Simple Summary
The TP53 mutation is one of the prevalent genetic alterations in human cancers and is often linked to a poor prognosis. While earlier studies have produced mixed results, they frequently involved small patient groups focused on specific breast cancer subtypes and treatments. To clarify these findings, we examined the clinical relevance of TP53 mutations in 650 patients across all subtypes, with consistent treatment based on subtype. In total, 172 (26.5%) had TP53 mutations, including 34 (19.8%) with missense hotspot mutations. Those with TP53 mutations had worse outcomes, with a 10-year recurrence-free survival rate of 83.5% compared to 86.6% for those without (p = 0.026), and a 10-year overall survival rate of 88.1% versus 91.0% (p = 0.003). However, the outcomes among patients with TP53 mutation did not differ significantly by mutation types or locations. Consequently, further research is necessary to explore the clinical relevance of the characteristics of TP53 mutation.
Keywords: TP53 mutation, missense mutation, missense hotspot, breast cancer, recurrence-free survival, overall survival
Abstract
Background: The TP53 mutation is one of the most frequently identified mutations in human cancers and is typically associated with a poor prognosis. However, there are conflicting findings regarding its impact. We aimed to clarify the clinical relevance of TP53 mutations across all breast cancer subtypes and treatments utilizing long-term follow-up data. Methods: We retrospectively identified the data of breast cancer patients who underwent TP53 mutation testing. Stratified log-rank tests and Cox regression analysis were performed to compare oncologic outcomes based on TP53 mutation status and the characteristics of these mutations, including types and locations. Mutations in exons 5-9 were identified using polymerase chain reaction—denaturing high-performance liquid chromatography (PCR-DHPLC) and direct sequencing. Results: Between January 2007 and December 2015, 650 breast cancer patients underwent TP53 mutation testing in Gangnam Severance Hospital. The TP53 mutations were identified in 172 patients (26.5%), with 34 (19.8%) exhibiting missense hotspot mutations. Patients with TP53 mutations (TP53-mutated group) had worse prognosis, demonstrated by a 10-year recurrence-free survival (RFS) rate of 83.5% compared to 86.6% in patients without mutations (HR, 1.67; p = 0.026) and a 10-year overall survival (OS) rate of 88.1% versus 91.0% (HR, 3.02; p = 0.003). However, subgroup analyses within the TP53-mutated group did not reveal significant differences in oncologic outcomes based on mutation types and locations. Conclusions: Our findings establish that TP53 mutations are linked to poorer oncologic outcomes in breast cancer across all subtypes. Yet, within the TP53-mutated group, the specific characteristics of TP53 mutations do not influence oncologic outcomes.
1. Introduction
The TP53 gene, which codes for the tumor-suppressor protein p53, is the most frequently mutated gene in human cancers [1]. Located on chromosome 17p13.1, TP53 consists of 11 exons, 10 introns, and 393 amino acid residues, and encodes the p53 protein, a transcription factor with distinct amino-terminal, DNA-binding, and carboxy-terminal domains [2]. The TP53-activated pathway exerts tumor-suppressive functions by regulating DNA repair, cell-cycle arrest, senescence, and apoptosis, thereby inhibiting early tumorigenesis, tumor growth, and progression [3,4,5]. As a result, the activation of p53 in normal tissues is critical for preventing tumorigenesis. However, tumors with TP53 mutations not only lose these tumor-suppressive functions but also often acquire gain-of-function mutations that promote tumor growth [6,7]. Consequently, TP53-mutated tumors typically exhibit rapid progression, resistance to treatment, and a poor prognosis [8,9,10].
According to the International Agency for Research on Cancer (IACR) database, over 75% of TP53 mutations were missense mutations, with approximately 97% located in exons encoding the DNA-binding domain (DBD, residue 98-292). Six codons (175, 220, 245, 248, 273, and 282) are recognized as well-known missense hotspots, each accounting for more than 2% of all missense mutations (https://www.cbioportal.org/, (accessed on 29 October 2024)). These single nucleotide substitutions disrupt the 3D structure of the p53 protein or impair its ability to bind DNA, leading to a loss-of-function [11].
TP53 mutations are identified in nearly 30% of all breast cancers [12,13]. Numerous preclinical and clinical studies have explored the clinical significance of TP53 mutations in breast cancer, with most associating them with poor prognosis [14,15,16,17]. However, studies that challenging the conventional understanding of the clinical relevance of TP53 mutations have also been published. Ostrowski et al. found that while tumors with p53 expression exhibited more aggressive clinicopathological features, there was no significant difference in survival outcomes compared to tumors without p53 expression [18]. In a study by Shiao et al., which evaluated the association between p53 gene alterations and survival in patients with TP53 mutations, differences in p53 gene alteration patterns were observed between Black and White patients. Among Black patients, TP53-mutated breast cancer was associated with poorer outcomes, whereas no such correlation was found in White patients [19]. Additionally, a meta-analysis including 26 studies with 3476 patients reported that patients with TP53 mutations had a better response to neoadjuvant chemotherapy [20]. However, studies investigating the association between TP53 mutations and breast cancer have generally been limited by small patient cohorts and prone to selection bias due to the varying prevalence of TP53 mutations across molecular subtypes. Moreover, the lack of standardization in TP53 mutation testing methods and treatment protocols, such as chemotherapy regimens, complicates the interpretation of findings. As a result, the clinical relevance of TP53 mutations in breast cancer remains controversial.
As breast cancer treatment becomes increasingly personalized, there is a growing need not only to access the presence of TP53 mutations but also to adopt a molecular approach to better understand these mutations. In breast tumors, gene sequencing revealed that missense mutations were dominant, accounting for about 80%, while other mutations, such as nonsense and frameshift mutations, made up approximately 20%. Additionally, the mutational events also differed from those observed in other cancers [21,22]. Moreover, TP53 mutations in breast cancer act through various mechanisms, including impairing DNA damage repair, promoting cancer stemness, and enhancing inflammatory responses, each of which may require different therapeutic strategies [23,24,25]. Therefore, more in-depth research on the specific types and locations of TP53 mutations is urgently needed. However, research in this area remains limited.
In light of these considerations, we investigated the association between TP53 mutations and prognosis in breast cancer patients using long-term follow-up data. Additionally, we explored the clinical relevance of the characteristics of TP53 mutations among patients harboring these mutations.
2. Materials and Methods
2.1. Data Collection
We retrospectively identified patients diagnosed with breast cancer who underwent TP53 mutation testing at Gangnam Severance Hospital from January 2007 to December 2015. Clinicopathological data were collected from electronic medical records including age at diagnosis, histologic subtype, histologic grade, estrogen receptor (ER) and progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) status, lymphovascular invasion (LVI), Ki67 index, T stage, N stage, and implementation of (neo)adjuvant chemotherapy. We also collected genetic information about TP53 mutation status and characteristics of TP53 mutation. Patients diagnosed with recurrent breast cancer and de novo metastatic breast cancer were excluded. We also excluded bilateral breast cancer to minimize bias from concomitant pathologies.
T stage and N stage were determined using surgical specimens according to the American Joint Committee on Cancer Guidelines (AJCC) (8th edition). Hormone receptor (HR), ER and PR status was determined from surgical specimen using immunohistochemistry (IHC). Positive for ER and PR were defined as those in which more than 1% of tumor nuclei in the sample were stained [26]. HER2 status was assessed following the recommendation of the 2013 American Society of Clinical Oncology (ASCO)/College of American Pathologist (CAP) [27]. Triple-negative breast cancer (TNBC) refers to tumors that are negative for ER and PR, and do not exhibit HER2 overexpression, as determined by IHC. In this study, we applied a 20% threshold, commonly used in luminal-like subtypes, to classify Ki67 status as high or low, establishing a broadly applicable standard across all breast cancer subtypes [28,29]. Neoadjuvant and adjuvant systemic therapies, including chemotherapy, radiotherapy and endocrine therapy, were administered in accordance with established guidelines based on the age at diagnosis, molecular subtype, and axillary lymph node status.
2.2. Mutational Analysis of TP53 Gene
Mutational analysis of exons 5-9 of the TP53 gene was performed using polymerase chain reaction—denaturing high performance liquid chromatography (PCR-DHPLC) and direct sequencing. Approximately 1 mg of samples from either biopsies or surgical specimens, freshly frozen of paraffin-embedded, were cut into pieces, and DNA was extracted using the Easy-DNATM kit (Invitrogen, Carlsbad, CA, USA) with 100 ng/µL of DNA used for each PCR reaction, where each PCR was performed in a 20 µL reaction mixture containing 100 ng of DNA, 20 µM of forward and reverse primers, 2 µL of Taq buffer (10×), 2.5 mM of deoxyribonucleotide triphosphates (dNTPs), 2.5 mM of MgCl2, and 0.7 U of Taq DNA polymerase, under conditions of 95 °C for 5 min, followed by 50 cycles of 94 °C for 10 s, 62 °C for 10 s, 72 °C for 15 s, and a final extension at 72 °C for 5 min in a DNA terminal cycler (Perkin-Elmer, GeneAmp PCR System 2400, Waltham, MA, USA), after which the PCR products were kept at 4 °C until further analysis, initially screened for mutations using DHPLC (WAVE; Transgenomic, Omaga, NE, USA), followed by sequence analysis if heteroduplex formation was detected, with DHPLC performed by mixing 20 µL of each exon PCR product with an equal amount of the corresponding wild-type PCR product, incubating at 95 °C for 5 min, and then at room temperature, and separating heteroduplex and homoduplex strands using triethylammonium acetate (TEAA) absorbed into the surface of the DNASep Cartridge (Transgenomic, USA) through an association with the negatively charged phosphate backbone of DNA, with elution using acetonitrile (ACN), in a gradient solution of buffer A (0.1 M TEAA solution, pH 7.0) and buffer B (0.1 M TEAA and 25% ACN, pH 7.0), with buffer C (8% ACN (syringe washing solution)) and buffer D (75% ACN (DNASep Cartridge Ultra-Clean and Storage Solution)) used for cleansing, while the stationary phase involved the DNASep Cartridge (Transgenomic, USA) column in an alkylated nonporous poly(styrene-divinylbenzene) form, washed with buffer D at 0.9 mL/min for 60 min, with the detection of separated DNA checked for purity by injecting 0.5 µL of the non-denatured specimen into the column at 0.9 mL/min at 50 °C, with the temperature elevated to 63 °C and the eluted DNA detected using an ultraviolet light detector at 260 nm, with analysis showing heteroduplexes eluted more rapidly than homoduplexes and appearing as separate forms in the chromatogram, and the DHPLC device operated per the manufacturer’s instruction, with denatured PCR products at 95 °C for 5 min, annealed at 55 °C for about 40 min, and monitored as a chromatogram, where heterogenous molecules typically displayed an additional peak compared to homozygous molecules, which had only one peak, and sequence analysis was performed using commercial reagents and an automated sequencer (ABI Prism BigDye Terminator v3.1 cycles sequencing kit and ABI 310 Genetic Analyzer; Applied Biosystems, Foster City, CA, USA), with both forward and reverse sequenced to confirm nucleotide alterations.
