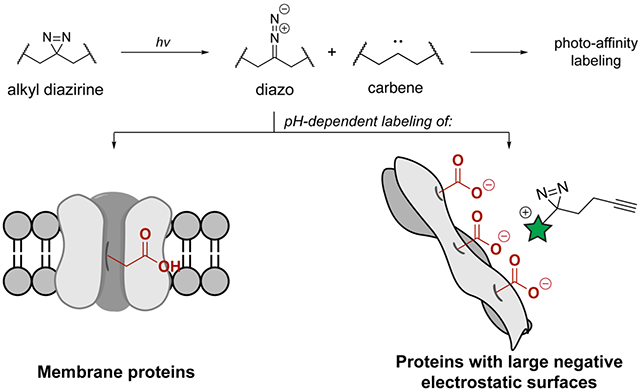
Abstract
Diazirines are widely used in photoaffinity labeling (PAL) to trap non-covalent interactions with biomolecules. However, design and interpretation of PAL experiments is challenging without a molecular understanding of the reactivity of diazirines with protein biomolecules. Here, we report a systematic evaluation of the labeling preferences of alkyl and aryl diazirines with individual amino acids, single proteins, and in the whole cell proteome. We find that alkyl diazirines exhibit preferential labeling of acidic amino acids in a pH-dependent manner that is characteristic of a reactive alkyl diazo intermediate, while aryl-fluorodiazirines labeling patterns reflect reaction primarily through a carbene intermediate. From a survey of 32 alkyl diazirine probes, we use this reactivity profile to rationalize why alkyl diazirine probes preferentially enrich highly acidic proteins or those embedded in membranes and why probes with a net positive-charge tend to produce higher labeling yields in cells and in vitro. These results indicate that alkyl diazirines are an especially effective chemistry for surveying the membrane proteome, and will facilitate design and interpretation of biomolecular labeling experiments with diazirines.
Graphical Abstract
INTRODUCTION
Since the development of photoaffinity labeling (PAL) in 1962,1 PAL has emerged as a key approach for measuring biomolecular interactions, protein structure, and the identification of small molecule targets in the cell. The combination of PAL with high-throughput sequencing2 and mass spectrometry-based proteomics3, 4 has significantly increased the depth of information measured from a single experiment and has enabled mapping of lipid,5 carbohydrate,6 and small molecule interactomes,7 broadly widening the scope of binding interactions with proteins in cells. In particular, the design of small tags embedded with the alkyl diazirine as the PAL functional group and a chemical enrichment handle has accelerated the use of PAL within large-scale biological measurements,8–10 and the combination of this technology with isotopically encoded enrichment tools has enabled the mapping of small molecule binding sites in cells.7, 11, 12
Despite the use of diazirines as probes to measure biomolecular interactions, the incomplete understanding of the labeling preferences of the diazirine limits the design and interpretation of these experiments. To overcome challenges in interpretation, the development of a diazirine “backgroundome”13 or methods to differentiate between selective and non-selective labeling,14 including the use of enantioprobes,12 or introduction of a scavenger,15 have been implemented. During photolysis, the diazirine primarily isomerizes to a diazo intermediate that further converts to the carbene for biomolecular conjugation,16, 17 which will either rapidly form a covalent bond with a nearby biomolecule or be quenched by the aqueous environment (Figure 1a).18 However, the generation of a reactive, electrophilic diazo intermediate during PAL may drive protein labeling through an alternate “pseudo-PAL” mechanism.18 For example, stabilized diazo compounds exhibit preferential reactivity with organic acids19 and selectively react with protein carboxylates to generate esters.20 Evidence for alkyl diazirine reactivity patterns that transition through the diazo isomer to preferentially react with acidic residues has been recently noted using in vitro peptide or protein model systems.21–23 Despite the investigation of reactivity preferences of other PAL functional groups (e.g., aryl azides,24, 25 benzophenones,26, 27 and aryl tetrazoles28–30) the chemical reactivity and resulting labeling preferences of the diazirine with amino acids have only been investigated via in vitro systems and peptide digests by mass spectrometry.31 Systematic establishment of how alkyl diazirines exhibit differential reactivity preferences from aryl diazirines, and the implications this has on biomolecular labeling, would facilitate interpretation of the measurements and conclusions from PAL experiments using diazirines.
Figure 1.
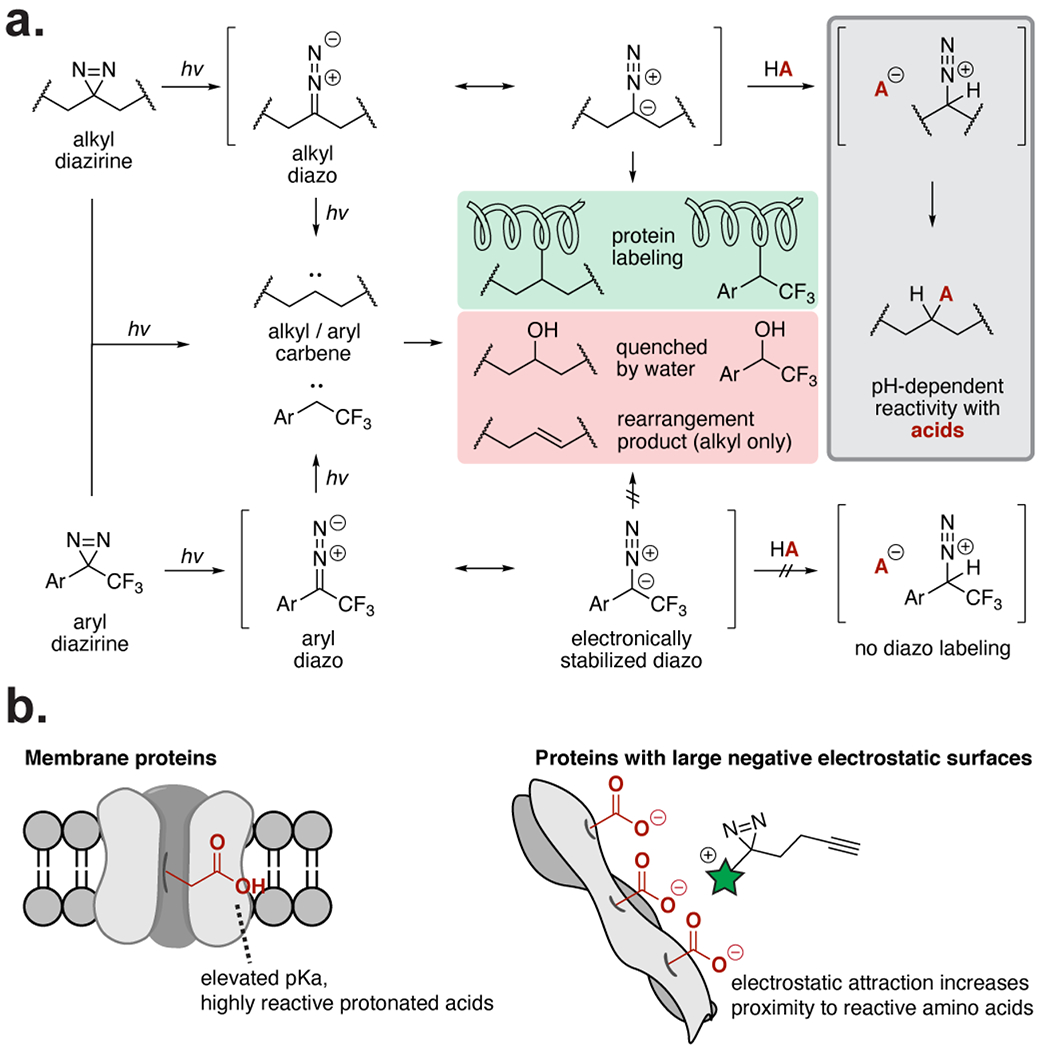
Overview of diazirine reactivity pathways. (a) Alkyl and aryl diazirines form carbene and diazo intermediates upon irradiation. Carbenes label nearby proteins, but are rapidly quenched if no protein substrate is nearby. Alkyl diazo intermediates react selectively with acids, while electronically stabilized aryl diazo intermediates do not. (b) The acid-selectivity of the alkyl diazo intermediates causes increased labeling of membrane proteins, which have more reactive protonated carboxylic acids, and proteins with large negative electrostatic surfaces. Labeling of these protein surfaces increases when the PAL probe (highlighted in green) is positively charged.
