Abstract
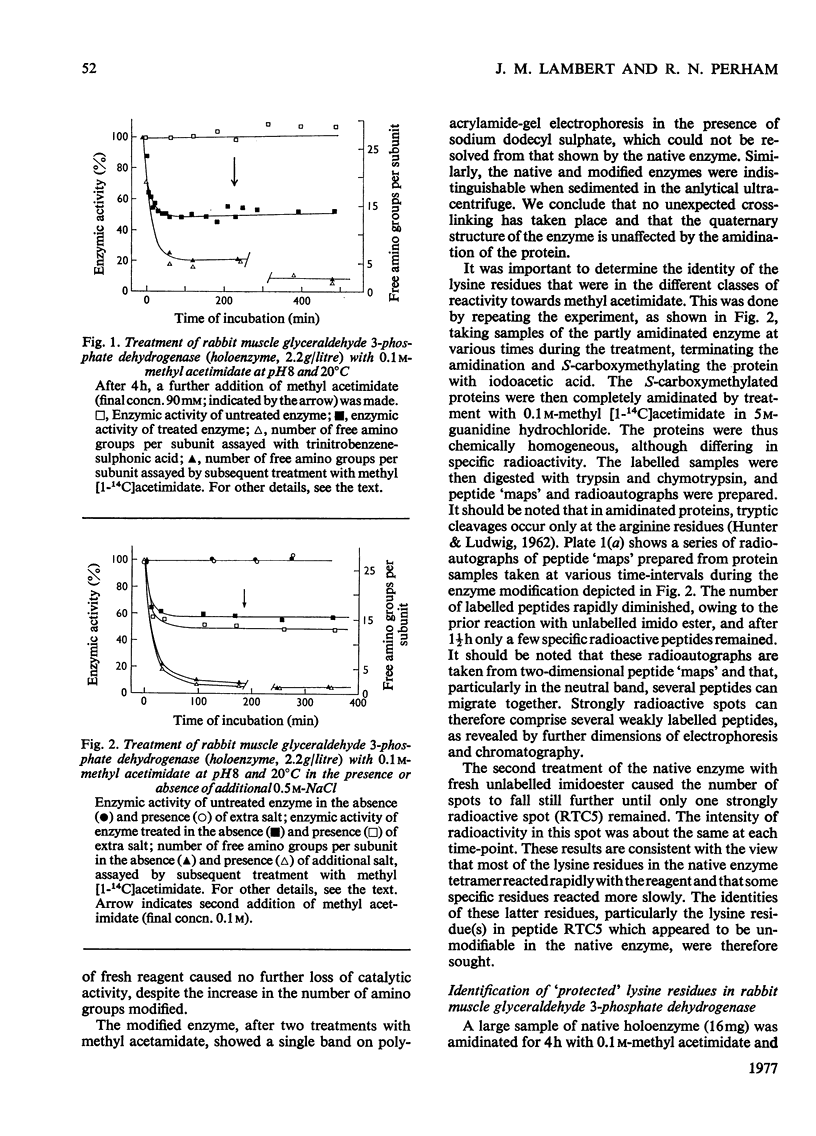
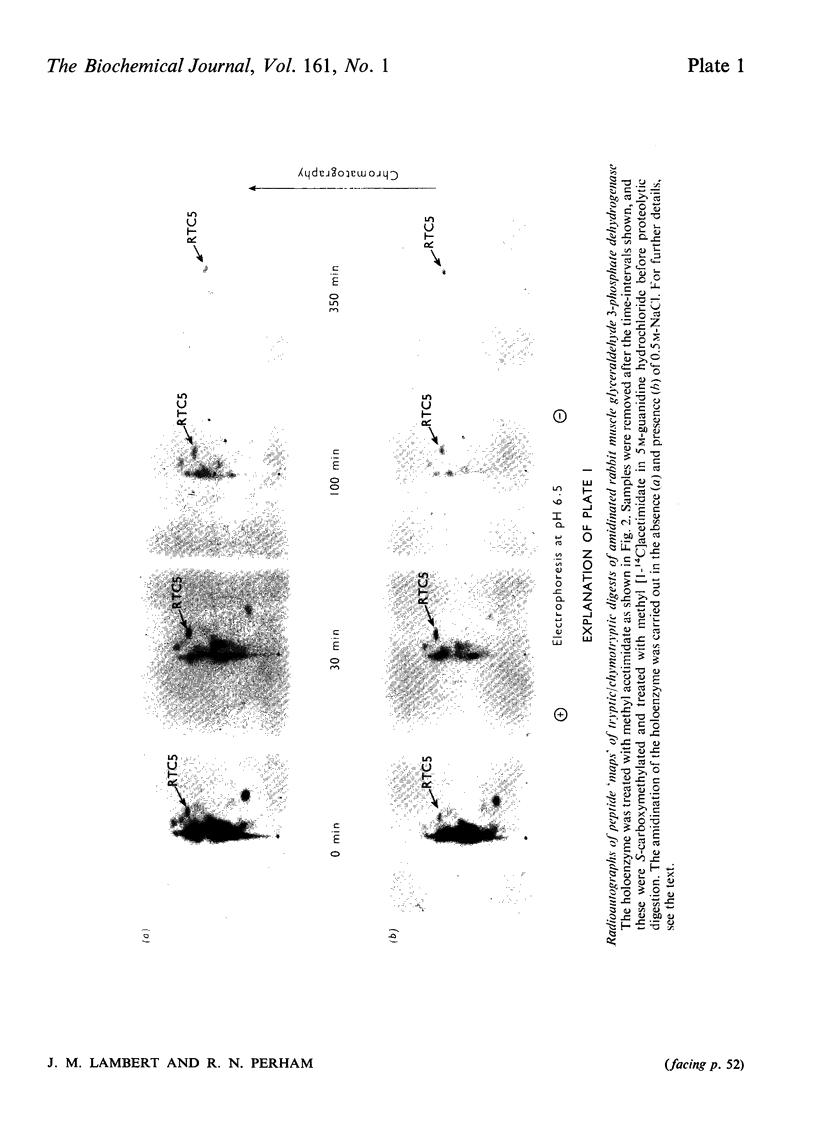
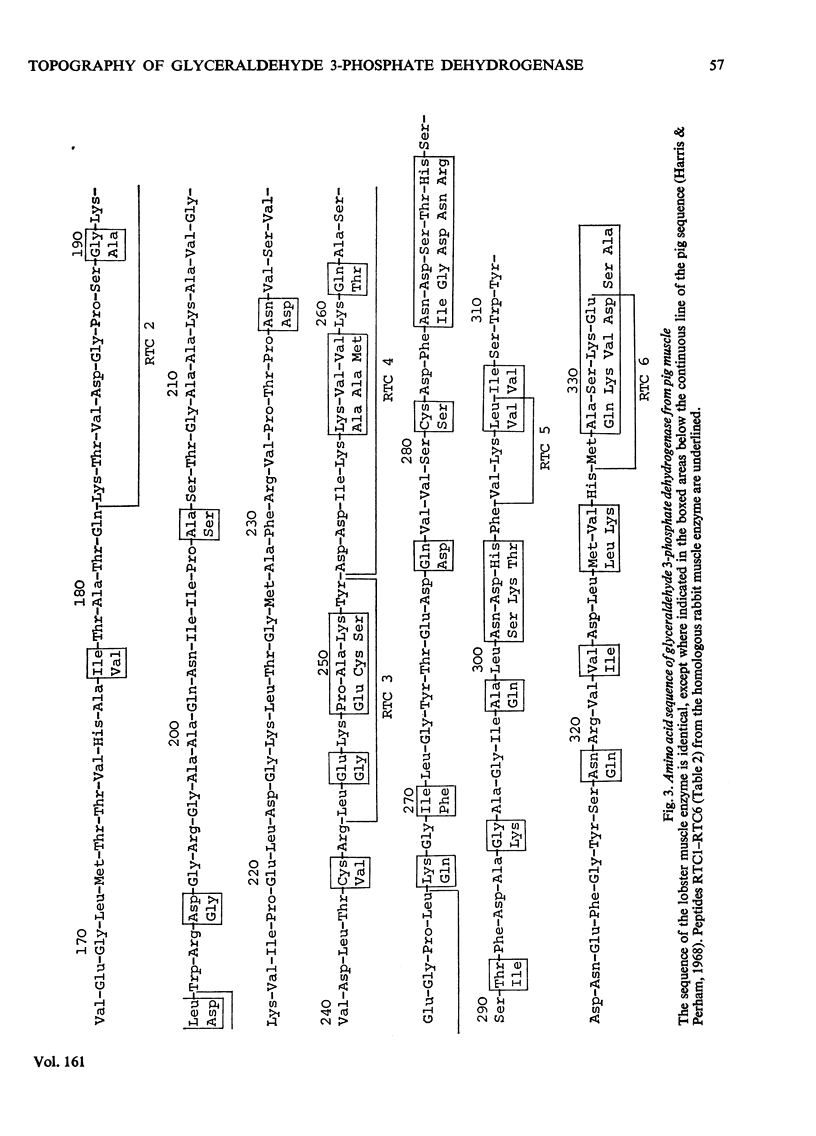
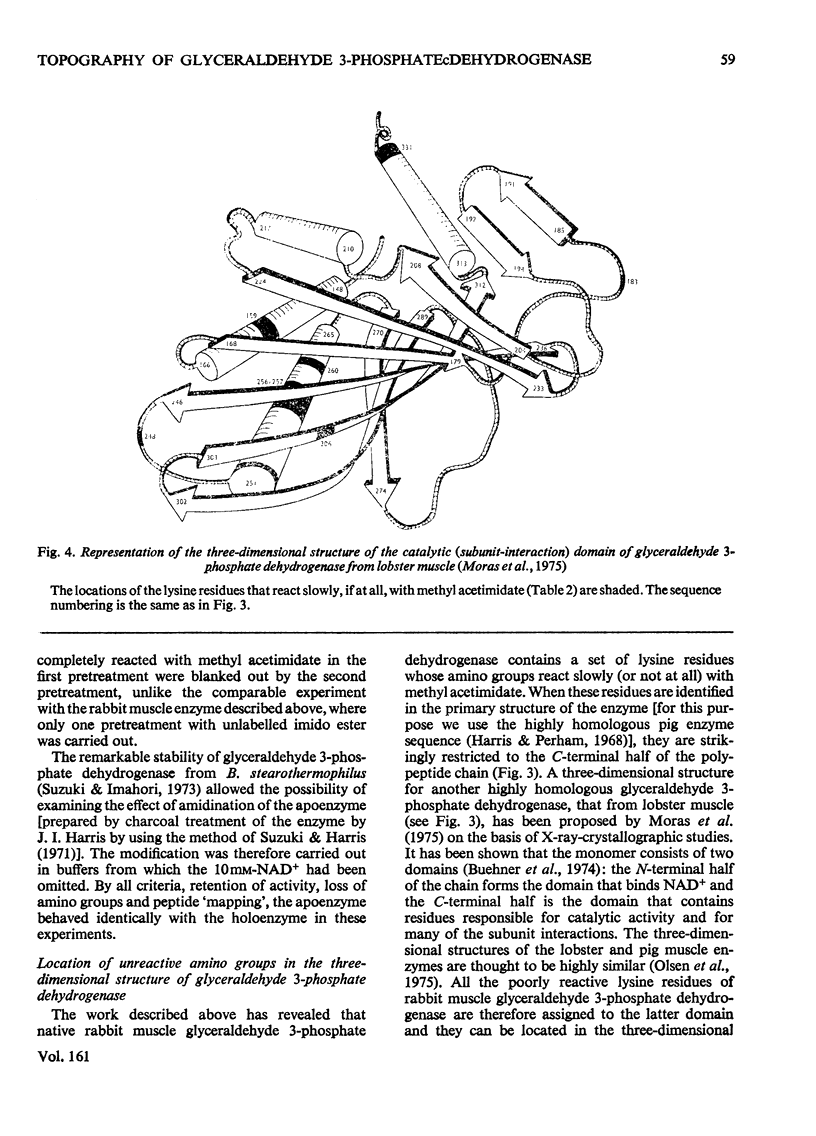
1. Treatment with methyl acetimidate was used to probe the topography of several tetrameric glyceraldehyde 3-phosphate dehydrogenases, in particular the holoenzymes from rabbit muscle and Bacillus stearothermophilus. During the course of the reaction with the rabbit muscle enzyme, the number of amino groups fell rapidly from the starting value of 27 per subunit to a value of approx. five per subunit. This number could be lowered further to values between one and two per subunit by a second treatment with methyl acetimidate. The enzyme remained tetrameric throughout and retained 50% of its initial catalytic activity at the end of the experiment. 