Abstract
Although antimicrobial peptides are considered one of the most promising alternatives to conventional antibiotics given the alarming increase in bacterial multidrug resistance, many aspects of their mechanism of action remain unclear, in particular the emergence and role of collective phenomena such as the spontaneous formation of nano-sized unstructured objects (clusters) and their effects on the biodynamics. We study this process using two novel peptides from the mucus of the garden snail Cornu aspersum as an example to reveal its dynamics and bioactivity implications through coordinated in silico and in vitro techniques — molecular dynamics simulations, UV–Vis and fluorescence spectroscopy, and antibacterial activity tests against two representative bacterial strains — one gram-negative (Escherichia coli 3458) and one gram-positive (Bacillus subtilis). The results obtained confirm the impact of the aggregation processes of the peptides on their biological activity and provide insight into possible synergies in their action.
Keywords: Antimicrobial peptides, Biological activity, Aggregation, Molecular dynamics, Fluorescence spectroscopy, Mechanism of action
Highlights
-
•
The emergence of collective phenomena and their role in peptide solutions is investigated.
-
•
As a testbed, two novel antimicrobial peptides from the Cornu aspersum mucus are employed.
-
•
Spontaneous formation of nano-sized clusters is demonstrated both in silico and experimentally.
-
•
A synergistic effect is observed in the activity of a multicomponent solution against gram-negative strains.
1. Introduction
Drug resistance is the basis of a number of serious problems, both in the treatment of infectious diseases directly caused by resistant strains, and in medical therapies such as complex surgical operations, chemotherapy, etc. [1], [2]. With multi-drug microbial resistance on the rise, there is an urgent need for the development of unconventional therapeutic options — new antimicrobial therapeutics with a novel mechanism of action and a lower potential for resistance development — to combat this major public health threat [3], [4]. Antimicrobial peptides (AMPs) are one such possibility. They are essential components of innate immunity in most multicellular organisms and represent an ancient non-specific host defense mechanism against infectious pathogens, including viruses, bacteria, and fungi, which complements the highly specific cell-mediated immune response [5], [6], [7], [8], [9]. In addition, they also have anticancer, anti-inflammatory, and immunomodulatory roles [10]. In recent years, AMPs have been actively explored as a platform for new antimicrobials with little potential for resistance development [11], [12], [13].
It is generally accepted that the target of most AMPs is the bacterial membrane, disrupting its integrity or function [14], [15], [16]. However, a number of questions about AMPs’ behaviour prior to the engagement with target membrane still remains open: when and how peptides acquire their biologically active conformation, whether they act individually or certain collective phenomena occur, and how their activity should be understood in relation to their low concentrations in bodily fluids [17] and generally what their mechanism of action is [18]. A common characteristic of AMPs is their propensity to form aggregated structures in solution. The aggregation process is understood as spontaneous and reversible association of molecular species to form complex supramolecular architectures [19]. Supramolecular assembly into various nanostructures, including -sheets and fibrils, heterogeneous particles, and branched networks is governed by the interplay among various noncovalent interactions, including electrostatics, Van der Waals interactions and interactions [20]. However, the implications of this phenomenon for peptide activity and selectivity have not been extensively studied [21], [22], and the results of experimental and theoretical studies in last 20 years on this topic are extremely contradictory [23], [24].
AMP aggregation is generally considered an undesirable property that affects the antimicrobial activity negatively [25], [26], [27]. However, numerous studies indicate a positive and even necessary role of this process. Peptide self-assembly at the bacterial membrane is necessary for the action of pore-forming AMPs [28] and self-associating of peptide monomers occurs at low concentrations [29], [30]. However, self-association and aggregation of cell-penetration peptides probably occur prior to the membrane interaction. Deca-arginine self-associates in solution which enhances its bioavailability [31]. Preassembly of cationic diastereomeric antimicrobial peptides is an essential factor in their membrane targeting to bacterial cells [32]. Melittin, like magainin, forms disordered toroidal pores in membranes, and peptide aggregation, either prior or after binding to the membrane surface, is a prerequisite to this pore formation [14]. Multimerization of the proline-rich Chex-Arg20 AMP changes its bacterial membrane interaction from non-lytic to disrupting [33]. The aggregation in solution of the killerFLIP peptide significantly enhances its selectivity for negatively charged membranes. This occurs by decreasing the effective hydrophobicity of the peptide, thereby reducing its affinity for membranes composed of neutral lipids, such as those found in the outer layer of healthy eukaryotic cell membranes [21]. Coaggregation in solution of magainin 2 and tachyplesin 1 into hetero-oligomers positively affects the membrane disruption in a synergistic manner [34].
In this study we explore the aggregation patterns of sample components in a substance with confirmed antibacterial properties — the mucus of garden snail Cornu aspersum, and, particularly, its low-molecular-weight fraction — in a search for correlations between the aggregation behaviour and the biological action. To this end, we explore in detail through both in silico (molecular dynamics simulations) and physicochemical (fluorescence spectroscopy) approaches the solvation behaviour and conformational stability of two novel peptides from the mucus fraction with MW below 3 kDa and their combination. Furthermore, we perform in vitro assays of the respective antimicrobial activities against one gram-negative (E. coli 3584) and one gram-positive (B. subtilis) bacterial strains. Our results suggest a new perspective on the mechanism of action of many small linear peptides and provide a basis for intelligent biologicals design.
2. Materials and methods
2.1. Dominant novel peptides in the LMW fraction of C. aspersum mucus
The mucus was collected from garden snails C. aspersum grown on Bulgarian farms by a technology not harming the snails — a patented device with a low-voltage electrical stimulation, as described in BG Utility model 2097/2015 [35], [36], [37]. After removing the coarse impurities from the crud extract, the supernatant was subjected to several cycles of filtration at 4 °C [38]. The obtained mucus extract was separated into two main fractions by ultrafiltration, using a membrane with pores sized 10 kDa (EMD Millipore Corporation, Billerica, USA). The mucus extract of compounds with MW 3 kDa was obtained from the mucus fraction with MW 10 kDa by additionally processing it at an Amicon® Ultra-15 centrifugal unit (centrifugation at 2500 × g, 4 °C, 20 min), with a 3 kDa membrane. The use of this non-invasive technique ensures the obtaining of fractions, which contain intact compounds.
2.2. Analysis of peptide fractions by mass spectrometric analysis
The peptides in the fraction with MW 3 kDa were analysed by MALDI-TOF-TOF mass spectrometry on AutoflexTM III (Bruker Daltonics), which uses a 200 Hz frequency-tripled Nd-YAG laser operating at a wavelength of 355 nm. The analysis was performed with of a mixture containing equal parts of the sample and of a matrix solution (7 mg/mL -cyano-4-hydroxyquinamic acid, CHCA) in 50% ACN containing 0.1% TFA on a target plate with 192 stainless steel wells. The mass spectrometer was calibrated with a standard mixture of angiotensin I, Glu-1-fibrinopeptide B, ACTH (1-17), and ACTH and MS/MS spectra were performed in reflector mode. The amino acid sequences of the peptides were identified by MALDI-MS/ MS using precursor ions from the MALDI-MS analysis. Amino acid sequences of two peptides (p1 and p3) were determined by de novo sequencing. Based on the primary structure so defined, for the subsequent experiments pure peptide samples were synthesized at GenScript Biotech (HK.1548).
2.3. 3D-structures of the peptides
The investigated peptides were recently isolated, and their three-dimensional structure is unknown. The experimentally determined amino acid sequences were generated in fully extended conformations and subjected to long-scale folding simulations using molecular dynamics (MD). Section 2.5 describes the simulation protocol. Based on the obtained results, three-dimensional molecular models were constructed as input structures for the subsequent computational studies.
2.4. Model systems for the in silico studies
To probe the actual behaviour of the novel peptides p1 and p3 after their secretion, we conducted MD simulations of monocomponent solutions of the two at concentrations of 10 mg/mL. For this, 27 monomers from each peptide were placed in a rectangular box with a minimal distance between each other and the box walls of 2 nm. This resulted in cubic simulation boxes with edges of 173.3 Å and 168915 water molecules, and 197 Å and 247632 water molecules for p1 and p3, respectively. Both simulations had a duration of .
As a toy model of the natural multicomponent substance, a bi-component system was constructed out of 21 p1-monomers and 14 p3-monomers, corresponding to a 1:1 mass ratio of the peptides and a concentration of 10 mg/mL. Similar to the monocomponent simulations, the peptides were uniformly placed in a cubic box, resulting in a box edge size of 158.7 Å and 127617 water molecules for each of the peptide species. The simulation time was 950 ns.
2.5. Molecular-dynamics simulations
All simulations were carried out using the MD simulation package GROMACS 2020.3 [39]. The CHARMM36 force field was used to parameterize the peptides [40], in conjunction with the modified TIP3P water model for the solvent. Under periodic boundary conditions, the peptides were solvated in cubic boxes with a minimal distance to the box walls of 1.2 nm. To neutralize the net charge and ensure physiological salinity, 0.15 mol/l sodium and chlorine ions were added to all systems. The energy of the systems was minimized by the steepest descent with a maximum force tolerance of 100 kJ/(mol nm), followed by short position-restraint simulations of 50 ps to equilibrate the solvent. The temperature was gradually increased to 310 K using a v-rescale thermostat [41] with a coupling constant of 0.1 ps and pressure was equilibrated at 1 atm using a Parrinello–Rahman barostat [42], [43] with a coupling constant of 2 ps in 10 ns simulations.