Here, we report a systematic study of alkyl and aryl diazirine labeling preferences with individual amino acids and single proteins, and use these data to interpret the protein binding partners visualized by a 32-membered alkyl diazirine library from the whole cell proteome. We demonstrate that the aryl-trifluorodiazirine primarily reacts through a carbene intermediate, while the alkyl diazirine transitions through a reactive diazo intermediate that preferentially reacts with acidic amino acids in a pH-dependent manner. Reaction of the alkyl diazirine through a diazo intermediate rationalizes the preferential labeling of membrane proteins and proteins with negatively charged electrostatic surfaces in cells (Figure 1b). PAL ligands that are protonated at physiological pH, but not ligands that are neutral or deprotonated at physiological pH, possess affinity for, and therefore heightened labeling of, these protein surfaces, which is reflected by global protein profiles and binding site maps. This understanding of diazirine labeling preferences improves the ability to design and interpret the results from biological experiments using diazirines.
RESULTS
Alkyl and aryl diazirine reactivity profile with individual amino acids under neat conditions
We initiated a systematic evaluation of diazirine chemistry by evaluating the photolysis products of the alkyl diazirine9 in aqueous conditions (20% CD3CN–D2O) and recovered a mixture of olefins (50%), a water-insertion product (35%), and a ketone (10–20%), presumably from reaction of the carbene with oxygen (Figure S1).32 We next examined the reactivity of the Fmoc-protected alkyl diazirine 1 with individual amino acids (Ac-AA-OMe) under neat conditions to represent the closest possible binding interaction between the diazirine and a protein. Neat mixtures of the Fmoc-diazirine 1 with each Ac-AA-OMe (4 equiv) were irradiated using a broadband lamp (280–400 nm) and analyzed by liquid chromatography–mass spectrometry (LC-MS) (Figure 2a). The Fmoc-diazirine 1 covalently labeled relatively acidic polar amino acids with the highest yields (40–60% yield), followed by a subset of the additional polar amino acids (<20%), and had no observable labeling of the remaining polar or aliphatic amino acids (<3%). For comparison, the aryl diazirine 2 was evaluated against individual amino acids (4 equiv) under neat conditions by 19F NMR. The aryl diazirine 2 produced insertion products with all 20 amino acids and afforded the highest yield with cysteine (Figure 2b, S2). While optimizing these reactions, we observed a transient 19F NMR peak at −57 ppm, matching the reported 19F NMR shift of similar aryl-trifluoromethyl diazo compounds.33 We found the intermediate did not independently react, and disappeared following longer irradiation times.
Figure 2.
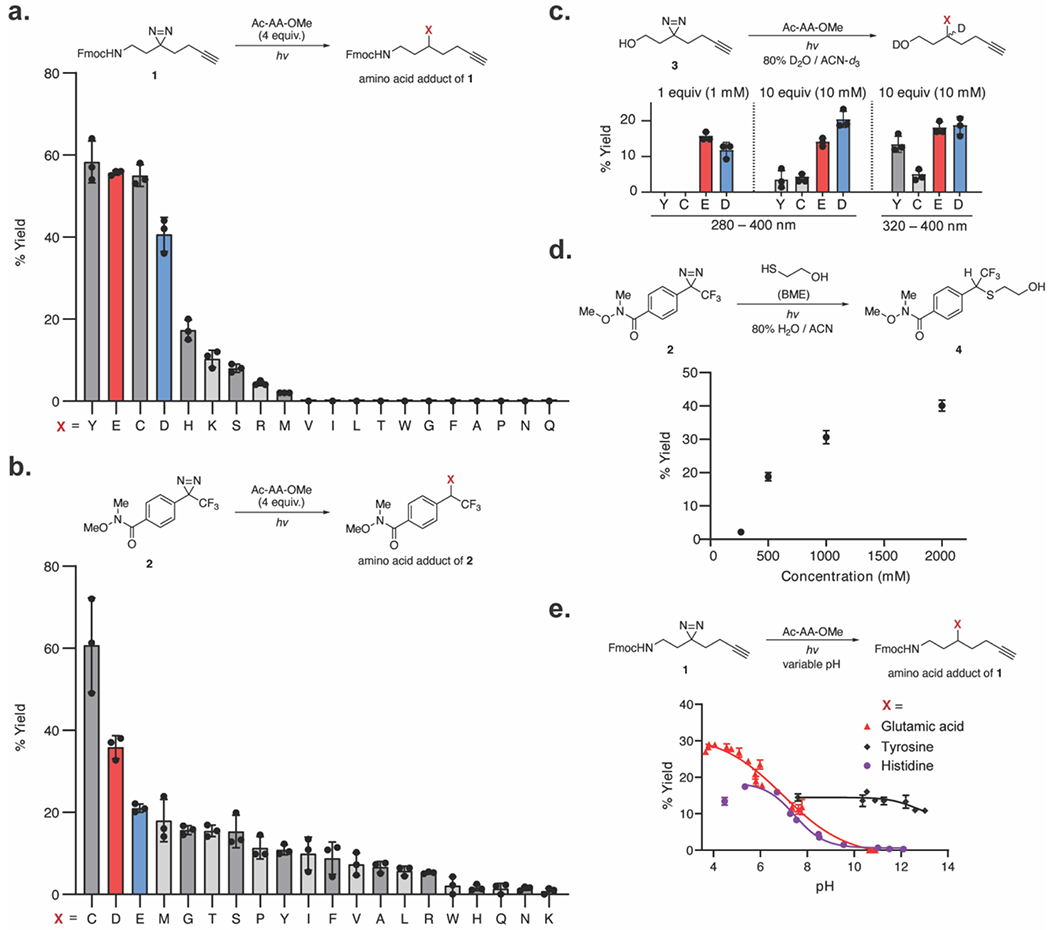
Reactivity of alkyl and aryl diazirines with the 20 natural amino acids. (a) Reactivity of alkyl diazirines with the 20 N-acetyl, O-Me protected amino acids (4 equiv) in neat conditions. Yields calculated against an internal standard by LC-MS. (b) Reactivity of aryl diazirines with the 20 amino acids (4 equiv) in neat conditions. Yields calculated against an internal standard by [isp]19F NMR. (c) Reactivity of alkyl diazirines with the 20 amino acids in solution (1 mM 3, 1 mM or 10 mM amino acid) with 280–400 nm or 320–400 nm excitation. Only reactive amino acids are displayed. Yields calculated against an internal standard by 1H NMR. (d) Reaction of aryl diazirine 2 with β-mercapto ethanol at varying concentrations in aqueous conditions. Yields calculated with relative integration by LC-MS. (e) Yield of alkyl diazirines with individual amino acids as a function of pH. Yields calculated against an internal standard by LC-MS. Plotted values show the mean with error bars representing the standard deviation with n = 3.
Alkyl and aryl diazirine reactivity in aqueous solution
We next evaluated the product distribution in aqueous conditions (80% D2O–CD3CN) at equimolar concentrations of the amino acid and the diazirine (1 mM). Under these conditions, we expected that the carbene intermediate generated by each probe would be rapidly quenched by D2O before reacting with the amino acid residue. Surprisingly, photo-irradiation of the alkyl diazirine 3 at equimolar amounts of the individual amino acids (1 mM) under aqueous conditions revealed selective conjugation with Glu and Asp (10–15% yield, Figure 2c, S3). The insertion of the alkyl diazirine 3 to Tyr and Cys was additionally detected in aqueous solution at elevated concentrations (10 mM, 10 equiv). In order to investigate the possibility that the alkyl diazo intermediate 3 was inducing the observed reactivity with acidic residues, analogous to the reactivity of bona fide diazo reagents with acidic amino acid residues,19 we filtered the broadband excitation wavelength to exclude wavelengths below 320 nm,34 and thus slowed the transition of the diazo to carbene at 300 nm. Indeed, filtration of the light source to a narrower range (320–400 nm) increased reactivity of the diazirine 3 with the weak acids, Tyr and Cys (Figure 2c).
