Abstract
Down syndrome (DS), caused by trisomy 21 (T21), results in immune and metabolic dysregulation. People with DS experience co-occurring conditions at higher rates than the euploid population. However, the interplay between immune and metabolic alterations and the clinical manifestations of DS are poorly understood. Here, we report an integrated analysis of immunometabolic pathways in DS. Using multi-omics data, we infered cytokine-metabolite relationships mediated by specific transcriptional programs. We observed increased mediation of immunometabolic interactions in those with DS compared to euploid controls by genes in interferon response, heme metabolism, and oxidative phosphorylation. Unsupervised clustering of immunometabolic relationships in people with DS revealed subgroups with different frequencies of co-occurring conditions. Across the subgroups, we observed distinct mediation by DNA repair, Hedgehog signaling, and angiogenesis. The molecular stratification associates with the clinical heterogeneity observed in DS, suggesting that integrating multiple omic profiles reveals axes of coordinated dysregulation specific to DS co-occurring conditions.
Multi-omic data integration reveals molecular mechanisms of immune and metabolic dysregulation in Down syndrome.
INTRODUCTION
Down syndrome (DS) is the most common human chromosomal disorder, with approximately 1 in 700 newborns in the United States having trisomy 21 (T21), the driver of DS (1, 2). The incidence of DS increased from 9.0 to 11.8 per 10,000 live births in the United States between 1979 and 2003 (3). In the same period, the average life span of individuals with DS increased from 30 to 60 years due to improved medical care (4). Despite this increased life span, individuals with DS and their families face ongoing health challenges from co-occurring conditions that are more prevalent among those with T21. People with DS are at a higher risk than the general population for developing disorders such as hearing and visual impairments, obesity, dyslipidemia, congenital heart defects, leukemias, diverse autoimmune disorders, Alzheimer’s disease, and neurological conditions like autism and epilepsy (5–13). In addition, individuals with DS experience more severe complications from viral respiratory infections, putting them at high risk of poor outcomes, as is the case for severe acute respiratory syndrome coronavirus 2 infections (14). Despite the known clinical conditions associated with DS, little is known about the mechanisms by which triplication of chromosome 21 alters molecular pathways, cellular function, and overall physiology to result in the observed pattern of co-occurring conditions.
DS is characterized as both an immune and metabolic disorder (4, 15). The cross-talk between cytokines, gene expression, and metabolites leads to coordinated regulation of metabolism and immune responses (16, 17). Inflammatory cytokine release is initially protective against infection, but extended and potentiated exposure is harmful (18). For example, elevated levels of acute phase proteins, such as C-reactive protein (CRP) and serum amyloid A (SAA), have been associated with mild cognitive impairment in individuals with DS (19, 20). Elevated levels of interleukin-6 (IL-6), CRP, and SAA have also been correlated with body mass index (BMI) and elevated fasting insulin, indicating their involvement in metabolic disorders (21). In a longitudinal study of individuals with Parkinson’s disease, CRP and SAA were correlated with tryptophan pathway metabolites (22). In a cohort of 165 people, Powers et al. (23) reported that 75 individuals with DS had higher circulating levels of kynurenine and quinolinic acid, a neurotoxic metabolite, compared to euploid controls. These previous studies, along with others, have focused on single-omic profiles and identified clear differences between individuals with T21 and disomic (D21) karyotypes (23–27). However, to our knowledge, no study has considered how immunometabolic cross-talk, inferred from cytokine-metabolite associations and their relations to gene expression, relates to disease co-occurrence in the context of T21.
Here, we identify the relationships between cytokine levels, metabolite profiles, and gene expression patterns to infer putative mechanisms of immunometabolic regulation in people with and without T21. Furthermore, by integrating cytokine and metabolite profiles, we define four distinct subgroups of individuals with DS that are enriched for different co-occurring conditions. This integrated analysis provides insights into the molecular underpinnings of co-occurring conditions in individuals with DS that are not apparent when using a single molecular profile.
RESULTS
The immunometabolic profile of DS differs from the general population
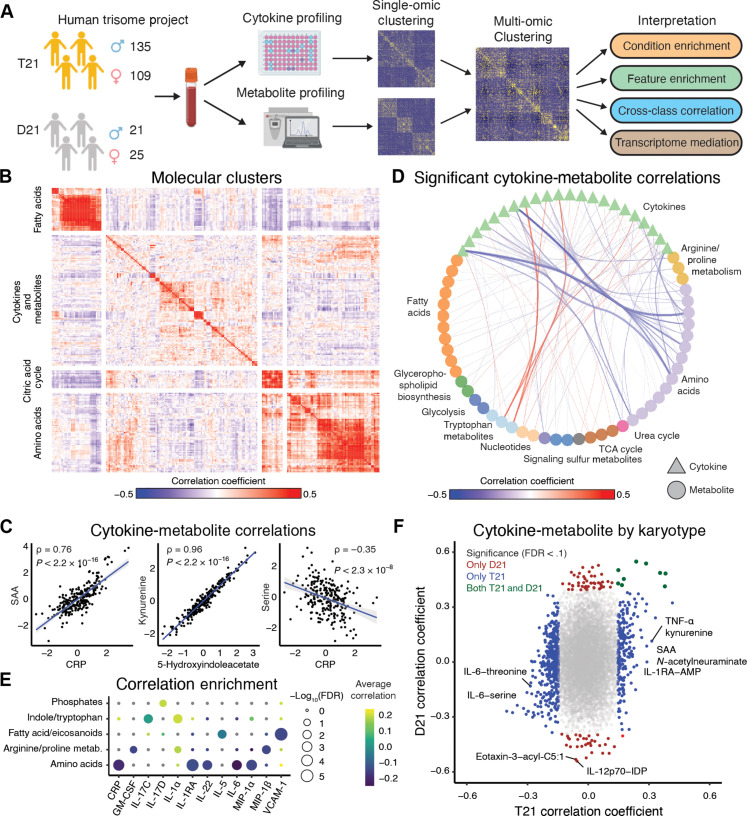
To investigate immunometabolic dysregulation in DS and its relationship to clinical features in this population, we analyzed datasets generated through the Human Trisome Project (HTP) cohort study [www.trisome.org; NCT02864108 (27)]. Multiple-omics datasets were generated from whole-blood samples, including 54 plasma immune markers using Meso Scale Discovery (MSD) assays, 174 plasma metabolites via ultrahigh performance liquid chromatography–mass spectrometry (UHPLC-MS), and 12,560 protein-coding gene transcript counts measured by RNA sequencing (fig. S1 and see Methods). Demographic data and information on co-occurring conditions were also collected through a combination of participant/caregiver surveys and review of medical records. The results presented here correspond to analysis from a subset of 290 individuals, of which 244 individuals were trisomic for chromosome 21 (T21), and 46 individuals were euploid controls (D21) (Fig. 1A).
Fig. 1. The immunometabolic profile of DS differs from the general population.
(A) Overview of analysis workflow. Whole-blood samples collected from individuals with T21 and D21 karyotypes were used to quantify plasma cytokines and metabolites. Individual-omic profiles were evaluated and an integrated analysis was performed. Identification of gene mediators of cytokine-metabolite relationships was performed, and immunometabolic subgroups were defined. Further analysis was performed on the subgroups. (B) Heatmap of Spearman correlation coefficients between standardized cytokine and metabolite abundances. Clustering with k-means defined four groups. (C) Representative scatterplots of standardized values between cytokines, metabolites, and across molecular assay type. Blue lines represent the lines of best fit between the features; gray areas represent the 95% confidence interval. Spearman correlation statistics are reported. (D) Statistically significant (FDR < 0.1) correlations between cytokines and metabolites, aggregated by metabolite class. The thickness of the edges between nodes represents the absolute value of aggregated correlations coefficients. (E) Overrepresentation enrichment of significant correlations between cytokines and metabolites (FDR < 0.1) annotated to metabolite classes using a Fisher’s exact test. (F) Spearman correlation coefficients between cytokines and metabolites in people with T21 and D21. Points are colored blue if the relationships are only significant (FDR < 0.1) in T21, red if they are only significant in D21, and dark green if they are significant in both T21 and D21 individuals. GM-CSF, granulocyte-macrophage colony-stimulating factor; FDR, false discovery rate; CRP, C-reactive protein; IL-17C, interleukin-17C; VCAM-1, vascular cell adhesion molecule–1; AMP, adenosine 5′-monophosphate; IDP, inosine 5′-diphosphate; SAA, serum amyloid A.
Consistent with previous reports, individuals with T21 significantly differed from euploid individuals in the occurrence of 9 of 17 clinical variables measured, including higher rates of obesity, autoimmune skin conditions, hypothyroidism, asthma, and recurrent otitis media, among others (table S1) (28–30). A greater portion of individuals with T21 were in their 20s and 30s, but we did not observe a statistically significant difference in age or sex by karyotype (fig. S2, A and B).
We performed differential abundance analysis for both immune markers and metabolites. In agreement with previous reports, on average, people with DS had increased cytokine abundances, including pro-inflammatory cytokines [e.g., thymic stromal lymphopoietin (TSLP), Interleukin (IL)–17D, IL-17C, IL-16, and IL-6] and chemokines [e.g., Monocyte chemoattractant protein-1 (MCP-1) and Interferon-γ inducible protein 10 (IP-10)], as well as acute phase proteins (e.g., CRP) (31). Anti-inflammatory cytokines (e.g., IL-10 and IL-1RA) and other immune mediators (vascular endothelial growth factor A, placenta growth factor, and fibroblast growth factor) were also elevated. Cytokines that significantly decreased in the population of people with DS included tumor necrosis factor–β (TNF-β) and IL-12/IL-23p40 (table S2). Increased metabolites in DS included markers of inflammation [e.g., sphingosine, urate, 5(S) hydroxyeicosatetraenoic acid (HETE), 12(S)-HETE, and 11-HETE], tryptophan pathway metabolites (e.g., kynurenine and 5-hydroxyindoleacetate), bile acids (e.g., taurolithocholic acid and ursodeoxycholic acid), and lysophospatidic acids (LPAs) (e.g., LPA 16:1) (32). Amino acids (e.g., glycine, histidine, serine, and tyrosine) were decreased in people with DS (table S3) (33). Overall, the metabolite and cytokine results are consistent with previously reported findings and provide support that the datasets are robust and represent the biology of individuals with DS (23–27, 31, 33).
