SUMMARY
The cardinal classes are a useful simplification of cortical interneuron diversity, but such broad subgroupings gloss over the molecular, morphological, and circuit specificity of interneuron subtypes, most notably among the somatostatin interneuron class. While there is evidence that this diversity is functionally relevant, the circuit implications of this diversity are unknown. To address this knowledge gap, we designed a series of genetic strategies to target the breadth of somatostatin interneuron subtypes and found that each subtype possesses a unique laminar organization and stereotyped axonal projection pattern. Using these strategies, we examined the afferent and efferent connectivity of three subtypes (two Martinotti and one non-Martinotti) and demonstrated that they possess selective connectivity with intratelecephalic or pyramidal tract neurons. Even when two subtypes targeted the same pyramidal cell type, their synaptic targeting proved selective for particular dendritic compartments. We thus provide evidence that subtypes of somatostatin interneurons form cell-type specific cortical circuits.
Keywords: cortex, interneurons, somatostatin, subtypes, pyramidal neurons, reciprocal connectivity, laminar specificity, optogenetics, monosynaptic rabies tracing, intratelencephalic, pyramidal tract, spatial transcriptomics
INTRODUCTION
The astonishing computational processing capacity of the mammalian cerebral cortex relies on the intricate connectivity between its two fundamental cell types, the glutamatergic excitatory neurons and GABAergic inhibitory interneurons. The layers of the cerebral cortex have long been recognized as being organized into a well-ordered circuitry comprised of distinct excitatory neuronal types across layers. It is less apparent whether cortical interneurons follow the same laminar organizational principles as the excitatory neurons, in part because of the absence of an obvious prescribed laminar distribution. Instead, diversity within cortical interneurons is primarily categorized by the expression of molecular markers and their targeting of distinct subcellular compartments1,2. Thus, cortical interneurons can be coarsely grouped into four major cardinal classes expressing parvalbumin (Pvalb), somatostatin (Sst), vasoactive intestinal peptide (Vip), or lysosome-associated membrane protein (Lamp5) genes3. These four classes show relatively stereotyped targeting of subcellular compartments, somas, dendrites, or axons, and are generally attributed to serving largely non-overlapping circuit functions: feedforward inhibition, feedback inhibition, disinhibition, and “bulk” slower inhibition2,4.
Here we focus on SST-expressing cortical interneurons that have previously been hypothesized to provide non-specific feedback inhibition to pyramidal neuron dendrites2,5–7. Despite this, emerging evidence suggests that diversity within SST interneurons allows them to function in a more specific manner. Previous work examining the biophysical properties, morphology, and molecular markers has described at least three SST interneuron subtypes. The majority of SST interneurons are Martinotti cells, defined by an axonal plexus in L1, which can be further divided based on their morphology into fanning-out Martinotti cells with axons that ramify in both L2/3 and L1, and T-shaped Martinotti cells that ramify in L1 alone8. In addition, there exists a population of non-Martinotti cells that target L4 instead of L18–11. Moreover, in vivo functional studies have shown that infragranular SST interneuron subtypes exert layer-specific control of sensory processing8,12. Finally, recent advances in single-cell genomics have rapidly expanded our knowledge of interneuron transcriptomic diversity and unraveled additional SST interneuron subtypes3,13–17. However, which properties best connote meaningful functional diversity is still a matter of debate. For example, in an effort to link transcriptionally-defined clusters (T) with historical classifications based on electrophysiology (E) and morphology (M), recent studies have utilized Patch-seq to collect and reconcile information on all three parameters from single neurons to define so-called MET types18–21. Although this work made a direct and concerted effort to unify the various parameters that distinguish interneuron subtypes, it is unclear how the three features used for MET analysis relate to the functionality of the cell types that emerge from these classifications.
In this study, using SST interneurons as an exemplar, we propose connectivity as an organizing principle that synthesizes noisy cellular features into meaningful cell types. To test this hypothesis, we divided SST interneurons into eight transcriptomic subtypes (nine if one includes the CHODL type, representing SST-expressing long-range projecting neurons) and designed various genetic strategies for selectively targeting these different SST subtypes. Using a combination of spatial transcriptomics, morphological reconstructions of sparse-labeled neurons, single-molecule fluorescent in situ hybridization (smFISH), and electrophysiology, we validated that these subtypes represent the totality of SST interneurons in primary somatosensory and visual cortices. Furthermore, these characterizations revealed that each SST subtype possesses a unique laminar organization and stereotyped axonal projection pattern. To test whether the subtype-specific organization reflected discrete circuit motifs within local cortical networks, we used optogenetics to map the efferent connectivity of three major SST subtypes that are distributed in different layers onto local excitatory neurons in V1. Intriguingly, each SST subtype had a distinct intralaminar or translaminar targeting pattern, as well as cell-type selective targeting of L5 pyramidal neurons and PV interneurons. Afferent mapping of two infragranular SST subtypes using monosynaptic rabies tracing provided further support that cell-type selective connectivity between different SST subtypes and excitatory neuron cell types is likely reciprocal. Finally, synaptic puncta analysis of two SST subtypes that innervate the same excitatory neuron cell type revealed that while they share a common efferent target, at the synaptic level, they likely gate distinct dendritic inputs. Our data demonstrate that SST interneurons can be divided into discrete subtypes that selectively contribute to cell-type specific circuits within the cortex. Taken together, this reveals an unanticipated precision of cortical interneurons in regulating the flow of excitation of cortical pyramidal cells.
RESULTS
SST interneurons subtypes are organized in layers
To assess the transcriptomic diversity of cortical SST interneurons, we took advantage of a single-nuclei RNA sequencing (snRNA-seq) dataset of cortical interneurons from mouse anterior lateral motor cortex (ALM) and primary visual cortex (V1) at postnatal day (P) 2822. The use of snRNA-seq prevents stress-sensitive artifacts in gene expression and the selective loss of particular cell types during fluorescence-activated cell sorting (FACS) that occur in whole-cell sequencing23. This dataset utilized a Dlx5/6-Cre driver line, which enriches for all cortical interneurons and allows for the collection of the breadth of interneuron subtypes in accordance with their relative abundances. Interneurons derived from the medial and caudal ganglionic eminences (MGE and CGE, respectively) are clearly separated into distinct branches as visualized in a Uniform Manifold Approximation and Projection (UMAP) plot (Figure 1A, inset). Based on our data, SST interneurons can be initially divided into nine different subtypes, and an additional CHODL type, which corresponds to nNos-expressing long-range projecting neurons15,16,24. Canonical correlation analysis (CCA) showed that this division closely matches with the supertypes of SST interneurons, as described in the recently published taxonomy of transcriptomic cell types of the isocortex and hippocampal formation 3. Therefore, with minor adjustments (e.g. combining two subtypes SST-Lpar1 and SST-Esm1 to SST-Nmbr; see additional information on https://fishelllab.hms.harvard.edu/publications), we were able to adhere to the current nomenclature utilized by Allen Institute in classifying SST interneurons into eight subtypes and CHODL type (Figure 1A), each of which possesses distinct marker genes (Figure 1B).
Figure 1. Spatial transcriptomic analysis reveals the laminar organization of eight SST interneuron subtypes.
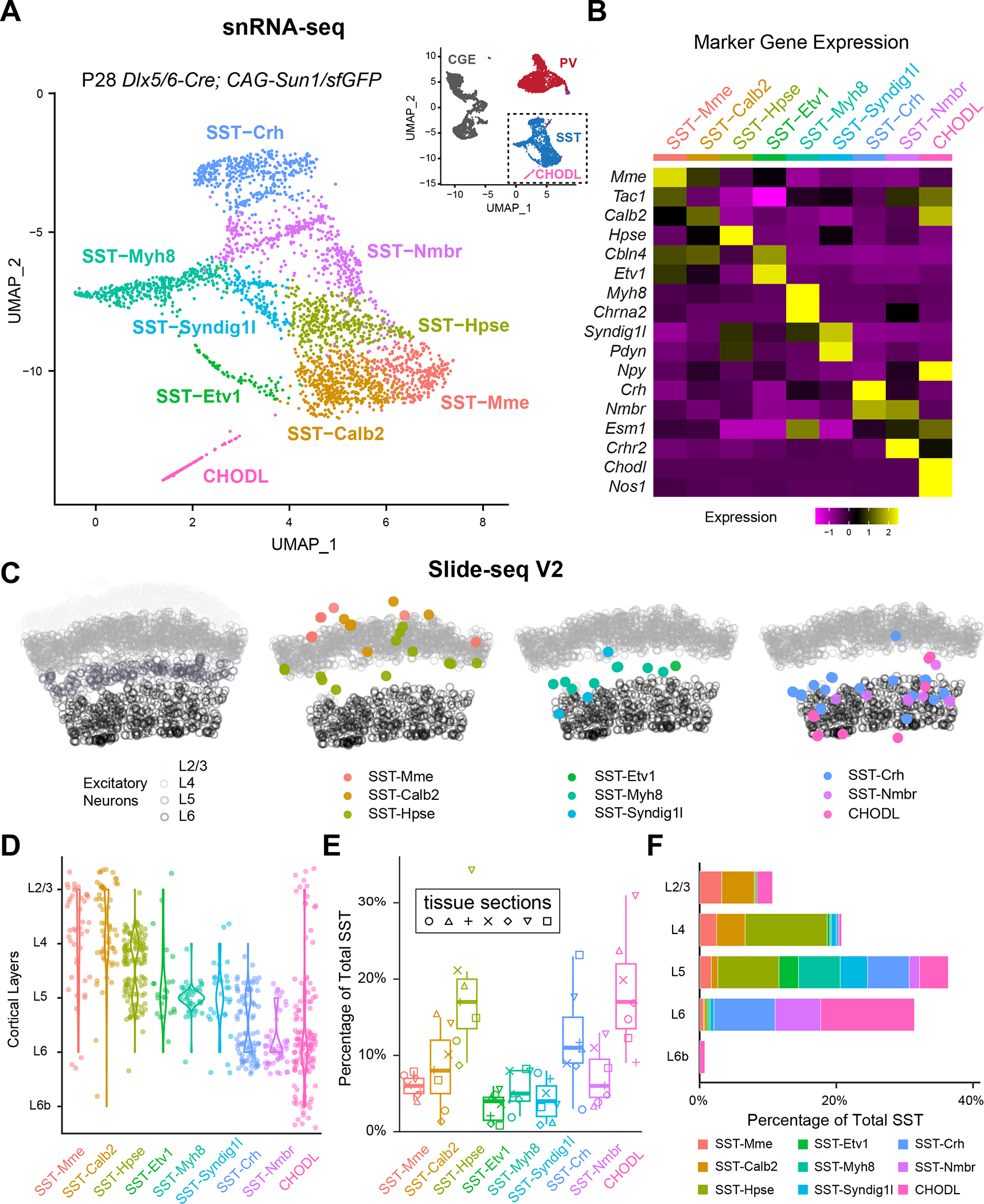
(A) UMAP visualization of snRNA-seq of P28 cortical interneurons22, illustrating eight SST subtypes and the CHODL subtype. Inset showing the UMAP of the entire dataset. CGE, caudal ganglionic eminence.
(B) Heatmap showing the scaled expression of marker genes for each SST subtype based on snRNA-seq data.
(C) Robust cell type decomposition (RCTD) assignment of spatial clusters to different SST subtypes on a representative Slide-seq V2 experiment based on a scRNA-seq reference (see Methods). Gray circles represent the location of excitatory neurons in different layers for reference.
(D) Violin plots demonstrate the laminar distribution of different SST subtypes identified in Slide-seqV2 experiments (n = 7 tissue sections, 4 mice).
(E) Boxplot showing the proportion of different SST subtypes out of 525 total SST interneurons identified.
(F) Bar plot showing the proportion of different SST subtypes identified across different cortical layers.
See also Figures S1–2, Tables S1–2.
To investigate the laminar distribution of these SST interneuron subtypes, we performed Slide-seq V2 experiments on the primary somatosensory cortex (S1) of ~1-month-old mice25. With reference to scRNA-seq data3, we used robust cell type decomposition (RCTD)26 to detect the spatial distribution of different excitatory neuron cell types (Figure S1A,B) and the locations of each SST interneuron subtype (See Methods). Interestingly, each SST subtype had a stereotyped laminar distribution: SST-Mme, SST-Calb2 are mainly found in upper layers; SST-Hpse reside in L4 and L5a; SST-Etv1, SST-Myh8, and SST-Syndig1l are all located in L5; and SST-Crh, SST-Nmbr, and CHODL are preferentially located within L6 (Figure 1C–D, S1C). To complement these observations, we performed single molecule fluorescent in situ hybridization (smFISH) against different marker genes for various SST subtypes in both S1 and V1 (Table S1). The laminar distribution of these marker genes confirms the Slide-seq V2 results, indicating that within these sensory cortices, specific SST subtypes reside in different cortical layers (Figure S2). These results also allowed us to estimate the relative proportion of different SST subtypes across different cortical layers (Figure 1E–F). In general, the results from Slide-seq V2 and smFISH agree well with each other. For instance, SST-Calb2 and SST-Crh subtypes were estimated to comprise ~10% of the total SST interneuron population in S1 by both Slide-SeqV2 (Figure 1E) and smFISH (Figure S2). However, we did notice that Slide-seq V2 over-estimated the proportion of the CHODL cells in S1, based on prior studies27. In addition, the proportion of different SST subtypes in V1 can be estimated by the relative abundance in our snRNA-seq dataset (Table S2) or by smFISH (Figure S2) and are in concordance. Most of the SST subtypes are similar across these two sensory cortices, except that V1 contains a higher proportion of SST-Calb2 than S1 (~19% in V1, ~9% in S1) as estimated by smFISH (Figure S2). This correlates with the observation that the majority of the L4 SST interneurons in S1 belongs to SST-Hpse subtype (~74% of SST-Hpse, ~9% of SST-Calb2), while L4 of V1 is comprised by both SST-Calb2 (~35%) and SST-Hpse (~50%) (Figure S2), which has been previously shown by Patch-Seq of L4 SST interneurons in S1 and V1 respectively28.
Genetic targeting of different SST subtypes reveals stereotyped axonal projection patterns
Based on marker gene expression, we designed direct and intersectional genetic strategies to target either one or a combination of multiple SST interneuron subtypes (Table S3). These genetic strategies revealed the distinct laminar organization and axonal projection patterns of different SST subtypes that were largely consistent between S1 and V1 (Figure 2A, S3A). One exception to this trend is the SST-Calb2 subtype. The intersectional strategy of Calb2Cre;SstFlpO primarily targets SST-Calb2 subtype in L2/3 and L5a, but not in L4 of S1 (Figure 2A). By comparison, the same genetic strategy showed that the SST-Calb2 subtype in V1 is distributed throughout L2/3 to L5a (Figure S3A), which is consistent with the smFISH results (Figure S2) and a previous publication (Scala et al., 2019). Instead, L4 of S1 is primarily populated by the SST-Hpse subtype, which can only be targeted by injection of recombinant adeno-associated viruses (AAVs) driving Cre recombinase-dependent (Cre-ON) expression of reporter protein under a Dlx enhancer, after the age of P7 (due to germline expression of Hpse gene and the postnatal onset of Hpse expression in SST interneurons) 29. SST-Hpse interneurons have extensive axons that arborize within L4, which results in the striking labeling of barrel fields in S1. Single-cell reconstruction confirms that the axons of SST-Hpse primarily target L4 in both S1 and V1, often with a collateral to L1. In S1, the axon of one SST-Hpse interneuron can fill an entire barrel field (Figure 2B, S3B). Therefore, the SST-Hpse subtype is an L4-targeting non-Martinotti cell that resides in L4 and L5a of both sensory cortices8,11,12,20,28. As an alternative to targeting with a viral strategy, the PdynCreER;NpyFlpO Cre-ON/Flp-ON intersectional strategy can also be used to target SST-Hpse subtype, although this strategy also labels a subset of SST-Calb2 interneurons. In addition, by crossing this compound allele with a Cre-ON/Flp-OFF reporter line, one can selectively target the SST-Syndig1l subtype, whose morphology corresponds to L5a T-shaped Martinotti cells (Figure 2, S3) 8,10. Another SST subtype that resides in L5a is the SST-Etv1 subtype, which resembles a fanning-out Martinotti shape and can be partially targeted using an Etv1CreER;SstFlpO intersectional strategy. Within L5b and 6, the Chrna2-Cre allele can be used to target the SST-Myh8 subtype, which also exhibits a T-Shaped Martinotti morphology (Figure 2, S3), as previously described30.
Figure 2. Genetically targeted SST subtypes showed stereotypical laminar distribution and morphology.
