Abstract
Concerted integration of retrovirus DNA termini into the host chromosome in vivo requires specific interactions between the cis-acting attachment (att) sites at the viral termini and the viral integrase (IN) in trans. In this study, reconstruction experiments with purified avian myeloblastosis virus (AMV) IN and retrovirus-like donor substrates containing wild-type and mutant termini were performed to map the internal att DNA sequence requirements for concerted integration, here termed full-site integration. The avian retrovirus mutations were modeled after internal att site mutations studied at the in vivo level with human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV). Systematic overlapping 4-bp deletions starting at nucleotide positions 7, 8, and 9 in the U3 terminus had a decreasing detrimental gradient effect on full-site integration, while more internal 4-bp deletions had little or no effect. This decreasing detrimental gradient effect was measured by the ability of mutant U3 ends to interact with wild-type U3 ends for full-site integration in trans. Modification of the highly conserved C at position 7 on the catalytic strand to either A or T resulted in the same severe decrease in full-site integration as the 4-bp deletion starting at this position. These studies suggest that nucleotide position 7 is crucial for interactions near the active site of IN for integration activity and for communication in trans between ends bound by IN for full-site integration. The ability of AMV IN to interact with internal att sequences to mediate full-site integration in vitro is similar to the internal att site requirements observed with MLV and HIV-1 in vivo and with their preintegration complexes in vitro.
The integration of the retrovirus linear DNA genome into the host chromosome is mediated by the retrovirus integrase (IN) (for reviews, see references 4, 17, 24, and 35). The integration process involves the concerted insertion of the viral DNA termini by IN into cellular DNA, here termed full-site integration. In vivo, the first ∼15 bp of the long terminal repeat (LTR) sequences at the viral DNA termini is required to various degrees for both 3′ OH trimming of the blunt-ended termini and subsequent insertion of the recessed termini into the host chromosome (3, 4, 8, 10, 16, 28, 32, 33). These ∼15-bp LTR end nucleotides are termed attachment (att) site sequences (4).
Several pathways have been used to investigate retrovirus full-site integration at the in vitro level. The most direct route has been by the characterization of preintegration complexes (PIC) isolated from the cytoplasm of newly virus-infected cells (3, 5, 8, 14, 25, 39). Studies with several retroviruses have suggested that the LTR ends in the PIC are held together by a protein bridge presumably promoted by IN (3, 31, 40), although one or more cellular proteins also may have similar bridging functions (13, 26, 39, 40). Two 3′ OH recessed termini interact to form a functional PIC for integration in vitro; this entity is termed an intasome (3, 8, 39, 40). Intasomes are characterized as having bacteriophage Mu-mediated PCR (MM-PCR)-protected footprints spanning several hundred base pairs at each end of the viral DNA and enhancements near the LTR ends, both requiring functional IN. The relationship of the relatively large MM-PCR-protected footprints (∼200 bp) observed in the PIC to the ∼15-bp attachment (att) site sequence requirement for integration in vivo is unknown (3, 8).
Reconstitution experiments with purified IN and linear retrovirus-like DNA substrates (∼500 bp in length) is another approach for investigating the full-site integration process at the in vitro level. Avian myeloblastosis virus (AMV) IN purified from virions (36–38) or recombinant Rous sarcoma virus (RSV) Prague A (PrA) IN (30) efficiently mediates concerted insertion of recessed LTR ends from two donor molecules (bimolecular reaction) into circular target DNA. The reaction products are characterized by direct physical assays—restriction enzyme analysis and agarose gel electrophoresis. Recombinant RSV Schmidt-Ruppin B IN also promotes full-site integration by inserting the termini of one donor substrate ∼300 bp in length (unimolecular reaction) into target DNA, and this reaction is enhanced by a cellular protein, HMG-I(Y) (1, 22). Recombinant simian immunodeficiency virus IN by itself (18) and human immunodeficiency virus type 1 (HIV-1) IN requiring the viral nucleocapsid (7) or HMG-1(Y) (22) for enhanced activities can also mediate full-site integration in vitro. In contrast, in the case of recombinant HIV-1 IN, HMG-1(Y) and several other DNA binding proteins do not enhance full-site integration (7).