To identify patterns of immunometabolic dysregulation in DS, we calculated pairwise Spearman correlations between all cytokine and metabolite pairs (fig. S1 and see Methods). In individuals with T21, we identified sets of cytokines and metabolites that share similar patterns of dysregulation using k-means clustering (k = 4) (Fig. 1, B and C; single dataset correlations are represented in fig. S3, A and B, and tables S4 and S5). Among cytokines, expectedly, factors in the IL-6/CRP/SAA signaling axis showed significant positive correlations with each other (Fig. 1C and fig. S3A). Interferon-γ (IFN-γ), IFNL1 (IL-29), and the IFN-inducible protein IP-10 were also positively correlated and associated with multiple monocyte-activating cytokines and chemokines, such as macrophage inflammatory protein-1α (MIP-1α), MCP-1, and MCP-4 (fig. S3C). Among metabolites, we observed strong correlations in tryptophan catabolites previously observed to be dysregulated in DS, such as kynurenine and 5-hydroxyindolacetate (Fig. 1C and fig. S1B) (23). Other examples of metabolites tightly correlated in individuals with DS included leukotriene B4 with hexadecenoic acid and resolvin D1/D2 with protectin D1 (fig. S3D).
Several interesting patterns were identified from the combined analyses of immune markers and metabolites. For example, CRP, SAA, and IL-6 were all negatively correlated with amino acid levels in DS, including serine, asparagine, and threonine (Fig. 1D and fig. S3E). As reported previously, TNF-α was positively correlated with the tryptophan catabolites kynurenine and 5-hydroxyindolacetate (23). We further evaluated the enrichment of significant correlations between cytokines and metabolites by metabolic class. Amino acids were enriched for negative correlations with several cytokines including CRP, IL-1RA, IL-22, IL-6, and MIP-1α (Fig. 1E). Enrichments of negative correlations were also observed between fatty acids/eicosanoids with IL-5 and VCAM-1. The cytokine IL-1α was enriched for positive correlations with arginine and proline metabolism as well as indole and tryptophan metabolites.
To assess the differences between karyotype, we compared the pairwise correlation coefficients between cytokines and metabolites in those with T21 versus euploid controls (D21). Overall, and consistent with the sample size of the groups, we observed more significant correlations between cytokines and metabolites [false discovery rate (FDR) < 0.1] in people with T21 (634 significant correlations) relative to D21 euploid controls (91 significant correlations) (Fig. 1F and table S6). To address the imbalance in sample size across karyotypes, we randomly subsampled the T21 cohort to 46 individuals to match the sample size of the D21 cohort. As with the full cohort, we observed more significant cytokine-metabolite correlations, both positive and negative, in those with T21 compared to euploid controls. Moreover, we observed a smaller range of correlation coefficients in euploids controls compared to the cohort with T21 (fig. S3F and table S6). For example, in those with T21, we observed more positive correlations between cytokines with kynurenine and N-acetylneuraminate compared to controls (Fig. 1F). N-acetylneuraminate is the predominant form of sialic acid in mammals and has been linked to immune signaling and recognition-memory impairment in mice (34). Together, these results reveal immunometabolic dysregulation in DS that differs from and is more heterogeneous than euploid individuals, which could contribute to the etiology of key co-occurring conditions in this population.
Mediation analysis reveals signaling pathways driving cytokine-metabolite relationships in DS
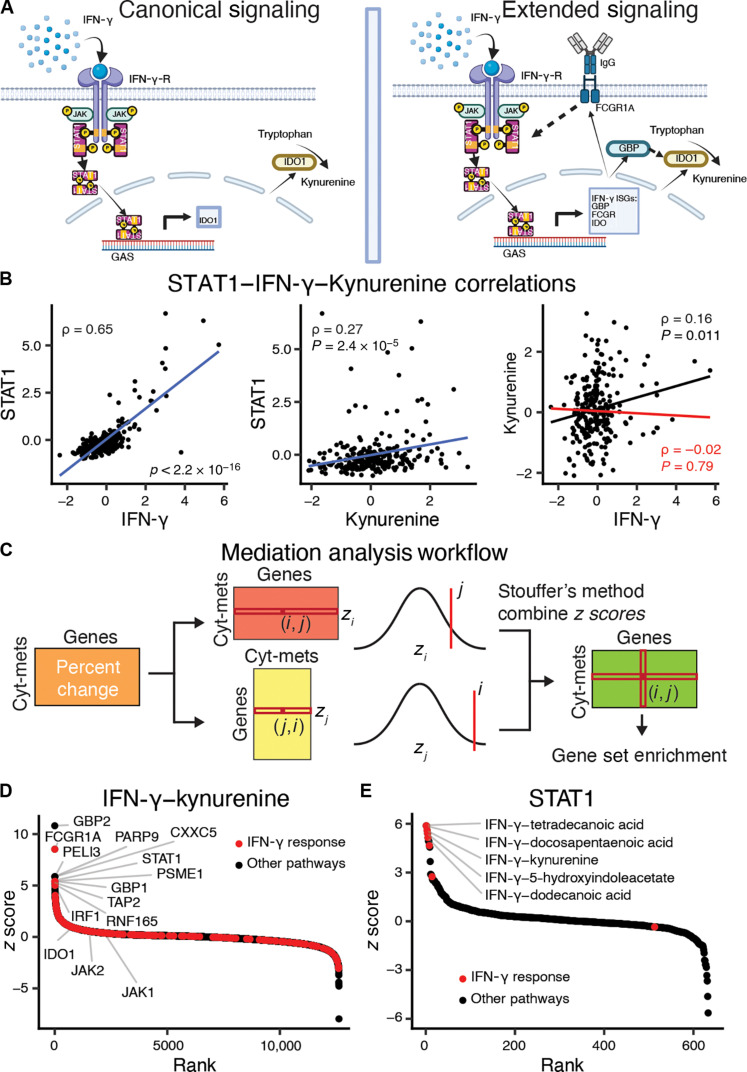
Cytokines can indirectly affect metabolite levels by altering the regulation and expression of enzymes involved in metabolic cascades (35). For example, IFN-γ signals through the IFN-γ receptors IFNGR1/IFNGR2 activate the Janus kinases 1 and 2 (JAK1/2) that, in turn, phosphorylate and activate the signal transducers and activators of transcription 1 (STAT1) transcription factor (Fig. 2A) (36). Upon translocation to the nucleus, STAT1 induces transcription of hundreds of IFN-stimulated genes (ISGs), including IDO1, which encodes indoleamine 2,3-dioxygenase 1, the rate-limiting enzyme in the conversion of tryptophan to kynurenine in the tryptophan catabolism pathway (37, 38). IDO1 expression is also induced by type I and type III IFNs through similar pathways (39, 40).
Fig. 2. Cytokines induce gene expression changes that mediate metabolite levels.
(A) Indoleamine 2,3-dioxygenase 1 (IDO1) is the rate-limiting enzyme in the conversion of tryptophan to kynurenine and is induced by IFN-γ signaling. IFN-γ signals through the IFN-γ receptor activating STAT1, which then up-regulates IDO1 via binding to the gamma interferon activation site (GAS). The right panel extends this signaling pathway based on the mediation analysis to include other ISGs. Created using BioRender.com. (B) Scatterplots for IFN-γ–STAT1, kynurenine-STAT1, and IFN-γ–kynurenine. Blue lines represent the lines of best fit between the features. The red line represents the linear fits of the partial correlation coefficient after adjustment for STAT1 expression. Spearman direct correlation (black) and partial correlation (red) statistics are reported. (C) Overview of the mediation analysis algorithm to identify immunometabolic relationships that are conditioned on gene expression. The partial Spearman correlation coefficients between cytokines and metabolites after adjusting for transcript abundances is compared to the direct Spearman correlations between cytokines and metabolites. The percentage changes between the partial and direct correlations over all cytokine-metabolite relationships and gene transcripts are calculated, and then both the cytokine-metabolite and gene axes are z-score normalized. Last, the z scores are combined using Stouffer’s method to combine z scores. (D) Gene mediation rankings for IFN-γ–kynurenine with IFN-γ response genes highlighted in red. (E) Cytokine-metabolite rankings for mediation by STAT1 with cytokine-metabolite relationships enriched for IFN-γ response genes are highlighted in red. FCGR, Fcγ receptor; GBP, guanylate-binding protein; IgG, immunoglobulin G; JAK, Janus kinase; PARP9, poly(ADP-ribose) polymerase.
To predict these kinds of mediating relationships, using IFN-γ (type II IFN) as a positive control, we used matched transcriptome data to identify genes that could regulate cytokine-metabolite relationships (fig. S1). Using the partial correlation (see Methods for technical details), we can show that STAT1 mediates the relationship between IFN-γ and kynurenine based on the 88% drop in correlation when the relationship is conditioned on STAT1 (Fig. 2B). This result demonstrates that analyzing cytokine, metabolite, and gene expression measurements through the partial correlation mediation analysis captures known biological regulation and can be used to prioritize genes as potential mediators of immunometabolic relationships.