2.3. Definition of TP53 Mutation Characteristics and Oncologic Outcomes
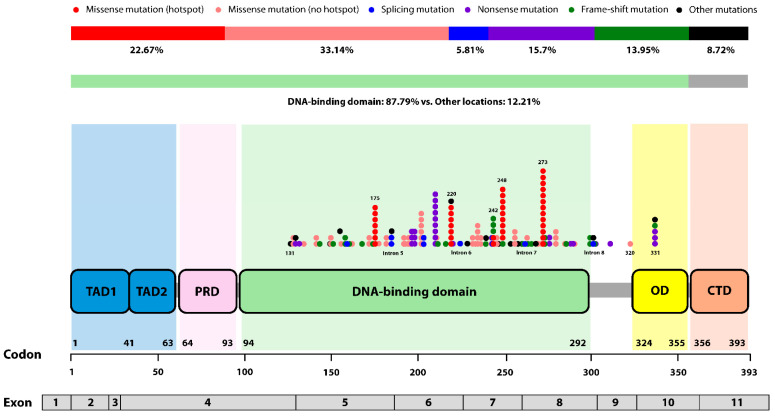
In this study, we classified cases with mutations identified in exons 5-9 through DNA sequencing, as previously described [30,31], into the TP53-mutated group and cases with no mutations detected into the TP53 wild-type group. To validate the clinical relevance of the characteristics of TP53 mutation, we subcategorized the TP53-mutated group into some categories. Since most TP53 mutations are missense mutations and are predominantly found in the DBD, we performed subgroup analyses by subdividing the TP53-mutated group into missense mutation vs. other mutations and DBD vs. other locations. Additionally, we distinguished and analyzed cases with missense hotspot mutations (missense mutations situated at codon 175, 220, 245, 248, 273, and 282) separately from other cases. The characteristics of TP53 mutations within the TP53-mutated group are visualized in Figure 1.
Figure 1.
Characteristics of TP53 mutations in patients within the TP53-mutated group. More than half of the identified TP53 mutations were missense mutations, with the majority occurring in the DNA-binding domain (DBD). Each circle represents a codon where a TP53 mutation occurred, with mutation type distinguishing by color. The number of circles indicate the total number of mutations occurring within specific codons. (Abbreviation, TAD; transactivation domain, PRD; proline-rich domain, OD; oligomerization domain, CTD; carboxy-terminal domain).
Recurrence-free survival (RFS) was defined as the time from treatment of breast cancer (surgery or neoadjuvant chemotherapy) to relapse or death from any cause. Tumor recurrence occurring in the parenchyma of the ipsilateral breast affected by the primary cancer was defined as local recurrence (LR), and metastasis to the ipsilateral axillary lymph node, internal mammary node, and supraclavicular node were classified as regional recurrence (RR). Metachronous breast cancer (recurrence affecting the contralateral breast diagnosed after 1 year from the first cancer diagnosis [32]) was also defined as regional recurrence in this study. Metastasis to all other organs was defined as distant metastasis (DM). Overall survival (OS) was defined as the time from the treatment to death from any cause.
2.4. Statistical Analysis
We utilized the chi-square test or Fisher’s exact test to compare the proportion of de-mographic and clinicopathological variables between the two groups based on TP53 mutation status. Comparisons among TP53-mutated subgroups, based on characteristics of TP53 mutation including mutation types and locations, were also conducted. Oncologic outcomes between the two groups, classified according to TP53 mutation status and characteristics, were compared using a stratified log-rank test at a two-sided significance level of 0.05. A stratified Cox regression analysis was performed to estimate hazard ratio (HR) and 95% confidence intervals (CIs) for oncologic outcome. To estimate the HR of each clinicopathological variable and TP53 mutation status for RFS and OS, we performed Cox proportional hazard model. Multivariable Cox analyses were performed using all variables with p-value (p) ≤ 0.05. Statistical significance was set as p ≤ 0.05. All data analysis was conducted with SPSS software version 26.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software version 10.0 (GraphPad software Inc., Boston, MA, USA).
3. Results
3.1. Baseline Patient Characteristics
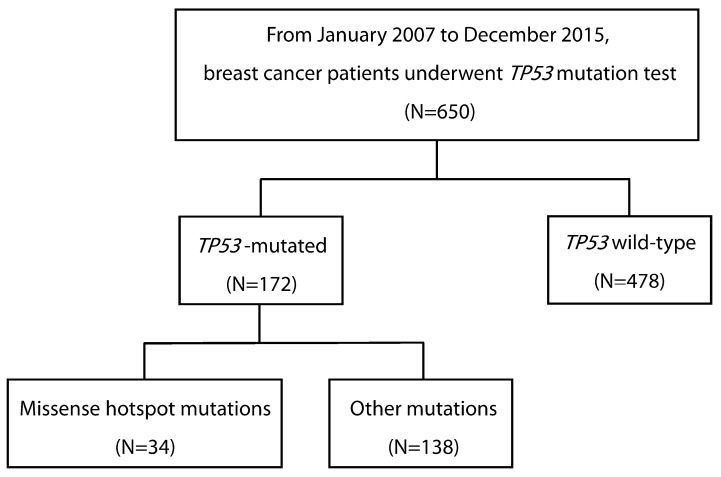
Between January 2007 and December 2015, 650 patients underwent TP53 mutation testing using preoperative biopsies or surgical specimens at Gangnam Severance Hospital. Among these, there were 172 patients (26.5%) who detected TP53 mutations. Of the patients with TP53 mutations, 34 (19.8%) had missense hotspot mutations (Figure 2).
Figure 2.
Consort diagram of this study.
Table 1 presents the demographic and clinicopathological characteristics of patients according to TP53 mutation status. The median age in both groups was 52 years. Compared to the TP53 wild-type group, the TP53-mutated group had a higher proportion of ductal-type breast cancer (86.0% vs. 76.6%; p = 0.016), more frequent histologic grade III tumors (61.6% vs. 28.9%, p < 0.001), an increased rate of LVI (34.5% vs. 17.4%, p < 0.001), and a higher Ki67 index (73.8% vs. 31.4%, p < 0.001). Additionally, the TP53-mutated group had a higher incidence of HR-negative tumor (64.7% vs. 35.9%, p < 0.001) and a greater frequency of HER2-positive tumors (44.2% vs. 26.8%, p < 0.001). When categorized by molecular subtype, the TP53-mutated group exhibited a lower proportion of HR-positive/HER2-negative tumors (18.0% vs. 50.9%) and higher proportions of HER2-positive (44.2% vs. 29.4%) and triple-negative tumors (37.8% vs. 19.7%) compared to the TP53 wild-type group (p < 0.001).
Table 1.
Baseline patients’ characteristics according to TP53 mutation status.
|
TP53-Mutated (N = 172) |
TP53 Wild-Type (N = 478) |
p-Value | |
|---|---|---|---|
| Age, median [IQR] | 52 [27–78] | 52 [51–87] | 0.284 |
| Histologic subtype | 0.016 | ||
| Ductal | 148 (86.0) | 366 (76.6) | |
| Lobular | 2 (1.2) | 23 (4.8) | |
| Others and Mixed | 22 (12.8) | 89 (18.6) | |
| Histologic grade | <0.001 | ||
| Grade III | 106 (61.6) | 138 (28.9) | |
| Grade I-II | 66 (38.4) | 340 (71.1) | |
| HR status # | <0.001 | ||
| Positive | 60 (35.3) | 261 (64.1) | |
| Negative | 110 (64.7) | 146 (35.9) | |
| HER2 status | <0.001 | ||
| Positive | 76 (44.2) | 128 (26.8) | |
| Negative | 96 (55.8) | 350 (73.2) | |
| Molecular subtype # | <0.001 | ||
| HR-positive/HER2-negative | 30 (17.6) | 209 (51.2) | |
| HER2-positive | 75 (44.1) | 113 (22.7) | |
| Triple-negative | 65 (38.2) | 86 (21.1) | |
| LVI # | <0.001 | ||
| Positive | 59 (34.5) | 83 (17.4) | |
| Negative | 112 (65.5) | 395 (82.6) | |
| Ki67 index (cutoff 20%) | <0.001 | ||
| High | 127 (73.8) | 150 (31.4) | |
| Low | 45 (26.2) | 328 (68.6) | |
| Neoadjuvant chemotherapy | 0.062 | ||
| Yes | 11 (6.4) | 15 (3.1) | |
| No | 161 (93.6) | 463 (96.9) | |
| T stage * | 0.035 | ||
| T1 | 69 (42.9) | 253 (54.6) | |
| T2 | 86 (53.4) | 195 (42.1) | |
| T3-4 | 6 (3.7) | 15 (3.2) | |
| N stage * | 0.826 | ||
| N0 | 97 (62.2) | 274 (60.8) | |
| N1 | 47 (30.1) | 135 (29.9) | |
| N2-3 | 12 (7.7) | 42 (9.3) | |
| Breast operation | 0.022 | ||
| BCS | 74 (43.0) | 159 (33.3) | |
| Mastectomy | 98 (57.0) | 319 (66.7) | |
| Axilla surgery | 0.152 | ||
| No approach | 6 (3.5) | 12 (2.5) | |
| SLNB | 135 (78.5) | 406 (84.9) | |
| ALND | 31 (18.0) | 60 (12.6) | |
| Adjuvant chemotherapy * | <0.001 | ||
| Yes | 140 (87.0) | 314 (67.8) | |
| No | 21 (13.0) | 149 (32.2) | |
| Post-operative radiotherapy | 0.007 | ||
| Yes | 96 (55.8) | 210 (43.9) | |
| No | 76 (44.2) | 268 (56.1) |
# Patients for whom accurate test values could not be confirmed were excluded. * Patients who received neoadjuvant chemotherapy or did not undergo surgery were excluded. Abbreviations, IQR; inter-quartile range, HR, hormone receptor, HER2; human epidermal growth factor receptor 2, LVI; lymphovascular invasion, BCS; breast-conserving surgery, SLNB; sentinel lymph node biopsy, ALND; axillary lymph node dissection.
