Abstract
Type 2 diabetes (T2D) is a common metabolic disorder characterized by dysregulation of glucose metabolism. Genome-wide association studies have defined hundreds of signals associated with T2D and related metabolic traits, predominantly in noncoding regions. While pancreatic islets have been a focal point given their central role in insulin production and glucose homeostasis, other metabolic tissues, including liver, adipose, and skeletal muscle, also contribute to T2D pathogenesis and risk. Here, we examined context-specific genetic regulation under basal and stimulated states. Using LHCN-M2 human skeletal muscle cells, we generated transcriptomic profiles and characterized regulatory activity of 327 metabolic trait-associated variants via a massively parallel reporter assay (MPRA). To identify condition-specific effects, we compared four different conditions: (1) undifferentiated, or (2) differentiated with basal media, (3) media supplemented with the AMP analog AICAR (to simulate exercise) or (4) media containing sodium palmitate (to induce insulin resistance). RNA-seq revealed these treatments extensively perturbed transcriptional regulation, with 498–3,686 genes showing significant differential expression between pairs of conditions. Among differentially expressed genes, we observed enrichment of relevant biological pathways including muscle differentiation (undifferentiated vs. differentiated), oxidoreductase activity (differentiated vs. AICAR), and glycogen binding (differentiated vs. palmitate). The results of our MPRA found broadly different levels of activity between all conditions. Our MPRA screen revealed a shared set of 7 variants with significant allelic activity across all conditions, along with a proportional number of variants showing condition-specific allelic bias and the total number of active oligos per condition. We found that a lead variant for serum triglyceride levels, rs490972, overlaps SP transcription factor motifs and has differential regulatory activity between conditions. Comparison of MPRA activity with paired gene expression data allowed us to predict that regulatory activity at this locus is mediated by SP1 transcription factor binding. While several of the MPRA variants have been previously characterized in other metabolic tissues, none have been studied in these stimulated states. Together, this work uncovers context-dependent transcriptomic and regulatory dynamics of T2D- and metabolic trait-associated variants in skeletal muscle cells, offering new insights into their functional roles in metabolic processes.
Keywords: transcription factors, enhancers, GWAS, genetic variation, metabolic traits, skeletal muscle
Introduction
Type 2 diabetes (T2D) is a common metabolic disorder characterized by dysregulation of glucose metabolism and is a major public health issue, affecting over 483 million individuals worldwide (90% of 537 million diabetes mellitus cases as of 2021) (IDF, 2021). T2D is a complex disease and results from a combination of both genetic and environmental factors. Previous genome-wide association studies (GWAS) have successfully mapped over 1,000 independent signals to roughly 600 loci in the genome (Suzuki et al. 2024; Mahajan et al. 2022; Vujkovic et al. 2020). Because the majority of these signals reside far from genes in noncoding regions, progress to translate them to specific disease mechanisms has proceeded slowly. This is due to several challenges including difficulty predicting the functional consequences of noncoding variants, tight linkage between neighboring genetic variants, and lack of certainty about which cell types and environmental contexts are casual to the disease (Cano-Gamez and Trynka 2020).
T2D research has focused on pancreatic islets due to their direct connection to glucose regulation by insulin secretion; however, metabolic disease pathogenesis and risk is distributed across other important peripheral metabolic tissues, including the liver, adipose, and, the tissue of focus for this study, skeletal muscle. Glucose levels are regulated in skeletal muscle through both insulin-independent and insulin-dependent mechanisms, modulating delivery, transport, and metabolism (Hulett, Scalzo, and Reusch 2022). Skeletal muscle is responsible for 80% of postprandial glucose uptake, and therefore is a tissue of high interest when studying metabolic disorders (DeFronzo and Tripathy 2009). Previous studies have used functional genomic data collected from primary skeletal muscle samples to link T2D GWAS signals and target genes (Scott et al. 2016; Orchard et al. 2021; Varshney et al. 2023), but few studies have further probed context-specific effects of these variants. Exploring the behavior of variants across stimulatory states is a crucial next step to reveal how dynamic environmental changes and the associated shifts in cell states affect the functional impact of disease-associated variants.
Here, we used the human skeletal muscle cell line LHCN-M2 (Zhu et al. 2007) across basal and stimulatory states to investigate cellular environmental effects on regulatory features of T2D and metabolic trait-associated variants (Figure 1). To examine developmental state changes, we differentiated LHCN-M2 proliferating myoblasts into terminally differentiated myotubes. To emulate different environmental stressors, we subsequently perturbed myotubes with either the AMP analog 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) to induce oxidative stress (Dolinar et al. 2018) and mimic exercise, or palmitate, a fatty acid known to cause insulin resistance (Mäkinen et al. 2017). We examined these stimulations’ effects on gene expression by performing differential expression analysis using bulk RNA-seq data from basal and perturbed cells. Then, to assess regulatory activity across a library of 327 metabolic trait-associated variants, we constructed and implemented a massively parallel reporter assay (MPRA) under the same conditions. Our study builds upon the current understanding of how metabolic disease-associated regulatory elements may be influenced by the cellular environment.
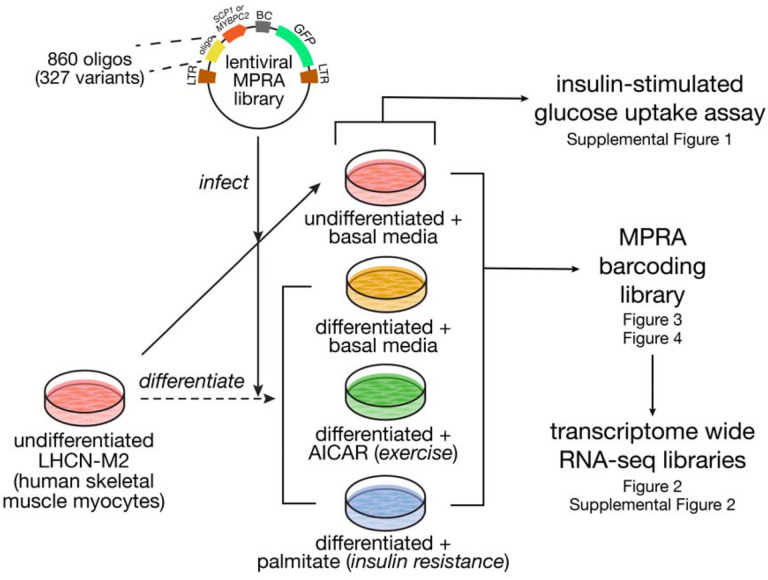
Figure 1. Study overview.
We synthesized a library of 860 oligos spanning 327 variants with either the super core (SCP1) or muscle-specific (MYBPC2) promoter. This library was used to infect undifferentiated LHCN-M2 human skeletal muscle myocytes, and subsequently, a portion of the cells were differentiated into myotubes. These were stimulated with AICAR to mimic exercise or palmitate to induce insulin resistance, or left untreated. Across these four conditions, we measured insulin-stimulated glucose uptake, gene expression (by bulk RNA-seq) and variant regulatory activity (by MPRA).
Results
Differentially expressed genes highlight developmental biology and cellular context
To model stimulation states relevant to metabolic disease, we cultured LHCN-M2 skeletal muscle cells in four conditions: undifferentiated with basal media, differentiated with basal media, differentiated with AICAR-supplemented media, and differentiated with palmitate-supplemented media. With cells across these conditions, we first examined insulin-stimulated glucose uptake, a physiological endpoint reflective of relevant muscle biology. As expected, we observed significant differences in baseline glucose uptake between undifferentiated cells in basal media compared to any of the other three conditions that used differentiated cells (Supplemental Figure 1; p = 0.028 for differentiated, p = 3.77 × 10−5 for AICAR-stimulated, and p = 3.88 × 10−5 for palmitate-stimulated, all compared to undifferentiated cells; t-test). Over the course of myogenesis, myocytes express higher levels of insulin receptors and thus are more responsive to insulin (Yun and Wold 1996).