By contrast, the aryl diazirine 2 formed no observed adducts with any amino acid up to 10 mM (10 equiv) in aqueous solution. To evaluate the limit of reactivity of the aryl diazirine 2 to conjugate with a molecule in aqueous solution, β-mercaptoethanol (BME) was used as a more soluble surrogate for the most-reactive amino acid, cysteine (Figure 2d, S4). The aryl diazirine 2 formed conjugated products with BME beginning at 250 mM concentrations (250 equiv). Notably, the aryl diazirine 2 formed comparable yields with BME at 500 mM to the alkyl diazirine 3 with 1 mM Glu. These data suggest that the aryl diazirine 2 primarily reacts through a short-lived carbene and that the aryl diazo intermediate is not driving alternate reaction pathways under the photolysis conditions used here.
Alkyl diazirine labeling of amino acids is pH-dependent
Reaction of acidic residues with the alkyl diazirine through the diazo intermediate requires a proton transfer from the acid to give a cationic intermediate prior to displacement of dinitrogen by the conjugate base (Figure 1a). If the proton transfer is the rate limiting step, the insertion yield will therefore be dependent on the protonation state of the amino acid. The pH-dependence of alkyl diazirine labeling was investigated by titrating aqueous solutions of Glu, His, Tyr, and Lys (10 equiv) with acid or base before addition of the diazirine 1 (1 equiv) and filtered UV irradiation (320–400 nm). Product yields were measured by LC-MS. Glu, His, and Tyr produced pH-dependent insertion products with the alkyl diazirine 1, while no observable insertion was observed with the fully deprotonated carboxylate and free base His (Figure 2e). Protonated Lys was also not reactive under these conditions, suggesting that the pKa of the conjugate acid is too high to react with the diazo intermediate generated by the diazirine 1. In sum, these data suggest that the alkyl diazirine 1 generates a reactive intermediate that leads to preferential reactivity with protonated acidic residues in aqueous solution.
In the course of these studies, we additionally found that a significant fraction of the photolyzed diazirine underwent rapid internal rearrangement to a mixture of olefins, which may be a major rearrangement product from the carbene intermediate.23 To minimize this elimination pathway, we drew on the kinetic isotope effect35 and synthesized a deuterated alkyl diazirine (Figure S5). Photolysis of the tetra-deuterated diazirine afforded a greater yield of the ketone (42%) and reduced the yield of the olefin products (34%) as compared
to the tetra-proton analogue. Thus, deuteration of the alkyl diazirine may improve covalent labeling yields in PAL experiments.
Alkyl diazirine labeling of single proteins is pH-dependent
To translate these observations to proteins, we next examined the labeling efficiency of alkyl and aryl diazirines36 in vitro on an individual protein, bovine serum albumin (BSA), over a 5.8–8.0 pH range in Tris buffer (Figure 3a). Analogous to the reactions with single amino acids, labeling from the alkyl diazirine 3 was higher at lower pH, while labeling from the aryldifluorodiazirine 5 was pH-independent over the range examined (Figure 3b, 3c). Interestingly, we found that performing a similar labeling experiment of BSA with the alkyl diazirine 3 in acidic phosphate buffer (pH = 5.8) resulted in concentration-dependent labeling of BSA by the alkyl diazirine 3, presumably via competitive reactivity with the phosphate buffer (Figure S7). Indeed, we observed formation of a phosphate adduct with 3 in a pH-dependent manner analogous to the aqueous reactivity of 3 with the protected amino acids, suggesting that acidic phosphate was reacting with the diazo isomer of 3. These data demonstrate that the differential reactive intermediates from the alkyl diazirine and the aryl diazirine predictably impact observed labeling patterns on individual substrates.
Figure 3.
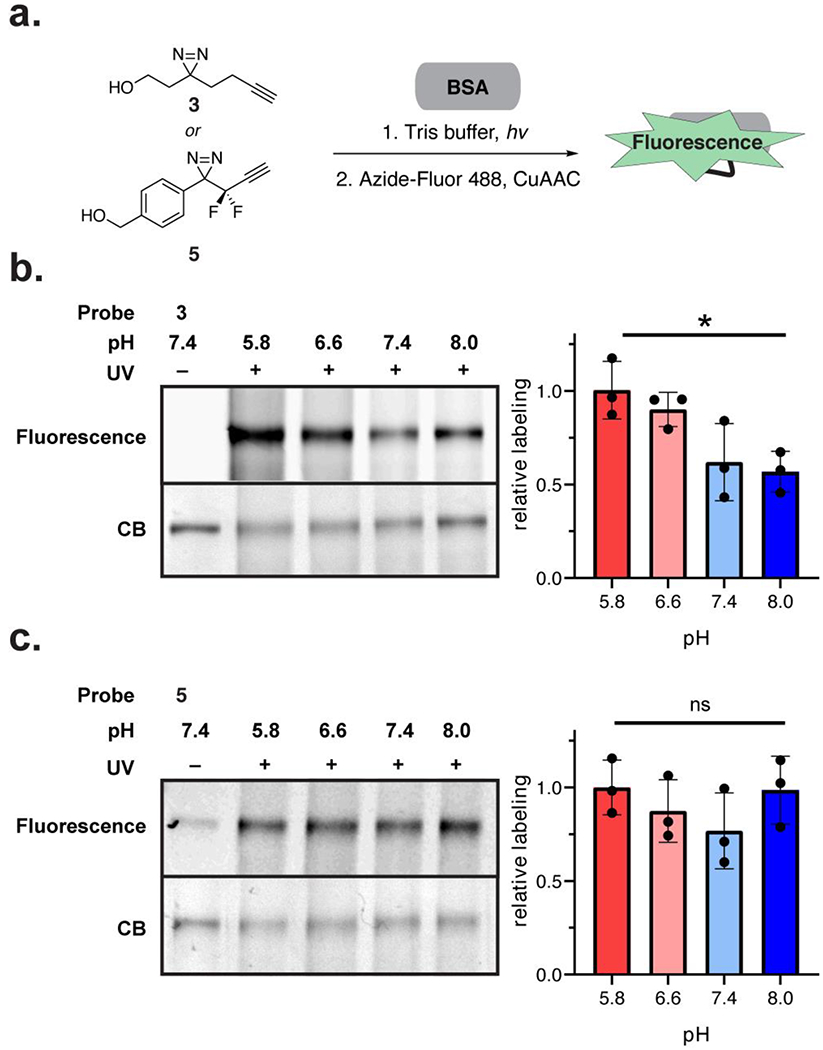
Photolabeling of single proteins as a function of pH. (a) Workflow schematic. Alkyl diazirine 3 or aryl-difluorodiazirine 5 were incubated with BSA in Tris buffer with varying pH (5.8, 6.6, 7.4, 8.0). The diazirines were crosslinked to the protein by UV irradiation and visualized by in-gel fluorescence following click chemistry with Azide-Fluor 488. (b) pH-dependent labeling of BSA by alkyl diazirine 3. (c) pH-independent labeling of BSA by aryl diazirine 5. Labeling yields were calculated with fluorescence signal normalized to coomassie blue (CB) stain signal. Plotted values show the mean with error bars representing the standard deviation with n = 3. Statistical significance was determined with a t-test between pH 5.8 and 8.0 samples (* = p-value < 0.05)
PAL probes with a net positive charge increase alkyl diazirine labeling in the cell
The pH dependence of alkyl diazirine chemistry in vitro implies that charge state will play a significant role in the capture of protein substrates and may explain the inherent labeling efficiency of different alkyl diazirine probes.7, 12 To evaluate the influence of alkyl diazirine reactivity on PAL experiments in cells, a set of alkyl diazirine probes with a distribution of net positive, negative, or neutral charges (representative structures in Figure 4a; for all structures see Figure S8). The PAL probes cover a diversity of small molecules including aromatic scaffolds (e.g., JN3, JN939, JN849, JN935), analogs of nucleotides (e.g., JN846, JN847, JN936), lipids (e.g., JN38), and other metabolites (e.g., JN247, JN835).
