Abstract
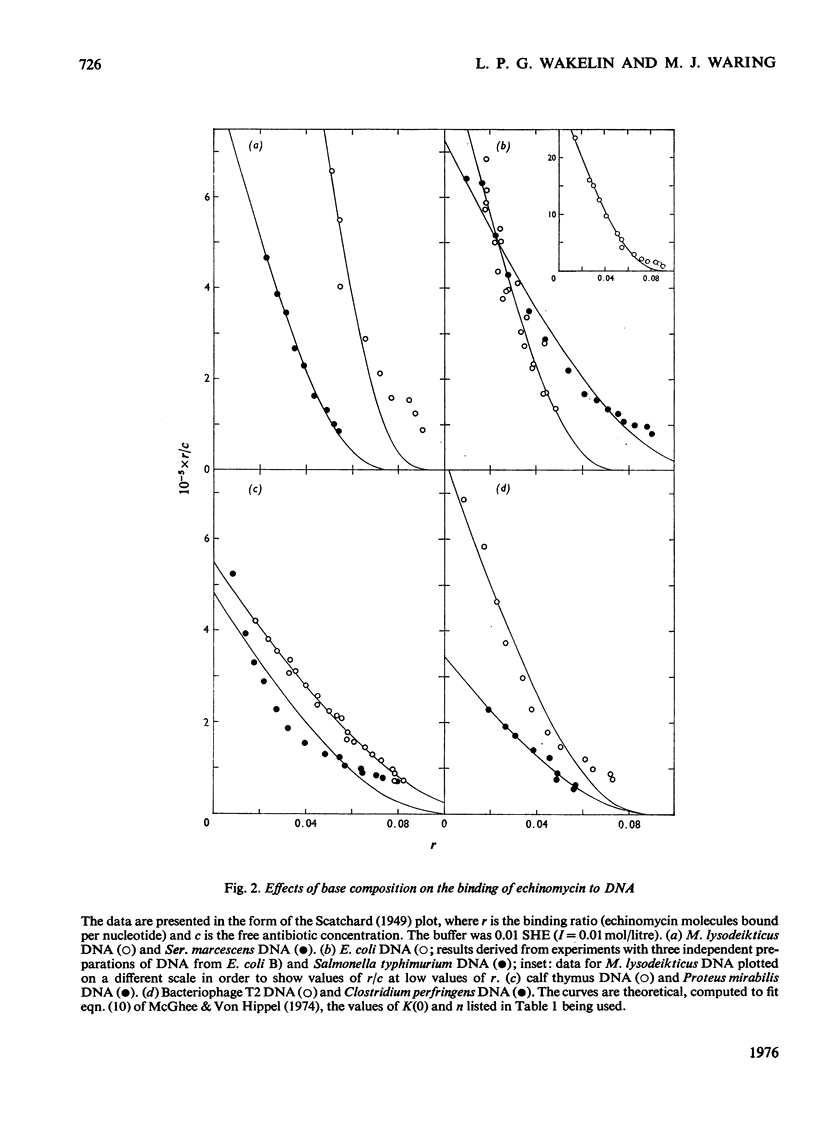
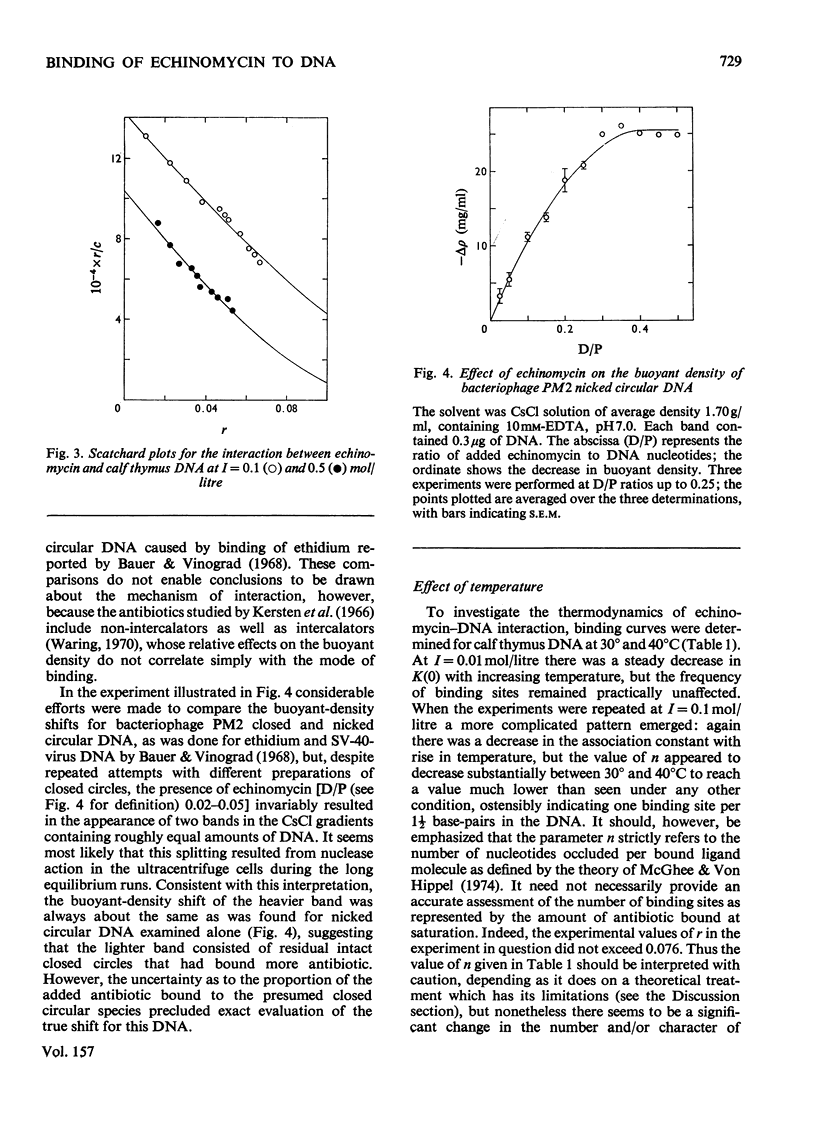
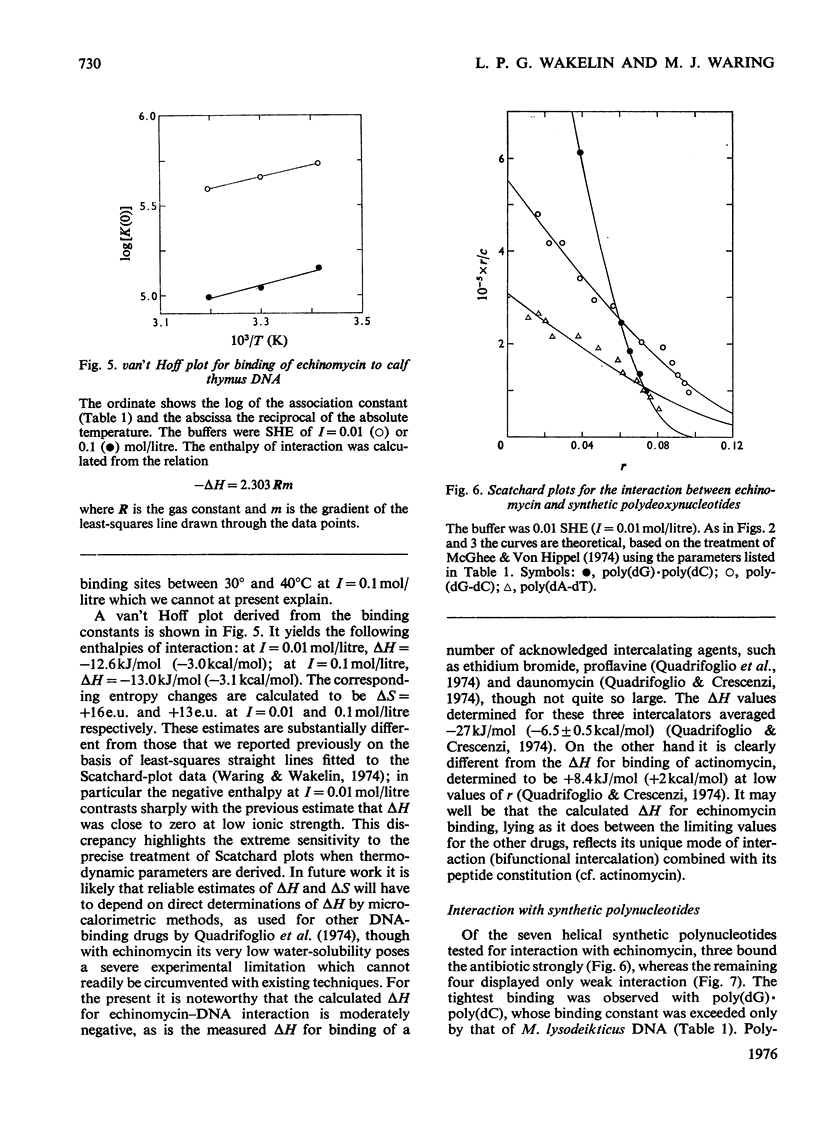
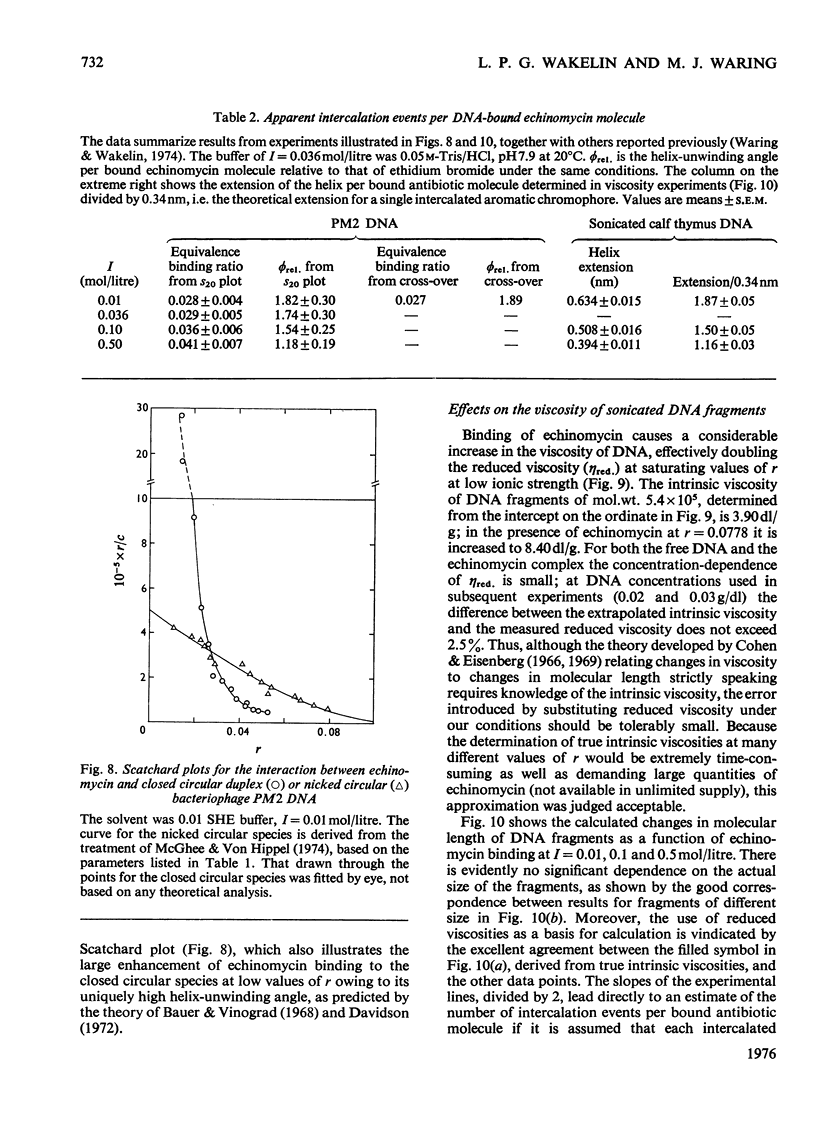
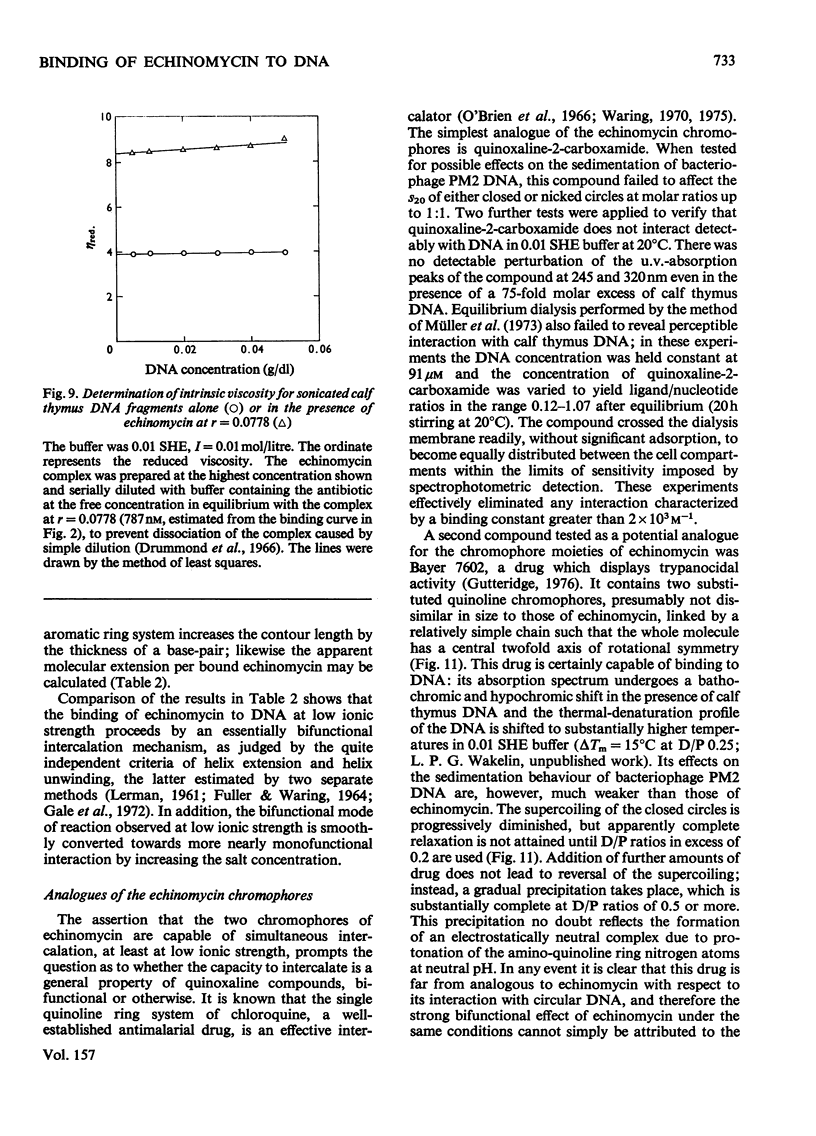
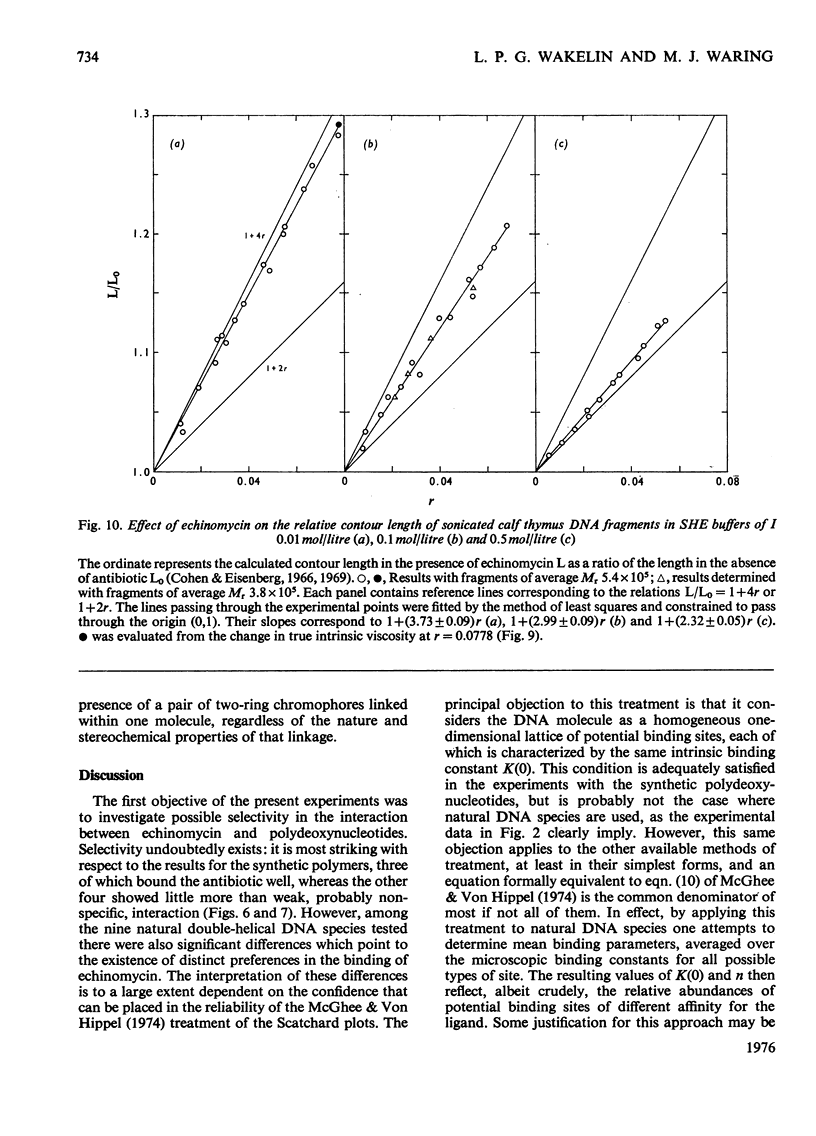
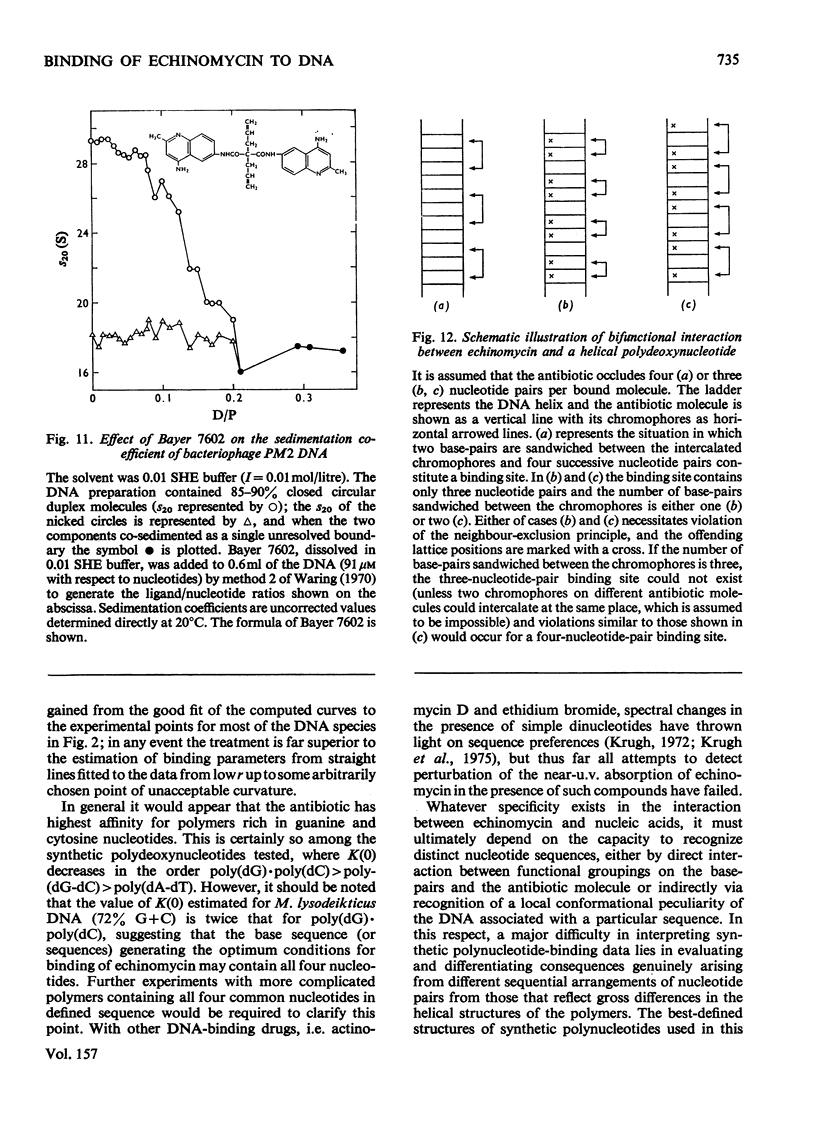
Echinomycin is a peptide antibiotic which binds strongly to double-helical DNA up to a limit of approximately one molecule per five base-pairs. There is no detectable interaction with rRNA and only extremely feeble non-specific interaction with poly(rA)-poly(rU). Heat denaturation of DNA greatly decreases the binding, and similarly limited interaction is observed with naturally occurring single-stranded DNA. Association constants for binding to nine double-helical DNA species from different sources are presented; they vary by a factor of approximately 10, but are not simply related to the gross base composition. The interaction with DNA is ionic-strength-dependent, the binding constant falling by a factor of 4 when the ionic strength is raised from 0.01 to 0.10mol/litre. From the effect of temperature on the association constant for calf thymus DNA, the enthalpy of interaction is calculated to be about -13kJ/mol (-3kcal/mol). Binding of echinomycin persists in CsCl gradients and the buoyant density of nicked bacteriophage PM2 DNA is decreased by 25 mg/ml. Echinomycin interacts strongly with certain synthetic poly-deoxynucleotides, the binding constant decreasing in the order poly(dG)-poly(dC) greater than poly(dG-dC) greater than poly(dA-dT). For the latter two polymers the number of base-pairs occluded per bound antibiotic molecule is calculated to be three, whereas for poly(dG)-poly(dC) it is estimated to be four to five. Poly(dA)-poly(dT) and poly(dI)-poly(dC) interact only very weakly with the