Abstract
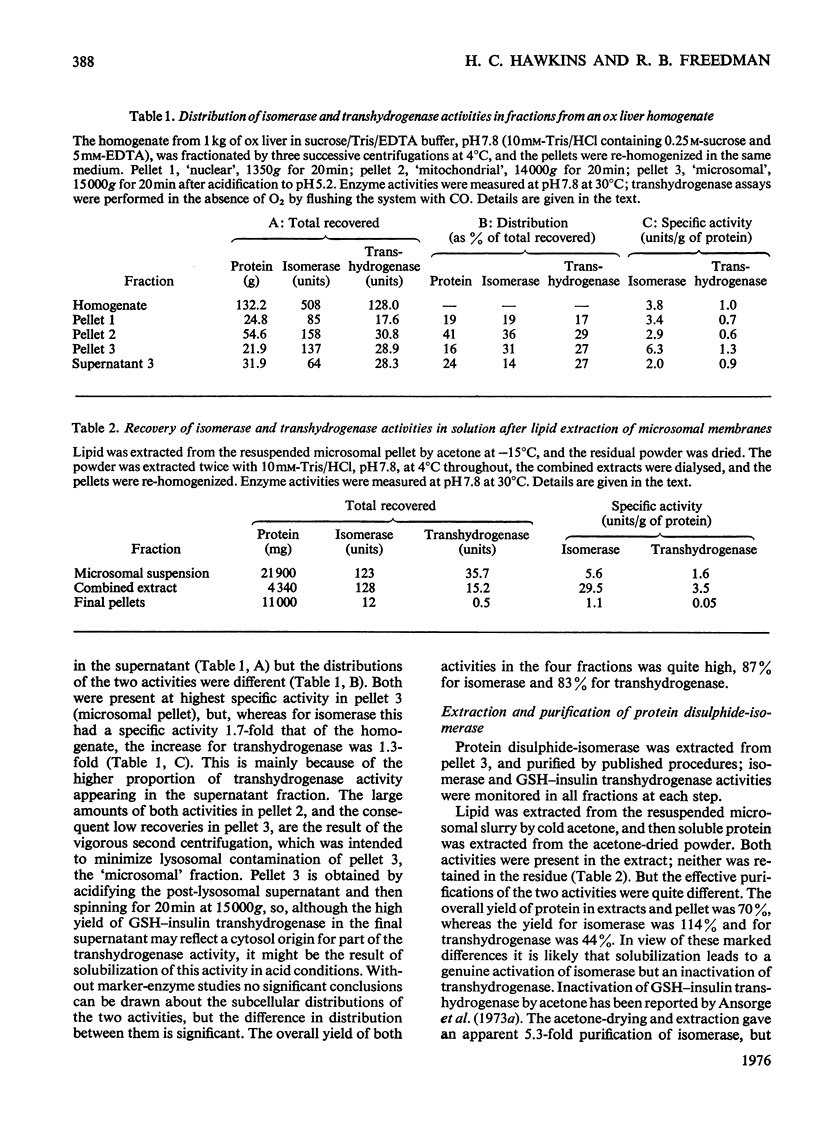
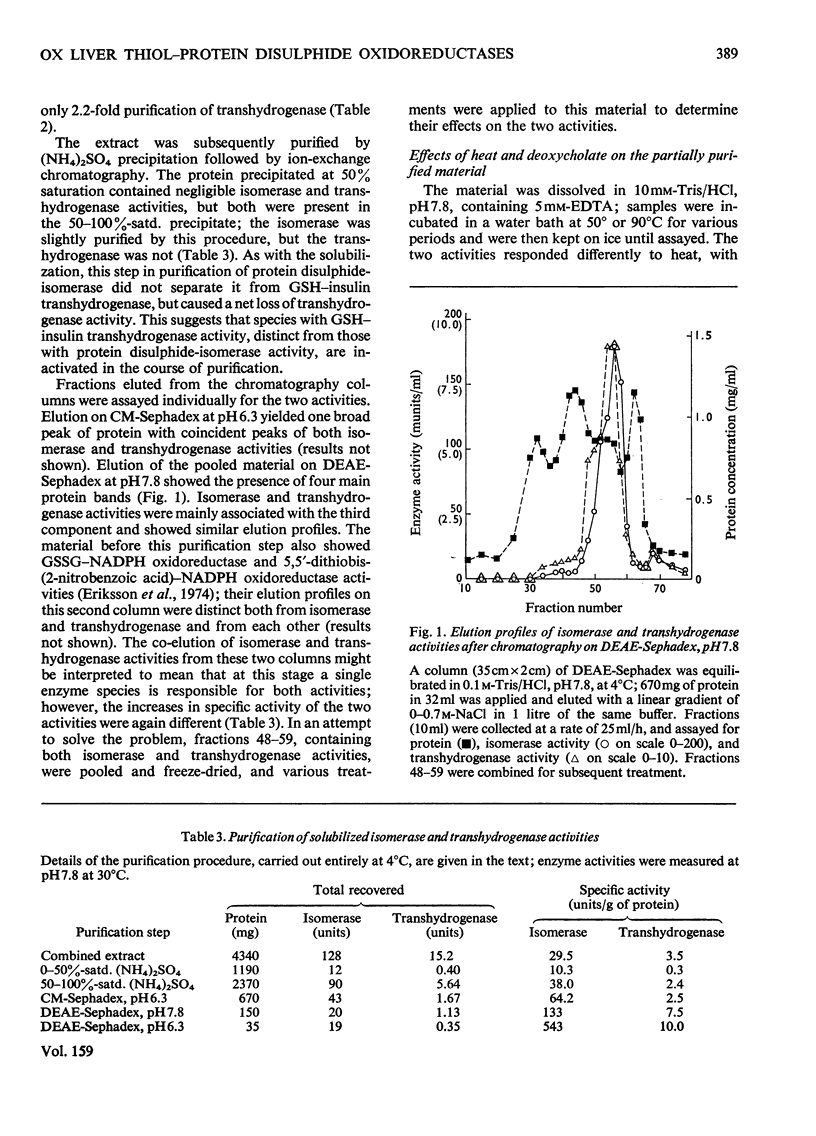
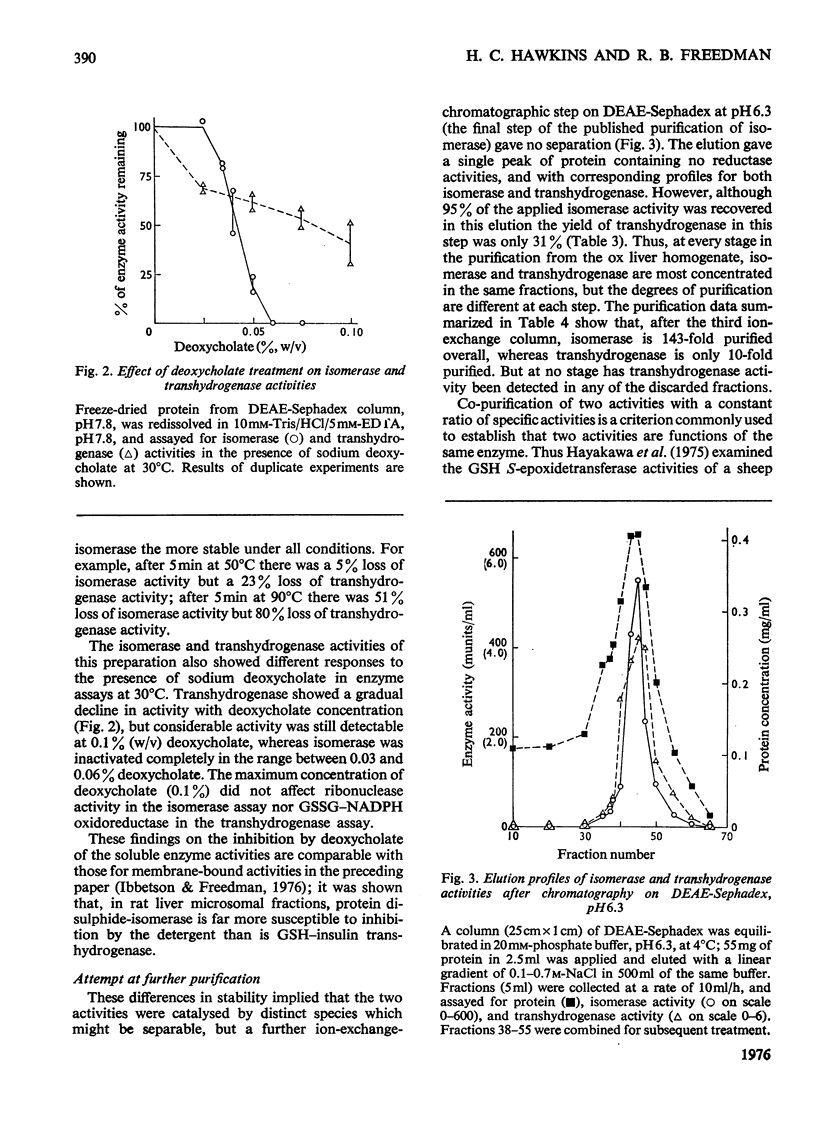
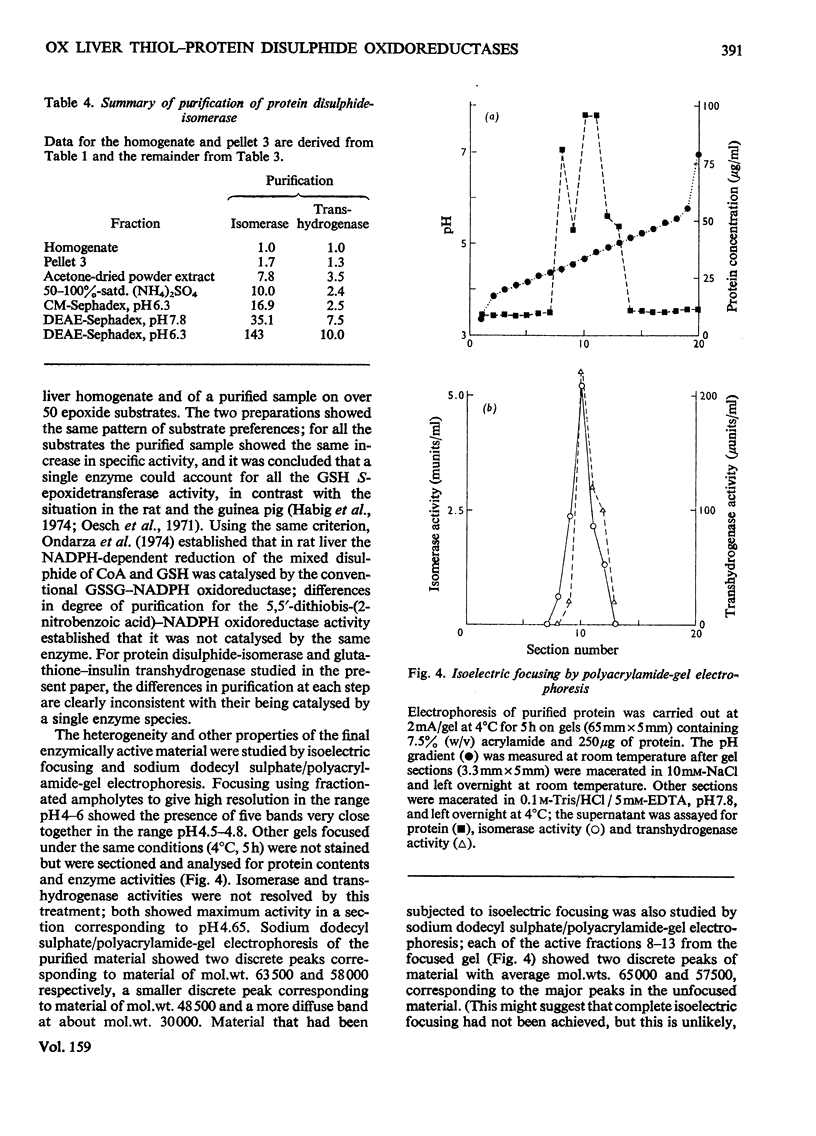
1. Protein disulphide-isomerase and glutathione-insulin transhydrogenase activities were assayed in parallel through a conventional purification of protein disulphide-isomerase from ox liver. 2. Throughout a series of purification steps (differential centrifugation, acetone extraction, (NH4)2SO4 precipitation and ion-exchange chromatography), the two activities appeared in the same fractions but were purified to different extents. 3. The final sample was 143-fold purified in protein disulphide-isomerase but only 10-fold purified in glutathione-insulin transhydrogenase; nevertheless the two activities in this preparation were not resolved by high-resolution isoelectric focusing and both showed pI4.65. 4. In a partially purified preparation containing both activities, glutathione-insulin transhydrogenase was far more sensitive to heat denaturation than was protein disulphide-isomerase; conversely protein disulphide-isomerase was more sensitive to inactivation by deoxycholate. 5. The data are inconsistent with a single enzyme being responsible for all the protein disulphide-isomerase and glutathione-insulin transhydrogenase activity of ox liver. It is suggested that several similiar thiol-protein disulphide oxidoreductases of overlapping specificities may better account for the data.