Figure 4.
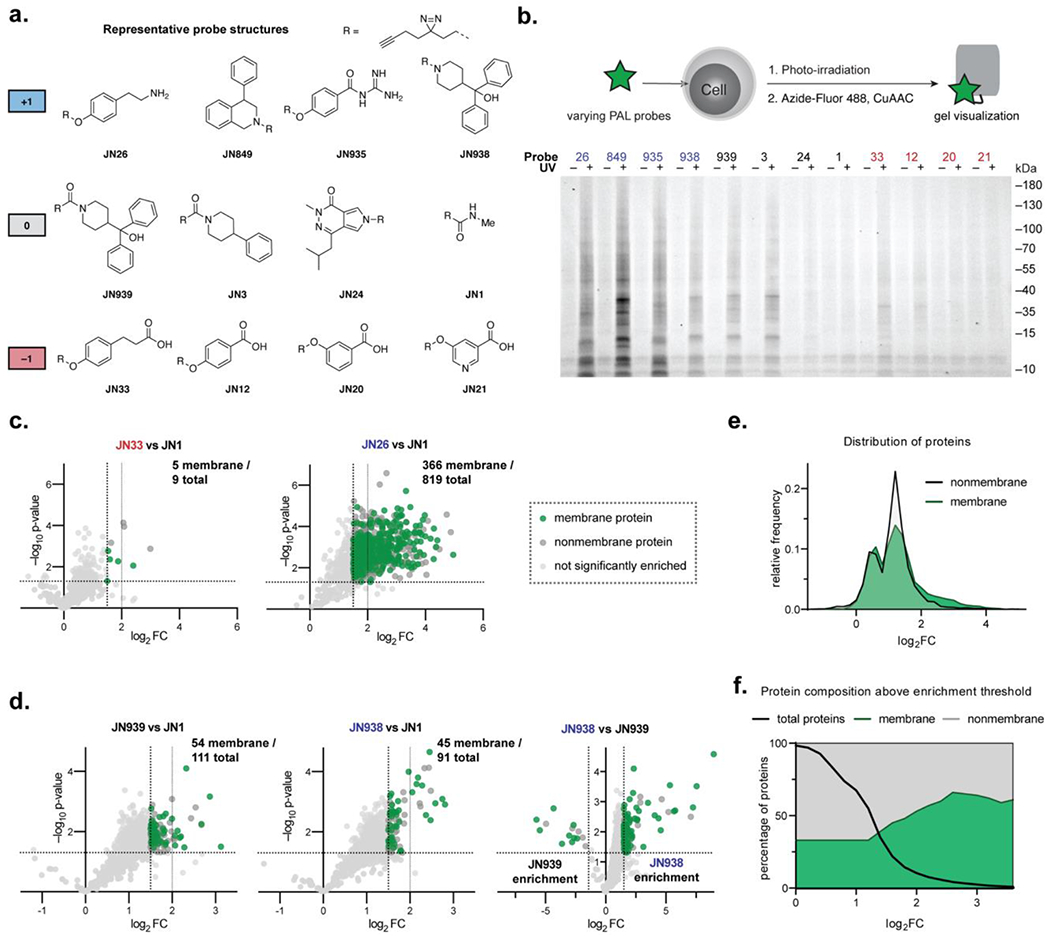
Whole proteome photolabeling in cells with PAL probes. (a) Representative probe structures (for a full list see Figure S8). (b) Photolabeling yields of compounds visualized by fluorescence signal. (c) Comparison of photolabeling profiles of carboxylic acid probe JN33 and primary amine JN26. Significantly enriched proteins are colored as membrane and nonmembrane proteins. (d) Photolabeling profiles of neutral amide probe JN939 and positively-charge amine analogue JN938. (e) Histogram showing the relative frequency of membrane and nonmembrane protein enrichments for all data in (c) and (d). (f) Percentage of membrane proteins above enrichment thresholds for all data in (c) and (d).
From this library, we selected representative probe structures based on their overall charge at physiological pH and evaluated their labeling efficiency by in-gel fluorescence. HEK293T cells were incubated with individual probes at 10 µM and photo-irradiated for 60 sec to crosslink the ligands to their protein binding partners. The cells were harvested, lysed, and visualized by in gel fluorescence following click chemistry with Azide-Fluor 488. As expected, probe labeling efficiency generally correlated with the net charge of the probe (Figure 4b). The alkyl diazirine probes with a net positive charge exhibited higher labeling than neutral or negatively charged probes (Figure 1b). To examine how factors such as cell permeability affected labeling efficiency, we performed PAL with the alkyl diazirine probes on HEK293T lysates (Figure S10). In general, the same trend of positively charged probes producing higher labeling yields than neutral or negatively charged probes were observed in lysates as in whole cells, but higher labeling from a subset of the negatively charged probes, most notably JN33, was found in lysates than from whole cells. These data suggest that in general positively charged probes have higher labeling propensities that may reflect increased interaction with negatively charged protein surfaces composed of acidic residues that react with alkyl diazirines, and that overall labeling yields from a particular probe reflect a composite of the diazirine chemistry and cellular permeability.
To directly examine the contribution of charge to the labeling efficiency in probe design, we compared the enriched proteome of two pairs of alkyl diazirine PAL probes with different net charge by chemical proteomics: first, a primary amine vs. a carboxylic acid (JN26 vs. JN33), and second, a tertiary amine vs. an amide (JN938 vs. JN939). SK-N-SH cells were incubated with individual PAL probes, photo-crosslinked, harvested, and lysed. Lysates were conjugated with biotin-azide using click chemistry and the biotinylated proteins were enriched with streptavidin-agarose. The enriched proteins were digested with trypsin for quantitative proteomics (TMT10plex). The positively charged JN26 exhibited a much higher degree of labeling compared to the negatively charged analogue JN33 (819 and 9 enriched proteins, respectively, Figure 4c). The effect from probe charge state was diminished, but still noticeable when comparing neutral versus positively charged probes with larger, more hydrophobic structures. The neutral amide probe JN939 significantly enriched slightly more proteins than the positively charged amine JN938 at a 3-fold enrichment threshold (111 and 91 enriched proteins, respectively), but the degree of enrichment was higher for the positive amine JN938 than the neutral amide JN939 when the enriched proteins from each probe were compared directly (JN938 vs JN939, Figure 4d). Taken together, these data imply that alkyl diazirine probes with a net positive charge will yield elevated labeling of the proteome, potentially due to increased localization to protein surfaces that are more readily crosslinked by alkyl diazirine chemistry, and that this effect is attenuated with larger hydrophobic probes.
PAL probes disproportionately label membrane proteins in cells
In line with prior reports,5, 7, 11–13, 37 we observed that alkyl diazirine probes possess an enrichment bias for membrane proteins (Figure 4e). While membrane proteins made up 33% of all proteins observed in our dataset, similar to the distribution of these proteins in the global proteome (27%),38 membrane proteins were over-represented after filtering for protein enrichment: 46% of proteins that had at least a 3-fold enrichment, and 56% of proteins with at least a 4-fold enrichment were membrane proteins (Figure 4f). These membrane proteins derived from throughout the cell, including the cell surface, ER/golgi, mitochondrion, and nuclear membranes. The preferential labeling of membrane proteins by alkyl diazirine probes may reflect the elevated pKa of Glu and Asp residues in lipid bilayers.39–41 Similarly, a recent study by Cravatt and co-workers examined the types proteins labeled by an assortment of alkyl diazirine probes and found that membrane proteins were indeed enriched (~20–40% of protein targets).7 Analysis of the most-enriched set of proteins in this dataset revealed that membrane proteins were disproportionately common (38–61% of proteins with a SILAC ratio ≥ 20 ) (Figure S12).