antibiotic. Poly(dI-dC) interacts to a slightly greater extent, but the binding curve is quite unlike that seen with the three strongly binding synthetic polynucleotides. Echinomycin affects the supercoiling of closed circular duplex bacteriophage PM2 DNA in the characteristic fashion of intercalating drugs. At low ionic strength the unwinding angle is almost twice that of ethidium. Likewise the extension of the helix, determined from changes in the viscosity of rod-like sonicated DNA fragments, is nearly double that expected for a simple (monofunctional) intercalation process. On this basis the interaction process is characterized as bifunctional intercalation. At higher ionic strength the unwinding angle relative to that of ethidium and the helix extension per bound echinomycin molecule fall, indicating a smooth progression towards more nearly monofunctional intercalation. Two simpler compounds which act as analogues of the quinoxaline chromophores of echinomycin, quinoxaline-2-carboxamide and the trypanocidal drug Bayer 7602, interact with DNA very much more weakly than does echinomycin, showing that the peptide portion of the antibiotic plays an essential role in determining the strength and specificity of the interaction.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen F. S., Gray D. M., Roberts G. P., Tinoco I., Jr The ultraviolet circular dichroism of some natural DNAs and an analysis of the spectra for sequence information. Biopolymers. 1972;11(4):853–879. doi: 10.1002/bip.1972.360110410. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hukins D. W., Smith P. J., Watts L. Structural details of double-helix observed for DNAs containing alternating purine and pyrimidine sequences. J Mol Biol. 1974 Sep 15;88(2):523–533. doi: 10.1016/0022-2836(74)90499-9. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Letter: The structure of polydeoxyguanylic acid with polydeoxycytidylic acid. J Mol Biol. 1974 Sep 15;88(2):551–552. doi: 10.1016/0022-2836(74)90502-6. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- BERGER J., LA SALA E. R., SCOTT W. E., MELTSNER B. R., STERNBACH L. H., KAISER S., TEITEL S., MACH E., GOLDBERG M. W. Antibiotics X-948 and X-1008. Experientia. 1957 Nov 15;13(11):434–436. doi: 10.1007/BF02157235. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Bram S. Secondary structure of DNA depends on base composition. Nat New Biol. 1971 Aug 11;232(2):174–176. doi: 10.1038/newbio232174a0. [DOI] [PubMed] [Google Scholar]
- Bram S. The secondary structure of DNA in solution and in nucleohistone. J Mol Biol. 1971 May 28;58(1):277–288. doi: 10.1016/0022-2836(71)90246-4. [DOI] [PubMed] [Google Scholar]
- Bram S., Tougard P. Polymorphism of natural DNA. Nat New Biol. 1972 Oct 4;239(92):128–131. doi: 10.1038/newbio239128a0. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Conformation studies on the sodium and cesium salts of calf thymus deoxyribonucleic acid (DNA). Biopolymers. 1966 Apr-May;4(4):429–440. doi: 10.1002/bip.1966.360040404. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Zimm B. H. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J Mol Biol. 1965 Jul;12(3):525–536. doi: 10.1016/s0022-2836(65)80310-2. [DOI] [PubMed] [Google Scholar]
- DAVIES D. R., BALDWIN R. L. X-ray studies on two synthetic DNA copolymers. J Mol Biol. 1963 Apr;6:251–255. doi: 10.1016/s0022-2836(63)80086-8. [DOI] [PubMed] [Google Scholar]
- Davidson N. Effect of DNA length on the free energy of binding of an unwinding ligand to a supercoiled DNA. J Mol Biol. 1972 May 14;66(2):307–309. doi: 10.1016/0022-2836(72)90482-2. [DOI] [PubMed] [Google Scholar]