To assess the specificity of potential gene regulation, we applied the mediation analysis across all other immunometabolic relationships (Fig. 2C and see Methods for more details). We identified several known genes as mediators of the IFN-γ–kynurenine relationship. Overall, STAT1 was the sixth highest ranked gene (Fig. 2D). Guanylate-binding proteins 2 and 1 (GBP2/1) were ranked first and eight, respectively. Bai et al. (41) found that knocking down GBPs inhibited IDO1 expression in human mesenchymal stromal cells. Another highly ranked potential mediator was Fcγ receptor I (FCGR1A). IFN-γ increases FCGR1A expression and the relative affinity of FCGR1A for immune complexes (42). Conversely, and consistent with known relationships, the immunometabolic relationships most significantly mediated by STAT1 were between IFN-γ and saturated fatty acids (e.g., dodecanoic acid, docosapentaenoic acid, and tretradecanoic acid) or tryptophan catabolites (e.g., kynurenine and 5-hydroxyindoleacetate) (Fig. 2E) (43). In summary, our approach of leveraging the difference between partial and direct correlations is effective for identifying genes that are most likely to mediate immunometabolic relationships, thus allowing us to expand and contextualize the known effectors as illustrated with IFN-γ signaling in Fig. 2A.
Distinct transcriptional programs mediate immunometabolic relationships
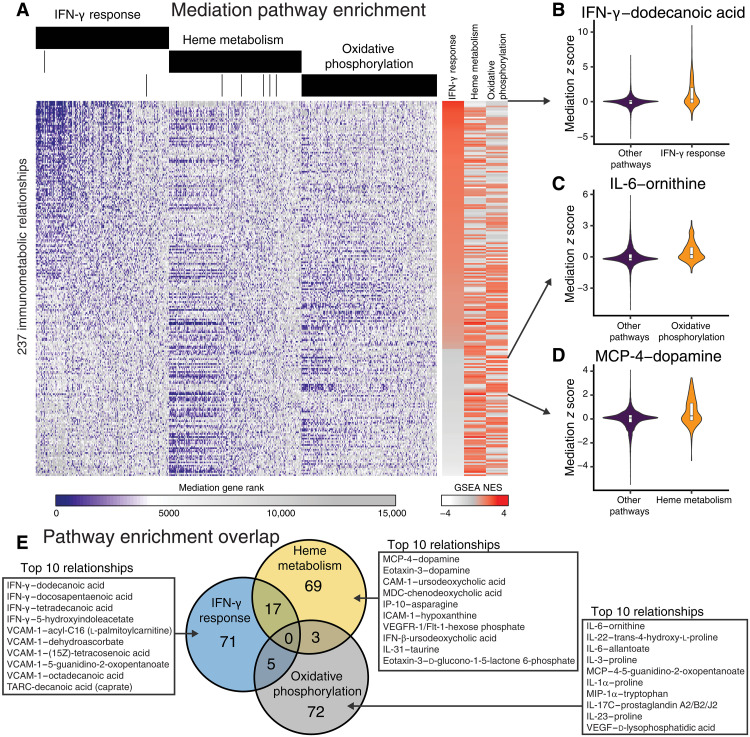
To understand the transcriptional mediation effect on cellular pathways and processes, we performed gene set enrichment analysis on the ranked list of mediator gene z scores for each cytokine-metabolite relationship (fig. S1) (44, 45). We previously identified T21-specific changes in gene expression in the whole-blood transcriptome including activation of the IFN-γ response, heme metabolism pathways, and oxidative phosphorylation (26, 27). Ranking the gene sets based on the variance of normalized enrichment scores (NESs) across cytokine-metabolite relationships and hierarchically clustering cytokine-metabolite relationships by pathway mediation NESs revealed that the IFN-γ and IFN-α response pathways varied widely across cytokine-metabolite relationships (fig. S4). The oxidative phosphorylation and heme metabolism pathways mediated sets of cytokine-metabolite relationships that are largely distinct from the IFN-γ and IFN-α sets. Together, these results demonstrate that specific cytokine-metabolite relationships are mediated by largely distinct transcriptional programs in people with DS.
IFN-γ response genes were significantly enriched (FDR < 0.05) for mediation in 93 cytokine-metabolite relationships (Fig. 3A). In ranking cytokine-metabolite relationships by IFN-γ response enrichment scores, we observed that the top result was IFN-γ and dodecanoic acid (or lauric acid) (Fig. 3B). Lauric acid has been shown to inhibit the effect of IFN-γ on intercellular adhesion molecule–1 (ICAM-1) and vascular cell adhesion molecule–1 (VCAM-1) expression in macrophages (46). The correlation coefficient between IFN-γ and dodecanoic acid is indeed negative (table S6), indicating that this relationship may be regulated by IFN-γ response genes. Overall, metabolite pathway enrichment analysis for IFN-γ revealed significant enrichment (FDR < 0.1) of sugars (e.g., maltose, mannitol, d-ribose, d-rhamnose, and d-arabitol) and saturated fatty acids [e.g., hexanoic acid (caproate), heptanoic acid, octanoic acid (caprylate), nonanoic acid (pelargonate), decanoic acid (caprate), dodecanoic acid, and tetradecanoic acid] among the immunometabolic interactions.
Fig. 3. Mediation analysis reveals signaling pathways driving cytokine-metabolite relationships in DS.
(A) Gene mediation rankings over all genes for cytokine-metabolite relationships. Genes (x axis) are ordered by aggregate ranking across immunometabolic relationships and separated by pathway annotation. Cytokine-metabolite relationships (y axis) are ordered by the normalized gene set enrichment scores IFN-γ response pathway genes. (B to D) Mediation scores for the immunometabolic relationship most enriched for mediation by (B) IFN-γ response, (C) oxidative phosphorylation, or (D) heme metabolism. (E) Overlap of cytokine-metabolite relationships with significant enrichments for gene mediation in IFN-γ response pathway genes, heme metabolism, and oxidative phosphorylation. The top 10 significant relationships specific to each pathway are listed in the insets. GSEA, gene set enrichment analysis; NES, normalized enrichment score.
Heme metabolism genes were significantly enriched for the mediation of 89 cytokine-metabolite relationships (Fig. 3A). Immunometabolic relationships between dopamine and the cytokines MCP-4 and eotaxin were most enriched for heme metabolism mediation (Fig. 3D). Other top-ranking cytokine-metabolite relationships were observed between cytokines ICAM-1, macrophage-derived cytokine (MDC), IFN-β, and VCAM-1 with bile acids (e.g., ursodeoxycholic acid and chenodeoxycholic acid). Metabolite pathway enrichment analysis on the immunometabolic relationships enriched for mediation by heme metabolism showed that the most represented metabolite classes included amino acids and bile acids (FDRs of 0.13 and 0.36, respectively).
Oxidative phosphorylation genes were significantly enriched for the mediation of 80 cytokine-metabolite relationships (Fig. 3A). The relationship between inflammatory cytokine IL-6 and urea cycle metabolite ornithine was most significantly enriched for mediation by oxidative phosphorylation (Fig. 3C). Moreover, we observed that oxidative phosphorylation mediated several of the immunometabolic relationships involving arginine and proline metabolism metabolites (e.g., ornithine, proline, trans-4-hydroxy-l-proline, 4-5-guanidino-2-oxopentanoate). Overall, we observed enrichment of oxidative phosphorylation mediation with relationships involving amino acids, phosphates, urea cycle, and arginine and proline metabolism at FDRs of 0.07, 011, 0.11, and 0.17, respectively.
Together, we observed that immunometabolic relationships were mostly regulated by distinct transcriptional programs dysregulated in DS, such as IFN-γ response, heme metabolism, and oxidative phosphorylation (Fig. 3E). These results suggest that dysregulated pathways in DS affect unique immunometabolic relationships.
Integrating immune markers and metabolites identifies clinical subgroups in DS
There is heterogeneity in co-occurring conditions in people with DS. For example, 78% of people with DS in this cohort have a history of sleep apnea, 65% have a history of autoimmune skin condition, and 42% are classified as obese (BMI ≥ 30), while only 29 and 11% have a history of asthma or depression, respectively. Given our observation that dysregulated pathways are related to distinct immunometabolic relationships, we performed an integrated clustering of the cytokine and metabolite profiles in DS to identify subgroups of individuals that were enriched for pre-existing, co-occurring conditions (fig. S1 and see Methods for details on clustering).
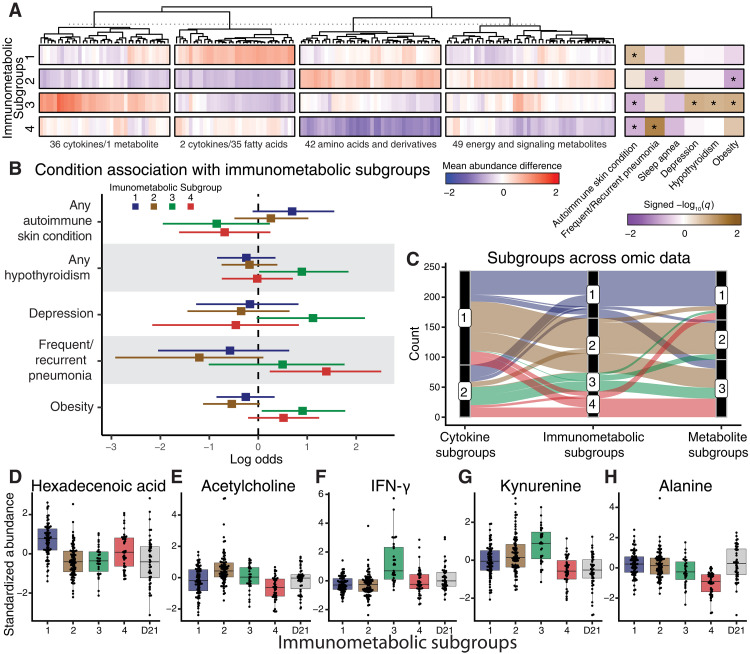
We identified an integrated immunometabolic cluster solution of four subgroups (ranging from 39 to 91 individuals) with distinct immunometabolic interactions and clinical profiles (Fig. 4, A and B; fig. S5A; and table S7). We compared the integrated subgroups to the subgroups based on individual cytokine or metabolite profiles, which consisted of two and three subgroups, respectively (fig. S5, B and C, and tables S8 and S9). The integrated solution stratified the cohort into unique groups with a combination of individuals from the single-omic clustering solutions (Fig. 4C and fig. S5D), suggesting that integrating the data identifies molecular patterns that are not captured in the single-omic profiles. Furthermore, there were more enriched co-occurring conditions across the clusters based on the integrated-omic data in comparison to the single-omic clusters (Fig. 4, A to C, and fig. S5, B and C and E and F). We refer to the integrated clusters as immunometabolic subgroups (IMSs) 1 to 4.