The same thermo- and barostat parameters were used for the production MD simulations in the NPT-ensemble. The leapfrog integrator [44] was used with a time step of 2 fs, while the PLINCS algorithm [45] was used to impose constraints on bonds between heavy atoms and hydrogens. Van der Waals interactions were turned off gradually from 1.0 nm away and truncated at 1.2 nm. The smooth PME method [46] was used to model electrostatic interactions, with a direct PME cut-off of 1.2 nm. Every 10 ps, a neighbourlist was created. Each production simulation lasted . Every 100 ps, trajectory frames were taken.
2.6. Synthetic data analysis
The MD trajectories were examined using standard GROMACS post-processing and analytic tools. The secondary structure was assigned by the STRIDE algorithm [47]. The visualization and manipulation program VMD [48] was used to create all structural figures.
2.7. UV–Vis absorption measurements
UV–Vis absorption measurements were carried out at a Shimadzu™ UVmini-1240 Model Spectrophotometer using asymmetric quartz cuvettes with 4/10 mm optical length at room temperature (22 °C). All absorption spectra were corrected by subtracting the absorption spectrum of the buffer solution in the same wavelength range (200–400 nm). From the two peptides and their mixture were freshly prepared solutions at concentrations from 0.25 mg/mL to 10 mg/mL. All spectroscopic experiments were carried out at pH 7.4 in phosphate buffer solutions.
2.8. Fluorescence analysis of the structure and stability of the peptides
Fluorescence spectra were recorded on a Shimadzu RF-6000 spectrofluorometer using a xenon lamp as the excitation source and fluorescence curette (Hellma, Macro, Suprasil quartz), limit 200–2500 nm spectral range, pathlength 10 × 10 mm. The p1 and p3 solutions with concentration of 10 mg/mL in a phosphate buffer with pH 7.4 were diluted with Milli-Q water to a final concentration of 0.286 mg/mL, which corresponds of 0.245 mM solution for p1 and 0.165 mM for p3. The concentration of the mixture containing p1 and p3 in a 1:1 ratio also was 0.286 mg/mL (p1 — 0.143 mg/mL and p3 — 0.143 mg/mL). All peptide solutions were investigated over a wide pH range by capillary microtitration. Small amounts of 0.5N HCl or 0.5N NaOH were added during the titration to the peptide samples. Emission spectra were recorded in the range 310–500 nm by exciting the samples at 295 nm at room temperature 25 °C. Fluorescence spectra were recorded after incubation for 5 min at each pH value.
2.9. Minimum inhibitory concentration (MIC)
The bacterial strains Bacillus subtilis and Escherichia coli 3458 were obtained from the National Bank for Industrial Microorganisms and Cell Cultures (Sofia, Bulgaria). The minimum inhibitory concentration (MIC) of the tested peptides was determined by broth microdilution method. To determine the lowest concentrations of the tested peptides required to inhibit visible growth of the microorganisms, they were dissolved in PBS (pH 7.4) to achieve 25 mg/mL. The peptides were serially diluted two-fold from 25 mg/mL to 0.195 mg/mL (the concentrations of each of the peptides in the solution of the p1+p3 combination were respectively from 25 to 0.0975 mg/mL). Bombinin, a renowned antimicrobial peptide, was used as a reference peptide with antibacterial properties. The bacterial strains used were cultured in sterile Muller-Hinton broth (MHB) (HiMedia Laboratories Pvt. Ltd., Mumbai, Maharashtra, India), incubated aerobically overnight, and diluted with fresh MHB media to reach 2.5 CFU/mL. Next, 50 L of the respective peptide concentrations, 50 L fresh MHB and 10 L bacterial suspension were added to wells of sterile 96-well round-bottom polypropylene microtiter plates. The plates were incubated at 37 °C for 20 h. MIC of the peptide capable of inhibiting visible bacterial growth was quantitatively determined by measuring OD600 for each well using an MRX A2000 Microplate Reader (KLAB Inc.). All MIC determinations were made in triplicate. Wells having bacteria, MHB and Gentamicin (Sopharma PLC) at a concentration of 0.032 mg/mL were used as negative control. Wells having bacteria and MHB without the peptides served as positive controls. Any resultant MIC was defined as the lowest concentration of peptide at which there was no bacterial growth upon juxtaposition with media control samples in the 96-well round-bottom microtiter plate. Any resultant MIC50 was defined as the lowest concentration of peptide at which there was at least 50% bacterial growth inhibition upon juxtaposition with media control samples in the plate.
3. Results
3.1. Isolation and characterization of peptides
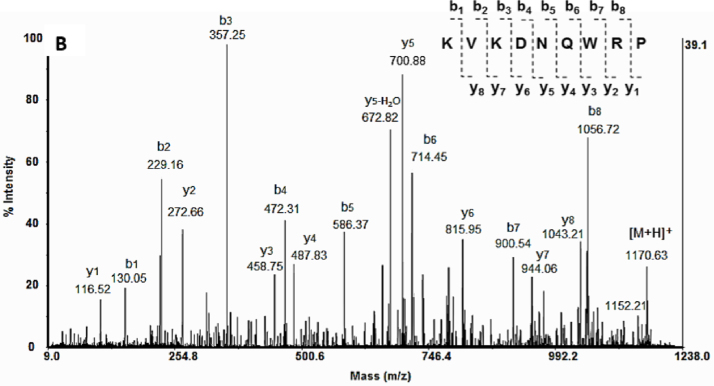
Individual peptide components in the C. aspersum mucus fraction with MW 3 kDa were identified by their molecular masses and amino acid sequences. The exact molecular weights of the peptides were determined as protonated molecular ions [M+H] by MALDI-TOF/MS analysis performed in positive ionization mode. The resulting MS spectrum revealed a group of six dominant peptides at the leftmost end of the mass spectrum (MW 1750 Da). Their primary structures were determined by means of tandem mass spectrometry, tracking the fragment y- and b- ions, as exemplified in Fig. 1 for the peptide represented as the molecular ion [M+H] at m/z 1170.63 Da.1
Fig. 1.
MS/MS and de novo sequence analysis of the peptide at m/z 1170, 63 [M+H].
In Table 1, the amino acid sequences, masses, and key physicochemical properties of these six peptides — net charge, isoelectric point (pI), grand average hydropathicity (GRAVY) index — as determined with the ExPASy ProtParam tool2 [50] are shown, as well as their antimicrobial activities predicted using the iAMPpred software3 [51].
Table 1.
The six dominant peptides in the C. aspersum mucus fraction below 1750 Da.
| No | Amino acid sequence | MALDI-TOF- | MW [Da] | pI | GRAVY | Charge | Antibac- | Anti- | Anti- |
|---|---|---|---|---|---|---|---|---|---|
| MS [M+H]+ | terial | viral | fungal | ||||||
| at m/z | [%] | [%] | [%] | ||||||
| p1 | KVKDNQWRP | 1170.63 | 1169.63 | 9.99 | −2.344 | +2 | 34 | 22 | 32 |
| p2 | VNVVGGGGGIVGGGIGGGGM | 1570.8 | 1569.79 | 5.49 | 0.9 | 0 | 51 | 55 | 29 |
| p3 | LFGGHQGGGLVGGLWRK | 1738.99 | 1737.94 | 11 | −0.024 | +2 | 76 | 41 | 78 |
| p4 | LGHDVH | 677.328 | 676.33 | 5.97 | −0.383 | −1 | 84 | 76 | 61 |
| p5 | LGLGNGGAGGGL | 942.49 | 941.49 | 5.52 | 0.575 | 0 | 90 | 54 | 67 |
| p6 | MLGGVLGGGPLK | 1098.63 | 1097.63 | 8.5 | 0.833 | +1 | 76 | 52 | 64 |
In the present study, we focus on two of the six dominant peptides in this fraction, p1 and p3. Our choice is motivated by two main arguments: (1) Most of the known AMPs are cationic, and (2) It is believed that tryptophan is essential for their antibacterial activity [52], [53]. The two selected peptides are both cationic and the only ones of the six containing tryptophan — one residue each, at positions 7 and 15, respectively, further referred to as Trp7 and Trp15. In the same time, they exhibit substantial differences in their length and molecular mass, charge distribution and relative hydrophobic content. As we aim to reveal the impact of these differences on the biological activity of the peptides, this selection fits best the objectives of our study.
3.2. Monomer-in-water simulations for structure resolution
The investigated peptides are novel, so there is no prior knowledge on their structure. The p1 peptide is a nonapeptide, so is not expected to adopt any stable secondary structure, as was confirmed by the conducted 500 ns molecular dynamics (MD) folding simulation. The p3 peptide is a 17-mer and could potentially collapse into a well-defined fold. The extensive MD simulations with a total duration of , though, proved the opposite — despite the temporary formation of different secondary-structure elements, no preferred conformation could be singled out. This is also substantiated by the fewer than two residues on average being involved in any secondary-structure element.