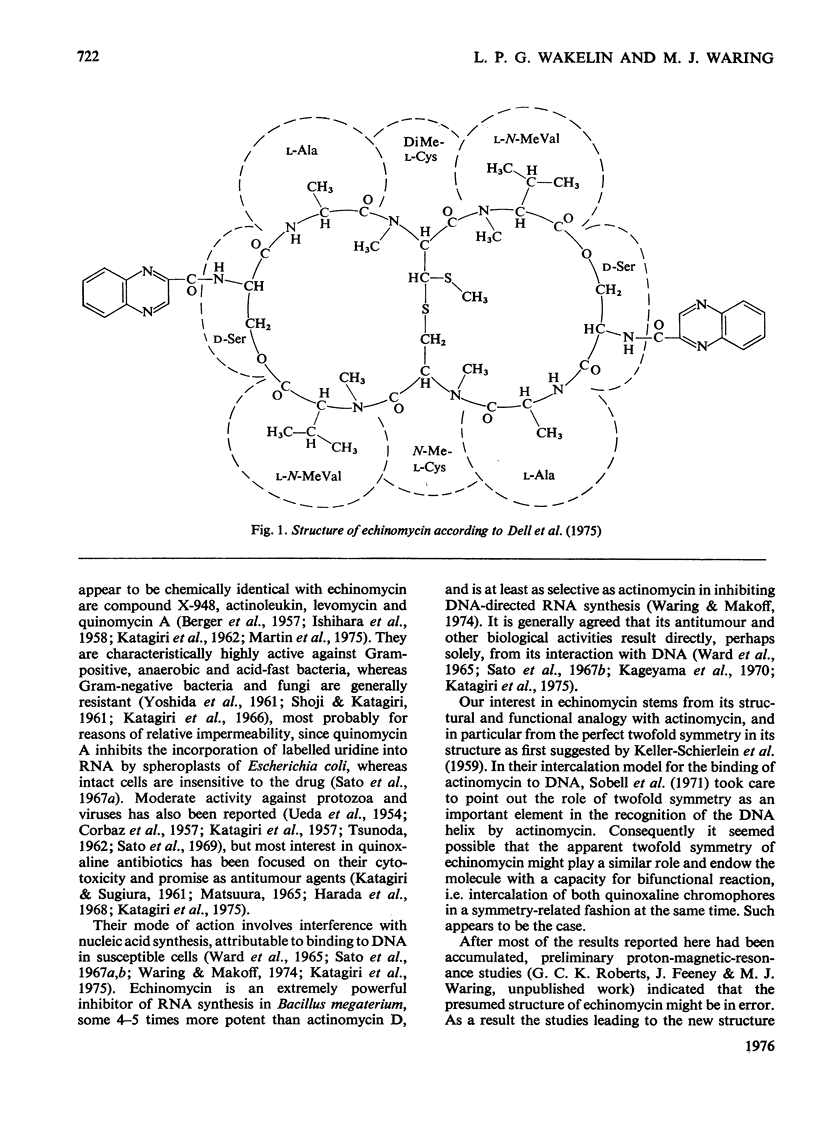
- Dell A., Williams D. H., Morris H. R., Smith G. A., Feeney J., Roberts G. C. Structure revision of the antibiotic echinomycin. J Am Chem Soc. 1975 Apr 30;97(9):2497–2502. doi: 10.1021/ja00842a029. [DOI] [PubMed] [Google Scholar]
- Drummond D. S., Pritchard N. J., Simpson-Gildemeister V. F., Peacocke A. R. Interaction of aminoacridines with deoxyribonucleic acid: viscosity of the complexes. Biopolymers. 1966 Oct-Nov;4(9):971–987. doi: 10.1002/bip.1966.360040903. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. C., Kodama M., Wells R. D. Enzymatic and physical studies on (dI-dC) n -(dI-dC) n and (dG-dC) n -(dG-dC) n . Biochemistry. 1972 Feb 29;11(5):805–815. doi: 10.1021/bi00755a020. [DOI] [PubMed] [Google Scholar]
- Harada Y., Sunagawa N., Katagiri K. Segregation of the nucleolar materials produced by quinoxaline antibiotics in JTC-13 cells. Comparison of effects among 4-nitro-quinoline 1-oxide, actinomycin-B, and quinoxaline antibiotics. Gan. 1968 Dec;59(6):513–522. [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. HELIX--RANDOM COIL TRANSITIONS IN DNA HOMOPOLYMER PAIRS. J Mol Biol. 1964 Apr;8:452–469. doi: 10.1016/s0022-2836(64)80003-6. [DOI] [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. Helix-random coil transitions in synthetic DNAs of alternating sequence. J Mol Biol. 1962 Aug;5:172–184. doi: 10.1016/s0022-2836(62)80082-5. [DOI] [PubMed] [Google Scholar]
- ISHIHARA S., UTAHARA R., SUZUKI M., OKAMI Y., UMEZAWA H. Studies on actinoleukin; relation to echinomycin and levomycin. J Antibiot (Tokyo) 1958 Jul;11(4):160–161. [PubMed] [Google Scholar]
- KATAGIRI K., SHOJI J., YOSHISA T. Identity of levomycin and quinomycin A (echimomycin). J Antibiot (Tokyo) 1962 Nov;15:273–273. [PubMed] [Google Scholar]