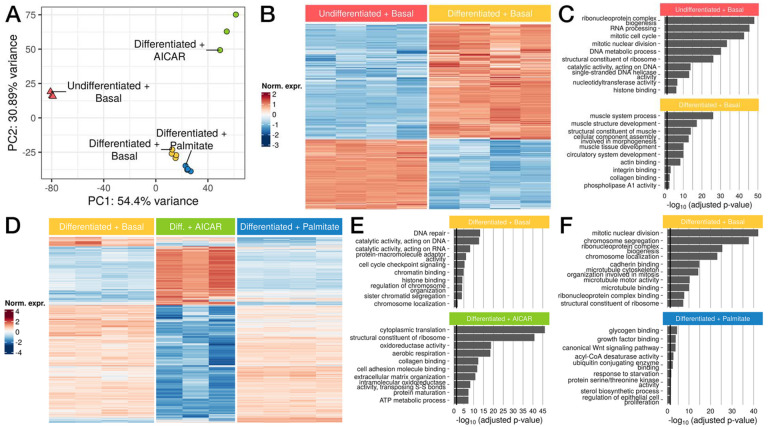
We performed bulk RNA-seq under the same conditions in quadruplicate, and clustered the resulting samples by principal component analysis (Figure 2A, Supplemental Table 1). Differentiation state was strongly separated along PC1 (54.40% of variance). The conditions using differentiated myotubes were further separated along PC2 (30.89% of variance) with AICAR-stimulated samples showing the greatest separation. We first compared the undifferentiated and basal differentiated conditions, and found 3,130 differentially expressed genes with DESeq2 (Figure 2B, Supplemental Figure 2A, Supplemental Table 2; log2FC > 1, FDR < 5%) (Love, Huber, and Anders 2014). Among the strongest changes was MYF5, a key myogenic fate determination factor which was highly downregulated (log2FC ~ −5; FDR-adjusted p < 10−300), consistent with its known expression pattern in vivo (Arnold and Braun 1999). Conversely, MYH3 was strongly induced during differentiation (log2FC ~ 5.22; FDR-adjusted p < 10−300). Enrichment analysis highlighted pathways involved in cell growth and division, such as DNA metabolism, cell division cycle, and ribosome biogenesis as highly expressed in undifferentiated samples (Figure 2C, Supplemental Table 3), consistent with myoblasts’ role as proliferating precursors of mature skeletal muscle cells (Vaughan and Lamia 2019), and with growth arrest due to permanent withdrawal from the cell cycle during differentiation (Myers, Andreuzza, and Franklin 2004). Conversely, pathways upregulated during differentiation included known in muscle cell development and morphogenesis gene sets, and actin filament based processes involved in myoblast elongation (Lehka and Rędowicz 2020). Taken together, these results highlight extensive transcriptional dynamics that direct the transition from proliferative myoblasts to specialized, mature myotubes.
Figure 2. Gene expression varies widely after myoblast to myotube differentiation and in response to AICAR and palmitate.
(A) Principal components analysis (PCA) of samples by gene expression. (B) Heatmap of genes differentially expressed between undifferentiated and differentiated samples (n=3,130) (C) Upregulated GO terms enriched in undifferentiated (top) or differentiated samples (bottom). (D) Heatmap of genes differentially expressed between differentiated LHCN-M2 myotubes in basal vs AICAR or palmitate-stimulated conditions (n=3,570) (E,F) Upregulated GO terms enriched in (E) differentiated basal (top) or AICAR-stimulated samples (bottom) and in (F) basal (top) or palmitate-stimulated samples (bottom). Black lines in (C), (E), and (F) correspond to p = 0.05.
To evaluate stimulus-specific effects, we performed pairwise comparisons between each perturbation (AICAR, palmitate) and the basal differentiated condition. First, we identified 3,686 differentially expressed genes between the AICAR-stimulated samples and differentiated samples (Figure 2D, Supplemental Figure 2B, Supplemental Table 4; log2FC > 1, FDR < 5%). In AICAR-stimulated cells, we observed a significant increase in MYC expression (log2FC ~ 2.58; DESeq2 FDR-adjusted p = 1.757 × 10−207). Several studies have reported significant increases in MYC expression after both resistance and endurance exercise in humans and mouse models (Jones et al. 2023; Broholm et al. 2011; Trenerry et al. 2007; Popov et al. 2019). By contrast, SIX1 was highly downregulated in AICAR-stimulated cells (log2FC ~ −2.13; DESeq2 FDR-adjusted p = 1.138 × 10−104). Previous studies have reported decreased SIX1 expression and activity in response to skeletal muscle overload (B. S. Gordon et al. 2012; Ramachandran et al. 2019; Kostek et al. 2007). Moreover, SIX1 is an important developmental transcription factor that regulates skeletal muscle differentiation and promotes myoblast fusion to generate myotubes (Niro et al. 2010; Bessarab et al. 2008; Ridgeway and Skerjanc 2001; Fougerousse et al. 2002). Pathway enrichment analysis identified DNA repair and cell cycle-associated genes as significantly downregulated following AICAR treatment (Figure 2E, Supplemental Table 5), consistent with previous studies showing that AICAR inhibits proliferation and promotes cell cycle arrest (Nagata et al. 2004; Igata et al. 2005). In contrast, genes more highly expressed in AICAR-perturbed cells were related to protein synthesis and cellular oxidative metabolism, with pathways such as cytoplasmic translation, maintenance of ribosome structure, and oxidoreductase activity. AICAR-perturbed cells likely undergo adaptive processes such as translation of stress-response genes and preservation of ribosome structures to combat the effects of oxidative damage (Shcherbik and Pestov 2019). Oxidoreductase activity is responsible for maintaining redox homeostasis, and introduction of oxidative stress by AICAR would cause an imbalance of reactive oxygen species, therefore requiring higher levels of redox activity (Sies, Berndt, and Jones 2017).
Response to palmitate identified 498 differentially expressed genes versus basal media (Figure 2D, Supplemental Figure 2C, Supplemental Table 6; log2FC > 1, FDR < 5%). Several lipid-associated genes were highly upregulated in palmitate-stimulated samples including SCD (log2FC ~ 1.66; DESeq2 FDR-adjusted p = 6.329 × 10−95) (Voss et al. 2005; Dziewulska et al. 2020), PLIN2 (log2FC ~ 1.97; DESeq2 FDR-adjusted p = 3.475 × 10−49) (Daemen, van Polanen, and Hesselink 2018; Cho and Kang 2015), FADS1 (log2FC ~ 1.18; DESeq2 FDR-adjusted p = 1.98 × 10−48) (Karjalainen et al. 2024; Selvaraj et al. 2022), and ANGPTL4 (log2FC ~ 4.55; DESeq2 FDR-adjusted p = 4.663 × 10−45) (Kuo et al. 2024; McCulloch et al. 2020), all of which are implicated in insulin resistance. Significantly downregulated pathways after palmitate treatment were related to mitotic processes such as nuclear division and chromosome segregation (Figure 2F, Supplemental Table 7). While this appears to contradict the growth arrest expected in differentiated myotubes, palmitate is known to further inhibit proliferation and induce differentiation (Grabiec et al. 2015). Genes highly expressed in palmitate-stimulated cells were associated with glycogen binding, growth factor binding and the Wnt signaling pathway. Increases in circulating fatty acids such as palmitate promote insulin resistance and modulate glycogen binding and synthesis (Samuel and Shulman 2016; Roden et al. 1996; Dimopoulos et al. 2006). Additionally, recent work implicates the Wnt signaling pathway as a mediator of insulin resistance (Suren Garg et al. 2023). Altogether, these results highlight the unique context-specific pathways that are enriched for each condition, demonstrating the importance of developmental and environmental conditions on gene expression.
Massively parallel reporter assay identifies allelic activity of oligos
We next used an MPRA to investigate the regulatory basis of the transcriptional differences between these stimulatory states. We constructed a library of 860 200-bp oligos, spanning 327 variants, including metabolic trait-associated variants that were previously characterized in luciferase assays (Orchard et al. 2021; Khetan et al. 2021; Roman et al. 2015; Pandey et al. 2024; Sinnott-Armstrong et al. 2021) and variants in tight linkage disequilibrium to measures of physical activity (Wang et al. 2022). To compare generic and tissue-specific promoter contexts, we cloned this oligo library into an MPRA vector with either the SCP1 synthetic housekeeping promoter or the promoter for the muscle-specific gene MYBPC2. Each cloned oligo fragment was tagged with a short randomized 20mer barcode such that each oligo could be associated with multiple barcodes (median = 927 barcodes/oligo) to provide internal replication. Subsequently, we delivered this library to LHCN-M2 human skeletal muscle cells by stable lentiviral transduction under all four conditions as mentioned above. After delivery, we deeply sequenced these MPRA barcodes from mRNA and genomic DNA (gDNA), with the ratio of these counts serving as a measure of each oligo’s activity in the MPRA.
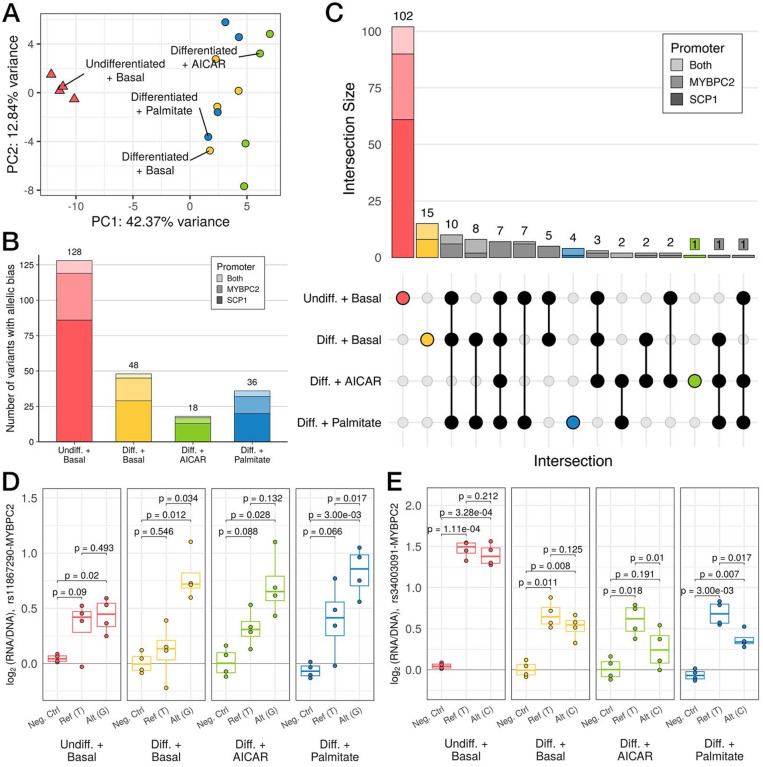
After filtering to oligos with at least 10 unique barcodes, we recovered 735 oligos (85.5% of total) spanning 295 variants (90.2% of total). Further separating these data by promoter context, we recovered 720 oligos (83.7%) spanning 294 variants (89.9%) paired with the MYBPC2 promoter and 721 oligos (83.8%) spanning 290 variants (88.7%) paired with the SCP1 promoter. Across the two promoters, we recovered 700 oligos (81.4%) in common spanning 288 variants (88.1%). Notably, the results of clustering on MPRA signal (Figure 3A) mirrored the clustering based upon bulk RNA-seq (Figure 2A), with the differentiation and treatment separating samples on the first two principal components (42.4% and 12.8% of variance, respectively). As with RNA-seq PCA, these differences were not driven by read coverage or barcode recovery (Supplemental Table 8). A similar grouping was observed by PCA restricted to oligos paired with the MYBPC2 promoter (Supplemental Figure 3A) or those paired with the SCP1 promoter (Supplemental Figure 3B), indicating that promoter context does not drive the clustering.