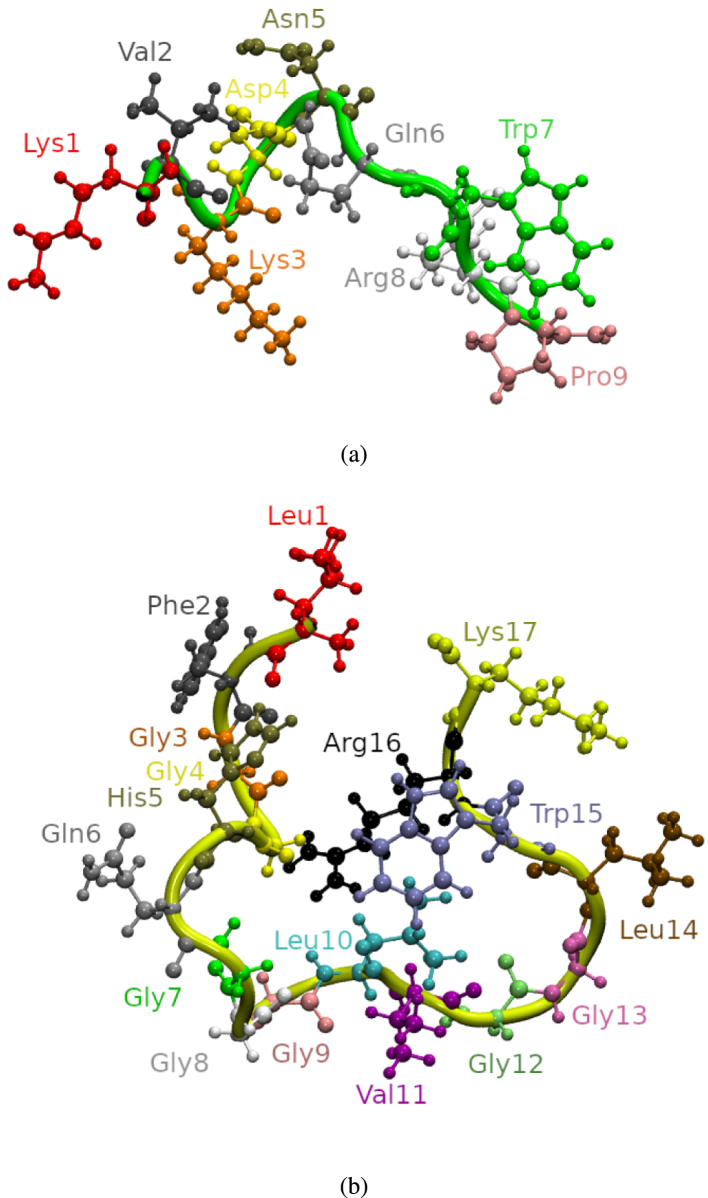
Based on the above observations, for the subsequent in silico investigations the randomly chosen conformations from the folding simulations shown in Fig. 2 were used as initial 3D structures for the peptides.
Fig. 2.
Structures of (a) p1 and (b) p3, as used in the subsequent MD simulations.
3.3. Computational modelling of mono- and multicomponent peptide solutions: Cluster formation and solvent exposure
3.3.1. Monocomponent p1 and p3 solutions
In the literature, there is no evidence supporting the single-peptide action mode [54], [55]. Based on studies of self-association patterns of different linear AMPs in water solution [56], a hypothesis was put forward about the role aggregation plays in the complex mechanism of antimicrobial action of bombinin H2 and, possibly, other small linear AMPs [57].
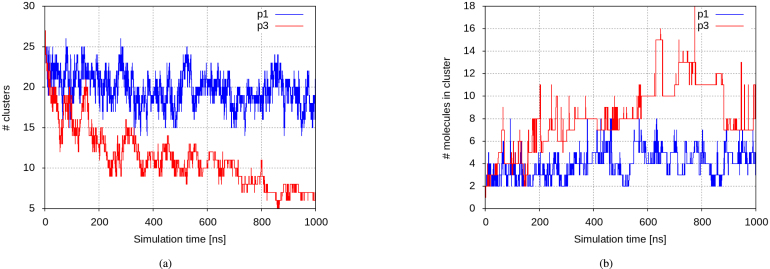
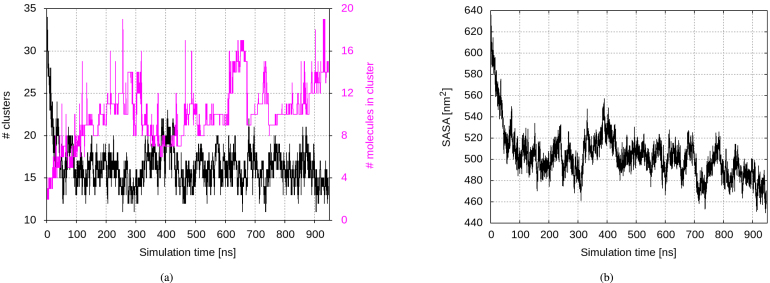
An insight into solvation behaviour of the two novel peptides after their secretion was gained through MD simulations with a duration of of monocomponent solutions of p1 and p3 at a concentration of 10 mg/mL. Both peptides show a clear tendency towards rapid oligomerization, yet at a different pace and scale, to be traced back to some key properties of the two: p1 is much shorter and with a much higher electric charge density, with four out of nine residues charged, while p3 has a substantially higher fraction of hydrophobic residues — 70% vs. 30% in p1. That way, while in both solutions the number of clusters decreases at the expense of their size, this process is more pronounced for p3. In the p1 solution the clusters remain rather small and by the end of the simulation their number decreases by 20%, the largest-cluster size averaged over the first vs. the last 200 ns of the simulation showing an increase of about 65%, up to 5 monomers. In the same time, in the p3 solution twice as big clusters are formed and within 1 microsecond their number decreases by 70%, coupled with an increase of the average largest-cluster size by 3.5 times (Fig. 3) . The same applies to the average cluster sizes — fluctuating around three monomers for p1 and more than doubling in size, from two to five monomers for p3 (Suppl. Figure S1).
Fig. 3.
(a) Number of clusters and (b) largest-cluster size over the aggregation process in monocomponent solutions at a concentration of 10 mg/mL of p1 (blue) and p3 (red) . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
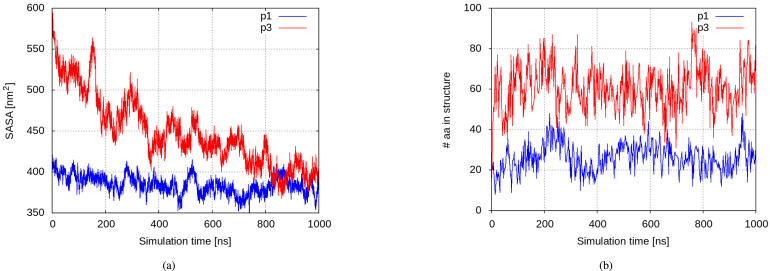
The aggregation process is further manifested through the evolution of the solvent-accessible surface areas (SASA) of the peptides and certain their residues. As expected, aggregate formation is associated with a decrease in all investigated SASAs, but again on different scales. In the p1 solution, the simulations reveal some 10% decrease in the SASA of all 27 monomers, with tryptophan residues buried in the clusters interior to a somewhat greater extent, the respective decrease being twice as high. In the p3 solution, the protein SASA drops by almost 30% (Fig. 4(a)). Since exposure to the solvent largely impacts the ability of a residue or domain to interact with other molecules, among them the bacterial membrane, it is important how it is influenced by the above aggregation process, specifically for the aromatic residues considered important for the biological function of the peptides — Trp7 in p1 and Trp15, Phe2, and His5 in p3. Indeed, two of the three aromatic residues demonstrate their hydrophobic nature, losing 31% (the hystidines) and 46% (the tryptophans) of their initial SASA (Suppl. Figure S2). Details of the SASA decrease according to residue types — hydrophobic, polar, and charged — are in accord with the amino acid content of the investigated peptides — the large portion of charged residues in p1 and the high hydrophobic content of p3 (Suppl. Figure S3).
Fig. 4.
(a) Evolution of the solvent-accessible surface area (SASA) and (b) number of amino acid residues participating in any secondary structure element in the aggregation process in the p1 (blue) and p3 (red) monocomponent solutions at a concentration of 10 mg/mL . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The largest clusters in the two monocomponent simulations are depicted in Suppl. Figure S4, residues coloured by their type. Interestingly, in the p3 simulation, there are two different structures of the same size and while the first one lines up well with the p1-cluster structure, resembling the one of a globular protein, the second one provides solvent exposure to a larger number of residues. Another observation concerns the cluster size. In the case of bombinin H2 [57] and of a very similar to p3 in size and molecular mass peptide, though charge-neutral (manuscript in preparation), the aggregates tend to be much bigger, gradually recruiting almost all monomers. The fact that this is not the case here raises even more acutely the question about the role of neutral peptides in natural multicomponent substances.