2. Use of methyl [1-14C]acetimidate and small-scale methods of protein chemistry showed that only one amino group per subunit, that of lysine-306, was completely unavailable for reaction with imido ester in the native enzyme. This results is consistent with the structure of the highly homologous glyceraldehyde 3-phosphate dehydrogenase of lobster muscle deduced from X-ray-crystallographic analysis, since lysine-306 can be seen to form an intrachain ion-pair with aspartic acid-241 in the hydrophobic environment of a subunit-subunit interface. 3. Several other amino groups in the rabbit muscle enzyme that reacted only slowly with the reagent were also identified chemically. These were found to be located entirely in the C-terminal half of the polypeptides chain, which comprises a folding domain associated with catalytic activity and subunit contact in the three-dimensional structure. Slow reaction of these 'surface' amino groups with methyl acetimidate is attributed to intramolecular ionic interactions of the amino groups with neighbouring side-chain carboxyl groups, a conclusion that is compatible with the reported three-dimensional structure and with the dependence of the reaction of ionic stength. 4. Very similar results were obtained with the enzymes from B. stearothermophilus and from ox muscle and ox liver, supporting the view that the ion-pair involving lysine-306 and aspartic acid-241 will be a common structural feature in glyceraldehyde-3-phosphate dehydrogenases. The B. stearothermophilus enzyme was fully active after modification. 5. No differences could be detected between the enzymes from ox muscle and ox liver, in accord with other evidence that points to the identify of these enzymes. 6. The pattern of slowly reacting amino groups in the enzyme from B. stearothermophilus, although similar to that of the mammalian enzymes, indicated one or two additional intramolecular ionic interactions of lysine residues that might contribute to the thermal stability of this enzyme.