In this study, we produced a series of 4-bp deletions and several point mutations near the LTR ends of linear 480-bp substrates to determine which internal att site sequences are essential for full-site integration in vitro. The design of the avian internal LTR mutations was based on LTR mutations produced in different retroviruses examined at the in vivo level (3, 8, 28, 32, 33, 39, 40) and on isolated PIC containing mutant viral DNA (3, 8, 40). Systematic 4-bp deletions starting at nucleotides 7, 8, and 9 had a decreasing detrimental gradient effect on full-site and half-site integration activities, while other, more internal 4-bp deletions had little effect on these activities. For reference, half-site integration is defined as the insertion of a single LTR end by IN into a target. The highly conserved pyrimidine nucleotide (C) at position 7 from the end is also essential for mediating efficient trans interactions between two LTR ends bound by IN for full-site integration. The LTR sequence requirements observed with avian IN for full-site integration in vitro appear similar to the LTR DNA requirements observed in vivo and with the purified PIC in vitro.
MATERIALS AND METHODS
Construction of LTR donor mutants.
A series of mutations (Fig. 1B and C) were introduced into one LTR end of a linear 480-bp DNA donor that contained wild-type (wt) U3 sequences at both ends of the DNA (Fig. 1A) (38). The parental wt U3-U3 donor fragment was located at the NdeI site of pUC19. Oligonucleotides containing the appropriate mutations were used as primers for PCR amplification of the U3-U3 donor. In all cases, the modified U3 end was located adjacent to the internal BglII site (Fig. 1B), allowing for uniform analysis of the donor-target recombinants following BglII digestion and agarose gel electrophoresis (see Fig. 4) (36–38). After amplification and insertion of the NdeI-digested PCR products into pUC19, the appropriate plasmids were screened and sequenced at both LTR ends to verify the modified and unmodified U3 LTR sequences. Following NdeI digestion, the 480-bp fragment was isolated from various mutants on agarose gels and purified.
FIG. 1.
Small deletions just distal to the LTR end affect full-site integration. (A) A 480-bp LTR donor containing wt U3 LTR sequences at both ends was produced by NdeI digestion, as indicated by the arrows at the CA dinucleotide. The internal region (indicated by dashes) of the donor contains the supF gene (37). (B) A series of 4-bp deletions were introduced only into the U3 LTR that maps closest to the unique BglII site in the donor. Only the catalytic strand is shown, and the recessed AC dinucleotide in 3′ OH recessed ends is underlined. The 4-bp deletions are indicated by dashes. The numbering of the positions starts from the blunt end of the donor. SupF, supF gene used for genetic selection. (C) Nucleotide 7 (C) was changed to an A or a T at one U3 terminus, while the other U3 LTR sequence was not modified. In the U3+A@P7 mutation, only an A nucleotide was inserted prior to the C nucleotide at position 7 while the other U3 LTR end was not modified.
FIG. 4.
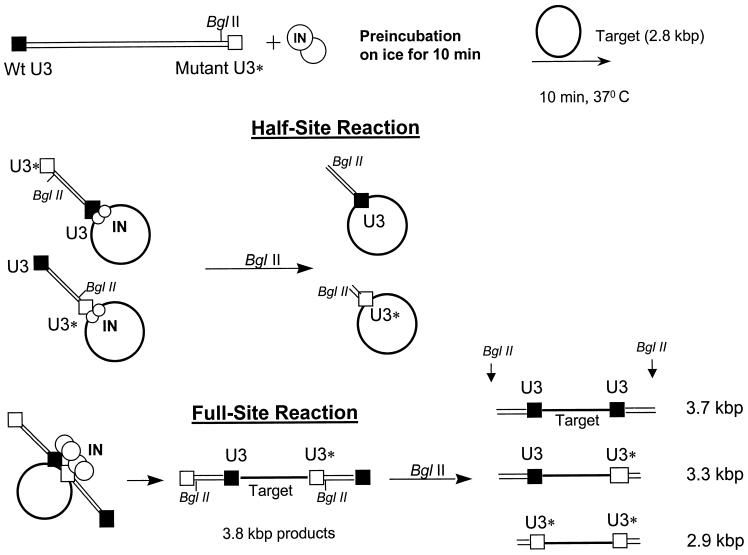
Diagram for half-site and full-site integration with BglII digestion of donor-target products. The LTR donor is depicted containing a wt U3 end (black box) and a mutant U3* end (open box). A unique BglII site is located ∼40 bp from the mutant U3* end. After preincubation with IN, the integration reaction is started by the addition of the target. The half-site reaction is depicted by the insertion of one LTR donor per target. The full-site reaction is depicted by the concerted insertion of two separate LTR donors per target, producing linear 3.8-kb products. BglII digestion of both donor-target products reveals the frequency of LTR end usage for both the half-site and the full-site integration reactions (37). The sizes of the fragments resulting from BglII digestion are depicted on the bottom right.