As discussed above, a single monomer — which is a reasonable approximation to the very low-concentration case — tends to remain unstructured, thus leaving open the question of when the peptides adopt their biologically active conformation. In [57] it was suggested that this happens at least partly within the aggregates thus formed, that is, a process of aggregation-driven folding takes place, bringing the potentially biologically active peptides into the optimal for the membrane impairment shape. Gradual structuring within the oligomers formed can be seen in the secondary-structure plots (Suppl. Figures S5, S6). In both monocomponent solutions, an increase in the number of amino acid residues involved in any structural element could be detected (Fig. 4(b)), a detailed account for the four main types — -helices, -bridges, -sheets, and turns — being presented in Suppl. Figures S7, S8. Despite the increase compared to the single-monomer simulations, the number of amino acid residues engaged in structural elements remains modest — roughly one residue per monomer in the p1 solution, and 2.4 residues per monomer in the p3 one. This might be due to the very short length of p1 and the high glycine content of p3, which is a known secondary structure breaker.
3.3.2. p1+p3 multicomponent solution
The examined peptides belong to the lightest fraction of the C. aspersum mucus, along with a number of other peptides, acids, salts, etc. Most likely, the biological activity of the whole complex substance stems from the synergy between two or more of its components. To test this assertion, a multicomponent solution of p1 and p3 was simulated for 950 ns. The resulting conformation is shown in Suppl. Figure S9. As can be seen, several larger clusters were formed, along with a number of smaller ones. At times, even larger constructs formed, which proved unstable and dissociated again into smaller aggregates. The reason for such dynamics is electrostatic repulsion, which overwhelms the hydrophobic effects. A close-up of the largest cluster formed is presented in Suppl. Figure S10.
This dynamics is also reflected in the cluster-formation data, shown in Fig. 5(a). Very rapidly, the monomers aggregate into dimers, trimers and higher oligomers, whose number then remains relatively constant, engaging between 10 and 15 monomers in the largest formation over the last 200 ns of the simulation.
Fig. 5.
Aggregation process in the p1+p3 multicomponent solution: (a) number of clusters (black) and maximal cluster size (magenta); (b) Total protein SASA . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The rapid and significant decrease of the protein SASA of the mixture is fully consistent with the dynamics of cluster formation (Fig. 5(b)): after a 20% decrease within the first 100 ns of the simulation, the SASA remains approximately the same, with only a further decrease of about 5%, reflecting a minor increase in cluster size. Focusing on the biologically important aromatic residues, we observe that in the mixture these residues from the different peptides behave differently. Suppl. Figure S11 demonstrates that p3 tryptophans and phenylalanines underwent a more severe decline in SASA, resp. of 55% and 37% (recall the corresponding values of 46% and 20% for the monocomponent solutions). At the same time, the SASA of p1 tryptophans registered only a slight decrease compared to the monocomponent solution — about 25% vs. 20%. Apparently, these residues remain exposed to the solvent, being located on the surface of the cluster (see Suppl. Figure S10b). Thus, in the mixture, the biological activity is likely to be due to the p1 monomers.
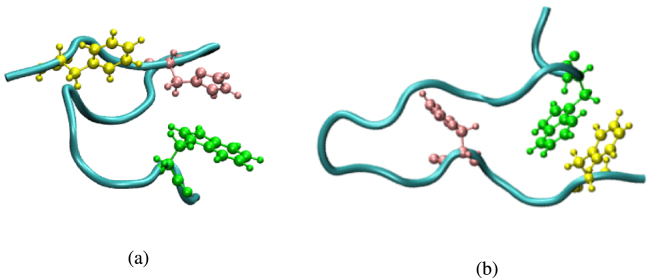
Due to the presence of several aromatic amino acids in the p3 monocomponent and in the multicomponent simulations, the possible impact of the - stacking on both the aggregation process in the solutions and the fluorescence emission merits attention. Such conformations already occur in the single-monomer simulation, as shown in Fig. 6. In the p3 monocomponent simulation there are 27 monomers and, correspondingly, 27 residues of each of the three aromatic amino acids present — tryptophan, phenylalanine and histidine — while in the peptide mixture there are 35 tryptophan residues and 14 residues of each phenylalanine and histidine. The analysis reveals that about 1% of all mutual orientations of the three aromatic amino acid residues and almost 6% of the respective paired conformations fall within the - stacking domain, as shown in Suppl. Figure S12. The influence of these conformations, despite their relatively small amount, will be addressed elsewhere.
Fig. 6.
Sample conformations for - stacking in the p3 monomer-in-water conformation. The aromatic amino acids are coloured as follows: Trp in green, Phe in yellow, and His in pink . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Self-association of peptides established by UV–Vis absorption spectroscopy
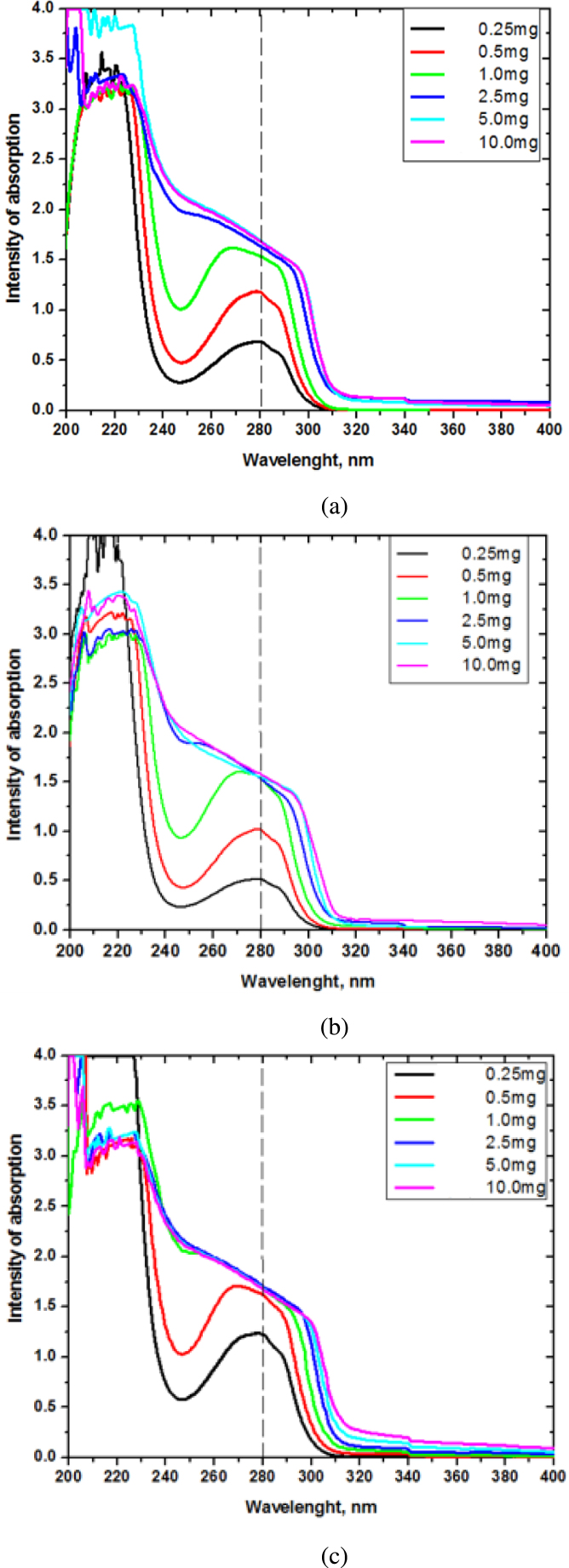
Absorption spectrum dependence on the peptide concentration for the two novel peptides and their mixture was studied in the range from 0.25 mg/mL to 10 mg/mL. At concentrations of 0.25 mg/mL and 0.5 mg/mL, one maximum at 280 nm was observed and one less intense peak at 292 nm, which are due to transitions in the indole group of tryptophan. At a concentration of 1 mg/mL, a shoulder was observed in the region 288–267 nm for both samples. Increasing the concentration to 10 mg/mL leads to significant differences in the absorption spectra of p1 and p3. For p1, an overlap of the emission spectra at concentrations of 5 mg/mL and 10 mg/mL and a shift of the observed shoulder to longer wavelengths around 300 nm was observed, which is due to the aggregation of the peptide. Thus, the self-association of p1 into aggregate structures takes place from 5 mg/mL onward, (Fig. 7(a)). For peptide 3, an overlap of absorbance intensity at 280 nm was observed at 1.0 mg/mL, 2.5 mg/mL, 5.0 mg/mL and 10 mg/mL. Moreover, at a concentration of 2.5 mg/mL, a red shift of the observed shoulder was also found, more pronounced at concentrations of 5.0 mg/mL and 10 mg/mL. These results are indicative of an onset of the oligomerization processes for p3 at 2.5 mg/mL (Fig. 7(b)).
Fig. 7.
UV–Vis absorption analyses at concentration of 0.25–10.0 mg/mL for: (a) p1; (b) p3; (c) mixture of p1 and p3, containing 0.25–10.0 mg/mL of each peptide in a 1:1 ratio.
The changes in the absorption spectra of the bi-component solution (p1 and p3 at the same concentrations in a 1:1 ratio) exhibit a similar pattern as p3. However, the overlap of the absorption spectra at concentrations from 1 mg/mL to 10 mg/mL suggests that the self-association processes here have already started at a concentration above 0.5 mg/mL (Fig. 7(c)).