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansorge S., Bohley P., Kirschke H., Langner J., Marquardt I., Wiederanders B., Hanson H. The identity of the insulin degrading thiol-protein disulfide oxidoreductase (glutathione-insulin transhydrogenase) with the sulfhydryl-disulfide interchange enzyme. FEBS Lett. 1973 Dec 1;37(2):238–240. doi: 10.1016/0014-5793(73)80468-5. [DOI] [PubMed] [Google Scholar]
- Ansorge S., Bohley P., Kirschke H., Langner J., Wiederanders B., Hanson H. Metabolism of insulin and glucagon. Glutathione-insulin transhydrogenase from microsomes of rat liver. Eur J Biochem. 1973 Jan 3;32(1):27–35. doi: 10.1111/j.1432-1033.1973.tb02574.x. [DOI] [PubMed] [Google Scholar]
- Chandler M. L., Varandani P. T. Insulin degradation. II. The widespread distribution of glutathione-insulin transhydrogenase in the tissues of the rat. Biochim Biophys Acta. 1972 Nov 24;286(1):136–145. [PubMed] [Google Scholar]
- De Lorenzo F., Goldberger R. F., Steers E., Jr, Givol D., Anfinsen B. Purification and properties of an enzyme from beef liver which catalyzes sulfhydryl-disulfide interchange in proteins. J Biol Chem. 1966 Apr 10;241(7):1562–1567. [PubMed] [Google Scholar]
- De Lorenzo F., Molea G. Relative levels of the disulfide-interchange enzyme in the microsomes of the bovine tissues. Biochim Biophys Acta. 1967;146(2):593–595. doi: 10.1016/0005-2744(67)90246-x. [DOI] [PubMed] [Google Scholar]