Full-site integration assay.
The conditions for the full-site integration assay were previously described (36). All of the reactions were performed in the linear range for strand transfer activities (∼40 to 50 nM IN for 10 min at 37°C). At 50 nM IN, the IN dimer/donor LTR end molar ratio is 12:1. In some experiments, the concentration of IN was varied. The donor/target molar ratio was 1:1. After strand transfer, the samples were processed and subjected to agarose gel electrophoresis. Digestion of the donor-target products by BglII was previously described (37). BglII digestion of the full-site products obtained with wt and mutant donors produced the same 3.7-, 3.3-, and 2.9-kbp fragments (see Fig. 4) (18, 29). U3* represents a modified wt U3 LTR sequence (see Fig. 3 to 5). The amount of 5′ 32P-labeled donor incorporated into target DNA as either a half-site or a full-site integration product was determined with a Molecular Dynamics STORM PhosphorImager (37).
FIG. 3.
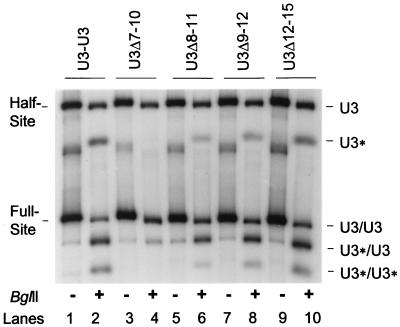
Defining internal LTR sequence requirements for communication in trans between LTR ends for full-site integration. The donor-target products from the indicated wt and mutant donor reactions were digested with BglII and subjected to 1.5% agarose gel electrophoresis. The five donors are identified at the top. The samples were not (−) or were (+) digested with BglII. The undigested half-site and full-site products are identified on the left, and the digested products are identified on the right. The products containing the various U3 LTR deletions are identified by U3*. U3/U3, U3*/U3, and U3*/U3* are 3.7, 3.3, and 2.9 kbp long, respectively (see Fig. 4). Note that only U3/U3 full-site products are possible with the wt-U3-U3 donor (lane 2). The donor-donor products and the donor substrates were run off of the 1.5% agarose gel.
FIG. 5.
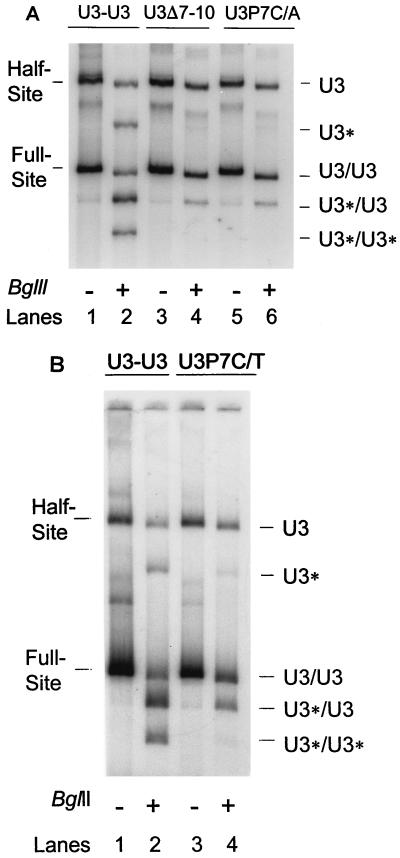
Conserved nucleotide 7 (C) of the U3 LTR is critical for trans interactions between LTR ends for full-site integration. (A) The U3-U3, U3Δ7-10, and U3P7C/A donors were used as substrates for insertion into pGEM with AMV IN (50 nM). The samples were not (−) or were (+) digested with BglII prior to electrophoresis on 1.5% agarose. The undigested half-site and full-site products are identified on the left, and the digested fragments are identified on the right. The donor-target products containing the various modified U3 LTR ends are identified by U3* (Fig. 4). (B) The U3-U3 and U3P7C/T donors were analyzed as described for panel A. The concentration of IN was 25 nM. The nomenclature is the same as that used in panel A.
Nitrocellulose filter DNA binding assays.