Figure 3. Variation in regulatory activity measured by MPRA recapitulates whole-transcriptome patterns.
(A) Principal components analysis (PCA) of MPRA activity across 860 oligos. (B) Stacked bar plot displaying the number of variants that display differences in regulatory activity across both alleles (i.e., allelic bias), as measured by MPRA (FDR < 5%). Bar plot shading corresponds to which promoter a variant was paired with when it demonstrated allelic bias (light shade: both, medium shade: MYBPC2, dark shade: SCP1). Total number of variants for each condition is listed above the corresponding bar. (C) UpSet plot shows counts of variants showing allelic bias in each subset of conditions (filled dots). (D,E) MPRA signals with log2(RNA/DNA) plotted vs alleles (Ref: reference, Alt: alternate) or a negative control, for (D) an example of allele-specific activity favoring the alternate allele primarily in differentiated conditions; and (E) a stimulation-specific example. (p-values in (C) and (D) from post hoc pairwise t-tests after passing mpralm 5% FDR threshold; N = 4 replicates per group)
We used mpralm (Myint et al. 2019) to estimate oligo activity in each individual condition (Supplemental Tables 9–12). We identified 303 active oligos in undifferentiated cells, 103 in basal differentiated cells, 38 in AICAR-stimulated cells, and 76 in palmitate-stimulated cells (FDR-adjusted p < 0.05; Supplemental Figure 3A). Across all four conditions, most active oligos were paired with the SCP1 promoter compared to MYBPC2 or both promoters, ranging from 57.2% (n = 59/103) in the differentiated condition and 71.1% (n = 27/38) in the AICAR-stimulated condition (Supplemental Table 13). Within these oligo sets, we also examined the number of corresponding variants with at least one active allele and identified 208 unique variants in undifferentiated cells, 76 in differentiated cells, 28 in AICAR-stimulated cells, and 59 in palmitate-stimulated cells (Supplemental Figure 4B). Next, we compared active oligo sets across conditions which revealed a shared set of 21 active oligos, the majority of which were paired with the MYBPC2 promoter (n = 20/21, 95.2%; Supplemental Figure 4C). The largest proportion of condition-specific active oligos was displayed by the undifferentiated group (276) with only 20, 3, and 3 uniquely active oligos in differentiated, palmitate-stimulated, and AICAR-stimulated groups, respectively.
We next tested for differential activity across alleles, focusing on variants where at least one allele was significantly active. (Supplemental Figure 4B, Supplemental Tables 14–17). We identified 128 variants with significant allelic bias in undifferentiated cells, 48 in differentiated cells, 18 in AICAR-stimulated cells, and 36 in palmitate-stimulated cells (FDR-adjusted p < 0.1; Figure 3B). As with the active oligo analysis, the majority of variants with allelic bias in each condition were paired with the SCP1 promoter, from 55.6% (n = 20/36) in palmitate-stimulated cells and 72.2% (n = 13/18) in AICAR-stimulated cells (Supplemental Table 18). We discovered a set of 7 variants with allelic bias across all four conditions, all of which displayed allelic bias when paired with the MYBPC2 promoter. A separate set of 122 variants showed condition-specific allelic bias (Figure 3C). Variants that displayed allelic bias across more than one condition did so in the same direction in all cases except one (rs4782722 paired with the SCP1 promoter; Supplemental Table 19).
Given the broad effects of differentiation upon gene expression and MPRA activity, we sought to identify examples where allelic effects were also influenced by differentiation state. One such example is rs11867290, which displays significant allelic bias in both basal differentiated and palmitate-stimulated cells, with nearly significant allelic bias in AICAR-stimulated cells, where the alternate allele (G) displays higher enhancer activity than the reference allele (T) (Figure 3D; p = 0.034, 0.132, and 0.017 in differentiated, AICAR-stimulated, and palmitate-stimulated cells, respectively; post hoc pairwise t-tests). By contrast, both alleles are active in undifferentiated cells, but without significant allelic bias. rs11867290 is located in the second intron of MARCHF10 and is a lead signal variant for height (Yengo et al. 2022), where the T allele is associated with decreased height. This variant overlaps an ENCODE-annotated enhancer for several metabolic tissues, including primary skeletal muscle and skeletal muscle-derived myoblasts and myotubes (ENCODE Project Consortium 2012; ENCODE Project Consortium et al. 2020; Luo et al. 2020).
We also identified variants that displayed allelic bias in a more restricted pattern, including rs34003091 (Figure 3E). While this variant enhances MPRA activity in both undifferentiated and differentiated cells, it displays allelic bias in neither condition. By contrast, the reference allele (T) is more active than the alternate allele (C) in both AICAR- and palmitate-stimulated cells (p = 0.01 and 0.017 in AICAR- and palmitate-stimulated, respectively; post hoc pairwise t-tests). rs34003091 is a lead signal variant for total cholesterol (Graham et al. 2021) and in high linkage with lead variants for LDL cholesterol (R2 = 1 with rs35081008 and R2 > 0.9 with rs34503352 across 1000 Genomes populations; Klimentidis et al. 2020; Sakaue et al. 2021; Hoffmann et al. 2018). The T allele is associated with decreased total cholesterol and on a shared haplotype with LDL cholesterol-raising alleles. This variant is located in a chromHMM-predicted weak promoter for ZNF329 in HepG2 hepatocytes and also colocalizes with e- and caQTLs for the same gene in liver tissue (Currin et al. 2021; Etheridge et al. 2020). The authors of this study also used a luciferase assay in HepG2 hepatocytes to evaluate enhancer activity at this variant and observed allelic effects with concordant direction of effect to what we observed here (Pandey et al. 2024). Similar to the liver, skeletal muscle is an important regulator of lipid metabolism. One previous study observed decreased ZNF329 expression in skeletal muscle biopsies taken from obese individuals before and after bariatric surgery, indicating that skeletal muscle may play a role in trait variation at this locus (Gancheva et al. 2019).
Regulatory features and condition-specific activity at metabolic trait-associated variants
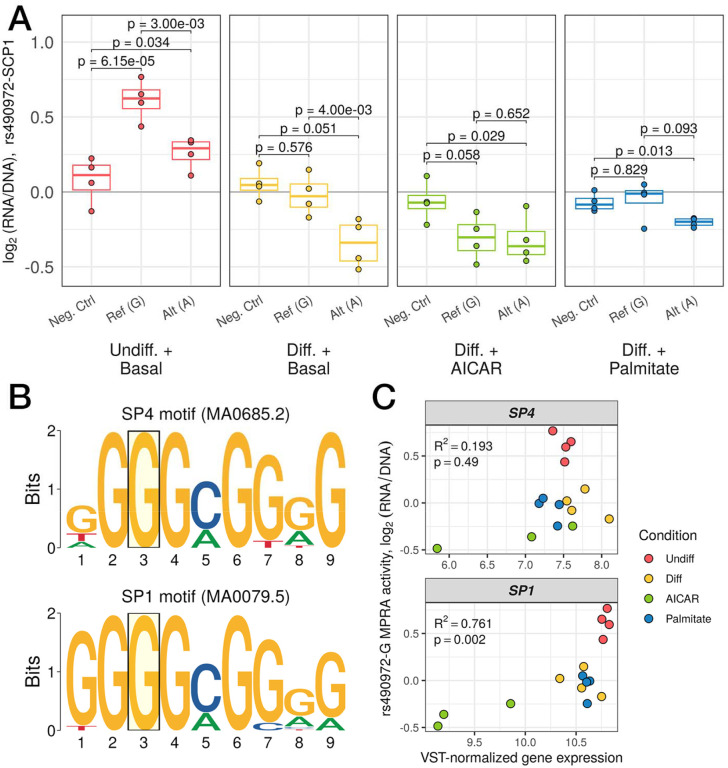
We next set out to identify upstream transcriptional regulators which may underlie the allelic effects observed in our MPRA. We focused on rs490972, a lead variant for serum triglyceride levels (DeForest et al. 2024; Benedet et al. 2024) and in tight linkage with a separate lead variant (rs1625595; R2 > 0.97 across 1000 Genomes populations) for both metabolic syndrome (Park et al. 2024) and self-reported moderate-to-vigorous physical activity (Wang et al. 2022). In most of the conditions assayed by MPRA, this variant displayed strong allelic bias with the alternate allele (A) showing lower activity in comparison to the reference allele (G) (Figure 4A; p = 0.003, 0.004, and 0.093 in undifferentiated, differentiated, and palmitate-stimulated cells, respectively; post hoc pairwise t-tests). rs490972 overlaps several motifs for SP family transcription factors, including SP1 and SP4 and the reference allele (G) of this variant matches the consensus base at a high-information content position (Figure 4B). The specificity protein (SP) family of transcription factors are zinc-finger proteins that play key roles in transcriptional regulation of foundational cellular functions, such as growth-regulation and developmental processes (Kaczynski, Cook, and Urrutia 2003), and recognize and bind to GC-rich promoter sites to regulate cellular function. To nominate which of these two TFs may underlie the allelic difference in activity at this site, we compared their mRNA expression with the MPRA activity of the motif-preserving reference allele. We observed significant positive correlation (Spearman R2 = 0.761; p = 0.002) only for SP1 but not SP4 (Figure 4C). We predict that the alternate allele A disrupts the SP motif leading to decreased binding and reporter activity which is further exaggerated when SP1 expression is decreased. This aligns with the observed reporter activity where this variant is only an enhancer in the undifferentiated group and is neither active nor displays allelic bias in the AICAR-stimulated group. Together, these data suggest that regulatory activity at the metabolic trait-associated variant rs490972 may be mediated by SP binding, restricted to conditions where the appropriate TF is expressed.