Fig. 4. Immunometabolic relationships define clinical subgroups in DS.
(A) Hierarchically clustered differences in mean feature abundance (IMS versus all others) separated by immunometabolic subgroups. Co-occurring conditions were tested for enrichment across subgroups (signed −log10 q values of one-sided Fisher’s exact tests; *FDR < 0.2). (B) Log odds ratios for reported co-occurring conditions in relation to each immunometabolic subgroup. (C) Alluvial plot demonstrating the distribution of individuals across subgroups based on cytokine profiles, metabolite profiles, or immunometabolic profiles. (D to H) Standardized abundances of individual cytokines or metabolites across immunometabolic subgroups and in relationship to individuals with D21 karyotype. TCA, tricarboxylic acid.
Immunometabolic subgroup 1 (IMS1) consists of 71 individuals. Compared to the rest of the individuals with T21, this group was enriched for people with a history of autoimmune skin conditions (Fig. 4B). Individuals in IMS1 have higher levels of fatty acids/eicosanoids. For example, hexadecanoic (palmitic) acid, a prevalent saturated fatty acid found in Western diets, was higher in this subgroup (Fig. 4D). Hexadecanoic acid has been associated with adaptive immunity and is known to enhance toll-like receptor–dependent inflammation by inducing ceramide metabolism (47).
IMS2 consists of 91 individuals. This subgroup was underrepresented for obese individuals and those with a history of frequent/recurrent pneumonia (Fig. 4B). Amino acids were more highly abundant in this subgroup, while fatty acids/eicosanoids had lower abundances. The feature most significantly elevated in IMS2 was acetylcholine (Fig. 4E). Acetylcholine inhibits the release of inflammatory cytokines and inflammation via α7 nicotinic acetylcholine receptors on immune cells (48), suggesting that acetylcholine may reduce inflammatory effects within this subgroup.
IMS3 consists of 31 individuals. This subgroup was enriched for individuals with a history of hypothyroidism, depression, and obesity (Fig. 4B). This subgroup also had fewer people with a history of autoimmune skin conditions. Individuals in this subgroup tend to have higher abundances of inflammatory cytokines (e.g., IFN-γ, SAA, and IL-29) (Fig. 4F) and tryptophan catabolites (e.g., kynurenine and 5-hydroxyindoleacetate) (Fig. 4G). This cluster was also enriched for lower abundances of fatty acids/eicosanoids.
IMS4 consists of 43 individuals. This group was enriched for people with a history of frequent or recurrent pneumonia and had fewer individuals with a history of autoimmune skin conditions (Fig. 4B). The individuals in this group had significantly lower levels of amino acids. For example, alanine was significantly lower in IMS4 compared to all other individuals with either T21 or D21 karyotypes (Fig. 4H).
To understand if there was a karyotype-specific immunometabolic effect, we investigated the euploid controls using the same approach as with the trisomic individuals. Within the euploid individuals, we identified two D21 IMSs (fig. S6A). On the molecular level, we found similarities in cytokine and metabolite clusters between T21 and D21 individuals (fig. S6, B and C). However, there was also variability in molecular clusters, indicating that there are karyotype-specific immunometabolic patterns.
Overall, the clustering of the integrated cytokine and metabolomic profiles revealed groupings in DS with enrichment and depletion for combinations of co-occurring conditions. Furthermore, the consistent molecular interactions within the stratifications implicate shared mediation of immunometabolic relationships and identify putative mechanisms related to the associated conditions.
Immunometabolic subgroups reveal differential mediation of cytokine-metabolite relationships
Given the distinct immunometabolic patterns of the four IMSs, we investigated the transcriptional programs associated with these patterns by applying our multi-omic mediation analysis within each subgroup (fig. S1). To create an IMS representation of pathway mediation, we compared the number of immunometabolic relationships with significant (FDR < 0.05) positive enrichments (NES > 0) across the hallmark gene sets. To identify shared and distinct patterns of pathway mediation, we normalized the counts of enriched gene sets within each IMS to the counts of enriched gene sets across all individuals with T21.
Comparatively, IMS1 (elevated history of autoimmune skin conditions) and IMS3 (elevated history of hypothyroidism, depression, and obesity) demonstrated increased immunometabolic mediation by the DNA repair gene set (Fig. 5A). In contrast, IMS4 (elevated history of frequent/recurrent pneumonia) showed increased mediation by genes involved in angiogenesis. IMS2, which included people with a healthier clinical profile, had comparatively higher enrichments and immunometabolic mediation by Hedgehog signaling. The rates of enriched immunometabolic mediation by IFN-γ, heme metabolism, and oxidative phosphorylation gene sets were similar across the four IMSs.
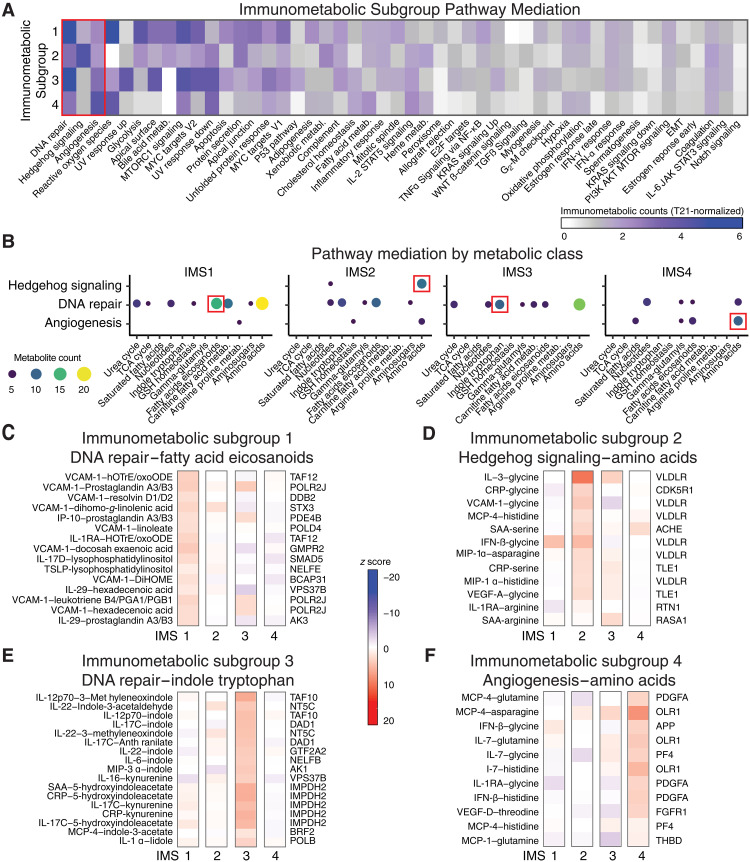
Fig. 5. Immunometabolic subgroups reveal differential mediation of cytokine-metabolite relationships.
(A) Number of immunometabolic relationships that were significantly (FDR < 0.05) and positively enriched (NES > 0) for mediation by genes in gene sets across each immunometabolic subgroup (IMS). Counts of enriched gene sets within each IMS were normalized to the counts of enriched gene sets across all individuals with T21. (B) Enrichment of metabolite classes for the cytokine-metabolite relationships mediated by the gene sets that were most differential across IMSs [red box in (A): DNA repair, Hedgehog signaling, and angiogenesis). (C to F) Top gene mediators and their relative strength of mediation (z scores) for cytokine-metabolite relationships within a selected metabolite class for each IMS [red boxes in (B)] are reported. (C) For IMS1, the top genes within DNA repair that mediate fatty acid/eicosaniod cytokine-metabolite relationships are shown. (D) For IMS2, the top genes within Hedgehog signaling that mediate amino acid cytokine-metabolite relationships are shown. (E) For IMS3, the top genes within DNA repair that mediate indole and tryptophan cytokine-metabolite relationships are shown. (F) For IMS4, the top genes within Angiogenesis that mediate amino acid cytokine-metabolite relationships are shown.
We next investigated the top three pathways that were most differential across the IMSs, specifically DNA repair, Hedgehog signaling, and angiogenesis (Fig. 5A). We compared the metabolite classes enriched for pathway mediation and found shared and distinct classes of immunometabolic mediation, even when the overall number of immunometabolic features was similarly enriched. For example, immunometabolic relationships involving amino acids were enriched for mediation by DNA repair genes in both IMS1 and IMS3; however, relationships with fatty acids/eicosanoids were distinctly mediated in IMS1, while relationships with indole and tryptophan metabolites were distinctly mediated in IMS3 (Fig. 5B).