After excluding patients who received neoadjuvant chemotherapy, the distribution of T stage in the TP53 wild-type group was 54.6% (253/463) for T1, 42.1% (195/463) for T2, and 3.2% (15/463) for T3-4. In the TP53-mutated group, the distribution was 42.9% (69/161) for T1, 53.4% (86/161) for T2, and 3.7% (6/161) for T3-4. There was no significant difference in the proportion of each N stage between the two groups (p = 0.922). In both groups, regardless of TP53 mutation status, the mastectomy rate was higher than the breast-conserving surgery (BCS) rate; however, the mastectomy rate was lower in the TP53-mutated group compared to the TP53 wild-type group (57.0% vs. 66.7%; p = 0.022). The majority of patients in both groups underwent sentinel lymph node biopsy (SLNB) alone, and while the axillary lymph node dissection (ALND) rate was higher in the TP53-mutated group, this difference was not statistically significant (18.0% vs. 12.6%; p = 0.152).
As previously mentioned, all patients included in the study received established standard treatment based on a comprehensive evaluation of their age at diagnosis, molecular subtype, and nodal metastasis. Patients in the TP53-mutated group were more likely to receive adjuvant chemotherapy (87.0% vs. 67.8%, p < 0.001) and post-operative radiotherapy (55.8% vs. 43.8%; p = 0.007) compared to those in the TP53 wild-type group. In HER2-positive subtype, a total of 29 patients (14.2%, 29/204) did not receive HER2-targeted therapy due to advanced age or comorbidities; however, the difference in the proportion of these patients between the TP53-mutated and TP53 wild-type groups was not statistically significant (12.0% in the TP53-mutated group vs. 17.7% in the TP53 wild-type group; p = 0.289) (data not shown).
At the time of data cut-off of this study, the median follow-up period was 86.2 months [IQR, 60.3–111.8] in the TP53-mutated group and 97.4 months [IQR, 63.6–134.4] in the TP53 wild-type group.
3.2. Oncologic Outcomes According to TP53 Mutation Status
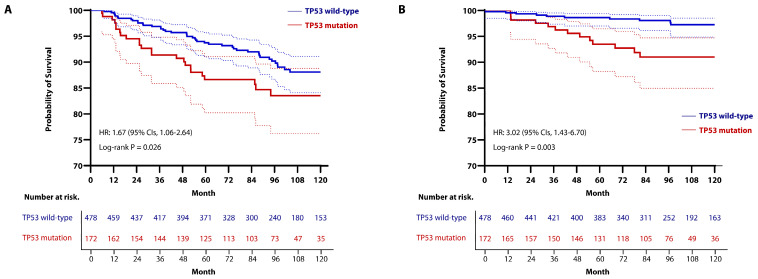
With an extended follow-up period, we assessed 5-year and 10-year oncologic out-comes by using Kaplan-Meier analysis and Cox regression analysis. The RFS rates at 5-year were 88.1% (95% CIs, 84.1–91.1) in the TP53 mutated-group, 93.7% (95% CIs, 91.0–95.7) in the TP53 wild-type group, and the 10-year RFS rates were 83.5% (95% CIs, 76.2–88.8) in the TP53-mutated group, 86.6% (95% CIs, 80.2–91.1) in the TP53 wild-type group, showing a statistically significant difference between the two groups (HR, 1.67; 95% CIs, 1.06–2.64; p = 0.026; Figure 3A).
Figure 3.
Kaplan-Meier curve for (A) RFS and (B) OS in patients stratified by TP53 mutation status. (A) Stratified log-rank test and Cox regression analysis showed a significant different between the two groups (The 5-year RFS rates: 88.1% (95% CIs, 84.1–91.1) in the TP53-mutated group vs. 93.7% (95% CIs, 91.0–95.7) in the TP53 wild-type group; the 10-year RFS rates; 83.5% (95% CIs, 76.2–88.8) in the TP53-mutated group vs. 86.6% (95% CIs, 80.2–91.1) in the TP53 wild-type group) (HR, 1.67; 95% CIs 1.06–2.64; p = 0.026). (B) Stratified log-rank test and Cox regression analysis showed a significant difference between the two groups (The 5-year OS rate: 89.8% (95% CIs, 83.8–93.6) in the TP53-mutated group vs. 95.3% (95% CIs, 92.8–97.0) in the TP53 wild-type group; the 10-year OS rate: 88.1% (95% CIs, 81.7–92.4) in the TP53-mutated group vs. 91.0% (95% CIs, 87.3–93.6) in the TP53 wild-type group) (HR, 3.02; 95% CIs, 1.43–6.70; p = 0.003).
The OS rates at 5-year were 89.8% (95% CIs, 83.8–93.6) in the TP53-mutated group and 95.3% (95% CIs, 92.8–97.0) in the TP53 wild-type group, while the 10-year OS rates were 88.1% (95% CIs, 81.7–92.4) in the TP53-mutated group and 91.0% (95% CIs, 87.3–93.6) in the TP53 wild-type group, indicating that the TP53-mutated group had a worse prognosis compared to the TP53 wild-type group (HR, 3.02; 95% CIs, 1.43–6.70; p = 0.003; Figure 3B). However, when recurrence events were analyzed by sites, there were no differences between the two groups in terms of local recurrence-free survival (LRFS), regional recurrence-free survival (RRFS), and distant metastasis-free survival (DMFS) (Figure S1).
We utilized a Cox regression model to explore predictive factors for RFS and OS. Univariable analysis showed that TP53 mutation was significantly associated with a shorter period of RFS (HR, 1.669; 95% CIs, 1.058–2.635; p = 0.028; Table 2) and OS (HR, 3.092; 95% CIs, 1.427–6.698; p = 0.004; Table 3). In multivariable analysis, which included all predictors with a p ≤ 0.05 from the univariable Cox analysis, TP53 mutation remained an independent predictor of worse RFS (HR, 1.29; 95% CIs, 1.008–1.832; p = 0.046; Table 2) and OS (HR, 2.488; 95% CIs, 1.407–3.788; p = 0.044; Table 3). Additionally, the multivariable Cox analysis indicated that the presence of LVI and a high Ki67 index were significantly associated with worse RFS (Table 2), and the presence of LVI was also an independent predictor of worse OS (Table 3). In the univariable analysis, large tumor size (more than 2 cm) was identified as a factor associated with worse RFS and OS, but it was not statistically significant in the multivariable analysis. In addition, factors such as young age at diagnosis, high histologic grade, HR and HER2 positivity, nodal involvement, breast preservation during surgery, and the use of HER2-targeted therapy were not significantly associated with survival outcomes in our study.
Table 2.
Univariable and multivariable analyses for RFS.
| Variable | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| HR | 95% CIs | p-Value | HR | 95% CIs | p-Value | |
| Age ≤ 50 years (ref. > 50 years) | 1.24 | 0.813–1.893 | 0.318 | |||
| TP53 mutation (ref. TP53 wild-type) | 1.669 | 1.058–2.635 | 0.028 | 1.29 | 1.008–1.832 | 0.046 |
| Histologic grade III (ref. HG I-II) | 1.123 | 0.730–1.730 | 0.597 | |||
| HR positive (ref. HR negative) # | 1.234 | 0.781–1.950 | 0.367 | |||
| HER2 positive (ref. HER2 negative) | 0.718 | 0.439–1.172 | 0.185 | |||
| LVI present (ref. LVI absent) # | 2.604 | 1.686–4.021 | <0.001 | 2.366 | 1.495–3.747 | <0.001 |
| Ki67 high (≥20%) (ref. Ki67 < 20%) | 1.829 | 1.198–2.790 | 0.005 | 1.607 | 1.030–2.506 | 0.037 |
| Tumor > 2 cm (ref. Tumor ≤ 2 cm) * | 1.743 | 1.115–2.724 | 0.015 | 1.355 | 0.843–2.178 | 0.209 |
| Nodal involvement (ref. Node-negative) * | 1.3 | 0.839–2.014 | 0.241 | |||
| BCS (ref. Mastectomy) | 1.383 | 0.903–2.117 | 0.136 | |||
| HER2-targeted therapy (ref. no treatment) | 0.774 | 0.455–1.316 | 0.343 | |||
# Patients without definite data were excluded. * Patients who underwent neoadjuvant chemotherapy were excluded. Abbreviations, HR, hazard ratio, CIs; confidence intervals, HR, hormone receptor, HER2; human epidermal growth factor receptor 2, LVI; lymphovascular invasion, BCS; breast-conserving surgery.
Table 3.