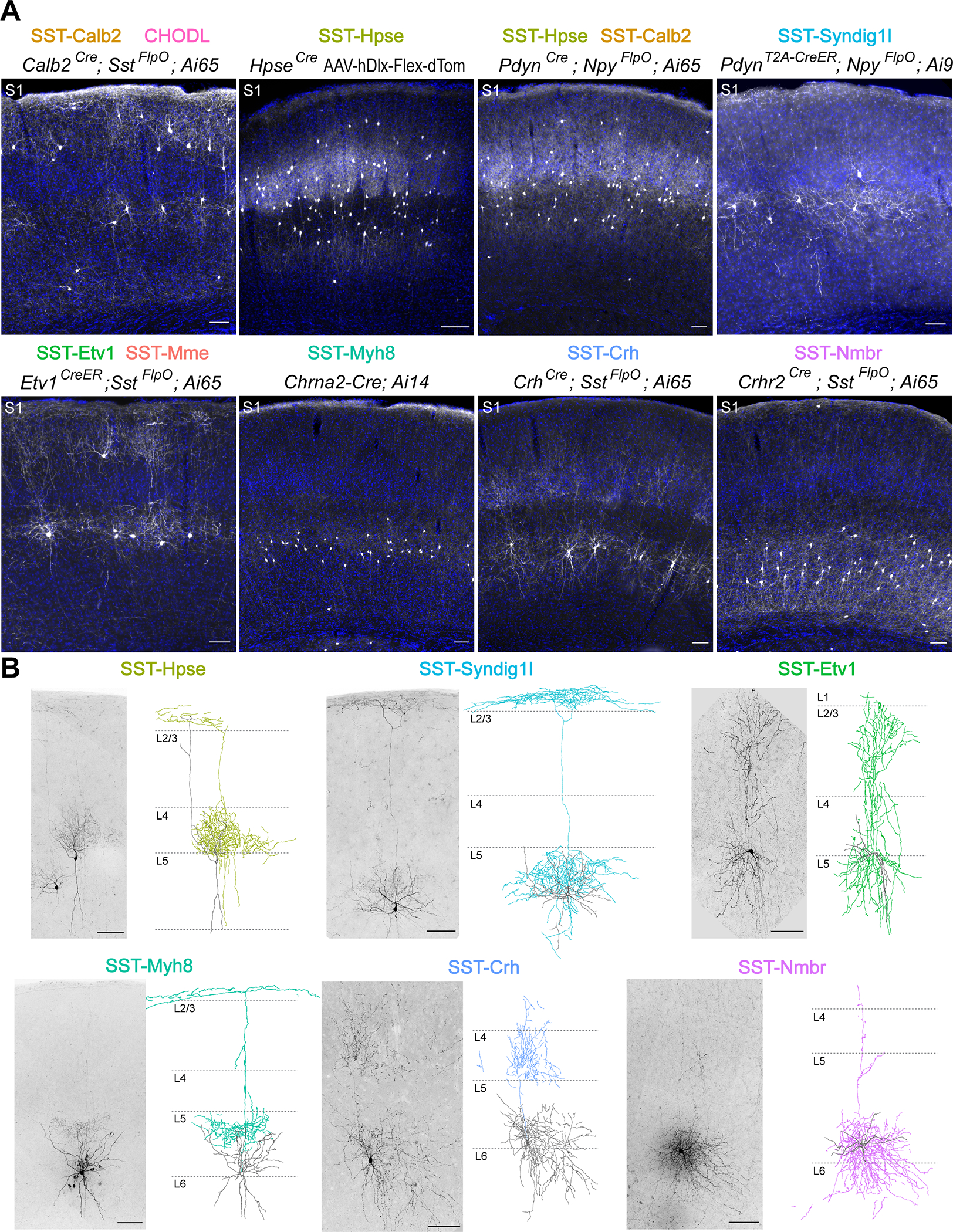
(A) Representative images of genetically targeted SST subtypes in S1, counterstained with DAPI for visualization of laminar distribution. All images were taken from 1–3 month old mice. Ai9 reporter line is used here as a Cre-ON/Flp-OFF strategy because the FRT sites flanking the LoxP cassette are retained in this mouse line52. SST-Hpse interneurons were occasionally observed in PdynT2A-CreER; NpyFlpO; Ai9 strategy, likely due to incomplete FlpO recombination, though not noted in this representative image. For labeling SST-Hpse subtype, rAAV9-hDlx-Flex-dTomato virus was stereotaxically injected in HpseCre mice in S1 at 1 month old and examined 13 days post-injection. Note that Etv1CreER; SstFlpO intersectional strategy may partially target SST-Calb2 subtype (Figure S5B) though not obvious in this example. Scale bars, 100 μm.
(B) Sparse labeling and Neurolucida reconstructions of selective SST subtypes in S1. Images of genetically labeled or biocytin-filled SST interneurons are shown to the left of the Neurolucida reconstruction of single-neuron morphology. SST-Etv1 interneurons are labeled by Etv1CreER; SstFlpO; RC::FPSit genetic strategy. SST-Hpse and SST-Syndig1l interneurons are both labeled by PdynT2A-CreER; Ai14 strategy and differentiated by their unique morphology. SST-Crh interneurons are labeled by CrhCre; SstFlpO; RC::FPSit. SST-Myh8 and SST-Nmbr are both labeled by biocytin-filling. All reconstructions were performed using P25–73 mice. Scale bars, 100 μm.
See also Figures S3–5, Tables S3–6.
Little has been previously reported about SST interneuron diversity in L6. We identified two strategies for targeting two L6 SST subtypes, each of which has distinct features. The CrhCre;SstFlpO intersectional strategy targets the SST-Crh subtype, which are L4-targeting non-Martinotti cells that reside within L5b and L612. The Crhr2Cre;SstFlpO intersectional strategy targets the SST-Nmbr subtype that resides almost exclusively in L6. The axons of these cells remain primarily in deep layers despite occasionally extending thin collateral towards L1, suggesting that they are also non-Martinotti cells (Figure 2, S3). This morphology was also observed in previously published single-cell reconstructions of L6 SST interneurons20. For SST subtypes not highlighted here, we have included a list of genetic targeting strategies (Table S4) and a summary of our current understanding of the putative SST subtypes targeted using each genetic approach (Table S4). Images of additional genetic strategies and raw images of sparse labeling of SST interneurons are available on public domains (https://doi.org/10.7910/DVN/NQDIPG). Based on the transcriptomic clusters (Table S4), our sparse labeling in general agrees with the published single-neuron reconstructions of the corresponding transcriptomic clusters20.
To assess the completeness and coverage of these genetic strategies, we performed smFISH against Sst mRNA for quantification of genetic labeling in S1 and V1 (Figure S4, S5). In general, most genetic strategies label the expected proportion of SST subtypes. For example, the Calb2Cre;SstFlpO strategy labels ~22% of total SST interneurons in V1 (Figure S4), which is consistent with the prevalence of the SST-Calb2 subtype as estimated by snRNA-seq (~20% of SST interneurons in V1, Table S2) and smFISH (~19%, Figure S2). In addition, consistent with that mentioned above, the Calb2Cre;SstFlpO strategy labels ~8% more SST interneurons in V1 compared to S1, in accordance with their greater abundance in the former. Similarly, the Crhr2Cre;SstFlpO strategy labels ~11% of total SST interneurons in V1 (Figure S4) and thus correlates well with the size of the SST-Nmbr population, which by snRNA-seq is estimated to be ~13% of total SST interneurons in this area (Table S2). However, the CrhCre;SstFlpO genetic targeting strategy labels only ~2–3% of total SST interneurons in both S1 and V1 (Figure S4), while the SST-Crh subtype is estimated to comprise >10% of total SST interneurons by Slide-seq V2 (Figure 1), snRNA-seq (Table S2) and smFISH (Figure S2). This is likely due to the ineffectiveness of genetic targeting caused by relative low expression of the Crh gene in SST-Crh subtype.
To evaluate the specificity of four of the genetic strategies, we performed smFISH experiments using selected marker genes. Overall, each genetic strategy showed the expected expression pattern of the selected marker genes in each of the targeted SST subtypes (Figure S5). For example, both Chrna2-Cre and Crhr2Cre;SstFlpO labeled SST interneurons with low levels of Calb2 and Hpse transcripts (Figure S5). However, the majority of the marker genes are not binary classifiers for each SST subtype. Instead, they showed graded expression across the examined SST subtypes (Figure 1B, Table S1). Notably, due to the high sensitivity of the smFISH method, a low level of transcripts is often detected. For example, Pdyn gene is expressed at a low level in the SST-Calb2 subtype (Table S1), resulting in a high percentage of Pdyn+ Calb2Cre;SstFlpO labeled SST interneurons (Figure S5A). Therefore, a thorough characterization of the specificity and coverage of each of the genetic strategies utilized would require a quantitative analysis involving smFISH multiplexing of upwards ~20 genes. A further caveat associated with strategies involving tamoxifen-dependent labeling is that different proportions of SST subtypes are labeled in specific experiments, depending on the recombination efficiency, as a result of the graded expression within the targeted populations. For example, immunostaining against Calretinin (the protein product of the Calb2 gene) suggested that the Etv1CreER;SstFlpO intersectional strategy, in addition to targeting SST-Etv1 subtype, also labels some SST-Mme interneurons (that expresses a low-level of Calb2) (Figure S5B), due to the graded expression of Etv1 gene in different SST subtypes.
A notable outcome of our genetic analysis was that it revealed that each SST transcriptomic subtype has a stereotypical laminar location and an associated axonal projection pattern (see also Gouwens et al., 202020). We likewise wondered whether they exhibited stereotyped electrophysiological properties. To test this, we decided to focus on three major SST interneuron subtypes that tiled the cortical column: the SST-Calb2, the SST-Myh8, and the SST-Nmbr subtypes, targeted with the Calb2Cre;SstFlpO, Chrna2-Cre, and Crhr2Cre;SstFlpO alleles, respectively. These three SST subtypes each reside in different cortical layers and have distinct morphologies. SST-Calb2 interneurons are fanning-out Martinotti cells found in L2/3 to L5, SST-Myh8 interneurons are T-shaped Martinotti cells concentrated in L5b, and SST-Nmbr interneurons are L6 non-Martinotti cells whose axons primarily arborize extensively in deep layers (Figure 2) 20,30. Previous studies have characterized three major electrophysiological profiles for SST interneurons: adapting regular spiking, quasi-fast spiking, and low-threshold spiking (LTS) 9–12,24. These electrophysiological types correlate with previously described SST subtypes in several transgenic lines (GIN, X94, and X98, respectively), as well as morphological parameters 9–11, but recent efforts to link transcriptomic clusters with electrophysiology have found significant variability across transcriptional types 20,28. To address whether these SST subtypes have particular biophysical identities, we analyzed 11 electrophysiological parameters from genetically labeled SST-Calb2, SST-Myh8, and SST-Nmbr interneurons (Table S5). To ensure our results are comparable with previous studies, we restricted our analysis to S1. Overall, all three subtypes displayed regular-spiking firing patterns with adaptation (Figure S3C). While SST-Myh8 interneurons displayed rebound burst firing, they did not have the characteristic high input resistance, low action potential (AP) threshold, or high adaptation index of low threshold spiking (LTS) cells. We trained a k-nearest neighbor classifier on our dataset and found that SST-Calb2 and SST-Nmbr interneurons were predicted with >80% accuracy, but SST-Myh8 interneurons were mixed with SST-Calb2, likely due to rebound firing in some SST-Calb2 cells (Figure S3B,C). SST-Nmbr interneurons were primarily distinguished by their higher firing frequency (Figure S3C). Notably, SST-Calb2 interneurons are found in both L2/3 and L5a in S1. To address whether SST-Calb2 interneurons are a continuum of one cell type or two distinct cell types in different layers, we compared the intrinsic electrophysiological properties of SST-Calb2 interneurons in these two layers. We found that across most parameters, SST-Calb2 interneurons in L2/3 were indistinguishable from those in L5a, with the exception that L5a SST-Calb2 interneurons were slightly more adapting (Table S6). In summary, despite their distinct electrophysiological properties, the three SST subtypes are better discerned by other features such as laminar location, morphology and axonal projection patterns.
Laminar positioning of SST subtypes partially predicts their output connectivity
Given that SST-Calb2, SST-Myh8, and SST-Nmbr are organized in distinct cortical layers, we hypothesized that they may form laminar-specific circuits. To test this, we used our genetic strategies (SstCre;SstFlpO for pan-SST cells, Calb2Cre;SstFlpO for SST-Calb2, Chrna2-Cre;SstFlpO for SST-Myh8, and Crhr2Cre;SstFlpO for SST-Nmbr) in combination with the intersectional reporter line Ai80, which allows for recombinase-mediated expression of a channelrhodopsin variant, CatCh, in all SST interneurons or a particular SST subtype, accordingly (Figure 3A) 31. Using this approach, we performed optogenetic-assisted circuit mapping experiments in V1.
Figure 3. Laminar positioning correlates with SST subtype innervation.
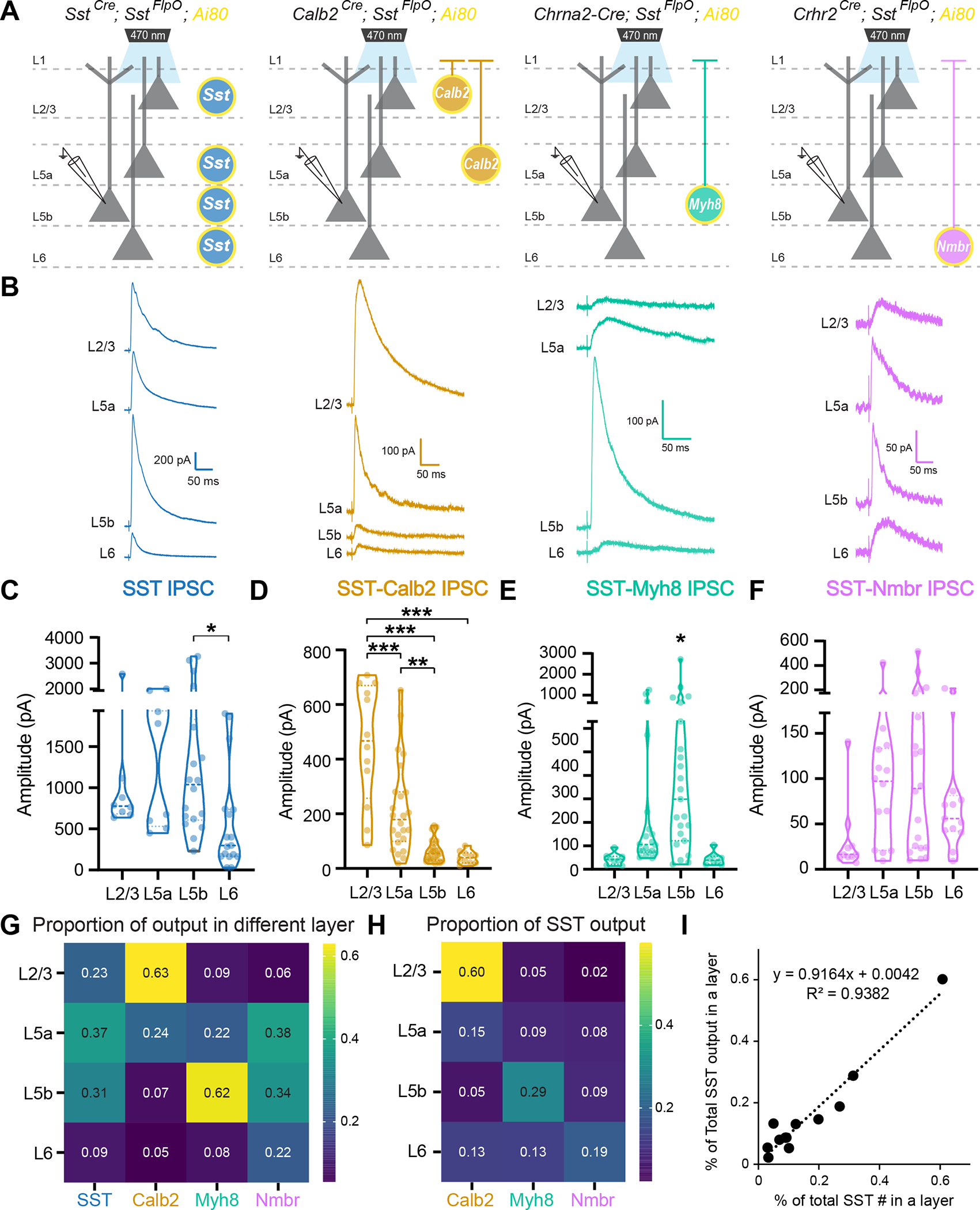
(A) Recording scheme. Pan-SST interneurons or three SST subtypes, SST-Calb2, SST-Myh8, SST-Nmbr, were genetically targeted to express CatCh by crossing with the Ai80 reporter line. Postsynaptic IPSCs were recorded from pyramidal neurons across layers in response to 1 ms light stimulation.