To investigate this further, we identified immunometabolic relationships and the top gene-mediating relationship within these metabolite clusters. For DNA repair genes in IMS1, POLR2J (RNA polymerase II subunit J) was the top mediator for several of the relationships between VCAM-1 and eicosanoids (e.g., prostaglandin A3/B3 and leukotriene B4/PGA1/PGB1) (Fig. 5C). IMS2 revealed pathway mediation of immunometabolic relationships involving amino acids by Hedgehog signaling, with very-low-density lipoprotein (VLDLR) receptor as the top mediator for several of the immunometabolic relationships involving amino acids (e.g., glycine) (Fig. 5D). In IMS3, Inosine monophosphate dehydrogenase 2 (IMPDH2) was the top mediator for relationships between inflammatory cytokines (e.g., CRP, SAA, and IL-17C) and tryptophan catabolites (kynurenine and 5-hydroxyindoleacetate) (Fig. 5E). In IMS4, which was enriched for people with a history of frequent/recurrent pneumonia, several of the immunometabolic relationships were mediated by platelet-derived growth factor subunit A (PDGFA) (Fig. 5F). The full set of mediator results across all clusters is available at 10.5281/zenodo.13370637.
To determine whether we captured different information in the mediation analysis compared to gene expression patterns across the IMSs, we performed differential expression analysis between each IMS and all other individuals with T21 (table S10) followed by gene set enrichment analysis. We then compared the gene set NES values based on differential expression to the IMS representation of gene set mediation (fig. S7, A to D). While some gene sets were enriched for both mediation and differential expression across all IMSs (e.g., IFN-γ response), several gene sets showed evidence of mediating immunometabolic relationships without induction or suppression. This implies that the gene sets enriched for mediation serve a functional role in immunometabolic regulation.
Together, IMS-specific mediation analyses revealed two important observations. First, the mechanisms and pathways previously identified as important to the disease pathology in DS, specifically interferon signaling, heme metabolism, and oxidative phosphorylation (26, 49–51) are ubiquitous and affect all IMSs equally. Second, there are patterns that indicate differential influence of pathways and processes in mediating immunometabolic relationships in an IMS-specific manner. Unraveling these interactions provides evidence of putative molecular interactions that may drive the heterogenous manifestation of co-occurring conditions in DS.
DISCUSSION
The triplication of chromosome 21 in DS causes an up-regulation of the expression of genes present on chromosome 21 and results in a wide array and severity of co-occurring conditions. J. Lejeune, who found T21 as the aneuploidy causing DS based on the slides of M. Gautier, hypothesized that DS is a “metabolic disease” (52, 53). It is only recently through efforts such as the HTP that molecular features in DS have been systematically and comprehensively measured. These data support J. Lejeune’s hypothesis where transcriptomic and metabolomic profiles have revealed dysregulation across metabolic pathways including insulin resistance, oxidative phosphorylation, lipid metabolism, homocysteine/folate/transulfuration pathways, heme metabolism, and many others (15, 26, 50, 54–56). As we have shown here, these data can further be used to identify putative molecular relationships using integrative computational methods.
Each blood plasma-omic profile used in this study provides a snapshot of the biological state of an individual. However, these molecular features provide complementary information, and we can gain different insights into potential mechanisms underlying DS and co-occurring conditions by studying the relationships between these molecular features. We showed that cytokine-metabolite relationships differ between euploid individuals (D21) and those with T21 (Fig. 1) and that gene expression data can be included to identify potential mediators of these relationships using a data-driven and systematic methodology (Fig. 2). We identified known relationships and characterized how gene sets, including IFN-γ signaling, heme metabolism, and oxidative phosphorylation, may affect the cytokine-metabolite relationships in DS (Fig. 3). Furthermore, we used these data to define immunometabolic subgroups of individuals with DS. These subgroups showed differential abundance of different cytokines and metabolites, as well as differential enrichment for the history of co-occurring conditions (Fig. 4). Last, we used mediation analysis to identify potential mechanisms that could explain why co-occurring conditions are associated with specific immunometabolic subgroups (Fig. 5).
An example of a potential immunometabolic mechanism specific to IMS2 is revealed by the pathway enrichment of cytokine-metabolite relationships involving amino acids and Hedgehog signaling (Fig. 5B). In IMS2, for which we observed comparatively lower levels of cytokines and increased abundances of amino acids (Fig. 4A), VLDLR was the top mediator for several of the immunometabolic relationships involving amino acids. Nguyen et al. (57) showed that VLDLR deficiency attenuates the inflammatory interaction between adipocytes and macrophages. As IL-3 regulates the proliferation of macrophages (58), and glycine is associated with decreased lipid accumulation in macrophages via attenuation of very-low-density lipoprotein) uptake (59), the relative increase in mediation by VLDLR is a potential explanation of the comparatively healthy profile of individuals in IMS2. Furthermore, previous studies have observed reduced Hedgehog signaling in people with DS, and this reduced signaling was associated with abnormalities in tissue development including craniofacial skeleton, heart, and the enteric nervous system tissues (60, 61). In T21 neural progenitor cells, treatment with a Sonic Hedgehog (SHH) agonist increased concentration of SHH and normalized T21-driven changes in gene expression and neuronal cell differentiation (62). In relation to immune signaling, IFN-γ has been shown to attenuate Hedgehog signaling in white adipose cells (63), and deficiency in the Hedgehog signaling gene VLDLR reduces inflammatory interaction between adipocytes and macrophages (57). Together, the increased mediation of immunometabolic relationships by Hedgehog signaling, specifically through VLDLR, in the blood of comparatively healthy individuals presents an interesting hypothesis for the role of Hedgehog signaling across tissues of people with DS; reduced Hedgehog signaling is associated with dysregulated neurogenesis yet healthier adipogenesis and inflammatory signaling. Future studies are necessary to model and further elucidate this mechanism in people with DS.
An expansion and more clinically focused translation of this work is in the stratification of individuals into groups with similar clinical characteristics to be leveraged for improved clinical care and potential prediction of drug response. As we have shown, the IMSs provide unique and molecularly defined groupings of individuals that are enriched for specific conditions. These groupings could help clinicians with defining common mechanisms of disease dysregulation. By incorporating the IMS-specific mediation analysis, we can reveal subgroups of people with T21 exhibiting more evidence of molecular regulation by specific genes and pathways (e.g., DNA repair in IMS1 and IMS3; angiogenesis in IMS4). Cross-referencing the evidence of mediation with gene and pathway drug targets may predict drug efficacy. For example, tofacitinib, a JAK/STAT inhibitor, is now in clinical trials for the treatment of autoimmune skin conditions and regression disorder in people with DS (NCT04246372 and NCT05662228). Our analysis suggested that IFN-γ response immunometabolic mediation is similar across IMSs. However, we hypothesize that differences in other interacting pathways across IMSs may predict the systemic response to this treatment within those groups of people; this will require follow-up studies.
This study also has distinct limitations. The cross-sectional nature of the data limits our ability to distinguish the molecular patterns that we identify as being causal or the result of co-occurring conditions. Although the data are deep (generating multiple-omic data on each sample), observations from a single time point must be considered in this context and present specific hypotheses to focus prospective analysis. In the mediation analysis, we assumed that cytokines induce transcriptional changes affecting metabolite levels. That is, we implied a directionality in the relationships based on biological knowledge, but the statistical model is undirected. Metabolites could also impact gene expression and then cytokine levels. Therefore, as with all associative analyses, causality cannot be proven without additional follow-up studies. We also note that our results should be interpreted in the context of the biospecimens and measurement approaches used, namely, blood samples and bulk-omic profiling. Last, here, we evaluated 244 samples from T21 and 46 samples for D21—any future multi-omic data analyses will benefit from increased sample size, particularly considering occurrence rates for each co-occurring condition. Future data generation can focus on specific conditions longitudinally, which will strengthen both the overall analysis and the condition-specific analysis. Despite these limitations, this is the first study to integrate cytokine, metabolite, and gene expression datasets sampled from the same individuals in DS, and we identified relationships for future studies.
In conclusion, we carried out a comprehensive study aiming to elucidate the mechanisms underlying immunometabolic regulation within subgroups of people with DS. Using this approach, we reproduced known mechanisms of regulation and prioritized hypotheses for future evaluation. We anticipate that this methodology and these results may aid DS researchers in elucidating the molecular causes of co-occurring conditions and prioritizing treatments with the highest likelihood of success.
METHODS
Study consent
The Crnic Institute’s HTP (www.trisome.org) enrolled participants under a study protocol approved by the Colorado Multiple Institutional Review Board (COMIRB #15-2170). Written informed consent was obtained from participants or their legally authorized representative; written consent was obtained from participants over 7 years old as allowed by cognitive ability. Study procedures were explained to all participants, regardless of age and/or cognitive ability.
Clinical histories
Clinical histories were recorded for study participants through a combination of participant/caregiver surveys and annotation of medical records. In the case of discrepancies, medical records took precedence. Demographic data collected at visit time include age at visit, sex, and BMI. All co-occurring conditions were recorded as “history of,” regardless of the status of the condition. Of the 17 recorded co-occurring conditions, we removed four conditions from the analysis with less than 15 case counts in people with DS, including history of autism spectrum disorder, cataracts, pulmonary hypertension, and regression. The conditions used in this study include history of anxiety, any autoimmune skin condition, hypothyroidism, hearing loss, sleep apnea, seizures, celiac disease, asthma or restrictive airway disease, frequent or recurrent pneumonia, obesity, and recurrent otits media.
Blood processing and molecular quantification
Waugh et al. (26) and Galbraith et al. (27) report detailed descriptions of blood processing and molecular quantification for omic profiling performed by the HTP. Briefly, peripheral blood was collected in Vacutainer K2 EDTA tubes (BD) and PAXgene RNA Tubes (QIAGEN). Plasma inflammatory markers (cytokines) were quantified using multiplex immunoassay platform V-PLEX Human Biomarker 54-Plex Kit (Meso Scale Diagnostics). Plasma metabolomic and lipidomic profiles were quantified using a Vanquish UHPLC coupled online to a Q Exactive high-resolution mass spectrometer (Thermo Fisher Scientific). Whole-blood paired-end RNA sequencing was performed using Illumina NovaSeq 6000 instrument (Novogene) on samples extracted from the PAXgene RNA tubes.
