high doses given over a long period may cause toxicity. The use of therapeutic concentrations of zinc has been considered beneficial for the reduction and prevention of postweaning diarrhea in weaners and postweaning scouring (20). However, the growth promotion effects are not well understood (12).
According to the risk assessments released at the end of 2016, the benefits of the therapeutic use of zinc compounds in animal production do not outweigh the environmental risks, and additionally, there is an acknowledged risk for coselection of antibiotic resistance; therefore the Committee for Veterinary Medicine Products has recommended the withdrawal of existing market authorizations for veterinary products containing zinc and refusal of approval for new formulations. This recommendation has been backed by the EU Commission, and phase-out is expected in the coming years. Therefore, such products will not be available for therapeutic purposes in the European Union member states in the future (14).
Other Metallic Compounds for Therapeutic Purposes
Arsenic is a metalloid compound that is naturally present in rocks but is present in low amounts in soil and water. The presence of arsenic in the environment may also occur due to human industrial activity (mining, burning nonferrous metals, burning fossil fuel, or use in fertilizers, pesticides, insecticides, etc.). Unlike copper and zinc, arsenic is not thought to be an essential trace element, and therefore supplementation in feed is not considered for nutritional purposes. Arsenic and inorganic arsenic compounds are considered toxic and carcinogenic, while other arsenic-derived compounds are considered either “possibly carcinogenic to humans” or “not classifiable as to their carcinogenicity to humans” (21). However, there have been reports of beneficial effects in cancer therapy, and therefore arsenic might have some unexpected beneficial effects (22). Additionally, exposure to inorganic arsenic in drinking water has been linked to a reduction in breast cancer (23).
Organic arsenical compounds are active against coccidia and other parasites, so they have been used in poultry production to reduce coccidial infection and promote growth. However, even though roxarsone, arsanilic acid, and nitarsone are organic compounds and are thought to be less toxic, it was noticed that their usage led to the increased occurrence of inorganic arsenic residues in liver detected by improved analytical methods, and they have therefore been under scrutiny. These compounds were widely used in the United States and have only recently been banned, starting with a ban on roxarsone, carbasone, and arsanilic acid in 2013, which was followed by a ban on the use of nitarsone for histomoniasis in turkeys in 2015. However, the widespread use and subsequent exposure to arsenic in humans was determined through a significant association between arsenic content in urine and poultry intake in a study performed in the United States between 2003 and 2010 (24). Because these compounds are now banned in the United States and the European Union, it is important to focus on other places in the world where this type of supplementation is still ongoing (25, 26), both to avoid consumption of arsenic by humans and to avoid pollution of the environment around farms (27).
Effect of Supplementation on Microflora and Organs
Copper
It has been generally assumed that copper does not affect the normal bacterial flora, but an extensive literature study found that in piglets and growing pigs, low copper concentrations (<50 mg/kg feed) affected the microbiota in the gastrointestinal tract. Similarly, 100 to 250 mg/kg copper sulfate was found to significantly change the microbial community structure. In poultry (broilers), supplementation with copper, even at low concentrations (<50 mg/kg feed), appeared to affect the microbiota in the gastrointestinal tract. In particular, the Clostridia population seemed to be affected even at low concentrations. At higher concentrations (>200 mg/kg feed), inorganic or organic bound copper also appeared to affect the population of lactobacilli and coliforms in broilers (28). Furthermore, the reduction of the pH of the gizzard content may cause severe gizzard erosion (4).
The risk assessments made for copper salt supplements (including copper carbonate) in relation to human consumption have concluded that a maximum residue level is not needed because it seems unlikely that copper salts, when used according to the specifications in veterinary medicine settings, are likely to cause harm in humans through consumption of residues contained in food of animal origin (29).
Zinc
The main consensus regarding zinc supplementation is that it does not appear to have major consequences on the microflora and organs. However, recent studies demonstrated that the effect observed might be due to the regulation of factors related to the immune response leading to a reduction of stem cell factor expression in the small intestine and causing a reduction in the number of mast cells and histamine release. These findings may have important implications for the prevention of weaning-associated diarrhea in piglets (30). However, a number of studies suggest that changes in the stress response may also indicate a more profound effect of therapeutic zinc supplementation that might affect the immune response (31).
The risk assessments made for zinc supplements in relation to human consumption have concluded that a maximum residue level is not needed because it seems unlikely that zinc salts, when used according to the specifications in veterinary medicine settings, are likely to cause harm in humans through consumption of residues contained in food of animal origin (32).
Usage Data and Distribution
High amounts of zinc additives are used for medical purposes in piglet production in Europe. As an example: European pig production was 248 million heads in 2008 (EURO-25 in 2008); 30% of these were produced in Denmark and Belgium. A percentage piglets were most likely given feed containing 2.5 g Zn/kg in the first 14 days after weaning. If daily feed consumption is 0.4 kg in the first week and 0.5 kg in the second week, the zinc consumed amounts to approximately 1,312 tons per year (13).
A similar scenario can be found in the United States. Here, as in Europe, zinc is fed to newly weaned pigs at high dietary levels of 2.0 to 3.0 g/kg feed (19). Assuming a mean of 2.5 g/kg and the same feed intake during the first 14 days after weaning as stated above for Europe, the 113.8 million pigs raised for slaughter in 2016 (33) would have consumed approximately 1,800 tons of zinc.
Taking Denmark as an example of a major pig-producing country using zinc by prescription, it was observed that the consumption of zinc oxide prescribed by Danish veterinarians has increased over the past 10 years, from ∼150 tons in 2005 to ∼500 tons in 2014. In this time period, the total consumption of zinc oxide tripled, while pig production increased by only 14% (15, 20). Another example of a European country using zinc oxide for therapeutic purposes is Belgium, where the use of zinc oxide in therapeutic doses in piglets administered for 2 weeks after weaning has been allowed only since September 2013. In Belgium, zinc oxide has become an alternative for colistin, which was previously used in weaned piglets. The data for 2014 and 2015 show a substantial increase in the use of this metal compound, with 81,964 kg and 87,199 kg of zinc oxide, respectively, consumed in Belgium. Simultaneously, the Belgian authorities noticed a decrease in the use of polymyxins (colistin) from 5.8 mg/PCU (population correction unit, which is defined as a measure of biomass, where 1 PCU is equivalent to 1 kg of biomass of livestock or slaughtered animals) in 2012, when zinc oxide was not available, to 3.3 mg/PCU in 2014, and a further 51% decrease was noticed in 2015 (16, 34). As mentioned above, the market authorizations for zinc products will be withdrawn for use for therapeutic purposes following the decision taken by European Medicines Agency, and therefore we expect a sharp drop in the consumption of this compound in the European Union. Because the use of zinc compounds is not exclusive to Europe and is quite widespread in North America (18) and other regions, some therapeutic use might remain, depending on local regulations. Reducing the amount of zinc to a dietary requirement of 100 mg/kg for newly weaned pigs would reduce the yearly zinc consumption in pig farming by over 90% (35).
It has been shown that high zinc supplementation after weaning until pigs reached 12 kg, followed by high copper supplementation until they reached about 25 kg in body weight was both cost-effective and promoted the best growth. Suggested therapeutic copper levels of 100 to 250 mg/kg are 95 to 98% above the dietary requirement for this age group (19, 35).
Environmental Concerns from Use of Metals in Agriculture: Metal Ecotoxicity
The extensive supplementation of feed with these compounds and the additional practices of therapeutic and/or growth promoter usage are not innocuous. Most of these compounds are excreted by the animals and end up in the environment, where their accumulation can have major consequences (36) because these are not degradable and will increase the concentration in the environment, especially in soils amended with slurry/manure from animal production (37). For example, Denmark, which is one of the largest pig-producing countries in the world and uses metals in animal production, has maintained a national monitoring program of heavy metals in the environment for the last 28 years to better understand the effects of these practices on the environment. The values and analyses published in 2016 indicate that the use of pig slurry has led to a significant increase in the concentrations of copper and zinc in soil (38).
Furthermore, the data show that 45% of the analyzed soils have reached zinc levels above the predicted no-effect concentrations (PNEC), while for copper, only one soil sample was above the PNEC. Continued agricultural use of zinc is expected to increase the levels, and larger areas will have soil exceeding the PNEC. Furthermore, the authors noted that the current situation raises concerns regarding the aquatic environment because the current use of zinc and copper in agriculture might lead to leaching of these metals into water and negatively affect aquatic species (37, 38).
In addition, at the European level, zinc is one of the compounds studied as part of risk assessments regarding the environmental pollution of heavy metals. These reports focus on aspects related to manufacturing of zinc products. A number of studies looking at the accumulation of zinc in soils due to agricultural use are also mentioned, and although these studies are relatively different in scope and methodology, they all agree that zinc will accumulate in soils in a significant manner, and increasing areas will exceed the critical concentrations. However, the time this will take varies and therefore needs to be assessed at the regional level (39). Taking into account these aspects and also acknowledging the risk for coselection, the European authorities have been alerted and are conducting ongoing collection of information for environmental risk assessment of heavy metals as part of veterinary medicinal products. The Committee for Veterinary Medicine Products has therefore concluded that the benefits do not outweigh the risks, and the risk for the environment justifies withdrawing the market authorizations for the therapeutic usage of zinc compounds (34).
Alternatives
As mentioned above, zinc and copper cannot be replaced since they are essential trace elements, but the concentrations can be reduced and adjusted to the essential requirements to avoid overuse. Similarly, in the future, the use of those metals at high concentrations for therapeutic purposes will be discontinued at least in the European Union, not only because of the added pressure leading to selection of bacterial resistance to copper and zinc and subsequently coselection of antibiotic-resistance genes, but mainly because of environmental concerns. Antimicrobials are alternatives for this therapeutic use but are not viable options since there are a number of ongoing efforts to reduce antimicrobial consumption in animals. Replacement should go hand in hand with improvement of management practices to reduce the need for the use of metals or other compounds during the weaning period.
DETECTION OF HEAVY METAL RESISTANCE
In recent years, the identification and/or characterization of strains of bacteria of animal origin that have reduced susceptibility or increased tolerance to heavy metals has gained a lot of attention, and therefore, an increasing number of studies include heavy metal susceptibility or tolerance testing. The Web of Science database was searched systematically for articles between January 2005 and December 2016 using the search terms “heavy+ metal+ resistance+ animal” and “heavy+ metal+ resistance+ bacteria+ MIC”. This resulted in the retrieval of 278 articles, of which only 23 reported MIC data on bacterial isolates of animal origin, including fish. These studies were reviewed and revealed a number of weaknesses, which can impact the quality of the results or the conclusions made. Here, we will highlight the major pitfalls that make comparisons of studies challenging.
Several methods, such as disc diffusion, agar dilution, and microbroth dilution, have been used in metal tolerance assays. Disc diffusion, the use of paper discs impregnated with a specific heavy metal salt concentration placed on agar plates spread with bacteria, was rarely used (only two studies [40, 41]). Agar and microbroth dilution were the most common methods to determine an isolate’s MIC to a particular heavy metal. Both methods are based on a dilution series of the heavy metal that is incorporated into either molten agar or liquid broth. The methodology is similar to that used for antibiotic susceptibility testing, where the dilution series is typically 2-fold dilutions rather an exponential serial dilution. While most studies use the lowest concentration at which there is inhibition of visible growth, a few studies use optical density measurements to determine the presence of bacterial growth (42, 43).
As previously described by Hasman et al. (44), the choice of media and the pH of that media are crucial factors that can influence the detection of metal resistance levels. For example, certain components in complex media can sequester free metal ions, reducing the concentration available for the bacteria. Therefore, it is advisable to use standardized media such as Mueller Hinton, which has been extensively validated for use in antibiotic susceptibility testing. Of the 23 studies included in this analysis and described using agar dilution (n = 18), up to six different media were used. While Mueller Hinton agar was the most common, brain heart infusion agar (45), Luria Bertani agar (46), nutrient agar (47, 48), and tryptic soy agar (49, 50) were also used. As previously mentioned, the pH of the medium can influence the detection of metal resistance. For example, pH values above 5.5 can cause zinc sulfate in complex media to precipitate, thereby reducing the concentration of the metal that the bacteria have to encounter. Furthermore, addition of metals into the medium can change the final pH, which can affect bacterial growth. Therefore, it is strongly recommended that after supplementation of the metal, the media is adjusted accordingly to ensure optimal bacterial growth and avoid misleading results. Only seven articles noted that pH adjustments were performed. However, even in studies where adjustments were reported, the pH value was not always consistent; e.g., for CuSO4 in Mueller-Hinton agar, a range of pH values were used by the different studies (pH 7, 7.2, and 7.4) (51–55). It is recommended that when using Mueller Hinton agar or broth, the adjusted pH should be as follows: zinc chloride, pH = 5.5; copper sulfate, pH = 7.2; sodium arsenate, cadmium acetate, and silver nitrate, pH = 7.4 (51, 56).