Figure 4.
(A) Regulatory activity of a negative control sequence compared to the reference, G, and alternate, A, alleles of rs490972. Each point represents a single replicate and groups are color-coded (p-values from post hoc pairwise t-tests after passing mpralm 5% FDR threshold). (B) Logo plots of the motifs for SP4 (top) and SP1 (bottom). The position corresponding to where rs490972 overlaps both motifs is highlighted by a transparent yellow box. The reference allele of this variant, G, is the consensus nucleotide for both motifs. (C) Scatterplots of variance stabilizing transformation-normalized RNA expression counts for SP4 (top) and SP1 (bottom) compared to log2(RNA/DNA) for the reference allele of rs490972. Spearman correlation coefficient and p-value are displayed in the top left corner for each comparison. (N = 3 replicates for AICAR-stimulated group, 4 replicates for other three groups for RNA-seq data; N = 4 replicates per group for MPRA data)
Discussion
In this study, we examined how transcriptional regulation and gene expression respond to differentiation and stimulation, and how those effects interact with allelic variation in a model of human skeletal muscle. We focused on skeletal muscle due to its involvement in metabolic disease processes, and investigated both developmental and environmental state differences using four different conditions. We differentiated LHCN-M2 proliferating myoblasts into terminally differentiated myotubes to assess developmental state differences, and we perturbed differentiated cells with either AICAR to mimic exercise, or palmitate to induce insulin resistance, models for different physiologically relevant cellular states.
A wide swath of genes showed differential expression between the conditions tested including the key muscle fate determination gene MYF5, which was induced during differentiation. AICAR treatment was marked by cellular responses to oxidative stress, while shifts after palmitate treatment included pathways involved in lipid metabolism. Enrichment for gene sets and pathways related to each of the chosen perturbations confirms that these treatments are appropriate models of the dynamic changes in state that muscle cells undergo during differentiation and disease processes.
Concurrently, we used an allelic MPRA to test metabolic-disease associated variants across the same four stimulatory states. Remarkably, clustering cells based on the MPRA measurements across 735 oligos recapitulated a qualitatively similar arrangement to PCA performed using whole-transcriptome RNA-seq, demonstrating that the selected conditions’ regulatory effects can be read out by MPRA. We noted that undifferentiated cells displayed the highest level of regulatory activity followed by differentiated cells in basal media, palmitate- and AICAR-stimulated cells. The basis for these differences is not clear, but they suggest that the undifferentiated, proliferative state may be globally more transcriptionally permissive, similar to what is observed during retinoic acid-induced differentiation of embryonic stem cells (Bulut-Karslioglu et al. 2018).
We also highlighted several variants in which our MPRA results demonstrated condition-specific allelic bias. One variant, rs11867290, displayed condition-specific effects reflecting differentiation state changes. This variant was recently uncovered as a lead signal variant for height (Yengo et al. 2022). rs11867290 demonstrated allelic bias in all conditions except for the basal undifferentiated cells. Another variant associated with total and LDL cholesterol (Graham et al. 2021; Klimentidis et al. 2020; Sakaue et al. 2021), rs34003091, only displayed allelic bias in AICAR- and palmitate-stimulated cells, illustrating a potential condition-specific effect. These instances of differentiation-state and environmental-induced differences on regulatory activity indicate that the function of regulatory variants are influenced by dynamic environmental changes and prompts further investigation of context-specific activity.
Lastly, to explore potential regulatory mechanisms functioning at disease-associated variants from our MPRA analysis, we investigated a specific variant, rs490972, which overlapped motifs for the SP family transcription factors SP1 and SP4. We hypothesize that regulatory activity at this loci may be mediated by SP1 transcription factor binding based on the differential regulatory activity between conditions. Taken together, these results reinforce the importance of examining cellular environments when assessing the functional impact of regulatory variation. While other studies have explored the effect of stimulatory states on regulatory variation as assessed in an MPRA, such as in pancreatic β cells through investigating steady state and endoplasmic reticulum stress conditions (Khetan et al. 2021), or at different time-points of neural differentiation (Kreimer et al. 2022), this study is the first that we are aware of to employ this framework in a model of skeletal muscle. As the primary site for insulin stimulated glucose uptake, skeletal muscle is an essential tissue in which to understand how regulatory variants affect metabolic traits. We explored regulatory activity across a small panel of variants, many of which were selected based on prior evidence of regulatory activity in a luciferase assay. Our results serve as a proof of principle for scaling up MPRAs to examine the thousands of variants across hundreds of loci linked to metabolic diseases, as well as saturation-scale efforts to mutationally dissect individual loci. to Moreover, while the use of an MPRA library provided robust initial findings, investigating these variants in their native genomic contexts (e.g., via CRISPR editing) is a crucial next step to fully characterize and understand their function. Finally, the regulatory inferences presented here lay the foundation for future work that integrates other functional genomic datasets to refine the regulatory mechanisms driving activity at disease-associated loci. For instance, ChIP-seq for specific transcription factors of interest (e.g., SP1, SP4) in LHCN-M2 cells across different perturbation states could provide further evidence for involvement at variants of interest.
Altogether, this study builds upon the current understanding of how metabolic disease-associated variants are regulated in skeletal muscle across developmental state changes and in response to relevant perturbations. Our findings provide evidence for widespread context-specific regulatory effects at metabolic disease-associated loci. Future studies should take this into consideration and assay an even broader set of tissues and contexts to eventually develop a comprehensive understanding of disease mechanisms.
Methods
Cell culture
We obtained LHCN-M2 human skeletal muscle myoblasts from Evercyte. We maintained LHCN-M2 cells on 0.1% gelatin-coated flasks in 4:1 DMEM (4.5 g/L glucose, glutamine, bicarbonate):Medium 199 (with bicarbonate) containing 15% FBS, 20 mM HEPES, 30 ng/mL zinc sulfate, 1.4 ug/mL vitamin B12, 55 ng/mL dexamethasone, 2.5 ng/mL recombinant human hepatocyte growth factor, 10 ng/mL basic fibroblast growth factor, and 600 U/mL penicillin/streptomycin. To differentiate the LHCN-M2 myoblasts to myotubes, we maintained cells in LHCN-M2 differentiation media containing 50:1 DMEM (1 g/L glucose):heat-inactivated horse serum for 7 days with daily media changes.
Insulin stimulated glucose uptake assay
Approximately two hours prior to applying perturbations, we exchanged LHCN-M2 media for unsupplemented DMEM (1 g/L glucose). Then, we incubated cells in basal culture media, basal differentiation media, or differentiation media supplemented with 2mM AICAR or 0.5mM palmitate for 24 hours followed by glucose starvation for 3.5 hours in modified Krebs Ringer buffer with BSA (pH 7.2) containing 1X SAB stock (10X SAB: 1.14M NaCl, 47mM KCl, 12mM KH2PO4, 11.6 mM MgSO4), 1M HEPES, 25 mM CaCl2, 0.2% BSA (w/v), and 25.5 mM NaHCO3. Afterward, we added fresh Krebs Ringer buffer with or without 100 nM insulin for 30 minutes. We measured 2-deoxyglucose uptake with the Glucose Uptake-Glo™ Assay (Promega, Fitchburg, WI, USA) following manufacturer’s instructions. We used a GloMax Multi+ Detection System (Promega, Fitchburg, WI, USA) to measure luminescence intensity (relative light units, RLU). We normalized raw data by subtracting an average of background RLU from eight empty wells.