- Eriksson S., Askelöf P., Axelsson K., Carlberg I., Guthenberg C., Mannervik B. Resolution of glutathione-linked enzymes in rat liver and evaluation of their contribution to disulfide reduction via thiol--disulfide interchange. Acta Chem Scand B. 1974;28(8):922–930. doi: 10.3891/acta.chem.scand.28b-0922. [DOI] [PubMed] [Google Scholar]
- Fujita T., Shoeman D. W., Mannering G. J. Differences in P-450 cytochromes from livers of rats treated with phenobarbital and with 3-methylcholanthrene. J Biol Chem. 1973 Mar 25;248(6):2192–2201. [PubMed] [Google Scholar]
- HIRAOKA T., GLICK D. Studies in histochemistry. LXXI. Measurement of protein in millimicrogram amounts by quenching of dye fluorescence. Anal Biochem. 1963 Jun;5:497–504. doi: 10.1016/0003-2697(63)90069-1. [DOI] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974 Nov 25;249(22):7130–7139. [PubMed] [Google Scholar]
- Hayakawa T., Udenfriend S., Yagi H., Jerina D. M. Substrates and inhibitors of hepatic glutathione-S-epoxide transferase. Arch Biochem Biophys. 1975 Oct;170(2):438–451. doi: 10.1016/0003-9861(75)90139-3. [DOI] [PubMed] [Google Scholar]
- Ibbetson A. L., Freedman R. B. Thiol-protein disulphide oxidoreductases. Assay of microsomal membrane-bound glutathione-insulin transhydrogenase and comparison with protein disulphide-isomerase. Biochem J. 1976 Nov;159(2):377–384. doi: 10.1042/bj1590377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen H. M., Tietze F. Studies on the specificity and mechanism of action of hepatic glutathione-insulin transhydrogenase. J Biol Chem. 1966 Aug 10;241(15):3561–3570. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lu A. Y., Levin W., West S. B., Jacobson M., Ryan D., Kuntzman R., Conney A. H. Reconstituted liver microsomal enzyme system that hydroxylates drugs, other foreign compounds, and endogenous substrates. VI. Different substrate specificities of the cytochrome P450 fractions from control and phenobarbital-treated rats. J Biol Chem. 1973 Jan 25;248(2):456–460. [PubMed] [Google Scholar]
- Oesch F., Jerina D. M., Daly J. W. Substrate specificity of hepatic epoxide hydrase in microsomes and in a purified preparation: evidence for homologous enzymes. Arch Biochem Biophys. 1971 May;144(1):253–261. doi: 10.1016/0003-9861(71)90476-0. [DOI] [PubMed] [Google Scholar]
- Ondarza R. N., Escamilla E., Gutiérrez J., De la Chica G. CoAS-Sglutathione and GSSG reductases from rat liver. Two disulfide oxidoreductase activities in one protein entity. Biochim Biophys Acta. 1974 Mar 21;341(1):162–171. doi: 10.1016/0005-2744(74)90076-x. [DOI] [PubMed] [Google Scholar]
- Peters T. J., Müller M., De Duve C. Lysosomes of the arterial wall. I. Isolation and subcellular fractionation of cells from normal rabbit aorta. J Exp Med. 1972 Nov 1;136(5):1117–1139. doi: 10.1084/jem.136.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti P., Drysdale J. W. Isoelectric focusing in polyacrylamide gels. Biochim Biophys Acta. 1971 Apr 27;236(1):17–28. doi: 10.1016/0005-2795(71)90144-9. [DOI] [PubMed] [Google Scholar]