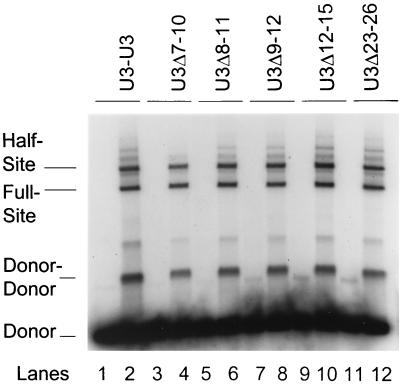
Each 480-bp donor was cut with HinfI, releasing three fragments, a wt U3 end (253 bp), a fragment derived from the middle of the donor and containing two HinfI ends (36 bp), and a mutant U3 end (191 bp). The fragments were 5′ end labeled with [γ-32P]ATP by use of polynucleotide kinase. Alternatively, the 480-bp LTR donors were labeled at the NdeI site prior to digestion with HinfI, eliminating the labeling of the internal 36-bp DNA fragment. In either case, the homogeneous mixture of labeled fragments was preincubated at 0°C for 10 min with various concentrations of IN under normal strand transfer conditions with 0.33 M NaCl. The IN-DNA complexes were immediately filtered at room temperature onto nitrocellulose filters equilibrated with 0.33 M NaCl buffer. The IN-DNA samples were washed with two independent 0.1-ml washes of the same buffer. The bound DNA was quantified by Cerenkov counting and subsequently eluted from the filters with a sodium dodecyl sulfate-containing buffer. After ethanol precipitation, the entire sample from each binding assay was loaded onto 2% agarose gels, which permitted separation of the above three fragments. The gels were dried, and the quantities of the retained fragments were defined with the PhosphorImager. The percentage of each fragment bound by IN was determined and compared to data for DNA controls not bound to IN. The controls were DNA in buffer alone and processed on agarose gels as described above or DNA alone placed on a filter without washing and then processed. Purified HinfI fragments containing only one LTR end were used in previous integration assays (29).
Purification of IN.
AMV IN (19) and recombinant RSV PrA IN (30) were purified as previously described.
RESULTS
Small deletions of nucleotides just distal to the LTR ends affect full-site integration activity.
We had established that nucleotides 5 and 6 from the blunt-ended LTR ends play a critical role in the preferential usage of wt U3 over wt U5 LTR ends by avian IN for 3′ OH processing and full-site integration in vitro (29, 38). To investigate the effect of LTR sequences distal to nucleotides 5 and 6 on full-site integration, a series of 4-bp deletions were constructed at one end of a U3-U3 donor while the wt U3 sequence was maintained at the other end (Fig. 1B). The LTR deletion mutations started at nucleotide position 7 and overlapped each other. In all deletion mutants, the modified sequences showed no resemblance to the wt U3 sequences (Fig. 1B). The numbering of the nucleotide positions started at the blunt-ended LTR end.
The wt U3-U3 donor and the donors containing a wt U3 end and a U3 end with a 4-bp deletion (Fig. 1B) were preincubated with AMV IN on ice for 10 min. Supercoiled DNA (2,867 bp) was added as a target, followed by immediate incubation for 10 min at 37°C. The total amounts of donor DNA incorporated into the target DNA for the wt U3-U3 and U3Δ7-10 donors were ∼12 and ∼8%, respectively, with the strand transfer activities of the other LTR deletion mutants occurring near or between these two values (Fig. 2). The percentages of donor molecules inserted into the products (half-site and full-site) (see Fig. 4) were generally equivalent. These results show that there is no major variation in the observed half-site and full-site products produced by the wt and mutant donors prior to BglII digestion.
FIG. 2.
Strand transfer activities of wt and mutant donors. The 480-bp LTR donors containing 4-bp deletions in one U3 LTR were preincubated with AMV IN (50 nM) on ice prior to strand transfer (10 min at 37°C). The donor substrates with only the modified LTR end showing are indicated at the top (Fig. 1). The samples were subjected to 1% agarose gel electrophoresis and exposed to X-ray film. The odd-numbered lanes contain no IN, while the even-numbered lanes contain IN. The half-site and full-site integration products as well as the donor-donor products and the donors are indicated on the left. The half-site reaction identifies the insertion of only one LTR end per target molecule (see Fig. 4).