Full text
PDF


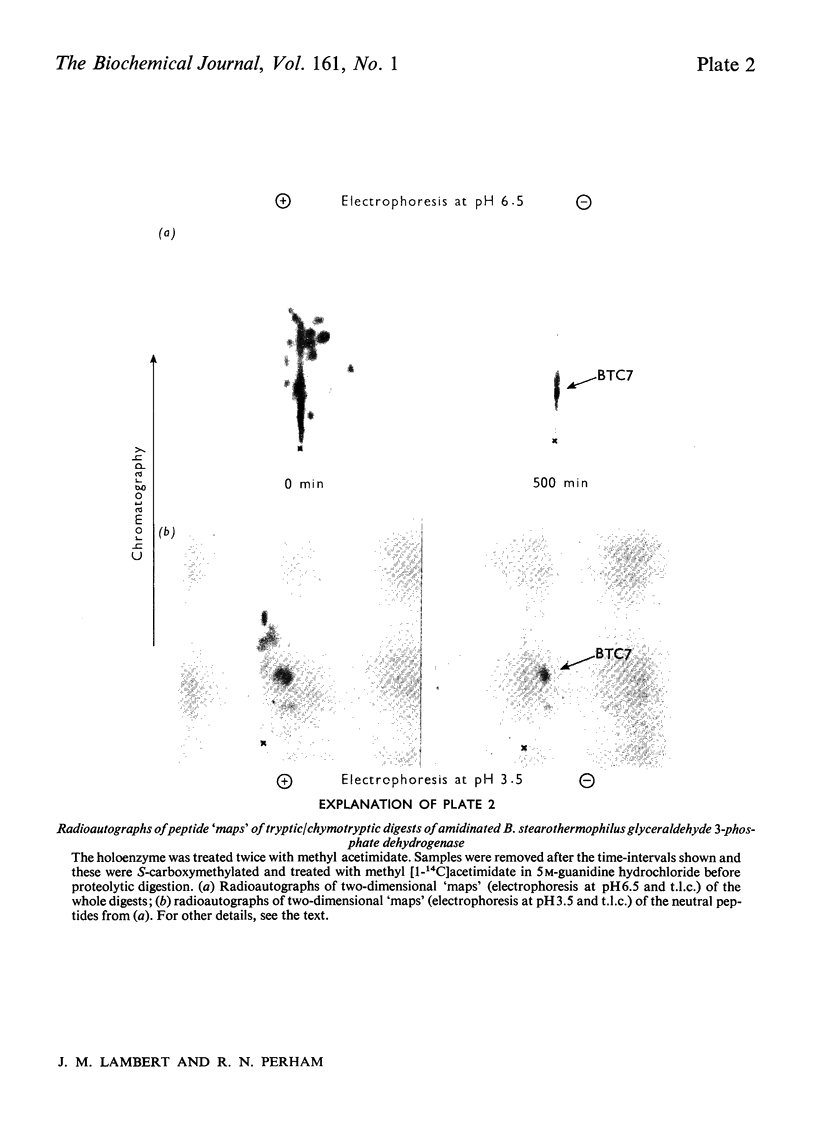
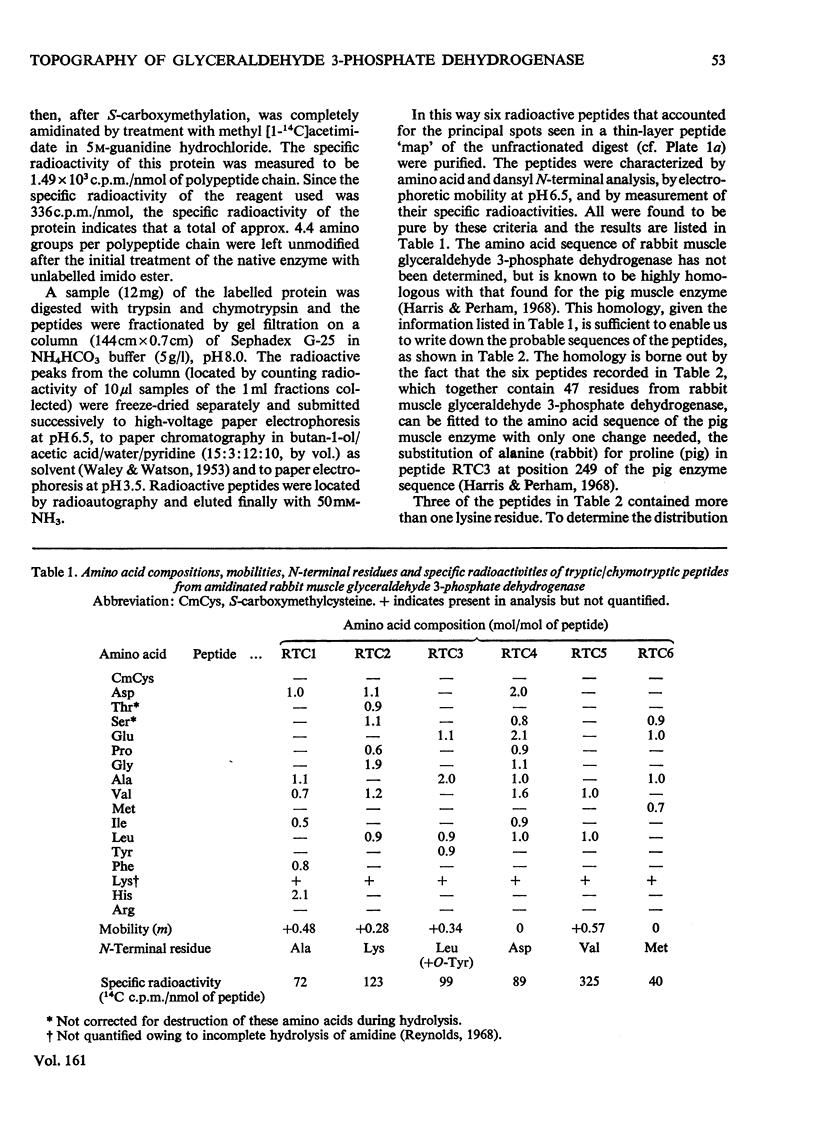

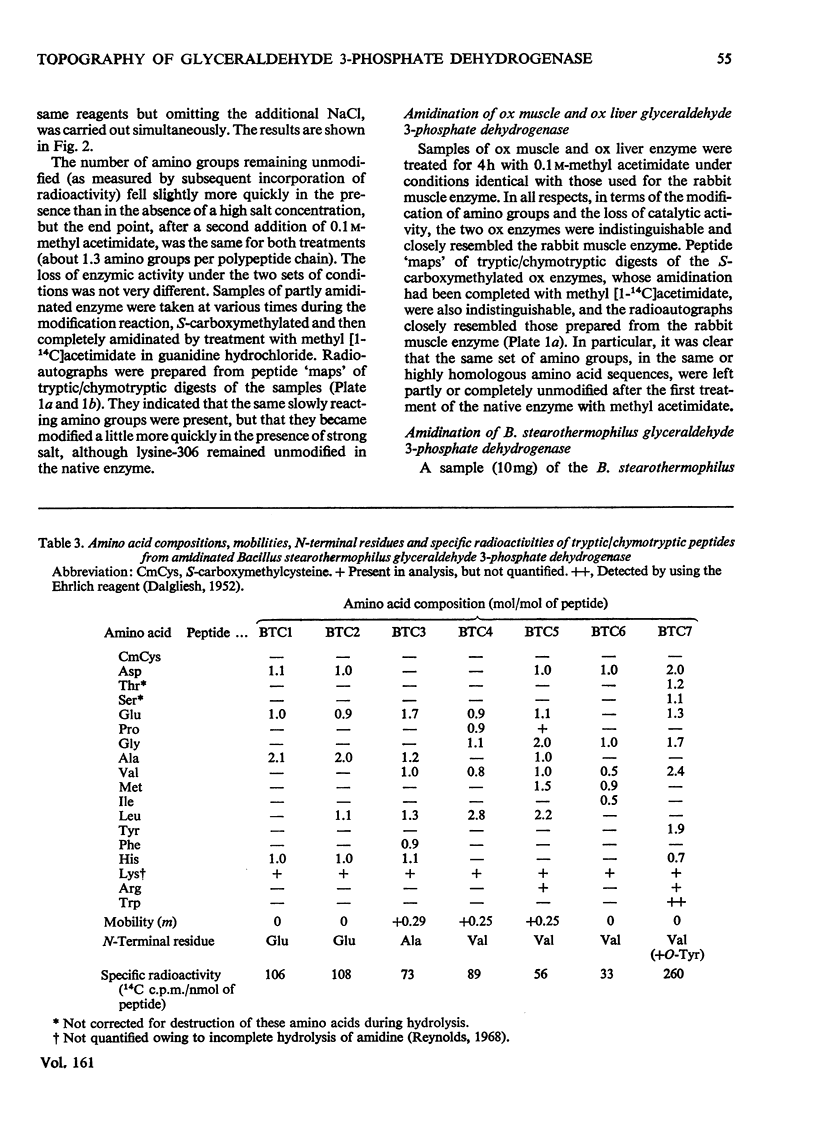
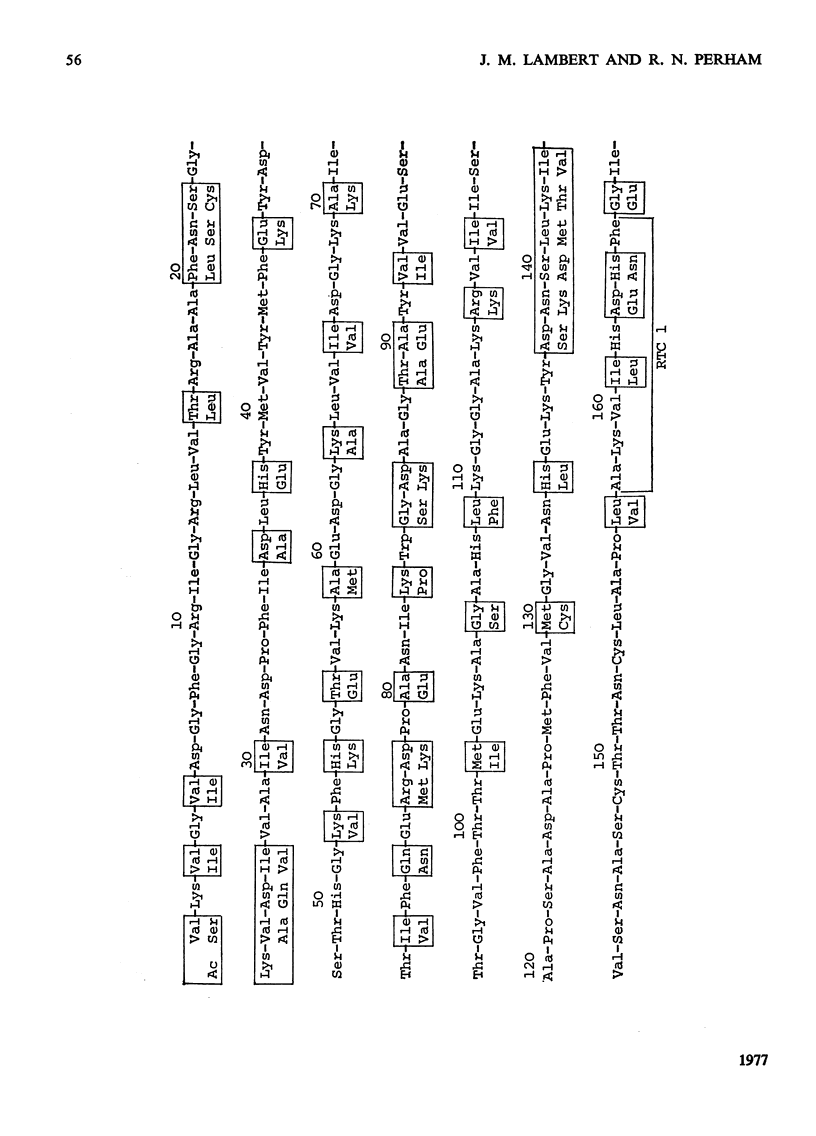




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLISON W. S., KAPLAN N. O. THE COMPARATIVE ENZYMOLOGY OF TRIOSEPHOSPHATE DEHYDROGENASE. J Biol Chem. 1964 Jul;239:2140–2152. [PubMed] [Google Scholar]
- Bates D. L., Perham R. N., Coggins J. R. Methods for obtaining peptide maps of proteins on a subnanomole scale. Anal Biochem. 1975 Sep;68(1):175–184. doi: 10.1016/0003-2697(75)90692-2. [DOI] [PubMed] [Google Scholar]
- Bell J. E., Dalziel K. Studies of coenzyme binding to rabbit muscle glyceraldehyde-3-phoshate dehydrogenase. Biochim Biophys Acta. 1975 Jun 24;391(2):249–258. doi: 10.1016/0005-2744(75)90248-x. [DOI] [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. An amino acid sequence in the active site of lipoamide dehydrogenase from pig heart. Biochem J. 1974 Mar;137(3):505–512. doi: 10.1042/bj1370505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne D. T., Kent S. B. Formation of non-amidine products in the chemical modification of horse liver alcohol dehydrogenase with imido esters. Biochem Biophys Res Commun. 1975 Nov 3;67(1):133–138. doi: 10.1016/0006-291x(75)90293-4. [DOI] [PubMed] [Google Scholar]
- Browne D. T., Kent S. B. Formation of non-amidine products in the reaction of primary amines with imido esters. Biochem Biophys Res Commun. 1975 Nov 3;67(1):126–132. doi: 10.1016/0006-291x(75)90292-2. [DOI] [PubMed] [Google Scholar]