Univariable and multivariable analyses for OS.
| Variable | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| HR | 95% CIs | p-Value | HR | 95% CIs | p-Value | |
| Age ≤ 50 years (ref. > 50 years) | 0.812 | 0.375–1.757 | 0.597 | |||
| TP53 mutation (ref. TP53 wild-type) | 3.092 | 1.427–6.698 | 0.004 | 2.488 | 1.407–3.788 | 0.044 |
| Histologic grade III (ref. HG I-II) | 0.909 | 0.405–2.038 | 0.816 | |||
| HR positive (ref. HR negative) # | 0.549 | 0.244–1.238 | 0.148 | |||
| HER2 positive (ref. HER2 negative) | 1.017 | 0.442–2.339 | 0.968 | |||
| LVI present (ref. LVI absent) # | 2.604 | 1.686–4.021 | <0.001 | 2.366 | 1.495–3.747 | <0.001 |
| Ki67 high (≥ 20%) (ref. Ki67 < 20%) | 2.419 | 1.096–5.340 | 0.029 | 2.35 | 0.966–5.717 | 0.06 |
| Tumor > 2 cm (ref. Tumor ≤ 2 cm) * | 2.781 | 1.079–7.170 | 0.034 | 2.061 | 0.765–5.551 | 0.153 |
| Nodal involvement (ref. Node-negative) * | 1.69 | 0.718–3.980 | 0.23 | |||
| BCS (ref. Mastectomy) | 0.927 | 0.413–2.085 | 0.855 | |||
| HER2-targeted therapy (ref. no treatment) | 0.914 | 0.367–2.276 | 0.846 | |||
# Patients without definite data were excluded. * Patients who underwent neoadjuvant chemotherapy were excluded. Abbreviations, OS; overall survival, HR, hazard ratio, CIs; confidence intervals, HR, hormone receptor, HER2; human epidermal growth factor receptor 2, LVI; lymphovascular invasion, BCS; breast-conserving surgery.
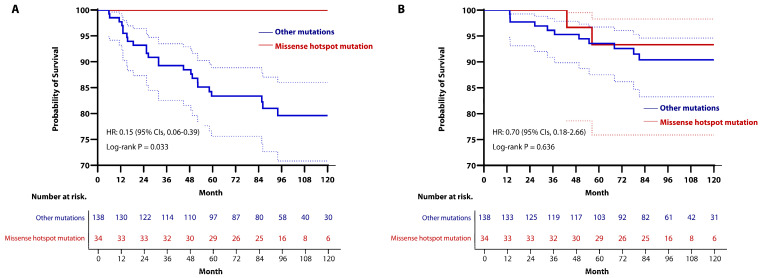
3.3. Subgroup Analysis Based on Mutation Types Within the TP53-Mutated Group
Since most TP53 mutations are known to be missense mutations, we conducted a subgroup analysis to examine potential differences in oncologic outcomes between missense mutation and other mutation types. Among the 172 cases with confirmed TP53 mutations, 96 (55.8%) were missense mutations, and 76 (44.2%) were other types of mutations.
After excluding patients who received neoadjuvant chemotherapy, the missense mutation subgroup had a higher proportion of tumors ≤ 2 cm, and a lower proportion of tumors > 2 cm compared to the other mutations subgroup (T1 tumors; 50.6% in the missense mutation subgroup vs. 33.3% in the other mutations subgroup; p = 0.026). Consequently, patients in the missense mutation subgroup underwent BCS more frequently (53.1% vs. 30.3%; p = 0.003) and were more likely to receive post-operative radiotherapy (63.5% vs. 46.1%; p = 0.022) than those in the other mutations subgroup. However, no differences were observed between the two groups in the proportion of other clinicopathologic variables with surgery and treatment implementation. Detailed information is presented in Table S1.
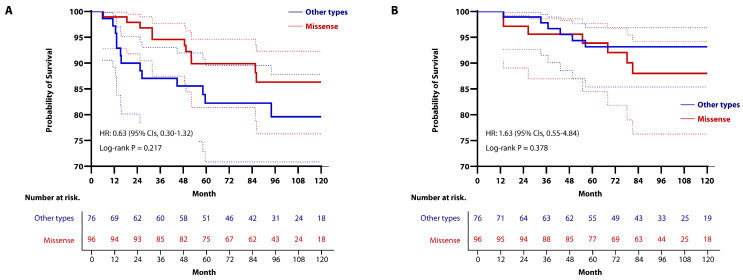
With a median follow-up period of 86.1 months (IQR, 54.1–110.8), there were no significant differences in RFS and OS between the two groups. The 5-year RFS rates were 89.9% (95% CIs, 81.4–94.6) in the missense mutation group and 82.3% (95% CIs, 70.8–89.5) in the other mutations group, whereas the rates of RFS at 10 years were 86.3% (95% CIs, 76.3–92.3) in the missense mutation group and 79.6% (95% CIs, 67.0–87.8) in the other mutations group (HR, 0.63; 95% CIs, 0.30–1.32; p = 0.217; Figure 4A). The 5-year OS rates were 93.9% (95% CIs, 84.5–97.7) in the missense hotspot mutation group and 93.2% (95% CIs, 85.4–96.9) in the other mutation group, while the 10-year OS rates were 88.0% (95% CIs, 76.3–94.2) in the missense mutation group and 93.2% (95% CIs, 85.4–96.9) in the other mutations group (HR, 1.63; 95% CIs, 0.55–4.84; p = 0.378; Figure 4B). Additionally, LRFS, RRFS, and DMFS did not differ significantly between the two groups (Figure S2).
Figure 4.
Kaplan-Meier curve for (A) RFS and (B) OS in the TP53-mutated group, stratified by type of mutation. To compare oncologic outcomes, we used stratified log-rank test and Cox regression analysis. (A) There was no significant difference between the two groups (the 5-year RFS rates: 89.9% (95% CIs, 81.4–94.6) in the missense mutation group vs. 82.3% (95% CIs, 70.8–89.5) in the other mutations group; the 10-year RFS rates: 86.3% (95% CIs, 76.3–92.3) in the missense mutation group vs. 79.6% (95% CIs, 67.0–87.8) in the other mutations group) (HR, 0.63; 95% CIs, 0.30–1.32; p = 0.217). (B) Similarly, there was no significant difference between the two groups (The 5-year OS rates: 93.9% (95% CIs, 84.5–97.7) in the missense mutation group vs. 93.2% (95% CIs, 85.4–96.9) in the other mutations group; the 10-year OS rates: 88.0% (95% CIs, 76.3–94.2) in the missense mutation group vs. 93.2% (95% CIs, 85.4–96.9) in the other mutations group) (HR, 1.63; 95% CIs, 0.55–4.84; p = 0.378).
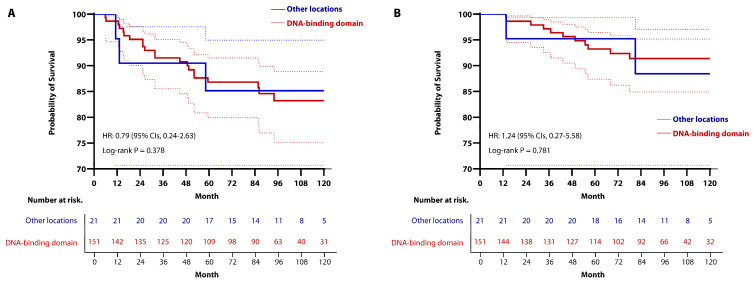
3.4. Subgroup Analysis Based on Locations of Mutation Within the TP53-Mutated Group
Next, focusing on the patient with TP53 mutation, we conducted a subgroup analysis to investigate oncologic outcomes based on the locations of TP53 mutations. First, we classified the location of TP53 mutations into the DBD and other locations. In total, there were 151 cases (87.8%) in the DBD subgroup and 21 cases (12.2%) in the other locations subgroup.
Compared to the other locations subgroup, the DBD subgroup had a lower proportion of HR-positive tumors (32.0% vs. 60.0%; p = 0.014). When tumors were classified by molecular subtype, the DBD subgroup exhibited a lower proportion of HR-positive/HER2-negative and HER2-positive tumors, and a higher proportion of triple-negative tumors (HR-positive/HER2-negative; 16.0% vs. 30.0%, HER2-positive; 42.0% vs. 60.0%, triple-negative; 42.0% vs. 10.0%; p = 0.018). However, there were no differences between the two groups in the proportion of the other collected variables (Table S3).
As with TP53 mutation type, there were no differences in oncologic outcomes between the two groups based on mutation locations. The 5-year RFS rates were 86.8% (95% CIs, 79.9–91.5) in the DBD group and 85.2% (95% CIs, 60.6–95.0) in the other locations group; and the 10-year RFS rates were 83.2% (95% CIs, 75.2–88.9) in the DBD group and 85.2% (95% CIs, 60.6–95.0) in the other locations group (HR, 0.79; 95% CIs, 0.24–2.63; p = 0.378; Figure 5A). The OS rates at 5 years were 93.2% (95% CIs, 87.4–96.4) in the DBD group and 95.2% (95% CIs, 70.7–99.3) in the other locations group, whereas the 10-year OS rates were 91.4% (95% CIs, 84.9–95.2) in the DBD group and 88.4% (95% CIs, 60.3–97.1) in the other locations group (HR, 0.24; 95% CIs, 0.27–5.58; p = 0.781; Figure 5B). LRFS, RRFS, and DMFS did not differ significantly between the two groups (Figure S3).
Figure 5.
Kaplan-Meier curve for (A) RFS and (B) OS in the TP53-mutated group, stratified by mutation locations. To compare oncologic outcomes, we used stratified log-rank test and Cox regression analysis. (A) There was no significant difference between the two groups (the 5-year RFS rates: 86.8% (95% CIs 79.9–91.5) in the DBD group vs. 85.2% (95% CIs, 60.6–95.0) in the other locations group; the 10-year RFS rates: 83.2% (95% CIs, 75.2–88.9) in the DBD group vs. 85.2% (95% CIs, 60.6–95.0) in the other locations group) (HR, 0.79; 95% CIs, 0.24–2.63; p = 0.378). (B) There was also no significant difference between the two groups (the 5-year OS rates: 93.2% (95% CIs, 87.4–96.4) in the DBD group vs. 95.2% (95% CIs, 70.7–99.3) in the other locations group; the 10-year OS rates: 91.4% (95% CIs, 84.9–95.2) in the DBD group vs. 88.4% (95% CIs, 60.3–97.1) in the other locations group) (HR, 1.24; 95% CIs, 0.27–5.58; p = 0.781).