Studies wanting to categorize bacterial isolates as susceptible or resistant to a particular heavy metal on the basis of their MICs or inhibition zone diameters require approved interpretive criteria. There are no clinical breakpoints for heavy metals. Therefore, epidemiological cutoff values are based on previous studies reporting the MIC distributions for a particular bacterial species. Five studies used a control strain, e.g., the laboratory strain Escherichia coli K12 or C600, and strains were considered resistant if the MIC values exceeded that of the control strain (57–61). While this could be a reasonable approach to characterize reduced heavy metal susceptibility among bacterial isolates, the control strain should belong to the same bacterial species as the test isolates. Akinbowale et al. (59) used E. coli K12 to describe resistance in Aeromonas spp. and Pseudomonas spp. While all isolates are Gram negative, there are intrinsic differences between the species that can affect the interpretation of the results.
Increased metal tolerance to zinc and copper has been described in many bacterial species. Initially, these resistant bacterial species were detected among environmental bacteria isolated from areas with high concentrations of heavy metals. Due to the use of heavy metals in food-producing animals and the association of heavy metal resistance with antimicrobial resistance as a result of co-selection, many studies have investigated the levels of reduced susceptibility (Table 3) and the associated genes in a number of bacterial species isolated from food-producing animals (Table 4).
TABLE 3.
Bacteria of animal origin for which copper and zinc resistance has been described
| Metal | Bacterial isolate(s) | Animal origin | References |
|---|---|---|---|
| Copper | Actinomycetes | Goat | 236 |
| Aeromonas hydrophilia | Fish including shellfish | 48 | |
| Citrobacter spp. | Fish, pigs | 47, 144 | |
| Enterococcus faecalis, E. faecium | Chicken, cattle, pigs | 56 | |
| Escherichia coli | Chicken, cattle, pigs | 56, 144, 145, 155 | |
| Klebsiella pneumoniae | Prawns | 237 | |
| Lactobacillus spp. | Pigs, cattle, poultry | 238 | |
| Salmonella spp. | Chicken, cattle, pigs | 56, 144, 156 | |
| Serratia marcescens | Fish including shellfish | 48 | |
| Staphylococcus aureus, S. hyicus | Chicken, cattle, pigs | 56 | |
| Streptomycetes | Goats | 236 | |
| Zinc | A. hydrophilia | Fish including shellfish | 48 |
| E. faecalis, E. faecium | Chicken, cattle, pigs | 56 | |
| E. coli | Chicken, cattle, pigs | 56 | |
| K. pneumoniae | Prawns | 237 | |
| Lactobacillus sp. | Pigs, cattle, poultry | 238 | |
| P. mirabilis | Fish including shellfish | 48 | |
| P. aeruginosa | Various | 49 | |
| Salmonella spp. | Chicken, cattle, pigs | 56 | |
| S. marcescens | Fish including shellfish | 48 | |
| S. aureus, S. hyicus | Chicken, cattle, pigs | 56 | |
| Vibrio cambelli, V. harveyi, V. mimicus, V. ordelli | Fish including shellfish | 48 |
TABLE 4.
Copper- and zinc-resistance genes identified in bacteria from livestock
| Heavy metal | Heavy metal-resistance gene | Bacterial species | Animal origin | References |
|---|---|---|---|---|
| Copper | pcoA, pcoD | Salmonella enterica serovar MbandakaS. enterica serovar DerbyS. enterica serovar HeidelbergS. enterica serovar TyphimuriumS. enterica serovar WorthingtonS. enterica serovar RissenS. enterica serovar AgonaS. enterica serovar SenftenbergS. enterica serovar LondonS. enterica serovar OhioEscherichia coliHistophilius somni | Pigs, cattle, poultry | 53, 146, 239–242 |
| copB | Staphylococcus aureus Staphylococcus epidermidis Staphylococcus haemolyticus Staphylococcus hominis Staphylococcus lentus Staphylococcus pasteuri Staphylococcus rostri Staphylococcus sciuri E. coli | Pigs, cattle, poultry | 199, 240, 243 | |
| cueO | E. coli | Poultry | 240 | |
| cusC | E. coli | Poultry | 240 | |
| mco | S. aureus | Pigs | 243 | |
| Zinc | czrC | Mainly associated with methicillin-resistant staphylococci:S. aureusStaphylococcus hyicusStaphylococcus epidermidisS. haemolyticusS. hominisS. lentus | Pigs, cattle, poultry, meat from these animal species | 51, 199–203, 244, 245 |
| czcD | S. TyphimuriumS. enterica serovar Infantis | Pigs | 53 | |
| zntA | E. coli | Poultry | 240 |
MECHANISMS OF METAL HOMEOSTASIS AND RESISTANCE
Metal homeostasis is a delicate balance of ensuring that a cell’s requirement for essential metals is met to ensure proper cell function while at the same time limiting the amount of metal in the cell to avoid toxicity. The ability to balance the influx and efflux of metals, metal homeostasis, is determined by the presence or absence of metal transport systems, which can depend on an organism’s living environment and therefore its exposure to metals.
Copper
As in other soft metals, the bioavailability of copper changed with the appearance of oxygen in the environment during the Great Oxidation Event. Copper became available due to the increased oxidation of copper sulfides and the higher solubility of Cu(II) (62, 63). With life initially evolving under anoxic conditions and the limited availability of copper for cells, prokaryotes utilized a limited number of copper-containing enzymes. Copper homeostasis mechanisms evolved to protect cells from increasing copper availability and its cytotoxicity due to Cu(I) attacking Fe-S clusters and the generation of reactive oxygen species under aerobic conditions (64–66).
Copper uptake
In contrast to eukaryotes, prokaryotes use a small number of copper-containing enzymes that are, with the exception of plastocyanin and caa3-type cytochrome oxidase, located in the periplasm and cytoplasmic membrane. Therefore, very few specific copper uptake systems have been identified in bacteria. In Bacillus subtilis, the protein YcnJ shows homology to CopCD, appearing to be a fusion of the two proteins, and was shown to be involved in copper uptake. Expression of ycnJ is regulated by the repressor YcnK (67, 68). While common in the genus Bacillus, ycnJ does not seem to be present in other members of the Firmicutes. Bacillus cereus produces a copper-chelator (coproporphyrin III) that might be involved in copper uptake (69).
Gram-negative bacteria face the additional challenge of an outer membrane to take up required nutrients. In Methylosinus trichosporium, E. coli and Pseudomonas aeruginosa porins have been found to play an important role in copper uptake, allowing the metal ion to enter the periplasm and therefore being able to interact with the transporters in the cytoplasmic membrane (70–72). From the periplasm, copper can enter the cell via several mechanisms. ZupT, a member of the ZIP family in E. coli, was shown to have a broad substrate spectrum also taking up Cu(II) (73, 74). Since members of this protein family are ubiquitously present, it can be assumed that they can also allow copper uptake in other species as well (75). Similar to YcnJ in B. subtilis, some members of the Proteobacteria have CopCD, with CopC being a periplasmic copper-binding protein and CopD a cytoplasmic membrane transporter. Overexpression of copCD increases the cells’ copper sensitivity, indicating an uptake system (76). In several Gram-negative bacteria, copper chelators (chalkophore) have been identified, and Cyanobacteria utilize siderophores for both copper and iron uptake (70, 77–82).
Chromosomally encoded copper defense systems
Due to the increased bioavailability of copper during Earth’s history and the capability of Cu(I) to damage iron-sulfur clusters and catalyze production of reactive oxygen species, organisms had to develop strategies to deal with the toxic effects of copper to cells. While the targets of copper toxicity are mainly restricted to the cytoplasm in Gram-positive bacteria, Gram-negative bacteria face the additional challenge of protecting their periplasm.
Since copper often enters the bacterial cell in an unspecific manner (utilizing other metal-uptake systems), bacteria cannot limit the amount of copper that enters the cytoplasm. Therefore, they had to develop mechanisms to expel excess cytoplasmic copper.
Widespread transporters utilized for copper efflux are members of the P1B-1-ATPase subfamily [Cu(I) transporters] of P1B-ATPases (formerly CPx-ATPases for the CPx-sequence motif in transmembrane helix VI) (83–90). In bacteria, the first copper-transporting ATPases were identified in Enterococcus hirae (91–95). E. hirae encodes two proteins, CopA and CopB, with one, CopB, belonging to the “classical” P1B-1-ATPases removing excess Cu(I) from the cytoplasm, while CopA belongs to the FixI/CopA2-like ATPases that are important for proper assembly of membrane-bound Cu-containing enzymes (85, 87, 96). P1B-ATPases have been identified to be involved in copper resistance in several Gram-positive bacteria (97, 98).
Similarly, in Gram-negative bacteria, P1B-1-ATPases have been shown to be involved in expelling excess copper from the cytoplasm. The best characterized are the chromosomally encoded copper homeostasis systems from E. coli, including copA encoding the P1B-ATPase CopA (99–101). In E. coli, copA is part of the CueR-regulon. The repressor CueR binds Cu(I) at zeptomolar concentrations and regulates the expression of copA and cueO (100, 102–105). While CopA in E. coli expels Cu(I) from the cytoplasm (106, 107), Cu(I) would now still be in the periplasm to cause damage to the cell. This necessitates CueO, which is a periplasmic multicopper oxidase that has been shown to oxidize Cu(I) into the less toxic Cu(II). CueO also oxidized enterobactin, enabling further sequestration of copper (108–116). The presence of genes encoding homologs of CopA and CueO is widespread among members of the Enterobacteriaceae, and this two-pronged approach appears to be a common strategy for copper detoxification (under “normal” aerobic circumstances) (49, 117–121). However, for CueO to be active, the organism requires the presence of oxygen. In the digestive tract of humans and/or animals, limited or no oxygen is available [and Cu(I) is also more stable than in an oxygenated environment]. Therefore, other strategies to keep the periplasm safe from copper toxicity need to be in place (99, 122). In E. coli, the CBA transport complex CusCBA is involved in Cu detoxification of the periplasm in the absence of CueO (99, 100, 123). Expression of cusCFBA is regulated by the two-component regulatory system CusRS, with CusS being a membrane-bound sensor kinase that detects periplasmic copper and relays the information to the cytoplasmic response regulator CusR, regulating cusCFBA expression (124, 125). It was found that in an anaerobic environment, expression was induced at much lower copper concentrations than in the presence of oxygen (100). The CusCBA complex is composed of the resistance nodulation cell division (RND)-protein CusA in the cytoplasmic membrane, the outer membrane factor (OMF) CusC in the outer membrane, and the membrane fusion protein CusB stabilizing the complex. Three molecules of CusA form a complex creating a “vestibule,” which forms a funnel to the channel formed by the CusC trimer. Cu(I) enters the funnel in the periplasm and via CusC is expelled into the surrounding medium. Cu(I) transport is energized via the proton gradient across the cytoplasmic membrane (101, 126–131). While CBA transport systems have been described for different types of heavy metals and organic compounds (including antibiotics), the presence of the periplasmic protein CusF is unique to transport systems involved in copper and silver transport (123). CusF could either act as a periplasmic chaperon delivering the metal ion to the CBA transport complex or function as a regulator of the transport (132–139). An alternative system for protection of the periplasm against copper-mediated toxicity under anaerobic conditions can be found in Salmonella. Here, the copper tolerance is linked to the presence of the periplasmic protein CueP. Expression of cueP in a cus-deletion strain of E. coli can partially restore its copper resistance under anaerobic conditions (120, 140–143). CueP seems to be confined to members of the genera Citrobacter and Salmonella.