- Buehner M., Ford G. C., Olsen K. W., Moras D., Rossman M. G. Three-dimensional structure of D-glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1974 Nov 25;90(1):25–49. doi: 10.1016/0022-2836(74)90254-x. [DOI] [PubMed] [Google Scholar]
- DALGLIESH C. E. The relation between pyridoxin and tryptophan metabolism, studied in the rat. Biochem J. 1952 Sep;52(1):3–14. doi: 10.1042/bj0520003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. E., Sajgò M., Noller H. F., Harris J. I. Amino-acid sequence of glyceraldehyde 3-phosphate dehydrogenase from lobster muscle. Nature. 1967 Dec 23;216(5121):1181–1185. doi: 10.1038/2161181a0. [DOI] [PubMed] [Google Scholar]
- Dworschack R., Tarr G., Plapp B. V. Identification of the lysine residue modified during the activation of acetimidylation of horse liver alcohol dehydrogenase. Biochemistry. 1975 Jan 28;14(2):200–203. doi: 10.1021/bi00673a002. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H., Colloms M. Reactivity of tobacco mosaic virus and its protein toward acetic anhydride. Biochemistry. 1967 Sep;6(9):2740–2745. doi: 10.1021/bi00861a014. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N. The reaction of aldolase with 2-methylmaleic anhydride. Biochem J. 1970 Mar;116(5):843–849. doi: 10.1042/bj1160843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeeb A. F. Determination of free amino groups in proteins by trinitrobenzenesulfonic acid. Anal Biochem. 1966 Mar;14(3):328–336. doi: 10.1016/0003-2697(66)90275-2. [DOI] [PubMed] [Google Scholar]
- Harris J. I., Perham R. N. Glyceraldehyde 3-phosphate dehydrogenase from pig muscle. Nature. 1968 Sep 7;219(5158):1025–1028. doi: 10.1038/2191025a0. [DOI] [PubMed] [Google Scholar]