3.5. Subgroup Analysis Based on the Presence of Missense Hotspot Mutations Within the TP53-Mutated Group
Lastly, we analyzed oncologic outcomes by distinguishing between cases with missense hotspot domains and those without within the patients with TP53 mutation. The majority of mutations identified at hotspot codons were missense mutations (39/44, 88.6%). However, no statistically significant differences in patients’ characteristics were observed between the two groups (Table S3).
The median follow-up period was 95.1 months (IQR, 81.9–98.8) in the missense hotspot mutations group and 83.7 months (IQR, 75.6–89.3) in the other mutations group. At 5 years, the RFS rates were 100% in the missense hotspot mutations group and 83.4% (95% CIs, 75.6–88.8) in the other mutations group. The Kaplan-Meier estimates of 10-year RFS rates were 100% in the missense hotspot mutations group and 79.6% (95% CIs, 70.8–86.0) in the other mutations group, indicating that the missense hotspot mutations group had a better oncologic outcome than the other mutations group (HR, 0.15; 95% CIs, 0.06–0.39; p = 0.033; Figure 6A). However, there was no significant difference in OS between the two groups (HR, 0.70; 95% CIs, 0.18–2.66; p = 0.636; Figure 6B). Additionally, the RFS rates stratified by recurrence sites were not significantly different between the two groups (Figure S4).
Figure 6.
Kaplan-Meier curve for (A) RFS and (B) OS in patients with TP53 mutation, stratified by the presence or absence of missense hotspot mutations. (A) By utilizing stratified log-rank test and Cox regression analysis, patients with missense hotspot mutations had a longer RFS period (the 5-year RFS rates: 100% in the missense hotspot mutations group vs. 83.4% (95% CIs, 75.6–88.8) in the other mutations group; the 10-year RFS rates: 100% in the missense hotspot mutations group vs. 79.6% (95% CIs, 70.8–86.0) in the other mutations group (HR, 0.15; 95% CIs, 0.06–0.39; p = 0.028). (B) Stratified log-rank test and Cox regression analysis showed that there was no significant difference between the two groups (the 5-year OS rates: 93.3% (95% CIs, 75.9–98.3) in the missense hotspot mutations group vs. 93.6% (95% CIs, 87.6–96.7) in the other mutations group; the 10-year OS rates; 93.3% (95% CIs, 75.9–98.3) in the missense hotspot mutations group vs. 90.4% (95% CIs, 83.3–94.6) in the other mutations group (HR, 0.70; 95% CIs, 0.18–2.66; p = 0.636).
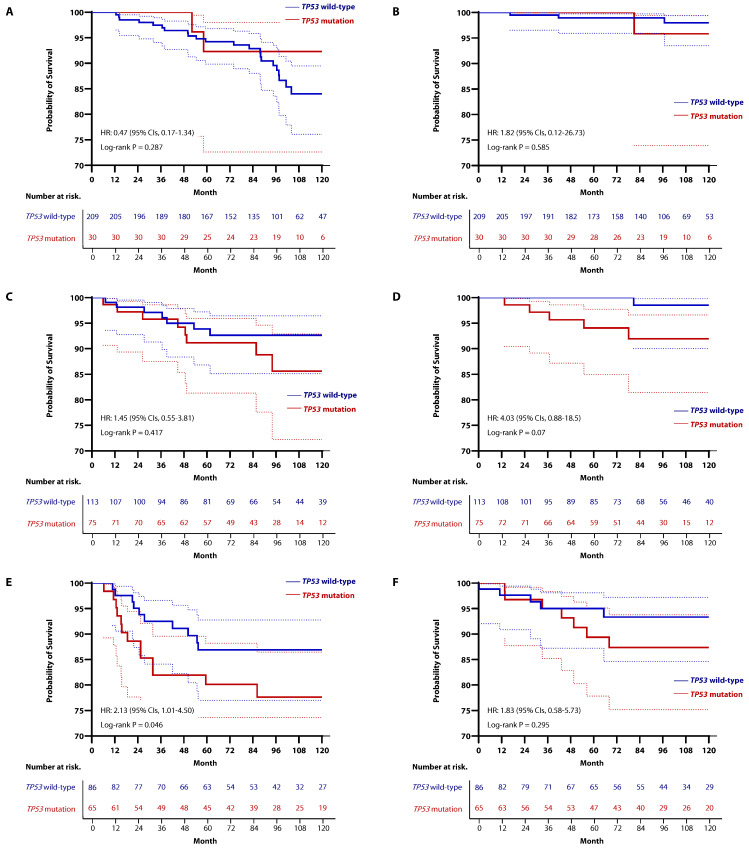
3.6. Clinical Relevance of TP53 Within Molecular Subtypes of Breast Cancer
As part of an exploratory analysis, we examined the clinical relevance of TP53 mutations within specific molecular subtypes. After excluding 72 patients for whom IHC-based HR status was unavailable, there were 239 patients (41.3%) with HR-positive/HER2-negative (HR+/HER2-) breast cancer, 188 patients (32.5%) with HER2-positive breast cancer, and 151 patients (26.1%) with TNBC. The proportion of patients with confirmed TP53 mutations in each subtype were 30 patients (12.6%) in the HR+/HER2- subtype, 75 patients (39.9%) in the HER2-positive subtype, and 65 patients (43.0%) in the TNBC group. When comparing survival outcomes based on TP53 mutation status within each subtype using the Kaplan-Meier estimated model, there were no differences in RFS or OS in HR+/HER2- (Figure 7A,B) and HER2-positive breast cancer (Figure 7C,D). However, in TNBC, patients with TP53 mutation had worse RFS compared to those with TP53 wild-type (HR, 2.13; 95% CIs, 1.01–4.50; p = 0.046; Figure 7E). Nevertheless, there was no statistical difference in OS according to TP53 mutation status in TNBC (HR, 1.83; 95% CIs, 0.58–5.73; p = 0.295; Figure 7F). In addition, when comparing survival outcomes based on HER2 overexpression status within the TP53-mutated group, no statistically significant differences were observed (Figure S5).
Figure 7.
Kaplan-Meier curve for RFS and OS according to TP53 mutation status in each subtype. In HR+/HER2-subtype, there was no difference in (A) RFS (HR, 0.47; 95% CIs, 0.17–1.34; p = 0.287) or (B) OS (HR, 1.82; 95% CIs; 0.12–26.73; p = 0.585) between TP53-muated group and TP53 wild-type group. Similarly, in the HER2-positive subtype, no differences were observed in (C) RFS (HR, 1.45; 95% CIs; 0.55–3.81; p = 0.417) or (D) OS (HR, 4.03; 95% CIs; 0.88–18.5; p = 0.07) based on TP53 mutation status. In the TNBC group, (E) TP53-mutated tumors showed worse RFS compared to the TP53 wild-type group (HR, 2.13; 95% CIs; 1.01–4.50; p = 0.046). However, (F) although there was a trend toward worse OS in TP53-mutated tumors, it was not statistically significant (HR, 1.83; 95% CIs, 0.58–5.73; p = 0.295).
4. Discussion
In this retrospective cohort study, we assessed the clinical relevance of TP53 mutations in breast cancer patients, including all subtypes and treatments, and conducted subgroup analyses based on the characteristics of TP53 mutations within the TP53-mutated group. TP53 mutations were more frequent in breast cancer with more aggressive clinicopathological variables, such as large tumors, tumors with LVI or high histologic grade, and overexpression of HER2. Patients with TP53 mutations had shorter RFS and OS compared to patients with TP53 wild-type tumors. However, within the TP53-mutated group, the oncologic outcomes did not significantly differ between subgroups based on the characteristics of the TP53 mutations. Missense mutation, mutation situated on DBD, and even missense mutation situated in hotspots, which are all well-known dominant characteristics of TP53 mutation, did not have clinical relevance compared to other types or locations of TP53 mutations. Although patients with missense hotspot mutations in the TP53-mutated group had a longer RFS period compared to other patients, there was no difference in OS rate. Therefore, the prognostic impact of missense hotspot mutations of TP53 gene remains questionable.
Although TP53 mutations are found in approximately 30% of all breast cancers [13], the proportion of these mutations varies by tumor subtypes. Furthermore, due to the differing mechanism of p53 protein among tumor subtypes and treatments, most studies on the clinical relevance of TP53 mutations in breast cancer have been conducted within specific subtypes or treatments. Given that p53 regulates cell response to DNA damage, there have been several studies investigating the role of TP53 mutations in patients undergoing chemotherapy or radiation, which induces tumor cell damage. Early preclinical trials indicated that p53 plays a role in regulating apoptosis or cell cycle arrest following cell damage such as radiation or systemic anticancer treatments [33,34,35,36]. Subsequent studies have shown that breast cancer patients with TP53 mutations often have higher pathologic complete response (pCR) rates following neoadjuvant chemotherapy [37,38,39,40]. Otherwise, there were studies showing neutral and negative results regarding the association between TP53 mutations and pCR rates following neoadjuvant chemotherapy [41,42,43]. Most of previous studies had small sample sizes and used different chemotherapy regimens and methods for detecting TP53 mutations, making it challenging to define the clinical relevance of TP53 mutations. Recently, a meta-analysis of 26 studies involving 3476 breast cancer patients who underwent neoadjuvant chemotherapy found that those with TP53 mutations had a higher pCR rate [20]. However, even though this study confirmed the clinical relevance of TP53 mutations through a sizable cohort, it also had the limitation of inconsistent TP53 mutation detecting methods across the included studies. Additionally, most cases receiving neoadjuvant chemotherapy were HER2-positive breast cancer or TNBC. Therefore, it is difficult to consider these studies as having a balanced representation of all breast tumor subtypes.