(B) Example average traces from pyramidal neurons across layers in response to pan-SST stimulation (left) and individual SST subtypes (three right panels).
(C-F) Violin plot of the evoked IPSC amplitude upon stimulation of pan-SST interneurons or SST-Calb2, SST-Myh8, SST-Nmbr interneurons.
(G) Heatmap of the ratio of median evoked IPSC amplitude for pan-SST interneurons or for individual SST subtypes across layers. Data were normalized across columns, where the value represents the ratio between the median evoked IPSC amplitude in a particular layer compared to the summed median IPSC amplitude of that SST subtype across layers.
(H) Heatmap of the proportion of inhibition from individual SST subtype as compared to the inhibition from pan-SST interneurons in different layers.
(I) Plot showing that percentage of individual SST subtype out of the total number of SST interneurons found in a particular layer (x-axis) is correlated with the proportion of the inhibitory output by individual SST subtype out of pan-SST interneuron response in that layer (y-axis).
We first examined the output of SST interneurons as a general class and found that SST interneurons strongly inhibit all layers, with the smallest response in L6 (Figure 3B–C). Note that the level of inhibition does not correlate with the abundance of SST interneurons found in each layer (Figure S4, S6E). Compared with the other layers, L6 seems to receive disproportionately less inhibition from SST interneurons (Figure S6E), suggesting that L6 excitatory neurons either receive less innervation or form weaker synapses with SST interneurons compared to excitatory neurons in other layers 32. We then tested whether individual SST subtypes also selectively target specific cortical layers. We found that corresponding with their laminar positioning, SST-Calb2 interneurons primarily innervate L2/3 and L5a (Figure 3B,D). SST-Myh8 interneurons likewise innervated their resident layer L5b, as well as pyramidal cells within the adjacent L5a layer (Figure 3B,E). Surprisingly, SST-Nmbr interneurons did not show preferential laminar targeting despite their cell bodies being mostly restricted to L6 (Figure 3B,F). The median evoked IPCS amplitude from SST-Nmbr subtype is larger in L5, as compared to L6 (Figure 3G), consistent with L6 excitatory cells receiving less SST-mediated inhibition than other layers. As a general trend, when we examined the contribution from each SST subtype in proportion to the pan-SST output, as measured in the soma, each subtype consistently contributes most to the overall inhibition of its resident layer (Figure 3H, Figure S6A–D). In fact, the percentage of each SST subtype found in each layer correlates well with the portion of their contribution to the total inhibitory output by SST interneurons in that layer (Figure 3I). However, the strength of inhibition was not distributed equally across each SST subtype. SST-Calb2 subtype tends to form stronger inhibition as compared to the other two subtypes (Figure S6F). This could be due to differences in the strength of the inhibitory synapses formed, the number of synapses per cell, or the receptors on the postsynaptic neuron.
Notably, compared to evoked IPSCs from pan-SST stimulation, all three subtypes evoked significantly lower responses (Figure S6A–D). To assess the combined contribution of these three SST subtypes to the pan-SST response, we compared simulated linear combinations of each subtype with the median pan-SST response for a particular cortical layer using a hierarchical bootstrapping method (see Methods). We found that the combined evoked IPSC amplitude from SST-Calb2, SST-Myh8, and SST-Nmbr was smaller than the pan-SST response in 80–85% of the trials across all layers (Figure S6E). To estimate the proportion of the pan-SST response that could be attributed to the output of these three subtypes, we took the ratio of a linear combination of each subtype compared to a pan-SST evoked amplitude (Figure S6F). The highest proportion was in L2/3 pyramidal neurons with a median ratio of 70.05%, followed by L6 at 58.39%, L5a at 46.41%, and L5b at 52.79%. This is not surprising given that the combination of these three SST subtypes constitutes ~46% of all SST interneurons, and depending upon the layer, varies from 40–60% in their relative abundance (Figure S6H).
Taken together, these results suggest that in aggregate each SST subtype contributes most to its resident layer, although some subtypes may target cells in other layers more strongly. It is particularly intriguing that Martinotti cells such as SST-Calb2 and SST-Myh8, despite their axons being largely restricted to L1, can still selectively target excitatory neurons in their resident layer. This suggests that there is a mechanism for SST interneurons to recognize the dendrites of pyramidal cells whose soma they are proximal to.
SST subtypes selectively target IT and PT neurons within L5
All three SST subtypes examined innervate L5, which contains two major types of pyramidal neurons: intratelencephalic (IT) neurons that project within the cortex and striatum; and pyramidal tract (PT) neurons that extend their axon subcerebrally to several targets including the tectum, brainstem, and spinal cord. We therefore wondered whether these SST subtypes selectively innervate specific pyramidal neuron types within the same layer. To address this, we injected rAAV2-retro-hSyn-mScarlet into either the ipsilateral retrosplenial cortex (Rs) or superior colliculus (SC) to retrogradely label IT and PT neurons (cortico-tectal neurons to be specific) in V1, respectively (Figure 4A). We then recorded optogenetically evoked IPSCs from virally labeled IT and PT neurons in L5 using the same genetic strategy for targeting SST subtypes (Figure 4B).
Figure 4. SST subtypes differentially target IT vs. PT pyramidal neurons in L5.
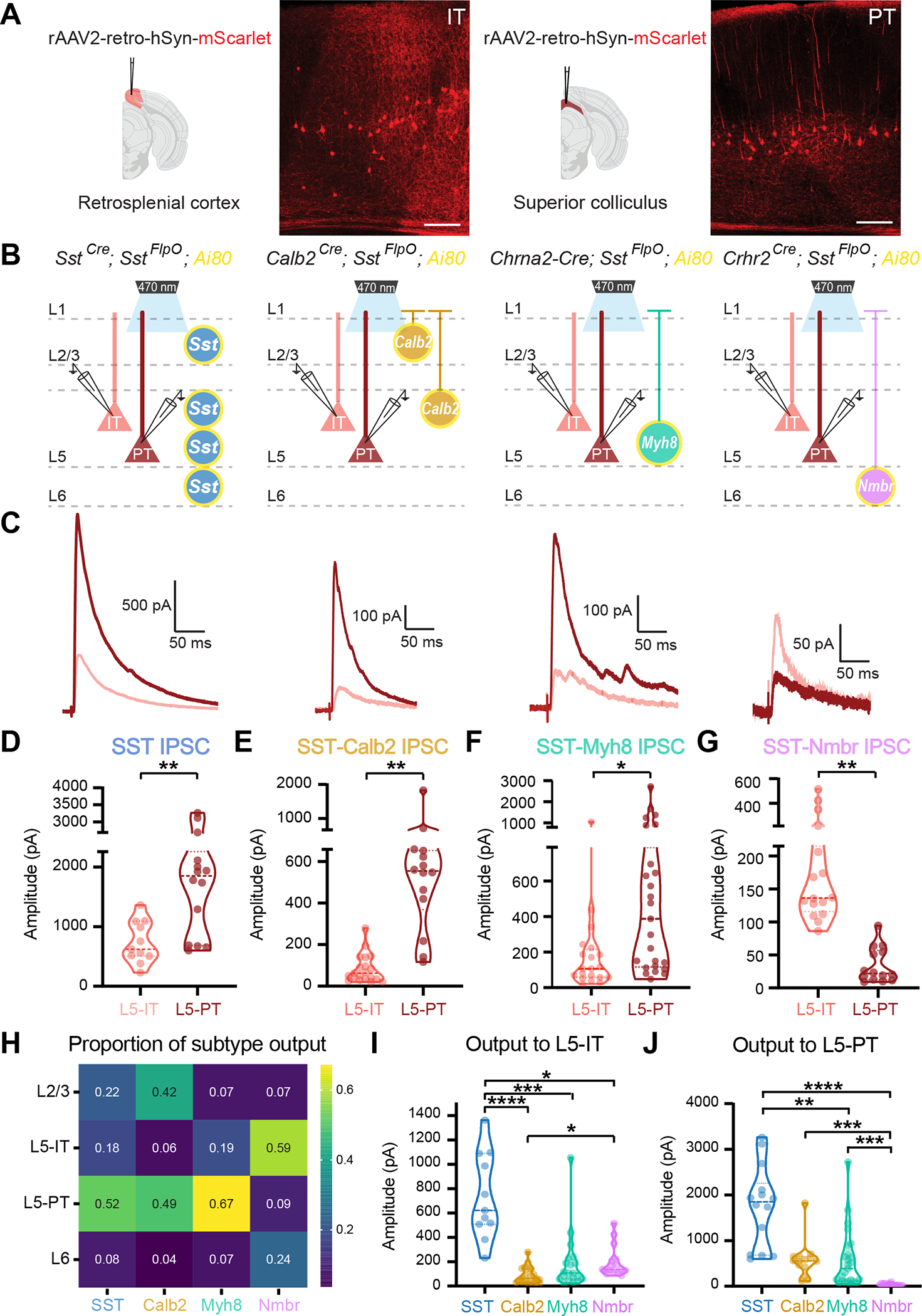
(A) Strategy for targeting IT and PT pyramidal neurons by injecting rAAV2-retro-hSyn-mScarlet into the retrosplenial cortex (Rs) or superior colliculus (SC), respectively. Representative images of mScarlet-labeled IT and PT neurons. Scale bars, 100 μm.
(B) Recording scheme. Pan-SST interneurons or three SST subtypes, SST-Calb2, SST-Myh8, SST-Nmbr, were genetically targeted to express CatCh by crossing with the Ai80 reporter line. Postsynaptic IPSCs are recorded from IT or PT neurons in response to 1 ms light stimulation.
(C) Representative average traces of evoked IPSC in IT (pink) and PT (red) neurons upon stimulation of pan-SST, SST-Calb2, SST-Myh8, and SST-Nmbr interneurons.
(D-G) Violin plot of evoked IPSC amplitude upon optogenetic stimulation of pan-SST interneurons, SST-Calb2, SST-Myh8 or SST-Nmbr interneurons in L5-IT and L5-PT neurons.
(H) Heatmap of the proportion of inhibition from individual SST subtype as compared to the inhibition from pan-SST interneurons in different layers and pyramidal neuron cell types.
(I-J) Violin plot of evoked IPSC amplitude in L5-IT or L5-PT pyramidal neurons.
We first tested the efferent connectivity of pan-SST interneurons to IT and PT neurons and found that SST interneurons strongly inhibit both types but have stronger output onto PT neurons (Figure 4C–D). Upon examination of individual SST subtypes, each one showed a clear innervation bias towards specific L5 pyramidal neuron cell types. Both SST-Calb2 and SST-Myh8 interneurons preferentially target L5-PT neurons, while SST-Nmbr interneurons preferentially inhibit L5-IT neurons (Figure 4C,E–G).
Compared across layers, this time with L5 separated into IT and PT neurons, pan-SST interneurons inhibited all layers and both cell types tested, although as noted above, significantly weaker in L6 (Figure 4H, Figure S6I). By comparison, clear subtype-specific patterns emerged for individual SST subtypes. SST-Calb2 strongly targeted L2/3 and L5-PT neurons, SST-Myh8 primarily targeted L5-PT neurons, and SST-Nmbr preferentially targeted L5-IT neurons (Figure 4H, Figure S6I–L). Note that SST-Calb2 and SST-Myh8 outputs to L5-PT neurons were not significantly different, showing that these two subtypes innervate L5-PT neurons with similar strength at a population level, while the inhibition from SST-Nmbr to L5-PT neurons was negligible (Figure 4J, Figure S6M–N). With regard to L5-IT neurons, outputs from these three SST subtypes are all relatively weak, although, amongst the three, SST-Nmbr was still the strongest (Figure 4I, Figure S6M–N).
To predict the contribution of these three types to the pan-SST inhibition, we repeated the hierarchical bootstrapping and linear combination simulations described above. We found that the combined IPSC simulated responses were 77.19% less in L5-IT cells and 79.49% less in L5-PT cells than observed upon pan-SST stimulation (Figure S6O). To estimate the proportion of the pan-SST inhibition that could be attributed to SST-Calb2, SST-Myh8, and SST-Nmbr outputs, we took the ratio of a linear combination of each subtype compared to a pan-SST evoked amplitude (Figure S6P). The median contribution for L5-IT neurons was 67.10%, and for L5-PT neurons the median contribution was 56.46%, indicating that these subtypes account for approximately two-thirds of the total SST inputs to both L5-IT and L5-PT neurons. However, in both bootstrapping analyses, our assumption that these inputs are linearly summated needs to be further investigated.
SST subtypes differentially inhibit PV interneurons across layers
As we observed a high degree of specificity between three SST subtypes and excitatory neurons, we wondered whether they formed specific connections with inhibitory neurons as well. Previous studies suggest that except for themselves, SST interneurons broadly inhibit all other cardinal classes of interneurons 33. There is already evidence that non-Martinotti SST interneurons innervate L4 PV interneurons more strongly than other SST interneurons 11. To test for selective outputs to PV interneurons from these three SST subtypes, we injected an AAV expressing GFP under the control of a PV-specific enhancer 34 into the various SST subtype-specific driver lines crossed with Ai80. We then proceeded to record from virally labeled PV interneurons in V1 (Figure 5A–B). We found that SST-Calb2 interneurons strongly targeted PV interneurons in the superficial layers, while SST-Myh8 and SST-Nmbr interneurons showed a trend of preferably targeting PV interneurons in infragranular layers but not at all in the superficial layers (Figure 5C–E). To compare this to previous studies demonstrating the innervation of L4 PV interneurons by non-Martinotti cells, we also tested the output of the SST interneurons labeled using the PdynCre;NpyFlpO strategy. This strategy primarily targets the L4-targeting non-Martinotti SST-Hpse subtype, as well as some SST-Calb2 cells. As expected, SST interneurons targeted using this genetic strategy strongly innervated L2/3 and L4 PV interneurons (see additional information on https://fishelllab.hms.harvard.edu/publications).
Figure 5. SST subtypes differentially innervate PV interneurons.
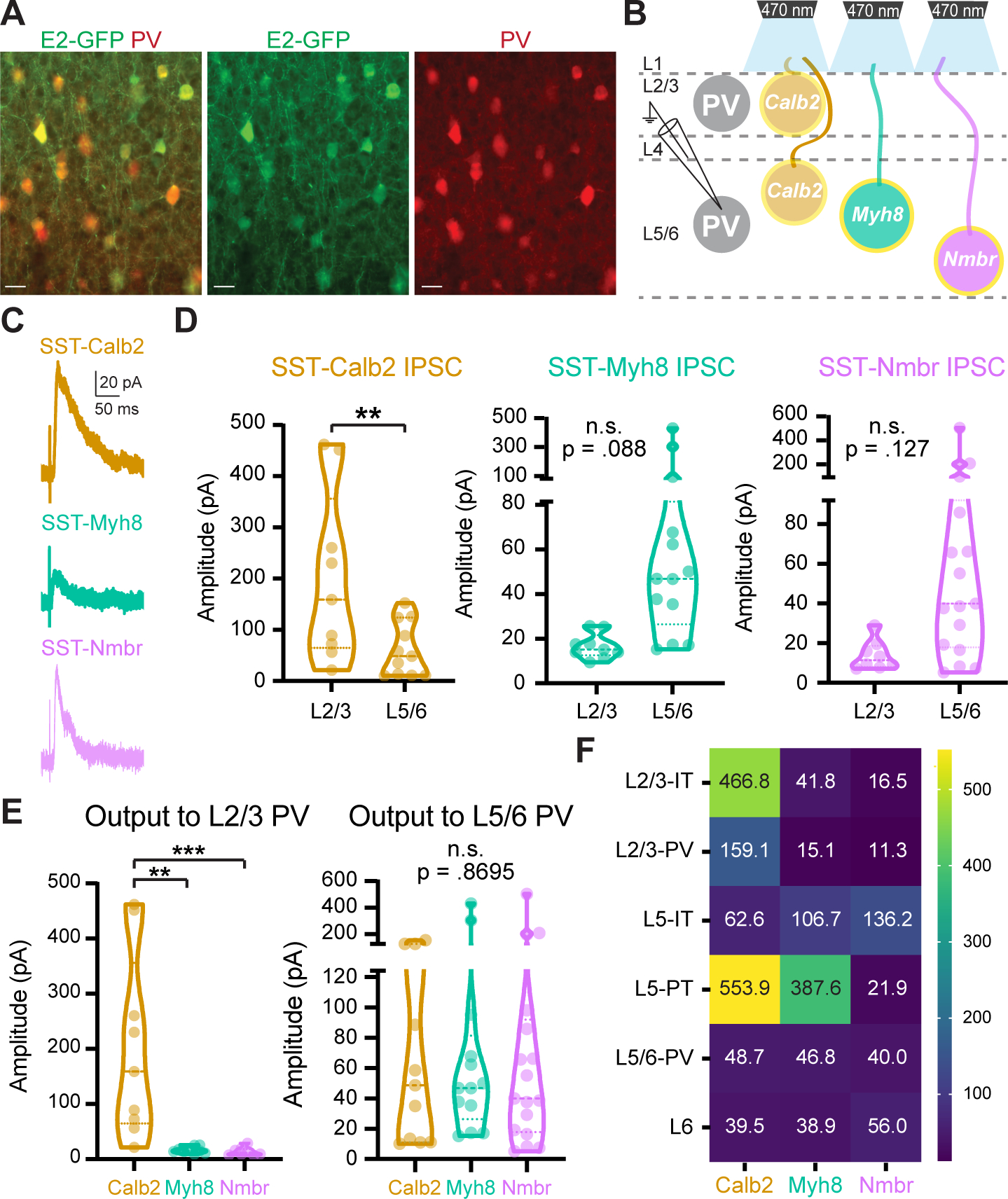
(A) Representative images of E2-GFP injection in V1 labeling PV interneurons. Scale bars, 5 μm.