3.5. Fluorescence analysis of peptide structure and stability
The presence of fluorophoric residues in both peptides — Trp7 in p1, respectively Trp15 and Phe5 in p3 — makes fluorescence emission spectroscopy a valuable tool in analysing their behaviour in solution. The fluorescence emission spectra obtained as described in Section 2.8 showed a single maximum at =350 nm (Suppl. Figure S13).
Fluorescence emission spectra of the peptides, their mixture, and the hydrophobic amino acid tryptophan (at uniform concentrations of 10 mg/mL) were obtained after excitation at = 295 nm. Significant differences are observed both in the position of the emission maxima and in their intensity. The fluorescence spectra of the two peptides and their mixture (p1 + p3) show a single maximum at = 350 nm, while tryptophan shows a single maximum at = 355 nm with the highest intensity (Suppl. Figure S13). The observed blue shift to lower wavelengths of the emission maxima and the decrease in their intensity for p1, p3 and their mixture compared to tryptophan indicate structural changes that are most likely related to the self-association of the peptides into oligomers. The results show that (p1+p3) and p3 have a higher tendency to form aggregates than p1.
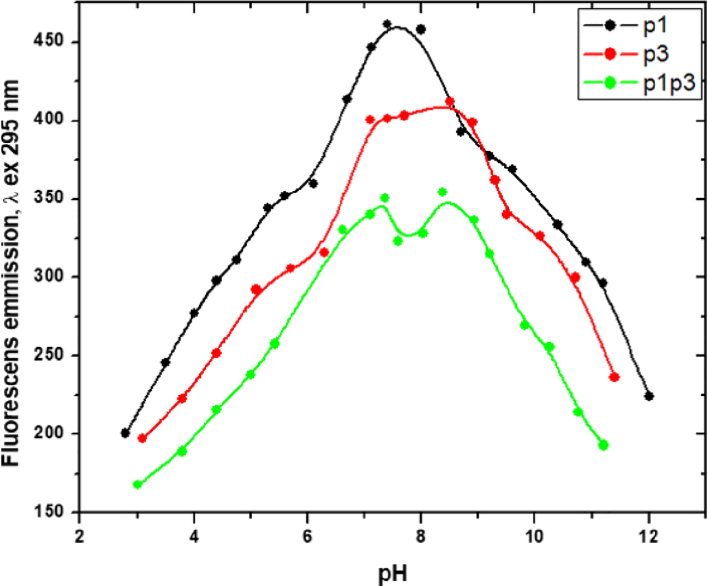
The pH dependence of the emission intensity was studied at solution pH values ranging from 3 to 12 in a phosphate buffer (Fig. 8). At this excitation wavelength the only fluorophores contributing to the emission are the tryptophan residues (Trp7 in p1 and Trp15 in p3, respectively). For both peptides, a shoulder was observed in the pH range 5.2–6.2, slightly less pronounced in the case of p1, followed by a steady intensity increase to the well-defined peak at pH 7.6 for p1 and an almost-plateau region at pH 7.0–9.3, with a minor increase in the emission intensity at pH 8.5 for p3. The p3 emission intensity was lower in the whole pH range, except for a narrow p3-dominant domain, pH 8.6–9.2.
Fig. 8.
pH dependence of the maximum emission intensity at nm of p1, p3, and p1+p3 combination in a phosphate buffer.
A significant difference was observed in the titration curve of the bi-component solution: it exhibits no shoulder in the acidic domain, but two maxima shortly above the physiological pH value, at pH 7.3 and at pH 8.4, and with a lower intensity than in the monocomponent solutions throughout the whole tested pH range.
3.6. Antibacterial activity
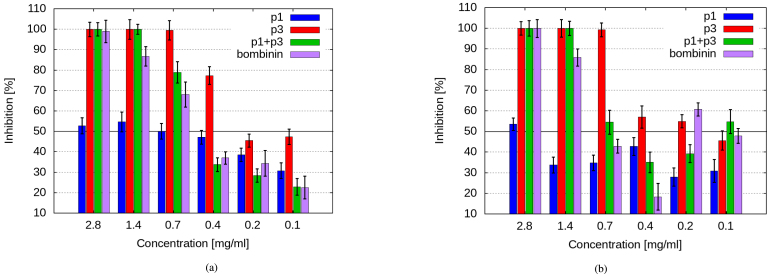
The antibacterial activity of p1, p3, and their mixture (p1+p3) against two representative bacterial strains — one gram-negative and one gram-positive. The chosen bacterial cultures, B. subtilis and E. coli, have previously been used for initial screening of antimicrobial peptides of different fractions from C. aspesum mucus [37], [38], as well as for studies of antibacterial activity of other AMPs [58], [59], [60], [61]. P1 and p3 antibacterial activity was tested using broth microdilution method (Section 2.9), with the known antimicrobial peptide bombinin as a reference (Fig. 9 and Suppl. Tables S1, S2). We observed an overall stronger antibacterial potential of p3 compared to p1 and even bombinin. In the case of B. subtilis, p3 fully inhibited its growth already from a concentration of 0.7 mg/mL, while even at the highest tested concentration (11.4 mg/mL) p1 achieved only about 63% inhibition. Complete growth inhibition after treatment with (p1+p3) combination and bombinin was found at concentrations of 1.4 mg/mL and 5.7 mg/mL, respectively. This trend was also manifested in the MIC50 values, with the lowest one for p3 — 0.4 mg/mL, 0.7 mg/mL for (p1+p3) combination and bombinin, and 1.4 mg/mL for p1.
Fig. 9.
MIC50 determination for p1, p3, and (p1+p3) combination against: (a) B. subtilis; (b) E. coli 3458.
In the case of E. coli 3458, a very interesting dependence of the inhibition rate on concentration and sample composition was observed. Full inhibition was registered starting from a concentration of 1.4 mg/mL for p3 and (p1+p3) mixture, 2.8 mg/mL for bombinin, and at 11.4 mg/mL for p1. However, the lowest MIC50 was detected for the bi-component sample (0.1 mg/mL, with almost 55% inhibition), suggesting a synergistic mode of action of the multicomponent natural substances in this very-low-concentration range. Another interesting low-concentration phenomenon is the Eagle-effect [62] resembling trend observed for the peptide mixture (p1+p3) and bombinin and somewhat less pronounced for p1 in the range 0.1 mg/mL – 0.7 mg/mL: increasing the concentration of these samples did not result in a proportional increase in bacterial growth inhibition. Thus, for (p1+p3) mixture a 50% inhibition was detected at 0.1 mg/mL, and then from 0.7 mg/mL onward, and for bombinin — at 0.2 mg/mL and then from 1.4 mg/mL, with a lower antibacterial activity of these samples at the intermediate concentrations.
4. Discussion
Intuitively, biological activity of the AMPs should be related to their affinity towards the target membrane. This also appears to be true for bacteriocins [63], [64], [65]. Most AMPs are cationic and this provides rationale for them targeting bacteria without harming the host: bacterial membranes are negatively charged, whereas the outer leaflet of the eukaryotic membranes is neutral [66], [67]. The two investigated peptides are cationic and exhibit a key similarity — the presence of a single tryptophan residue in each — but differ in a number of other important features. Thus, p1 is a nonapeptide, while p3 consists of 17 residues, but with seven of them glycines, its mass is only some 50% higher than that of the nonapeptide. Another difference is in their hydrophobic-amino-acid-residues content, 30% and 70%, respectively, which predictably affects their aggregation behaviour. Though both peptides have a net charge of +2, its distribution is substantially different. In fact, p1 contains four charged residues, but in the middle of its short chain one lysine (K3) and one aspartate (D4) mutually compensate their charges, so that the peptide is effectively left with one positive residue at each end. Unlike p1, p3 has only two charged residues, but they are located next to each other and complete its C-terminal domain. In both peptides, tryptophan residues are located at the C-termini, adjacent to arginine residues. This arrangement suggests a possible navigation role of arginine immediately after peptide secretion and an anchoring one once it reaches the target membrane. This effect is enhanced in p3 by the second positively charged residue — a lysine located next to the arginine, while the second positive charge in p1 is in the N-terminal, which depending on the adopted conformation can generate competing forces and, as a consequence, slower and/or weaker attraction between the peptide and the target membrane. The situation is further complicated by the presence of two more aromatic residues in p3 — phenylalanin and histidine — that necessitates the study of the possible - stacking effects. The above considerations suggest a higher biological activity for p3 compared to p1, in agreement with the predicted values in Table 1.
The self-association process in peptides is largely governed by the hydrophobic effect. Therefore, it is to be expected that p3 with its higher hydrophobic content will form more stable and larger clusters than p1 and will also influence the aggregation dynamics of the peptide mixture, consistent with the GRAVY indices of both peptides, 2.344 and 0.024 respectively. Our simulation results are well in line with these expectations.