ER-positive breast cancers account for about 70% of all breast cancers, making them the most prevalent subtype. In ER-positive breast tumors, the frequency of TP53 mutations is lower than in other subtypes [40]; however, when these mutations are present, they are associated with a poor prognosis. Many studies presented that TP53 mutations could lead to alterations in the p53 protein, potentially causing endocrine resistance [44,45,46]. However, the relationship between TP53 mutations and survival outcomes in patients receiving only hormone therapy has been controversial [40,45,47]. This is due to several factors such as the small sample size, the detection of TP53 mutations primarily through IHC, and the lack of information of additional treatments beyond hormone therapy. In a meta-analysis examining the clinical relevance of TP53 mutations in patients receiving only hormone therapy, it was found that patients with TP53 mutations had worse overall survival compared to those without TP53 mutations [48]. Although a different dataset with varying TP53 mutation detecting methods was utilized, we previously identified an association between TP53 mutations and high 21-gene recurrence score in ER+HER2- breast tumors [49]. This finding aligns with prior research indicating that TP53 mutations are associated with endocrine resistance in ER-positive breast tumors. Compared to ER-positive breast cancer, ER-negative breast cancer accounts for a smaller proportion of all breast tumors; however, the frequency of TP53 mutations is higher in ER-negative breast cancer. TP53 mutation rates are higher in HER2-positive and TNBC (also referred to as basal-like subtype) compared to luminal-type breast cancer, which are predominantly ER-positive tumors [13,50,51,52,53,54,55,56]. Some studies indicated that the presence of TP53 mutations is associated with poor prognosis and might confer resistance to chemotherapy in HER2-positive breast cancer and TNBC [54,57,58,59]. However, some studies showed no difference in oncologic outcomes based on TP53 mutation status in ER-negative tumors [60,61,62,63,64], or even suggested that TP53 mutations are associated with better prognosis [39,65,66]. This trend has become more pronounced in recent studies as chemotherapy regimens have continuously evolved and the clinical use of new drugs, such as dual HER2 blockade and immune checkpoint inhibitors, has increased. Consequently, determining the clinical significance of TP53 mutations in ER-negative breast cancer has become even more challenging. Given these circumstances, conducting studies to determine the clinical relevance of TP53 mutations across all subtypes and treatments involves many hurdles and interpreting the results is also challenging.
Therefore, the strength of our study is its ability to assess long-term oncologic outcomes using a large cohort that encompasses all breast cancer subtypes and treatments. Excluding 42 patients whose hormone receptor status was not clearly identified, the data for this study included 253 HR-positive, HER2-negative tumors (41.6%), 204 HER2-positive cases (33.6%), and 172 TNBC (28.3%), which means the distribution of tumor subtypes in the collected data was well-balanced. To date, few studies have investigated the clinical relevance of TP53 mutations using cohorts that included all breast cancer subtypes and treatments. In most of these studies, patients with TP53 mutations were found to have worse survival compared to the TP53 wild-type group [67,68,69,70]. However, these studies had limitations such as small sample size, lack of follow-up data, and inconsistent treatments even within the same subtype. This study, leveraging a large cohort from a single center, ensured consistent treatments according to tumor subtypes and stage, thereby minimizing bias from the data.
Another notable strength of our study is its focus on an Asian population, unlike most previous research on the link between TP53 mutations and poor prognosis in breast cancer, which has primarily included individuals of American and European descent. Large-scale retrospective cohort studies evaluating the clinical relevance of TP53 mutations in Asian breast cancer patients are still limited. Thus, our findings provide a unique opportunity to contribute evidence that could support cross-ethnic comparisons and validate the clinical implications of TP53 mutations in breast cancer. In addition, by collecting data from patients who underwent TP53 mutation testing between 2007 and 2015, we were able to secure comprehensive long-term follow-up data.
Furthermore, few studies have examined surgical outcomes based on the location and type of TP53 mutations in patients with confirmed mutations, underscoring the significance of our investigation. Given that mutations causing loss of DNA-binding can critically affect the biological activity of p53 [71], there is increasing interest in understanding the characteristics of different TP53 mutations. However, the clinical relevance of specific mutation types and locations remains underexplored. For instance, an analysis of the METABRIC cohort found that tumors harboring missense mutations in DNA-binding motifs (DBM) had a higher risk of breast cancer-specific mortality compared to tumors with non-missense mutations or missense mutations outside the DBM, though this difference did not reach statistical significance [64]. Similarly, a study in China demonstrated that metastatic breast cancer patients with TP53 mutations outside the DBD experienced poorer disease-free survival and OS compared to those with TP53 wild-type, with particularly poor outcomes observed in patients with non-missense mutations located within the DBD [72]. Pal, et al. assessed the 10 most common TP53 missense mutations using MCF10A cell lines for preclinical investigation and found that mutations such as R248W, R273C, R248Q, and Y220C were associated with the most aggressive tumor phenotypes [73]. Børresen, et al. reported that TP53 mutations in the zinc-binding domain were associated with worse prognosis compared to mutations outside this domain [74]. Meanwhile, Kucera, et al. observed no significant difference in survival outcomes between cases with mutations in the L2/3 domain and those without such mutations [75]. Data from the BIG 02-98 phase III trial also indicated that only truncating mutations were predictive of increased recurrence risk, while missense mutations showed no significant association [76]. Despite these findings, many studies have faced challenges in achieving statistical significance due to limited sample size, diverse patient populations, varying methods for analyzing TP53 mutations, and differences in study endpoints. In our study, we investigated the clinical relevance of several characteristics within the TP53-mutated group, including missense mutations and mutations located in the DBD, but did not find statistically significant results. Nonetheless, considering the limited research focused on the clinical implications of TP53 mutations in early breast cancer among Asian populations, we believe that our findings add valuable evidence to the existing literature and can help guide future research efforts.
Although our study allowed us to assess the clinical relevance of TP53 mutations and their characteristics within a large cohort encompassing all tumor subtypes and treatments, it still had inherent limitations. The first limitation is the sensitivity of TP53 mutations. In our study, we identified TP53 mutations in exons 5-9 using PCR-DHPLC and direct sequencing. Although most TP53 mutations occur within exon 5-9, this approach might lead to false-negative results for mutations occurring in other regions, particularly in exons 2-4 and 10-11 [77,78]. In addition, somatic mutations identified by PCR-DHPLC might not always be detectable by direct sequencing, because it has a threshold of detection of approximately 15–20% [79]. To overcome this limitation, NGS is now used for DNA sequencing in breast cancer [80]. However, since NGS was introduced at our institution in 2017, it was not applied for the patients retrospectively collected for this study. Another limitation of our study is the reliance on older data, which may have constrained our ability to control confounding variables effectively. Furthermore, although this study included a broad cohort covering all breast cancer subtypes, the overall number of patients and the subgroup sizes within the TP53 mutation category were limited, representing a notable study limitation. In our Cox regression analysis assessing associations between clinicopathological features and survival outcomes, several established prognostic and predictive markers did not reach statistical significance, likely due to the sample size constraints, which may have reduced the power to detect meaningful associations. Lastly, since the data were collected 10 years ago, the treatment protocols at that time may differ significantly from those currently used in clinical practice. Most patients in this study did not receive neoadjuvant chemotherapy, and treatments such as CDK4/6 inhibitors, immune checkpoint inhibitors, and dual HER2 blockade including pertuzumab were rarely administered at that time. Nevertheless, based on this study, we expected that we could conduct further research addressing a prognostic influence on the characteristics of TP53 mutations in breast cancer patients with advanced research methods and molecular studies.
5. Conclusions
Using consistently collected long-term follow-up data, we found that TP53 mutations are associated with worse prognosis in breast tumors encompassing all subtypes and treatments. Additionally, within the TP53-mutated group, there were no significant differences in surgical outcomes based on the characteristics of TP53 mutations such as mutation types and locations of mutation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16233899/s1, Table S1: Patients’ characteristics based on TP53 mutation type within the TP53-mutated group. Table S2: Patients’ characteristics based on location of TP53 mutations in the TP53-mutated group. Table S3: Patients’ characteristics between missense hotspot mutations and other mutations. Figure S1: Kaplan-Meier curve for (A) LRFS, (B) RRFS, and (C) DMFS in patients stratified by TP53 mutation. Figure S2: Kaplan-Meier curve for (A) LRFS, (B) RRFS, and (C) DMFS in patients with TP53 mutation, stratified by types of mutation. Figure S3: Kaplan-Meier curve for (A) LRFS, (B) RRFS, and (C) DMFS in patients with TP53 mutation, stratified by locations of mutation. Figure S4: Kaplan-Meier curve for (A) LRFS, (B) RRFS, and (C) DMFS in patients with TP53 mutation, stratified by the presence or absence of missense hotspot mutations. Figure S5: Kaplan-Meier curve for (A) RFS and (B) OS according to HER2-overexpression status within the TP53-mutated group.
Author Contributions
Conceptualization, S.H.H., S.H.B., S.G.A. and J.J.; formal analysis, S.H.H., S.H.B. and J.J.; investigation, S.H.H., S.H.B., M.J.L., Y.K., S.J.B., S.G.A. and J.J.; data curation, S.H.H., S.H.B., S.G.A. and J.J.; resources, S.H.H., S.H.B., M.J.L., Y.K., S.J.B., S.G.A. and J.J.; writing—original draft preparation, S.H.H. and S.H.B.; writing—review and editing, S.H.H., S.H.B., M.J.L., Y.K., S.J.B., S.G.A. and J.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Gangnam Severance Hospital, Yonsei University, Seoul, Korea (IRB no. 3-2024-0197).
Informed Consent Statement
Patient consent was waived due to retrospective design of this study.