Understanding alkyl diazirine reactivity improves interpretation of binding site mapping results
To further evaluate the labeling trends in cells, the entire 32-membered alkyl diazirine probe library was divided into groups according to similarities in the probe charge and structure and dosed to SK-N-SH cells for protein and binding site mapping (Figure 5a). In brief, SK-N-SH cells were dosed in biological duplicate with the alkyl diazirine probe library in six groups of 5–6 compounds at 10 µM concentrations of each compound for 2 h. The treated cells were photo-irradiated for 60 sec, collected, and lysed. Labeled proteins in the cellular lysates were tagged with an acid-cleavable biotin picolyl biotin azide42 for affinity enrichment and trypsin digestion on streptavidin–agarose beads. The labeled peptides representing binding interactions of the PAL probe with the protein target were subsequently eluted from the beads by acid cleavage and recovered for analysis by LC-MS, which were analyzed by two technical replicates. The probe-labeled peptide was assigned by database searching against the Swissprot human proteome, followed by filtration based on the embedded isotopic pattern,43 and manual validation. A total of 632 binding sites from 170 proteins were mapped in SK-N-SH cells across 4,200 peptide spectral matches (PSMs).
Figure 5.
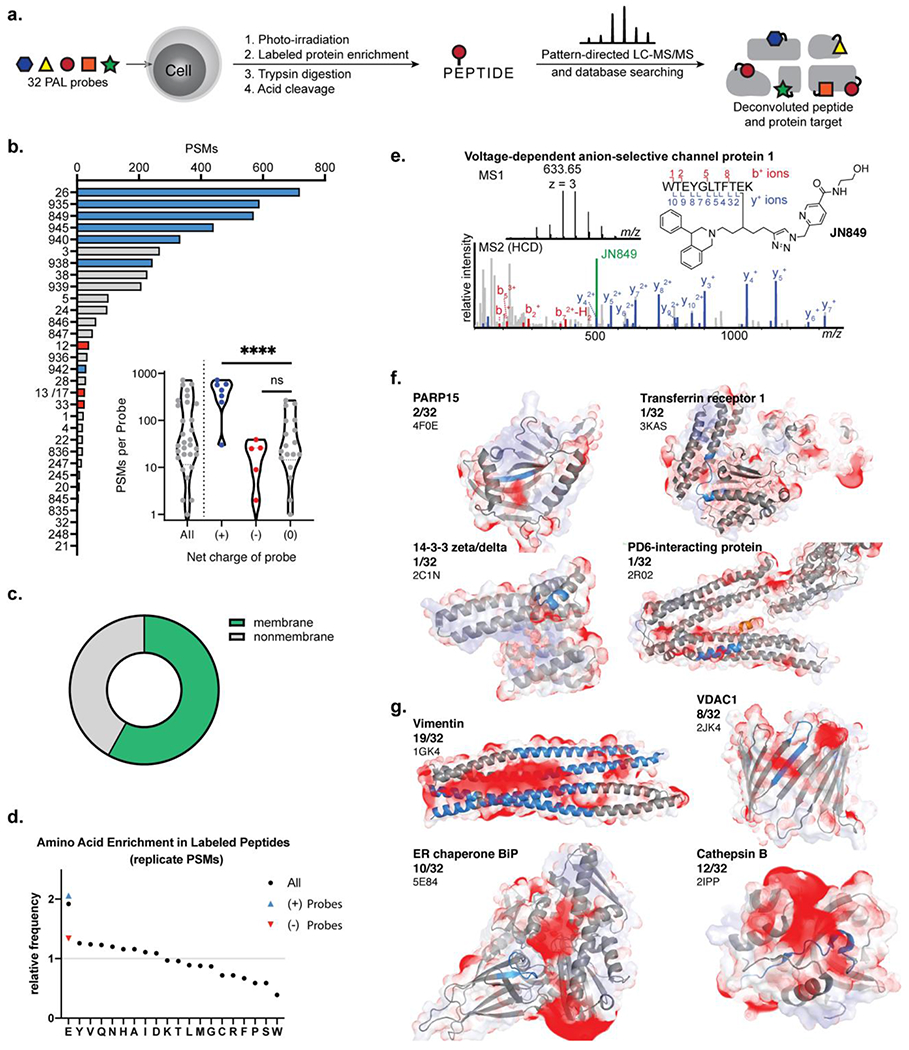
Binding site mapping with a 32 PAL probe library. (a) Cell cultures were treated simultaneously with 5 or 6 PAL probes which were photo-conjugated to protein binding partners. The conjugated proteins were enriched and the conjugated peptide representing the binding site of the PAL probe was isolated for analysis using isotope-targeted MS. (b) Count of peptide spectral matches (PSMs) assigned to each PAL probe. PAL probes with a net positive charge highlighted in blue, neutral probes in grey, and negatively charged probes in red. (c) Comparison of binding site PSMs from membrane and nonmembrane proteins. (d) Amino acid frequency in the conjugated peptides relative to frequency in human proteome. (e) Annotated mass spectra and assignment of a VDAC1 binding site. (f) Negative electrostatic maps (red) overlaid on the conjugated peptide (blue) for unique binding interactions. (g) Negative electrostatic maps (red) overlaid on conjugated peptide (blue) for frequently conjugated proteins.
Analysis of the mapped binding sites reflected the labeling trends observed on the proteome level. PSMs were used to approximate the relative abundance of labeling events with protein binding partners from each PAL probe (Figure 5b). Examination of the number of PSMs per probe revealed a bimodal distribution that clustered as a function of positively charged probes versus neutral or negatively charged probes (Figure 5b, inset). Positively charged probes produced on average 418 PSMs corresponding to 50 unique binding sites per probe whereas neutral probes produced an average of 66 PSMs (14 unique binding sites) and negatively-charge probes produced an average of 14 PSMs (5 unique binding sites). No statistical difference between the neutral or negatively charged PAL probes was observed, although neutral PAL probes with a larger hydrophobic surface area tended to yield a greater number of PSMs (e.g., JN3, JN939, JN38). Similar to observations from the proteome, we found that a majority of the binding sites were from membrane proteins (58% of PSMs) (Figure 5c). The wider structural diversity of the 32 PAL probes suggests the increased labeling of membrane proteins is a result of the labeling propensity of alkyl diazirine chemistry in contrast to the probes’ individual structures.
Although the positively charged members of the library were six of the top seven probes by PSM counting, the positively charged probe JN942 was an outlier at rank 21 of 32 probes. We investigated the cause of this outlier in vitro and observed unaffected labeling efficiencies on individual amino acids, and proteins (Figure S13), suggesting that in addition to the net charge, cellular accessibility of the small molecule to biomolecular targets should also be considered in probe design.
As further evidence of labeling preferences between alkyl diazirines and acidic amino acid residues in the whole proteome, we compared the occurrence of amino acids found within labeled peptides to their natural occurrence in the human proteome (Figure 5d). Glutamic acid was the most enriched amino acid in labeled peptides, increased specifically in peptides labeled by positively charged probes, and most of this enrichment was eliminated in binding sites measured from negatively charged probes. The limited enrichment of aspartate residues reflects previous trends observed from labeling propensities with bona fide diazo esterification reagents.20 This may be caused by differences in steric accessibility or the generally lower pKa of Asp relative to Glu, which would favor reactivity with Glu and disfavor reactivity with Asp. In sum, for alkyl diazirine PAL experiments in cells, glutamic acid has the highest propensity for labeling than other amino acids and the labeling frequency is influenced by the local pKa environment and the physical properties of the photoaffinity probe.