(B) Recording scheme. SST-Calb2, SST-Myh8, SST-Nmbr interneurons were genetically targeted to express CatCh by crossing with the Ai80 reporter line. IPSCs were recorded from PV neurons in response to 1 ms light stimulation.
(C-D) Representative traces of IPSCs and violin plots of IPSC amplitudes in PV interneurons in response to optogenetic stimulation of different SST subtypes.
(E) Comparison of different SST subtypes output to L2/3 (left) and L5/6 PV interneurons (right).
(F) Heatmap of median evoked IPSC amplitude (pA) from each SST subtype across pyramidal neurons and PV interneurons in different layers.
Statistics in Table S7.
A heatmap of median evoked IPSC amplitude summarizes the selective output patterns of the three SST subtypes across different layers and cell types (Figure 5F). The three SST subtypes we studied proved to have a combination of shared and distinct targets. SST-Calb2 interneurons targeted L2/3 and L5-PT pyramidal neurons and L2/3 PV interneurons and SST-Myh8 interneurons targeted L5-PT neurons. While SST-Nmbr interneurons targeted L5-IT neurons specifically, none of the three subtypes provided strong input to L5-IT or L6 pyramidal neurons as compared to the pan-SST response.
Two infragranular SST subtypes receive reciprocal selective excitatory neuron inputs
As our optogenetic experiments demonstrated that different SST subtypes had selective output connectivity, we wondered whether they also received selective input connectivity. To test this, we performed monosynaptic rabies tracing on two closely positioned infragranular SST subtypes, SST-Myh8 and SST-Nmbr. To restrict starter cells to specific SST subtypes, we utilized AAV-helper viruses that allow Cre-dependent expression of TVA receptor for the infection of EnvA-pseudotyped rabies virus (RV), and G protein for the replication and monosynaptic transport of RV. Specifically, for targeting SST-Myh8 interneurons, we injected AAV-helper viruses (AAV-hSyn-DIO-TVA-GFP-N2cG)35,36 in Chrna2-Cre mice at an early developmental age (P2–5), due to the decreased Chrna2-Cre expression around the third postnatal week. We subsequently injected N2c-RV-mCherry virus at P22–42 for S1 and P56–79 for V1 (Figure 6A). For targeting SST-Nmbr interneurons, we co-injected AAV-helper viruses (AAV-Dlx-DIO-TVA and AAV-Dlx-DIO-GFP-N2cG) with N2c-RV-mCherry in 1–3 month old Crhr2Cre mice within both S1 and V1 (Figure 6A). The use of the mDlx5/6 enhancer 29 in the AAV-helper viruses is necessary for selective targeting SST-Nmbr interneurons because a subset of L2/3 pyramidal neurons also express Crhr2 gene (see additional information on https://fishelllab.hms.harvard.edu/publications). Retrogradely traced presynaptic neurons were examined 10–14 days after RV injection. As expected, GFP-positive starter cells were mainly found in L5b for SST-Myh8 and L6 for SST-Nmbr (Figure S7A–B).
Figure 6. Monosynaptic rabies tracing from two different SST subtypes revealed cell type-specific afferent input.
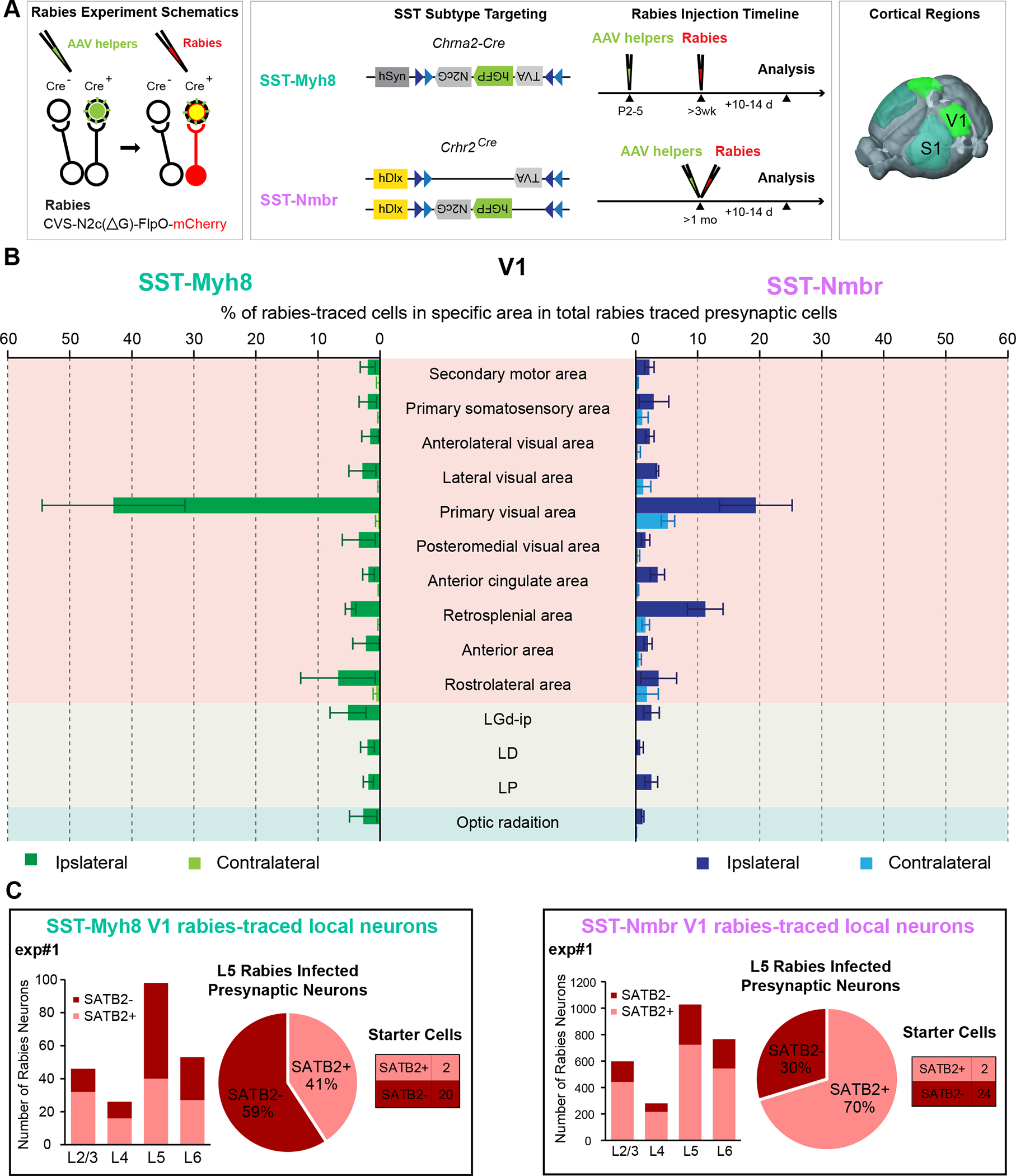
(A) Experimental design of rabies retrograde tracing from two SST subtypes. TVA and N2cG (green) are expressed via AAV helpers, followed by infection and retrograde tracing with rabies virus (red) (left panel). The design of AAV-DIO-helper viruses and the timeline of AAV-helpers and N2cRV injections for tracing from SST-Myh8 (top) and SST-Nmbr interneurons (bottom) using Chrna2-Cre and Crhr2Cre mouse lines, respectively. Rabies tracing patterns were analyzed 10–14 days post-infection (middle panel). The tracing was performed on both SST subtypes from two cortical regions, S1 and V1 (right panel).
(B) Presynaptic inputs to SST-Myh8 and SST-Nmbr interneurons in V1 quantified as the percentage of rabies traced cells in each regional category out of the total number of cells labeled in the brain. The top 10 input regional categories for either SST subtype are included in the plot. (n = 3 mice for each SST subtype). Abbreviations of thalamic regions: dorsal part of the lateral geniculate complex (LGd-ip), lateral dorsal nucleus of thalamus (LD), lateral posterior nucleus of the thalamus (LP).
(C) Quantification of rabies traced local presynaptic neurons in one representative experiment from SST-Myh8 (left) and SST-Nmbr interneurons (right), respectively. SATB2+ neurons are IT neurons, SATB2- neurons are either PT neurons or interneurons. For each experiment, a histogram of rabies traced neurons in each layer (left); a pie chart of the numbers of SATB2+ versus SATB2- rabies infected presynaptic neurons in L5 (middle), and a table shows the number of starter cells (right) are shown. Note that there are occasionally a small number of SATB2+ pyramidal neuron starter cells, due to the challenge of specifically targeting a small interneuron population that only constitutes ~2% of cortical neurons.
We quantified the retrogradely labeled presynaptic neurons and normalized the number to the total amount of rabies traced presynaptic cells. Overall, both SST subtypes primarily received input from local excitatory neurons, other cortical regions, and the corresponding sensory thalamus relative to the site of injection (albeit very few, correlating with weak inputs from the thalamus to adult SST interneurons) (Figure 6B, Figure S7C–D) 36,37. The top 10 brain regions for both SST subtypes combined, which contain almost exclusively cortical regions and the thalamus, could account for >70% of all afferent inputs identified (Figure 6B, Figure S7D). As expected, the topmost afferent region for both SST subtypes is the injection area, suggesting that SST interneurons primarily receive inputs from local excitatory neurons. Intriguingly, one difference noted was that SST-Nmbr interneurons seemed to receive more inputs from the contralateral cortex, while inputs to SST-Myh8 interneurons were almost exclusively from the ipsilateral side (n = 3 for SST-Myh8, n = 3 for SST-Nmbr in V1, Figure 6B; n = 3 for SST-Myh8, n = 2 for SST-Nmbr in S1; Figure S7D). Notably, different AAV-helper viruses were utilized for targeting SST-Myh8 and SST-Nmbr interneurons, and we observed many more rabies traced presynaptic neurons relative to the number of starter cells when tracing from SST-Nmbr interneurons as compared to SST-Myh8 interneurons (Figure S7C). As such, we wanted to confirm that the observed differences in contralateral versus ipsilateral inputs to these two SST subtypes were not experimental artifact. Specifically, we suspected that the larger amount of retrogradely traced presynaptic neurons in SST-Nmbr experiments was likely due to a higher level of G protein expression in SST-Nmbr starter cells, which utilized a more efficient rAAV construct. We therefore repeated one rabies tracing experiment from SST-Myh8 interneurons in S1, using the same AAV-helper viruses used for targeting SST-Nmbr (Figure S7E). Reassuringly, this experiment revealed few contralateral inputs to SST-Myh8 interneurons, despite yielding a larger number of retrogradely traced cells. Given that SST-Nmbr preferentially targets L5-IT neurons, while SST-Myh8 primarily targets L5-PT neurons, the larger fraction of contralateral inputs to SST-Nmbr interneurons could reflect preferred afferent connectivity from IT neurons.
To examine whether these two SST subtypes received inputs from distinct populations of local pyramidal neurons, we performed immunostaining against SATB2, a marker for IT neurons in the mature cortex 38, to determine the identity of the retrogradely traced local input neurons (Figure S8). The laminar distribution of the local inputs to both SST subtypes was very similar. The majority of the presynaptic neurons resided in the infragranular layers and most of them were found in L5 (Figure 6C, Figure S8). This result correlates with output mapping indicating that both SST subtypes preferentially target L5, despite SST-Nmbr interneurons residing primarily in L6 (Figure 3G). This also suggests that these two subtypes receive reciprocal innervations from L5 excitatory neurons. Furthermore, this distribution pattern is consistent between S1 and V1 (Figure S8), suggesting that the selective input and output connectivity we described might be stereotyped microcircuit properties intrinsic to different SST subtypes. Intriguingly, we found that the majority of L5 inputs to SST-Myh8 are SATB2-negative, indicating that they were either L5-PT neurons or interneurons (n = 4/5, S1 and V1 combined, Figure 6C and Figure S8). In contrast, the majority (70–80%) of L5 presynaptic neurons to SST-Nmbr interneurons are SATB2-positive, suggesting that they were L5-IT neurons (n = 4, S1 and V1 combined, Figure 6C and Figure S8). Again, this input connectivity seems to mirror the output connectivity, SST-Myh8 interneurons preferentially projected to L5-PT neurons, while SST-Nmbr interneurons preferentially connected to L5-IT neurons. This suggests that there are reciprocal selective connections between specific SST subtypes and excitatory neurons.
Two Martinotti SST subtypes showed distinct subcellular innervation of L5 PT dendrites
Our results have demonstrated that SST subtypes showed selective input/output connectivity that is both laminar and cell type specific. Of particular interest are L5-PT neurons, which received strong input from both SST-Calb2 and SST-Myh8 interneurons (Figure 4). We therefore wondered whether input from these SST subtypes was functionally redundant, or whether they provided qualitatively distinct forms of inhibition to a common target. Notably, these SST subtypes have distinct axonal morphologies: SST-Calb2 has a fanning-out shape, whereas SST-Myh8 has a T-shape (Figure 2). One possibility is therefore that they could impinge on different subcellular compartments of L5-PT dendrites.
To test this, we quantified the distribution of putative synaptic puncta from SST-Calb2 and SST-Myh8 interneurons onto virally labeled L5-PT basal, oblique, apical branch, and tuft dendrites in V1 (Figure 7A–E). Immunostaining for both the presynaptic marker Gad65 and the postsynaptic marker Gephyrin allowed us to identify putative inhibitory synaptic boutons at the intersection of four fluorescent channels (see Methods). We found that putative SST-Calb2 synapses were distributed across the apical dendritic arbors, with the greatest density situated on the tuft and apical branches (Figure 7F,H). Putative SST-Myh8 synapses, on the other hand, were concentrated solely on the tuft (Figure 7G–H). These results demonstrate that two SST subtypes impinging on the same excitatory population have distinct innervation patterns at a subcellular level, providing further support for the interesting possibility that they may gate different streams of information to a common target, as suggested in Muñoz et al.8
Figure 7. SST subtypes target distinct subcellular compartments of L5-PT dendrites.