MPRA library design, construction, and delivery
We designed, constructed and delivered the MPRA library as described in Tovar et al. 2023 based on methods in Tewhey et al. 2016 and Gordon et al. 2020. We gathered signal variants relevant to T2D and metabolic traits and designed 198-bp oligo sequences. We added 16-bp flanking adapter sequences for PCR amplification and cloning, and obtained a pool of 1,255 full-length 230-bp oligos from IDT. We added barcodes and a promoter cloning scaffold via PCR, and cloned amplified oligos into a modified pMPRA1 vector (a gift from Tarjei Mikkelsen; Addgene #49349) using Golden Gate assembly with PaqCI. We digested this assembly with SfiI to remove empty backbones then transformed into electrocompetent 10-beta cells. We inserted clonal promoter fragments using Golden Gate assembly and digested with AsiSI to remove promoterless constructs. We transformed this final assembly into electrocompetent 10-beta cells, expanded in 150 mL cultures, and isolated plasmid using the ZymoPURE Plasmid Maxiprep Kit.
MPRA barcode pairing
To create barcode-oligo pairs, we sequenced the promoterless constructs. We generated initial amplicons with primers specific to the regions immediately 5’ of the oligo and 3’ of the barcode. We amplified these pairing amplicon using P7 and P5 stubs, then added dual sequencing indexes. We sequenced the resulting libraries on an Illumina NovaSeq 6000 and received paired-end 150-bp reads.
MPRA/RNA-seq sample generation
To deliver the MPRA library to the LHCN-M2 human skeletal myoblasts, we ported the assembled MPRA block (oligo, barcode, promoter, GFP) to a lentiviral transfer vector (a gift from Nadav Ahituv; Addgene #137725) via restriction cloning (M. G. Gordon et al. 2020; Inoue et al. 2017). This lentiviral transfer vector was used by the University of Michigan viral vector core to produce infectious lentiviral particles. The undifferentiated cells were maintained and passaged while other samples per condition were differentiated for 5 days. On day 6, we infected 15 × 106 LHCN-M2 human skeletal myoblasts per replicate with our MPRA library at an MOI of ~10. On day 7, we exchanged media for fresh basal media (undifferentiated) or differentiation media and incubated cells for 24 hours. Subsequently, cells were incubated in basal media, basal differentiation media, or differentiation media supplemented with 2mM AICAR or 0.5mM palmitate for 24 hours. We isolated RNA and gDNA from each replicate using the Qiagen AllPrep DNA/RNA mini kit.
RNA-seq library generation
We used the NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA) to enrich poly(A)+ RNA transcripts from total RNA for RNA library preparation and sequencing. Afterward, we used the NEBNext® Ultra™ II Directional RNA Library Prep kit (New England BioLabs, Ipswich, MA) to prepare sequencing libraries following manufacturer’s instructions. At this stage, we excluded one sample (ATS0272, AICAR replicate 2) due to low library concentration. We sequenced libraries on an Illumina NovaSeqX and received paired-end 150-bp reads.
MPRA barcode library generation
After gDNA and RNA collection, we treated RNA with the TURBO DNA-free™ kit (Thermo Fisher Scientific, MA, USA) following the manufacturer’s instructions. We synthesized cDNA from the resulting cleaned RNA using the SuperScript™ III First-Strand Synthesis System (Thermo Fisher Scientific, MA, USA) with a GFP-specific primer containing a 15-bp UMI, and subsequently amplified cDNA via PCR. We attached sequencing adapters and a 13-bp UMI by PCR, then amplified libraries using P7 and P5 stubs. We added dual sequencing indexes to both cDNA and gDNA libraries in a final 6-cycle PCR reaction. We sequenced the resulting cDNA/gDNA libraries on an Illumina NovaSeq 6000 and received paired-end 150-bp reads.
RNA-seq data analysis
RNA-seq raw data was processed with a custom pipeline. We mapped reads to hg38 using STAR (v2.7.9a) (Dobin et al. 2013). We used SAMtools to filter high-quality read pairs (-q 255 -F 4 -F 256 -F 2048; v. 1.9 using htslib v. 1.9) (Li et al. 2009). We used FastQC to assess the quality of raw sequence data (v. 0.11.5) and MultiQC to summarize quality results (v. 1.11) (Andrews 2010; Ewels et al. 2016). To perform quality control on aligned reads, we used QoRTs (Quality of RNA-seq Tool-Set) (v. 1.3.6) (Hartley and Mullikin 2015). Given the dominant effects of differentiation state on gene expression (see Figure 2A), we chose to split the data into two subsets for differential expression analysis: (1) basal differentiated versus basal undifferentiated samples and (2) basal differentiated versus differentiated with AICAR or differentiated with palmitate samples. We conducted differential expression analysis using DESeq2 v.1.34.0 (Love, Huber, and Anders 2014) using a log2 fold change cutoff of 1 and a 5% false discovery rate. To perform pathway enrichment analysis, we split sets of differentially expressed genes based on log2FC to assign to each condition. For example, for the basal undifferentiated versus basal differentiated contrast, we assigned genes with a positive log2FC to the basal differentiated state and vice versa for the basal undifferentiated state. We then used g:Profiler v.0.2.3 (Raudvere et al. 2019) to perform pathway enrichment, subset results to the “Biological Process” and “Molecular Function” ontologies, and used rrvgo (Sayols 2023) with a threshold of 0.7 to cluster similar terms. To improve interpretability, we removed terms with fewer than 10 and more than 1000 member genes.
MPRA data analysis
To create the barcode-oligo pairing dictionary and process MPRA sequencing data, we used custom pipelines as described previously (Tovar et al. 2023). We used the R package mpra to estimate oligo activity and allelic bias (Myint et al. 2019). For our activity analysis, we used a 5% FDR threshold. For our allelic bias analysis, we required that at least one allele for a variant was active and to identify variants with differential allelic activity, we used a 10% FDR threshold.
Supplementary Material
Acknowledgements
The authors would like to acknowledge members of the Kitzman and Parker labs (University of Michigan) for their critical feedback. The authors would also like to thank the University of Michigan Viral Vector Core for producing lentivirus.
Funding
The authors received support from the National Institute of Diabetes and Digestive and Kidney Diseases, grants 1UM1 DK126185-01 (S.C.J.P.), R01 DK117960 (S.C.J.P.), Opportunity Pool Funding (A.T., S.C.J.P., J.O.K.), and T32 DK101357 (A.T.); National Institute of General Medical Sciences grant R35 GM153286 to J.O.K.; National Human Genome Research Institute K99/R00 Pathway to Independent Award HG013676 to A.T.; and the Burroughs Wellcome Fund Postdoctoral Diversity Enrichment Fellowship to A.T.
Funding Statement
The authors received support from the National Institute of Diabetes and Digestive and Kidney Diseases, grants 1UM1 DK126185-01 (S.C.J.P.), R01 DK117960 (S.C.J.P.), Opportunity Pool Funding (A.T., S.C.J.P., J.O.K.), and T32 DK101357 (A.T.); National Institute of General Medical Sciences grant R35 GM153286 to J.O.K.; National Human Genome Research Institute K99/R00 Pathway to Independent Award HG013676 to A.T.; and the Burroughs Wellcome Fund Postdoctoral Diversity Enrichment Fellowship to A.T.
Footnotes
Competing Interest Statement
The authors declare they have no conflicts of interest.
Data and code availability
Primer sequences used for MPRA library cloning and sequencing library generation are available in Supplemental Table 20. All processed RNA-seq counts and MPRA data are accessible through the Gene Expression Omnibus (GSE282191). Raw sequencing data are deposited on the NCBI Sequence Read Archive and accession numbers are listed in the GEO records. Complete results files from both sets of analyses are available as Supplemental Tables referenced in this article.
All code used to preprocess and analyze data presented here are available on Github. We used custom preprocessing pipelines for RNA-seq data (https://github.com/porchard/RNAseq-NextFlow) and MPRA barcode counting data (https://github.com/adelaidetovar/MPRA-Nextflow). Scripts used to analyze and visualize processed data are available as a separate repository at https://github.com/adelaidetovar/lhcn-perturb-mpra.