Copper resistance systems encoded on mobile elements
Genomic islands involved in copper resistance of Enterobacteria
A plasmid-bound copper resistance determinant in Enterobacteria was first described on plasmid pRJ1004 from E. coli (144, 145). Copper resistance encoded on this plasmid is linked to the presence of the pco system, which encodes a two-component regulatory system, PcoRS, as well as to the structural proteins involved in copper resistance: PcoA, a periplasmic multicopper oxidase, the outer membrane protein PcoB, the inner membrane protein PcoD, and two periplasmic proteins, PcoC and PcoE (146–149). Expression of pcoE is regulated via a chromosomally encoded system, CusRS (124, 149), while expression of pcoABCD is regulated by PcoRS (147). Periplasmic PcoE is able to sequester excess copper, giving the cell temporary protection from the toxic effects of copper (149). Periplasmic PcoC also binds copper but is able to transfer it to membrane-bound PcoD to catalyze Cu(I) uptake into the cell. Once in the cytoplasm, Cu(I) is incorporated into the multicopper oxidase PcoA, which is then exported into the periplasm via the twin-arginine translocation pathway. Once in the periplasm, PcoA detoxifies Cu(I) by either oxidation of catechol siderophores and subsequent sequestration of Cu(I) or oxidation of Cu(I) to Cu(II). Cu(II) might be removed from the periplasm via PcoB (Fig. 1) (101, 146, 148, 150, 151). Located adjacent to pco on pRJ1004 is a sil determinant (150, 151). The sil determinant was first identified on plasmid pMG101 from Salmonella enterica serovar Typhimurium and was shown to confer silver resistance (152, 153). Expression of the structural genes of the sil determinant is regulated by the two-component regulatory system SilRS. In the presence of silver, silCFBA is expressed forming the SilCBA efflux complex (similar to CusCBA), allowing for the export of Ag(I) and Cu(I) from the periplasm. SilF is a periplasmic protein (like CusF) that is able to bind Ag(I) and either shuttles Ag(I) to the transport complex as substrate for export or could act as a regulatory protein of the function of SilCBA. In addition to the RND-dependent transporter SilCBA, the P1B-1-ATPases SilP, the periplasmic protein SilE, and a periplasmic protein, SilG, of unknown function are encoded as part of the sil determinant. Like PcoE, SilE is able to bind metal ions [Ag(I) and Cu(I)] and protect the periplasm from short-term metal stress. The P1B-1-ATPase SilP transports silver ions from the cytoplasm into the periplasm, from where they can be removed via SilCBA (150, 151, 153, 154). In contrast to copper, silver ions cannot undergo redox reactions, and therefore oxidation of Ag(I) as a detoxification mechanism is not an option for the cell. However, the entire sil resistance determinant also confers resistance to Cu(I). The overall 20-gene cluster pco/sil has been referred to as a copper-pathogenicity island (150) or a copper homeostasis and silver resistance island (151) (Fig. 1). The entire gene cluster has been identified in isolates of E. coli and S. Typhimurium, including E. coli O104:H4 from pigs fed a high-copper diet (155, 156).
FIGURE 1.
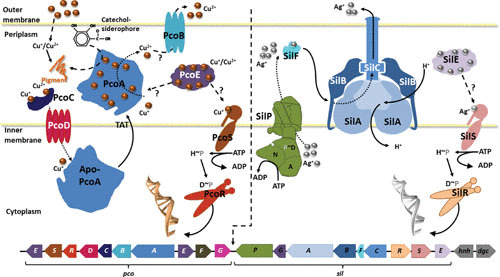
Copper fitness (or pathogenicity) island in Enterobacteriaceae. Genes and protein products of the enterobacterial copper fitness island composed of the pco- and sil-determinants. The genes, including their transcriptional/translational direction, are indicated below the illustration of the proposed or experimentally determined function of the proteins encoded by the pco/sil system. Refer to text for details. (Reprinted with permission [171].)
Analysis of the available completed genome and plasmid sequences of Enterobacteria (Table 5) revealed that the pco/sil gene cluster is frequently flanked by Tn7-like elements, allowing for transfer of the gene cluster (154, 157). Recent sequencing of E. coli J53 (pMG101) (NCTC 50110) showed that the pco/sil cluster has integrated from pMG101 (152, 153) into the bacterium’s genome (154). Interestingly, in some genera (Citrobacter, Kosakonia, and Raoultella), all of the identified pco/sil sequences are chromosomally encoded, while in others (Klyvera and Serratia), the sequences are carried only on plasmids. In most of the genera containing the pco/sil determinant, it can be carried on plasmids as well as on the bacterial chromosome, including strains that harbor a copy on each location. While in most cases the entire 20-gene cluster pco/sil is present, some strains of Enterobacter, Escherichia, Klebsiella, Pantoea, and Salmonella contain only one of the two resistance determinants, pco or sil (Table 5).
TABLE 5.
Distribution of pco/sil and yersiniabactin biosynthesis genes among Enterobacteriaceae
| Genusa | Number of sequences analyzedb | Occurrence of copper/silver tolerance determinants | ||||
|---|---|---|---|---|---|---|
| pco c | sil d | pco/sil e | Yersiniabactin synthesisf | pco/silP and yersiniabactin synthesisg | ||
| Citrobacter | 60 genomes | 10 | 10 | 10 | 3 | 0 |
| 50 plasmids | 0 | 0 | 0 | 0 | ||
| Cronobacter | 29 genomes | 1 | 1 | 1 | 0 | 0 |
| 22 plasmids | 6 | 6 | 6 | 0 | ||
| Enterobacter | 167 genomes | 22 | 28 | 21 | 0 | 0 |
| 121 plasmids | 6 | 7 | 6 | 0 | ||
| Escherichia | 946 genomes | 33 | 31 | 30 | 121 | 18 |
| 901 plasmids | 7 | 11 | 7 | 0 | ||
| Klebsiella | 674 genomes | 2 | 3 | 2 | 82 | 30 |
| 628 plasmids | 92 | 92 | 91 | 2 | ||
| Kluyvera | 6 genomes | 0 | 0 | 0 | 0 | 0 |
| 4 plasmids | 1 | 1 | 1 | 0 | ||
| Kosakonia | 10 genomes | 1 | 1 | 1 | 0 | 0 |
| 4 plasmids | 0 | 0 | 0 | 0 | ||
| Pantoea | 66 genomes | 0 | 0 | 0 | 0 | 0 |
| 61 plasmids | 0 | 1 | 0 | 0 | ||
| Raoultella | 9 genomes | 1 | 1 | 1 | 4 | 1 |
| 8 plasmids | 0 | 0 | 0 | 0 | ||
| Salmonella | 602 genomes | 14 | 13 | 13 | 5 | 0 |
| 369 plasmids | 4 | 6 | 4 | 7 | ||
| Serratia | 64 genomes | 0 | 0 | 0 | 0 | 0 |
| 27 plasmids | 2 | 2 | 2 | 0 | ||
| Yersinia | 192 genomes | 0 | 0 | 0 | 33 | 0 |
| 153 plasmids | 0 | 0 | 0 | 0 | ||
Genera of Enterobacteriaceae harboring pco, sil, and/or ybt.
Number of completed genomic and plasmid sequences of respective genera available for Microbial Genome BLAST (http://blast.ncbi.nlm.nih.gov; accessed 13 September 2017).
Analysis (blastn) using sil from pMG101 (GenBank accession number NG_035131.1 -[153]) as query.
Analysis (blastn) using pco (GenBank accession number X83541.1; [146]) and sil (accession number KC146966.1 [151]) from pRJ1004 as query.
Analysis (tblastn) using Ybt peptide/polyketide synthetase HMWP1 (GenBank accession number AAC69588.1 [246]) as query.
Number of strains harboring pco/sil and ybt with determinant being located on chromosome and/or plasmid, respectively.
Another strategy of Enterobacteriaceae to protect against the toxic effects of copper is the use of siderophores, especially yersiniabactin (158). The presence of the genes encoding the yersiniabactin synthesis pathway has previously been described as a virulence factor, but its presence in copper-resistant isolates indicates its importance in protection from copper toxicity. Yersiniabactin biosynthesis has been found in strains of E. coli, Salmonella, Yersinia pestis, and Klebsiella pneumoniae, including E. coli isolates from livestock fed high-copper diets (155, 159–163). Analysis of available completed genome and plasmid sequences of Enterobacteriaceae revealed the presence of the yersiniabactin biosynthesis pathways in the genera Citrobacter and Raoultella, in addition to the previously known genera. While commonly encoded on the bacterial chromosome, in some genera the yersiniabactin biosynthesis genes are located on plasmids as well (Table 5). Some strains of E. coli (including strains of enterohemorrhagic E. coli O104:H4), K. pneumoniae, and Raoultella have both the pco/sil gene cluster and the yersiniabactin biosynthesis determinant (Table 5).
Genomic islands involved in copper resistance of Enterococcus
In addition to Gram-negative bacteria, Gram-positive bacteria of the genus Enterococcus have frequently been isolated from livestock and shown to have high copper resistance (52, 121, 164, 165). Copper resistance in Enterococcus has been shown to be plasmid encoded and can be transferred via conjugation (121, 166–169). The first gene that could be linked to the plasmid-encoded copper resistance determinant in Enterococcus was tcrB. TcrB is a member of the P1B-ATPases of copper transporters, but due to the presence of a histidine-rich cytoplasmic N-terminus, a CPH-motif in transmembrane helix VI, and an MSXST-motif, it would be predicted to belong to the P1B-3-ATPase subfamily utilizing Cu(II) as substrate. TcrB is encoded as part of the tcrYAZB operon, which encodes TcrA, an additional P1B-ATPase of the P1B-1-ATPase subfamily with Cu(I) as the transported substrate, a cytoplasmic copper chaperon TcrZ, and TcrY, a copper-dependent regulator (150, 166, 170). Approximately 62% of Enterococcus strains that harbor transferrable copper resistance contained not only tcrYAZB but also cueO. This is in contrast to the strains analyzed in another study, where 22% only had tcrB and 16% encoded for CueO (121).
Sequence analysis of copper-resistant Enterococcus faecalis showed the presence of tcrZAYB and cueO as part of a larger gene cluster that also encoded a two-component regulatory system, CusRS, and an additional transcriptional regulator, CopY. Directly adjacent to copY, an additional P1B-1-ATPase named CopA is encoded, leading to the prediction of copA expression being regulated by CopY. Genes encoding several putative metal-binding proteins possibly serving as metal chaperones in the copper detoxification process are also in close proximity. The genes cusRS are directly downstream of cueO, suggesting regulation by this two-component regulatory system. Since the P1B-ATPases TcrA, TcrB, and CopA export copper ions from the cytoplasm, regulation of the respective genes by cytoplasmic copper-dependent regulators, CopY and TcrY, seems logical. In contrast, CueO, located just outside the cytoplasmic membrane, oxidizing Cu(I) to the less toxic Cu(II), appears to be regulated by CusRS sensing environmental copper concentrations (Fig. 2) (42, 150, 171). Other strains of E. faecalis and Enterococcus faecium contain related copper-resistance islands that vary slightly in the presence and arrangement of some of the genes in the copper-resistance island. These changes are probably at least in part due to the large number of putative transposons encoded in this region. BLAST analysis of the E. faecium HF50105 genome revealed a similar gene region in 5 of the 149 completed plasmid sequences of members of the genus Enterococcus, two belonging to Enterococcus durans, two to E. faecium, and one to Enterococcus gallinarum. No chromosomally encoded copper-resistance island could be identified. Further sequence analysis showed that this gene cluster is only present in the genus Enterococcus but not in other Firmicutes.
FIGURE 2.
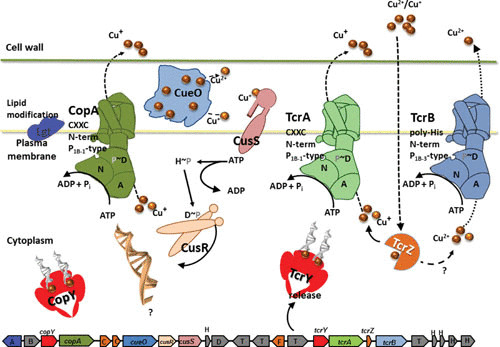
Copper fitness island in Enterococcus. Genes and proposed protein products of the copper island in Enterococcus faecium HF50105 (GenBank accession number AITS01000024). The genes, including their transcriptional/translational direction, are indicated below the illustration of the proposed function of the proteins. (Refer to text for details.) Adjacent to and separating the genes involved in copper resistance are genes encoding prolipoprotein diacylglyceryl transferase (A), integral membrane protein (B), predicted metal-binding protein/chaperone (C), hypothetical protein (H), transposase (T), and disrupted P-type ATPase (F) that have been identified. (Reprinted with permission [171].)
Zinc
Zinc bioavailability has changed drastically in course of Earth’s history. The greater bioavailability of zinc after the Great Oxidation Event is reflected in the increased use of zinc in eukaryotes (62, 63, 172). Zinc serves as a structural or catalytic component in hundreds of proteins and is one of the most abundant transition metals in a cell. The total amount of zinc in an E. coli cell was determined to be in a range similar to the cellular concentrations of calcium and iron (173). However, free Zn(II) in the cytoplasm is essentially nonexistent, since the Zn(II) regulators ZntR (efflux) and Zur (uptake) respond to free Zn(II) concentrations of 10−16 M in E. coli (173). This shows that bacteria very tightly control the uptake and efflux of Zn(II) to ensure proper cellular function while avoiding metal toxicity.