Across all the mapped probe-labeled binding sites, we found that a binding site on VDAC1 was frequently observed (representative assignment, Figure 5e). This peptide on VDAC1 is in close proximity to two negatively charged residues on the protein (Y67, E73). VDAC1 E73 is notable for its unusually high pKa (predicted membrane pKa = 7.4) and ability to sensitively mediate dimerization of VDAC1 in a pH-dependent manner.44 Protonation of VDAC1 E73 would increase the propensity for E73 to react with a diazo intermediate. VDAC1 and VDAC2, but not VDAC3, possess this glutamic acid residue, which explains why only VDAC1 and VDAC2 are frequently enriched targets in alkyl diazirine studies.13
Intrigued by this analysis with VDAC1, we next visualized a subset of the binding sites onto available protein structures (binding sites highlighted in dark blue, Figure 5f, 5g). The electrostatic potential of each protein surface was calculated using the adaptive Poisson-Boltzmann solver (APBS) and overlaid on the structure of the protein. For clarity, only the negative electrostatic surface map is displayed in red. For binding sites that were observed as labeled by only one or two PAL probes, the mapped peptide has a minor degree of negative electrostatic density nearby (Figure 5f). In some cases, the mapped peptide is within 9 Å21 in proximity to a known binding site for the natural protein ligand. For example, ADP-ribosyltransferase protein PARP15 has a nicotinamide binding cleft that is adjacent to the observed binding site.45 PD6-interacting protein (PDCD6IP) binds to a natural peptide ligand in close proximity to the observed small molecule binding site (peptide ligand highlighted in orange, Figure 5f).46 The 14–3-3 proteins are receptors for peptides in a cleft within 9 Å of the mapped binding site.47 Transferrin receptor 1 is a membrane-bound protein; the binding site is in the extracellular region.48
In contrast, binding sites on proteins that were mapped to multiple PAL probes exhibited significantly greater negative electrostatic densities around the labeled peptide (highlighted in dark blue, Figure 5g). Vimentin is a filamentous protein that is frequently identified in alkyl diazirine labeling experiments, likely due to the significant negative electrostatic density on vimentin (net charge = –27). Unsurprisingly, the 17 unique peptides conjugated across 19 different PAL probes identified from vimentin colocalize with the negative electrostatic density map. Likewise, 8–12 PAL probes conjugated to VDAC1, ER chaperone BiP, and cathepsin B, which each display similar negative electrostatic densities in close proximity to the mapped peptide binding site. Taken together, these data suggest that the subset of proteins that are labeled more frequently by alkyl diazirine chemistry typically have negative electrostatic regions that are characterized by a density of acidic residues or those with a relatively high localized pKa, resulting in an increase in the degree of protonation of the amino acid for labeling through the diazo intermediate during PAL experiments.
DISCUSSION
Despite the growing use and application of diazirine chemistry in PAL experiments, a systematic understanding of the labeling preferences of the diazirine in cells has yet to emerge. Here, we report a systematic analysis of diazirines and their preferential reactivity pattern with protein biomolecules in vitro and in cells. We find that alkyl diazirines preferentially react with acidic amino acid residues in a pH-dependent manner in vitro, which can be rationalized by the formation of a long-lived diazo intermediate that is intercepted by organic acids prior to formation of an alkyl carbene. By contrast, the aryl-trifluorodiazirine forms insertion products with all 20 amino acids, is less dependent on pH, and its labeling chemistry is readily quenched by water, which are characteristics of a reaction pathway through a carbene intermediate. Although reactions with individual amino acids in neat conditions is only an approximation of the PAL probe reactivity with a target protein in cells, the impact of the alkyl diazirine reactivity pattern translates to a predictable enhancement of cellular labeling with alkyl diazirine probes carrying a net positive charge from a library of 32 alkyl diazirine probes, which may arise from enhanced association of the probe with the matched acidic regions on protein surfaces. In line with this expectation, chemoproteomics and binding site data show that these probes preferentially enrich proteins with negative electrostatic surfaces or proteins embedded in membranes, which may be a consequence of the predicted elevation of pKa of acidic residues in the hydrophobic membrane environment.41
These data assist with the design and interpretation of PAL experiments using alkyl diazirine chemistry. Design of the probe can be tuned to enhance or reduce protein capture simply by altering the net overall charge of the probe molecule. Alkyl diazirine probes with a net neutral or negative charge may selectively visualize stronger binding interactions, while probes with a net positive charge may capture more transient interactions in the proteome. The charge state of the molecule is readily altered by the type of linker chemistry employed when embedding the alkyl diazirine to the probe molecule of interest (e.g., amide versus amine linker chemistry). Alkyl diazirine chemistry should also be selected if membrane proteins or those with large acidic patches are desired targets. The preference of alkyl diazirine chemistry for these protein types is reflected herein and prior studies.5, 7, 11–13, 37, 42, 49 This enhances the utility of diazirine chemistry in probes for visualizing binding over a range of affinities with membrane proteins, which are typically challenging proteins to target.
Although these results point to a significant contribution of alkyl diazirine PAL through the diazo intermediate, dissection of the exact ratio of labeling from the carbene and the diazo intermediate remains to be fully elucidated. Theoretically, products resulting from the carbene are more likely to arise from tight binding interactions with a lower binding constant, since the carbene is a short-lived intermediate that is rapidly quenched in aqueous media.17, 50 Conversely, products resulting from labeling through the diazo are more likely to represent weaker binding interactions, since the lifetime of the diazo is above the rate of diffusion.51 There are relatively few chemistries that have been used to selectively label carboxylic acids in whole cells,52–54 and fewer photoactivatable chemistries,30 suggesting that this chemistry could be intentionally utilized to profile carboxylic acids in cells as a method that senses the protonation state of the acids. Additionally, once a carbene intermediate is generated, the contribution from singlet or triplet states is another open question that may affect the amino acid labeling distribution, since the carbene is formed as a singlet, but can relax to the triplet state.55–59
In addition to the contribution of the chemical reactivity of the alkyl diazirine, the overall labeling trends of diazirine probes are also impacted by other factors, such as overall hydrophobicity of the probe, subcellular localization, or cell permeability, which must be additionally considered when designing PAL probes or interpreting data from PAL experiments. These factors likely contribute to the observed labeling trends, by either reducing cellular permeability60 or enhancing the cellular uptake and subcellular localization of positively charged probes to membranous organelles.61, 62 However, we observed higher labeling yields from probes with a positive charge in cells and cell lysates suggesting that increased cellular uptake of positive probes does not fully explain the increase in labeling. In addition, the increased membrane protein enrichment is a general trend observed here with alkyl diazirines, which is independent of the charge of the probe. Potentially, the subcellular localization of positively charged probes to the membrane is synergistic with the pH-dependent reactivity of alkyl diazirines. The influence of these factors can be distinguished by comparison of labeling trends of a given probe in vitro versus in cells, such as what we observed with JN942 and JN33.
CONCLUSION
In conclusion, the alkyl diazirine preferentially reacts with acidic amino acid residues due to the major contribution of a reactive diazo intermediate, while the aryl-trifluoro diazirine possesses a reactivity pattern in line with a reactive carbene intermediate. These reactivity patterns are consistent across individual amino acids, single proteins, and the whole proteome, and assist with the design and interpretation of PAL experiments using alkyl diazirine probes. Understanding the reactivity pattern of alkyl diazirines therefore facilitates design and interpretation of PAL experiments. Given these reactivity considerations for the alkyl diazirine, there is opportunity for innovation of new PAL chemistries with different labeling preferences. The design of new chemistries and the systematic examination of their reactivity patterns may provide additional opportunities for generation of an “ideal” PAL functional group or development of methods to selectively map non-covalent interactions with certain regions of the cell or types or proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Flaxman, N. Vallavoju, C. Chang, and A. D’Souza for their advice and helpful discussions, and M. Parsons for his assistance. Mass spectrometry data was collected at the Harvard University Mass Spectrometry and Proteomics Resource Laboratory (B. Budnik), and NMR data collected at the Laukin-Purcell Instrumentation Center (D. Cui). Support from NIH NIDA (DP1DA046586), Burroughs Welcome Fund, Sloan Foundation, Camille and Henry Dreyfus Foundation, and Harvard University are gratefully acknowledged.
Footnotes
SUPPORTING INFORMATION
A full description of the experimental methods used in this study can be found in the Supporting Information, along with supplementary figures, synthetic protocols, representative spectra for amino acid reactions, characterization of novel compounds, and representative binding site spectra.
CODE AVAILABILITY
The IsoStamp software used for analysis in this paper is available online at GitHub: https://github.com/harvardinformatics/quantproteomics/tree/master/Isostamp
COMPETING INTERESTS
The authors declare no competing interests
DATA AVAILABILITY
All proteomics data are available within this paper, in the Supporting Information, or the Supplementary Tables. The full mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD025140.