The solvent-accessible surface area (SASA) provides an adequate quantification of the aggregation dynamics. In all simulated systems we observed SASA decrease, however at a different rate, consistent with the respective cluster-formation dynamics. Our simulations confirmed the expected tendency for the aromatic residues to get buried in the hydrophobic core of the clusters. Notably, in the mixed sample the tryptophan residues of the two components experience different SASA decrease, stronger for those belonging to p3 than to p1. Thus, the tryptophan and phenylalanine residues in p3 lose about 9% and 17% more SASA than in the monocomponent solution, while for the p1 tryptophans this decrease amounts to only about 5%. Apparently, these residues remain solvent-exposed to a larger extent (see Suppl. Figure S10b, S11), becoming of greater importance for the biological activity of the mixture.
These computational findings were confirmed by the collected spectroscopic data. The absorption of light by the protein or peptide chromophores allows indirect determination of the changes in their structure during the aggregation process. The observed shifts of the maxima at 280 nm and 292 nm to longer wavelengths (red shifts) suggest that the tryptophan residues are in a more buried environment and less exposed to the solvent. In the case of aggregation of tryptophan-containing proteins or peptides, a shoulder around 290–300 nm is usually observed [68], as also seen in our results.
Following the approach in [69], we investigated the onset of the oligomerisation process by analysing the UV–Vis absorption spectra of the peptide solutions. The observed concentration threshold hierarchy — p1, followed by p3 and the (p1+p3) composition with, respectively, 5.0 mg/mL, 2.5 mg/mL, and 1.0 mg/mL — is in agreement with the simulation data, which shows that (p1+p3), followed by p3, self-assembles into larger and more stable oligomers much faster than p1. The results from the fluorescence study of p1, p3, and (p1+p3) compared to tryptophan (Suppl. Figure S13), also confirm the hypothesis of self-association of peptides in clusters in the mono- and multi-component solutions.
A potential influence of the environmental pH on peptides aggregation propensity may be of key importance due to its clinical significance and its impact on drug production and synthesis [70]. The observed differences in the pH dependence of the fluorescence intensity of the two peptides are most likely related to their aggregation specifics: while in the p1 solution only very small if any clusters may be formed, in the p3 solution much larger and more stable clusters are formed, consistent with the MD simulations data, which indicates the transient formation of a 3- to 5-monomer cluster in the first case against a stable 9-monomer cluster in the second. Moreover, as the simulation data suggests, for p3 there are two different maximal-cluster conformations (Suppl. Figure S4b), which might respond differently to the environmental pH changes in that the less compact one may keep intact, with its tryptophan residues exposed, in a wider pH range. Since the aggregation process results from the balance of the different forces involved — primarily the hydrophobic effect and the electrostatic interactions between the amino acid residues — these observations are consistent with the amino acid content of the investigated peptides. The lower emission of p3 may result from a possible interference on part of the histidine residues present therein — - stacking conformations may result in quenching of the emission of the Trp15 indole ring by the His5 imidazole ring, a phenomenon first reported in [71], [72]. In the case at hand, this role might be played by Phe2 and His5 of p3, as seen in the sample trajectory frames in Fig. 6.
The substantially different titration curve of the p1+p3 mixture is rather indicative of a different rate of unfolding of the pH dependence of the two ingredients and the different structure of the clusters in the mono- and multi-component solutions. Such behaviour might hint at a temporary counteraction to the natural emission decrease by fluorescence-emission promoting factors — the presence of a larger number of surface-exposed tryptophan residues on the one hand and a possible decrease in the quenching effect of histidine residues on the other — upon entering the alkaline pH range.
The self-association phenomenon appears to be of key importance for the biological activity of the AMPs. It is known that their antimicrobial action depends on a threshold concentration below which they cannot affect the target membrane. In recent years, however, a number of studies have pointed out the importance of the local concentration rather than the bulk (full-volume) one [14], [61]. Thus, in [14] it is reported that the peptide aggregation at or near the target membrane provides the critical local concentration necessary for the formation of transmembrane toroidal pores. Protegrin-1 forms oligomeric aggregates near a lipid bilayer [73].
In [57], a hypothesis was put forward based on in silico studies, that AMPs do not exist in solution as isolated monomers but a self-assembly process into nano-sized aggregates accompanied by aggregation-driven folding allows them to reach the target membrane in a sufficient concentration and fully functional state in order to effectively exert their antimicrobial action. The present study provides further support for this hypothesis: p3 reached the threshold local concentration at a much lower bulk concentration compared to the small and unstable cluster-forming p1. Moreover, with the observed over 50% growth inhibition of E. coli 3458 by the peptide mixture (p1+p3) at the lowest investigated concentration, exceeding by 20%, resp. 75% the inhibition achieved by the individual components, p3, resp. p1, our results are consistent with an additive or even synergistic effect in this concentration range as suggested in previous studies [37], [38]. Such a synergistic effect of different AMPs has also been observed for other peptides [74], [75], [76].
5. Conclusions
In this paper, we investigate the spontaneous formation of nano-sized unstructured objects (clusters) and their effects on the biodynamics of small linear peptides with putative antimicrobial properties, with two novel peptides from the mucus of the garden snail Cornu aspersum and their mixture as a test-bed. Using coordinated in silico and in vitro approaches we scrutinized the dynamics and bioactivity implications of this process when targeting two representative bacterial strains — one gram-negative (Escherichia coli 3458) and one gram-positive (Bacillus subtilis). The experimentally detected self-association processes were fully conform with the patterns and rate predicted by the computational modelling. The results of the in silico studies, the spectroscopic analyses, and in vitro antibacterial tests reveal the complex relation between peptide charge, concentration, aggregation patterns, and biological activity. In particular, they support the hypothesis about the important role of the formation of nano-sized clusters through spontaneous self-association in the complex mechanism of peptides’ antimicrobial action [57]. The observed in the case of gram-negative bacteria at low bulk concentrations (the real-life scenario) higher antibacterial activity of the multicomponent solution compared to its components is a strong indication about the synergy underlying the antimicrobial effect of snail mucus and other natural bioactive substances [37], [38].
The observed activity dependencies on the sample composition and concentration back the concept of a highly complex mechanism of the natural substances biological activity, their component complementarity, and critical concentration thresholds, encouraging further investigations into this topic in the context of biologics and biosimilars development.
CRediT authorship contribution statement
Dimitar Kaynarov: Investigation, Data curation. Karina Marinova: Investigation. Rossitsa Marinova: Writing – original draft, Investigation, Data curation. Peicho Petkov: Software, Methodology, Investigation, Formal analysis. Lyudmila Velkova: Writing – original draft, Methodology, Investigation, Formal analysis. Aleksandar Dolashki: Investigation. Petar Petrov: Investigation. Leandar Litov: Supervision, Resources, Investigation. Elena Lilkova: Visualization, Investigation, Formal analysis. Pavlina Dolashka: Writing – review & editing, Supervision, Investigation, Conceptualization. Nevena Ilieva: Writing – review & editing, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The computational resources for this study were provided by the Discoverer supercomputer – Sofia (Bulgaria) thanks to Discoverer PetaSC and EuroHPC JU, by the HPC cluster BioSim at the Physics Faculty of the Sofia University “St. Kl. Ohridski” (Bulgaria) and at the Tricity Academic Supercomputer & networK Centre (CI TASK) – Gdansk (Poland).
Funding
This research was supported in part by the Bulgarian Science Fund under Grant KP-06-OPR 03-10/2018.
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101753.
The following supporting information can be downloaded:
Figure S1 The average cluster size in the p1 and p3 monocomponent solutions.
Figure S2 SASA of the aromatic residues in the p1 and p3 monocomponent solutions.
Figure S3 SASA of the hydrophobic, polar, and charged residues in the p1,and p3 monocomponent solutions.
Figure S4 Conformations of the largest clusters in the p1 and p3 monocomponent solutions.
Figure S5 Evolution of the secondary structure of the p3 monocomponent solution.
Figure S6 Evolution of the secondary structure of the p1+p3 multicomponent solution.
Figure S7 Number of residues involved in the four main secondary-structure elements in the p1 solution.
Figure S8 Number of residues involved in the four main secondary-structure elements in the p3 solution.
Figure S9 Final conformation (clusters formed) by the end of the multicomponent simulation.
Figure S10 The largest cluster in the multicomponent p1+p3 simulation.
Figure S11 SASA of the Trp and Phe residues in the multicomponent solution.
Figure S12 Percentage of the - stacking conformations in the peptide mixture simulation.
Figure S13 Fluorescence emission spectra of p1, p3, and p1+p3 compared to hydrophobic amino acid tryptophan.
Table S1 Inhibition of B. subtilis growth by p1, p3, (p1+p3) combination, and bombinin at different concentrations.
Table S2 Inhibition of E. coli 3458 growth by p1, p3, (p1+p3) combination, and bombinin at different concentrations.
Contributor Information
Rossitsa Marinova, Email: rosie.marinova@gmail.com.
Lyudmila Velkova, Email: Lyudmila.Velkova@orgchm.bas.bg.
Appendix A. Supplementary data
The following is the Supplementary material related to this article. The following supporting information can be downloaded:
Figure S1 The average cluster size in the (a) p1, resp. (b) p3 monocomponent solutions at a concentration of 10 mg/mL.