To compare full-site integration activities between wt and mutant LTR donor ends, nearly equivalent quantities (counts per minute) of each reaction, similar to those shown in Fig. 2, were not or were subjected to BglII restriction digestion (Fig. 3). The undigested half-site and full-site products produced by each donor are shown in the odd-numbered lanes of Fig. 3. The full-site products containing the different LTR ends were subjected to BglII digestion, producing three possible bimolecular full-site integration products (Fig. 3, even-numbered lanes) (36–38). Note that the wt U3-U3 donor in Fig. 3 makes only U3/U3 (see below) full-site integration products which, upon BglII digestion, migrate at the same 3.7-, 3.3-, and 2.9-kbp positions as the three products produced by mutant donors (Fig. 4 and Table 1) (29). Within each mutant donor set, the integration reactions occurring between two wt termini (designated U3/U3), one mutant end with one wt end (U3*/U3), and two mutant ends (U3*/U3*) are shown on the right side of Fig. 3 and illustrated in Fig. 4.
TABLE 1.
Relative efficiencies of half-site and full-site integration reactions with 4-bp LTR deletion mutantsa
| Integration reaction | Relative efficiency of the reaction with the following donor substrate:
|
|||||
|---|---|---|---|---|---|---|
| U3-U3 | U3Δ7-10 | U3Δ8-11 | U3Δ9-12 | U3Δ12-15 | U3Δ23-26 | |
| Half-site | ||||||
| U3 | 1 | 1 | 1 | 1 | 1 | 1 |
| U3* | 0.70 | 0.06 | 0.23 | 0.19 | 0.39 | 0.68 |
| Full-site | ||||||
| U3/U3 | 1 | 1 | 1 | 1 | 1 | 1 |
| U3*/U3 | 1.6 | 0.2 | 0.8 | 1.0 | 1.5 | 1.7 |
| U3*/U3* | 0.70 | 0.03 | 0.16 | 0.24 | 0.49 | 0.67 |
The average of three independent experiments was used to calculate the relative efficiencies of the half-site and full-site integration reactions for each donor set. The activity of the wt U3 end was set to 1 for each reaction with each donor substrate because the observed wt U3/U3 full-site reactions vary significantly between donors (Fig. 3). The U3-U3 donor substrate set contains only wt U3 events in all five reactions. U3* represents the mutant LTR end reactions with wt U3 or with itself.
In comparison to the wt U3-U3 donor BglII digestion pattern (Fig. 3, lane 2), only the U3Δ7-10 donor (lane 4) showed a major loss of activity for the U3*/U3* and U3*/U3 full-site products and the U3* half-site product (Table 1). The adjacent U3 LTR deletion mutants, U3Δ8-11 (Fig. 3, lane 6) and U3Δ9-12 (lane 8), showed a less extensive loss of activity for these matching full-site reactions. The U3Δ12-15 deletion (Fig. 3, lane 10) and the U3Δ23-26 deletion (data not shown) (Table 1 and Fig. 1) had nearly wt activities for both their full-site and their half-site integration reactions. The results suggest that the critical interactions for IN to mediate full-site (or half-site) integration map to at least position 7 (C nucleotide) from the LTR end, with nucleotides 8 to 12 also influencing these reactions but to a lesser extent.
The same LTR donors (Fig. 1B) were used with purified recombinant RSV PrA IN (29) under standard assay conditions for strand transfer. The total amounts of full-site and half-site integration products (Fig. 2) as well as their BglII restriction products (Fig. 3 and 4) were essentially the same as those observed with AMV IN (data not shown). These results suggest recombinant RSV PrA IN has similar physical interactions and sequence recognition at the LTR ends as virion-derived AMV IN, as well as a high fidelity for producing the avian 6-bp host site duplication (38).
A quantitative comparison of reactions between wt U3-U3 and the 4-bp LTR deletion mutants (Fig. 3 and 4) is shown in Table 1. As visualized in Fig. 3, only the U3Δ7-10, U3Δ8-11, and U3Δ9-12 donor ends had appreciable effects on both the half-site and the full-site integration reactions in comparison to their respective wt donor ends in the same reaction mixture. Of particular interest, both the U3*/U3 and the U3*/U3* full-site reactions and the U3* half-site reaction recover catalytically at similar rates as the deletions map further from the LTR end (Fig. 3, even-numbered lanes). These results suggest that there is a close correlation between the capability for half-site integration and functionality for full-site integration (Table 1).