Data preprocessing
All data processing and analyses were performed in R (v4.2.1). This study included 290 individuals from the HTP cohort with matched immune, metabolite, and gene expression profiling, of which 244 individuals have DS. Before analysis, all the omic profiles were log2-transformed. To correct for confounding variables in clustering analyses, we adjusted for age, sex, and sample source in a random effects model using the “adjust” function from the “datawizard” R package (v0.7.1) (64). Age and sex were fixed effects, while sample source was included as a random effect. Data were z score–transformed for comparison across feature classes. Transcriptomic features were filtered to protein coding genes with a variance greater than 0 (12,624 features). Cytokine and metabolite profiles consisted of 54 and 174 features, respectively.
Molecular correlations
To evaluate the normality of cytokine and metabolite distributions, we performed Kolmogrov-Smirnov (KS) goodness of fit test (KS) and Shapiro-Wilk (SW) test for normality using the “ks.test” and “shapiro.test” functions, respectively, from the base “stats” package (v4.2.1) in R. For the 54 cytokines, we found that 23 (43%) and 43 (80%) for the KS and SW tests fell below 0.05, which rejects the null hypothesis that the data are sampled from a normal distribution. Similarly, for the 174 metabolites, we found that 54 (31%) and 122 (70%) for the KS and SW tests similarly fell below the 0.05 threshold. These analyses indicate that not all samples are normally distributed, so we selected the nonparametric Spearman correlation coefficient to be used for all correlation calculations.
Spearman rank-based correlations were calculated within and across data types using the “corr.test” function from the “psych” R package (v2.3.6). When comparing correlations, test results were corrected for multiple comparisons using the Benjamini-Hochberg procedure (65). When comparing correlations across karyotypes (i.e., T21 against D21), we addressed class imbalances by iteratively (1000 iterations) subsampling the T21 population to 46 individuals (matching the sample size of the D21 population) and recalculating Spearman correlation coefficients.
Differential abundance analysis
Given the large number of samples per group (minimum of 31 in IMS3), the statistical test to evaluate differential abundance for the cytokine and metabolomic data was the Wilcoxon ranked sum test, which we calculated using the “wilcox.test” function from the base stats package (v4.2.1) in R. Test results were corrected for multiple comparisons using the Benjamini-Hochberg procedure (65).
Differential gene expression analysis
We calculated the differential gene expression between each IMS compared to all other IMSs (e.g., IMS1 versus IMS2 to IMS4). Differential gene expression pipelines are well-established, and we used the “limma” package (v 3.52.4) with the RNA sequencing counts matrix as input and incorporated covariates into the model (i.e., age at visit, sex, and sample source) (66).
Metabolites class enrichment
We identified enriched metabolite classes within significantly correlated immunometablic relationships using an overrepresentation analysis testing the hypothesis that the proportion of significant metabolites from a particular metabolite class is higher than expected. We used the “fisher.test” function from the base stats package (v4.2.1) in R with the alternative hypothesis parameter set to “greater.” Test results were corrected for multiple comparisons using the Benjamini-Hochberg procedure (65).
Gene mediation analysis
We calculated the direct Spearman correlations between all significant (FDR < 0.1) combinations of cytokine abundance, metabolite abundance, and transcript counts (transformed and standardized as defined in the “Data pre-processing” section). We then calculated partial correlations between cytokine and metabolite abundances with adjustment for each gene count using the “pcor” function in the “ppcor” package (v1.1) (67). Equation 1 defines the partial correlation between cytokine (i) and metabolite (j) given transcript (k)
| (1) |
We then calculated the percent change between the direct and partial correlation coefficients for each cytokine-metabolite relationship across all genes using Eq. 2
| (2) |
To extend this partial correlation analysis systematically, we leveraged the context likelihood of relatedness (CLR) network inference method (68). The CLR approach aims to find relationships that are specific to the biological context tested. Therefore, to assess the specificity of the observed conditional change and prioritized potential mediators, we calculated the scores for the percent change for each cytokine-metabolite association across all genes (z1) and for the percent change for each gene across all cytokine-metabolite associations (z2). We then created a combined score for downstream analysis using Stouffer’s method for combining z scores (69) as defined in Eq. 3
| (3) |
Gene set enrichment analysis
We ranked gene expression transcripts based on differential mediation (either by karyotype or across clusters within people with DS) by the percent change in the mediation analysis. We then performed gene set enrichment analysis over the hallmark gene sets from the Molecular Signatures Database and using the“fgsea” package (version 1.22.0) (44, 45, 70).
Immunometabolic stratification
To identify immunometabolic subgroups, single- and multi-omic clustering was performed using neighborhood-based multi-omics (NEMO) clustering (71). First, interpatient similarity matrices were calculated for each molecular profile. The similarity measure used is based on the radial basis function kernel and incorporates a normalizing factor to control for the density of samples by averaging the squared distance between samples to their nearest neighbors and each other (71, 72). Next, the relative similarity matrix was calculated for each omic profile to account for the different distributions of features within each data type. For a single-omic profile, clustering was performed on the similarity matrix. For multi-omic profiles, the relative similarity matrices were combined into an average similarity matrix. Last, spectral clustering was performed using a variant based on the eigenvectors of the normalized Laplacian (73).
Determining the number of clusters and hyperparameters of NEMO
The minimum cluster size was set to 5% of the sample size for adequately powered statistics between clusters. The number of clusters was determined by evaluating several metrics, including the eigengap heuristic (74), the modified eigengap heuristic used in NEMO [i.e., the eigengap solution multiplied by the number of clusters (71)], the number of enriched co-occurring conditions, and the number of significantly differential molecular features. Of the evaluation criteria, we gave the most weight to the number of enriched conditions based on the goal of identifying molecular subgroups with shared biology in those with similar profiles of co-occurring conditions.
The NEMO algorithm is affected by the number of neighbors’ hyperparameter. The authors suggest that when the number of clusters is defined, then the number of neighbors should be equal to . However, the NEMO algorithm was developed for omic profiles with substantially more features (i.e., gene expression, methylation, and microRNA expression) than the cytokine and metabolite assays we tested. Therefore, we performed a grid search between 10 and 100 neighbors by increments of 5 to evaluate the number of enriched conditions per clustering solution.
We further validated the clusters using a bootstrapping approach (75), which randomly sampled the population with replacement, reclustered the random sample, and calculated the adjusted rand index (76) between the random clusters and the full cohort solution. The bootstrapping was performed for 1000 iterations, and the average adjusted rand index is reported.
Association testing with co-occurring conditions
One-sided Fisher’s exact tests were performed to test for independence within each immunometabolic subgroup. For any continuous phenotypes tested, Wilcoxon rank-sum tests were performed comparing the within cluster to the mean of all other subjects. Results were adjusted for multiple comparisons using the Benjamini-Hochberg procedure (65).
Acknowledgments
We would like to thank L. Feldman, C. Dieter, M. Orman, and S. Nandi for insightful comments during the early iterations of this analysis. We thank all self-advocates with DS and their families for participation in the HTP. We thank K. Jordan and her team at the Human Immune Monitoring Shared Resource for outstanding service in generation of the immune marker dataset and A. D’Alessandro and his team in the Metabolomics Shared Resource for generation of the metabolomics dataset. We are also grateful to the Colorado Translational and Sciences Institute and the Colorado Multiple Institutional Review Board for assistance in all clinical research projects involving the Crnic Institute. Special thanks to M. S. Whitten, the team at the Global Down Syndrome Foundation, J. Reilly, and R. Sokol for logistical support at multiple stages of the project.
Funding: This work was supported by grants from the National Institutes of Health R01AI150305 (to J.M.E.), HD109765 (to J.C.C.), GM007635-45S1 (to L.A.G.), P30CA046934, and UM1TR004399; the Linda Crnic Institute for Down Syndrome; the Global Down Syndrome Foundation; the Anna and John J. Sie Foundation; the GI & Liver Innate Immune Program; the Human Immunology and Immunotherapy Initiative, University of Colorado School of Medicine; and the Boettcher Foundation (to K.D.S.).
Author contributions: Conceptualization: L.A.G., M.D.G., J.C.C., and J.M.E. Formal analysis: L.A.G., M.D.G., and J.M.E. Funding acquisition: J.C.C. and J.M.E. Methodology: L.A.G., M.D.G., and J.C.C. Supervision: J.C.C., J.M.E., and M.D.G. Software: L.A.G. and J.M.E. Validation: L.A.G., J.C.C., M.D.G., and J.M.E. Resources: J.C.C., M.D.G., K.D.S., and J.M.E. Investigation: J.C.C., A.L.R., and J.M.E. Project administration: J.C.C., A.L.R., and J.M.E. Data curation: L.A.G., J.C.C., A.L.R., N.P.E., M.D.G., and J.M.E. Visualization: L.A.G., J.C.C., M.D.G., and J.M.E. Writing—original draft: L.A.G., J.C.C., J.M.E., and M.D.G. Writing—review and editing: L.A.G., J.C.C., K.D.S., N.P.E., and J.M.E.
Competing interests: J.M.E. has provided consulting services to Elli Lily, Gilead, Biohaven, and Perha Pharmaceuticals. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The demographics and clinical data for ~700 research participants in the HTP study are available on both the Synapse data sharing platform (https://doi.org/10.7303/syn31488784) and the INCLUDE Data Hub (https://portal.includedcc.org/). Whole blood transcriptome data for 400 research participants (26) can be accessed through Synapse (https://doi.org/10.7303/syn31488780), the INCLUDE Data Hub, and Gene Expression Omnibus (GSE190125). Transcriptome data from murine tissues can be accessed through the Gene Expression Omnibus under accession numbers GSE229762 and GSE229737. Targeted plasma proteomics data for inflammatory markers using MSD assays for 470+ research participants (25) can be accessed through Synapse (https://doi.org/10.7303/syn31475487) and the INCLUDE Data Hub. Plasma metabolomics data for 410+ participants can be accessed through Synapse (https://doi.org/10.7303/syn31488782), Metabolomics Workbench (study ID ST002200; https://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST002200&Access=UrlT3545), and the INCLUDE Data Hub. All code for running this analysis and generating figures is available at 10.5281/zenodo.13376529.