Zinc uptake
At least three systems (ABC-transporter, ZIP transporter, and phosphate-bound uptake) have been described to be involved in bacterial zinc uptake. Best understood are the systems involved in zinc uptake of E. coli. Under zinc-limiting conditions, E. coli utilizes the ABC (ATP-binding cassette) transporter ZnuABC. ZnuA is a zinc-binding protein located in the periplasm. ZnuB spans the cytoplasmic membrane and forms the channel to transport Zn(II) into the cytoplasm. ZnuC is bound to the cytoplasmic site on ZnuB, providing the energy for Zn(II)-transport via ATP hydrolysis (174, 175). Expression of the znu operon is regulated by Zur, which binds cytoplasmic Zn(II) and acts as repressor of znu by blocking the RNA polymerase from binding to the −10 region of the znuC promoter. Half-maximal repression by Zur in vitro has been shown to occur at a concentration of 2.0 (± 0.1) × 10−16 M free Zn(II), indicating that the presence of any free Zn(II) in the cell effectively turns off Zn(II) uptake by ZnuABC (173, 176–178). While genes encoding ZnuABC homologs have been identified in the genomes of many bacteria, it is absent in the genome of Cupriavidus metallidurans CH34, a bacterium originally isolated from a zinc decantation tank (179–181).
Under nonlimiting zinc conditions, E. coli utilizes a different uptake system, ZupT, the first identified bacterial member of the ZIP (ZRT-, IRT-like protein [ZRT, zinc-regulated transporter; IRT, iron-regulated transporter]) family of transport proteins (75, 182). Genes encoding ZupT have been identified in members of many bacterial phyla. In E. coli, zupT is expressed constitutively at a low level, and expression is therefore independent of external zinc availability. In contrast, in C. metallidurans, zupT expression is regulated by FurC, decreasing the amount of transporter under high zinc conditions and thereby limiting Zn(II) uptake (74, 181). It is not known if or how zupT expression is regulated in other microorganisms. While initially described as a zinc uptake system in E. coli, ZupT has broad substrate specificity and is also able to take up other metals, including Fe(II), Co(II), and Mn(II). Cadmium and copper also cross the cytoplasmic membrane via ZupT-mediated unspecific uptake (74). Several amino acids have been identified to contribute to ZupT’s substrate specificity (73). In uropathogenic E. coli as well as S. enterica, ZupT plays an important role in bacterial virulence, especially in the absence of znu (175, 183).
Under nonlimiting conditions, zinc can also enter E. coli as a metal-phosphate complex via PitA (inorganic phosphate uptake system) (184, 185). Magnesium uptake systems have been shown to have a broad substrate spectrum and are able to transport Zn(II) in addition to Mg(II) (180, 186–188). The presence of both zinc uptake systems is very common among bacteria.
Zinc efflux
Zinc resistance in bacteria is facilitated via efflux (189, 190). At least four systems (P1B-type ATPases, CDF transporters, 2-TM-GxN transporters, and CBA efflux systems) involved in transporting Zn(II) out of the cell have been identified. These systems can be encoded chromosomally or on plasmids.
The most effective transporters that export Zn(II) out of the cell are soft-metal P1B-type ATPases. Members of this family of membrane proteins contain six to eight transmembrane helices and cytoplasmic domains involved in ATP binding and hydrolysis as well as metal-binding domains. Based on structural features and the resulting substrate spectra, several subfamilies can be differentiated. Zn(II) is exported by members of the P1B-2-ATPase subgroup (83, 86). The best-characterized member of this group is ZntA from E. coli (191, 192). Members of this subgroup possess eight transmembrane helices, with a conserved CPC motif and Asp as transmembrane metal-binding sites. ZntA has a broad substrate specificity and can also transport Pb(II) and Cd(II), as can other members of this P1B-2-ATPase subgroup. Additionally, other metals such as Co(II), Ni(II), and Cu(II) bind to the transmembrane metal-binding domain, However, when these metals are bound, no ATP hydrolysis or transport takes place (193). Zn(II)-transporting ATPases can be chromosomally encoded, such as ZntA from E. coli, or encoded on plasmids, such as CadA from Staphylococcus aureus (191, 194, 195). Expression of zntA of E. coli is regulated by the MerR-like regulator ZntR. In the absence of Zn(II), apo-ZntR binds to the zntA promoter, repressing transcription. In the presence of Zn(II) in the cytoplasm, Zn-ZntR acts as a transcriptional activator and allows for transcription of zntA. In vitro analysis of ZntR affinity to Zn(II) revealed that concentration in the femtomolar range is required for ZntR to bind. However, in vivo studies identified ZntR responding to cytoplasmic free Zn(II) concentrations in the nanomolar range (173, 196). On the E. coli chromosome, zntR and zntA are not located in the same gene cluster (197). In contrast, on plasmid pI258 of S. aureus, the genes encoding the P1B-2-ATPase and its regulator are organized in an operon, cadCA. CadC is a member of the ArsR/SmtB-family and represses transcription in the absence of Zn(II), Cd(II), or Pb(II) (198). P1B-2-ATPases are highly efficient at removing Zn(II) from the cytoplasm, and deletion of the respective genes usually results in Zn(II) sensitivity of the organism (189, 191). Another P1B-2-ATPase of Staphylococcus is CzrC, which can be found in some methicillin-resistant strains encoded as part of SCCmec (staphylococcal cassette chromosome mec) (51, 199, 200). While often associated with SCCmec, czrC has also been identified in mecA-negative strains and on plasmids of S. aureus (201). Initially identified in S. aureus, czrC has also been found in isolates of other Staphylococcus species: S. haemolyticus, S. epidermitis, S. lentus, S. hominis, and S. hyicus (51, 199, 201–204). Out of the 489 completed Staphylococcus genomes in the NCBI database (as of 1 November 2017), 15 carry czrC (9 S. aureus, 1 S. condimenti, 3 S. epidermis, and 2 S. simulans), while the gene is not found on any of the 483 Staphylococcus plasmid sequences. While to date, the regulation of czrC expression has not yet been studied, it is likely regulated by an ArsR-like regulator encoded directly upstream of czrC.
A second family of membrane transporters linked to bacterial zinc export are cation diffusion facilitator (CDF) proteins, which contain six transmembrane helices with a C- and N-terminus located in the cytoplasm. E. coli contains two genes encoding CDF proteins. Both transporters, ZitB and FieF, have been shown to be involved in Zn(II) transport (205, 206). While deletion of zitB alone does not impact E. coli’s ability to handle elevated concentrations of Zn(II), a double mutant defective in both ZitB and the P1B-2-ATPase ZntA exhibits higher zinc sensitivity than a mutant strain defective only in ZntA (205). While no increase in zinc sensitivity was observed after deletion of fieF, everted membrane vesicles of E. coli GR362 (deficient in ZnuABC, ZupT, ZntA, ZntB, and ZitB) expressing fieF accumulated 65Zn(II). This transport was energized via the proton gradient, as has been shown for other CDF proteins (206). Using reporter gene fusion, expression of both genes, zitB and fieF, was found to be induced by Zn(II) and to a lesser extent by Cd(II). However, later analysis showed that mRNA levels of zitB remain constant after addition of Zn(II) and are elevated, but still independent of Zn(II) concentration, in a zntA deletion strain (196, 205, 206). It has been suggested that ZitB is involved in maintaining zinc homeostasis under “normal” conditions, while ZntA confers zinc resistance. CDF proteins involved in Zn(II) transport have been identified in many bacterial species. In S. aureus, the chromosomally encoded CDF transporter has been named ZntA and is encoded in an operon encoding ZntR in addition to ZntA. ZntA-null mutants were zinc sensitive, and expression of the operon was Zn(II) dependent and regulated by ZntR (207, 208).
First identified in S. Typhimurium, ZntB is a member of the 2-TM-GxN family of membrane transporters. This transporter family is widely found in bacteria, and ZntB-like proteins have been identified in many Proteobacteria. Deletion of zntB rendered S. Typhimurium more sensitive to Zn(II) and Cd(II). ZntB has two transmembrane helixes with the ∼270-amino acid N-terminus and the small C-terminus located in the cytoplasm. ZntB forms a pentamer, forming a cytoplasmic funnel. Each soluble domain monomer of ZntB binds three Zn2+, one in the funnel, while full-length ZntB binds four Zn2+ (209–213). In Agrobacterium tumefaciens, zntB is constitutively expressed but does not seem to contribute to the bacterium’s metal tolerance (214).
In contrast to the zinc efflux systems described so far, CBA efflux systems are protein complexes composed of a central transporter of the RND family, a membrane fusion protein, and an OMF. CBA efflux systems are involved in the transport of mono- and divalent heavy metals as well as organic compounds. The RND protein has 12 transmembrane helixes and two large periplasmic domains, which interact with the OMF. Both the RND and the OMF function as homotrimers, forming a channel starting in the periplasmic vestibule of the RND and reaching across the outer membrane via the OMF. The membrane fusion protein (hexamer) interacts with the RND protein and the OMF (190, 215, 216).
Of interest here are members of the heavy metal efflux (HME) family. Members of the HME-RND family are involved in the export of heavy metals, and several subfamilies can be differentiated based on sequence motifs and resulting metal substrates. Zn(II) transport has been credited to the HME1 subfamily (217). The best-characterized member of this family is CzcA from C. metallidurans CH34. The czc gene cluster is encoded on plasmid pMOL30 of C. metallidurans CH34 and allows growth in the presence of Co(II), Zn(II), and Cd(II), increasing the MIC by factors of 10, 25, and 100, respectively, compared to the plasmid-free strain AE104 (218). While export of the metal ions from the cytoplasm was assumed and data indicate transport by CzcA across the cytoplasmic membrane, the fact that a CDF protein as well as a P1B-4-ATPase are encoded as part of the czc gene cluster in addition to the two-component regulatory system CzcRS, which monitors the metal content within the periplasm, points toward export of the majority of metal ions from the periplasm into the surrounding environment by CzcCBA. As indicated, the CzcCBA system from C. metallidurans CH34 is the best-characterized zinc transporting CBA system to date. The structure of CzcCBA has not yet been solved (189, 215, 216, 219–224). The first structure of an RND protein was solved for AcrB, a multidrug transporter from E. coli (225, 226). To date, additional structures of RND transporters have been solved, including ZneA from C. metallidurans CH34. ZneA was shown to actively transport Zn(II) across the membrane via conformational changes within the three ZneA protomers. ZneA interacts with ZneB (membrane fusion protein) and ZneC (OMF) to form the membrane-spanning transport complex. The structure of ZneB was also shown to undergo conformational changes during export of the metal ions, reminiscent of CusB. However, the involvement of ZneCAB in zinc homeostasis of C. metallidurans CH34 is not understood (227, 228). Compared to CDF and P1B-type ATPases, CBA efflux systems are not as frequently present on bacterial genomes in bacterial handling of excess zinc.
Copper and Zinc Resistance Determinants: Link to Bacterial Virulence
In addition to these specific metal resistance determinants, there are numerous other genes that improve survival under elevated copper or zinc concentrations. This overview does not go into detail regarding these rather unspecific genes, but we refer the reader to a recent excellent study as an example (229).
The involvement of zinc and copper in bacterial killing is a recent important finding that is of relevance to understanding potential public health challenges arising from feeding livestock copper and zinc supplements. Professional phagocytes such as macrophages engulf pathogens and destroy them through elevation of Zn2+ and Cu+ concentrations in the phagosome, along with other mechanisms (oxidative burst, induction of Fe2+, and Mn2+ efflux) (230).
However, it is probable that the mechanisms macrophages use to kill infectious agents evolved in protozoa long before multicellular life arose and the need for macrophages appeared (231). This hypothesis was strengthened showing that copper (and probably zinc) poisoning is used in the model amoeba Dictyostelium discoideum to kill bacterial prey (232). Bacteria, in turn, were not just inert prey, and some succeeded in bypassing and resisting phagocytic cells by developing a number of strategies to avoid the protozoan killing mechanisms such as counteracting resistance systems specific for Zn and Cu and possibly other antimicrobial metals. This main concept of ongoing evolution driven by protozoan predation could explain the ongoing evolution of pathogens such as methicillin-resistant S. aureus, vancomycin-resistant Enterococcus, and extended spectrum beta-lactamase-producing E. coli, all of which pose a very important public health problem (233). Farmed animals such as pigs and poultry receive additional Zn and Cu in their diets due to supplementing elements in compound feed as well as medical remedies. Enteral bacteria in farmed animals have been shown to develop resistance to trace elements such as Zn and Cu. Resistance to Zn is often linked with resistance to methicillin in staphylococci, and Zn supplementation to animal feed may increase the proportion of multiresistant E. coli in the gut. Resistance to Cu in bacteria, enterococci in particular, is often associated with resistance to antimicrobial drugs such as macrolides and glycopeptides (e.g., vancomycin). Since Cu and Zn have an important role in protozoan predation, these metal resistances could make survival of these bacteria much more likely. These strains could then become pathogens after transfer to humans (234, 235).