Several other observations were apparent in the above 4-bp LTR deletion mutant studies. The quantity of the U3/U3 full-site reaction product (Fig. 4, 3.7-kbp product) was increased ∼3-fold with the Δ7-10 donor (Fig. 3, lane 4) relative to the quantity of the 3.7-kbp U3/U3 product derived from the wt U3-U3 donor (Fig. 3, lane 2). This result suggests that the affinity of IN is higher for the wt U3 LTR end than for the U3Δ7-10 LTR end, both present in the same mixture (1, 29). To account for the severely reduced quantity of the U3*/U3 product (Fig. 3, lane 4 and Fig. 4, 3.3-kbp products), the U3Δ7-10 mutation must have blocked the inclusion of the wt U3 end in nucleoprotein complexes capable of producing U3*/U3 products more than the other mutations did (Fig. 3, lanes 6, 8, and 10). This latter result is consistent with the in vivo observation that two good LTR ends are necessary for 3 ′ OH processing of both blunt-ended termini (32, 33); the same also appears to apply to the full-site integration reaction with 3′ OH recessed termini (Fig. 3) (29). However, the wt U3 end on the U3Δ7-10 donor is able to incorporate the U3* donor end to a small degree into nucleoprotein complexes producing the U3*/U3 product (Fig. 4, 3.3-kbp product), while the defective U3* half-site and the U3*/U3* full-site products (Fig. 3, lane 4, faint bottom fragment, and Fig. 4, 2.9-kbp products) are barely detectable. This “rescuing” effect for full-site integration by the wt U3 end suggests that IN must interact physically with the mutant U3* end, allowing communication between these ends to occur in trans. Therefore, IN must be binding to the defective U3* ends.
Nucleotide 7 (C) is critical for full-site integration.
The above LTR deletion studies suggest that position 7 plays a critical role in promoting full-site strand transfer. We modified the C nucleotide at position 7 (Fig. 1C) to determine whether it was necessary for full-site integration (Fig. 5). The single nucleotide change from C to A (U3P7C/A) at this position (Fig. 5A, lane 6) had the same effect on full-site and half-site integration reactions as the U3Δ7-10 mutation (Fig. 5A, lane 4). This pyrimidine-to-purine nucleotide substitution (U3P7C/A) effectively blocked the inclusion of wt U3 in nucleoprotein complexes producing the full-site U3*/U3 product. The insertion of a single A nucleotide prior to the C nucleotide at position 7 in another donor (Fig. 1C) produced results nearly identical to those observed with the U3P7C/A mutant (data not shown). Recombinant PrA IN displayed a response to the U3P7C/A mutant nearly identical to that of AMV IN (data not shown). The results suggest position 7 (C nucleotide) plays a critical role in the alignment of IN at the LTR ends, thereby allowing IN to communicate in trans between two LTR ends for full-site integration.
We investigated whether the other pyrimidine nucleotide (T) could substitute for the C nucleotide at position 7 (U3P7C/T) (Fig. 1C). Protein titration experiments with IN to measure strand transfer demonstrated that the T substitution in the U3-U3P7C/T donor resulted in an ∼40% decrease of activity relative to that obtained with the wt U3-U3 donor for the total observed half-site and full-site strand transfer reactions (data not shown) (Fig. 2). BglII restriction analysis showed that the U3P7C/T end was slightly more effective in communicating in trans with the wt U3 LTR end (Fig. 5B, the U3*/U3 reaction) than the U3P7C/A end (Fig. 5A) to mediate full-site integration. This slightly greater effect was observed by comparing quantitative full-site product data (U3*/U3 divided by U3/U3 within each set of reactions) (Fig. 5). By comparison of the average of four independent experiments with the U3P7C/T donor to results obtained with either the U3P7C/A or the U3Δ7-10 donor, it was determined that the C/T substitution was 2.5 times more effective than the other mutations at position 7 for promoting full-site reactions with wt U3. The results suggest that a pyrimidine nucleotide is preferred at position 7 (C>T) and that this nucleotide is critical for IN recognition, maximum strand transfer activity, and assembly of nucleoprotein complexes capable of full-site integration.
Retention of LTR DNA on nitrocellulose by IN does not correlate with full-site integration activities.
IN binds to DNA termini in a nonspecific fashion in vitro (2, 4, 41). We investigated whether DNA binding by IN under high-salt conditions for strand transfer to wt U3 LTR ends was significantly different from that to mutant U3 LTR ends, thereby accounting for some of the major differences observed at the strand transfer level. All of the donors were cut with HinfI, producing two LTR-containing fragments of 191 and 253 bp (29). The smaller fragment always contained the mutant LTR end. The donors were labeled either at the terminal NdeI sites or at both the NdeI and the HinfI sites. The latter technique allowed labeling of the internal 36-bp fragment containing only terminal HinfI sites.