- Steiner R. F., De Lorenzo F., Anfinsen C. B. Enzymically catalyzed disulfide interchange in randomly cross-linked soybean trypsin inhibitor. J Biol Chem. 1965 Dec;240(12):4648–4651. [PubMed] [Google Scholar]
- TOMIZAWA H. H., VARANDANI P. T. GLUTATHIONE-INSULIN TRANSHYDROGENASE OF HUMAN LIVER. J Biol Chem. 1965 Jul;240:3191–3194. [PubMed] [Google Scholar]
- Varandani P. T. Insulin degradation. V. Unmasking of glutathione-insulin transhydrogenase in rat liver microsomal membrane. Biochim Biophys Acta. 1973 May 28;304(3):642–659. doi: 10.1016/0304-4165(73)90210-9. [DOI] [PubMed] [Google Scholar]
- Varandani P. T. Insulin degradation. XII. The amino acid composition, amino-terminal, and carbohydrate content of beef pancreatic glutathione-insulin transhydrogenase. Biochim Biophys Acta. 1974 Dec 18;371(2):577–581. [PubMed] [Google Scholar]
- Varandani P. T., Nafz M. A., Chandler M. L. Interaction of insulin analogs, glucagon, growth hormone, vasopressin, oxytocin, and scrambled forms of ribonuclease and lysozyme with glytathione-insulin transhydrogenase (thiol: protein-disulfide oxidoreductase): dependence upon conformation. Biochemistry. 1975 May 20;14(10):2115–2120. doi: 10.1021/bi00681a011. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]