Data Availability Statement
The data that support the findings of this study contain clinical outcomes for which institutional review board (IRB) approval is required before analysis. Therefore, these data are not publicly available. The data will be provided to authorized researchers who have obtained IRB approval from their institution and Gangnam Severance Hospital, Yonsei University, Seoul, Korea. For data access requests, please contact the corresponding authors, namely Pf. J.J., email address: gsjjoon@yuhs.ac.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C., Nik-Zainal S., Martin S., Varela I., Bignell G.R., et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernández Borrero L.J., El-Deiry W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188556. doi: 10.1016/j.bbcan.2021.188556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley T., Sontag E., Chen P., Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 4.Walerych D., Napoli M., Collavin L., Del Sal G. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–2017. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.el-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 6.Kotler E., Shani O., Goldfeld G., Lotan-Pompan M., Tarcic O., Gershoni A., Hopf T.A., Marks D.S., Oren M., Segal E. A Systematic p53 Mutation Library Links Differential Functional Impact to Cancer Mutation Pattern and Evolutionary Conservation. Mol. Cell. 2018;71:178–190.e178. doi: 10.1016/j.molcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Alexandrova E.M., Mirza S.A., Xu S., Schulz-Heddergott R., Marchenko N.D., Moll U.M. p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis. 2017;8:e2661. doi: 10.1038/cddis.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X., Hao Q., Lu H. Mutant p53 in cancer therapy-the barrier or the path. J. Mol. Cell Biol. 2019;11:293–305. doi: 10.1093/jmcb/mjy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampath J., Sun D., Kidd V.J., Grenet J., Gandhi A., Shapiro L.H., Wang Q., Zambetti G.P., Schuetz J.D. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J. Biol. Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 11.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 12.Børresen-Dale A.L. TP53 and breast cancer. Hum. Mutat. 2003;21:292–300. doi: 10.1002/humu.10174. [DOI] [PubMed] [Google Scholar]
- 13.The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berns E.M., Foekens J.A., Vossen R., Look M.P., Devilee P., Henzen-Logmans S.C., van Staveren I.L., van Putten W.L., Inganäs M., Meijer-van Gelder M.E., et al. Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res. 2000;60:2155–2162. [PubMed] [Google Scholar]
- 15.Bonnefoi H., Diebold-Berger S., Therasse P., Hamilton A., Van De Vijver M., MacGrogan G., Shepherd L., Amaral N., Duval C., Drijkoningen R. Locally advanced/inflammatory breast cancers treated with intensive epirubicin-based neoadjuvant chemotherapy: Are there molecular markers in the primary tumour that predict for 5-year clinical outcome? Ann. Oncol. 2003;14:406–413. doi: 10.1093/annonc/mdg108. [DOI] [PubMed] [Google Scholar]
- 16.Hensel M., Schneeweiss A., Sinn H.P., Egerer G., Solomayer E., Haas R., Bastert G., Ho A.D. P53 is the strongest predictor of survival in high-risk primary breast cancer patients undergoing high-dose chemotherapy with autologous blood stem cell support. Int. J. Cancer. 2002;100:290–296. doi: 10.1002/ijc.10478. [DOI] [PubMed] [Google Scholar]
- 17.Malamou-Mitsi V., Gogas H., Dafni U., Bourli A., Fillipidis T., Sotiropoulou M., Vlachodimitropoulos D., Papadopoulos S., Tzaida O., Kafiri G., et al. Evaluation of the prognostic and predictive value of p53 and Bcl-2 in breast cancer patients participating in a randomized study with dose-dense sequential adjuvant chemotherapy. Ann. Oncol. 2006;17:1504–1511. doi: 10.1093/annonc/mdl147. [DOI] [PubMed] [Google Scholar]
- 18.Ostrowski J.L., Sawan A., Henry L., Wright C., Henry J.A., Hennessy C., Lennard T.J., Angus B., Horne C.H. p53 expression in human breast cancer related to survival and prognostic factors: An immunohistochemical study. J. Pathol. 1991;164:75–81. doi: 10.1002/path.1711640113. [DOI] [PubMed] [Google Scholar]
- 19.Shiao Y.H., Chen V.W., Scheer W.D., Wu X.C., Correa P. Racial disparity in the association of p53 gene alterations with breast cancer survival. Cancer Res. 1995;55:1485–1490. [PubMed] [Google Scholar]
- 20.Chen M.B., Zhu Y.Q., Xu J.Y., Wang L.Q., Liu C.Y., Ji Z.Y., Lu P.H. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: A meta-analysis. PLoS ONE. 2012;7:e39655. doi: 10.1371/journal.pone.0039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 22.Soussi T. p53 alterations in human cancer: More questions than answers. Oncogene. 2007;26:2145–2156. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- 23.Valenti F., Ganci F., Fontemaggi G., Sacconi A., Strano S., Blandino G., Di Agostino S. Gain of function mutant p53 proteins cooperate with E2F4 to transcriptionally downregulate RAD17 and BRCA1 gene expression. Oncotarget. 2015;6:5547–5566. doi: 10.18632/oncotarget.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Minin G., Bellazzo A., Dal Ferro M., Chiaruttini G., Nuzzo S., Bicciato S., Piazza S., Rami D., Bulla R., Sommaggio R., et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol. Cell. 2014;56:617–629. doi: 10.1016/j.molcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Chao C.H., Wang C.Y., Wang C.H., Chen T.W., Hsu H.Y., Huang H.W., Li C.W., Mai R.T. Mutant p53 Attenuates Oxidative Phosphorylation and Facilitates Cancer Stemness through Downregulating miR-200c-PCK2 Axis in Basal-like Breast Cancer. Mol. Cancer Res. 2021;19:1900–1916. doi: 10.1158/1541-7786.MCR-21-0098. [DOI] [PubMed] [Google Scholar]
- 26.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/jco.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/jco.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 28.Bustreo S., Osella-Abate S., Cassoni P., Donadio M., Airoldi M., Pedani F., Papotti M., Sapino A., Castellano I. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: A large case series study with a long-term follow-up. Breast Cancer Res. Treat. 2016;157:363–371. doi: 10.1007/s10549-016-3817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbeck N., Rastogi P., Martin M., Tolaney S.M., Shao Z.M., Fasching P.A., Huang C.S., Jaliffe G.G., Tryakin A., Goetz M.P., et al. Adjuvant abemaciclib combined with endocrine therapy for high-risk early breast cancer: Updated efficacy and Ki-67 analysis from the monarchE study. Ann. Oncol. 2021;32:1571–1581. doi: 10.1016/j.annonc.2021.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Ahn S.G., Yoon C.I., Lee J.H., Lee H.S., Park S.E., Cha Y.J., Cha C., Bae S.J., Lee K.A., Jeong J. Low PR in ER(+)/HER2(-) breast cancer: High rates of TP53 mutation and high SUV. Endocr. Relat. Cancer. 2019;26:177–185. doi: 10.1530/erc-18-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H.W., Lee H.M., Hwang S.H., Ahn S.G., Lee K.A., Jeong J. Patterns and Biologic Features of p53 Mutation Types in Korean Breast Cancer Patients. J. Breast Cancer. 2014;17:1–7. doi: 10.4048/jbc.2014.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim N.Y., Sroor M.Y., Darwish D.O. Impact of bilateral breast cancer on prognosis: Synchronous versus metachronous tumors. Asian Pac. J. Cancer Prev. 2015;16:1007–1010. doi: 10.7314/apjcp.2015.16.3.1007. [DOI] [PubMed] [Google Scholar]
- 33.Lowe S.W., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 34.Serrano M., Hannon G.J., Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 35.Chang B.D., Xuan Y., Broude E.V., Zhu H., Schott B., Fang J., Roninson I.B. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 36.Lowe S.W., Ruley H.E., Jacks T., Housman D.E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi S., Sato N., Kaneko K., Masuda N., Kawai M., Hirakawa H., Nomizu T., Iwata H., Ueda A., Ishikawa T., et al. TP53 signature predicts pathological complete response after neoadjuvant chemotherapy for breast cancer: Observational and confirmational study using prospective study cohorts. Transl. Oncol. 2024;48:102060. doi: 10.1016/j.tranon.2024.102060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertheau P., Plassa F., Espié M., Turpin E., de Roquancourt A., Marty M., Lerebours F., Beuzard Y., Janin A., de Thé H. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 2002;360:852–854. doi: 10.1016/s0140-6736(02)09969-5. [DOI] [PubMed] [Google Scholar]
- 39.Kim T., Han W., Kim M.K., Lee J.W., Kim J., Ahn S.K., Lee H.B., Moon H.G., Lee K.H., Kim T.Y., et al. Predictive Significance of p53, Ki-67, and Bcl-2 Expression for Pathologic Complete Response after Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer. J. Breast Cancer. 2015;18:16–21. doi: 10.4048/jbc.2015.18.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ungerleider N.A., Rao S.G., Shahbandi A., Yee D., Niu T., Frey W.D., Jackson J.G. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 2018;20:115. doi: 10.1186/s13058-018-1044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonnefoi H., Litière S., Piccart M., MacGrogan G., Fumoleau P., Brain E., Petit T., Rouanet P., Jassem J., Moldovan C., et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: A landmark and two-step approach analyses from the EORTC 10994/BIG 1-00 phase III trial. Ann. Oncol. 2014;25:1128–1136. doi: 10.1093/annonc/mdu118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandioler-Eckersberger D., Ludwig C., Rudas M., Kappel S., Janschek E., Wenzel C., Schlagbauer-Wadl H., Mittlböck M., Gnant M., Steger G., et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin. Cancer Res. 2000;6:50–56. [PubMed] [Google Scholar]
- 43.Bidard F.C., Matthieu M.C., Chollet P., Raoefils I., Abrial C., Dômont J., Spielmann M., Delaloge S., André F., Penault-Llorca F. p53 status and efficacy of primary anthracyclines/alkylating agent-based regimen according to breast cancer molecular classes. Ann. Oncol. 2008;19:1261–1265. doi: 10.1093/annonc/mdn039. [DOI] [PubMed] [Google Scholar]