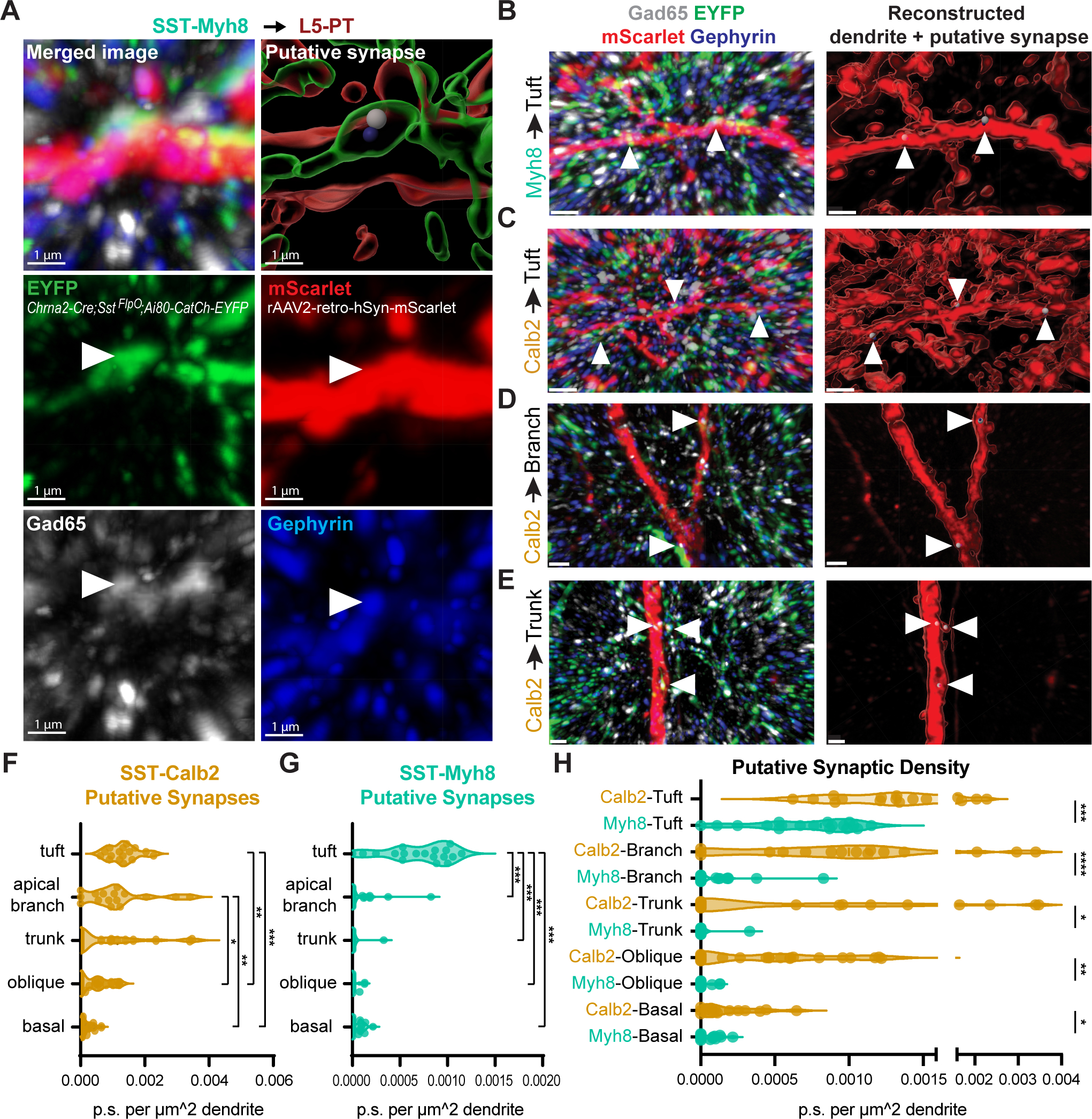
(A) Representative images of a putative synapse from SST-Myh8 interneurons onto a PT tuft dendrite. SST-Myh8 axons are labeled with Chrna2-Cre;SstFlpO;Ai80-CatCh-EYFP, L5-PT dendrites labeled with rAAV2-retro-hSyn-mScarlet, presynaptic puncta labeled with Gad65, and postsynaptic puncta labeled with Gephyrin. The top row shows the merged image with all four channels (left) and the 3D reconstruction in Imaris (right). Arrowheads indicate the location of the putative synapse identified by the colocalization of all four channels in Imaris. Scale bars, 1 μm.
(B) Representative images of putative synapses from SST-Myh8 interneurons onto PT tuft dendrites in lower magnification. Arrowheads indicate the location of putative synapses. Merged image (left) shows all four channels as in (A), and Imaris reconstruction (right) shows the locations of the putative synapses on the dendrite. Scale bars, 1 μm.
(C-E) Representative image of SST-Calb2 putative synapses on L5-PT tuft dendrites, dendritic apical branch, or dendritic trunk, as in (B). Scale bars, 1 μm.
(F) Quantification of SST-Calb2 puncta on L5-PT dendrites. The number of puncta is normalized by the surface area of the reconstructed dendrite. Each data point represents one ROI examined.
(G) Quantification of SST-Myh8 puncta on L5-PT dendrites. The number of puncta is normalized by the surface area of the reconstructed dendrite.
(H) Comparison of SST-Calb2 and SST-Myh8 puncta on L5-PT dendrites.
Statistics in Table S7.
DISCUSSION
The rapid expansion of single-cell transcriptomic analysis of cell type taxonomies in recent years has resulted in an unprecedented understanding of the molecular heterogeneity of cortical neurons39–41. However, despite recent efforts16–21,42–45, an understanding of how transcriptomic cell type correlates with other modalities including morphology, connectivity and in vivo functions is still largely lacking. In this study, we developed and characterized genetic strategies to target the breadth of transcriptomically identified SST subtypes. We then focused on three major SST subtypes and demonstrated that different transcriptomic subtypes form precise and partially reciprocal inhibitory microcircuits with excitatory and inhibitory neurons that are laminar, cell-type and subcellular specific. Previous studies also demonstrated that SST-Hpse and SST-Crh subtypes form specific reciprocal connections with L4 spiny stellate cells10,12. A schematic diagram summarizing the characterized and hypothesized local microcircuitry formed by individual SST subtypes in S1 and V1 is shown in Figure 8. Therefore, taking SST interneurons as an exemplar, we provide a roadmap for understanding interneuron subtypes, which emphasizes the previously underappreciated circuit specificity linking different subtypes of inhibitory and excitatory neurons.
Figure 8. Schematic drawing of the output circuitry of different SST subtypes in S1 and V1.
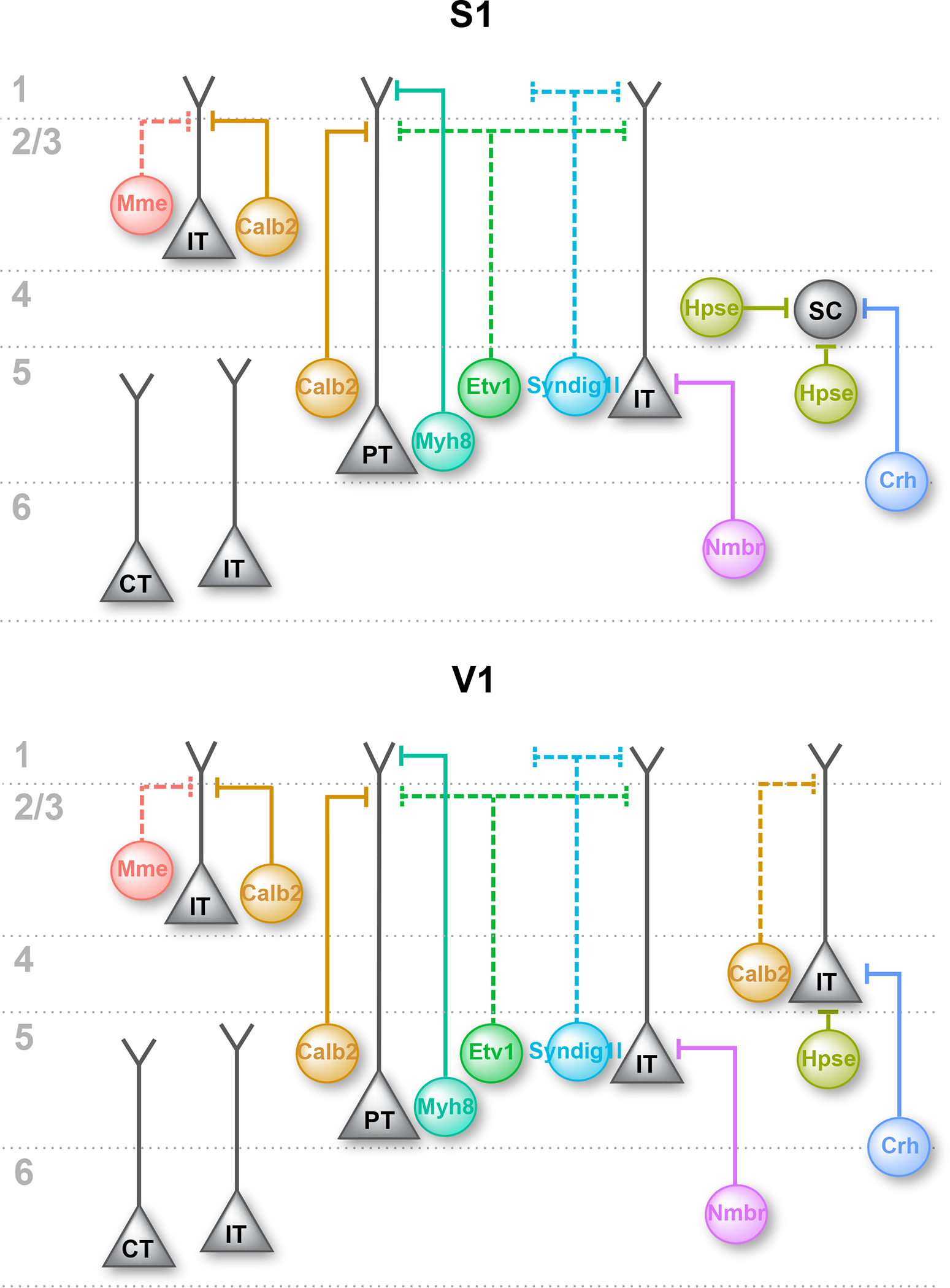
Summary of our current understanding of the innervation pattern of different SST subtypes in S1 and V1, showing the preferred postsynaptic excitatory neuron cell type of each SST subtype. Dashed lines showing hypothesized output circuitry for SST subtypes that have not been fully characterized. IT, intratelencephalic neuron; PT, pyramidal-tract neuron; SC, L4 spiny stellate cell; CT, corticothalamic neuron.
Inhibitory interneurons contribute to specific cortical microcircuits.
Although the spatial distribution of cortical interneurons does not strictly obey the laminar boundaries set by excitatory neurons, our results demonstrate that, like the local excitatory network, cortical inhibitory circuits are organized in both a layer and cell-type specific fashion. Therefore, the complexity of inhibitory circuitry is at least as multifaceted as local excitatory neuron networks. A complete description of them requires knowledge of the laminar position of their afferent and efferent targets. While this study begins to characterize the local inhibitory microcircuits of SST subtypes with selected excitatory neuron types, the inclusion of other neuronal types such as L5 near-projecting pyramidal neurons, L6 corticothalamic neurons, and VIP interneurons will be necessary to gain a complete understanding of these inhibitory circuits. Nevertheless, our results provide a first-pass look at the granularity of their multilayered specificity.
Given this unanticipated specificity, upon reflection, it is not surprising that cortical inhibitory circuits were hypothesized to primarily exert ‘blanket’ inhibition when analyzed at the cardinal class level 6,7. While SST interneurons as a class reside primarily in the infragranular layer, their overall inhibition to L2/3 excitatory neurons is equally as strong as to L5, resulting in a false impression of non-selective efferent targeting. By examining SST interneurons in terms of their different subtypes with respect to excitatory neurons, we observed that different layers of excitatory neurons receive inhibition roughly in proportion to the composition of SST subtypes found in their resident layer. Despite this, excitatory neurons clearly also receive a portion of SST inputs from populations residing in other layers46. For example, L5-IT neurons receive at least equally strong translaminar inhibition from the L6 SST-Nmbr subtype, as compared to SST-Myh8 and SST-Calb2 subtypes that both reside in L5. These results seem to suggest that the local inhibitory circuits have a hierarchical organization, whereby interneuron subtypes are distributed across layers to provide balanced laminar inhibition, while within each layer interneuron subtypes innervate specific postsynaptic inhibitory and excitatory neurons. The third layer of specificity is achieved subcellularly. SST-Myh8 and SST-Calb2 interneurons target L5-PT neurons with similar strength but innervate distinct dendritic domains. Thus, rather than inhibition being indiscriminate, specificity is achieved through the precise regulation of subtype number, subtype distribution, synaptic strength, and synaptic organization.
Do SST subtypes receive reciprocal cell-type specific excitatory inputs?
This study has focused on the efferent specificity of SST interneurons. This raises the question as to whether SST interneurons receive corresponding reciprocal afferent inputs from the local excitatory neurons that they target. Our retrograde rabies tracing experiments seem to support this conclusion: SST-Myh8 interneurons both target and receive connections from L5-PT neurons, while SST-Nmbr interneurons preferentially reciprocally target L5-IT neurons. Consistent with our findings, SST-Myh8 interneurons in A1 appear to reciprocally form connections with thick-tufted and therefore PT L5 pyramidal neurons 30. Additional evidence supporting reciprocal connections comes from previous studies that demonstrated that L4-targeting non-Martinotti cells in L5 form selective and reciprocal connections with L4 spiny stellate cells in S1. In addition, using double or triple patch-clamp recordings, two distinct L5 pyramidal neuron populations were found to form recurrent connections with two different SST populations47. Furthermore, L5 fanning-out Martinotti cells were found to primarily receive excitatory inputs from L2/3 in S1 and target L5 pyramidal neurons in S110,12. Our data suggest that SST-Calb2 interneurons, as a whole, strongly inhibit L5-PT pyramidal neurons and L2/3. Such translaminar inhibitory circuits have been described by previous studies, that are mediated through SST interneurons48–50.
Extrapolating from these findings, we hypothesize that molecularly diverse interneuron subtypes are embedded in highly specific circuit motifs that can be understood as functional units. This raises the possibility that sophisticated cortical neural networks exist comprised of combinations of computational modules, which can be understood as assemblies of distinct functional units, akin to those found in integrated circuits.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, Gord Fishell (gordon_fishell@hms.harvard.edu)
Materials availability
Plasmids and viruses created in this study are available upon request from the Lead Contact with a completed Materials Transfer Agreement.
This study did not generate new unique reagents.
Data and code availability
Slide-seqV2 data generated for this study are available at the Broad Institute Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP2082/cortical-somatostatin-interneuron-subtypes-form-cell-type-specific-circuits#study-summary. Sparse labeling images of SST interneurons have been deposited at Harvard Dataverse and can be accessed through the following link: https://doi.org/10.7910/DVN/NQDIPG. Additional relevant information is available on our lab website: https://fishelllab.hms.harvard.edu/publications.