CONCLUSIONS
Copper and zinc have been widely used in livestock feed both as growth promoters and as necessary supplements. In many countries the use of these metals appears to have increased due to a ban on the use of antibiotics. They are effective growth promoters, but it is not entirely clear how this is achieved. The potential negative impacts include zinc and copper contamination in the environment but more importantly the potential of coselecting antibiotic-resistance genes and possibly generating more pathogenic strains of medically relevant bacteria.
REFERENCES
- 1.Suttle NF. 2010. Mineral Nutrition of Livestock, 4th ed. CABI Publishing, Wallingford, United Kingdom. p 283–342. [Google Scholar]
- 2.European Commission Scientific Committee for Animal Nutrition (SCAN). 2003. Opinion of the Scientific Committee for Animal Nutrition on the use of copper in feedingstuffs.
- 3.European Commission. 2003. Commission Regulation (EC) no 1334/2003 of 25 July 2003. Amending the conditions for authorisation of a number of additives in feedingstuffs belonging to the group of trace elements.
- 4.EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). 2016. Revision of the currently authorised maximum copper content in complete feed. EFSA J 14:e04563. 10.2903/j.efsa.2016.4563. [DOI] [Google Scholar]
- 5.European Medicines Agency (EMA), Committee for Veterinary Medicinal Products (CVMP). 1998. Copper chloride, copper gluconate, copper heptanoate, copper oxide, copper methionate, copper sulfate and dicopper oxide: Summary Report. EMEA/MRL/431/98-FINAL.
- 6.Todd WR, Elvehjem CA, Hart EB. 1980. Nutrition classics. The American Journal of Physiology. Volume 107, 1934, pages 146–156. “Zinc in the nutrition of the rat” by W.R. Todd, C.A. Elvehjem and E.B. Hart. Nutr Rev 38:151–154 10.1111/j.1753-4887.1980.tb05879.x. [DOI] [PubMed] [Google Scholar]
- 7.Tucker HF, Salmon WD. 1955. Parakeratosis or zinc deficiency disease in the pig. Proc Soc Exp Biol Med 88:613–616 10.3181/00379727-88-21670. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.O’Dell BL, Newberne PM, Savage JE. 1958. Significance of dietary zinc for the growing chicken. J Nutr 65:503–518 10.1093/jn/65.4.503. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Chesters JK. 1983. Zinc metabolism in animals: pathology, immunology and genetics. J Inherit Metab Dis 6(Suppl 1):34–38 10.1007/BF01811321. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Jezyk PF, Haskins ME, MacKay-Smith WE, Patterson DF. 1986. Lethal acrodermatitis in bull terriers. J Am Vet Med Assoc 188:833–839. [PubMed] [PubMed] [Google Scholar]
- 11.Hambidge KM, Casey CE, Krebs NF. 1986. Zinc, p 1–137. In Mertz W (ed), Trace Elements in Human and Animal Nutrition, vol 2. Academic Press, San Diego, CA. 10.1016/B978-0-08-092469-4.50005-4 [DOI] [Google Scholar]
- 12.European Commission’s Scientific Committee for Animal Nutrition (SCAN). 2003. Opinion of the Scientific Committee for Animal Nutrition on the use of zinc in feedingstuffs.
- 13.EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). 2014. Scientific Opinion on the potential reduction of the currently authorised maximum zinc content in complete feed. EFSA J 12:3668. 10.2903/j.efsa.2014.3668. [DOI] [Google Scholar]
- 14.European Medicines Agency (EMA). 2016. Committee for Medicinal Products for Veterinary Use (CVMP) meeting 6–8 December 2016. http://www.ema.europa.eu/ema/index.jsp?curl=pages/news_and_events/events/2015/09/event_detail_001206.jsp&mid=WC0b01ac058004d5c3
- 15.DANMAP. 2014. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark.DANMAP, Copenhagen, Denmark. [Google Scholar]
- 16.BelVetSAC. 2015. Belgian Veterinary Surveillance of Antibacterial Consumption National consumption report.
- 17.BelVetSAC. 2014. Belgian Veterinary Surveillance of Antibacterial Consumption National consumption report.
- 18.Slifierz M. 2016. The effects of zinc therapy on the co-selection of methicillin-resistance in livestock-associated Staphylococcus aureus and the bacterial ecology of the porcine microbiota. PhD thesis. University of Guelph, Guelph, Canada. [Google Scholar]
- 19.Jacela JY, DeRouchey JM, Tokach MD, Goodband RD, Nelssen JL, Renter DG, Dritz SS. 2010. Feed additives for swine: fact sheets–high dietary levels of copper and zinc for young pigs, and phytase. J Swine Health Prod 18:87–91. [Google Scholar]
- 20.Poulsen HD. 1995. Zinc oxide for weanling piglets. Acta Agriculturae Scandinavica 45:159–167. [Google Scholar]
- 21.European Food Safety Authority. 2014. Dietary exposure to inorganic arsenic. EFSA J 12:3597. 10.2903/j.efsa.2014.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antman KH. 2001. Introduction: the history of arsenic trioxide in cancer therapy. Oncologist 6(Suppl 2):1–2 10.1634/theoncologist.6-suppl_2-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Smith AH, Marshall G, Yuan Y, Steinmaus C, Liaw J, Smith MT, Wood L, Heirich M, Fritzemeier RM, Pegram MD, Ferreccio C. 2014. Rapid reduction in breast cancer mortality with inorganic arsenic in drinking water. EBioMedicine 1:58–63 10.1016/j.ebiom.2014.10.005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nigra AE, Nachman KE, Love DC, Grau-Perez M, Navas-Acien A. 2017. Poultry consumption and arsenic exposure in the U.S. population. Environ Health Perspect 125:370–377. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldas D, Pestana IA, Almeida MG, Henry FC, Salomão MSMB, de Souza CMM. 2016. Risk of ingesting As, Cd, and Pb in animal products in north Rio de Janeiro state, Brazil. Chemosphere 164:508–515 10.1016/j.chemosphere.2016.08.130. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XY, Zhou MY, Li LL, Jiang YJ, Zou XT. 2017. Effects of arsenic supplementation in feed on laying performance, arsenic retention of eggs and organs, biochemical indices and endocrine hormones. Br Poult Sci 58:63–68 10.1080/00071668.2016.1216945. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Xi GF, Zhou SB, Ding HC, Yao CX, Kong JJ. 2014. Characteristics of arsenic content in the livestock farms’ surrounding environment in Shanghai suburbs. Huan Jing Ke Xue 35:1928–1932. (In Chinese.) [PubMed] [PubMed] [Google Scholar]
- 28.Jensen BB. 2016. Extensive literature search on the ‘effects of copper intake levels in the gut microbiota profile of target animals, in particular piglets’. EFSA Supporting Publications 13:1024E-n/a. 10.2903/sp.efsa.2016.EN-1024 [DOI] [Google Scholar]
- 29.European Medicines Agency (EMA), Committee for Veterinary Medicinal Products (CVMP). 2016. European public MRL assessment report (EPMAR): copper carbonate (all food producing species). EMA/CVMP/ 758734/2015.
- 30.Ou D, Li D, Cao Y, Li X, Yin J, Qiao S, Wu G. 2007. Dietary supplementation with zinc oxide decreases expression of the stem cell factor in the small intestine of weanling pigs. J Nutr Biochem 18:820–826 10.1016/j.jnutbio.2006.12.022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Schulte JN, Brockmann GA, Kreuzer-Redmer S. 2016. Feeding a high dosage of zinc oxide affects suppressor of cytokine gene expression in Salmonella Typhimurium infected piglets. Vet Immunol Immunopathol 178:10–13 10.1016/j.vetimm.2016.06.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.European Medicines Agency (EMA), Committee for Veterinary Medicinal Products (CVMP). 1996. Zinc salts. EMEA/MRL/113/96-FINAL:
- 33.United States Department of Agriculture (USDA). 2017. Livestock slaughter 2016 summary. USDA National Agricultural Statistics Service, Washington, DC.
- 34.European Medicines Agency (EMA), European Surveillance of Veterinary Antimicrobial Consumption (ESVAC). 2016. Sales of veterinary antimicrobial agents in 29 European countries in 2014. EMA/61769/2016.
- 35.National Research Council, Committee on Nutrient Requirements of Swine. 2012. Nutrient Requirements of Swine. National Academies Press, Washington, DC. [Google Scholar]
- 36.Stolte J, Tesfai M, Øygarden L, Kværnø S, Keizer J, Verheijen F, Panagos P, Ballabio C, Hessel R (ed). 2016. Soil Threats in Europe. EUR 27607 EN. doi:10.2788/488054 (print); 10.2788/828742 (online). [DOI] [Google Scholar]
- 37.Jensen J, Larsen MM, Bak J. 2016. National monitoring study in Denmark finds increased and critical levels of copper and zinc in arable soils fertilized with pig slurry. Environ Pollut 214:334–340 10.1016/j.envpol.2016.03.034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Bak JL, Jensen J, Larsen MM. 2015. Belysning af kobber- og zinkindholdet i jord. Indhold og udvikling i kvadratnettet og måling på udvalgte brugstyper. Aarhus Universitet, DCE – Nationalt Center for Miljø og Energi, 72 s. - Videnskabelig rapport fra DCE - Nationalt Center for Miljø og Energi nr. 159.
- 39.Joint Research Center (JRC) European Union. 2010. Risk assessment report. CAS: 7440-66-6; EINECS No: 231-175-3 ZINC METAL.