Supplementary Materials
The PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S10
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S10
REFERENCES AND NOTES
- 1.Mai C. T., Isenburg J. L., Canfield M. A., Meyer R. E., Correa A., Alverson C. J., Lupo P. J., Riehle-Colarusso T., Cho S. J., Aggarwal D., Kirby R. S., National Birth Defects Prevention Network , National population-based estimates for major birth defects, 2010–2014. Birth Defects Res. 111, 1420–1435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC, Facts about Down Syndrome (CDC, 2020); www.cdc.gov/ncbddd/birthdefects/downsyndrome.html.
- 3.Shin M., Besser L. M., Kucik J. E., Lu C., Siffel C., Correa A., Prevalence of Down syndrome among children and adolescents in 10 regions of the United States. Pediatrics 124, 1565–1571 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Bittles A. H., Glasson E. J., Clinical, social, and ethical implications of changing life expectancy in Down syndrome. Dev. Med. Child Neurol. 46, 282–286 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Hartley D., Blumenthal T., Carrillo M., DiPaolo G., Esralew L., Gardiner K., Granholm A.-C., Iqbal K., Krams M., Lemere C., Lott I., Mobley W., Ness S., Nixon R., Potter H., Reeves R., Sabbagh M., Silverman W., Tycko B., Whitten M., Wisniewski T., Down syndrome and Alzheimer’s disease: Common pathways, common goals. Alzheimers Dement. 11, 700–709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull M. J., Committee on Genetics , Health supervision for Children with Down syndrome. Pediatrics 128, 393–406 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Maloney K. W., Taub J. W., Ravindranath Y., Roberts I., Vyas P., Down syndrome preleukemia and leukemia. Pediatr. Clin. North Am. 62, 121–137 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ivarsson S. A., Ericsson U. B., Gustafsson J., Forslund M., Vegfors P., Annerén G., The impact of thyroid autoimmunity in children and adolescents with Down syndrome. Acta Paediatr. 86, 1065–1067 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Zachor D. A., Mroczek-Musulman E., Brown P., Prevalence of celiac disease in Down syndrome in the United States. J. Pediatr. Gastroenterol. Nutr. 31, 275–279 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Sureshbabu R., Kumari R., Ranugha S., Sathyamoorthy R., Udayashankar C., Oudeacoumar P., Phenotypic and dermatological manifestations in Down syndrome. Dermatol. Online J. 17, 3 (2011). [PubMed] [Google Scholar]
- 11.F. Guaraldi, R. Rossetto Giaccherino, F. Lanfranco, G. Motta, D. Gori, E. Arvat, E. Ghigo, R. Giordano, Endocrine Autoimmunity in Down’s syndrome in Frontiers of Hormone Research, W. Savino, F. Guaraldi, Eds. (S. Karger AG, 2017;), vol. 48, pp. 133–146. [DOI] [PubMed] [Google Scholar]
- 12.Ji N. Y., Capone G. T., Kaufmann W. E., Autism spectrum disorder in Down syndrome: Cluster analysis of Aberrant Behaviour Checklist data supports diagnosis. J. Intellect. Disabil. Res. 55, 1064–1077 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Wexler I. D., Abu-Libdeh A., Rn Y. K., Nimrodi A., Kerem E., Tenenbaum A., Optimizing health care for individuals with Down syndrome in Israel. Isr. Med. Assoc. J. 11, 655–659 (2009). [PubMed] [Google Scholar]
- 14.De Toma I., Dierssen M., Network analysis of Down syndrome and SARS-CoV-2 identifies risk and protective factors for COVID-19. Sci. Rep. 11, 1930 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dierssen M., Fructuoso M., Martínez de Lagrán M., Perluigi M., Barone E., Down syndrome is a metabolic disease: Altered insulin signaling mediates peripheral and brain dysfunctions. Front. Neurosci. 14, 670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao N., Nie M., Pang H., Wang B., Hu J., Meng X., Li K., Ran X., Long Q., Deng H., Chen N., Li S., Tang N., Huang A., Hu Z., Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 12, 1618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill L. A. J., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huggard D., Doherty D. G., Molloy E. J., Immune dysregulation in children with Down syndrome. Front. Pediatr. 8, 73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Bryant S. E., Zhang F., Silverman W., Lee J. H., Krinsky-McHale S. J., Pang D., Hall J., Schupf N., Proteomic profiles of incident mild cognitive impairment and Alzheimer’s disease among adults with Down syndrome. Alzheimers Dement. 12, e12033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manti S., Cutrupi M. C., Cuppari C., Ferro E., Dipasquale V., Di Rosa G., Chimenz R., La Rosa M. A., Valenti A., Salpietro V., Inflammatory biomarkers and intellectual disability in patients with Down syndrome. J. Intellect. Disabil. Res. 62, 382–390 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Helmersson J., Vessby B., Larsson A., Basu S., Association of type 2 diabetes with cyclooxygenase-mediated inflammation and oxidative stress in an elderly population. Circulation 109, 1729–1734 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Heilman P. L., Wang E. W., Lewis M. M., Krzyzanowski S., Capan C. D., Burmeister A. R., Du G., Escobar Galvis M. L., Brundin P., Huang X., Brundin L., Tryptophan metabolites are associated with symptoms and nigral pathology in Parkinson’s disease. Mov. Disord. 35, 2028–2037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers R. K., Culp-Hill R., Ludwig M. P., Smith K. P., Waugh K. A., Minter R., Tuttle K. D., Lewis H. C., Rachubinski A. L., Granrath R. E., Carmona-Iragui M., Wilkerson R. B., Kahn D. E., Joshi M., Lleó A., Blesa R., Fortea J., D’Alessandro A., Costello J. C., Sullivan K. D., Espinosa J. M., Trisomy 21 activates the kynurenine pathway via increased dosage of interferon receptors. Nat. Commun. 10, 4766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan K. D., Evans D., Pandey A., Hraha T. H., Smith K. P., Markham N., Rachubinski A. L., Wolter-Warmerdam K., Hickey F., Espinosa J. M., Blumenthal T., Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci. Rep. 7, 14818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araya P., Kinning K. T., Coughlan C., Smith K. P., Granrath R. E., Enriquez-Estrada B. A., Worek K., Sullivan K. D., Rachubinski A. L., Wolter-Warmerdam K., Hickey F., Galbraith M. D., Potter H., Espinosa J. M., IGF1 deficiency integrates stunted growth and neurodegeneration in Down syndrome. Cell Rep. 41, 111883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waugh K. A., Minter R., Baxter J., Chi C., Galbraith M. D., Tuttle K. D., Eduthan N. P., Kinning K. T., Andrysik Z., Araya P., Dougherty H., Dunn L. N., Ludwig M., Schade K. A., Tracy D., Smith K. P., Granrath R. E., Busquet N., Khanal S., Anderson R. D., Cox L. L., Estrada B. E., Rachubinski A. L., Lyford H. R., Britton E. C., Fantauzzo K. A., Orlicky D. J., Matsuda J. L., Song K., Cox T. C., Sullivan K. D., Espinosa J. M., Triplication of the interferon receptor locus contributes to hallmarks of Down syndrome in a mouse model. Nat. Genet. 55, 1034–1047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galbraith M. D., Rachubinski A. L., Smith K. P., Araya P., Waugh K. A., Enriquez-Estrada B., Worek K., Granrath R. E., Kinning K. T., Paul Eduthan N., Ludwig M. P., Hsieh E. W. Y., Sullivan K. D., Espinosa J. M., Multidimensional definition of the interferonopathy of Down syndrome and its response to JAK inhibition. Sci. Adv. 9, eadg6218 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonarakis S. E., Down syndrome and the complexity of genome dosage imbalance. Nat. Rev. Genet. 18, 147–163 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Capone G. T., Chicoine B., Bulova P., Stephens M., Hart S., Crissman B., Videlefsky A., Myers K., Roizen N., Esbensen A., Peterson M., Santoro S., Woodward J., Martin B., Smith D., Down Syndrome Medical Interest Group DSMIG-USA Adult Health Care Workgroup , Co-occurring medical conditions in adults with Down syndrome: A systematic review toward the development of health care guidelines. Am. J. Med. Genet. A. 176, 116–133 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Gensous N., Bacalini M. G., Franceschi C., Garagnani P., Down syndrome, accelerated aging and immunosenescence. Semin. Immunopathol. 42, 635–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rachubinski A. L., Wallace E., Gurnee E., Estrada B. A. E., Worek K. R., Smith K. P., Araya P., Waugh K. A., Granrath R. E., Britton E., Lyford H. R., Donovan M. G., Eduthan N. P., Hill A. A., Martin B., Sullivan K. D., Patel L., Fidler D. J., Galbraith M. D., Dunnick C. A., Norris D. A., Espinosa J. M., JAK inhibition decreases the autoimmune burden in Down syndrome. eLife 13, RP99323 (2024). [Google Scholar]
- 32.S. Kapoor, M. Fitzpatrick, E. Clay, R. Bayley, G. R. Wallace, S. P. Young, “Metabolomics in the Analysis of Inflammatory Diseases” in Metabolomics, U. Roessner, Ed. [InTech, Rijeka (HR), 2012; http://www.ncbi.nlm.nih.gov/books/NBK402338/]. [PubMed]
- 33.Donovan M. G., Eduthan N. P., Smith K. P., Britton E. C., Lyford H. R., Araya P., Granrath R. E., Waugh K. A., Enriquez Estrada B., Rachubinski A. L., Sullivan K. D., Galbraith M. D., Espinosa J. M., Variegated overexpression of chromosome 21 genes reveals molecular and immune subtypes of Down syndrome. Nat. Commun. 15, 5473 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzzi S., Croese T., Ravid A., Gold O., Clark A. R., Medina S., Kitsberg D., Adam M., Vernon K. A., Kohnert E., Shapira I., Malitsky S., Itkin M., Brandis A., Mehlman T., Salame T. M., Colaiuta S. P., Cahalon L., Slyper M., Greka A., Habib N., Schwartz M., N-acetylneuraminic acid links immune exhaustion and accelerated memory deficit in diet-induced obese Alzheimer’s disease mouse model. Nat. Commun. 14, 1293 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Carriere J., Lin X., Xie N., Feng P., Interplay between cellular metabolism and cytokine responses during viral infection. Viruses 10, 521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark G. R., Darnell J. E., The JAK-STAT Pathway at Twenty. Immunity 36, 503–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroder K., Hertzog P. J., Ravasi T., Hume D. A., Interferon-γ: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Taylor M. W., Feng G. S., Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5, 2516–2522 (1991). [PubMed] [Google Scholar]
- 39.MacMicking J. D., Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng C.-W., Shieh P.-C., Lin Y.-C., Chen Y.-J., Lin Y.-H., Kuo D.-H., Liu J.-Y., Kao J.-Y., Kao M.-C., Way T.-D., Indoleamine 2,3-dioxygenase, an immunomodulatory protein, is suppressed by (−)-epigallocatechin-3-gallate via Blocking of γ-interferon-induced JAK-PKC-δ-STAT1 signaling in human oral cancer cells. J. Agric. Food Chem. 58, 887–894 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Bai S., Mu Z., Huang Y., Ji P., Guanylate binding protein 1 inhibits osteogenic differentiation of human mesenchymal stromal cells derived from bone marrow. Sci. Rep. 8, 1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swisher J. F. A., Feldman G. M., The many faces of FcγRI: Implications for therapeutic antibody function. Immunol. Rev. 268, 160–174 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Sisler J. D., Morgan M., Raje V., Grande R. C., Derecka M., Meier J., Cantwell M., Szczepanek K., Korzun W. J., Lesnefsky E. J., Harris T. E., Croniger C. M., Larner A. C., The signal transducer and activator of transcription 1 (STAT1) inhibits mitochondrial biogenesis in liver and fatty acid oxidation in adipocytes. PLOS ONE 10, e0144444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.G. Korotkevich, V. Sukhov, A. Sergushichev, Fast gene set enrichment analysis. bioRxiv 060012 [Preprint] (2019). https://doi.org/10.1101/060012.