Original codes for clustering of snRNA-seq data and analysis of Slide-seq V2 experiments are available at https://github.com/gs512/slideseq-engine.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
All experiments were approved by and in accordance with Harvard Medical School IACUC protocol number IS00001269. Animals were group housed and maintained under standard, temperature-controlled laboratory conditions. Mice were kept on a 12:12 light/dark cycle and received water and food ad libitum. Both female and male animals were used indiscriminately for all experiments. Though a systematic analysis was not performed to assess whether there are sex-related differences, no obvious pattern was observed. Transgenic mouse lines used in this study are included in Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Polyclonal rabbit anti-DsRed | Clontech | Cat #632496; RRID:AB_10013483 |
| Polyclonal goat anti-GFP | Sicgen | Cat# AB0020-200; RRID:AB_2333099 |
| Rat anti-RFP | Chromotek | #5f8; RRID:AB_2336064 |
| Rabbit anti-somatostatin | Peninsula Laboratories | T4103; RRID:AB_518614 |
| Mouse anti-Calretinin | Millipore | Cat# MAB1568; RRID:AB_94259 |
| Rabbit anti-Calretinin | Swant | Cat# CR 7697; RRID:AB_2619710 |
| Rabbit anti-Satb2 | Abcam | Cat# ab34735; RRID:AB_2301417 |
| Mouse IgG1 anti-Gephyrin | Synaptic Systems | Cat# 147 011; RRID:AB_2810215 |
| Polyclonal chicken anti-GFP | Aves Labs | Cat# 1020; RRID:AB_10000240 |
| Mouse IgG2a anti-Gad65 | Millipore | Cat# MAB351R; RRID:AB_94905 |
| Alexa Fluor™ 488, Donkey anti-Goat | Thermo Fisher Scientific | Cat# A-11055, RRID:AB_2534102 |
| Alexa Fluor™ 594, Donkey anti-Rabbit | Thermo Fisher Scientific | Cat# A-21207, RRID:AB_141637 |
| Alexa Fluor™ 594, Donkey anti-Rat | Thermo Fisher Scientific | Cat# A-21209, RRID:AB_2535795 |
| Alexa Fluor™ 647, Donkey anti-Rabbit | Thermo Fisher Scientific | Cat# A-31573, RRID:AB_2536183 |
| Alexa Fluor™ 647, Goat anti-Mouse IgG1 | Thermo Fisher Scientific | Cat# A-21240, RRID:AB_2535809 |
| Alexa Fluor® 488 polyclonal Donkey anti-Chicken | Jackson ImmunoResearch Labs | Cat# 703-545-155, RRID:AB_2340375 |
| DyLightTM 405 Polyclonal Goat anti-Mouse IgG2a | Jackson ImmunoResearch Labs | Cat# 115-477-186, RRID:AB_2632530 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| rAAV9-hSyn-DIO-TVA-GFP-N2cG | This paper | RRID:Addgene_175439 |
| rAAV1/2-Dlx-DIO-TVA | This paper | N/A |
| rAAV1/2-Dlx-DIO-GFP-N2cG | This paper | N/A |
| rAAV9-Dlx-DIO-TVA | This paper | N/A |
| rAAV9-Dlx-DIO-GFP-N2cG | This paper | N/A |
| rAAV2-retro-hSyn-mScarlet | Dr. David Ginty This paper | N/A |
| rAAV PHP.eB-S5E2-GFP-fGFP | This paper | RRID:Addgene_135631 |
| rAAV9-hDlx-Flex-dTomato | This paper | RRID:Addgene_83894 |
| EnvA-CVS-N2C(DG)-FlpO-mCherry | K. Ritola, Janelia Pouchelon et al.37 PMID: 34758329 PMCID: PMC8832360 |
N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Corn Oil | Sigma-Aldrich | Cat# C8267 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay | Advanced Cell Diagnostics | Cat. No. 323100 |
|
| ||
| Deposited data | ||
|
| ||
| P28 cortical interneuron snRNA-seq data | Allaway et al.22 PMID: 34552240 PMCID: PMC9316417 |
GEO: GSE165233 |
| Mouse Whole Cortex and Hippocampus Smart-Seq | Yao et al.3 PMID: 34004146 PMCID: PMC8195859 |
GEO: GSE185862 |
| Slide-SeqV2 data | This paper | https://singlecell.broadinstitute.org/single_cell/study/SCP2082/cortical-somatostatin-interneuron-subtypes-form-cell-type-specific-circuits#study-summary |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| Mouse: B6 Ssttm2.1(cre)Zjh/J | Jackson Laboratories | RRID:IMSR_JAX:013044 |
| Mouse: B6J.Cg-Ssttm3.1(flpo)Zjh/AreckJ | Jackson Laboratories | RRID:IMSR_JAX:031629 |
| Mouse: B6;129S-Tac1tm1.1(cre)Hze/J | Jackson Laboratories | RRID:IMSR_JAX:021877 |
| Mouse: B6(Cg)-Etv1tm1.1(cre/ERT2)ZJh/J | Jackson Laboratories | RRID:IMSR_JAX:013048 |
| Mouse: B6;129S-Pdyntm1.1(cre)Mjkr/LowlJ | Jackson Laboratories | RRID:IMSR_JAX:027958 |
| Mouse: B6;129S-Pdyntm1.1(cre/ERT2)Hze/J | Jackson Laboratories | RRID:IMSR_JAX:030197 |
| Mouse: B6.Cg-Npytm1.1(flpo)Hze/J | Jackson Laboratories | RRID:IMSR_JAX:030211 |
| Mouse: B6.Cg-Hpseem1(cre)Ngai/TasicJ | Dr. David A. Stafford | RRID:IMSR_JAX:037334 |
| Mouse: Chrna2-Cre (Tg(Chrna2-cre)1Kldr) | Dr. Klas KullanderHilscher et al.30 PMID: 28182735 PMCID: PMC5300109 |
N/A |
| Mouse: CrhCre | Dr. Bradford Lowell Krashes et al.52 PMID: 24487620 PMCID: PMC3955843 |
N/A |
| Mouse: B6.129S4(SJL)-Crhr2tm1.1(cre)Lbrl/J | Jackson Laboratories | RRID:IMSR_JAX:033728 |
| Mouse: B6.Cg-Gt(ROSA) 26Sortm65.2(CAG-tdTomato)Hze/J; Ai65 | Jackson Laboratories | RRID:IMSR_JAX:021875 |
| Mouse: B6.Cg-Gt(ROSA) 26Sortm14(CAG-tdTomato)Hze/J; Ai14 | Jackson Laboratories | RRID:IMSR_JAX:007914 |
| Mouse: B6.Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze/J; Ai9 | Jackson Laboratories | RRID:IMSR_JAX:007909 |
| Mouse: B6;129S4-Gt(ROSA) 26Sortm3(CAG-tdTomato,-EGFP*)Zjh/J; IS reporter | Jackson Laboratories | RRID:IMSR_JAX:028582 |
| Mouse: B6.Cg-Gt(ROSA) 26Sortm32(CAG-COP4*H134R/EYFP)Hze/J; Ai32 | Jackson Laboratories | RRID:IMSR_JAX:024109 |
| Mouse: B6.Cg-Gt(ROSA) 26Sortm80.1(CAG-COP4*L132C/EYFP)Hze/J; Ai80 | Jackson Laboratories | RRID:IMSR_JAX:025109 |
| Mouse: B6;129S6-Gt(ROSA) 26Sortm10(CAG-Syp/EGFP*,-tdTomato)Dym/J; RC::FPSit | Jackson Laboratories | RRID:IMSR_JAX:030206 |
| Mouse: Gt(ROSA)26Sortm1.1(CAG-EGFP)Fsh/Mmjax; RCE:loxP | MMRRC | RRID:MMRRC_032037-JAX |
| Mouse: Gt(ROSA)26Sortm1.2(CAG-EGFP)Fsh/Mmjax; RCE:FRT | MMRRC | RRID:MMRRC_032038-JAX |
|
| ||
| Oligonucleotides | ||
|
| ||
| RNAscope® Probe- Mm-Gad1 | ACDBio | Cat#400951 |
| RNAscope® Probe- Mm-Sst | ACDBio | Cat#404631, 404631-C2, 404631-C3, 404631-C4 |
| RNAscope® Probe- Mm-Calb2 | ACDBio | Cat#313641-C3 |
| RNAscope® Probe- Mm-Hpse | ACDBio | Cat#412251 |
| RNAscope® Probe- Mm-Cbln4 | ACDBio | Cat#428471 |
| RNAscope® Probe- Mm-Pdyn | ACDBio | Cat#318771 |
| RNAscope® Probe- Mm-Crh | ACDBio | Cat#316091-C2 |
| RNAscope® Probe- Mm-Chodl | ACDBio | Cat#450211 |
| RNAscope® Probe- tdTomato | ACDBio | Cat#317041-C2 |
| HCR RNA-FISH probe Sst - B5 | Molecular Instruments | N/A |
| HCR RNA-FISH amplifier B5 - Alexa Fluor 647 | Molecular Instruments | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Code for snRNA-seq and Slide-SeqV2 analysis | This paper | https://github.com/gs512/slideseq-engine |
| Zen blue 2.6 | Zeiss | RRID:SCR_013672 |
| ImageJ | NIH | RRID:SCR_003070 |
| Fiji | http://fiji.sc | RRID:SCR_002285 |
| Adobe Photoshop CS6 | Adobe | RRID:SCR_014199 |
| Adobe Illustrator CS6 | Adobe | RRID:SCR_014198 |
| R Project for Statistical Computing | Open Source | RRID:SCR_001905 |
| RStudio | Open Source | RRID:SCR_000432 |
| Seurat | Rahul Satija Lab | https://satijalab.org/seurat/ |
| SPACEXR (formerlly RCTD) | Fei Chen Lab | https://github.com/dmcable/spacexr |
| ClampFit 11 | Molecular Devices | RRID:SCR_011323 |
| Neurolucida 360 | MBF Bioscience | RRID:SCR_016788 |
| NeuroInfo® | MBF Bioscience | https://www.mbfbioscience.com/neuroinfo |
| Prism 9.1.2 | Graphpad Software | RRID:SCR_002798 |
| IBM SPSS Statistics 26 | IBM | RRID:SCR_019096 |
| MATLAB | MathWorks | https://www.mathworks.com/ |
METHOD DETAILS
Tamoxifen Induction
Tamoxifen (Sigma-Aldrich, T5648) was dissolved in corn oil (Sigma-Aldrich) at 10 or 20 mg/ml) with agitation or sonication. Tamoxifen solution was either stored at RT and used within one week of preparation or stored long-term at −80°C and warmed up prior to injection. Tamoxifen solution was administrated to mice through oral gavage. A wide range of tamoxifen is administrated to achieve different levels of recombination. To achieve sparse labeling of SST interneurons for examining single-neuron morphology, a single dose of 0.5 – 2 mg of tamoxifen was administrated to PdynCreER; Ai14, PdynCreER; Ai32 or Etv1CreER; SstFlpO; RC::FPSit mice. To induce a higher level of recombination for assessing the specificity and coverage of different genetic targeting strategies, a varying dosage ranging from a single dose of 1 mg up to 5 doses of 2 mg of tamoxifen per mouse was administered to PdynCreER; Ai14, PdynCreER; NpyFlpO; Ai9 and Etv1CreER; SstFlpO; Ai65 mice. All Tamoxifen administrations were performed on mice aged from 2-week to 3-month.
Perfusion and Immunohistochemistry
For all histological experiments, mice were deeply anesthetized with sodium pentobarbital (Euthasol) by intraperitoneal injection and transcardially perfused with 1X PBS followed by 4% paraformaldehyde (PFA) in 1X PBS. Brains were dissected out and post-fixed overnight at 4°C.
To examine the expression pattern of transgenic mouse lines, immunofluorescence is routinely used to amplify the fluorescent signal of reporter protein labeling. For these experiments, fixed brain samples were then cryopreserved in 30% sucrose in 1X PBS. 40 μm brain sections were obtained through a Leica sliding microtome. For immunofluorescence, free-floating brain sections were incubated in primary antibodies diluted in antibody incubation solution (5% normal donkey serum, 0.25% Triton X-100 in 1X PBS) in coldroom overnight or up to three days. Secondary antibodies were diluted in antibody incubation solution at RT for 1–3 hrs, or in coldroom overnight.
For sparse labeling and single neuron morphology reconstruction, fixed brain samples were sectioned through a vibratome (Leica VT1200S) into 100–150 μm slices. Brain sections were incubated in primary antibodies diluted in antibody incubation solution in coldroom for 2–3 days. Secondary antibodies were diluted in antibody incubation solution (5% normal donkey serum, 0.25% Triton X-100 in 1X PBS) at RT for 1–3 hrs, or in coldroom overnight or up to 2 days.
For synaptic puncta staining, tissue was sectioned at 50 μm on a vibratome (Leica VT 1200S). Free-floating brain sections were stored in antifreeze solution until processing. Free-floating brain sections were blocked for one hour (0.1% Triton X-100, 3% Normal Donkey Serum and 3% Normal Goat Serum in 1X PBS) for 1 hour, followed by primary antibody incubation in the same solution overnight at 4°C. The following day, sections were rinsed in 0.1% Triton X-100 in 1X PBS for a minimum of 3 × 5 minutes, followed by secondary incubation in the same blocking solution for 2 hours at room temperature. Sections were then rinsed again for a minimum of 3 × 5 minutes in 1X PBS and mounted.
A list of primary antibodies used in this study can be found in Key Resources Table.
Slide-seq V2
Slide-seq V2 experiments were performed on 10 μm thick coronal sections from four different wild-type mice aged between P28–37. Experimental procedures were detailed previously 25. Samples were sequenced on an Illumina NovaSeq SP flow cell 100 cycle kit with 8 samples per run (four samples per lane). The Slide-seq tools (https://github.com/MacoskoLab/slideseq-tools) software was used to collect, demultiplex and sort reads across barcodes. Slide-seq V2 data generated in this study can be found at the Broad Institute Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP2082/cortical-somatostatin-interneuron-subtypes-form-cell-type-specific-circuits#study-summary. In addition, one published dataset from somatosensory cortex, Puck_200306_03, was included in the analysis, which can be accessed through https://singlecell.broadinstitute.org/single_cell/study/SCP815/sensitive-spatial-genome-wide-expression-profiling-at-cellular-resolution#study-summary.
Single Molecule Fluorescent In Situ Hybridization Histochemistry
For single molecule fluorescent in situ hybridization (smFISH) combined with immunohistochemistry, mice were perfused and brains were fixed overnight in 4% PFA in 1X PBS followed by cryoprotection in 30% sucrose in 1X PBS. Then, 16–20 μm (for RNAscope®) or 40–80 μm (for HCR-FISH) thick brain sections were obtained using a Leica cryostat or a sliding microtome. Brain slices sectioned using cryostat are directly mounted on glass slides (Fisherbrand Superfrost Plus) and preserved at −80 °C. Brain sections obtained using sliding microtome were preserved in Section Storage Buffer containing 28% (w/v) sucrose, 30% (v/v) ethylene glycol in 0.1M sodium phosphate buffer, pH 7.4 and stored at −80 °C, before smFISH experiments.
For RNAscope® experiments, samples were processed according to the ACDBio Multiplex Flourescent v2 Kit protocol (ACDBio #323100) for fixed frozen tissue. Briefly, tissue was pre-treated with a series of dehydration, H2O2, antigen retrieval, and protease III steps before incubation with the probe for 2 hours at 40 °C. Note here protease III incubation was performed at room temperature to better preserve protein for immunostaining. A list of probes purchased from ACDBio is included in Key Resources Table. Three amplification steps were carried out prior to developing the signal with Opal™ or TSA® Dyes (Akoya Biosciences). Immuostaining following RNAscope® experiment was performed according to Technical Note 323100-TNS from ACDBio. Samples were counterstained with DAPI and mounted using Prolong Gold antifade mounting medium (Molecular Probes #P369300).
HCR RNA-FISH experiments were performed with a modified protocol to the manufacturer’s recommendation (Molecular Instruments). Briefly, three to four 40 or 80 μm brain slices were placed in a single well of a 24-well plate. The brain slices then went through a series of pre-treatment including post-fixation, an optional ethanol dehydration step, and a mild proteinase K treatment (2 μg/ml, 15 min, RT), before incubating with 3.3–4.5 nM of HCR RNA-FISH probes at 37 °C overnight. After repeated wash with probe wash buffer and 5X SSCT, the signal is developed and amplified with 60 nM hairpin pairs at RT for 4–16 hrs. After the amplification step, the brain slices were washed with 5X SSCT for 1.5 hr with periodic buffer change. Immunostaining following the HCR RNA-FISH was performed by blocking the brain slices with 2% BSA/PBST for ~15 min, followed by overnight incubation with primary antibody diluted in 1% BSA/PBST at 4 °C overnight. After washing with 1X PBST, the brain slices are incubated with secondary antibodies diluted in 1% BSA/PBST at RT for 1–2 hrs. Brain slices were counterstained with DAPI (5 μM, Sigma #D9542) and mounted using Fluoromount-G (Invitrogen) or Prolong Gold antifade mounting medium (Molecular Probes #P369300). HCR RNA-FISH probes and amplifiers used in this study can be found in Key Resources Table.
Cell Culture, transfection and AAV production
HEK293FT cells (Thermo Fisher Scientific, #R70007) were cultured in Dulbecco’s Modified Eagle’s medium with high glucose and pyruvate, GlutaMAX Supplement, 10% fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml). The cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2. For AAV production, HEK293FT cells were seeded on 15-cm plates without antibiotics for 24 hours and co-transfected with the following plasmids using Polyethylenimine (100 μg/dish, Polysciences, #23966–1): pHGTI-helper (22 μg/dish), rAAV2-retro helper (Addgene plasmid #81070, 12 μg/dish), AAV9 helper (Addgene plasmid #112865, 12 μg/dish),. and the AAV expression vector (12 μg/dish). 72 hours after transfection, transfected cells were harvested and lysed (150 mM NaCl, 20 mM Tris pH 8.0) by three freeze-thaw cycles and Benzonase treatment (375 U/dish; Sigma, #E1014) for 15 minutes at 37 °C. The supernatants were cleared by centrifugation at 4000 RPM for 20 minutes at 4 °C, then transferred to Iodixanol gradients (OptiPrep Density Gradient Medium, Sigma, #D1556) for ultracentrifugation (VTi50 rotor, Beckman Coulter) at 50,000 RPM for 1.5 hours at 16 °C. The 40% iodixanol fraction containing the AAVs was collected, underwent ultrafiltration with PBS in Amicon Ultra (15 ml, 100K, Millipore, #UFC910024) for 4 times, aliquoted and stored at −80 °C. The number of genomic viral copies was determined by qPCR using the following primers against the WPRE sequence: Fw: AGC TCC TTT CCG GGA CTT TC and Rv: CAC CAC GGA ATT GTC AGT GC. A list of viral vectors used in this study can be found in Key Resources Table.
Viral labeling of IT/PT neurons and PV interneurons
Juvenile mice (P10–15) were head-fixed using soft tissue Zygoma ear cups (Kopf #921). rAAV2-retro-hSyn-mScarlet (Dr. David Ginty) was used for retrograde labeling. Viral aliquots were loaded into a Drummond Nanoinjector III. All coordinates are referenced from Lambda suture. For PT labeling, 150 nl was injected into the ipsilateral superior colliculus at AP 0.15, ML 0.38, DV −1.45. For L5-IT labeling, 100 nl was injected into the ipsilateral retrosplenial cortex at AP 1.1, ML 0.39, DV −.25 and 50 nl at AP 0.5, ML 0.31, DV −.25. For L6-IT labeling, 150 nl was injected into contralateral V1 at AP 0.2, ML 2.0, DV −.45. For PV labeling, 200 nl of rAAV PHP.eB-S5E2-GFP-fGFP (Addgene #135631, Titer: 9.4×1011 vg/mL) was injected into ipsilateral V1 at AP .2, ML 2.0, DV −.45. Coordinates were slightly adjusted based on the age of the mouse at the time of injection (+/− .2).