- 40.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2005. DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob Agents Chemother 49:4681–4688 10.1128/AAC.49.11.4681-4688.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salem AZM, Ammar H, Lopez S, Gohar YM, González JS. 2011. Sensitivity of ruminal bacteria isolates of sheep, cattle and buffalo to some heavy metals. Anim Feed Sci Technol 163:143–149 10.1016/j.anifeedsci.2010.10.017. [DOI] [Google Scholar]
- 42.Chen S, Li X, Sun G, Zhang Y, Su J, Ye J. 2015. Heavy metal induced antibiotic resistance in bacterium LSJC7. Int J Mol Sci 16:23390–23404 10.3390/ijms161023390. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriques I, Tacão M, Leite L, Fidalgo C, Araújo S, Oliveira C, Alves A. 2016. Co-selection of antibiotic and metal(loid) resistance in Gram-negative epiphytic bacteria from contaminated salt marshes. Mar Pollut Bull 109:427–434 10.1016/j.marpolbul.2016.05.031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Hasman H, Franke S, Rensing C. 2005. Resistance to metals used in agricultural production, p 99–114. In Aarestrup FM, Wegener HC (ed), Antimicrobial Resistance in Bacteria of Animal Origin. ASM Press, Washington, DC. [Google Scholar]
- 45.Jacob ME, Fox JT, Nagaraja TG, Drouillard JS, Amachawadi RG, Narayanan SK. 2010. Effects of feeding elevated concentrations of copper and zinc on the antimicrobial susceptibilities of fecal bacteria in feedlot cattle. Foodborne Pathog Dis 7:643–648 10.1089/fpd.2009.0401. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Bednorz C, Oelgeschläger K, Kinnemann B, Hartmann S, Neumann K, Pieper R, Bethe A, Semmler T, Tedin K, Schierack P, Wieler LH, Guenther S. 2013. The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int J Med Microbiol 303:396–403 10.1016/j.ijmm.2013.06.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Toroglu S, Dincer S. 2009. Heavy metal resistances of Enterobacteriaceae from Aksu River (Turkey) polluted with different sources. Asian J Chem 21:411–420. [Google Scholar]
- 48.Kumar PA, Joseph B, Patterson J. 2011. Antibiotic and heavy metal resistance profile of pathogens isolated from infected fish in Tuticorin, south-east coast of India. Indian J Fish 58:121–125. [Google Scholar]
- 49.Deredjian A, Colinon C, Brothier E, Favre-Bonté S, Cournoyer B, Nazaret S. 2011. Antibiotic and metal resistance among hospital and outdoor strains of Pseudomonas aeruginosa. Res Microbiol 162:689–700 10.1016/j.resmic.2011.06.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Wei LS, Musa N, Wee W. 2010. Bacterial flora from a healthy freshwater Asian sea bass (Lates calcarifer) fingerling hatchery with emphasis on their antimicrobial and heavy metal resistance pattern. Vet Arh 80:411–420. [Google Scholar]
- 51.Cavaco LM, Hasman H, Stegger M, Andersen PS, Skov R, Fluit AC, Ito T, Aarestrup FM. 2010. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob Agents Chemother 54:3605–3608 10.1128/AAC.00058-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fard RM, Heuzenroeder MW, Barton MD. 2011. Antimicrobial and heavy metal resistance in commensal enterococci isolated from pigs. Vet Microbiol 148:276–282 10.1016/j.vetmic.2010.09.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Medardus JJ, Molla BZ, Nicol M, Morrow WM, Rajala-Schultz PJ, Kazwala R, Gebreyes WA. 2014. In-feed use of heavy metal micronutrients in U.S. swine production systems and its role in persistence of multidrug-resistant salmonellae. Appl Environ Microbiol 80:2317–2325 10.1128/AEM.04283-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mourão J, Marçal S, Ramos P, Campos J, Machado J, Peixe L, Novais C, Antunes P. 2016. Tolerance to multiple metal stressors in emerging non-typhoidal MDR Salmonella serotypes: a relevant role for copper in anaerobic conditions. J Antimicrob Chemother 71:2147–2157 10.1093/jac/dkw120. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Amachawadi RG, Scott HM, Vinasco J, Tokach MD, Dritz SS, Nelssen JL, Nagaraja TG. 2015. Effects of in-feed copper, chlortetracycline, and tylosin on the prevalence of transferable copper resistance gene, tcrB, among fecal enterococci of weaned piglets. Foodborne Pathog Dis 12:670–678 10.1089/fpd.2015.1961. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Aarestrup FM, Hasman H. 2004. Susceptibility of different bacterial species isolated from food animals to copper sulphate, zinc chloride and antimicrobial substances used for disinfection. Vet Microbiol 100:83–89 10.1016/j.vetmic.2004.01.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Hacioglu N, Tosunoglu M. 2014. Determination of antimicrobial and heavy metal resistance profiles of some bacteria isolated from aquatic amphibian and reptile species. Environ Monit Assess 186:407–413 10.1007/s10661-013-3385-y. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Fang L, Li X, Li L, Li S, Liao X, Sun J, Liu Y. 2016. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci Rep 6:25312 10.1038/srep25312. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akinbowale OL, Peng H, Grant P, Barton MD. 2007. Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. Int J Antimicrob Agents 30:177–182 10.1016/j.ijantimicag.2007.03.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.He Y, Jin L, Sun F, Hu Q, Chen L. 2016. Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from fresh shrimps in Shanghai fish markets, China. Environ Sci Pollut Res Int 23:15033–15040 10.1007/s11356-016-6614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Q, Chen L. 2016. Virulence and antibiotic and heavy metal resistance of Vibrio parahaemolyticus isolated from crustaceans and shellfish in Shanghai, China. J Food Prot 79:1371–1377 10.4315/0362-028X.JFP-16-031. [DOI] [PubMed] [Google Scholar]
- 62.Williams RJ, Fraústo Da Silva JJ. 2003. Evolution was chemically constrained. J Theor Biol 220:323–343 10.1006/jtbi.2003.3152. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Hong Enriquez RP, Do TN. 2012. Bioavailability of metal ions and evolutionary adaptation. Life (Basel) 2:274–285 10.3390/life2040274. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA 106:8344–8349 10.1073/pnas.0812808106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dupont CL, Grass G, Rensing C. 2011. Copper toxicity and the origin of bacterial resistance: new insights and applications. Metallomics 3:1109–1118 10.1039/c1mt00107h. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol 192:2512–2524 10.1128/JB.00058-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chillappagari S, Miethke M, Trip H, Kuipers OP, Marahiel MA. 2009. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J Bacteriol 191:2362–2370 10.1128/JB.01616-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirooka K, Edahiro T, Kimura K, Fujita Y. 2012. Direct and indirect regulation of the ycnKJI operon involved in copper uptake through two transcriptional repressors, YcnK and CsoR, in Bacillus subtilis. J Bacteriol 194:5675–5687 10.1128/JB.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamada-Ankei T, Iwasaki H, Mori T. 1977. Production of copper coproporphyrin III by Bacillus cereus. I. Purification and identification of copper coproporphyrin III. J Biochem 81:835–842 10.1093/oxfordjournals.jbchem.a131547. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Balasubramanian R, Kenney GE, Rosenzweig AC. 2011. Dual pathways for copper uptake by methanotrophic bacteria. J Biol Chem 286:37313–37319 10.1074/jbc.M111.284984. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188:7242–7256 10.1128/JB.00837-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whiting GC, Rowbury RJ. 1995. Increased resistance of Escherichia coli to acrylic acid and to copper ions after cold-shock. Lett Appl Microbiol 20:240–242 10.1111/j.1472-765X.1995.tb00437.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Taudte N, Grass G. 2010. Point mutations change specificity and kinetics of metal uptake by ZupT from Escherichia coli. Biometals 23:643–656 10.1007/s10534-010-9319-z. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. 2005. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol 187:1604–1611 10.1128/JB.187.5.1604-1611.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guerinot ML. 2000. The ZIP family of metal transporters. Biochim Biophys Acta 1465:190–198 10.1016/S0005-2736(00)00138-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Cha JS, Cooksey DA. 1993. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl Environ Microbiol 59:1671–1674. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fitch MW, Graham DW, Arnold RG, Agarwal SK, Phelps P, Speitel GE Jr, Georgiou G. 1993. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol 59:2771–2776. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anttila J, Heinonen P, Nenonen T, Pino A, Iwaï H, Kauppi E, Soliymani R, Baumann M, Saksi J, Suni N, Haltia T. 2011. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim Biophys Acta 1807:311–318 10.1016/j.bbabio.2010.12.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD, Taylor A. 1998. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol 180:3606–3613. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PM. 2004. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305:1612–1615 10.1126/science.1098322. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Zahn JA, DiSpirito AA. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J Bacteriol 178:1018–1029 10.1128/jb.178.4.1018-1029.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicolaisen K, Hahn A, Valdebenito M, Moslavac S, Samborski A, Maldener I, Wilken C, Valladares A, Flores E, Hantke K, Schleiff E. 2010. The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochim Biophys Acta 1798:2131–2140 10.1016/j.bbamem.2010.07.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Argüello JM. 2003. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biol 195:93–108 10.1007/s00232-003-2048-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Argüello JM, González-Guerrero M. 2008. Cu+ -ATPases brake system. Structure 16:833–834 10.1016/j.str.2008.05.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Argüello JM, González-Guerrero M, Raimunda D. 2011. Bacterial transition metal P(1B)-ATPases: transport mechanism and roles in virulence. Biochemistry 50:9940–9949 10.1021/bi201418k. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Argüello JM, Eren E, González-Guerrero M. 2007. The structure and function of heavy metal transport P1B-ATPases. Biometals 20:233–248 10.1007/s10534-006-9055-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Raimunda D, González-Guerrero M, Leeber BW III, Argüello JM. 2011. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals 24:467–475 10.1007/s10534-010-9404-3. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenzweig AC, Argüello JM. 2012. Toward a molecular understanding of metal transport by P(1B)-type ATPases. Curr Top Membr 69:113–136 10.1016/B978-0-12-394390-3.00005-7. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Solioz M, Odermatt A, Krapf R. 1994. Copper pumping ATPases: common concepts in bacteria and man. FEBS Lett 346:44–47 10.1016/0014-5793(94)00316-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Solioz M, Vulpe C. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci 21:237–241 10.1016/S0968-0004(96)20016-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Odermatt A, Suter H, Krapf R, Solioz M. 1992. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann N Y Acad Sci 671(1 Ion-Motive AT):484–486 10.1111/j.1749-6632.1992.tb43836.x. [DOI] [PubMed] [Google Scholar]
- 92.Solioz M, Odermatt A. 1995. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem 270:9217–9221 10.1074/jbc.270.16.9217. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Solioz M, Stoyanov JV. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev 27:183–195 10.1016/S0168-6445(03)00053-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Odermatt A, Krapf R, Solioz M. 1994. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem Biophys Res Commun 202:44–48 10.1006/bbrc.1994.1891. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Odermatt A, Solioz M. 1995. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J Biol Chem 270:4349–4354 10.1074/jbc.270.9.4349. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.González-Guerrero M, Raimunda D, Cheng X, Argüello JM. 2010. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol 78:1246–1258 10.1111/j.1365-2958.2010.07402.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Gaballa A, Cao M, Helmann JD. 2003. Two MerR homologues that affect copper induction of the Bacillus subtilis copZA operon. Microbiology 149:3413–3421 10.1099/mic.0.26225-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Reyes A, Leiva A, Cambiazo V, Méndez MA, González M. 2006. Cop-like operon: structure and organization in species of the Lactobacillale order. Biol Res 39:87–93 10.4067/S0716-97602006000100010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Grass G, Rensing C. 2001. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol 183:2145–2147 10.1128/JB.183.6.2145-2147.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Outten FW, Huffman DL, Hale JA, O’Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677 10.1074/jbc.M104122200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Rensing C, Grass G. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27:197–213 10.1016/S0168-6445(03)00049-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Outten FW, Outten CE, Hale J, O’Halloran TV. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J Biol Chem 275:31024–31029 10.1074/jbc.M006508200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Petersen C, Møller LB. 2000. Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene 261:289–298 10.1016/S0378-1119(00)00509-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Stoyanov JV, Hobman JL, Brown NL. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol 39:502–511 10.1046/j.1365-2958.2001.02264.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, Mondragón A. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383–1387 10.1126/science.1085950. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA 97:652–656 10.1073/pnas.97.2.652. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fan B, Rosen BP. 2002. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J Biol Chem 277:46987–46992 10.1074/jbc.M208490200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Grass G, Rensing C. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286:902–908 10.1006/bbrc.2001.5474. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Kim C, Lorenz WW, Hoopes JT, Dean JF. 2001. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J Bacteriol 183:4866–4875 10.1128/JB.183.16.4866-4875.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, Tollin G, Rensing C, Montfort WR. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci USA 99:2766–2771 10.1073/pnas.052710499. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roberts SA, Wildner GF, Grass G, Weichsel A, Ambrus A, Rensing C, Montfort WR. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J Biol Chem 278:31958–31963 10.1074/jbc.M302963200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Grass G, Thakali K, Klebba PE, Thieme D, Müller A, Wildner GF, Rensing C. 2004. Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J Bacteriol 186:5826–5833 10.1128/JB.186.17.5826-5833.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Singh SK, Grass G, Rensing C, Montfort WR. 2004. Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol 186:7815–7817 10.1128/JB.186.22.7815-7817.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sakurai T, Kataoka K. 2007. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem Rec 7:220–229 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- 115.Djoko KY, Chong LX, Wedd AG, Xiao Z. 2010. Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J Am Chem Soc 132:2005–2015 10.1021/ja9091903. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Singh SK, Roberts SA, McDevitt SF, Weichsel A, Wildner GF, Grass GB, Rensing C, Montfort WR. 2011. Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence. J Biol Chem 286:37849–37857 10.1074/jbc.M111.293589. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lim SY, Joe MH, Song SS, Lee MH, Foster JW, Park YK, Choi SY, Lee IS. 2002. CuiD is a crucial gene for survival at high copper environment in Salmonella enterica serovar Typhimurium. Mol Cells 14:177–184. [PubMed] [PubMed] [Google Scholar]