- Harris J. I., Polgár L. Amino acid sequence around a reactive lysine in glyceraldehyde 3-phosphate dehydrogenase. J Mol Biol. 1965 Dec;14(2):630–633. doi: 10.1016/s0022-2836(65)80218-2. [DOI] [PubMed] [Google Scholar]
- Hartley B. S. Strategy and tactics in protein chemistry. Biochem J. 1970 Oct;119(5):805–822. doi: 10.1042/bj1190805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock S. E. Cross-linking of troponin with dimethylimido esters. Biochemistry. 1975 Nov 18;14(23):5162–5167. doi: 10.1021/bi00694a022. [DOI] [PubMed] [Google Scholar]
- Kaplan H., Stevenson K. J., Hartley B. S. Competitive labelling, a method for determining the reactivity of individual groups in proteins. The amino groups of porcine elastase. Biochem J. 1971 Sep;124(2):289–299. doi: 10.1042/bj1240289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. M., Perham R. N. A comparison of the glyceraldehyde 3-phosphate dehydrogenase from ox muscle and liver. FEBS Lett. 1974 Apr 1;40(2):305–308. doi: 10.1016/0014-5793(74)80250-4. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Perham R. N., Coggins J. R. Intramolecular ionic interactions of lysine residues and a possible folding domain in fructose diphosphate aldolase. Biochem J. 1977 Jan 1;161(1):63–71. doi: 10.1042/bj1610063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Olsen K. W., Sabesan M. N., Buehner M., Ford G. C., Rossmann M. G. Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1975 Dec 10;250(23):9137–9162. doi: 10.2210/pdb1gpd/pdb. [DOI] [PubMed] [Google Scholar]
- Offord R. E. Electrophoretic mobilities of peptides on paper and their use in the determination of amide groups. Nature. 1966 Aug 6;211(5049):591–593. doi: 10.1038/211591a0. [DOI] [PubMed] [Google Scholar]