REFERENCES
- 1.Singh A; Thornton ER; Westheimer FH, The Photolysis of Diazoacetylchymotrypsin. J. Biol. Chem 1962, 237 (9), PC3006–PC3008. [PubMed] [Google Scholar]
- 2.Reuter JA; Spacek DV; Snyder MP, High-throughput sequencing technologies. Mol. Cell 2015, 58 (4), 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y; Fonslow BR; Shan B; Baek MC; Yates JR 3rd, Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 2013, 113 (4), 2343–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pappireddi N; Martin L; Wühr M, A Review on Quantitative Multiplexed Proteomics. ChemBioChem 2019, 20 (10), 1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niphakis Micah J.; Lum Kenneth M.; Cognetta Armand B.; Correia Bruno E.; Ichu T-A; Olucha J; Brown Steven J.; Kundu S; Piscitelli F; Rosen H, et al. , A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell 2015, 161 (7), 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S-H; Boyce M; Wands AM; Bond MR; Bertozzi CR; Kohler JJ, Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl. Acad. Sci. U.S.A 2012, 109 (13), 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker CG; Galmozzi A; Wang Y; Correia BE; Sasaki K; Joslyn CM; Kim AS; Cavallaro CL; Lawrence RM; Johnson SR, et al. , Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell 2017, 168 (3), 527–541.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan S; Jang S-Y; Wang D; Liew SS; Li Z; Lee J-S; Yao SQ, A Suite of “Minimalist” Photo-Crosslinkers for Live-Cell Imaging and Chemical Proteomics: Case Study with BRD4 Inhibitors. Angew. Chem 2017, 129 (39), 11978–11983. [DOI] [PubMed] [Google Scholar]
- 9.Li Z; Hao P; Li L; Tan CYJ; Cheng X; Chen GYJ; Sze SK; Shen H-M; Yao SQ, Design and Synthesis of Minimalist Terminal Alkyne-Containing Diazirine Photo-Crosslinkers and Their Incorporation into Kinase Inhibitors for Cell- and Tissue-Based Proteome Profiling. Angew. Chem. Int. Ed 2013, 52 (33), 8551–8556. [DOI] [PubMed] [Google Scholar]
- 10.Chang C-F; Mfuh A; Gao J; Wu H-Y; Woo CM, Synthesis of an electronically-tuned minimally interfering alkynyl photo-affinity label to measure small molecule–protein interactions. Tetrahedron 2018, 74, 3273–3277. [Google Scholar]
- 11.Gao J; Mfuh A; Amako Y; Woo CM, Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs. J. Am. Chem. Soc 2018, 140 (12), 4259–4268. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y; Dix MM; Bianco G; Remsberg JR; Lee H-Y; Kalocsay M; Gygi SP; Forli S; Vite G; Lawrence RM, et al. , Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs. Nat. Chem 2019, 11 (12), 1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleiner P; Heydenreuter W; Stahl M; Korotkov VS; Sieber SA, A Whole Proteome Inventory of Background Photocrosslinker Binding. Angew. Chem. Int. Ed 2017, 56 (5), 1396–1401. [DOI] [PubMed] [Google Scholar]
- 14.Hagan B, Chapter 4 The photoaffinity labeling experiment. In Laboratory Techniques in Biochemistry and Molecular Biology, Work TS; Burdon RH, Eds. Elsevier: 1983; Vol. 12, pp 66–111. [Google Scholar]
- 15.Ruoho AE; Kiefer H; Roeder PE; Singer SJ, The Mechanism of Photoaffinity Labeling. Proc. Natl. Acad. Sci. U.S.A 1973, 70 (9), 2567–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platz MS, A perspective on physical organic chemistry. In J. Org. Chem., American Chemical Society: 2014; Vol. 79, pp 2341–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y; Burdzinski G; Kubicki J; Vyas S; Hadad CM; Sliwa M; Poizat O; Buntinx G; Platz MS, Study of the S1 Excited State of para-Methoxy-3-phenyl-3-methyl Diazirine by Ultrafast Time Resolved UV−Vis and IR Spectroscopies and Theory. J. Am. Chem. Soc. 2009, 131 (38), 13784–13790. [DOI] [PubMed] [Google Scholar]
- 18.Westheimer FH, Photoaffinity Labeling-Retrospect and Prospect. Ann. N. Y. Acad. Sci. 1980, 346 (1), 134–143. [Google Scholar]
- 19.Roberts JD; Watanabe W, The Kinetics and Mechanism of the Acid-Catalyzed Reaction of Diphenyldiazomethane with Ethyl Alcohol1,2. J. Am. Chem. Soc 1950, 72 (11), 4869–4879. [Google Scholar]
- 20.Mix KA; Lomax JE; Raines RT, Cytosolic Delivery of Proteins by Bioreversible Esterification. J. Am. Chem. Soc 2017, 139 (41), 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flaxman HA; Chang CF; Wu HY; Nakamoto CH; Woo CM, A Binding Site Hotspot Map of the FKBP12-Rapamycin-FRB Ternary Complex by Photoaffinity Labeling and Mass Spectrometry-Based Proteomics. J. Am. Chem. Soc 2019, 141 (30), 11759–11764. [DOI] [PubMed] [Google Scholar]
- 22.Iacobucci C; Götze M; Piotrowski C; Arlt C; Rehkamp A; Ihling C; Hage C; Sinz A, Carboxyl-Photo-Reactive MS-Cleavable Cross-Linkers: Unveiling a Hidden Aspect of Diazirine-Based Reagents. Anal. Chem 2018, 90 (4), 2805–2809. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien JGK; Jemas A; Asare-Okai PN; am Ende CW; Fox JM, Probing the Mechanism of Photoaffinity Labeling by Dialkyldiazirines through Bioorthogonal Capture of Diazoalkanes. Org. Lett 2020, 22 (24), 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith RAG; Knowles JR, Aryldiazirines. Potential Reagents for Photolabeling of Biological Receptor Sites. J. Amer. Chem. Soc 1972, 95. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka Y; Bond MR; Kohler JJ, Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst 2008, 4 (6), 473–480. [DOI] [PubMed] [Google Scholar]
- 26.Wittelsberger A; Thomas BE; Mierke DF; Rosenblatt M, Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 2006, 580 (7), 1872–1876. [DOI] [PubMed] [Google Scholar]
- 27.Deseke E; Nakatani Y; Ourisson G, Intrinsic Reactivities of Amino Acids towards Photoalkylation with Benzophenone − A Study Preliminary to Photolabelling of the Transmembrane Protein Glycophorin A. Eur. J. Org. Chem 1998, 1998 (2), 243–251. [Google Scholar]
- 28.Cheng K; Lee J-S; Hao P; Yao SQ; Ding K; Li Z, Tetrazole-Based Probes for Integrated Phenotypic Screening, Affinity-Based Proteome Profiling, and Sensitive Detection of a Cancer Biomarker. Angew. Chem. Int. Ed 2017, 56, 15044–15048. [DOI] [PubMed] [Google Scholar]
- 29.Tian Y; Jacinto MP; Zeng Y; Yu Z; Qu J; Liu WR; Lin Q, Genetically Encoded 2-Aryl-5-carboxytetrazoles for Site-Selective Protein Photo-Cross-Linking. J. Am. Chem. Soc 2017, 139, 6078–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bach K; Beerkens BLH; Zanon PRA; Hacker SM, Light-Activatable, 2,5-Disubstituted Tetrazoles for the Proteome-wide Profiling of Aspartates and Glutamates in Living Bacteria. ACS Cent. Sci 2020, 6 (4), 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziemianowicz DS; Bomgarden R; Etienne C; Schriemer DC, Amino Acid Insertion Frequencies Arising from Photoproducts Generated Using Aliphatic Diazirines. J. Am. Soc. Mass. Spectrom 2017, 28 (10), 2011–2021. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen PE; Buchardt O, Aryl Azides as Photoaffinity Labels. A Photochemical Study of some 4-substituted Aryl Azides. Photochem. Photobiol 1982, 35 (3), 317–323. [Google Scholar]