IN was preincubated at 0°C for 10 min with each set of HinfI fragments derived from wt U3-U3, U3Δ7-10, and U3P7C/A donors under standard 0.33 M NaCl integration conditions. The concentration of IN was varied from 5 to 50 nM, and the donor DNA concentration was kept at 10 ng. The samples were filtered onto nitrocellulose filters at room temperature. No major differences were observed between the different LTR-containing donors in the total quantity of DNA retained on the filters. The quantity of retained DNA was nearly proportional to the IN concentration, with ∼80% of the DNA of each donor set being retained at between 30 and 40 nM IN (data not shown).
To determine if IN was able to selectively retain one LTR fragment over another, the retained fragments were eluted from the filters and the samples were subjected to agarose gel electrophoresis. No major differences were observed between the quantities of any mutant U3 and wt U3 LTR fragments (data not shown) retained on the filters. We cannot exclude the possibility that IN binds selectively or nonselectively to the 5′ 3-base overhang associated with the HinfI end on each fragment (or to internal regions), resulting in the observed DNA binding data. However, the 36-bp DNA fragment containing two HinfI ends was not retained by IN on the nitrocellulose filters. These results suggest that AMV IN bound to all of the LTR donor ends in a similar quantitative but not qualitative fashion.
DISCUSSION
In this report, the LTR sequence requirements with either AMV IN or recombinant RSV PrA IN and wt and mutant LTR donor substrates mirror the DNA att site requirements for full-site integration observed in vivo (3, 17, 28, 32, 33) and with the PIC in vitro (3, 8, 39, 40). Earlier and more extensive mutational analyses of the LTR end sequences using oligonucleotides to define the requirements for 3′ OH processing of blunt-ended termini or half-site strand transfer activities (2, 4, 6, 9, 11, 12, 15, 23, 34, 41) also mirror the sequence requirements observed in our reconstitution experiments for full-site integration; that is, mutations at or near the LTR ends (<15 bp) control IN activities. Our studies suggest that mutations in one LTR end influence the ability of avian IN to mediate half-site as well as full-site integration activities with another LTR end (Fig. 2, 3, and 5) (1, 29, 39). The physical interactions that are observed between IN subunits bound to two LTR ends occur in trans. These apparent allosteric interactions regulate both the effective participation of LTR ends in nucleoprotein complexes and the observed full-site integration activity associated with the assembled nucleoprotein complexes.
Two active LTR ends are required for murine leukemia virus (MLV) IN to promote 3′ OH processing of both blunt-ended LTR ends in vivo (32) as well as for IN-mediated MM-PCR enhancements and protection of LTR sequences in isolated MLV PIC (39, 40) or HIV-1 PIC (3, 8). Concurring with MLV and HIV-1 models in vivo (3, 8, 28, 32, 33), deletions in or modifications of internal sequences (<12 nucleotides from the end) have major effects in wt and mutant avian LTR ends on the promotion of full-site integration (Fig. 2 and 3). A 22-bp deletion starting at position 12 from the MLV LTR end has little or no effect on IN-associated properties in the PIC (40). This lack of effect is also analogous to the effect of deletions more distal from the ends in the avian LTR (Fig. 2 and 3 and Table 1). In summary, although there surely are some differences between retroviruses, the ability of AMV IN to interact with the att sequences to mediate full-site integration in vitro appear similar to results observed in vivo with MLV and HIV-1 and their PIC in vitro.
IN binds to DNA ends in a nonspecific fashion (4). The nitrocellulose filter DNA binding studies with the U3 LTR deletion mutants and AMV IN revealed no specific preference toward wt or mutant ends. The DNA binding specificities of IN for LTR fragments in 0.33 M NaCl were nearly equivalent and were not related to strand transfer activities. However, it appears that the interactions of IN with several defective U3 LTR ends (Fig. 3 and 5, U3*/U3 reactions) are sufficient to allow communication in trans with the wt U3 end to mediate full-site integration, arbitrarily at a lower level. The ability of IN to bind defective LTR ends may be biologically significant because even with one defective end, IN would still have the ability to mediate integration in vivo at a lower level (3, 8, 28, 32, 33, 39, 40). The DNA binding and strand transfer data together suggest that the binding of IN to LTR ends is necessary but not sufficient to effectively mediate full-site integration. Specific interactions with IN must occur at the LTR sequence level.