- Kageyama M., Hasegawa M., Inagaki A., Egami F. Interaction of antibiotics with deoxyribonucleic acid. I. Sensitivity of the complexes to nucleolytic enzymes. J Biochem. 1970 Apr;67(4):549–557. doi: 10.1093/oxfordjournals.jbchem.a129280. [DOI] [PubMed] [Google Scholar]
- Kersten W., Kersten H., Szybalski W. Physicochemical properties of complexes between deoxyribonucleic acid and antibiotics which affect ribonucleic acid synthesis (actinomycin, daunomycin, cinerubin, nogalamycin, chormomycin, mithramycin, and olivomycin). Biochemistry. 1966 Jan;5(1):236–244. doi: 10.1021/bi00865a031. [DOI] [PubMed] [Google Scholar]
- Kohn K. W., Waring M. J., Glaubiger D., Friedman C. A. Intercalative binding of ellipticine to DNA. Cancer Res. 1975 Jan;35(1):71–76. [PubMed] [Google Scholar]
- Krugh T. R. Association of actinomycin D and deoxyribodinucleotides as a model for binding of the drug to DNA. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1911–1914. doi: 10.1073/pnas.69.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugh T. R., Wittlin F. N., Cramer S. P. Ethidium bromide-dinucleotide complexes. Evidence for intercalation and sequence preferences in binding to double-stranded nucleic acids. Biopolymers. 1975 Jan;14(1):197–210. doi: 10.1002/bip.1975.360140114. [DOI] [PubMed] [Google Scholar]
- LERMAN L. S. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961 Feb;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- Langridge R. Nucleic acids and polynucleotides. J Cell Physiol. 1969 Oct;74(2 Suppl):1–20. doi: 10.1002/jcp.1040740403. [DOI] [PubMed] [Google Scholar]
- Latt S. A., Sober H. A. Protein-nucleic acid interactions. II. Oligopeptide-polyribonucleotide binding studies. Biochemistry. 1967 Oct;6(10):3293–3306. doi: 10.1021/bi00862a040. [DOI] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- MATSUURA S. STUDIES ON QUINOXALINE ANTIBIOTICS. IV. SELECTIVE ANTITUMOR ACTIVITY OF EACH QUINOXALINE ANTIBIOTIC. J Antibiot (Tokyo) 1965 Jan;18:43–46. [PubMed] [Google Scholar]