References:
- International Diabetes Federation. IDF Diabetes Atlas 10th edn https://diabetesatlas.org/ (IDF, 2021). [Google Scholar]
- Andrews Simon. 2010. “Babraham Bioinformatics - FastQC A Quality Control Tool for High Throughput Sequence Data.” 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- Arnold Hans-Henning, and Braun Thomas. 1999. “4 Genetics of Muscle Determination and Development.” In Current Topics in Developmental Biology, edited by Ordahl Charles P., 48:129–64. Somitogenesis. Academic Press. 10.1016/S0070-2153(08)60756-5. [DOI] [PubMed] [Google Scholar]
- Benedet Patricia O., Safikhan Nooshin S., Pereira Maria J., Lum Bryan M., José Diego Botezelli Cheng-Hsiang Kuo, Wu Hua-Lin, et al. 2024. “CD248 Promotes Insulin Resistance by Binding to the Insulin Receptor and Dampening Its Insulin-Induced Autophosphorylation.” eBioMedicine 99 (January). 10.1016/j.ebiom.2023.104906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarab Dmitri A., Chong Shang-Wei, Srinivas Bhylahalli Purushottam, and Korzh Vladimir. 2008. “Six1a Is Required for the Onset of Fast Muscle Differentiation in Zebrafish.” Developmental Biology 323 (2): 216–28. 10.1016/j.ydbio.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Broholm Christa, Laye Matthew J., Brandt Claus, Vadalasetty Radhika, Pilegaard Henriette, Pedersen Bente Klarlund, and Scheele Camilla. 2011. “LIF Is a Contraction-Induced Myokine Stimulating Human Myocyte Proliferation.” Journal of Applied Physiology (Bethesda, Md.: 1985) 111 (1): 251–59. 10.1152/japplphysiol.01399.2010. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu Aydan, Macrae Trisha A., Oses-Prieto Juan A., Covarrubias Sergio, Percharde Michelle, Ku Gregory, Diaz Aaron, McManus Michael T., Burlingame Alma L., and Ramalho-Santos Miguel. 2018. “The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output.” Cell Stem Cell 22 (3): 369–383.e8. 10.1016/j.stem.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez Eddie, and Trynka Gosia. 2020. “From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases.” Frontiers in Genetics 11 (May). 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Kyung-Ah, and Kang Peter B.. 2015. “PLIN2 Inhibits Insulin-Induced Glucose Uptake in Myoblasts through the Activation of the NLRP3 Inflammasome.” International Journal of Molecular Medicine 36 (3): 839–44. 10.3892/ijmm.2015.2276. [DOI] [PubMed] [Google Scholar]
- Currin Kevin W., Erdos Michael R., Narisu Narisu, Rai Vivek, Vadlamudi Swarooparani, Perrin Hannah J., Idol Jacqueline R., et al. 2021. “Genetic Effects on Liver Chromatin Accessibility Identify Disease Regulatory Variants.” American Journal of Human Genetics 108 (7): 1169–89. 10.1016/j.ajhg.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemen Sabine, van Polanen Nynke, and Hesselink Matthijs K. C.. 2018. “The Effect of Diet and Exercise on Lipid Droplet Dynamics in Human Muscle Tissue.” The Journal of Experimental Biology 221 (Pt Suppl 1): jeb167015. 10.1242/jeb.167015. [DOI] [PubMed] [Google Scholar]
- DeForest Natalie, Wang Yuqi, Zhu Zhiyi, Dron Jacqueline S., Koesterer Ryan, Natarajan Pradeep, Flannick Jason, Amariuta Tiffany, Peloso Gina M., and Majithia Amit R.. 2024. “Genome-Wide Discovery and Integrative Genomic Characterization of Insulin Resistance Loci Using Serum Triglycerides to HDL-Cholesterol Ratio as a Proxy.” Nature Communications 15 (September):8068. 10.1038/s41467-024-52105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo Ralph A., and Tripathy Devjit. 2009. “Skeletal Muscle Insulin Resistance Is the Primary Defect in Type 2 Diabetes.” Diabetes Care 32 (Suppl 2): S157–63. 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos Nikolaos, Watson Maria, Sakamoto Kei, and Hundal Harinder S.. 2006. “Differential Effects of Palmitate and Palmitoleate on Insulin Action and Glucose Utilization in Rat L6 Skeletal Muscle Cells.” The Biochemical Journal 399 (3): 473–81. 10.1042/BJ20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin Alexander, Davis Carrie A., Schlesinger Felix, Drenkow Jorg, Zaleski Chris, Jha Sonali, Batut Philippe, Chaisson Mark, and Gingeras Thomas R.. 2013. “STAR: Ultrafast Universal RNA-Seq Aligner.” Bioinformatics 29 (1): 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinar Klemen, Jan Vid, Pavlin Mojca, Chibalin Alexander V., and Pirkmajer Sergej. 2018. “Nucleosides Block AICAR-Stimulated Activation of AMPK in Skeletal Muscle and Cancer Cells.” American Journal of Physiology-Cell Physiology 315 (6): C803–17. 10.1152/ajpcell.00311.2017. [DOI] [PubMed] [Google Scholar]
- Dziewulska Anna, Dobosz Aneta M., Dobrzyn Agnieszka, Smolinska Agnieszka, Kolczynska Katarzyna, Ntambi James M., and Dobrzyn Pawel. 2020. “SCD1 Regulates the AMPK/SIRT1 Pathway and Histone Acetylation through Changes in Adenine Nucleotide Metabolism in Skeletal Muscle.” Journal of Cellular Physiology 235 (2): 1129–40. 10.1002/jcp.29026. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. “An Integrated Encyclopedia of DNA Elements in the Human Genome.” Nature 489 (7414): 57–74. 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium, Moore Jill E., Purcaro Michael J., Pratt Henry E., Epstein Charles B., Shoresh Noam, Adrian Jessika, et al. 2020. “Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes.” Nature 583 (7818): 699–710. 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge Amy S., Gallins Paul J., Jima Dereje, Broadaway K. Alaine, Ratain Mark J., Schuetz Erin, Schadt Eric, et al. 2020. “A New Liver Expression Quantitative Trait Locus Map From 1,183 Individuals Provides Evidence for Novel Expression Quantitative Trait Loci of Drug Response, Metabolic, and Sex-Biased Phenotypes.” Clinical Pharmacology and Therapeutics 107 (6): 1383–93. 10.1002/cpt.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels Philip, Magnusson Måns, Lundin Sverker, and Käller Max. 2016. “MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report.” Bioinformatics 32 (19): 3047–48. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougerousse Françoise, Durand Muriel, Lopez Soledad, Suel Laurence, Demignon Josiane, Thornton Charles, Ozaki Hidenori, et al. 2002. “Six and Eya Expression during Human Somitogenesis and MyoD Gene Family Activation.” Journal of Muscle Research and Cell Motility 23 (3): 255–64. 10.1023/a:1020990825644. [DOI] [PubMed] [Google Scholar]
- Gancheva Sofiya, Ouni Meriem, Jelenik Tomas, Koliaki Chrysi, Szendroedi Julia, Toledo Frederico G. S., Markgraf Daniel F., et al. 2019. “Dynamic Changes of Muscle Insulin Sensitivity after Metabolic Surgery.” Nature Communications 10 (1): 4179. 10.1038/s41467-019-12081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon Bradley S., Delgado Díaz Diana C., White James P., Carson James A., and Kostek Matthew C.. 2012. “Six1 and Six1 Cofactor Expression Is Altered during Early Skeletal Muscle Overload in Mice.” The Journal of Physiological Sciences: JPS 62 (5): 393–401. 10.1007/s12576-012-0214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. Grace, Inoue Fumitaka, Martin Beth, Schubach Max, Agarwal Vikram, Whalen Sean, Feng Shiyun, et al. 2020. “lentiMPRA and MPRAflow for High-Throughput Functional Characterization of Gene Regulatory Elements.” Nature Protocols 15 (8): 2387–2412. 10.1038/s41596-020-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiec Kamil, Milewska Marta, Błaszczyk Maciej, Gajewska Małgorzata, and Grzelkowska-Kowalczyk Katarzyna. 2015. “Palmitate Exerts Opposite Effects on Proliferation and Differentiation of Skeletal Myoblasts.” Cell Biology International 39 (9): 1044–52. 10.1002/cbin.10477. [DOI] [PubMed] [Google Scholar]
- Graham Sarah E., Clarke Shoa L., Wu Kuan-Han H., Kanoni Stavroula, Zajac Greg J. M., Ramdas Shweta, Surakka Ida, et al. 2021. “The Power of Genetic Diversity in Genome-Wide Association Studies of Lipids.” Nature 600 (7890): 675–79. 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley Stephen W., and Mullikin James C.. 2015. “QoRTs: A Comprehensive Toolset for Quality Control and Data Processing of RNA-Seq Experiments.” BMC Bioinformatics 16 (1): 224. 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann Thomas J., Theusch Elizabeth, Haldar Tanushree, Ranatunga Dilrini K., Jorgenson Eric, Medina Marisa W., Kvale Mark N., et al. 2018. “A Large Electronic-Health-Record-Based Genome-Wide Study of Serum Lipids.” Nature Genetics 50 (3): 401–13. 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett Nicholas A., Scalzo Rebecca L., and Reusch Jane E. B.. 2022. “Glucose Uptake by Skeletal Muscle within the Contexts of Type 2 Diabetes and Exercise: An Integrated Approach.” Nutrients 14 (3): 647. 10.3390/nu14030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igata Motoyuki, Motoshima Hiroyuki, Tsuruzoe Kaku, Kojima Kanou, Matsumura Takeshi, Kondo Tatsuya, Taguchi Tetsuya, et al. 2005. “Adenosine Monophosphate-Activated Protein Kinase Suppresses Vascular Smooth Muscle Cell Proliferation through the Inhibition of Cell Cycle Progression.” Circulation Research 97 (8): 837–44. 10.1161/01.RES.0000185823.73556.06. [DOI] [PubMed] [Google Scholar]