Slice preparation and brain slice recording
Animals aged P25–35 were anesthetized with isoflurane followed by decapitation. The brain was quickly removed and immersed in ice-cold oxygenated sucrose cutting solution containing (in mM) 87 NaCl, 75 Sucrose, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 10 Glucose, 1 CaCl2, 2 MgCl2 (pH=7.4). 300 μm thick coronal slices were cut using a Leica VT 1200S vibratome through the primary visual cortex. Slices recovered in a holding chamber with ACSF containing (in mM) 124 NaCl, 20 Glucose, 3 KCl, 1.2 NaH2PO4, 26 NaHCO3, 2 CaCl2, 1 MgCl2 (pH=7.4) at 34 °C for 30 minutes and at room temperate for at least 45 minutes prior to recording. Note that one P33 mouse was sliced in NMDG solution as described in Ting et al., 201853.
Patch-clamp recordings were performed using two different electrophysiological rigs. The majority of the data were obtained with [rig A], containing an upright microscope (Scientifica) with oblique illumination Olympus optics. Cells were visualized using a 60x water immersion objective. Recordings were performed using a Multiclamp 700B amplifier (Molecular Devices) and digitized using a Digidata 1550A and the Clampex 10 program suite (Molecular Devices). A small set of experiments were obtained with [rig B] using an upright differential interference contrast microscope (BX51WI) with a 40 × water immersion objective (N.A. 0.9). Recordings were performed using a Multiclamp 700B amplifier and digitized using Digidata 1440A using a sampling rate of 20KHz.
Slices were perfused with ACSF in a recording chamber at 2 ml/min at room temperature. All slice preparation and recording solutions were oxygenated with carbogen gas (95% O2, 5% CO2, pH 7.4). Patch electrodes (3–7 MΩ) were pulled from borosilicate glass (1.5 mm OD, Harvard Apparatus). For current-clamp recordings, patch pipettes were filled with an internal solution containing (in mM): 130 K-Gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4 MgATP, 0.3 NaGTP, 5 Phosphocreatine and 0.4% biocytin, equilibrated with KOH CO2 to pH=7.3. For voltage-clamp recordings patch pipettes were filled with an internal solution containing (in mM): 125 Cs-gluconate, 2 CsCl, 10 HEPES, 1 EGTA, 4 MgATP, 0.3 Na-GTP, 8 Phosphocreatine-Tris, 1 QX-314-Cl, equilibrated with CsOH at pH=7.3. Voltage-clamp signals were filtered at 3 kHz and recorded with a sampling rate of 20 kHz. IPSCs were performed at a holding potential of 0 mV. Cells were only accepted for analysis if the initial series resistance was less than 40 MΩ and did not change by more than 20% during the recording period. The series resistance was compensated at least ~50% in voltage-clamp mode. No correction was made for the liquid junction potential. Experiments were performed at room temperature to ameliorate space clamp errors 54.
Optogenetic mapping
For output mapping, experiments were performed using mice express specific driver lines crossed with Ai80 for intersectional CatCh expression and injected with AAVs to label IT, PT neurons, and PV interneurons. Whole-cell patch-clamp recordings were obtained from virally labeled neurons or unlabeled putative pyramidal neurons across layers. Virally labeled excitatory neurons were included as L5a and L5b neurons in the analysis of outputs across layers, but PV interneurons were excluded.
For optogenetic stimulation on [rig A], 470 nm light was transmitted from a collimated LED (Mightex) attached to the epifluorescence port of the upright microscope. 1 ms pulses of light were directed to the slice in the recording chamber via a mirror coupled to the 60x objective (N.A. = 1.0). Flashes were delivered every 15 s over a total of 15 trials. The LED output was driven by a transistor-transistor logic output from the Clampex software. For optogenetic stimulation on [rig B], LED (ThorLabs LED4D021) is directed to the microscope via a 5mm liquid light guide and triggered by a LED controller (Mightex SLC-AA04-US). Optogenetically induced spikes in SST-Calb2 and SST-Nmbr interneurons were shown in additional figures available at https://fishelllab.hms.harvard.edu/publications.
Biocytin filling and staining
After recording with pipette solution containing 0.3–0.5% biocytin, the slices were fixed in 4% PFA overnight, then stored in 30% sucrose in 1X PBS till further processing. After washing out the PFA, the slices were incubated with ScaleCUBIC-1 solution for 2 days. After thorough washing with 1X PBS, the slices were incubated with Alexa-conjugated streptavidin in blocking solution (10% normal donkey or goat serum, 0.5% Triton X-100, 0.2% cold water fish gelatin in 1X PBS) overnight at room temperature. After thorough wash with 1X PBS, slices were transferred to ScaleCUBIC-2 solution and incubated for approximately 30 minutes before being mounted on a glass slide in ScaleCUBIC-2 solution for confocal microscopy imaging. Recipes for ScaleCUBIC-1 and ScaleCUBIC-2 can be found in Susaki et al., 201455.
Retrograde monosynaptic rabies tracing
For tracing afferent inputs to SST-Myh8 subtype, Chrna2-Cre mouse pups at P2–5 were anesthetized by hypothermia and stereotaxically micro-injected with AAV9-DIO-helper virus encoding N2c-G-P2A-TVA-P2A-eGFP (Addgene #170853; Titer of 9.5×1012 vg/mL) using Nanoject III at a rate of 1 nL/s. AAV9-DIO-helper virus was diluted 1:1 or 1:2 with 1X PBS and injected for a total of 10 nL (from Lambda: AP +1.5–2, ML +1.8–3, DV- 0.2–0.3 for S1; AP +0–0.4, ML −1.8–2.2, DV −0.05–0.3 for V1). EnvA-pseudotyped CVS-N2c(ΔG)-FlpO-mCherry (N2c-RV, Titer: 3.7E+09U/ml) was generously shared by K. Ritola at Janelia Farms Research Center as described in Pouchelon et al., 202136. N2c-RV was injected separately at P22-P42 (from Bregma: AP −1, ML +3, DV −0.85) for S1 or at P56–79 (from Bregma: AP −3, ML −2.5, DV −0.5) for V1. N2c-RV was diluted 1:10 with HBSS and stereotaxically injected for a total volume of 60–100 nL. Mice were sacrificed 10–15 days later for examination.
For tracing afferent inputs to SST-Nmbr subtype, Crhr2Cre mice at P34–79 were stereotaxically injected with AAV-Dlx-DIO-helpers and N2c-RV at the same time according to stereotaxic coordinates (from Bregma: AP −1, ML +3, DV-0.86 for S1; AP −3, ML −2.5, DV −0.50 for V1). AAV-Dlx-DIO-TVA, AAV-Dlx-DIO-GFP-N2cG, and 1:10 diluted N2c-RV were combined in 1 : 1 : 1–3 ratio for injection of a total 50–80 nL. For most of the rabies experiments, AAV9-Dlx-DIO-TVA (Titer: 6.89×1013 vg/mL) and AAV9-Dlx-DIO-GFP-N2cG (Titer: 5.46×1013 vg/mL) were used in combination, except for one experiment AAV1/2-Dlx-DIO-TVA (Titer: 3.5×1012 vg/mL) and AAV1/2-Dlx-DIO-GFP-N2cG (Titer: 2.9×1012 vg/mL) were used. Mice were sacrificed 13–14 days later for examination.
To confirm that the different afferent input patterns to these two SST subtypes were not caused by the use of different AAV-DIO-helper viruses, one test experiment was performed using AAV-Dlx-DIO-helpers from SST-Myh8 interneurons. Briefly, AAV9-Dlx-DIO-TVA and AAV9-Dlx-DIO-GFP-N2cG were stereotaxically injected in a P3 Chrna2-Cre mouse in 1:1 (v/v) ratio for a total of 10 nL (from Lambda: AP +1.8, ML +2.3, DV −0.25). N2c-RV was injected at P31. Mouse was sacrificed 15 days later for examination.
For all rabies tracing experiments, fixed brain samples were sectioned into 40 μm slices. Every third slice was collected for immunofluorescence experiments to examine the rabies tracing patterns.
Image acquisition
Images of transgenic mouse line labeling and rabies tracing were collected using a whole slide scanning microscope with a 10X objective (Olympus VS120 slide scanners) or using a motorized tiling scope (Zeiss Axio Imager A1) with a 5X or 10X objective.
Images of smFISH experiments were acquired with an upright confocal microscope (Zeiss LSM 800) with a 10X objective (Plan-Apochromat 10x/0.45 M27). For sparse labeling or biocytin filling experiments, images were acquired using the confocal microscope (Zeiss LSM 800) with a 20X objective (Plan-Apochromat 20x/0.8 M27). Images of synaptic puncta were acquired using an upright confocal microscope (Zeiss LSM800) with 40X oil immersion objective, 1.4 NA, 2.5 digital zoom, 1024 × 1024 pixels (~0.22 μm resolution using 510 nm emission) with 0.33 μm step size.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism, R, or IBM SPSS software. When appropriate, we have included nested design in our statistical analysis to account for potential correlation for data obtained from the same animal. A summary of the data number and statistical test results are included in Table S7.
snRNA-seq pre-processing, clustering and label transfer
snRNA-seq datasets of interneurons in ALM and V1 of P28 Dlx5/6-Cre; Sun1-eGFP mice (here on referred to as Fishell_P28) were previously published 22. Fishell_P28 and Allen mouse whole cortex and hippocampus Smart-seq dataset (https://portal.brain-map.org/atlases-and-data/rnaseq/mouse-whole-cortex-and-hippocampus-smart-seq) were pre-processed and aligned using Seurat (Satija lab). Supertype labels from the Allen dataset were transferred to Fishell_P28 dataset using Seurat integration. Briefly, Fishell_P28 was pre-processed as described in (https://github.com/gs512/slideseq-engine/), with the clustering resolution set to 2.1.
Allen dataset was filtered using region_label, retaining cells from ALM, SSp, VISp, SSs and VIS regions. Cells were then split on class_label: GABAergic, Glutamatergic and Non-Neuronal. Glutamatergic cells were then filtered to retain cells with region_label equal to SSp and SSs; subclass_label CR, DG, L2/3 IT PPP, L5/6 IT TPE-ENT, L6b/CT ENT were excluded. The remaining cells were then processed in Seurat using default parameters (https://github.com/gs512/slideseq-engine/blob/82ceaebdca62d541dd044667f030fff5fb08bcea/helper.R#L70).
GABAergic cells were then filtered on supertype_label, excluding interneurons found outside of the cortex: Meis2, Ntng1 HPF, Sst Ctsc HPF and Vip Cbln4 HPF. GABAergic interneurons in the Allen dataset were integrated with Fishell_P28 using Seurat (https://github.com/gs512/slideseq-engine/blob/82ceaebdca62d541dd044667f030fff5fb08bcea/helper.R#L89). Default parameters were applied, except for dims 1:100 set for IntegrateData, and resolution = 1.7 for FindClusters. For each SST positive Integrated cluster, the most represented Allen supertype_label was transferred to the Fishell_P28 dataset. Note that SST-Nts supertype defined in Yao et al., 20213 was considered part of PV/SST-Th subtype in our dataset, and therefore not considered a pure SST subtype. See additional information at https://fishelllab.hms.harvard.edu/publications.
Mapping SST subtypes from Allen Smart-seq dataset onto Slide-seq V2 with RCTD
The Allen mouse whole cortex and hippocampus Smart-seq dataset was processed as described above and used as the scRNA-seq reference for mapping SST subtypes in Slide-seq V2 data. We used the RCTD method 26, to integrate Allen Smart-seq data with spatial Slide-seq V2 data. Before running RCTD (now renamed as SPACEXR package), pucks were restricted to only include relevant cortical zones. SPACEXR was run in doublet mode, spots classified as doublet uncertain were not included in the downstream analysis. Each non-excitatory spot was then assigned to a layer [L2/3, L4, L5, L6] using a KNN graph where the majority of the n nearest excitatory cells layer determined the assigned layer. Codes can be found at https://github.com/gs512/slideseq-engine.
Quantification of marker gene expression and genetic labeling
Quantification of marker gene expression of smFISH experiments was performed by visual inspection of each genetically labeled neuron, or SST interneurons labeled by smFISH against Sst transcripts, in maximum orthogonal projection images of confocal image stacks. Neurons containing at least three puncta were considered positive for the gene. In a few experiments where background noise was higher, the threshold was adjusted to five puncta per neuron. Quantification of calretinin-immunopositive neurons within Etv1CreER; SstFlpO; Ai65 labeled neurons was also performed by visual inspection.
For quantification of the percentage of genetically labeled neurons out of the total SST interneuron population, we performed smFISH against Sst mRNA for labeling all SST interneurons. Because Sst mRNA is abundant in SST interneurons and usually labels the entire cell body, for the majority of the analysis, we instituted a semi-automated strategy using Fiji (Image J), which achieved similar effectiveness as compared to a small subset of images that were analyzed by manual inspection. Briefly, the maximum orthogonal projection image of the Z-stack confocal image was loaded in FiJi program and the channel containing smFISH of Sst mRNA was selected. The image then went through a routine of brightness/contrast adjustment, background subtraction, smooth, and Gaussian Blur filtering before automatic ROI detection. Automatically detected ROIs were further selected based on size using Analyze Particles function in Fiji. The outline of all the selected ROIs was then superimposed over the original images, and the final count of cell numbers was then performed manually using Cell Counter function in Fiji. A similar process is applied to the other channel containing genetically labeled neurons. ROIs from these two channels are then superimposed for identifying overlapping/non-overlapping neurons. DAPI channel was used to identify and quantify by cortical layer.
Neurolucida tracing
Z stacks of confocal images were loaded into Neurolucida 360 (MBF Biosciences). Cell body, dendrite, and axons were recognized and reconstructed in a semi-automated manner. Neuronal processes were traced using the ‘user guided’ option with Directional Kernels.
Intrinsic property recording and analysis
Passive and active membrane properties were measured in the current-clamp mode with a holding potential of −65 mV. Analysis was done in Clampfit 11 and Prism (GraphPad). The electrophysiological parameters were adapted10 and defined as follows:
Resting membrane potential (Vrest, in mV): membrane potential measured with no current applied, measured immediately after breaking into the cell;
Input resistance (IR; in MΩ): resistance measured from Ohm’s law from the peak of voltage responses to hyperpolarizing current injections (up to −40 or −50 pA);
Sag ratio (dimensionless): the ratio of voltage at the peak and voltage at steady-state in response to hyperpolarizing current injections, with the peak at approximately −90 mV;
AP threshold (APthre, in mV): measured from action potentials (APs) evoked at rheobase with 1-second current injections, as the membrane potential where the rise of the AP was 10 mV/ms;
AP amplitude (mV): The peak amplitude measured from APthre;
AP half-width (in ms): duration of the AP at half-amplitude from APthre;
AP maximum rate of rise (APrise, in mV/ms): measured from APs evoked at rheobase as the maximal voltage slope during the upstroke of the AP;
After hyperpolarization potential (AHP, in mV): measured as the difference between APthre and the peak of the fAHP.
Maximal firing frequency (HFF, in Hz): maximal firing frequency evoked with 1-s-long depolarizing current steps;
Adaptation (dimensionless): measured from trains of approximately 35 APs as [1 − (Ffirst/Flast)], where Ffirst and Flast are, respectively, the frequencies of the first and last ISI;
Rebound APs: the number of APs elicited at the end of a 1-s-long hyperpolarizing voltage deflection where the steady-state voltage response was approximately −90 mV.
Optogenetic experiment analysis
Data analysis was performed offline using the Clampfit module of pClamp (Molecular Devices) and Prism 9 (GraphPad). Individual waveforms from 15 trials per cell were averaged, and the averaged peak amplitude was recorded. To visualize the distribution of the data, for each graph the highest 75th percentile value of all plotted datasets was selected as a breakpoint for the y-axis, with 75% of the data below the breakpoint and 25% above the breakpoint.
Hierarchical bootstrapping and pan-SST response simulation
Analysis was done in Matlab. We addressed two levels of variability in our data: the variability of single-cell IPSCs across 15 sweeps of ChR2 stimulation, and the variability across cells within a given condition (layer or pyramidal cell type). First, we recomputed the average IPSC amplitude per cell by sampling with replacement across all 15 sweep amplitudes and taking a new average. Second, we recomputed the set of IPSC amplitudes per condition by sampling from the set of bootstrapped amplitudes with replacement. To simulate the linear combination of three SST subtypes, we added together one randomly selected amplitude per subtype (SST-Calb2, SST-Myh8, and SST-Nmbr). We repeated the bootstrapping procedure for the pan-SST IPSCs and subtracted the simulated linear combination IPSC from the median pan-SST IPSC to compare the two amplitudes (for example a difference of 0 means the amplitudes of the combined SST-Calb2, SST-Myh8, and SST-Nmbr equaled the pan-SST response, and a difference > 0 means that the combined SST-Calb2, SST-Myh8, and SST-Nmbr response was greater than the pan-SST response). This was repeated 100,000 times to determine the distribution of the difference between the simulated and measured pan-SST response.