- Martin D. G., Mizsak S. A., Biles C., Stewart J. C., Bacynsky L., Meulman P. A. Structure of quinomycin antibiotics. J Antibiot (Tokyo) 1975 Apr;28(4):332–336. doi: 10.7164/antibiotics.28.332. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Mitsui Y., Langridge R., Shortle B. E., Cantor C. R., Grant R. C., Kodama M., Wells R. D. Physical and enzymatic studies on poly d(I-C)-poly d(I-C), an unusual double-helical DNA. Nature. 1970 Dec 19;228(5277):1166–1169. doi: 10.1038/2281166a0. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M., Waring M. J. A non-intercalating proflavine derivative. Eur J Biochem. 1973 Nov 1;39(1):223–234. doi: 10.1111/j.1432-1033.1973.tb03120.x. [DOI] [PubMed] [Google Scholar]
- O'Brien R. L., Allison J. L., Hahn F. E. Evidence for intercalation of chloroquine into DNA. Biochim Biophys Acta. 1966 Dec 21;129(3):622–624. doi: 10.1016/0005-2787(66)90078-5. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pyeritz R. E., Schlegel R. A., Thomas C. A., Jr Hydrodynamic shear breakage of DNA may produce single-chained terminals. Biochim Biophys Acta. 1972 Jul 31;272(4):504–509. doi: 10.1016/0005-2787(72)90505-9. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Crescenzi V. On the binding of actinomycin and of daunomycin to DNA: a calorimetric and spectroscopic investigation. Biophys Chem. 1974 Jun;2(1):64–69. doi: 10.1016/0301-4622(74)80025-6. [DOI] [PubMed] [Google Scholar]
- RUBENSTEIN I., THOMAS C. A., Jr, HERSHEY A. D. The molecular weights of T2 bacteriophage DNA and its first and second breakage products. Proc Natl Acad Sci U S A. 1961 Aug;47:1113–1122. doi: 10.1073/pnas.47.8.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- SHOJI J. I., KATAGIRI K. Studies on quinoxaline antibiotics. II. New antibiotics, triostins A, B and C. J Antibiot (Tokyo) 1961 Nov;14:335–339. [PubMed] [Google Scholar]