- Inoue Fumitaka, Kircher Martin, Martin Beth, Cooper Gregory M., Witten Daniela M., McManus Michael T., Ahituv Nadav, and Shendure Jay. 2017. “A Systematic Comparison Reveals Substantial Differences in Chromosomal versus Episomal Encoding of Enhancer Activity.” Genome Research 27 (1): 38–52. 10.1101/gr.212092.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Ronald G., Andrea Dimet-Wiley Amin Haghani, da Silva Francielly Morena, Brightwell Camille R., Lim Seongkyun, Khadgi Sabin, et al. 2023. “A Molecular Signature Defining Exercise Adaptation with Ageing and in Vivo Partial Reprogramming in Skeletal Muscle.” The Journal of Physiology 601 (4): 763–82. 10.1113/JP283836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski Joanna, Cook Tiffany, and Urrutia Raul. 2003. “Sp1- and Krüppel-like Transcription Factors.” Genome Biology 4 (2): 206. 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karjalainen Minna K., Karthikeyan Savita, Clare Oliver-Williams Eeva Sliz, Allara Elias, Wing Tung Fung Praveen Surendran, et al. 2024. “Genome-Wide Characterization of Circulating Metabolic Biomarkers.” Nature 628 (8006): 130–38. 10.1038/s41586-024-07148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetan Shubham, Kales Susan, Kursawe Romy, Jillette Alexandria, Ulirsch Jacob C., Reilly Steven K., Ucar Duygu, Tewhey Ryan, and Stitzel Michael L.. 2021. “Functional Characterization of T2D-Associated SNP Effects on Baseline and ER Stress-Responsive β Cell Transcriptional Activation.” Nature Communications 12 (1): 5242. 10.1038/s41467-021-25514-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimentidis Yann C., Arora Amit, Newell Michelle, Zhou Jin, Ordovas Jose M., Renquist Benjamin J., and Wood Alexis C.. 2020. “Phenotypic and Genetic Characterization of Lower LDL Cholesterol and Increased Type 2 Diabetes Risk in the UK Biobank.” Diabetes 69 (10): 2194–2205. 10.2337/db19-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostek Matthew C., Chen Yi-Wen, Cuthbertson Daniel J., Shi Rongye, Fedele Mark J., Esser Karyn A., and Rennie Michael J.. 2007. “Gene Expression Responses over 24 h to Lengthening and Shortening Contractions in Human Muscle: Major Changes in CSRP3, MUSTN1, SIX1, and FBXO32.” Physiological Genomics 31 (1): 42–52. 10.1152/physiolgenomics.00151.2006. [DOI] [PubMed] [Google Scholar]
- Kreimer Anat, Ashuach Tal, Inoue Fumitaka, Khodaverdian Alex, Deng Chengyu, Yosef Nir, and Ahituv Nadav. 2022. “Massively Parallel Reporter Perturbation Assays Uncover Temporal Regulatory Architecture during Neural Differentiation.” Nature Communications 13 (1): 1504. 10.1038/s41467-022-28659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Chun-Heng, Wang Shu-Huei, Juan Hsien-Chia, Chen Szu-Chi, Kuo Ching-Hua, Kuo Han-Chun, Lin Shin-Yu, and Li Hung-Yuan. 2024. “Angiopoietin-like Protein 4 Induces Growth Hormone Variant Secretion and Aggravates Insulin Resistance during Pregnancy, Linking Obesity to Gestational Diabetes Mellitus.” BioFactors (Oxford, England), May. 10.1002/biof.2076. [DOI] [PubMed] [Google Scholar]
- Lehka Lilya, and Rędowicz Maria Jolanta. 2020. “Mechanisms Regulating Myoblast Fusion: A Multilevel Interplay.” Seminars in Cell & Developmental Biology 104 (August):81–92. 10.1016/j.semcdb.2020.02.004. [DOI] [PubMed] [Google Scholar]
- Li Heng, Handsaker Bob, Wysoker Alec, Fennell Tim, Ruan Jue, Homer Nils, Marth Gabor, Abecasis Goncalo, Durbin Richard, and 1000 Genome Project Data Processing Subgroup. 2009. “The Sequence Alignment/Map Format and SAMtools.” Bioinformatics 25 (16): 2078–79. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love Michael I., Huber Wolfgang, and Anders Simon. 2014. “Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2.” Genome Biology 15 (12): 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Yunhai, Hitz Benjamin C., Gabdank Idan, Hilton Jason A., Kagda Meenakshi S., Lam Bonita, Myers Zachary, et al. 2020. “New Developments on the Encyclopedia of DNA Elements (ENCODE) Data Portal.” Nucleic Acids Research 48 (D1): D882–89. 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan Anubha, Spracklen Cassandra N., Zhang Weihua, Ng Maggie C. Y., Petty Lauren E., Kitajima Hidetoshi, Yu Grace Z., et al. 2022. “Multi-Ancestry Genetic Study of Type 2 Diabetes Highlights the Power of Diverse Populations for Discovery and Translation.” Nature Genetics 54 (5): 560–72. 10.1038/s41588-022-01058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen Selina, Yen H Nguyen Paulina Skrobuk, and Koistinen Heikki A. 2017. “Palmitate and Oleate Exert Differential Effects on Insulin Signalling and Glucose Uptake in Human Skeletal Muscle Cells.” Endocrine Connections 6 (5): 331–39. 10.1530/EC-17-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch Laura J., Bramwell Laura R., Knight Bridget, and Kos Katarina. 2020. “Circulating and Tissue Specific Transcription of Angiopoietin-like Protein 4 in Human Type 2 Diabetes.” Metabolism: Clinical and Experimental 106 (May):154192. 10.1016/j.metabol.2020.154192. [DOI] [PubMed] [Google Scholar]
- Melnikov Alexandre, Zhang Xiaolan, Rogov Peter, Wang Li, and Mikkelsen Tarjei S.. 2014. “Massively Parallel Reporter Assays in Cultured Mammalian Cells.” Journal of Visualized Experiments: JoVE, no. 90 (August), 51719. 10.3791/51719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers Terri K., Andreuzza Sébastien E., and Franklin David S.. 2004. “p18INK4c and p27KIP1 Are Required for Cell Cycle Arrest of Differentiated Myotubes.” Experimental Cell Research 300 (2): 365–78. 10.1016/j.yexcr.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Myint Leslie, Avramopoulos Dimitrios G., Goff Loyal A., and Hansen Kasper D.. 2019. “Linear Models Enable Powerful Differential Activity Analysis in Massively Parallel Reporter Assays.” BMC Genomics 20 (1): 209. 10.1186/s12864-019-5556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Daisuke, Takeda Ryo, Sata Masataka, Satonaka Hiroshi, Suzuki Etsu, Nagano Tetsuo, and Hirata Yasunobu. 2004. “AMP-Activated Protein Kinase Inhibits Angiotensin II-Stimulated Vascular Smooth Muscle Cell Proliferation.” Circulation 110 (4): 444–51. 10.1161/01.CIR.0000136025.96811.76. [DOI] [PubMed] [Google Scholar]
- Niro Claire, Demignon Josiane, Vincent Stéphane, Liu Yubing, Giordani Julien, Sgarioto Nicolas, Favier Maryline, Isabelle Guillet-Deniau Alexandre Blais, and Maire Pascal. 2010. “Six1 and Six4 Gene Expression Is Necessary to Activate the Fast-Type Muscle Gene Program in the Mouse Primary Myotome.” Developmental Biology 338 (2): 168–82. 10.1016/j.ydbio.2009.11.031. [DOI] [PubMed] [Google Scholar]
- Orchard Peter, Manickam Nandini, Ventresca Christa, Vadlamudi Swarooparani, Varshney Arushi, Rai Vivek, Kaplan Jeremy, et al. 2021. “Human and Rat Skeletal Muscle Single-Nuclei Multi-Omic Integrative Analyses Nominate Causal Cell Types, Regulatory Elements, and SNPs for Complex Traits.” Genome Research 31 (12): 2258–75. 10.1101/gr.268482.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey Gautam K., Vadlamudi Swarooparani, Currin Kevin W., Moxley Anne H., Nicholas Jayna C., McAfee Jessica C., Broadaway K. Alaine, and Mohlke Karen L.. 2024. “Liver Regulatory Mechanisms of Noncoding Variants at Lipid and Metabolic Trait Loci.” HGG Advances 5 (2): 100275. 10.1016/j.xhgg.2024.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Sanghyeon, Kim Soyeon, Kim Beomsu, Dan Say Kim Jaeyoung Kim, Ahn Yeeun, Kim Hyejin, et al. 2024. “Multivariate Genomic Analysis of 5 Million People Elucidates the Genetic Architecture of Shared Components of the Metabolic Syndrome.” Nature Genetics 56 (11): 2380–91. 10.1038/s41588-024-01933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov Daniil V., Makhnovskii Pavel A., Shagimardanova Elena I., Gazizova Guzel R., Lysenko Evgeny A., Gusev Oleg A., and Vinogradova Olga L.. 2019. “Contractile Activity-Specific Transcriptome Response to Acute Endurance Exercise and Training in Human Skeletal Muscle.” American Journal of Physiology. Endocrinology and Metabolism 316 (4): E605–14. 10.1152/ajpendo.00449.2018. [DOI] [PubMed] [Google Scholar]