Monosynaptic rabies tracing analysis
Upon uploading all the images into NeuroInfo software, all sections were manually reordered from rostral to caudal of the brain. The software’s section detection parameters were adjusted to correctly recognize the borders of each brain section. Sections were aligned, first using the software’s Most Accurate alignment option, and adjusted manually if necessary. After specifying the distance (120 μm) between each section, the Section Registration function of the software would compare each section to an existing 3D model of the mouse brain to estimate the rostral-caudal location of each section. Non-linear registration was run on each section to account for the slight distortions that might happen during sectioning/mounting, and/or imperfections in the sectioning angle. In the Cell Detection function, parameters for cell size and distance from the background were adjusted, and then Neural Network with preset pyramidal-p64-c1-v15.pbx was used to automatically detect rabies-infected cells in the red channel. Detection results were reviewed manually to correct for any detection mistakes (false positives or negatives). Starter cells were manually marked and identified as GFP co-localized rabies-infected cells. The final results were exported to an Excel file for further analysis.
Synaptic puncta analysis
For analysis of synaptic density on dendritic compartments, the dendritic compartment was noted prior to image acquisition though in some cases a single image contained multiple compartments, requiring post-hoc segmentation.
To quantify the density of synaptic puncta on dendrites, 5 μm-thick image stacks were analyzed with IMARIS 9.5.0 or 9.7.0 using MATLAB scripts adapted from a previous study56. First, all channels underwent background subtraction and depth normalization. Then three-dimensional “surfaces” using the “Create Surface” tool were automatically reconstructed for mScarlet+ dendrites and GFP+ SST cells with a 2 μm2 area filter. The threshold was selected to include as much of the process as possible while minimizing background noise. Surfaces were manually edited to exclude artifacts, segment dendritic compartments, and remove the soma and dendrites of SST interneurons when necessary (clearly distinguishable from axons by size and brightness). Gad65+ and Gephyrin+ puncta were automatically reconstructed as “spots” of 0.6 and 0.3 μm diameter, respectively. To detect spots the built-in spot detection algorithm in Imaris first applies a 3D Mexican Hat filter using the spot size and then locates the spot centroid at the local maxima of the filtered image. Gad65+ spots located within the axon surface were identified using the “split into surface” tool using the radius of the spot as the threshold distance, and the same was done for Gephyrin+ spots in the dendrite surface. Finally, the presynaptic and postsynaptic boutons identified in the previous step were colocalized using their radii as a threshold to identify the number of puncta per image. That number was then normalized by the surface area of the reconstructed dendrite in that image.
To control for noisy signals, we reflected the Gad65 channel on the y-axis and repeated our analysis to determine how many puncta are detected by chance without the biologically correlated signal. As SST-Calb2 puncta were distributed across the dendritic arbor we included all images, but for SST-Myh8 interneurons we only included images of tuft dendrites as the other images contained little to no puncta. We found that for both SST-Calb2 and SST-Myh8, our analysis detected significantly more puncta in the original images (https://fishelllab.hms.harvard.edu/publications), confirming that we can detect synaptic puncta above a noise threshold.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marian Fernandez-Otero and Nusrath Yusuf for their technical assistance. Dr. David Ginty and Dr. Mark Springel (Harvard Medical School) for sharing with us the viral construct of pAAV-hSyn-mScarlet-WPRE. N2c-RV was generously shared by K. Ritola at Janelia Farms Research Center. Chrna2-Cre (Tg(Chrna2-cre)1Kldr) mice were kindly provided by Dr. Klas Kullander (Uppsala University, Sweden). Crhr2Cre mice were kindly provided by Dr. Stephen Liberles (Harvard Medical School). Hpse-IRES-Cre mice were made and kindly provided by Dr. David A. Stafford (University of California, Berkeley). Crh-IRES-Cre_BL mice were kindly provided by Dr. Bard Lowell (Beth Israel Deaconess Medical Center)51. Pdyn-CreER mice were kindly provided by Allen Institute. This work was supported by grants from the National Institutes of Health (NIH) R01NS081297, R37MH071679, and UG3MH120096 to G.F., the Simons Foundation SFARI to G.F., the Leonard and Isabelle Goldenson fellowship (FY19) to S.J.W., the William Randolph Hearst Fund (FY20) to S.J.W., F32 fellowship from National Institute of Mental Health F32MH125464 to S.J.W., F31 fellowship from National Institute of Neurological Disorders and Stroke F31NS110120 to E.S., and the BRAIN Initiative grant U19MH114830 to D.A.S. We thank the Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported this work. Finally, we are grateful for the comments and suggestions of Drs. Chris Harvey, Emilia Favuzzi, Elisabetta Furlanis, Leena A. Ibrahim on the manuscript. This facility is supported in part by the Neural Imaging Center as part of an NINDS P30 Core Center grant #NS072030.
INCLUSION AND DIVERSITY
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community.
Footnotes
DECLARATION OF INTERESTS
Gord Fishell is a founder of Regel Therapeutics, which has no competing interests with the present manuscript. Gord Fishell is an advisor for Neuron and Annual Review of Neuroscience.
REFERENCES
- 1.Fishell G, and Kepecs A (2020). Interneuron Types as Attractors and Controllers. Annu Rev Neurosci 43, 1–30. 10.1146/annurev-neuro-070918-050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tremblay R, Lee S, and Rudy B (2016). GABAergic interneurons in the neocortex: From cellular properties to circuits. Neuron 91, 260–292. 10.1016/j.neuron.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, et al. (2021). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26. 10.1016/j.cell.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kepecs A, and Fishell G (2014). Interneuron cell types are fit to function. Nature 505, 318–326. 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fino E, and Yuste R (2011). Dense Inhibitory Connectivity in Neocortex. Neuron 69, 1188–1203. 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fino E, Packer AM, and Yuste R (2013). The Logic of Inhibitory Connectivity in the Neocortex. Neuroscientist 19, 228–237. 10.1177/1073858412456743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karnani MM, Agetsuma M, and Yuste R (2014). A blanket of inhibition: functional inferences from dense inhibitory connectivity. Curr Opin Neurobiol 26, 96–102. 10.1016/j.conb.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñoz W, Tremblay R, Levenstein D, and Rudy B (2017). Layer-specific modulation of neocortical dendritic inhibition during active wakefulness. Science 355, 954–959. 10.1126/science.aag2599. [DOI] [PubMed] [Google Scholar]
- 9.Ma Y, Hu H, Berrebi AS, Mathers PH, and Agmon A (2006). Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci 26, 5069–5082. 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nigro MJ, Hashikawa-Yamasaki Y, and Rudy B (2018). Diversity and Connectivity of Layer 5 Somatostatin-Expressing Interneurons in the Mouse Barrel Cortex. J Neurosci 38, 1622–1633. 10.1523/JNEUROSCI.2415-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, Jeong H-Y, Tremblay R, and Rudy B (2013). Neocortical somatostatin-expressing GABAergic interneurons disinhibit the thalamorecipient layer 4. Neuron 77, 155–167. 10.1016/j.neuron.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naka A, Veit J, Shababo B, Chance RK, Risso D, Stafford D, Snyder B, Egladyous A, Chu D, Sridharan S, et al. (2019). Complementary networks of cortical somatostatin interneurons enforce layer specific control. eLife 8, e43696. 10.7554/eLife.43696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer C, Hafemeister C, Bandler RC, Machold R, Brito RB, Jaglin X, Allaway K, Butler A, Fishell G, and Satija R (2018). Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462. 10.1038/nature25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi D, Li Z, Lim L, Li M, Moissidis M, Yang Y, Gao T, Hu TX, Pratt T, Price DJ, et al. (2018). Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85. 10.1126/science.aar6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul A, Crow M, Raudales R, He M, Gillis J, and Huang ZJ (2017). Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539.e20. 10.1016/j.cell.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346. 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol 34, 199–203. 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, and Harkany T (2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol 34, 175–183. 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouwens NW, Sorensen SA, Baftizadeh F, Budzillo A, Lee BR, Jarsky T, Alfiler L, Baker K, Barkan E, Berry K, et al. (2020). Integrated Morphoelectric and Transcriptomic Classification of Cortical GABAergic Cells. Cell 183, 935–953.e19. 10.1016/j.cell.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scala F, Kobak D, Bernabucci M, Bernaerts Y, Cadwell CR, Castro JR, Hartmanis L, Jiang X, Laturnus S, Miranda E, et al. (2021). Phenotypic variation of transcriptomic cell types in mouse motor cortex. Nature 598, 144–150. 10.1038/s41586-020-2907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allaway KC, Gabitto MI, Wapinski O, Saldi G, Wang C-Y, Bandler RC, Wu SJ, Bonneau R, and Fishell G (2021). Genetic and epigenetic coordination of cortical interneuron development. Nature 597, 693–697. 10.1038/s41586-021-03933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, et al. (2018). Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLOS ONE 13, e0209648. 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. (2016). Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron 91, 1228–1243. 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, and Chen F (2021). Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol 39, 313–319. 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cable DM, Murray E, Zou LS, Goeva A, Macosko EZ, Chen F, and Irizarry RA (2022). Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol 40, 517–526. 10.1038/s41587-021-00830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomioka R, Okamoto K, Furuta T, Fujiyama F, Iwasato T, Yanagawa Y, Obata K, Kaneko T, and Tamamaki N (2005). Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. European Journal of Neuroscience 21, 1587–1600. 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- 28.Scala F, Kobak D, Shan S, Bernaerts Y, Laturnus S, Cadwell CR, Hartmanis L, Froudarakis E, Castro JR, Tan ZH, et al. (2019). Layer 4 of mouse neocortex differs in cell types and circuit organization between sensory areas. Nat Commun 10, 4174. 10.1038/s41467-019-12058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi G-A, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 19, 1743–1749. 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilscher MM, Leão RN, Edwards SJ, Leão KE, and Kullander K (2017). Chrna2-Martinotti Cells Synchronize Layer 5 Type A Pyramidal Cells via Rebound Excitation. PLOS Biology 15, e2001392. 10.1371/journal.pbio.2001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480.e22. 10.1016/j.cell.2018.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campagnola L, Seeman SC, Chartrand T, Kim L, Hoggarth A, Gamlin C, Ito S, Trinh J, Davoudian P, Radaelli C, et al. (2022). Local connectivity and synaptic dynamics in mouse and human neocortex. Science 375, eabj5861. 10.1126/science.abj5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeffer CK, Xue M, He M, Huang ZJ, and Scanziani M (2013). Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci 16, 1068–1076. 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, Allaway K, Sakopoulos S, Schneider G, Stevenson O, et al. (2020). Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat Neurosci 23, 1629–1636. 10.1038/s41593-020-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouchelon G, Vergara J, McMahon J, Gorissen BL, Lin JD, Vormstein-Schneider D, Niehaus JL, Burbridge TJ, Wester JC, Sherer M, et al. (2022). A versatile viral toolkit for functional discovery in the nervous system. Cell Reports Methods 2, 100225. 10.1016/j.crmeth.2022.100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pouchelon G, Dwivedi D, Bollmann Y, Agba CK, Xu Q, Mirow AMC, Kim S, Qiu Y, Sevier E, Ritola KD, et al. (2021). The organization and development of cortical interneuron presynaptic circuits are area specific. Cell Reports 37, 109993. 10.1016/j.celrep.2021.109993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sermet BS, Truschow P, Feyerabend M, Mayrhofer JM, Oram TB, Yizhar O, Staiger JF, and Petersen CC (2019). Pathway-, layer- and cell-type-specific thalamic input to mouse barrel cortex. eLife 8, e52665. 10.7554/eLife.52665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna WL, Ortiz-Londono CF, Mathew TK, Hoang K, Katzman S, and Chen B (2015). Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proceedings of the National Academy of Sciences 112, 11702–11707. 10.1073/pnas.1504144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callaway EM, Dong H-W, Ecker JR, Hawrylycz MJ, Huang ZJ, Lein ES, Ngai J, Osten P, Ren B, Tolias AS, et al. (2021). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102. 10.1038/s41586-021-03950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, and Zeng H (2017). The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 96, 542–557. 10.1016/j.neuron.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngai J (2022). BRAIN 2.0: Transforming neuroscience. Cell 185, 4–8. 10.1016/j.cell.2021.11.037. [DOI] [PubMed] [Google Scholar]
- 42.Berg J, Sorensen SA, Ting JT, Miller JA, Chartrand T, Buchin A, Bakken TE, Budzillo A, Dee N, Ding S-L, et al. (2021). Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158. 10.1038/s41586-021-03813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugeon S, Duffield J, Dipoppa M, Ritoux A, Prankerd I, Nicoloutsopoulos D, Orme D, Shinn M, Peng H, Forrest H, et al. (2022). A transcriptomic axis predicts state modulation of cortical interneurons. Nature 607, 330–338. 10.1038/s41586-022-04915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condylis C, Ghanbari A, Manjrekar N, Bistrong K, Yao S, Yao Z, Nguyen TN, Zeng H, Tasic B, and Chen JL (2022). Dense functional and molecular readout of a circuit hub in sensory cortex. Science 375, eabl5981. 10.1126/science.abl5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EJ, Zhang Z, Huang L, Ito-Cole T, Jacobs MW, Juavinett AL, Senturk G, Hu M, Ku M, Ecker JR, et al. (2020). Extraction of Distinct Neuronal Cell Types from within a Genetically Continuous Population. Neuron 107, 274–282.e6. 10.1016/j.neuron.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yetman MJ, Washburn E, Hyun JH, Osakada F, Hayano Y, Zeng H, Callaway EM, Kwon H-B, and Taniguchi H (2019). Intersectional monosynaptic tracing for dissecting subtype-specific organization of GABAergic interneuron inputs. Nat Neurosci 22, 492–502. 10.1038/s41593-018-0322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morishima M, Kobayashi K, Kato S, Kobayashi K, and Kawaguchi Y (2017). Segregated Excitatory-Inhibitory Recurrent Subnetworks in Layer 5 of the Rat Frontal Cortex. Cereb Cortex 27, 5846–5857. 10.1093/cercor/bhx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapfer C, Glickfeld LL, Atallah BV, and Scanziani M (2007). Supralinear increase of recurrent inhibition during sparse activity in the somatosensory cortex. Nat Neurosci 10, 743–753. 10.1038/nn1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otsuka T, and Kawaguchi Y (2009). Cortical Inhibitory Cell Types Differentially Form Intralaminar and Interlaminar Subnetworks withExcitatory Neurons. J. Neurosci. 29, 10533–10540. 10.1523/JNEUROSCI.2219-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pluta SR, Telian GI, Naka A, and Adesnik H (2019). Superficial Layers Suppress the Deep Layers to Fine-tune Cortical Coding. J. Neurosci. 39, 2052–2064. 10.1523/JNEUROSCI.1459-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, et al. (2014). An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242. 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ting JT, Lee BR, Chong P, Soler-Llavina G, Cobbs C, Koch C, Zeng H, and Lein E (2018). Preparation of Acute Brain Slices Using an Optimized N-Methyl-D-glucamine Protective Recovery Method. Journal of Visualized Experiments : JoVE. 10.3791/53825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams SR, and Mitchell SJ (2008). Direct measurement of somatic voltage clamp errors in central neurons. Nat Neurosci 11, 790–798. 10.1038/nn.2137. [DOI] [PubMed] [Google Scholar]
- 55.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. (2014). Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell 157, 726–739. 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 56.Favuzzi E, Deogracias R, Marques-Smith A, Maeso P, Jezequel J, Exposito-Alonso D, Balia M, Kroon T, Hinojosa AJ, F. Maraver E, et al. (2019). Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science 363, 413–417. 10.1126/science.aau8977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Slide-seqV2 data generated for this study are available at the Broad Institute Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP2082/cortical-somatostatin-interneuron-subtypes-form-cell-type-specific-circuits#study-summary. Sparse labeling images of SST interneurons have been deposited at Harvard Dataverse and can be accessed through the following link: https://doi.org/10.7910/DVN/NQDIPG. Additional relevant information is available on our lab website: https://fishelllab.hms.harvard.edu/publications.
Original codes for clustering of snRNA-seq data and analysis of Slide-seq V2 experiments are available at https://github.com/gs512/slideseq-engine.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