- Sato K., Niinomi Y., Katagiri K., Matsukage A., Minagawa T. Prevention of phage multiplication by quinomycin A. Biochim Biophys Acta. 1969 Jan 21;174(1):230–238. doi: 10.1016/0005-2787(69)90246-9. [DOI] [PubMed] [Google Scholar]
- Sato K., Shiratori O., Katagiri K. The mode of action of quinoxaline antibiotics. Interaction of quinomycin A with deoxyribonucleic acid. J Antibiot (Tokyo) 1967 Sep;20(5):270–276. [PubMed] [Google Scholar]
- Sato K., Yoshida T., Katagiri K. Inhibition of RNA synthesis in E. coli protoplasts by quinomycins A and C. J Antibiot (Tokyo) 1967 Jul;20(3):188–189. [PubMed] [Google Scholar]
- Sobell H. M., Jain S. C., Sakore T. D., Nordman C. E. Stereochemistry of actinomycin--DNA binding. Nat New Biol. 1971 Jun 16;231(24):200–205. doi: 10.1038/newbio231200a0. [DOI] [PubMed] [Google Scholar]
- TSUNODA A. Chemoprophylaxis of polimyelitis in mice with quinomycin. (Studies on the antibiotic substances from Actinomycetes. XLVI). J Antibiot (Tokyo) 1962 Mar;15:60–66. [PubMed] [Google Scholar]
- TUBBS R. K., DITMARS W. E., Jr, VANWINKLE Q. HETEROGENEITY OF THE INTERACTION OF DNA WITH ACRIFLAVINE. J Mol Biol. 1964 Aug;9:545–557. doi: 10.1016/s0022-2836(64)80226-6. [DOI] [PubMed] [Google Scholar]
- UEDA M., TANIGAWA Y., OKAMI Y., UMEZAWA H. A new toxic antibiotic, actinoleukin, produced by a streptomycete. J Antibiot (Tokyo) 1954 Aug;7(4):125–126. [PubMed] [Google Scholar]
- Wakelin L. P., Waring M. J. The unwinding of circular deoxyribonucleic acid by phenanthridinium drugs: structure-activity relations for the intercalation reaction. Mol Pharmacol. 1974 May;10(3):544–561. [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]