Several models describing specific interactions of IN with viral LTR DNA ends and target DNA within nucleoprotein complexes have been described in detail (12, 20, 21). The first three to four nucleotides from the HIV-1 LTR ends are photo-cross-linked to several specific regions of the catalytic core domain (residues 50 to 200) of HIV-1 IN (12, 20, 21). Mutational analysis of the carboxyl terminus of IN (residues 220 to 270) showed that residues L234 and R262 are critical for DNA binding (27). Residues R262 and K263 reside within a peptide photo-cross-linked to nucleotides at position 6 or 7 from the 3′ end of the viral DNA (20, 21). Nucleotide 7 was also found to be highly photo-cross-linked to this same carboxyl-terminal region of IN (12). In this report, position 7 (a C nucleotide) from the avian U3 LTR end coincides with this notion and appears to contribute significantly to the ability of AMV IN to mediate communication between two LTR ends bound by IN for full-site integration activity as well as for half-site strand transfer activity in vitro (Fig. 5A and B). In addition to nucleotide 7, the more distal nucleotides 8 to 12 also appear to influence full-site integration activity (Fig. 3 and Table 1) to various degrees, a result which is also consistent with IN interacting with these specific nucleotides (12).
In summary, our reconstitution experiments using purified IN and retrovirus-like DNA substrates closely mirrored the internal LTR sequence requirements observed either for integration in vivo or for the integration activity of purified PIC in vitro. Although not fully investigated, the avian LTR sequence requirements for full-site integration activity in vitro have allowed us to establish parameters to further investigate the assembly requirements for nucleoprotein complexes capable of mediating full-site integration. Molecular footprinting of these nucleoprotein complexes may allow us to determine whether other LTR sequences, besides those necessary for integration activity and communication in trans, are also necessary for the assembly process. Comparisons of molecular footprinting studies with assembled components for full-site integration to footprinting studies with the PIC capable of full-site integration should be enlightening.
ACKNOWLEDGMENTS
This work was supported by National Cancer Institute grant CA16312.
We thank Zhong-Ning Yang for discussions regarding the importance of a purine or pyrimidine at nucleotide position 7 from the LTR end.
REFERENCES
- 1.Aiyar N, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan M, Jonsson C B. Functional identification of nucleotides conferring substrate specificity of retroviral integrase reactions. J Virol. 1997;71:1025–1035. doi: 10.1128/jvi.71.2.1025-1035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown H E, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown P O. Integration. In: Coffin J M, Hughes S J, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 161–203. [Google Scholar]
- 5.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 6.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV-1 integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 7.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Wei S Q, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 9.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang J, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison V, Adams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG 1(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 14.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 17.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 18.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D P. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandgenett D P, Vora A C, Schiff R. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 20.Heuer T S, Brown P O. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 21.Heuer T S, Brown P O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 22.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 24.Kulkosky J, Skalka A M. Molecular mechanism of retroviral DNA integration. Pharmacol Ter. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y M H, Coffin J M. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutzke R A P, Plasterk R H. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vitro. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCord M, Chiu R, Vora A C, Grandgenett D P. Retrovirus DNA termini bound by integrase communicates in trans for full-site integration in vitro. Virology. 1999;259:392–401. doi: 10.1006/viro.1999.9782. [DOI] [PubMed] [Google Scholar]
- 30.McCord M, Stahl S J, Mueser T C, Hyde C C, Vora A C, Grandgenett D P. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Purification Expression. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- 31.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;65:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reicin A S, Kalpana G, Paik S, Marmon S, Goff S. Sequences in the human immunodeficiency virus type 1 U3 region required for in vivo and in vitro integration. J Virol. 1995;69:5904–5907. doi: 10.1128/jvi.69.9.5904-5907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scottline B P, Chow S, Ellison V, Brown P O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 35.Varmus H E, Brown P O. Retrovirus. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 36.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vora A C, McCord M, Fitzgerald M L, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vora A C, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 39.Wei S Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshinaga T, Fujiwara T. Different roles of bases within the integration signal sequence of human immunodeficiency virus type 1 in vitro. J Virol. 1995;69:3233–3236. doi: 10.1128/jvi.69.5.3233-3236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]