- 118.Kosman DJ. 2010. Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J Biol Inorg Chem 15:15–28 10.1007/s00775-009-0590-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Rademacher C, Moser R, Lackmann JW, Klinkert B, Narberhaus F, Masepohl B. 2012. Transcriptional and posttranscriptional events control copper-responsive expression of a Rhodobacter capsulatus multicopper oxidase. J Bacteriol 194:1849–1859 10.1128/JB.06274-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nies DH, Herzberg M. 2013. A fresh view of the cell biology of copper in enterobacteria. Mol Microbiol 87:447–454 10.1111/mmi.12123. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Silveira E, Freitas AR, Antunes P, Barros M, Campos J, Coque TM, Peixe L, Novais C. 2014. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. J Antimicrob Chemother 69:899–906 10.1093/jac/dkt479. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Padilla-Benavides T, George Thompson AM, McEvoy MM, Argüello JM. 2014. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J Biol Chem 289:20492–20501 10.1074/jbc.M114.577668. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185:3804–3812 10.1128/JB.185.13.3804-3812.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Munson GP, Lam DL, Outten FW, O’Halloran TV. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol 182:5864–5871 10.1128/JB.182.20.5864-5871.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Franke S, Grass G, Nies DH. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965–972 10.1099/00221287-147-4-965. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. 2007. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem 282:35695–35702 10.1074/jbc.M703937200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Su CC, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. 2009. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol 393:342–355 10.1016/j.jmb.2009.08.029. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467:484–488 10.1038/nature09395. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kulathila R, Kulathila R, Indic M, van den Berg B. 2011. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS One 6:e15610 10.1371/journal.pone.0015610. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. 2011. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562 10.1038/nature09743. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim EH, Nies DH, McEvoy MM, Rensing C. 2011. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 193:2381–2387 10.1128/JB.01323-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loftin IR, Franke S, Roberts SA, Weichsel A, Héroux A, Montfort WR, Rensing C, McEvoy MM. 2005. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44:10533–10540 10.1021/bi050827b. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Kittleson JT, Loftin IR, Hausrath AC, Engelhardt KP, Rensing C, McEvoy MM. 2006. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both CuI and AgI. Biochemistry 45:11096–11102 10.1021/bi0612622. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, Focia P, Spiro TG, Penner-Hahn JE, O’Halloran TV. 2008. Cu(I) recognition via cation-pi and methionine interactions in CusF. Nat Chem Biol 4:107–109 10.1038/nchembio.2007.57. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. 2008. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47:11408–11414 10.1021/bi801638m. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bersch B, Derfoufi KM, De Angelis F, Auquier V, Ekendé EN, Mergeay M, Ruysschaert JM, Vandenbussche G. 2011. Structural and metal binding characterization of the C-terminal metallochaperone domain of membrane fusion protein SilB from Cupriavidus metallidurans CH34. Biochemistry 50:2194–2204 10.1021/bi200005k. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Mealman TD, Bagai I, Singh P, Goodlett DR, Rensing C, Zhou H, Wysocki VH, McEvoy MM. 2011. Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry 50:2559–2566 10.1021/bi102012j. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chacón KN, Mealman TD, McEvoy MM, Blackburn NJ. 2014. Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins. Proc Natl Acad Sci USA 111:15373–15378 10.1073/pnas.1411475111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meir A, Natan A, Moskovitz Y, Ruthstein S. 2015. EPR spectroscopy identifies Met and Lys residues that are essential for the interaction between the CusB N-terminal domain and metallochaperone CusF. Metallomics 7:1163–1172 10.1039/C5MT00053J. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Pontel LB, Soncini FC. 2009. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol Microbiol 73:212–225 10.1111/j.1365-2958.2009.06763.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J Biol Chem 285:25259–25268 10.1074/jbc.M110.145953. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yun BY, Piao S, Kim YG, Moon HR, Choi EJ, Kim YO, Nam BH, Lee SJ, Ha NC. 2011. Crystallization and preliminary X-ray crystallographic analysis of Salmonella Typhimurium CueP. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:675–677 10.1107/S1744309111010645. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yoon BY, Yeom JH, Kim JS, Um SH, Jo I, Lee K, Kim YH, Ha NC. 2014. Direct ROS scavenging activity of CueP from Salmonella enterica serovar Typhimurium. Mol Cells 37:100–108 10.14348/molcells.2014.2238. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Williams JR, Morgan AG, Rouch DA, Brown NL, Lee BT. 1993. Copper-resistant enteric bacteria from United Kingdom and Australian piggeries. Appl Environ Microbiol 59:2531–2537. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tetaz TJ, Luke RK. 1983. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol 154:1263–1268. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brown NL, Barrett SR, Camakaris J, Lee BT, Rouch DA. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol Microbiol 17:1153–1166 10.1111/j.1365-2958.1995.mmi_17061153.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Rouch DA, Brown NL. 1997. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology 143:1191–1202 10.1099/00221287-143-4-1191. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Lee SM, Grass G, Rensing C, Barrett SR, Yates CJ, Stoyanov JV, Brown NL. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem Biophys Res Commun 295:616–620 10.1016/S0006-291X(02)00726-X. [DOI] [PubMed] [Google Scholar]
- 149.Zimmermann M, Udagedara SR, Sze CM, Ryan TM, Howlett GJ, Xiao Z, Wedd AG. 2012. PcoE: a metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J Inorg Biochem 115:186–197 10.1016/j.jinorgbio.2012.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 150.Hao X, Lüthje FL, Qin Y, McDevitt SF, Lutay N, Hobman JL, Asiani K, Soncini FC, German N, Zhang S, Zhu YG, Rensing C. 2015. Survival in amoeba: a major selection pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island”. Appl Microbiol Biotechnol 99:5817–5824 10.1007/s00253-015-6749-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Staehlin BM, Gibbons JG, Rokas A, O’Halloran TV, Slot JC. 2016. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in Enterobacteria. Genome Biol Evol 8:811–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McHugh GL, Moellering RC, Hopkins CC, Swartz MN. 1975. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet 1:235–240 10.1016/S0140-6736(75)91138-1. [DOI] [PubMed] [Google Scholar]
- 153.Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat Med 5:183–188 10.1038/5545. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Randall CP, Gupta A, Jackson N, Busse D, O’Neill AJ. 2015. Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J Antimicrob Chemother 70:1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lüthje FL, Hasman H, Aarestrup FM, Alwathnani HA, Rensing C. 2014. Genome sequences of two copper-resistant Escherichia coli strains isolated from copper-fed pigs. Genome Announc 2:e01341 10.1128/genomeA.01341-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qin Y, Hasman H, Aarestrup FM, Alwathnani HA, Rensing C. 2014. Genome sequences of three highly copper-resistant Salmonella enterica subsp. I serovar Typhimurium strains isolated from pigs in Denmark. Genome Announc 2:e01334-14 10.1128/genomeA.01334-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Peters JE, Fricker AD, Kapili BJ, Petassi MT. 2014. Heteromeric transposase elements: generators of genomic islands across diverse bacteria. Mol Microbiol 93:1084–1092. [DOI] [PubMed] [Google Scholar]
- 158.Chaturvedi KS, Henderson JP. 2014. Pathogenic adaptations to host-derived antibacterial copper. Front Cell Infect Microbiol 4:3 10.3389/fcimb.2014.00003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schubert S, Dufke S, Sorsa J, Heesemann J. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol 51:837–848 10.1046/j.1365-2958.2003.03870.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 160.Rakin A, Schneider L, Podladchikova O. 2012. Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front Cell Infect Microbiol 2:151/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, Grassl GA, Gal-Mor O. 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol 16:977–994 10.1111/1462-2920.12351. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Chaturvedi KS, Hung CS, Giblin DE, Urushidani S, Austin AM, Dinauer MC, Henderson JP. 2014. Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem Biol 9:551–561 10.1021/cb400658k. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fodah RA, Scott JB, Tam HH, Yan P, Pfeffer TL, Bundschuh R, Warawa JM. 2014. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS One 9:e107394 10.1371/journal.pone.0107394. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, Donabedian S, Jensen LB, Francia MV, Baquero F, Peixe L. 2011. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol 49:925–931 10.1128/JCM.01750-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang S, Wang D, Wang Y, Hasman H, Aarestrup FM, Alwathnani HA, Zhu YG, Rensing C. 2015. Genome sequences of copper resistant and sensitive Enterococcus faecalis strains isolated from copper-fed pigs in Denmark. Stand Genomic Sci 10:35-015-0021-1. 10.1186/s40793-015-0021-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hasman H, Aarestrup FM. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob Agents Chemother 46:1410–1416 10.1128/AAC.46.5.1410-1416.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Amachawadi RG, Shelton NW, Jacob ME, Shi X, Narayanan SK, Zurek L, Dritz SS, Nelssen JL, Tokach MD, Nagaraja TG. 2010. Occurrence of tcrB, a transferable copper resistance gene, in fecal enterococci of swine. Foodborne Pathog Dis 7:1089–1097 10.1089/fpd.2010.0540. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Amachawadi RG, Scott HM, Alvarado CA, Mainini TR, Vinasco J, Drouillard JS, Nagaraja TG. 2013. Occurrence of the transferable copper resistance gene tcrB among fecal enterococci of U.S. feedlot cattle fed copper-supplemented diets. Appl Environ Microbiol 79:4369–4375 10.1128/AEM.00503-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pasquaroli S, Di Cesare A, Vignaroli C, Conti G, Citterio B, Biavasco F. 2014. Erythromycin- and copper-resistant Enterococcus hirae from marine sediment and co-transfer of erm(B) and tcrB to human Enterococcus faecalis. Diagn Microbiol Infect Dis 80:26–28 10.1016/j.diagmicrobio.2014.06.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Hasman H. 2005. The tcrB gene is part of the tcrYAZB operon conferring copper resistance in Enterococcus faecium and Enterococcus faecalis. Microbiology 151:3019–3025 10.1099/mic.0.28109-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 171.Rensing C, Alwathnani HA, McDevitt SF. 2016. The copper metallome in prokaryotic cells, p 161–173. In de Bruijn FJ (ed), Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. John Wiley & Sons, Inc., Hoboken, NJ 10.1002/9781119004813.ch13 [DOI] [Google Scholar]
- 172.Hedges SB, Chen H, Kumar S, Wang DY, Thompson AS, Watanabe H. 2001. A genomic timescale for the origin of eukaryotes. BMC Evol Biol 1:4 10.1186/1471-2148-1-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Outten CE, O’Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492 10.1126/science.1060331. [PubMed] [DOI] [PubMed] [Google Scholar]
- 174.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210 10.1046/j.1365-2958.1998.00883.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 175.Sabri M, Houle S, Dozois CM. 2009. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect Immun 77:1155–1164 10.1128/IAI.01082-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332 10.1074/jbc.M001775200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.Sigdel TK, Easton JA, Crowder MW. 2006. Transcriptional response of Escherichia coli to TPEN. 188:6709–6713. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gilston BA, Wang S, Marcus MD, Canalizo-Hernández MA, Swindell EP, Xue Y, Mondragón A, O’Halloran TV. 2014. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol 12:e1001987 10.1371/journal.pbio.1001987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, Nies DH. 2011. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J Bacteriol 193:4652–4663 10.1128/JB.05293-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Herzberg M, Bauer L, Nies DH. 2014. Deletion of the zupT gene for a zinc importer influences zinc pools in Cupriavidus metallidurans CH34. Metallomics 6:421–436 10.1039/c3mt00267e. [PubMed] [DOI] [PubMed] [Google Scholar]