- Ward D. C., Reich E., Goldberg I. H. Base specificity in the interaction of polynucleotides with antibiotic drugs. Science. 1965 Sep 10;149(3689):1259–1263. doi: 10.1126/science.149.3689.1259. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Henley S. M. Stereochemical aspects of the interaction between steroidal diamines and DNA. Nucleic Acids Res. 1975 Apr;2(4):567–586. doi: 10.1093/nar/2.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J. Stablilzation of two-standard ribohomopolymer helices and destabilzation of a three-stranded helix by ethidium bromide. Biochem J. 1974 Nov;143(2):483–486. doi: 10.1042/bj1430483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J. The effects of antimicrobial agents on ribonucleic acid polymerase. Mol Pharmacol. 1965 Jul;1(1):1–13. [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P., Lee J. S. A solvent-partition method for measuring the binding of drugs to DNA. Application to the quinoxaline antibiotics echinomycin and triostin A. Biochim Biophys Acta. 1975 Oct 1;407(2):200–212. doi: 10.1016/0005-2787(75)90285-3. [DOI] [PubMed] [Google Scholar]
- Waring M., Makoff A. Breakdown of pulse-labeled ribonucleic acid and polysomes in Bacillus megaterium: actions of streptolydigin, echinomycin, and triostins. Mol Pharmacol. 1974 Mar;10(2):214–224. [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- YOSHIDA T., KATAGIRI K., YOKOZAWA S. Studies on quinoxaline antibiotics. II. Isolation and properties of quinomycins A, B and C. J Antibiot (Tokyo) 1961 Nov;14:330–334. [PubMed] [Google Scholar]
- Zasedatelev A. S., Gurskii G. V., Vol'kenshtein M. V. Theory of one-dimensional adsorption. I. Adsorption of small molecules on a homopolymer. Mol Biol. 1971 Mar-Apr;5(2):194–198. [PubMed] [Google Scholar]