- Olsen K. W., Moras D., Rossmann M. G. Sequence variability and structure of D-glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1975 Dec 25;250(24):9313–9321. [PubMed] [Google Scholar]
- Park J. H., Agnello C. F., Mathew E. S-N transfer and dual acetylation in the S-acetylation and N-acetylation of 3-phosphoglyceraldehyde dehydrogenase by substrates. J Biol Chem. 1966 Feb 10;241(3):769–771. [PubMed] [Google Scholar]
- Perham R. N., Anderson P. J. The reactivity of thiol groups in aldolase. Biochem Soc Symp. 1970;31:49–58. [PubMed] [Google Scholar]
- Perham R. N., Richards F. M. Reactivity and structural role of protein amino groups in tobacco mosaic virus. J Mol Biol. 1968 May 14;33(3):795–807. doi: 10.1016/0022-2836(68)90320-3. [DOI] [PubMed] [Google Scholar]
- Perham R. N. The reactivity of functional groups as a probe for investigating the topography of tobacco mosaic virus. The use of mutants with additional lysine residues in the coat protein. Biochem J. 1973 Jan;131(1):119–126. doi: 10.1042/bj1310119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M. F., Raidt H. Stereochemical basis of heat stability in bacterial ferredoxins and in haemoglobin A2. Nature. 1975 May 15;255(5505):256–259. doi: 10.1038/255256a0. [DOI] [PubMed] [Google Scholar]
- Plapp B. V. Enhancement of the activity of horse liver alcohol dehydrogenase by modification of amino groups at the active sites. J Biol Chem. 1970 Apr 10;245(7):1727–1735. [PubMed] [Google Scholar]