- 33.Hyde S; Veliks J; Ascough DMH; Szpera R; Paton RS; Gouverneur V, Enantioselective rhodium-catalysed insertion of trifluorodiazoethanes into tin hydrides. Tetrahedron 2019, 75 (1), 17–25. [Google Scholar]
- 34.Pezacki JP; Pole DL; Warkentin J; Chen T; Ford F; Toscano JP; Fell J; Platz MS, Laser Flash and Dual Wavelength Photolysis of 3,4-Diaza-2,2-dimethoxy-1-oxa[4.5]spirooct-3-ene. Migration of Hydrogen and Carbon in Cyclobutylidene and in the Excited State of Its Precursor. J. Am. Chem. Soc 1997, 119 (13), 3191–3192. [Google Scholar]
- 35.Albu TV; Lynch BJ; Truhlar DG; Goren AC; Hrovat DA; Borden WT; Moss RA, Dynamics of 1,2-Hydrogen Migration in Carbenes and Ring Expansion in Cyclopropylcarbenes. J. Phys. Chem. A 2002, 106 (21), 5323–5338. [Google Scholar]
- 36.Kumar NS; Young RN, Design and synthesis of an all-in-one 3-(1,1-difluoroprop-2-ynyl)-3H-diazirin-3-yl functional group for photo-affinity labeling. Bioorg. Med. Chem 2009, 17, 5388–5395. [DOI] [PubMed] [Google Scholar]
- 37.Parker CG; Kuttruff CA; Galmozzi A; Jørgensen L; Yeh C-H; Hermanson DJ; Wang Y; Artola M; McKerrall SJ; Josyln CM, et al. , Chemical Proteomics Identifies SLC25A20 as a Functional Target of the Ingenol Class of Actinic Keratosis Drugs. ACS Cent. Sci 2017, 3 (12), 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almén MS; Nordström KJV; Fredriksson R; Schiöth HB, Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panahi A; Brooks CL 3rd, Membrane environment modulates the pKa values of transmembrane helices. J. Phys. Chem 2015, 119 (13), 4601–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcoline Frank V.; Bethel N; Guerriero Christopher J.; Brodsky Jeffrey L.; Grabe M, Membrane Protein Properties Revealed through Data-Rich Electrostatics Calculations. Structure 2015, 23 (8), 1526–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teixeira VH; Vila-Viçosa D; Reis PBPS; Machuqueiro M, pKa Values of Titrable Amino Acids at the Water/Membrane Interface. J. Chem. Theory Comput 2016, 12 (3), 930–934. [DOI] [PubMed] [Google Scholar]
- 42.Miyamoto DK; Flaxman HA; Wu H-Y; Gao J; Woo CM, Discovery of a Celecoxib Binding Site on Prostaglandin E Synthase (PTGES) with a Cleavable Chelation-Assisted Biotin Probe. ACS Chem. Biol 2019, 14 (12), 2527–2532. [DOI] [PubMed] [Google Scholar]
- 43.Woo CM; Felix A; Byrd WE; Zuegel DK; Ishihara M; Azadi P; Iavarone AT; Pitteri SJ; Bertozzi CR, Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes. J. Proteome Res. 2017, 16 (4), 1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergdoll LA; Lerch MT; Patrick JW; Belardo K; Altenbach C; Bisignano P; Laganowsky A; Grabe M; Hubbell WL; Abramson J, Protonation state of glutamate 73 regulates the formation of a specific dimeric association of mVDAC1. Proc. Natl. Acad. Sci. U.S.A 2018, 115 (2), E172–E179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersson CD; Karlberg T; Ekblad T; Lindgren AEG; Thorsell A-G; Spjut S; Uciechowska U; Niemiec MS; Wittung-Stafshede P; Weigelt J, et al. , Discovery of Ligands for ADP-Ribosyltransferases via Docking-Based Virtual Screening. J. Med. Chem 2012, 55 (17), 7706–7718. [DOI] [PubMed] [Google Scholar]
- 46.Zhai Q; Fisher RD; Chung H-Y; Myszka DG; Sundquist WI; Hill CP, Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol 2008, 15 (1), 43–49. [DOI] [PubMed] [Google Scholar]
- 47.Macdonald N; Welburn JPI; Noble MEM; Nguyen A; Yaffe MB; Clynes D; Moggs JG; Orphanides G; Thomson S; Edmunds JW, et al. , Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14–3-3. Mol. Cell 2005, 20 (2), 199–211. [DOI] [PubMed] [Google Scholar]
- 48.Abraham J; Corbett KD; Farzan M; Choe H; Harrison SC, Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol 2010, 17 (4), 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gertsik N; Am Ende CW; Geoghegan KF; Nguyen C; Mukherjee P; Mente S; Seneviratne U; Johnson DS; Li YM, Mapping the Binding Site of BMS-708163 on gamma-Secretase with Cleavable Photoprobes. Cell Chem. Biol 2017, 24 (1), 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modarelli DA; Morgan S; Platz MS, Carbene formation, hydrogen migration, and fluorescence in the excited states of dialkyldiazirines. J. Am. Chem. Soc 1992, 114 (18), 7034–7041. [Google Scholar]
- 51.Procacci B; Roy SS; Norcott P; Turner N; Duckett SB, Unlocking a Diazirine Long-Lived Nuclear Singlet State via Photochemistry: NMR Detection and Lifetime of an Unstabilized Diazo-Compound. J. Am. Chem. Soc 2018, 140 (48), 16855–16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mix KA; Raines RT, Optimized Diazo Scaffold for Protein Esterification. Org. Lett 2015, 17 (10), 2358–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian Y; Schürmann M; Janning P; Hedberg C; Waldmann H, Activity-Based Proteome Profiling Probes Based on Woodward’s Reagent K with Distinct Target Selectivity. Angew. Chem. Int. Ed 2016, 55 (27), 7766–7771. [DOI] [PubMed] [Google Scholar]
- 54.Martín-Gago P; Fansa EK; Winzker M; Murarka S; Janning P; Schultz-Fademrecht C; Baumann M; Wittinghofer A; Waldmann H, Covalent Protein Labeling at Glutamic Acids. Cell Chem. Biol 2017, 24 (5), 589–597.e5. [DOI] [PubMed] [Google Scholar]
- 55.Arenas JF; López-Tocón I; Otero JC; Soto J, Carbene Formation in Its Lower Singlet State from Photoexcited 3H-Diazirine or Diazomethane. A Combined CASPT2 and ab Initio Direct Dynamics Trajectory Study. J. Am. Chem. Soc 2002, 124 (8), 1728–1735. [DOI] [PubMed] [Google Scholar]
- 56.Geise CM; Wang Y; Mykhaylova O; Frink BT; Toscano JP; Hadad CM, Computational and Experimental Studies of the Effect of Substituents on the Singlet−Triplet Energy Gap in Phenyl(carbomethoxy)carbene. J. Org. Chem 2002, 67 (9), 3079–3088. [DOI] [PubMed] [Google Scholar]
- 57.Matzinger S; Bally T; Patterson EV; McMahon RJ, The C7H6 Potential Energy Surface Revisited: Relative Energies and IR Assignment. J. Am. Chem. Soc 1996, 118 (6), 1535–1542. [Google Scholar]
- 58.Admasu A, D. Gudmundsdóttir A, S. Platz M, S. Watt D, Kwiatkowski S, J. Crocker P, A laser flash photolysis study of p-tolyl(trifluoromethyl)carbene. J. Chem. Soc., Perkin Trans 2 1998, (5), 1093–1100. [Google Scholar]
- 59.Hirai K; Itoh T; Tomioka H, Persistent Triplet Carbenes. Chem. Rev 2009, 109 (8), 3275–3332. [DOI] [PubMed] [Google Scholar]
- 60.Yang NJ; Hinner MJ, Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. In Site-Specific Protein Labeling: Methods and Protocols, Gautier, A.; Hinner, M. J, Eds. Springer; New York: New York, NY, 2015; pp 29–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng N; Tsai HN; Zhang X; Shedden K; Rosania GR, The Subcellular Distribution of Small Molecules: A Meta-Analysis. Mol. Pharm 2011, 8 (5), 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richter MF; Drown BS; Riley AP; Garcia A; Shirai T; Svec RL; Hergenrother PJ, Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, 545 (7654), 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomics data are available within this paper, in the Supporting Information, or the Supplementary Tables. The full mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD025140.