Figure S2 SASA of the aromatic residues in the (a) p1, resp. (b) p3 monocomponent solutions at a concentration of 10 mg/mL.
Figure S3 SASA of the hydrophobic, polar, and charged residues in the (a) p1, resp. (b) p3 monocomponent solutions at a concentration of 10 mg/mL.
Figure S4 Conformations of the largest clusters in the (a) p1 and (b) p3 monocomponent solutions.
Figure S5 Evolution of the secondary structure of the p3 monocomponent solution.
Figure S6 Evolution of the secondary structure of the p1+p3 multicomponent solution.
Figure S7 Number of residues involved in the four main secondary-structure elements in the p1 solution.
Figure S8 Number of residues involved in the four main secondary-structure elements in the p3 solution.
Figure S9 Final conformation (clusters formed) by the end of the multicomponent simulation.
Figure S10 The largest cluster in the multicomponent p1+p3 simulation, in surface representation: (a) coloured by residue type; and (b) coloured by peptide type.
Figure S11 SASA of the Trp and Phe residues in the multicomponent solution.
Figure S12 Percentage of the - stacking conformations in the peptide mixture simulation: (a) by kind per aromatic pairs; (b) by kind per amino acid type pairs.
Figure S13 Fluorescence emission spectra of p1, p3, and p1+p3 compared to hydrophobic amino acid tryptophan (concentration of 10 mg/mL) at immediately after the digestion of the peptides in a phosphate buffer (asymmetric quartz cuvette with 4/10 mm optical length at 22 °C).
Table S1 Inhibition of B. subtilis growth by p1, p3, (p1+p3) combination, and bombinin at different concentrations.
Table S2 Inhibition of E. coli 3458 growth by p1, p3, (p1+p3) combination, and bombinin at different concentrations.
.
Data availability
The synthetic data underlying the present study will be made available upon request.
References
- 1.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization C. World Health Organization; 2014. Antimicrobial Resistance: Global Report on Surveillance; p. xxii. 232. [Google Scholar]
- 4.World Health Organization C. World Health Organization; 2018. Monitoring Global Progress on Addressing Antimicrobial Resistance. [Google Scholar]
- 5.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020;19:311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q.-Y., Yan Z.-B., Meng Y.-M., Hong X.-Y., Shao G., Ma J.-J., Cheng X.-R., Liu J., Kang J., Fu C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Milit. Med. Res. 2021;8:48. doi: 10.1186/s40779-021-00343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fjell C.D., Hiss J.A., Hancock R.E.W., Schneider G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 8.Kang H.-K., Kim C., Seo C.H., Park Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017;55:1–12. doi: 10.1007/s12275-017-6452-1. [DOI] [PubMed] [Google Scholar]
- 9.Beutler B. Innate immunity: An overview. Mol. Immunol. 2004;40(12):845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Hoskin D.W., Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochimica et Biophys. Acta (BBA) - Biomembr. 2008;1778(2):357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P., Kizhakkedathu J.N., Straus S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in Vivo. Biomolecules. 2018;8 doi: 10.3390/biom8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luong H.X., Thanh T.T., Tran T.H. Antimicrobial peptides – advances in development of therapeutic applications. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theuretzbacher U., Outterson K., Engel A., Karlén A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2019;18:275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengupta D., Leontiadou H., Mark A.E., Marrink S.-J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochimica et Biophys. Acta (BBA) - Biomembr. 2008;1778(10):2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Mahlapuu M., Håkansson J., Ringstad L., Björn C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cellular Infect. Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haney E.F., Mansour S.C., Hancock R.E. Antimicrobial peptides: An introduction. Methods Mol. Biol. 2017;1548:3–22. doi: 10.1007/978-1-4939-6737-7_1. [DOI] [PubMed] [Google Scholar]
- 17.Ronc̆ević T., Puizina J., Tossi A. Antimicrobial peptides as anti-infective agents in pre-post-antibiotic era? Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vignoli Muniz G.S., De la Torre L.I., Duarte E.L., Lorenzón E.N., Cilli E.M., Balan A., Lamy M.T. Interaction of synthetic antimicrobial peptides of the Hylin a1 family with models of eukaryotic structures: Zwitterionic membranes and DNA. Biochem. Biophys. Rep. 2020;24 doi: 10.1016/j.bbrep.2020.100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forlano N., Bucci R., Contini A., Venanzi M., Placidi E., Gelmi M.L., Lettieri R., Gatto E. Non-conventional peptide self-assembly into a conductive supramolecular rope. Nanomaterials. 2023;13(2):333. doi: 10.3390/nano13020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W., Liu M., Yu L., Mo S., Deng Z., Liu S., Yang Y., Wang C., Wang C. Perturbation effect of single polar group substitution on the self-association of amphiphilic peptide helices. J. Colloid Interface Sci. 2022;610:1005–1014. doi: 10.1016/j.jcis.2021.11.154. [DOI] [PubMed] [Google Scholar]
- 21.Vaezi Z., Bortolotti A., Luca V., Perilli G., Mangoni M.L., Khosravi-Far R., Bobone S., Stella L. Aggregation determines the selectivity of membrane-active anticancer and antimicrobial peptides: The case of killerFLIP. Biochimica et Biophys. Acta (BBA) - Biomembr. 2020;1862(2) doi: 10.1016/j.bbamem.2019.183107. [DOI] [PubMed] [Google Scholar]
- 22.Lin B., Hung A., Singleton W., Darmawan K.K., Moses R., Yao B., Wu H., Barlow A., Sani M.A., Sloan A.J., Hossain M.A., Wade J.D., Hong Y., O’Brien-Simpson N.M., Li W. The effect of tailing lipidation on the bioactivity of antimicrobial peptides and their aggregation tendency. Aggregate. 2023;4 doi: 10.1002/agt2.329. [DOI] [Google Scholar]
- 23.Rotem S., Mor A. Antimicrobial peptide mimics for improved therapeutic properties. Biochimica et Biophys. Acta (BBA) - Biomembr. 2009;1788(8):1582–1592. doi: 10.1016/j.bbamem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Malekkhaiat Häffner S., Malmsten M. Influence of self-assembly on the performance of antimicrobial peptides. Curr. Opin. Colloid Interf. Sci. 2018;38:56–79. doi: 10.1016/j.cocis.2018.09.002. [DOI] [Google Scholar]
- 25.Feder R., Dagan A., Mor A. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 2000;275(6):4230–4238. doi: 10.1074/jbc.275.6.4230. [DOI] [PubMed] [Google Scholar]
- 26.Farrotti A., Conflitti P., Srivastava S., Ghosh J.K., Palleschi A., Stella L., Bocchinfuso G. Molecular dynamics simulations of the host defense peptide temporin l and its Q3K derivative: An atomic level view from aggregation in water to bilayer perturbation. Molecules. 2017;22(7) doi: 10.3390/molecules22071235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zai Y., Xi X., Ye Z., Ma C., Zhou M., Chen X., Siu S.W.I., Chen T., Wang L., Kwok H.F. Aggregation and its influence on the bioactivities of a novel antimicrobial peptide, temporin-PF, and its analogues. Int. J. Mol. Sci. 2021;22(9) doi: 10.3390/ijms22094509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goki H.N., Tehranizadeh A.Z., Saberi R.M., Khameneh B., Bazzaz S.F.B. Structure, function, and physicochemical properties of pore-forming antimicrobial peptides. Curr. Pharmaceut. Biotechnol. 2024;25(8):1041–1057. doi: 10.2174/0113892010194428231017051836. [DOI] [PubMed] [Google Scholar]
- 29.Syryamina V.N., De Zotti M., Toniolo C., Formaggio F., Dzuba S.A. Alamethicin self-assembling in lipid membranes: Concentration dependence from pulsed EPR of spin labels. Phys. Chem. Chem. Phys. 2018;20:3592–3601. doi: 10.1039/C7CP07298H. [DOI] [PubMed] [Google Scholar]
- 30.Afanasyeva E.F., Syryamina V.N., De Zotti M., Formaggio F., Toniolo C., Dzuba S.A. Peptide antibiotic trichogin in model membranes: Self-association and capture of fatty acids. Biochimica et Biophys. Acta (BBA) - Biomembr. 2019;1861(2):524–531. doi: 10.1016/j.bbamem.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Tesei G., Vazdar M., Jensen M.R., Cragnell C., Mason P.E., Heyda J., Skepö M., Jungwirth P., Lund M. Self-association of a highly charged arginine-rich cell-penetrating peptide. Proc. Natl. Acad. Sci. 2017;114(43):11428–11433. doi: 10.1073/pnas.1712078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sal-Man N., Oren Z., Shai Y. Preassembly of membrane-active peptides is an important factor in their selectivity toward target cells. Biochemistry. 2002;41(39):11921–11930. doi: 10.1021/bi0260482. [DOI] [PubMed] [Google Scholar]
- 33.Li W., O’Brien-Simpson N.M., Tailhades J., Pantarat N., Dawson R.M., Otvos L., Reynolds E.C., Separovic F., Hossain M.A., Wade J.D. Multimerization of a proline-rich antimicrobial peptide, Chex-Arg20, alters its mechanism of interaction with the escherichia coli membrane. Chem. Biol. 2015;22(9):1250–1258. doi: 10.1016/j.chembiol.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Remington J.M., Liao C., Sharafi M., Ste.Marie E.J., Ferrell J.B., Hondal R.J., Wargo M.J., Schneebeli S.T., Li J. Aggregation state of synergistic antimicrobial peptides. J. Phys. Chem. Lett. 2020;11(21):9501–9506. doi: 10.1021/acs.jpclett.0c02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velkova L., Nissimova A., Dolashki A., Daskalova E., Dolashka P., Topalova Y. Glycine-rich peptides from Cornu aspersum snail with antibacterial activity. Bulg. Chem. Commun. 2018;50:169–175. [Google Scholar]