- Ramachandran Krithika, Senagolage Madhavi D., Sommars Meredith A., Futtner Christopher R., Omura Yasuhiro, Allred Amanda L., and Barish Grant D.. 2019. “Dynamic Enhancers Control Skeletal Muscle Identity and Reprogramming.” PLoS Biology 17 (10): e3000467. 10.1371/journal.pbio.3000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere Uku, Kolberg Liis, Kuzmin Ivan, Arak Tambet, Adler Priit, Peterson Hedi, and Vilo Jaak. 2019. “G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update).” Nucleic Acids Research 47 (W1): W191–98. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgeway A. G., and Skerjanc I. S.. 2001. “Pax3 Is Essential for Skeletal Myogenesis and the Expression of Six1 and Eya2.” The Journal of Biological Chemistry 276 (22): 19033–39. 10.1074/jbc.M011491200. [DOI] [PubMed] [Google Scholar]
- Roden M., Price T. B., Perseghin G., Petersen K. F., Rothman D. L., Cline G. W., and Shulman G. I.. 1996. “Mechanism of Free Fatty Acid-Induced Insulin Resistance in Humans.” The Journal of Clinical Investigation 97 (12): 2859–65. 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman Tamara S., Marvelle Amanda F., Fogarty Marie P., Vadlamudi Swarooparani, Gonzalez Arlene J., Buchkovich Martin L., Huyghe Jeroen R., et al. 2015. “Multiple Hepatic Regulatory Variants at the GALNT2 GWAS Locus Associated with High-Density Lipoprotein Cholesterol.” American Journal of Human Genetics 97 (6): 801–15. 10.1016/j.ajhg.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue Saori, Kanai Masahiro, Tanigawa Yosuke, Karjalainen Juha, Kurki Mitja, Koshiba Seizo, Narita Akira, et al. 2021. “A Cross-Population Atlas of Genetic Associations for 220 Human Phenotypes.” Nature Genetics 53 (10): 1415–24. 10.1038/s41588-021-00931-x. [DOI] [PubMed] [Google Scholar]
- Samuel Varman T., and Shulman Gerald I.. 2016. “The Pathogenesis of Insulin Resistance: Integrating Signaling Pathways and Substrate Flux.” Journal of Clinical Investigation 126 (1): 12–22. 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayols Sergi. 2023. “Rrvgo: A Bioconductor Package for Interpreting Lists of Gene Ontology Terms.” microPublication Biology 2023 (April): 10.17912/micropub.biology.000811. https://doi.org/10.17912/micropub.biology.000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott Laura J., Erdos Michael R., Huyghe Jeroen R., Welch Ryan P., Beck Andrew T., Wolford Brooke N., Chines Peter S., et al. 2016. “The Genetic Regulatory Signature of Type 2 Diabetes in Human Skeletal Muscle.” Nature Communications 7 (1): 11764. 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj Margaret Sunitha, Paruchuri Kaavya, Haidermota Sara, Bernardo Rachel, Rich Stephen S., Peloso Gina M., and Natarajan Pradeep. 2022. “Genome-Wide Discovery for Diabetes-Dependent Triglycerides-Associated Loci.” PloS One 17 (10): e0275934. 10.1371/journal.pone.0275934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik Natalia, and Pestov Dimitri G.. 2019. “The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation.” Cells 8 (11): 1379. 10.3390/cells8111379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies Helmut, Berndt Carsten, and Jones Dean P.. 2017. “Oxidative Stress.” Annual Review of Biochemistry 86 (Volume 86, 2017): 715–48. 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- Sinnott-Armstrong Nasa, Sousa Isabel S., Laber Samantha, Elizabeth Rendina-Ruedy Simon E. Nitter Dankel, Ferreira Teresa, Mellgren Gunnar, et al. 2021. “A Regulatory Variant at 3q21.1 Confers an Increased Pleiotropic Risk for Hyperglycemia and Altered Bone Mineral Density.” Cell Metabolism 33 (3): 615–628.e13. 10.1016/j.cmet.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg Suren, Sourbh Kriti Kushwaha, Dubey Rupal, and Gupta Jeena. 2023. “Association between Obesity, Inflammation and Insulin Resistance: Insights into Signaling Pathways and Therapeutic Interventions.” Diabetes Research and Clinical Practice 200 (June):110691. 10.1016/j.diabres.2023.110691. [DOI] [PubMed] [Google Scholar]
- Suzuki Ken, Hatzikotoulas Konstantinos, Southam Lorraine, Taylor Henry J., Yin Xianyong, Lorenz Kim M., Mandla Ravi, et al. 2024. “Genetic Drivers of Heterogeneity in Type 2 Diabetes Pathophysiology.” Nature 627 (8003): 347–57. 10.1038/s41586-024-07019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey Ryan, Kotliar Dylan, Park Daniel S., Liu Brandon, Winnicki Sarah, Reilly Steven K., Andersen Kristian G., et al. 2016. “Direct Identification of Hundreds of Expression-Modulating Variants Using a Multiplexed Reporter Assay.” Cell 165 (6): 1519–29. 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar Adelaide, Kyono Yasuhiro, Nishino Kirsten, Bose Maya, Varshney Arushi, Parker Stephen C. J., and Kitzman Jacob O.. 2023. “Using a Modular Massively Parallel Reporter Assay to Discover Context-Specific Regulatory Grammars in Type 2 Diabetes.” bioRxiv: The Preprint Server for Biology, October, 2023.10.08.561391. 10.1101/2023.10.08.561391. [DOI] [Google Scholar]
- Trenerry Marissa K., Carey Kate A., Ward Alister C., and Cameron-Smith David. 2007. “STAT3 Signaling Is Activated in Human Skeletal Muscle Following Acute Resistance Exercise.” Journal of Applied Physiology (Bethesda, Md.: 1985) 102 (4): 1483–89. 10.1152/japplphysiol.01147.2006. [DOI] [PubMed] [Google Scholar]
- Varshney Arushi, Manickam Nandini, Orchard Peter, Tovar Adelaide, Zhang Zhenhao, Feng Fan, Erdos Michael R., et al. 2023. “Population-Scale Skeletal Muscle Single-Nucleus Multi-Omic Profiling Reveals Extensive Context Specific Genetic Regulation.” bioRxiv: The Preprint Server for Biology, December, 2023.12.15.571696. 10.1101/2023.12.15.571696. [DOI] [Google Scholar]
- Vaughan Megan, and Lamia Katja A.. 2019. “Isolation and Differentiation of Primary Myoblasts from Mouse Skeletal Muscle Explants.” Journal of Visualized Experiments: JoVE, no. 152 (October), 10.3791/60310. https://doi.org/10.3791/60310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss M. D., Beha A., Tennagels N., Tschank G., Herling A. W., Quint M., Gerl M., Metz-Weidmann C., Haun G., and Korn M.. 2005. “Gene Expression Profiling in Skeletal Muscle of Zucker Diabetic Fatty Rats: Implications for a Role of Stearoyl-CoA Desaturase 1 in Insulin Resistance.” Diabetologia 48 (12): 2622–30. 10.1007/s00125-005-0025-2. [DOI] [PubMed] [Google Scholar]
- Vujkovic Marijana, Keaton Jacob M., Lynch Julie A., Miller Donald R., Zhou Jin, Tcheandjieu Catherine, Huffman Jennifer E., et al. 2020. “Discovery of 318 New Risk Loci for Type 2 Diabetes and Related Vascular Outcomes among 1.4 Million Participants in a Multi-Ancestry Meta-Analysis.” Nature Genetics 52 (7): 680–91. 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Zhe, Emmerich Andrew, Pillon Nicolas J., Moore Tim, Hemerich Daiane, Cornelis Marilyn C., Mazzaferro Eugenia, et al. 2022. “Genome-Wide Association Analyses of Physical Activity and Sedentary Behavior Provide Insights into Underlying Mechanisms and Roles in Disease Prevention.” Nature Genetics 54 (9): 1332–44. 10.1038/s41588-022-01165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo Loïc, Vedantam Sailaja, Marouli Eirini, Sidorenko Julia, Bartell Eric, Sakaue Saori, Graff Marielisa, et al. 2022. “A Saturated Map of Common Genetic Variants Associated with Human Height.” Nature 610 (7933): 704–12. 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K., and Wold B.. 1996. “Skeletal Muscle Determination and Differentiation: Story of a Core Regulatory Network and Its Context.” Current Opinion in Cell Biology 8 (6): 877–89. 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- Zhu Chun-Hong, Mouly Vincent, Cooper Racquel N., Mamchaoui Kamel, Bigot Anne, Shay Jerry W., Di Santo James P., Butler-Browne Gillian S., and Wright Woodring E.. 2007. “Cellular Senescence in Human Myoblasts Is Overcome by Human Telomerase Reverse Transcriptase and Cyclin-Dependent Kinase 4: Consequences in Aging Muscle and Therapeutic Strategies for Muscular Dystrophies.” Aging Cell 6 (4): 515–23. 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primer sequences used for MPRA library cloning and sequencing library generation are available in Supplemental Table 20. All processed RNA-seq counts and MPRA data are accessible through the Gene Expression Omnibus (GSE282191). Raw sequencing data are deposited on the NCBI Sequence Read Archive and accession numbers are listed in the GEO records. Complete results files from both sets of analyses are available as Supplemental Tables referenced in this article.
All code used to preprocess and analyze data presented here are available on Github. We used custom preprocessing pipelines for RNA-seq data (https://github.com/porchard/RNAseq-NextFlow) and MPRA barcode counting data (https://github.com/adelaidetovar/MPRA-Nextflow). Scripts used to analyze and visualize processed data are available as a separate repository at https://github.com/adelaidetovar/lhcn-perturb-mpra.