- 182.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. 2002. ZupT is a Zn(II) uptake system in Escherichia coli. J Bacteriol 184:864–866 10.1128/JB.184.3.864-866.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cerasi M, Liu JZ, Ammendola S, Poe AJ, Petrarca P, Pesciaroli M, Pasquali P, Raffatellu M, Battistoni A. 2014. The ZupT transporter plays an important role in zinc homeostasis and contributes to Salmonella enterica virulence. Metallomics 6:845–853 10.1039/C3MT00352C. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jackson RJ, Binet MR, Lee LJ, Ma R, Graham AI, McLeod CW, Poole RK. 2008. Expression of the PitA phosphate/metal transporter of Escherichia coli is responsive to zinc and inorganic phosphate levels. FEMS Microbiol Lett 289:219–224 10.1111/j.1574-6968.2008.01386.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Beard SJ, Hashim R, Wu G, Binet MR, Hughes MN, Poole RK. 2000. Evidence for the transport of zinc(II) ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol Lett 184:231–235 10.1111/j.1574-6968.2000.tb09019.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Webb M. 1970. Interrelationships between the utilization of magnesium and the uptake of other bivalent cations by bacteria. Biochim Biophys Acta 222:428–439 10.1016/0304-4165(70)90133-9. [DOI] [PubMed] [Google Scholar]
- 187.Krom BP, Huttinga H, Warner JB, Lolkema JS. 2002. Impact of the Mg(2+)-citrate transporter CitM on heavy metal toxicity in Bacillus subtilis. Arch Microbiol 178:370–375 10.1007/s00203-002-0465-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Knoop V, Groth-Malonek M, Gebert M, Eifler K, Weyand K. 2005. Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol Genet Genomics 274:205–216 10.1007/s00438-005-0011-x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 189.Legatzki A, Grass G, Anton A, Rensing C, Nies DH. 2003. Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J Bacteriol 185:4354–4361 10.1128/JB.185.15.4354-4361.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nies DH. 2007. How cells control zinc homeostasis. Science 317:1695–1696 10.1126/science.1149048. [PubMed] [DOI] [PubMed] [Google Scholar]
- 191.Rensing C, Mitra B, Rosen BP. 1998. A Zn(II)-translocating P-type ATPase from Proteus mirabilis. Biochem Cell Biol 76:787–790 10.1139/o98-071. [PubMed] [DOI] [PubMed] [Google Scholar]
- 192.Rensing C, Ghosh M, Rosen BP. 1999. Families of soft-metal-ion-transporting ATPases. J Bacteriol 181:5891–5897. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sharma R, Rensing C, Rosen BP, Mitra B. 2000. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem 275:3873–3878 10.1074/jbc.275.6.3873. [PubMed] [DOI] [PubMed] [Google Scholar]
- 194.Smith K, Novick RP. 1972. Genetic studies on plasmid-linked cadmium resistance in Staphylococcus aureus. J Bacteriol 112:761–772. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nucifora G, Chu L, Misra TK, Silver S. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA 86:3544–3548 10.1073/pnas.86.10.3544. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang D, Hosteen O, Fierke CA. 2012. ZntR-mediated transcription of zntA responds to nanomolar intracellular free zinc. J Inorg Biochem 111:173–181 10.1016/j.jinorgbio.2012.02.008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 10.1126/science.277.5331.1453. [PubMed] [DOI] [PubMed] [Google Scholar]
- 198.Yoon KP, Silver S. 1991. A second gene in the Staphylococcus aureus cadA cadmium resistance determinant of plasmid pI258. J Bacteriol 173:7636–7642 10.1128/jb.173.23.7636-7642.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Argudín MA, Butaye P. 2016. Dissemination of metal resistance genes among animal methicillin-resistant coagulase-negative staphylococci. Res Vet Sci 105:192–194 10.1016/j.rvsc.2016.02.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Slifierz MJ, Park J, Friendship RM, Weese JS. 2014. Zinc-resistance gene CzrC identified in methicillin-resistant Staphylococcus hyicus isolated from pigs with exudative epidermitis. Can Vet J 55:489–490. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 201.Vandendriessche S, Vanderhaeghen W, Larsen J, de Mendonça R, Hallin M, Butaye P, Hermans K, Haesebrouck F, Denis O. 2014. High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCCmec remnant DNA in MSSA CC398. J Antimicrob Chemother 69:355–362 10.1093/jac/dkt366. [PubMed] [DOI] [PubMed] [Google Scholar]
- 202.Vandendriessche S, Vanderhaeghen W, Soares FV, Hallin M, Catry B, Hermans K, Butaye P, Haesebrouck F, Struelens MJ, Denis O. 2013. Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J Antimicrob Chemother 68:1510–1516 10.1093/jac/dkt047. [PubMed] [DOI] [PubMed] [Google Scholar]
- 203.Agersø Y, Hasman H, Cavaco LM, Pedersen K, Aarestrup FM. 2012. Study of methicillin resistant Staphylococcus aureus (MRSA) in Danish pigs at slaughter and in imported retail meat reveals a novel MRSA type in slaughter pigs. Vet Microbiol 157:246–250 10.1016/j.vetmic.2011.12.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 204.Nair R, Thapaliya D, Su Y, Smith TC. 2014. Resistance to zinc and cadmium in Staphylococcus aureus of human and animal origin. Infect Control Hosp Epidemiol 35(Suppl 3):S32–S39 10.1086/677834. [PubMed] [DOI] [PubMed] [Google Scholar]
- 205.Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. 2001. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol 183:4664–4667 10.1128/JB.183.15.4664-4667.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D. 2005. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch Microbiol 183:9–18 10.1007/s00203-004-0739-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Xiong A, Jayaswal RK. 1998. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J Bacteriol 180:4024–4029. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Singh VK, Xiong A, Usgaard TR, Chakrabarti S, Deora R, Misra TK, Jayaswal RK. 1999. ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol Microbiol 33:200–207 10.1046/j.1365-2958.1999.01466.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 209.Van Pham ST, Engman H, Dahlgren LG, Cornvik T, Eshaghi S. 2010. A systematic approach to isolate mono-disperse membrane proteins: purification of zinc transporter ZntB. Protein Expr Purif 72:48–54 10.1016/j.pep.2010.02.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 210.Papp-Wallace KM, Maguire ME. 2007. Bacterial homologs of eukaryotic membrane proteins: the 2-TM-GxN family of Mg(2+) transporters. Mol Membr Biol 24:351–356 10.1080/09687680701441883. [PubMed] [DOI] [PubMed] [Google Scholar]
- 211.Caldwell AM, Smith RL. 2003. Membrane topology of the ZntB efflux system of Salmonella enterica serovar Typhimurium. J Bacteriol 185:374–376 10.1128/JB.185.1.374-376.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wan Q, Ahmad MF, Fairman J, Gorzelle B, de la Fuente M, Dealwis C, Maguire ME. 2011. X-ray crystallography and isothermal titration calorimetry studies of the Salmonella zinc transporter ZntB. Structure 19:700–710 10.1016/j.str.2011.02.011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Worlock AJ, Smith RL. 2002. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J Bacteriol 184:4369–4373 10.1128/JB.184.16.4369-4373.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chaoprasid P, Nookabkaew S, Sukchawalit R, Mongkolsuk S. 2015. Roles of Agrobacterium tumefaciens C58 ZntA and ZntB and the transcriptional regulator ZntR in controlling Cd2+/Zn2+/Co2+ resistance and the peroxide stress response. Microbiology 161:1730–1740 10.1099/mic.0.000135. [PubMed] [DOI] [PubMed] [Google Scholar]
- 215.Nies DH, Silver S. 1995. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol 14:186–199 10.1007/BF01569902. [PubMed] [DOI] [PubMed] [Google Scholar]
- 216.Legatzki A, Franke S, Lucke S, Hoffmann T, Anton A, Neumann D, Nies DH. 2003. First step towards a quantitative model describing Czc-mediated heavy metal resistance in Ralstonia metallidurans. Biodegradation 14:153–168 10.1023/A:1024043306888. [PubMed] [DOI] [PubMed] [Google Scholar]
- 217.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 218.Nies D, Mergeay M, Friedrich B, Schlegel HG. 1987. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol 169:4865–4868 10.1128/jb.169.10.4865-4868.1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Saier MH Jr, Tam R, Reizer A, Reizer J. 1994. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol 11:841–847 10.1111/j.1365-2958.1994.tb00362.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 220.Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. 1995. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol 14:142–153 10.1007/BF01569896. [PubMed] [DOI] [PubMed] [Google Scholar]
- 221.Grosse C, Anton A, Hoffmann T, Franke S, Schleuder G, Nies DH. 2004. Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch Microbiol 182:109–118 10.1007/s00203-004-0670-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 222.Zoropogui A, Gambarelli S, Covès J. 2008. CzcE from Cupriavidus metallidurans CH34 is a copper-binding protein. Biochem Biophys Res Commun 365:735–739 10.1016/j.bbrc.2007.11.030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 223.Scherer J, Nies DH. 2009. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol Microbiol 73:601–621 10.1111/j.1365-2958.2009.06792.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 224.von Rozycki T, Nies DH. 2009. Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie van Leeuwenhoek 96:115–139 10.1007/s10482-008-9284-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 225.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA 106:7173–7178 10.1073/pnas.0900693106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179 10.1038/nature05076. [PubMed] [DOI] [PubMed] [Google Scholar]
- 227.De Angelis F, Lee JK, O’Connell JD III, Miercke LJ, Verschueren KH, Srinivasan V, Bauvois C, Govaerts C, Robbins RA, Ruysschaert JM, Stroud RM, Vandenbussche G. 2010. Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc Natl Acad Sci USA 107:11038–11043 10.1073/pnas.1003908107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pak JE, Ekendé EN, Kifle EG, O’Connell JD III, De Angelis F, Tessema MB, Derfoufi KM, Robles-Colmenares Y, Robbins RA, Goormaghtigh E, Vandenbussche G, Stroud RM. 2013. Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter. Proc Natl Acad Sci USA 110:18484–18489 10.1073/pnas.1318705110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vaccaro BJ, Lancaster WA, Thorgersen MP, Zane GM, Younkin AD, Kazakov AE, Wetmore KM, Deutschbauer A, Arkin AP, Novichkov PS, Wall JD, Adams MW. 2016. Novel metal cation resistance systems from mutant fitness analysis of denitrifying Pseudomonas stutzeri. Appl Environ Microbiol 82:6046–6056 10.1128/AEM.01845-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Djoko KY, Ong CL, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961 10.1074/jbc.R115.647099. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.German N, Doyscher D, Rensing C. 2013. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol 8:1257–1264 10.2217/fmb.13.100. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.Hao X, Lüthje F, Rønn R, German NA, Li X, Huang F, Kisaka J, Huffman D, Alwathnani HA, Zhu YG, Rensing C. 2016. A role for copper in protozoan grazing: two billion years selecting for bacterial copper resistance. Mol Microbiol 102:628–641 10.1111/mmi.13483. [PubMed] [DOI] [PubMed] [Google Scholar]
- 233.Poole K. 2017. At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol 25:820–832 10.1016/j.tim.2017.04.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 234.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71:20–28 10.1128/AEM.71.1.20-28.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chandrangsu P, Rensing C, Helmann JD. 2017. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15:338–350 10.1038/nrmicro.2017.15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tan H, Deng Z, Cao L. 2009. Isolation and characterization of actinomycetes from healthy goat faeces. Lett Appl Microbiol 49:248–253 10.1111/j.1472-765X.2009.02649.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 237.Choudhury P, Kumar R. 1998. Multidrug- and metal-resistant strains of Klebsiella pneumoniae isolated from Penaeus monodon of the coastal waters of deltaic Sundarban. Can J Microbiol 44:186–189 10.1139/w97-144. [PubMed] [DOI] [PubMed] [Google Scholar]
- 238.Dutta GN, Devriese LA. 1981. Sensitivity and resistance to growth promoting agents in animal lactobacilli. J Appl Bacteriol 51:283–288 10.1111/j.1365-2672.1981.tb01243.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 239.Siddaramappa S, Challacombe JF, Duncan AJ, Gillaspy AF, Carson M, Gipson J, Orvis J, Zaitshik J, Barnes G, Bruce D, Chertkov O, Detter JC, Han CS, Tapia R, Thompson LS, Dyer DW, Inzana TJ. 2011. Horizontal gene transfer in Histophilus somni and its role in the evolution of pathogenic strain 2336, as determined by comparative genomic analyses. BMC Genomics 12:570. 10.1186/1471-2164-12-570. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Deus D, Krischek C, Pfeifer Y, Sharifi AR, Fiegen U, Reich F, Klein G, Kehrenberg C. 2017. Comparative analysis of the susceptibility to biocides and heavy metals of extended-spectrum β-lactamase-producing Escherichia coli isolates of human and avian origin, Germany. Diagn Microbiol Infect Dis 88:88–92 10.1016/j.diagmicrobio.2017.01.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 241.Agga GE, Scott HM, Amachawadi RG, Nagaraja TG, Vinasco J, Bai J, Norby B, Renter DG, Dritz SS, Nelssen JL, Tokach MD. 2014. Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Prev Vet Med 114:231–246 10.1016/j.prevetmed.2014.02.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 242.Campos J, Cristino L, Peixe L, Antunes P. 2016. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill 21:30270 10.2807/1560-7917.ES.2016.21.26.30270. [PubMed] [DOI] [PubMed] [Google Scholar]
- 243.Gómez-Sanz E, Kadlec K, Feßler AT, Zarazaga M, Torres C, Schwarz S. 2013. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother 57:3275–3282 10.1128/AAC.00171-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Argudín MA, Lauzat B, Kraushaar B, Alba P, Agerso Y, Cavaco L, Butaye P, Porrero MC, Battisti A, Tenhagen BA, Fetsch A, Guerra B. 2016. Heavy metal and disinfectant resistance genes among livestock-associated methicillin-resistant Staphylococcus aureus isolates. Vet Microbiol 191:88–95 10.1016/j.vetmic.2016.06.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 245.Cavaco LM, Hasman H, Aarestrup FM, Wagenaar JA, Graveland H, Veldman K, Mevius D, Fetsch A, Tenhagen B-A, Concepcion Porrero M, Dominguez L, Granier SA, Jouy E, Butaye P, Kaszanyitzky E, Dán A, Zmudzki J, Battisti A, Franco A, Schwarz S, Gutierrez M, Weese JS, Cui S, Pomba C, Members of MRSA-CG. 2011. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet Microbiol 150:344–348 10.1016/j.vetmic.2011.02.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 246.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol 5:573–586 10.1016/S1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]