- 36.Vassilev N.G., Simova S.D., Dangalov M., Velkova L., Atanasov V., Dolashki A., Dolashka P. An 1H NMR- and MS-based study of metabolites profiling of garden snail Helix aspersa Mucus. Metabolites. 2020;10(9) doi: 10.3390/metabo10090360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolashki A., Velkova L., Daskalova E., Zheleva N., Topalova Y., Atanasov V., Voelter W., Dolashka P. Antimicrobial activities of different fractions from Mucus of the garden snail Cornu aspersum. Biomedicines. 2020;8(9) doi: 10.3390/biomedicines8090315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topalova Y., Belouhova M., Velkova L., Dolashki A., Zheleva N., Daskalova E., Kaynarov D., Voelter W., Dolashka P. Effect and mechanisms of antibacterial peptide fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785. Biomedicines. 2022;10(3) doi: 10.3390/biomedicines10030672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 40.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2016;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126(1) doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 42.Parrinello M., Rahman A. Crystal structure and pair potentials: A molecular-dynamics study. Phys. Rev. Lett. 1980;45:1196. doi: 10.1103/physrevlett.45.1196. [DOI] [Google Scholar]
- 43.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 44.Hockney R., Goel S., Eastwood J. Quiet high-resolution computer models of a plasma. J. Comput. Phys. 1974;14(2):148–158. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 45.Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4(1):116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 46.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 47.Frishman D., Argos P. Knowledge-based secondary structure assignment. Prot.: Struct. Funct. Genet. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 48.Humphrey W., Dalke A., Schulten K. VMD – visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.Ilieva N., Petkov P., Lilkova E., Lazarova T., Dolashki A., Velkova L., Dolashka P., Litov L. In silico study on the structure of novel natural bioactive peptides. In: Lirkov I., Margenov S., editors. Large-Scale Scientific Computing; LSSC 2019; Cham: Springer International Publishing; 2020. pp. 332–339. [DOI] [Google Scholar]
- 50.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. In: The Proteomics Protocols Handbook. Walker J.M., editor. Humana Press; Totowa, NJ: 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [DOI] [Google Scholar]
- 51.Meher P.K., Sahu T.K., Saini V., Rao A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017;7:42362. doi: 10.1038/srep42362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Jesus A.J., Allen T.W. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochimica et Biophys. Acta (BBA) - Biomembr. 2013;1828(2):864–876. doi: 10.1016/j.bbamem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Clark S., Jowitt T.A., Harris L.K., Knight C.G., Dobson C.B. The lexicon of antimicrobial peptides: A complete set of arginine and tryptophan sequences. Commun. Biol. 2021;4:605. doi: 10.1038/s42003-021-02137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J., Mura M., Zhou Y., Pinna M., Zvelindovsky A.V., Dennison S.R., Phoenix D.A. The cooperative behaviour of antimicrobial peptides in model membranes. Biochimica et Biophys. Acta (BBA) - Biomembr. 2014;1838(11):2870–2881. doi: 10.1016/j.bbamem.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Huang H.W. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochimica et Biophys. Acta (BBA) - Biomembr. 2006;1758(9):1292–1302. doi: 10.1016/j.bbamem.2006.02.001. Membrane Biophysics of Antimicrobial Peptides. [DOI] [PubMed] [Google Scholar]
- 56.Oren Z., Shai Y. Mode of action of linear amphipathic -helical antimicrobial peptides. Peptide Sci. 1998;47(6):451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 57.Petkov P., Lilkova E., Ilieva N., Litov L. Self-association of antimicrobial peptides: A molecular dynamics simulation study on Bombinin. Int. J. Mol. Sci. 2019;20(21):5450. doi: 10.3390/ijms20215450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu G., Ding J., Li H., Li L., Zhao R., Shen Z., Fan X., Xi T. Effects of cations and PH on antimicrobial activity of Thanatin and s-Thanatin against Escherichia coli ATCC25922 and B. subtilis ATCC 21332. Nat. Preced. 2008;57(6):552–557. doi: 10.1038/npre.2008.2006.1. [DOI] [PubMed] [Google Scholar]
- 59.Abdou A.M., Higashiguchi S., Aboueleinin A., Kim M., Ibrahim H.R. Antimicrobial peptides derived from hen egg lysozyme with inhibitory effect against Bacillus species. Food Control. 2007;18(2):173–178. doi: 10.1016/j.foodcont.2005.09.010. [DOI] [Google Scholar]
- 60.Lu Y., Tian H., Chen R., Liu Q., Jia K., Hu D.-L., Chen H., Ye C., Peng L., Fang R. Synergistic antimicrobial effect of antimicrobial peptides CATH-1, CATH-3, and PMAP-36 with erythromycin against bacterial pathogens. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.953720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laadhari M., Arnold A.A., Gravel A.E., Separovic F., Marcotte I. Interaction of the antimicrobial peptides caerin 1.1 and aurein 1.2 with intact bacteria by 2H solid-state NMR. Biochimica et Biophys. Acta (BBA) - Biomembr. 2016;1858(12):2959–2964. doi: 10.1016/j.bbamem.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Eagle H., Musselman A.D. The rate of bactericidal action of penicillin in vitro as a function of its concentration, and its paradoxically reduced activity at high concentrations against certain organisms. J. Exp. Med. 1948;88:99–131. doi: 10.1084/jem.88.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shentu H., Ye P., Zhou Q., Li P., Gu Q. Purification, characterization, and mode of action of Sakacin ZFM225, a novel bacteriocin from Lactobacillus sakei ZFM225. Biochem. Biophys. Rep. 2023;35 doi: 10.1016/j.bbrep.2023.101494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sang Y., Blecha F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008;9(2):227–235. doi: 10.1017/S1466252308001497. [DOI] [PubMed] [Google Scholar]
- 65.Simons A., Alhanout K., Duval R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. 2020;8(5) doi: 10.3390/microorganisms8050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He Y., Lazaridis T. Activity determinants of helical antimicrobial peptides: A large-scale computational study. PLOS ONE. 2013;8(6):1–13. doi: 10.1371/journal.pone.0066440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochimica et Biophys. Acta (BBA) - Biomembr. 2009;1788(8):1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 68.Pignataro M.F., Herrera M.G., Dodero V.I. Evaluation of peptide/protein self-assembly and aggregation by spectroscopic methods. Molecules. 2020;25(20) doi: 10.3390/molecules25204854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venanzi M., Savioli M., Cimino R., Gatto E., Palleschi A., Ripani G., Cicero D., Placidi E., Orvieto F., Bianchi E. A spectroscopic and molecular dynamics study on the aggregation process of a long-acting lipidated therapeutic peptide: The case of semaglutide. Soft Matter. 2020;16(44):10122–10131. doi: 10.1039/D0SM01011A. [DOI] [PubMed] [Google Scholar]
- 70.Wang W. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 2005;289(1):1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 71.Shinitzky M., Goldman R. Fluorometric detection of histidine-tryptophan complexes in peptides and proteins. Eur. J. Biochem. 1967;3(2):139–144. doi: 10.1111/j.1432-1033.1967.tb19508.x. [DOI] [PubMed] [Google Scholar]
- 72.Loewenthal R., Sancho J., Fersht A.R. Fluorescence spectrum of barnase: Contributions of three tryptophan residues and a histidine-related pH dependence. Biochemistry. 1991;30(27):6775–6779. doi: 10.1021/bi00241a021. [DOI] [PubMed] [Google Scholar]
- 73.Buffy J.J., Waring A.J., Lehrer R.I., Hong M. Immobilization and aggregation of the antimicrobial peptide protegrin-1 in lipid bilayers investigated by solid-state NMR. Biochemistry. 2003;42(46):13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- 74.Bechinger B., Juhl D.W., Glattard E., Aisenbrey C. Revealing the mechanisms of synergistic action of two Magainin antimicrobial peptides. Front. Med. Technol. 2020;2 doi: 10.3389/fmedt.2020.615494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G., Baeder D.Y., Regoes R.R., Rolff J. Combination effects of antimicrobial peptides. Antimicrob. Agents. Chemother. 2016;60(3):1717–1724. doi: 10.1128/aac.02434-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao S., Zhai X., Cheng Y., Zhang R., Wang W., Hou H. Starch/PBAT blown antimicrobial films based on the synergistic effects of two commercial antimicrobial peptides. Int. J. Biol. Macromol. 2022;204:457–465. doi: 10.1016/j.ijbiomac.2022.01.183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
Data Availability Statement
The synthetic data underlying the present study will be made available upon request.