- 44.Liu W., Konduri S.D., Bansal S., Nayak B.K., Rajasekaran S.A., Karuppayil S.M., Rajasekaran A.K., Das G.M. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J. Biol. Chem. 2006;281:9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- 45.Konduri S.D., Medisetty R., Liu W., Kaipparettu B.A., Srivastava P., Brauch H., Fritz P., Swetzig W.M., Gardner A.E., Khan S.A., et al. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. USA. 2010;107:15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grote I., Bartels S., Kandt L., Bollmann L., Christgen H., Gronewold M., Raap M., Lehmann U., Gluz O., Nitz U., et al. TP53 mutations are associated with primary endocrine resistance in luminal early breast cancer. Cancer Med. 2021;10:8581–8594. doi: 10.1002/cam4.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Archer S.G., Eliopoulos A., Spandidos D., Barnes D., Ellis I.O., Blamey R.W., Nicholson R.I., Robertson J.F. Expression of ras p21, p53 and c-erbB-2 in advanced breast cancer and response to first line hormonal therapy. Br. J. Cancer. 1995;72:1259–1266. doi: 10.1038/bjc.1995.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shahbandi A., Nguyen H.D., Jackson J.G. TP53 Mutations and Outcomes in Breast Cancer: Reading beyond the Headlines. Trends Cancer. 2020;6:98–110. doi: 10.1016/j.trecan.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji J.H., Bae S.J., Kim K., Chu C., Lee K.A., Kim Y., Kim J.H., Jeong J., Ahn S.G. Association between TP53 mutation and high 21-gene recurrence score in estrogen receptor-positive/HER2-negative breast cancer. NPJ Breast Cancer. 2022;8:19. doi: 10.1038/s41523-022-00384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langerød A., Zhao H., Borgan Ø., Nesland J.M., Bukholm I.R., Ikdahl T., Kåresen R., Børresen-Dale A.L., Jeffrey S.S. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fedorova O., Daks A., Shuvalov O., Kizenko A., Petukhov A., Gnennaya Y., Barlev N. Attenuation of p53 mutant as an approach for treatment Her2-positive cancer. Cell Death Discov. 2020;6:100. doi: 10.1038/s41420-020-00337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan D.S., Marchió C., Jones R.L., Savage K., Smith I.E., Dowsett M., Reis-Filho J.S. Triple negative breast cancer: Molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res. Treat. 2008;111:27–44. doi: 10.1007/s10549-007-9756-8. [DOI] [PubMed] [Google Scholar]
- 54.Chae B.J., Bae J.S., Lee A., Park W.C., Seo Y.J., Song B.J., Kim J.S., Jung S.S. p53 as a specific prognostic factor in triple-negative breast cancer. Jpn. J. Clin. Oncol. 2009;39:217–224. doi: 10.1093/jjco/hyp007. [DOI] [PubMed] [Google Scholar]
- 55.Dumay A., Feugeas J.P., Wittmer E., Lehmann-Che J., Bertheau P., Espié M., Plassa L.F., Cottu P., Marty M., André F., et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int. J. Cancer. 2013;132:1227–1231. doi: 10.1002/ijc.27767. [DOI] [PubMed] [Google Scholar]
- 56.Bertheau P., Lehmann-Che J., Varna M., Dumay A., Poirot B., Porcher R., Turpin E., Plassa L.F., de Roquancourt A., Bourstyn E., et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast. 2013;22((Suppl. 2)):S27–S29. doi: 10.1016/j.breast.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Nakopoulou L.L., Alexiadou A., Theodoropoulos G.E., Lazaris A.C., Tzonou A., Keramopoulos A. Prognostic significance of the co-expression of p53 and c-erbB-2 proteins in breast cancer. J. Pathol. 1996;179:31–38. doi: 10.1002/(SICI)1096-9896(199605)179:1<31::AID-PATH523>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 58.Geisler S., Lønning P.E., Aas T., Johnsen H., Fluge O., Haugen D.F., Lillehaug J.R., Akslen L.A., Børresen-Dale A.L. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61:2505–2512. [PubMed] [Google Scholar]
- 59.Li J.P., Zhang X.M., Zhang Z., Zheng L.H., Jindal S., Liu Y.J. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine. 2019;98:e15449. doi: 10.1097/MD.0000000000015449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darb-Esfahani S., Denkert C., Stenzinger A., Salat C., Sinn B., Schem C., Endris V., Klare P., Schmitt W., Blohmer J.U., et al. Role of TP53 mutations in triple negative and HER2-positive breast cancer treated with neoadjuvant anthracycline/taxane-based chemotherapy. Oncotarget. 2016;7:67686–67698. doi: 10.18632/oncotarget.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abubakar M., Guo C., Koka H., Sung H., Shao N., Guida J., Deng J., Li M., Hu N., Zhou B., et al. Clinicopathological and epidemiological significance of breast cancer subtype reclassification based on p53 immunohistochemical expression. NPJ Breast Cancer. 2019;5:20. doi: 10.1038/s41523-019-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu B., Yi Z., Guan Y., Ouyang Q., Li C., Guan X., Lv D., Li L., Zhai J., Qian H., et al. Molecular landscape of TP53 mutations in breast cancer and their utility for predicting the response to HER-targeted therapy in HER2 amplification-positive and HER2 mutation-positive amplification-negative patients. Cancer Med. 2022;11:2767–2778. doi: 10.1002/cam4.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coutant C., Rouzier R., Qi Y., Lehmann-Che J., Bianchini G., Iwamoto T., Hortobagyi G.N., Symmans W.F., Uzan S., Andre F., et al. Distinct p53 gene signatures are needed to predict prognosis and response to chemotherapy in ER-positive and ER-negative breast cancers. Clin. Cancer Res. 2011;17:2591–2601. doi: 10.1158/1078-0432.CCR-10-1045. [DOI] [PubMed] [Google Scholar]
- 64.Silwal-Pandit L., Vollan H.K., Chin S.F., Rueda O.M., McKinney S., Osako T., Quigley D.A., Kristensen V.N., Aparicio S., Børresen-Dale A.L., et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. 2014;20:3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 65.Kim J.Y., Lee E., Park K., Park W.Y., Jung H.H., Ahn J.S., Im Y.H., Park Y.H. Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes. Oncotarget. 2017;8:27997–28007. doi: 10.18632/oncotarget.15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bae S.Y., Lee J.H., Bae J.W., Jung S.P. Differences in prognosis by p53 expression after neoadjuvant chemotherapy in triple-negative breast cancer. Ann. Surg. Treat. Res. 2020;98:291–298. doi: 10.4174/astr.2020.98.6.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergh J., Norberg T., Sjögren S., Lindgren A., Holmberg L. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat. Med. 1995;1:1029–1034. doi: 10.1038/nm1095-1029. [DOI] [PubMed] [Google Scholar]
- 68.Rossner P., Jr., Gammon M.D., Zhang Y.J., Terry M.B., Hibshoosh H., Memeo L., Mansukhani M., Long C.M., Garbowski G., Agrawal M., et al. Mutations in p53, p53 protein overexpression and breast cancer survival. J. Cell. Mol. Med. 2009;13:3847–3857. doi: 10.1111/j.1582-4934.2008.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olivier M., Langerød A., Carrieri P., Bergh J., Klaar S., Eyfjord J., Theillet C., Rodriguez C., Lidereau R., Bièche I., et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 70.Meric-Bernstam F., Zheng X., Shariati M., Damodaran S., Wathoo C., Brusco L., Demirhan M.E., Tapia C., Eterovic A.K., Basho R.K., et al. Survival Outcomes by TP53 Mutation Status in Metastatic Breast Cancer. JCO Precis. Oncol. 2018;2018:1–15. doi: 10.1200/PO.17.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho Y., Gorina S., Jeffrey P.D., Pavletich N.P. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 72.Bai H., Yu J., Jia S., Liu X., Liang X., Li H. Prognostic Value of the TP53 Mutation Location in Metastatic Breast Cancer as Detected by Next-Generation Sequencing. Cancer Manag. Res. 2021;13:3303–3316. doi: 10.2147/CMAR.S298729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pal A., Gonzalez-Malerva L., Eaton S., Xu C., Zhang Y., Grief D., Sakala L., Nwekwo L., Zeng J., Christensen G., et al. Multidimensional quantitative phenotypic and molecular analysis reveals neomorphic behaviors of p53 missense mutants. NPJ Breast Cancer. 2023;9:78. doi: 10.1038/s41523-023-00582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Børresen A.L., Andersen T.I., Eyfjörd J.E., Cornelis R.S., Thorlacius S., Borg A., Johansson U., Theillet C., Scherneck S., Hartman S., et al. TP53 mutations and breast cancer prognosis: Particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995;14:71–75. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- 75.Kucera E., Speiser P., Gnant M., Szabo L., Samonigg H., Hausmaninger H., Mittlböck M., Fridrik M., Seifert M., Kubista E., et al. Prognostic significance of mutations in the p53 gene, particularly in the zinc-binding domains, in lymph node- and steroid receptor positive breast cancer patients. Austrian Breast Cancer Study Group. Eur. J. Cancer. 1999;35:398–405. doi: 10.1016/S0959-8049(98)00400-6. [DOI] [PubMed] [Google Scholar]
- 76.Fernández-Cuesta L., Oakman C., Falagan-Lotsch P., Smoth K.S., Quinaux E., Buyse M., Dolci M.S., Azambuja E.D., Hainaut P., Dell’orto P., et al. Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: Results from the BIG 02-98 phase III trial. Breast Cancer Res. 2012;14:R70. doi: 10.1186/bcr3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez K.D., Noltner K.A., Buzin C.H., Gu D., Wen-Fong C.Y., Nguyen V.Q., Han J.H., Lowstuter K., Longmate J., Sommer S.S., et al. Beyond Li Fraumeni Syndrome: Clinical characteristics of families with p53 germline mutations. J. Clin. Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 78.Robles A.I., Harris C.C. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb. Perspect. Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rohlin A., Wernersson J., Engwall Y., Wiklund L., Björk J., Nordling M. Parallel sequencing used in detection of mosaic mutations: Comparison with four diagnostic DNA screening techniques. Hum. Mutat. 2009;30:1012–1020. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- 80.Hainaut P., Pfeifer G.P. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb. Perspect. Med. 2016;6:a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study contain clinical outcomes for which institutional review board (IRB) approval is required before analysis. Therefore, these data are not publicly available. The data will be provided to authorized researchers who have obtained IRB approval from their institution and Gangnam Severance Hospital, Yonsei University, Seoul, Korea. For data access requests, please contact the corresponding authors, namely Pf. J.J., email address: gsjjoon@yuhs.ac.