- Price N. C., Radda G. K. The binding of NAD+ to rabbit muscle glyceraldehyde-3-phosphate dehydrogenase studied by protein fluorescence quenching. Biochim Biophys Acta. 1971 Apr 14;235(1):27–31. doi: 10.1016/0005-2744(71)90029-5. [DOI] [PubMed] [Google Scholar]
- Reynolds J. H. Acetimidation of bovine pancreatic ribonuclease A. Biochemistry. 1968 Sep;7(9):3131–3135. doi: 10.1021/bi00849a016. [DOI] [PubMed] [Google Scholar]
- Shapiro A. L., Maizel J. V., Jr Molecular weight estimation of polypeptides by SDS-polyacrylamide gel electrophoresis: further data concerning resolving power and general considerations. Anal Biochem. 1969 Jun;29(3):505–514. doi: 10.1016/0003-2697(69)90335-2. [DOI] [PubMed] [Google Scholar]
- Smith C. M., Velick S. F. The glyceraldehyde 3-phosphate dehydrogenases of liver and muscle. Cooperative interactions and conditions for functional reversibility. J Biol Chem. 1972 Jan 10;247(1):273–284. [PubMed] [Google Scholar]
- Suzuki K., Imahori K. Glyceraldehyde 3-phosphate dehydrogenase of Bacillus stearothermophilus. Kinetics and physicochemical studies. J Biochem. 1973 Nov;74(5):955–970. [PubMed] [Google Scholar]
- Suzuki Koichi, Ieuan Harris J. Glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. FEBS Lett. 1971 Mar 16;13(4):217–220. doi: 10.1016/0014-5793(71)80539-2. [DOI] [PubMed] [Google Scholar]
- WALEY S. G., WATSON J. The action of trypsin on polylysine. Biochem J. 1953 Sep;55(2):328–337. doi: 10.1042/bj0550328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vijlder J. J., Slater E. C. The reaction between NAD+ and rabbit-muscle glyceraldehydephosphate dehydrogenase. Biochim Biophys Acta. 1968 Aug 27;167(1):23–34. doi: 10.1016/0005-2744(68)90274-x. [DOI] [PubMed] [Google Scholar]