- 46.Lim W.-S., Gan M.-S.-Y., Ong M.-H.-L., Chew C.-H., Lauric acid abolishes interferon-gamma (IFN-γ)-induction of intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) expression in human macrophages. Asian Pac. J. Reprod. 4, 217–221 (2015). [Google Scholar]
- 47.Seufert A. L., Napier B. A., A new frontier for fat: Dietary palmitic acid induces innate immune memory. Immunometabolism 5, e00021 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang E. H., Chavan S. S., Pavlov V. A., Cholinergic control of inflammation, metabolic dysfunction, and cognitive impairment in obesity-associated disorders: Mechanisms and novel therapeutic opportunities. Front. Neurosci. 13, 263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan K. D., Lewis H. C., Hill A. A., Pandey A., Jackson L. P., Cabral J. M., Smith K. P., Liggett L. A., Gomez E. B., Galbraith M. D., DeGregori J., Espinosa J. M., Trisomy 21 consistently activates the interferon response. eLife 5, e16220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayona-Bafaluy M. P., Garrido-Pérez N., Meade P., Iglesias E., Jiménez-Salvador I., Montoya J., Martínez-Cué C., Ruiz-Pesini E., Down syndrome is an oxidative phosphorylation disorder. Redox Biol. 41, 101871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambert K., Moo K. G., Arnett A., Goel G., Hu A., Flynn K. J., Speake C., Wiedeman A. E., Gersuk V. H., Linsley P. S., Greenbaum C. J., Long S. A., Partridge R., Buckner J. H., Khor B., Deep immune phenotyping reveals similarities between aging, Down syndrome, and autoimmunity. Sci. Transl. Med. 14, eabi4888 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lejeune J., On the mechanism of mental deficiency in chromosomal diseases. Hereditas 86, 9–14 (1977). [DOI] [PubMed] [Google Scholar]
- 53.Elisabeth Pain “After more than 50 years, a dispute over Down syndrome discovery” Science, 11 February 14; www.science.org/content/article/after-more-50-years-dispute-over-down-syndrome-discovery. [DOI] [PubMed]
- 54.Pogribna M., Melnyk S., Pogribny I., Chango A., Yi P., James S. J., Homocysteine metabolism in children with Down syndrome: In vitro modulation. Am. J. Hum. Genet. 69, 88–95 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Convertini P., Menga A., Andria G., Scala I., Santarsiero A., Castiglione Morelli M. A., Iacobazzi V., Infantino V., The contribution of the citrate pathway to oxidative stress in Down syndrome. Immunology 149, 423–431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buonuomo P. S., Bartuli A., Mastrogiorgio G., Vittucci A., Di Camillo C., Bianchi S., Pires Marafon D., Villani A., Valentini D., Lipid profiles in a large cohort of Italian children with Down syndrome. Eur. J. Med. Genet. 59, 392–395 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Nguyen A., Tao H., Metrione M., Hajri T., Very low density lipoprotein receptor (VLDLR) expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J. Biol. Chem. 289, 1688–1703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen B. D., Clark C. R., Interleukin 3 (IL 3) regulates the in vitro proliferation of both blood monocytes and peritoneal exudate macrophages: Synergism between a macrophage lineage-specific colony-stimulating factor (CSF-1) and IL 3. J. Immunol. 137, 563–570 (1986). [PubMed] [Google Scholar]
- 59.Rom O., Villacorta L., Zhang J., Chen Y. E., Aviram M., Emerging therapeutic potential of glycine in cardiometabolic diseases: Dual benefits in lipid and glucose metabolism. Curr. Opin. Lipidol. 29, 428–432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Currier D. G., Polk R. C., Reeves R. H., A Sonic hedgehog (Shh) response deficit in trisomic cells may be a common denominator for multiple features of Down syndrome. Prog. Brain Res. 197, 223–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. J. Moyer, F.-X. Fernandez, Y. Li, D. K. Klinedinst, L. D. Florea, Y. Kazuki, M. Oshimura, R. H. Reeves, Overexpression screen of chromosome 21 genes reveals modulators of Sonic hedgehog signaling relevant to Down syndrome. bioRxiv 492735 [Preprint] (2022). https://doi.org/10.1101/2022.05.19.492735. [DOI] [PMC free article] [PubMed]
- 62.Klein M., Thomas M., Hofmann U., Seehofer D., Damm G., Zanger U. M., A systematic comparison of the impact of inflammatory signaling on absorption, distribution, metabolism, and excretion gene expression and activity in primary human hepatocytes and HepaRG cells. Drug Metab. Dispos. 43, 273–283 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Todoric J., Strobl B., Jais A., Boucheron N., Bayer M., Amann S., Lindroos J., Teperino R., Prager G., Bilban M., Ellmeier W., Krempler F., Müller M., Wagner O., Patsch W., Pospisilik J. A., Esterbauer H., Cross-talk between interferon-γ and hedgehog signaling regulates adipogenesis. Diabetes 60, 1668–1676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patil I., Makowski D., Ben-Shachar M. S., Wiernik B. M., Bacher E., Lüdecke D., Datawizard: An R package for easy data preparation and statistical transformations. J. Open Source Softw. 7, 4684 (2022). [Google Scholar]
- 65.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995). [Google Scholar]
- 66.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S., Ppcor: An R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22, 665–674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faith J. J., Hayete B., Thaden J. T., Mogno I., Wierzbowski J., Cottarel G., Kasif S., Collins J. J., Gardner T. S., Large-scale mapping and validation of escherichia coli transcriptional regulation from a compendium of expression profiles. PLOS Biol. 5, e8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. A. Stouffer, E. A. Suchman, L. C. Devinney, S. A. Star, R. M. Williams Jr., The American Soldier: Adjustment during Army Life. (Studies in Social Psychology in World War II) (Princeton Univ. Press, 1949), vol. 1. [Google Scholar]
- 70.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rappoport N., Shamir R., NEMO: Cancer subtyping by integration of partial multi-omic data. Bioinformatics 35, 3348–3356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. D. Buhmann, Radial Basis Functions: Theory and Implementations (Cambridge Univ. Press, ed. 1, 2003). [Google Scholar]
- 73.A. Ng, M. Jordan, Y. Weiss, On spectral clustering: Analysis and an algorithm, in Advances in Neural Information Processing Systems (MIT Press, 2001), vol. 14. [Google Scholar]
- 74.von Luxburg U., A tutorial on spectral clustering. Stat. Comput. 17, 395–416 (2007). [Google Scholar]
- 75.Fang Y., Wang J., Selection of the number of clusters via the bootstrap method. Comput. Stat. Data Anal. 56, 468–477 (2012). [Google Scholar]
- 76.J. M. Santos, M. Embrechts, On the use of the adjusted rand index as a metric for evaluating supervised classification in Artificial Neural Networks – ICANN 2009, C. Alippi, M. Polycarpou, C. Panayiotou, G. Ellinas, Eds. (Springer, 2009), vol. 5769. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S10
Tables S1 to S10